Abstract
Most cancer vaccines target peptide antigens, necessitating personalization owing to the vast inter-individual diversity in major histocompatibility complex (MHC) molecules that present peptides to T cells. Furthermore, tumours frequently escape T cell-mediated immunity through mechanisms that interfere with peptide presentation1. Here we report a cancer vaccine that induces a coordinated attack by diverse T cell and natural killer (NK) cell populations. The vaccine targets the MICA and MICB (MICA/B) stress proteins expressed by many human cancers as a result of DNA damage2. MICA/B serve as ligands for the activating NKG2D receptor on T cells and NK cells, but tumours evade immune recognition by proteolytic MICA/B cleavage3,4. Vaccine-induced antibodies increase the density of MICA/B proteins on the surface of tumour cells by inhibiting proteolytic shedding, enhance presentation of tumour antigens by dendritic cells to T cells and augment the cytotoxic function of NK cells. Notably, this vaccine maintains efficacy against MHC class I-deficient tumours resistant to cytotoxic T cells through the coordinated action of NK cells and CD4+ T cells. The vaccine is also efficacious in a clinically important setting: immunization following surgical removal of primary, highly metastatic tumours inhibits the later outgrowth of metastases. This vaccine design enables protective immunity even against tumours with common escape mutations.
We developed a conceptually new cancer vaccine that targets a tumour immune escape mechanism. The vaccine targets the MICA and MICB (MICA/B) stress proteins that are upregulated in response to DNA damage in many types of human cancers but are expressed at low or undetectable levels by healthy cells2,3,5,6. Engagement of the activating NKG2D receptor by membrane-bound MICA/B triggers the cytotoxicity programme in natural killer (NK) cells and co-stimulatory signalling in CD8+ T cells7-9. However, many human tumours evade this important immune recognition pathway by proteolytic shedding of MICA/B from the cell surface4,10-13. Shedding substantially reduces the surface density on tumour cells of these immunostimulatory ligands for the NKG2D receptor12,14. Shed MICA/B proteins have also been reported to induce NKG2D receptor internalization and inhibit NK cell function12,14-16. Patients with melanoma responding to an autologous cell-based cancer vaccine (GVAX) plus anti-CTLA-4 were found to develop anti-MICA antibodies, and the presence of these antibodies correlated with reduced serum levels of shed MICA and augmented CD8+ T cell and NK cell responses17,18.
Design of the MICA and MICB α3 domain vaccine
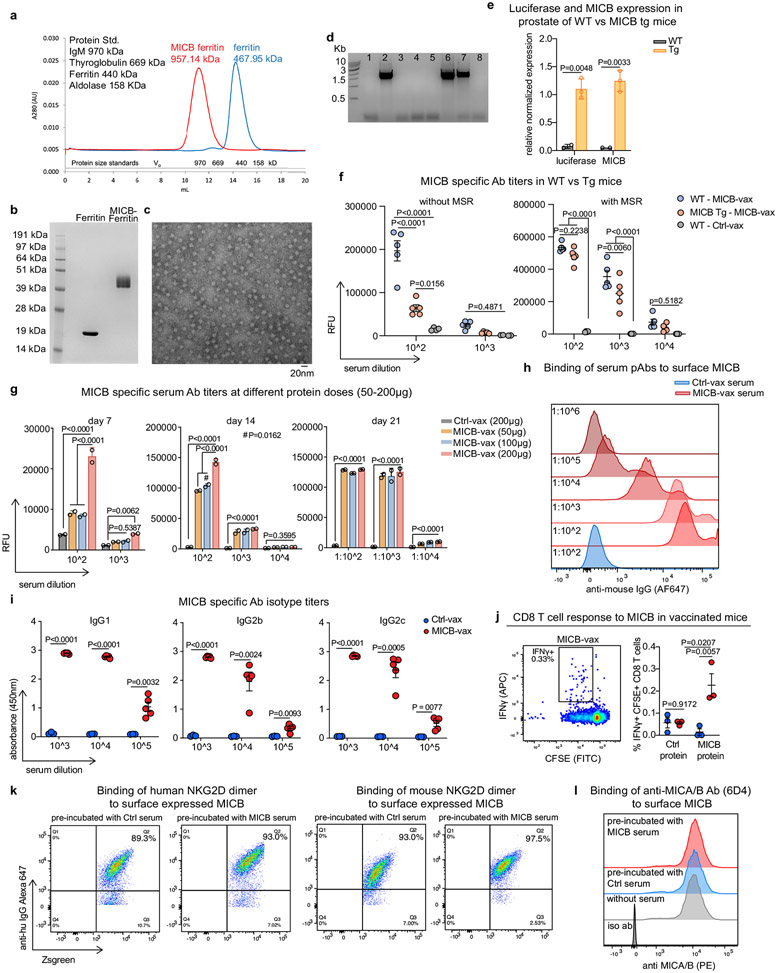
Our vaccine targeted the highly conserved α3 domain in MICA/B, the site of proteolytic shedding, and was designed to induce tumour immunity by T cells and NK cells (Fig. 1a)19. We intentionally omitted the α1–α2 domains to avoid induction of antibodies that could block NKG2D receptor binding20. Multivalent display of vaccine antigens greatly enhances immunogenicity21, and we therefore fused the α3 domain of MICB or MICA to the N terminus of ferritin from Heliobacter pylori, which forms particles composed of 24 subunits22 (Extended Data Fig. 1a-c). Ferritin was used as a control antigen in all experiments. A recently developed mesoporous silica rod (MSR) biodegradable scaffold formulated with granulocyte-macrophage colony-stimulating factor (GM-CSF; for dendritic cell (DC) recruitment) and CpG ODN 1826 (adjuvant) was used for vaccine delivery23.
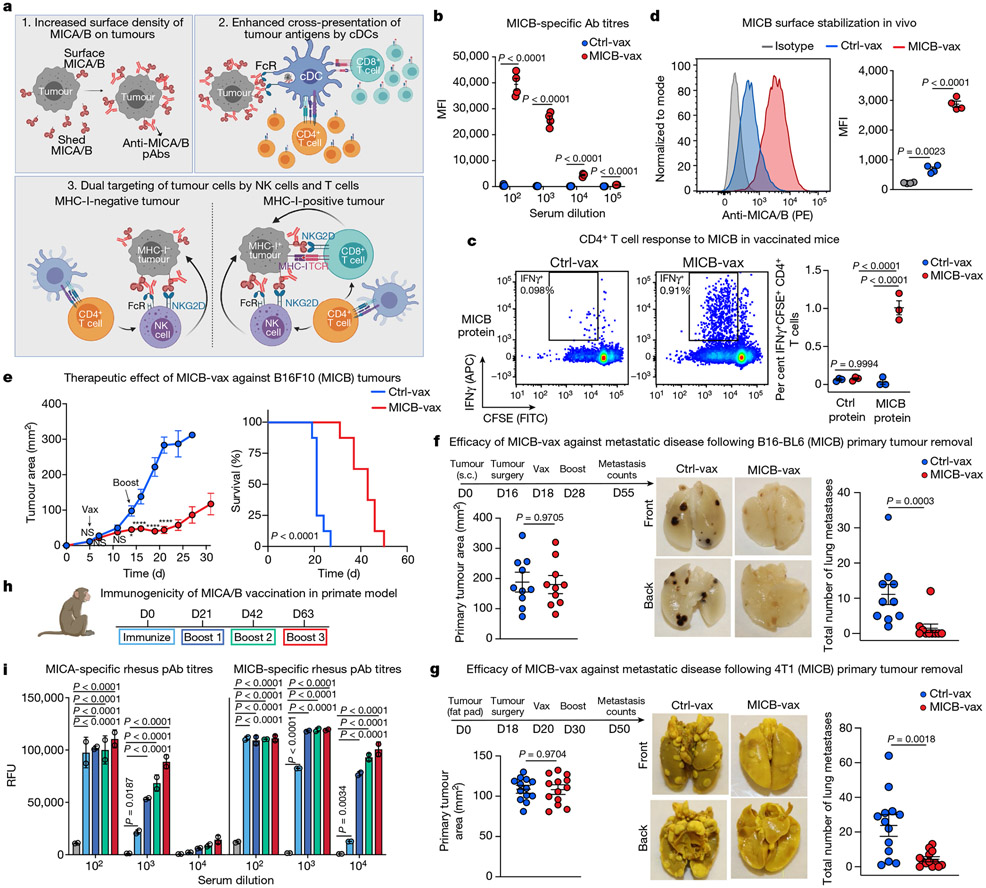
Fig. 1 ∣. Efficacy of the MICA/B α3 domain cancer vaccine.
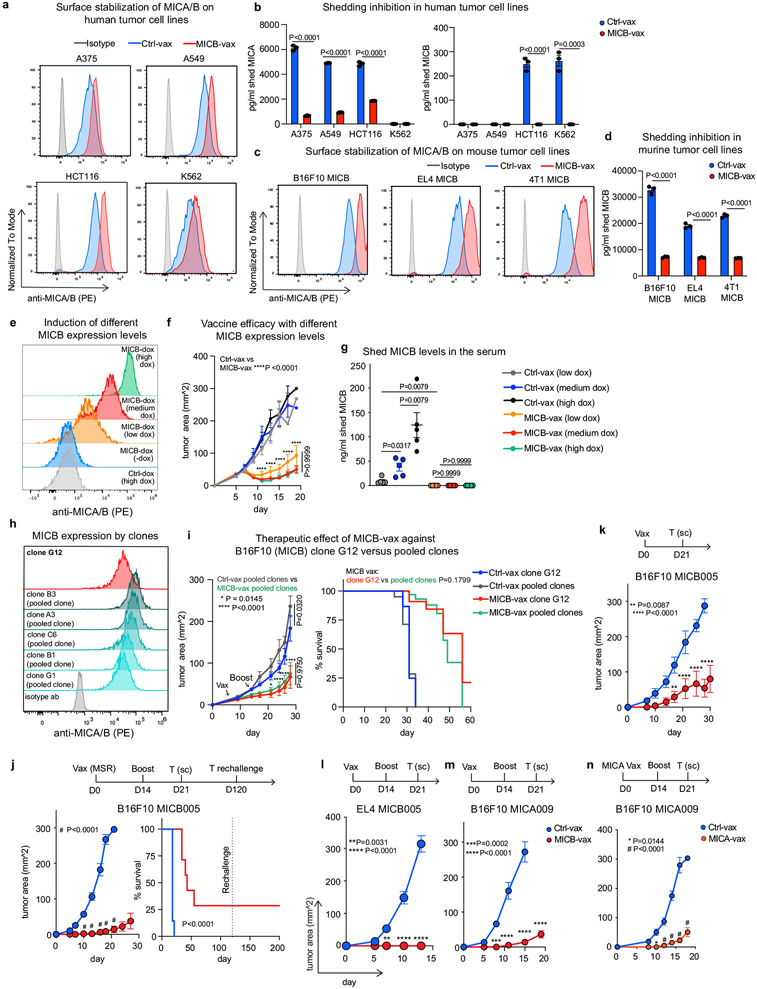
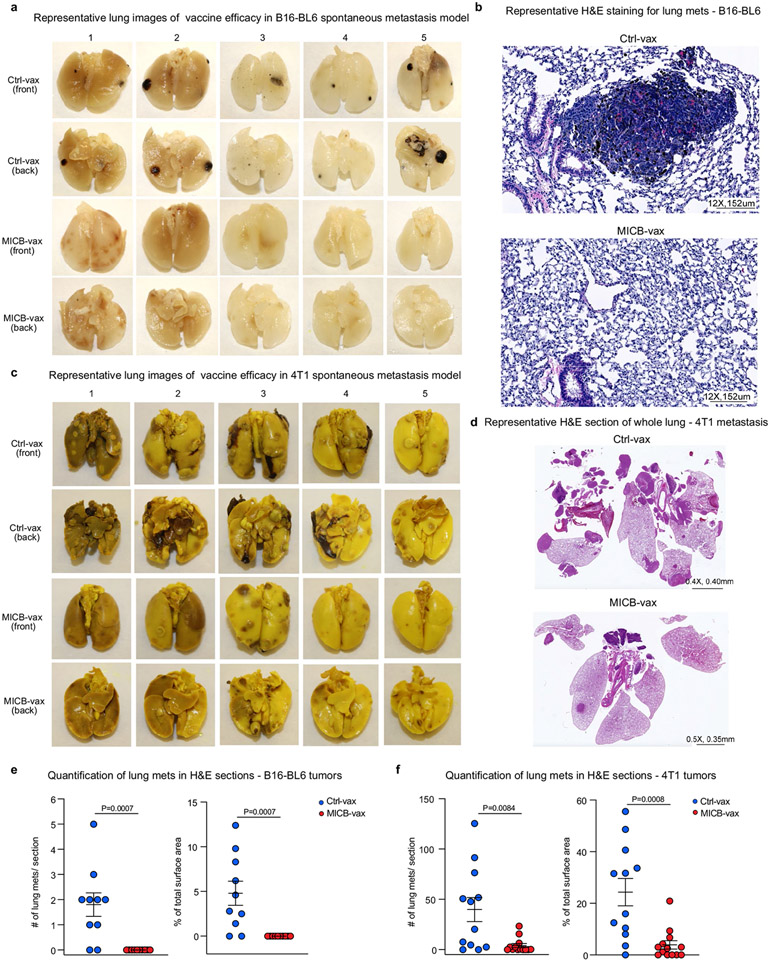
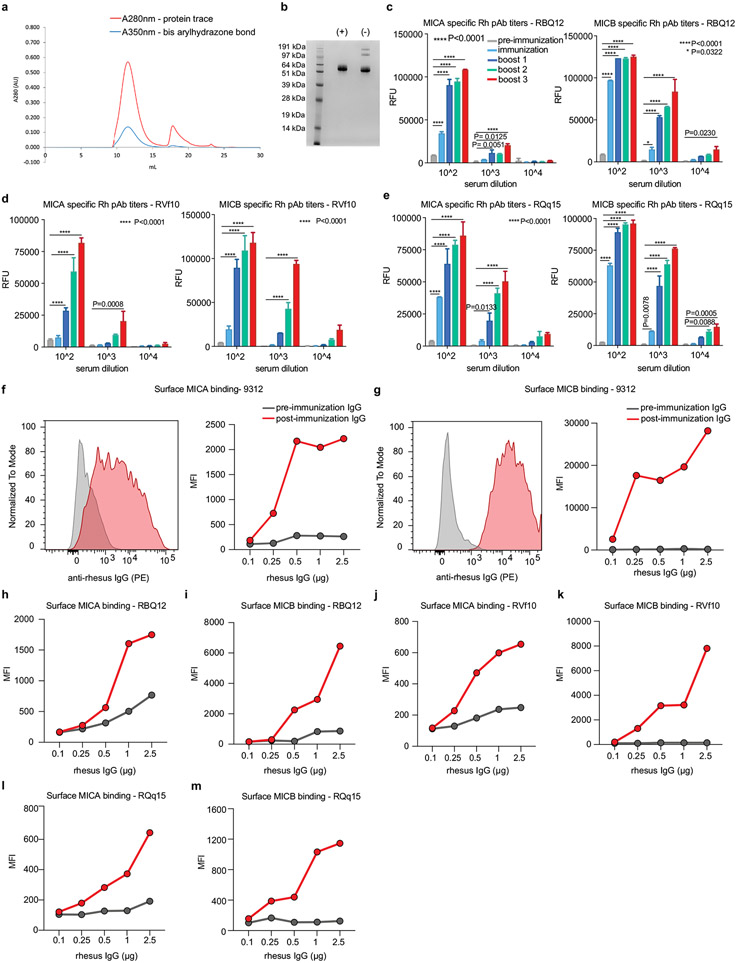
a, Design of the MICA/B vaccine. pAbs, polyclonal serum IgG. b, MICB-specific serum antibody (Ab) titres quantified by flow cytometry (n = 4 mice per group) in MICB-transgenic mice immunized with Ctrl-vax (blue) or MICB-vax (red). MFI, median fluorescence intensity. c, MICB-specific CD4+ T cell responses following immunization with MICB-vax or Ctrl-vax; CFSE dilution of splenocytes stimulated with MICB or control protein (ovalbumin (OVA)); shown are representative flow cytometry plots (left) and quantification for three mice per group (right). d, Cell-surface levels of MICB on B16F10 (MICB) tumours from mice immunized with MICB-vax or Ctrl-vax (n = 4 mice per group); staining of tumour cells with isotype-control monoclonal antibody (grey) or anti-MICA/B monoclonal antibody (specific for the α1–α2 domains, monoclonal antibody not blocked by vaccine-induced antibodies). e, Therapeutic efficacy of MICB-vax (red) or Ctrl-vax (blue) in mice with established B16F10 (MICB) tumours immunized at the indicated time points (n = 7 mice per group). Vax, vaccination. *P = 0.0137, ****P <0.0001; NS (not significant), P > 0.999. f, g, Vaccine efficacy in two models of spontaneous metastasis. Mice were immunized with Ctrl-vax (blue) or MICB-vax (red) following surgical removal of primary tumours using the B16-B6 melanoma (f; 10 mice per group) or 4T1 breast cancer (g; 13 mice per group) models. Shown are the size of primary tumours at the time of surgery (left), representative images of lung metastases (middle) and quantification of the total number of lung surface metastases (right). D, day; s.c., subcutaneous. h, i, Immunogenicity of the rhesus MICA/B α3 domain vaccine in the rhesus macaque model. h, Timeline of vaccination; blood was drawn 24 h before indicated immunization or boost. i, Serum titres of antibody to rhesus MICA/B for animal ID 9312. Representative data are shown from at least three (b) or two (c–g) independent experiments. Data from a single experiment with technical replicates for each time point are shown in i. Statistical significance was assessed by two-tailed unpaired Student’s t test (b), two-way ANOVA with Sidak’s multiple-comparison test (c), one-way ANOVA with Tukey’s multiple-comparison test (d), two-way ANOVA with Bonferroni’s post hoc test (left) and the log-rank (Mantel–Cox) test (right) (e), two-tailed Mann–Whitney test (f, g) and two-way ANOVA with Tukey’s multiple-comparison test (i). Data are depicted as the mean ± s.e.m. (b–g) or mean±s.d. (i).
Efficacy against metastatic disease
The mouse NKG2D receptor binds human MICA/B proteins24,25, yet mouse NKG2D ligands have only limited homology to human MICA/B. We therefore expressed human MICB or MICA proteins in mouse cancer cell lines for in vivo experiments. Human MICA/B proteins are shed by these mouse cancer cell lines, similarly to human cancer cells25. Vaccination experiments were performed in MICB-transgenic mice in which MICB and a luciferase reporter were expressed under the control of the prostate-specific probasin (Pbsn) promoter14. Expression of MICB and luciferase mRNA in the prostate was confirmed by reverse-transcription qPCR (Extended Data Fig. 1d, e). Significantly lower antibody titres were measured in MICB-transgenic mice than in wild-type (WT) mice following immunization with MICB–ferritin in the absence of the MSR scaffold, providing evidence for partial tolerance towards MICB in these transgenic mice. Inclusion of the MSR scaffold substantially increased antibody titres in both strains, enabling induction of high-titre antibodies in partially tolerant MICB-transgenic mice (Extended Data Fig. 1f).
The MICB α3 domain vaccine (MICB-vax) induced high-titre antibodies that strongly labelled B16F10 (MICB) tumour cells (expressing human MICB) but not control B16F10 tumour cells (Fig. 1b and Extended Data Fig. 1g-i). The vaccine also induced CD4+ T cell and CD8+ T cell responses against MICB (Fig. 1c and Extended Data Fig. 1j). As expected, MICB α3 domain-specific antibodies did not interfere with MICB binding by human or mouse NKG2D receptor (Extended Data Fig. 1k, l). Small quantities of sera from MICB-vaccinated mice significantly increased cell-surface MICA/B protein levels on human and mouse cancer cell lines by inhibiting MICA/B shedding (Extended Data Figs 2a-d). In vivo, MICB-vax inhibited MICB shedding to undetectable levels and substantially increased the cell-surface density of MICB on B16F10 tumours (Fig. 1d). This vaccine showed substantial efficacy in controlling subcutaneous B16F10 and EL4 tumours that expressed either MICB or MICA (Fig. 1e and Extended Data Fig. 2e-n). To evaluate immunological memory induced by MICB-vax, mice that remained tumour free were re-challenged with B16F10 (MICB) tumour cells 4 months after the initial immunization and were found to be fully protected (Extended Data Fig. 2j). Vaccine efficacy was observed over a wide range of MICB expression levels induced in isogenic tumours using a doxycycline-inducible promoter (Extended Data Fig. 2e-g), including in tumour cells with low MICB expression. Additionally, the vaccine showed similar efficacy against B16F10 (MICB) cell lines composed of either a single clone or multiple clones (Extended Data Fig. 2h, i).
Both NK cells and T cells have a critical role in the control of metastases26,27. One of the most challenging problems in oncology is that many patients with locally advanced tumours already have established micrometastases that cause recurrence despite successful removal of the primary tumour. We therefore evaluated the efficacy of MICB-vax in this challenging setting by immunizing mice following surgical removal of primary tumours with a high propensity for early metastatic dissemination. We used two models of spontaneous metastasis, the B16-BL6 (MICB) melanoma model and the 4T1 (MICB) triple-negative breast cancer model. Following surgical removal of primary tumours, mice received either MICB-vax or Ctrl-vax (ferritin). In this therapeutic setting, MICB-vax greatly reduced the number of lung metastases detected in both models more than 1 month after surgery (Fig. 1f, g). Histological analysis of lung sections further demonstrated that MICB-vax resulted in a substantial reduction in metastasis number and size in comparison to Ctrl-vax (Extended Data Fig. 3a-f).
Immunogenicity in non-human primates
We next examined vaccine safety and immunogenicity in rhesus macaques, which endogenously express MICA/B proteins28. Macaque MICA/B α3 domains were used for vaccination, rather than those from the human proteins, to enable full assessment of vaccine immunogenicity. Immunization with both MICA/B α3 domains provided better coverage of the limited number of polymorphic positions in the MICA and MICB alleles in this outbred species. MICA/B α3 domains were displayed on ferritin, and CpG ODN 2395 was conjugated to nanoparticles because the MSR scaffold had not been evaluated in non-human primates (Extended Data Fig. 4a, b). All four macaques developed anti-MICA and anti-MICB antibodies following immunization, and titres increased 100- to 1,000-fold with subsequent booster immunization (Fig. 1h, i and Extended Data Fig. 4c-e). Differences in purified serum IgG titres among these outbred animals could be due to allelic differences in immune-related genes (such as major histocompatibility complex class II (MHC-II) genes) or other factors. Purified IgG from vaccinated animals strongly labelled macaque MICA and MICB proteins on transfected cells (Extended Data Fig. 4f-m). There were no clinical side effects or changes in blood chemistry following immunization, providing preliminary evidence of vaccine safety.
Recruitment of T cells and NK cells
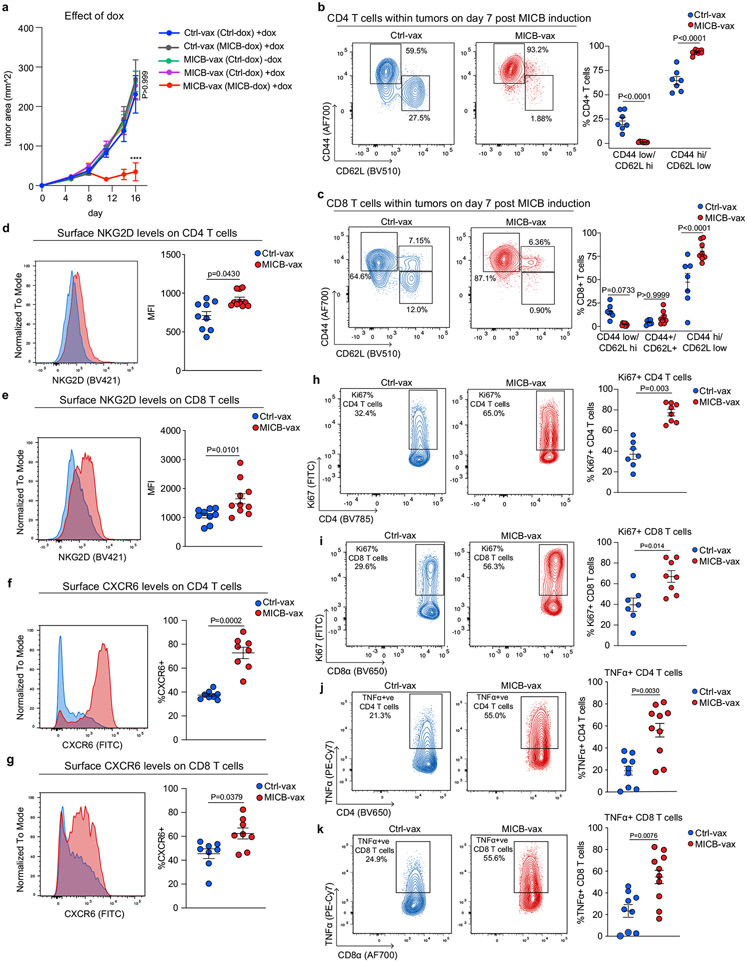
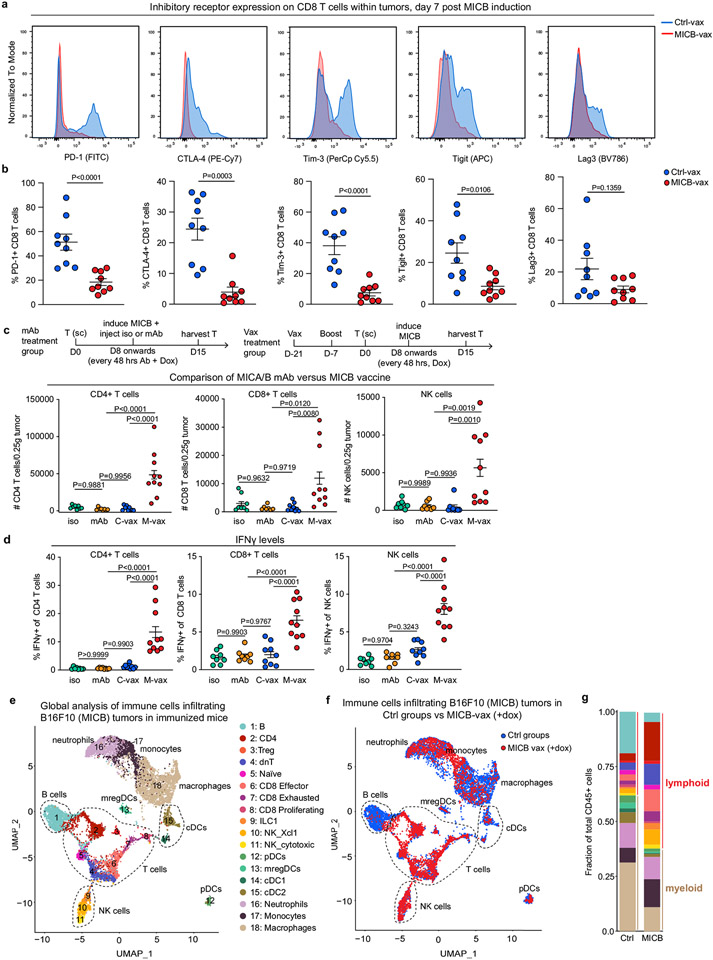
We used the aggressive B16F10 (MICB) melanoma model to study the mechanisms of vaccine efficacy. Owing to the efficacy of MICB-vax, tumours in experimental mice were substantially smaller or undetectable when tumours in control mice had already reached the endpoint. To enable better comparison of the tumour microenvironment between vaccination groups, MICB expression was induced in B16F10 (MICB-dox) tumour cells by doxycycline treatment once mice had been vaccinated and tumours were established (Fig. 2a); MICB expression was undetectable in B16F10 (MICB-dox) tumours before doxycycline administration (Extended Data Fig. 2e). Moreover, doxycycline did not affect tumour growth in mice receiving Ctrl-vax (Extended Data Fig. 5a). Multiple effector lymphocyte populations were strongly enriched in tumours from mice immunized with MICB-vax, with a 29.3-fold enrichment in CD4+ T cells, a 17.9-fold enrichment in CD8+ T cells and a 38.9-fold enrichment in NK cells in these mice compared with mice receiving Ctrl-vax (Fig. 2b-e and Extended Data Fig. 5b, c). By contrast, the percentage of FoxP3+ regulatory T cells was reduced among total CD4+ T cells in tumours from mice immunized with MICB-vax (Fig. 2c). Higher NKG2D and CXCR6 receptor levels were present on tumour-infiltrating CD4+ and CD8+ T cells in mice immunized with MICB-vax relative to those receiving Ctrl-vax (Extended Data Fig. 5d-g). Furthermore, both CD4+ and CD8+ T cells showed enhanced functionality with larger percentages of interferon-γ (IFNγ)+, tumour necrosis factor-α (TNFα)+ and Ki67+ cells in mice immunized with MICB-vax (Fig. 2f, g and Extended Data Fig. 5h-k). Expression of the PD-1, CTLA-4, Tim-3, Tigit and Lag3 inhibitory receptors was significantly lower on CD8+ T cells from mice immunized with MICB-vax compared with those receiving Ctrl-vax (Extended Data Fig. 6a, b). In contrast to MICB-vax, administration of an anti-MICA/B monoclonal antibody directed against the α3 domain did not increase T cell recruitment into tumours (Extended Data Fig. 6c, d).
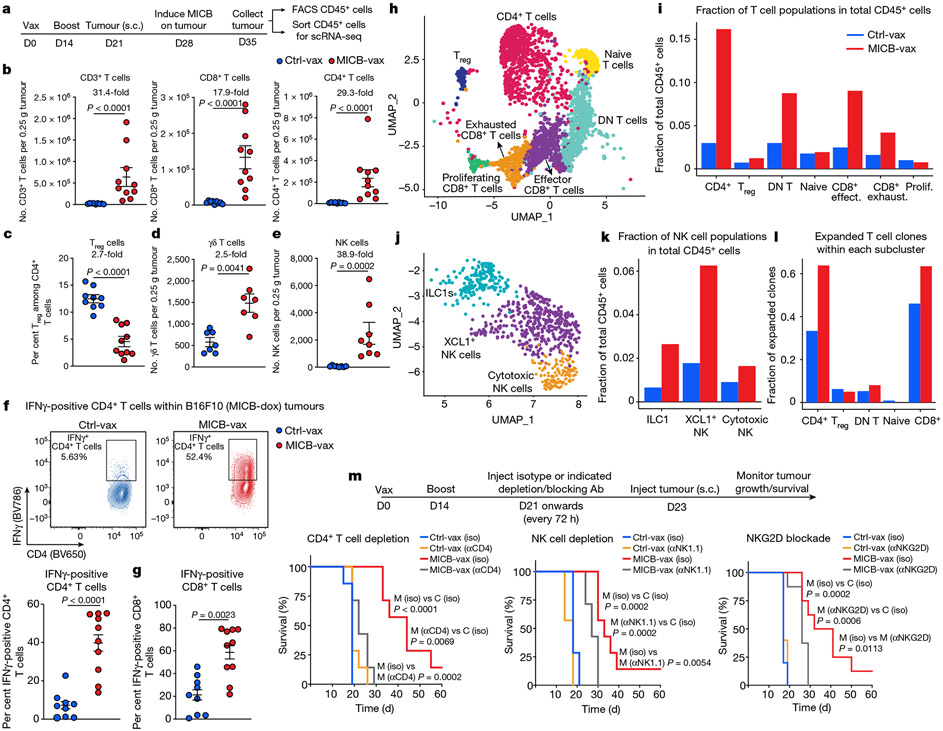
Fig. 2 ∣. Vaccine induces T cell and NK cell recruitment into tumours.
a, Strategy for characterization of tumour-infiltrating immune cells by flow cytometry (FACS) and scRNA-seq. Mice received two immunizations (days 0 and 14), B16F10 (MICB-dox) tumour cells were implanted (day 21), MICB expression was induced on tumour cells by doxycycline treatment (day 28) and tumour-infiltrating immune cells were analysed 7 d later. b–d, Tumour-infiltrating T cell populations following immunization with Ctrl-vax (blue) or MICB-vax (red) (Ctrl-vax, n = 9 mice per group; MICB-vax, n = 10 mice per group; n = 7 mice per group for the γδ T cell panel). Treg cells, regulatory T cells. e, Tumour-infiltrating NK cells (n = 8 mice per group). f, g, Quantification of IFNγ-positive CD4+ and CD8+ T cells (Ctrl-vax, n = 9 mice per group; MICB-vax, n = 10 mice per group). h, i, UMAP representation of all T cell clusters from scRNA-seq data (h) and the fraction of each T cell subpopulation among total CD45+ cells from the experimental group (MICB-vax + doxycycline; red) and three combined control groups (Ctrl-vax±doxycycline and MICB-vax without doxycycline; blue) (i). DN, double negative. j, k, UMAP representation of NK and ILC1 cells from all experimental groups (j) and the fraction of these subpopulations among total CD45+ cells from experimental (red) and combined control (blue) groups (k). l, Fraction of T cells representing expanded clones based on TCR sequence analysis for the experimental group (red) and combined control groups (blue). m, Contribution of CD4+ T cells, NK cells and NKG2D receptor to vaccine efficacy. Mice were first immunized with MICB-vax (M) or Ctrl-vax (C) (days 0 and 14) and treated with isotype-control monoclonal antibody (iso), depleting monoclonal antibody (targeting CD4+ T cells or NK cells (αCD4 and αNK1.1, respectively)) or NKG2D receptor-blocking monoclonal antibody (αNKG2D) starting on day 21, followed by implantation of B16F10 (MICB) tumour cells (n = 7 mice per group). Representative data from three independent experiments are shown in b–g. scRNA-seq data from a single experiment with sorted CD45+ cells pooled from five mice per group are shown in h–l. Representative data from two independent experiments are shown in m. Statistical significance was assessed by two-tailed Mann–Whitney test (b–g) or log-rank (Mantel–Cox) test (m). Data are depicted as the mean±s.e.m.
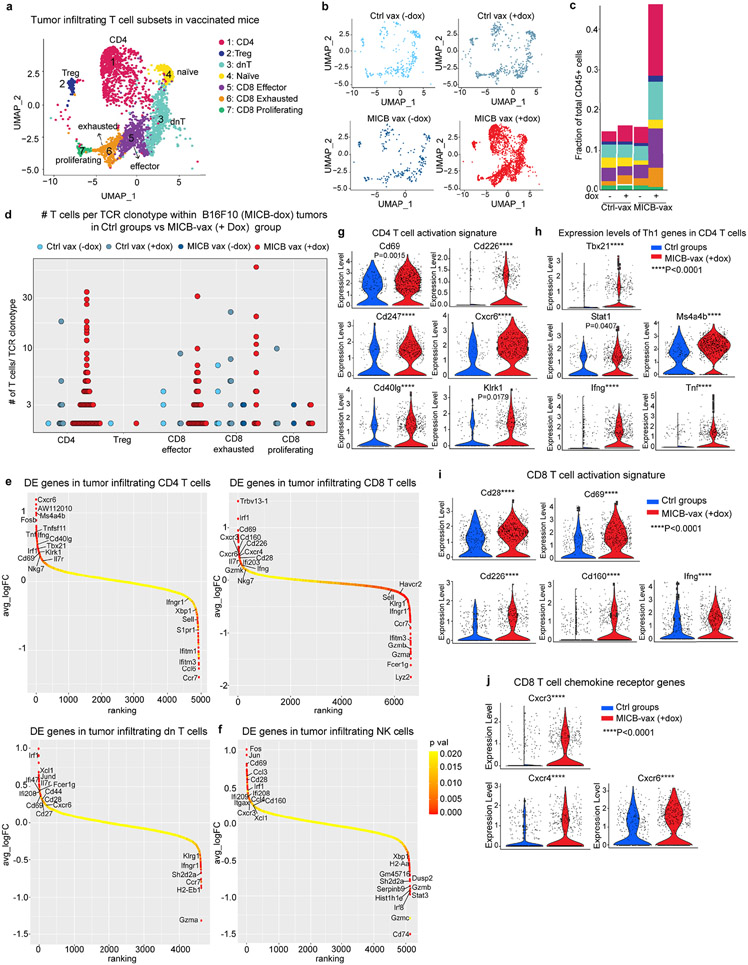
Single-cell RNA sequencing (scRNA-seq) analysis demonstrated marked differences in the composition of tumour-infiltrating immune cells between the experimental group (MICB-vax + doxycycline) and three control groups (Ctrl-vax ± doxycycline, MICB-vax without doxycycline). While myeloid cells dominated the immune infiltrate in all control groups, the majority of CD45+ immune cells in the experimental group receiving MICB-vax + doxycycline were T cells and NK cells (Extended Data Figs. 6e-g and 7a-c, and Fig. 2h-l). T cell receptor (TCR) sequence analysis provided evidence for substantial clonal expansion of both CD4+ and CD8+ T cell populations in the experimental group (Fig. 2l and Extended Data Fig. 7d). CD4+ T cells in the experimental group showed significant upregulation of activation-related genes, including Cd69, Cd226, Cd40lg and Klrk1; a similar activation signature was also observed for CD8+ T cells. CD4+ T cells also had a strong T helper type 1 signature with significantly higher expression of Tbx21 and Ifng in the experimental group than in the control groups (Extended Data Fig. 7e-j).
Chemokine receptors and their ligands are critical for recruitment of T cells and NK cell populations to tumours. Both CD8+ T cells and NK cells showed increased expression of the Cxcr3 chemokine receptor and CD4+ and CD8+ T cells had higher levels of Cxcr6 in the group receiving MICB-vax + doxycycline compared with the control groups (Extended Data Fig. 7e, f, j). Three clusters of group 1 innate lymphoid cells (ILCs) were identified: ILC1s, Xcl1+ NK cells and cytotoxic NK cells (Fig. 2j). Cells in all three of these clusters were enriched in the experimental group compared with the control groups, with Xcl1+ NK cells corresponding to the predominant cluster in the experimental group (Fig. 2k). XCL1 is important for recruiting XCR1+ conventional DC type 1 (cDC1) cells to tumours29, and the corresponding gene was significantly upregulated in NK cells and double-negative T cells in the experimental group compared with the control groups (Extended Data Fig. 7e, f). These data demonstrate that MICB-vax induced efficient recruitment of multiple effector T cell and NK cell populations to tumours that were in a highly activated state.
Immunity against resistant tumours
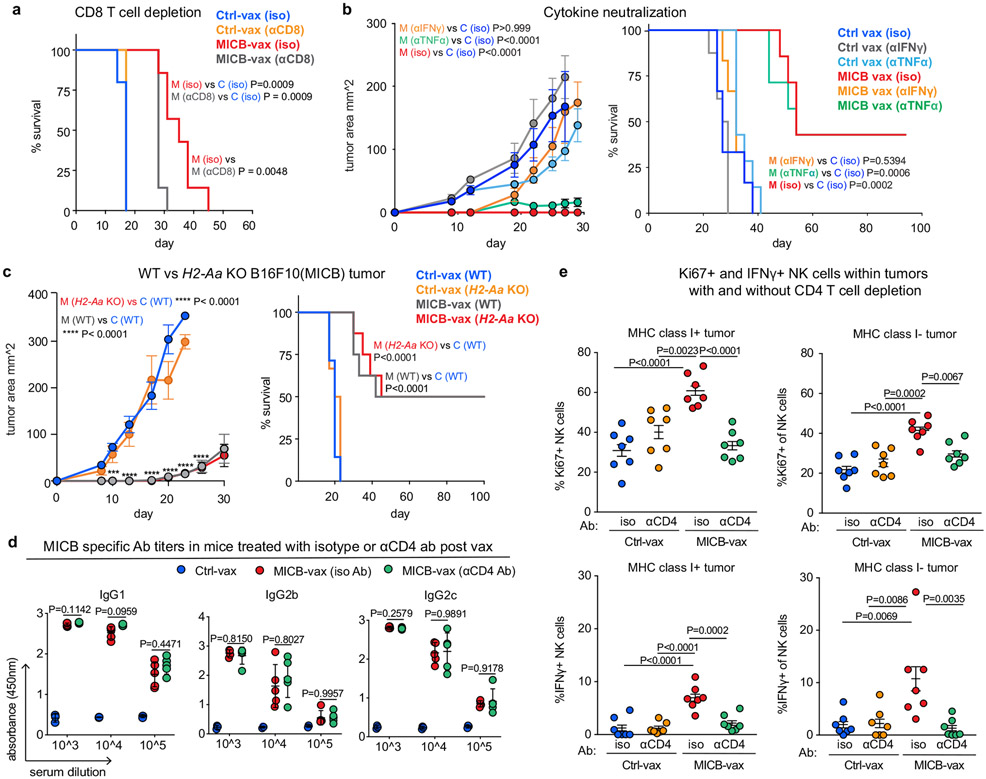
Cell depletion experiments demonstrated that CD4+ T cells were critical for the efficacy of the MICB vaccine; depletion of CD8+ T cells or NK cells as well as NKG2D receptor blockade also reduced the survival advantage conferred by MICB-vax (Fig. 2m and Extended Data Fig. 8a). Neutralization of IFNγ abrogated the protective effect of the MICB vaccine against B16F10 (MICB) tumours (Extended Data Fig. 8b). Thus, multiple effector cell populations and effector mechanisms substantially contributed to the efficacy of the MICB vaccine.
Resistance to current immunotherapies is frequently mediated by inactivating mutations (or downregulation) of genes in the MHC-I antigen presentation or IFNγ signalling pathways that greatly impair CD8+ T cell-mediated tumour immunity30-32. We theorized that MICB-vax could be effective against such resistant tumours because it induces the recruitment of multiple effector cell populations, including substantial numbers of NK cells. Indeed, we found that MICB-vax was efficacious against B16F10 (MICB) tumours with mutations resulting in loss of MHC-I (B2m−/−), MHC-II (H2-Aa−/−) or IFNγ receptor (Ifngr1−/−) expression, with 50–75% of mice remaining tumour free beyond day 100 (Fig. 3a, b and Extended Data Fig. 8c). Cellular depletion experiments demonstrated a requirement for both CD4+ T cells and NK cells for effective control of B2m−/− tumours in mice immunized with MICB-vax (Fig. 3c). These mice already had high titres of anti-MICB antibodies when the CD4+ T cell-depleting monoclonal antibody was administered, and antibody titres were unaffected by CD4+ T cell depletion (Extended Data Fig. 8d). Purified serum IgG from mice immunized with MICB-vax enabled strong NK cell-mediated killing of B2m−/− B16F10 (MICB) tumours in vitro (Fig. 3d). Both B2m−/− B16F10 (MICB) and B16F10 (MICB) tumours showed significantly increased accumulation of NK cells, including IFNγ+ and Ki67+ cells, in mice immunized with MICB-vax relative to those receiving Ctrl-vax. CD4+ T cell depletion resulted in a marked reduction in intratumoral NK cell numbers (Fig. 3e and Extended Data Fig. 8e). These data demonstrated that the efficacy of the MICB vaccine against resistant tumours (B2m−/− and Ifngr1−/−) was mediated by both NK cells and CD4+ T cells and that CD4+ T cells had an essential role in NK cell recruitment to tumours.
Fig. 3 ∣. Vaccine retains efficacy against MHC-I-deficient tumours.
a, b, Comparison of vaccine efficacy against B16F10 (MICB) WT tumours and tumours with resistance mutations in the B2m (a) or Ifngr1 (b) gene. Mice received MICB-vax or Ctrl-vax and were then challenged with tumours of the indicated genotype (n = 7 mice per group). KO, knockout. c, Effect of CD4+ T cell and NK cell depletion on immunity to B2m-knockout tumours. Mice were immunized with MICB-vax or Ctrl-vax; treatment with depleting or isotype-control monoclonal antibody was started 2 d before injection of B2m-knockout B16F10 (MICB) tumour cells (n = 7 mice per group). d, Contribution of vaccine-induced anti-MICB antibodies to NK cell-mediated cytotoxicity against B2m-knockout B16F10 (MICB) tumour cells. CFSE-labelled B2m-knockout B16F10 (MICB) tumour cells were pre-incubated with 10 μg per well of purified serum IgG from mice immunized with MICB-vax or Ctrl-vax before the addition of NK cells at different effector to target (E:T) ratios as indicated. The percentage of dead target cells was assessed by flow cytometry. e, Effect of CD4+ T cell depletion on vaccine-induced NK cell infiltration into tumours. Flow cytometry quantification of total NK cell numbers is shown in WT (left) and B2m-knockout (right) tumours for the following treatment groups: Ctrl-vax+isotype-control monoclonal antibody (blue), Ctrl-vax + anti-CD4 (orange), MICB-vax+isotype-control monoclonal antibody (red) and MICB-vax + anti-CD4 (green) (n = 7 mice per group). Representative data from two independent experiments are shown in a–e. Statistical significance was assessed by two-way ANOVA with Bonferroni’s post hoc test (left) and log-rank (Mantel–Cox) test (right) (a–c), two-way ANOVA with Sidak’s multiple-comparison test (d) and one-way ANOVA with Tukey’s multiple-comparison test (e). Data are depicted as the mean±s.e.m.
Mechanisms of NK cell recruitment
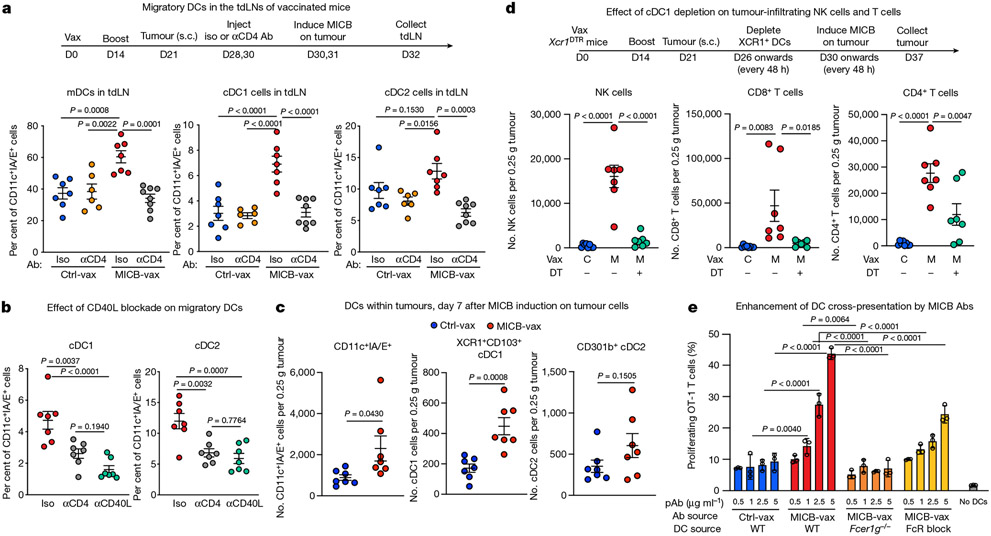
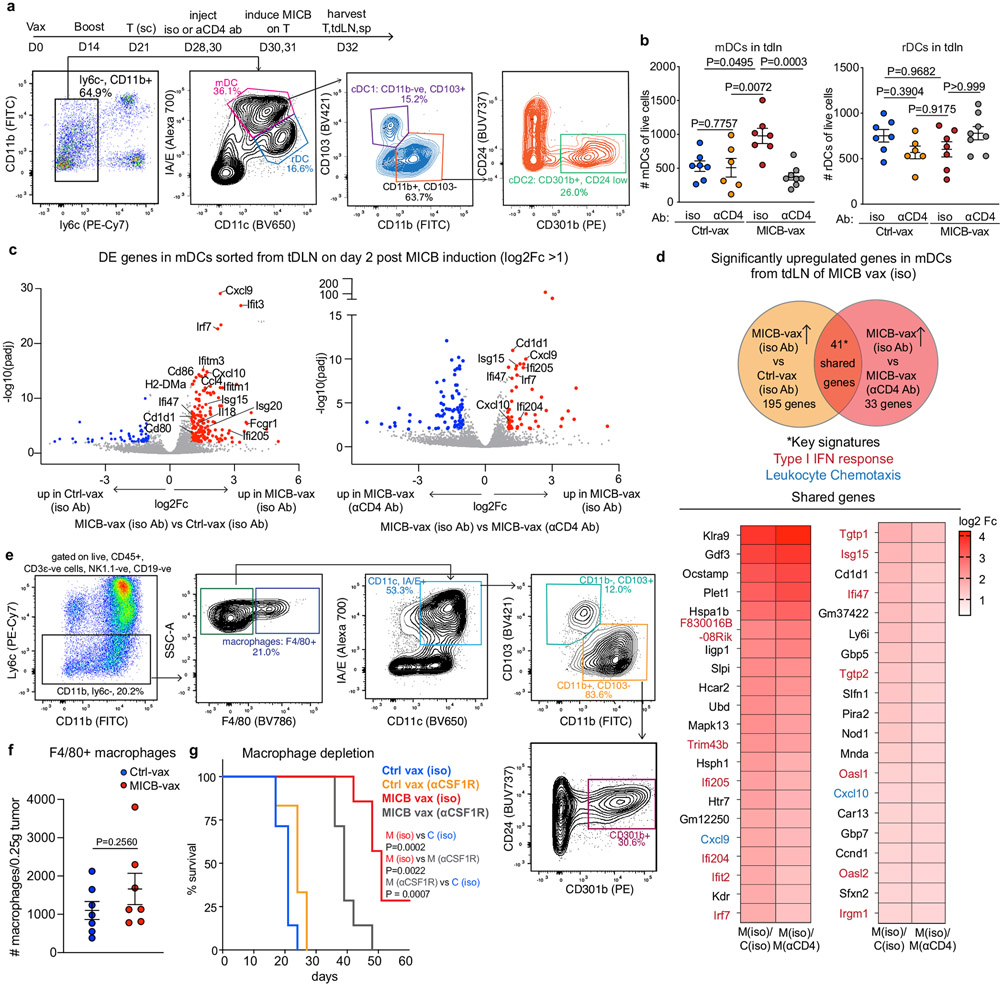
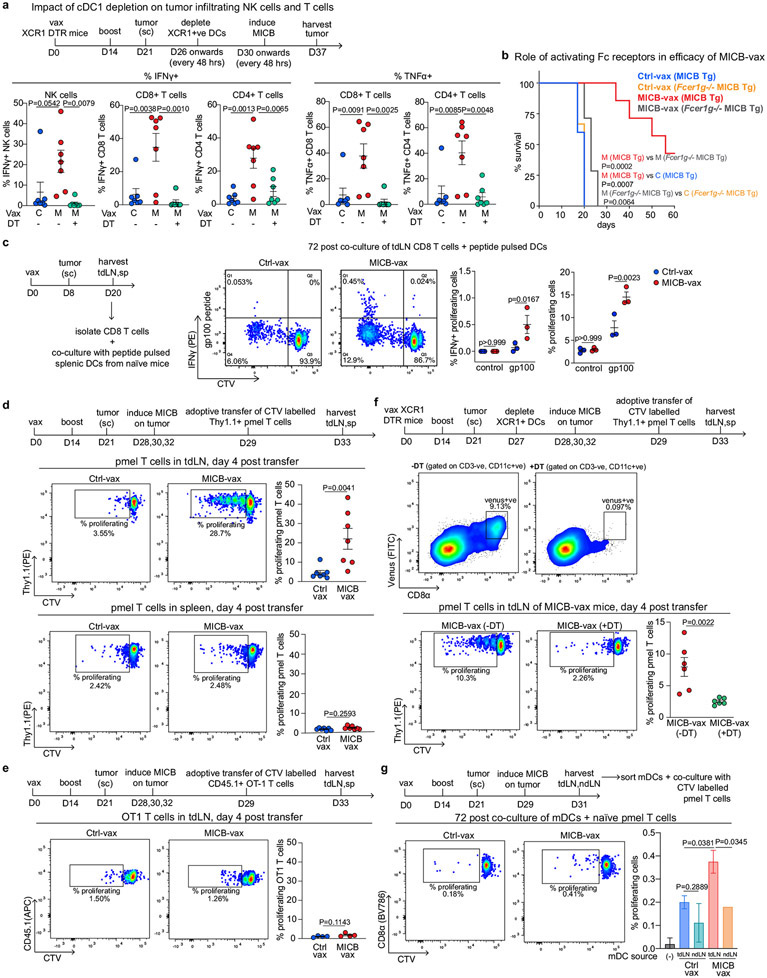
We investigated the mechanisms by which CD4+ T cells enabled recruitment of NK cells and other effector populations to tumours. cDCs (cDC1 and cDC2) have an important role in transporting tumour antigens to tumour draining lymph nodes (tdLNs), where they prime anti-tumour T cells33-35. cDC1 cells are particularly important for priming of CD8+ and CD4+ T cells with cell-associated antigens34-37. We observed a significant increase in the numbers of migratory cDC1 and cDC2 cells in the tdLNs of mice immunized with MICB-vax relative to those receiving Ctrl-vax at 48 h following doxycycline-induced expression of MICB on tumour cells (Fig. 4a and Extended Data Fig. 9a, b). This increase in migratory cDCs was abrogated by CD4+ T cell depletion or CD40 ligand (CD40L) blockade (Fig. 4a, b). RNA-seq analysis of migratory DCs sorted from the tdLN showed significant upregulation of interferon response genes (Isg15, Ifi47 and Irf7), lymphocyte chemotaxis genes (Cxcl9, Cxcl10 and Ccl4) and genes associated with lymphocyte activation (Cd80, Cd86 and Il18) in mice immunized with MICB-vax compared with those receiving Ctrl-vax (Extended Data Fig. 9c, d). Increased numbers of cDC1 cells were also observed in the tumours of mice immunized with MICB-vax compared with control mice, but macrophage numbers were not different (Fig. 4c and Extended Data Fig. 9e-g).
Fig. 4 ∣. Role of CD4+ T cells and cDC1 cells in NK cell recruitment to tumours.
a, Effect of CD4+ T cells on migratory DC populations in the tdLNs of mice immunized with MICB-vax versus Ctrl-vax. Total migratory DCs as well as cDC1 and cDC2 cells were quantified 2 d after induction of MICB expression in tumour cells by doxycycline treatment (n = 7 mice per group, except n = 6 for Ctrl-vax without anti-CD4). b, Migratory DC subsets within the tdLN of MICB-vax-immunized mice treated following immunization (days 28 + 30) with isotype-control, CD4-depleting or CD40L-blocking monoclonal antibody (n = 7 mice per group). c, Quantification of DC populations within the tumours of mice immunized with Ctrl-vax (blue) or MICB-vax (red) on day 7 following induction of MICB expression with doxycycline (n = 7 mice per group). d, Effect of cDC1 depletion on MICB vaccine-induced T cell and NK cell accumulation within tumours in Xcr1DTR mice. Mice were treated with DT or left untreated starting on day 26 following immunization with Ctrl-vax or MICB-vax (days 0 + 14) and B16F10 (MICB-dox) tumour implantation (day 21). Immune cells were analysed in tumours 7 d after induction of MICB expression on tumours with doxycycline (day 37) (n = 7 mice per group). e, Contribution of vaccine-induced anti-MICB antibodies to DC-mediated cross-presentation of tumour antigens to CD8+ T cells. Bone marrow-derived DCs (BMDCs) were pre-incubated with B2m-knockout B16F10 (MICB-OVA) tumour cells in the presence of affinity-purified serum IgG from mice immunized with Ctrl-vax or MICB-vax at the indicated concentrations. DCs were co-cultured with CFSE-labelled OT-1 CD8+ T cells with T cell proliferation as the readout. The role of activating Fc receptor (FcR) was assessed using BMDCs from Fcer1g−/− mice (orange) or pre-incubation of BMDCs with FcR-blocking antibody (yellow) before tumour cell addition. Representative data from two independent experiments are shown in a–e. Statistical significance was assessed by one-way ANOVA with Tukey’s multiple-comparison test (a, b, d), two-tailed Mann–Whitney test (c) and two-way ANOVA with Tukey’s multiple-comparison test (e). Data are depicted as the mean ± s.e.m. (a–d) or mean ± s.d. (e).
Many studies have highlighted the importance of cDC1 cells for recruitment of CD8+ T cells to tumours38-41. We assessed the contribution of cDC1 cells to NK cell recruitment to tumours using Xcr1DTR mice in which cDC1 cells could be specifically depleted by diphtheria toxin (DT) administration41. cDC1 depletion substantially decreased the number of intratumoral NK cells, CD8+ T cells and CD4+ T cells in mice immunized with MICB-vax (Fig. 4d). Similarly, the percentages of tumour-infiltrating IFNγ+ NK cells, CD8+ T cells and CD4+ T cells were substantially reduced following cDC1 depletion in mice immunized with MICB-vax (Extended Data Fig. 10a). Thus, MICB-vax induced cDC1 migration to tdLNs by a CD4+ T cell-dependent mechanism, and cDC1 cells were required for accumulation of NK cells and CD8+ T cells in tumours of mice immunized with MICB-vax.
We next investigated whether vaccine-induced anti-MICB antibodies could enhance the function of DCs. DCs were co-cultured with irradiated B16F10 (Ova) tumour cells with a B2m mutation such that they could not directly present the antigen to OT-1 CD8+ T cells. Addition of purified serum IgG from mice immunized with MICB-vax compared with Ctrl-vax substantially enhanced CD8+ T cell proliferation in these co-cultures. Enhanced DC function in the presence of anti-MICB antibodies was dependent on activation of Fc receptors on DCs, as shown in co-culture experiments with Fcer1g-knockout DCs (Fig. 4e). Additionally, the efficacy of MICB-vax was greatly diminished in Fcer1g-knockout mice even though MICB-specific antibody titres were not affected (Extended Data Fig. 10b).
These results suggested that MICB-vax could enhance T cell responses against melanoma tumour antigens not encoded by the MICB vaccine. CD8+ T cells from MICB-vax-immunized mice with B16F10 (MICB) melanomas showed a stronger proliferative response to the gp100 melanoma peptide not included in the vaccine than CD8+ T cells from mice receiving Ctrl-vax (Extended Data Fig. 10c). Furthermore, pmel-1 T cells specific for the gp100 melanoma antigen proliferated robustly in the tdLNs of mice immunized with MICB-vax but not Ctrl-vax, in line with increased presentation of this melanoma antigen in the tdLNs of MICB-vax-immunized mice (Extended Data Fig. 10d, e). Depletion of cDC1 cells in Xcr1DTR mice41 just before transfer of pmel-1 T cells greatly reduced the proliferation of the transferred T cells in the tdLNs of MICB-vax-immunized mice (Extended Data Fig. 10f). Finally, migratory DCs from the tdLNs of mice immunized with MICB-vax but not Ctrl-vax induced proliferation of naive pmel-1 CD8+ T cells in vitro in the absence of exogenous antigen, demonstrating enhanced in vivo uptake of this melanoma antigen by DCs in MICB-vax-immunized mice (Extended Data Fig. 10g).
Conclusions
The presented vaccine design enabled tumour immunity by multiple effector cell populations, including diverse T cell and NK cell populations. Vaccine-induced antibodies inhibited proteolytic MICA/B shedding by tumour cells, enhanced the cytotoxic function of NK cells and increased cDC1-mediated cross-presentation of tumour antigens to CD8+ T cells. The vaccine also induced CD4+ and CD8+ T cell responses against MICA/B as well as epitope spreading to melanoma antigens not included in the vaccine. Engagement of multiple effector cell populations enabled induction of protective immunity against tumours with common escape mechanisms, including loss of MHC-I expression or IFNγ receptor signalling by tumour cells. Vaccine-induced immunity against MHC-I-deficient tumours was mediated by the coordinated action of CD4+ T cells and NK cells. A limitation of this work was that human MICA/B proteins had to be expressed in mouse tumour cells because mouse NKG2D ligands have only limited homology to MICA/B. This approach will require evaluation in a clinical trial in patients with cancer, and a first-in-human clinical trial is being planned. Serum levels of shed MICA/B and tumour cell expression of MICA/B will serve as major biomarkers for patient selection. This vaccine may also be of interest in combination with local radiation therapy because DNA damage enhances MICA/B expression by cancer cells2.
Methods
Mice
All mice were maintained under specific pathogen-free conditions, and experiments were performed in compliance with federal laws and were approved by the Dana-Farber Cancer Institute Institutional Animal Care and Use Committee (IACUC, protocol 04-103). Transgenic mice that expressed MICB under the control of the prostate-specific Pbsn promoter were generated as previously reported14. The cDNA for the protein-coding sequence of the full-length human MICB*005:02 allele was cloned downstream of the minimal rat Pbsn promoter (−426 bp to +28 bp), followed by an IRES and a luciferase cDNA sequence. This MICB-IRES insert was synthesized by Genscript and cloned into the pGL4.20 Luc2/Puro vector using the 5′ NheI and 3′ XhoI restriction sites. The expression cassette (rPB-MICB-IRES-luciferase-SV40 poly(A)) was excised with NheI and BamHI restriction enzymes, purified by agarose gel electrophoresis and recovered using the QIAEXII Gel Extraction kit (Qiagen). The linearized transgene construct was microinjected into fertilized C57BL/6J embryos by the Dana-Farber/Harvard Cancer Center Mouse Engineering Core Facility. Transgenic founders were identified by PCR analysis of genomic DNA extracted from tail biopsies using 5′-ACAAGTGCATTTAGCCTCTCCAGTA-3′ forward and 5′-TGTGTCTTGGTCTTCATGGC-3′ reverse primers. RNA expression of MICB and luciferase reporter genes in the prostate was confirmed by real-time qPCR. Total RNA was extracted using the RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. Extracted RNA (1 μg) was transcribed into cDNA using SuperScript IV VILO master mix with ezDNase enzyme according to the manufacturer’s protocol (ThermoFisher). The cDNA samples were diluted and used for real-time qPCR. Powerup SYBR Green master mix (ThermoFisher) and gene-specific primers were used for PCR amplification and detection using a Quant-Studio 6 Flex Real-Time PCR System (ThermoFisher). The real-time qPCR data were normalized to mouse Hprt (a housekeeping gene) and are presented as the fold change in gene expression in transgenic mice compared with WT C57BL/6J mice.
Homozygous breeding pairs were generated by breeding MICB-transgenic male and female mice. MICB-heterozygous males used for the vaccination studies were generated by crossing MICB-homozygous males with C57BL/6J females. C57BL/6J (B6, stock 000664), B6.SJL-Ptprca Pepcb/BoyJ (B6 Cd45.1, stock 002014), BALB/cJ (stock 000651), C57BL/6-Tg(TcraTcrb)1100Mjb/J (OT-1; stock 003831), B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J (pmel-1; stock 005023) and B6;129P2-Fcer1gtm1Rav/J (Fcer1g−/−; stock 002847) mice were purchased from the Jackson Laboratory. B6.Cg-Xcr1<tm2(HBEGF/Venus)Ksho>(Xcr1DTR) mice were previously reported41 and obtained from the RIKEN BioResource Research Center, Japan. OT-1, pmel-1, Fcer1g−/− and Xcr1DTR mice were all maintained as homozygous breeding pairs. Fcer1g−/− mice were crossed to B6 mice for six generations to generate Fcer1g−/− mice on the B6 background. Fcer1g−/− MICB-transgenic mice were generated and maintained by crossing Fcer1g−/− homozygous females with MICB-transgenic males. Strain-specific genotyping primers standardized by Jackson Laboratory was used for genotyping. Mice were age and sex matched for all experiments, and mice between 8 and 10 weeks of age were used for all experiments. Sample size was determined on the basis of our previous studies and/or pilot studies. For therapeutic and spontaneous metastasis experiments, mice were assigned into two groups prior to immunization on the basis of tumour size to have an equal distribution of tumour sizes across both experimental and control groups. No formal blinding was used for in vivo experiments, but various steps in key studies were performed by different investigators, who were each independent of the steps performed by the other investigator.
Cell lines
B16F10, 4T1, EL4, A375, A549, HCT116, K562 and HEK293T cell lines were purchased from the American Type Culture Collection (ATCC). Sf9 cells were purchased from Invitrogen. The B16-BL6 cell line was obtained from the National Institutes of Health. Cell culture media and supplements were purchased from Gibco. B16F10, EL4 and HEK293T cells were grown in DMEM supplemented with 10% (vol/vol) FBS, 1× GlutaMax and 100 U ml−1 penicillin-streptomycin. 4T1, A375 and B16-BL6 cells were grown in RPMI 1640 supplemented with 10% (vol/vol) FBS, 1× GlutaMax and 100 U ml−1 penicillin-streptomycin. The K562 cell line was grown in IMDM supplemented with 10% (vol/vol) FBS, 1× GlutaMax and 100 U ml−1 penicillin-streptomycin All primary cells were grown in RPMI 1640 supplemented with 10% (vol/vol) FBS, 1× GlutaMax, 100 U ml−1 penicillin-streptomycin, 20 mM HEPES, 1mM sodium pyruvate and 50 μM 2-mercaptoethanol. All mammalian cells were cultured at 37 °C in a humidified incubator with 5% CO2. Sf9 cells were grown in Sf900 II serum-free medium (Invitrogen) on an orbital shaker platform (125–150 r.p.m.) at 28°C in a non-humidified incubator without CO2. ATCC cell lines were authenticated using STR profiling by ATCC. None of the commonly misidentified cell lines were used in this study. Parental lines and their derivatives were confirmed to be mycoplasma negative using the ATCC Universal Mycoplasma Detection kit.
The B16F10 MICA*009 cell line was generated as described previously25. B16F10, B16-BL6, 4T1 and EL4 cell lines overexpressing human MICB*005:02 were generated using a GeneHero Mouse Rosa26 Safe-Harbour Knock-in kit (SH054, Genecopoeia). The cell lines were co-transfected with (1) Rosa26 donor plasmid (SH150, Genecopoeia) containing human MICB*005:02 cDNA (synthesized by IDT) under the control of the mPGK promoter and (2) a Rosa26 sgRNA/CRISPR–Cas9 plasmid with an mCherry reporter (SH250, Genecopoeia) at a 1:1 ratio using Lipofectamine 3000 transfection reagent (Invitrogen). One week after transfection, MICB-positive cells were single-cell sorted into 96-well plates using a FACSAria IIIu (BD Biosciences); mCherry-positive cells were excluded from the sorted cells to avoid potential immunogenicity due to Cas9 expression. Single-cell clones were grown for 2–3 weeks, genomic DNA was extracted and integration of the donor plasmid at the Rosa26 locus was confirmed by performing junction PCR using forward and reverse primers that bound at the SV40 poly(A) region of the plasmid and the 3′ Rosa26 homology arm, respectively. To generate a metastatic derivative of B16-BL6 (MICB) cells, C57BL/6J mice were injected subcutaneously with B16-BL6 (MICB) cells and the whole tumour mass was surgically removed on day 21, leaving a clean margin without residual tumour cells. Thirty days after surgical removal of the primary tumour, lungs were excised and metastatic clones colonizing the lungs were carefully picked, dissociated into a single-cell suspension and grown in complete RPMI for 2 weeks before validating MICB expression by these cells by flow cytometry using a BD LSRFortessa X-20 instrument (BD Biosciences).
The doxycycline-inducible B16F10 (MICB-dox) cell line was generated using Lenti-X Tet-ON 3G inducible expression vectors (631363, Clontech). Human MICB*005:02 cDNA (synthesized by IDT) was cloned into the pLVX-Tre3G plasmid using the BamHI and MluI restriction sites. Separate regulator (pLVX-pEF1a-Tet3G) and response (pLVX-Tre3G-MICB) lentiviral supernatants were produced using HEK293T cells and Lenti-X Packaging Single Shots (631277, Clontech) according to the manufacturer’s protocol. Supernatants containing viral particles were collected 48 h after transfection, centrifuged at 300g for 5 min to remove cellular debris and filtered through 0.45-μm sterile syringe filters (Millipore). Viral particles were concentrated by centrifuging the supernatants at 112,000g for 30 min at 4 °C. B16F10 cells were co-transduced with pLVX-pEF1a-Tet3G and pLVX-Tre3G-MICB lentiviral stocks combined at a a 1:1 ratio in the presence of polybrene (4 μg ml−1). Virus-containing medium was removed after 8 h and replaced with fresh growth medium containing doxycycline (25 ng ml−1). MICB-expressing cells were double sorted 1 week after transduction using a BD FACSAria IIIu instrument. The B16F10 cell line co-transduced with pLVX-pEF1a-Tet3G and empty pLVX-Tre3G vector was used as a negative control.
B16F10 (MICB) B2m−/−, H2-Aa−/− and Ifngr1−/− tumour cells were generated by electroporation with ribonucleoprotein (RNP) complexes consisting of Cas9 protein with bound guide RNAs (gRNAs). Respective gRNAs (B2m−/−, 5-′ATTTGGATTTCAATGTGAGG-3′; H2-Aa−/−, 5′-CTTGCCAATTGGCCAAACTC-3′; Ifngr1−/−, 5′-GGTATTCCCAGCATACGACA-3′) (20 μM) were mixed with Cas9 protein (Cas9-NLS, University of California, Berkeley) at an equimolar ratio. Gene editing was performed by electroporation of 2 × 105 tumour cells with RNP complexes using an SF Cell Line 96-Well Nucleofector kit (Lonza). Editing was validated by performing a genomic cleavage assay using T7 endonuclease I (NEB) following the manufacturer’s protocol. Control (lacz gRNA) and knockout B16F10 (MICB) cells were treated with 10 ng ml−1 IFNγ (for B2m−/− and Ifngr1−/− cells) or with 10 ng ml−1 IFNγ + 5 ng ml−1 TNFα (for H2-Aa−/− cells) for 24 h and then stained with APC-conjugated anti-mouse H2-Kb/H2-Db (clone 28-8-6, BioLegend), PE-conjugated anti-mouse IFNγ receptor β chain (clone MOB-47, BioLegend) or Alexa Fluor 488-conjugated anti-mouse IA/E (clone M5/144.15.2, BioLegend) antibodies. Cells were washed and analysed using a BD LSRFortessa X-20 instrument to confirm the absence of MHC-I expression on B2m−/− cells, MHC-I and IFNγ receptor expression on Ifngr1−/− cells and MHC-II expression on H2-Aa−/− cells.
Codon-optimized rhesus macaque MICA*01-T2A-eGFP and MICB*01-T2A-eGFP gBlocks were synthesized by IDT and cloned into the UCOE Hu-P vector (504867, EMD Millipore) containing a puromycin selection cassette. HEK293T cells were transfected with the expression plasmids using Lipofectamine 3000 (Invitrogen) transfection reagent. Cells were grown in medium containing 1 μg ml−1 puromycin to select for transfected cells and sorted for purity on the basis of eGFP expression using a FACSAria IIIu instrument (BD Biosciences).
Protein production and purification
Proteins used for immunization were produced in Expi293F cells using cDNA constructs that were codon optimized for mammalian expression and synthesized by Genscript. A point mutation (Asn19Gly) was introduced into the ferritin cDNA from H. pylori to remove a site for N-linked glycosylation. All constructs contained an N-terminal signal peptide to target the protein to the secretory pathway, a Ser-Lys-Ser linker and an HA epitope tag for downstream protein purification. The α3 domain encoded by human MICA*009 or MICB*005 was linked to the N terminus of ferritin using a Gly-Ser-Gly linker. The rhesus macaque constructs contained linked MICA*01 and MICB*01 α3 domains, again attached to ferritin with a Gly-Ser-Gly linker. A ferritin construct without linked MICA or MICB α3 domain was generated as a control. The constructs were cloned into the UCOE Hu-P expression vector and transfected into Expi293F mammalian cells following the manufacturer’s protocol (ThermoFisher Scientific). Proteins were produced with transiently transfected cells; alternatively, stable cell lines were generated using puromycin selection (50 μg ml−1 for initial selection and 10 μg ml−1 after scaling up). Supernatants were collected 4 d after transfection (or 4 d after scaling up in the case of stable lines), centrifuged at 6,000 r.p.m. for 30 min and filtered using Nalgene rapid-flow disposable filter units with PES membrane (0.45 μm). Proteins were affinity purified using anti-HA affinity columns, eluted with 50 mM CAPS buffer (pH 11.5) and neutralized immediately by addition of 2 M Tris, pH 7.5. Eluted proteins were buffer exchanged into PBS and concentrated to 2 mg ml−1 using Amicon Ultra-15 centrifugal filters (EMD Millipore) with a 30-kDa molecular weight cut-off (MWCO). Concentrated proteins were purified further by HPLC size-exclusion chromatography using the following columns: Superose 6 Increase 10/300 GL (for human MICA α3–ferritin and MICB α3–ferritin), Superdex 200 (for ferritin) or tandem Superose 6 Increase 10/300 GL (for macaque MICA/B α3–ferritin) (GE Health-care) with PBS (fusion proteins) or HBS (ferritin) as running buffer at a flow rate of 0.8 ml min−1. Proteins were concentrated to 1 mg ml−1 using Amicon Ultra-15 centrifugal filter units with a 30-kDa MWCO and stored at −80 °C until use.
Proteins for measuring anti-MICA/B antibody titres were produced in insect cells using the baculovirus system. Separate constructs were generated for the full-length extracellular domains encoded by human MICA*009, human MICB*005, rhesus macaque MICA*01 and rhesus macaque MICB*01. These constructs contained an N-terminal signal peptide followed by a Ser-Lys-Ser linker and a PC tag for protein purification. Codon-optimized cDNAs were synthesized and cloned into the pAcDB3 baculovirus expression vector, and proteins were expressed in Sf9 cells infected with recombinant baculovirus at a multiplicity of infection of 10 (BestBac 2.0, v-cath/chiA Deleted Baculovirus Cotransfection Kit, Expression Systems). Supernatants were collected 3 d later, clarified by centrifugation at 6,000 r.p.m. for 30 min and buffer exchanged into 100 mM Tris (pH 7.5) and 150 mM NaCl. Proteins were purified using anti-Protein C affinity columns, followed by elution and size-exclusion chromatography using a Superose 6 column (GE Healthcare) as described above. Protein purity was assessed using 4–12% Novex Tris-glycine gels (ThermoFisher Scientific) stained with Coomassie Brilliant Blue.
Mouse vaccine formulation
MSRs were fabricated as described previously23. In brief, 4g of P123 surfactant (average Mn of about 5,800; Sigma-Aldrich) were dissolved in 150 ml of 1.6 M HCl solution and stirred with 8.6 g of tetraethyl-orthosilicate (TEOS, 98%; Sigma-Aldrich) at 40 °C for 20 h, followed by ageing at 100 °C for 24 h. TEOS was extracted in 1% ethanol in HCl at 80 °C for 18 h. To prepare the MSR vaccines, 2 mg of the MSR was adsorbed with 100 μg of murine class B CpG ODN 1826 (sequence 5′-TCCATGACGTTCCTGACGTT-3′; IDT) and 2 mg of MSR was loaded with an equimolar amount of ferritin (control, 120 μg) or 200 μg MICB–ferritin protein. Loading was carried out for 6h at room temperature under shaking, and MSRs were subsequently lyophilized. Separately, 1 mg of the MSRs was loaded with mouse GM-CSF (Peprotech) for 1 h at 37 °C under shaking. MSR vaccines for subsequent boosts were prepared by halving the amounts of protein, CpG and GM-CSF adsorbed to the MSR. The MSRs were combined and resuspended in 150 μl cold PBS before immunization and boost. MSR vaccines were injected via an 18-gauge needle, subcutaneously in the flank region with the mouse under brief isoflurane anaesthesia.
Rhesus macaque vaccine formulation and immunization studies
Macaque MICA/B α3–ferritin protein was conjugated to 5′-amine-modified CpG-2396 (5′-NH2-CpG, 5′-TCGTCGTTTTCGGCGCGCGCCG-3′) using a Solulink Protein-Oligo Conjugation kit (S-9011, Trilink Biotechnologies) following the manufacturer’s protocol. In brief, CpG oligonucleotide and MICA/B α3–ferritin protein were desalted and modified with S-4B cross-linker and S-HyNic, respectively. HyNic–MICA/B α3–ferritin protein and 4FB–CpG were mixed at a 1:10 molar ratio for 1 h, resulting in the formation of a stable bis-aryl hydrazone bond. Excess unreacted oligonucleotide in the reaction mixture was removed by size-exclusion chromatography using a Superose 6 Increase 10/300 GL column. The protein–oligonucleotide conjugate was concentrated to 1 mg ml−1 using Amicon Ultra-15 centrifugal filters with a 30-kDa MWCO. Absorbance was measured at 354 nm using a DS-11 spectrophotometer/fluorometer (Denovix), and the molar substitution ratio of CpG to MICA/B–ferritin protein was calculated using the manufacturer’s protocol.
Rhesus macaque immunization studies were performed at the Yerkes National Primate Research Center (YNPRC). YNRPC’s animal care facilities are accredited by the US Department of Agriculture (USDA) and by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All animal procedures were performed in line with institutional regulations and guidelines set forth by the NIH’s Guide for the Care and Use of Laboratory Animals, 8th edition. Macaque immunization experiments were reviewed and approved by Emory University’s IACUC (protocol number PROTO201700713). Animals assigned to the study were requested to be more than 4 years old without preference for sex or MHC haplotype. The assigned macaques were all males between 5 and 14 years of age.
For each macaque to be immunized, 500 μg of MICA/B α3–ferritin protein conjugated to 25 μg CpG-2395 (1 mg ml−1 in PBS) was mixed with 10 μg of rhesus macaque GM-CSF (10 μg μl−1; Cell Sciences). Vaccines were prepared by mixing 500 μl of GM-CSF-loaded protein–CpG conjugate with 500 μl of Addavax (squalene in oil emulsion; Invivogen) and injected intramuscularly using a 21-gauge needle. Macaques received a total of four injections, spaced 3 weeks apart. Serum samples were collected 2 d before initial immunization and 3 weeks after each immunization or boost. Complete blood count and blood chemistry were measured at the YNPRC from whole blood samples collected 2 d before initial immunization and 3 weeks after the final boost. These animals had been enrolled in prior vaccine and treatment studies (with interim rest periods), and prior treatment history could have been a variable affecting anti-MICA/B antibody titres in individual animals.
Measurement of antibody titres by ELISA or DELFIA
Serum samples collected from immunized mice (on days–1, 7, 14 and 28) by retro-orbital bleed were processed and stored at −80 °C until analysis. Nunc Maxisorp 96-well plates (Thermo Scientific) were coated with 100 μl of PC-tagged human MICA or MICB extracellular domain (2 μg ml−1 in PBS) by incubating plates overnight at 4 °C. All incubation steps for ELISAs were performed on a rocker platform with gentle agitation. Following incubation, wells were washed twice with 300 μl wash buffer (0.05% Tween-20 in PBS) and blocked with 250 μl blocking buffer (2% BSA in PBS) for 2 h at room temperature. Serum samples serially diluted in PBS were added to the wells (100 μl per well) in technical duplicates or triplicates, and plates were incubated for 2 h at room temperature and washed three times; then, 100 μl of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgM, IgG1, IgG2b, IgG2c or IgG3 (cross-adsorbed antibodies; Southern Biotech) diluted 1:2,000 in 0.5% BSA in PBS was added to the wells and plates were incubated for 1 h at room temperature. The wells were washed three times and incubated with 100 μl of 1-Step Ultra TMB-ELISA substrate (ThermoFisher Scientific) for 10 min. The reaction was stopped by adding 100 μl of 1 N sulfuric acid, and plates were read at 450 nm using an EnVision Multimode plate reader (PerkinElmer).
Dissociation-enhanced lanthanide fluorescent immunoassay (DELFIA) was used to measure antibody titres in immunized rhesus macaques. DELFIA 96-well plates (PerkinElmer) were coated with 100 μl of PC-tagged macaque MICA or MICB extracellular domain (2 μg ml−1 in carbonate buffer, pH 9.4) and plates were incubated overnight at 4 °C. Following two washes, 300 μl DELFIA assay buffer (PerkinElmer) was added to wells followed by incubation for 2 h at room temperature. The wells were washed three times and incubated with serum samples as described above; then, 100 μl of a 1:10,000 dilution (in 0.5% BSA in PBS) of goat anti-monkey IgA/IgM/IgG–biotin (cross-adsorbed; Novus Biologicals) was added to wells and plates were incubated for 1 h at room temperature. Wells were washed four times and incubated with 100 μl streptavidin–europium substrate (PerkinElmer) diluted 1:2,000 in assay buffer. Following five washes, 100 μl europium enhancement solution was added to the wells and plates were incubated in the dark for 15 min at room temperature. Fluorescence was measured using TRF settings (excitation, 340 nm; emission, 615 nm) on an EnVision Multimode plate reader.
Measurement of antibody titres by flow cytometry
B16F10 cells expressing full-length human MICA or MICB were collected, counted and plated at a density of 2 × 105 cells per well in a 96-well V-bottom plate. All centrifugation steps were performed at 300g for 5 min. Cells were washed twice with PBS, pelleted and stained with 100 μl Zombie Green dead cell exclusion dye (diluted 1:800 in PBS; BioLegend) for 20 min at room temperature. Cells were washed with PBS containing 2% FBS and 2 mM EDTA (FACS buffer) and incubated with 100 μl of serum samples serially diluted in FACS buffer for 1 h at room temperature. Following two washes, cells were stained with 100 μl of Alexa Fluor 647-conjugated goat anti-mouse IgG (1 μg ml−1 in FACS buffer; BioLegend) for 30 min at room temperature. Samples were washed twice with FACS buffer. Samples were analysed using an LSRFortessa X-20 instrument (BD Biosciences) and FlowJo software. Sera from control immunized mice, parental cells without MICA or MICB expression and respective secondary antibodies were used as negative controls for flow cytometry.
A similar staining procedure was followed for assays using macaque serum samples with the following modifications. HEK293T cells expressing rhesus macaque MICA or MICB protein were stained with serum samples, and 100 μl of diluted (1 μg ml−1 in FACS buffer) PE-conjugated goat anti-rhesus macaque IgG (Southern Biotech) was used for detection.
Purification of serum IgG antibodies from immunized mice
Polyclonal serum IgG was purified using Pierce Protein A/G magnetic agarose beads (Thermo Scientific) following the manufacturer’s protocol. Following the elution and neutralization steps, IgG was buffer exchanged into PBS and concentrated to 1 mg ml−1 using 0.5-ml Amicon Ultracentrifugation filters with a 30-kDa MWCO.
Assays assessing stabilization of MICA/B surface proteins
Stabilization of MICA or MICB surface protein by serum antibodies was assessed following incubation for 24 h. Human A375, A549, HCT116 and K562 cells were seeded at a density of 5 × 104 cells per well in a 96-well flat-bottom plate, and 10 μl of serum samples from mice vaccinated with ferritin control or MICB α3–ferritin was added to the wells. The final volume in each well was adjusted to 200 μl, and plates were incubated at 37 °C in a humidified incubator with 5% CO2 for 24 h. Following incubation, plates were centrifuged at 300g for 5 min, and supernatants were collected for quantification of shed MICA and/or MICB by ELISA. Cells were detached using Versene (50 μl per well; Gibco), washed twice with 250 μl PBS and incubated briefly with human TrueStain FcX (BioLegend) before addition of PE-conjugated anti-human MICA/B antibody (6D4, BioLegend) or corresponding isotype-control antibody (1:100 dilution in FACS buffer). Cells were stained with antibody for 30 min at room temperature and washed twice with FACS buffer before data acquisition using an LSRFortessa X-20 flow cytometer.
The B16F10 (MICB-dox) cell line was used to assess stabilization of MICB protein on the surface of tumour cells in vivo. Mice vaccinated with ferritin control or MICB α3–ferritin were challenged subcutaneously with 0.3 × 106 B16F10 (MICB-dox) cells 14 d after immunization. MICB expression was induced on tumour cells by intraperitoneal injection of doxycycline (5 mg kg−1 in PBS) every 48 h. Serum samples were collected from mice before induction of MICB expression and 5 d after doxycycline treatment and frozen at −80 °C for quantification of shed MICB by ELISA. Tumours were collected 5 d after induction of MICB expression and dissociated into single cells by mechanical disruption using 70-μm cell strainers. Surface MICB protein levels on tumour cells were quantified by flow cytometry following staining with PE-conjugated anti-human MICB antibody (6D4, BioLegend) as described above. The 6D4 monoclonal antibody binds to the α1–α2 domains of MICA/B, and its binding is not inhibited by antibodies directed against the α3 domain (Extended Data Fig. 1l).
ELISA for quantification of shed MICA/B
Shed MICA/B levels in serum samples or cell culture supernatants were measured using the Human MICA ELISA (ab100592, Abcam) and Human MICB DuoSet ELISA (DY1599, R&D Systems) kits following the manufacturers’ protocols.
Binding of NKG2D–Fc fusion proteins to cell-surface MICA/B
These assays were performed to determine whether MICA/B α3-specific purified serum IgG antibodies interfered with human or mouse NKG2D binding to the MICA/B α1–α2 domains. Cells were pre-incubated with sera from mice immunized with Ctrl-vax, MICA-vax or MICB-vax, followed by staining of cells with NKG2D–Fc fusion protein. Control transduced B16F10 cells (negative control) and human MICA- or MICB-expressing B16F10 tumour cells (1 × 105) were pre-incubated with 10 μl of sera from immunized mice for 30 min at room temperature. Cells were then incubated with 100 μl recombinant human NKG2D–Fc chimera protein (10 μg ml−1 in FACS buffer; 1299-NK-050, R&D Systems) or with recombinant mouse NKG2D–Fc chimera protein (10 μg ml−1 in FACS buffer; 139-NK-050, R&D Systems) for 1 h on ice. Cells were washed twice with FACS buffer and stained with 100 μl Alexa Fluor 647-conjugated goat anti-human IgG cross-adsorbed secondary anti-body (Invitrogen, A-21445; 1:10,000 dilution in FACS buffer) for 30 min at room temperature. Cells were washed twice with FACS buffer, and NKG2D–Fc fusion protein binding was analysed using an LSRFortessa X-20 flow cytometer.
Prophylactic and therapeutic vaccine experiments using subcutaneous tumour models
Tumour cells were thawed and passaged twice before injection. Cells were detached using 0.05% trypsin-EDTA, washed twice with PBS, passed through 70-μm cell strainers and prepared at a density of 0.25 × 106 cells in 50 μl PBS. In a prophylactic setting, MICB-transgenic male mice were immunized with ferritin (control) or with MICB α3–ferritin or MICA α3–ferritin (experimental) vaccine; booster immunizations were administered on day 14 where indicated. Mice were injected subcutaneously with 0.25 × 106 MICA- or MICB-expressing tumour cells 21 d after the initial immunization. Tumour-free mice in the MICB α3–ferritin group were re-challenged with the same dose of tumour cells on the opposite flank 120 d after primary tumour challenge, and tumour growth was monitored. To determine the efficacy of the vaccine over a wide range of MICB expression levels, the doxycycline-inducible B16F10 (MICB-dox) cell line was used. Mice immunized (day 0) and boosted (day 14) with Ctrl-vax or MICB-vax were injected subcutaneously with 0.25 × 106 B16F10 (MICB-dox) tumour cells. MICB expression was induced on tumour cells by intraperitoneal injection of doxycycline (low dox group, 2.5 mg kg−1; medium dox group, 5 mg kg−1; high dox group, 10 mg kg−1) every 48 h once the tumours were palpable (day 4 after tumour cell inoculation).
In therapy experiments, MICB-transgenic males were injected subcutaneously with 0.25 × 106 MICA- or MICB-expressing tumour cells, followed by immunization with Ctrl-vax or MICB-vax when tumours were palpable, as indicated. Tumour size was measured every 3 d using digital callipers, and tumour area was calculated by measuring the longest length and width of the tumour. Mice were killed when the longest side of the tumour reached 18–20 mm or when the tumour area reached 350 mm2.
Vaccination after surgical removal of primary tumours
Primary tumours were surgically removed, and mice were then immunized to assess vaccine efficacy against metastatic disease. Two models of spontaneous metastasis were used, the B16-BL6 melanoma model and the 4T1 breast cancer model. For the B16-BL6 melanoma model, MICB-transgenic mice were injected subcutaneously with 0.3 × 106 B16-BL6 (MICB) cells in 50 μl PBS. Tumours were resected on day 16 when they reached ~250–300 mm2. Mice were placed under brief iso-flurane anaesthesia, and the incision site was closed using medical clips. Two days after surgical removal of the primary tumour, mice were immunized with Ctrl-vax or MICB-vax on the opposite flank; a booster vaccine was administered 10 d later. Mice were killed on day 55 after primary tumour challenge and lungs were collected, washed in PBS and fixed immediately in 10% neutral-buffered formalin solution. For the metastatic 4T1 breast cancer model, 0.4 × 106 4T1 (MICB) cells in 50 μl PBS were injected into the fourth mammary fat pad of BALB/cJ mice. Tumours were resected on day 18 when they reached about 200 mm2, and mice were immunized as described above. Mice were killed on day 50 after primary tumour inoculation, and lungs were collected, washed in PBS and placed immediately into Bouin’s fixative. Metastatic nodules in fixed lung tissue were counted manually under a stereomicroscope. Fixed tissue was also embedded in paraffin, and one section per lung was stained with haematoxylin and eosin. The number of metastases per section was counted under a microscope, and metastasis surface area was quantified using ImageJ software version 1.53b.
In vivo treatments
To assess the contribution of individual immune cell populations to vaccine efficacy against B16F10 (MICB) tumours, mice were injected intraperitonially with 200 μg (in 100 μl PBS) of anti-mouse CD4 (clone GK1.5, BioXcell), CD8β (clone 53-5.8, BioXcell), NK1.1 (clone PK136, BioXcell) or CSF1R (clone AFS98, BioXcell) antibodies to deplete CD4+ T cells, CD8+ T cells, NK cells and macrophage/monocytes, respectively. The following isotype-control antibodies were used: rat IgG2b (clone LTF-2, BioXcell; control for anti-CD4), rat IgG1 (clone TNP6A7, BioXcell; control for anti-CD8β), mouse IgG2a (clone C1.18.4, BioXcell; control for anti-NK1.1) and rat IgG2a (clone 2A3, BioXcell; control for anti-CSF1R). All depletion antibodies were injected 2 d before tumour inoculation and then every third day until the study endpoint. IFNγ and TNFα cytokines were neutralized by injecting immunized mice intraperitoneally with 200 μg (in 100 μl PBS) anti-mouse IFNγ (clone XMG1.2, BioXcell) and TNFα (clone XT3.11, BioXcell) antibodies 1 d before tumour challenge and then every 48 h until the study endpoint. Rat IgG1 was used as a control for the cytokine-neutralizing antibodies. Depletion of CD4+ T cells, CD8+ T cells and NK cells was confirmed by staining peripheral blood mononuclear cells (PBMCs) isolated from depleted or control groups 7 d after antibody treatment with BioLegend anti-mouse CD4 (RM4-4), CD8α (53-6.7) and NK1.1 (PK136) antibodies, respectively, and the stained cells were analysed by flow cytometry.
In NKG2D blocking experiments, mice were injected with 200 μg (in 100 μl PBS) NKG2D blocking antibody (clone CX5, eBioscience) or rat IgG1 isotype control (clone eBRG1, eBioscience) starting 1 d before tumour inoculation and then every 48 h until the study end-point. For CD40L blocking experiments, mice were treated with 200 μg anti-CD40L (clone MR-1, BioXcell) or rat IgG2b isotype-control antibody (clone LTF-2, BioXcell) on days–1, 0 and 2 with respect to induction of MICB expression on tumour cells by doxycycline treatment.
XCR1+ cDC1 cells were depleted by injecting Xcr1DTR mice with 500 ng un-nicked DT (150, List Biologicals) intraperitoneally in 100 μl PBS. DT was typically administered 2 d before induction of MICB expression on tumour cells by doxycycline treatment and then every 48 h until the study endpoint. Depletion of XCR1+ DCs was confirmed by checking for Venus reporter expression by flow cytometry in splenocytes collected from mock-treated or DT-injected mice; DCs were stained with BioLegend anti-mouse CD11c (clone N418), IA/E (clone M5/114.15.2) and XCR1 (clone S15046E) antibodies.
Doxycycline-inducible expression of MICB on tumour cells for analysis of tumour-infiltrating immune cells by flow cytometry and scRNA-seq
Because of the efficacy of the MICA/B α3 domain vaccine, a substantial fraction of mice in the experimental group did not have tumours when control immunized mice were at the study endpoint. To enable better comparison of the effect of the vaccine on the tumour microenvironment, MICB expression was induced on tumour cells by doxycycline treatment once mice had received experimental or control vaccine. Tumour-infiltrating immune cells were then analysed 7 d after induction of MICB expression on tumour cells. C57BL/6J mice immunized (day 0) and boosted (day 14) with Ctrl-vax or MICB-vax were injected subcutaneously with 0.3 × 106 B16F10 (MICB-dox) cells in 50 μl PBS. MICB expression was induced in tumours by intraperitoneal injection of doxycycline (5 mg kg−1 in 100 μl PBS every 48 h) when they reached a size of about 50 mm2. Surface MICB expression was undetectable in vivo before doxycycline treatment. Surface MICB expression on tumour cells after doxycycline treatment was confirmed by preparing a single-cell suspension of excised tumour cells and labelling cells with a PE-conjugated anti-human MICA/B antibody (clone 6D4, BioLegend) or a corresponding isotype-control antibody (clone MOPC-173, BioLegend). Cells were washed twice and analysed by flow cytometry using an LSRFortessa X-20 instrument.
Flow cytometry analysis
Tumours were excised, weighed and dissected into small fragments using surgical scissors. Tumour fragments were transferred into 50-ml Falcon tubes with RPMI (5 ml digestion medium per 0.25 g tumour) containing 160 U ml−1 collagenase type IV (Gibco) and 80 U ml−1 DNase I (Sigma). Samples were incubated at 37 °C on a shaker platform (250 r.p.m.) for 30 min. Following enzymatic digestion, the reaction was quenched by washing cells with FACS buffer and centrifugation at 300g for 6 min to pellet the cells. The cells were resuspended in FACS buffer and passed through 70-μm cell strainers. CD45+ cells were isolated using CD45 TIL Microbeads (Miltenyi Biotec) following the manufacturer’s protocol.
Lymph nodes (LNs) were digested in 2 ml RPMI containing 80 U ml−1 collagenase type IV and 40 U ml−1 DNase I at 37 °C for 30 min. LNs were pipetted vigorously several times during the incubation step to facilitate cell dissociation. Following enzymatic digestion, LNs were mechanically dissociated using 70-μm cell strainers, and cells were washed with FACS buffer. tdLNs included axillary and brachial LNs. Single-cell suspensions of mouse spleens were prepared by pressing the tissue through a 70-μm cell strainer followed by red blood cell lysis for 90 s using ACK lysis buffer (ThermoFisher Scientific). The reaction was terminated by washing the cells with an excess of FACS buffer.
For flow cytometry analysis, cells were stained for 30 min with 100 μl of the indicated dead cell exclusion dye (from Tonbo or Bio-Legend) diluted 1:800 in PBS. Cells were washed once with FACS buffer and pre-incubated with TruStain FcX anti-mouse CD16/32 antibody (clone 93, BioLegend) at 1 μg per 106 cells in 100 μl PBS for 5–10 min on ice before immunostaining. For flow cytometry analysis of T cells and NK cells, cells were incubated with a combination of fluorochrome-conjugated antibodies to the following surface markers (from BioLegend unless otherwise indicated): CD3ε (17A2, BioLegend/BD Bioscience), TCRβ (H57-597, BioLegend/BD Bioscience), CD4 (RM4-5), CD8α (53-6.7), TCRγ/δ (GL3), CD44 (1M7), CD45 (30-F11), CD62L (MEL14), CD69 (H1.2F3), NK1.1 (PK13), CD49b (HMα2), NKG2D (CX5), PD-1 (29F.1A12), Tim3 (RMT3-23), Lag3 (C9B7W), CTLA-4 (UC10-4B9), Tigit (4D4/mTIGIT) and CXCR6 (SA051D1). Cells were stained in FACS buffer (total volume of 100 μl) for 30 min at room temperature, washed twice with FACS buffer and subsequently fixed in Fixation Buffer (BD Biosciences) or fixed and permeabilized using the FoxP3/Transcription Factor Staining Buffer Set (ThermoFisher Scientific) when intracellular staining was required. Fluorochrome-conjugated antibodies directed against the following intracellular markers were used: FoxP3 (FJK-16s, eBioscience), granzyme B (NGZB, eBioscience), Eomes (DAN11MAG, eBioscience), IL-2 (JES6-5H4, BioLegend), IFNγ (XMG1.2, BioLegend), Ki67 (SolA15, eBioscience), perforin (eBioOMAK-D, eBioscience) and TNFα (MP6-XT22, BioLegend). For analysis of intracellular cytokines in T cells, LN and tumour single-cell suspensions were incubated with 1× Leukocyte Activation Cocktail with BD GolgiPlug (BD Bioscience) for 4 h in complete RPMI before surface and intracellular staining.
For flow cytometry analysis of myeloid cells, cells were incubated with a combination of fluorochrome-conjugated antibodies to the following surface markers (from BioLegend unless otherwise indicated): CD3ε (clone 17A2), CD19 (clone 6D5), TCRβ (clone H57-597), NK1.1 (clone PK136; dump channel markers), F4/80 (clone BM8), Ly6c (clone HK1.4), CD11b (M1/70), CD11c (clone N418), IA/E (clone M5/114.15.2), CD103 (clone 2E7), CD8α (53-6.7), PD-L1 (clone 10F.9G2), XCR1 (clone S15046E), CD24 (clone M1/69, BD Biosciences) and CD301b (clone URA-1).
All antibodies for surface markers were used at a 1:300 dilution except anti-IA/E (1:500 dilution); antibodies for intracellular proteins were used at a dilution of 1:200. Stained and fixed cells were resuspended in 300 μl FACS buffer and stored at 4 °C protected from light until analysis within 48 h. Samples were acquired using an LSRFortessa X-20 flow cytometer, and FACS DIVA software was used for data acquisition. Compensation for fluorescence overlap was set up using OneComp eBeads (ThermoFisher) stained with the identical set of antibodies used for analysis. Collected data were analysed using FlowJo (Tree Star) version 10.7.1.
Adoptive T cell transfer experiments
Spleens collected from naive Thy1.1+ pmel-1 or CD45.1+ OT-1 TCR-transgenic mice were processed into single-cell suspensions, and CD8+ T cells were isolated by negative selection (EasySep Mouse CD8+ T Cell Isolation Kit, Stemcell Technologies). Enriched CD8+ T cells were labelled with CellTrace-Violet (CTV, ThermoFisher) by incubating cells (1 × 107 cells per ml in PBS) for 3 min with 1 μM CTV in a 37-°C water bath. The reaction was quenched by washing the cells once with an excess of prewarmed RPMI (Gibco) containing 10% FBS followed by two washes with PBS (without additives). Labelled CD8+ T cells (2 × 106 in 250 μl PBS) were adoptively transferred via tail vein 16 h after MICB expression had been induced on tumour cells by doxycycline treatment. For adoptive transfer experiments using Xcr1DTR recipient mice, 500 ng un-nicked DT (150, List Biologics) was injected intraperitoneally every other day beginning 2 d before adoptive T cell transfer until the experimental endpoint. Tumours, spleens and tdLNs were collected 4 d after T cell transfer and processed into a single-cell suspension as described above. Cells were stained with Zombie Green dead cell exclusion marker (BioLegend), followed by surface staining using anti-mouse Thy1.1 (OX-7; for pmel-1 T cells) or CD45.1 (A20; for OT-1 T cells), CD3ε (17A2) and CD8α (53-6.7) antibodies. T cell proliferation was measured by determining CTV dilution in transferred T cells using an LSRFortessa X-20 flow cytometer. Data were analysed using FlowJo software.
In vitro T cell assays
Epitope spreading of CD8+ T cell responses to other tumour antigens was analysed in mice that had received Ctrl-vax or MICB-vax. C57BL/6J mice were injected subcutaneously with 0.3 × 106 B16F10 (MICB) tumour cells. When the tumours were palpable (approximately day 9), mice were immunized with Ctrl-vax or MICB-vax. Spleens and tdLNs were collected 12 d after immunization, and CD8+ T cells were isolated and labelled with CTV as described above. Spleens were collected from naive mice and subjected to enzymatic digestion using a Spleen Dissociation kit (Miltenyi Biotec). DCs were isolated from digested spleens using mouse CD11c MicroBeads UltraPure (Miltenyi Biotec) according to the manufacturer’s protocol. DCs were seeded into 96-well U-bottom plates (2 × 104 cells per well) with complete RPMI and pulsed with 10 μg ml−1 (final concentration for each well) of the melanoma gp100 peptide (EGSRNQDWL) or a control peptide (SIINFEKL) (synthesized by 21st Century Biochemicals). Labelled T cells and peptide-loaded DCs were co-cultured at a 5:1 ratio (1 × 105 T cells and 2 × 104 DCs in 200 μl medium) for 72 h. On the day of analysis, cells were incubated with 1× Leukocyte Activation Cocktail with BD GolgiPlug for 4 h before staining for surface and intracellular markers. T cell proliferation and cytokine secretion were measured using an LSRFortessa X-20 instrument and analysed using FlowJo software.
NK cell cytotoxicity assays
NK cells were isolated from the spleens of WT C57BL/6 mice by magnetic separation using the EasySep Mouse NK Cell Isolation kit (Stemcell Technologies) according to the manufacturer’s instructions. Isolated NK cells were cultured in complete RPMI (supplemented with 5 ng ml−1 IL-2 (Peprotech, AF-212–12) and cultured for 72 h. B2m−/− B16F10 (MICB) tumour cells were labelled with CFSE (Life Technologies, C34554) and seeded in a 96-well U-bottom plate at a density of 0.25 × 105 cells per well in 100 μl complete RPMI. Ten micrograms of purified polyclonal antibodies from mice immunized with Ctrl-vax or MICB-vax was added to the wells containing tumour cells, and plates were incubated for 2 h. In parallel, NK cells were collected and washed with PBS, and the pellet was resuspended at a density of 2 × 106 cells per ml. Following incubation of tumour cells with polyclonal antibodies, NK cells were added to the wells at different NK cell to tumour cell ratios (0.25–2.5 to 1), and the total volume in each well was adjusted to 200 μl with complete RPMI. Wells containing target cells alone without the addition of NK cells were used as controls to determine spontaneous cell death during the assay. Tumour cells lysed with 2% Triton X-100 were used as a positive control for complete lysis. Plates were incubated for 6 h at 37 °C (5% CO2, humidified atmosphere). Following incubation, cells were collected and stained with Zombie NIR (BioLegend), followed by surface staining using anti-mouse CD45 (BioLegend, clone 30-F 11) and NK1.1 (BioLegend, clone PK136) antibodies. Samples were analysed using an LSRFortessa X-20 flow cytometer, and NK cell cytotoxicity was measured by evaluating the percentage of dead target cells at different effector to target ratios.
Ex vivo cross-presentation assays using polyclonal antibodies generated in vaccinated mice
BMDCs were differentiated in vitro from bone marrow precursors. Bone marrow cells were collected from the femurs and tibias of WT C57BL/6 mice and Fcer1g−/− mice (JAX strain 002847). Red blood cells were removed by adding ACK lysis buffer (Life Technologies). Cells were resuspended in complete medium supplemented with 20 ng ml−1 GM-CSF (BioLegend, 576306) and 5 ng ml−1 IL-4 (Peprotech, 10780-142), and half of the medium was replaced every 2 d. On day 8, non-adherent and loosely adherent cells in the culture supernatant were collected for assays.
BMDCs were co-cultured with apoptotic tumour cells before co-culture with T cells. MICB-expressing B2m−/− B16F10 Ova-Zsgreen cells were irradiated at 10,000 rads; 48 h after irradiation, purified polyclonal antibodies (0.5–5 μg ml−1) from mice immunized with Ctrl-vax or MICB-vax were added to the wells containing tumour cells and plates were incubated for 2 h. In parallel, BMDCs were collected and seeded into 96-well U-bottom plates at a density of 10,000 cells per well. After 2 h of incubation with polyclonal antibodies, tumour cells were washed to remove excess antibodies and added to the wells with BMDCs at a 1:1 ratio. For Fc blocking conditions, Fc blocking antibodies were added to the BMDCs and plates were incubated for 30 min before the addition of tumour cells. CD8+ T cells from the spleens and LNs of OT-1 mice were enriched by magnetic separation using the EasySep Mouse CD8+ T Cell Isolation kit (Stemcell Technologies) according to the manufacturer’s instructions. Enriched CD8+ T cells were then labelled with CFSE (Life Technologies, C34554). CFSE-labelled CD8+ T cells were added to the plate containing BMDCs and tumour cells and co-cultured at a 5:1 ratio of T cells to BMDCs. Three days later, cells were collected and stained with Zombie UV viability dye (BioLegend), followed by staining with BioLegend mouse antibodies directed against CD11c (clone N418) and CD8 (53-6.7). Cells were washed twice and analysed by flow cytometry using an LSRFortessa X-20 instrument. T cell proliferation was evaluated by CFSE dilution.
Ex vivo co-culture of migratory DCs with pmel-1 T cells
DCs were isolated from Ctrl-vax- and MICB-vax-immunized mice to investigate whether DCs from MICB-vax-immunized mice had enhanced in vivo uptake of endogenous melanoma antigens. MICB expression was induced on B16F10 (MICB-dox) tumour cells 2 d before DC isolation by doxycycline treatment. Axillary and brachial tdLNs and control mesenteric LNs were collected, and single-cell suspensions were stained with Ghost Violet dead cell exclusion dye (Tonbo Biosciences), followed by Fc blocking with anti-CD16/32 (BioLegend) and surface staining with anti-bodies directed against CD11c (clone N418) and IA/E (clone M5/114.15.2) as well as a panel of dump channel markers, including CD3ε (clone 17A2), CD19 (clone 6D5), TCRβ (clone H57-597), NK1.1 (clone PK136), F4/80 (clone BM8) and Ly6c (clone HK1.4) (all from BioLegend). Migratory DCs (defined as CD3−TCRβ−CD19−NK1.1−Ly6c−F4/80−CD11c+IA/Ehl) were sorted from the stained samples using a FACSAria IIIu instrument (BD Biosciences). CD8+ T cells were isolated from the spleens of naive pmel-1 TCR-transgenic mice, labelled with CTV and co-cultured with sorted migratory DCs at a 5:1 ratio for 72 h. T cell proliferation was measured on the basis of CTV dilution using an LSRFortessa X-20 instrument.
Bulk RNA-seq analysis of migratory DCs
The effect of MICB-vax versus Ctrl-vax on migratory DCs in LNs was investigated, both with and without prior depletion of CD4+ T cells. C57BL/6J mice were immunized twice, and B16F10 (MICB-dox) tumour cells were injected subcutaneously on day 21 after the initial immunization. When tumours were palpable (about 50 mm2), mice were randomized to two groups that received an CD4-depleting monoclonal antibody (0.2 mg in 0.1 ml PBS; clone GK1.5, BioXcell) or a rat IgG2b isotype-control antibody (clone LTF-2, BioXcell). MICB expression was induced on tumour cells by intraperitoneal injection of doxycycline (5 mg kg−1 in PBS) 2 d after injection with anti-CD4 or control monoclonal antibody. tdLNs were collected 48 h after induction of MICB expression, and single-cell suspensions were prepared as described earlier. Fc receptors were blocked before staining cells with a PE-conjugated anti-mouse CD11c antibody (clone N418, BioLegend). Labelled cells were enriched using an EasySep Mouse PE Positive Selection Kit II (Stem-cell Technologies) following the manufacturer’s protocol. Enriched cells were stained with Ghost Violet dead cell exclusion dye (Tonbo Biosciences), followed by surface staining with antibodies directed against IA/E (clone M5/114.15.2) and NK1.1 (clone PK136) as well as a cocktail of monoclonal antibodies specific for CD3ε (clone 17A2), CD19 (clone 6D5), TCRβ (clone H57-597), F4/80 (clone BM8) and Ly6c (clone HK1.4) (dump channel markers). Migratory DCs (defined as CD3−TCRβ−CD19−NK1.1−Ly6c−F4/80−CD11c+IA/Ehi) were sorted directly into 200 μl TRIzol using a FACSAria IIIu instrument. The collection tubes were spun down briefly and snap frozen at −80 °C. Samples were submitted to Genewiz for RNA isolation, library preparation and data analysis. The HiSeq 2 × 150-bp paired-end high-output platform was used for sequencing. Sequence reads were trimmed to remove adaptor sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Mus musculus GRCm38 reference genome available on Ensembl using STAR aligner v.2.5.2b. Extracted gene hit counts were used for downstream differential expression analysis. Using DESeq2, migratory DC gene expression levels were compared between the MICB-vax and Ctrl-vax groups as well as the MICB-vax (isotype-control monoclonal antibody) and MICB-vax (CD4+ T cell depletion) groups. P values and log2-transformed fold change values were determined using the Wald test. Genes with an adjusted P value of <0.05 and absolute log2-transformed fold change of >1 were considered to be differentially expressed in these datasets.
scRNA-seq analysis
C57BL/6J mice immunized and boosted with MICB-vax or Ctrl-vax were injected subcutaneously with 0.3 × 106 B16F10 (MICB-dox) tumours. Once the tumours were palpable (about 50 mm2), mice immunized with MICB-vax or Ctrl-vax were injected intraperitonially with either 100 μl doxycycline (5 mg kg−1 in PBS) or PBS, resulting in four groups, including three control groups (Ctrl-vax + PBS, Ctrl-vax + doxycycline, MICB-vax + PBS) and one experimental group (MICB-vax + doxycycline). Tumours were collected 7 d later, and single-cell suspensions were prepared and stained with antibodies as described in the mouse tissue digestion and flow cytometry sections. In brief, dead cells were excluded by staining with Zombie Green viability dye (BioLegend) and Fc receptors were blocked before the addition of PE-conjugated anti-mouse CD45 (clone 30-F11, BioLegend) antibody. Then, 5,000 live CD45+ cells were sorted from each tumour sample using a FACSAria IIIu instrument. Equal numbers of sorted CD45+ cells from five mice per group were pooled, pelleted and resuspended at 1 × 103 cells per μl in PBS containing 0.04% RNase-free BSA. For each of the four groups, 5,000 cells were used for the 10x Genomics 5′ v2 single-cell assay. Reverse transcription, cDNA amplification and library preparation were performed according to the manufacturer’s protocol. Completed libraries were sequenced on the Illumina HiSeq 2500 instrument with rapid-run mode, yielding > 40,000 reads per cell. Amplified cDNA was also used for TCR V(D)J targeted enrichment using the Chromium Single Cell Mouse TCR Amplification kit. TCR V(D)J libraries were sequenced on the Illumina HiSeq 2500 platform with a 150-bp paired-end configuration. A description of scRNA-seq data processing is available in the Supplementary Information.
Statistical analyses
Statistical analyses were performed using GraphPad Prism 9 software. Each experiment was repeated as indicated in the respective figure legend. An unpaired Student’s t test, one-way ANOVA with Sidak’s multiple-comparison or Tukey’s multiple-comparison test, two-way ANOVA with Tukey’s multiple-comparison test or unpaired two-sided Mann–Whitney test was used as indicated for comparisons between the groups. Tumour growth kinetics were analysed using two-way ANOVA with Bonferroni’s post hoc test. For comparison of mouse survival curves, a log-rank (Mantel–Cox) test was used. All P values are two sided, and statistical significance was evaluated at the 0.05 level.
Extended Data
Extended Data Fig. 1 ∣. Characterization of vaccine targeting MICA/B vaccine α3 domain.
a, HPLC gel filtration analysis of affinity-purified ferritin (immunogen for Ctrl-vax) and MICB-ferritin (immunogen for MICB-vax) proteins. The proteins formed nanoparticles of ~957 kDa (MICB-ferritin) and ~468 kDa (ferritin); molecular weight standards are indicated. b, SDS-PAGE analysis of purified ferritin and MICB-ferritin proteins under reducing conditions. c, Electron microscopy image showing MICB-ferritin protein assembled into nanoparticles (98,000x magnification). d, Identification of MICB transgenic (Tg) mice by PCR amplification using genomic DNA extracted from tail biopsies (lanes 2,6, MICB Tg; lane 7, positive control amplification from plasmid with the transgene cassette; lane 8, negative control reaction). e, Quantitative RT-PCR analysis of luciferase and MICB mRNA in the prostate of WT (grey) and MICB-tg (orange) mice (n = 2 WT, n = 3 MICB-Tg mice) normalized to mouse Hprt mRNA. f, Quantification of day 14 MICB-specific serum Ab titers (fluorescence-based immunoassay, DELFIA) in WT and MICB-tg mice immunized with Ctrl-vax (n = 4 mice/group) (grey) (120μg ferritin protein, 100μg CpG, 1μg GM-CSF) or MICB-vax (n = 5 mice/group) (200μg MICB-ferritin, 100μg CpG, 1μg GM-CSF) without MSR (left) or with MSR (right) scaffold. g, Quantification of MICB-specific serum Ab titers (fluorescence-based immunoassay, DELFIA) in mice immunized with Ctrl-vax (grey) or MICB-vax using differing protein doses, 50μg (yellow), 100μg (blue) or 200μg (red) (n = 2 mice/group). h, Representative flow cytometry plots showing binding of polyclonal serum Abs from mice immunized with Ctrl-vax (blue) or MICB-vax (red) to cell surface MICB on B16F10 (MICB) tumor cells; serum dilutions are indicated for each condition. i, Titers of MICB Ab isotypes assessed by ELISA (n = 5 mice/group) in MICB transgenic mice immunized with Ctrl-vax (blue) or MICB-vax (red). j, Analysis of MICB-specific CD8 T-cell responses in the spleen of mice immunized with Ctrl-vax (blue) or MICB-vax (red). Intracellular cytokine staining (IFNγ) and CFSE dilution is shown in representative flow cytometry plots (left); data are quantified for T-cells from both vaccine groups (3 mice/group, right). k, Analysis of human NKG2D dimer (left) and mouse NKG2D dimer (right) binding to cell surface MICB on B16F10 (MICB-ZsGreen) tumor cells pre-incubated with sera (5μl) from Ctrl-vax or MICB-vax mice. l, Representative flow cytometry plots showing binding of anti-human MICA/B antibody (6D4, specific for MICA/B α1-α2 domains) to cell surface MICB on B16F10 (MICB) tumor cells pre-incubated with sera from mice immunized with Ctrl-vax (blue) or MICB-vax (red). Representative data from >3 independent experiments (a, b). Data from a single experiment with technical replicates (c). Representative data from three experiments (d, e, g, h, i). Representative data from two experiments (j, k, l). Two-tailed unpaired Student’s t-test (e); Two-way ANOVA with Tukey’s multiple comparison test (f, g); Two-way ANOVA with Sidak’s multiple comparison test (i, j). Data depict mean +/− SD (e,g) or mean +/− SEM (f, i,j).
Extended Data Fig. 2 ∣. Characterization of vaccine-induced immune responses.
a, b, Inhibition of MICA/B shedding and surface stabilization by vaccine-induced Abs. Flow cytometric analysis of cell surface MICA/B levels (a) and shed MICA/B in supernatants (b) for human A375 melanoma, A549 lung carcinoma, HCT116 colon carcinoma and K562 myelogenous leukemia cell lines 24 h following incubation with 10 μl of sera from Ctrl-vax (blue) or MICB-vax (red) immunized mice; isotype control Ab staining shown in grey (a). c, d, Inhibition of MICB shedding and surface stabilization by vaccine-induced Abs on mouse tumor cell lines. Flow cytometric analysis of cell surface MICB levels (c) and shed MICB in supernatants (d) for mouse B16F10 (MICB) melanoma, EL4 (MICB) lymphoma and 4T1 (MICB) triple negative breast cancer cell lines 24 h following incubation with 10 μl of sera from Ctrl-vax (blue) or MICB-vax (red) immunized mice. Isotype control Ab staining shown in grey (c). e–g. Vaccine efficacy at different MICB expression levels induced in tumor cells using a doxycycline (dox) inducible promoter. Representative flow cytometry histograms showing MICB surface expression levels on B16F10 (MICB-dox) tumors in vivo in mice treated with PBS (blue histogram) or different concentrations of doxycycline (dox): low dox (2.5mg/kg, orange), medium dox (5mg/kg, red) or high dox (10mg/kg, green) or control B16F10 (Ctrl-dox) tumors treated with high dox (10mg/kg, grey) (e). Analysis of B16F10 (MICB-dox) tumor growth kinetics at different MICB expression levels by tumor cells. Mice received Ctrl-vax (grey, blue, black) or MICB-vax (orange, red, green) on day 0 and a boost on day 14. B16F10 (MICB-dox) tumor cells were implanted on day 21, and MICB expression was induced on tumor cells on day 25 when tumors were palpable by treating mice with different concentrations of doxycycline as indicated (f) (n=7 mice/group). Quantification of serum levels of shed MICB in mice immunized with Ctrl-vax (grey, blue, black) or MICB-vax (orange, red, green) 96 h post dox-mediated induction of MICB on B16F10 (MICB-dox) tumor cells. Serum levels of shed MICB were analyzed in 5 randomly selected mice in each group (g). h–i, Representative flow cytometry histogram showing surface MICB levels on B16F10 (MICB) clone G12 (red) or indicated pooled clones (gradient of turquoise). Grey histogram represents isotype antibody staining of B16F10 (MICB) clone G12 (h). Assessment of therapeutic efficacy of MICB-vax (red, green) or Ctrl-vax (blue, grey) in mice with tumors established with B16F10 (MICB) clone G12 or pooled clones (B3, A3, C6, B1, G1) (n=8 mice/group) (i). j–n, Assessment of vaccine efficacy targeting MICA or MICB α3 domains. The MSR scaffold was formulated with antigens, GM-CSF and CpG. Mice received one or two doses of Ctrl-vax, MICB-vax (j–m) or MICA-vax (n) and were then challenged with B16F10 tumor cells expressing MICB (allele 005) (j, k) or MICA (allele 009) (m, n) or EL4 tumor cells expressing MICB (allele 005) (l). Vaccination and tumor challenge schedule is illustrated above each experiment; n = 7 mice/group (j–l), n = 6 mice in Ctrl-vax, n = 8 mice in MICB-vax (m) and n = 6 mice/group (n). For experiments shown in (j), tumor-free mice were rechallenged on day 120 using the same dose of B16F10 (MICB) tumor cells as in the initial inoculation. Representative data from two experiments (a–d, j–n). Data from single experiment (e–g, h–i). Two-tailed unpaired Student’s t-test (b, d); one-way ANOVA with Tukey’s multiple comparison test (g); two-way ANOVA with Bonferroni’s post hoc test (f, i (left), j (left), k. l, m, n); log-rank (Mantel-Cox) test (i (right), j (right). Data depict mean +/− SD (b, d) or mean +/− SEM (f, g, i–n).
Extended Data Fig. 3 ∣. Protection from metastatic disease by vaccination following surgical removal of primary tumors.
a, c, Images of lung metastases from five representative mice per group immunized with Ctrl-vax (top two rows) or MICB-vax (bottom two rows). Mice were immunized following surgical removal of primary B16-BL6 (MICB) (n = 10 mice/group) (a) and 4T1 (MICB) (c) tumors (n = 13 mice/group), as described in Fig. 1f, g. b, d, Representative bright field images of H&E stained histological sections of lung metastases from mice with B16-BL6 (MICB) (b) or 4T1 (MICB) (d) tumors. e–f, Quantification of the number of metastases per H&E stained section (left) and percentage of area of lung section occupied by metastases (right) for mice immunized with Ctrl-vax (blue) or MICB-vax (red) following surgical removal of primary B16-BL6 (MICB) tumors (e) or 4T1 (MICB) tumors (f). Representative data from two experiments (a-f). Representative images of 5 histological lung sections per mouse (b, d), Two-tailed Mann Whitney test (e, f). Data depict mean+/− SEM.
Extended Data Fig. 4 ∣. Immunogenicity of MICA/B α3 domain vaccine in non-human primates.
a, Characterization of the MICA/B α3 immunogen used in the primate study. The α3 domains of rhesus macaque MICA and MICB were expressed as a fusion protein with ferritin to generate nanoparticles that displayed both α3 domains on the surface. Nanoparticles formed by this fusion protein were conjugated using click chemistry to CpG ODN 2935 as the adjuvant and characterized by HPLC gel filtration chromatography. Shown are HPLC traces of the protein following conjugation to the CpG oligonucleotide (red: 280 nm trace for detection of protein; blue: 350 nm trace for detection of bis-aryl hydrazone bond). b, SDS-PAGE analysis of purified macaque MICA/B α3–ferritin protein under reducing (+) and non-reducing (−) conditions following CpG conjugation and final purification using a HPLC gel filtration column. c–e, Characterization of serum antibody responses to rhesus macaque MICA (left) and MICB (right) proteins (full-length extracellular domains without ferritin fusion partner) at different steps in the immunization process. Antibody responses were investigated in three animals (RBQ12, RVf10 and RQq15) at multiple timepoints (pre-immunization; three weeks following initial immunization and boosts 1-3, as illustrated in Fig. 1h) using a fluorescence-based ELISA (RFU, relative fluorescence units) at multiple serum dilutions (1:102 to 1:104). f–m, Binding of purified polyclonal serum IgG to cell surface MICA (left) and MICB (right) using HEK293T transfectants that displayed rhesus macaque MICA (Mamu-A*01) or MICB (Mamu-B*01) proteins. Preimmune sera were used as a control (grey) for sera obtained following immunization (red). Representative histograms (left in f, g) and graphical summaries of flow cytometry data (right in f, g, h–m) are shown for the four immunized macaques (9312, RBQ12, RVf10 and RQq15). Representative data from two experiments (a–b). Data from a single macaque immunization experiment with technical replicates for each macaque analyzed (c–m). Two-way ANOVA with Tukey’s multiple comparison test (c-e). Data depict mean+/− SD.
Extended Data Fig. 5 ∣. Impact of vaccine on functional programs of tumor-infiltrating CD4 and CD8 T-cells.
a, Effect of doxycycline on tumor growth in vaccinated mice. Mice received Ctrl-vax (blue, grey) or MICB-vax (green, magenta, red) on day 0 and a boost on day 14. B16F10 cells transduced with Ctrl-dox or MICB-dox lentiviral vectors as indicated were implanted on day 21, and mice were treated with doxycycline (or PBS as control) starting on day 25 to induce MICB expression on tumor cells (n=5 mice/group). b–k, Analysis of tumor-infiltrating T-cell populations. Tumor-infiltrating T-cells were analyzed 7 days following induction of MICB expression by tumor cells (n = 7 mice/group). b–c, Analysis of CD62L and CD44 expression by tumor-infiltrating CD4 (b) and CD8 (c) T-cells. d–e, Representative flow plots (left) and quantification (right) of NKG2D receptor expression by CD4 (d) and CD8 (e) T-cells. f–g, Representative flow plots and quantification of CXCR6 receptor expression by CD4 (f) and CD8 (g) T-cells. h–i, Quantification of proliferating Ki67+ CD4 (h) and CD8 (i) T-cells. j–k, Quantification of TNFα positive CD4 (j) and CD8 (k) T-cells. Two-way ANOVA with Bonferroni’s post hoc test (a); two-way ANOVA with Tukey’s multiple comparison test (b–c); two-tailed Mann Whitney test (d–k). Representative data from two independent experiments (b–i); representative data from three independent experiments (j–k). Data depict mean+/− SEM.
Extended Data Fig. 6 ∣. Flow cytometric and scRNA-seq analysis of changes in tumor-infiltrating immune cells induced by the vaccine.
a–b, Representative histograms (a) and quantification (b) of PD-1, CTLA-4, Tim-3, Tigit and Lag3 expression by tumor-infiltrating CD8 T-cells from Ctrl-vax (blue) or MICB-vax (red) mice (n = 9 mice/group). c–d, Comparison of T-cell and NK cell populations in B16F10 (MICB) tumors following treatment with a MICA/B mAb or the MICB vaccine. In the vaccine arm, mice received Ctrl-vax (C-vax) (n = 9 mice) or MICB-vax (M-vax) (n = 10 mice) on days 0 and 14, while mice in the mAb treatment group (n = 8 mice/ group) received two buffer injections. Mice were implanted with B16F10 (MICB-dox) tumor cells on day 21. MICB expression was induced on tumor cells by doxycycline treatment starting on day 28, and mice in the mAb treatment group received either mouse IgG2a isotype control mAb (iso) or MICA/B mAb (mAb) treatment every 48 h starting on day 28. Tumor-infiltrating immune cells were analyzed in all groups on day 35. Total numbers of tumor-infiltrating CD4+ T-cells, CD8+ T-cells and NK cells were quantified by flow cytometry (c), and intracellular staining was performed for IFNγ (d) in all four treatment groups. e–g, scRNA-seq analysis of changes in tumor-infiltrating immune cells induced by the vaccine. CD45+ immune cells in B16F10 (MICB-dox) tumors were investigated by scRNA-seq under four experimental conditions: the MICB-vax (+dox) experimental group and the three control groups, Ctrl-vax (−dox), Ctrl-vax (+dox) and MICB-vax (−dox). Doxycycline was administered to mice for seven days prior to scRNA-seq analysis to induce MICB expression on tumor cells in two of these groups (+dox). For each of the four groups, CD45+ immune cells were pooled from five mice to reduce variation from individual tumors. e, UMAP projection of CD45+ immune cells combined from all experimental groups. Major immune cell populations are annotated based on differentially expressed genes. f, Comparison of immune subpopulations across all clusters for the experimental MICB-vax (+dox) group (red) versus the three combined control groups (blue). g, Distribution of CD45+ cells across individual clusters (color-coded as in e) for the experimental MICB-vax (+dox) group (MICB) and the three combined control groups (Ctrl). Representative data of two experimental repeats (a–b). Data from a single experiment (c–d). ScRNA-seq data from a single experiment with sorted CD45+ cells pooled from 5 mice/group (e–g). Two-tailed Mann Whitney test (b); one-way ANOVA with Tukey’s multiple comparison test (c–d). Data depict mean+/− SEM.
Extended Data Fig. 7 ∣. Gene expression programs of tumor-infiltrating T-cell and NK cell populations.
a–d, scRNA-seq analysis of T-cell clusters among CD45+ tumor-infiltrating cells in B16F10 (MICB-dox) tumors. UMAP representation of T-cell subclusters (a) and visualization of T-cell populations for the experimental MICB-vax (+dox) group (MICB) and the three control groups (b). c, Contribution of each T-cell subcluster to the total CD45+ immune population for each of the four treatment groups. d, Quantification of expanded TCR clonotypes for CD4, Treg, CD8 effector, CD8 exhausted and CD8 proliferating clusters shown for all four treatment groups. e, f, Ranking of differentially expressed genes in scRNA-seq data from the indicated T-cell subpopulations (e) and NK cells (f) comparing cells from the experimental MICB-vax (+dox) group to cells from the three combined control groups, Ctrl-vax (−dox), Ctrl-vax (+dox) and MICB-vax (−dox). g–j, Violin plots showing expression levels of activation-related genes (g) and Th1-related genes (h) in CD4 T-cells as well as activation-related genes (i) and chemokine receptor genes (j) in CD8 T-cells from the experimental MICB-vax (+dox) group compared to cells from the three combined control groups (Ctrl). ScRNA-seq data from a single experiment with sorted CD45+ cells pooled from 5 mice/group (a–j). Pairwise Wilcoxon rank sum test (g–j).
Extended Data Fig. 8 ∣. T cell and NK cell responses in MHC-I expressing and MHC-I deficient tumors.
a, Contribution of CD8 T cells to vaccine efficacy. Mice were first immunized with MICB-vax or Ctrl-vax (d0, d14), treated with either isotype control mAb, depleting mAb targeting CD8 T-cells starting on day 21, followed by implantation of B16F10 (MICB) tumor cells (n = 7 mice/group). b, Impact of IFNγ versus TNFα neutralization on the efficacy of the MICB α3 domain vaccine. MICB-transgenic mice received IFNγ or TNFα neutralizing mAbs or an isotype control mAb every 48 h, starting two days prior to subcutaneous injection of B16F10 (MICB) tumor cells on day 21 following immunization (n = 7 mice/group). Tumor growth (left) and survival analysis (right) are shown. c, Comparison of vaccine efficacy against B16F10 (MICB) wild-type tumors and tumors with resistance mutation in H2-Aa gene. Mice received MICB-vax (n=8 mice/ group) or Ctrl-vax (n = 7 mice/ group) and were then challenged with tumors of the indicated genotypes. d, Quantification of MICB-specific serum Ab titers in mice immunized with Ctrl-vax (blue) or MICB-vax followed by treatment with CD4 T-cell depletion (green) or control (red) mAbs for 3 weeks, starting on day 28 following immunization (n=5 mice/group). e, Impact of CD4 T-cell depletion on vaccine-induced NK cell infiltration into tumors. Flow cytometric quantification of the percentage of Ki67+ NK cells (top) and IFNγ+ NK cells (bottom) in WT (left) and B2m-KO (right) tumors for the following treatment groups: Ctrl-vax + isotype mAb (blue), Ctrl-vax + anti-CD4 (orange), MICB-vax + isotype mAb (red) and MICB-vax+anti-CD4 (green); (n = 7 mice/group). Representative data from two independent experiments (a-c, e). Data from a single experiment with technical triplicates (d). Log-rank (Mantel-Cox) test (a); two-way ANOVA with Bonferroni’s post hoc test (b left, c left) and Log-rank (Mantel-Cox) test (b right, c right); two-way ANOVA with Tukey’s multiple comparison test (d); one-way ANOVA with Tukey’s multiple comparison test (e).
Extended Data Fig. 9 ∣. Characterization of dendritic cells in the tumor draining lymph nodes of vaccinated mice.
a, Experimental outline and gating strategy for identification of migratory DC (mDC) subsets within the tumor draining lymph node (tdLN) of immunized mice by flow cytometry. b, Impact of CD4 T-cell depletion on mDC and resident (rDC) populations in tdLN of vaccinated mice. DC populations were analyzed in tdLN on day 32, two days following induction of MICB expression on tumor cells with dox (n = 7 mice per group, except in control-αCD4 ab (n = 6 mice). c, Differentially expressed genes of mDCs (bulk RNA-seq) sorted from tdLN of MICB-vax and Ctrl-vax mice treated with CD4 depleting (αCD4) or isotype control (iso) mAb. d, Upregulated genes in mDCs from the MICB-vax (isotype control mAb, iso) group compared to either MICB-vax plus CD4 T-cell depletion (αCD4) or the Ctrl-vax (isotype mAb) groups. Venn diagram and heatmap of genes with higher expression in MICB-vax (iso) compared to both other groups. e–g, Characterization of myeloid cells within the tumors of vaccinated mice. Mice were immunized (d0 and 14), B16F10 (MICB-dox) tumor cells were implanted on day 21 and MICB expression was induced on tumor cells on day 28 by doxycycline (dox) treatment. Tumor-infiltrating myeloid cells were analyzed 7 days later. e, Gating strategy used for identification of tumor-infiltrating CD103+ cDC1, CD301b+ cDC2 and F4/80+ macrophages in vaccinated mice by flow cytometry. f, Quantification of macrophage population within tumors of Ctrl-Vax (blue) and MICB-vax (red) mice on day 7 following MICB induction on tumors with dox (n = 7 mice/group). g, Impact of αCSF1R treatment on immunity to B16F10 (MICB) tumors. Mice were immunized with Ctrl-vax or MICB-vax; treatment with αCSF1R or isotype control Ab was started two days prior to subcutaneous injection of B16F10 (MICB) tumor cells; mAb treatment was continued every third day; n=7 mice/group. Data representative of two independent experiments (a–b, e–g). Data from one experiment with three biological replicates per sample. DCs from 3 mice were pooled for each biological replicate (n = 9 mice/group). Significance was determined using thresholds of −log10 >2 (adjusted P value), and log2 >1 (fold change) (c–d). One-way ANOVA with Tukey’s multiple comparison test (b); Two-tailed Mann Whitney test (f); log-rank (Mantel-Cox) test (g). Data represent mean +/− SEM.
Extended Data Fig. 10 ∣. Cross-presentation of endogenous melanoma antigen by dendritic cells from MICB-vax mice.
a, Impact of cDC1 depletion on MICB vaccine-induced T-cell and NK accumulation within tumors in Xcr1DTR mice. Mice were treated +/− diphtheria toxin (DT) starting on day 26 following immunization with Ctrl-vax (C) or MICB-vax (M) (days 0 and 14) and B16F10 (MICB-dox) tumor implantation (day 21). Immune cells were analyzed in tumors 7 days following MICB induction on tumors with dox (day 37) (n = 7 mice/group). b, Contribution of activating Fc receptors to efficacy of MICB-vax. Survival curves of FceR1g−/− MICB-Tg versus MICB-Tg mice immunized with Ctrl-vax (blue) and MICB-vax (red) (n = 7 mice/group). c, Analysis of endogenous gp100 specific CD8 T-cell responses. CD8 T-cells were isolated from tdLN of mice immunized with MICB-vax or Ctrl-vax, labeled with the CTV cell proliferation dye and then co-cultured for 72 h with DCs pulsed with control (Ova) or gp100 peptide (10μg/ml). Intracellular cytokine staining (IFNγ) and CTV dilution are shown in representative flow cytometry plots (left); data are quantified for T-cells from both vaccine groups (3 mice/group, right). d, Proliferation of transferred CD8 T-cells specific for the gp100 melanoma antigen (from Pmel-1 transgenic mice) in tumor-draining lymph nodes of mice immunized with MICB-vax compared to Ctrl-vax. Mice were vaccinated twice (days 0 and 14) and B16F10 (MICB-dox) tumor cells were implanted subcutaneously on day 21. Doxycycline treatment was initiated on day 28 to induce MICB expression on tumor cells, one day prior to transfer of Thy1.1+ Pmel-1 CD8 T-cells (2x106 cells/mouse). Proliferation of CTV-labeled Pmel-1 T-cells was analyzed in tumor-draining LN (top) and spleen (bottom, control organ) four days following T-cell transfer. Cells were gated based on CD3, CD8 and Thy1.1 markers; shown is Thy1.1 marker of transferred Pmel-1 T-cells (Y-axis) and CTV dye dilution in proliferating T-cells (X-axis). Proliferating T-cell populations are indicated in representative flow plots (left) and quantification is shown (right) across the entire cohort of Ctrl-vax (blue) versus MICB-vax (red) mice (n = 7 mice/group). e, Control experiment for (d) with CD8 T-cells of irrelevant specificity (OT-1 T-cells, n = 4 mice/group). f, Role of XCR1+ DCs in the activation of transferred gp100-specific pmel-1 CD8 T-cells. Xcr1DTR mice were immunized with MICB-vax (days 0 and 14), and B16F10 (MICB-dox) melanoma cells were implanted on day 21. XCR1+ DCs were depleted by injection of diphtheria toxin (+DT, green) or solvent as a control (−DT, red) one day prior to induction of MICB expression by tumor cells with doxycycline. Thy1.1+ Pmel-1 CD8 T-cells were transferred and proliferation of these T-cells was analyzed in tdLN four days later by dilution of the CTV dye. Top flow cytometry plots show depletion of XCR1+ cells (Venus fluorescent reporter protein) in diphtheria toxin (+DT) treated mice (right) compared to control mice (−DT, left), six days following initiation of DT treatment. Bottom flow cytometry plots show proliferation of transferred pmel-1 CD8 T-cells based on dilution of the CTV dye (X-axis); data are quantified on the right (n=6 mice/group). g, Presentation of gp100 peptide by migratory DCs from MICB-vax mice. Naïve Pmel-1 CD8 T-cells were co-cultured for 72 h with migratory DCs (mDC, CD11c+, IA/E high) isolated from tdLN or non-tumor draining LN of Ctrl-vax or MICB-vax mice (pooled from 10 mice/group) implanted with B16F10 (MICB) tumor cells. CD8 T-cell proliferation was assessed based on CTV dilution. Data are representative of two independent experiments (a-d, f-g). Data from a single experiment (e). One-way ANOVA with Tukey’s multiple comparison test (a); log rank (Mantel-Cox) test (b); two-tailed Mann Whitney test (c-f); one-way ANOVA with Dunnett’s multiple comparison test (g). Data depict mean +/− SEM (a, c–f) or mean +/− SD (g).
Supplementary Material
Acknowledgements
We thank the Dana-Farber/Harvard Cancer Center Transgenic Core Facility for generating MICB-transgenic mice; the Dana-Farber/Harvard Cancer Center Rodent Histopathology Core facility and B. Roderick for histological analysis of lung metastases; J. Agudo, H. Dong and M. Raundhal for helpful discussions; and D. Pan for providing B2m and Ifngr gRNAs. Schematic illustrations were created using BioRender.com (licence numbers XU22LPSR1O and RJ23SQGA27). This work was supported by James and Tania McCann (to K.W.W.), the Parker Institute for Cancer Immunotherapy (PICI; to K.W.W.), the Ludwig Center at Harvard Medical School (to K.W.W.), NIH grants R01 CA238039, R01 CA251599, P01 CA163222 and P01 CA236749 (to K.W.W.), R01 CA234018 (to K.W.W.) and R01 CA223255 (to D.J.M.), and a sponsored research agreement with Novartis (to K.W.W.). S.B. was supported by a US Department of Defense fellowship (DOD CA150776) and was the 2018 Baruj Benacerraf Fellow in Immunology.
Footnotes
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Competing interests K.W.W. serves on the scientific advisory board of TCR2 Therapeutics, TScan Therapeutics, SQZ Biotech and Nextechinvest, and he receives sponsored research funding from Novartis. He is a co-founder of Immunitas Therapeutics, a biotech company.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-04772-4.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-04772-4.
Data availability
Raw RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession code GSE181997. Source data are provided with this paper.
References
- 1.Sharma P, Hu-Lieskovan S, Wargo JA & Ribas A Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gasser S, Orsulic S, Brown EJ & Raulet DH The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436, 1186–1190 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier LL NKG2D receptor and its ligands in host defense. Cancer Immunol. Res 3, 575–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaiser BK et al. Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature 447, 482–486 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Bahram S, Bresnahan M, Geraghty DE & Spies T A second lineage of mammalian major histocompatibility complex class I genes. Proc. Natl Acad. Sci. USA 91, 6259–6263 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raulet DH, Gasser S, Gowen BG, Deng W & Jung H Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol 31, 413–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer S et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Groh V et al. Costimulation of CD8αβ T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol 2, 255–260 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Ogasawara K & Lanier LL NKG2D in NK and T cell-mediated immunity. J. Clin. Immunol 25, 534–540 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Holdenrieder S et al. Soluble MICA in malignant diseases. Int. J. Cancer 118, 684–687 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Raffaghello L et al. Downregulation and/or release of NKG2D ligands as immune evasion strategy of human neuroblastoma. Neoplasia 6, 558–568 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salih HR, Rammensee HG & Steinle A Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J. Immunol 169, 4098–4102 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Wu JD et al. Prevalent expression of the immunostimulatory MHC class I chain-related molecule is counteracted by shedding in prostate cancer. J. Clin. Invest 114, 560–568 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G et al. Perturbation of NK cell peripheraL homeostasis accelerates prostate carcinoma metastasis. J. Clin. Invest 123, 4410–4422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doubrovina ES et al. Evasion from NK cell immunity by MHC class I chain-related molecules expressing colon adenocarcinoma. J. Immunol 171, 6891–6899 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Groh V, Wu J, Yee C & Spies T Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 419, 734–738 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Hodi FS et al. Immunologic and clinical effects of antibody blockade of cytotoxic T lymphocyte-associated antigen 4 in previously vaccinated cancer patients. Proc. Natl Acad. Sci. USA 105, 3005–3010 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinushi M, Hodi FS & Dranoff G Therapy-induced antibodies to MHC class I chain-related protein A antagonize immune suppression and stimulate antitumor cytotoxicity. Proc. Natl Acad. Sci. USA 103, 9190–9195 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X et al. An six-amino acid motif in the α3 domain of MICA is the cancer therapeutic target to inhibit shedding. Biochem. Biophys. Res. Commun 387, 476–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P et al. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol 2, 443–451 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Ungaro F et al. VLPs and particle strategies for cancer vaccines. Expert Rev. Vaccines 12, 1173–1193 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Kanekiyo M et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J et al. Injectable, spontaneously assembling, inorganic scaffolds modulate immune cells in vivo and increase vaccine efficacy. Nat. Biotechnol 33, 64–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diefenbach A, Jamieson AM, Liu SD, Shastri N & Raulet DH Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat. Immunol 1, 119–126 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Ferrari de Andrade L et al. Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell-driven tumor immunity. Science 359, 1537–1542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Demaria S et al. Immune-mediated inhibition of metastases after treatment with local radiation and CTLA-4 blockade in a mouse model of breast cancer. Clin. Cancer Res 11, 728–734 (2005). [PubMed] [Google Scholar]
- 27.Krasnova Y, Putz EM, Smyth MJ & Souza-Fonseca-Guimaraes F Bench to bedside: NK cells and control of metastasis. Clin. Immunol 177, 50–59 (2017). [DOI] [PubMed] [Google Scholar]
- 28.de Groot NG, Blokhuis JH, Otting N, Doxiadis GG & Bontrop RE Co-evolution of the MHC class I and KIR gene families in rhesus macaques: ancestry and plasticity. Immunol. Rev 267, 228–245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottcher JP et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1037 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167, 397–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitt JM et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity 44, 1255–1269 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Zaretsky JM et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N. Engl. J. Med 375, 819–829 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binnewies M et al. Unleashing type-2 dendritic cells to drive protective antitumor CD4+ T cell immunity. Cell 177, 556–571 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merad M, Sathe P, Helft J, Miller J & Mortha A The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol 31, 563–604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts EW et al. Critical role for CD103+/CD141+ dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell 30, 324–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broz ML et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 638–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferris ST et al. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature 584, 624–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ataide MA et al. BATF3 programs CD8+ T cell memory. Nat. Immunol 21, 1397–1407 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Hildner K et al. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spranger S, Dai D, Horton B & Gajewski TF Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31, 711–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamazaki C et al. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J. Immunol 190, 6071–6082 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO series accession code GSE181997. Source data are provided with this paper.