Abstract
The genetic base of soybean cultivars (Glycine max (L.) Merr.) has been narrowed through selective domestication and specific breeding improvement, similar to other crops. This presents challenges in breeding new cultivars with improved yield and quality, reduced adaptability to climate change, and increased susceptibility to diseases. On the other hand, the vast collection of soybean germplasms offers a potential source of genetic variations to address those challenges, but it has yet to be fully leveraged. In recent decades, rapidly improved high-throughput genotyping technologies have accelerated the harness of elite variations in soybean germplasm and provided the important information for solving the problem of a narrowed genetic base in breeding. In this review, we will overview the situation of maintenance and utilization of soybean germplasms, various solutions provided for different needs in terms of the number of molecular markers, and the omics-based high-throughput strategies that have been used or can be used to identify elite alleles. We will also provide an overall genetic information generated from soybean germplasms in yield, quality traits, and pest resistance for molecular breeding.
Keywords: Soybean germplasm, Core collection, Gene mapping, Omics, Elite allele, Molecular breeding
Introduction
Crop domestication is an expansive process that occurs in four stages including management, purposeful cultivation, geographic expansion of the domesticate, and deliberate breeding (Gaut et al. 2018). There are three evolutionary types related to domestication including the wild relatives of crop species, local domesticates called landraces, and modern varieties, collectively called the crop germplasms. Over the course of around 10,000 years of artificial and natural selection, these germplasms have stored rich biodiversity, which is fundamental in modern crop breeding (Jeong et al. 2019). However, only a small fraction of the available biodiversity has been utilized. For example, less than 1% of soybean germplasms conserved had been used for breeding (Qiu et al. 2013b). This means that only a few improved cultivars coming from a few excellent parents have been planted, which has resulted in a narrow genetic basis, reduced diversity, and enhanced consistency (Schoener and Fehr 1979). The limited biodiversity has slowed the progress in conventional crop breeding and has increased the risk of disease and pest outbreak. Abundant germplasm resources are the prerequisite for continuously developing new varieties with high yield and quality to ensure food security.
It is time to explore and use new variations conserved in the gene banks for agriculture as genotyping approaches, such as SNPs array and re-sequencing, are rapidly developing. Biodiversity in germplasms could be explored by starting with genotyping representative germplasm populations followed by phenotyping them and testing their regional differences. Then, the genetic constitution of the germplasm for both whole genome and individual traits, as well as the corresponding markers for the detected genes/alleles with their regional differentiation, can be analyzed. Finally, the identified elite accessions, along with phenotypic and genetic knowledge, can fuel advances in improving cultivars by breeding programs to meet the food and nutritional demand of the growing population of the world (Susan et al. 2013). Soybean is a major legume crop providing a significant amount of edible oil and protein for human and animal feed around the world. In this article, we review the conservation of soybean germplasm and the situation of the mining the favorable variations in this wealth of genetic resources.
The conservation of soybean germplasms
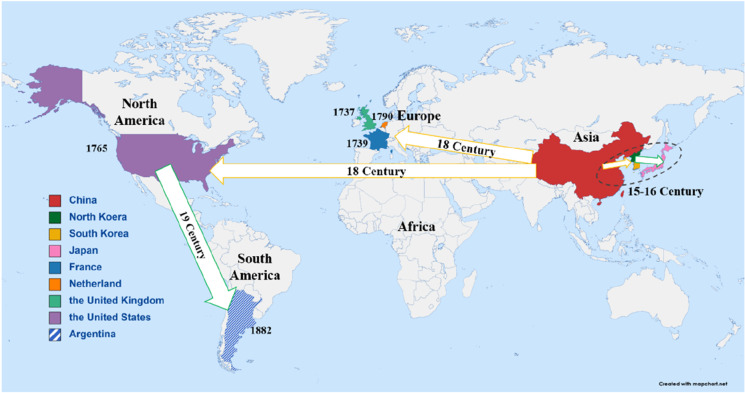
The genus Glycine is composed of two subgenera: Glycine and Soja. Subgenus Glycine contains around 30 perennial species which are mainly originated from Australia and distributed there (Chung and Singh 2008). Tetraploid species are distributed as far north as southeast coast of China. The subgenus Soja contains annual wild (Glycine soja Sieb. & Zucc.) and cultivated soybean Glycine max (L.) Merr. G. soja is native to East Asia and mainly distributed in China, Korea, Japan, and Russia (Sherman-Broyles et al. 2014). G. max originated from central China and domesticated from its ancestor annual wild soybean G. soja about 6000 to 9000 years ago (Zhuang et al. 2022; Li et al. 2023b). Soybean from China was first introduced into neighboring Korea during the Chinese Warring States Period and then spread from Korea to Japan. Soybean was known in Europe after the eighteenth century, and was introduced to the Netherlands in 1737, and then planted in France and the UK between 1739 and 1790. The earliest soybean planting in the USA dates back to 1765. Samuel Bowen, a seaman from the East India Company, brought Chinese soybean back to Savannah, Georgia. The first introduction of soybean into South America was to Argentina in 1882 (Chang 1989; Fig. 1). Since then, soybeans have been gradually planted almost all over the world and have acted as one of the most important legume crops due to their richness in protein and oil.
Fig. 1.
The transportation route of soybean. Information from Chang (1989). The yellow and green arrows on the map represent direct or indirect transportation from China separately, with the occurrence time marked near the respective country
Collection and conservation of soybean germplasm
The purpose of crop germplasm conservation is to preserve them before losing the existing genetic variability due to the widespread use of modern cultivars. According to rough statistics, there are more than 200 thousand soybean accessions conserved in more than 70 countries worldwide, but only 30% of them could be unique. As the origin and domestication site, China has rich soybean germplasm. The germplasm collection was initialized in early 20th country (Qiu et al. 2011). In 1913, germplasm from northeast China was collected and evaluated by researchers in Gongzhuling. In the meantime, the southern China germplasm was also collected by researchers in Nanjing Agricultural University (former Jinling University). Since 1949, Chinese government has organized experts to conduct three national surveys of agricultural collections during 1956 ~ 1957, 1979 ~ 1983, and 2015 ~ present, respectively (http://www.gov.cn/zhengce/zhengceku/2021-03/25/content_5595469.htm). Until now, around 42 thousand soybean accessions, including ~ 10 thousand G. soja and ~ 32 thousand G. max, were collected in China (Wang 1982; Chang and Sun 1991; Chang et al. 1996; Qiu et al. 2013a; Guo et al. 2022). The Chinese soybean germplasm collection has played a key role, not only in the domestic soybean breeding, but also in world soybean breeding and production such as cultivar development in the USA, Brazil, Argentina, Korea, Japan, Europe, and Australia (Qiu et al. 2011). For example, it is estimated that the northern US cultivars contain 50% nuclear DNA and 83% cytoplasmic DNA from China ancestors (Gizlice et al. 1994). In addition, 3.6 thousand cultivated soybeans were introduced from abroad and conserved in Chinese National Soybean GeneBank (CNSGB). Until now, China has conserved the largest number (~ 46,000) of soybean germplasm in the world.
The USDA Soybean Collection was established in 1895 but formally started to keep records in 1898. It was not until 1949 that the government realized the importance of conservation and utilization of the important crop germplasm, introduced soybean germplasm from China, carefully conserved since then (Nelson 2011). The USA conserved the third largest soybean germplasm, which includes approximately 1.2 thousand G. soja and 21 thousand G. max accessions, fall behind Korea with around 26 thousand accessions. The conservation situation of soybean germplasm in other countries is shown in Table 1.
Table 1.
The conservation of soybean germplasm resources
| Country | Institution | Accessions (G. max/G. soja) | Ref. or website |
|---|---|---|---|
| China | Chinese National Soybean GeneBank (CNSGB) | 36,000/9862 | Wang (1982), Chang and Sun (1991), Chang et al. (1996), Qiu et al. (2013a), Guo et al. (2022) |
| USA | USDA Soybean Germplasm Collection | 20,224/1179 | https://npgsweb.ars-grin.gov/gringlobal/search |
| Korea | National Agrobiodiversity Center (NAC) | ~ 26,000/NA | Kim et al. (2022) |
| Japan | National Agricuture and Food Research Organization (NARO) | 8523/2346 | https://www.gene.affrc.go.jp/ |
| Ukraine | Institute of Agroecology and Biotechnology | 7000/NA | Boerma and Specht (2004) |
| Russia | N. I. Vavilov Research Institute of Plant Industry | 6126/310 | Boerma and Specht (2004) |
| Brazil | Centro Nacional de Pesquisa De Recursos Genéticos e Biotec. (CENARGEN) | 4693/NA | Boerma and Specht (2004) |
| India | India National Genebank (NGB) | 3702/NA | Babu et al. (2018) |
| Germany | Genebank, Institute for Plant Genetics and Crop Plant Res. (IPK) | 3063/2 | Boerma and Specht (2004) |
| France | G. I. E. Amelioration Fourragere | 1582/NA | Boerma and Specht (2004) |
| Indonesia | Sukamandi Research Institute for Food Crops (SURIF) | 2194/4 | Boerma and Specht (2004) |
| Canada | Plant Gene Resources of Canada (PGRC) | 1031/NA | Fu et al. (2021) |
Construction of soybean core collection
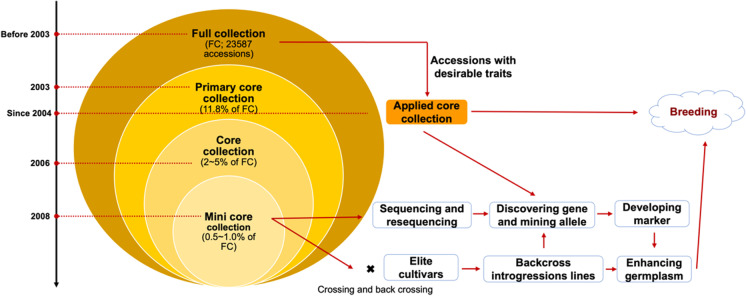
The collection and conservation of crop germplasm resources to maintain genetic diversity have long been an important priority worldwide. However, as the size of collections increases, the cost of conservation and evaluation has risen (Wang et al. 2006). To deal with this challenge, Frankel (1984) first proposed sampling the collections to yield a manageable sample called “core collection.” A core collection is a subset of accessions that represents the genetic diversity of a species and its relatives. Before 2000, the construction of core germplasm worldwide mainly depended on phenotypic data in the field, such as plant height, leaf size, maturity, and disease resistance performance. Because different countries and regions paid attention to different traits, there were great difficulties in the consistency, comparability, and compatibility of collected data. The rapid development and application of DNA molecular marker technology promoted the exploration of technology and methods to establish core collections for major crops, including wheat, rice, maize, and soybean (Li et al. 2004; Balfourier et al. 2007; Hao et al. 2008; Zhang et al. 2011). Based on the DNA molecular fingerprint data of accessions, through cluster analysis, representative accessions are selected for core collections, which has been widely accepted by the academic community as a popular strategy to establish a core collection. In China, in order to accelerate the evaluation and utilization of the germplasm, the projects of “Establishment of soybean core collection” (1998–2003) and “Gene diversity of mini core collection in soybean” (2004–2008) were carried out by the continuous support from National Basic Research Program (973 project). Taking a total of 23,587 soybean accessions as full collection (FC), serial core collections including primary core, core, and mini-core collections had been constructed (Qiu et al. 2003, 2009, 2011, 2013b; Wang et al. 2006; Fig. 2). The primary core collection consisting of 2794 cultivars (11.8% of FC) was constructed using single sequence repeats (SSR) molecular markers and phenotypic data analysis, which account for 100% and 73.6% of phenotypic and genetic diversity separately. This primary core collection represents the achievement of soybean core research, encompassing theory and application, with an emphasis on evaluating phenotype and gene diversity. Based on the core collection, a cultivated soybean mini-core collection (MCC, 1% of FC) was further constructed with 94.5% and 63.5% representativeness of the overall resource phenotype and molecular level, respectively. These applied core collections could fulfill the needs of adaptation to different eco-regions and broadening the genetic base of germplasm for breeding programs. A succession of applied core collections has been constructed for different desirable traits. For resistance to soybean cyst nematode (SCN), Ma et al. (2006) constructed a 28 accessions core collection from 432 immune or highly resistant Chinese accessions. Using a GIS-assisted approach, Zhao et al. (2004) constructed a phosphorus efficiency of soybean applied core collection. However, the sample size could increase suddenly if all the applied core collections for each desirable trait are merged, which goes against the principle of the core collection. Thus, an integrated applied core collection (IACC) with cold, drought, salt tolerances, and resistances to SCN, SMV, high protein, and fat content has been developed (Guo et al. 2014). Simultaneously, an annual Chinese wild soybean’s core collection was also developed (Zhao et al. 2005). These core collections have played good leading roles in constructing core germplasm for other crops and promoted the research of crop germplasm resources nationwide and even globally.
Fig. 2.
The pipeline of the development and utilization of soybean core collections in China. Information from Qiu et al. (2009, 2013b)
The construction of soybean core germplasm has also been carried out in other countries. Gizlice et al. (1994) identified twenty-eight ancestors and seven first progeny, which would be a useful core collection. Oliveira et al. (2010) developed a core collection of 1600 accessions from around 16,000 accessions in USDA Soybean germplasm collection using the multivariate proportional sampling strategy. Priolli et al. (2013) developed a 31 entries Brazilian core set from 435 soybean cultivars. Kaga et al. (2012) developed mini core collections from around 1600 accessions of NIAS in Japan. A core set of Korean landraces was developed from around 2800 accessions conserved in the National Genebank of Rural Development Administration (RDA-Genebank; Cho et al. 2008).
The evaluation of the genotypes of soybean germplasms
Genotyping is used to identify and track the genetic diversity for soybean germplasms. This polymorphism information could be used for identification, selection, and genetic basis dissection of traits that are desirable for specific growing conditions, end-use applications, and addressing specific biotic and abiotic stresses. The knowledge enables the development of new soybean varieties through breeding and genetic modification and broadens the genetic basis of current soybean-improved cultivars. There are various genotyping solutions available that can be chosen based on the desired throughput and number of genomic molecular markers (Fig. 3A).
Fig. 3.
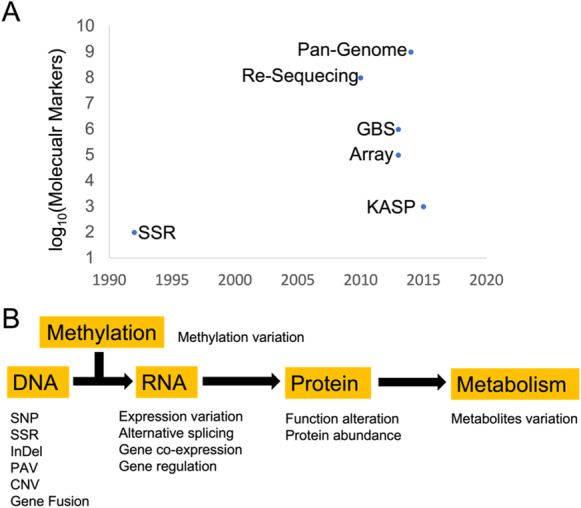
The genic and multi-omics variation for genetic basis dissection of trait. A Different genotyping platforms or technologies. The y-axis represents number of molecular markers, and x-axis indicates the time applied in soybean studies. B Omics data and the revealed polymorphisms for association studies
Genotyping strategies for different molecular markers
Two decades ago, molecular makers were developed as the main technique for genotyping via utilizing the DNA polymorphism, such as restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), randomly amplified polymorphic DNA (RAPD), SSR, and single nucleotide polymorphism (SNP). SSR is the polymorphism of DNA length (Akkaya et al. 1992) and one of the most widely markers used for linkage mapping (Cregan et al. 1999) and population study (Abe et al. 2003) in soybean. Another important type of marker, SNPs, exists at a higher frequency and is more stable. It is widely used with different strategies (Hyten et al. 2006, 2010; Shu et al. 2011; Shi et al. 2015), such as the kompetitive allele specific PCR (KASP) technology, which is cost-effective and can be applied in high-throughput systems.
In the last decade, tremendous progress has been achieved in genotyping methods, particularly in the development of high-throughput and cost-effective methods, such as SNP arrays and next-generation sequencing platforms. SNP arrays are a fast and efficient way for researchers to genotype a large number of samples, and they are usually less expensive than other sequencing-based genotyping methods. Soybean SNP arrays (Song et al. 2013; Lee et al. 2015; Wang et al. 2016; Li et al. 2022d; Sun et al. 2022; Ma et al. 2023) have been designed with a range of sizes, from 20 to 618 K, to meet different research goals. Song et al. (2013) developed an array of 50,000 SNPs with a high minor allele frequency among landraces, elite cultivars, and wild soybeans. They used this array to identify genetic regions associated with the signature of domestication and selection during soybean breeding. Lee et al. (2015) selected 180,961 SNPs for creation from millions of resequencing SNPs. This array provided a higher quality of genotyping for diversity analyses, linkage mapping, and association studies. Wang et al. (2016) constructed a 355,595 SNPs array for population genetic analysis to track the evolutionary history and identify the genetic regions for improvement. Sun et al. (2022) brought out a functional SNPs array aimed for cost-effective genome selection that included 158,959 SNPs covering 91% of soybean genes and known QTLs of agronomy traits. Li et al. (2022d) and Ma et al. (2023) both designed array with more than 600 K SNPs, which has a high-resolution for germplasm studies and breeding programs.
Those solid chip-based SNPs arrays have the advantage of being able to be customized to focus on specific regions of interest. This, however, might potentially be a drawback because it necessitates prior information and prevents the genotyping of novel SNPs. A more flexible liquid SNP array, a technique genotyping by target sequencing, was applied in soybean (Liu et al. 2022). This technique allows for easy updating of target SNPs and is more cost-effective than solid chip-based SNP arrays. In addition, genotyping by sequencing (GBS) is another approach that uses restriction enzymes to simplify the genome and requires no prior knowledge (Elshire et al. 2011; Sonah et al. 2013). Compared to re-sequencing, GBS is a much less expensive genotyping solution for species with a large genome size. GBS was widely used for genetic dissection of agronomy traits (Sonah et al. 2015), germplasm screening (Lemay et al. 2019), and geographic adaptation (Li et al. 2020b) in soybean.
Genome re-sequencing is the most comprehensive method for genome-wide genotyping, and it allows for the detection of a wider range of genetic variations, like insertion and deletion (InDel), presence-absence variation (PAV), copy number variation (CNV), and gene fusion. Lam et al. (2010) first demonstrated in-depth understandings of variety and selection of soybean from re-sequencing of 31 wild soybean and cultivars. After that, hundreds to thousands soybean lines were genotyped by re-sequencing (Zhou et al. 2015; Liu et al. 2020b; Torkamaneh et al. 2021; Bayer et al. 2022; Yang et al. 2022; Li et al. 2023b), of which gave higher resolution on domestication, improvement, adaptation, and genetic basis dissection of traits. Millions of SNPs and short InDels were typically identified and genotyped in those re-sequencing studies. Liu et al. (2020b) and Li et al. (2023b) both genotyped over 2000 soybean accessions, providing better insight into diversity, genetic basis of agronomic traits, and domestication and improvement of soybean. Liu et al. (2020b) identified large structural variations and linked them with gene expressions and traits. Li et al. (2023b) proposed soybean evolutionary route with four geographic paths, expansion of annual wild soybean G. soja from Southern China, domestication in Central China, expansion of landrace, and local breeding.
Development of soybean pan-genome
In 2010, the first soybean reference genome was made public (Schmutz et al. 2010). A 1.1-gigabase chromosome-scale draft sequence assembly for the Williams 82, a northern USA improved cultivar, was built using a combination of whole genome shotgun sequencing and BAC-by-BAC sequencing. Long reads sequencing technology and algorithms were becoming mature and more accurate in last 5 years, of which made the chromosome level assembly with a reasonable cost on sequencing, labors, and computing resource (Sohn and Nam 2018). Soybean-improved cultivars from represent growing areas were assembled: Enrei from Japan (Shimomura et al. 2015), Zhonghuang 13 from China (Yang and Huang 2018; Shen et al. 2019), and Lee from southern USA (Valliyodan et al. 2019). Wild relative of soybean cultivars was also assembled: annual wild soybean PI 483,463 (Valliyodan et al. 2019), W05 (Xie et al. 2019), and a perennial wild soybean (Glycine latifolia) PI 559,298 (Liu et al. 2018). Soybean pan-genome studies (Li et al. 2014; Liu et al. 2020b; Torkamaneh et al. 2021; Bayer et al. 2022) were reported to further investigate the diversity of core genome and complete set of genes. Liu et al. (2020b) combined the PacBio SMRT, Bionano optical maps, Hi-C scaffolding, and Illumina next-generation sequencing error correction for de novo assembly of 26 soybean genomes and created a graph-based genome. Recently, the Telomere-to-Telomere (T2T) reference chromosome or genome with completed sequence was achieved in human (Nurk et al. 2022) and plants (Song et al. 2021; Deng et al. 2022; Wang et al. 2022), but not in soybean yet. In the near future, a more comprehensive and complete reference genome will enable more accurate genotyping for soybean germplasm studies.
The identification of elite functional alleles in soybean germplasms
Strategies for gene mapping
The identification of elite genes and functional alleles is the foundation of crop improvement of yield and quality, crop development resistant to climate changes and stress tolerance, and the understanding of plant evolution and diversity. It is a critical area of study in the field that has benefitted from developments in genotyping technologies and reference genome, and considerably contributed to our understanding of genetics and genomics in soybean. Meanwhile, there are challenges and limitations for gene mapping that soybean is self-pollinating leading with a low recombination rate and a slow LD decay (Zhou et al. 2015). There are two classical mapping approaches in soybean: linkage mapping for the bi-parental genetic populations (Keim et al. 1990) and association study for diversity panels (Hao et al. 2012).
Linkage mapping is an analysis to the examination of Mendelian inheritance patterns between genotyped markers and phenotype to identify the location of causal genes for qualitative and quantitative traits. Bulked segregant analysis (BSA) (Michelmore et al. 1991) is a linkage mapping method used to rapidly identify molecular markers linked to the trait, which normally is qualitative. The next-generation sequencing (NGS) based BSA offers genome-wide fast gene mapping via high-density molecular markers. Multiple NGS-based BSA methods were developed in plants, such as SHOREmap in Arabidopsis (Schneeberger et al. 2009) and MutMap in rice (Abe et al. 2012). A method named BSR-Seq (Liu et al. 2012b) utilized the RNA-Seq, instead of genome re-sequencing, for genotyping that only focus on the genic region of genome and capture gene expression information. The NGS-based BSA methods were widely used for gene mapping in soybean traits, such as gnarled trichome (Campbell et al. 2016), high sucrose fast neutron (Dobbels et al. 2017), and cotyledon color (Song et al. 2017). However, quantitative traits are more common, especially agronomy traits, in plants. Quantitative trait loci (QTLs) mapping was applied to identify the genetic basis of soybean flowering time (Orf et al. 1999), height (Zhang et al. 2004), yield (Yuan et al. 2002), quality (Reinprecht et al. 2006), drought tolerance (Specht et al. 2001), and disease resistance (Yuan et al. 2002).
Genome-wide association studies (GWAS) conduct association analysis between the variation of genetic markers and variation of phenotype on diversity panels composed of unrelated individuals, rather than bi-parental populations. A nested association panel named SoyNAM was built from crossing between 40 important soybean varieties with the common hub parent, IA3023. GWASs using the genotyped SoyNAM have been used to identify genetic variants associated with yield-related traits (Wen et al. 2015), canopy coverage (Xavier et al. 2017), resistance to soybean cyst nematode (Cook et al. 2014) and Phytophthora sojae (Scott et al. 2019), water use efficiency (Lopez et al. 2019), and environmental interactions (Jarquin et al. 2018). Several important domestication genes have been identified in soybean through GWAS. These genes are involved in the regulation of various traits related to soybean domestication, including flowering time (Li et al. 2020a; Dong et al. 2021), yield (Duan et al. 2022; Li et al. 2022a), quality (Gao et al. 2021; Goettel et al 2022), and other agronomy traits (Liu et al. 2020a).
Omics data facilitates gene mapping
Both linkage mapping and association study identify genetic regions surrounding dozens or hundreds of candidate genes, but the causal genes are often unknown, making it challenging and time-consuming to identify the functional gene underlying a trait. On the other hand, the identified loci from either linkage mapping or association studies only explain a proportion of the total heritability of complex trait, of which is referred as “missing heritability” (Manolio et al. 2009). Meanwhile, omics data, including transcriptomics (RNA), proteomics (protein), epigenomics (methylation), and metabolomics (metabolism), are producing using high-throughput and cost-effective cutting-edge technologies. These omics data can be used to identify functional genes or be integrated with GWAS to prioritize candidate genes (Fig. 3B).
Transcriptomics sequencing could provide not only genetic variation but more importantly, gene expression. Transcriptome-wide association studies (TWAS) were a method to study the association between gene expression and phenotype variation. The gene expression could either be integrated with GWAS as eQTLs (Gusev et al. 2016; Li et al. 2020c; Ma et al. 2021) or used as measured expression independently (Lin et al. 2017; Zheng et al. 2020; Li et al. 2021; Tang et al. 2021). Lin et al. (2017) developed the first plant TWAS method named eRD-GWAS performed association between the variation of the measured gene expression and phenotype variation. The genes identified by TWAS are complementary with loci reveal by GWAS in maize, of which further be confirmed in maize (Zheng et al. 2020), Brassica napus (Tang et al. 2021) and Arabidopsis (Li et al. 2021). Li et al. (2021) reported that TWAS can reduce effects of slow linkage disequilibrium in soybean, which may make gene-level association mapping alliable in soybean. The gene co-expression networks were also used to identify genetic variation associated with soybean trait, such as seed development (Gao et al. 2018).
Proteome-wide association study (PWAS) links the gene proteome variation and phenotype variation. To our knowledge, PWAS was reported in human studies but not in plant yet. There were two PWAS strategies: the first one is to predict protein function alteration via DNA sequence, which hypothesized that causal variants in coding regions of certain gene affect phenotypes by altering the biochemical functions of the genes’ protein products (Brandes et al. 2020); the second one is to measure the abundance of genes’ protein products genome-widely, and integrated with independent GWAS results (Wingo et al. 2021). Besides that, epigenomics (Huan et al. 2019) and metabolomics (Lord et al. 2021) were also reported to be used to integrate with GWAS results to identify DNA methylations or metabolites involved in human diseases via Mendelian randomization analysis. The integration, interpretation, and application of omics data in soybean research has the potential to accelerate our understanding of the genetic basis of traits and offers promising opportunities for both genetic research and breeding programs.
Soybean elite alleles identified
Adaption related
Soybean is a classic short-day crop that is particularly photoperiod sensitive. Individual varieties or germplasm resources are generally only suitable for planting in restricted geographic areas with small latitude spans. The genetic dissection study of flowering/maturity time could give us insights into the regulation of geographic adaptation and broaden the adaption of varieties. Currently, hundreds of loci associated with flowering/maturity time have been identified through linkage mapping and association studies and are available in SoyBase (https://www.soybase.org/; Grant et al. 2010). Functional gene of some major QTLs was cloned, including E1 to E4, E9, J, Tof5, Tof11, GmRPP3b, GmGBP1, and GmCDPK38 (Xia et al. 2012; Tsubokura et al. 2013; Zhao et al. 2016, 2018; Lu et al. 2017a, 2020; Li et al. 2020a, 2022c; Dong et al. 2022). Three distinct e1 mutant alleles, comprising a non-synonymous SNP, a one-base insertion, and a deletion of the entire gene, were identified, all resulting in an earlier flowering time (Xia et al. 2012). A major mutant e2 allele (Tsubokura et al. 2014) caused by a stop-gained SNP also induced earlier flowering time. Similarly, the natural e3, e4, tof11, gmprr3b, and gmcdpk38 mutant alleles resulted in earlier flowering/maturity time. Conversely, the e9, j, tof5, and gmgbp1 mutant alleles induced later flowering/maturity time, which enhanced adaptation in higher latitudes. The inferred flowering regulatory network was reviewed recently (Hou et al. 2022; Du et al. 2023). Since flowering/maturity time is the key factor for geographic adaption, some of those identified flowering/maturity time loci undergone selection either during domestication or improvement (Li et al. 2020a; Lu et al. 2020). The elite alleles of some of those loci in germplasms were already effectively utilized to expand the suitable growing areas for soybeans in breeding programs. The nature mutant alleles of J cause long juvenile trait, and breed soybean varieties adapt from temperate to tropic environment, such as Brezil (Dong et al. 2021). The mutant alleles of photoperiod sensitivity genes were selected by breeders in the high latitude (Jiang et al. 2014; Lu et al. 2020).
Yield related
High yield is always the ultimately goal of soybean breeding. Plant height, seed size, and seed weight are all important factors in determining the final yield. Dt1 (Liu et al. 2010) and Dt2 (Ping et al. 2014; Zhang et al. 2019a) are two major genes regulating the soybean stem growth habit, which were categorized into three types: determinate (dt1dt1), semi-determinate (Dt1Dt1/Dt2Dt2) and indeterminate (Dt1Dt1/dt2dt2). dt1 allele is the dominant allele of Dt1 locus and has an epistatic effect on Dt2 (Ping et al. 2014). Benefiting from the development of genotyping and gene editing, the seed size-related genes were mapped and functionally proved: PP2C (Lu et al. 2017b), GmKIX8-1 (Nguyen et al. 2021), POWR1 (Goettel et al. 2022), and GmGA3ox1 (Hu et al. 2022). The PP2C allele from wild soybean was found that could increase seed size (Lu et al. 2017b). gmkix8-1 mutant alleles increase seed size (Nguyen et al. 2021). powr1 allele with transposable element insertion is the major allele in cultivated soybean and increased the seed weight (Goettel et al. 2022). gmga3ox1 mutant alleles have lower seed weight but increased seed number (Hu et al. 2022). The cloned seed weight-regulated genes included GmSWEET10a (Wang et al. 2020b), GmSSS1 (Zhu et al. 2022), ST1 (Li et al. 2022a), and GmST05 (Duan et al. 2022). The wild soybean dominant alleles of GmSWEET10a have a decreased frequency in cultivars, while the cultivar alleles display varying degrees of increased seed weight and oil content (Wang et al. 2020b). The natural gmsss1 mutant allele is induced by a non-synonymous SNP and has an increased seed weight (Zhu et al. 2022). ST1 influences seed weight and exhibits high levels of surrounding polymorphisms, including SNPs and InDels, resulting in a wide range of seed weight variation (Li et al. 2022a). Two major alleles of GmST05 were identified, displaying a geographical distribution and expression levels that positively correlated with seed weight. GmST05 also affects protein and oil content, with the low-protein allele being selected during domestication and improvement (Duan et al. 2022).
Seed quality related
Soybean is a vital leguminous crop that plays a crucial role in providing a global source of vegetable protein and oil. The quality of the seed, specifically in terms of oil and protein content, is a key trait to consider. GmOLEO1 (Zhang et al. 2019b) and GmSWEET39 (Miao et al. 2020) both regulate seed oil content. The alleles contributing to higher oil were both artificially selected during the domestications. And alleles increased oil negatively impacts on protein content. Pleiotropic genes GmST05 (Duan et al. 2022) and GmSWEET10a (Wang et al. 2020b) not only contribute the seed size, but also the oil and protein contents. POWR1 (Goettel et al. 2022) also regulated seed quality as a pleiotropic gene that its mutant had increased oil content and seed weight but decreased protein content. Besides that, GmCCD4 (Gao et al. 2021) regulates seed carotenoid that mutant alleles resulted in increasing of carotenoid content. GmZFP7 (Feng et al. 2022) and GmMYB29 (Chu et al. 2017) involve in isoflavone biosynthesis regulation of soybean. Variations within the promoters of GmZFP7 were found to affect isoflavone accumulation, which is positively correlated with the expression of GmZFP7 (Feng et al. 2022). A single exon SNP in GmMYB29 accounts for 14.91% of the variation in total isoflavone content (Chu et al. 2017).
Biotic and abiotic stress related
The resistance to biotic and abiotic stress is an important factor to ensure yield and quality of final production. SCN, as a represented biotic stress, is a pathogen that causes worldwide significant damage to soybean crops with reducing growth and yield. SCN is difficult to control, as the nematode is able to survive in the soil for long periods of time. The most effective method of control is through the use of resistant soybean varieties. Two major SCN resistance QTLs Rhg1 and Rhg4 were mapped (Concibido et al. 2004) and cloned (Cook et al. 2012; Liu et al. 2012a). Copy number variation of Rhg1 is positively correlated with the SCN resistance (Cook et al. 2012). Two naturally occurring SNPs were identified that alter the function of the Rhg4 enzyme, resulting in a change from resistance to susceptibility (Liu et al. 2012a). Developing the ability of varieties to tolerate drought and salt conditions is crucial in addressing the challenges posed by climate change. Pd1-Ps-P1 feedback loop was discovered to regulate pubescence formation affecting insect and drought tolerance (Liu et al. 2020a). GmSALT3 affects soybean salt tolerance with five haplotypes, and four haplotypes have salt-sensitive phenotype, while the salt tolerance haplotype widely distributes in saline areas (Guan et al. 2014; Qu et al. 2021).
With the rapid development in high-throughput phenotyping platforms (Yang et al. 2020; Jin et al. 2021), it is possible to phenotype time-series, instead of single-point, data of trait in larger population. After that, the time-series data could introduce the time dimension to reveal new trait and increase the statistic power for the genetic dissection of traits (Li and Sillanpaa 2015). For example, a novel trait, speed of canopy closure, was inferred from the time-series canopy coverage in soybean (Li et al. 2023a). GWAS were combined with the time-series data to identify the elite alleles of plant height (Chang et al. 2018), canopy coverage (Xavier et al. 2018; Li et al. 2023a) with the time-series data in soybean. Li et al. (2023a) identified 35 genetic loci associated with canopy coverage that displayed dynamic regulation, which were grouped into three categories based on their function in “Earlier,” “Later,” or “All” of soybean developmental stages. And geographically favored alleles from two different canopy coverage-associated loci were identified with potential applications for breeding programs.
Conclusion
Soybean germplasm studies were dramatically accelerated with developing genotyping technologies in the last decades; however, there were both opportunities and challenges to obtain genetic basis of desired traits from germplasms for future genetic research and breeding. The “missing heritability” was a normal phenomenon in genetic basis dissection of complex quantitative traits, while limited number of major QTLs were repeatedly be identified from independent studies. “Missing heritability” was partially induced by the rare alleles, population size, and the power of statistic model. The cost of genotyping has been continuously decreasing with the development of sequencing platforms, while the quality of reference genomes and pan-genomes is improving. The identification of genetic variation will be more cost-effectively and accurate. High-throughput phenotyping (HTP) methods were widely developed for phenotyping of soybeans and are expected to be improved with advances in sensors, image-processing algorithms, and deep learning (Wang et al. 2020a; Watt et al. 2020; Jin et al. 2021). However, to our knowledge, there have been a limited number of studies (Xavier et al. 2017; Bai et al. 2022; Li et al. 2023a) that have applied HTP platforms to identify elite materials and alleles in diverse populations. These platforms will enable large population sizes in greenhouses or fields, which will further observe higher phenotype diversity. The combination of high-throughput genotyping and phenotyping offers more possibilities with increased population size together with more rare alleles. On the other hand, the statistic models (Liu et al. 2016; Huang et al. 2019; Li et al. 2022b) were developed to addressing the challenges in GWAS: (1) the computational efficiency was increased by addressing increased markers number and samples size; (2) statistic power was increased to explained more phenotype variation; (3) the gene by environment interaction was estimated to handle multiple environment data. Those advanced achievements on genotyping, phenotyping, and statistics will enable us to identify more elite materials and QTLs on desired traits.
Identifying the causal genes within QTLs (quantitative trait loci) is crucial for understanding the genes and regulatory mechanisms that are responsible for desirable traits such as high yield, disease resistance, and drought tolerance. Those information can aid in more precise breeding efforts. The omics data were reported that could prioritize candidate genes and provide complementary results with genetic results in other plants (Lin et al. 2017; Li et al. 2020c, 2021; Zheng et al. 2020; Tang et al. 2021), and multi-omics data were integrated as one network map in maize (Han et al. 2023). However, there is no public omics data for a diverse population in soybean yet. The omics data will certainly facilitate the candidate gene analysis. Moreover, gene editing has been widely used for gene functional validation but still presents challenges. The process of transformation, introducing new genetic material into soybean, is the bottleneck in soybean gene editing, as it is not as advanced as it is in other crops. And soybean is an ancient polyploid with many redundant genes, which affect not only the precision of target gene, but also the phenotype of edited loss-of-function gene. The combination of advanced high-throughput phenotyping and genotyping can expand the range of traits available for selection and increase genetic diversity, resulting in the discovery of novel elite alleles from germplasms. This broadens the genetic base for developing better improved cultivars, advancing genetic research and breeding towards a “green revolution” of soybean.
Acknowledgements
We thank Dr. Bo Zhang (Virginia Tech) for her assistance in editing the manuscript.
Author contribution
YHL, LJQ, DL, and ZZ carried out the literature review and wrote the manuscript, with help from XG, HZ, DB, QW, and TZ. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2021YFD1201601), the earmarked fund for CARS (CARS-04-PS01), and the Agricultural Science and Technology Innovation Program (ASTIP).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors provided the consent for publication.
Competing interests
The authors declare no competing interests.
Footnotes
This article is part of the Topical Collection on Soybean Functional Genomics.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Delin Li and Zhengwei Zhang contributed equally to this work.
Contributor Information
Ying-Hui Li, Email: liyinghui@caas.cn.
Li-Juan Qiu, Email: qiulijuan@caas.cn.
References
- Abe J, Xu D, Suzuki Y, Kanazawa A, Shimamoto Y. Soybean germplasm pools in Asia revealed by nuclear SSRs. Theor Appl Genet. 2003;106(3):445–453. doi: 10.1007/s00122-002-1073-3. [DOI] [PubMed] [Google Scholar]
- Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol. 2012;30(2):174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- Akkaya MS, Bhagwat AA, Cregan PB. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics. 1992;132(4):1131–1139. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu PK, Radhamani J, Aravind J, Varghese E, Tyagi RK. Field performance of 30-year-old soybean germplasm conserved in Indian National genebank. Indian J Plant Genet Resour. 2018;31(2):152–163. doi: 10.5958/0976-1926.2018.00018.9. [DOI] [Google Scholar]
- Bai D, Li D, Zhao C, Wang Z, Shao M, Guo B, Liu Y, Wang Q, Li J, Guo S. Estimation of soybean yield parameters under lodging conditions using RGB information from unmanned aerial vehicles. Front Plant Sci. 2022;13:1012293. doi: 10.3389/fpls.2022.1012293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balfourier F, Roussel V, Strelchenko P, Exbrayat-Vinson F, Sourdille P, Boutet G, Koenig J, Ravel C, Mitrofanova O, Beckert M, Charmet G. A worldwide bread wheat core collection arrayed in a 384-well plate. Theor Appl Genet. 2007;114(7):1265–1275. doi: 10.1007/s00122-007-0517-1. [DOI] [PubMed] [Google Scholar]
- Bayer PE, Valliyodan B, Hu H, Marsh JI, Yuan Y, Vuong TD, Patil G, Song Q, Batley J, Varshney RK, Lam H-M, Edwards D, Nguyen HT (2022) Sequencing the USDA core soybean collection reveals gene loss during domestication and breeding. Plant Genome 15(1):e20109. 10.1002/tpg2.20109 [DOI] [PubMed]
- Boerma HR, Specht JE. Soybeans: improvement, production, and uses. Wisconsin: American Society of Agronomy; 2004. [Google Scholar]
- Brandes N, Linial N, Linial M. PWAS: proteome-wide association study-linking genes and phenotypes by functional variation in proteins. Genome Biol. 2020;21(1):173. doi: 10.1186/s13059-020-02089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BW, Hofstad AN, Sreekanta S, Fu F, Kono TJ, O’Rourke JA, Vance CP, Muehlbauer GJ, Stupar RM. Fast neutron-induced structural rearrangements at a soybean NAP1 locus result in gnarled trichomes. Theor Appl Genet. 2016;129(9):1725–1738. doi: 10.1007/s00122-016-2735-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang R. Chinese soybean genetic resources’ utilization abroad. World Agric. 1989;3:20–21. [Google Scholar]
- Chang R, Sun J. Catalogues of Chinese soybean germplasm and resources: continuation I. Beijing: China Agricultural Press; 1991. [Google Scholar]
- Chang R, Sun J, Qiu L, Chen Y. Catalogues of Chinese soybean germplasm and resources: continuation II. Beijing: China Agricultural Press; 1996. [Google Scholar]
- Chang F, Guo C, Sun F, Zhang J, Wang Z, Kong J, He Q, Sharmin RA, Zhao T. Genome-wide association studies for dynamic plant height and number of nodes on the main stem in summer sowing soybeans. Front Plant Sci. 2018;9:1184. doi: 10.3389/fpls.2018.01184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho G, Yoon M, Lee J, Baek H, Kang J, Kim T, Paek N (2008) Development of a core set of korean soybean landraces [Glycine max(L.) Merr.]. J Crop Sci Biotech 11(3):157–162
- Chu S, Wang J, Zhu Y, Liu S, Zhou X, Zhang H, Wang C-e, Yang W, Tian Z, Cheng H (2017) An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PLoS Genet 13(5):e1006770. 10.1371/journal.pgen.1006770 [DOI] [PMC free article] [PubMed]
- Chung G, Singh RJ. Broadening the genetic base of soybean: a multidisciplinary approach. CRC Crit Rev Plant Sci. 2008;27(5):295–341. doi: 10.1080/07352680802333904. [DOI] [Google Scholar]
- Concibido VC, Diers BW, Arelli PR. A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 2004;44(4):1121–1131. doi: 10.2135/cropsci2004.1121. [DOI] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, Diers BW, Jiang J, Hudson ME, Bent AF. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in Soybean. Science. 2012;338(6111):1206–1209. doi: 10.1126/science.1228746. [DOI] [PubMed] [Google Scholar]
- Cook DE, Bayless AM, Wang K, Guo X, Song Q, Jiang J, Bent AF. Distinct copy number, coding sequence, and locus methylation patterns underlie Rhg1-mediated soybean resistance to soybean cyst nematode. Plant Physiol. 2014;165(2):630–647. doi: 10.1104/pp.114.235952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J, Specht JE. An integrated genetic linkage map of the soybean genome. Crop Sci. 1999;39(5):1464–1490. doi: 10.2135/cropsci1999.3951464x. [DOI] [Google Scholar]
- Deng Y, Liu S, Zhang Y, Tan J, Li X, Chu X, Xu B, Tian Y, Sun Y, Li B, Xu Y, Deng XW, He H, Zhang X. A telomere-to-telomere gap-free reference genome of watermelon and its mutation library provide important resources for gene discovery and breeding. Mol Plant. 2022;15(8):1268–1284. doi: 10.1016/j.molp.2022.06.010. [DOI] [PubMed] [Google Scholar]
- Dobbels AA, Michno J-M, Campbell BW, Virdi KS, Stec AO, Muehlbauer GJ, Naeve SL, Stupar RM (2017) An induced chromosomal translocation in soybean disrupts a KASI ortholog and is associated with a high-sucrose and low-oil seed phenotype. G3: Genes Genom Genet 7(4):1215–1223. 10.1534/g3.116.038596 [DOI] [PMC free article] [PubMed]
- Dong L, Fang C, Cheng Q, Su T, Kou K, Kong L, Zhang C, Li H, Hou Z, Zhang Y, Chen L, Yue L, Wang L, Wang K, Li Y, Gan Z, Yuan X, Weller JL, Lu S, Kong F, Liu B. Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat Commun. 2021;12(1):5445. doi: 10.1038/s41467-021-25800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L, Cheng Q, Fang C, Kong L, Yang H, Hou Z, Li Y, Nan H, Zhang Y, Chen Q, Zhang C, Kou K, Su T, Wang L, Li S, Li H, Lin X, Tang Y, Zhao X, Lu S, Liu B, Kong F. Parallel selection of distinct Tof5 alleles drove the adaptation of cultivated and wild soybean to high latitudes. Mol Plant. 2022;15(2):308–321. doi: 10.1016/j.molp.2021.10.004. [DOI] [PubMed] [Google Scholar]
- Du H, Fang C, Li Y, Kong F, Liu B. Understandings and future challenges in soybean functional genomics and molecular breeding. J Integr Plant Biol. 2023;65(2):468–495. doi: 10.1111/jipb.13433. [DOI] [PubMed] [Google Scholar]
- Duan Z, Zhang M, Zhang Z, Liang S, Fan L, Yang X, Yuan Y, Pan Y, Zhou G, Liu S. Natural allelic variation of GmST05 controlling seed size and quality in soybean. Plant Biotechnol J. 2022;20(9):1807–1818. doi: 10.1111/pbi.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLOS One 6(5):e19379. 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed]
- Feng Y, Zhang S, Li J, Pei R, Tian L, Qi J, Azam M, Agyenim-Boateng KG, Shaibu AS, Liu Y. Dual-function C2H2-type zinc-finger transcription factor GmZFP7 contributes to isoflavone accumulation in soybean. New Phytol. 2022;237(5):1794–1809. doi: 10.1111/nph.18610. [DOI] [PubMed] [Google Scholar]
- Frankel OH. Genetic manipulation: impact on man and society. Cambridge: Cambridge University Press; 1984. Genetic perspectives of germplasm conservation; pp. 161–170. [Google Scholar]
- Fu YB, Cober ER, Morrison MJ, Marsolais F, Peterson GW, Horbach C. Patterns of genetic variation in a soybean germplasm collection as characterized with genotyping-by-sequencing. Plants. 2021;10(8):1611. doi: 10.3390/plants10081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Wang Y, Li W, Gu Y, Lai Y, Bi Y, He C. Transcriptomic comparison reveals genetic variation potentially underlying seed developmental evolution of soybeans. J Exp Bot. 2018;69(21):5089–5104. doi: 10.1093/jxb/ery291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Yang S, Tang K, Li G, Gao X, Liu B, Wang S, Feng X. GmCCD4 controls carotenoid content in soybeans. Plant Biotechnol J. 2021;19(4):801–813. doi: 10.1111/pbi.13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Seymour DK, Liu Q, Zhou Y. Demography and its effects on genomic variation in crop domestication. Nat Plants. 2018;4(8):512–520. doi: 10.1038/s41477-018-0210-1. [DOI] [PubMed] [Google Scholar]
- Gizlice Z, Carter T, Burton J. Genetic base for North American public soybean cultivars released between 1947 and 1988. Crop Sci. 1994;34:1143–1151. doi: 10.2135/cropsci1994.0011183X003400050001x. [DOI] [Google Scholar]
- Goettel W, Zhang H, Li Y, Qiao Z, Jiang H, Hou D, Song Q, Pantalone VR, Song B-H, Yu D. POWR1 is a domestication gene pleiotropically regulating seed quality and yield in soybean. Nat Commun. 2022;13(1):1–11. doi: 10.1038/s41467-022-30314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D, Nelson RT, Cannon SB, Shoemaker RC (2010) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38 (Database issue):D843–D846. 10.1093/nar/gkp798 [DOI] [PMC free article] [PubMed]
- Guan R, Qu Y, Guo Y, Yu L, Liu Y, Jiang J, Chen J, Ren Y, Liu G, Tian L, Jin L, Liu Z, Hong H, Chang R, Gilliham M, Qiu L. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014;80(6):937–950. doi: 10.1111/tpj.12695. [DOI] [PubMed] [Google Scholar]
- Guo Y, Li Y, Hong H, Qiu LJ. Establishment of the integrated applied core collection and its comparison with mini core collection in soybean (Glycine max) Crop J. 2014;2(1):38–45. doi: 10.1016/j.cj.2013.11.001. [DOI] [Google Scholar]
- Guo B, Sun L, Ren H, Sun R, Wei Z, Hong H, Luan X, JunWang WX, Xu D, Li W, Qiu LJ. Soybean genetic resources contributing to sustainable protein production. Theor Appl Genet. 2022;135(11):4095–4121. doi: 10.1007/s00122-022-04241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, Sullivan PF, Nikkola E, Alvarez M, Civelek M, Lusis AJ, Lehtimaki T, Raitoharju E, Kahonen M, Seppala I, Raitakari OT, Kuusisto J, Laakso M, Price AL, Pajukanta P, Pasaniuc B. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zhong W, Qian J, Jin M, Tian P, Zhu W, Zhang H, Sun Y, Feng JW, Liu X, Chen G, Farid B, Li R, Xiong Z, Tian Z, Li J, Luo Z, Du D, Chen S, Jin Q, Li J, Li Z, Liang Y, Jin X, Peng Y, Zheng C, Ye X, Yin Y, Chen H, Li W, Chen LL, Li Q, Yan J, Yang F, Li L. A multi-omics integrative network map of maize. Nat Genet. 2023;55(1):144–153. doi: 10.1038/s41588-022-01262-1. [DOI] [PubMed] [Google Scholar]
- Hao CY, Dong YC, Wang LF, You GX, Zhang HN, Gai HM, Jia JZ, Zhang XY. Establishment and genetic analysis of a wheat core collection in China. Chin Sci Bull. 2008;53:908–915. doi: 10.1007/s11434-008-0212-x. [DOI] [Google Scholar]
- Hao D, Cheng H, Yin Z, Cui S, Zhang D, Wang H, Yu D. Identification of single nucleotide polymorphisms and haplotypes associated with yield and yield components in soybean (Glycine max) landraces across multiple environments. Theor Appl Genet. 2012;124(3):447–458. doi: 10.1007/s00122-011-1719-0. [DOI] [PubMed] [Google Scholar]
- Hou Z, Liu B, Kong F (2022) Chapter Two - Regulation of flowering and maturation in soybean. In: Lam H-M, Li M-W (eds) Adv Bot Res 102:43–75. 10.1016/bs.abr.2022.02.007
- Hu D, Li X, Yang Z, Liu S, Hao D, Chao M, Zhang J, Yang H, Su X, Jiang M, Lu S, Zhang D, Wang L, Kan G, Wang H, Cheng H, Wang J, Huang F, Tian Z, Yu D. Downregulation of a gibberellin 3beta-hydroxylase enhances photosynthesis and increases seed yield in soybean. New Phytol. 2022;235(2):502–517. doi: 10.1111/nph.18153. [DOI] [PubMed] [Google Scholar]
- Huan T, Joehanes R, Song C, Peng F, Guo Y, Mendelson M, Yao C, Liu C, Ma J, Richard M, Agha G, Guan W, Almli LM, Conneely KN, Keefe J, Hwang S-J, Johnson AD, Fornage M, Liang L, Levy D. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun. 2019;10(1):4267. doi: 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Liu X, Zhou Y, Summers RM, Zhang Z (2019) BLINK: a package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 8(2):giy154. 10.1093/gigascience/giy154 [DOI] [PMC free article] [PubMed]
- Hyten DL, Song Q, Zhu Y, Choi I-Y, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci U S A. 2006;103(45):16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Choi I-Y, Song Q, Specht JE, Carter TE, Shoemaker RC, Hwang E-Y, Matukumalli LK, Cregan PB. A high density integrated genetic linkage map of soybean and the development of a 1536 universal soy linkage panel for quantitative trait locus mapping. Crop Sci. 2010;50(3):960–968. doi: 10.2135/cropsci2009.06.0360. [DOI] [Google Scholar]
- Jarquin D, Howard R, Xavier A, Das Choudhury S. Increasing predictive ability by modeling interactions between environments, genotype and canopy coverage image data for soybeans. Agronomy. 2018;8(4):51. doi: 10.3390/agronomy8040051. [DOI] [Google Scholar]
- Jeong N, Kim K-S, Jeong S, Kim J-Y, Park S-K, Lee JS, Jeong S-C, Kang S-T, Ha B-K, Kim D-Y (2019) Korean soybean core collection: genotypic and phenotypic diversity population structure and genome-wide association study. PLOS One 14(10):e0224074. 10.1371/journal.pone.0224074 [DOI] [PMC free article] [PubMed]
- Jiang B, Nan H, Gao Y, Tang L, Yue Y, Lu S, Ma L, Cao D, Sun S, Wang J (2014) Allelic combinations of soybean maturity loci E1, E2, E3 and E4 result in diversity of maturity and adaptation to different latitudes. PLOS ONE 9(8):e106042. 10.1371/journal.pone.0106042 [DOI] [PMC free article] [PubMed]
- Jin X, Zarco-Tejada P, Schmidhalter U, Reynolds MP, Hawkesford MJ, Varshney RK, Yang T, Nie C, Li Z, Ming B, Xiao Y, Xie Y, Li S. High-throughput estimation of crop traits: a review of ground and aerial phenotyping platforms. IEEE Trans Geosci Remote Sens. 2021;9(1):200–231. doi: 10.1109/mgrs.2020.2998816. [DOI] [Google Scholar]
- Kaga A, Shimizu T, Watanabe S, Tsubokura Y, Katayose Y, Harada K, Vaughan DA, Tomooka N. Evaluation of soybean germplasm conserved in NIAS genebank and development of mini core collections. Breed Sci. 2012;61(5):566–592. doi: 10.1270/jsbbs.61.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Diers BW, Olson TC, Shoemaker RC. RFLP mapping in soybean: association between marker loci and variation in quantitative traits. Genetics. 1990;126(3):735–742. doi: 10.1093/genetics/126.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Jo JW, Wang X, Shin M-J, Hur OS, Ha B-K, Hahn B-S. Diversity characterization of soybean germplasm seeds using image analysis. Agronomy. 2022;12(5):1004. doi: 10.3390/agronomy12051004. [DOI] [Google Scholar]
- Lam H-M, Xu X, Liu X, Chen W, Yang G, Wong F-L, Li M-W, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SS-M, Zhang G. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42(12):1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- Lee YG, Jeong N, Kim JH, Lee K, Kim KH, Pirani A, Ha BK, Kang ST, Park BS, Moon JK. Development, validation and genetic analysis of a large soybean SNP genotyping array. Plant J. 2015;81(4):625–636. doi: 10.1111/tpj.12755. [DOI] [PubMed] [Google Scholar]
- Lemay M-A, Torkamaneh D, Rigaill G, Boyle B, Stec AO, Stupar RM, Belzile F. Screening populations for copy number variation using genotyping-by-sequencing: a proof of concept using soybean fast neutron mutants. BMC Genom. 2019;20(1):634. doi: 10.1186/s12864-019-5998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sillanpaa MJ. Dynamic quantitative trait locus analysis of plant phenomic data. Trends Plant Sci. 2015;20(12):822–833. doi: 10.1016/j.tplants.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Li Y, Shi Y, Cao Y, Wang T. Establishment of a core collection for maize germplasm preserved in Chinese National Genebank using geographic distribution and characterization data. Genet Resour Crop Evol. 2004;51:845–852. doi: 10.1007/s10722-005-8313-8. [DOI] [Google Scholar]
- Li YH, Zhou G, Ma J, Jiang W, Jin LG, Zhang Z, Guo Y, Zhang J, Sui Y, Zheng L, Zhang SS, Zuo Q, Shi XH, Li YF, Zhang WK, Hu Y, Kong G, Hong HL, Tan B, Song J, Liu ZX, Wang Y, Ruan H, Yeung CKL, Liu J, Wang H, Zhang LJ, Guan RX, Wang KJ, Li WB, Chen SY, Chang RZ, Jiang Z, Jackson SA, Li R, Qiu LJ. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat Biotechnol. 2014;32(10):1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- Li C, Li YH, Li Y, Lu H, Hong H, Tian Y, Li H, Zhao T, Zhou X, Liu J, Zhou X, Jackson SA, Liu B, Qiu LJ. A domestication-associated gene GmPRR3b regulates the circadian clock and flowering time in soybean. Mol Plant. 2020;13(5):745–759. doi: 10.1016/j.molp.2020.01.014. [DOI] [PubMed] [Google Scholar]
- Li YH, Li D, Jiao YQ, Schnable JC, Li YF, Li HH, Chen HZ, Hong HL, Zhang T, Liu B, Liu ZX, You QB, Tian Y, Guo Y, Guan RX, Zhang LJ, Chang RZ, Zhang Z, Reif J, Zhou XA, Schnable PS, Qiu LJ. Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnol J. 2020;18(2):389–401. doi: 10.1111/pbi.13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang P, You C, Yu J, Zhang X, Yan F, Ye Z, Shen C, Li B, Guo K, Liu N, Thyssen GN, Fang DD, Lindsey K, Zhang X, Wang M, Tu L. Combined GWAS and eQTL analysis uncovers a genetic regulatory network orchestrating the initiation of secondary cell wall development in cotton. New Phytol. 2020;226(6):1738–1752. doi: 10.1111/nph.16468. [DOI] [PubMed] [Google Scholar]
- Li D, Liu Q, Schnable PS. TWAS results are complementary to and less affected by linkage disequilibrium than GWAS. Plant Physiol. 2021;186(4):1800–1811. doi: 10.1093/plphys/kiab161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Y, Ma R, Huang W, Hou J, Fang C, Wang L, Yuan Z, Sun Q, Dong X. Identification of ST1 reveals a selection involving hitchhiking of seed morphology and oil content during soybean domestication. Plant Biotechnol J. 2022;20(6):1110–1121. doi: 10.1111/pbi.13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhang YW, Zhang ZC, Xiang Y, Liu MH, Zhou YH, Zuo JF, Zhang HQ, Chen Y, Zhang YM. A compressed variance component mixed model for detecting QTNs and QTN-by-environment and QTN-by-QTN interactions in genome-wide association studies. Mol Plant. 2022;15(4):630–650. doi: 10.1016/j.molp.2022.02.012. [DOI] [PubMed] [Google Scholar]
- Li X, Hu D, Cai L, Wang H, Liu X, Du H, Yang Z, Zhang H, Hu Z, Huang F. CALCIUM-DEPENDENT PROTEIN KINASE38 regulates flowering time and common cutworm resistance in soybean. Plant Physiol. 2022;190(1):480–499. doi: 10.1093/plphys/kiac260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Li YH, Su SS, Reif JC, Qi ZM, Wang XB, Wang X, Tian Y, Li DL, Sun RJ, Liu ZX, Xu ZJ, Fu GH, Ji YL, Chen QS, Liu JQ, Qiu LJ. SoySNP618K array: a high-resolution single nucleotide polymorphism platform as a valuable genomic resource for soybean genetics and breeding. J Integr Plant Biol. 2022;64(3):632–648. doi: 10.1111/jipb.13202. [DOI] [PubMed] [Google Scholar]
- Li D, Bai D, Tian Y, Li YH, Zhao C, Wang Q, Guo S, Gu Y, Luan X, Wang R, Yang J, Hawkesford MJ, Schnable JC, Jin X, Qiu LJ. Time series canopy phenotyping enables the identification of genetic variants controlling dynamic phenotypes in soybean. J Integr Plant Biol. 2023;65:117–132. doi: 10.1111/jipb.13380. [DOI] [PubMed] [Google Scholar]
- Li YH, Qin C, Wang L, Jiao C, Hong H, Tian Y, Li Y, Xing G, Wang J, Gu Y, Gao X, Li D, Li H, Liu Z, Jing X, Feng B, Zhao T, Guan R, Guo Y, Liu J, Yan Z, Zhang L, Ge T, Li X, Wang X, Qiu H, Zhang W, Luan X, Han Y, Han D, Chang R, Guo Y, Reif JC, Jackson SA, Liu B, Tian S, Qiu LJ. Genome-wide signatures of the geographic expansion and breeding of soybean. Sci China Life Sci. 2023;66:350–365. doi: 10.1007/s11427-022-2158-7. [DOI] [PubMed] [Google Scholar]
- Lin HY, Liu Q, Li X, Yang J, Liu S, Huang Y, Scanlon MJ, Nettleton D, Schnable PS. Substantial contribution of genetic variation in the expression of transcription factors to phenotypic variation revealed by eRD-GWAS. Genome Biol. 2017;18(1):192. doi: 10.1186/s13059-017-1328-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J. The soybean stem growth habit gene Dt1 Is an ortholog of arabidopsis TERMINAL FLOWER1. Plant Physiol. 2010;153(1):198–210. doi: 10.1104/pp.109.150607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kandoth PK, Warren SD, Yeckel G, Heinz R, Alden J, Yang C, Jamai A, El-Mellouki T, Juvale PS, Hill J, Baum TJ, Cianzio S, Whitham SA, Korkin D, Mitchum MG, Meksem K. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature. 2012;492(7428):256–260. doi: 10.1038/nature11651. [DOI] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS (2012b) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLOS One 7(5):e36406. 10.1371/journal.pone.0036406 [DOI] [PMC free article] [PubMed]
- Liu Q, Chang S, Hartman GL, Domier LL. Assembly and annotation of a draft genome sequence for Glycine latifolia, a perennial wild relative of soybean. Plant J. 2018;95(1):71–85. doi: 10.1111/tpj.13931. [DOI] [PubMed] [Google Scholar]
- Liu S, Fan L, Liu Z, Yang X, Zhang Z, Duan Z, Liang Q, Imran M, Zhang M, Tian Z. A Pd1–Ps–P1 feedback loop controls pubescence density in soybean. Mol Plant. 2020;13(12):1768–1783. doi: 10.1016/j.molp.2020.10.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou GA, Zhang H, Liu Z, Shi M, Huang X, Li Y, Zhang M, Wang Z, Zhu B, Han B, Liang C, Tian Z. Pan-Genome of wild and cultivated soybeans. Cell. 2020;182(1):162–176.e13. doi: 10.1016/j.cell.2020.05.023. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12(2):e1005767. 10.1371/journal.pgen.1005767 [DOI] [PMC free article] [PubMed]
- Liu Y, Liu S, Zhang Z, Ni L, Chen X, Ge Y, Zhou G, Tian Z (2022) GenoBaits Soy40K: a highly flexible and low-cost SNP array for soybean studies. Sci China Life Sci 65(9):1898–1901. 10.1007/s11427-022-2130-8 [DOI] [PubMed]
- Lopez MA, Xavier A, Rainey KM (2019) Phenotypic variation and genetic architecture for photosynthesis and water use efficiency in soybean (Glycine max L. Merr). Front Plant Sci 10:680. 10.3389/fpls.2019.00680 [DOI] [PMC free article] [PubMed]
- Lord J, Jermy B, Green R, Wong A, Xu J, Legido-Quigley C, Dobson R, Richards M, Proitsi P (2021) Mendelian randomization identifies blood metabolites previously linked to midlife cognition as causal candidates in Alzheimer’s disease. Proc Natl Acad Sci U S A 118(16):e2009808118. 10.1073/pnas.2009808118 [DOI] [PMC free article] [PubMed]
- Lu S, Zhao X, Hu Y, Liu S, Nan H, Li X, Fang C, Cao D, Shi X, Kong L. Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet. 2017;49(5):773–779. doi: 10.1038/ng.3819. [DOI] [PubMed] [Google Scholar]
- Lu X, Xiong Q, Cheng T, Li QT, Liu XL, Bi YD, Li W, Zhang WK, Ma B, Lai YC. A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol Plant. 2017;10(5):670–684. doi: 10.1016/j.molp.2017.03.006. [DOI] [PubMed] [Google Scholar]
- Lu S, Dong L, Fang C, Liu S, Kong L, Cheng Q, Chen L, Su T, Nan H, Zhang D. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat Genet. 2020;52(4):428–436. doi: 10.1038/s41588-020-0604-7. [DOI] [PubMed] [Google Scholar]
- Ma Y, Wang W, Wang L, Ma F, Wang P, Chang R, Qiu L. Genetic diversity of soybean and the establishment of a core collection focused on resistance to soybean cyst nematode. J Integr Plant Biol. 2006;48(6):722–731. doi: 10.1111/j.1744-7909.2006.00256.x. [DOI] [Google Scholar]
- Ma Y, Min L, Wang J, Li Y, Wu Y, Hu Q, Ding Y, Wang M, Liang Y, Gong Z, Xie S, Su X, Wang C, Zhao Y, Fang Q, Li Y, Chi H, Chen M, Khan AH, Lindsey K, Zhu L, Li X, Zhang X. A combination of genome-wide and transcriptome-wide association studies reveals genetic elements leading to male sterility during high temperature stress in cotton. New Phytol. 2021;231(1):165–181. doi: 10.1111/nph.17325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Fan L, Zhang Z, Yang X, Liu Y, Ma Y, Pan Y, Zhou G, Zhang M, Ning H, Kong F, Ma J, Liu S, Tian Z. Global dissection of the recombination landscape in soybean using a high-density 600K SoySNP array. Plant Biotechnol. 2023;J21:606–620. doi: 10.1111/pbi.13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Yang S, Zhang K, He J, Wu C, Ren Y, Gai J, Li Y. Natural variation and selection in GmSWEET39 affect soybean seed oil content. New Phytol. 2020;225(4):1651–1666. doi: 10.1111/nph.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore RW, Paran I, Kesseli RV. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci U S A. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RL. Managing self-pollinated germplasm collections to maximize utilization. Plant Genet Resour. 2011;9(01):123–133. doi: 10.1017/s147926211000047x. [DOI] [Google Scholar]
- Nguyen CX, Paddock KJ, Zhang Z, Stacey MG. GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol. 2021;229(2):920–934. doi: 10.1111/nph.16928. [DOI] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen N-C, Cheng H, Chin C-S, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O’Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM. The complete sequence of a human genome. Science. 2022;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MF, Nelson RL, Geraldi IO, Cruz CD, de Toledo JFF. Establishing a soybean germplasm core collection. Field Crops Res. 2010;119(2–3):277–289. doi: 10.1016/j.fcr.2010.07.021. [DOI] [Google Scholar]
- Orf JH, Chase K, Jarvik T, Mansur LM, Cregan PB, Adler FR, Lark KG (1999) Genetics of soybean agronomic traits: I. comparison of three related recombinant inbred populations. Crop Sci 39(6):1642–1651. 10.2135/cropsci1999.3961642x
- Ping J, Liu Y, Sun L, Zhao M, Li Y, She M, Sui Y, Lin F, Liu X, Tang Z. Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell. 2014;26(7):2831–2842. doi: 10.1105/tpc.114.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolli R, Wysmierski P, Cunha C, Pinheiro J, Vello N. Genetic structure and a selected core set of Brazilian soybean cultivars. Genet Mol Biol. 2013;36(3):382–390. doi: 10.1590/S1415-47572013005000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu LJ, Li YH, Guan RX, Liu ZX, Wang LX, Chang RZ. Establishment, representative testing and research progress of soybean core collection and mini core collection. Acta Agron Sin. 2009;35(4):571–579. doi: 10.3724/sp.J.1006.2009.00571. [DOI] [Google Scholar]
- Qiu LJ, Chen PY, Liu ZX, Li YH, Guan RX, Wang LH, Chang RZ. The worldwide utilization of the Chinese soybean germplasm collection. Plant Genet Resour. 2011;9(01):109–122. doi: 10.1017/s1479262110000493. [DOI] [Google Scholar]
- Qiu LJ, Chang RZ, Liu ZX. Catalogues of Chinese soybean germplasm and resources: continuation III. Beijing: China Agricultural Press; 2013. [Google Scholar]
- Qiu LJ, Xing LL, Guo Y, Wang J, Jackson SA, Chang RZ. A platform for soybean molecular breeding: the utilization of core collections for food security. Plant Mol Biol. 2013;83(1–2):41–50. doi: 10.1007/s11103-013-0076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Cao Y, Chang R, Zhou X, Wang G, Sun J, Xie H, Zhang B, Li X, Xu Z, Liu L (2003) Establishment of Chinese soybean (G. max) core collection I: sampling strategy. Sci Agric Sin 36(12):1442–1449
- Qu Y, Guan R, Bose J, Henderson SW, Wege S, Qiu L, Gilliham M. Soybean CHX-type ion transport protein GmSALT3 confers leaf Na+ exclusion via a root derived mechanism, and Cl− exclusion via a shoot derived process. Plant Cell Environ. 2021;44(3):856–869. doi: 10.1111/pce.13947. [DOI] [PubMed] [Google Scholar]
- Reinprecht Y, Poysa VW, Yu K, Rajcan I, Ablett GR, Pauls KP (2006) Seed and agronomic QTL in low linolenic acid, lipoxygenase-free soybean (Glycine max (L.) Merrill) germplasm. Genome 49(12):1510–1527. 10.1139/g06-112 [DOI] [PubMed]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6(8):550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Schoener CS, Fehr WR (1979) Utilization of plant introductions in soybean breeding populations. Crop Sci 19(2):cropsci1979.0011183X001900020003x. 10.2135/cropsci1979.0011183X001900020003x
- Scott K, Balk C, Veney D, McHale LK, Dorrance AE. Quantitative disease resistance loci towards Phytophthora sojae and three species of Pythium in six soybean nested association mapping populations. Crop Sci. 2019;59(2):605–623. doi: 10.2135/cropsci2018.09.0573. [DOI] [Google Scholar]
- Shen Y, Du H, Liu Y, Ni L, Wang Z, Liang C, Tian Z. Update soybean Zhonghuang 13 genome to a golden reference. Sci China Life Sci. 2019;62(9):1257–1260. doi: 10.1007/s11427-019-9822-2. [DOI] [PubMed] [Google Scholar]
- Sherman-Broyles S, Bombarely A, Powell AF, Doyle JL, Egan AN, Coate JE, Doyle JJ. The wild side of a major crop: soybean’s perennial cousins from Down Under. Am J Bot. 2014;101(10):1651–1665. doi: 10.3732/ajb.1400121. [DOI] [PubMed] [Google Scholar]
- Shi Z, Liu S, Noe J, Arelli P, Meksem K, Li Z. SNP identification and marker assay development for high-throughput selection of soybean cyst nematode resistance. BMC Genom. 2015;16(1):314. doi: 10.1186/s12864-015-1531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura M, Kanamori H, Komatsu S, Namiki N, Mukai Y, Kurita K, Kamatsuki K, Ikawa H, Yano R, Ishimoto M (2015) The Glycine max cv. Enrei genome for improvement of Japanese soybean cultivars. Int J Genomics 2015:358127. 10.1155/2015/358127 [DOI] [PMC free article] [PubMed]
- Shu Y, Li Y, Zhu Z, Bai X, Cai H, Ji W, Guo D, Zhu Y. SNPs discovery and CAPS marker conversion in soybean. Mol Biol Rep. 2011;38(3):1841–1846. doi: 10.1007/s11033-010-0300-2. [DOI] [PubMed] [Google Scholar]
- Sohn JI, Nam JW. The present and future of de novo whole-genome assembly. Brief Bioinform. 2018;19(1):23–40. doi: 10.1093/bib/bbw096. [DOI] [PubMed] [Google Scholar]
- Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J. 2015;13(2):211–221. doi: 10.1111/pbi.12249. [DOI] [PubMed] [Google Scholar]
- Sonah H, Bastien M, Iquira E, Tardivel A, Légaré G, Boyle B, Normandeau É, Laroche J, Larose S, Jean M (2013) An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLOS One 8(1):e54603. 10.1371/journal.pone.0054603 [DOI] [PMC free article] [PubMed]
- Song J, Li Z, Liu Z, Guo Y, Qiu LJ. Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Front Plant Sci. 2017;8:919. doi: 10.3389/fpls.2017.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JM, Xie WZ, Wang S, Guo YX, Koo DH, Kudrna D, Gong C, Huang Y, Feng JW, Zhang W. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol Plant. 2021;14(10):1757–1767. doi: 10.1016/j.molp.2021.06.018. [DOI] [PubMed] [Google Scholar]
- Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLOS One 8(1):e54985. 10.1371/journal.pone.0054985 [DOI] [PMC free article] [PubMed]
- Specht J, Chase K, Macrander M, Graef G, Chung J, Markwell J, Germann M, Orf J, Lark K. Soybean response to water: a QTL analysis of drought tolerance. Crop Sci. 2001;41(2):493–509. doi: 10.2135/cropsci2001.412493x. [DOI] [Google Scholar]
- Sun R, Sun B, Tian Y, Su S, Zhang Y, Zhang W, Wang J, Yu P, Guo B, Li H, Li Y, Gao H, Gu Y, Yu L, Ma Y, Su E, Li Q, Hu X, Zhang Q, Guo R, Chai S, Feng L, Wang J, Hong H, Xu J, Yao X, Wen J, Liu J, Li Y, Qiu L. Dissection of the practical soybean breeding pipeline by developing ZDX1, a high-throughput functional array. Theor Appl Genet. 2022;135(4):1413–1427. doi: 10.1007/s00122-022-04043-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susan M, Baute J, James B, Paula B, Peter K, Edward B, Burke M, David C, Sylvie C, Glenn CJN. Agriculture: feeding the future. Nature. 2013;499:23–24. doi: 10.1038/499023a. [DOI] [PubMed] [Google Scholar]
- Tang S, Zhao H, Lu S, Yu L, Zhang G, Zhang Y, Yang QY, Zhou Y, Wang X, Ma W, Xie W, Guo L. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant. 2021;14(3):470–487. doi: 10.1016/j.molp.2020.12.003. [DOI] [PubMed] [Google Scholar]
- Torkamaneh D, Lemay MA, Belzile F. The pan-genome of the cultivated soybean (PanSoy) reveals an extraordinarily conserved gene content. Plant Biotechnol J. 2021;19(9):1852–1862. doi: 10.1111/pbi.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubokura Y, Matsumura H, Xu M, Liu B, Nakashima H, Anai T, Kong F, Yuan X, Kanamori H, Katayose Y. Genetic variation in soybean at the maturity locus E4 is involved in adaptation to long days at high latitudes. Agronomy. 2013;3(1):117–134. doi: 10.3390/agronomy3010117. [DOI] [Google Scholar]
- Tsubokura Y, Watanabe S, Xia Z, Kanamori H, Yamagata H, Kaga A, Katayose Y, Abe J, Ishimoto M, Harada K. Natural variation in the genes responsible for maturity loci E1, E2, E3 and E4 in soybean. Ann Bot. 2014;113(3):429–441. doi: 10.1093/aob/mct269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B, Cannon SB, Bayer PE, Shu S, Brown AV, Ren L, Jenkins J, Chung CYL, Chan TF, Daum CG. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019;100(5):1066–1082. doi: 10.1111/tpj.14500. [DOI] [PubMed] [Google Scholar]
- Wang G. Catalogues of Chinese soybean germplasm and resources. Beijing: China Agricultural Press; 1982. [Google Scholar]
- Wang L, Guan Y, Guan R, Li Y, Ma Y, Dong Z, Liu X, Zhang H, Zhang Y, Liu Z, Chang R, Xu H, Li L, Lin F, Luan W, Yan Z, Ning X, Zhu L, Cui Y, Piao R, Liu Y, Chen P, Qiu L. Establishment of Chinese soybean Glycine max core collections with agronomic traits and SSR markers. Euphytica. 2006;151(2):215–223. doi: 10.1007/s10681-006-9142-3. [DOI] [Google Scholar]
- Wang J, Chu S, Zhang H, Zhu Y, Cheng H, Yu D. Development and application of a novel genome-wide SNP array reveals domestication history in soybean. Sci Rep. 2016;6:20728. doi: 10.1038/srep20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cimen E, Singh N, Buckler E. Deep learning for plant genomics and crop improvement. Curr Opin Plant Biol. 2020;54:34–41. doi: 10.1016/j.pbi.2019.12.010. [DOI] [PubMed] [Google Scholar]
- Wang S, Liu S, Wang J, Yokosho K, Zhou B, Yu Y-C, Liu Z, Frommer WB, Ma JF, Chen L-Q. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl Sci Rev. 2020;7(11):1776–1786. doi: 10.1093/nsr/nwaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang X, Jia Y, Xu Y, Jia P, Dang N, Wang S, Xu T, Zhao X, Gao S, Dong Q, Ye K. High-quality Glycine max genome assembly with nanopore and HiFi Long reads. Genomics Proteomics Bioinformatics. 2022;20(1):4–13. doi: 10.1016/j.gpb.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt M, Fiorani F, Usadel B, Rascher U, Muller O, Schurr U. Phenotyping: new windows into the plant for breeders. Annu Rev Plant Biol. 2020;71:689–712. doi: 10.1146/annurev-arplant-042916-041124. [DOI] [PubMed] [Google Scholar]
- Wen Z, Boyse JF, Song Q, Cregan PB, Wang D. Genomic consequences of selection and genome-wide association mapping in soybean. BMC Genom. 2015;16:671. doi: 10.1186/s12864-015-1872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo AP, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM, Dammer EB, Robins C, Beach TG, Reiman EM, Epstein MP, De Jager PL, Lah JJ, Bennett DA, Seyfried NT, Levey AI, Wingo TS. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer’s disease pathogenesis. Nat Genet. 2021;53(2):143–146. doi: 10.1038/s41588-020-00773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A, Hall B, Hearst AA, Cherkauer KA, Rainey KM. Genetic architecture of phenomic-enabled canopy coverage in Glycine max. Genetics. 2017;206(2):1081–1089. doi: 10.1534/genetics.116.198713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A, Jarquin D, Howard R, Ramasubramanian V, Specht JE, Graef GL, Beavis WD, Diers BW, Song Q, Cregan PB (2018) Genome-wide analysis of grain yield stability and environmental interactions in a multiparental soybean population. G3: Genes Genom Genet 8(2):519–529. 10.1534/g3.117.300300 [DOI] [PMC free article] [PubMed]
- Xia Z, Watanabe S, Yamada T, Tsubokura Y, Nakashima H, Zhai H, Anai T, Sato S, Yamazaki T, Lu S, Wu H, Tabata S, Harada K. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc Natl Acad Sci U S A. 2012;109(32):E2155–2164. doi: 10.1073/pnas.1117982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Chung CY-L, Li M-W, Wong F-L, Wang X, Liu A, Wang Z, Leung AK-Y, Wong T-H, Tong S-W, Xiao Z, Fan K, Ng M-S, Qi X, Yang L, Deng T, He L, Chen L, Fu A, Ding Q, He J, Chung G, Isobe S, Tanabata T, Valliyodan B, Nguyen HT, Cannon SB, Foyer CH, Chan T-F, Lam H-M. A reference-grade wild soybean genome. Nat Commun. 2019;10(1):1216. doi: 10.1038/s41467-019-09142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Huang X. A new high-quality genome sequence in soybean. Sci China Life Sci. 2018;61(12):1604. doi: 10.1007/s11427-018-9431-8. [DOI] [PubMed] [Google Scholar]
- Yang W, Feng H, Zhang X, Zhang J, Doonan JH, Batchelor WD, Xiong L, Yan J. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol Plant. 2020;13(2):187–214. doi: 10.1016/j.molp.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Yang C, Yan J, Jiang S, Li X, Min H, Wang X, Hao D. Resequencing 250 soybean accessions: new insights into genes associated with agronomic traits and genetic networks. Genomics Proteomics Bioinformatics. 2022;20(1):29–41. doi: 10.1016/j.gpb.2021.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Njiti V, Meksem K, Iqbal M, Triwitayakorn K, Kassem MA, Davis G, Schmidt M, Lightfoot D. Quantitative trait loci in two soybean recombinant inbred line populations segregating for yield and disease resistance. Crop Sci. 2002;42(1):271–277. doi: 10.2135/cropsci2002.2710. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Wang M, Sun J, Qi Y, Li J, Wei X, Han L, Qiu Z, Tang S, Li Z. A core collection and mini core collection of Oryza sativa L. in China. Theor Appl Genet. 2011;122(1):49–61. doi: 10.1007/s00122-010-1421-7. [DOI] [PubMed] [Google Scholar]
- Zhang D, Wang X, Li S, Wang C, Gosney MJ, Mickelbart MV, Ma J. A post-domestication mutation, Dt2, triggers systemic modification of divergent and convergent pathways modulating multiple agronomic traits in soybean. Mol Plant. 2019;12(10):1366–1382. doi: 10.1016/j.molp.2019.05.010. [DOI] [PubMed] [Google Scholar]
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY (2004) QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet 108(6):1131–1139. 10.1007/s00122-003-1527-2 [DOI] [PubMed]
- Zhang D, Zhang H, Hu Z, Chu S, Yu K, Lv L, Yang Y, Zhang X, Chen X, Kan G (2019b) Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet 15(7):e1008267. 10.1186/s12870-016-0704-9 [DOI] [PMC free article] [PubMed]
- Zhao J, Fu J, Liao H, He Y, Nian H, Hu Y, Qiu L, Dong Y, Yan X. Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. Chin Sci Bull. 2004;49(15):1611–1620. doi: 10.1360/04wc0142. [DOI] [Google Scholar]
- Zhao L, Dong Y, Liu B, Hao S, Wang K, Li X. Establishment of a core collection for the Chinese annual wild soybean (Glycine soja) Chin Sci Bull. 2005;50:989–996. doi: 10.1360/982004-657. [DOI] [Google Scholar]
- Zhao C, Takeshima R, Zhu J, Xu M, Sato M, Watanabe S, Kanazawa A, Liu B, Kong F, Yamada T, Abe J. A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 2016;16:20. doi: 10.1186/s12870-016-0704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Li M, Xu C, Yang X, Li D, Zhao X, Wang K, Li Y, Zhang X, Liu L. Natural variation in GmGBP1 promoter affects photoperiod control of flowering time and maturity in soybean. Plant J. 2018;96(1):147–162. doi: 10.1111/tpj.14025. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Hey S, Jubery T, Liu H, Yang Y, Coffey L, Miao C, Sigmon B, Schnable JC, Hochholdinger F, Ganapathysubramanian B, Schnable PS. Shared genetic control of root system architecture between Zea mays and Sorghum bicolor. Plant Physiol. 2020;182(2):977–991. doi: 10.1104/pp.19.00752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, Fang C, Shen Y, Liu T, Li C, Li Q, Wu M, Wang M, Wu Y, Dong Y, Wan W, Wang X, Ding Z, Gao Y, Xiang H, Zhu B, Lee S-H, Wang W, Tian Z. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol. 2015;33(4):408–414. doi: 10.1038/nbt.3096. [DOI] [PubMed] [Google Scholar]
- Zhu W, Yang C, Yong B, Wang Y, Li B, Gu Y, Wei S, An Z, Sun W, Qiu L. An enhancing effect attributed to a nonsynonymous mutation in SOYBEAN SEED SIZE 1, a SPINDLY-like gene, is exploited in soybean domestication and improvement. New Phytol. 2022;236(4):1375–1392. doi: 10.1111/nph.18461. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Wang X, Li X, Hu J, Fan L, Landis JB, Cannon SB, Grimwood J, Schmutz J, Jackson SA, Doyle JJ, Zhang XS, Zhang D, Ma J. Phylogenomics of the genus Glycine sheds light on polyploid evolution and life-strategy transition. Nat Plants. 2022;8(3):233–244. doi: 10.1038/s41477-022-01102-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.