Summary
Genome-wide association studies (GWASs) have identified more than 200 genomic loci for breast cancer risk, but specific causal genes in most of these loci have not been identified. In fact, transcriptome-wide association studies (TWASs) of breast cancer performed using gene expression prediction models trained in breast tissue have yet to clearly identify most target genes. To identify candidate genes, we performed a GWAS analysis in a breast cancer dataset from UK Biobank (UKB) and combined the results with the GWAS results of the Breast Cancer Association Consortium (BCAC) by a meta-analysis. Using the summary statistics from the meta-analysis, we performed a joint TWAS analysis that combined TWAS signals from multiple tissues. We used expression prediction models trained in 11 tissues that are potentially relevant to breast cancer from the Genotype-Tissue Expression (GTEx) data. In the GWAS analysis, we identified eight loci distinct from those reported previously. In the TWAS analysis, we identified 309 genes at 108 genomic loci to be significantly associated with breast cancer at the Bonferroni threshold. Of these, 17 genes were located in eight regions that were at least 1 Mb away from published GWAS hits. The remaining TWAS-significant genes were located in 100 known genomic loci from previous GWASs of breast cancer. We found that 21 genes located in known GWAS loci remained statistically significant after conditioning on previous GWAS index variants. Our study provides insights into breast cancer genetics through mapping candidate target genes in a large proportion of known GWAS loci and discovering multiple new loci.
Keywords: transcriptome-wide association study, multiple tissues, genome-wide association study, meta-analysis, breast cancer, susceptibility genes, genetic epidemiology
Gao and colleagues identify eight loci of breast cancer susceptibility in a genome-wide association study. By leveraging gene expression data from multiple tissues, they also discover 309 breast cancer susceptibility genes in a transcriptome-wide association study. Known genetic risk variants could act on these genes to cause breast cancer.
Introduction
Breast cancer is the most common malignancy among women in most countries around the world, accounting for one-quarter of all cancer cases in women.1 In the past 15 years, genome-wide association studies (GWASs) have identified over 200 loci significantly associated with breast cancer.2,3,4 Although some of these findings have yielded functional insights into breast cancer,4 these genetic variants account for a relatively small proportion of heritability, suggesting that more genetic variants have yet to be identified. Because the vast majority of risk variants identified in GWASs are located in intergenic regions and are not nonsynonymous coding variants, the putative genes on which these risk variants act to cause breast cancer remain unclear for most GWAS-identified loci.
To further elucidate the role of genetic variants in complex traits, transcriptome-wide association studies (TWASs) have been conducted to quantify the relationship between a predicted level of genetically regulated gene expression and the phenotype of interest.5,6 TWASs of breast cancer have identified dozens of genes whose expression is significantly associated with breast cancer and its subtypes.4,7,8,9 However, these genes account for only a small proportion of known GWAS loci of breast cancer. These TWASs were performed primarily by associating a cis-regulated level of gene expression with breast cancer in single tissues (breast tissue or whole blood). Our recent study demonstrated that integrating information from multiple tissues in a TWAS could improve association detection.10 In addition, existing TWASs used gene expression prediction models trained on data from older versions of the Genotype-Tissue Expression (GTEx) project, such as v.6 or v.7. The recent v.8 of GTEx has much larger sample sizes compared to the older versions, so the expression models trained in GTEx v.8 will be more accurate than those trained in older versions in prediction of expression levels.11,12 By using GTEx v.8, one can explore heritability for expression more efficiently, and more genes can pass the filtering threshold and be used for TWAS analysis.12 Therefore, the prediction models trained in GTEx v.8 have the potential to increase the power of TWAS in detecting susceptibility genes.
In this study, we aimed to identify candidate genes for breast cancer by performing joint TWAS analyses of breast cancer by combining TWAS information from multiple tissues. We applied our TWAS method to the summary statistics from a meta-analysis of data from 122,977 breast cancer cases and 105,974 controls in the Breast Cancer Association Consortium (BCAC)3 and 10,534 breast cancer cases and 185,116 controls in UK Biobank (UKB).13
Subjects and methods
GWAS summary statistics on women of European ancestry from the BCAC
In our meta-analysis, we used the summary statistics data from the GWAS of breast cancer in 122,977 cases and 105,974 controls of European ancestry from the BCAC. The details of the BCAC have been described previously.3,14 Briefly, the BCAC included: (1) 61,282 female cases with breast cancer and 45,494 female controls of European ancestry that were genotyped using the OncoArray, including 570,000 SNPs; (2) 46,785 breast cancer cases and 42,892 controls of European ancestry from Collaborative Oncological Gene-environment Study (iCOGS) that were genotyped using a custom Illumina iSelect genotyping array containing ∼211,155 variants; and (3) 11 other breast cancer GWASs (14,910 cases and 17,588 controls). Genotype data from iCOGS, OncoArray, and GWASs were imputed using the October 2014 release of the 1000 Genomes Project data as a reference. Genetic association results for breast cancer risk were combined using inverse-variance fixed-effect meta-analyses.3
GWAS analysis using data from UK Biobank
The UK Biobank project recruited approximately 500,000 participants, ages 40–69, between 2007 and 2010, across 22 study centers in the United Kingdom. The project collected detailed demographic, lifestyle, and disease histories at baseline, as well as disease occurrences through prospective follow-up and database linkages.13 Whole-genome genotyping was conducted using UK Biobank Axiom Arrays for 488,377 participants, and imputation was performed using the Haplotype Reference Consortium and 1000 Genomes phase 3 as reference panels to obtain >90 million genetic markers.13 In this study, we selected female individuals with both phenotypic and genotypic data available. Unrelated individuals with European ancestry were selected using principal-component analysis (PCA). We further filtered out samples with genotyping call rates <95%. After these exclusions, the analysis included 10,534 breast cancer cases (including 6,055 prevalent cases before enrollment and 4,479 incident cases during a median of 7 years follow-up) and 185,116 controls (Table S1). We performed GWAS analysis using logistic regression, comparing breast cancer cases with controls, adjusting for age at enrollment and top ten eigenvectors from PCA of genotypes with software package PLINK 2.0.15 As a sensitivity analysis, we performed a case-case GWAS analysis, comparing incident cases with prevalent cases. In the logistic regression models adjusting for age and top ten eigenvectors, we found that no variants were genome-wide significant (alpha = 5 × 10−8), suggesting that it is reasonable to combine incident and prevalent cases in the primary analysis.
Gene expression prediction models
Gene expression prediction models were built with the genotype and RNA-seq data in 49 tissues of European ancestry from the GTEx project (v.8).12 Specifically, building prediction models for a gene includes the following steps. (1) Across all tissues, cis-expression quantitative trait loci (cis-eQTLs) were discovered with a false discovery rate of 5% per tissue. Only genes with cis-eQTLs were selected. (2) Fine mapping was performed in each tissue in the corresponding cis-gene region by the DAP-G method16,17 to select variants with minor allele frequency >0.01 and posterior inclusion probabilities (PIPs) >0.01 and to select genes with at least one credible set of PIP >0.1 (where the credible-set PIP is the sum of PIPs of variants in the set). Then, in each credible set, only the variant with the highest PIP was kept. For the 49 tissues, a union of selected variants across 49 tissues was obtained, and linkage disequilibrium (LD) pruning was applied to the union of variants to remove redundant variants. (3) The multivariate adaptive shrinkage (MASH) method was applied to the marginal eQTL effects across the 49 tissues at the union of variants to jointly estimate effects of eQTLs, allowing sparse effects (that is, with many zero effects) and accounting for correlation among non-zero effects in different tissues.18 (4) The predicted expression of the gene in each tissue was calculated as the linear combination of genotypes multiplying by their estimated effect sizes. In this study, we used the prediction models for 11 tissues, including female tissues (breast, ovary, uterus, and vagina), tissues that resemble connective and fat tissues in the breast (subcutaneous adipose, visceral adipose, and cultured fibroblasts), tissues related to immune cells (spleen, EBV-transformed lymphocytes, and whole blood), and liver. These tissues are potentially relevant to breast cancer development or carcinogen metabolism.
Summary statistic-based imputation
For variants included in the GTEx prediction models but not in the GWAS summary statistics, we imputed Z scores with the method ImpG-Summary.19 The ImpG-Summary method assumes that, under null hypothesis, the vector of Z scores at all SNPs in a locus is approximately distributed as a Gaussian distribution, , with being the correlation matrix among all pairs of SNPs induced by LD. We estimated posterior mean of Z scores at unobserved SNPs. We used the GWAS summary statistics and correlation matrix estimated from the genotype data in the GTEx samples as the input for the ImpG-Summary method.
Joint TWAS across multiple tissues
The joint TWAS analysis includes two steps: (1) performing a traditional TWAS analysis in each of the 11 tissues with the software S-PrediXcan20 to obtain the p values and (2) constructing test statistics using the aggregated Cauchy association test (ACAT) method21 that combined p values for each gene from the single-tissue TWAS analyses across the 11 tissues. Specifically, the ACAT test statistic is , where is a nonnegative weight. We used . The p value of the ACAT test statistic is approximated by /π. We noticed that, for some genes, expression prediction models were only available for K of the 11 tissues (K < 11), then the ACAT test statistic was calculated using the S-PrediXcan p value from the K tissues, with a weight .
Conditional joint TWAS
To test if the signals at the 309 genes detected by the TWAS are independent of a set of published GWAS index SNPs and newly identified index SNPs in our GWAS analysis, we performed a TWAS conditional on these index SNPs that were genome-wide significant (p < 5 × 10−8). At each gene, we considered two sets of SNPs: the target set of SNPs used for predicting gene expression and the conditioning set of significant index SNPs from GWASs within ±2 Mb of the transcription start or stop sites of the gene. For the target set of SNPs, we calculated adjusted effects (beta) on breast cancer risk and their variances, after conditioning on the index SNPs using the conditional and joint multiple-SNP (COJO) analysis method of Yang et al.22 We then ran S-PrediXcan20 on these conditional summary statistics in single tissues and performed the joint TWAS analysis that combines p values from the single-tissue analyses using the ACAT method.21
Colocalization analysis
For genes identified in the TWAS, we calculated regional colocalization probabilities (RCPs) using the method ENLOC.17 ENLOC divides the genome into roughly independent LD blocks using the approach described in Berisa and Pickrell.23 For a gene located in a specific LD block, ENLOC calculates the colocalization probability of causal GWAS hits and causal eQTLs in the LD block. Variant-level fine-mapping analysis was done to reveal possible causal eQTLs and GWAS hits. To calculate RCP for a gene in an LD block, we used the GTEx (v.8) eQTLs for the gene and the meta-analysis GWAS summary statistics in the LD block. Because ENLOC can calculate RCP only for single tissues, we calculated RCP in each of the 11 tissues and then calculated maximum RCP (Max RCP) among the 11 tissues. An RCP at a gene greater than a threshold (such as 0.5) provides supportive information that the gene identified by the TWAS has a high probability of colocalizing with a nearby GWAS variants, which strengthens the association signal.
Gene-based fine-mapping
We performed a gene-based statistical fine-mapping over the gene-trait association signals from our TWAS using the software FOCUS (fine-mapping of causal gene sets).24 For an LD block, we identified a credible set of genes that contain the causal genes at a predefined confidence level of 90%. We also computed the marginal PIP in the target tissue (breast tissue) for each gene in the region to be causal given the observed TWAS statistics. FOCUS accounts for the correlation structure induced by LD and prediction weights used in the TWAS and controls for certain pleiotropic effects. We used 11 tissues and related expression prediction weights from the GTEx v.8 and assigned the breast tissue as the target tissue. When the expression prediction model for a gene in the breast tissue was unavailable (18% of the genes), we randomly selected an alternative tissue with a prediction model as a proxy for the gene.
Gene set enrichment and functional annotation
For the set of significant genes identified by our TWAS, we conducted enrichment of protein-coding and long non-coding RNA (lncRNA) genes against gene sets from multiple biological pathways, functional categories, and databases using the FUMA package.25 Specifically, we used the GENE2FUNC module of FUMA and specified 33,527 protein-coding and lncRNA genes as the background genes for enrichment testing. Multiple testing correction was performed per data source of tested gene sets (e.g., canonical pathways, GWAS catalog categories) using Benjamini-Hochberg false discovery rate adjustment. We reported pathways/categories with adjusted p values ≤0.05 and at least two genes that overlapped with the gene set of interest.
Results
GWAS in BCAC and UKB
We performed a GWAS analysis in a breast cancer dataset from UKB (genomic control λ = 1.02) and then combined the UKB GWAS results with the previously published GWAS results of the BCAC data3 by a meta-analysis using the software METAL26 (Figure S1). We identified eight GWAS loci that were not reported by previous studies (Table 1; Figure S2). In six loci, the sentinel variants are located at least 2 Mb away from any of the risk variants identified by previous GWASs, and in two loci, the sentinel variants are located at least 500 kb away (rs9833726 and rs62483813), but none of these index variants are in LD with previous GWAS signals. Each of the index variants showed the same association direction in BCAC and UKB GWASs. No significant heterogeneity was observed in the meta-analysis at any sentinel variants. All the sentinel variants are common and are located in the introns of nearby genes.
Table 1.
The lead breast cancer GWAS risk variants at eight previously unreported locia
| rsid | Position (hg38) | Locus | Nearest geneb | Allelesc | EAF | Data source | OR (95% CI) | p value |
|---|---|---|---|---|---|---|---|---|
| rs707475 | 7857016 | 1p36.23 | UTS2 | A/G | 0.393 | UKB | 0.97 (0.94–1.00) | 2.05E−02 |
| BCAC | 0.97 (0.95–0.98) | 1.29E−07 | ||||||
| Meta | 0.97 (0.95–0.98) | 7.17E−09 | ||||||
| rs60504827 | 168127440 | 1q24.2 | GPR161 | T/C | 0.126 | UKB | 0.96 (0.92–1.00) | 4.03E−02 |
| BCAC | 0.95 (0.93–0.97) | 1.76E−07 | ||||||
| Meta | 0.95 (0.93–0.97) | 1.90E−08 | ||||||
| rs9833726 | 86154659 | 3p12.1 | LOC102723364 | T/G | 0.135 | UKB | 0.93 (0.89–0.97) | 8.21E−04 |
| CADM2 | BCAC | 0.96 (0.94–0.98) | 5.98E−06 | |||||
| Meta | 0.95 (0.94–0.97) | 4.02E−08 | ||||||
| rs35016840 | 150318622 | 4q31.3 | LRBA | T/C | 0.650 | UKB | 1.03 (1.00–1.06) | 3.55E−02 |
| BCAC | 1.03 (1.02–1.05) | 3.73E−07 | ||||||
| Meta | 1.03 (1.02–1.05) | 4.25E−08 | ||||||
| rs62483813 | 102478605 | 7q22.1 | POLR2J | T/C | 0.369 | UKB | 1.04 (1.01–1.07) | 7.79E−03 |
| BCAC | 1.04 (1.02–1.05) | 1.89E−07 | ||||||
| Meta | 1.04 (1.02–1.05) | 5.34E−09 | ||||||
| rs77457752 | 13942941 | 9p23 | LINC00583 | A/G | 0.127 | UKB | 0.91 (0.87–0.96) | 1.41E−04 |
| BCAC | 0.95 (0.94–0.97) | 7.95E−07 | ||||||
| Meta | 0.95 (0.93–0.97) | 1.70E−09 | ||||||
| rs3235 | 13907609 | 10p13 | FRMD4A | A/G | 0.656 | UKB | 1.04 (1.01–1.07) | 8.68E−03 |
| BCAC | 1.03 (1.02–1.05) | 9.34E−07 | ||||||
| Meta | 1.03 (1.02–1.05) | 3.37E−08 | ||||||
| rs71063528 | 113863643 | 11q23.2 | USP28 | (A)17/delA | 0.275 | UKB | 0.96 (0.93–0.99) | 1.29E−02 |
| BCAC | 0.96 (0.95–0.98) | 7.91E−07 | ||||||
| Meta | 0.96 (0.95–0.98) | 3.15E−08 |
EAF, effect allele frequency; OR, odds ratio; CI, confidence interval; GWAS, genome-wide association study; UKB, UK Biobank; BCAC, Breast Cancer Association Consortium; meta, meta-analysis.
The heterogeneity tests comparing effects in BCAC and UKB were not significant in any of the loci
Variants are located in the introns of nearby genes
Effect allele/reference allele
Joint TWAS combining information across multiple tissues
We used expression prediction models trained in 11 tissues of European ancestry (with sample sizes ranging from 129 to 670 and a median of 227) from the GTEx v.8 data using the MASH method.11,12,18 In total, 19,274 genes across the 11 tissues with prediction models, including 14,613 genes expressed in breast tissue, were tested in our TWAS analysis. Using the meta-analysis summary statistics, we performed a single-tissue TWAS analysis in each tissue with S-PrediXcan20 and then a joint TWAS analysis using ACAT21 to combine p values of single-tissue TWASs across the 11 tissues for each gene.
The results of the joint TWAS analysis are summarized in the Manhattan plots against the variant-based GWAS analysis results (Figure S1). Of the 19,274 genes tested in our joint TWAS analysis, we identified 299 genes whose predicted expression was associated with breast cancer risk at the Bonferroni-corrected threshold of p < 2.59 × 10−6 (Table S2). Only 141 genes were identified when TWAS analysis used only breast tissue, i.e., a conventional single-tissue TWAS approach20 (Table S3). Of these 141 genes, 131 genes were also identified in the joint, multi-tissue TWAS. The remaining 10 genes identified only in the breast-tissue TWAS analysis were also marginally significant in the joint TWAS (p < 1.62 × 10−5), so we described the 309 genes from either TWAS in further analysis. Table S4 shows the detailed single-tissue TWAS results for the 309 genes in the analysis of two databases (BCAC and UKB) pooled and separately. We found that Z scores across tissues were moderately concordant on average, with an intraclass correlation coefficient of 0.561, but the agreement between tissues as well as the strongest association signals varied across genes. These findings suggest that the multi-tissue joint TWAS could provide additional information compared to a traditional single-target-tissue TWAS and address the possibility that the target tissue(s) could vary for different genes. We also found that TWAS results using the BCAC and UKB databases were very consistent, with a Pearson r = 0.871 (Figure S3).
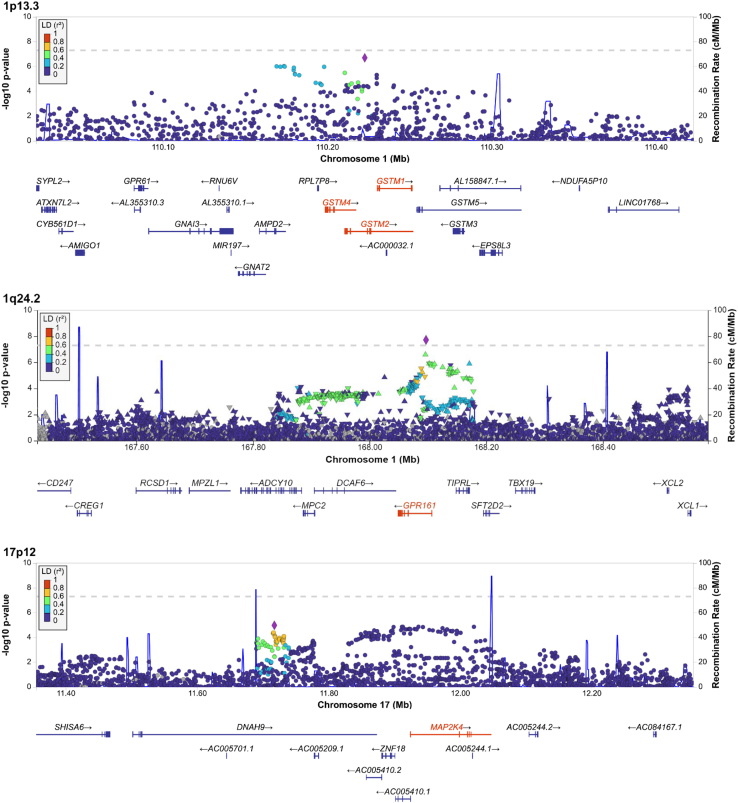
Of the 309 genes identified in our TWAS, 108 genes have been reported in previous TWASs (Tables S2 and S5), and multiple genes have been implicated in previous GWASs, such as FGFR2, TOX3, and ESR1. The 309 genes identified in our TWAS are distributed among 108 genomic loci (Figure 1). Based on NHGRI-EBI GWAS Catalog27 and literature review, we curated 226 GWAS loci of breast cancer susceptibility (Table S6). Including the eight GWAS loci discovered in the current study (Table 1), there are a total of 234 GWAS breast cancer susceptibility loci (Figure 1). Our TWAS identified 292 significant genes that are located in 100 known GWAS loci. The remaining 17 genes are located in eight TWAS loci that are at least 1.4 Mb away from any risk variant identified in previous GWASs and are not in LD with risk variants (Table 2). Of the 17 genes found in the eight TWAS loci, 10 genes in six loci were significant in the breast-tissue-based TWAS at the Bonferroni threshold. For example, we found MAP2K4 in the 17p12 locus was significant in both the multi-tissue joint TWAS and the breast-tissue-based TWAS, although there was no reported GWAS signal in this locus (Figure 2). Notably, the 1q24.2 locus was identified both in our GWAS and in our TWAS, and the GWAS index variant rs60504827 is located in an intron of GPR161 (Figure 2).
Figure 1.
Ideogram of the 309 TWAS-identified genes in the context of known breast cancer GWAS loci
Table 2.
The 17 genes identified by TWAS located at 8 genomic loci at least 1 Mb away from previous GWAS hits
| Locusa | Gene symbol | Position (hg38) |
Joint ACAT p value |
Breast p value |
PIPb | In credible set | Max RCPc | RCP in breast |
|---|---|---|---|---|---|---|---|---|
| 1q21.1, L1 | H3-2 | chr1: 143,894,544–143,905,977 | 1.13E−10 | 9.13E−01 | – | – | 0.000 | 0.000 |
| FAM72C | chr1: 143,955,287–143,971,986 | 2.77 E−12 | – | 0.419 | Yes | 0.000 | 0.000 | |
| 1q24.2 | GPR161 | chr1: 168,079,542–168,137,667 | 1.04 E−06 | 2.30E−01 | 0.000 | No | 0.513 | 0.000 |
| 6q24.1 | TXLNB | chr6: 139,240,061–139,291,998 | 8.21 E−06 | 2.24E−06 | 0.761 | Yes | 0.681 | 0.681 |
| 7q22.1, L1 | SPDYE3 | chr7: 100,307,702–100,322,196 | 1.91 E−07 | – | 0.253 | Yes | 0.127 | 0.040 |
| PILRB | chr7: 100,352,176–100,367,831 | 2.04 E−07 | 1.71E−07 | 0.412 | Yes | 0.505 | 0.505 | |
| PILRA | chr7: 100,367,530–100,400,096 | 2.69 E−07 | 3.47E−07 | 0.016 | No | 0.386 | 0.386 | |
| ZCWPW1 | chr7: 100,400,826–100,428,992 | 6.72 E−07 | 8.53E−06 | 0.005 | No | 0.261 | 0.202 | |
| MEPCE | chr7: 100,428,322–100,434,126 | 1.74 E−07 | – | 0.219 | Yes | 0.259 | 0.057 | |
| C7orf61 | chr7: 100,456,620–100,464,260 | 7.19 E−07 | 2.92E−07 | 0.079 | Yes | 0.155 | 0.088 | |
| TSC22D4 | chr7: 100,463,359–100,479,232 | 1.08 E−06 | 1.38E−06 | 0.001 | No | 0.228 | 0.228 | |
| NYAP1 | chr7: 100,483,927–100,494,802 | 1.15 E−06 | 5.41E−02 | 0.000 | No | 0.086 | 0.000 | |
| 10p12.1 | YME1L1 | chr10: 27,110,111–27,155,266 | 4.99 E−06 | 4.99E−07 | 0.920 | Yes | 0.249 | 0.084 |
| 17p12 | MAP2K4 | chr17: 12,020,829–12,143,830 | 9.59 E−07 | 7.28E−07 | 0.893 | Yes | 0.630 | 0.630 |
| 17q23.1 | RPS6KB1 | chr17: 59,893,046–59,950,574 | 3.97 E−06 | 1.80E−06 | 0.823 | Yes | 0.144 | 0.095 |
| 20q13.33 | RGS19 | chr20: 64,073,181–64,079,988 | 9.27 E−07 | 1.76E−06 | 0.166 | Yes | 0.922 | 0.862 |
| – | OPRL1 | chr20: 64,080,082–64,100,643 | 4.37 E−07 | 2.81E−07 | 0.792 | Yes | 0.925 | 0.791 |
“L1” and “L2” denote the first and second locus defined by LD block in the same cytoband, respectively
Posterior inclusion probability (PIP) calculated by FOCUS
Maximum marginal posterior inclusion probability (RCP) in all tissues calculated in colocalization analysis
Figure 2.
Manhattan plots of exemplar genes in three loci identified by the joint TWAS analysis
The genes highlighted in red are significant genes identified in the joint transcriptome-wide association study.
Conditional joint TWAS on known GWAS index variants
To determine whether the associations for the genes identified by the joint TWAS were independent of GWAS association signals, we performed conditional analyses adjusting for nearby GWAS index risk variants. We found 21 genes located in 15 known GWAS loci that were conditionally significant (Table 3). This suggests that additional genetic variants, which are neither genome-wide significant nor in LD with GWAS-significant variants, may account for the association between expression of these genes and breast cancer risk at these loci.
Table 3.
The 21 genes identified by TWAS in the 15 known loci that were significant after adjusting for known GWAS index variants
| Locus | Gene symbol | Position (hg38) | Joint ACAT p value | Conditional p valuea | Closest index SNPb | Dist (kb)c | PIPd | In Credible set | Max RCPe |
|---|---|---|---|---|---|---|---|---|---|
| 1p13.3 | GSTM1 | chr1: 109,687,814–109,709,039 | 2.91E−08 | 1.10E−06 | rs5776993 | 7.5 | 0.491 | Yes | 0.996 |
| 2q35 | DIRC3 | chr2: 217,284,019–217,756,593 | 1.67E−16 | 2.31E−10 | rs6436017 | 102.1 | 1.000 | Yes | 0.001 |
| 3p24.1, L1 | NEK10 | chr3: 27,106,484–27,369,460 | <5.0E−17 | 1.77E−07 | rs4973768 | 5.1 | 1.000 | Yes | 0.209 |
| 5p15.33, L1 | AHRR | chr5: 321,714–438,291 | 1.22E−07 | 4.78E−07 | rs62641919 | 0 | 0.000 | No | 0.379 |
| EXOC3 | chr5: 443,175–471,937 | 2.14E−06 | 1.87E−06 | rs62641919 | 98.2 | 0.317 | Yes | 0.065 | |
| 6q22.33 | RSPO3 | chr6: 127,118,671–127,199,481 | 2.21E−06 | 2.57E−06 | rs2180341 | 80 | 0.055 | No | 0.324 |
| 8q21.13 | HNF4G | chr8: 75,407,914–75,566,834 | 1.14E−12 | 2.18E−07 | rs72658071 | 14.4 | 1.000 | Yes | 0.069 |
| 11p15.5, L2 | LSP1 | chr11: 1,850,904–1,892,267 | <5.0E−17 | 7.24E−07 | rs576603 | 0.6 | 0.000 | No | 0.478 |
| 11q13.1 | OVOL1 | chr11: 65,787,063–65,797,214 | 2.08E−12 | 2.23E−06 | rs3903072 | 18.4 | 0.982 | Yes | 0.165 |
| 11q13.3 | RP11-554A11.8 | chr11: 69,147,228–69,171,564 | 1.58E−08 | 6.02E−09 | rs72932540 | 0 | 0.004 | No | 0.000 |
| 12p11.22, L1 | CCDC91 | chr12: 28,133,249–28,581,511 | 2.22E−16 | 2.90E−11 | rs7297051 | 111.4 | 0.005 | No | 0.000 |
| 12p11.22, L2 | OVCH1 | chr12: 29,412,474–29,497,686 | 1.43E−06 | 1.63E−06 | rs1027113 | 425.1 | 0.000 | No | 0.000 |
| 16q12.1–q12.2 | TOX3 | chr16: 52,436,417–52,547,802 | <5.0E−17 | <5.0E−17 | rs3803662 | 4.6 | 1.000 | Yes | 0.333 |
| 19q13.32 | GIPR | chr19: 45,668,221–45,683,722 | 2.31E−08 | 8.44E−07 | rs61373376 | 0 | 0.001 | No | 0.353 |
| 20q11.23 | PHF20 | chr20: 35,771,974–35,950,370 | 8.79E−07 | 2.33E−06 | rs112208395 | 0 | 0.330 | Yes | 0.322 |
| CNBD2 | chr20: 35,954,564–36,030,700 | 2.38E−06 | 3.66E−06 | rs112208395 | 21.5 | 0.251 | Yes | 0.038 | |
| 22q12.1–q12.2 | TTC28 | chr22: 27,978,014–28,679,840 | 1.14E−08 | 9.40E−12 | rs62235681 | 4.9 | 0.000 | No | 0.000 |
| CHEK2 | chr22: 28,687,743–28,742,422 | 9.02E−13 | 4.39E−15 | rs62235681 | 3.0 | 1.000 | Yes | 0.001 | |
| HSCB | chr22: 28,742,039–28,757,515 | 1.36E−12 | 6.49E−15 | rs17879961 | 16.9 | 0.000 | No | 0.000 | |
| CCDC117 | chr22: 28,772,674–28,789,301 | 1.46E−07 | 1.62E−09 | rs17879961 | 47.6 | 0.000 | No | 0.000 | |
| XBP1 | chr22: 28,794,555–28,800,597 | 6.35E−09 | 1.26E−10 | rs17879961 | 69.5 | 0.003 | No | 0.000 |
Conditional ACAT p value after adjusting for adjacent index SNPs
SNPs that were identified to be significant in previous genome-wide association studies
Distance from the gene to closest index SNP (kb)
Posterior inclusion probability (PIP) calculated by the FOCUS method
Maximum marginal posterior inclusion probability (RCP) in all tissues calculated in colocalization analysis
Colocalization analysis and gene-based fine-mapping
In colocalization analysis using ENLOC,17 we found 45 of 309 genes with RCP values greater than 0.5 using breast tissue and 75 genes with maximum RCP values greater than 0.5 (Table S2). The colocalization RCP value for breast tissue was inversely correlated with the p value from breast-tissue TWAS (Spearman r = −0.287), suggesting that genes with stronger TWAS significance are more likely to colocalize with GWAS causal variants.
As the LD among variants can induce significant gene-trait associations for non-causal genes, we conducted gene-level fine-mapping analysis using the package FOCUS.24 We found that 141 genes are in credible sets that contain causal genes at the confidence level of 90%; for these genes, the median PIP was 0.792 (Table S2). As the fine-mapping analysis in FOCUS mainly used breast tissue as the target tissue (82%), genes identified in the breast-tissue TWAS were more likely in the credible sets (94 genes).
As candidate causal genes, we selected 114 genes if they are in the credible sets with gene PIPs greater than 0.15 and located in regions in which the null model is not a possible outcome (Table S7). These 114 genes are located in 83 loci. For most loci (n = 61), fine-mapping identified only one causal gene candidate, eliminating many TWAS-identified genes; for example, CHEK2 (PIP = 1.0) was identified as the possible causal gene in locus 22q12.1–q12.2 (out of six TWAS-identified genes). For fewer loci, multiple candidate genes were identified after fine-mapping analysis; for example, GSTM1, GSTM2, and GSTM4, three members of the glutathione S-transferase multigene family, were suggested to be possible causal genes in locus 1p13.3 (Table S7; Figure 2).
Gene set enrichment and functional annotation
Of the 309 TWAS-identified genes, 272 are protein-coding genes, 34 are lncRNA genes, and 3 are pseudogenes. We tested the enrichment of this set of protein-coding and lncRNA genes against background gene sets from multiple databases using the FUMA software package.25 We found that these TWAS-identified genes were significantly enriched in several biological pathways, such as the Trail signaling pathway, Fas signaling pathway, apoptosis pathway, biosynthesis, and cell cycle regulation; all of these pathways are important in cancer development or a hallmark of cancer, which further warrants efforts into studying how the genes identified in our TWAS may contribute to breast cancer etiology (Table S8). Interestingly, we found significant enrichment in the genes underlying several breast cancer risk factors, including body fat distribution, mammographic density, alcohol use, and body height,28 suggesting that the TWAS-identified genes may indirectly contribute to breast cancer susceptibility through their impacts on known lifestyle/environmental risk factors. We also found strong enrichment for other diseases, such as inflammatory bowel disease, diabetes, and other cancers (including melanoma, chronic myeloid leukemia, and cancers of the esophagus, pancreas, bladder, and prostate), suggesting that some of breast cancer genes have pleiotropic effects. These results were consistent with the notion that there are shared genetic components between various cancer sites29 and suggest that further research into collating TWAS results across cancers may be beneficial to understanding their shared genetic etiologies. Lastly, differential gene expression analysis in GTEx showed that the TWAS-identified genes had strong tissue specificity, although our joint TWAS weighted each tissue similarly; the most up-expressed tissues of these genes were uterus, ovary, vagina, and breast (Figure S4).
Enrichment of genetic variants in prediction models
Fachal et al.30 identified a set of 11,289 credible causal variants (CCVs) for breast cancer risk in a fine-mapping study of 150 breast cancer risk regions. We performed a hypergeometric test to examine whether genetic variants included in the expression prediction models for the 292 significant genes identified by our TWAS in 100 known GWAS loci were enriched in the set of CCVs. In 100 known GWAS loci, there were 1,160,671 genetic variants with minor allele frequency >0.01 in the GTEx v.8 data. Among these variants, 7,537 were shared with the set of CCVs identified by Fachal et al.30 The prediction models for the 292 genes used 1,702 unique genetic variants; of these variants, 114 variants were CCVs (Table S9). Therefore, there was an approximately 10-fold over-representation of CCVs among our genetic variants of prediction models (hypergeometric p value 3.17 × 10−75). Although this analysis cannot take into account LD among genetic variants, the strong enrichment is unlikely to be due to confounders.
Fachal et al.30 also classified GWAS signals as strong and moderate signals. We examined whether CCVs in regions with strong GWAS signals are more likely to be genetic variants of our prediction models, compared with CCVs in regions with moderate GWAS signals. Of the 5,117 CCVs in regions with strong GWAS signals, 83 (1.6%) were genetic variants in our gene expression prediction models. In contrast, 30 of 5,973 (0.5%) CCVs in regions with moderate GWAS signals were genetic variants in our gene expression prediction models, so there was a >3-fold enrichment (p = 4.81 × 10−9). In short, genetic variants used in gene expression models are more likely to be variants identified from fine-mapping of breast cancer GWAS, and there is a stronger enrichment for regions with strong GWAS signals.
Discussion
In this study, we performed a breast cancer TWAS analysis that leveraged the genetically predicted gene expression levels across multiple tissues. We identified 309 significant genes at the Bonferroni threshold, including 17 genes located in eight loci not reported in previous studies and 292 genes located in 100 known GWAS loci. In about 43% of known breast cancer GWAS loci, our study was able to identify possible susceptibility gene(s). We also found 21 genes in known GWAS loci that were independent of previously reported GWAS risk variants, suggesting potentially additional breast cancer susceptibility signals. Generally, our study findings are consistent with previous TWASs; of the 368 genes reported in previous TWASs,4,7,8,9,31,32,33,34,35,36 108 genes were replicated in our study.
The number of TWAS-significant genes identified in our study is similar to the number identified in all previous studies combined, possibly because of several notable differences in methodologies. First, we used GWAS data from a large number of breast cancer cases (n = 133,511) and controls (n = 291,090) from BCAC and UKB. This large sample size provided high statistical power in the association analysis and helped to identify eight GWAS loci that were not reported previously. Second, we aggregated TWAS signals across 11 tissues. This multi-tissue approach resulted in more genes being identified compared to the TWAS using breast tissue alone. This suggests that while breast tissue is an important tissue to be utilized when conducting breast cancer TWASs, other tissues can contribute additional information for gene discovery. For example, our multi-tissue approach identified FGFR2, a gene with strong evidence in breast cancer etiology, but this gene has not been identified in previous TWASs and would have been missed if we had utilized only the TWAS exclusive to breast tissue. Expression of FGFR2 in fibroblasts, ovary, vagina, and liver were associated with breast cancer risk (Table S4). Third, we used expression prediction models trained in GTEx v.8 with the MASH method based on fine-mapping that selected possible causal eQTLs as predictors for each gene. The expression models trained in GTEx v.8 can be more accurate than those trained in older versions of GTEx for three reasons: (1) The sample sizes for tissues in GTEx v.8 are larger than those in older versions of GTEx (for example, we used 329 samples from GTEx v.8 to build prediction models of breast tissue in European ancestry individuals, while Wu et al.8 used 67 samples from v.6); (2) selecting possibly causal eQTL through fine-mapping can reduce the probability that non-causal eQTLs were used in the prediction models12; and (3) MASH accounts for eQTL correlation across tissues and provides more accurate estimates of beta coefficients of eQTLs used as final weights in the prediction models. By using the prediction models trained in GTEx (v.8) data, we were able to perform this joint TWAS analysis on 19,274 genes with prediction models of good performance (i.e., with eQTL signals). In contrast, two previously published large TWASs that relied on breast tissues in older versions of GTEx and traditional methods could evaluate a smaller subset of genes.7,8 Wu et al.8 evaluated 8,597 genes in their TWAS and commented that several highly implicated breast cancer susceptibility genes, such as ESR1, TERT, and MRPS30, could not be investigated because of poor performance of prediction models. Our study was able to identify these three genes as significant at the Bonferroni threshold. Similarly, Feng et al.7 investigated 901 genes in their TWAS.
We identified 17 genes in eight loci that are at least 1 Mb away from any risk variants and not in LD with risk variants reported in previous GWASs. This finding suggests that transcriptome-based association studies are able to discover cancer susceptibility signals, extending the capacity of variant-based association studies. These genes and loci are plausibly important in breast cancer susceptibility, based on evidence from previous studies in other cancers or known cancer pathways. For example, MAP2K4 in the 17p12 locus has not been implicated in breast cancer susceptibility. We found that the predicted expression of MAP2K4 in multiple tissues, including breast tissue, was positively associated with breast cancer risk. Both colocalization analysis (RCP = 0.63) and fine-mapping analysis (PIP = 0.893 in breast tissue) suggested that MAP2K4 is a possible causal breast cancer gene, and the effect was driven by multiple weak variants. MAP2K4 (also known as MKK4) is a member of the MAPK family, which act as integration points for multiple biochemical signals and are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development. MAP2K4 has been found to be a metastasis suppressor gene in ovarian carcinoma.37 Furthermore, MAP2K4 was identified as a driver gene mutated in both early and metastatic breast cancer.38,39 Taken together, it is possible that MAP2K4 was a breast cancer susceptibility gene in the 17p12 locus.
Most of our TWAS-identified genes are located in known GWAS susceptibility loci. Interestingly, we are able to identify possible susceptibility genes in a large proportion of the known breast cancer GWAS loci. In these scenarios, our TWAS revealed possible target genes that GWAS-identified risk variants act on to cause breast cancer. Interestingly, genetic variants used in our expression prediction models and TWAS analysis were highly enriched in the set of CCVs identified in a previous fine-mapping study of breast cancer,30 and the enrichment was even stronger for loci with strong GWAS signals. Using eQTL analyses, Guo et al.40 inferred 101 target genes in known breast cancer GWAS loci. We re-discovered 51 of these 101 genes in our TWAS.
One interesting GWAS locus is 1p13.3, a region containing >20 genes within 400 kb (Figure 2). Although none of the SNPs in this locus reached GWAS significance in BCAC, this locus was recently reported to be associated with breast cancer in a cross-ancestry study.41 Of the genes in this region, it is unclear which ones are breast cancer susceptibility genes simply based on GWAS signals. Our TWAS found that the predicted expression of GSTM1, GSTM2, and GSTM4 in multiple tissues, including breast tissue, was inversely associated with breast cancer risk. After adjusting for GWAS index SNPs in conditional analysis, the three genes were no longer significant, suggesting that GWAS risk SNPs may be responsible for the observed TWAS signals. Both colocalization analysis (RCP >0.99) and fine-mapping analysis suggested that all three genes are possible breast cancer candidate genes. GSTM1, GSTM2, and GSTM4 encode members of the glutathione S-transferase multigene family, which can detoxify xenobiotics, including carcinogenic compounds, and thus were proposed as cancer susceptibility genes.42 The GSTM1 null genotype has been associated with risk of several cancers.43,44,45,46,47,48,49 Therefore, there exists evidence that all three genes are possible cancer suppressors responsible for GWAS signals in 1p13.3 through carcinogen metabolism.
Another interesting example is GPR161 at 1q24.2 (Figure 2). We discovered this locus in our meta-analysis GWAS, and its expression in adipose and fibroblasts was inversely associated with breast cancer risk in our TWAS. This gene is overexpressed in triple-negative breast cancer, promotes cell proliferation, stimulates migration and invasion, and disrupts E-cadherin localization in vitro.50 Overexpression of GPR161 in fibroblasts has also been experimentally shown to increase cAMP signaling, ultimately resulting in decreased signaling of the Sonic hedgehog (Shh) pathway,51 which plays an essential role in embryonic development and tumorigenesis.52 Germline GPR161 mutations have been associated with pediatric medulloblastoma.53 Taken together, there is some evidence that GPR161 may play a role in breast cancer etiology.
Determining causality of TWAS-identified genes remains challenging because these genes may be associated with disease phenotypes through their correlation with disease-causal gene(s) in the same LD region. Based on gene-based fine-mapping, we proposed 114 genes in 83 loci to have a high probability of being causal genes. Still, these genes need to be investigated in future functional experiments. Guo et al.40 used luciferase reporter assays to study functional target genes, and they found a significant difference between alternative and reference alleles in promoter activity for five genes (DCLRE1B, SSBP4, MRPS30, ATG10, and PAX9) but failed to show functional activity for ARRDC3. These findings are consistent with ours: we found that all five genes were TWAS significant, and three (DCLRE1B, SSBP4, and PAX9) are in our proposed list of causal genes (Table S7). Consistent with Guo et al.,40 we did not find ARRDC3 to be TWAS significant.
The current study has several limitations. First, although the joint TWAS identified more genes than single-tissue TWAS, it may generate more false-positive hits because it utilizes other tissues that may not be causal to breast cancer risk,54 and it uses a large number of prediction models from multiple tissues. This may increase the chance of poor/unreliable prediction models being used for downstream association analysis and may result in identification of non-causal genes. We tried to control the type I error using a stringent Bonferroni alpha level and conducted gene-based fine-mapping analysis to suggest causal genes. Furthermore, the target tissue for cancer development might not be distinct, and gene expression across multiple tissues could be partially correlated.10,11 We also observed moderate consistency between the results of single-tissue TWASs. For instance, breast tissue is presumably the target tissue for breast cancer, but gene expression in liver might better reflect carcinogen metabolism. Fortunately, the ACAT method used in our joint TWAS analysis calculates a weighted average of p values from multiple tissues and is relatively conservative in identifying significant genes. Strikingly, the top tissues in which the joint TWAS-identified genes were upregulated were all female tissues (breast, uterus, ovary, and vagina), suggesting that our joint TWAS method was able to automatically prioritize target tissues. One focus of our future methods research is to develop more efficient methods to combine TWAS signals across tissues by effectively accounting for the correlation of the signals across tissues or giving high weights to potential target tissues.
Second, the current study analyzed only data from individuals with European ancestry and focused on overall breast cancer risk. Breast cancer is a heterogeneous disease consisting of several molecular subtypes. The genetic architecture of estrogen receptor (ER)-negative breast cancer may be different from the ER-positive subtype. In the BCAC consortium, 76% of patients had ER-positive breast cancer,3 so the current study may mainly identify genes for susceptibility of ER-positive breast cancer. Future TWASs that focus on ER-negative breast cancer or in other racial/ethnic populations are highly desirable.
Third, the current study examined only overall expression of genes but did not consider the effect of RNA splicing on disease etiology. Li et al.55 reported that RNA splicing is another primary link between genetic variation and complex diseases. Therefore, TWASs evaluating associations of genetically predicted splicing with breast cancer have great promise for identifying novel putative candidate disease genes. We are currently working on a splicing-based TWAS of breast cancer. Last, our study focused on cis-eQTL effects when constructing the expression prediction models. We did not consider trans-eQTL due to limited sample sizes in the GTEx data, but this is an interesting topic for future studies.
In conclusion, our joint TWAS identified more than 300 breast cancer genes for further functional investigation. Our approach has discovered susceptibility loci not reported previously and mapped out candidate genes in multiple known susceptibility loci. Future studies in diverse populations and with a focus on homogeneous phenotypes of breast cancer using innovative TWAS methodology are warranted. There is potential to map out most candidate genes in GWAS loci of breast cancer, the most common malignancy affecting women across the world.
Acknowledgments
This work was supported by the National Cancer Institute (R01-CA242929, R01-CA228198, P20CA233307), Breast Cancer Research Foundation (BCRF-22-071), and the NIDDK (P30 DK20595). For BCAC data, the breast cancer genome-wide association analyses were supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research; the ‘Ministère de l’Économie, de la Science et de l’Innovation du Québec’ through Génome Québec and grant PSR-SIIRI-701; the National Institutes of Health (U19 CA148065, X01HG007492); Cancer Research UK (C1287/A10118, C1287/A16563, C1287/A10710); and the European Union (HEALTH-F2-2009-223175 and H2020 633784 and 634935). All studies and funders are listed in Michailidou et al.3 This research has been conducted using the UK Biobank under application number 49564. The authors thank the participants, investigators, and staff of the UK Biobank for providing them with the resources to pursue this research. We thank Sarah Sumner for help editing the paper.
Author contributions
G.G., H.K.I., and D.H. conceived the study and contributed to the study design. P.N.F., J.M., A.N.B., G.G., and D.H. performed statistical analyses. G.G., P.N.F., J.M., A.N.B., J.L.L., O.I.O., H.K.I, and D.H. wrote and revised the manuscript.
Declaration of interests
O.I.O. reported receiving grants from Tempus (scientific advisory board) during the study, being cofounder of CancerIQ, serving as a member of the board of directors for 54gene, and receiving grants from Color Genomics (research support) and from Roche (clinical trial support for IIT) outside the submitted work.
Published: May 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.04.005.
Contributor Information
Hae Kyung Im, Email: haky@uchicago.edu.
Dezheng Huo, Email: dhuo@health.bsd.uchicago.edu.
Web resources
BCAC summary statistics, https://bcac.ccge.medschl.cam.ac.uk/bcacdata/oncoarray/oncoarray-and-combined-summary-result
COJO (GCTA), https://yanglab.westlake.edu.cn/software/gcta/
FUMA, http://fuma.ctglab.nl
GTEx Portal, https://gtexportal.org/home/
PLINK 2.0, https://www.cog-genomics.org/plink/2.0/
PrediXcan GTEx v.8 MASHR models, https://predictdb.org/
S-PrediXcan, https://github.com/hakyimlab/MetaXcan and https://github.com/hakyimlab/summary-gwas-imputation
UK Biobank, http://ukbiobank.ac.uk
Supplemental information
Data and code availability
In this study, we used only existing datasets that are publicly available (see web resources). The code pipeline and results for our joint TWAS analysis are available at https://zenodo.org/record/7814694#.ZDaspXbMK5d (DOI: 10.5281/zenodo.7814694). For specific method code, we made minor modifications to S-PrediXcan to combine results with ACAT (https://github.com/shugamoe/MetaXcan/tree/catch_up). We also made minor modifications to FOCUS to accommodate PrediXcan GTEx v.8 MASHR models (https://github.com/shugamoe/focus).
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H., Ahearn T.U., Lecarpentier J., Barnes D., Beesley J., Qi G., Jiang X., O'Mara T.A., Zhao N., Bolla M.K., et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 2020;52:572–581. doi: 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira M.A., Gamazon E.R., Al-Ejeh F., Aittomäki K., Andrulis I.L., Anton-Culver H., Arason A., Arndt V., Aronson K.J., Arun B.K., et al. Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat. Commun. 2019;10:1741. doi: 10.1038/s41467-018-08053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., GTEx Consortium. Nicolae D.L., et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H., Gusev A., Pasaniuc B., Wu L., Long J., Abu-Full Z., Aittomäki K., Andrulis I.L., Anton-Culver H., Antoniou A.C., et al. Transcriptome-wide association study of breast cancer risk by estrogen-receptor status. Genet. Epidemiol. 2020;44:442–468. doi: 10.1002/gepi.22288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu L., Shi W., Long J., Guo X., Michailidou K., Beesley J., Bolla M.K., Shu X.O., Lu Y., Cai Q., et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet. 2018;50:968–978. doi: 10.1038/s41588-018-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao G., Pierce B.L., Olopade O.I., Im H.K., Huo D. Trans-ethnic predicted expression genome-wide association analysis identifies a gene for estrogen receptor-negative breast cancer. PLoS Genet. 2017;13:e1006727. doi: 10.1371/journal.pgen.1006727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbeira A.N., Pividori M., Zheng J., Wheeler H.E., Nicolae D.L., Im H.K. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 2019;15:e1007889. doi: 10.1371/journal.pgen.1007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguet F., Anand S., Ardlie K.G., Gabriel S., Getz G.A., Graubert A., Hadley K., Handsaker R.E., Huang K.H., Kashin S., et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbeira A.N., Melia O.J., Liang Y., Bonazzola R., Wang G., Wheeler H.E., Aguet F., Ardlie K.G., Wen X., Im H.K. Fine-mapping and QTL tissue-sharing information improves the reliability of causal gene identification. Genet. Epidemiol. 2020;44:854–867. doi: 10.1002/gepi.22346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J., et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M., et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wen X., Lee Y., Luca F., Pique-Regi R. Efficient Integrative Multi-SNP Association Analysis via Deterministic Approximation of Posteriors. Am. J. Hum. Genet. 2016;98:1114–1129. doi: 10.1016/j.ajhg.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen X., Pique-Regi R., Luca F. Integrating molecular QTL data into genome-wide genetic association analysis: Probabilistic assessment of enrichment and colocalization. PLoS Genet. 2017;13:e1006646. doi: 10.1371/journal.pgen.1006646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urbut S.M., Wang G., Carbonetto P., Stephens M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet. 2019;51:187–195. doi: 10.1038/s41588-018-0268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasaniuc B., Zaitlen N., Shi H., Bhatia G., Gusev A., Pickrell J., Hirschhorn J., Strachan D.P., Patterson N., Price A.L. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics. 2014;30:2906–2914. doi: 10.1093/bioinformatics/btu416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Xie J. Cauchy combination test: a powerful test with analytic p-value calculation under arbitrary dependency structures. J. Am. Stat. Assoc. 2020;115:393–402. doi: 10.1080/01621459.2018.1554485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Ferreira T., Morris A.P., Medland S.E., Genetic Investigation of ANthropometric Traits GIANT Consortium. DIAbetes Genetics Replication And Meta-analysis DIAGRAM Consortium. Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berisa T., Pickrell J.K. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe K., Taskesen E., van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J., et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brinton L., Gaudet M., Gierach G. In: Schottenfeld and Fraumeni Cancer Epidemiology and Prevention. Thun M., Linet M., Cerhan J., Haiman C., Schottenfeld D., editors. Oxford University Press; 2018. Breast Cancer. [Google Scholar]
- 29.Rashkin S.R., Graff R.E., Kachuri L., Thai K.K., Alexeeff S.E., Blatchins M.A., Cavazos T.B., Corley D.A., Emami N.C., Hoffman J.D., et al. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat. Commun. 2020;11:4423. doi: 10.1038/s41467-020-18246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fachal L., Aschard H., Beesley J., Barnes D.R., Allen J., Kar S., Pooley K.A., Dennis J., Michailidou K., Turman C., et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat. Genet. 2020;52:56–73. doi: 10.1038/s41588-019-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffman J.D., Graff R.E., Emami N.C., Tai C.G., Passarelli M.N., Hu D., Huntsman S., Hadley D., Leong L., Majumdar A., et al. Cis-eQTL-based trans-ethnic meta-analysis reveals novel genes associated with breast cancer risk. PLoS Genet. 2017;13:e1006690. doi: 10.1371/journal.pgen.1006690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia G., Ping J., Shu X., Yang Y., Cai Q., Kweon S.S., Choi J.Y., Kubo M., Park S.K., Bolla M.K., et al. Genome- and transcriptome-wide association studies of 386,000 Asian and European-ancestry women provide new insights into breast cancer genetics. Am. J. Hum. Genet. 2022;109:2185–2195. doi: 10.1016/j.ajhg.2022.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen W., Chen Z., Bao J., Long Q., Shu X.O., Zheng W., Guo X. Genetic variations of DNA bindings of FOXA1 and co-factors in breast cancer susceptibility. Nat. Commun. 2021;12:5318. doi: 10.1038/s41467-021-25670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J., Wen W., Beeghly A., Chen Z., Cao C., Shu X.O., Zheng W., Long Q., Guo X. Integrating transcription factor occupancy with transcriptome-wide association analysis identifies susceptibility genes in human cancers. Nat. Commun. 2022;13:7118. doi: 10.1038/s41467-022-34888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song X., Ji J., Rothstein J.H., Alexeeff S.E., Sakoda L.C., Sistig A., Achacoso N., Jorgenson E., Whittemore A.S., Klein R.J., et al. MiXcan: a framework for cell-type-aware transcriptome-wide association studies with an application to breast cancer. Nat. Commun. 2023;14:377. doi: 10.1038/s41467-023-35888-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kar S.P., Considine D.P.C., Tyrer J.P., Plummer J.T., Chen S., Dezem F.S., Barbeira A.N., Rajagopal P.S., Rosenow W.T., Moreno F., et al. Pleiotropy-guided transcriptome imputation from normal and tumor tissues identifies candidate susceptibility genes for breast and ovarian cancer. HGG Adv. 2021;2:100042. doi: 10.1016/j.xhgg.2021.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamada S.D., Hickson J.A., Hrobowski Y., Vander Griend D.J., Benson D., Montag A., Karrison T., Huo D., Rutgers J., Adams S., Rinker-Schaeffer C.W. Mitogen-activated protein kinase kinase 4 (MKK4) acts as a metastasis suppressor gene in human ovarian carcinoma. Cancer Res. 2002;62:6717–6723. [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lefebvre C., Bachelot T., Filleron T., Pedrero M., Campone M., Soria J.C., Massard C., Lévy C., Arnedos M., Lacroix-Triki M., et al. Mutational Profile of Metastatic Breast Cancers: A Retrospective Analysis. PLoS Med. 2016;13:e1002201. doi: 10.1371/journal.pmed.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X., Lin W., Bao J., Cai Q., Pan X., Bai M., Yuan Y., Shi J., Sun Y., Han M.R., et al. A Comprehensive cis-eQTL Analysis Revealed Target Genes in Breast Cancer Susceptibility Loci Identified in Genome-wide Association Studies. Am. J. Hum. Genet. 2018;102:890–903. doi: 10.1016/j.ajhg.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adedokun B., Du Z., Gao G., Ahearn T.U., Lunetta K.L., Zirpoli G., Figueroa J., John E.M., Bernstein L., Zheng W., et al. Cross-ancestry GWAS meta-analysis identifies six breast cancer loci in African and European ancestry women. Nat. Commun. 2021;12:4198. doi: 10.1038/s41467-021-24327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebbeck T.R. Molecular epidemiology of the human glutathione S-transferase genotypes GSTM1 and GSTT1 in cancer susceptibility. Cancer Epidemiol. Biomarkers Prev. 1997;6:733–743. [PubMed] [Google Scholar]
- 43.Liu X., Li Z., Zhang Z., Zhang W., Li W., Xiao Z., Liu H., Jiao H., Wang Y., Li G. Meta-analysis of GSTM1 null genotype and lung cancer risk in Asians. Med. Sci. Monit. 2014;20:1239–1245. doi: 10.12659/MSM.890490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai X., Yang L., Chen H., Wang C. An updated meta-analysis of the association between GSTM1 polymorphism and colorectal cancer in Asians. Tumour Biol. 2014;35:949–953. doi: 10.1007/s13277-013-1125-0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X.L., Cui Y.H. GSTM1 null genotype and gastric cancer risk in the Chinese population: an updated meta-analysis and review. OncoTargets Ther. 2015;8:969–975. doi: 10.2147/OTT.S79099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H., Yang S., Liu J., Shao F., Wang H., Wang Y. The association of GSTM1 deletion polymorphism with lung cancer risk in Chinese population: evidence from an updated meta-analysis. Sci. Rep. 2015;5:9392. doi: 10.1038/srep09392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu J., Zou H., Zheng L., Li X., Chen S., Zhang L. GSTM1 null genotype is associated with increased risk of gastric cancer in both ever-smokers and non-smokers: a meta-analysis of case-control studies. Tumour Biol. 2014;35:3439–3445. doi: 10.1007/s13277-013-1454-z. [DOI] [PubMed] [Google Scholar]
- 48.Economopoulos K.P., Choussein S., Vlahos N.F., Sergentanis T.N. GSTM1 polymorphism, GSTT1 polymorphism, and cervical cancer risk: a meta-analysis. Int. J. Gynecol. Cancer. 2010;20:1576–1580. doi: 10.1111/IGC.0b013e3181ca1dfc. [DOI] [PubMed] [Google Scholar]
- 49.Zubair H., Aurangzeb J., Zubair B., Imran M. Association of GSTM1 and GSTT1 genes insertion/deletion polymorphism with colorectal cancer risk: A case-control study of Khyber Pakhtunkhwa population, Pakistan. J. Pak. Med. Assoc. 2022;72:457–463. doi: 10.47391/JPMA.1393. [DOI] [PubMed] [Google Scholar]
- 50.Feigin M.E., Xue B., Hammell M.C., Muthuswamy S.K. G-protein-coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. Proc. Natl. Acad. Sci. USA. 2014;111:4191–4196. doi: 10.1073/pnas.1320239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S.J., Jackson P.K. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 52.Carballo G.B., Honorato J.R., de Lopes G.P.F., Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun. Signal. 2018;16:11. doi: 10.1186/s12964-018-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Begemann M., Waszak S.M., Robinson G.W., Jäger N., Sharma T., Knopp C., Kraft F., Moser O., Mynarek M., Guerrini-Rousseau L., et al. Germline GPR161 Mutations Predispose to Pediatric Medulloblastoma. J. Clin. Oncol. 2020;38:43–50. doi: 10.1200/JCO.19.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K., et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In this study, we used only existing datasets that are publicly available (see web resources). The code pipeline and results for our joint TWAS analysis are available at https://zenodo.org/record/7814694#.ZDaspXbMK5d (DOI: 10.5281/zenodo.7814694). For specific method code, we made minor modifications to S-PrediXcan to combine results with ACAT (https://github.com/shugamoe/MetaXcan/tree/catch_up). We also made minor modifications to FOCUS to accommodate PrediXcan GTEx v.8 MASHR models (https://github.com/shugamoe/focus).