Abstract
Aims
We conducted observational and Mendelian randomization (MR) analyses to explore the associations between blood proteins and risk of peripheral artery disease (PAD).
Methods and results
The observational cohort analyses included data on 257 proteins estimated in fasting blood samples from 12 136 Swedish adults aged 55–94 years who were followed up for incident PAD via the Swedish Patient Register. Mendelian randomization analyses were undertaken using cis-genetic variants strongly associated with the proteins as instrumental variables and genetic association summary statistic data for PAD from the FinnGen study (11 924 cases and 288 638 controls) and the Million Veteran Program (31 307 cases and 211 753 controls). The observational analysis, including 86 individuals diagnosed with incident PAD during a median follow-up of 6.6-year, identified 13 proteins [trefoil factor two, matrix metalloproteinase-12 (MMP-12), growth differentiation factor 15, V-set and immunoglobulin domain-containing protein two, N-terminal prohormone brain natriuretic peptide, renin, natriuretic peptides B, phosphoprotein associated with glycosphingolipid-enriched microdomains one, C-C motif chemokine 15, P-selectin, urokinase plasminogen activator surface receptor, angiopoietin-2, and C-type lectin domain family five member A] associated with the risk of PAD after multiple testing correction. Mendelian randomization analysis found associations of T-cell surface glycoprotein CD4, MMP-12, secretoglobin family 3A member 2, and ADM with PAD risk. The observational and MR associations for T-cell surface glycoprotein CD4 and MMP-12 were in opposite directions.
Conclusion
This study identified many circulating proteins in relation to the development of incident PAD. Future studies are needed to verify our findings and assess the predictive and therapeutic values of these proteins in PAD.
Keywords: Cohort, Mendelian randomization, Protein, Peripheral artery disease
Graphical Abstract
Graphical abstract.
Introduction
Peripheral artery disease (PAD) is a common vascular disease with a high incidence1 and causes a large disease burden, especially in high-income countries.2 Several modifiable factors, such as smoking, diabetes, hypertension, and concomitant cardiovascular diseases (CVDs), have been associated with the development of PAD.2,3 There are some treatments for this disease, like statin therapy, P2Y12 inhibitors, low-dose rivaroxaban, vorapaxar, and cilostazol, supervised exercise therapy, and revascularization for PAD patients with severe pain. Proteins principally regulating molecular pathways have been highlighted for drug development. With multiplex methods, circulating proteins can be efficiently measured in a large sample with high accuracy. Several studies have investigated the associations of blood proteins with risk of coronary atherosclerosis.4–7 However, there is a scarcity of data on the associations between blood proteins and the risk PAD.
Employing genetic variants as an instrument for the exposure, Mendelian randomization (MR) analysis can strengthen the causal inference by minimizing confounding and reverse causation.8 This approach resembles the design of randomized controlled trials. In detail, given that genetic variants are randomly assorted at conception, MR based on genetic variants as the instrumental variable for the exposure can assign the participants into groups by natural randomization. The exposure proxied by genetic instruments is not associated with confounders. This process mimics randomization of randomized controlled trials and thus minimizes confounding. In addition, since the germline genotype is fixed and cannot be modified by onset or progression of disease, MR design can also diminish the influence of reverse causation. This method has been widely employed to assess the causality of associations and has been found to be valid as randomized controlled trials.9–11 For MR investigations on a protein, one or more cis-genetic variants (i.e. a genetic variant within the gene region that encodes the protein) is usually utilized as the instrumental variable. Compared to using trans-genetic variants, using cis-variants as an instrumental variable is more likely to reflect protein-specific effects and thus satisfy the three key assumptions of MR that are (i) the selected genetic variant strongly associated with the protein, (ii) the used genetic variant for the protein are not associated with confounders, and (iii) the genetic variant affects the outcome only via the exposure.12 Here, we conducted cohort and MR analyses to explore the associations of cardiovascular and cardiometabolic proteins with risk of PAD.
Methods
Study design
We explored the observational associations between 257 blood proteins and the risk of incident PAD in two Swedish cohorts. To strengthen the causality of the protein-PAD associations, MR analyses were conducted using data from large-scaled genetic studies.
Observational analysis
Study population
The observational analysis was based on data from two clinical sub-cohorts of the Swedish Infrastructure for Medical Population-Based Life-Course and Environmental Research (SIMPLER) that includes the Swedish Mammography Cohort (SMC) and Cohort of Swedish Men (COSM). Fasting blood samples were collected in clinical examinations by trained nurses during 2003–2009 and 2010–2019 for sub-cohorts of SMC and COSM, respectively. Meanwhile, participants were asked to fill in some questionnaires on diet, health status, and lifestyle. In total, 12 314 participants were recruited in two sub-cohorts. After removing 178 people with baseline PAD diagnosed before the day of blood sample collection, we included 12 136 participants in the analysis. Detailed information on cohorts and used questionnaires can be found on the SIMPLER website (https://www.simpler4health.se/).
Proteomic profiling
Venous blood samples were collected after an overnight fast and immediately centrifuged and stored at −80°C until analysis. In total, 276 plasma protein biomarkers were analysed using three high-throughput multiplex immunoassays: the Olink Proseek Multiplex CVD II, CVD III, and Metabolism (Olink Bioscience, Uppsala, Sweden), where the levels of protein expression were normalized on a log2 scale standardized per analysis plate. Values below limit of detection (LOD) were provided by the manufacturer and used as a protein selection criterium. The analyses were performed at SciLifeLab, Uppsala University, Sweden.13 In this analysis, 19 proteins with more than 50% samples below the LOD were excluded (see Supplementary material online, Table S1). A small portion of specimens was set to missing given the analysed sample did not pass the manufacturer’s quality control (3.6%, 0.8%, and 0.7% for CVD II, CVD III, and Metabolism panels, respectively). Included 257 proteins can be found in the Supplementary material online, Table S2.
Case ascertainment and follow-up
Incident PAD cases were ascertained by a medical diagnosis of PAD as the primary or contributing causes with diagnostic data obtained from the Swedish National Patient Register. The diagnostic codes used are shown in Supplementary material online, Table S3. The Swedish National Patient Register covers nearly all information on hospital-based inpatient and outpatient care.14 We obtained date of death from the Swedish Death Registry. Participants were followed up from the baseline until the date of diagnosis of PAD, date of death, or end of follow-up (i.e. 31 December 2019), whichever came first.
Measurements of covariates
We obtained data on age, sex, education attainment, smoking, alcohol consumption, physical activity, and dietary intake from self-reported questionnaires. Diet quality was assessed by a modified Dietary Approaches to Stop Hypertension (mDASH) score.15 Weight, height, estimated glomerular filtration rate (eGFR), levels of blood lipids and glucose, and blood pressure were measured by trained nurses in the health exam. Body mass index (BMI) was calculated by weight (in kg) divided by square of height (in m). Baseline diagnosis of cardiovascular disease (CVD) including coronary artery disease, heart failure, stroke, and atrial fibrillation was extracted from the Swedish National Patient Register. Detailed information of covariates is shown in Supplementary material online, Table S4.
Cox regression analyses
We used multiple imputation by chained equations (20 imputation cycles) to impute missing values of protein and covariates. The associations between circulating proteins and the risk of incident PAD were calculated by Cox proportional hazards regression model with age as the underlying time scale. The assumption of proportionality was examined by Schoenfeld residuals and found to be met. Two models were used: Model 1 adjusted for sex and plate, and Model 2 adjusted for sex, plate, BMI,16 educational attainment,17 baseline CVD,18 smoking status,19 alcohol consumption,3 physical activity,3 mDASH score,3 eGFR,20 low- and high-density lipoprotein cholesterol (LDLC and HDLC),21 triglycerides,21 systolic blood pressure,21 and blood glucose levels.22 Traits included in the Model 2 have been associated with PAD; however, it remains unclear whether all these are associated with blood proteins. In a conservative way of minimizing confounding, we adjusted for these factors in the Model 2. To examine the robustness of the associations, we conducted a sensitivity analysis among individuals with (n = 2617) and without (n = 9519) baseline CVD separately. The statistical tests were two-sided, and the analyses performed in Stata/SE (version 15.0; StataCorp, Texas, USA). The false discovery rate (FDR) based on Benjamini–Hochberg method was used to account for multiple testing. False discovery rate < 0.05 was considered statistically significant.
Mendelian randomization analysis
Peripheral artery disease data sources
Summary-level genetic data on PAD were obtained from the FinnGen R7 data release that included 11 924 participants with and 288 638 participants without PAD23 and from a genome-wide association analysis in the Million Veteran Program (MVP) comprised of 31 307 participants with and 211 753 participants without PAD.24 Detailed information (e.g. case definition and covariate adjustment) of the data sources is presented in Supplementary material online, Table S5.
Genetic instrument selection
To examine the causality of the association between proteins and PAD and reduce the possibility of Type 2 error caused by a heavy multiple-testing burden in cohort analysis, we included proteins associated with PAD at the nominal significance level in the cohort analysis to MR analysis. We used cis-genetic variants associated with proteins at the genome-wide significance threshold (P < 5 × 10−8) as instrumental variables for circulating proteins. The genetic associations (β coefficients and standard errors) with proteins were obtained from six genome-wide association studies, where protein profiles were analysed by SomaLogic assay in four studies25–28 and Olink in two studies.29,30 Lead cis-genetic variants provided by five studies25–28,30 and single nucleotide polymorphisms (SNPs) in the protein-coding gene region with the pruning threshold of R2 < 0.01 from Folkersen et al. study29 were used. Single nucleotide polymorphisms for the identical protein from different studies were treated as separate instruments and independently used in MR analysis with the aim of mutual verification. Missing SNP was replaced by a proxy SNP with strong linkage disequilibrium (r2 ≥ 0.8), and proteins without genetic instruments were removed from the analysis. Detailed information on used studies is presented in Supplementary material online, Table S6. Instrument variables are listed in Supplementary material online, Table S7.
Reverse Mendelian randomization analysis for matrix metalloproteinase-12
In this analysis, genetic liability to PAD was deemed the exposure and levels of matrix metalloproteinase-12 (MMP-12) the outcome. We selected genetic variants associated with PAD at the genome-wide significance level in the MVP comprised of 31 307 participants with and 211 753 participants without PAD.24 After pruning genetic variants with linkage disequilibrium r2 < 0.01, we selected 18 SNPs as an instrumental variable for PAD. Summary-level data on MMP-12 were extracted from the SCALLOP consortium (Folkersen et al. study29) where blood proteins were measured using the Olink platform in > 30 000 participants.
Mendelian randomization statistical analysis
We calculated F statistic to assess the strength of the genetic instrument. The F statistic >10 indicates a good strength of minimal weak instrument bias.31 Mendelian randomization associations were estimated by the Wald ratio method (i.e. ratio estimate equals the β coefficient for the effect of the SNP on PAD divided by the β coefficient for the effect of the SNP on the protein).32 The standard error of the ratio estimate was estimated using the delta method.32 For proteins with two or more SNPs from Folkersen et al. study,29 the inverse variance weighted method was used to estimate MR associations with PAD. Mendelian randomization estimates for each protein from the FinnGen and the MVP studies were combined using the fixed-effect meta-analysis method. We performed two sensitivity analyses, the weighted median and MR–Egger methods, for proteins with greater than or equal to three cis-SNPs as instrumental variables to examine the consistency of the results and detect potential horizontal pleiotropy. The weighted median method assumes that more than half of the weight is from valid SNPs and thus generate consistent MR estimates. The MR–Egger regression method can detect horizontal pleiotropy by its intercept test and provide estimates after correction for potential horizontal pleiotropy; however, the method is usually underpowered. The analyses were conducted using TwoSampleMR and MendelianRandomization packages in R software 4.1.1.
Results
Observational analysis of proteins and peripheral artery disease
During a median follow-up of 6.4 (the interquartile range of 8.5) years, 86 participants developed incident PAD diagnosed. Baseline characteristics of participants by incident PAD status are shown in Table 1. In brief, compared to individuals without incident PAD diagnosis, those with incident PAD were more likely to be men and had more baseline traditional CVD risk factors.
Table 1.
Baseline characteristics of 12 136 Swedish adults by incident peripheral artery disease (PAD) status during follow up
| Characteristic | Without incident PAD (n = 12 050) |
Incident PAD (n = 86) |
P for difference |
|---|---|---|---|
| Age in year | 71.5 ± 6.8 | 74.5 ± 6.7 | <0.001 |
| Male, n (%) | 4593 (38.1) | 44 (51.2) | 0.013 |
| Body mass index in kg/m2 | 26.4 ± 4.2 | 25.9 ± 3.1 | 0.083 |
| Education attainment ≥12 years, n (%) | 3443 (28.6) | 17 (20.0) | 0.072 |
| Baseline cardiovascular disease, n (%) | 2583 (21.4) | 34 (39.5) | <0.001 |
| Coronary artery disease, n (%) | 70 (0.6) | 16 (1.6) | <0.001 |
| Heart failure, n (%) | 74 (0.7) | 12 (1.5) | 0.008 |
| Stroke, n (%) | 84 (0.7) | 2 (0.5) | 0.608 |
| Atrial fibrillation, n (%) | 70 (0.7) | 16 (1.2) | 0.026 |
| Current smoker, n (%) | 4181 (34.7) | 54 (62.8) | <0.001 |
| Excessive alcohol consumption, n (%) | 1406 (11.8) | 11 (12.8) | 0.773 |
| Physical activity, n (%) | 0.023 | ||
| <10 min/day | 685 (5.7) | 5 (5.8) | |
| 10–30 min/day | 1391(11.5) | 19 (22.1) | |
| 30–60 min/day | 5385 (44.7) | 35 (40.7) | |
| >60 min/day | 4589 (38.1) | 27 (31.4) | |
| mDASH score | 18.1 ± 3.7 | 17.8 ± 3.5 | 0.327 |
| eGFR in mL/min/1.73m2 | 81.0 ± 15.2 | 75.6 ± 17.9 | <0.001 |
| LDLC in mmol/L | 3.3 ± 1.0 | 3.4 ± 1.0 | 0.303 |
| HDLC in mmol/L | 1.5 ± 0.4 | 1.4 ± 0.3 | 0.025 |
| Triglyceride in mmol/L | 1.3 ± 0.7 | 1.5 ± 0.7 | 0.021 |
| Glucose in mmol/L | 5.7 ± 1.4 | 6.1 ± 1.6 | 0.011 |
| Systolic blood pressure in mmHg | 138.9 ± 17.6 | 146.1 ± 20.3 | <0.001 |
eGFR, estimated glomerular filtration rate; HDLC, high-density lipoprotein cholesterol; LDLC, low-density lipoprotein cholesterol; mDASH, modified Dietary Approaches to Stop Hypertension; PAD, peripheral artery disease; continuous and categorical variables were expressed in mean (standard deviation) and n (%), respectively.
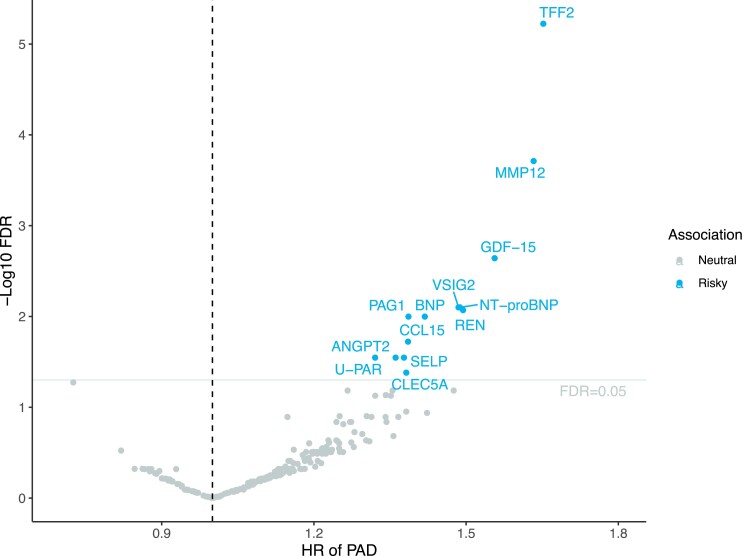
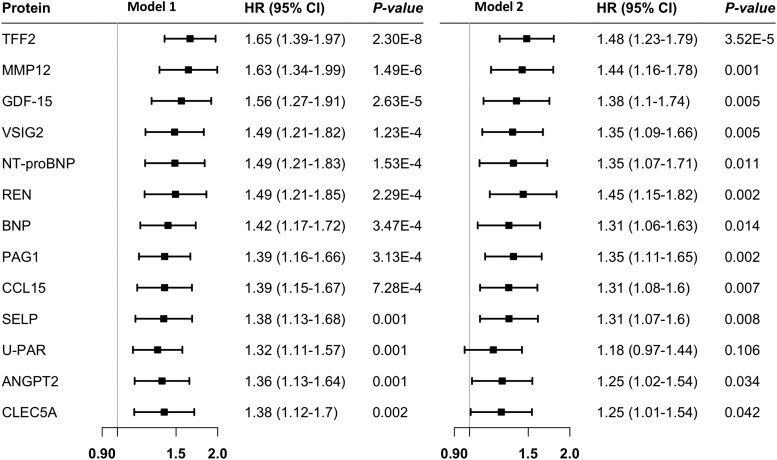
In total, plasma levels of 13 out of 257 proteins were associated with the risk of incident PAD after FDR correction in Model 1 adjusted for age, sex, and plate (Figure 1 and Figure 2). Per standard deviation (SD) increase in circulating protein levels, the hazard ratio (HR) of PAD was 1.65 [95% confidence interval (CI) 1.39–1.97] for TFF2 (trefoil factor 2), 1.63 (95% CI 1.34–1.99) for MMP-12, 1.56 (95% CI 1.27–1.91) for GDF-15 (growth differentiation factor-15), 1.49 (95% CI 1.21–1.82) for VSIG2 (v-set and immunoglobulin domain-containing protein 2), 1.49 (95% CI 1.21–1.83) for NT-proBNP (N-terminal prohormone brain natriuretic peptide), 1.49 (95% CI 1.21–1.85) for REN (renin), 1.42 (95% CI 1.17–1.72) for BNP (natriuretic peptides B), 1.39 (95% CI 1.16–1.66) for PAG1 (phosphoprotein associated with glycosphingolipid-enriched microdomains 1), 1.39 (95% CI 1.15–1.67) for CCL15 (C-C motif chemokine 15), 1.38 (95% CI 1.13–1.68) for SELP (P-selectin), 1.38 (95% CI 1.12–1.7) for CLEC5A (C-type lectin domain family 5 member A), 1.36 (95% CI 1.13–1.64) for ANGPT2 (angiopoietin-2), and 1.32 (95% CI 1.11–1.57) for U-PAR (urokinase plasminogen activator surface receptor). After further adjustment for lifestyle and clinical factors in Model 2, the associations for all proteins except for U-PAR remained significant (Figure 2). These associations remained overall consistent in the sensitivity analysis in individual with and without baseline CVD albeit with larger CIs (see Supplementary material online, Table S8). There were 33 associations with the nominal P < 0.05 and FDR-adjusted P > 0.05 (see Supplementary material online, Table S9).
Figure 1.
Volcano plot of the observational associations between 257 proteins and the risk of incident peripheral artery disease (PAD) in the Swedish cohort analysis. FDR, false discovery rate; HR, hazard ratio. The associations were derived from the model adjusted for age, sex, and plate. Full name of proteins can be found in Supplementary material online, Table S2.
Figure 2.
The observational associations between 13 proteins and the risk of incident peripheral artery disease with false discovery rate <0.05. CI, confidence interval; HR, hazard ratio. Model 1 adjusted for age, sex, and plate; Model 2 adjusted for age, sex, body mass index, plate, education attainment, baseline cardiovascular disease, smoking status, alcohol consumption, physical activity, mDASH score, eGFR, low- and high-density lipoprotein cholesterol, triglycerides, blood pressure, and blood glucose levels. Full name of proteins can be found in Supplementary material online, Table S2.
Mendelian randomization analysis of the protein–peripheral artery disease association
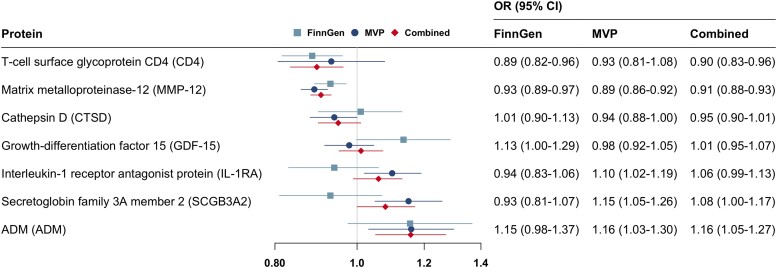
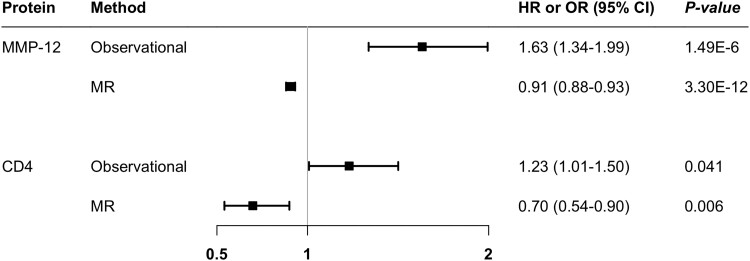
We conducted MR analyses for 46 proteins associated with PAD at the nominal significant level in the Cox regression analysis adjusted for age, sex, and plate. Seven proteins were removed from the analysis due to without suitable instrumental variables, and one protein (LEG9, galectin-9) was excluded from the analysis in FinnGen due to missing without a proxy SNP. All F statistics for used genetic instruments were >10. Genetically predicted levels of MMP-12, CD4 (T-cell surface glycoprotein CD4), and GDF-15 were associated with PAD risk in FinnGen (Figure 3). For genetically predicted per SD increase, the odds ratio (OR) of PAD was 0.93 (95% CI 0.89–0.97) for MMP-12, 1.13 (95% CI 1.00–1.29) for GDF-15, and 0.89 (95% CI 0.82–0.96) for CD4. In the MVP study, there were additional associations for IL-1ra (interleukin-1 receptor antagonist protein), ADM, CTSD (cathepsin D), and SCGB3A2 (secretoglobin family 3A member 2) (Figure 3). In the combined analysis of MR estimate in FinnGen and MVP studies, genetically predicted levels of CD4 (OR 0.90; 95% CI 0.83–0.96), MMP-12 (OR 0.91; 95% 0.88–0.93), SCGB3A2 (OR 1.08; 95% CI 1.00–1.17), and ADM (OR 1.16; 95% CI 1.05–1.27) were associated with the risk of PAD (Figure 3). These associations were generally consistent using genetic instrumental variables from different studies (see Supplementary material online, Table S10). For proteins with SNPs greater than or equal to three, the associations were consistent in the sensitivity analysis, and we detected no indication of horizontal pleiotropy in MR–Egger intercept test (P > 0.05; Supplementary material online, Table S11). There were no associations of genetically predicted levels of other studied proteins with PAD risk (see Supplementary material online, Table S10). Of note, the observational and MR associations for MMP-12 and CD4 were in the opposite direction (Figure 4).
Figure 3.
Potential causal associations of circulating proteins with risk of peripheral artery disease in Mendelian randomization analysis. CI, confidence interval; MVP, the Million Veteran Program; OR, odds ratio.
Figure 4.
Contradictory associations of MMP-12 and CD4 with peripheral artery disease risk in observational and Mendelian randomization analyses. The observational associations were adjusted for age, sex, and plate. CI, confidence interval; CD 4, T-cell surface glycoprotein CD4; HR, hazard ratio; MMP-12, matrix metalloproteinase-12; OR, odds ratio; MR, Mendelian randomization.
Reverse Mendelian randomization analysis for matrix metalloproteinase-12
Genetic liability to PAD was not associated with levels of MMP-12 (see Supplementary material online, Table S12). The null association was observed in sensitivity analyses. There was heterogeneity among SNPs’ estimates but no horizontal pleiotropy (P for MR–Egger intercept = 0.89).
Discussion
The present study identified 13 circulating proteins that were observationally associated with PAD incidence in Swedish adults. Mendelian randomization analysis in two populations of Finnish and US adults, including a total of 43 231 PAD cases and 500 391 controls, revealed associations of CD4, MMP-12, SCGB3A2, and ADM with PAD risk. However, the associations for CD4 and MMP-12 were in opposite directions in the observational and MR analyses. The reverse MR found limited data in support of an association between genetic liability to PAD and MMP-12 levels.
A study including data from six US community-based cohorts estimated the residual lifetime risk of PAD in relation to many traditional risk factors for CVD.33 The direction of associations for identified proteins in this study were comparable to that of most risk factor for PAD, like diabetes, SBP, and hyperlipidaemia.33 This comparison implies the vital roles of identified circulating proteins in the development of PAD, which warrants more explorations for PAD prevention and treatment using these protein targets.
High levels of MMP-12 have been associated with an increased risk of several atherosclerotic cardiovascular conditions, such as atherosclerotic media destruction and ectasia,34 intima-media thickness in the common carotid artery (IMT-CCA) and in the bulb (IMT-bulb),5 and incident cardiovascular events4,35 in some but not all36 observational studies. Our observational analysis for the first time revealed a positive association between MMP-12 and PAD in which atherosclerosis plays a vital role. The mechanisms in support of this positive association may be related to lesion formation, plaque stability, reduced coagulation, and perivascular fibrosis.37,38 However, the MR analysis identified a strong albeit opposite effect of MMP-12 on PAD, which is in line with previous MR findings of inverse associations of MMP-12 with the risk of coronary artery disease28 and ischaemic stroke.4,39 Even though the direction of the association was discordant between observational and MR analyses, these findings pointed out a possible causal role of MMP-12 in the development of atherosclerotic outcomes. The reason of this discrepancy is unclear but possibly related to the feedback mechanism,4 which means that MMP-12 may be upregulated to compensate for proatherogenic processes during the early stage of atherogenesis40 and thus individuals with reduced capacity to synthesize this protein have an increased cardiovascular risk. To test this hypothesis, we conducted an inverse MR analysis on the association of genetic liability to PAD with MMP-12 levels. However, the reverse MR analysis did not detect a significant association instead of an inverse trend in the primary MR analysis even though the analysis might be underpowered. Thus, if anything, this inverse link partly supports this hypothesis. Of note, confounding may also contribute to this disagreement. However, we measured and adjusted for many important risk factors in observational study although we could not completely rule out confounding caused by unmeasured factors.
Our observational and MR associations were also contradictive for the association between CD4 and PAD risk. This disagreement was also observed in previous studies. In some observational studies, a decline in CD4 count was associated with an elevated risk of endothelial dysfunction41 and CVD.42 However, lack of CD4 was shown to substantially decreased the development of atherosclerosis in apolipoprotein E knockout mice.43 As for the discrepancy in our study, the difference in the content actually measured in observational and MR analysis may be the reason. In observational study, serum CD4 protein was measured, and this protein in the circulation may be majorly derived from the membrane particles after CT4 T-cell death. However, CD 4 protein levels proxied by genetic variants may reflect more the amount of CD4+ T cells. Although there are no robust explanation for the disagreement, these findings suggest the complex roles of the adaptive immune response in the development of the atherosclerosis,44 which may provide attractive targets for atherosclerotic prevention.
Evidence on the positive association between GDF-15 and atherosclerosis risk is consistent between studies.45,46 Our study found a positive between GDF-15 and PAD in a generally healthy middle-aged population for the first time, and this association was partly strengthened in MR analysis in FinnGen instead of in the MVP study. Likewise, IL-1RA was positively associated with PAD risk in the cohort analysis and MR analysis in the MVP study, which is in line with previous findings.47,48 Given that these associations were not consistently observed in our analyses using different sources, more studies are needed to verify these findings. In addition, the study found possible associations of CTSD and SCGB3A2 with PAD risk, which are novel findings that need confirmation. Our cohort analysis identified the association of previously established atherosclerosis-related proteins, such as NT-proBNP and U-PAR,4 as well as novel proteins, such as VSIG2, REN, CCL15, and SELP, with incident PAD, but the MR analysis was not able to provide evidence for a causal relationship between these proteins and PAD. Nonetheless, these proteins may have utility as predictive biomarkers for PAD.
The strengths of our study include a large sample size with protein data, nearly complete follow-up information, inclusion of important covariates, and combination of traditional observational and MR analyses. At the same time, there are limitations that deserve to be emphasized when interpreting our results. First, the number of PAD cases in the observational analysis was limited despite the large overall cohort and a long follow-up. Thus, some weak associations might be missed due to inadequate power, and multiple correction might inflate the rate of Type 2 error. Second, our cohort findings among patients with baseline CVD might be affected by residual confounding from use of certain medications, like statins. However, we were able to adjust for baseline CVD diagnosis, which tightly correlated with corresponding medications. In addition, these associations remained overall consistent in individuals without baseline CVD who usually use few medications. Another confounding source is lipoprotein(a), an important risk factor for PAD,49 which was not measured in the cohorts and thus could not be adjusted for in the analysis, even though whether lipoprotein(a), which is largely determined by variations in the LPA gene, is associated with studied blood proteins is unknown. Third, our observational and MR analyses were based on individual of European populations. Whether our findings can be generalized to other populations needs verification. Fourth, the proteomic platform used for the cohort study was targeted and incomplete in its coverage of potential proteins of interest, undoubtedly missing important potential protein biomarkers. Fifth, we could not replicate the results of the cohort analysis in an independent sample. However, the potential protein–PAD associations were tested by MR analyses in two populations with generally consistent findings although FinnGen population may have a different genetic background from the traditional European populations. Sixth, blood samples in the cohort analysis were collected at different timepoints and time in freezer might affect the levels of proteins, which might influence the results. However, an inflated variation in protein levels would attenuate the association towards to a null hypothesis in a conservative way. Seventh, horizontal pleiotropy could not be examined in MR analysis due to the lack of summary-level data even though this bias should be minimal when using cis-SNPs with a clear function as instrumental variables.
In summary, this study discovered many circulating proteins in relation to the development of PAD. These population-level findings may provide clues for future molecular exploration of mechanistical insights of PAD development. In addition, more studies are needed to assess the predictive and therapeutic values of these PAD-associated proteins.
Supplementary Material
Acknowledgements
We want to acknowledge the participants and investigators of SIMPLER for provisioning of facilities and experimental support. SIMPLER receives funding through the Swedish Research Council under grant number 2017-00644. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project simpl2020002. We also want to acknowledge the participants and investigators of the VA Million Veteran Program and the FinnGen study. S.M.D. is supported by IK2-CX001780. This publication does not represent the views of the Department of Veterans Affairs or the United States Government.
Contributor Information
Shuai Yuan, Unit of Cardiovascular and Nutritional Epidemiology, Institute of Environmental Medicine, Karolinska Institutet, Nobels väg 13, 171 65 Solna, Stockholm, Sweden.
Olga E Titova, Unit of Medical Epidemiology, Department of Surgical Sciences, Uppsala University, Uppsala, Sweden.
Ke Zhang, Key Laboratory of Growth Regulation and Translational Research of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou, China; Westlake Intelligent Biomarker Discovery Lab, Westlake Laboratory of Life Sciences and Biomedicine, Hangzhou, China.
Jie Chen, Department of Big Data in Health Science School of Public Health, Center of Clinical Big Data and Analytics of The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China.
Xue Li, Department of Big Data in Health Science School of Public Health, Center of Clinical Big Data and Analytics of The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China.
Derek Klarin, VA Palo Alto Healthcare System, 4951 Arroyo Rd, Livermore, CA 94550, USA; Department of Surgery, Stanford University School of Medicine, Stanford University, Palo Alto, CA, USA.
Agneta Åkesson, Unit of Cardiovascular and Nutritional Epidemiology, Institute of Environmental Medicine, Karolinska Institutet, Nobels väg 13, 171 65 Solna, Stockholm, Sweden.
Scott M Damrauer, Corporal Michael J. Crescenz VA Medical Center, Philadelphia, PA, USA; Department of Surgery & Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
Susanna C Larsson, Unit of Cardiovascular and Nutritional Epidemiology, Institute of Environmental Medicine, Karolinska Institutet, Nobels väg 13, 171 65 Solna, Stockholm, Sweden; Unit of Medical Epidemiology, Department of Surgical Sciences, Uppsala University, Uppsala, Sweden.
Lead author biography
 Shuai Yuan is a PhD candidate at Unit of Cardiovascular and Nutritional Epidemiology, Institute of Environmental Medicine, Karolinska Institutet. He is interested in exploring the roles of lifestyle and nutritional factors in the development of venous thrombosis and PAD using population-based cohort and MR analyses. He also conducts research to identify protein biomarkers and drug targets for common diseases by integrating human plasma proteome with genome.
Shuai Yuan is a PhD candidate at Unit of Cardiovascular and Nutritional Epidemiology, Institute of Environmental Medicine, Karolinska Institutet. He is interested in exploring the roles of lifestyle and nutritional factors in the development of venous thrombosis and PAD using population-based cohort and MR analyses. He also conducts research to identify protein biomarkers and drug targets for common diseases by integrating human plasma proteome with genome.
Data availability
De-identified SIMPLER data are available for researchers upon application (http://www.simpler4health.se/). Summary-level data from the FinnGen study of PAD can be obtained via https://finngen.gitbook.io/documentation/. Summary data from the MVP GWAS of PAD can be obtained via dbGAP, accession code no. phs001672.v2.p1.
Supplementary material
Supplementary material is available at European Heart Journal Open online.
Funding
Funding for this study came from the Karolinska Institutet Grants (Grant number 2020-01842), the Swedish Research Council (Vetenskapsrådet; Grant Number 2019-00977), the Swedish Research Council for Health, Working Life and Welfare (Forte; 2018-00123), and the Swedish Heart-Lung Foundation (Hjärt-Lungfonden; Grant number 20190247).
Author contributions
S.Y. and S.C.L. conceived and designed the study. S.Y. and O.E.T undertook the statistical analyses. S.Y. wrote the first draft of the manuscript. All authors provided important comments to the manuscript and approved the final version of the manuscript.
References
- 1. Sundaram V, Bloom C, Zakeri R, Halcox J, Cohen A, Bowrin K, Briere JB, Banerjee A, Simon DI, Cleland JGF, Rajagopalan S, Quint JK. Temporal trends in the incidence, treatment patterns, and outcomes of coronary artery disease and peripheral artery disease in the UK, 2006–2015. Eur Heart J 2020;41:1636–1649. [DOI] [PubMed] [Google Scholar]
- 2. Song P, Rudan D, Zhu Y, Fowkes FJI, Rahimi K, Fowkes FGR, Rudan I. Global, regional, and national prevalence and risk factors for peripheral artery disease in 2015: an updated systematic review and analysis. Lancet Glob Health 2019;7:e1020–e1030. [DOI] [PubMed] [Google Scholar]
- 3. Yuan S, Damrauer SM, Håkansson N, Åkesson A, Larsson SC. A prospective evaluation of modifiable lifestyle factors in relation to peripheral artery disease risk. Eur J Vasc Endovasc Surg 2022;64:83–91. [DOI] [PubMed] [Google Scholar]
- 4. Lind L, Gigante B, Borné Y, Feldreich T, Leppert J, Hedberg P, Östgren CJ, Nyström FH, Sundström J, Ärnlöv J, Baldassarre D, Tremoli E, Veglia F, Hamsten A, O'Donnell CJ, Franceschini N, Orho-Melander M, Nilsson J, Melander O, Engström G, Mälarstig A. Plasma protein profile of carotid artery atherosclerosis and atherosclerotic outcomes: meta-analyses and Mendelian randomization analyses. Arterioscler Thromb Vasc Biol 2021;41:1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lind L, Gigante B, Borne Y, Mälarstig A, Sundström J, Ärnlöv J, Ingelsson E, Baldassarre D, Tremoli E, Veglia F, Hamsten A, Orho-Melander M, Nilsson J, Melander O, Engström G. The plasma protein profile and cardiovascular risk differ between intima-media thickness of the common carotid artery and the bulb: A meta-analysis and a longitudinal evaluation. Atherosclerosis 2020;295:25–30. [DOI] [PubMed] [Google Scholar]
- 6. Ferrannini G, Manca ML, Magnoni M, Andreotti F, Andreini D, Latini R, Maseri A, Maggioni AP, Ostroff RM, Williams SA, Ferrannini E. Coronary artery disease and type 2 diabetes: a proteomic study. Diabetes Care 2020;43:843–851. [DOI] [PubMed] [Google Scholar]
- 7. Prentice RL, Zhao S, Johnson M, Aragaki A, Hsia J, Jackson RD, Rossouw JE, Manson JE, Hanash SM. Proteomic risk markers for coronary heart disease and stroke: validation and mediation of randomized trial hormone therapy effects on these diseases. Genome Med 2013;5:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burgess S, Thompson SG. Mendelian Randomization: methods for using genetic variants in causal estimation. London, UK: Chapman and Hall/CRC; 2015. [Google Scholar]
- 9. Ference BA, Robinson JG, Brook RD, Catapano AL, Chapman MJ, Neff DR, Voros S, Giugliano RP, Davey Smith G, Fazio S, Sabatine MS. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med 2016;375:2144–2153. [DOI] [PubMed] [Google Scholar]
- 10. Yuan S, Wang L, Zhang H, Xu F, Zhou X, Yu L, Sun J, Chen J, Ying H, Xu X, Yu Y, Spiliopoulou A, Shen X, Wilson J, Gill D, Theodoratou E, Larsson SC, Li X. Mendelian Randomization and clinical trial evidence supports TYK2 inhibition as a therapeutic target for autoimmune diseases. EBioMedicine 2023;89:104488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bennett DA, Holmes MV. Mendelian randomisation in cardiovascular research: an introduction for clinicians. Heart 2017;103:1400–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Warensjö Lemming E, Byberg L, Stattin K, Ahmad S, Lind L, Elmståhl S, Larsson SC, Wolk A, Michaëlsson K. Dietary pattern specific protein biomarkers for cardiovascular disease: A cross-sectional study in 2 independent cohorts. J Am Heart Assoc 2019;8:e011860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ludvigsson JF, Andersson E, Ekbom A, Feychting M, Kim JL, Reuterwall C, Heurgren M, Olausson PO. External review and validation of the Swedish national inpatient register. BMC Public Health 2011;11:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan S, Bruzelius M, Håkansson N, Åkesson A, Larsson SC. Lifestyle factors and venous thromboembolism in two cohort studies. Thromb Res 2021;202:119–124. [DOI] [PubMed] [Google Scholar]
- 16. Larsson SC, Bäck M, Rees JMB, Mason AM, Burgess S. Body mass index and body composition in relation to 14 cardiovascular conditions in UK Biobank: a Mendelian randomization study. Eur Heart J 2020;41:221–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nash D, McClure G, Mastracci TM, Anand SS. Social deprivation and peripheral artery disease. Can J Cardiol 2022;38:612–622. [DOI] [PubMed] [Google Scholar]
- 18. Aday AW, Matsushita K. Epidemiology of peripheral artery disease and polyvascular disease. Circ Res 2021;128:1818–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan S, Titova OE, Damrauer SM, Åkesson A, Larsson SC. Swedish Snuff (snus) dipping, cigarette smoking, and risk of peripheral artery disease: a prospective cohort study. Sci Rep 2022;12:12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bourrier M, Ferguson TW, Embil JM, Rigatto C, Komenda P, Tangri N. Peripheral artery disease: its adverse consequences with and without CKD. Am J Kidney Dis 2020;75:705–712. [DOI] [PubMed] [Google Scholar]
- 21. Fowkes FG, Housley E, Riemersma RA, Macintyre CC, Cawood EH, Prescott RJ, Ruckley CV. Smoking, lipids, glucose intolerance, and blood pressure as risk factors for peripheral atherosclerosis compared with ischemic heart disease in the Edinburgh artery study. Am J Epidemiol 1992;135:331–340. [DOI] [PubMed] [Google Scholar]
- 22. Gao JW, Hao QY, Gao M, Zhang K, Li XZ, Wang JF, Vuitton DA, Zhang SL, Liu PM. Triglyceride-glucose index in the development of peripheral artery disease: findings from the atherosclerosis risk in communities (ARIC) study. Cardiovasc Diabetol 2021;20:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, Loukola A, Lahtela E, Mattsson H, Laiho P, Parolo P Della Briotta, Lehisto AA, Kanai M, Mars N, Rämö J, Kiiskinen T, Heyne HO, Veerapen K, Rüeger S, Lemmelä S, Zhou W, Ruotsalainen S, Pärn K, Hiekkalinna T, Koskelainen S, Paajanen T, Llorens V, Gracia-Tabuenca J, Siirtola H, Reis K, Elnahas AG, Sun B, Foley CN, Aalto-Setälä K, Alasoo K, Arvas M, Auro K, Biswas S, Bizaki-Vallaskangas A, Carpen O, Chen CY, Dada OA, Ding Z, Ehm MG, Eklund K, Färkkilä M, Finucane H, Ganna A, Ghazal A, Graham RR, Green EM, Hakanen A, Hautalahti M, Hedman ÅK, Hiltunen M, Hinttala R, Hovatta I, Hu X, Huertas-Vazquez A, Huilaja L, Hunkapiller J, Jacob H, Jensen JN, Joensuu H, John S, Julkunen V, Jung M, Junttila J, Kaarniranta K, Kähönen M, Kajanne R, Kallio L, Kälviäinen R, Kaprio J, FinnGen, Kerimov N, Kettunen J, Kilpeläinen E, Kilpi T, Klinger K, Kosma VM, Kuopio T, Kurra V, Laisk T, Laukkanen J, Lawless N, Liu A, Longerich S, Mägi R, Mäkelä J, Mäkitie A, Malarstig A, Mannermaa A, Maranville J, Matakidou A, Meretoja T, Mozaffari SV, Niemi MEK, Niemi M, Niiranen T, Donnell CJ O, Obeidat ME, Okafo G, Ollila HM, Palomäki A, Palotie T, Partanen J, Paul DS, Pelkonen M, Pendergrass RK, Petrovski S, Pitkäranta A, Platt A, Pulford D, Punkka E, Pussinen P, Raghavan N, Rahimov F, Rajpal D, Renaud NA, Riley-Gillis B, Rodosthenous R, Saarentaus E, Salminen A, Salminen E, Salomaa V, Schleutker J, Serpi R, Shen HY, Siegel R, Silander K, Siltanen S, Soini S, Soininen H, Sul JH, Tachmazidou I, Tasanen K, Tienari P, Toppila-Salmi S, Tukiainen T, Tuomi T, Turunen JA, Ulirsch JC, Vaura F, Virolainen P, Waring J, Waterworth D, Yang R, Nelis M, Reigo A, Metspalu A, Milani L, Esko T, Fox C, Havulinna AS, Perola M, Ripatti S, Jalanko A, Laitinen T, Mäkelä TP, Plenge R, McCarthy M, Runz H, Daly MJ, Palotie A.. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 2023;613:508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klarin D, Busenkell E, Judy R, Lynch J, Levin M, Haessler J, Aragam K, Chaffin M, Haas M, Lindström S, Assimes TL, Huang J, Min Lee K, Shao Q, Huffman JE, Kabrhel C, Huang Y, Sun YV, Vujkovic M, Saleheen D, Miller DR, Reaven P, DuVall S, Boden WE, Pyarajan S, Reiner AP, Trégouët DA, Henke P, Kooperberg C, Gaziano JM, Concato J, Rader DJ, Cho K, Chang KM, Wilson PWF, Smith NL, O'Donnell CJ, Tsao PS, Kathiresan S, Obi A, Damrauer SM, Natarajan P. Genome-wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat Genet 2019;51:1574–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, Hoover H, Gudmundsdottir V, Horman SR, Aspelund T, Shu L, Trifonov V, Sigurdsson S, Manolescu A, Zhu J, Olafsson Ö, Jakobsdottir J, Lesley SA, To J, Zhang J, Harris TB, Launer LJ, Zhang B, Eiriksdottir G, Yang X, Orth AP, Jennings LL, Gudnason V. Co-regulatory networks of human serum proteins link genetics to disease. Science 2018;361:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yao C, Chen G, Song C, Keefe J, Mendelson M, Huan T, Sun BB, Laser A, Maranville JC, Wu H, Ho JE, Courchesne P, Lyass A, Larson MG, Gieger C, Graumann J, Johnson AD, Danesh J, Runz H, Hwang SJ, Liu C, Butterworth AS, Suhre K, Levy D. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat Commun 2018;9:3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, Sarwath H, Thareja G, Wahl A, DeLisle RK, Gold L, Pezer M, Lauc G, El-Din Selim MA, Mook-Kanamori DO, Al-Dous EK, Mohamoud YA, Malek J, Strauch K, Grallert H, Peters A, Kastenmüller G, Gieger C, Graumann J. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 2017;8:14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Genomic atlas of the human plasma proteome. Nature 2018;558:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Folkersen L, Gustafsson S, Wang Q, Hansen DH, Hedman Å K, Schork A, Page K, Zhernakova DV, Wu Y, Peters J, Eriksson N, Bergen SE, Boutin TS, Bretherick AD, Enroth S, Kalnapenkis A, Gådin JR, Suur BE, Chen Y, Matic L, Gale JD, Lee J, Zhang W, Quazi A, Ala-Korpela M, Choi SH, Claringbould A, Danesh J, Davey Smith G, de Masi F, Elmståhl S, Engström G, Fauman E, Fernandez C, Franke L, Franks PW, Giedraitis V, Haley C, Hamsten A, Ingason A, Johansson Å, Joshi PK, Lind L, Lindgren CM, Lubitz S, Palmer T, Macdonald-Dunlop E, Magnusson M, Melander O, Michaelsson K, Morris AP, Mägi R, Nagle MW, Nilsson PM, Nilsson J, Orho-Melander M, Polasek O, Prins B, Pålsson E, Qi T, Sjögren M, Sundström J, Surendran P, Võsa U, Werge T, Wernersson R, Westra HJ, Yang J, Zhernakova A, Ärnlöv J, Fu J, Smith JG, Esko T, Hayward C, Gyllensten U, Landen M, Siegbahn A, Wilson JF, Wallentin L, Butterworth AS, Holmes MV, Ingelsson E, Mälarstig A. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab 2020;2:1135–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun BB, Chiou J, Traylor M, Benner C, Hsu Y-H, Richardson TG, Surendran P, Mahajan A, Robins C, Vasquez-Grinnell SG, Hou L, Kvikstad EM, Burren OS, Cule M, Davitte J, Ferber KL, Gillies CE, Hedman ÅK, Hu S, Lin T, Mikkilineni R, Pendergrass RK, Pickering C, Prins B, Raj A, Robinson J, Sethi A, Ward LD, Welsh S, Willis CM, Burkitt-Gray L, Black MH, Fauman EB, Howson JMM, Kang HM, McCarthy MI, Melamud E, Nioi P, Petrovski S, Scott RA, Smith EN, Szalma S, Waterworth DM, Mitnaul LJ, Szustakowski JD, Gibson BW, Miller MR, Whelan CD. Genetic regulation of the human plasma proteome in 54,306 UK Biobank participants. bioRxiv 2022;doi: 10.1101/2022.06.17.496443. [DOI] [Google Scholar]
- 31. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res 2017;26:2333–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsushita K, Sang Y, Ning H, Ballew SH, Chow EK, Grams ME, Selvin E, Allison M, Criqui M, Coresh J, Lloyd-Jones DM, Wilkins JT. Lifetime risk of lower-extremity peripheral artery disease defined by ankle-brachial index in the United States. J Am Heart Assoc 2019;8:e012177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luttun A, Lutgens E, Manderveld A, Maris K, Collen D, Carmeliet P, Moons L. Loss of matrix metalloproteinase-9 or matrix metalloproteinase-12 protects apolipoprotein E-deficient mice against atherosclerotic media destruction but differentially affects plaque growth. Circulation 2004;109:1408–1414. [DOI] [PubMed] [Google Scholar]
- 35. Goncalves I, Bengtsson E, Colhoun HM, Shore AC, Palombo C, Natali A, Edsfeldt A, Dunér P, Fredrikson GN, Björkbacka H, Östling G, Aizawa K, Casanova F, Persson M, Gooding K, Strain D, Khan F, Looker HC, Adams F, Belch J, Pinnoli S, Venturi E, Kozakova M, Gan LM, Schnecke V, Nilsson J. Elevated plasma levels of MMP-12 are associated with atherosclerotic burden and symptomatic cardiovascular disease in subjects with type 2 diabetes. Arterioscler Thromb Vasc Biol 2015;35:1723–1731. [DOI] [PubMed] [Google Scholar]
- 36. Lamblin N, Bauters C, Hermant X, Lablanche JM, Helbecque N, Amouyel P. Polymorphisms in the promoter regions of MMP-2, MMP-3, MMP-9 and MMP-12 genes as determinants of aneurysmal coronary artery disease. J Am Coll Cardiol 2002;40:43–48. [DOI] [PubMed] [Google Scholar]
- 37. Stawski L, Haines P, Fine A, Rudnicka L, Trojanowska M. MMP-12 deficiency attenuates angiotensin II-induced vascular injury, M2 macrophage accumulation, and skin and heart fibrosis. PLoS One 2014;9:e109763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnson JL, George SJ, Newby AC, Jackson CL. Divergent effects of matrix metalloproteinases 3, 7, 9, and 12 on atherosclerotic plaque stability in mouse brachiocephalic arteries. Proc Natl Acad Sci U S A 2005;102:15575–15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cárcel-Márquez J, Cullell N, Muiño E, Gallego-Fabrega C, Lledós M, Ibañez L, Krupinski J, Montaner J, Cruchaga C, Lee JM, Gill D, Paré G, Mola-Caminal M, Roquer J, Jimenez-Conde J, Martí-Fàbregas J, Fernandez-Cadenas I. Causal effect of MMP-1 (matrix metalloproteinase-1), MMP-8, and MMP-12 levels on ischemic stroke: a Mendelian randomization study. Stroke 2021;52:e316–e320. [DOI] [PubMed] [Google Scholar]
- 40. Chelluboina B, Nalamolu KR, Klopfenstein JD, Pinson DM, Wang DZ, Vemuganti R, Veeravalli KK. MMP-12, a promising therapeutic target for neurological diseases. Mol Neurobiol 2018;55:1405–1409. [DOI] [PubMed] [Google Scholar]
- 41. Mogadam E, King K, Shriner K, Chu K, Sondergaard A, Young K, Naghavi M, Kloner RA. The association of nadir CD4-T cell count and endothelial dysfunction in a healthy HIV cohort without major cardiovascular risk factors. SAGE Open Med 2020;8:2050312120924892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Helleberg M, Kronborg G, Larsen CS, Pedersen G, Pedersen C, Obel N, Gerstoft J. CD4 Decline is associated with increased risk of cardiovascular disease, cancer, and death in virally suppressed patients with HIV. Clin Infect Dis 2013;57:314–321. [DOI] [PubMed] [Google Scholar]
- 43. Zhou X, Robertson AK, Rudling M, Parini P, Hansson GK. Lesion development and response to immunization reveal a complex role for CD4 in atherosclerosis. Circ Res 2005;96:427–434. [DOI] [PubMed] [Google Scholar]
- 44. Saigusa R, Winkels H, Ley K. T cell subsets and functions in atherosclerosis. Nat Rev Cardiol 2020;17:387–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Echouffo-Tcheugui JB, Daya N, Ndumele CE, Matsushita K, Hoogeveen RC, Ballantyne CM, Coresh J, Shah AM, Selvin E. Diabetes, GDF-15 and incident heart failure: the atherosclerosis risk in communities study. Diabetologia 2022;65:955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Echouffo-Tcheugui JB, Daya N, Matsushita K, Wang D, Ndumele CE, Al Rifai M, Hoogeveen RC, Ballantyne CM, Selvin E. Growth differentiation factor (GDF)-15 and cardiometabolic outcomes among older adults: the atherosclerosis risk in communities study. Clin Chem 2021;67:653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herder C, de Las Heras Gala T, Carstensen-Kirberg M, Huth C, Zierer A, Wahl S, Sudduth-Klinger J, Kuulasmaa K, Peretz D, Ligthart S, Bongaerts BWC, Dehghan A, Ikram MA, Jula A, Kee F, Pietilä A, Saarela O, Zeller T, Blankenberg S, Meisinger C, Peters A, Roden M, Salomaa V, Koenig W, Thorand B. Circulating levels of interleukin 1-receptor antagonist and risk of cardiovascular disease: meta-analysis of six population-based cohorts. Arterioscler Thromb Vasc Biol 2017;37:1222–1227. [DOI] [PubMed] [Google Scholar]
- 48. Merhi-Soussi F, Kwak BR, Magne D, Chadjichristos C, Berti M, Pelli G, James RW, Mach F, Gabay C. Interleukin-1 plays a major role in vascular inflammation and atherosclerosis in male apolipoprotein E-knockout mice. Cardiovasc Res 2005;66:583–593. [DOI] [PubMed] [Google Scholar]
- 49. Laschkolnig A, Kollerits B, Lamina C, Meisinger C, Rantner B, Stadler M, Peters A, Koenig W, Stöckl A, Dähnhardt D, Böger CA, Krämer BK, Fraedrich G, Strauch K, Kronenberg F. Lipoprotein (a) concentrations, apolipoprotein (a) phenotypes, and peripheral arterial disease in three independent cohorts. Cardiovasc Res 2014;103:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified SIMPLER data are available for researchers upon application (http://www.simpler4health.se/). Summary-level data from the FinnGen study of PAD can be obtained via https://finngen.gitbook.io/documentation/. Summary data from the MVP GWAS of PAD can be obtained via dbGAP, accession code no. phs001672.v2.p1.