Abstract
Xylose is the second most abundant monomeric sugar in plant biomass. Consequently, xylose catabolism is an ecologically important trait for saprotrophic organisms, as well as a fundamentally important trait for industries that hope to convert plant mass to renewable fuels and other bioproducts using microbial metabolism. Although common across fungi, xylose catabolism is rare within Saccharomycotina, the subphylum that contains most industrially relevant fermentative yeast species. The genomes of several yeasts unable to consume xylose have been previously reported to contain the full set of genes in the XYL pathway, suggesting the absence of a gene–trait correlation for xylose metabolism. Here, we measured growth on xylose and systematically identified XYL pathway orthologs across the genomes of 332 budding yeast species. Although the XYL pathway coevolved with xylose metabolism, we found that pathway presence only predicted xylose catabolism about half of the time, demonstrating that a complete XYL pathway is necessary, but not sufficient, for xylose catabolism. We also found that XYL1 copy number was positively correlated, after phylogenetic correction, with xylose utilization. We then quantified codon usage bias of XYL genes and found that XYL3 codon optimization was significantly higher, after phylogenetic correction, in species able to consume xylose. Finally, we showed that codon optimization of XYL2 was positively correlated, after phylogenetic correction, with growth rates in xylose medium. We conclude that gene content alone is a weak predictor of xylose metabolism and that using codon optimization enhances the prediction of xylose metabolism from yeast genome sequence data.
Keywords: codon optimization, yeasts, metabolic evolution, genome evolution, xylose
Introduction
Xylose is the most abundant pentose sugar and the second most abundant monomeric sugar in plant biomass, second only to glucose. Xylose occurs in xylan polymers in hemicellulose; therefore, the ability to hydrolyze xylan and oxidize xylose for energy is a common trait in saprophytic fungi (Polizeli et al. 2005). Metabolic conversion of xylose is also a key process in the efficient conversion of lignocellulosic biomass into biofuels and other bioproducts via fermentation by industrially leveraged yeast species. Unlike filamentous fungi, native xylose assimilation appears to be a somewhat rare trait within budding yeasts. Saccharomyces cerevisiae is the choice microbe for the industrial production of the vast majority of biofuels due to its high ethanol tolerance, high glycolytic and fermentative capacity, and amenability to genetic engineering (Hong and Nielsen 2012). However, S. cerevisiae requires genetic engineering to metabolize xylose, and even engineered strains are often inefficient in the fermentation of lignocellulosic xylose (Osiro et al. 2019; Lee, Tremaine et al. 2021; Lee, Yook et al. 2021; Sun and Jin 2021). This has led to the suggestion that cost-effective industrial conversion of xylose would be better achieved using native pentose-fermenting yeast species. One successful approach to identifying xylolytic species is the isolation of yeasts from xylose-rich environments, such as rotting logs and the guts of wood-boring beetles (Nguyen et al. 2006; Cadete et al. 2012; Urbina et al. 2013). Given that budding yeast genomes are increasingly available (Riley et al. 2016; Shen et al. 2018), a simpler means of identifying xylolytic yeasts through genome sequence data would facilitate the discovery of additional xylose-metabolizing yeasts.
The budding yeast xylose catabolism pathway was first described in Cyberlindnera jadinii and Candida albicans (Chiang and Knight 1960; Veiga et al. 1960; Chakravorty et al. 1962), but most subsequent characterization has focused on xylose-fermenting genera, including Scheffersomyces and, more recently, Spathaspora (Verduyn et al. 1985; Kötter et al. 1990; Cadete et al. 2016). The native enzymatic pathway consists of three genes: XYL1, XYL2, and XYL3. XYL1 and XYL2 encode a xylose reductase (XR) and xylitol dehydrogenase (XDH), respectively, which function in the oxidoreductive conversion of xylose to xylulose with xylitol as an intermediate. XYL3 encodes a xylulokinase, which phosphorylates xylulose to xylulose-5-phosphate to be fed into the nonoxidative branch of the pentose phosphate pathway (PPP). The identification of yeasts with complete pathways that were nonetheless unable to grow on xylose in previous surveys suggests a weak or absent gene–trait association between complete XYL pathways and xylose assimilation traits (Wohlbach et al. 2011; Riley et al. 2016).
In addition to a complete XYL pathway, other genetic and regulatory features may be important in determining xylose metabolic traits. Most studies have focused on the role of redox imbalance, which is thought to be produced by the different cofactor preferences of XR and XDH due to their preferences for NADPH and NAD+, respectively (Bruinenberg et al. 1983). This hypothesis is supported by the observation that some well-studied yeasts that efficiently metabolize xylose have evolved XR enzymes able to use NADH in addition to or in lieu of NADPH (Bruinenberg et al. 1984; Schneider et al. 1989; Cadete et al. 2016). Recently, it has been suggested that changes to cofactor preference in methylglyoxal reductase (encoded by GRE2) may also alleviate redox imbalance in xylofermentative yeasts (Borelli et al. 2019). Additional properties, such as transporter presence or copy number and the expression of other metabolic genes, have also been implicated in xylose utilization (Wohlbach et al. 2011). It is difficult to say how broadly applicable any of these explanations may be because the presence of XYL genes in the absence of xylose catabolism has only been studied in a handful of related yeast species. Thus, we do not know the extent of this lack of association across budding yeasts and whether other genome characteristics would enhance predictions concerning xylose metabolism.
The identification of some yeasts with complete XYL pathways that lack xylose assimilation suggests that xylose utilization may be much more difficult to predict based on gene content than many other metabolic traits, such as galactose utilization (Riley et al. 2016; Shen et al. 2018). An alternative strategy to predicting metabolic traits from gene content is evaluating specific metabolic genes for evidence of selection. Measuring selection on codon usage is one such approach. Among metrics developed to measure codon usage bias (Bennetzen and Hall 1982; Sharp and Li 1987; Wright 1990), codon optimization captures how well matched individual codons are to their respective tRNA copy numbers in a given genome (dos Reis et al. 2004). Accordingly, a codon with a low-copy corresponding tRNA is less optimized than a codon with a high-copy corresponding tRNA. The codon optimization index of a gene therefore measures the concordance between its transcript and the cellular tRNA pool and has repeatedly been shown to correlate with gene expression levels (Gouy and Gautier 1982; Duret and Mouchiroud 1999; Zhou et al. 2016). Recent work has shown that codon usage is under translational selection in most fungal species (Wint et al. 2022), including within budding yeasts (Labella et al. 2019). Studies examining the relationship between codon usage and metabolism in fungi have found that codon bias is elevated in genes encoding important metabolic pathways (Gonzalez et al. 2020), and, further, that codon optimization of metabolic genes is predictive of growth in corresponding conditions (LaBella et al. 2021). Codon optimization of xylolytic genes has not been studied, but we hypothesize that it may be more useful than gene content in predicting which budding yeast species are well adapted to xylose metabolism.
Here, we measure growth on xylose and systematically identify XYL pathway orthologs across 332 publicly available budding yeast genomes (Shen et al. 2018). In agreement with previous work, we find that an intact XYL pathway often does not confer xylose assimilation. We find multicopy XYL1 and XYL2 lineages to be common, and we find support for the hypothesis that XYL gene copy number is important by showing that XYL1 copy number coevolves with the ability to consume xylose. We then generate codon optimization indices for all XYL homologs and show that XYL3 codon optimization is significantly correlated with the ability to consume xylose, whereas codon optimization of XYL2 is significantly positively correlated with kinetic growth rates on xylose. Collectively, our analyses reveal two genomic properties, copy number of XYL1 and codon optimization of XYL2 and XYL3, that correlate with xylose metabolism and can be used as novel means of predicting xylolytic traits from genome sequence alone.
Results
Identification of XYL Homologs Across 332 Budding Yeast Species
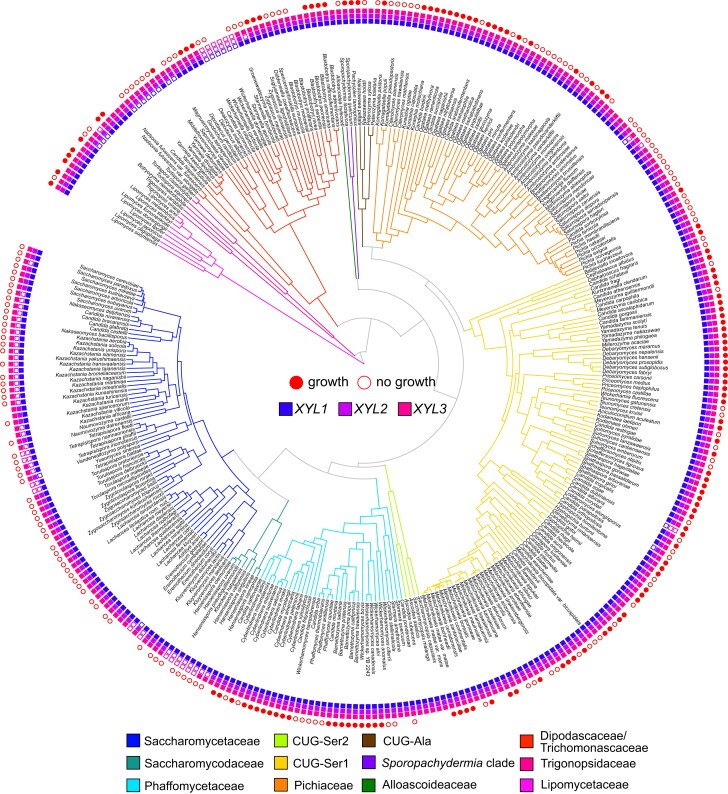
We detected at least one of the three XYL pathway genes in 325 of 332 species (fig. 1). Complete pathways were found in 270 species. We were unable to detect any XYL genes in seven species. Six of the seven species with no detected XYL homologs were the six representative species of the Wickerhamiella/Starmerella (W/S) clade, so it appears that the entire XYL pathway has been lost in this clade. XYL1 and XYL2 have evidence of gene duplications, losses, horizontal transfers, and multiple origins prior to the origin of Saccharomycotina, as well as within the budding yeasts. However, due to the sheer breadth of evolutionary distance in this group, confident elucidation of the complete gene history for these genes is intractable with current taxon sampling.
Fig. 1.
XYL pathway presence and xylose growth across a representative set of 332 Saccharomycotina species. Major yeast clades are depicted by branch color and are described in Shen et al. (2018). Presence of XYL homologs is indicated by filled boxes at tips; the innermost ring depicts XYL1 presence, followed by XYL2, then XYL3. Complete pathways of XYL1, XYL2, and XYL3 were found in 270 species. Species with nonzero growth rates in xylose medium are indicated by a filled red circle, and species unable to assimilate xylose are indicated by an empty red circle. Species without circles were not assayed for growth. Time-calibrated phylogeny from Shen et al. (2018).
The phylogenies of XYL1 and XYL2 homologs were able to resolve previously ambiguous S. cerevisiae orthology (supplementary figs. S1–S3, Supplementary Material online). GRE3 has known XR activity, but it has been annotated as a nonspecific aldo–keto reductase and believed to be distinct from the XR-encoding genes of xylose-fermenting yeasts (Kuhn et al. 1995; Träff et al. 2002; Toivari et al. 2004). We found definitive phylogenetic evidence that GRE3 is a member of the XR-encoding gene family and is orthologous to the XYL1 genes of more distantly related yeasts (supplementary fig. S1, Supplementary Material online). In contrast, S. cerevisiae is known to contain a XYL2 homolog, but the function of XYL2 has remained unclear given the inability of most S. cerevisiae strains to metabolize xylose. The nearly identical S. cerevisiae paralogs SOR1 and SOR2 also fell within the XYL2 clade of the family Saccharomycetaceae. SOR1 and SOR2 are annotated as encoding sorbitol dehydrogenases and are upregulated in response to sorbose and xylose (Toivari et al. 2004) (supplementary fig. S2, Supplementary Material online).
The XYL2 gene phylogeny showed more evidence of gene diversification and retention than was expected, given that species of the family Saccharomycetaceae are generally not able to use xylose as a carbon source. To further clarify XYL2 evolution within the Saccharomycetaceae, we generated a maximum likelihood tree of the XYL2 homologs within the Saccharomycetaceae and included S. cerevisiae XDH1, a gene encoding a XDH present in some wine strains (but not the S288C reference strain) that was previously identified as being sufficient for weak xylose utilization (Wenger et al. 2010). The resulting tree supports an ancestral duplication of XYL2, which produced two distinct paralogous lineages that we name the SOR lineage and the XYL2 lineage based on the S. cerevisiae paralogs contained therein (supplementary fig. S3, Supplementary Material online). The XYL2 lineage homolog was preferentially retained by most Saccharomycetaceae species, whereas a handful retained only the SOR paralog, and a few retained both. The tree also supported a few subsequent duplications, including the lineage-specific duplication of SOR1/SOR2 in S. cerevisiae. The phylogeny also showed that the XDH1 gene identified in some wine strains of S. cerevisiae by Wenger et al. (2010) is orthologous to S. cerevisiae SOR1/SOR2, not to S. cerevisiae XYL2. The protein sequence is identical to the Torulaspora microellipsoides SOR homolog, further corroborating a known 65-kb transfer from T. microellipsoides to the S. cerevisiae EC1118 wine strain and its relatives (Marsit et al. 2015).
A Complete XYL Pathway Is Necessary, but Not Sufficient, for Xylose Catabolism
The XYL pathway has been repeatedly shown to underlie xylose catabolism in focal budding yeasts, and no alternative pathways are known. Nonetheless, previous genomic surveys have turned up multiple taxa that possess complete pathways but are unable to catabolize xylose (Wohlbach et al. 2011; Riley et al. 2016). In agreement with these previous studies, we measured maximum growth rates in a minimal medium containing xylose as the sole carbon source for 282 of the 332 species examined and found that only 52% of species with complete pathways were able to grow on xylose (123/236, fig. 1). To explicitly test for an evolutionary relationship between XYL pathway presence and xylose utilization, we used Pagel's (1994) method to test for a correlation between the two binary traits and found strong support for the coevolution of complete XYL pathways and xylose metabolism (P = 1.1 × 10−5, supplementary table S1, Supplementary Material online). Indeed, 235 of 236 species that exhibited growth in xylose medium contained complete pathways. Only Candida sojae appeared able to catabolize xylose although lacking a complete pathway, but this is likely attributed to an incomplete C. sojae genome, rather than true pathway absence (Shen et al. 2018). These data collectively demonstrate that a complete XYL pathway is necessary, but not sufficient, for xylose catabolism, which suggests that there may be other quantifiable genomic features that would enhance predictions of xylose catabolism.
XYL1 Copy Number Is Correlated with Xylose Metabolism
Duplications and losses of enzyme-encoding genes are well-documented evolutionary modulators of metabolic activities (Kliebenstein 2008; Wolfe et al. 2015). XYL1 and XYL2 were frequently found as multicopy in our data set, so we next tested for a relationship between increased copy number and xylose metabolism. We scored yeast taxa as either multicopy or single-copy and again used Pagel's (1994) method to look for a correlation between xylose catabolism and copy number. Copy number of XYL1 was significantly correlated with the ability to grow on xylose (P = 1.5 × 10−4, supplementary fig. S4, Supplementary Material online). The coevolutionary model with the most support assumed that the two traits were interdependent (weighted Akaike information criterion [AIC] = 0.51, supplementary table S2, Supplementary Material online), but a model in which growth depended on XYL1 copy number was almost as strongly supported (weighted AIC = 0.48). Contrary to XYL1, coevolution between XYL2 copy number and growth on xylose was not supported (P = 0.60, supplementary table S3, Supplementary Material online). We did not test for a correlation with XYL3 copy number because only four species had multiple copies of this gene. As with gene content, the correlation between XYL1 duplication and growth in xylose medium was not perfect; indeed, 43% (20/46) of multicopy lineages were unable to metabolize xylose. Whereas these data point to a significant role of XYL1 duplication in some taxa, we conclude that XYL1 copy number alone is insufficient to explain yeast variation in xylose metabolic traits.
XYL1 and XYL2 Are Highly Codon Optimized
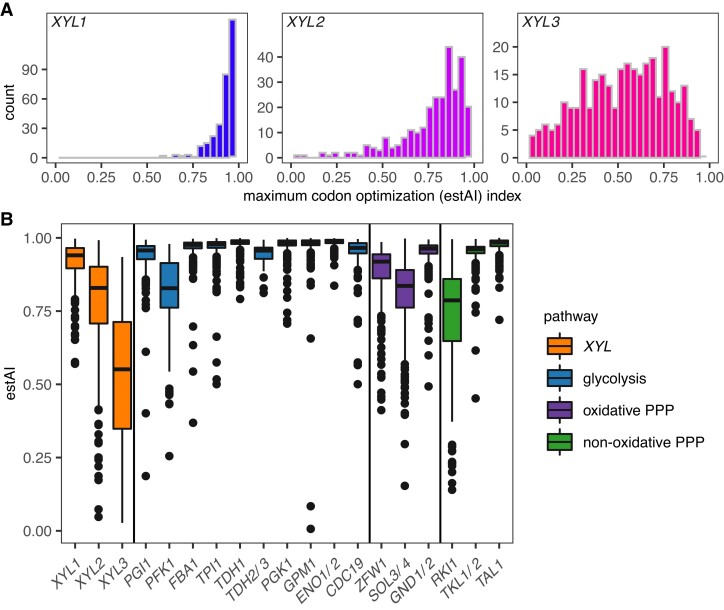
We next examined whether codon optimization of the XYL pathway genes would be useful in predicting metabolic capabilities. Codon optimization indices (estAI values) of XYL pathway homologs were calculated for 320 of the 325 species in which a XYL1, XYL2, or XYL3 gene was detected. XYL1 and XYL2 estAI distributions were both heavily skewed with median estAI values of 0.94 and 0.83, which means these genes have a higher optimization than 94% and 83% of the coding genome of an individual species, respectively. XYL3 estAI values were more variable with a lower median optimization index of 0.55 (fig. 2A).
Fig. 2.
Distribution of codon optimization indices (estAI values). (A) Histograms of the distribution of maximum estAI values among 320 of the 325 species for XYL1, XYL2, and XYL3 are shown. XYL1 genes were skewed towards highly optimized (blue), XYL2 estAI values were somewhat less skewed (violet), and XYL3 estAI values were broadly distributed (magenta). Median estAI values of 0.94, 0.83, and 0.55 were calculated for XYL1, XYL2, and XYL3, respectively. (B) XYL gene estAI distributions were compared with other carbon metabolism pathways related to xylose metabolism. The XYL pathway (orange), in general, was less optimized than glycolysis (blue) or either branch of the PPP (purple/green). Specifically, the XYL1 distribution was significantly lower than the estAI distributions of highly expressed glycolytic genes (FBA1, TPI1, TDH1, PGK1, GPM1, and ENO1/ENO2), but it was similar to PGI1. XYL2 genes had estAI values similar to the rate-limiting steps in glycolysis (PFK1) and the oxidative PPP (ZWF1). XYL3 was less optimized on average than genes involved in glycolysis or the PPP.
To provide context to codon optimization index distributions for XYL genes, we compared them to the optimization indices of genes that function in glycolysis and the PPP (fig. 2B). The XYL1 distribution was lower than the estAI distributions of highly expressed glycolytic genes (FBA1, TPI1, TDH1, PGK1, GPM1, and ENO1/ENO2), but it was similar to PGI1, which encodes the glycolysis-initiating enzyme phosphoglucose isomerase. XYL2 genes were less codon optimized than most glycolytic genes, but interestingly, the XYL2 estAI distribution was similar to the rate-limiting steps in glycolysis (PFK1) and the oxidative PPP (ZWF1). XYL3 was clearly less codon optimized on average than genes involved in glycolysis or the PPP.
Codon Optimization of XYL3 Predicts Xylose Growth Abilities
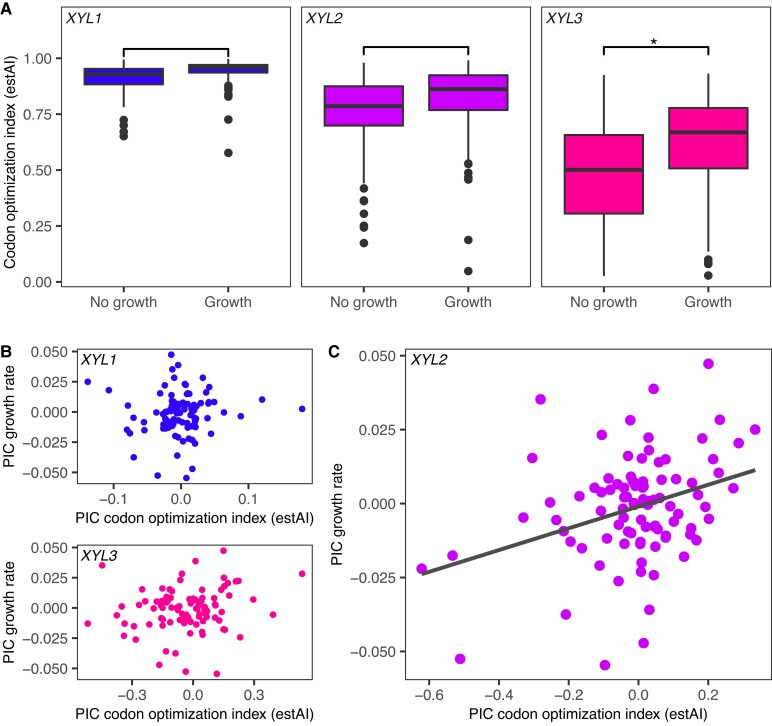
The distributions of codon optimization indices for the three XYL genes in species able to grow in xylose medium were higher than the distributions of species showing no growth (fig. 3A). Because this difference could also be due to shared ancestry, we tested whether codon optimization of XYL genes was correlated with xylose utilization by using a Bayesian phylogenetic linear mixed model (GLMM) to control for shared evolutionary history. Using this model, only codon optimization of XYL3 was significantly correlated with the ability to metabolize xylose (pMCMC = 0.039), whereas codon optimizations of XYL1 and XYL2 were not (supplementary table S4, Supplementary Material online).
Fig. 3.
XYL3 codon optimization predicts the ability to metabolize xylose. (A) Boxplots showing the distribution of estAI values for species unable to use xylose (left) compared with those that can (right) for XYL1 (blue), XYL2 (violet), and XYL3 (magenta). Asterisk denotes significant as assessed by a Bayesian phylogenetic linear mixed model (GLMM) (supplementary table S4, Supplementary Material online). (B, C) PIC analyses of XYL1, XYL2, and XYL3 estAI in relation to xylose growth. Kodamaea laetipori and Blastobotrys adeninivorans were removed as outliers prior to analyses. (B) Codon optimizations of XYL1 and XYL3 did not correlate with xylose growth rates. (C) Codon optimization of XYL2 was significantly correlated with growth rate in xylose medium (P = 9 × 10−4, r = 0.34).
Codon Optimization of XYL2 Correlates with Xylose Growth Rates
We have shown previously that codon optimization indices of specific genes involved in galactose metabolism not only predict whether a budding yeast species can utilize galactose, but can also be used to predict the rates of growth on galactose (LaBella et al. 2021). We similarly compared XYL gene codon optimization to growth rates measured in medium containing xylose as the sole carbon source to determine whether this trait would be useful in predicting yeast growth rates when consuming xylose. Phylogenetically independent contrasts (PICs) were used to compare estAI values and growth rates for the 93 species with complete pathways and for which there was previously published evidence of selection on codon usage (Labella et al. 2019). Of the three genes examined, only XYL2 had a significant correlation between codon optimization and growth rate (P = 9 × 10−4, r = 0.34; fig. 3B and C).
Discussion
Xylose fermentation is an ecologically important trait of immense biotechnological value for the conversion of sustainable plant feedstocks into biofuels. This study identifies systematically XYL pathway homologs across a wide breadth of Saccharomycotina that includes representative species from all 12 major clades. Whereas most genomes examined contain complete pathways, less than half of those species were able to assimilate xylose under laboratory conditions. This stands in contrast to other metabolic traits that have been investigated in yeasts that exhibit strong gene–trait associations (Riley et al. 2016; Shen et al. 2018). For example, a survey of galactose metabolism across the same extensive collection of budding yeast species found that 89% of species with complete GAL pathways were able to use galactose as a carbon source in the laboratory (LaBella et al. 2021). The poor ability of gene content to predict xylose–metabolism traits has been noted before in surveys of a small number of biotechnologically important yeasts (Wohlbach et al. 2011; Riley et al. 2016), but it was unclear whether this limited gene–trait association would apply broadly across budding yeasts. Whereas complete pathways are found in all major yeast clades, xylose metabolism is variable; most CUG-Ser1 species are able to utilize xylose, assimilation shows up sporadically in most other clades, and it is completely absent in the Saccharomycetaceae. These patterns are consistent with previous observations (reviewed in Ruchala and Sibirny (2021)).
One limitation of this study and a possible explanation for the poor correlation between genotype and phenotype is that xylose catabolism requires specific conditions. We analyzed only growth data generated in our assay under a single laboratory condition. For some species, our data conflict with data aggregated from species descriptions (Opulente et al. 2018). For other species, conflicting data also exist elsewhere in the literature. For example, Kluyveromyces marxianus did not grow in our 96-well plate assay but has been found to consume xylose in shake flasks (Margaritis and Bajpai 1982). Oxygenation, base media, and temperature have all been documented as affecting xylose metabolism in different yeast species (Signori et al. 2014; Osiro et al. 2019). Beyond condition dependence, intraspecific metabolic heterogeneity, such as is known to occur in Kluyveromyces lactis and Torulaspora delbrueckii, could also produce inconsistencies (Lyutova et al. 2021; Silva et al. 2023). A final reason why our data may conflict with preexisting descriptions is historical human errors in species typing and identification (Haase et al. 2017). Our choice to confine our analysis to the data we directly collected from taxonomic type strains may have obscured growth in a few species, but in general, it eliminated the effects of inconsistent conditions and taxonomical error.
Whereas it remains unclear why XYL pathway presence is not sufficient to confer xylose catabolism, the finding that most yeast species do, in fact, have intact XYL pathways has implications for industrial strain development at a time when researchers are actively searching for new candidate species. The first of these is that engineering xylose consumption in nonutilizing species will likely be more difficult than the simple heterologous expression of XYL gene cassettes. A second, more promising, implication is that most yeast species already have the genetic potential for xylose metabolism and could perhaps be coaxed into xylose utilization with adaptive laboratory evolution, mutagenesis, or a combination thereof.
Although we find pathway completeness alone to be insufficient for xylose assimilation, each of the three genes was found to have a property correlated with xylose metabolism. Increased copy number of XYL1 and increased codon optimization of XYL3 are important for determining whether a species will consume xylose, whereas codon optimization of XYL2 determines how efficiently xylose is converted to biomass. Of these, copy number has known relevance based on the observations that duplications and functional divergences of XYL1 are consequential in xylose-fermenting yeasts (Bruinenberg et al. 1984; Mayr et al. 2000; Cadete et al. 2016), and that amplification of heterologous XYL1 is a frequent mode of adaptation in engineered yeast populations evolved for xylose consumption in the lab (Li and Alper 2016; Peris et al. 2017). The present study confirms a statistically significant phylogenetic coevolutionary relationship between XYL1 copy number and xylose metabolism. The relationship between XYL1 amplification and xylose metabolism is unlikely to be a matter of simple flux; XYL2, not XYL1, is thought to be the rate-limiting step in xylose catabolism (Kim et al. 2012; Zha et al. 2012; Ryu et al. 2016). Instead, detailed studies of XYL1 paralog pairs within the CUG-Ser1 clade show divergence in cofactor preferences between paralogs (Bruinenberg et al. 1984; Cadete et al. 2016), which provides an attractive hypothesis in which duplicate XYL1 genes resolve redox imbalance.
Both the XYL1 and XYL2 phylogenies generated show evidence of widespread duplication and loss. Despite evidence of xylitol oxidation to xylulose being the rate-limiting step in xylose degradation, XYL2 copy number was not associated with xylose catabolism. The phylogenetic distribution of retained XYL2 paralogs is curious. Given the seeming ecological irrelevance of xylose utilization in the Saccharomycetaceae, the diversification and retention of XYL2 genes in this group lack a clear explanation unless the primary function of XYL2 homologs in this family is not in xylose catabolism. Several lines of evidence in the literature support this notion: 1) there is ample evidence that budding yeast XDH enzymes are promiscuous across polyols (Ko et al. 2006; Biswas et al. 2010, 2013; Sukpipat et al. 2017); 2) the Xyl2 reverse reaction (reduction of xylulose to xylitol) is more energetically favorable by an order of magnitude (Rizzi et al. 1989); and 3) the strongest phylogenetic signal of XYL gene loss we observed was in the W/S clade of yeasts, which is a group of fructose-specializing yeasts that have evolved a novel means of reducing fructose to maintain redox balance (Gonçalves et al. 2019). Taken together, these data are suggestive of an alternative role of the XYL pathway and XYL2 in particular. Instead of supporting xylose utilization, XDH activity in these yeasts may be important for regenerating oxidized NAD+ in certain growth conditions through the reduction of sugars, including xylulose, fructose, and mannose, to the polyols xylitol, sorbitol, and mannitol, respectively. Additional experimental work in the family Saccharomycetaceae is needed to determine if XDH activity plays a role in redox balance as hypothesized above, or perhaps functions in a yet-to-be-discovered process.
It was initially surprising to find that XYL2 copy number does not covary with qualitative xylose consumption because XDH is considered a rate-limiting step, and overexpression often increases xylose fermentation rates in engineered strains (Jeppsson et al. 2003; Karhumaa et al. 2007). Instead, we found that XYL2 codon optimization positively correlates with growth rates on xylose. The correlation between codon optimization and growth that we report supports the hypothesis that endogenous XYL2 expression levels affect rates of xylose consumption in natively xylose-consuming yeasts. This optimization could be partly to overcome the unfavorable reaction kinetics and subpar substrate specificity mentioned above. Interestingly, the XYL2 estAI distribution we observed was highly similar to that of rate-limiting steps of glycolysis (PFK1) and the oxidative PPP (ZWF1), perhaps pointing to a general trend in genes encoding enzymes with rate-limiting or regulatory roles.
The codon optimization distribution of XYL3 was much broader than the other two genes in the XYL pathway. There is little evidence that increasing xylulose kinase activity alone increases xylose pathway flux, and so the broad distribution we observe may simply reflect a lack of selection on XYL3 gene expression. Nonetheless, only XYL3 codon optimization was correlated with the actual ability to consume xylose. The finding that XYL3 codon optimization is correlated with qualitative growth, but not quantitative growth rate, coupled with the broad distribution of codon optimization across species, suggests that there may be an important threshold of XYL3 expression or that the phylogenetically corrected signal was simply not as strong as for XYL2 in this data set. The different distributions observed between the XYL genes could also be related to other correlates of codon usage selection, such as the evolutionary ages of the genes (Prat et al. 2009). Indeed, XYL1 and XYL2 are members of large and ancient gene families of aldo–keto reductases and medium-chain dehydrogenases, respectively, whereas XYL3 does not appear to belong to a large fungal gene family.
Xylose metabolism cannot be predicted by gene content alone in budding yeasts. Here, we show that there is a significant predictive value of codon optimization in the detection of native xylose-metabolizing yeasts for two of the three genes required for xylose degradation. Xylose fermentation is a trait of great ecological and biotechnological interest, whereas being exceedingly rare. Instead of expending resources testing large sets of yeasts or their synthesized genes, copy number and codon optimization could be used to filter for candidate yeasts with a higher probability of containing highly xylolytic pathways. We also show that XYL2 optimization has a linear relationship with growth rates on xylose. In the absence of growth or metabolic data, XYL2 sequences can be used to predict which species are likely to catabolize xylose especially well. This work presents a novel framework of leveraging signatures of selection, specifically codon optimization, for understanding weak and variable gene–trait associations and could be a valuable tool for understanding trait variation in other systems.
Materials and Methods
Identification of XYL1, XYL2, and XYL3 Homologs
We identified homologs of XYL1, XYL2, and XYL3 across 332 published budding yeast genome assemblies (Shen et al. 2018) using hidden Markov model (HMMER) sequence similarity searches (v3.3, http://hmmer.org). HMM profiles were built using sequences retrieved from a BLASTp search using Spathaspora passalidarum XYL1.1, XYL2.1, and XYL3. Hits were manually curated to retain an alignment of 14 sequences representing a phylogenetically diverse taxon set. HMMER searches were performed on protein annotations generated with ORFfinder (NCBI RRID:SCR_016643) using default settings, which include nonconventional start codons. Sequences were later manually curated to confirm probable start sites (see below). We did not account for modified translation tables found in some yeast clades (CUG-Ser1, CUG-Ser2, and CUG-Ala clades; Shen et al. 2018) because this codon is known to be rare (Labella et al. 2019).
HMMER searches for XYL1 and XYL2 both identified large gene families of aldose reductases and medium-chain dehydrogenases, respectively. To identify the XYL orthologous sequences, HMMER hits were assigned KEGG orthology with BLASTKoala (Kanehisa et al. 2016), and approximate maximum likelihood trees of KEGG-annotated hits were built with FastTree v2.1.10 (Price et al. 2009) (supplementary figs. S5 and S6, Supplementary Material online). Subclades containing XYL gene homologs based on KEGG orthology (XYL1 - K17743 and XYL2 - K05351) were identified for XYL1 and XYL2.
Coding sequences of homologs for all three genes were then manually curated. Probable start sites were identified using TranslatorX (Abascal et al. 2010), and sequences were trimmed or expanded accordingly. A combination of alignment visualization and collapsed tree inspection was used to identify highly divergent sequences that were then examined via BLAST; likely bacterial contaminants were removed. Maximum likelihood phylogenies of protein sequences for each of the three genes were built with IQTree (Trifinopoulos et al. 2016) using ModelFinder (Kalyaanamoorthy et al. 2017) automated model selection (Xyl1- LG+F+I+G4, Xyl2- LG+I+G4, and Xyl3- LG+F+I+G4, supplementary figs. S1, S2, and S7, Supplementary Material online) based on 1,000 bootstrap replications. An independent maximum likelihood tree of Xyl2 protein sequences in the family Saccharomycetaceae with the addition of S. cerevisiae Xdh1 originating from a wine strain (Wenger et al. 2010) was generated using IQ tree with an LG+I+G4 substitution model and node support based on 1,000 bootstrap replications. Trees were visualized and annotated in iTOL (Letunic and Bork 2021).
Growth Assays
All yeast strains used in growth experiments were first plated on yeast extract peptone dextrose (YPD) agar plates and grown until single colonies were visible. The plates were then stored at 4 °C for up to a month. Single colonies were then cultured in liquid YPD for a week at room temperature on a culture wheel. After a week of growth, yeast strains were subcultured in 96-well plates containing minimal medium with 1% glucose or 1% xylose and allowed to grow for a week at room temperature. The 96-well plates contained a four-quadrant moat around the edge of the plate where 2 mL of water was added to each quadrant. The addition of water to the plate prevents evaporation in the edge and corner wells, allowing for the whole plate to be utilized. After the initial week of growth on the treatments, all yeasts were transferred into fresh 1% glucose or 1% xylose minimal medium and placed on a plate reader and stacker (BMG FLUOstar Omega). Plates were read every 2 h for a week at OD600. All growth experiments were replicated three times. In each replicate, both the order of yeasts on the plate and order of sugars on the plate were randomized to alleviate plate effects. Growth rates were quantified in R using the package grofit (Kahm et al. 2010). Average growth rates were calculated across replicates for each species.
Codon Optimization
Codon optimization indices of XYL1, XYL2, and XYL3 homologs were determined as in LaBella et al. (2021). Species-specific codon optimization values (wi values) for all codons were retrieved from Labella et al. (2019). For each ortholog analyzed, each codon was identified and assigned its species-specific wi value. The codon optimization index (stAI) for each ortholog was then calculated as the geometric mean of wi values for each gene. Five species in our data set do not have corresponding wi values due to software issues (LaBella et al. 2019) and were dropped from codon optimization analyses (Middelhovenomyces tepae, Nadsonia fulvescens var. fulvescens, Spencermartinsiella europaea, Botryozyma nematodophila, and Martiniozyma abiesophila). To compare codon optimization values between species, the gene-specific stAI value of each gene was normalized to the genome-wide distribution of stAI values for the respective species using the empirical cumulative distribution function. The resulting normalized codon optimization index (estAI value) is an estimate of the genome-wide percentile of codon optimization for each gene (e.g., an estAI value of 0.95 indicates a gene that is more optimized than 95% of genes in the genome). For species with multiple paralogs, including those derived from the whole genome duplication, only the gene with the highest estAI value was considered in further analysis.
Orthologs of glycolysis pathway genes (CDC19, ENO1/ENO2, FBA1, GPM1, PFK1, PGI1, PGK1, TDH1, TDH2/TDH3, and TPI1) and PPP genes (GND1/GND2, RKI1, SOL3/SOL4, TAL1, TKL1/TKL2, and ZWF1) were identified using HMMER searches as described above with the exception of manual curation. Codon optimization for each gene was measured as described above. For species with multiple paralogs, only the maximum estAI value per gene per species was retained for analysis.
Statistical Analyses of Growth Data and Codon Optimization
Pagel's (1994) tests were used to test for correlated evolution between binary growth traits and the binary traits of pathway completeness or multicopy genes. Growth was scored as present in all species exhibiting nonzero growth in xylose media and absent in species without detectable growth. XYL pathways were scored as complete in all taxa possessing at least one copy of XYL1, XYL2, and XYL3 and incomplete when any of the three genes was absent. Taxa with two or more copies of XYL1 or XYL2 were scored as multicopy, whereas taxa with only one copy were scored as single copy. Tests were performed using the R package phytools (Revell 2012).
A Bayesian phylogenetic linear mixed model was used to test the effect of codon optimization and binary growth traits using MCMCglmm with family set to “categorical” (Hadfield 2010). Quantitative codon optimization indices were scaled to have a mean of 0 and standard deviation of 1. All three genes were combined in a single model with phylogeny as a random effect. Priors were set with an inverse-gamma prior with shape and scale equal to 0.001. The model was run with 4 × 107 iterations, a burn-in of 105 iterations, and a thinning interval of 104. Chains were visually inspected and model convergence was assessed using Heidelberger and Welch’s convergence diagnostic.
The effect of codon optimization on quantitative growth rates was tested separately for each gene using PICs. To compare xylose growth rates to estAI values, we first retained data for only those species previously found to have evidence of genome-wide selection on codon usage (Labella et al. 2019). Two species had extremely high growth rates that did not appear to be artifactual (supplementary fig. S8, Supplementary Material online). Since phylogenetic independent contrasts are highly sensitive to outlier data, we removed these two species. For the remaining 93 species, growth rate was compared with codon optimization by fitting a linear model to PIC values to account for phylogenetic relatedness. PIC values were generated using the ape package in R (Paradis and Schliep 2019). All other statistical analyses were performed using R stats v3.6.2.
Supplementary Material
Acknowledgments
We thank members of the Hittinger and Rokas groups for helpful discussions. This work was supported by the National Science Foundation under Grant Nos. DEB-1442148, DEB-2110403, DEB-1442113, and DEB-2110404; in part by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science DE-SC0018409); and the USDA National Institute of Food and Agriculture (Hatch Project 1020204). C.T.H. is an H. I. Romnes Faculty Fellow, supported by the Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. Research in A.R.'s lab is also supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R56 AI146096 and R01 AI153356) and the Burroughs Wellcome Fund. K.J.F. was a Morgridge Metabolism Interdisciplinary Fellow, supported by the Morgridge Institute for Research - Metabolism Theme.
Contributor Information
Rishitha L Nalabothu, Laboratory of Genetics, J. F. Crow Institute for the Study of Evolution, Wisconsin Energy Institute, Center for Genomic Science Innovation, University of Wisconsin-Madison, Madison, WI; DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI.
Kaitlin J Fisher, Laboratory of Genetics, J. F. Crow Institute for the Study of Evolution, Wisconsin Energy Institute, Center for Genomic Science Innovation, University of Wisconsin-Madison, Madison, WI; Department of Biological Sciences, State University of New York at Oswego, Oswego, NY.
Abigail Leavitt LaBella, Department of Biological Sciences, Vanderbilt University, Nashville, TN; Evolutionary Studies Initiative, Vanderbilt University, Nashville, TN; Department of Bioinformatics and Genomics, University of North Carolina at Charlotte, Charlotte, NC.
Taylor A Meyer, Laboratory of Genetics, J. F. Crow Institute for the Study of Evolution, Wisconsin Energy Institute, Center for Genomic Science Innovation, University of Wisconsin-Madison, Madison, WI; DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI.
Dana A Opulente, Laboratory of Genetics, J. F. Crow Institute for the Study of Evolution, Wisconsin Energy Institute, Center for Genomic Science Innovation, University of Wisconsin-Madison, Madison, WI; DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI; Department of Biology, Villanova University, Villanova, PA.
John F Wolters, Laboratory of Genetics, J. F. Crow Institute for the Study of Evolution, Wisconsin Energy Institute, Center for Genomic Science Innovation, University of Wisconsin-Madison, Madison, WI; DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI.
Antonis Rokas, Department of Biological Sciences, Vanderbilt University, Nashville, TN; Evolutionary Studies Initiative, Vanderbilt University, Nashville, TN.
Chris Todd Hittinger, Laboratory of Genetics, J. F. Crow Institute for the Study of Evolution, Wisconsin Energy Institute, Center for Genomic Science Innovation, University of Wisconsin-Madison, Madison, WI; DOE Great Lakes Bioenergy Research Center, University of Wisconsin-Madison, Madison, WI.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
K.J.F., A.L.L., A.R., and C.T.H. conceived of the project. R.L.N., T.A.M., K.J.F., and A.L.L. performed bioinformatic analyses. J.F.W. wrote a custom bioinformatic pipeline for sequence similarity searches. D.A.O. collected and analyzed growth rate data. R.L.N., K.J.F., and A.L.L. performed statistical analyses. R.L.N., K.J.F., and C.T.H. wrote the paper with input from all authors. K.J.F., A.L.L., A.R., and C.T.H. provided mentorship throughout the study.
Data Availability
Analyses were performed on the 332 published and publicly available assemblies analyzed in Shen et al. (2018). Codon optimization values were obtained from the figshare repository from LaBella et al. (2019) (https://doi.org/10.6084/m9.figshare.c.4498292). All data generated in this project, including curated XYL gene sequences, are available in the figshare associated with this manuscript (https://doi.org/10.6084/m9.figshare.c.6011956.v1).
Conflict of interest statement. A.R. is a scientific consultant for LifeMine Therapeutics, Inc.
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38(suppl_2):W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. 1982. Codon selection in yeast. J Biol Chem. 257(6):3026–3031. [PubMed] [Google Scholar]
- Biswas D, Datt M, Aggarwal M, Mondal AK. 2013. Molecular cloning, characterization, and engineering of xylitol dehydrogenase from Debaryomyces hansenii. Appl Microbiol Biotechnol. 97(4):1613–1623. [DOI] [PubMed] [Google Scholar]
- Biswas D, Datt M, Ganesan K, Mondal AK. 2010. Cloning and characterization of thermotolerant xylitol dehydrogenases from yeast Pichia angusta. Appl Microbiol Biotechnol. 88(6):1311–1320. [DOI] [PubMed] [Google Scholar]
- Borelli G, Fiamenghi MB, Dos Santos LV, Carazzolle MF, Pereira GAG, José J. 2019. Positive selection evidence in xylose-related genes suggests methylglyoxal reductase as a target for the improvement of yeasts’ fermentation in industry. Genome Biol Evol. 11(7):1923–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinenberg PM, de Bot PHM, van Dijken JP, Scheffers WA. 1983. The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J Appl Microbiol Biotechnol. 18(5):287–292. [Google Scholar]
- Bruinenberg PM, de Bot PHM, van Dijken JP, Scheffers WA. 1984. NADH-linked aldose reductase: the key to anaerobic alcoholic fermentation of xylose by yeasts. Appl Microbiol Biotechnol. 19(4):256–260. [Google Scholar]
- Cadete RM, de las Heras AM, Sandström AG, Ferreira C, Gírio F, Gorwa-Grauslund M-F, Rosa CA, Fonseca C. 2016. Exploring xylose metabolism in Spathaspora species: XYL1.2 from Spathaspora passalidarum as the key for efficient anaerobic xylose fermentation in metabolic engineered Saccharomyces cerevisiae. Biotechnol Biofuels. 9(1):167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadete RM, Melo MA, Dussan KJ, Rodrigues RCLB, Silva SS, Zilli JE, Vital MJS, Gomes FCO, Lachance M-A, Rosa CA. 2012. Diversity and physiological characterization of D-xylose-fermenting yeasts isolated from the Brazilian Amazonian Forest. PLoS ONE. 7(8):e43135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty M, Veiga LA, Bacila M, Horecker BL. 1962. Pentose metabolism in Candida: II. The diphosphopyridine nucleotide-specific polyol dehydrogenase of Candida utilis. J Biol Chem. 237(4):1014–1020. [PubMed] [Google Scholar]
- Chiang C, Knight SG. 1960. Metabolism of D-xylose by moulds. Nature 188(4744):79–81. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. 2004. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 32(17):5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci. 96(8):4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves C, Ferreira C, Gonçalves LG, Turner DL, Leandro MJ, Salema-Oom M, Santos H, Gonçalves P. 2019. A new pathway for mannitol metabolism in yeasts suggests a link to the evolution of alcoholic fermentation. Front Microbiol. 10:2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Corsini G, Lobos S, Seelenfreund D, Tello M. 2020. Metabolic specialization and codon preference of lignocellulolytic genes in the white rot basidiomycete Ceriporiopsis subvermispora. Genes (Basel) 11(10):1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Gautier C. 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10(22):7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase MAB, Kominek J, Langdon QK, Kurtzman CP, Hittinger CT. 2017. Genome sequence and physiological analysis of Yamadazyma laniorum fa sp. nov. and a reevaluation of the apocryphal xylose fermentation of its sister species, Candida tenuis. FEMS Yeast Res. 17(3):fox019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J Stat Softw. 33:1–22.20808728 [Google Scholar]
- Hong K-K, Nielsen J. 2012. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci. 69(16):2671–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppsson M, Träff K, Johansson B, Hahn-Hägerdal B, Gorwa-Grauslund MF. 2003. Effect of enhanced xylose reductase activity on xylose consumption and product distribution in xylose-fermenting recombinant Saccharomyces cerevisiae. FEMS Yeast Res. 3(2):167–175. [DOI] [PubMed] [Google Scholar]
- Kahm M, Hasenbrink G, Lichtenberg-Fraté H, Ludwig J, Kschischo M. 2010. Grofit: fitting biological growth curves. Nat Preced. 1:1. [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 428(4):726–731. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Fromanger R, Hahn-Hägerdal B, Gorwa-Grauslund M-F. 2007. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 73(5):1039–1046. [DOI] [PubMed] [Google Scholar]
- Kim SR, Ha S-J, Kong II, Jin Y-S. 2012. High expression of XYL2 coding for xylitol dehydrogenase is necessary for efficient xylose fermentation by engineered Saccharomyces cerevisiae. Metab Eng. 14(4):336–343. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ. 2008. A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One 3(3):e1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko BS, Jung HC, Kim JH. 2006. Molecular cloning and characterization of NAD+-dependent xylitol dehydrogenase from Candida tropicalis ATCC 20913. Biotechnol Prog. 22(6):1708–1714. [DOI] [PubMed] [Google Scholar]
- Kötter P, Amore R, Hollenberg CP, Ciriacy M. 1990. Isolation and characterization of the Pichia stipitis xylitol dehydrogenase gene, XYL2, and construction of a xylose-utilizing Saccharomyces cerevisiae transformant. Curr Genet. 18(6):493–500. [DOI] [PubMed] [Google Scholar]
- Kuhn A, van Zyl C, van Tonder A, Prior BA. 1995. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl Environ Microbiol. 61(4):1580–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBella AL, Opulente DA, Steenwyk JL, Hittinger CT, Rokas A. 2019. Variation and selection on codon usage bias across an entire subphylum. PLoS Genet. 15(7):e1008304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBella AL, Opulente DA, Steenwyk JL, Hittinger CT, Rokas A. 2021. Signatures of optimal codon usage in metabolic genes inform budding yeast ecology. PLoS Biol. 19(4):e3001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-B, Tremaine M, Place M, Liu L, Pier A, Krause DJ, Xie D, Zhang Y, Landick R, Gasch AP, et al. 2021. Crabtree/Warburg-like aerobic xylose fermentation by engineered Saccharomyces cerevisiae. Metab Eng. 68:119–130. [DOI] [PubMed] [Google Scholar]
- Lee JW, Yook S, Koh H, Rao CV, Jin Y-S. 2021. Engineering xylose metabolism in yeasts to produce biofuels and chemicals. Curr Opin Biotechnol. 67:15–25. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49(W1):W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Alper HS. 2016. Enabling xylose utilization in Yarrowia lipolytica for lipid production. Biotechnol J. 11(9):1230–1240. [DOI] [PubMed] [Google Scholar]
- Lyutova LV, Naumov GI, Shnyreva AV, Naumova ES. 2021. Molecular polymorphism of β-galactosidase LAC4 genes in dairy and natural strains of Kluyveromyces yeasts. Mol Biol. 55(1):66–74. [DOI] [PubMed] [Google Scholar]
- Margaritis A, Bajpai P. 1982. Direct fermentation of D-xylose to ethanol by Kluyveromyces marxianus strains. Appl Environ Microbiol. 44(5):1039–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit S, Mena A, Bigey F, Sauvage F-X, Couloux A, Guy J, Legras J-L, Barrio E, Dequin S, Galeote V. 2015. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol Biol Evol. 32(7):1695–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr P, Brüggler K, Kulbe KD, Nidetzky B. 2000. D-Xylose metabolism by Candida intermedia: isolation and characterisation of two forms of aldose reductase with different coenzyme specificities. J Chromatogr B Biomed Sci Appl. 737(1–2):195–202. [DOI] [PubMed] [Google Scholar]
- Nguyen NH, Suh S-O, Marshall CJ, Blackwell M. 2006. Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol Res. 110(10):1232–1241. [DOI] [PubMed] [Google Scholar]
- Opulente DA, Rollinson EJ, Bernick-Roehr C, Hulfachor AB, Rokas A, Kurtzman CP, Hittinger CT. 2018. Factors driving metabolic diversity in the budding yeast subphylum. BMC Biol. 16(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osiro KO, Borgström C, Brink DP, Fjölnisdóttir BL, Gorwa-Grauslund MF. 2019. Exploring the xylose paradox in Saccharomyces cerevisiae through in vivo sugar signalomics of targeted deletants. Microb Cell Fact. 18(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc Lond B: Biol Sci. 255(1342):37–45. [Google Scholar]
- Paradis E, Schliep K. 2019. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35(3):526–528. [DOI] [PubMed] [Google Scholar]
- Peris D, Moriarty RV, Alexander WG, Baker E, Sylvester K, Sardi M, Langdon QK, Libkind D, Wang QM, Bai FY, et al. 2017. Hybridization and adaptive evolution of diverse Saccharomyces species for cellulosic biofuel production. Biotechnol Biofuels. 10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizeli M, Rizzatti ACS, Monti R, Terenzi HF, Jorge JA, Amorim DS. 2005. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 67(5):577–591. [DOI] [PubMed] [Google Scholar]
- Prat Y, Fromer M, Linial N, Linial M. 2009. Codon usage is associated with the evolutionary age of genes in metazoan genomes. BMC Evol Biol. 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 26(7):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3(2):217–223. [Google Scholar]
- Riley R, Haridas S, Wolfe KH, Lopes MR, Hittinger CT, Göker M, Salamov AA, Wisecaver JH, Long TM, Calvey CH. 2016. Comparative genomics of biotechnologically important yeasts. Proc Natl Acad Sci. 113(35):9882–9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi M, Harwart K, Bui-Thanh N-A, Dellweg H. 1989. A kinetic study of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J Ferment Bioeng. 67(1):25–30. [Google Scholar]
- Ruchala J, Sibirny AA. 2021. Pentose metabolism and conversion to biofuels and high-value chemicals in yeasts. FEMS Microbiol Rev. 45(4):fuaa069. [DOI] [PubMed] [Google Scholar]
- Ryu S, Hipp J, Trinh CT. 2016. Activating and elucidating metabolism of complex sugars in Yarrowia lipolytica. Appl Environ Microbiol. 82(4):1334–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Lee H, de FS Barbosa M, Kubicek CP, James AP. 1989. Physiological properties of a mutant of Pachysolen tannophilus deficient in NADPH-dependent D-xylose reductase. Appl Environ Microbiol. 55(11):2877–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li W-H. 1987. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15(3):1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, et al. 2018. Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175(6):1533–1545.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signori L, Passolunghi S, Ruohonen L, Porro D, Branduardi P. 2014. Effect of oxygenation and temperature on glucose-xylose fermentation in Kluyveromyces marxianus CBS712 strain. Microb Cell Fact. 13(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M, Pontes A, Franco-Duarte R, Soares P, Sampaio JP, Sousa MJ, Brito PH. 2023. A glimpse at an early stage of microbe domestication revealed in the variable genome of Torulaspora delbrueckii, an emergent industrial yeast. Mol Ecol. 32(10):2396–2412. [DOI] [PubMed] [Google Scholar]
- Sukpipat W, Komeda H, Prasertsan P, Asano Y. 2017. Purification and characterization of xylitol dehydrogenase with L-arabitol dehydrogenase activity from the newly isolated pentose-fermenting yeast Meyerozyma caribbica 5XY2. J Biosci Bioeng. 123(1):20–27. [DOI] [PubMed] [Google Scholar]
- Sun L, Jin YS. 2021. Xylose assimilation for the efficient production of biofuels and chemicals by engineered Saccharomyces cerevisiae. Biotechnol J. 16(4):2000142. [DOI] [PubMed] [Google Scholar]
- Toivari MH, Salusjärvi L, Ruohonen L, Penttilä M. 2004. Endogenous xylose pathway in Saccharomyces cerevisiae. Appl Environ Microbiol. 70(6):3681–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Träff KL, Jönsson LJ, Hahn-Hägerdal B. 2002. Putative xylose and arabinose reductases in Saccharomyces cerevisiae. Yeast 19(14):1233–1241. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina H, Schuster J, Blackwell M. 2013. The gut of Guatemalan passalid beetles: a habitat colonized by cellobiose-and xylose-fermenting yeasts. Fungal Ecol. 6(5):339–355. [Google Scholar]
- Veiga LA, Bacila M, Horecker BL. 1960. Pentose metabolism in Candida albicans. I. The reduction of D-xylose and L-arabinose. Biochem Biophys Res Commun. 2(6):440–444. [Google Scholar]
- Verduyn C, Van Kleef R, Frank J, Schreuder H, Van Dijken JP, Scheffers WA. 1985. Properties of the NAD (P) H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem J. 226(3):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger JW, Schwartz K, Sherlock G. 2010. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 6(5):e1000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wint R, Salamov A, Grigoriev IV. 2022. Kingdom-wide analysis of fungal transcriptomes and tRNAs reveals conserved patterns of adaptive evolution. Mol Biol Evol. 39(2):msab372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, LaButti KM, Sun H, Clum A, Pangilinan JL, Lindquist EA. 2011. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc Natl Acad Sci. 108(32):13212–13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Armisen D, Proux-Wera E, OhEigeartaigh SS, Azam H, Gordon JL, Byrne KP. 2015. Clade-and species-specific features of genome evolution in the Saccharomycetaceae. FEMS Yeast Res. 15(5):fov035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. 1990. The ‘effective number of codons’ used in a gene. Gene 87(1):23–29. [DOI] [PubMed] [Google Scholar]
- Zha J, Hu M, Shen M, Li B, Wang J, Yuan Y. 2012. Balance of XYL1 and XYL2 expression in different yeast chassis for improved xylose fermentation. Front Microbiol. 3:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Dang Y, Zhou M, Li L, Yu C, Fu J, Chen S, Liu Y. 2016. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc Natl Acad Sci. 113(41):E6117–E6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Analyses were performed on the 332 published and publicly available assemblies analyzed in Shen et al. (2018). Codon optimization values were obtained from the figshare repository from LaBella et al. (2019) (https://doi.org/10.6084/m9.figshare.c.4498292). All data generated in this project, including curated XYL gene sequences, are available in the figshare associated with this manuscript (https://doi.org/10.6084/m9.figshare.c.6011956.v1).
Conflict of interest statement. A.R. is a scientific consultant for LifeMine Therapeutics, Inc.