Abstract
During mRNA 3′ end formation, cleavage stimulation factor (CstF) binds to a GU-rich sequence downstream from the polyadenylation site and helps to stabilise the binding of cleavage-polyadenylation specificity factor (CPSF) to the upstream polyadenylation sequence (AAUAAA). The 64 kDa subunit of CstF (CstF-64) contains an RNA binding domain and is responsible for the RNA binding activity of CstF. It interacts with CstF-77, which in turn interacts with CPSF. The Drosophila suppressor of forked gene encodes a homologue of CstF-77, and mutations in it affect mRNA 3′ end formation in vivo. A Drosophila homologue for CstF-64 has now been isolated, both through homology with the human protein and through protein–protein interaction in yeast with the suppressor of forked gene product. Alignment of CstF-64 homologues shows that the proteins have a conserved N-terminal 200 amino acids, the first half of which is the RNA binding domain with the second half likely to contain the CstF-77 interaction domain; a central region variable in length and rich in glycine, proline and glutamine residues and containing an unusual degenerate repeat motif; and then a conserved C-terminal 50 amino acids. In Drosophila, the CstF-64 gene has a single 63 bp intron, is transcribed throughout development and probably corresponds to l(3)91Cd.
INTRODUCTION
The 3′ ends of eukaryotic mRNAs are generated by processing of pre-mRNA, which in most cases occurs by endonucleolytic cleavage followed by addition of poly(A) to the new 3′ end (1,2). The mechanism for this process has been studied using extracts of human tissue culture cells with pre-formed RNA substrates (e.g. 3). Two multi-subunit complexes have been defined which interact with the RNA, and with each other, to define the site where processing will occur. Cleavage polyadenylation specificity factor (CPSF) consists of subunits of 160, 100, 73 and 30 kDa (4,5), while cleavage stimulation factor (CstF) consists of subunits of 77, 64 and 50 kDa (6). Some of the roles for the different subunits of CPSF and CstF are understood, and some of their interactions have been described (7–14). CPSF binds through its 160 kDa subunit (CPSF-160) to the polyadenylation signal (usually AAUAAA) 10–30 bases upstream from the site of cleavage/polyadenylation while CstF binds through its 64 kDa subunit (CstF-64) to a GU-rich sequence usually situated downstream from the site of cleavage/polyadenylation. The complexes interact via CPSF-160 and CstF-77 (14).
Mammalian CstF-64 has an N-terminal domain that includes an RNA recognition motif of the RNP class (11), and was originally identified in crude extracts of tissue culture cells from its binding to RNA in a polyadenylation signal-dependent manner (15,16). The RNA binding domain (RBD) on its own does bind RNA containing GU-rich sequences, although the CstF complex seems to bind more effectively (17). CstF-64 interacts with CstF-77 but not with CstF-50 (13). In human CstF-64 there is a region of 12 contiguous repeats of an amino-acid motif related to MEARA/G, embedded within a region rich in proline and glycine (11). The repeats are highly conserved in mouse and chicken, although in chicken the middle A is often P (18). They are not well conserved in a Xenopus homologue (19), and their function is not known.
The Drosophila homologue of CstF-77 is encoded by the suppressor of forked [su(f)] gene (13,20). Viable mutants of su(f) appear to have less efficient mRNA 3′ end formation, so that promoter-proximal sites are used less often, thereby allowing the transcribing RNA polymerase to reach more distal sites for processing (21). We describe here the cloning and characterisation of a Drosophila homologue of CstF-64 though its homology to the human sequence, and by protein–protein interaction in a yeast two-hybrid screen using the Drosophila homologue of CstF-77 encoded by su(f) as ‘bait’. Our results provide insights into the structure and function of CstF-64 and its conservation during evolution.
MATERIALS AND METHODS
Drosophila
Flies were raised at 25°C on cornmeal-yeast-sugar-agar medium. Df(3R)ChaM5 and Df(3R)148.5-1 stocks were from J. Hall and A. Villella (University of Brandeis) and Df(3R)fruW24 was from B. S. Baker and L. Ryner (Stanford University). These deficiencies are described in (22). The P-lacZ third chromosome balancer strain was from G. Tear (King’s College, University of London). Preparation of DNA from single embryos for analysis by PCR was as described in (23). Preparation of poly(A)-containing RNA from different stages of Drosophila development and RNA blotting was as described in (20).
PCR techniques
Part of the RBD of CstF-64 was amplified from Drosophila using redundant oligonucleotide primers based upon the human CstF-64 sequence. The reaction contained as template 0.5 µg of phage DNA from a Drosophila ovary cDNA library in Lambda ZAPII (a gift from Tulle Hazelrigg) with 1 µg each of the oligonucleotides RBD-5 (5′-GGNAAC/TATA/C/TCCNT-AC/TGAA/GGC-3′, 384-fold redundant) and RBD-3 (5′-TG-A/GT-CC/TTGA/GTAC/TTCA/GCAA/GAA-3′, 64-fold redundant), and ran for 35 cycles.
Deficiency mapping using PCR on single embryos was as described in (24). Stocks were made where the deficiency chromosomes were maintained using a balancer chromosome that carried a molecular marker, a P-lacZ transgene. Embryos from such stocks that are homozygous for the deficiency chromosome can then be identified as it is not possible to amplify a lacZ fragment from their DNA. Other embryos are either homozygous or heterozygous for the balancer chromosome and do amplify the lacZ fragment. DNA preparations from homozygous deficiency embryos are then assessed by PCR for the presence or absence of the dCstF-64 gene using a control gene from another chromosome. The following oligonucleotide pairs were used: 5′-CGACTGATCCACCCAGTCCC-3′ and 5′-GCGATGTCGGTTTCCGCGAG-3′ for lacZ giving a 739 bp product; 5′-CGATGACACTATCGCAGTTACATCC-3′ and 5′-CTGGTTTTAAGTTGGAATTTAGAAAGAAC-3′ for the X chromosome control gene su(f) giving a 1119 bp product; 5′-ATGCAGCAGCTGCTTCAGGG-3′ and 5′-CAATCTGTTCG-TCGGACAGC-3′ for Drosophila CstF-64 giving an 886 bp product. The reaction contained 2 µl of embryo DNA in a total of 40 µl with 20 ng of each lacZ primer, 100 ng of each su(f) primer and 40 ng of each Drosophila CstF-64 primer. After an initial denaturation at 95°C for 3 min, 16 cycles were ran with denaturation at 95°C for 0.5 min, annealing for 1 min starting at 55°C but dropping by 0.25°C per cycle and extension at 72°C for 2.5 min. A further 24 cycles were then run with a constant annealing temperature of 51°C.
Other recombinant DNA procedures
A 140 bp long PCR product from a Drosophila ovary cDNA library corresponding to residues 21–67 of human CstF-64 was generated as described above and cloned into pBluescript. The DNA insert was labelled and used to screen 106 plaques from the cDNA library. Positive clones were plaque-purified and phagemid DNAs were analysed by restriction enzyme digestion. The longest cDNA (pZd64-19) was used to isolate the corresponding gene from a Drosophila genomic library. Positive clones were purified and their inserts mapped by restriction enzyme digestion. A 4.3 kb EcoRI fragment that included the region where the cDNA hybridised was subcloned in pBluescript. DNA sequences of the insert in pZd64-19 and part of the insert in the genomic subclone were determined using Sequenase (USB) or T7 DNA polymerase (Pharmacia).
Yeast two-hybrid screen
The Drosophila SU(F) protein, homologous to human CstF-77, was fused downstream from the GAL4 DNA binding domain in the vector pGBT9 (Clonetech) that carries the TRP1 marker. The 5′ end of the fusion with respect to su(f) was within the 5′ UTR of a su(f) cDNA so that the hybrid protein has 18 residues that are encoded by the 5′ UTR. The 3′ end of the fusion was just before the C-terminal end of the SU(F) protein so that the hybrid protein lacks the C-terminal 23 (out of 733) residues of su(f). This ‘bait’ construct was transformed into the Saccharomyces cerevisiae strain HF7c (LEU2–HIS3–TRP1–) selecting for TRP1+ transformants. This strain was then transformed with a library (a gift from Susan Parkhurst) of Drosophila cDNAs from 0–4 h embryos (25). The library was in the vector pVP16 (26) which contains the LEU2 gene as a marker and has cDNA-encoded proteins fused with a nuclear-localised acidic transcriptional activation domain from VP16 of Herpes Simplex Virus. Interaction of a cDNA-encoded protein fused to VP16 with the GAL4-SU(F) fusion ‘bait’ protein leads to activation of transcription of the chromosomal HIS3 gene in HF7c as its transcription is under the control of the GAL4 upstream activating sequence, UASG Colonies that grew in the absence of added histidine, leucine and tryptophan were tested for expression of the Escherichia coli β-galactosidase gene (lacZ) also present in the chromosomes of HF7c under UASG control. Other yeast procedures were as described in the Clonetech Matchmaker manual.
RESULTS
Isolation of a Drosophila homologue of human CstF-64 by sequence homology
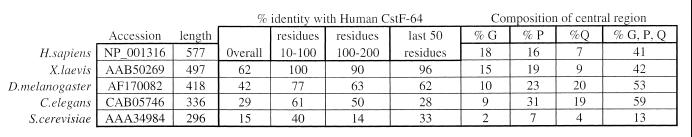
A Drosophila cDNA for a homologue of CstF-64 was isolated by first generating a 140 bp fragment using PCR on an ovary cDNA library with redundant oligonucleotide primers corresponding to the RBD region of human CstF-64. This fragment was then used to screen the same ovary cDNA library by hybridisation and the positive clone with the largest insert, pZd64-19, was characterised by DNA sequencing. The genomic sequence corresponding to this cDNA (see below) has been given the accession number AF170082. The cDNA insert of 1.4 kb in pZd64-19 appears to be complete with a 67 base 5′ untranslated region followed by an open reading frame of 1257 bases and then a 3′ untranslated region of 98 bases that includes an AATAAA polyadenylation signal close to the 3′ end. The protein encoded by the open reading frame is 418 amino acids long and is 42% identical to the human sequence. There are two regions of considerably better conservation: the N-terminal 200 amino acids in the Drosophila protein are 67% identical to human and the C-terminal 40 amino acids are 68% identical.
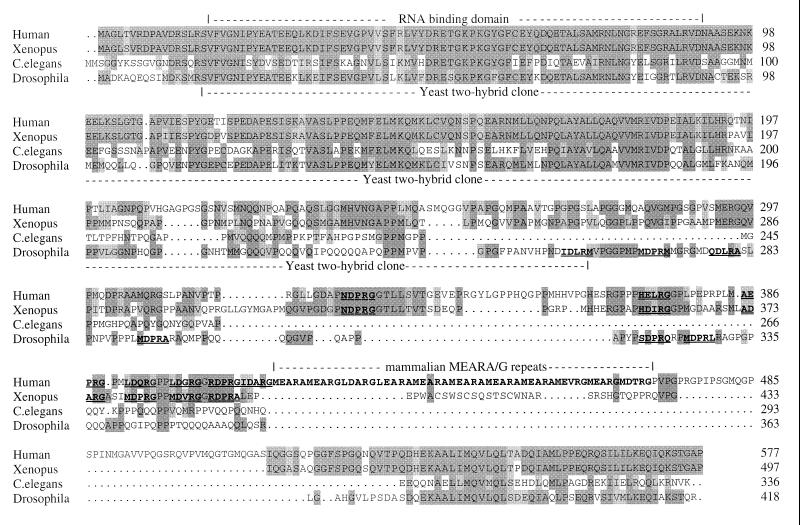
The Drosophila and human sequences (11) are aligned in Figure 1 with the sequence of a Xenopus homologue (19) and a predicted homologue (accession number 2414209 corresponding to CE16126 in WormPep) from the nematode worm, Caenorhabditis elegans (27,28). This four-way comparison confirms that the N- and C-terminal regions of CstF-64 are the most conserved regions, with the four sequences being 46% identical for the N-terminal 200 amino acids and 26% identical for the C-terminal 50 amino acids. The first half of the conserved N-terminal domain (up to around position 90) corresponds to a single copy of the RNA recognition motif found, often as multiple copies, in many RNA-binding proteins. The second half of the conserved N-terminal and the conserved C-terminal region appear to be conserved only in CstF-64. The difference in length of the proteins is due to the poorly conserved central region. This includes the 12 tandem copies of MEARA/G in the human sequence which, although well conserved in the mouse and chicken homologues (18), are at best poorly conserved in Drosophila and Xenopus laevis, and not at all in C.elegans (see Fig. 1 and Discussion). Much of the rest of this central region is made up of glycine, proline and glutamine residues (41, 42, 53 and 59% for human, Xenopus, Drosophila and C.elegans respectively; see Table 1).
Figure 1.
Alignment of amino acid sequences for the human, Xenopus, Drosophila and C.elegans 64 kDa subunits of CstF. Dark grey boxes show positions where the amino acids are conserved in all four sequences while grey boxes show where at least two of the sequences are conserved. The RBD, the region in the cDNA isolated through protein–protein interaction with su(f) and the human-specific MEARA/G repeats are shown. Degenerate copies of this sequence are underlined and in bold.
Table 1. Comparison of CstF-64 homologues.
Isolation of a Drosophila homologue of human CstF-64 by protein–protein interaction in yeast with the protein encoded by su(f)
A ‘bait’ construct was made where the Drosophila SU(F) protein, homologous to human CstF-77 (13), was fused downstream from the yeast GAL4 DNA binding domain. This hybrid protein lacks the C-terminal 23 (out of 733) residues of su(f). Around 8 × 106 embryonic cDNAs were assessed, and 12 were isolated that activated transcription of both HIS3 and lacZ. After confirmation that transcriptional activation required the presence of both the cDNA and the su(f) ‘bait’ plasmid, the cDNA inserts were characterised by partial DNA sequencing and database searching.
Two cDNAs were found to contain fragments from different parts of the Drosophila mitochondrial 16S rRNA. This RNA is enriched in poly(A)-containing RNA preparations from Drosophila (presumably because it contains A-rich regions) and other yeast two-hybrid screens have reported finding ribosomal RNA clones as false positives. However, other HIS3 and lacZ positive clones did contain cDNAs that encoded proteins, and one was found to encode part of a Drosophila homologue of human CstF-64. The DNA sequence of this partial cDNA matches perfectly that described above. The insert corresponds to amino acids 17–257 of Drosophila CstF-64. It includes the region of the RNA recognition domain, but also the rest of the conserved N-terminal domain (Fig. 1). We suggest that this well-conserved region, particularly around residues 120–200, is the domain where the 64 kDa subunit of CstF interacts with the 77 kDa subunit. This interaction in yeast between a Drosophila CstF-77 homologue encoded by su(f), and a Drosophila CstF-64 homologue, does not require the conserved C-terminal domain of CstF-64 (absent from the isolated cDNA), nor the C-terminal 23 amino acids of CstF-77 (missing from the ‘bait’ construct).
Mapping the gene for Drosophila CstF-64
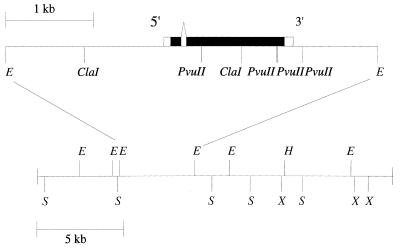
Genomic DNA hybridising to the full-length cDNA, pZd64-19, was isolated by screening a genomic λ library. Two overlapping phage were mapped and the 4.3 kb EcoRI fragment where the cDNA hybridised was subcloned (Fig. 2). The DNA sequence of the region of the genomic subclone corresponding to the cDNA was determined (accession number AF170082). A comparison of the genomic DNA sequence with the cDNA sequence shows that the gene has a single 63 bp intron within the region encoding the RBD. Putative TATA sequences for initiation of transcription, and a GU-rich sequence for mRNA 3′ end formation, are present upstream and downstream respectively from the positions corresponding to the 5′ and 3′ ends of the cDNA.
Figure 2.
Physical map of the Drosophila CstF-64 gene. Above is shown the 4.3 kb EcoRI interval where the full-length cDNA hybridises. The deduced structure of the gene is shown with black boxes to indicate translated regions of exons and white boxes to indicate untranslated regions. Below is the map for the region cloned from a λ genomic library. E, EcoRI; S, SalI; X, XhoI. The orientation of this interval with respect to the chromosome is not known.
In situ hybridisation of the genomic DNA to salivary gland polytene chromosomes from third instar larvae identified a single hybridising locus on the right arm of chromosome 3, around 91B–C (J.K.Lim, personal communication). This region (Fig. 3) includes the fruitless (fru) gene at 91B1-2 (29–31) and the Choline acetyltransferase (Cha) gene at 91C7 (32). Genomic DNA from our walk does not cross-hybridise with either the fru walk (L.Ryner and B.S.Baker, personal communication) or the Cha walk (T.Kitamoto, personal communication). Deficiency mapping using the plasmid subclone to probe DNA blots of genomic DNA from deficiency stocks for the region suggested that the gene was towards the distal end of 91B–C (data not shown). However, as no rearrangement specific fragments were identified, this assignment depended upon quantitation of the hybridisation signal as being double or single dose with respect to controls.
Figure 3.
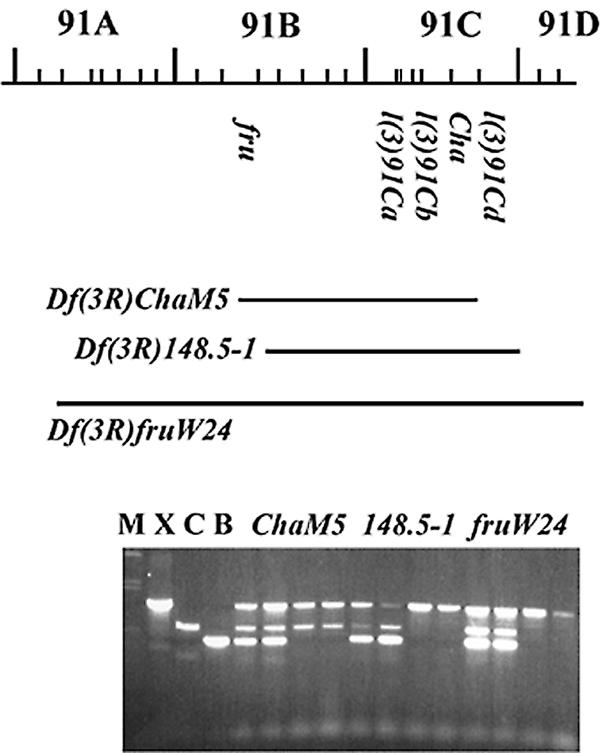
Cytogenetic mapping the Drosophila CstF-64 gene. The extent of several deficiencies and the locations of genes in the 91B–D region on the right arm of chromosome 3 are shown. Sets of four single embryos from Df(3R)148.5-1, Df(3R)Cha5 and Df(3R)fruW24 analysed simultaneously with the three sets of primers are shown. Embryos from Df(3R)148.5-1 that lack the balancer chromosome do amplify CstF-64 while embryos from Df(3R)Cha5 and Df(3R)fruW24 that lack the balancer chromosome do not amplify CstF-64. M, molecular weight marker; C, X and B are reactions using a single set of primers for the control gene su(f), CstF-64 and the balancer-specific lacZ gene respectively.
To more precisely map the gene we used PCR on single embryos laid by deficiency stocks (24). Figure 3 shows an analysis of this sort, which shows that Drosophila CstF-64 is absent from Df(3R)148.5-1, but present on Df(3R)ChaM5. These two small deficiencies are reported to have identical cytology (91B3;91D1) although Df(3R)ChaM5 has its proximal break within fru while Df(3R)148.5-1 leaves fru intact (29,30). This suggests that Drosophila CstF-64 maps in 91D1 before the distal end of Df(3R)148.5-1 but after the distal end of Df(3R)ChaM5. The only gene known in this interval is l(3)91Cd but as the mutants of l(3)91Cd are no longer extant (W.Gelbart, personal communication), it is not possible to test if l(3)91Cd does encode Drosophila CstF-64. The close proximity of the Drosophila CstF-64 gene to Cha [also known as l(3)91Cc] is further supported by the observation that part of our genomic sequence and part of Cha (accession number M63724) overlap the currently incomplete sequence for the Drosophila genomic BAC clone BACR01F15 (accession number AC007812).
Transcription of Drosophila CstF-64
Transcription of Drosophila CstF-64 was assessed during development by probing a blot of poly(A)-containing RNA from different stages with pZd64-19 (Fig. 4). A single RNA ~1.5 kb in size is present throughout development. The RNA is most abundant in early embryos, and there is more in adult females than adult males, suggesting that much of this early embryo RNA may be maternally contributed. It is present throughout larval and pupal development, and appears to be up-regulated in early pupae. This profile is very similar to that for su(f) (20) and for the 30 and 160 kDa subunits of CPSF in Drosophila (33,34), and is similar to that of many other genes for proteins (RNA polymerase subunits, splicing factors etc.) required to make mRNAs.
Figure 4.
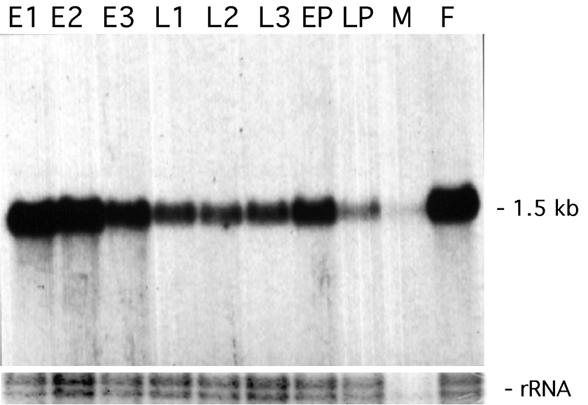
Transcription of Drosophila CstF-64 during development. An RNA blot of poly(A)-containing RNA from stages of Drosophila development was hybridised with a probe made form the full-length cDNA for Drosophila CstF-64. As a loading control, the rRNAs as revealed by ethidium bromide staining are shown. Note that in Drosophila mature 28S rRNA is cleaved, and on denaturing gels the larger part migrates with the 18S rRNA as a doublet of around 1950 bases while the smaller part migrates at around 1750 bases. E1, 0–4 h embryos; E2, 4–8 h embryos; E3, 8–24 h embryos; L1, first instar larvae; L2, second instar larvae; L3, third instar larvae; EP, early pupae; LP, late pupae; M, adult males; F, adult females.
The two cDNAs analysed here, one from 0–4 h embryos and one from ovaries, and an EST from an adult head cDNA (accession number AI514198) all show the same pattern of splicing with elimination of a single small intron. There is no evidence that this gene undergoes alternate splicing or uses alternate 5′ or 3′ ends.
DISCUSSION
Structure and function of CstF-64
The Drosophila gene that we have isolated encodes a protein that is 42% identical overall to human CstF-64. In addition to very highly conserved CstF-64 homologues from mouse and chicken (18), a X.laevis homologue has been characterised that is 62% identical to human CstF-64 (19) while the closest homologue in the yeast S.cerevisiae, RNA15, is 15% identical to human CstF-64 (11,35). However, the conservation is not uniform along the length of the protein with the most conserved regions being the N-terminal 200 and C-terminal 50 amino acids (Fig. 1 and Table 1).
The N-terminal region includes the RBD (approximately residues 10–100). The sequence specificity of binding to RNA of this domain in isolation (17) and in the CstF complex (17,36) have been studied. GU- and U-rich sequences similar to those found downstream of natural cleavage/polyadenylation sites are specifically bound, and these sequences function in vitro as cleavage/polyadenylation signals (17,36). Although this domain on its own may be sufficient to bind RNA, it does not bind as effectively or as specifically as the intact CstF heterotrimer (16,17). In the overall cleavage/polyadenylation reaction, binding of CstF to RNA containing the appropriate sequences occurs in a co-operative fashion with CPSF (14). CstF-64 was first identified in crude extracts after UV cross-linking to RNA that contained the AAUAAA sequence where CPSF interacts (15,16,37). The binding of CPSF to the RNA and protein–protein interaction between CPSF and CstF presumably helped stabilise the binding of CstF to the RNA, allowing CstF-64 to be cross-linked to the RNA.
The high degree of conservation of this region is striking (Table 1) suggesting that this part of the protein from different species has the same structure. The structures of several RBDs have been determined, and this domain of CstF-64 is likely to adopt the same fold (38). Homology (39) and structure prediction programmes (40) suggest that the RBD of Drosophila CstF-64 is most similar to the second RBD of Sex-lethal, which, like CstF-64, binds preferentially to U-rich sequences (41).
In mammals, the AAUAAA sequence where CPSF binds upstream from the site of cleavage is more highly conserved than the GU-rich sequence where CstF binds downstream. Polyadenylation sequences from mammals, for example rabbit β-globin (42) and SV40 early region (43), have been used in vectors for expressing proteins in Drosophila cells in culture and in transgenic flies, respectively. Although the 3′ ends of the mRNAs are rarely examined in any detail in such experiments, successful production of proteins suggests that Drosophila CstF and CPSF do recognise mammalian polyadenylation sequences.
The second half of the N-terminal 200 amino acids of CstF-64 is well conserved, although not as highly as the RNA binding domain (Table 1). This region is within the protein encoded by the Drosophila CstF-64 cDNA selected by interaction with Drosophila CstF-77 in yeast. We propose that this is where CstF-64 binds to CstF-77, and this is supported by in vitro experiments with the human protein (Y.Takagaki and J.L.Manley, submitted). Comparing this region of the homologues (residues 100–200) with human CstF-64, the degree of conservation in different species is similar to the degree of conservation of CstF-77 from those species with human CstF-77. In Drosophila this region of CstF-64 is 63% identical to human CstF-64 while Drosophila SU(F) is 57% identical to human CstF-77 (17), in the C.elegans CstF-64 homologue this region is 50% identical to human CstF-64 while the C.elegans CstF-77 homologue is 49% identical to human CstF-77 (44,45), and in yeast this region of RNA15 is 14% identical to human CstF-64 while the closest yeast homologue of CstF-77, RNA14, is 24% identical to human CstF-77 (17,35). This is consistent with co-evolution of this region of CstF-64 with CstF-77.
The conserved C-terminal domain is not present in the cDNA isolated in our yeast two-hybrid screen, so it cannot be necessary for CstF-64 to interact with CstF-77. Interestingly, the conservation of this region between human and yeast is higher than that between human and C.elegans (Table 1). Homologues do exist for all three CstF subunits in C.elegans (unpublished observations; C.J.Williams and T.Blumenthal, personal communication), suggesting that a nematode complex similar to mammalian CstF exists. However, the organisation of a quarter of C.elegans genes into operons (46) and the coupling of upstream cleavage/polyadenylation with downstream trans-splicing (47) may have resulted in the evolution of a mechanism for mRNA 3′ end formation significantly different to that described for human genes. In yeast, although RNA14 is similar to CstF-77 and RNA15 is similar to CstF-64 (35), there is no good homologue in the yeast genome for CstF-50. Moreover, the protein complex in yeast (CF I) that includes RNA14 and RNA15 has other additional subunits not present in CstF (2,48,49). Further work will be needed to define the role of the C-terminal region of CstF-64 in mRNA 3′ end formation.
The central region is not well conserved, with many proline, glycine and glutamine residues, and variability in this central region is the major reason why the homologues differ in overall length. It includes the 12 tandem copies related to MEARA/G in the human protein that are perfectly conserved in mouse and conserved as 11 tandem copies related to L/MEPRG in chicken (18). Six sequences with some similarity to these repeats can be found in both the Drosophila and Xenopus homologues (underlined in Fig. 1), although they are not contiguous and are more varied in sequence. Some of these degenerate repeats align with similar sequences in the human protein (see Fig. 1). No sequences with any similarity to the repeat motif occur in the worm and yeast homologues. Given the lack of conservation of this region and its amino acid composition, it seems likely that the structure of this part of the protein is not necessary for the essential function of CstF in mRNA 3′ end formation, although the repeats may contribute in some way to CstF function.
Expression and regulation of CstF-64 and mRNA 3′ end formation
In vertebrates, regulation of CstF-64 expression and activity seem to be an important aspect of the regulation of gene expression at the level of mRNA 3′ end formation. CstF-64 activity (50) and protein (18) increase during B cell activation, and manipulation of the level of expression of CstF-64 affects the switch from membrane bound to secreted forms of IgM as well as the total amount of IgM heavy chain (18,51). CstF-64 levels appear to vary during the cell cycle, and manipulation of the level of expression of CstF-64 affects progression through the cell cycle (51). Drosophila CstF-64 mRNA is most abundant during development in stages where cell division is rapid—embryos and early pupae. This profile matches that of other genes required for mRNA production including other subunits of CstF and CPSF, so it is not clear if this reflects a specific requirement for CstF-64 during cell proliferation. More detailed studies may reveal if changes in expression of Drosophila CstF-64 are responsible for, or correlated with, developmental changes in mRNA 3′ end formation.
Alternate forms of CstF-64 exist in vertebrates. A bovine isoform of 70 kDa (36) and a murine testis-specific form have been described (52). The mouse and chicken genes each contain 14 exons and mouse cDNAs have been identified that correspond to alternatively spliced mRNAs (Y.Takagaki and J.L.Manley, unpublished). We have found no evidence for such complexity for Drosophila CstF-64.
Null mutants of the gene for a Drosophila CstF-77 homologue, su(f), are lethal and their phenotypes indicate that the gene is required at many different times and places during development (see 22 for details). This presumably reflects the requirement for CstF in the production of many genes’ mRNAs. Null mutants for the genes of the other subunits of CstF are also likely to be lethal, and chicken CstF-64 is known to be required for viability of cultured cells (53). Weak mutations in the su(f) gene lead to suppression of the bristle phenotype of some insertion mutants of the forked gene. This is due to changes in mRNA 3′ end formation (21), and it has been suggested that in viable su(f) mutants, the interaction of CstF with CPSF is less effective (14). As this interaction occurs between the CstF-77 and CPSF-160 subunits, this rather specific phenotype need not necessarily be produced by mutations in the genes for the CstF-64 or CstF-50 subunits, as they would only indirectly, if at all, affect the CstF–CPSF interaction.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Susan Parkhurst (Fred Hutchison Cancer Center, WA) for providing the library used in the two-hybrid screen, and Tulle Hazelrigg (Columbia University, NY) for the ovary cDNA library. Jeff Hall and Adriana Villella (Brandeis University, MA), Bruce Baker and Lisa Ryner (Stanford University, CA), Kevin Moses (Emory University, GA) and Guy Tear (King’s College, London) kindly provided Drosophila stocks and Johng Lim (University of Wisconsin, Eau Claire) performed the in situ hybridisation that located the Drosophila CstF-64 gene to the 91B–C region. Work in London was supported by grant 040633 from the Wellcome Trust and in New York by NIH grant GM 28983. L.F. was an ERASMUS exchange student from the University of Oporto.
DDBJ/EMBL/GenBank accession no. AF170082
REFERENCES
- 1.Colgan D.F. and Manley,J.L. (1997) Genes Dev., 11, 2755–2766. [DOI] [PubMed] [Google Scholar]
- 2.Keller W. and Minvielle-Sebastia,L. (1997) Curr. Opin. Cell Biol., 9, 329–336. [DOI] [PubMed] [Google Scholar]
- 3.Takagaki Y., Ryner,L.C. and Manley,J.L. (1989) Genes Dev., 3, 1711–1724. [DOI] [PubMed] [Google Scholar]
- 4.Bienroth.S, Wahle,E., Suter-Crazzolara,C. and Keller,W. (1991) J. Biol. Chem., 266, 19768–19776. [PubMed] [Google Scholar]
- 5.Murthy K.G. and Manley,J.L. (1992) J. Biol. Chem., 267, 14804–14811. [PubMed] [Google Scholar]
- 6.Takagaki Y., Manley,J.L., MacDonald,C.C., Wilusz,J. and Shenk,T. (1990) Genes Dev., 4, 2112–2120. [DOI] [PubMed] [Google Scholar]
- 7.Jenny A., Hauri,H.P. and Keller,W (1994) Mol. Cell. Biol., 14, 8183–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenny A. and Keller,W. (1995) Nucleic Acids Res., 23, 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenny A., Minvielle-Sebastia,L., Preker,P.J. and Keller,W. (1996) Science, 274, 1514–1517. [DOI] [PubMed] [Google Scholar]
- 10.Barabino S.M., Hubner,W., Jenny,A., Minvielle-Sebastia,L. and Keller,W. (1997) Genes Dev., 11, 1703–1716. [DOI] [PubMed] [Google Scholar]
- 11.Takagaki Y., MacDonald,C.C., Shenk.T. and Manley,J.L. (1992) Proc. Natl Acad. Sci. USA, 89, 1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takagaki Y. and Manley,J.L. (1992) J. Biol. Chem., 267, 23471–23474. [PubMed] [Google Scholar]
- 13.Takagaki Y. and Manley,J.L. (1994) Nature, 372, 471–474. [DOI] [PubMed] [Google Scholar]
- 14.Murthy K.G. and Manley,J.L. (1995) Genes Dev., 9, 2672–2683. [DOI] [PubMed] [Google Scholar]
- 15.Wilusz J. and Shenk,T. (1988) Cell, 52, 221–228. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald C.C., Wilusz,J. and Shenk,T. (1994) Mol. Cell. Biol., 14, 6647–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takagaki Y. and Manley,J.L. (1997) Mol. Cell. Biol., 17, 3907–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takagaki Y., Seipelt,R.L., Peterson,M.L. and Manley,J.L. (1996) Cell, 87, 941–952. [DOI] [PubMed] [Google Scholar]
- 19.Barbaux S., Seery,L.T., Schoenberg,D.R., Sellar,G.C. and Whitehead,A.S. (1996) Comp. Biochem. Physiol. B Biochem. Mol. Biol., 114, 313–315. [DOI] [PubMed] [Google Scholar]
- 20.Mitchelson A., Simonelig,M., Williams,C. and O’Hare,K. (1993) Genes Dev., 7, 241–249. [DOI] [PubMed] [Google Scholar]
- 21.O’Hare K. (1995) Trends Genet., 11, 255–257. [DOI] [PubMed] [Google Scholar]
- 22.Lindsley D.L. and Zimm,G.G. (1992) The Genome of Drosophila. Academic Press, New York, NY.
- 23.Gloor G.B., Nassif,N.A., Johnson-Schlitz,D.M., Preston,C.R. and Engels,W.R. (1991) Science, 253, 1110–1117. [DOI] [PubMed] [Google Scholar]
- 24.Hatton L.S. and O’Hare,K. (1999) Technical Tips Online, T01816 http://tto.trends.com
- 25.Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vojtek A.B., Hollenberg,S.M. and Cooper,J.A. (1993) Cell, 74, 205–214. [DOI] [PubMed] [Google Scholar]
- 27.McMurray A. (1996) Accession number CAB05746.
- 28.Wilson R., Ainscough,R., Anderson,K., Baynes,C., Berks,M., Bonfield,J., Burton,J., Connell,M., Copsey,T., Cooper,J. et al. (1994) Nature, 368, 32–38. [DOI] [PubMed] [Google Scholar]
- 29.Gailey D.A. and Hall,J.C. (1989) Genetics, 121, 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryner L.C., Goodwin,S.F., Castrillon,D.H., Anand,A., Villella,A., Baker,B.S., Hall,J.C., Taylor,B.J. and Wasserman,S.A. (1996) Cell, 87, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 31.Ito H., Fujitani,K., Usui,K., Shimizu-Nishikawa,K., Tanaka,S. and Yamamoto,D. (1996) Proc. Natl Acad. Sci. USA, 93, 9687–9692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamoto T., Wang,W. and Salvaterra,P.M. (1998) J. Biol. Chem., 273, 2706–2713. [DOI] [PubMed] [Google Scholar]
- 33.Salinas C.A., Sinclair,D.A., O’Hare,K. and Brock,H.W. (1998) Mol. Gen. Genet., 257, 672–680. [DOI] [PubMed] [Google Scholar]
- 34.Bai C. and Tolias,P.P. (1996) Mol. Cell. Biol., 16, 6661–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minvielle-Sebastia L., Winsor,B., Bonneaud,N. and Lacroute,F. (1991) Mol. Cell. Biol., 11, 3075–3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyer K., Dandekar,T. and Keller,W. (1997) J. Biol. Chem., 272, 26769–26779. [DOI] [PubMed] [Google Scholar]
- 37.Wilusz J., Shenk,T., Takagaki,Y. and Manley,J.L. (1990) Mol. Cell. Biol., 10, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai K., Oubridge,C., Ito,N., Avis,J. and Evans,P. (1995) Trends Biochem. Sci., 20, 235–240. [DOI] [PubMed] [Google Scholar]
- 39.Corpet F., Gouzy,J. and Kahn,D. (1998) Nucleic Acids Res., 26, 323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rost B. and Sander,C. (1993) J. Mol. Biol., 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 41.Crowder S.M., Kanaar,R., Rio,D.C. and Alber,T. (1999) Proc. Natl Acad. Sci. USA, 96, 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourouis M. and Jarry,B. (1983) EMBO J., 2, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brand A.H. and Perrimon,N. (1993) Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 44.O’Hare K. and Williams,C. (1995) Accession number L39893.
- 45.Tanaka Y., Ohta,A., Matsuo,M. and Sakamoto,H. (1995) DNA Res., 2, 143–146. [DOI] [PubMed] [Google Scholar]
- 46.Blumenthal T. and Spieth,J. (1996) Curr. Opin. Genet. Dev., 6, 692–698. [DOI] [PubMed] [Google Scholar]
- 47.Kuersten S., Lea,K., MacMorris,M., Spieth,J. and Blumenthal,T. (1997) RNA, 3, 269–278. [PMC free article] [PubMed] [Google Scholar]
- 48.Minvielle-Sebastia L., Preker,P.J. and Keller,W. (1994) Science, 266, 1702–1705. [DOI] [PubMed] [Google Scholar]
- 49.Kessler M.M., Zhao,J. and Moore,C.L. (1996) J. Biol. Chem., 271, 27167–27175. [DOI] [PubMed] [Google Scholar]
- 50.Edwalds-Gilbert G. and Milcarek,C. (1995) Mol. Cell. Biol., 15, 6420–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martincic K., Campbell,R., Edwalds-Gilbert,G., Souan,L., Lotze,M.T. and Milcarek,C. (1998) Proc. Natl Acad. Sci. USA, 95, 11095–11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace A.M., Dass,B., Ravnik,S.E., Tonk,V., Jenkins,N.A., Gilbert,D.J., Copeland,N.G. and MacDonald,C.C. (1999) Proc. Natl Acad. Sci. USA, 96, 6763–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takagaki Y. and Manley,J.L. (1998) Mol. Cell, 2, 761–771. [DOI] [PubMed] [Google Scholar]