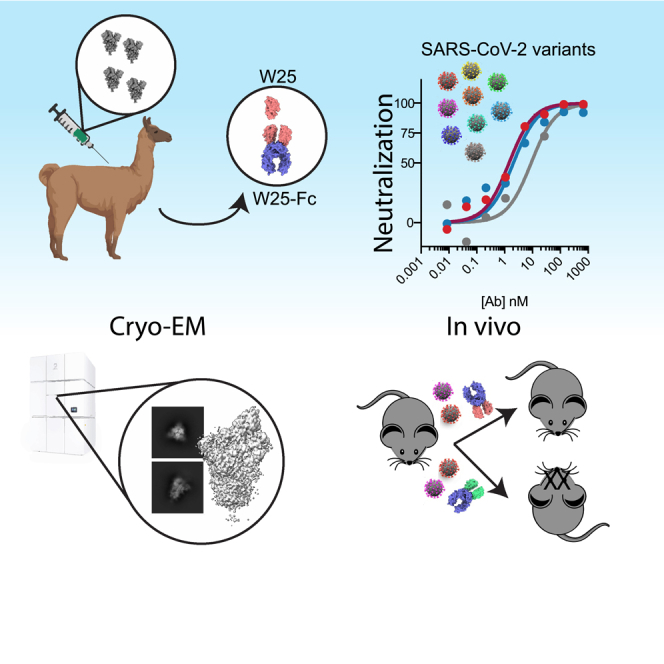
Summary
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) Omicron variant sub-lineages spread rapidly worldwide, mostly due to their immune-evasive properties. This has put a significant part of the population at risk for severe disease and underscores the need for effective anti-SARS-CoV-2 agents against emergent strains in vulnerable patients. Camelid nanobodies are attractive therapeutic candidates due to their high stability, ease of large-scale production, and potential for delivery via inhalation. Here, we characterize the receptor binding domain (RBD)-specific nanobody W25 and show superior neutralization activity toward Omicron sub-lineages in comparison to all other SARS-CoV2 variants. Structure analysis of W25 in complex with the SARS-CoV2 spike glycoprotein shows that W25 engages an RBD epitope not covered by any of the antibodies previously approved for emergency use. In vivo evaluation of W25 prophylactic and therapeutic treatments across multiple SARS-CoV-2 variant infection models, together with W25 biodistribution analysis in mice, demonstrates favorable pre-clinical properties. Together, these data endorse W25 for further clinical development.
Subject areas: Public health, Information system model, Decision science
Graphical abstract
Highlights
-
•
The nanobody W25 efficiently neutralizes SARS-CoV2 omicron sub-lineages
-
•
A dimeric W25-Fc fusion shows enhanced neutralization activity
-
•
W25 binds a unique conserved SARS-CoV2 spike RBD epitope
-
•
W25 protects K18-huACE2 mice from Wuhan and Omicron SARS-CoV-2 infections
Public health; Information system model; Decision science
Introduction
Since emergence in late 2019, the highly transmissible coronavirus—severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)—has infected more than 300 million individuals and claimed more than 6.5 million lives (WHO, updated September 2022). Despite the rapid development of vaccine strategies and preventive measures that help curb disease progression, viral transmission persists, and viral variants with altered virulence and antigenic sites have emerged in successive waves across the globe. Understanding the antigenic nature of emerging variants and the development of broad-spectrum therapeutics and vaccines remain an urgent priority.
SARS-CoV-2 expresses a surface spike (S) glycoprotein, which consists of S1 and S2 subunits that form a homotrimeric viral spike which serves as both viral attachment protein and membrane fusogen.1 Cell receptor interaction is mediated by the S1 receptor-binding domain (RBD), which binds the peptidase domain of human angiotensin-converting enzyme 2 (hACE2). Structural studies have revealed different conformations of the spike from both in vitro and in situ.2,3,4 In the prefusion stage, the RBD switches between a closed and an open conformation to facilitate hACE2 interaction.5 Subsequently, the S2 undergoes a substantial conformational change which drives the fusion of viral and cellular membranes.1
Investigations of sera from COVID-19 convalescent patients enabled isolation of potent neutralizing antibodies (NAbs), the majority of which target the RBD.6,7,8,9,10,11,12,13 Some of these have received Emergency Use Authorization (EUA) from the U.S. Food & Drug Administration for SARS-CoV2 treatment or pre-exposure prophylaxis and have shown to be effective in treating certain patients with COVID-19 (https//ww.fda.gov). However, virus evolution causes ongoing emergence of novel SARS-CoV2 variants of concern (VOCs), concomitant with reduction of vaccine14,15 and therapeutic antibody efficacy.16,17,18
The latest SARS-CoV-2 VOC B.1.1.529, designated Omicron (O), was first reported to WHO from South Africa at the end of November 2021 and is rapidly spreading across many countries, thereby replacing the already highly transmissible SARS-CoV-2 Delta variant (B.1.617.2).19 SARS-CoV-2 Omicron accumulated a large number of mutations compared to its earlier pandemic variants, of which >30 substitutions, deletions, or insertions are located in the spike protein. Scientists all over the world are unanimously reporting significantly reduced efficacies of vaccine-elicited sera against Omicron.20,21,22 To the best of our knowledge, the majority of potent monoclonal NAbs, including EUA NAbs, showed strongly reduced or no detectable neutralization activity toward Omicron. A notable exception is EUA NAb sotrovimab (S309), which however still exhibits 2- to 3-fold reduced neutralizing activity.23 Accordingly, the identification of novel, potent anti-Omicron NAbs would be highly desirable to strengthen the therapeutic repertoire.
Nanobodies are considered the smallest antigen-binding entity, representing the variable heavy-chain fragment (VHH) derived from heavy-chain-only antibodies found in camelids (llamas, alpacas, guanacos, vicuñas, dromedary, and camels). Because of their small size (2.5 nm by 4 nm; 12–15 kDa) and unique binding domains, nanobodies offer many advantages over conventional antibodies including the ability to bind cryptic epitopes not accessible to bulky conventional antibodies, high tissue permeability, ease of production, and thermostability. Due to their superior stability, nanobodies are highly suited for development as potential bio-inhaled therapies against respiratory diseases. ALX-0171, an inhaled anti-respiratory syncytial virus (RSV) nanobody, demonstrated robust antiviral effect, reduced symptoms of virus infection in animal models,24,25 and displayed promising result in reducing RSV viral load.26
We recently described the neutralizing RBD-specific nanobody,27 W25, derived from an alpaca immunized with the S protein from the ancestral SARS-CoV-2 strain (Wuhan, Wu). Here, we extend these early findings and show that unlike all hitherto described NAbs, W25 neutralizes the Omicron variant even more potently than the ancestral isolate. In order to understand the molecular basis of neutralization activity and breadth of W25, we determined the cryoelectron microscopy (cryo-EM) structures of W25 in complex with SARS-CoV-2 spike from both ancestral and Omicron SARS-CoV-2, revealing that the vast majority of mutations in the Omicron variant RBD are outside of the W25-RBD binding interface. In addition, W25-binding and live virus neutralization assays demonstrated activity against a broad range of SARS-CoV-2 VOCs. Functional assays reveal that W25 triggers fusion and that this mechanism is conserved across VOCs, including the less fusogenic Omicron variant. Furthermore, we demonstrate a protection and robust reduction of viral burden and prevention of lung pathology in the K18-hACE2 mouse model of SARS-CoV-2 challenged with VOC Beta from both prophylactic and therapeutic administration of human immunoglobulin G (IgG) Fc-nanobody fusion, W25-Fc. Importantly, W25-Fc treatment results in survival of K18-hACE2 mouse challenged with lethal dose of Beta SARS-CoV-2. Finally, we analyzed the pharmacodynamics of radiolabeled W25-Fc in mice to gain a better understanding of the pharmacological characteristics of W25-Fc to inform further drug development efforts.
Results
W25 ultra-potently neutralizes the SARS-CoV2 omicron variant
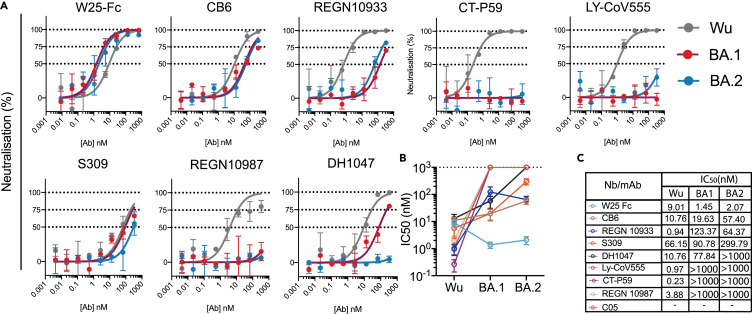
We have previously reported the neutralization activity of W25-Fc against SARS-CoV-2 Wu and D614G.27 Given the number of substitutions in the Omicron BA.1 spike protein, including G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H and BA.2 spike including G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, N501Y, and Y505H in the RBD, neutralization activity of W25-Fc was tested against SARS-CoV-2 Omicron BA.1 and BA.2 live virus on Vero E6 cells.28 Notably, W25-Fc completely and potently neutralized the Omicron BA.1 and BA.2 variant, with an IC50 (half maximal inhibitory concentration) of 1.45 nM ± 0.31 nM and 2.07 nM ± 0.66 nM, respectively, which is ∼7-fold more potent compared to the ancestral strain (Figures 1A–1C). In comparison, EUA monoclonal antibodies (mAbs) including REGN10933, REGN10987, LY-CoV555, and CT-P59 significantly or completely lost neutralizing activity against Omicron BA.1 and BA.2. Other RBD-specific mAbs including CB6 and DH1047 displayed a significant reduction of neutralizing activity against Omicron BA.1 variant, by ∼4- and 7-fold compared to the Wu strain, respectively (Figures 1A–1C), and DH1047 completely lost neutralizing activity against BA.2. Interestingly, the neutralization profiles demonstrated that while W25-Fc exhibited complete inhibition of Wu-1 and Omicron infection, S309, a broadly neutralizing mAb, did not reach 100% neutralization, as seen previously29 (Figure 1A).
Figure 1.
Authentic virus neutralization of SARS-CoV-2 variants by W25, EUA, and other mAbs
(A) Neutralization curves comparing the sensitivity of SARS-CoV-2 strains Wu (gray line) and Omicron BA.1 (red line) and Omicron BA.2 (blue line) to antibody as indicated including W25-Fc, C05, CB6, REGN10933, CT-P59, LY-CoV555, S309, REGN10987, and DH1047. The data were analyzed and plotted using nonlinear regression (curve fit, three parameters).
(B and C) Comparison of IC50 values across mAbs tested between Wu and Omicron. IC50 was calculated from the neutralization curve by Graphpad Prism 8 software. Color represents different mAbs as indicated. Data are generated from two independent experiments, each performed in technical triplicate. Data are represented as mean ± SEM.
Structural analysis of W25 binding to SARS-CoV-2 spike
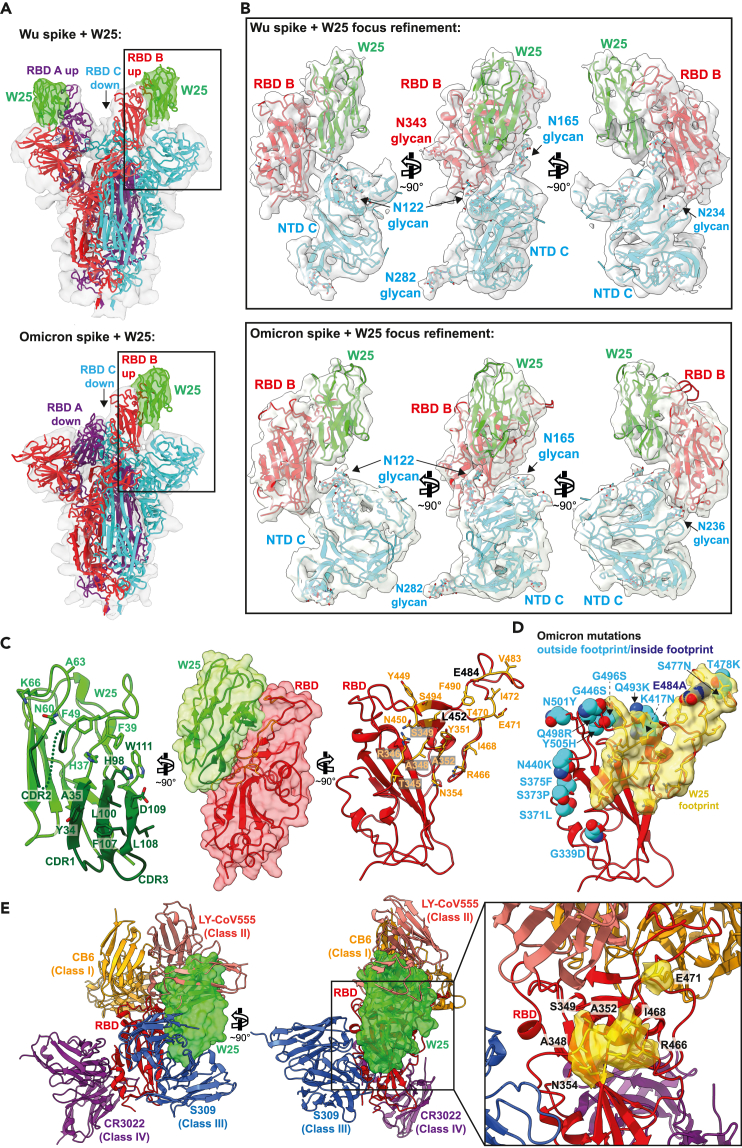
To gain mechanistic understanding of how W25 binds SARS-CoV-2 Wu and Omicron spikes, we determined their cryo-EM structures in complex with W25 (Table S1). After extensive 2D and 3D classification, two major conformational states of the Wu spike/W25 complex were identified (Figure S1). Class 1 spike trimers displayed two RBDs in an upward-facing position, with the third one in “down” position, while class 2 trimers possessed one “up” RBD and two “down” RBDs (Figure S1). For both classes, cryo-EM density maps showed additional density adjacent to the “up” RBDs, not covered by the structural model of the spike trimer, strongly suggesting that these density portions correspond to W25 (Figure 2A, upper panel, and S2). For the Omicron spike/W25 complex, multiple rounds of 2D and 3D classification identified “3 down”, “2 up, 1 down”, and “1 up, 2 down” RBD classes (Figure S3). The latter state displayed clear extra density corresponding to W25 in similar position as for the Wu spike (Figure 2A, lower panel).
Figure 2.
Structure analysis of the W25/spike interaction
(A) Cryo-EM densities of Wu (upper panel) and Omicron (lower panel) spike/W25 complexes (gray semi-transparent surfaces) (map 1, Figure S1 and map 4, Figure S3). Fitted trimeric SARS-COV2 spike protein structures (PDB: 6ZXN (Wu) and PBD: 7WG6 (Omicron)30,31 are shown in cartoon representation, with protomer A colored purple, protomer B red, and protomer C cyan. W25 is shown as green cartoon. Densities corresponding to W25 are colored green.
(B) Three views of a focused refinement of the regions illustrated in A. Proteins are shown in cartoon representation, and N-linked glycans as sticks, in the same colors as in A. Cryo-EM densities (map 3, Figure S1, and map 5, Figure S4) are shown as semi-transparent light-gray surfaces.
(C) Detailed “open book” view of the W25/RBD interaction interface between RBD and W25. Amino acids involved in intermolecular interaction are shown as sticks.
(D) Visualization of the RBD surface in contact with W25 (yellow). The RBD is shown as red cartoon. Residues mutated in the Omicron RBD are shown as spheres, and colored light blue when located outside of the W25 contact surface, or dark blue when inside.
(E) Two views of a superposition of selected RBD-bound NABs with the RBD/W25 complex, in cartoon representation (CB6 – PDB: 7C01, LY-CoV555 – PDB: 7l3N, S309 – PDB: 7R6W, CR3022 – PDB: 6YLA). W25 is shown as semi-transparent green molecular surface. For clarity, only the W25-bound RBD is shown as red cartoon. The inset shows an enlarged view of the unique portion of the W25 epitope as yellow surface.
Focused refinement of the Wu and Omicron spike RBDs, the N-terminal domain (NTD) regions of the adjoining spike protomer, and W25 resulted in contiguous density maps of sufficient quality for flexible fitting and refinement of a W25 structure model generated by AlphaFold232,33 (Figure 2B, Table S1 and Video S1). W25 binds to the side and upper edge of the RBD, with the downward-facing side of W25 surrounded by N-linked glycans arising from spike protomer B NTD N122 and N165 (Figure 2B). Since W25 positions relative to Wu or Omicron RBDs were equivalent and the map quality and resolution of the Wu spike/W25 complex were slightly better (Figures 2B and S1 and S3), the Wu spike/W25 model was subjected to further structural analysis. Superposition of the RBD/W25 complex structure with the Wu spike class 1 “down” RBD indicates that W25 binding would lead to a slight overlap with the adjacent spike NTD, rationalizing why W25 preferentially binds to RBDs in the “up” conformation (Figure S4A).
Detailed inspection of the RBD/W25 contact surface reveals an interface area of 847 Å2 with mostly hydrophobic character, involving W25 β-sheet 3 CDR1 residues Y34 and A35 and downstream amino acids H37 and F39 (Figure 2C). In addition, there are interfacing residues in the loop region connecting β-sheets 3 and 4, which include CDR2 (M41, R47, F49, F53, G54, N60, A62, Y61, A63, K66), and ultimately residues in β-sheet 7 (CDR3): H98, L100, and L105-W111. From the RBD side, amino acid residues in an N-terminal loop region contribute to W25 binding (T345-S349, Y351, A352, N354), as well as from the loop downstream of β-sheet 3 (Y449-L452) and from a large loop preceding β-sheet 4 (R466, I468, T470-I472, G482-E484, F490, S494). In addition, there are hydrogen bonds between W25 side chains D109 and W111 and RBD side chain S349 and the backbone carbonyl group of N450, as well as between RBD side chains N354 and T470 and backbone carbonyls of W25 F53 and L105. Twenty-two out of 23 epitope residues are conserved between Wu and Omicron. Only one interfacing residue is non-conservatively exchanged between Wu and Omicron SARS-CoV2 (E484A) (Figure 2D, S4B and Video S1).
Comparison with RBD binding modes of other nanobodies, NAbs, and ACE2
In order to put the present structure analysis in context, PDB entries containing camelid or synthetic SARS-CoV2-specific nanobodies were collated and structurally aligned on their RBDs, together with the RBD/W25 complex (Figure S4C and Table S2). The vast majority of nanobodies attach either to one side of the RBD (“side-1” or cluster 1 in34) or to the upper RBD surface (“top” or cluster 2). Two of the nanobodies bind to an RBD region opposite of cluster 1 (“side-2”). Interestingly, the W25 binding mode seems to incorporate characteristics of two of these clusters by occupying a surface area overlapping with epitopes of both “top” and “side-2” binders.
In addition, structures of the variable fragments of selected NAbs, representing distinct RBD binding modes (classes 1–4),35 were superimposed on the W25/RBD complex structure (Figure 2E). W25 partially overlaps with the VH fragment of class 2 NAb LY-CoV555, resulting in partially shared epitopes involving RBD amino acid residues 351, 449–453, and 480–490. Furthermore, W25 minimally clashes with loop regions of class 3 NAb S309 VH (residues 102–109) and VL (residue 94); their respective epitopes are adjacent but do not overlap. Importantly, a significant portion of the W25 epitope, including surface-exposed RBD amino acid stretches 348–354 and 466–471, is unique and not covered by the CDRs of the representative class 1–4 NAbs analyzed here (Figure 2E, right panel).
Subsequently, structural alignment of the RBD moieties of the RBD/W25 complex presented here with the structure of the RBD bound to its human cell surface receptor ACE2 was performed. The analysis demonstrates that there is a slight overlap of the molecular surfaces (Figure S4D). In this hypothetical model, W25 side chain E46 directly opposes ACE2 residue E35, which might induce electrostatic repulsion and lead to obstruction of the ACE2-binding site or ACE2 displacement from the RBD (Figure S4E).
W25 binds multiple VOCs of SARS-CoV-2 and retains binding to BA.2 Omicron sublineage
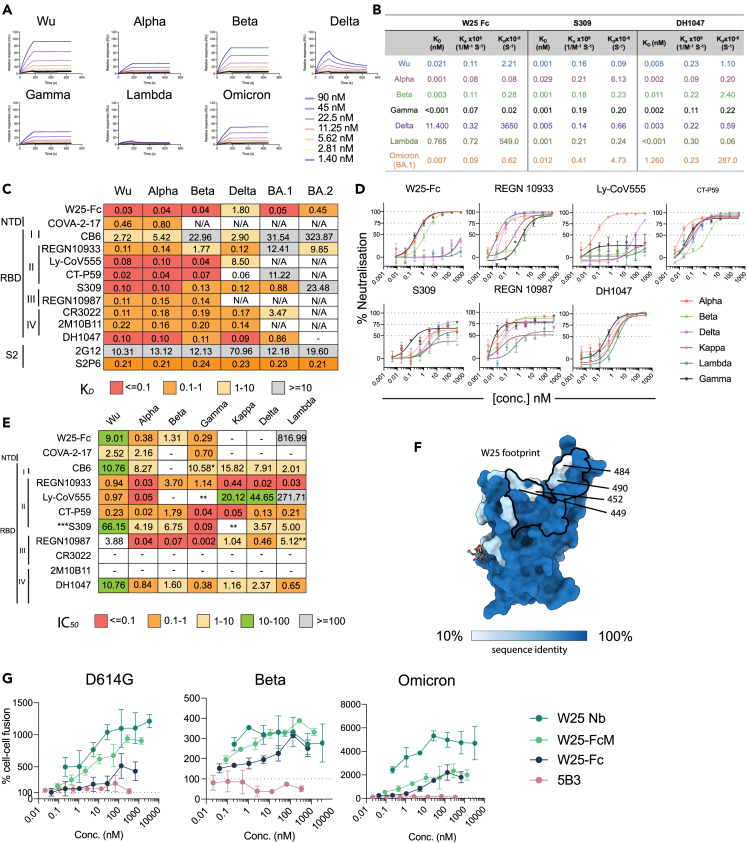
To evaluate W25 binding to previous SARS-CoV-2 VOCs and the rapidly expanding BA.2 Omicron sublineage, we expressed and purified Wu and VOC spikes (bearing mutations indicated in Figure S5A) and validated spike purity and trimer formation by SDS-PAGE (Figure S5B) and size exclusion chromatography (SEC) (Figure S5C). Binding kinetics of W25-Fc to the spike variant proteins were assessed by surface plasmon resonance (SPR) (Figure 3A). Subnanomolar dissociation constants (KD) were observed for Wu, Alpha, Beta, Gamma, and Omicron BA.1 using a 1:1 kinetic model (Figure 3B). The KD values of W25-Fc for Delta and Lambda variants were higher compared to other variants, at 11.4 and 0.7 nM, respectively (Figures 3A and 3B). As control, S309 and DH1047 NAbs also yielded subnanomolar KD values (Figures 3B and S6). ELISA was consistent with these findings (Figures 3C and S7A), showing that W25 binds all variants with similar KD (0.033–0.048 nM), except for the Delta variant (1.8 nM). W25 still retains binding affinity to XBB1.5 spike (Kd value = 0.1 nM) (Figure S7D). In agreement with recent reports,36,37 and our neutralization assays, EUA RBD-specific mAbs including REGN10933, LY-CoV555, and CT-P59 showed significantly reduced binding affinity (>10-fold reduction of KD) for Omicron BA.1, with the exception of S309 (Figure 3C). We observed no binding of REGN10987 to Omicron BA.1 variant. Structurally defined cross-reactive mAbs including DH1047 (RBD-specific NAb), CR3022, and 2M10B11 (non-neutralizing cryptic RBD epitopes) showed significantly reduced binding to Omicron BA.1 variant (∼5-fold).
Figure 3.
Broad reactivity of W25-Fc is conferred by conserved patch on SARS-CoV-2 RBD and potential inhibition mechanism of W25
(A) SPR sensograms of W25-Fc. W25-Fc was immobilized on SPR protein A chips. Various concentrations of SARS-CoV-2 spike variant proteins as indicated were injected for 30 s, at 30 μL/min followed by dissociation for 600 s. Dissociation constants (KD) were determined on the basis of fits, applying a 1:1 interaction model. Similar experiments were conducted for S309, DH1047, and C05 (as control, Figure S6).
(B) Summary table showing KD, Ka,Kd of indicated Nb/mAbs.
(C) Kd values from ELISA binding curves of W25-Fc, EUA mAbs, and other epitope-specific mAbs to SARS-CoV-2 spikes variants.
(D) Neutralization curves comparing the sensitivity of live SARS-CoV-2 viruses including Alpha, Beta, Delta, Kappa, Lambda, and Gamma to W25-Fc, EUA, and other RBD-specific mAbs as indicated. The data were analyzed and plotted using nonlinear regression (curve fit, three parameters), and IC50 value was calculated from neutralization curves by Graphpad Prism 8 software.
(E) Summary of IC50 values (nM) of neutralistion of SARS-CoV-2 variants performed in Vero E6 cells. Values that approached 50% neutralization were estimated from (D). Data are generated from two independent experiments, each performed in technical duplicate.
(F) Molecular surface conservation of RBD VOCs. (G) W25 and its derivatives enhance spike-mediated cell fusion. Cell-cell fusion assay was performed. Stable cells expressing the rLuc-GFP components (effector cells) were transiently transfected SARS-CoV-2 spike proteins bearing spike mutations for D614G (top left), Beta (top right), Omicron (Bottom left), or control plasmid (no envelope control). W25 Nb, W25 Dimeric Fc (W25-Fc), or W25 monomeric Fc (W25-FcM) were incubated with effector cells for 1 h prior to co-culturing with target cells (huACE2-HEK293T cells). After 24–48 h, Renilla luciferase was read and analyzed by subtraction with control plasmid treatment. Data are generated from at least three biological replicates. Data are represented as mean ± SEM.
Continuing surveillance of Omicron evolution revealed the newly emerged BA.2 Omicron sublineage, containing 8 unique spike mutations while lacking 13 spike mutations found in BA.1.29 Of the anti-RBD panel we tested, only W25-Fc retained sub-nanomolar affinity to BA.2 with a KD value of ∼0.4 nM. Conversely, all EUA mAbs, including S309, showed significantly impaired binding to BA.2 spike protein (Figures 3C and S7B). S2-specific mAbs S2P6 and 2G12 bound all variants with KD’s of approximately 0.2 nM and 10–12 nM, respectively (Figure 3C). Additionally, a direct comparison with other previously characterized engineered Nb-Fc fusions demonstrated that W25-Fc exhibits superior binding affinity for Omicron BA.1 (Figure S7C).
W25 neutralizes multiple VOCs of SARS-CoV-2
As SARS-CoV-2 continues to evolve, the dominant variants are continuously changing. To explore and compare the efficacy of W25 for neutralization of SARS-CoV-2 VOCs, we performed live virus neutralization on Vero E6 cells against major VOCs including Alpha, Beta, Gamma, Delta, Kappa, and Lambda and found that W25-Fc neutralized Alpha, Beta, and Gamma variants (Figures 3D and 3E). W25-Fc showed complete and potent inhibition of SARS-CoV-2 Alpha, Beta, and Gamma variant infections with IC50 values ranging from 0.38 to 1.31 nM. The monomeric W25 also neutralized similar VOCs with IC50 values ranging from 0.42 to 2.12 nM (Figures S8A and S8B). Interestingly, both monovalent and divalent forms exhibited significantly weaker neutralization for Kappa, Lambda, and Delta variants. Of note, monomeric W25 was effective against Beta and Gamma, two of the most resistant variants leading to first-generation RBD-associated antibodies.15 Similar to other reports, we demonstrated that introducing bivalency through Fc fusion further improved efficacy of W25-Fc (∼10-fold for Gamma variant). Overall, the neutralization data are consistent with the W25 binding mode observed in the cryo-EM analysis since key residues within the W25 binding interface, with the exception of L452 and E484, were highly conserved across all VOCs examined in this work (Figure 3F and S9).
W25 stimulates spike-dependent cell-cell fusion
We furthermore explored the modulation of spike-mediated membrane fusion as a potential neutralization mechanism, using established cell-cell fusion assays.38 W25 and its derivatives (Fc fusion and monomeric Fc fusion [FcM]) caused extensive cell-cell fusion for SARS-CoV2 D614G-, Beta-, and Omicron-infected cells, demonstrated by luciferase activity (Figure 3G), when compared to a control antibody (anti-Nipah virus Fusion protein, 5B3). This was most striking for Omicron, which in the absence of W25 did not cause significant cell-cell fusion. Overall, W25-Fc induced less spike-mediated fusion, followed by W25-FcM and W25 (W25 Nb) in all variants tested. No enhancement was observed for control antibody treatment. Additionally, live cell GFP-reporter IncuCyte assays were performed in parallel and showed consistent results, where W25 addition greatly enhanced the GFP signal (Figures S10 and 11). Thus, W25 may additionally act through premature fusion activation, causing irreversible inactivation of coronavirus particles.
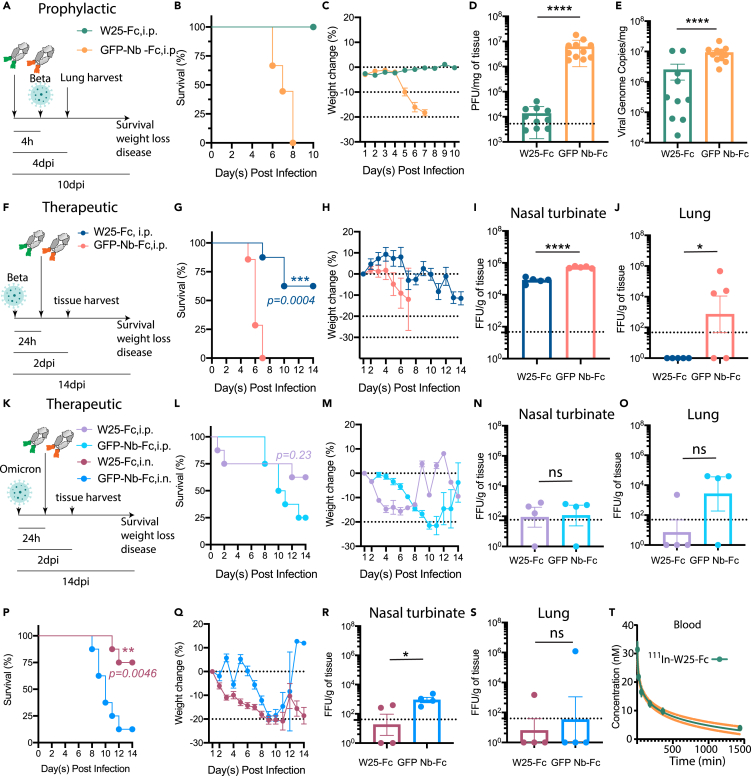
W25 prophylactically and therapeutically reduces disease burden and protects from SARS-CoV-2 infection mortality in mice
Since nanobody affinity and function have shown to be limited by temperature,39 thermostability of W25 was tested and showed W25 retains its antigen-binding function after heat treatment at 90°C (Figures S12A and S12B). In addition, full stability of the monomeric W25 after nebulization was observed, suggesting potential stability for airway administration (Figures S12C and S12D). To assess the use of W25-Fc for pre- and post-exposure therapy to counter SARS-CoV-2 infection, the K18-hACE2 transgenic murine model was employed. W25-Fc or GFP-Nb-Fc (control) were administered by intraperitoneal (i.p.) injection 4 h prior to or 24 h after intranasal (i.n.) challenge of K18-hACE2 mice with SARS-CoV-2 Beta variant (Figures 4A and 4F). It has been shown previously that SARS-CoV-2 Beta is significantly more lethal and causes more severe organ damage in K18-hACE2 mice than both the Wu ancestral strain and the D614G variant, mimicking severe SARS-CoV-2 infection in humans.40 Infected mice were monitored for 10 or 14 days after infection. All SARS-CoV-2-infected animals from the prophylactical W25-Fc treatment group survived, as opposed to control-treated group (Figure 4B). Symptoms, including ruffled fur/piloerection and accelerated shallow breathing, were observed in control-treated animals at 4–5 days after infection and progressed rapidly. Some non-treated, infected mice reached the humane endpoint state by day 6 post infection. Prophylactical W25-Fc treatment prevented all respiratory disease (Figure S13) and weight loss (Figure 4D) caused by SARS-CoV-2 Beta infection. In a separate experiment, lungs were collected after 4 days of infection. W25-Fc treatment reduced infectious virus level up to 103-fold when compared to the control group, and viral RNA also showed a significant reduction (Figures 4D and 4E). Therapeutic treatment (24 h post infection) of SARS-CoV-2 Beta-infected mice (Figure 4F) with W25-Fc significantly improved survival (60%) (Figure 4G) and weight loss (Figure 4H) and lead to lower viral load in nasal turbinate (Figure 4I) and lung (Figure 4J), compared to GFP-Nb-Fc-treated infected mice.
Figure 4.
W25-Fc protects mice from lethal Beta and Omicron SARS-CoV-2 infection and W25-Fc pharmacodynamic analysis
(A) Experimental schematic: eight- to twelve-week-old male and female K18 transgenic mice were inoculated via the intranasal route with 1 × 103 PFU of SARS-CoV-2 (Isolate B.1.351). W25-Fc or GFP Nb-Fc were administered intraperitoneally 4 h prior to infection.
(B) Survival and (C) weight change were monitored and scored. Two experiments were performed (n = 9–12, Log rank (Mantel-Cox) test ∗∗∗∗p < 0.0001).
(D) Viral burdens were determined in lung tissues 4 day post infection via plaque forming assays for infectious virus and (E) RT-qPCR for viral genome copy number. Two experiments were performed (N = 10–11).
(F) Experimental schematic: five- to six-week-old female K18 transgenic mice were inoculated via the intranasal route with 1 × 104 FFU of SARS-CoV-2 (Beta, Isolate B.1.351) N = 8, and N = 4–5 each group for survival and tissue harvest experiment, respectively. For therapeutic treatment, W25-Fc or GFP-Nb-Fc were administered intraperitoneally 24 h after SARS-CoV-2 Beta infection at 5 × 103 FFU/mouse.
(G) Survival and (H) weight change were monitored. Viral loaded were determined in (I) nasal turbinate and (J) lung tissues 2 day post infection by immuoplaque assays for infectious virus.
(K) Experimental schematic: for Omicron infection, five- to six-week-old female K18 transgenic mice were inoculated via the intranasal route with 1 × 105 FFU of SARS-CoV-2 (Isolate B.1.1.529). W25-Fc or GFP-Nb-Fc were administered intraperitoneally or intranasally 24 h after infection.
(L and P) Survival and (M and Q) weight changes were monitored and scored. Viral loaded were determined in (N and R) nasal turbinate and (O and S) lung tissues 2 day post infection by immunoplaque assays for infectious virus. Bars represent medians, dots are individual animals, and dotted horizontal lines indicate the limit of detection. Data are represented as mean ± SEM.
(T) The pharmacodynamics of W25-Fc in blood was determined by conjugation of W25-Fc to radioactive 111Indium. One mg/kg were injected intravenously through the tail to 6 groups of mice (group 1: 5 min, group 2: 20 min, group 3: 60 min, group 4: 3 h, group 5: 5 h and group 6: 24 h). The mice were dissected, and concentration of 111In W25-Fc was measured in the blood using an Auto-Gamma Counter.
The efficacy of W25-Fc against Omicron BA.1 was also tested. Omicron virus appears less pathogenic in K18-ACE2 mice41 compared to earlier isolates. In this work, we inoculated mice with a lethal dose (105 FFU/mouse), and W25-Fc was administrated 24 h after infection via i.p. or i.n. delivery (Figure 4K). In parallel experiments, tissues were harvested 2 days post infection. For i.p. delivery, mice treated with W25-Fc had slight improvement of survival rate as compared to those which received control treatment (62.5% vs. 25%, p=0.23). Some mice treated with W25-Fc reached human endpoint by 2 days post-treatment (Figure 4L). Mice treated with W25 via i.p. displayed weight loss between day 2–8 post infection (Figure 4M). No significant reduction of viral load in nasal turbinate (Figure 4N) and lung was observed (Figure 4O). Notably, i.n. delivery of W25-Fc to Omicron-infected mice significantly improved survival (75%) compared to control-treated group (Figure 4P). Mice that received W25-Fc i.n. had lower weight compared to control-treated mice (Figure 4Q) but also lower viremia in nasal turbinate compared to controls (Figure 4R). No significant difference between W25-Fc- and control-treated mice was observed for viremia in lung (Figure 4S). Consistent with other studies, we observed low level of viremia in lung from Omicron-infected mice compared to SARS-CoV-2 Beta variant infection.41,42 Finally, we determined the pharmacokinetic profile of single dose intravenously injected W25-Fc radiolabeled with 1.2–1.5 MBq/mouse (1 mg/kg) of indium-111 in C57BL/6 mice (Figure S14A). Ex vivo studies were carried out measuring radioactivity in several tissues including blood, lungs, liver, kidneys, spleen, brain, muscle, and tail after 5 min, 20 min, 1 h, 3 h, 6 h, and 24 h (Figures S14B–S14H). We found that the half-life of W25-Fc in blood is 20.6 h, and after 24 h, the concentration of W25-Fc in blood was 3.8 ± 03 nM, well above the in vitro neutralization IC50 values for most variants (Figures 3D, 3E, and 4T).
Discussion
COVID-19 vaccines have proven to be highly effective in reducing the number of severe cases43,44 and are associated with lower transmissibility,45 but SARS-CoV-2 continues to evolve. The Delta variant has been dominant around Sep–Dec 2021, but more recently Omicron and its sub-lineages prevailed due to their unparalleled transmissibility. Certain therapeutic antibody cocktails have been approved for post-exposure treatment to reduce severe illness. However, the potency and breadth of protection by the majority of therapeutic antibodies are compromised by emerging SARS-CoV-2 immune-escape mutations, underlining an urgent need for developing, preparing, and improving antiviral avenues.
The COVID-19 pandemic highlighted the importance of the rapid discovery of antiviral drugs. In this effort, a growing list of neutralizing antibodies, nanobodies, and synthetic antivirals have been developed using tools such as human/mouse B cells or immunized camelids.8,9,10,11,46 In contrast to conventional antibodies that require post-translation modifications from mammalian cells, nanobodies can be expressed in prokaryotic cells and, hence, are cheaper to produce. Benefiting from their small size, nanobodies may bind with higher affinity, target epitopes not accessible to conventional antibodies, and are amenable to nasal administration to directly reach infected mucosal tissues and lungs.47 However, due to their low molecular weight, nanobodies may have very short half-lives in the bloodstream. Our work and previous reports48 showed that dimerization via Fc fusion significantly improved neutralization potency, potentially through simultaneous engagement of two adjacent “up” RBDs (Figures 3D and 3E).
The majority of NAbs raised against Wu SARS-CoV-2 are not effective against SARS-CoV-2 Omicron. In this work, we demonstrate that W25 and W25-Fc display potent inhibitory activity in vitro against the SARS-CoV-2 Omicron variant, as well as other VOCs including Alpha, Beta, and Gamma (Figures 1, 3D, and 3E) in live virus neutralization assays. This is consistent with structure-based sequence alignments (Figure S9), demonstrating that the majority of W25 residues involved in RBD binding are highly conserved across SARS-CoV-2 VOCs (Figure 3F), with the exception of L452, which is centrally located in the binding interface. Neutralization of Kappa, Lambda, and Delta variants by W25-Fc is strongly impeded (Figures 3D and 3E). These variants share the L452R/Q mutation, which may lead to steric clashes with W25 residues F39 and/or W111 (Figures 2D, 3F, and S9), thus rationalizing weaker binding affinity and neutralization activity of W25 toward these variants (Figures 3C–3E). This also suggests that W25 might not be effective against Omicron BA4/5. Unlike other antibodies,49,50,51,52,53 mutations at RBD position 484 (E484 K/A/Q), presented on Beta, Gamma, Kappa. and Omicron spike variants, do not impede the binding and neutralizing activity of W25. These mutations might be tolerated since E484 is only peripherally situated in the W25/RBD interface (Figures 2D and 3F). Intriguingly, W25, but not S309, retains a relatively high affinity to BA2.1 spike bearing mutation at amino acid 371 which is thought to be the key change due to introduction of a bulky phenylalanine side chain, causing loss of efficacy for several EUA mAbs and nanobodies.53,54 This is also in accordance with our structure analysis, showing no involvement of residue 371 in W25 binding. Importantly, BA.2 derivative strains recently emerged; among them is the XBB.1.5 lineage. XBB.1.5 contains RBD amino acid exchanges R346T and F490S, which are located in the W25/RBD binding interface. F490S is also found in the Lambda variant, which is not sensitive to W25. However, W25 retains significant higher binding affinity to the XBB1.5. spike (Kd = 0.1 nM, Figure S7) than to the Lambda spike (Kd = 0.765, Figures 3A and 3B), suggesting that the Lambda L452R substitution may be the main determinant of W25 inefficiency. Ultimately, however, how and if these affinity differences translate into neutralizing activity of W25 against these variants remains to be tested.
Several SARS-CoV-2 NAbs have been reported targeting NTD and S2 epitopes; the most effective antibody classes however appear to target the RBD due to its natural role in binding the hACE2 receptor. Prior structural studies have classified these SARS-CoV2 NAbs according to their RBD binding modes35,55 (Figure 2E). Class 1 and 2 NAbs obstruct the ACE2-binding site on top of the RBD. However, they are highly susceptible to VOCs escape mutations including K417N, G446S, E484A, and Q493R. Classes 3 and 4 bind largely outside of the ACE2 interaction interface, to opposite surfaces on the side of the RBD. Our structure analysis showed that W25 targets a unique RBD epitope, partially overlapping with class 2 and 3 NAbs, not affected by Omicron mutations. Accordingly, W25 might be suitable as therapeutic agent in combination with class 1 and/or 4 NAbs to improve treatment efficacy against SARS-CoV2 Omicron strains or newly emerging VOCs.
Most N-glycosylation sites are highly conserved across multiple SARS-CoV-2 variants, particularly in the NTD (N61, N122, N165, and N234). Our structural analysis found that W25 is surrounded by N-linked glycans at N122 and N165 from NTD of the neighboring spike protomer (Figure 2B). Interestingly, beyond its shielding function, N165 act as a molecular switch for RBD conformation by helping to maintain RBD in the “up” or ACE2-accessible state.56,57 Accordingly, RBD/W25-interaction might interfere with these glycan-mediated RBD opening processes. In addition, the binding of a class 2 NAb, BD-23, is also facilitated by this glycan.58
The mode of action of W25 in activating fusion mechanism may have clinical implications. Our structural analysis indicates that W25 only associates with RBD-up state and slightly overlaps with the ACE2-binding surface. This is in line with our fusion assay demonstrating that W25 triggers fusion activity of transfected spike protein in the presence of the cognate receptor ACE2 on the target cell population. Thus W25 may promote RBD-up conformation facilitating ACE2 engagement and enhance fusion. However, no infectivity enhancement was observed in authentic virus neutralization assay (Figures 3D and 3E); thus W25 may trigger premature fusogenic conformational changes on spike leading to viral inactivation prior to cellular engagement. A similar mechanism has been reported for RBD-specific mAbs including antibodies CR302259 and S23060 and nanobodies such as VHH E, W, and U.53 However, the mode of engagement of W25 to spike is unique to these previously reported binders, and it incorporates the outward face of RBD and is largely non-overlapping with the ACE2 site (Figure S15). Interestingly, induction of fusion was reduced using the dimeric W25-Fc relative to the monomeric form, although dimeric W25-Fc showed increased antiviral activity; thus multiple modes of action may underpin the potency of W25.
In K18-hACE2 mouse model, W25-Fc administrated before infection provided full protection to all treated animals from a fatal SARS-CoV-2 challenge dose. Mice that received W25 prophylactic treatment exhibited no sickness and weight loss following the challenge with SARS-CoV-2 Beta variant (Figures 4A–4E). Single dose of therapeutic treatment of W25 after Beta and Omicron challenge provided improvement in survival (Figures 4F–4S). Interestingly, i.n. delivery of W25 conferred a better outcome compared to i.p. delivery This could be attributed to virus tropism where Omicron is reported to have higher replication in ex vivo bronchial epithelium as compared to other VOCs and Wu.61 This suggests that i.n. delivery of W25 may directly block the key infection site for Omicron. Nebulization, being more efficient at targeting the deep and local pulmonary structures, could be tested for W25 efficacy in preclinical setting. W25 in combination with other Nbs could be used as novel cocktail treatment that could be rapidly generated to block virus mutational escape. Of note, the efficiency of delivery methods for each VOC may need to be considered and assessed. In conclusion, our results show that the W25 nanobody, raised against the RBD of the ancestral SARS-CoV-2 strain, has highly potent neutralizing activity against multiple SARS-CoV-2 variants including Omicron in vitro. Our structural analyses reveal that the binding interface of W25 on the RBD is unique and highly conserved across multiple VOCs. In vivo mouse model SARS-CoV-2 infection studies demonstrated that W25 confers protection when administrated prophylactically or therapeutically, suggesting its therapeutic potential as a passive immunotherapy. The COVID-19 pandemic is still ongoing, with SARS-CoV-2 constantly evolving, prompting speculation that there will be new VOCs driving a resurgence of infections. Although vaccination remains the main measure against the pandemic, it is not effective in or possible for all patients. Accordingly, there is still an urgent need to provide additional means for infection prevention and/or treatment of high-risk patients and those in mid- to low-income countries. Because of its high efficiency, remarkable stability, and resilience to nebulization, W25 has high therapeutic potential in SARS-CoV-2-infected individuals.
Limitations of the study
We also recognize that there are some limitations in our present study. Firstly, our experiments analyzing the neutralization profile and cell-to-cell fusion were performed in Vero E6 cells and in HEK293 cells overexpressing ACE2. While these cells are widely used and validated in preclinical settings, future investigations using primary nasal and/or lung epithelial cells may offer insight into the observed in vivo therapeutic activity. Secondly, as nebulization was only performed in uninfected mice, nebulization in an infection model would need to be performed to access the efficacy. Relevant to this, however, i.n. delivery of W25-Fc was shown to be therapeutically beneficial for Omicron infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| W25 | Valenzuela Nieto et al.27 | N/A |
| W270 | Valenzuela Nieto et al.27 | N/A |
| NB3 | Schoof et al.52 | N/A |
| NB11 | Schoof et al.52 | N/A |
| NB20 | Schoof et al.52 | N/A |
| MR17 | Li et al.62 | N/A |
| SB14 | N/A | |
| SB42 | N/A | |
| SB4 | N/A | |
| Ty1 | Hanke et al.31 | N/A |
| H11 | Huo et al.63 | N/A |
| Nb against GFP | Kubala et al.64 | N/A |
| S309 | Pinto et al.9 | N/A |
| REGN10987 | Hansen et al.10 | N/A |
| REGN10933 | Hansen et al.10 | N/A |
| Ly-CoV555 | Jones et al.11 | N/A |
| CT-P59 | Kim et al.12 | N/A |
| CB6 | Shi et al.8 | N/A |
| DH1047 | Martinez et al.13 | N/A |
| 5B3 | Dang et al.65 | N/A |
| C05 | Ekiert et al.66 | N/A |
| Anti-human IgG conjugated with an IRDye 800CW | Millennium Scientific | Cat#LCR-926-32232 |
| Goat Anti-Human IgG Fc Highly Cross-Adsorbed Secondary Antibody | Thermofisher | Cat#A18829 |
| Bacterial and virus strains | ||
| StellarTM Competent cells | Takara | Cat#636763 |
| hCoV-19/Australia/QLD02/20200 | This paper | GISAID accession ID, EPI_ISL_407896 |
| hCoV-19/Australia/QLD1517/2021 | This paper | GISAID accession ID EPI_ISL_944644 |
| hCoV-19/Australia/QLD1520/2020 | This paper | GISAID accession ID EPI_ISL_968081 |
| hCoV-19/Australia/QLD1893C/2021 | This paper | GISAID accession ID EPI_ISL_2433928 |
| hCoV-19/Australia/NSW4318/2021 | This paper | GISAID accession ID EPI_ISL_1121976 |
| hCoV-19/Australia/NSW4431/2021 | This paper | GISAID accession ID EPI_ISL_1494722 |
| hCoV-19/Australia/NSW-RPAH-1933/2021 | This paper | GISAID accession ID EPI_ISL_6814922 |
| Chemicals, peptides, and recombinant proteins | ||
| Spike proteins (Wuhan, Alpha, Beta, Gamma, Delta Lambda, BA.1, BA.2) | This paper | N/A |
| Critical commercial assays | ||
| KPL milk diluent/blocking solution concentrate | SeraCare | Cat#5140-0011 |
| 3,3,5,5′-Tetramethylbenzidine | Thermofisher | Cat#002023 |
| Experimental models: Cell lines | ||
| ExpiCHO-STMcells | Thermofisher | Cat#29127 |
| Flp-In™ T-REx™ 293 cells | Thermofisher | Cat#R78007 |
| HEK293T | ATCC | CRL-3216 |
| VeroE6 | ATCC | CRL-1586 |
| VeroE6-TMPRSS2 | This paper | N/A |
| VeroE6-ACE2-TMPRSS2 | BEI resource | NR-54970 |
| Experimental models: Organisms/strains | ||
| Transgenic hACE2 mice (2B6.Cg-Tg(K18-ACE2)2Prlmn/J) | Jackson laboratory | RRID:IMSR_JAX:034860 |
| Oligonucleotides | ||
| primers for SARS-CoV-2 N gene Forward 5′- TTACAAACATTGGCCGCAAA-3′ | This paper | N/A |
| primers for SARS-CoV-2 N gene Reverse 5′-GCGCGACATTCCGAAGAA-3′ | This paper | N/A |
| primers for HPRT1 Forward 5′- GTTGGATACAGGCCAGACTTTGTTG-3′ | This paper | N/A |
| primers for HPRT1 Reverse 5′-GAGGGTAGGCTGGCCTATTGGCT-3′ | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.01 | GraphPad Software Inc. |
https://www.graphpad.com/scientific-software/prism/" https://www.graphpad.com/scientific-software/prism/ |
| MotionCorr2 | Zheng et al.67 | N/A |
| CTFFind4 | Rohou et al.68 | N/A |
| Relion 3.0.7 | Zivanov et al.69 | N/A |
| Relion 3.1.3 | Zivanov et al.69 | N/A |
| cryoSPARC 3.3.1 | Punjani et al.70 | N/A |
| AlphaFold2 | Jumper et al.,71 Mirdita et al.33 | N/A |
| Coot 0.9.5 | Emsley et al.72 | N/A |
| Phenix 1.19.2–4158 | Liebschner et al.73 | N/A |
| ChimeraX | Petterson et al.84 | N/A |
| Biacore Insight Evaluation | Cytiva | https://www.cytivalifesciences.com/en/us/shop/protein-analysis/spr-label-free-analysis/software/biacore-insight-evaluation-software-p-23528 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel Watterson (d.watterson@uq.edu.au).
Material availability
Mabs plasmids and mAbs generated in this study are available from the authors upon request.
Experimental model and study participant details
Cell lines
HEK293T (ATCC), Vero-E6 (ATCC) Vero-Ace2-Tmprss2 cells (BEI resources) were maintained in DMEM supplemented with 10% heat-inactivated (HI) Fetal Calf Serum (FCS, Bovogen), penicillin (100 IU/mL)/streptomycin (100 μg/mL) (P/S, Gibco) and L-glutamine (2 mM) (Life Technologies). Cells were cultured at 37°C and 5% CO2. ExpiCHO cells were maintained in ExpiCHO Expression media as per the manufacturer’s protocol. Flp-In T-REx 293 cells were cultured according to the manufacturer’s protocol. All cell lines used in this study were routinely tested for mycoplasma and found to be mycoplasma-free (MycoAlert Mycoplasma Detection Kit MycoAlert, Lonza).
Evaluating the efficacy of W25 in K18-huACE2 mice
Six to 8 weeks old female transgenic hACE2 mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were purchased from The Jackson Laboratory and Animal Resources Center and/or bred in-house. C57BL/6J female mice weighing 20–23 g were purchased from Janvier, France, La Rochelle. All mice were kept in a climate-controlled facility with a 12 h light/dark cycle.
Study approval
All work with infectious virus was performed in a biosafety level 3 laboratory. In vitro work and protocol were approved by the University of Queensland Biosafety committee (IBC/447B/SCMB/2021) and the Penn Institutional Biosafety Committee and Environmental Health and Safety. All the animals infected by SARS-CoV-2 were handled in a biosafety level 3 animal facilities in accordance with the recommendations for care and use of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the Guidelines to promote the wellbeing of animals used for Scientific purposes. The protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania (protocol #807017) and the University of Queensland animal ethics committee (2021/AE000929 and 2021/AE001119). All the authors declare their compliance with publishing ethics. C57BL/6J mice were conducted in accordance with the European Commission’s Directive 2010/63/EU, FELASA and ARRIVE guidelines for animal research and, with approval from The Danish Council for Animal Ethics (license numbers 2017-15-0201-01283).
Method details
Transient expression of S protein, mAbs and Nbs
Expression and purification of SARS-CoV-2 Spike
Soluble, trimeric spikes (residue 1–1204 amino acid) of SARS-CoV-2/human/China/Wuhan-Hu-1/2019 (referred to as Wu) (GenBank: MN908947), SARS-CoV-2 Beta variant (B.1.351) (L18F, D80A, D215G, Δ242–244, K417N, E484K, N501Y, D614G and A701V), Alpha variant (B.1.1.7) (Δ69-70, ΔY144–145, N501Y, A570D, D614G, P681H, T716I, S982A and D1118H), Gamma variant (P.1) L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, D655Y, T1027I and V1176F), Lambda variant (C.37) (G75V, T76I, R246N, Δ247–253, L452Q, F490S, D614G and T859N), Delta variant (B.1.617) (T19R, E156G, Δ157–158, L452R, T478K, D614G, P681R and D950N), Omicron (BA.1) (A67V, Δ69–70, T95I, G142D, Δ143-145, Δ211, L212I, 214EPEinsert, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N796Y, N856K, Q954H, N969K, L981F) and Omicron (BA.2) (T19I, L24S, Δ25–26, A67V, Δ69-70, G142D, V213G, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H and N969K) spike mutations were added in silico into the codon-optimised Wuhan reference stain and were cloned to pNBF plasmid. Spike proteins contain 6 proline mutations (F817P, A892P, A899P, A942P, K986P and V987P) and substituted at the furin cleavage site (residues 682–685).74 Cells were harvested at 5 days post transfection and purified using 0.1M NaAc pH3 before buffer exchanged to PBS pH7.4. Purified proteins were subjected to size exclusion chromatography (SEC) using Superose6 Increase 10/300 (Cytiva) in 10 mM Tris-HCl pH 7.8, 150 mM NaCl. A stable Flp-In T-REx 293 cell line (Thermo Fisher) expressing the prefusion-stabilized HexaPro variant of the Wu Spike75 was generated according to the manufacturer’s protocol (polyclonal selection). Cells were seeded in T-300 flasks (0.3 × 106 cells/mL), and after 24 h the medium was exchanged against fresh medium containing 1 μg/mL tetracycline to induce protein expression. After 4 days, cell culture supernatant was dialyzed against binding buffer (50 mM Tris-HCl pH 7.8, 500 mM NaCl), and spike was purified using Strep-Tactin XT beads (IBA Lifesciences), followed by SEC using a Superdex200 Increase 10/300 column (Cytiva), equilibrated in 10 mM Tris-HCl pH 7.8, 150 mM NaCl. SEC peak fractions containing spike protein were pooled, concentrated to 1 mg/mL, flash-frozen in liquid nitrogen and stored at −80°C.
For antibodies and nanobodies, DNA encoding antibody variable domains including W25, W270,27 NB3, Nb11, Nb20,52 MR17,62 SB14, SB42, SB4,76 Ty131 and H1163 and Nb against GFP protein67 were cloned in-frame with an upstream IgG leader sequence and downstream human IgG1 Fc open reading frame into the mammalian expression vector pNBF. For monoclonal antibodies, heavy and light chains of S309,9 REGN10987, REGN10933,10 Ly-CoV555,11 CT-P59,12 CB6,8 DH1047,13 5B3,65 and C0566 were cloned into a human IgG1 expression vector as described previously.77 After validation of the cloning by Sanger sequencing, the plasmids were transfected into ExpiCHO cells (Thermo Scientific) according to the manufacturer’s protocol (1 μg DNA/mL of cells; 2:3 ratio of heavy chain to light chain). After 7 days, cell culture media were clarified by centrifugation and the IgG captured using Protein A resin (GE Healthcare). Proteins were eluted from the resin using citrate buffer pH 3, eluate was buffer exchanged to PBS pH7.4. For nanobody production, W25 and W270 were expressed as previously described.27 The pHen6-W25 vector inoculated in 20 mL of liquid LB-medium containing 100 μg/mL ampicillin and 1% glucose. The bacteria were cultured at 37°C with agitation for 16 h. The bacteria were then diluted into 1L Terrific Broth (TB) medium containing 100 μg/mL ampicillin, 2 mM MgCl2, 0.1% glucose and incubated at 37°C to an OD600 of 0.6–0.9. The expression of the nanobodies was induced by adding 1 mM of IPTG (isopropyl-β-d-1-thiogalactopyranoside) for 20 h at 28°C. Bacteria were collected by centrifugation at 8000 rpm for 8 min at 4°C. The harvested bacteria were resuspended in a 12 mL TES buffer (0.2 M Tris pH 8.0, 0.5 mM EDTA, 0.5 M sucrose) and incubated for 1 h on ice, then incubated for another hour on ice in a 18 mL TES buffer, diluted 4 times and centrifuged at 8000 rpm at 4°C to pellet down cell debris. The supernatant was loaded on 5 mL of HisPur Ni–NTA agarose resin which was pre-equilibrated with binding buffer (Tris 50 mM, NaCl 500 mM, imidazole 10 mM pH 7.5). The lysed cells containing His- and myc-tagged nanobodies were added to the column followed by adding the column’s volume in binding buffer for a total of eight times. The column was washed by adding 8-fold the columnś volume with wash buffer (Tris 50 mM, NaCl 500 mM, imidazole 30 mM pH a 7.5), and eluted with 15 mL of elution buffer (Tris 50 mM, NaCl 150 mM, 150 mM imidazole, 1 mM DTT pH 7.5). Purified antibodies, and nanobodies were verified by SDS-PAGE Coomassie staining analysis.
Cryo-EM sample preparation, imaging and processing
Wu spike (0.5 μM trimer) and W25 (1.7 μM) were incubated for 2 h on ice in a volume of 50 μL buffer containing 10 mM Tris-HCl pH 7.8 and 150 mM NaCl. 3.5 μL sample were adsorbed for 45 s on glow-discharged 400 mesh R1.2/R1.3 holey carbon grids (Quantifoil) and plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher) with 1 s blotting time at 4°C and 99% humidity. Data were collected on a 300 kV Tecnai Polara cryo-EM (Thermo Fisher) equipped with a K2 Summit direct electron detector (Gatan) in super-resolution mode, at a nominal magnification of 31000×, with a pixel size of 0.63 Å/px and the following parameters: defocus range of −1 to −2.5 μm, 50 frames per movie, 200 ms exposure per frame, electron dose of 6.2 e/Å2/s, leading to a total dose of 62 e/Å2 per movie. Data were collected using Leginon.78 Movies were aligned and dose-weighted using MotionCor2.67 Initial estimation of the contrast transfer function (CTF) was performed with the CTFFind4 package via the Relion 3.0.7 GUI.68,69 Power spectra were manually inspected to remove ice contamination and astigmatic, weak, or poorly defined spectra. Subsequent image processing procedures are shown in Figure S1. Particles were picked initially using the Relion 3.0.7 LoG autopicker. After 2D and 3D classification, the best defined 3D class was used as template for a second round of autopicking in Relion 3.0.7. 2D and 3D classification yielded 141.257 clean particles, which were further processed using Relion 3.0.7 or cryoSPARC 3.3.170 as indicated in Figure S1. Particles were subjected to an additional ab-initio reconstruction (3 classes, class similarity = 0), followed by a consensus refinement, exhibiting a 2 RBD-up, 1 RBD-down conformation. Subsequently, particles were locally refined to one RBD in the “up” conformation using a stalk/RBD/W25 mask, followed by a focused classification without alignment using an RBD/W25 mask for the identification of the most defined WuRBD/W25 complex subset. This particle subset was subjected to another local refinement using the RBD/NTD mask. Locally refined maps were used as reference in a heterogeneous refinement. This revealed that particles were partially in a 1 RBD-up, 2 RBD-down conformation. Clean particles were re-assigned to both global conformations, followed by non-uniform refinement, resulting in 3.8 Å map for the 2 RBD-up, 1 RBD-down class (map 1, Figure S1) and a 3.81 Å map for the 1 RBD-up, 2 RBD-down class (map 2). Both particle subsets were pooled again and locally refined to the shared RBD in “up” conformation, resulting in a 5.92 Å map.
Omicron spike and W25 were mixed in 1:6 M ratio and incubated on 37°C for 45 min. Omicron spike-W25 complex were adsorbed for 10 s onto glow discharged Quantifoil grids (Q1.2/1.3) and plunge frozen into liquid ethane using an EMGP2 system (Leica). Cryo-EM data collection was performed with Serial EM software (Version 3.1) using CRYO ARM 300 (JEOL) at the Center of Microscopy and microanalysis UQ. Movies were acquired in super resolution and CDS mode with a slit width of 10 eV using a K3 detector (Gatan) for 3.3 s during which 40 movie frames were collected with a 1 e−/Å2 per frame. Data were collected at a magnification of 60,000x, corresponding to a calibrated pixel size of 0.4 Å/pixel. Movies were binned 2x during motion Correction.67 A total of 9,258 micrographs were collected, 8,845 micrographs were selected based on an estimated resolution cutoff of 4.5 Å. Particles were extracted using Relion 3.0.7. All following processing steps were performed in Cryosparc 3.3.170 as indicated in Figure S3. Extracted particles were cleaned up from ice and false-positive picks using 2D classification. After 2D classification, most defined classes with high-resolution features were retained (164.806 particles) and used for an ab-initio 3D reconstruction (3 classes, class similarity = 0). Particles were further processed using heterogeneous refinement and previously generated maps, followed by a re-assignment to both global conformations from the Wuhan dataset (maps 1 and 2, Figure S1). A 3D variability analysis was performed for particles in 1 RBD-up, 2 RBD down conformation using a mask for the RBD in the “up” conformation. This revealed that particles were partially in a 3 RBD-down conformation. Using a less strict 2D class selection (222237 particles), the particle subset were then cleaned again using heterogeneous refinement, followed by re-assignment to the three identified global conformations. Particles subsets in the 2 RBD-up, 1 RBD-down and 1 RBD-up, 2 RBD-down conformation were cleaned in an additional round of heterogeneous refinement. The 1 RBD-up, 2 RBD-down particle subset showed the most defined RBD/W25 density and was therefore refined using non-uniform refinement (4.97 Å, map 4, Figure S3), followed by local refinement using an RBD-up/NTD mask, resulting in a 6.04 Å map (map 5).
Model building and refinement
The trimeric base, and the individual RBDs and NTDs of a Wu spike model (PDB 6ZXN,31) were fitted as rigid bodies in map 1 (Figure S1). One RBD and the adjacent NTD, as well as an AlphaFold233,71 model of the W25 nanobody were then fitted as rigid bodies in focus map 3 (Figure S1), trimmed manually using Coot 0.9.5,72 subjected to molecular dynamics flexible fitting using the Namdinator server79 with standard parameters, followed by real space and ADP refinement in Phenix 1.19.2–415873 and manual adjustment using Coot 0.9.5. For Omicron spike model building, a similar strategy was pursued, however using PDB 7WG630 as starting model, and map 4 and 5 for fitting (Figure S3). Molecular models and maps were visualized using the ChimeraX software.80
Surface plasmon resonance (SPR)
SPR measurements were performed using a Biacore 8K+ instrument (Cytiva). Purified W25-Fc, S309, DH1047 or C05 antibody were captured on a Protein A Series S Sensor Chip (Cytiva). Binding of SARS-CoV-2 spike variants were tested using a Multi-Cycle High Performance Kinetics assay. The antibodies were all prepared at 1 μg/mL in running buffer (HBS-P+, pH 7.4, Cytiva) and injected for 30 s at 30 μL/min over Fc2 on all 8 channels. Multiple concentrations of SARS-CoV-2 spike variants were included: 0 nM, 1.40625 nM, 2.8125 nM, 5.625 nM, 11.25 nM, 22.5 nM, 45 nM and 90 nM. The proteins were injected over both Fc1 and Fc2 of Channels 1–8 in series for 180 s at 30 μL/min, followed by a dissociation period of 600 s. The chip surface was regenerated between each cycle using 10 mM glycine pH 1.5. The data was double reference subtracted; reference cell subtraction (Fc1) from the active cell (Fc2), and zero concentration subtraction for each analyte-antibody pair. Sensorgrams for the association and dissociation phases were fitted to a 1:1 binding model, using Biacore Insight Evaluation software (Cytiva).
ELISA
To test nanobody and antibodies, SARS-CoV-2 spike variant proteins in PBS pH7.4 were immobilised on Maxisorb ELISA (Nunc) plates at a concentration of 2 μg/mL overnight. Serial 5-fold dilutions of Fc-fusion nanobodies or antibodies in 1X KPL in blocking buffer (Seracare) were incubated with the immobilized antigen, followed by incubation with HRP-coupled anti-human IgG (MilleniumScience) and the chromogenic substrate TMB (ThermoFisher). Reactions were stopped with 2 M H2SO4 and absorption measured at 450 nm.
Viruses
The SARS-CoV-2 Ancestral (Wu) variant, hCoV-19/Australia/QLD02/2020 (GISAID accession ID, EPI_ISL_407896), Alpha variant, hCoV-19/Australia/QLD1517/2021, (GISAID accession ID EPI_ISL_944644), Beta variant hCoV-19/Australia/QLD1520/2020, (GISAID accession ID EPI_ISL_968081), Delta: hCoV-19/Australia/QLD1893C/2021 (GISAID accession ID EPI_ISL_2433928) were kindly provided by Dr Alyssa Pyke (Queensland Health Forensic & Scientific Services, Queensland Department of Health, Brisbane, Australia). For Omicron BA.1 variant (hCoV-19/Australia/NSW-RPAH-1933/2021), Gamma variants, hCoV-19/Australia/NSW4318/2021, Lambda variants hCoV-19/Australia/NSW4431/2021 were kindly provided by A/Prof. Stuart Turville (University of New South Wales, Australia). Omicron BA.2 variant was kindly provided by Prof. Andreas Suhrbier (QIMR Berghofer Medical Research Institute). The passage 2 of SARS-CoV-2 variants (except Gamma and Lambda) was received, and passage 3 were propagated in TMPRSS2-expressing VeroE6 and used as virus stock. Passage 3 of and passage 4 of Gamma, Lambda and BA.2 variants were obtained and passage 4 was propagated in TMPRSS2-expressing VeroE6 and used as virus stock, viruses were stored at −80°C. All working virus stocks were sequenced and validated using either Nanopore sequencing or Sanger sequencing on spike genes. For SARS-CoV-2 Beta variant (B.1.351) used in the animal experiment, was obtained from Andy Pekosz (Center for Emerging Viruses and Infectious Diseases (CEVID)). Infectious stocks were grown in Vero-Ace2-TMPRSS2 cells (BEI Resources) and stored at −80°C. All work with infectious virus was performed in a biosafety level 3 laboratory and approved by the University of Queensland Biosafety committee (IBC/447B/SCMB/2021) and the Penn Institutional Biosafety Committee and Environmental Health and Safety.
Plaque reduction neutralisation test (PRNT)
The levels of neutralising antibodies were assessed using our established PRNT protocol28 and supplementary materials and methods. Briefly, purified mAbs were 5-fold serially diluted in DMEM containing 2% HI-FCS and 1% P/S (Gibco). Subsequently, serial diluted antibodies were incubated with 50–100 immuno-plaques of SARS-CoV-2 and incubated at 37°C for 1 h. Then, 50 μL of mixture (virus/antibody complex) was added onto pre-seeded VeroE6 cells in 96-well plates at 6 × 104 cells/well and incubated at 37°C for 30 min. Following, overlay medium (containing 1% CMC, 1X M199, 2% HI-FCS and 1% (P/S)) was added on top of the inoculum and incubated at 37°C with 5% of CO2. Twenty-four hours after infection, the overlay was removed, and the monolayer was fixed with cold fixative solution (80% acetone and 20% phosphate buffered saline (PBS)) and incubated at −20°C for 30min. Fixative reagent was removed, and the monolayer was fully dried. Monolayer was then treated with 100 μL of 1X milk blocking solution (KPL, Seracare) diluted in 1X-PBS containing 0.05% Tween 20 (PBS-T) and incubated at 4°C for overnight. Next, the blocking buffer was removed, and immuno-plaques were stained using anti-spike antibody (mouse CR3022) as the primary antibody for the following variants: SARS-CoV-2 Wu, Alpha, Gamma, Beta, Lambda, Kappa and Delta variants. Anti-M SARS-CoV-2 antibody was used as a primary antibody to stain the immune-plaques for Omicron variant. The monolayer was incubated at 37°C for 1 h with the primary antibody and the unbounded antibody was removed by five consecutive washes with 5min incubation between each washed. To reveals the immuno-plaques, an infra-red dye conjugated secondary antibody (IRDye 800CW Goat anti-Mouse or Goat anti-streptavidin, MillenniumScience) was added and incubated at 37°C for 1 h, followed by five consecutive washes with 5min incubation in between. Finally, plate was fully dried avoiding long light exposure and scanned using the LI-COR Biosciences Odyssey Infrared Imaging System (Odessey CLx, Li-COR, USA). Immuno-plaques were analyzed and counted using an automated foci counter program, Viridot.81 A stock of primary and secondary antibodies at 1 mg/mL were diluted 1/1000 and 1/2500 in blocking buffer solution, respectively and 50 μL/well was used for the staining.
Cell-cell fusion assay
Cell-cell fusion assay was conducted as previously described.38 HEK293T Lenti rLuc-GFP 1–7 (effector cells) and HEK293T Lenti rLuc-GFP 8–11 (target cells) were seeded separately at 7.5×105 per well in a six-well dish in 3 mL of PRF-DMEM-10% and incubated overnight at 37°C, 5% CO2. Transfection mixes were set up in 200 μL Opti-MEM (Gibco) with the TransIT-X2 Dynamic Delivery System as per the manufacturer’s recommendations (Mirus). SARS-CoV-2 Spike glycoproteins of D614G, Beta and Omicron and human ACE2 plasmids were transfected into effector cells. A mock-transfected (pcDNA3.1 empty plasmid, - vGP) and positive transfection control (250 ng rLuc-GFP 8–11 plasmid) was also set up. W25, its derivatives and control antibody were diluted to specific concentration in sterile 1.5 mL tubes using serum-free PRF-DMEM and plated at 25 μL/well in a white-bottomed, sterile 96-well plate (Corning), including no antibody controls. The antibodies were incubated with 2×104 effector cells in 50 μL at 37°C, 5% CO2 for 1 h, after which target cells were co-cultured to corresponding wells and incubated for 18–24 h, after which GFP-positive syncytia and Renilla luciferase were quantified. Negative controls (effector cells only, target cells only) and positive transfection controls (HEK293T Lenti rLuc-GFP 1–7 cells transfected with rLuc-GFP 8–11 plasmid) were always included. The assay was conducted separately for Renilla luciferase and GFP readout.
To quantify Renilla luciferase expression in fusion assays media were replaced with 100 μL of phosphate-buffered saline (PBS) followed by 60 μL of diluted substrate, Coelenterazine-H, 1 μM (Promega) 1: 400 with PBS. The plate was incubated in the dark for 2 min then read on the GloMax Multi+ Detection System (Promega). To quantify GFP expression, cells were plated in clear flat-bottomed 96-well plates (Nunc) and imaged every hour using the IncuCyte S3 live cell imaging system (Essen BioScience). Five fields of view were taken per well at 10× magnification, and GFP expression was determined using the total integrated intensity metric included in the IncuCyte S3 software (Essen BioScience). To analyze images generated on the IncyCyte S3, a collection of representative images is first taken to set fluorescence and cellular thresholds, which allows for the removal of background fluorescence, and selection of cell boundaries (‘objects’) by creating ‘masks’. Following this, the total integrated intensity metric can be accurately calculated by the software, which takes the total sum of objects’ fluorescent intensity in the image, expressed as green count units (GCU) μm.
Mouse infections
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and the Guidelines to promote the wellbeing of animals used for Scientific purposes. The protocols were approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania (protocol #807017) and the University of Queensland animal ethics committee (2021/AE000929 and 2021/AE001119). Heterozygous K18-hACE C57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory and/or bred in-house. Animals were housed in groups and fed standard chow diets. Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering. For prophylactic treatment study, mice of different ages and both sexes were infected with SARS-CoV-2 Beta variant at 1 × 103 plaque-forming units (PFU) via intranasal administration 4 h after intraperitoneal administration of 100 μg W25-Fc or GFP-Nb-Fc in 100 μl PBS. For therapeutic treatment study, 5–6 weeks old female mice were infected with SARS-CoV-2 Beta (5 × 103 FFU/mouse) or Omicron (1 × 105 FFU/mouse) variants via intranasal administration. Infected mice were administrated with 100 μg W25-Fc or GFP-Nb-Fc via intraperitoneal (100 μL) or intranasal administration (20 μL) 24 h post infection. The dose of W25 Fc was based on previous report.82 Mice were monitored daily for weights and clinical respiratory disease scores were obtained. Mice were sacrificed when exhibiting greater than 20% weight loss or after reaching a respiratory disease score of 5 for longer than 2 days or greater than 5. Respiratory disease was scored based on activity, ruffled fur/piloerection, accelerated shallow breathing, labored breathing, tremor/tension/paralysis and/or weight loss of greater than 7.5% with a point added for each identified. Lungs and Nasal turbinates were collected on 2 or 4 days post-infection and viral titers were determined using plaque-forming assay, immunoplaque-forming assay and RT-PCR.
Plaque-forming assay and immunoplaque-forming assay
Tissues were weighed and homogenized with zirconia beads in a FastPrep-24 instrument (MP Biomedicals) in 0.6 mL of DMEM media supplemented with 10% heat-inactivated FBS. Tissue homogenates were clarified by centrifugation at 10,000 r.p.m. for 5 min and stored at −80°C. Vero-Ace2-Tmprss2 cells (BEI Resources) were seeded at a density of 2.5 × 106 cells per plate in flat-bottom 6-well tissue culture plates. The following day, media was removed and replaced with 200 μL of 10-fold serial dilutions of the tissue homogenate to be titered, diluted in DMEM. One hour later, seaplaque/seakem agarose was added. Plates were incubated for 72 h, then fixed with 4% paraformaldehyde in phosphate-buffered saline for at least 1 h. Plaques were visualized with 0.05% (w/v) crystal violet in 20% methanol and washed with water prior to enumeration of plaques.
Viral load via RT-qPCR or immuno-plaque assay (iPA)
For RT-qPCR, tissue homogenate was added to Trizol at 1:3 ratio and extracted using Zymo Direct-zol RNA kit following manufactures protocol. RNA was reverse transcribed and amplified using iScript cDNA Synthesis Kit (Biorad). Gene-specific primers for SARS-CoV-2 N gene (F:TTACAAACATTGGCCGCAAA & R:GCGCGACATTCCGAAGAA) and mouse HPRT1 (F: GTTGGATACAGGCCAGACTTTGTTG & R: GAGGGTAGGCTGGCCTATTGGCT) with Power SYBR Green PCR Master Mix (Applied Biosystems) were used to amplify viral and cellular RNA by QuantStudio 3 Flex Real-Time PCR Systems (Applied Biosystems). The relative expression levels of target genes were calculated using the standard curve method using quantitative synthetic RNA (ATCC) and normalized to HPRT1 RNA as an internal control.
For iPA, organs such as lung and nasal turbinates were collected in preweighted tubes and homogenised using a tissue homogenizer (TissueLyser LT, Qiagen). Homogenates were then clarified by centrifugation at 10000 × g, 4°C for 5 min. Supernatants were collected and stored at −80°C. Viral loads were determined by iPA on VeroE6 following our optimised protocol for viral titration. Viral titers were expressed in FFU per grams of tissue (FFU/g).
Radiochemistry
[111In]InCl3 was purchased from Curium. The analytical-HPLC system consists of a 170U UVD detector, a Scansys radiodetector and a Dionex system connected to a P680A pump. The system was run by Chromeleon software. The radiochemical conversion (RCC) of the radiolabeled compounds was determined by analyzing an aliquot of the crude reaction mixture by radio-HPLC analysis integrating the radioactive peaks of the chromatogram.83 Radiolabeled products were characterized by associating the UV-HPLC traces of the authentic cold compounds with the radio-HPLC chromatogram of the reaction mixtures. The radiochemical yield (RCY) was determined using the initial activity at the beginning of the synthesis and that of the formulated product at the end of thes, corrected for decomposition and corrected for decay.
111In-labeling DOTA-tetrazine
1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) -PEG11-tetrazine was dissolved (2 mg/mL) in metal-free water and stored at −80°C before use. In general, an aliquot of 50–100 μL (10–30 MBq) of [111In]indium chloride in 0.1M HCl was combined with 2 μL DOTA-PEG11-tetrazine and 1M NH4OAc buffer (pH 5.5) at a volume ratio of 1:10 was added. The mixture was shaken at 600 rpm for 5 min at 60°C in an Eppendorf ThermoMixer C. Then, 10 mM diethylenetriamine-pentaacetic acid (DTPA, volume ratio 1:10) and 2 μL 10 mg/mL gentisic acid in saline was added and the solution was shaken for an additional 5 min at 60°C in an Eppendorf ThermoMixer C. Quantitative labeling yield and a radiochemical purity reater than 95% were obtained with this method, as confirmed by radio-HPLC.
Trans–cyclooctene (TCO)-modifications W25-Fc
W25-Fc (2.0 mg/mL) in PBS (pH 7.4) was aliquoted in five vials of 100 μL. To each aliquot, 25 equiv TCO-PEG4-NHS (Broadpharm, BP-22418) and sodium carbonate buffer (1 M, 3.1 μL, pH 8.0) was added. The mixture was shaken at 600 rpm for 2 h at room temperature in the dark. Unreacted TCO-PEG4-NHS was removed by purification with Zeba spin desalting columns (7K MWCO, 0.5 mL, 89882, Thermofisher) and eluted in PBS (pH 7.4), with >95% protein recovery after purification. Final protein concentration was 1.7 mg/mL, measured with NanoDrop (NanoDrop 2000, ThermoScientific). Titration experiments were conducted to quantify the amount of reactive TCOs per protein-conjugate. 111In-labeled Tz stock was diluted accordingly to add 5 μL an excess of 111In-labeled Tz (2 equiv of Tz per protein-conjugate) to 5 μL of TCO-HSA and the mixture was shaken at 600 rpm for 1 h at 37°C. 3 μL NuPAGE LDS Sample Buffer (NP0007, Invitrogen) was added and the mixture was shaken for 10 min at 70°C. Samples were applied to NuPAGE 4 to 12%, Bis-Tris, 1.0 mm, Mini Protein Gel, 12-well (NP0322BOX, Invitrogen) SDS-PAGE gels. SDS-PAGE gels were exposed to phosphor storage screens and read by a Cyclone Storage Phosphor System (PerkinElmer Inc.). Quantification of plate readings was done with Optiquant software (version 5.00, PerkinElmer Inc.). Quantification by radioactive SDS-PAGE revealed presence of approximately 1.6 reactive TCO/protein.
Radiolabeling of TCO-W25-Fc with 111In-tetrazine
To a 5 mL Eppendorf vial was added TCO-W25-Fc (465 μL, 10.16 nmol protein, 16.26 nmol TCO) and 111In-Tz (68 MBq, 95 μL, 1.6 nmol, 0.1 TCO/Tz eq.). The mixture was shaken at 600 rpm for 1 h at 37°C, giving a radiochemical conversion of 68%. Ultrafiltration (Vivaspin 500, 5,000 MWCO PES, Sartorius) yielded in 40 MBq 111In-W25-Fc with a radiochemical purity of 96%, determined by radio-HPLC. Protein concentration after purification was measured with NanoDrop (NanoDrop One, ThermoScientific). The 111In-labeled protein was formulated in PBS to a final concentration of 15 MBq/mL.
Biodistribution of 111In-labelled W25-Fc in vivo
C57BL/6J female mice weighing 20–23 g (Janvier, France, La Rochelle) were housed in cages of 7–8 mice per cage. All mice were kept in a climate-controlled facility with a 12 h light/dark cycle. The cages contained a biting stick, fed with commercial breeding diet ad libitum (1310 FORTI- Avlsfoder, Brogaarden, Altromin International) and had access to water. All procedures were conducted in accordance with the European Commission’s Directive 2010/63/EU, FELASA and ARRIVE guidelines for animal research and, with approval from The Danish Council for Animal Ethics (license numbers 2017-15-0201-01283) as well as the Department of Experimental Medicine, University of Copenhagen. Mice were evenly divided into six different groups. All mice were weighted prior to injection of 111In-W25Fc (1.2–1.5 MBq/mouse) through a tail vein. The animals were sacrificed under anesthesia (3% sevoflurane) by decapitation at the designated time points after tracer administration (group 1: 5 min, group 2: 20 min, group 3: 60 min, group 4: 3 h, group 5: 5 h and group 6: 24 h). The mice were dissected and of each mouse the blood, lungs, liver, kidney, spleen, brain, muscle and tail were collected and placed into preweighed gamma counting tubes (Polypropylene, 5 mL 75 × 12 mm Ø, round base, Hounisens). The tubes with collected tissue were weighed and radioactivity measurements were conducted on a Cobra II Auto-Gamma Counter (Model D5005, Packard BioScience Company). The measurements were corrected for radioactivity decay and background. The ex vivo biodistribution results were expressed as a percentage of the injected radioactivity dose per gram of tissue (% ID/g) and nmol protein per tissue volume. For these calculations, tissue weight was converted to volume based on different tissue density for different organs. Radioactive counts in the tail were used as a quality control for the injections, which led to exclusion of one mouse in group 4 that had >20% ID in remaining in the tail.
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism 8.01 software (GraphPad) ANOVA or Mann-Whitney test was performed for group comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001 was considered as statistically significant with mean ± SEM. All of the statistical details of experiments can be found in the figure legends.
Acknowledgments
We thank Drs Matthias Flotenmeyer and Lou Brillault from Center for Microscopy and Microanalysis (CMM), the University of Queensland, for facilitating cryo-EM work to be carried out and for scientific and technical assistance. We thank UQBR animal staff (Maya Patrick and Barb Arnts) and UQBR facility at the Australian Institute for Bioengineering and Nanotechnology (AIBN). We thank Research Computing Center (RCC), UQ, for partially providing computational resources. We thank Queensland Health Forensic and Scientific Services, Queensland, and the Kirby Institute University of New Souths Wales for providing SARS-CoV-2 virus isolates. We thank the National Biologics Facility, AIBN, for access to the Biacore 8K+ instrument for the SPR assay. We thank Prof. Andreas Suhrbier for providing Omicron BA.2 virus isolate. We thank Dr. Michael Landsberg for advice and helpful discussion. We thank Ian Mortimer (ITS Infrastructure Operations, UQ) for IT support. We thank the Max-Planck-Institut für molekulare Genetik (MPI-MG) for granting access to the TEM instrumentation of the microscopy and cryo-EM service group. N.M would like to thank Alpaca ‘Budda’ whose nanobody was isolated and Dzongsar Jamyang Khyentse Rinpoche for generous contribution. Funding. This work was supported by NHMRC MRFF Coronavirus Research Response grant APP1202445 to D.W., K.C., P.R.Y., and T.M. Cryo-EM equipment at CMM is supported by ARC The Linkage Infrastructure, Equipment and Facilities (LIEF) scheme. N.M. was supported by UQ Research Stimulus. D. S. was supported by the German Research Foundation (DFG) Emmy Noether Programme (SCHW1851/1-1) and by an EMBO Advanced grant (aALTF-1650). The Chilean platform for the generation and characterization of Camelid Nanobodies to A.R.F is funded by ANID-FONDECYT No. 1200427; the regional Council of the “Los Rios region” projects FICR19-20, FICR21-01; FICR20-49 to R.J; the Bio & Medical Technology Development Program of the National Research Foundation (NRF) of the Korean government (MSIT) (NRF-2020M3A9H5112395); the ANID-MPG MPG190011 and ANID-STINT CS2018-7952 grants; and the EU-LAC T010047. DB is funded by UKRI Biotechnology and Biological Sciences Research Council (UKRI - BBSRC, https://www.ukri.org/) Institute Strategic Program Grant (ISPG) to The Pirbright Institute (BBS/E/I/00007034, BBS/E/I/00007030 and BBS/E/I/00007039).
Author contributions
N.M.: Experimental design; conceptualized; designed, generated, purified spike proteins, monoclonal antibodies, W25-Fc and control Fc; characterized spike proteins and nanobodies; generated W25/Omicron spike dataset including sample preparation, optimization and determination of optimal freezing conditions, image acquisition, performed single particle analysis (SPA); performed PRNTs; performed K18-huACE2 mouse challenge experiments; generated figures; drafted and edited manuscript. S.M.L.: Performed Wu spike Hexapro/W25 and Omicron spike Hexapro/W25 cryo-EM single particle and structure analysis. A.A.A.: Propagated live virus stock, performed PRNTs, performed K18-huACE2 mouse challenge experiments and data analysis. P.W.: Performed K18-huACE2 mouse challenge experiment. S.I.L.B.: Experimental design for the biodistribution studies, performed them with the help of J.T.J. and I.V.A. Y.S.L: Sample optimization for W25/Omicron spike cryo-EM, determination optimal freezing conditions, data acquisition, data analysis. N.T.: Performed cell-cell fusion experiments. B.L.: Assisted in cloning and purification of spike proteins and antibodies; assisted in K18-huACE2 mouse challenge experiments. G.V.N.: Identification of W25 cloning of W25 and performed thermostability and nebulization stability assays of W25. J.J.: Performed SPA analysis and focuses classification for W25/Omicron dataset. D.P.: Cloned, expressed, and purified published nanobodies. A.I.: Reformatted published nanobodies. J.D.S.: Generated live virus stock and generated VeroE6-TMPRSS6 cells for virus propagation. D.S.: Performed K18-huACE2 mouse challenge experiment. J.T.J.: Performed the biodistribution study with S.I.L.B and I.V.A. Y.C.: Performed optimization and purification of W25. J.B.: Assisted in Wu spike Hexapro/W25 cryo-EM single particle and structure analysis. I.V.A.: Performed the biodistribution study with S.I.L.B and J.T.J. J.H.: Performed optimization and purification of W25 for biodistribution assays. R.J.: Supervised vibrating mesh nebulizer stability assays. R.M.: Supervised vibrating mesh nebulizer stability assays. Z.M.C.: Performed optimization of W25. P.C.C.: supervised optimization of purification of W25 for cryo-EM and biodistribution. V.K.: Experimentally designed the biodistribution studies. T.M. and C.M.T.S.: Supervised Wu spike Hexapro/W25 cryo-EM single particle and structure analysis. A.A.K.: Obtained SARS-CoV-2 ancestral, Alpha, Beta isolates. Chief Investigator for BSL3 biocontainment facility at School of Chemistry and Molecular Biosciences. M.L.J.: Performed and analyzed SPR experiment. A.K.: Designed the biodistribution studies, supervised the work of J.T.J. M.M.H.: Designed the labeling and biodistribution studies, supervised the work of I.V.A. and S.I.L.B. K.A.J.: Supervised K18-huACE2 prophylactic mouse work. D. Sc.: Prepared and purified Wu Hexapro spike, prepared Wu spike Hexapro/W25 complex cryo-EM sample, performed and supervised Wu spike Hexapro/W25 cryo-EM and Omicron spike Hexapro/W25 single particle and structure analysis, generated figures, wrote initial manuscript draft. D.W.: Performed SPA of W25/Omicron dataset. The overall project was supervised, conceptualized, and edited by D.W., A.R.F., and D.S.
Declaration of interests
D.W., K.C., and P.R.Y are listed as inventors of ‘Molecular Clamp’ patent, US 2020/0040042.
Published: June 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107085.
Contributor Information
David Schwefel, Email: david.schwefel@charite.de.
Alejandro Rojas-Fernandez, Email: alejandro.rojas@uach.cl.
Daniel Watterson, Email: d.watterson@uq.edu.au.
Supplemental information
Data and code availability
All data are available in the main text or the supplementary materials. Cryo-EM maps and molecular models have been deposited in the Electron Microscopy DataBank (EMDB) and the Protein DataBank (PDB), respectively, with accession codes EMD-15994 (map 1), EMD-15997 (map 2), EMD-16010 (map 3), EMD-16028 (map 4), EMD-16030 (map 5) and PDB: 8BEV (Wu NTD/RBD/W25) and PDB: 8BGG (Omicron NTD/RBD/W25). The accession numbers and more detailed information are also available in Supplementary information. This paper does not report original code. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 2.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schön A., Bimela J., et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879.e5. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 9.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 10.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim C., Ryu D.K., Lee J., Kim Y.I., Seo J.M., Kim Y.G., Jeong J.H., Kim M., Kim J.I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez D.R., Schaefer A., Gobeil S., Li D., De la Cruz G., Parks R., Lu X., Barr M., Manne K., Mansouri K., et al. A broadly neutralizing antibody protects against SARS-CoV, pre-emergent bat CoVs, and SARS-CoV-2 variants in mice. bioRxiv. 2021 doi: 10.1101/2021.04.27.441655. Preprint at. [DOI] [Google Scholar]
- 14.Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 18.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 19.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 21.Yu J., Collier A.R.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheward D.J., Kim C., Ehling R.A., Pankow A., Castro Dopico X., Dyrdak R., Martin D.P., Reddy S.T., Dillner J., Karlsson Hedestam G.B., et al. Neutralisation sensitivity of the SARS-CoV-2 omicron (B.1.1.529) variant: a cross-sectional study. Lancet Infect. Dis. 2022;22:813–820. doi: 10.1016/S1473-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larios Mora A., Detalle L., Gallup J.M., Van Geelen A., Stohr T., Duprez L., Ackermann M.R. Delivery of ALX-0171 by inhalation greatly reduces respiratory syncytial virus disease in newborn lambs. mAbs. 2018;10:778–795. doi: 10.1080/19420862.2018.1470727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., et al. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunningham S., Piedra P.A., Martinon-Torres F., Szymanski H., Brackeva B., Dombrecht E., Detalle L., Fleurinck C., RESPIRE study group Nebulised ALX-0171 for respiratory syncytial virus lower respiratory tract infection in hospitalised children: a double-blind, randomised, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2021;9:21–32. doi: 10.1016/S2213-2600(20)30320-9. [DOI] [PubMed] [Google Scholar]
- 27.Valenzuela Nieto G., Jara R., Watterson D., Modhiran N., Amarilla A.A., Himelreichs J., Khromykh A.A., Salinas-Rebolledo C., Pinto T., Cheuquemilla Y., et al. Potent neutralization of clinical isolates of SARS-CoV-2 D614 and G614 variants by a monomeric, sub-nanomolar affinity nanobody. Sci. Rep. 2021;11:3318. doi: 10.1038/s41598-021-82833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amarilla A.A., Modhiran N., Setoh Y.X., Peng N.Y.G., Sng J.D.J., Liang B., McMillan C.L.D., Freney M.E., Cheung S.T.M., Chappell K.J., et al. An optimized high-throughput immuno-plaque assay for SARS-CoV-2. Front. Microbiol. 2021;12:625136. doi: 10.3389/fmicb.2021.625136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of SARS-CoV-2 BA.2 variant. bioRxiv. 2022 doi: 10.1101/2022.02.14.480335. Preprint at. [DOI] [Google Scholar]
- 30.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., et al. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., Bridgland A., Cowie A., Meyer C., Laydon A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. ColabFold - making protein folding accessible to all. bioRxiv. 2022 doi: 10.1101/2021.08.15.456425. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Q., Owens R.J., Naismith J.H. Structural biology of nanobodies against the spike protein of SARS-CoV-2. Viruses. 2021;13:2214. doi: 10.3390/v13112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28:490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakur N., Conceicao C., Isaacs A., Human S., Modhiran N., McLean R.K., Pedrera M., Tan T.K., Rijal P., Townsend A., et al. Micro-fusion inhibition tests: quantifying antibody neutralization of virus-mediated cell-cell fusion. J. Gen. Virol. 2021;102:jgv001506. doi: 10.1099/jgv.0.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esparza T.J., Martin N.P., Anderson G.P., Goldman E.R., Brody D.L. High affinity nanobodies block SARS-CoV-2 spike receptor binding domain interaction with human angiotensin converting enzyme. Sci. Rep. 2020;10:22370. doi: 10.1038/s41598-020-79036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radvak P., Kwon H.J., Kosikova M., Ortega-Rodriguez U., Xiang R., Phue J.N., Shen R.F., Rozzelle J., Kapoor N., Rabara T., et al. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat. Commun. 2021;12:6559. doi: 10.1038/s41467-021-26803-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suryawanshi R.K., Chen I.P., Ma T., Syed A.M., Brazer N., Saldhi P., Simoneau C.R., Ciling A., Khalid M.M., Sreekumar B., et al. Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination. Nature. 2022;607:351–355. doi: 10.1038/s41586-022-04865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dagan N., Barda N., Kepten E., Miron O., Perchik S., Katz M.A., Hernán M.A., Lipsitch M., Reis B., Balicer R.D. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eyre D.W., Taylor D., Purver M., Chapman D., Fowler T., Pouwels K.B., Walker A.S., Peto T.E.A. Effect of Covid-19 vaccination on transmission of alpha and Delta variants. N. Engl. J. Med. 2022;386:744–756. doi: 10.1056/NEJMoa2116597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Heeke G., Allosery K., De Brabandere V., De Smedt T., Detalle L., de Fougerolles A. Nanobodies(R) as inhaled biotherapeutics for lung diseases. Pharmacol. Ther. 2017;169:47–56. doi: 10.1016/j.pharmthera.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., VIB-CMB COVID-19 Response Team. Hoffmann M., et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pymm P., Adair A., Chan L.J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O'Neill M.T., Tan L.L., et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2101918118. e2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huo J., Mikolajek H., Le Bas A., Clark J.J., Sharma P., Kipar A., Dormon J., Norman C., Weckener M., Clare D.K., et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021;12:5469. doi: 10.1038/s41467-021-25480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370:1479–1484. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;370:1473–1479. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koenig P.A., Das H., Liu H., Kümmerer B.M., Gohr F.N., Jenster L.M., Schiffelers L.D.J., Tesfamariam Y.M., Uchima M., Wuerth J.D., et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:eabe6230. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iketani S., Liu L., Guo Y., Liu L., Chan J.F.W., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shrestha L.B., Tedla N., Bull R.A. Broadly-neutralizing antibodies against emerging SARS-CoV-2 variants. Front. Immunol. 2021;12:752003. doi: 10.3389/fimmu.2021.752003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Kopp M., Haynes B.F., Acharya P. Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. bioRxiv. 2020 doi: 10.1101/2020.06.26.173765. Preprint at. [DOI] [Google Scholar]
- 58.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walls A.C., Xiong X., Park Y.J., Tortorici M.A., Snijder J., Quispe J., Cameroni E., Gopal R., Dai M., Lanzavecchia A., et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e15. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hui K.P.Y., Ho J.C.W., Cheung M.C., Ng K.C., Ching R.H.H., Lai K.L., Kam T.T., Gu H., Sit K.Y., Hsin M.K.Y., et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 62.Li T., Cai H., Yao H., Zhou B., Zhang N., van Vlissingen M.F., Kuiken T., Han W., GeurtsvanKessel C.H., Gong Y., et al. A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection. Nat. Commun. 2021;12:4635. doi: 10.1038/s41467-021-24905-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 64.Kubala M.H., Kovtun O., Alexandrov K., Collins B.M. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010;19:2389–2401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dang H.V., Chan Y.P., Park Y.J., Snijder J., Da Silva S.C., Vu B., Yan L., Feng Y.R., Rockx B., Geisbert T.W., et al. An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections. Nat. Struct. Mol. Biol. 2019;26:980–987. doi: 10.1038/s41594-019-0308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ekiert D.C., Kashyap A.K., Steel J., Rubrum A., Bhabha G., Khayat R., Lee J.H., Dillon M.A., O'Neil R.E., Faynboym A.M., et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 71.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Watterson D., Wijesundara D.K., Modhiran N., Mordant F.L., Li Z., Avumegah M.S., McMillan C.L., Lackenby J., Guilfoyle K., van Amerongen G., et al. Preclinical development of a molecular clamp-stabilised subunit vaccine for severe acute respiratory syndrome coronavirus 2. Clin. Transl. Immunol. 2021;10:e1269. doi: 10.1002/cti2.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahmad J., Jiang J., Boyd L.F., Zeher A., Huang R., Xia D., Natarajan K., Margulies D.H. Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction. J. Biol. Chem. 2021;297:101202. doi: 10.1016/j.jbc.2021.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Modhiran N., Song H., Liu L., Bletchly C., Brillault L., Amarilla A.A., Xu X., Qi J., Chai Y., Cheung S.T.M., et al. A broadly protective antibody that targets the flavivirus NS1 protein. Science. 2021;371:190–194. doi: 10.1126/science.abb9425. [DOI] [PubMed] [Google Scholar]
- 78.Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 79.Kidmose R.T., Juhl J., Nissen P., Boesen T., Karlsen J.L., Pedersen B.P. Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ. 2019;6:526–531. doi: 10.1107/S2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katzelnick L.C., Coello Escoto A., McElvany B.D., Chávez C., Salje H., Luo W., Rodriguez-Barraquer I., Jarman R., Durbin A.P., Diehl S.A., et al. Viridot: an automated virus plaque (immunofocus) counter for the measurement of serological neutralizing responses with application to dengue virus. PLoS Negl. Trop. Dis. 2018;12:e0006862. doi: 10.1371/journal.pntd.0006862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schepens B., van Schie L., Nerinckx W., Roose K., Van Breedam W., Fijalkowska D., Devos S., Weyts W., De Cae S., Vanmarcke S., et al. An affinity-enhanced, broadly neutralizing heavy chain-only antibody protects against SARS-CoV-2 infection in animal models. Sci. Transl. Med. 2021;13:eabi7826. doi: 10.1126/scitranslmed.abi7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herth M.M., Ametamey S., Antuganov D., Bauman A., Berndt M., Brooks A.F., Bormans G., Choe Y.S., Gillings N., Häfeli U.O., et al. On the consensus nomenclature rules for radiopharmaceutical chemistry - reconsideration of radiochemical conversion. Nucl. Med. Biol. 2021;93:19–21. doi: 10.1016/j.nucmedbio.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 84.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Cryo-EM maps and molecular models have been deposited in the Electron Microscopy DataBank (EMDB) and the Protein DataBank (PDB), respectively, with accession codes EMD-15994 (map 1), EMD-15997 (map 2), EMD-16010 (map 3), EMD-16028 (map 4), EMD-16030 (map 5) and PDB: 8BEV (Wu NTD/RBD/W25) and PDB: 8BGG (Omicron NTD/RBD/W25). The accession numbers and more detailed information are also available in Supplementary information. This paper does not report original code. Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.