Abstract
DNA-dependent protein kinase (DNA-PK) is utilized in both DNA double-strand break repair (DSBR) and V(D)J recombination, but the mechanism by which this multiprotein complex participates in these processes is unknown. To evaluate the importance of DNA-PK-mediated protein phosphorylation in DSBR and V(D)J recombination, we assessed the effects of the phosphatidyl inositol 3-kinase inhibitor wortmannin on the repair of ionizing radiation-induced DNA double-strand breaks and V(D)J recombination in the V(D)J recombinase inducible B cell line HDR37. Wortmannin radiosensitized HDR37, but had no affect on V(D)J recombination despite a marked reduction in DNA-PK activity. On the other hand, studies with mammalian expression vectors for wild-type human DNA-PK catalytic subunit (DNA-PKcs) and a kinase domain mutant demonstrated that only the kinase active form of DNA-PKcs can reconstitute DSBR and V(D)J recombination in a DNA-PKcs-deficient cell line (Sf19), implying that DNA-PKcs kinase activity is essential for both DSBR and V(D)J recombination. These apparently contradictory results were reconciled by analyses of cell lines varying in their expression of recombinant wild-type human DNA-PKcs. These studies establish that minimal DNA-PKcs protein levels are sufficient to support V(D)J recombination, but insufficient to confer resistance to ionizing radiation.
INTRODUCTION
Double-strand breaks (DSBs) in DNA are induced by a number of DNA-damaging agents including ionizing radiation and radiomimetic drugs. If left unrepaired, they can lead to chromosome loss and cell death. The predominant pathway for the repair of DSBs in mammalian cells is non-homologous end joining (NHEJ). To date, three factors, DNA-dependent protein kinase (DNA-PK), XRCC4 and DNA ligase IV, have been shown to be involved (1).
DNA-PK is a multiprotein complex with serine/threonine protein kinase activity that must be physically associated with DNA to be active (reviewed in 2). The complex is comprised of two subunits: the ∼465 kDa catalytic subunit (DNA-PKcs), which possesses DNA binding and protein kinase activities (3–5), and the regulatory subunit Ku, which directs DNA-PKcs to DNA ends and stabilizes its binding so that it is efficiently activated (6,7). Ku binds to DNA ends in a sequence-independent manner and then translocates to internal sites (8). XRCC4 is a 37 kDa DNA-binding protein that has been shown to interact with and stimulate the catalytic activity of DNA ligase IV (9–11). An XRCC4–DNA ligase IV complex carries out the final step of joining DNA ends (12,13).
These three components of the NHEJ machinery have also been shown to be involved in the site-specific recombination process [V(D)J recombination] which assembles functional immune receptor genes from their disparate component gene segments during lymphocyte development in all jawed vertebrates (12,14–16). Recombination is initiated by a lymphocyte-specific endonuclease (the products of recombination activating genes I and II; 15,16), whereas components of the NHEJ pathway function in resolution of recombination intermediates.
Despite advances in the characterization of NHEJ and the establishment of a link between NHEJ and V(D)J recombination, the precise mechanisms by which repair proteins participate in these two processes remains unclear. Here we address the role of DNA-PKcs in NHEJ and V(D)J recombination and, in particular, investigate whether the kinase activity of DNA-PKcs is important for its function. DNA-PKcs is a member of the phosphatidyl inositol kinase (PIK)-related subfamily of phosphatidyl inositol 3-kinases (PI 3-kinases) (3,17–20). It has generally been presumed that DNA-PKcs will function as a protein kinase in these processes (21–23). In fact, DNA-PK has been shown to phosphorylate numerous proteins in vitro, including Ku, DNA-PKcs, XRCC4, p53, various transcription factors, histone proteins, topoisomerases I and II, the RNA polymerase II C-terminal domain and the 34 kDa subunit of DNA replication factor A (2,24–26).
On the other hand, the in vivo substrates of DNA-PK phosphorylation have yet to be identified (1). DNA-PKcs is an extremely large protein and others have suggested that its role in NHEJ and V(D)J recombination might be structural, aligning DNA termini to promote their ligation, or architectural, acting as a scaffold to which other proteins may be recruited (21,27). To investigate the importance of DNA-PK-mediated protein phosphorylation in NHEJ and V(D)J recombination, we have (i) explored the effect of the PI 3-kinase inhibitor wortmannin (28–30) on both of these processes, (ii) assessed the ability of a recombinantly expressed DNA-PKcs kinase domain mutant to reconstitute radioresistance and V(D)J recombination defects in a DNA-PKcs-deficient cell line and (iii) studied radioresistance and V(D)J recombination in cell lines varying in their expression of recombinant wild-type DNA-PKcs. Our results support the view that while DNA-PKcs kinase activity is essential for both NHEJ and V(D)J recombination, establishing normal radioresistance requires higher DNA-PKcs kinase activity levels.
MATERIALS AND METHODS
Cell lines and culture
The V(D)J recombinase inducible mouse B cell line HDR37, a generous gift of Dr Fred Alt (Harvard Medical School, Boston, MA), was propagated in a humidified 5% CO2 atmosphere in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, 57.6 µM 2-mercaptoethanol and 500 U/ml penicillin/streptomycin. HDR37 cells were routinely grown at 37°C and heat shocked at 43°C for 1.5 h to induce V(D)J recombination activity (31).
NS47, a wild-type mouse fibroblast cell line generously provided by Dr Kiyoshi Ariizumi (University of Texas Southwestern Medical Center, Dallas, TX), Sf19, a scid (severe combined immune deficiency) mouse fibroblast cell line generously provided by Dr Mel Bosma (Fox Chase Cancer Center, Philadelphia, PA), and 100E, a mouse scid fibroblast cell line containing a small fragment of human chromosome 8 (22) generously provided by Dr Cordula Kirchgessner (Stanford University School of Medicine, Stanford, CA), were propagated in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 500 U/ml penicillin/streptomycin. NS47, Sf19 and 100E were routinely grown at 37°C in a humidified 5% CO2 atmosphere.
Murine cell lines expressing wild-type and mutant human DNA-PKcs (hDNA-PKcs) were derived by stably transfecting the DNA-PKcs mutant cell line Sf19 with the wild-type (32) and mutant (described below) hDNA-PKcs expression constructs, respectively. FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals, Indianapolis, IN) was used to perform all transfections. Transfections were performed according to the manufacturer’s instructions in 60 mm tissue culture dishes utilizing 25 µg PvuI-linearized hDNA-PKcs expression plasmid (wild-type or mutant), 1 µg NotI-linearized pcDNAI/Neo plasmid (Invitrogen, Carlsbad, CA) to allow for drug resistance to neomycin and 35 µl FuGENE 6. Forty-eight hours after each transfection, cells were placed under selection conditions (200 µg/ml G418) and grown for 8 days, at which time cells were cloned by limiting dilution. Individual colonies were screened for DNA-PKcs expression by immunoblot analysis and further cultured. Cell lines 2-2, 2-7 and 2-9 are Sf19 cells transfected with the wild-type hDNA-PKcs expression vector. Cell line 18-6 is Sf19 transfected with the K3752M mutant hDNA-PKcs. Cell line 6-1 is Sf19 transfected with the pCMV6 eukaryotic expression vector. All of these Sf19-derived cell lines were propagated at 37°C in a humidified 5% CO2 atmosphere in the Dulbecco’s modified Eagle’s medium described above, with the addition of 200 µg/ml G418.
Plasmids
The V(D)J recombination substrates pJH201, pJH289 and pJH290 (33,34) and the RAG-1 and RAG-2 expression plasmids pJH548 and pJH549 (35) were a gift from Dr M. Gellert (National Institutes of Health, Bethesda, MD). The wild-type hDNA-PKcs expression vector was described earlier (32). The mutant DNA-PKcs construct was generated by overlap extension using PCR (36). Two PCR fragments were amplified and subcloned using the following pairs of primers (coding mutation underlined): K>M top (5′-CACCCTTTCCTGGTCATGGGAGGCGAGGACCTG-3′) and 3′ BbvC1 (5′-CTTTATATTCACACGGCGGTGC-3′); K>M bottom (5′-CAGGTCCTCGCCTCCCATGACCAGGAAAGGGTG-3′) and 5′ Eco721 (5′-TATGACGGTAGGGGAAAGCCATTG-3′). The resulting two fragments were ‘Soed’ together generating a fragment in which Lys3752 was mutated to Met. This fragment was subcloned into a fragment of human DNA-PKcs spanning nt 7911–12358 of the coding sequence via unique Eco721 and BbvC1 restriction endonuclease sites in the DNA-PKcs cDNA. At this point, the sequence of each fragment was confirmed using the dideoxy termination method (37) with the ABI PRISM Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer, Foster City, CA). Sequences were primed with the 5′ Eco721 and 3′ BbvC1 oligonucleotides. The resulting fragment spanning nt 7911–12358 with a point mutation at codon 3752 was subcloned into the previously described expression vector encoding wild type human DNA-PKcs via the unique Eco721 site and a unique XmaI site in the cloning cassette.
Assessment of irradiation sensitivity
For experiments with HDR37 cells, 800 HDR37 cells, suspended in 2 ml of culture medium, were treated with 20 µM wortmannin or 0.2% dimethyl sulfoxide (DMSO, the solvent) for 30 min at 37°C and then exposed to either 2 or 4 Gy ionizing radiation, using a 137Ce source calibrated at 761.7 R/min, or left unirradiated (0 Gy value). Afterwards, cell suspensions were placed in 150 mm2 tissue culture dishes containing 18 ml of culture medium with 20 µM wortmannin or 0.2% DMSO and incubated at 37°C for 6–8 days. During the incubation, the wortmannin was neither removed nor replaced. Following the incubation, cell colonies were fixed with 2% formaldehyde followed by 100% methanol, stained with trypan blue and counted. Percent survival was calculated by dividing the clonogenic survival at a given dose of irradiation by the clonogenic survival of unirradiated cells. Within an experiment irradiation sensitivities were determined in triplicate and all experiments were repeated at least three times.
Experiments with Sf19 transfectants were performed similarly except that 1000 cells were used for each radiation dose, no wortmannin or DMSO pretreatment occurred and the radiation doses were 0.5, 1, 2 and 4 Gy.
V(D)J recombination assay
Extrachromosomal recombination assays were performed essentially as described by Lieber et al. (38). Briefly, to assess V(D)J recombination in HDR37 cells, 20 × 106 cells, grown at 37°C, were transfected with 20 µg pJH289 or pJH290 by electroporation using an Electro cell manipulator 600 (BTX, San Diego, CA) set at 950 FF and 280–290 V. Transfected HDR37 cells were cultured for 17–19 h at 37°C, pretreated with wortmannin (20 or 50 µM) or the solvent DMSO (0.2%) for 30 min, heat shocked at 43°C for 1.5 h and then returned to 37°C for 7 h. At 1 and 4 h after heat shock, the culture medium was changed by precipitation of the cells and resuspension in fresh wortmannin- or DMSO-containing medium, since wortmannin has a half-life of <5 h in tissue culture medium (39,40). To facilitate maintenance of a constant wortmannin concentration, the recombination assay was shortened from that described (48 h; 31) resulting in somewhat lower recombination rates than reported previously for the HDR37 cell line. After 7 h at 37°C, plasmid DNA was recovered from the cells by alkaline lysis, treated with DpnI and a portion of the recovered plasmid was used to transform competent Escherichia coli (Library or Max efficiency DH5″‘; Life Technologies, Gaithersburg, MD) according to the manufacturer’s instructions. Approximately 2% of each transformation mix was spread onto each of two LB agar plates containing 100 µg/ml ampicillin (Amp); the remainder was spread onto three LB agar plates containing 100 µg/ml Amp and 55 µg/ml chloramphenicol (Cam). Plates were incubated for 14–48 h at 37°C and then the number of colonies growing on each type of plate was determined.
To assess V(D)J recombination in Sf19 cells, RAG-1 and RAG-2 expression constructs (6 µg each), wild-type or mutant hDNA-PKcs expression plasmid or the pCMV6 vector control (6 µg) and recombination substrate (1 µg pJH201 or pJH290) were transiently introduced into Sf19 cells by liposome transfection using FuGENE 6 Transfection Reagent according to the manufacturer’s instructions. Forty-eight hours later plasmid substrates were rescued from the cells by alkaline lysis and processed as above. However, transformed bacteria were spread onto two LB agar plates containing 100 µg/ml Amp and two LB agar plates containing 100 µg/ml Amp and 22 µg/ml Cam.
Analysis of recombinants
For all V(D)J recombination assays, at least 20% of the putative recombinants were examined for authenticity by restriction enzyme digestion or hybridization with an oligonucleotide from the oop terminator (5′-GACGACGACATGGCTCGATTG-3′); authentic recombinants fail to hybridize to the oop oligonucleotide.
Sequence analysis of recombinant plasmids was performed using the dideoxy termination method as above. Two oligonucleotides, CAT (5′-CCTCAAAATGTTCTTTA-3′) and REV (5′-CAGGAAACAGCTATGAC-3′), were used as sequencing primers.
The fidelity of pJH289 signal joints was examined by ApaLI digestion of recombinant plasmids and electrophoresis on 0.8% agarose gels. A novel ApaLI site is created by a perfectly head-to-head fused pJH289 signal joint.
Protein extract preparation and western blotting
Cells were harvested, washed and resuspended in 50 mM Tris pH 7.4, 1% NP40, 0.25% deoxycholate, 150 mM NaCl and 1 mM EGTA with the addition of a cocktail of protease inhibitors (Complete, EDTA-free; Roche Molecular Biochemicals, Indianapolis, IN). The cell suspension was frozen in liquid nitrogen and thawed at 37°C three times. The extract was microcentrifuged at 15 000 r.p.m. for 7 min at 4°C; concentrations of the resulting whole cell extract were determined by Bradford analysis using bovine serum albumin (BSA) as the standard. Indicated amounts of extracts were electrophoresed in SDS–5% polyacrylamide gels and transferred to poly(vinylidene difluoride) membranes. The mouse monoclonal anti-DNA-PKcs antibody 42psc, which recognizes the C-terminal 150 kDa of DNA-PKcs (a gift of Dr T. Carter, St John’s University, Jamaica, NY; 22) was used as the primary antibody (1:300) and a goat anti-mouse IgG conjugated to horseradish peroxidase as the secondary antibody. Membranes were then incubated with a chemiluminescent substrate (ECL; Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer’s recommendations.
DNA-PK microfractionation and measurement of kinase activity
Whole cell extracts were prepared by a modification of the method of Finnie et al. (41). Briefly, 20 × 106 cells were harvested, washed three times in phosphate-buffered saline and cell pellets were frozen at –80°C. Frozen cell pellets were resuspended in 20 µl of extraction buffer (50 mM NaF, 20 mM HEPES pH 7.8, 450 mM NaCl, 25% v/v glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 0.5 µg/ml leupeptin, 0.5 µg/ml protease inhibitor and 1.0 µg/ml trypsin inhibitor), subjected to three freeze/thaw cycles (liquid nitrogen/37°C), and centrifuged at 8160 g for 7 min at 4°C. Supernatants were stored at –80°C prior to use and concentrations were determined by Bradford analysis using BSA as the standard.
The SignaTECT DNA-PK assay system (Promega Corp., Madison, WI) was used to assay DNA-PK activity with the following modifications. Ten microliters of extract (280 µg) was diluted 1:3 in buffer A (25 mM HEPES pH 7.9, 50 mM KCl, 10 mM MgCl2, 10% v/v glycerol, 1 mM EDTA, 1 mM EGTA and 1 mM DTT) and incubated with 20 µl of pre-swollen double-stranded DNA–cellulose beads (Amersham Pharmacia Biotech) for 30 min at 4°C. The double-stranded DNA–cellulose was then washed three times with 1 ml of buffer A each time, before it was resuspended in 20 µl of DNA-PK reaction buffer containing 100 µg/ml BSA. Kinase reactions were conducted with 10 µl aliquots of the resuspended DNA-PK absorbed on cellulose beads and were performed in both the presence and absence of a biotinylated DNA-PK p53-derived substrate peptide; no activator was used. Terminated reactions were analyzed by spotting onto SAM2‘ membrane, washing and counting in a scintillation counter as per the manufacturer’s instructions. All assays were performed in duplicate with at least two different extract preparations.
RESULTS
Wortmannin radiosensitizes, but does not inhibit, V(D)J recombination in a recombinase inducible cell line
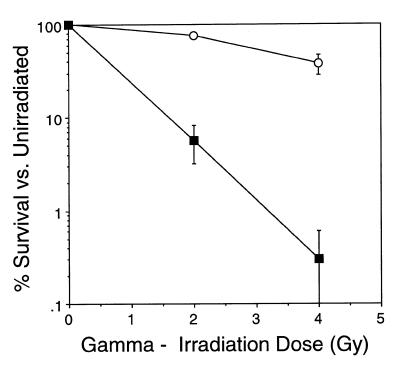
To assess the possible role of a specific phosphorylation event catalyzed by DNA-PK during either the repair of DSBs or during V(D)J recombination, we utilized the fungal metabolite wortmannin, which inhibits PI 3-kinase at nanomolar concentrations (28–30) and DNA-PKcs at higher concentrations (3). Wortmannin covalently and irreversibly binds a lysine residue within the kinase domain of PI 3-kinase that is crucial to the phosphate transfer reaction and is shared by all PI 3-kinase family members (42). Initially, we determined whether wortmannin sensitizes the murine V(D)J recombinase inducible B cell line HDR37 to ionizing radiation (Fig. 1). Earlier studies have shown that wortmannin acts as a radiosensitizer in cultured cell lines (39,43,44); consistent with these studies, exposure of HDR37 cells to 20 µM wortmannin for 30 min decreased the surviving fraction of cells at both 2 and 4 Gy.
Figure 1.
Wortmannin sensitizes HDR37 cells to radiation. HDR37 cells were exposed to 20 µM wortmannin or 0.2% DMSO (the solvent) for 30 min, irradiated at the indicated dose and assessed for colony formation as described in Materials and Methods, 6–8 days later. Open circles, 0.2% DMSO; filled squares, 20 µM wortmannin. Survival curves are the mean of three independent experiments and bars indicate the SD.
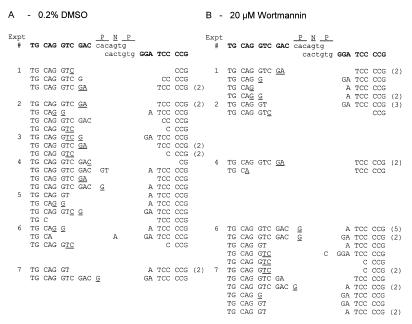
To assess the function of wortmannin-sensitive kinases during V(D)J recombination, HDR37 cells were treated with 20 (Table 1) or 50 µM (data not shown) wortmannin and assayed for ability to rearrange the V(D)J recombination substrates, pJH289 and pJH290, which allow for evaluation of signal and coding joint formation, respectively (33,34). In these experiments, constant wortmannin concentrations were maintained throughout the culture period. No defects in the efficiency of either signal or coding end joining were observed in wortmannin-treated HDR37 cells. Furthermore, signal (Table 1) and coding (Fig. 2) junctions recovered from wortmannin-treated HDR37 cells were indistinguishable from those recovered from DMSO-treated HDR37 cells. Signal joints were precise, as indicated by digestion with ApaLI enzyme (Table 1) and coding junctions showed characteristics of normal joints, including small nucleotide deletions from one or both coding ends (<10 bp) and P elements (Fig. 2).
Table 1. Analysis of signal and coding joint formation in 20 µM wortmannin-treated HDR37 cells by transient V(D)J recombination assay.
| Treatment | Experiment | Signal (pJH289) |
|
Fidelityb | Coding (pJH290) |
|
|---|---|---|---|---|---|---|
| AmpR + CamR/AmpR | Ratea (%) | AmpR + CamR/AmpR | Ratea (%) | |||
| 0.2% DMSO | 1 | 87/13 356 | 0.651 | 100.0 (10/10) | 66/14 196 | 0.465 |
| 2 | 65/8932 | 0.728 | 100.0 (6/6) | 34/11 676 | 0.291 | |
| 3 | 34/8316 | 0.409 | 100.0 (6/6) | 52/13 664 | 0.381 | |
| 4 | 17/17 603 | 0.097 | 100.0 (6/6) | 23/24 752 | 0.093 | |
| 5 | 47/26 824 | 0.175 | 83.3 (5/6) | 20/28 056 | 0.071 | |
| 6 | 126/23 946 | 0.526 | 100.0 (12/12) | 100/27 762 | 0.360 | |
| 7 | 284/80 310 | 0.354 | 100.0 (10/10) | 98/36 221 | 0.271 | |
| 0.420 ± 0.234c | 97.6 ± 6.3d | 0.276 ± 0.147c | ||||
| 20 µM Wortmannin | 1 | 24/6188 | 0.388 | 90.5 (19/21) | 50/9201 | 0.543 |
| 2 | 16/4452 | 0.359 | 100.0 (10/10) | 5/3164 | 0.158 | |
| 3 | 10/4480 | 0.223 | 88.9 (8/9) | ND | ND | |
| 4 | 6/9781 | 0.061 | 100.0 (5/5) | 12/12 003 | 0.100 | |
| 5 | 7/9053 | 0.077 | 100.0 (6/6) | 2/11 891 | 0.017 | |
| 6 | 129/31 807 | 0.406 | 94.4 (17/18) | 63/17 983 | 0.350 | |
| 7 | 82/29 121 | 0.282 | 81.8 (9/11) | 42/18 874 | 0.223 | |
| 0.257 ± 0.143c | 93.7 ± 7.0d | 0.232 ± 0.190c |
aRate is the percentage of replicated substrates which underwent V(D)J recombination and is derived from the formula [AmpR + CamR/AmpR] × 100.
bFidelity is expressed as the percentage of recovered signal joints that were susceptible to ApaLI digestion.
cAverage recombination rate ± SD.
dAverage fidelity ± SD.
Figure 2.
Nucleotide sequences of coding joints formed in DMSO- or 20 µM wortmannin-treated HDR37 cells. (A) Junctions formed in the presence of 0.2% DMSO. (B) Junctions formed in the presence of 20 µM wortmannin. The sequences of coding ends and heptamers, as they are in the pJH290 substrate, are shown above the sequences of the recombinant junctions. Junctional sequences are grouped according to transfection and the experiment numbers are consistent with the experiment numbers in Table 1. Bracketed numbers to the right of sequences indicate the number of observations of that sequence. N and P denote nucleotides added to each junction. N nucleotides are random bases, while P nucleotides form a palindromic sequence with the neighboring coding element. Nucleotides that cannot unequivocally be assigned to a particular coding end, or classified as a P nucleotide, are underlined and listed in the more 5′ location.
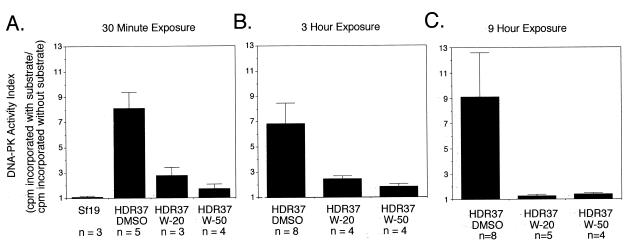
To confirm that DNA-PK activity was inhibited under these conditions, HDR37 cells were exposed to wortmannin for 0.5, 3 and 9 h, washed extensively and cell extracts were assayed for DNA-PK activity (Fig. 3). Aliquots of 20 and 50 µM wortmannin markedly inhibited DNA-PK in HDR37 cells after only 30 min exposure. However, detectable levels of DNA-PK activity remained in HDR37 cells following exposure to either 20 or 50 µM wortmannin for 9 h. This low level of DNA-PK activity may be sufficient to support V(D)J recombination, although it is insufficient to confer resistance to ionizing radiation.
Figure 3.
Inhibition of DNA-PK by wortmannin. HDR37 cells were pretreated for 30 min with 20 (W-20) or 50 µM wortmannin (W-50) or 0.2% DMSO, heat shocked for 1.5 h and then cultured for another 7 h. At 1 and 4 h after heat shock treatment, fresh medium containing wortmannin or DMSO was supplied. Whole cell extracts were prepared after the first 30 min exposure to wortmannin or DMSO (left) and 1 (middle) and 7 (right) h after heat shock (3 and 9 h exposure to wortmannin or DMSO, respectively). Whole cell extracts were also prepared from the DNA-PKcs mutant cell line Sf19, as a control. DNA-PK activity in 280 µg whole cell extract was assayed as described in Materials and Methods. Extracts were tested in duplicate and the number of independent extracts examined for any condition (n) is listed below a column. Bars indicate the SD.
Only a kinase active form of DNA-PKcs reconstitutes NHEJ and V(D)J recombination in murine scid cells
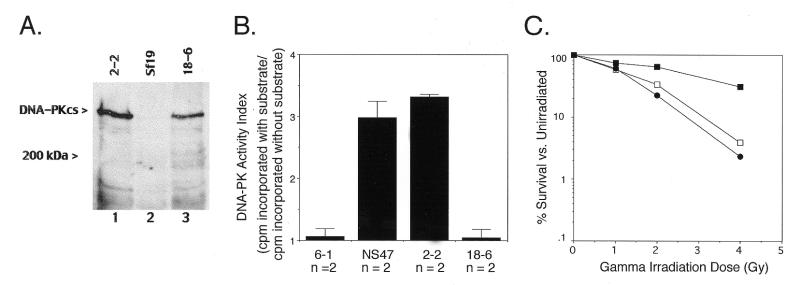
To more directly address the role of DNA-PKcs kinase activity in NHEJ and V(D)J recombination, we generated mammalian expression vectors for wild-type human DNA-PKcs (32) and a kinase domain mutant and tested their ability to reconstitute NHEJ and V(D)J recombination defects in a DNA-PKcs-deficient cell line. The hDNA-PKcs kinase domain mutant had Lys3752 changed to Met (K3752M). This residue is conserved within the kinase catalytic domains of all PI 3-kinases and protein kinases (42,45,46) and studies have shown that it is important for ATP phosphate donor binding and orientation (46) and interacts with wortmannin (42). Methionine was chosen as the replacement residue since it is similar in both size and flexibility to lysine. Expression vectors were stably transfected into the DNA-PKcs-deficient cell line Sf19, a murine scid fibroblast cell line, and individual neomycin-resistant clones were isolated from the bulk transfected cultures [cell lines 2-2 (wild-type hDNA-PKcs) and 18-6 (hDNA-PKcsK3752M mutant)]. Expression of recombinant protein was confirmed by western blot analysis with the mouse monoclonal anti-DNA-PKcs antibody 42psc (22; Fig. 4A). Levels of recombinant wild-type and mutant DNA-PKcs observed in 2-2 and 18-6 transfectants, respectively, were comparable; no DNA-PKcs was detected in the DNA-PKcs-deficient Sf19 cell line.
Figure 4.
(A) Expression of recombinant wild-type and PI 3-kinase domain mutant hDNA-PKcs in the scid mouse fibroblast cell line Sf19. Two hundred micrograms each of protein extract from untransfected Sf19 cells (lane 2) or Sf19 cells stably transfected with a wild-type (lane 1, cell line 2-2) or the K3752M mutant hDNA-PKcs cDNA expression vector (lane 3, cell line 18-6) were separated on a 5% SDS–polyacrylamide gel under reducing conditions and immunoblotted with 42psc (22). The molecular weight standard position, in kDa, is indicated on the left, as is the ∼465 kDa recombinant hDNA-PKcs protein. (B) Sf19 cells expressing recombinant PI 3-kinase domain mutant hDNA-PKcs lack DNA-PK activity. Whole cell extracts (280 µg) derived from normal mouse fibroblasts (NS47) and the Sf19 transfectants 2-2, 18-6 and 6-1 (pCMV6 eukaryotic expression vector alone transfection control) were assayed for DNA-PK activity as described in Materials and Methods. Cell extracts were tested in duplicate and the number of independent extracts analyzed (n) is listed below a column. Bars indicate the SD. (C) Radiation sensitivity of wild-type and mutant hDNA-PKcs-expressing Sf19 cell lines. Sf19 cells stably transfected with the wild-type (2-2, closed squares) or K3752M mutant hDNA-PKcs cDNA expression vector (18-6, open squares) or the pCMV6 vector alone (6-1, closed circles) were irradiated at the indicated dose and assessed for colony formation as described in Materials and Methods, 8 days later. Survival curves are the average of triplicates from one experiment.
Confirmation that the K3752M replacement mutation in hDNA-PKcs abolished the protein serine/threonine kinase activity of the enzyme was obtained by assaying the 18-6 and 2-2 whole cell extracts utilized in the western analysis for DNA-PK activity (Fig. 4B). Extracts of 2-2 cells contained DNA-PK activity at a level comparable to that found in the wild-type mouse fibroblast cell line NS47. In contrast, Sf19 and 18-6 cell extracts lacked DNA-PK activity completely.
Cell lines derived from mice homozygous for the scid mutation exhibit a profound hypersensitivity to agents that cause DNA DSBs, including ionizing radiation (47–49). Evaluation of the γ-ray sensitivity of transfected cell lines 2-2 and 18-6 showed that only the kinase active form of hDNA-PKcs restored radioresistance to Sf19 cells (Fig. 4C). Transfectant 2-2 was significantly less radiosensitive than Sf19 cells transfected with the eukaryotic expression vector pCMV6 (cell line 6-1). In contrast, 18-6 cells showed radiation sensitivity levels similar to cell line 6-1. Thus, the kinase activity of DNA-PKcs is required in the end joining reactions following ionizing radiation.
DNA-PKcs-deficient scid-derived cell lines also show an impairment in V(D)J recombination; V(D)J coding end joining is severely reduced while signal joining formation is relatively normal. To test whether catalytically inactive hDNA-PKcs can overcome this V(D)J coding end resolution defect in Sf19 cells, we assayed V(D)J recombination in Sf19 cells transiently co-transfected with hDNA-PKcs expression plasmids (wild-type or mutant), extrachromosomal V(D)J recombination substrates [coding (pJH290) or signal joint (pJH201); 33] and RAG-1 and RAG-2 expression vectors (35), which provide the specific V(D)J recombination functions to the non-lymphoid cells. Two or three independent transfection experiments were carried out (Table 2). As expected, coding joint formation was undetectable in the control transfection of Sf19 cells with the pCMV6 expression vector and RAG expression constructs. Furthermore, as we have shown previously (32), introduction of wild-type hDNA-PKcs into Sf19 cells generated coding joint formation in the reporter plasmid pJH290 (32). However, introduction of the K3752M kinase domain mutant hDNA-PKcs into Sf19 cells did not yield detectable coding joint formation with pJH290. Rates of signal joint formation varied little between Sf19 cells and Sf19 cells expressing wild-type or kinase domain mutant hDNA-PKcs. Significant levels of signal joint formation were detected in Sf19 cells with the reporter plasmid pJH201 and the introduction of wild-type or mutant hDNA-PKcs had no effect on this rate. Thus, these V(D)J recombination studies demonstrate that the kinase activity of DNA-PKcs is essential for V(D)J coding joint formation.
Table 2. Analysis of signal and coding joint formation in murine scid fibroblasts by transient V(D)J recombination assay.
| Expression vectors | Experiment | Signal (pJH201) | Coding (pJH290) | ||
|---|---|---|---|---|---|
| AmpR + CamR/AmpR | Ratea (%) | AmpR + CamR/AmpR | Ratea (%) | ||
| PCMV6 | 1 | 8/10 368 | 0.078 | 0/21 120 | <0.005 |
| RAG1 + RAG2 | 2 | 15/12 960 | 0.116 | 0/8928 | <0.011 |
| 3 | ND | ND | 0/20 736 | <0.005 | |
| Wild-type DNA-PKcs | 1 | 29/16 704 | 0.174 | 76/69 888 | 0.109 |
| RAG1 + RAG2 | 2 | 17/26 784 | 0.078 | 8/24 192 | 0.033 |
| 3 | ND | ND | 8/27 456 | 0.020 | |
| K3752M DNA-PKcs | 1 | 17/25 248 | 0.032 | 0/34 464 | <0.003 |
| RAG1 + RAG2 | 2 | 25/27 456 | 0.091 | 0/7872 | <0.013 |
| 3 | ND | ND | 0/60 768 | <0.002 |
aRate is the percentage of replicated substrate which underwent V(D)J recombination and is derived from the formula [AmpR + CamR/AmpR] × 100.
Minimal DNA-PKcs is required to reconstitute coding end resolution in murine scid cells
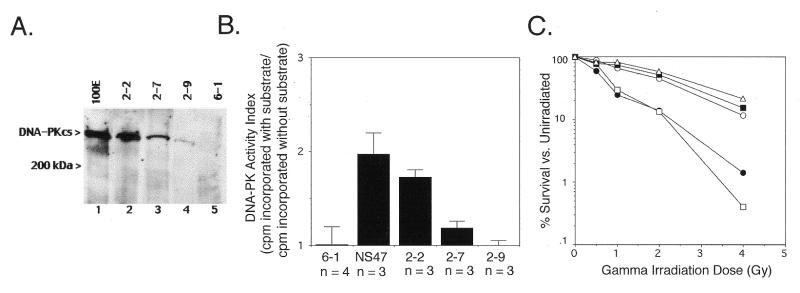
While DNA-PKcs kinase activity is required for both radioresistance and V(D)J coding joint formation, the wortmannin studies above suggest that V(D)J recombination requires only very low levels of DNA-PKcs kinase activity. To examine this issue further, during the generation of the wild-type hDNA-PKcs-transfected Sf19 line 2-2 we selected two other clonal Sf19 transfectants that express hDNA-PKcs at lower levels (cell lines 2-7 and 2-9) and analyzed their ability to repair ionizing radiation-induced DSBs and rearrange transiently transfected V(D)J recombination constructs. Figure 5A shows the expression of hDNA-PKcs within the three Sf19 transfectants. Western blot analysis with monoclonal anti-DNA-PKcs antibody 42psc (22) was performed. Cell line 2-2 expresses the highest level of wild-type hDNA-PKcs and this level is somewhat less than the hDNA-PKcs detected in a mouse scid fibroblast cell line containing a fragment of human chromosome 8 encoding DNA-PKcs, 100E. Cell line 2-7 expresses ∼5-fold lower hDNA-PKcs than cell line 2-2 and cell line 2-9 expresses minimally detectable DNA-PKcs (∼10-fold lower than cell line 2-2).
Figure 5.
(A) Expression analysis of recombinant wild-type hDNA-PKcs in scid mouse fibroblast Sf19 stable transfectants. Two hundred micrograms each of protein extract from the hDNA-PKcs transfectants 2-2, 2-7 and 2-9 (lanes 2–4), the vector only transfectant 6-1 (lane 5) and the cell line 100E, a murine scid fibroblast cell line harboring a fragment of human chromosome 8 encoding DNA-PKcs (lane 1), were separated on a 5% SDS–polyacrylamide gel under reducing conditions and immunoblotted with 42psc (22). The molecular weight standard position, in kDa, is indicated on the left, as is the ∼465 kDa recombinant hDNA-PKcs protein. (B) DNA-PK activity of wild-type hDNA-PKcs-expressing Sf19 cell lines. Whole cell extracts (280 µg) derived from normal mouse fibroblasts (NS47), the hDNA-PKcs transfectants 2-2, 2-7 and 2-9 and the vector-only transfectant 6-1 were assayed for DNA-PK activity as described in Materials and Methods. Cell extracts were tested in duplicate and the number of independent extracts analyzed (n) is listed below a column. Bars indicate the SD. (C) Radiation sensitivity of wild-type hDNA-PKcs transfectants. Normal mouse fibroblasts (NS47, open triangles) and Sf19 cell lines stably transfected with a wild-type hDNA-PKcs cDNA expression vector (2-2, filled squares; 2-7, open circles; 2-9, open squares) or the pCMV6 vector alone (6-1, filled circles) were irradiated at the indicated dose and assessed for colony formation as indicated in Materials and Methods, 8 days later. Survival curves are the average of triplicates from one experiment.
Measurement of the DNA-PK activity within whole cell extracts of these three transfected cell lines confirmed that DNA-PK kinase activity correlates with expression of DNA-PKcs. Extracts of 2-2 cells contained DNA-PK activity at a level comparable to that found in normal mouse fibroblasts. Extracts of 2-7 cells contained ∼1.5-fold lower DNA-PK activity than 2-2 extracts and DNA-PK activity was undetectable in 2-9 cell extracts.
Cell survival assays following acute dose irradiation showed that the level of DNA-PKcs expression in Sf19 transfectants 2-2 and 2-7 is sufficient to repair DSBs induced by these dosages of ionizing radiation, but the level of DNA-PKcs expressed in 2-9 is not (Fig. 5C). Transfected cell lines 2-2 and 2-7 showed radiation sensitivity levels similar to wild-type mouse fibroblasts, whereas 2-9 showed radiation sensitivity levels similar to Sf19 transfected with the eukaryotic expression vector pCMV6 (cell line 6-1).
V(D)J coding end resolution in Sf19 transfectants was analyzed following transient co-transfection of the extrachromosomal V(D)J recombination substrate pJH290 and the RAG-1 and RAG-2 expression vectors. Six independent transfection experiments were carried out (Table 3). As with the parental Sf19 cell line, the vector-only transfectant 6-1 did not support coding end resolution. However, transfectant 2-2, which expresses hDNA-PKcs at levels similar to DNA-PKcs in normal mouse fibroblasts, supported substantial coding end resolution. In transfectant 2-9, which expresses minimal DNA-PKcs, the rate of coding joint formation was ∼5-fold less than transfectant 2-2, but 30-fold higher than the vector-only transfectant, suggesting that coding joint formation is partially restored in transfectant 2-9. Sequence analysis of representative recombinants isolated from each of the transfectants revealed that the fidelity of coding resolutions is normal in transfectants 2-2 and 2-9 (Fig. 6). No large deletions or long P segments were observed in coding joints isolated from transfectants 2-2 or 2-9. In contrast, the few rearrangements isolated from the vector-only transfectant 6-1 exhibited large deletions, a characteristic of coding joints isolated from scid mice (50,51). Thus, we conclude that minimal DNA-PK activity can substantially restore the defect in coding end resolution in murine scid cells.
Table 3. Analysis of coding joint formation in stable scid transfectants by transient V(D)J recombination assay.
| Sf19-derived cell line | Expression vector stably introduced | Experiment | Coding (pJH290) |
|
|---|---|---|---|---|
| (AmpR + CamR/AmpR) | Ratea (%) | |||
| 6-1 | pCMV6 alone | 1 | 0/30 528 | <0.003 |
| 2 | 0/66 336 | <0.002 | ||
| 3 | 1/36 672 | 0.003 | ||
| 4 | 0/19 453 | <0.005 | ||
| 5 | 3/68 600 | 0.004 | ||
| 6 | 0/48 216 | <0.002 | ||
| 2-2 | Wild-type DNA-PKcs | 1 | 97/42 720 | 0.227 |
| 2 | 136/83 520 | 0.163 | ||
| 3 | 40/63 072 | 0.063 | ||
| 4 | 51/50 372 | 0.101 | ||
| 5 | 39/18 816 | 0.207 | ||
| 6 | 53/23 422 | 0.226 | ||
| 2-9 | Wild-type DNA-PKcs | 1 | 20/46 944 | 0.043 |
| 2 | 16/51 072 | 0.025 | ||
| 3 | 23/41 520 | 0.058 | ||
| 4 | 16/72 128 | 0.022 | ||
| 5 | 8/30 331 | 0.026 | ||
| 6 | 4/16 360 | 0.031 |
aRate is the percentage of replicated substrate which underwent V(D)J recombination and is derived from the formula [AmpR + CamR/AmpR] × 100.
Figure 6.
Nucleotide sequences of coding joints formed in Sf19 transfectants expressing varying levels of recombinant wild-type hDNA-PKcs. Sequences are displayed as indicated in Figure 2.
DISCUSSION
Two potential roles have been proposed for DNA-PKcs in DNA DSBR and V(D)J recombination. One is a structural or scaffolding role, due to the large size of the molecule, and the other is for DNA-PKcs to act as a protein kinase. This study has examined only the importance of the kinase activity of DNA-PKcs in the repair of ionizing radiation-induced DNA DSBs and V(D)J recombination. Our results with kinase-inactive hDNA-PKcs transfectants confirm that protein phosphorylation mediated by DNA-PK is critical for both of these processes. Two other groups have also recently provided evidence that the kinase activity of hDNA-PKcs is essential for the repair of DNA DSBs in mammalian cells (52,53). Baumann and West demonstrated that wortmannin inhibits an in vitro end joining system that involves DNA-PK, XRCC4 and DNA ligase IV (52). Then, using an expression approach similar to ours, Kurimasa et al. showed that different mutations within the kinase domain of hDNA-PKcs abolish the ability of the protein to complement the DSB and V(D)J recombination defects in the DNA-PKcs mutant cell line V3 (53).
The function of the protein kinase activity of DNA-PKcs in DSB rejoining and V(D)J recombination is currently unclear. The in vivo substrates of DNA-PK phosphorylation have not been identified, but it is known that DNA-PK phosphorylates proteins bound to the same DNA molecule most effectively (6). DNA-PK kinase activity could regulate the activities of components of the DNA repair and recombination apparatus (21,54). Alternatively, a signal transduction pathway could be activated to alert the cell to DNA damage (21,54,55). Finally, components of the transcription apparatus might be phosphorylated and inactivated by DNA-PK to repress transcription in the vicinity of a DSB (21,54).
A new finding in this study is that radioresistance requires higher DNA-PKcs protein levels than does V(D)J recombination. An Sf19 transfectant expressing minimal levels of hDNA-PKcs was found to support substantial V(D)J coding joint formation (30-fold higher than the vector-only transfectant), but lacked any complementation for the radiation sensitivity of Sf19 cells, whereas two other Sf19 transfectants expressing higher levels of DNA-PKcs supported higher frequencies of V(D)J coding joint formation and showed significant correction of the γ-ray sensitivity characteristic of Sf19 cells using the standard high dose irradiation procedure. Furthermore, the minimally detectable DNA-PK activity which remains in wortmannin-treated HDR37 cells is also sufficient to support wild-type levels of V(D)J recombination as tested in a standard transient assay. In sum, these results are consistent with the hypothesis that DNA-PKcs expression correlates with the number of DNA breaks which can be resolved in a given cell line and that the number of DNA breaks introduced during the V(D)J recombination assay are lower than the number introduced by the ionizing radiation doses we used.
The number of DSBs introduced by ionizing radiation has been estimated to be 10–40 DSBs/Gy/cell (reviewed in 56). Though the number of DNA breaks introduced during the transient V(D)J recombination assay has not been established, the minimal number of breaks can be estimated based on recombination frequency and transfection efficiency. In these experiments, we routinely isolated 100–500 recombinants in transfections of 20 000 000 cells. After compensating for transfection efficiency, this translates into 1 recombination event in every 100–1000 cells transfected. Thus, in these experiments, it is likely that the number of DSBs/cell introduced during the recombination assay is significantly lower than the number of DSBs/cell introduced with the radiation doses used here. Perhaps a more relevant question is whether extremely low levels of DNA-PK activity could suffice to support V(D)J recombination in a living organism. In developing lymphocytes, V(D)J recombination is temporally controlled such that rearrangement occurs at only a single receptor locus (and probably at only one allele) at once. This predicts the presence of only 2 DSBs/cell (or 4 DSBs, if both alleles rearrange concurrently). Thus, our data predict that low levels of DNA-PK might suffice for V(D)J recombination in vivo. In fact, Riballo et al. have recently described another example of a defect in NHEJ which results in marked radiosensitivity but normal V(D)J recombination (57). In this instance, a mutation in the active site of DNA ligase IV was discovered in a cell line derived from a leukemia patient who was highly sensitive to radiotherapy. Though the mutant DNA ligase IV was severely deficient in its capacity to generate a stable enzyme–adenylate intermediate, explaining the marked radiosensitivity, the residual activity of the mutant protein was sufficient to support normal rates of V(D)J recombination (58). Thus, this previous report and the data we present here are consistent with the following hypothesis. Minimal levels of the factors involved in NHEJ are sufficient to repair the relatively few DSBs introduced during V(D)J recombination, but normal levels are required to repair the abundant DNA strand breaks generated by γ-ray irradiation. There are also qualitative differences in the DSBs introduced by ionizing radiation and during V(D)J recombination; it is also possible that qualitative differences in the DSBs generated contribute to the more stringent NHEJ requirements necessary to maintain normal radioresistance.
Earlier studies using YAC fusion hybrids or human chromosome 8–mouse hybrids also showed only a partial complementation of the radiosensitivity in DNA-PKcs-deficient rodent cells, but full complementation of the V(D)J recombination defect (21,59–62). DNA-PKcs was therefore assumed to function not simply as a kinase, but also to mediate interaction(s) with other proteins; the partial complementation was presumed to be because hDNA-PKcs could not function as well as rodent DNA-PKcs in this role. Notably, in these YAC fusion hybrids, both DNA-PKcs protein levels and DNA-PK activity were elevated above the levels expressed in normal rodent cells. However, two recent studies using hDNA-PKcs expression vectors demonstrate that hDNA-PKcs can fully complement the radiosensitivity of both V3 and murine scid cells (32,53). Thus, the partial complementation of the radiosensitivity in DNA-PKcs-deficient rodent cells by YACs cannot be explained by species differences in the DNA-PKcs protein itself. One potential explanation for the difference in complementation with hDNA-PKcs expressed from plasmid vectors versus YACs is that the higher levels of DNA-PKcs expressed in the YAC fusion hybrids may be in some way deleterious to radioresistance in rodent cells.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Public Health Service grants AI32600 and AI32600 from the National Institutes of Health, the Harold C. Simmons Research Center and the American Heart Association, Texas Affiliate (K.M.).
REFERENCES
- 1.Jeggo P.A. (1998) Adv. Genet., 38, 185–218. [DOI] [PubMed] [Google Scholar]
- 2.Lees-Miller S.P. (1996) Biochem. Cell Biol., 74, 503–512. [DOI] [PubMed] [Google Scholar]
- 3.Hartley K.O., Gell,D., Smith,G.C., Zhang,H., Divecha,N., Connelly,M.A., Admon,A., Lees-Miller,S.P., Anderson,C.W. and Jackson,S.P. (1995) Cell, 82, 849–856. [DOI] [PubMed] [Google Scholar]
- 4.Yaneva M., Kowalewski,T. and Lieber,M.R. (1997) EMBO J., 16, 5098–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammarsten O. and Chu,G. (1998) Proc. Natl Acad. Sci. USA, 95, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlieb T.M. and Jackson,S.P. (1993) Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 7.Suwa A., Hirakata,M., Takeda,Y., Jesch,S.A., Mimori,T. and Hardin,J.A. (1994) Proc. Natl Acad. Sci. USA, 91, 6904–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dynan W.S. and Yoo,S. (1998) Nucleic Acids Res., 26, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z., Otevrel,T., Gao,Y., Cheng,H.L., Seed,B., Stamato,T.D., Taccioli,G.E. and Alt,F.W. (1995) Cell, 83, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 10.Modesti M., Hesse,J.E. and Gellert,M. (1999) EMBO J., 18, 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 12.Grawunder U., Zimmer,D., Kulesza,P. and Lieber,M.R. (1998) J. Biol. Chem., 273, 24708–24714. [DOI] [PubMed] [Google Scholar]
- 13.Grawunder U., Zimmer,D., Fugmann,S., Schwarz,K. and Lieber,M.R. (1998) Mol. Cell, 2, 477–484. [DOI] [PubMed] [Google Scholar]
- 14.Grawunder U., West,R.B. and Lieber,M.R. (1998) Curr. Opin. Immunol., 10, 172–180. [DOI] [PubMed] [Google Scholar]
- 15.Lewis S.M. (1994) Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- 16.Gellert M. (1997) Adv. Immunol., 64, 39–64. [DOI] [PubMed] [Google Scholar]
- 17.Poltoratsky V.P., Shi,X., York,J.D., Lieber,M.R. and Carter,T.H. (1995) J. Immunol., 155, 4529–4533. [PubMed] [Google Scholar]
- 18.Hunter T. (1995) Cell, 83, 1–4. [DOI] [PubMed] [Google Scholar]
- 19.Jackson S.P. (1995) Curr. Biol., 5, 1210–1212. [DOI] [PubMed] [Google Scholar]
- 20.Keith C.T. and Schreiber,S.L. (1995) Science, 270, 50–51. [DOI] [PubMed] [Google Scholar]
- 21.Blunt T., Finnie,N.J., Taccioli,G.E., Smith,G.C., Demengeot,J., Gottlieb,T.M., Mizuta,R., Varghese,A.J., Alt,F.W., Jeggo,P.A. et al. (1995) Cell, 80, 813–823. [DOI] [PubMed] [Google Scholar]
- 22.Kirchgessner C.U., Patil,C.K., Evans,J.W., Cuomo,C.A., Fried,L.M., Carter,T., Oettinger,M.A. and Brown,J.M. (1995) Science, 267, 1178–1183. [DOI] [PubMed] [Google Scholar]
- 23.Peterson S.R., Kurimasa,A., Oshimura,M., Dynan,W.S., Bradbury,E.M. and Chen,D.J. (1995) Proc. Natl Acad. Sci. USA, 92, 3171–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Critchlow S.E., Bowater,R.P. and Jackson,S.P. (1997) Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- 25.Anderson C.W. (1993) Trends Biochem. Sci., 18, 433–437. [DOI] [PubMed] [Google Scholar]
- 26.Leber R., Wise,T.W., Mizuta,R. and Meek,K. (1998) J. Biol. Chem., 273, 1794–1801. [DOI] [PubMed] [Google Scholar]
- 27.Finnie N.J., Gottlieb,T.M., Blunt,T., Jeggo,P.A. and Jackson,S.P. (1996) Phil. Trans. R. Soc. Lond. B Biol. Sci., 351, 173–179. [DOI] [PubMed] [Google Scholar]
- 28.Arcaro A. and Wymann,M.P. (1993) Biochem. J., 296, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Powis G., Bonjouklian,R., Berggren,M.M., Gallegos,A., Abraham,R., Ashendel,C., Zalkow,L., Matter,W.F., Dodge,J., Grindey,G. et al. (1994) Cancer Res., 54, 2419–2423. [PubMed] [Google Scholar]
- 30.Okada T., Sakuma,L., Fukui,Y., Hazeki,O. and Ui,M. (1994) J. Biol. Chem., 269, 3563–3567. [PubMed] [Google Scholar]
- 31.Oltz E.M., Alt,F.W., Lin,W.C., Chen,J., Taccioli,G., Desiderio,S. and Rathbun,G. (1993) Mol. Cell. Biol., 13, 6223–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin E.K., Rijkers,T., Pastink,A. and Meek,K. (2000) J. Immunol., 164, 1416–1424. [DOI] [PubMed] [Google Scholar]
- 33.Hesse J.E., Lieber,M.R., Gellert,M. and Mizuuchi,K. (1987) Cell, 49, 775–783. [DOI] [PubMed] [Google Scholar]
- 34.Lewis S.M., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Cell, 55, 1099–1107. [DOI] [PubMed] [Google Scholar]
- 35.Sadofsky M.J., Hesse,J.E. and Gellert,M. (1994) Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F., Nicklen,S. and Coulson,A.R. (1977) Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lieber M.R., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1987) Genes Dev., 1, 751–761. [DOI] [PubMed] [Google Scholar]
- 39.Rosenzweig K.E., Youmell,M.B., Palayoor,S.T. and Price,B.D. (1997) Clin. Cancer Res., 3, 1149–1156. [PubMed] [Google Scholar]
- 40.Kimura K., Hattori,S., Kabuyama,Y., Shizawa,Y., Takayanagi,J., Nakamura,S., Toki,S., Matsuda,Y., Onodera,K. and Fukui,Y. (1994) J. Biol. Chem., 269, 18961–18967. [PubMed] [Google Scholar]
- 41.Finnie N.J., Gottlieb,T.M., Blunt,T., Jeggo,P.A. and Jackson,S.P. (1995) Proc. Natl Acad. Sci. USA, 92, 320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wymann M.P., Bulgarelli-Leva,G., Zvelebil,M.J., Pirola,L., Vanhaesebroeck,B., Waterfield,M.D. and Panayotou,G. (1996) Mol. Cell. Biol., 16, 1722–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price B.D. and Youmell,M.B. (1996) Cancer Res., 56, 246–250. [PubMed] [Google Scholar]
- 44.Boulton S., Kyle,S., Yalcintepe,L. and Durkacz,B.W. (1996) Carcinogenesis, 17, 2285–2290. [DOI] [PubMed] [Google Scholar]
- 45.Anderson C.W. and Carter,T.H. (1996) Curr. Top. Microbiol. Immunol., 217, 91–111. [DOI] [PubMed] [Google Scholar]
- 46.Hanks S.K. and Hunter,T. (1995) FASEB J., 9, 576–596. [PubMed] [Google Scholar]
- 47.Fulop G.M. and Phillips,R.A. (1990) Nature, 347, 479–482. [DOI] [PubMed] [Google Scholar]
- 48.Biedermann K.A., Sun,J.R., Giaccia,A.J., Tosto,L.M. and Brown,J.M. (1991) Proc. Natl Acad. Sci. USA, 88, 1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendrickson E.A., Qin,X.Q., Bump,E.A., Schatz,D.G., Oettinger,M. and Weaver,D.T. (1991) Proc. Natl Acad. Sci. USA, 88, 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuler W., Weiler,I.J., Schuler,A., Phillips,R.A., Rosenberg,N., Mak,T.W., Perry,R.P., Bosma,M.J., Malynn,B.A., Blackwell,T.K., Fulop,G.M., Rathbun,G.A., Furley,A.J., Ferrier,P., Yancopoulos,G.D. and Alt,F.W. (1988) Cell, 54, 453–460. [DOI] [PubMed] [Google Scholar]
- 51.Malynn B.A., Blackwell,T.K., Fulop,G.M., Rathbun,G.A., Furley,A.J., Ferrier,P., Phillips,R.A., Yancopoulos,G.D. and Alt,F.W. (1988) Cell, 54, 453–460. [DOI] [PubMed] [Google Scholar]
- 52.Baumann P. and West,S.C. (1998) Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurimasa A., Kumano,S., Boubnov,N.V., Story,M.D., Tung,C.S., Peterson,S.R. and Chen,D.J. (1999) Mol. Cell. Biol., 19, 3877–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeggo P.A., Taccioli,G.E. and Jackson,S.P. (1995) Bioessays, 17, 949–957. [DOI] [PubMed] [Google Scholar]
- 55.Critchlow S.E. and Jackson,S.P. (1998) Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 56.Prise K.M., Ahnstrom,G., Belli,M., Carlsson,J., Frankenberg,D., Kiefer,J., Lobrich,M., Michael,B.D., Nygren,J., Simone,G. and Stenerlow,B. (1998) Int. J. Radiat. Biol., 74, 173–184. [DOI] [PubMed] [Google Scholar]
- 57.Riballo E., Critchlow,S.E., Teo,S.H., Doherty,A.J., Priestley,A., Broughton,B., Kysela,B., Beamish,H., Plowman,N., Arlett,C.F., Lehmann,A.R., Jackson,S.P. and Jeggo,P.A. (1999) Curr. Biol., 9, 699–702. [DOI] [PubMed] [Google Scholar]
- 58.Badie C., Goodhardt,M., Waugh,A., Doyen,N., Foray,N., Calsou,P., SingletonB., Gell,D., Salles,B., Jeggo,P., Arlett,C.F. and Malaise,E.P. (1997) Cancer Res., 57, 4600–4607. [PubMed] [Google Scholar]
- 59.Priestley A., Beamish,H.J., Gell,D., Amatucci,A.G., Muhlmann-Diaz,M.C., Singleton,B.K., Smith,G.C., Blunt,T., Schalkwyk,L.C., Bedford,J.S., Jackson,S.P., Jeggo,P.A. and Taccioli,G.E. (1998) Nucleic Acids Res., 26, 1965–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banga S.S., Hall,K.T., Sandhu,A.K., Weaver,D.T. and Athwal,R.S. (1994) Mutat. Res., 315, 239–247. [DOI] [PubMed] [Google Scholar]
- 61.Kirchgessner C.U., Tosto,L.M., Biedermann,K.A., Kovacs,M., Araujo,D., Stanbridge,E.J. and Brown,J.M. (1993) Cancer Res., 53, 6011–6016. [PubMed] [Google Scholar]
- 62.Zdzienicka M.Z., Jongmans,W., Oshimura,M., Priestley,A. and Whitmore,G.F. (1995) Radiat. Res., 143, 238–244. [PubMed] [Google Scholar]