Abstract
Second messengers transfer signals from changing intra- and extracellular conditions to a cellular response. Over the last few decades, several nucleotide-based second messengers have been identified and characterized in especially bacteria and eukaryotes. Also in archaea, several nucleotide-based second messengers have been identified. This review will summarize our understanding of nucleotide-based second messengers in archaea. For some of the nucleotide-based second messengers, like cyclic di-AMP and cyclic oligoadenylates, their roles in archaea have become clear. Cyclic di-AMP plays a similar role in osmoregulation in euryarchaea as in bacteria, and cyclic oligoadenylates are important in the Type III CRISPR–Cas response to activate CRISPR ancillary proteins involved in antiviral defense. Other putative nucleotide-based second messengers, like 3′,5′- and 2′,3′-cyclic mononucleotides and adenine dinucleotides, have been identified in archaea, but their synthesis and degradation pathways, as well as their functions as secondary messengers, still remain to be demonstrated. In contrast, 3′-3′-cGAMP has not yet been identified in archaea, but the enzymes required to synthesize 3′-3′-cGAMP have been found in several euryarchaeotes. Finally, the widely distributed bacterial second messengers, cyclic diguanosine monophosphate and guanosine (penta-)/tetraphosphate, do not appear to be present in archaea.
Keywords: archaea, signaling, second messenger, cyclic diadenylate, cyclic oligoadenylate
Signaling in archaea
Archaea constitute one of the three domains of life, next to the bacteria and eukaryotes. They are found in a wide variety of environments, including extreme conditions such as hydrothermal vents, acidic pools, and high-salt lakes, but also in more moderate environments such as the oceans, soil, and human body. Similar to bacterial and eukaryotic cells, archaeal cells respond to changing conditions and extracellular signals. The transfer of information encoded within DNA to a protein (e.g. replication, transcription, and translation) in archaea resembles these processes in eukaryotes (Grohmann and Werner 2011, Ausiannikava and Allers 2017, Jenal et al. 2017, White and Allers 2018, Schmitt et al. 2020, Wenck and Santangelo 2020). However, the manner in which environmental changes are sensed is more similar to their bacterial counterparts.
Both in bacteria and in archaea, signal transduction occurs most via one-component systems, which are a direct fusion of an input domain to an output domain in a single protein, like repressors and activators that contain ligand-binding and DNA-binding domains (Ulrich et al. 2005). In bacteria, signaling also often occurs via two-component systems. Two-component systems seem absent from Crenarchaeota, Nanoarchaeota, and Korarchaeota but are found in Haloarchaea and Thaumarchaea (Galperin et al. 2018, Krell 2018). Whereas the most observed output of bacterial two-component systems is the regulation of transcription, this seems less the case for archaea, where only a small number of two-component systems have a DNA-binding output domain (Galperin et al. 2018, Krell 2018). Indeed, the well-studied Che chemosensory pathway is a more sophisticated version of a two-component system and plays an important role in regulating the rotational direction of the archaellum, the archaeal motility structure, in response to external stimuli (Quax et al. 2018, Altegoer et al. 2022). Reversible phosphorylation of proteins by protein kinases and phosphatases in response to external stimuli can also be used to transduce signals (Esser et al. 2016). For example, phosphorylation and dephosphorylation of regulatory components in the archaellum regulatory network are important for regulating the expression of the archaellum (Reimann et al. 2012, Hoffmann et al. 2019, Ye et al. 2020). A further method to transfer signals is second messengers, which are well studied in both bacteria and eukaryotes.
Second messengers are small molecules that are involved in intracellular signaling pathways and mediate the effects of first messengers, which are stimuli such as chemical and biochemical signaling molecules or physical cues. First messengers cause a conformational change in specific receptors that trigger the production, release, import, or degradation of a second messenger. The second messenger then interacts with the specific target or other intermediate stages of the signaling pathway, ultimately leading to a cellular response. There are various types of second messengers. Currently known second messengers can be categorized into four distinct groups: (i) ions that signal within and between different compartments; (ii) gases and free radicals that can diffuse through the cell and even to other cells; (iii) lipid-based messengers that signal within cell membranes; and (iv) nucleotide-based messengers and other soluble molecules that signal within the cytosol (Newton et al. 2016).
Nucleotide-based second messengers are involved in many different processes. Their presence and function have been studied in detail in eukaryotes and bacteria (Newton et al. 2016, Thompson and Malone 2020), but significantly less is known about their role in archaea. Isomers of the 3′,5′- and 2′,3′-cyclic mononucleotide-based messengers, as well as the dinucleotide-based messengers c-di-AMP, 5′-phosphoadenylyl-3′,5′-adenosine (5′-pApA), diadenosine tetraphosphate (Ap4A), and oligonucleotide-based cyclic oligoadenylate messengers, have been identified in archaea, whereas the presence of the widely distributed bacterial second messengers cyclic diguanosine monophosphate (3′,5′-c-di-GMP) and guanosine (penta-)/tetraphosphate [(p)ppGpp] has not yet been described. In this review, we focus on the current knowledge of the occurrence and role of nucleotide-based second messengers in archaea.
Mononucleotide-based second messengers
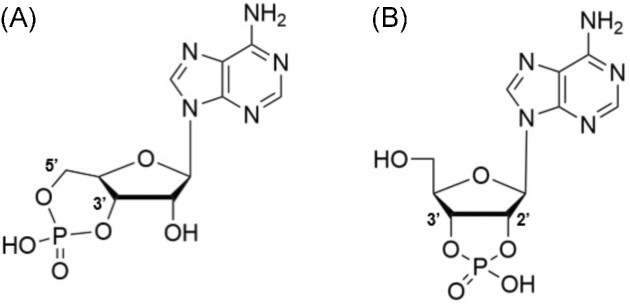
Currently, the best-characterized mononucleotide-based second messengers are 3′,5′- and 2′,3′-cyclic nucleotides (see Fig. 1), and guanine penta- and tetraphosphate (p)ppGpp.
Figure 1.
Structures of mononucleotide-based second messengers: (A) 3′,5′-cyclic nucleotides and (B) 2′,3′-cyclic nucleotides using 3′,5′-cyclic adenosine monophosphate and 2′,3′-cyclic adenosine monophosphate as examples.
3′,5′-cAMP
3′,5′-cAMP was the first mononucleotide-based second messenger discovered (Sutherland and Rall 1958). In animals, it activates protein kinase A (PKA) to regulate cell growth, metabolism, and stress resistance through the phosphorylation of various protein substrates (Turnham and Scott 2016). It also targets a family of cyclic nucleotide-gated cation channels (Yau 1994) and nucleotide exchange factors, known as EPAC (de Rooij et al. 1998). The combination of EPAC- and PKA-dependent pathways determines the overall effects of 3′,5′-cAMP signaling (Cheng et al. 2008). In plants, 3′,5′-cAMP is involved in the regulation of cell-cycle progression and plant resistance to bacterial pathogens (Sabetta et al. 2019). 3′,5′-cAMP is also the first second messenger described in bacteria (Makman and Sutherland 1965) and is primarily associated with the regulation of carbon utilization (Görke and Stülke 2008, Green et al. 2014) as well as other central traits such as biofilm formation (Liu et al. 2020) and virulence (McDonough and Rodriguez 2011).
In 1986, using a radioimmunodetection assay, 3′,5′-cAMP was identified as the first nucleotide-based second messenger in archaea. 3′,5′-cAMP was shown to be present in the crenarchaea Saccharolobus solfataricus (formerly named Sulfolobus solfataricus), the euryarchaea Haloferax volcanii, and Methanothermobacter thermautotrophicus (Leichtling et al. 1986). The presence of 3′,5′-cAMP was also demonstrated in Halobacterium salinarium using an immunoassay kit (Baumann et al. 2007). Recently, the concentrations of 3′,5′-cAMP were determined in Haloferax volcanii and Sulfolobus acidocaldarius using mass spectrometry and were approximately three to five times higher during exponential growth than in the stationary phase (Braun et al. 2021) (see Table 1). The detected levels of 3′,5′-cAMP in Haloferax volcanii remained unchanged when the temperature was lowered from its optimal temperature of 42°C to 25°C. These levels determined by mass spectrometry were significantly lower than those initially observed for Haloferax volcanii (Leichtling et al. 1986, Baumann et al. 2007), and the levels observed for S. acidocaldarius were much lower than those observed for the closely related Sa. solfataricus (Leichtling et al. 1986). The observed discrepancies most likely originated from the different detection methods used. The cytosolic 3′,5′-cAMP concentration determined by mass spectrometry would be 1 µM or lower in both H. volcanii and S. acidocaldarius (see Table 1). This is close to the levels observed in eukaryotes, where the intracellular concentration of cAMP in unstimulated conditions is typically around 1 µM in eukaryotic cells (Börner et al. 2011), but 20–200-fold lower than in Escherichia coli, where the 3′,5′-cAMP concentration increases from ∼20 to ∼180 μm when the growth of E. coli is switched from growth on low levels of glucose to growth on excess glucose (Notley-McRobb et al. 1997). Several possible functions have been assigned to 3′,5′-cAMP in Archaea. Initially, it was observed that in M. thermautotrophicus, 3′,5′-cAMP levels were higher upon starvation for H2, which suggested that 3′,5′-cAMP is involved in the regulation of central metabolic processes (Leichtling et al. 1986). It was further shown that H. salinarum exhibits cell-cycle-dependent fluctuations in its 3′,5′-cAMP levels (Baumann et al. 2007). This observation indicated a potential role for 3′,5′-cAMP in the regulation of the cell cycle of H. salinarum, an effect that was, by then, only known in certain eukaryotes (Baumann et al. 2007). Finally, a characterization of SSO3182, one of the three typical eukaryotic protein kinases encoded on the genome of Sa. solfataricus, showed that the isolated kinase domain of SSO3182 was inhibited by 3’,5’-cAMP, with an estimated Ki of ∼23 µM (Ray et al. 2015), suggesting a regulatory role for 3′,5′-cAMP in the level of protein phosphorylation.
Table 1.
Concentration of (putative) nucleotide-based messengers detected in Braun et al. (2021) after nucleotide extraction and mass spectrometry, converted to micromolar concentrations assuming a cytosolic content of 200 mg/mL (Ray et al. 2015).
| Organims | Haloferax volcanii | Sulfolobus acidocaldarius | ||
|---|---|---|---|---|
| Molecule | Levels during exponential growth (µM) | Levels during stationary growth (µM) | Levels during exponential growth (µM) | Levels during stationary growth (µM) |
| 3′,5′-cCMP | 0.52 | 0.13 | n.d. | n.d. |
| 3′,5′-cUMP | 0.38 | 0.20 | n.d. | n.d. |
| 2′,3′-cAMP | 30 | 20 | 1.0 | 0.69 |
| 2′,3′-cGMP | 22 | 13 | 0.56 | 0.52 |
| 2′,3′-cCMP | 15 | 11 | 0.22 | 0.32 |
| 2′,3′-cUMP | 0.51 | 0.40 | n.d. | ≤ 0.02 |
| 3′,5′-c-di-AMP | 8.2 | 9.1 | n.d. | n.d. |
| 5′-pApA | ≤ 0.6 | 0.22 | n.d. | ≥ 0 |
| Ap4A | 1.9 | 0.29 | 125 | 8.1 |
n.d., not detected.
3′,5′-cAMP is synthesized by adenylate cyclases, which convert ATP to 3′,5′-cAMP and pyrophosphates. Adenylate cyclases are divided into six distinct classes, all of which are present in bacteria, whereas eukaryotes encode only Class III adenylate cyclases (Khannpnavar et al. 2020). Several archaeal genomes encode a protein that is often annotated as CyaB-like Class IV adenylyl cyclase. These proteins are members of the CYTH-like domain superfamily (IPR033469), which includes Class IV adenylyl cyclases, thiamine triphosphatases, and inorganic tri- or polyphosphatases (also known as triphosphatase tunnel metalloenzymes or TTMs). Recently, using structural and functional approaches, it was demonstrated that the Saci_0718 protein of S. acidocaldarius, which was also previously annotated as an adenylate cyclase, does not have this function, but functions as a triphosphatase (Vogt et al. 2021). Molecular determinants that discriminate the adenylyl cyclase from the different phosphatase activities in the CYTH domain family and a systematic sequence similarity network analysis demonstrated that archaeal CYTH enzymes are functionally divergent from CyaB-like Class IV adenylyl cyclases (Vogt et al. 2021). 3′,5′-cAMP is degraded by 3′,5′-cAMP-specific phosphodiesterases (Matange 2015). A search of the NCBI databases identified some phosphodiesterases annotated as 3′,5′-cyclic-AMP phosphodiesterases; however, for none of these, this function has been demonstrated. Thus, although 3′,5′-cAMP is present in archaea, proteins that either synthesize, degrade, or interact with 3′,5′-cAMP have not been characterized, and a more general function of 3′,5′-cAMP in archaea is currently unknown.
3′,5′-cNMPs
In addition, other cyclic nucleotides such as 3′,5′-cGMP, 3′,5′-cCMP, 3′,5′-cUMP, and 3′,5′-cIMP have been detected in several organisms, but these cyclic nucleotides are found to be less widespread than 3′,5′-cAMP. 3′-5′-cGMP is generated by a guanylate cyclase and might function as a second messenger in bacteria (Rauch et al. 2008, Linder 2010, Ryu et al. 2015). Recently, it was demonstrated that 3′,5′-cCMP and 3′,5′-cUMP cyclases function as part of the Pycsar (pyrimidine cyclase system for antiphage resistance) family of bacterial antiphage defense systems. The production of these cyclic pyrimidine molecules is triggered specifically by phage infection after which effector proteins are activated that execute abortive infection through membrane impairment or depletion of cellular NAD+ (Tal et al. 2021). These 3′,5′-cCMP and 3′,5′-cUMP cyclases are highly homologous to the adenylate cyclases, but mutations in the active site result in high selectivity for pyrimidines. They are widespread in bacteria and found in some euryarchaea (Tal et al. 2021). However, with the exception of 3′,5′-cAMP, no other 3′,5′-cNMPs were detected in archaea until recently; 3′,5′-cGMP, 3′,5′-cCMP, and 3′,5′-cUMP were detected using nucleotide extraction followed by Liquid-Chromatography-Massspectroscopy/Massspectroscopy (LC-MS/MS) in H. volcanii (Braun et al. 2021). The levels of these three 3′,5′-cNMPs and 3′,5′-cAMPs were higher in exponentially growing cells than in stationary cells, but no cyclases could be identified in H. volcanii. Similar experiments in S. acidocaldarius detected only 3′,5′-cAMP and very low levels of 3′,5′-cGMP (Braun et al. 2021). Currently, no enzymes involved in 3′,5′-cGMP, 3′,5′-cCMP, or 3′,5′-cUMP synthesis or degradation in archaea have been characterized.
2′,3′-cNMPs
The 2′,3′-cNMPs have been identified in eukaryotes, bacteria, and archaea. 2′,3′-Cyclic phosphate termini are produced during RNA cleavage by many endoribonucleases, either as intermediates or final products (Filipowicz 2016). In eukaryotes, they originate from trans-phosphorylation (Thompson et al. 1994) or RNA cyclase activity (Shigematsu et al. 2018), and in E. coli, they have been shown to originate from RNase I-dependent RNA degradation (Fontaine et al. 2018) or RNA cyclase activity (Genschik et al. 1997). RNA 3′-phosphate cyclases are found in most archaea. No function has been established for 2′,3′-cNMPs. In addition to the cyclic 3′,5′-isomers, mass spectrometry of the cell extracts of H. volcanii and S. acidocaldarius identified cyclic 2′,3′-AMP, cyclic 2′,3′-GMP, cyclic 2′,3′-CMP, and cyclic 2′,3′-UMP (Braun et al. 2021). In H. volcanii, the concentrations of the 2′,3′-cNMPs were ∼30 times higher than those of the corresponding 3′,5′-cNMPs, and the levels detected in H. volcanii were also significantly higher than those detected in S. acidocaldarius. The levels of these nucleotides were higher in the exponential phase than in the stationary phase. The levels of 2′,3′-cUMP were lower than those of the other 2′,3′-cNMPs in both organisms, possibly indicating that 2′,3′-cUMP is less stable. 2′,3′-cNMPs may be derived from RNA degradation, as this is a common source of 2′,3′-cNMPs in eukaryotes and bacteria (Fontaine et al. 2018, Thompson and Malone 2020). It is not known whether any of the detected 2′,3′-cNMPs are specifically synthesized and/or function as second messengers in these organisms.
pppGpp and ppGpp
Guanine penta- and tetraphosphate [(p)ppGpp] were first identified in 1969 in bacteria (Cashel and Gallant 1969) and have been shown to play an important role in the bacterial stringent response (Irving et al. 2021). Analysis of the cell extracts from S. acidocaldarius and H. volcanii for the presence of the alarmone ppGpp and its precursor pppGpp did not detect any (p)ppGpp in the extracts from either species (Braun et al. 2021). This is consistent with previous studies that also found that these alarmones were not produced in these organisms, even under stress conditions (Scoarughi et al. 1995, Cellini et al. 2004). A study on the distribution of (p)ppGpp synthetases and hydrolases across the tree of life also suggests that these enzymes are generally rare in archaea (Atkinson et al. 2011).
Dinucleotide-based second messengers
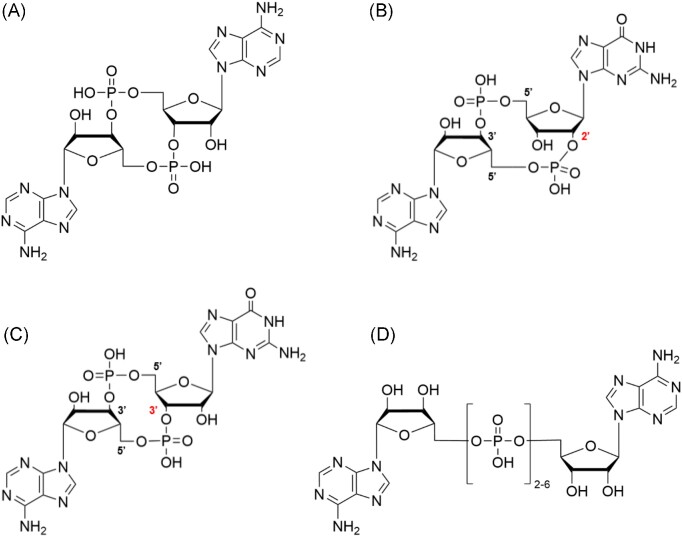
Several nucleotide-based second messengers that are built from two nucleotides have been identified in prokaryotes. They can either have a cyclic structure like cyclic 3′,5′-di-GMP (c-di-GMP), 3′,5′-cyclic di-AMP (c-di-AMP), 3′-3′-cGAMP, and 2′-3′-cGAMP, or have a linear structure like dinucleotide polyphosphates (NpnN) (see Fig. 2).
Figure 2.
Structures of dinucleotide-based second messengers: (A) 3′,5′-cyclic diadenosine monophosphate (c-di-AMP); (B) 2′,3′-cyclic guanosine monophosphate-adenosine monophosphate (2’,3’-cGAMP); (C) 3’,3’-cyclic guanosine monophosphate-adenosine monophosphate (3′,3′-cGAMP); and (D) diadenosine oligo (n = 2–6) phosphate (Ap2–6A). For 2’,3’-cGAMP and 3’,3’-cGAMP, the number of the isomer-determining carbon atom is highlighted in red.
3′,5′-c-di-GMP
3′,5′-c-di-GMP (c-di-GMP) has been extensively studied in many bacteria and is involved in the transitions between motile and sessile lifestyles (Hengge 2009, Römling et al. 2013, Jenal et al. 2017), but has also been found to regulate various processes such as virulence (Tischler and Camilli 2005), predation (Hobley et al. 2012), cellular development (Tschowri et al. 2014), DNA repair (Fernandez et al. 2018), and cell shape regulation (Fernandez et al. 2018). c-di-GMP is synthesized by diguanylate cyclases, which contain a conserved GGDEF motif and are degraded by phosphodiesterases with specific EAL or HD-GYP motifs (Tchigvintsev et al. 2010, Galperin and Chou 2022). c-di-GMP levels can be sensed by different c-di-GMP binding transcription factors, riboswitches, and protein complexes (Hengge 2009, Jenal et al. 2017, Römling and Galperin 2017), and bacteria often contain multiple diguanylate cyclases, c-di-GMP phosphodiesterases, and c-di-GMP binding output domains, which allow for the integration of many different signals. Although very common in bacteria, c-di-GMP and its (intermediate) degradation product 5′-pGpG could not be detected by mass spectrometric analysis of the cell extracts from S. acidocaldarius and H. volcanii (Braun et al. 2021) and have also not been detected in other archaea. Indeed, diguanylate cyclases appear to be almost exclusively found in bacteria, similar to proteins with all other domains associated with 3′,5′-c-di-GMP signaling (e.g. EAL- or PilZ-domain) (Römling et al. 2013).
3′,5′-c-di-AMP
3′,5′-c-di-AMP (c-di-AMP) is essential for the maintenance of cellular osmolarity, and its main function is to regulate the import and export of potassium and other osmoprotective molecules (Bai et al. 2014, Schuster et al. 2016, Gundlach et al. 2017, Pham et al. 2018, Zarrella et al. 2018). Therefore, both low and high levels of c-di-AMP are detrimental to the cells (Gundlach et al. 2015, Huynh and Woodward 2016, Commichau et al. 2019). c-di-AMP is also involved in various cellular processes and pathways such as DNA integrity sensing (Bejerano-Sagie et al. 2006, Witte et al. 2008, Gándara and Alonso 2015, Gundlach et al. 2016, Torres et al. 2019), antibiotic resistance (Chin et al. 2015), control of cell size and cell wall homeostasis (Corrigan et al. 2011, Luo and Helmann 2012), regulation of fatty acid synthesis (Zhang et al. 2013), and biofilm formation (Peng et al. 2016, Townsley et al. 2018). c-di-AMP is synthesized by diadenylate cyclases (Corrigan and Gründling 2013, He et al. 2020, Yin et al. 2020) and degraded by phosphodiesterases, which are often characterized by a DHH-DHHA1 domain, although other c-di-AMP degrading phosphodiesterases have also been identified (Commichau et al. 2019, He et al. 2020). Degradation of c-di-AMP often occurs via a 5′-pApA intermediate. In contrast to the presence of several diguanylate cyclases in many bacteria, in general only one diadenylate cyclase is found in most bacteria. Similar to c-di-GMP, c-di-AMP can bind to transcription factors, riboswitches, and protein complexes to regulate their activities (Corrigan et al. 2013, Commichau et al. 2019). c-di-AMP binds diverse proteins, such as proteins with RCK_C (regulator of conductance of K+, C-terminal domain) (Corrigan et al. 2013, Chin et al. 2015, Kim et al. 2015), cystathionine beta synthase (CBS) (Sureka et al. 2014, Huynh et al. 2016, Schuster et al. 2016), and universal stress protein (USP) (Corrigan et al. 2013, Moscoso et al. 2016) domains. These proteins then function as signal transduction and regulatory proteins or directly regulate transport proteins. Genomic data analysis predicted the presence of diadenylate cyclases in several Euryarchaeots (Shenroy and Visweswariah 2004, Corrigan and Gründling 2013) and in several recently proposed candidate archaeal phyla (Galperin 2023). Similar to bacterial diadenylate cyclases of the DacY/CdaZ class, several archaeal diadenylate cyclases have an N-terminal domain with a fold similar to the C-terminal alpha/beta domain of pyruvate kinase (Braun et al. 2019, Yin et al. 2020). However, the function of this domain remains unknown. Recently, the N-terminal domains of different diadenylate cyclases were analyzed in more detail, and a revised classification of diadenylate cyclases was proposed (Galperin 2023).
After it was shown that the purified Methanocaldococcus jannaschii CdaZ exhibited diadenylate cyclase activity (Kellenberger et al. 2015), H. volcanii was the first archaeon where the presence of c-di-AMP in cell extracts was demonstrated (Braun et al. 2019). H. volcanii has a single diadenylate cyclase gene (dacZ) that is responsible for c-di-AMP production (Braun et al. 2019). The dacZ gene is essential, and its overexpression leads to cell death, suggesting the need for tight regulation of c-di-AMP levels. A strain with decreased c‐di‐AMP levels exhibited an increased cell area in a hyposalt medium, suggesting a similar function in osmoregulation as shown in bacteria (Braun et al. 2019). The concentration of c-di-AMP was ∼8 µM during both exponential growth and stationary stages (Braun et al. 2019, 2021) (see Table 1). A similar analysis of S. acidocaldarius cells did not identify any c-di-AMP (Braun et al. 2021). Indeed, proteins with diadenylate cyclase (DAC) domains (COG1624) are frequently found in euryarchaeotes but are absent in crenarchaeota (Römling 2008, Witte et al. 2008, He et al. 2020). It is currently unknown which proteins degrade c-di-AMP in archaea. Possible candidates include nanoRNases/aPa phosphatases containing a DHH-DHHA1 domain (arCOG01566), which are found in euryarchaeotes and may hydrolyze c-di-AMP and/or pApA (Lee et al. 2022). No archaeal proteins that bind c-di-AMP have been identified, but many euryarchaeotes contain proteins with C-terminal TrkA domains that are linked to potassium transporters, which are excellent candidates for c-di-AMP binding.
3′-3′-cGAMP and 2′-3′-cGAMP
3′-3′-cGAMP is a more recently discovered dinucleotide-based second messenger that has been shown to control diverse processes, such as chemotaxis in Vibrio cholerae (Davies et al. 2012), anaerobic respiration in Geobacter metallireducens (Nelson et al. 2015), biofilm formation and motility in E. coli (Li et al. 2019), predation in Bdellovibrio bacteriovorus (Lowry et al. 2022, Rangarajan and Waters 2022), and functions in a prokaryotic anti-phage defense mechanism (Cohen et al. 2019, Athukoralage and White 2022, Duncan-Lowey and Kranzusch 2022). In prokaryotes, 3′,3′-cGAMP is produced by cGAS/DncV-like nucleotidyltransferases (called CD-NTases or GMP-AMP cyclases (cGASs) (Kranzusch et al. 2014) or by a more recently discovered class of cGASs that are structurally related to the GGDEF family of diguanylate cyclases but have ‘Hypr’ GGDEF domains that favor 3′-3′-cGAMP production (Hallberg et al. 2016). The 3′-3′-cGAMPs are degraded by both EAL and HD-GYP motif-containing phosphodiesterases (Hallberg et al. 2016, Yadav et al. 2019, Wright et al. 2020). The cGAS proteins have also been identified in animals. Mammalian cGAS proteins are structural and functional homologs of bacterial CD-NTases (Kranzusch et al. 2014). In animals, the binding of cGAS to double-stranded DNA allosterically activates its catalytic activity and leads to the production of 2′-3′-cGAMP, which functions as a potent agonist of the stimulator of interferon genes (STING) pathway. The cGAS-STING pathway serves as a crucial element of immunity in many organisms (Sun et al. 2013, Decout et al. 2021).
In bacteria, cGAS proteins are involved in the widespread cyclic oligonucleotide-based anti-phage signaling systems (CBASSs), a family of defense systems against bacteriophages (Cohen et al. 2019, Millman et al. 2020, Athukoralage and White 2022, Duncan-Lowey and Kranzusch 2022). Larger cyclic oligonucleotides consisting of three to six AMP molecules and their role in Type III CRISPR–Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated genes)-based CBASSs are described below. Central to dinucleotide-based CBASSs are cGAS proteins, which generate a variety of cyclic dinucleotides in response to phage infection, and effector proteins, which are activated by these cyclic dinucleotides (Whiteley et al. 2019). These CBASSs are mainly found in bacteria and have been divided into four classes (Millman et al. 2020). Clusters of genes belonging to the Type IV CBASS class, which is a rare and currently uncharacterized class, have been found in the genomes of several euryarchaea (Millman et al. 2020), suggesting the presence of 3′-3′-cGAMP in archaea. However, 3′-3′-cGAMP and 2′-3′-cGAMP could not be detected by mass spectrometric analysis of the cell extracts from S. acidocaldarius and H. volcanii, which correlated with the absence of dinucleotide-based CBASS systems in the genomes of these two organisms (Braun et al. 2021). Thus, although it is likely that 3′-3′-cGAMP is present in euryarchaeotes encoding cGAS-based CBASSs, its presence still needs to be confirmed.
Adenine dinucleotides
Adenine dinucleotides (ApnA) are formed by two adenosines bridged at their 5′ ends by several phosphates. Diadenosine 5′,5′′′-P1, P4-tetraphosphate (Ap4A) is the best-characterized ApnA and is mostly produced under stress conditions, such as pH or heat shock, treatment with heavy metal ions or antibiotics, DNA damage, and oxidative stress; thus, Ap4A has been proposed to be an alarmone that signals cellular stress (Lee et al. 1983, Bochner et al. 1984, Johnstone and Farr 1991, Despotović et al. 2017, Ferguson et al. 2020). Ap4A levels can increase >100-fold under stress conditions to concentrations >300 µM (Varshavsky 1983, Bochner et al. 1984). Several mechanisms have been proposed by which Ap4A functions in response to stress have been proposed (Ferguson et al. 2020). Under stress conditions that elevate cellular Np4N concentrations, diverse mRNAs and sRNAs acquire cognate Np4 caps (Luciano et al. 2019, Hudeček et al. 2020, Luciano and Belasco 2020). This cap can be methylated and can protect mRNA from degradation, thus stabilizing mRNA (Hudeček et al. 2020). In Bacillus subtilis, Ap4A regulates inosine-5′-monophosphate dehydrogenase, thus affecting nucleotide metabolite homoeostasis (Altegoer et al. 2022). Ap4A also binds to other ATP-binding proteins; however, it has been questioned whether it only binds to an ATP analog (Despotović et al. 2017). In eukaryotes, Ap4A is involved in activating the microphthalmia-associated transcription factor during allergic response IgE (Lee et al. 2004, Yannay-Cohen et al. 2009) and inhibiting the cGAS-STING pathway (Guerra et al. 2020). Ap4N furthermore functions as an extracellular messenger through the activation of purinoceptors (Jankowski et al. 2009), and the activation of the posttranslational modifiers ubiquitin and ubiquitin-like proteins results in the formation of Ap4A and Ap3A (Götz et al. 2019). Ap4A is produced by many different enzymes that involve an acyl-adenylate and/or enzyme-adenylate intermediate, like acyl-coenzyme A synthetase, DNA and RNA ligases, ubiquitin and ubiquitin-like E1 activating enzymes, and non-ribosomal peptide synthetases (Ferguson et al. 2020), but most commonly by aminoacyl-transfer RNA (tRNA) synthetases (Zamecnik et al. 1966, Giammarinaro et al. 2022). Several hydrolase families have been identified that degrade Ap4A (Ferguson et al. 2020).
Ap4A was found in all three kingdoms. In S. acidocaldarius, the concentration of Ap4A, as determined by mass spectrometry after nucleotide extraction, was ∼125 µM Ap4A in the exponential stage and decreased ∼18-fold in the stationary stage (Braun et al. 2019, 2021) (see Table 1). In H. volcanii, the concentration of Ap4A was ∼2 µM Ap4A in the exponential stage and decreased ∼6-fold in the stationary stage (see Table 1). As the enzymes that synthesize Ap4A are diverse and widespread, the specific proteins responsible for their synthesis in archaea are difficult to identify. However, it has been shown that phenylalanyl-tRNA synthetase from Methanosarcina barkeri can produce Ap4A (Rauhut et al. 1985), suggesting that tRNA synthetases are a source of Ap4A in archaea. Whether they act as second messengers, possibly via stabilization of mRNAs via 5′ capping, remains unknown.
Oligonucleotide-based second messengers
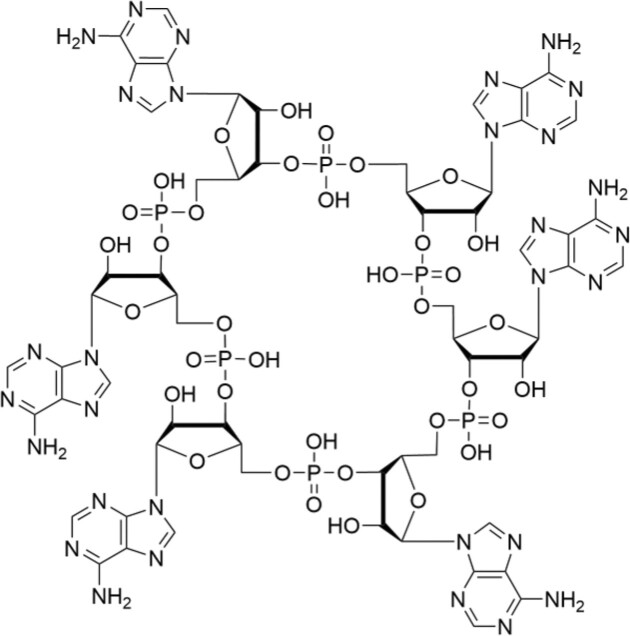
Cyclic oligonucleotides of three to six AMP molecules (see Fig. 3) have been shown to function as second messengers in Type III CRISPR–Cas-based CBASSs (Kazlauskiene et al. 2017, Niewoehner et al. 2017, Athukoralage and White 2022). CRISPR–Cas systems provide immunity against invading genetic elements, such as bacteriophages or plasmids, and have been divided into Class I and Class II systems, with both classes further divided into different types (Makarova et al. 2015). The Type III CRISPR–Cas systems of Class I are known for their ability to target both invader RNA and DNA and are characterized by the presence of a Cas10 protein, which has two enzymatic functions (Elmore et al. 2016, Estrella et al. 2016, Kazlauskiene et al. 2016, Tamulaitis et al. 2017). The first function is to degrade single-stranded DNA, whereas the second function is the synthesis of cyclic oligoadenylates from ATP. Upon recognition of a target RNA, cyclic oligoadenylates are generated (Kazlauskiene et al. 2017, Niewoehner et al. 2017), which activate CRISPR ancillary nucleases that indiscriminately degrade both host and invader RNA. To control cyclic oligoadenylate levels, RNA nucleases often containing a CRISPR-associated Rossman fold domain that cleaves cyclic oligoadenylates are expressed or activated (Athukoralage et al. 2018, 2020, Samolygo et al. 2020, Molina et al. 2021).
Figure 3.
Structure of an oligonucleotide-based second messenger: structure of cyclic hexa-adenylate.
Although S. acidocaldarius encodes a functional Type III CRISPR system that contains a Cas10 subunit (Zink et al. 2021) and cyclic oligoadenylates were detected in the closely related Sa. solfataricus (Rouillon et al. 2018), no cyclic oligoadenylates could be detected by mass spectrometry after nucleotide extraction. Possibly, this oligonucleotide can be detected only when cells are infected with a virus. Also, no cyclic oligoadenylates were detected in H. volcanii, but this was expected because it lacks a Type III CRISPR system (Maier et al. 2013).
Conclusions and future perspectives
Nucleotide-based second messengers play important roles in the signaling processes in both bacteria and eukaryotes. The observations that (i) cyclic di-AMP plays a similar role in osmoregulation as in bacteria; (ii) cyclic oligoadenylate messengers play a role in the bacterial and archaeal Type III CRISPR–Cas response to activate CRISPR ancillary proteins involved in antiviral defense; and (iii) homologs of enzymes that synthesize 3′-3′-cGAMP are found in CBASS systems found in both archaea and bacteria suggest that nucleotide-based second messengers in archaea resemble these systems in bacteria. The role that cyclic oligoadenylate messengers play in the bacterial and archaeal Type III CRISPR–Cas response to activate CRISPR ancillary proteins has been studied in quite some detail, but little is still known about the other putative nucleotide-based second messengers and will require further studies. Remarkably, contrary to the nucleotide-based secondary messengers involved in the antiviral defense systems, the widely distributed bacterial second messengers cyclic diguanosine monophosphate, (p)ppGpp, and possibly cAMP, which are involved in general cellular regulation systems, do not appear to play an important role in archaea. Most likely, the antiviral defense systems are more easily transmitted via lateral gene transfer.
Contributor Information
Chris van der Does, Molecular Biology of Archaea, Institute of Biology, University of Freiburg, 79104 Freiburg, Germany.
Frank Braun, Molecular Biology of Archaea, Institute of Biology, University of Freiburg, 79104 Freiburg, Germany.
Hongcheng Ren, Molecular Biology of Archaea, Institute of Biology, University of Freiburg, 79104 Freiburg, Germany.
Sonja-Verena Albers, Molecular Biology of Archaea, Institute of Biology, University of Freiburg, 79104 Freiburg, Germany.
Conflict of interest statement
None declared.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) within the scope of SPP 1879: Nucleotide Second Messenger Signaling in Bacteria. HR was supported by a CSC fellowship (Chinese Science Council).
References
- Altegoer F, Quax TEF, Weiland P et al. Structural insights into the mechanism of archaellar rotational switching. Nat Commun. 2022;13:2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, McMahon SA, Zhang C et al. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020;577:572–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, Rouillon C, Graham S et al. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018;562:277–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukoralage JS, White MF. Cyclic nucleotide signaling in phage defense and counter-defense. Annu Rev Virol. 2022;9:451–68. [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausiannikava D, Allers T. Diversity of DNA replication in the archaea. Genes. 2017;8:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang J, Zarrella TM et al. Cyclic di-AMP impairs potassium uptake mediated by a cyclic di-AMP binding protein in Streptococcus pneumoniae. J Bacteriol. 2014;196:614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A, Lange C, Soppa J. Transcriptome changes and cAMP oscillations in an archaeal cell cycle. BMC Cell Biol. 2007;8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I et al. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–90. [DOI] [PubMed] [Google Scholar]
- Bochner BR, Lee PC, Wilson SW et al. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell. 1984;37:225–32. [DOI] [PubMed] [Google Scholar]
- Börner S, Schwede F, Schlipp A et al. FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells. Nat Protoc. 2011;6:427–38. [DOI] [PubMed] [Google Scholar]
- Braun F, Recalde A, Bähre H et al. Putative nucleotide-based second messengers in the archaeal model organisms Haloferax volcanii and Sulfolobus acidocaldarius. Front Microbiol. 2021;12:779012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F, Thomalla L, van der Does C et al. Cyclic nucleotides in archaea: cyclic di-AMP in the archaeon Haloferax volcanii and its putative role. Microbiol Open. 2019;8:e00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashel M, Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–41. [DOI] [PubMed] [Google Scholar]
- Cellini A, Scoarughi GL, Poggiali P et al. Stringent control in the archaeal genus Sulfolobus. Res Microbiol. 2004;155:98–104. [DOI] [PubMed] [Google Scholar]
- Cheng X, Ji Z, Tsalkova T et al. Epac and PKA: a tale of two intracellular cAMP receptors. Acta Biochim Biophys Sin. 2008;40:651–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K-H, Liang J-M, Yang J-G et al. Structural insights into the distinct binding mode of cyclic di-AMP with SaCpaA_RCK. Biochemistry. 2015;54:4936–51. [DOI] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman A et al. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature. 2019;574:691–5. [DOI] [PubMed] [Google Scholar]
- Commichau FM, Heidemann JL, Ficner R et al. Making and breaking of an essential poison: the cyclases and phosphodiesterases that produce and degrade the essential second messenger cyclic di-AMP in bacteria. J Bacteriol. 2019;201:e00462–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne H et al. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Campeotto I, Jeganathan T et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110:9084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Gründling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–24. [DOI] [PubMed] [Google Scholar]
- Davies BW, Bogard RW, Young TS et al. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Zwartkruis FJ, Verheijen MH et al. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474–7. [DOI] [PubMed] [Google Scholar]
- Decout A, Katz JD, Venkatraman S et al. The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21:548–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despotović D, Brandis A, Savidor A et al. Diadenosine tetraphosphate (Ap4A)—an E. coli alarmone or a damage metabolite?. FEBS J. 2017;284:2194–215. [DOI] [PubMed] [Google Scholar]
- Duncan-Lowey B, Kranzusch PJ. CBASS phage defense and evolution of antiviral nucleotide signaling. Curr Opin Immunol. 2022;74:156–63. [DOI] [PubMed] [Google Scholar]
- Elmore JR, Sheppard NF, Ramia N et al. Bipartite recognition of target rnas activates DNA cleavage by the type III-B CRISPR–Cas system. Genes Dev. 2016;30:447–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser D, Hoffmann L, Pham TK et al. Protein phosphorylation and its role in archaeal signal transduction. FEMS Microbiol Rev. 2016;40:625–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo F-T, Bailey S. RNA-activated DNA cleavage by the type III-B CRISPR–Cas effector complex. Genes Dev. 2016;30:460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson F, McLennan AG, Urbaniak MD et al. Re-evaluation of diadenosine tetraphosphate (Ap4A) from a stress metabolite to bona fide secondary messenger. Front Mol Biosci. 2020;7:606807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez NL, Srivastava D, Ngouajio AL et al. Cyclic di-GMP positively regulates DNA repair in vibrio cholerae. J Bacteriol. 2018;200:e00005–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W. RNA 3’-terminal phosphate cyclases and cyclase-like proteins. Postepy Biochem. 2016;62:327–34. [PubMed] [Google Scholar]
- Fontaine BM, Martin KS, Garcia-Rodriguez JM et al. RNase I regulates Escherichia coli 2’,3’-cyclic nucleotide monophosphate levels and biofilm formation. Biochem J. 2018;475:1491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Chou S-H. Sequence conservation, domain architectures, and phylogenetic distribution of the HD-GYP type c-di-GMP phosphodiesterases. J Bacteriol. 2022;204:e0056121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI et al. Phyletic distribution and lineage-specific domain architectures of archaeal two-component signal transduction systems. J Bacteriol. 2018;200:e00681–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. All dacs in a row: domain architectures of bacterial and archaeal diadenylate cyclases. J Bacteriol. 2023;205:e0002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gándara C, Alonso JC. DisA and c-di-AMP act at the intersection between DNA-damage response and stress homeostasis in exponentially growing Bacillus subtilis cells. DNA Repair (Amst). 2015;27:1–8. [DOI] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz M et al. The human RNA 3’-terminal phosphate cyclase is a member of a new family of proteins conserved in eucarya, bacteria and archaea. EMBO J. 1997;16:2955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammarinaro PI, Young MKM, Steinchen W et al. Diadenosine tetraphosphate regulates biosynthesis of GTP in Bacillus subtilis. Nat Microbiol. 2022;7:1442–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–24. [DOI] [PubMed] [Google Scholar]
- Götz KH, Mex M, Stuber K et al. Formation of the alarmones diadenosine triphosphate and tetraphosphate by ubiquitin- and ubiquitin-like-activating enzymes. Cell Chem Biol. 2019;26:1535–43. [DOI] [PubMed] [Google Scholar]
- Green J, Stapleton MR, Smith LJ et al. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr Opin Microbiol. 2014;18:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann D, Werner F. Recent advances in the understanding of archaeal transcription. Curr Opin Microbiol. 2011;14:328–34. [DOI] [PubMed] [Google Scholar]
- Guerra J, Valadao A-L, Vlachakis D et al. Lysyl-tRNA synthetase produces diadenosine tetraphosphate to curb STING-dependent inflammation. Sci Adv. 2020;6:eaax3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach J, Herzberg C, Kaever V et al. Control of potassium homeostasis is an essential function of the second messenger cyclic di-AMP in Bacillus subtilis. Sci Signal. 2017;10:eaal3011. [DOI] [PubMed] [Google Scholar]
- Gundlach J, Mehne FMP, Herzberg C et al. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol. 2015;197:3265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundlach J, Rath H, Herzberg C et al. Second messenger signaling in Bacillus subtilis: accumulation of cyclic di-AMP inhibits biofilm formation. Front Microbiol. 2016;7:804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg ZF, Wang XC, Wright TA et al. Hybrid promiscuous (Hypr) GGDEF enzymes produce cyclic AMP-GMP (3’,3’-cGAMP). Proc Natl Acad Sci USA. 2016;113:1790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yin W, Galperin MY et al. Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 2020;48:2807–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–73. [DOI] [PubMed] [Google Scholar]
- Hobley L, Fung RKY, Lambert C et al. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 2012;8:e1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann L, Anders K, Bischof LF et al. Structure and interactions of the archaeal motility repression module ArnA–ArnB that modulates archaellum gene expression in Sulfolobus acidocaldarius. J Biol Chem. 2019;294:7460–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudeček O, Benoni R, Reyes-Gutierrez PE et al. Dinucleoside polyphosphates act as 5’-RNA caps in bacteria. Nat Commun. 2020;11:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Choi PH, Sureka K et al. Cyclic di-AMP targets the cystathionine beta-synthase domain of the osmolyte transporter OpuC. Mol Microbiol. 2016;102:233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh TN, Woodward JJ. Too much of a good thing: regulated depletion of c-di-AMP in the bacterial cytoplasm. Curr Opin Microbiol. 2016;30:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving SE, Choudhury NR, Corrigan RM. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat Rev Microbiol. 2021;19:256–71. [DOI] [PubMed] [Google Scholar]
- Jankowski V, van der Giet M, Mischak H et al. Dinucleoside polyphosphates: strong endogenous agonists of the purinergic system. Br J Pharmacol. 2009;157:1142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–84. [DOI] [PubMed] [Google Scholar]
- Johnstone DB, Farr SB. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 1991;10:3897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č et al. A cyclic oligonucleotide signaling pathway in type III CRISPR–Cas systems. Science. 2017;357:605–9. [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G et al. Spatiotemporal control of type III-A CRISPR–Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell. 2016;62:295–306. [DOI] [PubMed] [Google Scholar]
- Kellenberger CA, Chen C, Whiteley AT et al. RNA-based fluorescent biosensors for live cell imaging of second messenger cyclic di-AMP. J Am Chem Soc. 2015;137:6432–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khannpnavar B, Mehta V, Qi C et al. Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr Opin Struct Biol. 2020;63:34–41. [DOI] [PubMed] [Google Scholar]
- Kim H, Youn S-J, Kim SO et al. Structural studies of potassium transport protein KtrA regulator of conductance of K+ (RCK) C domain in complex with cyclic diadenosine monophosphate (c-di-AMP). J Biol Chem. 2015;290:16393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee ASY, Wilson SC et al. Structure-guided reprogramming of human cGAS dinucleotide linkage specificity. Cell. 2014;158:1011–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krell T. Exploring the (almost) unknown: archaeal two-component systems. J Bacteriol. 2018;200:e00774–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Bochner BR, Ames BN. AppppA, heat-shock stress, and cell oxidation. Proc Natl Acad Sci USA. 1983;80:7496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee VT, Sondermann H, Winkler WC. Nano-rnases: oligo- or dinucleases?. FEMS Microbiol Rev. 2022;46:fuac038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-N, Nechushtan H, Figov N et al. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity. 2004;20:145–51. [DOI] [PubMed] [Google Scholar]
- Leichtling BH, Rickenberg HV, Seely RJ et al. The occurrence of cyclic AMP in archaebacteria. Biochem Biophys Res Commun. 1986;136:1078–82. [DOI] [PubMed] [Google Scholar]
- Li F, Cimdins A, Rohde M et al. DncV synthesizes cyclic GMP-AMP and regulates biofilm formation and motility in Escherichia coli ECOR31. Mbio. 2019;10:e02492–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder J.U. cGMP production in bacteria. Mol Cell Biochem. 2010;334:215–9. [DOI] [PubMed] [Google Scholar]
- Liu C, Sun D, Zhu J et al. The regulation of bacterial biofilm formation by cAMP-CRP: a mini-review. Front Microbiol. 2020;11:802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry RC, Hallberg ZF, Till R et al. Production of 3’,3’-cGAMP by a Bdellovibrio bacteriovorus promiscuous GGDEF enzyme, Bd0367, regulates exit from prey by gliding motility. PLoS Genet. 2022;18:e1010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Belasco JG. Np4A alarmones function in bacteria as precursors to RNA caps. Proc Natl Acad Sci USA. 2020;117:3560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Levenson-Palmer R, Belasco JG. Stresses that raise Np4A levels induce protective nucleoside tetraphosphate capping of bacterial RNA. Mol Cell. 2019;75:957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83:623–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier LK, Lange SJ, Stoll B et al. Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biol. 2013;10:865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makman RS, Sutherland EW. Adenosine 3’,5’-phosphate in Escherichia coli. J Biol Chem. 1965;240:1309–14. [PubMed] [Google Scholar]
- Matange N. Revisiting bacterial cyclic nucleotide phosphodiesterases: cyclic AMP hydrolysis and beyond. FEMS Microbiol Lett. 2015;362:fnv183. [DOI] [PubMed] [Google Scholar]
- McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat Rev Microbiol. 2011;10:27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman A, Melamed S, Amitai G et al. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat Microbiol. 2020;5:1608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina R, Jensen ALG, Marchena-Hurtado J et al. Structural basis of cyclic oligoadenylate degradation by ancillary type III CRISPR–Cas ring nucleases. Nucleic Acids Res. 2021;49:12577–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso JA, Schramke H, Zhang Y et al. Binding of cyclic di-AMP to the Staphylococcus aureus sensor kinase KdpD occurs via the universal stress protein domain and downregulates the expression of the Kdp potassium transporter. J Bacteriol. 2016;198:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JW, Sudarsan N, Phillips GE et al. Control of bacterial exoelectrogenesis by c-AMP-GMP. Proc Natl Acad Sci USA. 2015;112:5389–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton AC, Bootman MD, Scott JD. Second messengers. Cold Spring Harb Perspect Biol. 2016;8:a005926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–8. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, Death A, Ferenci T The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiol Read Engl. 1997;143:1909–18. [DOI] [PubMed] [Google Scholar]
- Peng X, Zhang Y, Bai G et al. Cyclic di-AMP mediates biofilm formation. Mol Microbiol. 2016;99:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham HT, Nhiep NTH, Vu TNM et al. Enhanced uptake of potassium or glycine betaine or export of cyclic-di-AMP restores osmoresistance in a high cyclic-di-AMP Lactococcus lactis mutant. PLoS Genet. 2018;14:e1007574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quax TEF, Albers S-V, Pfeiffer F. Taxis in archaea. Emerg Top Life Sci. 2018;2:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan AA, Waters CM. Double take: a dual-functional Hypr GGDEF synthesizes both cyclic di-GMP and cyclic GMP-AMP to control predation in Bdellovibrio bacteriovorus. PLoS Genet. 2022;18:e1010263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Leipelt M, Russwurm M et al. Crystal structure of the guanylyl cyclase Cya2. Proc Natl Acad Sci USA. 2008;105:15720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R, Gabius HJ, Engelhardt R et al. Archaebacterial phenylalanyl-tRNA synthetase. Accuracy of the phenylalanyl-tRNA synthetase from the archaebacterium Methanosarcina barkeri, Zn(II)-dependent synthesis of diadenosine 5’,5’’’-P1,P4-tetraphosphate, and immunological relationship of offnylalanyl-tRNA synthetases from different urkingdoms. J Biol Chem. 1985;260:182–7. [PubMed] [Google Scholar]
- Ray WK, Potters MB, Haile JD et al. Activation of SsoPK4, an archaeal eIF2α kinase homolog, by oxidized CoA. Proteomes. 2015;3:89–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann J, Lassak K, Khadouma S et al. Regulation of archaella expression by the FHA and von Willebrand domain-containing proteins ArnA and ArnB in Sulfolobus acidocaldarius. Mol Microbiol. 2012;86:24–36. [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77:1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U, Galperin MY. Discovery of the second messenger cyclic di-GMP. Methods Mol Biol Clifton NJ. 2017;1657:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U. Great times for small molecules: c-di-AMP, a second messenger candidate in bacteria and archaea. Sci Signal. 2008;1:pe39. [DOI] [PubMed] [Google Scholar]
- Rouillon C, Athukoralage JS, Graham S et al. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. Elife. 2018;7:e36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Youn H, Kang IH et al. Identification of bacterial guanylate cyclases. Proteins. 2015;83:799–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabetta W, Vandelle E, Locato V et al. Genetic buffering of cyclic AMP in Arabidopsis thaliana compromises the plant immune response triggered by an avirulent strain of Pseudomonas syringae pv. tomato. Plant J Cell Mol Biol. 2019;98:590–606. [DOI] [PubMed] [Google Scholar]
- Samolygo A, Athukoralage JS, Graham S et al. Fuse to defuse: a self-limiting ribonuclease-ring nuclease fusion for type III CRISPR defence. Nucleic Acids Res. 2020;48:6149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Coureux P-D, Kazan R et al. Recent advances in archaeal translation initiation. Front Microbiol. 2020;11:584152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CF, Bellows LE, Tosi T et al. The second messenger c-di-AMP inhibits the osmolyte uptake system OpuC in Staphylococcus aureus. Sci Signal. 2016;9:ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoarughi GL, Cimmino C, Donini P. Lack of production of (p)ppGpp in Halobacterium volcanii under conditions that are effective in the eubacteria. J Bacteriol. 1995;177:82–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenroy AR, Visweswariah SS. Class III nucleotide cyclases in bacteria and archaebacteria: lineage-specific expansion of adenylyl cyclases and a dearth of guanylyl cyclases. FEBS Lett. 2004;561:11–21. [DOI] [PubMed] [Google Scholar]
- Shigematsu M, Kawamura T, Kirino Y. Generation of 2’,3’-cyclic phosphate-containing rnas as a hidden layer of the transcriptome. Front Genet. 2018;9:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F et al. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureka K, Choi PH, Precit M et al. The cyclic di-nucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014;158:1389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland EW, Rall TW. Fractionation and characterization of a cyclic adenine ribonucleotide formed by tissue particles. J Biol Chem. 1958;232:1077–91. [PubMed] [Google Scholar]
- Tal N, Morehouse BR, Millman A et al. Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cell. 2021;184:5728–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Venclovas Č, Siksnys V. Type III CRISPR–Cas immunity: major differences brushed aside. Trends Microbiol. 2017;25:49–61. [DOI] [PubMed] [Google Scholar]
- Tchigvintsev A, Xu X, Singer A et al. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J Mol Biol. 2010;402:524–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Malone JG. Nucleotide second messengers in bacterial decision making. Curr Opin Microbiol. 2020;55:34–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Venegas FD, Raines RT. Energetics of catalysis by ribonucleases: fate of the 2’,3’-cyclic phosphodiester intermediate. Biochemistry. 1994;33:7408–14. [DOI] [PubMed] [Google Scholar]
- Tischler AD, Camilli A. Cyclic diguanylate regulates vibrio cholerae virulence gene expression. Infect Immun. 2005;73:5873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Carrasco B, Gándara C et al. Bacillus subtilis DisA regulates RecA-mediated DNA strand exchange. Nucleic Acids Res. 2019;47:5141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley L, Yannarell SM, Huynh TN et al. Cyclic di-AMP acts as an extracellular signal that impacts Bacillus subtilis biofilm formation and plant attachment. Mbio. 2018;9:e00341–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschowri N, Schumacher MA, Schlimpert S et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control streptomyces development. Cell. 2014;158:1136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnham RE, Scott JD. Protein kinase A catalytic subunit isoform PRKACA; history, function and physiology. Gene. 2016;577:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005;13:52–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. Diadenosine 5’, 5"’-P1, P4-tetraphosphate: a pleiotropically acting alarmone?. Cell. 1983;34:711–2. [DOI] [PubMed] [Google Scholar]
- Vogt MS, Ngouoko Nguepbeu RR, Mohr MKF et al. The archaeal triphosphate tunnel metalloenzyme SaTTM defines structural determinants for the diverse activities in the CYTH protein family. J Biol Chem. 2021;297:100820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenck BR, Santangelo TJ. Archaeal transcription. Transcription. 2020;11:199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF, Allers T. DNA repair in the archaea—an emerging picture. FEMS Microbiol Rev. 2018;42:514–26. [DOI] [PubMed] [Google Scholar]
- Whiteley AT, Eaglesham JB, de Oliveira Mann CC et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature. 2019;567:194–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Hartung S, Büttner K et al. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–78. [DOI] [PubMed] [Google Scholar]
- Wright TA, Jiang L, Park JJ et al. Second messengers and divergent HD-GYP phosphodiesterases regulate 3’,3’-cGAMP signaling. Mol Microbiol. 2020;113:222–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Pal K, Sen U. Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication. Biochem J. 2019;476:3333–53. [DOI] [PubMed] [Google Scholar]
- Yannay-Cohen N, Carmi-Levy I, Kay G et al. LysRS serves as a key signaling molecule in the immune response by regulating gene expression. Mol Cell. 2009;34:603–11. [DOI] [PubMed] [Google Scholar]
- Yau KW. Cyclic nucleotide-gated channels: an expanding new family of ion channels. Proc Natl Acad Sci USA. 1994;91:3481–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Vogt MS, van der Does C et al. The phosphatase PP2A interacts with ArnA and ArnB to regulate the oligomeric state and the stability of the ArnA/B complex. Front Microbiol. 2020;11:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Cai X, Ma H et al. A decade of research on the second messenger c-di-AMP. FEMS Microbiol Rev. 2020;44:701–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamecnik PC, Stephenson ML, Janeway CM et al. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem Biophys Res Commun. 1966;24:91–7. [DOI] [PubMed] [Google Scholar]
- Zarrella TM, Metzger DW, Bai G. Stress suppressor screening leads to detection of regulation of cyclic di-AMP homeostasis by a Trk family effector protein in Streptococcus pneumoniae. J Bacteriol. 2018;200:e00045–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li W, He Z-G. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288:3085–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink IA, Fouqueau T, Tarrason Risa G et al. Comparative CRISPR type III-based knockdown of essential genes in hyperthermophilic Sulfolobales and the evasion of lethal gene silencing. RNA Biol. 2021;18:421–34. [DOI] [PMC free article] [PubMed] [Google Scholar]