Abstract
Breast cancer is the most common type of cancer in women worldwide, with the luminal subtype being the most widespread. Although characterized by better prognosis compared with other subtypes, luminal breast cancer is still considered a threatening disease due to therapy resistance, which occurs via both cell- and non–cell-autonomous mechanisms. Jumonji domain-containing 6, arginine demethylase and lysine hydroxylase (JMJD6) is endowed with a negative prognostic value in luminal breast cancer and, via its epigenetic activity, it is known to regulate many intrinsic cancer cell pathways. So far, the effect of JMJD6 in molding the surrounding microenvironment has not been explored.
Here, we describe a novel function of JMJD6 showing that its genetic inhibition in breast cancer cells suppresses lipid droplet formation and ANXA1 expression, via estrogen receptor alpha and PPARα modulation. Reduction of intracellular ANXA1 results in decreased release in the tumor microenvironment (TME), ultimately preventing M2-type macrophage polarization and tumor aggressiveness.
Implications:
Our findings identify JMJD6 as a determinant of breast cancer aggressiveness and provide the rationale for the development of inhibitory molecules to reduce disease progression also through the remodeling of TME composition.
Introduction
Breast cancer is the most common cancer in women worldwide and it represents the third cause of cancer-related deaths in Europe (1, 2). Breast cancer is highly heterogeneous, making therapy decision and prognosis extremely variable depending on the molecular landscape of the tumor along with its tumor size, morphology, and presence of lymph node metastases (3–5). Breast cancer is classified in different subtypes based on the expression of estrogen receptor alpha (ERα), progesterone receptor and HER2 (6). In ER-positive breast cancer, hormone therapy, mostly administered in adjuvant settings, represents the standard treatment, although about 50% of patients experience local or distant relapse.
Jumonji domain-containing 6 (JMJD6) is an epigenetic regulator which demethylates arginine and hydroxylates lysine on both histone and nonhistone proteins, including ERα (7). Expression of JMJD6 has been linked to progression and poor prognosis of several tumor types, including oral cancer (8), colon cancer (9), and hepatocellular carcinoma (10). Interestingly, the highest rate of gene amplification is observed in breast cancer (pan-TCGA, cBioportal) and several studies confirmed an association between JMJD6 expression and breast cancer aggressiveness and poor prognosis (11–13). The vast majority of these investigations focused on cell-intrinsic role of JMJD6, also highlighting its capability to regulate ERα (14), but whether JMJD6 impacts the clinical outcome also by affecting the surrounding microenvironment still remains largely unexplored.
ANXA1 is a Ca2+-dependent phospholipid-binding protein endowed with an anti-inflammatory potential which has been described both in tumor-related and unrelated studies (15, 16). ANXA1 has been shown to affect the phenotype of macrophages and to promote their polarization towards M2 (17). In physiologic settings, this process favors inflammation resolution (18), but in cancer it skews macrophages to a pro-tumor state (17). ANXA1 is largely stored in lipid droplets (LD; refs. 19, 20) whose formation has been shown to be controlled by hormones, through the regulation of adiposity and lipid homeostasis (21), and recently also by JMJD6 (22).
In our work, we sought to dissect the link between ERα, JMJD6 and ANXA1, and the possible involvement of LDs in this axis, demonstrating that JMJD6 can affect the aggressiveness of breast cancer, not only in a cell-intrinsic fashion, but also through the regulation of macrophage activity. We provide evidence that JMJD6 supports high levels of ANXA1 via transcriptional expression, and likely also via LD accumulation, through a mechanism which depends on ERα and PPARα, a nuclear receptor important for the maintenance of lipid homeostasis (23) and LD formation (24), and which is suppressed by ERα (25, 26). High levels of ANXA1 eventually trigger the polarization of macrophages towards a pro-tumor M2-like state and promote the growth of breast cancer in vivo.
Materials and Methods
Cell lines and siRNAs
The human luminal A-like MCF7, BT474 and triple-negative MDA-MB231 breast cancer cell lines were purchased from the ATCC together with the murine macrophage RAW264.7 cell line. The mouse PyMT-41C mammary cancer cell line was derived from MMTV-PyMT mice and already described (27). MCF7 and PyMT-41C cells were cultured in DMEM (Gibco, Thermo Fisher), while MDA-MB231 and RAW264.7 cell lines were cultured in RPMI1640 medium (Gibco, Thermo Fisher) supplemented with 10% FBS (EuroClone), 2 mmol/Ll-glutamine, sodium pyruvate (NaPyr), HEPES and nonessential amino acids (NEAA, all from LONZA). Mycoplasma screening was performed every month by TaKaRa PCR Mycoplasma Detection Set (Catalog no. 6601). Experiments were generally performed one week after cell thawing. For repetitive experiments, cells were maintained in culture for 6 weeks at most. MDA-MB231 and MCF7 were authenticated by Eurofins Genomics on January 16, 2020. Lipofectamine RNAiMAX transfection reagent (13778075, Thermo Fisher) was used for silencing experiments according to the manufacturer's protocol. The following siRNAs were purchased from ThermoFisher: PPARα (AM51331, ID: 5348), PPARα (AM16708, ID: 142804), ERα (4392420, ID: s4823), ERα (AM16708, ID: 4004), and JMJD6 (AM16708, ID: 172187 and 108684).
Generation of knockout cell lines with CRISPR-Cas9 technology
Amaxa cell line nucleofector kit V (VCA-1003, Lonza) was used for electroporation-based transfection of MCF7 cells. Lipofectamine 3000 transfection reagent (L3000015, Thermo Fisher Scientific) was used to transfect PyMT-41C cells and MDA-MB231 cells according to the manufacturer's protocol. All cell lines were transfected with the pSpCas9(BB)-2A-GFP plasmid (ID 48138; a gift from F. Zhang, Addgene, Cambridge, MA), in which the following gRNAs targeting Jmjd6 were cloned: CACCgGAGCAAGAAGCGCATCCGCG (1B1), CACCgGCTCTCGTAGTAGTTGTGCC (1A2). Control clones were obtained by transfection with the scramble vector. Three days after transfection, GFP-positive cells were sorted with the BD FACS Aria III sorter (BD Biosciences). After 2 to 3 days, sorted cells were seeded at single cell (0.7 cell/well) in 96-well plates and cultured at 37°C until confluence was reached. After expansion, clones were tested for JMJD6 expression by Western blot.
Cell lysis and Western blot
For Western blot assay, cells were harvested, washed with PBS, boiled for 10 minutes in lysis buffer (125 mmol/L Tris HCl pH 6.8, 5% of SDS) and supplemented with protease inhibitors (11873580001, cOmplete, Roche). Samples were then sonicated for 20 seconds, followed by centrifugation for 15 minutes at 13,000 rpm and quantified with micro BCA Assay kit reagent (23235, Thermo Fisher Scientific) using the Spark microplate reader (TECAN). Proteins were separated by SDS-PAGE on precast 4% to 12% Bis-Tris NuPAGE gels (NP0321PK2, Thermo Fisher Scientific) and blotted to Immobilon PVDF membranes (IPVH00010, Merck Millipore) using the XCell II blot module (EI9051, Thermo Fisher Scientific). Membranes were then saturated for 1 hour in Tris-buffered saline containing 5% BSA and incubated overnight with the antihuman primary antibodies recognizing: JMJD6 (ab64575, Abcam and P1495, Sigma-Aldrich), Actin-β (A1978, Sigma-Aldrich), ERα (sc-544, G20, Santa Cruz Biotechnology and MA1–80216, Invitrogen), ANXA1 (ab214486, Abcam) and Vinculin (V9131, Sigma-Aldrich). After hybridization with the appropriate secondary antibody (anti-rabbit and anti-mouse IgG, GE HealthCare), proteins were detected by chemiluminescence (EuroClone).
Immunofluorescence
For immunofluorescence staining, cells cultured on coverslips or in black optical 96-well plates were washed with PBS followed by fixation with 4% paraformaldehyde in PBS for 10 minutes at room temperature. Cells were then permeabilized with 0.1% Triton X-100 in PBS and 0.5% BSA for 5 minutes and blocked with 3% BSA in PBS for 1 hour at room temperature, prior to primary antibody incubation. The following antibodies were used: anti-ERα (MA1–80216, Invitrogen) for 2 hours at room temperature and anti-ANXA1 (ab214486, Abcam) overnight at 4°C. After extensive washes, cells were incubated with secondary antibodies Alexa Fluor 555 anti-mouse and anti-rabbit (Thermo Fisher Scientific) for 1 hour at room temperature in the dark, followed by DAPI staining for nuclei. Fluorescence was detected using a microscope (DFC450C, Leica Biosystems).
LD staining and ANXA1 IHC
For Oil Red O analysis (LD), cells were seeded in 24-well plates and stained with Oil Red O staining kit (ab150678, Abcam) according to the manufacturer's protocol. When Oil Red O staining was combined with ANXA1 IHC, cells were cultured in coverslips and washed with PBS before endogenous peroxidase blockade with 3% of dH2O2 for 10 minutes at room temperature in the dark. Cells were then washed with PBS and nonspecific bindings were blocked 3% of BSA in PBS for 1 hour at room temperature. ANXA1 antibody + 1% BSA in PBS (ab214486, Abcam) was then incubated overnight at 4°C. After extensive washes with PBS, cells were incubated with HRP-tagged anti-rabbit secondary antibody (GE HealthCare) for 1 hour at room temperature. Signal was developed for 3 to 5 minutes of DAB staining enhancer (AR9432, Leica Biosystems) incubation, followed by washes with tap water. Cells were then stained with the Oil Red O stain kit according to the manufacturer's protocol, and coverslips were mounted on slides using an aqueous mounting medium, and analyzed with the microscope (DFC450C, Leica Biosystems). For HCS LipidTox Red neutral lipid staining (LD), cells were cultured on coverslips or black optical 96 well plates, and fixed with 4% PFA for 10 minutes at room temperature, after that HCS LipidTox Red neutral lipid staining kit (H34476, ThermoFisher) was used according to the manufacturer's instructions (1 hour at room temperature). After PBS washes, nuclei were stained with DAPI and coverslips were mounted on slides with a mounting medium. Fluorescence signal was observed at 550/640 nm with a fluorescence microscope (DFC450C, Leica Biosystems).
ANXA1 ELISA assay
To detect ANXA1 in conditioned medium (CM) of wild-type (WT), scramble or ERα and/or PPARα silenced MCF7 cells, the human Annexin A1 ELISA kit was used (ab222868, Abcam). Briefly, cells seeded in equal numbers in 12-well plates and silenced as previously described. CM was collected after 72 hours and used for ELISA assay. Absorbance was read at 450 nm with the Spark microplate reader (TECAN). The standard curve was used to define the protein concentration.
RNA isolation and real-time PCR
Before stimulation, cells were cultured in medium with 10% FBS. After 24 hours culture medium was changed to phenol red-free DMEM containing 1% of dextran-coated charcoal serum (DCC). Then, 10 nmol/L β2-estradiol (E2) was added for 6 hours. Cells were then washed with PBS and harvested, and pellet used for RNA isolation and real-time PCR. RNA was isolated with miRNeasy Minikit (217004, Qiagen) according to the manufacturer's protocol. For cDNA synthesis (from 500–1,000 ng RNA), the high capacity cDNA reverse transcription kit (4374967, Thermo Fisher Scientific) was used according to the manufacturer's protocol. Real-time PCR was carried out using the TaqMan Fast universal PCR Master Mix (4352042, Thermo Fisher Scientific) and results were analyzed with QuantStudio 3 (Thermo Fisher Scientific). Transcript levels were quantified by using the dCt method or ddCt (Fig. 1E), and normalized to GAPDH expression. The following TaqMan assays were used for human cells: Hs00947536 (PPARA), Hs01046816 (ESR1), Hs00167549 (ANXA1), Hs04986394_s1 (BCL2), Hs01556702 (PGR), Hs00940446 (RARA), Hs00231936_m1 (XBP1), Hs99999905 (GAPDH), all from ThermoFisher, and Hs.PT.56a.2905156 (BCL2) from IDT. Mouse probes Mm00466679 (Jmjd6), Mm00441891 (Cd40), Mm00475988 (Arg1), and Mm99999915 (Gapdh) were purchased from ThermoFisher, Mm.PT.58.43705194 (Nos2) and Mm.PT.58.42560062 (Mrc1) from IDT.
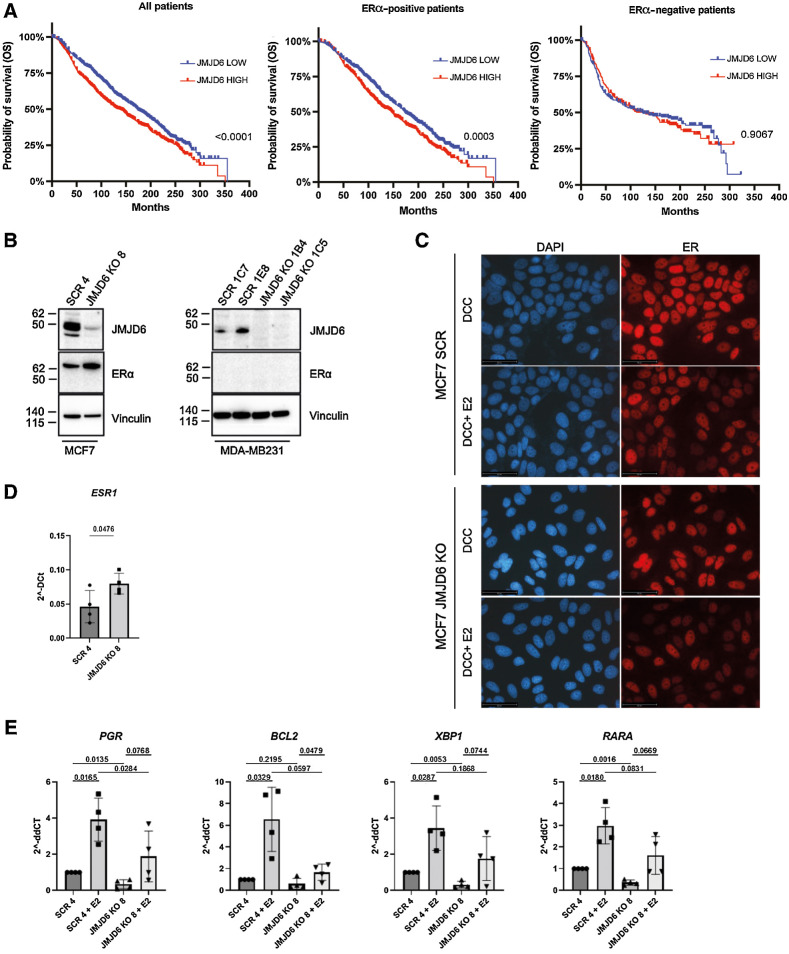
Figure 1.
JMJD6 regulates ERα activity and affects prognosis in luminal patients with breast cancer. A, Kaplan–Meier analysis of patients with breast cancer included in the METABRIC dataset (available on cBioportal) showing the OS of all (left, n = 1,884), ERα-positive (middle, n = 1,440) or ERα-negative patients (right, N = 444). ‘High’ (red) and ‘low’ (blue) JMJD6 expression is shown according to median. P value was calculated with Log-rank (Mantel-Cox) test. B, Western blot showing the expression of JMJD6 and ERα in control (SCR) and JMJD6 KO MCF7 and MDA-MB231 clones. Vinculin is shown as loading control. C, Immunofluorescence staining of ERα (red) in scramble or JMJD6 KO MCF7 cells stimulated or not with E2. Nuclei were stained with DAPI (blue), bars = 50 μmol/L. D, Real Time PCR to evaluate the levels of ESR1 and (E) its target genes (PGR, BCL2, XBP1 and RARA) in control (SCR) of JMJD6 KO MCF7 stimulated or not with E2. Statistical significance was calculated by paired two-tailed t test comparing four independent experiments.
Generation of bone marrow–derived macrophages
WT C57BL/6 female mice were sacrificed between 6 and 8 weeks, and bone marrow was flushed from femurs with PBS containing 5% FBS into a 50 mL tube by using a 27-gauge needle. Cell suspension was centrifuged at 1,300 rpm for 5 minutes and the supernatant discarded. Cells were treated with 1X ACK lysis buffer (Ammonium-Chloride-Potassium) for 5 minutes at 4°C to lyse erythrocytes, and reaction was then blocked with PBS containing 5% of FBS. Cell suspension was centrifuged at 1,300 rpm for 5 minutes and cells were seeded in 6-well plates in DMEM containing 10% FBS and 20 ng/mL of M-CSF (Peprotech 315–02). Medium containing M-CSF was then changed every 2 days. After 7 days, M1 macrophages were differentiated for 24 hours with 20 ng/mL IFNγ (Peprotech, 315–05) and 10 ng/mL LPS (Sigma, 916374), while M2 macrophages were differentiated with 40 ng/mL IL4 (Peprotech, 214–14). The experiment was repeated 3 times and 2 mice were used in each experiment.
Macrophages in vitro culture with breast cancer cell CM
CM of WT, scramble or Jmjd6 knockout (KO) PyMT-41C cells was collected and used 1:3 with fresh normal medium (10% FBS in RPMI) to culture RAW264.7 macrophage cell line for 24 hours, thereafter, RAW264.7 cells were harvested and used for expression profiling (ClariomS analysis) or further analyses. For BMDMs, macrophages were first differentiated towards M1 or M2 phenotypes as described above, and then, CM was added 1:4 to M0, M1 or M2 macrophages for 24 hours. BMDMs were harvested and used for flow cytometry analysis. Alternatively, CM of WT, scramble or Jmjd6-KO PyMT-41C cells, with or without the Annexin-1 protein mimetic peptide Ac2–26 (R&D Systems, catalog no. 1845, 10 ng/mL) was used to culture M0, M1, and M2 macrophages for 24 hours. Macrophages were then harvested and used for further experiments.
In vivo experiments
WT C57BL/6 female mice were purchased from Charles River Italy and housed in a specific pathogen-free animal facility and treated in accordance with the European Community guidelines. The study was approved by the Institutional Ethical Committee and by the Italian Ministry of Health (authorization number 297/2020-PR). For tumor growth experiments, 6- to 8-week-old C57BL/6 mice were injected in the mammary fat pad with 1 × 106 JMJD6 scramble (n = 8) or JMJD6 KO PyMT-41C cells (n = 8) in 100-μL saline solution. Two different clones were used for each condition. Tumor length and width were measured once a week, and tumor volume was estimated by (length × width2)/2. At the experimental endpoint, mice were euthanized and the tumor was stored in formalin for further experiments or used for flow cytometry analysis.
Flow cytometry analysis
For the analysis of myeloid cell infiltration, tumors generated from scramble or Jmjd6 KO PyMT-41C breast cancer cells were collected, minced and filtered through a 40-μm pores cell strainer (BD Biosciences). Red blood cells were removed using 1X ACK lysis buffer (ammonium chloride potassium). Cells were Fc-blocked using CD16/32 antibody (eBioscience) before staining. Antibodies used were: CD45 (clone 30-F11) BV510 (BD Biosciences, catalog no. 563891), CD11b (clone M1/70) PE (TONBO Biosciences, catalog no. 50–0112), F4/80 (clone BM8), PE-Cy7 (eBioscience, catalog no. 25–4801–82), Gr1 (clone RB6–8C5) APC (eBioscience, catalog no. 17–5931–82), Ly6c (clone AL-21) BV421 (BD Biosciences, catalog no. 562727), CD206 (clone C068C2) Alexa Fluor 488 (BioLegend, catalog no. 141704), PD-L1 (clone MIH5) BV786 BD (Biosciences, catalog no. 741014). All samples were analyzed in single; 6 samples were analyzed for each group. For analysis of BMDMs cultured in CM of scramble or Jmjd6 KO 41C cells, M0, M1, and M2-differentiated macrophages were detached with TrypLE Express Enzyme 1X without phenol red (Gibco, 12604013), washed with PBS and stained for 15 minutes at 4°C with the following antibodies: MRC1 (clone C068C2) BV650 (BD Biosciences, catalog no. 141723), CD40 (clone 3/23) PerCP/Cy5.5 (BD Biosciences, catalog no. 124624), PDL1 (clone 10F.942) BV711 (BD Biosciences, catalog no. 124319), PDL2 (clone TY25) APC (BD Biosciences, catalog no. 107210), I-A/I-E (HLA-II) (clone M5/114.15.2) BV510 (BD Biosciences, catalog no. 107635), F4/80 (clone BM8) PE/Cyanine 7 (BD Biosciences, catalog no. 123114), CD11b (clone M1/70) PE (TOMBO, catalog no. 50–0112), CD86 (clone GL1) FITC (eBioscience, catalog no. 11–0862–85). Samples were acquired using a BD LSR II Fortessa instrument and analyzed with FlowJo software (TreeStar).
Histology and IHC
After formalin fixation, scramble or Jmjd6 KO PyMT-41C primary tumors were formalin-fixed, paraffin-embedded (FFPE). FFPE sections of 4 μmol/L were stained with Mayer-hematoxylin and eosin (H&E). Alternatively, after re-hydratation and antigen retrieval, IHC was performed using the streptavidin–biotin–peroxidase complex method, and 3,3′-Diaminobenzidine tetrahydrochloride as chromogenic substrate, according to standard protocols. Anti-ANXA1 (Genetex, 113329) was used as the primary antibody.
Clariom array and exon array analysis
Total RNA was extracted using Qiazol reagent and purified with the RNeasy kit on a QIAcube automated station (Qiagen) according to the manufacturer's instructions. The quantity and quality of the extracted RNA were determined with 4200 TapeStation system (Agilent) and Qubit Fluorometer (ThermoFisher), respectively. Clariom S assay human (for MCF7 and MDA-MB231 cells) and Clariom D mouse assay (for PyMT-41C cells; Affymetrix) were used for gene expression profiling. Briefly, total RNA was amplified, labeled, and hybridized following the manufacturer's instructions. The Affymetrix GeneChip WT Pico Kit was used for cDNA preparation, biotin labeling, and cRNA synthesis. The arrays were subsequently incubated for 16 hours in an Affymetrix GeneChip 645 hybridization oven at 45°C with rotation at 60 rpm and processed with the GeneChip Wash and Stain kit. The chips were scanned with an Affymetrix GeneChip Scanner 3000 with default manufacturers’ settings and raw data were acquired with the AGCC scan control v4.0.0. Raw data were imported in R software and a RMA normalization (28) was performed exploiting the oligo package. After quality control, multiple probes matching the same gene were collapsed with function collapseRows from the WGCNA package. Differentially expressed gene (DEG) were identified with the limma package (29) and P value was corrected with Benjamini–Hochberg method; genes were considered significant with FDR < 0.05. From class comparison results, pre-ranked gene set enrichment analysis (GSEA) was performed to calculate which hallmark pathways were significantly up- or down-modulated (FDR < 0.05).
In silico analyses
The list of genes identified in the GSEA of ‘Hallmark of Adipogenesis’ obtained from ClariomS performed on MCF7 breast cancer cells were run on the free, open-source, curated, and peer-reviewed ‘Reactome Pathway Database’, and can be found at the following link: https://reactome.org/PathwayBrowser/#/ANALYSIS = MjAyMTEyMjcxNjU0MTVfMzk0.
Graphs and statistical analysis
Graphs were analyzed and plotted with Graphpad Prism 9. All experiments have been performed at least in 3 independent experiments and plotted as average ± SE% or SD. P values are shown in each graph. If asterisks are shown *, P < 0.005; **, P < 0.01; ***, P < 0.001.
Data availability
The transcriptome data generated in this study have been deposited in the National Center for Biotechnology Information in the Gene Expression Omnibus database under the accession number GSE164399, GSE216234, and GSE216235.
Results
JMJD6 depletion increases ERα expression without enhancing its activity
We investigated the potential clinical value of JMJD6 in patients with breast cancer included in the METABRIC dataset (30) by correlating the expression levels of JMJD6 with patients’ overall survival (OS). High levels of JMJD6 were associated with significantly shorter OS (Fig. 1A), suggesting that JMJD6 might support breast cancer development and progression. However, such correlation was significant only in ERα-positive patients, while no impact on OS was found in ERα-negative patients. This suggests that JMJD6 effects might dependent on ERα expression and activity. Because patients with ERα-positive tumors can co-express HER2 receptor, we checked if these patients also showed different HER2 expression levels based on JMJD6 classification, which might bias the analysis. As shown in Supplementary Table S1 and as expected, HER2-positive patients were few within the two groups (62 HER2+ vs. 661 HER2− in JMJD6 high patients and 42 HER2+ vs. 675 HER2− in JMJD6 low), hence slightly impacting the survival analysis.
To further explore the correlation between JMJD6 and ERα, we generated JMJD6 KO breast cancer MCF7 and MDA-MB231 cells (ERα-positive and ERα-negative cell line, respectively) with CRISPR-Cas9 technology. As shown in Fig. 1B, KO of JMJD6 in MCF7 cells leads to upregulation of ERα protein levels. This evidence was also confirmed in another human cell line, BT474, in which, in mass cultures and at least in one clone, ERα was increased by JMJD6 KO (Supplementary Fig. S1A). No expression of ERα was detected in MDA-MB231 independently of JMJD6 expression (Fig. 1B).
Because the levels of ERα are upregulated in the presence of reduced JMJD6 levels, we asked whether ERα is also more active. To this end, we investigated its localization in unstimulated conditions and upon β2-estradiol (E2) stimulation, in cells cultured in phenol red–deprived medium supplemented with DCC (Fig. 1C) or in normal medium. In any condition tested, JMJD6 depletion did not affect ERα localization. Moreover, although ERα is more expressed at protein (Fig. 1B) and transcription (Fig. 1D) level, its activity is not enhanced. In fact, the expression levels of a number of genes, which are known to be transcribed by ERα, are significantly reduced in unstimulated conditions (PGR, XBP1, and RARA; Fig. 1E) and are not upregulated more than control Scramble cells upon E2 stimulation. Hence, despite the fact that ERα is more expressed in JMJD6 KO cells (Fig. 1D), its transcriptional activity is not increased (Fig. 1E). Altogether, these results show that JMJD6 is able to tune ERα expression and function, and its depletion increases ERα levels without augmenting its transactivation activity.
JMJD6 controls different biological pathways in MCF7 and MDA-MB231 cells
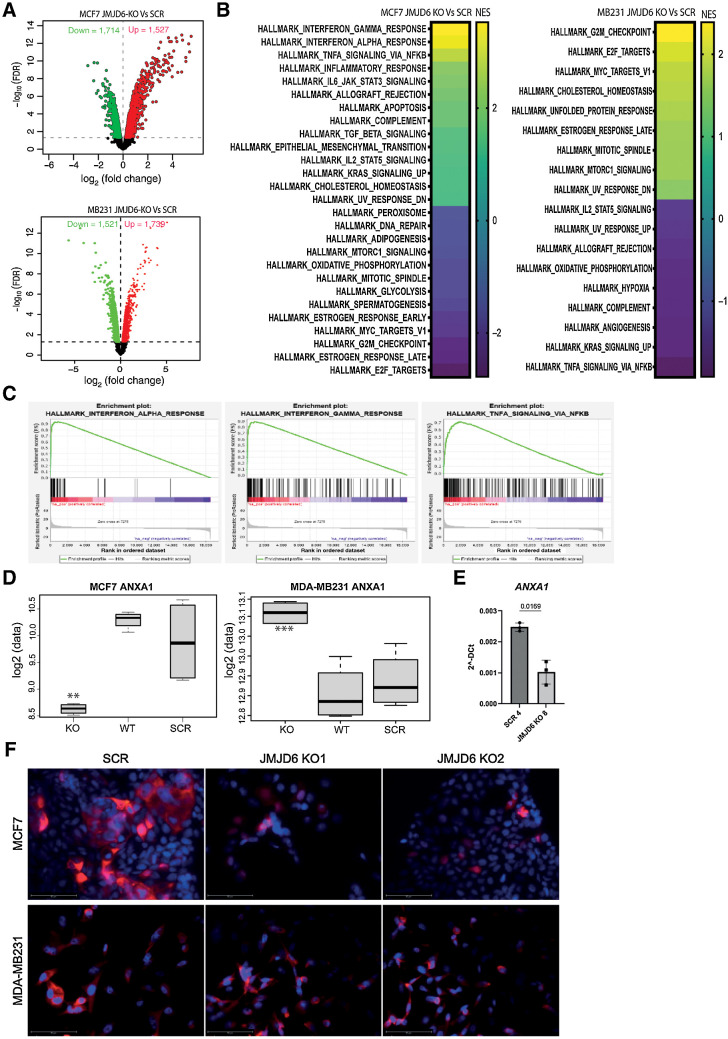
To better explore the role of JMJD6 expression in MCF7 and MDA-MB231 cells, we performed microarray profiling of both cell lines. Two different clones were used for each scramble and KO conditions. As shown in the volcano plot in Fig. 2A and listed in Supplementary Table S2A, 3241 genes were found differentially expressed between JMJD6 KO and scramble MCF7 cells, while 3,260 genes were differentially expressed in MDA-MB231 pair comparison. DEGs were also analyzed by comparing WT (parental) and scramble MCF7 and MDA-MB231 cells as an additional control (Supplementary Fig. S1B). DEGs emerging from JMJD6 KO versus scramble in either MCF7 or MDA-MB231 were used to perform GSEA and the most significant up- or downregulated biological pathways are listed on the basis of normalized enrichment score (NES) in Fig. 2B. Complete list of significant (FDR < 0.05) GSEA pathways is provided in Supplementary Table S2. The vast majority of the enriched biological pathways in JMJD6 KO MCF7 involved inflammatory mechanisms, including IFNα and IFNγ response, and TNFα signaling via NF-κB (Fig. 2C), suggesting that loss of JMJD6 could affect the immune composition of tumor microenvironment (TME) and therefore the inflammatory response. Contrarily, IFN pathways were not affected by JMJD6 KO in MDA-MB231 cells.
Figure 2.
Transcriptome profile of scramble and JMJD6 KO MCF7 and MDA-MB231 cells. A, Volcano plot showing statistical significance values against fold-change and highlighting the most upregulated (red) and downregulated (green) genes between JMJD6 KO and scramble MCF7 (top), and MDA-MB231 (bottom) cells. B, GSEA analysis of the most significantly up- or downregulated hallmark pathways in JMJD6 KO MCF7 (left) and MDA-MB231 (right) cells compared with scramble control. C, GSEA plots showing enrichment of genes associated with inflammatory pathways in JMJD6 KO MCF7 cells compared with scramble control. Significant GSEA pathways are defined on the basis of FDR < 0.05. D, mRNA expression of ANXA1 in WT, scramble and JMJD6 KO MCF7 and MDA-MB231 cells. t test was performed between JMJD6 KO MCF7 and scramble cells, and between JMJD6 KO and scramble cells. **, P < 0.01; ***, P < 0.001. E, Real Time PCR showing the levels of ANXA1 in control and JMJD6 KO clones. Statistical significance was calculated by paired two-tailed t test. F, Immunofluorescence staining showing the expression of ANXA1 (red) in scramble or JMJD6 KO MCF7 and MDA-MB231 cell clones. Nuclei were stained with DAPI (blue), bars = 50 μmol/L.
On the basis of a publicly available dataset (12) and further validated in our own dataset, ANXA1 was found downregulated in JMJD6 KO MCF7 compared with both scramble and WT MCF7 cells. On the contrary, in JMJD6 KO MDA-MB231 cells, increased levels of ANXA1 were detected compared with controls (Fig. 2D). The reduction of ANXA1 in JMJD6 depleted cells was further validated by Real Time PCR (Fig. 2E). As ANXA1 is involved in several inflammatory processes (31, 32), we focused on its regulation by JMJD6. We found that ANXA1 is generally localized in the cytoplasm and membranes of WT and scramble MCF7 cells, whereas the cytoplasmic accumulation was massively reduced in the JMJD6 KO counterpart (Fig. 2F).
JMJD6 promotes LD formation
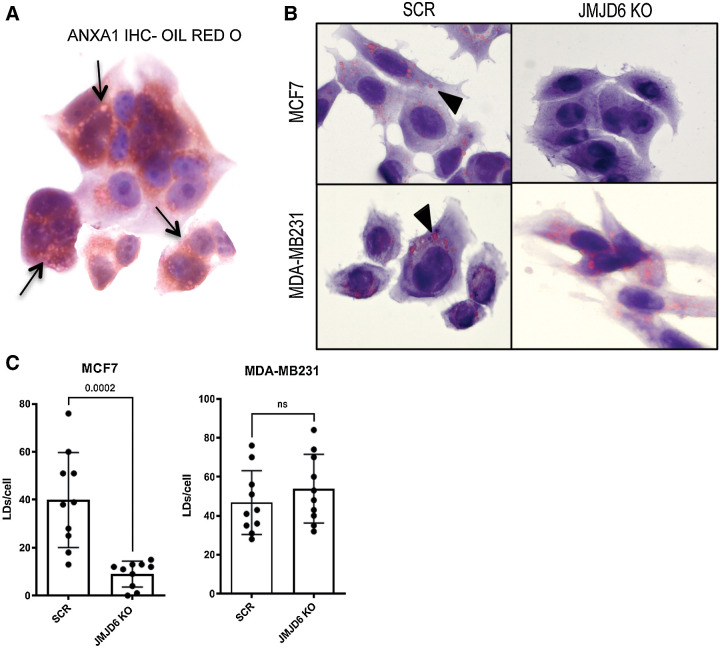
ANXA1 has been shown to be stored in LD, whose formation has recently been shown to be promoted by JMJD6 (22). Therefore, we explored the possibility that JMJD6 might affect LD formation and consequent ANXA1 levels also in MCF7 cells. LDs were stained with Oil Red O and anti-ANXA1 antibody showing their co-localization (Fig. 3A; Supplementary Fig. S2A).
Figure 3.
ANXA1 is accumulated in LDs. A, IHC staining of ANXA1 combined with Oil Red O staining of WT MCF7 cells. Black arrows show colocalization areas. B, Oil Red O stained scramble or JMJD6 KO MCF7 and MDA-MB231 cells. Black arrows show LDs. C, Quantification of LDs shown in panel B. P values were calculated by unpaired two-tailed t test.
These data indicate that JMJD6 loss could impair ANXA1 levels not only through direct transcriptional downregulation, but also through impaired LD formation. Notably, the reduced LD formation, occurring in JMJD6 KO MCF7 cells, was not observed in JMJD6 KO MDA-MB231 cells (Fig. 3B and C), suggesting that the effect of JMJD6 in LD formation could be cell-specific and likely ERα-dependent. To identify possible determinants of LD formation regulated by JMJD6, we analyzed the DEGs obtained by comparing scramble and JMJD6 KO MCF7, and we focused on genes belonging to the ‘Hallmark of Adipogenesis’ (Supplementary Fig. S2B) which we found significantly modulated and which is enriched by genes related to lipid metabolism. By using the Reactome browser, we identified PPARα-associated pathways among the most affected (Supplementary Fig. S2C).
JMJD6 promotes ANXA1 expression in an ERα/PPARα-dependent manner
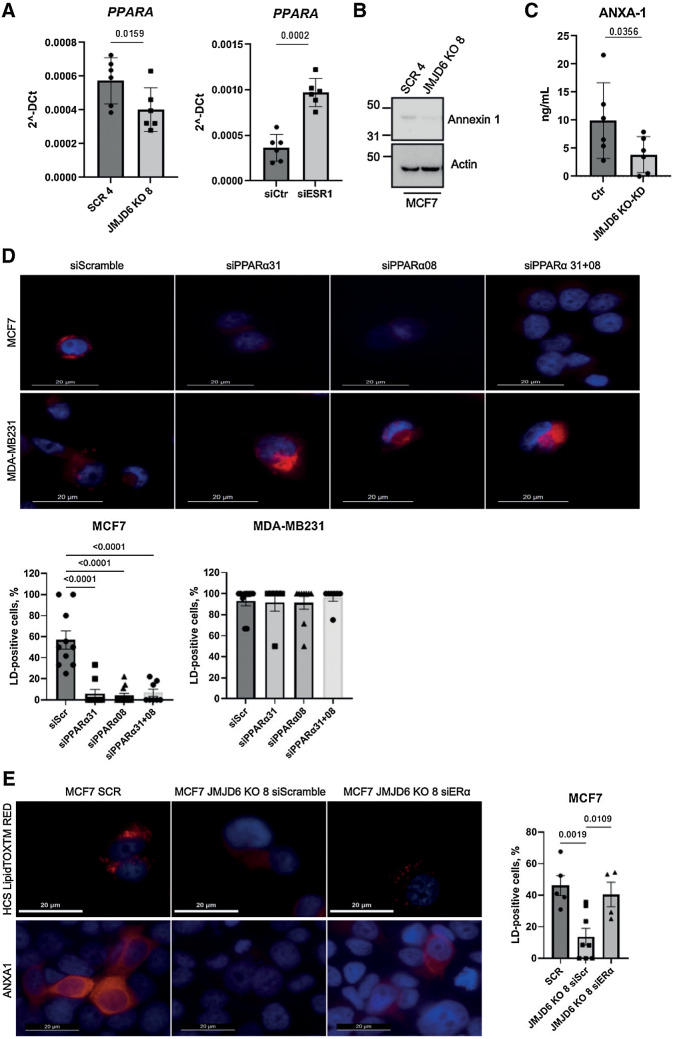
Having shown that JMJD6 is necessary for LD formation and that this could affect ANXA1 levels and localization, we investigated the mechanisms underlying the observed LD reduction in JMJD6 KO MCF7 cells. Because the PPARα pathway resulted to be affected by JMJD6 (Supplementary Fig. S2C), we tested whether JMJD6 depletion results in a reduction of PPARα expression, confirming a reduction at a transcription level (Fig. 4A, left). We have previously shown that JMJD6 reduction results in an increase of ERα expression and protein levels (Fig. 1B and D), we then asked whether this could be responsible for PPARα reduction. In agreement with our hypothesis, we found that the silencing of ERα indeed increases PPARα expression (Fig. 4A, right). The genetic inhibition of JMJD6 results in reduction of ANXA1 protein levels (Fig. 4B), as already show at a transcriptional levels (Fig. 2E), and release in the medium (Fig. 4C). To investigate the involvement of ERα in PPARα-dependent LD formation (and consequent ANXA1 accumulation and release), we compared MCF7 and MDA-MB231 cells, finding that PPARα silencing leads to loss of LDs in MCF7, but not in MDA-MB231 cells (Fig. 4D). Moreover, being ERα upregulated in MCF7 JMJD6 KO cells, which display defective LD formation (Fig. 4D) and reduced PPARα expression (Fig. 4A, left), we tested whether ERα silencing could restore LD formation. In agreement with our hypothesis, silencing of ERα restored LD formation and ANXA1 expression in MCF7 depleted for JMJD6 (Fig. 4E).
Figure 4.
PPARα regulates LD droplet formation and ANXA1 levels via ERα signaling. A, Real time PCR showing the expression of PPARα in control and JMJD6 KO MCF7 clones (left) and in MCF7 silenced for ERα (right). Graphs show the average of six independent experiments. P values were calculated by paired two-tailed t test. B, Western blot showing the levels of ANXA1 in MCF7 control or JMJD6 KO clones. Actin-β was used as loading control. C, ANXA1 ELISA performed with CM from control (n = 6) and JMJD6 KD (n = 3) and KO (n = 3) collected in independent experiments. P values were calculated by paired two-tailed t test. D, Fluorescence images showing the presence of LDs (HCS LipidTOX RED) in MCF7 and MDA-MB231 cells silenced for PPARα. Nuclei were stained with DAPI (blue), bars = 20 μmol/L. Graphs show the percentage of LD-positive cells/field; cells from at least six independent fields were counted. Statistical significance was evaluated by ordinary one-way Anova, multiple comparisons test. E, Fluorescence images showing the presence of LDs and ANXA1 (red) in scramble and JMJD6 KO MCF7 cells silenced for ERα. Nuclei were stained with DAPI (blue), bars = 20 μmol/L. Graph shows the percentage of LD-positive cells/field; cells from at least four independent fields were counted. Statistical significance was evaluated by ordinary one-way Anova, multiple comparisons test.
JMJD6-mediated ANXA1 affects macrophage differentiation
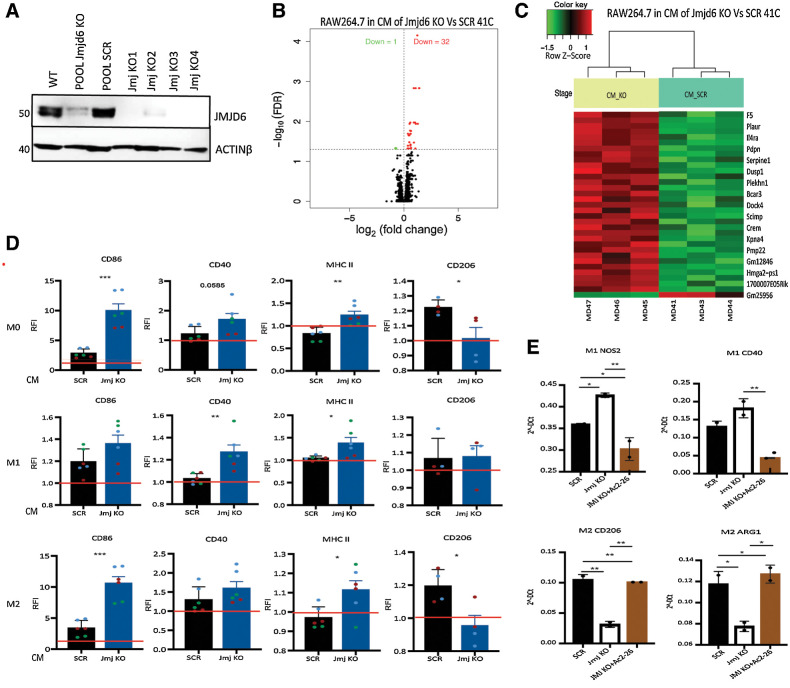
Having shown that JMJD6 controls ANXA1 levels via ERα and knowing the role of ANXA1 in inflammation and macrophage differentiation, we tested whether JMJD6 may represent a regulator of the TME. To this end, we employed the PyMT-41C (41C) cell line, which was derived from a spontaneous mammary tumor of a MMTV-PyMT transgenic mouse and hence allows to study this mammary tumor in a syngeneic immunocompetent context. Cells either scramble or KO for Jmjd6 were used and characterized to verify the depletion of JMJD6 (Fig. 5A). Moreover, by silencing, we confirmed that JMJD6 reduction correlates with reduced ANXA1 cytoplasmic accumulation and secretion, and hampers LD formation (Supplementary S3A–S3D). CM from 41C cells was added to RAW264.7 macrophages, which were subsequently profiled using microarray gene expression technique. As shown in the volcano plot in Fig. 5B, 32 genes were upregulated and 1 gene downregulated in RAW264.7 cells cultured in CM from Jmjd6 KO 41C versus scramble cells (adj. value < 0.05). The heatmap in Fig. 5C shows the most significant DEGs between the two groups. GSEA highlighted several inflammatory pathways upregulated in RAW264.7 cultured with CM of Jmjd6 KO cells, suggesting that loss of JMJD6 in tumor cells might promote the differentiation of pro-inflammatory macrophages, thus, potentially affecting tumor growth.
Figure 5.
ANXA1 released by tumor cells affects macrophage differentiation in vitro. A, Western blot showing the expression of JMJD6 in PyMT-41C cells in different KO clones. Actin-β was used as housekeeping protein expression. B, Volcano plot of statistical significance against fold-change between RAW264.7 macrophages cultured in CM of Jmjd6 KO or scramble PyMT-41C cells showing the significantly upregulated (32, red) and downregulated (1, green) genes. C, Heatmap shows the degree of clustering between biological replicates and distinct pattern of gene expression level between RAW264.7 macrophages cultured in CM of Jmjd6 KO (yellow) and scramble PyMT-41C cells (light green) cells. Color coding green to red shows the z-score value from -1.5 to 1. D, Relative fluorescence intensity (RFI) plots showing the expression level of M1 and M2 markers in M0, M1, and M2 BMDMs in CM of scramble and Jmjd6 KO PyMT-41C cells. Data were normalized on macrophages cultured without CM (red bar). Datapoints show the biological replicates of three independent experiments (green, blue and red) with two biological replicates in each experiment. Error bars show the SEM. P value is calculated with one-way ANOVA, Tukey multiple comparison test. E, Real-time PCR showing the expression of M1 markers (Cd40 and Nos2) and M2 markers (Cd206 and Arg1) in M1 and M2 differentiated BMDMs cultured in CM of scramble and Jmjd6 KO PyMT-41C cells, with or without the ANXA1 mimetic peptide Ac2–26.
To further explore this hypothesis, bone marrow-derived macrophages (BMDM) were generated from C57BL/6 female mice and differentiated into M0, M1, and M2 phenotypes. Macrophages were then cultured with CM of Jmjd6 KO 41C cells, scramble or normal medium and then characterized by FACS analysis for changes in expression of common M1 (CD86, CD40, and MHC-II) or M2 markers (CD206; Fig. 5D; Supplementary Fig. S4A and S4B). We found that, while CM from scramble tumor cells stimulated the expression of M2 marker CD206, CM from Jmjd6 KO cells impeded M2 differentiation and enhanced the expression of M1 makers CD86, CD40, and MHC II in M0 macrophages. CM medium from Jmjd6 KO cells given to previously M1 differentiated cells further increased M1 markers without modifying CD206 expression, whereas, when given to M2 macrophages, a certain degree of conversion to M1 was observed according to the decrease expression of CD206 and increase of M1 markers.
To test the hypothesis that Jmjd6 KO-dependent ANXA1 reduction is indeed responsible for the increase of M1 polarization, we added the ANXA1 agonist peptide Ac2–26 to the CM of Jmjd6 KO 41C cells. As expected, the presence of Ac2–26 could successfully revert M1 to M2 polarization, analyzed by evaluating the mRNA expression of classical M1 and M2 markers, including Cd40 and Nos2 for M1 and Cd206 and Arginase-1 (Arg1) for M2. Addition of Ac2–26 into CM of Jmjd6-KO 41C cells successfully restored the expression of M1 and M2 markers in M1 and M2 macrophages respectively, to comparable levels of CM of scramble 41C cells (Fig. 5E). These results support a role for tumor expressing JMJD6 in conditioning macrophage polarization towards a pro-tumoral M2-type via ANXA1 release.
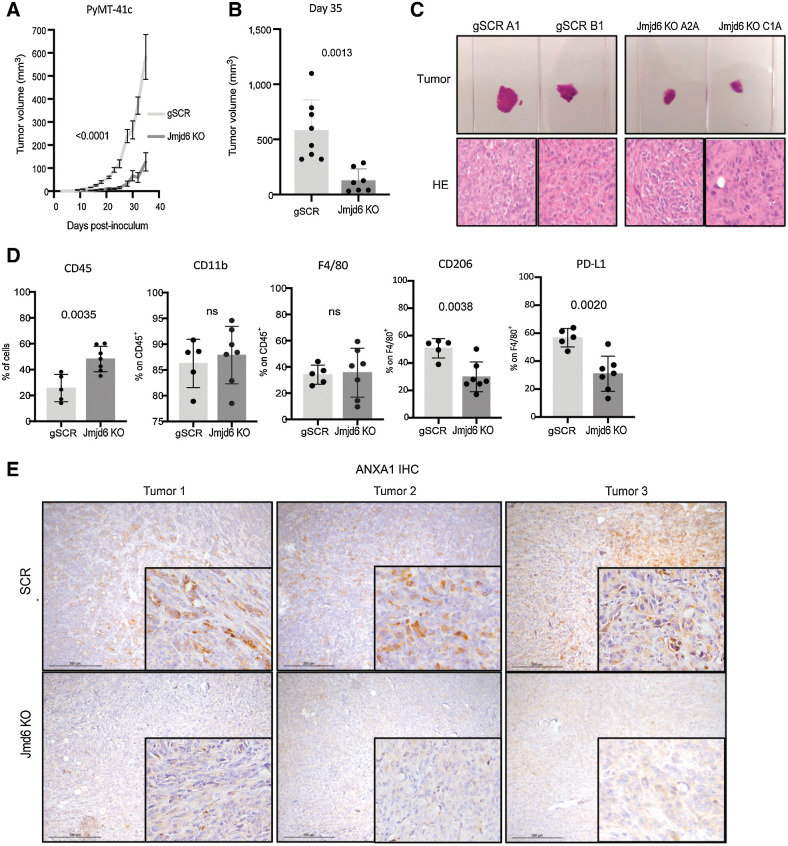
JMJD6 affects macrophage differentiation in vivo and promotes tumor growth
To validate our findings in vivo, scramble or Jmjd6 KO 41C cells were inoculated into syngeneic C57BL/6 female mice. Tumors generated from scramble 41C cells showed faster growth rate and increased volume compared with Jmjd6 KO 41C counterpart (Fig. 6A–C). On the basis of our in vitro data, this result might, at least in part, depend on the different macrophage subtypes infiltrating the two groups of tumor. Flow cytometry performed on these tumors showed a higher percentage of CD45+ leukocytes in Jmjd6-KO 41C tumors than scramble controls. The fraction of CD11b+F4/80+ macrophages was similar in the two groups. However, the percentage of CD206+F4/80+ and PD-L1+F4/80+ macrophages was significantly reduced in tumors from Jmjd6 KO 41C cells compared with scramble control, suggesting a reduced skewing toward an M2-like, immune suppressive phenotype (Fig. 6D). Flow cytometry gating strategy is shown in Supplementary Fig. S4C.
Figure 6.
Tumor cell JMJD6 supports M2 macrophage differentiation and tumor growth in breast cancer mouse model. A, Tumor growth after inoculation of scramble (light gray) and Jmjd6 KO (dark gray) PyMT-41C cells in C57BL/6 mice. P value is calculated with two-way ANOVA, Sidak multiple comparison test. Error bars shows S.D. B, Tumor volume of scramble (light gray) and Jmjd6 KO (dark gray) PyMT-41C cells in C57BL/6 mice at the experimental endpoint of 35 days. P value is calculated with two-way ANOVA, Sidak multiple comparison test. Each dot represents a different mouse. Error bars shows SD among samples. C, Top, Representative images of two scramble PyMT-41C cells-derived tumors (clone A1 and clone B1) and two Jmjd6-KO PyMT-41C cells-derived tumors (clone A2A and clone C1A). Below, H&E staining of the different tumors. D, Percentage of CD45+ immune cells, CD11b+ myeloid cells, F4/80+ monocytes, macrophages, CD206+ M2-like macrophages over F4/80+ macrophages and PD-L1+ immunosuppressive macrophages over F4/80+ macrophages in tumors derived from scramble or Jmjd6 KO PyMT-41C cells. P value is calculated with unpaired t test. Each dot represents a different mouse. Error bars show SD among samples. E, IHC analysis of ANXA1 expression in tumors derived from scramble and Jmjd6 KO PyMT-41C cells injected in C57BL/6 mice.
Furthermore, expression of ANXA1 in Jmjd6 KO 41C tumors was profoundly diminished compared with scramble 41C tumors (Fig. 6E), confirming that also in vivo the Jmjd6 genetic deletion in tumor cells leads to ANXA1 reduction. The latter event could in turn be responsible for augmented pro-inflammatory M1 macrophage portion in the tumors which reduces their growth.
Discussion
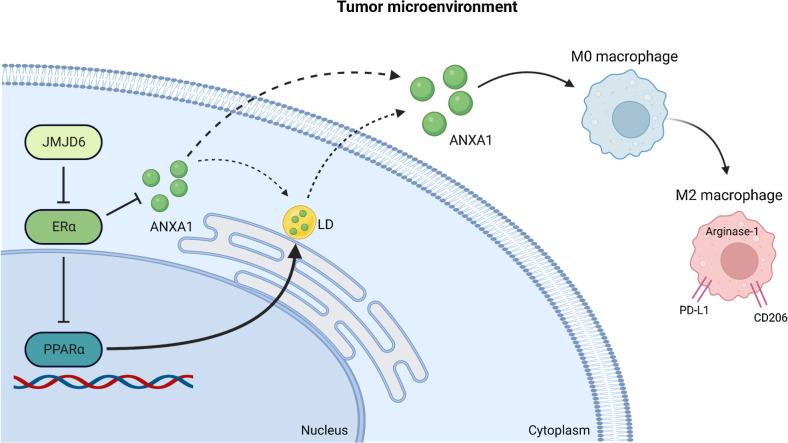
Our work supports the notion that JMJD6 affects breast cancer prognosis not only by affecting cancer cell-intrinsic mechanisms, but also by molding the TME. In fact, we show that JMJD6 favors ANXA1 release by cancer cells triggering the conversion of macrophages to a pro-tumor M2-like state. Our data suggests that JMJD6, on the one hand, promotes the expression of ANXA1, on the other, it promotes its protein accumulation via LD formation, through a pathway involving PPARα and ERα (Fig. 7). The latter evidence suggests that JMJD6 could regulate these pathways in breast cancer via different mechanisms according to the expression of ERα, which is the crucial marker for defining breast cancer subtypes.
Figure 7.
JMJD6 molds TME through ANXA1-dependent macrophages polarization. JMJD6 favors ANXA1 expression, intracellular accumulation, through a pathway involving PPARα and ERα, and its increase in the TME. ANXA1 released by cancer cells triggers the conversion of macrophages to a pro-tumor M2-like state which favors tumor development.
This observation could contribute to explain why JMJD6 is endowed with a negative prognostic factor mainly in luminal, ERα-positive, breast cancer. Notably, JMJD6 has already been proposed as a regulator of ERα transactivation activity by forming a complex with it (14) and by regulating ERα posttranslational modification, via methylation (7). JMJD6 is usually associated to breast cancer aggressiveness due to cell-intrinsic mechanisms by promoting cell proliferation and motility (12), cell transformation and tumor progression (13), DNA damage response (33, 34) and cell cycle (35). All these activities have usually been linked to its enzymatic arginine demethylase and lysine hydroxylase activity. In few cases, we and others have showed also enzymatic-independent activities (34, 36).
In our previous studies, we found that JMJD6 can be secreted into the extracellular space, where it binds with collagen contributing to tissue fibrosis and stiffness (36). Antibody-mediated inhibition of JMJD6-Collagen interaction reduces fibrosis and leads to increased CD45+ leukocyte tumor-infiltration, suggesting that JMJD6 might suppress the inflammatory response. In the current study, we describe another mechanism by which JMJD6 regulates the immune infiltrate and, in particular, macrophage polarization. We show that JMJD6 promotes the expression of ANXA1 in a LD-dependent manner and its release. Although we do not provide direct evidence that LD loss caused by JMJD6 depletion is responsible for the reduced secretion of ANXA1, several lines of evidence support our hypothesis. In fact, it has previously been shown that ANXA1 is accumulated in LDs (20) and also our findings confirm a co-localization between LD and ANXA1. Moreover, in our settings, the defect of LD formation found upon PPARα silencing recapitulates JMJD6 KO and results in reduction of intracellular ANXA1 accumulation.
Notably, during the revision of this manuscript, another group demonstrated that JMJD6 regulates LD formation in other tumor types (22) by promoting the expression of DGAT1. In our cell lines, we did not confirm a JMJD6-dependent regulation of DGAT1, but we identified PPARα as a crucial mediator of JMJD6-dependent LD formation. This discrepancy could be due to ERα expression that as we observed by comparing ER-positive MCF7 and MDA-MB231 cells and through genetic inhibition of ERα, could represent a discriminating factor which determines the pathways actually controlled by JMJD6.
The latter notion is not surprising considering that many pieces of evidence support a relation between ANXA1, lipid metabolism and hormones. For instance, ANXA1 expression was found increased in adipose tissue in mice fed with a high-fat diet (37), in obese children (38), and in the subcutaneous fat of overweight patients (39). At the same time, the correlation between estrogens, their receptors and lipid metabolism have been widely described. Treatment of female mice with β2-estradiol leads to decreased expression of lipogenic genes in adipose tissue (40), and ERα-null female mice show increased fat, adipocyte number, and cholesterol levels (21). ER activation reduces lipid synthesis in pancreatic islets and hepatic cells (41, 42). Postmenopausal women have increased body adiposity and are more at risk of developing atherosclerosis in association with reduced estrogen levels (43). Hence, due to its crucial role in these mechanisms, ERα could discriminate how and if JMJD6 regulates LDs and, in turn, ANXA1 metabolism.
In conclusion, our results uncover a novel function of JMJD6 in molding the TME and provide a rationale for developing inhibitory molecules against JMJD6 to reduce disease progression. These first-in-class compounds could be effective in breast cancer ERα-positive therapy not only by direct targeting of cancer cells, but also by molding the immune infiltrate and skewing macrophages towards an antitumor, pro-inflammatory state.
Supplementary Material
List of BC patients with ER-positive tumors included in the METABRIC dataset
Effect of JMJD6 KO in ER positive and negative cell lines. A) Western blot showing the expression of JMJD6 and ERα in wild-type, scramble or JMJD6 KO BT474 cells both in mass cultures (Left panel) and in single clones (right panel). Vinculin is shown as loading control. B) Volcano plot showing the most upregulated (red) and downregulated (green) genes between scramble and wild-type (parental) MCF7 (left) and MDA-MB231 (right) cells.
JMJD6 regulates lipid metabolism in MCF7 cells. A) Immunohistochemistry staining for ANXA1 (brown) combined with Oil red-O staining (pink) for lipid droplets visualization in wild-type MCF7 cells B) Gene set enrichment plot for 'Hallmark of Adipogenesis' found to be significantly downregulated in JMJD6 KO MCF7 cells compared to scramble control. C) List of 'Reactome Pathways' found to be regulated by the genes included in the gene set enrichment pathway of 'Hallmark of Adipogenesis' and present in the DEGs list of JMJD6 KO versus scramble MCF7 cells.
Significant enriched pathways in JMJD6-KO MCF7 and MDA-MB231 cells
Effect of Jmjd6 silencing in PyMT41C-cells. A) Real-Time PCR showing the levels of Jmjd6 in PyMT-41C cells transfected with non-targeting siRNA (siCtr) or specific for Jmjd6. Graph is the average of two independent experiments. Two-tailed unpaired t student test was applied to calculate statistical significance. B) Western blot performed on conditioned medium collected from PyMT-41C transfected as in A). Red Ponceau is shown as loading control. C) IF to detect ANXA1 levels and D) lipid droplet formation in cell transfected as in A).
Jmjd6-mediated ANXA1 secretion promotes M2 macrophage differentiation in vitro and ex vivo. A) Mean fluorescence intensity (MFI) of M1 and M2 markers in BMDM differentiated into M0, M1 and M2.** p<0,001, *** means p<0,0001, **** p<0,00001 without conditioned medium. B) Real time PCR showing the expression M1 (CD40 and NOS2) and M2 (CD206 and ARG1) markers in BMDMs differentiated into M0, M1 and M2 macrophages without CM. Datapoints show technical replicates.** p<0,001, *** p<0,0001. C) Gating strategies for FACS analysis on ex vivo mouse samples of Figure 6D. CD45+ leukocytes were gated on live cells, excluding dead cells and debris. In CD45+ population, myeloid cells were analyzed by the expression of CD11b, Gr-1, Ly6C and F4/80 markers. Monocyte-like myleloid-derived immunosuppressive cells (M-MDSC) were identified as Ly6Chigh Gr-1int expression. Granulocyte-like myeloid-derived immunosuppressive cells (G-MDSC) were identified as Gr-1high Ly6Cint cells. M2 macrophages were identified as CD206+ cells in the F4/80 gate. G-MDSC were identified as Gr-1high Ly6Cint cells. M2 macrophages were identified as CD206+ cells in the F4/80 gate.
Acknowledgments
The authors thank Lorena Ventura for helping with IHC and Dr. Elena Jachetti for providing antibodies for flow cytometry analysis and for helping with experimental settings. Enrico Fontanella, Nicolò Mercurio, and Roberta Bongiorno provided technical support during the revision of the manuscript.
B. Cioni is supported by the iCARE-2 fellowship program funded by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 800924, project code 23866. M.P. Colombo is supported by AIRC grant IG2020 no. 24363 and from Italian Ministry of Health “Ricerca Corrente”.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
B. Cioni: Conceptualization, investigation, methodology, writing–original draft, project administration. S. Ratti: Validation, investigation. A. Piva: Data curation, software. I. Tripodi: Data curation, software. M. Milani: Data curation, software. F. Menichetti: Investigation. T. Langella: Investigation. L. Botti: Resources, methodology. L. De Cecco: Data curation. C. Chiodoni: Conceptualization, writing–review and editing. D. Lecis: Conceptualization, supervision, writing–review and editing. M.P. Colombo: Conceptualization, supervision.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, 2019. CA Cancer J Clin 2019;69:438–51. [DOI] [PubMed] [Google Scholar]
- 3. Carter CL, Allen C, Henson DE. Relation of tumor size, lymph node status, and survival in 24,740 breast cancer cases. Cancer 1989;63:181–7. [DOI] [PubMed] [Google Scholar]
- 4. Andre F, Pusztai L. Molecular classification of breast cancer: implications for selection of adjuvant chemotherapy. Nat Clin Pract Oncol 2006;3:621–32. [DOI] [PubMed] [Google Scholar]
- 5. Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod Pathol 2010;23:S60–4. [DOI] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumors. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poulard C, Rambaud J, Hussein N, Corbo L, Le Romancer M. JMJD6 regulates ERalpha methylation on arginine. PLoS One 2014;9:e87982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang J, Ni SS, Zhao WL, Dong XC, Wang JL. High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol 2013;34:2397–401. [DOI] [PubMed] [Google Scholar]
- 9. Wang F, He L, Huangyang P, Liang J, Si W, Yan R, et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol 2014;12:e1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wan J, Liu H, Yang L, Ma L, Liu J, Ming L. JMJD6 promotes hepatocellular carcinoma carcinogenesis by targeting CDK4. Int J Cancer 2019;144:2489–500. [DOI] [PubMed] [Google Scholar]
- 11. Poulard C, Rambaud J, Lavergne E, Jacquemetton J, Renoir JM, Tredan O, et al. Role of JMJD6 in breast tumorigenesis. PLoS One 2015;10:e0126181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee YF, Miller LD, Chan XB, Black MA, Pang B, Ong CW, et al. JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res 2012;14:R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aprelikova O, Chen K, El Touny LH, Brignatz-Guittard C, Han J, Qiu T, et al. The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-myc to enhance cellular transformation, tumor progression, and metastasis. Clin Epigenetics 2016;8:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao WW, Xiao RQ, Zhang WJ, Hu YR, Peng BL, Li WJ, et al. JMJD6 licenses ERalpha-dependent enhancer and coding gene activation by modulating the recruitment of the CARM1/MED12 co-activator complex. Mol Cell 2018;70:340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Y, Cai L, Wang H, Wu P, Gu W, Chen Y, et al. Pleiotropic regulation of macrophage polarization and tumorigenesis by formyl peptide receptor-2. Oncogene 2011;30:3887–99. [DOI] [PubMed] [Google Scholar]
- 16. Ferlazzo V, D'Agostino P, Milano S, Caruso R, Feo S, Cillari E, et al. Anti-inflammatory effects of annexin-1: Stimulation of IL10 release and inhibition of nitric oxide synthesis. Int Immunopharmacol 2003;3:1363–9. [DOI] [PubMed] [Google Scholar]
- 17. Moraes LA, Kar S, Foo SL, Gu T, Toh YQ, Ampomah PB, et al. Annexin-A1 enhances breast cancer growth and migration by promoting alternative macrophage polarization in the tumor microenvironment. Sci Rep 2017;7:17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McArthur S, Juban G, Gobbetti T, Desgeorges T, Theret M, Gondin J, et al. Annexin A1 drives macrophage skewing to accelerate muscle regeneration through AMPK activation. J Clin Invest 2020;130:1156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Khor VK, Ahrends R, Lin Y, Shen WJ, Adams CM, Roseman AN, et al. The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One 2014;9:e105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kalinec F, Webster P, Maricle A, Guerrero D, Chakravarti DN, Chakravarti B, et al. Glucocorticoid-stimulated, transcription-independent release of annexin A1 by cochlear Hensen cells. Br J Pharmacol 2009;158:1820–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barros RP, Gustafsson JA. Estrogen receptors and the metabolic network. Cell Metab 2011;14:289–99. [DOI] [PubMed] [Google Scholar]
- 22. Zhou J, Simon JM, Liao C, Zhang C, Hu L, Zurlo G, et al. An oncogenic JMJD6-DGAT1 axis tunes the epigenetic regulation of lipid droplet formation in clear cell renal cell carcinoma. Mol Cell 2022;82:3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lau C, Abbott BD, Corton JC, Cunningham ML. PPARs and xenobiotic-induced adverse effects: relevance to human health. PPAR Res 2010;2010:954639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stojanovic O, Altirriba J, Rigo D, Spiljar M, Evrard E, Roska B, et al. Dietary excess regulates absorption and surface of gut epithelium through intestinal PPARα. Nat Commun 2021;12:7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma H, Sprecher HW, Kolattukudy PE. Estrogen-induced production of a peroxisome proliferator-activated receptor (PPAR) ligand in a PPARgamma-expressing tissue. J Biol Chem 1998;273:30131–8. [DOI] [PubMed] [Google Scholar]
- 26. Faddy HM, Robinson JA, Lee WJ, Holman NA, Monteith GR, Roberts-Thomson SJ. Peroxisome proliferator-activated receptor alpha expression is regulated by estrogen receptor alpha and modulates the response of MCF-7 cells to sodium butyrate. Int J Biochem Cell Biol 2006;38:255–66. [DOI] [PubMed] [Google Scholar]
- 27. Majorini MT, Cancila V, Rigoni A, Botti L, Dugo M, Triulzi T, et al. Infiltrating mast cell-mediated stimulation of estrogen receptor activity in breast cancer cells promotes the luminal phenotype. Cancer Res 2020;80:2311–24. [DOI] [PubMed] [Google Scholar]
- 28. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–64. [DOI] [PubMed] [Google Scholar]
- 29. Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 2016;10:946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumors reveals novel subgroups. Nature 2012;486:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugimoto MA, Vago JP, Teixeira MM, Sousa LP. Annexin A1 and the resolution of inflammation: modulation of neutrophil recruitment, apoptosis, and clearance. J Immunol Res 2016;2016:8239258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han PF, Che XD, Li HZ, Gao YY, Wei XC, Li PC. Annexin A1 involved in the regulation of inflammation and cell signaling pathways. Chin J Traumatol 2020;23:96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y, Long YH, Wang SQ, Zhang YY, Li YF, Mi JS, et al. JMJD6 regulates histone H2A.X phosphorylation and promotes autophagy in triple-negative breast cancer cells via a novel tyrosine kinase activity. Oncogene 2019;38:980–97. [DOI] [PubMed] [Google Scholar]
- 34. Huo D, Chen H, Cheng Y, Song X, Zhang K, Li MJ, et al. JMJD6 modulates DNA damage response through downregulating H4K16ac independently of its enzymatic activity. Cell Death Differ 2020;27:1052–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biswas A, Mukherjee G, Kondaiah P, Desai KV. Both EZH2 and JMJD6 regulate cell cycle genes in breast cancer. BMC Cancer 2020;20:1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miotti S, Gulino A, Ferri R, Parenza M, Chronowska A, Lecis D, et al. Antibody-mediated blockade of JMJD6 interaction with collagen I exerts antifibrotic and antimetastatic activities. FASEB J 2017;31:5356–70. [DOI] [PubMed] [Google Scholar]
- 37. Akasheh RT, Pini M, Pang J, Fantuzzi G. Increased adiposity in annexin A1-deficient mice. PLoS One 2013;8:e82608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aguilera CM, Gomez-Llorente C, Tofe I, Gil-Campos M, Canete R, Gil A. Genome-wide expression in visceral adipose tissue from obese prepubertal children. Int J Mol Sci 2015;16:7723–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alfadda AA, Benabdelkamel H, Masood A, Moustafa A, Sallam R, Bassas A, et al. Proteomic analysis of mature adipocytes from obese patients in relation to aging. Exp Gerontol 2013;48:1196–203. [DOI] [PubMed] [Google Scholar]
- 40. Bryzgalova G, Lundholm L, Portwood N, Gustafsson JA, Khan A, Efendic S, et al. Mechanisms of antidiabetogenic and body weight-lowering effects of estrogen in high-fat diet-fed mice. Am J Physiol Endocrinol Metab 2008;295:E904–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tiano JP, Delghingaro-Augusto V, Le May C, Liu S, Kaw MK, Khuder SS, et al. Estrogen receptor activation reduces lipid synthesis in pancreatic islets and prevents beta cell failure in rodent models of type 2 diabetes. J Clin Invest 2011;121:3331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiu S, Vazquez JT, Boulger E, Liu H, Xue P, Hussain MA, et al. Hepatic estrogen receptor alpha is critical for regulation of gluconeogenesis and lipid metabolism in males. Sci Rep 2017;7:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faulds MH, Zhao C, Dahlman-Wright K, Gustafsson JA. The diversity of sex steroid action: Regulation of metabolism by estrogen signaling. J Endocrinol 2012;212:3–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of BC patients with ER-positive tumors included in the METABRIC dataset
Effect of JMJD6 KO in ER positive and negative cell lines. A) Western blot showing the expression of JMJD6 and ERα in wild-type, scramble or JMJD6 KO BT474 cells both in mass cultures (Left panel) and in single clones (right panel). Vinculin is shown as loading control. B) Volcano plot showing the most upregulated (red) and downregulated (green) genes between scramble and wild-type (parental) MCF7 (left) and MDA-MB231 (right) cells.
JMJD6 regulates lipid metabolism in MCF7 cells. A) Immunohistochemistry staining for ANXA1 (brown) combined with Oil red-O staining (pink) for lipid droplets visualization in wild-type MCF7 cells B) Gene set enrichment plot for 'Hallmark of Adipogenesis' found to be significantly downregulated in JMJD6 KO MCF7 cells compared to scramble control. C) List of 'Reactome Pathways' found to be regulated by the genes included in the gene set enrichment pathway of 'Hallmark of Adipogenesis' and present in the DEGs list of JMJD6 KO versus scramble MCF7 cells.
Significant enriched pathways in JMJD6-KO MCF7 and MDA-MB231 cells
Effect of Jmjd6 silencing in PyMT41C-cells. A) Real-Time PCR showing the levels of Jmjd6 in PyMT-41C cells transfected with non-targeting siRNA (siCtr) or specific for Jmjd6. Graph is the average of two independent experiments. Two-tailed unpaired t student test was applied to calculate statistical significance. B) Western blot performed on conditioned medium collected from PyMT-41C transfected as in A). Red Ponceau is shown as loading control. C) IF to detect ANXA1 levels and D) lipid droplet formation in cell transfected as in A).
Jmjd6-mediated ANXA1 secretion promotes M2 macrophage differentiation in vitro and ex vivo. A) Mean fluorescence intensity (MFI) of M1 and M2 markers in BMDM differentiated into M0, M1 and M2.** p<0,001, *** means p<0,0001, **** p<0,00001 without conditioned medium. B) Real time PCR showing the expression M1 (CD40 and NOS2) and M2 (CD206 and ARG1) markers in BMDMs differentiated into M0, M1 and M2 macrophages without CM. Datapoints show technical replicates.** p<0,001, *** p<0,0001. C) Gating strategies for FACS analysis on ex vivo mouse samples of Figure 6D. CD45+ leukocytes were gated on live cells, excluding dead cells and debris. In CD45+ population, myeloid cells were analyzed by the expression of CD11b, Gr-1, Ly6C and F4/80 markers. Monocyte-like myleloid-derived immunosuppressive cells (M-MDSC) were identified as Ly6Chigh Gr-1int expression. Granulocyte-like myeloid-derived immunosuppressive cells (G-MDSC) were identified as Gr-1high Ly6Cint cells. M2 macrophages were identified as CD206+ cells in the F4/80 gate. G-MDSC were identified as Gr-1high Ly6Cint cells. M2 macrophages were identified as CD206+ cells in the F4/80 gate.
Data Availability Statement
The transcriptome data generated in this study have been deposited in the National Center for Biotechnology Information in the Gene Expression Omnibus database under the accession number GSE164399, GSE216234, and GSE216235.