Abstract
Mitochondrial ribosomal proteins (MRPs) assemble as specialized ribosome to synthesize mtDNA‐encoded proteins, which are essential for mitochondrial bioenergetic and metabolic processes. MRPs are required for fundamental cellular activities during animal development, but their roles beyond mitochondrial protein translation are poorly understood. Here, we report a conserved role of the mitochondrial ribosomal protein L4 (mRpL4) in Notch signaling. Genetic analyses demonstrate that mRpL4 is required in the Notch signal‐receiving cells to permit target gene transcription during Drosophila wing development. We find that mRpL4 physically and genetically interacts with the WD40 repeat protein wap and activates the transcription of Notch signaling targets. We show that human mRpL4 is capable of replacing fly mRpL4 during wing development. Furthermore, knockout of mRpL4 in zebrafish leads to downregulated expression of Notch signaling components. Thus, we have discovered a previously unknown function of mRpL4 during animal development.
Keywords: Drosophila, mitochondrial ribosomal protein L4, Notch, wap, zebrafish
Subject Categories: Development, Metabolism, Signal Transduction
The mitochondrial ribosome component mRpL4 regulates Notch pathways, which is likely independent from its role in mitochondrial protein synthesis. The regulatory role of mRpL4 in Notch signaling is conserved during fly and zebrafish development.

Introduction
Mitochondria are best known as the powerhouse of cells as they generate the majority of cellular ATP through coupled reactions carried out by five oxidative phosphorylation (OXPHOS) protein complexes (Quirós et al, 2016). Most OXPHOS proteins are generated from nuclear genes and imported into mitochondria, with the exception of 13 OXPHOS proteins encoded by the mitochondrial genome (Richter‐Dennerlein et al, 2015). The mitochondrial protein synthesis depends on the mitochondrial ribosomal proteins (MRPs), which assemble to form a specialized form of ribosome (Kummer & Ban, 2021). Disruption of MRPs function leads to deficiency in OXPHOS protein synthesis and mitochondrial activity, which in turn impacts a wide variety of cellular processes (Kummer & Ban, 2021).
Systemic mutagenesis analysis in Drosophila (Marygold et al, 2007) and mice (Cheong et al, 2020) have demonstrated that MRPs are crucial for animal development. Mutations of MRPs are associated with a number of developmental disorders and fatal diseases in humans (Huang et al, 2020; Ferrari et al, 2021). The impacts of MRPs on fundamental developmental events such as cell cycle and cell growth have been extensively investigated (Galloni, 2003; Frei et al, 2005; Mandal et al, 2005; Tselykh et al, 2005; Liao et al, 2006; Ohsawa et al, 2012; Wang et al, 2012; Chen et al, 2018a). These findings highlight the importance of MRPs for mitochondrial activity and fit well with current view that mitochondria function not only as the power generator but also as a signaling hub (Quirós et al, 2016). Recent studies have begun to reveal the diversified roles of MRPs, some of which are independent of mitochondrial protein synthesis (Amikura et al, 2001; Zhang et al, 2015; Han et al, 2020; Huang et al, 2020). However, the functions of MRPs outside of mitochondrial ribosome during animal development are not fully understood.
The highly conserved Notch signaling pathway functions to distinguish adjacent cells and is required for various developmental processes (Bray, 2006). In Drosophila, the Notch gene encodes a transmembrane receptor, which is activated by Delta or Serrate presented on the membrane of signal‐sending cell (Henrique & Schweisguth, 2019). The receptor–ligand engagement triggers a series of proteolytic cleavage of the Notch protein and releases the Notch intracellular domain (NICD). NICD is translocated into the nucleus, where it forms a transcription activation complex with the DNA‐binding protein Suppressor of Hairless [Su(H)] to drive the expression of downstream target genes (Guruharsha et al, 2012). In the absence of Notch activation, Su(H) recruits co‐repressors to suppress the expression of Notch targets (Henrique & Schweisguth, 2019).
The Notch signaling cascade regulates mitochondrial homeostasis and activity in both fly and vertebrates (Thörig et al, 1981a,b; Vilkki & Portin, 1987; Landor et al, 2011; Basak et al, 2014; Ludikhuize et al, 2020; Dubal et al, 2022), but only a few mitochondrial proteins are directly regulated by Notch signaling at the transcriptional level (Xu et al, 2015; Lee & Long, 2018; Kung‐Chun Chiu et al, 2019). Interestingly, NICD also functions through Su(H)‐independent pathways to regulate mitochondria activity (Perumalsamy et al, 2010; Xu et al, 2015; Chen et al, 2018b; Zhou et al, 2019; Dai et al, 2020). In this noncanonical pathway, NICD is found to localize in the mitochondria and interact with various mitochondrial proteins, including the OXPHOS components (Lee et al, 2013; Ojha et al, 2022). The cross talk between mitochondria and Notch signaling is bidirectional, with studies in Drosophila follicle cells showing for the first time that mitochondria fission activates Notch signaling (Mitra et al, 2012). Subsequent studies reveal that mitochondria modulate Notch activity through signaling molecules such as calcium (Kasahara et al, 2013) and reactive oxygen species (ROS; Hamanaka et al, 2013; Cao et al, 2016; Khacho et al, 2016; Perez‐Gomez et al, 2020). However, our understanding about the reciprocal regulatory relationship between mitochondria and Notch signaling is still incomplete.
We have isolated an MRP gene, mRpL4, as a positive regulator of Notch signaling during Drosophila wing development. We found that mRpL4 functions in the Notch signal‐receiving cells to permit transcription of target genes. Likely independent of its role in OXPHOS protein synthesis, mRpL4 interacts with wap to facilitate the recruitment of Su(H) to the chromatin. We further demonstrate that knockout of mRpL4 in zebrafish leads to decreased Notch signaling activity. Our findings reveal a previously unknown function of MRP during animal development and emphasize the complexity of Notch signaling regulation.
Results
mRpL4 regulates Notch signaling activity in the Drosophila wing
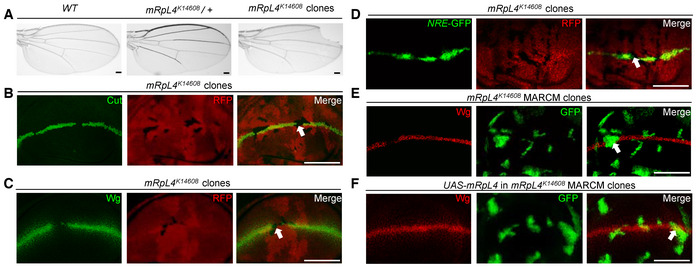
During a somatic mosaic screen (Mo et al, 2022), one of the Bruinfly mutant stocks, mRpL4 K14608 , was found to cause marginal defects when homozygous clones were generated in the wings (Fig 1A). As this phenotype is reminiscent of impaired Notch signaling, we examined the expression level of Notch target genes Cut and Wingless (Wg). In the wild‐type wing imaginal disk, Cut (Appendix Fig S1A) and Wg (Appendix Fig S1B) were produced in cells located at the dorsal/ventral (D/V) boundary. In mRpL4 K14608 homozygous clones located at the D/V boundary, the expression of Cut and Wg was abolished (Fig 1B and C). The expression of Notch activity reporter NRE‐GFP (Saj et al, 2010) was also reduced in mRpL4 K14608 mutant cells (Fig 1D). The expression of Notch (Appendix Fig S1C and D) and Dl (Appendix Fig S1E and F) was not significantly affected in mRpL4 mutant cells. Importantly, when a mRpL4 transgene was expressed in mRpL4 K14608 mutant cells using the MARCM technique (Lee & Luo, 2001), the Notch signaling defect was rescued (Fig 1E and F; Appendix Fig S1G and H).
Figure 1. mRpL4 mutant leads to Notch signaling defects in the Drosophila wing.
-
ARepresentative image of wings (n > 20 wings) from control adult flies, mRpL4 K14608 heterozygous flies and flies harboring mRpL4 K14608 homozygous mutant clones.
-
B–DRepresentative image of wing imaginal disks (n > 15 wing disks) stained for Cut and Wg, and wing disk expressing Notch signaling reporter NRE‐GFP, respectively.
-
E, FRepresentative images of wing disks (n > 10 wing disks) bearing MARCM clones stained for Wg. In (F), UAS‐mRpL4 are expressed under the control of tub‐Gal4 in the MARCM clones.
Data information: The mRpL4 K14608 mutant clones in these images are marked by the absence of RFP (B–D), while the MARCM clones are marked by GFP (E, F). Representative clones are marked by white arrows in (B–F). Scale bars = 100 μm.
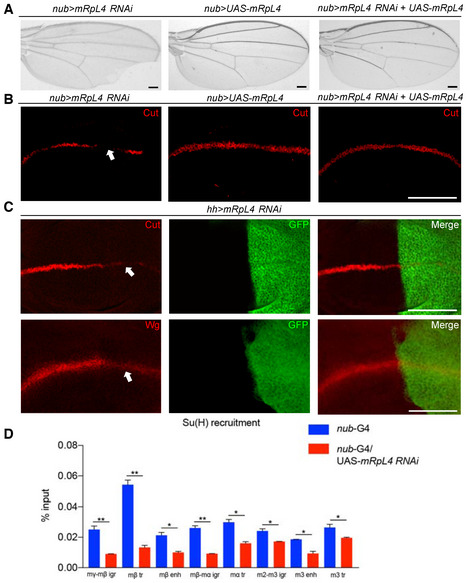
Consistent with the mutant phenotypes, inhibiting the expression of mRpL4 by RNAi also led to marginal nicks in the adult wing (Fig 2A). The mRpL4 RNAi resulted in moderate wing margin defects, while RNAi knockdown of two other MRP genes (mRpS28 and mRpL24) showed little effect on wing margin integrity (Appendix Fig S2A–D). The mRpL4 transgene was sufficient to rescue both the adult wing margin defect (Fig 2A) and downregulation of Cut in the wing disk (Fig 2B) when co‐expressed with the RNAi construct. When mRpL4 RNAi was driven by hh‐Gal4 in the posterior compartment of wing disk, the expression of Cut and Wg was clearly dampened within the posterior region (Fig 2C; Appendix Fig S2E).
Figure 2. mRpL4 regulates Notch signaling in the Drosophila wing.
-
ARepresentative image of wings (n > 20 wings) from flies expressing mRpL4 RNAi, UAS‐mRpL4 and both under the control of nub‐Gal4.
-
BRepresentative image of wing imaginal disks (n > 15 wing disks) stained for Cut, from flies expressing mRpL4 RNAi, UAS‐mRpL4 and both under the control of nub‐Gal4.
-
CRepresentative images of wing disks (n > 15 wing disks) stained for Cut and Wg from flies expressing UAS‐mRpL4‐RNAi under the control of hh‐Gal4 (marked by GFP).
-
DThe level of Su(H) occupancy at E(spl)mβ gene family regions as assessed by qPCR following ChIP, from wild‐type and UAS‐mRpL4‐RNAi‐expressing wing disks. Data are presented as mean ± SEM, two biological replicates for each genotype and three technical replicates for each sample. Statistical significance was tested using two‐tailed unpaired t‐test. *P < 0.05, **P < 0.01.
Data information: Representative regions showing Notch activity defects are marked by white arrows in (B, C). Scale bars = 100 μm.
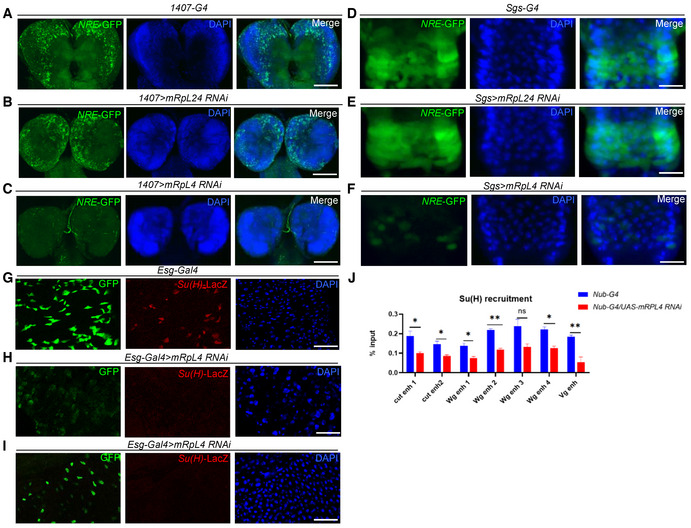
Notch signaling plays important roles during the development of numerous tissues, and whether mRpL4 is broadly involved in the regulation of Notch signaling was further investigated. Notch signal activity could be readily monitored by reporter lines such as NRE‐GFP in larval neuroblasts (Liu et al, 2017) and salivary gland imaginal rings (Yang & Deng, 2018), as well as by Su(H)‐LacZ in adult midgut (Zhao et al, 2022). RNAi knockdown of mRpL4 but not mRpL24 was able to attenuate the expression of NRE‐GFP in larval neuroblasts (Fig EV1A–C) and salivary gland imaginal rings (Fig EV1D–F). In adult midgut, the expression level of Su(H)‐LacZ was also reduced by mRpL4 RNAi (Fig EV1G–I). These observations indicate that mRpL4 might modulate Notch signaling in various developmental events.
Figure EV1. Effects of mRpL4 RNAi on Notch signal activity.
-
A–CRepresentative image showing the expression of NRE‐GFP in larval neuroblasts (n > 10 larvae) of control (A), mRpL24 RNAi (B) and mRpL4 RNAi (C) larvae.
-
D–FRepresentative image showing the expression of NRE‐GFP in salivary gland imaginal rings (n > 10 larvae) of control (D), mRpL24 RNAi (E) and mRpL4 RNAi (F) larvae.
-
G–IRepresentative image showing the expression of Su(H)‐LacZ in midgut cells (n > 10 flies) of control (G) and mRpL4 RNAi (H, I) adult flies.
-
JThe level of Su(H) occupancy at Wg, Cut and Vg genomic regions as assessed by qPCR following ChIP, from wild‐type and UAS‐mRpL4‐RNAi‐expressing wing disks. Data are presented as mean ± SEM, two biological replicates for each genotype and three technical replicates for each sample. Statistical significance was tested using two‐tailed unpaired t‐test. *P < 0.05, **P < 0.01, ns means “not significant”.
Data information: Scale bars = 100 μm.
Activation of Notch signaling relies on binding of Su(H) at the enhancer region of target genes (Krejcí & Bray, 2007; Gomez‐Lamarca et al, 2018). When examined by chromatin immunoprecipitation (ChIP) in wing disk cells, mRpL4 knockdown by RNAi was found to decrease the occupancy of Su(H) at regulatory regions of the Enhancer of split Complex family genes (Fig 2D), as well as at enhancer regions of Cut, Wg, and Vestigial (Fig EV1J). Collectively, these findings demonstrate that mRpL4 positively regulates Notch signaling activity.
mRpL4 functions in signal‐receiving cells
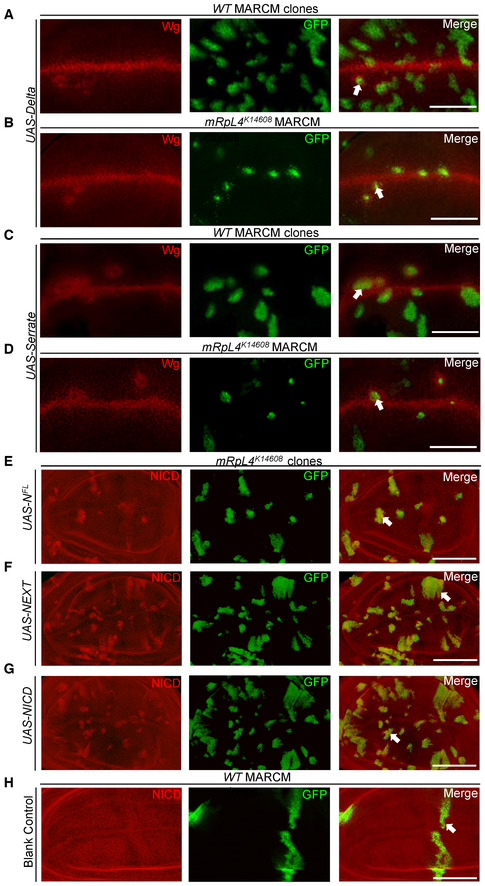
Previous studies have shown that mRpL4 is involved in fly eye and ovary development (Mandal et al, 2005; Ohsawa et al, 2012; Wang et al, 2012), but its role in Notch signaling has not been reported. Therefore, we performed genetic analysis to further dissect the role of mRpL4 in Notch signal transduction. To distinguish whether mRpL4 functions in the signal‐sending or receiving cells, the MARCM system was employed to overexpress Dl, Ser, and Notch proteins in mRpL4 K14608 mutant cells. Expression of Dl in wild‐type cells led to the induction of Wg expression along the border of MARCM clones (Fig EV2A). Overexpression of Dl in mRpL4 K14608 clones also induced Wg expression in the surrounding cells (Fig EV2B). Similarly, overexpression of Ser led to the induction of Wg along the border of MARCM clones in both wild‐type (Fig EV2C) and mRpL4 K14608 mutant (Fig EV2D) cells. These results suggest that mRpL4 is dispensable in the signal‐sending cells.
Figure EV2. Effects of overexpressing Notch signaling components.
-
A–DRepresentative image of wing imaginal disks (n > 10 wing disks) containing MARCM clones stained for Wg. Notch signaling ligand Dl is overexpressed in wild‐type (A) or mRpL4 K14608 mutant (B) cells. Notch signaling ligand Ser is overexpressed in wild‐type (C) or mRpL4 K14608 mutant (D) cells.
-
E–HRepresentative image of wing imaginal disks (n > 10 wing disks) containing MARCM clones stained for NICD. Full‐length Notch protein (NFL) (E), NEXT (F) and NICD (G) are overexpressed in mRpL4 K14608 mutant cells. Blank MARCM clones are generated in the wild‐type wing disks (H).
Data information: The MARCM clones are marked by GFP and representative clones are marked by white arrows. Scale bars = 100 μm.
Similar experiment with full‐length Notch (NFL) was performed to investigate the role of mRpL4 in the signal‐receiving cells. In wild‐type cells, NFL induced Cut expression in proximity of the wing margin (Fig 3A). However, NFL failed to induce Cut expression in mRpL4 K14608 clones (Fig 3B). These observations indicate a requirement for mRpL4 in the signal‐receiving cells to activate Notch targets.
Figure 3. mRpL4 is required in Notch signal‐receiving cells.
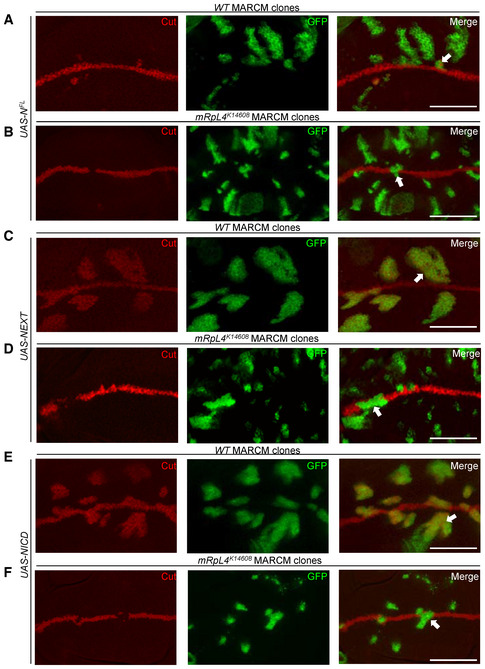
-
A–FRepresentative image of wing disks (n > 10 wing disks) bearing MARCM clones stained for Cut. NFL (A, B), NEXT (C, D) and NICD (E, F) are overexpressed in wild‐type (A, C, E) or mRpL4 K14608 mutant (B, D, F) cells. The MARCM clones are marked by GFP and representative clones are marked by white arrows. Scale bars = 100 μm.
In the signal‐receiving cells, an active membrane‐bound form of Notch (NEXT) is generated through proteolytic cleavage by the metalloprotease Kuzbanian after binding with the ligands. NEXT is further cleaved by Presenilin to form NICD, which interacts with Su(H) to regulate target gene expression (Bray, 2006). To refine in which step mRpL4 is required for Notch processing, we overexpressed NEXT and NICD in mRpL4 K14608 mutant cells. In wild‐type cells, NEXT was sufficient to induce the expression of downstream target Cut (Fig 3C). NEXT failed to induce the expression of Cut in mRpL4 K14608 clones (Fig 3D). Furthermore, we found that NICD robustly induced Cut expression in wild‐type cells (Fig 3E), but failed to do so in mRpL4 K14608 mutant cells (Fig 3F). The expression of NFL (Fig EV2E), NEXT (Fig EV2F), and NICD (Fig EV2G and H) in MARCM clones was confirmed by immunostaining using antibody raised against NICD. These genetic data place the function of mRpL4 downstream of NICD production to regulate target gene expression.
mRpL4 regulates OXPHOS activity and Notch signal through parallel pathways
Mitochondrial ribosomal proteins are required for optimal mitochondrial activity, and ROS, a major metabolite of mitochondria, has been shown to modulate Notch signaling activity in various developmental contexts (Hamanaka et al, 2013; Cao et al, 2016; Khacho et al, 2016; Perez‐Gomez et al, 2020). The cellular ROS level was indeed reduced in both mRpL4 K14608 mutant clones and mRpL4 RNAi cells (Fig 4A and B), implying that the effect of mRpL4 on Notch signal transduction might be relayed by ROS. However, several findings are inconsistent with this simplified model. Mutations of two other MRP genes, mRpS28 and mRpL24, led to reduction of ROS without affecting Cut expression (Fig 4C and D). In addition, RNAi knockdown of mRpS2 and mRpS12 impaired ROS production but not Cut expression (Fig 4E and F). We further examined the role of Cytochrome c oxidase Va (CoVa) during wing development. CoVa is a component of the OXPHOS complex IV, which functions downstream of mRpL4 to regulate cell cycle progression (Mandal et al, 2005; Mitra et al, 2012). The production of ROS was inhibited in CoVa tend mutant clones (Fig 4G) and CoVa RNAi cells (Figs 4H and EV3A), but Cut was expressed normally (Figs 4G and H and EV3B). Unlike CoVa and mRpL4 mutations, inhibition of other OXPHOS complexes led to accumulation of ROS (Fig EV3C–F), which could induce oxidative stress response and trigger changes in multiple signaling pathways (Owusu‐Ansah et al, 2008; Ohsawa et al, 2012; Wang et al, 2012; Perez‐Gomez et al, 2020). Taken together, we conclude that reduction of cellular ROS is not the cause of Notch signaling defects during fly wing development. We hypothesize that additional factors are involved in the regulation of Notch signal transduction by mRpL4.
Figure 4. Subset of MRPs regulate ROS production but not Notch activity.
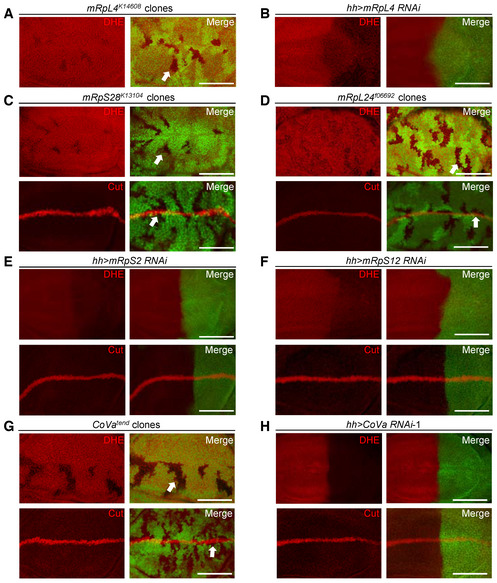
-
A, BRepresentative images of wing disks (n > 15 wing disks) stained with DHE. The mRpL4 K14608 mutant clones are marked by the absence of GFP (A), while UAS‐mRpL4‐RNAi are expressed in the posterior region (B).
-
C, DRepresentative images of wing disks (n > 10 wing disks) bearing mRpS28 k13104 (C) and mRpL24 f06692 (D) mutant clones stained with DHE and Cut.
-
E, FRepresentative images of wing disks (n > 15 wing disks) stained with DHE and Cut. The hh‐Gal4 (marked by GFP) is used to drive the expression of RNAi against mRpS2 (E) and mRpS12 (F).
-
GRepresentative images of wing disks (n > 10 wing disks) bearing CoVa tend mutant clones stained with DHE and Cut.
-
HRepresentative images of wing disks (n > 15 wing disks) stained with DHE and Cut. UAS‐CoVa‐RNAi are expressed in the posterior region under the control of hh‐Gal4 (marked by GFP).
Data information: The clones are marked by the absence of GFP and representative clones are marked by white arrows (A, C, D, G). Scale bars = 100 μm.
Figure EV3. mRpL4 may regulate OXPHOS activity and Notch signaling through branched pathways.
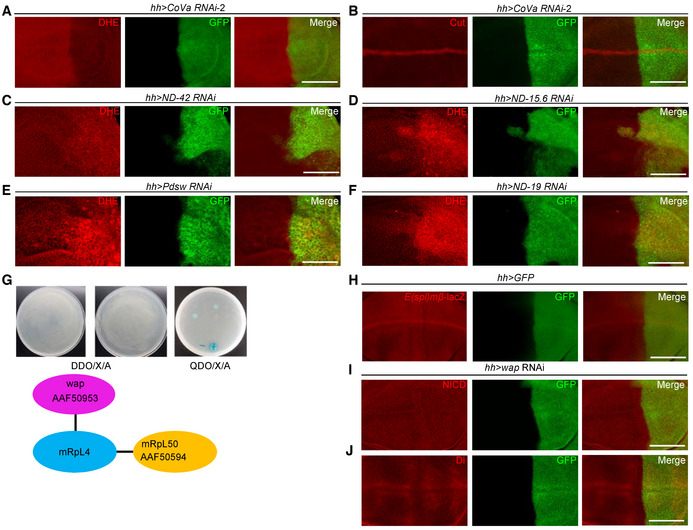
-
A, BRepresentative images of wing imaginal disks (n > 15 wing disks) expressing CoVa RNAi under the control of hh‐gal4 (marked by GFP) that are stained for DHE (A) and Cut (B).
-
C–FRepresentative images of wing imaginal disks (n > 20 wing disks) stained by DHE. The expression of ND‐42 (C), ND‐15.6 (D), Pdsw (E) and ND‐19 (F) RNAi are under the control of hh‐Gal4 (marked by GFP).
-
GTwo proteins, mRpL50 and wap are isolated as mRpL4 interaction partners through yeast two‐hybridization. The GenBank accession numbers assigned to mRpL50 and wap are AfAF50594 and AAF50953.
-
HRepresentative image of wing imaginal disks (n > 15 wing disks) showing E(spl)mβ‐LacZ expression pattern, and cells in the posterior compartment are marked by GFP.
-
I, JRepresentative image of wing imaginal disks (n > 15 wing disks) stained for NICD (I) and Dl (J). In these wing disks, wap RNAi are expressed under the control of hh‐Gal4 (marked by GFP).
Data information: Scale bars = 100 μm.
mRpL4 interacts with wap to regulate Notch signaling
In order to understand how mRpL4 regulates Notch pathway, we screened for mRpL4 interacting proteins by yeast two‐hybridization and found that wings apart (wap, also known as Riquiqui) and mRpL50 physically interact with mRpL4 (Fig EV3G). The interaction between mRpL4 and mRpL50 fits with the fact that they are both components of the large subunit of mitochondrial ribosome. As a WD40‐repeat protein, wap regulates Hippo and extracellular signal‐regulated kinase (ERK) pathways during fly wing development (Degoutin et al, 2013; Yang et al, 2016), but its interaction with MRPs and Notch signaling has not been reported. To confirm that wap is an mRpL4‐interacting protein, we performed immunoprecipitation experiments with wing disk cell lysates. Using an antibody developed against fly mRpL4 protein (Fig 5A), a physical association between wap and mRpL4 was detected (Fig 5B). The potential role of wap in Notch signal transduction was further investigated. RNAi knockdown of wap led to wing margin notches (Fig 5C). In the wing disk, the expression of Cut (Fig 5D) and Notch activity reporter E(spl)mβ‐LacZ (Figs 5E and EV3H) was attenuated in wap RNAi cells, while ROS production was largely unaffected (Fig 5F). Similar as that for mRpL4, knockdown of wap showed little effect on Notch or Dl (Fig EV3I and J). Importantly, wap was sufficient to restore the expression of Wg in mRpL4 K14608 mutant cells (Fig 5G). These results suggest that wap acts as a downstream factor of mRpL4 to regulate Notch activation in the wing.
Figure 5. mRpL4 physically and genetically interacts with wap.
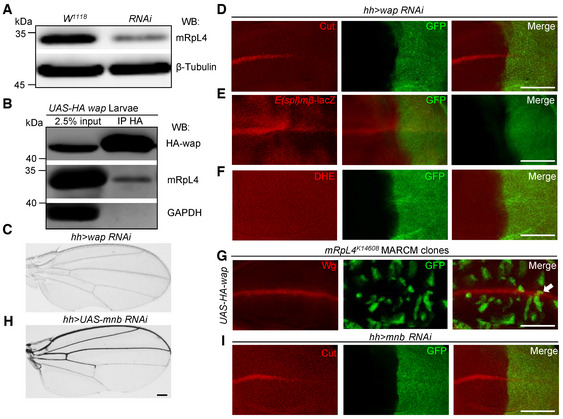
-
ARepresentative western blotting (n = 3 biological repeats) of lysates from wild‐type and UAS‐mRpL4‐RNAi‐expressing wing disks. The custom mRpL4 antibody was used and the β‐tubulin protein was included as loading control.
-
BRepresentative immunoprecipitation analysis (n = 3 biological repeats) using lysates from wing disks overexpressing HA‐tagged wap. Anti‐HA antibodies were used for immunoprecipitation. Western blotting was performed using anti‐HA and anti‐mRpL4 antibodies to reveal wap and mRpL4, respectively. GAPDH was used as control.
-
CRepresentative image of wings (n > 15 wings) from flies expressing UAS‐wap‐RNAi under the control of hh‐Gal4.
-
D–FRepresentative images of wing disks (n > 15 wing disks) from flies expressing UAS‐wap‐RNAi under the control of hh‐Gal4 that have been stained for Cut (D), E(spl)mβ‐LacZ (E) and DHE (F).
-
GRepresentative image of wing disks (n > 10 wing disks) bearing MARCM clones (marked by GFP) stained for Wg. In the mRpL4 K14608 MARCM clones, UAS‐HA‐wap are expressed under the control of tub‐Gal4.
-
HRepresentative image of wings (n > 15 wings) from flies expressing UAS‐mnb‐RNAi under the control of hh‐Gal4.
-
IRepresentative image of wing disks (n > 15 wing disks) from flies expressing UAS‐mnb‐RNAi under the control of hh‐Gal4 that have been stained for Cut.
Data information: Representative clone is marked by white arrow in (G). Scale bars = 100 μm.
Source data are available online for this figure.
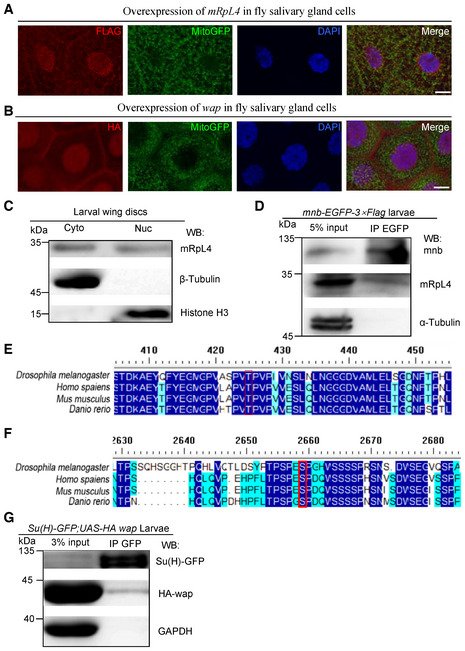
Mitochondrial ribosomal proteins are believed to function within mitochondria to synthesize OXPHOS proteins (Kummer & Ban, 2021). However, mRpL4 (Fig EV4A) and wap (Fig EV4B) were found in both cytoplasm and nucleus when overexpressed in fly salivary gland cells. Fractionation assays using wing disk cell lysates confirmed that endogenous mRpL4 protein was present in both the cytoplasmic and nucleus fractions (Fig EV4C). The presence of mRpL4 in cell nucleus is of particular interest, as we have shown that mRpL4 functions downstream of NICD to regulate Notch signal transduction. It has been reported that the Ser/Thr protein kinase minibrain (mnb) forms a heterodimer with wap, which phosphorylates key signaling components during fly wing development (Degoutin et al, 2013; Yang et al, 2016). A physical interaction between mnb and mRpL4 was detected by immunoprecipitation using wing disk cell lysates (Fig EV4D). RNAi knockdown of mnb in the developing wing resulted in wing margin nicks (Fig 5H) and reduction of Cut expression (Fig 5I). Searching of the mnb targeting sequence (Degoutin et al, 2013) identified residues [T426 in Su(H) and S2659 in Notch] that could be potentially recognized and phosphorylated by the wap‐mnb heterodimer (Fig EV4E and F). In wing disk cells, Su(H) was found to interact with wap when examined by immunoprecipitation (Fig EV4G). Taken together, we propose a model that mRpL4 interacts with wap‐mnb to regulate Notch signaling activity, probably acting on Su(H) to modulate the transcriptional output.
Figure EV4. mRpL4 and wap may function through Su(H) to regulate Notch signaling.
-
A, BRepresentative images of third instar larvae salivary glands (n > 10 larvae) from flies expressing FLAG‐tagged mRpL4 (A) and HA‐tagged wap (B). Immunostaining was performed using anti‐FLAG and anti‐HA antibodies to reveal mRpL4 and wap, respectively. Mitochondria are marked by GFP and cell nuclei are labeled by DAPI.
-
CRepresentative western blotting (n = 3 biological replicates) of mRpL4 protein distribution in cytoplasmic (Cyto) and nuclear (Nuc) fractions from wing disks lysates.
-
DRepresentative immunoprecipitation analysis (n = 3 biological replicates) using lysates from wing disks expressing GFP‐tagged mnb. Anti‐GFP antibodies were used for immunoprecipitation. Western blotting was performed using anti‐GFP and anti‐mRpL4 antibodies to reveal mnb and mRpL4, respectively. α‐Tubulin was used as control.
-
EAlignment of Su(H) protein sequences from fly, human, mice and zebrafish. The region covering the mnb phosphorylation consensus sequence is shown, and the conserved Thr residue (T426) is labeled by red box.
-
FAlignment of Notch protein sequences from fly, human, mice and zebrafish. The region covering the mnb phosphorylation consensus sequence is shown, and the conserved Ser residue (S2659) is labeled by red box.
-
GRepresentative immunoprecipitation analysis (n = 2 biological replicates) using lysates from wing disks expressing GFP‐tagged Su(H) and HA‐tagged wap. Anti‐GFP antibodies were used for immunoprecipitation. Western blotting was performed using anti‐GFP and anti‐HA antibodies to reveal Su(H) and wap, respectively. GAPDH was used as control.
Data information: Scale bars = 50 μm in (A and B).
Source data are available online for this figure.
The role of mRpL4 in Notch signaling regulation is conserved
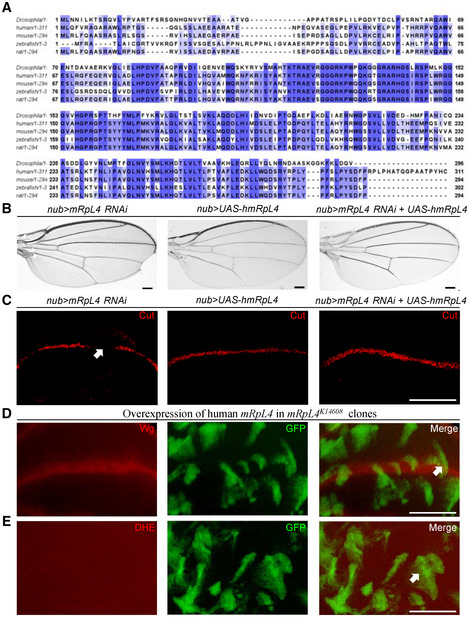
MRPs are known to play highly conserved roles for mitochondrial protein synthesis (Richter‐Dennerlein et al, 2015). Our findings indicate a moonlighting role of fly mRpL4 in Notch signaling regulation, whether such additional function is a common feature across different species or specifically acquired by Drosophila is an intriguing question. Amino acid sequence alignment reveals that mRpL4 is highly conserved from fly to human (Fig 6A). A transgenic fly expressing human mRpL4 protein was constructed to examine the biological activity of the homologous protein during fly wing development. The human mRpL4 transgene was sufficient to rescue both the adult wing margin defect (Fig 6B) and downregulation of Cut in the wing disk (Fig 6C) when co‐expressed with the RNAi construct. Importantly, the human mRpL4 protein was sufficient to restore both Wg expression (Fig 6D) and ROS production (Fig 6E) in mRpL4 K14608 mutant cells. These results indicate that mRpL4 might play a conserved role in Notch signal regulation.
Figure 6. mRpL4 regulates Notch signaling in the Drosophila wing.
-
AAlignment of mRpL4 protein sequences from fly, human, mice, zebrafish and rat. The conserved residues are labeled with blue shadow.
-
BRepresentative images of wings (n > 15 wings) from flies expressing mRpL4 RNAi, UAS‐hmRpL4 and both under the control of nub‐Gal4.
-
CRepresentative images of wing imaginal disks (n > 10 wing disks) stained for Cut from flies expressing mRpL4 RNAi, UAS‐hmRpL4 and both under the control of nub‐Gal4. Representative regions showing Notch activity defects are marked by white arrow.
-
D, ERepresentative images of wing disks (n > 10 wing disks) bearing MARCM clones stained for Wg (D) and DHE (E). The MARCM clones are marked by GFP, and UAS‐hmRpL4 are expressed under the control of tub‐Gal4 in these clones.
Data information: Representative clones are marked by white arrows (D, E). Scale bars = 100 μm.
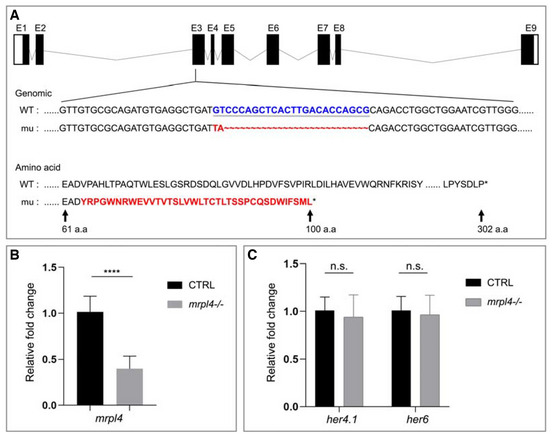
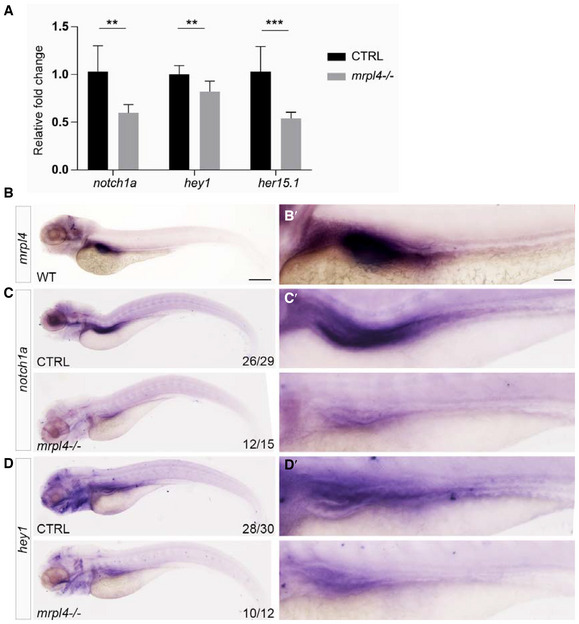
To further investigate whether mRpL4 modulates Notch signaling in vertebrates, we analyzed the role of mRpL4 homolog in zebrafish, an excellent vertebrate model for developmental studies (Zhao et al, 2021). The zebrafish mRpL4 (zmRpL4) null allele was generated using CRISPR‐Cas9‐mediated genome editing system. The isolated mutant allele, zmRpL4 mu , carries an indel (24‐bp deletion and 2‐bp insertion) that leads to frameshifting and premature translation termination. As a result, the mutant allele produces a small hybrid protein that retains the first 63 amino acids of wild‐type zmRpL4, followed by 37 irrelevant amino acids (Fig EV5A). In the homozygous zmRpL4 mu mutant larvae, the mRNA level of zmRpL4 was significantly decreased (Fig EV5B), indicating that the mutation inhibits transcript production and/or impairs the mRNA stability. Using quantitative real‐time PCR analysis, we surveyed the expression level of several members of the Notch signaling pathway. In 5‐day postfertilization (dpf) larvae, the expression level of Notch signaling receptor gene, notch1a, and target genes, hey1 and her15.1, was significantly reduced in the zmRpL4 mu mutant (Fig 7A). The expression of two other Notch target genes, her4.1 and her6, was not affected (Fig EV5C). These observations suggest that Notch signaling lies genetically downstream of zmRpL4 during zebrafish development. Next, we visualized the expression pattern of Notch signaling components by in situ hybridization. At 5 dpf, zmRpL4 was highly expressed in the digestive system, mainly in the liver and intestinal bulb (Fig 7B). In consistent with previous studies (Lorent et al, 2004; Crosnier et al, 2005), notch1a (Fig 7C) and hey1 (Fig 7D) were also expressed in the digestive system. When examined by whole‐mount RNA hybridization, notch1a (Fig 7C) and hey1 (Fig 7D) expression was found to be severely decreased in the zmRpL4 mu mutant larvae at 5 dpf. Taken together, these data demonstrate that Notch signaling is also modulated by mRpL4 during zebrafish development.
Figure EV5. Function of mRpL4 in zebrafish.
-
ASchematic diagrams of wild‐type (WT) and the zmRpL4 mutant allele (mu) generated by the CRISPR‐Cas9 genome editing system. The mutant allele contains a 24‐bp deletion (GTCCCAGCTCACTTGACACCAGCG, in blue) and a 2‐bp insertion (TA, in red) in the third exon. As a result, the mutant allele encodes a small polypeptide of 100 amino acid residues containing part of the wild‐type residues and a disordered Carbon‐terminal tail due to frame shifting (amino acid, in red).
-
BThe mRNA levels of zmRpL4 in wild‐type control and mrpl4‐null larvae at 5 dpf as measured by quantitative PCR (three biological replicates for each genotype and three technical replicates for each sample). Statistical significance was tested using two‐tailed unpaired t‐test. Error bars represent ± SD, ****P < 0.00001.
-
CBar graph showing relative levels of Notch signaling target genes her4.1 and her6 in mrpl4‐null larvae comparing to wild‐type control at 5 dpf as measured by quantitative PCR (n = 3 biological replicates per group). Statistical significance was tested using two‐tailed unpaired t‐test. Error bars represent ± SD; n.s., not significant.
Figure 7. Zebrafish mRpL4 modulates expression of Notch signaling components during development.
-
AExpression of notch1a, hey1, and her15.1 mRNA in mrpl4‐null larvae comparing to wild‐type control at 5 dpf as measured by quantitative PCR (three biological replicates for each genotype and three technical replicates for each sample). Statistical significance was tested using two‐tailed unpaired t‐test. Error bars represent ± SD; **P < 0.01, ***P < 0.001.
-
BRepresentative bright field image of 5 dpf wild‐type larva (n > 30) illustrated by in situ hybridization with a mrpl4 riboprobe.
-
CRepresentative bright field image of 5 dpf CTRL (n = 26/29) and mrpl4 mutant (n = 12/15) illustrated by in situ hybridization with a notch1a riboprobe.
-
DRepresentative bright field images of 5 dpf CTRL (n = 28/30) and mrpl4 mutant (n = 10/12) illustrated by in situ hybridization with a hey1 riboprobe.
Data information: Scale bars: 250 μm in (B–D); 100 μm in (B′–D′).
Discussion
Mitochondrial ribosomal proteins are essential for animal development and have been implicated in developmental defects and genetic disorders, but it is unknown whether and how MRPs could regulate Notch signaling. Here, we report that mRpL4 regulates Notch signaling activation downstream of NICD production during fly wing development. The observations that human mRpL4 protein rescues fly wing developmental defects and that zebrafish Notch signaling is also impacted in zmRpL4 mutant indicate that this regulatory mode might be conserved in vertebrates. The WD40 repeat protein wap and the Ser/Thr protein kinase mnb provide a potential link between mRpL4 and Su(H). Given that the only annotated domain of mRpL4 (ribosomal L4) functions to bind rRNA, it is difficult to interpret how mRpL4 exerts regulatory function on Su(H) protein. mRpL4 protein might possess undiscovered biochemical activity, but several clues suggest that the mnb kinase might be involved to modify Notch signaling. We found that, similar as mRpL4 and wap, mnb also positively regulates Notch signaling during fly wing development.
Both Su(H) and NICD bear conserved mnb phosphorylation consensus sequences, and the potential functional significance is discussed below. Phosphorylation at various sites impacts Su(H) protein stability and affinity with DNA, as well as the formation and dynamics of repressor or activator complex (Auer et al, 2015; Nagel et al, 2017; Frankenreiter et al, 2021; Fechner et al, 2022). The presumptive mnb target residue (T426) lies in the C‐terminal domain (CTD) of Su(H), a region that is not involved in DNA binding (Kovall & Hendrickson, 2004; Wilson & Kovall, 2006). Thus, although mRpL4 and wap might recruit mnb to phosphorylate Su(H), such modification will unlikely alter its affinity with chromosome. The T426 phosphorylation site resides in the conserved β‐strand motif that interacts with the ankyrin repeats domain of NICD (Nam et al, 2006; Choi et al, 2012) and the transcription repressor Hairless (Yuan et al, 2016). It is attempting to speculate that T426 phosphorylation could potentially affect the interaction of Su(H) with NICD and Hairless, which in turn may modulate the composition, stability, activity, and turnover of Su(H) transcription regulatory complexes. These hypotheses could help to explain the reduced occupation of Su(H) on Notch targets observed in mRpL4 RNAi wing disk cells. Interestingly, the mitogen‐activated protein kinase (MAPK) phosphorylates Su(H) at P424 to attenuate Notch signaling (Auer et al, 2015; Fechner et al, 2022). The mnb and MAPK phosphorylation sites are in such close proximity, making it hard to ignore the potential antagonistic effect between them. At present, we could not rule out the possibility that mnb may also target less conserved consensus sites in other domains of Su(H) to modify its activity. Alternatively, unknown kinases that interact with wap could contribute to modification and regulation of Su(H). The vertebrate orthologs of mnb are known to phosphorylate NICD and attenuate Notch signaling (Fernandez‐Martinez et al, 2009; Hämmerle et al, 2011; Morrugares et al, 2020); whether mnb could phosphorylate NICD during fly development is still illusive. Further investigations are needed to reveal how mRpL4 and wap‐mnb regulate Su(H) and Notch signaling activity.
Apart from the co‐factors of transcription regulatory complexes, the heterogeneous nuclear ribonucleoprotein Hrb27/Hrp48 is a recently identified nuclear protein that regulates Notch signaling during fly wing development. Hrb27 utilizes at least two separate pathways to modulate Notch signaling. In female flies, Hrb27 represses the expression of the sex determination master gene Sex‐lethal (Sxl) to ensure a proper amount of Notch during wing development (Suissa et al, 2010). Sxl protein binds Notch mRNA and inhibits Notch protein translation in ovary cells (Penn & Schedl, 2007), but whether similar mode of action exists in wing disk cells has not been directly tested yet. In both males and females, Hrb27 interacts with the ubiquitin ligase Deltex (Dx) to attenuate Notch signaling activity in a Sxl‐independent manner (Dutta et al, 2017, 2020). Epistasis assays demonstrate that Hrb27 functions upstream of NICD in both pathways (Suissa et al, 2010; Dutta et al, 2017). In our hands, mRpL4 regulates Notch signaling activity in both sexes and likely functions downstream of NICD. We believe that Hrb27 and mRpL4 might not directly collaborate with each other to regulate Notch signaling.
Recent studies have begun to reveal that mitochondria and related proteins could regulate Notch signaling through various pathways. Mitochondrial fission factor Drp1 promotes Notch activation in fly ovariole follicle cells (Mitra et al, 2012) and human breast cancer cells (Chen et al, 2018b). Mitochondrial fusion proteins inhibit Notch activity in fly lymph gland (Ray et al, 2021) and mouse embryonic heart (Cao et al, 2016), but act to promote Notch activation in mouse neural stem cells (Khacho et al, 2016). In fly neuroblasts, depletion of mitochondrial fusion protein Opa1, but not the other fusion protein Marf, leads to reduction of Notch pathway activity (Dubal et al, 2022). These studies indicate that mitochondria morphology and Notch signaling might be generally associated during animal development, but the mode of action varies significantly among different tissues. In several developmental events, the effects of mitochondria on Notch signaling are mediated by one of its major metabolites, ROS. Mitochondrial transcription factor A (TFAM) maintains cellular ROS production to activate Notch signaling during mouse keratinocytes differentiation (Hamanaka et al, 2013). On the contrary, ROS inhibits Notch activity by triggering autophagic degradation of NICD in mouse hematopoietic stem cells (Cao et al, 2016) and by inducing expression of inhibitory gene in mouse neural stem cells (Khacho et al, 2016). In fly wing disk cells, burst of ROS stimulates TOR activity, which activates Notch signaling as a secondary response (Perez‐Gomez et al, 2020). Thus, ROS also impacts Notch signaling activity in a developmental context‐dependent manner. Unlike in mouse keratinocytes, reduction of cellular ROS is insufficient to inhibit Notch activation in fly wing imaginal disk. We demonstrated that fly mRpL4 regulates Notch signaling through a separate route which is likely independent of OXPHOS protein synthesis, thus adding another layer to the complex regulatory network between mitochondria and Notch signaling.
Based on our findings and previous studies, we argue that the functional diversity of MRPs might be seriously underestimated. Increasing evidence indicates that MRPs perform common as well as separate functions during animal development. For the five MRPs we examined, all of them are required for ROS production in wing disk cells, but only mRpL4 regulates Notch signaling activity. During fly eye development, mRpL4 and mRpL17, but not mRpS15 and mRpL12, are required for cell cycle progressing (Galloni, 2003; Frei et al, 2005; Mandal et al, 2005). Instead, mRpL12 possesses a unique ability to regulate cell growth in larval eye disk, wing disk, and fat body cells (Frei et al, 2005). mRpL55 is essential for eye cell survival in the pupal stage, without affecting cell cycle nor cell growth in the larvae (Tselykh et al, 2005). We noticed that mutation of mRpL4 leads to the accumulation of cellular ROS in fly ovary follicle cells (Wang et al, 2012), but results in reduction of ROS in wing disk (Fig 3A) and eye disk cells (Mandal et al, 2005). Given that the same mRpL4 allele is used, it is likely that mRpL4 regulates mitochondrial OXPHOS activity in a tissue‐specific manner. The underlying mechanism and the biological significance of tissue‐specific as well as component‐specific functions of fly MRPs demand further investigation. We also looked into the ProteinAtlas database (www.proteinatlas.org; Thul et al, 2017) to assess the expression pattern of vertebrate MRPs. The subcellular localization status of 58 human MRPs has been examined by immunofluorescence analysis, and about half of them (27/58) are found in cellular compartments other than mitochondria. Interestingly, around one third of these MRPs (19/58) are present within or nearby cell nucleus. As for mRpL4, no classical nuclear localization signal (NLS) motifs are found in fly and vertebrate proteins. However, a pat‐7 type NLS (PDKRFRL) was identified in wap using the PSORT predication program (Horton et al, 2007). It is possible that mRpL4 and other MRPs might be localized to distinct cellular compartments by their interacting partners. Therefore, in contrast to the general thoughts of being confined in the mitochondria to synthesis OXPHOS proteins, many MRPs may have “part‐time jobs” in other cellular organelles. Comprehensive investigations of how MRPs regulate cell cycle, cell growth, and tissue development would help us to better understand their diverse functions.
Materials and Methods
Drosophila genetics
The Gal4‐UAS system was utilized for tissue‐specific gene expression. The Hh‐Gal4, Sgs‐Gal4 (Du et al, 2016), nub‐Gal4 (Bloomington Drosophila Stock Center, BDSC 86108), and 1407‐Gal4 (BDSC 8751) stocks were used to drive transgene and RNAi expression. To induce mosaic clones, the following stocks were used: Ubx‐FLP;Ubi‐mRFP, FRT40A; Ubx‐FLP;Ubi‐GFP, FRT40A and Ubx‐FLP; FRT82B, Ubi‐mRFP. The MARCM experiments were performed with the hs‐FLP, tub‐GAL4, UAS‐GFP; tub‐GAL80, FRT40A stock. We used mRpL4 K14608 , FRT40A (Kyoto Stock Center, KSC 111435), FRT82B, CoVa tend (BDSC 33839), FRT42D, mRpS28 k13104 (KSC 111025) and mRpL24 f06692 , FRT40A (KSC 114548) for genetic analysis. The NRE‐GFP (BDSC 30728) and E(spl)mβ‐LacZ (Yu et al, 2013) reporters were used to visualize Notch signaling activity. The RNAi lines used in this work are as follows: UAS‐mRpL4 RNAi (Vienna Drosophila Resource Center, VDRC 101351), UAS‐wap RNAi (VDRC 107076), UAS‐mnb RNAi (VDRC 107066), UAS‐CoVa RNAi (VDRC 109070), UAS‐CoVa RNAi‐2 (BDSC 58282), UAS‐mRpS2 RNAi (National Institute of Genetics, Japan, 2937R‐1), UAS‐mRpS12 RNAi (BDSC 38251), UAS‐ND‐42 RNAi (BDSC 58282), UAS‐mRpL24 RNAi (VDRC 103782), UAS‐mRpS28 RNAi (VDRC 107181), UAS‐ND‐15.6 RNAi (Tsinghua Fly Center, THFC 1766), UAS‐Pdsw RNAi (THFC 3175), and UAS‐ND‐19 RNAi (THFC 4895). The transgenic lines used in this work are as follows: UAS‐Delta (BDSC 26695), UAS‐Serrate (BDSC 5815), UAS‐N FL (BDSC 26820), UAS‐NEXT (BDSC 63220), and UAS‐NICD (Go et al, 1998). The Su(H)‐GFP and mnb‐GFP stocks carried BAC transgenes that express GFP‐tagged fusion proteins under the control of their genomic regulatory sequence (Sarov et al, 2016). The Su(H)‐lacZ; Esg‐Gal4, UAS‐mCD8‐GFP, tub‐Gal80TS stock was used to monitor Notch signal activity in adult midgut cells (Zhao et al, 2022).
The somatic mosaic screening was performed by crossing the Bruinfly mutant stocks (Chen et al, 2005) with virgin females bearing Ubx‐Flp and corresponding FRT site. The wing morphology of the progenies was examined, and mutants that resulted in wing margin nicks were selected for secondary screen. The Notch signaling activity was monitored in third instar larval wing imaginal disks in the secondary screen (Ren et al, 2018). For MARCM analysis, virgin females of mRpL4 K14608 , FRT40A were first crossed with desirable transgenic strain and males of the progeny were picked to mate with the FRT40A MARCM stock. Three‐day‐old larval from the second cross were then heat‐shocked at 37°C for 1 h using water bath to induce clones.
Generation of transgenic flies
Total RNAs were extracted from third instar larvae with TRIeasy reagent (Yeasen Biotech, Shang Hai, China) and followed by cDNA synthesis with Hifair First Strand cDNA Synthesis Kit (Yeasen Biotech). Sequence‐specific primers (Appendix Table S1) were designed to amplify cDNA fragments of fly mRpL4 and wap by High‐Fidelity Master Mix (Molecular Cloning Laboratories, MCLAB). Human mRpL4 ortholog was amplified from a cDNA clone provided by Dr. Jiahuai Han. The cDNAs were subcloned into pUAST‐attB vector using EcoR I and Kpn I sites by ClonExpress II One Step Cloning Kit (Vazyme). The transgenic flies were generated by phiC31 integrase‐mediated site‐specific insertion at the left arm of third chromosome with cytogenetic location at 68A4. The embryonic injections were performed by Core Facility of Drosophila Resource and Technology at the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (Shanghai, China) and Fungene Biotechnology (Qidong, Jiangshu Province, China).
Immunofluorescence staining
Third instar larvae were dissected in ice‐cold PBS and fixed in 4% paraformaldehyde for 15 min and then washed in PBST (PBS containing 0.1% Triton X‐100) for 15 min. The larvae were blocked in PBST with 0.2% BSA for 1 h before incubating with primary antibodies overnight at 4°C. The following primary antibodies were used: mouse anti‐Cut (1:200; 2B10; Developmental Studies Hybridoma Bank, DSHB), mouse anti‐Wingless (1:200; 4D4; DSHB), mouse anti‐Notch intracellular domain (1:200; C17.9C6; DSHB), mouse anti‐Delta (1:200; C594.9B; DSHB), rabbit anti‐LacZ (1:4000; Cappel), and rabbit anti‐GFP (1:50; G10362; Thermo Fisher). Alexa flour‐conjugated secondary antibodies (1:400; Invitrogen) were used. Wing disks and adult midguts were dissected and mounted in 40% glycerol for imaging.
Dihydroethidium staining
We monitored cellular ROS level in wing disks by DHE staining (Robinson et al, 2006). Third instar larvae were incubated for 10 min at 22°C in SFX‐Insect medium (HyClone) containing 8 μM DHE (Sigma) and then rinsed twice in SFX‐Insect medium before fixed in 4% paraformaldehyde for 5 min. The wing disks were rinsed once with PBS and mounted in 40% glycerol.
Generation of mRpL4 antibody
A rabbit polyclonal antibody was generated against a synthetic peptide (DYTDCLPVSRNTARQAW) corresponding to amino acids 52–68 of the Drosophila mRpL4 protein (ABclonal, Wuhan, China). The specificity of rabbit antisera was examined by immunoblotting (1:2,000) using protein lysates extracted from wild‐type and mRpL4 RNAi‐expressing wing disks (Fig 5A). Note that this antibody is not suitable for immunofluorescence.
Biochemistry
Third instar larval brain and imaginal disks were lysed in RIPA buffer (150 mM sodium chloride [NaCl], 1.0% NP‐40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris–HCl, pH 8.0) supplemented with protease inhibitor cocktail (20124ES10; Yeasen Biotech). Immunoblotting analyses were carried out using standard protocols. The following antibodies were used for immunoblotting: mouse anti‐α‐Tubulin (1:5,000; A11126; Thermo Fisher), mouse anti‐β‐Tubulin (1:1,000; E7; DSHB), rabbit anti‐GAPDH (1:2,000; sc25778; Santa Cruz), rabbit anti‐HA (1:5,000; 3,724; Cell Signaling), mouse anti‐FLAG (1:5,000; F1804; Thermo Fisher), rabbit anti‐GFP (1:400; G10362; Thermo Fisher), rabbit anti‐mRpL4 (1: 2,000; E6023; ABclonal), and mouse anti‐Histone3 (1:20,000; BE3015; EasyBio, Beijing, China).
Immunoprecipitations were performed using anti‐HA agarose (E6779; Sigma) and magnetic anti‐GFP beads (GNM‐25‐1000; Lablead) according to the manufacturer's instructions. Third instar larval brain and imaginal disks were lysed in NP‐40 buffer (150 mM sodium chloride [NaCl], 1.0% NP‐40, 50 mM Tris–HCl, pH 8.0) supplemented with protease inhibitor cocktail (20124ES10; Yeasen Biotech).
Subcellular fractionations were separated as previously described with minor modification (Du et al, 2016). Briefly, larval brain and imaginal wing disks were collected at 1,250 g for 10 min and washed in PBS before lysed in Buffer I (15 mM Hepes pH 7.4, 10 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT) for 30 min on ice to generate the cytoplasmic fraction. Nuclei were pelleted at 3,000 g for 10 min and washed twice in Buffer I before resuspended in Buffer II (50 mM Hepes, pH 7.6, 110 mM KCl, 150 mM NaCl, 1% Triton X‐100, 10% glycerol, 1 mM DTT, 0.1 mM EDTA) for nuclear protein extraction. The precipitate is removed by centrifugation at 5,000 g for 10 min. All lysis buffers were supplemented with protease inhibitor cocktail (20124ES10; Yeasen Biotech).
Mounting of adult fly wings
Adult flies with correct genotypes were collected and fixed overnight in isopropanol. Dissected adult wings were mounted in Euparal mounting media (BioQuip).
Yeast two‐hybridization
Yeast two‐hybridization was performed with the Matchmaker Gold Yeast Two‐Hybrid System (Takara, 630489). The mRpL4 cDNAs were cloned into pGBKT7 vector using Nde I and EcoR I sites by ClonExpress II One Step Cloning Kit (Vazyme) and expressed as a fusion to the Gal4 DNA‐binding domain as bait protein. We confirmed that the mRpL4 bait does not autonomously activate the reporter genes in the absence of a prey protein. A concentrated overnight culture of the bait strain was combined with the Mate & Plate Library—Universal Drosophila (Takara, 630485) strain and incubated at 30°C for 20–24 h with slowly shaking (30 rpm) to allow mating between the two strains. Presence of yeast zygotes was examined under a phase contrast microscope (40×), and the mated culture was spread on agar plates with double dropout media containing 40 μg/ml X‐alpha‐Gal and 200 ng/ml Aureobasidin A (DDO/X/A). All the blue colonies that grew on DDO/X/A were transferred to higher stringency agar plates with quadruple dropout media containing 40 μg/ml X‐alpha‐Gal and 200 ng/ml Aureobasidin A (QDO/X/A). The QDO/X/A positive clones were further analyzed to verify the interactions and identify the insert in the prey plasmid. Two candidate preys were isolated, and the cDNA inserts were sequenced.
Chromatin immunoprecipitation (ChIP)
The ChIP experiments form wing imaginal disks were performed based on previously described protocols (Krejcí & Bray, 2007; Du et al, 2016). Roughly, 2000 wing disks were dissected in ice‐cold SFX‐insect medium for each genotype. After fixation in 1.8% formaldehyde on a rotating wheel at room temperature for 15 min, disks were divided into two parts for fragmentation by sonication (ON = 30 s, OFF = 1 min, high power, 4 times). The size of bulk DNA fragments ranges from 100 to 300 bp. Mouse monoclonal antibody against Su(H) (sc398453, Santa Cruz Biotechnology) was used for immunoprecipitation. The Perfect Start Green qPCR Super Mix (TransGen Biotech) was used for real‐time quantitative PCR experiments conducted on an ABI QuantStudio 6 Flex System (Thermo Fisher, USA). Primers are designed to cover the Su(H) binding sites in the regulatory regions of cut, Wg, vestigial, and E(Spl) family genes (Bailey & Posakony, 1995; Lecourtois & Schweisguth, 1995; Klein & Arias, 1999; Krejcí & Bray, 2007). The sequences of primers are shown in Appendix Table S1. The value of % Input was calculated by the following formula:
Generation of zebrafish zmRpL4 mutant allele
The zebrafish zmRpL4 mutant line was generated using the CRISPR/Cas9 system. Potential CRISPR target sites were screened on the CRISPRscan website (https://www.crisprscan.org/). To generate the F0 generation of mutants, 200 pg zmRpL4 guide RNA (targeting sequence 5′‐GGGCTGGTGTCAAGTGAGCT‐3′, located in the 3rd exon) and 200 pg Cas9 mRNA were co‐injected into wild‐type embryos at one‐cell stage. The F1 generation was genotyped by PCR amplifying a genomic DNA fragment harbored the zmRpL4 target site and subsequent sequencing. The zmRpL4 mutant allele was identified, containing a 24‐base pair (bp) deletion and a 2‐bp insertion at exon 3 (Fig EV5A).
qRT–PCR
Wing imaginal disks were dissected from third instar larvae, and total RNAs were extracted by TRIeasy reagent (Yeasen Biotech), followed by cDNA synthesis with Hifair First Strand cDNA Synthesis Kit (Yeasen Biotech). Around 200 wing disks were collected for each genotype. The Perfect Start Green qPCR Super Mix (TransGen) was used for real‐time quantitative PCR experiments conducted on an ABI QuantStudio 6 Flex System (Thermo Fisher). For zebrafish experiments, 5 dpf wild‐type and zmRpL4 mutant larvae were collected for each biological replicate, and total RNA was isolated using the TRNzol Universal (TIANGEN, China), following reverse transcription into cDNA using TransScript® Uni All‐in‐One First‐Strand cDNA Synthesis SuperMix system (TransGen) according to the manufacturer's protocol. The quantitative PCR was performed using PerfectStart® Green qPCR SuperMix (TransGen) with a QuantStudio™ 3 Real‐Time PCR Instrument (Applied Biosystems). The expression of β‐actin was used for normalization to calculate fold differences in selected transcripts between experimental groups. The sequence of qPCR primers used in this study is provided in Appendix Table S1.
Zebrafish whole‐mount in situ hybridization
At 5 dpf, zebrafish larvae were fixed in 4% paraformaldehyde (PFA) at 4°C overnight and rinsed in PBST (0.1% Tween‐20 in PBS) following subsequently dehydrated in methanol and stored at −20°C. The whole‐mount in situ hybridization was performed as described previously (Thisse & Thisse, 2008). To generate antisense riboprobes of mrpl4 and hey1, cDNA fragments were amplified (the sequence of primers was provided in Appendix Table S1) and cloned into pEASY‐T3 (TransGen Biotech) and TOPO (Life Technologies) vectors, respectively. The plasmids containing zmRpL4, hey1, and notch1a (Zhao et al, 2014) cDNA fragments were linearized, and the digoxigenin‐labeled antisense RNA riboprobes were in vitro transcribed using a DIG RNA Labeling Kit (Roche). The hybridized probes were detected with alkaline‐phosphatase‐conjugated anti‐digoxigenin antibody (Roche), and the signal was developed with BM purple (Roche).
Image capture and processing
The fluorescence images were acquired with a Leica SP8 confocal microscope and processed in Photoshop and ImageJ. The following detection wavelengths were used: 510–530 nm for Alexa 488, 525–550 nm for GFP, 580–600 nm for RFP, and 590–610 nm for Alexa 568. The images of adult wings were acquired with a Leica DMIL inverted microscope equipped with a QImaging QICAM Fast 1394 digital camera. Minor image adjustments (overall brightness and/or contrast) were performed in Photoshop and Microsoft PowerPoint.
Statistics
All genetic experiments were performed independently at least two times, and independent but genetically identical samples were used. For adult wing phenotypes, at least 30 flies were analyzed. For wing imaginal disk staining, at least 10 disks were examined for each genotype. For qRT–PCR analysis, three biological repeats were performed. Error bars show mean ± standard error of measurement. Boxplots show median (thick line in the box) and maximum values (whiskers). Statistical significance was tested using two‐tailed unpaired t‐test. P‐value was indicated as follows: *P < 0.05, N.S., not significant.
Author contributions
Dongqing Mo: Data curation; formal analysis; investigation; writing – original draft. Chenglin Liu: Data curation; formal analysis; investigation. Yao Chen: Data curation; formal analysis; validation. Xinkai Cheng: Data curation; formal analysis; investigation. Jie Shen: Conceptualization; supervision; funding acquisition; project administration; writing – review and editing. Long Zhao: Conceptualization; funding acquisition; project administration; writing – review and editing. Junzheng Zhang: Conceptualization; supervision; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
PDF+
Source Data for Figure 5
Acknowledgments
We would like to thank Drs. Alan Jian Zhu, Renjie Jiao, Zhouhua Li, Wei Wu, the Bloomington Stock Center, the Kyoto Fly Stock Center, the Tsinghua Fly Stock Center, the Vienna Drosophila Resource Center, National Institute of Genetics (NIG), and the Developmental Studies Hybridoma Bank (DSHB) for fly stocks and antibodies. We thank Dr. Jiahuai Han for providing human mRpL4 cDNA clone. We thank Haomiao Li for helping with adult fly gut experiments. We thank Core Facility of Drosophila Resource and Technology, CEMCS, CAS, and Fungene Biotechnology (Qidong, Jiangshu Province, China) for generation of transgenic fly strains. This work was supported by the National Natural Science Foundation of China (31970478 and 31772526 to J.Z; 32030012 and 31872295 to J.S.; 31970506 and 32170541 to L.Z.).
EMBO reports (2023) 24: e55764
Data availability
This study includes no data deposited in external repositories.
References
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S (2001) Presence of mitochondria‐type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci USA 98: 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer JS, Nagel AC, Schulz A, Wahl V, Preiss A (2015) MAPK‐dependent phosphorylation modulates the activity of suppressor of hairless in Drosophila . Cell Signal 27: 115–124 [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW (1995) Suppressor of hairless directly activates transcription of Enhancer of split complex genes in response to Notch receptor activity. Genes Dev 9: 2609–2622 [DOI] [PubMed] [Google Scholar]
- Basak NP, Roy A, Banerjee S (2014) Alteration of mitochondrial proteome due to activation of Notch1 signaling pathway. J Biol Chem 289: 7320–7334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ (2006) Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Bio 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Cao Y, Fang Y, Cai J, Li X, Xu F, Yuan N, Zhang S, Wang J (2016) ROS functions as an upstream trigger for autophagy to drive hematopoietic stem cell differentiation. Hematology 21: 613–618 [DOI] [PubMed] [Google Scholar]
- Chen J, Call GB, Beyer E, Bui C, Cespedes A, Chan A, Chan J, Chan S, Chhabra A, Dang P et al (2005) Discovery‐based science education: functional genomic dissection in Drosophila by undergraduate researchers. PLoS Biol 3: e59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Tiosano D, Guran T, Baris HN, Bayram Y, Mory A, Shapiro‐Kulnane L, Hodges CA, Akdemir ZC, Turan S et al (2018a) Mutations in the mitochondrial ribosomal protein MRPS22 lead to primary ovarian insufficiency. Hum Mol Genet 27: 1913–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhang J, Lyu Z, Chen Y, Ji X, Cao H, Jin M, Zhu J, Yang J, Ling R et al (2018b) Positive feedback loop between mitochondrial fission and notch signaling promotes survivin‐mediated survival of TNBC cells. Cell Death Dis 9: 1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong A, Archambault D, Degani R, Iverson E, Tremblay KD, Mager J (2020) Nuclear‐encoded mitochondrial ribosomal proteins are required to initiate gastrulation. Development 147: dev188714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Wales TE, Nam Y, O'Donovan DJ, Sliz P, Engen JR, Blacklow SC (2012) Conformational locking upon cooperative assembly of Notch transcription complexes. Structure 20: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Vargesson N, Gschmeissner S, Ariza‐McNaughton L, Morrison A, Lewis J (2005) Delta‐notch signalling controls commitment to a secretory fate in the zebrafish intestine. Development 132: 1093–1104 [DOI] [PubMed] [Google Scholar]
- Dai SH, Wu QC, Zhu RR, Wan XM, Zhou XL (2020) Notch1 protects against myocardial ischaemia‐reperfusion injury via regulating mitochondrial fusion and function. J Cell Mol Med 24: 3183–3191 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Degoutin JL, Milton CC, Yu E, Tipping M, Bosveld F, Yang L, Bellaiche Y, Veraksa A, Harvey KF (2013) Riquiqui and minibrain are regulators of the hippo pathway downstream of Dachsous. Nat Cell Biol 15: 1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhang J, He T, Li Y, Su Y, Tie F, Liu M, Harte PJ, Zhu AJ (2016) Stuxnet facilitates the degradation of Polycomb protein during development. Dev Cell 37: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubal D, Moghe P, Verma RK, Uttekar B, Rikhy R (2022) Mitochondrial fusion regulates proliferation and differentiation in the type II neuroblast lineage in Drosophila . PLoS Genet 18: e1010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Paul MS, Singh A, Mutsuddi M, Mukherjee A (2017) Regulation of notch signaling by the heterogeneous nuclear ribonucleoprotein Hrp48 and Deltex in Drosophila melanogaster . Genetics 206: 905–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Mutsuddi M, Mukherjee A (2020) Regulation of notch signaling in Drosophila melanogaster: the role of the heterogeneous nuclear ribonucleoprotein Hrp48 and Deltex. Adv Exp Med Biol 1227: 95–105 [DOI] [PubMed] [Google Scholar]
- Fechner J, Ketelhut M, Maier D, Preiss A, Nagel AC (2022) The binding of CSL proteins to either co‐activators or co‐repressors protects from proteasomal degradation induced by MAPK‐dependent phosphorylation. Int J Mol Sci 23: 12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Martinez J, Vela EM, Tora‐Ponsioen M, Ocaña OH, Nieto MA, Galceran J (2009) Attenuation of notch signalling by the Down‐syndrome‐associated kinase DYRK1A. J Cell Sci 122: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Ferrari A, Del'Olio S, Barrientos A (2021) The diseased mitoribosome. FEBS Lett 595: 1025–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenreiter L, Gahr BM, Schmid H, Zimmermann M, Deichsel S, Hoffmeister P, Turkiewicz A, Borggrefe T, Oswald F, Nagel AC (2021) Phospho‐site mutations in transcription factor suppressor of hairless impact notch signaling activity during hematopoiesis in Drosophila . Front Cell Dev Biol 9: 658820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C, Galloni M, Hafen E, Edgar BA (2005) The Drosophila mitochondrial ribosomal protein mRpL12 is required for cyclin D/Cdk4‐driven growth. EMBO J 24: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M (2003) Bonsaï, a ribosomal protein S15 homolog, involved in gut mitochondrial activity and systemic growth. Dev Biol 264: 482–494 [DOI] [PubMed] [Google Scholar]
- Go MJ, Eastman DS, Artavanis‐Tsakonas S (1998) Cell proliferation control by notch signaling in Drosophila development. Development 125: 2031–2040 [DOI] [PubMed] [Google Scholar]
- Gomez‐Lamarca MJ, Falo‐Sanjuan J, Stojnic R, Abdul Rehman S, Muresan L, Jones ML, Pillidge Z, Cerda‐Moya G, Yuan Z, Baloul S et al (2018) Activation of the notch signaling pathway in vivo elicits changes in CSL nuclear dynamics. Dev Cell 44: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Kankel MW, Artavanis‐Tsakonas S (2012) The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet 13: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, Getsios S, Gottardi CJ, DeBerardinis RJ, Lavker RM et al (2013) Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal 6: ra8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämmerle B, Ulin E, Guimera J, Becker W, Guillemot F, Tejedor FJ (2011) Transient expression of Mnb/Dyrk1a couples cell cycle exit and differentiation of neuronal precursors by inducing p27KIP1 expression and suppressing Notch signaling. Development 138: 2543–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, An O, Hong H, Chan THM, Song Y, Shen H, Tang SJ, Lin JS, Ng VHE, Tay DJT et al (2020) Suppression of adenosine‐to‐inosine (A‐to‐I) RNA editome by death associated protein 3 (DAP3) promotes cancer progression. Sci Adv 6: eaba5136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Schweisguth F (2019) Mechanisms of notch signaling: a simple logic deployed in time and space. Development 146: dev172148 [DOI] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams‐Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35: W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Li H, Zhang H (2020) Abnormal expression of mitochondrial ribosomal proteins and their encoding genes with cell apoptosis and diseases. Int J Mol Sci 21: 8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A, Cipolat S, Chen Y, Dorn GW 2nd, Scorrano L (2013) Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science 342: 734–737 [DOI] [PubMed] [Google Scholar]
- Khacho M, Clark A, Svoboda DS, Azzi J, MacLaurin JG, Meghaizel C, Sesaki H, Lagace DC, Germain M, Harper ME et al (2016) Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Cell Stem Cell 19: 232–247 [DOI] [PubMed] [Google Scholar]
- Klein T, Arias AM (1999) The vestigial gene product provides a molecular context for the interpretation of signals during the development of the wing in Drosophila . Development 126: 913–925 [DOI] [PubMed] [Google Scholar]
- Kovall RA, Hendrickson WA (2004) Crystal structure of the nuclear effector of notch signaling, CSL, bound to DNA. EMBO J 23: 3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejcí A, Bray S (2007) Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev 21: 1322–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer E, Ban N (2021) Mechanisms and regulation of protein synthesis in mitochondria. Nat Rev Mol Cell Biol 22: 307–325 [DOI] [PubMed] [Google Scholar]
- Kung‐Chun Chiu D, Pui‐Wah Tse A, Law CT, Ming‐Jing XI, Lee D, Chen M, Kit‐Ho Lai R, Wai‐Hin Yuen V, Wing‐Sum Cheu J, Wai‐Hung Ho D et al (2019) Hypoxia regulates the mitochondrial activity of hepatocellular carcinoma cells through HIF/HEY1/PINK1 pathway. Cell Death Dis 10: 934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landor SK, Mutvei AP, Mamaeva V, Jin S, Busk M, Borra R, Grönroos TJ, Kronqvist P, Lendahl U, Sahlgren CM (2011) Hypo‐ and hyperactivated notch signaling induce a glycolytic switch through distinct mechanisms. Proc Natl Acad Sci USA 108: 18814–18819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F (1995) The neurogenic suppressor of hairless DNA‐binding protein mediates the transcriptional activation of the Enhancer of split complex genes triggered by notch signaling. Genes Dev 9: 2598–2608 [DOI] [PubMed] [Google Scholar]
- Lee SY, Long F (2018) Notch signaling suppresses glucose metabolism in mesenchymal progenitors to restrict osteoblast differentiation. J Clin Invest 128: 5573–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24: 251–254 [DOI] [PubMed] [Google Scholar]
- Lee KS, Wu Z, Song Y, Mitra SS, Feroze AH, Cheshier SH, Lu B (2013) Roles of PINK1, mTORC2, and mitochondria in preserving brain tumor‐forming stem cells in a noncanonical notch signaling pathway. Genes Dev 27: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao TS, Call GB, Guptan P, Cespedes A, Marshall J, Yackle K, Owusu‐Ansah E, Mandal S, Fang QA, Goodstein GL et al (2006) An efficient genetic screen in Drosophila to identify nuclear‐encoded genes with mitochondrial function. Genetics 174: 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Shen D, Shen J, Gao SM, Li B, Wong C, Feng W, Song Y (2017) The super elongation complex drives neural stem cell fate commitment. Dev Cell 40: 537–551 [DOI] [PubMed] [Google Scholar]
- Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M (2004) Inhibition of jagged‐mediated notch signaling disrupts zebrafish biliary development and generates multi‐organ defects compatible with an Alagille syndrome phenocopy. Development 131: 5753–5766 [DOI] [PubMed] [Google Scholar]
- Ludikhuize MC, Meerlo M, Gallego MP, Xanthakis D, Burgaya Julià M, Nguyen NTB, Brombacher EC, Liv N, Maurice MM, Paik JH et al (2020) Mitochondria define intestinal stem cell differentiation downstream of a FOXO/notch axis. Cell Metab 32: 889–900 [DOI] [PubMed] [Google Scholar]
- Mandal S, Guptan P, Owusu‐Ansah E, Banerjee U (2005) Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila . Dev Cell 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC et al (2007) The ribosomal protein genes and minute loci of Drosophila melanogaster . Genome Biol 8: R216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Rikhy R, Lilly M, Lippincott‐Schwartz J (2012) DRP1‐dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis. J Cell Biol 197: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo D, Shen J, Zhang J (2022) Use of FLP/FRT system to screen for notch signaling regulators in the Drosophila wing. Methods Mol Biol 2472: 39–48 [DOI] [PubMed] [Google Scholar]
- Morrugares R, Correa‐Sáez A, Moreno R, Garrido‐Rodríguez M, Muñoz E, de la Vega L, Calzado MA (2020) Phosphorylation‐dependent regulation of the Notch1 intracellular domain by dual‐specificity tyrosine‐regulated kinase 2. Cell Mol Life Sci 77: 2621–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel AC, Auer JS, Schulz A, Pfannstiel J, Yuan Z, Collins CE, Kovall RA, Preiss A (2017) Phosphorylation of suppressor of hairless impedes its DNA‐binding activity. Sci Rep 7: 11820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC (2006) Structural basis for cooperativity in recruitment of MAML coactivators to notch transcription complexes. Cell 124: 973–983 [DOI] [PubMed] [Google Scholar]
- Ohsawa S, Sato Y, Enomoto M, Nakamura M, Betsumiya A, Igaki T (2012) Mitochondrial defect drives non‐autonomous tumour progression through hippo signalling in Drosophila . Nature 490: 541–551 [DOI] [PubMed] [Google Scholar]
- Ojha R, Tantray I, Rimal S, Mitra S, Cheshier S, Lu B (2022) Regulation of reverse electron transfer at mitochondrial complex I by unconventional notch action in cancer stem cells. Dev Cell 57: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu‐Ansah E, Yavari A, Mandal S, Banerjee U (2008) Distinct mitochondrial retrograde signals control the G1‐S cell cycle checkpoint. Nat Genet 40: 356–361 [DOI] [PubMed] [Google Scholar]
- Penn JK, Schedl P (2007) The master switch gene sex‐lethal promotes female development by negatively regulating the N‐signaling pathway. Dev Cell 12: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Gomez R, Magnin V, Mihajlovic Z, Slaninova V, Krejcí A (2020) Downregulation of respiratory complex I mediates major signalling changes triggered by TOR activation. Sci Rep 10: 4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumalsamy LR, Nagala M, Sarin A (2010) Notch‐activated signaling cascade interacts with mitochondrial remodeling proteins to regulate cell survival. Proc Natl Acad Sci USA 107: 6882–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós PM, Mottis A, Auwerx J (2016) Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol 17: 213–226 [DOI] [PubMed] [Google Scholar]
- Ray A, Kamat K, Inamdar MS (2021) A conserved role for Asrij/OCIAD1 in progenitor differentiation and lineage specification through functional interaction with the regulators of mitochondrial dynamics. Front Cell Dev Biol 9: 643444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Mo D, Li Y, Liu T, Yin H, Jiang N, Zhang J (2018) A genetic mosaic screen identifies genes modulating notch signaling in Drosophila . PLoS One 13: e0203781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter‐Dennerlein R, Dennerlein S, Rehling P (2015) Integrating mitochondrial translation into the cellular context. Nat Rev Mol Cell Biol 16: 586–592 [DOI] [PubMed] [Google Scholar]
- Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS (2006) Selective fluorescent imaging of superoxide in vivo using ethidium‐based probes. Proc Natl Acad Sci USA 103: 15038–15043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T, Dürrenberger M, Paro R, Boutros M, Merdes G (2010) A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev Cell 18: 862–876 [DOI] [PubMed] [Google Scholar]
- Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, K J VV, Krishnan RT et al (2016) A genome‐wide resource for the analysis of protein localisation in Drosophila . Elife 5: e12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suissa Y, Kalifa Y, Dinur T, Graham P, Deshpande G, Schedl P, Gerlitz O (2010) Hrp48 attenuates Sxl expression to allow for proper notch expression and signaling in wing development. Proc Natl Acad Sci USA 107: 6930–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B (2008) High‐resolution in situ hybridization to whole‐mount zebrafish embryos. Nat Protoc 3: 59–69 [DOI] [PubMed] [Google Scholar]
- Thörig GE, Heinstra PW, Scharloo W (1981a) The action of the notch locus in Drosophila melanogaster. II. Biochemical effects of recessive lethals on mitochondrial enzymes. Genetics 99: 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thörig GE, Heinstra PW, Scharloo W (1981b) The action of the notch locus in Drosophila melanogaster. I. Effects of the notch8 deficiency on mitochondrial enzymes. Mol Gen Genet 182: 31–38 [DOI] [PubMed] [Google Scholar]
- Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Björk L, Breckels LM et al (2017) A subcellular map of the human proteome. Science 356: eaal3321 [DOI] [PubMed] [Google Scholar]
- Tselykh TV, Roos C, Heino TI (2005) The mitochondrial ribosome‐specific MrpL55 protein is essential in Drosophila and dynamically required during development. Exp Cell Res 307: 354–366 [DOI] [PubMed] [Google Scholar]
- Vilkki J, Portin P (1987) Fine structure of flight muscles in different notch mutants of Drosophila melanogaster reared at different temperatures. Rouxs Arch Dev Biol 196: 12–15 [DOI] [PubMed] [Google Scholar]
- Wang ZA, Huang J, Kalderon D (2012) Drosophila follicle stem cells are regulated by proliferation and niche adhesion as well as mitochondria and ROS. Nat Commun 3: 769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA (2006) Crystal structure of the CSL‐notch‐mastermind ternary complex bound to DNA. Cell 124: 985–996 [DOI] [PubMed] [Google Scholar]
- Xu J, Chi F, Guo T, Punj V, Lee WN, French SW, Tsukamoto H (2015) NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J Clin Invest 125: 1579–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SA, Deng WM (2018) Serrate/notch signaling regulates the size of the progenitor cell pool in Drosophila imaginal rings. Genetics 209: 829–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Paul S, Trieu KG, Dent LG, Froldi F, Forés M, Webster K, Siegfried KR, Kondo S, Harvey K et al (2016) Minibrain and wings apart control organ growth and tissue patterning through down‐regulation of Capicua. Proc Natl Acad Sci USA 113: 10583–10588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Wu H, Chen H, Wang R, Liang X, Liu J, Li C, Deng WM, Jiao R (2013) CAF‐1 promotes notch signaling through epigenetic control of target gene expression during Drosophila development. Development 140: 3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Praxenthaler H, Tabaja N, Torella R, Preiss A, Maier D, Kovall RA (2016) Structure and function of the Su(H)‐hairless repressor complex, the major antagonist of notch signaling in Drosophila melanogaster . PLoS Biol 14: e1002509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gao X, Coots RA, Conn CS, Liu B, Qian SB (2015) Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat Struct Mol Biol 22: 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Borikova AL, Ben‐Yair R, Guner‐Ataman B, MacRae CA, Lee RT, Burns CG, Burns CE (2014) Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci USA 111: 1403–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Gao F, Gao S, Liang Y, Long H, Lv Z, Su Y, Ye N, Zhang L, Zhao C et al (2021) Biodiversity‐based development and evolution: the emerging research systems in model and non‐model organisms. Sci China Life Sci 64: 1236–1280 [DOI] [PubMed] [Google Scholar]
- Zhao H, Ren X, Kong R, Shi L, Li Z, Wang R, Ma R, Zhao H, Liu F, Chang HC et al (2022) Auxilin regulates intestinal stem cell proliferation through EGFR. Stem Cell Rep 17: 1120–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XL, Wu X, Xu QR, Zhu RR, Xu H, Li YY, Liu S, Huang H, Xu X, Wan L et al (2019) Notch1 provides myocardial protection by improving mitochondrial quality control. J Cell Physiol 234: 11835–11841 [DOI] [PubMed] [Google Scholar]


