Abstract
Current combined challenges of rising food demand, climate change and farmland degradation exert enormous pressure on agricultural production. Worldwide soil salinization, in particular, necessitates the development of salt-tolerant crops. Soybean, being a globally important produce, has its genetic resources increasingly examined to facilitate crop improvement based on functional genomics. In response to the multifaceted physiological challenge that salt stress imposes, soybean has evolved an array of defences against salinity. These include maintaining cell homeostasis by ion transportation, osmoregulation, and restoring oxidative balance. Other adaptations include cell wall alterations, transcriptomic reprogramming, and efficient signal transduction for detecting and responding to salt stress. Here, we reviewed functionally verified genes that underly different salt tolerance mechanisms employed by soybean in the past two decades, and discussed the strategy in selecting salt tolerance genes for crop improvement. Future studies could adopt an integrated multi-omic approach in characterizing soybean salt tolerance adaptations and put our existing knowledge into practice via omic-assisted breeding and gene editing. This review serves as a guide and inspiration for crop developers in enhancing soybean tolerance against abiotic stresses, thereby fulfilling the role of science in solving real-life problems.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-023-01383-3.
Keywords: Soybean, Salt stress, Osmotic regulation, Ion homeostasis, Transcription regulation, Oxidative stress response
Introduction
The forecasted population growth in the 21st century will not only bring about increased demand in food production, but also accelerated urbanization. The current agricultural output will no longer be sufficient to support the increasing population, while urbanization will shrink the area of available farmland, worsening the situation. Although breeders and scientists are dedicated to increasing crop yield, the increased yield will not be sufficient to satisfy the demand. Exploring ways of utilizing salt-affected land for crop production appears to be an attractive option for increasing crop production (Shrivastava and Kumar 2015). To exacerbate the situation, agricultural irrigation is a major driving force of farmland salinization (Hassani et al. 2021). At the same time, changes in precipitation patterns and global warming due to climate change have been predicted to massively alter the distribution of salinized soil (Hassani et al. 2021). To better utilize saline soil and prevent yield loss due to farmland salinization, researchers are determined to gain a better understanding of the salt stress responses in crops to improve their salt tolerance through molecular breeding or genetic engineering.
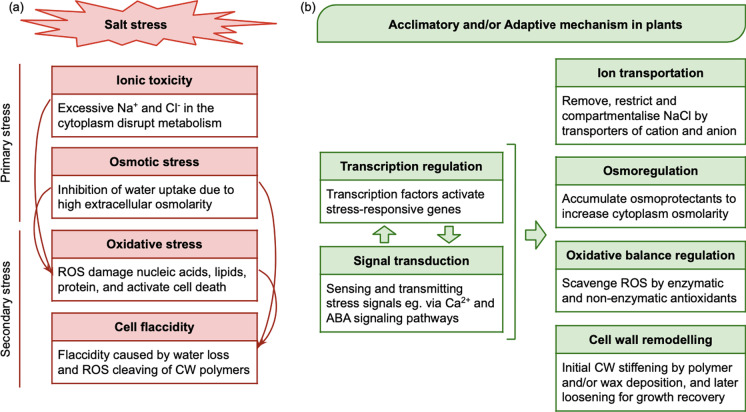
Salt stress is caused by the presence of excessive salt in the soil, which hampers the normal physiology of plants by inducing multiple stressors simultaneously (Fig. 1a). In turn, plants evolved acclimatory or adaptive responses to salinity (Fig. 1b). Salt stress includes the primary stresses of osmotic and ionic imbalance, and secondary stresses caused by reactive oxygen species (Ashraf 1994) and cell flaccidity (Yang and Guo 2018). The excessive salt content in the soil lowers the osmotic potential, and thus reduces the availability of water for uptake by the plant root. This causes physiological drought in the plant, where, despite the presence of water in the soil, the plant cannot uptake enough water through the roots to compensate for the water loss through transpiration. In some cases, the ionic salt is absorbed concurrently with the soil water, then transported and built up in the aerial parts of the plant. The accumulation of the toxic ionic salt causes ionic stress and disrupts cellular functions. The osmotic and ionic stresses then trigger the production of ROS, which damages macromolecules such as DNA and lipid, leading to cell death. Furthermore, net water loss causes cells to collapse from the reduced turgor pressure, which is acerbated by cell wall impairment caused by ROS. The ability to eliminate or tolerate these stress components is hence the key to salt tolerance. Plants have evolved mitigation towards each and different components of salt stress, which have been described in detail by many well written reviews (Deinlein et al. 2014; Hanin et al. 2016; Phang et al. 2008; van Zelm et al. 2020). Briefly, at the molecular level, ion transportation by specialised membrane proteins can restrict the negative impacts of excess salt ions. Water uptake is enhanced, and water loss is reduced by accumulating osmoprotectants to increase the osmolarity inside the cytoplasm. Antioxidants that scavenge and neutralise excessive ROS are produced to restore the oxidative balance. The loss of cell structural support is compensated by cell wall strengthening, and later loosening to enable growth and elongation. Meanwhile, all these responses are regulated and enabled by the interplay between transcription regulation and signal transduction.
Fig. 1.
Salt stress and molecular tolerance mechanisms in plants. Each mechanism and its genetic components in soybeans are discussed in detail in individual sections. Abbreviations: NaCl, sodium chloride; ROS, reactive oxygen species; CW, cell wall; ABA, abscisic acid
Soybean (Glycine max) is a relatively sustainable crop. It could acquire nitrogen through symbiotic nitrogen fixation in the nodules. The surplus of organic nitrogen can also replenish soil fertility. Therefore, compared to other major crops, soybean has a lower dependency on synthetic nitrogen fertilizer, of which production and utilization are energy-demanding and polluting to the environment. Soybean seeds contain roughly 40% protein and 20% oil, making it a good alternative to animal protein and lipid. With soybean being a primary producer, its consumption minimizes the energy and material loss during trophic transfer. Owing to its importance, soybean is also predicted to become a dominant crop in Africa in the future (Foyer et al. 2019).
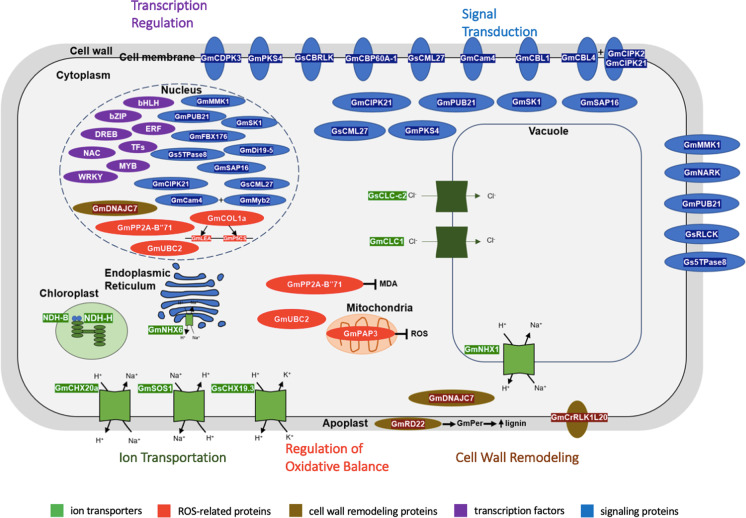
Soybean is regarded as a moderately salt-tolerant crop (Ashraf 1994), and thus its production and acreage expansion are also hampered by soil salinization. However, unlike the model plant Arabidopsis, there is a lack of mutant collection for soybean. Screening soybean mutants for salt-tolerance genes is, hence, not a viable strategy. Twenty years ago, salt tolerance research in soybean was confined to physiological characterization and some low-resolution mapping. There were limited identification and characterization of functional genes. However, in the past twenty years, the advances in sequencing technologies have enabled the genome-wide identification and characterization of gene families with well-annotated reference genomes. Moreover, sequencing-based genotyping methods, such as genotyping-by-sequencing and genome resequencing, have generated high-density markers for precision mapping of genes related to salt tolerance. Transcriptomic studies have also been able to detect global gene expression changes under salt stress. We have previously reviewed the soybean salt tolerance mechanisms before the genomic age (Phang et al. 2008). In this review, we will explore the progress made in the identification and functional characterization of salt tolerance genes from soybean in the past twenty years (Fig. 2). From that, we will discuss the selection priority of various salt-tolerance mechanisms and suggest methods to incorporate our knowledge into field application.
Fig. 2.
Overview of subcellular localization of soybean salt tolerance genes at cellular level. Light and dark green boxes indicate cation and anion transporters. respectively. CLC, chloride/proton exchanger or chloride channel; CHX, cation/H+ exchanger; NDH, subunit of NAD(P)H dehydrogenase complex; NHX, SOS (Salt Overly Sensitive), Na+/H+ exchanger. Genes involve in regulation of oxidative balance are colored in red. GmCOL1a, CONSTANS-LIKE 1a protein; GmLEA and GmP5CS are downstream target genes of GmCOL1a. GmPAP3, purple acid phosphatase 3; GmPP2A-B’71, B” subunit of phosphatase 2A; GmUCB2, ubiquitin-conjugating enzyme. Genes involve in cell wall remodeling are shown in brown. GmCrRLK1L20, Catharanthus roseus RLK1-like protein; GmDNAJC7, co-chaperone DNAJ protein; GmRD22, BURP-domain protein. Transcription factors are indicated in purple. bHLH, basic/helix-loop-helix protein; bZIP, basic leucine-zipper; DREB, dehydration responsive element-binding proteins; ERF, ethylene-responsive factors; MYB, MYB transcription factors; NAC, NAC (NAM, ATAF, CUC) transcription factors; WRKY, WRKY transcription factors; TFs, other transcription factors. Blue boxes indicate genes involve in signal transduction. GmCam4, calmodulin; GmCBL1, GmCBL4, calcineurin B-like protein; GmCBP60A-1, calmodulin-binding protein; GsCBRLK, calcium-dependent calmodulin-binding receptor-like kinase; GmCDPK3, calcium-dependent protein kinase; GmCIPK2, GmCIPK21, GmPKS4, calcineurin B-like protein interacting kinase; GmCML27, calmodulin-like protein. GmDi19-5, drought-induced protein; GmFBX176, F-box protein; GmMMK1, mitogen-activated kinase; GmNARK, nodule autoregulation receptor kinase; GmPUB21, U-box E3-ubiquitin ligase; GsRLCK, receptor-like cytoplasmic serine/threonine protein kinase; GmSAP16, stress associated protein; GmSK1, S-phase kinase-associated protein 1; Gs5PTase8, inositol polyphosphate 5-phosphatase
Ion transportation
Sodium (Na+) is not an essential nutrient for plants. However, Na+ can serve as a subpar substitute of potassium (K+), an essential macronutrient of plant, when K+ is scarce (Maathuis 2014). Meanwhile, chloride (Cl−) is a micronutrient of higher plants that is involved in vital functions such as photosynthesis, growth and development etc. (Geilfus 2018). Although sodium and chloride are beneficial to plants at low dosage, excessive amount of either one could be toxic. Therefore, to survive salt stress, soybean plant would need to remove excessive NaCl. Ion transportation is the major mechanism for soybean plants to maintain ion homeostasis. Different classes of ion transporters are localized in different subcellular membrane to alleviate salt stress through removing excessive NaCl from the cell, compartmentalization of NaCl into vacuole, and restricting the movement of NaCl from root to shoot (Table S1). Furthermore, it is reported that under salinity, H+-ATPase and H+-PPase located in tonoplast are more active, which provide the proton gradient to drive the active transport of ions from cytoplasm into vacuoles (Yu et al. 2005).
Cation transporters
In the past two decades, the major breakthrough for salt tolerance mechanisms in soybean is the identification of the cation/proton antiporter (CPA)-encoding gene, GmCHX1, which is the major determinant of the salt tolerance level of soybean (Guan et al. 2014; Qi et al. 2014; Qu et al. 2021). A salt tolerance-conferring major quantitative trait locus (QTL) has been mapped repeatedly to chromosome 3 of the soybean genome since 2004 (Ha et al. 2013; Hamwieh et al. 2011; Hamwieh and Xu 2008; Lee et al. 2004; Qi et al. 2014), but it was not until 2014 that the causal gene within this QTL was cloned.
GmCHX1 was first cloned through QTL mapping using a wild-cultivated soybean recombinant inbred population (Qi et al. 2014). It was later further confirmed by map-based cloning using other recombinant populations and genome-wide association studies (GWAS) with different nomenclatures, GmNcl/GmSALT3 (Guan et al. 2014; Patil et al. 2016). To prevent confusions in nomenclature, we will use the gene name reported in the first literature, GmCHX1 in the following sections. The protein sequence of the salt tolerance allele is largely conserved in salt-tolerant soybean varieties, while natural variations of this gene, either in the promoter region or in the coding region, have led to salt sensitivity (Guan et al. 2014; Qi et al. 2014). GmCHX1 is predominantly expressed in the tissue associated with the root vasculature to restrict the loading of Na+ into the shoot, protecting the shoot from salt damage (Guan et al. 2014; Qi et al. 2014; Qu et al. 2021). It is known that GmCHX1 is localized in the ER membrane, but the molecular mechanism of how this protein restricts the loading of Na+ into the shoot is still largely unknown. Interestingly, although GmCHX1 serves primarily as a CPA, transporting Na+ and K+ through the exchange of H+ in the root (Jia et al. 2021; Qu et al. 2021), a functional GmCHX1 is also needed for Cl− exclusion in the shoot in a root-independent manner (Qu et al. 2021).
Besides GmCHX1, the salt tolerance function of another CPA-encoding gene, GmCHX20a, which is located adjacent to GmCHX1 in the soybean genome, was also investigated. Although both GmCHX1 and GmCHX20a are homologs of AtCHX20 in Arabidopsis, the functions of GmCHX1 and GmCHX20a have diverged from those of AtCHX20 (Jia et al. 2021). First of all, the expression of GmCHX1 and GmCHX20a showed a negative correlation in the root upon salt treatment (Jia et al. 2021). That is, the expression of GmCHX1 was suppressed by the initial salt shock and then recovered after prolonged treatment while that of GmCHX20a was highly induced upon salt shock but was slowly reduced after prolonged treatment (Jia et al. 2021). When these genes were overexpressed in tobacco Bright-Yellow 2 (BY-2) cells, the plasma membrane-localized GmCHX20a enhanced the uptake of Na+ while GmCHX1 enhanced its exclusion (Jia et al. 2021). Consistent with this observation, the ectopic expression of GmCHX20a in transgenic Arabidopsis and soybean hairy root led to higher salt sensitivity. On the contrary, the transgenic BY-2 cells expressing GmCHX20a showed higher osmotic stress tolerance (Jia et al. 2021). One possible explanation is that GmCHX20a may function as an osmotic regulator by recruiting Na+ to combat osmotic stress during the early phase of salt stress before GmCHX1 kicks in. A later study demonstrated that the level of histone 3 lysine 9 acetylation (H3K9ac) at the promoter of GmCHX20a was decreased upon mild salt stress priming treatment, implying that switching off GmCHX20a by salt stress priming may contribute to the enhanced salt tolerance in subsequent salt stresses (Yung et al. 2022).
Indeed, another major clade in the CPA family is the Na+/H+ exchanger (NHX). There are 9–10 NHX-encoding genes in the soybean genome (Chen et al. 2015b; Joshi et al. 2021). For example, GmSOS1 is the homolog of the well-characterized AtSOS1/AtNHX7 (Arabidopsis Salt Overly Sensitive 1/Arabidopsis Na+/H+ exchanger 7), which has played crucial roles in salt tolerance in Arabidopsis (Qiu et al. 2002). However, unlike AtSOS1, which has two copies in the Arabidopsis genome, GmSOS1 is a single-copy gene in the soybean genome (Zhang et al. 2022c). The expression of GmSOS1 is salt-responsive and in a dosage-dependent manner in the root (Zhang et al. 2022c). The ectopic overexpression of GmSOS1 can complement the salt overly sensitive phenotype of atsos1 mutant, suggesting that the function of the SOS1 homologs is essentially conserved (Nie et al. 2015). A loss-of-function mutation of GmSOS1 in Williams 82 and Jack backgrounds increased the sensitivity of the plant toward salt stress in terms of the survivability in 160 mM NaCl treatment (Zhang et al. 2022c). It was observed that the gmsos1 mutants had lower net Na+ fluxes but higher K+ fluxes in the root meristem zone. The impaired ion fluxes significantly increased and decreased the Na+/K+ ratio in the root and leaf, respectively, of the mutants (Zhang et al. 2022c). The imbalance in monovalent ions may be the reason for the salt sensitivity. This imbalance of ions also subsequently perturbed the expression of ion transporter-encoding genes such as KEA2, GmCHX20b, NCX1, and CIPK9 (Zhang et al. 2022c).
GmNHX1 is highly induced by NaCl or polyethylene glycol (PEG) treatment. It is localized in the tonoplast membrane and responsible for the compartmentalization of Na+ into vacuoles under salt stress (Li et al. 2006). The ectopic expression of GmNHX1 enhanced the salt tolerance of BY-2 cells (Li et al. 2006), Arabidopsis (Sun et al. 2019a), soybean hairy root (Wang et al. 2011a), and Lotus corniculatus L. (Sun et al. 2006). GmNHX5 was responsive to NaCl treatment in the salt-tolerant cultivar Jidou-7, but not in the salt-sensitive cultivar Mustang (Sun et al. 2021). The overexpression of GmNHX5 in transgenic soybean alleviated the salt stress symptoms and significantly reduced Na+ accumulation while enhancing K+ accumulation in the leaf and root (Sun et al. 2021). Nevertheless, it is still unclear how GmNHX5 alters the accumulation of these ions.
GsCHX19.3 (Jia et al. 2017) and GmNHX6 are two CPA-encoding genes that are responsive to salt-alkaline stress (Jin et al. 2022a). Although GsCHX19.3 was localized to the plasma membrane while GmNHX6 was localized to the Golgi, they both facilitated the uptake of K+ and reduced Na+ accumulation under salt-alkaline stress in transgenic Arabidopsis to maintain a low Na+/K+ ratio (Jia et al. 2017), which may be responsible for the higher salt tolerance of the transgenic Arabidopsis (Jia et al. 2017; Jin et al. 2022a).
The high-affinity K+ transporter (HKT) family is another important class of ion transporters conferring salt tolerance in plants (Singh and Lone 2022). There are six HKT-encoding genes identified in the soybean genome, namely GmHKT1;1-GmHKT1;6 (Chen et al. 2014). Thus far, only the function of GmHKT1;1 (GmHKT1) and GmHKT1;4 have been characterized. The ectopic expression of either GmHKT1;1 or GmHKT1;4 was sufficient to confer salt tolerance in transgenic tobacco plants (Chen et al. 2014, 2011). Like other salt tolerance-conferring ion transporters, both of these HKTs reduced Na+ and improved K+ accumulation in the transgenic plants upon salt treatment when compared to the wild type (Chen et al. 2014, 2011).
Through QTL mapping using a recombinant inbred population of Kefeng No. 1 and Nannong 1138–2, QTLs related to salt tolerance at the germination stage, in terms of imbibition rate, germination index, germination potential and germination rate, were mapped to chromosome 8 in the soybean genome (Zhang et al. 2019a). Seventeen single nucleotide polymorphisms (SNPs) significantly associated with the salt tolerance of 211 soybean accessions at the germination stage were also identified in the same regions (Zhang et al. 2019a). Amongst the polymorphic genes between the Kefeng No. 1 and Nannong 1138–2 within the region, only GmCDF1 (soybean cation diffusion factor 1; Glyma.08g102000) was induced by salt stress and was highly differentially expressed between the two parental lines under salt stress (Zhang et al. 2019a). In general, a higher expression of GmCDF1 is associated with a lower salt tolerance, as illustrated by the expression study on soybean accessions with different salt tolerance, and by transgenic composite soybean plants with roots either overexpressing or underexpressing GmCDF1 (Zhang et al. 2019a). Although the expression of GmCDF1 was positively associated with the accumulation of Na+ in the root and affected the expression of other salt tolerance-related genes, the direct link between GmCDF1 and salt sensitivity is still unknown.
Some ion transporters involved in salt tolerance may not be directly involved in NaCl transportation. One example is the calcium/proton exchangers (CAXs). A stress-responsive soybean transcript, GmCAX1, was found to be differentially expressed upon PEG, CaCl2, NaCl, ABA and LiCl treatments. Overexpression of GmCAX1 could alleviate the salt stress symptoms of transgenic Arabidopsis (Luo et al. 2005), probably through increasing the accumulation of Ca2+ and reducing the accumulation of monovalent ions (Na+, K+, and Li+, if present) (Luo et al. 2005). Whether GmCAX1 alters the accumulation of monovalent ions through direct transportation or regulates ion transportation indirectly through calcium signaling is still unknown.
Auto-inhibited Ca2+-ATPases (Rodrigues et al.) regulate the cytosolic Ca2+ concentration that contributes to salt tolerance. A NaHCO3- and NaCl-responsive gene, GsACA1, was cloned from wild soybean (Glycine soja) (Sun et al. 2016). Transgenic alfalfa overexpressing GsACA1 showed better salt tolerance and higher chlorophyll content than the wild type plants upon salt treatment. Moreover, compared to the wild type, transgenic plants showed lower membrane permeability and a lower malondialdehyde (MDA) content but a higher content of proline, which is both an osmolyte and ROS scavenger (Sun et al. 2016). GsACA1 probably regulates salt tolerance through calcium signaling, but that requires further investigation.
Anion transporters
The chloride channel protein (CLCs) family is one of the crucial classes of chloride transporters involved in salt tolerance in plants (Wu and Li 2019). There are eight CLC-encoding genes in the soybean genome (Wei et al. 2019), while only two of them have been demonstrated to be involved in soybean salt tolerance. It was found that GmCLC1, CLC-b1, CLC-b2, CLC-c1, and CLC-c2 were significantly induced by salt treatment in both the salt-sensitive cultivar N23674 and the salt-tolerant wild soybean BB52 (Wei et al. 2019). As the only member containing non-synonymous SNP between N23674 and BB52, GsCLC-c2 from the wild soybean BB52 was able to confer salt tolerance in composite soybean plants with hairy roots overexpressing GsCLC-c2 (Wei et al. 2019). The tonoplast-localized GsCLC-c2 could compartmentalize NaCl in the root and promote the accumulation of NO3− and K+ in the shoot to counteract the damages caused by salt treatment (Wei et al. 2019). An electrophysiology experiment using oocytes injected with GsCLC-c2 cRNA demonstrated that GsCLC-c2 could function as a pH-independent Cl− channel with a higher affinity towards Cl− and NO3− (Wei et al. 2019).
GmCLC1 is another CLC-encoding gene. Although its expression in soybean was highly induced by both NaCl and PEG, its ectopic expression in BY-2 cells could only confer tolerance to salt but not PEG (Li et al. 2006). Lucigenin staining of isolated vacuoles demonstrated that GmCLC1 could compartmentalize Cl− into the vacuoles upon NaCl treatment (Li et al. 2006). An electrophysiology study as well as sequence homology implied that GmCLC1 is a Cl−/H+ antiporter (Wong et al. 2013). The ectopic expression of GmCLC1 could also confer salt tolerance to transgenic Arabidopsis, transgenic soybean hairy roots and transgenic hairy root composite plants (Wei et al. 2016). An in planta experiment suggested that GmCLC1 was able to compartmentalized Cl− in the root to prevent the accumulation of Cl− in the shoot (Wei et al. 2016).
Other transporters
Besides ion transporters, other kinds of transporters were shown to be involved in salt stress responses. Plasma membrane intrinsic proteins (PIPs) govern the water transport across the plasma membrane (Chaumont and Tyerman 2014). The expression of GmPIP1;6 was initially suppressed within the first few hours of salt treatment and then the gene was induced in the following days (Zhou et al. 2014), probably due to its differential functions during the osmotic stress and ionic stress phases. Transgenic soybean plants overexpressing GmPIP1;6 showed better salt tolerance than the wild type in terms of growth, photosynthetic performance, and higher root hydraulic conductance under salt treatment conditions (Zhou et al. 2014). Furthermore, the transgenic plants also bore lower Na+ contents than the wild type (Zhou et al. 2014). Although it is still unsure how GmPIP1;6 confers salt tolerance, the major hypothesis suggests that GmPIP1;6 serves as a filter for the soil water, by allowing water to enter the cell while blocking the uptake of NaCl to maintain the water status of the plant.
Apart from managing material transfer or serving as components of signal transduction pathways, transporters are also essential for maintaining the energy status of the plant under salt stress. Active ion transportation is an energy-intensive process. A collapse of energy production mechanism such as photosynthesis upon salt stress is usually used as an indicator of salt sensitivity. ndhB and ndhH are genes encoding the subunits of NAD(P)H dehydrogenase (NDH), which drives the cyclic electron flow for ATP production (He et al. 2015). The expression of both ndhB and ndhH were induced by salt treatment at both the transcript and the protein levels. Furthermore, the expressions of both genes were higher in both the salt-tolerant soybean S111-9 and the salt-sensitive Melrose in both normal and salt-treated conditions. Interestingly, upon salt treatment, Na+ was mainly compartmentalized in the vacuole of S111-9 and the chloroplast of Melrose, which could be the cause of the difference in salt sensitivity (He et al. 2015). Although direct evidence is lacking, with these observations, it is proposed that the active NDH-dependent cyclic electron flow might be important for providing ATP for active Na+ sequestration into the vacuole to enhance the salt tolerance of the plant (He et al. 2015).
Osmoregulation
Upon the early stage of salt stress, due to the sudden presence of high solute concentration in the soil, the water potential around the root is lowered, and water is thus unavailable to the plant. The water loss through transpiration supersedes the water uptake through the root, leading to water deficit. It is reflected in drooping leaves within the first hour of salt treatment due to the loss of turgor pressure in the cells. In general, the soybean plant is able to cope with this osmotic stress within a few hours through different mechanisms.
Accumulation of osmoprotectants in the cell is one way to balance the osmolarity between the inside and outside of the cell. As mentioned in the previous section, pumping Na+ into the cells as a readily available osmolyte through active transportation could be a quick way to reduce the cellular water potential, though this strategy will likely result in subsequent ion toxicity (Jia et al. 2021). In addition to the ionic salt, the plant also produces and accumulates compactible metabolites such as onium compounds, polyols/sugars, amino acids, and alkaloids (Phang et al. 2008). Since this mechanism has been intensively discussed (Hasegawa et al. 2000; Phang et al. 2008), it will not be covered in this review.
Osmoregulation by late embryogenesis abundant proteins
Late embryogenesis abundant (LEA) proteins have been investigated broadly with respect to salt tolerance. The expressions of LEA-encoding genes were highly induced under different stress conditions such as drought, heat, cold and salt stresses (Bhardwaj et al. 2013; Hasegawa et al. 2000). The LEA proteins might function by (i) participating as antioxidant to safeguard against ROS (Sobhanian et al.); (ii) stabilizing the plasma membrane and proteins; (iii) filling the space and maintain the shape of cells upon dehydration (Bhardwaj et al. 2013; Tunnacliffe and Wise 2007). Given that LEA proteins are in general highly hydrophilic, the accumulation of LEA proteins in cells upon desiccation is proposed to be an important mechanism for osmoregulation. LEA proteins can be divided into four groups according to their structures, with each group serving a different function (Bhardwaj et al. 2013; Tunnacliffe and Wise 2007; Wise and Tunnacliffe 2004).
The functions of a few soybean LEA proteins, such as PM11 and Em belonging to group I LEAs, have been studied (Lan et al. 2005; Cai et al. 2006). Transgenic Escherichia coli expressing PM11 underwent a short lag phase when growing in a medium containing 800 mM NaCl when compared to the vector-only control (Lan et al. 2005). Apart from improving the salt tolerance of transgenic E. coli, Em could also confer salt tolerance in transgenic tobacco plants (Cai et al. 2006). Besides, group II LEA proteins such as GmLEA2-1 can also enhance salt tolerance. Overexpressing GmLEA2-1 in Arabidopsis showed improved tolerance under mannitol and NaCl treatment, in which the expression might be regulated by DREB-, DBF-, CBF-, MYC- or MYB-like transcription factors (Wang et al. 2018b).
The functions of PM30 and PM2, belonging to group III LEA proteins, have also been studied using transgenic E. coli. Similar to PM11, PM30 was able to confer tolerance to transgenic E. coli treated with 800 mM NaCl (Lan et al. 2005). Transgenic E. coli expressing PM2 did not gain any advantage over the control under 1100 mM sorbitol treatment, but it showed improvement in salt tolerance when treated with 500 mM NaCl and 500 mM KCl (Liu and Zheng 2005). Furthermore, the expression of the 22-mer repeating region of PM2 in E. coli demonstrated a similar protective effect as the full-length PM2, suggesting that the 22-mer repeating region is responsible for the tolerance (Liu and Zheng 2005).
Regulation of oxidative balance
Reactive oxygen species (ROS) are by-products of redox reactions in aerobic respiration and photosynthesis (Dixon et al. 2010). Integral to plant metabolic pathways, ROS exhibit dual effects depending on their cellular concentrations (Das and Roychoudhury 2014). At low levels, they act as signaling molecules or substrates for metabolism (Czarnocka and Karpinski 2018), whereas at high concentrations, they cause oxidative damage to nucleic acids, proteins, lipids and pigments (Gill and Tuteja 2010). Salinity induces secondary oxidative stress by perturbing ROS homeostasis, by increasing ROS production while slowing their removal (Chawla et al. 2013). The overaccumulation of ROS and their downstream products cause damage to biomolecules and cell tissue, and eventually lead to cell death (Czarnocka and Karpinski 2018; Gill and Tuteja 2010). Therefore, key adaptations in salinity tolerance usually entail enhanced ROS scavenging to restore oxidative balance at the transcription, translation and post-translational modification levels (Lv et al. 2014; Matsuura et al. 2010; Schmidt et al. 2013; Yu et al. 2010). It is thus critical for crop improvement research to investigate the genes responsible for ROS scavenging during salt stress.
To restore oxidative balance under salt stress, soybean employs both antioxidant enzymes and secondary metabolites (Das and Roychoudhury 2014), of which synthesis is activated by high concentrations of ROS and malondialdehyde (MDA), a toxic electrophile produced from the peroxidation of polyunsaturated fatty acid (Dixon et al. 2010). Enzymatic antioxidants catalyze direct ROS scavenging or reactions that replenish ROS decomposition substrates (Das and Roychoudhury 2014), while non-enzymatic antioxidants serve as scavenging substrates or direct scavengers (Dogan 2011). These two groups complement each other in ROS elimination. Thus far, a number of genes underpinning the production of antioxidants and their ROS scavenging pathways in soybean have been discovered and functionally verified (Table S2).
In general, superoxide dismutase (SOD) catalyzes the conversion of superoxide radicals into less reactive hydrogen peroxide (H2O2) and oxygen molecules. Hydrogen peroxide is normally detoxified by catalase (CAT) (Rodrigues et al.), peroxidase (POD) or ascorbate peroxidase (APX) coupled with the ascorbate–glutathione cycle (Das and Roychoudhury 2014). A class III peroxidase-encoding gene, GsPRX9, was cloned from a salt-tolerant wild soybean (Jin et al. 2019). The expression of GsPRX9 was significantly induced by salt stress and the induction was more prominent in salt-tolerant wild soybeans, LY01-10 and LY16-08, than in the salt-sensitive cultivated soybean, Tianlong1, and the salt-sensitive wild soybean, LY01-06 (Jin et al. 2019). Composite soybean plants with roots overexpressing GsPRX9 apparently grew better than those transformed with empty vector, with higher SOD and POD activities and glutathione concentration (Jin et al. 2019). GsPRX9 overexpression also led to the upregulation of seven genes involved in the phenylpropanoid pathway (Jin et al. 2019), which is the major pathway producing secondary metabolites with antioxidant activities.
Glutathione S-transferases (GSTs) are another class of potent antioxidants that catalyze the conjugation of the nucleophilic glutathione (GSH) to lipid hydroperoxides, hence preventing the downstream generation of the toxic MDA (Sharma et al. 2004). Under acute salt stress at 200 mM NaCl, GsGST-overexpression in tobacco led to a sixfold increase in GST activities and a significantly higher survivorship and root elongation compared to the wild type (Ji et al. 2010). Meanwhile, under salt treatment, GmGSTL1-overexpression in tobacco BY-2 cells and Arabidopsis reduced ROS accumulation and leaf chlorosis (Chan and Lam 2014), which is a typical symptom of ROS toxicity (Lim et al. 2007). Chan and Lam (2014) elucidated the anti-oxidation mechanism of soybean GST through its conjugation with the secondary metabolites, quercetin and kaempferol, and demonstrated the interactions between quercetin and GST via exogenous applications.
ROS detoxification also entails MDA scavenging by aldehyde dehydrogenases (ALDHs), which catalyze the conversion of aldehydes into carboxylic acids (Singh et al. 2013). Under salinity stress, the ectopic expression in Arabidopsis and tobacco of Glycine max turgor-responsive 55 kDa protein (GmTP55), which encodes an antiquitin-like ALDH, resulted in higher germination efficiency and seedling development than the wild-type plants (Rodrigues et al. 2006). Notably, transgenic Arabidopsis under 100 mM NaCl retained the same germination rate as in unstressed condition (Rodrigues et al. 2006). The elevated salt tolerance was achieved by a higher anti-oxidation efficiency as evidenced by GmTP55-overexpressing transgenic plants exhibiting lower MDA levels and less severe oxidative stress symptoms under peroxide and paraquat treatments than wild-type plants (Rodrigues et al. 2006).
Alternative salt stress response pathways involving purple acid phosphatase 3 have also been uncovered (Liao et al. 2003). GmPAP3 is localized in mitochondria, a primary site of ROS generation (Li et al. 2008). Under salt and oxidative stress treatments, the overexpression of GmPAP3 in tobacco BY-2 cells resulted in reduced ROS levels and increased ascorbic acid-like antioxidation pathway activities, while GmPAP3-overexpressing Arabidopsis experienced reduced lipid peroxidation (Li et al. 2008). The ectopic expression of GmPAP3 in rice also yielded increased SOD, POD and CAT activities, a higher proline content, and a reduced MDA content under salt treatment compared to wild type (Deng et al. 2014). Since the ROS-scavenging effects from GmPAP3 overexpression were diminished when an ion chelator was present, it is possible that the redox reactions of GmPAP3 play a role in ROS scavenging (Li et al. 2008). However, whether GmPAP3 acts on ROS directly or through interfering Fenton or Heiber-Weiss reactions is still inconclusive and awaits further investigations.
Increasing the generation of secondary metabolites involved in ROS catabolism can also contribute to the restoration of oxidative balance in soybean. L-ascorbic acid (AA), a prominent antioxidant in photosynthetic eukaryotes, reacts with peroxides to form the nontoxic docosahexaenoic acid (Wheeler et al. 2015). AA synthesis, catalyzed by GDP-D-mannose pyrophosphorylase (GMP), hence acts as a universal defence against oxidative stress (Wheeler et al. 2015). Indeed, under salt stress, overexpressing GmGMP1 in transgenic Arabidopsis conferred higher salt tolerance via significantly elevated AA levels and reduced levels of superoxide radicals and lipid peroxidation (Xue et al. 2018). Other important non-enzymatic antioxidants in soybean include flavonoids and proline. Flavonoids, a class of polyphenolic compounds, can directly scavenge peroxides, singlet oxygen and hydroxyl radicals (Brunetti et al. 2013). Similarly, proline, which also acts as an osmolyte, scavenges singlet oxygen and hydroxyl radicals, and prevents damages from lipid peroxidation (Dogan 2011). In transgenic Arabidopsis, GmMYB12 overexpression improved salt stress tolerance during seed germination, root development, and in the vegetative stage by increasing flavonoid and proline productions and upregulating genes involved in their biosynthesis, alongside increased SOD and POD activities (Wang et al. 2019b).
Some gene products may not act directly on ROS but regulate the downstream expressions of genes that mitigate the salt-induced oxidative stress. Xiong et al. (2022) examined the interactions between ROS, salt tolerance and protein phosphatase 2A (PP2A), an enzyme family that is known to modulate oxidative stress in plants. Specifically, PP2A-B”71 was shown to mediate the stress-induced abscisic acid (ABA) signaling (Yang et al. 2020a), and its expression was responsive to salt stress (Xiong et al. 2022). Under salt treatment, GmPP2A-B”71-overexpressing soybean hairy roots had increased levels of chlorophyll, proline, CAT and POD, and lower MDA content, while GmPP2A-B”71-RNA-interference plants exhibited the opposite phenotype (Xiong et al. 2022). Crucially, GmPP2A-B”71 overexpression markedly upregulated genes responsible for the synthesis of CATs (GmCAT1 and GmCAT2) and POD (GmPOD1), and two genes with putative roles in stress-responsive antioxidation (GmLEA15 and GmERF115) (Xiong et al. 2022). Similarly, CONSTANS-LIKE 1a (GmCOL1a), a flowering repressor gene in soybean, was found to participate in oxidative stress alleviation by promoting the accumulation of stress-responsive proteins under salinity treatment (Xu et al. 2022). GmCOL1a proteins, of which expressions are highly induced by NaCl, are localized in the nucleus and promote the transcriptional activation of the stress-tolerance genes, GmLEA and GmP5CS, by binding to their promoters (Xu et al. 2022). Compared to the knockout mutants and wild type, GmCOL1a-overexpressing and GmP5CS-overexpressing soybean hairy roots had much more effective antioxidation under salt treatment via reduced ROS levels, and significantly elevated enzymatic antioxidant activities and proline concentrations (Xu et al. 2022). Both studies attest to the importance of characterizing the regulation of downstream salt-tolerance genes by the target genes under salt stress.
Besides directly participating in the restoration of oxidative balance, signal transduction (which will be discussed in detail in a later section) is crucial for plants to detect salt stress and acclimatize accordingly. Genes encoding substrate or receptor proteins involved in signal transduction can mediate salinity stress responses including ROS scavenging. The ectopic expression experiments of such soybean genes have implied their potential roles in salt-induced oxidative stress responses. The phytohormone, abscisic acid (ABA), is an important messenger in stress signaling pathways in plants. In both maize and wheat, exogenous ABA applications at low doses increased the activities of SOD, CAT, APX and glutathione reductase (Agarwal et al. 2005; Jiang and Zhang 2001). By increasing ABA sensitivity, a lower threshold is required to activate stress responses, thereby allowing plants to mitigate salinity stress more quickly and preventing the overaccumulation of intracellular ABA, which can be toxic at high concentrations (Agarwal et al. 2005; Jiang and Zhang 2001). GmSAP16, which encodes stress-associated proteins (SAPs) of the zinc-finger protein family, increased ABA sensitivity and proline level, and reduced MDA level when overexpressed in Arabidopsis and soybean hairy roots (Zhang et al. 2019b). Similarly, in Arabidopsis under salt treatment, the ectopic expression of GmST1, of which function has yet to be characterized, increased ABA sensitivity and reduced peroxide level (Ren et al. 2016). The expressions of lectin receptor protein kinases (LecRLK), another important group of receptors to external stress signals in plants, were induced in wild soybean roots under salt treatment (Zhang et al. 2022d). Under salt stress, GmLecRlk-overexpressing soybean hairy roots had increased proline content and CAT activities, and reduced peroxide and MDA levels compared to the wild-type control (Zhang et al. 2022d). The exact functional mechanism of GmLecRlk in soybean oxidative stress reduction is unknown, but the expression patterns of its homologs in Arabidopsis, rice and other legumes under salinity stress suggested its involvement in mediating the ABA signaling pathway (Joshi et al. 2010; Li et al. 2014; Sun et al. 2013b). The exact physiological mechanisms of these genes in soybean oxidative stress reduction are not as well characterized as their roles in improving ion homeostasis or signal transduction. As a result, their roles in restoring oxidative balance are only implicated. The improved ROS scavenging due to the expressions of these genes is likely due to their downstream regulation of the antioxidation regulatory network, which awaits verification by future research.
Besides salinity, secondary oxidative stress and the subsequent antioxidation metabolism in plants are also inducible by other biotic and abiotic stresses (Cheng et al. 2015; Li et al. 2021a; Mira et al. 2021). In particular, multiple soybean genes responsible for oxidative balance restoration have been functionally verified with drought stress, an agricultural challenge that often occurs concurrently with salinity (Li et al. 2018a, 2013; Wang et al. 2019b). Furthermore, genome-wide identification has been applied to characterize the soybean gene families associated with the enzymatic antioxidants, SOD and glutathione peroxidase (Aleem et al. 2022), while proteomic and metabolomic analyses have uncovered indirect pathways that can improve soybean antioxidation (Pi et al. 2016). Future studies could verify the functions of these genes with salt treatment, thereby expanding the genetic avenues available for improving oxidation–reduction responses in soybean under salinity stress.
Cell wall remodeling
In plants, cell wall (CW) provides structural support by maintaining cell stiffness (Houston et al. 2016), and the apoplast serves as an important site for reactions and signal transduction (Farvardin et al. 2020). The physical and biochemical properties of CW render it indispensable to plant survival under abiotic stress (Farvardin et al. 2020; Le Gall et al. 2015; Tenhaken 2015). Salinity impairs CW functioning and arrests plant growth by inducing osmotic imbalance that reduces cell turgidity (Liu et al. 2021a), and by causing secondary oxidative stress, where excessive ROS results in cell wall loosening (Gigli-Bisceglia et al. 2020). There is increasing evidence suggesting CW remodeling to be a critical adaptation to salt stress (Liu et al. 2021a). To survive under high salinity, plants constantly modulate CW structure and composition to optimize stress signal detection, CW integrity repair, and subsequent loosening to allow for growth under sustained stress (Houston et al. 2016; Le Gall et al. 2015; Liu et al. 2021a; Tenhaken 2015). Exploring the genes underlying the adaptive remodeling of soybean CW under salt stress offers insights into improving crop tolerance against salinity.
CW is a dynamic and complex matrix consisting of polysaccharides, proteins, and other compounds that help maintain or modulate CW functions. The cellulose microfibril scaffold is interconnected by hemicellulose and pectin (Lampugnani et al. 2018; Loix et al. 2017; Somerville et al. 2004). The depolymerization of these polysaccharides via enzymatic or ROS cleavage, their strengthening by increased cross-linkages, or changes in their biosynthetic rates, all combine to modulate CW plasticity and tensile strength as plants respond to abiotic stress (Houston et al. 2016; Le Gall et al. 2015; Liu et al. 2021a; Tenhaken 2015). Cell wall proteins, which can either be embedded in the CW or in soluble forms in the apoplast, include enzymes that catalyze CW component biosynthesis and modifications, as well as those involved in stress signal reception and transduction (Jamet et al. 2006; Tenhaken 2015). Mediating the abundance, composition and distribution of aforementioned CW components are integral to plant survival and development under abiotic stress.
The immediate challenge faced by plants under high salinity is osmotic stress, which causes the loss of cell turgor (Liu et al. 2021a). Furthermore, the ROS generated as the secondary stress response cleave polysaccharide polymer linkages, hence lowering CW tensile strength and aggravating the water loss-induced flaccidity (Tenhaken 2015). Therefore, the first line of CW defense involves increasing mechanical strength by polymer biosynthesis and deposition. In soybean, a co-chaperone DNAJ protein, GmDNAJC7, was found to upregulate and co-express with genes involved in cellulose biosynthesis, where under salt treatment, GmDNAJC7-overexpressing Arabidopsis had a higher germination rate, a higher cotyledon greening rate and greater root length compared to wild type (WT) (Jin et al. 2022b). The production of lignin, a phenolic polysaccharide of which deposition at secondary CW confers tensile strength, is often increased to stiffen CW amidst stresses (Cesarino 2019). GmRD22 encodes a BURP-domain protein localized in the apoplast that interacts with a cell wall peroxidase to increase CW lignification (Wang et al. 2012). Under salt treatment, its overexpression in tobacco BY-2 cells, Arabidopsis and rice resulted in higher survivorship while reducing the negative effects of NaCl on Arabidopsis root elongation (Wang et al. 2012). In both Arabidopsis and rice transgenic systems, GmRD22 overexpression markedly elevated lignin contents, evincing its protective properties in soybean under salt stress (Wang et al. 2012).
Besides CW strengthening, the hydrophobic cuticle is also an important barrier against dehydration, as its waterproofing property could reduce transpirational water loss during salinity stress (Ziv et al. 2018). EARLI (early Arabidopsis aluminium-induced gene1) is a CW-localized protein that contains lipid transfer protein (LTP) motifs (Bubier and Schlappi 2004), which are involved in cutin biosynthesis and membrane formation (Kader 1996). In soybean, GsEARLI17 encodes a hybrid proline-rich protein (HyPRP) (Liu et al. 2015), which is involved in CW reinforcement (Jose-Estanyol and Puigdomenech 2000). Under salt treatment and compared to WT, its overexpression in Arabidopsis led to higher germination rates and healthier cotyledons, while GsEARLI17-overexpressing seedlings had higher rates of leaf opening and greening (Liu et al. 2015). Remarkably, the transgenic line had cuticles up to 167% thicker than those of WT (Liu et al. 2015). This corroborates the role of proline-rich proteins in altering soybean CW structure under salt stress, as proposed by He et al. (2002), where the expression of soybean SbPRP3, encoding a HyPRP with unknown function, was found to be inducible by salinity stress (He et al. 2002).
The growth and functionality of plant cells are constrained by the CW architecture (Lampugnani et al. 2018; Somerville et al. 2004), which is partly underpinned by the cortical microtubule array that directs the arrangement of cellulose and other CW components (Oda 2015). Coumarin, a phytotoxin typically produced by plants to combat pathogens and herbivory (Gnonlonfin et al. 2012; Prats et al. 2007; Sun et al. 2014), has been found to mediate plant growth and development (Lupini et al. 2014). Specifically, it exhibits a dose-dependent effect on Arabidopsis root morphology by indirectly altering the cortical microtubule organization, and hence, the architecture of the root apical meristem (Bruno et al. 2021; Lupini et al. 2014). In soybean, the expression of GmF6′H1, which encodes the enzyme for coumarin biosynthesis, was highly induced by salt stress (Duan et al. 2019). The GmF6′H1-overexpressing transgenic Arabidopsis had higher salt tolerance, along with a higher germination rate and chlorophyll content, produced more siliques and had less growth impairment under NaCl than the WT plants (Duan et al. 2019). Coumarin might play a role in altering the soybean cortical microtubule array, thereby causing adaptive changes in the CW structure to confer salt tolerance. However, since the role of coumarin in soybean CW modification is only implied, future verification is required to characterize the actual changes in CW composition and organization following increased coumarin biosynthesis under salt stress.
Achieving efficient signal transduction to enable rapid acclimation is as crucial as the direct modulation of the CW structure. For instance, the Catharanthus roseus receptor-like kinase (CrRLK1L) subfamily is crucial in regulating cell expansion via spatially and temporally controlled downstream signal transduction when plants undergo development or respond to stresses (Nissen et al. 2016). In soybean, GmCrRLK1L20 encodes a cell membrane-localized FERONIA receptor kinase that mediates Ca2+ signaling (Feng et al. 2018; Wang et al. 2021b). Under salt treatment, soybean hairy roots overexpressing GmCrRLK1L20 had delayed leaf wilt, longer roots, and higher chlorophyll content compared to wild type (Wang et al. 2021b). Additionally, elevated contents of enzymatic and secondary metabolite antioxidants were coupled with a lower MDA level and less cell membrane damage, as indicated by lower membrane permeability in the transgenic line (Wang et al. 2021b). Importantly, CrRLK1L-mediated Ca2+ signaling is activated by changes in pectin structure and polymerization (Lin et al. 2022; Nissen et al. 2016), which are a common feature of both adaptive and maladaptive salt responses in plants (Feng et al. 2018), evincing the critical role of apoplastic modifications in salt stress responses.
After overcoming the salinity-induced osmotic stress, secondary responses at later stages typically involve CW loosening and cell elongation for plants to continue to grow under prolonged salt stress (Voxeur and Hofte 2016). The brassinosteroid (BR) signaling pathway is involved in cellulose deposition (Planas-Riverola et al. 2019), and is activated as a protective response against the stress-induced loss of CW integrity due to imbalanced pectin modification (Wolf et al. 2012). Meanwhile, BIN2 (brassinosteroid-insensitive 2), a negative regulator of BR signaling (Li et al. 2020c), acts as a molecular switch from immediate salt stress responses to growth recovery (Li et al. 2020c). In soybean, GmBIN2 encodes glycogen synthase kinase 3 (GSK3), and its expression is induced by exposure to NaCl (Wang et al. 2018a). Under salt treatment, GmBIN2-overexpressing Arabidopsis had a higher germination rate and Ca2+ content, longer root length, and a reduced Na+ content than WT, while GmBIN2 overexpression in soybean hairy roots resulted in lower cell membrane permeability (Wang et al. 2018a). Given that BIN2 inhibits the cellulose biosynthesis required for early salt responses, future studies could examine the temporal control mechanism of GmBIN expressions and verify the downstream gene regulation cascade in soybean CW remodeling.
Past researches on soybean CW remodeling have mainly focused on growth and development (Hong et al. 1987; Ye and Varner 1991), and responses to pests and pathogens (Borkowska et al. 1998; Liu et al. 2017). Proteomic studies have uncovered enzymes that are key to adaptive CW remodeling and responsive to salinity (Rehman et al. 2022; Sobhanian et al. 2010), but the underlying genes have yet to be determined. Moreover, CW-modifying protein gene families, including those involved in CW loosening, cellulose biosynthesis and pectin modification, have been identified and characterized in genome-wide analyses (Feng et al. 2022; Nawaz et al. 2017; Wang et al. 2021b). Given that CW remodelling is closely tied to salt-induced growth regulation (Julkowska and Testerink 2015), future study could first finely characterize the relative growth rate of different soybean organs and the salt-induced sequential changes in cell wall structure, then investigate the underlying genetic components using functional verification.
Transcription regulation
Plants, when subjected to salt stress, undergo extensive transcriptome reprogramming to make physiological and metabolic adjustments to survive the damage caused by the salt (Liu et al. 2019). In the past 20 years, a number of transcriptomic studies have been conducted to identify the differentially expressed genes (DEGs) in soybean under salt stress (Table S3). The global transcriptomic view allows researchers to determine transcriptionally the most affected cellular processes or pathways during salt stress.
Transcription factors (TFs) play a key role in salt stress-induced transcriptome reprogramming by activating or repressing their target genes. A number of TFs belonging to various families, including bHLH, bZIP, AP2/ERF, MYB, NAC, and WRKY, have been reported to participate in plant tolerance against abiotic stress such as drought and salinity (Golldack et al. 2011; Khan et al. 2018). In soybean, in the salt-induced DEGs belonging to these same families of transcription factors are also identified in response to salt stress. For instance, 862 TFs clustering mostly in the WRKY, NAC, AP2-EREBP, ZIM, and C2C2 (Zn) CO-like families were identified among the DEGs in salt-treated soybean (Belamkar et al. 2014). For the remaining 1,235 DEGs under salt stress, 117 were identified as TFs and 17 of them were putative members of the MYB family of TFs (Liu et al. 2021b). A single TF can regulate a series of downstream stress-responsive genes and induce comprehensive phenotypic adjustments for salt tolerance (Khan et al. 2018). Therefore, manipulating the expressions of a few salt stress-related TFs could lead to significant improvements in salt tolerance. Summarized in Table 1 are the functional analyses using transgenic plants to characterize the roles of selected TFs that are differentially expressed under salt stress.
Table 1.
A list of functionally verified soybean transcription factors involved in salt tolerance
| Gene | Phenotype | Functions | References |
|---|---|---|---|
| bHLH transcription factor | |||
| GmbHLH3 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | GmbHLH3 regulates the expression of GmCLC1, which in turn retains NaCl in the root to prevent salt damage in the shoot | (Liu et al. 2022) |
| bZIP transcription factors | |||
| GmbZIP1 | Overexpression in Arabidopsis, wheat and tobacco enhances salt tolerance | GmbZIP1 regulates the expressions of ABA- and stress-related genes to mediate the ABA response | (Gao et al. 2011) |
| GmbZIP2 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | Ectopic expression of GmbZIP2 can enhance the expressions of stress-responsive genes under normal conditions, salt stress, and mannitol treatment | (Yang et al. 2020b) |
| GmbZIP15 | Overexpression in Arabidopsis and soybean results in increased salt sensitivity | GmbZIP15 positively regulates the transcription of GmSAHH1 and negatively regulates GmWRKY12 and GmABF1 under abiotic stress | (Zhang et al. 2020a) |
| GmbZIP19 | Overexpression in Arabidopsis and soybean results in increased salt sensitivity | GmbZIP19 upregulates ABA-, JA-, ET-, and SA-inducible marker genes by interacting with their promoters but downregulates salt- and drought-responsive genes | (He et al. 2020a) |
| GmbZIP44, GmbZIP62, GmbZIP78 | Overexpression in Arabidopsis enhances salt tolerance | GmbZIP44, GmbZIP62, and GmbZIP78 mediate ABA signaling by upregulating ABI1 and ABI2 which regulate downstream stress-responsive genes | (Liao et al. 2008c) |
| GmbZIP110 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | GmbZIP110 alters the expressions of stress-related genes in the ABA-signaling pathway and those related to ion transportation | (Xu et al. 2016) |
| GmbZIP132 | Overexpression in Arabidopsis enhances salt tolerance at germination stage | GmbZIP132 promotes the transcription of stress-responsive genes including RD29B, DREB2A, and P5CS | (Liao et al. 2008a) |
| GmbZIP152 | Overexpression in Arabidopsis enhances salt tolerance | GmbZIP152 activates antioxidant enzymes and the transcription of biotic and abiotic stress-related marker genes when stressed. GmbZIP152 can bind directly to the promoters of ABA-, JA-, ET-, and SA-inducible genes | (Chai et al. 2022) |
| GmFDL19 | Overexpression in soybean enhances salt tolerance | GmFDL19 reduces Na+ accumulation and enhances expression of ABA- and stress-responsive genes | (Li et al. 2017b) |
| GmTGA13 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | GmTGA13 mediates ion homeostasis by facilitating K+ and Ca2+ absorption | (Ke et al. 2022) |
| GmTGA17 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
GmTGA17 upregulates ABA-responsive genes and regulates ABA signaling under salt and drought stress | (Li et al. 2019a) |
| Dehydration-responsive element-binding (DREB) proteins | |||
| GmDREB1 | Overexpression in alfalfa enhances salt tolerance | Transgenic alfalfa exhibits a higher level of P5CS transcripts, which is consistent with a higher level of free proline | (Jin et al. 2010) |
| GmDREB2 | Overexpression in Arabidopsis enhances salt tolerance | GmDREB2 can induce the expressions of downstream genes RD29A and COR15a | (Chen et al. 2007) |
| GmDREB3 | Overexpression in Arabidopsis and tobacco enhances salt tolerance | GmDREB3 can bind to DREs and activate downstream genes | (Chen et al. 2009) |
| GmDREB6 | Overexpression in soybean enhances salt tolerance | GmDREB6 can enhance the expression of GmP5CS and increase the proline content when under salt stress | (Nguyen et al. 2019) |
| Ethylene-responsive factors (ERFs) | |||
| GmERF3 | Overexpression in tobacco enhances salt tolerance | GmERF3 can interact with the GCC box and DRE/CRT and activate the associated genes | (Zhang et al. 2009) |
| GmERF4 | Overexpression in tobacco enhances salt tolerance | GmERF4 possesses GCC-box- and DRE/CRT-binding specificities and inhibits the expressions of GCC-box-containing genes | (Zhang et al. 2010) |
| GmERF7 | Overexpression in tobacco enhances salt tolerance | GmERF7 functions as a transcription activator through binding to the GCC box of its target genes | (Zhai et al. 2013) |
| GmERF057, GmERF089 | Overexpression in tobacco enhances salt tolerance | GmERF057 and GmERF089 are induced by salt, drought, ET, SA, JA, and ABA, suggesting that they may be involved in various stress signaling pathways | (Zhang et al. 2008) |
| GmERF75 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | GmERF75 is induced in SA, JA, and ET treatments, suggesting its role in integrating the signals from SA and ET/JA pathways | (Zhao et al. 2019b) |
| GmERF135 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | GmERF135 regulates ET and ABA signaling by activating ET-producing genes, AtACO4, AtACS2, and ABA biosynthesis genes, ABA1, ABA2 | (Zhao et al. 2019c) |
| GRAS transcription factor | |||
| GmGRAS37 |
Overexpression in soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
GmGRAS37-overexpressing plants show upregulated GmDREB1, GmNCED3, GmCLC1, GmSOS1, and GmSODs under salt stress, so it can regulate ABA biosynthesis, ion transportation and ROS scavenging | (Wang et al. 2020) |
| Early Flowering 3 | |||
| GmJ |
Overexpression in soybean hairy roots enhances salt tolerance NIL-j soybean hairy roots show increased salt sensitivity |
J is involved in the regulation of downstream salt-responsive genes, including GmWRKY12, GmWRKY27, GmWRKY54, GmNAC11, and GmSIN1 | (Cheng et al. 2020) |
| MYB transcription factors | |||
| GmMYB12 | Overexpression in Arabidopsis enhances salt tolerance | GmMYB12 activates gene expressions and enzymatic activities of P5CS, SOD, and POD upon salt stress, and therefore helps maintain the proper levels of proline and ROS | (Wang et al. 2019b) |
| GmMYB12B2 | Overexpression in Arabidopsis enhances salt tolerance | GmMYB12B2 can increase the expressions of salt stress-responsive genes including DREB2A and RD17 | (Li et al. 2016b) |
| GsMYB15 | Overexpression in Arabidopsis enhances salt tolerance | GsMYB15 reduces the transcript levels of ABA marker genes (ABIs) and activates several TFs, conferring salt tolerance | (Shen et al. 2018a) |
| GmMYB46 | Overexpression in Arabidopsis enhances salt tolerance | GmMYB46 induces the expressions of salt-responsive genes, P5CS1, SOD, POD, and NCED3 to turn on salt tolerance mechanisms | (Liu et al. 2021b) |
| GmMYB68 |
Overexpression in soybean enhances salt tolerance RNAi soybean shows increased salt sensitivity |
GmMYB68 activates FBP and P5CR to promote the accumulation of soluble sugars and free proline | (He et al. 2020b) |
| GmMYB76, GmMYB92, GmMYB177 | Overexpression in Arabidopsis enhances salt tolerance | All three genes lead to the activation of DREB2A, RD17, and P5CS. In addition, GmMYB76 and GmMYB177 can enhance the expressions of RD29B, ERD10, and COR78 | (Liao et al. 2008b) |
| GmMYB81 | Overexpression in Arabidopsis enhances salt tolerance at germination stage | GmMYB81 regulates stress tolerance together with abiotic stress regulator GmSGF14l | (Bian et al. 2020) |
| GmMYB84 | Overexpression in Arabidopsis and soybean enhances salt tolerance | GmMYB84 can bind to the cis-regulatory sequence of GmAKT1 to mediate K+ transport and in turn establish a higher [K+]/[Na+] ratio | (Zhang et al. 2020b) |
| GmMYB118 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance CRISPR-edited soybean shows increased salt sensitivity |
GmMYB118 regulates the expressions of stress-associated genes to reduce ROS and MDA levels under salt and drought stress | (Du et al. 2018) |
| GmMYB133 | Overexpression in Arabidopsis enhances salt tolerance | GmMYB133 confers salt tolerance through inducing the expression of salt tolerance-related genes including EARL1, MPK3 and AZI1 | (Shan et al. 2021) |
| GmMYB173 |
Overexpression in soybean roots enhances salt tolerance RNAi soybean roots show increased salt sensitivity |
Phosphorylated GmMYB173 has a higher affinity to the promoter of GmCHS5 and facilitates the accumulation of dihydroxy B-ring flavonoids | (Pi et al. 2018) |
| GmMYB183 |
Overexpression in soybean hairy roots shows increased salt sensitivity RNAi soybean hairy roots have enhanced salt tolerance |
GmMYB183 regulates GmCYP81E11 and enhances the accumulation of ononin, which has negative effects on soybean salt tolerance | (Pi et al. 2019) |
| NAC transcription factors | |||
| GmNAC06 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance CRISPR-edited soybean hairy roots show increased salt sensitivity |
GmNAC06 promotes the expressions of GmUBC2 and GmHKT1 to maintain ion homeostasis and mediate oxidative stress responses | (Li et al. 2021b) |
| GmNAC15 | Overexpression in soybean hairy roots enhances salt tolerance | GmNAC15 regulates osmolyte biosynthesis to eliminate osmotic stress and induces the expressions of stress-responsive genes including GmERF3 and GmWRKY54 | (Li et al. 2018b) |
| GmNAC11 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | GmNAC11 is involved in the regulation of DREB1A and other stress-associated genes | (Hao et al. 2011) |
| GmNAC20 | Overexpression in Arabidopsis enhances salt tolerance | GmNAC20 activates the DREB/CBF-COR pathway | (Hao et al. 2011) |
| GmNAC085 | Overexpression in soybean enhances salt tolerance | GmNAC085 enhances the expressions of stress-related genes involved in proline biosynthesis, Na+ transport, and dehydrin accumulation | (Hoang et al. 2021) |
| GmNAC109 | Overexpression in Arabidopsis enhances salt tolerance | GmNAC109 increases the transcript levels of stress-responsive genes such as DREBs, AREBs, RD29A, and COR15A. ABA-responsive genes, ABIs, and auxin-related genes, AIR3 and ARF2, are also induced | (Yang et al. 2019) |
| GmSIN1 |
Overexpression in soybean enhances salt tolerance RNAi soybeans show increased salt sensitivity |
GmSIN1 enhances the expressions of GmNCED3s and GmRbohBs, forming a positive feed-forward loop to amplify the initial salt stress signal by rapidly accumulating ABA and ROS | (Li et al. 2019b) |
| NF-YA transcription factors | |||
| GmNFYA |
Overexpression in soybean enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
Stress-induced GmNFYA destabilizes the GmFVE-GmHDA13 complex by interacting with GmFVE, resulting in the activation of salt stress-responsive genes by histone acetylation | (Lu et al. 2021) |
| GmNFYA13 |
Overexpression in Arabidopsis and soybean enhances salt tolerance RNAi soybeans show increased salt sensitivity |
GmNFYA13 can bind to and regulate the transcription of GmSALT3, GmMYB84, GmNCED3, and GmRbohB | (Ma et al. 2020) |
| Plant homeodomain finger protein | |||
| GmPHD2 | Overexpression in Arabidopsis enhances salt tolerance | GmPHD2 may downregulate several negative regulators of stress response-related genes such as CBF2, STRS1, and STRS2 | (Wei et al. 2009) |
| RAV transcription factor | |||
| GmRAV-03 | Overexpression in Arabidopsis enhances salt tolerance | (Zhao et al. 2017) | |
| Trihelix transcription factors | |||
| GmGT-2A, GmGT-2B | Overexpression in Arabidopsis enhances salt tolerance | The two GmGTs induce the stress tolerance genes, STZ/ZAT and DREB2A, and activate antioxidative mechanisms. Furthermore, GmGT-2B can enhance the expressions of ABA-dependent genes including NCED3, LTP3, LTP4 and PAD3 | (Xie et al. 2009) |
| VOZ transcription factor | |||
| GmVOZ1G |
Overexpression in soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
GmVOZ1G enhances the activities of POD and SOD under drought and salt stress | (Li et al. 2020a) |
| WRKY transcription factors | |||
| GmWRKY12 | Overexpression in soybean hairy roots enhances salt tolerance | The promoter of GmWRKY12 contains cis-acting elements including ABER4, MYB, MYC, GT-1, W-box, GARE, and DPBF, suggesting the role of GmWRKY12 in various stress and hormone responses | (Shi et al. 2018) |
| GmWRKY13 | Overexpression in Arabidopsis leads to increased salt sensitivity | GmWRKY13 upregulates ARF6 to promote lateral root development and ABI1 to mediate ABA response | (Zhou et al. 2008) |
| GmWRKY16 | Overexpression in Arabidopsis enhances salt tolerance | GmWRKY16 enhances the expressions of WRKY8, KIN1, and RD29A in salt stress | (Ma et al. 2019) |
| GmWRKY27 |
Overexpression in soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
GmWRKY27 interacts with GmMYB174 and inhibits the expression of GmNAC29, which negatively regulates stress tolerance | (Wang et al. 2015) |
| GmWRKY45 | Overexpression in Arabidopsis enhances salt tolerance | (Li et al. 2020b) | |
| GmWRKY49 | Overexpression in Arabidopsis and soybean enhances salt tolerance | GmWRKY49 can bind directly to the W-box and possibly modulates the expressions of downstream stress-related genes | (Xu et al. 2018) |
| GmWRKY54 | Overexpression in Arabidopsis enhances salt tolerance | GmWRKY54 regulates the expressions of DREB2A and STZ/Zat10 by binding to the W-boxes in their promoter region | (Zhou et al. 2008) |
| GmWRKY111 | Overexpression in soybean hairy roots enhances salt tolerance | (Xu et al. 2014) | |
| C2H2-type zinc finger protein | |||
| GmZAT4 | Overexpression in Arabidopsis enhances salt tolerance | GmZAT4 regulates the expressions of ABA-responsive marker genes including RD29A, RD29B, RAB, and ABI | (Sun et al. 2019b) |
Among the 10 groups of GmbZIPs identified in soybean (131 in total), the group A genes were reported to be involved in ABA-dependent stress signaling (Liao et al. 2008c). For example, the overexpression of GmbZIP1 in Arabidopsis upregulated ABA-regulated genes such as ABA INSENSITIVE 1 (ABI1), ABI2, RESPONSIVE TO DESICCATION 29B (RD29B), and RESPONSIVE TO ABA 18 (Rab18), and downregulated POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 1 (KAT1) and KAT2, to promote stomatal closure upon salt stress (Gao et al. 2011). The overexpression of GmFDL19 upregulated another subset of stress-responsive genes including the ion transporters GmCHX1 and GmNHX1, and some ABA-responsive TFs such as GmbZIP1, GmNAC11, and GmNAC29 to activate salt tolerance mechanisms (Li et al. 2017b). Thus, these two groups of genes may enhance salt tolerance in an ABA-dependent manner. On the other hand, some non-group A members were determined to be negative regulators of ABA signaling in salt stress. Transgenic plants overexpressing a group D member GmbZIP132, a group S member GmbZIP44, a group C member GmbZIP62, or a group G member GmbZIP78 showed reduced ABA sensitivity but enhanced salt tolerance (Liao et al. 2008a, 2008c). Similar observations were also reported in three MYB genes GmMYB76, GmMYB92, and GmMYB177, which also negatively regulate ABA signaling for soybean salt tolerance (Liao et al. 2008c). Besides, GsMYB15-overexpressing Arabidopsis repressed the expressions of the marker genes in the ABA pathway, AtABI1 and AtABI2, and showed improved salt tolerance, so GsMYB15 may also participate in the ABA-dependent pathway in salt stress responses (Shen et al. 2018a).
There are around 180 members in the NAM/ATAF/CUC (NAC) family of TF-encoding genes in the soybean genome (Melo et al. 2018). A number of them have been characterized to play roles in salt stress tolerance (Table 1). Some members in the NAC family also play roles in the ABA pathway. For example, the overexpression of GmNAC109 resulted in the upregulation of ABA-responsive genes, ABI1 and ABI5, conferring increased sensitivity to ABA and greater tolerance against salt stress (Yang et al. 2019). SALT INDUCED NAC1 (GmSIN1) regulates the ABA biosynthesis genes, 9-cis-epoxycarotenoid dioxygenases (GmNCED3s), which in turn leads to the rapid accumulation of ABA and enhances salt tolerance by amplifying the initial salt stress signal (Li et al. 2019b).
At the same time, the ethylene responsive factor (ERF) family of TFs are also involved in ABA and multiple phytohormone signaling pathways upon salt stress. The induction of GmERF75 by exogenous salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) suggested that GmERF75 may integrate signals form the SA and ET/JA pathways and positively regulate salt stress responses (Zhao et al. 2019b). GmERF057 and GmERF089 were also induced by salt, drought, ET, SA, JA, and ABA, implying their participation in various phytohormone-mediated signaling pathways in enhancing salt tolerance (Zhang et al. 2008). Meanwhile, the ET biosynthesis genes, AtACO4 and AtACS2, and ABA biosynthesis genes, ABA1 and ABA2, were upregulated in transgenic Arabidopsis overexpressing GmERF135, indicating that GmERF135 modulates salt tolerance by regulating both ET and ABA signaling pathways (Zhao et al. 2019c).
The WRKY TFs are characterized by the core amino acid sequence, WRKYGQK, which binds specifically to the W-box sequence (T)TTGAC(C/T) in the promoters of their target genes (Zhou et al. 2008). GmWRKY54 can regulate the expressions of DREB2A and STZ/Zat10 and induce stress tolerance mechanisms. GmWRKY49 can also bind directly to the W-box in its target gene promoters and possibly modulate the expressions of downstream stress-related genes, leading to enhanced salt tolerance in GmWRKY49-overexpressing plants (Xu et al. 2018).
Signal transduction
When plants sense salt stress, multiple signal transduction pathways, such as the phytohormone-mediated, Ca2+-dependent, and phosphatidylinositol signals, are initiated (Liu et al. 2019). Regulators such as transcription factors are activated and the gene expressions of downstream stress-responsive genes are then changed to induce salt tolerance mechanisms and help plants combat salt stress. In soybean, the Ca2+-mediated as well as ABA-dependent pathways related to salt stress are most extensively documented. Components in the pathways involved in soybean salt tolerance and their functional studies are listed in Table 2.
Table 2.
A list of functionally verified components of the Ca2+-mediated and ABA-dependent pathways involved in salt tolerance in soybean
| Putative function | Gene | Role in salt tolerance | References |
|---|---|---|---|
| Ca2+-mediated pathway | |||
| P-type II Ca2+ ATPase | GsACA1 | Overexpression in alfalfa enhances salt tolerance | (Sun et al. 2016) |
| Calmodulin | GmCam4 |
Overexpression in soybean enhances salt tolerance Silencing in soybean leads to increased salt sensitivity |
(Rao et al. 2014) |
| Calcineurin B-like protein | GmCBL1 | Overexpression in Arabidopsis enhances salt tolerance | (Li et al. 2012) |
| Calcineurin B-like protein | GmCBL4 | Overexpression in soybean hairy roots enhances salt tolerance | (Li et al. 2022a) |
| Calmodulin-binding protein | GmCBP60A-1 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
(Yu et al. 2021) |
| Calcium-dependent calmodulin-binding receptor-like kinase | GsCBRLK | Overexpression in Arabidopsis and soybean enhances salt tolerance | (Ji et al. 2016b; Yang et al. 2010) |
| Calcium-dependent protein kinase | GmCDPK3 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
(Wang et al. 2019a) |
| Calcineurin B-like protein interacting protein kinase | GmCIPK2 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
(Li et al. 2022a) |
| Calcineurin B-like protein interacting protein kinase | GmCIPK21 |
Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance RNAi soybean hairy roots show increased salt sensitivity |
(Wang et al. 2019a) |
| Calmodulin-like protein | GsCML27 | Overexpression in Arabidopsis leads to increased salt sensitivity | (Chen et al. 2015a) |
| Calcineurin B-like protein-interacting protein kinase | GmPKS4 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | (Ketehouli et al. 2021) |
| ABA-mediated pathway | |||
| Drought-induced protein | GmDi19-5 | Overexpression in Arabidopsis shows increased salt and ABA sensitivities | (Feng et al. 2015) |
| F-box protein | GmFBX176 | Overexpression in Arabidopsis shows increased salt sensitivity and reduced ABA sensitivity | (Yu et al. 2020) |
| Glucose-6-phosphate dehydrogenase | GmG6PH7 | Overexpression in Arabidopsis enhances salt tolerance | (Jin et al. 2010) |
| Mitogen-activated protein kinase | GmMMK1 | Overexpression in Arabidopsis and soybean hairy roots leads to increased salt sensitivity | (Liao et al. 2021) |
| Nodule autoregulation receptor kinase | GmNARK | Overexpression in Arabidopsis leads to increased salt and ABA sensitivities | (Cheng et al. 2018) |
| U-box E3-ubiquitin ligase | GmPUB21 |
Overexpression in tobacco leads to increased salt sensitivity Knock-down soybean shows increased salt sensitivity |
(Yang et al. 2022) |
| Receptor-like cytoplasmic serine/threonine protein kinase | GsRLCK | Overexpression in Arabidopsis enhances salt tolerance and reduces ABA sensitivity | (Sun et al. 2013a) |
| Stress-associated protein | GmSAP16 | Overexpression in Arabidopsis enhances salt tolerance and ABA sensitivity | (Zhang et al. 2019b) |
| S-phase kinase-associated protein 1 | GmSK1 | Overexpression in tobacco enhances salt tolerance | (Chen et al. 2018b) |
| Salt tolerance 1 | GmST1 | Overexpression in Arabidopsis enhances salt tolerance and ABA sensitivity | (Ren et al. 2016) |
| With No Lysine serine-threonine kinase | GmWNK1 | Overexpression in Arabidopsis enhances salt and ABA tolerance | (Wang et al. 2011b) |
| Inositol polyphosphate 5-phosphatase | Gs5PTase8 | Overexpression in Arabidopsis and soybean hairy roots enhances salt tolerance | (Jia et al. 2020) |
Ca2+-mediated pathway
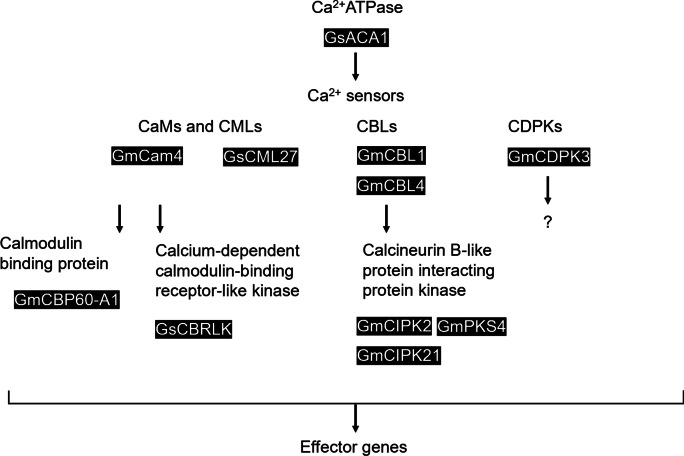
When salt stress is detected, Ca2+ serves as one of the secondary messengers to trigger a series of sequential responses (Phang et al. 2008). Its cytosolic concentration is controlled tightly by Ca2+ transporters such as Ca2+ ATPase. One good example is GsACA1 detailed in a previous section (Sun et al. 2016) (Fig. 3).
Fig. 3.
Schematic representation of Ca2+-mediated pathway involve in salt stress in soybean. Upon salt stress, Ca2+ATPase regulate cytosolic Ca2+ level. Ca2+ signals are amplified and transmitted by Ca2+ sensors including calmodulins (CaMs), calmodulin-like proteins (CMLs), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinase (CDPKs). Through interacting with their associated interacting proteins, downstream effectors genes are subsequentially activated to give physiological response countering salt stress. Black boxes indicate functionally verified genes in the Ca2+-mediated pathway involve in salt tolerance in soybean
Following the rapid changes in cytosolic Ca2+ level in response to environmental stimuli, Ca2+ signals are amplified and transmitted through Ca2+ sensors. There are three main classes of EF-hand Ca2+ sensors in plants, including calmodulins (CaMs) and CaM-like proteins (CMLs), calcium-dependent protein kinases (CDPKs), and calcineurin B-like protein (CBLs) (DeFalco et al. 2010). While CaMs, CMLs, and CBLs are non-catalytic relay sensors regulating downstream signaling, CDPKs are direct responders possessing catalytic activities that can transduce a signal (DeFalco et al. 2010). Among 17 drought- and salt-induced CDPK genes in soybean, GmCDPK3 demonstrated a relatively high differential expression level and was subjected to further functional analyses (Wang et al. 2019a). Soybean hairy roots with GmCDPK3 overexpression had increased protein, chlorophyll and decreased MDA contents, while the opposite results were obtained in GmCDPK3-RNA-interference soybean hairy roots. The overexpression of GmCDPK3 also enhanced salt tolerance in Arabidopsis, with shortened primary roots and more lateral roots, indicating that GmCDPK3 can regulate drought and salt stress responses as well as root growth and development (Wang et al. 2019a). Despite confirming the role of GmCDPK3 in salt tolerance by functional analysis, further investigation would be required to study the direct downstream effects of GmCDPK3 in transducing the salt stress signal.
CaMs are prominent non-enzymatic Ca2+ sensors that interact and alter the activities of other proteins upon the binding of Ca2+ in response to environmental stress. GmCaM4 has been shown to directly interact with the transcription factor, Myb2, which is involved in the regulation of stress-responsive genes (Rao et al. 2014). The overexpression of GmCam4 in Arabidopsis can induce AtMyb2-regulated genes, including P5CS1, which causes the accumulation of proline and improves salt tolerance. Moreover, the interaction between GmCaM2 and a CaM-binding reporter-like kinase, GmCBRLK, was also confirmed. Therefore, a GmCaM4-Myb2-mediated and GmCaM2-GmCBRLK-mediated mechanism may be involved in salt tolerance in soybean. GsCML27, which contains four conserved calcium-binding EF-hand motifs, was isolated from a salt-alkali-resistant wild soybean (Chen et al. 2015a). It shows Ca2+-binding affinity and has a higher expression under bicarbonate, salt, and osmotic stresses. Interestingly, the ectopic expression of GsCML27 promoted seed germination under bicarbonate stress but inhibited it under salt and osmotic stresses (Chen et al. 2015a), implying the complex function of GsCML27 in various stress signaling pathways and its negative regulatory role in salt stress during early growth stages.
CBLs are Ca2+ sensors unique to plants that decode Ca2+ signals by interacting with CBL-interacting protein kinases (CIPKs). GmCBL1-overexpression conferred salt tolerance in Arabidopsis by activating several stress-responsive genes in response to high salinity, including DREB1A, DREB2A, RD29A, and KIN1 (Li et al. 2012). Salt tolerance was also enhanced in GmCBL4-overexpressing soybean hairy roots and Arabidopsis, resulting in lower levels of MDA and H2O2 detected (Li et al. 2022a, b). GmCIPK2 and GmCIPK21 were identified as interacting partners of GmCBL4. The overexpression of either GmCIPK2 or GmCIPK21 can promote the activities of antioxidant-related enzymes such as POD, GST, and CAT, consistent with the lower MDA and H2O2 contents and the greater salt tolerance phenotype observed. Thus, the GmCBL4-GmCIPK2 and GmCBL4-GmCIPK21 complexes participate in salt tolerance in soybean by enhancing ROS scavenging. GmPKS4 is another CIPK that could regulate salt stress responses through activating ROS scavenging systems and promoting the transcription of stress-related genes (Ketehouli et al. 2021). In Arabidopsis and soybean hairy roots overexpressing GmPKS4, lower MDA, higher proline, and lower ROS levels as well as less membrane damage were observed. Moreover, the expression levels of stress-related genes, such as SNF4, CBL1, and NHX1, were also higher (Ketehouli et al. 2021).
The roles of the calcium-dependent calmodulin-binding receptor-like kinase (CBRLK) and calmodulin-binding protein 60 (CBP60) in soybean salt tolerance were also reported. Isolated from Glycine soja, GsCBRLK was a novel CaM-binding protein kinase containing Ca2+-dependent activities and a CaM-binding site within the kinase domain (Yang et al. 2010). Enhanced salt tolerance was observed in Arabidopsis and soybean overexpressing GsCBRLK (Ji et al. 2016b; Yang et al. 2010). Proteomic studies also revealed that GsCBRLK plays a role in regulating ROS scavenging and photosynthesis by affecting the abundance of signaling, photosynthetic, and metabolic salt-responsive proteins (Ji et al. 2016b). Among 19 CBP60 members identified in soybean, GmCBP60A-1 was induced significantly under high salt and dehydration (Yu et al. 2021). Soybean hairy roots overexpressing GmCBP60A-1 demonstrated better salt tolerance and RNAi plants showed increased sensitivity to salt stress, corroborating the role of GmCBP60A-1 in enhancing salt tolerance in soybean (Yu et al. 2021).
ABA-dependent pathway
Abscisic acid (ABA) is a plant hormone that plays a vital role in stress responses due to its rapid accumulation upon stress and participation in various stress signaling pathways (Zhang et al. 2006). The ABA signaling pathway generally encompasses the initial stress signal perception, cellular signal transduction and regulation of the expressions of genes involved in ABA biosynthesis and catabolism, which in turn regulate the accumulation of ABA and trigger salt stress responses. Besides the TFs involved in ABA signaling mentioned in the previous section, some genes were also reported to play roles in the ABA signaling pathway by altering the expression of ABA-responsive genes.
Various kinases are found to be involved in the ABA pathway through altering the activities of ABA-responsive genes. Arabidopsis overexpressing an ABA-induced nodule autoregulation receptor kinase GmNARK was more sensitive to salt stress and had significantly upregulated ABA-responsive genes, ABI3, ABI4, ABI5, RAB18, RD29A, and RD29B, suggesting the role of GmNARK in salt stress through the ABA pathway (Cheng et al. 2018). The role of the mitogen-activated protein kinase (MAPK) cascade in salt stress signaling is well-known in model plants. In soybean, a salt stress-related MAPK, GmMMK1, was identified (Liao et al. 2021). GmMMK1-overexpressing plants demonstrated hypersensitivity to salinity (Liao et al. 2021). In particular, the transcript levels of PYLs and PP2Cs, which are the core components in ABA signaling, increased significantly in transgenic Arabidopsis overexpressing GmMMK1. Therefore, GmMMK1 may regulate salt stress response in an ABA-dependent manner (Liao et al. 2021). On the contrary, the ectopic expression of With No Lysine serine-threonine kinase, GmWNK1, and receptor-like cytoplasmic serine/threonine protein kinase, GsRLCK, in Arabidopsis conferred improved tolerance against salt stress (Sun et al. 2013a; Wang et al. 2011b). GmWNK1 regulates the ABA level by altering the expressions of ABA-responsive genes. The downregulation of ABA-catabolic genes, CYP707As, coupled with an increased level of endogenous ABA, in GmWNK1-overexpressing Arabidopsis indicated that GmWNK1 regulates ABA homeostasis and catabolism (Wang et al. 2011b). Receptor protein kinases could work as sensors to trigger a signaling cascade. Plants overexpressing GsRLCK had altered expressions of ABA-regulating genes, PYR1, ABI5, ABF4, and ABI2, and stress-responsive genes induced by ABA, such as COR47, RAB18, KIN1, NCED3, COR15A, and RD29A, supporting the role of GsRLCK in controlling ABA levels and salt sensitivity in soybean (Sun et al. 2013a). Notably, GmSK1 was induced by various phytohormones including ABA, JA, and SA, indicating that GmSK1 may participate in multiple stress-signaling pathways (Chen et al. 2018b).
Other proteins are also involved in the ABA pathway during salt stress responses. For instance, the overexpression of GmDi19-5 and GmFBX176 caused the transgenic Arabidopsis to be more sensitive to NaCl (Feng et al. 2015; Yu et al. 2020). GmDil19-5 and GmFBX176 regulate the expressions of genes related to ABA-signaling. The expressions of ABA-related genes, such as CYP707A3, ABF3, ABF4, ABI1, ABI5, RAB18, and SOS2, were altered in transgenic Arabidopsis, suggesting these genes are also involved in the regulation of ABA response under stress. Meanwhile, enhanced salt tolerance was observed in Arabidopsis overexpressing GmSAP16, Gs5PTase8, or GmG6PD7, with alterations in the transcript levels of ABA-responsive genes (Jia et al. 2020; Jin et al. 2010; Zhang et al. 2019b). Specifically, Arabidopsis overexpressing GmG6PD7 exhibited downregulated AtPYL8, AtABIs, and AtSnKRs, and ABA-biosynthesis genes, AtNCEDs, and upregulated ABA-catabolic genes, AtCYP707As, which implies that GmG6PD7 functions by lowering the ABA level under salt stress, and resulting in increased germination rate (Jin et al. 2010).
Perspectives
In the past few decades, great strides have been taken in identifying and validating soybean salt tolerance genes, and in applying such knowledge to germplasm generation (Tran and Mochida 2010; Zhang et al. 2022b). Progress has been accelerated by multiple factors, including the release of reference-grade genomes for cultivated and wild soybeans (Liu et al. 2020; Schmutz et al. 2010; Shen et al. 2018b; Valliyodan et al. 2019; Xie et al. 2019) and advances in next-generation sequencing that increase the study resolution from the genic to the nucleotide level (Zhang et al. 2022b). Functional studies, which examine how the overexpression or mutation of salt-responsive genes impacts plant phenotypes under salinity, are effective in verifying the functions of these genes that are potentially useful for improving soybean salt tolerance (Tran and Mochida 2010; Zhang et al. 2022b). Genome-wide association studies (GWAS), whole-genome scanning, and functional annotation have also assisted in characterizing genes with putative salt tolerance functions (Zhou et al. 2015).
In this section, we aim to discuss how to use the systemic approach of multi-omics to identify potential genes and pathways to complement the knowledge gap in genomic studies. Then, we lay out the strategic roadmap to incorporate current knowledge into molecular breeding, producing salt-tolerant soybeans that brings actual societal benefits.
Filling knowledge gaps with multi-omics
Since salt tolerance involves the simultaneous interactions of various phenotypic traits with multiple salt-associated stressors, relying solely on genomics is insufficient to resolve the complex mechanisms by which soybean exhibits salt tolerance. Therefore, there have been calls for taking the multi-omic approach—combining genomic, transcriptomic, proteomic and epigenomic techniques—to obtain holistic insights into the mechanisms underpinning salt stress resistance and to expand the toolkit for developing salt-tolerant soybeans (Zhang et al. 2022b).
For some gene products, their physiological and metabolic mechanisms in conferring salt tolerance in soybean remain elusive. Deciphering the pathways by which these proteins activate salt tolerance require proteomics and metabolomics. Some proteomic studies have uncovered protein families and biochemical pathways that are integral to salt stress mitigation in soybean, including ROS scavenging (Pi et al. 2016; Xu et al. 2011), cell wall remodeling (Rehman et al. 2022) and signal transduction (Ji et al. 2016a). Meanwhile, the metabolomic profiling of salt-tolerant versus salt-sensitive soybeans has revealed metabolites and metabolic pathways that play significant roles in mediating salt tolerance (Das et al. 2017; Jiao et al. 2018; Li et al. 2017a; Zhang et al. 2016). Since the genes that underlie the synthesis or modulation of these proteins, secondary metabolites and their systems have yet to be pinpointed, combining genomic, proteomic and metabolomic approaches could unlock a wealth of genetic resources for improving soybean salt tolerance.
While proteomics and metabolomics deepen our understanding of the downstream effects of genome modifications, epigenomics elucidates the mechanisms that control the expressions of stress tolerance genes in crops. Epigenetic features, including chromatin architecture and small RNA expressions, are emergent key players in crop tolerance to abiotic stress by regulating the expressions of stress-related genes. Chromatin architecture is a major epigenetic gateway governing the plant’s response to external stresses (Bhadouriya et al. 2021). Its alteration includes DNA methylation and histone modifications, where genomic reprogramming could manifest in the form of gene silencing or upregulation (Bhadouriya et al. 2021). Most fascinatingly, nuclear re-organizations have been shown to be functionally linked with changes in stress conditions, suggesting a mechanistic basis of stress memories within the individual plant’s soma and across generations (Bhadouriya et al. 2021). Adaptive epigenetic memory is manifested in priming effects, where an initial exposure to a specific stress induces the plant to acquire improved tolerance in subsequent exposures (Conrath et al. 2015). Indeed, priming was shown to induce histone modifications that modulate the transcriptional responses in soybean to salt stress (Yung et al. 2022). Given that stress memories might be inherited at the somatic, intergenerational and transgenerational scales, priming could help ensure a stable soybean production in the changing climate (Wang et al. 2017). Another epigenetic feature involves the expression of miRNAs—short, non-coding RNAs that modulate post-transcriptional gene-silencing by repressing translation or inducing cleavage in their target mRNAs (Chaudhary et al. 2021). They have been shown to regulate many biotic and abiotic stress responses in plants, and could be useful for producing stress-resistant crops (Zhang 2015; Zhang et al. 2022a). For example, miR172a and miR172c, of which expressions are induced by salt treatment, have been functionally verified to improve soybean salt tolerance through the regulation of thiamine production. While miR172a regulates long-distance signalling between root and shoot (Pan et al. 2016), miR172c controls ABA signalling (Li et al. 2016a). In their review, Chaudhary et al. (Chaudhary et al. 2021) have explored the functional studies of miRNAs that regulate stress responses in crops, and summarized the methodologies for mining functional miRNAs, which could be adopted in soybean bioengineering.
Multi-omic approaches have been applied to crop improvement against abiotic stresses in several major cereal crops (Farooqi et al. 2022; Kaur et al. 2021). Online databases specifically for multi-omic system analyses have also been constructed (Scossa et al. 2021; Yang et al. 2021). With a plethora of high-throughput technologies and publicly available databases such as the Soybean Proteome Database (Sakata et al. 2009) and SoyNet (Kim et al. 2017), future studies could focus on functional omics analyses, and not just on identifying the specific members of different classes of biological molecules.
Priority in selecting salt tolerance genes in soybean
As explored above, the genetic control and effects of salt-tolerance mechanisms are highly variable. Despite having been functionally verified, genes differ in their usefulness for real-life molecular breeding.
Given that ion homeostasis is the primary mechanism for soybean salt tolerance and its major effect gene has been confirmed repeatedly, the most straightforward way to develop or screen for salt-tolerant cultivars is by targeting the selection of ion transportation. After discovering the major-effect salt-tolerant gene GmCHX1 in the soybean genome, marker-assisted breeding has been applied to cross-breed salt tolerant soybean cultivars with high-yield soybean cultivars in China, which yielded three non-transgenic cultivars, Longhuang #1–3, which exhibit dual tolerance towards salt and drought (Li et al. 2020d). Such applications will be most useful in a locale where salt stress is the primary cause of crop penalty.
When soybean is met with a multitude of stressors other than salinity, gene components of osmoprotectants and antioxidation have the potential of conferring multi-stress tolerance. Osmolytes like prolines increase plant tolerance against drought and heat stress, and also act as a ROS scavenger. Antioxidant metabolism, particularly, is crucial in honing overall crop resilience as oxidative imbalance is incited by a wide range of biotic and abiotic stresses. Targeting these two components of salt tolerance, which is rarely done in crop improvement studies, might produce cultivars that can adapt to multiple stressors that occur concurrently with salinity.
The potential of utilising cell wall remodelling as a salt-tolerant strategy pales in comparison due to the unresolved spatiotemporal complexity of this mechanism. We currently lack high-resolution temporal characterisation of how plant organs and tissues differentially respond to salt stress, and studies on their genetic control are scattered and unsystematic. As mentioned, plasticity is the key to cell wall modification during the course of salt stress. The immediate response of salt-induced cell wall reinforcement is antagonistic to the subsequent loosening for continuous growth under salt stress. A genetic change that permanently stiffen cell wall might confer salt tolerance for a short time span, which is often the case for lab experiments, but compromise later growth. Crucially, the genetic switch governing the response transition is still poorly understood. The extent of cell wall remodelling’s contribution to overall salt tolerance is also unknown. Future studies will have to answer these glaring mechanistic questions before application.
The pleiotropic nature of transcriptional factors and signalling controls render their modification for crop improvement a double-edged sword. On one hand, being able to unlock a stream of salt stress adaptations by modifying a single gene might be efficient for gene editing. For instance, signal transduction involving stress hormones regulate not only molecular defences, but also anatomical adjustments like root system architecture to enhance water and nutrient acquisition under salinity (Julkowska and Testerink 2015). On the flip side, changes in pleiotropic genes could also lead to unintended consequences that may harm crop performance. To resolve this, examining both mechanisms could conduct higher resolution characterisation of their downstream pathways, and to conduct realistic field trials. For transcription regulation, that is to verify the functions of their target genes in soybean physiology, not just their roles in conferring salt tolerance. For tuning plant signal transduction to survive salt stress, (Julkowska and Testerink 2015) et al. laid out the outstanding questions, particularly in relation to plant’s structural acclimation to high salinity. If the mechanisms are well understood, transcriptional and signalling modifications can best be applied to crop improvement by gene editing tools, which will be discussed below in more detail.
Thus far, the salt tolerance mechanisms covered in this review are universal among different crop species as these adaptations have been studied extensively. Exploring adaptive traits unique to soybean, namely its symbiosis with Rhizobia in root nodules, will prove crucial to developing salt-tolerant cultivars. Rhizobia-legume symbiosis has long been recognised for its agronomic and environmental significance. Salt stress hampers this interaction by reducing nodulation success and nitrogen-fixing abilities of Rhizobia (Singleton and Bohlool 1984), hence handicapping nutrient acquisition and productivity in soybean. Recently, salt-induced expression of the transcription factor GmNAC181, renamed from GmNAC11 in Li et al. (Li et al. 2017b), was found to promote soybean nodulation under salt stress by binding to and activating the GmNINa promoter (Wang et al. 2022). Future studies could continue unravelling the interaction between salinity and soybean nodulation genetic components, thereby providing useful insights into improving Rhizobia recruitment efficiency under salt stress.
It might be tempting to combine multiple salt tolerance traits in a single cultivar. However, salt tolerance mechanisms, such as ion transportation and ROS scavenging, are known to incur an energetic cost to plants and might affect crop yield (Munns et al. 2020; Zorb et al. 2019). Future studies could rate the salinity threshold of soybeans and measure their yield to quantify and compare the effectiveness of different improvement strategies while taking into account the energetic trade-off.
Toolkits to improve salt tolerance in soybean
Marker-assisted selection (MAS) and genomic selection (GS) are conventional methods of applying genomic knowledge to crop development (Ashraf et al. 2012; Chen et al. 2018a). Thanks to the advancement in artificial intelligence, MAS and GS can now be enhanced by machine learning, which leverages the big data of soybean phenotypes and genotypes (Tong and Nikoloski 2021). Computational modeling offers several advantages over conventional MAS and GS. First, it can incorporate multi-omic datasets to describe the intermediate phenotypes of crops (Tong and Nikoloski 2021). Environmental factors, such as soil type and local climate, can also be incorporated into the model as genotype-by-environment (G × E) interactions (Burgueno et al. 2012), which yield much higher trait predictability than models that omit G × E effects (Lopez-Cruz et al. 2015). Tools developed for crop phenotypic data collection and phenomic data analysis have been reviewed and shown great potential (Zhao et al. 2019a). A model constructed from soybean phenomic data has integrated agro-management practices in predicting yield, which takes a step forward to informing and optimizing crop production (Parmley et al. 2019). All these show that machine learning-facilitated breeding could overcome the difficulties in applying ex situ experimental knowledge in the field. Tong and Nikoloski (2021) have evaluated past applications of machine learning in crop trait improvement via GS, and highlighted the potential of integrating high-throughput multi-omic phenotypic data to improve the efficacy of crop breeding, serving as a reference point for future improvements in soybean salt tolerance.
Genome editing refers to a suite of molecular techniques that induce precise and targeted modifications in genomes (Zhang et al. 2018), offering effective tools in generating new germplasms with controlled mutagenesis. Since genome editing does not involve the transfer of foreign genes, i.e. transgenesis, gene-edited crops side-step the legal limitations of, or ethical concerns for, conventional genetically modified organisms (GMOs) (Rahman et al. 2022). In soybean, a breakthrough occurred when transcription activator-like effector nuclease (TALENs) was applied to create a high-oleic acid soybean cultivar by knocking out two fatty acid desaturase 2 genes (GmFAD2-1A and GmFAD2-1B) (Haun et al. 2014), which has become the first-ever gene-edited crop to be commercialised (Calyxt 2019). The oil produced from this new soybean contains 80% oleic acids and 20% less saturated fat, thereby significantly improving its health benefits, heat stability and shelf life.
Among various gene editing methods, CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9) stands out for being rapid and cost-effective. It involves inducing mutations in target genes by cleaving specific DNA strands via the action of the Cas9 enzyme guided by the CRISPR sequence (Zafar et al. 2020), offering an effective tool in the molecular breeding of new germplasms with controlled mutagenesis. In soybean, CRISPR/Cas9 has been applied in different forms of mutation, namely targeted addition, deletion, replacement and correction (Lu and Tian 2022; Xu et al. 2020). Most notably, Do et al. (Do et al. 2019) were able to reproduce the high-oleic acid phenotype by using CRISPR/Cas 9 instead of TALEN and yielded a higher dual-mutation efficiency. However, in relation to soybean salt tolerance, the application of CRISPR/Cas 9 has mostly been limited to performing functional analyses on target genes (Sun et al. 2021; Wang et al. 2021b). The potential, application, challenges and key to the success of utilizing CRISPR/Cas9 to develop stress-resistant crops have been discussed in several reviews (Rahman et al. 2022; Shelake et al. 2022; Zafar et al. 2020; Mao et al. 2019).
Beyond the above methods, an excellent review by Varshney et al. (2021) has presented a holistic, strategic pipeline to fast-forward crop improvement. It starts from developing system-level understanding of crop phenotype via multi-omics and machine learning, to genomic prediction and speed-breeding approaches. We encourage future studies to reference this roadmap when developing salt-tolerant soybeans if we were to move on from laboratory experiments to field application.
From lab to field: strategies to apply current knowledge
Given the socio-economic importance of soybean, the value of genomic research can only be fully realized by applications. The goal of improving salt tolerance in soybean is to optimise crop production in spite of the saline farmland, not maximising salt tolerance per se. As agronomic viability is the prerequisite of applying lab-based knowledge to field production, newly developed soybean lines must be evaluated in the field while considering the local production context.
Laboratory or greenhouse settings are unrealistic, hence, inadequate in testing crop performance. The screening for soybean salt-tolerance is often limited to seedlings, with the assumption that higher survival or vigor during the vegetative stage is a good proxy for its crop performance. This is, however, not guaranteed as too high a salinity could cause plants to divert most photosynthetic energy to stress tolerance rather than growth and reproduction (Zorb et al. 2019). Interestingly, salt-tolerant soybeans might even exhibit the same level of early vigor as salt-sensitive lines under salt stress despite performing better in field (Liu et al. 2016). Tolerant soybeans are not immune to salt-induced crop penalty, which might occur in form of reduced number pods per plant, seed multiplication ratio, hundred-seed weight et cetera. All these point to the necessity of field trials. Studies that improve soybean salt tolerance on a molecular basis seldom quantify the yield performance of their product, but examples include those developed from the introgression of GmCHX1 into salt sensitive cultivars with MAS (Do et al. 2016; Liu et al. 2016) and CRISPR/Cas9-induced mutagenesis of GmAITR, which increased ABA sensitivity (Wang et al. 2021a). All three studies demonstrated that the salt-tolerant line outperformed salt-sensitive ones in terms of yield-related traits. Future studies could consider quantifying the raise in salinity threshold of improved soybeans (Zorb et al. 2019).
Same as the selection process of other agronomic traits, the evaluation of salt tolerance should consider the local environment and the need of producers. While most lab-based soybean salt tolerance experiments use NaCl for salt treatment, salt-affected land worldwide vary in their chemical characteristics like pH and the presence of other types of salt (Rengasamy 2010). Developing salt-tolerant soybeans, whether by breeding or gene editing, can be based off landraces that are adapted to local conditions. These include, but are not limited to, soil physicochemical properties, climate, irrigation regime, pest and disease. Despite having undergone domestication, cultivated soybeans are reasonably high in genetic diversity (Hyten et al. 2006). This corroborates the well-recognised importance of preserving local seed resources, of which genetic diversity could improve crop resilience, avoid genetic bottleneck and inbreeding depression.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from the Guangdong Provincial Department of Science and Technology 2020 Key Areas Research and Development Programs: Breeding for High-Yield and High-Quality New Soybean Varieties for Tropics and Subtropics (2020B020220008), the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16) and the Lo Kwee-Seong Biomedical Research Fund. Ms. Jee Yan Chu copy-edited the manuscript. Any opinions, findings, conclusions, or recommendations expressed in this publication do not reflect the views of the Government of the Hong Kong Special Administrative Region or the Innovation and Technology Commission.
Author contributions
H-ML conceptualized the manuscript. L-YC, C-HL, and H-SL co-wrote the first draft. L-YC, C-HL, H-SL, M-WL and H-ML reviewed and edited the manuscript.
Funding
The work was supported by the Guangdong Provincial Department of Science and Technology 2020 Key Areas Research and Development Programs: Breeding for High-Yield and High-Quality New Soybean Varieties for Tropics and Subtropics (2020B020220008), the Hong Kong Research Grants Council Area of Excellence Scheme [AoE/M-403/16] and Lo Kwee-Seong Biomedical Research Fund awarded to H-ML.
Data availability
Not applicable.
Declarations
Not applicable.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
This article is part of the Topical Collection on Soybean Functional Genomics.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hoi-Sze Leung, Long-Yiu Chan and Cheuk-Hin Law contributed equally.
References
- Agarwal S, Sairam RK, Srivastava GC, Meena RC. Changes in antioxidant enzymes activity and oxidative stress by abscisic acid and salicylic acid in wheat genotypes. Biol Plant. 2005;49(4):541–550. doi: 10.1007/s10535-005-0048-z. [DOI] [Google Scholar]
- Aleem M, Aleem S, Sharif I, Wu ZY, Aleem M, Tahir A, Atif RM, Cheema HMN, Shakeel A, Lei S, Yu DY, Wang H, Kaushik P, Alyemeni MN, Bhat JA, Ahmad P. Characterization of SOD and GPX Gene Families in the Soybeans in Response to Drought and Salinity Stresses. Antioxidants. 2022;11(3):460. doi: 10.3390/antiox11030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M. Breeding for salinity tolerance in plants. Crit Rev Plant Sci. 1994;13(1):17–42. doi: 10.1080/713608051. [DOI] [Google Scholar]
- Ashraf M, Akram NA, Mehboob Ur R, Foolad MR. Marker-assisted selection in plant breeding for salinity tolerance. Methods Mol Biol. 2012;913:305–333. doi: 10.1007/978-1-61779-986-0_21. [DOI] [PubMed] [Google Scholar]
- Belamkar V, Weeks NT, Bharti AK, Farmer AD, Graham MA, Cannon SB. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics. 2014;15:950. doi: 10.1186/1471-2164-15-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhadouriya SL, Mehrotra S, Basantani MK, Loake GJ, Mehrotra R. Role of chromatin architecture in plant stress responses: an update. Front Plant Sci. 2021;11:603380. doi: 10.3389/fpls.2020.603380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj R, Sharma I, Kanwar M, Sharma R, Handa N, Kaur H, Kapoor D, Poonam . LEA proteins in salt stress tolerance. In: Ahmad P, Azooz MM, Prasad MNV, editors. Salt Stress in plants: signalling, omics and adaptations. New York: Springer New York; 2013. pp. 79–112. [Google Scholar]
- Bian SM, Jin DH, Sun GQ, Shan BH, Zhou HN, Wang JY, Zhai LL, Li XY. Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene. 2020;753:144803. doi: 10.1016/j.gene.2020.144803. [DOI] [PubMed] [Google Scholar]
- Borkowska M, Krzymowska M, Talarczyk A, Awan MFM, Yakovleva L, Kleczkowski K, Wielgat B. Transgenic potato plants expressing soybean ß-1,3-Endoglucanase gene exhibit an increased resistance to Phytophthora infestans. Zeitschrift für Naturforschung C. 1998;53(11–12):1012–1016. doi: 10.1515/znc-1998-11-1212. [DOI] [PubMed] [Google Scholar]
- Brunetti C, Di Ferdinando M, Fini A, Pollastri S, Tattini M. Flavonoids as antioxidants and developmental regulators: relative significance in plants and humans. Int J Mol Sci. 2013;14(2):3540–3555. doi: 10.3390/ijms14023540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L, Talarico E, Cabeiras-Freijanes L, Madeo ML, Muto A, Minervino M, Lucini L, Miras-Moreno B, Sofo A, Araniti F. Coumarin interferes with polar auxin transport altering microtubule cortical array organization in Arabidopsis thaliana (L.) Heynh. Root apical meristem. International Journal of Molecular Sciences. 2021;22(14):7305. doi: 10.3390/ijms22147305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier J, Schlappi M. Cold induction of EARLI1, a putative Arabidopsis lipid transfer protein, is light and calcium dependent. Plant Cell Environ. 2004;27(7):929–936. doi: 10.1111/j.1365-3040.2004.01198.x. [DOI] [Google Scholar]
- Burgueno J, de los Campos G, Weigel K, Crossa J. Genomic prediction of breeding values when modeling genotype x environment interaction using pedigree and dense molecular markers. Crop Sci. 2012;52(2):707–719. doi: 10.2135/cropsci2011.06.0299. [DOI] [Google Scholar]
- Cai D, Zheng Y, Lan Y. Expression of Em gene (LEA1) from soybean immature seeds confers salt tolerance to Escherichia coli and tobacco plants. J Shenzhen Univ Sci Eng. 2006;23(3):235. [Google Scholar]
- Calyxt I (2019) First commercial sale of calyxt high oleic soybean oil on the U.S. Market. Calyxt, Inc. https://calyxt.com/first-commercial-sale-of-calyxt-high-oleic-soybean-oil-on-the-u-s-market/. Accessed 12/03/2023 2023
- Cesarino I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr Opin Biotechnol. 2019;56:209–214. doi: 10.1016/j.copbio.2018.12.012. [DOI] [PubMed] [Google Scholar]
- Chai MN, Fan RB, Huang YM, Jiang XH, Wai MH, Yang Q, Su H, Liu KC, Ma SZ, Chen ZT, Wang FJ, Qin Y, Cai HY. GmbZIP152, a soybean bZIP transcription factor, confers multiple biotic and abiotic stress responses in plant. Int J Mol Sci. 2022;23(18):10935. doi: 10.3390/ijms231810935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Lam HM. A putative lambda class glutathione S-Transferase enhances plant survival under salinity stress. Plant Cell Physiol. 2014;55(3):570–579. doi: 10.1093/pcp/pct201. [DOI] [PubMed] [Google Scholar]
- Chaudhary S, Grover A, Sharma PC. MicroRNAs: potential targets for developing stress-tolerant crops. Life. 2021;11(4):289. doi: 10.3390/life11040289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Tyerman SD. Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol. 2014;164(4):1600–1618. doi: 10.1104/pp.113.233791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla S, Jain S, Jain V. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryza sativa L.) J Biochem Biotechnol. 2013;22(1):27–34. doi: 10.1007/s13562-012-0107-4. [DOI] [Google Scholar]
- Chen M, Wang QY, Cheng XG, Xu ZS, Li LC, Ye XG, Xia LQ, Ma YZ. GmDREB2, a soybean DRE-binding transcription factor, conferred drought and high-salt tolerance in transgenic plants. Biochem Biophys Res Commun. 2007;353(2):299–305. doi: 10.1016/j.bbrc.2006.12.027. [DOI] [PubMed] [Google Scholar]
- Chen M, Xu ZS, Xia LQ, Li LC, Cheng XG, Dong JH, Wang QY, Ma YZ. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.) J Exp Bot. 2009;60(1):121–135. doi: 10.1093/jxb/ern269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HT, He H, Yu DY. Overexpression of a novel soybean gene modulating Na+ and K+ transport enhances salt tolerance in transgenic tobacco plants. Physiol Plant. 2011;141(1):11–18. doi: 10.1111/j.1399-3054.2010.01412.x. [DOI] [PubMed] [Google Scholar]
- Chen HT, Chen X, Gu HP, Wu BY, Zhang HM, Yuan XX, Cui XY. GmHKT1;4, a novel soybean gene regulating Na+/K+ ratio in roots enhances salt tolerance in transgenic plants. Plant Growth Regul. 2014;73(3):299–308. doi: 10.1007/s10725-014-9890-3. [DOI] [Google Scholar]
- Chen C, Sun XL, Duanmu HZ, Zhu D, Yu Y, Cao L, Liu AL, Jia BW, Xiao JL, Zhu YM. GsCML27, a gene encoding a calcium-binding Ef-Hand protein from Glycine soja, plays differential roles in plant responses to bicarbonate, salt and osmotic stresses. Plos One. 2015;10(11):e0141888. doi: 10.1371/journal.pone.0141888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-t, Chen X, Wu B-y, Yuan X-x, Zhang H-m, Cui X-y, Liu X-q. Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J Integr Agric. 2015;14(6):1171–1183. doi: 10.1016/S2095-3119(14)60918-7. [DOI] [Google Scholar]
- Chen HT, Liu XQ, Zhang HM, Yuan XX, Gu HP, Cui XY, Chen X. Advances in salinity tolerance of soybean: Genetic diversity, heredity, and gene identification contribute to improving salinity tolerance. J Integr Agric. 2018;17(10):2215–2221. doi: 10.1016/S2095-3119(17)61864-1. [DOI] [Google Scholar]
- Chen YP, Chi Y, Meng QC, Wang XL, Yu DY. GmSK1, an SKP1 homologue in soybean, is involved in the tolerance to salt and drought. Plant Physiol Biochem. 2018;127:25–31. doi: 10.1016/j.plaphy.2018.03.007. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Li N, Dong L, Zhang D, Fan S, Jiang L, Wang X, Xu P, Zhang S. Overexpression of Soybean Isoflavone Reductase (GmIFR) enhances resistance to Phytophthora sojae in Soybean. Front Plant Sci. 2015;6:1024. doi: 10.3389/fpls.2015.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CH, Li CM, Wang DD, Zhai LF, Cai ZM. The soybean GmNARK affects ABA and salt responses in transgenic Arabidopsis thaliana. Front Plant Sci. 2018;9:514. doi: 10.3389/fpls.2018.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Gan ZR, Wang YP, Lu SJ, Hou ZH, Li HY, Xiang HT, Liu BH, Kong FJ, Dong LD. The soybean gene J contributes to salt stress tolerance by up-regulating salt-responsive genes. Front Plant Sci. 2020;11:272. doi: 10.3389/fpls.2020.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Langenbach CJG, Jaskiewicz MR. Priming for enhanced defense. Annu Rev Phytopathol. 2015;53(53):97–119. doi: 10.1146/annurev-phyto-080614-120132. [DOI] [PubMed] [Google Scholar]
- Czarnocka W, Karpinski S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radical Biol Med. 2018;122:4–20. doi: 10.1016/j.freeradbiomed.2018.01.011. [DOI] [PubMed] [Google Scholar]
- Das K, Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front Environ Sci. 2014;2:53. doi: 10.3389/fenvs.2014.00053. [DOI] [Google Scholar]
- Das A, Rushton PJ, Rohila JS. Metabolomic profiling of soybeans (Glycine max L.) Reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants. 2017;6(2):21. doi: 10.3390/plants6020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco TA, Chiasson D, Munro K, Kaiser BN, Snedden WA. Characterization of GmCaMK1, a member of a soybean calmodulin-binding receptor-like kinase family. FEBS Lett. 2010;584(23):4717–4724. doi: 10.1016/j.febslet.2010.10.059. [DOI] [PubMed] [Google Scholar]
- Deinlein U, Stephan AB, Horie T, Luo W, Xu GH, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014;19(6):371–379. doi: 10.1016/j.tplants.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng LX, Chen F, Jiang LP, Lam HM, Xiao GY. Ectopic expression of GmPAP3 enhances salt tolerance in rice by alleviating oxidative damage. Plant Breed. 2014;133(3):348–355. doi: 10.1111/pbr.12171. [DOI] [Google Scholar]
- Dixon DP, Skipsey M, Edwards R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry. 2010;71(4):338–350. doi: 10.1016/j.phytochem.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Do TD, Chen HT, Vu HTT, Hamwieh A, Yamada T, Sato T, Yan YL, Cong H, Shono M, Suenaga K, Xu DH. Ncl Synchronously regulates Na+, K+, and Cl- in soybean and greatly increases the grain yield in saline field conditions. Sci Rep-Uk. 2016;6:19147. doi: 10.1038/srep19147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do PT, Nguyen CX, Bui HT, Tran LTN, Stacey G, Gillman JD, Zhang ZJ, Stacey MG. Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and alpha-linolenic acid phenotype in soybean. BMC Plant Biol. 2019;19(1):311. doi: 10.1186/s12870-019-1906-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan M. Antioxidative and proline potentials as a protective mechanism in soybean plants under salinity stress. Afr J Biotech. 2011;10(32):5972–5977. doi: 10.5897/AJB10.2114. [DOI] [Google Scholar]
- Du YT, Zhao MJ, Wang CT, Gao Y, Wang YX, Liu YW, Chen M, Chen J, Zhou YB, Xu ZS, Ma YZ. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018;18:320. doi: 10.1186/s12870-018-1551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CL, Mao TT, Sun SQ, Guo XJ, Guo LX, Huang LL, Wang ZX, Zhang Y, Li M, Sheng YT, Yi YJ, Liu JY, Zhang HX, Zhang J. Constitutive expression of GmF6 ' H1 from soybean improves salt tolerance in transgenic Arabidopsis. Plant Physiol Biochem. 2019;141:446–455. doi: 10.1016/j.plaphy.2019.06.027. [DOI] [PubMed] [Google Scholar]
- Farooqi MQU, Nawaz G, Wani SH, Choudhary JR, Rana M, Sah RP, Afzal M, Zahra Z, Ganie SA, Razzaq A, Reyes VP, Mahmoud EA, Elansary HO, El-Abedin TKZ, Siddique KHM. Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.) Front Plant Sci. 2022;13:965878. doi: 10.3389/fpls.2022.965878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farvardin A, Gonzalez-Hernandez AI, Llorens E, Garcia-Agustin P, Scalschi L, Vicedo B. The apoplast: A key player in plant survival. Antioxidants. 2020;9(7):604. doi: 10.3390/antiox9070604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng ZJ, Cui XY, Cui XY, Chen M, Yang GX, Ma YZ, He GY, Xu ZS. The soybean GmDi19–5 interacts with GmLEA31 and increases sensitivity of transgenic plants to abiotic stresses. Front Plant Sci. 2015;6:179. doi: 10.3389/fpls.2015.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan QH, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, Yvon R, Kudla J, Wu HM, Cheung AY, Dinneny JR. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol. 2018;28(5):666-+. doi: 10.1016/j.cub.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Li CT, He FM, Xu YQ, Li L, Wang X, Chen QS, Li FL. Genome-wide identification of Expansin genes in wild soybean (Glycine soja) and functional characterization of Expansin B1 (GsEXPB1) in soybean hair root. Int J Mol Sci. 2022;23(10):5407. doi: 10.3390/ijms23105407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Siddique KHM, Tai APK, Anders S, Fodor N, Wong FL, Ludidi N, Chapman MA, Ferguson BJ, Considine MJ, Zabel F, Prasad PVV, Varshney RK, Nguyen HT, Lam HM. Modelling predicts that soybean is poised to dominate crop production across Africa. Plant Cell Environ. 2019;42(1):373–385. doi: 10.1111/pce.13466. [DOI] [PubMed] [Google Scholar]
- Gao SQ, Chen M, Xu ZS, Zhao CP, Li LC, Xu HJ, Tang YM, Zhao X, Ma YZ. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol Biol. 2011;75(6):537–553. doi: 10.1007/s11103-011-9738-4. [DOI] [PubMed] [Google Scholar]
- Geilfus CM. Chloride: from nutrient to toxicant. Plant Cell Physiol. 2018;59(5):877–886. doi: 10.1093/pcp/pcy071. [DOI] [PubMed] [Google Scholar]
- Gigli-Bisceglia N, Engelsdorf T, Hamann T. Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell Mol Life Sci. 2020;77(11):2049–2077. doi: 10.1007/s00018-019-03388-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48(12):909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Gnonlonfin GJB, Sanni A, Brimer L. Review Scopoletin - A Coumarin Phytoalexin with medicinal properties. Crit Rev Plant Sci. 2012;31(1):47–56. doi: 10.1080/07352689.2011.616039. [DOI] [Google Scholar]
- Golldack D, Luking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 2011;30(8):1383–1391. doi: 10.1007/s00299-011-1068-0. [DOI] [PubMed] [Google Scholar]
- Guan RX, Qu Y, Guo Y, Yu LL, Liu Y, Jiang JH, Chen JG, Ren YL, Liu GY, Tian L, Jin LG, Liu ZX, Hong HL, Chang RZ, Gilliham M, Qiu LJ. Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J. 2014;80(6):937–950. doi: 10.1111/tpj.12695. [DOI] [PubMed] [Google Scholar]
- Ha BK, Vuong TD, Velusamy V, Nguyen HT, Shannon JG, Lee JD. Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463. Euphytica. 2013;193(1):79–88. doi: 10.1007/s10681-013-0944-9. [DOI] [Google Scholar]
- Hamwieh A, Xu DH. Conserved salt tolerance quantitative trait locus (QTL) in wild and cultivated soybeans. Breed Sci. 2008;58(4):355–359. doi: 10.1270/jsbbs.58.355. [DOI] [Google Scholar]
- Hamwieh A, Tuyen DD, Cong H, Benitez ER, Takahashi R, Xu DH. Identification and validation of a major QTL for salt tolerance in soybean. Euphytica. 2011;179(3):451–459. doi: 10.1007/s10681-011-0347-8. [DOI] [Google Scholar]
- Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci. 2016;7:1787. doi: 10.3389/fpls.2016.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68(2):302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hassani A, Azapagic A, Shokri N. Global predictions of primary soil salinization under changing climate in the 21st century. Nat Commun. 2021;12(1):6663. doi: 10.1038/s41467-021-26907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, Mathis L, Voytas DF, Zhang F. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J. 2014;12(7):934–940. doi: 10.1111/pbi.12201. [DOI] [PubMed] [Google Scholar]
- He CY, Zhang JS, Chen SY. A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet. 2002;104(6–7):1125–1131. doi: 10.1007/s00122-001-0853-5. [DOI] [PubMed] [Google Scholar]
- He Y, Fu JL, Yu CL, Wang XM, Jiang QS, Hong J, Lu KX, Xue GP, Yan CQ, James A, Xu LG, Chen JP, Jiang DA. Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J Exp Bot. 2015;66(21):6877–6889. doi: 10.1093/jxb/erv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Cai HY, Bai MY, Zhang M, Chen FQ, Huang YM, Priyadarshani SVGN, Chai MN, Liu LP, Liu YH, Chen HH, Qin Y. A soybean bZIP transcription factor GmbZIP19 confers multiple biotic and abiotic stress responses in plant. Int J Mol Sci. 2020;21(13):4701. doi: 10.3390/ijms21134701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YX, Dong YS, Yang XD, Guo DQ, Qian XY, Yan F, Wang Y, Li JW, Wang QY. Functional activation of a novel R2R3-MYB protein gene, GmMYB68, confers salt-alkali resistance in soybean (Glycine max L.) Genome. 2020;63(1):13–26. doi: 10.1139/gen-2018-0132. [DOI] [PubMed] [Google Scholar]
- Hoang XLT, Chuong NN, Hoa TTK, Doan H, Van PHP, Trang LDM, Huyen PNT, Le DT, Tran LSP, Thao NP. The drought-mediated soybean GmNAC085 functions as a positive regulator of plant response to salinity. Int J Mol Sci. 2021;22(16):8986. doi: 10.3390/ijms22168986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JC, Nagao RT, Key JL. Characterization and sequence-analysis of a developmentally regulated putative cell-wall protein gene isolated from soybean. J Biol Chem. 1987;262(17):8367–8376. doi: 10.1016/S0021-9258(18)47573-4. [DOI] [PubMed] [Google Scholar]
- Houston K, Tucker MR, Chowdhury J, Shirley N, Little A. The plant cell wall: a complex and dynamic structure as revealed by the responses of genes under stress conditions. Front Plant Sci. 2016;7:984. doi: 10.3389/fpls.2016.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyten DL, Song QJ, Zhu YL, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB. Impacts of genetic bottlenecks on soybean genome diversity. P Natl Acad Sci USA. 2006;103(45):16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E, Canut H, Boudart G, Pont-Lezica RF. Cell wall proteins: a new insight through proteomics. Trends Plant Sci. 2006;11(1):33–39. doi: 10.1016/j.tplants.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Ji W, Zhu YM, Li Y, Yang LA, Zhao XW, Cai H, Bai X. Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotech Lett. 2010;32(8):1173–1179. doi: 10.1007/s10529-010-0269-x. [DOI] [PubMed] [Google Scholar]
- Ji W, Cong R, Li S, Li R, Qin ZW, Li YJ, Zhou XL, Chen SX, Li J. Comparative proteomic analysis of soybean leaves and roots by iTRAQ provides insights into response mechanisms to short-term salt stress. Front Plant Sci. 2016;7:573. doi: 10.3389/fpls.2016.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Koh J, Li S, Zhu N, Dufresne CP, Zhao XW, Chen SX, Li J. Quantitative proteomics reveals an important role of GsCBRLK in salt stress response of soybean. Plant Soil. 2016;402(1–2):159–178. doi: 10.1007/s11104-015-2782-0. [DOI] [Google Scholar]
- Jia B, Sun M, DuanMu H, Ding X, Liu B, Zhu Y, Sun X. GsCHX19.3, a member of cation/H+ exchanger superfamily from wild soybean contributes to high salinity and carbonate alkaline tolerance. Sci Rep-Uk. 2017;7(1):9423. doi: 10.1038/s41598-017-09772-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Sun S, Kong DF, Song JL, Wu LM, Yan Z, Zuo L, Yang YJ, Liang KJ, Lin WX, Huang JW. Ectopic expression of Gs5PTase8, a soybean inositol polyphosphate 5-Phosphatase, enhances salt tolerance in plants. Int J Mol Sci. 2020;21(3):1023. doi: 10.3390/ijms21031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Li MW, Zheng CW, Xu YY, Sun S, Li Z, Wong FL, Song JL, Lin WW, Li QH, Zhu YB, Liang KJ, Lin WX, Lam HM. The soybean plasma membrane-localized cation/H+ exchanger GmCHX20a plays a negative role under salt stress. Physiol Plant. 2021;171(4):714–727. doi: 10.1111/ppl.13250. [DOI] [PubMed] [Google Scholar]
- Jiang MY, Zhang JH. Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell Physiol. 2001;42(11):1265–1273. doi: 10.1093/pcp/pce162. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Bai ZZ, Xu JY, Zhao ML, Khan Y, Hu YJ, Shi LX. Metabolomics and its physiological regulation process reveal the salt tolerant mechanism in Glycine soja seedling roots. Plant Physiol Biochem. 2018;126:187–196. doi: 10.1016/j.plaphy.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Jin TC, Chang Q, Li WF, Yin DX, Li ZJ, Wang DL, Liu B, Liu LX. Stress-inducible expression of GmDREB1 conferred salt tolerance in transgenic alfalfa. Plant Cell Tissue Organ Cult. 2010;100(2):219–227. doi: 10.1007/s11240-009-9628-5. [DOI] [Google Scholar]
- Jin T, Sun YY, Zhao RR, Shan Z, Gai JY, Li Y. Overexpression of peroxidase gene GsPRX9 confers salt tolerance in soybean. Int J Mol Sci. 2019;20(15):3745. doi: 10.3390/ijms20153745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, An JX, Xu HD, Chen J, Pan L, Zhao RR, Wang N, Gai JY, Li Y. A soybean sodium/hydrogen exchanger GmNHX6 confers plant alkaline salt tolerance by regulating Na+/K+ homeostasis. Front Plant Sci. 2022;13:938635. doi: 10.3389/fpls.2022.938635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Shan Z, Zhou S, Yang QQ, Gai JY, Li Y. GmDNAJC7 from soybean is involved in plant tolerance to alkaline-salt, salt, and drought stresses. Agronomy-Basel. 2022;12(6):1419. doi: 10.3390/agronomy12061419. [DOI] [Google Scholar]
- Jose-Estanyol M, Puigdomenech P. Plant cell wall glycoproteins and their genes. Plant Physiol Biochem. 2000;38(1–2):97–108. doi: 10.1016/S0981-9428(00)00165-0. [DOI] [Google Scholar]
- Joshi A, Dang HQ, Vaid N, Tuteja N. Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconj J. 2010;27(1):133–150. doi: 10.1007/s10719-009-9265-6. [DOI] [PubMed] [Google Scholar]
- Joshi S, Kaur K, Khare T, Srivastava AK, Suprasanna P, Kumar V. Genome-wide identification, characterization and transcriptional profiling of NHX-type (Na+/H+) antiporters under salinity stress in soybean. 3 Biotech. 2021;11(1):16. doi: 10.1007/s13205-020-02555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska MM, Testerink C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015;20(9):586–594. doi: 10.1016/j.tplants.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Kader JC. Lipid-transfer proteins in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- Kaur B, Sandhu KS, Kamal R, Kaur K, Singh J, Roeder MS, Muqaddasi QH. Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: applications, challenges, and prospects. Plants. 2021;10(10):1989. doi: 10.3390/plants10101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D, He Y, Fan L, Niu R, Cheng L, Wang L, Zhang Z. The soybean TGA transcription factor GmTGA13 plays important roles in the response to salinity stress. Plant Biol. 2022;24(2):313–322. doi: 10.1111/plb.13360. [DOI] [PubMed] [Google Scholar]
- Ketehouli T, Zhou YG, Dai SY, Carther KFI, Sun DQ, Li Y, Nguyen QVH, Xu H, Wang FW, Liu WC, Li XW, Li HY. A soybean calcineurin B-like protein-interacting protein kinase, GmPKS4, regulates plant responses to salt and alkali stresses. J Plant Physiol. 2021;256:153331. doi: 10.1016/j.jplph.2020.153331. [DOI] [PubMed] [Google Scholar]
- Khan SA, Li MZ, Wang SM, Yin HJ. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int J Mol Sci. 2018;19(6):1634. doi: 10.3390/ijms19061634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Hwang S, Lee I. SoyNet: a database of co-functional networks for soybean Glycine max. Nucleic Acids Res. 2017;45(D1):D1082–D1089. doi: 10.1093/nar/gkw704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Khan GA, Somssich M, Persson S. Building a plant cell wall at a glance. J Cell Sci. 2018;131(2):jcs207373. doi: 10.1242/jcs.207373. [DOI] [PubMed] [Google Scholar]
- Lan Y, Cai D, Zheng YZ. Expression in Escherichia coli of three different soybean late embryogenesis abundant (LEA) genes to investigate enhanced stress tolerance. J Integr Plant Biol. 2005;47(5):613–621. doi: 10.1111/j.1744-7909.2005.00025.x. [DOI] [Google Scholar]
- Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C. Cell wall metabolism in response to abiotic stress. Plants. 2015;4(1):112–166. doi: 10.3390/plants4010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GJ, Boerma HR, Villagarcia MR, Zhou X, Carter TE, Li Z, Gibbs MO. A major QTL conditioning salt tolerance in S-100 soybean and descendent cultivars. Theor Appl Genet. 2004;109(8):1610–1619. doi: 10.1007/s00122-004-1783-9. [DOI] [PubMed] [Google Scholar]
- Li WY, Wong FL, Tsai SN, Phang TH, Shao G, Lam HM. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 2006;29(6):1122–1137. doi: 10.1111/j.1365-3040.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- Li WYF, Shao G, Lam HM. Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytol. 2008;178(1):80–91. doi: 10.1111/j.1469-8137.2007.02356.x. [DOI] [PubMed] [Google Scholar]
- Li ZY, Xu ZS, He GY, Yang GX, Chen M, Li LC, Ma YZ. Overexpression of soybean GmCBL1 enhances abiotic stress tolerance and promotes hypocotyl elongation in Arabidopsis. Biochem Biophys Res Commun. 2012;427(4):731–736. doi: 10.1016/j.bbrc.2012.09.128. [DOI] [PubMed] [Google Scholar]
- Li YJ, Zhang JC, Zhang J, Hao L, Hua JP, Duan LS, Zhang MC, Li ZH. Expression of an Arabidopsis molybdenum cofactor sulphurase gene in soybean enhances drought tolerance and increases yield under field conditions. Plant Biotechnol J. 2013;11(6):747–758. doi: 10.1111/pbi.12066. [DOI] [PubMed] [Google Scholar]
- Li CH, Wang G, Zhao JL, Zhang LQ, Ai LF, Han YF, Sun DY, Zhang SW, Sun Y. The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell. 2014;26(6):2538–2553. doi: 10.1105/tpc.114.125187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WB, Wang T, Zhang YH, Li YG. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J Exp Bot. 2016;67(1):175–194. doi: 10.1093/jxb/erv450. [DOI] [PubMed] [Google Scholar]
- Li XW, Wang Y, Yan F, Li JW, Zhao Y, Zhao X, Zhai Y, Wang QY. Overexpression of soybean R2R3-MYB transcription factor, GmMYB12B2, and tolerance to UV radiation and salt stress in transgenic Arabidopsis. Genet Mol Res. 2016;15(2):15026573. doi: 10.4238/gmr.15026573. [DOI] [PubMed] [Google Scholar]
- Li MX, Guo R, Jiao Y, Jin XF, Zhang HY, Shi LX. Comparison of salt tolerance in Soja based on metabolomics of seedling roots. Front Plant Sci. 2017;8:1101. doi: 10.3389/fpls.2017.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Chen QZ, Nan HY, Li XM, Lu SJ, Zhao XH, Liu BH, Guo CH, Kong FJ, Cao D. Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. Plos One. 2017;12(6):e0179554. doi: 10.1371/journal.pone.0179554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DH, Chen FJ, Li HY, Li W, Guo JJ. The soybean GmRACK1 gene plays a role in drought tolerance at vegetative stages. Russ J Plant Physiol. 2018;65(4):541–552. doi: 10.1134/S1021443718040155. [DOI] [Google Scholar]
- Li M, Hu Z, Jiang QY, Sun XJ, Guo Y, Qi JC, Zhang H. GmNAC15 overexpression in hairy roots enhances salt tolerance in soybean. J Integr Agric. 2018;17(3):530–538. doi: 10.1016/S2095-3119(17)61721-0. [DOI] [Google Scholar]
- Li B, Liu Y, Cui XY, Fu JD, Zhou YB, Zheng WJ, Lan JH, Jin LG, Chen M, Ma YZ, Xu ZS, Min DH. Genome-wide characterization and expression analysis of soybean TGA transcription factors identified a novel TGA gene involved in drought and salt tolerance. Front Plant Sci. 2019;10:549. doi: 10.3389/fpls.2019.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Wang N, Ji DD, Zhang WX, Wang Y, Yu YC, Zhao SZ, Lyu MH, You JJ, Zhang YY, Wang LL, Wang XF, Liu ZH, Tong JH, Xiao LT, Bai MY, Xiang FN. A GmSIN1/GmNCED3s/GmRbohBs feed-forward loop acts as a signal amplifier that regulates root growth in soybean exposed to salt stress. Plant Cell. 2019;31(9):2107–2130. doi: 10.1105/tpc.18.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zheng JC, Wang TT, Min DH, Wei WL, Chen J, Zhou YB, Chen M, Xu ZS, Ma YZ. Expression analyses of soybean VOZ transcription factors and the role of GmVOZ1G in drought and salt stress tolerance. Int J Mol Sci. 2020;21(6):2177. doi: 10.3390/ijms21062177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liu XY, Ruan H, Zhang JY, Xie FB, Gai JY, Yang SP. GmWRKY45 enhances tolerance to phosphate starvation and salt stress, and changes fertility in transgenic Arabidopsis. Front Plant Sci. 2020;10:1714. doi: 10.3389/fpls.2019.01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Zhou HP, Zhang Y, Li Z, Yang YQ, Guo Y. The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev Cell. 2020;55(3):367–380. doi: 10.1016/j.devcel.2020.08.005. [DOI] [PubMed] [Google Scholar]
- Li M-W, Wang Z, Jiang B, Kaga A, Wong F-L, Zhang G, Han T, Chung G, Nguyen H, Lam H-M. Impacts of genomic research on soybean improvement in East Asia Abstract. Theor Appl Genet. 2020;133(5):1655–1678. doi: 10.1007/s00122-019-03462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KP, Wong CH, Cheng CC, Cheng SS, Li MW, Mansveld S, Bergsma A, Huang TF, van Eijk MJT, Lam HM. GmDNJ1, a type-I heat shock protein 40 (HSP40), is responsible for both Growth and heat tolerance in soybean. Plant Direct. 2021;5(1):e00298. doi: 10.1002/pld3.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen R, Jiang QY, Sun XJ, Zhang H, Hu Z. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol Biol. 2021;105(3):333–345. doi: 10.1007/s11103-020-01091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu ZN, Li Q, Zhu WL, Wang XH, Xu P, Cao X, Cui XY. CBL-Interacting protein kinase 2 improves salt tolerance in soybean (Glycine max L.) Agronomy. 2022;12(7):1595. doi: 10.3390/agronomy12071595. [DOI] [PubMed] [Google Scholar]
- Li H, Wang XH, Li Q, Xu P, Liu ZN, Xu M, Cui XY. GmCIPK21, a CBL-interacting protein kinase confers salt tolerance in soybean (Glycine max. L) Plant Physiol Biochem. 2022;184:47–55. doi: 10.1016/j.plaphy.2022.05.027. [DOI] [PubMed] [Google Scholar]
- Liao H, Wong FL, Phang TH, Cheung MY, Li WY, Shao G, Yan X, Lam HM. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene. 2003;318:103–111. doi: 10.1016/s0378-1119(03)00764-9. [DOI] [PubMed] [Google Scholar]
- Liao Y, Zhang JS, Chen SY, Zhang WK. Role of soybean GmbZIP132 under abscisic acid and salt stresses. J Integr Plant Biol. 2008;50(2):221–230. doi: 10.1111/j.1744-7909.2007.00593.x. [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS, Chen SY. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008;18(10):1047–1060. doi: 10.1038/cr.2008.280. [DOI] [PubMed] [Google Scholar]
- Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228(2):225–240. doi: 10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- Liao XL, Shi MQ, Zhang W, Ye Q, Li YL, Feng XZ, Bhat JA, Kan GZ, Yu DY. Association analysis of GmMAPKs and functional characterization of GmMMK1 to salt stress response in soybean. Physiol Plant. 2021;173(4):2026–2040. doi: 10.1111/ppl.13549. [DOI] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu Rev Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- Lin WW, Tang WX, Pan X, Huang AB, Gao XQ, Anderson CT, Yang ZB. Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling. Curr Biol. 2022;32(3):497–507. doi: 10.1016/j.cub.2021.11.030. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zheng YZ. PM2, a group 3 LEA protein from soybean, and its 22-mer repeating region confer salt tolerance in Escherichia coli. Biochem Biophys Res Commun. 2005;331(1):325–332. doi: 10.1016/j.bbrc.2005.03.165. [DOI] [PubMed] [Google Scholar]
- Liu AL, Yu Y, Li RT, Duan XB, Zhu D, Sun XL, Duanmu HZ, Zhu YM. A novel hybrid proline-rich type gene GsEARLI17 from Glycine soja participated in leaf cuticle synthesis and plant tolerance to salt and alkali stresses. Plant Cell Tissue Organ Cult. 2015;121(3):633–646. doi: 10.1007/s11240-015-0734-2. [DOI] [Google Scholar]
- Liu Y, Yu LL, Qu Y, Chen JL, Liu XX, Hong HL, Liu ZX, Chang RZ, Gilliham M, Qiu LJ, Guan RX. GmSALT3, which confers improved soybean salt tolerance in the field, increases leaf Cl- exclusion prior to Na+ Exclusion but does not improve early vigor under salinity. Front Plant Sci. 2016;7:1485. doi: 10.3389/fpls.2016.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SM, Kandoth PK, Lakhssassi N, Kang JW, Colantonio V, Heinz R, Yeckel G, Zhou Z, Bekal S, Dapprich J, Rotter B, Cianzio S, Mitchum MG, Meksem K. The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat Commun. 2017;8:14822. doi: 10.1038/ncomms14822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AL, Xiao ZX, Li MW, Wong FL, Yung WS, Ku YS, Wang QW, Wang X, Xie M, Yim AKY, Chan TF, Lam HM. Transcriptomic reprogramming in soybean seedlings under salt stress. Plant Cell Environ. 2019;42(1):98–114. doi: 10.1111/pce.13186. [DOI] [PubMed] [Google Scholar]
- Liu YC, Du HL, Li PC, Shen YT, Peng H, Liu SL, Zhou GA, Zhang HK, Liu Z, Shi M, Huang XH, Li Y, Zhang M, Wang Z, Zhu BG, Han B, Liang CZ, Tian ZX. Pan-genome of wild and cultivated soybeans. Cell. 2020;182(1):162-+. doi: 10.1016/j.cell.2020.05.023. [DOI] [PubMed] [Google Scholar]
- Liu JW, Zhang W, Long SJ, Zhao CZ. Maintenance of cell wall integrity under high salinity. Int J Mol Sci. 2021;22(6):3260. doi: 10.3390/ijms22063260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang XX, Zhang B. Transcriptome analysis and functional identification of GmMYB46 in soybean seedlings under salt stress. Peerj. 2021;9:e12492. doi: 10.7717/peerj.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Pi BY, Du ZY, Yang T, Gu MY, Sun SL, Yu BJ. The transcription factor GmbHLH3 confers Cl-/salt tolerance to soybean by upregulating GmCLC1 expression for maintenance of anion homeostasis. Environ Exp Bot. 2022;194:104755. doi: 10.1016/j.envexpbot.2021.104755. [DOI] [Google Scholar]
- Loix C, Huybrechts M, Vangronsveld J, Gielen M, Keunen E, Cuypers A. Reciprocal interactions between cadmium-induced cell wall responses and oxidative stress in plants. Front Plant Sci. 2017;8:1867. doi: 10.3389/fpls.2017.01867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cruz M, Crossa J, Bonnett D, Dreisigacker S, Poland J, Jannink JL, Singh RP, Autrique E, de los Campos G. Increased prediction accuracy in wheat breeding trials using a marker x environment interaction genomic selection model. G3-Genes Genomes Genet. 2015;5(4):569–582. doi: 10.1534/g3.114.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QSM, Tian LN. An efficient and specific CRISPR-Cas9 genome editing system targeting soybean phytoene desaturase genes. BMC Biotechnol. 2022;22(1):7. doi: 10.1186/s12896-022-00737-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Wei W, Tao JJ, Lu X, Bian XH, Hu Y, Cheng T, Yin CC, Zhang WK, Chen SY, Zhang JS. Nuclear factor Y subunit GmNFYA competes with GmHDA13 for interaction with GmFVE to positively regulate salt tolerance in soybean. Plant Biotechnol J. 2021;19(11):2362–2379. doi: 10.1111/pbi.13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, Wang HW, Huang J, Tian AG, Wang YJ, Zhang JS, Chen SY. A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol Biol. 2005;59(5):809–820. doi: 10.1007/s11103-005-1386-0. [DOI] [PubMed] [Google Scholar]
- Lupini A, Araniti F, Sunseri F, Abenavoli MR. Coumarin interacts with auxin polar transport to modify root system architecture in Arabidopsis thaliana. Plant Growth Regul. 2014;74(1):23–31. doi: 10.1007/s10725-014-9893-0. [DOI] [Google Scholar]
- Lv DW, Subburaj S, Cao M, Yan X, Li XH, Appels R, Sun DF, Ma WJ, Yan YM. Proteome and phosphoproteome characterization reveals new response and defense mechanisms of Brachypodium distachyon leaves under salt stress. Mol Cell Proteomics. 2014;13(2):632–652. doi: 10.1074/mcp.M113.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QB, Xia ZL, Cai ZD, Li L, Cheng YB, Liu J, Nian H. GmWRKY16 enhances drought and salt tolerance through an ABA-Mediated pathway in Arabidopsis thaliana. Front Plant Sci. 2019;9:1979. doi: 10.3389/fpls.2018.01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XJ, Fu JD, Tang YM, Yu TF, Yin ZG, Chen J, Zhou YB, Chen M, Xu ZS, Ma YZ. GmNFYA13 improves salt and drought tolerance in transgenic soybean plants. Front Plant Sci. 2020;11:587244. doi: 10.3389/fpls.2020.587244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM. Sodium in plants: perception, signalling, and regulation of sodium fluxes. J Exp Bot. 2014;65(3):849–858. doi: 10.1093/jxb/ert326. [DOI] [PubMed] [Google Scholar]
- Mao YF, Botella JR, Liu YG, Zhu JK. Gene editing in plants: progress and challenges. Natl Sci Rev. 2019;6(3):421–437. doi: 10.1093/nsr/nwz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura H, Ishibashi Y, Shinmyo A, Kanaya S, Kato K. Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol. 2010;51(3):448–462. doi: 10.1093/pcp/pcq010. [DOI] [PubMed] [Google Scholar]
- Melo BP, Fraga OT, Silva JCF, Ferreira DO, Brustolini OJB, Carpinetti PA, Machado JPB, Reis PAB, Fontes EPB. Revisiting the soybean GmNAC superfamily. Front Plant Sci. 2018;9:1864. doi: 10.3389/fpls.2018.01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MM, Huang SL, Hill RD, Stasolla C. Tolerance to excess moisture in soybean is enhanced by over-expression of the Glycine max Phytoglobin (GmPgb1) Plant Physiol Biochem. 2021;159:322–334. doi: 10.1016/j.plaphy.2020.12.033. [DOI] [PubMed] [Google Scholar]
- Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, Bose J, Byrt CS, Chen ZH, Foster KJ, Gilliham M, Henderson SW, Jenkins CLD, Kronzucker HJ, Miklavcic SJ, Plett D, Roy SJ, Shabala S, Shelden MC, Soole KL, Taylor NL, Tester M, Wege S, Wegner LH, Tyerman SD. Energy costs of salt tolerance in crop plants. New Phytol. 2020;225(3):1072–1090. doi: 10.1111/nph.15864. [DOI] [PubMed] [Google Scholar]
- Nawaz MA, Rehman HM, Imtiaz M, Baloch FS, Lee JD, Yang SH, Lee SI, Chung G. Systems identification and characterization of cell wall reassembly and degradation related genes in Glycine max (L.) Merill, a bioenergy Legume. Sci Rep-Uk. 2017;7:10862. doi: 10.1038/s41598-017-11495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen QH, Vu LTK, Nguyen LTN, Pham NTT, Nguyen YTH, Le SV, Chu MH. Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci Rep-Uk. 2019;9:19663. doi: 10.1038/s41598-019-55895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie WX, Xu L, Yu BJ. A putative soybean GmSOS1 confers enhanced salt tolerance to transgenic Arabidopsis sos1-1 mutant. Protoplasma. 2015;252(1):127–134. doi: 10.1007/s00709-014-0663-7. [DOI] [PubMed] [Google Scholar]
- Nissen KS, Willats WGT, Malinovsky FG. Understanding CrRLK1L Function: cell walls and growth control. Trends Plant Sci. 2016;21(6):516–527. doi: 10.1016/j.tplants.2015.12.004. [DOI] [PubMed] [Google Scholar]
- Oda Y. Cortical microtubule rearrangements and cell wall patterning. Front Plant Sci. 2015;6:236. doi: 10.3389/fpls.2015.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WJ, Tao JJ, Cheng T, Bian XH, Wei W, Zhang WK, Ma B, Chen SY, Zhang JS. Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol Plant. 2016;9(9):1337–1340. doi: 10.1016/j.molp.2016.05.010. [DOI] [PubMed] [Google Scholar]
- Parmley KA, Higgins RH, Ganapathysubramanian B, Sarkar S, Singh AK. Machine learning approach for prescriptive plant breeding. Sci Rep-Uk. 2019;9:17132. doi: 10.1038/s41598-019-53451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil G, Do T, Vuong TD, Valliyodan B, Lee JD, Chaudhary J, Shannon JG, Nguyen HT. Genomic-assisted haplotype analysis and the development of high-throughput SNP markers for salinity tolerance in soybean. Sci Rep-Uk. 2016;6:19199. doi: 10.1038/srep19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang TH, Shao GH, Lam HM. Salt tolerance in soybean. J Integr Plant Biol. 2008;50(10):1196–1212. doi: 10.1111/j.1744-7909.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- Pi EX, Qu LQ, Hu JW, Huang YY, Qiu LJ, Lu H, Jiang B, Liu C, Peng TT, Zhao Y, Wang HZ, Tsai SN, Ngai SM, Du LQ. Mechanisms of soybean roots' tolerances to salinity revealed by proteomic and phosphoproteomic comparisons between two cultivars. Mol Cell Proteomics. 2016;15(1):266–288. doi: 10.1074/mcp.M115.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi EX, Zhu CM, Fan W, Huang YY, Qu LQ, Li YY, Zhao QY, Ding F, Qiu LJ, Wang HZ, Poovaiah BW, Du LQ. Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol Cell Proteomics. 2018;17(6):1209–1224. doi: 10.1074/mcp.RA117.000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi EX, Xu J, Li HH, Fan W, Zhu CM, Zhang TY, Jiang JC, He LT, Lu HF, Wang HZ, Poovaiah BW, Du LQ. Enhanced Salt Tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis. Mol Cell Proteomics. 2019;18(11):2225–2243. doi: 10.1074/mcp.RA119.001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegon-Putze I, Bosch N, Ibanes M, Cano-Delgado AI. Brassinosteroid signaling in plant development and adaptation to stress. Development. 2019;146(5):dev151894. doi: 10.1242/dev.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats E, Llamas MJ, Jorrin J, Rubiales D. Constitutive coumarin accumulation on sunflower leaf surface prevents rust germ tube growth and appressorium differentiation. Crop Sci. 2007;47(3):1119–1124. doi: 10.2135/cropsci2006.07.0482. [DOI] [Google Scholar]
- Qi XP, Li MW, Xie M, Liu X, Ni M, Shao GH, Song C, Yim AKY, Tao Y, Wong FL, Isobe S, Wong CF, Wong KS, Xu CY, Li CQ, Wang Y, Guan R, Sun FM, Fan GY, Xiao ZX, Zhou F, Phang TH, Liu X, Tong SW, Chan TF, Yiu SM, Tabata S, Wang J, Xu X, Lam HM. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun. 2014;5:4340. doi: 10.1038/ncomms5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. P Natl Acad Sci USA. 2002;99(12):8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Guan RX, Bose J, Henderson SW, Wege S, Qiu LJ, Gilliham M. Soybean CHX-type ion transport protein GmSALT3 confers leaf Na+ exclusion via a root derived mechanism, and Cl- exclusion via a shoot derived process. Plant Cell Environ. 2021;44(3):856–869. doi: 10.1111/pce.13947. [DOI] [PubMed] [Google Scholar]
- Rahman MU, Zulfiqar S, Raza MA, Ahmad N, Zhang BH. Engineering abiotic stress tolerance in crop plants through CRISPR genome editing. Cells. 2022;11(22):3590. doi: 10.3390/cells11223590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, El-Habbak MH, Havens WM, Singh A, Zheng DM, Vaughn L, Haudenshield JS, Hartman GL, Korban SS, Ghabrial SA. Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol Plant Pathol. 2014;15(2):145–160. doi: 10.1111/mpp.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman HM, Chen SJ, Zhang SD, Khalid M, Uzair M, Wilmarth PA, Ahmad S, Lam HM. Membrane proteomic profiling of soybean leaf and root tissues uncovers salt-stress-responsive membrane proteins. Int J Mol Sci. 2022;23(21):13270. doi: 10.3390/ijms232113270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren SX, Lyle C, Jiang GL, Penumala A. Soybean salt tolerance 1 (GmST1) reduces ROS production, enhances ABA sensitivity, and abiotic stress tolerance in Arabidopsis thaliana. Front Plant Sci. 2016;7:445. doi: 10.3389/fpls.2016.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengasamy P. Soil processes affecting crop production in salt-affected soils. Funct Plant Biol. 2010;37(7):613–620. doi: 10.1071/Fp09249. [DOI] [Google Scholar]
- Rodrigues SM, Andrade MO, Gomes APS, DaMatta FM, Baracat-Pereira MC, Fontes EPB. Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J Exp Bot. 2006;57(9):1909–1918. doi: 10.1093/jxb/erj132. [DOI] [PubMed] [Google Scholar]
- Sakata K, Ohyanagi H, Nobori H, Nakamura T, Hashiguchi A, Nanjo Y, Mikami Y, Yunokawa H, Komatsu S. Soybean proteome database: a data resource for plant differential omics. J Proteome Res. 2009;8(7):3539–3548. doi: 10.1021/pr900229k. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, Fisahn J, Segundo BS, Guiderdoni E, Schippers JHM, Mueller-Roeber B. SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell. 2013;25(6):2115–2131. doi: 10.1105/tpc.113.113068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Scossa F, Alseekh S, Fernie AR. Integrating multi-omics data for crop improvement. J Plant Physiol. 2021;257:153352. doi: 10.1016/j.jplph.2020.153352. [DOI] [PubMed] [Google Scholar]
- Shan BH, Wang W, Cao JF, Xia SQ, Li RH, Bian SM, Li XY. Soybean GmMYB133 inhibits hypocotyl elongation and confers salt tolerance in arabidopsis. Front Plant Sci. 2021;12:764074. doi: 10.3389/fpls.2021.764074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Yang Y, Sharma A, Awasthi S, Awasthi YC. Antioxidant role of glutathione S-transferases: Protection against oxidant toxicity and regulation of stress-mediated apoptosis. Antioxid Redox Signal. 2004;6(2):289–300. doi: 10.1089/152308604322899350. [DOI] [PubMed] [Google Scholar]
- Shelake RM, Kadam US, Kumar R, Pramanik D, Singh AK, Kim J-Y. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 2022;3(6):100417. doi: 10.1016/j.xplc.2022.100417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XJ, Wang YY, Zhang YX, Guo W, Jiao YQ, Zhou XA. Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa Armigera in Transgenic Arabidopsis. Int J Mol Sci. 2018;19(12):3958. doi: 10.3390/ijms19123958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YT, Liu J, Geng HY, Zhang JX, Liu YC, Zhang HK, Xing SL, Du JC, Ma SS, Tian ZX. De novo assembly of a Chinese soybean genome. Sci China-Life Sci. 2018;61(8):871–884. doi: 10.1007/s11427-018-9360-0. [DOI] [PubMed] [Google Scholar]
- Shi WY, Du YT, Ma J, Min DH, Jin LG, Chen J, Chen M, Zhou YB, Ma YZ, Xu ZS, Zhang XH. The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int J Mol Sci. 2018;19(12):4087. doi: 10.3390/ijms19124087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava P, Kumar R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci. 2015;22(2):123–131. doi: 10.1016/j.sjbs.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Lone RA. Chapter 3 - High-affinity K+ transporters and their functions in plants. In: Upadhyay SK, editor. Cation transporters in plants. Academic Press; 2022. pp. 49–61. [Google Scholar]
- Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89–101. doi: 10.1016/j.freeradbiomed.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton PW, Bohlool BB. Effect of salinity on nodule formation by soybean. Plant Physiol. 1984;74(1):72–76. doi: 10.1104/pp.74.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhanian H, Razavizadeh R, Nanjo Y, Ehsanpour AA, Jazii FR, Motamed N, Komatsu S. Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Science. 2010;8:19. doi: 10.1186/1477-5956-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Bauer S, Brininstool G, Facette M, Hamann T, Milne J, Osborne E, Paredez A, Persson S, Raab T, Vorwerk S, Youngs H. Toward a systems approach to understanding plant-cell walls. Science. 2004;306(5705):2206–2211. doi: 10.1126/science.1102765. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wang D, Bai Y, Wang N, Wang Y. Studies on the overexpression of the soybean GmNHX1 in Lotus corniculatus: The reduced Na+ level is the basis of the increased salt tolerance. Chin Sci Bull. 2006;51(11):1306–1315. doi: 10.1007/s11434-006-1306-y. [DOI] [Google Scholar]
- Sun XL, Sun MZ, Luo X, Ding XD, Ji W, Cai H, Bai X, Liu XF, Zhu YM. A Glycine soja ABA-responsive receptor-like cytoplasmic kinase, GsRLCK, positively controls plant tolerance to salt and drought stresses. Planta. 2013;237(6):1527–1545. doi: 10.1007/s00425-013-1864-6. [DOI] [PubMed] [Google Scholar]
- Sun XL, Yu QY, Tang LL, Ji W, Bai X, Cai H, Liu XF, Ding XD, Zhu YM. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J Plant Physiol. 2013;170(5):505–515. doi: 10.1016/j.jplph.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Sun HH, Wang L, Zhang BQ, Ma JH, Hettenhausen C, Cao GY, Sun GL, Wu JQ, Wu JS. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J Exp Bot. 2014;65(15):4305–4315. doi: 10.1093/jxb/eru203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MZ, Jia BW, Cui N, Wen YD, Duanmu HZ, Yu QY, Xiao JL, Sun XL, Zhu YM. Functional characterization of a Glycine soja Ca2+ATPase in salt-alkaline stress responses. Plant Mol Biol. 2016;90(4–5):419–434. doi: 10.1007/s11103-015-0426-7. [DOI] [PubMed] [Google Scholar]
- Sun T-J, Fan L, Yang J, Cao R-Z, Yang C-Y, Zhang J, Wang D-M. A Glycine max sodium/hydrogen exchanger enhances salt tolerance through maintaining higher Na+ efflux rate and K+/Na+ ratio in Arabidopsis. BMC Plant Biol. 2019;19(1):469. doi: 10.1186/s12870-019-2084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZX, Liu RH, Guo B, Huang KS, Wang L, Han YH, Li HY, Hou SY. Ectopic expression of GmZAT4, a putative C2H2-type zinc finger protein, enhances PEG and NaCl stress tolerances in Arabidopsis thaliana. 3 Biotech. 2019;9(5):166. doi: 10.1007/s13205-019-1673-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TJ, Ma N, Wang CQ, Fan HF, Wang MX, Zhang J, Cao JF, Wang DM. A Golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front Plant Sci. 2021;12:638340. doi: 10.3389/fpls.2021.638340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenhaken R. Cell wall remodeling under abiotic stress. Front Plant Sci. 2015;5:771. doi: 10.3389/fpls.2014.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Nikoloski Z. Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data. J Plant Physiol. 2021;257:153354. doi: 10.1016/j.jplph.2020.153354. [DOI] [PubMed] [Google Scholar]
- Tran LSP, Mochida K. Functional genomics of soybean for improvement of productivity in adverse conditions. Funct Integr Genomics. 2010;10(4):447–462. doi: 10.1007/s10142-010-0178-z. [DOI] [PubMed] [Google Scholar]
- Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94(10):791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- Valliyodan B, Cannon SB, Bayer PE, Shu SQ, Brown AV, Ren LH, Jenkins J, Chung CYL, Chan TF, Daum CG, Plott C, Hastie A, Baruch K, Barry KW, Huang W, Patil G, Varshney RK, Hu HF, Batley J, Yuan YX, Song QJ, Stupar RM, Goodstein DM, Stacey G, Lam HM, Jackson SA, Schmutz J, Grimwood J, Edwards D, Nguyen HT. Construction and comparison of three reference-quality genome assemblies for soybean. Plant J. 2019;100(5):1066–1082. doi: 10.1111/tpj.14500. [DOI] [PubMed] [Google Scholar]
- van Zelm E, Zhang YX, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020;71:403–433. doi: 10.1146/annurev-arplant-050718-100005. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Bohra A, Roorkiwal M, Barmukh R, Cowling WA, Chitikineni A, Lam HM, Hickey LT, Croser JS, Bayer PE, Edwards D, Crossa J, Weckwerth W, Millar H, Kumar A, Bevan MW, Siddique KHM. Fast-forward breeding for a food-secure world. Trends Genet. 2021;37(12):1124–1136. doi: 10.1016/j.tig.2021.08.002. [DOI] [PubMed] [Google Scholar]
- Voxeur A, Hofte H. Cell wall integrity signaling in plants: "To grow or not to grow that's the question". Glycobiology. 2016;26(9):950–960. doi: 10.1093/glycob/cww029. [DOI] [PubMed] [Google Scholar]
- Wang M-J, Hou W-S, Wang Q-Y, Lam H-M, Han T-F. Enhancing salt tolerance of soybean roots by overexpression of GmNHX1. Soybean Science. 2011;30:889–894. [Google Scholar]
- Wang YX, Suo HC, Zhuang CX, Ma H, Yan XL. Overexpression of the soybean GmWNK1 altered the sensitivity to salt and osmotic stress in Arabidopsis. J Plant Physiol. 2011;168(18):2260–2267. doi: 10.1016/j.jplph.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Wang HM, Zhou L, Fu YP, Cheung MY, Wong FL, Phang TH, Sun ZX, Lam HM. Expression of an apoplast-localized BURP-domain protein from soybean (GmRD22) enhances tolerance towards abiotic stress. Plant Cell Environ. 2012;35(11):1932–1947. doi: 10.1111/j.1365-3040.2012.02526.x. [DOI] [PubMed] [Google Scholar]
- Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, Ma B, Bi YD, Lai YC, Liu XL, Man WQ, Zhang JS, Chen SY. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 2015;83(2):224–236. doi: 10.1111/tpj.12879. [DOI] [PubMed] [Google Scholar]
- Wang X, Liu FL, Jiang D. Priming: A promising strategy for crop production in response to future climate. J Integr Agric. 2017;16(12):2709–2716. doi: 10.1016/S2095-3119(17)61786-6. [DOI] [Google Scholar]
- Wang LS, Chen QS, Xin DW, Qi ZM, Zhang C, Li SN, Jin YM, Li M, Mei HY, Su AY, Wu XX. Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots. J Integr Agric. 2018;17(9):1959–1971. doi: 10.1016/S2095-3119(17)61863-X. [DOI] [Google Scholar]
- Wang ZK, Yang Q, Shao YP, Zhang BB, Feng AY, Meng FL, Li WB. GmLEA2–1, a late embryogenesis abundant protein gene isolated from soybean (Glycine Max (L.) Merr.), confers tolerance to abiotic stress. Acta Biologica Hungarica. 2018;69(3):270–282. doi: 10.1556/018.68.2018.3.4. [DOI] [PubMed] [Google Scholar]
- Wang D, Liu YX, Yu Q, Zhao SP, Zhao JY, Ru JN, Cao XY, Fang ZW, Chen J, Zhou YB, Chen M, Ma YZ, Xu ZS, Lan JH. Functional Analysis of the Soybean GmCDPK3 Gene Responding to Drought and Salt Stresses. Int J Mol Sci. 2019;20(23):5909. doi: 10.3390/ijms20235909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang FB, Ren XQ, Zhang F, Qi MY, Zhao HY, Chen XH, Ye YX, Yang JY, Li SG, Zhang Y, Niu Y, Zhou Q. A R2R3-type MYB transcription factor gene from soybean, GmMYB12, is involved in flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis. Plant Biotechnology Reports. 2019;13(3):219–233. doi: 10.1007/s11816-019-00530-7. [DOI] [Google Scholar]
- Wang TT, Yu TF, Fu JD, Su HG, Chen J, Zhou YB, Chen M, Guo J, Ma YZ, Wei WL, Xu ZS. Genome-wide analysis of the GRAS gene family and functional identification of GmGRAS37 in drought and salt tolerance. Front Plant Sci. 2020;11:604690. doi: 10.3389/fpls.2020.604690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TY, Xun HW, Wang W, Ding XY, Tian HA, Hussain S, Dong QL, Li YY, Cheng YX, Wang C, Lin R, Li GM, Qian XY, Pang JS, Feng XZ, Dong YS, Liu B, Wang SC. Mutation of GmAITR Genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front Plant Sci. 2021;12:598. doi: 10.3389/fpls.2021.779598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Yu TF, Sun GZ, Zheng JC, Chen J, Zhou YB, Chen M, Ma YZ, Wei WL, Xu ZS. Genome-wide analysis of the Catharanthus roseus RLK1-like in soybean and GmCrRLK1L20 responds to drought and salt stresses. Front Plant Sci. 2021;12:614909. doi: 10.3389/fpls.2021.614909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Chen K, Zhou MM, Gao YK, Huang HM, Liu C, Fan YY, Fan ZH, Wang YN, Li X. GmNAC181 promotes symbiotic nodulation and salt tolerance of nodulation by directly regulating GmNINa expression in soybean. New Phytol. 2022;236(2):656–670. doi: 10.1111/nph.18343. [DOI] [PubMed] [Google Scholar]
- Wei W, Huang J, Hao YJ, Zou HF, Wang HW, Zhao JY, Liu XY, Zhang WK, Ma B, Zhang JS, Chen SY. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. Plos One. 2009;4(9):e7209. doi: 10.1371/journal.pone.0007209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei PP, Wang LC, Liu AL, Yu BJ, Lam HM. GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front Plant Sci. 2016;7:1082. doi: 10.3389/fpls.2016.01082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei PP, Che BN, Shen LK, Cui YQ, Wu SY, Cheng C, Liu F, Li MW, Yu BJ, Lam HM. Identification and functional characterization of the chloride channel gene, GsCLC-c2 from wild soybean. BMC Plant Biol. 2019;19:121. doi: 10.1186/s12870-019-1732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler G, Ishikawa T, Pornsaksit V, Smirnoff N. Evolution of alternative biosynthetic pathways for vitamin C following plastid acquisition in photosynthetic eukaryotes. Elife. 2015;4:e06369. doi: 10.7554/eLife.06369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A. POPP the question: what do LEA proteins do? Trends Plant Sci. 2004;9(1):13–17. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Wolf S, Mravec J, Greiner S, Mouille G, Hofte H. Plant cell wall homeostasis is mediated by Brassinosteroid feedback signaling. Curr Biol. 2012;22(18):1732–1737. doi: 10.1016/j.cub.2012.07.036. [DOI] [PubMed] [Google Scholar]
- Wong TH, Li MW, Yao XQ, Lam HM. The GmCLC1 protein from soybean functions as a chloride ion transporter. J Plant Physiol. 2013;170(1):101–104. doi: 10.1016/j.jplph.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Wu HH, Li ZH. The importance of Cl- Exclusion and vacuolar Cl- Sequestration: revisiting the role of Cl- Transport in plant salt tolerance. Front Plant Sci. 2019;10:1418. doi: 10.3389/fpls.2019.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZM, Zou HF, Lei G, Wei W, Zhou QY, Niu CF, Liao Y, Tian AG, Ma B, Zhang WK, Zhang JS, Chen SY. Soybean trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic arabidopsis. Plos One. 2009;4(9):e6898. doi: 10.1371/journal.pone.0006898. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xie M, Chung CYL, Li MW, Wong FL, Wang X, Liu AL, Wang ZL, Leung AKY, Wong TH, Tong SW, Xiao ZX, Fan KJ, Ng MS, Qi XP, Yang LF, Deng TQ, He LJ, Chen L, Fu AS, Ding Q, He JX, Chung G, Isobe S, Tanabata T, Valliyodan B, Nguyen HT, Cannon SB, Foyer CH, Chan TF, Lam HM. A reference-grade wild soybean genome. Nat Commun. 2019;10:1216. doi: 10.1038/s41467-019-09142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Fan XH, Wang Q, Yin ZG, Sheng XW, Chen J, Zhou YB, Chen M, Ma YZ, Ma J, Xu ZS. Genomic analysis of soybean PP2A-B’’ family and its effects on drought and salt tolerance. Front Plant Sci. 2022;12:784038. doi: 10.3389/fpls.2021.784038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XY, Fan R, Zheng R, Li CM, Yu DY. Proteomic analysis of seed germination under salt stress in soybeans. J Zhejiang Univ Sci B. 2011;12(7):507–517. doi: 10.1631/jzus.B1100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZL, Ali Z, Yi JX, Xu L, Liu XQ, He XL, Huang YH, Karim I, Rasul M, Ma HX, Zhang DY. Over-expression of GmWRKY111 Enhances NaCl Tolerance of Salt-Sensitive Genotype of Glycine max. Int J Agric Biol. 2014;16(1):153–159. [Google Scholar]
- Xu ZL, Ali Z, Xu L, He XL, Huang YH, Yi JX, Shao HB, Ma HX, Zhang DY. The nuclear protein GmbZIP110 has transcription activation activity and plays important roles in the response to salinity stress in soybean. Sci Rep-Uk. 2016;6:20366. doi: 10.1038/srep20366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZL, Raza Q, Xu L, He XL, Huang YH, Yi JX, Zhang DY, Shao HB, Ma HX, Ali Z. GmWRKY49, a Salt-Responsive Nuclear Protein, Improved Root Length and Governed Better Salinity Tolerance in Transgenic Arabidopsis. Front Plant Sci. 2018;9:809. doi: 10.3389/fpls.2018.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhang LX, Zhang K, Ran YD. Progresses, Challenges, and Prospects of Genome Editing in Soybean (Glycine max) Front Plant Sci. 2020;11:571138. doi: 10.3389/fpls.2020.571138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Shan J, Liu T, Wang Q, Ji Y, Zhang Y, Wang M, Xia N, Zhao L. CONSTANS-LIKE 1a positively regulates salt and drought tolerance in soybean. Plant Physiol. 2022 doi: 10.1093/plphys/kiac573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue CC, Xu JY, Wang C, Guo N, Hou JF, Xue D, Zhao JM, Xing H. Molecular cloning and functional characterization of a soybean GmGMP1 gene reveals its involvement in ascorbic acid biosynthesis and multiple abiotic stress tolerance in transgenic plants. J Integr Agric. 2018;17(3):539–553. doi: 10.1016/S2095-3119(17)61727-1. [DOI] [Google Scholar]
- Yang YQ, Guo Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018;217(2):523–539. doi: 10.1111/nph.14920. [DOI] [PubMed] [Google Scholar]
- Yang LA, Ji W, Zhu YM, Gao P, Li Y, Cai H, Bai X, Guo DJ. GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J Exp Bot. 2010;61(9):2519–2533. doi: 10.1093/jxb/erq084. [DOI] [PubMed] [Google Scholar]
- Yang XF, Kim MY, Ha J, Lee SH. Overexpression of the soybean NAC Gene GmNAC109 increases lateral root formation and abiotic stress tolerance in transgenic Arabidopsis plants. Front Plant Sci. 2019;10:1036. doi: 10.3389/fpls.2019.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang QS, Yang B, Li JZ, Wang Y, Tao RY, Yang F, Wu XY, Yan XH, Ahmad M, Shen JQ, Bai SL, Teng YW. ABA-responsive ABRE-BINDING FACTOR3 activates DAM3 expression to promote bud dormancy in Asian pear. Plant Cell Environ. 2020;43(6):1360–1375. doi: 10.1111/pce.13744. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yu TF, Ma J, Chen J, Zhou YB, Chen M, Ma YZ, Wei WL, Xu ZS. The soybean bZIP transcription factor gene GmbZIP2 Confers drought and salt resistances in transgenic plants. Int J Mol Sci. 2020;21(2):670. doi: 10.3390/ijms21020670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YD, Saand MA, Huang LY, Abdelaal WB, Zhang J, Wu Y, Li J, Sirohi MH, Wang FY. Applications of multi-omics technologies for crop improvement. Front Plant Sci. 2021;12:563953. doi: 10.3389/fpls.2021.563953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Karthikeyan A, Yin JL, Jin TT, Ren R, Fang F, Cai H, Liu MZ, Wang DG, Li K, Zhi HJ. The E3 Ligase GmPUB21 negatively regulates drought and salinity stress response in soybean. Int J Mol Sci. 2022;23(13):6893. doi: 10.3390/ijms23136893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH, Varner JE. Tissue-specific expression of cell-wall proteins in developing soybean tissues. Plant Cell. 1991;3(1):23–37. doi: 10.2307/3869197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BJ, Lam HM, Shao GH, Liu YL. Effects of salinity on activities of H+ -ATPase, H+-PPase and membrane lipid composition in plasma membrane and tonoplast vesicles isolated from soybean (Glycine max L.) seedlings. J Environ Sci. 2005;17(2):259–262. [PubMed] [Google Scholar]
- Yu LJ, Nie JN, Cao CY, Jin YK, Yan M, Wang FZ, Liu J, Xiao Y, Liang YH, Zhang WH. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 2010;188(3):762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- Yu YH, Wang P, Bai YC, Wang Y, Wan HN, Liu C, Ni ZY. The soybean F-box protein GmFBX176 regulates ABA-mediated responses to drought and salt stress. Environ Exp Bot. 2020;176:104056. doi: 10.1016/j.envexpbot.2020.104056. [DOI] [Google Scholar]
- Yu Q, Liu YL, Sun GZ, Liu YX, Chen J, Zhou YB, Chen M, Ma YZ, Xu ZS, Lan JH. Genome-wide analysis of the soybean calmodulin-binding protein 60 family and identification of GmCBP60A-1 responses to drought and salt stresses. Int J Mol Sci. 2021;22(24):13501. doi: 10.3390/ijms222413501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung WS, Wang QW, Huang MK, Wong FL, Liu AL, Ng MS, Li KP, Sze CC, Li MW, Lam HM. Priming-induced alterations in histone modifications modulate transcriptional responses in soybean under salt stress. Plant J. 2022;109(6):1575–1590. doi: 10.1111/tpj.15652. [DOI] [PubMed] [Google Scholar]
- Zafar SA, Zaidi SSEA, Gaba Y, Singla-Pareek SL, Dhankher OP, Li XY, Mansoor S, Pareek A. Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J Exp Bot. 2020;71(2):470–479. doi: 10.1093/jxb/erz476. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Wang Y, Li YJ, Lei TT, Yan F, Su LT, Li XW, Zhao Y, Sun X, Li JW, Wang QY. Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene. 2013;513(1):174–183. doi: 10.1016/j.gene.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Zhang BH. MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot. 2015;66(7):1749–1761. doi: 10.1093/jxb/erv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Jia WS, Yang JC, Ismail AM. Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res. 2006;97(1):111–119. doi: 10.1016/j.fcr.2005.08.018. [DOI] [Google Scholar]
- Zhang GY, Chen M, Chen XP, Xu ZS, Guan S, Li LC, Li AL, Guo JM, Mao L, Ma YZ. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.) J Exp Bot. 2008;59(15):4095–4107. doi: 10.1093/jxb/ern248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GY, Chen M, Li LC, Xu ZS, Chen XP, Guo JM, Ma YZ. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60(13):3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GY, Chen M, Chen XP, Xu ZS, Li LC, Guo JM, Ma YZ. Isolation and characterization of a novel EAR-motif-containing gene GmERF4 from soybean (Glycine max L.) Mol Biol Rep. 2010;37(2):809–818. doi: 10.1007/s11033-009-9616-1. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang DS, Li MX, Shi LX. Metabolic profiles reveal changes in wild and cultivated soybean seedling leaves under salt stress. Plos One. 2016;11(7):e0159622. doi: 10.1371/journal.pone.0159622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Massel K, Godwin ID, Gao C. Applications and potential of genome editing in crop improvement. Genome Biol. 2018;19:1–11. doi: 10.1186/s13059-018-1586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liao XL, Cui YM, Ma WY, Zhang XN, Du HY, Ma YJ, Ning LH, Wang H, Huang F, Yang H, Kan GZ, Yu DY. A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. Plos Genetics. 2019;15(1):e1007798. doi: 10.1371/journal.pgen.1007798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XZ, Zheng WJ, Cao XY, Cui XY, Zhao SP, Yu TF, Chen J, Zhou YB, Chen M, Chai SC, Xu ZS, Ma YZ. Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and soybean. Front Plant Sci. 2019;10:1453. doi: 10.3389/fpls.2019.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu YH, Cai HY, Guo ML, Chai MN, She ZY, Ye L, Cheng Y, Wang BR, Qin Y. The bZIP transcription factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int J Mol Sci. 2020;21(20):7778. doi: 10.3390/ijms21207778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WX, Wang N, Yang JT, Guo H, Liu ZH, Zheng XJ, Li S, Xiang FN. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020;291:110326. doi: 10.1016/j.plantsci.2019.110326. [DOI] [PubMed] [Google Scholar]
- Zhang FY, Yang JW, Zhang N, Wu JH, Si HJ. Roles of microRNAs in abiotic stress response and characteristics regulation of plant. Front Plant Sci. 2022;13:919243. doi: 10.3389/fpls.2022.919243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Liu SL, Wang Z, Yuan YQ, Zhang ZF, Liang QJ, Yang X, Duan ZB, Liu YC, Kong FJ, Liu BH, Ren B, Tian ZX. Progress in soybean functional genomics over the past decade. Plant Biotechnol J. 2022;20(2):256–282. doi: 10.1111/pbi.13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MH, Cao JF, Zhang TX, Xu T, Yang LY, Li XY, Ji FD, Gao YX, Ali S, Zhang QZ, Zhu JH, Xie LA. A putative plasma membrane Na+/H+ antiporter GmSOS1 is critical for salt stress tolerance in Glycine max. Front Plant Sci. 2022;13:870695. doi: 10.3389/fpls.2022.870695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, Fang QW, Zheng JQ, Li ZY, Li Y, Feng Y, Han YP, Li YG. GmLecRlk, a Lectin Receptor-like Protein Kinase, contributes to salt stress tolerance by regulating salt-responsive genes in soybean. Int J Mol Sci. 2022;23(3):1030. doi: 10.3390/ijms23031030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SP, Xu ZS, Zheng WJ, Zhao W, Wang YX, Yu TF, Chen M, Zhou YB, Min DH, Ma YZ, Chai SC, Zhang XH. Genome-wide analysis of the RAV family in soybean and functional identification of GmRAV-03 involvement in salt and drought stresses and exogenous ABA treatment. Front Plant Sci. 2017;8:905. doi: 10.3389/fpls.2017.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CJ, Zhang Y, Du JJ, Guo XY, Wen WL, Gu SH, Wang JL, Fan JC. Crop phenomics: current status and perspectives. Front Plant Sci. 2019;10:714. doi: 10.3389/fpls.2019.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MJ, Yin LJ, Liu Y, Ma J, Zheng JC, Lan JH, Fu JD, Chen M, Xu ZS, Ma YZ. The ABA-induced soybean ERF transcription factor gene GmERF75 plays a role in enhancing osmotic stress tolerance in Arabidopsis and soybean. BMC Plant Biol. 2019;19(1):506. doi: 10.1186/s12870-019-2066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MJ, Yin LJ, Ma J, Zheng JC, Wang YX, Lan JH, Fu JD, Chen M, Xu ZS, Ma YZ. The roles of GmERF135 in improving salt tolerance and decreasing ABA sensitivity in soybean. Front Plant Sci. 2019;10:940. doi: 10.3389/fpls.2019.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6(5):486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- Zhou L, Wang C, Liu RF, Han Q, Vandeleur RK, Du J, Tyerman S, Shou HX. Constitutive overexpression of soybean plasma membrane intrinsic protein GmPIP1;6 confers salt tolerance. BMC Plant Biol. 2014;14:181. doi: 10.1186/1471-2229-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZK, Jiang Y, Wang Z, Gou ZH, Lyu J, Li WY, Yu YJ, Shu LP, Zhao YJ, Ma YM, Fang C, Shen YT, Liu TF, Li CC, Li Q, Wu M, Wang M, Wu YS, Dong Y, Wan WT, Wang X, Ding ZL, Gao YD, Xiang H, Zhu BG, Lee SH, Wang W, Tian ZX. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol. 2015;33(4):408–U125. doi: 10.1038/nbt.3096. [DOI] [PubMed] [Google Scholar]
- Ziv C, Zhao ZZ, Gao YG, Xia Y. Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci. 2018;9:1088. doi: 10.3389/fpls.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorb C, Geilfus CM, Dietz KJ. Salinity and crop yield. Plant Biol. 2019;21:31–38. doi: 10.1111/plb.12884. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.