Abstract
Owing to their superior agronomic performance, the hybrids of vegetable crops are currently applied extensively. However, effective hybrid production requires a laborious manual emasculation to ensure the purity of hybrid seeds in tomato because of the lack of an effective male sterility system. Here, we created two types of tomato nuclear male-sterile lines with different screening markers in a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system. Co-knockouts of male sterile 1035 (Ms1035) and glutathione S-transferase (GSTAA) created a male-sterile line marked by a green hypocotyl. The Ms1035 biallelic mutation was introduced into the woolly tomato background, resulting in the linkage of male sterility and a non-woolly phenotype. Two types of male-sterile lines were easily selected at the seedling stage by hypocotyl color or trichome density and further showed high seed purity during hybrid seed production. Our work established the procedure for a rapid transfer of the male-sterile phenotype to the parents of hybrids without extra-modification by the CRISPR/Cas9 system that can be practically applied to hybrid seed production in tomato. This method will be the basis and example for sterile parent creation of multiple crops for hybrid production with the CRISPR/Cas9 system.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01215-2.
Keywords: Tomato, CRISPR/Cas9, Male-sterile, Ms1035, GSTAA
Introduction
The use of hybrid seeds in most commercial horticultural and field crops is universal, which increases crop production and improves the quality of agriculture products. The introduction of male sterility can reduce the process of artificial emasculation or chemical hybridiziation during hybrid seed production and improve seed purity (Chen and Liu 2014; Song et al. 2015; Kim and Zhang 2018). Tomato is a vital vegetable crop with heterosis and the use of male sterility has been receiving attention in tomato breeding. Since the discovery of the first male-sterile material, various genes controlling male sterility have been reported, including male sterile 1035 (Ms1035), Ms32, positional sterile-2 (PS-2), and 7B-1, and few mutants have been applied in hybrid seed production (Crane 1915; Atanassova 1999, 2000; Jeong et al. 2014; Pucci et al. 2017; Liu et al. 2019). Currently, commercial varieties of tomato are mainly hybrid seeds that require efficient cross-pollination between parental inbred lines and to avoid self-pollination of the maternal parent. The commercial production of tomato hybrids mainly employs artificial emasculation, which is laborious and expensive work. If emasculation during the hybridization process is incomplete, the purity of hybrid seed purity is reduced. Therefore, creating a superior male-sterile line with a screening marker to simplify tomato hybrid production is a very meaningful work.
It is not only laborious and time consuming to establish tomato male sterile lines through traditional breeding methods, but many male sterile lines cannot be used for practical breeding because there are no suitable seedling screening markers or molecular markers to screen male-sterile plants. ms-1035aa is a fully fertile male-sterile mutation in tomato that is linked to the locus anthocyanin absent (aa) and exhibits a green hypocotyl at the seedling stage (Mutschler et al. 1987). Thus, it is easy to select male-sterile plants via the color of the hypocotyl and cotyledon during early developmental stages (Mutschler et al. 1987). Previous studies indicated that Ms1035 encodes a basic helix-loop-helix transcription factor to regulate another development, such as through meiosis and programmed cell death of the tapetum (Jeong et al. 2014). GSTAA encodes a putative glutathione S-transferase that participates in the sequestration of anthocyanins in vacuoles (Jeong et al. 2014). In double mutant ms-1035aa, a 398-bp insertion into the promoter of the Ms1035 gene reduces the transcription expression of Ms1035, thereby leading to male sterility, while deletion of the GSTAA gene caused the absence of anthocyanins (Zhang et al. 2016).
Trichomes are prevalent biological structures on the epidermis of almost all terrestrial plants and play vital roles in plant adaptation to stress, including resistance to insects, drought, and heat (Valverde et al. 2001; Yang et al. 2015). Woolly (Wo) and its allele are spontaneous mutations in tomato that exhibit increased trichome density and is thus named the woolly phenotype (Shilling, 1959). The woolly phenotype is caused by a single amino acid substitution in the Wo gene that encodes a HD-ZipIV transcription factor containing a START domain (Yang et al. 2011). A nucleotide substitution at 1904 bp in Wo was shown to cause embryo lethality when homozygous, and the self-pollinated offspring from this plant type were always divided into two phenotypes, non-woolly and woolly (Yang et al. 2011). Plants with a woolly phenotype can be easily distinguished by observing the trichome density on the leaves or hypocotyls at the seedling stage. Thus, the woolly characteristic has the potential to become a visual marker for male sterile line screening and cultivation.
The CRISPR/Cas9 system is a new type of gene editing tool developed after zinc-finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN) (Maeder et al. 2008; Mussolino et al. 2011; Cong et al. 2013). Due to its simplicity and reliability, the CRISPR/Cas9 system became popular all over the world within a short time. With the wide application of the CRISPR/Cas9 system in plant genome editing, researchers can use its rapid and effective orientation to change the function of target genes and cultivate new germplasm resources. The CRISPR/Cas9 system has been shown to be highly efficient in vegetable crops and has created tomatoes that are tolerant to storage and transportation, diploid potatoes that are self-compatible, and cucumbers with broad virus resistance by targeted mutagenesis of the PL, S-RNase, and eIF4E genes, respectively (Chandrasekaran et al. 2016; Uluisik et al. 2016; Ye et al. 2018). However, there are no studies that have used CRISPR/Cas9 technology to generate a marked male-sterile line in vegetable crops.
Here, we used CRISPR/Cas9 technology for directed mutagenesis of the Ms1035 gene in heterozygous woolly phenotype tomato and created a tomato male-sterile line linked to the non-woolly phenotype. Additionally, the Ms1035 and GSTAA double mutant was also generated in purple hypocotyl tomato and exhibited complete male sterility with a green hypocotyl in the first generation. These two types of male-sterile lines were easily identified during the early developmental stage and showed favorable application prospects in hybrid seed production. Our results demonstrate the utility of the CRISPR/Cas9 system for the rapid generation of male-sterile lines with a selection marker in tomato could be an example for generations of male-sterile lines for large-scale application in crop breeding.
Materials and methods
Plant materials, growth conditions, and generation of transgenic plants
CR-ms1035, CR-tpd, and CR-cyp704b1 were derived from Micro-Tomd+ (a functional DWARF gene was introduced in Micro-Tom) by the CRISPR/Cas9 system. Both 6Q71 and 5N21 are tomato inbred lines obtained by self-crossing through six generations or more. CR-ms1035/Wo was derived from 6Q71 by the transformation of the pCRI-Ms1035 vector. CR-ms1035/f3h and CR-ms1035/gstaa were derived from the transformation of the pCRI-GSTAA-Ms1035 and pCRI-F3H-Ms1035 vectors, respectively, using 5N21 as a host. Transgenic plants were generated using Agrobacterium-mediated transformation as previously described (Park et al. 2003). Unless indicated, all experimental plants were grown in the field at the Northwest A & F University, Yangling (34_N, 108_E), during the normal tomato growing season.
Vector construct
CRISPR/Cas9 was constructed as previously described (Xing et al. 2014). Briefly, according to the sequences and accession number of the target genes, the appropriate targets were selected as targets through online target predicted software RGEN Tools (http://www.rgenome.net/cas-designer/). Equivalent amounts of forward and reverse primers (100 μM) for Ms1035 as a single target were mixed and incubated at 65 °C for 5 min, then gradually cooled to room temperature to form the Ms1035 target adaptor. For the dual-target adaptor, we paired the target primers of Ms1035 with the GSTAA and F3H target primers, and carried out nested PCR using the vector pDT1T2 as a template to obtain the dual-target adaptor Ms1035+GSTAA and Ms1035+F3H, respectively. Approximately 500 ng each of the dual-target adaptor was digested using 10 U Bsa I (NEB). Three target adaptors, Ms1035, Ms1035+GSTAA, and Ms1035+F3H, were ligated into the linearized pKSE401 vector (digested with 10 U of Bsa I). The CRISPR/Cas9 constructs for TPD1 and CYP704B1 genes were the same as for the pCRI-Ms1035 vector, only the targets were different. The relevant PCR primers are listed in Supplementary Table 3.
Mutation detection and analysis of transgenic plants
To determine the mutation of target sites, genomic DNA from the leaves of transgenic tomato plants was extracted using the sodium dodecyl sulfate method (Dellaporta et al. 1983). Primers were designed according to the position of the target to amplify the fragments more than 300 bp upstream and downstream of the target. The PCR products were directly sequenced or cloned into pMD19-T (TAKARA, #6028) for sequencing.
For identification of “T-DNA free” plants, genomic DNA from the T1 progeny was analyzed by PCR using gRNA expression cassette specific primers. The pKSE401 plasmids and wild-type (WT) plant DNA were selected as positive and negative controls, respectively. The relevant PCR primers are listed in Supplementary Table 3.
Pollen viability assay
Pollen viability was assessed by Alexander staining (Solarbio, #G3050). Fresh anthers from mature flower were collected, put into a 0.5-ml centrifuge tubes containing 200 μL staining solution, and mashed to release the pollen. After instantaneous centrifugation, the supernatant containing pollen grains was mounted on glass microscope slides and observed with a microscope (Olympus, Japan, BX63).
Restriction enzyme site loss assay for genotyping
Genomic DNA was extracted from T2 and T3 plants as described above. The genomic DNA fragment containing the target site was amplified by PCR using Es Taq MasterMix (Cwbio, CW0718M) and the pair of Ms1035 or F3H-specific primers (Table S3). PCR cycling conditions were as follows: initial denaturation at 94°C for 4 min, followed by 31 cycles of denaturation at 94 °C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 30 s, and final extension at 72°C for 2 min. The PCR product was run on an agarose gel to confirm the presence of a single band of the expected size (Ms1035, 557 bp; F3H, 564bp). Four microliters of unpurified amplicons was digested with 3 U of Hph I (NEB, #R0158S) or Xba I (NEB, #R0145L) in a 6 μL reaction mix and then detected by 1% agarose gel electrophoresis.
Performance of hybrid seeds obtained from edited plants
The edited male sterile lines and its WT were crossed with another tomato inbred line, 5N11 or 6N55 (multigenerational inbred lines in our lab). Hybrid seeds were collected after fruit maturity and the number of seeds per pollinated fruit was analyzed. The harvested hybrid seeds were cultivated in seedling-raising plates, and each hybrid combination was randomly selected. To determine seed purity, 120 plants were used to extract genomic DNA. Since 6Q71 and 5N21 did not carry the tomato yellow flower curly leaf virus resistance gene Ty-3a, and 5N11/6N55 carries it, the Ty-3a molecular marker primers were used to detect seed purification (Ji et al. 2007).
Results
Identification of tomato male-sterility genes and its linkage marker genes
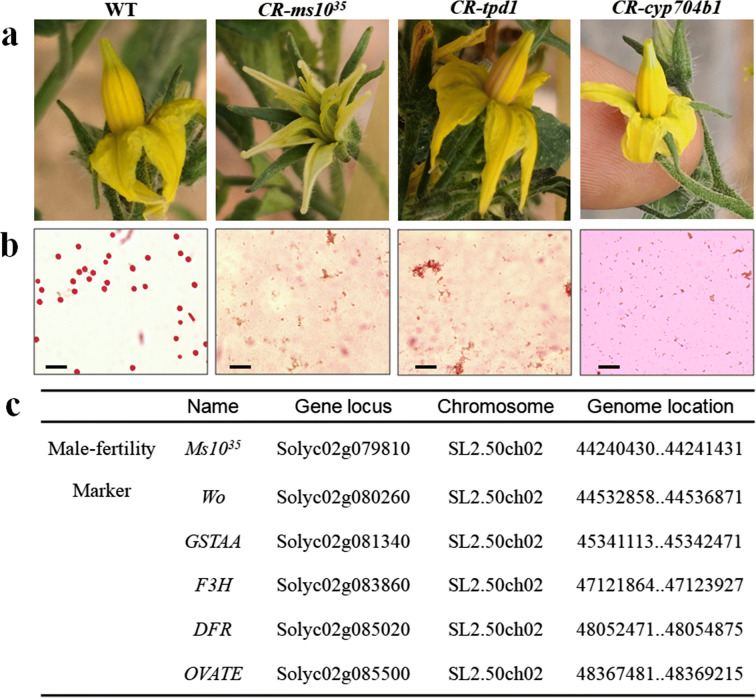
To identify fertility gene suitable for creating male sterile lines in tomato, we performed gene editing on fertility genes Ms1035, TPD1 (homologous genes of AtTPD1), and CYP704B1 (homologous genes of AtCYP704B1) with the CRISPR/Cas9 system. There were no significant differences in growth and development between the WT and edited plants until the flowering stage. At the flowering stage, the whole flower phenotypes of edited plants CR-tpd and CR-cyp704b1 resembled those of WT plants (Fig. 1a); however, CR-ms1035 plants had loosely spread anther cones (Fig. 1a). To determine the fertility of these edited plants, an Alexander’s staining assay, which distinguished aborted and non-aborted pollen grains, was performed with mature pollen grains released from anthers of fully opened flowers. Pollen from WT flowers exhibited a red spherical shape, whereas no staining signals were observed in edited plants pollen grains (Fig. 1b). According to the above results, CR-ms1035, CR-tpd, and CR-cyp704b1 were infertile, and only CR-ms1035 could be hand pollinated without artificial emasculation because of the anther cones. Therefore, Ms1035 has application prospects for male sterility in tomato.
Fig. 1.
Identification of tomato male-sterility genes and their linkage marker genes. a Morphological comparison of wild type (WT), CR-ms1035, CR-tpd1, and CR-cyp704b1 flowers. b Alexander staining assay of pollen viability of the WT, CR-ms1035, CR-tpd1, and CR-cyp704b1; scale bar: 100 μm. c Information on the male-sterility gene Ms1035 and its linkage marker genes
Previous investigation found that Ms1035 is an important fertility gene that is localized to chromosome 2 of tomato and linked to the gene of GSTAA (Jeong et al. 2014; Zhang et al. 2016). Based on the research of existing tomato functional genes, five visible marker trait genes near the region of Ms1035 were selected, which including the anthocyanin synthesis genes dihydroflavonol 4-reductase (DFR) and flavonoid 3-hydroxylase (F3H), the anthocyanin transporter gene GSTAA, the trichome developmental regulatory gene Wo, and the fruit shape-related gene OVATE (Fig. 1c) (Goldsbrough et al. 1994; Liu et al. 2002; Yang et al. 2011; Maloney et al. 2014). Since GSTAA, F3H, and Wo are close to Ms1035 on the chromosome, we chose these genes as visible markers for further gene editing.
Using the CRISPR/Cas9 system for knockout of Ms1035 and its marker genes
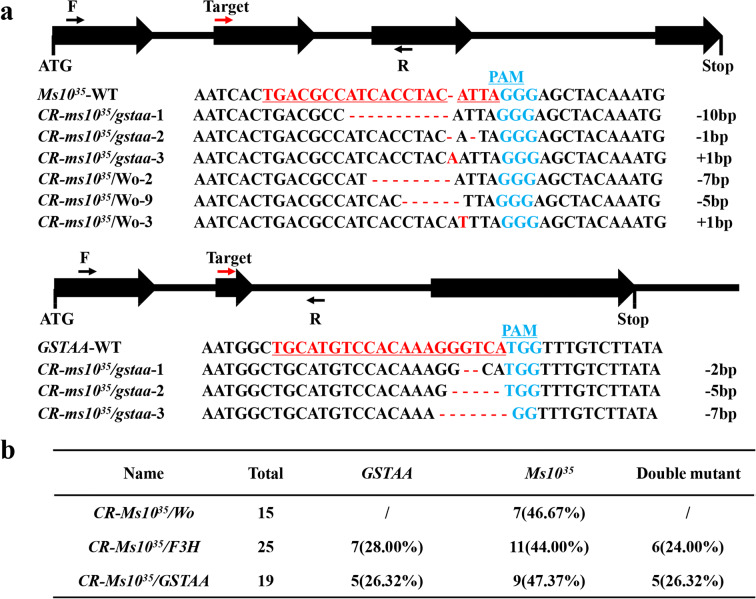
To achieve edited mutants, three targets were designed using an online analysis software that targeted exon 2 of Ms1035, GSTAA, and F3H separately. Three CRISPR/Cas9 vectors, single-target vector CR-Ms1035, and two double-target vectors CR-GSTAA-Ms1035 and CR-F3H-Ms1035 were constructed and transformed into tomato by Agrobacterium-mediated transformation. We produced a total of 59 T0 independent transgenic lines carrying CR-Ms1035 (15 lines), CR-GSTAA-Ms1035 (25 lines), or CR-F3H-Ms1035 (19 lines). To evaluate the efficiency of mutations, the targets of all T0 plants derived from the three constructs were analyzed through sequencing. The three targets were successfully mutated and the mutation sites were all biallelic mutants, which caused a frameshift mutation in the target genes (Fig. 2a and Fig. S3a). The editing efficiency of Ms1035 was 46.67, 44, and 47.37% in CR-Ms1035, CR-GSTAA-Ms1035, and CR-F3H-Ms1035, respectively, and the efficiency of GSTAA and F3H were 28 and 26.32%, respectively. Because the two targets sometimes cannot work at the same time, the efficiency of double mutants was only 24 and 26.32% in the CR-GSTAA-Ms1035 and CR-F3H-Ms1035 transgenic plants, respectively (Fig. 2b). Due to uncertainty about whether the target had an off-target effect, five T0 edited plants were randomly chosen, and the two most probable off-target sites were selected for each target site. The results indicated that no mutations were found at any of the potential off-target loci (Table S1).
Fig. 2.
Using the CRISPR/Cas9 system for knockout of Ms1035 and its marker genes. a The second exon of Ms1035 and its marker genes were targeted by the CRISPR/Cas9 system using single-guide RNAs (red arrows indicate target). Black arrows indicate the forward (F) and reverse (R) primers used for PCR genotyping and sequencing. The target sequences in Ms1035 and GSTAA are underlined, and minus symbols represent deletions. The numbers on the right show the type of mutation and how many nucleotides are involved, with “−”or “+” indicating the deletion or insertion of the given number of nucleotides, respectively. b Editing efficiency of CR-Ms1035/Wo and CR-Ms1035/GSTAA in T0 transgenic plants
Morphological analysis of the CR-ms1035/Wo
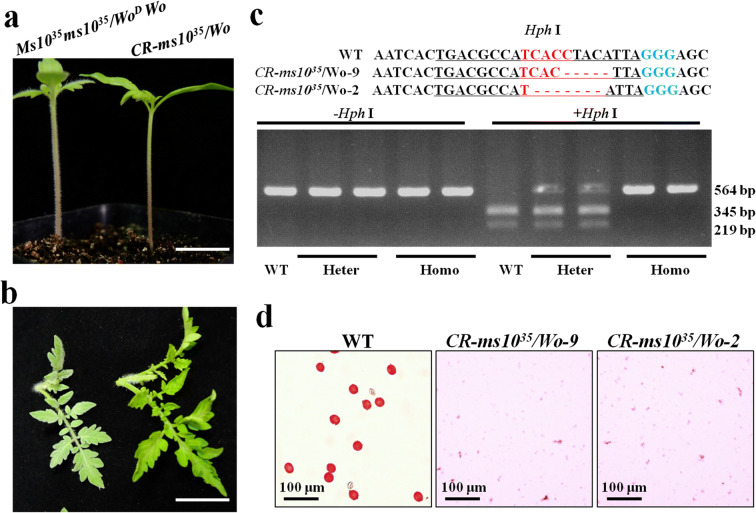
The higher trichome density of the tomato inbred line 6Q71 was caused by one amino acid substitution from Pro-635 to Arg in the Wo gene (WoD), and the embryo was lethal when the mutation was homozygous (Yang et al. 2011). Therefore, non-woolly phenotype plants that were isolated from 6Q71 were selected as the transformation material. Edited plants of CR-ms1035/Wo were infertile and had the woolly phenotype. Plants in the background of 6Q71 were used to pollinate CR-ms1035/Wo. In the T2 generation, the CR-ms1035/Wo plants were selected as the male-sterile lines marked by non-woolly phenotype, and Ms1035ms1035/WoDWo plants were selected as maintainer lines to form the 1:1 line. Woolly and non-woolly phenotype plants from the 1:1 line were easily identified at the seedling stage through the trichome density of the hypocotyls and leaves (Fig. 3a–3b). As the nucleotide deletion of the Ms1035 target missed an Hph I restriction site, we were able to easily detect the mutant allele via an Hph I restriction enzyme assay (Fig. 3c). Moreover, a competitive allele-specific PCR (Semagn et al. 2013) marker based on the mutation in the target sequence region could be also applied to the genotyping of WT and CR-ms1035 plants.
Fig. 3.
Analysis of phenotypes and genotypes of plants derived from selfed Ms1035ms1035/WoDWo . a Seven-day-old tomato seedlings from Ms1035ms1035/WoDWo selfed progeny; scale bar: 1 cm. b The phenotype of T2 plants from Ms1035ms1035/WoDWo selfed progeny before planting; scale bar: 1 cm. c Editing analysis of Ms1035 using restriction enzyme site Hph I loss. The two bands and three bands indicate the WT and heterozygous line, respectively. d Alexander staining assay of pollen grains at the dehiscence stage
With regard to the plants grown in the field during the normal growing season, no discernible phenotypic differences between the CR-ms1035/Wo homozygote and heterozygote were observed before flowering (Fig. S1a). As reported previously (Jung et al. 2020), flowers of the CR-ms1035/Wo homozygote were slightly smaller (Fig. S1b), and the stigma was exposed due to anther deformation (Fig. S1c). Otherwise, mature anther cones of the CR-ms1035/Wo homozygote were not observed in normal pollen by Alexander’s staining (Fig. 3d).
Morphological features of the CR-ms1035/gstaa and CR-ms1035/f3h
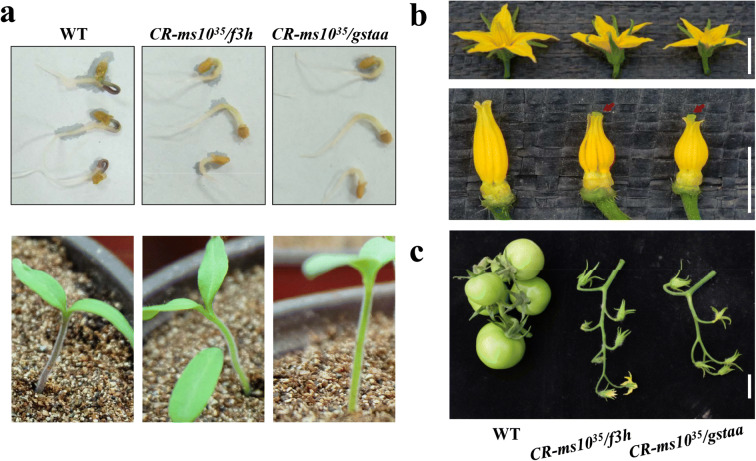
To confirm the morphological features of knockout plants CR-ms1035/gstaa and CR-ms1035/f3h, we tracked the phenotype of these mutants for more than one generation. Owing to the complete male sterility of CR-ms1035/gstaa and CR-ms1035/f3h plants, the next generation of edited plants was obtained by artificial pollination and the non-transgenic plants were selected by PCR. The hypocotyls of CR-ms1035/gstaa and CR-ms1035/f3h plants appeared to have anthocyanin deficiency at the seedling stage (Fig. 4a). The phenotype of flowers of CR-ms1035/gstaa and CR-ms1035/f3h plants tended to be consistent with CR-ms1035/Wo (Fig. 4b), which did not set fruit containing normal seeds unless artificial pollination occurred planting in the field (Fig. 4c and Fig. S2). Thus, CR-ms1035/gstaa and CR-ms1035/f3h plants identified in the T2 generation were male-sterile lines marked by a green hypocotyl; meanwhile, Ms1035ms1035/GSTAAgstaa and Ms1035ms1035/F3Hf3h plants were the maintainer lines. Knocking out F3H caused anthocyanin synthesis to be blocked, resulting in green hypocotyls (Maloney et al. 2014). Surprisingly, three hypocotyls colors, purple, lavender, and green, appeared in T2 generation, which was isolated from Ms1035ms1035/F3Hf3h plants (Fig. S3b). With the restriction endonuclease Xba I deletion assay, the result indicated that the lavender hypocotyl plants were F3H heterozygous mutations, while the purple hypocotyl plants did not contain an edited F3H (Fig. S3c).
Fig. 4.
Morphology of 5N21 plants with CR-ms1035/f3h and CR-ms1035/gstaa at multiple stages. a Representative example for distinguishing male-sterile plants (green hypocotyl) and WT (purple hypocotyl) at the seedling stages. Top: seedlings 3 days after germination. Bottom: seedlings 5 days after germination. b Morphological comparison of WT, CR-ms1035/f3h, and CR-ms1035/gstaa flowers. Top: full-opened flower. Bottom: stamen and stigma, red arrows indicate exerted stigmata. c CR-ms1035/f3h and CR-ms1035/gstaa plants did not set fruit following self-pollination; scale bar: 1 cm
Linkage analysis between Ms1035 and its markers
Natural mutant ms-1035aa was not completely linked in natural isolation, which means Ms1035 and its marker genes were genetically distant on chromosome 2. We found that Ms1035 was not completely linked to its marker genes because of the separation of Ms1035 from its marker genes in T2 and T3 plants. In particular, the CR-ms1035/f3h plants were directly discarded because of the excess separation ratio of Ms1035 and F3H (Table S2). To determine whether the linkage between Ms1035 and its markers was stable, we selected two independent Ms1035ms1035/WoDWo lines as a male parent to pollinate CR-ms1035/Wo plants in T3 generation. To test the linkage between ms1035 and Wo, two independent Ms1035ms1035/WoDWo lines were crossed with ms1035CR/Wo plants to construct two populations for detection. The ratio of woolly phenotype plants to non-woolly phenotype plants in these populations was close to 1:1. By genotyping ms1035 in non-woolly phenotype plants to confirm fertility, we identified one fertile plant in each of the two ms-Wo populations, the proportion of which was less than 1% in non-woolly phenotype plants (Table 1).
Table 1.
Linkage analysis between ms1035 and Wo
| Population | Non-woolly | No. of non-woolly plants | No. of woolly plants | |
|---|---|---|---|---|
| Male sterile | Male fertility | |||
| ms-Wo #1 | 122 (99.19%) | 1 (0.81%) | 124 | 126 |
| ms-Wo #2 | 107 (99.07%) | 1 (0.93%) | 108 | 106 |
Two independent lines of the Ms1035ms1035/WoDWo crossed with CR-ms1035/Wo plants to construct populations for testing
Meanwhile, two independent CR-ms1035/gstaa lines were selected as female parent and crossed with Ms1035ms1035/GSTAAgstaa plants in the T3 generation. Seeds harvested from the two independent CR-ms1035/gstaa plants were divided into two populations. The ratio of purple hypocotyl plants to green hypocotyl plants in these populations was close to 1:1 (Table 2). However, the green hypocotyl was not completely linked to male infertility; approximately 5% of the plants with a green hypocotyl plants were male fertile. Since the stamen was opened and the stigma was exposed after Ms1035 loss of function, it was y to select the mingled fertile plants during the hybrid seed production process.
Table 2.
Linkage analysis between ms1035 and gstaa
| Population | Green hypocotyl | No. of green hypocotyl plants | No. of purple hypocotyl plants | |
|---|---|---|---|---|
| Male sterile | Male fertility | |||
| ms-g #1 | 121 (94.53%) | 7 (5.47%) | 128 | 125 |
| ms-g #2 | 113 (95.76%) | 5 (4.24%) | 118 | 120 |
Two independent lines of the Ms1035ms1035/GSTAAgstaa as male parents crossed with CR-ms1035/gstaa plants to construct populations for testing
Application of edited plants in tomato hybrid breeding
Excellent male sterile lines must have not only visual screening markers but the quality of the seeds also cannot be reduced. Although it has been reported that the simultaneous mutation of Ms1035 and GSTAA had no effect on plant and fruit characteristics, ms1035, which is a natural mutation, is not completely knocked out in ms-1035aa mutant; therefore, whether it has negative effects on the growth and development of tomato after being completely knocked out has not yet been confirmed. According to the tomato hybrid seed production process, CR-ms1035/Wo and CR-ms1035/gstaa female parents were crossed with inbred lines 5N11 (in 2018) and 6N55 (in 2019), respectively. The fruit of the sterile lines after pollination did not differ significantly in the whole fruit growth and development cycle compared with the WT, especially in the number of seeds per pollinated fruit and seed germination rate (Fig. S4 and Table 3). The maturity of the seeds was not significantly different from that of the WT and the results of molecular marker identification showed that the seed purity of male-sterile lines was better than that of control groups (Table 3). As recessive genes, Ms1035 and GSTAA heterozygous mutations may have no adverse effects on the growth and development of hybrid F1 plants.
Table 3.
Performance of 6Q71, 5N21, CR-ms1035/Wo, and CR-ms1035/gstaa in tomato seed production
| Crossa | Fruit set (fruits developed/No. of hand pollination) | Seeded fruits (fruits with seeds/No. of fruits developed) | Seeds (No. per fruit) | Purification of F1 hybrid seeds |
|---|---|---|---|---|
| ♀6Q71×♂5N11 | 38/40 | 38/38 | 103±5.3a | 98.3% (118/120) |
| ♀CR-ms1035/Wo×♂5N11 | 38/40 | 38/38 | 99±4.7a | 100% (120/120) |
| ♀5N21×♂5N11 | 39/40 | 39/39 | 76±5.2b | 95.8% (115/120) |
| ♀CR-ms1035/gstaa×♂5N11 | 39/40 | 39/39 | 74±4.6b | 100% (120/120) |
| ♀6Q71×♂6N55 | 30/30 | 30/30 | 54±5.3d | 99.2% (119/120) |
| ♀CR-ms1035/Wo×♂6N55 | 30/30 | 30/30 | 64±7.4c | 100% (120/120) |
| ♀5N21×♂6N55 | 30/30 | 30/30 | 72±8.1b | 97.5% (117/120) |
| ♀CR-ms1035/gstaa×♂6N55 | 30/30 | 30/30 | 61±9.0c | 100% (120/120) |
aCR-ms1035/Wo, CR-ms1035/gstaa, and their background materials as female parents were crossed with inbred lines 5N11 (in 2018) and 6N55 (in 2019), respectively
Successful fruit set and number of fruits with seeds versus total number of hand pollination are recorded. Values are means ± SE, which differ at the 0.05 significance level are labeled with different letters (ANOVA and Holm-Sidak test)
Discussion
The CRISPR/Cas9 system is a powerful tool for gene function research and generating genetic diversity in crops. This system is relatively simple to operate and can induce directed mutations in vegetable crops, such as tomato, potato, and lettuce (Brooks et al. 2014; Wang et al. 2015; Woo et al. 2015). According to recent reports, the CRISPR/Cas9 system has been successfully applied for the generation of male-sterile rice, tomato, sorghum, and wheat lines (Cigan et al. 2017; Barman et al. 2019; Okada et al. 2019). By this approach, male-sterile lines can be easily generated in excellent tomato breeding material by targeted mutagenesis of pollen development genes.
Previous studies have reported a series of genes that determine pollen development. Of them, only Ms1035 can be artificially pollinated without removing the stamens through the evaluation of sterile materials in tomato. Therefore, Ms1035 was selected as a candidate gene to create male-sterile lines with the CRISPR/Cas9 system. Our goal was to demonstrate the utility of the CRISPR/Cas9 system for the rapid generation of male-sterile lines and verify the characteristics of Ms1035, GSTAA, and F3H in tomato. As expected, co-knockouts of Ms1035 and GSTAA or F3H in the purple hypocotyl tomato 5N21 resulted in complete male sterility and obtained male-sterile lines marked by a green hypocotyl in T0 generation. About 20% of green hypocotyl plants from the CR-ms1035/f3h heterozygous self-crossing progeny were identified as male fertile plants via genotyping of Ms1035. Therefore, F3H was not suitable as a visible marker gene for Ms1035, but it can be used as a visible marker at the seedling stage for F1 hybrid seed purity identification. However, 90% of progeny with a green hypocotyl from CR-ms1035/gstaa heterozygous self-crossing plants were identified as male-sterile plants. This result was consistent with previous reports of male sterility in ms-1035aa double mutant (Zhang et al. 2016). Although the obtained sterile line contained about 5% fertile plants with a green hypocotyl in hybrid seed production, fertile plants could be removed based on the phenotypes of the stamen cones, stigma, and flower size. Even if the fertile plants with a green hypocotyl were not completely removed, less F1 generation plants without hybridization could be removed according to the hypocotyl color at the seedling stage. Another type of male-sterile line was created in the woolly phenotype tomato 6Q71. Non-woolly phenotype plants separated from 6Q71 were selected for edited Ms1035 to obtain completely male-sterile plants in the T0 generation. Offspring separation testing confirmed that linkage between Wo and Ms1035 was close. When the male-sterile line was separated, the number of mixed fertile plants was less than 1%. Like male-sterile line marked by a green hypocotyl, mixed fertile plants can be selected according to the phenotype of the flower. These two types of sterile lines could be crossed with their heterozygous mutant to form a 1:1 line or crossed with other tomato male parent materials for the production of hybrid seeds.
Our tested method can be applied to the purple hypocotyl or woolly phenotype (the Wo mutation type of homozygous lethal) tomato plants, but the absence of anthocyanin and other woolly phenotypes was limited. Additionally, the linkage between male sterility traits and marker traits was not 100% satisfactory. In recent years, some studies showed that fertility restorer genes and marker genes can be co-transformed into the male-sterile line to create an artificial restorer line (Chang et al. 2016). To conquer this difficulty, the artificial maintainer line can be created by co-overexpression of the Ms1035 native expression box and anthocyanin synthesis gene (or seed-specific expression of the red fluorescent protein) under the background of CR-ms1035 (Du et al., 2020). Moreover, the visible marker gene can be inserted into the region that is completely linked to the fertility genes through the CRISPR/Cas9 system mediated by homologous recombination (Čermák et al. 2015).
In the breeding of common tomatoes, the advantages of male sterile lines are not very obvious, but the advantages in cherry tomato and processing tomato are quite obvious. Due to the small fruit, cherry tomatoes are not only easy to pollinate but also have fewer seeds per fruit. The value of processing tomato is relatively low. However, it is impossible to invest too much in seed cost during tomato cultivation. Therefore, the production of cherry tomato and processing tomato are still dominated by conventional species. At present, we have obtained cherry tomato and processing tomato male-sterile lines marked with a green hypocotyl via this technology but have not yet tested them.
In summary, our work provides a platform to construct male-sterile lines with a screening marker for tomato breeding with the CRISPR/Cas9 system. Using this technology to create marked tomato male-sterile lines can effectively reduce the labor intensity and cost of tomato hybrid production, and significantly improve seed purity. Our work will promote the application of male-sterile lines with screening marker in tomato breeding, especially for the selection of new varieties of cherry and processing tomatoes, which will facilitate the development of hybrid tomato breeding. Certainly, this method can be applied for not only hybrid tomato breeding but also the breeding of other hybrid horticultural crops. Moreover, a new generation of male-sterile lines can be developed on this basis.
Supplementary information
Morphological analysis of Ms1035ms1035/WoDWo and CR-ms1035/Wo. a Two-month-old plants in the field, woollyand non-woolly phenotype plants from two dependent male-sterile lines. b Morphological comparison of Ms1035ms1035/WoDWo and CR-ms1035/Wo flowers; scale bar: 1 cm. c Morphology of Ms1035ms1035/WoDWo and CR-ms1035/Wo anther cones, red arrow indicates exerted stigmata; scale bar: 1 cm. (PNG 2262 kb)
CR-Ms035/Wo and CR-Ms035/gstaa can set normal fruits by artificial pollination. The red arrow indicates the fruit set by artificial pollination. (TIF 6068 kb) (PNG 2090 kb)
Analysis of hypocotyl color and genotype in Ms1035ms1035/F3Hf3h self-crossed progeny. a The second exon of F3H was targeted by the CRISPR/Cas9 system using single-guide RNAs (red arrows indicate target). Black arrows indicate forward (F) and reverse (R) primers used for PCR genotyping and sequencing. The target sequences in F3H are underlined, minus symbols represent deletions. The numbers on the right show the type of mutation and how many nucleotides are involved, with “−” indicating deletion of the given number of nucleotides. b Morphological comparison of wild-type (WT) and CR-ms1035/f3h hypocotyls; scale bar: 1 cm. c Editing analysis of F3H using restriction enzyme site Xba I loss. The two bands and three bands indicate WT and heterozygous, respectively. (PNG 4642 kb)
a Morphology of fruits and seeds in 6Q71, 5N21, CR-ms1035/Wo and CR-ms1035/gstaa during tomato seed production. b Germination rate of hybrid F1 seeds from 6Q71, 5N21, CR-ms1035/Wo and CR-ms1035/gstaa. Per plug, 128 seeds were sowing, and the germination rate was counted seven days after sowing. Three plugs were sown for each type of seed as three independent biological repeats. Statistically significant differences were test by Student’s t-test (P < 0.05). (PNG 1007 kb)
Detection of mutations in potential off-target sites in T0 edited plants. (PDF 184 kb)
Linkage between ms1035 and f3h in the T2 and T3 generations. (PDF 246 kb)
Primers used for this study. (PDF 255 kb)
(DOCX 151 kb)
Acknowledgements
We thank Prof Qijun Chen (China Agricultural University, Beijing) for kindly providing the plant genome editing vector pKSE401 and pCBC1-DT1T2 plasmids.
Author contribution
J.L., S.W., H.W., Y.Z., and X.W. designed the studies. J.L., S.W., H.W., B.L., Y.C., and X.L. performed the experiments. J.L., S.W., H.W., Y.Z., and X.W. wrote the manuscript. All the authors gave final approval for submission of the manuscript.
Funding
This work was financially supported by NSFC (No. 31972430) and Research Collaborative Innovation of Yangling Demonstration Zone (2018CXY-09) to Xiaofeng Wang, “Light of the West” Talent Training and Introduction Program of Chinese Academy of Sciences (XAB2018AW16) to Yanfeng Zhang.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jianwei Liu, Shufen Wang and Hao Wang contributed equally to this work.
Contributor Information
Yanfeng Zhang, Email: zhangyfcl@126.com.
Xiaofeng Wang, Email: wangxff99@nwsuaf.edu.cn.
References
- Atanassova B. Functional male sterility (ps-2) in tomato (Lycopesicon esculentum Mill.) and its application in breeding and hybrid seed production. Euphytica. 1999;107:13–21. doi: 10.1023/A:1003527714805. [DOI] [Google Scholar]
- Atanassova B. Functional male sterility in tomato (Lycopersicon esculentum Mill.) and its application in hybrid seed production. Acta Physiol Plant. 2000;22:221–225. doi: 10.1007/s11738-000-0015-4. [DOI] [Google Scholar]
- Barman HN, et al. Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 2019;19:109. doi: 10.1186/s12870-019-1715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C, Nekrasov V, Lippman ZB, Van Eck J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 system. Plant Physiol. 2014;166:1292–1297. doi: 10.1104/pp.114.247577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran J, et al. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17:1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, et al. Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci U S A. 2016;113:14145–14150. doi: 10.1073/pnas.1613792113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu Y. Male sterility and fertility restoration in crops. In: Merchant SS, editor. Annual Review of Plant Biology. 2014. pp. 579–606. [DOI] [PubMed] [Google Scholar]
- Cigan AM, et al. Targeted mutagenesis of a conserved anther-expressed P450 gene confers male sterility in monocots. Plant Biotechnol J. 2017;15:379–389. doi: 10.1111/pbi.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane MB. Heredity of types of inflorescence and fruits in tomato. J Genet. 1915;5:1–11. doi: 10.1007/BF02982149. [DOI] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: Version II. Plant Mol Biol Report. 1983;1:19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- Du M, et al. A biotechnology-based male-sterility system for hybrid seed production in tomato. Plant J. 2020;102:1090–1100. doi: 10.1111/tpj.14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsbrough A, Belzile F, Yoder JI. Complementation of the tomato anthocyanin without (aw) mutant using the dihydroflavonol 4-reductase gene. Plant Physiol. 1994;105:491–496. doi: 10.1104/pp.105.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, et al. Tomato male sterile 10(35) is essential for pollen development and meiosis in anthers. J Exp Bot. 2014;65:6693–6709. doi: 10.1093/jxb/eru389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, et al. Co-dominant SCAR markers for detection of the Ty3 and Ty3a loci from Solanum chilense at 25 cM of chromosome 6 of tomato. Tomato Genet Cooper. 2007;57:25–29. [Google Scholar]
- Jung YJ et al (2020) Knockout of SlMS10 gene (Solyc02g079810) encoding bHLH transcription factor using CRISPR/Cas9 system confers male sterility phenotype in tomato. Plants (Basel) 9. 10.3390/plants9091189 [DOI] [PMC free article] [PubMed]
- Kim Y, Zhang D. Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 2018;23:53–65. doi: 10.1016/j.tplants.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Liu J, Van Eck J, Cong B, Tanksley SD. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc Natl Acad Sci U S A. 2002;99:13302–13306. doi: 10.1073/pnas.162485999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X et al (2019) A putative bHLH transcription factor is a candidate gene for male sterile 32, a locus affecting pollen and tapetum development in tomato. Hortic Res-England 6. 10.1038/s41438-019-0170-2 [DOI] [PMC free article] [PubMed]
- Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney GS, DiNapoli KT, Muday GK. The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 2014;166:254–614. doi: 10.1104/pp.114.240507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschler M, Tanksley S, Rick CM (1987) Linkage maps of the tomato (Lycopersicon esculentum) Vol 37.,
- Okada A, et al. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J. 2019;17:1905–1913. doi: 10.1111/pbi.13106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Morris JL, Park JE, Hirschi KD, Smith RH. Efficient and genotype-independent Agrobacterium-mediated tomato transformation. J Plant Physiol. 2003;160:1253–1257. doi: 10.1078/0176-1617-01103. [DOI] [PubMed] [Google Scholar]
- Pucci A, Picarella ME, Mazzucato A. Phenotypic, genetic and molecular characterization of 7B-1, a conditional male-sterile mutant in tomato. Theor Appl Genet. 2017;130:2361–2374. doi: 10.1007/s00122-017-2964-7. [DOI] [PubMed] [Google Scholar]
- Semagn K, Babu R, Hearne S, Olsen M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol Breed. 2013;33:1–14. doi: 10.1007/s11032-013-9917-x. [DOI] [Google Scholar]
- Shilling, Richard P (1959) Investigation of the hereditary character, woolly, in the tomato
- Song Y, Wang J, Zhang G, Zhao X, Zhang P, Niu N, Ma S. Microspore abortion and abnormal tapetal degeneration in a male-sterile wheat line induced by the chemical hybridizing agent SQ-1. Crop Sci. 2015;55:1117–1128. doi: 10.2135/cropsci2014.08.0538. [DOI] [Google Scholar]
- Uluisik S, et al. Genetic improvement of tomato by targeted control of fruit softening. Nat Biotechnol. 2016;34:950–952. doi: 10.1038/nbt.3602. [DOI] [PubMed] [Google Scholar]
- Valverde P, Fornoni J, Núñez-Farfán J. Defensive role of leaf trichome in resistance to herbivorous in Datura stramonium. J Evol Biol. 2001;14:424–432. doi: 10.1046/j.1420-9101.2001.00295.x. [DOI] [Google Scholar]
- Wang S, Zhang S, Wang W, Xiong X, Meng F, Cui X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015;34:1473–1476. doi: 10.1007/s00299-015-1816-7. [DOI] [PubMed] [Google Scholar]
- Woo JW, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33:1156–1162. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- Xing H et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327 doi:10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed]
- Yang C, Li H, Zhang J, Wang T, Ye Z. Fine-mapping of the woolly gene controlling multicellular trichome formation and embryonic development in tomato. Theor Appl Genet. 2011;123:625–633. doi: 10.1007/s00122-011-1612-x. [DOI] [PubMed] [Google Scholar]
- Yang C, Gao Y, Gao S, Yu G, Xiong C, Chang J, Li H, Ye Z. Transcriptome profile analysis of cell proliferation molecular processes during multicellular trichome formation induced by tomato Wo (v) gene in tobacco. BMC Genomics. 2015;16:868. doi: 10.1186/s12864-015-2099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, et al. Generation of self-compatible diploid potato by knockout of S-RNase. Nat Plants. 2018;4:651–654. doi: 10.1038/s41477-018-0218-6. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang Z, Wang X, Gao J, Guo Y, Du Y, Hu H (2016) Fine mapping and molecular marker development of anthocyanin absent, a seedling morphological marker for the selection of male sterile 10 in tomato. Mol Breed. 36 doi:10.1007/s11032-016-0531-6
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological analysis of Ms1035ms1035/WoDWo and CR-ms1035/Wo. a Two-month-old plants in the field, woollyand non-woolly phenotype plants from two dependent male-sterile lines. b Morphological comparison of Ms1035ms1035/WoDWo and CR-ms1035/Wo flowers; scale bar: 1 cm. c Morphology of Ms1035ms1035/WoDWo and CR-ms1035/Wo anther cones, red arrow indicates exerted stigmata; scale bar: 1 cm. (PNG 2262 kb)
CR-Ms035/Wo and CR-Ms035/gstaa can set normal fruits by artificial pollination. The red arrow indicates the fruit set by artificial pollination. (TIF 6068 kb) (PNG 2090 kb)
Analysis of hypocotyl color and genotype in Ms1035ms1035/F3Hf3h self-crossed progeny. a The second exon of F3H was targeted by the CRISPR/Cas9 system using single-guide RNAs (red arrows indicate target). Black arrows indicate forward (F) and reverse (R) primers used for PCR genotyping and sequencing. The target sequences in F3H are underlined, minus symbols represent deletions. The numbers on the right show the type of mutation and how many nucleotides are involved, with “−” indicating deletion of the given number of nucleotides. b Morphological comparison of wild-type (WT) and CR-ms1035/f3h hypocotyls; scale bar: 1 cm. c Editing analysis of F3H using restriction enzyme site Xba I loss. The two bands and three bands indicate WT and heterozygous, respectively. (PNG 4642 kb)
a Morphology of fruits and seeds in 6Q71, 5N21, CR-ms1035/Wo and CR-ms1035/gstaa during tomato seed production. b Germination rate of hybrid F1 seeds from 6Q71, 5N21, CR-ms1035/Wo and CR-ms1035/gstaa. Per plug, 128 seeds were sowing, and the germination rate was counted seven days after sowing. Three plugs were sown for each type of seed as three independent biological repeats. Statistically significant differences were test by Student’s t-test (P < 0.05). (PNG 1007 kb)
Detection of mutations in potential off-target sites in T0 edited plants. (PDF 184 kb)
Linkage between ms1035 and f3h in the T2 and T3 generations. (PDF 246 kb)
Primers used for this study. (PDF 255 kb)
(DOCX 151 kb)