Abstract
The concept of probiotics is witnessing increasing attention due to its benefits in influencing the host microbiome and the modulation of host immunity through the strengthening of the gut barrier and stimulation of antibodies. These benefits, combined with the need for improved nutraceuticals, have resulted in the extensive characterization of probiotics leading to an outburst of data generated using several ‘omics’ technologies. The recent development in system biology approaches to microbial science is paving the way for integrating data generated from different omics techniques for understanding the flow of molecular information from one ‘omics’ level to the other with clear information on regulatory features and phenotypes. The limitations and tendencies of a ‘single omics’ application to ignore the influence of other molecular processes justify the need for ‘multi-omics’ application in probiotics selections and understanding its action on the host. Different omics techniques, including genomics, transcriptomics, proteomics, metabolomics and lipidomics, used for studying probiotics and their influence on the host and the microbiome are discussed in this review. Furthermore, the rationale for ‘multi-omics’ and multi-omics data integration platforms supporting probiotics and microbiome analyses was also elucidated. This review showed that multi-omics application is useful in selecting probiotics and understanding their functions on the host microbiome. Hence, recommend a multi-omics approach for holistically understanding probiotics and the microbiome.
Subject terms: Bacterial genetics, Genomics
Introduction
The gastrointestinal tract (GIT) is an intricate ecosystem harboring the microbiome consisting of fungi, bacteria, viruses and archaebacteria living symbiotically with the host1. The microbiome benefits the host through selective carbohydrates and polyphenols fermentation2 to produce bioactive metabolites3. However, age, diet, antimicrobials, and stress may cause dysbiosis contributing to chronic diseases. Probiotics, the preparations of microorganisms with health benefits when consumed by the host in adequate amounts4, have been used to modulate and restore the gut microbiome, stimulate immune response and enhance the host’s resistance to diseases5,6. Increasing awareness regarding the microbiome has greatly improved the consideration of microbes beyond disease-causing pathogens to a better understanding of their beneficial effects on human and animal health, thereby expanding the use of probiotics7,8. Formerly, the knowledge of probiotics was limited to elementary microbiology and food processes. However, this has changed in the postgenomic period of biomedicine as an important area for developing functional nutraceuticals, gastroenterology, allergology, skin care, cancer therapy, psycho-neuroendocrinology, and veterinary applications9. Probiotics have received increased attention in the scientific, healthcare settings, and the larger society10. Lactic acid bacteria (LAB), Bifidobacterium, Escherichia coli Nissle1917, and yeasts (Saccharomyces cerevisiae and S. boulardii) are among the species of microbes regarded as safe status with the ability to express heterologous genes encoding anti-inflammatory and antimicrobial biomolecules9. Other potential bacteria, such as Akkermansia muciniphila, with the ability to improve the host metabolic function and immunity during cancer treatment11, and Faecalibacterium prausnitzii with benefits, including the improvement of liver health by regulating fat build-up12 and alleviates atopic dermatitis in experimental animals were identified and termed next-generation probiotics (NGPs)13. The growing awareness of the application of beneficial microbes and the increase in data generation due to the characterization of these microbes have necessitated a clearer understanding of probiotics for precision therapy because they are strain14, disease15, and host specificity in their actions16. Therefore, the application of multi-omics will grant a holistic understanding of the mechanism of probiotic action at the system biology level for efficient applications.
The biological system has many regulatory features, including DNA, mRNA, proteins, metabolites, and epigenetic components such as DNA methylation and histone post-translational modifications (PTMs). These features can influence the signaling cascades and phenotypes when affected by physiological or pathological changes. Furthermore, the microbiome can influence the host’s genome, protein expression, and PTMs17. Hence, the advent of high-throughput technologies, including whole genome, transcriptome, and reduced representation bisulfite sequencing, and liquid/gas chromatography-mass spectrometry (LC-MS, GC-MS) have greatly enhanced the comprehensive studies of molecular features at different omics levels18. A system biology approach permits the integration of large datasets from genomics, epigenomics, transcriptomics, proteomics, metabolomics, and lipidomic (the so-called “multi-omics”) analyses from experimental and theoretical models19. Technological advancement, including DNA and RNA sequencing, proteomics, metabolomics, lipidomics, and the initiation of microbiome studies and computational collation of clinical and research data in the past two decades, has increased biological data generation. These developments require advanced analytical tools to derive useful biological information for meaningful inferences19. Omics tools are continually becoming powerful methods of obtaining an impartial and integrated view of complex biological processes such as the course of diseases and treatment efficiency20,21. Multi-omics data analysis derives valuable information about cellular functions21 and grants an understanding of the complex biology22 with a clear picture of the endotypes23. This review describes the growing aspects of system biology using multi-omics related to probiotic studies while elucidating its effects on the host and the gut microbiome.
Omics techniques used in studying probiotics and microbiome
Genomics and metagenomics
The development of ‘omics’ technologies has changed the perspective in research by generating high-throughput genomic data, bridging genome and transcription to the phenome24. An organism’s genome assembly is necessary for understanding its biology. The genomics revolution in microbiology has enabled the sequencing of diverse strains of microbial species with clear information on the pangenome for protein-coding sequences25. For example, accurate genome sequencing demonstrates several differences between microbial reference sequences of a species26 with considerable physiological effects that can influence experimental results with downstream consequences on bioprocess designs27. The use of whole-genome sequencing to decipher the activities of Limosilactobacillus reuteri PNW1 (formerly; Lactobacillus reuteri PNW1) showed several genes in the genome assembly that are important for its action as a probiotic28. Alayande et al.28 further revealed the presence of several important genes, including those associated with lactic acid production, mucosal adhesion, stress tolerance, and therapeutically useful peptides (Table 1). Data from genomic analyses are also useful in tracing the origin of probiotic bacteria and their relationship with the gut microbiome. Pasolli et al.29 applied comparative genomics analyses to show the diversity and relationship of the LAB strains present in food and gut microbiome, with fermented food as the frequent niche of LAB in nature. Another comparative genomics study of the lactobacilli genomes revealed extensive gene loss and acquisitions via horizontal transfer during co-evolution in their habitats30.
Table 1.
‘Omics’ techniques applied in studying probiotic and microbiome interaction.
| Tool | Probiotic bacteria | Application | Findings | References |
|---|---|---|---|---|
| Whole-genome sequencing | Limosilactobacillus reuteri PNW1 | Determination of probiotic potential |
• Genes encoding D and L-lactate dehydrogenases. • Genes responsible for adhesion to epithelial tissues, including antiadhesion Pls, Sortase A, exopolysaccharide cluster, • Genes encoding bioactive peptides (S-ribosyl homocysteine lyase, Autoinducer-2 production protein LuxS • Four coding regions associated with enhancing host metabolism and enzymes such as Poly (glycerol-phosphate) alpha-glucosyltransferase. |
28 |
| Metagenomics (Sequencing of V3-V4 region of the bacterial 16S rRNA) | Lactobacillus rhamnosus, Enterococcus faecalis | Determination of the effects of direct-fed microbes on the rumen microbiome of goats | • Showed the influence of direct-fed Lactic acid bacteria on the microbiota in goats | 35 |
| Metagenomics (Sequencing of V3-V4 region of the bacterial 16S rRNA) | L. rhamnosus PT9, L. rhamnosus PT10 | Determination of the effects of direct-fed microbes on the rumen microbiome of sheep | • Revealed the effects of the lactic acid bacterial administration on the ruminal microbiome of sheep | 36 |
| Metagenomics (Sequencing of V3-V4 region of the bacterial 16S rRNA) | Saccharomyces cerevisiae boulardii CNCM I-1079 Lactobacillus acidophilus BT1386 | Determination of the effects of directly fed microbes on calves’ ruminal microbiome |
• Decreased in pathogenic and increased in beneficial bacterial populations, respectively. • Impacts on pathways include cell cycle, bile secretion, proteasome, cAMP signaling, thyroid hormone synthesis, and dopaminergic synapse pathways. |
34 |
| Transkingdom network analysis | Lactobacillus johnsonii and Lactobacillus gasseri) | Determination of probiotics on the liver functions | • Attenuation of western diet-induced diabetes through the improvement of lipid metabolism and enhanced mitochondrial health. | 45 |
| Transcriptomics | Lactiplantibacillus plantarum LIP-1 | Determination of Lactiplantibacillus plantarum LIP-1 responses to different Ph | • Unraveled the microbe’s responses and enhanced the means to improve survival in a lyophilized state. | 46 |
| Transcriptomics | Bifidobacterium breve UCC2003 | Influence of B. breve UCC2003 on the intestinal barrier | • Described the functions of B. breve UCC2003 in intestinal epithelial homeostasis during early life in neonatal murine intestinal cells. | 47 |
| Metatranscriptomics | yeasts and Lactobacillus species | The interplay of the microbes during the fermentation of | • Revealed the role of the probiotic microbes in pyruvate metabolism | 49 |
| Metatranscriptomics | Oral microbiota | Interactions of the oral microbiome in biofilm biomass | • Unraveled the complex interactions of the oral microbiome in biofilm assembly | 50 |
| Peptidomics and Metagenomics | Lactobacillus helvetius NS8 and Lactobacillus fermentum NS9 | Determine the effects of the probiotics on the peptidome of specific pathogen-free mice |
• Alters the hippocampus peptidome by acting on the gut-brain axis. • No substantial alteration on the gut microbiome |
58 |
| Shotgun metaproteomics | Gut microbiota | Revealed the response of the host to the microbiota | • Revealed human proteins and antimicrobial peptides | 60 |
| Metabolomics and 16S RNA sequencing | Lactobacillus casei Zhang | Determined changes in the metabolic profile of L. casei Zhang culture |
• Revealed changes in several metabolic pathways including amino acid and carbohydrate metabolism. • Showed the metabolic changes associated with glucose restriction to strain. |
69 |
| Metabolomics | Lactobacillus plantarum MLK 14-2, L. plantarum KCCM 11322 | Determined the variations in the metabolic changes in kimchi during fermentation with different strains |
• Revealed the applications of metabolomics to monitor the fermentation characteristics of the strain. • Showed that the metabolites vary with the starter culture strain used |
71 |
| Metabolomics | Lactobacillus plantarum NCU116 | Determined the effects of ingesting L. plantarum NCU116 on the metabolite profile of hyperlipidemic rat model fed high fat diet |
• Revealed biomarkers associated with high-fat diet in the serum of the model experimental animal. • Showed the biological pathways and functions associated with the metabolites. • Improvement in the hyperlipidemic condition of the rat via biosynthetic and metabolic pathways. |
75 |
| Metabolomics | Lactobacillus paracasei subsp. paracasei SM20 and Propionibacterium jensenii SM11 | Studied the antiyeast activities of the strains against Candida pulcherrima and Rhodotorula mucilaginosa |
• Antiyeast compounds resistant to proteinase K and pronase E treatments were detected. • Other low molecular weight compounds that play role in the complex antiyeast activities were also identified |
76 |
| Metabolomics | Lactobacillus rhamnosus GG | Determine the effects of supplementing L. rhamnosus GG as a probiotic in alcoholic liver disease induced rats |
• Normalized the level of fatty acids in the liver and feces of alcoholic liver disease induced rats. • Prevents alcoholic liver disease in the treated rats. • Modifies the gut microbiome to stimulate the synthesis of long chin fatty acids. • Elevates the level of some essential amino acids through amino acid biosynthesis. |
77 |
| Lipidomics coupled with RT-PCR gene expression | Multi-strain probiotics VSL#3 and IT-3 (containing different strains of lactobacilli, Bifidobacterium and Streptococcus thermophilus) | Determine the lipidemic response of Caenorhabditis elegans to different probiotics preparations. |
• Variation in the lipid contents C. elegans fed VSL#3 compared to IT-3. • Positive correlation between the genes encoding the fatty acid and the levels of the respective lipids. |
82 |
| Lipidomics | Lactobacillus plantarum APsulloc 331261 | study the extracellular vesicle phospholipid of the strain |
• Variation in the phospholipid level between the extracellular vesicle and the parent cells • Revealed high level of phosphatidylcholine synthase and lipopolysaccharide which could be used as biomarkers for clinical applications |
83 |
The invention of high throughput metagenomic sequencing has aided the analyses of microbes, including non-culturable bacteria, at strain levels31. Metagenomics and metatranscriptomics have improved the development of new generations of probiotics. These techniques have also increased our understanding of the human microbiome and its contribution to gut physiology by reducing disease risks32. An important aspect of microbiome study is the use of metagenomics to generate testable hypotheses on the mechanisms of disease evasion by the host33. In a study to analyze the effects of directly fed microbes on the rumen microbiome of cattle, Fomenky et al.34 applied 16S RNA gene sequencing of the V3 to V5 region of the entire microbial community to decipher the effect of Saccharomyces cerevisiae boulardii CNCM I-1079 (SCB) and Lactobacillus acidophilus BT1386 (LA)), and an antibiotic growth promoter (ATB) on the gut microbiota. These authors further showed the roles of the direct-fed microbial culture on the gut microbial structures with consequent inhibition of pathogens through metagenomics sequencing. The mechanisms of the host-microbe interactions, including effects on pathways like cell signaling, bile secretion, proteasome, cAMP signaling, thyroid hormone synthesis, and dopaminergic synapse pathways, were elucidated34 (Table 1). Similarly, Maake et al.35 and Mani et al.36 showed the influence of direct-fed microbes on the microbiome of goats and sheep, respectively, and the resultant modulation of the host health (Table 1). Furthermore, Mani et al.36 revealed that probiotics could improve the diversity of the gut microbiome while decreasing the level of pathogenic microbes such as Pseudomonas species. Maake et al.35 also showed the inhibition of pathogenic microbes, including Chlamydia species, in goat’s rumen by direct-fed lactic acid bacteria (LAB) using metagenomics analysis. The advancement in genomic sequencing technology has greatly facilitated research in microbial ecology, microbial interactions within the commensal community, and host-probiotics-microbiome interactions through 16S rRNA and shotgun-metagenomic sequencing37–39. These studies indicate the potential of metagenomic analyses to interpret microbial structures and diversity in their ecological niches. They revealed the interactions between probiotics on the microbiome and the host37,40.
Transcriptomics and metatranscriptomics
Transcriptomics studies the whole ‘transcriptome’ of a cell, tissue, organ, or organism under defined conditions41. The term transcriptome, first attributed to Charles Auffray, is regarded as the whole set of ribonucleic acid (RNA) expressed in a cell, tissue, or organism42. Transcriptomics includes everything relating to RNAs, such as transcription and expression levels, functions, locations, trafficking, and degradation41. It also encompasses the structures of transcripts and their parent genes regarding starting sites, 5′ and 3′ end sequences, splicing patterns, and post-transcriptional modifications43. Transcriptome covers all types of transcripts, including messenger RNAs (mRNAs), microRNAs (miRNAs), and different types of long noncoding RNAs (lncRNAs)41. It employs advanced methods to analyze the expression of multiple transcripts under different physiological or pathological conditions, thereby rapidly expanding the understanding of the relationship between the transcriptome and phenotypes across a wide range of living entities41. RNA sequencing data has enabled the quantification of gene expression, non-coding RNAs (ncRNAs), and post-transcriptional regulations in an organism44.
Gene expression studies coupled with electron microscopy by Rodrigues et al.45 showed the ability of Lactobacillus johnsonii and Lactobacillus gasseri strains to modulate the activity of the liver with resultant improvement in lipid metabolism through enhancing mitochondrial health in type-2 diabetes-induced mice models. Transcriptomics was used to study the internal response of cells or organisms to physical perturbations. For example, Jingjing et al.46 applied transcriptomics analysis to unravel the responses of Lactiplantibacillus plantarum LIP-1 to different pH conditions. These authors revealed the mechanisms of survival of the organism in a lyophilized state. Transcriptomics has also been applied to decipher host-microbe interactions in understanding the influence of probiotics on host immune modulation. Kiu et al.47 described the effects of Bifidobacterium breve UCC2003 in strengthening the intestinal barrier by modulating the intestinal epithelial cells. They showed the impact of the microbiome on the intestinal epithelial cells through global RNA sequencing, differentially expressed genes and metabolic pathways. These authors further revealed the central role of B. breve UCC2003 in maintaining intestinal epithelial homeostasis in neonatal murine intestinal cells.
Metatranscriptomics is a powerful tool for studying the microbial community’s structure and transcriptional regulation of active genes through gene expression in response to environmental perturbations48. Like metagenomics, metatranscriptomics has been applied to reveal microbial structure and diversity from different ecological niches. Song et al.49 described the interplay between the fermentation bacteria during solid-state fermentation in producing Chinese Mao-Tai-flavored liquor using high-throughput 16S rRNA gene amplicon sequencing, internal transcribed space amplicon sequencing, and metatranscriptomics sequencing. These authors showed the involvement of yeasts (genera Pichia, Schizosaccharomyces, Saccharomyces, and Zygosaccharomyces) and lactic acid bacteria (genus Lactobacillus) in pyruvate metabolism through metatranscriptomics analysis. Similarly, the complex interactions of the oral microbiome in biofilm assembly were elucidated by Edlund et al.50 using metatranscriptomics analysis. Apart from applying metatranscriptomics in studying microbial structure through gene expression, this high throughput technique was also used to screen targets in disease conditions such as bacterial vaginosis51. Metatranscriptome analysis normally sequences the entire transcriptome from the microbial community and unravels the complex interactions between probiotics, the microbiome, and the host. It can give a clear picture of specific targets and precision application of probiotics, especially when coupled with other omics techniques such as proteomics and metabolomics.
Proteomics and metaproteomics
The proteome is the total protein complement of a cell or subcellular fraction of an organism in a specified growth phase and physiological condition52,53. Proteomics allows the identification of proteins that participate in cellular processes such as catalysis and stress responses and quantifying the complete proteins present in a cell and tissue under defined conditions53. Proteomics permits high-throughput identification of important proteins for probiotic interactions with their environment, including the food and host’s gut53. The application of proteomics also provides a unique framework for identifying post-translational modifications (methylation, phosphorylation, or glycosylation) that may strongly affect protein functions commonly overlooked by other “omics” techniques54. Currently, there is an increase in the amount of proteomics data in public repositories, which has continued to grow at an unprecedented rate55,56. Proteomics has been combined with metabolomics to characterize the functions of probiotics originating from different means of production and determine the influence of production processes on the probiotic’s potential57. A previous study employing low molecular weight proteomic analysis (peptidomics) by Zhang et al.58 revealed the ability of probiotics to modulate the gut-brain axis through a dynamic landscape of the peptidome across multiple regions. This study indicates the potential of proteomics to unravel probiotic interactions with the host beyond the gut to the nervous system.
In-depth Characterization of the microbiome composition, structure, and functions is usually achieved by sequencing the total DNA and RNA59. However, the gene/transcripts do not necessarily denote complete protein expression. Therefore, metaproteomics which measures the total expressed proteins, has been considered to provide exact functional information60. Additionally, mass spectrometry (MS) based proteomics allows for the simultaneous measurement of proteins of both the host and microbiome61 and is a useful tool that aids in unraveling host-microbe interactions in complex intestinal ecosystems62. Verberkmoes et al.60 demonstrated metaproteomics as an important tool in understanding the host’s response to the gut microbiota by revealing thousands of proteins, including antimicrobial peptides. Thus, showing the mechanisms of the complex interplay between the host and the microbiome. These studies indicate the applications of proteomics in understanding probiotics interactions with the gut microbiome and the host’s responses to such modulations. Metaproteomics is a powerful tool for functional microbiome analysis and can reveal complex host-microbe interactions61. Metaproteomics focuses directly on the total proteins expressed in a microbial community and therefore provides an understanding of the community phenotypes63. Additionally, proteins constitute the bulk of a cell; hence metaproteomics estimates the contribution of individual community members to the community biomass64.
Metabolomics and lipidomics
Metabolomics is an important technique that simultaneously detects hundreds of small molecules in a biological system65,66. The metabolites reveal the organism’s health by acquiring information about the biological system and providing a clear picture of the phenotype67. Metabolomics identifies the putative substances from probiotics, such as reduced glutathione, beneficial to the host45. Metabolomics quantitively and qualitatively analyze low molecular weight compounds, including peptides, carbohydrates, amino acids, nucleic acid metabolites, vitamins, organic acids, and minerals produced by microbial metabolism68. A previous study combining 16S rRNA gene sequencing with proton nuclear magnetic resonance analysis revealed that metabolomics correlates specific biological functions with taxonomy, providing an understanding of the mechanisms underlying the inhibition of pathogens by some probiotic bacteria69. Moreover, studies have revealed that bioactive secondary metabolites produced by many probiotic agents affect bacterial community interaction and attenuate the virulent markers on several pathogens70. Metabolomics has also been applied in studying the changes that occur during the fermentation of foods and dairy products in the presence of lactic acid bacteria71–74. Several studies have applied metabolomics to elucidate metabolic changes resulting from the administration of probiotic bacteria, such as the alleviation of hyperlipidemia in rats by Lactobacillus plantarum NCU11675, Characterization of antimicrobial metabolites by Lactobacillus and Propionibacterium coculture76, alleviation and the prevention of alcohol-induced liver disease in experimental rat models77, and the determination of probiotic function through metabolic Characterization78. Metabolomics can be used in probiotics to identify the precise bioactive substances with specific functions for desirable applications, thereby aiding precision therapy. Lipidomics encompasses a broad range of mass spectrometry (MS) workflows that aim to identify and quantify various lipid classes, including their molecular species in the biological systems79,80. Lipidomic technologies become useful to characterize the lipid content of an organism through the analysis of the structure, function, or interaction of cellular lipids, which also play an essential role in nutritional research81. Lipidomics has been used to show probiotics’ impacts on the host’s physiology. A recent study combining gene expression and lipidomics (using mass-spectrometry) revealed the response of Caenorhabditis elegans (C. elegans) to multi-strain probiotics; VSL#3 and IT-3 (containing four lactobacilli, three bifidobacterial, and S. thermophilus strains) with a resultant variation in the level of the fatty acids based on dietary interventions82. The lipidomic analysis of the extracellular vesicles of Lactobacillus plantarum APsulloc 331261 probiotic revealed a variation in the phospholipid contents secreted in the media compared to that of the parent cell. Hence giving an insight into the mechanism of lipid biogenesis and lipid-mediated cell-to-cell interactions between or within species83. Considering the powerful applications of lipidomics in understanding the biochemistry of the cell membrane84, integrating lipidomics data with other ‘omics’ techniques would be a great means of understanding probiotics functions and their influence on the host and gut microbiome.
The need for integrative ‘multi-omics’ in studying biological systems
A single ‘omics’ technique is insufficient to understand biological processes clearly. For instance, metagenomics sequencing alone is fundamentally limited due to its inability to account for the functional activities of the microbial community directly. Individual omics techniques/data provide important information in understanding several biological processes. However, each type of study (genomics, transcriptomics, proteomics) neglects the influence of the other domains and their interplay. Hence, resulting in limited information on the biological processes85. Furthermore, the collective studies and integration of omics data from the host and microbiome domains (a concept known as holo-omics) are imperative for understanding the mechanisms of host-microbe interactions86. While multi-omics employs data from one domain (host or microbe), holo-omics integrates data from the host and the microbiome. For instance, to accurately model the health-associated outcomes of bacterial configuration, omics datasets covering several covariables from the host and microbial domains are integrated to reveal the nature of interactions87. As a result, additional omics data are needed for a full description of microbial activities, such as the abundances of RNA (metatranscriptome), proteins (metaproteome), and metabolites (metabolome), preferably in an integrated fashion88.
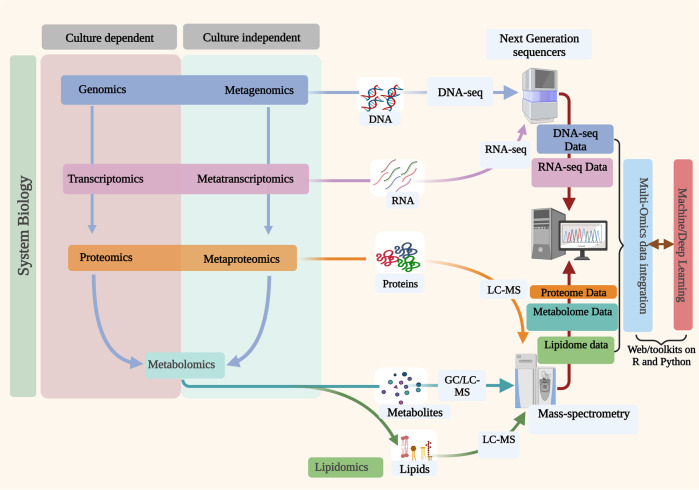
Integrative multi-omics has also paved the way for understanding complex disease interplay and gives a clear route for applying therapeutic regimens89. The growing interest in the relationships between the host and their associated microorganisms has changed the perceptions of the biology underlying the host’s genetic properties based on their interactions in several biological processes85. These microbes play important roles in the host, including nutrient acquisition90, immune modulation91 and development92, biomolecule synthesis93, and influence on the host’s behavior94. Furthermore, the advent of meta-omics techniques such as metagenomics95,96, metatranscriptomics97, and metaproteomics98 enables understanding of microbial behaviors in their natural habitat where they are part of communities frequently dominated by as-yet unculturable populations99,100. A summary of the different omics techniques applicable for both culture-dependent and culture-independent microbial studies and the other various biological levels for multi-omics data integration for system biology is presented in Fig. 1.
Fig. 1. Multi-omics applications in studying biological systems.
Omics techniques have applications in both culture-dependent and culture-independent studies of bacteria. The data generated from each technique can be integrated for a holistic understanding of the biological system.
Multi-omics application at different biological levels and domains in probiotics and microbiomics
Studying the host genomics revealed the host genome’s influence on the diversity of the microbiome and how it affects the host phenotypes101. Similarly, metabolomics reveals the metabolic activities of the gut microbiome and its influence on the host gut metabolome102. Hence, an integrative multi-omics approach employing metagenomics and untargeted metabolomics of the host and microbiome at both the intra- and inter-domain levels revealed the host-microbiome metabolites interactions with insight into the effects of microbiota on ageing102. Furthermore, genomics aids in mining probiotic potentials from commensal microbes103, while metagenomics can decipher probiotics-microbiome interactions104. Integration of transcriptomics, proteomics and metabolomics datasets profiles host-microbe crosstalk and the influence of probiotics supplementation on the host105. The immunomodulatory characteristics of probiotics are determined using transcriptomics by applying RNA-sequencing or microarray gene expression. The proteome changes are determined using protein chips with antibodies, nucleic acid or other proteins that bind to protein targets106. The application of multi-omics enables the purification of antimicrobial peptides from microbes using liquid-chromatography tandem mass-spectrometry and the characterization of the genes encoding these compounds through integrated genomics and proteomics analysis107. Thus, enabling an evolutionary insight using comparative genomics and discovering novel antimicrobial compounds. This is because mass-spectrometry can identify antimicrobial peptides sequence and masses from a sample mixture, revealing their primary, secondary, and tertiary structures and functions108,109.
‘Omics’ data integration to elucidate probiotics action
Biological data integration describes the analytical methods that combine information from multiple sources into a single biological inference110. Multi-omics data integration provides information on biomolecules from different biological layers for the systematic and holistic understanding of complex biology22. Integrated approaches combine individual omics data sequentially or simultaneously to elucidate the complex interplay of molecules111. This help in assessing information flow from one omics level to the other, thus closing the gap from genotypes to phenotypes21. Approaches to profile cellular characteristics and processes from the genome112, epigenome113, RNA114, RNA isoforms115, and proteins116 are increasingly applied for understanding cellular activities, especially in single-cell studies117. Multi-omics data integration combines individual omics datasets sequentially to understand molecular interactions111. Analyzing two or more datasets is necessary for understanding the relationships between different biological functional levels. It is becoming evident that the integration of ‘omics’ data, such as transcriptomics, proteomics, and metabolomics, provides a better understanding of the biological system21,118. Recent work by Lee et al.119 revealed the integration of metagenomics, genomics, and transcriptomics of bacteria and the analysis of mouse intestinal transcriptome and serum metabolome data to show the mechanisms by which bacteria determine the efficacy of cancer therapeutics where Bifidobacterium bifidum (a probiotic bacteria) was found to be influential in patients responsive to therapy. Their result further showed the ability of B. bifidum to potentiate the production of interferon-γ through the enhancement of immune-stimulating molecules and metabolites. A longitudinal multi-omics integration of data from the gut microbiome, metabolome, host epigenome, and transcriptome of the mechanisms behind irritable bowel syndrome (IBS) revealed purine metabolism as a novel host-microbial metabolic pathway in IBS with translational potential120. Another study by Rasmussen et al.121 applied 16 S metagenomic sequencing and untargeted metabolomics to reveal probiotics’ immunomodulatory and growth-promoting actions in rainbow trout. Their studies further elaborated on the association of the gut microbial diversity with the microbial metabolites when fed the probiotics preparation. Hence, revealing the complex mechanisms of microbiome-host interactions. Studies highlighting multi-omics applications in understanding probiotic actions, especially in-vivo, are rare. However, several studies have shown the applications of integrative multi-omics in host-microbiome interactions85,122–124. Therefore, the need for integrative multi-omics studies in understanding the actions of probiotics on the host and the host microbiome is crucial.
Integrative ‘multi-omics’ data platforms and tools
Previous studies have revealed large-scale improvements in data coverage and measurement fidelity to track dynamic changes in RNA transcripts, ribosome profiling, proteins, and metabolites quantitatively in unprecedented detail125–127. Multi-omics studies provide the potential for a more holistic picture enabling a comprehensive understanding of complex diseases and biological processes128,129. This has led to increased bioinformatics and statistical tools to aid the integration of multiple omics datasets130. A previous study by Huang et al.131 described several tools that integrate multi-omics datasets for deriving useful biological inferences. Therefore, more recently developed platforms, including web tools for multi-omics data integration, are discussed in this study. A study by Zhou et al.132 introduced OmicsAnalyst (https://www.omicsanalyst.ca/), a web-based platform for integrating and visualizing multi-omics data. OmicsAnalyst is user-friendly and aided by three main visual analytic tracks: feature correlation network, cluster heatmap, and dimension reduction analyses. OmicsAnalyst supports three multi-view clustering algorithms: spectral and perturbation clustering and similarity network fusion. Another multi-omics data integration platform is the PaintsOmics 3 (https://www.paintomics.org/), a free web-based interphase that allows integrated analyses of multi-omics datasets, visualization, and network analysis onto the KEGG pathway133. PaintsOmics is interactive in its usage and permits extensive exploration of multi-omics data in addition to comprehensive pathway analysis, automatic feature name/identifier conversion, multi-layered feature matching, pathway enrichment, network analysis, interactive heatmaps, trends charts, and accepts a variety of omics data types including transcriptomics, proteomics and metabolomics, and region-based approaches such as ATACseq or ChIP-seq data. The increase in the need for a comprehensive understanding of biological systems has prompted the development of integrative network analysis platforms like OmicsNet134 (https://www.omicsnet.ca/) and MOPED135 (Multi-Omics Profiling Expression Database, http://moped.proteinspire.org), which are also web-based multi-omics platforms. OmicsNet allows data integration from different sources, including humans and the microbiome and is enhanced with several network analytics functions and interactive 3D charts. MOPED was designed to enhance the integration of genomic and protein expression data into pathway analysis. It also has meta-data detailing functions to ensure data quality, consistency, and reuse136,137.
In conclusion, this article reviewed the applications of different omics techniques in studying probiotics and the gut microbiome in culture-dependent and culture-independent scenarios. The need for a systematic approach to understanding the biological system also motivated this work to highlight the integration of multi-omics data in studying probiotics and the gut microbiome. In this study, we elaborated on the applications of different multi-omics data integration platforms available for studying probiotics and gut microbiomes. The different multi-omics data integrating platforms applicable in the study of probiotics and the gut microbiome have the potential to integrate and interpret ‘omics’ data from different biological levels and take various impute data sources. The studies of probiotics at the different biological levels, and the integration of the derived omics data, can unravel the important trends in their activities and the specific mechanisms by which they exert their health benefits. Hence, the room for careful selection and applications in precision therapy.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors acknowledge the College of Agriculture, Engineering and Sciences, University of KwaZulu-Natal, South Africa, for granting the first author tuition remission for his PhD program. We also acknowledge the Agricultural Research Council, Animal Production Institute, Gastrointestinal Microbiology, and Biotechnology platform, where the authors conceived this project.
Author contributions
M.A.A., O.A.A., M.O. and I.D.K. – Conceptualization; I.D.K. – Literature search, writing – drafted the original manuscript; M.A.A., O.A.A. and M.O. – supervised and edited the manuscript. All the authors read and approved the final draft for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41538-023-00199-x.
References
- 1.Westfall S, et al. Optimization of probiotic therapeutics using machine learning in an artificial human gastrointestinal tract. Sci. Rep. 2021;11:1–15. doi: 10.1038/s41598-020-79947-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yin R, et al. Gut microbiota, dietary phytochemicals, and benefits to human health. Curr. Pharmacol. Rep. 2019;5:332–344. doi: 10.1007/s40495-019-00196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westfall S, Pasinetti GM. The gut microbiota links dietary polyphenols with management of psychiatric mood disorders. Front. Neurosci. 2019;13:1196. doi: 10.3389/fnins.2019.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill C, et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 5.Westfall S, et al. Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell. Mol. Life Sci. 2017;74:3769–3787. doi: 10.1007/s00018-017-2550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Toole P, Marchesi J, Hill C. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017;2:17057. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 7.Chin-Lee B, Curry WJ, Fetterman J, Graybill MA, Karpa K. Patient experience and use of probiotics in community-based health care settings. Patient Prefer. Adherence. 2014;8:1513. doi: 10.2147/PPA.S72276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson SA, et al. Improving end-user trust in the quality of commercial probiotic products. Front. Microbiol. 2019;10:739. doi: 10.3389/fmicb.2019.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh B, Mal G, Marotta F. Designer probiotics: paving the way to living therapeutics. Trends Biotechnol. 2017;35:679–682. doi: 10.1016/j.tibtech.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Cunningham M, et al. Shaping the future of probiotics and prebiotics. Trends Microbiol. 2021;29:667–685. doi: 10.1016/j.tim.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhang T, Li Q, Cheng L, Buch H, Zhang F. Akkermansia muciniphila is a promising probiotic. Microb. Biotechnol. 2019;12:1109–1125. doi: 10.1111/1751-7915.13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munukka E, et al. Faecalibacterium prausnitzii treatment improves hepatic health and reduces adipose tissue inflammation in high-fat fed mice. ISME J. 2017;11:1667–1679. doi: 10.1038/ismej.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y, et al. Oral administration of Faecalibacterium prausnitzii and Akkermansia muciniphila strains from humans improves atopic dermatitis symptoms in DNCB induced NC/Nga mice. Sci. Rep. 2022;12:1–15. doi: 10.1038/s41598-022-11048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleerebezem M, et al. Understanding mode of action can drive the translational pipeline towards more reliable health benefits for probiotics. Curr. Opin. Biotechnol. 2019;56:55–60. doi: 10.1016/j.copbio.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 15.McFarland LV, Evans CT, Goldstein EJ. Strain-specificity and disease-specificity of probiotic efficacy: a systematic review and meta-analysis. Front. Med. 2018;5:124. doi: 10.3389/fmed.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, et al. Autochthonous Lactobacillus spp. isolated from Murrah buffalo calves show potential application as probiotic. Curr. Res. Biotechnol. 2021;3:109–119. doi: 10.1016/j.crbiot.2021.04.002. [DOI] [Google Scholar]
- 17.Graw, S. et al. Multi-omics data integration considerations and study design for biological systems and disease. Mol. Omics. 17, 170–185 (2021). [DOI] [PMC free article] [PubMed]
- 18.Qiu C, et al. Multi-omics data integration for identifying osteoporosis biomarkers and their biological interaction and causal mechanisms. iScience. 2020;23:100847. doi: 10.1016/j.isci.2020.100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi A, Rienks M, Theofilatos K, Mayr M. Systems biology in cardiovascular disease: a multiomics approach. Nat. Rev. Cardiol. 2021;18:313–330. doi: 10.1038/s41569-020-00477-1. [DOI] [PubMed] [Google Scholar]
- 20.Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat. Rev. Genet. 2018;19:299–310. doi: 10.1038/nrg.2018.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights. 2020;14:1177932219899051. doi: 10.1177/1177932219899051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief. Bioinforma. 2018;19:1370–1381. doi: 10.1093/bib/bbx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyler SR, Bunyavanich S. Leveraging-omics for asthma endotyping. J. Allergy Clin. Immunol. 2019;144:13–23. doi: 10.1016/j.jaci.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromer M, et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monk JM, et al. Multi-omics quantification of species variation of Escherichia coli links molecular features with strain phenotypes. Cell Syst. 2016;3:238–251.e12. doi: 10.1016/j.cels.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freddolino PL, Amini S, Tavazoie S. Newly identified genetic variations in common Escherichia coli MG1655 stock cultures. J. Bacteriol. 2012;194:303–306. doi: 10.1128/JB.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nahku R, et al. Stock culture heterogeneity rather than new mutational variation complicates short-term cell physiology studies of Escherichia coli K-12 MG1655 in continuous culture. Microbiology. 2011;157:2604. doi: 10.1099/mic.0.050658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alayande KA, Aiyegoro OA, Nengwekhulu TM, Katata-Seru L, Ateba CN. Integrated genome-based probiotic relevance and safety evaluation of Lactobacillus reuteri PNW1. PLoS One. 2020;15:e0235873. doi: 10.1371/journal.pone.0235873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasolli E, et al. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-16438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova K, et al. Comparative genomics of the lactic acid bacteria. Proc. Natl Acad. Sci. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sybesma W, Kort R, Lee Y-K. Locally sourced probiotics, the next opportunity for developing countries? Trends Biotechnol. 2015;33:197–200. doi: 10.1016/j.tibtech.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Bisanz JE, et al. A genomic toolkit for the mechanistic dissection of intractable human gut bacteria. Cell Host Microbe. 2020;27:1001–1013.e9. doi: 10.1016/j.chom.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fomenky BE, et al. Direct-fed microbial supplementation influences the bacteria community composition of the gastrointestinal tract of pre-and post-weaned calves. Sci. Rep. 2018;8:1–21. doi: 10.1038/s41598-018-32375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maake TW, Aiyegoro OA, Adeleke MA. Effects of Lactobacillus rhamnosus and Enterococcus faecalis Supplementation as Direct-Fed Microbials on Rumen Microbiota of Boer and Speckled Goat Breeds. Vet. Sci. 2021;8:103. doi: 10.3390/vetsci8060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mani S, Aiyegoro OA, Adeleke MA. Characterization of rumen microbiota of two sheep breeds supplemented with direct-fed lactic acid bacteria. Front. Vet. Sci. 2021;7:1199. doi: 10.3389/fvets.2020.570074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasc C, Peyret P. Hybridization capture reveals microbial diversity missed using current profiling methods. Microbiome. 2018;6:1–9. doi: 10.1186/s40168-018-0442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alcon-Giner C, et al. Microbiota supplementation with Bifidobacterium and Lactobacillus modifies the preterm infant gut microbiota and metabolome: an observational study. Cell Rep. 2020;1:100077. doi: 10.1016/j.xcrm.2020.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breitwieser FP, Lu J, Salzberg SL. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2019;20:1125–1136. doi: 10.1093/bib/bbx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F, Zou Q, Gao B. GutBalance: a server for the human gut microbiome-based disease prediction and biomarker discovery with compositionality addressed. Brief. Bioinform. 2021;22:436. doi: 10.1093/bib/bbaa436. [DOI] [PubMed] [Google Scholar]
- 41.Milward E, et al. Transcriptomics. Encyclo. Cell Biol. 2016;4:160–165. doi: 10.1016/B978-0-12-394447-4.40029-5. [DOI] [Google Scholar]
- 42.Morozova O, Hirst M, Marra MA. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genom. Hum. Genet. 2009;10:135–151. doi: 10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y, Ye Y, Zhang Z, Han L. Maximizing the utility of cancer transcriptomic data. Trends Cancer. 2018;4:823–837. doi: 10.1016/j.trecan.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Rodrigues RR, et al. Transkingdom interactions between Lactobacilli and hepatic mitochondria attenuate western diet-induced diabetes. Nat. Commun. 2021;12:1–15. doi: 10.1038/s41467-020-20313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jingjing E, et al. Effects of different initial pH values on freeze-drying resistance of Lactiplantibacillus plantarum LIP-1 based on transcriptomics and proteomics. Food Res. Int. 2021;149:110694. doi: 10.1016/j.foodres.2021.110694. [DOI] [PubMed] [Google Scholar]
- 47.Kiu R, et al. Bifidobacterium breve UCC2003 induces a distinct global transcriptomic program in neonatal murine intestinal epithelial cells. iScience. 2020;23:101336. doi: 10.1016/j.isci.2020.101336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitra S, et al. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinform. 2011;12:1–8. doi: 10.1186/1471-2105-12-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song Z, Du H, Zhang Y, Xu Y. Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing. Microbiol. 2017;8:1294. doi: 10.3389/fmicb.2017.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edlund A, et al. Uncovering complex microbiome activities via metatranscriptomics during 24 h of oral biofilm assembly and maturation. Microbiome. 2018;6:1–22. doi: 10.1186/s40168-018-0591-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Twin J, et al. The potential of metatranscriptomics for identifying screening targets for bacterial vaginosis. PLoS One. 2013;8:e76892. doi: 10.1371/journal.pone.0076892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Görg A, Boguth G, Obermaier C, Weiss W. Two‐dimensional electrophoresis of proteins in an immobilized pH 4–12 gradient. Electrophoresis. 1998;19:1516–1519. doi: 10.1002/elps.1150190850. [DOI] [PubMed] [Google Scholar]
- 53.Ruiz L, et al. Tackling probiotic and gut microbiota functionality through proteomics. J. Proteom. 2016;147:28–39. doi: 10.1016/j.jprot.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 54.Koponen J, et al. Effect of acid stress on protein expression and phosphorylation in Lactobacillus rhamnosus GG. J. Proteom. 2012;75:1357–1374. doi: 10.1016/j.jprot.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 55.Deutsch EW, et al. The ProteomeXchange consortium in 2020: enabling ‘big data’approaches in proteomics. Nucleic Acids Res. 2020;48:D1145–D1152. doi: 10.1093/nar/gkz984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai C, et al. A proteomics sample metadata representation for multiomics integration, and big data analysis. Nat. Commun. 2021;12:5854. doi: 10.1038/s41467-021-26111-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bianchi L, et al. A combined proteomics, metabolomics and in vivo analysis approach for the characterization of probiotics in large-scale production. Biomolecules. 2020;10:157. doi: 10.3390/biom10010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang P, et al. A dynamic mouse peptidome landscape reveals probiotic modulation of the gut-brain axis. Sci. Signal. 2020;13:eabb0443. doi: 10.1126/scisignal.abb0443. [DOI] [PubMed] [Google Scholar]
- 59.Fraher MH, O’toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat. Rev. Gastroenterol. Hepatol. 2012;9:312–322. doi: 10.1038/nrgastro.2012.44. [DOI] [PubMed] [Google Scholar]
- 60.Verberkmoes NC, et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X, et al. Deep metaproteomics approach for the study of human microbiomes. Anal. Chem. 2017;89:9407–9415. doi: 10.1021/acs.analchem.7b02224. [DOI] [PubMed] [Google Scholar]
- 62.Zhang X, et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-05357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kleikamp HB, et al. Database-independent de novo metaproteomics of complex microbial communities. Cell Syst. 2021;12:375–383.e5. doi: 10.1016/j.cels.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Kleiner M, et al. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 2017;8:1–14. doi: 10.1038/s41467-017-01544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiehn, O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol.48, 155–171 (2002). [PubMed]
- 66.Jain A, Li XH, Chen WN. An untargeted fecal and urine metabolomics analysis of the interplay between the gut microbiome, diet and human metabolism in Indian and Chinese adults. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-45640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gromski PS, et al. A tutorial review: Metabolomics and partial least squares-discriminant analysis–a marriage of convenience or a shotgun wedding. Anal. Chim. Acta. 2015;879:10–23. doi: 10.1016/j.aca.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 68.Jewett MC, Hofmann G, Nielsen J. Fungal metabolite analysis in genomics and phenomics. Curr. Opin. Biotechnol. 2006;17:191–197. doi: 10.1016/j.copbio.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 69.Pan L, et al. Metabolomic analysis of significant changes in Lactobacillus casei Zhang during culturing to generation 4,000 under conditions of glucose restriction. J. Dairy Sci. 2019;102:3851–3867. doi: 10.3168/jds.2018-15702. [DOI] [PubMed] [Google Scholar]
- 70.Salminen S, et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021;18:649–667. doi: 10.1038/s41575-021-00440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park S-E, et al. GC–MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. LWT Food Sci. Technol. 2016;68:313–321. doi: 10.1016/j.lwt.2015.12.046. [DOI] [Google Scholar]
- 72.Jeong SH, Lee SH, Jung JY, Choi EJ, Jeon CO. Microbial succession and metabolite changes during long‐term storage of kimchi. J. Food Sci. 2013;78:M763–M769. doi: 10.1111/1750-3841.12095. [DOI] [PubMed] [Google Scholar]
- 73.Shim S-M, et al. Profiling of fermentative metabolites in kimchi: volatile and non-volatile organic acids. J. Korean Soc. Appl. Biol. Chem. 2012;55:463–469. doi: 10.1007/s13765-012-2014-8. [DOI] [Google Scholar]
- 74.Mozzi F, Ortiz ME, Bleckwedel J, De Vuyst L, Pescuma M. Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Res. Int. 2013;54:1152–1161. doi: 10.1016/j.foodres.2012.11.010. [DOI] [Google Scholar]
- 75.Li C, et al. Serum metabolomics analysis for biomarker of Lactobacillus plantarum NCU116 on hyperlipidaemic rat model feed by high fat diet. J. Funct. Foods. 2018;42:171–176. doi: 10.1016/j.jff.2017.12.036. [DOI] [Google Scholar]
- 76.Schwenninger SM, et al. Characterization of low-molecular-weight antiyeast metabolites produced by a food-protective Lactobacillus-Propionibacterium coculture. J. Food Prot. 2008;71:2481–2487. doi: 10.4315/0362-028X-71.12.2481. [DOI] [PubMed] [Google Scholar]
- 77.Shi X, et al. Hepatic and fecal metabolomic analysis of the effects of Lactobacillus rhamnosus GG on alcoholic fatty liver disease in mice. J. Proteome Res. 2015;14:1174–1182. doi: 10.1021/pr501121c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, Y. et al. Metabolism characteristics of lactic acid bacteria and the expanding applications in food industry. Front. Bioeng. Biotechnol. 9, 612285 (2021). [DOI] [PMC free article] [PubMed]
- 79.Shevchenko A, Simons K. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 2010;11:593–598. doi: 10.1038/nrm2934. [DOI] [PubMed] [Google Scholar]
- 80.Hsu F-F. Mass spectrometry-based shotgun lipidomics–a critical review from the technical point of view. Anal. Bioanal. Chem. 2018;410:6387–6409. doi: 10.1007/s00216-018-1252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung H-J, Sim J-H, Min T-S, Choi H-K. Metabolomics and lipidomics approaches in the science of probiotics: a review. J. Med. Food. 2018;21:1086–1095. doi: 10.1089/jmf.2017.4175. [DOI] [PubMed] [Google Scholar]
- 82.Schifano E, et al. In vitro and in vivo lipidomics as a tool for probiotics evaluation. Appl. Microbiol. Biotechnol. 2020;104:8937–8948. doi: 10.1007/s00253-020-10864-w. [DOI] [PubMed] [Google Scholar]
- 83.Kim H, et al. Comparative lipidomic analysis of extracellular vesicles derived from Lactobacillus plantarum APsulloc 331261 living in green tea leaves using liquid chromatography-mass spectrometry. Int. J. Mol. Sci. 2020;21:8076. doi: 10.3390/ijms21218076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 2011;80:301–325. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nyholm L, et al. Holo-omics: integrated host-microbiota multi-omics for basic and applied biological research. iScience. 2020;23:101414. doi: 10.1016/j.isci.2020.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Limborg MT, et al. Applied hologenomics: feasibility and potential in aquaculture. Trends Biotechnol. 2018;36:252–264. doi: 10.1016/j.tibtech.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 87.Liwinski, T., Leshem, A. & Elinav, E. Breakthroughs and bottlenecks in microbiome research. Trends Mol. Med. 27, 298–301 (2021). [DOI] [PubMed]
- 88.Franzosa EA, et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat. Rev. Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tyler SR, et al. Merged Affinity Network Association Clustering: Joint multi-omic/clinical clustering to identify disease endotypes. Cell Rep. 2021;35:108975. doi: 10.1016/j.celrep.2021.108975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Falcinelli S, et al. Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep09336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu H-J, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rudman SM, et al. Microbiome composition shapes rapid genomic adaptation of Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2019;116:20025–20032. doi: 10.1073/pnas.1907787116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 94.Liang S, Wu X, Jin F. Gut-brain psychology: rethinking psychology from the microbiota–gut–brain axis. Front. Integr. Neurosci. 2018;12:33. doi: 10.3389/fnint.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 96.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 97.Shi Y, Tyson GW, DeLong EF. Metatranscriptomics reveals unique microbial small RNAs in the ocean’s water column. Nature. 2009;459:266–269. doi: 10.1038/nature08055. [DOI] [PubMed] [Google Scholar]
- 98.Wilmes P, Bond PL. Metaproteomics: studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006;14:92–97. doi: 10.1016/j.tim.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 99.Puspita, I. D., Kamagata, Y., Tanaka, M., Asano, K. & Nakatsu, C. H. Are uncultivated bacteria really uncultivable? Microbes Environ. 27, 356–366 (2012). [DOI] [PMC free article] [PubMed]
- 100.Delogu F, et al. Integration of absolute multi-omics reveals dynamic protein-to-RNA ratios and metabolic interplay within mixed-domain microbiomes. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-18543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Henry LP, Bruijning M, Forsberg SK, Ayroles JF. The microbiome extends host evolutionary potential. Nat. Commun. 2021;12:1–13. doi: 10.1038/s41467-021-25315-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen L, et al. Integrative multiomics analysis reveals host-microbe-metabolite interplays associated with the aging process in Singaporeans. Gut Microbes. 2022;14:2070392. doi: 10.1080/19490976.2022.2070392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salvetti E, O’Toole PW. The genomic basis of lactobacilli as health-promoting organisms. Microbiol. Spectr. 2017;5:22. doi: 10.1128/microbiolspec.BAD-0011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gueimonde M, Collado M. Metagenomics and probiotics. Clin. Microbiol. Infect. 2012;18:32–34. doi: 10.1111/j.1469-0691.2012.03873.x. [DOI] [PubMed] [Google Scholar]
- 105.Bottacini F, van Sinderen D, Ventura M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem. J. 2017;474:4137–4152. doi: 10.1042/BCJ20160756. [DOI] [PubMed] [Google Scholar]
- 106.Kiousi DE, Rathosi M, Tsifintaris M, Chondrou P, Galanis A. Pro-biomics: Omics technologies to unravel the role of probiotics in health and disease. Adv. Nutr. 2021;12:1802–1820. doi: 10.1093/advances/nmab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ngashangva N, Mukherjee PK, Sharma C, Kalita MC, Sarangthem I. Integrated genomics and proteomics analysis of Paenibacillus peoriae IBSD35 and insights into its antimicrobial characteristics. Sci. Rep. 2022;12:1–18. doi: 10.1038/s41598-022-23613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability‐based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 109.Ma B, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 110.Miao, Z., Humphreys, B. D., McMahon, A. P. & Kim, J. Multi-omics integration in the age of million single-cell data. Nat. Rev. Nephrol. 17, 710–724 (2021). [DOI] [PMC free article] [PubMed]
- 111.Bersanelli M, et al. Methods for the integration of multi-omics data: mathematical aspects. BMC Bioinform. 2016;17:167–177. doi: 10.1186/s12859-015-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mallory XF, Edrisi M, Navin N, Nakhleh L. Methods for copy number aberration detection from single-cell DNA-sequencing data. Genome Biol. 2020;21:1–22. doi: 10.1186/s13059-020-02119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baek S, Lee I. Single-cell ATAC sequencing analysis: from data preprocessing to hypothesis generation. Comput. Struct. Biotechnol. J. 2020;18:1429–1439. doi: 10.1016/j.csbj.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Erhard F, et al. scSLAM-seq reveals core features of transcription dynamics in single cells. Nature. 2019;571:419–423. doi: 10.1038/s41586-019-1369-y. [DOI] [PubMed] [Google Scholar]
- 115.Hu Y, et al. Single-cell RNA cap and tail sequencing (scRCAT-seq) reveals subtype-specific isoforms differing in transcript demarcation. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Specht H, et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. 2021;22:1–27. doi: 10.1186/s13059-021-02267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rautenstrauch, P., Vlot, A. H. C., Saran, S. & Ohler, U. Intricacies of single-cell multi-omics data integration. Trends Genet. 38, 128–139 (2021). [DOI] [PubMed]
- 118.Lê Cao K-A, González I, Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lee S-H, et al. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021;6:277–288. doi: 10.1038/s41564-020-00831-6. [DOI] [PubMed] [Google Scholar]
- 120.Mars RA, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182:1460–1473.e17. doi: 10.1016/j.cell.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rasmussen JA, et al. A multi-omics approach unravels metagenomic and metabolic alterations of a probiotic and synbiotic additive in rainbow trout (Oncorhynchus mykiss) Microbiome. 2022;10:1–19. doi: 10.1186/s40168-021-01221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Heintz-Buschart A, et al. Integrated multi-omics of the human gut microbiome in a case study of familial type 1 diabetes. Nat. Microbiol. 2016;2:1–13. doi: 10.1038/nmicrobiol.2016.180. [DOI] [PubMed] [Google Scholar]
- 123.Eloe-Fadrosh EA, et al. The National Microbiome Data Collaborative Data Portal: an integrated multi-omics microbiome data resource. Nucleic Acids Res. 2022;50:D828–D836. [Google Scholar]
- 124.Yan X, et al. Integrated multi-omics of the gastrointestinal microbiome and ruminant host reveals metabolic adaptation underlying early life development. Microbiome. 2022;10:1–18. doi: 10.1186/s40168-022-01396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fuhrer T, Zamboni N. High-throughput discovery metabolomics. Curr. Opin. Biotechnol. 2015;31:73–78. doi: 10.1016/j.copbio.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 126.Kahn SD. On the future of genomic data. Science. 2011;331:728–729. doi: 10.1126/science.1197891. [DOI] [PubMed] [Google Scholar]
- 127.Zhang Z, Wu S, Stenoien DL, Paša-Tolić L. High-throughput proteomics. Annu. Rev. Anal. Chem. 2014;7:427–454. doi: 10.1146/annurev-anchem-071213-020216. [DOI] [PubMed] [Google Scholar]
- 128.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:1–15. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Integrative, H. The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe. 2014;16:276–289. doi: 10.1016/j.chom.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chong J, Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites. 2017;7:62. doi: 10.3390/metabo7040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front. Genet. 2017;8:84. doi: 10.3389/fgene.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou G, Ewald J, Xia J. OmicsAnalyst: a comprehensive web-based platform for visual analytics of multi-omics data. Nucleic Acids Res. 2021;49:W476–W482. doi: 10.1093/nar/gkab394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hernández-de-Diego R, et al. PaintOmics 3: a web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res. 2018;46:W503–W509. doi: 10.1093/nar/gky466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou G, Xia J. OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic Acids Res. 2018;46:W514–W522. doi: 10.1093/nar/gky510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Montague E, et al. Beyond protein expression, MOPED goes multi-omics. Nucleic Acids Res. 2015;43:D1145–D1151. doi: 10.1093/nar/gku1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou G, Pang Z, Lu Y, Ewald J, Xia J. OmicsNet 2.0: a web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 2022;50:W527–W533. doi: 10.1093/nar/gkac376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Allendes Osorio RS, et al. Panomicon: A web-based environment for interactive, visual analysis of multi-omics data. Heliyon. 2020;6:e04618. doi: 10.1016/j.heliyon.2020.e04618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.