Abstract
SMART (a Simple Modular Architecture Research Tool) allows the identification and annotation of genetically mobile domains and the analysis of domain architectures (http://SMART.embl-heidelberg.de ). More than 400 domain families found in signalling, extracellular and chromatin-associated proteins are detectable. These domains are extensively annotated with respect to phyletic distributions, functional class, tertiary structures and functionally important residues. Each domain found in a non-redundant protein database as well as search parameters and taxonomic information are stored in a relational database system. User interfaces to this database allow searches for proteins containing specific combinations of domains in defined taxa.
INTRODUCTION
The explosion of sequence data increases the need for computational sequence analysis tools that annotate novel genes with predicted functions. Function prediction, however, is fraught with potential pitfalls such as considerable sequence divergence, non-equivalent functions of homologues and non-identical multi-domain architectures (1). Detecting non-enzymatic regulatory domains is essential to predict a protein’s cellular role, binding partners and subcellular localisation. Such domains are usually divergent in sequence and occur in contrasting multi-domain contexts. This leads to difficulties unravelling the evolution and function of multi-domain proteins. These problems are addressed by the SMART Web tool, which was first described by Schultz et al. (2) as a database for signalling domains. Here we report on the expansion of SMART’s domain coverage, its relational database system and the development of new Web tools for the analysis of mobile domains.
THE SMART ALIGNMENT SET
Domain detection in SMART relies on multiple sequence alignments of representative family members (2). In the past year, we have improved the alignment construction method to achieve higher levels of reproducibility and have increased the number of domain families detectable by SMART. Older alignments have been updated to integrate new homology and structural findings. As a consequence, SMART alignments are of high quality and have been exploited in recent comparative genomics studies (e.g., 3–5).
Alignment construction protocol
The starting point for the construction of a multiple sequence alignment that optimally represents a domain family, is an alignment of divergent family members based on known tertiary structures, where possible, or from homologues identified in a PSI-BLAST (6) analysis. These alignments are optimised manually and, following construction of a hidden Markov model (HMM) (7), used to search current sequence databases. Each sequence of the alignment is also used as a query in a PSI-BLAST search. All sequences that are significantly similar [as detected by HMM (E < 0.01) or PSI-BLAST (E < 0.001) searches] are added to the alignment using the sequence versus HMM alignment method of HMMer. Alignments are checked manually for potential false positives or misassembled protein sequences derived from genomic sources. From this alignment, one of each sequence pair sharing >67% identity is deleted to reduce redundancy. The resulting alignment is used as a starting point for a subsequent round of searches. This iterative procedure is pursued until no new homologues are detected.
Increased coverage
Originally, SMART was intended as a tool for the analysis of domains involved in eukaryotic signal transduction (2) but was expanded to detect domains of extracellular proteins and bacterial two-component regulatory systems (8). In 1999, domains associated with DNA, RNA, chromatin and actin cytoskeleton functions have been added (see http://SMART.embl-heidelberg. de/changes.shtml for a list of all new added domains). In addition, new reported domain families that fall within the categories covered by SMART have been incorporated. These include extracellular GPS (9) and PSI (10) domains, intracellular signalling domains as ENTH (11) and GoLoco (12) as well as domains in splicing factors [e.g., FF (13) and PWI (14)]. During this process, additional, previously undetected members are often recognized, as for example ENTH domains in Saccharomyces cerevisiae and mammalian huntingtin interacting proteins or PWI domains in fungal proteins. As a result of this improvement in coverage, SMART now includes >400 domains.
Updating of alignments
In 1999 more than 40 alignments have been updated (see http://SMART.embl-heidelberg.de/changes.shtml for a list). For instance, in cases where the tertiary structure of a domain has been solved, we ensured that domain boundaries derived from sequence analysis are consistent with the three dimensional structure. The histidine kinase structures (15–17), for example, revealed two structurally independent domains, namely A, which contains the phosphorylation site, and B, the catalytic core. The previous SMART histidine kinase alignment was therefore split into two domains, HisKA and HATPase_c, the latter includes heat shock protein 90 and DNA gyrase B homologues (18). Updates were also undertaken to ensure that newly-deposited sequences are suitably represented in the current SMART alignments. Recent identification of distant domain homologues such as SH3 (19) and VWA domains in prokaryotes (4) and VWA domains in integrin β-subunits (20), have been incorporated into the SMART database. Updates of domain families have resulted in unexpected structural or functional predictions. For example, revisiting the SET domain family resulted in the prediction that some plant N-methyltransferases (21) contain this domain (unpublished data). This suggests that SET domain proteins may possess methyltransferase activities.
DATA ACQUISITION AND INTEGRATION INTO A RELATIONAL DATABASE SYSTEM
SMART was designed to facilitate the study of domain evolution and multi-domain architectures by correlations with phyletic distributions. Consequently, it was essential that all members of a domain family complete with associated taxonomic information were recorded in an easy-to-retrieve format.
The SMART database
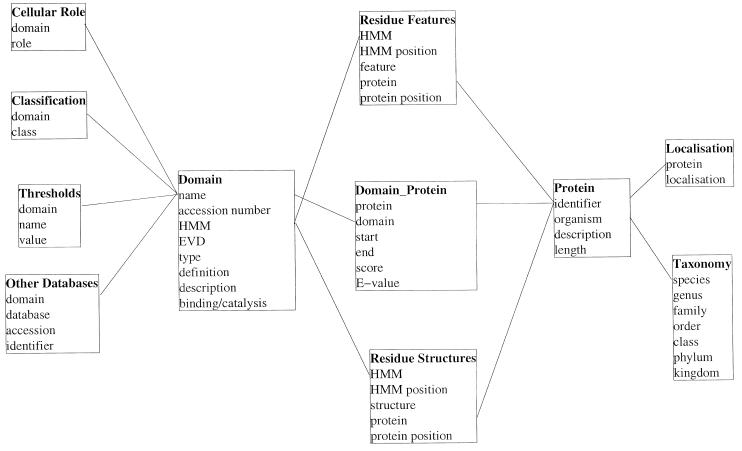
Information on >400 domain types in >54 000 different proteins is stored in SMART using a relational database management system (RDBMS; see http://www.PostgreSQL.org ). For each domain hit, boundaries, raw bit score and E-value are recorded. The protein accession code, description line, the sequence length and the species name are stored. To allow phylogenetic analyses, the full taxonomic description for each species derived from the NCBI Taxonomy database (see http://www.ncbi.nlm.nih.gov/Taxonomy/tax.html ) is also recorded. Each SMART domain is identified by a unique accession number, thus providing stable references for other domain databases and is linked to corresponding domains in Pfam (22) and PROSITE (23). By including into the database annotation, search parameters (see below) and cross-references to other domain databases, SMART has been converted into a relational database scheme, resulting also in increased system stability and easier maintenance (see Fig. 1 for the structure of the database).
Figure 1.
Structure of the relational database underlying SMART. Table names are in bold and lines indicate the relationships between tables in the RDBMS.
New searching method
To improve sensitivity of domain and repeat detection, SMART’s searching method has been changed to HMMs using the implementation of the HMMer2 program (7) (see http://hmmer. wustl.edu ). HMMer2 provides statistically sound E-values, giving a robust estimate of the significance of a domain hit. From a database search with a HMM derived from the SMART alignment, the highest per protein E-value of identified true positives (Ep) and the lowest per protein E-value of predicted true negatives (En) are stored within the SMART database. Similarly, for two or more repeats in a protein, the lowest E-value of a false positive repeat (Er) is stored. To ensure that the E-value thresholds are independent from the database size, the size of the protein database used when deriving the thresholds is also recorded. SMART will predict a domain homologue within any sequence, that has an E-value <Ep or else where Ep < E-value < En and E-value < 1.0. In cases where no repeat threshold is defined, all hits in a protein are reported, otherwise only those with E-values < Er are shown.
THE SMART WEB SERVER: NEW FEATURES
SMART offers different Web interfaces to query the underlying RDBMS for particular domain architectures. This query can be limited to specific taxonomic groups. In addition, we have improved the output of basic SMART searches, to present results in a more coherent and concise format.
Architecture SMART and Alert SMART
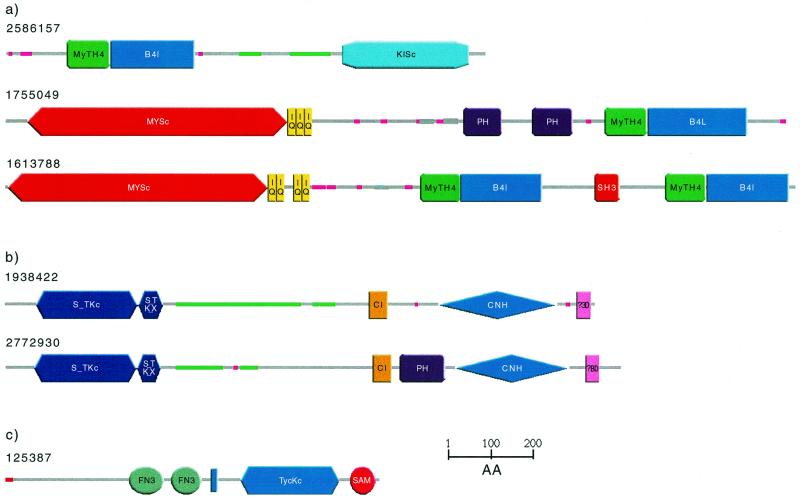
Architecture SMART allows users to search for specific domain architectures using an AND/NOT logic. Searches can be restricted to any taxonomic group. Selecting for plant proteins with B41 domains, for example, reveals a single domain architecture consisting of MyTH4, B41 and C-terminal kinesin motor (KISc) domains (Fig. 2a). In metazoa, by contrast, B41 domains can be found in combination with 18 other domains. Restricting the search to metazoan proteins with both B41 and MyTH4 domains reveals two distinct domain architectures (Fig. 2a) both of which contain an N-terminal myosin-like ATPase motor domain (MYSc). Thus, in plants and in metazoans, the B41/MyTH4 domain pair is combined with motor domains, but in contrasting domain architectures.
Figure 2.
Schematic representations, produced using SMART, of the domain architecture of proteins discussed in the text. Green/grey lines indicate predicted coiled coil regions and pink lines show low complexity segments. Regions of proteins without any predicted features are marked with grey bars and can be subjected individually to gapped BLAST searches. (a) Selective SMART. Domain architecture of plant (Arabidopsis thaliana; gi: 2586157) and metazoan (gi:1755049 Bos taurus; gi:1613788 Homo sapiens) proteins with MyTH4 and B41 domains. (b) Finding proteins with similar domain architecture. SMART detects no PH domain in the C.elegans protein K08B12.5 (gi:1938422), though all thus far sequenced proteins with an identical domain architecture show a PH domain between the C1 and the CNH domain (representative gi:2772930 Drosophila melanogaster). (c) Improved representation of results. SMART merges domain prediction and intrinsic features into a single line output. The red line indicates a signal sequence, the blue bar a transmembrane helix.
Users wishing to be kept informed by Email of sequences newly deposited in databases, that contain particular domains, should register their requirements using the alert SMART facility.
Finding proteins with similar domain architecture
SMART can search for all proteins that have an identical domain architecture as the query (having all the domains of the query protein in the same collinear order) or an identical domain composition (at least one of all domain types of the query protein, irrespective of order). Identification of proteins with identical, or near-identical, domain architectures as the query may improve predictions of protein, as opposed to domain, functions. This feature also reveals, using a taxonomic breakdown, the phyletic distribution of the architecture. In addition, it allows the detection of very divergent members of domain families that are not detectable by standard sequence searching methods. The Caenorhabditis elegans protein K08B12.5 (gi 1938422), for example, is predicted by SMART to contain the following domains: S_TKc, S_TK_X, C1, CNH and PBD (Fig. 2b). Searching for proteins that contain each of these domains in identical order demonstrates, that all such proteins possess a PH domain between the C1 and CNH domains (Fig. 2b). This suggests, that further investigation might also reveal a divergent PH domain in K08B12.5.
Improved representation of results
SMART analysis of a query sequence reveals not only domains, but also intrinsic features such as signal sequences (24), transmembrane helices (25), coiled coil regions (26) and compositionally biased regions (27). In the last year, methods for the prediction of GPI anchors (28) and for improved repeat detection (M.A.Andrade, EMBL, Heidelberg, unpublished data) have been added. To provide a comprehensive overview of these features, all predictions are merged into a single line output (Fig. 2c). The following priority list is used to resolve overlapping predictions based on the perceived prediction accuracy: Domain > Signal > TM > Coils > Seg. All predictions are also provided in a tabular format.
FUTURE PERSPECTIVES
SMART detects domains from sequences with relatively high selectivity and specificity. Domain families that contain extremely divergent representatives are deliberately targeted for inclusion in this database due to problems in their detection using other methods. Future work will focus on increasing the types of mobile domains detected and on improved functional predictions within single families.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank colleagues from the EMBL group for lively discussions and help, in particular B. Eisenhaber for linking the GPI prediction and M. A. Andrade for providing repeater to SMART. J.S., T.D. and P.B. are supported by the DFG and by the EC (grant 01KW9602/6) as well as by the BMBF grants MEDSEQ and TARGID. C.P.P. is supported by the Medical Research Council, UK.
REFERENCES
- 1.Doerks T., Bairoch,A. and Bork,P. (1998) Trends Genet., 14, 248–250. [DOI] [PubMed] [Google Scholar]
- 2.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chervitz S.A., Aravind,L., Sherlock,G., Ball,C.A., Koonin,E.V., Dwight,S.S., Harris,M.A., Dolinski,K., Mohr,S. et al. (1998) Science, 282, 2022–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting C.P., Aravind,L., Schultz,J., Bork,P. and Koonin,E.V. (1999) J. Mol. Biol., 18, 729–745. [DOI] [PubMed] [Google Scholar]
- 5.Copley R.R., Schultz,J., Ponting,C.P. and Bork,P. (1999) Curr. Opin. Struct. Biol., 9, 408–415. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbin R., Eddy,S., Krogh,A. and Mitchison,G. (1998) Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge, UK.
- 8.Ponting C.P., Schultz,J., Milpetz,F. and Bork,P. (1999) Nucleic Acids Res., 27, 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponting C.P., Hofmann,K. and Bork,P. (1999) Curr. Biol., 9, R585–R588. [DOI] [PubMed] [Google Scholar]
- 10.Bork P., Doerks,T., Springer,T.A. and Snel,B. (1999) Trends Biochem. Sci., 24, 261–263. [DOI] [PubMed] [Google Scholar]
- 11.Kay B.K., Yamabhai,M., Wendland,B. and Emr,S.D. (1999) Protein Sci., 8, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siderovski D.P., Diverse-Pierluissi,M. and De Vries,L. (1999) Trends Biochem. Sci., 24, 340–334. [DOI] [PubMed] [Google Scholar]
- 13.Bedford M.T. and Leder,P. (1999) Trends Biochem. Sci., 24, 264–265. [DOI] [PubMed] [Google Scholar]
- 14.Blencowe B.J. and Ouzounis,C.A. (1999) Trends Biochem. Sci., 24, 179–180. [DOI] [PubMed] [Google Scholar]
- 15.Park H., Saha,S.K. and Inouye,M. (1998) Proc. Natl Acad. Sci. USA, 95, 6728–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T., Saha,S.K., Tomomori,C., Ishima,R., Liu,D., Tong,K.I., Park,H., Dutta,R., Qin,L., Swindells,M.B. et al. (1998) Nature, 396, 88–92. [DOI] [PubMed] [Google Scholar]
- 17.Tomomori C., Tanaka,T., Dutta,R., Park,H., Saha,S.K., Zhu,Y., Ishima,R., Liu,D., Tong,K.I., Kurokawa,H. et al. (1999) Nature Struct. Biol., 6, 729–734. [DOI] [PubMed] [Google Scholar]
- 18.Mushegian A.R., Bassett,D.E.,Jr, Boguski,M.S., Bork,P. and Koonin,E.V. (1997) Proc. Natl Acad. Sci. USA, 94, 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whisstock J.C. and Lesk,A.M. (1999) Trends Biochem. Sci., 24, 132–133. [DOI] [PubMed] [Google Scholar]
- 20.Ponting C.P., Schultz,J., Copley,R.R., Andrade,M.A., and Bork,P. (2000) Adv. Protein Chem., in press. [DOI] [PubMed] [Google Scholar]
- 21.Klein R.R. and Houtz,R.L. (1995) Plant. Mol. Biol., 27, 249–261. [DOI] [PubMed] [Google Scholar]
- 22.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L.L. (2000) Nucleic Acids Res., 28, 263–266 (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 25.von Heijne G. (1992) J. Mol. Biol., 225, 487–494. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A., Van Dyke,M. and Stock,J. (1991) Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 27.Wootton J.C. and Federhen,S. (1996) Methods Enzymol., 266, 554–573. [DOI] [PubMed] [Google Scholar]
- 28.Eisenhaber B., Bork,P. and Eisenhaber,F. (1998) Protein Eng., 11, 1155–1161. [DOI] [PubMed] [Google Scholar]