Highlights
-
•
Biliary tract cancers (BTCs), including intrahepatic, perihilar, and distal cholangiocarcinoma as well as gallbladder cancer are rare but aggressive malignancies with few effective therapies.
-
•
Integrative clinical sequencing of 124 BTC patients at University of Michigan identified actionable/ potentially actionable aberrations in a majority of cases.
-
•
Patients treated with molecularly matched targeted therapies showed a significantly improved survival as compared to those who could not be treated with matched therapy.
Keywords: Cholangiocarcinoma, Next-generation sequencing, Precision oncology
Abstract
Background
Biliary tract cancers (BTCs) including intrahepatic, perihilar, and distal cholangiocarcinoma as well as gallbladder cancer, are rare but aggressive malignancies with few effective standard of care therapies.
Methods
We implemented integrative clinical sequencing of advanced BTC tumors from 124 consecutive patients who progressed on standard therapies (N=92 with MI-ONCOSEQ and N=32 with commercial gene panels) enrolled between 2011-2020.
Results
Genomic profiling of paired tumor and normal DNA and tumor transcriptome (RNA) sequencing identified actionable somatic and germline genomic alterations in 54 patients (43.5%), and potentially actionable alterations in 79 (63.7%) of the cohort. Of these, patients who received matched targeted therapy (22; 40.7%) had a median overall survival of 28.1 months compared to 13.3 months in those who did not receive matched targeted therapy (32; P < 0.01), or 13.9 months in those without actionable mutations (70; P < 0.01). Additionally, we discovered recurrent activating mutations in FGFR2, and a novel association between KRAS and BRAF mutant tumors with high expression of immune modulatory protein NT5E (CD73) that may represent novel therapeutic avenues.
Conclusions
Overall, the identification of actionable/ potentially actionable aberrations in a large proportion of cases, and improvement in survival with precision oncology supports molecular analysis and clinical sequencing for all patients with advanced BTC.
Introduction
Biliary tract cancers (BTCs) arise from the epithelial lining of the biliary ducts and comprise of intrahepatic and extrahepatic (perihilar and distal) cholangiocarcinoma (CCA), and gallbladder cancer. The incidence of these cancers is rising, driven predominantly by intrahepatic CCA [1], [2], [3]. A majority of these tumors are advanced and unresectable at diagnosis [1]. Prognosis of patients with advanced BTC remains poor with median overall survival (OS) from diagnosis of less than 12 months [4,5], and five-year survival rate of about 5% despite therapy [6]. Current systemic chemotherapy options for patients with advanced BTCs remain nonspecific and suboptimal, thus it is imperative to further our understanding of the molecular biology of this disease and to define more targeted and effective therapeutic options.
Molecular profiling of BTCs has identified many common drivers [7], [8], [9], [10], [11], as well as molecular aberrations associated with specific anatomical subgroups, for example, FGFR2 fusions, and mutations in IDH1/2, BAP1, ARID1A, and KRAS predominantly seen in intrahepatic CCA (iCCA) [12], [13], [14]; KRAS, TP53, and ARID1A mutations or amplification of ERBB2 or ERBB3 in extrahepatic CCA; and TP53, ERBB2/3, CDKN2A/B, and ARID1A in gallbladder cancer [13,15]. Among these, targeted therapy options have recently become available for BTC patients with FGFR2 and IDH1 aberrations. Pan-FGFR inhibitors pemigatinib and infigratinib received accelerated FDA approval for use in patients with FGFR2 fusion or translocation who had progressed on first-line therapy, following multiple clinical trials that demonstrated clinical benefit in refractory BTC patients with objective response rates (ORR) ranging from 25% to 36%, and disease control rates as high as 70% to 80% [16], [17], [18]. Ivosidenib, an IDH1 inhibitor, demonstrated a statistically significant improvement in median progression-free survival (PFS) from 1.4 months to 2.7 months and disease control rate from 28% to 53% in patients as compared to placebo which led to its FDA approval in 2021 [19]. Other promising targets in BTC under clinical investigation include BRAF V600E [20] and ERBB2/HER-2 amplification [21]. Additionally, application of immune checkpoint inhibitors (ICI) in BTCs has yielded variable benefit across several trials evaluating role of ICI as single agent or dual therapy, and combination with chemotherapy. In the frontline setting, the combination of chemotherapy with ICI resulted in response rates ranging from 27-73% and median PFS of 4.3–11.0 months and median OS of 10.6–20.7 months [22], [23], [24], [25]. In the refractory population, single agent or combination immunotherapy demonstrated an ORR ranging from 5.8 to 23% with median PFS of 1.5–3.6 months and median OS of 4.3–14.23 months [24,[26], [27], [28], [29], [30]. In many of these trials, the median duration of response had not been reached suggesting a subset of patients had durable response. Fewer than 5% of patients with BTC have underlying microsatellite instability /deficient mismatch repair or high tumor mutational burden for which ICI has received FDA approval in a tissue-type agnostic manner.
Herein, we summarize findings from clinical sequencing of 124 BTC patients with a focus on defining the spectrum of molecularly matched therapeutic options for this rare cancer and assess their impact on the clinical management of patients.
Materials and methods
Sequencing was performed via the MI-ONCOSEQ program using standard protocols under Institutional Review Board (IRB HUM00046018, HUM00067928, HUM00056496) approved studies at Michigan Center for Translational Pathology a Clinical Laboratory Improvement Amendments (CLIA) compliant sequencing lab at University of Michigan [31], [32], [33], [34]. Patients enrolled in the MI-ONCOSEQ study provided written informed consent to perform comprehensive molecular profiling of tumor/germline exomes and tumor transcriptome on either fresh tumor biopsies or (FFPE) tissue blocks. In addition, patient data was collected from the electronic medical records under IRB application HUM00165244.
Next generation sequencing library preparation
Sample details, including age, gender, and disease stage are summarized in Table 1 and Supplementary Table S1. Tissue acquisition, pathology review and sequencing of matched pair (tumor/normal DNA) exome, and tumor only transcriptome libraries were prepared using previously described protocols [32]. Samples with low tumor content were macro-dissected to enrich for tumor tissue based on pathologist assessment. “Human All Exon v4” Agilent exon probes and a selected target capture panel probes were used to capture tumor DNA and enriched following manufacturer's protocol (Agilent/Roche). DNA/RNA paired-end sequencing libraries were sequenced using the Illumina HiSeq 2000 or HiSeq 2500 (2 × 100 nucleotide read length) (Illumina Inc. San Diego, CA).
Table 1.
Patient Characteristics.
| All | Actionable Matched Treated |
Actionable Matched Untreated |
Non-actionable | |
|---|---|---|---|---|
| Total, N (%) | 124 (100) | 22 (17.7) | 32 (25.8) | 70 (56.5) |
| Age, years | ||||
| Median | 59 | 56 | 58 | 61 |
| Range | 17-80 | 27-75 | 17-72 | 20-80 |
| Sex, N (%) | ||||
| Female | 65 (52) | 13 (59.1) | 19 (59.4) | 33 (47.1) |
| Male | 59 (48) | 9 (40.1) | 13 (40.6) | 37 (52.9) |
| Race, N (%) | ||||
| White or Caucasian | 111 (89.5) | 21 (95.5) | 30 (93.8) | 59 (84.3) |
| Asian or Asian American | 5 (4.0) | 1 (3.1) | 3 (4.3) | |
| Black or African American | 4 (3.2) | 1 (3.1) | 5 (7.1) | |
| American Indian | 2 (1.6) | 1 (4.5) | 1 (1.4) | |
| Other or missing | 2 (6) | 2 (2.9) | ||
| Primary tumor site, N (%) | ||||
| Intrahepatic | 88 (71) | 21 (95.4) | 27 (84.4) | 40 (57.1) |
| Extrahepatic perihilar | 10 (8) | 1 (3.1) | 9 (12.9) | |
| Extrahepatic distal | 8 (6) | 2 (6.3) | 6 (8.6) | |
| Gallbladder | 13 (10) | 1 (3.1) | 12 (17.1) | |
| Mixed hepatocellular/ cholangiocarcinoma | 5 (4) | 1 (4.5) | 1 (3.1) | 3 (4.3) |
| First-line systemic therapy, N (%) | ||||
| Gemcitabine/platinum +/- agent | 90 (72.6) | 16 (72.7) | 24 (75.0) | 50 (71.4) |
| Gemcitabine/taxane | 4 (3.2) | 3 (9.4) | 1 (1.4) | |
| 5-fluorouracil based regimen | 15 (12.1) | 2 (9.1) | 1 (3.1) | 12 (17.1) |
| Immunotherapy only | 5 (4.0) | 1 (4.5) | 1 (3.1) | 3 (4.3) |
| Other | 4 (3.2) | 3 (13.6) | 1 (1.4) | |
| Unknown or none | 6 (4.8) | 3 (9.4) | 3 (4.3) | |
| NGS Platform, N (%) | ||||
| MI-ONCOSEQ | 92 (74.2) | 14 (63.6) | 27 (84.4) | 51 (81.4) |
| Other | 32 (25.8) | 8 (36.4) | 5 (15.6) | 19 (27.1) |
| Stage at biopsy, N (%) | ||||
| Resectable | 16 (12.9) | 2 (9.1) | 3 (9.4) | 11 (15.7) |
| Locally advanced unresectable | 11 (8.9) | 1 (4.5) | 4 (12.5) | 6 (8.6) |
| Metastatic | 97 (78.2) | 19 (86.4) | 25 (78.1) | 53 (75.7) |
| Biopsy specimen, N (%) | ||||
| Primary | 61 (49) | 13 (59.1) | 20 (62.5) | 29 (41.4) |
| Metastatic | 63 (51) | 9 (40.9) | 12 (37.5) | 41 (58.6) |
| Biopsy in relation to chemotherapy, N (%) | ||||
| Pre-chemotherapy | 41 (33) | 5 (22.7) | 13 (40.6) | 23 (32.9) |
| Post-chemotherapy | 83 (67) | 17 (77.3) | 19 (59.4) | 47 (67.1) |
NGS, next-generation sequencing; MI-ONCOSEQ, Michigan Oncology Sequencing
Exome sequencing analysis
Whole Exome paired end Fastq sequence files were aligned to GRCh37 genome build using Novoalign multithreaded (version 2.08.02, Novocraft). Novosort and Picard (version 1.93) were used to sort, index and remove duplicates from the aligned bam files. Mutation analysis was carried out on matched normal–tumor pairs using freebayes (version 1.0.1) and pindel (version 0.2.5b9) as previously described [31,32,35]. Somatic SNV and Indel files from freebayes and pindel were postfiltered using at least 5% variant allelic fraction, minimum six variant reads, <2% variant allelic fraction in normal with at least 20X coverage. The indel thresholds were optimized using a pool of hundreds of matched normal samples sequenced using the same protocol and platform as described [35]. Germline mutation analysis was performed using at least 10 variant reads in normal sample, with >=20% allelic fraction and, <1% population frequency in 1000 Genomes and ExAC. Variant annotation was performed using snpEff and snpSift (version 4.1g) based on refseq (from UCSC genome browser, retrieved on 8/22/2016), COSMIC v79, dbSNP v146, ExAC v0.3, and 1000 Genomes phase 3 databases.
Copy number aberration analysis was performed on exome data using DNAcopy (version 1.48.0) to get CBS segments, regions were normalized for GC content, and log2-transformed exon coverage ratio between tumor/normal samples across the targeted regions were calculated as previously described [32,35]. Cohort wise copy number analysis was performed by merging all the segment files used as input to gistic version 2, and maftools was used to generate cumulative copy number plot.
RNA sequencing data analysis
Strand-specific RNA sequencing (RNA-seq) libraries were used for gene expression and fusion analysis. Gene expression quantification was performed using kallisto version 0.43.1, transcript per million (TPM) values were used as input for qlucore omics (https://www.qlucore.com/) software for downstream expression analysis. Genes with transcripts with <1 TPM in at least 95% of the cohort were removed and the data was transformed to log2. The expression data was normalized for preservation method (FFPE/Fresh Frozen), biopsy sites and tumor content. Unsupervised hierarchical clustering was performed for the 69 immune marker genes, including 66 genes recently evaluated (Cancer Genome Atlas Research Network) plus IFN-γ responsive chemokines (CXCL9-11). Fusion calling was performed using a combination of CRISP, CODAC MI-ONCOSEQ pipeline [32,35], fusioncatcher_v1.10 [36] and arriba_v1.1.0. [37] The fusions calls were compiled and reported in Supplementary Table S8.
Mutation burden estimation
Freebaye's mutation calls were used for the mutation burden estimation. Mutations were filtered for coverage (>=10x) and variant allelic fraction (>=6%). Mutation burden was expressed as (number of mutations/ total covered bases) × 106. Varscan2 processed VCF files from TCGA CCA cohort (N=51) were downloaded from the GDC data portal and lifted-over from the GRCh38 to GRCh37 reference genome using CrossMap for comparison with the MI-ONCOSEQ cohort.
Pathogenic germline variant analysis
Pathogenicity of germline variants were determined through review of the published literature, public databases including but not limited to ClinVar, the Human Genome Mutation Database, Leiden Open Variation Databases, and variant-specific databases. Only cancer-relevant germline variants that had been previously categorized as pathogenic or likely pathogenic in ClinVar, or adjudicated at the precision molecular tumor board as pathogenic were included in the study.
Survival analysis
Subject efficacy data was manually extracted from review of electronic medical records. OS was defined as the duration of time from the date of advanced unresectable or metastatic disease until death from any cause. Follow-up time was censored at the date of last disease evaluation. The survival analysis was estimated using the product-limit method of Kaplan and Meier (GraphPad Prism 8, San Diego, CA). The analyzes should be considered post hoc, and the results herein exploratory with the intention to guide further definitive studies. A significance threshold for P value was arbitrarily set to 0.05 for all statistical tests.
In order to associate patient outcome with reported molecular alterations, we included all consecutive subjects with BTC with targeted gene panel analysis completed using alternative CLIA platforms at our institution. Gene alterations predictive of response to an FDA approved drug(s) were classified as ‘actionable (Tier 1)’, and aberrations associated with potential responsiveness to experimental drugs based on emerging data from ongoing clinical trials or compelling pre-clinical evidence were designated as ‘potentially actionable (Tier 2)’, and frequent aberrations noted in this cohort for which currently no therapeutic approach is available were deemed non-actionable (Supplementary Table S9).
Results
Somatic aberration landscape of advanced BTCs presents diverse therapeutic avenues
Clinical sequencing data was obtained from a total of 124 consecutive patients with advanced BTC (from a total of 239 patients enrolled between September 2011 and February 2020 (Fig. S1). The sequencing cohort was comprised of 52% women, median age of 59 (range, 17–80) years), including intrahepatic (N=88), perihilar (N=10), and distal (N=8) CCA, mixed hepatocellular/CCA (N=5), and gallbladder cancer (N=13), with 83 (67%) cases being post-chemotherapy and 63 (51%) metastatic (Table 1 and Supplementary Table S1). We obtained high quality exome sequencing data from 92 tumor/normal samples at MI-ONCOSEQ as indicated by the 94% median alignment rate (range, 53-97%), mean coverage of 203X for whole exome (WXS) and 506X for target capture panel, and overall low PCR duplication rate averaging 8% (range 0.6–79%) (Supplementary Table S2). In parallel, high quality capture transcriptome sequencing data from 85 tumor tissues was analyzed for gene fusions (Supplementary Table S7), and gene expression (Supplementary Table S8). Additionally, tumors from 32 cases were analyzed through other CLIA-approved gene panels from commercial vendors including 30 from Foundation Medicine, and 2 from Guardant Health) (Supplementary Table S4).
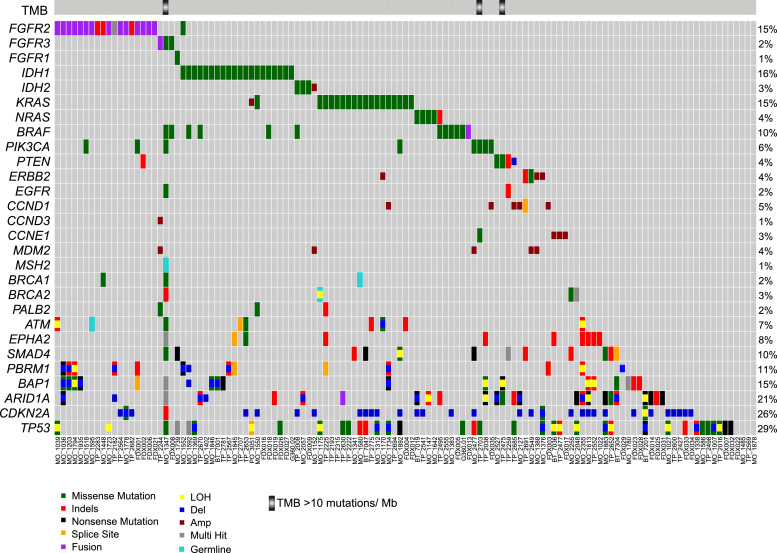
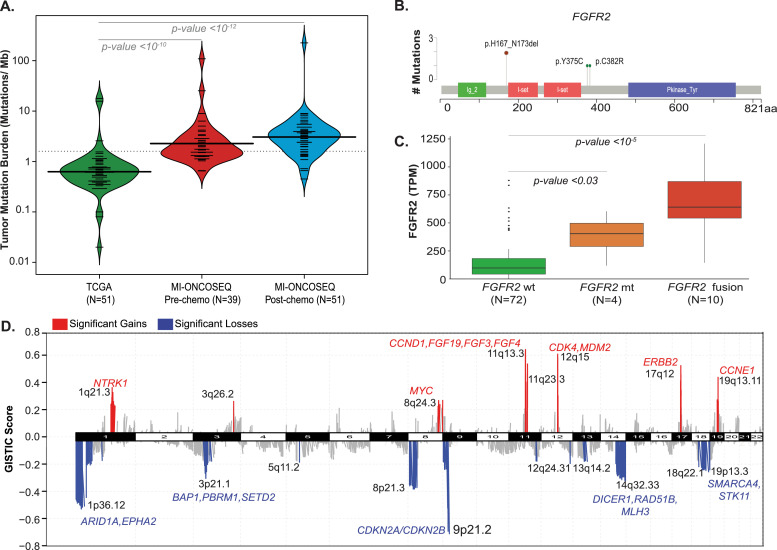
Somatic mutations (Supplementary Tables S3 and S4), copy number aberrations (Supplementary Table S6), gene fusions (Supplementary Table S7), and tumor mutation burden (Supplementary Table S5) from all the cases were assessed for potential clinical relevance and summarized in executive reports returned to the treating oncologists. The most frequently altered genes in the cohort included TP53 (N=43, 34.7%) and CDKN2A (N=36, 29%) followed by ARID1A (N=25, 20.2%), IDH1/2 (N=23, 18%), BAP1 (N=19, 15.3%), FGFR2/3 (N=25 20.2%), KRAS (N=19, 15%), and PBRM1 (N=13, 10%) (Fig. 1). Among driver aberrations, FGFR2, KRAS and IDH1 were largely mutually exclusive except for one patient with concurrent mutations in IDH1 and KRAS. Actionable genomic alterations (Tier 1), defined as predictive of response to FDA approved drug(s) in any cancer, were noted in a total of 54 cases (43.5%), and included hotspot activating/hotspot mutations in IDH1/2 (N=23), gene fusions in FGFR2/3 (N=15), BRAF V600E mutation (N=4), ERBB2 amplification (N=4), deleterious mutations in BRCA1/BRCA2 (N=4), and KRAS G12C (N=3). Apart from mutations, high mutation burden in tumors also defines an actionable aberration, that can be potentially matched with checkpoint blockade immunotherapy. Enumeration of mutation burden in the MI_Oncoseq cohort identified three cases with high tumor mutation burden, defined as >10 mutations/Mb (Supplementary Table S5). These included MO_1347, a 46-year-old male with metastatic CCA (and a history of ampullary carcinoma) previously treated with gemcitabine and cisplatin; a lymph node biopsy from this case, histologically seen as poorly differentiated high-grade adenocarcinoma admixed with prominent inflammation, was found to harbor 225 mutations/MB and high microsatellite instability (MSI-high) score, consistent with a biallelic loss of function of the mismatch repair deficiency gene MSH2 (with truncating germline mutation MSH2 c.2494G>T; p.Glu832Ter; dbSNP: rs863225396), coupled with the somatic loss of heterozygosity through the splice acceptor mutation, MSH2 c.1662-1G>A. No specific mutation or extrinsic etiology could be associated with the high mutation burden of 109 mutations/MB in the tumor from TP_2475, a 62-year-old female with stage IV metastatic CCA, “mixed” subtype (CMS-HCC) previously treated with CDDP/gemcitabine. The third case with high mutation burden, TP_2703 with 25.3 mutations/MB displayed mutation signature 4 (associated with tobacco smoking [18,19]), consistent with the patient's 30 pack-year history of smoking. The average mutation burden in the MI_Oncoseq cohort, after excluding 2 cases with low tumor content and one, MO_1347 with MSI high associated outlier mutation burden (Supplementary Table S5), was calculated as 4.3 mutations/Mb (range 0.45 to 108.9 mutations/Mb). This mutation burden in the cohort of advanced, metastatic tumors was found to be significantly higher than that of the TCGA-BTC cohort comprised of primary tumors (Wilcoxon rank test p-value 0.05*, Fig. 2A), consistent with similar observations across tumor types (for example, Robinson et al [32]). No significant difference was noted in the mutation burden of tumor samples post-chemotherapy (N=52), compared to advanced tumors prior to chemotherapy (N=39).
Fig. 1.
Integrative landscape of molecular alterations in biliary cancer. Landscape of molecular alterations in biliary tract cancer cohort (n=124). Each column represents a sample, identified at the bottom, each row represents a gene shown on the left, arranged according to functional groups as indicated. The bar on top displays cases with tumor mutation burden (TMB >10 mutations/Mb). Mutation types are color coded: missense (green), indels (red), nonsense (black), splice site (orange), pathogenic germline (cyan), fusion (purple), copy number deletion (blue), copy number amplification (dark red), multiple hits (grey), and promotor mutation (dark pink). Respective percentages of cases with gene aberrations are shown on the right.
Fig. 2.
Frequent genomic aberrations seen in biliary cancer. (A) Violin plots showing mutation burden across cholangiocarcinoma cohorts: TCGA-CHOL, MI-ONCOSEQ pre- and post-chemotherapy BTC cohort. The overall mutation burden is significantly higher in MI-ONCOSEQ advanced biliary tract cohort relative to the primary tumors in the TCGA cohort. (B) A summary of mutations in chromatin modifier genes in the BTC cohort. Individual samples represented in columns, genes in rows. The bar graphs on the right summarize the mutation frequency and number of samples mutated for each gene. Molecular alterations are color coded as described in Fig. 1. (C) Copy number landscape of the BTC cohort (GISTIC2, Q < 0.05), significant losses or gains with interesting genes are represented in blue and red color, respectively.
Gene fusion analysis using RNA-seq data identified known [12] and novel translocation events in 12 (9.7%) patients, including FGFR2, FGFR3 and YAP1 fused in frame with known and novel partners (Fig. S2). FGFR translocations were enriched in iCCA subtype (N=9) with three cases of FGFR2-BICCI, and one each with FGFR2-KIAA1967, FGFR2-AFF4, FGFR2-AHCYL1 and FGFR2-CCDC6 fusion, and one each with novel partners including, FGFR2-TAX1BP1 and MATN4-FGFR2. One gallbladder patient was identified to have FGFR3-TACC3 fusion, and one patient with mixed hepatocellular and CCA subtype had FGFR2-BICCI fusion (Fig. S1). All the FGFR rearrangements were found to retain the kinase domain and all the FGFR fusion partners potentially exhibited oligomerization capability, suggesting a shared mode of kinase activation as noted previously [12].
In addition to FGFR2 gene fusions, two samples had hotspot activating mutations p.Y375C (also reported as p.Y276C) and p.C382R (also reported as p.C383R), and two cases had a novel in-frame indel p.H167_N173del located in the extracellular domain (Figs. 2B and S3). Importantly, a significant upregulation of FGFR2 gene expression (P= 0.029) was noted in the FGFR2 mutants (N=4) as compared to the wild type cases (Fig. 2C), suggesting that the two patients with the novel indels represent a potentially activating aberration. Moreover, the median OS of patients in the fusion cohort (N=12; 21.3 months) and mutation cohort was similar (N=4; 21.5 months).
Patient MO_1778 with perihilar CCA exhibited two known, recurrent driver oncogenic fusions: FGFR2- BICC1 and YAP1-MAML2. While the FGFR2 fusion is a known driver in iCCA, recurrent YAP1-MAML2 fusion associated with aberrant Hippo pathway signaling has not been reported in CCA, but has been previously identified in other cancers [38], [39], [40], [41]. The YAP1-MAML2 fusion encodes TEAD1, WW1 and WW2 domains from YAP1 and loss of Notch interaction domain in MAML2, associated with transactivation of TEAD target genes leading to dedifferentiation or proliferation [39,42,43].
Potentially actionable aberrations and novel avenues for targeted therapies in advanced BTCs
A total of 79 (63.7%) cases harbored one or more potentially actionable (Tier 2) aberrations for which preliminary clinical/ preclinical rationale is available to match with experimental targeted therapeutics in ongoing clinical trials (Supplementary Table S9). Among the most common aberrations in this category were 35 patients with homozygous deletion or biallelic loss of Cyclin Dependent Kinase Inhibitor 2A, CDKN2A (p16INK4), associated with potential sensitivity to CDK4/6 inhibitors.; 21 patients with activating mutations in KRAS/NRAS (and one case with deleterious mutation in NF1), that may be considered for treatment with novel KRAS and/or MEK inhibitors; and 23 patients with truncating mutations in ARID1A, a SWI-SNF pathway regulator, potentially associated with synthetic lethality to PARPi, ATRi or EZH2i. Additional cases with potentially actionable aberrations included tumors with amplification of MDM2 (with wild type TP53); CDK4 and CCND1 (with wild type RB1), NTRK1, MYC and CCNE1 (Fig. 2D), supported by outlier expression of these genes (data not shown). We identified recurrent in frame indels in FGFR2 that may represent gain of function mutations responsive to FGFR inhibitors as recently shown, and non-BRAF V600 mutations (class II and class III) [44] that may respond to MEK inhibitors. Overall, 105 out of 124 (84.7%) cases analyzed, were determined to harbor one or more actionable or potentially actionable aberrations that could be matched with FDA approved or experimental therapies in ongoing clinical trials.
Germline alterations
Pathogenic germline mutations were noted in 6 patients in the MI_Oncoseq cohort (6.5%) with majority in DNA damage repair pathway genes (2 cases of MUTYH, one each of BRCA1, BRCA2, ATM and MSH2), and one case with germline mutation in FH, an essential gene in the tricarboxylic acid cycle (Fig. 1; Supplementary Tables S3 and S4). Three patients with pathogenic germline mutations in MSH2, MUTYH and BRCA2 were found to harbor a second somatic aberration in the tumor resulting in biallelic loss.
Targeted therapies and survival
The median OS for the expanded cohort (MI-ONCOSEQ and other CLIA platforms) from date of diagnosis of advanced unresectable or metastatic disease was 15.2 months (range, 1.5–96.9), and from date of diagnosis was 19.2 months (range, 1.3–166). The median follow-up from date of release of genomic analysis report was 8.6 months (range, -1.4–61.3). We observed no significant imbalances in the baseline characteristics and treatment variables between the actionable matched and unmatched cohorts, including gender, age, ECOG performance status, FGFR status, exposure to platinum therapy, or number of lines of therapy, or distance from cancer center (P > 0.05 by Fisher's exact test or Wilcoxon test; data not shown).
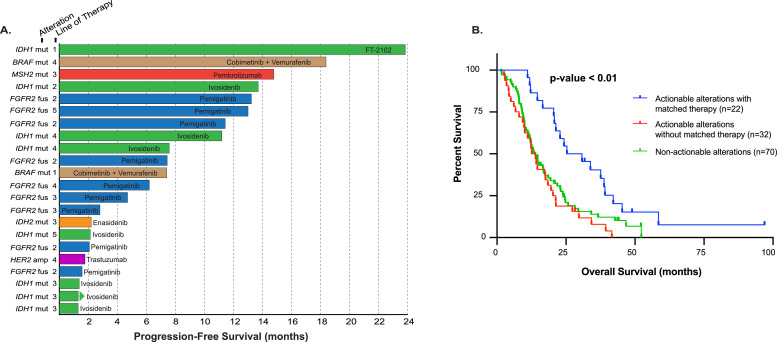
In the actionable cohort (N=54; 43.5%), defined by the presence of a molecular aberration that can be matched with an FDA approved therapeutic, 22 (40.7%) subjects received a molecularly matched therapy (matched treated cohort) off-label or on clinical trials (Table 2), while 32 (59.3%) patients did not receive molecularly matched therapy (matched untreated cohort). The remaining patients were defined as non-actionable for this analysis (70; 56.5%). In the matched treated group, patients received matched therapy after failure of systemic chemotherapy with the exception of one subject who received cobimetinib and vemurafenib off-label as first-line therapy for BRAF V600E mutation (Table 2, Fig. 3A). Patients with actionable mutations who received a matched therapy (N=22) had significantly longer OS than the 70 patients in the non-actionable group, or 32 patients in the matched untreated group (28·1 months, 13.3 months and 13.9 months, respectively; P < 0.01). The median OS between the matched treated and untreated arms of the actionable cohort had a hazard ratio of 0·33 (95% CI, 0·18-0·60, P < 0·01). However, median OS did not differ between the untreated actionable group and non-actionable group (HR 1.13, 95% CI, 0·80-2.00, P=0·31) (Fig. 3B).
Table 2.
Patients in the study who underwent treatment with experimental agents.
| Case ID | Gender | Age (years) | Anatomic subtype | Stage at diagnosis | Pre-biopsy treatment(s) for advanced disease | Actionable Alterations | Potentially Actionable Alterrations | Other Notable Alterations | Post-biopsy Treatment(s) |
|---|---|---|---|---|---|---|---|---|---|
| MO_1022 | M | 49 | iCCA | Localized resectable | Capecitabine/ oxaliplatin | Gemcitabine/capecitabine/bevacizumab (OL); Tivantinib/gemcitabine; Erlotinib/bevacizumab; MEK inhibitor (CT); cabozantinib (OL) | |||
| MO_1175 | M | 66 | CUP (iCCA) | Metastatic | Gemcitabine/carboplatin; FOLFOX/ bevacizumab | gBRCA2 E13fs | KRAS G12D, TP53 R273C, GNAS R201H | MEK inhibitor (CT) | |
| MO_1203 | F | 46 | iCCA | Localized resectable | Gemcitabine/ cisplatin; FOLFOX; IAP antagonist (CT) | FGFR2-CCAR2 fusion | BAP1 Y173C, PBRM1 E991fs | SMAC mimetic (CT); EGFR antibody (CT) | |
| MO_1338 | M | 42 | iCCA | Metastatic | Gemcitabine/ cisplatin | STK11 deletion | SMAD4 deletion, KEAP1 deletion, GNA11 deletion, TP53 P98fs, gFH K477dup- likely pathogenic/ VUS | 5FU (CT); mTOR inhibitor (OL) | |
| MO_1347 | M | 45 | iCCA | Metastatic | Gemcitabine/cisplatin | Hypermutation (TMB>10), gMSH2 E832*+sMSH2 c.1662-1G>A splA | EGFR Q787R, BRCA2 T3033fs, CDKN2A P105fs, ARID1A P1326fs | TP53 R174G and R158H, BAP1 V27fs, TET2 K534* | MEK inhibitor (CT) |
| MO_1369 | F | 61 | iCCA | Localized resectable | Gemcitabine/ cisplatin; RFA; Capecitabine/RT; SBRT; Capecitabine/ oxaliplatin; Capecitabine/ gemcitabine | IDH1 R132C | TP53 R248Q | Ivosidenib (CT) | |
| MO_1388 | M | 44 | iCCA | Metastatic | Gemcitbaine/ cisplatin | ERBB2 amplification & outlier expression | MDM2 amplification + outlier expression | FOLFIRI; FOLFOX; Trastuzumab (OL) | |
| MO_1518 | F | 63 | CUP (iCCA) | Metastatic | None | FGFR2-CCDC6 fusion | PIK3CA E545G | Gemcitabine/cisplatin; Pemigatinib (CT); Anti-LAG3 antibody (CT) | |
| MO_1595 | M | 50 | iCCA | Metastatic | Gemcitabine/ cisplatin; FOLFOX | FGFR2-MATN4 fusion | gATM R2832C | FOLFIRI; Pemigatinib (CT) | |
| MO_1613 | M | 70 | iCCA | Locally advanced | Gemcitabine/ nab-paclitaxel (CT); Gemcitabine/ cisplatin; FOLFOX | ARID1A p.Q177*+LOH | BAP1 R252fs+LOH, CDKN2A deletion | FOLFIRI; CDK4/6 inhibitor (CT) | |
| MO_1642 | M | 53 | iCCA | Metastatic | Gemcitabine/ cisplatin; Gemcitabine/ carboplatin | NRAS G12D | Anti-PD-L1 antibody/TAK-659 (CT) | ||
| MO_1723 | F | 50 | Mixed | Locally advanced | Gemcitabine/ carboplatin; SIRT; FOLFOX; TACE | FGFR2-AHCYL1 fusion | Pemigatinib (CT) | ||
| MO_1778 | M | 27 | CUP (iCCA) | Metastatic | Cisplatin/ etoposide; FOLFIRINOX | FGFR2-BICC1 fusion | CDKN2A p.H83Y+LOH | Pan FGFR inhibitor (CT) | |
| MO_1780 | M | 71 | Mixed | Metastatic | FOLFOX/ bevacizumab; Capecitabine/ bevacizumab; Sorafenib | BAP1 K421fs+c.375+1G>T splice donor | Anti-PD-1 antibody (OL) | ||
| MO_1794 | F | 51 | iCCA | Metastatic | Gemcitabine/ cisplatin | FGFR2-BICC1 fusion | BAP1 T203K, PBRM1 R690fs | FOLFOX; Pemigatinib (CT); FOLFIRI; SIRT; Gemcitabine/paclitaxel; Sunitinib (CT) | |
| MO_1883 | F | 57 | GB | Metastatic | Gemcitabine/ cisplatin | CDKN2A deletion, ARID1A Q479* | TP53 R175H+LOH, SMAD4 G386D+LOH | Gemcitabine/capecitabine; CTLA-4 antibody/PD-1 antibody (CT) | |
| MO_2057 | F | 63 | CUP (iCCA) | Locally advanced | Carboplatin/ etoposide; Capecitabine/ temozolomide | IDH2 R172W | ARID1A D2178fs+LOH | RASA1 p.L870fs+LOH | Gemcitabine/oxaliplatin; Enasidenib (OL) |
| MO_2127 | M | 20 | iCCA | Metastatic | Gemcitabine/ cisplatin/anti-PD-1 antibody (CT) | CCND1 amplification+outlier expression, ARID1A Q335*+LOH | TGFBR1 I109fs+LOH, APC P1594fs+LOH, CDKN2A deletion | FOLFOX | |
| MO_2549 | F | 52 | iCCA | Metastatic | Gemcitabine/ cisplatin | ERBB2 amplification+outlier expression | MDM2 amplification+outlier expression | CDKN2A/B deletion | PARP inhibitor/PD-1 antibody (CT) |
| TP_2475 | F | 62 | Mixed | Locally advanced | None | Hypermutation (TMB >10) | ARID1A P153A, PTEN S129G | BAP1 p.E31*+LOH, RB1 p.S567*+LOH | Gemcitabine/cisplatin; Anti-CTLA-4 antibody/anti-PD-1 antibody (CT); Capecitabine/oxaliplatin |
| TP_2495 | F | 67 | iCCA | Metastatic | None | BRAF D594G, ARID1A p.E1836fs, NRAS p.V14_Gdel in frame | gMUTYH G393D, TP53 N29fs+Splice Donor | Gemcitabine/cisplatin; FOLFOX; Regorafenib (CT); FOLFIRI | |
| TP_2541 | M | 59 | iCCA | Localized resectable | None | NRAS G13R | CDKN2A deletion | Gemcitabine/cisplatin/anti-PD-1 antibody (CT) | |
| TP_2670 | F | 65 | iCCA | Metastatic | None | IDH1 R132G | ARID1A P120fs | RASA1 I859fs | PARP inhibitor/anti-PD-1 antibody (CT) |
| TP_2682 | M | 43 | iCCA | Localized resectable | Gemcitabine/ cisplatin | TSC1 R786*+LOH | MAX R33*+LOH, NF2 K44*+LOH, CDKN2A deletion | Capecitabine/gemcitabine; 5-FU/nal-irinotecan/anti-PD-1 antibody (CT) | |
| TP_2694 | F | 55 | iCCA | Metastatic | None | FOXQ1 E147*+ E107K, KRAS G12V | Gemcitabine/cisplatin; 5-FU/nal-irinotecan/anti-PD-1 antibody (CT); FOLFOX; Gemcitabine/nab-paclitaxel | ||
| BT_7001 | M | 57 | iCCA | Metastatic | None | IDH1 R132G | BAP1 L100R+LOH | Gemcitabine/cisplatin/anti-PD-1 antibody (CT); Ivosidenib (CT) | |
| BT_7019 | M | 51 | iCCA | Metastatic | Anti-CTLA-4 antibody/anti-PD-1 antibody (CT) | ARID1A Q67*+LOH | NRAS G12D, CDKN2A deletion | Gemcitabine/cisplatin/nab-paclitaxel; Capecitabine/gemcitabine | |
| BT_7036 | M | 59 | iCCA | Metastatic | Anti-CTLA-4 antibody/anti-PD-1 antibody (CT) | CCNE1 amplification+moderate expression | TP53 p.R175H | Gemcitabine/cisplatin; FOLFOX | |
| BT_7047 | M | 61 | iCCA | Locally Advanced | Gemcitabine/ cisplatin/ anti-PD-1 antibody (CT) | TP53 H178fs+LOH, SMAD4 p.E49*+LOH, CDKN2A deletion, KRAS G12D, KDM6A deletion | SIRT; resection followed by Gemcitabine/cisplatin, SBRT | ||
| BT_7201 | F | 54 | iCCA | Metastatic | None | ARID1A p.R1528*+LOH | TP53 p.R306*+LOH, CDKN2A p.H83Y+LOH | Gemcitabine/cisplatin; PARP inhibitor/anti-PD-1 antibody (CT); Capecitabine/oxaliplatin; Gemcitabine/nab-paclitaxel | |
| BT_7304 | F | 66 | GB | Metastatic | Capecitabine/ gemcitabine | ARID1A E2250fs+LOH, AKT2 amplification | BAP1 D672G+LOH, SMAD4 splice acceptor+LOH | 5-FU/nal-irinotecan/anti-PD-1 antibody (CT); Capecitabine/oxaliplatin | |
| G360.01 | M | 75 | iCCA | Metastatic | None | BRAF V600E | TP53 G279E | Cobimetinib + vemurafenib (OL) | |
| FDX001 | M | 54 | iCCA | Metastatic | FOLFIRI plus nab-paclitaxel (CT) | FGFR2-SLMAP fusion | PIK3CA H1047L | BAP1 splice site 255+2T>G, PBRM1 L848fs*16 | Pemigatinib (CT) |
| FDX002 | F | 60 | iCCA | Metastatic | Gemcitabine/ cisplatin | FGFR2-SHROOM3 fusion | PTEN D268fs*30, PIK3CA K593fs*8 | CDKN2A A17_G23>GR, TERT promoter -146C>T | Pemigatinib (CT) |
| G360.02 | F | 57 | iCCA | Metastatic | None | IDH1 R132C | MYC A299V | Ivosidenib (CT) | |
| FDX005 | F | 55 | iCCA | Metastatic | Capecitabine | BRAF V600E | Cobimetinib + vemurafenib (OL) | ||
| FDX006 | F | 52 | iCCA | Metastatic | None | VCL-FGFR2 fusion | Pemigatinib (CT) | ||
| FDX026 | M | 64 | iCCA | Metastatic | Anti-CTLA-4 antibody/anti-PD-1 antibody (CT) | IDH1 R132C | TP53 K132N | Ivosidenib (CT) |
Bold denotes matched treatment to actionable or potentially actionable mutation; iCCA, intrahepatic cholangiocarcinoma; GB, gall bladder carcinoma; mixed, mixed hepatocellular/ cholangiocarcinoma; CUP, carcinoma of unknown primary; OL, off-label; CT, clinical trial
Fig. 3.
Targeted therapies and survival. (A) Swimmers plot highlighting actionable alterations and progression-free survival for molecularly matched therapies (n=22). Arrow denotes ongoing therapy. (B) Kaplan-Meier plot showing overall survival (OS) from diagnosis of advanced cancer in patients with actionable alterations treated with matched targeted therapy (blue; n=22 in Fig. 3C), actionable alterations without matched therapy (red; n=32), and patients with no actionable alterations (green; n=70) with an overall P < 0.01.
A novel association between BRAF/ KRAS mutations and immune-modulator NT5E
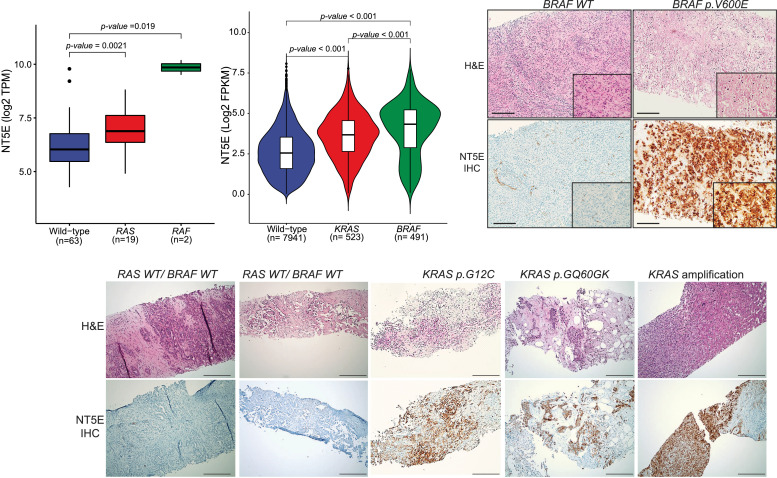
Apart from somatic mutations or copy number aberrations, we used RNAseq data (in MI_Oncoseq cohort) to help inform precision oncology avenues. This included sensitive detection of FGFR2 gene fusions in a partner-agnostic manner, and corroboration of outlier expression in cases with amplification of targetable genes such as ERBB2, CCND1, CCNE1, and MDM2. Additionally, querying individual driver aberrations for therapeutically informative gene expression correlates, we discovered a remarkable association between tumors with RAS/RAF mutations and expression of 5′-Nucleotidase Ecto, NT5E (CD73), a membrane protein that converts extracellular nucleotides to membrane-permeable nucleosides, associated with promotion of tumor immunosuppression. Fig. 4A shows a significantly higher level of NT5E in BTC cases in the MI_Oncoseq cohort with activating mutations in KRAS and BRAF, the latter being significantly higher than KRAS. To assess this correlation in an external dataset, we accessed TCGA pan-cancer dataset from cBioportal, and compared NT5E expression in tumors with (1) BRAF V600E mutation, (2) KRAS G12/13 or Q61 substitutions, and (3) wild-type BRAF and KRAS. Tumors with other mutations in BRAF or KRAS, and cases with mutations in NRAS or HRAS, as well as cases with amplification or deletion of NT5E were excluded from this analysis to ensure relatively discreet comparisons. As seen in Fig. 4B, tumors with activating KRAS/BRAF mutations showed significantly higher levels of NT5E expression, with BRAF mutated tumors showing relatively higher expression than KRAS mutated. In the context of BTCs, we corroborated the association between BRAF/KRAS mutations and NT5E expression level by IHC staining of select tumor tissue sections, as indicated (Fig. 4C-D). This association suggests follow up investigations for combination therapy with MEK and NT5E inhibitors in KRAS/BRAF mutant cases.
Fig. 4.
Biliary cancers with KRAS/ BRAF mutations show increased expression of immune-modulatory membrane protein CD73. (A) Box-plot representation of NT5E expression in MI-ONCOSEQ BTC cases with indicated status of KRAS and BRAF mutation. The y-axis shows NT5E expression levels (RPKM, reads per kb per million reads). (B) Violin-plot representation of NT5E expression in TCGA pan-cancer cohort with indicated status of KRAS and BRAF mutation. The y-axis shows NT5E expression levels (RPKM, reads per kb per million reads). (C) NT5E overexpression in BRAF/ KRAS mutated cholangiocarcinoma by immunohistochemistry (IHC), as indicated. Scale bar represents 50 uM (3B) and 10 uM (3C) respectively.
Discussion
Recent large-scale sequencing efforts like TCGA, ICGC and TARGET have provided insights into underlying molecular mechanisms in variety of cancer types. In this study, we analyzed a cohort of 124 patients with advanced BTC and subjected data to integrative clinical sequencing. Overall, a sizeable 43% of BTC patients harbored actionable mutations, of which, the 40.7% that received matched therapy had significantly longer OS by approximately 15 months compared to the cohort with actionable mutations that did not receive matched therapy. This suggests that patients with well-defined actionable molecular alterations derive considerable benefit in survival from receiving matched targeted therapy.
Admittedly, the definition of actionability varies in literature but we used a common and perhaps stringent interpretation to include only FDA approved therapies for specific molecular alterations in any cancer unless BTC-specific data suggested lack of benefit, such as palbociclib monotherapy in cases with CDKN2A deletion [45]. Unfortunately, only 40.7% of the actionable cohort received matched targeted therapy. The most common reason was lack of available early phase clinical trial (n=15), but other reasons included, the molecular analysis report preceded clinical trial investigation/availability (n=9), inability to obtain off-label targeted therapy for those who did not meet trial eligibility (n=5), decline in functional status or demise of the patient prior to release of the molecular analysis report (n=2), or patient refusal to participate in clinical trial (n=1). These outcomes suggest that precision oncology has a substantial clinical impact in patients with biliary cancer and warrants consideration of genomic analysis in all patients particularly earlier in their treatment course, and continued investigation of novel biomarkers and therapeutics in this rare cancer.
We found the mutational burden in our cohort to be significantly higher compared to the TCGA cohort perhaps since majority of the patients in our cohort had sequencing on tissue obtained at advanced disease (89%), biopsies mostly included metastatic sites (55%), and patients had had prior exposure to chemotherapy (56%). In comparison the TCGA cohort includes tissue obtained at primary resection. This finding supports the hypothesis that tumor mutational burden may worsen with prior exposure to chemotherapy and perhaps during the natural progression of the cancer. The overall tumor mutational burden is still low, however, compared to other cancers [46], and only a small percentage of tumors have a high enough mutational burden (3% with ≥10 mutations/Mb in our cohort) to leverage potential therapeutic benefit from immune checkpoint blockade [47,48].
Tumors with DDR gene mutations have been associated with sensitivity to DNA damaging chemotherapy, including platinum agents, as well as PARP inhibition [49], [50], [51]. Germline mutants of BRCA1 and 2 without defined locus specific loss of heterozygosity (LOH) in tumors have been associated with functional homologous recombination deficiency as the wild-type allele may be inactivated via alternative mechanisms, such as promoter methylation. However, in absence of LOH inactivation they may lack sensitivity to DNA damaging agents [52]. Results from the phase 3 POLO trial showed significant improvement in median PFS when patients with germline BRCA1 or 2 mutated metastatic pancreatic adenocarcinoma were treated with olaparib as maintenance therapy after platinum-based chemotherapy in patients compared to placebo [53]. It is worthwhile to hypothesize a similar benefit in BRCA-mutated BTC treated with PARP inhibitors, and indeed multiple clinical trials with PARP inhibitors alone (NCT04298021, NCT04042831), or in combination with anti-PD1 antibody (NCT03639935) are accruing patients with BTC. In addition to BRCA1 or 2 (incidence of 3–5%) [54] in BTC, other DDR mutated genes have also been identified including ATM and PALB2 (5 patients in our cohort; 4%) that may also benefit with PARP inhibitors [55,56]. Furthermore, patients with IDH1 or IDH2 hotspot mutations (10–15%) may also be susceptible to PARP inhibitors due to production of (R)-2-hydroxyglutarate (2HG), an oncometabolite that may impair homologous recombination by inhibiting the function of histone demethylases [57]. Thus, there is a potential for significant benefit in up to 30% of patients with BTC with PARP inhibitors.
A significantly high frequency (34.6%) of deleterious mutations in epigenetic modifiers, ARID1A, BAP1 and PBRM1 in the SWI/SNF accessory subunit highlights the role of dysregulated chromatin remodeling in BTC. ARID1A, BAP1 and PBRM1 encode subunits of the SWI/SNF chromatin-remodeling genes and were mutated in 20%, 15% and 10% of the samples, respectively; these have previously been shown to be drivers of progression in iCCA [15]. ARID1A and BAP1 have been shown to impair homologous repair in vitro [58,59] and therefore increase susceptibility to PARP inhibitors; clinical trials to test this hypothesis are ongoing (e.g. NCT03207347). In addition, epigenetic inhibitors such as HDAC and EZH2 inhibitors [60], proteolysis targeting chimera (PROTAC) degraders [61], anti PD-1 antibodies [62], and Aurora kinase A inhibitors [63] may also hold promise in targeting these mutations in BTC.
In addition to the mutations in the SWI/SNF complex, other epigenetic regulators such as IDH1 (and less commonly IDH2, FH) hotspot mutations have been described in iCCA [64,65]. As noted above, the 2-HG oncometabolite is a byproduct of the IDH1 mutation and is known to dysregulate the function of the histone methylases [57]. Recently an IDH1 inhibitor, ivosidenib showed significant improvement in median PFS and OS compared to placebo in a phase 3 clinical trial [66]. Interestingly, 6 out of 23 (26%) patients with IDH1/2 mutations had concurrent mutations in either ARID1A, BAP1 or PBRM1 thus suggesting potential benefit from a combination of an IDH inhibitor and histone modifying agents such as HDAC or demethylating inhibitors in this subset, similar to AML [67,68,69].
ERBB2 amplification was identified in 4% of our cohort consistent with other studies with fluke-negative BTCs [70]. Data from in vitro experiments [71], retrospective case series [72], and prospective phase 1/2 trial [73] support the ongoing investigation of ERBB2 targeted therapies in clinical trials in BTC (NCT02693535, NCT03613168, NCT01953926, NCT04466891). We also identified amplifications in CCND1 [74], MDM2 [75] and NTRK [76,77] and when targeted have shown modest preliminary clinical data noted in other cancers, and are under further investigation in integral biomarker trials.
Other molecular alterations with less than 5% incidence in BTC that have shown promising activity include BRAF V600E mutation [20]. We identified 12 (9.7%) patients with BRAF mutation in our cohort of which 7 patients had non-V600E activating mutations, including class III (D594N, D594E, D594G), and undefined kinase domain mutations, K483E, M693V, G466E as well as N661K. Cells with BRAF class III mutations have been shown to be responsive to MEK inhibitors [44]. BRAF K483E is a recurrent mutation, shown to be transforming in culture [78], and thus may represent a therapeutic target. Additionally, one case had a gene TRIM24-BRAF fusion, previously reported in a case of melanoma, sensitive to MEK inhibitor [79].
The MI-ONCOSEQ study first described the FGFR2 fusions across diverse cancers in 2013. Herein, we describe that FGFR2 activating mutations also lead to upregulation of gene expression similar to the fusions. Moreover, the median OS in the FGFR fusion cohort was similar to the FGFR activating mutation cohort (21.3 versus 21.5 months, respectively; data not shown) although the cohort sizes are small (N=15 and 4, respectively). The median OS of patients in the FGFR cohort (fusions or activating mutations) was higher compared to the FGFR wild type (21.3 versus 14.0 months, respectively; p value 0.07; data not shown). Of the 19 patients in the FGFR fusion/activating mutation cohort, 8 patients were treated with pan FGFR inhibitors and had a median OS of 22.8 months compared to 17.3 months in the untreated arm (p value of 0.31; data not shown). These data suggest that FGFR fusions (and potentially activating mutations) are both prognostic and predictive biomarkers in this rare cancer. We also identified a FGFR3-TACC3 fusion in a patient with gallbladder cancer, and to our knowledge this is the first report of a FGFR3 fusion in gallbladder cancer. Multiple FGFR fusion partners have been previously identified of which BICC1 is the most commonly noted [16]. Herein, we describe additional novel fusion partners, specifically the FGFR2-TAX1BP1, and MATN4-FGFR2. Clinical sequencing efforts like MI-ONCOSEQ which incorporate transcriptome analysis for gene fusions are important to identify targetable FGFR fusions due to the combinatorial possibilities of FGFR family fusion to a variety of oligomerization partners, as well as other rare fusions [80,81].
The discovery of novel association between BRAF/KRAS mutations and the expression of immunomodulatory target NT5E may define dual-precision therapeutic targets in a subset of cancers including the relatively intractable KRAS driven cancers. Notably, CD73 inhibitors are under intense clinical investigation for therapy across various cancers, wherein some exciting results were noted in pancreatic cancer, a predominantly KRAS driven malignancy. In a Phase I ARC-8 trial (NCT04104672), treatment with small-molecule CD73 inhibitor AB680 in combination with gemcitabine and nab-paclitaxel and PD-1 inhibitor zimberelimab, in previously untreated patients with metastatic pancreatic adenocarcinoma demonstrated effectiveness, with ORR 41%. Tumors reportedly shrank or stabilized in 11 of 13 patients who received the treatment for at least 16 weeks [82], spurring dose-expansion and placebo-controlled phase II trials.
We acknowledge the limitations of sample resources including neoplastic cellularity which reduced the sample size in RNA-seq and immune cluster analysis. Our study also merged data from different sequencing platforms (whole exome and targeted sequencing), thus limiting our analysis across the cohort to genomic regions common across the platforms. However, a uniform MI-ONCOSEQ analysis pipeline was used to ensure consistency and concordance across samples. These results may not be applicable in the community for multiple reasons, including the use of a more inclusive genomic analysis platform such as MI-ONCOSEQ, lack of clinical trials at many non-academic sites, patient willingness to travel to an academic institution which may represent a more motivated sub-group (preserved performance status, younger age), and use of non-MI-ONCOSEQ genomic analysis reports in our expanded cohort includes a biased group of patients referred specifically for open clinical trials.
Conclusion
This study highlights the importance of integrative clinical sequencing in defining molecularly matched targeted therapy options for biliary tract cancer, a rare yet anatomically and molecularly diverse malignancy with an aggressive clinical course, poor long-term prognosis due to limited therapeutic options. We observed significant improvement in survival when patients with actionable targets can receive matched therapies, and also enumerate several potentially actionable targets that provide a basis for matching with investigational drugs in ongoing clinical trials. Furthermore, we describe novel FGFR activating mutations and novel FGFR2 fusion partners which are likely to have direct impact on patient care, and diagnostic and therapeutic investigation. The novel association between KRAS/BRAF mutant tumors and the immunomodulatory target NT5E merits further investigation as a potential dual targeting modality in subsets of BTCs (as well as other cancers). These data provide evidence to strongly consider molecular analysis of tumors in patients with this rare cancer and the role of investigational therapies.
CRediT authorship contribution statement
Chandan Kumar-Sinha: Conceptualization, Data curation, Formal analysis, Writing – review & editing, Writing – original draft. Pankaj Vats: Conceptualization, Data curation, Formal analysis, Writing – review & editing. Nguyen Tran: Conceptualization, Data curation, Formal analysis, Writing – review & editing, Writing – original draft. Dan R. Robinson: Data curation, Formal analysis, Writing – original draft. Valerie Gunchick: Data curation, Writing – original draft. Yi-Mi Wu: Data curation, Writing – original draft, Formal analysis, Writing – original draft. Xuhong Cao: Data curation, Writing – original draft. Yu Ning: Data curation, Writing – original draft. Rui Wang: Data curation, Writing – original draft. Erica Rabban: Data curation, Writing – original draft. Janice Bell: Data curation, Writing – original draft. Sunita Shankar: Data curation, Writing – original draft. Rahul Mannan: Data curation, Writing – original draft. Yuping Zhang: Data curation, Writing – original draft. Mark M. Zalupski: Data curation, Writing – original draft. Arul M. Chinnaiyan: Data curation, Formal analysis, Writing – original draft. Vaibhav Sahai: Conceptualization, Data curation, Formal analysis, Writing – review & editing, Writing – original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Grant Support: This work was supported by grants from the NCI Early Detection Research Network (U01CA214170) and NIH/NCI Outstanding Investigator Award (R35CA231996)
Disclosures: MMZ – Institutional grant funding from AstraZeneca, MedImmune and Seattle Genetics. VS – Institutional grant funding from Agios, Bristol-Myers Squibb, Celgene, Clovis, Cornerstone, Exelixis, Fibrogen, Incyte, Ipsen, Medimmune, Merck, NCI, Rogel Cancer Center, Repare, Relay, Servier, Syros and Transthera; and consultant fees from AstraZeneca, Autem, Cornerstone, Delcath Systems, GlaxoSmithKline, Helsinn, Histosonics, Incyte, Ipsen, Kinnate, Lynx Group, QED, Servier and Taiho.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2023.100910.
Contributor Information
Arul M. Chinnaiyan, Email: arul@umich.edu.
Vaibhav Sahai, Email: vsahai@umich.edu.
Appendix. Supplementary materials
References
- 1.Patel N., Benipal B. Incidence of cholangiocarcinoma in the USA from 2001 to 2015: a US cancer statistics analysis of 50 states. Cureus. 2019;11:e3962. doi: 10.7759/cureus.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao K.J., Jabbour S., Parekh N., et al. Increasing mortality in the United States from cholangiocarcinoma: an analysis of the national center for health statistics database. BMC Gastroenterol. 2016;16:117. doi: 10.1186/s12876-016-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valle J.W., Kelley R.K., Nervi B., et al. Biliary tract cancer. Lancet. 2021;397:428–444. doi: 10.1016/S0140-6736(21)00153-7. [DOI] [PubMed] [Google Scholar]
- 4.Sahai V., Catalano P.J., Zalupski M.M., et al. Nab-paclitaxel and gemcitabine as first-line treatment of advanced or metastatic cholangiocarcinoma: a phase 2 clinical trial. JAMA Oncol. 2018 doi: 10.1001/jamaoncol.2018.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valle J., Wasan H., Palmer D., et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010;4:395–397. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 6.Nathan H., Pawlik T.M., Wolfgang C.L., et al. Trends in survival after surgery for cholangiocarcinoma: a 30-year population-based SEER database analysis. J. Gastrointest. Surg. 2007;11:1488–1497. doi: 10.1007/s11605-007-0282-0. [DOI] [PubMed] [Google Scholar]
- 7.Sia D., Hoshida Y., Villanueva A., et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Churi C.R., Shroff R., Wang Y., et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Javle M., Bekaii-Saab T., Jain A., et al. Biliary cancer: Utility of next-generation sequencing for clinical management. Cancer. 2016;122:3838–3847. doi: 10.1002/cncr.30254. [DOI] [PubMed] [Google Scholar]
- 10.Chan-On W., Nairismagi M.L., Ong C.K., et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 2013;45:1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 11.Simbolo M., Fassan M., Ruzzenente A., et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget. 2014;5:2839–2852. doi: 10.18632/oncotarget.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y.M., Su F., Kalyana-Sundaram S., et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farshidfar F., Zheng S., Gingras M.C., et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell Rep. 2017;19:2878–2880. doi: 10.1016/j.celrep.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montal R., Sia D., Montironi C., et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J. Hepatol. 2020;73:315–327. doi: 10.1016/j.jhep.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y., Pawlik T.M., Anders R.A., et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat. Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abou-Alfa G.K., Sahai V., Hollebecque A., et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–684. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Javle M., Lowery M., Shroff R.T., et al. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J. Clin. Oncol. 2018;36:276–282. doi: 10.1200/JCO.2017.75.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner N.C., Krop I.E., Bardia A., et al. Abstract OT2-07-01: a phase 2 study of futibatinib (TAS-120) in metastatic breast cancers harboring fibroblast growth factor receptor (<em>FGFR</em>) amplifications (FOENIX-MBC2) Cancer Res. 2020;80 OT2-07-01-OT2-07-01. [Google Scholar]
- 19.Abou-Alfa G.K., Macarulla T., Javle M.M., et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21:796–807. doi: 10.1016/S1470-2045(20)30157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wainberg Z.A., Lassen U.N., Elez E., et al. Efficacy and safety of dabrafenib (D) and trametinib (T) in patients (pts) with BRAF V600E–mutated biliary tract cancer (BTC): a cohort of the ROAR basket trial. J. Clin. Oncol. 2019;37 187-187. [Google Scholar]
- 21.Javle M., Churi C., Kang H.C., et al. HER2/neu-directed therapy for biliary tract cancer. J. Hematol. Oncol. 2015;8:58. doi: 10.1186/s13045-015-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X., Wu X., Wu H., et al. SHR-1210 plus GEMOX as first line treatment in biliary tract cancer: results from a single-arm exploratory study. J. Clin. Oncol. 2019;37 4092-4092. [Google Scholar]
- 23.Sahai V., Griffith K.A., Beg M.S., et al. A multicenter randomized phase II study of nivolumab in combination with gemcitabine/cisplatin or ipilimumab as first-line therapy for patients with advanced unresectable biliary tract cancer (BilT-01) J. Clin. Oncol. 2020;38 4582-4582. [Google Scholar]
- 24.Ueno M., Ikeda M., Morizane C., et al. Nivolumab alone or in combination with cisplatin plus gemcitabine in Japanese patients with unresectable or recurrent biliary tract cancer: a non-randomised, multicentre, open-label, phase 1 study. Lancet Gastroenterol. Hepatol. 2019;4:611–621. doi: 10.1016/S2468-1253(19)30086-X. [DOI] [PubMed] [Google Scholar]
- 25.Oh D.Y., Lee K.H., Lee D.W., et al. Phase II study assessing tolerability, efficacy, and biomarkers for durvalumab (D) ± tremelimumab (T) and gemcitabine/cisplatin (GemCis) in chemo-naïve advanced biliary tract cancer (aBTC) J. Clin. Oncol. 2020;38 4520-4520. [Google Scholar]
- 26.Bang Y.J., Ueno M., Malka D., et al. Pembrolizumab (pembro) for advanced biliary adenocarcinoma: results from the KEYNOTE-028 (KN028) and KEYNOTE-158 (KN158) basket studies. J. Clin. Oncol. 2019;37 4079-4079. [Google Scholar]
- 27.Floudas C.S., Xie C., Brar G., et al. Combined immune checkpoint inhibition (ICI) with tremelimumab and durvalumab in patients with advanced hepatocellular carcinoma (HCC) or biliary tract carcinomas (BTC) J. Clin. Oncol. 2019;37 336-336. [Google Scholar]
- 28.Kim R.D., Chung V., Alese O.B., et al. A phase 2 multi-institutional study of nivolumab for patients with advanced refractory biliary tract cancer. JAMA Oncol. 2020;6:1–8. doi: 10.1001/jamaoncol.2020.0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang J., Yoo C., Jeong J.H., et al. Efficacy and safety of pembrolizumab in patients with PD-L1 positive advanced biliary tract cancer (BTC): a prospective cohort study. J. Clin. Oncol. 2019;37 4082-4082. [Google Scholar]
- 30.Klein O., Kee D., Nagrial A., et al. Combination immunotherapy with ipilimumab and nivolumab in patients with advanced biliary tract cancers. J. Clin. Oncol. 2020;38 doi: 10.1001/jamaoncol.2020.2814. 4588-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson D., Van Allen E.M., Wu Y.M., et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;162:454. doi: 10.1016/j.cell.2015.06.053. [DOI] [PubMed] [Google Scholar]
- 32.Robinson D.R., Wu Y.M., Lonigro R.J., et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mody R.J., Wu Y.M., Lonigro R.J., et al. Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA. 2015;314:913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roychowdhury S., Iyer M.K., Robinson D.R., et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci. Transl. Med. 2011;3 doi: 10.1126/scitranslmed.3003161. 111ra121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y.M., Cieslik M., Lonigro R.J., et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell. 2018;173:1770–1782. doi: 10.1016/j.cell.2018.04.034. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicorici D., Şatalan M., Edgren H., et al. <strong>FusionCatcher</strong>– a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014 [Google Scholar]
- 37.Arriba. We used Arriba https://github.com/suhrig/arriba/ to detect gene fusions from RNA-Seq data.
- 38.Agaimy A., Stoehr R., Tögel L., et al. YAP1-MAML2-rearranged poroid squamous cell carcinoma (squamoid porocarcinoma) presenting as a primary parotid gland tumor. Head Neck Pathol. 2020 doi: 10.1007/s12105-020-01181-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vivero M., Davineni P., Nardi V., et al. Metaplastic thymoma: a distinctive thymic neoplasm characterized by YAP1-MAML2 gene fusions. Mod. Pathol. 2020;33:560–565. doi: 10.1038/s41379-019-0382-x. [DOI] [PubMed] [Google Scholar]
- 40.Picco G., Chen E.D., Alonso L.G., et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat. Commun. 2019;10:2198. doi: 10.1038/s41467-019-09940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valouev A., Weng Z., Sweeney R.T., et al. Discovery of recurrent structural variants in nasopharyngeal carcinoma. Genome Res. 2014;24:300–309. doi: 10.1101/gr.156224.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tonon G., Modi S., Wu L., et al. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat. Genet. 2003;33:208–213. doi: 10.1038/ng1083. [DOI] [PubMed] [Google Scholar]
- 43.Wu L., Liu J., Gao P., et al. Transforming activity of MECT1-MAML2 fusion oncoprotein is mediated by constitutive CREB activation. EMBO J. 2005;24:2391–2402. doi: 10.1038/sj.emboj.7600719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao Z., Yaeger R., Rodrik-Outmezguine V.S., et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017 doi: 10.1038/nature23291. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baghdadi T.A., Halabi S., Garrett-Mayer E., et al. Palbociclib in patients with pancreatic and biliary cancer with CDKN2A alterations: results from the targeted agent and profiling utilization registry study. JCO Precis. Oncol. 2019:1–8. doi: 10.1200/PO.19.00124. [DOI] [PubMed] [Google Scholar]
- 46.Chalmers Z.R., Connelly C.F., Fabrizio D., et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowery M.A., Ptashkin R., Jordan E., et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin. Cancer Res. 2018 doi: 10.1158/1078-0432.CCR-18-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spizzo G., Puccini A., Xiu J., et al. Molecular profile of BRCA-mutated biliary tract cancers. ESMO Open. 2020;5 doi: 10.1136/esmoopen-2020-000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audeh M.W., Carmichael J., Penson R.T., et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 50.Ledermann J., Harter P., Gourley C., et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 51.Gelmon K.A., Tischkowitz M., Mackay H., et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 52.Maxwell K.N., Wubbenhorst B., Wenz B.M., et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat. Commun. 2017;8:319. doi: 10.1038/s41467-017-00388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Golan T., Hammel P., Reni M., et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spizzo G., Puccini A., Xiu J., et al. Frequency of BRCA mutation in biliary tract cancer and its correlation with tumor mutational burden (TMB) and microsatellite instability (MSI) J. Clin. Oncol. 2019;37 4085-4085. [Google Scholar]
- 55.O'Reilly E.M., Lee J.W., Zalupski M., et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J. Clin. Oncol. 2020;38:1378–1388. doi: 10.1200/JCO.19.02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radhakrishnan S.K., Bebb D.G., Lees-Miller SP. Targeting ataxia-telangiectasia mutated deficient malignancies with poly ADP ribose polymerase inhibitors. Transl. Cancer Res. 2013;2:155–162. [Google Scholar]
- 57.Sulkowski P.L., Corso C.D., Robinson N.D., et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu H., Pak H., Hammond-Martel I., et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Natl. Acad. Sci. 2014;111:285–290. doi: 10.1073/pnas.1309085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park J.H., Park E.J., Lee H.S., et al. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 2006;25:3986–3997. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bitler B.G., Fatkhutdinov N., Zhang R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin. Ther. Targets. 2015;19:1419–1422. doi: 10.1517/14728222.2015.1062879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farnaby W., Koegl M., Roy M.J., et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 2019;15:672–680. doi: 10.1038/s41589-019-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen J., Ju Z., Zhao W., et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018;24:556–562. doi: 10.1038/s41591-018-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu C., Lyu J., Yang E.J., et al. Targeting AURKA-CDC25C axis to induce synthetic lethality in ARID1A-deficient colorectal cancer cells. Nat. Commun. 2018;9:3212. doi: 10.1038/s41467-018-05694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaelin W.G., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang P., Dong Q., Zhang C., et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abou-Alfa G.K., Macarulla Mercade T., Javle M., et al. ClarIDHy: a global, phase III, randomized, double-blind study of ivosidenib (IVO) vs placebo in patients with advanced cholangiocarcinoma (CC) with an isocitrate dehydrogenase 1 (IDH1) mutation. Ann. Oncol. 2019;30 v872-v873. [Google Scholar]
- 67.Dinardo C.D., Stein A.S., Stein E.M., et al. Mutant IDH1 inhibitor ivosidenib (IVO; AG-120) in combination with azacitidine (AZA) for newly diagnosed acute myeloid leukemia (ND AML). 2019;37:7011–7011.
- 68.Kim G.H., Choi S.Y., Oh T.I., et al. IDH1(R132H) Causes Resistance to HDAC Inhibitors by Increasing NANOG in Glioblastoma Cells. Int. J. Mol. Sci. 2019;20:2679. doi: 10.3390/ijms20112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiNardo C.D., Stein A.S., Fathi A.T., et al. Mutant isocitrate dehydrogenase (mIDH) inhibitors, enasidenib or ivosidenib, in combination with azacitidine (AZA): preliminary results of a phase 1b/2 study in patients with newly diagnosed acute myeloid leukemia (AML) Blood. 2017;130 639-639. [Google Scholar]
- 70.Jusakul A., Cutcutache I., Yong C.H., et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discovery. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nam A.R., Kim J.W., Cha Y., et al. Therapeutic implication of HER2 in advanced biliary tract cancer. Oncotarget. 2016;7:58007–58021. doi: 10.18632/oncotarget.11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Javle M., Churi C., Kang H.C., et al. HER2/neu-directed therapy for biliary tract cancer. J. Hematol. Oncol. 2015;8 doi: 10.1186/s13045-015-0155-z. 58-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B.T., Li T., Johnson M.L., et al. Safety and efficacy of pyrotinib in patients with NSCLC and other advanced solid tumors with activating HER2 alterations: A phase I basket trial. J. Clin. Oncol. 2020;38 3510-3510. [Google Scholar]
- 74.Edelman M.J., Redman M.W., Albain K.S., et al. SWOG S1400C ( NCT02154490)-a phase II study of palbociclib for previously treated cell cycle gene alteration-positive patients with stage IV squamous cell lung cancer (lung-MAP substudy) J. Thorac. Oncol. 2019;14:1853–1859. doi: 10.1016/j.jtho.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meric-Bernstam F., Somaiah N., DuBois S.G., et al. 475P - A phase IIa clinical trial combining ALRN-6924 and palbociclib for the treatment of patients with tumours harboring wild-type p53 and MDM2 amplification or MDM2/CDK4 co-amplification. Ann. Oncol. 2019;30 v179-v180. [Google Scholar]
- 76.Hong D.S., Bauer T.M., Lee J.J., et al. Larotrectinib in adult patients with solid tumours: a multi-centre, open-label, phase I dose-escalation study. Ann. Oncol. 2019;30:325–331. doi: 10.1093/annonc/mdy539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hempel D., Wieland T., Solfrank B., et al. Antitumor activity of larotrectinib in esophageal carcinoma with NTRK gene amplification. Oncologist. 2020;25:e881–e886. doi: 10.1634/theoncologist.2019-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ng P.K., Li J., Jeong K.J., et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell. 2018;33:450–462. doi: 10.1016/j.ccell.2018.01.021. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hutchinson K.E., Lipson D., Stephens P.J., et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin. Cancer Res. 2013;19:6696–6702. doi: 10.1158/1078-0432.CCR-13-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borad M.J., Gores G.J., Roberts L.R. Fibroblast growth factor receptor 2 fusions as a target for treating cholangiocarcinoma. Curr. Opin. Gastroenterol. 2015;31:264–268. doi: 10.1097/MOG.0000000000000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arai Y., Totoki Y., Hosoda F., et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59:1427–1434. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 82.Blocking CD73 can shrink pancreatic tumors. Cancer Discov. 2021;11 doi: 10.1158/2159-8290.CD-NB2021-0313. OF4-OF4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.