Abstract
More than 1.1 billion tonnes of maize grain were harvested across 197 million hectares in 2019 (FAOSTAT 2020). The vast global productivity of maize is largely driven by denser planting practices, higher yield potential per area of land, and increased yield potential per plant. Shoot architecture, the three-dimensional structural arrangement of the above-ground plant body, is critical to maize grain yield and biomass. Structure of the shoot is integral to all aspects of modern agronomic practices. Here, the developmental genetics of the maize vegetative shoot is reviewed. Plant architecture is ultimately determined by meristem activity, developmental patterning, and growth. The following topics are discussed: shoot apical meristem, leaf architecture, axillary meristem and shoot branching, and intercalary meristem and stem activity. Where possible, classical and current studies in maize developmental genetics, as well as recent advances leveraged by “-omics” analyses, are highlighted within these sections.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01208-1.
Keywords: Maize, Shoot architecture, Shoot apical meristem, Axillary meristem, Blade/sheath boundary, Tiller, Leaf, Stem
Introduction
An urgent challenge facing plant scientists and breeders is how to feed the world’s growing population on limited arable land (Tilman et al. 2011). The cereal maize (Zea mays ssp. mays) is one of the most cultivated crops, making up over 38% of the world’s cereal production (FAOSTAT 2020). Increased global production is attributed, in part, to adapting maize shoot architecture to dense planting (Duvick 2005). Shoot architecture is defined as the three-dimensional structural arrangement of the above-ground plant body (Sussex and Kerk 2001; Reinhardt and Kuhlemeier 2002) and is a major factor contributing to unit area grain yield (Russell 1991). Structure of the maize shoot is integral to all aspects of productivity and modern agronomic practices: row cropping, light interception, photosynthetic capacity, planting density, canopy shape, synchronized flowering, yield, and mechanized harvestability (Crosbie 1982; Troyer 1996; Tuberosa and Salvi 2009; Duvick 2005; Wang et al. 2018a). Maize stands out among the cereals as a crop with an impressive history of genetic investigation, owing largely to its amenability for forward, reverse, and modifier genetic screens (Candela and Hake 2008; Schnable and Freeling 2011; Nannas and Dawe 2015). Additionally, advances in new methodologies such as single cell technology (Efroni and Birnbaum 2016; Libault et al. 2017; Nelms and Walbot 2019; Satterlee et al. 2020), plant transformation (Lowe et al. 2016; Altpeter et al. 2016; Kaush et al. 2019), image processing (Clark et al. 2020; Guo et al. 2020), genomics (Kremling et al. 2018; Yang et al. 2019; Ricci et al. 2019; Franco et al. 2020; Soyk et al. 2020; Wang et al. 2020a; Hufford et al. 2021), metabolomics (Xu et al. 2019; Zhou et al. 2019), proteomics (Walley et al. 2016), phenomics (Pauli et al. 2016; Yang et al. 2017; Gage et al. 2019), genome editing (Kelliher et al. 2019; Gao et al. 2020; Liu et al. 2020; Atkins and Voytas 2020; Taagen et al. 2020), machine learning (Wang et al. 2020b; Washburn et al. 2019, 2020), and synthetic biology (Leydon et al. 2020) have deepened the toolboxes of plant scientists and breeders for more efficient and comprehensive strategies toward maize improvement.
Domestication of maize from teosinte reshaped shoot architecture
Maize was domesticated from the teosinte Z. mays ssp. parviglumis about 9000 years ago through a single domestication event in the Balsas region of southwest Mexico (Matsuoka et al. 2002; Piperno et al. 2009). Vegetative and reproductive morphologies of domesticated maize differ drastically compared with teosinte (Stitzer and Ross-Ibarra 2018). The domestication of maize resulted in a conspicuous increase in apical dominance of the shoot and morphological transformations of the pistillate inflorescence, or ear (Doebley and Stec 1991; Doebely et al. 1997). Maize has relatively few, if any, basal axillary branches, or tillers. A tiller originates from an actively growing axillary bud that nests in the axil between the attachment point of the leaf sheath and the stem node. Growth of the axillary bud is suppressed in maize shortly after it produces a whorl of protective leaves. About midway up the adult maize shoot, 2–3 aerial lateral branches (also called ear shanks or ear shoots) originate singly at each node in the sheath axil, each of which is terminated by a large ear. By comparison, the teosinte shoot has multiple, long tillers and lateral branches that originate basally and from aerial mid-shoot nodes, each of which terminates with a staminate inflorescence, or tassel. Teosinte tillers are prolific, bearing clusters of reduced ears at each node (Iltis 1983). Maize geneticists and breeders use “degree of prolificacy” to describe the potential for ear production (Liable and Dirks 1968; Hallauer 1974); prolificacy is reduced in modern maize and remains high in teosinte (Wills et al. 2013). Thus, domestication of maize from teosinte, with its strong selection for tiller suppression at the base of the shoot and elongation of lateral ear shoots in the middle of the plant, impacted several agronomically important aspects of plant architecture.
Maize domestication also conditioned more subtle effects on the shoot architecture through changes in leaf morphology. In maize and teosinte, leaves alternate at each node. Each leaf has a stem clasping proximal sheath and a distal blade that angles away from the shoot axis. Leaf sheath and blade are separated by a hinge-like auricle and epidermally derived ligule. In teosinte, each long lateral branch is akin to the maize shoot in that leaf attachment alternates at each node, with each leaf having sheath, auricle, ligule, and pronounced blade. In maize, the shorter internodes of the ear shank are wrapped entirely in a whorl of leaves of predominantly sheath tissue, though auricle, ligule, and blade are present, albeit highly reduced (Iltis 1983; Doebley 2004). Leaf traits, such as blade width and length, sheath length, size, and total estimated area, show significant quantitative variation between maize and teosinte, with each showing a proportional increase in maize (Milla and Matesanz 2017; Fu et al. 2019). Additionally, leaf blades along the teosinte main axis can be slightly more upright compared to blade angle for some maize inbred lines (Tian et al. 2019).
Shoot architecture as a readout of developmental patterning and growth
Plant development lies at the heart of shaping shoot architecture through the size, identity, timing, and duration of maturation schedules of active, pluripotent stem cell tissues called meristems. As plant stem cells, meristems give rise to daughter cells that form new organs as well as undergo self-renewal (Steeves and Sussex 1989). Meristem tissues of the maize shoot include the shoot apical meristem (SAM), axillary meristem, intercalary meristem, and meristems of the inflorescences. Meristems subsequently produce leaves, lateral branches, stem (discussed herein), and, upon floral transition, tassel and ears (discussed elsewhere).
Shoot apical meristem
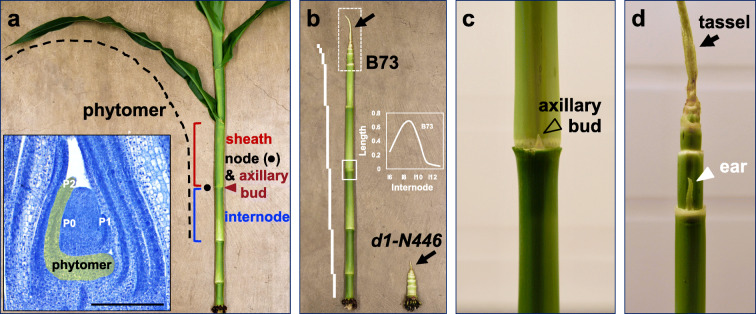
The capacity for continuous postembryonic shoot development requires establishment and maintenance of shoot meristems (Steeves and Sussex 1989). In maize, shoot meristematic tissue develops during embryogenesis approximately a week after pollination (Randolph 1936; Nardmann and Werr 2009; Takacs et al. 2012). Subsequently, SAM initiates a leaf primordium singly at its flank in a distichous (alternate) phyllotaxy (Kiesselbach 1949; see Jackson 2009). Concomitant with cells becoming fated to a leaf primordium, cells from the meristem also are recruited to establish an axillary meristem in the axil of the initiating leaf and the internode section of stem. Collectively, these vegetative organs form a repeating unit of the shoot called the phytomer (Fig. 1a) (Galinat 1959). To describe the relatively uniform time interval between successive initiation of a primordium and its associated phytomer, plant biologists use the term plastochron. Plastochron index, abbreviated as “P” and a number, denotes the position of the primordia relative to an apical meristem actively producing primordia (Lamoreaux et al. 1978). For example, P2 is the second youngest primordium produced by the meristem, P1 is the most recently initiated primordium, and P0 is the incipient primordium at the meristem periphery (Fig. 1a, inset). Meristem size is dynamic in between plastochrons and during shoot maturation: the SAM is smallest in the embryo and increases as each successive primordium is initiated until floral transition (Abbe et al. 1951; Ledin 1954; Bassiri et al. 1992; Thompson et al. 2014). The geometry and size of the SAM is influenced by genetic modifier loci in various maize inbred backgrounds (Vollbrecht et al. 2000; Thompson et al. 2014, 2015; Leiboff et al. 2015). SAM traits such as size, height, and volume can be predicative of adult plant phenotypes (Thompson et al. 2015; Leiboff et al. 2015).
Fig. 1.
The maize shoot. a Representation of the B73 phytomer consisting of internode, node with axillary bud, and leaf with sheath attached at the node and blade. Inset, longitudinal section of maize shoot apex depicting development of the phytomer pseudocolored in yellow. P0–P2, leaf primordia of plastochron stages. b Same plant shown in (a) but with all leaves removed. For comparison, a d1-N446 mutant with leaves dissected to expose short internodes. White bars represent internode length plotted in graph for adult internodes 8–12 (inset), showing sigmoidal distribution of length across the shoot. Arrows denote tassels; solid outline box around an axillary bud shown in (c); dashed outline box around an ear shown in (d). c Axillary bud. d Tassel (arrow) and ear (arrowhead)
Histology of the SAM captures the beauty of its internal organization and cell division patterns. Longitudinal sections through the SAM expose cells that divide almost exclusively anticlinal (perpendicular to the surface) as clearly marked outer cell layer termed the L1 or tunica (Esau 1965). Beneath the L1/tunica is an inner mass of cells collectively termed L2 (subepidermal) or corpus, where cells division is in all orientations (Abbe et al. 1951; Ledin 1954). The central zone of the SAM houses initial cells akin to stem cells; cells of this zone are less dense in cytoplasm with infrequent divisions (Jackson 2009). The subjacent peripheral zone cups the central zone and is characterized by cells that are more concentrated in cytoplasm engaged in more frequent division, which relates to this zone’s function in organogenesis. Underneath the girth of the peripheral zone is the centrally positioned rib zone, a region of more regular cell files that produce stem tissues.
Cell fate in the SAM is determined according to position, as demonstrated by numerous clonal analyses (Poethig 1987). Poethig et al. (1986) demonstrated that phytomers are clonally iterated, and others have shown that ~200 “founder” cells are recruited in the SAM for development of a phytomer (Steffensen 1968; Poethig 1984, 1987; McDaniel and Poethig 1988; Poethig and Szymkowiak 1995). Additionally, SAMs can be reprogrammed by responding to signals originating from lateral primordia (Irish and Karlen 1998; Je et al. 2016).
Leaves and the blade/sheath boundary
Maize leaves develop at the SAM flank. Leaf initiation is first detected by periclinal cell divisions in the L1 and L2 cell layers that correspond positionally to the emergent midrib along the medial axis of the incipient primordium (Sharman 1942; Esau 1965). Cell divisions spread bidirectionally and laterally around the entire circumference of the meristem flank to define an ensheathing, crescent-shaped ridge of founder cells known as the disc of insertion (Sharman 1942; Poethig 1984; Johnston et al. 2015). Additionally, lateral domains of the eventual distal blade and the proximal sheath are initiated during founder cell recruitment (Scanlon et al. 1996; Johnston et al. 2015). With initiation of each primordium, the sheathing leaf base surrounds the meristem and all younger leaves with characteristic overlapping lateral margins. Leaf margins trace back to overlapping domains of founder cells in the disc of insertion (Poethig and Szymkowiak 1995; Scanlon 2000). Delineation of the distinct founder cell population is a fundamental step in forming a boundary between the SAM and the developing leaf primordium (Johnston et al. 2015). Establishing and maintaining a boundary between the meristem and primordium is crucial to support stem cell homeostasis in the meristem and to ensure proper leaf morphology (Richardson and Hake 2019). From the disc of insertion, the primordium grows upward along the curvature of the meristem to produce its distinctive hood shape (Sharman 1942). Uniform cell divisions throughout the primordium account for growth that is continuous from the circumference of the meristem flank with the base of the primordium (Poethig 1984; Sylvester et al. 1990; Poethig and Szymkowiak 1995).
As the primordium grows from the meristem periphery, the three axes that define the mature maize leaf—proximal-distal, medial-lateral, and adaxial-abaxial—become apparent (Fig. 2a, b). Immature leaf primordia can be separated into proximal-distal regions soon after primordium emergence, although regions of the sheath and blade are morphologically indistinguishable (Sylvester et al. 1990). The plastochron can be used to track early developmental events. For example, a primordium staged between P1 and P3 consists solely of cells fated to become distal blade (Poethig and Szymkowiak 1995). The sheath margins that originate from overlapping regions in the disc of insertion become fated as early as P3/P4 (Poethig and Szymkowiak 1995; Johnston et al. 2015). A pre-ligule inductive signal is thought to initiate from the medial plane of the primordium at the midrib and move laterally to each margin (Becraft and Freeling 1991). This signal sets up the formation of the pre-ligule band, the first morphological indicator of a boundary between the blade and sheath (Sylvester et al. 1990). The pre-ligule band becomes apparent around P6 due to higher rates of localized anticlinal cell division and a decrease in cell expansion (Sylvester et al. 1990; Moon et al. 2013). At about P7, epidermal cells at the base of the pre-ligule band subsequently undergo periclinal divisions to give rise to the emerging ligule (Fig. 2c) (Sylvester et al. 1990). Much less is known about auricle development; however, it has been suggested that the cells of the pre-ligule band and overlying cells, positioned immediately distal to the emergent ligule, differentiate to form auricle (Sylvester et al. 1996). In summary, leaf development is defined by three crucial stages that occur between P0 and P7: (1) formation of the crescent-shaped disc of insertion between the meristem and incipient primordium that will eventually produce a boundary between successive leaf whorls; (2) separation of sheath lateral margins at overlapping domains within the disc of insertion; and (3) delineation of the sheath, ligule, auricle, and blade along the proximal-distal axis.
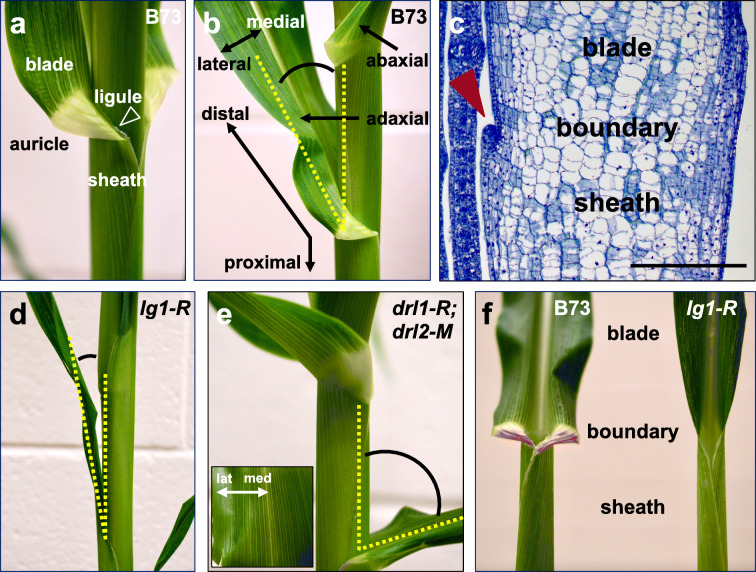
Fig. 2.
Basic trait units that contribute to maize leaf architecture. a Blade/sheath boundary of a mature B73 leaf. b Side view of a mature B73 leaf denoting major axes and blade angle. c Longitudinal section through the P7 blade/sheath boundary with arrowhead denoting the adaxial ligule. Scale bar, 200 μm. d Side view of a mature lg1-R mutant leaf with blade angle labeled. Note the more upright blade with reduced angle. e Side view of a mature drl1-R; drl2-M double mutant leaf with blade angle labeled. Note the more horizontal blade with increased angle. Inset, adaxial view of drl1-R; drl2-M double mutant leaf blade showing reduced midrib. f Front view of B73 and lg1-R leaf showing deletion of the ligule and auricle and narrow blade in lg1. Images in a, b, and d–f are not to scale
At P8 and beyond, a developing maize leaf exhibits a gradient of differentiation until the leaf is fully mature: the distal tip consists of dividing cells that will produce specific cell types, whereas concomitantly cells at the proximal base are still acquiring identity of unique cell types (Marjeran et al. 2010; Wang et al. 2013). Patterning along the adaxial-abaxial domains leads to differentiation of specific cell types on the dorsal and ventral sides of the leaf, respectively (Foster and Timmermans 2009). The adaxial epidermis is characterized by the ligule at the blade/sheath boundary and files of bulliform cells and macrohairs in the adult blade (Timmermans et al. 1998). Patterning of bulliform cell files, which may function in leaf rolling and blade angle, occurs in a region spanning 25–28 ml above the ligule during leaf expansion (Qiao et al. 2019). Adaxial-abaxial features are also expressed in internal tissues within polarized vascular bundles of xylem adaxially positioned to abaxial phloem and lignified sclerenchyma hypodermal cells that differentiate and connect to the adaxial epidermis at the midvein (Sharman 1942; Russell and Evert 1985). Along the medial-lateral axis of the blade, bundle sheath and mesophyll cells form rings around veins such that vascular bundles are separated in an ordered pattern of a bundle sheath cell, two mesophyll cells, and a bundle sheath cell (Hughes et al. 2019). Parallel to the midrib and leaf margins, densely arrayed lateral veins are separated from one another by two ranks of intermediate veins (Sharman 1942; Russell and Evert 1985; Hughes et al. 2019).
In the mature maize leaf, the proximal leaf sheath wraps around the stem and younger leaves to provide structural support (Kiesselbach 1949). The distal leaf blade projects away from the sheath at an angle, which maximizes light capture and gas exchange for photosynthesis. Sheath and blade tissues are joined at the ligule and auricle (Sharman 1942). The articulated auricle consists of a pair of triangular-shaped tissues that are widest at the leaf margin and relatively tapered at the midrib. The fringe-like ligule grows out from the epidermis at the base of the auricle (Fig. 2a) and may function in plant defense (Anderson et al. 2019; Kolkman et al. 2020). Interactions between the sheath, ligule, auricle, blade, and midrib significantly impact morphology of the shoot, primarily at the level of blade angle (Fig. 2b) (Sylvester et al. 1990; Becraft and Freeling 1991; Fowler and Freeling 1996; Harper and Freeling 1996; Strable et al. 2017; Tian et al. 2019). Because leaves are fundamental units of the canopy (Stewart et al. 2003), which is a key component of defining optimal plant density in row crops (Pepper et al. 1977; Lambert and Johnston 1978), regions of the leaf have undergone intense selection over the past century of maize breeding to alter its architecture (Tian et al. 2011; Tian et al. 2019). As a result, commercial maize hybrids exhibit a leaf architecture that is more upright in the upper nodes (Fig. 3) (Duvick 2005). Collectively, the constellation of traits that underlie leaf architecture has led to significant gains in grain yield in the last century of maize breeding (Pendleton et al. 1968; Duvick 2005; Tian et al. 2019).
Fig. 3.
Canopy and leaf structure in field-grown maize. a Maize hybrid field showing the difference between lower (more sprawling) and upper (more upright) leaf canopies separated by dotted line. b Three commonly used maize inbred lines, B73, Mo17, and W22, denoting the position of the upper ear (arrow) and the upper three leaves (arrowhead). Note the shift in blade angle of the upper leaves in B73
Axillary meristems
Axillary meristems initiate in the axil of the leaf sheath at the stem node (Fig. 1b, c). Once formed, the meristem initiates a protective whorl of unexpanded leaves, producing an axillary bud. After the bud forms, its growth largely arrests when the bud enters a state of quiescence (Figs. 1a and 4a–c). In maize, an axillary bud can either remain dormant or grow based on its position along the shoot (Doebley et al. 1990). Axillary buds from the middle to upper nodes of the maize shoot grow in response to floral transition to produce a lateral branch that bears an ear. Buds of the basal nodes may remain dormant or grow into tillers. Tillers are akin to long branches that recapitulate phytomers of the main shoot. The capacity for a bud to remain dormant or become active is highly regulated by both internal and external stimuli (Whipple et al. 2011; Dong et al. 2019a). Maize concentrates resources to the primary shoot axis, resulting in the suppression of axillary bud growth. When overall apical dominance of the shoot is reduced, buds grow into tillers and compete with the shoot for resources (Fig. 4a) (Doebely et al. 1997).
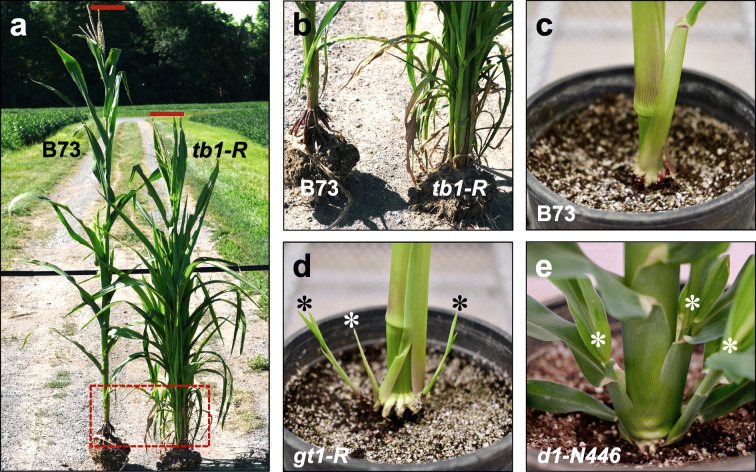
Fig. 4.
Tiller growth in maize. a Field-grown B73 and tb1-R mutant (backcrossed in B73). Red bars denote plant height. Boxed region shown in (b). b High tiller number from outgrowth of basal axillary buds in tb1-R, whereas B73 axillary bud growth is suppressed. c B73 with suppressed axillary bud growth. d gt1-R mutant with active axillary bud growth (leaves have been removed). e d1-N446 mutant with active axillary bud growth (leaves have been removed)
Intercalary meristems
The maize stem is a cylindrical structure that provides mechanical support below the shoot apex (Peiffer et al. 2013). The stem is comprised of repeating units of node, where a leaf is attached, and internode, where stem growth occurs (Kiesselbach 1949). Shoot development is divided into juvenile and adult vegetative phases and a reproductive phase (Poethig 2003). During the juvenile phase in the first few weeks of shoot growth, juvenile internodes elongate very little, and as a result, node spacing is compact and the leaf whorl is tight. During transition to the adult phase and at the onset of the reproductive phase, adult internodes undergo substantial elongation, and spacing between nodes and leaves lengthens (Morrison et al. 1994; Poethig 2003). As a result, adult internodes constitute a significant proportion of overall plant height (Peiffer et al. 2014). Collectively, the variable individual growth rates and the differential elongation of each internode produce an overall sigmoidal pattern (Fig. 1b). This growth pattern importantly separates photosynthetic leaves along the shoot axis, improving efficient light capture and gas exchange, as well as elevating the tassel and ear to promote anemophily, i.e., wind pollination (Ku et al. 2015).
Internode elongation is controlled by the intercalary meristem positioned immediately above the node at the base of each internode (Tsuda et al. 2017). Node and internode tissues are derived from a shared cell population from the shoot apex (Jorhi and Coe 1983). Intercalary meristems regulate internode elongation independent of the SAM; additionally, growth is not synchronous for all internodes (Morrison et al. 1994). Vascular tissue develops in association with internodes and, in transverse section, appears separate and dispersed throughout the stem (Tsuda et al. 2017).
Genes that shape shoot architecture of maize: an overview
Maize geneticists have been identifying, collecting, and maintaining mutants with altered shoot architecture for over 100 years (Fig. 5) (Neuffer et al. 1997). Hundreds of mutations affect nearly every organ and cell type in maize; cloning numerous loci has provided a wealth of information about the functional and agronomic properties of maize. Some of the genes described in maize are unique to the grasses and therefore underscore the importance of maize as a model for discovering new developmental genes with no orthologs in eudicots. A current, albeit far from comprehensive, survey of the characterized loci that impact maize shoot architecture are summarized in Table S1 (in the supplementary material). Of the cloned genes, approximately 73% have significant associations with polymorphisms that explain natural variation in vegetative traits (Table S1). A common theme that emerges from mutant analyses is that many of the genes are pleiotropic (Table S1). Untangling the individual phenotypic components of pleiotropy is crucial if developmental genes are to be utilized to improve shoot architecture in maize. The following sections highlight several mutant and gene characterization studies with the primary focus on components of shoot architecture: shoot meristem, leaf, axillary bud, and stem.
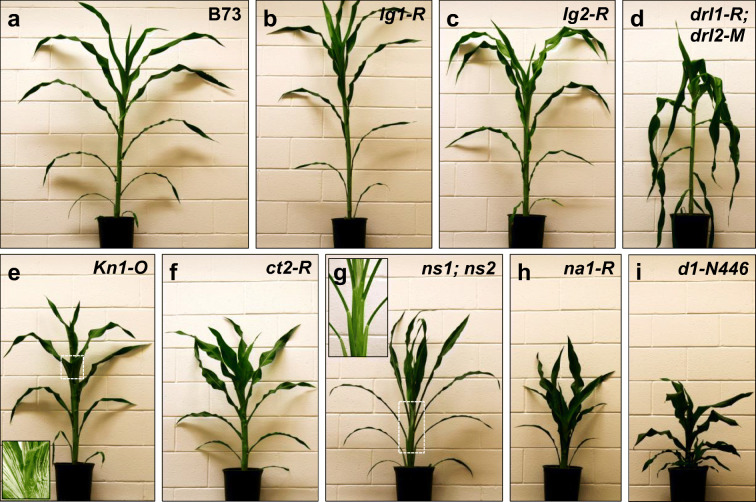
Fig. 5.
Variation in shoot architecture caused by mutations in key developmental regulators. All mutants are backcrossed in B73, were greenhouse grown, and were imaged at the same stage of maturity. a B73. b liguleless1-R (lg1-R). c liguleless2-R (lg2-R). d drooping leaf1-R; drooping leaf2-M (drl1-R; drl2-M). e Knotted1-O (Kn1-O); inset, boxed region showing knots on Kn1-O leaves. f compact plant2-R (ct2-R). g narrow sheath1; narrow sheath2 (ns1; ns2); inset, boxed region showing narrow sheaths and lower blade of ns1; ns2 h nana plant1-R (na1-R) i dwarf plant1-N446 (d1-N446)
Shoot apical meristem establishes and maintains shoot development
Shoot apical meristem maintenance
One of the best characterized plant transcription factors that regulates meristem activity is KNOTTED1 (KN1), the founding member of the plant KN1-LIKE HOMEOBOX (KNOX) gene family (Hake et al. 1989; Vollbrecht et al. 1991). Some dominant, gain-of-function alleles of KN1 produce “knots” by ectopically expressing KNOX genes along lateral veins in the blade (Fig. 5e, inset) (Freeling and Hake 1985; Sinha and Hake 1990). These knots are reprogrammed cells that continue to divide as if they were meristematic and eventually differentiate into sheath and auricle rather than blade. In contrast, strong loss-of-function kn1 mutants show failure to initiate or maintain the SAM (Kerstetter et al. 1997). Loss-of-function alleles of KN1 are sensitive to genetic modifier loci in various maize backgrounds (Vollbrecht et al. 2000). In permissive backgrounds, vegetative development is mostly normal, but development of the tassel and ear is severely compromised; in restrictive backgrounds, the SAM cannot be maintained, resulting in a limited (abortive) shoot (Vollbrecht et al. 2000). KN1 transcripts accumulate throughout the SAM and developing stem, but not in the incipient primordium (P0), developing primordia (>P1), or the L1 layer of the meristem proper (Jackson et al. 1994). KN1 protein, however, traffics cell-to-cell via plasmodesmata and is found in the base of young leaf primordia and in the L1 (Lucas et al. 1995; Jackson 2002; Kim et al. 2002; Xu et al. 2011). By leveraging mRNA sequencing (RNA-seq) in kn1 loss-of-function mutant immature ears and tassels and SAMs with chromatin immunoprecipitation paired sequencing (ChIP-seq) of normal developing ears, Bolduc et al. (2012b) found that KN1 binds to over 5000 loci in the maize genome and modulates the expression of around 650 genes. Many of KN1 target genes encode transcription factors and components of phytohormone pathways, which include auxin and gibberellin (Bolduc and Hake 2009; Bolduc et al. 2012b). Such candidate genes could be well-suited for improving maize shoot architecture.
Genetic analysis can be complicated by functional redundancy (Kafri et al. 2009). KNOX genes exhibit redundancy that is hierarchical: KNOX family members cannot substitute for KN1 function, but knox mutants enhance kn1 in a dose-dependent manner (Bolduc et al. 2014). A loss-of-function mutation in the KNOX gene ROUGH SHEATH1 (RS1) does not condition a single mutant phenotype. However, in combinations with a kn1 loss-of-function allele, rs1 heterozygous or homozygous mutants interact synergistically, resulting in a limited shoot (Bolduc et al. 2014). KN1 and RS1 transcripts accumulate in overlapping domains in the shoot apex (Jackson et al. 1994), and KN1 and RS1 proteins bind and modulate the expression of similar genes (Bolduc et al. 2014). Furthermore, KN1 can heterodimerize with transcription factors that co-localize in planta, such as BELL1-like homoeobox12 (BLH12) and BLH14 (Tsuda et al. 2017). Neither blh12 nor blh14 single mutants display an obvious meristem phenotype; however, blh12; blh14 double mutants have reduced SAM height and width, as well as suppressed axillary meristem maintenance (Tsuda et al. 2017), similar to kn1 loss-of-function alleles (Kerstetter et al. 1997; Vollbrecht et al. 2000). Whether meristem phenotypes of blh12; blh14 double mutants are sensitive to modifier loci in diverse genetic backgrounds remains to be explored.
Limited shoot phenotypes also arise from mutations in genes that participate in small RNA, phytohormone, metabolite, and nutrient pathways. Small RNAs (sRNAs) are crucial regulators of shoot development and have been linked to SAM establishment and maintenance (Klesen et al. 2020). Strong alleles of RAGGED SEEDLING2/ARGONAUT7 and RNA-DEPENDENT RNA POLYMERASE6 machinery components of sRNA production give rise to seedlings with limited shoots that terminate shortly after germination (Douglas et al. 2010; Petsch et al. 2015). Additional shoot abortive phenotypes can be teased out by generating double mutants between sRNA biosynthesis components, such as by stacking mutant alleles in LEAFBLADELESS1/SUPPRESSOR-OF-GENE-SILENCING3 and DICER-LIKE4 (Petsch et al. 2015). Plant hormones regulate development of specific regions of the SAM (Hepworth and Pautot 2015). The phytohormone cytokinin promotes proliferation of meristematic cells (Giulini et al. 2004; Kurakawa et al. 2007). The maize cytokinin biosynthesis gene LONELY GUY7 (ZmLOG7) is expressed in the meristem tip, and Zmlog7 mutants display a limited shoot (Knauer et al. 2019). Additionally, SAM maintenance is promoted by certain metabolites. BLADEKILLER1 (BLK1) encodes a THIAMINE BIOSYNTHESIS2 (THI2) enzyme; blk1/thi2 mutants display a progressive reduction in meristem height that leads to an abortive adult shoot (Woodward et al. 2010). blk1/thi2 mutant shoots can be rescued with exogenous thiamine, highlighting the importance of thiamine in meristem maintenance (Woodward et al. 2010).
Shoot apical meristem size
A core pathway at the heart of stem cell homeostasis in plants is the CLAVATA (CLV)-WUSCHEL (WUS) feedback loop, which consists of peptide ligands, receptors, and transcription factors that operate in different regions of the meristem (Somssich et al. 2016). Briefly, stem cell fate is promoted by WUS, a homeodomain transcription factor. WUS transcripts accumulate in cells situated between the central and rib zones, but WUS protein moves into the central zone where activates the expression of CLV3, which encodes a secreted peptide. The central zone-derived CLV3 peptide ligand binds to the leucine-rich repeat receptor-like kinase (LRR-RLK) CLV1. CLV1 is expressed beneath the central zone, whereas transcripts for the LRR-RL protein (LRR-RLP) encoding CLV2 gene accumulate throughout the SAM. Both CLV1 and CLV2 are required to repress WUS expression. The CLV-WUS pathway is conserved in maize, as highlighted by genetic analyses that have mainly focused on meristems in inflorescences (Wu et al. 2018). Mutations in the CLV2 in ortholog FASCIATED EAR2 (FEA2) significantly enlarged the SAM (Taguchi-Shiobara et al. 2001; Je et al. 2018), as do mutations in a CLV3 ortholog, maize CLV3/EMBRYO SURROUNDING REGION-RELATED7 (ZmCLE7; Rodriguez-Leal et al. 2019). Interestingly, a mutant allele of the maize CLV1 ortholog THICK TASSEL DWARF1 (TD1; Bommert et al. 2005) reduces SAM size, which is consistent with the observation that td1; kn1 double mutants cannot rescue the limited shoot of kn1 loss-of-function mutants (Lunde and Hake 2009). Maize WUS1 (ZmWUS1) and ZmWUS2 are putative WUS co-orthologs (Nardmann and Werr 2006); however, laser microdissection, single-cell RNA sequencing, and RT-PCR analyses of the maize vegetative shoot apex did not detect cells that accumulated ZmWUS1/2 transcripts, suggesting that the canonical CLV-WUS pathway appears to have been bypassed in the maize vegetative SAM (Knauer et al. 2019; Satterlee et al. 2020).
Enlargement of the SAM results from mutations in several key signaling components of stem cell homeostasis: the LRR-RLP encoding FEA3 (Je et al. 2016), the PERIANTHIA (PAN) ortholog FEA4 (Pautler et al. 2015), the α-subunit (Gα) of a heterotrimeric GTP binding protein encoded by COMPACT PLANT2 (CT2; Bommert et al. 2013), the β-subunit (Gβ) of a heterotrimeric GTP binding protein (Wu et al. 2020), the maize ortholog of rice FON2-LIKE CLE PROTEIN1 (ZmFCP1; Je et al. 2016; Knauer et al. 2019), the CORYNE ortholog ZmCRN (Je et al. 2018), and a cytokinin type A response regulator encoding gene ABERRANT PHYLLOTAXY1 (ABPH1; Giulini et al. 2004). Additionally, a subset of genes that regulate meristem size also impact adult shoot architecture. Mutant abph1 plants additionally display decussate phyllotaxy due to broadening of the SAM (Giulini et al. 2004). In certain genetic backgrounds, fea4 mutants have reduced shoot stature (Pautler et al. 2015). FEA4 interacts with MALE STERILE CONVERTED ANTHER1 (MSCA1), a glutaredoxin protein (Yang et al. 2015). Mutant mcsa1 plants exhibit reduced SAM size, whereas mis-expression of MCSA1 in the dominant Abph2 mutant increases SAM size and results in decussate phyllotaxy (Yang et al. 2015), similar to abph1 mutants. In addition to a larger SAM, ct2 mutants display an overall reduced shoot stature, and leaf blades are relatively wider and more upright compared to non-mutants (Fig. 5f) (Bommert et al. 2013; Wu et al. 2018).
Besides core signaling pathways that operate within stem cells of the SAM, signaling feedback to the SAM from leaf primordia is crucial for balancing stem cell proliferation. The ZmFCP1 peptide is produced in lateral primordia and signals to its receptor FEA3, which is localized to cells that subtend meristem central and core regions (Je et al. 2016). Maize YABBY (YAB)/CRABS CLAW co-orthologous genes DROOPING LEAF1 (DRL1/YAB2) and DRL2/YAB7 are expressed in the incipient and initiating leaf primordia, but not in the SAM or subtending stem (Strable et al. 2017). However, meristem size is reduced in drl1 and drl2 single mutants and in a dose-dependent manner in drl1; drl2 double mutants (Strable et al. 2017). These observations indicate the DRL genes function non-cell autonomously to regulate stem cell proliferation and suggest that activity levels and/or interactions among gene products from the loci are important to their function (Strable et al. 2017).
Micronutrients can impact SAM size. Mutations in an aquaporin encoding gene, TASSEL-LESS1 (TLS1), confer a progressive reduction of SAM size that can lead to complete meristem consumption (Durbak et al. 2014). TLS1 is necessary for boron transport during shoot development, as tls1 mutants have low boron content. Furthermore, tls1 meristem defects can be rescued by boron fortification, underlining the importance of the bioavailability of this micronutrient in soils (Durbak et al. 2014).
Shoot apical meristem determinacy
Maize shoots become determinate, or limited in growth, when the SAM differentiates into the tassel (Fig. 1b, d). Determinate meristems produce a limited number of organs before vegetative growth terminates and reproductive growth is acquired. Indeterminate activity of a meristem, on the other hand, results in prolonged growth due to sustained self-renewal of stem cells and continuous initiation of new organs. For a majority of maize lines, the SAM produces a predictable range of leaf primordia during vegetative growth prior to the reproductive phase. In temperate maize inbred lines, the number of leaves initiated before the SAM converts to a tassel is background dependent and can range from 13 to 22 leaves, in either short- or long-day photoperiods (Russell and Stuber 1983; Meng et al. 2011). By comparison, early-flowering Gaspé Flint maize produces 6–8 leaves before flowering in either short- or long-day photoperiods, whereas photoperiod-sensitive tropical maize CML436 produces 26–30 leaves under long days and 18–20 leaves in short days (Meng et al. 2011).
Leaf number is a major component of shoot architecture in maize. Leaves along the shoot can be divided into two groups: those below the uppermost (primary) ear, where the canopy is relatively more sprawling, and those above the uppermost ear where the canopy is relatively more upright (Fig. 4a) (Vlăduţu et al. 1999; Flint-Garcia et al. 2005; Li et al. 2016). The proportion of lower and upper canopy leaves importantly enhances photosynthetic efficiency across pre- and post-anthesis stages of shoot development, respectively (Duncan 1971; Zhang et al. 2017). Maize is rich in natural variation for total leaf number (Russell and Stuber 1983; Flint-Garcia et al. 2005; Li et al. 2016). For example, in a set of 100 diverse inbred lines, total leaf number ranged from 7 to 19, leaf number below the primary ear ranged from 3 to 10, and leaf number above the primary ear ranged from 3 to 8 (Flint-Garcia et al. 2005). Most modern, temperate maize lines are photoperiod insensitive (Colasanti and Muszynski 2009), and developmental (i.e., endogenous) programs regulate vegetative to floral transition (Irish and Nelson 1991; Irish and Jegla 1997). Genetic studies on leaf number are largely deduced from variation in flowering time. This intricate that link between flowering time and total leaf number is due to the fact that floral transition converts the vegetative SAM into a tassel; thus, floral transition marks an end to leaf production.
Some genes regulate leaf initiation rate and total leaf number without significantly changing flowering time. Mutations in the VIVIPAROUS8 (VP8) gene increase leaf initiation rate especially during early shoot growth, such that at any same stage of growth, vp8 mutants display more leaves when compared with normal siblings (Evans and Poethig 1997). Interestingly, vp8 have more leaves at maturity compared with normal sibling plants, and yet both genotypes flower simultaneously (Evans and Poethig 1997). VP8 encodes a membrane-localized peptidase related to the Arabidopsis ALTERED MERISTEM PROGRAM1, which may be important for abscisic acid (ABA) homeostasis (Suzuki et al. 2008). Similarly, terminal ear1 (te1) mutants initiate leaves more frequently compared with normal sibling plants, as well as alters phyllotaxy (Veit et al. 1998). TE1 encodes an RNA binding protein (Veit et al. 1998), for which the target RNAs remain uncharacterized. The SQUAMOSA PROMOTER BINDING (SBP) transcription factor encoding UNBRANCHED2 (UB2), UB3, and TASSELSHEATH4 genes function redundantly in the central domain of the SAM to balance the rate of cell differentiation during lateral primordia initiation (Chuck et al. 2014). This balance is lost in ub2; ub3; tsh4 triple mutants, where an abbreviated plastochron index increases total leaf number without altering flowering time (Chuck et al. 2014). Mutants with shifts in total leaf number and a subsequent shift in flowering time also greatly alter shoot architecture and are covered elsewhere (Colasanti and Muszynski 2009).
Natural variation of the SAM morphology
Exploring natural diversity of SAM morphology and understanding its underlying genetic architecture requires phenotypic analysis at microscopic scale. A handful of studies in maize have utilized biparental recombinant inbred line (RIL) and maize diversity panels to quantify SAM width, height, arch length, midpoint width, height of P1, plastochron internode length, and cell number of the L1 layer along the length of meristem arc (Thompson et al. 2014, 2015) or to model the SAM as a paraboloid (Leiboff et al. 2015). Interestingly, candidate genes in quantitative trait locus (QTL) regions that associate with SAM size and morphology do not represent genes that have been characterized previously to regulate SAM activity, suggesting that genes crucial to SAM maintenance do not necessarily contribute to natural variation of SAM morphology (Thompson et al. 2014, 2015; Leiboff et al. 2015). Curiously, SAM size and flowering time are correlative, such that maize lines with large SAMs associate with earlier flowering (Thompson et al. 2015; Leiboff et al. 2015). As expected, maize lines with larger SAMs also correlate with fewer nodes and reduced plant height (Leiboff et al. 2015). The height of the SAM relates to stem diameter, whereas SAM radius is associated with the height of the primary ear along the plant axis (Leiboff et al. 2015). These SAM “microphenotypes” were recently utilized in genomic prediction models as a way to optimize trait selection in breeding programs (Yu et al. 2020). Furthermore, laser microdissection coupled with transcriptomic analysis of SAM functional domains uncovered gene regulatory circuits that also regulate adult shoot traits (Knauer et al. 2019).
Leaf architecture is governed by multiple interacting traits
Blade/sheath boundary and blade angle
Maize leaves play a significant role in defining overall shoot architecture. The blade/sheath boundary, articulated by auricle and ligule tissues, impacts blade angle and is a crucial feature of the maize leaf. Dominant, gain-of-function mutations in KNOX genes, such as in the transcriptional regulators KN1, RS1, GNARLEY1 (GN1), LIGULELESS3 (LG3), and LG4, have highlighted a key role for these genes in specifying proximal cues during the establishment of the blade/sheath boundary (reviewed in Bolduc et al. 2012a). Leaves of neomorphic Knox mutants produce ectopic blade/sheath boundaries due to the mis-regulation of KNOX genes during leaf development (Fig. 5e). Cells in the blade that mis-express KNOX genes take on a sheath-like identity; the juxtaposition of blade and ectopic sheath-like cells produces an ectopic boundary, where ligule-/auricle-like tissues subsequently develop (Smith et al. 1992; Jackson et al. 1994; Freeling and Hake 1985; Becraft and Freeling 1994; Schneeberger et al. 1995; Fowler et al. 1996; Muehlbauer et al. 1999; Foster et al. 1999). A dominant, gain-of-function allele of the WAVY AURICLE IN BLADE1 (WAB1) gene similarly displaces auricle-like tissue distally into the blade due to WAB1 mis-expression (Hay and Hake 2004; Lewis et al. 2014). Interestingly, ligule tissue does not develop with the ectopic auricle of Wab1 mutants, and kn1 loss-of-function mutants have no effect on Wab1, suggesting WAB1 functions independent of the KNOX pathway (Hay and Hake 2004). WAB1 encodes a TCP (TEOSINTE BRANCHED1, CYCLOIDEA, PCF) transcription factor that is not normally expressed during leaf development (Lewis et al. 2014). The HAIRY SHEATH FRAYED1 (HSF1) gene encodes a maize HISTIDINE KINASE1 receptor for cytokinin (Muszynski et al. 2020). Semi-dominant Hsf1 mutants initiate proximal sheath-like prongs along the distal blade margin due to cytokinin hypersignaling, suggesting that ectopic activation of cytokinin response is sufficient for activating at least some leaf patterning programs (Bertrand-Garcia and Freeling 1991; Muszynski et al. 2020). Deciphering the downstream genetic networks for WAB1 and HSF1 will be important next steps in understanding how they regulate leaf patterning.
Once the blade/sheath boundary is established, the ligule and auricle region is elaborated (Fig. 2a, b). A recessive, loss-of-function class of liguleless (lg) mutants has illuminated the importance of specifying the pre-ligule band (~P6) and proper ligule outgrowth (~P7/P8) (Sylvester et al. 1990; Moreno et al. 1997; Walsh et al. 1998). Mutant lg1 plants were originally identified by their acutely upright leaves (Figs. 2d and 5b) (Emerson 1912a). Ligule and auricle tissues are not established in lg1 mutants, and yet despite lacking these tissues, the blade and sheath are separated by a clear boundary (Fig. 2d, f) (Becraft et al. 1990; Sylvester et al. 1990; Becraft and Freeling 1991; Moreno et al. 1997; Li et al. 2017). Genetic mosaic analysis indicates LG1 functions cell autonomously in an early step of ligule and auricle initiation (Becraft and Freeling 1994). LG1 encodes a grass-specific SBP transcription factor, and LG1 transcripts and LG1 protein accumulate in the pre-ligule band and throughout the initiating ligule (Moreno et al. 1997; Moon et al. 2013; Johnston et al. 2014; Lewis et al. 2014). LG1 is mis-expressed in dominant, gain-of-function Wab1 mutants in domains of the leaf that also ectopically express WAB1 (Lewis et al. 2014). However, in wab1 loss-of-function mutants, expression of LG1 at the pre-ligule band is unaltered, suggesting that although WAB1 is sufficient to regulate LG1 expression, it is not required (Lewis et al. 2014). This observation leaves open the possibility that LG1 is regulated by a different TCP transcription factor. Additional insight into LG1 regulation was provided through fine mapping and cloning a QTL for leaf angle, qLA2. Cloning qLA2 uncovered a maize gene orthologous to rice INCREASED LEAF INCLINATION1 (ZmILI1; Ren et al. 2020). ZmILI1 is a helix-loop-helix (HLH) protein that has affinity for the LG1 promoter (Ren et al. 2020). Because HLH proteins lack a basic domain required for DNA binding (Ruzinova and Benezra 2003; Zhang et al. 2009), but form heterodimers with bHLH proteins, it will be important to understand the mechanism for which ZmILI1 associates with target promoters. One such mechanism could be through heterodimerization with maize ILI1 BINDING bHLH1-1 (ZmIBH1-1), a gene that underlies the qLA2-1 leaf angle QTL and encodes a bHLH protein with a basic DNA binding domain (Cao et al. 2020). However, whether ZmILI1 and ZmIBH1-1 physically interact to regulate LG1 expression currently remains to be reported.
The recessive, loss-of-function lg2 mutant has upright leaves due to incomplete patterning of the auricle and ligule (Fig. 5c). In contrast with lg1 mutants, a reduced ligule and auricle develop at the margin but not at the midrib in lg2 mutants, and the blade/sheath boundary is shifted along the proximal/distal axis (Harper and Freeling 1996). Although the expression pattern of LG2 has not been reported by RNA in situ hybridization, LG2 is expressed broadly in the SAM and young primordia (i.e., ≤P4), earlier than LG1 expression, according to transcriptomic analyses of laser microdissected sections of the maize shoot apex (Knauer et al. 2019; Leiboff et al. 2020). Genetic mosaic analysis suggests that LG2 functions non-cell autonomously (Harper and Freeling 1996). LG2 encodes a grass-specific TGA-class bZIP transcription factor (Walsh et al. 1998). Genetic interaction analysis between lg1 and lg2 mutants indicates the two genes work in the same pathway, with LG2 potentially functioning earlier than LG1 to establish the position of the blade/sheath boundary (Harper and Freeling 1996). In support, LG2 is expressed early and is bound by KN1, whereas LG1 is expressed late and is not a KN1 target (Bolduc et al. 2012b). The Arabidopsis TGA-class bZIP protein PAN can interact with organ boundary proteins BLADE-ON-PETIOLE1 (BOP1) and BOP2 in yeast and in planta (Hepworth et al. 2005; Xu et al. 2010). It is tempting to speculate that LG2 may similarly form heterodimers with other transcriptional regulators to establish and/or pattern the blade/sheath boundary.
Connecting developmental events, such as patterning with growth, along the proximodistal, mediolateral, and adaxial-abaxial axes of the leaf is critical to understanding leaf architecture. The semi-dominant Liguleless narrow (Lgn) mutant links development of the blade/sheath boundary with medial/lateral growth (Moon et al. 2013). Leaf width is significantly reduced in Lgn mutant leaves, and the ligule develops only at the midrib along the medial plane of an otherwise poorly patterned blade/sheath boundary (Moon et al. 2013). LGN encodes a serine/threonine kinase, in which the kinase activity is severely compromised in Lgn mutants (Moon et al. 2013). Expression levels of LG1 and LG2 are low in Lgn mutants; moreover, genetic interactions between lg1 and lg2 with Lgn are synergistic (Moon et al. 2013). Similar to the early expression pattern of the KN1-target LG2, LGN transcripts accumulate early and ubiquitously in the leaf, and LGN is bound by KN1 (Bolduc et al. 2012b; Moon et al. 2013). Thus, a LGN-mediated phosphorylation cascade may regulate a ligule signal from the medial midrib laterally to the leaf margin. Because TGA-class bZIP activity can be regulated post-translationally by phosphorylation (Schütze et al. 2008), it will be important to know if LG2 and LGN physically interact and if LGN can phosphorylate LG2. Lgn is sensitive to genetic modifier loci, as noted by its variable expressivity in various genetic backgrounds and response to temperature (Buescher et al. 2014). Cloning a large effect modifier QTL for Lgn, Sympathy for the ligule (Sol), uncovered a homolog of Arabidopsis ENHANCED DISEASE RESISTANCE4 (EDR4), suggesting Lgn may trigger a MAP kinase signaling cascade, whereas Sol/EDR4 may suppress this cascade (Anderson et al. 2019).
Cloning of the semi-dominant Semidwarf3 (Sdw3) mutant revealed the importance of the phytohormone ethylene in regulating growth at the blade/sheath boundary (Li et al. 2020a). SDW3 encodes a maize ethylene biosynthesis enzyme, ZmACS7, and the Sdw3 mutant overproduces ethylene (Li et al. 2020a). Mutant Sdw3 plants have a wide leaf angle due to growth-related expansion of the auricle at the margin (Li et al. 2020a). The phytohormone brassinosteroid plays a key role in regulating blade/sheath boundary development in maize. RNA interference (RNAi) downregulation of maize BRASSINOSTEROID INSENSITIVE1 (ZmBRI1) homologs gave rise to leaves with highly reduced ligule and auricle tissues (Kir et al. 2015). Developing ligule and auricle cells accumulate a fluorescent protein marker of brassinosteroid activity, and Zmbri1-RNAi lines have reduced accumulation of this fluorescent protein marker (Kir et al. 2015). Mutations in the brassinosteroid biosynthesis genes BRASSINOSTEROID DEPENDENT1 (BRD1; Makarevitch et al. 2012) and NANA PLANT1 (NA1; Hartwig et al. 2011) disrupt margin-to-margin expansion of the ligule and result in acute involution of auricle tissue (Makarevitch et al. 2012; J. Strable, unpublished data). Furthermore, na1 mutants have markedly upright leaves (Fig. 5h). Laser microdissection of the developing ligule and auricle region coupled with RNA sequencing of these tissues found that BRD1 and a maize homolog of Dwarf11 were both upregulated in pre-ligule tissue, further implicating brassinosteroid biosynthesis in patterning the blade/sheath boundary (Johnston et al. 2014). Interestingly, the rice HOMEOBOX1 (OSH1) transcription factor dampens the brassinosteroid pathway by upregulating the expression of CYTOCHROME P450 (CYP) brassinosteroid catabolic genes. Furthermore, cyp-RNAi lines have patterning defects of the blade/sheath boundary (Tsuda et al. 2014). In maize, CYP genes are direct targets of KN1 and are upregulated in Kn1 dominant, gain-of-function mutants (Bolduc et al. 2012b). These observations suggest that KNOX proteins regulate CYP genes to maintain low levels of brassinosteroid in some cells of the developing leaf, which may be necessary to establish the blade/sheath boundary in the cereals (Tsuda et al. 2014).
The maize DRL genes regulate leaf development along all three axes (Strable et al. 2017). The syntenic paralogous DRL1 and DRL2 genes are co-orthologous to Arabidopsis CRABS CLAW and rice DROOPING LEAF genes (Bowman and Smyth 1999; Yamaguchi et al. 2004; Strable et al. 2017). The moderate droopy leaves of drl1 single mutants are enhanced in drl1; drl2 double mutants (Figs. 2e and 5d). Not only is the midrib markedly reduced by mutations in the DRL genes, auricle tissue is distally expanded at the presumptive midrib (Fig. 2e, inset), and hypodermal sclerenchyma cells are underdeveloped (Strable et al. 2017). The blade angle traits in drl mutants, along with narrower blades and elongated internodes, were noted previously (Strable et al. 2017) as reminiscent of rice brassinosteroid mutants (Yamamuro et al. 2000; Zhang et al. 2009; Sun et al. 2015), which either have elevated brassinosteroid levels or enhanced brassinosteroid signaling. In summary, the interaction of midrib, auricle, and supportive sclerenchyma cells are key factors that control blade angle, and all are under the control of the DRL genes (Strable et al. 2017).
Leaf morphometrics: length, width, and size
Leaf morphology and size greatly impact photosynthetic capacity. Leaves are the primary photosynthetic organ of the maize shoot and play crucial roles in shoot growth and grain filling (Duncan 1971; Stewart et al. 2003; Lee and Tollenaar 2007; Ma et al. 2014; Zhao et al. 2015; Zhang et al. 2017). Genetic analysis of leaf development has identified several key regulators of blade length, blade width, and sheath length. Many of the genes encode transcription factors that also regulate hormone biosynthesis, signaling, or response pathways. Dominant Kn1-N mutants have wider and shorter leaf blades and shorter sheaths. This is due in part to KN1 directly activating the gibberellin catabolic gene GA2 OXIDASE1 (GA2ox1; Bolduc and Hake 2009). Mutations in a gibberellin biosynthesis gene DWARF PLANT3 (D3) result plants with short stature and wide blades (Winkler and Helentjaris 1995), and d3 mutants have low levels of bioactive gibberellin (Phinney and Spray 1982; Fujioka et al. 1988; Winkler and Helentjaris 1995). Transgenic maize that overexpress a gibberellin anabolic-related gene, GA20ox5 homolog, has increased blade length along with increased plant height (Nelissen et al. 2015). Analysis of KN1 target genes by ChIP-seq found that KN1 preferentially binds adjacent to genes in the gibberellin pathway, including GA2ox1 and GA20ox5, among other phytohormone pathways (Bolduc et al. 2012b). In Rs1 and Gn1 mutants, sheath length is similarly reduced, and blade length tends to be unchanged (Becraft and Freeling 1994; Foster et al. 1999). GA20ox5 is mis-expressed in recessive kn1 mutants, and GA2ox1 is mis-expressed in dominant Kn1, Rs1, Gn1, and Lg3 mutant leaves, where enrichment of KNOX binding overlaps KN1 target loci (Bolduc and Hake 2009; Bolduc et al. 2014). These observations suggest that KNOX genes have redundant functions in regulating hormonal response. In support, loss-of-function mutations in RS2, a negative regulator of KNOX genes during leaf development, produce short stature mutant plants with wide blades (Schneeberger et al. 1998). Mutant rs2 plants ectopically accumulate KNOX proteins in developing leaves (Schneeberger et al. 1998; Tsiantis et al. 1999; Timmermans et al. 1999); however, the gibberellin levels or responsiveness have not been reported in rs2 mutants. Collectively, these studies suggest that KNOX-mediated control of gibberellin levels at the base of newly initiating leaves is important in coordinating cell expansion and elongation during leaf development.
Studies of other phytohormones and growth regulators have expanded the framework for understanding the regulation of maize leaf morphology and size. In addition to critical roles in patterning the blade/sheath boundary, brassinosteroid also plays an important role in determining leaf size. Mutations in the rice DWARF1 homolog NA2, a brassinosteroid biosynthesis gene, result in reduced brassinosteroid metabolites and short stature plants with shorter and narrower leaf blades (Best et al. 2016). Dampening brassinosteroid signaling in Zmbri1-RNAi knockdown lines also conditions reduced blade and sheath lengths largely through decreasing the number and rate of dividing cells (Kir et al. 2015). Mis-regulation of cytokinin signaling through the semi-dominant Hsf1 mutant decreases blade and sheath length, as well as blade width (Muszynski et al. 2020). Similarly, the recessive abph1 mutant, a negative regulator of cytokinin signaling, results in narrower blades (Jackson and Hake 1999; Giulini et al. 2004). Treating non-mutant seedlings with exogenous cytokinin phenocopies the effects of Hsf1 on leaf growth (Muszynski et al. 2020). The ethylene pathway functions largely to restrict growth (Dubois et al. 2018). Increased stability of the ZmACS7 biosynthesis enzyme in the semi-dominant Sdw3 mutant leads to increased ethylene production in sheath tissues and a subsequent reduction in sheath length (Li et al. 2020a). Exogenous application of the ethylene precursor ACC also reduces sheath length (Li et al. 2020a), as well as leaf blade length (J. Strable, unpublished data).
Further insight into the regulation of maize leaf morphology and size is provided by genetic analyses of transcription factors that pattern the blade/sheath boundary. In lg1 and drl1 single mutants, leaf blades are narrower and shorter, and sheaths are longer (Foster et al. 2004; Strable et al. 2017). These traits are enhanced in drl1; drl2 double mutants (Strable et al. 2017). Furthermore, a genome wide association study (GWAS) identified rare polymorphisms with large phenotypic effects for leaf width within QTL intervals for DRL1/YAB2 and DRL2/YAB7 (Strable et al. 2017). The duplicate NARROW SHEATH genes (NS1 and NS2) encode WUS-HOMEOBOX transcription factors and are required to recruit founder cells from the SAM into marginal domains of the primordium, mediating mediolateral growth of the sheath and blade (Scanlon et al. 1996; Nardmann et al. 2004). The NS gene products accumulate in a lateral domain of the primordium (Nardmann et al. 2004; Conklin et al. 2020). Double mutant ns1; ns2 plants have narrow sheaths and proximal blades that lack morphological markers of margin identity (Fig. 5g) (Scanlon et al. 1996). Blade width is reduced in a dose-dependent manner by the Wab1 mutation (Hay and Hake 2004). The narrow leaves observed in Wab1 versus those found in ns1; ns2 double mutants differ in telling ways: Wab1 distal blade width is predominantly reduced, whereas the proximal blade and sheath are narrow in ns1; ns2 (Scanlon et al. 1996; Hay and Hake 2004), suggesting that WAB1 promotes growth in the blade lateral domain. Indeed, genetic interactions between Wab1 and ns1; ns2 mutants indicate WAB1 regulates growth of a leaf domain distinct from the NS domain.
Coordination between cell division, expansion, and differentiation is crucial to determining mature leaf morphology and size. The AP2-EREB171 transcription factor DWARF & IRREGULAR LEAF1 (DWIL1) importantly regulates cell size, shape, and arrangement in the leaf that correlates with the shorter and wider blades in dwil1 mutants (Jiang et al. 2012). Maize SCARECROW1 (ZmSCR1) co-orthologs encode GRAS transcription factors, and Zmscr1; Zmscrh double mutants display significant reductions in blade length and width, along with overall stunted shoot growth (Hughes et al. 2019). The narrow blades of Zmscr1; Zmscrh double mutants have defects in mesophyll cell divisions that increase vein density, indicating these genes function to regulate cell-type patterning during leaf development (Hughes et al. 2019). In transgenic studies, constitutive (i.e., driven by the UBIQUITIN [UBI] promoter) or localized ectopic expression (i.e., driven by ZmGA2ox promoter) of the maize PLASTOCHRON1 (PLA1) gene gives rise to longer leaves due to an increase in the duration of leaf elongation, which is also correlated with patterns of auxin accumulation (Sun et al. 2017). These results indicate that PLA1 promotes leaf growth by increasing the duration of cell division and that auxin may play a key role in determining the period of growth (Sun et al. 2017). Similarly, constitutive expression of the maize ARGOS1 (AUXIN REGULATED GENE INVOLVED IN ORGAN SIZE1; ZAR1) gene under the UBI promoter increased leaf size in field-grown transgenic plants largely by increasing cell number but not cell size (Guo et al. 2013). ARGOS proteins also negatively regulate ethylene signaling (Shi et al. 2015), suggesting that crosstalk between hormone pathways could play a role in regulating both cellular dynamics and leaf size.
Cell division and elongation defects that culminate in narrower and shorter leaf blades and shorter sheaths characterize asceapen1 (asc1; Brooks et al. 2009), narrow odd dwarf (nod; Rosa et al. 2017), and cellulose synthase-like D1 (csld1; Hunter et al. 2012) mutants. Natural variation in the CSLD1 gene, which is important for cell wall development in maize, was found to underlie a QTL for leaf width (Li et al. 2018). NOD encodes the CELL NUMBER REGULATOR13/MID-COMPLEMENTING ACTIVITY membrane-localized protein that is enriched in dividing leaf tissues (Rosa et al. 2017), and ASC1 is a D4-CYCLIN (Brooks et al. 2009), emphasizing the importance of coordinating cell division during leaf morphogenesis. However, mis-regulation of cell division and cell shape is not always correlated with altered leaf shape. For example, leaves of tangled-1 mutant plants are reduced proportionally in size largely due to much slower growth rate (Smith et al. 1996).
Natural variation of leaf morphology
Maize is rich in natural variation for leaf traits, such as blade width and length, sheath length, blade angle of middle and upper canopies, auricle area and length, and total leaf area (Flint-Garcia et al. 2005; Kong et al. 2017; Cui et al. 2017; Dzievit et al. 2018). As a result, numerous QTL mapping analyses have harnessed variation in leaf morphology to identify underlying genetic factors (Mickelson et al. 2002; Ding et al. 2015; Li et al. 2015; Ku et al. 2010, 2012a, 2012b; Tian et al. 2011; Wei et al. 2016; Pan et al. 2017; Cui et al. 2017; Dzievit et al. 2018; Li et al. 2018). The maize nested association mapping (NAM) population, in which 25 diverse founders were crossed to a common parent to generate 5000 RILs (Yu et al. 2008; Gage et al. 2020), was utilized to dissect the genetic architecture of leaf width, length, and angle (Tian et al. 2011). By conducting a GWAS, Tian et al. (2011) found that these three classes of leaf traits are governed by myriad QTL with small effects that display little epistasis, environmental interaction, or pleiotropy. Linkage mapping in bi- and quad-parental populations has also been leveraged to identify leaf trait QTL (Mickelson et al. 2002; Ding et al. 2015; Li et al. 2015; Ku et al. 2010, 2012a, 2012b; Wei et al. 2016; Cui et al. 2017; Dzievit et al. 2018; Li et al. 2018). Recent dissection of leaf angle in three historically important US maize heterotic groups isolated 12 QTL that, when merged into a meta-analysis, identified 58 genomic regions that harbored 33 candidate genes (Dzievit et al. 2018). This analysis identified several cloned genes with significant roles in leaf architecture, as well as paralogs of these genes. These include LG1 (Moreno et al. 1997) and its paralog SISTER OF LG1/SBP28, LG2 (Walsh et al. 1998), and its paralog LG2 RELATED SEQUENCE1/bZIP90, and DRL1/YAB2 and its paralog DRL2/YAB7 (Strable et al. 2017). Genetic associations at LG1 and LG2 (Tian et al. 2011) and DRL1 and DRL2 (Strable et al. 2017) were found to underlie significant QTL for blade angle and width traits. However, despite the large number of QTL that have been identified and mapped, a majority of the underlying causative loci remain to be cloned and characterized.
A biparental maize-teosinte population that generated 866 RILs (Shannon 2012; Shannon et al. 2019) has proven to be a powerful resource for uncovering novel genetic variants for leaf architecture (Fu et al. 2019; Tian et al. 2019). This population has great potential (1) for allele discovery, such as beneficial genetic variants that were lost during domestication, and (2) as a source for alleles to improve maize shoot architecture (Tian et al. 2019). Uncovering genetic variation in wild ancestors not only provides insights into domestication, it also paves the way for future maize improvement. Tian et al. (2019) demonstrated the power of utilizing the maize-teosinte RILs to discover leaf architecture QTL and improve yield potential under dense planting. A screen for leaf angle variants important for high density planting uncovered two large effect QTL for leaf blade angle: Upright Plant Architecure1 (UPA1) and UPA2 (Tian et al. 2019). Cloning UPA2 revealed a regulatory sequence 9.5 kb upstream of the maize gene RELATED TO ABI3/VP1 (RAV)-LIKE1 (ZmRAVL1), which encodes a B3-domain transcriptional activator (Tian et al. 2019). The UPA2 regulatory sequence carries a DRL1 binding motif (Tian et al. 2019). This DRL1 binding motif is complete in most teosinte lines, whereas in all maize lines, two nucleotides are missing, which weakens DRL1 binding affinity (Tian et al. 2019). As a result, near isogenic lines (NILs) that retain the teosinte allele displayed slightly more upright leaves. Interestingly, the ZmRAVL1 promoter carries a LG1 binding motif important for LG1-mediated activation of ZmRAVL1 expression (Tian et al. 2019). DRL1 and LG1 proteins physically interact in vitro and in vivo, and DRL1 attenuates the activity of LG1 (Tian et al. 2019). Thus, in modern maize, ZmRAVL1 levels are higher due to the reduced binding of DRL1 to UPA2 that subsequently weakens its repressive interaction with LG1 (Tian et al. 2019).
In parallel, Tian et al. (2019) also cloned the large effect UPA1 leaf angle QTL. UPA1 QTL was found to be controlled by the brassinosteroid biosynthesis gene BRD1, and NILs with the teosinte UPA1/BRD1 allele have a wider blade angle, due to an increase in auricle size and a reduction in abaxial hypodermal sclerenchyma (Tian et al. 2019). The BRD1 promoter contains ZmRAVL1 binding motifs, which provides a mechanism for regulating brassinosteroid levels during development of the blade/sheath boundary (Tian et al. 2019). Therefore, in modern maize, DRL1-LG1-mediated activation of ZmRAVL1 upregulates BRD1/UPA1 and increases brassinosteroid levels at the developing ligule/auricle region (Tian et al. 2019). Understanding the mechanism by which DRL1 and LG1 proteins interact in planta to regulate ZmRAVL1 expression will be an important area of future research.
Maize-teosinte RILs, additionally, have been leveraged to dissect genome-wide expression QTL to detect shifts in gene expression that may associate with domestication (Wang et al. 2018b). Additionally, maize-teosinte RIL populations with diverse maize common parents have been used to hunt for QTL that explain natural variation in leaf traits (Fu et al. 2019; Liu et al. 2019a). Fu et al. (2019) noted that large effect QTL underlie leaf length, whereas conversely numerous small effect loci control blade width and sheath length. This study implicated several cloned genes known to regulate these processes, such as DRL1 and DRL2 (blade width and sheath length, respectively; Strable et al. 2017), microRNA396a (sheath length; Nelissen et al. 2015), DWIL1 (blade width; Jiang et al. 2012), and LGN1 (leaf length; Moon et al. 2013). Recently, a teosinte NAM (TeoNAM) population was developed, in which 5 diverse teosinte inbred lines were crossed with a common maize parent to generate 1257 RILs, to facilitate genetic mapping of domestication and agronomically useful QTL (Chen et al. 2019; Chen et al. 2020). TeoNAM carries with it the potential for identifying beneficial alleles from teosinte that could be then introgressed into maize breeding lines and programs.
Axillary branching is regulated by external and internal cues
Axillary meristem initiation and maintenance
An axillary bud is initiated by an axillary meristem located at the node within the leaf axil. These lateral buds will either enter dormancy or remain active and grow into branches. Auxin and downstream transcription factors serve as key regulators of axillary meristem initiation. SPARSE INFLORESENCE1 (SPI1) encodes a maize YUCCA-like flavin monooxygenase involved in local auxin biosynthesis (Gallavotti et al. 2008). The serine/threonine kinase BARREN INFLORESENCE2 (BIF2) is co-orthologous to Arabidopsis PINOID (McSteen et al. 2007) and functions in polar auxin transport (Skirpan et al. 2009). Transcripts for both SPI1 and BIF2 accumulate in axillary meristems, and spi1; bif2 double mutants produce fewer vegetative phytomers compared with either single mutant (McSteen et al. 2007; Gallavotti et al. 2008). Mutations in SPI1 and BIF2 reduce tiller number in highly branched teosinte branched1 (tb1; Doebely et al. 1997) mutants, indicating that these genes importantly regulate axillary meristem initiation (McSteen et al. 2007; Gallavotti et al. 2008). BARREN STALK1 (BA1) encodes a bHLH transcription factor orthologous to rice LAX PANICLE1 (LAX1) (Gallavotti et al. 2004). Additionally, BA1 is regulated post-translationally by BIF2 phosphorylation (Skirpan et al. 2008). ba1 mutants fail to initiate all vegetative and reproductive axillary meristems, and, as expected, ba1; tb1 double mutants display complete tiller suppression (Ritter et al. 2002). BA1 levels are under the control of BARREN STALK FASTIGIATE1 (BAF1), a transcriptional regulator with an AT-hook DNA binding motif (Gallavotti et al. 2011). Mutant baf1 plants fail to initiate axillary buds that are fated to become lateral ear shoots; as a result, baf1 mutants are earless (Gallavotti et al. 2011). BA1 and BAF1 are co-expressed in boundary regions flanking the axillary meristem stem cell niche, indicating expression of these genes represents one of the first committed steps of axillary meristem initiation (Gallavotti et al. 2004; Gallavotti et al. 2011). The BA2 gene is required for axillary meristem initiation, much like BA1. ba2 mutants fail to initiate vegetative and reproductive axillary meristems, and ba2; tb1 double mutants display complete tiller suppression (Yao et al. 2019). BA2 encodes a nuclear localized protein orthologous to rice LAX2 (Yao et al. 2019). BA1 and BA2 physically interact to form a protein complex, which likely functions downstream of auxin signaling (Yao et al. 2019). Whether BA2 is regulated transcriptionally by BAF1 or whether BIF2 phosphorylates BA2 remains to be explored.
Once formed, maintenance of axillary meristems is equally critical for shoot branching. Genetic analysis of maize BLH genes has shed some light on this process. Vegetative shoots of blh12; blh14 double mutants lack tillers and ears (Tsuda et al. 2017). However, cells in the axils of blh12; blh14 double mutant leaves are histologically dense staining, and these cells accumulate KN1 protein (Tsuda et al. 2017). These hallmark features of meristematic cells indicate that axillary meristem initiation is unaffected by mutations in BLH12 and BLH14 at early plastochron stages. However, around P7, histological staining of cells weakens and KN1 accumulation lessens to the extent of eventual undetectable levels, suggesting that axillary meristem maintenance fails in the absence of BLH12 and BLH14 (Tsuda et al. 2017).
Axillary bud potential
Shoot branching of modern maize is largely at the level of lateral ears and not so much by tillers. This is due to growth arrest of axillary buds and bud dormancy, not a lack of axillary meristem initiation, at basal nodes. Key transcription factors expressed within basal buds regulate their dormancy (Dong et al. 2019a). TB1 encodes a TCP transcription factor that acts as a growth repressor to promote apical dominance in modern maize (Doebely et al. 1997). Mutant tb1 plants resemble teosinte in that the mutants are highly tillered and overproduce aerial lateral branches from mid-shoot nodes (Fig. 4a, b) (Doebley et al. 1995). Modern maize contains a hopscotch retrotransposon inserted ~58 kb upstream of the TB1 promoter that predates maize domestication (Clark et al. 2006; Studer et al. 2011). This hopscotch element has transcriptional enhancer activity (Studer et al. 2011; Ricci et al. 2019), which conditions overexpression of TB1 in domesticated maize (Doebely et al. 1997). Mutations in GRASSY TILLERS1 (GT1), a class I HOMEODOMAIN LEUCINE ZIPPER transcription factor encoding gene, increases tiller growth (Fig. 4d) (Whipple et al. 2011). TB1 binds the GT1 promoter to activate its expression (Whipple et al. 2011; Dong et al. 2019b). TB1 also binds to the domestication QTL prolificacy1.1 (prol1.1; Wills et al. 2013) located 7 kb upstream of GT1 to repress secondary branching in the ear (Dong et al. 2019b). Additionally, the TASSELS REPLACE UPPER EARS1 (TRU1) domestication locus participates in a TB1 module to regulate shoot architecture. tru1 mutants have long lateral ear shanks from mid-shoot nodes that terminate with a tassel (Sheridan 1988). TRU1 encodes a BTB/POZ ankyrin repeat protein that accumulates in axillary buds, similar to TB1 (Dong et al. 2017). TRU1 is a direct target of TB1 and is overexpressed in maize relative to teosinte in a tissue-specific manner (Dong et al. 2017). The TB1 module highlights how overexpression was selected to produce a suite of domestication and agronomic traits (Dong et al. 2019a)
External factors, such as light, and internal signals, such as photohormones, metabolites, sugar status, and light, are key regulators of axillary bud activity. Suppression of bud growth is a component of the shade avoidance response and PHYTOCHROME (PHY)-mediated perception of red to far-red light ratios (R:FR; Dubois and Brutnell 2009). The TB1-GT1 regulatory module is sensitive to R:FR ratios, where a decrease in R:FR is perceived as shade (Whipple et al. 2011). Furthermore, FR light reduces tillering in teosinte and sorghum, which are two grass species where phytochrome signaling activates the shade avoidance response (Whipple et al. 2011). In sorghum, the red-light receptor PHYB signals to repress TB1 expression in axillary buds, whereas high R:FR ratio suppresses PHYB activity and promotes shade avoidance (Kebrom et al. 2006). In maize buds, TB1 regulates metabolite levels involved in sugar sensing, as well as numerous phytohormone pathways that do not include auxin (Dong et al. 2019b). Levels of the phytohormone jasmonic acid are significantly reduced in tb1 and gt1 mutant buds; furthermore, TB1 directly targets the jasmonic acid biosynthesis gene OXOPHYTODIENOATE REDUCTASE8 (OPR8), which is downregulated in tb1 and gt1 mutant buds (Dong et al. 2019b). opr7; opr8 double mutants have many more nodes with de-repressed buds, and lateral ear shanks are longer relative to non-mutant plants (Yan et al. 2012), further indicating a role for jasmonic acid in suppressing axillary buds and branch growth.
Bud activity can be regulated independently of TB1. A QTL that increases tiller number in sweet maize was identified as TILLER NUMBER1 (TIN1; Zhang et al. 2019a). TIN1 encodes a C2H2 ZINC FINGER transcriptional regulator, where a splice site variant in sweet maize causes intron retention that enhances TIN1 expression and bud growth (Zhang et al. 2019a). TIN1 physically interacts with TOPLESS (TPL) co-repressor proteins and represses GT1 expression (Zhang et al. 2019a). Mutations in the TPL member RAMOSA ENHANCER LOCUS2 (REL2) result in earless shoots in some maize backgrounds due to the loss of lateral ear buds (Liu et al. 2019b). Interestingly, rel2; tb1 double mutants only slightly impact tiller number compared with tb1 single mutants, suggesting that basal axillary buds retain growth potential independent of REL2 (Liu et al. 2019b). The phytohormone strigolactone controls lateral branching through TB1 homologs in other plant species (Domagalska and Leyser 2011). In maize, however, strigolactone and TB1 pathways work independently, as mutations in the maize strigolactone biosynthesis gene CAROTENOID CLEAVAGE DIOXYGENASE8 (CCD8) have only a modest effect on axillary bud growth and do not impact TB1 expression (Guan et al. 2012).
Genetic and hormonal regulation of intercalary meristem and stem traits
Regulators of cell size and number have key roles in internode elongation, a few of which have been referred to previously in this review (e.g., NOD, ASC1, DWIL1). Phytohormone pathways also play an integral role in the control of plant height. In fact, reduction of plant height in wheat drove the Green Revolution, and subsequent characterization of the genes responsible for this trait placed them in the gibberellin pathway (Hedden 2003). Short stature maize mutants have long been recognized by maize geneticists (Emerson 1912b; Emerson and Emerson 1922). Some of the short stature mutants that piqued Emerson’s curiosity were later found to have genetic lesions in the gibberellin pathway. For example, recessive mutants d1, d3, d5, and anther ear1 (an1) all respond to exogenous gibberellin (Figs. 1b and 5i) (Fujioka et al. 1988), and all four genes encode gibberellin biosynthesis enzymes (Benson et al. 1995; Winkler and Helentjaris 1995; Chen et al. 2014; Fu et al. 2016). Dominant D8 and D9 mutants are insensitive to exogenous gibberellin (Winkler and Freeling 1994; Peng et al. 1999) and encode DELLA proteins involved in gibberellin signaling (Cassani et al. 2009; Lawitt et al. 2010). Additionally, D1 was found to underlie a major QTL for plant height (Teng et al. 2013), and GWAS for plant height traits using the maize NAM RILs found several significant associations with hormone genes that regulate internode elongation, including D1 and BRD1 (Peiffer et al. 2014). The genetic architecture of maize height is highly polygenic (Peiffer et al. 2014), and many candidate loci remain uncharacterized.
Several examples of hormonal regulation of internode elongation have been mentioned in previous sections of this review. These include brassinosteroid- (e.g., BRD1, NA1, NA2, BRI1), cytokinin- (e.g., HSF1, ABPH1), ethylene- (e.g., SDW3), strigolactone- (e.g., CCD8), abscisic acid- (e.g., VP8), and auxin (e.g., BIF2, SPI1)-related genes, all of which control plant height across all nodes of the shoot. Similarly, mutations in BREVIS PLANT1 (BV1) significantly reduce plant height by decreasing stem elongation across all nodes (Avilla et al. 2016). BV1 encodes an inositol phosphate phosphatase involved in auxin signaling (Avilla et al. 2016). Curiously, in bracytic2 (br2) mutants, only lower internodes are reduced, while upper internode length remains relatively normal (Multani et al. 2003; Pilu et al. 2007; Xing et al. 2015; Li et al. 2020b). BR2 encodes an MDR class of P-glycoproteins necessary for polar auxin transport (Multani et al. 2003), whereas others have suggested that BR2 is more closely related to an ATP binding cassette type B protein that exports auxin from intercalary meristems of lower nodes (Knöller et al. 2010). Given the lower versus upper node differences in br2 mutants, it would be interesting to know whether BR2 levels are regulated by a program related to the juvenile phase.
In addition to regulating plant height, auxin transport also plays a critical role in promoting negative gravitropism. The prostrate stem1 mutant fails to maintain an upright shoot, which becomes recumbent as an adult (Dong et al. 2013). Genetic complementation tests between prostrate stem1 and the maize mutant lazy plant1 (la1; Jenkins and Gerhardt 1931; van Overbeek 1936) indicated the two are allelic (Dong et al. 2013). ZmLA1 is functionally orthologous to rice and Arabidopsis LAZY genes. Interestingly, the ZmLA1 protein localizes to both the plasma membrane and the nucleus where it interacts with auxin transport and singling proteins, respectively (Dong et al. 2013; Howard et al. 2014). In future work, it will be important to know the mechanism by which ZmLA1 regulates both auxin transport and signaling proteins.
Transcriptional regulators of internode development have also been characterized. Double mutant drl1; drl2 plants have longer adult internodes compared with non-mutant sibling plants (Strable et al. 2017). Additionally, a GWAS identified rare polymorphisms with large phenotypic effects for average internode length within QTL intervals for DRL1/YAB2 and DRL2/YAB7 (Strable et al. 2017). Through a mechanism that may involve partnering with KN1, BLH12 and BLH14 proteins prevent precocious differentiation of the internode by maintaining intercalary meristems, as well as balance anastomosis of provascular bundles in early stem development (Tsuda et al. 2017). Intercalary meristem activity is also governed by GROWTH REGULATING FACTOR (GRF)-INTERACTING FACTOR1 (GIF1; Zhang et al. 2018), a transcriptional regulator that forms complexes with GRF transcription factors (Kim and Tsukaya 2015). Mutant gif1 plants have short, asymmetric (kinked) internodes and show decreased expression of gibberellin-related genes (Zhang et al. 2018). In rice, GRF1 activity in the intercalary meristem is induced by gibberellin (van der Knaap et al. 2000), suggesting that hormonal regulation of GIF1 and GRF binding partners may be similar in maize. GRF1 levels are regulated by microRNA396 (miR396; Rodriguez et al. 2010); maize plants that overexpress a miR396-resistant GRF1 transgene have short internodes and wide leaves (Nelissen et al. 2015), phenotypes that are reminiscent of mutants with altered gibberellin levels. GRF1, along with twelve other GRFs, physically interacts with GIF1, which is enriched in meristematic tissues, suggesting that the spatiotemporal context and modularity of GRF-GIF1 complexes are likely a key aspect of their function during stem development (Nelissen et al. 2015; Zhang et al. 2018).
Conclusions and outlook
Plant developmental genetics provide a framework for translating information about gene function into breeding programs to improve agronomic traits. Maize has a long history as a genetic and breeding model and is an indispensable global crop. Positional cloning of genes has revealed the importance of structural variation (e.g., Kn1, Veit et al. 1990; Ramirez et al. 2009; Abph2, Yang et al. 2015) and transposable elements (e.g., Corngrass1, Chuck et al. 2007; Kn1, Hake et al. 1989; Ramirez et al. 2009; tru1, Dong et al. 2017; drl1, Strable et al. 2017; Sdw3, Li et al. 2020a) in generating novel shoot phenotypes. Genome mining of diversity panels and RILs has uncovered natural variants of cloned genes, which have given rise to a plethora of quantitative shoot phenotypes (qPH3.1/D1, Teng et al. 2013; qPH1/BR2, Xing et al. 2015; DRL2, Strable et al. 2017; qLW10/ZmCSLD1, Li et al. 2018; UPA1/BRD1, Tian et al. 2019; qLA2-1/ZmIBH1, Cao et al. 2020; qLA2/ZmILI1, Ren et al. 2020). A major step forward will be to functionally characterize candidate genes that underlie QTL more efficiently, such as demonstrated with quantitative trait gene sequencing (Zhang et al. 2019b). Given the tremendous diversity found in maize (Tenaillon et al. 2001; Romay et al. 2013), more high-quality inbred line genome assemblies are needed to fully catalog genetic variation, such as those captured by the (Hufford et al. 2021). High-quality genome assemblies are also needed for teosintes, landraces, and tropical maize lines, as illustrated by the assembly of a tropical maize landrace genome, which uncovered thousands of structural variations when compared with either the genomes of B73 or Mo17, both temperate maize inbred lines (Yang et al. 2019). The previously mentioned implementation of UPA1 and UPA2 regions in maize to increase yield under dense planting nicely demonstrates the power of utilizing teosinte-maize populations to find rare and beneficial genetic variants and incorporating pre-domesticate alleles into elite maize inbred lines (Tian et al. 2019). Recent advances including the construction of a practical haplotype graph in maize and a pangenome representative of haplotype diversity and structural diversity should prove to be valuable resources for modern breeding programs by improving the effectiveness of deciphering the genetic basis of phenotypic variation (Franco et al. 2020).
Current sequence databases catalog a wealth of genetic variation; however, a vast number of genes in maize are yet to be functionally characterized, providing scant resolution to the traits they may regulate. Plant transformation has been a historical roadblock for in maize functional genetics, where high cost, low efficiency, and a limited number of amenable transformable genetic backgrounds have hampered progress. Recent advances in transformation technology have circumvented two of these obstacles by increasing transformation efficiency and making the process independent of genotype (Lowe et al. 2016, 2018). Dovetailing with advances in transformation, CRISPR/Cas9 genome engineering has allowed for the generation of multiple alleles of a single gene, simultaneous targeting multiple genes of a functionally redundant gene family, or broadly targeting many independent genes. Genetic redundancy has challenged maize geneticists for decades; however, multiplex gene knockout with CRISPR/Cas9 is a method to overcome functional redundancy by generating higher-order mutants in a single experiment (Wu et al. 2018). Therefore, scalability of maize CRISPR/Cas9 targets has recently become less of an obstacle. In an astounding tour de force, high-throughput usage of multiplex CRISPR/Cas9 achieved editing of >740 candidate genes in maize that generated >410 alleles in ~190 genes with discernable phenotypes (Liu et al. 2020). Considering the recent identification of 160 loci with adaptive agronomic potential, as well as >1800 loci associated with trait improvement in 350 elite inbred lines (Wang et al. 2020a), multiplex CRISPR/Cas9 provides an efficient platform to discover and characterize loci in QTL regions and provides opportunities to accelerate precision maize breeding.
Many of the developmental mutants mentioned in the previous sections display pleiotropy, which can have a negative effect on yield. Pleiotropy that is often uncovered in developmental studies can arise from functional null mutations due to lesions in coding sequences and/or the inherent nature of phytomer, where genes that regulate phytomer development often have pleiotropic effects. Genetic variation in cis-regulatory elements (CRE) located in promoters and other regulatory regions are often less pleiotropic because transcript levels and/or spatiotemporal accumulation patterns are shifted versus completely removed (Wittkopp and Kalay 2012). Thousands of CREs in the maize genome function over long ranges (>20 kb) to regulate gene activity (Ricci et al. 2019). Untangling the individual phenotypic components of pleiotropy is one way to utilize developmental genes to improve yield. Mutagenesis of CREs through CRISPR/Cas9 has thus far proven to be an effective method of modulating expression levels and/or patterns, as well as generating quantitative phenotypes in crops by inducing dosage variation within a genetic circuit that regulates meristem size (Rodriguez-Leal et al. 2017, 2019). In recent efforts to regulate gene expression in maize, promoter sequences have been inserted, albeit at low efficiency, at endogenous loci using defined CRISPR/Cas9 target sites and homology directed repair (Svitashev et al. 2015; Shi et al. 2016). CRISPR/Cas9 could be used to overcome pleiotropic effects if the abundance of binding partners or downstream targets within a network could be modulated in a developmental or tissue-specific context. Equally promising is the implementation of CRISPR/Cas9 to generate complex trait loci to enable trait stacking, as demonstrated in multi-trait maize hybrids (Gao et al. 2020). Chromosomal regions engineered with a site-specific insertion landing pad are utilized to accept transgene(s) through recombinase-mediated cassette exchange, thus providing an effective method for overcoming linkage issues (Gao et al. 2020). Collectively, approaches that harness CRISPR/Cas9 technologies, along with rapid advances in transformation, genomics, and machine learning, demonstrate the transformative utility ahead for functional genetics and applied breeding programs.
Supplementary information
(XLSX 30 kb)
Acknowledgements
This manuscript benefited from the insightful comments and helpful critique provided by China Lunde, Jack Satterlee, Zhaobin Dong, María Jazmín Abraham-Juárez, and Kevin Ahern, as well as two anonymous reviewers—to all of you, many thanks for improving clarity of this manuscript. Bret Timmons and Thomas Paul are gratefully acknowledged for excellent plant care.
Code availability
Not applicable
Author contribution
Manuscript preparation (JS)
Funding
JS was supported by the National Science Foundation Plant Genome Postdoctoral Research Fellowship (IOS-1710973)
Data Availability
Not applicable
Declarations
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The author declares no competing interests.
Footnotes
This article is part of the Topical Collection on Maize Genetics, Genomics and Sustainable Improvement
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abbe EC, Phinney BO, Baer DF. The growth of the shoot apex of maize: internal features. Am J Bot. 1951;38:744–751. doi: 10.1002/j.1537-2197.1951.tb14887.x. [DOI] [Google Scholar]
- Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, Lemaux PG, Medford JI, Orozco-Cádenas ML, Tricoli DM, Van Eck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart CN. Advancing crop transformation in the era of genome editing. Plant Cell. 2016;28:1510–1520. doi: 10.1105/tpc.16.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, St Aubin B, Abraham-Juarez MJ, Leiboff S, Shen Z, Briggs S, Brunkard JO, Hake S (2019) The second site modifier, Sympathy for the ligule, encodes a homolog of Arabidopsis ENHANCED DISEASE RESISTANCE4 and rescues the Liguleless narrow maize mutant. Plant Cell 31:1829–1844 [DOI] [PMC free article] [PubMed]
- Atkins PAP, Voytas DF. Overcoming bottlenecks in plant gene editing. Curr Opin Plant Biol. 2020;54:79–84. doi: 10.1016/j.pbi.2020.01.002. [DOI] [PubMed] [Google Scholar]
- Avilla LM, Cerrudo D, Swanton C, Lukens L. Brevis plant1, a putative inositol polyphosphate 5-phosphatase, is required for internode elongation in maize. J Exp Bot. 2016;67(5):1577–1588. doi: 10.1093/jxb/erv554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassiri A, Irish EE, Poethig RS. Heterchronic effects of Teopod 2 on growth and photosensitivity of the maize shoot. Plant Cell. 1992;4:497–504. doi: 10.2307/3869450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Freeling M. Sectors of liguleless-1 tissue interrupt an inductive signal during maize leaf development. Plant Cell. 1991;3:801–807. doi: 10.1105/tpc.3.8.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Freeling M. Genetic analysis of Rough sheath1 developmental mutants of maize. Genetics. 1994;136:295–311. doi: 10.1093/genetics/136.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft PW, Bongard-Pierce DK, Sylvester AW, Poethig RS, Freeling M (1990) The liguleless-1 gene acts tissue specifically in maize leaf development. Dev Biol 141:220–232 [DOI] [PubMed]
- Benson RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP. Cloning and characterization of the maize an1 gene. Plant Cell. 1995;7:75–84. doi: 10.1105/tpc.7.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand-Garcia R, Freeling M. Hairy-sheath frayed #1-O: a systemic, heterochronic mutant of maize that specifies slow developmental stage transitions. Am J Bot. 1991;78:747–765. doi: 10.1002/j.1537-2197.1991.tb14477.x. [DOI] [Google Scholar]
- Best NB, Hartwig T, Budka J, Fujioka S, Johal G, Schulz B, Dilkes BP. nana plant2 encodes a maize ortholog of the Arabidopsis brassinosteroid biosynthesis gene DWARF1, identifying developmental interactions between brassinosteroid and gibberellins. Plant Physiol. 2016;171:2633–2647. doi: 10.1104/pp.16.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Hake S. The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell. 2009;21:1647–1658. doi: 10.1105/tpc.109.068221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, O’Connor D, Moon J, Lewis M, Hake S. How to pattern a leaf. Cold Spring Harb Symp Quant Biol. 2012;77:47–51. doi: 10.1101/sqb.2012.77.014613. [DOI] [PubMed] [Google Scholar]
- Bolduc N, Tyers RG, Freeling M, Hake S. Unequal redundancy in maize knotted1 homeobox genes. Plant Physiol. 2014;164:229–238. doi: 10.1104/pp.113.228791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012;26:1685–1690. doi: 10.1101/gad.193433.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P, Je BI, Goldschmidt A, Jackson D. The maize Gα gene COMPACT PLANT2 functions in CLAVATA signaling to control shoot meristem size. Nature. 2013;502:555–558. doi: 10.1038/nature12583. [DOI] [PubMed] [Google Scholar]
- Bommert P, Lunde C, Nardmann J, Vollbrecht E, Running M, Jackson D, Hake S. thick tassel dwarf1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2005;132:1235–1245. doi: 10.1242/dev.01671. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development. 1999;126:2387–2396. doi: 10.1242/dev.126.11.2387. [DOI] [PubMed] [Google Scholar]
- Brooks L, Strable J, Zhang X, Ohtsu K, Zhou R, Sarkar A, Hargreaves S, Elshire RJ, Eudy D, Pawlowska T, Ware D, Janick-Buckner D, Buckner B, Timmermans MCP, Schnable PS, Nettleton D, Scanlon MJ. Microdissection of shoot meristem functional domains. PLoS Genet. 2009;5(5):e1000476. doi: 10.1371/journal.pgen.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher EM, Moon J, Runkel A, Hake S, Dilkes DP. Natural variation at sympathy for the ligule controls penetrance of the semidominant Liguleless narrow-R mutation in Zea mays. G3. 2014;4:2297–2306. doi: 10.1534/g3.114.014183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H, Hake S. The art and design of genetic screens: maize. Nat Rev Genet. 2008;9:192–203. doi: 10.1038/nrg2291. [DOI] [PubMed] [Google Scholar]
- Cao Y, Zeng H, Ku L, Ren Z, Han Y, Su H, Dou D, Liu H, Dong Y, Zhu F, Li T, Zhao Q, Chen Y. ZmIBH1-1 regulates plant architecture in maize. J Exp Bot. 2020;71:2943–2955. doi: 10.1093/jxb/eraa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassani E, Bertolini E, Badone FC, Landoni M, Gavina D, Sirizzotti A, Pilu R. Characterization of the first dominant dwarf maize mutant carring a single amino acid insertion in the VHYNP domain of the dwarf8 gene. Mol Breed. 2009;24:375–385. doi: 10.1007/s11032-009-9298-3. [DOI] [Google Scholar]
- Chen Q, Samoyoa LF, Yang CJ, Bradbury PJ, Olukolu BA, Neumeyer MA, Romay MC, Sun Q, Lorant A, Buckler ES, Ross-Ibarra J, Holland JB, Doebley JF. The genetic architecture of the maize progenitor, teosinte, and how it was altered during maize domestication. PLoS Genet. 2020;16:e10088791. doi: 10.1371/journal.pgen.1008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yang CJ, York AM, Xue W, Daskalska LL, Devalk CA, Krueger KW, Lawton SB, Spiegelberg BG, Schnell J, Neumeyer MA, Perry JS, Peterson AC, Kim B, Bergstrom L, Yang L, Barber IC, Tian F, Doebley JF. TeoNAM: a nested association mapping population for domestication and agronomic trait analysis in maize. Genetics. 2019;213:1065–1078. doi: 10.1534/genetics.119.302594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hou M, Liu L, Wu S, Shen Y, Ishiyama K, Kobayashi M, McCarty DR, Tan BC. The maize dwarf1 encodes a gibberellin 3-oxidase and is dual localized to the nucleus and cytosol. Plant Physiol. 2014;166:2028–2039. doi: 10.1104/pp.114.247486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Brown PJ, Meeley R, Hake S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc Natl Acad Sci U S A. 2014;111:18775–18780. doi: 10.1073/pnas.1407401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Clark NM, Van den Broeck L, Guichard M, Stager A, Tanner HG, Blilou I, Grossman G, Iyer-Pascuzzi AS, Maizel A, Sparks EE, Sozzani R. Novel imaging modalities shedding light on plant biology: start small and grow big. Annu Rev Plant Biol. 2020;71:789–816. doi: 10.1146/annurev-arplant-050718-100038. [DOI] [PubMed] [Google Scholar]
- Clark RM, Wagler TN, Quijada P, Doebley J. A distinct upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescence architecture. Nat Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- Colasanti J, Muszynski MG. Handbook of Maize: Its Biology. New York: Springer; 2009. The maize floral transition; pp. 41–55. [Google Scholar]
- Conklin PA, Johnston R, Conlon BR, Shimizu R, Scanlon MJ. Plant homeodomain proteins provide a mechanism for how leaves grow wide. Development. 2020;147:dev193623. doi: 10.1242/dev.193623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosbie T. Changes in physiological traits associated with long-term breeding efforts to improve grain yield in maize. In: Loden H, Wilkinson D, editors. Proceedings of the 37th Annual Corn and Sorghum Research Conference. Washington, DC: American Seed Trade Association; 1982. pp. 206–223. [Google Scholar]
- Cui T, He K, Chang L, Zhang X, Xue J, Liu J. QTL mapping for leaf area in maize (Zea mays L.) under multi-environments. J Integr Agric. 2017;16:800–808. doi: 10.1016/S2095-3119(16)61524-1. [DOI] [Google Scholar]
- Ding J, Zhang L, Chen J, Li X, Li Y, Cheng H, Huang R, Zhou B, Li Z, Wang J, Wu J. Genomic dissection of leaf angle in maize (Zea mays L.) using a four-way cross mapping population. PLoS One. 2015;10:e0141619. doi: 10.1371/journal.pone.0141619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A. Genetic analysis of the morphological differences between maize and teosinte. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Gustus C. teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebely J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Wendel J, Edwards M. Genetic and morphological analysis of a maize-teosinte F2 populations: implications for the origin of maize. Proc Natl Acad Sci U S A. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska MA, Leyser O. Signal integration in the control of shoot branching. Nat Rev Mol Biol. 2011;12:211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
- Dong Z, Alexander M, Chuck G. Understanding grass domestication through maize mutants. Trends Genet. 2019;35:118–128. doi: 10.1016/j.tig.2018.10.007. [DOI] [PubMed] [Google Scholar]
- Dong Z, Jiang C, Chen X, Zhang T, Ding L, Song W, Luo H, Lai J, Chen H, Liu R, Zhang X, Jin W. Maize LAZY1 mediates shoot gravitropism and inflorescence development through regulating auxin transport, auxin signaling, and light response. Plant Physiol. 2013;163:1306–1322. doi: 10.1104/pp.113.227314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Li W, Unger-Wallace E, Yang J, Vollbrecht E, Chuck G. Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc Natl Acad Sci U S A. 2017;114:E8656–E8664. doi: 10.1073/pnas.1714960114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Xiao Y, Govindarajulu R, Feil R, Siddoway ML, Nielsen T, Lunn JE, Hawkins J, Whipple C, Chuck G. The regulatory landscape of a core maize domestication module controlling bud dormancy and growth repression. Nat Commun. 2019;10:1–15. doi: 10.1038/s41467-019-11774-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RN, Wiley D, Sarkar A, Springer N, Timmermans MCP, Scanlon MJ. ragged seedling2 encodes and ARGONAUTE7-like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell. 2010;22:1441–1451. doi: 10.1105/tpc.109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Van den Broeck L, Inzé D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018;23:311–323. doi: 10.1016/j.tplants.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois PG, Brutnell TP. Handbook of Maize: Its Biology. N.Y. pp: Springer; 2009. Light signal transduction networks in maize; pp. 205–227. [Google Scholar]
- Duncan WG. Leaf angle, leaf area, and canopy photosynthesis. Crop Sci. 1971;11:482–485. doi: 10.2135/cropsci1971.0011183X001100040006x. [DOI] [Google Scholar]
- Durbak AR, Phillips KA, Pike S, O’Neill MA, Mares J, Gallavotti A, Malcomber ST, Gassmann W, McSteen P. Transport of boron by the tassel-less1 aquaporin is critical for vegetative and reproductive development in maize. Plant Cell. 2014;26:2978–2995. doi: 10.1105/tpc.114.125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN. Genetic progress in yield of United States maize (Zea mays L.) Maydica. 2005;50:193–202. [Google Scholar]
- Dzievit MJ, Li X, Yu J. Dissection of leaf angle variation in maize through genetic mapping and meta-analysis. Plant Genome. 2018;12:180024. doi: 10.3835/plantgenome2018.05.0024. [DOI] [PubMed] [Google Scholar]
- Efroni I, Birnbaum KD. The potential of single-cell profiling in plants. Genome Biol. 2016;17:65. doi: 10.1186/s13059-016-0931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RA. The inheritance of the ligule and auricle of corn leaves. Neb Agric Exp Sta An Rep. 1912;25:81–85. [Google Scholar]
- Emerson RA. The inheritance of certain abnormalities in maize. Am Breeders Assoc Rep. 1912;8:385–399. [Google Scholar]
- Emerson RA, Emerson SE. Genetic interrelations of two andromonecious types of maize, dwarf and anther ear. Genetics. 1922;7:203–227. doi: 10.1093/genetics/7.3.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Plant Anatomy. 2. New York: Wiley; 1965. [Google Scholar]
- Evans MMS, Poethig RS. The viviparous8 mutation delays vegetative phase change and accelerates the rate of seedling growth in maize. Plant J. 1997;12:769–779. doi: 10.1046/j.1365-313X.1997.12040769.x. [DOI] [Google Scholar]
- Flint-Garcia SA, Thuillet AC, Yu J, Pressoir G, Romero SM, Mithcell SE, Doebley J, Kresovich S, Goodman MM, Buckler ES. Maize association population: a high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- FAOSTAT (2020). Food and Agricultural Organization of the United Nations Agriculture Databases (FAOSTAT, December 22, 2020); http://www.fao.org/faostat/en/#data/QC
- Foster T, Hay A, Johnston R, Hake S. The establishment of axial patterning in the maize leaf. Development. 2004;131:3921–3929. doi: 10.1242/dev.01262. [DOI] [PubMed] [Google Scholar]
- Foster T, Yamaguchi J, Wong BC, Veit B, Hake S (1999) Gnarley1 is a dominant mutation in the knox4 homeobox gene affecting cell shape and identity. Plant Cell 11:1239–1252 [DOI] [PMC free article] [PubMed]
- Foster TM, Timmermans MCP. Handbook of Maize: Its Biology. New York: Springer; 2009. Axial patterning of the maize leaf; pp. 161–178. [Google Scholar]
- Fowler JE, Freeling M. Genetic analysis of mutations that alter cell fates in maize leaves: dominant Liguleless mutations. Dev Genet. 1996;18:198–222. doi: 10.1002/(SICI)1520-6408(1996)18:3<198::AID-DVG2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Fowler JE, Muehlbauer GJ, Freeling M. Mosaic analysis of Liguleless3 mutant phenotype in maize by coordinate suppression of Mutator-insertion alleles. Genetics. 1996;143:489–503. doi: 10.1093/genetics/143.1.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco JAV, Gage JL, Bradbury PJ, Johnson LC, Miller ZR, Buckler ES, Romay MC. bioRxiv. 2020. A maize practical haplotype graph leverages diverse NAM assemblies. [Google Scholar]
- Freeling M, Hake S. Developmental genetics of mutants that specify knotted leaves in maize. Genetics. 1985;111:617–634. doi: 10.1093/genetics/111.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Ren F, Lu X, Mao H, Xu M, Degenhardt J, Peters RJ, Wang Q. A tandem array of ent-kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol. 2016;170:742–751. doi: 10.1104/pp.15.01727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Xu G, Chen H, Wang X, Chen Q, Huang C, Li D, Xu D, Tian J, Wu W, Lu S, Li C, Tian F. QTL mapping for leaf morphology traits in a large maize-teosinte population. Mol Breed. 2019;39:103. doi: 10.1007/s11032-019-1012-5. [DOI] [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Gaskin P, Macmillan J, Phinney BO, Takahashi Qualitative and quantitative analyses of gibberellins in vegetative shoots of normal, dwarf-1, dwarf-2, dwarf-3, and dwarf-5 seedlings of Zea mays L. Plant Physiol. 1988;88:1367–1372. doi: 10.1104/pp.88.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, Monier B, Giri A, Buckler ES. Ten years of the maize nested association mapping population: Impact, limitations, and future directions. Plant Cell. 2020;32:2083–2093. doi: 10.1105/tpc.19.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, Richards E, Lepak N, Kaczmar N, Soman C, Chowdhary G, Gore MA, Buckler ES. In-field whole-plant maize architecture characterized by subcanopy rovers and latent space phenotyping. Plant Phe J. 2019;2:1–21. doi: 10.2135/tppj2019.07.0011. [DOI] [Google Scholar]
- Galinat W. The phytomer in relation to the floral homologies in the American Maydea. Bot Mus Lealf Harv Univ. 1959;19:1–32. [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P. sparse infloresence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci U S A. 2008;105:15196–15201. doi: 10.1073/pnas.0805596105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Malcomber S, Gaines C, Stanfield S, Whipple C, Kellogg E, Schmidt RJ. BARREN STALK FASTIGIATE1 is an AT-hook protein required for the formation of maize ears. Plant Cell. 2011;23:1756–1771. doi: 10.1105/tpc.111.084590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter MK, Doebley JF, Pé ME, Schmidt RJ. The role of barren stalk1 in the architecture of maize. Nature. 2004;432:630–635. doi: 10.1038/nature03148. [DOI] [PubMed] [Google Scholar]
- Gao H, Mutti J, Young JK, Yang M, Schroder M, Lenderts B, Wang L, Peterson D, St. Clair G, Jones S, Feigenbutz L, Marsh W, Zeng M, Wagner S, Farrell J, Snopek K, Scelonge C, Sopko X, Sander JD, Betts S, Cigan AM, Chilcoat ND. Complex trait loci in maize enabled by CRISPR-Cas9 mediated gene insertion. Front Plant Sci. 2020;11:535. doi: 10.3389/fpls.2020.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulini A, Wang J, Jackson D. Control of phyllotaxy by the cytokinin inducible response regulator homologue ABPHYL1. Nature. 2004;430:1031–1034. doi: 10.1038/nature02778. [DOI] [PubMed] [Google Scholar]
- Guan JC, Koch KE, Suzuki M, Wu S, Latshaw S, Petruff T, Goulet C, Klee HJ, McCarty DR. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 2012;160:1303–1317. doi: 10.1104/pp.112.204503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Rupe MA, Wei J, Winkler C, Goncalves-Butruille M, Weers BP, Cerwick SF, Dieter JA, Duncan KE, Howard RJ, Hou Z, Löffler CM, Cooper M, Simmons CR. Maize ARGOS1 (ZAR1) transgenic alleles increase hybrid maize yield. J Exp Bot. 2013;65:249–260. doi: 10.1093/jxb/ert370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Qiu Y, Nettleton D, Yeh CT, Zheng Z, Hey S, Schnable PS. bioRxiv. 2020. Automatic traits extraction and fitting for field high-throughput phenotyping systems. [Google Scholar]
- Hake S, Vollbrecht E, Freeling M. Cloning Knotted, the dominant morphological mutant in maize using Ds2 as a transposon tag. EMBO J. 1989;8:15–22. doi: 10.1002/j.1460-2075.1989.tb03343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallauer AR. Heritability of prolificacy in maize. J Hered. 1974;65:163–168. doi: 10.1093/oxfordjournals.jhered.a108490. [DOI] [Google Scholar]
- Harper L, Freeling M. Interactions of liguleless1 and liguleless2 function during ligule induction in maize. Genetics. 1996;144:1871–1882. doi: 10.1093/genetics/144.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig T, Chuck GS, Fujioka S, Klempien A, Weizbauer R, Potluri DPV, Choe S, Johal GS, Schulz B. Brassinosteroid control of sex determination in maize. Proc Natl Acad Sci U S A. 2011;108:19814–19819. doi: 10.1073/pnas.1108359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Hake S. The dominant mutant Wavy auricle in blade1 disrupts patterning in a lateral domain of the maize leaf. Plant Physiol. 2004;135:300–308. doi: 10.1104/pp.103.036707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/S0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Pautot VA. Beyond the divide: boundaries for patterning and stem cell regulation in plants. Front Plant Sci. 2015;6:1052. doi: 10.3389/fpls.2015.01052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell. 2005;17:1434–1448. doi: 10.1105/tpc.104.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TP, Hayward AP, Tordillos A, Fragoso C, Moreno MA, Tohme J, Kausch AP, Mottinger JP, Dellaporta SL. Identification of the maize gravitropism gene lazy plant1 by transposon-tagging genome resequencing strategy. PLoS One. 2014;9:e87053. doi: 10.1371/journal.pone.0087053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, Seetharam AS, Woodhouse MR, Chougule KM, Ou S, Liu J, Ricci WA, Guo T, Olson A, Qiu Y, Della Coletta R, Tittes S, Hudson AI, Marand AP, Wei S, Lu Z, Wang B, Tello-Ruiz MK, Piri RD, Wang N, won Kim D, Zeng Y, O’Connor CH, Li X, Gilbert AN, Baggs E, Krasileva KV, Portwood JL, Cannon EKS, Andorf CM, Manchanda N, Snodgrass SJ, Hufnagel DE, Jiang Q, Pedersen S, Syring ML, Kudrna DA, Llaca V, Fengler K, Schmitz RJ, Ross-Ibarra J, Yu J, Gent JI, Hirsch CN, Ware D, Dawe RK (2021) De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes. bioRxiv. 10.1101/2021.01.14.426684
- Hughes TE, Sedelnikova OV, Wu H, Becraft PW, Langdale JA. Redundant SCARECROW genes pattern distinct cell layers in roots and leaves of maize. Development. 2019;146:e177543. doi: 10.1242/dev.177543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CT, Kirienko DH, Sylvester AW, Peter GF, McCarty DR, Koch KE. Cellulose synthase-like D1 is integral to normal cell division, expansion and leaf development in maize. Plant Physiol. 2012;158:708–724. doi: 10.1104/pp.111.188466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iltis HH. From teosinte to maize: the catastrophic sexual transmutation. Science. 1983;222:886–894. doi: 10.1126/science.222.4626.886. [DOI] [PubMed] [Google Scholar]
- Irish EE, Jegla D. Regulation of extent of vegetative development of the maize shoot meristem. Plant J. 1997;11:63–71. doi: 10.1046/j.1365-313X.1997.11010063.x. [DOI] [Google Scholar]
- Irish EE, Karlen S. Restoration of juvenility in maize shoots by meristem culture. Int J Plant Sci. 1998;159:695–701. doi: 10.1086/297587. [DOI] [Google Scholar]
- Irish EE, Nelson T. Identification of multiple stages in the conversion of vegetative to floral development. Development. 1991;112:891–898. doi: 10.1242/dev.112.3.891. [DOI] [Google Scholar]
- Jackson D. Double labeling of KNOTTED1 mRNA and protein reveals multiple potential sites of protein trafficking in the shoot apex. Plant Physiol. 2002;129:1423–1429. doi: 10.1104/pp.006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. Handbook of Maize: Its Biology. New York: Springer; 2009. Vegetative shoot meristems; pp. 1–12. [Google Scholar]
- Jackson D, Hake S. Control of phyllotaxy in maize by the abphy1 gene. Development. 1999;126:315–323. doi: 10.1242/dev.126.2.315. [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–413. doi: 10.1242/dev.120.2.405. [DOI] [Google Scholar]
- Je BI, Gruel J, Lee YK, Bommert P, Demesa-Arevalo E, Eveland AL, Wu Q, Goldshmidt A, Meeley R, Bartlett M, Komatsu M, Sakai H, Jönsson H, Jackson D. Signaling from maize primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet. 2016;48:785–791. doi: 10.1038/ng.3567. [DOI] [PubMed] [Google Scholar]
- Je BI, Xu F, Wu Q, Liu L, Meeley R, Gallagher JP, Corcilius L, Payne RJ, Bartlett ME, Jackson D. The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. Elife. 2018;7:e35673. doi: 10.7554/eLife.35673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins MT, Gerhardt F. A gene influencing the composition of the culm in maize. Iowa Agric Exp Stn Res Bull. 1931;138:121–151. [Google Scholar]
- Jiang F, Guo M, Yang F, Duncan K, Jackson D, Rafalski A, Wang S, Li B. Mutations in an AP2 transcription factor-like gene affect internode length and leaf shape in maize. PLoS One. 2012;7(5):e37040. doi: 10.1371/journal.pone.0037040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston R, Leiboff S, Scanlon MJ. Ontogeny of the sheathing leaf base in maize (Zea mays) New Phytol. 2015;205:306–315. doi: 10.1111/nph.13010. [DOI] [PubMed] [Google Scholar]
- Johnston R, Wang M, Sun Q, Sylvester AW, Hake S, Scanlon MJ. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell. 2014;26:4718–4732. doi: 10.1105/tpc.114.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorhi MM, Coe EH. Clonal analysis of corn plant development. I. The development of the tassel and the ear. Dev Biol. 1983;97:154–172. doi: 10.1016/0012-1606(83)90073-8. [DOI] [PubMed] [Google Scholar]
- Kafri R, Springer M, Pilpel Y. Genetic redundancy: new tricks for old genes. Cell. 2009;136:389–392. doi: 10.1016/j.cell.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Kaush AP, Nelson-Vasilchik K, Hague J, Mookkan M, Quemada H, Dellaporta S, Fragoso C, Zhang ZJ. Edit at will: genotype independent plant transformation in the ear of advanced genomics and genome editing. Plant Sci. 2019;281:186–205. doi: 10.1016/j.plantsci.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA. Phytochrome B represses teosinte branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006;140:1109–1117. doi: 10.1104/pp.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher T, Starr D, Su X, Tang G, Chen Z, Carter J, Wittich PE, Dong S, Green J, Burch E, McCuiston J, Gu W, Sun Y, Strebe T, Robers J, Bate NJ, Que Q. One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol. 2019;37:287–292. doi: 10.1038/s41587-019-0038-x. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- Kiesselbach T. The Structure and Reproduction of Corn. Lincoln, NE: University of Nebraska College of Agriculture; 1949. [Google Scholar]
- Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot. 2015;66:6093–6107. doi: 10.1093/jxb/erv349. [DOI] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Cilia M, Khalfan-Jagani Z, Jackson D. Intercellular trafficking of a KNOTTED1 green fluorescent protein fusion in the leaf and shoot meristem of Arabidopsis. Proc Natl Acad Sci U S A. 2002;99:4103–4108. doi: 10.1073/pnas.052484099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kir G, Ye H, Nelissen H, Neelakandan AK, Kusnandar AS, Luo A, Inzé D, Sylvester AW, Yin Y, Becraft PW. RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 2015;169:826–839. doi: 10.1104/pp.15.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klesen S, Hill K, Timmermans MCP. Small RNAs as plant morphogens. Curr Opin Dev Biol. 2020;137:455–480. doi: 10.1016/bs.ctdb.2019.11.001. [DOI] [PubMed] [Google Scholar]
- Knauer S, Javelle M, Li L, Li X, Ma X, Wimalanathan K, Kumari S, Johnston R, Leiboff S, Meeley R, Schnable PS, Ware D, Lawrence-Dill C, Yu J, Muehlbauer GJ, Scanlon MJ, Timmermans MCP. A high-resolution gene expression atlas links dedicated meristems genes to key architectural traits. Genome Res. 2019;29:1962–1973. doi: 10.1101/gr.250878.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knöller AS, Blakeslee JJ, Richards EL, Peer WA, Murphy AS. Bracytic2/ZmABCB1 functions in IAA export from intercalary meristems. J Exp Bot. 2010;61:3689–3696. doi: 10.1093/jxb/erq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolkman JM, Strable J, Harline K, Kroon DE, Wiesner-Hanks T, Bradbury PJ, Nelson RJ. Maize introgression library provides evidence for the involvement of liguleless1 in resistance to Northern Leaf Blight. G3. 2020;10:3611–3622. doi: 10.1534/g3.120.401500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Zhang T, Liu J, Heng S, Shi Q, Zhang H, Wang Z, Ge L, Li P, Lu X, Li G. Regulation of leaf angle by auricle development in maize. Mol Plant. 2017;10:516–519. doi: 10.1016/j.molp.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Kremling KAG, Chen SY, Su MH, Lepak NK, Romay MC, Swarts KL, Lu F, Lorant A, Bradbury PJ, Buckler ES. Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature. 2018;555:520–523. doi: 10.1038/nature25966. [DOI] [PubMed] [Google Scholar]
- Ku L, Cao L, Wei X, Su H, Tian Z, Guo S, Zhang L, Ren Z, Wang X, Zhu Y, Li G, Wang Z, Chen Y (2015) Genetic dissection of internode length above the uppermost ear in four RIL populations of maize (Zea mays L.). G3 5:281–289 [DOI] [PMC free article] [PubMed]
- Ku L, Zhang J, Zhang JC, Guo S, Liu H, Zhao R, Yan Q, Chen Y. Genetic dissection of leaf area by jointing two F2:3 populations in maize (Zea mays L.) Plant Breed. 2012;131:591–599. doi: 10.1111/j.1439-0523.2012.01993.x. [DOI] [Google Scholar]
- Ku LX, Zhang J, Guo SL, Liu HY, Zhan RF, Chen YH. Integrated multiple population analysis of leaf architecture traits in maize (Zea mays L.) J Exp Bot. 2012;63:261–274. doi: 10.1093/jxb/err277. [DOI] [PubMed] [Google Scholar]
- Ku LX, Zhao WM, Zhang J, Wu LC, Wang CL, Wang PA, Zhang WQ, Chen YH. Quantitative trait loci mapping of leaf angle and leaf orientation value in maize (Zea mays L.) Theor Appl Genet. 2010;121:951–959. doi: 10.1007/s00122-010-1364-z. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Lambert RJ, Johnston RR. Leaf angle, tassel morphology, and the performance of maize hybrids. Crop Sci. 1978;18:499–502. doi: 10.2135/cropsci1978.0011183X001800030037x. [DOI] [Google Scholar]
- Lamoreaux RJ, Chaney WR, Brown KM. The plastochron index: a review after two decades of use. Am J Bot. 1978;65:586–593. doi: 10.1002/j.1537-2197.1978.tb06113.x. [DOI] [Google Scholar]
- Lawitt SJ, Wych HM, Xu D, Kundu S, Tomes DT. Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 2010;51:1854–1868. doi: 10.1093/pcp/pcq153. [DOI] [PubMed] [Google Scholar]
- Ledin R. The vegetative shoot apex of Zea mays. Am J Bot. 1954;41:11–17. doi: 10.1002/j.1537-2197.1954.tb14298.x. [DOI] [Google Scholar]
- Lee EA, Tollenaar M. Physiological basis of successful breeding strategies for maize grain yield. Crop Sci. 2007;47:S-202–S-215. doi: 10.2135/cropsci2007.04.0010IPBS. [DOI] [Google Scholar]
- Leiboff S, Li X, Hu HC, Todt N, Yang J, Li X, Yu X, Muehlbauer GJ, Timmermans MCP, Yu J, Schnable PS, Scanlon MJ. Genetic control of morphometric diversity in the maize shoot apical meristem. Nat Commun. 2015;6:8974. doi: 10.1038/ncomms9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiboff S, Strable J, Johnston R, Federici S, Sylvester AW, Scanlon MJ (2020) Network analyses identify a transcriptomic proximodistal prepattern in the maize leaf primordium. New Phytol. 10.1111/nph.171132 [DOI] [PubMed]
- Lewis MW, Bolduc N, Hake K, Htike Y, Hay A, Candela H, Hake S. Gene regulatory interactions at lateral organ boundaries in maize. Development. 2014;141:4590–4597. doi: 10.1242/dev.111955. [DOI] [PubMed] [Google Scholar]
- Leydon AR, Gala HP, Guiziou S, Nemhauser JL. Engineering synthetic signaling in plants. Annu Rev Plant Biol. 2020;71:767–788. doi: 10.1146/annurev-arplant-081519-035852. [DOI] [PubMed] [Google Scholar]
- Li C, Li Y, Shi Y, Song Y, Zhang D, Buckler ES, Zhang Z, Wang T, Li Y. Genetic control of the leaf angle and leaf orientation value as revealed by ultra-high density maps in three connected maize populations. PLoS One. 2015;10:e0121624. doi: 10.1371/journal.pone.0121624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liu C, Qi X, Wu Y, Fei X, Mao L, Cheng B, Li X, Xie C. RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol J. 2017;15:1566–1576. doi: 10.1111/pbi.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Tang J, Hu Z, Wang J, Yu T, Yi H, Cao M. A novel maize dwarf mutant generated by Ty1-copia LTR-retrotransposon insertion in Brachytic2 after spaceflight. Plant Cell Rep. 2020;39:393–408. doi: 10.1007/s00299-019-02498-8. [DOI] [PubMed] [Google Scholar]
- Li D, Wang X, Zhang X, Chen Q, Xu G, Xu D, Wang C, Liang Y, Wu L, Huang C, Tian J, Wu Y, Tian F. The genetic architecture of leaf number and its genetic relationship to flowering time in maize. New Phytol. 2016;210:256–268. doi: 10.1111/nph.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang L, Liu M, Dong Z, Li Q, Fei S, Xiang H, Liu B, Jin W. Maize plant architecture is regulated by the ethylene biosynthetic gene ZmACS7. Plant Physiol. 2020;183:1184–1199. doi: 10.1104/pp.19.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang Z, Yao J, Li J, Song W, Yang X. Cellulose synthase-like D1 controls organ size in maize. BMC Plant Biol. 2018;18:239. doi: 10.1186/s12870-018-1453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liable CA, Dirks VA. Genetic variance and selection value of ear number of corn (Zea mays L.) Crop Sci. 1968;8:540–543. doi: 10.2135/cropsci1968.0011183X000800050010x. [DOI] [Google Scholar]
- Libault M, Pingault L, Zogli P, Schiefelbein J. Plant systems biology at the single-cell level. Trends Plant Sci. 2017;22:949–960. doi: 10.1016/j.tplants.2017.08.006. [DOI] [PubMed] [Google Scholar]
- Liu HJ, Jian L, Xu J, Zhang Q, Zhang M, Jin M, Peng Y, Yan J, Han B, Liu J, Gao F, Liu X, Huang L, Wei W, Ding Y, Yan g X, Li Z, Zhang M, Sun J, Bai M, Song W, Chen H, Sun X, Li W, Lu Y, Liu Y, Zhao J, Qian Y, Jackson D, Fernie AR, Yan J. High-throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell. 2020;32:1397–1413. doi: 10.1105/tpc.19.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Huang J, Ne L, Liu N, Du Y, Hou R, Du H, Qiu F, Zhang Z. Dissecting the genetic architecture of important traits that enhance wild germplasm resource usage in modern maize breeding. Mol Breed. 2019;39:157. doi: 10.1007/s11032-019-1061-9. [DOI] [Google Scholar]
- Liu X, Galli M, Camehl I, Gallavotti A. RAMOSA1 ENHANCER LOCUS2-mediated transcriptional repression regulates vegetative and reproductive architecture. Plant Physiol. 2019;179:348–363. doi: 10.1104/pp.18.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cellular Dev Biol – Plant. 2018;54:240–252. doi: 10.1007/s11627-018-9905-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlain M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco C, Register J, Zhao ZY, Xu D, Jones T, Kamm-Gordon W. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell. 2016;28:1998–2015. doi: 10.1105/tpc.16.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science. 1995;270:1980–1983. doi: 10.1126/science.270.5244.1980. [DOI] [PubMed] [Google Scholar]
- Lunde C, Hake S. The interaction of knotted1 and thick tassel dwarf1 in vegetative and reproductive meristems of maize. Genetics. 2009;181:1693–1697. doi: 10.1534/genetics.108.098350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DL, Xie RZ, Niu XK, Li SK, Long HL, Liu YE. Changes in the morphological traits of maize genotypes in China between the 1950s and 2000s. Eur J Agron. 2014;58:1–10. doi: 10.1016/j.eja.2014.04.001. [DOI] [Google Scholar]
- Makarevitch I, Thompson A, Muehlbauer GJ, Springer NM. Brd1 gene in maize encodes a brassinosteroid C-6 oxidase. PLoS One. 2012;7:e30798. doi: 10.1371/journal.pone.0030798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjeran W, Friso G, Ponnala L, Connolly B, Huang M, Reidel E, Zhang C, Asakura Y, Bhuiyan NH, Sun Q, Turgeon R, van Wijk KJ. Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell. 2010;22:3509–3542. doi: 10.1105/tpc.110.079764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez J, Buckler E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci U S A. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel CN, Poethig RS. Cell lineage patterns in the shoot apical meristem of the germinating maize embryo. Planta. 1988;175:13–22. doi: 10.1007/BF00402877. [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S. barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 2007;144:1000–1011. doi: 10.1104/pp.107.098558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. The FT-like ZCN8 gene functions as a floral activation and is involved in photoperiod sensitivity in maize. Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelson SM, Stuber CS, Senior L, Kaeppler SM. Quantitative trait loci controlling leaf and tassel traits in a B73 x Mo17 population of maize. Crop Sci. 2002;42:1902–1909. doi: 10.2135/cropsci2002.1902. [DOI] [Google Scholar]
- Milla R, Matesanz S. Growing larger with domestication: a matter of physiology, morphology or allocation? Plant Biol. 2017;19:475–483. doi: 10.1111/plb.12545. [DOI] [PubMed] [Google Scholar]
- Moon J, Candela H, Hake S. The Liguleless narrow mutation affects proximal-distal signaling and leaf growth. Development. 2013;140:405–412. doi: 10.1242/dev.085787. [DOI] [PubMed] [Google Scholar]
- Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997;11:616–628. doi: 10.1101/gad.11.5.616. [DOI] [PubMed] [Google Scholar]
- Morrison TA, Kessler JR, Buxton DR. Maize internode elongation patterns. Crop Sci. 1994;34:1055–1060. doi: 10.2135/cropsci1994.0011183X003400040040x. [DOI] [Google Scholar]
- Muehlbauer GJ, Fowler JE, Girard L, Tyers R, Harper L, Freeling M. Ectopic expression of the maize homeobox gene liguleless3 alters cell fates in the maize leaf. Plant Physiol. 1999;119:651–662. doi: 10.1104/pp.119.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Moss-Taylor L, Chudalayandi S, Cahill J, Del Valle-Echevarria AR, Alvarez-Castro I, Petefish A, Sakakibara H, Krivosheev DM, Lomin SN, Romanov GA, Thamotharan S, Dam T, Li B, Brugiére The maize Hairy sheath frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell. 2020;32:1501–1518. doi: 10.1105/tpc.19.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannas NJ, Dawe RK. Genetic and genomic toolbox of Zea mays. Genetics. 2015;199:655–669. doi: 10.1534/genetics.114.165183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardmann J, Werr W. The shoot stem cell niche in angiosperms: expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol Biol Evol. 2006;23:2492–2504. doi: 10.1093/molbev/msl125. [DOI] [PubMed] [Google Scholar]
- Nardmann J, Werr W. Handbook of Maize: Its Biology. New York: Springer; 2009. Patterning of the maize embryo and the perspective of evolutionary developmental biology; pp. 105–119. [Google Scholar]
- Nardmann J, Ji J, Werr W, Scanlon MJ (2004) The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131:2827–2839 [DOI] [PubMed]
- Nelissen H, Eeckhout D, Demuynck K, Persiau G, Walton A, van Bel M, Vervoort M, Canadese J, De Block J, Aesaert S, Van Lijsebettens M, Goormachtig S, Vandepoele K, Van Leene J, Muszynski M, Gevaert K, Inzé D, De Jaeger G (2015) Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf. Plant Cell 27:1605–1619 [DOI] [PMC free article] [PubMed]
- Nelms B, Walbot V. Defining the developmental program leading to meiosis in maize. Science. 2019;364:52–56. doi: 10.1126/science.aav6428. [DOI] [PubMed] [Google Scholar]
- Neuffer MG, Coe EH, Wessler SR. Mutants of Maize. 2. Plainview: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Pan Q, Xu Y, Li K, Peng Y, Zhan W, Li W, Li L, Yan J. The genetic basis of plant architecture in 10 maize recombinant inbred line populations. Plant Physiol. 2017;175:858–873. doi: 10.1104/pp.17.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli D, Chapman SC, Bart R, Topp CN, Lawrence-Dill CJ, Poland J, Gore MA. The quest for understanding phenotypic variation via integrated approaches in the field environment. Plant Physiol. 2016;172:622–634. doi: 10.1104/pp.16.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautler M, Eveland AL, LaRue T, Yang F, Weeks R, Je BI, Meeley R, Komatsu M, Vollbrecht E, Sakai H, Jackson D. FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell. 2015;27:104–120. doi: 10.1105/tpc.114.132506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Flint-Garcia S, de Leon N, McMullen M, Kaeppler SM, Buckler ES. The genetic architecture of maize stalk strength. PLoS One. 2013;8:e67066. doi: 10.1371/journal.pone.0067066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CA, McMullen M, Holland JB, Bradbury JP, Buckler ES. The genetic architecture of maize height. Genetics. 2014;196:1337–1356. doi: 10.1534/genetics.113.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ. Field investigations of the relationships of leaf angle in corn (Zea mays L.) to grain yield and apparent photosynthesis. Agron J. 1968;60:422–424. doi: 10.2134/agronj1968.00021962006000040027x. [DOI] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP. ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Pepper GE, Pearce RB, Mock JJ. Leaf orientation and yield of maize. Crop Sci. 1977;17:883–886. doi: 10.2135/cropsci1977.0011183X001700060017x. [DOI] [Google Scholar]
- Petsch K, Manzotti PS, Tam OT, Meeley R, Hammell M, Consonni G, Timmermans MCP. Novel DICER-LIKE1 siRNAs bypass the requirement for DICER-LIKE4 in maize development. Plant Cell. 2015;27:2163–2177. doi: 10.1105/tpc.15.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phinney BO, Spray CR. Plant growth substances 1982. London: Academic Press; 1982. Biochemical genetics and the gibberellin biosynthetic pathway in Zea mays L; pp. 101–110. [Google Scholar]
- Pilu R, Cassani E, Villa D, Curiale S, Panzeri D, Badone FC, Landoni M. Isolation and characterization of a new mutant allele of brachytic 2 maize gene. Mol Breed. 2007;20:83–91. doi: 10.1007/s11032-006-9073-7. [DOI] [Google Scholar]
- Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci U S A. 2009;106:5019–5024. doi: 10.1073/pnas.0812525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig RS. Contemporary Problems in Plant Anatomy. Cambridge: Academic Press; 1984. Cellular parameters of leaf development in maize and tobacco. [Google Scholar]
- Poethig RS. Clonal analysis of cell lineage patterns in plant development. Am J Bot. 1987;74:581–594. doi: 10.1002/j.1537-2197.1987.tb08679.x. [DOI] [Google Scholar]
- Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- Poethig RS, Coe EH, Johri MM. Cell linage patterns in maize embryogenesis: a clonal analysis. Dev Biol. 1986;117:392–404. doi: 10.1016/0012-1606(86)90308-8. [DOI] [Google Scholar]
- Poethig RS, Szymkowiak EJ. Clonal analysis of leaf development in maize. Maydica. 1995;40:67–76. [Google Scholar]
- Qiao P, Lin M, Vasquez M, Matschi S, Chamness J, Baseggio M, Smith LG, Sabuncu MR, Gore MA, Scanlon MJ. Machine learning enables high-throughput phenotyping for analyses of the genetic architecture of bulliform cell patterning in maize. G3. 2019;9:4235–4243. doi: 10.1534/g3.119.400757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, Bolduc N, Lisch D, Hake S. Distal expression of knotted1 in maize leaves leads to reestablishment of proximal/distal patterning and leaf dissection. Plant Physiol. 2009;151:1878–1888. doi: 10.1104/pp.109.145920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph LF. Developmental morphology of the caryopsis in maize. J Agric Res. 1936;53:881–917. [Google Scholar]
- Reinhardt D, Kuhlemeier C. Plant architecture. EMBO Rep. 2002;3:846–851. doi: 10.1093/embo-reports/kvf177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Wu L, Ku L, Wang H, Zeng H, Su H, Wei L, Dou D, Liu H, Cao Y, Zhang D, Han S, Chen Y. ZmILI1 regulates leaf angle by directly affecting liguleless1 expression in maize. Plant Biotechnol J. 2020;18:881–883. doi: 10.1111/pbi.13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerrra MK, Colomé-Tatché M, Johannes F, Rowley MJ, Corces VG, Zhai J, Scanlon MJ, Buckler ES, Gallavotti A, Springer NM, Schmitz RJ, Zhang X. Widespread long-range cis-regulatory elements in the maize genome. Nat Plants. 2019;5:1237–1249. doi: 10.1038/s41477-019-0547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AE, Hake S. Drawing a line: grasses and boundaries. Plants. 2019;8:4. doi: 10.3390/plants8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter MW, Padilla CM, Schmidt RJ. The maize mutant barren stalk1 is defective in axillary meristem development. Am J Bot. 2002;89:203–210. doi: 10.3732/ajb.89.2.203. [DOI] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–480e8. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Leal D, Xu C, Kwon CT, Soyars C, Demesa-Arevalo E, Man J, Liu L, Lemmon ZH, Jones DS, Van Eck J, Jackson DP, Bartlett ME, Nimchuk ZL, Lippman ZB. Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat Genet. 2019;51:786–792. doi: 10.1038/s41588-019-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romay MC, Millard MJ, Glaubitz JC, Peiffer JA, Swarts KL, Casstevents TM, Elshire RJ, Acharya CB, Mitchell SE, Flint-Garcia SA, McMullen MD, Holland JB, Buckler ES, Gardner CA. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 2013;14:R55. doi: 10.1186/gb-2013-14-6-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa M, Abraham-Juarez MJ, Lewis MW, Fonesca JP, Tian W, Ramirez V, Luan S, Pauly M, Hake S. The maize MID-COMPLEMENTING ACTIVITY homolog CELL NUMBER REGULATOR13/NARROW ODD DWARF coordinates organ growth and tissue patterning. Plant Cell. 2017;29:474–490. doi: 10.1105/tpc.16.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WA. Genetic improvements of maize yields. Adv Agron. 1991;46:245–298. doi: 10.1016/S0065-2113(08)60582-9. [DOI] [Google Scholar]
- Russell SH, Evert RF. Leaf vasculature in Zea mays L. Planta. 1985;164:448–458. doi: 10.1007/BF00395960. [DOI] [PubMed] [Google Scholar]
- Russell WK, Stuber CW. Effects of photoperiod and temperatures on the duration of vegetative growth in maize. Crop Sci. 1983;23:847–850. doi: 10.2135/cropsci1983.0011183X002300050008x. [DOI] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/S0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Satterlee JW, Strable J, Scanlon MJ. Plant stem cell organization and differentiation at single-cell resolution. Proc Natl Acad Sci U S A. 2020;117:33689–33699. doi: 10.1073/pnas.2018788117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ. NARROW SHEATH1 functions from two meristematic foci during founder-cell recruitment in maize leaf development. Development. 2000;127:4573–4585. doi: 10.1242/dev.127.21.4573. [DOI] [PubMed] [Google Scholar]
- Scanlon MJ, Schneeberger RG, Freeling M. The maize mutant narrow sheath fails to establish leaf margin identity in the meristematic domain. Development. 1996;122:1683–1691. doi: 10.1242/dev.122.6.1683. [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M. Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One. 2011;6:e17855. doi: 10.1371/journal.pone.0017855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger RG, Becraft PW, Hake S, Freeling M. Ectopic expression of the knox homeo box gene rough sheath1 alters cell fate in the maize leaf. Genes Dev. 1995;9:2292–2304. doi: 10.1101/gad.9.18.2292. [DOI] [PubMed] [Google Scholar]
- Schneeberger RG, Tsiantis M, Freeling M, Langdale JA. The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development. 1998;125:2857–2865. doi: 10.1242/dev.125.15.2857. [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaben C. Post-translational regulation of plant bZIP factors. Trends Plant Sci. 2008;13:247–255. doi: 10.1016/j.tplants.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Shannon LM. The genetic architecture of maize domestication and range expansion. PhD thesis. Madison: The University of Wisconsin-Madison; 2012. [Google Scholar]
- Shannon LM, Chen Q, Doebley JF (2019) A BC2S3 maize-teosinte RIL population for QTL mapping. Maize Genet Coop News Lett 93
- Sharman BC. Developmental anatomy of the shoot of Zea mays L. Ann Bot (Lond) 1942;6:245–282. doi: 10.1093/oxfordjournals.aob.a088407. [DOI] [Google Scholar]
- Sheridan WF. Maize developmental genetics: genes of morphogenesis. Annu Rev Genet. 1988;22:353–385. doi: 10.1146/annurev.ge.22.120188.002033. [DOI] [PubMed] [Google Scholar]
- Shi J, Gao H, Wang H, Lafitte HR, Archiblad RL, Yang M, Hakimi SM, Mo H, Habben JE. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2016;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Habben JE, Archibald RL, Drummond BJ, Chamberlin MA, Williams RW, Lafitte HR, Weers BP. Overexpression of ARGOS gene modifies plant sensitivity to ethylene, leading to improved drought tolerance in both Arabidopsis and maize. Plant Physiol. 2015;169:266–282. doi: 10.1104/pp.15.00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N, Hake H. Mutant characters of Knotted maize leaves are determined in the innermost tissue layers. Dev Biol. 1990;141:203–210. doi: 10.1016/0012-1606(90)90115-Y. [DOI] [PubMed] [Google Scholar]
- Skirpan A, Culler AH, Gallavotti A, Jackson D, Cohen JD, McSteen P. BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol. 2009;50:652–657. doi: 10.1093/pcp/pcp006. [DOI] [PubMed] [Google Scholar]
- Skirpan A, Wu X, McSteen P. Genetic and physical interaction suggest that BARREN STALK1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 2008;55:787–797. doi: 10.1111/j.1365-313X.2008.03546.x. [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. A dominant mutation in the maize homeobox, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development. 1992;116:21–30. doi: 10.1242/dev.116.1.21. [DOI] [PubMed] [Google Scholar]
- Smith LG, Hake S, Sylvester AW. The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development. 1996;122:481–489. doi: 10.1242/dev.122.2.481. [DOI] [PubMed] [Google Scholar]
- Somssich M, Je BI, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- Soyk S, Benoit M, Lippman ZB. Epistasis in crop quantitative trait variation. Annu Rev Genet. 2020;54:13.1–13.21. doi: 10.1146/annurev-genet-050720-122916. [DOI] [PubMed] [Google Scholar]
- Steeves T, Sussex I. Patterns in plant development. Cambridge, UK: University Presse; 1989. [Google Scholar]
- Steffensen DM. A reconstruction of cell development in the shoot apex of maize. Am J Bot. 1968;55:354–369. doi: 10.1002/j.1537-2197.1968.tb07387.x. [DOI] [Google Scholar]
- Stewart DW, Costa C, Dwyer LM, Smith DL, Hamilton RI, Ma BL. Canopy structure, light interception, and photosynthesis in maize. Agron J. 2003;95:1465–1474. doi: 10.2134/agronj2003.1465. [DOI] [Google Scholar]
- Stitzer MC, Ross-Ibarra J. Maize domestication and gene interaction. New Phytol. 2018;220:395–408. doi: 10.1111/nph.15350. [DOI] [PubMed] [Google Scholar]
- Strable J, Wallace JG, Unger-Wallace E, Briggs S, Bradbury PJ, Buckler ES, Vollbrecht E. Maize YABBY genes drooping leaf1 and drooping leaf2 regulate plant architecture. Plant Cell. 2017;29:1622–1641. doi: 10.1105/tpc.16.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Chen D, Li X, Qiao S, Shi C, Li C, Shen H, Wang X. Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev Cell. 2015;34:220–228. doi: 10.1016/j.devcel.2015.05.019. [DOI] [PubMed] [Google Scholar]
- Sun X, Cahill J, Van Hautegem T, Feys K, Whipple C, Novák O, Delbare S, Versteele C, Demuynck K, De Block J, Storme V, Claeys H, Van Lijsebettens M, Coussens G, Ljung K, De Vliegher A, Muszynski M, Inzé D, Nelissen H. Altered expression of maize PLASTOCHRON1 enhances biomass and seed trait by extending cell division duration. Nat Commun. 2017;8:14752. doi: 10.1038/ncomms14752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM. The evolution of plant architecture. Curr Opin Plant Biol. 2001;4:33–37. doi: 10.1016/S1369-5266(00)00132-1. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Latshaw S, Sato Y, Settles AM, Koch KE, Hannah LC, Kojima M, Sakakibara H, McCarty DR. The maize Viviparaous8 locus, encoding a putative ALTERED MERISTEM PROGRAM1-like peptidase, regulates abscisic acid accumulation and coordinates embryo and endosperm development. Plant Physiol. 2008;146:1193–1206. doi: 10.1104/pp.107.114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015;169:931–945. doi: 10.1104/pp.15.00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvester AW, Cande WZ, Freeling M. Division and differentiation during normal and liguleless-1 maize leaf development. Development. 1990;110:985–1000. doi: 10.1242/dev.110.3.985. [DOI] [PubMed] [Google Scholar]
- Sylvester AW, Smith LG, Freeling M. Acquisition of identity in the developing leaf. Annu Rev Cell Dev Biol. 1996;12:257–304. doi: 10.1146/annurev.cellbio.12.1.257. [DOI] [PubMed] [Google Scholar]
- Taagen E, Bogdanove AJ, Sorrells ME. Counting on crossovers: controlled recombination for plant breeding. Trends Plant Sci. 2020;25:455–465. doi: 10.1016/j.tplants.2019.12.017. [DOI] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Yuan Z, Hake S, Jackson D. The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 2001;15:2755–2766. doi: 10.1101/gad.208501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takacs EM, Li J, Du C, Ponnala L, Janick-Buckner D, Yu J, Muehlbauer GJ, Schnable PS, Timmermans MCP, Sun Q, Nettleton D, Scanlon MJ. Ontogeny of the maize shoot apical meristem. Plant Cell. 2012;24:3219–3234. doi: 10.1105/tpc.112.099614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proc Natl Acad Sci U S A. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Zhai L, Liu R, Bai W, Wang L, Huo D, Tao Y, Zheng Y, Zhang Z. ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J. 2013;73:405–416. doi: 10.1111/tpj.12038. [DOI] [PubMed] [Google Scholar]
- Thompson AM, Crants JE, Schnable PS, Yu J, Timmermans MCP, Springer NM, Scanlon MJ, Muehlbauer GJ (2014) Genetic control of maize shoot apical meristem architecture. G3 4:1327–1337 [DOI] [PMC free article] [PubMed]
- Thompson AM, Yu J, Timmermans MCP, Schnable PS, Crants JE, Scanlon MJ, Muehlbauer GJ. Diversity of maize shoot apical meristem architecture and its relationship to plant morphology. G3. 2015;5:819–827. doi: 10.1534/g3.115.017541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- Tian J, Wang C, Xia J, Wu L, Xu G, Wu W, Li D, Qin W, Han X, Chen Q, Jin W, Tian F. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science. 2019;365:658–664. doi: 10.1126/science.aax5482. [DOI] [PubMed] [Google Scholar]
- Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci U S A. 2011;108:20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MCP, Hudson A, Becraft PW, Nelson T. ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science. 1999;284:151–153. doi: 10.1126/science.284.5411.151. [DOI] [PubMed] [Google Scholar]
- Timmermans MCP, Schultes NP, Jankovsky JP, Nelson T. Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development. 1998;125:2813–2823. doi: 10.1242/dev.125.15.2813. [DOI] [PubMed] [Google Scholar]
- Troyer AF. Breeding widely adapted, popular maize hybrids. Euphytica. 1996;92:163–174. doi: 10.1007/BF00022842. [DOI] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science. 1999;284:154–156. doi: 10.1126/science.284.5411.154. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Abraham-Juarez MJ, Maeno A, Dong Z, Aromdee D, Meeley R, Shiroishi T, Nonomura K, Hake S. KNOTTED1 cofactors, BLH12 and BLH14, regulate internode patterning and vein anastomosis in maize. Plant Cell. 2017;29:1105–1118. doi: 10.1105/tpc.16.00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Kurata N, Ohyanagi H, Hake S. Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell. 2014;26:3488–3500. doi: 10.1105/tpc.114.129122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuberosa R, Salvi S. Handbook of Maize – Volume II: Genetics and Genomics. New York: Springer; 2009. QTL for agronomic traits in maize plants; pp. 501–541. [Google Scholar]
- van der Knaap E, Kim JH, Kende H. A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 2000;122:695–704. doi: 10.1104/pp.122.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek J. “Lazy” an a-geotropic form of maize. “Gravitational indifference” rather than structural weakness accounts for prostrate growth habit of this form. J Hered. 1936;27:93–96. doi: 10.1093/oxfordjournals.jhered.a104191. [DOI] [Google Scholar]
- Veit B, Briggs SP, Schmidt RJ, Yanofsky MF, Hake S. Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature. 1998;393:166–168. doi: 10.1038/30239. [DOI] [PubMed] [Google Scholar]
- Veit B, Vollbrecht E, Mathern J, Hake S. A tandem duplication causes the Kn1-O allele of Knotted, a dominant morphological mutant of maize. Genetics. 1990;125:623–631. doi: 10.1093/genetics/125.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlăduţu C, McLaughlin J, Phillips RL. Fine mapping and characterization of linked quantitative trait loci involved in the transition of the maize apical meristem from vegetative to generative structures. Genetics. 1999;153:993–1007. doi: 10.1093/genetics/153.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E, Reiser L, Hake S. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development. 2000;127:3161–3172. doi: 10.1242/dev.127.14.3161. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Walley JW, Sartor RC, Shen Z, Schmitz RJ, Wu KJ, Urich MA, Nery JR, Smith LG, Schnable JC, Ecker JR, Briggs SP. Integration of omic networks in a developmental atlas of maize. Science. 2016;353:814–818. doi: 10.1126/science.aag1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh J, Waters CA, Freeling M. The maize gene liguleless2 encodes a basic-leucine zipper protein involved in the establishment of the blade-sheath boundary. Genes Dev. 1998;12:208–218. doi: 10.1101/gad.12.2.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Lin Z, Li X, Zhao Y, Zhao B, Wu G, Ma X, Wang H, Xie Y, Li Q, Song G, Kong D, Zheng Z, Wei H, Shen R, Wu H, Chen C, Meng Z, Wang T, Li Y, Li X, Chen Y, Lai J, Hufford MB, Ross-Ibarra J, He H, Wang H. Genome-wide selection and genetic improvement during modern maize breeding. Nat Genet. 2020;52:565–571. doi: 10.1038/s41588-020-0616-3. [DOI] [PubMed] [Google Scholar]
- Wang B, Smith SM, Li J. Genetic regulation of shoot architecture. Annu Rev Plant Biol. 2018;69:437–468. doi: 10.1146/annurev-arplant-042817-040422. [DOI] [PubMed] [Google Scholar]
- Wang H, Cimen E, Singh N, Buckler E. Deep learning for plant genomics and crop improvement. Curr Opin Plant Biol. 2020;54:34–41. doi: 10.1016/j.pbi.2019.12.010. [DOI] [PubMed] [Google Scholar]
- Wang P, Kelly S, Fouracre JP, Langdale JA. Genome-wide transcript analysis of early maize leaf development reveals gene cohorts associated with the differentiation of C4 Kranz anatomy. Plant J. 2013;75:656–670. doi: 10.1111/tpj.12229. [DOI] [PubMed] [Google Scholar]
- Wang X, Chen Q, Wu Y, Lemmon ZH, Xu G, Huang C, Liang Y, Xu D, Li D, Doebley JF, Tian F. Genome-wide analysis of transcriptional variability in a large maize-teosinte population. Mol Plant. 2018;11:443–459. doi: 10.1016/j.molp.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Washburn JD, Burch MB, Franco JAV. Predictive breeding for maize: making use of molecular phenotypes, machine learning and physiological crop models. Crop Sci. 2020;60:622–638. doi: 10.1002/csc2.20052. [DOI] [Google Scholar]
- Washburn JD, Mejía-Guerrra MK, Ramstein G, Kremling KA, Valluru R, Buckler ES, Wang H. Evolutionary informed deep learning methods for predictive relative transcript abundance from DNA sequence. Proc Natl Acad Sci U S A. 2019;116:5542–5549. doi: 10.1073/pnas.1814551116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Wang X, Guo S, Zhou J, Shi Y, Wang H, Dou D, Song X, Li G, Ku L, Cheng Y. Epistatic and QTLxenvironment interaction effects on leaf area-associated traits in maize. Plant Breed. 2016;135:671–676. doi: 10.1111/pbr.12422. [DOI] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci U S A. 2011;108:E506–E512. doi: 10.1073/pnas.1102819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills DM, Whipple CJ, Takuno S, Kursel LE, Shannon LM, Ross-Ibarra J, Doebley JF. From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 2013;9:e1003604. doi: 10.1371/journal.pgen.1003604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Freeling M. Physiological genetics of the dominant gibberellin-non-responsive maize dwarfs, Dwarf8 and Dwarf9. Planta. 1994;193:341–348. doi: 10.1007/BF00201811. [DOI] [Google Scholar]
- Winkler RG, Helentjaris T. The maize dwarf3 gene encodes a cytochrome P450-mediated early step in gibberellin biosynthesis. Plant Cell. 1995;7:1307–1317. doi: 10.1105/tpc.7.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Woodward JB, Abeydeera ND, Paul D, Phillips K, Rapala-Kozik M, Freeling M, Begley TP, Ealick SE, McSteen P, Scanlon MJ. A maize thiamine auxotroph is defective in shoot meristem maintenance. Plant Cell. 2010;22:3305–3317. doi: 10.1105/tpc.110.077776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Regan M, Furukawa H, Jackson D. Role of heterotrimeric Gα proteins in maize development and enhancement of agronomic traits. PLoS Genet. 2018;14:e1007374. doi: 10.1371/journal.pgen.1007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Xu F, Liu L, Char SN, Ding Y, Je BI, Schmelz E, Yang B, Jackson D. The maize heterotrimeric G protein β subunit controls shoot meristem development and immune response. Proc Natl Acad Sci U S A. 2020;117:1799–1805. doi: 10.1073/pnas.1917577116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A, Gao Y, Ye L, Zhang W, Cai L, Ching A, Llaca V, Johnson B, Liu L, Yang X, Kang D, Yan J, Li J. A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J Exp Bot. 2015;66:3791–3802. doi: 10.1093/jxb/erv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Cao J, Wang X, Chen Q, Jin W, Li Z, Tian F. Evolutionary metabolomics identifies substantial metabolic divergence between maize and its wild ancestor, teosinte. Plant Cell. 2019;31:1990–2009. doi: 10.1105/tpc.19.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, McKim SM, Murmu J, Haughn GW, Hepworth SR. Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 2010;63:974–989. doi: 10.1111/j.1365-313X.2010.04299.x. [DOI] [PubMed] [Google Scholar]
- Xu XM, Wang J, Xuan Z, Goldschmidt A, Borrill PGM, Hariharan N, Kim JY, Jackson D. Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science. 2011;333:1141–1144. doi: 10.1126/science.1205727. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell. 2004;16:500–509. doi: 10.1105/tpc.018044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Christensen S, Isakeit T, Engelberth J, Meeley R, Hayward A, Emery RJN, Kolomiets MV. Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. Plant Cell. 2012;24:1420–1436. doi: 10.1105/tpc.111.094151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Bui HT, Pautler M, Llaca V, Johnston R, Lee BH, Kolbe A, Sakai H, Jackson D. A maize glutaredoxin gene, Abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell. 2015;27:121–131. doi: 10.1105/tpc.114.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Liu J, Zhao C, Li Z, Huang Y, Yu H, Xu B, Yang X, Zhu D, Zhang X, Zhang R, Feng H, Zhao X, Li Z, Li H, Yang H. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: current status and perspectives. Front Plant Sci. 2017;8:1111. doi: 10.3389/fpls.2017.01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Liu J, Gao Q, Gui S, Chen L, Yang L, Huang J, Deng T, Luo J, He L, Wang Y, Xu P, Peng Y, Shi Z, Lan L, Ma Z, Yang X, Zhang Q, Bai M, Li S, Li W, Liu L, Jackson D, Yan J. Genome assembly of a tropical maize inbred line provides insights into structural variation and crop improvement. Nat Genet. 2019;51:1052–1059. doi: 10.1038/s41588-019-0427-6. [DOI] [PubMed] [Google Scholar]
- Yao H, Skirpan A, Wardell B, Matthes MS, Best NB, McCubbin T, Durbak A, Smith T, Malcomber S, McSteen P. The barren stalk2 gene is required for axillary meristem development in maize. Mol Plant. 2019;12:374–389. doi: 10.1016/j.molp.2018.12.024. [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Leiboff S, Li X, Guo T, Ronning N, Zhang X, Muehlbauer GJ, Timmermans MCP, Schnable PS, Scanlon MJ, Yu J (2020) Genomic prediction of maize microphenotypes provides insights for optimizing selection and mining diversity. Plant Biotechnol J. 10.1111/pbi.13420 [DOI] [PMC free article] [PubMed]
- Zhang D, Sun W, Singh R, Zheng Y, Cao Z, Li M, Hake S, Zhang Z. GRF-interacting factor1 regulates shoot architecture and meristem determinacy in maize. Plant Cell. 2018;30:360–374. doi: 10.1105/tpc.17.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang X, Pan Q, Li P, Liu Y, Lu X, Zhong W, Li M, Han L, Li J, Wang P, Li D, Liu Y, Li Q, Yang F, Zhang YM, Wang G, Li L. QTG-seq accelerates QTL fine mapping through QTL partitioning and whole-genome sequencing of bulked segregant samples. Mol Plant. 2019;12:426–437. doi: 10.1016/j.molp.2018.12.018. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, Yang H, Xu Y, Kim SH, Fujioka S, Lin WH, Chong K, Lu T, Wang ZY. Antagonistic HLH.bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huang C, Wu D, Qiao F, Li W, Duan L, Wang K, Xiao Y, Chen G, Liu Q, Xiong L, Yang W, Yan J. High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol. 2017;173:1554–1564. doi: 10.1104/pp.16.01516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin Z, Wang J, Liu H, Zhou L, Zhong S, Li Y, Zhu C, Liu J, Lin Z. The tin1 gene retains the function of promoting tillering in maize. Nat Commun. 2019;10:5608. doi: 10.1038/s41467-019-13425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yang X, Lin X, Sassenrath GF, Dai S, Lv S, Chen X, Chen F, Mi G. Radiation interception and use efficiency contributes to higher yields of newer maize hybrids in Northeast China. Agron J. 2015;107:1473–1480. doi: 10.2134/agronj14.0510. [DOI] [Google Scholar]
- Zhou S, Kremling KA, Bandillo N, Ritcher A, Zhang YK, Ahern KR, Artyukhin AB, Hui JX, Younkin GC, Schroeder FC, Buckler ES, Jander G. Metabolome-scale genome-wide association studies reveal chemical diversity and genetic control of maize specialized metabolites. Plant Cell. 2019;31:937–955. doi: 10.1105/tpc.18.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 30 kb)
Data Availability Statement
Not applicable