Abstract
COVID-19 has accounted for more than six million deaths worldwide. Bacillus Calmette-Guérin (BCG), the existing tuberculosis vaccine, is known to induce heterologous effects over other infections due to trained immunity and has been proposed to be a potential strategy against SARS-CoV-2 infection. In this report, we constructed a recombinant (r) BCG expressing domains of the SARS-CoV-2 nucleocapsid and spike proteins (termed rBCG-ChD6), recognized as major candidates for vaccine development. We investigated whether rBCG-ChD6 immunization followed by a boost with the recombinant nucleocapsid and spike chimera (rChimera) together with alum provided protection against SARS-CoV-2 infection in K18-hACE2 mice. A single dose of rBCG-ChD6 boosted with rChimera associated with alum elicited the highest anti-Chimera total IgG and IgG2c antibody titers with neutralizing activity against SARS-CoV-2 Wuhan strain when compared to control groups. Importantly, following SARS-Cov2 challenge, this vaccination regimen induced IFN-γ and IL-6 production in spleen cells and reduced viral load in the lungs. In addition, no viable virus was detected in rBCG-ChD6 boosted with rChimera immunized mice which was associated with decreased lung pathology when compared to BCG WT-rChimera/alum or rChimera/alum control groups. Overall, our study demonstrates the potential of a prime-boost immunization system based on a rBCG expressing a chimeric protein derived from SARS-CoV-2 to protect mice against viral challenge.
Keywords: rBCG, COVID-19, Chimera, K18-hACE2 mice, vaccine, protection
Introduction
Since the outbreak of the Coronavirus Disease-19 (COVID-19) in late 2019, massive loss has been experienced around the globe and current death counts exceed six million worldwide (1, 2). First identified in Wuhan, China (3), a tremendous effort has been put forward by the scientific community to contain the spread of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) as governments struggle to deal with exhausted health-care services (4, 5). Designated a pandemic in March 2020 by the World Health Organization (WHO), several actions have been taken to manage this critical situation (6). Lockdown countermeasures, physical distancing, home-office practices, financial aid packages and drug repositioning studies (7, 8) are among the many strategies that have been implemented over the past two years.
The prompt availability of viral genome sequencing (9) was instrumental for the development of accurate diagnoses and experimental treatments (10). In that context, genomic surveillance programs have been playing critical roles in supporting direct governmental measures and epidemiological strategies (11). The SARS-CoV-2 genome consists of a single-stranded positive sense RNA that encodes among its structural proteins, two major candidates for vaccine development: the nucleocapsid protein (N) (12, 13) and the spike glycoprotein (S) (14, 15). The latter is the primary vaccine candidate as it is essential for viral attachment to the angiotensin-converting enzyme 2 (ACE2) receptor and subsequent entry of the virus into the host (16, 17). This human respiratory disease presents clinical manifestations ranging from asymptomatic to severe. After an incubation period of 4–14 days, the most common symptoms are characterized by chills or fever, cough, headache and fatigue, although intense pneumonia, ageusia and anosmia (loss of taste and smell) have also been reported in some cases. Depending on the prognosis, severe cases may present cytokine storm followed by respiratory failure and death (16, 18).
As a highly transmittable viral infection, large-scale mass vaccination has become a critical management tool. According to WHO, about 160 vaccine candidates are currently in clinical evaluation (1–4) (19). Intervention strategies are in its majority based on inducing neutralizing antibodies against S or other structural proteins; but vaccine designs aiming the inhibition of proteases or RNA-dependent polymerases are also being evaluated (20–22). Multiple platforms have been assessed since December 2020 in order to develop a protective vaccine against SARS-CoV-2. So far, it is still unclear which vaccine methodology is the most effective. The mRNA-based vaccines (such as BNT162 from Pfizer/BioNTech and mRNA-1273 from Moderna Inc. (23, 24)), adenovirus-based vaccines (such as AZD1222/chAdOx1 developed by the University of Oxford and AstraZeneca and Sputnik V by the Gamaleya Research Institute of Epidemiology and Microbiology (25, 26)), whole-virus-based vaccines (such as CoronaVac developed by Sinovac Research and Development and BBV152/Covaxin from Bharat Biotec (27, 28)) are among the current methodologies with approved vaccine candidates for population use. More than 11 billion doses of vaccines have been administered to the worldwide population (2). However, despite the mass-production and distribution of vaccines, new variants of concern (VOCs) continue to emerge (6), causing worries about escape from the vaccine-induced immune protection. Several studies have shown key mutations on the viral proteins compromise antibody neutralization (24, 29–31), jeopardizing population-coverage and therefore threatening COVID-19 control. This highlights the feasibility of using other viral structural proteins as candidates (32) or even a combination of them (33). Therefore, it is of the utmost importance to better understand the long-term protective mechanisms of currently used vaccines, as well as to invest in full characterization of new vaccine candidates in order to contain the spread of SARS-CoV-2 and its VOCs in a new wave of global pandemic.
Another candidate that has been investigated due to its beneficial heterologous effects is the Bacillus Calmette-Guérin (BCG). This attenuated strain of Mycobacterium bovis is protective against Tuberculosis and nowadays it is the most applied vaccine in the world (34). A growing body of evidence indicates that BCG vaccination also provides different levels of protection against non-specific pathogens (35–37), the result of a phenomenon known as “trained immunity” (38). This so-called heterologous protection induced by BCG immunization has been reported against viral infections, such as herpes simplex virus, respiratory syncytial virus, yellow fever virus, human papillomavirus, hepatitis B and influenza A (34, 39). These findings raised the possibility of BCG vaccination as a promising weapon against SARS-CoV-2 infection (40). Multiple randomized trials and retrospective cohort studies are being carried out worldwide to assess if BCG vaccination provides any level of protection against SARS-CoV-2 infection (41–43). A number of studies have suggested an association of prior BCG vaccination with lower incidence of COVID-19 disease (42, 44). Nevertheless, this concept remains controversial. Currently more than 20 clinical trials are recorded in Clinicaltrials.gov database (Clinicaltrials.gov) from which results are expected to shed some light into this question. In addition, recent preclinical studies have demonstrated the use of BCG as an adjuvant enhances the COVID-19 vaccine protection in the murine model (45–47).
Beyond any heterologous protective effects, this live bacterial vaccine can also be used as a powerful adjuvant-like tool to elicit strong cellular and humoral immune responses, enhancing vaccine responses against viral infections (35, 36, 48, 49). These immunological features laid the foundation for the development of BCG-based platforms to deliver heterologous antigens (also known as, recombinant BCG – rBCG) (50, 51). To date, several strategies for stable and controlled expression of foreign antigens by rBCG strains have been investigated with promising results, including viral, bacterial and parasitic proteins (52–54). Considering these studies, a rBCG strain expressing SARS-CoV-2 antigens merits investigation as a vaccine candidate against COVID-19 (55). This strategy could provide a powerful combination of the beneficial effects elicited by the BCG upon the innate immune system (i.e. trained immunity) and also specific cellular and humoral responses against SARS-CoV-2 elicited by the specific recombinant antigens vectorized by this system. Herein, we describe a recombinant BCG expressing a chimeric protein consisting of nucleocapsid- and spike-derived immunogenic epitopes from SARS-CoV-2 (termed here rBCG-ChD6), previously identified by our team (12, 14). Furthermore, we demonstrated the protective effects of rBCG-ChD6 against SARS-CoV-2 infection in K18-hACE2 mice. A single administration of rBCG-ChD6 followed by a booster dose of the recombinant chimera protein with alum elicited a strong cellular and humoral immune responses that resulted in protection against severe SARS-CoV-2 infection in the lungs of K18-hACE2 mouse model.
Methods
Ethics statement and mice
All animal experiments were conducted in accordance with the Brazilian Federal Law number 11.794, which regulates the scientific use of animals, and IACUC guidelines, to minimize discomfort and suffering of animals. All protocols involving animals used in this study were approved by the Ethics Committee for Animal Experimentation at the Federal University of Minas Gerais (UFMG), under permit 93/2022. K18-hACE2 mice from the Jackson Laboratory (strain #034860 - B6.Cg-Tg(K18-hACE2)2Prlmn/J) were housed in a ventilated rack system with standard 12-hour light/dark cycles. In vivo experiments were carried out using six- to 8-week-old mice for each group (n = 5–7), including male and female animals homogenously distributed among experimental groups. Standardized environmental enrichment (red mouse igloo and foraging) was used for animal welfare. Mice were properly transported to a BSL-3 facility at the University of São Paulo (Ribeirão Preto, Brazil) for SARS-CoV-2 infection and further experiments. All experiments not involving SARS-CoV-2 infected mice were conducted at an animal facility at the Federal University of Minas Gerais (Belo Horizonte, Brazil).
Peptides, bacteria, virus and chemicals
All peptides used in this work (termed Spep and Npep) were purchased from GenScript® (Nanjing, China). Escherichia coli strains DH5α and CodonPlus™ (Invitrogen, Waltham, USA) were used for the cloning and expression protocols, respectively. The Mycobacterium bovis BCG Danish strain was used to generate the recombinant BCG strains (namely rBCG-ChHsp and rBCG-ChD6) and also as wild type control for in vitro and in vivo immune experiments. The SARS-CoV-2 Wuhan strain was used for in vivo and in vitro assays. This wild type strain (HIAE-02, SARS-CoV-2/SP02/human/2020/BRA) was isolated from a nasopharyngeal specimen of a COVID-19 positive patient from Hospital Israelita Albert Einstein, São Paulo, Brazil, which presents 99.9993% similarity to the original strain isolated in Wuhan, China (9)). All reagents were purchased from Sigma-Aldrich, CO (St. Louis, USA) unless otherwise stated.
Accession numbers
SARS-CoV-2 HIAE-02 (GenBank MT126808.1 - https://www.ncbi.nlm.nih.gov/nuccore/MT126808), Spike (NCBI reference YP_009724390.1 - https://www.ncbi.nlm.nih.gov/protein/YP_009724390.1) and Nucleocapsid proteins (NCBI reference YP_009724397.2 - https://www.ncbi.nlm.nih.gov/protein/YP_009724397.2).
Chimera cloning for E. coli and BCG expression systems
The Chimera coding sequence (CDS) was designed as part of a functional cassette for direct cloning into Escherichia coli and Mycobacterium bovis specific expression vectors. The cassette contains the Chimera CDS, restriction sites for molecular handling alongside BCG regulatory regions (including a transcription terminator and a Shine-Dalgarno sequence). This cassette, herein termed Chimera-cassette, was cloned by GenScript® (Nanjing, China) into the pET-28a expression vector between NdeI and EcoRI restriction sites and the coding sequences were optimized for E. coli and M. bovis codon usage. The plasmid herein termed pET-28a::Chimera was designed in a way Chimera CDS is under the control of the T7 promoter and in frame with a poli-Histidine-tag for further affinity chromatographic recovery. The Chimera-cassete was in-house subcloned into BCG expression vectors (pJ-Hsp60 and pJK-D6) with specific promoters described elsewhere (56). Subcloning was performed using NdeI and EcoRI restriction sites so that the Chimera CDS was under the control of each promoter for constitutive expression and in frame with the transcription terminator, so that other regulatory genetic regions would not disturb BCG expression fit. The Shine-Dalgarno sequence was inserted after the transcription terminator and in frame with an auxotrophic mark (present in plasmids pJ-Hsp60 and pJK-D6) for further experiments which would not require antibiotic resistance. E. coli DH5α was transformed with each recombinant plasmid individually (herein termed pJ-Hsp60::Chimera and pJK-D6::Chimera) and screened on LB agar plates containing kanamycin (50 μg/mL). Clones were confirmed by Sanger Method sequencing at Fiocruz (Minas Gerais, Brazil).
Biophysical characterization of antigenic peptides by NMR spectroscopy
NMR measurements were performed at 2 mM peptide in a DMSO-d6 solution. NMR experiments were recorded using Bruker Avance III HD 800 MHz spectrometer (UFRJ, Rio de Janeiro, Brazil) at 300 K. Two-dimensional (2D) experiments were recorded for sequential NMR assignment (57) and distance restriction derivation. These included 1H-1H TOCSY using the DIPSI-2 pulse sequence (58) with water suppression. The spectral width was 7812.5 Hz, and 512 t1 increments with 40 transients of 1024 points were collected. Nuclear Overhauser spectroscopy (NOESY) spectra were acquired using a pulse sequence with watergate suppression (noesyesgpph) and a mixing time of 150 ms (59). The spectral width was 7812.5 Hz, 512 t1 increments were collected with 40 transients with 4096 points for each FID. 1H-13C heteronuclear single quantum coherence (HSQC) spectra were collected in the processed mode with phase-sensitive, multiple-processed HSQC using the echo-antiecho sequence (hsqcedetgp) with F1 and F2 spectral widths of 72,010 Hz and 9615 Hz, respectively (60). A total of 128 t1 increments with 300 transients with 2048 points were acquired. Chemical shift reference was performed with the internal control. All data were processed using TopSpin® software 4.1 (academic license, Bruker, Germany), assigned using CCPNMR analysis software (61) 3D structures manipulation was performed using Chimera (UCSF, USA) (62). The software DANGLE was used to derive dihedral constraints (63). Aria2.3 software was used for structure calculation (64). An ensemble of 50,000 structures was generated using a simulated annealing protocol as proposed by Krishnan et al. 2021 (65).
rChimera antigen preparation
The pET-28a::Chimera construct was transformed into E. coli CodonPlus™ (Agilent Technologies, California, USA) cells. For protein expression, it was cultured in one liter of LB medium supplemented with kanamycin (50 μg/mL) in 3 L Erlenmeyer flasks on a rotary shaker at 180 rpm at 37 °C until an OD600nm of 0.8 was achieved. Gene expression was induced by adding isopropyl-β-D-thiogalactoside (IPTG) at a final concentration of 1 mM and expression was carried out for 4 hours at 37 °C at 180 rpm. Cells were harvested by centrifugation at 3000 g at 4 °C for 15 minutes and the pelleted cells gently suspended in lysis buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 0.5 M NaCl and 10 mM imidazole). Mechanical disruption was achieved by four cycles of sonication (Branson Sonifier SLPe – Emerson Electric Co) with pulses of thirty seconds with amplitude of 60% and intervals of 30 seconds and the lysate was centrifuged at 3000 g for 15 minutes. Since rChimera was recovered in inclusion bodies, it was resuspended in denaturing buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 0.5 M NaCl, 40 mM imidazole and 8 M urea) for solubilization. Protein recovery was achieved by affinity chromatography on a Ni-Sepharose column (Hitrap chelating 5mL) using an AKTA explorer chromatography system (GE Healthcare, São Paulo, Brazil) previously loaded with appropriate buffer (Na2HPO4 10 mM, NaH2PO4 10 mM, NaCl 0.5 M, urea 8M, imidazole 40 mM, pH 7.5). Isocratic elution was performed with buffer containing Na2HPO4 10 mM, NaH2PO4 10 mM, NaCl 0.5 M, urea 8M, imidazole 0.5 M, pH 7.5. Purified proteins were dialyzed against PBS pH 7.0 at 4 °C using a Spectra/Por® membrane (MWCO 6–8.000 kDa; Spectrum Medical Industries, Inc., Laguna Hills, CA). Protein concentration was determined using the BCA™ Protein Assay Kit (Thermo Fisher Scientific, Walhtam, USA) according to manufacturer’s instructions.
ELISA assays with human sera
Human sera were used in ELISA to measure specific IgG antibodies against rChimera, Spep or Npep. The protocol was adjusted from a previously report from our group (66). Briefly, microtiter plates (Sarstedt, Nümbrecht, Germany) were coated with 1–3 μg/mL of each antigen individually. Plates were incubated with sera diluted 1:40 or 1:100 in appropriate buffer. For the unpaired analysis, sera from healthy individuals harvested before the COVID-19 pandemic (year 2015) were used as negative individuals (n = 35) and sera from SARS-CoV-2 infected individuals prior mass vaccination (year 2020; positive by molecular diagnosis – qPCR) were used as positive individuals (n = 35). For the paired analysis, sera from 13 individuals (all negative by qPCR and ELISA) were assessed before and after CoronaVac immunization (three weeks after the second vaccine dose). Plate-bound antibodies were detected using peroxidase-conjugated anti-human IgG (Promega, Fitchburg, USA). Colorimetric reaction was induced by 3,3’,5,5’-Tetramethylbenzidine and was stopped with H2SO4 solution. Plates were read at 450 nm in a microplate reader (BioRad, Hercules, USA).
Wild type and recombinant BCG preparation
The Mycobacterium bovis BCG strain Danish (herein termed BCG WT) was cultured in supplemented Middlebrook 7H9 broth (0.05% Tween 80, 0.1% glycerol and 10% oleic acid-albumin-dextrose-catalase [OADC]) to mid-exponential phase as previously described (67). Cells were then harvested by centrifugation and suspended in 10% cold glycerol, and this procedure was performed twice for cell sensitization. BCG cells were aliquoted and stored at −80 °C for in vitro and in vivo use and for electroporation of recombinant plasmids.
In order to transfect pJ-Hsp60::Chimera or pJK-D6::Chimera, 0.1 μg of each purified and dialyzed recombinant plasmid was individually electroporated into a single BCG WT aliquot with the following parameters. The recombinant plasmids and BCG WT were incubated together in 2 mm cuvettes on ice for 15 minutes prior to electroporation using a Gene Pulser Xcell™ Electroporation System (Bio-Rad, Hercules, USA) exponential wave, a resistance of 1000 Ω, 2500 V voltage and 25 μF capacitance. Immediately after the pulse, cells were transferred to 2 mL supplemented Middlebrook 7H9 and incubated for sixteen hours at 37 °C and 5% CO2 atmosphere. The culture was later seeded on supplemented selective Middlebrook 7H11 (0.5% glycerol, 10% OADC and 50 μg/mL kanamycin) agar plates and incubated at 37 °C and 5% CO2 atmosphere. A single colony for each transformant was cultured on selective supplemented Middlebrook 7H9 medium until mid-exponential phase. Cells were then harvested by centrifugation and suspended in saline buffer containing 10% glycerol and stored at −80 °C until use.
SDS-PAGE and immunoblotting
rBCG lysate supernatant preparations were obtained as it follows. Late-log phase cultures of BCG WT and rBCG were disrupted on ice with twenty sonication cycles (Branson Sonifier SLPe – Emerson Electric Co) with pulses of ten seconds, amplitude of 50% and intervals of ten seconds in lysis buffer (Tris 10 mM, EDTA 1 mM and SDS 1%). The lysate was submitted to centrifugation at 3000 g for twenty minutes at 4 °C and the supernatant was collected for total protein concentration determination using a NanoDrop® ND-1000 spectrophotometer. Purified rChimera and rBCG lysate supernatant preparations were analyzed by SDS-PAGE as previously described (68). Gels were submitted to electroblotting onto a nitrocellulose membrane using a semidry system (69). The membranes were blocked with TBST (tris-buffered saline plus 0.05% Tween 20, pH 7.2) containing 5% non-fat dry milk for 2 hours at room temperature. The membranes were then incubated with sera containing polyclonal anti-rChimera antibodies (1:200) from mice in TBST for one hour at 4 °C. Membranes were later incubated with goat anti-mouse IgG conjugated to horseradish peroxidase (HRP) (1:2000) at room temperature for one hour. Reaction was developed using ECL western blotting detection reagents (Amershan Biosciences, Piscataway, USA) according to the manufacturer’s instructions and visualized in an Amershan Imager 600 device (GE Healthcare, Chicago, USA). Densitometry analyses were performed as needed using ImageJ software.
Generation of BMDMs, in vitro assays and CFU analysis
After C57BL/6 mice were sacrificed, femurs and tibias were aseptically removed and polished free from adherent tissues. Collected bones had their ends cut and bone marrow was flushed out with PBS using a 23G needle. Bone marrow derived macrophages (BMDMs) were generated by culturing the bone marrow cells in supplemented media in Petri dishes for 7days as previously described (67). Briefly, culture medium was changed at day four and, at the seventh day of cell differentiation, cells were obtained completely differentiated in macrophages. BMDMs (5×105 cells/well) were seeded in 24-wells plates (Thermo Fisher Scientific, Waltham, USA) for 24 hours in supplemented media without antibiotics prior to BCG infection. Before use, BCG WT, rBCG-ChHsp and rBCG-ChD6 aliquots were thawed and passed through a 27G needle ten times to disrupt bacteria clumps. Infection was carried out with BCG WT, rBCG-ChHsp or rBCG-ChD6 at a 5:1 multiplicity of infection (MOI) ratio. After four hours post infection (hpi), the monolayer of cells was washed with fresh medium to remove extracellular bacteria and then supplemented medium was added. Infected cells were either lysed (4 hpi analysis) either incubated at 37 °C with 5% CO2 atmosphere for 72 hours (72 hpi analysis). Lysis was performed with saponin 0,1% for twenty minutes and cell lysates were plated onto Middlebrook 7H11 agar plates serially diluted. Colony-forming units (CFUs) were determined within 2–3 weeks.
Immunization and challenge of mice
Groups of 6- to 8-week-old K18-hACE2 mice (n = 5–7) were immunized subcutaneously in the nape of the neck by the following prime-boost regime. At day 0, mice were administered PBS, BCG WT or rBCG-ChD6 (1×106 CFU / animal). For weeks later, mice were boosted with 25 μg rChimera associated with 100 μg Al(OH)3 (Alum - Alhydrogel® adjuvant 2%, Invivogen, San Diego, USA), boosted with PBS formulations containing 100 μg of Alum or boosted with rBCG-ChD6 (1×106 CFU / animal). Twenty days after the second immunization, mice were (1) either challenged with SARS-CoV-2 (2×104 PFU) intranasally at a BSL-3 facility, (2) or sacrificed and the spleens were harvested for cytokine analysis. Independent experiments were carried out for both outcomes (SARS-CoV-2 challenge or euthanasia for spleen collection). Challenged animals were observed daily for body weight assessment and at day six post-infection were sacrificed for blood and lung collection. Lungs were harvested and processed for viral load determination.
Measurement of anti-rChimera antibodies
Sera were collected from all mice in each experimental group at day 50 before euthanasia by submandibular bleeding. In order to assess the levels of anti-rChimera antibodies, indirect ELISA was performed as previously described (66). Briefly, microtiter plates (Sarstedt, Nümbrecht, Germany) were coated with rChimera (10 μg/mL). Serum samples were serially diluted and incubated at room temperature for one hour. Plate-bound antibodies were detected using peroxidase-conjugated anti-mouse total IgG (1:2000) or its isotypes IgG1 (1:2000) and IgG2c (1:5000) (Promega, Fitchburg, USA) diluted in PBS. The colorimetric reaction was induced by 3,3’,5,5’-Tetramethylbenzidine and sulfuric acid was used to stop the reaction. Absorbance was detected at 450 nm in a microplate reader (BioRad, Hercules, USA). Isotype switch was calculated by IgG2c/IgG1 ratio using detected absorbances at 1:100 dilution. Endpoint titers were estimated by detecting the dilution of each sample that reached the average of the control sera ± 2 standard deviation units.
Assessment of neutralizing antibodies
Plasmids and HEK293T/ACE2 cells for generation of the pseudotyped virus neutralization assay were kindly provided by Drs. Paul Bieniasz and Frauke Muecksch at Rockefeller University (New York, NY, USA) (70). Sera from the immunized mice were incubated for one hour with an HIV-1-based pseudovirus at different dilutions. Pseudovirus-sera solutions were used to infect 293T/ACE2 cells in 96-wells plate and NanoLuc (RLU) activity was detected. Neutralization rate was calculated using the formula [1 - (MRS - MRNC) / (MRPC - MRNC)] × 100; where MRS = means RLU of the samples, MRNC = means RLU of the negative control (cells without virus inoculation), MRPC = means RLU from the positive control (cells inoculated with virus without any serum). The readings from MRPC were used in order to assess the limit of detection of the experiment. Results were plotted as neutralizing rate and compared to the control group (samples incubated with sera from mice previously immunized with CoronaVac vaccine). Neutralizing assays were performed using Wuhan wildtype strain as well as Gamma, Delta and Omicron variants.
Spleen cell culture and cytokine concentration determination
Spleens of mice were harvested 20 days after boost with rChimera or PBS in formulation with alum and spleen cells culture was performed as previously described (66). Briefly, spleens were mechanically disrupted, treated with ammonium-chloride-potassium buffer, washed and resuspended in RPMI supplemented with 10% FBS and 1% penicillin-streptomycin. Ninety-six-wells plates were seeded with 1×106 cells/well and cultures were treated with Polymyxin B (30 μg/mL). Spleen cells were further stimulated with rChimera (25 μg/mL) and with Concanavalin A (ConA - 5 μg/mL) or LPS (1 μg/mL) as positive controls. Cytokine levels were assessed using the Duoset ELISA kit (R&D Diagnostic, Minneapolis, USA) according to the manufacturer’s directions. Readings from the analytical standard curve were used for assessment of the experimental limits of detection. Spleen cell culture supernatants were assayed for IL-6 and IFN-γ (after 24 and 72 hours of stimulus, respectively).
RT-qPCR analysis
Lungs were harvested at day six post-infection and homogenized in TRIzol (Thermo Fisher Scientific, USA) reagent using a TissueLyzer LT (QIAgen, Hilden, Germany) to isolate total RNA according to the manufacturer instructions. Real time quantitative PCR was conducted for viral load determination using TaqMan Fast Virus 1-Step Master Mix kit (Applied Biosystems, Waltham, USA) according to manufacturer recommendations. The reaction was performed as previously described (71) and primers and probes are described in Table I. Samples were analyzed in duplicate and compared to a standard relative curve using viral stocks from cell culture of SARS-CoV-2, the serial 10-fold dilution starting from 100 ng of viral RNA, and the limit of detection was stablished at CT 40 equivalent to 0.0001 ng of total RNA. PCR reaction was performed in a QuantStudio 3 Real-Time PCR System (Thermo Fischer Scientific, Waltham, USA). Viral load CT values were interpolated to the standard curve and presented as log10 of RNA nanogram.
Table I.
Primers and probe used for real time quantitative PCR in this study.
| Primers and probe used for SARS-CoV-2 viral load determination | |
|---|---|
| Probe | 5’-FAM-CAGGTGGAACCTCATCAGGAGATGC-BHQ-1–3’ |
| Forward | 5’-GTGAAATGGTCATGTGTGGCGG-3’ |
| Reverse | 5’-CAAATGTTAAAAACACTATTAGCATA-3’ |
Analysis of viable viral titers analysis (PFU)
At six days post-infection, lungs were harvested and weighed, being disrupted in a TissueLyzer LT (QIAgen, Hilden, Germany) after appropriate weight/volume standardization. Homogenates were centrifuged at 14000 g for 10 minutes at 4 °C and supernatant containing virus was collected and serially diluted in order to infect VERO-E6 cells previously seeded on 48-well plates (5×104 cells/well) as previously described (71). Briefly, infection was carried out for 1 hour and supernatant was discarded. Carboxi-metil-celulose (CMC) 4% was added to each well and plates were incubated for 4 days at 37 °C with 5% CO2 atmosphere for plaque formation. Cells were fixed with 4% formaldehyde solution and stained with crystal violet. Plaque forming units (PFUs) were counted in all dilutions and applied to the formula (PFU mean counts) / (dilution × used virus volume). Viable virus titer results are represented as PFU/g and the limit of detection was the maximum dilution in which PFUs could be counted in the positive control wells.
Histopathological analysis
At six days post-infection, lungs were harvested and fixed in phosphate-buffered 10% formaldehyde for 48 hours being later dehydrated, diaphanized, and embedded in paraffin. Three-four micrometers tissue sections were stained with hematoxylin and eosin (H&E) for further histological analyses. Overall lung architecture was analysed in the slides and 15–20 representative images for each slide were captured using an optical microscope Olympus CX31 attached to a microcamera JVC 1270/RGB using 10x or 20x magnification lens. Cell infiltration was depicted in representative pictures. The free alveolar space was measured using ImageJ software. Briefly, captured digital images were used to create binary images and pixels were later converted into μm2. Free area was measured and expressed in relation to the total area of each section for percentage calculation. For free alveolar space determination, vascular vessels, bronchi and bronchioles were excluded from the analysis.
Statistical analysis
All statistical analysis were performed using the GraphPad Prism software package (La Jolla, Irvine, USA). Kolmogorov-Smirnov normality tests were applied. Analysis with two groups were performed using Student’s t-test for parametric data or Mann-Whitney test for nonparametric data. Analysis with more than two groups were performed with OneWay or TwoWay ANOVA (Tukey or Bonferroni adjustments were included for multiple comparisons). P-values obtained by these methods were considered significant when less than 0.05. For humoral assays, ELISA data were submitted to ROC curve analysis in order to determine the cut-off points, when necessary.
Results
Characterization of immunogenic SARS-CoV-2-derived peptides
The rational design of an immunogenic vaccine against SARS-CoV-2 by immunoinformatic analysis of viral structural proteins has been previously reported by our group (12, 14). The nucleocapsid (N) and spike (S) proteins were screened for major B and T cell epitopes for potential targets and the most immunodominant epitopes were selected for further characterization. We selected two N- and S-derived peptides for characterization based on previous analysis. Peptide #1 (Figure 1) comprises epitopes identified in the NTD region of the N protein. This region is highly conserved and has high potential for T cell cross-reactivity with Nucleocapsid proteins from other human coronaviruses. Another N-derived peptide (peptide #2 in Figure 1) comprises epitopes from an unstructured serine-arginine-rich region between the NTD and CTD regions and represents a strong immunodominant B cell epitope. The selected S-derived peptides (#3 and #4 in Figure 1), are both epitopes identified in the Receptor Binding Domain (RBD) region with high binding affinity for multiple human MHC I and II alleles. This region is also predicted to have several B cell epitopes that are likely to elicit a strong antibody response mediated by T cell help. Taken together, these immune features predict highly immunogenic potential for the use of these peptides in a chimeric multi-epitope vaccine.
Figure 1. Immunodominant B and T cell epitopes identified in the Nucleocapsid and Spike proteins for vaccine formulation.
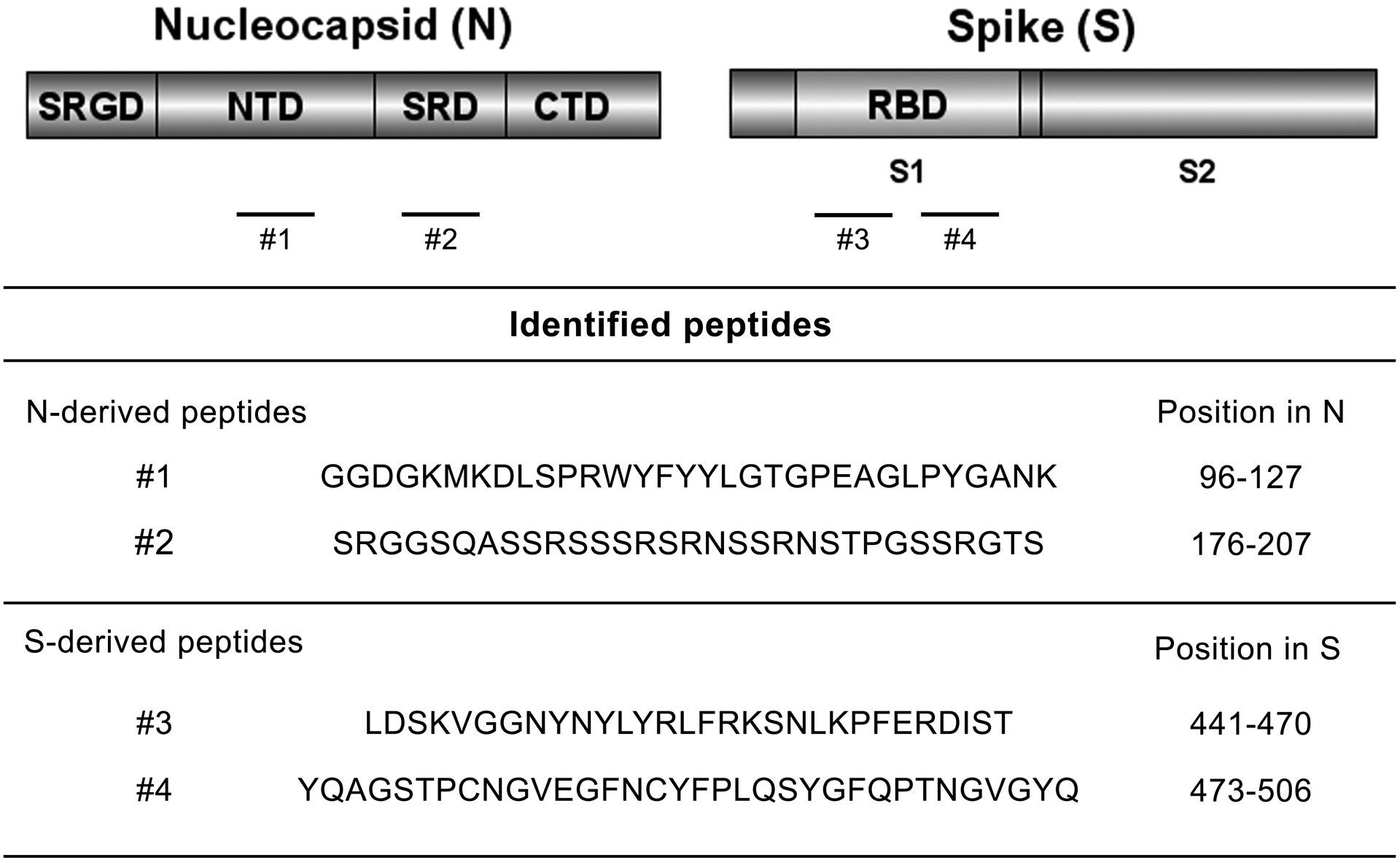
Schematic representation of the Nucleocapsid (N) and Spike (S) structural proteins from SARS-CoV-2. Protein domains are represented by N- and C-terminal domains (NTD and CTD, respectively) for the Nucleocapsid, including the serine-glycine-arginine rich domain (SGRD) and the serine-arginine rich domain (SRD); and by S1, S2 and the Receptor Binding Domain (RBD) for Spike. Immunogenic peptides previously identified by our group are represented as numbered dashes. Identified N- and S-derived amino acid sequence and position are also shown. The complete immunoinformatic analysis is described elsewhere (12, 14). Figures were prepared using DOG illustrator of protein domains structures (88). N and S accession numbers are YP_009724397.2 and YP_009724390.1, respectively.
In order to validate our immune predictions and also gain further insights into preferential structural conformations, peptides #1 and #3 (Figure 1), herein termed Npep and Spep, respectively, were chemically synthesized and used in further analyses (Figure 2). The 3D structures were solved in DMSO in order to evaluate the peptide preferred conformations in a medium able to mimic the cellular environment. Both peptides were found to lack mid and long-range constraints. The Npep forms a concavity in the peptide center, exposing residues 11P, 12R, 13W, 14Y and 16Y and seems to be highly flexible in the extremities (Figure 2A). Similarly, Spep structures appear to form a concavity in this region, exposing some residues (14R, 16F, 17R and 18R), while the N- and C- terminal regions are more flexible (Figure 2B). The best Spep RMSD values are in the region of residues 9 to 10. The Spep concavity exposes an aromatic residue surrounded by highly positively charged residues, while the Npep exposes a positive large arginine surrounded by aromatic tyrosines and a tryptophan.
Figure 2. N- and S-derived peptides are stable and reactive to SARS-CoV-2 infected sera.
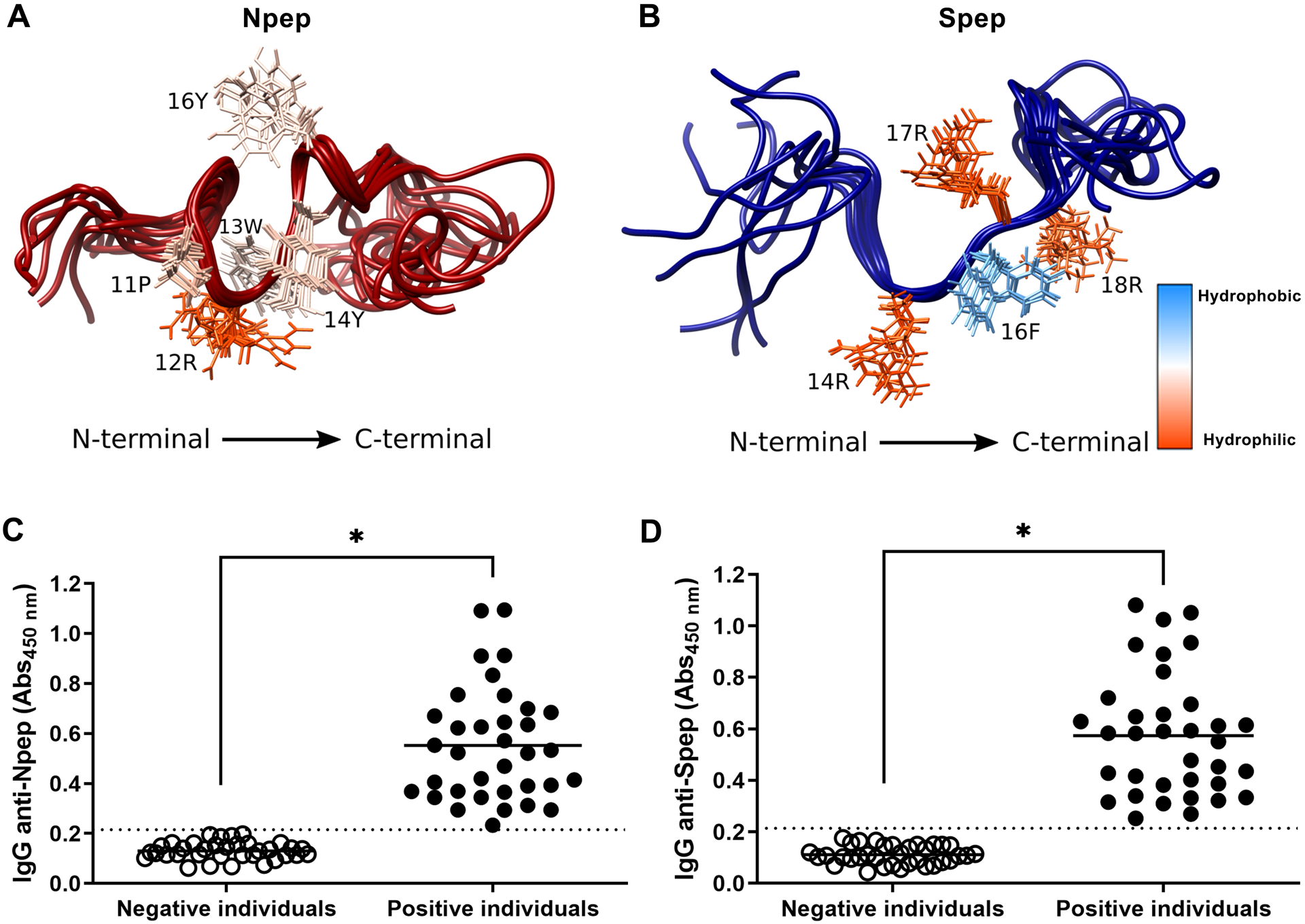
NMR 3D structures for peptides (A) Npep and (B) Spep. The ten structures of lowest energy are represented with the superposition of the backbone traces. The residues’ side chains are colored by the Kyte-DoLittle hydrophobicity scale, labeled at right, being dodger blue the most hydrophobic residues and orange red the most hydrophilic ones. The highlighted side chains are in the peptide’s concavities. Furthermore, Npep (C) and Spep (D) were tested against human sera by ELISA. Sera from individuals infected with SARS-CoV-2 prior to mass vaccination and positive by molecular diagnosis (qPCR) were used as Positive individuals (n = 35). Samples from healthy people collected prior to 2015 were used as Negative individuals (n = 35). Sera were diluted 1:100 for the assay and total IgG detected values are plotted. Graphs are representative of two independent experiments. Statistical analysis were performed using Student’s t-test and * represents p value < 0.05. Dotted lines represent the cut-off between the samples and were obtained by ROC curve analyses.
Predicted immunogenicity of Npep and Spep was assessed by indirect ELISA using human sera. Sera from individuals with positive qPCR diagnosis for SARS-CoV-2 before mass vaccination program (i.e., collected in June, 2020, so individuals with no vaccine-induced antibodies) were used as positive controls and sera from healthy individuals (obtained before COVID-19 pandemics) were used as negative controls. Both Npep (Figure 2C) and Spep (Figure 2D) revealed to be reactive to sera from SARS-CoV-2 infected patients and non-reactive to sera from negative healthy individuals, validating our previous immunoinformatics analysis and highlighting their potential use in vaccine formulations.
Expression of the chimeric protein derived from SARS-CoV-2 containing immunodominant peptides in E. coli and BCG
The immunogenic peptides characterized in the N and S proteins from SARS-CoV-2 were used to guide a rational design of a chimeric recombinant protein suitable for use in vaccine formulation, herein termed rChimera. This recombinant protein was designed to contain two domains (Figure 3A). Its N-terminal domain (consisting of Nucleocapsid-derived peptides, herein termed N-domain) is thought to elicit strong humoral and cellular responses against SARS-CoV-2 and potentially to evoke cross-reactivity against other human coronaviruses (including peptides #1 and #2 listed in Figure 1). This first domain comprises 161 amino acids, incorporating the NTD and also a very flexible, nonuniform serine-arginine-rich region from SRD (peptide #2) as a linker to the second domain (Spike-domain) of the rChimera. Its C-terminal domain (consisting of Spike-derived peptides, herein termed S-domain) exhibits several B cell epitopes and it is designed to induce neutralizing antibodies against RBD from Spike, impairing viral attachment and subsequent genomic RNA delivery. This second domain comprises 294 amino acids including peptides #3 and #4 listed in Figure 1. The rChimera protein was designed to have 455 amino acids in length (52.2 kDa).
Figure 3. Recombinant Chimera is reactive to human sera from SARS-CoV-2 infected patients and also from CoronaVac immunized individuals.
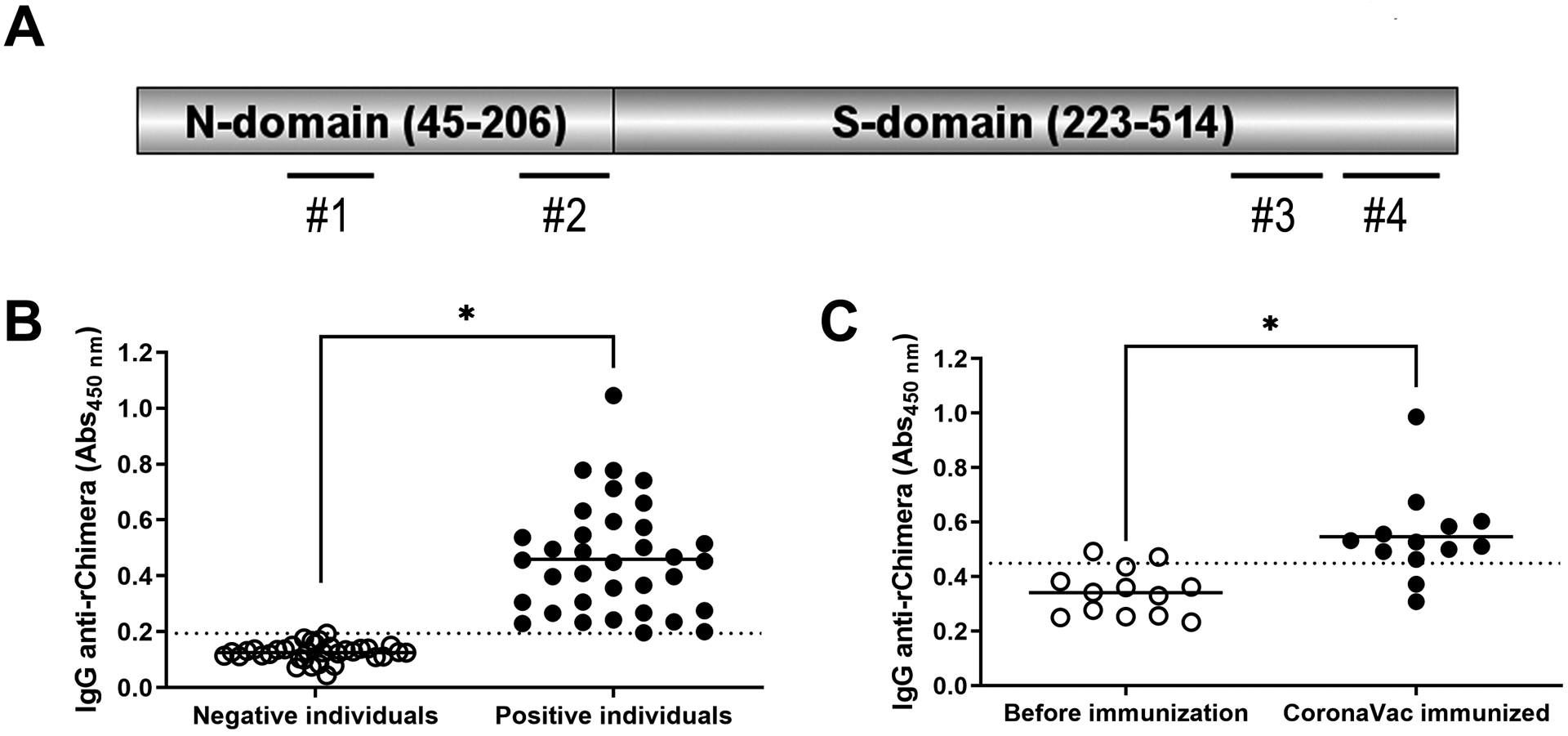
A) Schematic representation of rChimera. The N-domain is composed of 45–206 amino acids from N protein (YP_009724397.2) while the S-domain incorporates 223–514 amino acids from the S protein (YP_009724390.1). The positions of peptides #1, 2, 3 and 4 are shown by dashed lines. B) Reactivity of rChimera against human sera was assessed by ELISA. Sera from SARS-CoV-2 infected individuals (from year 2020, prior to mass vaccination and positive by molecular diagnosis) and sera from healthy individuals (from year 2015, prior to the pandemic) were used as positive and negative controls of the assay (n = 35). Sera were diluted 1:100 for the assay. C) Sera from non-SARS-CoV-2-infected individuals (negative by molecular and serological diagnosis) were assessed to rChimera’s reactivity before and after CoronaVac immunization. Samples were collected at day zero (before immunization) and three weeks after CoronaVac booster dose (n = 13). Both results are representative of total IgG antibodies against rChimera detected in the samples. Graphs are representative of two independent experiments. Statistical analysis were performed using Student’s t-test and * represents p value < 0.05. Dotted lines represent the cut-off between the samples and were obtained by ROC curve analyses.
The coding sequence of rChimera was optimized for heterologous expression in Escherichia coli and Mycobacterium bovis systems as described in the Methods. After heterologous expression in E. coli and affinity chromatographic recovery, the reactivity of rChimera against human sera was demonstrated by indirect ELISA (Figure 3B), and was clearly able to distinguish SARS-CoV-2 naturally infected individuals from healthy donors. Interestingly, it was also reactive to the sera from CoronaVac-immunized individuals (which had not been previously infected), demonstrating the rChimera protein is also recognized by antibodies generated from inactivated whole-virus immunization (Figure 3C). Once rChimera was recognized by anti-SARS-CoV-2 antibodies, its coding sequence (CDS) was subcloned into suitable plasmids for expression in BCG.
The Chimera CDS was cloned in two different plasmids, pJ-Hsp60 and pJK-D6, under the control of two promoters: PHsp60 or PD6, both previously described elsewhere (56) and details are described in the methods section. These constructs (Supplementary Figure 1A) were designed with regulatory sites after Chimera CDS, containing a transcription terminator site (TSS) and an auxotrophic marker (LysA). These molecular elements were added so in further experiments, the TSS could be enzymatically removed, and the plasmids would be able to be maintained by auxotrophic complementation in an appropriate BCG strain, without requiring antibiotic selection. Recombinant plasmids were transformed into BCG Danish strain (herein termed BCG WT) and recombinant BCGs were handled accordingly. Two recombinant strains were generated and cultured for further experiments: rBCG-ChHsp and rBCG-ChD6, containing the recombinant plasmids with Chimera CDS under the control of PHsp60 and of a high strength promoter (PD6, obtained through error-prone PCR of the strong PL5 promoter, originally from mycobacteriophage L5), respectively. Late-log phase cultures were disrupted by sonication in appropriate lysis buffer and lysate supernatants containing soluble rChimera were analyzed by Western blotting (Supplementary Figure 1B). Densitometry analysis demonstrates rBCG-ChD6 strain expresses 1.8-fold higher levels of rChimera, when compared to rBCG-ChHsp, observed at approximately 52 kDa. As expected, BCG WT control did not reveal a protein band at this molecular weight under the incubation with sera from rChimera-immunized mice. The differential growth of the recombinant strains was assessed in vitro by incubation with bone-marrow-derived macrophages (BMDMs) at a multiplicity of infection of 5:1 (Supplementary Figure 1C). Bacterial growth was monitored at 4- and 72-hours post-infection (hpi). Despite a slight significant increase in the uptake of rBCG strains by the macrophages when compared to BCG WT at 4 hpi, this phenotype was not observed at 72 hpi. Altogether, these results indicate both rBCG strains replicate well in macrophages. As rBCG-ChD6 expresses higher levels of rChimera, this strain was selected as a suitable candidate for in vivo analysis and further use in vaccine formulation.
Prime/boost strategy with rBCG-ChD6 and rChimera induces neutralizing antibodies against SARS-CoV-2 Wuhan strain
In order to assess the efficacy of our designed chimeric protein for vaccine formulation, a prime/boost immunization strategy was performed (Figure 4A). K18-hACE2 mice were immunized subcutaneously with BCG WT or rBCG-ChD6, or with PBS as control. Four weeks later, mice were boosted with a formulation containing rChimera or PBS as control (both with alum). Twenty days later serum was harvested to measure total IgG responses, IgG2c/IgG1 ratio, neutralizing antibodies and cytokine (IFN-γ and IL-6) responses.
Figure 4. Immunization of K18-hACE2 mice with rBCG-ChD6 and boosted with rChimera protein induces IgG and IgG2c/IgG1 isotype responses, neutralizing antibodies, IFN-γ and IL-6 production.
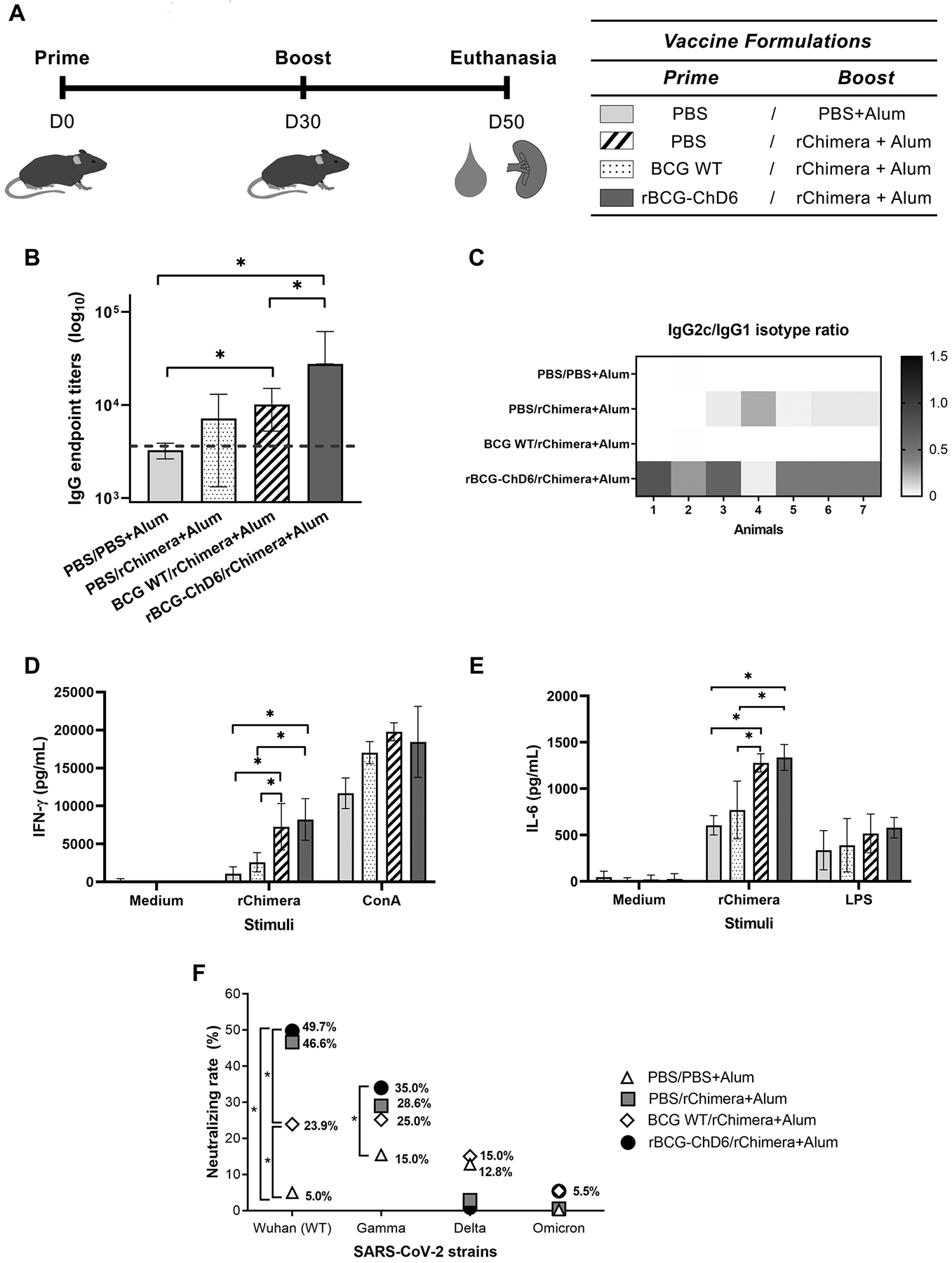
A) Schematic representation of K18-hACE2 mice vaccination. Mice (n = 5–7 mice) were immunized at day 0 with PBS, BCG WT or rBCG-ChD6 (s.c.). At day 30, mice were boosted with formulations containing rChimera+Alum or PBS+Alum. At day 50, blood was collected and spleens were harvested. Immunization groups are indicated next to the vaccination schedule. Sera collected at day 50 were used to determine antibodies against rChimera by ELISA. B) Sera was diluted serially and IgG endpoint-titers were determined by ELISA. The dilution of each sample that reached the average of the control sera (± 2 standard deviation units) was represented in bars by mean ± S.D and the dotted line indicates the limit of detection. C) Sera diluted 1:100 were assayed for IgG1 and IgG2c antibody isotype determination against rChimera. IgG2c/IgG1 ratio is represented by the heatmap and the color scale ranges from 0.0 in black to 1.5 in white, indicating the IgG2c/IgG1 ratio of detected absorbances. D-E) Spleens were harvested 20 days after booster dose. Culture cell supernatant was assayed for (D) IFN-γ and (E) IL-6 cytokine productions in response to stimulation with rChimera, LPS or ConcanavalinA. Bars represent mean cytokine concentration ± SD for each group. F) Neutralizing rates were assessed by incubating the mice sera for one hour with an HIV-1-based pseudovirus at different dilutions. Pseudovirus-sera solutions were used to infect 293T/ACE2 cells in 96-well plate and NanoLuc (RLU) activity was detected. The neutralization rate against wildtype and variant viruses is obtained by the formula described in the methods section and final results are plotted as the observed percentage of neutralization. Statistical analyses were performed using 1-way ANOVA (for B and F) and 2-way ANOVA (for D and E) followed by the Bonferroni post-hoc test. * represents p value < 0.05. Results are representative of two independent experiments. Limits of detections are described in the methods section.
Sera from immunized mice were assayed by ELISA (Figure 4B) and a strong humoral response was detected (at day 50) in the animals immunized with rBCG-ChD6 and boosted with purified rChimera associated with alum when compared to the other studied groups. Mice previously immunized with BCG WT or PBS before rChimera boost presented less anti-rChimera antibody titers when compared to the rBCG-ChD6 group. Significant antibody levels were not detected prior to booster dose for all groups at day 30 (data not shown). We also assessed the antibody isotype profile measuring IgG2c/IgG1 ratio in vaccinated mice. We observed an increased IgG2c/IgG1 ratio in rBCG-ChD6 immunized compared to the other vaccinated groups (Figure 4C). As the IgG2c antibody isotype correlates with a Th1-type of immunity, an early IgG2c response could suggest an effective neutralizing antibody and cellular responses against SARS-CoV-2 in rBCG-ChD6 immunized animals. In parallel, we observed higher IFN-γ (Figure 4D) and IL-6 (Figure 4E) production by splenocytes of rBCG-ChD6/rChimera and BCG WT vaccinated groups when compared to rChimera or PBS with alum administered animals. Even though alum is known as an adjuvant that primes host immune responses to Th2, BCG has the ability to induce a strong Th1 immunity. It has been previously demonstrated that T cell responses in recovering COVID-19 patients are predominantly Th1-driven (72). Furthermore, rBCG-ChD6/rChimera+Alum vaccinated group presented increased levels of neutralizing antibodies against SARS-CoV-2 Wuhan strain when compared to BCG WT/rChimera group (Figure 4F). Alternatively, the same sera were assayed against SARS-CoV-2 variants of concern but resulted in decreased neutralizing rate for Gamma and failed to neutralize Delta and Omicron variants (Figure 4F). Collectively, these results demonstrate the rBCG-ChD6/rChimera+Alum vaccinated animals engendered a type of immune response that correlates with protective potential against COVID-19, but further investigation might be required regarding viral variants of concern.
Vaccination with rBCG-ChD6 followed by rChimera booster dose elicits protective immunity against SARS-CoV-2 infection in K18-hACE2 mice
Next, we evaluated the protective potential of the rBCG-ChD6/rChimera+Alum immunization regimen employing a murine model of SARS-Cov2 infection (Figure 5A). Twenty days after the boost immunization, K18-hACE2 mice were intranasally inoculated with 2×104 SARS-CoV-2 PFU. Control groups, comprising PBS/PBS+Alum and PBS/rChimera+Alum, demonstrated severe clinical deterioration beginning at day 4 post-infection and animals were very weak at the sixth day of observation (Figure 5B). This is suggestive that a single dose of rChimera protein might have not been sufficient to elicit a protective response against SARS-CoV-2 infection. A similar phenotype was also observed in the animals immunized with BCG WT/rChimera+Alum, suggesting the immune response elicited by BCG immunization was also not enough to protect against the infection, even with a rChimera protein booster dose (Figure 5B). Remarkably, rBCG-ChD6 immunized mice demonstrated no apparent weight loss when observed for a week (Figure 5B), indicating the immune response induced by this vaccine played an important role in the protection observed.
Figure 5. Prime-boost immunization regimen with rBCG-ChD6 and rChimera reduced weight loss in SARS-CoV-2 infected K18-hACE2 murine model.
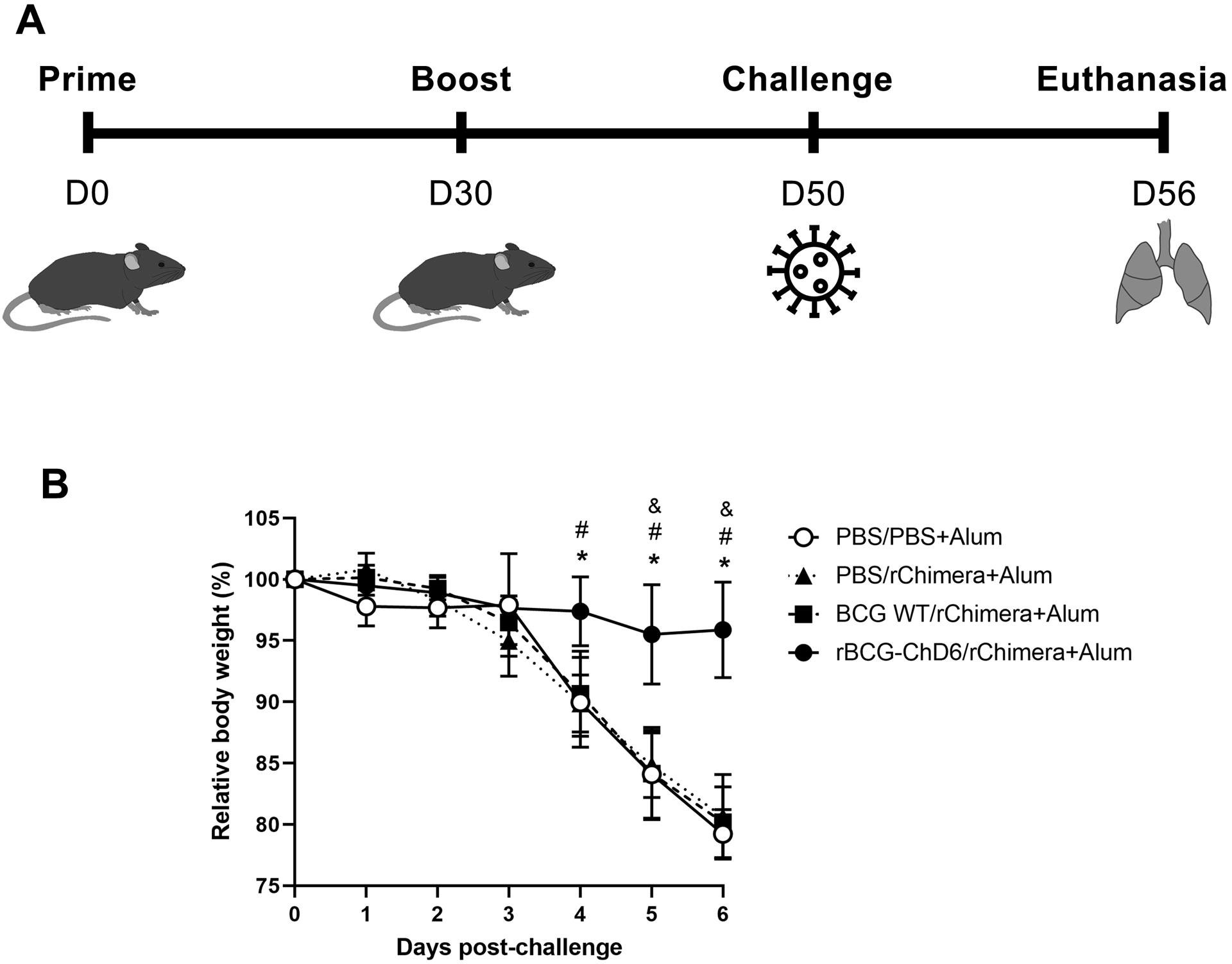
A) Schematic representation of K18-hACE2 mice (n = 5–7) prime-boost vaccination. Mice were immunized subcutaneously at day 0 with PBS, BCG WT or rBCG-ChD6 (s.c.). At day 30, mice were boosted with formulations containing PBS or rChimera with Alum. SARS-CoV-2 challenge was performed intranasally at day 50 and lungs were harvested at day 56. Results are representative of two independent experiments. B) Mice were observed for the following six days and relative body weight is represented. Statistical analyses were performed using 2-way ANOVA followed by the Bonferroni post-hoc test. An asterisk represents statistical difference when rBCG-ChD6/rChimera+Alum group (indicated by black circles connected by straight lines) was compared to PBS/PBS+Alum (indicated by white circles connected by straight lines), # when it is compared to PBS/rChimera+Alum (indicated by black triangles connected by dotted lines) and & when it is compared to BCG WT/rChimera+Alum (indicated by black squares connected by dashed lines) (p value < 0.05).
At day six of SARS-CoV-2 post-infection, mice were euthanized, and lungs harvested for further analysis. Bulk-RNA extracted from the tissues was used as template for RT-qPCR determination of the viral load (Figure 6A) using specific primers for SARS-CoV-2 structural proteins. rBCG-ChD6 immunized group presented a marked decrease in the amount of detected viral RNA in the tissues from infected lungs and statistically significant differences were observed for all assessed groups. Whereas the other experimental groups apparently presented lesser RNA viral load, no significant difference was detected (p value = 0.2247 when BCG WT/rChimera+Alum immunized group was compared to the PBS control group; and p value = 0.234 when PBS/rChimera+Alum immunized group was compared to the PBS control group). Additionally, the PFU analysis revealed all three vaccinated groups demonstrated reduced viral viability when compared to the PBS control group (Figure 6B). However, full viral impairment in infected cells was observed in the rBCG-ChD6 immunized group, where no PFU was detected. Regarding lung pathology associated with infection, rBCG-ChD6/rChimera+Alum immunized mice presented overall pathologic amelioration at 6 days post-infection, with reduced inflammatory infiltrates in the alveoli (Figure 7 A–D), leading to alveolar space preservation in a greater extent (Figure 7E). Alternatively, we observed that mice which received two doses of rBCG-ChD6 as a homologous prime-boost strategy failed to be protected against viral infection. Body weight loss (Supplementary Figure 2A) and viral load of rBCG-ChD6 vaccinated mice were not statistically significant (Supplementary Figures 2B-C) when compared to mice immunized with PBS, highlighting the importance of a rChimera boost for the development of an effective and protective immune response. Collectively, these findings demonstrate that mice immunized with a single dose of rBCG-ChD6 and boosted with purified rChimera associated with alum are protected against COVID-19 clinical disease manifestations due to an early and rapid viral control and reduced pathology.
Figure 6. rBCG-ChD6/rChimera+Alum immunization provides protection against SARS-CoV-2 infection in the lungs of K18-hACE2 mice.
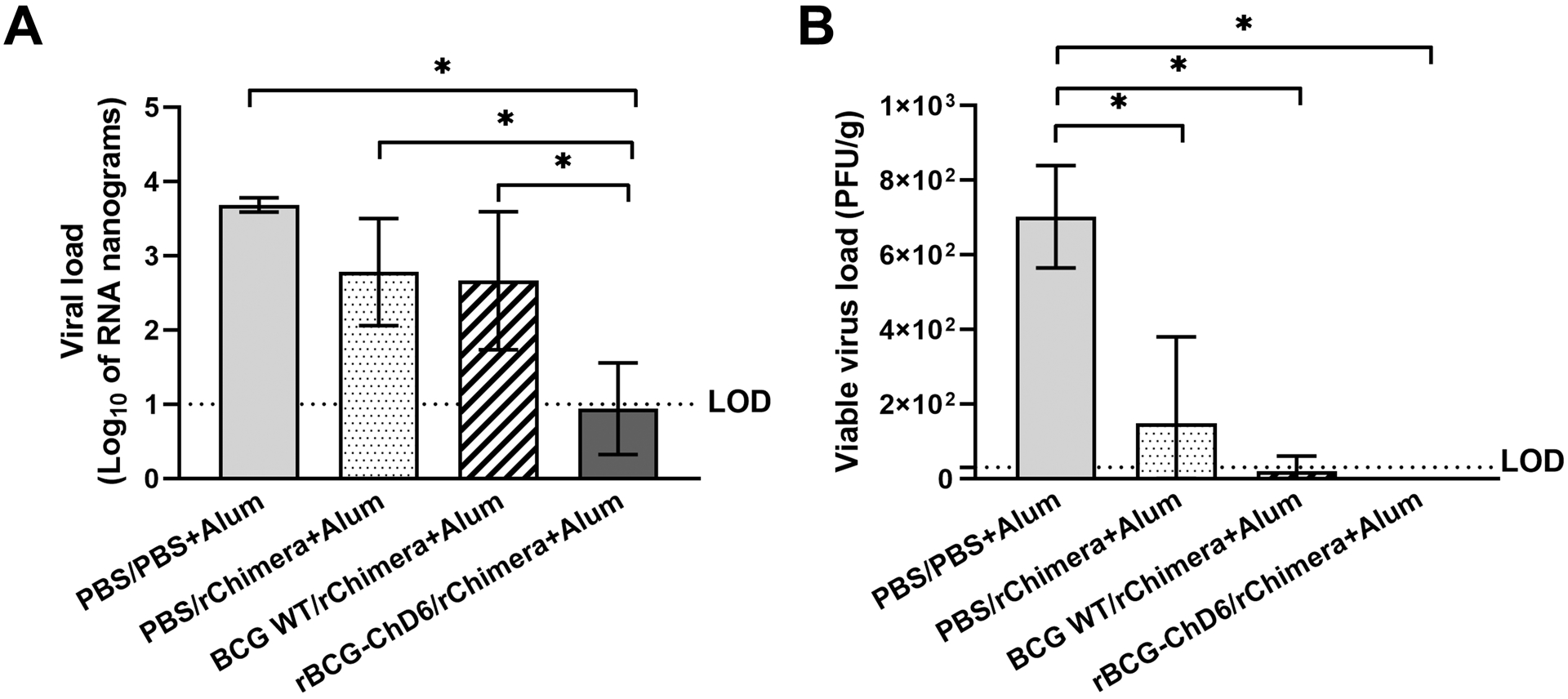
Harvested lungs from SARS-CoV-2 infected mice were processed after 6 days. A) RNA extraction followed by RT-qPCR using 100 ng of total RNA determined the viral load of each sample and is presented as Log10 of nanograms of RNA. B) Supernatant from lung homogenates containing viable viruses were serially diluted and used to infect VERO-E6 cells for 1 hour. After applying CMC and letting plaque formation, cells were fixed with formol and stained with crystal violet. PFUs were counted and viable viral titers are represented as PFU/g. Statistical analysis were performed using 1-way ANOVA followed by the Bonferroni post-hoc test and * represents p value < 0.05. Results are representative of two independent experiments. LOD represents the limit of detection for each experiment.
Figure 7. rBCG-ChD6 priming followed by rChimera+Alum boosting ameliorates lung pathology in K18-hACE2 mice infected with SARS-CoV-2.
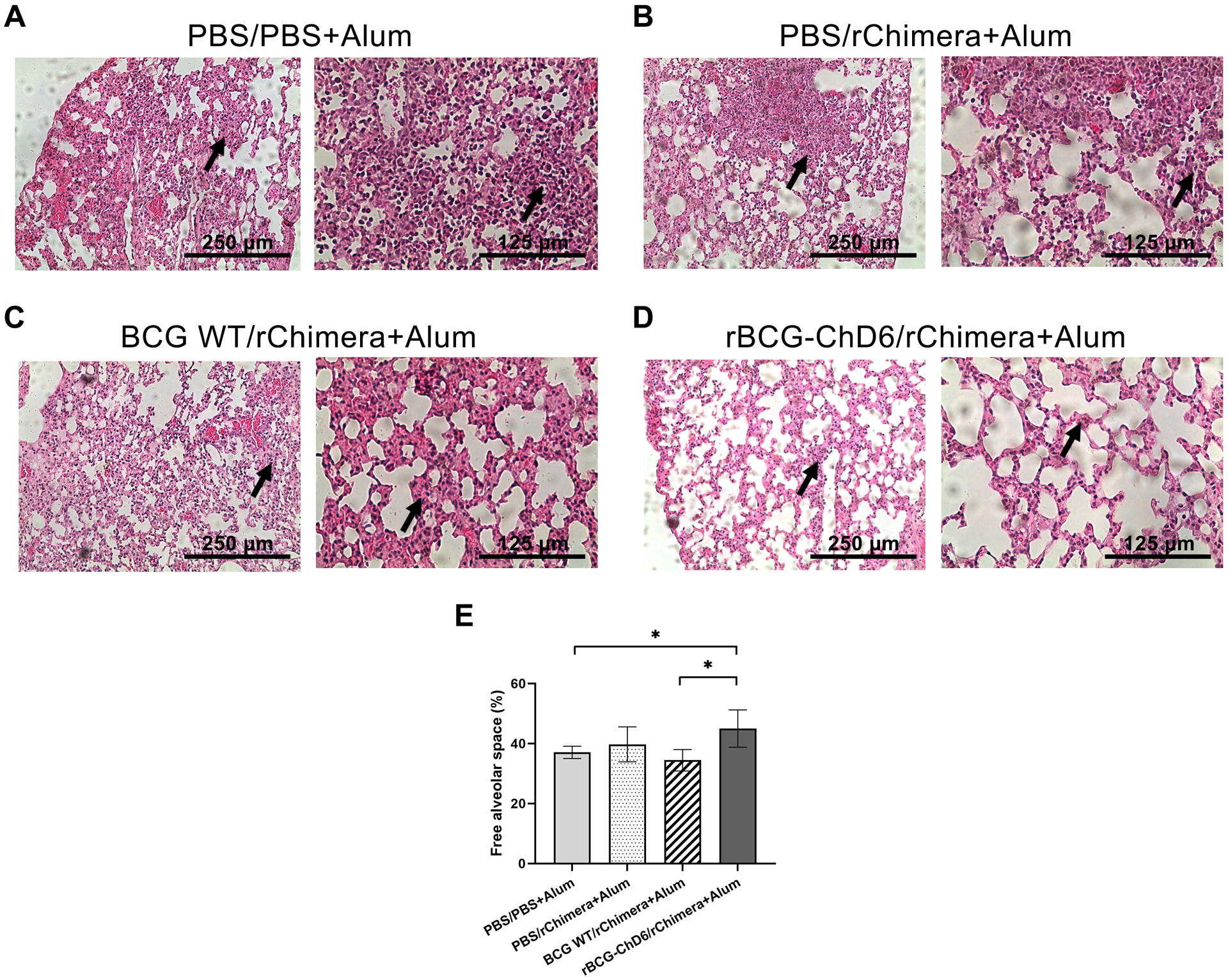
The lungs from previously immunized K18-hACE2 mice were harvested 6 days post-infection with 2×104 PFU SARS-CoV-2 for histopathological analyses. Panels A-D indicate representative images from lungs of each immunized group analysed at 10- and 20-times magnification with a micrometer scale depicted at each picture for comparison. Black arrows indicate inflammatory infiltrates. Panels A-C represent diffuse pneumonia while panel D represents overall preservation of pulmonary alveoli spaces. Panel E demonstrates alveoli wall thickening analyses using ImageJ software plotted as free alveolar space percentage. Twenty images were analysed for each lung and 5–7 mice were used for each group for the whole histopathological analysis. Statistical analyses were performed using 1-way ANOVA followed by the Bonferroni post-hoc test and * represents p value < 0.05. Results are representative of two independent experiments.
Discussion
The COVID-19 pandemic has had a tremendous impact on society, with a high number of deaths and economic loss. A critical issue is ensuring the adequate supply of vaccines to low and middle-income countries. The mRNA vaccines, for instance, require maintenance at low temperatures and have complex logistical requirements. An interesting strategy is to reposition existing licensed vaccines for the use against COVID-19. In this context, the Mycobacterium bovis Calmette-Guerin (BCG), the most frequently applied vaccine worldwide has drawn particular interest (73). Despite its original purpose as a tuberculosis vaccine, numerous studies have demonstrated the potential of BCG to provide effective protection against heterologous infections. Particularly, BCG vaccination is known to provide protection against other respiratory infections by inducing trained immunity (39). Our hypothesis was that recombinant BCG expressing immunodominant peptides from SARS-CoV-2 could be an effective strategy to induce protection against COVID-19, by combining the adjuvant-like properties of BCG, its ability to induce strong cellular and trained immunity, and anti-SARS-CoV-2 specific neutralizing antibodies.
In this study, we describe the construction of a recombinant BCG-based vaccine which enables complete elimination of viable SARS-CoV-2 in the lungs of K18-hACE2 mice. We have designed a Nucleocapsid/Spike chimeric protein based on highly immunogenic peptides identified by bioinformatic analysis (12, 14). These peptides were predicted to elicit strong humoral and cellular immune responses against two key structural proteins from SARS-CoV-2. The structure of these peptides were defined by NMR and our results show that the N-peptide exhibits the typical random shape of spectra previously observed for longer β-sheet-forming peptides (74). These preferred conformations of the N-peptide correlate with the immunogenic character of T cell epitopes, which were generally found to be disordered structures with preferred conformations in specific regions (75, 76). On the other hand, we observed that the S-peptide appears to have a conformational folded preference that resembles those found in peptides bound to antibody molecules (77). The interaction between peptide and antibody is guided by many complex factors, and the structure and stability of the peptide in solution is one of the most important features, demonstrating the importance of having these peptide domains in the chimeric recombinant protein.
The strategy here was not only to target the spike protein but also key regions from the N protein, which could also confer broader cross-recognition of immunogenic peptides with other beta-coronaviruses (12). A recent study demonstrates that cross-protective immunity could be of interest when assessing SARS-CoV-2 vaccines (78). As the Spike protein is highly mutated in different viral variants and may account for loss of antibody-mediated protection as new variants appear (79), incorporating N-derived peptides in our vaccine design was an important strategy. The N-epitopes selected in this study predicts to elicit T-cell mediated responses, activating another arm of the immune system and not solely relying on neutralizing antibodies. The rChimera protein was recognized by the sera of SARS-CoV-2 infected patients and by CoronaVac-immunized individuals demonstrating its antigenic properties. K18-hACE2 mice were immunized with the rBCG-ChD6 or BCG WT as control and boosted with rChimera protein associated with alum, aiming to induce strong humoral and cellular immune responses to N- and S- peptides. Recently, proteomic studies demonstrated that Mycobacterium bovis BCG and SARS-CoV-2 have significant peptide-homology (80, 81). Eggenhuizen and colleagues, reported that CD4+ and CD8+ T-cells in vitro primed with BCG-derived-peptides present cross-reactivity to SARS-CoV-2-peptide homologues, what could partially account to the epidemiological observations of BCG-vaccinated individuals being partially protected against SARS-CoV-2 infection. Furthermore, BCG induces nonspecific heterologous protection due to “trained immunity” by reprogramming innate immune responses (39). These findings reinforce the use of a rBCG-system for the delivery of SARS-CoV-2 derived antigens to elicit efficient immune responses.
We observed that immunization with rBCG-ChD6/rChimera+Alum elicited the highest anti-rChimera total IgG and IgG2c antibody titers with neutralizing activity to SARS-CoV-2 Wuhan strain in pre-challenge experiments when compared to BGC WT or with rChimera vaccinated groups. Additionally, rBCG-ChD6 or BCG WT-immunized animals presented higher levels of IFN-γ and IL-6 by spleen cells when compared to rChimera with alum, highlighting the importance of the BCG-based system in evoking strong cellular responses. A similar profile has been seen in mice vaccinated with a recombinant BCG expressing an HIV-1 p24 Gag (82) which led to a Th1-like cellular response and higher IgG2a/IgG1 ratio (83, 84). Recently, another recombinant BCG expressing SARS-CoV-2 N protein was reported (85) to induce a Th1-cell response when spleen cells from immunized mice were stimulated with the nucleocapsid protein. However, these investigators showed that a single dose of rBCG expressing N antigen was insufficient to elicit a strong humoral response. This reinforces the need of a booster dose to enhance antibody titers with neutralizing activity in the rBCG-platform vaccine. We also observed that a homologous prime-boost protocol based only on rBCG-ChD6 failed to protect mice from SARS-CoV-2 infection, which corroborates the importance of a purified rChimera protein booster dose in order to induce a more robust and effective immune response.
In addition to several clinical trials being conducted to evaluate the impact of BCG-vaccination against COVID-19, several groups have been assessing the heterologous protective effects of BCG vaccination upon SARS-CoV-2 infection in K18-hACE2 murine model, but divergent findings have been reported. Kaufmann and colleagues (86) have observed BCG vaccination fails to protect against SARs-CoV-2. At their work, K18-hACE2 mice were subcutaneously or intravenously immunized with BCG before challenge but no differences in morbidity or mortality were observed when compared to the BCG non-vaccinated group. The same phenotype was observed in Syrian Golden Hamster model, which naturally expresses the ACE2 receptor. In contrast, Hilligan and colleagues (87) observed a different outcome for intravenous inoculation of BCG. In their work, they observed that mice intravenously immunized with BCG (but not subcutaneously) were protected against lethal SARS-CoV-2 challenge with viral load being significantly reduced in the lungs from K18-hACE2 mice. They suggest this outcome was due to changes in both composition and function of pulmonary cellular components induced by BCG, as intravenous inoculation allows the Mycobacterium to reach the lungs, inducing innate immune alterations. Similar findings were reported by Zhang and collaborators (47), where protection was achieved by intravenous inoculation of BCG. Despite the use of different BCG strains (Tice, Pasteur and Tokyo, respectively) or different SARS-CoV-2 doses (1×103 TCID50, 4×103 TCID50, 104 TCID50 or 1.25×104 PFU) employed in these studies, the key feature that accounted for protection was the intravenous inoculation of BCG; however, it is not a clinically accepted practice in humans.
In our study, immunizations were performed subcutaneously, as this route is similar to intradermal BCG vaccination performed in humans what makes this model more suitable for ready-implementation in a clinical setting. Here, post-challenge analysis demonstrates rBCG-ChD6/rChimera+Alum immunized mice showed no apparent clinical manifestation of COVID-19, featured by no weight loss at day 6 pos-infection and overall pulmonary alveoli preservation at histopathologic analysis, but also diminished viral load detected in the lungs by qPCR and no viable virus detected by PFU analysis when compared to BCG WT and rChimera control groups. These data altogether point to a prompt neutralizing response against SARS-CoV-2 elicited in rBCG-ChD6/rChimera+Alum vaccinated animals which led to full eradication of viable virus in the lungs of the mice. The neutralizing activity to SARS-CoV-2 Wuhan strain from antibodies derived from rChimera’s S-domain could have had a significant impact on the early control of the infection. Together with a combination with a Th1-type of cellular response induced by vaccination (presumably by both N- and S-domains from rChimera) lead to the protective response observed against viral challenge; however, further investigation is required to define the mechanisms involved in protective immunity. Interestingly, although the BGC WT-immunized group has presented significant decrease in viable virus in the lungs, mice had lost 20% of their body weight at day six post-infection. It is important to mention that, despite this group had been boosted with rChimera with alum, they showed a severe deteriorated phenotype comparable to the PBS control group. Similar results have been reported by Counoupas and collaborators (45). In their study, K18-hACE2 mice were subcutaneously immunized with a single formulation containing PBS or BCG and later challenged with SARS-CoV-2. Their findings revealed both these vaccinated groups presented body weight loss and elevated viral titers in the lungs, with no statistical difference. Protection was only achieved by immunizing mice with a formulation containing a mix of BCG WT, full-length trimeric spike and alum.
In summary, our study revealed that a prime-boost immunization system based on a rBCG-ChD6 expressing a chimeric protein (rChimera) derived from SARS-CoV-2 Wuhan strain followed by vaccination with rChimera protein plus alum fully protects mice against viral challenge. A booster dose of rChimera protein with alum provided efficient neutralizing antibody rates against SARS-CoV-2 Wuhan strain which led to prompt and early control of infection. This immunization system relies on the BCG platform and alum adjuvant, both clinically approved for to use in human practice, which would confer rapid incorporation into health care systems where BCG is currently employed. Despite failing to neutralize Delta and Omicron variants and with inferior neutralization rate to Gamma strain, further investigation is required upon the effectiveness of this BCG-based system against SARS-CoV-2 most recent VOCs, which could culminate in updating the rChimera protein by incorporating VOCs immunogenic epitopes into the construct.
Supplementary Material
Key points:
S- and N-predicted epitopes are antigenic for sera of infected patients
Vaccination with rBCG leads to neutralization against virus and cytokine responses.
rBCG immunization protects K18-hACE2 mice against SARS-CoV-2 challenge.
Acknowledgments
This study was supported by grants funded by CNPq (grants#303044/2020–9, 401209/2020–2 and 465229/2014–0), CAPES grants #88887.506612/2020–00 and #88887.504421/2020–00, FAPESP (grants# 2017/24832–6 and 2023/02577–5), Pro-Reitoria de Pesquisa-Universidade de São Paulo (Pro-Reitoria de Pesquisa USP), Howard Hughes Medical Institute (grant# 55007412) and FAPEMIG Rede Mineira de Imunobiologicos grant #REDE-00140–16.
Abbreviations
- BCG
Bacillus Calmette-Guérin
- COVID-19
Coronavirus Disease 19
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- rBCG
Recombinant BCG
- rChimera; Ch
Recombinant Chimera
- N
Nucleocapsid protein
- S
Spike glycoprotein
- ACE2
Angiotensin-Converting Enzyme 2
- WHO
World Health Organization
- VOCs
Variants of Concern
- BSL-3
Biosafety Level 3
- CDS
Coding sequence
- NMR
Nuclear magnetic resonance
- BMDMs
Bone marrow derived macrophages
- OADC
Oleic acid-albumin-dextrose-catalase
- s.c.
subcutaneously
- CFU
Colony-forming units
- CMC
Carboxi-metil-celulose
- PFU
Plaque-forming units
Footnotes
Conflict of interest
The authors declare no conflict of interest with the contents of this article.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
- 1.JHU. 2022. COVID-19 Dashboard by the Center for Systems Science and Engineering at Johns Hopkins University.
- 2.WHO. 2022. WHO Coronavirus (COVID-19) Dashboard. [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus I, and Research T. 2020. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma A, Ahmad Farouk I, and Lal SK. 2021. COVID-19: A Review on the Novel Coronavirus Disease Evolution, Transmission, Detection, Control and Prevention. Viruses 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chams N, Chams S, Badran R, Shams A, Araji A, Raad M, Mukhopadhyay S, Stroberg E, Duval EJ, Barton LM, and Hajj Hussein I. 2020. COVID-19: A Multidisciplinary Review. Front Public Health 8: 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadj Hassine I 2021. Covid-19 vaccines and variants of concern: A review. Rev Med Virol: e2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinobe RT, and Owens L. 2021. A systematic review of experimental evidence for antiviral effects of ivermectin and an in silico analysis of ivermectin’s possible mode of action against SARS-CoV-2. Fundam Clin Pharmacol 35: 260–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yousefi H, Mashouri L, Okpechi SC, Alahari N, and Alahari SK. 2021. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: A review describing drug mechanisms of action. Biochem Pharmacol 183: 114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araujo DB, Machado RRG, Amgarten DE, Malta FM, de Araujo GG, Monteiro CO, Candido ED, Soares CP, de Menezes FG, Pires ACC, Santana RAF, Viana AO, Dorlass E, Thomazelli L, Ferreira LCS, Botosso VF, Carvalho CRG, Oliveira DBL, Pinho JRR, and Durigon EL. 2020. SARS-CoV-2 isolation from the first reported patients in Brazil and establishment of a coordinated task network. Mem Inst Oswaldo Cruz 115: e200342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asselah T, Durantel D, Pasmant E, Lau G, and Schinazi RF. 2021. COVID-19: Discovery, diagnostics and drug development. J Hepatol 74: 168–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Candido DS, Claro IM, de Jesus JG, Souza WM, Moreira FRR, Dellicour S, Mellan TA, du Plessis L, Pereira RHM, Sales FCS, Manuli ER, Theze J, Almeida L, Menezes MT, Voloch CM, Fumagalli MJ, Coletti TM, da Silva CAM, Ramundo MS, Amorim MR, Hoeltgebaum HH, Mishra S, Gill MS, Carvalho LM, Buss LF, Prete CA Jr., Ashworth J, Nakaya HI, Peixoto PS, Brady OJ, Nicholls SM, Tanuri A, Rossi AD, Braga CKV, Gerber AL, de CGAP, Gaburo N Jr., Alencar CS, Ferreira ACS, Lima CX, Levi JE, Granato C, Ferreira GM, Francisco RS Jr., Granja F, Garcia MT, Moretti ML, Perroud MW Jr., Castineiras T, Lazari CS, Hill SC, de Souza Santos AA, Simeoni CL, Forato J, Sposito AC, Schreiber AZ, Santos MNN, de Sa CZ, Souza RP, Resende-Moreira LC, Teixeira MM, Hubner J, Leme PAF, Moreira RG, Nogueira ML, Brazil-Uk DG Centre for Arbovirus Discovery, N. Epidemiology Genomic, Ferguson NM, Costa SF, Proenca-Modena JL, Vasconcelos ATR, Bhatt S, Lemey P, Wu CH, Rambaut A, Loman NJ, Aguiar RS, Pybus OG, Sabino EC, and Faria NR. 2020. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science 369: 1255–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira SC, de Magalhaes MTQ, and Homan EJ. 2020. Immunoinformatic Analysis of SARS-CoV-2 Nucleocapsid Protein and Identification of COVID-19 Vaccine Targets. Front Immunol 11: 587615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Z, Cao Y, Liu W, and Li J. 2021. The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation. Viruses 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Queiroz N, Marinho FV, Chagas MA, Leite LCC, Homan EJ, de Magalhaes MTQ, and Oliveira SC. 2020. Vaccines for COVID-19: perspectives from nucleic acid vaccines to BCG as delivery vector system. Microbes Infect 22: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, and Veesler D. 2020. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181: 281–292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chilamakuri R, and Agarwal S. 2021. COVID-19: Characteristics and Therapeutics. Cells 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beyerstedt S, Casaro EB, and Rangel EB. 2021. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis 40: 905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu B, Huang S, and Yin L. 2021. The cytokine storm and COVID-19. J Med Virol 93: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. 2022. Landscape of COVID-19 candidate vaccines.
- 20.Ita K 2021. Coronavirus Disease (COVID-19): Current Status and Prospects for Drug and Vaccine Development. Arch Med Res 52: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F, Kream RM, and Stefano GB. 2020. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med Sci Monit 26: e924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salian VS, Wright JA, Vedell PT, Nair S, Li C, Kandimalla M, Tang X, Carmona Porquera EM, Kalari KR, and Kandimalla KK. 2021. COVID-19 Transmission, Current Treatment, and Future Therapeutic Strategies. Mol Pharm 18: 754–771. [DOI] [PubMed] [Google Scholar]
- 23.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Khetan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, and Group CS. 2021. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med 384: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou J, Xie X, Fontes-Garfias CR, Swanson KA, Kanevsky I, Tompkins K, Cutler M, Cooper D, Dormitzer PR, and Shi PY. 2021. The effect of SARS-CoV-2 D614G mutation on BNT162b2 vaccine-elicited neutralization. NPJ Vaccines 6: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, Avanzato VA, Bushmaker T, Flaxman A, Ulaszewska M, Feldmann F, Allen ER, Sharpe H, Schulz J, Holbrook M, Okumura A, Meade-White K, Perez-Perez L, Edwards NJ, Wright D, Bissett C, Gilbride C, Williamson BN, Rosenke R, Long D, Ishwarbhai A, Kailath R, Rose L, Morris S, Powers C, Lovaglio J, Hanley PW, Scott D, Saturday G, de Wit E, Gilbert SC, and Munster VJ. 2020. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 586: 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logunov DY, Dolzhikova IV, Zubkova OV, Tukhvatullin AI, Shcheblyakov DV, Dzharullaeva AS, Grousova DM, Erokhova AS, Kovyrshina AV, Botikov AG, Izhaeva FM, Popova O, Ozharovskaya TA, Esmagambetov IB, Favorskaya IA, Zrelkin DI, Voronina DV, Shcherbinin DN, Semikhin AS, Simakova YV, Tokarskaya EA, Lubenets NL, Egorova DA, Shmarov MM, Nikitenko NA, Morozova LF, Smolyarchuk EA, Kryukov EV, Babira VF, Borisevich SV, Naroditsky BS, and Gintsburg AL. 2020. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet 396: 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Z, Hu Y, Xu M, Chen Z, Yang W, Jiang Z, Li M, Jin H, Cui G, Chen P, Wang L, Zhao G, Ding Y, Zhao Y, and Yin W. 2021. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis 21: 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ella R, Vadrevu KM, Jogdand H, Prasad S, Reddy S, Sarangi V, Ganneru B, Sapkal G, Yadav P, Abraham P, Panda S, Gupta N, Reddy P, Verma S, Kumar Rai S, Singh C, Redkar SV, Gillurkar CS, Kushwaha JS, Mohapatra S, Rao V, Guleria R, Ella K, and Bhargava B. 2021. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBV152: a double-blind, randomised, phase 1 trial. Lancet Infect Dis 21: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forchette L, Sebastian W, and Liu T. 2021. A Comprehensive Review of COVID-19 Virology, Vaccines, Variants, and Therapeutics. Curr Med Sci 41: 1037–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Oosterhout C, Hall N, Ly H, and Tyler KM. 2021. COVID-19 evolution during the pandemic - Implications of new SARS-CoV-2 variants on disease control and public health policies. Virulence 12: 507–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu L, Mok BW, Chen LL, Chan JM, Tsang OT, Lam BH, Chuang VW, Chu AW, Chan WM, Ip JD, Chan BP, Zhang R, Yip CC, Cheng VC, Chan KH, Jin DY, Hung IF, Yuen KY, Chen H, and To KK. 2021. Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients. Clin Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matchett WE, Joag V, Stolley JM, Shepherd FK, Quarnstrom CF, Mickelson CK, Wijeyesinghe S, Soerens AG, Becker S, Thiede JM, Weyu E, O’Flanagan SD, Walter JA, Vu MN, Menachery VD, Bold TD, Vezys V, Jenkins MK, Langlois RA, and Masopust D. 2021. Cutting Edge: Nucleocapsid Vaccine Elicits Spike-Independent SARS-CoV-2 Protective Immunity. J Immunol 207: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dangi T, Class J, Palacio N, Richner JM, and Penaloza MacMaster P. 2021. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Rep 36: 109664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh AK, Netea MG, and Bishai WR. 2021. BCG turns 100: its nontraditional uses against viruses, cancer, and immunologic diseases. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moorlag S, Arts RJW, van Crevel R, and Netea MG. 2019. Non-specific effects of BCG vaccine on viral infections. Clin Microbiol Infect 25: 1473–1478. [DOI] [PubMed] [Google Scholar]
- 36.Pasco ST, and Anguita J. 2020. Lessons from Bacillus Calmette-Guerin: Harnessing Trained Immunity for Vaccine Development. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez-Perez M, Sanchez-Tarjuelo R, Shor B, Nistal-Villan E, and Ochando J. 2021. The BCG Vaccine for COVID-19: First Verdict and Future Directions. Front Immunol 12: 632478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netea MG, Quintin J, and van der Meer JW. 2011. Trained immunity: a memory for innate host defense. Cell Host Microbe 9: 355–361. [DOI] [PubMed] [Google Scholar]
- 39.Arts RJW, Moorlag S, Novakovic B, Li Y, Wang SY, Oosting M, Kumar V, Xavier RJ, Wijmenga C, Joosten LAB, Reusken C, Benn CS, Aaby P, Koopmans MP, Stunnenberg HG, van Crevel R, and Netea MG. 2018. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe 23: 89–100 e105. [DOI] [PubMed] [Google Scholar]
- 40.Netea MG, Giamarellos-Bourboulis EJ, Dominguez-Andres J, Curtis N, van Crevel R, van de Veerdonk FL, and Bonten M. 2020. Trained Immunity: a Tool for Reducing Susceptibility to and the Severity of SARS-CoV-2 Infection. Cell 181: 969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koneru G, Batiha GE, Algammal AM, Mabrok M, Magdy S, Sayed S, AbuElmagd ME, Elnemr R, Saad MM, Abd Ellah NH, Hosni A, Muhammad K, and Hetta HF. 2021. BCG Vaccine-Induced Trained Immunity and COVID-19: Protective or Bystander? Infect Drug Resist 14: 1169–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivas MN, Ebinger JE, Wu M, Sun N, Braun J, Sobhani K, Van Eyk JE, Cheng S, and Arditi M. 2021. BCG vaccination history associates with decreased SARS-CoV-2 seroprevalence across a diverse cohort of health care workers. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelleni MT 2021. BCG vaccination potential for COVID-19: an analytical approach. Hum Vaccin Immunother 17: 2448–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Escobar LE, Molina-Cruz A, and Barillas-Mury C. 2020. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19). Proc Natl Acad Sci U S A 117: 17720–17726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Counoupas C, Johansen MD, Stella AO, Nguyen DH, Ferguson AL, Aggarwal A, Bhattacharyya ND, Grey A, Hutchings O, Patel K, Siddiquee R, Stewart EL, Feng CG, Hansbro NG, Palendira U, Steain MC, Saunders BM, Low JKK, Mackay JP, Kelleher AD, Britton WJ, Turville SG, Hansbro PM, and Triccas JA. 2021. A single dose, BCG-adjuvanted COVID-19 vaccine provides sterilising immunity against SARS-CoV-2 infection. NPJ Vaccines 6: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hilligan KL, Namasivayam S, Clancy CS, O’Mard D, Oland SD, Robertson SJ, Baker PJ, Castro E, Garza NL, Lafont BAP, Johnson R, Ronchese F, Mayer-Barber KD, Best SM, and Sher A. 2022. Intravenous administration of BCG protects mice against lethal SARS-CoV-2 challenge. J Exp Med 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang BZ, Shuai H, Gong HR, Hu JC, Yan B, Yuen TT, Hu YF, Yoon C, Wang XL, Hou Y, Lin X, Huang X, Li R, Au-Yeung YM, Li W, Hu B, Chai Y, Yue M, Cai JP, Ling GS, Hung IF, Yuen KY, Chan JF, Huang JD, and Chu H. 2022. Bacillus Calmette-Guerin-induced trained immunity protects against SARS-CoV-2 challenge in K18-hACE2 mice. JCI Insight. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stensballe LG, Nante E, Jensen IP, Kofoed PE, Poulsen A, Jensen H, Newport M, Marchant A, and Aaby P. 2005. Acute lower respiratory tract infections and respiratory syncytial virus in infants in Guinea-Bissau: a beneficial effect of BCG vaccination for girls community based case-control study. Vaccine 23: 1251–1257. [DOI] [PubMed] [Google Scholar]
- 49.Ritz N, Mui M, Balloch A, and Curtis N. 2013. Non-specific effect of Bacille Calmette-Guerin vaccine on the immune response to routine immunisations. Vaccine 31: 3098–3103. [DOI] [PubMed] [Google Scholar]
- 50.Bastos RG, Borsuk S, Seixas FK, and Dellagostin OA. 2009. Recombinant Mycobacterium bovis BCG. Vaccine 27: 6495–6503. [DOI] [PubMed] [Google Scholar]
- 51.Mouhoub E, Domenech P, Ndao M, and Reed MB. 2021. The Diverse Applications of Recombinant BCG-Based Vaccines to Target Infectious Diseases Other Than Tuberculosis: An Overview. Front Microbiol 12: 757858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goulart C, Rodriguez D, Kanno AI, Converso TR, Lu YJ, Malley R, and Leite LCC. 2017. A Combination of Recombinant Mycobacterium bovis BCG Strains Expressing Pneumococcal Proteins Induces Cellular and Humoral Immune Responses and Protects against Pneumococcal Colonization and Sepsis. Clin Vaccine Immunol 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanekiyo M, Matsuo K, Hamatake M, Hamano T, Ohsu T, Matsumoto S, Yamada T, Yamazaki S, Hasegawa A, Yamamoto N, and Honda M. 2005. Mycobacterial codon optimization enhances antigen expression and virus-specific immune responses in recombinant Mycobacterium bovis bacille Calmette-Guerin expressing human immunodeficiency virus type 1 Gag. J Virol 79: 8716–8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varaldo PB, Leite LC, Dias WO, Miyaji EN, Torres FI, Gebara VC, Armoa GR, Campos AS, Matos DC, Winter N, Gicquel B, Vilar MM, McFadden J, Almeida MS, Tendler M, and McIntosh D. 2004. Recombinant Mycobacterium bovis BCG expressing the Sm14 antigen of Schistosoma mansoni protects mice from cercarial challenge. Infect Immun 72: 3336–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta PK 2020. New disease old vaccine: Is recombinant BCG vaccine an answer for COVID-19? Cell Immunol 356: 104187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanno AI, Goulart C, Rofatto HK, Oliveira SC, Leite LCC, and McFadden J. 2016. New Recombinant Mycobacterium bovis BCG Expression Vectors: Improving Genetic Control over Mycobacterial Promoters. Appl Environ Microbiol 82: 2240–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wüthrich K 1986. NMR with Proteins and Nucleic Acids. Europhys. News 17: 11–13. [Google Scholar]
- 58.Griesinger C, Otting G, Wuethrich K, and Ernst RR. 1988. Clean TOCSY for proton spin system identification in macromolecules. J. Am. Chem. Soc 110: 7870–7872. [Google Scholar]
- 59.Kumar A, Ernst RR, and Wuthrich K. 1980. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochem Biophys Res Commun 95: 1–6. [DOI] [PubMed] [Google Scholar]
- 60.Willker W, Leibfritz D, Kerssebaum R, and Bermel W. 1993. Gradient selection in inverse heteronuclear correlation spectroscopy. Magnetic Ressonance in Chemistry 31. [Google Scholar]
- 61.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, and Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696. [DOI] [PubMed] [Google Scholar]
- 62.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 63.Cheung MS, Maguire ML, Stevens TJ, and Broadhurst RW. 2010. DANGLE: A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J Magn Reson 202: 223–233. [DOI] [PubMed] [Google Scholar]
- 64.Bardiaux B, Malliavin T, and Nilges M. 2012. ARIA for solution and solid-state NMR. Methods Mol Biol 831: 453–483. [DOI] [PubMed] [Google Scholar]
- 65.Krishnan VV, Bentley T, Xiong A, and Maitra K. 2021. Conformational Ensembles by NMR and MD Simulations in Model Heptapeptides with Select Tri-Peptide Motifs. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mambelli FS, Figueiredo BC, Morais SB, Assis NRG, Fonseca CT, and Oliveira SC. 2020. Recombinant micro-exon gene 3 (MEG-3) antigens from Schistosoma mansoni failed to induce protection against infection but show potential for serological diagnosis. Acta Trop 204: 105356. [DOI] [PubMed] [Google Scholar]
- 67.Marinho FV, Fahel JS, de Araujo A, Diniz LTS, Gomes MTR, Resende DP, Junqueira-Kipnis AP, and Oliveira SC. 2020. Guanylate binding proteins contained in the murine chromosome 3 are important to control mycobacterial infection. J Leukoc Biol 108: 1279–1291. [DOI] [PubMed] [Google Scholar]
- 68.Laemmli UK 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 69.Towbin H, Staehelin T, and Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76: 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, and Bieniasz PD. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fumagalli MJ, Castro-Jorge LA, Fraga-Silva TFC, de Azevedo PO, Capato CF, Rattis BAC, Hojo-Souza NS, Floriano VG, de Castro JT, Ramos SG, da Fonseca BAL, Bonato VLD, Gazzinelli RT, and Figueiredo LTM. 2021. Protective Immunity against Gamma and Zeta Variants after Inactivated SARS-CoV-2 Virus Immunization. Viruses 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, Marrama D, de Silva AM, Frazier A, Carlin AF, Greenbaum JA, Peters B, Krammer F, Smith DM, Crotty S, and Sette A. 2020. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181: 1489–1501 e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lobo N, Brooks NA, Zlotta AR, Cirillo JD, Boorjian S, Black PC, Meeks JJ, Bivalacqua TJ, Gontero P, Steinberg GD, McConkey D, Babjuk M, Alfred Witjes J, and Kamat AM. 2021. 100 years of Bacillus Calmette-Guerin immunotherapy: from cattle to COVID-19. Nat Rev Urol 18: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dyson HJ, and Wright PE. 1995. Antigenic peptides. FASEB J 9: 37–42. [DOI] [PubMed] [Google Scholar]
- 75.Yanaka S, Ueno T, Shi Y, Qi J, Gao GF, Tsumoto K, and Sugase K. 2014. Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex. J Biol Chem 289: 24680–24690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boulat B, Emsley L, Muller N, Corradin G, Maryanski JL, and Bodenhausen G. 1991. NMR studies of an oligoproline-containing peptide analogue that binds specifically to the H-2Kd histocompatibility molecule. Biochemistry 30: 9429–9434. [DOI] [PubMed] [Google Scholar]
- 77.Saito NG, and Paterson Y. 1996. Nuclear Magnetic Resonance Spectroscopy for the Study of B-Cell Epitopes. Methods 9: 516–524. [DOI] [PubMed] [Google Scholar]
- 78.Dangi T, Palacio N, Sanchez S, Park M, Class J, Visvabharathy L, Ciucci T, Koralnik IJ, Richner JM, and Penaloza-MacMaster P. 2021. Cross-protective immunity following coronavirus vaccination and coronavirus infection. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahardika GN, Mahendra NB, Mahardika BK, Suardana IBK, and Pharmawati M. 2022. Annotating Spike Protein Polymorphic Amino Acids of Variants of SARS-CoV-2, Including Omicron. Biochem Res Int 2022: 2164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eggenhuizen PJ, Ng BH, Chang J, Fell AL, Cheong RMY, Wong WY, Gan PY, Holdsworth SR, and Ooi JD. 2021. BCG Vaccine Derived Peptides Induce SARS-CoV-2 T Cell Cross-Reactivity. Front Immunol 12: 692729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Urban S, Paragi G, Burian K, McLean GR, and Virok DP. 2020. Identification of similar epitopes between severe acute respiratory syndrome coronavirus-2 and Bacillus Calmette-Guerin: potential for cross-reactive adaptive immunity. Clin Transl Immunology 9: e1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim BJ, Kim BR, Kook YH, and Kim BJ. 2018. Development of a Live Recombinant BCG Expressing Human Immunodeficiency Virus Type 1 (HIV-1) Gag Using a pMyong2 Vector System: Potential Use As a Novel HIV-1 Vaccine. Front Immunol 9: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finkelman FD, Holmes J, Katona IM, Urban JF Jr., Beckmann MP, Park LS, Schooley KA, Coffman RL, Mosmann TR, and Paul WE. 1990. Lymphokine control of in vivo immunoglobulin isotype selection. Annu Rev Immunol 8: 303–333. [DOI] [PubMed] [Google Scholar]
- 84.Barr TA, Brown S, Mastroeni P, and Gray D. 2009. B cell intrinsic MyD88 signals drive IFN-gamma production from T cells and control switching to IgG2c. J Immunol 183: 1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soto JAD, Retamal-Díaz FE, Gálvez A, Melo-González NMS, Piña-Iturbe F, Ramírez A, Bohmwald MA, González K, Bueno PA, Kalergis SM, A. M. 2022. BCG-Based Vaccines Elicit Antigen-Specific Adaptive and Trained Immunity against SARS-CoV-2 and Andes orthohantavirus. Vaccines 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaufmann E, Khan N, Tran KA, Ulndreaj A, Pernet E, Fontes G, Lupien A, Desmeules P, McIntosh F, Abow A, Moorlag S, Debisarun P, Mossman K, Banerjee A, Karo-Atar D, Sadeghi M, Mubareka S, Vinh DC, King IL, Robbins CS, Behr MA, Netea MG, Joubert P, and Divangahi M. 2022. BCG vaccination provides protection against IAV but not SARS-CoV-2. Cell Rep 38: 110502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hilligan KL, Namasivayam S, Clancy CS, O’Mard D, Oland SD, Robertson SJ, Baker PJ, Castro E, Garza NL, Lafont BAP, Johnson R, Ronchese F, Mayer-Barber KD, Best SM, and Sher A. 2021. Intravenous administration of BCG protects mice against lethal SARS-CoV-2 challenge. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ren J, Wen L, Gao X, Jin C, Xue Y, and Yao X. 2009. DOG 1.0: illustrator of protein domain structures. Cell Res 19: 271–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request.


