Abstract
Lysine malonylation is a recently characterized post-translational modification involved in the regulation of energy metabolism and gene expression. One unique feature of this post-translational modification is its potential susceptibility to decarboxylation, which poses possible challenges to its study. As a step towards addressing these challenges, we report the synthesis and evaluation of a stable isostere of malonyllysine. First, we find that synthetic substitution of the malonyl group with a tetrazole isostere results in amino acid’s resistant to thermal decarboxylation. Next, we demonstrate that protected variants of this amino acid are readily incorporated into peptides. Finally, we show that tetrazole isosteres of malonyllysine can be recognized by anti-malonyllysine antibodies and histone deacylases, validating their ability to mimic features of the endogenous lysine modification. Overall, this study establishes a new chemical strategy for stably mimicking a metabolite-derived post-translational modification, providing a foothold for tool development and functional analyses.
Keywords: acetylation, isosteres, malonylation, post-translational modification, unnatural amino acids
Introduction
Reversible acetylation plays a critical role in many important cellular functions, including the regulation of metabolism and gene expression.[1] This paradigm has been further expanded by the identification of several related post-translational modifications, collectively referred to as lysine acylations.[2] Similar to acetylation, the majority of lysine acylations are derived from acyl-CoA metabolites and have been found to be enzymatically removed. However, many of these modifications are physiochemically distinct from acetylation, allowing them to exert unique effects on the properties of modified proteins. One of the most distinct is lysine malonylation (Figure 1a). Lysine malonylation is a post-translational modification first discovered in 2011 via LC-MS/MS analysis of bacterial and cancer cell proteomes.[3] Malonylation possesses a larger steric footprint than acetylation and also exerts a more profound electrostatic effect, replacing the positively charged epsilon amine of protein lysine residues with a negatively charged carboxylate. Lysine malonylation is derived from malonyl-CoA, a precursor for anabolic fatty acid synthesis whose production is highly regulated by the metabolic state of the cell.[4] In addition to being intrinsically sensitive to fed/fasted state, levels of malonyl-CoA can be altered by cellular transformation, administration of fatty acid synthase inhibitors, and inborn errors of metabolism known as malonic acidurias.[5] Removal of lysine malonylation is catalyzed by SIRT5, an NAD-dependent deacylase enzyme which primarily localizes to the mitochondria but can also be found in the cytosol and nucleus.[3,6] In contrast, protein acetyltransferase enzymes that catalyze lysine malonylation at appreciable rates have yet to be identified. Building on early observations of non-enzymatic malonylation, our group recently applied a reactivity-based approach to define malonyl-CoA as a hyperreactive metabolite that drives non-enzymatic lysine malonylation in the cytosol and nucleus.[7] Non-enzymatic malonylation was found to functionally alter glycolytic enzyme activities, consistent with the biological observations of others.[8] These findings hint at the potential of lysine malonylation to influence signaling via effects on protein structure and activity, as well as significantly reflect an organism’s history of environmental exposures and metabolic state.
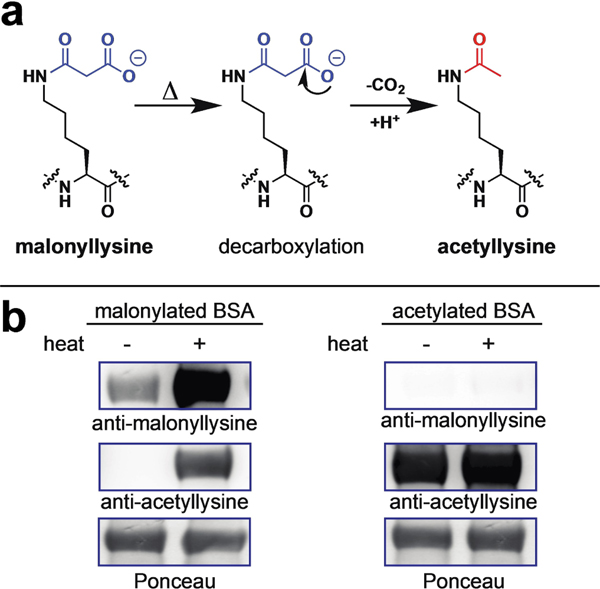
Figure 1.
(a) Decarboxylation of malonyllysine to acetyllysine. (b) No decarboxylation of malonyllysine is seen at room temperature. However, heat can cause chemical decarboxylation of malonyllysine-containing proteins to form acetyllysine, a cross-reactive epitope. Note that higher malonyllysine signal is also observed after heating, likely due to increased reaction of proteins with malonyl-NAC.
One relatively uncharacterized property of lysine malonylation is its potential chemical lability. Upon heating malonyl amides are known to undergo decarboxylation to acetamides in solution, which has the effect of converting malonyllysine to acetyllysine within proteins (Figure 1a). This reaction was first noted in early studies of protein malonylation, which used the facile decarboxylation of malonyllysine-containing peptides in the gas-phase as a characteristic signature for identification of malonyllysine in multi-stage peptide mass spectrometry experiments.[3] This reactivity is unique to lysine malonylation versus other acylations, and is conceptually similar to labile phosphorylations such as phosphohistidine, phosphoarginine, and phospholysine, whose instability has been found to hinder their detection and investigation.[9] In contrast, the ability to detect and raise antibodies against malonyllysine suggests its reactivity may be relatively modest compared to labile phosphorylations, and may only become relevant under specific conditions.
In order to better characterize the potential chemical lability of malonyllysine, we first set out to generate a malonylated protein standard for use in model reactions. In previous studies, we have applied acyl-CoAs as well as N-acetyl-cysteamine (NAC) esters as non-enzymatic acylation reagents.[7] To prepare malonylated bovine serum albumin (BSA) for use in model studies, we thus compared three non-enzymatic malonylation agents: malonyl-CoA, malonyl-NAC, and Meldrum’s acid, which can malonylate proteins via a decarboxylative acylation reaction (Figure S1a). Reagents were individually incubated with BSA overnight, and analyzed for malonylation via SDS-PAGE and anti-malonyllysine immunoblotting. The relative intensity of malonylation in these treated samples was found to follow the order: malonyl-NAC>malonyl-CoA ⪢ Meldrum’s acid (Figure S1b). These studies indicate malonyl-NAC as an effective reagent for in vitro non-enzymatic protein malonylation. Next, we explored our samples for evidence of decarboxylation. In contrast to its sensitive detection by anti-malonyllysine Western blot, malonylated BSA possessed no observable cross-reactivity with acetyllysine antibodies (Figure 1b), indicating it is relatively stable at room temperature. However, heating of malonylated BSA under conditions commonly employed for SDS-PAGE loading (90 °C, 5 min) resulted in production of an intense acetylation signal (Figure 1b). These studies suggest malonyllysine is a stable post-translational modification under physiological conditions which demonstrates “cryptic” chemical lability upon heating.
Studies of labile post-translational modifications such as phosphohistidine have greatly benefited from the development of stable analogues of these modifications.[9] In this approach, synthetic amino acids are developed that mimic the chemical properties of the native post-translational modification but which possess sufficient stability to allow for incorporation into proteins and peptides for functional analysis. Access to such amino acids can facilitate antibody generation as well as the interrogation of receptor-binding interactions dependent upon the modified residue, enabling fine-tuning of affinity and selectivity. This notion inspired us to ask the question: could one design an isostere of malonyllysine? As an initial proof-of-concept, we explored the effect of replacing the terminal carboxy- group of malonyllysine with 5-substituted-1H-tetrazoles.[10] 5-substituted tetrazoles (also referred to herein as tetrazolic acids) are a class of heterocyclic bioisosteres that exhibit similar physical properties to carboxylic acids. These include a planar structure and nearly identical pKa values (carboxylate pKa 4.5; tetrazole pKa 4.9) that lead to similar ionization at physiological pH. Distinct chemical features of tetrazoles include the delocalization of negative charge around the ring, as well as relatively increased size and lipophilicity (Figure 2a). Most importantly, unlike the terminal carboxy-group found in malonyllysine, tetrazoles cannot decompose via entropically favorable release of carbon dioxide. These considerations led us to evaluate 5-substituted tetrazoles as a stable class of malonyllysine isosteres.
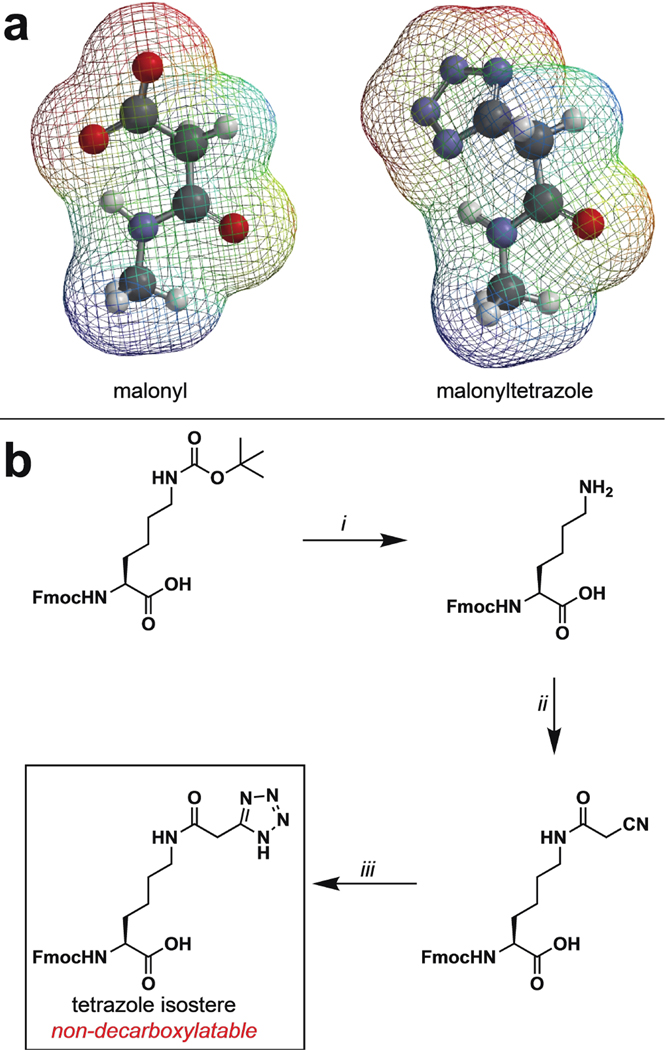
Figure 2.
(a) Electrostatic potential map comparing malonyl and a malonyltetrazole isostere. (b) Synthesis of a tetrazole isostere of malonyllysine amino acid. i) 4 M HCl, THF, 99%; ii) Cyanoacetic acid, DCC, DCM, 20%; iii) NaN3, ZnBr2, H2O/iPrOH (2:1), reflux, 30%.
Tetrazole mimics of malonyllysine were prepared via a facile three-step synthesis from the commercially available Fmoc-protected lysine amino acid (Figure 2b). Following deprotection, cyanoacetic acid was coupled to the epsilon amine of Fmoc-Lys-OH using standard carbodiimide-mediated coupling conditions. Pilot studies found that omitting bases such as diisopropylethylamine from this reaction led to greatly improved yields, possibly by limiting decomposition of the highly acidic O-acylisourea of cyanoacetic acid. An alternative route, in which lysine cyanoacetamide was produced via sequential bromoacetic acid coupling and cyanide displacement, was also explored and found to provide product, albeit with reduced yields. The lysine cyanoacetamide was further subjected to zinc bromide-catalyzed cycloaddition with sodium azide to furnish the 5-substituted tetrazole amino acid. These studies establish a straightforward synthetic route to tetrazole isosteres of a malonyllysine.
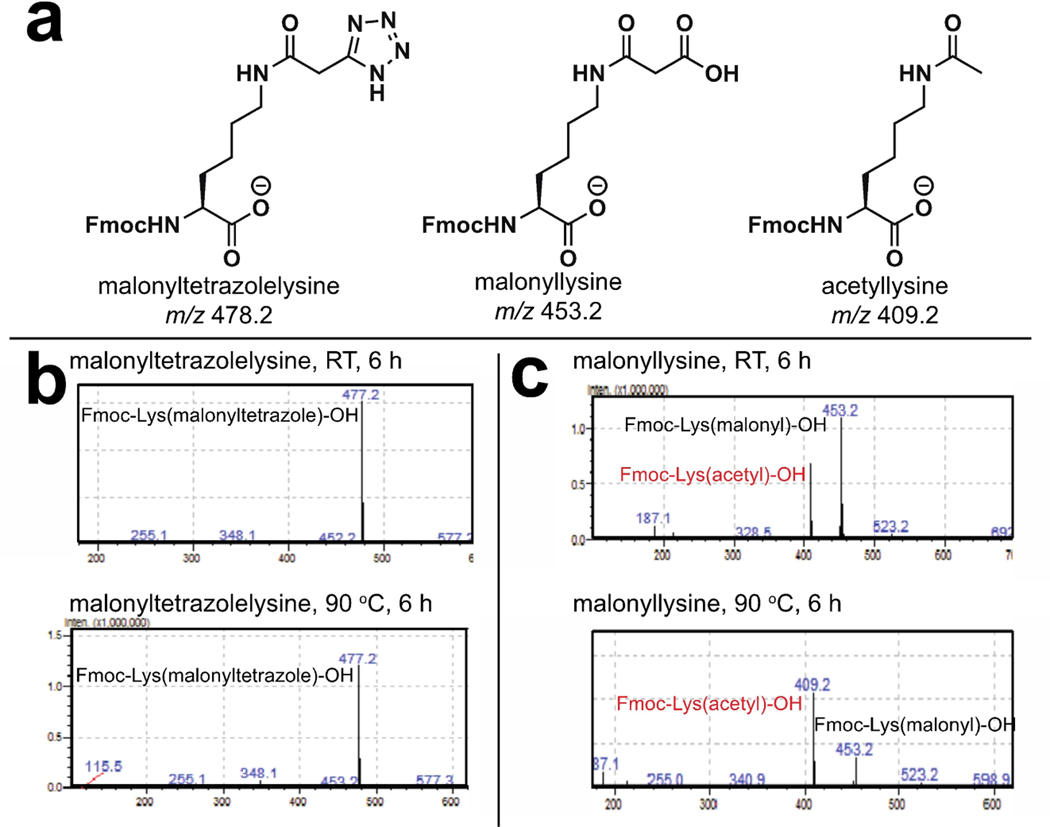
In drug design, useful isosteres mimic the parent functional group while simultaneously imbuing a biomolecule with novel properties, such as increased binding affinity or metabolic stability. To establish the distinct chemical properties of tetrazole and malonyllysine amino acids, we compared their susceptibility to the aforementioned thermally-induced decarboxylative decomposition. For these studies, Fmoc-protected malonyllysine and malonyltetrazole amino acids were incubated in neutral PBS (pH 7) for 0–12 hours at two different temperatures, 23°C and 90°C. As anticipated from our studies of malonylated BSA, incubation of the malonyllysine amino acid at elevated temperatures resulted in increased decarboxylation, as assessed by the relative ion intensity of malonylated versus acetylated amino acids (Figure 3). Due to the non-quantitative nature of our LC-MS method we must note these results are consistent with, but do not represent unambiguous proof of heat-accelerated decarboxylation. In contrast, no similar decomposition was observed with the malonyltetrazole amino acid. These observations indicate 5-substituted malonyltetrazoles may serve as chemically stable alternatives to cryptically labile malonylated amino acids.
Figure 3.
(a) Calculated masses of modified lysine amino acids. (b) Malonyltetrazolelysine is stable to prolonged heating at 90°C. (c) Malonyllysine decarboxylates upon ionization, and increasing levels of the decarboxylated mass are observed upon analysis of amino acids which have been heated at 90°C.
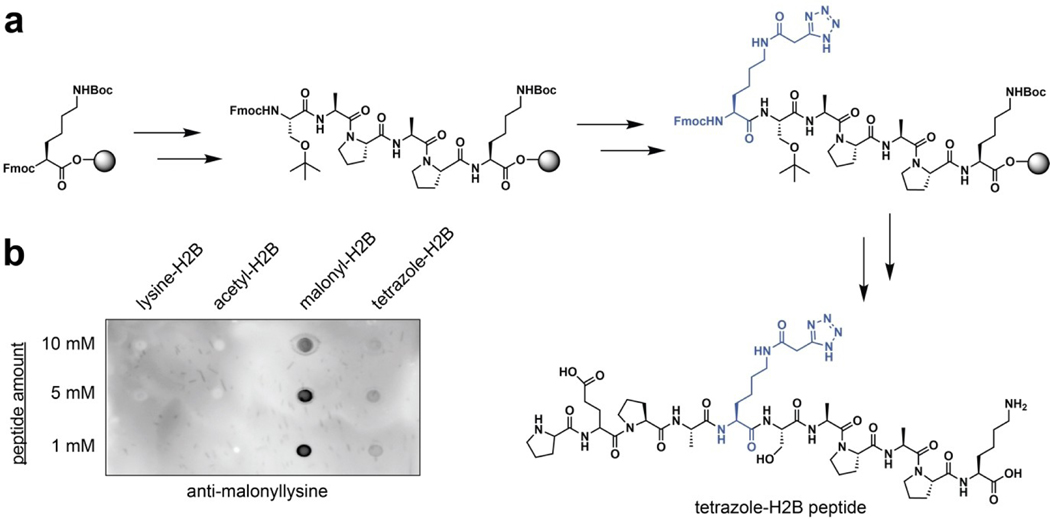
Next, we explored whether malonyltetrazole amino acids could be incorporated into biologically-relevant sites of malonylation using conventional solid-phase peptide synthesis. As a target for these studies, we chose a peptide based on the N-terminus of the nuclear structural protein histone H2B (Figure 4a). Recent proteomic studies found that malonylation of lysine 5 in histone H2B undergoes a large increase in abundance upon knockout of SIRT5, suggesting it is highly regulated.[8] Coupled with the known role of histone acylation in chromatin structure and transcription,[1] this suggests H2BK5 may be an important node linking malonylation with gene expression in the nucleus. Considering building blocks for the introduction of malonyl isostere, we opted to utilize the unprotected tetrazole amino acid. We reasoned the lack of an additional protection/deprotection step would be preferable from a technical perspective, as it may facilitate the production of sufficient quantities of amino acid to allow its utilization in excess during solid-phase peptide synthesis reactions. Previous studies have found unprotected 5-substituted tetrazoles to be compatible with peptide coupling reagents,[11] suggesting the synthetic feasibility of this strategy. Accordingly, peptides based on histone H2B (residues 1–11) containing either malonyltetrazole or native lysine at K5 were synthesized according to standard Fmoc solid-phase peptide synthesis methods on Wang resin (Figure 4a). Cleavage from resin with TFA followed by reverse-phase HPLC purification provided the cognate peptides in multi-milligram amounts. These studies demonstrate the compatibility of malonyltetrazole amino acids with solid-phase synthesis strategies.
Figure 4.
Incorporation of tetrazole isosteres of malonyllysine into peptides by solid-phase peptide synthesis. (a) Synthetic scheme. (b) Peptides containing tetrazole isosteres are recognized by anti-malonyllysine antibodies.
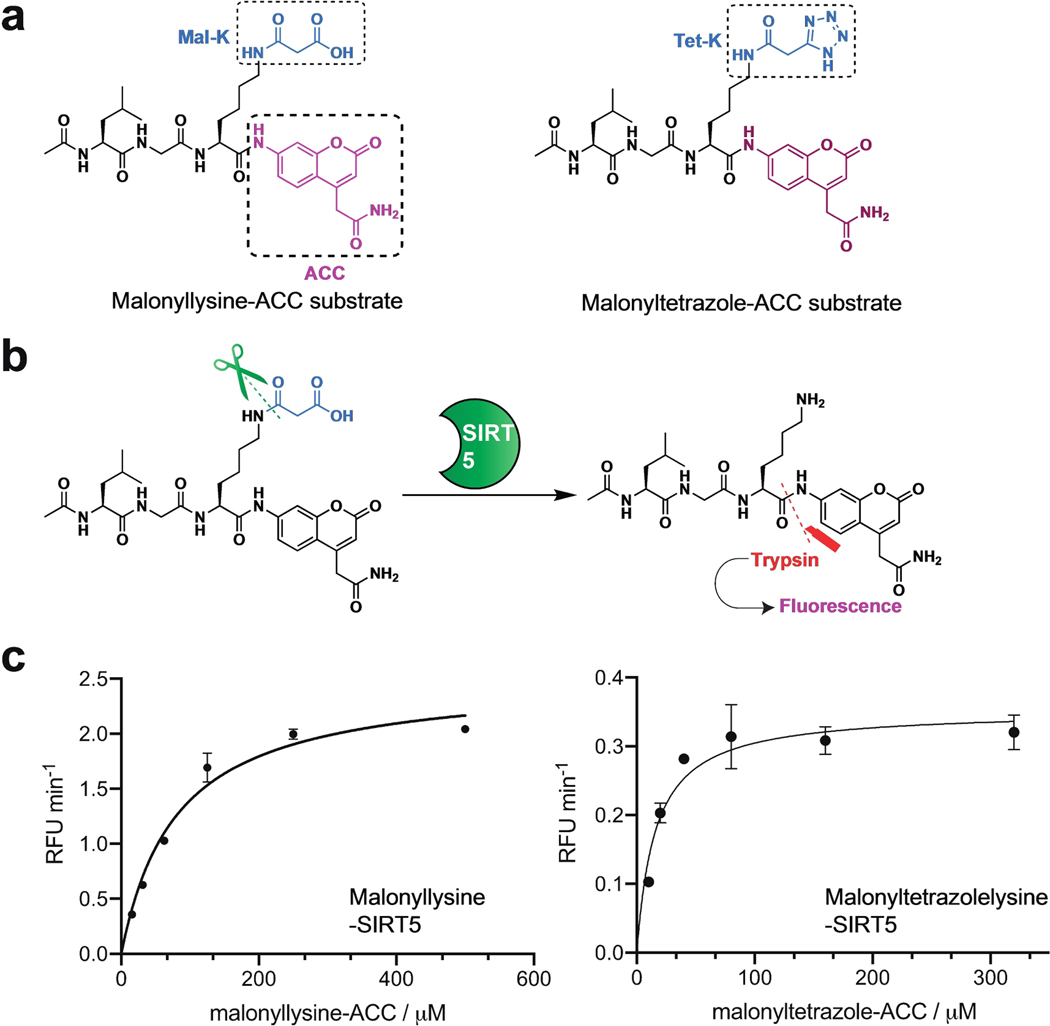
Having established a unique chemical property of malonyltetrazole, as well as the ability to incorporate it into peptides, one final question was whether this non-native amino acid could mimic the interaction of malonyllysine with biomolecules. To assess this, we tested the recognition of these two amino acids using anti-malonyllysine antibodies by dot blot. These studies utilized the aforementioned H2B (1–11) peptides derivatized at position 5 with either lysine, acetyllysine, malonyllysine, or malonyltetrazole. Peptides were spotted onto nitrocellulose membranes, blocked, and probed with a polyclonal anti-malonyllysine antibody (PTM Biolabs #PTM-901) followed by secondary detection.[3] These dot blot experiments revealed the polyclonal antibody readily detects malonyllysine and malonyltetrazole, but not acetyllysine or unmodified lysine, in the H2B peptide context (Figure 4b). This suggests malonyltetrazole- and malonyllysine-containing peptides can present similar molecular interaction surfaces for receptor recognition. Interestingly, a significantly less intense signal was observed for the malonyltetrazole H2B peptide compared to the analogous malonyllysine species. This suggests that within the polyclonal anti-malonyllysine antibody population, individual antibodies exist that recognize conformations of malonyllysine not mimicked by the tetrazole analogue. Consistent with this, using dot blot analysis we identified one monoclonal anti-malonyllysine antibody (CST #14942S) that is able to distinguish malonyllysine and malonyltetrazole (Figure S2).[6] Distinct chemical features of malonyltetrazole that may preclude detection by this antibody include equilibration to the 2H-5-substituted tetrazole isomer, as well as increased size, lipophilicity, and delocalization of negative charge. To further extend our understanding of how the recognition properties of these amino acids overlap we synthesized a known substrate of the human deacylase SIRT5 and compared its turnover upon substitution of a malonyllysine residue with a malonyltetrazole (Figure 5, Table 1). Steady-state kinetic analyses revealed the tetrazole-modified substrate was turned over by SIRT5, albeit with significantly reduced (~4-fold) catalytic efficiency (kcat/KM). This reflected the combined effect of an ~20-fold reduction in the rate of turnover of the malonyltetrazole peptide, as well as its 5-fold higher KM. Since KM is conceptually correlated with binding, we tested the tetrazole-containing peptide as a SIRT5 inhibitor, and found it could inhibit demalonylase activity with an IC50 value around 77.4 μM (Figure S3). Overall, these findings indicate the receptor and enzyme recognition landscape presented by malonyltetrazole- and malonyllysine- is characterized both by overlapping, as well as distinct, elements.
Figure 5.
Deacylase activity of SIRT5 on malonyllysine-ACC and malonyltetrazole-ACC substrate. (a) Chemical structure of malonyllysine-ACC and malonyltetrazole-ACC substrate. (b) Scheme for the enzymatic assay based on the fluorogenic peptide substrate, (c) Michaelis-Menten kinetic curves of SIRT5 on malonyllysine-ACC (15.625 μM–500 μM) and malonyltetrazole-ACC (10 μM–320 μM) substrate.
Table 1.
Michaelis-Menten kinetic parameters for malonyllysine-ACC and malonyltetrazole-ACC substrate hydrolysis by SIRT5.
| Ac-LGK(Mal)-ACC | Ac-LGK(Tet)-ACC | |
|---|---|---|
|
| ||
| kcat (s−1) | (3.83 ± 0.15) × 10 −3 | (1.79 ± 0.08) × 10 −4 |
| KM (μM) | 80.26 ± 9.18 | 15.73 ± 2.87 |
| Vmax (μM S−1) | (3.83 ± 0.15) × 10 −3 | (5.38 ± 0.23) × 10 −4 |
| kcat/KM(M−1 S−1) | 47.76 ± 16.11 | 11.40 ± 2.66 |
To summarize, here we have reported the design and characterization of a stable bioisostere of malonyllysine. Lysine amino acids incorporating a 5-substituted malonyltetrazole at the epsilon amino group serve as synthetically accessible analogues of malonyllysine. Model studies indicate this analogue possesses reduced thermal lability relative to the native malonyl post-translational modification, and can be readily incorporated into peptides via solid-phase peptide synthesis. Then, we utilized antibody-based dot blotting to demonstrate the ability of malonyltetrazole-containing peptides to present a molecular interaction surface which overlaps with that of malonyllysine. Finally, we performed fluorescence-based in vitro SIRT5 deacylase assays to reveal that malonyltetrazole-containing substrate can be turned over by SIRT5 with lower catalytic efficiency compared with native malonyllysine, and malonyltetrazole-containing peptide can function as a SIRT5 inhibitor. The knowledge generated from this proof-of-concept study should find several applications. First, the cryptic chemical lability of malonyllysine has to date been only sparingly characterized in the literature.[3] Our results using malonylated BSA indicate that malonyllysine is quite stable at room temperature and can decompose when heated. This decarboxylation of malonyllysine has the potential not only to limit detection of this malonyllysine, but also to cause artifactual detection of acetyllysine at authentic malonylation sites. This suggests mild, rather than extreme, heating should be used when preparing malonylated samples for experiments such as denaturing polyacrylamide gel electrophoresis. Second, our studies suggest that in cases where the terminal carboxy- functional group of malonyllysine hinders applications, malonyltetrazoles may be employed as alternative bioisostere that is easily incorporated into peptides and engage in similar receptor binding interactions as the natural modified amino acid. One caveat to this strategy stems from our identification of a monoclonal antibody that distinguishes malonyllysine and malonyltetrazole, which suggests malonyltetrazole may only partially mimic the molecular interaction landscape presented to the immune system by malonyllysine. Whether the converse is true, i.e. can malonyltetrazole elicit the production of antibodies that do not recognize malonyllysine, remains unknown but will be important to explore if this amino acid is employed as an immunogen. A multitude of carboxylate isosteres are known in the synthetic literature, and an interesting future question is whether replacing the tetrazole with one of these alternatives may more effectively mimic malonyllysine and circumvent this limitation. It is important to note that, as we explicitly note in the text, unlike phosphoamino acids the generation of malonyllysine antibodies is not necessarily limited by their cryptic lability, and further motivation for asking whether effective isosteres of this modification can be generated lies in their potential to enable additional applications. The ability to incorporate malonyltetrazole into peptides, as well as possibly into proteins via native chemical ligation,[9] may enable the identification of proteins which interact with this modification with high affinity as ‘readers.’ Relatedly, our kinetic studies indicate malonyltetrazole is turned over by SIRT5 but with relatively low efficiency, which may increase its stability for chemoproteomics applications. The increased lipophilicity, cell permeability, and potential “cageability” of tetrazoles relative to carboxylates may also usefully incorporate this analogue into proteins via unnatural amino acid mutagenesis approaches.[12] Finally, these studies open the door to the development of bioisostere of additional dicarboxylate metabolite-derived lysine acylations, including lysine succinylation and glutarylation.[5] In particular it will be interesting to evaluate whether (and how) these analogues influence turnover by SIRT-family deacylases. By expanding the methodological arsenal available to study reversible acylation, these new tools may facilitate a greater understanding of how post-translational modifications function as key mediators of biology and disease.
Supplementary Material
Acknowledgements
We thank Arissa Bavari for contributing synthetic procedures for this study. This work was supported by National Institutes of Health, National Cancer Institute, Center for Cancer Research (ZIABC011488).
Footnotes
A previous version of this manuscript has been deposited on a preprint server (https://doi.org/10.1101/2020.08.23.263285).
Supporting information for this article is available on the WWW under https://doi.org/10.1002/cbic.202100491
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Ali I, Conrad RJ, Verdin E, Ott M, Chem. Rev. 2018, 118, 1216–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Lin H, Su X, He B, ACS Chem. Biol. 2012, 7, 947–960; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Olsen CA, Angew. Chem. Int. Ed. 2012, 51, 3755–3756; Angew. Chem. 2012, 124, 3817–3819. [DOI] [PubMed] [Google Scholar]
- [3].Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y, Mol. Cell. Proteomics 2011, 10, M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Martin DB, Vagelos PR, J. Biol. Chem. 1962, 237, 1787–1792. [PubMed] [Google Scholar]
- [5].Hirschey MD, Zhao Y, Mol. Cell. Proteomics 2015, 14, 2308–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H, Science 2011, 334, 806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kulkarni RA, Worth AJ, Zengeya TT, Shrimp JH, Garlick JM, Roberts AM, Montgomery DC, Sourbier C, Gibbs BK, Mesaros C, Tsai YC, Das S, Chan KC, Zhou M, Andresson T, Weissman AM, Linehan WM, Blair IA, Snyder NW, Meier JL, Cell Chem. Biol. 2017, 24, 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, Verdin E, Mol. Cell 2015, 59, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hauser A, Penkert M, Hackenberger CPR, Acc. Chem. Res. 2017, 50, 1883–1893. [DOI] [PubMed] [Google Scholar]
- [10].Herr RJ, Bioorg. Med. Chem. 2002, 10, 3379–3393. [DOI] [PubMed] [Google Scholar]
- [11].Sureshbabu VV, Venkataramanarao R, Naik SA, Chennakrishnareddy G, Tetrahedron Lett. 2007, 48, 7038–7041. [Google Scholar]
- [12].Neumann H, Peak-Chew SY, Chin JW, Nat. Chem. Biol. 2008, 4, 232–234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.