Abstract
Tools for on-demand protein activation enable impactful gain-of-function studies in biological settings. Thus far, however, proteins have been chemically caged at primarily Lys, Tyr, and Sec, typically through the genetic encoding of unnatural amino acids. Here, we report that the preferential reactivity of diazo compounds with protonated acids can be used to expand this toolbox to solvent-accessible carboxyl groups with an elevated pKa. As a model protein, we employed lysozyme (Lyz), which has an active-site Glu35 with a pKa of 6.2. A diazo compound with a bioorthogonal self-immolative handle esterified Glu35 selectively, inactivating Lyz. The hydrolytic activity of the caged Lyz on bacterial cell walls was restored with two small-molecule triggers. The decaging was more efficient by small molecules than by esterases. This simple chemical strategy was also applied to a hemeprotein and an aspartyl protease, setting the stage for broad applicability.
Keywords: bioorthogonal chemistry, diazo compounds, enzymology, lysozyme, protein caging, HIV-1 protease, cytochrome c
Entry for the Table of Contents
The caging of amino acid side chains can provide exquisite control of protein function. As nature’s catalysts, enzymes are high-priority targets for caging. We report on the first chemical caging of carboxyl groups (Asp and Glu) in enzymic active sites. Further, we demonstrated that caging of a heme propionate in a protein is also feasible. Our caging approach is based solely on the esterification of a carboxyl group with a tuned diazo compound. Decaging is achieved with small-molecule reagents that elicit a Staudinger reaction or 1,3-dipolar cycloaddition.
Introduction
Spatiotemporal control over protein activation has emerged as a powerful tool for studying and manipulating biological processes. The advantage of these tools over strategies for protein inactivation is that they can probe the sufficiency, rather than necessity, of a protein in a signaling pathway.[1] In the last two decades, these protein activation tools have prompted numerous advances, ranging from temporal proteomics and protein-based prodrugs[2] to the discovery of precise intracellular roles of several proteins involved in disease[3,4] and cell motility.[5]
Partly due to the rapid development of bioorthogonal cleavage reactions, the use of chemically caged unnatural amino acids (UAAs) for on-demand protein activation has drawn considerable attention.[6] In this approach, genetic code expansion is used to incorporate caged UAAs in active sites, which can be deprotected bioorthogonally by triggers such as metal ions or small molecules. Compared to photocaged UAAs, chemically caged UAAs are more tunable, versatile, and compatible with applications in living systems that are not penetrant to light.[6] Traceless rescue of the native protein distinguishes UAA-based approaches from other systems, such as split[7] or switchable[8] proteins. Further, the caging of a specific residue enables a more nuanced control over protein activity than does the caging of an entire protein via steric regulator molecules.[9]
Currently, the rescue strategy based on chemically caged UAAs is limited primarily to three types of amino acids and, for this reason, is applicable to just a subset of proteins.[6,10] This shortfall is consequential. For example, the three most common catalytic residues in enzymic active sites are His, Asp, and Glu; and carboxyl groups are the most prevalent functional group for effecting enzymatic catalysis.[11] Yet, only the less common Lys, Tyr, and Sec residues have been exploited for on-demand protein activation with bioorthogonal chemical triggers.[ 6,10] The gap in the coverage of acidic residues has been partially filled by photocaged Asp[12,13] and Glu[14] residues, but examples of genetic encoding of unnatural Asp and Glu are sparse.[15,16] Although a variety of caged carboxyl groups have been incorporated into small-molecule[17,18] or peptide-based[19] prodrugs and turn-on probes,[20] to the best of our knowledge, chemically-caged carboxyl groups have not been incorporated into proteins to control their activity.
We reasoned that such a cage could be installed on carboxyl groups via diazo compounds. Recently, we showed that α-aryl-α-diazoamides with optimized basicity can esterify carboxyl groups in proteins under mild aqueous conditions.[21,22] In addition to their chemoselectively, key advantages of these diazo-based scaffolds over other reagents that modify carboxyl groups are their tunable reactivity, modularity, and facile synthesis.[23]
The first step in productive esterification by diazo compounds is protonation by an acid (Figure S1),[21,24] Accordingly, we suspected that pKa differences between acids could be exploited to control esterification selectivity. Several important protein targets have carboxyl groups with an elevated pKa, including the T-cell surface antigen CD2 (Glu41, pKa = 6.7),[25] anion channel VDAC1 (Glu73, pKa = 7.4),[26] ribonuclease H1 (Asp10, pKa = 6.1),[27] xylanase (Glu172, pKa = 6.7),[28] ketosteroid isomerase (Asp99, pKa > 9),[29] aromatase (Asp309, pKa = 8.2),[30] and many others.[31,32] This strategy has gained support from Gillingham and coworkers, who demonstrated that diazo compounds can selectively label phosphoryl groups in peptides that contain competing carboxyl groups.[33] Further, Woo and coworkers have shown that diazirine probes, which transition through a reactive diazo intermediate, preferentially label the membrane proteins in living cells—a phenomenon that may reflect the elevated pKa of carboxyl groups in lipid bilayers.[34] Also, our group has found that diazo compounds preferentially esterify Glu over Asp residues in the green fluorescent protein[35] for steric reasons or their higher intrinsic pKa.[36] These observations provide evidence that diazo compounds exhibit pKa-driven selectivity. Hence, we sought to expand the toolbox of on-demand activation tools and develop chemically-cleavable diazo-based cages for proteins that contain functional[37] solvent-accessible carboxyl groups. In comparison to UAA-based caging approaches, our strategy is purely chemical and simple in execution.
Results and Discussion
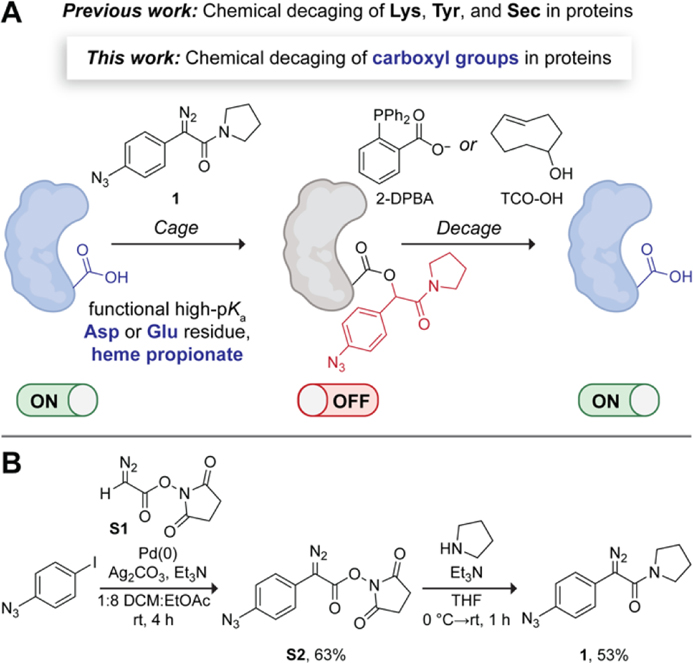
We envisioned a bioorthogonal chemical strategy for decaging esterified carboxyl groups in proteins—enabling ester release through a 1,6-type quinone-methide elimination.[38] Specifically, we decided to modify the aryl ring of the α-aryl-α-diazoamide scaffold with a trigger-responsive handle—an azido group. This versatile functional group has been used in caged UAAs[6,39,40] and can be released with a variety of chemical triggers, including phosphines, strained alkenes, and metal complexes.[6,41] We chose to create the amide moiety within the α-aryl-α-diazoamide with a pyrrolidine, thereby increasing the synthetic yield of these scaffolds.[42] Our rationally designed diazo compound (1) and proposed on-demand protein activation strategy are depicted in Figure 1A. We hypothesized that 1 would inactivate a protein of interest by esterifying a functional solvent-accessible carboxyl group (e.g., Asp or Glu residue, or a heme propionate) with an elevated pKa. Treatment of the caged protein with small-molecule triggers, 2-(diphenylphosphino)benzoic acid (2-DPBA)[43] or trans-cyclooctenol (TCO-OH),[39] was then expected to release the native protein with restored activity through a mechanism that involves the elimination of an imino-quinone methide (IQM).
Figure 1.
(A) Proposed chemical strategy for on-demand activation of proteins. (B) Synthetic route to diazo compound 1. DCM, dichloromethane; THF, tetrahydrofuran.
To effect our strategy, we first synthesized diazo compound 1 by adapting a recently developed two-step route to α-aryl-α-diazoamides[42] (Figure 1B). We began the route by cross-coupling the commercially available 1-azido-4-iodobenzene to N-succimidyl 2-diazoacetate (S1). Although aryl azides can be susceptible to reduction by phosphines,[44] the use of 20 mol% tri(furan-2-yl)phosphine as a ligand for the palladium catalyst still resulted in substantial product formation (63% yield). In the second step, we subjected the diazoester S2 to aminolysis with pyrrolidine, obtaining diazo compound 1 in 53% overall yield.
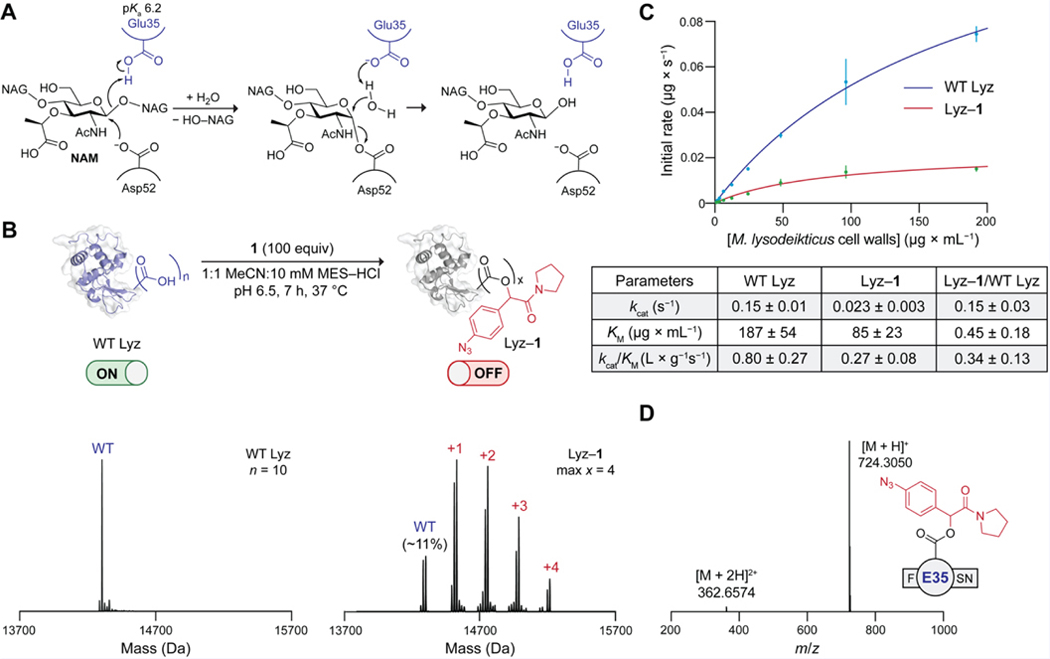
To validate the approach in Figure 1A, we chose Lyz from chicken egg white as a model for an initial proof of concept. Lyz has a catalytic Glu35 with an elevated pKa of 6.2 (Table S1),[32] and both its chemistry and biology are well-established. For example, Lyz was the first enzyme whose three-dimensional structure was determined by X-ray crystallography[45,46] and has an accepted enzymatic reaction mechanism.[48] In animals, Lyz is known to be a critical component of innate immunity against bacteria.[49] The enzyme kills bacteria by cleaving the cell-wall peptidoglycan that protects bacterial cells from osmotic stress.[49] Specifically, Lyz catalyzes the hydrolysis of the β−1,4 glycosidic bond between N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) residues. Catalysis relies on the active-site carboxyl groups of Glu35, which acts as a Brønsted acid, and Asp52, which acts as a nucleophile (Figure 2A).[50] An E35Q substitution completely inactivates Lyz.[51] Taking advantage of these properties of Lyz, we wanted to esterify Glu35 selectively with diazo compound 1 and then re-activate the caged enzyme with small molecules.
Figure 2.
(A) Catalytic mechanism of Lyz. (B) Top: Optimal conditions for the esterification of WT Lyz with diazo compound 1. Bottom: Deconvoluted Q-TOF mass spectra of WT Lyz and Lyz–1. N = total number of carboxyl groups in WT Lyz; max x = maximum number of ester labels. Structures of Lyz are based on PDB entry 1hew.[47] The ion intensity of intact protein spectra was normalized (norm.) so that the ordinate of the highest point is equal to 1.0. (C) Top: Fitted initial rates of the hydrolytic activity of WT Lyz and Lyz–1 on Micrococcus lysodeikticus cell walls. Bottom: Table of kinetic parameters. Values are the mean ± 95% confidence interval (n = 4). (D) Q-TOF mass spectrum of the most abundant esterified peptide in the pepsin digest of Lyz–1. MES, 2-(N-morpholino)ethanesulfonic acid.
We proceeded to screen esterification conditions for selectively caging Lyz at Glu35. Specifically, we sought a condition that would result in approximately one ester label on the protein with minimal remaining unlabeled Lyz. Imposing this constraint would later simplify the identification of the preferentially esterified carboxyl group. The pH of the reaction medium is a key parameter for controlling esterification by diazo compounds.[33] Hence, we incubated wild-type (WT) Lyz with diazo compound 1 (10 equiv per carboxyl group) in buffers of varying pH and analyzed the resultant conjugates (Lyz–1) by Q-TOF mass spectrometry (MS) (Figure S2). After 3 h, the conjugates esterified at pH 5.5 and 6.0 contained mainly 2–4 labels (Figure S2A). In contrast, conjugates esterified at pH 6.5 and 7.0 for 3 h contained primarily 1–2 labels along with some unlabeled Lyz (Figure S2A), which was indicative of higher selectivity. This result is consistent with most carboxyl groups in Lyz being carboxylates at pH 6.5 or above, whereas a large fraction of Glu35 remains protonated (Table S2). Note that incubating Lyz with a lower excess of diazo compound 1 (2.5 equiv per carboxyl group) resulted in insufficient labeling even at pH 5.5 (Figure S2B). When we extended the esterification time to 19 h, unknown byproducts began to appear (Figure S2C). Since esterification at pH 7 proceeded rather slowly, we ultimately opted for caging Lyz at pH 6.5 for 7 h (Figures 2B and S3). By ionization intensities, the optimized Lyz–1 conjugate contained primarily 1–2 labels and only ~11% WT Lyz (Figure 2B).
To determine whether Lyz–1 (Figure 2B) is inactivated through a caged catalytic residue, we assessed the impact of esterification on enzymatic catalysis. Specifically, we compared the activities of WT Lyz and Lyz–1 on authentic Micrococcus lysodeikticus cell walls. In this assay,[52] the bacterial cell walls are labeled with fluorescein to such a degree that fluorescence is quenched. Active Lyz catalyzes the cleavage of the β−1,4 glycosidic bonds in this substrate, yielding an increase in fluorescence that is proportional to enzymatic activity. Using a Michaelis–Menten analysis based on initial rates, we obtained kinetic parameters for catalysis by WT Lyz and Lyz–1 (Figure 2C). Because even conservative substitutions of Glu35 (and, to a lesser extent, Asp52) inactivate WT Lyz,[51] we hypothesized that esterification of one of these residues would inactive Lyz–1 at the catalytic step. This scenario would result in a decrease in the value of kcat that is proportional to the fraction of Lyz that has a caged active site, with little effect on the value of KM. Our results support this hypothesis (Figure 2C). We found that the kcat of Lyz–1 is only 15% that of WT Lyz, which aligns with the kcat value we would expect if only the residual ~11% of unlabeled Lyz in Lyz–1 (Figure 2B) were active. The large decrease in kcat and smaller change in KM also suggests that the esterification of a catalytic residue, rather than a residue important for substrate binding, inactivates the enzyme. Given that the activity of Lyz is remarkably stable to non-active site substitutions,[53] the observed 3-fold drop in the catalytic efficiency of Lyz–1 (Figure 2C) is further indicative of labeling within the active site. Changes in the structure of Lyz are also unlikely to account for the observed decrease in activity because Lyz is a remarkably robust protein (e.g., Tm ~75 °C).[54] Overall, our kinetic data support the conclusion that 1 esterifies a catalytic carboxyl group in Lyz.
Next, we probed whether Glu35, which has a higher pKa than does Asp52 (Table S1), is the major esterified catalytic residue in Lyz–1 (Figure 2B). We digested the caged enzyme with pepsin and analyzed the resultant peptides by intact MS (Figure S4). Gratifyingly, out of all the detected esterified peptides, peptides esterified at the Glu35 residue were the most abundant by peak area, and the most abundant esterified peptide was F(E35)SN (Figure 2D). Although we also detected peptides esterified at Asp52 (Figure S4), they were at least 7-fold less abundant than peptides esterified at Glu35, which suggests that Glu35 is the primary target of esterification. Note that, whereas intact mass spectrometry cannot be used to make absolute comparisons between peptide abundances, peak areas serve as a valid proxy for the overall trend.[55,56] To confirm that our observations are in line with the assumption that pH impacts esterification selectivity, we also inspected the digestion products (Figure S5) of Lyz–1 synthesized at pH 5.5 (Figure S2). In this dataset, Glu35 was no longer the top esterified residue. These results agree with the findings of Gillingham and coworkers,[33] who reported that a pH at or above the pKa of the target acid maximizes selectivity.[57] Altogether, our data suggest that the esterification of Glu35 is predominantly responsible for the inactivation of Lyz–1.
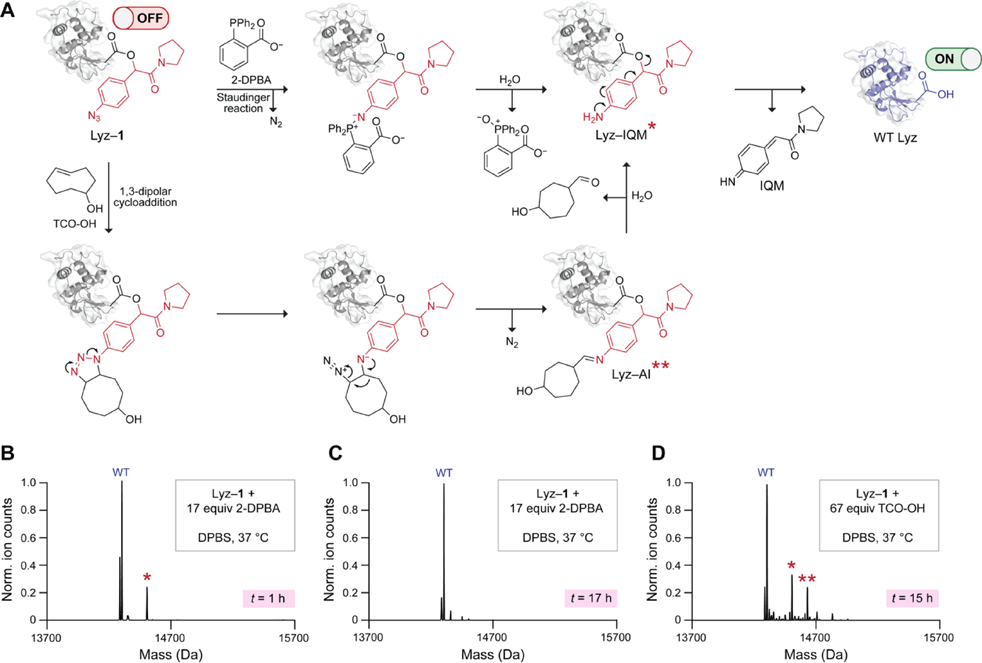
We proceeded to test whether the treatment of Lyz–1 with two commercially available small-molecule triggers, 2-DPBA and TCO-OH, can release the native protein. We expected that 2-DPBA would convert the aryl azide in Lyz–1 to an amine via a Staudinger reduction (Figure 3A). This ortho-substituted phosphine accelerates iminophosphorane hydrolysis through neighboring-group participation, leading to rapid 1,6-elimination.[44] Inside living cells, 2-DPBA deprotects caged UAAs with a t1/2 of less than 2 h.[43] As a complementary trigger to 2-DPBA, we selected the strained alkene TCO-OH, which also reduces aryl azides, albeit more slowly than phosphines.[58] Drawing on previous studies,[39] we anticipated that TCO-OH would trigger 1,6-elimination of the aryl azide in Lyz–1 by engaging in a release mechanism (Figure 3A) that begins with a strain-promoted 1,3-dipolar cycloaddition.
Figure 3.
(A) Putative mechanism of Lyz–1 decaging with 2-DPBA and TCO-OH. A single asterisk refers to the Lyz–IQM byproduct. Two asterisks refer to the Lyz–AI byproduct. (B) Deconvoluted Q-TOF mass spectrum of Lyz–1 decaged with 2-DPBA and spontaneous hydrolysis for 1 h. (C) Deconvoluted Q-TOF mass spectrum of Lyz–1 decaged with 2-DPBA and spontaneous hydrolysis for 17 h. (D) Deconvoluted Q-TOF mass spectrum of Lyz–1 decaged with TCO-OH and spontaneous hydrolysis for 15 h. DPBS, Dulbecco’s phosphate-buffered saline.
We began by optimizing the conditions for Lyz–1 decaging with 2-DPBA. We found that a 1-h Lyz–1 incubation with 2-DPBA (17 equiv per protein) led to deprotection of the esters and release of WT Lyz (Figures 3B and S7). We also noticed the formation of a minor byproduct that corresponds to the incomplete elimination of the aniline intermediate from a carboxyl group. We designate this byproduct: Lyz–IQM (Figure 3A). Some esters and quinone-methide adducts on proteins[59] and DNA[60] are cleaved only slowly. To assess the stability of Lyz–IQM, we extended the incubation time to 17 h. Under these conditions, the Lyz–IQM byproduct vanished, and WT Lyz was released tracelessly (Figures 3B and S7).
After settling on two optimal Lyz–1 decaging conditions with 2-DPBA, we turned to TCO-OH. As this reagent unmasks aryl azides more slowly than do phosphines, we first ensured that Lyz–1 is relatively stable to background hydrolysis (Figure S8). We found that a 15-h incubation of Lyz–1 with TCO-OH (67 equiv per protein) was required for complete enzyme decaging (Figures 3D and S8). Although WT Lyz was the major product in this reaction, we also detected Lyz–IQM and a byproduct of incomplete hydrolysis of the aldimine intermediate (AI) formed upon triazoline degradation on Lyz (Lyz-AI) (Figure 3A).
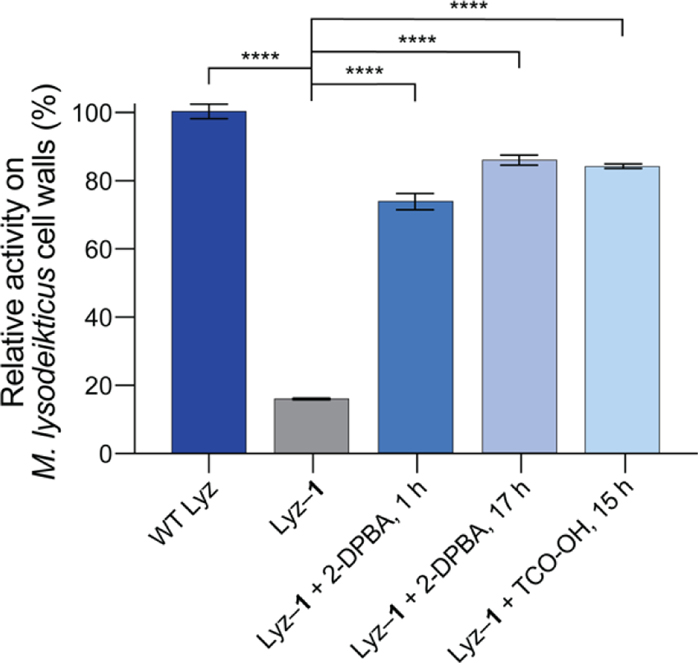
With the three optimized decaging conditions identified (Figures 3B–D), we tested their suitability for on-demand activation of Lyz–1. In these assays, we took endpoint measurements. The relative hydrolytic activities of 0.125 μM WT Lyz, Lyz–1, and decaged Lyz on M. lysodeikticus cell walls are shown in Figures 4 and S9A. All three decaging conditions resulted in efficient enzyme reactivation: decaged Lyz regained about 80% of the activity of WT Lyz. Moreover, at 0.125 μM, decaged Lyz was about 5-fold more active than Lyz–1. At higher concentrations, the change in activity upon decaging was slightly less pronounced (Figure S9B). This experiment demonstrates the feasibility of on-demand Lyz activation through small molecule-mediated decaging of the catalytic Glu35 residue.
Figure 4.
Relative hydrolytic activities on Micrococcus lysodeikticus cell walls of WT Lyz, Lyz–1, and Lyz–1 decaged under optimized conditions. Values are the mean ± SD (n = 3). ****p < 0.0001.
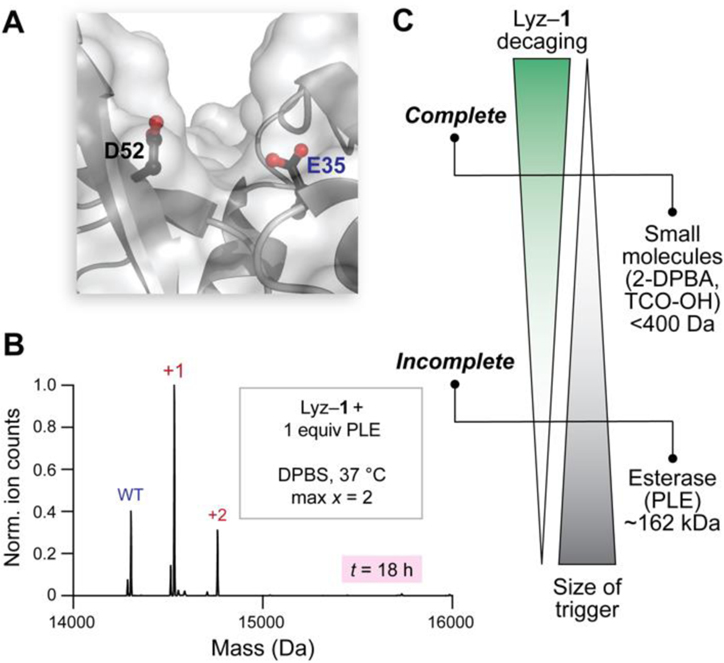
One future application of the strategy presented herein is to manipulate proteins within living cells. Accordingly, we compared esterases to small molecules in terms of their ability to cleave the esters in Lyz–1. Because mammalian esterases are much larger than Lyz, they would encounter steric hindrance upon approaching esterified residues. The most challenging residues for esterases to decage would likely be those found in grooves within Lyz, such as the active-site Glu35 (Figure 5A). To evaluate the stability of Lyz–1 to cleavage by esterases, we incubated this conjugate with pig liver esterase (PLE), which is a model esterase, or lysates prepared from human M21 melanoma cells, which contain a variety of human esterases. After 4 h, Lyz–1 incubated with PLE or lysates retained one major ester label on average, which was indicative of slow cleavage (Figure S10). Extending the cleavage time to 18 h did not alter these results, as the cleavage of ester labels was still incomplete (Figures 5B, S11, and S12). In contrast, TCO-OH and 2-DPBA were able to remove all of the ester labels on Lyz–1 on similar and much shorter time scales (i.e., ~1 h) (Figures 3B–D, S7, and S8). We wondered whether these differences could be explained by small molecules being able to reach esterified residues in the active site of Lyz more easily than do esterases. To answer this question, we compared the kcat of WT Lyz to that of Lyz–1 cleaved by PLE for 18 h. The kcat of the cleaved Lyz dropped by about twofold (Figure S13), which indicated that some active-site residues remained caged, even upon prolonged exposure to PLE. Intact MS analysis (Figure S14) of the cleaved Lyz (Figure 5B) confirmed that Glu35 was the predominant remaining esterified residue. Thus, we conclude that sterics hinder esterases from reaching an esterified carboxyl group within the active site of Lyz. Collectively, our data show that esterases decage ester labels in Lyz–1 much less efficiently than do small molecules (Figure 5C). If additional resistance to intracellular esterases is required, the stability of the pendant esters to esterase-mediated hydrolysis could be increased even further. For example, steric bulk around the aryl ring of diazo compound 1 could be increased, as has been implemented in esterase-resistant auxins caged with a photolabile ester bond.[17]
Figure 5.
(A) Steric hindrance in the active site of Lyz. (B) Deconvoluted Q-TOF mass spectrum of Lyz–1 subjected to cleavage with PLE. (C) Relationship between the size of the trigger and the degree of Lyz–1 decaging.
To validate further the preferential reactivity of diazo compound 1 with high-pKa carboxyl groups and showcase the versatility of our approach beyond glycosidases, we explored the caging and on-demand activation of two additional proteins—cytochrome c (Cyt c), a hemeprotein that plays a central role in mammalian apoptosis,[61] and HIV-1 protease (HIVPR), an aspartyl protease that is necessary for the replication of HIV-1.[62]
We began to probe the scope of our strategy by using horse Cyt c. This protein is covalently linked to a heme c prosthetic group, iron-protoporphyrin IX.[61] One of the two propionates in this heme, HP6, is solvent-accessible[63,64] and has an anomalously high pKa > 9.[65] To test whether our approach could be used to cage HP6 selectively, we esterified Cyt c with diazo compound 1 (6.7 equiv per carboxyl group) at pH 5.5, digested the fully caged protein (Cyt c–1) (Figure S15) with Glu-C, and examined the resulting peptides via intact MS. By peak area, a heme propionate (most likely HP6) was the most abundant esterified carboxyl group (Figures S16 and S17). To quantify the amount of Cyt c–1 esterified at the heme propionate, we analyzed the absorbance of heme c-containing digests at 410 nm[66] and obtained a lower-bound estimate of 39% (Figures S18). This estimate represents the minimal amount of heme propionate esterified in Cyt c–1 because a fraction of the esters hydrolyzed (Figure S19) during the incubation (15 h, 37 °C) with Glu-C. Because HP6 is solvent-accessible and has a pKa > 9, the selectivity observed at pH 5.5 is not surprising. Next, we wondered whether caging and subsequent decaging of the heme propionate would engender control over intrinsic apoptosis. In this pathway, Cyt c binds to Apaf-1, commencing a proteolytic cascade of caspases.[61] We decaged Cyt c–1 with 2-DPBA (Figure S20) and used immunoblot- and luciferin-based assays to measure the ability of various Cyt c samples to activate caspase-3/7 (Figures S21–S23). These experiments were performed in cytosolic fractions that contain esterases. Gratifyingly, we found that, compared to WT and decaged Cyt c, Cyt c–1 was less capable of activating caspases. A possible explanation for this result is that the caging of HP6 disrupts the assembly of the apoptosome. Support for this hypothesis comes from HP6 residing in an exposed heme edge[67] that is involved in the Cyt c·Apaf-1 interaction.[68] Further, HP6 forms a hydrogen bond with Thr49 of Ω-loop C (residues 40–57), which also interacts with Apaf-1.[69] Our work with Cyt c creates opportunities for developing novel tools for manipulating intrinsic apoptosis.
Next, we turned our attention to HIVPR. An inherent complexity is that this aspartyl protease is an obligate homodimer (Kd = 23 pM).[70] Solvent-accessible residues D25 and D25′, one donated by each monomer, form the catalytic dyad of HIVPR.[71] Although there is disagreement regarding the exact pKa values of the catalytic aspartic acids, the pKa of one (nominally, D25) is depressed and that of the other (D25′) is elevated to be 4.9–7.3.[71-75] HIVPR is prone to autodegradation,[76] and we were curious if this challenging target would withstand our caging–decaging protocol. We expressed a stabilized variant of HIVPR[77] that contains substitutions that obviate cysteine oxidation and restrict autoproteolysis, though not fully.[78,79] Aiming to minimize autodegradation of caged HIVPR (HIVPR–1) by the native protein, we esterified HIVPR at room temperature (instead of 37 °C) and accelerated the reaction by adding a large excess of diazo compound 1 (22 equiv) at pH 5.5, as lower pH results in faster caging. By ionization intensities, the resultant conjugate contained primarily one ester label and 29% of WT HIVPR (Figure S24). Intact MS analysis of pepsin digests of HIVPR–1 suggested that the most abundant singly esterified peptide was GQLK(Glu21)ALL(D25) (Figure S25). Although we could not determine which of the two carboxyl groups was esterified, this experiment supported the conjecture that D25′ was caged. (According to one computational source, the pKa of E21/E21′ is 5.03/5.06, and the pKa of D25/D25′ is 1.54/6.89.[71]) As an ultimate test for the compatibility of our approach with sensitive enzymes, we compared the relative rates of catalysis by WT HIVPR, HIPVR–1, and HIPVR–1 decaged with 2-DPBA (Figure S26) using a FRET-based assay. As seen in Figure S27, HIVPR–1 was about 4-fold less active than WT HIVPR, and the relative kcat/KM of HIVPR–1 (0.26 ± 0.07) roughly corresponded to the amount of WT HIVPR (29%) in the caged protein (Figure S24). Further, decaging restored catalytic activity. (For a more in-depth analysis, see Figures S27 and S28.) Together, our results provide evidence that D25′ was caged selectively and suggest that our method is general. We anticipate that spatiotemporal control over HIVPR activity will be useful for both virological[80] and medicinal[62] studies.
Conclusion
We have developed the first method for on-demand protein activation through chemically caged carboxyl groups. Our simple, purely chemical strategy does not require protein engineering (such as UAA incorporation) and makes the activity of a new class of targets—proteins that contain a functional, solvent-accessible carboxyl group with an elevated pKa (e.g., Asp, Glu, and heme propionate)—amendable to spatiotemporal control. As an initial proof of concept, we showed that a diazo compound with a self-immolative handle can be used to cage Lyz, which could later be switched back “ON” with small molecules. To validate the generality of our strategy, we demonstrated its applicability to the hemeprotein Cyt c and the aspartyl protease HIVPR, creating opportunities for future research.
We also demonstrated that the esterified Lyz is cleaved more efficiently by small molecules than by esterases, opening the door for the utility of our strategy both outside and inside of living cells. Because the esterification reaction would not be specific in complex biological environments, proteins would first need to be caged and then delivered into cells using a method (chosen from many available options)[81,82] suitable for the specific user-defined application. For instance, if delivery into many cells is required, caged proteins could be conjugated with cell-penetrating peptides,[83] encapsulated into nanoparticles,[84] or co-incubated with endosomolytic reagents.[85] Alternatively, if delivery to a small group of live cells (<100) would suffice, microinjection would be the method of choice.[81] This approach has been successfully applied in a variety of human cell lines,[86] Xenopus oocytes,[87] and zebrafish embryos.[88] In the latter example,[88] Deiters and coworkers microinjected embryos with the machinery for caged UAA incorporation and showed that the resultant proteins can be activated by adding a phosphine to the embryo water. Efforts towards exploring our carboxyl group caging strategy in zebrafish embryos are currently underway in our laboratory. Applications in the extracellular space (which has a low concentration of esterases[89,90]) are also promising, especially in the context of a growing field of activatable antibodies.[91]
Finally, we anticipate that the concept of exploiting pKa differences between acids to reversibly cage functional groups of interest could be translatable to many systems including phosphorylated and pyrophosphorylated proteins, nucleic acids (e.g., selective caging of 5′ phosphoryl groups in mRNA[92]), and stimuli-responsive biomaterials.
Supplementary Material
Acknowledgements
This work was supported by Grants R01 GM044783 and R35 GM148220 (NIH). Y.D.P. was supported by NSF Graduate Research Fellowship 4000143422. C.S.G. was supported by an NDSEG Fellowship sponsored by the Air Force Research Laboratory. We are grateful to Dr. Eric Spooner from the Whitehead Proteomics Facility for general MS advice and MS/MS analysis. We thank Dr. Mohanraja Kumar from the Department of Chemistry Instrumentation Facility at the Massachusetts Institute of Technology for HRMS analysis of diazo compounds. The M21 cell line was a kind gift from Dr. Oscar Ortiz (Merck KGaA, Darmstadt, Germany), and we thank Professor Norbert Sewald (Universität Bielefeld) for advice about that cell line. We are grateful to Dr. Joomyung V. Jun and JoLynn B. Giancola for helpful discussions. We are also grateful to Dr. Aniekan M. Okon, Dr. Brian J. Graham, Nile S. Abularrage, and Evans C. Wralstad for their comments on the manuscript. We thank Professor Matthew D. Shoulders and Jessica E. Patrick for help with preparative ultracentrifugation. Figure 1 was created with BioRender.com.
Footnotes
Supporting Information for this article is given via a link at the end of the document.
References
- [1].Zorn JA, Wells JA, Nat. Chem. Biol 2010, 6, 179–188. [DOI] [PubMed] [Google Scholar]
- [2].Wang J, Liu Y, Liu Y, Zheng S, Wang X, Zhao J, Yang F, Zhang G, Wang C, Chen PR, Nature 2019, 569, 509–513. [DOI] [PubMed] [Google Scholar]
- [3].Qiao Y, Molina H, Pandey A, Zhang J, Cole PA, Science 2006, 311, 1293–1297. [DOI] [PubMed] [Google Scholar]
- [4].Li J, Yu J, Zhao J, Wang J, Zheng S, Lin S, Chen L, Yang M, Jia S, Zhang X, Chen PR, Nat. Chem 2014, 6, 352–361. [DOI] [PubMed] [Google Scholar]
- [5].Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS, Science 2004, 304, 743–746. [DOI] [PubMed] [Google Scholar]
- [6].Wang J, Wang X, Fan X, Chen PR, ACS Cent. Sci 2021, 7, 929–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pratt MR, Schwartz EC, Muir TW, Proc. Natl. Acad. Sci. U.S.A 2007, 104, 11209–11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM, Nature 2000, 407, 395–401. [DOI] [PubMed] [Google Scholar]
- [9].Takamori S, Yamaguchi S, Ohashi N, Nagamune T, Chem. Commun 2013, 49, 3013–3015. [DOI] [PubMed] [Google Scholar]
- [10].Liu J, Zheng F, Cheng R, Li S, Rozovsky S, Wang Q, Wang L, J. Am. Chem. Soc 2018, 140, 8807–8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ribeiro AJM, Tyzack JD, Borkakoti N, Holliday GL, Thornton JM, J. Biol. Chem 2020, 295, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mendel D, Ellman JA, Schultz PG, J. Am. Chem. Soc 1991, 113, 2758–2760. [Google Scholar]
- [13].Short GF, Lodder M, Laikhter AL, Arslan T, Hecht SM, J. Am. Chem. Soc 1999, 121, 478–479. [Google Scholar]
- [14].Ling X, Zuo Y, Chen H, Ji D, Wang J, Chang L, Liu T, CCS Chem. 2023, 0, 1–7. [Google Scholar]
- [15].Dumas A, Lercher L, Spicer CD, Davis BG, Chem. Sci 2015, 6, 50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yang X, Miao H, Xiao R, Wang L, Zhao Y, Wu Q, Ji Y, Du J, Qin H, Xuan W, Chem. Sci 2021, 12, 9778–9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kusaka N, Maisch J, Nick P, Hayashi K.-i., Nozaki H, ChemBioChem 2009, 10, 2195–2202. [DOI] [PubMed] [Google Scholar]
- [18].Davies S, Qiao L, Oliveira BL, Navo CD, Jiménez-Osés G, Bernardes GJL, ChemBioChem 2019, 20, 1541–1546. [DOI] [PubMed] [Google Scholar]
- [19].Sainlos M, Iskenderian-Epps WS, Olivier NB, Choquet D, Imperiali B, J. Am. Chem. Soc 2013, 135, 4580–4583. [DOI] [PubMed] [Google Scholar]
- [20].Chyan W, Raines RT, ACS Chem. Biol 2018, 13, 1810–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Mix KA, Raines RT, Org. Lett 2015, 17, 2358–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Caution! Exposure to heat, light, pressure, or shock can effect the exothermic decomposition of some diazo compounds. The diazo compounds used in this work are, however, predicted to be safe for use in the contexts of chemical biology. See: Green SP, Wheelhouse KM, Payne AD, Hallett JP, Miller PW, W. P, Bull JA, Org. Process Res. Dev 2020, 24, 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mix KA, Aronoff MR, Raines RT, ACS Chem. Biol 2016, 11, 3233–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roberts JD, Watanabe W, McMahon RE, J. Am. Chem. Soc 1951, 73, 760–765. [Google Scholar]
- [25].Chen HA, Pfuhl M, McAlister MSB, Driscoll PC, Biochemistry 2000, 39, 6814–6824. [DOI] [PubMed] [Google Scholar]
- [26].Bergdoll LA, Lerch MT, Patrick JW, Belardo K, Altenbach C, Bisignano P, Laganowsky A, Grabe M, Hubbell WL, Abramson J, Proc. Natl. Acad. Sci. U.S.A 2018, 115, E172–E179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Oda Y, Yamazaki T, Nagayama K, Kanaya S, Kuroda Y, Nakamura H, Biochemistry 1994, 33, 5275–5284. [DOI] [PubMed] [Google Scholar]
- [28].McIntosh LP, Hand G, Johnson PE, Joshi MD, Körner M, Plesniak LA, Ziser L, Wakarchuk WW, Withers SG, Biochemistry 1996, 35, 9958–9966. [DOI] [PubMed] [Google Scholar]
- [29].Thornburg LD, Hénot F, Bash DP, Hawkinson DC, Bartel SD, Pollack RM, Biochemistry 1998, 37, 10499–10506. [DOI] [PubMed] [Google Scholar]
- [30].Di Nardo G, Breitner M, Bandino A, Ghosh D, Jennings GK, Hackett JC, Gilardi G, J. Biol. Chem 2015, 290, 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Harris TK, Turner GJ, IUBMB Life 2002, 53, 85–98. [DOI] [PubMed] [Google Scholar]
- [32].Grimsley GR, Scholtz JM, Pace CN, Protein Sci. 2009, 18, 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fei N, Sauter B, Gillingham D, Chem. Commun 2016, 52, 7501–7504. [DOI] [PubMed] [Google Scholar]
- [34].West AV, Muncipinto G, Wu H-Y, Huang AC, Labenski MT, Jones LH, Woo CM, J. Am. Chem. Soc 2021, 143, 6691–6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mix KA, Lomax JE, Raines RT, J. Am. Chem. Soc 2017, 139, 14396–14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Thurlkill RL, Grimsley GR, Scholtz JM, Pace CN, Protein Sci. 2006, 15, 1214–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Casari G, Sander C, Valencia A, Nat. Struct. Biol 1995, 2, 171–178. [DOI] [PubMed] [Google Scholar]
- [38].Gnaim S, Shabat D, Acc. Chem. Res 2014, 47, 2970–2984. [DOI] [PubMed] [Google Scholar]
- [39].Ge Y, Fan X, Chen PR, Chem. Sci 2016, 7, 7055–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wesalo JS, Luo J, Morihiro K, Liu J, Deiters A, ChemBioChem 2020, 21, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lozhkin B, Ward TR, Bioorg. Med. Chem 2021, 45, 116310. [DOI] [PubMed] [Google Scholar]
- [42].Jun JV, Raines RT, Org. Lett 2021, 23, 3110–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Luo J, Liu Q, Morihiro K, Deiters A, Nat. Chem 2016, 8, 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lukasak B, Morihiro K, Deiters A, Sci. Rep 2019, 9, 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Blake CCF, Koenig DF, Mair GA, North ACT, Phillips DC, Sarma VR, Nature 1965, 206, 757–761. [DOI] [PubMed] [Google Scholar]
- [46].Phillips DC, Proc. Natl. Acad. Sci. U.S.A 1967, 57, 484–495. [Google Scholar]
- [47].Cheetham JC, Artymiuk PJ, Phillips DC, J. Mol. Biol 1992, 224, 613–628. [DOI] [PubMed] [Google Scholar]
- [48].Vocadlo DJ, Davies GJ, Laine R, Withers SG, Nature 2001, 412, 835–838. [DOI] [PubMed] [Google Scholar]
- [49].Ragland SA, Criss AK, PLoS Pathog. 2017, 13, e1006512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kirby AJ, Nat. Struct. Biol 2001, 8, 737–739. [DOI] [PubMed] [Google Scholar]
- [51].Malcolm BA, Rosenberg S, Corey MJ, Allen JS, de Baetselier A, Kirsch JF, Proc. Natl. Acad. Sci. U.S.A 1989, 86, 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Maeda H, Biochem J. 1980, 88, 1185–1191. [DOI] [PubMed] [Google Scholar]
- [53].Kunichika K, Hashimoto Y, Imoto T, Protein Eng. Des. Sel 2002, 15, 805–809. [DOI] [PubMed] [Google Scholar]
- [54].Knubovets T, Osterhout JJ, Connolly PJ, Klibanov AM, Proc. Natl. Acad. Sci. U.S.A 1999, 96, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Coombes KR, Koomen JM, Baggerly KA, Morris JS, Kobayashi R, Cancer Inform. 2005, 1, 117693510500100103. [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang G, Ueberheide BM, Waldemarson S, Myung S, Molloy K, Eriksson J, Chait BT, Neubert TA, Fenyö D, in Computational Biology, Humana Press, Totowa, NJ, 2010, pp. 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].We also attempted to corroborate our findings with tandem MS (MS/MS) but obtained low coverage of esterified residues (Figure S6). A possible explanation for this result is that our digestion protocol produced many singly charged peptides, which do not fragment well. See: König SJ, Mass Spectrom. 2021, 56, e4616. [Google Scholar]
- [58].Matikonda SS, Orsi DL, Staudacher V, Jenkins IA, Fiedler F, Chen J, Gamble AB, Chem. Sci 2015, 6, 1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rose DA, Treacy JW, Yang ZJ, Ko JH, Houk KN, Maynard HD, J. Am. Chem. Soc 2022, 144, 6050–6058. [DOI] [PubMed] [Google Scholar]
- [60].Weinert EE, Dondi R, Colloredo-Melz S, Frankenfield KN, Mitchell CH, Freccero M, Rokita SE, J. Am. Chem. Soc 2006, 128, 11940–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Alvarez-Paggi D, Hannibal L, Castro MA, Oviedo-Rouco S, Demicheli V, Tórtora V, Tomasina F, Radi R, Murgida DH, Chem. Rev 2017, 117, 13382–13460. [DOI] [PubMed] [Google Scholar]
- [62].Ghosh AK, Osswald HL, Prato G, J. Med. Chem 2016, 59, 5172–5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Timkovich R, Biochem. J 1980, 185, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Moore GR, FEBS Lett. 1983, 161, 171–175. [DOI] [PubMed] [Google Scholar]
- [65].Hartshorn RT, Moore GR, Biochem. J. 1989, 258, 595–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Butt WD, Keilin D, Proc. R. Soc. B: Biol. Sci 1997, 156, 429–458. [DOI] [PubMed] [Google Scholar]
- [67].Deng Y, Zhong F, Alden SL, Hoke KR, Pletneva EV, Biochemistry 2018, 57, 5827–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Purring-Koch C, McLendon G, Proc. Natl. Acad. Sci 2000, 97, 11928–11931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gu J, Shin D-W, Pletneva EV, Biochemistry 2017, 56, 2950–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Todd MJ, Semo N, Freire E, J. Mol. Biol 1998, 283, 475–488. [DOI] [PubMed] [Google Scholar]
- [71].McGee TD Jr., Edwards J, Roitberg AE, J. Phys. Chem. B 2014, 118, 12577–12585. [DOI] [PubMed] [Google Scholar]
- [72].Hyland LJ, Tomaszek TA, Meek TD, Biochemistry 1991, 30, 8454–8463. [DOI] [PubMed] [Google Scholar]
- [73].Ido E, Han HP, Kezdy FJ, Tang J, J. Biol. Chem 1991, 266, 24359–24366. [PubMed] [Google Scholar]
- [74].Smith R, Brereton IM, Chai RY, Kent SBH, Nat. Struct. Biol 1996, 3, 946–950. [DOI] [PubMed] [Google Scholar]
- [75].Hofer F, Kraml J, Kahler U, Kamenik AS, Liedl KR, J. Chem. Inf. Model 2020, 60, 3030–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hui JO, Tomasselli AG, Reardon IM, Lull JM, Brunner DP, Tomich C-SC, Heinrikson RL, Protein Chem J. 1993, 12, 323–327. [DOI] [PubMed] [Google Scholar]
- [77].Windsor IW, Raines RT, Sci. Rep 2015, 5, 11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT, Proteins: Struct. Funct. Genet 2001, 43, 455–464. [DOI] [PubMed] [Google Scholar]
- [79].Louis JM, Deshmukh L, Sayer JM, Aniana A, Clore GM, Biochemistry 2015, 54, 5414–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yang H, Nkeze J, Zhao RY, Cell Biosci. 2012, 2, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Stewart MP, Langer R, Jensen KF, Chem. Rev 2018, 118, 7409–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mann G, Sadhu P, Brik A, Acc. Chem. Res 2022, 55, 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jun JV, Petri YD, Erickson LW, Raines RT, J. Am. Chem. Soc 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mout R, Ray M, Tay T, Sasaki K, Yesilbag Tonga G, Rotello VM, ACS Nano 2017, 11, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Akishiba M, Takeuchi T, Kawaguchi Y, Sakamoto K, Yu H-H, Nakase I, Takatani-Nakase T, Madani F, Gräslund A, Futaki S, Nat. Chem 2017, 9, 751–761. [DOI] [PubMed] [Google Scholar]
- [86].Brustugun OT, Fladmark KE, Døskeland SO, Orrenius S, Zhivotovsky B, Cell Death Differ. 1998, 5, 660–668. [DOI] [PubMed] [Google Scholar]
- [87].Saxena SK, Rybak SM, Winkler G, Meade HM, McGray P, Youle RJ, Ackerman EJ, J. Biol. Chem 1991, 266, 21208–21214. [PubMed] [Google Scholar]
- [88].Brown W, Wesalo J, Tsang M, Deiters A, J. Am. Chem. Soc 2023, 145, 2395–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Bahar FG, Ohura K, Ogihara T, Imai T, J. Pharm. Sci 2012, 101, 3979–3988. [DOI] [PubMed] [Google Scholar]
- [90].Wang D, Zou L, Jin Q, Hou J, Ge G, Yang L, Acta Pharm. Sin. B 2018, 8, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Lucchi R, Bentanachs J, Oller-Salvia B, ACS Cent. Sci 2021, 7, 724–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gampe CM, Hollis-Synynkywicz M, Zécri F, Angew. Chem. Int. Ed 2016, 55, 10283–10286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.