Abstract
Cannabinoids, natural or synthetic, have antidepressant, anxiolytic, anticonvulsant, and anti-psychotic properties. Cannabidiol (CBD) and delta-9-tetrahydrocannabinol (Δ9-THC) are the most studied cannabinoids, but recently, attention has turned towards minor cannabinoids. Delta-8-tetrahydrocannabinol (Δ8-THC), an isomer of Δ9-THC, is a compound for which, to date, there is no evidence of its role in the modulation of synaptic pathways. The aim of our work was to evaluate the effects of Δ8-THC on differentiated SH-SY5Y human neuroblastoma cells. Using next generation sequencing (NGS), we investigated whether Δ8-THC could modify the transcriptomic profile of genes involved in synapse functions. Our results showed that Δ8-THC upregulates the expression of genes involved in the glutamatergic pathway and inhibits gene expression at cholinergic synapses. Conversely, Δ8-THC did not modify the transcriptomic profile of genes involved in the GABAergic and dopaminergic pathways.
Keywords: delta-8-tetrahydrocannabinol, SH-SY5Y cells, transcriptomic analysis, glutamatergic synapses, cholinergic synapses, dopaminergic synapses, GABAergic synapses
1. Introduction
Cannabis sativa L. is a dioecious plant belonging to the Cannabaceae family known for its beneficial properties, even if some of its phytocompounds have a psychoactive ac-tion [1,2]. The chemical composition of its phytocompounds can be summarized in carbohydrates, terpenes, amides, amines, phytosterols, fatty acids and their esters, but the most famous are certainly cannabinoids [3].
Several studies have demonstrated the efficacy of cannabinoids in psychiatric and neurodegenerative diseases [1,2].
The most studied cannabinoid is delta-9-tetrahydrocannabinol (Δ9-THC), but the side effects of being psychoactive have led to its limited use. For this reason, researchers have focused their attention on cannabinoids with minor psychoactive effects such as delta-8-tetrahydrocannabinol (Δ8-THC). Δ8-THC is a double bond isomer of Δ9-THC. It derives from the cyclization of cannabidiol (CBD), differing from Δ9-THC for the position of the double bond [4,5].
It has been widely reported that cannabinoids exert their effects by targeting different receptors such as cannabinoid receptor 1 (CB1) and cannabinoid receptor 2 (CB2) included in the endocannabinoid system (ECS) [4].
Both Δ8-THC and Δ9-THC are reported to be partial agonists of CB1. However, the lower affinity of Δ8-THC for CB1 could explain the less psychotropic effect of Δ8-THC [5].
∆8-THC may be associated with beneficial effects, including decreased adverse effects of chemotherapy, decreased tumor cell proliferation, and analgesic properties [6].
Furthermore, in a previous study, our group demonstrated that Δ8-THC can exert protective effects against amyloid-β toxicity in an in vitro model of Alzheimer’s disease [7].
Cannabinoid receptors (CBs) belong to the G protein-coupled receptor family and are widely distributed in the central and peripheral nervous systems. Cannabinoids are compounds that interact with CBs; therefore, they can influence ion channels and synaptic transmission by influencing neurotransmitter release [8,9].
Synaptic transmission and plasticity are important cellular processes that enable the nervous system to process information and respond to changes in the environment and internal environment [10].
In a previous study, we showed that the phytocannabinoids CBD and Cannabigerol (CBG) are able to affect the transcription of genes involved in glutamate, GABA and do-pamine signaling [11].
Glutamate is the primary excitatory neurotransmitter of the central nervous system (CNS) and is involved in several neuronal functions including synaptic transmission, long-term potentiation (LTP), long-term depression (LTD), plasticity and excitability [12,13]. Once released into the synaptic cleft, glutamate can bind to various classes of receptors including glutamatergic receptors (GluRs), which are divided into ionotropic (iGluRs) and metabotropic (mGluRs). N-methyl-d-aspartate receptors (NMDARs), AMPA receptors (AMPARs) and kainate receptors (KARs) are classified among iGluRs that act as mediators in the fast part of excitatory transmission. Instead, mGluRs include G protein coupled receptors (GPCRs) involved both in neuronal plasticity and cognitive functions [14,15].
Through glutamate transmission, cannabinoids could indirectly act on dopaminergic signaling. Dopamine (4-(2-aminoethyl)-1,2-benzenediol) is produced by dopaminergic neurons and regulates several physiological functions such as cognition, mood, sleep, emotion, motor functions, memory and learning [16,17,18].
Cannabinoids are also able to exert their effect on cholinergic and GABAergic synapses [19,20]. Acetylcholine (ACh), one of main neurotransmitters involved in synaptic plasticity, behavior and movement, is hydrolyzed by acetylcholinesterase (AChE) present both in the synaptic cleft and neuromuscular junctions [21].
Instead, gamma-aminobutyric acid (GABA) is an inhibitory neurotransmitter that modulates synaptic transmission and neuronal development, as well as learning and depression [22,23].
The aim of this study was to investigate, using Next Generation Sequencing (NGS) technology, the transcriptomic changes on the synaptic pathways induced by Δ8-THC in all-trans Retinoic acid (RA)-differentiated SH-SY5Y cells.
2. Results
2.1. Differentiation of SH-SY5Y Cells with RA
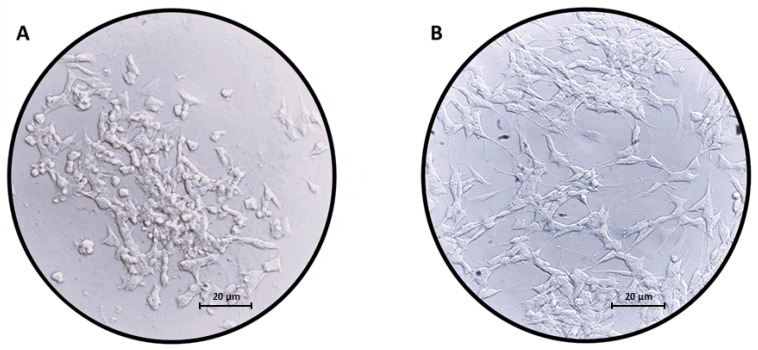
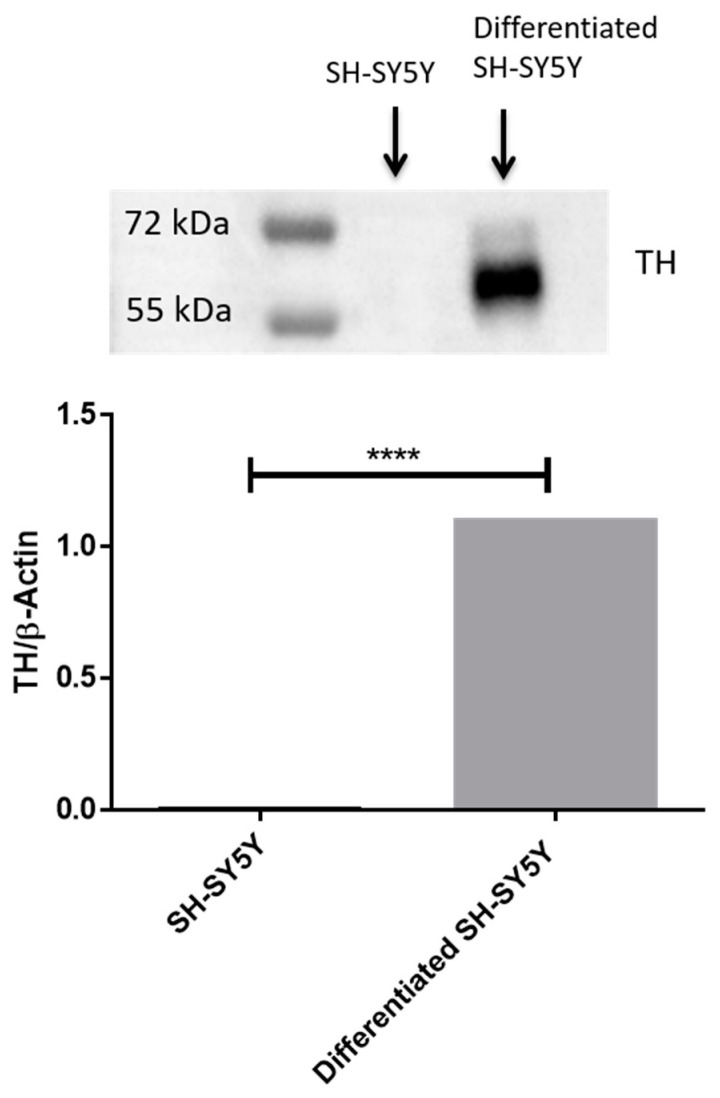
Differentiated SH-SY5Y (Figure 1B) cells acquired a neuronal like morphology compared to undifferentiated cells (Figure 1A). Our results obtained by Western blot analysis show that only RA-differentiated SH-SY5Y cells express TH (Figure 2) compared to undifferentiated cells.
Figure 1.
Images of (A) undifferentiated SH-SY5Y neuroblastoma cells and (B) SH-SY5Y neuroblastoma cells differentiated with RA for 5 days. The differentiation leads to changes in morphology; indeed, SH-SY5Y cells acquire a neuron-like phenotype when treated with RA. Scale bar: 20 µm.
Figure 2.
Western blot for TH. RA treatment caused an increase in TH protein levels. In order to evaluate that blots were loaded with equal amounts of protein lysates; we incubated the blot with an antibody for β-Actin. **** p < 0.0001. The blot for β-Actin is available in the Supplementary Figure S2.
2.2. Transcriptomic Analysis
After treatment of differentiated SH-SY5Y cells with Δ8-THC, we performed a NGS transcriptomic analysis and we focused on differentially expressed genes (DEGs) between Δ8-THC (treated) and CTRL (non-treated) groups.
Δ8-THC exerts its effects on differentiated SH-SY5Y cells, as demonstrated by transcriptomic analysis. In detail, bioinformatics analysis highlighted 8819 DEGs, among which 4307 are upregulated and 4512 are downregulated DEGs. Specifically, upregulated genes are more expressed in the Δ8-THC group, whereas downregulated genes are more expressed in the CTRL group.
We performed a Gene Ontology analysis on the whole transcriptomic using the Panther database (http://pantherdb.org/ (accessed on 5 April 2023)), a web page that allows us to make classification and enrichment tests of DEGs.
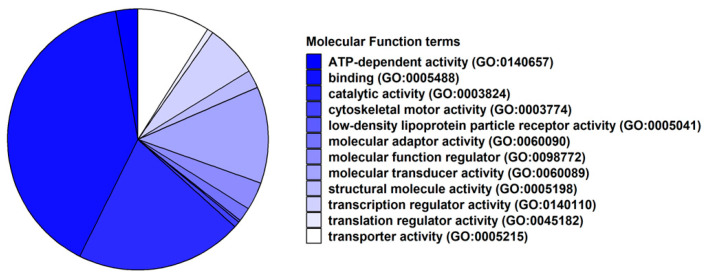
In Figure 3, we inspected how our DEGs are classified on the basis of their Molecular Function terms of the Gene Ontology. It is interesting to note that “ATP-dependent activity” (GO:0140657) and “binding” (GO:0005488) are the most observed terms in the classification.
Figure 3.
Pie chart of the classification of Gene Ontology terms related to Molecular Function. It is interesting to note that Panther classified most of the included DEGs in “ATP-dependent activity” (GO:0140657) and “binding” (GO:0005488) terms.
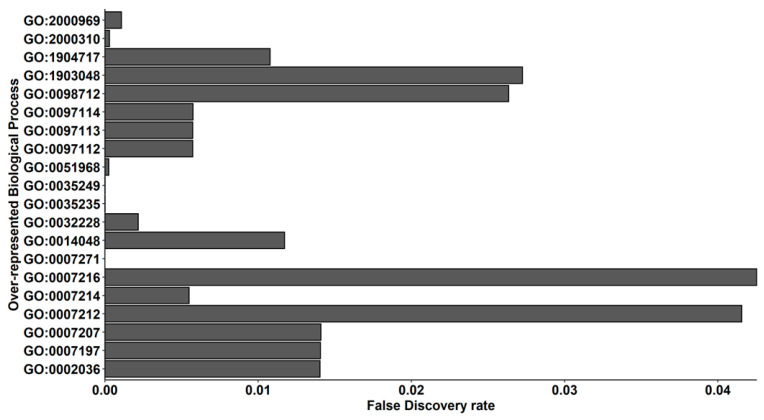
In order to understand the Biological Process term in which our DEGs were expressed in, we also performed an over-representation test of our DEGs that shows the 348 terms in Table S1. Herein, the aim of our work was to observe how Δ8-THC is able to modulate synaptic pathways. For this reason, in Figure 4, we plotted the 20 terms that are related to synaptic activity. Of note, most terms are related to the glutamatergic pathway: “NMDA glutamate receptor clustering” (GO:0097114), “regulation of NMDA receptor activity” (GO:2000310), “AMPA glutamate receptor clustering” (GO:0097113), “positive regulation of AMPA receptor activity” (GO:2000969), “regulation of AMPA glutamate receptor clustering” (GO:1904717), “gamma-aminobutyric acid receptor clustering” (GO:0097112), “regulation of L-glutamate import across plasma membrane” (GO:0002036), “ionotropic glutamate receptor signaling pathway” (GO:0035235), “L-glutamate import across plasma membrane” (GO:0098712), “G protein-coupled glutamate receptor signaling pathway” (GO:0007216), “regulation of glutamate secretion” (GO:0014048), “positive regulation of synaptic transmission, glutamatergic” (GO:0051968), “synaptic transmission, glutamatergic” (GO:0035249), “gamma-aminobutyric acid signaling pathway” (GO:0007214), “regulation of synaptic transmission, GABAergic” (GO:0032228). Remaining terms are associated to cholinergic terms: “regulation of acetylcholine-gated cation channel activity” (GO:1903048), “phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway” (GO:0007207), “adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway” (GO:0007197), “synaptic transmission, cholinergic” (GO:0007271). Only “dopamine receptor signaling pathway” (GO:0007212) is linked to dopaminergic terms.
Figure 4.
Barplot of over-represented terms in the Biological Process category of Gene Ontology related to synapses. On the x-axis, the False Discovery Rate for each term in the y-axis is represented. Even if acetylcholine (GO:1903048, GO:0007207, GO:0007197, GO:0007271) and dopaminergic (GO:0007212) terms are included, most of the terms (GO:0097114, GO:2000310, GO:0097113, GO:2000969, GO:1904717, GO:0097112, GO:0002036, GO:0035235, GO:0098712, GO:0007216, GO:0014048, GO:0051968, GO:0035249, GO:0007214, GO:0032228) are associated to glutamatergic synapsis.
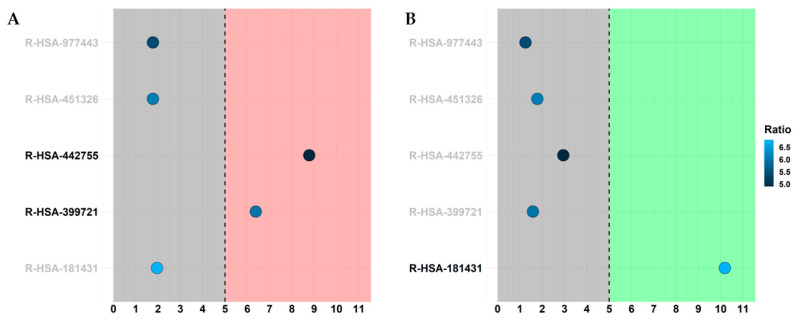
Next, we took advantage of the Amigo2 project. Specifically, Amigo2 is a web page (http://amigo.geneontology.org/amigo (accessed on 5 April 2023)) that allows us to perform queries in the Gene Ontology database that, in turn, collects all the knowledge about genes characterized for biological process terms. For this reason, we used Amigo2 to retrieve all the genes that are included in the biological process term “nervous system process” (GO:0050877) and we kept all DEGs that fall in the ontology. In detail, 366 DEGs are in the GO:0050877, among which 168 are upregulated and 198 are downregulated. Thus, we searched in the Reactome database, in which the sub-pathways of “Neuronal System” (R-HSA-112316) were enriched. Indeed, Reactome returns the False Discovery Rate (FDR) for each pathway, showing the significance of the pathway itself. We found that the only enriched pathway (FDR < 0.05) was “Neurotransmitter receptors and postsynaptic signal transmission” (R-HSA-112314). Thus, we inspected all the sub-pathways: “Activation of NMDA receptors and postsynaptic events” (R-HSA-442755), “Glutamate binding, activation of AMPA receptors and synaptic plasticity” (R-HSA-399721), “Acetylcholine binding and downstream events” (R-HSA-181431), “GABA receptor activation” (R-HSA-977443), “Activation of kainate receptors upon glutamate binding” (R-HSA-451326). In detail, in both figures, each sub-pathway in the y axis is highlighted by its significance in the x-axis with a score computed by −log2(FDR). In this line, only the pathways in the red or green regions that have a score higher than 5 are significative because they have an FDR lower than 0.05, and they are highlighted in bold. Additionally, each pathway was filled by a color palette that shows the ratio of DEGs we found in the pathway over the total number of entities that define that pathway in Reactome. In particular, we define a score computed as -log2(entities ratio) so that pathways with brighter filler have more DEGs in spite of the number of entities in the pathway itself. Thus, in Figure 5A, the only enriched pathways are glutamatergic pathways related to NMDA (R-HSA-442755) and AMPA (R-HSA-399721) so that they are upregulated. In contrast, in Figure 5B, the “Acetylcholine binding and downstream event” (R-HSA-181431) pathway is the only enriched and downregulated pathway. No significance was observed for GABAergic synapses. Of note, not even pathways related to dopaminergic synapses were enriched, but it is not included in the plot because it is not included in any enriched pathway mentioned above. All the data obtained by Reactome that were plotted are in Table S2.
Figure 5.
Bubbleplot of the Reactome pathways for glutamatergic, cholinergic and gabaergic synapses. The x-axis represents a score obtained by −log2(FDR), so that pathways with score higher than 5 are significantly upregulated (red region in panel (A)) or downregulated (green region in panel (B)). Each point is filled with the color obtained with a score computed as −log2(entities ratio).
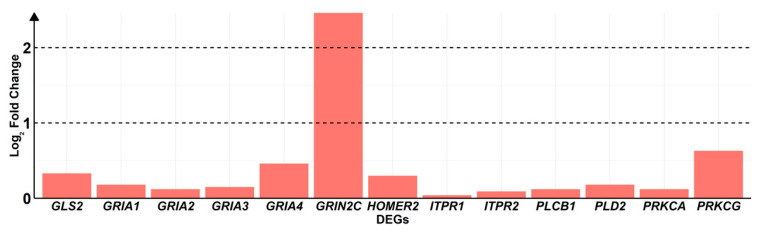
Particularly, we focus our attention on the KEGG “Glutamatergic synapse pathway” (hsa04724). As shown in Figure 6, Δ8-THC upregulates the following 13 genes: GLS2, GRIA1, GRIA2, GRIA3, GRIA4, GRIN2C, HOMER2, ITPR1, ITPR2, PLCB1, PLD2, PRKCA, PRKCG.
Figure 6.
Barplot of DEGs highlighted in KEGG “Glutamatergic synapse pathway” (hsa04724). Each bar in the x-axis highlights the order of magnitude of log2 fold changes obtained by log2(∆8-THC/CTRL) in the y-axis. All DEGs taken into consideration are upregulated; thus, they are more expressed in ∆8-THC than CTRL.
In this line, Table 1 highlights that Δ8-THC is able to modulate transcripts that are associated with the translation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPA receptor) (GRIA1, GRIA2, GRIA3, GRIA), N-methyl-D-aspartate receptor (NMDA receptor) (GRIN2C), IP3R protein involved in calcium signaling (ITPR1 and ITPR2), and genes involved in the downstream of metabotropic glutamate receptors (mGluR) activity (PLCB1, PLD2, PRKCA, PRKCG).
Table 1.
Differentially expressed genes involved in KEGG Glutamatergic synapse pathway (hsa04724).
| Gene | CTRL | Δ8-THC | log2 Fold Change | q-Value | Protein |
|---|---|---|---|---|---|
| GLS2 | 120.39 | 151.17 | 0.33 | 2.38 × 10−2 | GLS |
| GRIA1 | 494.82 | 561.49 | 0.18 | 1.19 × 10−2 | AMPA receptor |
| GRIA2 | 1928.70 | 2098.12 | 0.12 | 8.32 × 10−4 | |
| GRIA3 | 1727.64 | 1915.39 | 0.15 | 8.36 × 10−5 | |
| GRIA4 | 541.77 | 745.06 | 0.46 | 1.46 × 10−13 | |
| GRIN2C | 2.41 | 13.29 | 2.46 | 4.22 × 10−3 | NMDA receptor |
| HOMER2 | 816.27 | 1007.53 | 0.30 | 6.95 × 10−9 | Homer |
| SHANK3 | 1790.25 | 1910.40 | 0.09 | 1.52 × 10−2 | SHANK |
| ITPR1 | 9393.09 | 9664.99 | 0.04 | 1.53 × 10−2 | IP3R |
| ITPR2 | 1542.24 | 1645.44 | 0.09 | 2.57 × 10−2 | |
| PLCB1 | 1167.82 | 1269.17 | 0.12 | 1.16 × 10−2 | PLC |
| PLD2 | 1390.54 | 1577.33 | 0.18 | 1.25 × 10−5 | PLD |
| PRKCA | 8513.01 | 9226.42 | 0.12 | 1.58 × 10−12 | PKC |
| PRKCG | 87.89 | 136.22 | 0.63 | 6.35 × 10−5 |
For each DEG, the difference in the level of expression computed by log2(∆8-THC/CTRL) results in the log2 fold change. The post hoc correction of the p-value is highlighted in q-Value column. All values were rounded to the second decimal digit.
In order to highlight that the role of the glutamatergic pathway is biologically considerable in spite of gabaergic, cholinergic and dopaminergic ones, Table 2 highlights key DEGs of these synapses.
Table 2.
Differentially expressed genes involved in gabaergic, cholinergic or dopaminergic synapsis.
| Gene | CTRL | Δ8-THC | log2 Fold Change | q-Value | Synapsis |
|---|---|---|---|---|---|
| GABRA3 | 551.40 | 616.31 | 0.16 | 2.03 × 10−2 | Gabaergic |
| GABRG3 | 50.57 | 98.84 | 0.97 | 1.12 × 10−6 | Gabaergic |
| GABRR2 | 30.10 | 17.44 | −0.79 | 2.27 × 10−2 | Gabaergic |
| CHRNA3 | 3521.51 | 3364.80 | −0.07 | 2.01 × 10−2 | Cholinergic |
| CHRNA6 | 15.65 | 1.66 | −3.24 | 1.60 × 10−4 | Cholinergic |
| CHRNA7 | 2073.17 | 1844.79 | −0.17 | 2.26 × 10−6 | Cholinergic |
| CHRNB2 | 9219.72 | 8367.57 | −0.14 | 5.31 × 10−18 | Cholinergic |
| PRKACB | 3247.01 | 3042.53 | −0.09 | 1.17 × 10−3 | Cholinergic |
| MAPK1 | 1856.47 | 1716.04 | −0.11 | 3.28 × 10−3 | Cholinergic |
| CREB3L3 | 13.24 | 4.98 | −1.41 | 1.67 × 10−2 | Cholinergic |
| CREB3L4 | 1438.70 | 1281.63 | −0.17 | 1.24 × 10−4 | Cholinergic |
| CREB5 | 2677.55 | 2544.16 | −0.07 | 2.34 × 10−2 | Cholinergic |
| SLC18A3 | 280.52 | 213.47 | −0.39 | 1.11 × 10−4 | Cholinergic |
| DRD4 | 108.35 | 54.82 | −0.98 | 3.20 × 10−8 | Dopaminergic |
| CALY | 7.22 | 1.66 | −2.12 | 2.62 × 10−2 | Dopaminergic |
| DDC | 13,066.29 | 12,497.37 | −0.06 | 4.46 × 10−6 | Dopaminergic |
For each DEG, the difference in the level of expression computed by log2(∆8-THC/CTRL) results in the log2 fold change. The post hoc correction of the p-value is highlighted in q-Value column. All values were rounded to the second decimal digit.
3. Discussion
In our study, we used RA-differentiated SH-SY5Y cells and treated them with (20 µM) Δ8-THC. The aim was to verify, through transcriptomic analysis, whether this cannabinoid modifies the expression of genes belonging to the synaptic pathways.
RA has powerful growth-inhibiting and cellular differentiation-promoting properties [24,25].
As shown in Figure 1, the SH-SY5Y cells treated with RA assumed a neuronal phenotype compared to the undifferentiated ones. Furthermore, to support the differentiation process, we analyzed the expression levels of the protein tyrosine hydroxylase (TH). This protein is a neuronal enzyme that catalyzes the conversion of the amino acid L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA) [26,27]. As shown in Figure 2 (Figure S1), TH was expressed only in RA-treated SH-SY5Y and not in the control, thus indicating that SH-SY5Y cells are differentiated. As housekeeping protein we have used β-Actin (Figure S2).
In a previous paper, we demonstrated that Δ8-THC showed no toxicity in a range of concentrations 5–20 µM. Moreover, the dose 20 µM Δ8-THC was able to restore cell viability after 10 µM Amyloid Beta treatment [7], suggesting that Δ8-THC may exert important neuroprotective effects. Considering this background, we studied how Δ8-THC affects the transcriptomic profile of genes involved in synaptic pathways, since there is no documented evidence about its role on the modulation of synaptic pathways.
Chemical signals are transferred between neurons in the nervous system through the neuro-transmitters released at presynaptic sites, followed by the diffusion across the synaptic boundaries, and the activation of post-synaptic receptors. There are several ways of communication among neurons, regulated by the release of specific neurotransmitters and subsequent responses given by their specific receptors in the post-synaptic neurons. As was previously reported in the literature, exposure to cannabinoids affects synaptic pathways, changing the outcome of various neurobiological signals [11,28,29,30,31]. For this reason, in this study, we wanted to focus our attention on the effect of Δ8-THC on the transcriptomic profile of genes involved in synaptic pathways related to synaptic transmission.
At first, we performed a GO analysis and classification of molecular functions and biological processes. Notably, “ATP-dependent activity” and “binding” were the most observed molecular functions (Figure 3 and Figure 4 and Tables S1 and S2). This result can be explained by the neuronal physiology, given that they represent high energy-demanding cells, consuming about 20% of the resting energy of the body.
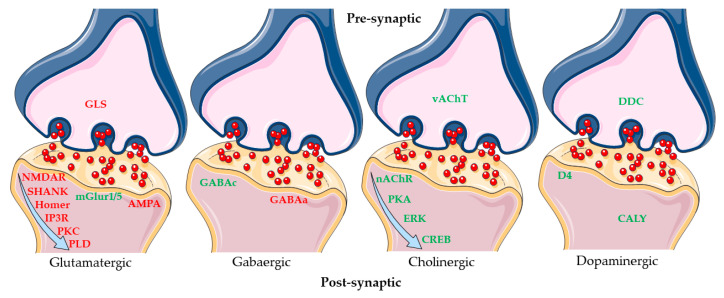
Our transcriptomic analysis demonstrated that Δ8-THC influenced several genes involved in “Glutamatergic Synapse” and “Cholinergic synapse”. Instead, it is not able to exert an action on “GABAergic synapse” and “Dopaminergic synapse” (Figure 7).
Figure 7.
Mechanism modulated in SH-SY5Y cells differentiated with RA after Δ8-THC treatment. The figure represents how the different synapses are influenced by Δ8-THC treatment and the genes that take place. The red genes are upregulated; the green genes are downregulated. The results of transcriptomic analysis suggest that Δ8-THC influenced the Glutamatergic synapse more than others synapse types. The figure was created using Servier Medical Art by Servier (http://smart.servier.com/ (accessed on 9 April 2023)), licensed under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/ (accessed on 9 April 2023)).
Glutamate and GABA are important neurotransmitters in the excitatory and inhibitory balance that neurons exhibit [32] because many aspects of the physiological brain function, such as cognition memory and learning, are directly or indirectly affected by them [18,33,34,35].
Our analysis showed that Δ8-THC influenced the expression of genes involved in glutamatergic pathways related to NMDA (R-HSA-442755) and AMPA (R-HSA-399721) but not in the kainate receptor (R-HSA-451326) signaling (Figure 5). In particular, Δ8-THC promoted the expression of several genes of Glutamatergic Synapse pathway such as GLS2, GRIA1, GRIA2, GRIA3, GRIA4, GRIN2C, SHANK3, HOMER, ITPR1, ITPR2 (Table 1 and Figure 6).
Glutaminase 2, encoded by the GLS2 gene, is an enzyme belonging to the glutaminase family. This protein catalyzes the synthesis of glutamate by converting glutamine inside the mitochondrion. Once glutamate is released from presynaptic neurons, the neuro-transmitter predominantly binds AMPA and NMDA receptors. These are the main receptors of glutamatergic synapse and they are important for plasticity and neuronal transmission [36,37]. AMPA receptors are predominantly expressed in postsynaptic neurons and can be composed by four different types of subunits (from GluA1 to GluA4) [14]. Our transcriptomic analysis showed that Δ8-THC induced the upregulation of gene encoding for all the subunits of AMPA receptors (GRIA1, GRIA2, GRIA3, GRIA4). The AMPA receptor is important for memory, thus the deletion of a single AMPA receptor subunit, such as GluA1, results in alteration of the short-term memory [38].
Furthermore, we have also shown that NMDA receptor subunit epsilon-3 (GRIN2C) expression is influenced by Δ8-THC. This receptor is permeable to sodium and potassium ions and also to calcium, an important second messenger, known as an activator of several forms of intracellular signaling [39]. AMPA and NMDA receptors interact with each other during neurotransmission; in fact, the NMDA channel is activated due to release of Mg2+ ions blockade, which occurs after cation influx into the neuron via the AMPA receptor activity [40]. Considering these results, we suppose that Δ8-THC could modulate the AMPA and NMDA functions in the glutamatergic activity.
Moreover, we discovered up-regulation of other important genes involved in AMPA and NMDA signaling. SHANK3 and HOMER proteins, which are respectively encoded by SHANK3 and HOMER, are scaffold proteins that permit the co-localization of AMPA and NMDA receptors in post-synaptic neurons, also known as postsynaptic density (PSD), to induce neuronal depolarization [41,42]. The SHANK3 gene is involved in glutamatergic synaptic transmission and long-term potentiation in the hippocampus [43]. HOMER contributes to form a physical link among signaling molecules in glutamatergics. PSD also interacts with inositol triphosphate receptor (IP3R) and mGluRs [44,45]. IP3R is a receptor involved in calcium signaling, which is important for the glutamatergic synapse [46]. Interestingly, our results showed an upregulation of ITPR1 and ITPR2 genes, both encoding for IP3R. Indeed, SHANK3, HOMER and IP3R upregulation support the hypothesis that there is an activation of the glutamatergic synapse after Δ8-THC treatment.
Furthermore, another set of genes that were upregulated in our analysis are PLCB1, PRKCA, PRKCG, PLD2 (Table 1 and Figure 6). The proteins encoded by these genes are involved in mGluRs signaling in glutamatergic synapses. In fact, when activated, the beta-type phospholipase C (PLC-β) enzymes (PLCB1) [47] synthetize diacyl glycerol (DAG) and inositol trisphosphate (IP3) through the hydrolyzation of phosphatidylinositol 4,5-bisphosphate (PIP2). DAG and IP3R activity are important for Protein-kinase C (PKC), encoded by PRKCA and PRKCG. PKC plays an important role in the regulation of phospholipase D (PLD) activity [48], which is encoded by PLD2 and in turn synthesizes Phosphatidic acid (PtdOH), an anionic phospholipid involved in neuro-transmission [49]. These results also suggest that the mGluRs pathway could be influenced by Δ8-THC.
Furthermore, our findings suggest how Δ8-THC does not exert an action on the other types of synapses, or it is able to induce inhibitory effects. Unlike the GLS2 gene, our analyses do not show a deregulation of the GAD gene, encoding for glutamic acid decarboxylase (GAD), which plays a role in the synthesis of GABA from glutamate [50].
There are several GABA receptors, such as GABAA, GABAB and GABAC [51,52]. GABAA is an ionotropic receptor, composed of pentameric combinations of different subunits [53]. Using KEGG and inspecting the “Glutamatergic synapse pathway”, there are 16 genes encoding for GABAA subunits, but only two were deregulated in our analysis (GABRA3, GABRG3). On the counterpart, our analyses have highlighted that GABRR2 gene, encoding for a subunit of GABAC, is downregulated while the genes encoding for GABAB are not influenced by Δ8-THC. As can be seen in Figure 5 (R-HSA-977443), these results suggest that Δ8-THC does not affect the GABAergic synapse pathway and that there could be a higher concentration of glutamate in treated cells than in the control. For these reasons, we suppose that Δ8-THC treatment could induce stronger glutamatergic action compared to the inhibitory action of GABA.
Our results suggest that Δ8-THC exerts an inhibitory action in “Acetylcholine binding and downstream events” (R-HSA-181431) (Figure 5). Especially, in our work, we show that Δ8-THC treatment inhibits the transcription of genes involved in cholinergic synapse pathways such as SLC18A3, CHRNA3, CHRNA6, CHRNA7, CHRNB2, MAPK1, CREB3L3, CREB3L4, CREB5, DDC, DDR4 and CALY (Table 2).
Ach is a neurotransmitter that supports cognitive functions and activates muscle activity and contraction. Moreover, it is the major neurotransmitter in the autonomic nervous system. Neurons that are able to secrete acetylcholine are known as cholinergic [54]. Once this neurotransmitter is released into the synaptic cleft, it is able to bind the Nicotinic acetylcholine receptors (nAChRs) [55].
We noticed that the SLC18A3 gene, which encodes for the vesicular acetylcholine transporter (VAChT), is downregulated. Its role in the presynaptic terminal is related to the transport of acetylcholine (Ach) into vesicles for eventual release into synapses [56].
Its downregulation supports the idea that there is a reduction of acetylcholine release in the synaptic cleft.
Our analysis showed that the genes encoding for nAChRs subunits (CHRNA3, CHRNA6, CHRNA7 and CHRNB2) are mostly downregulated. These receptors are composed of five subunits, arranged around a water-filled pore [57]. nAChRs are permeable to small monovalent and divalent cations, such as sodium and calcium. The latter generates specific and complex signals involving adenylyl cyclase and protein kinase A (PKA) [58,59]. The subsequent activation of PKA determines the activation of ERK (MAPK1), which exerts downstream signaling through CREBs (CREB3L3, CREB3L4, CREB5). ERK plays a role in a number of cellular processes, including cholinergic synapse pathways [59,60]. The downregulation of genes encoding for these proteins suggest once again that the Δ8-THC treatment could have an inhibitory action on cholinergic synapse activity.
In addition, our results showed that Δ8-THC treatment does not have an effect on dopaminergic synapses. Dopamine is a neurotransmitter belonging to the catecholamine family. Among the functions that are controls, there are: movement, working memory, pleasure, reward, sleep regulation and cognitive functions [61]. DOPA decarboxylase (DDC) is a lyase enzyme that is able to regulate the transformation of L-DOPA to dopamine [62]. The DDC gene, which encodes for DDC, is downregulated in our analyses, suggesting a low dopamine production. Moreover, in order to exert its action, dopamine interacts with its receptors, which are G protein-coupled receptors [63]. Table 2 shows the DRD4 gene, encoding for Dopamine receptor D4 (DRD4), which is down-regulated, thus supporting the hypothesis that dopaminergic activity is low in treated cells. Except DRD4, in our analyses, there are no deregulations of other genes encoding for dopamine receptors. Moreover, our results show downregulation of the gene expression of CALY, encoding for Neuron-specific vesicular protein calcyon (Calcyon). Calcyon plays an important role in DRD1 signaling and it is involved in neuropsychiatric disorders such as schizophrenia, bipolar disorder and attention disorder [64]. These results suggest that Δ8-THC treatment does not increase dopaminergic synapse activity. Instead, it could inhibit dopaminergic synapse activity through inhibition of dopaminergic receptor expression.
In conclusion, our analysis demonstrated that Δ8-THC is able to influence the transcriptomic profile of genes involved in synaptic functionality.
4. Materials and Methods
4.1. Synthesis and Purification of Δ8-THC
All details about synthesis and purification were already published by Gugliandolo, A et al. [7]. Supplementary Materials presented all the details.
4.2. Cell Culture, Differentiation and Treatment with Δ8-THC
The SH-SY5Y cell line was acquired from American Type Culture Collection (ATCC) (Manassas, VA, USA). We grew the human neuroblastoma cell line SH-SY5Y in a monolayer at 37 °C in a 5% CO2 humidified atmosphere using Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham (DMEM/F12) medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich), 1% glutamine and 1% penicillin-streptomycin (100 U-100 µg/mL). To induce neuronal differentiation of SH-SY5Y cells, we incubated them for 5 days with 10 µM of all-trans RA (Sigma-Aldrich) [7,65]. We dissolved Δ8-THC in DMSO and then diluted in PBS. We added it in the medium at the final concentration (the final DMSO concentration was <0.1%). We treated the differentiated SH-SY5Y cells with 20 µM Δ8-THC for 24 h.
4.3. Western Blot Analysis for Tyrosine Hydroxylase
Proteins were extracted from undifferentiated and RA-differentiated SH-SY5Y cells using RIPA Buffer and quantified using Bradford Assay (Bio-Rad, Hercules, CA, USA). After denaturation at 95 °C, 25 µg of protein for each sample was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto PVDF membranes (Immobilon–P, Millipore, Burlington, MA, USA). Membranes were blocked with 5% skimmed milk in TBS for 1 h. The blots were incubated at 4 °C overnight with Anti-Tyrosine Hydroxylase (1:1000, Millipore, Temecula, CA, USA) and Anti-β-Actin (1:1000, Santa Cruz Biotechnology, Dallas, TX, USA). The blots were incubated for 1 h at room temperature with the following secondary antibodies: mouse anti-rabbit IgG-HRP (1:850, Santa Cruz Biotechnology, Dallas, TX, USA) and Chicken anti-Mouse IgG Secondary Antibody, HRP (1:1000, ThermoFisher, Rockgord, IL, USA). ChemiDocTM MP System (Bio-Rad) acquired bands after exposure to an enhanced chemiluminescence system (Luminata Western HRP Substrates, Millipore, Burlington, MA, USA).
4.4. RNA Extraction and Library Preparation
Cells were harvested and RNA was extracted using Maxwell® RSC simplyRNA Cells Kit (Promega, Madison, WI, USA) following the protocol. The library was prepared using the TruSeq RNA Exome protocol (Illumina, San Diego, CA, USA) following the manufacturer’s instruction. Experiments were performed in triplicate.
4.5. Sequencing Analysis
We used the fastqc tool version 0.11.4 from the Babraham Institute (Cambridge, UK) to evaluate the raw data obtained from the NextSeq 550 Dx instrument of Illumina. We then eliminated adapters and low-quality bases with Trimmomatic version 0.38 (Usadel Lab, Aachen, Germany) [66]. In detail, we used the paired-end option (PE), the -phred33 encoding, 2 seed mismatches, 30 palindrome clips and 10 simple clip thresholds for the ILLUMINACLIP, 20 of both for LEADING and TRAILING, 4 in window size and 15 for the quality of the window in SLIDINGWINDOW, with 75 minimum length. In the next step, we aligned the cleaned reads to the human reference genome (GRCh38) with the STAR RNA-seq aligner 2.7.3a (New York, NY, USA) [67]. We used the primary assembly annotation file v39 of gencode for -sjdbGTFfile option, the -outFilterIntronMotifs RemoveNoncanonical and the -quantMode GeneCounts. In the last step, we computed the expression levels of the transcripts with the htseq-count python package version 0.6.1p1 (European Molecular Biology Laboratory (EMBL), Heidelberg, Germany) [68] using the -s reverse option.
4.6. Comparative Analysis and In Silico Inspection
Once the expression was obtained, we identified differentially expressed genes (DEGs) with the DESeq2 library in R version 3.6.3 (R Core Team) [69]. The full script (script.R) is included the supplementary materials. In detail, we performed a Wald test for each gene by DESeq2, returning a p-value. Because of the multiple Wald tests performed, we defined the q-value as the p-value corrected by the Benjamini–Hochberg procedure to drop the number of false positives. Thus, a gene was marked as DEG only if the q-value was lower than 0.05. Panther web page (current release 17.0) was used to perform classification and enrichment analysis with default parameters [70]. From Amigo2, we retrieved all protein coding genes included in the “nervous system process” (GO:0050877). Then, DEGs in the GO:0050877 were enriched for pathways using Reactome web page database. In detail, we used the analysis tool including default parameters. Thus, we kept the “project to human” option, allowing us to include information related to association coming from other species about orthologous genes in the analysis; we did not include interactors. We define all pathways with a p-value corrected with the Benjamini–Hochberg post-hoc procedure lower than 0.05 as significative. Then, based on the aim of our work, we inspected the “Neurotransmitter receptors and postsynaptic signal transmission” (R-HSA-112314) pathway included in the “Neuronal System” category [71,72].
5. Conclusions
Our analysis shows that Δ8-THC is able to upregulate the expression of genes involved in glutamate signaling involving AMPA and NMDA. In parallel, treatment with Δ8-THC reduces the expression of genes belonging to the cholinergic synapse pathway and it do not seem to have effect on the expression of genes involved in GABAergic and dopaminergic signaling. Based on our transcriptomic results, Δ8-THC could be considered as a potential compound that is useful to improve glutamatergic transmission in neurons. In this regard, further studies are needed to validate these results.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24119486/s1. References [7,73,74] are cited in Supplementary Materials.
Author Contributions
Conceptualization, E.M.; methodology, A.G.; software, L.C.; validation, E.M.; formal analysis, L.C.; investigation, I.A. and G.S.; resources, F.P., S.S. and D.C.; data curation, L.C.; writing—original draft preparation, I.A. and G.S.; writing—review and editing, E.M.; visualization, L.C.; supervision, E.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in the NCBI Sequence Read Archive at BioProject accession numbers PRJNA973106.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by Ministry of Health, Italy, Current Research Funds 2023.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Pellati F., Borgonetti V., Brighenti V., Biagi M., Benvenuti S., Corsi L. Cannabis sativa L. and Nonpsychoactive Cannabinoids: Their Chemistry and Role against Oxidative Stress, Inflammation, and Cancer. BioMed Res. Int. 2018;2018:1691428. doi: 10.1155/2018/1691428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pertwee R.G. Pharmacological actions of Cannabinoids. Handb. Exp. Pharmacol. 2005;168:1–51. doi: 10.1007/3-540-26573-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Appendino G., Chianese G., Taglialatela-Scafati O. Cannabinoids: Occurrence and medicinal chemistry. Curr. Med. Chem. 2011;18:1085–1099. doi: 10.2174/092986711794940888. [DOI] [PubMed] [Google Scholar]
- 4.Hanus L.O., Meyer S.M., Munoz E., Taglialatela-Scafati O., Appendino G. Phytocannabinoids: A unified critical inventory. Nat. Prod. Rep. 2016;33:1357–1392. doi: 10.1039/C6NP00074F. [DOI] [PubMed] [Google Scholar]
- 5.Tagen M., Klumpers L.E. Review of delta-8-tetrahydrocannabinol (Δ8-THC): Comparative pharmacology with Δ9-THC. Br. J. Pharmacol. 2022;179:3915–3933. doi: 10.1111/bph.15865. [DOI] [PubMed] [Google Scholar]
- 6.LoParco C.R., Rossheim M.E., Walters S.T., Zhou Z., Olsson S., Sussman S.Y. Delta-8 tetrahydrocannabinol: A scoping review and commentary. Addiction. 2023;118:1011–1028. doi: 10.1111/add.16142. [DOI] [PubMed] [Google Scholar]
- 7.Gugliandolo A., Blando S., Salamone S., Caprioglio D., Pollastro F., Mazzon E., Chiricosta L. Delta(8)-THC Protects against Amyloid Beta Toxicity Modulating ER Stress In Vitro: A Transcriptomic Analysis. Int. J. Mol. Sci. 2023;24:6598. doi: 10.3390/ijms24076598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo B., Schlicker E. Effects of Cannabinoids on neurotransmission. Handb. Exp. Pharmacol. 2005;168:327–365. doi: 10.1007/3-540-26573-2_11. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman A.F., Lupica C.R. Synaptic targets of Delta9-tetrahydrocannabinol in the central nervous system. Cold Spring Harb. Perspect. Med. 2013;3:a039743. doi: 10.1101/cshperspect.a012237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennedy M.B. Synaptic Signaling in Learning and Memory. Cold Spring Harb. Perspect. Biol. 2013;8:a016824. doi: 10.1101/cshperspect.a016824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gugliandolo A., Silvestro S., Chiricosta L., Pollastro F., Bramanti P., Mazzon E. The Transcriptomic Analysis of NSC-34 Motor Neuron-Like Cells Reveals That Cannabigerol Influences Synaptic Pathways: A Comparative Study with Cannabidiol. Life. 2020;10:227. doi: 10.3390/life10100227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moretto E., Murru L., Martano G., Sassone J., Passafaro M. Glutamatergic synapses in neurodevelopmental disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;84:328–342. doi: 10.1016/j.pnpbp.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Pittenger C., Bloch M.H., Williams K. Glutamate abnormalities in obsessive compulsive disorder: Neurobiology, pathophysiology, and treatment. Pharmacol. Ther. 2011;132:314–332. doi: 10.1016/j.pharmthera.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., Dingledine R. Glutamate receptor ion channels: Structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miladinovic T., Nashed M.G., Singh G. Overview of Glutamatergic Dysregulation in Central Pathologies. Biomolecules. 2015;5:3112–3141. doi: 10.3390/biom5043112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi K. Role of catecholamine signaling in brain and nervous system functions: New insights from mouse molecular genetic study. J. Investig. Dermatol. Symp. Proc. 2001;6:115–121. doi: 10.1046/j.0022-202x.2001.00011.x. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Ruiz J., Hernandez M., Ramos J.A. Cannabinoid-dopamine interaction in the pathophysiology and treatment of CNS disorders. CNS Neurosci. Ther. 2010;16:e72–e91. doi: 10.1111/j.1755-5949.2010.00144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teleanu R.I., Niculescu A.G., Roza E., Vladacenco O., Grumezescu A.M., Teleanu D.M. Neurotransmitters-Key Factors in Neurological and Neurodegenerative Disorders of the Central Nervous System. Int. J. Mol. Sci. 2022;23:5954. doi: 10.3390/ijms23115954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domino E.F. Cannabinoids and the cholinergic system. J. Clin. Pharmacol. 1981;21:249S–255S. doi: 10.1002/j.1552-4604.1981.tb02602.x. [DOI] [PubMed] [Google Scholar]
- 20.Vanin A.P., Tamagno W.A., Alves C., Mesacasa L., Santin L.F., Sutorillo N.T., Bilibio D., Muller C., Galon L., Kaizer R.R. Neuroprotective potential of Cannabis sativa-based oils in Caenorhabditis elegans. Sci. Rep. 2022;12:15376. doi: 10.1038/s41598-022-19598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pundir C.S., Chauhan N. Acetylcholinesterase inhibition-based biosensors for pesticide determination: A review. Anal. Biochem. 2012;429:19–31. doi: 10.1016/j.ab.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Chattopadhyaya B. Molecular mechanisms underlying activity-dependent GABAergic synapse development and plasticity and its implications for neurodevelopmental disorders. Neural Plast. 2011;2011:734231. doi: 10.1155/2011/734231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu W., Bromley-Coolidge S., Li J. Regulation of GABAergic synapse development by postsynaptic membrane proteins. Brain Res. Bull. 2017;129:30–42. doi: 10.1016/j.brainresbull.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotan R. Retinoids in cancer chemoprevention. FASEB J. 1996;10:1031–1039. doi: 10.1096/fasebj.10.9.8801164. [DOI] [PubMed] [Google Scholar]
- 25.Melino G., Thiele C.J., Knight R.A., Piacentini M. Retinoids and the control of growth/death decisions in human neuroblastoma cell lines. J. Neuro-Oncol. 1997;31:65–83. doi: 10.1023/A:1005733430435. [DOI] [PubMed] [Google Scholar]
- 26.Khwanraj K., Phruksaniyom C., Madlah S., Dharmasaroja P. Differential Expression of Tyrosine Hydroxylase Protein and Apoptosis-Related Genes in Differentiated and Undifferentiated SH-SY5Y Neuroblastoma Cells Treated with MPP+ Neurol. Res. Int. 2015;2015:734703. doi: 10.1155/2015/734703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daubner S.C., Le T., Wang S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011;508:1–12. doi: 10.1016/j.abb.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerdeman G., Lovinger D.M. CB1 cannabinoid receptor inhibits synaptic release of glutamate in rat dorsolateral striatum. J. Neurophysiol. 2001;85:468–471. doi: 10.1152/jn.2001.85.1.468. [DOI] [PubMed] [Google Scholar]
- 29.Morera-Herreras T., Ruiz-Ortega J.A., Ugedo L. Two opposite effects of Delta(9)-tetrahydrocannabinol on subthalamic nucleus neuron activity: Involvement of GABAergic and glutamatergic neurotransmission. Synapse. 2010;64:20–29. doi: 10.1002/syn.20701. [DOI] [PubMed] [Google Scholar]
- 30.Kelley B.G., Thayer S.A. Delta 9-tetrahydrocannabinol antagonizes endocannabinoid modulation of synaptic transmission between hippocampal neurons in culture. Neuropharmacology. 2004;46:709–715. doi: 10.1016/j.neuropharm.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 31.Bara A., Ferland J.N., Rompala G., Szutorisz H., Hurd Y.L. Cannabis and synaptic reprogramming of the developing brain. Nat. Rev. Neurosci. 2021;22:423–438. doi: 10.1038/s41583-021-00465-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petroff O.A. GABA and glutamate in the human brain. Neuroscientist. 2002;8:562–573. doi: 10.1177/1073858402238515. [DOI] [PubMed] [Google Scholar]
- 33.Filpa V., Moro E., Protasoni M., Crema F., Frigo G., Giaroni C. Role of glutamatergic neurotransmission in the enteric nervous system and brain-gut axis in health and disease. Neuropharmacology. 2016;111:14–33. doi: 10.1016/j.neuropharm.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 34.Panther E.J., Dodd W., Clark A., Lucke-Wold B. Gastrointestinal Microbiome and Neurologic Injury. Biomedicines. 2022;10:500. doi: 10.3390/biomedicines10020500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y., Danbolt N.C. GABA and Glutamate Transporters in Brain. Front. Endocrinol. 2013;4:165. doi: 10.3389/fendo.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luscher C., Xia H., Beattie E.C., Carroll R.C., von Zastrow M., Malenka R.C., Nicoll R.A. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24:649–658. doi: 10.1016/S0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 37.Gambrill A.C., Storey G.P., Barria A. Dynamic regulation of NMDA receptor transmission. J. Neurophysiol. 2011;105:162–171. doi: 10.1152/jn.00457.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanderson D.J., Sprengel R., Seeburg P.H., Bannerman D.M. Deletion of the GluA1 AMPA receptor subunit alters the expression of short-term memory. Learn. Memmory. 2011;18:128–131. doi: 10.1101/lm.2014911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam M.S. Calcium Signaling: From Basic to Bedside. Adv. Exp. Med. Biol. 2020;1131:1–6. doi: 10.1007/978-3-030-12457-1_1. [DOI] [PubMed] [Google Scholar]
- 40.Lipton S.A. Paradigm shift in neuroprotection by NMDA receptor blockade: Memantine and beyond. Nat. Rev. Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- 41.Naisbitt S., Kim E., Tu J.C., Xiao B., Sala C., Valtschanoff J., Weinberg R.J., Worley P.F., Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/S0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 42.Ha H.T.T., Leal-Ortiz S., Lalwani K., Kiyonaka S., Hamachi I., Mysore S.P., Montgomery J.M., Garner C.C., Huguenard J.R., Kim S.A. Shank and Zinc Mediate an AMPA Receptor Subunit Switch in Developing Neurons. Front. Mol. Neurosci. 2018;11:405. doi: 10.3389/fnmol.2018.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang M., Bozdagi O., Scattoni M.L., Wohr M., Roullet F.I., Katz A.M., Abrams D.N., Kalikhman D., Simon H., Woldeyohannes L., et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J. Neurosci. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castelli V., Brancato A., Cavallaro A., Lavanco G., Cannizzaro C. Homer2 and Alcohol: A Mutual Interaction. Front. Psychiatry. 2017;8:268. doi: 10.3389/fpsyt.2017.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petralia R.S., Wang Y.X., Sans N., Worley P.F., Hammer J.A., 3rd, Wenthold R.J. Glutamate receptor targeting in the postsynaptic spine involves mechanisms that are independent of myosin Va. Eur. J. Neurosci. 2001;13:1722–1732. doi: 10.1046/j.0953-816x.2001.01553.x. [DOI] [PubMed] [Google Scholar]
- 46.Maher B.J., Mackinnon R.L., 2nd, Bai J., Chapman E.R., Kelly P.T. Activation of postsynaptic Ca(2+) stores modulates glutamate receptor cycling in hippocampal neurons. J. Neurophysiol. 2005;93:178–188. doi: 10.1152/jn.00651.2004. [DOI] [PubMed] [Google Scholar]
- 47.Neyman S., Manahan-Vaughan D. Metabotropic glutamate receptor 1 (mGluR1) and 5 (mGluR5) regulate late phases of LTP and LTD in the hippocampal CA1 region in vitro. Eur. J. Neurosci. 2008;27:1345–1352. doi: 10.1111/j.1460-9568.2008.06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J.S., Exton J.H. Regulation of phospholipase D2 activity by protein kinase C alpha. J. Biol. Chem. 2004;279:22076–22083. doi: 10.1074/jbc.M311033200. [DOI] [PubMed] [Google Scholar]
- 49.Raben D.M., Barber C.N. Phosphatidic acid and neurotransmission. Adv. Biol. Regul. 2017;63:15–21. doi: 10.1016/j.jbior.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yogeswara I.B.A., Maneerat S., Haltrich D. Glutamate Decarboxylase from Lactic Acid Bacteria-A Key Enzyme in GABA Synthesis. Microorganisms. 2020;8:1923. doi: 10.3390/microorganisms8121923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wisden W., Yu X., Franks N.P. GABA Receptors and the Pharmacology of Sleep. Handb. Exp. Pharmacol. 2019;253:279–304. doi: 10.1007/164_2017_56. [DOI] [PubMed] [Google Scholar]
- 52.Zhang D., Pan Z.H., Awobuluyi M., Lipton S.A. Structure and function of GABA(C) receptors: A comparison of native versus recombinant receptors. Trends Pharm. Sci. 2001;22:121–132. doi: 10.1016/S0165-6147(00)01625-4. [DOI] [PubMed] [Google Scholar]
- 53.Ghit A., Assal D., Al-Shami A.S., Hussein D.E.E. GABA(A) receptors: Structure, function, pharmacology, and related disorders. J. Genet. Eng. Biotechnol. 2021;19:123. doi: 10.1186/s43141-021-00224-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Picciotto M.R., Higley M.J., Mineur Y.S. Acetylcholine as a neuromodulator: Cholinergic signaling shapes nervous system function and behavior. Neuron. 2012;76:116–129. doi: 10.1016/j.neuron.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogg R.C., Raggenbass M., Bertrand D. Nicotinic acetylcholine receptors: From structure to brain function. Rev. Physiol. Biochem. Pharmacol. 2003;147:1–46. doi: 10.1007/s10254-003-0005-1. [DOI] [PubMed] [Google Scholar]
- 56.O’Grady G.L., Verschuuren C., Yuen M., Webster R., Menezes M., Fock J.M., Pride N., Best H.A., Benavides Damm T., Turner C., et al. Variants in SLC18A3, vesicular acetylcholine transporter, cause congenital myasthenic syndrome. Neurology. 2016;87:1442–1448. doi: 10.1212/WNL.0000000000003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dani J.A. Neuronal Nicotinic Acetylcholine Receptor Structure and Function and Response to Nicotine. Int. Rev. Neurobiol. 2015;124:3–19. doi: 10.1016/bs.irn.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarnawski L., Reardon C., Caravaca A.S., Rosas-Ballina M., Tusche M.W., Drake A.R., Hudson L.K., Hanes W.M., Li J.H., Parrish W.R., et al. Adenylyl Cyclase 6 Mediates Inhibition of TNF in the Inflammatory Reflex. Front. Immunol. 2018;9:2648. doi: 10.3389/fimmu.2018.02648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen J.X., Yakel J.L. Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharmacol. Sin. 2009;30:673–680. doi: 10.1038/aps.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Resende R.R., Adhikari A. Cholinergic receptor pathways involved in apoptosis, cell proliferation and neuronal differentiation. Cell Commun. Signal. 2009;7:20. doi: 10.1186/1478-811X-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klein M.O., Battagello D.S., Cardoso A.R., Hauser D.N., Bittencourt J.C., Correa R.G. Dopamine: Functions, Signaling, and Association with Neurological Diseases. Cell. Mol. Neurobiol. 2019;39:31–59. doi: 10.1007/s10571-018-0632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guenter J., Lenartowski R. Molecular characteristic and physiological role of DOPA-decarboxylase. Postep. Hig. Med. Dosw. 2016;70:1424–1440. doi: 10.5604/17322693.1227773. [DOI] [PubMed] [Google Scholar]
- 63.Beaulieu J.M., Gainetdinov R.R. The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol. Rev. 2011;63:182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 64.Ha C.M., Park D., Han J.K., Jang J.I., Park J.Y., Hwang E.M., Seok H., Chang S. Calcyon forms a novel ternary complex with dopamine D1 receptor through PSD-95 protein and plays a role in dopamine receptor internalization. J. Biol. Chem. 2012;287:31813–31822. doi: 10.1074/jbc.M112.370601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalinovskii A.P., Osmakov D.I., Koshelev S.G., Lubova K.I., Korolkova Y.V., Kozlov S.A., Andreev Y.A. Retinoic Acid-Differentiated Neuroblastoma SH-SY5Y Is an Accessible In Vitro Model to Study Native Human Acid-Sensing Ion Channels 1a (ASIC1a) Biology. 2022;11:167. doi: 10.3390/biology11020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S., Pyl P.T., Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mi H., Lazareva-Ulitsky B., Loo R., Kejariwal A., Vandergriff J., Rabkin S., Guo N., Muruganujan A., Doremieux O., Campbell M.J., et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., Ami G.O.H., Web Presence Working G. AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gillespie M., Jassal B., Stephan R., Milacic M., Rothfels K., Senff-Ribeiro A., Griss J., Sevilla C., Matthews L., Gong C., et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D692. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Choi Y.H., Hazekamp A., Peltenburg-Looman A.M., Frederich M., Erkelens C., Lefeber A.W., Verpoorte R. NMR assignments of the major cannabinoids and cannabiflavonoids isolated from flowers of Cannabis sativa. Phytochem. Anal. PCA. 2004;15:345–354. doi: 10.1002/pca.787. [DOI] [PubMed] [Google Scholar]
- 74.Gaoni Y., Mechoulam R. The isolation and structure of delta-1-tetrahydrocannabinol and other neutral cannabinoids from hashish. J. Am. Chem. Soc. 1971;93:217–224. doi: 10.1021/ja00730a036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are openly available in the NCBI Sequence Read Archive at BioProject accession numbers PRJNA973106.