Summary
Background
Pretreatment identification of pathological extranodal extension (ENE) would guide therapy de-escalation strategies for in human papillomavirus (HPV)-associated oropharyngeal carcinoma but is diagnostically challenging. ECOG-ACRIN Cancer Research Group E3311 was a multicentre trial wherein patients with HPV-associated oropharyngeal carcinoma were treated surgically and assigned to a pathological risk-based adjuvant strategy of observation, radiation, or concurrent chemoradiation. Despite protocol exclusion of patients with overt radiographic ENE, more than 30% had pathological ENE and required postoperative chemoradiation. We aimed to evaluate a CT-based deep learning algorithm for prediction of ENE in E3311, a diagnostically challenging cohort wherein algorithm use would be impactful in guiding decision-making.
Methods
For this retrospective evaluation of deep learning algorithm performance, we obtained pretreatment CTs and corresponding surgical pathology reports from the multicentre, randomised de-escalation trial E3311. All enrolled patients on E3311 required pretreatment and diagnostic head and neck imaging; patients with radiographically overt ENE were excluded per study protocol. The lymph node with largest short-axis diameter and up to two additional nodes were segmented on each scan and annotated for ENE per pathology reports. Deep learning algorithm performance for ENE prediction was compared with four board-certified head and neck radiologists. The primary endpoint was the area under the curve (AUC) of the receiver operating characteristic.
Findings
From 178 collected scans, 313 nodes were annotated: 71 (23%) with ENE in general, 39 (13%) with ENE larger than 1 mm ENE. The deep learning algorithm AUC for ENE classification was 0·86 (95% CI 0·82–0·90), outperforming all readers (p<0·0001 for each). Among radiologists, there was high variability in specificity (43–86%) and sensitivity (45–96%) with poor inter-reader agreement (κ 0·32). Matching the algorithm specificity to that of the reader with highest AUC (R2, false positive rate 22%) yielded improved sensitivity to 75% (+ 13%). Setting the algorithm false positive rate to 30% yielded 90% sensitivity. The algorithm showed improved performance compared with radiologists for ENE larger than 1 mm (p<0·0001) and in nodes with short-axis diameter 1 cm or larger.
Interpretation
The deep learning algorithm outperformed experts in predicting pathological ENE on a challenging cohort of patients with HPV-associated oropharyngeal carcinoma from a randomised clinical trial. Deep learning algorithms should be evaluated prospectively as a treatment selection tool.
Funding
ECOG-ACRIN Cancer Research Group and the National Cancer Institute of the US National Institutes of Health.
Introduction
Head and neck cancer incidence is rising, driven by an increase in human papillomavirus (HPV)-associated oropharyngeal carcinoma with approximately 15 000 incidents annually in the USA.1,2 Standard treatment frameworks for HPV-associated oropharyngeal carcinoma include upfront surgical or non-operative (ie, chemoradiation) approaches, both of which can be associated with substantial morbidity. HPV-associated oropharyngeal carcinoma is associated with favourable prognosis compared with tobacco and alcohol-associated head and neck cancers,3,4 and therefore the long-term treatment sequelae in survivors can be more apparent and detrimental.5 The high morbidity of treatment has led to investigations of therapy de-escalation strategies to maintain favourable oncological outcomes while reducing toxicity,6,7 including the use of upfront transoral robotic surgery.8–11
The ECOG-ACRIN Cancer Research Group E3311 (NCT01898494) was a multicentre phase 2 trial studying primary transoral robotic surgery for HPV-associated oropharyngeal carcinoma followed by risk-adapted post-operative therapy dependent on pathological risk factors. Protocol details have been published previously.9 Patients in the intermediate pathological risk group were randomly assigned to de-escalated versus standard dose post-operative radiotherapy (at 50 Gy or 60 Gy), which was the experimental focus of the trial. Patients were deemed high-risk, and not eligible for radiation alone, if surgical pathology showed more than four malignant lymph nodes, positive margin, or more than 1 mm of extranodal extension (ENE).9
ENE, when malignant cells infiltrate beyond the lymph node into surrounding tissue, is both a poor prognostic factor for oropharyngeal carcinoma and an indication for therapy intensification with postoperative chemoradiation (ie, trimodality therapy),12 which increases toxicity and health-care costs.13–15 ENE is only definitively diagnosed on pathology, and attempts to identify or predict ENE via human interpretation of pretreatment imaging have generally shown poor results.16–21 Accordingly, there are high rates of incidental ENE following surgery for oropharyngeal carcinoma.21,22 For example, although the E3311 protocol specifically excluded patients with radiographically overt ENE, 31% of patients were allocated to the high-risk, trimodality arm, with 77% of these due to an ENE greater than 1 mm.9 Additionally, patients receiving trimodality therapy had poorer toxicity and quality of life outcomes. Therefore, tools are needed to better predict ENE in the pretreatment setting to help select the ideal treatment de-escalation or intensification trials for patients and avoid unnecessary trimodality therapy.
Deep learning algorithms within the field of artificial intelligence synthesise large amounts of data and make predictions based on learned features of a training dataset.23 Deep learning has transformed the field of computer vision and is now used in several US Food and Drug Administration-approved imaging applications.24–26 In previous work, some of the present authors developed, externally validated, and benchmarked a CT-based deep learning algorithm for identifying ENE in patients with head and neck cancers,27,28 finding that it outperformed head and neck radiologists.28 In this study, we rigorously evaluated the algorithm as an ENE screening tool for transoral surgery in the E3311 cohort, a difficult litmus test in a prospectively accrued patient population already screened for readily identifiable ENE, where pretreatment identification of ENE would be particularly impactful to management decisions.
Methods
Study design
This retrospective evaluation of deep learning algorithm performance used data from the multicentre, randomised de-escalation trial E3311. This study was reviewed by the institutional review boards at the participating institutions and granted exemption and waiver of informed consent. The study was done with support of ECOG-ACRIN in providing de-identified imaging and pathology data. The report follows the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis guidelines.29
Participants
All enrolled patients on E3311 required pretreatment and diagnostic head and neck imaging; patients with radiographically overt ENE were excluded per study protocol. E3311 was held in the USA across 58 participating medical centres. All surgical specimens underwent centralised pathology review and ENE extent in millimeters was recorded. We retrospectively included all intent-to-treat E3311 patients with readily available pretreatment, diagnostic quality CT with intravenous contrast enhancement, and corresponding lymph node dissection pathology report with annotated ENE status and extent of ENE in millimeters. E3311 allowed pretreatment CT or MRI, and those without iodinated, intravenous contrast-enhanced CT were excluded. Generally, surgical pathology reports included ENE diagnosis at the nodal level by including description of the nodal station and including diameter information. Corresponding lymph node radiographic annotations were made in accordance with the previous study protocol27 and assigned a level of certainty (appendix p 2). Only nodes with a high degree of certainty were included in the study dataset. Given the known association between lymph node size and ENE,30 we included the node with longest short-axis diameter on each scan along with 1–2 additional smaller nodes at random that had certain ENE correlations (appendix p 2).
Procedures
We used DualNet, a three-dimensional (3D) convolutional neural network algorithm developed and validated in previous studies28 for all analyses. DualNet is a visual geometry group-inspired architecture that receives and merges two 3D representations of lymph node regions of interest: one that retains original volume information, and one that is rescaled to be size invariant. Algorithm architecture and hyperparameters were all identical to those implemented in previous work.27 Before doing the present validation study, several modifications to image acquisition and preprocessing workflows were made a priori (appendix pp 2–3). Given the fundamental knowledge that increasing training data improves neural network performance, we retrained our initial model for this study, which was developed from a single institution,27 with two additional datasets that had been previously used as external validators.28 Therefore, a total of three data sources were made use of for model training and tuning (appendix p 6). The model was then locked for testing on the E3311 dataset (figure 1).
Figure 1: E3311 validation and benchmarking study framework.
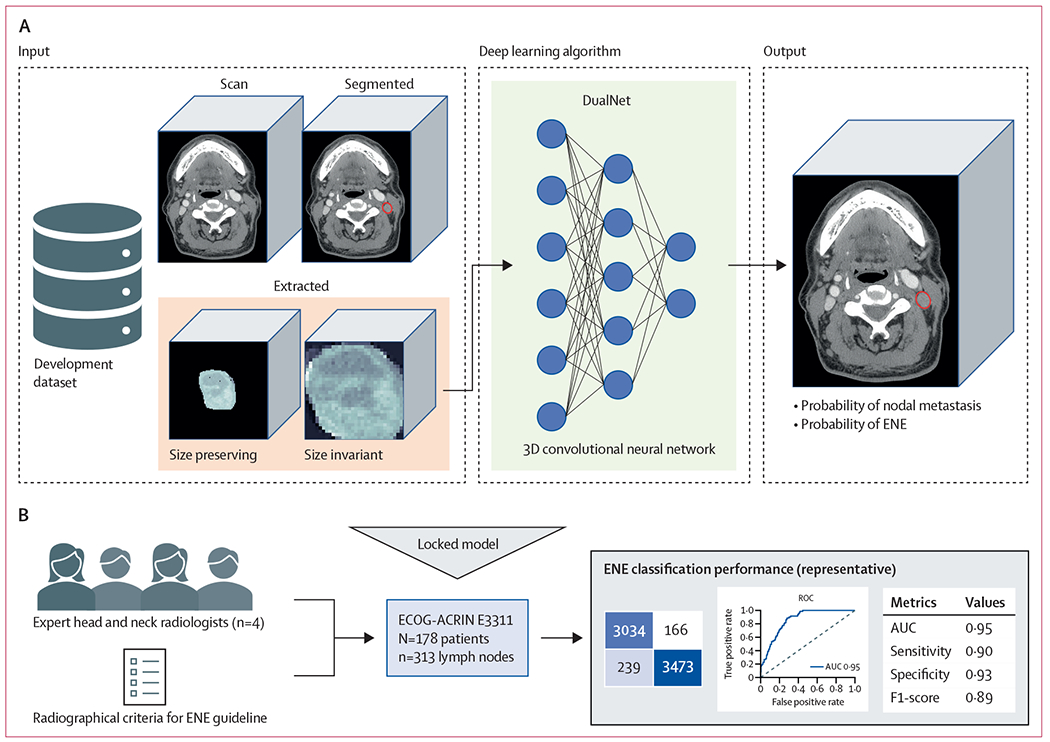
(A) The previously developed deep learning algorithm was retrained on a combined, multi-institutional dataset from three sources to predict probability of nodal metastasis and ENE on a node-by-node basis. (B) The model was locked and tested on a curated dataset of 313 lymph nodes from 178 patients enrolled on the E3311 de-escalation trial for patient withs human papillomavirus-associated oropharyngeal carcinoma, a trial that specifically excluded radiographic matted nodes or overt clinical ENE. Four expert head and neck radiologists from National Comprehensive Cancer Network comprehensive cancer centers, and with access to a validated educational guideline for radiographic ENE criteria, individually reviewed the lymph nodes, and made a prediction of node positivity or ENE on a forced Likert scale. ENE classification performance was compared between the deep learning algorithm and the radiologists, with a primary endpoint of area under the receiver operating characteristic curve. 3D=three dimensional. AUC=area under curve. ECOG-ACRIN=Eastern Cooperative Oncology Group and the American College of Radiology Imaging Network. ENE=extranodal extension. ROC=receiver operating characteristic.
Four board-certified, fellowship-trained neuroradiologists who specialise in head and neck cancer (median experience 11 years in practice post-residency, range 4–17 years) from three National Cancer Institute-designated Comprehensive Cancer Centers were recruited as expert ENE readers (labelled as R1–4). No readers were involved in the development of the algorithm. Each reader independently reviewed all study CT scans, one at a time, via the open-source software, 3D Slicer (version 4). The CT scan was viewable in the axial, sagittal, and coronal planes, with overlaid lymph node segmentations, which could be hidden at the reader’s discretion. Reviews were done in isolation and readers were masked to the segmentation labels. Readers were instructed to use best judgement in measuring ENE likelihood for the segmented nodes and were additionally provided with an educational tool to assist with ENE diagnosis containing visual descriptions of CT features previously found to be associated with ENE (appendix p 21).31 Readers documented likelihood of malignant involvement and ENE on a 4-point forced-Likert scale (1 meaning very unlikely, 2 meaning somewhat unlikely, 3 meaning somewhat likely, and 4 meaning very likely) for each segmented lymph node and for the scan overall.
Statistical analysis
Statistical analyses were done in Python (version 3.8.5) using the scikit-learn package (version 0.24), unless otherwise specified. The primary endpoint of the study was the area under the curve (AUC) of the receiver operating characteristic for ENE prediction, which measures discriminatory performance and reflects a combination of sensitivity and specificity. From previous work,28 we anticipated a minimum of 155 lymph nodes would be needed to detect a type I error of 5% with 80% power for the expected AUC improvement of 0·85 (+ 15%) over the expected benchmark of 0·70 for expert controls (appendix p 13). 95% CIs were calculated, and algorithm AUC was compared with that of each reader via the Delong method,32 with a p value of less than 0·05 indicating statistical significance. For primary analyses, reader Likert scores were dichotomised (1–2 vs 3–4) to mimic a real-world decision to indicate negative or positive ENE prediction. Secondary endpoints were sensitivity (1 – false negative rate), specificity (1 – false positive rate), positive predictive value, negative predictive value, and raw accuracy. Deep learning algorithm secondary endpoints were calculated using four probability thresholds: a threshold that optimised Youden index (sensitivity + specificity – 1) on internal validation and thresholds that yielded false positive rates at clinically meaningful values: 10%, 20%, and 30%. Performance metrics were evaluated for ENE overall and ENE more than 1 mm and in the subgroup of lymph nodes with short-axis diameter of 10 mm or more, which we believe would be the most relevant for real-world use as this threshold is used clinically to denote lymph nodes at risk of metastasis. Calibration was assessed via reliability diagrams and expected calibration error.33 Post-hoc calibration using temperature scaling33,34 was then done on the internal validation set to improve uncertainty estimation without affecting discriminatory performance (appendix p 4).
We did analyses to identify the deep learning algorithm’s resilience to adversarial attacks and enhance interpretability and trust. Given the reliance of the algorithm on segmented lymph nodes and known interoperator variability in segmentation tasks,35 we analysed the algorithm test-time tolerance to the injection of random segmentation perimeter perturbations, from 1 mm to 10 mm in the axial planes. A custom script was created to mimic potential human variations in node segmentation (appendix p 5) and AUC, range, and standard deviation were also analysed. Additionally, scanner noise on the order of around 5 Hounsfield units (HUs) has been recently implicated as adversarial images that could mislead neural networks,36 so we did sensitivity analyses by injecting random Gaussian noise to images at test time. We did ten iterations of each type of sensitivity analysis. For interpretability analysis, we implemented gradient-weighted class activation maps, highlighting image regions most important for algorithm prediction.37 Finally, we did an analysis of failures and compared lymph-node characteristics between correct and incorrect ENE predictions using the algorithm probability threshold that maximised the Youden index.
Role of the funding source
ECOG-ACRIN supported data collection for this work. The funders of the study had no role in study design, data analysis, data interpretation, or writing of the report.
Results
Of 360 eligible and treated patients in E3311, we obtained complete data meeting our inclusion criteria for 178 (49%; appendix p 24). The remaining patients had either CT or pathology data that could not be retrieved from the participating institution. Included scans were from 46 participating institutions and various scanner models (appendix p 7). From these, 313 lymph-node segmentations were manually generated and annotated for malignancy and ENE. There were 105 (34%) benign lymph nodes, 137 (44%) malignant lymph nodes without ENE, and 71 (23%) malignant lymph nodes with ENE. Of nodes with ENE, 20 (28%) were 1 mm or smaller, 39 (55%) were larger than 1 mm, and 12 (17%) were unspecified per the obtained pathology report. For further analysis, we labelled unspecified ENE as more than 1 mm in size, as these ENE could not be excluded in these cases. We also did sensitivity analyses excluding patients with unspecified ENE. Median short-axis diameter was 7 mm (range 4–14) for benign lymph nodes, 20 mm (6–37) for malignant lymph nodes without ENE, and 24 mm (12–42) for lymph nodes with ENE, and 204 (65%) of the 313 nodes had a short-axis diameter of 10 mm or more (appendix p 9).
Deep learning algorithm performance for ENE identification was superior to each of the four readers (algorithm 0·857, 95% CI 0·82–0·90; R1 0·66, 0·60–0·73; R2 0·71, 0·64–0·77; R3 0·70, 0·66–0·73; R4 0·63, 0·56–0·69; p<(1 × 10−5) for each; figure 2; table). For an ENE of more than 1 mm, the algorithm yielded superior performance compared with the readers (0·859, 0·82–0·90; p<0·0001 for R1–4). For the subgroup of nodes with a short-axis diameter of 10 mm or more (n=204), the algorithm also had improved performance (AUC 0·74, 0·67–0·81), compared with readers, with reader performance particularly low in this subgroup (AUC range 0·55–0·62, mean 0·58, p<0·006, figure 2; appendix pp 15–16). Inter-reader agreement for ENE was modest overall (Fleiss-κ 0·32) and low for nodes with a short-axis diameter of 10 mm or more (Fleiss-κ 0·16). Initial algorithm calibration was adequate, with probabilities tending to underestimate the likelihood of ENE.
Figure 2: Predictive performance of DLA and radiologists for ENE prediction in E3311.
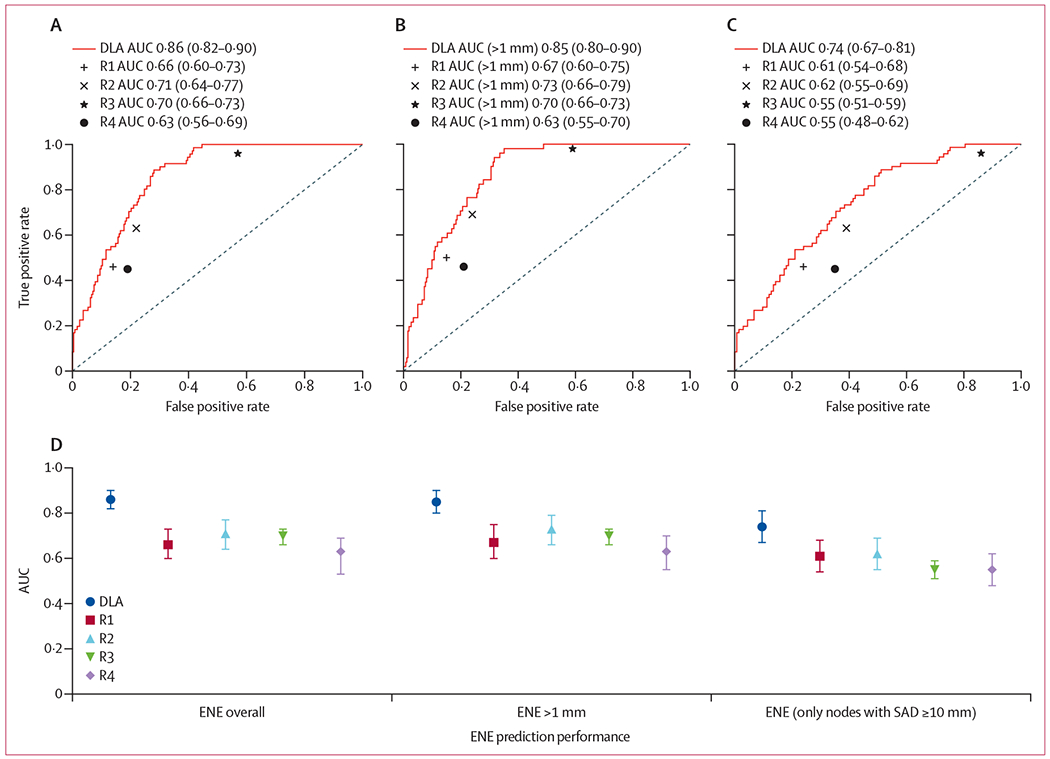
(A) Overall, (B) for ENE larger than 1 mm in extent (n=178 patients, 313 lymph nodes), and (C) in nodes with a short-axis diameter of 10 mm or more (n=204 lymph nodes). ROC curves are displayed with corresponding area under the curve (AUC). Comparative AUC with confidence intervals are shown (D) with the addition of performance in the subgroup of nodes with SAD ≥10 mm. p<0·001 for each comparison between DLA and R1–4. AUC=area under curve. DLA=deep learning algorithm. ENE=extranodal extension. R1–4=radiologists 1–4. ROC=receiver operating characteristic. SAD=short-axis diameter.
Table:
ENE predictive performance for deep learning algorithm and radiologists for pathological ENE overall and ENE larger than 1 mm in extent
| Overall ENE |
ENE >1 mm |
|||||||
|---|---|---|---|---|---|---|---|---|
| AUC (95% CI) | Sensitivity | Specificity | Accuracy | AUC | Sensitivity | Specificity | Accuracy | |
| DLA | 0·857 (0·82–0·90) | .. | .. | .. | 0·859 (0·82–0.90) | .. | .. | .. |
|
| ||||||||
| Best YI | NA | 0·89 | 0·72 | 0·76 | NA | 0·94 | 0·68 | 0·73 |
|
| ||||||||
| 30% FPR | NA | 0·90 | 0·70 | 0·74 | NA | 0·84 | 0·70 | 0·72 |
|
| ||||||||
| 20% FPR | NA | 0·72 | 0·80 | 0·78 | NA | 0·69 | 0·80 | 0·78 |
|
| ||||||||
| 10% FPR | NA | 0·49 | 0·90 | 0·81 | NA | 0·49 | 0·90 | 0·83 |
|
| ||||||||
| R1 | 0·66 (0·60–0·73) | 0·46 | 0·86 | 0·77 | 0·68 (0·61–0·75) | 0·51 | 0·85 | 0·79 |
|
| ||||||||
| R2 | 0·71 (0·64–0·77) | 0·63 | 0·78 | 0·74 | 0·73 (0·66–0·80) | 0·71 | 0·76 | 0·75 |
|
| ||||||||
| R3 | 0·70 (0·66–0·73) | 0·96 | 0·43 | 0·55 | 0·71 (0·67–0·74) | 1·0 | 0·41 | 0·51 |
|
| ||||||||
| R4 | 0·63 (0·56–0·69) | 0·45 | 0·81 | 0·73 | 0·63 (0·56–0·70) | 0·47 | 0·79 | 0·74 |
AUC=area under curve. ENE=extranodal extension. FPR=false positive rate. NA=not applicable. R1–4=radiologists 1–4. YI=Youden index.
Deep learning algorithm sensitivity at the optimised Youden index threshold was 0·89 and specificity was 0·72. Sensitivity dropped to 0·72 when setting a threshold to allow no more than 20% false positive rate. There was considerable variation between reader sensitivities (range 0·45–0·96) and specificities (0·43–0·86), with the highest performing reader (R2) having a sensitivity of 0·63 and specificity of 0·78. Except for R3, specificity was higher than sensitivity, and no reader had both sensitivity and specificity greater than 70%. Matching the algorithm specificity to that of the reader with highest AUC (R2, false positive rate 22%) yielded improved sensitivity to 75% (+13%). Positive predictive value was uniformly lower than negative predictive value for all readers and the algorithm, probably owing to the lower prevalence of ENE, and the F1 score was higher for the algorithm than the readers (appendix p 14). Following post-hoc calibration with temperature scaling, calibration on the test set improved (expected calibration error 0·426 vs 0·246; figure 3), without an effect on the AUC (appendix p 4).
Figure 3: Calibration curves for DLA ENE classifier for E3311.

Plots are binned in intervals of 10%, with predicted probability in the x axis, compared with the actual rate of ENE within that prediction interval on the y axis. Raw probabilities after testing on E3311 indicate mild underestimation of actual ENE rates. To develop a calibrated model whose probabilities would indicate real-world certainty, we did temperature scaling on the raw model outputs using the internal validation set from the combined Yale–Sinai–TCIA dataset. We then applied this mapping to the output probabilities for the E3311 dataset to yield a calibrated model, which maps very well to actual ENE rates, with improved expected calibration error. DLA=deep learning algorithm. ECE=expected calibration error. ENE=extranodal extension. TCIA=The Cancer Imaging Archive.
Using algorithm prediction on the largest lymph node as a surrogate for patient-level ENE yielded improved performance compared to all readers’ predictions of patient-level ENE based on their scan-level review (algorithm AUC 0·68 vs R1–4 AUC range 0·54–0·62; appendix p 16). Augmenting uncertain reader predictions with algorithm prediction resulted in overall improved discriminatory performance, with substantial improvements in sensitivity and inter-rater agreement (appendix p 17).
Studies of random peripheral segmentation variance for lymph nodes between 1 mm and 10 mm did not affect algorithm performance (mean AUC 0·860, range 0·856–0·867; appendix pp 19–20). Studies of adversarial images with Gaussian noise of −5 HU to 5 HU minimally degraded performance (0·853, 0·851–0·854). Gradient-weighted class activation map visualisations highlighted the importance of peripheral nodal regions in classifying ENE (appendix p 21).
Confusion matrices yielded eight (11%) of 71 false negative predictions and 68 (28%) of 242 false positive predictions for the algorithm with a threshold balanced for the optimal Youden index (figure 4; appendix p 17). Of false negatives, only one (13%) had an ENE of more than 1 mm (3 mm in extent). Of false positives, none had a calibrated probability of more than 85%, and only 12 (18%) had a predicted probability of greater than 70%. Lymph node mean short-axis diameter was higher for false positive predictions compared with the overall lymph node mean short-axis diameter (24 mm vs 17 mm, p<0·001), but lymph node mean short-axis diameter was not sigificantly different between false negative predictions and the overall cohort (21 mm vs 17 mm, p=0·16). Regarding readers, both false positive predictions and false negative predictions were more likely in larger nodes (p<0·001 for each, R1–4), with the sole exception being false negative predictions for R3 (p=0·97). Scanner parameters, such as pixel spacing, slice thickness, and scanner manufacturer were not significantly associated with the algorithm or reader failure (appendix p 19).
Figure 4: Analysis of successes and failures of ENE classification by DLA and radiologists by comparison with representative cases.
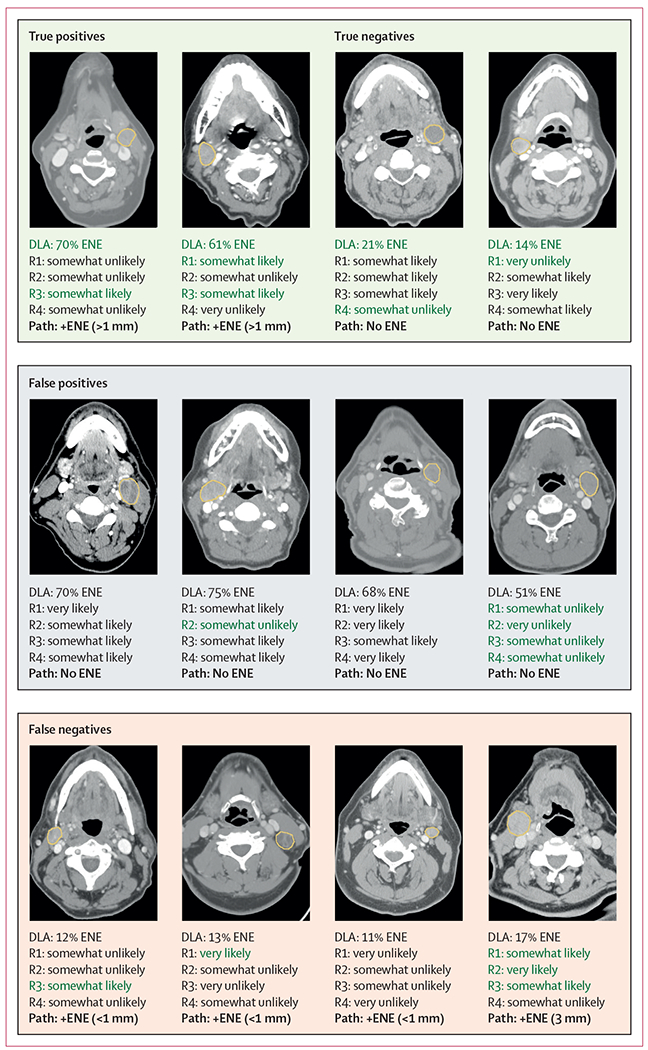
For the DLA, calibrated probabilities are shown. For the radiologists, predictions are shown via the forced Likert scale. Images are shown at varying soft tissue window widths and levels. The ground truth pathological ENE status is shown in bold font. Green font is used to denote a correct prediction. There was variable concordance between the DLA and radiologists, as well as radiologists among themselves. DLA=deep learning algorithm.
ENE=extranodal extension. R1–4=radiologists 1–4. +ENE=positive ENE.
Discussion
This study shows that imaging-based deep learning can improve the identification of ENE, including those larger than 1 mm, for HPV-associated oropharyngeal carcinoma in the pretreatment setting and is positioned for use as a screening tool to help treatment decision-making and selecting the optimal de-escalation strategy. Deep learning algorithm performance generalises to a diagnostically challenging set of HPV-associated cases where pretreatment knowledge of ENE would be clinically impactful. Furthermore, the algorithm performs this task with higher discriminatory performance, and particularly improved sensitivity, compared with experienced head and neck radiologists from tertiary comprehensive cancer centres. The improved sensitivity of the algorithm would translate to fewer missed diagnoses of incidental ENE and less subsequent trimodality therapy. In addition to the substantial performance gains compared with experts, the algorithm also has the benefit of producing standardised predictions, an adjustable threshold to fit the end user’s preferences, and calibrated probabilities that align with real-world ENE likelihoods and could enhance clinical usability.
Deep learning algorithm results on this prescreened, diagnostically challenging cohort show stable performance, with an AUC of 0·86, comparable to our previous work.28 The study shows that an algorithm trained largely on HPV-negative head and neck cancers generalises well to the HPV-associated setting. We hypothesise that this generalisability is because ENE is a morphological, local phenomenon, with the algorithm detecting subtle imaging signals in the node periphery that are universally indicative of ENE across molecular subtypes. Given that this cohort was prescreened for overt ENE or matted nodes, we expect it to perform even better in unscreened populations.
To our knowledge, there has been only one other deep learning classifier developed for pathological ENE detection, in a study of 51 patients with oral carcinoma, which did not have external validation, limiting direct comparison to our study.18 By contrast, there have been numerous studies of radiological diagnosis of ENE using a combination of radiologist clinical judgment and traditional imaging criteria.16,17,38–40 These studies show a wide range of sensitivities and specificities for ENE, with high inter-reader variability, and AUCs generally less than 0·70, consistent with the performance of the experienced head and neck radiologists in this study. Notably, we found extreme inter-reader variability in this dataset, and wide-ranging in sensitivities (46% to 96%) and specificities (43% to 86%), despite optional use of an educational guideline intended to help standardise predictions. We hypothesise that uncertainty in ENE prediction led to overestimation or underestimation depending on the readers’ individual tendencies that might derive from personal or institutional experiences,45,46 and this was reflected in our post-study survey results (appendix p 23). For example, R3 reported a tendency to overestimate ENE, which reflected their high sensitivity and low specificity. Specific ENE detection training and improved guidelines might improve interobserver variability and accuracy in the future, though current strategies do not appear sufficient. However, the algorithm is agnostic to these biases, which might be a strength of an algorithm-based detection strategy.
In positioning the algorithm for clinical use, an acceptable tradeoff between sensitivity and specificity of ENE identification is important to consider, while understanding that the ideal threshold is specific to the clinical user. Allowing a false positive ENE rate of 30% yielded 90% sensitivity, meaning that applying this algorithm at protocol screening could have substantially reduced allocation to the E3311 high-risk arm, which required trimodality therapy. In practice, however, clinicians might prefer a false positive rate of no more than 10% to 20%, which compromises sensitivity. Findings from several trials, including E3311,9 DART,43 and ORATOR,11,44 suggest that, in current frameworks, patients with ENE might be better served with non-operative definitive therapy. Conversely, patients without ENE are currently the most suitable candidates for surgically based treatment de-escalation, with reduced trimodality therapy and reduced long-term sequelae, as shown in E3311.9 The relevance of ENE could depend on its extent and, reassuringly, the algorithm performed comparably in discriminating ENE by size (ie, >1 mm or ≤1 mm). The algorithm operating point can be adapted to various scenarios, though will always require expert input to optimise its value in clinical context. Notably, aside from ENE, there are other important considerations in determining suitability for operative management, such as primary tumour location, extent and number of positive nodes, and patient preferences that must be accounted for in decision-making. Beyond selection for de-escalation strategies, more accurate ENE prediction via the algorithm could also be helpful in selecting patients appropriate for treatment escalation, including addition of chemotherapy to radiation in earlier stage patients, and clinical trials of systemic therapy intensification. Given the limitations of radiological ENE detection, this study suggests that algorithm-enhanced ENE prediction could contribute to decision-making in newly diagnosed patients with HPV-associated oropharyngeal carcinoma.
Deep learning medical applications are still nascent, and there are barriers to clinical translation.45,46 To address some of these barriers, we did test-time experiments to ensure that the network would be robust to adversarial images and generalise to various clinical scenarios, including variations in lymph-node segmentations and scanner parameters. To test for every possible scenario is not feasible and so, for this algorithm, as with any other deep learning application, we would recommend a run-in period of local testing at an institution to identify possible dataset or performance drift.47 The algorithm (and readers) performed worse on larger lymph nodes, and we are actively investigating ways to improve performance in this subgroup.
There are several other limitations to this study. Due to logistical and archiving issues, we could not procure the entire E3311 dataset for analysis, instead receiving a random sample of roughly half of the trial population. Additionally, validating the algorithm requires node-level annotations for ENE, which increase the manual resource needs for independent testing. We annotated at least the largest lymph node on each scan to ensure the algorithm analysed the node at highest risk of ENE but recognise that there might be exceptions to this, so other nodes with clear pathological ENE correlations were annotated as well. There is also the possibility that some node labels were misidentified and that pathological confirmation could be subject to interobserver variability, though the granular reporting and centralised pathological review of E3311 probably minimised these risks. Patient-level ENE prediction would promote translation to clinical use without reliance on manual segmentation, and work is ongoing to develop autosegmentation tools to facilitate this,48,49 although there are inherent benefits of a node-based model. Above all, node-level prediction greatly denoises the imaging framework space and allows the algorithm to focus on the region where ENE is present. ENE is generally a subtle, localised radiographical phenomenon, and therefore, a node-by-node approach to detection is more scientifically plausible than scan-level prediction in the absence of datasets much larger than are currently available. Additionally, our study shows that the algorithm is robust to variations in node segmentation, so performance would not be expected to decline due to interuser segmentation variability. Incorporation of imaging methods such as MRI and PET might improve the identification of ENE, both from a radiologist and algorithmic perspective, however studies thus far have been inconclusive.50–53
With intensifying interest in de-escalation paradigms for HPV-associated oropharyngeal carcinoma, tools are needed to optimise patient selection, including improved pretreatment ENE identification.54 We show the utility of CT-based deep learning to predict pathological ENE in a diagnostically challenging cohort of patients from a prospective, randomised de-escalation trial for HPV-associated oropharyngeal carcinoma. The algorithm shows high sensitivity and specificity for ENE identification, and substantially outperforms expert head and neck radiologists on direct comparison. Algorithm predictions could be provided to radiologists during the scan review or addended to a radiology report and given to oncology providers during the decision-making process. The deep learning algorithm should be prospectively tested in a randomised trial to determine its effect on treatment decision-making, quality of life, and disease control.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for articles published from Jan 1, 1990, until April 5, 2022, for diagnostic performance for extranodal extension (ENE) for head and neck cancers, using the terms: ((extranodal extension) OR (extracapsular extension)) AND ((computed tomography) OR (magnetic resonance imaging)) AND ((deep learning) or (machine learning) or (radiologist)) AND ((head and neck cancer) OR (oropharynx)). Reviews were excluded, yielding 12 articles, ten of which pertained to head and neck cancer lymph nodes. Eight articles qualified the diagnostic accuracy of radiologists in identifying ENE and universally showed subpar performance or high inter-reader variability using various radiographical criteria. Our group previously published the first internal and externally validated CT-based, deep learning algorithm for ENE identification, showing superior performance when compared directly with two human experts. One other deep learning algorithm for ENE identification in patients with oral cancer has been since published, trained on a small, single-institution dataset without external validation. We found no articles investigating deep learning for ENE identification in the setting of human papillomavirus (HPV)-associated oropharyngeal carcinoma.
Added value of this study
This is the first study evaluating deep learning as a screening tool for the pretreatment identification of ENE in HPV-associated oropharyngeal carcinoma and does so in the context of a multinational, randomised treatment deescalation trial. This study was done on a large cohort of patients who were, per protocol, excluded if overt radiographic ENE was suspected. This population thus represents a difficult litmus test for the algorithm in a cohort of patients where pretreatment ENE identification would be consequential. Additionally, the cohort had centralised pathology review for ENE, which included extent of ENE. The algorithm is directly benchmarked by four expert radiologists and shows that deep learning has a clear superiority in identifying ENE compared with the current standard.
Implications of all the available evidence
This study shows that deep learning can predict ENE in a prospectively accrued cohort of patients with HPV-associated oropharyngeal carcinoma in the pretreatment setting with high accuracy, outperforming diagnostic experts. Given the increasing interest in therapeutic de-escalation strategies for HPV-associated oropharyngeal carcinoma, the algorithm could be used to screen for ENE and help to select patients for operative versus non-operative management, thereby improving treatment personalisation and minimising the use of trimodality therapy.
Acknowledgments
This study was supported by the ECOG-ACRIN Cancer Research Group (Peter J O’Dwyer and Mitchell D Schnall, Group Co-Chairs, University of Pennsylvania School of Medicine) and the National Cancer Institute of the US National Institutes of Health under the following award numbers: U10CA180794, U10CA180820, UG1CA233180, UG1CA233184, UG1CA233337, UG1CA233253, and UG1CA232760. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interests
BHK reports research support from the US National Institutes of Health (NIH; grant number K08DE030216-01) and the Radiological Society of North America, honoraria from Dedham Group, and stock options in Monogram Orthopedics. JDS reports research support paid to their institution from Merck, Bristol Myers Squibb (BMS), Regeneron, Debiopharm, and Merck KGA; consulting, scientific advisory board, and travel fees from Castle Biosciences, Genentech, Immunitas, Debiopharm, BMS, Nanobiotix, Tilos, AstraZeneca, LEK Consulting, Catenion, ACI Clinical, Astellas, Stimit, and Merck KGA; expert witness fees; stock options in Immunitas; and equity in Doximity. RH reports payment for consulting from Celgene, Nanobiotix, ISA, Merck, Eisai, BMS, AstraZeneca, Pfizer, Loxo, Genentech, Immunomic Therapeutics, GlaxoSmithKline, Gilead Sciences, Vaccinex, Emmanuel Merck Darmstadt (EMD), Serono, BioNTech, Achilles Therapeutics, Bayer, Coherus Biosciences, Boehringer Ingelheim, and Mirati Therapeutics; and research funding from Boehringer Ingelheim, Merck, BMS, Celgene, AstraZeneca, Genentech, Pfizer, Kura Oncology, EMD, and Serono. SA reports research support from the Agency for Healthcare Research and Quality, National Cancer Institute, National Science Foundation, American Cancer Society, American Society of Clinical Oncology, Amazon, and Patterson Trust; fees from American Society of Clinical Oncology, HighCape Capital, Emmerson Collective; and stock options in Prophet Brand Strategy. HJWLA reports research support from NIH (grant numbers U24CA194354, U01CA190234, U01CA209414, and R35CA22052), and the European Union—European research Council (grant number 866504) and stock and other ownership interests in Onc.Ai, Love Health, Health-AI, and Bayerische Motoren Werke AG. BAB reports a consulting or advisory role with Merck, Debiopharm Group, CUE Biopharma, Maverick Therapeutics, Rakuten Medical, Nanobiotix, Macrogenics, ALX Oncology, IO Biotech, Ipsen, Genentech/Roche, Kura Oncology, Merck KGA, Pharmaceutical Product Development Global, and Exelixis; research funding to their institution from Merck, Aduro Biotech, Formation Biologics, BMS, and CUE Biopharma; and travel, accommodations, or expenses from Merck and Debiopharm Group. RLF reports stock and other ownership interests in Novasenta; a consulting or advisory role with Merck, Pfizer, EMD Serono, Numab, Macrogenics, Aduro Biotech, Novasenta, Sanofi, Zymeworks, and BMS; and research funding from Bristol Myers Squibb, AstraZeneca/MedImmune, Merck, Tesaro, and Novasenta. All other authors declare no competing interests.
Footnotes
See Online for appendix
Data sharing
Data used for this study was acquired via data user agreement with ECOG-ACRIN and the E3311 study investigators. Individual participant data are not publically available because this requirement was not anticipated in the study protocol. The study source code and model can be found at https://github.com/bhkann/DualNet-ENE/.
References
- 1.Ellington TD, Henley SJ, Senkomago V, et al. Trends in incidence of cancers of the oral cavity and pharynx—United States 2007–2016. MMWR Morb Mortal Wkly Rep 2020; 69: 433–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damgacioglu H, Sonawane K, Zhu Y, et al. Oropharyngeal cancer incidence and mortality trends in all 50 states in the US, 2001–2017. JAMA Otolaryngol Head Neck Surg 2022; 148: 155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010; 363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strojan P, Hutcheson KA, Eisbruch A, et al. Treatment of late sequelae after radiotherapy for head and neck cancer. Cancer Treat Rev 2017; 59: 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langendijk JA, Doornaert P, Verdonck-de Leeuw IM, Leemans CR, Aaronson NK, Slotman BJ. Impact of late treatment-related toxicity on quality of life among patients with head and neck cancer treated with radiotherapy. J Clin Oncol 2008; 26: 3770–76. [DOI] [PubMed] [Google Scholar]
- 6.Noor A, Mintz J, Patel S, et al. Predictive value of computed tomography in identifying extracapsular spread of cervical lymph node metastases in p16 positive oropharyngeal squamous cell carcinoma. J Med Imaging Radiat Oncol 2019; 63: 500–09. [DOI] [PubMed] [Google Scholar]
- 7.Marur S, Lee J-W, Cmelak A, et al. ECOG 1308: a phase II trial of induction chemotherapy followed by cetuximab with low dose versus standard dose IMRT in patients with HPV-associated resectable squamous cell carcinoma of the oropharynx (OP). J Clin Oncol 2012; 30: 5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstein GS, O’Malley BW Jr, Magnuson JS, et al. Transoral robotic surgery: a multicenter study to assess feasibility, safety, and surgical margins. Laryngoscope 2012; 122: 1701–07. [DOI] [PubMed] [Google Scholar]
- 9.Ferris RL, Flamand Y, Weinstein GS, et al. Phase II randomized trial of transoral surgery and low-dose intensity modulated radiation therapy in resectable p16+ locally advanced oropharynx cancer: an ECOG-ACRIN Cancer Research Group trial (E3311). J Clin Oncol 2022; 40: 138–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma DJ, Price KA, Moore EJ, et al. Phase II Evaluation of aggressive dose de-escalation for adjuvant chemoradiotherapy in human papillomavirus-associated oropharynx squamous cell carcinoma. J Clin Oncol 2019; 37: 1909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nichols AC, Theurer J, Prisman E, et al. Randomized trial of radiotherapy versus transoral robotic surgery for oropharyngeal squamous cell carcinoma: long-term results of the ORATOR trial. J Clin Oncol 2022; 40: 866–75. [DOI] [PubMed] [Google Scholar]
- 12.Bernier J, Cooper JS, Pajak TF, et al. Defining risk levels in locally advanced head and neck cancers: a comparative analysis of concurrent postoperative radiation plus chemotherapy trials of the EORTC (#22931) and RTOG (# 9501). Head Neck 2005; 27: 843–50. [DOI] [PubMed] [Google Scholar]
- 13.Ling DC, Chapman BV, Kim J, et al. Oncologic outcomes and patient-reported quality of life in patients with oropharyngeal squamous cell carcinoma treated with definitive transoral robotic surgery versus definitive chemoradiation. Oral Oncol 2016; 61: 41–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sher DJ, Fidler MJ, Tishler RB, Stenson K, al-Khudari S. Cost-effectiveness analysis of chemoradiation therapy versus transoral robotic surgery for human papillomavirus-associated, clinical N2 oropharyngeal cancer. Int J Radiat Oncol Biol Phys 2016; 94: 512–22. [DOI] [PubMed] [Google Scholar]
- 15.de Almeida JR, Moskowitz AJ, Miles BA, et al. Cost-effectiveness of transoral robotic surgery versus (chemo)radiotherapy for early T classification oropharyngeal carcinoma: a cost-utility analysis. Head Neck 2016; 38: 589–600. [DOI] [PubMed] [Google Scholar]
- 16.Carlton JA, Maxwell AW, Bauer LB, et al. Computed tomography detection of extracapsular spread of squamous cell carcinoma of the head and neck in metastatic cervical lymph nodes. Neuroradiol J 2017; 30: 222–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Almulla A, Noel CW, Lu L, et al. Radiologic-pathologic correlation of extranodal extension in patients with squamous cell carcinoma of the oral cavity: implications for future editions of the TNM classification. Int J Radiat Oncol Biol Phys 2018; 102: 698–708. [DOI] [PubMed] [Google Scholar]
- 18.Ariji Y, Sugita Y, Nagao T, et al. CT evaluation of extranodal extension of cervical lymph node metastases in patients with oral squamous cell carcinoma using deep learning classification. Oral Radiol 2020; 36: 148–55. [DOI] [PubMed] [Google Scholar]
- 19.Kann BH, Buckstein M, Carpenter TJ, et al. Radiographic extracapsular extension and treatment outcomes in locally advanced oropharyngeal carcinoma. Head Neck 2014; 36: 1689–94. [DOI] [PubMed] [Google Scholar]
- 20.Faraji F, Aygun N, Coquia SF, et al. Computed tomography performance in predicting extranodal extension in HPV-positive oropharynx cancer. Laryngoscope 2020; 130: 1479–86. [DOI] [PubMed] [Google Scholar]
- 21.McMullen CP, Garneau J, Weimar E, et al. Occult nodal disease and occult extranodal extension in patients with oropharyngeal squamous cell carcinoma undergoing primary transoral robotic surgery with neck dissection. JAMA Otolaryngol Head Neck Surg 2019; 145: 701–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian HE, Park HS, Barbieri A, et al. Pretreatment predictors of adjuvant chemoradiation in patients receiving transoral robotic surgery for squamous cell carcinoma of the oropharynx: a case control study. Cancers Head Neck 2016; 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–44. [DOI] [PubMed] [Google Scholar]
- 24.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature 2020; 577: 89–94. [DOI] [PubMed] [Google Scholar]
- 26.Benjamens S, Dhunnoo P, Meskó B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. NPJ Digit Med 2020; 3: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kann BH, Aneja S, Loganadane GV, et al. Pretreatment identification of head and neck cancer nodal metastasis and extranodal extension using deep learning neural networks. Sci Rep 2018; 8: 14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kann BH, Hicks DF, Payabvash S, et al. Multi-institutional validation of deep learning for pretreatment identification of extranodal extension in head and neck squamous cell carcinoma. J Clin Oncol 2020; 38: 1304–11. [DOI] [PubMed] [Google Scholar]
- 29.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 2015; 162: 55–63. [DOI] [PubMed] [Google Scholar]
- 30.An Y, Park HS, Kelly JR, et al. The prognostic value of extranodal extension in human papillomavirus-associated oropharyngeal squamous cell carcinoma. Cancer 2017; 123: 2762–72. [DOI] [PubMed] [Google Scholar]
- 31.Huang SH, Chernock R, O’Sullivan B, Fakhry C. Assessment criteria and clinical implications of extranodal extension in head and neck cancer. Am Soc Clin Oncol Educ Book 2021; 41: 265–78. [DOI] [PubMed] [Google Scholar]
- 32.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–45. [PubMed] [Google Scholar]
- 33.Guo C, Pleiss G, Sun Y, et al. On calibration of modern neural networks. 2017. http://arxiv.org/abs/1706.04599 (accessed Feb 5, 2020).
- 34.Küppers F, Kronenberger J, Shantia A, et al. Multivariate confidence calibration for object detection. 2020. https://openaccess.thecvf.com/content_CVPRW_2020/papers/w20/Kuppers_Multivariate_Confidence_Calibration_for_Object_Detection_CVPRW_2020_paper.pdf (accessed March 1, 2022).
- 35.Vinod SK, Jameson MG, Min M, Holloway LC. Uncertainties in volume delineation in radiation oncology: a systematic review and recommendations for future studies. Radiother Oncol 2016; 121: 169–79. [DOI] [PubMed] [Google Scholar]
- 36.Joel MZ, Umrao S, Chang E, et al. Using adversarial images to assess the robustness of deep learning models trained on diagnostic images in oncology. JCO Clin Cancer Inform 2022; 6: e2100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaraju RR, Cogswell M, Das A, et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. 2016. http://arxiv.org/abs/1610.02391 (accessed March 1, 2022).
- 38.Maxwell JH, Rath TJ, Byrd JK, et al. Accuracy of computed tomography to predict extracapsular spread in p16-positive squamous cell carcinoma. Laryngoscope 2015; 125: 1613–18. [DOI] [PubMed] [Google Scholar]
- 39.Patel MR, Hudgins PA, Beitler JJ, et al. Radiographic imaging does not reliably predict macroscopic extranodal extension in human papilloma virus-associated oropharyngeal cancer. ORL J Otorhinolaryngol Relat Spec 2018; 80: 85–95. [DOI] [PubMed] [Google Scholar]
- 40.Chai RL, Rath TJ, Johnson JT, et al. Accuracy of computed tomography in the prediction of extracapsular spread of lymph node metastases in squamous cell carcinoma of the head and neck. JAMA Otolaryngol Head Neck Surg 2013; 139: 1187–94. [DOI] [PubMed] [Google Scholar]
- 41.Shinagare AB, Lacson R, Boland GW, et al. Radiologist preferences, agreement, and variability in phrases used to convey diagnostic certainty in radiology reports. J Am Coll Radiol 2019; 16: 458–64. [DOI] [PubMed] [Google Scholar]
- 42.Cochon LR, Kapoor N, Carrodeguas E, et al. Variation in follow-up imaging recommendations in radiology reports: patient, modality, and radiologist predictors. Radiology 2019; 291: 700–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma DJ, Price K, Eric MJ, et al. Long-term results for MC1273, a phase II evaluation of de-escalated adjuvant radiation therapy for human papillomavirus associated oropharyngeal squamous cell carcinoma (HPV+ OPSCC). Int J Radiat Oncol Biol Phys 2021; 111: S61. [Google Scholar]
- 44.Palma DA, Prisman E, Berthelet E, et al. Assessment of toxic effects and survival in treatment deescalation with radiotherapy vs transoral surgery for HPV-associated oropharyngeal squamous cell carcinoma: the ORATOR2 phase 2 randomized clinical trial. JAMA Oncol 2022; 8: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kann BH, Hosny A, Aerts HJWL. Artificial intelligence for clinical oncology. Cancer Cell 2021; 39: 916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Glocker B, McCradden MM, Ghassemi M, Denniston AK, Oakden-Rayner L. The medical algorithmic audit. Lancet Digit Health 2022; 4: e384–97. [DOI] [PubMed] [Google Scholar]
- 47.Finlayson SG, Subbaswamy A, Singh K, et al. The clinician and dataset shift in artificial intelligence. N Engl J Med 2021; 385: 283–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plana D, Guntaka PK, Qian JM, et al. Deep learning and harmonization of multi-institutional data for automated gross tumor and nodal segmentation for oropharyngeal cancer. Int J Radiat Oncol Biol Phys 2021; 111: e97–98. [Google Scholar]
- 49.Oreiller V, Andrearczyk V, Jreige M, et al. Head and neck tumor segmentation in PET/CT: the HECKTOR challenge. Med Image Anal 2022; 77: 102336. [DOI] [PubMed] [Google Scholar]
- 50.Kubicek GJ, Champ C, Fogh S, et al. FDG-PET staging and importance of lymph node SUV in head and neck cancer. Head Neck Oncol 2010; 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yousem DM, Som PM, Hackney DB, Schwaibold F, Hendrix RA. Central nodal necrosis and extracapsular neoplastic spread in cervical lymph nodes: MR imaging versus CT. Radiology 1992; 182: 753–59. [DOI] [PubMed] [Google Scholar]
- 52.Lodder WL, Lange CAH, van Velthuysen M-LF, et al. Can extranodal spread in head and neck cancer be detected on MR imaging. Oral Oncol 2013; 49: 626–33. [DOI] [PubMed] [Google Scholar]
- 53.Abdel-Halim CN, Rosenberg T, Dyrvig A-K, et al. Diagnostic accuracy of imaging modalities in detection of histopathological extranodal extension: a systematic review and meta-analysis. Oral Oncol 2021; 114: 105169. [DOI] [PubMed] [Google Scholar]
- 54.Price KAR, Nichols AC, Shen CJ, et al. Novel strategies to effectively de-escalate curative-intent therapy for patients with HPV-associated oropharyngeal cancer: current and future directions. Am Soc Clin Oncol Educ Book 2020; 40: 1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used for this study was acquired via data user agreement with ECOG-ACRIN and the E3311 study investigators. Individual participant data are not publically available because this requirement was not anticipated in the study protocol. The study source code and model can be found at https://github.com/bhkann/DualNet-ENE/.


