Abstract
Several unnatural, predominantly hydrophobic nucleobases that pack efficiently in duplex DNA without hydrogen bonding functionalities are reported to circumvent the hydrogen bonding-based specificity, both during oligonucleotide hybridization and enzymatic DNA synthesis. The reported nucleoside analogs are efficient ‘universal bases’ for hybridization, template directed DNA synthesis and chain termination. Moreover, several of the universal bases function in their biological role, hybridization or replication, with an efficiency not significantly reduced relative to their natural counterparts.
INTRODUCTION
DNA duplex stability results from a combination of interstrand hydrogen bonds and intrastrand hydrophobic interactions between nucleobases (1). However, sequence specificity is determined solely by the specific interstrand hydrogen bonding patterns of the natural bases (1). Hydrophobic base analogs without hydrogen bonding groups (2–4) that pack efficiently in duplex DNA can show little selectivity in pairing with native bases (5). Such ‘universal bases’ have attracted much attention due to their potential utility in the design of oligonucleotide primers or hybridization probes where the identity of one or more bases in the target sequence is unknown (6–14). In practice, sequence ambiguities occur frequently due to the degeneracies in the genetic code. In cases where sequence data is available, ambiguities can still remain due to polymorphic or species-dependent sequence differences. Therefore, for any given gene, it would be of value to have an oligonucleotide probe that is capable of selective hybridization even in the presence of polymorphisms.
There has been some success in the design of nucleobase analogs that can hybridize non-selectively to each of the native bases (6–14), but analogs that can universally hybridize without significant duplex destabilization are rare. Deoxyoligonucleotides containing either 3-nitropyrrole or 5-nitroindole bases have received the most attention (6,10–13). These nucleoside analogs pair non-discriminantly opposite each natural base. Duplex DNA containing 5-nitroindole paired opposite each natural base was shown to melt with only a 3°C range of Tm values. Although these universal bases do not strongly discriminate the native bases, their incorporation destabilizes duplex DNA by at least 4.0–7.0°C per analog base. Significant destabilization limits the number of these analogs that can be incorporated in any given oligonucleotide, thus limiting their practical utility.
In addition to a base for universal hybridization, an unnatural base that is non-discriminantly recognized by a DNA polymerase in the template (‘universal template base’) (15–19) or as a triphosphate (‘universal triphosphate’) (20–22) would also be useful for a variety of recombinant DNA techniques, such as library generation, random oligonucleotide labeling and random chain termination. When in the template, the universal nucleobase would direct the polymerase to incorporate each dNTP indiscriminately, and as triphosphate it would be indiscriminately incorporated opposite each native base in the DNA template. The practicality of a universal triphosphate will require that it be randomly incorporated into the priming oligonucleotide in the presence of the natural triphosphates. This competitive insertion will require the unnatural nucleoside triphosphate to be incorporated into DNA with a rate approaching that of native base pair synthesis. A shortcoming of the traditional dideoxy method of chain termination is that four different dideoxynucleotide triphosphates, one corresponding to each base, must be used in great excess to compete with more efficient incorporation of natural nucleoside triphosphates. Excess chain dideoxy terminators are required because they are not incorporated into the growing oligonucleotide strand competitively with the natural substrates. For example, the Klenow fragment of Escherichia coli DNA polymerase I prefers the natural dNTP substrates by several thousand-fold relative to the chain terminator dideoxy analogs (23).
Universal bases for hybridization, replication or random chain termination would be valuable tools for nucleic acid manipulation and analysis, but have proven difficult to design. We have been interested in the design and characterization of unnatural hydrophobic nucleobase analogs which when present in a given biological context (oligonucleotide hybridization or DNA replication) behave universally. Herein we report nucleobase analogs that act as universal bases in each biological context. We describe a base analog capable of pairing stably, but non-specifically, with each natural base. We also describe a base analog which when present in a DNA template directs a DNA polymerase to randomly insert a natural nucleobase triphosphate. Finally, an analog triphosphate is described which is efficiently inserted opposite any natural base in template with a rate approaching that of correct natural triphosphate insertion and subsequently causes chain termination.
MATERIALS AND METHODS
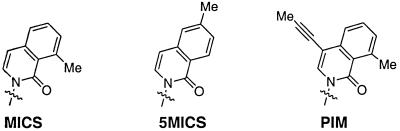
The nucleoside of 3-methyl 7-propynyl isocarbostyril (PIM) (Fig. 1) was synthesized as reported previously (24). Synthesis of the 3-methyl isocarbostyril (MICS), and 5-methyl isocarbostyril (5MICS) nucleosides (Fig. 1) will be reported elsewhere (accepted for publication in Journal of the American Chemical Society). The nucleosides were converted into the triphosphate (3,25) and phosphoramidite (3) by literature methods. The thermal stability of the base pairs was evaluated by determining the melting temperature (Tm) of duplex DNA containing MICS or 5MICS paired with dG, dC, dA or dT (Table 1). The melting experiments were done with 3 µM duplex in 10 mM PIPES, 10 mM MgCl2, 100 mM NaCl, pH 7, using a Cary 300 Bio UV/Vis spectrophotometer. The heating rate was 0.5°C/min between 16 and 80°C. Melting temperatures were obtained by the derivative method utilizing the Cary Win UV thermal application software. MICS and PIM were evaluated as substrates for the exonuclease-deficient Klenow fragment of E.coli DNA polymerase I (KF) purchased from Amersham Pharmacia Biotech (Piscataway, NJ). Initial velocities were determined during extension of a γ-32P-labeled primer with varying concentrations of nucleoside triphosphates (26). The reactions were analyzed by polyacrylamide gel electrophoresis; a Phosphorimager (Molecular Dynamics, Sunnyvale, CA) was used to quantify gel band intensities corresponding to the extended primer. The measured velocities were plotted against the concentration of dNTP and subsequently fitted to the Michaelis–Menten equation. Unnatural nucleobases were assayed both in template DNA and as incoming nucleoside triphosphates. Steady-state kinetic parameters for single incorporation of dGTP, dATP, dTTP or dCTP opposite MICS are reported in Table 2. Steady-state kinetic parameters for single incorporation of dPIMTP opposite dG, dA, dT or dC are reported in Table 3.
Figure 1.
Hydrophobic nucleobases.
Table 1. Tm values for duplex containing MICS and 5MICSa.
| Duplex | N | X | Tm (°C) |
|---|---|---|---|
| 5′-GCGTACXCATGCG | A | MICS | 53.6 |
| 3′-CGCATGNGTACGC | T | MICS | 55.1 |
| C | MICS | 54.8 | |
| G | MICS | 54.8 | |
| MICS | Average = 54.6 ± 0.7°C | ||
| A | 5MICS | 55.7 | |
| T | 5MICS | 55.5 | |
| C | 5MICS | 54.3 | |
| G | 5MICS | 55.5 | |
| 5MICS | Average = 55.3 ± 0.6°C | ||
| 5′-GCGTACNCATGCG | A | MICS | 54.2 |
| 3′-CGCATGXGTACGC | T | MICS | 51.9 |
| C | MICS | 55.2 | |
| G | MICS | 51.2 | |
| MICS | Average = 53.1± 1.9 °C | ||
| A | 5MICS | 54.6 | |
| T | 5MICS | 51.6 | |
| C | 5MICS | 54.2 | |
| G | 5MICS | 49.9 | |
| 5MICS | Average = 52.6± 2.2 °C |
aExperiments were run in triplicate. See text for experimental details.
Table 2. Steady-state kinetic parameters for KF exo–-mediated synthesis of DNA with MICS in the templatea.
| Template (X) | Nucleoside triphosphate | kcat (min–1) | KM (µM) | kcat/KM (M–1 min–1) |
|---|---|---|---|---|
| MICS | dATP | 0.36 ± 0.02 | 15 ± 2 | 2.4 × 104 |
| dGTP | 0.28 ± 0.02 | 48 ± 6 | 5.8 × 103 | |
| dCTP | 0.27 ± 0.03 | 43 ± 15 | 6.3 × 103 | |
| dTTP | 1.6 ± 0.1 | 80 ± 7 | 2.0 × 104 |
5′-dTAATACGACTCACTATAGGGAGA
3′-dATTATGCTGAGTGATATCCCTCTXGTCA
aExperiments were run in triplicate. See text for experimental details.
Table 3. Steady-state kinetic parameters for KF exo–-mediated insertion of dPIMTP opposite dA, dG, dC and dT in the templatea.
| Template (X) | kcat (min–1) | KM (µM) | kcat/KM (M–1 min–1) |
|---|---|---|---|
| dA | 5.3 ± 0.2 | 3.1 ± 0.5 | 1.7 × 106 |
| dG | 1.33 ± 0.02 | 4.1 ± 0.3 | 3.2 × 105 |
| dC | 3.2 ± 0.2 | 4.8 ± 0.6 | 6.7 × 105 |
| dT | 11.0 ± 0.5 | 6.6 ± 1.0 | 1.7 × 106 |
5′-dTAATACGACTCACTATAGGGAGA
3′-dATTATGCTGAGTGATATCCCTCTXGTCA
aExperiments were run in triplicate. dNTP concentrations were varied from 0.1 to 100 µM. See text for experimental details.
RESULTS
We have investigated a wide variety of unnatural hydrophobic nucleobases that do not have hydrogen bonding functionality (5,24,27). We systematically evaluated the stability of duplex DNAs containing these unnatural bases paired opposite native bases. Several of the unnatural bases show little thermal discrimination against the different natural bases and do not compromise duplex stability. In addition, we have evaluated the ability of several polymerases to synthesize DNA containing the unnatural bases both in template and as triphosphates. Several of these base analogs are efficiently, but non-discriminantly, accepted as DNA polymerase substrates.
Universal bases for oligonucleotide hybridization
In addition to interstrand hydrogen bonding between pairing bases, duplex DNA is stabilized by intrastrand base interactions, including dipole–dipole interactions as well as nucleobase polarizability (28–31). An unnatural nucleobase, lacking hydrogen bond donor or acceptor functionalities, which can sufficiently stabilize duplex DNA by intrastand base interactions may act as a universal base, without destabilizing the duplex. A universal base must also pair indiscriminately in any sequence context. For example, there are large effects on natural and unnatural base pair stability when the flanking bases are changed from purines to pyrimidines and the universal bases must behave indiscriminately in each sequence context.
The MICS and 5MICS nucleobases (Fig. 1) have polarizable N-glycosidic linkages with a hydrophilic minor groove carbonyl group and are expected to present a large and polarizable hydrophobic surface for intrabase packing. The data in Table 1 demonstrates that these predominantly hydrophobic bases form stable base pairs with each of the natural bases when incorporated into the central position of either oligodeoxyribonucleotide strand of a 13mer duplex (Table 1). When incorporated into one oligonucleotide strand and paired opposite each of the four natural bases, the average Tm values for MICS and 5MICS are 54.6 ± 0.7 and 55.3 ± 0.6°C, respectively (Table 1). This compares favorably with the stability of the duplex with a native dT·dA pair (Tm = 58.7°C). The spread in measured Tm values for MICS and 5MICS opposite each natural base is the same (1.4°C). A single MICS or 5MICS paired opposite any natural base is significantly more stable than a mispair in the same sequence context (ΔTm = 3.8–17°C for mispairs among the native bases in the same sequence context; data not shown). These nucleoside analogs are both more universal and less destabilizing than any universal base reported in the literature. For example, in a different sequence context, 5-nitroindole destabilized duplex DNA by 4.0–7.0°C (12,13). When MICS and 5MICS were examined in the opposite strand, position Y, paired opposite natural bases, the measured ΔTm values were slightly larger (ΔTm = 1.9 and 2.2°C for MICS and 5MICS, respectively). In this sequence context, the stability of the MICS and 5MICS base pairs with dT and dG are lower than the pairs with dA and dC, which fall into the same range as that observed in the opposite strand context. This illustrates the sensitivity of the universal behavior to sequence context.
A universal template nucleobase
Few studies have been reported that describe universal bases for DNA replication (15–19). For example, no base analog is known that universally templates the incorporation of each natural nucleoside triphosphate. Such bases would have important applications. The ability to synthesize DNA with mutations focused to certain regions of a gene might provide alternatives to the currently available, but arduous, techniques of creating genetic diversity, such as error-prone PCR or gene shuffling. Mutations might be introduced by including a base analog in the template, which directs a polymerase to incorporate each of the native triphosphates indiscriminately.
We have identified a variety of predominantly hydrophobic nucleobases that efficiently direct KF to incorporate native triphosphates. Despite a compromise in absolute rate, when present in template MICS directs the insertion of each native triphosphate with approximately equal efficiency (Table 2). Each natural triphosphate is inserted by KF opposite MICS with an efficiency between 5.8 × 103 and 2.4 × 104 M–1 min–1, with dATP and dTTP inserted slightly faster than dCTP and dGTP. KF inserts each native triphosphate opposite MICS with only a 4-fold variation in efficiency, and with high concentrations of dNTPs KF synthesizes full-length DNA (Fig. 2). The additional major band seen in Figure 2 results from single nucleotide extension of the primer through incorporation of a native triphosphate opposite MICS in the template. The accumulation of this intermediate implies that the continued synthesis after incorporation opposite MICS is slow. The ability of MICS to direct KF to insert triphosphates with no bias demonstrates that predominantly hydrophobic bases are capable of bypassing the nucleobase hydrogen bonding specificities during DNA replication.
Figure 2.
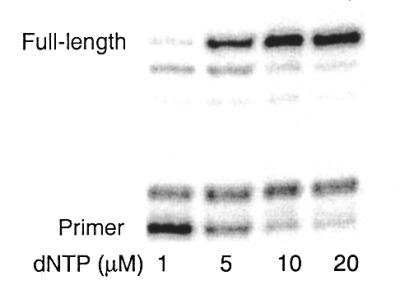
Full-length synthesis past MICS in the template. Assay conditions were as follows: 40 nM DNA duplex, 24 nM KF exo– and 1–20 µM dNTPs. Reactions were incubated at room temperature for 60 min.
A universal nucleobase triphosphate chain terminator
Several predominantly hydrophobic nucleobase triphosphates that are largely incapable of forming hydrogen bonds have been shown to be surprisingly good substrates for KF (5,27,32,33). These hydrophobic bases bear no obvious shape complementarity to the native bases, but are incorporated with rates that in some cases are equal to those observed for correct pair synthesis. We recently reported the synthesis and thermal characterization of the unnatural nucleobase PIM (Fig. 1; 24). We were interested in examining PIM as a substrate for KF. Steady-state kinetic experiments were conducted with exonuclease-deficient KF (26) and the rates for the single nucleotide primer extension are reported in Table 3. The universal behavior of dPIMTP in this sequence context is evident from the relative efficiencies (kcat/KM) for incorporation opposite each natural base (Table 3), which vary by only 5-fold. The insertion of dPIMTP opposite dA and dT is slightly more efficient than insertion opposite dC or dG. Remarkably, the average rate of dPIMTP insertion is only 40-fold reduced relative to the insertion of dTTP opposite dA (kcat/KM = 4.7 × 107 M–1 min–1) in the same sequence context.
After incorporation of dPIMTP, KF is unable to continue DNA synthesis. Even under forcing conditions (0.5 h incubation time and 1 mM dNTPs) no band corresponding to extension of the PIM-terminated primer was detectable. Therefore, PIM inserts efficiently, but randomly into the growing oligonucleotide strand and then terminates synthesis. The average length of synthesized oligonucleotide may be tuned by choice of chain terminator concentration (Fig. 3). The efficient insertion of dPIMTP opposite any native template results in the efficient generation of random length oligonucleotides, with low concentrations of a single chain terminator. This strongly contrasts with the traditional method of dideoxy based termination, as discussed above. In this regard, dPIMTP may have practical applications, for example in the generation of random length oligonucleotides for sequencing by mass spectrometry (34–36).
Figure 3.
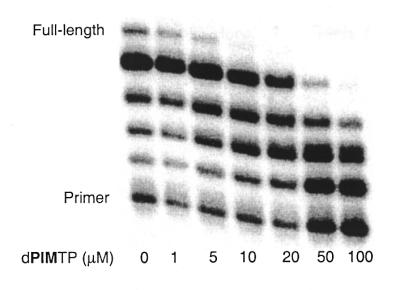
Random chain termination with dPIMTP. Assay conditions were as follows: 40 nM DNA duplex, 1.34 nM KF exo–, 0–100 µM dPIMTP and 1 µM dNTPs. Reactions were incubated at room temperature for 1 min.
This study demonstrates that hydrophobicity may act as an ambiguous but sufficiently strong force to mediate the interbase interactions involved in hybridization and replication. We are currently exploring derivatives of these bases, which may be even more universal with regard to kinetic or thermodynamic behavior. The ability of oligonucleotides to form relatively stable duplexes with a variety of predominantly hydrophobic bases paired opposite native bases as well as the remarkable ability of DNA polymerases to accept unnatural bases bearing little or no hydrogen bond or shape complementarity to native bases demonstrates the potential of the approach. Moreover, the hydrophobicity-based approach is not limited to the isocarbostyril ring structure common to PIM, MICS and 5MICS and we are continuing to examine alternative scaffoldings. Experiments are underway to evaluate the ability of oligonucleotides containing multiple MICS or 5MICS substitutions to hybridize to target sequences with multiple degeneracies. We are also evaluating the utility of oligonucleotides containing these base analogs in PCR-based applications.
Acknowledgments
ACKNOWLEDGEMENTS
Funding was provided by the National Institutes of Health (GM 60005 to F.E.R.) and the Skaggs Institute for Chemical Biology (F.E.R. and P.G.S.) and a National Institutes of Health post-doctoral fellowship (F32 GM19833-01 to A.K.O.)
REFERENCES
- 1.Kornberg A. and Baker,T.A. (1992) DNA Replication, 2nd Edn. W.H. Freeman and Co., New York, NY.
- 2.Millican T.A., Mock,G.A., Chauncey,M.A., Patel,T.P., Eaton,M.A., Gunning,J., Cutbush,S.D., Neidle,S. and Mann,J. (1984) Nucleic Acids Res., 12, 7435–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweitzer B.A. and Kool,E.T. (1995) J. Am. Chem. Soc., 117, 1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matray T.J. and Kool,E.T. (1999) Nature, 399, 704–708. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa A.K., Wu,Y., McMinn,D.L., Liu,J., Schultz,P.G. and Romesberg,F.E. (2000) J. Am. Chem. Soc., 122, 3274–3287. [Google Scholar]
- 6.Van Aerschot A., Rozenski,J., Loakes,D., Pillet,N., Schepers,G. and Herdewijn,P. (1995) Nucleic Acids Res., 23, 4363–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang P., Johnson,W.T., Klewer,D., Paul,N., Hoops,G., Davisson,V.J. and Bergstrom,D.E. (1998) Nucleic Acids Res., 26, 2208–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seela F. and Mittelbach,C. (1999) Nucleosides Nucleotides, 18, 425–441. [Google Scholar]
- 9.Bergstrom D.E., Zhang,P. and Johnson,W.T. (1997) Nucleic Acids Res., 25, 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amosova O., George,J. and Fresco,J.R. (1997) Nucleic Acids Res., 25, 1930–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergstrom D.E., Zhang,P., Toma,P.H., Andrews,P.C. and Nichols,R. (1995) J. Am. Chem. Soc., 117, 1201–1209. [Google Scholar]
- 12.Loakes D., Brown,D.M., Linde,S. and Hill,F. (1995) Nucleic Acids Res., 23, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loakes D. and Brown,D.M. (1994) Nucleic Acids Res., 22, 4039–4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichols R., Andrews,P.C., Zhang,P. and Bergstrom,D.E. (1994) Nature, 369, 492–493. [DOI] [PubMed] [Google Scholar]
- 15.Hill F., Loakes,D. and Brown,D.M. (1997) Nucleosides Nucleotides, 16, 1507–1511. [Google Scholar]
- 16.Hoops G.C., Zhang,P., Johnson,W.T., Paul,N., Bergstrom,D.E. and Davisson,V.J. (1997) Nucleic Acids Res., 25, 4866–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamiya H., Murata-Kamiya,N., Kong Thoo Lin,P., Brown,D.M. and Ohtsuka,E. (1994) Nucleosides Nucleotides, 13, 1483–1492. [Google Scholar]
- 18.Hill F., Loakes,D. and Brown,D.M. (1998) Proc. Natl Acad. Sci. USA, 95, 4258–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loakes D., Van Aerschot,A., Brown,D.M. and Hill,F. (1996) Nucleosides Nucleotides, 15, 1891–1904. [Google Scholar]
- 20.Sala M., Pezo,V., Pochet,S. and Wain-Hobson,S. (1996) Nucleic Acids Res., 24, 3302–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beaussire J.-J. and Pochet,S. (1999) Nucleosides Nucleotides, 18, 403–410. [Google Scholar]
- 22.Smith C.L., Simmonds,A.C., Felix,I.R., Hamilton,A.L., Kumar,S., Nampalli,S., Loakes,D., Hill,F. and Brown,D.M. (1998) Nucleosides Nucleotides, 17, 541–554. [Google Scholar]
- 23.Astatke M., Grindley,N.D.F. and Joyce,C.M. (1998) J. Mol. Biol., 278, 147–165. [DOI] [PubMed] [Google Scholar]
- 24.Berger M., Ogawa,A.K., McMinn,D.L., Wu,Y., Schultz,P.G. and Romesberg,F.E. (2000) Angew. Chem. Int. Ed. Engl., in press. [DOI] [PubMed] [Google Scholar]
- 25.Kovacs T. and Otvos,L. (1988) Tetrahedron Lett., 29, 4525–4528. [Google Scholar]
- 26.Goodman M.F., Creighton,S., Bloom,L.B. and Petruska,J. (1993) Crit. Rev. Biochem. Mol. Biol., 28, 83–126. [DOI] [PubMed] [Google Scholar]
- 27.McMinn D.L., Ogawa,A.K., Wu,Y., Liu,J., Schultz,P.G. and Romesberg,F.E. (1999) J. Am. Chem. Soc., 121, 11585–11586. [Google Scholar]
- 28.Bergman E.D. and Weiler-Feilchenfeld. (1971) In Bergman,E.D. and Pullman,B. (eds), The Purines. Theory and Experiment. The Israel Academy of Sciences and Humanities, Jerusalem, Israel, pp. 21–28.
- 29.Lister J.H. (1996) In Taylor,E.C. (ed.), The Purines. Supplement 1. The Chemistry of Heterocyclic Compounds. John Wiley & Sons, New York, NY, p. 54.
- 30.Hurst D.T. (1980) An Introduction to the Chemistry and Biochemistry of Pyrimidines, Purines and Pteridines. Page Bros, Norwich, UK.
- 31.Bugg C.E. (1971) In Bergmann,E.D. and Pullman,B. (eds), The Purines. Theory and Experiment. The Israel Academy of Sciences and Humanities, Jerusalem, Israel, pp. 178–204.
- 32.Morales J.C. and Kool,E.T. (1998) Nature Struct. Biol., 5, 950–954. [DOI] [PubMed] [Google Scholar]
- 33.Kool E.T. (1998) Biopolymers, 48, 3–17. [DOI] [PubMed] [Google Scholar]
- 34.Griffin T.J., Tang,W. and Smith,L.M. (1997) Nature Biotechnol., 15, 1368–1372. [DOI] [PubMed] [Google Scholar]
- 35.Gut I.G. and Beck,S. (1995) Nucleic Acids Res., 23, 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray K.K. (1996) J. Mass Spectrom., 31, 1203–1215. [DOI] [PubMed] [Google Scholar]