Abstract
Many examples of enzymes that have lost their catalytic activity and perform other biological functions are known. The opposite situation is rare. A previously unnoticed structural similarity between the λ integrase family (Int) proteins and the AraC family of transcriptional activators implies that the Int family evolved by duplication of an ancient DNA-binding homeodomain-like module, which acquired enzymatic activity. The two helix–turn–helix (HTH) motifs in Int proteins incorporate catalytic residues and participate in DNA binding. The active site of Int proteins, which include the type IB topoisomerases, is formed at the domain interface and the catalytic tyrosine residue is located in the second helix of the C-terminal HTH motif. Structural analysis of other ‘tyrosine’ DNA-breaking/rejoining enzymes with similar enzyme mechanisms, namely prokaryotic topoisomerase I, topoisomerase II and archaeal topoisomerase VI, reveals that the catalytic tyrosine is placed in a HTH domain as well. Surprisingly, the location of this tyrosine residue in the structure is not conserved, suggesting independent, parallel evolution leading to the same catalytic function by homologous HTH domains. The ‘tyrosine’ recombinases give a rare example of enzymes that evolved from ancient DNA-binding modules and present a unique case for homologous enzymatic domains with similar catalytic mechanisms but different locations of catalytic residues, which are placed at non-homologous sites.
The wealth of biochemical, sequence and structural information accumulated over the years of molecular biology provides examples of proteins that change function in the course of evolution (1–4). Enzymes having a chemical requirement for invariant amino acids in the active site are particularly vulnerable to selection pressure. Using sequence similarity, one can detect proteins evolutionarily related to enzymes but lacking catalytic activity due to disruption of their active sites. These proteins may function, for example, as transcription regulators (4). Given that an overwhelming majority of homologs to such proteins are indeed enzymes and that the non-catalytic variants are uncommon (4), there is little doubt about the direction of evolution in these cases: the enzyme has lost its activity/acquired a new function. The reverse path of evolution is rather rare. There are few examples of normally non-enzymatic domains that gain catalytic activity (5,6), particularly for transcription regulators. One such example is discussed here.
The helix–turn–helix (HTH) DNA-binding motif is ubiquitous and detected in many transcription regulators (7–9). HTH transcription factors are diversified across a variety of orthologous families and the HTH motif is incorporated into several structural scaffolds (9). The most common of these scaffolds, hereafter referred to as homeodomain-like (HHTH), has a hydrophobic core of two α-helices (helices B and C) completed by another, usually N-terminal, α-helix (helix A). This structure can be described as a right-handed three-helical bundle (Fig. 1b). Some examples of HHTH proteins are homeodomains, AraC-type transcriptional activators and members of the winged HTH family (HHTHw), typified by the C-terminal domain of catabolite gene activator protein (CAP) (7). HTH bundles can usually be distinguished from other three-helical structures by a sequence signal in the HTH motif (8–11). Very divergent representatives with known spatial structure can be recognized by the characteristic packing of α-helices B and C at nearly a right angle to each other (Fig. 1b, helices B1–C1 and B2–C2, Fig. 1c). The turn between α-helices B and C offsets α-helix C so that the N-terminal part of C is packed against the middle of B. α-Helix B is usually short (two or three turns) and C, which binds to the DNA major groove, is longer (12). A monophyletic origin for most HHTH proteins has been proposed (8).
Figure 1.
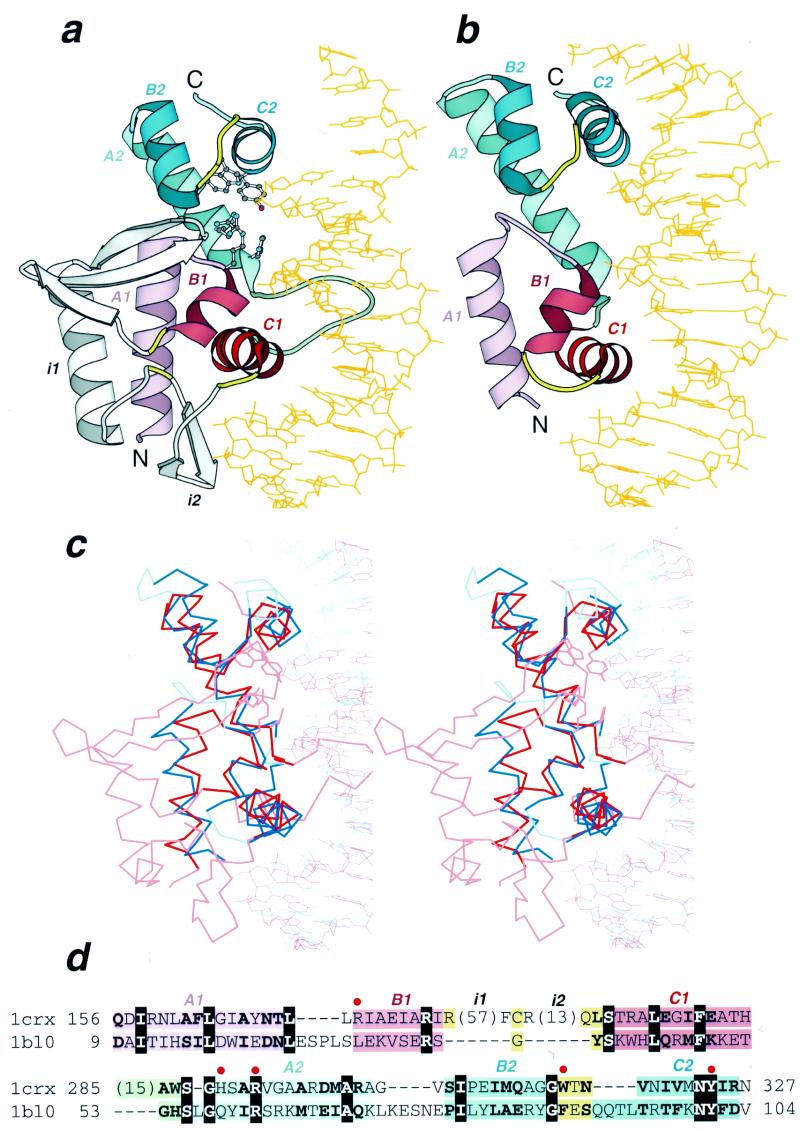
Structural similarity between Cre recombinase and MarA. Ribbon diagrams of (a) Cre recombinase from bacteriophage P1 (pdb entry 1crx, residues A154–A330) and (b) MarA transcription regulator from E.coli (pdb entry 1bl0, residues A9–A106) in complex with DNA drawn by Bobscript (48), a modified version of Molscript (49). The structures were superimposed and then separated for clarity. N- and C-termini are labeled. The spatially equivalent structural elements are colored correspondingly in the two structures. N- and C-terminal HHTH domains are colored red and blue, respectively. α-Helices of the HTH motifs are in darker color. The turns in the HTH motifs are yellow and the loop connecting two HHTH domains is green. Long insertions (i1 and i2) in the first HHTH domain of Cre recombinase are shown in gray. DNA chains are orange. α-Helices are labeled A, B and C followed by a domain index (1 or 2). Side chains of active site residues in Cre recombinase are shown in ball-and-stick presentation. (c) The stereodiagram of Cre recombinase (red) and MarA (blue) superposition. The Cα traces of protein and DNA segments are shown. The regions used in r.m.s.d. minimization are outlined in darker colors. Superposition was performed using the InsightII package (MSI Inc) according to the DALI alignment (34). (d) Structure-based sequence alignment of Cre recombinase (1crx) and MarA (1bl0) generated by DALI (34). The starting and ending residues are numbered and the segments are labeled with the same letters as in (a) and (b). Color shading of the regions is the same as in (a) and (b). Invariant residues are shown in bold white letters boxed with black and conserved substitutions are shown in bold. The number of residues omitted from the alignment are shown in parentheses. The active site residues are marked with a red dot above the alignment and their side chains are displayed in (a).
Site-specific recombination allows living organisms to rearrange and redistribute their genetic content by cutting and rejoining DNA segments at specific sequences. Recombinases catalyze DNA breakage, strand exchange and ligation. One of the two major recombinase types, the λ integrase family (Int), uses a tyrosine nucleophile in a reaction that proceeds through a stable 3′-phosphotyrosine DNA–enzyme intermediate (13,14). The structures of several family members, namely bacteriophage λ integrase (15), bacteriophage HP1 integrase (16), XerD from Escherichia coli (17) and Cre recombinase from bacteriophage P1 (Fig. 1a) (18), have recently been solved. The most extensive structural information obtained concerns the DNA-binding mode and mechanism of Cre enzyme (19,20). X-ray crystallography revealed that type IB topoisomerases (21), which include eukaryotic (22,23) and viral (24) enzymes, also belong to the Int family due to extensive conservation of the structural core, active site arrangement and the catalytic mechanism (25,26).
The Int family has always been treated as a unique fold without much structural similarity to other proteins (27–29). SCOP (30,31) groups Int family structures into the fold named ‘DNA-breaking/rejoining enzymes’ of α+β class. CATH (32) places them in the ‘mainly α’ class with non-bundle architecture. However, structure similarity searches with such programs as DALI and VAST initiated with Cre recombinase coordinates (18) (pdb entry 1crx, Fig. 1a) reveal a highly significant and striking match that spans the entire length of the MarA transcriptional activator molecule (33) (pdb entry 1bl0, Fig. 1b). DALI (34,35) superimposes 88 Cα atoms of 1crx (322 residues) and 1bl0 (116 residues) with a Z score of 4.0, r.m.s.d. of 3.3 Å and 17% identity in the resulting sequence alignment (Fig. 1d). VAST (36) aligns 78 Cα atoms of these proteins with a P value of 0.0002, r.m.s.d. of 2.5 Å and a sequence identity of 16.7%. Additionally, superposition of Cα traces of Cre recombinase and MarA results in an almost perfect superposition of DNA molecules present in the crystals (Fig. 1a–c) despite the fact that DNA coordinates were not used in r.m.s.d. minimization. Thus the modes of DNA binding are essentially identical for Cre recombinase and MarA. Such an extensive structural resemblance combined with similar substrate binding and non-random sequence identity (18%, Fig. 1c) argues for homology (3,37) between DNA-breaking/rejoining enzymes and MarA. Surprisingly, similarity between the two proteins remained unnoticed to date.
MarA is a member of the AraC family of transcription activators that control expression of a variety of genes (33). The MarA structure consists of two HHTH modules with a unique mutual arrangement, previously unrecognized for multi-HTH proteins, in which two HHTH domains are approximately related by a translation (33) (Fig. 1b). This arrangement results in tight packing of the two domains and places two almost parallel DNA-binding helices in the major groove at a separation of one DNA double helix turn (Fig. 1b). Both MarA domains have structural counterparts in the Cre recombinase–DNA complex and all six MarA α-helices are superimposable between the two proteins (Fig. 1). The homology of Cre and MarA suggested by structural, functional and sequence similarity implies that the catalytic segment of Int proteins consists of two consecutive HHTH domains. However, it is difficult to determine at present if the common ancestor of Int and MarA already contained two HHTH domains or if duplications in these proteins occurred in parallel. Interestingly, among the four articles describing different independently solved Int protein structures (15–18), only one discusses the structural similarity of the first HHTH domain in Int proteins with the HTH motif of the catabolite activator protein DNA-binding domain (17). X-ray crystallography revealed that the second HHTH domain, which contains a catalytic tyrosine residue, is conformationally variable between different representatives of the family, as well as between different DNA complexes of the same Cre protein, and thus might fold into the HHTH structure upon DNA binding only (15–18,27–29). For example, in λ integrase the catalytic tyrosine is modeled in a flexible β-strand-like region. Such flexibility might be necessary for proper functioning of the enzymatic HHTH domain. It is well known that the active sites of many enzymes include regions of higher flexibility to accommodate changes in the substrate during catalysis. Therefore, it is likely that the second HHTH domain, which contains most of the active site residues (Fig. 1a and c), acquired some structural flexibility while the first HHTH domain, which is used mostly for DNA binding in a standard HTH-like manner, remained rigid.
Thus the Int family fold has likely evolved by a duplication of an ancient HHTH protein (Fig. 1a, red and blue). The first HHTH domain was elaborated with long insertions (Fig. 1a, gray) placed in the ‘turn’ region (Fig. 1a, yellow) of the HTH motif. These insertions are structured in subdomains that contain small β-sheets (Fig. 1a, gray). It is not unusual for HTH proteins to incorporate insertions in ‘turn’ regions, found for example in the endonuclease FokI (38). The presence of these subdomains disrupting the HTH motif masks the sequence signal and prevents motif detection in Int proteins by sequence analysis. The first HHTH domain of Int proteins is used primarily for DNA binding while the second HHTH domain is adapted to a catalytic role.
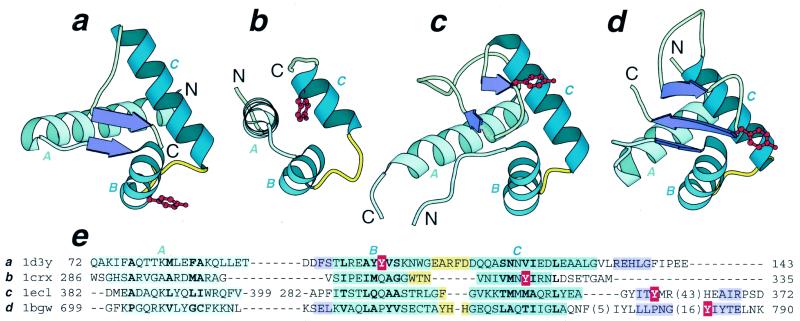
The following question arises: are there other examples of HTH domains that are not only present in an enzyme as nucleotide-binding modules but possess enzymatic activity (i.e. carry at least some of the catalytic residues)? PDB (39,40) searches by DALI (34,35) and VAST (36,41) reveal domains of different topoisomerases that contain catalytic tyrosine residues as members of the HHTH fold. The presence of HHTH domains in type IA, II and VI topoisomerases (42–44) has been detected previously (44–46). Topoisomerase IA, II and VI HHTH domains contain a small amount of β-sheet and should be classified as CAP-like ‘winged’ HTH domains (Fig. 2a, c and d). Notably, all of these enzymes possess a catalytic mechanism similar to the one established for Int proteins, i.e. tyrosine is utilized as a nucleophile and found in an HHTH domain. The Int family includes type IB topoisomerases. Thus an evolutionary connection exists between all ‘tyrosine’ DNA-breaking/rejoining enzymes with known structure, namely type IA, IB, II and VI topoisomerases, which all contain an enzymatic HHTH module. Structure superpositions of these domains in the four enzymes reveal that the position of the catalytic tyrosine residue is not structurally conserved (Fig. 2e). In the topoisomerase VI structure (44) Tyr103 is placed in α-helix B (Fig. 2a); in the Int family, including topoisomerase IB (21,22,24,47) and Cre recombinase (18), Tyr324 (Cre numbering) is incorporated in α-helix C (Fig. 2b); in topoisomerase IA (42) Tyr319 is at the C-terminal end of the first β-stand in the ‘wing’ segment of the HHTH domain (Fig. 2c); in topoisomerase II (43) Tyr782 is located after a long loop at the beginning of the second β-strand in the ‘wing’ (Fig. 2d). The sites in homologous HTH domains where catalytic tyrosines are located are not homologous; therefore, the catalytic properties of HTH domains in DNA-breaking/rejoining enzymes are likely to have evolved independently in parallel. Thus catalytic HHTH domains provide a unique example of homologous enzymes with a similar mechanism but different location of active site residues which are placed at non-homologous sites.
Figure 2.
Catalytic HHTH domains with a topoisomerase-like mechanism. Ribbon diagrams of (a) DNA topoisomerase VI A subunit from Methanococcus jannaschii (pdb entry 1d3y, residues A72–A142), (b) Cre recombinase from bacteriophage P1 (pdb entry 1crx, residues A286–A330), (c) topoisomerase I from E.coli (pdb entry 1ecl, residues 279–405), and (d) topoisomerase II from yeast Saccharomyces cerevisiae (pdb entry 1bgw, residues 699–789) were drawn by Bobscript (48), a modified version of Molscript (49). Corresponding α-helices are labeled A, B and C. α-Helices of the HTH motifs are in a darker color. The turns in the HTH motifs are yellow. β-Strands are shown as purple arrows. The side chains of catalytic tyrosine residues are shown in ball-and-stick presentation and are colored red. Dots in (c) replace a long partially disordered insertion. (e) Structure-based sequence alignment of domains shown in (a)–(d). The starting and ending residues are numbered and the three helices are labeled. The number of residues omitted from the alignment are shown in brackets. Color shading of these regions matches that in (a)–(d). Residues at conserved hydrophobic positions are shown in bold. The catalytic tyrosine residues are shown in white and are boxed with red. The HHTH domain of 1ecl (c) is circularly permuted, which is reflected in the residue numbering of the segment.
Acknowledgments
ACKNOWLEDGEMENTS
The author is grateful to Yuri Wolf for fruitful discussions and to Hong Zhang and Monica Horvath for critical reading of the manuscript and helpful comments.
REFERENCES
- 1.Murzin A.G. (1993) Trends Biochem. Sci., 18, 403–405. [DOI] [PubMed]
- 2.Artymiuk P.J., Poirrette,A.R., Rice,D.W. and Willett,P. (1997) Nature, 388, 33–34. [DOI] [PubMed]
- 3.Murzin A.G. (1998) Curr. Opin. Struct. Biol., 8, 380–387. [DOI] [PubMed]
- 4.Aravind L. and Koonin,E.V. (1998) Curr. Biol., 8, R111–R113. [DOI] [PubMed]
- 5.Lorick K.L., Jensen,J.P., Fang,S., Ong,A.M., Hatakeyama,S. and Weissman,A.M. (1999) Proc. Natl Acad. Sci. USA, 96, 11364–11369. [DOI] [PMC free article] [PubMed]
- 6.Boerner R.J., Consler,T.G., Gampe,R.T.,Jr, Weigl,D., Willard,D.H., Davis,D.G., Edison,A.M., Loganzo,F.,Jr, Kassel,D.B., Xu,R.X. et al. (1995) Biochemistry, 34, 15351–15358. [DOI] [PubMed]
- 7.Wintjens R. and Rooman,M. (1996) J. Mol. Biol., 262, 294–313. [DOI] [PubMed]
- 8.Rosinski J.A. and Atchley,W.R. (1999) J. Mol. Evol., 49, 301–309. [DOI] [PubMed]
- 9.Aravind L. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed]
- 10.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- 11.Altschul S.F. and Koonin,E.V. (1998) Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed]
- 12.Wintjens R.T., Rooman,M.J. and Wodak,S.J. (1996) J. Mol. Biol., 255, 235–253. [DOI] [PubMed]
- 13.Nunes-Duby S.E., Kwon,H.J., Tirumalai,R.S., Ellenberger,T. and Landy,A. (1998) Nucleic Acids Res., 26, 391–406. [DOI] [PMC free article] [PubMed]
- 14.Gopaul D.N. and Duyne,G.D. (1999) Curr. Opin. Struct. Biol., 9, 14–20. [DOI] [PubMed]
- 15.Kwon H.J., Tirumalai,R., Landy,A. and Ellenberger,T. (1997) Science, 276, 126–131. [DOI] [PMC free article] [PubMed]
- 16.Hickman A.B., Waninger,S., Scocca,J.J. and Dyda,F. (1997) Cell, 89, 227–237. [DOI] [PubMed]
- 17.Subramanya H.S., Arciszewska,L.K., Baker,R.A., Bird,L.E., Sherratt,D.J. and Wigley,D.B. (1997) EMBO J., 16, 5178–5187. [DOI] [PMC free article] [PubMed]
- 18.Guo F., Gopaul,D.N. and van Duyne,G.D. (1997) Nature, 389, 40–46. [DOI] [PubMed]
- 19.Gopaul D.N., Guo,F. and Van Duyne,G.D. (1998) EMBO J., 17, 4175–4187. [DOI] [PMC free article] [PubMed]
- 20.Guo F., Gopaul,D.N. and Van Duyne,G.D. (1999) Proc. Natl Acad. Sci. USA, 96, 7143–7148. [DOI] [PMC free article] [PubMed]
- 21.Redinbo M.R., Champoux,J.J. and Hol,W.G. (1999) Curr. Opin. Struct. Biol., 9, 29–36. [DOI] [PubMed]
- 22.Redinbo M.R., Stewart,L., Kuhn,P., Champoux,J.J. and Hol,W.G. (1998) Science, 279, 1504–1513. [DOI] [PubMed]
- 23.Redinbo M.R., Stewart,L., Champoux,J.J. and Hol,W.G. (1999) J. Mol. Biol., 292, 685–696. [DOI] [PubMed]
- 24.Cheng C., Kussie,P., Pavletich,N. and Shuman,S. (1998) Cell, 92, 841–850. [DOI] [PubMed]
- 25.Sherratt D.J. and Wigley,D.B. (1998) Cell, 93, 149–152. [DOI] [PubMed]
- 26.Wigley D.B. (1998) Structure, 6, 543–548. [DOI] [PubMed]
- 27.Grindley N.D. (1997) Curr. Biol., 7, R608–R612. [DOI] [PubMed]
- 28.Lilley D.M. (1997) Chem. Biol., 4, 717–720. [DOI] [PubMed]
- 29.Yang W. and Mizuuchi,K. (1997) Structure, 5, 1401–1406. [DOI] [PubMed]
- 30.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) J. Mol. Biol., 247, 536–540. [DOI] [PubMed]
- 31.Hubbard T.J., Ailey,B., Brenner,S.E., Murzin,A.G. and Chothia,C. (1999) Nucleic Acids Res., 27, 254–256. [DOI] [PMC free article] [PubMed]
- 32.Orengo C.A., Michie,A.D., Jones,S., Jones,D.T., Swindells,M.B. and Thornton,J.M. (1997) Structure, 5, 1093–1108. [DOI] [PubMed]
- 33.Rhee S., Martin,R.G., Rosner,J.L. and Davies,D.R. (1998) Proc. Natl Acad. Sci. USA, 95, 10413–10418. [DOI] [PMC free article] [PubMed]
- 34.Holm L. and Sander,C. (1995) Trends Biochem. Sci., 20, 478–480. [DOI] [PubMed]
- 35.Holm L. and Sander,C. (1997) Nucleic Acids Res., 25, 231–234. [DOI] [PMC free article] [PubMed]
- 36.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed]
- 37.Russell R.B., Saqi,M.A., Bates,P.A., Sayle,R.A. and Sternberg,M.J. (1998) Protein Eng., 11, 1–9. [DOI] [PubMed]
- 38.Wah D.A., Hirsch,J.A., Dorner,L.F., Schildkraut,I. and Aggarwal,A.K. (1997) Nature, 388, 97–100. [DOI] [PubMed]
- 39.Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.F.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) Eur. J. Biochem., 80, 319–324. [DOI] [PubMed]
- 40.Abola E.E., Sussman,J.L., Prilusky,J. and Manning,N.O. (1997) Methods Enzymol., 277, 556–571. [DOI] [PubMed]
- 41.Marchler-Bauer A., Addess,K.J., Chappey,C., Geer,L., Madej,T., Matsuo,Y., Wang,Y. and Bryant,S.H. (1999) Nucleic Acids Res., 27, 240–243. [DOI] [PMC free article] [PubMed]
- 42.Lima C.D., Wang,J.C. and Mondragon,A. (1994) Nature, 367, 138–146. [DOI] [PubMed]
- 43.Berger J.M., Gamblin,S.J., Harrison,S.C. and Wang,J.C. (1996) Nature, 379, 225–232. [DOI] [PubMed]
- 44.Nichols M.D., DeAngelis,K., Keck,J.L. and Berger,J.M. (1999) EMBO J., 18, 6177–6188. [DOI] [PMC free article] [PubMed]
- 45.Murzin A.G. (1994) Curr. Opin. Struct. Biol., 4, 441–449.
- 46.Berger J.M., Fass,D., Wang,J.C. and Harrison,S.C. (1998) Proc. Natl Acad. Sci. USA, 95, 7876–7881. [DOI] [PMC free article] [PubMed]
- 47.Shuman S. (1998) Biochim. Biophys. Acta, 1400, 321–337. [DOI] [PubMed]
- 48.Esnouf R. (1997) J. Mol. Graph. Model., 15, 133–138. [DOI] [PubMed]
- 49.Kraulis P. (1991) J. Appl. Crystallogr., 24, 946–950.