Abstract
Genome editing approaches have transformed our ability to make user-defined changes to genomes in both ex vivo and in vivo contexts. Despite the abundant development of technologies that permit the installation of nucleotide-level changes, until recently, larger-scale sequence edits via technologies independent of DNA double-strand breaks (DSBs) had remained less explored. Here we review recent advances toward DSB-free technologies that enable kilobase-scale modifications including insertions, deletions, inversions, replacements, and others. These technologies provide new capabilities for users, while offering hope for the simplification of putative therapeutic strategies by moving away from small mutation-specific edits and towards generalizable kilobase-scale approaches.
Graphical Abstract
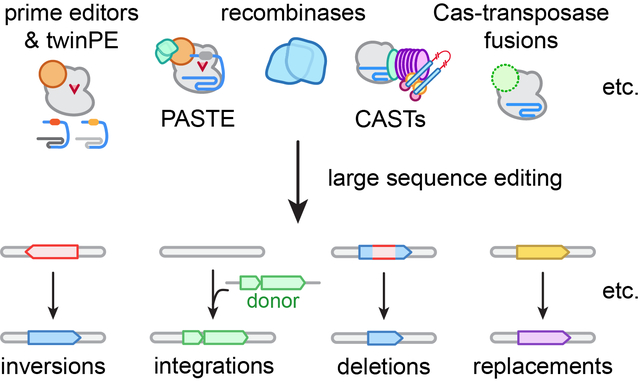
Towards large(r) genetic edits. New classes of genome editing technologies are transforming our ability to make large sequence edits ranging from dozens of base pairs to several kilobases.
The continued development of CRISPR-Cas technologies has expanded our ability to make customizable modifications to the human genome, revolutionizing the pursuit of permanent therapeutic edits. Typically, RNA-programmed CRISPR-Cas enzymes initiate genome editing events by catalyzing locus-specific DNA double-stranded breaks (DSBs) in a genome of interest. Subsequent repair of the DSBs by cellular processes can result in gene knockouts or targeted deletions via non-homologous end-joining or microhomology-mediated end-joining (NHEJ and MMEJ, respectively), or result in knock-in of small or large desired edits that are encoded on donor DNA molecules via homology-directed repair1 (HDR; Fig. 1a). Despite these capabilities, the precise installation or deletion of small and large DNA sequences in various cells and organisms via nuclease mediated DSBs is challenging and/or can lead to unwanted side effects. For instance, DSB-based methods suffer from heterogeneous and sometimes undesired insertion or deletion mutations (indels) at the on-target site2,3, unpredictable large-scale deletions3, chromosomal alterations due to DSBs that co-occur at on- and/or off-target sites4,5, toxicity resulting from cellular DSB-response6,7, and a reliance on certain cellular factors or DNA repair pathways that may not be expressed in the target cell type. Thus, next-generation technologies that produce targeted DNA modifications directly on the sequence of interest without DSBs are critical to overcoming these caveats.
Fig. 1. CRISPR-based technologies for small and large genome edits.
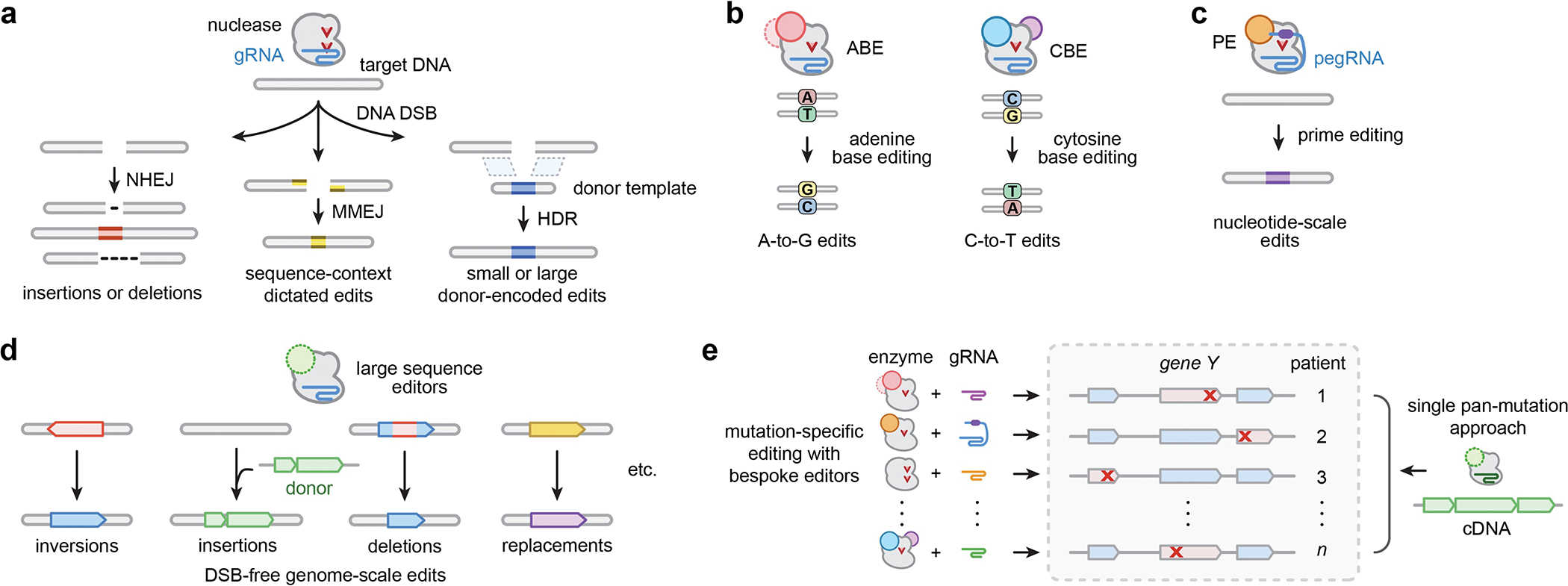
a, Summary of major edit outcomes following nuclease-mediated editing. CRISPR-Cas enzymes paired with guide RNAs (gRNAs) generate DNA double-strand breaks (DSBs) to initiate editing events, which are generally repaired by non-homologous end joining (NHEJ), micro-homology-mediated end joining (MMEJ), or homology-directed repair (HDR). b, Adenine and cytosine base editors (ABEs and CBEs) generate A-to-G and C-to-T changes, respectively, without intentionally causing DSBs or requiring HDR. c, Prime editors (PEs) generate short insertion, deletion, and/or substitution edits that are encoded on a prime editing guide RNA (pegRNA). d, Large sequence editors can generate targeted inversions, insertions, and deletions of multi-kb sequences, which would enable novel editing approaches for previously inaccessible classes of diseases. e, Compared to the requisite suite of mutation-specific gene editing approaches for diseases caused by numerous heterogeneous mutations, multi-kilobase integration and replacement technologies could act as single pan-mutation therapies for all genotypes.
There has been a recent expansion in the breadth of DSB-free technologies that generate nucleotide-level changes with higher precision, versatility, and programmability compared to prior approaches8. For example, base editors (BEs) typically facilitate the installation of A-to-G9,10 or C-to-T changes11,12 (ABEs and CBEs, respectively; Fig. 1b) within short sequence windows, as directed by the guide RNA (gRNA). BEs are comprised of fusions of adenine or cytosine deaminases to catalytically inactive or nicking variants of SpCas9 (nCas9) or other Cas orthologs, and they have been shown to mediate high levels of single nucleotide edits in primary human cells and in vivo for the treatment of human diseases13–17. More recently, prime editors (PEs) have been developed to permit user-defined sequence modifications via the fusion of nCas9 to a reverse transcriptase (RT), enabling the genetic writing of small edits that are pre-programmed on prime editor guide RNAs18 (pegRNAs; Fig. 1c). PEs can insert, substitute, or delete short sequences18, and have been shown to function in various cell types ex vivo and organisms in vivo, though with varying efficiencies19–23. Recent efforts to develop optimized prime editors21,24,25 and pegRNAs26, combined with future advances (e.g. understanding determinants of activity24,27,28, cell-specific optimizations, etc.) may lead to efficient prime editing in a variety of contexts. The development of BEs and PEs highlights a concentrated effort to engineer editors capable of small sequence edits, with applicability to correct a range of disease-causing nucleotide-level mutations using bespoke enzymes and gRNAs.
Despite the promise of small sequence editors, the fact that most diseases are caused by heterogenous mutations is an obstacle for clinical translation given the time and resources required to optimize safe and effective editing approaches. One potential solution to this bottleneck is large sequence editors capable of precisely inserting, deleting, inverting, translocating, or replacing kilobases of DNA without DSBs (Fig. 1d). In contrast to nuclease-, BE-, and canonical PE-based approaches that necessitate the design and optimization of new enzyme and gRNA combinations for treating any mutation (Fig. 1e), programmable integration would enable insertion of wild-type genes or cDNAs at endogenous genetic locations (concomitantly eliminating expression of the mutant gene) or direct replacement of mutated sequences, respectively. Such capabilities could act as genotype agnostic pan-mutation genetic therapies for individual diseases caused by various heterogenous mutations (Fig. 1e) including cystic fibrosis, Duchenne muscular dystrophy, amyotrophic lateral sclerosis, neurofibromatosis, Leber congenital amaurosis, and primary immunodeficiencies. Moreover, they would facilitate insertion of engineered genes, genetic elements, or circuits at specified locations for cell engineering applications (e.g. CAR-T cells). Targeted deletions and inversions (Fig. 1d) could treat diseases caused by duplications, nucleotide expansions (e.g. Huntington’s Disease, Fragile X syndrome) and certain cases of Hunter’s syndrome and Hemophilia, while targeted translocations could treat certain cases of Down’s syndrome and cancers. Thus, large sequence editors could obviate the burden of designing and optimizing custom enzyme and gRNA combinations for the vast diversity of heterogeneous pathogenic substitutions (Fig. 1e). Moreover, these technologies would create new basic research applications by enabling predictable cell engineering of structural variants as well as facilitating new screening and library-based approaches (including in situ saturation mutagenesis and deletion screens).
An ideal large-sequence editor would optimally have certain properties, including being: (1) able to generate a wide array of multi-kilobase edits at high efficiency and specificity without DSBs, (2) independent of HDR to improve cell-type applicability, (3) scarless without leaving residual sequences at the target locus, (4) compact in coding sequence for delivery via viruses, (5) tunable to user-defined parameters for a variety of applications, etc. Towards these ambitious goals, a suite of new technologies has begun to emerge, including programmable approaches and systems for precise deletion, integration, and inversion of genetic sequences.
Recent developments of next-generation deletion and short sequence replacement technologies have thus far largely been based on adapted prime editing methods. Instead of using a single pegRNA as done in the canonical approach, the use of paired pegRNAs offers additional versatility to encode and generate edits. For example, in the methods PrimeDel29 and twinPE30, the paired pegRNAs are utilized to install 3’ DNA flaps homologous to target site DNA or to each other, respectively, whose annealing and resolution leads to the programmed deletion or replacement of the intervening DNA sequence between the two nicks (Fig. 2a). Variations on this methodology have either relied on longer reverse transcription templates (RTTs) and substantial polymerase-mediated gap filling to increase insertion size31, or utilized nuclease Cas9 to promote repair of the flap-target DNA (though inherently introducing DSBs)32. Despite the promise of these approaches, their efficiency decreases as the deletion or replacement sizes increase since their mechanisms rely on 3’ DNA flap localization to a homologous segment of DNA or another 3’ flap located at the opposite end of the intended deletion/replacement site.
Fig. 2. Emerging approaches for kilobase-scale edits.
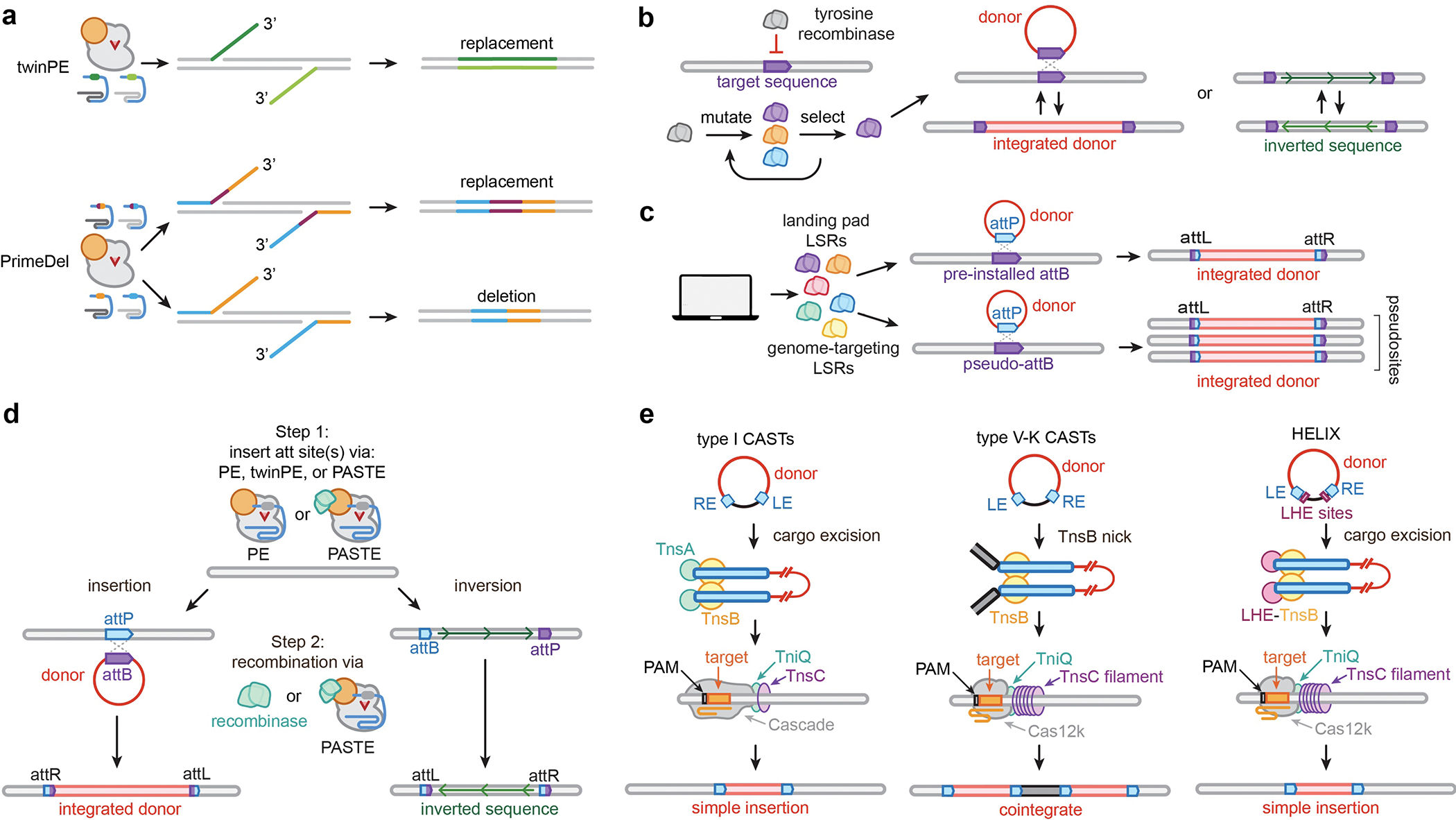
a, Paired pegRNA prime editing approaches, such as twinPE and PrimeDel, enable precise deletion and replacement of target sequences. b, Tyrosine recombinases can be evolved to recognize new therapeutically relevant target sites for site-specific donor recombination or inversion, which is a reversible process. c, Novel large serine recombinases (LSRs) have been discovered and characterized for use with pre-installed genomic landing pads (landing pad LSRs) or to integrate into predicted pseudosites (genome targeting LSRs). d, Two-step large sequence edits by combining PE or twinPE with site-specific serine recombinases (directly fused to the PE or co-expressed separately) can install programmable targeted integrations or inversions. e, RNA-guided DNA integration with type I or V-K CRISPR-associated transposases (CASTs), or engineered versions, such as HELIX.
Targeted integration and inversion technologies have largely focused on recombinases and transposases, though other recombineering-like approaches have also recently been developed33. Tyrosine recombinases have been evolved to recognize new target sites for targeted inversion and integration (Fig. 2b), expanding their therapeutic potential34–37. However, their reversible recombination mechanisms can limit their activity and utility. In contrast, site-specific serine recombinases act through an irreversible mechanism, renewing interest in their discovery and application in single-step reactions to integrate donor-encoded sequences into the human genome38 (Fig. 2c). However, reprogramming the target specificity of recombinases to defined loci is laborious, and there are specificity concerns about intentionally targeting predicted pseudosites given the number of times these occur in the genome39,40. Furthermore, dCas9-recombinase fusions are less efficient, and the recombinase catalytic domains can retain moderate sequence preferences, limiting reprogrammability to any user-defined site41. Despite these potential limitations, the discovery and characterization of additional recombinases has begun to yield new enzymes with unique and useful properties. Leveraging these datasets could lead to a machine-learning framework that can ab initio predict high probability recombinase enzyme variants with user-defined sequence specificities42.
In another recent approach for generating large sequence insertions, site-specific serine recombinases have been coupled with PEs in a multi-step process (Fig. 2d). PEs first are utilized to install recombinase attachment sites at defined genomic locations, and then site-specific recombinases are co-expressed or directly fused to the PE to recombine desired sequences into the genome30,43. While these PE-based approaches hold promise for targeted DNA integration (as well as targeted inversions and potentially replacements and translocations), these experiments require the extensive design and optimization of multiple parameters (i.e. target site, pegRNA components, recombinase, donor, etc.), the coding sequences of current enzyme complexes are very large (>8 kb coding size), the edits are not scarless, and current integrases remains less efficient compared to generating small sequence edits with other leading-edge technologies.
Transposons are another class of technology that offer unique properties for large DNA insertions (Fig. 2e). CRISPR-associated transposases (CASTs) are Tn7 or Tn5053-like transposons that have co-opted type I or type V CRISPR-Cas systems, respectively, for genetic element mobilization44–47. CASTs have garnered attention due to their high-efficiency RNA-guided DNA integration in bacteria. Beyond the expanded exploration of the aphylogenetic diversity of CASTs to uncover useful new characteristics48,49, derivative systems have been engineered that offer more streamlined use and optimal editing properties and capabilities50,51. For example, HELIX employs a nicking homing endonuclease fusion to TnsB of type V-K CASTs, enabling these systems to achieve comparable simple insertion product purity, specificity, and efficiency to type I systems51 (Fig. 2e). Furthermore, recent structural studies have provided unique insights into mechanisms of CASTs52–57, potentially motivating new engineering approaches to enhance their properties. While CAST-based technologies hold tantalizing potential to enable facile and efficient transposition as human therapeutics and for cell engineering, translation into eukaryotic cells has yet to be demonstrated. Alternatively, transposase enzymes have been directly fused to Cas DNA binding domains to localize transposition events to defined target sites. While the initial development of piggybac-dCas9 or sleeping-beauty-dCas9 fusions displayed significant off-target editing and lower efficiency58,59, recent adaptations have shown promise by fusing evolved piggybac variants with a Cas9 nuclease60 though this approach generates DSBs. Still, there remains a major need to optimize the efficiency, deliverability, and translatability of transposase-based technologies in therapeutically relevant contexts.
Despite this progress towards developing optimal next-generation kilobase editors, there exist several challenges for DSB-free technologies including: their relatively low editing efficiency, their complex design determinants, poorly characterized mechanisms, that certain types of kilobase-scale changes remain nascent or largely unexplored (e.g. translocations, replacements, etc), that most approaches leave undesirable sequence scars from recombined sites or transposon ends in gene insertion products, the large sizes of current-generation machinery, and the necessity to co-deliver donor molecules encoding genetic cargoes. To overcome these challenges, there exists potentially transformative opportunities for technological advances. For example, retrotransposases, which naturally integrate a reverse-transcribed RNA template, could be harnessed for direct insertion of RNA-encoded sequences into the genome61. However, some challenges include the fidelity of some retrotransposase RTs62,63, the potential for integration of 5’ truncated templates resulting from insufficient RT processivity and/or transcript degradation64, the programmability of retrotransposase specificity65–67, and others. Together, the discovery and engineering of other integration enzymes or methods with capabilities for scarless gene products and facile and efficient multiplexing would increase utility and create new approaches to cell engineering and gene therapy.
Similar to other genome editing technologies, the delivery of kilobase-scale editors for in vivo applications represents a challenge for the field68. Many of these nascent technologies editors have very large coding sequences and can require multiple sgRNAs or pegRNAs, which together can constrain delivery by size-constrained viral vectors69. Moreover, codelivery of donor molecules is required for edits involving integration or replacement of large sequences. Continued innovation in viral, nanoparticle, exosome, and virus-like particle technologies that package DNA, RNA, and RNP cargos, enable selective targeting to diseased tissues, and minimize immunogenicity will be crucial for all in vivo genome editing approaches68, particularly kilobase-scale alterations. Advances in the composition and size of the editing technologies themselves will also facilitate more effective delivery strategies.
Together, kilobase-scale DSB-free and HDR-independent technologies represent an exciting new frontier of genome modification. These tools hold promise for high impact as research reagents and for various applications. Continued optimization of these technologies might unlock their potential as blanket therapies to treat diseases caused by dispersed heterogeneous mutations within patient populations (Fig. 1e), as treatment strategies for currently intractable structural variants, as methods to facilitate engineering of therapeutic cells, and as tools for biological studies through variant modelling and screening approaches. Further metagenomic discovery will continue to reveal the vast diversity of enzymes that can be harnessed as kilobase-scale editors either on their own (e.g. new classes of RNA-guided systems), as more optimal components in current technologies (e.g. novel types of RNA-guided nucleases70–74), and/or in tandem with new approaches (e.g. PE-based systems). The continued interest in and development of these diverse but nascent technologies will refine and simplify our ability to precisely edit genomes at the kilobase scale.
Acknowledgements
We thank C.R.R. Alves, L.T. Hille, L. Ma, and J. Ferreira da Silva for suggestions about the manuscript. C.J.T. was supported by a National Science Foundation Graduate Research Fellowship Grant No. 2020295403. B.P.K was supported by a Mass General Hospital Howard M. Goodman Fellowship, the Gilbert Family Foundation’s Gene Therapy Initiative Grant No. 521004, and National Institutes of Health (NIH) awards R00-CA218870 and P01-HL142494.
Footnotes
Declaration of Interest
C.J.T. and B.P.K are inventors on patents and/or patent applications filed by Mass General Brigham that describe genome engineering technologies. B.P.K. is a consultant for EcoR1 Capital and is an advisor to Acrigen Biosciences, Life Edit Therapeutics, and Prime Medicine.
References
- 1.Yeh CD, Richardson CD & Corn JE Advances in genome editing through control of DNA repair pathways. Nature Cell Biology 21, 1468–1478 (2019). [DOI] [PubMed] [Google Scholar]
- 2.van Overbeek M, Capurso D, Carter MM, Thompson MS, Frias E, Russ C, Reece-Hoyes JS, Nye C, Gradia S, Vidal B, Zheng J, Hoffman GR, Fuller CK & May AP DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Molecular Cell 63, 633–646 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kosicki M, Tomberg K & Bradley A Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leibowitz ML, Papathanasiou S, Doerfler PA, Blaine LJ, Sun L, Yao Y, Zhang C-Z, Weiss MJ & Pellman D Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat Genet 53, 895–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alanis-Lobato G, Zohren J, McCarthy A, Fogarty NME, Kubikova N, Hardman E, Greco M, Wells D, Turner JMA & Niakan KK Frequent loss of heterozygosity in CRISPR-Cas9–edited early human embryos. PNAS 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enache OM, Rendo V, Abdusamad M, Lam D, Davison D, Pal S, Currimjee N, Hess J, Pantel S, Nag A, Thorner AR, Doench JG, Vazquez F, Beroukhim R, Golub TR & Ben-David U Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nature Genetics 52, 662–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgens DW, Wainberg M, Boyle EA, Ursu O, Araya CL, Tsui CK, Haney MS, Hess GT, Han K, Jeng EE, Li A, Snyder MP, Greenleaf WJ, Kundaje A & Bassik MC Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat Commun 8, 15178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nature Biotechnology 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI & Liu DR Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, Doudna JA & Liu DR Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nature Biotechnology 38, 883–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koblan LW, Doman JL, Wilson C, Levy JM, Tay T, Newby GA, Maianti JP, Raguram A & Liu DR Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nature Biotechnology 36, 843–846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webber BR, Lonetree C, Kluesner MG, Johnson MJ, Pomeroy EJ, Diers MD, Lahr WS, Draper GM, Slipek NJ, Smeester BA, Lovendahl KN, McElroy AN, Gordon WR, Osborn MJ & Moriarity BS Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nature Communications 10, 5222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy JM, Yeh W-H, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q & Liu DR Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nature Biomedical Engineering 4, 97–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong Z-M, Tavarez UL, Davison LM, Gete YG, Mao X, Newby GA, Doherty SP, Narisu N, Sheng Q, Krilow C, Lin CY, Gordon LB, Cao K, Collins FS, Brown JD & Liu DR In vivo base editing rescues Hutchinson–Gilford progeria syndrome in mice. Nature 1–7 (2021). doi: 10.1038/s41586-020-03086-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothgangl T, Dennis MK, Lin PJC, Oka R, Witzigmann D, Villiger L, Qi W, Hruzova M, Kissling L, Lenggenhager D, Borrelli C, Egli S, Frey N, Bakker N, Walker JA, Kadina AP, Victorov DV, Pacesa M, Kreutzer S, Kontarakis Z, Moor A, Jinek M, Weissman D, Stoffel M, van Boxtel R, Holden K, Pardi N, Thöny B, Häberle J, Tam YK, Semple SC & Schwank G In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nature Biotechnology 1–9 (2021). doi: 10.1038/s41587-021-00933-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musunuru K, Chadwick AC, Mizoguchi T, Garcia SP, DeNizio JE, Reiss CW, Wang K, Iyer S, Dutta C, Clendaniel V, Amaonye M, Beach A, Berth K, Biswas S, Braun MC, Chen H-M, Colace TV, Ganey JD, Gangopadhyay SA, Garrity R, Kasiewicz LN, Lavoie J, Madsen JA, Matsumoto Y, Mazzola AM, Nasrullah YS, Nneji J, Ren H, Sanjeev A, Shay M, Stahley MR, Fan SHY, Tam YK, Gaudelli NM, Ciaramella G, Stolz LE, Malyala P, Cheng CJ, Rajeev KG, Rohde E, Bellinger AM & Kathiresan S In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021). [DOI] [PubMed] [Google Scholar]
- 18.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A & Liu DR Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petri K, Zhang W, Ma J, Schmidts A, Lee H, Horng JE, Kim DY, Kurt IC, Clement K, Hsu JY, Pinello L, Maus MV, Joung JK & Yeh J-RJ CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat Biotechnol 40, 189–193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang H, Jo DH, Cho CS, Shin JH, Seo JH, Yu G, Gopalappa R, Kim D, Cho S-R, Kim JH & Kim HH Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat Biomed Eng 6, 181–194 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Liu P, Liang S-Q, Zheng C, Mintzer E, Zhao YG, Ponnienselvan K, Mir A, Sontheimer EJ, Gao G, Flotte TR, Wolfe SA & Xue W Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat Commun 12, 2121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böck D, Rothgangl T, Villiger L, Schmidheini L, Matsushita M, Mathis N, Ioannidi E, Rimann N, Grisch-Chan HM, Kreutzer S, Kontarakis Z, Kopf M, Thöny B & Schwank G In vivo prime editing of a metabolic liver disease in mice. Science Translational Medicine 14, eabl9238 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newby GA & Liu DR In vivo somatic cell base editing and prime editing. Molecular Therapy (2021). doi: 10.1016/j.ymthe.2021.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen P-F, Chen C, Nelson JW, Newby GA, Sahin M, Osborn MJ, Weissman JS, Adamson B & Liu DR Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 184, 5635–5652.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu B, Dong X, Cheng H, Zheng C, Chen Z, Rodríguez TC, Liang S-Q, Xue W & Sontheimer EJ A split prime editor with untethered reverse transcriptase and circular RNA template. Nat Biotechnol 1–6 (2022). doi: 10.1038/s41587-022-01255-9 [DOI] [PubMed] [Google Scholar]
- 26.Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An M, Newby GA, Chen JC, Hsu A & Liu DR Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol 1–9 (2021). doi: 10.1038/s41587-021-01039-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HK, Yu G, Park J, Min S, Lee S, Yoon S & Kim HH Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol 39, 198–206 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Ferreira da Silva J, Oliveira GP, Arasa-Verge EA, Kagiou C, Moretton A, Timelthaler G, Jiricny J & Loizou JI Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat Commun 13, 760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi J, Chen W, Suiter CC, Lee C, Chardon FM, Yang W, Leith A, Daza RM, Martin B & Shendure J Precise genomic deletions using paired prime editing. Nat Biotechnol 1–9 (2021). doi: 10.1038/s41587-021-01025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anzalone AV, Gao XD, Podracky CJ, Nelson AT, Koblan LW, Raguram A, Levy JM, Mercer JAM & Liu DR Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol 40, 731–740 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, He Z, Wang G, Zhang R, Duan J, Gao P, Lei X, Qiu H, Zhang C, Zhang Y & Yin H Efficient targeted insertion of large DNA fragments without DNA donors. Nat Methods 19, 331–340 (2022). [DOI] [PubMed] [Google Scholar]
- 32.Jiang T, Zhang X-O, Weng Z & Xue W Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol 1–8 (2021). doi: 10.1038/s41587-021-01026-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Qu Y, Cheng JKW, Hughes NW, Zhang Q, Wang M & Cong L dCas9-based gene editing for cleavage-free genomic knock-in of long sequences. Nat Cell Biol 24, 268–278 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarkar I, Hauber I, Hauber J & Buchholz F HIV-1 Proviral DNA Excision Using an Evolved Recombinase. Science 316, 1912–1915 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Karpinski J, Hauber I, Chemnitz J, Schäfer C, Paszkowski-Rogacz M, Chakraborty D, Beschorner N, Hofmann-Sieber H, Lange UC, Grundhoff A, Hackmann K, Schrock E, Abi-Ghanem J, Pisabarro MT, Surendranath V, Schambach A, Lindner C, van Lunzen J, Hauber J & Buchholz F Directed evolution of a recombinase that excises the provirus of most HIV-1 primary isolates with high specificity. Nat Biotechnol 34, 401–409 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Lansing F, Paszkowski-Rogacz M, Schmitt LT, Schneider PM, Rojo Romanos T, Sonntag J & Buchholz F A heterodimer of evolved designer-recombinases precisely excises a human genomic DNA locus. Nucleic Acids Research 48, 472–485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lansing F, Mukhametzyanova L, Rojo-Romanos T, Iwasawa K, Kimura M, Paszkowski-Rogacz M, Karpinski J, Grass T, Sonntag J, Schneider PM, Günes C, Hoersten J, Schmitt LT, Rodriguez-Muela N, Knöfler R, Takebe T & Buchholz F Correction of a Factor VIII genomic inversion with designer-recombinases. Nat Commun 13, 422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durrant MG, Fanton A, Tycko J, Hinks M, Chandrasekaran SS, Perry NT, Schaepe J, Du PP, Lotfy P, Bassik MC, Bintu L, Bhatt AS & Hsu PD Large-scale discovery of recombinases for integrating DNA into the human genome. 2021.11.05.467528 Preprint at 10.1101/2021.11.05.467528 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS & Calos MP Site-Specific Genomic Integration in Mammalian Cells Mediated by Phage φC31 Integrase. Molecular and Cellular Biology 21, 3926–3934 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Z, Chen L, Jia C, Zhu H, Wang W & Zhong J Screening of potential pseudo att sites of Streptomyces phage ΦC31 integrase in the human genome. Acta Pharmacol Sin 34, 561–569 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaikind B, Bessen JL, Thompson DB, Hu JH & Liu DR A programmable Cas9-serine recombinase fusion protein that operates on DNA sequences in mammalian cells. Nucleic Acids Research 44, 9758–9770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt LT, Paszkowski-Rogacz M, Jug F & Buchholz F Prediction of designer-recombinases for DNA editing with generative deep learning. 2022.04.01.486669 Preprint at 10.1101/2022.04.01.486669 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ioannidi EI, Yarnall MTN, Schmitt-Ulms C, Krajeski RN, Lim J, Villiger L, Zhou W, Jiang K, Roberts N, Zhang L, Vakulskas CA, Walker JA, Kadina AP, Zepeda AE, Holden K, Gootenberg JS & Abudayyeh OO Drag-and-drop genome insertion without DNA cleavage with CRISPR-directed integrases. 2021.11.01.466786 (2021). doi: 10.1101/2021.11.01.466786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters JE, Makarova KS, Shmakov S & Koonin EV Recruitment of CRISPR-Cas systems by Tn7-like transposons. PNAS 114, E7358–E7366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klompe SE, Vo PLH, Halpin-Healy TS & Sternberg SH Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV & Zhang F RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saito M, Ladha A, Strecker J, Faure G, Neumann E, Altae-Tran H, Macrae RK & Zhang F Dual modes of CRISPR-associated transposon homing. Cell 184, 2441–2453.e18 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rybarski JR, Hu K, Hill AM, Wilke CO & Finkelstein IJ Metagenomic discovery of CRISPR-associated transposons. Proceedings of the National Academy of Sciences 118, e2112279118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klompe SE, Jaber N, Beh LY, Mohabir JT, Bernheim A & Sternberg SH Evolutionary and mechanistic diversity of Type I-F CRISPR-associated transposons. Molecular Cell (2022). doi: 10.1016/j.molcel.2021.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vo PLH, Ronda C, Klompe SE, Chen EE, Acree C, Wang HH & Sternberg SH CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nature Biotechnology 1–10 (2020). doi: 10.1038/s41587-020-00745-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tou CJ, Orr B & Kleinstiver BP Cut-and-Paste DNA Insertion with Engineered Type V-K CRISPR-associated Transposases. 2022.01.07.475005 Preprint at 10.1101/2022.01.07.475005 (2022) [DOI] [PubMed] [Google Scholar]
- 52.Halpin-Healy TS, Klompe SE, Sternberg SH & Fernández IS Structural basis of DNA targeting by a transposon-encoded CRISPR–Cas system. Nature 577, 271–274 (2020). [DOI] [PubMed] [Google Scholar]
- 53.Park J-U, Tsai AW-L, Mehrotra E, Petassi MT, Hsieh S-C, Ke A, Peters JE & Kellogg EH Structural basis for target site selection in RNA-guided DNA transposition systems. Science 373, 768–774 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Querques I, Schmitz M, Oberli S, Chanez C & Jinek M Target site selection and remodelling by type V CRISPR-transposon systems. Nature 599, 497–502 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen Y, Gomez-Blanco J, Petassi MT, Peters JE, Ortega J & Guarné A Structural basis for DNA targeting by the Tn7 transposon. Nat Struct Mol Biol 29, 143–151 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Schmitz M, Querques I, Oberli S, Chanez C & Jinek M Structural basis for RNA-mediated assembly of type V CRISPR-associated transposons. 2022.06.17.496590 Preprint at 10.1101/2022.06.17.496590 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park J-U, Tsai AW-L, Chen TH, Peters JE & Kellogg EH Mechanistic details of CRISPR-associated transposon recruitment and integration revealed by cryo-EM. Proceedings of the National Academy of Sciences 119, e2202590119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hew BE, Sato R, Mauro D, Stoytchev I & Owens JB RNA-guided piggyBac transposition in human cells. Synthetic Biology 4, ysz018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kovač A, Miskey C, Menzel M, Grueso E, Gogol-Döring A & Ivics Z RNA-guided retargeting of Sleeping Beauty transposition in human cells. eLife 9, e53868 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pallarès-Masmitjà M, Ivančić D, Mir-Pedrol J, Jaraba-Wallace J, Tagliani T, Oliva B, Rahmeh A, Sánchez-Mejías A & Güell M Find and cut-and-transfer (FiCAT) mammalian genome engineering. Nat Commun 12, 7071 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bothmer AH, COTTA-RAMUSINO CGS, Salomon WE, RUBENS JR, Citorik RJ, WANG ZJ, Kim K, KOTLAR RM, RAY A, ALTSHULER RC, Kumar S, Roquet N & Steinberg BE Methods and compositions for modulating a genome. (2021). at <https://patents.google.com/patent/WO2021178720A2/en> [Google Scholar]
- 62.Boutabout M, Wilhelm M & Wilhelm F-X DNA synthesis fidelity by the reverse transcriptase of the yeast retrotransposon Ty1. Nucleic Acids Research 29, 2217–2222 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jamburuthugoda VK & Eickbush TH The Reverse Transcriptase Encoded by the Non-LTR Retrotransposon R2 Is as Error-Prone as That Encoded by HIV-1. Journal of Molecular Biology 407, 661–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D & Boeke JD Molecular archeology of L1 insertions in the human genome. Genome Biology 3, research0052.1 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grandi FC & An W Non-LTR retrotransposons and microsatellites. Mobile Genetic Elements 3, e25674 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fujiwara H Site-specific non-LTR retrotransposons. Microbiology Spectrum 3, 3.2.09 (2015). [DOI] [PubMed] [Google Scholar]
- 67.Sultana T, Essen D. van, Siol O, Bailly-Bechet M, Philippe C, Aabidine AZE, Pioger L, Nigumann P, Saccani S, Andrau J-C, Gilbert N & Cristofari G The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-insertion Sequence Biases and Post-insertion Selection. Molecular Cell 74, 555–570.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Raguram A, Banskota S & Liu DR Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li C & Samulski RJ Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 21, 255–272 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF & Doudna JA CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369, 333–337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pausch P, Soczek KM, Herbst DA, Tsuchida CA, Al-Shayeb B, Banfield JF, Nogales E & Doudna JA DNA interference states of the hypercompact CRISPR–CasΦ effector. Nat Struct Mol Biol 28, 652–661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Altae-Tran H, Kannan S, Demircioglu FE, Oshiro R, Nety SP, McKay LJ, Dlakić M, Inskeep WP, Makarova KS, Macrae RK, Koonin EV & Zhang F The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science 374, 57–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karvelis T, Druteika G, Bigelyte G, Budre K, Zedaveinyte R, Silanskas A, Kazlauskas D, Venclovas Č & Siksnys V Transposon-associated TnpB is a programmable RNA-guided DNA endonuclease. Nature 599, 692–696 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schuler G, Hu C & Ke A Structural basis for RNA-guided DNA cleavage by IscB-ωRNA and mechanistic comparison with Cas9. Science 0, eabq7220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]


