Abstract
The explosion of microbiome research over the past decade has shed light on the various ways that external factors interact with the human microbiome to drive health and disease. Each individual is exposed to more than 300 environmental chemicals every day. Accumulating evidence indicates that the microbiome is involved in the early response to environmental toxicants and biologically mediates their adverse effects on human health. However, few review articles to date provided a comprehensive framework for research and translation of the role of the gut microbiome in environmental health science. This review summarizes current evidence on environmental compounds and their effect on the gut microbiome, discusses the involved compound metabolic pathways, and covers environmental pollution-induced gut microbiota disorders and their long-term outcomes on host health. We conclude that the gut microbiota may crucially mediate and modify the disease-causing effects of environmental chemicals. Consequently, gut microbiota needs to be further studied to assess the complete toxicity of environmental exposures. Future research in this field is required to delineate the key interactions between intestinal microbiota and environmental pollutants and further to elucidate the long-term human health effects.
Keywords: Gut microbiome, Environmental chemicals, Human host, Mediation role
1. Introduction: the problem
Every year, humans are exposed to more than 600 pounds of potentially toxic chemicals that are released into the air, ground, and water throughout the world. The U.S. Environmental Protection Agency (EPA) estimates that there are 85,000 compounds currently in use, with the average of 2000 newer chemicals on average are added to the market annually (U.S. Department of Health and Human Services, Centers for Disease Control and Prevention., 2021). Environmental health researchers announced that the environment has been extensively contaminated by not only synthetic but also natural chemicals and the impacts of this chemical pollution on human health is a growing concern worldwide (Chiu et al., 2020a). These chemicals are found in common everyday stuffs, such as containers, plastic bottles, and food boxes, and are destined to be more and more abundant in coming years and decades (Rosenfeld, 2017). Increasing research is focusing on the gut microbiome as a possible target of this toxicity because of its sensitivity to chemical pollution. New evidence shows that gut microbiome has been increasingly involved in an early response system to environmental toxics and a potential but likely critical mediation of their adverse effects on human health (Cho & Blaser, 2012; Claus et al., 2016(a), 2016(b); E. and M. National Academies of SciencesE., 2018; Rude, Keogh, et al., 2019).
Indeed, emergent research indicates that environmental chemicals affect gut microbial communities and, the gut microbiome may influence, directly or indirectly, xenobiotic metabolisms and subsequent impact on human health (Lu et al., 2015). This cycle may contribute to adverse health consequences caused both by environmental exposures and gut microbiome disruption. While both exposure to environmental pollutants and gut microbiome disturbances have been associated with various diseases, including but not limited to obesity, inflammatory bowel diseases, and colon cancer (Claus et al., 2016a; Tsiaoussis et al., 2019), the interaction between these pollutants and the GI (gastrointestinal) microbiota and its role in both health and disease have not been clearly elucidated.
The gut microbiome is “term that embraces all the microorganisms present in the human gastrointestinal system, so all the bacteria, archaea, viruses, and fungi and their overall genetic information” (E. and M. National Academies of Sciences, 2018). Of note, the gut microbiota’s genome contains over 3.3 million genes, more than 100 times higher than the genome of a typical human cell (Rowland et al., 2018). Therefore, the gut microbiome has been slowly recognized as a possible “new organ system” that significantly affects human physiology. In healthy conditions, there is constant crosstalk between the host and the microbiota, in order to keep a healthy symbiosis and avoid the over-growth of potentially pathogenic bacteria. However, this equilibrium may be interrupted if any insult, such as an environmental toxin, produce an imbalance among the beneficial and opportunistic bacteria, exposing the gut microbiome to possible pathogenic and prolonged variations (DeGruttola et al., 2016). Consequently, gut dysbiosis, that may be then correlated with disease (Eggers et al., 2018; Tamboli et al., 2004; Wang et al., 2019), can occur for three principal causes: (1) shortage of beneficial flora; (2) decreased diversity; (3) competition of the opportunistic and commensal flora (Humphreys, 2020).
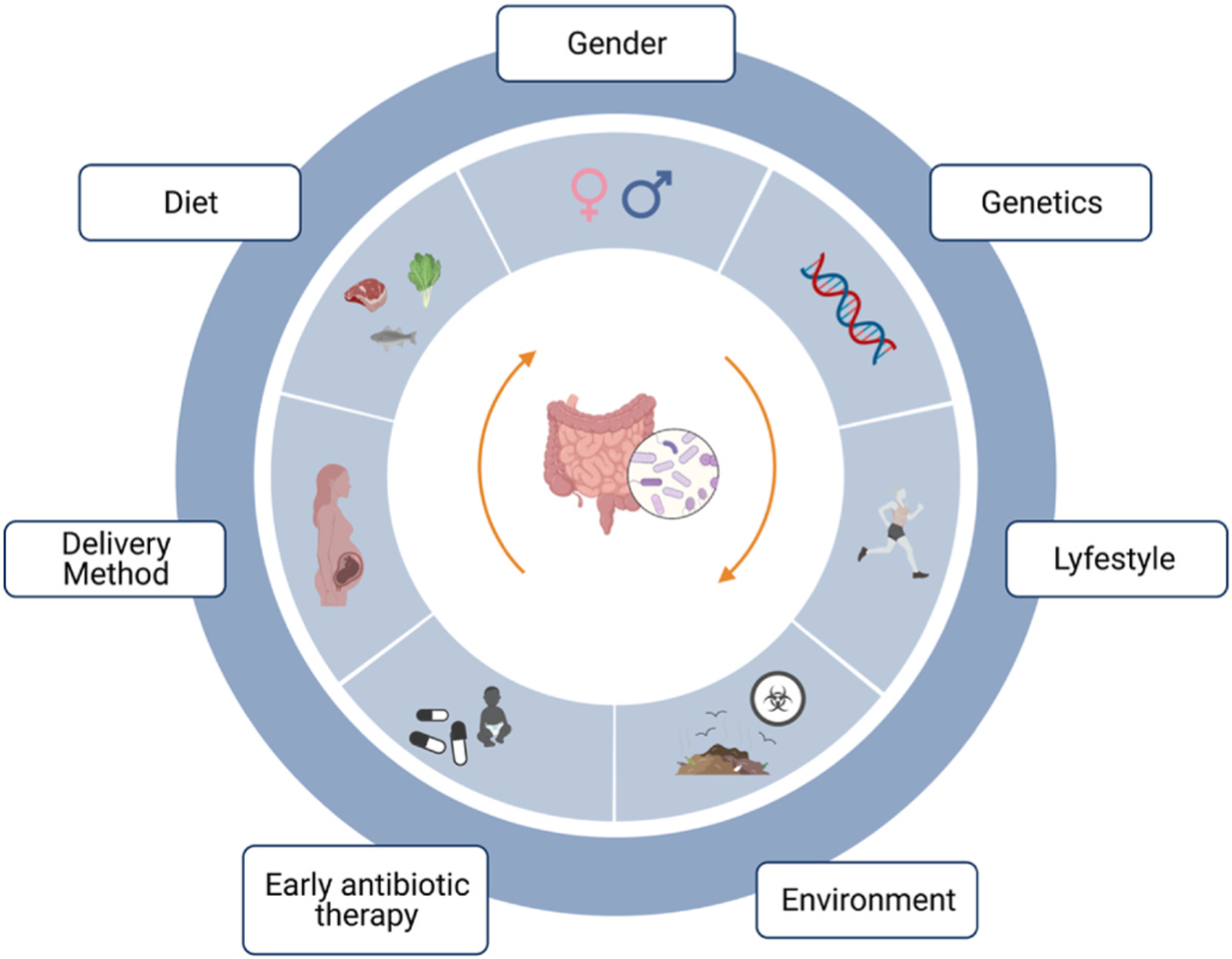
Gut microbial diversity is shaped very early in life and remains relatively stable in adulthood. However, throughout one’s lifetime, the gut microbiome is modulated by various endogenous (e.g., genotype, gender and age) and exogenous factors (e.g., diet composition, socio-economic status, mode of delivery, early antibiotic use, and exposure to environmental xenobiotics) (Fig. 1) (Chu et al., 2016; Dethlefsen & Relman, 2011; Gacesa et al., 2020; Gibson et al., 2016; Tsiaoussis et al., 2019). Each human’s gut microbiome has a distinct, much like a “bacterial fingerprint” (Tsiaoussis et al., 2019). In spite of such diversity, only a little fraction of the variation among people is explained by the known factors (Schmidt et al., 2018; Tsiaoussis et al., 2019).
Fig. 1. Factors that contribute to variation of human gut microbiota: genotype, early antibiotic therapy, diet composition, lifestyle, gender, delivery method and environmental exposure.a.
Although there are published review on gut microbiome and xenobiotics’ metabolism (Chiu et al., 2020a; N. Li et al., 2021; Tsiaoussis et al., 2019), these topics have not been extensively discussed, with few current articles on the role of the microbiome as a mediator of environmental exposures on human health. This review article intends to fill these gaps and offer up-to-date information on the metabolic pathways and human health disorders that can be mediated by disrupted gut microbiome in response to toxicants exposure. We discuss current knowledge and discuss potential interactions between human microbiome and overall human health. Finally, we review biological mechanisms, compound metabolic pathological pathways, and health outcomes associated with most common environmental exposures.
For this review, we used the PubMed database to search for relevant studies published between 2004 and 2022 using the following keywords: “gut microbiota”, “microbiome”, “chemicals”, “xenobiotics”, “human diseases”, “obesity”, “diabetes”, “cardiovascular diseases”, “neurologic/al disorders”, “biological pathways”, “metabolomics”, “metagenomics”. Both animal and human studies were included. Search results were excluded if there were not correlated to the subject of the study or not written in English.
1.1. Beyond traditional techniques: new tools for microbiome analysis
One of the earliest techniques to better understand the role of gut microbiome in response to environmental exposures is amplicon sequencing (Hamady & Knight, 2009). However, next-generation sequencing platforms are the newest and most promising technologies for high-resolution microbiome studies. Decreased average costs also have allowed more common use of the un-targeted method of shotgun metagenome sequencing, currently enabling sequencing of millions of reads per sample. High throughput methods have also recently been developed to comprehensively detect longer reads from metagenomic sequencing to RNA meta transcriptomes (Franzosa et al., 2014). All these technological advances have opened new paths to determine the function of gut microbiome in the toxicity of chemicals, developing the field of genomic toxicology to a larger metagenomic toxicology research area (Mesnage et al., 2018). More recently, other remarkable evolutions have been made on metabolomic and meta-proteomic practices. These cutting-edge methodologies permit a more accurate quantification and identification of cellular proteins and their post-translational alterations (Soufi & Soufi, 2016), and also provide valuable information on the microbiome characteristics and interaction of the microbial community with the host environment. Ideally, the study of metabolomics can help to develop biomarkers of environmental stressors (Aguiar-Pulido et al., 2016; Franzosa et al., 2015)
2. Principal biological mechanisms mediated by gut microbiota in response to environmental chemicals exposure
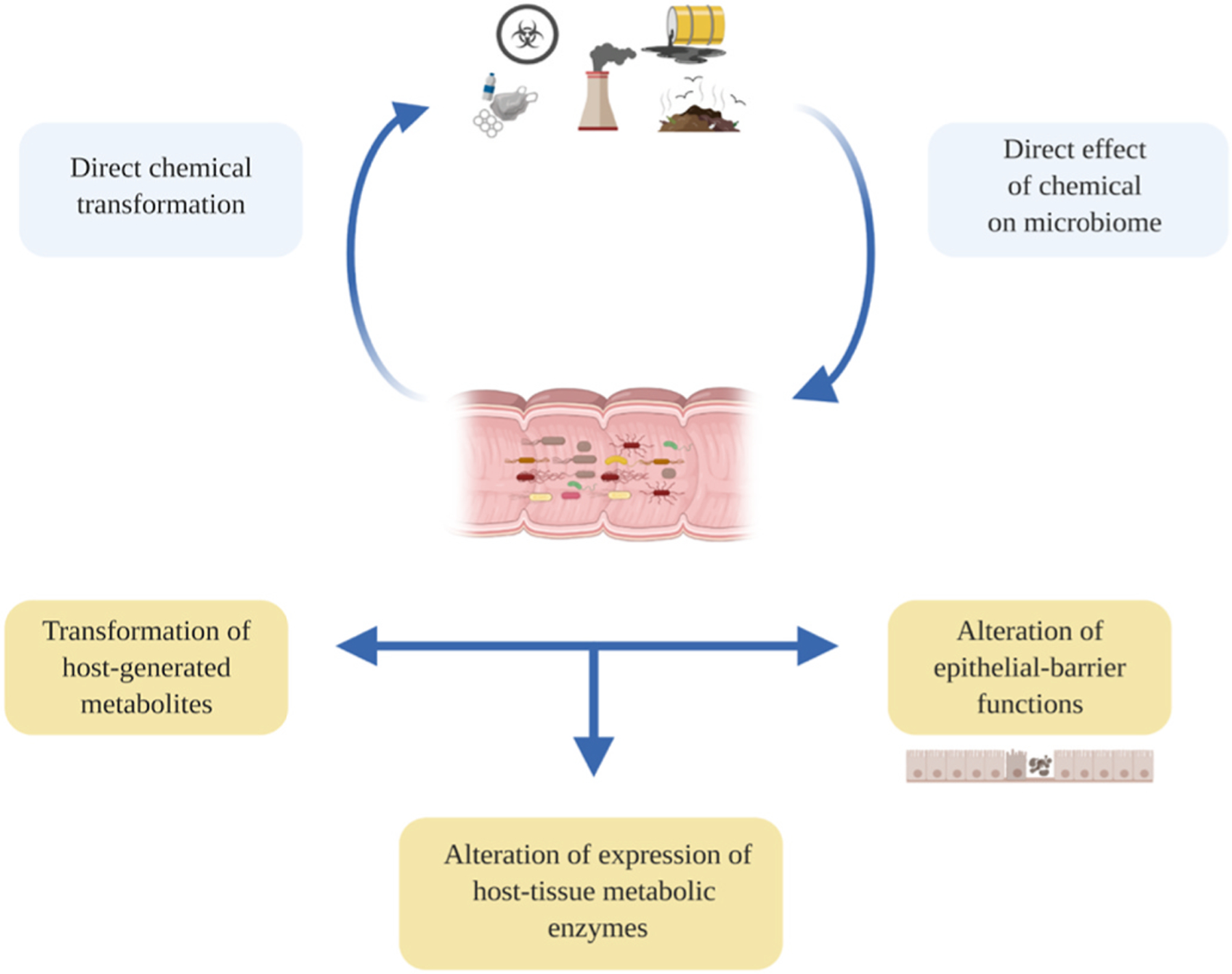
There are various mechanisms by which gut microbiota may modulate, directly or indirectly, the interaction between chemical exposures and host response. (Fig. 2). Several studies indicate that environmental chemicals exposure could modify microbes affecting their function and composition. Alternatively, microbial metabolism of chemicals may favorably or unfavorably affect the host through metabolic byproducts or modulation of compound toxicity. These processes include both direct metabolic transformations and secondary transformations including deconjugation of metabolites generated by the host, regulation of epithelial permeability, and control of the expression of key metabolic pathways. Notably, the first mechanism may interfere with other indirect mechanisms that are arbitrated down the line by the microbiome.
Fig. 2. Principal mechanisms by which the microbiome may directly or indirectly modulate the toxicants. Source: Adapted from National Academies Press (2018).
For example, the gut microbiome may first metabolically transform environmental chemicals directly - a mouse study shows that exposure to arsenic from drinking water can robustly and broadly disrupt gut microbes and dysregulate both bile acid and indole levels, which are usually directly created or modified by gut bacteria (Lu et al., 2014). Second, chemical reactions that involve the biotransformation of toxicants from gut bacteria are possible. They include both reduction and hydrolysis reactions (Claus et al., 2016a). In fact, some human and animal microbiome gut data indicated that gut microbiome modifies the biotoxicity of toxicants, like heavy metals, in several ways (Diaz-Bone & van de Wiele, 2009). For instance, whole methylation of inorganic arsenic to dimethyl arsenic in humans is catalyzed by methyltransferase activity. However, studies that used human bacteria revealed that after reduction and methylation, inorganic arsenic can be alternatively converted to intermediate forms, some of which are even more noxious than the original metalloid (Van de Wiele et al., 2010).
Secondary transformations can involve disruption of the gut microbiome epithelial barrier. Some studies showed that after with either gnotobiotic nurturing, antibiotic or probiotic treatment, gut microbiome has altered intestinal permeability similar to exposure to stress, infection, and diet changes (Everard et al., 2013; Leclercq et al., 2014). Indeed, introduction of a new bacterial species called Akk. muciniphila in a mouse study increased the expression of both some crucial tight-junctions proteins that are correlated with decreased intestinal absorbency capacity (Everard et al., 2013; Plovier et al., 2017). These findings were confirmed by another study using a mouse model of autism spectrum disorder, in which administration of the Bacteroides fragilis species produced higher genomic expression of two claudins proteins (8 and 15) linked with declining GI barrier integrity (Hsiao et al., 2013). Using metabolomics analysis in the urine mice, Zhao et al. investigated the toxic effects of Chlorpyrifos and concluded they it leads to a sustained level of intestinal inflammation and disrupted permeability of GI cells (Zhao et al., 2016). Proof of the role of the gut microbiota in regulating chemical metabolic pathways is instead provided in some recent RNA sequencing analyses of the intestinal and liver epithelia (C. Y. Li et al., 2016; Selwyn et al., 2016). For instance, the first evaluation of gene expression of livers among control mice and bacterial-free mice disclosed significant divergences in the expression of genes involved in the liver metabolism of chemicals (Selwyn et al., 2015). Importantly, CYP3 expression was drastically decreased under bacterial-free conditions, and subsequent microbes was able to increase expression levels comparable to control mice (E. and M. National Academies of SciencesE., 2018; Selwyn et al., 2016). Those observations are relevant because regulation of CYP3a expression arises via the Pregnane X signaling, a crucial receptor, since it is considered one of the key biochemical mediators among the microbiome and the host (Björkholm et al., 2009).
Secondary transformations include conjugation reactions (e.g., deconjugation) intermediated by host liver enzymes. These reactions that contribute to detoxification of environmental toxicants but can be reversed by microbial enzymes. A prominent example of deconjugation of metabolites generated by the host is the β-glucuronidase enzyme activity, which was already linked to metabolism of nitrated PAHs (E. and M. National Academies of Sciences, 2018). After inhalation exposure, the PAH 2-nitrofluorene (2-NF) is metabolized to hydroxylated nitrofluorenes (OH-NFs) (Claus et al., 2016b). After they enter into body, OH-NFs can be further detoxified and excreted as a glucuronide conjugate, but a microbial beta-glucuronidase enzyme is able to restimulate the synthesis of OH-NFs, increasing risk intestine cancer. Interestingly, the same compound is acetylated after oral exposure, and after further hydroxylation by gut microbiota, is transformed to metabolites that have, instead, fewer mutagenic potential and are ultimately fully expelled (E. National Academies of Sciences and Medicine, 2018). Also, a metagenome-wide detection approach described by Das et al. confirmed the occurrence of several homologues conjugation genes, such as glutathione S-transferases and N-acetyltransferases in human gut bacteria, reinforcing a potential role for these microbial enzymes in detoxification pathways (Das et al., 2016). In summary, we are only beginning to understand the extensive influence of microbes’ metabolic activity on environmental chemicals. While environmental chemicals can directly affect the microbiome itself, disturbances in the composition or richness of a bacterial species also can potentially affect all other pathways influenced by the microbiome. Conceptually, every single interaction can be favorable or unfavorable. Thus, it is perhaps not surprising that the role of the gut microbiome in altering toxicity of environmental exposures remain controversial.
3. Metabolic pathways underlying environmental pollutants and GI microbiota, and human diseases interaction
Here we provide some biological insights about how environmental toxicants may contribute to gut microbiome-associated diseases, including their potential metabolic pathways (Fig. 3). Although not every change in the microbiome associated with chemical exposure is harmful by definition, many may contribute to the onset or development of human-associated diseases (Tu et al., 2020). Understanding of xenobiotic bacteria-induced metabolism is essential to evaluate the potential effects on environmental pollutants through several interconnected pathways (Fig. 3). For example, environmental exposures that reach the gut through ingestion can alter gut microbiota composition, supporting growth of more opportunistic bacteria while harming beneficial ones. Lu et al. (2014) reported that exposure to arsenic from water alters the composition of gut microbiota of mice by decreasing some Firmicutes families. Further, male fish exposed to PFBS show increased abundance of Cetobacterium spp. Compared with controls (Chen et al., 2018). In an additional animal study, rats exposed to PFOS though their diet showed marked modulations in the richness of Firmicutes, Bacteroidetes, Proteobacteria, and Cyanobacteria bacteria phyla (Lai et al., 2018). In humans, Shen et el. (2022) recently reported that higher childhood blood Cadmium was positively associated with Flavonifractor plautii, a potential pathogen, already linked to worse Social Responsiveness Scale scores in young children (Laue et al., 2020; Ogita et al., 2020; Shen et al., 2022). Further, a human study from the Norwegian Microbiota Cohort showed that PFOS exposure in the breast milk is associated with both lower alpha diversity of the infant gut microbiome and greater relative abundance of Enterococcus ssp., underlining that PFOS exposure may influence infant gut microbial composition and functioning also during a critical developmental window (Iszatt et al., 2019). Another population analysis showed that continuous use of a TCS-based toothpaste is associated with a notable rise in the abundance of Proteobacteria spp. in the stools, together with a subsequent higher concentration in the urine of infants included in the cohort, probably through breast milk exposure (Ribado et al., 2017). The gut microbiome of these infants also showed decreased alpha diversity after their mother started using TCS-based toothpaste in their daily routine (N. Li et al., 2021). This evidence highlights the critical role of microbiome in chemicals exposure and its harmful effects on GI bacteria and their function. Bacterial imbalance and dysbiosis can disturb bacterial metabolisms, which can produce reduced production and circulation of SCFAs and bile acids, sustained by the increase prevalence of bacterial toxins and their metabolites. For instance, Nicholson at al. (2012) reported decreased biosynthesis of SCFAs in response to arsenic exposure (Nicholson et al., 2012), while Liu et al. used metagenomic sequencing to show reduced SCFA levels in the colon after cadmium administration in mice. Another mouse study showed that administration of Parabacteroides distasonis species is associated with alteration of the bile acid metabolism, specifically elevated Lithocholic acids and Ursodeoxycholic acids, and succinate levels in the gut. Interestingly, increased levels of these metabolites can improve obesity, metabolic dysfunction, and tumor formation in mice fed with high-fat diet (Koh et al., 2018; Wang et al., 2019). Similarly, bacterial fermentation of some proteins produce metabolites such as N-nitroso that can induce mutagenic effects, especially colorectal tumor development (Tu et al., 2020). In fact, in another animal study has shown that TCS exposure significantly interferes with microbiome, consequently increasing inflammation of the colon and even colon cancer risk (Gao et al., 2017; N. Li et al., 2021; Yang et al., 2018) (see Fig. 4).
Fig. 3. Effects of environmental contaminants on gut microbiome and their subsequent consequences on the human host. Dotted arrows indicate that the chemicals can altern gut microbiota composition, barrier and production of metabolites such as TMA (Trimethylamine), BA (Bile Acids), TMAO (Trimethylamine oxide), LPS (Lipopolysaccharide), SCFA (Short Chain Fatty acids), IL-6 (Interleukin-6), IL-1β (Interleukin-1 beta), and TNF-α (Tumor Necrosis Factor alpha). Further, both the inflammation and the dysbiosis on microbiome and the actions of the metabolites could produce adverse health outcomes such as cardiovascular, metabolic, and neurobehavioral disorders (solid arrows).
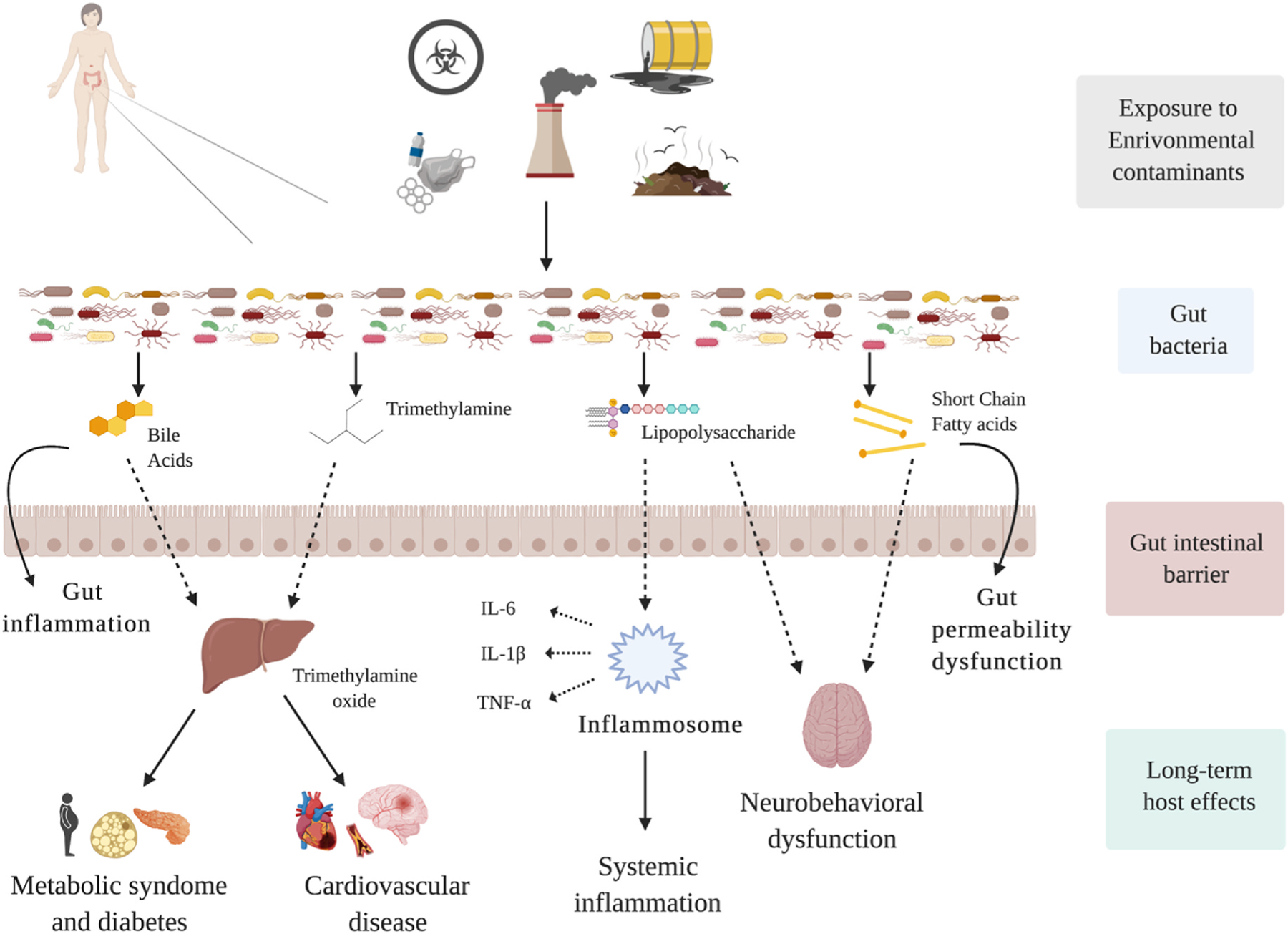
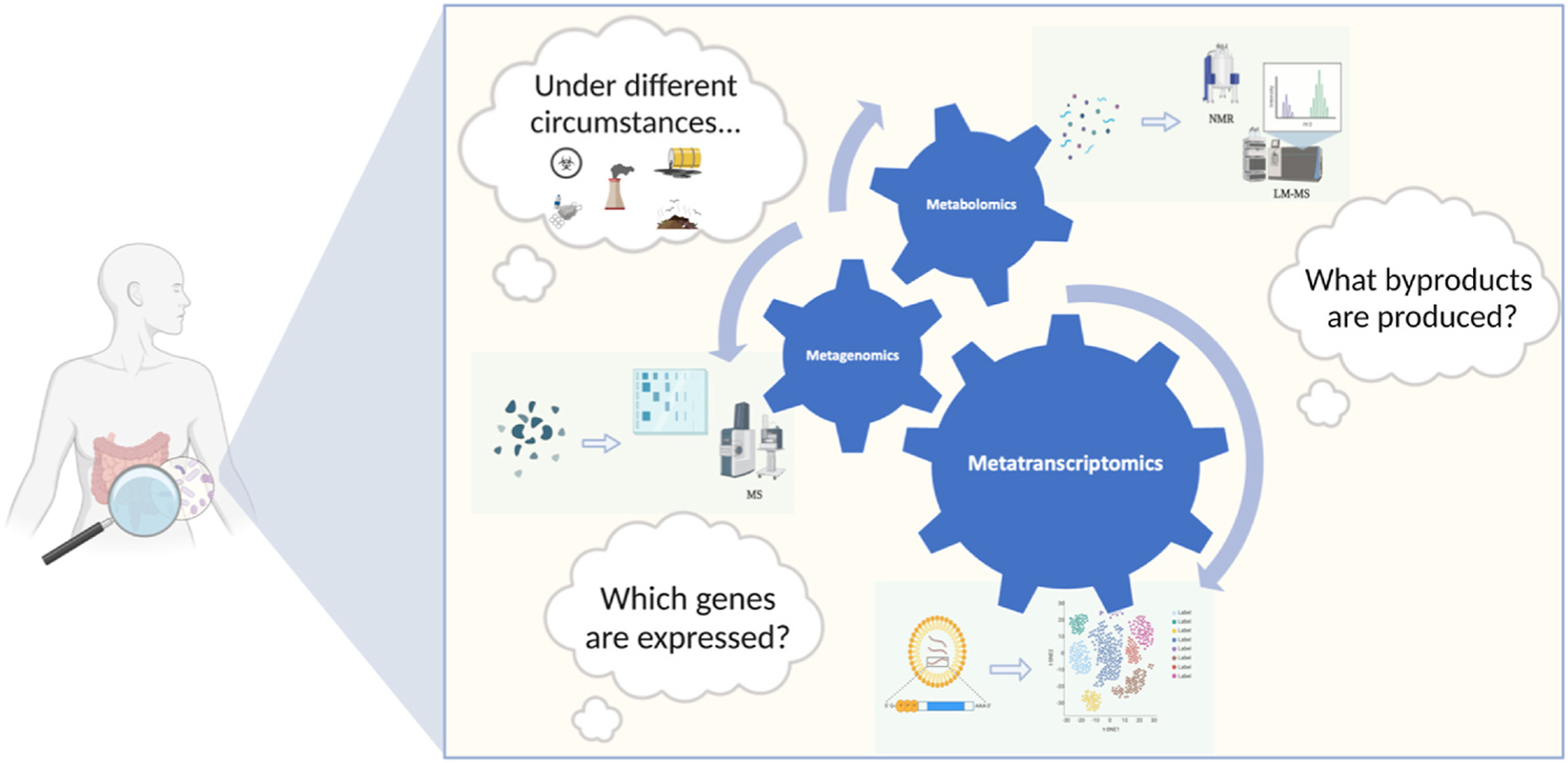
Fig. 4. Molecular methods to study microbiome-host interfaces from DNA and RNA-based approaches, to emergent metaproteomics and metabolomics in order to evaluate microbial gene expression, taxonomic profiles, and deeper-level genomes. Newer approaches include both mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectrometry, which are the most used non-sequencing-based methods for molecular-level investigation of the human gut microbiome. Source: Adapted from Ilhan (2016).
Moreover, chemicals triggers may impair the functions of GI epithelial cells by upregulation of TLR-4, which can activate inflmmatory paths with cytokine storms and production of harmful ROS (Yuan et al., 2019a). Indeed, Gao et al. recently used mice to explore the effects of lead on the microbiome via both whole-genome metagenomics and metabolomics sequencing, indicating that lead exposure can affect the uniformity of the microbiome and of several associated biochemical pathways including oxidative stress and detoxification functions (Assefa & Köhler, 2020; Gao et al., 2017) Due to inflammation activation, the lipo-polysaccharide (LPS), a gram-negative bacterial endotoxin, can be then increasingly released into the gut and act on the TLR-4 of innate immune cells (Gillois et al., 2018). The host immune is subsequently triggered by induction of both metalloproteases synthesis, proliferation and activation of differentiated T-cells, and consequently increased secretion of potent inflammatory cytokines such as IL-1β and TNF-α. (Tsiaoussis et al., 2019; Yuan et al., 2019b). Ultimately, this modulation of gut immune response together with the massive activation of the inflammation pathways could contribute to development of several systemic inflammatory-mediated conditions such as autoimmune diseases and hepatic inflammation (Claus et al., 2016a; Yuan et al., 2019b). For instance, an in vitro study of cultured human fecal suspensions with TCDF showed that TCDF exposure increases levels of IL-8 produced from the GI epithelial cells, which may indicate that TCDF can further stimulate immunotoxicity in response to microbiome disturbances (L. Zhang et al., 2015a). These alterations due to TCDF exposure can cause also potential liver dysfunction, such as proliferations in bile acids and reduced activity of the Farnesoid X receptor pathway (Chiu et al., 2020a; P. Zhang et al., 2017).
Further, systemic inflammation could eventually lead to dementia and other neurodegenerative diseases due to neuroinflammation triggered by chemical exposure (Bendorius et al., 2018). For example, low but prolonged exposure to di (2-ethylhexyl)phthalate (DEHP) in mice can increase synthesis of a substance called p-cresol, which usually reduces butyrate, a metabolite produced by bacteria that is critical for neurological function development (Lei et al., 2019).
Additionally, tight junctions’ proteins may be injured, automatically increasing GI cell permeability and potential infiltration of the lamina propria by bacterial derivates and toxins (Gillois et al., 2018; Groh et al., 2017).
Beneficial bacteria are a critical source of essential vitamins and nutrients within human body. Therefore, reduced bacterial production of beneficial metabolites could be detrimental to human health. Of note, the described disruption of bile acids’ cycles subsequently impairs a critical hormone, the glucagon-like peptide-1 (GLP-1) (D’Onofrio et al., 2010) which long-term may significantly affect insulin secretion and increase the risk of type-2 diabetes and metabolic disorders. In addition, tryptophan metabolites (e.g., indole 3-propionic acid) may be disrupted as well. Further, animal models showed that lead seems both to reduce diversity and altered metabolism of many pathways including vitamin E, bile acids and nitrogen metabolism (Assefa & Köhler, 2020; Gao et al., 2017). These compounds are usually secreted via commensal bacteria and are helpful to control the barrier function through the activation of aryl hydrocarbon receptor (AHR) (Dada et al., 2018; Dahan et al., 2018; Dai et al., 2018; Dempsey et al., 2019). AHR is deeply involved in inflammatory bowel disease (IBD). Consequently, disruption in bacterial-based tryptophan secretion, as may happen in response to xenobiotics, may play a role into IBD onset in susceptible individuals (Dahan et al., 2018).
Gut bacteria can modify some common aminoacidic nutritional components. For example, choline e L-carnitina that are converted to trimethylamine (TMA), which is then transformed into trimethylamine oxide (TMAO). Increased concentrations of bacterial-derived TMAO has positively associated with increased risk of cardiovascular disease (Aizawa et al., 2016). Results from a human study in Bangladesh showed that arsenic exposure from water intake is associated with increased abundance of Citrobacter spp., which are correlated with increased thickness of intima-media of the carotid vessels, a well-known predictor of cardiovascular risk (Tsiaoussis et al., 2019). This suggests that Citrobacter spp. may mediate the development of atherosclerosis resulting from arsenic exposure. Metabolites derived from gut microbiome also play a role in neurologic function through what is known as the gut–brain axis (Tsiaoussis et al., 2019). Specifically, gut microbes may influence brain structure and function through metabolic byproducts or innervation of the vagal nerve. Bacteria also contribute to the production of many of known neurotransmitters and their precursors, such as serotonin and gamma-aminobutyric acid. Of note, a significant pathway by which gut bacteria communicate with central nervous system involves enteroendocrine cells in the gut. Enteroendocrine cells can produce different types of molecules that can go into the systemic circulation and have an incidence on the nervous system regulatory pathways. For instance, serotonin (5-HT) is released by enterochromaffin cells starting from the metabolism of food derivate as tryptophan that is mediated by gut bacteria. Dysbiosis in the gut microbiome may impact the 5-HT homeostasis, especially in more vulnerable windows like pregnancy, where its disruption can cause impairment of forebrain development in the fetus. Indeed, Gonkowski et al. (2020) reported that prenatal BPA exposure may impact the synthesis of 5-HT directly inhibiting the serotonin release from the enterochromaffin cells (Gonkowski, 2020).
Based on all these biological interactions, the gut microbiome may be a central player in mediating toxicity from environmental contaminants as well as a potential biomarker reflecting the exposure to and action by environmental chemicals (Claus et al., 2016a; Yuan et al., 2019b). This evidence emphasizes that the gut microbiome is likely a marker of decreased health in addition to a reflection of external exposures and thus making it a new promising nexus between the environment toxicity and human diseases that, through such environmental-inducted changes, trigger the development and progression of prevalent human diseases.
4. Current challenges and future directions
This review presents the diverse effect of single compound on gut microbiota and the mechanisms driving interactions between environmental pollutants, the microbiome and human diseases. Gut bacteria have wide-ranging properties and can differently metabolize environmental chemicals, enhancing or decreasing their toxicity to the host. Equally, these chemicals can also affect both the structure and function of the microbiome. Up-to-date evidence indicates that the microbiota are fully tangled in the toxicity of environmental compounds. However, although some solid associations have already been recognized between gut microbiome changes and environmental exposures, several research gaps and challenges remain to be addressed.
For instance, experimental animal studies have been supported using standard organism models such as zebrafish and drosophila (Tu et al., 2020). More germ-free (e.g. fish) (Tlaskalova-Hogenova et al., 2014) and humanized animal models (Chiu et al., 2020b) with better and well-defined microbiome communities are needed to reduce measurement and experimental microbiome variability among experimental studies (Douglas, 2019). Moreover, future human studies will need to carefully control for diet and other known factors that may markedly perturb the microbiome (David et al., 2014).
Another recent challenge is the difficulty of capturing the impact of all chemicals at the same time. Indeed, contaminants are omnipresent and they can act synchronously as a mixture (Hernandez et al., 2019). Many studies examining the relationship between chemicals, microbiome and human outcomes have handled environmental exposures by considering one chemical at a time, but this methodology does not provide enough knowledge about the human health effects of the variety of potentially hazardous environmental factors that people are concurrently exposed to. In addition, it is still unclear if and how the reaction of gut microbiota changes when facing multiple detrimental stressors (e.g., chemicals) in association with one or more beneficial exposures (e.g., micronutrients). In this context, evaluating the combined effect of multiple chemicals or pollutants as a whole comprehensive mixture, rather than single exposures, would help accurately assess and identify risk factors and interactions and ultimately answer real-world public health questions.
Additionally, another methodology challenge introduced by whole genome sequencing platforms is elucidating the numerous, dynamic and convoluted interactions and networks possible among individual ‘omics datasets (Franzosa et al., 2015). Thus, a more integrative approach to this analysis is imperative. One example of integrative analysis is presented by Lu et al. (2014) who observed significant effects on both microbiome composition and metabolite production after introduction of arsenic into the mouse gut environment (Lu et al., 2014). A similar study was conducted by Zhang et al. (2015b)thought introduction of disinfection byproducts from drinking water which confirmed that the different omics are interdependent and that a combined approach can lead to more useful findings (Zhang et al., 2015b).
However, further studies are required to integrate all ‘omics data to provide a complete picture of the genotype to phenotype response to chemical insults. Further, several epidemiological studies have discovered discrepancies in the composition and diversity of the early-life microbiome that then do not persist later in life, indicating that perhaps this stage represents a “critical frame” of life throughout which individuals are more susceptible to environment-microbiome modulation (Tanaka & Nakayama, 2017). Environmental insults during this period, such as food, infections and antibiotic therapies can easily disrupt this optimal microbial progression. However, at the same time, it may offer a window of opportunity for modulating the microbiota through pointed interventions (diet, probiotics, prebiotics etc.) to promote a healthy gut microbiome growth and development (Sbihi et al., 2019). Therefore, it is critical to consider the impact of an intervention or perturbation according to the context, whether in a healthy adult or a developing infant.
In addition, future work should necessarily involve a more shotgun metagenomics-oriented approach, not only for higher resolution, but also because to explore a larger range of microorganisms present in the human gut microbiome ecosystem like fungi, viruses, and small eukaryotes. One example is provided by Eggers et al. who found associations between prenatal lead exposure and some gut fungal community composition at one month of age (Sitarik et al., 2020). Given the paucity of research examining these associations in humans, particularly for fungal child microbiota, further investigation is needed. Looking at the type and modality of relationship that these underappreciated microorganisms communities in the gut microbiome establish with environmental factors is a promising area of research (Zheng et al., 2020).
In conclusion, the gut microbiome is surely becoming an attractive and promising target but many studies to date have only reported the fluctuations in the microbiome and have not elucidated the existent mechanisms underlying these changes. Future studies are necessary to characterize these mechanisms and identify opportunities to modulate the microbiome (e.g., fecal microbiota transplantation, probiotics, and prebiotics etc.). To better address this modulation, the role of healthy gut microbiome needs to be primarily clarified.
Advances in laboratory technologies while incorporating multiple ‘omics, such as metabolomics, transcriptomics, and genomics will provide exciting opportunities to both obtain a multidimensional picture of the environmental exposure effects and also disentangle the health effects intermediated by microbes from the impacts resulting from direct exposures (Ahn & Hayes, 2021). Given the convolution of the human-microbiome symbiosis, only these “real life” data will return the necessary information for more realistic predictive models.
Only integration of new and more advanced gut microbiome human studies into environmental toxicology will ultimately offer a deeper and more comprehensive knowledge of the multifaced interactions among xenobiotics, gut microbiome and individuals. This idea towards precision medicine will accelerate progress of novel diagnostic biomarkers and targeted therapies and will help introduce new public health policies needed to protect individuals from these harmful environmental exposures.
Abbreviations
- BPA
bisphenol A
- CYP3
cytochrome P450-3
- DEHP
di (2-ethylhexyl) phthalate
- PAHs
polycyclic aromatic hydrocarbons
- PFOS/PFBS
perfluorooctyl Sulfonate
- TLR4
toll-like receptors-4
- TMA
trimethylamine
- TMAO
trimethylamine oxide
- TCS
triclosan
- TCDD
2,3,7,8 -tetrachlorodibenzo-p-dioxin
- SCFA
short chain fatty acids
Footnotes
This paper has been recommended for acceptance by Mingliang Fang.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- Aguiar-Pulido V, Huang W, Suarez-Ulloa V, Cickovski T, Mathee K, Narasimhan G, 2016. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evolutionary Bioinformatics Online 12 (Suppl. 1), 5–16. 10.4137/EBO.S36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Hayes RB, 2021. Environmental influences on the human microbiome and implications for noncommunicable disease. Annu. Rev. Publ. Health 42 (1), 277–292. 10.1146/annurev-publhealth-012420-105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa E, Tsuji H, Asahara T, Takahashi T, Teraishi T, Yoshida S, Ota M, Koga N, Hattori K, Kunugi H, 2016. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord 202, 254–257. 10.1016/j.jad.2016.05.038. [DOI] [PubMed] [Google Scholar]
- Assefa S, Köhler G, 2020. Intestinal microbiome and metal toxicity. Current Opinion in Toxicology 19, 21–27. 10.1016/j.cotox.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendorius M, Po C, Muller S, Jeltsch-David H, 2018. From systemic inflammation to neuroinflammation: the case of neurolupus. Int. J. Mol. Sci 19 (11), E3588. 10.3390/ijms19113588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S, 2009. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4 (9), e6958. 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang W, Hua J, Hu C, Lok-Shun Lai N, Qian P-Y, Lam PKS, Lam JCW, Zhou B, 2018. Dysregulation of intestinal health by environmental pollutants: involvement of the estrogen receptor and aryl hydrocarbon receptor. Environ. Sci. Technol 52 (4), 2323–2330. 10.1021/acs.est.7b06322. [DOI] [PubMed] [Google Scholar]
- Chiu K, Warner G, Nowak RA, Flaws JA, Mei W, 2020a. The impact of environmental chemicals on the gut microbiome. Toxicol. Sci. : An Official Journal of the Society of Toxicology 176 (2), 253–284. 10.1093/toxsci/kfaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu K, Warner G, Nowak RA, Flaws JA, Mei W, 2020b. The impact of environmental chemicals on the gut microbiome. Toxicol. Sci 176 (2), 253–284. 10.1093/toxsci/kfaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ, 2012. The human microbiome: at the interface of health and disease. Nat. Rev. Genet 13 (4), 260–270. 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM, 2016. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med 8 (1) 10.1186/s13073-016-0330-z, 77–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Guillou H, Ellero-Simatos S, 2016a. The gut microbiota: a major player in the toxicity of environmental pollutants? Npj Biofilms and Microbiomes 2. 10.1038/npjbiofilms.2016.3, 16003–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Guillou H, Ellero-Simatos S, 2016b. The gut microbiota: a major player in the toxicity of environmental pollutants? Npj Biofilms and Microbiomes 2 (1), 1–11. 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dada N, Sheth M, Liebman K, Pinto J, Lenhart A, 2018. Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci. Rep 8 (1), 2084. 10.1038/s41598-018-20367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan D, Jude BA, Lamendella R, Keesing F, Perron GG, 2018. Exposure to arsenic alters the microbiome of larval zebrafish. Front. Microbiol 9, 1323. 10.3389/fmicb.2018.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Yan Z, Ma S, Yang Y, Wang Q, Hou C, Wu Y, Liu Y, Diao Q, 2018. The herbicide glyphosate negatively affects midgut bacterial communities and survival of honey bee during larvae reared in vitro. J. Agric. Food Chem 66 (29), 7786–7793. 10.1021/acs.jafc.8b02212. [DOI] [PubMed] [Google Scholar]
- Das A, Srinivasan M, Ghosh TS, Mande SS, 2016. Xenobiotic metabolism and gut microbiomes. PLoS One 11 (10), e0163099. 10.1371/journal.pone.0163099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ, 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 (7484), 559–563. 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E, 2016. Current understanding of dysbiosis in disease in human and animal models. Inflamm. Bowel Dis 22 (5), 1137–1150. 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey JL, Little M, Cui JY, 2019. Gut microbiome: an intermediary to neurotoxicity. Neurotoxicology 75, 41–69. 10.1016/j.neuro.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA, 2011. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A 108 (Suppl. 1), 4554–4561. 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Bone RA, van de Wiele TR, 2009. Biovolatilization of metal(loid)s by intestinal microorganisms in the simulator of the human intestinal microbial ecosystem. Environ. Sci. Technol 43 (14), 5249–5256. 10.1021/es900544c. [DOI] [PubMed] [Google Scholar]
- D’Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K, 2010. Siderophores from neighboring organisms promote the growth of uncultured bacteria. Chem. Biol 17 (3), 254–264. 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE, 2019. Simple animal models for microbiome research. Nat. Rev. Microbiol 17 (12), 764–775. 10.1038/s41579-019-0242-1. [DOI] [PubMed] [Google Scholar]
- Eggers S, Malecki KM, Peppard P, Mares J, Shirley D, Shukla SK, Poulsen K, Gangnon R, Duster M, Kates A, Suen G, Sethi AK, Safdar N, 2018. Wisconsin microbiome study, a cross-sectional investigation of dietary fibre, microbiome composition and antibiotic-resistant organisms: rationale and methods. BMJ Open 8 (3), e019450. 10.1136/bmjopen-2017-019450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, Vos W. M. de, Cani PD, 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 110 (22), 9066–9071. 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Hsu T, Sirota-Madi A, Shafquat A, Abu-Ali G, Morgan XC, Huttenhower C, 2015. Sequencing and beyond: integrating molecular “omics” for microbial community profiling. Nat. Rev. Microbiol 13 (6), 360–372. 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J, Garrett WS, Chan AT, Huttenhower C, 2014. Relating the metatranscriptome and metagenome of the human gut. Proc. Natl. Acad. Sci. U. S. A 111 (22), E2329–E2338. 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gacesa R, Kurilshikov A, Vila AV, Sinha T, Klaassen M.a.Y., Bolte LA, Andreu-Sanchez S, Chen L, Collij V, Hu S, Dekens J.a.M., Lenters VC, Björk JR, Swarte JC, Swsertz MA, Jansen BH, Gelderloos-Arends J, cohort Study L., Hofker M, Weersma RK, 2020. The Dutch Microbiome Project Defines Factors that Shape the Healthy Gut Microbiome, 11.27.401125, p. 2020. 10.1101/2020.11.27.401125.bioRxiv. [DOI]
- Gao B, Chi L, Mahbub R, Bian X, Tu P, Ru H, Lu K, 2017. Multi-omics reveals that lead exposure disturbs gut microbiome development, key metabolites, and metabolic pathways. Chem. Res. Toxicol 30 (4), 996–1005. 10.1021/acs.chemrestox.6b00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MK, Wang B, Ahmadi S, Burnham CAD, Tarr PI, Warner BB, Dantas G, 2016. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nature Microbiology 1. 10.1038/nmicrobiol.2016.24, 16024–16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillois K, Lévêque M, Théodorou V, Robert H, Mercier-Bonin M, 2018. Mucus: an underestimated gut target for environmental pollutants and food additives. Microorganisms 6 (2), E53. 10.3390/microorganisms6020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonkowski S, 2020. Bisphenol A (BPA)-Induced changes in the number of serotonin-positive cells in the mucosal layer of porcine small intestine—the preliminary studies. Int. J. Mol. Sci 21 (3), 1079. 10.3390/ijms21031079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh KJ, Geueke B, Muncke J, 2017. Food contact materials and gut health: implications for toxicity assessment and relevance of high molecular weight migrants. Food Chem. Toxicol 109, 1–18. 10.1016/j.fct.2017.08.023. [DOI] [PubMed] [Google Scholar]
- Hamady M, Knight R, 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19 (7), 1141–1152. 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez AF, Buha A, Constantin C, Wallace DR, Sarigiannis D, Neagu M, Antonijevic B, Hayes AW, Wilks MF, Tsatsakis A, 2019. Critical assessment and integration of separate lines of evidence for risk assessment of chemical mixtures. Arch. Toxicol 93 (10), 2741–2757. 10.1007/s00204-019-02547-x. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK, 2013. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155 (7), 1451–1463. 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys C, 2020. 19—intestinal permeability. In: Pizzorno JE, Murray MT (Eds.), Textbook of Natural Medicine, fifth ed. Churchill Livingstone, pp. 166–177. 10.1016/B978-0-323-43044-9.00019-4. e4). [DOI] [Google Scholar]
- Iszatt N, Janssen S, Lenters V, Dahl C, Stigum H, Knight R, Mandal S, Peddada S, González A, Midtvedt T, Eggesbø M, 2019. Environmental toxicants in breast milk of Norwegian mothers and gut bacteria composition and metabolites in their infants at 1 month. Microbiome 7 (1), 34. 10.1186/s40168-019-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KP, Ng AH-M, Wan HT, Wong AY-M, Leung CC-T, Li R, Wong CK-C, 2018. Dietary exposure to the environmental chemical, PFOS on the diversity of gut microbiota, associated with the development of metabolic syndrome. Front. Microbiol 9, 2552. 10.3389/fmicb.2018.02552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue HE, Moroishi Y, Jackson BP, Palys TJ, Madan JC, Karagas MR, 2020. Nutrient-toxic element mixtures and the early postnatal gut microbiome in a United States longitudinal birth cohort. Environ. Int 138, 105613 10.1016/j.envint.2020.105613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K, de Timary P, Delzenne NM, 2014. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. U. S. A 111 (42), E4485–E4493. 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Menon R, Manteiga S, Alden N, Hunt C, Alaniz RC, Lee K, Jayaraman A, 2019. Environmental chemical diethylhexyl phthalate alters intestinal microbiota community structure and metabolite profile in mice. mSystems 4 (6). 10.1128/mSystems.00724-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Renaud HJ, Klaassen CD, Cui JY, 2016. Age-specific regulation of drug-processing genes in mouse liver by ligands of xenobiotic-sensing transcription factors. Drug Metabol. Dispos.: The Biological Fate of Chemicals 44 (7), 1038–1049. 10.1124/dmd.115.066639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Li J, Zhang Q, Gao S, Quan X, Liu P, Xu C, 2021. Effects of endocrine disrupting chemicals in host health: three-way interactions between environmental exposure, host phenotypic responses, and gut microbiota. Environmental Pollution (Barking, Essex: 1987) 271, 116387. 10.1016/j.envpol.2020.116387. [DOI] [PubMed] [Google Scholar]
- Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG, 2014. Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect 122 (3), 284–291. 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Mahbub R, Fox JG, 2015. Xenobiotics: interaction with the intestinal microflora. ILAR J 56 (2), 218–227. 10.1093/ilar/ilv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage R, Antoniou MN, Tsoukalas D, Goulielmos GN, Tsatsakis A, 2018. Gut microbiome metagenomics to understand how xenobiotics impact human health. Current Opinion in Toxicology 11 (12), 51–58. 10.1016/j.cotox.2019.02.002. [DOI] [Google Scholar]
- National Academies of Sciences, E. and M., 2018. Environmental Chemicals, the Human Microbiome, and Health Risk: A Research Strategy National Academies Press, p. 109. 10.17226/24960. [DOI] [PubMed] [Google Scholar]
- National Academies of Sciences, E., and Medicine, 2018. Environmental Chemicals, the Human Microbiome, and Health Risk: A Research Strategy The National Academies Press, Washington, DC. 10.17226/24960. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S, 2012. Host-gut microbiota metabolic interactions. Science (New York, N.Y.) 336 (6086), 1262–1267. 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Ogita T, Yamamoto Y, Mikami A, Shigemori S, Sato T, Shimosato T, 2020. Oral administration of flavonifractor plautii strongly suppresses Th2 immune responses in mice. Front. Immunol 11. 10.3389/fimmu.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas M-E, Maiter D, et al. , 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med 23 (1), 107–113. 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- Ribado JV, Ley C, Haggerty TD, Tkachenko E, Bhatt AS, Parsonnet J, 2017. Household triclosan and triclocarban effects on the infant and maternal microbiome. EMBO Mol. Med 9 (12), 1732–1741. 10.15252/emmm.201707882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld CS, 2017. Gut dysbiosis in animals due to environmental chemical exposures. Front. Cell. Infect. Microbiol 7, 396. 10.3389/fcimb.2017.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K, 2018. Gut microbiota functions: metabolism of nutrients and other food components. Eur. J. Nutr 57 (1), 1–24. 10.1007/s00394-017-1445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rude KM, Keogh CE, Gareau MG, 2019. The role of the gut microbiome in mediating neurotoxic outcomes to PCB exposure. Neurotoxicology 75, 30–40. 10.1016/j.neuro.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbihi H, Boutin RCT, Cutler C, Suen M, Finlay BB, Turvey SE, 2019. Thinking bigger: how early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease. Allergy 74 (11), 2103–2115. 10.1111/all.13812. [DOI] [PubMed] [Google Scholar]
- Schmidt TSB, Raes J, Bork P, 2018. The human gut microbiome: from association to modulation. Cell 172 (6), 1198–1215. 10.1016/j.cell.2018.02.044. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY, 2015. Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci.: An Official Journal of the Society of Toxicology 147 (1), 84–103. 10.1093/toxsci/kfv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Klaassen CD, Cui JY, 2016. Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metabol. Dispos.: The Biological Fate of Chemicals 44 (2), 262–274. 10.1124/dmd.115.067504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Laue HE, Shrubsole MJ, Wu H, Bloomquist TR, Larouche A, Zhao K, Gao F, Boivin A, Prada D, Hunting DJ, Gillet V, Takser L, Baccarelli AA, 2022. Associations of childhood and perinatal blood metals with children’s gut microbiomes in a Canadian gestation cohort. Environ. Health Perspect 130 (1), 017007 10.1289/EHP9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitarik AR, Arora M, Austin C, Bielak LF, Eggers S, Johnson CC, Lynch SV, Kyun Park S, Hank Wu K-H, Yong GJM, Cassidy-Bushrow AE, 2020. Fetal and early postnatal lead exposure measured in teeth associates with infant gut microbiota. Environ. Int 144, 106062 10.1016/j.envint.2020.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi Y, Soufi B, 2016. Mass spectrometry-based bacterial proteomics: focus on dermatologic microbial pathogens. Front. Microbiol 10.3389/fmicb.2016.00181. [DOI] [PMC free article] [PubMed]
- Tamboli CP, Neut C, Desreumaux P, Colombel JF, 2004. Dysbiosis in inflammatory bowel disease. Gut 53 (1), 1–4. 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Nakayama J, 2017. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int 66 (4), 515–522. 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Vannucci L, Klimesova K, Stepankova R, Krizan J, Kverka M, 2014. Microbiome and colorectal carcinoma: insights from germ-free and conventional animal models. Cancer J 20 (3), 217–224. 10.1097/PPO.0000000000000052. [DOI] [PubMed] [Google Scholar]
- Tsiaoussis J, Antoniou MN, Koliarakis I, Mesnage R, Vardavas CI, Izotov BN, Psaroulaki A, Tsatsakis A, 2019. Effects of single and combined toxic exposures on the gut microbiome: current knowledge and future directions. Toxicol. Lett 312, 72–97. 10.1016/j.toxlet.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Tu P, Chi L, Bodnar W, Zhang Z, Gao B, Bian X, Stewart J, Fry R, Lu K, 2020. Gut microbiome toxicity: connecting the environment and gut microbiome-associated diseases. Toxics 8 (1), 19. 10.3390/toxics8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- March U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2021. Centers for Disease Control and Prevention, Fourth Report on Human Exposure to Environmental Chemicals, Updated Tables. Atlanta, GA: [https://www.cdc.gov/exposurereport/. [Google Scholar]
- Van de Wiele T, Gallawa CM, Kubachk KM, Creed JT, Basta N, Dayton EA, Whitacre S, Laing GD, Bradham K, 2010. Arsenic metabolism by human gut microbiota upon in vitro digestion of contaminated soils. Environ. Health Perspect 118 (7), 1004–1009. 10.1289/ehp.0901794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Shen M, Zhou J, Jin Y, 2019. Chlorpyrifos disturbs hepatic metabolism associated with oxidative stress and gut microbiota dysbiosis in adult zebrafish. Comparative Biochemistry and Physiology. Toxicology & Pharmacology: CB (Curr. Biol.) 216, 19–28. 10.1016/j.cbpc.2018.11.010. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang W, Romano KA, Gu M, Sanidad KZ, Kim D, Yang J, Schmidt B, Panigrahy D, Pei R, Martin DA, Ozay EI, Wang Y, Song M, Bolling BW, Xiao H, Minter LM, Yang G-Y, Liu Z, Zhang G, 2018. A common antimicrobial additive increases colonic inflammation and colitis-associated colon tumorigenesis in mice. Sci. Transl. Med 10 (443) 10.1126/scitranslmed.aan4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Pan Z, Jin C, Ni Y, Fu Z, Jin Y, 2019a. Gut microbiota: an underestimated and unintended recipient for pesticide-induced toxicity. Chemosphere 227, 425–434. 10.1016/j.chemosphere.2019.04.088. [DOI] [PubMed] [Google Scholar]
- Yuan X, Pan Z, Jin C, Ni Y, Fu Z, Jin Y, 2019b. Gut microbiota: an underestimated and unintended recipient for pesticide-induced toxicity. Chemosphere 227, 425–434. 10.1016/j.chemosphere.2019.04.088. [DOI] [PubMed] [Google Scholar]
- Zhang L, Nichols RG, Correll J, Murray IA, Tanaka N, Smith PB, Hubbard TD, Sebastian A, Albert I, Hatzakis E, Gonzalez FJ, Perdew GH, Patterson AD, 2015a. Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ. Health Perspect 123 (7), 679–688. 10.1289/ehp.1409055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhu W, Wang D, Yan J, Wang Y, Zhou Z, He L, 2017. A combined NMR- and HPLC-MS/MS-based metabolomics to evaluate the metabolic perturbations and subacute toxic effects of endosulfan on mice. Environ. Sci. Pollut. Res. Int 24 (23), 18870–18880. 10.1007/s11356-017-9534-z. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhao F, Deng Y, Zhao Y, Ren H, 2015b. Metagenomic and metabolomic analysis of the toxic effects of trichloroacetamide-induced gut microbiome and urine metabolome perturbations in mice. J. Proteome Res 14 (4), 1752–1761. 10.1021/pr5011263. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang Y, Wang G, Han R, Xie X, 2016. Effects of chlorpyrifos on the gut microbiome and urine metabolome in mouse (Mus musculus). Chemosphere 153, 287–293. 10.1016/j.chemosphere.2016.03.055. [DOI] [PubMed] [Google Scholar]
- Zheng D, Liwinski T, Elinav E, 2020. Interaction between microbiota and immunity in health and disease. Cell Res 30 (6), 492–506. 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.


