Abstract

Viruses can elicit varying types and severities of symptoms during plant host infection. We investigated changes in the proteome and transcriptome of Nicotiana benthamiana plants infected by grapevine fanleaf virus (GFLV) with an emphasis on vein clearing symptom development. Comparative, time-course liquid chromatography tandem mass spectrometry and 3′ ribonucleic acid sequencing analyses of plants infected by two wildtype GFLV strains, one symptomatic and one asymptomatic, and their asymptomatic mutant strains carrying a single amino acid change in the RNA-dependent RNA polymerase (RdRP) were conducted to identify host biochemical pathways involved in viral symptom development. During peak vein clearing symptom display at 7 days post-inoculation (dpi), protein and gene ontologies related to immune response, gene regulation, and secondary metabolite production were overrepresented when contrasting wildtype GFLV strain GHu and mutant GHu-1EK802GPol. Prior to the onset of symptom development at 4 dpi and when symptoms faded away at 12 dpi, protein and gene ontologies related to chitinase activity, hypersensitive response, and transcriptional regulation were identified. This systems biology approach highlighted how a single amino acid of a plant viral RdRP mediates changes to the host proteome (∼1%) and transcriptome (∼8.5%) related to transient vein clearing symptoms and the network of pathways involved in the virus–host arms race.
Keywords: transcriptomics, proteomics, virus, grapevine, symptomology, Nicotiana benthamiana, time-course, nepovirus, RNA-dependent RNA polymerase
Introduction
Viruses induce targeted changes to host cells to facilitate replication, movement, and propagation. Plant viruses elicit host responses that primarily occur through protein–protein (i.e., pattern-recognition receptors and pathogen-associated molecular pattern), RNA-protein (i.e., host RNA binding proteins and viral silencing pathways), or protein-lipid interactions (i.e., membrane-anchored receptors and cross-membrane signaling).1−4 Selection has favored the evolution of a network of pathways for viral detection and suppression, typically starting with immune receptors and co-receptors.5,6 Likewise, selection has favored the evolution of viral proteins to combat these defense mechanisms by interfering at the host RNA or protein level to diminish plant responses.7 Common host targets by viruses are regulators of transcription and translation, small molecule/hormone pathways, and the antiviral pathways of RNA silencing. Host changes induced by viruses can be quantified at various levels, with most studies examining the differential abundance of transcripts or proteins within biological samples.
Multi-layered systems biology approaches have been performed to decipher virus–host interactions at the levels of genome, transcriptome, proteome, and the metabolome. For example, coupled proteomic–transcriptomic analyses have been used to study the infection of human and zoonotic viruses such as influenza A virus (H1N1)8 and severe acute respiratory syndrome coronavirus-2.9 Similarly, comparative proteomics and transcriptomics have been used to advance our understanding of virus infections in plant hosts such as mungbean yellow mosaic India virus (MYMIV) in soybean,10 potato virus Y (PVY) in potato,11 sugarcane mosaic virus (SCMV) in sugarcane,12 and turnip mosaic virus (TuMV) in cabbage.13 For MYMIV, host genes related to stress response, metabolism, and resistance were found to be critical for symptomatic infection.10 For PVY, photosynthesis-related genes were most impacted at both protein and transcript levels.11 Viruses have elegant mechanisms to reprogram their host for replication, and under certain conditions, these changes result in disease symptom development. The genome of a virus is significantly smaller than that of their host (approximately 500–5000 times over), indicating that multi-functional virus proteins are efficiently encoded for optimal infection and survival.14 Furthermore, viruses encode proteins to combat host defense pathways aimed to clear viral infection. Despite tremendous progress, our understanding of viral–host interactive pathways and downstream effects in plants is still limited.
Using two or more omics techniques rather than a single omics approach has higher confirmatory value for the identification of host factors involved in functional virus–host interactions. Respectively, mass spectrometry (MS) and ribonucleic acid sequencing (RNA-Seq) are quantitative methodologies for providing insights into the changes that occur within the plant host proteome and transcriptome in response to infection. Perturbations in the relative abundance of proteins and transcripts can allude to specific host pathways and networks behind biological processes that are involved, ultimately deepening our understandings of viral infections.15 Of note are key steps of viral pathogenesis and gene regulatory networks that contribute to disease symptom development negatively affecting plant health. For example, many pathways utilized by viruses for replication are well-documented to contribute to symptomology, but the exact mechanisms that perturb plant cellular functions remain unresolved.16 Using multiple omics techniques to quantify differentially abundant genes/proteins is a powerful approach to determine how these changes are critical for permissive host–pathogen interactions and disease development. If a mechanism is defined, efforts can then be tailored to disrupt the molecular interactions, thereby avoiding disease symptoms and curbing the negative impacts of viruses and other pathogens. In addition, tools such as genome assemblies, reverse genetics, more advanced sequencing technologies, and computational power have been integral to explore large datasets across multi-omics approaches for informing viral infections.17
Grapevine fanleaf virus (GFLV) is the most devastating virus of grapevine.18,19 This virus causes severe yield losses (up to 80%) and substantially reduces the productive lifespan of vineyards. GFLV is a representative member of the species NepovirusGFLV in the genus Nepovirus of the family Secoviridae.20 Its genome consists of two RNA molecules, each encoding a polyprotein that is proteolytically cleaved by the RNA1-encoded proteinase 1DPro into eight mature proteins21 (Figure 1A). GFLV can be mechanically transferred to herbaceous hosts such as Nicotiana benthamiana on which distinct symptoms can develop.22 Host responses and cellular reprogramming that are induced during GFLV infection, particularly for symptom development, remain largely unknown. Current hypotheses attribute symptom eliciting responses to the GFLV RNA1-encoded RNA-dependent RNA polymerase (RdRP, protein 1EPol) interacting with host components via protein–protein interactions.23 Previous attempts at identifying host proteins associated with GFLV 1EPol were performed by affinity-purification MS23 using infectious clones carrying a V5 epitope-tagged 1EPol. Several host protein candidates were found to complex with GFLV 1EPol; however, a large gap remains for understanding how GFLV mechanistically elicits symptoms in infected plant hosts.23
Figure 1.
GFLV genetic composition and symptom expression in N. benthamiana. (A) Simplified schematic of the structure and expression of RNA1 of wildtype GFLV strains F13 (blue) and GHu (red) and RNA2 of wildtype GFLV strain GHu (purple). The viral genome-linked protein VPg is shown at the 5′ end of the RNA molecules and the polyadenylated tail at the 3′ end. Untranslated 5′ and 3′ regions are shown as horizontal gray bars. Vertical gray lines represent proteolytic cleavage sites as processed by RNA1-encoded proteinase 1DPro. The nucleotide identity of 87.78% between the two GFLV RNA1s is depicted in gray (Jalview v2.11.2.2). Critical amino acid residue 1E802Pol for vein clearing symptoms is indicated by an arrow and colored vertical line on RNA1 molecules. (B) GFLV-infected N. benthamiana plants at 7 days post-inoculation (dpi) with asymptomatic wildtype strain F13 (top) and symptomatic wildtype strain GHu (bottom) causing a stark vein clearing phenotype. Plants infected with mutant GFLV-GHu 1EK802GPol appear identical to asymptomatic plants infected with GFLV-F13 (image editing: individual plants are shown after background removal). (C) Symptom expression of wildtype GFLV-GHu in infected N. benthamiana over time. Apical leaves observed across three cohorts of plants (n1 = 23, n2 = 48, and n3 = 15) for vein clearing symptoms were rated on a binary scale (1 = symptomatic, 0 = asymptomatic). GFLV symptoms emerged in a few inoculated plants as early as 4–6 days post-inoculation (dpi); most plants expressed symptoms at 7–10 dpi, and all plants fully recovered by 17 dpi. The average percent of symptomatic plants in three trials is shown with standard deviations as error bars. Severity of symptoms is diagramed as representative leaf drawings above the plotted line. The three time points used for tissue collection for transcriptomics and proteomics work are indicated with a star.
Powerful biological tools are available to study how GFLV elicits symptoms in plants using ‘omics approaches. First, N. benthamiana sustains systemic GFLV infection with different virus strains and mutants that vary in their ability to elicit symptoms.22,24 For example, GFLV strain GHu induces vein clearing symptoms on apical leaves of N. benthamiana, while strain F13 remains asymptomatic (Figure 1B).25 A single amino acid in position 802 of GFLV-GHu 1EPol is critical for manifestation of vein clearing symptoms, which are transient and accompanied by a progressive host recovery within 2 weeks or less without clearing viral infection.25 Second, the genome of N. benthamiana is sequenced and well-annotated;26 thus, this model herbaceous plant is ideal for conducting transcriptomics and proteomics studies to identify host factors involved in virus infection.26 We hypothesized that contrasting transcript/protein expression profiles between wildtype GFLV strain GHu and its single amino acid mutant GHu 1EK802GPol in the RdRP coding region24 would allow us to pinpoint key cellular pathways putatively involved in vein clearing symptom development. In this study, we leveraged the unique features of GFLV in planta to test this hypothesis using parallel tandem liquid chromatography MS (LC–MS/MS) and 3′RNA-Seq analyses for profiling changes in the proteome and transcriptome of infectedN. benthamiana over time. Our analyses revealed a short list of gene candidates that were cross-validated through both proteomics and transcriptomics, showing unique interactions of GFLV strain GHu with host components to elicit vein clearing symptoms, different from gene-for-gene or phytohormone-activated pathways most identified in plant–pathogen interactions. Furthermore, these data showed that the change in regulatory networks was most prevalent at peak symptom development, not before nor after.
Experimental Section
Plant Material
Seeds of N. benthamiana were sown in “Cornell” LM-2 potting mix and transplanted 16 days after sowing into individual 3″ diameter pots. Plants were maintained in a controlled growth chamber at 25 °C and a 16:8 light/dark cycle at 70% relative humidity.
GFLV Strains
Four GFLV strains were used in this study: wildtype strains GHu and F13 and mutant strains GHu-1EK802GPol and F13-1EG802KPol.24 The bipartite genomic RNAs of wildtype strains GHu and F13 are sequenced. They share 87.78% nucleotide identity (Figure 1A). The mutant strains used in this study, GHu-1EK802GPol and F13-1EG802KPol, differ from their wildtype strains at a single amino acid of the RdRP in position 802 with a K to G mutation for mutant GHu and a G to K mutation for mutant F1324 (Figure 1A, arrow). Infectious cDNA clones of these four GFLV strains in binary plasmid pCLEAN-G181 for Agrobacterium tumefaciens-mediated delivery in planta were used as previously described.13
Mechanical Inoculation of N. benthamiana with GFLV
For mechanical inoculation, leaf tissue of N. benthamiana plants infected with one of the four GFLV strains was macerated in inoculation buffer (0.15 M KH2PO4, 0.35 M Na2HPO4, pH 7.0) with carborundum powder by mortar and pestle. The crude sap inoculum was rubbed onto the two primary leaves of the plants at the six-leaf developmental stage (4 weeks post-germination), and carborundum was immediately rinsed off with ddH2O.
A cohort of 75 N. benthamiana plants was divided into 5 groups of 15 plants each and inoculated with one of the following viral treatments: wildtype GFLV strain GHu, wildtype GFLV strain F13, GFLV mutant GHu 1EK802GPol, GFLV-F13 mutant 1EG802KPol, and inoculation buffer supplemented with carborundum as the mock control. Mock control plants were used to account for induced wounding response of the host following mechanical inoculation. These plant groups served as the five treatment categories for the proteomics and transcriptomics analysis.
GFLV-GHu Symptomology Observations
Three cohorts of N. benthamiana plants (n1 = 23, n2 = 48, and n3 = 15) were inoculated with wildtype GFLV strain GHu, group n3 belonging to the transcriptomics and proteomics experiments. Vein clearing symptoms (Figure 1B, bottom) were observed daily in the apical leaves over a 16-day period until plants fully recovered. Symptoms were rated on a binary scale (0 = asymptomatic, 1 = symptomatic). All inoculated plants were tested in duplicate 17 days post-inoculation (dpi) via double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) with specific GFLV antibodies following the manufacturer’s recommendations (Bioreba, Reinach, Switzerland). Samples were tested with the positive control, buffer control, and GFLV-free samples in duplicate per 96-well microtiter plate. Only plants that tested positive for GFLV in DAS-ELISA were considered for symptomology recordings.
Leaf Tissue Collection for Proteomics and Transcriptomics Analyses
Preliminary pilot studies documented detectable GFLV in apical tissue at the protein and transcript levels using LC–MS/MS of protein tissue extracts and reverse transcription (RT) polymerase chain reaction (PCR) with specific primers,24 respectively, by the fourth dpi (data not shown). Based on these results, apical tissue of N. benthamiana was collected at three time points following inoculation (4, 7, and 12 dpi). One leaf was removed per plant at each collection, and five 1.0 cm hole punches were taken per leaf from three leaves of the same treatment and time point. The resulting 15 leaf discs were pooled into a 2.0 mL Eppendorf tube. This was repeated five times for each treatment, resulting in five biological replicates per treatment for each time point. This was mirrored for the two ‘omics techniques, as leaf discs were taken from the left or right side of leaves and placed into a microfuge tube for protein or RNA extraction, respectively. This resulted in 150 leaf tissue collections in total, including 75 for RNA isolation and 75 for protein isolation (Figure S1). All tissue was flash-frozen in liquid nitrogen and kept at −80 °C until further processing.
Total RNA Purification and 3′-RNA Sequencing
Total RNA was purified from N. benthamiana leaf tissue using an adapted protocol from the Omega E.Z.N.A Plant RNA Mini Kit (cat no. R6827), treated with DNAse I and filtered using RNA Clean & Concentrator (Zymo Research, cat no. R1013). An elution volume of 90 μL provided RNA concentrations above 200 ng/μL. RNA quantity and integrity were analyzed using an Agilent 2100 Bioanalyzer with RNA Nano Kits (part no. 5067-1511). Library sequencing was performed at the Cornell Institute of Biotechnology (Ithaca, NY, USA) using Illumina NextSeq500 technology (Illumina, CA, USA) with single end, 1 × 86 bp reads.
Transcriptomics Data Processing
Following Illumina sequencing, adapter sequences were trimmed using Trimmomatic (v0.39)27 and the quality of reads was analyzed using FastQC (v0.11.9).28 Sequence alignments were performed by creating a reference library using HiSat229 and the N. benthamiana (v1.0.1)26 draft genome assembly from Sol Genomics at the Boyce Thompson Institute (Ithaca, NY). Annotated sequences that contained the descriptor “gene” were extracted using FeatureCounts (v0.11.9)30 from Subread (v2.0.1).31 Sequence alignments were also performed against a curated genome collection of GFLV isolates and manually entered mutations for strains that were used in this study to capture polyadenylated viral RNA. Mapped sequence reads were then extracted and exported as a data matrix.
The following data processing steps were performed in RStudio v4.1.3. Count files were parsed and merged into a single count matrix, and a corresponding metadata file was created to perform differential sequence analyses with the package DESeq2 (v1.32.0).32 DESeq2 model construction followed the methodical and classical approach of using the treatment and time point as explanatory factors. However, this attempt resulted in poor explanation of variance. Therefore, a new factor for the model that considers treatment, time, and their interaction was created and named “group”. This new model served for full-scale experiment-wide comparisons. Treatment comparisons were thereafter generated using only models per time point and never included samples from other time points. A matrix containing the counts per gene before normalization can be found on the GitHub page. Raw data files can be accessed through the NCBI database, Bioproject: PRJNA838211.
Total Protein Isolation and Liquid Chromatography Tandem MS Analysis
Total protein extractions were performed on mirrored N. benthamiana leaf samples to RNA extractions (Figure S1). This protocol was adapted from original methods previously described33,34 with the following two modifications: First, N. benthamiana tissue was macerated with steel BBs in 2 mL centrifuge tubes following flash freezing in liquid nitrogen. Second, addition of 1 mL of working solution (10% trichloroacetic acid—TCA, 2% β-mercaptoethanol in acetone) to each sample was followed by probe sonication for cell lysis and membrane disruption. Samples were then incubated in an ethanol dry ice bath for at least 1 h and centrifuged to isolate insoluble proteins. The supernatant was removed and discarded, and the pellet was washed with 500 μL of pure acetone three times followed by centrifugation. The pellets were dried completely for 30 min and then were stored at −80 °C for further processing.
The protein-containing pellets were resuspended in 150 μL of 7 M urea/100 mM ammonium bicarbonate and kept on ice. Micromagnetic stir bars (Fisherbrand, Cat. no. 14-513-57) were added, and pellets were left to stir overnight at 4 °C. Resuspended proteins were subject to a Bradford assay using bovine serum albumin (CAS: 9048-46-8) to obtain a standard curve. Equal protein concentrations were utilized to standardize protein loading amounts for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on gradient (4–20%) Mini-PROTEAN© TGX (BIO-RAD, no. 4561096) precast protein gels for visualization of protein integrity and concentration uniformity.
Based on calculated concentrations from the Bradford analysis and gel electrophoresis, 100 μg of protein per sample was used for further processing. Reduction and cysteine blocking of proteins were performed with tris(2-carboxyethyl)phosphine and methyl methanethiosulfonate, for 1 h at 30 °C and 1 h at room temperature, respectively. Proteins were digested using ≥99.9% sequencing-grade modified trypsin (Promega, no. V5117) for 16 h at 30 °C while being agitated at 450 rpm. Peptides were subject to reverse-phase silica Sep-Pak column clean up (Waters, SKU: 186004618) and then dried via VacuFuge (Labconco Centrivap, Cat. no. 78100-00-C) evaporation. One sample was lost during the digestion step due to a handling error (wildtype GFLV-F13 at 12 dpi, biological replicate no. 1).
The nano-LC–MS/MS analysis was carried out using a Q Exactive (Thermo Fisher Scientific, San Jose, CA) mass spectrometer equipped with a nanospray ion source and coupled with the EASY-nLC (Thermo Fisher Scientific, San Jose, CA). Each peptide sample was reconstituted in 100 μL of 0.1% formic acid. A total of 4 μL for each reconstituted sample was injected onto a NanoViper C-18 RP trap column (3 μm, 75 μm × 20 mm, Thermo Fisher) at a 12 μL/min flow rate for on-line desalting and separated on a NanoViper C-18 RP trap column (2 μm, 0.075 mm × 250 mm, Thermo Fisher). The peptides were eluted in a 90 min gradient of 5–42% of 80% acetonitrile in 0.1% formic acid (elution B) at 300 nL/min, followed by a 2 min ramping to 100% elution B and then being held for 18 min. The Q Exactive is operated in the positive ion mode with nano-spray voltage set at 2.9 kV and source temperature at 250 °C. The top 10 precursor ions with >2 changes were selected for the MS2 scan. MS survey scans were taken at a resolving power of 70,000 with a mass range of m/z 350–1550, AGC = 5 × 105, and Max IT = 50 ms and MS2 scans at 17,500 resolution with AGC = 1 × 105, Max IT = 110 ms, and with isolation window 2.0 m/z. All data were acquired under Xcalibur (v4.2.47) operation software and Q Exactive Tune (v2.11 QF1 Build 3006) (Thermo Fisher Scientific). A blank was run between each sample to prevent any carry-over of peptide. One sample did not return any peptide reads (mock inoculated control at 4 dpi, biological replicate no. 3), ultimately providing a total of 73 output files. Each sample was analyzed twice as analytical replicates (A and B) culminating in 146 runs and output files.
Proteomics Data Processing
Raw files obtained from LC–MS/MS (73 samples/146 files total) were converted to .dat files with MSConvert from ProteoWizard.35 Peptide identification was then performed via Mascot Daemon (v2.5.1)36 against the previously defined “NibenNepo” database23 with N. benthamiana, GFLV, related nepoviruses, potential contaminants, and decoys to return protein abundance for analysis, as performed previously.23 Spectral counts were extracted by uploading search results of the 146 outputs into Scaffold Q+ Quantification software (v4.11.1)37 through MudPIT experiments that combined the two technical replicates of each sample into single biological samples to ease computational constraints. A count matrix was constructed containing gene names, descriptions, and counts for each peptide match to that respective gene per biological replicate for a total of 73 samples (Table S2, File S2). A false discovery rate of 1.0% at both protein and peptide levels, a minimum of two peptides per protein, and standard protein cluster analysis were utilized across all samples. Given that the same amount of trypsin was added to each protein digest, all samples were normalized to the trypsin enzyme spectral counts through the E chain peptide hits [36.3% coefficient of variation (CV)] as in the NibenNepo database (GenBank identifier: 3318722). This matrix was then utilized for differential expression analysis through the “DEP: differential enrichment analysis of proteomics data” pipeline (v1.16.0)38,39 in RStudio. After normalization and imputation, data was subject to k-means clustering and principal component analysis (PCA) to understand the interrelatedness of samples between time points and treatment groups. The designed summarized experiment object was broken into three smaller summarized experiments for differential protein abundance analysis and downstream analysis per time point, as done with the transcriptomics data. The MS proteomics data (reference FASTA, .raw, .dat, MudPIT combined technical mzIdent, and results) have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PCD037185 and 10.6019/PXD037185.
Statistical Analyses of Transcriptomics and Proteomics Data
Transcripts were defined as differentially abundant through the DeSeq2 default Wald Test, where only transcripts meeting adjusted p-value <0.05 and |log2(fold-change)| ≥ 1 were considered. Proteins were considered differentially abundant after processing with the DEP pipeline using protein-wise linear models and empirical Bayes statistics, meeting the qualifications for highly differentially abundant proteins (DEPs) [p-value <0.05, |log2(fold-change)| ≥ 1] and medium DEPs [p-value <0.05, |log2(fold-change)| ≥ 0.5].
Statistical analysis of the viral titer in transcriptomics data was performed using a Kruskal–Wallis ANOVA with a post-hoc Dunnet’s test. The viral titer analysis of MS-acquired reads used a Student’s t-test with a Bonferroni multiple correction40 after normalization to trypsin chain E.
Gene set enrichment analysis (GSEA) was performed on transcripts and proteins passing thresholds of padj/pvalue <0.05 and |log2(fold-change)| ≥ 1 and 0.5, respectively. Briefly, a reference was created in gprofiler2 (v0.2.1)41,42 with the entire annotated N. benthamiana genome .gff file and gene list comparisons being analyzed for enrichment. The comparison between transcripts for wildtype GFLV strain GHu at 7 dpi and transcripts of all other groups was executed in GSEA (v4.1.0)43,44 and network analysis in Cytoscape (v3.8.2).45 This analysis was performed to identify transcript gene ontologies (GOs) unique to only plants infected with wildtype GFLV-GHu during symptom expression. Further GSEA contrasts were executed considering time and treatment contrasts of interest, namely, wildtype GFLV-GHu to other treatments at 7 dpi and the two GFLV-F13 strains to other treatments at 4 dpi.
Weighted gene co-expression network analysis (WGCNA) was performed using the WGCNA package (v1.71)46,47 to observe gene clusters with similar expression patterns over time between treatments. Parameters outside default were maxBlockSize = 10,000; mergeCutHeight = 0.25; minKMEtoStay = 0.3; minModuleSize = 200; and power = 7. The 28 modules created were then assigned to colors and were referred to as ME′color′. Select modules were then exported and analyzed for GO overrepresentation. Specifically, gprofiler2 was used to group and report significant ontology overrepresentation per WGCNA module, representing a unique expression profile over time between treatments. All scripts and required files can be found in the provided GitHub page and can be clarified upon request.
Quantitative Real-Time Polymerase Chain Reaction
To confirm the expression of a gene detected by RNA-Seq but not LC–MS/MS, total RNA samples used for transcriptome analysis were partially thawed on ice and used in the Luna Universal One-Step RT-qPCR Kit (New England Biolabs Inc., no. E3005) with two primer pairs (Nb01478-FWD/REV and FBOX-F/R, Table S1). Samples were loaded in triplicate and analyzed using a BIO-RAD CFX96 Real-Time System/C1000 Touch Thermocycler qPCR with the following cycles: 55 °C for 10 min; 95 °C for 1 min; 40 cycles of 95 °C for 10 s and 60 °C for 30 s; plate read; melt curve from 65–95 °C in increments of 0.5 °C for 5 s at each level; and end incubation at 8 °C. SYBR green fluorescence was measured and compared to that of the housekeeping gene FBOX48 (Table S1). Target gene expression was normalized through the standard ΔΔCt method from collected Ct data and plotted for each sample using RStudio. Script and data files are available on the GitHub page.
Results
GFLV Infection for Symptom Observation in N. benthamiana and Tissue Collection
Apical leaves of all N. benthamiana plants (100%, 60 of 60) that were selected for transcriptomics and proteomics analyses tested positive for GFLV in DAS-ELISA using specific antibodies when mechanically inoculated with one of the four virus strains used in this study [Figure S1(1)], revealing systemic infection. None (0%, 0 of 15) of the mock control plants tested positive for GFLV in DAS-ELISA, as expected. Vein clearing symptoms (Figure 1B) emerged in apical leaves of plants infected with wildtype GFLV strain GHu at 5–6 dpi (average 44 and 48% of plants, respectively). Symptoms were apparent, showing strong contrast of vein clearing (Figure 1B, bottom) in nearly all of the plants (94%) by 7–10 dpi with full plant recovery occurring thereafter (Figure 1C). The percentage of plants displaying symptoms from 7 to 10 dpi was on average 81 and 93%, respectively (Figure 1C). Consistent with previous reports, vein clearing symptoms never appeared in new foliage after recovery, and none of the other GFLV strains elicited disease symptoms.23−25 To capture the events leading up to vein clearing, peak symptomology, and the start of recovery, three time points of interest were selected at 4, 7, and 12 dpi (Figure 1C). Preliminary LC–MS/MS data (not shown here) suggested that GFLV does not replicate to detectable levels in apical tissue until at least 4 dpi and therefore was the earliest time point for consideration.
Proteomic Analysis of N. benthamiana Leaf Tissue
Total proteins from the mock control and GFLV-infected N. benthamiana leaves were isolated at 4, 7, and 12 dpi and analyzed by LC–MS/MS (Figure S1). A total report output from Scaffold Q+ was generated to display spectral counts across the entire experiment (Table S2A,B). A summary of the proteome dataset obtained with 73 samples showed an average of 2176 spectral counts per sample (Figure S2A). An average of 1928 spectral counts per sample were obtained after low-count filtering for values missing completely at random. The CV of raw data prior to filtering or normalization was 5.14 and 5.41% after low-count filtering. Most proteins were detected in all samples across the entire experiment, with less proteins unique per sample and a left-handed distribution of intensities (Figure S2B–D). Finally, after imputation and normalization, the CV was 0.78% across all samples.
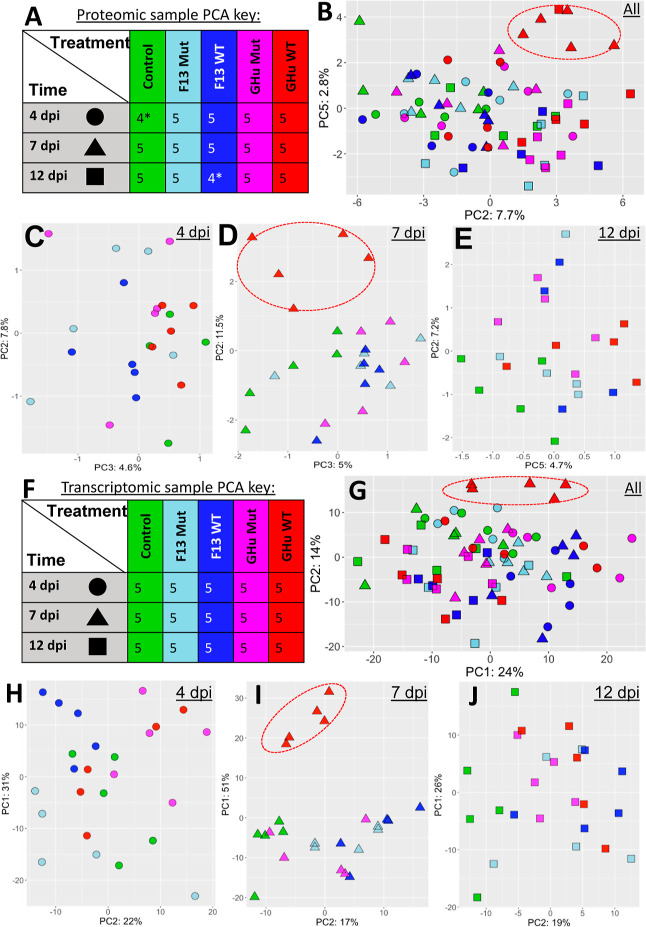
Low-count filtering removed proteins with low abundance (Figure S2A,B) and reduced near-zero distribution (Figure S3C). Variance-stabilized normalization (vsn) was performed as suggested by the DEP pipeline. This resulted in the dataset shifting from a left-centered distribution to a normalized curve (Figure S3E,F). A large proportion of missing protein identifications was present toward the lower end of spectral counts, indicating that the instrument’s detection limit was preventing total coverage of the dataset. This is generally considered to be missing values with a combination of these missing at random (MAR) and missing not at random (MNAR) (Figure S3C,D). Several instances of MNAR appeared when observing the missing values (Figure S4). For this reason, we tested a mixed imputation method after vsn to correct for both MNAR and MAR. We considered the most efficient and comprehensive imputation methods reported in the recent literature,49,50 including the biplot for principal component analysis (bpca) method for MAR and the zero method for MNAR (Figure S5). These methods did not drastically outperform simple bpca MNAR imputations in terms of hierarchal clustering and Pearson correlation. Therefore, we considered only bpca MNAR to simplify the data processing scheme. The missing values of protein identification were extracted for subsequent analyses. The simplified imputation technique resulted in an average of 2165 protein gene IDs per sample. Results showed the proteomic samples grouping by treatment and time point in hierarchal clustering. Pearson’s correlation matrices and relation by Gower’s matrices (Figure S6) were nuanced due to the relative similarity of treatment groups besides wildtype GFLV strain GHu at 7 dpi. The overlap and mixed clustering observed at each time point for the other groups was expected from previous observations and the absence of vein clearing outside of wildtype GFLV strain GHu at the middle time point. The datasets of plants infected by wildtype GFLV strain GHu at 7 dpi (symptomatic) and 12 dpi (asymptomatic) were more distinct than those at 4 dpi (asymptomatic). Viral treatments, regardless of the GFLV strain, were distinct from mock controls by PCA loadings during late infection (12 dpi). PCA revealed lower principal components (i.e., PC2, PC3, and PC5) more likely to be discriminated by the treatment group rather than PC1 (Figure 2A–E). PCA also showed wildtype GFLV strain GHu at 7 dpi being distinct from all other treatments and groupings. The proteome of plants infected with wildtype GFLV strain GHu at 7 dpi segregated from all other time points and treatments by PC5 and PC2 (Figure 2B,D). Small differences in tissue maturity and offset of the infection state could explain the observed discrepancies of proteome profiles, as revealed by correlation matrices (Figure S6) and in PCA (Figure 2).
Figure 2.
PCA for unbiased observation of acquired proteomics and transcriptomics data. Proteomics data utilizes top 100 proteins after vsn and bpca imputation. Transcriptomics data utilizes top 500 varying genes after vsn and low-count filtering. (A) Key for biological replicates displaying colors for treatments and shape used for three time points (4, 7, and 12 dpi as circles, triangles, and squares, respectively) of N. benthamiana inoculated with various GFLV strains and controls for (B) proteomics analysis of 73* samples for which grouping of samples is minorly apparent, especially for the wildtype GFLV strain GHu treatment (red triangles) in the upper-right quadrant. (C) Breakdown of samples at 4 dpi, (D) 7 dpi with large separation of wildtype GFLV strain GHu symptomatic plants in red (D), and (E) 12 dpi with cluster effects, suggesting subtle variations of the proteome upon these treatment groups. (F) Key displaying colors for treatments and shape used for the three time points (4, 7, and 12 dpi) following inoculation of N. benthamiana with various GFLV strains for (G) all 75 samples from DESeq2 analysis, normalization, and transformation plotted in PCA where PC2 separates out wildtype GHu 7 dpi, (H) 4 dpi with minor grouping of treatment groups, (I) 7 dpi with a large separation PC1 for symptomatic wildtype GFLV strain GHu, and (J) 12 dpi with minor grouping of treatment groups, although PC2 separates control from viral treatment groups.*Two samples were lost during sample processing resulting in 73 samples rather than the designed 75 sample experiment. This did not impact the ability of contrasts with a larger sample size taken and no more than one sample lost in a treatment group or time point.
Transcriptomic Analysis of N. benthamiana Leaf Tissue
Total RNA from the mock control and GFLV-infected N. benthamiana plants was isolated and analyzed by 3′RNA-Seq (Figure S1). All files passed quality control metrics (mean Phred score ∼33) and were deemed acceptable for downstream analysis. On average, 79.8% of reads mapped to the host reference genome with 2,148,843 reads per sample (Table S3 and Figure S7A). After normalization and low-count thresholds were placed, an average of 136,809 reads were attributed per sample (Figure S7B). No distinct trends were associated with any treatment or time point for count data indicating little to no technical bias from tissue processing steps, purification efforts, and sequencing. Analysis through HiSat2 returned 59,814 gene identities matching the assembly across all samples. Only 31,394 of these genes contained ≥2 counts per entry, which was used as a low-count filter threshold. A range of 10,000 to over 400,000 viral RNA2 reads were acquired in GFLV-infected plant samples, corresponding to polyadenylated ssRNA captured in 3′RNA-Seq. GFLV RNA1 reads were acquired with a range from 61 to over 60,000 reads but less consistently for the two F13 strains. The difference in abundance between GFLV RNA2 and GFLV RNA1 was 10–100-fold.
Model construction and variance stabilization normalization steps supported further analyses, as shown by dispersion estimate graphs with typical profiles (Figure S8), which showed dispersion estimates centered around the fitted gene estimate line. A hierarchal clustering using the pair-wise McQuitty method51 revealed grouping of treatments within time points with minimal mixing (Figure S9). PCAs on the 75 samples and groupings by time points (Figure 2F–J) revealed distinct separation for wildtype GFLV strain GHu at 7 dpi by both PC1 and PC2. Overall, the host transcriptome remained unresolved by other GFLV treatments and time points, with a maximum of 10.2% variance explained by the top 500 genes (Figure S10A). Both PCA and hierarchal clustering showed a distinct transcriptome profile for wildtype GFLV-GHu at 7 dpi from all the other datasets, with the major source of variation explained by the developmental or infection stage rather than from viral treatment groups (Figures 2B and S10B,C). The individual PCA plots per time point revealed more resolution among samples than the treatment group across the entire experiment (Figure 2H–J). The time-based groupings were then used for further analysis to avoid possible temporal bias in the transcriptome.
Detection of GFLV RNA and Protein Abundance across Treatments and Time Is Independent of Symptom Development
Across the four GFLV strains used in this study, the RNA-Seq data suggested little viral presence in apical leaf tissue at 4 dpi with an increase at 7 dpi coinciding with vein clearing and a reduced average number of reads at 12 dpi, as measured by GFLV RNA2 mapping transcripts normalized to the total read number per million or simply transcripts per million (Figure S11A). No significant difference in the virus titer among GFLV treatments was observed at any time point through paired Student’s t-test and post-hoc Levene’s test (P > 0.05). Wildtype GFLV strain GHu and mutant GFLV-F13 1EG802KPol contained elevated levels of RNA2 in comparison with wildtype GFLV-F13 and mutant GFLV-GHu 1EK802GPol, but differences were not statistically significant (P > 0.05). This result is consistent with the viral titer not determining symptom development, as previously reported.24
Peptides from GFLV RNA1- (1EPol) and RNA2-encoded (2BMP and 2CCP) proteins were consistently detected within the proteomics dataset (Figure S11B–D). Only samples within the mutant GFLV-F13 1EG802KPol treatment group showed spectra typically at 4 dpi (Figure S11C,D). This is consistent with a slightly faster replication or systemic infection rate for this GFLV strain compared to that for the other strains, which is also consistent with the RNA-Seq data (Figure S11A) in addition to the trend of elevated levels (and variability) of protein 2BMP for GFLV-F13 1EG802KPol at 7 dpi. Mock controls returned no spectra matching to viral peptides at any time point, as expected. Peptide spectra matching to GFLV protein 2BMP were detected in the highest relative abundance as compared to other viral proteins across all treatments with our extraction protocol and showed a similar trend to strain specific abundance, as with the RNA-Seq data corresponding to RNA2 reads. Treatments with wildtype GFLV-GHu and mutant GFLV-F13-1EG802KPol returned the most reads at 7 dpi, while wildtype GFLV-F13 and mutant GFLV-GHu 1EK802GPol returned lower counts; however, these were not statistically different from each other (P > 0.05).
GFLV-GHu Symptom Development Is Associated with Changes in the Plant Host Transcriptome and Proteome
We set out to identify genes/proteins whose change could be correlated to vein clearing symptom development by wildtype GFLV-GHu. Changes in abundance of transcripts (Figure 3) and proteins (Figure 4) were plotted using viral treatment contrasts, and the total number of differentially abundant transcripts (DEGs) (Table 1) and proteins (DEPs) (Table 2) was calculated.
Figure 3.
Select transcriptomics contrasts at 4, 7, and 12 dpi of N. benthamiana infected with various GFLV strains. Thresholds of |log2FoldChange| > 1 and p-adjusted value > 0.05 were used to identify DEGs. Data points meeting both requirements (green), significant reads but insufficient expression differences (blue), expression differences but not statistical significance (gray), and neither requirement (black) are shown. Contrasts between wildtype GFLV strain GHu and mutant GFLV-GHu 1EK802GPol at (A) 4 dpi with little difference in transcript expression, (B) 7 dpi with many DEGs, and (C) 12 dpi with few DEGs. Contrasts between wildtype GFLV strains GHu and F13 show a similar pattern at (D) 4 dpi with little difference in DEGs, (E) 7 dpi with many DEGs, and (F) 12 dpi with a reduced number of DEGs. Contrasts between wildtype GFLV strain F13 and mutant GFLV-F13 1EK802GPol show a different pattern at (G) 4 dpi with more DEGs than at (H) 7 dpi and (I) 12 dpi. (B) and (E) share 62.5% identity of significant DEGs, showing that most changes in the transcriptome are the result of a single amino acid and not the background of all other genomic differences between wildtype GFLV strains.
Figure 4.
Select proteomics contrasts at 4, 7, and 12 dpi of N. benthamiana with various GFLV strains. Thresholds of |log2FoldChange| > 1 (high confidence), |log2FoldChange| > 0.5 (medium confidence), and p-value <0.05 were used to identify DEPs. Data points meeting requirements for high confidence (purple), medium confidence (pink), significant reads but insufficient expression differences (blue), and neither requirement (black) are shown. Contrasts between wildtype GFLV strain GHu and mutant GFLV-GHu 1EK802GPol at (A) 4 dpi with very limited DEPs, (B) 7 dpi with several DEPs identified with medium and high confidence, and (C) 12 dpi with limited DEPs. Contrasts between wildtype GFLV-GHu and F13 show a similar pattern at (D) 4 dpi with limited DEPs, (E) 7 dpi with larger differences in protein expression, and (F) 12 dpi with limited DEPs. Contrasts between wildtype GFLV-F13 and mutant GFLV-F13 1EK802GPol with few DEPs at (G) 4 dpi, (H) 7 dpi, and (I) 12 dpi. The illustration of these time-resolved volcano plots demonstrates that the proteome is mainly disrupted at 7 dpi only for the wildtype GFLV-GHu treatment, corresponding to vein clearing symptom expression.
Table 1. Summary of DEGs between GFLV Treatment Groups at 4, 7, and 12 Days Post-Inoculation (dpi) of N. benthamianaa.
| wildtype GHu vs mutant GHuc |
wildtype GHu vs wildtype F13 |
wildtype F13 vs mutant F13 |
wildtype F13 vs mutant GHu |
wildtype GHu vs mutant F13 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| contrasts (dpi)b | UP | DOWN | UP | DOWN | UP | DOWN | UP | DOWN | UP | DOWN |
| 4 | 12 | 0 | 9 | 33 | 227 | 450 | 229 | 27 | 347 | 587 |
| 7 | 1838 | 869 | 1944 | 679 | 0 | 0 | 2 | 1 | 1579 | 688 |
| 12 | 16 | 2 | 51 | 11 | 0 | 2 | 1 | 1 | 85 | 66 |
The total number of unique transcripts detected total 31,396 genes after low-count filtering and normalization of data. The contrasts shown here are representative of the RNA-Seq data.
Contrasts are shown at 4, 7, and 12 dpi of N. benthamiana with wildtype GFLV-GHu and F13 and mutants GFLV-GHu 1EK802GPol and F13 1EG802KPol.
Upregulated (UP) and downregulated (DOWN) transcripts based on abundance and significance thresholds are indicated.
Table 2. Summary of DEPs Identified with Medium or High Confidence between GFLV Treatment Groups at Three Time Points Post-Inoculation of N. benthamianaa.
| wild type
GHu vs mutant GHuc |
wild type GHu vs wildtype F13 |
wild type F13 vs mutant F13 |
wild type F13 vs mutant GHu |
wild type GHu vs mutant F13 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| contrasts (dpi)b | UP | DOWN | UP | DOWN | UP | DOWN | UP | DOWN | UP | DOWN |
| 4 | 0 (0)d | 1 (0) | 1 (0) | 3 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (0) |
| 7 | 21 (0) | 7 (1) | 12 (0) | 18 (5) | 3 (0) | 2 (0) | 8 (0) | 1 (0) | 21 (0) | 10 (0) |
| 12 | 0 (0) | 0 (0) | 3 (0) | 2 (0) | 0 (0) | 2 (0) | 0 (0) | 3 (0) | 0 (0) | 1 (0) |
These contrasts are the result of 2152 common total proteins detected through LC–MS/MS analysis, averaging ∼10,800 spectral counts per sample.
Contrasts are shown at 4, 7, and 12 dpi of N. benthamiana with wildtype GFLV strains GHu and F13 and mutant GFLV strains GHu 1EK802GPol and F13 1EG802KPol.
Upregulated (UP) and downregulated (DOWN) proteins based on relative abundance and significance thresholds are indicated for each contrast.
Protein identifications with moderate and high (in parentheses) dysregulation are indicated for each contrast per time point. Thresholds of |log2FoldChange| > 1 (high dysregulation), |log2FoldChange| > 0.5 (medium dysregulation), and p-value <0.05 (confidence value for both) were used to identify DEPs.
The symptomatic wildtype GFLV-GHu provided the largest amount of contrast to other viral treatments during symptom expression at 7 dpi (Figures 3 and S12). Of interest were over 1800 upregulated and over 800 downregulated host transcripts for the wildtype and mutant GFLV-GHu comparison (Table 1) and 22 upregulated and 7 downregulated proteins at 7 dpi (Table 2).
Wildtype GFLV-GHu provided by far the most drastic difference in transcript expression levels when compared with any other treatments, suggesting that protein 1EPol residue 802 has a disproportionate impact on host response relative to the overall genetic composition of the viral genome. The difference of one amino acid between the two GFLV-F13 strains did not have a similar impact on the transcript expression levels at 7 dpi, as expected from phenotypic observations.
Contrast of the transcriptomic profiles of host plants infected with wildtype strain compared to mutant GFLV-GHu, over time, indicated that host gene expression was majorly affected at 7 dpi by the wildtype strain, but no substantial differences were observed at 4 and 12 dpi between the two (Figure 3A–C). This is reflected once more when contrasting wildtype GFLV-GHu with the mock control (Supporting Information Figure S12). A total of 2707 of 31,934 (8.48%) transcripts were classified as differentially abundant at 7 dpi between the two GFLV-GHu strains that differ in a single amino acid in protein 1EPol (Figure 3B). A vast difference in significant genes with both large fold changes and adjusted P-values was observed at 7 dpi (Figure 3B,E). This pattern was also obtained when contrasting wildtype GFLV-F13 and GHu (Figure 3D–F) with 2623 genes identified as differentially abundant at 7 dpi. It was notable that 62.5% of the classified DEGs from 7 dpi when comparing wildtype and mutant GFLV-GHu 1EK802GPol to wildtype strains GHu and F13 were identical (Figure 3B,E), suggesting that residue 802 of protein 1EPol is primarily responsible for underlying changes in the transcriptome. This means that a total of 1692 detected host genes could potentially be dysregulated due to a lysine in position 802 of the RdRP upon infection by wildtype GFLV-GHu. Remarkably, a disruption of transcript abundance between wildtype and mutant GFLV-F13 1EG802KPol was observed at 4 dpi (1042 genes) but not at later time points (Figure 3G–I). This may indicate initial response of the host to the presence of GFLV-F13, as previously mentioned for viral titer analyses, but these differences were reverted thereafter in the 7 dpi and 12 dpi contrasts. The wildtype and mutant GFLV-F13 had a larger host response at earlier and later time points than wildtype GFLV-GHu, all compared against mock-inoculated control plants (Figure S12).
Beyond these transcriptomic contrasts, treatment differences revealed dysregulation of genes from 0 (0.00%) to 2267 genes (7.10%) of the detected transcriptome (Table 1). Contrasting wildtype and mutant GFLV-F13 resulted in 677, 0, and 2 differentially abundant transcripts (DEGs) at 4, 7, and 12 dpi, respectively. Contrasting wildtype GFLV-F13 and mutant GFLV-GHu 1EK802GPol resulted in 256, 3, and 2 DEGs at 4, 7, and 12 dpi, respectively. Additionally, contrasting wildtype GFLV-GHu and mutant GFLV-F13 1EG802KPol resulted in 934, 2267, and 151 DEGs at 4, 7 dpi, and 12 dpi, respectively.
Analyses of the proteome revealed DEPs identified with high or moderate dysregulation (Figures 4 and S13). The number of proteins in each of these categories per treatment contrast and time point was determined out of over 2000 proteins per time point (Table 2). Additionally, spectral counts for each time point were documented as averages across samples to be 10,612 at 4 dpi, 11,028 at 7 dpi, and 10,661 at 12 dpi. For wildtype GFLV-GHu and mutant GFLV-GHu 1EK802GPol at 7 dpi, 28 (1.3%) DEPs were detected. Of these, 27 (1.3%) were moderately dysregulated proteins (P < 0.05 and |log2FoldChange| > 0.5), and 1 (<0.1%) was a highly dysregulated protein (P < 0.05 and |log2FoldChange| > 1). Similar trends of wildtype GFLV-GHu at 7 dpi reached the most pronounced contrast against other treatment groups with 30 DEPs against wildtype GFLV-F13 and 31 against mutant GFLV-F13 1EG802KPol both at 7 dpi (Table 2).
A single DEP was found at 4 dpi and none at 12 dpi when contrasting wildtype and mutant GFLV-GHu strains (Table 2). Nuances associated with GFLV F13 strains from transcriptomics data were not reflected in the proteomics contrasts (Figure S13). Additional proteomics contrasts revealed 0, 5, and 2 DEPs between wild and mutant GFLV-F13 at 4, 7, and 12 dpi, respectively (Table 2 and Figures 4 and S13). Contrasting wildtype GFLV-F13 with mutant GFLV-GHu 1EK802GPol found 0, 9, and 2 proteins of differential abundance at 4, 7, and 12 dpi, respectively. The consistent dysregulation of proteins between wildtype GFLV-GHu and other treatments at peak vein clearing and less DEPs at the early and late time points provide additional confidence in these findings.
GO Analyses of the Transcriptome and Proteome Indicative of Disrupted Host Pathways
GO analysis revealed GFLV- and GFLV strain-specific patterns of functional gene expression changes during infection. Contrasts for transcriptomics DEGs resulted in enriched gene functions for the wildtype GFLV-GHu against mutant GHu at 7 dpi: nucleic acid binding (GO: 0000496), nucleotide binding (GO: 0000166), RNA binding (GO: 0003723), structural constituent of the ribosome (GO: 0003735), photosynthesis (GO: 0015979), inositol-3-phosphate synthase activity (GO: 0004512), mitochondrial outer membrane (GO: 0005741), mitochondrial outer membrane translocase complex (GO: 0005742), translation elongation factor activity (GO: 0003746), and ssDNA binding (GO: 0003698) (Figure 5). Contrasting wildtype GFLV-GHu with the mock control at 7 dpi resulted in a similar output with seven identical ontologies, two new GO terms (translation initiation factor activity (GO: 0003743) and glycerol ether metabolic process (GO: 0006662), and two absent GO terms (translation elongation factor activity (GO: 0003746) and nucleic acid binding (GO: 0000496)) (Figure 5). Contrasting wildtype GFLV strains GHu and F13 at 7 dpi showed additional trends with differences of the GHu strain contrast being of three additional GO terms [rRNA processing (GO: 0006365), ribonucleoprotein complex (GO: 0030529), and nucleus (GO: 0005634)] and three GO terms removed [ssDNA binding (GO: 0003698), mitochondrial outer membrane translocase complex (GO: 0005742), and mitochondrial outer membrane (GO: 0005741)] (Figure 5).
Figure 5.
Gene set enrichment analysis of overrepresented genes present in the transcriptome analyses. GeneIDs were extracted from individual genes in contrast with a |log2FoldChange| > 1 and p-value <0.05. GeneID was inserted into “gprofiler2” package v0.2.1 for matching identity of functional annotation of N. benthamiana genes. Size of the dot represents relative abundance, and the color from blue to red signifies increasing significance (p-value range of 0.05 to <0.01). The upper panel shows phenotypes obtained following inoculation of N. benthamiana by various GFLV strains above each contrast for ease of comparing the visual phenotype to the transcriptome profile.
Other transcriptome contrasts are not reported here due to low numbers of DEGs, which did not return any significant GO terms (P < 0.05), mainly at early and late time points outside of symptom expression. Contrasts for the proteome of plants infected with wildtype GFLV-GHu showed similar trends but only resulted in three major ontologies with significance across the entire experiment: nucleotide binding (GO: 0000166), isopentenyl diphosphate biosynthetic process (GO: 0009240), and thiamine biosynthetic process (GO: 0009228) (Figure 6A).
Figure 6.
Overrepresented GOs for proteomic analysis. (A) Gene IDs were extracted from individual genes in multiple treatment-wise contrasts with a |log2FoldChange| > 0.5 and p-value <0.05 (medium confidence). The gene ID was inserted into “gprofiler2” package v0.2.1 for matching identity of functional annotation of N. benthamiana genes and returned GO terms overrepresented in the dysregulated proteome across the entire experiment. Contrasts were included from all time points of the experiment and from the following treatment contrasts: wildtype GFLV strain GHu vs mutant GHu 1EK802GPol, wildtype GFLV strain F13 vs mutant F13 1EK802GPol, wildtype GFLV-GHu vs wildtype GFLV-F13, and wildtype GFLV-F13 vs mutant GHu 1EK802GPol. The relative ratio of genes is shown by the x-axis scale, and the adjusted p-value (padj) is shown by the color of red to blue for most to least significant. Adjusted p-values indicated as significant by * = padj <0.1, ** = padj <0.05, and *** = padj<0.01. (B) GOs extracted from MNAR proteins across the entire 73 protein samples. Only significant ontologies are shown with the adjusted p-value increasing from blue to red (0.025 to <0.005).
GSEA and network analysis were performed from the significant transcriptomic DEGs for the wildtype GFLV-GHu treatment at 7 dpi versus all other treatments, providing a network of overrepresented, characteristic ontologies. The produced gene network displays three over-arching gene functions of immune response, metabolism, and gene regulation, most being up-regulated (Figure 7). For immune response-related genes, three main GO terms were apparent: response to oxidative stress (GO: 0006979), endopeptidase activity (GO: 0004175), and protein peptidyl–prolyl isomerization (GO: 0000413). Additional annotated genes related to host gene regulation and host immune response included gene silencing by RNA (GO: 0031047) which can act for both host and viral RNAs. Furthermore, RNA binding (GO: 0003723), nucleus (GO: 0005634), nuclear pore (GO: 0005643), transcription/DNA-templated (GO: 0006351), transcription coregulator activity (GO: 0001104), translation initiation factor activity (GO: 0003743), translation elongation factor activity (GO: 0003746), and NAD+ ADP-ribosyltransferase activity (GO: 0003950) all were upregulated and overrepresented terms for wildtype GFLV-GHu at 7 dpi compared with all other treatments (Figure 7).
Figure 7.
Differential gene set enrichment analysis network of wildtype GFLV strain GHu at 7 days post-inoculation of N. benthamiana contrasted against all other treatments (wildtype GFLV-GHu at 7 dpi was contrasted against the profile of all other treatments in GSEA with parameters of |log2FoldChange| > 1 and an adjusted p-value <0.05). Gene clusters upregulated (blue circle) and downregulated (red circle) are shown and are colored by saturation according to their respective calculated p-values. The size of a circle represents the relative number of genes present within a cluster ranging from 15 to 359. Lines represent the potential interactant connection network between GO terms. Black lined circles encompassing several clusters represent a general classification that was manually annotated; this analysis confirmed three main categories related to metabolism, immunity, and gene regulatory processes in the event of vein-clearing symptom development.
Extrapolating the proteomics dataset revealed several proteins to be MNAR (Figure S4, red brackets), for which the bulk might be relevant to treatment-specific phenotypes and representative of GFLV symptoms (i.e., downregulated beyond detection capacities through LC–MS/MS). A list containing these gene IDs was extracted from the DEP pipeline and was subject to GSEA. The resultant significant GOs were consistent with observations in the transcriptome to be structural constituent of the ribosome (GO: 0003735), ATP binding (GO: 0005524), RNA binding (GO: 0003723), calcium ion binding (GO: 0005509), nucleosome (GO: 0000786), intramolecular transferase activity (GO: 0016866), and inositol-3-phosphate synthase activity (GO: 0004512) (Figure 6B). This result suggested that MNAR proteins in this dataset are associated with the vein clearing phenotype, signifying that their abundance is too low for detection in treatment-specific samples through our protocol. This difference in protein presence or absence was observed between GFLV-GHu strains against GFLV-F13 strains and mock controls. This finding was reinforced by the fact that the transcriptome shows similar dysregulated GOs.
The largest change in the host transcriptome and proteome was observed with plants infected with wildtype GFLV-GHu. The top five proteins/genes based on fold change were for (1) proteomics; NUCLEOLAR PROTEIN 56, RNA-BINDING PROTEIN-LIKE, 4-HYDROXY-3-METHYLBUT-2-ENYL DIPHOSPHATE REDUCTASE [twice], and INOSITOL-3-PHOSPHATE SYNTHASE and (2) transcriptomics; 60S ACIDIC RIBOSOMAL PROTEIN P1 [twice], GLUTATHIONE S-TRANSFERASE U9, 30S RIBOSOMAL PROTEIN S19, and TETRATRICOPEPTIDE REPEAT (TPR)-LIKE SUPERFAMILY PROTEIN (Table S5). These genes represent top candidates for future investigation into their function during GFLV infection and symptom development.
Weighted Gene Co-Expression Network Analysis Further Demonstrates Time- and GFLV Strain-Specific Events
Gene clusters were constructed using WGCNA, resulting in 28 eigengene modules constructed from the transcriptomics dataset (Figure S14). Four expression modules contained 6985 downregulated genes only for wildtype GFLV-GHu at 7 dpi (MEbrown, MEyellow, MEblack, and MEpurple), while MEblue contained 3582 upregulated genes only for wildtype GFLV-GHu at 7 dpi (Figure 8A). All other treatments had relatively constant expression profiles in comparison to wildtype GFLV-GHu. A gene list extracted from all five of these modules and subject to GSEA revealed the same GOs as those displayed in the GSEA contrast of wildtype and mutant GFLV-GHu at 7 dpi, including nucleic acid binding, structural constituent of the ribosome, GTPase activity (GO: 0061745), protein folding (GO: 0006457), and translation initiation factor activity (Figure 8B). Similarly, a separate analysis of eigengene module MEblue showed many similar GOs with nucleic acid binding (GO: 0000496), nucleotide binding (GO: 0000166), RNA binding (GO: 0003723), structural constituent of the ribosome (GO: 0003735), and GTPase activity (GO: 0003924) being the top five GOs (Figure 8C). The same analysis was performed on MEmagenta, MEwhite, and MEgreenyellow for genes down- or up-regulated by mutant GFLV-F13 1EG802KPol at 4 dpi (Figure S15A), revealing GOs related to metabolic processes (GO: 0008152), nuclease activity (GO: 0004518), and cytoskeleton organization (GO: 0007010) (Figure S15B). Since wildtype GFLV-F13 does not manifest symptoms, these regulated genes are most likely unrelated to the vein clearing phenotype but may relate to virus–plant interactions early in the infection cycle of N. benthamiana. Other WGCNA modules were grouped based on their relative changes in expression for other treatments (i.e., mutant GFLV-F13 1EG802KPol at 12 dpi with MEorange and MEdarkorange), but no significant resulting GOs were found (data not shown). This was most likely due to the robust selective nature of our model and the low number of genes fitting into each module (<400 genes). All other unique gene profile expression modules were unsuccessful in fulfilling the minimum requirements and P-value thresholds to construct GSEA after extraction.
Figure 8.
Functional categorization of WGCNA eigengene modules unique to infection of N. benthamiana by GFLV wildtype strain GHu at 7 dpi. (A) Selected modules with unique expression patterns. Four of five modules (MEbrown, MEyellow, MEblack, and MEpurple) display downregulation of genes, while the MEblue module shows upregulation of genes. The consensus eigengene-based connectivity (kME) value was set to 0.3 and mergeCutHeight to 0.25 for selecting closely related gene expression profiles. (B) GOs overrepresented in all five modules subjected to GSEA. GOs unique to downregulated pathways are indicated by gray down arrows. Bars are colored by the p-value as indicated by the in-plot legend. (C) GOs overrepresented in MEblue subjected to GSEA.
Cross-Examination of Transcriptomics and Proteomics Datasets Shows Substantial Overlap
Overlap of differentially abundant genes from the transcriptomics and proteomics datasets was assessed by extracting gene IDs from each analysis and finding duplicates, after loosening our initial fold-change parameters of transcriptomics to cover a similar range for the proteomic values (Figure 9A).
Figure 9.
(A) Contrast of differentially abundant gene candidates identified by LC–MS/MS (purple) and 3′RNA-Seq (green) between wildtype GFLV strain GHu and mutant GFLV-GHu 1EK802GPol. From left to right, Venn diagrams show the number of differentially abundant genes identified by proteomics (|log2FoldChange| > 0.5 and p-value < 0.05), transcriptomics (adjusted p-value <0.05), and both (overlap) at 4, 7, and 12 dpi, respectively. The overlap between the two techniques shows 82% of the DEPs also being identified within the transcriptomics pipeline for this contrast at 7 dpi. No overlap was seen at 4 dpi and 12 dpi, perhaps indicative of differences in data acquired or biological timing differences in the transcriptome and proteome upon viral infection. (B) Comparative bar plots for Log2FoldChange values in proteomics (purple) and transcriptomics (green) data when contrasting wildtype GFLV-GHu and mutant GFLV-GHu 1EK802GPol. The 23 genes were pulled from the overlapping gene IDs seen in the central Venn diagram from A. All expression levels are in the same absolute value between LC–MS/MS- and 3′RNA-Seq-acquired data.
The range of differently abundant genes/proteins between the two techniques was large with 30 proteins compared to over 2500 transcripts during a single contrast but showed some overlap. Contrasting wildtype GFLV-GHu vs mutant GFLV-GHu 1EK802G revealed that most proteins classified as differentially abundant (82%, 23 of 28) were found within the transcriptomics data at 7 dpi (Figure 9A). Five proteins did not appear as DEGs within this contrast at 7 dpi, further stressing the need for a multi-omics approach where gene/protein candidates of interest could have been missed by one technique alone. Little overlap was found at the early and late time points, which was not surprising given the small number of DEPs in these contrasts. Additionally, the major GOs that overlapped between the two methods included biosynthetic processes, catalytic activity, GTPase activity, inositol-3 phosphate synthase, isoprenoid biosynthesis, nucleotide binding, and RNA binding. These genes followed identical trends for differential abundance with, for instance, a downregulation of INOSITOL-3 PHOSPHATE SYNTHASE in the transcriptome and proteome analyses for wildtype GFLV-GHu at 7 dpi (Figure 9B). Other contrasts with respective time points displayed similar trends (Table S4).
Identification of Networks of Host Pathways for GFLV Symptom Development
Previous virus–plant studies provided a springboard for profiling the host transcriptome and proteome and biological pathway in GFLV–N. benthamiana interactions.10−13,23−25 Then, information from closely related plant viruses with similar phenotypes was considered to provide the following list of cellular pathways to probe: viral silencing,52 viral receptors,5,6,53 kinase cascades,54 plant hormone synthesis,10,55 integrated stress response,56,57 ribosomal complexes,58,59 and photosynthesis-related genes.12,60 Searching relative terms to these seven pathways in the annotated N. benthamiana genome generated three pathways of interest for wildtype GFLV-GHu: (1) host defense, (2) ribosome filter hypothesis, and (3) post-transcriptional gene silencing (Figure 10).
Figure 10.
Hypothesized major networks for symptom development in N. benthamiana upon infection by GFLV strain GHu deduced from profiling proteome and transcriptome changes. Highly represented at 4 dpi and during peak symptom expression at 7 dpi were genes related to basal host defense responses including chitinases, ROS modulators, and a senescence-related protein, which were all upregulated. Other basal defense systems suppressed the expression of GLUTAMINE SYNTHASE PATHOGEN RELATED-1. Additionally, ribosomal subunits were upregulated at 7 dpi, related to changes of major translational networks for protein production. An increased abundance of certain ribosomal subunits could modify translational products and reprogram the cellular structure to be more conducive for the expression of the viral genome. Silencing mechanisms of miRNA- and RNAi-mediated defense involved the upregulation of SUPPRESSOR OF GENE SILENCING and DOUBLE-STRANDED RNA BINDING PROTEIN transcripts at 7 dpi. Other genes of the RNA silencing pathways such as DICER-LIKE 4 returned results with conflicting expression levels or very low counts (ARGONAUTE) and, therefore, may contribute in minor but non-critical capacities to the vein clearing phenotype. ROS = reactive oxygen species, GS-PR1 = glutamine synthase pathogenesis related-1, eIF = eukaryotic translation initiation factor, SGS = suppressor of gene silencing, DRB = double-stranded RNA binding protein, DCL = dicer-like protein, and AGO = argonaute protein.
The normalized RNA-Seq counts of the genes involved in these three cellular pathways were calculated (Table S5 and File S1). Several genes had increased abundance in plants infected with wildtype GFLV-GHu at 7 dpi, such as SCENESCENCE-RELATED PROTEIN, ROS modulators (CHITINASE 8/9, ROS MODULATOR 1), viral silencing components (RNA-DEPENDENT RNA POLYMERASE 2, DICER-LIKE PROTEIN 4, SUPPRESSOR OF GENE SILENCING, and SERRATE EFFECTOR PROTEIN), and ribosomal components (60S RIBOSOMAL PROTEIN L13A-1, L13A-4) (Figure 10). Several other virus-related pathways that were downregulated in plants infected with wildtype GFLV-GHu included GLUTAMINE SYNTHASE and many photosynthetic components (PHOTOSYSTEM REACTION CENTER SUBUNITS PSAK1/2, PHOTOSYSTEM II 10 kDa POLYPEPTIDE, CARBONIC ANHYDRASE, and RUBISCO 1/2). Differences in isoprenoid production pathways were observed for which wildtype-GHu increased the expression of isoprenoid synthesis-related genes, while the other three GFLV strains and mock control had lower levels of these respective genes. It remains unclear how these pathways are triggered for modification in response to GFLV-GHu 1E802KPol.
Additional Transcriptomic Treatment Contrasts Reveal Host Regulatory Genes during Early GFLV Infection
Both GFLV-F13 strains modified the transcriptome at a faster rate than that of wildtype GFLV-GHu (Figure S12D, G, J) The effect of the relative transcript abundance for wildtype GFLV-F13 at 4 dpi against the mock treatment was apparent with 132 genes upregulated and 259 genes downregulated (Figure S12G). Comparing mutant GFLV-F13 1EG802KPol and mock control treatments resulted in 605 upregulated and 481 downregulated genes (Figure S12J). In contrast, when comparing wildtype GFLV-GHu and mock control at 4 dpi, only two genes were upregulated, and three genes were downregulated, while a tremendous difference in gene expression was observed at 7 dpi (Figure S12D,E). Other GFLV treatments varied more drastically to the control than between viral treatments at 4, 7, and 12 dpi (Figure S12A–L). For treatment groups other than wildtype GFLV-GHu, differences were more drastic at 4 and 12 dpi (Figure S12D–L). Since no virus symptoms are observed for the other treatment groups, observing changes in the transcriptome at early and/or late time points is perplexing and warrants follow-up work.
Both proteome and transcriptome techniques showed that nucleotide binding is the most overrepresented GO for wildtype GFLV-GHu at 7 dpi and that several other metabolic functions could be resolved (Figures 5 and 6). Unique to wildtype and mutant GFLV-F13 were the overrepresented gene functions of DNA binding (Figure 5) and perturbations in metabolic processes, nuclease activity, and cytoskeleton organization (Figure S15).
Confirmation of the DEG Niben101Scf01478g00014
We examined the DEG encoding 60S ACIDIC RIBOSOMAL PROTEIN P1, as it (Niben101Scf01478g00014) had a Log2FoldChange of 8.53 and an adjusted P-value of 6.09 × 10–21 between wildtype GFLV-GHu and mutant GFLV-GHu 1EK802GPol at 7 dpi (Table S5). The 3′RNA-Seq results only found this transcript in high abundance in plants infected with wildtype GFLV-GHu at 7 dpi (average normalized counts ∼550) as compared to basal levels in all other treatments and time points (average normalized counts ∼10) (Figure S16). The corresponding protein was not present in the proteomics data likely because its expression was below the detection threshold for LC–MS/MS. The expression of Niben101Scf01478g00014 was confirmed in plants infected with GFLV-GHu by RT-qPCR at 7 dpi with a significantly higher relative expression (2–ΔΔCT) in plants infected with the wildtype strain (p < 0.01) when normalized to housekeeping gene FBOX but not in plants infected by mutant GFLV-GHu 1EK802GPol and mock inoculated control plants (Figure S16). This analysis validated the specific expression level of a host transcript for which the corresponding protein was undetectable through LC–MS/MS.
Discussion
We characterized the proteome and transcriptome of N. benthamiana upon GFLV infection with four distinct virus strains, one strain (GHu) eliciting transient vein clearing symptoms and three others (GHu-1EK802GPol, F13, and F13-1EG802KPol) causing asymptomatic infections. A time course experiment analyzed through 3′RNA-Seq and LC–MS/MS revealed distinct host responses to GFLV infection and strain- and single amino acid-specific perturbations to the host transcript and protein expression. Changes in the transcriptome and proteome of N. benthamiana were prominent at peak vein clearing symptom expression (∼7 dpi, Figure 1C) compared with earlier (4 dpi) or later (12 dpi) time points for wildtype GFLV-GHu (Figures 3 and 4). Additionally, comparative analyses of transcriptome and proteome datasets obtained with wildtype GFLV-GHu and mutant GFLV-GHu 1EK802GPol revealed residue 802 of the RdRP highly influencing host gene and protein expression profiles (Figures 3A–C and 4B). This work yielded a short list of five host protein and gene candidates correlated and unique to symptom expression when contrasted with other GFLV-infected plants (Table S5). In total, 23 gene candidates will be selected for future validation of function in addition to leveraging the proteomics and transcriptomics datasets to examine host pathways influenced by GFLV infection (Table S4).
A rigorous quality control determined that the samples after proteome and transcriptome acquisition were acceptable for the rest of the bioinformatic pipeline. LC–MS/MS resolved over 1500 proteins, while transcriptomics identified over 30,000 transcripts matching to the host genome per sample. Despite a differential resolution and range of these methodologies, both aligned with our hypotheses for wildtype GFLV-GHu related to immune response, metabolism, and host reprogramming genes for symptom development with considerable overlap at peak vein clearing expression (Figures 5–7). The proteome showed only a handful of proteins that were differentially abundant in the contrasts; however, additional information was gained by looking at proteins MNAR, which we suspected to be mainly identifying phenotype-specific functions (Table 2, Figure 6B). The most drastic changes in the transcriptome and proteome were observed at 7 dpi between wildtype GFLV strains GHu and F13 (Figures 3 and 4, Tables 1 and 2). This result was expected given the difference in genetic makeup between the two GFLV strains and their phenotypic differences on N. benthamiana. Furthermore, the wildtype and mutant GFLV-GHu contrast shared many DEGs in comparison with the wildtype GFLV strains GHu and F13 contrast (62.5%, 1692 of 2707 genes) (Table 1, Figure 3B,E). This finding suggested that the lysine (K) to glycine (G) mutation in the RdRP of GFLV-GHu has more impact on the transcriptome and proteome of N. benthamiana for symptom development than the other >10% genetic differences at the nucleotide level between wildtype GFLV strains GHu and F13. Previous and current phenotypic observations remain consistent with this observation.24,25
From the proteome analyses, plant host protein candidates potentially critical for symptom expression were identified. These included proteins involved in nucleotide binding, inositol-3-phosphate synthase activity, isopentenyl diphosphate biosynthesis, and thiamine biosynthesis (Figure 6A). This list was consistent with the results of transcriptomic GSEA, as analysis of the full transcriptome through DEG identification and subsequent over-enriched gene analysis showed nucleic acid binding, ribosomal component, and biosynthesis-related genes characteristic of the vein clearing phenotype (Figure 5). This was also confirmed through WGCNA, a gene grouping analysis across multiple treatments and time points, where these same GOs were overrepresented at 7 dpi for wildtype GFLV-GHu, and distinct from all other treatments and time points (Figure 8). Five gene clusters represented the unique gene expression profiles related to vein clearing symptom expression including hypersensitive response and defense-related genes being dysregulated only at 7 dpi for wildtype GFLV-GHu (Figure 8A). Other similar GOs were overrepresented as with previous gene set enrichment analysis (Figure 8B,C). These combined findings provided confidence that gene functions related to immune response (RNA binding, response to oxidative stress, and gene silencing), translation regulation (structural constituent of the ribosome, RNA binding, and translation elongation factor activity), and metabolism (inositol-3-phosphate synthase activity, pseudouridine synthesis, and thiamine biosynthetic process) are unique to vein clearing symptom expression at 7 dpi, as they were not observed prior to (4 dpi) or after (12 dpi) symptom development. In addition, examining the GOs for MNAR proteins was equally informative to the phenotype.
A network analysis following GSEA for wildtype GFLV-GHu at 7 dpi against all other treatment groups provided the most condensed and comprehensive form of which GO is associated with vein clearing symptom expression (Figure 7). By adding manual group annotations to the network analysis, three key gene categories were most associated with this phenotype: gene regulation, immune response, and metabolism (Figure 7). These major gene functions were found again when temporal analysis through WGCNA isolated gene expression profiles specific to wildtype GFLV-GHu at 7 dpi (Figure 8). Together, these gene categories suggested that a hypersensitive response is induced when GFLV first enters apical tissue (4–7 dpi), and this signal is not sustained later into infection (12 dpi), yet the exact mechanism remains unclear. Nevertheless, the overlap of GSEA and WGCNA in identifying differentially abundant genes was confirmatory to other network analyses.
At peak observation of the vein clearing phenotype at 7 dpi, the list of differentially abundant host genes obtained by contrasting the proteomic and transcriptomic datasets between the wildtype GFLV-GHu and its single-amino acid mutant was consistent (Figure 9A). These two techniques complemented each other to provide a list of 23 protein and gene candidates associated with virus-induced symptom development, although the proteome complexity of plant tissues and stochasticity of LC–MS/MS acquisition may have prevented lower-abundance proteins from being identified (Table S6). These 23 candidates once again belonged to 3 main gene host functions: translational regulation, nucleotide binding, and metabolism. All 23 candidates showed the same absolute expression values with correlated fold change values (Figure 9B and Table S8). Even in the instance of a gene product not being present in the proteomics dataset, confidence in our analysis was obtained by performing RT-qPCR on the top gene candidate from 3′RNA-Seq, a 60S ACIDIC RIBOSOMAL PROTEIN P1 (Niben101Scf01478g00014). A large abundance of this transcript was documented in plant samples infected only by wildtype GFLV-GHu at 7 dpi with basal levels detected in any other sample infected by one of the other three GFLV strains or the control (Figure S16). We were surprised to observe only cases of overlap where the absolute value of expression (positive change in abundance or negative) was the same across both techniques with no discrepancies. This may only be reflective of thresholds we placed to obtain the 23 genes of overlap between GFLV GHu strains at 7 dpi, but we interpreted that active mRNA transcripts captured through 3′RNA-Seq are complementary to shotgun proteomics approaches such as LC–MS/MS. The complementary nature of these two techniques we found to be as, if not more satisfactory, as RT-qPCR validation of candidate genes.
The overlap of proteomics and transcriptomics revealed tRNA ligase activity to be upregulated, suggesting a fine tuning of translational regulation and amino acid availability for transcript selection.61 Plant tRNA ligases are non-homologous to other RNA ligases and able to perform three different catalytic functions; polynucleotide kinase activity, cyclic phosphodiesterase activity, and adenylyltransferase/ligase activity.62 Lysine, proline, and threonine tRNA ligases were upregulated in both the proteomics and transcriptomics data (Table S7). Changing the expression pattern of these enzymes was correlated with the disruption of many genes in GFLV–N. benthamiana symptomatic interactions, as expected from such global regulators of host gene expression.
Secondary metabolites play major roles in plant defense against pathogens and abiotic stress.63,64 Regulating enzymes that control secondary metabolite pathways helps maintain plant growth patterns in response to biotic and abiotic factors. Not only did two wildtype GFLV strains show drastically different host responses with GOs related to biosynthetic pathways, but mutant GFLV strain GHu-1EK802GPol showed a significant dysregulation of isoprenoid related enzymes. In addition, gene enrichment analysis between two treatments and network analysis showed that wildtype GFLV-GHu increased the expression of genes and proteins involved in secondary metabolite production. In fact, isoprenoids and similar biosynthetic components play major roles in viral defense in all eukaryotic organisms.65,66 It is possible that this metabolite pathway identified in N. benthamiana is unique to wildtype GFLV-GHu, as a direct metabolite profile was documented in infected grapevines.67 Genes related to oxidative burst were also differentially expressed in symptomatic plants, suggesting that host immune response could be involved in the development of vein clearing symptoms caused by wildtype GFLV-GHu. Future work focusing on collecting quantitative data to measure oxidative stress metabolites in infected plants (i.e., peroxide, O3–) would be more informative to this end.
DEGs identified in this study showed parallels and juxtaposition to other virus–host plant interaction studies. MYMV, an ssDNA begomovirus, causes differential expression of proteasome subunits, carbonic anhydrase, glutathione S transferase, and photosynthetic components;10 many of these appeared in contrasts of symptomatic and asymptomatic GFLV strains. In the case of PVY, an ssRNA potyvirus, symptomology and response from infection were shown to be heavily regulated and initiated by salicylic acid.11 Photosynthetic components in addition to ribosomal proteins were also majorly dysregulated in PVY.11 Our study on GFLV–N. benthamiana interactions revealed little to no involvement of plant hormone pathways but shared similarity in the downregulation of photosynthesis genes. For SCMV, an ssRNA potyvirus, infection led to major dysregulation in photosynthetic pathway components12 but to a broader and deeper degree than that observed in our system. A discrepancy between SCMV and GFLV with regard to the magnitude of dysregulated photosynthesis relates perhaps to distinct symptom types resulting from infection with a transient vein clearing for GFLV and a strong and stable mosaic for SCMV.
Many reports associate recovery of virus symptoms with RNA silencing components, often when virus–host interactions induce post-transcriptional gene silencing (PTGS) and shuttle viral small interfering RNAs through vasculature tissue ahead of virus movement.68 With the recent report of two viral silencing suppressors identified in the GFLV genome, proteins 1A and 1BHel, and the previously noted interaction of protein 1EPol with 1BHel, we sought out genes related to PTGS.23,69 We documented dysregulation of RNA silencing pathways regardless of the GFLV strains, suggesting little, if any, involvement of RNA silencing components in GFLV-GHu vein clearing symptom induction in N. benthamiana (Table S9 and File S1). Slightly elevated expression levels were observed for DICER-LIKE PROTEIN 4 and SUPPRESSOR OF GENE SILENCING; to the same extent, the virus titer was detected in samples, suggesting that RNA silencing components are active during infection but diminish after symptoms subside. However, the counts on many of these genes were relatively low for robust analyses and definite conclusions.
Based on our results and consistent with the literature,23,24 the display of vein clearing symptoms was not correlated with the GFLV titer. Both the abundance of GFLV RNA2 in the transcriptomics data and three viral proteins (1EPol, 2BMP, and 2CCP) in the proteomics data support a statistically similar GFLV titer in infected N. benthamiana plants, regardless of the virus strain (Figure S11). Therefore, we suspect that the overall burden of viral replication and abundance has little to no impact on symptom development in this instance. Overall lower reads were acquired for RNA1 compared with those for RNA2 for all GFLV strains in the transcriptomics data, and no reads were identified for GFLV RNA1 for the two strains F13, perhaps due to a limitation of 3′RNA-Seq or a specific feature in this strain’s replication. The reason behind a lower RNA1 titer relative to RNA2 remains unresolved but is consistent with the proteomics work for which RNA2-encoded proteins were more abundant than RNA1-encoded proteins.70 The disparate ratio of GFLV RNA molecules and departure of the viral titer related to symptomology were previously documented for Vitis vinifera cv. “Chardonnay” in a vineyard setting.71 This RNA ratio was also recently documented for tobacco ringspot virus and tomato ringspot virus (ToRSV), two nepoviruses related to GFLV, in pawpaw, another fruit crop.72
Some parallels can be made between our findings on GFLV and other nepoviruses such as ToRSV73,74 and artichoke Italian latent virus (AILV). A previous study used microarray analysis to identify DEGs in N. benthamiana infected with ToRSV, along with two other fruit crop viruses.75 This study examined plant host gene expression levels after plant recovery from symptoms. Many GOs similar to those identified in this study were reported including induced or upregulated genes related to stress/pathogenesis, protein synthesis components, gene regulators, proteolysis, and developmental regulators. Additionally, repressed genes related to photosynthesis, protein synthesis, and unknown components were documented for ToRSV at a greater magnitude than for the other two viruses.75 These same gene functions were found dysregulated in our study using 3′-RNA-Seq and LC–MS/MS, especially with an improved annotation of the N. benthamiana genome and a more extensive read volume for genes. Another study revealed recovery of N. benthamiana from ToRSV in association with reduced RNA2 translation and decreased AGO1 levels.76 Similar recovery aspects with decreased RNA2 levels were reported for AILV.77 No major correlations with AGO proteins or GFLV RNA2 levels were found in our study when contrasting the proteome and transcriptome of symptomatic and asymptomatic GFLV infection (Figures 10 and S11). Differences in virus–host interactions among nepoviruses add to the diversity of this plant virus genus78 and brings additional merit to the uniqueness of GFLV and its protein 1EPol for symptom expression in N. benthamiana.
Our findings related to metabolic pathways in addition to immune response and translational regulation suggest an entanglement of the metabolome, transcriptome, and proteome contributing to the observed phenotype in N. benthamiana. By initial perception of GFLV-GHu, the host response likely triggers an oxidative burst and an overall disruption of metabolic pathways as an incomplete hypersensitive reaction to infection. The way other GFLV strains avoid this pattern is unclear because the expression of very few genes was altered by the reverse mutant GFLV F13-1EG802KPol treatment compared with the control when considered against the symptomatic wildtype GFLV-GHu carrying the same amino acid residue in position 802 of protein 1EPol. Additional approaches will need to be considered to delve into not only plant–virus interactions but also virus–virus interactions and host–host interactions upon GFLV infection to address additional questions for mechanistic understandings of GFLV symptom development in planta. Perhaps pull-downs of tagged proteins beyond previous studies with protein 1EPol could be implemented to more precisely determine cell components involved in vein clearing symptoms. To focus on the tissue type, a more optimized protein elution or a longer run time on LC–MS/MS could bring further resolution to virus–host protein complexes that give rise to symptom development.
Upon systemic infection, it appears that wildtype GFLV-GHu has a delayed reaction in establishing full infection and largely avoids detection by the host, indicated by low changes in the host transcriptome and proteome at 4 dpi (Figure S12D,G,J) and lower virus presence at 4 dpi (Figure S11). These data suggested that N. benthamiana recognizes the invasion of wildtype GFLV-GHu later than other GFLV strains and induces a hypersensitive response as an attempt to clear out or restrict the virus. The differential expression of pathogen- and immune-response-related genes at 4 dpi is unique in this instance. Host genes activated during the initial stages of viral replication might initiate signaling cascades for a robust yet short-lived oxidative burst. Some aspect of protein interactions through the GFLV 1EPol C-terminal end could be responsible for these major differences. Furthermore, unique gene networks of ribosomal filtering and host modification were induced by the virus. Both translation efficiency induced by viral replication and host defense were altered, as shown by translation initiation factors, translation elongation factors, tRNA ligases, and ribosomal proteins being in differential abundance analyses. Finally, vein clearing symptoms were shown to be correlated with metabolism and photosynthetic components of N. benthamiana. Biosynthetic and photosynthetic pathway-related genes were consistently dysregulated in plants infected with wildtype GFLV-GHu and to a much higher degree than the other GFLV strains. The transcriptome and proteome analyses were mostly consistent with this network of pathogen detection and overcoming disease symptomology. A subcellular resolution of GFLV protein 1EPolin planta or future targeted co-immunoprecipitation studies could further explore these mechanisms, particularly those triggering the cascade of events by tracking specific host interactants of protein 1EPol.
Conclusions
Our study provided insights into host proteome and transcriptome changes and distinct GOs and pathways related to GFLV infection with an emphasis on vein clearing symptom development in N. benthamiana. Several differentially abundant gene candidates were identified in the proteome and transcriptome of N. benthamiana via bioinformatic and computational analyses for symptom development following GFLV-GHu infection. Contrasting two different omics techniques validated changes in the transcriptome and proteome with 3′RNA-Seq providing additional candidates that were in lower abundance compared with LC–MS/MS. In the arms race between GFLV and its plant host, many layers of transcriptional and translational regulation were hypothesized and subsequently identified within each dataset generated. These two techniques generated consistent results that were valuable in examining GFLV–N. benthamiana interactions. Ultimately, our findings suggested gene expression patterns unique to the GFLV-GHu symptom determinant residue 802 of protein 1EPol. A functional analysis of the DEG candidates may yield exciting new information on the molecular mechanisms of symptom development during plant virus infection.
Glossary
Abbreviations
- 1EPol
GFLV-RNA1-encoded protein “E”, also referred to as viral polymerase or RNA-dependent RNA polymerase
- 3′RNA-Seq
three prime ribonucleic acid sequencing
- AGO
argonaute protein
- AILV
artichoke Italian latent virus
- ANOVA
analysis of variance
- AP-MS
affinity purification-mass spectrometry
- CV
coefficient of variation
- DAS-ELISA
double-antibody sandwich enzyme-linked immunosorbent assay
- DPI
days post inoculation
- GFF
general feature format
- GFLV
grapevine fanleaf virus
- GO
gene ontology
- GSEA
gene set enrichment analysis
- LC–MS/MS
liquid chromatography tandem mass spectrometry
- MAR
missing at random
- ME
module eigengene
- MMTS
methyl methanethiosulfonate
- MYM(I)V
mungbean yellow mosaic (India) virus
- NMAR
not missing at random
- PCA
principal component analysis
- PTGS
post-transcriptional gene silencing
- PVY
potato virus Y
- RdRP
RNA-dependent RNA polymerase
- ROS
reactive oxygen species
- RT-qPCR
real time-quantitative polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SCMV
soybean chlorotic mosaic virus
- TCA
trichloroacetic acid
- TCEP
tris(2-carboxyethyl) phosphine
- ToRSV
tomato ringspot virus
- TRSV
tobacco ringspot virus
- WGCNA
weighted gene co-expression network analysis
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.3c00069.
Sequence of primers, summary of proteomics mapping, statistics, overlap, top five proteins and genes by fold change, gene names, DEGs, overlapping genes, gene count matrix, schematic representation, summary analysis of full proteome data, distribution of protein counts, missing value patterns, summary statistics, Euclidian heatmaps, dispersion estimates, hierarchical clustering, PCA, boxplots, additional transcriptomics data, volcano plots, eigengene modules, functional categorization, and RNA expression levels (PDF)
Boxplot figures of targeted pathway analysis in transcriptomics data using normalized reads of transcripts aligned to the Niben v1.0.1 assembly (ZIP)
Relevant data, code, and files associated with protein and transcript analysis (ZIP)
Author Contributions
B.G.R., S.D., Y.Y., T.T., M.H., and M.F. contributed to experimental design. B.G.R. wrote the initial draft that was edited by S.D., Y.Y., T.T., M.H., and M.F. Formal analysis of transcriptomics was performed by B.G.R., and proteomics analysis was performed by B.G.R. with assistance from S.D., Y.Y., T.T., and M.H. Data curation was performed by B.G.R. and Y.Y. Visualization was performed by B.G.R. Project administration and supervision were executed by T.T., M.H., and M.F. Funding acquisition was performed by M.H. and M.F.
B.G.R. was supported by a Cornell University Graduate School Fellowship and the National Science Foundation Digital Plant Science Research Traineeship award no. 1922551. Transcriptomics analysis was partially funded by Cornell AgriTech Venture Funds. MS and protein analysis were partially funded by USDA grants 8062-22410-007-000-D and 8062-21000-047-000-D. This work was partially supported by the California Department of Food and Agriculture Pierce’s Disease & Glassy-Winged Sharpshooter Board Research & Outreach Program (awards 21-0269-000-SA and 22-0550-000-SA).
The authors declare no competing financial interest.
Supplementary Material
References
- Xu K.; D Nagy P. Dissecting Virus-Plant Interactions through Proteomics Approaches. Curr. Proteomics 2010, 7, 316. 10.2174/157016410793611792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carli M.; Benvenuto E.; Donini M. Recent Insights into Plant–Virus Interactions through Proteomic Analysis. J. Proteome Res. 2012, 11, 4765–4780. 10.1021/pr300494e. [DOI] [PubMed] [Google Scholar]
- Osterbaan L. J.; Fuchs M. Dynamic Interactions between Plant Viruses and Their Hosts for Symptom Development. J. Plant Pathol. 2019, 101, 885–895. 10.1007/s42161-019-00323-5. [DOI] [Google Scholar]
- Zanardo L. G.; de Souza G. B.; Alves M. S. Transcriptomics of Plant–Virus Interactions: A Review. Theor. Exp. Plant Physiol. 2019, 31, 103–125. 10.1007/s40626-019-00143-z. [DOI] [Google Scholar]
- Calil I. P.; Fontes E. P. B. Plant Immunity against Viruses: Antiviral Immune Receptors in Focus. Ann. Bot. 2017, 119, 711–723. 10.1093/aob/mcw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia B. C.; Calil I. P.; Machado J. P. B.; Santos A. A.; Fontes E. P. B. Immune Receptors and Co-Receptors in Antiviral Innate Immunity in Plants. Front. Microbiol. 2017, 7, 2139. 10.3389/fmicb.2016.02139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivalkar-Mehla S.; Vakharia J.; Mehla R.; Abreha M.; Kanwar J. R.; Tikoo A.; Chauhan A. Viral RNA Silencing Suppressors (RSS): Novel Strategy of Viruses to Ablate the Host RNA Interference (RNAi) Defense System. Virus Res. 2011, 155, 1–9. 10.1016/j.virusres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Zhang X.; Ma Q.; Ma F.; Zhou H. Transcriptomics and Proteomics in the Study of H1N1 2009. Genomics, Proteomics Bioinf. 2010, 8, 139–144. 10.1016/S1672-0229(10)60016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A. D.; Williamson M. K.; Lewis S.; Shoemark D.; Carroll M. W.; Heesom K. J.; Zambon M.; Ellis J.; Lewis P. A.; Hiscox J. A.; Matthews D. A. Characterisation of the Transcriptome and Proteome of SARS-CoV-2 Reveals a Cell Passage Induced in-Frame Deletion of the Furin-like Cleavage Site from the Spike Glycoprotein. Genome Med. 2020, 12, 68. 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan Kumar B. K.; Kanakala S.; Malathi V. G.; Gopal P.; Usha R. Transcriptomic and Proteomic Analysis of Yellow Mosaic Diseased Soybean. J. Plant Biochem. Biotechnol. 2017, 26, 224–234. 10.1007/s13562-016-0385-3. [DOI] [Google Scholar]
- Stare T.; Stare K.; Weckwerth W.; Wienkoop S.; Gruden K. Comparison between Proteome and Transcriptome Response in Potato (Solanum Tuberosum L.) Leaves Following Potato Virus Y (PVY) Infection. Proteomes 2017, 5, 14. 10.3390/proteomes5030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar S.; Yao W.; Yu K.; Qin L.; Ruan M.; Powell C. A.; Chen B.; Zhang M. Photosynthetic Characterization and Expression Profiles of Sugarcane Infected by Sugarcane Mosaic Virus (SCMV). Photosynth. Res. 2021, 150, 279–294. 10.1007/s11120-019-00706-w. [DOI] [PubMed] [Google Scholar]
- Lyu S.; Gao L.; Zhang R.; Zhang C.; Hou X. Correlation Analysis of Expression Profile and Quantitative ITRAQ-LC-MS/MS Proteomics Reveals Resistance Mechanism Against TuMV in Chinese Cabbage (Brassica Rapa Ssp. Pekinensis). Front. Genet. 2020, 11, 963. 10.3389/fgene.2020.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez J. D.; Cilia M.; Weisbrod C. R.; Ju H.-J.; Eng J. K.; Gray S. M.; Bruce J. E. Cross-Linking Measurements of the Potato Leafroll Virus Reveal Protein Interaction Topologies Required for Virion Stability, Aphid Transmission, and Virus–Plant Interactions. J. Proteome Res. 2012, 11, 2968–2981. 10.1021/pr300041t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y.; Seldin M.; Lusis A. Multi-Omics Approaches to Disease. Genome Biol. 2017, 18, 83. 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M. M.; Cilia M. A Molecular Tug-of-War: Global Plant Proteome Changes during Viral Infection. Curr. Plant Biol. 2016, 5, 13–24. 10.1016/j.cpb.2015.10.003. [DOI] [Google Scholar]
- Tang Z.; Fan W.; Li Q.; Wang D.; Wen M.; Wang J.; Li X.; Zhou Y. MVIP: Multi-Omics Portal of Viral Infection. Nucleic Acids Res. 2022, 50, D817–D827. 10.1093/nar/gkab958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andret-Link P.; Laporte C.; Valat L.; Ritzenthaler C.; Demangeat G.; Vigne E.; Laval V.; Pfeiffer P.; Stussi-Garaud C.; Fuchs M. Grapevine Fanleaf Virus: Still a Major Threat to the Grapevine Industry. J. Plant Pathol. 2004, 86, 183–195. [Google Scholar]
- Maliogka V. I.; Martelli G. P.; Fuchs M.; Katis N. I.. Chapter Six - Control of Viruses Infecting Grapevine. In Advances in Virus Research; Loebenstein G., Katis N. I., Eds.; Control of Plant Virus Diseases; Academic Press, 2015; Vol. 91, pp 175–227. [DOI] [PubMed] [Google Scholar]
- Fuchs M.; Hily J.-M.; Petrzik K.; Sanfaçon H.; Thompson J. R.; van der Vlugt R.; Wetzel T. ICTV Virus Taxonomy Profile: Secoviridae 2022. J. Gen. Virol. 2022, 103, 001807. 10.1099/jgv.0.001807. [DOI] [PubMed] [Google Scholar]
- Morris-Krsinich B. A. M.; Forster R. L. S.; Mossop D. W. The Synthesis and Processing of the Nepovirus Grapevine Fanleaf Virus Proteins in Rabbit Reticulocyte Lysate. Virology 1983, 130, 523–526. 10.1016/0042-6822(83)90105-8. [DOI] [PubMed] [Google Scholar]
- Roy B. G.; Fuchs M. Herbaceous Plant Hosts as Supermodels for Grapevine Viruses: A Historical Perspective. J. Plant Pathol. 2022, 1–30. 10.1007/s42161-022-01267-z. [DOI] [Google Scholar]
- Osterbaan L. J.; Hoyle V.; Curtis M.; DeBlasio S.; Rivera K. D.; Heck M.; Fuchs M.. Identification of Protein Interactions of Grapevine Fanleaf Virus RNA-Dependent RNA Polymerase during Infection of Nicotiana Benthamiana by Affinity Purification and Tandem Mass Spectrometry. J. Gen. Virol. 2021, 102( (5), ). 10.1099/jgv.0.001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterbaan L. J.; Choi J.; Kenney J.; Flasco M.; Vigne E.; Schmitt-Keichinger C.; Rebelo A. R.; Heck M.; Fuchs M. The Identity of a Single Residue of the RNA-Dependent RNA Polymerase of Grapevine Fanleaf Virus Modulates Vein Clearing in Nicotiana Benthamiana. Mol. Plant-Microbe Interact. 2019, 32, 790–801. 10.1094/MPMI-12-18-0337-R. [DOI] [PubMed] [Google Scholar]
- Vigne E.; Gottula J. W.; Schmitt-Keichinger C.; Komar V.; Ackerer L.; Belval L.; Rakotomalala L.; Lemaire O.; Ritzenthaler C.; Fuchs M. A strain-specific segment of the RNA-dependent RNA polymerase of grapevine fanleaf virus determines symptoms in Nicotiana species. J. Gen. Virol. 2013, 94, 2803–2813. 10.1099/vir.0.057646-0. [DOI] [PubMed] [Google Scholar]
- Bombarely A.; Rosli H. G.; Vrebalov J.; Moffett P.; Mueller L. A.; Martin G. B. A Draft Genome Sequence of Nicotiana Benthamiana to Enhance Molecular Plant-Microbe Biology Research. Mol. Plant-Microbe Interact. 2012, 25, 1523–1530. 10.1094/MPMI-06-12-0148-TA. [DOI] [PubMed] [Google Scholar]
- Bolger A. M.; Lohse M.; Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S.FastQC: A Quality Control Tool for High Throughput Sequence Data, 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Kim D.; Paggi J. M.; Park C.; Bennett C.; Salzberg S. L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.; Smyth G. K.; Shi W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liao Y.; Smyth G. K.; Shi W. The Subread Aligner: Fast, Accurate and Scalable Read Mapping by Seed-and-Vote. Nucleic Acids Res. 2013, 41, e108 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilia M.; Fish T.; Yang X.; McLaughlin M.; Thannhauser T. W.; Gray S. A Comparison of Protein Extraction Methods Suitable for Gel-Based Proteomic Studies of Aphid Proteins. J. Biomol. Tech. 2009, 20, 201–215. [PMC free article] [PubMed] [Google Scholar]
- Ramsey J. S.; Chin E. L.; Chavez J. D.; Saha S.; Mischuk D.; Mahoney J.; Mohr J.; Robison F. M.; Mitrovic E.; Xu Y.; Strickler S. R.; Fernandez N.; Zhong X.; Polek M.; Godfrey K. E.; Giovannoni J. J.; Mueller L. A.; Slupsky C. M.; Bruce J. E.; Heck M. Longitudinal Transcriptomic, Proteomic, and Metabolomic Analysis of Citrus Limon Response to Graft Inoculation by Candidatus Liberibacter Asiaticus. J. Proteome Res. 2020, 19, 2247–2263. 10.1021/acs.jproteome.9b00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adusumilli R.; Mallick P.. Data Conversion with ProteoWizard MsConvert. In Proteomics; Comai L., Katz J. E., Mallick P., Eds.; Methods in Molecular Biology; Springer New York: New York, NY, 2017; Vol. 1550, pp 339–368. [DOI] [PubMed] [Google Scholar]
- Perkins D. N.; Pappin D. J. C.; Creasy D. M.; Cottrell J. S. Probability-Based Protein Identification by Searching Sequence Databases Using Mass Spectrometry Data. Electrophoresis 1999, 20, 3551–3567. . [DOI] [PubMed] [Google Scholar]
- Searle B. C. Scaffold: A Bioinformatic Tool for Validating MS/MS-Based Proteomic Studies. Proteomics 2010, 10, 1265–1269. 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Smits A. H.; van Tilburg G. B.; Ovaa H.; Huber W.; Vermeulen M. Proteome-Wide Identification of Ubiquitin Interactions Using UbIA-MS. Nat. Protoc. 2018, 13, 530–550. 10.1038/nprot.2017.147. [DOI] [PubMed] [Google Scholar]
- Smits A.; Huber W.. DEP: Differential Enrichment Analysis of Proteomics Data, 2022.
- Bonferroni C. E.Teoria statistica delle classi e calcolo delle probabilita; Seeber, 1936. [Google Scholar]
- Raudvere U.; Kolberg L.; Kuzmin I.; Arak T.; Adler P.; Peterson H.; Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolberg L.; Raudvere U.; Kuzmin I.; Vilo J.; Peterson H. Gprofiler2 -- an R Package for Gene List Functional Enrichment Analysis and Namespace Conversion Toolset g:Profiler. F1000Research 2020, 9, 709. 10.12688/f1000research.24956.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.; Tamayo P.; Mootha V. K.; Mukherjee S.; Ebert B. L.; Gillette M. A.; Paulovich A.; Pomeroy S. L.; Golub T. R.; Lander E. S.; Mesirov J. P. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V. K.; Lindgren C. M.; Eriksson K.-F.; Subramanian A.; Sihag S.; Lehar J.; Puigserver P.; Carlsson E.; Ridderstråle M.; Laurila E.; Houstis N.; Daly M. J.; Patterson N.; Mesirov J. P.; Golub T. R.; Tamayo P.; Spiegelman B.; Lander E. S.; Hirschhorn J. N.; Altshuler D.; Groop L. C. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P.; Horvath S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinf. 2008, 9, 559. 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B.; Horvath S.. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4 ( (1), ), 17. 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
- Liu D.; Shi L.; Han C.; Yu J.; Li D.; Zhang Y. Validation of Reference Genes for Gene Expression Studies in Virus-Infected Nicotiana Benthamiana Using Quantitative Real-Time PCR. PLoS One 2012, 7, e46451 10.1371/journal.pone.0046451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Välikangas T.; Suomi T.; Elo L. L. A Systematic Evaluation of Normalization Methods in Quantitative Label-Free Proteomics. Briefings Bioinf. 2016, 19, 1–11. 10.1093/bib/bbw095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L.; Bi Y.; Hu C.; Qu J.; Shen S.; Wang X.; Tian Y. A Comparative Study of Evaluating Missing Value Imputation Methods in Label-Free Proteomics. Sci. Rep. 2021, 11, 1760. 10.1038/s41598-021-81279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuitty L. L.; Clark J. A. Clusters from Iterative, Intercolumnar Correlational Analysis. Educ. Psychol. Meas. 1968, 28, 211–238. 10.1177/001316446802800201. [DOI] [Google Scholar]
- Wang M.-B.; Masuta C.; Smith N. A.; Shimura H. RNA Silencing and Plant Viral Diseases. Mol. Plant-Microbe Interact. 2012, 25, 1275–1285. 10.1094/MPMI-04-12-0093-CR. [DOI] [PubMed] [Google Scholar]
- Teixeira R. M.; Ferreira M. A.; Raimundo G. A. S.; Loriato V. A. P.; Reis P. A. B.; Fontes E. P. B. Virus Perception at the Cell Surface: Revisiting the Roles of Receptor-like Kinases as Viral Pattern Recognition Receptors. Mol. Plant Pathol. 2019, 20, 1196–1202. 10.1111/mpp.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.; Wang C.; Wang H.; Li L.; Wang C. The Function of MAPK Cascades in Response to Various Stresses in Horticultural Plants. Front. Plant Sci. 2020, 11, 952. 10.3389/fpls.2020.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem M.; Lin N. Roles of Plant Hormones in the Regulation of Host–Virus Interactions. Mol. Plant Pathol. 2014, 16, 529–540. 10.1111/mpp.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof M.; Wang L.; Cogan J. Z.; Lawrence R. E.; Boone M.; Wuerth J. D.; Frost A.; Walter P. Viral Evasion of the Integrated Stress Response through Antagonism of EIF2-P Binding to EIF2B. Nat. Commun. 2021, 12, 7103. 10.1038/s41467-021-26164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Zhang Z.; Li Y.; Li Y. The Regulation of Integrated Stress Response Signaling Pathway on Viral Infection and Viral Antagonism. Front. Microbiol. 2022, 12, 814635. 10.3389/fmicb.2021.814635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.-S.; Himmelbach A.; Browning K. S.; Hohn T.; Ryabova L. A. A Plant Viral “Reinitiation” Factor Interacts with the Host Translational Machinery. Cell 2001, 106, 723–733. 10.1016/S0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Hashimoto M.; Neriya Y.; Yamaji Y.; Namba S. Recessive Resistance to Plant Viruses: Potential Resistance Genes Beyond Translation Initiation Factors. Front. Microbiol. 2016, 7, 1695. 10.3389/fmicb.2016.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y.; Zhang Y.; Wang C.; Lei R.; Wu Y.; Li X.; Zhu S. Cucumber Mosaic Virus Coat Protein Induces the Development of Chlorotic Symptoms through Interacting with the Chloroplast Ferredoxin I Protein. Sci. Rep. 2018, 8, 1205. 10.1038/s41598-018-19525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J.; Walter P. TRNA Ligase Structure Reveals Kinetic Competition between Non-Conventional MRNA Splicing and MRNA Decay. eLife 2019, 8, e44199 10.7554/eLife.44199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M.; Beier H. Plant TRNA Ligases Are Multifunctional Enzymes That Have Diverged in Sequence and Substrate Specificity from RNA Ligases of Other Phylogenetic Origins. Nucleic Acids Res. 2005, 33, 388–399. 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink M. Evolution of Secondary Metabolites from an Ecological and Molecular Phylogenetic Perspective. Phytochemistry 2003, 64, 3–19. 10.1016/S0031-9422(03)00300-5. [DOI] [PubMed] [Google Scholar]
- Akula R.; Ravishankar G. A. Influence of Abiotic Stress Signals on Secondary Metabolites in Plants. Plant Signaling Behav. 2011, 6, 1720–1731. 10.4161/psb.6.11.17613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K. The Biosynthesis of Isoprenoids and the Mechanisms Regulating It in Plants. Biosci., Biotechnol., Biochem. 2011, 75, 1219–1225. 10.1271/bbb.110228. [DOI] [PubMed] [Google Scholar]
- Fraga B. M. Natural sesquiterpenoids. Nat. Prod. Rep. 2005, 22, 465–486. 10.1039/B501837B. [DOI] [PubMed] [Google Scholar]
- Martin I. R.; Vigne E.; Velt A.; Hily J.-M.; Garcia S.; Baltenweck R.; Komar V.; Rustenholz C.; Hugueney P.; Lemaire O.; Schmitt-Keichinger C. Severe Stunting Symptoms upon Nepovirus Infection Are Reminiscent of a Chronic Hypersensitive-like Response in a Perennial Woody Fruit Crop. Viruses 2021, 13, 2138. 10.3390/v13112138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal B.; Sanfaçon H. Symptom Recovery in Virus-Infected Plants: Revisiting the Role of RNA Silencing Mechanisms. Virology 2015, 479–480, 167–179. 10.1016/j.virol.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Choi J.; Pakbaz S.; Yepes L. M.; Cieniewicz E. J.; Schmitt-Keichinger C.; Labarile R.; Minutillo S. A.; Heck M.; Hua J.; Fuchs M. Grapevine Fanleaf Virus RNA1-Encoded Proteins 1A and 1BHel Suppress RNA Silencing. Mol. Plant-Microbe Interact. 2023, 10.1094/MPMI-01-23-0008-R. [DOI] [PubMed] [Google Scholar]
- Pinck L.; Fuchs M.; Pinck M.; Ravelonandro M.; Walter B. A Satellite RNA in Grapevine Fanleaf Virus Strain F13. J. Gen. Virol. 1988, 69, 233–239. 10.1099/0022-1317-69-1-233. [DOI] [PubMed] [Google Scholar]
- Kubina J.; Hily J.-M.; Mustin P.; Komar V.; Garcia S.; Martin I. R.; Poulicard N.; Velt A.; Bonnet V.; Mercier L.; Lemaire O.; Vigne E. Characterization of Grapevine Fanleaf Virus Isolates in ‘Chardonnay’ Vines Exhibiting Severe and Mild Symptoms in Two Vineyards. Viruses 2022, 14, 2303. 10.3390/v14102303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.; Osatuke A. C.; Erich G.; Stevens K.; Hwang M. S.; Al Rwahnih M.; Fuchs M. High-Throughput Sequencing Reveals Tobacco and Tomato Ringspot Viruses in Pawpaw. Plants 2022, 11, 3565. 10.3390/plants11243565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel J.; Walker M.; Sanfaçon H. Salicylic Acid-Dependent Restriction of Tomato Ringspot Virus Spread in Tobacco Is Accompanied by a Hypersensitive Response, Local RNA Silencing, and Moderate Systemic Resistance. Mol. Plant-Microbe Interact. 2011, 24, 706–718. 10.1094/MPMI-09-10-0224. [DOI] [PubMed] [Google Scholar]
- Bitterlin M. W.; Gonsalves D. Serological Grouping of Tomato Ringspot Virus Isolates: Implications for Diagnosis and Cross-Protection. Phytopathology 1988, 78, 278–285. 10.1094/phyto-78-278. [DOI] [Google Scholar]
- Dardick C. Comparative Expression Profiling of Nicotiana Benthamiana Leaves Systemically Infected with Three Fruit Tree Viruses. Mol. Plant-Microbe Interact. 2007, 20, 1004–1017. 10.1094/MPMI-20-8-1004. [DOI] [PubMed] [Google Scholar]
- Ghoshal B.; Sanfaçon H. Temperature-Dependent Symptom Recovery in Nicotiana Benthamiana Plants Infected with Tomato Ringspot Virus Is Associated with Reduced Translation of Viral RNA2 and Requires ARGONAUTE 1. Virology 2014, 456–457, 188–197. 10.1016/j.virol.2014.03.026. [DOI] [PubMed] [Google Scholar]
- Santovito E.; Mascia T.; Siddiqui S. A.; Minutillo S. A.; Valkonen J. P. T.; Gallitelli D. Infection Cycle of Artichoke Italian Latent Virus in Tobacco Plants: Meristem Invasion and Recovery from Disease Symptoms. PLoS One 2014, 9, e99446 10.1371/journal.pone.0099446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H. Re-Examination of Nepovirus Polyprotein Cleavage Sites Highlights the Diverse Specificities and Evolutionary Relationships of Nepovirus 3C-like Proteases. Arch. Virol. 2022, 167, 2529–2543. 10.1007/s00705-022-05564-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.