Abstract
Cardiolipins are phospholipids that are central to proper mitochondrial functioning. Because mitochondria play crucial roles in differentiation, development, and maturation, we would also expect cardiolipin to play major roles in these processes. Indeed, cardiolipin has been implicated in the mechanism of three human diseases that affect young infants, implying developmental abnormalities. In this paper, we will: 1) Review the biology of cardiolipin; 2) Outline the evidence for essential roles of cardiolipin during organismal development, including embryogenesis and cell maturation in vertebrate organisms; 3) Place the role(s) of cardiolipin during embryogenesis within the larger context of the roles of mitochondria in development; and 4) Suggest avenues for future research.
Keywords: Mitochondria, Oxidative phosphorylation, Phospholipids, Signaling, Tafazzin
Introduction
Cardiolipins (CL’s) comprise a class of phospholipids that play diverse and important roles in mitochondria.1-3 Given that mitochondria play crucial roles in embryogenesis and organogenesis, we would expect CL, as the so-called “signature” phospholipid of mitochondria, to also play major roles in these developmental processes.
We currently know very little about the roles of mitochondrial phospholipids during developmental processes; thus, CL may serve as an illustrative model. Here, we will review the biology of CL, and the evidence for roles of CL during organismal development, including embryogenesis and cell maturation. We will then discuss these roles of CL during embryogenesis within the larger context of the roles of mitochondria in development to draw additional inferences. We focus on post-gastrulation development, which includes cellular differentiation and organogenesis. In addition to studies in cellular model systems, we will establish that CL plays a role in vertebrate development and then outline the evidence for possible underlying mechanisms. We then suggest questions and avenues for future research.
Lipids, Phospholipids, and Cardiolipin: General Biology
What might be termed “developmental lipid biology” is an understudied research area, considering the importance of lipids in multiple cell processes and functions. As a major component of cell membranes and playing essential roles in energy storage, cell signaling and transport, lipids also play fundamental roles in development, including mammalian embryo preimplantation development and cell fate. We now know that lipid metabolites influence cellular events including differentiation, proliferation, migration, and organogenesis.4-7 Inherited disorders of lipid metabolism currently include almost 150 Mendelian diseases, with over 80 involving complex lipid biosynthesis and remodeling7; many of these may be considered developmental disorders with pleiotropic effects on multiple organ systems in the newborn infant, such as Barth syndrome (see below) and MEGDEL syndrome.
Phospholipids are “complex lipids” consisting of two fatty acid tails and a phosphate-containing head group joined by a glycerol molecule.7 They are critical not only to membrane structure and function but also to protein interactions, including in mitochondria8, cell signaling, autophagy, cell division, and oxidative phosphorylation (OXPHOS).7 Disrupting phospholipid biosynthesis can lead to severe developmental consequences.9
CL is a dimeric phospholipid, diphosphatidylglycerol (1,3-bis(sn-3’-phosphatidyl)-sn-glycerol) (Figure 1).2,3,10 Among its diverse and important roles in mitochondrial functioning and morphology, CL is particularly critical to mitochondrial OXPHOS via the assembly of high-order oligomers of respiratory chain complexes.11-15 Less known but no less important, is CL’s role in stabilizing the lateral organization of protein-rich membranes and mitochondrial dynamics.16-17 CL biosynthesis starts from phosphatidic acid, and nascent CL is formed from phosphatidylglycerol (PG) via cardiolipin synthase (CRLS1), along the matrix-facing side of the inner mitochondrial membrane. Nascent CL then undergoes post-synthetic exchange of acyl groups, also known as remodeling (Figure 2). Catalyzed by the transacylase tafazzin, CL is remodeled via acyl exchange between phospholipids and monolyso-cardiolipin to its mature form found in the inner membrane of the mitochondria.18 Other acyltransferases -namely MLCLAT1 and ALCAT1 – may also be involved in CL remodeling.19,20 Most commonly, remodeled (mature) cardiolipin contains a higher number of unsaturated acyl chains, but a notable exception is the extramitochondrial cardiolipin, tetrapalmitoyl cardiolipin.21 Of note, tafazzin does not exhibit the usual enzyme specificity; instead, tafazzin relies on the surrounding membrane biophysical milieu (e.g., membrane curvature and membrane protein density) to remodel nascent CL into many different possible “mature” CL species.18 It is therefore important to emphasize that CL is not a single molecule: remodeling of its acyl chains results in an enormous diversity of acyl chain combinations, and consequently, many possible species of CL (Figure 1). Individual CL species together form a fluid with characteristic physical properties whose differences may be required in different cellular milieu.22,23 Specific CL species profiles are tissue- and species-dependent and as we will outline, even developmentally stage-dependent.
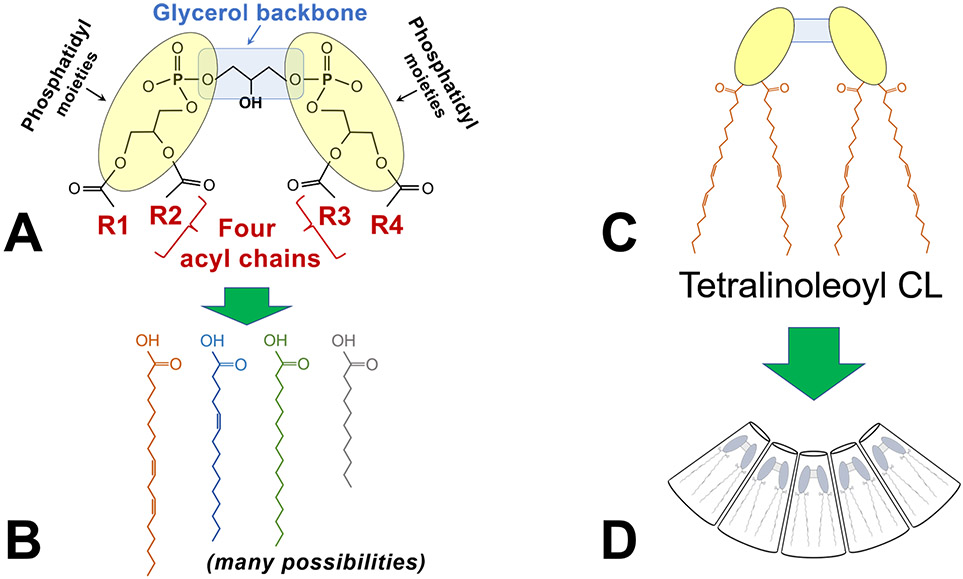
FIGURE 1. Cardiolipin structure, an overview.
A) The schematic shows the key features of cardiolipin: A glycerol backbone and two phosphatidyl moieties that together form the dimeric phosphatidylglycerol head group; and 4 acyl chains (designated R1-R4).
B) Each of the four acyl chains may vary by length, degrees of saturation, and positions of double bonds, resulting in an enormous possible diversity of acyl chain combinations.
C) The most abundant form of cardiolipin in the mammalian heart is tetralinoleoyl cardiolipin (CL), in which the four acyl chains are linoleic acid.
D) Tetralinoleoyl CL has a unique conical shape that is critical to its biophysical properties within the lipid membrane.
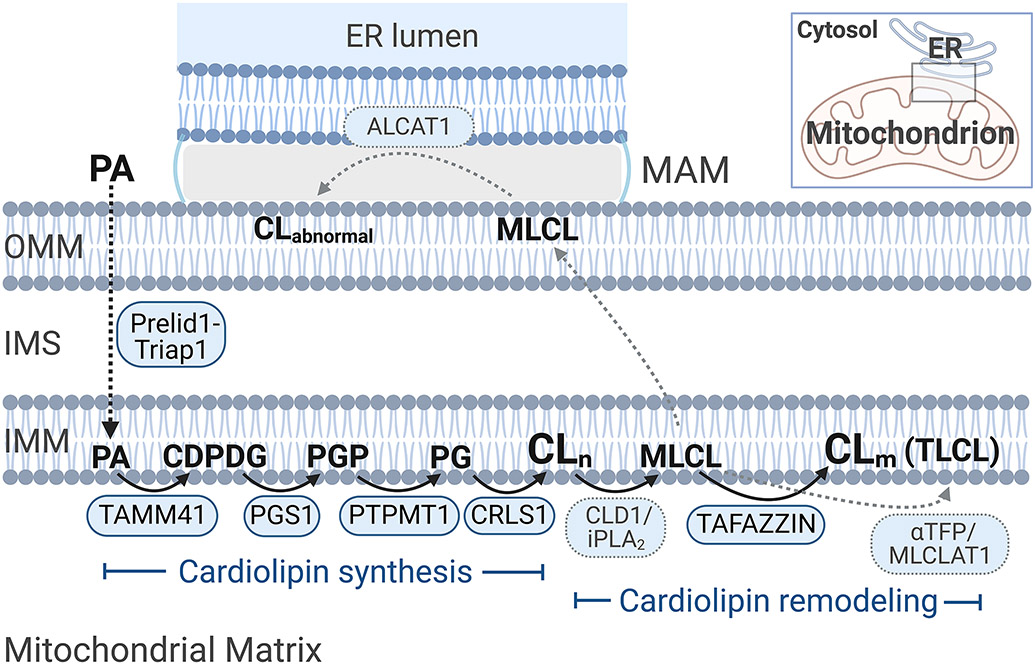
FIGURE 2. Cardiolipin biosynthetic and remodeling pathways.
Cardiolipin synthesis: Phosphatidic acid (PA) is the central precursor for the biosynthesis of glycerolipids and glycerophospholipids. PA is transported by Prelid1-Triap1 from the endoplasmic reticulum (ER) membranes to the inner mitochondrial membrane (IMM), where sequential reactions lead to the biosynthesis of cardiolipin (CL). The first reaction is catalyzed by TAMM41, by which PA is converted to cytidine diphosphate-diacylglycerol (CDPDG). The enzyme PGS1 then transfers a phosphatidyl group from CDPDG to glycerol-3-phosphate to form phosphatidylglycerophosphate (PGP). The third reaction is catalyzed by PTPMT1, which removes the terminal phosphate group from PGP to form phosphatidylglycerol (PG). The fourth and final reaction of CL biosynthesis is catalyzed by cardiolipin synthase (CRLS1), which actually uses two phospholipid substrates, PG and CDPDG, to form so-called “nascent” CL (CLn).
Cardiolipin remodeling: Following its synthesis, nascent CL undergoes a process known as remodeling, acquiring a new set of fatty acids (acyl chains). Nascent CL is first deacylated to monolysocardiolipin (MLCL); in yeast, the enzyme has been identified as CLD1, although in mammals, the calcium-independent phospholipase A2 (iPLA2) specifically responsible for this step has not been definitively identified.218,219 Tafazzin is the best-known and most well-characterized remodeling enzyme, a transacylase that transfers an acyl chain to MLCL from a donor lipid, forming “mature” CL (CLm); the most dominant mature CL species in the mammalian heart is tetralinoleoyl cardiolipin (TLCL). The most recent data localize tafazzin to the matrix side of the inner mitochondrial membrane220, although previous experiments had indicated localization to the intermembrane space (IMS)-facing leaflet of the IMM.221,222 Two additional postulated remodeling enzymes are ALCAT1 and MLCLAT1, both acyl-CoA-dependent lysocardiolipin acyltransferases. ALCAT1 has been localized to the ER, specifically the mitochondrial-associated membrane (MAM) space – a contact site between the ER and the mitochondrion, where lipids are exchanged. ALCAT1 catalyzes acylation of lysocardiolipin back to CL in vitro19, but has also been implicated in the production of “abnormal” CL (CLabnormal) with deficiency of TLCL in vivo (for a review, see Zhang and Shi223); moreover, this enzyme’s primary role is the remodeling of phosphoinositol rather than CL in vivo.224,225 Thus, the in vivo physiological role of ALCAT1 in CL remodeling is unclear. MLCLAT1 was first identified to acylate MLCL to CL20, and is identical to the α-subunit of trifunctional protein (αTFP, also known as HADHA) without the first N-terminal 191 residues70; MLCLAT1 may be a splice variant of trifunctional protein, which itself plays key roles in fatty acid beta-oxidation.69,70 Recent evidence suggests that HADHA does not remodel MLCL to any significant extent.69 Thus, there remain questions about the exact role of MLCLAT1 in remodeling CL.
Note: The pathway depicted is for mammals but has also been well-characterized in yeast, which exhibit slight differences and also slightly different notations. For the purposes of this review, mammalian enzymes are shown.
Abbreviations: αTFP, α-subunit of mitochondrial trifunctional protein; ALCAT1, acyl-CoA:lysocardiolipin acyltransferase 1; CDPDG, CDP-diacylglycerol; CLabnormal, “abnormal” cardiolipin; CLm, mature (remodeled) cardiolipin; CLn, nascent (non-remodeled) cardiolipin; CLD1/iPLA2, cardiolipin-specific deacylase / calcium-independent phospholipase A2; CRLS1, cardiolipin synthase; ER, endoplasmic reticulum; IMM, inner mitochondrial membrane; IMS, intermembrane space; MAM, mitochondrial-associated membrane (of the endoplasmic reticulum); MLCL, monolysocardiolipin; MLCLAT1, monolysocardiolipin acyltransferase 1; OMM, out mitochondrial membrane; PA, phosphatidic acid; PG, phosphatidylglycerol; PLA2, phospholipase A2; PGP, phosphatidylglycerophosphate; PTPMT1, protein tyrosine phosphatase mitochondrial 1; TAMM41, TAM41 mitochondrial translocator assembly and maintenance homolog (phosphatidate cytidylyltransferase, mitochondrial); TLCL, tetralinoleoyl cardiolipin. (Created with BioRender.com)
Cardiolipin Profiles Change During Normal Development
Phospholipid biochemical profiles, including CL species and profiles, change during normal development. In this review, we consider not only metazoan “development” (e.g., organogenesis) but also ongoing maturation, such as that which occurs in the early postnatal period in mammals. As recently reviewed by Alvarez-Dominguez and Melton24, how specialized cells “mature” is important for cell and developmental biology, and this should be considered a dynamic continuum of adaptive phenotypic changes that involve form, gene circuitry, interconnectivity and function, rhythms, and proliferation. Alterations in this developmental trajectory and maturation can potentially lead to developmental abnormalities. Although CL profiles continue to mature even during early postnatal development25, in this section we will focus primarily on prenatal development and review the data on the normal developmental trajectory of CL.
In a study of normal myocardial development in chick embryos, there were stage-dependent increases in CL levels, as well as phosphatidylethanolamine (PE) and phosphatidylcholine (PC) plasmalogens; CL was present in only minute amounts in the yolk sac membranes, however, demonstrating tissue-specificity of its distribution.26 In addition to the embryo proper, the yolk sac in vertebrates plays important roles in hematopoiesis, germ cell development, and nutrition supply.27 Meng and colleagues studied chick yolk sac lipidomics during embryogenesis and determined several changes in both amount and CL composition.27 However, it is not clear whether the changes in CL abundance could simply have been attributed to changes in the abundance of mitochondria. Moreover, the “yolk sac” in this study included the yolk sac membrane as well as the yolk sac contents, making it difficult to make any conclusions about CL in the embryo proper. Nevertheless, both studies suggest temporal- and tissue-specificity of CL changes during vertebrate development.
Chen and colleagues recently published quantitative lipidomic data on CL profiles in the mouse heart at different stages, during both embryonic development and adulthood.28 They found CL in embryonic hearts exhibited more diverse acyl chain compositions, while the predominant form of CL in the adult heart was tetralinoleoyl CL. CL, monolysocardiolipin (MLCL), and dilysocardiolipin (DLCL) all showed minor changes between embryonic day (E)11.5 and E15.5 but dramatic changes when compared to the 2- and 10-month-old postnatal adult mouse. The investigators suggested that pathways involved in CL biosynthesis and metabolism may have different functions between embryonic and adult hearts.28 However, we feel it is more likely that the function of the pathways remains the same, namely to synthesize and degrade CL, but different CL compositions are required because of functional differences between embryonic mitochondria and adult mitochondria.
Cheng and colleagues examined the potential role of temporal changes in CL content and/or its molecular composition during the development of the mammalian brain.29 During the perinatal period in mammalian species, active neuronal remodeling occurs in the cortex via apoptosis. The investigators first used a shotgun lipidomics approach to analyze lipid extracts from mouse cortex, as well as from human, rabbit, and rat brains; they revealed a highly diversified profile of CL molecular species across all examined mammals. Total CL levels in mouse cortex were then found to be significantly increased during development in utero, transiently decreased at birth, and then increased in the postnatal period. To determine the mechanisms underlying the diversification of the CL profile, as well as the temporal variations in CL content, the investigators also analyzed the expression levels of mRNAs encoding CRLS1, phospholipases A2, and tafazzin. They found that all these enzymes, which are critical to the biosynthesis and remodeling of CL, were abundantly expressed during the embryonic period and changes in their mRNA expression levels paralleled the age-dependent alterations in CL composition. Finally, they demonstrated that the CL profile remained highly diversified throughout adult life yet was distinct from that present during embryonic development. Their results suggest that changes in the CL profile during the perinatal period are temporally correlated to the alterations in neuronal remodeling and apoptosis that occur immediately after birth.29
In another study, Keilhoff and colleagues investigated the role of CL in neuronal differentiation using a model cell line, NSC-34.30 NSC-34 cells are produced by the fusion of neuroblastoma cells and primary spinal cord motor neurons. The investigators demonstrated a correlation between the content of distinct CL species and the differentiation status of NSC-34 cells. In fact, CL composition manipulation, via fatty acid supplementation, stimulated cell differentiation in the NSC-34 cells, suggesting a link between CL and normal neuronal cell differentiation.30 However, it should be noted that fatty acid supplementation changes the fatty acid composition of all lipids not just CL, and therefore has multiple other effects, which would need to be investigated to isolate CL’s specific roles. Nevertheless, we will see that the potential roles played by CL in cellular differentiation will be a recurrent theme.
Our knowledge of CL’s developmental trajectories and particularly the developmental biology of tafazzin31, is far from complete, but there is clear evidence of dynamic changes in gene expression of the cardiolipin biosynthetic enzymes throughout development and in different tissues (Figure 3). One unanswered question is whether the changes in the amounts of CL track with mitochondrial mass or are entirely independent. Still, limited available data indicate that CL changes during development, both in amount and in its acyl chain composition and diversity of species (Figure 4). The CL profiles are also tissue-specific. In vertebrate and mammalian models, the prenatal CL profiles are dramatically different from the mature, adult organism. These developmental trajectories strongly suggest important and perhaps changing, roles for CL throughout development.
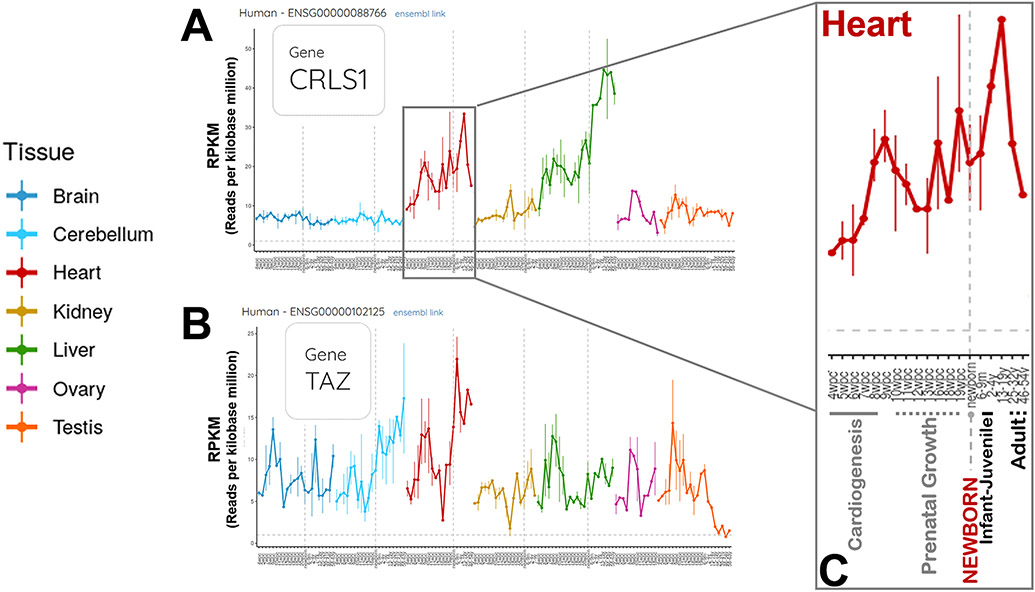
FIGURE 3. Gene expression profiles across developmental stages for human cardiolipin synthase and tafazzin. (https://apps.kaessmannlab.org/evodevoapp/, accessed 02/24/2022).
A & B) Panel A shows data for cardiolipin synthase (CRLS1). Panel B shows data for tafazzin (TAZ, now known as TAFAZZIN). The human developmental stages include 4 weeks post-conception through adulthood. Panels A & B illustrate key features of these two key enzymes in the cardiolipin biosynthetic pathway: Gene expression profiles that are both stage- and tissue-dependent.
C) The developmental changes in CRLS1 expression in the human heart are enlarged in Panel C, to illustrate more clearly the time scale during prenatal development and postnatal maturation. Major developmental milestones are shown: Cardiogenesis, prenatal growth, newborn, infant-juvenile, and adult.
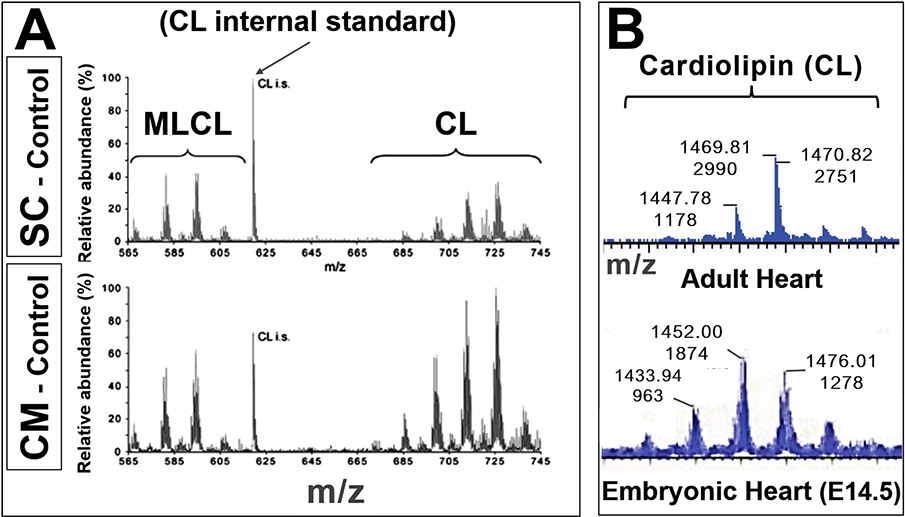
FIGURE 4. Cardiolipin species change during different developmental stages.
A) Cardiolipin (CL) and monolysocardiolipin (MLCL) in mouse embryonic stem cells (SC) and differentiated cardiomyocytes (CM). Lipid extracts of control (wildtype) cells were analyzed by mass spectrometry: Mass spectra of CL and MLCL are shown. The specific CL peaks at different m/z demonstrate differences in CL species between undifferentiated (SC) and differentiated (CM) cells, not simply a shift in MLCL:CL ratios. (Adapted from Acehan et al.64, with permission)
B) Mass spectra from wildtype mouse hearts: Adult (top) and E14.5 embryo (bottom: x-axes, or m/z axes, are aligned). The cardiolipin profiles (shifts in m/z [top numbers] and peak heights [bottom numbers]) indicate different cardiolipin species at different developmental stages. (Unpublished data, Phoon and Schlame labs)
These data illustrate the power of mass spectrometry to determine developmental lipidomics.
Disruption of Cardiolipin Synthesis Alters Developmental Trajectories
Disturbances at any of the steps along their complex biosynthetic pathway can lead to CL abnormalities. Because the CL biosynthetic pathway is conserved across different species, different model organisms have been utilized to study the consequences of CL deficiency and to better understand CL’s role in various functions. Major clues to the critical importance of CL in normal embryogenesis and its roles in cellular differentiation come from CL-deficient cellular and animal models that lead to lethality at various stages (knockouts of TAMM41, PGS1, PTPMT1, CRLS1, and TAFAZZIN). The following subsections take a closer look at disruptions in the CL biosynthetic pathway and the impact that the resulting CL deficiency has on normal development.
TAMM41
Residing at the inner mitochondrial membrane, TAMM41 encodes a protein with cytidine diphosphate-diacylglycerol synthase activity, an essential early step in the biosynthesis of PG and CL.32 TAMM41 appears to be required for heart valve differentiation by regulating the PINK1-PARK2-dependent mitophagy pathway; moreover, a genetic screen suggested an association of TAMM41 heterozygous variants with loss of full-length transcripts in the congenital heart defect known as atrioventricular septal defect.33 Very recently, Thompson and colleagues reported three unrelated individuals harboring biallelic variants in TAMM41, with clinical features of mitochondrial disease, including lethargy at birth, hypotonia, developmental delay, myopathy, and ptosis.34 Skeletal muscle samples from two of these individuals showed severe loss of subunits of complexes I-IV, a decrease in fully assembled OXPHOS complexes I-V, and decreased TAMM41 protein levels. CL levels were also significantly decreased in the skeletal muscle of these affected individuals. Interestingly, unlike families with pathogenic TAFAZZIN variants (Barth syndrome) or CRLS1 variants (see below), individuals with TAMM41 deficiency have shown no evidence of cardiomyopathy. Investigators noted the need for further research to clarify why skeletal muscle is severely affected but cardiac muscle is not, despite both being striated muscle.34
We caution that TAMM41 is several enzymatic steps upstream from the final steps in CL biosynthesis; therefore, there are almost certainly metabolic derangements in addition to cardiolipin deficiency. Moreover, there is an alternate pathway that contributes to CL synthesis through which CDPDG may also be transported from the ER membranes to the mitochondria (see review35); we speculate this alternate pathway is stage- or tissue-dependent and may sidestep TAMM41, which may then explain differential effects on skeletal and cardiac muscle.
PGS1
PGS1 is a phosphatidylglycerol phosphate synthase that catalyzes the first committed step in the CL biosynthetic pathway by converting CDP-DG to phosphatidylglycerol phosphate (PGP).22,36 While there are no published data on the roles of PGS1 in vertebrate development, the loss of PGS1 has been shown to adversely affect embryonic development in Arabidopsis. However, we acknowledge that PG is a major lipid in plants, not just an intermediate on the CL pathway: PG biosynthesis is essential for the development of embryos and normal membrane structures of chloroplasts and mitochondria.37 In mammalian development, information from online databases such as Evo-Devo Mammalian Organs (https://apps.kaessmannlab.org/evodevoapp/) and the International Mouse Phenotyping Consortium (https://www.mousephenotype.org/data/genes/MGI:1921701) suggest important roles of PGS1 in normal embryogenesis since PGS1 expression changes in a tissue-specific manner (data not shown) and loss of PGS1 results in complete pre-weaning lethality (i.e., no homozygote PGS1−/− pups). However, embryonic viability has not yet been tested.
PTPMT1
PTPMT1 is a protein tyrosine phosphatase localized to the mitochondrion that dephosphorylates the glycerophospholipid PGP and is therefore essential for mitochondrial function by regulating CL biosynthesis.38,39 Zhang and colleagues determined that all Ptpmt1 homozygotes died before the E8.5 gestational stage.39 To further investigate the physiological functions of Ptpmt1 during development, these researchers generated Cre-treated KO mouse embryonic fibroblasts (MEFs). KO MEFs were found to have a proliferation rate reduced by 25% compared to the control cells and overall, had fewer and smaller colonies. The KO MEFs also had a marked decrease in CL content, alterations in both the content and the acyl chain composition of CL, and distortion and deficiency of mitochondrial cristae. These results emphasize the role of the Ptpmt1 gene during embryonic development and suggest that PTPMT1 might be required for the increased mitochondrial demand that occurs during organogenesis in post-implantation embryos.39
To circumvent early embryonic lethality and to investigate specific roles in heart development, Chen and colleagues studied cardiac-specific deletion of PTPMT1 in mice, using a cTnT-Cre driver that constitutively expresses Cre DNA recombinase in cardiomyocytes starting at E7.5.28 While pre-organogenesis lethality was avoided, the mouse embryos died between E16.5 and E18.5, with cardiac morphological changes beginning around E12.5 following reduced cardiac cell proliferation in the compact myocardial zone at E11.5. Both respiration and expression of respiratory complex proteins were impaired at E11.5 and E12.5.28 Similarly, Zheng and colleagues investigated the effects of neural cell-specific deletion of PTPMT1 in mice, using a Nestin-Cre driver that constitutively expresses Cre DNA recombinase in neural precursor cells starting at E10.5.40 In these mice, cerebellar development was blocked and cerebral development was compromised, and all mice died by P12. Additional timed knockout experiments using a tamoxifen-inducible model verified that the role of PTPMT1 in cerebellar development is developmental stage-specific and that defective Bergmann glia development is responsible for the block of cerebellar development in PTPMT1 knockout (PTPMT1fl/fl/Nestin-Cre+) mice. The loss of PTPMT1 decreased mitochondrial aerobic metabolism by limiting utilization of pyruvate that affected only neural precursor or stem cells (not progenitor or mature cells), leading to cell cycle arrest through activation of the AMPK-p19/p21 pathway.40 However, CL itself was not studied.
In addition to vertebrates, PTPMT1 also plays a crucial role in early silkworm development.41 PTPMT1 KO silkworms demonstrate embryonic lethality, developmental arrest, and third instar lethality. Some PTPMT1 KO silkworms lack total enzyme activity, while others lack mitochondrial translocation signals. PTPMT1 has also been implicated in the biosynthesis or secretion of juvenile hormone, an insect hormone important in development, reproduction, and polyphenism.41
PTPMT1 may also play a role during T cell differentiation. Previous studies have shown that changes in mitochondrial shape, cristae morphology, and function impact CD8+ T cell activation, differentiation, and functional memory T cell development (see review42). Using T cells deficient for PTPMT1, Corrado et al.43 found that de novo PTPMT1-dependent CL synthesis is necessary to increase mitochondrial metabolic capacity and allow activation-induced cell death resistance during memory T cell differentiation. Importantly, it has been previously shown that induction of activation-induced cell death, or autophagy, sustains memory T cell development, suggesting that CL synthesis supports memory T cell development.43 While these studies do not directly investigate roles of CL during development, they do suggest that CL plays important roles in cellular differentiation.
We note that PTPMT1 is still one enzymatic step upstream from the final step in CL synthesis. And as a protein tyrosine phosphatase, PTPMT1 may dephosphorylate other mitochondrial signaling molecules.44 Therefore, it is not known if additional metabolic consequences of PTPMT1 ablation, not just CL deficiency, contribute to abnormal development and early lethality.
CRLS1
Cardiolipin synthase (CRLS1) catalyzes the fourth and final reaction of CL biosynthesis by using two phospholipid substrates, PG and CDP-DG, to form CL.45 Kasahara and colleagues found that deletion of the CRLS1 gene in mice led to early embryonic lethality, particularly at the peri-implantation stage.46 The investigators postulated that CRLS1 deletion interfered with cell division. To bypass the problems specific to cell division and neuronal proliferation during development, they generated postmitotic state-specific and neuron-specific CRLS1 conditional knockout (cKO) mice and found that disruption of the CRLS1 gene reduced CL levels and caused abnormalities in inner membrane structures of surviving hippocampal neurons, decreased respiratory supercomplex assembly in the hippocampus and cerebral cortex, and altered mitochondrial calcium dynamics.46
Very recently, deleterious variants in CRLS1 have been shown to cause an autosomal recessive mitochondrial disease, presenting as a severe infantile encephalopathy with multisystemic involvement.47 Biallelic CRLS1 variants were described in 4 patients with severe developmental anomalies. Three affected individuals had a similar infantile presentation of progressive encephalopathy, bull’s eye maculopathy, auditory neuropathy, diabetes insipidus, autonomic instability, cardiac defects, and early death. The fourth affected individual presented with chronic encephalopathy with neurodevelopmental regression, congenital nystagmus with decreased vision, sensorineural hearing loss, failure to thrive and acquired microcephaly. CRLS1 variants resulted in impaired CL biosynthesis, altered CL acyl-chain profile, abnormal cristae morphology, reduction in complex IV-stimulated respiration and a compensatory increase in OXPHOS protein levels and mtDNA copy number, and concerted endoplasmic reticulum and mitochondrial stress responses.47 This is the first time human developmental abnormalities have been associated with CRLS1.
TAFAZZIN
As a phospholipid-lysophospholipid transacylase, tafazzin plays a crucial role in CL remodeling by catalyzing both the removal and re-attachment of fatty acids.48,49 Tafazzin loss-of-function leads to decreased amounts of CL and increased amounts of MLCL, with loss of remodeling of CL and abnormal acyl chain composition.50,51 Barth syndrome is a rare X-linked mitochondrial disease that results from mutated tafazzin and leads to CL deficiency.50,52-56 Patients with Barth syndrome most commonly display cardiomyopathy, skeletal myopathy, growth retardation in early life (with later catch-up growth), and neutropenia57,58; the early clinical manifestations, including the lesser-known neurobiological manifestations59, suggest an abnormal developmental trajectory. In one study, Wang et al. developed iPSC-derived cardiomyocytes harboring tafazzin mutations from patients with Barth syndrome and defined metabolic, structural, and functional abnormalities. Such abnormalities included defects in sarcomerogenesis, which could be mitigated with ROS scavengers; thus, cardiolipin’s role in mediating ROS was implicated in tissue maturation.60 We note that any study employing iPSC-derived cardiomyocytes is essentially investigating a “fetal” phenotype and therefore likely looking at some aspect of cardiomyocyte development.
In a morphant zebrafish model, Khuchua and colleagues found that blocking mRNA translation or expression of tafazzin in the zebrafish resulted in severe developmental abnormalities, with the heart and tail as the most affected structures.61 These tafazzin morphant embryos developed marked edema with large pericardial effusions, dysmorphic and slowly beating hearts, shortened curved tails, and absent blood circulation. Importantly, the investigators also demonstrated that tafazzin is ubiquitously expressed in early stages of development (10 to 30 hours post-fertilization), suggesting it is essential for normal zebrafish development of multiple tissues and organs, not just of skeletal and cardiac muscle.61 Phoon and colleagues utilized an inducible, short hairpin RNA tafazzin-knockdown mouse model, knocking down tafazzin at different stages.62 In this model, several lines of evidence pointed to important developmental roles of tafazzin and CL function. First, significant embryonic lethality suggested a crucial role for tafazzin during development. Second, tafazzin knockdown could recapitulate noncompaction cardiomyopathy only when tafazzin was knocked down early, suggesting an important developmental window for tafazzin function (as also highlighted by Chin and Conway31). Moreover, abnormalities in cardiomyocyte structure and mitochondrial ultrastructure in the embryonic heart, leading to impaired myocardial development, indicated a disturbance in normal development of the cardiac muscle and sarcomerogenesis.62 However, it is worth noting the possible confounding effect of doxycycline in the tafazzin-KD mouse model of this study.63
The effects of tafazzin ablation – with consequent CL deficiency – also impact cellular differentiation. To compare the effects of tafazzin deletion in undifferentiated versus differentiated cells, Acehan et al. generated tafazzin-deficient mouse embryonic stem cells, which were then differentiated into cardiomyocytes. They found significant alterations of the mitochondrial ultrastructure in specific differentiated tissues but not in embryonic stem cells. These data suggest that normal concentration and composition of CL is essential only in certain types of mitochondria. More specifically, mitochondria with high cristae density, such as in skeletal and heart muscle tissue, are those most susceptible to tafazzin deletion.64 In fact, myogenic differentiation has been shown to be accompanied by major changes in mitochondrial metabolism, mitochondrial energy production, and mitochondria-mediated activation of apoptotic pathways.65 Given this prominent role of mitochondria in myogenic differentiation, Lou and colleagues developed a tafazzin-KO C2C12 myocyte cell line to determine the effect of defective CL remodeling on myogenic determination.66 C2C12 is a myoblast cell line with a high metabolic demand, like skeletal muscle cells. Strikingly, tafazzin-KO myoblast cells showed impaired phenotypic differentiation into myotubes, as well as defects in the metabolic transition from glycolysis to mitochondrial oxidative metabolism. Moreover, in contrast to WT cells, tafazzin-KO cells exhibited an increased reliance on glycolysis and decreased flux to mitochondrial oxidative metabolism following the switch to myocyte differentiation media. These data demonstrate that the loss of CL leads to defective skeletal myocyte differentiation.66 Finally, Seneviratne et al.67 assessed the effects of tafazzin knockdown on acute myeloid leukemia (AML) stem and progenitors and differentiation. Their study demonstrated that tafazzin knockdown reduces AML propagation by reducing AML stemness both in vivo and in vitro. Interestingly, however, reduction of the stemness of leukemia cells was attributed to an increase in phosphatidylserine levels and activation of toll-like receptor signaling, in contrast to the expected disruption of mitochondrial structure and impairment of OXPHOS by tafazzin knockdown. Additionally, tafazzin knockdown increased the differentiation of AML cells, demonstrating that tafazzin-mediated phospholipid production regulates both AML stemness and differentiation.67
Taken together, the published data strongly support the notion that tafazzin plays a crucial role in tissue development and differentiation.
Additional evidence implicating CL in development
Additional acyltransferases involved in CL-remodeling may also play roles during developmental processes. Acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) (alternatively known as lysocardiolipin acyltransferase or lycat) encodes an acyl-CoA: lysocardiolipin acyltransferase 1 that possesses both acyl-CoA: monolysocardiolipin acyltransferase and acyl-CoA: dilysocardiolipin acyltransferase activities.19 The lycat gene in zebrafish has been shown to play a necessary role in the generation of both endothelial and hematopoietic lineages, and the mouse orthologue of this gene plays a crucial role in the early specification of hematopoietic and endothelial cells. Overexpression of the lycat gene increases both hematopoietic and endothelial gene expression and increases the formation of both cell types. In fact, siRNA-mediated gene knockdown of lycat decreased hematopoietic and endothelial gene expression during embryonic cell differentiation into embryoid bodies, suggesting that this lycat gene is important for endothelial and hematopoietic development in mouse embryonic stem cells.68 The link to cardiolipin, however, was not clear in this study.
Mutations in the alpha-subunit of mitochondrial trifunctional protein (TFPα/HADHA) in iPSC-derived human cardiomyocytes lead to deficiency in tetralinoleoyl cardiolipin, similar to the biochemical phenotype of Barth syndrome69; TFPα/HADHA is felt to be, essentially, MLCLAT1.69-72 However, we are not aware of any human disease stemming from mutations in MLCLAT1 specifically that do not also affect mitochondrial trifunctional protein.
Finally, a Barth-like, autosomal recessive syndrome characterized by dilated cardiomyopathy with ataxia (DCMA) has been described in the Canadian Dariusleut Hutterite population, now attributed to homozygous variants in DNAJC19.73 While not an acyltransferase or directly involved in cardiolipin synthesis, DNAJC19 is a mitochondrial co-chaperone that forms a complex with prohibitins (PHB) to regulate CL remodeling.74 This syndrome affects infants and young children, with early-onset cardiomyopathy, cerebellar ataxia, and developmental delays. Investigators have observed impaired cell growth and defective cristae morphogenesis with the loss of PHB/DNAJC19 complexes, affecting CL acylation and leading to the accumulation of CL species with altered acyl chains.74 However, the precise function(s) of DNAJC19 are not well-understood. Recently, Rohani et al. studied peripheral blood mononuclear cells from two patients with DCMA and controls, quantifying multiple CL species in iPSCs and iPSC-derived cardiomyocytes (iPSC-CMs) using mass spectrometry.75 Several small but statistically significant differences were seen between the undifferentiated control and patient iPSCs. Fewer CL species were detectable in the iPSCs compared with the iPSC-CMs, and, importantly, the mature form, CL(18:2)4, was not present in significant amounts (<1%) in the undifferentiated iPSCs. Thus, there was greater CL diversity in the iPSC-CMs. In the iPSCs, CL(18:2)(18:1)3 predominated, comprising approximately 40% of the total CL. In the differentiated iPSC-CMs, no consistent and significant differences between the patients with DCMA and the control strain were seen. "Mature" (tetralinoleoyl) CL contributed between 6.5% and 14.4% of the total—a lower proportion than would be expected in mature cardiac tissue. Patient iPSC-CMs demonstrated highly fragmented and abnormally shaped mitochondria associated with an imbalanced isoform ratio of the mitochondrial protein OPA1, an important regulator of mitochondrial fusion. These abnormalities were reversible by incubation with SS-31, which is purported to interact with and stabilize CL.75
As summarized by Chin and Conway31, invertebrates appear to be far less susceptible to loss of CL. In Drosophila, inactivation of either CRLS176 or tafazzin77 failed to lead to any developmental consequences, including embryonic lethality. In C. elegans, inactivation of CRLS1 utilizing two different deletion alleles of CRLS1 led only to abnormal mitochondrial morphology (including disrupted cristae) and function, with reduced germ cell proliferation.78 Thus, it appears CL becomes increasingly important as organisms move to a higher order.
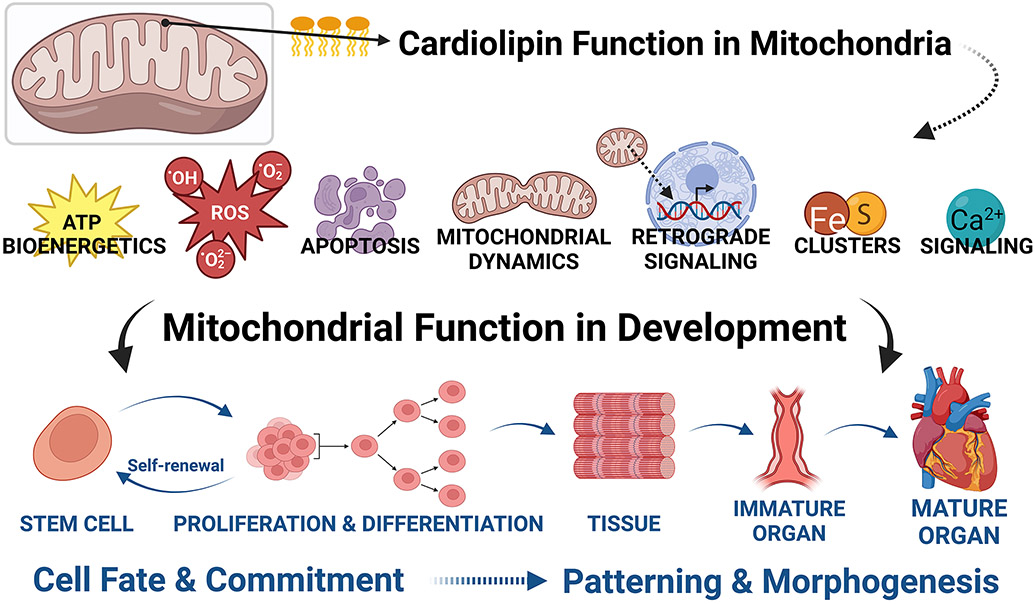
Cardiolipin Functions Can Be Linked to Mitochondria and Developmental Processes
CL’s known roles in normal mitochondrial functioning align with what we know about the overall importance of normal mitochondrial functioning in organismal development. In this section, we will attempt to connect known functions of CL to proposed roles in developmental pathways and processes, even when (as of yet) there are no direct data linking the specific CL function to the specific developmental process.
Mitochondria: General Statements
Development, organogenesis, and maturation require the tightly spatial and temporal coordination of many different processes. Such processes include regulation of gene expression, translation, and post-translational modifications; cellular fate specification and differentiation from stem cells; multi-level control of cell lineage progression; intra- and intercellular signaling events; cellular proliferation; programmed cell death (apoptosis); cell migration; regulation of cell shape and polarity; cellular coalescence and assembly into tissues; cellular and tissue responses to environmental cues, including paracrine and exocrine signals, and extrinsic physical forces; and ultimately, patterning.79-87 We now know that normal mitochondrial function is essential to many, if not all, of these processes.
Teleologically, mitochondria are well-suited to play important roles in cellular differentiation and organismal development. As extremely “plastic” organelles88, their form and function are tissue-specific and may even vary within cells. During the development of a mammalian embryo, changes occur in both mitochondrial structure89,90 and function.91-95 Clues to the potentially critical roles of mitochondria during prenatal development and embryogenesis also come from numerous studies of their critical roles in the cellular differentiation and fate of stem cells (see review96). More than “just” energy generators, mitochondria are active metabolic signaling organelles, and the mechanisms by which mitochondria control stem cell fate involve reactive oxygen species (ROS), calcium signaling, tricarboxylic acid cycle metabolites, the NAD+/NADH ratio, pyruvate metabolism, and mitochondrial dynamics (i.e., fission and fusion). In other words, mitochondrial metabolic signaling can control transcriptional networks, independently of energy production.96-98 Thus, mitochondria act as important “hubs” in coordinating cell differentiation.99 The reader is directed to selected reviews on cardiogenesis100, hematopoiesis96,101 and cellular-mediated immunity102, myogenesis103-105, osteogenesis, adipogenesis106-108, and various other mesenchymal tissues106,109, intestinal stem cells110,111, germ cell stem cells112, and adult neurogenesis113, as broad examples.
Bioenergetics
During vertebrate development, large amounts of energy are required for cell growth, division, and differentiation. Metabolism encompasses all the biochemical reactions that are involved in energy conversion pathways in an organism, including glycolysis, OXPHOS, the Krebs cycle, and fatty acid oxidation. The rapidly changing anabolic and catabolic demands in the developing embryo are reflected in an evolving metabolic profile, with an embryonic energy metabolism that is tuned to the cell’s state of differentiation, proliferation, and specific energy requirements including changes in oxygen availability. Metabolic pathways are thus spatiotemporally regulated and lineage-specific.114,115 In general, the transition from glycolysis to OXPHOS and fatty acid oxidation signals terminal differentiation and termination of the proliferative phase.24,116-118 This lineage-specific metabolic rewiring has been implicated in cellular differentiation and cell fate in many tissues.103,106,119 As but two examples, mitochondrial functioning is required for zebrafish somite maturation and embryonic development120, while dramatic changes are required in mitochondria to activate complex I and for the formation of respiratory chain supercomplexes during the increase in OXPHOS.121
CL is most closely identified with its roles in bioenergetics and specifically, OXPHOS. At the most fundamental level, CL is required for the mitochondrial inner membrane to achieve proper protein density and function, including membrane docking of mitochondrial ribosomes and protein synthesis122,123, protein “crowding” in the mitochondrial inner membrane124, and stabilization of proteins for proper activities, including the assembly of respiratory chain supercomplexes.11-15,125 CL deficiency leads to disruption of supercomplexes’ stability and results in overall decreased energy production during aerobic metabolism.3,125-127
ROS
Under normal physiological conditions, ROS are natural byproducts of aerobic metabolism (OXPHOS) in mitochondria and play numerous roles in maintaining cellular homeostasis by modulating cell death, cell survival, and cell signaling.3,128,129 Redox signaling is fundamental to developmental processes, including proliferation, differentiation, apoptosis, and left-right asymmetry.130,131 During organogenesis, stem cells’ self-renewal and fate decisions are governed and regulated by ROS levels.99 In many adult stem cells, including spermatogonial, airway basal, hematopoietic, keratinocyte, mesenchymal, skeletal muscle and neural stem cells, a low level of mitochondrial ROS is required for self-renewal and quiescence, while a moderate physiological increase in ROS is necessary for normal stem cell proliferation and differentiation (see reviews96,98,106,119). In late organogenesis and particularly in cardiogenesis, it has been shown that cardiomyocytes at different embryonic stages display distinct levels of ROS, further implicating a regulatory role for ROS during developmental phases.65 Cardiomyocyte differentiation was shown to be inhibited and enhanced with oxidant and antioxidant treatment, respectively, suggesting that redox signaling pathways regulate differentiation in these cells.65 ROS production is spatially and temporally regulated by the cell, and indeed, ROS levels have been shown to differ at specific events during embryonic development, starting as early as fertilization and including such events as embryonic cleavage and cell cycling.132,133 Possible active roles by which ROS regulates these developmental processes include regulation of protein phosphorylation, regulation of transcription factor activity, and transcriptional activation of specific genes involved in oxidative stress.130,131,134 Mitochondrial dysfunction may lead to excessive ROS and oxidative stress, which may, in turn, lead to developmental consequences. Disruptions to ROS levels can affect the differentiation of stem cells; decreased ROS levels can lead to undesired increased quiescence and defects in differentiation, while aberrantly high mitochondrial ROS production can trigger stem cell exhaustion.96,131
Data from several CL-deficient model systems have demonstrated that reduced CL leads to elevated mitochondrial ROS60,127,135-138, although other studies have questioned this association in models of Barth syndrome.139,140 Abnormally increased mitochondrial ROS causes a disruption in the balance between ROS and antioxidant enzymes, leading to mitochondrial ultrastructural changes in mtDNA, proteins, and lipids, and ultimately resulting in mitochondrial dysfunction (see reviews3,141). ROS may be intertwined with other mitochondrial processes, including mitochondrial dynamics and retrograde signaling (see elsewhere). Khacho and colleagues found that mitochondrial dynamics could dictate neural stem cell fate by driving a physiological ROS-mediated process that triggers a transcriptional program to suppress self-renewal and promote differentiation via NRF2 (nuclear factor erythroid 2-related factor 2)-mediated retrograde signaling.142 Given the potential role of CL in the regulation of ROS levels, it can be hypothesized that CL may also be at play in this retrograde pathway.
Apoptosis
Apoptosis is an organized and controlled process of cell death that occurs in organisms to control growth and allow for normal development.29,143,144 During development, apoptosis is necessary to counterbalance cell proliferation and regulate tissue growth. The central role of mitochondria in cell apoptosis also contributes to embryonic size, shape, and growth, as well as culling abnormal cells from this rapidly developing organism145 and tissue remodeling.144,146
In the overall mitochondrial-mediated apoptotic pathway, stress conditions lead to the translocation of normal CL to the outer mitochondrial membrane. Once translocated, CL recruits intrinsic apoptotic factors that contribute to the permeabilization of the outer mitochondrial membrane. Externalized CL interacts with cytochrome c, a mitochondrial protein, to form a cytochrome c/CL complex with peroxidase activity that can target and oxidize CL; a number of oxygenated CL species undergo phospholipase A2-catalyzed hydrolysis to generate multiple oxygenated fatty acids, including well-known lipid mediators, involved in apoptosis and other biological functions.147 The lower affinity of cytochrome c toward oxidized CL results in its release from mitochondria to the cytosol, initiating the apoptotic cascade pathway (see reviews3,148-150). Using a shotgun lipidomics approach, Cheng and colleagues found that changes in brain CL content during the perinatal period were temporally correlated to the amount of apoptosis that occurred during this period of development.29 Changes in CL composition, particularly the increased peroxidation of CL and a reduction in CL levels, can therefore lead to accelerated and deregulated levels of apoptosis.141,151-155
Mitochondrial Dynamics
Mitochondrial dynamics play an essential role during embryonic development. Mitochondrial dynamics refers to coordinated cycles of fusion/fission events, which contribute to the dynamic nature of mitochondria including the movement of mitochondria along the cytoskeleton and the regulation of mitochondrial morphology and distribution.156,157 Fission is the process by which mitochondria divide to form two separate mitochondrial organelles, while fusion is the physical content mixing between two originally distinct mitochondria.158 The importance of mitochondrial fusion during embryogenesis was demonstrated in murine embryonic fibroblasts (MEFs) lacking either Mfn (mitofusin)1 or Mfn2, two core proteins essential for mitochondrial fusion.159 Mfn1- and Mfn2- knockout mice display distinct types of fragmented mitochondria and die in utero at mid-gestation, while Mfn1/Mfn2 double knockout mice die earlier in development. Interestingly, MEFs from the double knockout mice can survive in culture despite the lack of mitochondrial fusion, suggesting that mitochondrial fusion is not essential for cell survival but is instead required for embryonic development and cell survival at later stages.159,160 In neural stem and progenitor cells, cell fate is also determined by mitochondrial fusion-fission dynamics.142,161 Disruption of the genes that promote mitochondrial fusion, such as Opa1 (optic atrophy 1), leads to an increase in neuronal differentiation, whereas the disruption of genes that promote mitochondrial fission, such as Drp1 (dynamin-related protein 1), leads to decreased neurogenesis.162 Finally, mitochondrial dynamics have also been implicated in the regulation of apoptosis, whereby inhibition of mitochondrial fission-mediating proteins can reduce apoptosis, while inhibition of mitochondrial fusion-mediating proteins can enhance apoptosis, both potentially disrupting normal development.163
CL is critical in mitochondrial fusion/fission events via its interaction with and recruitment of, fusion and fission core mediating proteins, including Opa1 and Drp1.164-166 CL deficiency leads to reduced levels of Opa1 and Drp1, resulting in defects in mitochondrial morphology.158,167 Moreover, changes in CL composition, mainly in the length and level of saturation of CL acyl chains, disrupt the balance between fission/fusion events.150,168,169 CL appears to be critical in modulating multiple Opa1 functions in the silkworm larva, thus regulating mitochondrial fusion, morphology, and quality control during development.168 Although specific CL remodeling by tafazzin appeared dispensable in this same silkworm model170, another group demonstrated that tafazzin deficiency in a conditional (heart-specific) tafazzin knockout mouse model led to dramatically decreased Mfn1 and Mfn2, while the long isoform of Opa1 was slightly decreased; the mitochondrial fission protein Drp1 and its phosphorylation status were unchanged.125 However, this study did not directly examine mitochondrial dynamics to confirm a cellular phenotype.125
Mitochondrial Retrograde Signaling
Development is of course guided by transcription, and retrograde signaling is one of the ways to regulate transcription. In retrograde signaling, nuclear gene expression can be modulated in response to functional changes in downstream components of a pathway, such as changes in organelles. All but 13 mitochondrial proteins are encoded by nuclear genes (nDNA, anterograde signaling), but because mitochondria also house their own genome (mtDNA), the two genomes (nDNA and mtDNA) need to coordinate with anterograde and retrograde signaling to generate the normal mitochondrial proteome.171-175 Molecules and processes that have been implicated in retrograde signaling during development include ROS (see above), HIF, and the mitochondrial unfolded protein response.
In low-oxygen, or hypoxic, environments, altered gene expression is initiated by the dimeric transcription factor, hypoxia-inducible factor (HIF).114,176 When oxygen levels are low, HIF1α is activated and upregulates the transcription of a group of genes that either promote glycolytic metabolism to reduce oxygen consumption or act to extend or modify the surrounding vasculature, including altering the composition and organization of the respiratory chain.177 While no study has directly elucidated the role that CL plays in this HIF-retrograde pathway, it has been shown that CL-deficiency in tafazzin-KO cells disrupts the transcription of HIF1α and hence leads to a significant reduction in the HIF1α protein, along with reduced expression of the hypoxia-specific isoforms of complex IV subunits of the respiratory chain.178 Thus, CL deficiency is implicated in an abnormal cascade of cellular events that include decreased HIF1α with potential disruptions in retrograde signaling.
As placental vertebrates, mammals develop in a relatively hypoxic environment. While the HIF transcriptional system is most often discussed in the context of pathological hypoxia, HIF also plays a crucial regulatory role in the normal hypoxic environment of normal mammalian development.178-180 HIF activity is required for the overall normal development of the placenta and the development of specific organ tissue, including lung181, bone and cartilage.114,182 HIF activity is also required for normal heart morphogenesis. Loss of HIF1α or HIF1β leads to arrested morphogenesis at multiple stages from the cardiac crescent stage to chamber formation (reviewed by Dunwoodie114).
When proteins fail to undergo proper protein folding and maturation in the mitochondria, the resulting stress on the organelle leads to the activation of a nuclear transcriptional program termed the mitochondrial unfolded protein response (mtUPR) that aims to increase the transcription of mitochondrial chaperones and proteases to counteract mitochondrial dysfunction.183 The mtUPR plays an important regulatory role in stem cell proliferation, cellular differentiation, aging184, oocyte and embryo development, and oocyte mitochondrial function and dynamics.185 In the early phase of mtUPR, there is a rapid remodeling of CL, resulting in a changed lipid microenvironment that helps to stabilize protein import machinery upon mtUPR induction.183
Mitochondrial Fe-S Clusters
Normal iron-sulfur (Fe-S) machinery is also required for normal mammalian embryonic viability and development.186 Fe-S clusters are important protein cofactors that can be found in the nucleus, cytosol, and mitochondria. Within mitochondria, Fe-S clusters are involved in the citric acid cycle, the electron transfer chain, fatty acid oxidation, and in lipoate and biotin biosynthesis.187 We now know that iron content and stores, including brain iron content (which includes mitochondrial iron content, stored primarily in the Fe-S clusters), change during development.188 Studies of other proteins involved in Fe-S clusters, such as NARFL189, have also demonstrated a critical role in embryonic development.
Given the roles of Fe-S clusters in electron transport chain functioning and mitochondrial OXPHOS190, it is no surprise that they also mediate the production of mitochondrial ROS and may underlie the pathogenesis of disease. In fact, in both TAZKO cells (modeling Barth syndrome) and CL-depleted yeast cells, researchers have demonstrated reduced catalytic activity of enzymes requiring Fe-S cofactors, overall defective Fe-S biogenesis and iron homeostasis, and consequent hypersensitivity to oxidative stress.191 Moreover, these TAZKO cells and CL-depleted yeast cells demonstrate decreased levels of frataxin, an essential protein of the Fe-S biogenesis pathway, and it has been shown that there is a direct interaction between CL and frataxin, which seems to suggest that the absence of this interaction in CL-deficient mitochondria may explain the defects in iron biogenesis and iron homeostasis.191
Calcium
Calcium (Ca2+) and calcium signaling pathways are important in many facets of embryonic development192-197, including the development of the nervous system, cardiac system, immune system, and the development of kidney and muscle.192,193 Mitochondria are both involved in and are affected by, intracellular and intraorganellar Ca2+ homeostasis. Two important mechanisms by which mitochondria regulate Ca2+ levels and/or use Ca2+ for cellular processes are the mitochondrial Ca2+ uniporter (MCU) and the mitochondrial permeability transition pore (mPTP), a non-specific conductance channel.
Within the mitochondria, the main role of Ca2+ is in the stimulation of OXPHOS. The MCU helps maintain overall Ca2+ homeostasis, specifically via Ca2+ uptake across the mitochondrial membrane.198-204 Recent studies have alluded to the importance of specific lipids for the function and formation of the MCU. One study showed a specific requirement of CL for MCU stability and activity, where the loss of CL triggered rapid turnover of MCU homologs and impaired Ca2+ transport.205 Moreover, tafazzin mutations, and by extension CL deficiencies, have been linked to abnormal Ca2+ handling and consequent decreased cardiomyocyte contractility.206 When Ca2+ is increased intracellularly, the mPTP opens and allows the free passage of small metabolites and ions into the mitochondria.207 The opening of the mPTP is also often accompanied by enhanced ROS production. 208,209 CL peroxidation, which occurs in the presence of high ROS levels, has been shown to participate, together with Ca2+, in the opening of the mPTP.148,210 It is thought that CL may first associate with adenine nucleotide translocator (ANT), a protein that facilitates the exchange of ADP and ATP across the mitochondrial membrane, which then allows Ca2+ to bind and activate mPTP opening.207 Although the prolonged opening of mPTP is linked to disease, the transient opening of mPTP has been implicated in both cardiac and brain development, particularly at the level of cell differentiation. In cardiomyocytes, closure of the mPTP leads to maturation of mitochondrial structure and function, decreasing ROS, increasing membrane potential and, ultimately, accelerating cell differentiation.65,211 In neuronal development, cell differentiation also appears to be mediated by the opening of the mPTP.211,212
Conclusions and Future Directions
While much of this evidence remains circumstantial, we have summarized here the available evidence for evolving and essential roles of CL during organismal development, including organogenesis and tissue maturation (Figure 5). Furthermore, normal mitochondrial functioning is known to contribute to normal developmental processes. As a key mitochondrial phospholipid, CL should prove to have even more detailed roles, from early pre-implantation embryogenesis through cellular differentiation, organogenesis, and tissue maturation. Here is a sampling of the questions we would pose to the scientific community:
FIGURE 5. Specific functions of cardiolipin are linked to developmental processes.
Cardiolipin’s roles during development include maintenance of cell stemness, and roles in cell fate and commitment (differentiation) and patterning and morphogenesis of tissues and organs. While many of these roles have not been demonstrated mechanistically, the importance of mitochondria in development via their involvement in bioenergetics, the generation and movement of reactive oxygen species, apoptosis, fission/fusion events, retrograde signaling, iron-sulfur clusters, and calcium signaling all strongly implicate direct mechanistic roles for cardiolipin. (Created with BioRender.com)
How does CL change throughout development and organismal maturation? Do the developmental changes in mitochondria (ultrastructure) lead to changes in CL, or does CL change primarily, thereby contributing to mitochondrial maturation? Data point to the requirement of the biophysical milieu for tafazzin specificity.18,213 Therefore, if the biophysical milieu determines tafazzin specificity, then the evolution of mitochondrial membrane morphology during prenatal development (especially early development) will in part determine what tafazzin does and the specifics of CL remodeling. The role(s) of additional remodeling enzymes, such as ALCAT1 and MLCLAT1, also require clarification, including during development.
Why do mitochondria need CL at each developmental stage? How do the changes in CL affect lipid bilayer functioning, and how do these, in turn, contribute to the development and maturation of different tissues? Assembly of OXPHOS complexes induces CL remodeling, which, in turn, leads to CL stabilization. Data from our laboratory suggest that protein crowding in the OXPHOS system imposes packing stress on the lipid bilayer, which is relieved by CL remodeling to form tightly packed lipid-protein complexes.214 Because the cristae are rather poorly developed in the early embryo, the requirement for relief of protein crowding stress may not be apparent; we do not know yet the developmental trajectory of protein density in mitochondrial membranes.
How does the relatively hypoxic environment of the mammalian fetus affect CL and mitochondrial function? Mammalian development occurs in the low oxygen environment of the uterus. Organogenesis is affected by hypoxia-inducible factor (HIF) transcription factors that are sensors of hypoxia215,216, and we know that mitochondrial biology, CL, and HIF-1α are linked.178 Changes in oxygen levels in the developing mammalian organism (such as that which occurs with some forms of congenital heart defects or with placental insufficiency, for example) may therefore influence transcriptional regulation via CL, or conversely, abnormal CL may impact HIF-1α178 and therefore organogenesis.
In which organs is CL critical to development, and how and why? Are there non-mitochondrial roles of CL during development? While CL is traditionally considered to be the “signature” phospholipid of mitochondria, recent work from our lab has revealed a novel, extramitochondrial role for CL, specifically TPCL as critical to normal spermatogenesis and acrosome formation.21,217 Thus, there may be roles for CL species not related to mitochondrial function, yet related to cellular differentiation and perhaps other developmental processes.
Finally, CL appears to assume more importance in higher-order organisms than in simple eukaryotes. Increasing systems complexity may require CL to be “fine-tuned” across the life span and different tissues. Investigations of cardiolipin biology in model systems should keep this caveat in mind. Three human mitochondrial diseases now implicate CL primarily in their pathogenesis, affecting young infants and therefore displaying developmental abnormalities: Barth syndrome, and recently-reported biallelic variants in TAMM41 and CRLS1. Thus, a deeper understanding of CL biology during organogenesis and tissue maturation in complex metazoans is likely to lead to the identification of therapeutic targets and approaches in such diseases of developmental CL deficiency.
Acknowledgments
This review was based in part on a talk by CKLP given at the conference, “Cardiolipin as Key Lipid of Mitochondria in Health and Disease,” 4th Biennial Conference, Bologna, Italy, on September 10, 2019 (Scientific Session: Role of Cardiolipin in Physiopathological States). This work was supported by the Dean’s Undergraduate Research Fund (MOV) and grants from the Barth Syndrome Foundation (CKLP, MR) and National Institutes of Health (R01GM115593, to MS).
Footnotes
Conflict of interest disclosure: The authors declare they have no conflicts of interest to disclose.
References
- 1.Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends Biochem Sci. 2012;37(1):32–41. doi: 10.1016/j.tibs.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlame M, Greenberg ML. Biosynthesis, remodeling, and turnover of mitochondrial cardiolipin. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862(1):3–7. doi: 10.1016/j.bbalip.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zegallai HM, Hatch GM. Barth syndrome: cardiolipin, cellular pathophysiology, management, and novel therapeutic targets. Mol Cell Biochem. 2021;476(3):1605–1629. doi: 10.1007/s11010-020-04021-0 [DOI] [PubMed] [Google Scholar]
- 4.Ferreira CR, Jarmusch AK, Pirro V, et al. Ambient ionisation mass spectrometry for lipid profiling and structural analysis of mammalian oocytes, preimplantation embryos and stem cells. Reprod Fertil Dev. 2015;27(4):621–637. doi: 10.1071/RD14310 [DOI] [PubMed] [Google Scholar]
- 5.Poulsen RR, McClaskey CM, Rivkees SA, Wendler CC. The Sphingosine-1-phospate receptor 1 mediates S1P action during cardiac development. BMC Dev Biol. 2011;11:37. doi: 10.1186/1471-213X-11-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukacs M, Roberts T, Chatuverdi P, Stottmann RW. Glycosylphosphatidylinositol biosynthesis and remodeling are required for neural tube closure, heart development, and cranial neural crest cell survival. elife. 2019;8:e45248. doi: 10.7554/eLife.45248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao C, Rossignol F, Vaz FM, Ferreira CR. Inherited disorders of complex lipid metabolism: A clinical review. J Inherit Metab Dis. 2021;44(4):809–825. doi: 10.1002/jimd.12369 [DOI] [PubMed] [Google Scholar]
- 8.Acoba MG, Senoo N, Claypool SM. Phospholipid ebb and flow makes mitochondria go. J Cell Biol. 2020;219(8):e202003131. doi: 10.1083/jcb.202003131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vance DE, Vance JE. Physiological consequences of disruption of mammalian phospholipid biosynthetic genes. J Lipid Res. 2009;50 Suppl(Suppl):S132–137. doi: 10.1194/jlr.R800048-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mejia EM, Hatch GM. Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr. 2016;48(2):99–112. doi: 10.1007/s10863-015-9601-4 [DOI] [PubMed] [Google Scholar]
- 11.Pfeiffer K, Gohil V, Stuart RA, et al. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278(52):52873–52880. doi: 10.1074/jbc.M308366200 [DOI] [PubMed] [Google Scholar]
- 12.Bazán S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem. 2013;288(1):401–411. doi: 10.1074/jbc.M112.425876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnarez C, Marrink SJ, Periole X. Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem Sci. 2016;7(7):4435–4443. doi: 10.1039/c5sc04664e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker CD, Basu Ball W, Pryce EN, Gohil VM. Specific requirements of nonbilayer phospholipids in mitochondrial respiratory chain function and formation. Mol Biol Cell. 2016;27(14):2161–2171. doi: 10.1091/mbc.E15-12-0865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennington ER, Sullivan EM, Fix A, et al. Proteolipid domains form in biomimetic and cardiac mitochondrial vesicles and are regulated by cardiolipin concentration but not monolyso-cardiolipin. J Biol Chem. 2018;293(41):15933–15946. doi: 10.1074/jbc.RA118.004948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennington ER, Funai K, Brown DA, Shaikh SR. The role of cardiolipin concentration and acyl chain composition on mitochondrial inner membrane molecular organization and function. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(7):1039–1052. doi: 10.1016/j.bbalip.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlame M, Xu Y. The function of tafazzin, a mitochondrial phospholipid-lysophospholipid acyltransferase. J Mol Biol. 2020;432(18):5043–5051. doi: 10.1016/j.jmb.2020.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao J, Liu Y, Lockwood J, Burn P, Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum- associated acyl-CoA:lysocardiolipin acyltrans- ferase (ALCAT1) in mouse. J Biol Chem. 2004;279:31727–31734. doi: 10.1074/jbc.M402930200 [DOI] [PubMed] [Google Scholar]
- 20.Taylor WA, Hatch GM. Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1). J Biol Chem. 2009;284(44):30360–30371. doi: 10.1074/jbc.M109.048322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren M, Xu Y, Erdjument-Bromage H, et al. Extramitochondrial cardiolipin suggests a novel function of mitochondria in spermatogenesis. J Cell Biol. 2019;218(5):1491–1502. doi: 10.1083/jcb.201808131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren M, Phoon CKL, Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 23.Bozelli JC Jr, Epand RM. Determinants of lipids acyl chain specificity: A tale of two enzymes. Biophys Chem. 2020;265:106431. doi: 10.1016/j.bpc.2020.106431 [DOI] [PubMed] [Google Scholar]
- 24.Alvarez-Dominguez JR, Melton DA. Cell maturation: Hallmarks, triggers, and manipulation. Cell. 2022; 185(2):235–249. doi: 10.1016/j.cell.2021.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiebish MA, Yang K, Sims HF, et al. Myocardial regulation of lipidomic flux by cardiolipin synthase: setting the beat for bioenergetic efficiency. J Biol Chem. 2012;287(30):25086–25097. doi: 10.1074/jbc.M112.340521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmy FM, Aikins A, Hughes J, Belfield C, Juracka A. Studies on the endogenous phospholipids of chick embryo myocardium and their in vitro hydrolysis by endogenous phospholipases during embryogenesis. Cell Biochem Funct. 2007;25(5):571–579. doi: 10.1002/cbf.1422 [DOI] [PubMed] [Google Scholar]
- 27.Meng Y, Qiu N, Mine Y, Keast R. Comparative lipidomics of chick yolk sac during the embryogenesis provides insight into understanding the development-related lipid supply. J Agric Food Chem. 2021;69(26):7467–7477. doi: 10.1021/acs.jafc.1c01728 [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Zhu S, Wang H, et al. PTPMT1 Is required for embryonic cardiac cardiolipin biosynthesis to regulate mitochondrial morphogenesis and heart development. Circulation. 2021;144(5):403–406. doi: 10.1161/CIRCULATIONAHA.121.054768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng H, Mancuso DJ, Jiang X, et al. Shotgun lipidomics reveals the temporally dependent, highly diversified cardiolipin profile in the mammalian brain: temporally coordinated postnatal diversification of cardiolipin molecular species with neuronal remodeling. Biochemistry. 2008;47(21):5869–5880. doi: 10.1021/bi7023282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keilhoff G, Mbou RP, Lucas B, Schild L. The differentiation of spinal cord motor neurons is associated with changes of the mitochondrial phospholipid cardiolipin. Neuroscience. 2019;400:169–183. doi: 10.1016/j.neuroscience.2019.01.005 [DOI] [PubMed] [Google Scholar]
- 31.Chin MT, Conway SJ. Role of Tafazzin in mitochondrial function, development and disease. J Dev Biol. 2020;8(2):10; doi: 10.3390/jdb8020010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kutik S, Rissler M, Guan XL, et al. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008;183(7):1213–1221. doi: 10.1083/jcb.200806048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang RM, Tao J, Zhan M, et al. TAMM41 is required for heart valve differentiation via regulation of PINK-PARK2 dependent mitophagy. Cell Death Differ. 2019;26(11):2430–2446. doi: 10.1038/s41418-019-0311-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson K, Bianchi L, Rastelli F, et al. Biallelic variants in TAMM41 are associated with low muscle cardiolipin levels, leading to neonatal mitochondrial disease. HGG Adv. 2022;3(2):100097. doi: 10.1016/j.xhgg.2022.100097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blunsom NJ, Cockcroft S. CDP-diacylglycerol synthases (CDS): Gateway to phosphatidylinositol and cardiolipin synthesis. Front Cell Dev Biol. 2020;8:63. doi: 10.3389/fcell.2020.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998;273(16):9829–9836. doi: 10.1074/jbc.273.16.9829 [DOI] [PubMed] [Google Scholar]
- 37.Tanoue R, Kobayashi M, Katayama K, Nagata N, Wada H. Phosphatidylglycerol biosynthesis is required for the development of embryos and normal membrane structures of chloroplasts and mitochondria in Arabidopsis. FEBS Lett. 2014;588(9):1680–1685. doi: 10.1016/j.febslet.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 38.Xiao J, Engel JL, Zhang J, Chen MJ, Manning G, Dixon JE. Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc Natl Acad Sci U S A. 2011;108(29):11860–11865. doi: 10.1073/pnas.1109290108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Guan Z, Murphy AN, et al. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13(6):690–700. doi: 10.1016/j.cmet.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng H, Yu WM, Shen J, et al. Mitochondrial oxidation of the carbohydrate fuel is required for neural precursor/stem cell function and postnatal cerebellar development. Sci Adv. 2018;4(10):eaat2681. doi: 10.1126/sciadv.aat2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homma Y, Toga K, Daimon T, Shinoda T, Togawa T. A mitochondrial phosphatase PTPMT1 is essential for the early development of silkworm, Bombyx mori. Biochem Biophys Res Commun. 2020;530(4):713–718. doi: 10.1016/j.bbrc.2020.07.124 [DOI] [PubMed] [Google Scholar]
- 42.Faas MM, de Vos P. Mitochondrial function in immune cells in health and disease. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165845. doi: 10.1016/j.bbadis.2020.165845 [DOI] [PubMed] [Google Scholar]
- 43.Corrado M, Edwards-Hicks J, Villa M, et al. Dynamic cardiolipin synthesis is required for CD8+ T cell immunity. Cell Metab. 2020;32(6):981–995.e7. doi: 10.1016/j.cmet.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papakyrikos AM, Kim MJ, Wang X. Drosophila PTPMT1 has a function in tracheal air filling. iScience. 2020;23(7):101285. doi: 10.1016/j.isci.2020.101285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houtkooper RH, Akbari H, van Lenthe H, et al. Identification and characterization of human cardiolipin synthase. FEBS Lett. 2006;580(13):305930–64. doi: 10.1016/j.febslet.2006.04.054 [DOI] [PubMed] [Google Scholar]
- 46.Kasahara T, Kubota‐Sakashita M, Nagatsuka Y, et al. Cardiolipin is essential for early embryonic viability and mitochondrial integrity of neurons in mammals. FASEB J. 2020;34(1):1465–1480. doi: 10.1096/fj.201901598r [DOI] [PubMed] [Google Scholar]
- 47.Lee RG, Balasubramaniam S, Stentenbach M, et al. Deleterious variants in CRLS1 lead to cardiolipin deficiency and cause an autosomal recessive multi-system mitochondrial disease. Hum Mol Genet. 2022;31(21):3597–3612. doi: 10.1093/hmg/ddac040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, Kelley RI, Blanck TJ, Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J Biol Chem. 2003;278(51):51380–51385. doi: 10.1074/jbc.M307382200 [DOI] [PubMed] [Google Scholar]
- 49.Xu Y, Malhotra A, Ren M, Schlame M. The enzymatic function of tafazzin. J Biol Chem. 2006;281(51):39217–39224. doi: 10.1074/jbc.M606100200 [DOI] [PubMed] [Google Scholar]
- 50.Vreken P, Valianpour F, Nijtmans LG, et al. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279(2):378–382. doi: 10.1006/bbrc.2000.3952 [DOI] [PubMed] [Google Scholar]
- 51.Vaz FM, Houtkooper RH, Valianpour F, Barth PG, Wanders RJ. Only one splice variant of the human TAZ gene encodes a functional protein with a role in cardiolipin metabolism. J Biol Chem. 2003;278(44):43089–43094. doi: 10.1074/jbc.M305956200 [DOI] [PubMed] [Google Scholar]
- 52.Barth PG, Scholte HR, Berden JA, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci. 1983;62(1–3):327–355. doi: 10.1016/0022-510x(83)90209-5 [DOI] [PubMed] [Google Scholar]
- 53.Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12(4):385–389. doi: 10.1038/ng0496-385 [DOI] [PubMed] [Google Scholar]
- 54.Schlame M, Towbin JA, Heerdt PM, Jehle R, DiMauro S, Blanck TJ. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann Neurol. 2002;51(5):634–637. doi: 10.1002/ana.10176 [DOI] [PubMed] [Google Scholar]
- 55.Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580(23):5450–5455. doi: 10.1016/j.febslet.2006.07.022 [DOI] [PubMed] [Google Scholar]
- 56.Miller PC, Ren M, Schlame M, Toth MJ, Phoon CKL. A Bayesian analysis to determine the prevalence of Barth syndrome in the pediatric population. J Pediatr. 2020;217:139–144. doi: 10.1016/j.jpeds.2019.09.074 [DOI] [PubMed] [Google Scholar]
- 57.Clarke SLN, Bowron A, Gonzalez IL, et al. Barth syndrome. Orphanet J Rare Dis. 2013;8:23. doi: 10.1186/1750-1172-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor C, Rao ES, Pierre G, et al. Clinical presentation and natural history of Barth Syndrome: An overview. J Inherit Metab Dis. 2022;45(1):7–16. doi: 10.1002/jimd.12422 [DOI] [PubMed] [Google Scholar]
- 59.Olivar-Villanueva M, Ren M, Phoon CKL. Neurological and psychological aspects of Barth syndrome: Clinical manifestations and potential pathogenic mechanisms. Mitochondrion. 2021;61:188–195. doi: 10.1016/j.mito.2021.06.011 [DOI] [PubMed] [Google Scholar]
- 60.Wang G, McCain ML, Yang L, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20(6):616–623. doi: 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khuchua Z, Yue Z, Batts L, Strauss AW. A zebrafish model of human Barth syndrome reveals the essential role of tafazzin in cardiac development and function. Circ Res. 2006;99(2):201–208. doi: 10.1161/01.RES.0000233378.95325.ce [DOI] [PubMed] [Google Scholar]
- 62.Phoon CKL, Acehan D, Schlame M, et al. Tafazzin knockdown in mice leads to a developmental cardiomyopathy with early diastolic dysfunction preceding myocardial noncompaction. J Am Heart Assoc. 2012;1(2):jah3–e000455. doi: 10.1161/JAHA.111.000455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren M, Miller PC, Schlame M, Phoon CKL. A critical appraisal of the tafazzin knockdown mouse model of Barth syndrome: what have we learned about pathogenesis and potential treatments? Am J Physiol Heart Circ Physiol. 2019a;317(6), 183–193. doi: 10.1152/ajpheart.00504.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Acehan D, Khuchua Z, Houtkooper RH, et al. Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion. 2009;9(2):86–95. doi: 10.1016/j.mito.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hom JR, Quintanilla RA, Hoffman DL, et al. The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Dev Cell. 2011;21(3):469–478. doi: 10.1016/j.devcel.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lou W, Reynolds CA, Li Y, et al. Loss of tafazzin results in decreased myoblast differentiation in C2C12 cells: A myoblast model of Barth syndrome and cardiolipin deficiency. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(8):857–865. doi: 10.1016/j.bbalip.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seneviratne AK, Xu M, Henao JJA, et al. The mitochondrial transacylase, tafazzin, regulates for AML stemness by modulating intracellular levels of phospholipids. Cell Stem Cell. 2019;24(4):621–636.e16. doi: 10.1016/j.stem.2019.02.020 (Erratum in: Cell Stem Cell. 2019;24(6):1007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, Faloon PW, Tan Z, et al. Mouse lysocardiolipin acyltransferase controls the development of hematopoietic and endothelial lineages during in vitro embryonic stem-cell differentiation. Blood. 2007;110(10):3601–3609. doi: 10.1182/blood-2007-04-086827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miklas JW, Clark E, Levy S, et al. TFPa/HADHA is required for fatty acid beta-oxidation and cardiolipin re-modeling in human cardiomyocytes. Nat Commun. 2019;10(1):4671. doi: 10.1038/s41467-019-12482-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Taylor WA, Mejia EM, Mitchell RW, Choy PC, Sparagna GC, Hatch GM. Human trifunctional protein alpha links cardiolipin remodeling to beta-oxidation. PLoS One. 2012;7(11):e48628. doi: 10.1371/journal.pone.0048628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mejia EM, Ibdah JA, Sparagna GC, Hatch GM. Differential reduction in cardiac and liver monolysocardiolipin acyltransferase-1 and reduction in cardiac and liver tetralinoleoyl-cardiolipin in the α-subunit of trifunctional protein heterozygous knockout mice. Biochem J. 2015;471(1):123–129. doi: 10.1042/BJ20150648 [DOI] [PubMed] [Google Scholar]
- 72.Xia C, Fu Z, Battaile KP, Kim JJP. Crystal structure of human mitochondrial trifunctional protein, a fatty acid β-oxidation metabolon. Proc Natl Acad Sci U S A. 2019;116(13):6069–6074. doi: 10.1073/pnas.1816317116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Davey KM, Parboosingh JS, McLeod DR, et al. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet. 2006;43(5):385–393. doi: 10.1136/jmg.2005.036657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Richter-Dennerlein R, Korwitz A, Haag M, et al. DNAJC19, a mitochondrial cochaperone associated with cardiomyopathy, forms a complex with prohibitins to regulate cardiolipin remodeling. Cell Metab. 2014;20(1):158–171. doi: 10.1016/j.cmet.2014.04.016 [DOI] [PubMed] [Google Scholar]
- 75.Rohani L, Machiraju P, Sabouny R, et al. Reversible mitochondrial fragmentation in iPSC-derived cardiomyocytes from children with DCMA, a mitochondrial cardiomyopathy. Can J Cardiol. 2020;36(4):554–563. doi: 10.1016/j.cjca.2019.09.021 [DOI] [PubMed] [Google Scholar]
- 76.Acehan D, Malhotra A, Xu Y, Ren M, Stokes DL, Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys J. 2011; 100(9):2184–2192. doi: 10.1016/j.bpj.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Y, Condell M, Plesken H, et al. A Drosophila model of Barth syndrome. Proc Natl Acad Sci U S A. 2006;103(31):11584–11588. doi: 10.1073/pnas.0603242103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sakamoto T, Inoue T, Otomo Y, et al. Deficiency of cardiolipin synthase causes abnormal mitochondrial function and morphology in germ cells of Caenorhabditis elegans. J Biol Chem. 2012;287(7):4590–4601. doi: 10.1074/jbc.M111.314823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buckingham M Tissue differentiation: A personal account of research on myogenesis and cardiogenesis. Curr Top Dev Biol. 2016;116:135–151. doi: 10.1016/bs.ctdb.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 80.Donadon M, Santoro MM. The origin and mechanisms of smooth muscle cell development in vertebrates. Development. 2021;148(7):dev197384. doi: 10.1242/dev.197384 [DOI] [PubMed] [Google Scholar]
- 81.McCauley HA, Wells JM. Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development. 2017;144(6): 958–962. doi: 10.1242/dev.140731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miquerol L, Kelly RG. Organogenesis of the vertebrate heart. Wiley Interdiscip Rev Dev Biol. 2013;2(1): 17–29. doi: 10.1002/wdev.68 [DOI] [PubMed] [Google Scholar]
- 83.Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res. 2011;317(19):2702–2710. doi: 10.1016/j.yexcr.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sivakumar A, Kurpios NA. Transcriptional regulation of cell shape during organ morphogenesis. J Cell Biol. 2018;217(9):2987–3005. doi: 10.1083/jcb.201612115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20(4):327–348. doi: 10.1007/s11065-010-9148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Udan RS, Culver JC, Dickinson ME. Understanding vascular development. Wiley Interdiscip Rev Dev Biol. 2013;2(3):327–346. doi: 10.1002/wdev.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vilches-Moure JG. Embryonic children (Gallus gallus domesticus) as a model of cardiac biology and development. Comp Med. 2019;69(3):184–203. doi: 10.30802/AALAS-CM-18-000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bahat A, Gross A. Mitochondrial plasticity in cell fate regulation. J Biol Chem. 2019;294(38):13852–13863. doi: 10.1074/jbc.REV118.000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shepard TH, Muffley LA, Smith LT. Ultrastructural study of mitochondria and their cristae in embryonic rats and primate (N. nemistrina). Anat Rec. 1998;252:383–392. doi: [DOI] [PubMed] [Google Scholar]
- 90.Shepard TH, Muffley LA, Smith LT. Mitochondrial ultrastructure in embryos after implantation. Hum Reprod. 2000;15 Suppl 2:218–228. doi: 10.1093/humrep/15.suppl_2.218 [DOI] [PubMed] [Google Scholar]
- 91.Marin-Garcia J, Ananthakrishnan R, Goldenthal MJ. Heart mitochondrial DNA and enzyme changes during early human development. Mol Cell Biochem. 2000;210(1–2):47–52. doi: 10.1023/a:1007031919298 [DOI] [PubMed] [Google Scholar]
- 92.Bass A, Stejskalová M, Stieglerová A, Ostádal B, Samánek M. Ontogenetic development of energy-supplying enzymes in rat and guinea-pig heart. Physiol Res. 2001;50:237–245. [PubMed] [Google Scholar]
- 93.Mando’ C, Marino MA, Miriam F, et al. OS048 Mitochondrial content and function in placental cells and tissues of preeclampsia and IUGR. Pregnancy Hypertens. 2012;2(3):203. doi: 10.1016/j.preghy.2012.04.049 [DOI] [PubMed] [Google Scholar]
- 94.Baker CN, Ebert SN. Development of aerobic metabolism in utero: Requirement for mitochondrial function during embryonic and fetal periods. OA Biotechnology. 2013;2(2):16 [Google Scholar]
- 95.Kolarova H, Krizova J, Hulkova M, et al. Changes in transcription pattern lead to a marked decrease in COX, CS and SQR activity after the developmental point of the 22(nd) gestational week. Physiol Res. 2018;67(1):79–91. doi: 10.33549/physiolres.933542 [DOI] [PubMed] [Google Scholar]
- 96.Chakrabarty RP, Chandel NS. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28(3):394–408. doi: 10.1016/j.stem.2021.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rossmann MP, Dubois SM, Agarwal S, Zon LI. Mitochondrial function in development and disease. Dis Model Mech. 2021;14(6):dmm048912. doi: 10.1242/dmm.048912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tiwari SK, Mandal S. Mitochondrial control of stem cell state and fate: Lessons from Drosophila. Front Cell Dev Biol. 2021;9:606639. doi: 10.3389/fcell.2021.606639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Noguchi M, Kasahara A. Mitochondrial dynamics coordinate cell differentiation. Biochem Biophys Res Commun. 2018;500(1):59–64. doi: 10.1016/j.bbrc.2017.06.094 [DOI] [PubMed] [Google Scholar]
- 100.Zhao Q, Sun Q, Zhou L, Liu K, Jiao K. Complex regulation of mitochondrial function during cardiac development. J Am Heart Assoc. 2019;8(13):e012731. doi: 10.1161/JAHA.119.012731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bonora M, Kahsay A, Pinton P. Mitochondrial calcium homeostasis in hematopoietic stem cell: Molecular regulation of quiescence, function, and differentiation. Int Rev Cell Mol Biol. 2021;362:111–140. doi: 10.1016/bs.ircmb.2021.05.003 [DOI] [PubMed] [Google Scholar]
- 102.Liu X, Peng G. Mitochondria orchestrate T cell fate and function. Nat Immunol. 2021;22(3):276–278. doi: 10.1038/s41590-020-00861-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wagatsuma A, Sakuma K. Mitochondria as a potential regulator of myogenesis. ScientificWorldJournal. 2013;2013:593267. doi: 10.1155/2013/593267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goody MF, Henry CA. A need for NAD+ in muscle development, homeostasis, and aging. Skelet Muscle. 2018;8(1):9. doi: 10.1186/s13395-018-0154-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bhattacharya D, Scimè A. Mitochondrial function in muscle stem cell fates. Front Cell Dev Biol. 2020;8:480. doi: 10.3389/fcell.2020.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Q, Gao Z, Chen Y, Guan MX. The role of mitochondria in osteogenic, adipogenic and chondrogenic differentiation of mesenchymal stem cells. Protein Cell. 2017;8(6):439–445. doi: 10.1007/s13238-017-0385-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mohammadalipour A, Dumbali SP, Wenzel PL. Mitochondrial transfer and regulators of mesenchymal stromal cell function and therapeutic efficacy. Front Cell Dev Biol. 2020;8:603292. doi: 10.3389/fcell.2020.603292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Morganti C, Bonora M, Marchi S, et al. Citrate mediates crosstalk between mitochondria and the nucleus to promote human mesenchymal stem cell in vitro osteogenesis. Cells. 2020; 9(4):1034. doi: 10.3390/cells9041034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ren L, Chen X, Chen X, Li J, Cheng B, Xia J. Mitochondrial dynamics: Fission and fusion in fate determination of mesenchymal stem cells. Front Cell Dev Biol. 2020;8:580070. doi: 10.3389/fcell.2020.580070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morris O, Jasper H. Reactive oxygen species in intestinal stem cell metabolism, fate and function. Free Radic Biol Med. 2021; 166:140–146. doi: 10.1016/j.freeradbiomed.2021.02.015 [DOI] [PubMed] [Google Scholar]
- 111.Ludikhuize MC, Meerlo M, Gallego MP, et al. Mitochondria define intestinal stem cell differentiation downstream of a FOXO/Notch axis. Cell Metab. 2020;32(5):889–900.e7. doi: 10.1016/j.cmet.2020.10.005 [DOI] [PubMed] [Google Scholar]
- 112.Garcez M, Branco-Santos J, Gracio PC, Homem CCF. Mitochondrial dynamics in the Drosophila ovary regulates germ stem cell number, cell fate, and female fertility. Front Cell Dev Biol. 2021;8:596819. doi: 10.3389/fcell.2020.596819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maffezzini C, Calvo-Garrido J, Wredenberg A, Freyer C. Metabolic regulation of neurodifferentiation in the adult brain. Cell Mol Life Sci. 2020;77(13):2483–2496. doi: 10.1007/s00018-019-03430-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Dev Cell. 2009;17(6):755–773. doi: 10.1016/j.devcel.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 115.Miyazawa H, Aulehla A. Revisiting the role of metabolism during development. Development. 2018;145(19):dev131110. doi: 10.1242/dev.131110 [DOI] [PubMed] [Google Scholar]
- 116.Agathocleous M, Love NK, Randlett O, et al. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14(8):859–864. doi: 10.1038/ncb2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11(5):596–606. doi: 10.1016/j.stem.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Folmes CD, Terzic A. Metabolic determinants of embryonic development and stem cell fate. Reprod Fertil Dev. 2014;27(1):82–88. doi: 10.1071/RD14383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Khacho M, Harris R, Slack RS. Mitochondria as central regulators of neural stem cell fate and cognitive function. Nat Rev Neurosci. 2019;20(1):34–48. doi: 10.1038/s41583-018-0091-3 [DOI] [PubMed] [Google Scholar]
- 120.Arribat Y, Grepper D, Lagarrigue S, et al. Mitochondria in embryogenesis: An organellogenesis perspective. Front Cell Dev Biol. 2019;7:282. doi: 10.3389/fcell.2019.00282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Beutner G, Eliseev RA, Porter GA Jr. Initiation of electron transport chain activity in the embryonic heart coincides with the activation of mitochondrial complex 1 and the formation of supercomplexes. PLoS One. 2014;9(11):e113330. doi: 10.1371/journal.pone.0113330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee RG, Gao J, Siira SJ, et al. Cardiolipin is required for membrane docking of mitochondrial ribosomes and protein synthesis. J Cell Sci. 2020;133(14):jcs240374. doi: 10.1242/jcs.240374 [DOI] [PubMed] [Google Scholar]
- 123.Itoh Y, Andréll J, Choi A, et al. Mechanism of membrane-tethered mitochondrial protein synthesis. Science. 2021;371(6531):846–849. doi: 10.1126/science.abe0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xu Y, Erdjument-Bromage H, Phoon CKL, Neubert TA, Ren M, Schlame M. Cardiolipin remodeling enables protein crowding in the inner mitochondrial membrane. EMBO J. 2021;40(23):e108428. doi: 10.15252/embj.2021108428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhu S, Chen Z, Zhu M, et al. Cardiolipin remodeling defects impair mitochondrial architecture and function in a murine model of Barth syndrome cardiomyopathy. Circ Heart Fail. 2021;14(6):e008289. doi: 10.1161/CIRCHEARTFAILURE.121.008289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361(3):462–469. doi: 10.1016/j.jmb.2006.06.057 [DOI] [PubMed] [Google Scholar]
- 127.Dudek J, Cheng IF, Balleininger M, et al. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 2013;11(2):806–819. doi: 10.1016/j.scr.2013.05.005 [DOI] [PubMed] [Google Scholar]
- 128.Geldon S, Fernández-Vizarra E, Tokatlidis K. Redox-mediated regulation of mitochondrial biogenesis, dynamics, and respiratory chain assembly in yeast and human cells. Front Cell Dev Biol. 2021;9:720656. doi: 10.3389/fcell.2021.720656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuznetsov AV, Margreiter R, Ausserlechner MJ, Hagenbuchner J. The complex interplay between mitochondria, ROS and entire cellular metabolism. Antioxidants (Basel). 2022;11(10):1995. doi: 10.3390/antiox11101995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Covarrubias L, Hernández-García D, Schnabel D, Salas-Vidal E, Castro-Obregón S. Function of reactive oxygen species during animal development: passive or active? Dev Biol. 2008;320(1):1–11. doi: 10.1016/j.ydbio.2008.04.041 [DOI] [PubMed] [Google Scholar]
- 131.Timme-Laragy AR, Hahn ME, Hansen JM, Rastogi A, Roy MA. Redox stress and signaling during vertebrate embryonic development: Regulation and responses. Semin Cell Dev Biol. 2018;80:17–28. doi: 10.1016/j.semcdb.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lopes AS, Lane M, Thompson JG. Oxygen consumption and ROS production are increased at the time of fertilization and cell cleavage in bovine zygotes. Hum Reprod. 2010;25(11):2762–2773. doi: 10.1093/humrep/deq221 [DOI] [PubMed] [Google Scholar]
- 133.Han Y, Ishibashi S, Iglesias-Gonzalez J, Chen Y, Love NR, Amaya E. Ca2+-induced mitochondrial ROS regulate the early embryonic cell cycle. Cell Rep. 2018;22(1):218–231. doi: 10.1016/j.celrep.2017.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Milkovic L, Cipak Gasparovic A, Cindric M, Mouthuy PA, Zarkovic N. Short overview of ROS as cell function regulators and their implications in therapy concepts. Cells. 2019;8(8):793. doi: 10.3390/cells8080793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen S, He Q, Greenberg ML. Loss of tafazzin in yeast leads to increased oxidative stress during respiratory growth. Mol Microbiol. 2008;68(4):1061–1072. doi: 10.1111/j.1365-2958.2008.06216.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Szczepanek K, Allegood J, Aluri H, Hu Y, Chen Q, Lesnefsky EJ. Acquired deficiency of tafazzin in the adult heart: Impact on mitochondrial function and response to cardiac injury. Biochim Biophys Acta. 2016;1861(4):294–300. doi: 10.1016/j.bbalip.2015.12.004 [DOI] [PubMed] [Google Scholar]
- 137.Cole LK, Kim JH, Amoscato AA, et al. Aberrant cardiolipin metabolism is associated with cognitive deficiency and hippocampal alteration in tafazzin knockdown mice. Biochim Biophys Acta Mol Basis Dis. 2018;1864(10):3353–3367. doi: 10.1016/j.bbadis.2018.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cole LK, Mejia EM, Sparagna GC, et al. Cardiolipin deficiency elevates susceptibility to a lipotoxic hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2020;144:24–34. doi: 10.1016/j.yjmcc.2020.05.001 [DOI] [PubMed] [Google Scholar]
- 139.Huang Z, Jiang J, Tyurin VA, et al. Cardiolipin deficiency leads to decreased cardiolipin peroxidation and increased resistance of cells to apoptosis. Free Radic Biol Med. 2008;44(11):1935–1944. doi: 10.1016/j.freeradbiomed.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goncalves RLS, Schlame M, Bartelt A, Brand MD, Hotamışlıgil GS. Cardiolipin deficiency in Barth syndrome is not associated with increased superoxide/H2 O2 production in heart and skeletal muscle mitochondria. FEBS Lett. 2021;595(3):415–432. doi: 10.1002/1873-3468.13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dudek J Role of cardiolipin in mitochondrial signaling pathways. Front Cell Dev Biol. 2017;5:90. doi: 10.3389/fcell.2017.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khacho M, Clark A, Svoboda DS, et al. Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell. 2016;19(2):232–247. doi: 10.1016/j.stem.2016.04.015 [DOI] [PubMed] [Google Scholar]
- 143.Zucker RM, Rogers JM. Confocal laser scanning microscopy of morphology and apoptosis in organogenesis-stage mouse embryos. Methods Mol Biol. 2019;1965:297–311. doi: 10.1007/978-1-4939-9182-2_20 [DOI] [PubMed] [Google Scholar]
- 144.Voss AK, Strasser A. The essentials of developmental apoptosis. F1000Res. 2020;9:F1000 Faculty Rev-148. doi: 10.12688/f1000research.21571.1 [DOI] [Google Scholar]
- 145.Lima A, Burgstaller J, Sanchez-Nieto JM, Rodríguez TA. The mitochondria and the regulation of cell fitness during early mammalian development. Curr Top Dev Biol. 2018;128:339–363. doi: 10.1016/bs.ctdb.2017.10.012 [DOI] [PubMed] [Google Scholar]
- 146.Kumar Sharma R, Chafik A, Bertolin G. Mitochondrial transport, partitioning, and quality control at the heart of cell proliferation and fate acquisition. Am J Physiol Cell Physiol. 2022;322(2):C311–C325. doi: 10.1152/ajpcell.00256.2021 [DOI] [PubMed] [Google Scholar]
- 147.Tyurina YY, Poloyac SM, Tyurin VA, et al. A mitochondrial pathway for biosynthesis of lipid mediators. Nat Chem. 2014;6(6):542–552. doi: 10.1038/nchem.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Role of cardiolipin peroxidation and Ca2+ in mitochondrial dysfunction and disease. Cell Calcium. 2009;45(6):643–650. doi: 10.1016/j.ceca.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 149.Pointer CB, Klegeris A. Cardiolipin in central nervous system physiology and pathology. Cell Mol Neurobiol. 2017;37(7):1161–1172. doi: 10.1007/s10571-016-0458-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Paradies G, Paradies V, Ruggiero FM, Petrosillo G. Role of cardiolipin in mitochondrial function and dynamics in health and disease: Molecular and pharmacological aspects. Cells. 2019;8(7):728. doi: 10.3390/cells8070728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Shidoji Y, Hayashi K, Komura S, Ohishi N, Yagi K. Loss of molecular interaction between cytochrome c and cardiolipin due to lipid peroxidation. Biochem Biophys Res Commun. 1999;264(2):343–347. doi: 10.1006/bbrc.1999.1410 [DOI] [PubMed] [Google Scholar]
- 152.Petrosillo G, Casanova G, Matera M, Ruggiero FM, Paradies G. Interaction of peroxidized cardiolipin with rat-heart mitochondrial membranes: induction of permeability transition and cytochrome c release. FEBS Lett. 2006;580(27):6311–6316. doi: 10.1016/j.febslet.2006.10.036 [DOI] [PubMed] [Google Scholar]
- 153.Vlasova II, Tyurin VA, Kapralov AA, et al. Nitric oxide inhibits peroxidase activity of cytochrome c.cardiolipin complex and blocks cardiolipin oxidation. J Biol Chem. 2006;281(21):14554–14562. doi: 10.1074/jbc.M509507200 [DOI] [PubMed] [Google Scholar]
- 154.Makaryan V, Kulik W, Vaz FM, et al. The cellular and molecular mechanisms for neutropenia in Barth syndrome. Eur J Haematol. 2012;88(3):195–209. doi: 10.1111/j.1600-0609.2011.01725.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ji J, Greenberg ML. Cardiolipin function in the yeast S. cerevisiae and the lessons learned for Barth syndrome. J Inherit Metab Dis. 2022;45(1):60–71. doi: 10.1002/jimd.12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Giacomello M, Pyakurel A, Glytsou C, Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat Rev Mol Cell Biol. 2020;21(4):204–224. doi: 10.1038/s41580-020-0210-7 [DOI] [PubMed] [Google Scholar]
- 157.Yapa NMB, Lisnyak V, Reljic B, Ryan MT. Mitochondrial dynamics in health and disease. FEBS Lett. 2021;595(8):1184–1204. doi: 10.1002/1873-3468.14077 [DOI] [PubMed] [Google Scholar]
- 158.Frohman MA. Role of mitochondrial lipids in guiding fission and fusion. J Mol Med (Berl). 2015;93(3):263–269. doi: 10.1007/s00109-014-1237-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160(2):189–200. doi: 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rangaraju V, Lewis TL Jr, Hirabayashi Y, et al. Pleiotropic mitochondria: The influence of mitochondria on neuronal development and disease. J Neurosci. 2019;39(42):8200–8208. doi: 10.1523/JNEUROSCI.1157-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Iwata R, Vanderhaeghen P. Regulatory roles of mitochondria and metabolism in neurogenesis. Curr Opin Neurobiol. 2021;69: 231–240. doi: 10.1016/j.conb.2021.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125(7):1241–1252. doi: 10.1016/j.cell.2006.06.010 [DOI] [PubMed] [Google Scholar]
- 164.DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol. 2009;186(6):793–803. doi: 10.1083/jcb.200906098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ban T, Heymann JA, Song Z, Hinshaw JE, Chan DC. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum Mol Genet. 2010;19(11):2113–2122. doi: 10.1093/hmg/ddq088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandran R. Cardiolipin's propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell. 2015;26(17):3104–3116. doi: 10.1091/mbc.E15-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Joshi AS, Thompson MN, Fei N, Hüttemann M, Greenberg ML. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J Biol Chem. 2012;287(21):17589–17597. doi: 10.1074/jbc.M111.330167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ban T, Ishihara T, Kohno H, et al. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol. 2017;19(7):856–863. doi: 10.1038/ncb3560 [DOI] [PubMed] [Google Scholar]
- 169.Kameoka S, Adachi Y, Okamoto K, Iijima M, Sesaki H. Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 2018;28(1):67–76. doi: 10.1016/j.tcb.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ban T, Kohno H, Ishihara T, Ishihara N. Relationship between OPA1 and cardiolipin in mitochondrial inner-membrane fusion. Biochim Biophys Acta Bioenerg. 2018;1859(9): 951–957. doi: 10.1016/j.bbabio.2018.05.016 [DOI] [PubMed] [Google Scholar]
- 171.Parikh VS, Morgan MM, Scott R, Clements LS, Butow RA. The mitochondrial genotype can influence nuclear gene expression in yeast. Science. 1987;235(4788):576–580. doi: 10.1126/science.3027892 [DOI] [PubMed] [Google Scholar]
- 172.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14(1):1–15. doi: 10.1016/s1097-2765(04)00179-0 [DOI] [PubMed] [Google Scholar]
- 173.Tan JX, Finkel T. Mitochondria as intracellular signaling platforms in health and disease. J Cell Biol. 2020; 219(5):e202002179. doi: 10.1083/jcb.202002179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Feng Y, Huang W, Paul C, et al. Mitochondrial nucleoid in cardiac homeostasis: bidirectional signaling of mitochondria and nucleus in cardiac diseases. Basic Res Cardiol. 2021;116(1):49. doi: 10.1007/s00395-021-00889-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Walker BR, Moraes CT. Nuclear-mitochondrial interactions. Biomolecules. 2022;12(3):427. doi: 10.3390/biom12030427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang GL, Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A. 1993;90(9):4304–4308. doi: 10.1073/pnas.90.9.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129(1):111–122. doi: 10.1016/j.cell.2007.01.047 [DOI] [PubMed] [Google Scholar]
- 178.Chowdhury A, Aich A, Jain G, et al. Defective mitochondrial cardiolipin remodeling dampens HIF-1α expression in hypoxia. Cell Rep. 2018;25(3):561–570.e6. doi: 10.1016/j.celrep.2018.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Simsek T, Kocabas F, Zheng J, et al. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7(3):380–390. doi: 10.1016/j.stem.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Krock BL, Eisinger-Mathason TS, Giannoukos DN, et al. The aryl hydrocarbon receptor nuclear translocator is an essential regulator of murine hematopoietic stem cell viability. Blood. 2015;125(21):3263–72. doi: 10.1182/blood-2014-10-607267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Saini Y, Harkema JR, LaPres JJ. HIF1alpha is essential for normal intrauterine differentiation of alveolar epithelium and surfactant production in the newborn lung of mice. J Biol Chem. 2008;283(48):33650–33657. doi: 10.1074/jbc.M805927200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Provot S, Zinyk D, Gunes Y, et al. Hif-1alpha regulates differentiation of limb bud mesenchyme and joint development. J Cell Biol. 2007;177(3):4514–64. doi: 10.1083/jcb.200612023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Poveda-Huertes D, Taskin AA, Dhaouadi I, et al. Increased mitochondrial protein import and cardiolipin remodelling upon early mtUPR. PLoS Genet. 2021;17(7):e1009664. doi: 10.1371/journal.pgen.1009664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shen G, Liu W, Xu L, Wang LL. Mitochondrial unfolded protein response and its roles in stem cells. Stem Cells Dev. 2020;29(10):627–637. doi: 10.1089/scd.2019.0278 [DOI] [PubMed] [Google Scholar]
- 185.Wang T, Babayev E, Jiang Z, et al. Mitochondrial unfolded protein response gene Clpp is required to maintain ovarian follicular reserve during aging, for oocyte competence, and development of pre-implantation embryos. Aging Cell. 2018;17(4):e12784. doi: 10.1111/acel.12784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nordin A, Larsson E, Thornell LE, Holmberg M. Tissue-specific splicing of ISCU results in a skeletal muscle phenotype in myopathy with lactic acidosis, while complete loss of ISCU results in early embryonic death in mice. Hum Genet. 2011;129(4):371–378. doi: 10.1007/s00439-010-0931-3 [DOI] [PubMed] [Google Scholar]
- 187.Stehling O, Lill R. The role of mitochondria in cellular iron-sulfur protein biogenesis: mechanisms, connected processes, and diseases. Cold Spring Harb Perspect Biol. 2013;5(8):a011312. doi: 10.1101/cshperspect.a011312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Holmes-Hampton GP, Chakrabarti M, Cockrell AL, et al. Changing iron content of the mouse brain during development. Metallomics. 2012;4(8):761–770. doi: 10.1039/c2mt20086d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Luo J, Zhang X, He S, et al. Deletion of narfl leads to increased oxidative stress mediated abnormal angiogenesis and digestive organ defects in zebrafish. Redox Biol. 2020;28:101355. doi: 10.1016/j.redox.2019.101355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Read AD, Bentley RE, Archer SL, Dunham-Snary KJ. Mitochondrial iron-sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox Biol. 2021;47:102164. doi: 10.1016/j.redox.2021.102164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li Y, Lou W, Grevel A, et al. Cardiolipin-deficient cells have decreased levels of the iron-sulfur biogenesis protein frataxin. J Biol Chem. 2020;295(33):11928–11937. doi: 10.1074/jbc.RA120.013960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cao X, Chen Y. Mitochondria and calcium signaling in embryonic development. Semin Cell Dev Biol. 2009;20(3):337–345. doi: 10.1016/j.semcdb.2008.12.014 [DOI] [PubMed] [Google Scholar]
- 193.Paudel S, Sindelar R, Saha M. Calcium signaling in vertebrate development and its role in disease. Int J Mol Sci. 2018;19(11):3390. doi: 10.3390/ijms19113390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Mammano F, Bortolozzi M. Ca2+ signaling, apoptosis and autophagy in the developing cochlea: Milestones to hearing acquisition. Cell Calcium. 2018;70:117–126. doi: 10.1016/j.ceca.2017.05.006 [DOI] [PubMed] [Google Scholar]
- 195.Martinho-Dias D, Leite-Moreira A, Castro-Chaves P. Calreticulin in the heart: From embryological development to cardiac pathology. Curr Mol Med. 2016;16(1):12–22. doi: 10.2174/1566524016666151222142816 [DOI] [PubMed] [Google Scholar]
- 196.Yoshiba S, Hamada H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 2014;30(1):10–17. doi: 10.1016/j.tig.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 197.Webb SE, Miller AL. Visualization of Ca2+ signaling during embryonic skeletal muscle formation in vertebrates. Cold Spring Harb Perspect Biol. 2011;3(2):a004325. doi: 10.1101/cshperspect.a004325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427(6972):360–364. doi: 10.1038/nature02246 [DOI] [PubMed] [Google Scholar]
- 199.De Stefani D, Raffaello A, Teardo E, Szabò I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476(7360):336–340. doi: 10.1038/nature10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bick AG, Calvo SE, Mootha VK. Evolutionary diversity of the mitochondrial calcium uniporter. Science. 2012;336(6083):886. doi: 10.1126/science.1214977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Marchi S, Pinton P. The mitochondrial calcium uniporter complex: molecular components, structure and physiopathological implications. J Physiol. 2014;592(5):829–839. doi: 10.1113/jphysiol.2013.268235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liu JC, Parks RJ, Liu J, et al. The in vivo biology of the mitochondrial calcium uniporter. Adv Exp Med Biol. 2017;982:49–63. doi: 10.1007/978-3-319-55330-6_3 [DOI] [PubMed] [Google Scholar]
- 203.Murphy E, Steenbergen C. Regulation of mitochondrial Ca2+ uptake. Annu Rev Physiol. 2021;83:107–126. doi: 10.1146/annurev-physiol-031920-092419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Garbincius JF, Elrod JW. Mitochondrial calcium exchange in physiology and disease. Physiol Rev. 2022;102(2):893–992. doi: 10.1152/physrev.00041.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ghosh S, Basu Ball W, Madaris TR, et al. An essential role for cardiolipin in the stability and function of the mitochondrial calcium uniporter. Proc Natl Acad Sci U S A. 2020;117(28):16383–16390. doi: 10.1073/pnas.2000640117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu X, Wang S, Guo X, et al. Increased reactive oxygen species-mediated Ca2+/calmodulin-dependent protein kinase II activation contributes to calcium handling abnormalities and impaired contraction in Barth syndrome. Circulation. 2021;143(19):1894–1911. doi: 10.1161/CIRCULATIONAHA.120.048698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ferko M, Andelová N, Szeiffová Bačová B, Jašová M. Myocardial adaptation in pseudohypoxia: Signaling and regulation of mPTP via mitochondrial connexin 43 and cardiolipin. Cells. 2019;8(11):1449. doi: 10.3390/cells8111449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Odagiri K, Katoh H, Kawashima H, et al. Local control of mitochondrial membrane potential, permeability transition pore and reactive oxygen species by calcium and calmodulin in rat ventricular myocytes. J Mol Cell Cardiol. 2009;46(6):989–997. doi: 10.1016/j.yjmcc.2008.12.022 [DOI] [PubMed] [Google Scholar]
- 209.Seidlmayer LK, Juettner VV, Kettlewell S, Pavlov EV, Blatter LA, Dedkova EN. Distinct mPTP activation mechanisms in ischaemia-reperfusion: contributions of Ca2+, ROS, pH, and inorganic polyphosphate. Cardiovasc Res. 2015;106(2):237–248. doi: 10.1093/cvr/cvv097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Petrosillo G, Ruggiero FM, Pistolese M, Paradies G. Reactive oxygen species generated from the mitochondrial electron transport chain induce cytochrome c dissociation from beef-heart submitochondrial particles via cardiolipin peroxidation. Possible role in the apoptosis. FEBS Lett. 2001;509(3):435–438. doi: 10.1016/s0014-5793(01)03206-9 [DOI] [PubMed] [Google Scholar]
- 211.Pérez MJ, Quintanilla RA. Development or disease: duality of the mitochondrial permeability transition pore. Dev Biol. 2017;426(1):1–7. doi: 10.1016/j.ydbio.2017.04.018 [DOI] [PubMed] [Google Scholar]
- 212.Namba T, Dóczi J, Pinson A, et al. Human-specific ARHGAP11B acts in mitochondria to expand neocortical progenitors by glutaminolysis. Neuron. 2020;105(5):867–881.e9. doi: 10.1016/j.neuron.2019.11.027 [DOI] [PubMed] [Google Scholar]
- 213.Schlame M Protein crowding in the inner mitochondrial membrane. Biochim Biophys Acta Bioenerg. 2021;1862(1):148305. doi: 10.1016/j.bbabio.2020.148305 [DOI] [PubMed] [Google Scholar]
- 214.Xu Y, Anjaneyulu M, Donelian A, et al. Assembly of the complexes of oxidative phosphorylation triggers the remodeling of cardiolipin. Proc Natl Acad Sci U S A. 2019;116(23):11235–11240. doi: 10.1073/pnas.1900890116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tomanek RJ, Lund DD, Yue X. Hypoxic induction of myocardial vascularization during development. Adv Exp Med Biol. 2003;543:139–149. doi: 10.1007/978-1-4419-8997-0_10 [DOI] [PubMed] [Google Scholar]
- 216.Imanirad P, Dzierzak E. Hypoxia and HIFs in regulating the development of the hematopoietic system. Blood Cells Mol Dis. 2013;51(4):256–263. doi: 10.1016/j.bcmd.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ren M, Xu Y, Phoon CKL, et al. Condensed mitochondria assemble into the acrosomal matrix during spermiogenesis. Front Cell Dev Biol. 2022;10:867175. doi: 10.3389/fcell.2022.867175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mancuso DJ, Sims HF, Han X, et al. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J Biol Chem. 2007;282(48):34611–34622. doi: 10.1074/jbc.M707795200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Malhotra A, Edelman-Novemsky I, Xu Y, et al. Role of calcium-independent phospholipase A2 in the pathogenesis of Barth syndrome. Proc Natl Acad Sci U S A. 2009;106(7):2337–2341. doi: 10.1073/pnas.0811224106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Schiller J, Laube E, Wittig I, Kühlbrandt W, Vonck J, Zickermann V. Insights into complex I assembly: Function of NDUFAF1 and a link with cardiolipin remodeling. Sci Adv. 2022;8(46):eadd3855. doi: 10.1126/sciadv.add3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol. 2006;174(3):379–390. doi: 10.1083/jcb.200605043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Falabella M, Vernon HJ, Hanna MG, Claypool SM, Pitceathly RDS. Cardiolipin, mitochondria, and neurological disease. Trends Endocrinol Metab. 2021;32(4):224–237. doi: 10.1016/j.tem.2021.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang J, Shi Y. In search of the Holy Grail: Toward a unified hypothesis on mitochondrial dysfunction in age-related diseases. Cells. 2022;11(12):1906. doi: 10.3390/cells11121906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Imae R, Inoue T, Nakasaki Y, et al. LYCAT, a homologue of C. elegans acl-8, acl-9, and acl-10, determines the fatty acid composition of phosphatidylinositol in mice. J Lipid Res. 2012;53(3):335–347. doi: 10.1194/jlr.M018655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Valentine WJ, Yanagida K, Kawana H, et al. Update and nomenclature proposal for mammalian lysophospholipid acyltransferases, which create membrane phospholipid diversity. J Biol Chem. 2022;298(1):101470. doi: 10.1016/j.jbc.2021.101470 [DOI] [PMC free article] [PubMed] [Google Scholar]