Abstract
Mammalian genomes possess multiple enhancers spanning an ultralong distance (>megabases) to modulate important genes, yet it is unclear how these enhancers coordinate to achieve this task. Here, we combine multiplexed CRISPRi screening with machine learning to define quantitative enhancer-enhancer interactions. We find that the ultralong distance enhancer network possesses a nested multi-layer architecture that confers functional robustness of gene expression. Experimental characterization reveals that enhancer epistasis is maintained by three-dimensional chromosomal interactions and BRD4 condensation. Machine learning prediction of synergistic enhancers provides an effective strategy to identify non-coding variant pairs associated with pathogenic genes in diseases beyond Genome-Wide Association Studies (GWAS) analysis. Our work unveils nested epistasis enhancer networks, which can better explain enhancer functions within cells and in diseases.
One-Sentence Summary:
A nested synergistic and additive enhancer network confers robustness of gene expression, which can model disease risks.
Disease-associated genes including oncogenes are frequently associated with many remote enhancers spanning across a long genomic distance (>megabases, Mb) (1-4). Genome-wide association studies (GWAS) reveal that non-coding variants of the regulatory elements, including enhancers, account for >90% of the variants in diseases and can spread over a long distance (5-8). While individual enhancer variants may present modest clinical risks (9), there are examples showing that a combination of multiple variants may greatly amplify the effects in traits and diseases (10-12). Like gene interaction (13), these enhancers may interact as an epistatic network wherein the effect of an enhancer is dependent on other enhancers to regulate gene dosage and confer robustness. Aside from these observations, it remains largely unknown why multiple ultralong-distance enhancers exist for important genes and how their interactions modulate gene regulation and diseases.
Enhancer interactions were previously studied within a single enhancer cluster. For example, super-enhancers were defined as a dense cluster, which contains adjacent enhancers within tens of kilobases (kb) (14, 15, 16). Other enhancer clusters like super-enhancers were also reported, including ‘stretch enhancers’ and ‘enhancer clusters’ (17, 18). A few examples by perturbing local enhancers within these enhancer clusters showed they may interact additively or synergistically for regulatory roles (19-25). However, these short-range enhancers organized in a cluster cannot explain the prevalence of ultralong-distance enhancers in the human genome.
It remains unknown how multiple enhancers interact with one another over long genomic distances to confer regulatory roles in gene expression and disease risks. Here, we hypothesize that by using ultralong-distance enhancers ( >1Mb), disease-associated genes have evolved high robustness to disruptive effects from genetic variations. These interactions likely occur via an elaborate network on the three-dimensional (3D) genome organization level.
High-resolution multiplexed perturbation of enhancers reveals a nested two-layer epistasis network
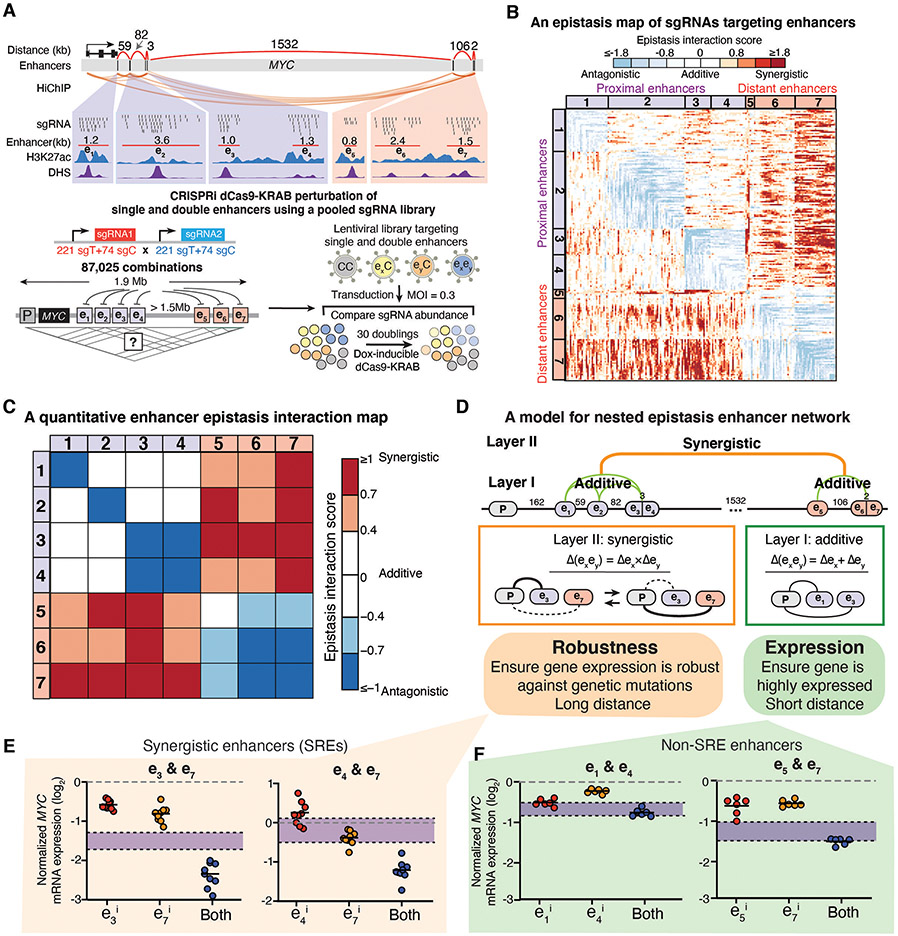
To gain insights into the ultralong-distance enhancer network for disease-relevant genes, we adopted a high-resolution approach to quantitatively analyze enhancer interactions in gene regulation. We chose the endogenous MYC locus as a model system. As an important oncogene governing cancer cell proliferation, the MYC locus encompasses 7 enhancers (e1-e7) spanning a 1.9 Mb region in K562 erythroleukemia cells (26). The reported linear correlation between MYC expression and cellular growth supports its use as a model system to quantitatively dissect the enhancer epistatic network in ultralong distance (26, 27). We conducted a multiplexed CRISPR interference (CRISPRi) screen (28, 29, 30), using a pooled library consisting of 87,025 pairs of single guide RNAs (sgRNAs) tiling all single and pairwise combinations of 7 enhancers (Fig. 1A, fig. S1A-B, table S1). We transduced the pooled sgRNA library into K562 cells stably expressing a doxycycline-inducible nuclease-dead dCas9-KRAB fusion and cultured cells for 30 doublings.
Fig. 1. High-resolution multiplexed CRISPRi perturbation of ultralong-distance enhancers at the MYC locus reveals a nested two-layer epistasis network.
A, Top: The MYC locus regulated by multiple enhancers distributed over an ultralong distance (~1.9Mb). Bottom: Diagram showing the multiplexed CRISPRi screening for high-resolution dissection of enhancer interactions. K562 cells expressing the doxycycline (Dox)-inducible dCas9-KRAB are transduced by a pooled sgRNA library targeting single or double MYC enhancers. Cells are harvested to sequence the pairwise sgRNA enrichment before and after 30 doublings. sgT, targeting sgRNA; sgC, control sgRNA.
B, A quantitative epistasis map of sgRNA pairs targeting all enhancer combinations in the MYC locus. Each dot represents the epistasis interaction score of a pair of sgRNAs smoothed by adjacent sgRNAs.
C, A quantitative enhancer epistasis map at the MYC locus.
D, A nested two-layer model for the enhancer epistasis network.
E,F, qRT-PCR of MYC mRNA expression for perturbing SREs e3&e7 or e4&e7 (E), or non-SREs e1&e4 or e5&e7 (F). P=0.02, 1.13E-05, 0.13, 0.61, for e3&e7, e4&e7, e1&e4, e5&e7, respectively. Data are represented as individual biological replicates (dots) and the mean value (black bar). The purple area indicates the expected additive effect by plotting mean ± one standard derivation. P values are calculated by t-test.
We calculated the depletion score of each sgRNA pair by comparing the relative abundance before and after cell culture (Fig. 1A, fig. S2A and B, table S2, see Methods). Using the depletion scores to fit a linear additive model, we calculated enhancer interaction scores to identify epistasis interactions and generated a high-density quantitative epistasis map of enhancer-targeting sgRNAs (Fig. 1B, fig. S2C-E, see Methods). We confirmed the epistasis interaction scores were reproducible across biological replicates and different sgRNA pairs targeting the same enhancer pair (fig. S2F-I). We observed clusters of sgRNAs targeting the same pairs of enhancers showing similar patterns of synergistic or additive interactions, suggesting an epistatic interaction relationship between enhancer pairs (Fig. 1B, fig. S2E, fig. S3A).
We computed the epistasis interaction scores for each enhancer pair by averaging the epistasis interaction scores of the top 25% sgRNA pairs (Fig. 1C, fig. S3B, see Methods). We observed synergistic epistasis when perturbing distant enhancer pairs (>1Mb), with all four proximal enhancers (e1-e4) showing strong synergistic interactions with the other three distant enhancers (e5-e7) upon perturbation. In contrast, perturbation of enhancer pairs within the proximal or distant group mostly showed additive interactions (Fig. 1C).
Our data suggested a nested two-layer architecture of the enhancer epistasis network in regulating genes with large-scale landscapes (Fig. 1D). In the first layer (Layer I), enhancer pairs (<100kb at MYC locus) behave additively after perturbation, suggesting individual enhancers contribute independently to gene expression. In the second layer (Layer II), distant enhancer pairs showed non-linear synergistic effects after perturbation, which is speculated to function as compensatory regulatory elements for one another to maintain the robustness of gene expression upon perturbation. These synergistic enhancers are distributed over long genomic distances, which likely reduces the chance of co-mutation and thus confers robustness of gene expression against mutations or chromosome perturbations. We define synergistic regulatory enhancers (SREs) as a pair of distant enhancers with synergistic effects on gene expression upon perturbation.
We experimentally validated SREs and non-SRE pairs by examining whether they can combinatorically perturb MYC expression and cellular growth. Using different sgRNA pairs targeting the same SREs (e3&e7; e4&e7), we observed synergistically decreased MYC expression as well as cell proliferation (Fig. 1E, fig. S4A-C, table S1). In comparison, inhibiting enhancers within the same proximal or distant groups led to additive repression effects (Fig. 1F, table S1).
We performed H3K9me3 and H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) to characterize the resolution of using dCas9-KRAB for enhancer perturbation. We confirmed no spreading effects of KRAB on adjacent enhancers (fig. S5A and B, fig. S6A and B). We also knocked out pairs of enhancers by transducing sgRNAs to K562 cells that stably expressed the nuclease Cas9 (see Methods). We confirmed consistent synergistic and additive interactions between e3&e7 and e1&e4, respectively (fig. S7A and B). However, we also observed deletions of large chromatin regions when knocking out pairs of enhancers (fig. S7C-F). This observation was consistent with reports that gene editing at multiple sites on the same DNA can induce megabase-scale chromosome deletions, which potentially confounds the study of enhancer interactions (31, 32). These results together confirm that dCas9-KRAB is a high-resolution approach for studying multiple enhancer interactions without unwanted large DNA deletions.
Machine learning modeling reveals determinants of SRE synergy
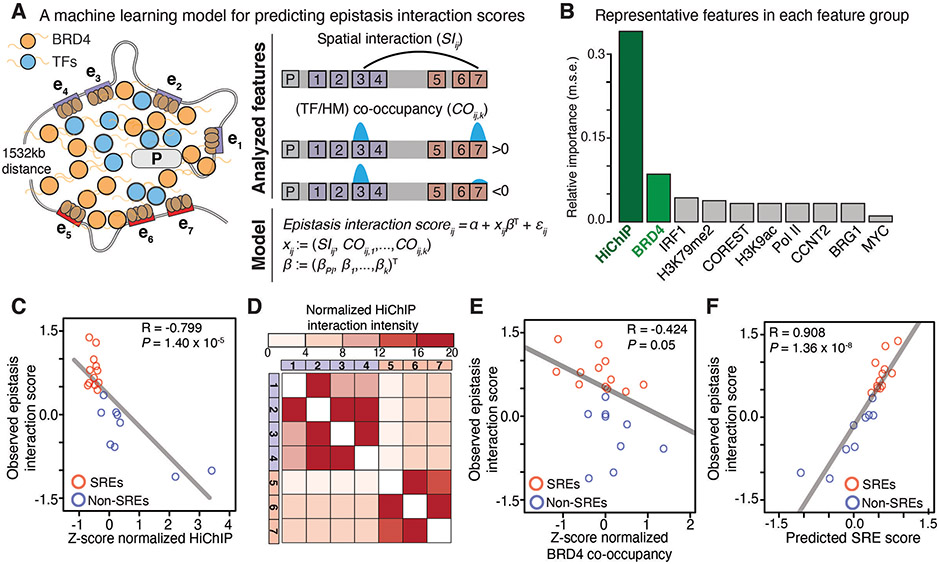
We next developed a machine learning model based on an elastic-net regularized generalized linear model to analyze the determinants of SRE synergy (33) (Fig. 2A). We examined publicly available transcription factor (TF) binding profiles, histone modification (HM) profiles, and H3K27ac HiChIP datasets that capture DNA-DNA spatial contacts in K562 cells (table S3, see Methods) (5). Among all features, spatial DNA contact is the most relevant feature, and was inversely correlated with calculated epistasis interaction scores (Fig. 2B and C, fig. S8A). We found that the spatial contacts between SREs were weaker than non-SREs, which displayed an inverse pattern with the enhancer epistasis map (Fig. 2D vs Fig. 1C). In addition, the co-occupancy of bromodomain-containing protein 4 (BRD4), a key chromatin-associated coactivator, showed a strong anticorrelation with epistasis interaction scores (Fig. 2B and E, fig. S8A and B).
Fig. 2. A machine learning model for analyzing determinants of the SRE synergy.
A, An elastic net regularized linear regression model for predicting epistasis interaction scores. We selected features including the chromatin spatial interaction (SIij) and co-occupancy (COij,k) of 38 TFs and 8 HM profiles.
B, The relative importance of each feature group for predicting epistasis interaction scores. The representative feature has the highest correlation in that group (fig. S8A). m.s.e., mean squared error.
C-F, Correlation between epistasis interaction scores and Z-scores normalized spatial contact (C) and BRD4 co-occupancy (E). D, Heatmap of normalized HiChIP interaction intensity between enhancers. F, Correlation between predicted SRE scores and observed epistasis interaction scores. In C, D, F: red, SREs; blue, non-SREs. The Pearson correlation coefficient (R) and P value are shown.
The elastic net regression model performed better for predicting SREs compared to simple linear models using individual representative features (fig. S8C and D). Predicted scores of all enhancer pairs were correlated with observed epistasis interaction scores assessed from the CRISPRi screen (Fig. 2F). Altogether, our machine learning model suggests that spatial DNA contacts and BRD4 co-occupancy are two major determinants for predicting SREs.
The SRE model can predict synergistic enhancer interactions at other genomic loci
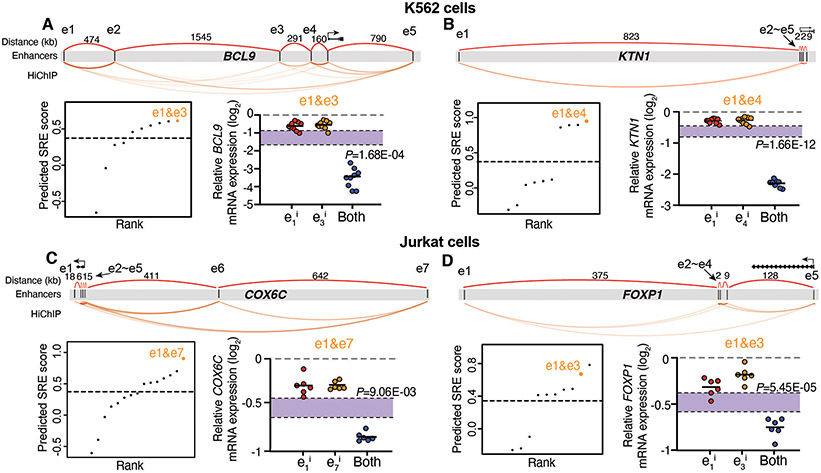
We next verified whether the SRE prediction model can be generalized to study other genes that possess multiple enhancers spanning an ultralong distance in different cell types (fig. S9A, see Methods). We examined the enhancer profiles of four disease-relevant genes: BCL9 and KTN1 in K562 cells, and COX6C and FOXP1 in Jurkat cells, all of which possess multiple enhancers spreading over a large genomic distance (3.3Mb, 0.8Mb, 1.1Mb, and 0.5Mb, respectively) (Fig. 3A-D, see Methods). We used the SRE prediction model to calculate putative SREs and non-SREs and designed sgRNA pairs to target each SRE and non-SRE.
Fig. 3. Experimental validation of predicted SREs at other genomic loci in different cell types.
A-D, Prediction and validation of SREs at BCL9 (A) and KTN1 loci (B) in K562 cells, and COX6C (C) and FOXP1 loci (D) in Jurkat cells. Top: Diagram showing multiple enhancers spanning an ultralong distance at each genomic locus. Bottom left: Rank of predicted SREs using the model. Dashed line represents the empirical threshold from the MYC locus. Orange dots indicate the validated SREs. Bottom right: qRT-PCR of mRNA expression for each gene when perturbing the predicted SREs. Data are represented as individual biological replicates (dots) and the mean value (black bar). The purple area indicates the expected additive effect by plotting mean ± one standard derivation. P values are calculated by t-test.
We observed synergistic changes of gene expression when targeting the predicted SRE pairs (Fig. 3A-D, fig. S9B-E), as well as additive effects when targeting the non-SRE pairs (fig. S9B and C). These data suggested that our machine learning model can predict functional interaction between enhancers (SRE or non-SRE) that regulate different genes spanning an ultralong distance in different cell types. We further developed a website (http://enhancer.stanford.edu/) by exploring all 4,835 putative networks of ultralong distance enhancers (>=5 enhancers; >200kb inter-distance) across 6 cell types (GM12878, K562, Jurkat, A549, HUVEC, and HCT116), which reports many predicted SREs and associated epistasis interaction scores.
Inhibition of SREs leads to synergistic reduction of local spatial contacts and BRD4 condensation
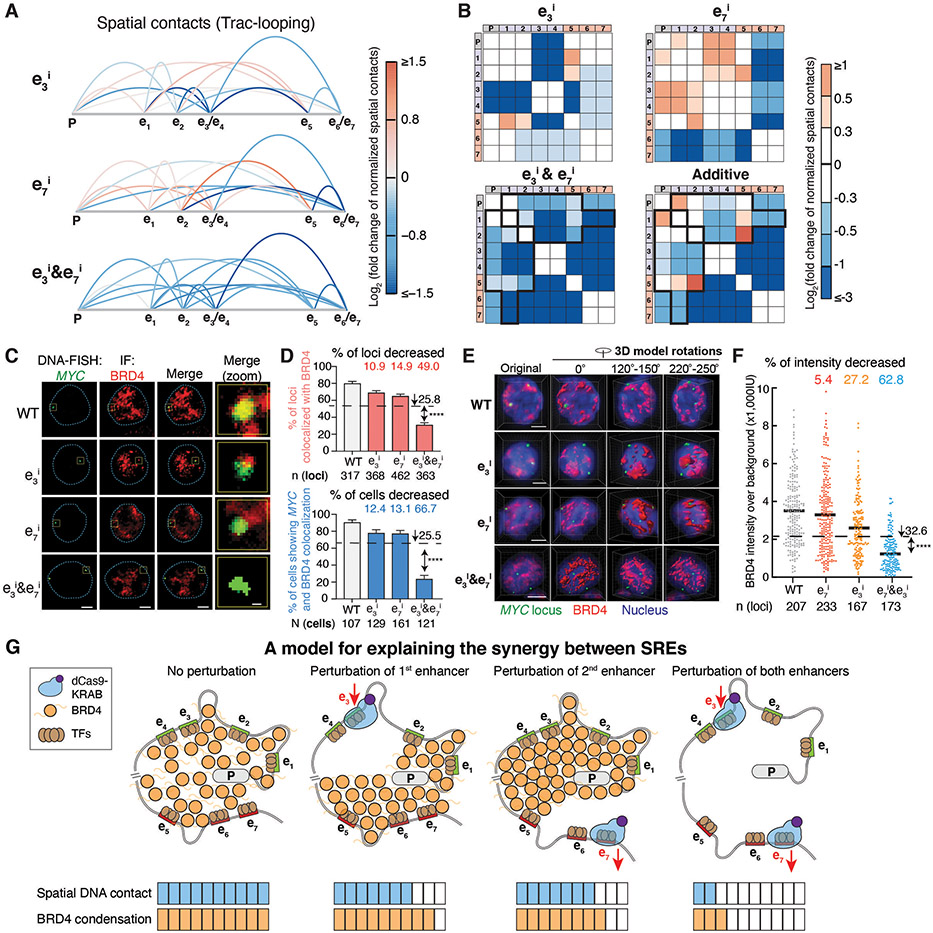
To experimentally examine the predicted determinants of the SRE model, we performed Trac-looping assays on CRISPRi-perturbed samples targeting a SRE pair e3&e7 to measure both spatial contacts and chromatin accessibility (fig. S10A) (34). We observed inhibition of individual enhancers decreased spatial contacts only between the targeted enhancer and other elements, while simultaneous inhibition of e3&e7 led to synergistic reduction of the spatial contacts at the MYC locus (Fig. 4A and B), which is consistent with the observed epistatic effects on MYC expression and cell growth (Fig. 1E, fig. S4A and B). In comparison, simultaneous inhibition of a non-SRE pair e1&e4 led to additive reduction of spatial contacts (fig. S10B). We also observed that inhibition of SREs showed no significant difference from the additive effects on chromatin accessibility (fig. S10C-E), suggesting that chromatin accessibility is less involved in synergistic interactions.
Fig. 4. Perturbation of SREs leads to synergistic reduction of spatial contacts and BRD4 condensation at the genomic locus.
A-B, Spatial contacts between the promoter and enhancers measured by Trac-looping for the MYC locus upon perturbation of e3, e7, and e3&e7. Colors represent the log2 fold change of spatial contacts normalized to the wildtype cells. Black boxes in (B) indicate synergistically decreased (more than additive) spatial contacts of e3&e7 pair perturbation.
C-F, DNA-FISH colocalization between BRD4 and the MYC locus of representative K562 cells for 2D (C-D) and 3D image analysis (E-F) upon perturbation of e3, e7, and e3&e7. In (C&E), red, BRD4 immunofluorescence (IF) staining; green, DNA-FISH at the MYC locus; blue dashed line, nuclear periphery determined by DAPI staining (not shown); scale bars, 5μm. The rightmost column in (C) shows insets in the yellow boxes. Scale bars, 500 nm. Quantification of BRD4 and the MYC locus colocalization are shown for 2D (D) and 3D image analysis (F). In (D), percentage of loci with colocalization is shown on the top and percentage of cells (≥ 2 colocalization loci) is shown on the bottom; data is represented as mean ± standard error of the mean. In (F), each dot represents an individual locus. n = total loci, N = total cells. **** P < 0.0001 in Fisher’s exact test (D) or t-test (F) versus the expected additive effect (dashed line).
G, A model to explain the synergy between SREs.
Perturbation of the distant enhancer e7 increased spatial contacts among the proximal enhancers and the promoter (e.g., e1-e3, e1-e4, e2-e3, e2-e4, e3-promoter and e4-promoter) (Fig. 4A and B). Similarly, perturbing e3 or e4 led to increased spatial contacts among the distant enhancers (Fig. 4A and B, fig. S10B). These observations imply a possible compensation mechanism on the spatial DNA contact between the SREs, which likely confers robustness of gene expression upon genome disruption (e.g., mutations or loss of DNA-TF interactions).
We next investigated the relationship between enhancer interactions and BRD4 localization. Clustered coactivator condensates mediated by BRD4 can assemble the transcription apparatus at enhancers to drive robust gene expression (35-37). Our machine learning model predicted that the SREs were associated with distinct BRD4 clusters (Fig. 2E, fig. S11A). We examined this relationship by studying BRD4 colocalization at the MYC locus via immunostaining and fluorescence in situ hybridization (FISH) confocal imaging.
Compared to wildtype K562 cells, inhibiting individual enhancers (e3 or e7) resulted in a small reduction of colocalization between BRD4 and MYC loci, whereas simultaneous inhibition of e3&e7 synergistically decreased the colocalization (49.0%) and the percentage of cells showing colocalization (66.7%) (Fig. 4C and D). Similar results were observed for another SRE pair e4&e7 (fig. S11B and C). We also performed 3D FISH to better quantify the fluorescent intensity of the BRD4 condensate at the MYC locus. While individual enhancer perturbation slightly decreased the BRD4 intensity (27.2% and 5.4% for e3 and e7, respectively), simultaneous perturbation led to synergistic BRD4 reduction (62.8%) (Fig. 4E and F, Movie S1-S4). In comparison, simultaneous inhibition of non-SRE e1&e4 led to additive decrease of colocalization between BRD4 and MYC loci (fig. S11D). We further used a BRD4 inhibitor, JQ1, to investigate whether BRD4 condensation was involved in maintaining the synergistic interaction of SRE (38). Consistently, with increasing JQ1 concentrations, the synergistic effect from SRE perturbation decreased and then disappeared, implying the importance of BRD4 condensation for enhancer synergy (fig. S11E and F).
These results together confirmed that SRE perturbation synergistically reduced spatial DNA contact and BRD4 condensation at the target genomic locus, which led to synergistic changes of gene expression (Fig. 4A-F, Fig. 1E). Based on computational and experimental analysis, we propose a speculative model (Fig. 4G): while perturbing individual enhancers modestly reduces spatial contacts and BRD4 condensation, perturbation of two distant enhancers dramatically alters the 3D chromosome organization and BRD4 condensation to confer synergistic regulatory roles.
Synergistic interactions between predicted SRE variants influence gene expression and disease risk
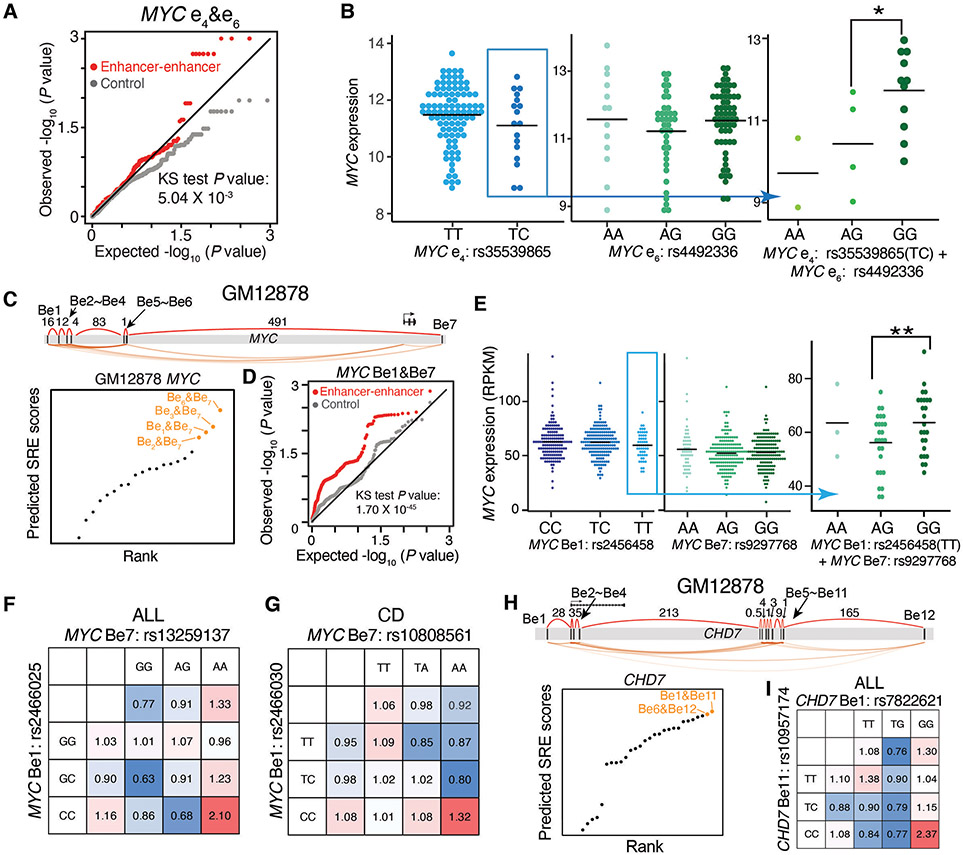
We evaluated whether SRE genetic variants spanning the ultralong distance can alter gene regulation and disease risks in an epistatic manner (fig. S12A). We examined the effect of our validated SREs within the MYC locus using an acute myeloid leukemia (LAML) patient database containing genomic and transcriptomic data. In LAML patients, we observed that e4&e6 SRE variants interacted more frequently to alter MYC expression than expected by chance, additive effect, and non-SRE variants (Fig. 5A, fig. S12B-D, see Methods, Supplementary Text). A large difference in MYC expression levels was observed in two patient groups stratified by the genotype combinations of e4&e6 SRE variants, while there were no dynamic changes when considering the genotypes of individual SRE variants (Fig. 5B).
Fig. 5. Synergistic interactions between predicted SRE variants influence gene expression and disease risk in an epistatic manner.
A-B, Analysis of predicted SRE variants at the MYC locus in K562 cells for influence on gene expression. A, quantile-quantile (QQ) plot showing the distribution of P values for the epistasis influence on MYC expression between e4&e6 variants (red) in LAML patients, compared to random permutations (grey); P value in Kolmogorov–Smirnov (KS) test. B, MYC expression in LAML patients stratified by e4&e6 SRE variants. * P < 0.05 in Wilcoxon test.
C-G, Analysis of predicted SRE variants at the MYC locus in GM12878 cells for influence on gene expression and associated disease risk. C, Diagram showing the rank of predicted SREs; orange dots show top SREs. D, QQ plot showing the distribution of P values for the epistasis influence of Be1&Be7 variants (red) on MYC expression in the B lymphoblasts of 373 European individuals, compared to random permutations (grey). P value in KS test. E, MYC expression in the B lymphoblasts from individuals stratified by Be1&Be7 variant. ** P < 0.01 in Wilcoxon test. F-G, Calculated odds ratio on the relapse risk in acute lymphoblastic leukemia (ALL) (F) and Crohn's disease (CD) (G). Odds ratios are calculated by considering the genotypes of individual variants or both SRE variants. Colors represent the odds ratios.
H-I, Analysis of predicted SRE variants at the CHD7 locus in GM12878 cells for influence on ALL. H, Diagram showing the rank of predicted SREs; orange dots show top SREs. I, Calculated odds ratio on the relapse risk in ALL. Odds ratios are calculated by considering the genotypes of individual variants or both SRE variants. Colors represent the odds ratios.
We further examined the epistatic effect of MYC SRE variants on gene regulation in B-lymphoblastic cells. We named the enhancers in GM12878 B lymphoblastoid cells as Be and used the SRE model to predict the interaction network among seven enhancers and rank SREs (Fig. 5C). We examined the interactions of variants across predicted SREs in a database of B-lymphoblast genomic variants and transcriptomes (39). While there was no difference of MYC expression by looking at the genotypes of single enhancer variants, a significant difference of MYC expression was observed when combinatorically considering the genotypes of SRE variants at Be1&Be7 (Fig. 5D and E, fig. S12E-G, Supplementary Text), or Be6&Be7 (fig. S12H and I).
Next, we applied the predicted SREs to investigate the association of MYC SRE variants in B-cell-associated diseases, acute lymphoblastic leukemia (ALL) and Crohn's disease (CD) patients (40-44). In the top four predicted SRE pairs, we identified two SRE instances, Be1&Be7 and Be2&Be7, where the SRE variant pairs can synergistically influence the clinical risk, including ALL relapse risk and CD disease risk (fig. S13A-E, Supplementary Text). Particularly, when we stratified case and control population based on SRE variants, the odds ratio was significantly higher than the odds ratio determined by individual SRE variants alone or additively (Fig. 5F and G, fig. S13F and G, Supplementary Text).
We also predicted SREs in other gene loci in GM12878 cells and observed the epistatic influence of SRE variants in gene expression and clinical risks, including the leukemogenesis-associated CHD7 locus and B-cell antigen CD180 locus (45) (Fig. 5H and I, fig. S13G, fig. S14, fig. S15, see Methods), which both possess enhancer networks spreading ultralong genomic distance (0.4Mb and 1.2Mb, respectively).
The SRE model better identifies epistatic influence of genome-wide non-coding variants on disease risk
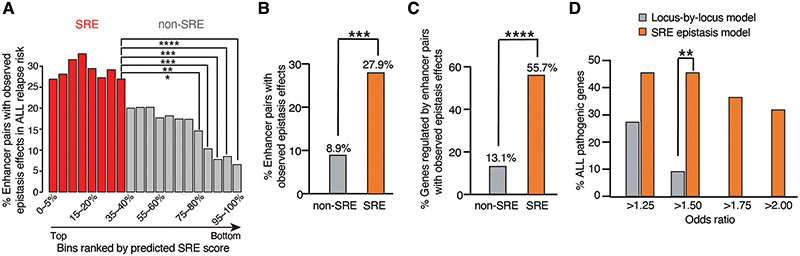
Finally, we applied the SRE prediction model to the genome-wide analysis in GM12878 cells to link multiple enhancer variants to disease risk. Among >900 genes containing ultralong distance enhancers networks, we focused on 70 immune- or cancer-related and highly expressed genes (fig. S16A-B, fig. S17, Supplementary Text). Notably, the predicted SRE scores correlated well with the epistatic effects of non-coding variants on the clinical risk for ALL relapse patients (Fig. 6A, Supplementary Text). Specifically, 27.9% of predicted SREs targeting 55.7% of genes showed epistatic effects on ALL relapse risk via our SRE model, which is significantly higher than the non-SRE pairs (Fig. 6B and C). Furthermore, the SRE model also identified significantly more ALL-associated pathogenic genes compared to the traditional locus-by-locus model (Fig. 6D). For example, among 22 literature reported ALL-associated pathogenic genes (table S4, Supplementary Text), our SRE model recovered 10 genes, whereas the locus-by-locus model showed only 2 genes (Fig. 6D, Supplementary Text). Therefore, the SRE prediction model can effectively elucidate the epistatic influence of multiple non-coding variants on associated clinical risk.
Fig. 6. Genome-wide analysis of epistatic influence of SRE variants on disease risk.
A, Percentage of enhancer pairs with observed epistatic effects on ALL relapse risk for predicted SREs and non-SREs.
B-C, Percentage of enhancer pairs (B) and genes (C) exhibiting interactive effects on ALL relapse risk. SRE pairs: enhancer pairs with top 40% SRE predicted score; non-SRE pairs: enhancer pairs with bottom 10% SRE predicted score.
D, Comparison of identified ALL pathogenic genes between the SRE model and the traditional locus-by-locus model at different odds ratio levels.
In all figures, *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001 in Fisher’s exact test.
Discussion
Our work is different from previous studies on interactions (<100kb) within enhancer clusters (e.g., super-enhancer) (19-25, 46). While small-scale perturbations revealed additive (21, 22, 46) or synergistic (23, 25) interactions within these enhancer clusters, it remained unknown if enhancers distributed on a very large scale (>1Mb) play interactive roles for gene regulation. Our results demonstrate that the observed nested synergistic interactions over the long distance and additive interactions in the short distance are important for an integrated function in the enhancer network: while the additive effects ensure a high expression level, the synergistic effects confer robustness against perturbations. Additional quantitative interaction mapping at more genomic loci in more cell types (e.g., diploid cells to rule out aneuploidy effects) should allow for the derivation of distance requirements for ultralong distance enhancer networks and a universal prediction model for enhancer networks. It should also help elucidate whether strong versus weak inhibition effects of individual enhancers determine if they are SREs or non-SREs.
Our analysis showed that SREs are prevalent in the mammalian genome. The identification of SREs is consistent with evidence from studies in the 1000 Genomes Project, which showed that enhancer regions can be deleted without obvious phenotypic alterations (47, 48). Theoretically, long-distance enhancers are less likely to be mutated at the same time, which avoids co-mutagenesis and thus provide compensation effects on important gene expression against mutations. Our website that comprehensively explores genome-wide SREs provides a resource to study enhancer interactions for gene regulation and multiple non-coding variants for diseases.
Since perturbations of individual enhancers may exhibit modest effects on gene expression, multiplexed perturbation of enhancers in the native chromatin context is crucial to fully elucidate their roles. we observed clusters of sgRNA pairs showing similar patterns of synergistic or additive interactions within an enhancer (fig. S3A), suggesting a high-resolution (~300bp) sub-enhancer interaction mapping capability. We note that due to dCas9-KRAB spreading effects (500bp~1kb estimated by H3K9me3 peaks) (fig. S5A and B), results from dCas9-KRAB should be validated using the Cas9 nuclease knockout for very close enhancers (<1kb). Nevertheless, our analysis among 15 cell lines showed >90% of enhancers have inter-distance >1kb. On the other hand, since the Cas9 nuclease may induce unwanted DNA deletions when perturbing multiple enhancers (fig. S7C-F) (31, 32), dCas9-KRAB offer a technology for high-throughput study of enhancer interactions with a high resolution and minimal side effects.
We provided a speculative model that links the 3D genome and BRD4 interaction to the ultralong distance enhancer network (Fig. 4G). In this model, large BRD4 condensates are formed by smaller distinct BRD4 clusters at individual enhancers (49), which connects these enhancers across ultralong distances to create ‘weak’ 3D spatial contacts (50). This model is consistent with our quantitative mapping of enhancer networks that showed an inverse correlation between spatial contacts and synergistic interactions. Although the inverse correlation may be partly derived from the genomic distance, our experimental validation demonstrated the 3D genome organization at SREs is casually linked to the synergistic interactions.
With more whole-genome DNA sequencing data available in patients, the SRE model can be applied to infer the biological roles of SRE variants in cancer and other diseases and interpret the interactive influence of non-coding elements on disease risk to aid diagnosis and therapy.
Supplementary Material
Acknowledgments:
The authors thank all members from Lei Stanley Qi lab for useful comments and help on experiments and manuscript preparation. The authors thank Youqiong Ye and Leng Han for helping with the LAML linear model. The authors thank Stephen Shang and Xinyi Chen for helping with the fluorescence-activated cell sorting. The authors thank Mengting Han for helping with the imaging. The authors thank Jens Magnusson for comments. We acknowledge the data generated by the TCGA Research Network (https://www.cancer.gov/tcga), which allowed us to generate the results of variant interaction. We acknowledge the CD GWAS dataset generated by the Wellcome Trust Case Control Consortium. The ALL Relapse GWAS dataset used for the analyses described in this manuscript were obtained from dbGaP at phs000638.v1.p1. The ALL Relapse GWAS dataset was generated at St. Jude Children's Research Hospital and by the Children's Oncology Group, supported by NIH grants CA142665, CA21765, CA158568, CA156449, CA36401, CA98543, CA114766, CA140729, and U01GM92666, Jeffrey Pride Foundation, the National Childhood Cancer Foundation, and by ALSAC.
Funding:
X.Z. acknowledges supports by the Stein Fellowship from Stanford University and Institute for Computational and Data Sciences Seed Grant from the Pennsylvania State University. W.H.W acknowledges support from NIH R01 HG010359. L.S.Q. acknowledges support from the Li Ka Shing Foundation and National Science Foundation. The project is supported by the Li Ka Shing Foundation and a National Science Foundation CAREER award (L.S.Q., award no. 2046650). L.S.Q. is a Chan Zuckerberg Biohub investigator.
Footnotes
Competing interests: L.S.Q. is a founder and scientific advisor of Epicrispr Biotechnologies, and a scientific advisor of Laboratory of Genomics Research. The roles are unrelated to this study.
Data and materials availability: The CRISPRi functional tiling screen, Trac-looping data, ChIP-seq data, and ATAC-seq data have been deposited in the Gene Expression Omnibus under the accession ID GSE160768. The codes for the analysis of CRISPRi screen and the SRE prediction model are publicly accessible at Zenodo (51,52). The CRISPRi double sgRNA library and key plasmids will be available on Addgene (https://www.addgene.org/Stanley_Qi/).
References and Notes
- 1.Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, Shih DJH, Hovestadt V, Zapatka M, Sturm D, Jones DTW, Kool M, Remke M, Cavalli FMG, Zuyderduyn S, Bader GD, VandenBerg S, Esparza LA, Ryzhova M, Wang W, Wittmann A, Stark S, Sieber L, Seker-Cin H, Linke L, Kratochwil F, Jäger N, Buchhalter I, Imbusch CD, Zipprich G, Raeder B, Schmidt S, Diessl N, Wolf S, Wiemann S, Brors B, Lawerenz C, Eils J, Warnatz H-J, Risch T, Yaspo M-L, Weber UD, Bartholomae CC, von Kalle C, Turányi E, Hauser P, Sanden E, Darabi A, Siesjö P, Sterba J, Zitterbart K, Sumerauer D, van Sluis P, Versteeg R, Volckmann R, Koster J, Schuhmann MU, Ebinger M, Grimes HL, Robinson GW, Gajjar A, Mynarek M, von Hoff K, Rutkowski S, Pietsch T, Scheurlen W, Felsberg J, Reifenberger G, Kulozik AE, von Deimling A, Witt O, Eils R, Gilbertson RJ, Korshunov A, Taylor MD, Lichter P, Korbel JO, Wechsler-Reya RJ, Pfister SM, Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 511, 428–434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren W-J, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung W-K, Liu ET, Wei C-L, Cheung E, Ruan Y, An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 462, 58–64 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X, Chen S, Flynn E, Wu S, Wintner D, Shen Y, Distinct epigenomic patterns are associated with haploinsufficiency and predict risk genes of developmental disorders. Nat. Commun 9, 2138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Goldstein DB, Enhancer Domains Predict Gene Pathogenicity and Inform Gene Discovery in Complex Disease. Am. J. Hum. Genet 106, 215–233 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ENCODE Project Consortium et al. , An integrated encyclopedia of DNA elements in the human genome. Nature. 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, Integrative analysis of 111 reference human epigenomes. Nature. 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA, Systematic localization of common disease-associated variation in regulatory DNA. Science. 337, 1190–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kellis M, Wold B, Snyder MP, Bernstein BE, Kundaje A, Marinov GK, Ward LD, Birney E, Crawford GE, Dekker J, Dunham I, Elnitski LL, Farnham PJ, Feingold EA, Gerstein M, Giddings MC, Gilbert DM, Gingeras TR, Green ED, Guigo R, Hubbard T, Kent J, Lieb JD, Myers RM, Pazin MJ, Ren B, Stamatoyannopoulos JA, Weng Z, White KP, Hardison RC, Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. U. S. A 111, 6131–6138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM, Finding the missing heritability of complex diseases. Nature. 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee S, Kapoor A, Akiyama JA, Auer DR, Lee D, Gabriel S, Berrios C, Pennacchio LA, Chakravarti A, Enhancer Variants Synergistically Drive Dysfunction of a Gene Regulatory Network In Hirschsprung Disease. Cell. 167, 355–368.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corradin O, Cohen AJ, Luppino JM, Bayles IM, Schumacher FR, Scacheri PC, Modeling disease risk through analysis of physical interactions between genetic variants within chromatin regulatory circuitry. Nat. Genet 48, 1313–1320 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Factor DC, Barbeau AM, Allan KC, Hu LR, Madhavan M, Hoang AT, Hazel KEA, Hall PA, Nisraiyya S, Najm FJ, Miller TE, Nevin ZS, Karl RT, Lima BR, Song Y, Sibert AG, Dhillon GK, Volsko C, Bartels CF, Adams DJ, Dutta R, Gallagher MD, Phu W, Kozlenkov A, Dracheva S, Scacheri PC, Tesar PJ, Corradin O, Cell Type-Specific Intralocus Interactions Reveal Oligodendrocyte Mechanisms in MS. Cell. 181, 382–395.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phillips PC, Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet 9, 855–867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA, Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA, Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA, Super-enhancers in the control of cell identity and disease. Cell. 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, NISC Comparative Sequencing Program, Black BL, Visel A, Pennacchio LA, F. S. Collins, National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors, NISC Comparative Sequencing Program Authors, Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. U. S. A 110, 17921–17926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman İ, Tena JJ, Morán I, Gómez-Marín C, van de Bunt M, Ponsa-Cobas J, Castro N, Nammo T, Cebola I, García-Hurtado J, Maestro MA, Pattou F, Piemonti L, Berney T, Gloyn AL, Ravassard P, Skarmeta JLG, Müller F, McCarthy MI, Ferrer J, Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet 46, 136–143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA, Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol. Cell 58, 362–370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J, Liu X, Li D, Shao Z, Cao H, Zhang Y, Trompouki E, Bowman TV, Zon LI, Yuan G-C, Orkin SH, Xu J, Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev. Cell 36, 9–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hay D, Hughes JR, Babbs C, Davies JOJ, Graham BJ, Hanssen L, Kassouf MT, Marieke Oudelaar AM, Sharpe JA, Suciu MC, Telenius J, Williams R, Rode C, Li P-S, Pennacchio LA, Sloane-Stanley JA, Ayyub H, Butler S, Sauka-Spengler T, Gibbons RJ, Smith AJH, Wood WG, Higgs DR, Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet 48, 895–903 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, Zeisberger P, Benk AS, Amit I, Zandstra PW, Lupien M, Dick JE, Trumpp A, Spitz F, A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 553, 515–520 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Shin HY, Willi M, HyunYoo K, Zeng X, Wang C, Metser G, Hennighausen L, Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet 48, 904–911 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Li K, Cai W, Liu X, Zhang Y, Orkin SH, Xu J, Yuan G-C, Dissecting super-enhancer hierarchy based on chromatin interactions. Nat. Commun 9, 943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry MW, Boettiger AN, Levine M, Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U. S. A 108, 13570–13575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM, Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM, Identification and characterization of essential genes in the human genome. Science. 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thakore PI, D’Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA, Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS, Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosicki M, Tomberg K, Bradley A, Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol 36, 765–771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L, CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun 8, 15464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman J, Hastie T, Tibshirani R, Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw 33, 1 (2010). [PMC free article] [PubMed] [Google Scholar]
- 34.Lai B, Tang Q, Jin W, Hu G, Wangsa D, Cui K, Stanton BZ, Ren G, Ding Y, Zhao M, Liu S, Song J, Ried T, Zhao K, Trac-looping measures genome structure and chromatin accessibility. Nat. Methods 15, 741–747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA, A Phase Separation Model for Transcriptional Control. Cell. 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, Cisse II, Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA, Coactivator condensation at super-enhancers links phase separation and gene control. Science. 361 (2018), doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE, Selective inhibition of BET bromodomains. Nature. 468 (2010), pp. 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lappalainen T, Sammeth M, Friedländer MR, ’t Hoen PAC, Monlong J, Rivas MA, Gonzàlez-Porta M, Kurbatova N, Griebel T, Ferreira PG, Barann M, Wieland T, Greger L, van Iterson M, Almlöf J, Ribeca P, Pulyakhina I, Esser D, Giger T, Tikhonov A, Sultan M, Bertier G, MacArthur DG, Lek M, Lizano E, Buermans HPJ, Padioleau I, Schwarzmayr T, Karlberg O, Ongen H, Kilpinen H, Beltran S, Gut M, Kahlem K, Amstislavskiy V, Stegle O, Pirinen M, Montgomery SB, Donnelly P, McCarthy MI, Flicek P, Strom TM, Geuvadis Consortium, Lehrach H, Schreiber S, Sudbrak R, Carracedo A, Antonarakis SE, Häsler R, Syvänen A-C, van Ommen G-J, Brazma A, Meitinger T, Rosenstiel P, Guigó R, Gut IG, Estivill X, Dermitzakis ET, Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 501, 506–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Craddock NJ, Consortium, Wellcome Trust Case Control, Jones IR, Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447, 661–678 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang JJ, Cheng C, Devidas M, Cao X, Fan Y, Campana D, Yang W, Neale G, Cox NJ, Scheet P, Borowitz MJ, Winick NJ, Martin PL, Willman CL, Bowman WP, Camitta BM, Carroll A, Reaman GH, Carroll WL, Loh M, Hunger SP, Pui C-H, Evans WE, Relling MV, Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat. Genet 43, 237–241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang JJ, Cheng C, Devidas M, Cao X, Campana D, Yang W, Fan Y, Neale G, Cox N, Scheet P, Borowitz MJ, Winick NJ, Martin PL, Bowman WP, Camitta B, Reaman GH, Carroll WL, Willman CL, Hunger SP, Evans WE, Pui C-H, Loh M, Relling MV, Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood. 120, 4197–4204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vijayakrishnan J, Kumar R, Henrion MYR, Moorman AV, Rachakonda PS, Hosen I, da Silva Filho MI, Holroyd A, Dobbins SE, Koehler R, Thomsen H, Irving JA, Allan JM, Lightfoot T, Roman E, Kinsey SE, Sheridan E, Thompson PD, Hoffmann P, Nöthen MM, Heilmann-Heimbach S, Jöckel KH, Greaves M, Harrison CJ, Bartram CR, Schrappe M, Stanulla M, Hemminki K, Houlston RS, A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 31, 573–579 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijayakrishnan J, Qian M, Studd JB, Yang W, Kinnersley B, Law PJ, Broderick P, Raetz EA, Allan J, Pui C-H, Vora A, Evans WE, Moorman A, Yeoh A, Yang W, Li C, Bartram CR, Mullighan CG, Zimmerman M, Hunger SP, Schrappe M, Relling MV, Stanulla M, Loh ML, Houlston RS, Yang JJ, Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat. Commun 10, 5348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhen T, Kwon EM, Zhao L, Hsu J, Hyde RK, Lu Y, Alemu L, Speck NA, Liu PP, Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood. 130, 2431–2442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam DD, de Souza FSJ, Nasif S, Yamashita M, López-Leal R, Otero-Corchon V, Meece K, Sampath H, Mercer AJ, Wardlaw SL, Rubinstein M, Low MJ, Partially Redundant Enhancers Cooperatively Maintain Mammalian Pomc Expression Above a Critical Functional Threshold. Plos Genetics. 11 (2015), p. e1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu D, Gokcumen O, Khurana E, Loss-of-function tolerance of enhancers in the human genome. Plos Genet. 16, e1008663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE, Visel A, Pennacchio LA, Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young RA, Chakraborty AK, Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 75, 549–561.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA, Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 76, 473–484.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin X, Qi LS, Code and processed data for: Nested Epistasis Enhancer Networks for Robust Genome Regulation. Zenodo; 10.5281/zenodo.6823833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin X, Qi LS, SRE predictor for: Nested Epistasis Enhancer Networks for Robust Genome Regulation. Zenodo; 10.5281/zenodo.6823807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li G-W, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B, Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 155 (2013), pp. 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Yu C, Daley TP, Wang F, Cao WS, Bhate S, Lin X, Still C 2nd, Liu H, Zhao D, Wang H, Xie XS, Ding S, Wong WH, Wernig M, Qi LS, CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell. 23, 758–771.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takei Y, Shah S, Harvey S, Qi LS, Cai L, Multiplexed Dynamic Imaging of Genomic Loci by Combined CRISPR Imaging and DNA Sequential FISH. Biophys. J 112, 1773–1776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robinson JT, Turner D, Durand NC, Thorvaldsdóttir H, Mesirov JP, Aiden EL, Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Systems. 6 (2018), pp. 256–258.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mumbach MR, Satpathy AT, Boyle EA, Dai C, Gowen BG, Cho SW, Nguyen ML, Rubin AJ, Granja JM, Kazane KR, Wei Y, Nguyen T, Greenside PG, Corces MR, Tycko J, Simeonov DR, Suliman N, Li R, Xu J, Flynn RA, Kundaje A, Khavari PA, Marson A, Corn JE, Quertermous T, Greenleaf WJ, Chang HY, Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet 49, 1602–1612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A, NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 41, D991–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Shirley Liu X, Model-based Analysis of ChIP-Seq (MACS). Genome Biology. 9 (2008), p. R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lyu J, Li JJ, Su J, Peng F, Chen YE, Ge X, Li W, DORGE: Discovery of Oncogenes and tumoR suppressor genes using Genetic and Epigenetic features. Science Advances. 6, eaba6784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fulco CP, Nasser J, Jones TR, Munson G, Bergman DT, Subramanian V, Grossman SR, Anyoha R, Doughty BR, Patwardhan TA, Nguyen TH, Kane M, Perez EM, Durand NC, Lareau CA, Stamenova EK, Aiden EL, Lander ES, Engreitz JM, Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat. Genet 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Y, Liu S, Ren G, Tang Q, Zhao K, cLoops2: a full-stack comprehensive analytical tool for chromatin interactions. Nucleic Acids Res. 50, 57–71 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Phanstiel DH, Boyle AP, Heidari N, Snyder MP, Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics. 31, 3092–3098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC, PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet 81 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST, The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet 39, 1181–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goldman M, Craft B, Brooks A, Zhu J, Haussler D, The UCSC Xena Platform for cancer genomics data visualization and interpretation. bioRxiv (2018), p. 326470. [Google Scholar]
- 70.Gong J, Mei S, Liu C, Xiang Y, Ye Y, Zhang Z, Feng J, Liu R, Diao L, Guo A-Y, Miao X, Han L, PancanQTL: systematic identification of cis-eQTLs and trans-eQTLs in 33 cancer types. Nucleic Acids Res. 46, D971–D976 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stegle O, Parts L, Piipari M, Winn J, Durbin R, Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc 7, 500–507 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS, A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.