Abstract
Historical and demographical human cohorts of populations exposed to famine, as well as animal studies, revealed that exposure to food deprivation is associated to lasting health-related effects for the exposed individuals, as well as transgenerational effects in their offspring that affect their diseases’ risk and overall longevity. Autophagy, an evolutionary conserved catabolic process, serves as cellular response to cope with nutrient starvation, allowing the mobilization of an internal source of stored nutrients and the production of energy. We review the evidence obtained in multiple model organisms that support the idea that autophagy induction, including through dietary regimes based on reduced food intake, is in fact associated to improved health span and extended lifespan. Thereafter, we expose autophagy-induced chromatin remodeling, such as DNA methylation and histone posttranslational modifications that are known heritable epigenetic marks, as a plausible mechanism for transgenerational epigenetic inheritance of hunger.
Subject terms: Autophagy, Epigenetics
Facts
Human cohorts revealed that fetal, and prepubertal childhood exposure to famine are linked to transgenerational effects on health span and longevity in offspring.
Animal studies demonstrated that postnatal dietary restriction is associated with extended lifespan for the affected organism and its successive generations.
Autophagy induction, including upon nutrient deprivation, is associated with improved health span and longevity in multiple model organisms.
Autophagy induction is associated to epigenetic chromatin changes, including DNA methylation and histone posttranslational modifications.
Heritable chromatin information, such as DNA methylation and histone posttranslational modifications, contribute to transgenerational epigenetic inheritance.
Open Questions
Are histone posttranslational modifications contributing to famine-induced transgenerational epigenetic inheritance in human, as observed in animals?
Why are prenatal versus postnatal exposure to nutrient deprivation, or autophagy induction, leading to contrasting effects on health span and longevity?
Most important, are autophagy-induced epigenetic modifications the drivers in nutrient starvation-induced transgenerational effects on offspring?
A memory of hunger and its impact on offspring longevity
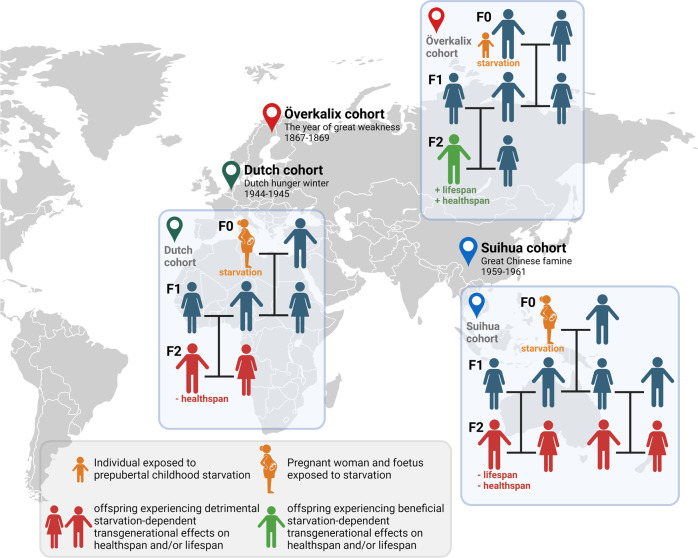
In 2010, the TIME Magazine highlighted findings by Lars Olov Bygrov and colleagues describing how hunger can not only affect your own lifespan but also the lifespan of your children and grandchildren [1]. While it is long known that famine can have acute detrimental health-related effects, the use of historical demographical data, the Överkalix cohort, from an isolated parish in the remote northernmost part of Sweden, where a bad harvest (referred as the year of great weakness, “storsvagåret”) led to famine in the mid-nineteenth century (1867–1869) showed that long-term adverse, as well as more unexpectedly favorable transgenerational effects can also be observed (Fig. 1). In their pioneering work, they unveiled a significant connection between the availability, or lack thereof, of food during the prepubertal slow growth period (5–12 years of age) and lifetime expectancy of the second-generation offspring (i.e. grandparents to grandchild transgenerational response) [2, 3]. Boys enjoying food availability had grandsons which lived on average 6 years shorter than the grandsons of starved boys. Taking socioeconomic variations into account, the difference in lifetime jumped to 32 years [3]. The increase in longevity of the grandsons was accompanied by a reduction in the risk of cardiovascular disease and diabetes mortality [4, 5]. These results were surprising considering that in clear contrast maternal malnutrition during mid-childhood was linked to increased granddaughters cardiovascular mortality [4–6]. The Uppsala birth cohort multigeneration study (UBCos Multigen), based on much larger Swedish population data set than the Överkalix cohort, confirmed that paternal grandfather’s food access in pre-puberty predicts grandsons’, but not granddaughters all-cause and cancer mortality [7]. Further study performed on the UBCos Multigen cohort established that this male-line transgenerational response to malnutrition is observed for several types of cancers [8]. A study performed in mice showed that paternal malnutrition is associated with an epigenetic and metabolic reprogramming of the mammary tissue of their daughters that in turns higher rates of mammary cancer, however this cancer risk association was not observed in the above human cohort [8, 9]. Hence, in the second-offspring generation, it appears that the mortality rate of men was linked exclusively to their paternal grandfather’s food supply during the prepubertal slow growth period, whereas the mortality rate of women was associated instead to the food supply of their paternal grandmothers, suggesting a sex-specific transgenerational response to starvation during mid-childhood, operating through a paternal line.
Fig. 1. Historical and demographical human cohorts illustrate the impact of famine on human health and longevity, including transgenerational effects up to 2 generations.
The Swedish Överkalix cohort (famine 1867–1869), based on individuals exposed to food deprivation during their prepubertal period (5–12 years of age) showed beneficial transgenerational effects in term of health span and lifespan in the grandsons of exposed grandfathers. The Dutch cohort (famine 1944–1945), and the Chinese Suihua cohort (famine 1959–1961) based on individuals, whose pregnant mothers and themselves in utero as foetuses were exposed to nutrients deprivation showed detrimental transgenerational effects in term of health span in the offspring of prenatally exposed fathers (but not mothers) for the Dutch cohort and in term of decreased health and lifespan in the offspring of parents exposed in utero to famine for the Chinese cohort. The image was created with BioRender.com.
Beside children in their prepubertal slow growth period, pregnant women and indirectly their foetuses are also known to represent sensitive groups during a period of famine. Indeed, evidence suggests that the nutritional status during fetal development, reflection of the maternal diet during pregnancy, leads to health outcomes not only on a person as an adult but also on their offspring (reviewed in [10, 11]). An early study on individuals conceived during the Dutch Hunger Winter from 1944 to 1945 suggested that famine exposure in utero was associated with offspring’s poor health in later life with increased chronic disease, however this conclusion was later challenged in a subsequent study performed by the same research team [12, 13]. It remained that adult offspring of prenatally exposed fathers (but not mothers) to famine had higher weights and body mass index than the offspring of prenatally unexposed fathers [13]. Population-based cohort studies based on participants recruited from the Suihua rural area that was affected by the great Chinese famine that occurred between the spring of 1959 and the end of 1961, may offer better understanding of the effect of maternal under-nutrition during gestation. Indeed, these demographical data revealed that prenatal exposure to starvation was associated with elevated risks of developing hyperglycemia, type 2 diabetes, renal dysfunction, and chronic kidney disease in adulthood across two consecutive generations [14–17]. According to the date of birth, F1 subjects were classified as fetal exposed and nonexposed (where the F0 mothers experience famine during their pregnancies). The F2 subjects were classified as having no parents exposed in utero to famine, maternal famine exposure, paternal famine exposure, or parental famine exposure. In the F1 parental generation, prenatal exposure to malnutrition was significantly associated with greater risks of having hyperglycemia and type 2 diabetes [14], as well as of renal dysfunction (as measured by estimated glomerular filtration rate) and chronic kidney disease [15]. One generation down, in the F2 offspring generation, participants from exposed parents, especially those from both exposed parents, still exhibited an increased risk for hyperglycemia and renal dysfunction during adulthood [14–17]. Additional human cohort also based on the great Chinese famine further revealed that parents experience famine in utero or early in life was associated with decreased of their cognitive function. In their offspring, father’s fetal famine exposure was associated with increased depression risk, whereas maternal infant and adolescent famine exposure was associated with decreased cognitive ability [18].
Hence, collectively these human population-based historical cohort studies revealed that in utero fetal, and prepubertal childhood exposure to food deprivation are both linked to long-term health-related effects for the exposed individuals but as well as transgenerational effects in their offspring that affect their diseases’ risk and overall longevity. However, it is important to note the striking difference in the observed transgenerational effects in the offspring based on the time of exposure of the grandparents to nutrient deprivation, with in utero fetal-exposure reported to be linked to detrimental effects whereas in contrast prepubertal childhood exposure was associated to beneficial effects. Collectively, these discoveries infer the existence of “a heritable memory of starvation/hunger”. Nevertheless, by their nature, the human cohorts present clear limitations when it comes to the investigation and possible understanding of the molecular and cellular mechanisms behind nutrient-starvation-induced beneficial effects on longevity. In addition, it could also be argued that an obvious caveat is that some of the observed effects in human populations that exhibit large degree of variation, might be purely due to survival of the fittest, hence starvation could remove the “weak” ones and led to stronger offspring.
More than a century ago, stunted female rats from poor nutrition were reported to live longer, as compared to control stock rat [19]. Later studies in rats showed that a reduction in maternal food intake during pregnancy produced female offspring with shorter lifespan [20], hence replicating some of the observation made with the human cohorts. Since, multiple organism models, ranging from yeast (Saccharomyces cerevisiae), fruit flies and sawflies (Drosophila melanogaster and Athalia rosae), nematodes (Caenorhabditis elegans), rodent (Mus musculus, and Rattus norvegicus), as well as non-human primate such as rhesus monkeys (Macaca mulatta) and gray mouse lemurs (Microcebus murinus) have been developed that reproduce the observation that a postnatal dietary restriction history is associated with an extended lifespan for the organism itself and for some of these models even its offspring [21–26]. However, the beneficial effects of nutrient restriction on longevity is not universal, as its effects vary between mouse strains (i.e. C57BL/6, DBA/2, and B6D2F1) and it is not observed in houseflies (Musca domestica) [27, 28].
Role for autophagy, cellular response to starvation, in lifespan expansion
An essential ability of single cells and by extension of multicellular organisms is to sense nutrient fluctuations in their environment and to adjust their consumption and response accordingly. This adaptation enables cells to survive during periods of nutrient deficiency, and to grow and proliferate when nutrients are plentiful. The capability of cells to sense and respond to nutrient availability and lack thereof in their environment is a prerequisite for life. In fact, nutrient shortage is a selective pressure that has shaped the evolution of most cellular processes [29].
Cells have developed an evolutionary conserved catabolic process, termed macroautophagy (hereafter referred as autophagy), to cope with stress conditions such as nutrient starvation [30–32]. During starvation-induced autophagy, non-essential cellular components are packed into double membrane vesicles, known as autophagosomes, to be thereafter broken down when these autophagosomes fuse with lysosomes, which are rich in proteolytic enzymes; the resulting metabolites can then be reused for core biosynthetic processes or energy production. Proteins constitute a reservoir of amino acids that can be mobilized upon autophagy induction to be recycled and used to sustain new protein synthesis required under starvation conditions. In addition, under those periods of starvation, amino acids are catabolized for the production of energy required to fuel the particular needs of certain vital organs. Hence, autophagy serves as an internal source of stored nutrients under conditions of nutrient limitation. The autophagic catabolic process is also involved in the sequestration and dismiss of protein aggregates, damaged organelles, or pathogenic organisms. Under physiological conditions, a basal level of autophagy is required to maintain cellular homeostasis [33]. Under stress conditions, initiated by a range of extra- or intracellular stress stimuli, autophagy can be enhanced to protect the organisms [34]. Consequently, disturbances in this biological process have been linked to many human diseases, including neurodegenerative diseases, metabolic disorders, psychiatric disorders, cardiovascular diseases, and cancers [35]. Thus, autophagy is divided into mechanistically distinct steps, including induction, cargo recognition and selection, autophagosome formation and fusion with the lysosome and breakdown of the cargo. At the molecular level, the membrane-rearrangements and cargo recognition are tightly regulated and mediated by a core set of Autophagy-related (ATG) proteins [35]. The autophagic core machinery is out of the scope of the present article, hence for details we refer to reviews on the topic, as well as the illustration in Fig. 2 [36, 37].
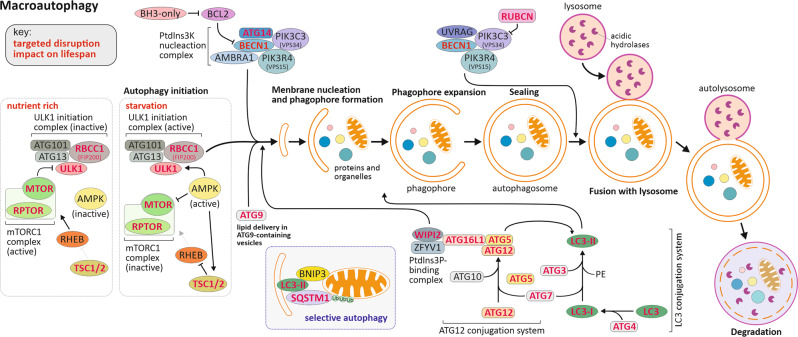
Fig. 2. The autophagy core machinery with indication of which components manipulation have been shown to impact on longevity in various organisms.
Bulk autophagy starts with the stepwise engulfment of cytoplasmic material by the phagophore, which matures into a double-layered vesicle named an autophagosome. AMP-activated protein kinase (AMPK/PRKAA2) and MTOR-containing mTORC1 complex promote and repress autophagy induction, respectively, through phosphorylation of ULK1 (unc-51 like autophagy activating kinase-1) at distinct residues. Under nutrient rich conditions, AMPK is inactive but mTORC1 is active and phosphorylates and inactivates ULK1. When nutrient starvation occurs, AMPK is activated and mTORC1 is inhibited by AMPK through the phosphorylation of TSC1/2 (TSC complex subunit 1/2) and RPTOR/RAPTOR (Regulator-associated protein of MTOR). Subsequently, ULK1 can interact with and be activated by AMPK-mediated phosphorylation, thereby initiating autophagy. Activation of the ULK1-containing initiation complex triggers phagophore formation by phosphorylating components of the BECN1 (Beclin-1) and ATG14-containing class III phosphatidylinositol 3-kinase (Ptdlns3K) nucleation complex. The activated Ptdlns3K nucleation complex generates PtdIns3P, which leads to the recruitment of the effector proteins WIPI2 (WD repeat domain phosphoinositide-interacting protein 2) and ZFYV1 (Zinc finger FYVE domain-containing protein 1). The expansion of the phagophore requires two ubiquitin-like conjugation systems. The ATG12-conjugation system that supports in the formation of ATG12–ATG5-ATG16L1 ternary complex, which in turn promotes the second conjugation reaction. The second system, the LC3 conjugation system, involves the conjugation of phosphatidylethanolamine (PE) to MAP1LC3/LC3 (microtubule associated protein 1 light chain 3, in mammals). Lipid conjugation converts the soluble form of LC3-I into a phagophore membrane-bound LC3-II form that functions in phagophore expansion, and in cargo recognition of ubiquitinated proteins and organelles, including upon selective autophagy with the involvement of autophagy receptors, e.g. BNIP3 (BCL2/adenovirus E1B 19 kDa protein-interacting protein 3), and Ub-dependent autophagy receptors, e.g. SQSTM1/p62 (Sequestosome-1). As a result of membrane expansion and sealing, the autophagic cargo becomes sequestered within the autophagosome. Autophagy completion involves the fusion of the mature autophagosome with a vacuole or lysosome. An alternative Ptdlns3K complex containing UVRAG (UV radiation resistance-associated gene protein), negatively regulated by RUBCN/RUBICON (Run domain Beclin-1-interacting cysteine-rich domain-containing protein), has been reported to regulate the processes of fusion between autophagosomes and lysosomes. Docking and fusion of the outer autophagosomal membrane with that of the lysosome exposes the inner vesicle to the lysosomal lumen, where acidic hydrolases degrade and recycle the macromolecular components for cellular use. Key: Component of the autophagic machinery whose alterations are reported to impact on lifespan are highlighted with red bold text.
At the molecular level, an evolutionary conserved serine/threonine protein kinase, the mechanistic target of rapamycin (MTOR), when part of mTOR complex 1 (MTORC1), functions as a primary nutrient sensor, in particular for amino acids, linking the activation versus repression of the cellular processes to the nutritional supply [29]. When nutrients are abundant, active MTORC1 promotes numerous anabolic processes, such as protein, nucleotide and lipid biosynthesis, while catabolic processes, including autophagy, are repressed. Upon amino acids limitation, MTORC1 is rapidly inactivated, and autophagy induced, allowing the cells to cope with the occurring nutrients starvation (Fig. 2) [38]. Hence, MTOR has emerged as a nutrient sensor of a plethora of extracellular and intracellular cues, and one of the central regulators of autophagy induction. Nutrient-sensing pathways, such as insulin/insulin-like growth factor 1 signaling and mTOR inhibition are thought to act as a determinant of increasing longevity in fruit flies [39, 40]. In C. elegans, inhibition of TOR, known to lead to autophagy induction, is associated to a strikingly doubling of lifespan for the nematode [41]. Likewise, in D. melanogaster the modulation of TOR, by the overexpression of TSC1 and TSC2 (tuberous sclerosis complex genes 1 and 2) that act together to inhibit TOR, or by the expression of dominant-negative forms of TOR all cause lifespan extension of the fruit flies (Fig. 2) [40].
Supporting evidence suggests that autophagy declines during aging, which might contribute to increased cellular stress and accumulation of damaged organelles that could lead to the development of age-related diseases and reduced lifespan expectancy [42]. In that regard, centenarians are excellent subjects to uncover potential mechanisms involved in a healthy aging and consequently human longevity. Remarkably, the genome wide analysis of the transcriptome by RNA sequencing of centenarians, centenarian-children, as well as their spouses revealed that, among the differentially expressed genes, the autophagy pathway was found to be significantly maintained by Centenarians. Remarkably the overexpression of one of these centenarians differentially expressed autophagy-related genes, namely WIPI1 (Atg18a in the fly) in D. melanogaster was shown to extend their lifespan [43]. The analysis of the expression levels of 40 mTOR pathway genes in a unique cohort of Dutch families with extended survival across generations, showed that RPTOR (Raptor) gene was found to be expressed at a lower level in the long-lived individuals, a differential expression that was conserved in their offspring, suggesting an association with familial longevity [44]. The expression of RUBCN/Rubicon (run domain Beclin-1-interacting and cysteine-rich domain-containing protein), a negative regulator of autophagy, increases in aged worm, fly and mouse, suggesting that an age-dependent increase in RUBCN could reduce autophagy activity over time [45]. Knockdown of RUBCN extends worm and fly lifespan and ameliorates several age-associated phenotypes. RUBCN was also found to be suppressed in several long-lived worms and calorie restricted mice [46]. In addition, studies report that the expression levels of a plethora of core component for the autophagic machinery, such as ULK1, ATG7, MAP1LC3, LAMP2, SQSTM1/p62, or BECN1 are found to be significantly affected in aged organisms [47–49]. Moreover, there is evidence that artificial maintenance of the basal autophagy activity level upon aging, for example via the overexpression of ATG5 in mice, or Atg8a, Atg1 or SQSTM1 in fruit flies or SQST-1 in nematodes increase lifespan [50–53]. A knock-in gain-of-function point mutation in Becn1 which disrupt BECN1-BCL2 interaction and constitutively activates autophagy has also been shown to extend lifespan in mice [38].
In contrast, the targeted disruption of core autophagy-related genes (e.g. ULK1, ATG5, ATG7, ATG9, BECN1, or their homologs) in yeast, flies, worms and mice results in a block of starvation-induced autophagy which is linked to reduced viability [54–58]. It appears that prolonged lifespan upon starvation, as well as other autophagy stimuli, is only observed in autophagy-proficient organisms. D. melanogaster lacking the core autophagy regulator Atg7, that is required for starvation-induced autophagy in the flies, are viable but exhibit reduced longevity. These flies are hypersensitive to nutrient and oxidative stress and accumulate ubiquitin-positive aggregates in degenerating neurons [56]. In C. elegans, atg7 is required for longevity in the dietary restriction eat-2 mutant, but not in the insulin-resistant daf-2 longevity model [58, 59] Depletion of another core autophagy-related gene, Beclin-1 (bec-1) in C. elegans, abolished the beneficial effects on longevity of resveratrol or calories restriction [57]. Taken together, these results suggest that suppression of autophagic activity is one of the signatures of aging and that counteracting this decrease can contribute to longevity in multiple organisms.
The direct effect of autophagy modulation on one individual’s cancer onset risk remains controversial (reviewed in [60, 61]). As an illustration, taking advantage of short-hairpin RNA targeting Atg5 expression placed under the control of a doxycycline-dependent promoter allowing the conditional and inducible silencing of this core autophagy gene in mouse, it was shown that the systemic autophagy inhibition from the age of 2 months accelerated aging and aged-associated pathologies, hence reduced lifespan. In contrast, the transient inhibition of autophagy, achieved by the restoration of Atg5 expression at the age of 4 months age in the mice, provided a near complete recovery from the aging phenotype and extended lifespan. However, autophagy-restored mice still succumb earlier than the control mice due to an increase in spontaneous tumor formation [62].
The popular dietary regimes based on reduced food intake, i.e. exposing ourselves voluntarily to hunger, thought to promote longevity, such as calorie restriction, or intermittent time-restricted feeding, also known as intermittent fasting, are all prominent inducers of autophagy leading to the assumption that autophagy is the key pathway in the induction of longevity. Calorie restriction, referred to as a reduction in dietary calorie intake without malnutrition, has been demonstrated to be the most physiological inducer of autophagy to extend longevity across animal models, Epidemiologically, calorie restriction in humans moderates intrinsic processes of aging through cellular and metabolic adaptations and decrease the risk factors in age-related diseases in humans. Therefore, autophagy is an integral mediator of calorie-restriction-induced lifespan extension with the potential to be inherited since the beneficial effect of this dietary regime on longevity fail if autophagy is inactivated.
As an illustration, the inactivation of two essential autophagy genes, bec-1 and Ce-atg7, which are orthologs of human BECN1 (Beclin-1) and ATG7, respectively, barred the longevity phenotype of the C. elegans dietary restriction mutant (eat-2(ad1113) animals [59]. Likewise, the silencing of SIRT1 (Situin-1) expression in the nematode, which is required for autophagy induction upon dietary restriction, abrogated the lifespan extension in the worm [57]. The effect of calorie restriction has been also studied in non-human primates, but the results are conflicting. A 20-year longitudinal adult-onset calorie-restriction study in rhesus monkeys reported a positive impact of this diet on health, age-related survival, and all-cause survival; hence an overall increased longevity [63]. However, another longitudinal study also conducted on rhesus monkeys failed to report an effect of calorie restriction on longevity [64]. Reason for this discrepancy has been suggested to rely in the source of the monkeys, the feeding practices and diet composition, as well as the age of onset of calorie restriction [24]. The established links between calorie restriction, autophagy, longevity and healthy aging has brought interest of the public and scientific communities into caloric-restriction mimetics (e.g. aspirin, resveratrol, spermidine) that could activate autophagy, and prolong life- and health span without the need to cut calories [65]. Intermittent fasting that restricts food intake to specific hours of the day, has also gained interest as a potential anti-ageing treatment [66]. In fruit flies, such dietary regime has been shown to significantly extend the health span and lifespan (by up to 25%) [67, 68]. The beneficial effects of this dietary regime rely on a circadian rhythm-dependent activation of autophagy [68]. In fact, autophagy has long been known to be regulated by the circadian clock such that it peaks at night [69, 70]. Considering that both circadian regulation and autophagy are highly conserved processes, this study provides the interesting possibility that behavioral or pharmaceutical interventions that promote circadian-regulated autophagy could be used to promote healthy aging and increase longevity even in human.
Inhibition of MTOR/TOR kinase by its name defining inhibitory drug, rapamycin (also known as Sirolimus, isolated from Streptomyces hygroscopicus, and certainly one of the best-characterized pharmacological inducers of autophagy) or its analogs (rapalogs), lead to an extension of lifespan in several organisms, seemingly mimicking the effect of dietary restriction. The effects of both chronic treatment and brief pulse of the drug have been considered. Feeding rapamycin to adult Drosophila or adult C. elegans reproduces the lifespan extension observed with some TOR-related mutants (4E-BP (Eukaryotic translation initiation factor 4E-binding protein) null mutant flies, daf-15 (abnormal Dauer Formation-15 and RPTOR homolog) and rheb-1 (RHEB (Ras homolog enriched in brain) homolog) deficient worms) [71, 72]. In mice, chronic exposure to rapamycin using dietary encapsulated rapamycin extends lifespan [73–75]. Additionally, rapamycin administration in food, starting at ~9 months of age, increased the lifespan of females by 18% and male by 10% [74]. Rapamycin even fed late in life, starting at ~20 months of age, was able to increase the lifespan of females by 15% and male by 9% [73]. In mice, rapamycin treatment can delay several age-related diseases, such as cognitive decline, cardiac dysfunction, and cancer [75–77]. However, even at low doses, long-term rapamycin administration can cause adverse effects, and therefor transient administration of the drugs has been considered. In female D. melanogaster, a brief, early rapamycin treatment of adults extended lifespan to the same degree as lifelong dosing [78]. Transient rapamycin treatment, for a 3 months period, is sufficient to increase life expectancy by up to 60% and improve measures of health span in middle-aged mice [78, 79]. The long-term effects on longevity of a transient induction of autophagy by rapamycin, and potentially other autophagy inducers, once more supporting the existence of a memory of this biological process.
Additional natural compounds which are inducers of autophagy, and considered to be caloric-restriction mimetics, have been reported to promote longevity. For example, spermidine a natural polyamide, originally isolated from semen, but also found to be enriched in wheat germ, grapefruit and soybean, induces autophagy and promote longevity in autophagy-proficient model systems [80–82]. Lifelong access to drinking water supplemented with spermidine prolonged the median lifespan of female mice by ~10% [81]. Another example, resveratrol, a polyphenol found in red wine, and a potent activator of Sirtuin-1 (SIRT1), induces SIRT1-dependent autophagy and improve longevity in several organisms. Early study, revealed in the budding yeast S. cerevisiae, that resveratrol treatment stimulating the SIRT1 ortholog Sir2, was able to significantly extend lifespan by a striking ~70% [83]. These observations of increased lifespan and Sir2/SIRT1 dependency were further extended to the metazoans C. elegans and D. melanogaster [84]. Thereafter, autophagy was shown to mediate the lifespan expansion of C. elegans by resveratrol, as this compound, as well as other Sir2/SIRT1 activators were only able to prolong the lifespan of autophagy-proficient nematodes [57]. However, multi-centers comparative studies failed to show that resveratrol treatment can impact on the lifespan of genetically heterogeneous male and female mice [74, 85], suggesting that the longevity promoting effect of resveratrol may not be conserved in higher organisms. Even the widely administrated drug aspirin, also known as acetylsalicylic acid, a nonsteroidal anti-inflammatory drug used to reduce pain, fever, and inflammation, can recapitulate features of caloric restriction, and stimulate the induction of autophagy by virtue of its ability to inhibit the acetyltransferase activity of EP300/p300 [86, 87]. The induction of autophagy by aspirin, or its active metabolite salicylate is observed in both mice and nematodes, but not in the EP300 ortholog cpb1-deficient nematodes [87]. it should be noted that whether treatment with aspirin, beyond its health promoting effects, acting as a caloric-restriction mimetic can promote longevity, remains to be formally established. However, a multi-centers study showed that aspirin led to increase lifespan of genetically heterogeneous male (but not female) mice [88]. In addition, the Finnish centenarian study, a population-based survey of subjects over 100 years old, suggests that subjects taking aspirin had a statistically significant increase in survival as compared to subjects who did not take the drug [89]. As illustrated in Table 1, beyond the above-mentioned examples, the list of natural and synthetic compounds activating autophagy and increasing longevity in various model organisms is increasing by the day.
Table 1.
List of compounds that regulate both autophagy and longevity.
| Pharmacological agent/Compounds | Mechanism of action/Impact on autophagy | Effect on health and lifespan | References |
|---|---|---|---|
| Acetylsalicylic acid and derivatives (C8-SA) | Anti-inflammation effect, autophagy induction by inhibition of EP300/p300; Activation of mitochondria unfolded protein response |
Increase lifespan of genetically heterogeneous male mice; Increase lifespan in worms |
[86, 87] |
| Alpha-ketoglutarate (AKG) | Reduce mTOR activation by inhibiting ATP synthase | Prolong lifespan in worms and flies, but reduces reproduction | [144] |
| Cannabidiol | SIRT1-dependent autophagy induction | Prolong lifespan and increased neuronal health in worms | [145] |
| Flavonoid 4,4′-dimethoxychalcone (DMC) | Induces autophagy in an mTOR-independent manner, but depends on GATA transcription factors | Increase lifespan of yeast, worm and flies | [146] |
| Ganoderma lucidum, dietary supplements | Induce autophagy in an mTOR-dependent manner and stress resistance by reducing the levels of Fibrilarin 1(FIB-1) and LGG-1 | Increase longevity in worms and human cells | [147] |
| Glucosamine (GlcN) | Promotes autophagy via an mTOR-independent manner | Extend lifespan in nematodes | [148, 149] |
| Nordihydroguaiaretic acid (NDGA) | Inhibition of EP300/p300 and activation of autophagy | Increase lifespan in flies and mice | [150] |
| Pyrroloquinoline quinone (PQQ) | Activation of autophagy via Insulin/IGF1 signaling pathway | Extends lifespan in worms by 31% | [151] |
| Metformin | Promote autophagy via AMPK | Extends lifespan in nematodes and mice | [152] |
| Nicotinamide riboside (NR) and nicotinamide mononucleotide (NMN), dietary supplements | Increase autophagy and mitophagy | Increase lifespan in worms, flies and mice | [153] |
| Rapamycin | Autophagy induction by direct inhibition of mTOR | Extends lifespan in worms, flies and mice | [154] |
| Resveratrol | SIRT1-dependent autophagy induction | Improve longevity in yeast, worms, flies and mice | [155] |
| SGLT2 Inhibitors | Mimics calorie restriction by inducing glycosuria. Promotes upregulation of AMPK, SIRT1 and mTOR inhibition | Appear to be a promising treatment extending longevity and reduce oxidative stress | [156] |
| Shatavarin IV | Via eat-2 activation promotes autophagy by promoting the expression of autophagy-related genes expression in an mTOR-dependent manner | On dietary restriction prolonged lifespan | [157] |
| Spermidine | Autophagy induction, reduce inflammation, lipid metabolism, regulation of cell growth, proliferation and cell death | Prolong median lifespan of female mice | [158] |
| Verapamil | Inhibition of calcineurin activity and activation of autophagy | Prolong lifespan, improve health span and delay senescence in worms | [159] |
Does autophagy induction, including upon nutrient starvation, always promotes longevity? This is certainly a matter for debates. Indeed, as mentioned earlier on treatments with autophagy inducers, often caloric-restriction mimetics, do not necessarily lead to increased lifespan across species. For example, whereas rapamycin/rapalogs appear to promote longevity in all the tested model organisms, resveratrol was unable to increase lifespan in mice [74]. The observed effect may also be sex-dependent, as illustrated by the beneficial effect of aspirin on male but not female mice lifespan [88]. Finally, even the prevailing notion that autophagy is beneficial for longevity has even been challenged. Indeed, a study presented data indicating that autophagy induction coupled with increased mitochondrial permeability is in fact detrimental to the lifespan of nematode [90, 91]. Elevated autophagy unexpectedly shortens the lifespan of C. elegans lacking sgk-1 (serum/glucocorticoid regulated kinase-1, and homolog of SGK) or rict-1 (rapamycin-insensitive companion of TOR, and homolog of RICTOR), two negative regulators of autophagy, because of a concomitant increase in mitochondrial permeability in these mutants [91, 92]. Likewise, overexpression of vdac-1 (homolog of voltage-dependent anion channel VDAC1), increase mitochondria permeability by regulating the opening of the mitochondrial permeability transition pore in promoted autophagy but decreased lifespan in C. elegans. Inhibition of autophagy, targeting bec-1 or lgg-1 expression, or even silencing vdac-1, restored the normal lifespan of sgk-1 mutant [91]. However, it should be highlighted that it remains unclear from these studies whether the decrease in lifespan observed in the nematode manipulating genes that affect both autophagy and mitochondrial functions is the result of a decreased longevity or increased toxicity. In fact, mice lacking liver Sgk1 gene expression are more sensitive to liver ischemia/reperfusion injury and thus would live shorter as a result.
The missing link: an epigenetic memory of autophagy
To summarize, (i) exposure to famine, i.e. prolonged nutrient starvation, in human is associated to effects on both health span and longevity for the affected individuals as well as their offspring; and (ii) autophagy induction, the cellular response to nutrient starvation, is likewise associated to similar effects on these parameters for the affected organisms. However, the question that remains to be addressed is how autophagy induction could lead to long-term, as well as transgenerational effects? Noteworthy, for an extended period of time, cytoplasmic events were considered of sole importance for this biological process as enucleated cells are still able to show signs of autophagy [57, 93]. However less than a decade ago, this view was been challenged with establishment that the nucleus, with both transcriptomic and epigenetic events, plays central roles in both the regulation and execution of autophagy [94, 95]. Epigenetic modifications refer to both mitotically and meiotically heritable alterations in gene expression that occur without changes in the underlying DNA sequence. Acquired epigenetic modifications, including DNA methylation, histone modifications and non-coding RNAs, can be propagated through mitotic cell divisions. In addition, these non-DNA sequence-based epigenetic information can be inherited across several generations, via transgenerational epigenetic inheritance [96]. A consensus is established that biological factors transmitted from parent to offspring include not only genetic but also epigenetic contribution [97]. Of interest for the current review, in C. elegans, as well as in D. melanogaster, alterations in several of these epigenetic mechanisms are reported to impact on longevity [96, 98–102]. Hence, an attractive potential mediator of the response to autophagy induction by dietary restriction leading to transgenerational longevity is epigenetics.
Posttranslational modification of core histone proteins, including the acetylation, and methylation of histone tails, is key mechanism of epigenetic regulation. Important groups of histone modifying enzymes in this regulatory network are the histone acetyltransferases (HATs/ KATs for lysine specificity), the histone deacetylases (HDACs), histone methyltransferases (HMTs) and histone lysine demethylases (KDMs). The histone posttranslational modifications can affect the overall chromatin structure and thereby either activate or repress transcription, by modulating the accessibility and binding of transcription factors, and coregulators to the chromatin. The identification of an increasing number of histone marks associated with the regulation and execution of the autophagic process offers an additional attractive conceptual framework to understand the potential long-term response to autophagy (Fig. 3) [94, 103, 104]. Among these autophagy histone marks, histone H4 lysine 16 acetylation (H4K16ac), is found to be reduced upon autophagy induction, through downregulation of the histone acetyltransferase KAT8 (also known hMOF). Indeed, H4K16 deacetylation upon autophagy induction is associated with the downregulation of autophagy-related genes that provides a negative regulatory feedback loop, which in turn serves as a key determinant for survival versus death responses [105]. In yeast, H4K16 acetylation regulate cellular lifespan, due to a decrease of the H4K16 deacetylase Sir2 (SIRT1 homolog) expression. Antagonizing the activities of Sir2 or Sas2 (KAT8 homolog), respectively shortened and extended the replicative yeast lifespan through regulation of H4K16ac levels [106, 107]. Duplication of the sir-2.1 gene, in C. elegans extends the lifespan of the worm by 50% [108]. In D. melanogaster, Sir2 was found to be directly involved in the calorie-restriction lifespan-extending pathway, as an increase in dSir2 extends lifespan, whereas a decrease in dSir2 blocks the lifespan-extending effect of calorie reduction [109]. From a transgenerational viewpoint, maternally inherited hMOF/KAT8-mediated H4K16 acetylation provides a transcriptomic instruction to the offspring, priming future zygotic gene activation in D. melanogaster. The maintenance of H4K16ac from oocytes to fertilized embryos was found to be conserved in mouse [110]. While histone acetylation at lysine residues is generally associated with increased gene expression, methylation of arginine or lysine residues, activate or represses gene expression depending on which residue methylated. Among the different methylated histone marks, two repressive marks, histone H3 lysine 9 trimethylation (H3K9me3), and H3K27me3, and one activating mark, H3K4me3, are reported to be involved in transgenerational epigenetic inheritance in the fruit fly, nematode and mouse [96, 99]. Interestingly, in starved zebrafish myotubes, the transcriptional state of genes involved on the autophagy process (such as atg4b, sqstm1 and lc3b) has been shown to be under the strict epigenetic control of all three-histone methylation marks [111]. In mammalian cells, under nutrient rich conditions, the H3K9 methyltransferase EHMT2/G9a binds to the promoter regions of ATG genes including MAP1LC3 and WIPI and represses their expression, whereas during starvation-induced autophagy,, EHMT2/G9a chromatin displacement leads to a reduction in H3K9me3 levels and the transcriptional activation of ATG genes [112]. In human and mouse, the age-associated decrease of the H3K9 histone methyltransferase SUV39H1, which in turn limit the ability of hematopoietic stem cells to generate B lymphocytes, contributes to a decrease in immune function [113]. However, in a progeria (premature aging) mouse model, depletion of SUV39H1 improves DNA repair capacity and extends lifespan of the mice by ~60% [114]. In C. elegans, the most commonly used animal model for transgenerational epigenetic inheritance and longevity, worms with increased H3K9me2 levels have a longer lifespan that can be passed down to twenty generations [115]. H3K27me3, another transgenerational heritable repressive methylation mark, is established by the H3K27 methyltransferase EZH2 containing polycomb-repressive complex 2 (PRC2). Interestingly, EZH2 has been shown to be potent regulator of autophagy, where the downregulation of TSC2 by EZH2 elicits MTOR activation, which in turn inhibits autophagy [116]. In the fruit flies, increased E(z)(EZH2 in D. melanogaster)-dependent histone H3K27 trimethylation was found to mediate the transgenerational programming on longevity after early-life dietary restriction with shortened lifespan of F0 and F2 offspring [117]. Mutations in E(z) or the H3 binding protein esc, reduce levels of H3K27me3 and promote longevity in D. melanogaster [102]. Likewise in C. elegans, downregulation of the H3K27 demethylase UTX-1 increased global levels of H3K27me3 and improved lifespan [118]. Exposure of pregnant rats to environmental toxicants reported to induce epigenetic transgenerational inheritance of phenotype variations or diseases, such as the fungicide vinclozolin or the pesticide DDT (dichlorodiphenyltrichloroethane), resulted in altered H3K27me3 levels at so-called differential methylated histone retention sites that were conserved in sperm of the F3 generation [119, 120]. H3K4me3, the third histone methylation mark associated to transgenerational epigenetic inheritance, decreases with age and has been shown to be linked to regulation of autophagy in yeast and human cells [105, 121]. In nematodes, deficiencies in the H3K4 methyltransferase SET-2 containing ASH-2 trithorax complex, which establish H3K4me3, extend lifespan, while the H3K4 demethylase RBR-2 is required for normal lifespan, supporting that H3K4me3 associated active chromatin is detrimental for longevity [98]. Likewise in the fruit flies, decrease of Lid, RBR-2 orthologue, increase H3K4me3 levels and decrease lifespan [100]. Transgenic mice overexpressing the H3K4 demethylase KDM1A/LSD1, with a resulting decreased in H3K4me2 level in sperm, impairs their offspring health and survival transgenerationnaly [122]. A fourth histone methylation mark, H4K20me3 is localized in constitutive heterochromatin regions and associated with transcriptional repression and is reported to increase with age in rat liver and kidney [123], as well as in human cells derived from patients with Hutchinson-Gilford progeria, a premature aging syndrome [124]. Serum starvation, a prominent inducer of autophagy, has a marked impact on the global level of H4K20me3 in various mouse cell types [125]. In addition, radiation-induced autophagy promotes H4K20 trimethylation on GABARAPL1 gene leading to autophagy induction in non-small cell lung cancer patients [126]. H4K20me3 is tightly linked to several of the above-mentioned histone marks associated to both the regulation of autophagy and longevity, e.g. H4K16ac and H3K9me3. Indeed, H4K20me is deposited through a preceding deacetylation of H4K16ac, and linked to the presence of H3K9me3, thus determining the levels of H4K20me3 throughout the genome [127]. summary, a role for histone posttranslational modifications, i.e. histone marks, including those reported to be modulated upon autophagy induction, in mediating epigenetic inheritance is well established in invertebrates but only suggested in the germ cells of vertebrates. DNA methylation is regulated by DNA methyltransferase (DNMTs) enzymes that catalyze the transfer of a methyl group to the fifth carbon of a cytosine ring in cytosine-guanine dinucleotide dinucleotides generating 5-methylcytosine. A role for DNA methylation in the long-term transcriptional control of autophagy was recently uncovered (Fig. 3). In fact, cell exposure even to brief autophagic stimuli, including amino acid starvation, is associated with an upregulation of the DNMT3A (DNA methyltransferase 3 alpha) gene expression [128]. The serine/threonine kinase ULK3 (unc-51 like kinase 3)-dependent activation of GLI1 (GLI family zinc finger 1) contributes to the observed transcriptional upregulation of DNMT3A gene expression [129]. DNMT3A protein was found to be recruited to and promote the methylation of the promoter region of MAP1LC3 genes in vitro. Transient autophagy induction was also found to lead to persistent downregulation of map1lc3 genes expression in zebrafish (Danio rerio). In mammals at birth, following the sudden termination of the trans-placental nutrient supply, some organs suffer temporary but severe starvation, which triggers a transient autophagic response [16, 31]. Longitudinal transcriptomic data available for murine lung tissues affected by the early neonatal starvation period revealed that Map1lc3b exhibited a significant and sustained decrease in gene expression, whereas Dnmt3a gene expression showed over time significant increase in gene expression [128]. A recent demonstration of transgenerational epigenetic inheritance was obtained in methylation-edited mammal, where an engineered epigenetic mutation, i.e. DNA methylation of promoter-associated CpG islands, in mice was shown to be inherited across four generations of offspring. The acquired CpG islands methylation was subjected to demetylation in parental primordial germ cells, the gamete precursors, but the heritable epigenetic memory was found to be subsequently re-established in the next generation at the post-implantation epiblast E6.5 embryonic stage [130]. Hence, these recent observations provide a concrete step toward demonstrating DNA methylation based transgenerational epigenetic inheritance in mammals, which may have implications in our understanding of the long-term and heritable effects of autophagy induction.
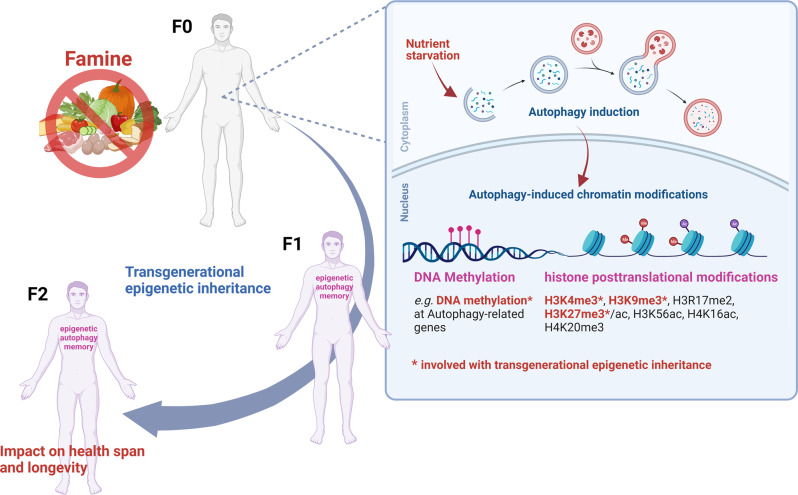
Fig. 3. Proposed contribution of autophagy-induced chromatin modifications to transgenerational epigenetic inheritance in response to famine, i.e. nutrient starvation.
Exposure of human to famine, i.e. nutrient deprivation, is linked to transgenerational effects on health span and longevity in their offspring. Nutrient starvation-induced autophagy is associated to modifications of the chromatin, including DNA methylation and histone posttranslational modifications (including H3K4me3, H3K9me3, H3R17me2, H3K27me3/ac, H3K56ac, H4K16ac and H4K20me3), some of which are reported to contribute to transgenerational epigenetic inheritance. Hence, an epigenetic memory of autophagy could contribute to food deprivation induced effects across successive generations. The image was created with BioRender.com.
Looking at the human population-based historical cohort studies referred above, it appears that DNA methylation-mediated transgenerational epigenetic inheritance also occurs in human upon exposure to famine. Indeed, in the population-based cohort study from Suihua China, blood DNA methylomes performed on 138 subjects across two generations revealed 961 and 503 differentially methylated sites, in the F1 and F2 generations respectively, between the famine and non-famine exposed groups. Remarkably, 19 differentially methylated sites, including in the loci of CUX1, PPARGC1A, ELMO1 and AGTR1, that have been linked to autophagy regulation, were shown to be conserved across generations suggesting that DNA methylation modifications occurring after nutrient restriction can be subject to transgenerational transmission [16]. It should be noted that whereas conservation of DNA methylation profiles across generations has been reported in human cohorts exposed to hunger, as well as in human cohort of long-lived individuals such as nonagenarian and centenarian, conservation of histone marks reported in animal model to contribute to the regulation of autophagy as well as transgenerational epigenetic inheritance remain to be demonstrated.
Conclusion and perspective
Overall, the discovery that autophagy induction is associated to epigenetic alterations, including DNA methylation and histone posttranslational modifications, and that some of this epigenetic information can be inherited by future generations, offer a conceptual frame for the understanding of the long-term effect of food deprivation including in the context of human famine (Fig. 3). Additional epigenetic modifications induced by nutrient starvation and autophagy induction, beyond the above-described ones, will certainly emerge as further epigenetic regulator of health span and lifespan. For example, in C. elegans, small RNAs induced gene silencing has been shown to be able to persist over several generations via transgenerational inheritance [131]. In fact, a set of starvation-induced small RNAs are transmitted transgenerationally, providing a mean for starved worms to control the expression of relevant genes in consecutive generations [132]. Evidence from human famines, as well as animal studies indicates that nutrient starvation affect the health and lifespan of the famished individuals as well as their progeny. However, these studies also indicate that these effects depend on (1) the sex of both the individuals exposed to nutrient deprivations as well as the sex of the offspring and (2) the time of hunger exposure, prenatal (in utero) versus postnatal exposure to nutrient deprivations. The exact mechanisms behind these observed differences in the response to starvation and associated transgenerational epigenetic inheritance remain to be established. However, it should be noted that sex-dependent differences in adaptation to famine have long been appreciated [133, 134]. Autophagy shows sex-dependent differences in both physiological and pathological processes, with the potential impact of sex steroid hormones and a role for the X chromosome (reviewed in [135, 136]). Even constitutive autophagy exhibits a sex bias, as autophagy levels investigated in spinal cord and skeletal muscle tissues of wild-type and unchallenged mice at 40–120 days of age was shown to exhibit significant sex and tissue specific differences [137].
Moreover, sex can be influenced by epigenetic changes promoted by the environment that can make one sex more adaptable to a new environment than the other, and thus promote its survival [138]. The first studies on environmental sex determination (ESD) linked elevated temperature levels led to an increase of males in European sea bass due to an increase of DNA methylation levels at the promoter of the enzyme responsible for estrogen synthesis, cyp19a1a [139]. Half-smooth tongue sole (Cynoglossus semilaevis), a marine fish that has both chromosomal genetic sex determinant and temperature-dependent ESD, shows DNA methylation as the responsible mechanisms for the transition from GSD to ESD. Studies in C. semilaevis demonstrate that ESD it is not limited to cyp19a1a gene rather across genes involved on the sex determination network, with an enrichment of differential DNA methylation patterns. Moreover, transgenerational epigenetic sex inheritance studies performed in that model shows that offspring from a normal male (ZZ) and a female (ZW) exposed to 28 °C during development induced genetic females (ZW) into pseudomales. Pseudomales was subsequently crossed with one normal female to produce F1 pseudomales and females. Interestingly, offspring of pseudomales can spontaneously develop into a functional pseudo-male without the environmental factor involved, due to novel DNA methylation patterns during the juvenile fish at high temperature alters the developmental fate of the gonadal cells. These findings suggest that these new environmental-dependent DNA methylation patterns are imprinted in the genome and transferred to the offspring [140]. In mice, Jmjd1a, the histone methylase responsible for H3K9me2 mark controls the expression of the mammalian Y chromosome sex-determining gene (SRY). Interestingly, XY mice with non-mutated Sry gene have developed as female which suggest the role of epigenetics modifications in mammalian sex determination [141].
Regarding the dramatic difference in observed effects of the transgenerational epigenetic inheritance of the response to nutrient starvation exposure on both health span and longevity in human depending on whether there was a fetal or prepubertal childhood exposure, developmental epigenetic events can be brought forward as possible explanation. Indeed, the fetal period in human, as well as numerous organisms, is a period associated to robust epigenetic (re-)programming events that contribute to proper development. In fact, prenatal exposure to environmental factors, beyond nutrient deprivation, such as stress, infections, toxins, can disrupt gene expression programming in the fetus, resulting in long-term epigenome alterations and developmental deficits that can affect the individual later in life [142, 143]. One can speculate that the effects observed in in utero exposed individuals, as well as their offspring, is the result of the sum of autophagy-induced epigenetic modifications and perturbation of developmental epigenetic programs. In contrast, the long-term effects of postnatal hunger could be the sole or predominant result of autophagy-induced transgenerational epigenetic inheritance. Interestingly, detailed analysis of the effect on lifespan extension of targeting various autophagy-related genes in C. elegans using RNA interference revealed that their manipulations had contrasting detrimental or beneficial effects on worm longevity depending on whether the gene silencing was performed maternally or adult. These results suggest that autophagy induction may not always be beneficial to longevity but may also function to restrict lifespan in the nematode [58]. In summary, one can envision that autophagy associated epigenetic changes in DNA methylation, RNA silencing and histones posttranslational modifications are passed down from one generation to the next, affecting the epigenetic age and lifespan not only of the exposed but also subsequent generations. As a possible illustration of this concept in human, as previously highlighted nonagenarians and centenarians have been shown to exhibit both differentially expressed autophagy genes, as well as unique heritable DNA methylation profiles across generations which could contribute to longevity [43, 44].
Supplementary information
Author contributions
BJ, PGR and JF wrote and edited the manuscript. BJ conceptualized and generated Figs. 1–3, and PGR generated Table 1.
Funding
Research in the authors laboratories is supported by grants from the Swedish Research Council, the Swedish Brain Foundation, the Swedish Cancer Foundation, the Swedish Cancer Society, the Karolinska Institutet Foundation, the Swedish Childhood Cancer Foundation (to BJ), and the Wenner-Gren Foundation (to PGR). We apologize to the many authors of important original research articles that could not be cited for lack of space. Open access funding provided by Karolinska Institute.
Competing interests
JF is current employee of Cambridge Epigenetix and holds stock options. BJ is co-founder of CERVO Therapeutics AB.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-023-01159-4.
References
- 1.Cloud J. Why your DNA isn’t your destiny? TIME Magzine; 2010.
- 2.Bygren LO, Kaati G, Edvinsson S. Longevity determined by paternal ancestors’ nutrition during their slow growth period. Acta Biotheor. 2001;49:53–59. doi: 10.1023/A:1010241825519. [DOI] [PubMed] [Google Scholar]
- 3.Kaati G, Bygren LO, Pembrey M, Sjöström M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–90. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 4.Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet. 2002;10:682–8. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- 5.Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–66. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 6.Bygren LO, Tinghög P, Carstensen J, Edvinsson S, Kaati G, Pembrey ME, et al. Change in paternal grandmothers’ early food supply influenced cardiovascular mortality of the female grandchildren. BMC Genet. 2014;15:12. doi: 10.1186/1471-2156-15-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vågerö D, Pinger PR, Aronsson V, van den Berg GJ. Paternal grandfather’s access to food predicts all-cause and cancer mortality in grandsons. Nat Commun. 2018;9:5124. doi: 10.1038/s41467-018-07617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vågerö D, Cederström A, van den Berg GJ. Food abundance in men before puberty predicts a range of cancers in grandsons. Nat Commun. 2022;13:7507. doi: 10.1038/s41467-022-35217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Cruz RS, Carney EJ, Clarke J, Cao H, Cruz MI, Benitez C, et al. Paternal malnutrition programs breast cancer risk and tumor metabolism in offspring. Breast Cancer Res. 2018;20:99. doi: 10.1186/s13058-018-1034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aiken CE, Tarry-Adkins JL, Ozanne SE. Transgenerational effects of maternal diet on metabolic and reproductive ageing. Mamm Genome. 2016;27:430–9. doi: 10.1007/s00335-016-9631-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moraru A, de Almeida MM, Degryse JM. PALTEM: what parameters should be collected in disaster settings to assess the long-term outcomes of famine? Int J Environ Res Public Health. 2018;15:857. doi: 10.3390/ijerph15050857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Painter RC, Osmond C, Gluckman P, Hanson M, Phillips DI, Roseboom TJ. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG. 2008;115:1243–9. doi: 10.1111/j.1471-0528.2008.01822.x. [DOI] [PubMed] [Google Scholar]
- 13.Veenendaal MV, Painter RC, de Rooij SR, Bossuyt PM, van der Post JA, Gluckman PD, et al. Transgenerational effects of prenatal exposure to the 1944-45 Dutch famine. BJOG. 2013;120:548–53. doi: 10.1111/1471-0528.12136. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Liu S, Li S, Feng R, Na L, Chu X, et al. Prenatal exposure to famine and the development of hyperglycemia and type 2 diabetes in adulthood across consecutive generations: a population-based cohort study of families in Suihua, China. Am J Clin Nutr. 2017;105:221–7. doi: 10.3945/ajcn.116.138792. [DOI] [PubMed] [Google Scholar]
- 15.Wang N, Ning Z, Xia F, Chen C, Cheng J, Chen Y, et al. Exposure to famine in early life and chronic kidney diseases in adulthood. Nutr Diabetes. 2018;8:4. doi: 10.1038/s41387-017-0014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Han T, Duan W, Dong Q, Hou W, Wu H, et al. Prenatal famine exposure and estimated glomerular filtration rate across consecutive generations: association and epigenetic mediation in a population-based cohort study in Suihua China. Aging. 2020;12:12206–21. doi: 10.18632/aging.103397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan S, Ruan J, Wang Y, Xu J, Sun C, Niu Y. Association of prenatal famine exposure with inflammatory markers and its impact on adulthood liver function across consecutive generations. Front Nutr. 2021;8:758633. doi: 10.3389/fnut.2021.758633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Lu Y, Li J, He S. Association of parental famine exposure with offspring depression and cognition function. Front Psychiatry. 2022;13:812805. doi: 10.3389/fpsyt.2022.812805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborne TB, Mendel LB, Ferry EL. The effect of retardation of growth upon the breeding period and duration of life of rats. Science. 1917;45:294–5. doi: 10.1126/science.45.1160.294. [DOI] [PubMed] [Google Scholar]
- 20.Aihie Sayer A, Dunn R, Langley-Evans S, Cooper C. Prenatal exposure to a maternal low protein diet shortens life span in rats. Gerontology. 2001;47:9–14. doi: 10.1159/000052764. [DOI] [PubMed] [Google Scholar]
- 21.Swindell WR. Dietary restriction in rats and mice: a meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev. 2012;11:254–70. doi: 10.1016/j.arr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 23.Paul SC, Putra R, Müller C. Early life starvation has stronger intra-generational than transgenerational effects on key life-history traits and consumption measures in a sawfly. PLoS ONE. 2019;14:e0226519. doi: 10.1371/journal.pone.0226519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, et al. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun. 2017;8:14063. doi: 10.1038/ncomms14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–29. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 26.Pifferi F, Terrien J, Marchal J, Dal-Pan A, Djelti F, Hardy I, et al. Caloric restriction increases lifespan but affects brain integrity in grey mouse lemur primates. Commun Biol. 2018;1:30. doi: 10.1038/s42003-018-0024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forster MJ, Morris P, Sohal RS. Genotype and age influence the effect of caloric intake on mortality in mice. FASEB J. 2003;17:690–2. doi: 10.1096/fj.02-0533fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper TM, Mockett RJ, Sohal BH, Sohal RS, Orr WC. Effect of caloric restriction on life span of the housefly, Musca domestica. FASEB J. 2004;18:1591–3. doi: 10.1096/fj.03-1464fje. [DOI] [PubMed] [Google Scholar]
- 29.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–10. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeifer U. Cellular autophagy and cell atrophy in the rat liver during long-term starvation. A quantitative morphological study with regard to diurnal variations. Virchows Arch B Cell Pathol. 1973;12:195–211. doi: 10.1007/BF02893998. [DOI] [PubMed] [Google Scholar]
- 31.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 32.He C. Balancing nutrient and energy demand and supply via autophagy. Curr Biol. 2022;32:R684–R696. doi: 10.1016/j.cub.2022.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryter SW, Cloonan SM, Choi AM. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013;36:7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–36. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J. 2021;40:e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang W, Chen X, Ji C, Zhang W, Song J, Li J, et al. Key regulators of autophagosome closure. Cells. 2021;10:2814. doi: 10.3390/cells10112814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandes SA, Demetriades C. The multifaceted role of nutrient sensing and mTORC1 signaling in physiology and aging. Front Aging. 2021;2:707372. doi: 10.3389/fragi.2021.707372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papadopoli D, Boulay K, Kazak L, Pollak M, Mallette F, Topisirovic I, et al. mTOR as a central regulator of lifespan and aging. F1000Res. 2019;8:F1000 Faculty Rev-998. [DOI] [PMC free article] [PubMed]
- 40.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–90. doi: 10.1016/j.cub.2004.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Müller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- 42.Barbosa MC, Grosso RA, Fader CM. Hallmarks of aging: an autophagic perspective. Front Endocrinol. 2018;9:790. doi: 10.3389/fendo.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao FH, Chen XQ, Yu Q, Ye Y, Liu YW, Yan D, et al. Transcriptome evidence reveals enhanced autophagy-lysosomal function in centenarians. Genome Res. 2018;28:1601–10. doi: 10.1101/gr.220780.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Passtoors WM, Beekman M, Deelen J, van der Breggen R, Maier AB, Guigas B, et al. Gene expression analysis of mTOR pathway: association with human longevity. Aging Cell. 2013;12:24–31. doi: 10.1111/acel.12015. [DOI] [PubMed] [Google Scholar]
- 45.Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–91. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura S, Oba M, Suzuki M, Takahashi A, Yamamuro T, Fujiwara M, et al. Suppression of autophagic activity by Rubicon is a signature of aging. Nat Commun. 2019;10:847. doi: 10.1038/s41467-019-08729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tóth ML, Sigmond T, Borsos E, Barna J, Erdélyi P, Takács-Vellai K, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4:330–8. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Shi S, Gu Z, Du Y, Liu M, Yan S, et al. Impaired autophagic function in rat islets with aging. Age. 2013;35:1531–44. doi: 10.1007/s11357-012-9456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalil H, Tazi M, Caution K, Ahmed A, Kanneganti A, Assani K, et al. Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics. 2016;11:381–8. doi: 10.1080/15592294.2016.1144007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–84. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- 51.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aparicio R, Rana A, Walker DW. Upregulation of the autophagy adaptor p62/SQSTM1 prolongs health and lifespan in middle-aged Drosophila. Cell Rep. 2019;28:1029–1040.e1025. doi: 10.1016/j.celrep.2019.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumsta C, Chang JT, Lee R, Tan EP, Yang Y, Loureiro R, et al. The autophagy receptor p62/SQST-1 promotes proteostasis and longevity in C. elegans by inducing autophagy. Nat Commun. 2019;10:5648. doi: 10.1038/s41467-019-13540-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–34. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 56.Juhász G, Erdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–6. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hashimoto Y, Ookuma S, Nishida E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes Cells. 2009;14:717–26. doi: 10.1111/j.1365-2443.2009.01306.x. [DOI] [PubMed] [Google Scholar]
- 59.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007;3:597–9. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 60.Galluzzi L, Kroemer G. Transient autophagy inhibition precipitates oncogenesis: a red flag for pharmacological autophagy inhibitors? Trends Cell Biol. 2020;30:339–40. doi: 10.1016/j.tcb.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 61.Ahmadi-Dehlaghi F, Mohammadi P, Valipour E, Pournaghi P, Kiani S, Mansouri K. Autophagy: a challengeable paradox in cancer treatment. Cancer Med. 2023. [DOI] [PMC free article] [PubMed]
- 62.Cassidy LD, Young ARJ, Young CNJ, Soilleux EJ, Fielder E, Weigand BM, et al. Temporal inhibition of autophagy reveals segmental reversal of ageing with increased cancer risk. Nat Commun. 2020;11:307. doi: 10.1038/s41467-019-14187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Colman RJ, Beasley TM, Kemnitz JW, Johnson SC, Weindruch R, Anderson RM. Caloric restriction reduces age-related and all-cause mortality in rhesus monkeys. Nat Commun. 2014;5:3557. doi: 10.1038/ncomms4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–21. doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hofer SJ, Davinelli S, Bergmann M, Scapagnini G, Madeo F. Caloric restriction mimetics in nutrition and clinical trials. Front Nutr. 2021;8:717343. doi: 10.3389/fnut.2021.717343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattson MP, Longo VD, Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Res Rev. 2017;39:46–58. doi: 10.1016/j.arr.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Catterson JH, Khericha M, Dyson MC, Vincent AJ, Callard R, Haveron SM, et al. Short-term, intermittent fasting induces long-lasting gut health and tor-independent lifespan extension. Curr Biol. 2018;28:1714–1724.e1714. doi: 10.1016/j.cub.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulgherait M, Midoun AM, Park SJ, Gatto JA, Tener SJ, Siewert J, et al. Circadian autophagy drives iTRF-mediated longevity. Nature. 2021;598:353–8. doi: 10.1038/s41586-021-03934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfeifer U. [Circadian rhythm of cellular autophagy] Naturwissenschaften. 1971;58:152. doi: 10.1007/BF00593114. [DOI] [PubMed] [Google Scholar]
- 70.Ma D, Li S, Molusky MM, Lin JD. Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol Metab. 2012;23:319–25. doi: 10.1016/j.tem.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, et al. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 2012;15:713–24. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, et al. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle. 2011;10:4230–6. doi: 10.4161/cc.10.24.18486. [DOI] [PubMed] [Google Scholar]
- 76.Halloran J, Hussong SA, Burbank R, Podlutskaya N, Fischer KE, Sloane LB, et al. Chronic inhibition of mammalian target of rapamycin by rapamycin modulates cognitive and non-cognitive components of behavior throughout lifespan in mice. Neuroscience. 2012;223:102–13. doi: 10.1016/j.neuroscience.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flynn JM, O’Leary MN, Zambataro CA, Academia EC, Presley MP, Garrett BJ, et al. Late-life rapamycin treatment reverses age-related heart dysfunction. Aging Cell. 2013;12:851–62. doi: 10.1111/acel.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Juricic P, Lu Y-X, Leech T, Drews LF, Paulitz J, Lu J, et al. Long-lasting geroprotection from brief rapamycin treatment in early adulthood by persistently increased intestinal autophagy. Nat Aging. 2022;2:824–36. doi: 10.1038/s43587-022-00278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bitto A, Ito TK, Pineda VV, LeTexier NJ, Huang HZ, Sutlief E, et al. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife. 2016;5:e16351. doi: 10.7554/eLife.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–14. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 81.Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016;22:1428–38. doi: 10.1038/nm.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Filfan M, Olaru A, Udristoiu I, Margaritescu C, Petcu E, Hermann DM, et al. Long-term treatment with spermidine increases health span of middle-aged Sprague-Dawley male rats. Geroscience. 2020;42:937–49. doi: 10.1007/s11357-020-00173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 84.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–9. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 85.Strong R, Miller RA, Astle CM, Baur JA, de Cabo R, Fernandez E, et al. Evaluation of resveratrol, green tea extract, curcumin, oxaloacetic acid, and medium-chain triglyceride oil on life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2013;68:6–16. doi: 10.1093/gerona/gls070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aguas AP, Soares JO, Nunes JF. Autophagy in mouse hepatocytes induced by lysine acetylsalicylate. Experientia. 1978;34:1618–9. doi: 10.1007/BF02034711. [DOI] [PubMed] [Google Scholar]
- 87.Pietrocola F, Castoldi F, Markaki M, Lachkar S, Chen G, Enot DP, et al. Aspirin recapitulates features of caloric restriction. Cell Rep. 2018;22:2395–407. doi: 10.1016/j.celrep.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strong R, Miller RA, Astle CM, Floyd RA, Flurkey K, Hensley KL, et al. Nordihydroguaiaretic acid and aspirin increase lifespan of genetically heterogeneous male mice. Aging Cell. 2008;7:641–50. doi: 10.1111/j.1474-9726.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agüero-Torres H, Viitanen M, Fratiglioni L, Louhija J. The effect of low-dose daily aspirin intake on survival in the Finnish centenarians cohort. J Am Geriatr Soc. 2001;49:1578–80. doi: 10.1046/j.1532-5415.2001.4911264.x. [DOI] [PubMed] [Google Scholar]
- 90.Savini M, Wang MC. Does autophagy promote longevity? It depends. Cell. 2019;177:221–2. doi: 10.1016/j.cell.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 91.Zhou B, Kreuzer J, Kumsta C, Wu L, Kamer KJ, Cedillo L, et al. Mitochondrial permeability uncouples elevated autophagy and lifespan extension. Cell. 2019;177:299–314.e216. doi: 10.1016/j.cell.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen AT, Guo C, Dumas KJ, Ashrafi K, Hu PJ. Effects of Caenorhabditis elegans sgk-1 mutations on lifespan, stress resistance, and DAF-16/FoxO regulation. Aging Cell. 2013;12:932–40. doi: 10.1111/acel.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joseph B. Accidentally enucleating autophagy. Nat Rev Mol Cell Biol. 2015;16:4. doi: 10.1038/nrm3917. [DOI] [PubMed] [Google Scholar]
- 94.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 95.Shi Y, Shen HM, Gopalakrishnan V, Gordon N. Epigenetic regulation of autophagy beyond the cytoplasm: a review. Front Cell Dev Biol. 2021;9:675599. doi: 10.3389/fcell.2021.675599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fitz-James MH, Cavalli G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat Rev Genet. 2022;23:325–41. doi: 10.1038/s41576-021-00438-5. [DOI] [PubMed] [Google Scholar]
- 97.Adrian-Kalchhauser I, Sultan SE, Shama LNS, Spence-Jones H, Tiso S, Keller Valsecchi CI, et al. Understanding ‘non-genetic’ inheritance: insights from molecular-evolutionary crosstalk. Trends Ecol Evol. 2020;35:1078–89. doi: 10.1016/j.tree.2020.08.011. [DOI] [PubMed] [Google Scholar]
- 98.Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–7. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–71. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS Genet. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kaletsky R, Moore RS, Vrla GD, Parsons LR, Gitai Z, Murphy CT. C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance. Nature. 2020;586:445–51. doi: 10.1038/s41586-020-2699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siebold AP, Banerjee R, Tie F, Kiss DL, Moskowitz J, Harte PJ. Polycomb repressive complex 2 and trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci USA. 2010;107:169–74. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Füllgrabe J, Heldring N, Hermanson O, Joseph B. Cracking the survival code: autophagy-related histone modifications. Autophagy. 2014;10:556–61. doi: 10.4161/auto.27280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baek SH, Kim KI. Epigenetic control of autophagy: nuclear events gain more attention. Mol Cell. 2017;65:781–5. doi: 10.1016/j.molcel.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 105.Füllgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–71. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–7. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kozak ML, Chavez A, Dang W, Berger SL, Ashok A, Guo X, et al. Inactivation of the Sas2 histone acetyltransferase delays senescence driven by telomere dysfunction. EMBO J. 2010;29:158–70. doi: 10.1038/emboj.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 109.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Samata M, Alexiadis A, Richard G, Georgiev P, Nuebler J, Kulkarni T, et al. Intergenerationally maintained histone H4 Lysine 16 acetylation is instructive for future gene activation. Cell. 2020;182:127–144.e123. doi: 10.1016/j.cell.2020.05.026. [DOI] [PubMed] [Google Scholar]
- 111.Biga PR, Latimer MN, Froehlich JM, Gabillard JC, Seiliez I. Distribution of H3K27me3, H3K9me3, and H3K4me3 along autophagy-related genes highly expressed in starved zebrafish myotubes. Biol Open. 2017;6:1720–5. doi: 10.1242/bio.029090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, Mann AO, Taoda Y, Gorman JA, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–93. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Djeghloul D, Kuranda K, Kuzniak I, Barbieri D, Naguibneva I, Choisy C, et al. Age-associated decrease of the histone methyltransferase SUV39H1 in HSC perturbs heterochromatin and B lymphoid differentiation. Stem Cell Rep. 2016;6:970–84. doi: 10.1016/j.stemcr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu B, Wang Z, Zhang L, Ghosh S, Zheng H, Zhou Z. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat Commun. 2013;4:1868. doi: 10.1038/ncomms2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee TW, David HS, Engstrom AK, Carpenter BS, Katz DJ. Repressive H3K9me2 protects lifespan against the transgenerational burden of COMPASS activity in C. elegans. Elife. 2019;8:e48498. doi: 10.7554/eLife.48498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei FZ, Cao Z, Wang X, Wang H, Cai MY, Li T, et al. Epigenetic regulation of autophagy by the methyltransferase EZH2 through an MTOR-dependent pathway. Autophagy. 2015;11:2309–22. doi: 10.1080/15548627.2015.1117734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xia B, Gerstin E, Schones DE, Huang W, Steven de Belle J. Transgenerational programming of longevity through E(z)-mediated histone H3K27 trimethylation in. Aging. 2016;8:2988–3008. doi: 10.18632/aging.101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–90. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ben Maamar M, Sadler-Riggleman I, Beck D, Skinner MK. Epigenetic transgenerational inheritance of altered sperm histone retention sites. Sci Rep. 2018;8:5308. doi: 10.1038/s41598-018-23612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Skinner MK, Ben Maamar M, Sadler-Riggleman I, Beck D, Nilsson E, McBirney M, et al. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin. 2018;11:8. doi: 10.1186/s13072-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, et al. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 2012;8:e1002473. doi: 10.1371/journal.pgen.1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350:aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 123.Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 124.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci. 2004;117:2491–501. doi: 10.1242/jcs.01238. [DOI] [PubMed] [Google Scholar]
- 126.Lee TG, Kim SY, Kim HR, Kim H, Kim CH. Radiation induces autophagy. Anticancer Res. 2020;40:2537–48. doi: 10.21873/anticanres.14224. [DOI] [PubMed] [Google Scholar]
- 127.Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27:639–53. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.González-Rodríguez P, Cheray M, Füllgrabe J, Salli M, Engskog-Vlachos P, Keane L, et al. The DNA methyltransferase DNMT3A contributes to autophagy long-term memory. Autophagy. 2021;17:1259–77. doi: 10.1080/15548627.2020.1816664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.González-Rodríguez P, Cheray M, Keane L, Engskog-Vlachos P, Joseph B. ULK3-dependent activation of GLI1 promotes DNMT3A expression upon autophagy induction. Autophagy. 2022;18:2769–80. doi: 10.1080/15548627.2022.2039993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takahashi Y, Morales Valencia M, Yu Y, Ouchi Y, Takahashi K, Shokhirev MN, et al. Transgenerational inheritance of acquired epigenetic signatures at CpG islands in mice. Cell. 2023;186:715–731.e719. doi: 10.1016/j.cell.2022.12.047. [DOI] [PubMed] [Google Scholar]
- 131.Houri-Ze’evi L, Korem Y, Sheftel H, Faigenbloom L, Toker IA, Dagan Y, et al. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 132.Rechavi O, Houri-Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ, et al. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–87. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Widdowson EM. The response of the sexes to nutritional stress. Proc Nutr Soc. 1976;35:175–80. doi: 10.1079/PNS19760030. [DOI] [PubMed] [Google Scholar]
- 134.Hoyenga KB, Hoyenga KT. Gender and energy balance: sex differences in adaptations for feast and famine. Physiol Behav. 1982;28:545–63. doi: 10.1016/0031-9384(82)90153-6. [DOI] [PubMed] [Google Scholar]
- 135.Shang D, Wang L, Klionsky DJ, Cheng H, Zhou R. Sex differences in autophagy-mediated diseases: toward precision medicine. Autophagy. 2021;17:1065–76. doi: 10.1080/15548627.2020.1752511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lista P, Straface E, Brunelleschi S, Franconi F, Malorni W. On the role of autophagy in human diseases: a gender perspective. J Cell Mol Med. 2011;15:1443–57. doi: 10.1111/j.1582-4934.2011.01293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oliván S, Calvo AC, Manzano R, Zaragoza P, Osta R. Sex differences in constitutive autophagy. Biomed Res Int. 2014;2014:652817. doi: 10.1155/2014/652817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Govender P, Ghai M, Okpeku M. Sex-specific DNA methylation: impact on human health and development. Mol Genet Genomics. 2022;297:1451–66. doi: 10.1007/s00438-022-01935-w. [DOI] [PubMed] [Google Scholar]
- 139.Navarro-Martín L, Viñas J, Ribas L, Díaz N, Gutiérrez A, Di Croce L, et al. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 2011;7:e1002447. doi: 10.1371/journal.pgen.1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shao C, Li Q, Chen S, Zhang P, Lian J, Hu Q, et al. Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 2014;24:604–15. doi: 10.1101/gr.162172.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sánchez A, Marchal JA, Romero-Fernández I, Pinna-Senn E, Ortiz MI, Bella JL, et al. No differences in the Sry gene between males and XY females in Akodon (Rodentia, Cricetidae) Sex Dev. 2010;4:155–61. doi: 10.1159/000309780. [DOI] [PubMed] [Google Scholar]
- 142.Kundakovic M, Jaric I. The epigenetic link between prenatal adverse environments and neurodevelopmental disorders. Genes. 2017;8:104. doi: 10.3390/genes8030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cao-Lei L, de Rooij SR, King S, Matthews SG, Metz GAS, Roseboom TJ, et al. Prenatal stress and epigenetics. Neurosci Biobehav Rev. 2020;117:198–210. doi: 10.1016/j.neubiorev.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 144.Su Y, Wang T, Wu N, Li D, Fan X, Xu Z, et al. Alpha-ketoglutarate extends. Aging. 2019;11:4183–97. doi: 10.18632/aging.102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Z, Zheng P, Chen X, Xie Y, Weston-Green K, Solowij N, et al. Cannabidiol induces autophagy and improves neuronal health associated with SIRT1 mediated longevity. Geroscience. 2022;44:1505–24. doi: 10.1007/s11357-022-00559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Carmona-Gutierrez D, Zimmermann A, Kainz K, Pietrocola F, Chen G, Maglioni S, et al. The flavonoid 4,4’-dimethoxychalcone promotes autophagy-dependent longevity across species. Nat Commun. 2019;10:651. doi: 10.1038/s41467-019-08555-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Peng HH, Wu CY, Hsiao YC, Martel J, Ke PY, Chiu CY, et al. Ganoderma lucidum stimulates autophagy-dependent longevity pathways in Caenorhabditis elegans and human cells. Aging (Albany NY) 2021;13:13474–95. doi: 10.18632/aging.203068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shintani T, Yamazaki F, Katoh T, Umekawa M, Matahira Y, Hori S, et al. Glucosamine induces autophagy via an mTOR-independent pathway. Biochem Biophys Res Commun. 2010;391:1775–9. doi: 10.1016/j.bbrc.2009.12.154. [DOI] [PubMed] [Google Scholar]
- 149.Shintani T, Kosuge Y, Ashida H. Glucosamine extends the lifespan of Caenorhabditis elegans via autophagy induction. J Appl Glycosci (1999) 2018;65:37–43. doi: 10.5458/jag.jag.JAG-2018_002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tezil T, Chamoli M, Ng CP, Simon RP, Butler VJ, Jung M, et al. Lifespan-increasing drug nordihydroguaiaretic acid inhibits p300 and activates autophagy. NPJ Aging Mech Dis. 2019;5:7. doi: 10.1038/s41514-019-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang L, Ye Q, Zhang X, Li K, Liang X, Wang M, et al. Pyrroloquinoline quinone extends. Food Funct. 2021;12:11319–30. doi: 10.1039/D1FO02128A. [DOI] [PubMed] [Google Scholar]
- 152.Parekh VV, Pabbisetty SK, Wu L, Sebzda E, Martinez J, Zhang J, et al. Autophagy-related protein Vps34 controls the homeostasis and function of antigen cross-presenting CD8α. Proc Natl Acad Sci USA. 2017;114:E6371–E6380. doi: 10.1073/pnas.1706504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lautrup S, Sinclair DA, Mattson MP, Fang EF. NAD+ in brain aging and neurodegenerative disorders. Cell Metab. 2019;30:630–55. doi: 10.1016/j.cmet.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lu YX, Regan JC, Eßer J, Drews LF, Weinseis T, Stinn J, et al. A TORC1-histone axis regulates chromatin organisation and non-canonical induction of autophagy to ameliorate ageing. Elife. 2021;10:e62233. doi: 10.7554/eLife.62233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hoong CWS, Chua MWJ. SGLT2 inhibitors as calorie restriction mimetics: insights on longevity pathways and age-related diseases. Endocrinology. 2021;162:bqab079. doi: 10.1210/endocr/bqab079. [DOI] [PubMed] [Google Scholar]
- 157.Smita SS, Trivedi S, Pandey T, Trivedi M, Pandey R. A bioactive compound shatavarin IV-mediated longevity as revealed by dietary restriction-induced autophagy in Caenorhabditis elegans. Biogerontology. 2020;21:827–44. doi: 10.1007/s10522-020-09897-5. [DOI] [PubMed] [Google Scholar]
- 158.Madeo F, Eisenberg T, Pietrocola F, Kroemer G. Spermidine in health and disease. Science. 2018;359:eaan2788. doi: 10.1126/science.aan2788. [DOI] [PubMed] [Google Scholar]
- 159.Liu W, Lin H, Mao Z, Zhang L, Bao K, Jiang B, et al. Verapamil extends lifespan in Caenorhabditis elegans by inhibiting calcineurin activity and promoting autophagy. Aging (Albany NY) 2020;12:5300–17. doi: 10.18632/aging.102951. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.