Abstract
Yeast telomeres consist of ~300 nt of degenerate repeats with the consensus sequence G2–3(TG)1–6. We developed a method for the amplification of a genetically marked telomere by PCR, allowing precise length and sequence determination of the G-rich strand including the 3′ terminus. We examined wild-type cells, telomerase RNA deficient cells and a strain deleted for YKU70, which encodes for a protein involved in telomere maintenance and DNA double strand break repair. The 3′ end of the G-rich strand was found to be at a variable position within the telomeric repeat. No preference for either thymine or guanine as the 3′ base was detected. Comparison of telomere sequences from clonal populations revealed that telomeres consist of a centromere-proximal region of stable sequence and a distal region with differing degenerate repeats. In wild-type as well as yku70-Δ cells, variation in the degenerate telomeric repeats was detected starting 40–100 nt from the 3′ end. Sequence divergence was abolished after deletion of the telomerase RNA gene. Thus, this region defines the domain where telomere shortening and telomerase-mediated extension occurs. Since this domain is much larger than the number of nucleotides lost per generation in the absence of telomerase, we propose that telomerase does not extend a given telomere in every cell cycle.
INTRODUCTION
Linear eukaryotic chromosomes end in a special chromatin structure called telomere. Telomeres protect chromosomes from end-to-end fusion (1–3) and act as a buffer zone against loss of DNA due to incomplete replication (4,5). Telomere maintenance requires the ribonucleoprotein enzyme telomerase, which adds telomeric repeats to the 3′ ends of chromosomes (6). The sequence of the repeats, which is specified by the telomerase RNA subunit, is GT-rich in the strand that contains the 3′ end. At the distal end, telomeres have a single-stranded 3′ overhang (7).
In the budding yeast Saccharomyces cerevisiae mutations in the genes coding for the RNA or the protein subunits of telomerase (TLC1 and EST1-3, respectively) cause a delayed lethal phenotype after about 75 generations, which is accompanied by progressive telomere shortening (8–10). Whereas telomerase synthesises the G-rich strand, the complementary strand is copied by DNA polymerases α and δ (11). Many other gene products are involved in telomere replication, affecting telomere length when mutated. Certain factors appear to regulate telomerase activity negatively (Rap1p, Rif1p, Rif2p) (12–16) or positively (Tel2p) (17). Some have dual roles at telomeres and in double strand break repair (Rad50p, Mre11p, Xrs2p, Yku70p, Yku80p) (18). Others may link telomere length control and checkpoint function (Tel1p, Set1p) (19–22). Clearly, further analysis of telomere replication factors and telomerase RNA mutants would be facilitated by an accurate and efficient method to determine the length and sequence of telomeres.
Unlike mammalian and most ciliate telomeres, yeast telomeric tracts do not have a precisely defined repeat sequence. They consist of degenerate copies of the TLC1 template region and can be described with the consensus G2–3(TG)1–6 (23,24). However, the exact roles of telomerase activity, lagging strand synthesis, recombination events and nucleolytic processing in the generation of repeat divergence have remained unclear. Also, the sequence of the telomeric 3′ end had thus far not been determined.
We have developed a terminal transferase-mediated PCR method to amplify a genetically marked S.cerevisiae telomere, allowing for rapid analysis of telomere length and sequence. Unlike available methods for telomere cloning (23–25), telomere PCR produces clones of complete chromosomal telomeres including the single-stranded 3′ end. Using this approach, we show that yeast telomeres do not end at a defined position within the telomeric repeat. Analysis of telomeres cloned from tlc1-Δ and yku70-Δ strains shows that the sequence variability at the 3′ terminus is not solely due to telomerase activity or Yku70p-dependent processes.
While all yeast telomeric repeats are degenerate (within the consensus), only the distal 40–100 nt of telomeres diverge during the growth of a colony. This repeat variation is dependent on telomerase activity.
MATERIALS AND METHODS
Yeast strains
GA426 is MATa ade2 trp1 can1 his3-11 leu2 ura3-52 DIA V-I. DIAV-I refers to a functional ADE2 gene placed at the telomere of the right arm of chromosome V (9). Deletion of TLC1 and YKU70 in strain GA426 was performed by PCR-based gene deletion (26). The disruption cassette for TLC1 was generated by PCR with primers TLC1KOsense (5′-CAATTAAAAGACCTTCTTTGTAGCTTTTAGTGTGATTTTTCTGGTTTGAGCGGATCCCCGGGTTAATTAA-3′) and TLC1KOantisense (5′-TATTTTTATTTGTATATTGTATATTCTAAAAAGAAGAAGCCATTTGGTGGGAATTCGAGCTCGTTTAAAC-3′) using pFA6a-His3MX6 as a template (26).
The disruption cassette for YKU70 was generated with primers deltaKUsense (5′-AATTTTGCTATGATTTGTTAAGTGACTCTAAGCCTGATTT-3′) and deltaKUantisense (5′-TACATCAAATACCCTACCCTACCTACATATTTTATGTAC-3′).
DNA isolation
Yeast genomic DNA was prepared according to Hoffman and Winston (27) with slight modifications. Single yeast colonies were picked and used to inoculate 2 ml of YPD. After overnight growth, the cultures were harvested by centrifugation and resuspended in 200 µl of lysis buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA). Glass beads (200 µl) and 200 µl of phenol/chloroform/isoamyl alcohol (25:24:1) were added and the mixture was vortexed for 5 min at 4°C. Then, 200 µl of TE were added to the cell lysate and the samples were centrifuged for 15 min at 10 000 g. The aqueous phase was transferred to a new tube and extracted with 500 µl of chloroform. After centrifugation, the supernatant was transferred to a new tube and precipitated with 500 µl of isopropanol. The DNA was centrifuged for 15 min at 10 000 g and washed once with 70% ethanol. The samples were vacuum dried and resuspended in 50 µl of 5 mM Tris–HCl, pH 8.0.
Terminal transferase mediated tailing
100 ng of genomic DNA (estimated from ethidium bromide stained agarose gels) were heat denatured and tailed in 10 µl of 20 mM Tris–HCl pH 7.8, 50 mM KAc, 10 mM MgAc2, 1 mM dCTP with 1 U of terminal deoxynucleotidyl transferase (Amersham Pharmacia Biotech, Uppsala, Sweden) for 30 min at 37°C. Following tailing, the enzyme was inactivated by incubating the samples for 10 min at 65°C and 5 min at 94°C. The tailing reaction was performed in 0.2 ml PCR-tubes using a thermal cycler with a heated lid.
Telomere PCR
After heat inactivation, the tailing reactions were supplemented with 30 µl of PCR-mix resulting in a final volume of 40 µl with the end concentrations: 67 mM Tris–HCl pH 8.8, 16 mM (NH4)2SO4, 5% glycerol, 0.01% Tween-20, 200 µM each dNTP, 2.5 mM MgAc2 (from tailing reaction), 2.5 U Taq-polymerase (Eurobio, Les Ullis Cedex B, France), 40 nM (each) of primers dG18-BamHI (5′-CGGGATCCG18-3′) and DIA5-1 (5′-GTGAGCGGATAACAATTTCACACAGTCTAGATGTCCGAATTGATCCCAGAGTAG-3′). Samples were denatured for 2 min at 94°C, then cycled 45 times with 20 s denaturation (94°C), 12 s annealing (62°C) and 20 s extension (72°C) followed by a final extension step of 5 min at 72°C. The amplified product contains on one end exactly 45 bp of subtelomeric non-TG1–3 sequence, 25 bp derived from primer DIA5-1 and, on the other end, 26 bp of the tail and a BamHI site introduced by the primer. The tail length recovered generally corresponded to the length of the dG-tract in the primer.
Cloning and sequencing of telomeres
Following gel electrophoresis, telomeric bands were excised, purified and cloned using a T/A-strategy (28). Plasmids were sequenced using the M13 forward/reverse primers and the SequiTherm EXCELTMII DNA Sequencing kit (Epicentre Technologies, Madison, WI) on a LiCor DNA-Sequencer.
Southern blotting
DNA samples (1 µg) were digested with 10 U of XhoI for 36 h at 37°C, electrophoresed on 0.7% agarose gels, denatured and blotted. The blot was hybridised with a Y′-probe according to Ritchie et al. (22).
RESULTS
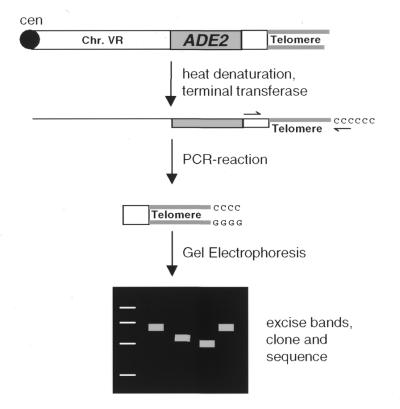
In budding yeast a variable number of subtelomeric X and Y′ elements is present near most chromosome ends which complicates the analysis of individual telomeres (29). Using a strain that contains an ADE2-marked telomere at the right arm of chromosome V (9), we developed a terminal transferase-mediated PCR protocol (referred to as ‘telomere PCR’, outlined in Fig. 1) that allows length measurement and cloning of a specific telomere.
Figure 1.
Principle of telomere PCR. Genomic DNA is isolated, denatured and tailed with dCTP in the presence of terminal deoxynucleotidyl transferase. The G-rich telomere strand is specifically amplified with a primer complementary to the dC tail and a primer specific for a sequence distal of the subtelomeric ADE2 gene. The PCR products are analysed for their length on agarose gels, then cloned and sequenced.
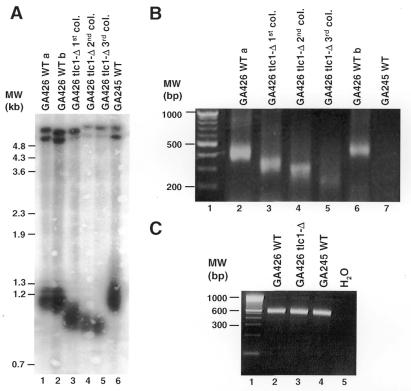
Upon deletion of essential genes of the telomere-maintenance pathway (e.g. EST1-3, TLC1), yeast cells show a progressive shortening of the telomeres and eventually cease dividing. This state is referred to as replicative senescence and occurs after 60–80 generations (three successive restreaks of a colony) (8). To compare the results of telomere PCR with length analysis of terminal restriction fragments by Southern blotting, we chose to follow this telomere shortening after deletion of the telomerase RNA component (tlc1-Δ). As shown in Figure 2A and B, both techniques consistently show a shortening of the telomeres by 200 bp during a growth period of 60–80 generations. The generation of a PCR product was dependent on the presence of a telomeric ADE2 gene (Fig. 2B, compare lanes 2 and 6 to lane 7) and the tailing reaction (not shown). The capacity of each DNA preparation to function as a template for PCR was confirmed by amplifying a portion of the internal ade2 gene (Fig. 2C). Thus, telomere PCR allows specific amplification of the ADE2-marked telomere.
Figure 2.
Comparison of telomere length analysis by Southern blotting and telomere PCR. (A) Southern blot of genomic DNA digested with XhoI and electrophoresed on a 0.7% agarose gel. The blot was hybridised with a Y′-element probe that corresponds to a subtelomeric repeat present at most yeast telomeres. Lanes 1 and 2 are independent DNA preparations from the same strain. (B) Telomere PCR of the same DNA samples as in (A), electrophoresed on 4% agarose gels. DNA from strain GA245 (no telomeric ADE2 gene) does not give a PCR product (lane 7). (C) Control reaction for the integrity of the genomic DNA. A primer pair amplifying the 3′ end of the internal ade2 gene yields a product of constant size in all yeast strains used in (B).
The PCR products were excised, cloned and sequenced. The length of the G-rich telomeric strand was determined to be 291 ± 30.6 nt (average length ±SD, n = 16) in wild-type cells and 181 ± 21.4 nt (n = 7) in yku70-Δ cells (Fig. 3 and data not shown). To confirm that the observed length variation was not introduced during the PCR reaction, we re-amplified a plasmid containing a tailed telomere under identical conditions as the tailed genomic DNA. The resulting band had the same size as the original insert. Sequencing of four cloned PCR products revealed no differences to the original clone (data not shown) and the telomere–tail junction could be unambiguously determined in all sequences. Thus telomere PCR does not introduce sequence variations.
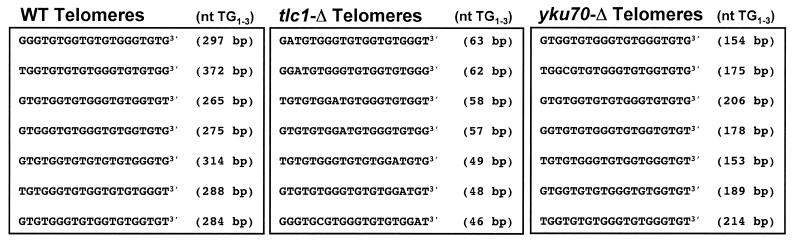
Figure 3.
The 3′ terminus of yeast telomeres has an irregular sequence. The last 20 nt of telomere sequence are shown. The total length of the TG1–3 tract recovered is indicated in parentheses. No preference for a guanine or thymine base at the 3′ position is evident (left, seven representative telomeres are shown). This heterogeneity is also present at telomeres isolated from yeast cells lacking telomerase RNA (centre, the telomeres shown were recovered at the point of senescence). Deletion of the yeast Ku70 subunit (yku70-Δ) does not affect the variability of the telomeric end (right). The sequenced telomeres of all strains contained some non-consensus adenine and cytosine bases (e.g. centre). They are stably maintained in the centromere-proximal part (see also Fig. 4C) and therefore not introduced during PCR amplification.
To ascertain that the DNA including the 3′ overhang remained intact during DNA isolation and tailing, a KpnI restricted plasmid (4 nt 3′ overhang) was added to yeast cells prior to preparing the genomic DNA. When tailed with dC and amplified with a plasmid-specific primer, the original KpnI site was reconstituted. Therefore, no degradation was detected during DNA isolation and tailing.
We compared the DNA sequences found at the 3′ end of cloned telomeres. As shown in Figure 3 (left), there was no preference for either G or T at the 3′ position [10× G, 6× T in wild-type (WT) cells; 6× G, 10× T in tlc1-Δ cells; 3× G, 4× T in yku70-Δ cells, Fig. 3 and data not shown], nor could we detect a conserved sequence motif within the last 20 bp.
Telomere length is determined by an equilibrium of shortening activities (incomplete leading and lagging strand synthesis, nucleolytic processing) and telomerase mediated extension. It is unclear which of these activities contributes to the sequence variability at the 3′ end. Since telomeres cloned from a tlc1-Δ strain showed no conserved sequence motifs at the 3′ end (Fig. 3, centre), telomerase is not the only activity responsible for the 3′ end sequence variability.
The yeast Ku heterodimer is involved in telomere maintenance and DNA double strand break repair (18). Loss of Ku function leads to persistence of long 3′ overhangs throughout the cell cycle, possibly due to altered nucleolytic processing of the telomere end (30). We cloned telomeres from a yku70-Δ strain and analysed the 3′ end sequence (Fig. 3, right). As for wild-type and tlc1-Δ strains, no defined sequences were found at the telomere ends. Thus, the variability of the 3′ terminus is also not solely due to YKU70-dependent activities in non-synchronised yeast cultures.
To examine sequence variation arising in a single telomere during clonal expansion, multiple telomeres were amplified and cloned from a single colony of yeast cells. After alignment of the sequences, two domains of the telomeric tract could be distinguished. First, a centromere-proximal part that was identical in all telomeres and second a distal region of 40–100 nt that showed divergence of the degenerate repeats (Fig. 4A). A similar organisation had been found in studies on telomeres of linear plasmids (24). As telomeres of yku70-Δ cells have long 3′ overhangs (30), more sequence loss should occur during leading strand replication of the telomere (5). However, distal repeat divergence did not extend further into the telomeric tract in yku70-Δ cells (Fig. 4B). Also, the fact that telomeres in these cells are shortened by 100 bp did not influence the extent of the diverging zone. In contrast, no distal repeat divergence was detected in tlc1-Δ cells (Fig. 4C). Therefore, telomerase activity is required for the divergence of distal telomeric repeats, while lagging strand synthesis of telomeric sequences is faithful. No evidence was found for a major role of recombination in the generation of telomere repeat degeneracy.
Figure 4.
Ungapped alignment of telomere 3′ ends. (A) Telomeres from a clonal population of wild-type cells show a divergence of the TG1–3 repeat sequence starting 40–100 bp from the 3′ end. (B) In yku70-Δ cells, telomeric repeat sequences diverge to approximately the same extent as in wild-type cells. (C) Divergence of the 3′ end repeat sequences is lost completely in tlc1-Δ cells (shown are the shortest telomeres recovered; distal repeat divergence was also undetectable in earlier generations).
DISCUSSION
In this study we present a PCR-based method (telomere PCR) for the amplification and length analysis of a marked telomere. The measured telomere lengths are consistent with those obtained by Southern blots of terminal restriction fragments. However, telomere PCR can be performed in 1 day including DNA isolation and requires much less genomic DNA. Most importantly, the method allows the analysis of telomere length and sequence in the same experiment. Although placing genetic markers at the chromosome ends results in artificial subtelomeric sequence, the steady-state lengths of the terminal G2–3(TG)1–6 tract correspond to those of wild-type telomeres. Thus, telomere homeostasis appears to be unperturbed (11,31,32). The principle of telomere PCR should be applicable to any telomere, as long as a unique subtelomeric sequence is known. This could for example facilitate the analysis of telomere dynamics during the cell cycle (11,31).
Yeast telomeres do not end in a defined sequence as is the case for ciliate macronuclear telomeres (33). Which models of 3′ end chromatin structure are compatible with a variable 3′ end sequence? Macronuclear telomeres are capped at their 3′ end with telomere binding proteins, which require a defined telomere end structure and sequence (34). For mammalian telomeres, a different chromosome end structure has been proposed recently (35). According to the T-loop model, the 3′ single-stranded portion of the telomere loops back and invades the duplex region, base pairing with the C-rich strand. This structure is not predicted to require a defined 3′ end sequence as long as base pairing is possible. Interestingly, T-loops also exist in the micronuclei of ciliates (36). In certain organisms, these two telomere structures may therefore represent alternative strategies to protect the chromosomal 3′ end. Since we detected no conserved sequence motif within the last 20 nt of the telomere, a T-loop or similar structure would fit our results better than a model involving telomere binding proteins similar to those found in ciliate macronuclei. Alternatively, Est1p and Cdc13p which bind to single-strand telomeric DNA without the need for a defined 3′ terminal sequence (37–39) may contribute to the 3′ end chromatin structure.
Since telomere PCR amplifies a specific telomere, we could follow the evolution of a single telomere during the growth of a colony. We examined telomere sequence divergence in wild-type cells and in strains lacking either Yku70p or telomerase RNA. While in both wild-type and yku70-Δ cells the last 40–100 nt of the telomere diverged during the growth of one colony, no sequence divergence was detected in tlc1-Δ cells even after the growth of three successive colonies (60–80 generations). Therefore, the domain of distal repeat divergence represents a dynamic region where telomeres have shortened and then been extended by telomerase. The size of this dynamic zone is considerably larger than what would be predicted from the rate of telomere shortening in the absence of telomerase [3–5 nt per generation (8)], if telomerase would act on every telomere in every cell cycle. This together with the observed length heterogeneity suggests a model in which a constant telomere shortening rate (31) is counterbalanced by a more stochastic process of telomere elongation by telomerase. At the steady state, a given telomere is not extended in every cell cycle. Rather, telomeres shorten during several generations before becoming extended by telomerase in vivo. The extent of telomere shortening could be limited to ~100 bp because short telomeres are elongated by telomerase more efficiently than longer ones (31). Our model is also consistent with the recent observation that incorporation of a mutant telomere sequence, specified by a mutant template RNA, is stochastic and does not immediately occur at every telomere (40).
Acknowledgments
ACKNOWLEDGEMENTS
We thank N. Simon-Vermot for the yku70-disrupted strain, M. Nabholz for discussion and C. Kelleher as well as other members of the Lingner laboratory for critical reading of the manuscript. This work was supported by grants from the Swiss National Science Foundation, the Human Frontier Science Program and a Boehringer-Ingelheim-Fonds Ph.D.-fellowship awarded to K.F.
REFERENCES
- 1.Muller H.J. (1938) Collect. Net. – Woods Hole, 13, 181–198. [Google Scholar]
- 2.McClintock B. (1941) Genetics, 26, 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Steensel B., Smogorzewska,A. and de Lange,T. (1998) Cell, 92, 401–413. [DOI] [PubMed] [Google Scholar]
- 4.Olovnikoff A.M. (1973) J. Theor. Biol., 41, 181–190.4754905 [Google Scholar]
- 5.Lingner J., Promisel Cooper,J. and Cech,T.R. (1995) Science, 269, 1533–1534. [DOI] [PubMed] [Google Scholar]
- 6.Nugent C.I. and Lundblad,V. (1998) Genes Dev., 12, 1073–1085. [DOI] [PubMed] [Google Scholar]
- 7.Wellinger R.J. and Sen,D. (1997) Eur. J. Cancer, 33, 735–749. [DOI] [PubMed] [Google Scholar]
- 8.Lundblad V. and Szostak,J.W. (1989) Cell, 57, 633–643. [DOI] [PubMed] [Google Scholar]
- 9.Singer M.S. and Gottschling,D.E. (1994) Science, 266, 404–409. [DOI] [PubMed] [Google Scholar]
- 10.Lendvay T.S., Morris,D.K., Sah,J., Balasubramanian,B. and Lundblad,V. (1996) Genetics, 144, 1399–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diede S.J. and Gottschling,D.E. (1999) Cell, 99, 723–733. [DOI] [PubMed] [Google Scholar]
- 12.Lustig A., Kurtz,S. and Shore,D. (1990) Science, 250, 549–553. [DOI] [PubMed] [Google Scholar]
- 13.Kyrion G., Boake,K. and Lustig,J. (1992) Mol. Cell. Biol., 12, 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardy C.F., Sussel,L. and Shore,D. (1992) Genes Dev., 6, 801–814. [DOI] [PubMed] [Google Scholar]
- 15.Wotton D. and Shore,D. (1997) Genes Dev., 11, 748–760. [DOI] [PubMed] [Google Scholar]
- 16.Marcand S., Gilson,E. and Shore,D. (1997) Science, 275, 986–990. [DOI] [PubMed] [Google Scholar]
- 17.Runge K.W. and Zakian,V.A. (1996) Mol. Cell. Biol., 16, 3094–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertuch A. and Lundblad,V. (1998) Trends Cell Biol., 8, 339–342. [DOI] [PubMed] [Google Scholar]
- 19.Greenwell P.W., Kronmal,S.L., Porter,S.E., Gassenhuber,J., Obermaier,B. and Petes,T.D. (1995) Cell, 82, 823–829. [DOI] [PubMed] [Google Scholar]
- 20.Nislow C., Ray,E. and Pillus,L. (1997) Mol. Biol. Cell, 8, 2421–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corda Y., Schramke,V., Longhese,M.P., Smokvina,T., Paciotti,V., Brevet,V., Gilson,E. and Geli,V. (1999) Nature Genet., 21, 204–208. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie K.B., Mallory,J.C. and Petes,T.D. (1999) Mol. Cell. Biol., 19, 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szostak J. and Blackburn,E. (1982) Cell, 29, 245–255. [DOI] [PubMed] [Google Scholar]
- 24.Wang S. and Zakian,V. (1990) Mol. Cell. Biol., 10, 4415–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prescott J. and Blackburn,E.H. (1997) Genes Dev., 11, 528–540. [DOI] [PubMed] [Google Scholar]
- 26.Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman C. and Winston,F. (1987) Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- 28.Kovalic D., Kwak,J.-H. and Weisblum,B. (1991) Nucleic Acids Res., 19, 4560–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louis E. and Borts,R. (1995) Genetics, 139, 125–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravel S., Larrivée,M., Labrecque,P. and Wellinger,R.J. (1998) Science, 280, 741–744. [DOI] [PubMed] [Google Scholar]
- 31.Marcand S., Brevet,V. and Gilson,E. (1999) EMBO J., 18, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray A. and Runge,K.W. (1999) Mol. Cell. Biol., 19, 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klobutcher L.A., Swanton,M.T., Donini,P. and Prescott,D.M. (1981) Proc. Natl Acad. Sci. USA, 78, 3015–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horvath M.P., Schweiker,V.L., Bevilacqua,J.M., Ruggles,J.A. and Schultz,S.C. (1998) Cell, 95, 963–974. [DOI] [PubMed] [Google Scholar]
- 35.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]
- 36.Murti K.G. and Prescott,D.M. (1999) Proc. Natl Acad. Sci. USA, 96, 14436–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin J.J. and Zakian,V.A. (1996) Proc. Natl Acad. Sci. USA, 93, 13760–13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virta-Pearlman V., Morris,D.K. and Lundblad,V. (1996) Genes Dev., 10, 3094–3104. [DOI] [PubMed] [Google Scholar]
- 39.Nugent C.I., Hughes,T.R., Lue,N.F. and Lundblad,V. (1996) Science, 274, 249–252. [DOI] [PubMed] [Google Scholar]
- 40.Prescott J.C. and Blackburn,E.H. (2000) Mol. Cell. Biol., 20, 2941–2948. [DOI] [PMC free article] [PubMed] [Google Scholar]