Abstract
Background
Studies have proven that as competing endogenous RNAs (ceRNAs), long non-coding RNAs (lncRNAs) play vital roles in regulating RNA transcripts in ischemic stroke. It has been reported that TTTY15, a lncRNA, is dysregulated in cardiomyocytes after ischemic injury. We intended to explore the potential regulating mechanism of TTTY15 in ischemic stroke.
Methods
TTTY15 and miR-520a-3p levels in vivo were measured in the cerebral ischemia/reperfusion (I/R) model. Cell apoptosis was measured by flow cytometry. To manifest TTTY15 functions in I/R injury, Neuro 2a (N2a) cells were exposed to oxygen-glucose deprivation/reoxygenation (OGD/R) and treated with si-NC, pcDNA3.1-NC, si-TTTY15 or pcDNA3.1-TTTY15.
Results
TTTY15 expression was elevated and miR-520a-3p expression was declined in mouse brains exposed to I/R and in N2a cells exposed to OGD/R. Bioinformatics analyses predicted the binding sites of miR-520a-3p in the 3′-UTRs of interferon regulatory factor 9 (IRF9) and TTTY15. Luciferase reporter assay exhibited that TTTY15 bound to miR-520a-3p directly and IRF9 was targeted by miR-520a-3p. MiR-520a-3p overexpression diminished N2a cell apoptosis caused by OGD/R. TTTY15 overexpression antagonized the inhibitory impacts of miR-520a-3p on IRF9 expression and apoptosis after OGD/R, while TTTY15 knockdown enhanced the inhibitory impacts of miR-520a-3p. Additionally, TTTY15 knockdown alleviated brain damages and neurological deficits induced by I/R in vivo. Our results revealed that TTTY15 modulated IRF9 via acting as a ceRNA for miR-520a-3p.
Conclusion
The study revealed the roles of TTTY15/miR-520a-3p/IRF9 signaling pathway in regulating cerebral ischemia/reperfusion injury.
Keywords: TTTY15, miR-520a-3p, IRF9, Apoptosis, Cerebral ischemia/reperfusion
At a glance commentary
Scientific background on the subject
Recently, long non-coding RNAs (lncRNAs) are considered as critical regulators in pathogenesis progression of ischemic stroke. The present study aimed to explore the potential effect of lncRNA TTTY15 and to investigate the underlying mechanism in ischemia/reperfusion (I/R) mode and N2a cells.
What this study adds to the field
We demonstrate TTTY15 targets miR-520a-3p, which accelerates cerebral ischemia/reperfusion injury. Our findings provide molecular mechanism by which TTTY15 enhanced cerebral I/R injury and facilitate development of therapeautical strategies for treating ischemic injury.
Introduction
Ischemic stroke is the most common life-threatening neurological disease and the major cause of serious long-term disability due to severe neural cell injuries [1,2]. Numerous pathological mechanisms are involved in the development of cerebral ischemia stroke. For example, diverse apoptosis initiation and regulation pathways are triggered in stroke, leading to cell death and brain disorders [3,4]. In parallel, researchers have focused on the mechanisms of excitotoxicity, oxidative and nitrosative stress, and inflammation induced by ischemic stroke [5]. Accordingly, it is of great importance to investigate novel regulatory factors to diminish neural injuries induced by ischemic stroke.
Long Non-coding (lncRNAs, >200 nt) have been widely considered crucial players in physiological processes in organisms and pathological progression in diseases [6]. Additionally, studies have proven that lncRNAs are vital factors in regulating neural apoptosis of many brain diseases, including ischemic stroke [7]. For example, lncRNA-N1LR facilitates cell cycle progression and proliferation and reduces cell apoptosis by inhibiting p53 phosphorylation after ischemic brain injuries [8]. The small nucleolar RNA host gene 1 (SNHG1) enhances brain microvascular endothelial cell (BMEC) survival as a ceRNA for miR-18a and regulates hypoxia inducible factor-1α (HIF-1α) [9]. LncRNA testis-specific transcript Y-linked 15 (TTTY15) is increased in human umbilical vein endothelial cells (HUVECs) subjected to hypoxia [10]. TTTY15 regulates cardiomyocyte apoptosis after ischemic injuries [11], but whether TTTY15 participates in ischemia stroke is still obscure.
MicroRNAs (miRNAs), a category of non-coding RNAs with lengths of approximately 20–22 nt play critical roles in ischemia/reperfusion (I/R)-induced cerebral damages [[12], [13], [14]]. For instance, miR-124 injection enhance neuronal survival and anti-inflammatory consequence [15]. MiR-223–5p inhibition improves ischemic injuries and ameliorates neurological outcomes by preventing the K+-dependent Na+/Ca2+ exchanger (NCKX2) downregulation after ischemia in rats [16]. MiRNA-520a-3p regulates cell proliferation, apoptosis, invasion, and migration in numerous cancers, such as non-small cell lung cancer (NSCLC) and papillary thyroid carcinoma (PTC) [17,18]. However, it remains to be determined whether miRNA-520a-3p is involved in ischemic stroke.
Interferon (IFN) regulatory factor 9 (IRF9) is a member of interferon regulatory factors (IRFs), which constitute nine members (IRF1-9) and are first identified as transcriptional factors of IFNs and IFN inducible genes [19,20]. Several studies have demonstrated that IRF9 participates in pathophysiological events in the central nervous system [21,22]. In addition, studies have verified the pro-neuronal death effect of IRF9. For instance, IRF9 knockdown protects the heart against cardiomyocyte death caused by I/R by ameliorating the downstream apoptosis-related signaling cascade of the histone deacetylase Sirt1 [23]. However, whether the effects of IRF9 on neural apoptosis could be regulated by ncRNAs, such as lncRNAs or miRNAs, is unclear, especially in cerebra ischemia injury.
Recently, noncoding RNAs, including lncRNAs and miRNAs, are considered therapeutic targets or diagnostic biomarkers of ischemic stroke-associated brain disorders due to their regulatory functions on cerebral I/R-induced cell damages [24]. To systematically demonstrate whether TTTY15, miRNA-520a-3p, and IRF9 participate in the neural damages following ischemia stroke, in this study, we detected the function of TTTY15/MiRNA-520a-3p/IRF9 in mice model and in N2a cells exposed to I/R or OGD/R. Furthermore, the potential regulation network among TTTY15, miRNA-520a-3p, and IRF9 were also investigated.
Materials and methods
Focal cerebral I/R
Adult C57BL/6 J male mice 8–10 weeks of age were from the Laboratory Animal Center of Harbin Medical University (Harbin, China) and housed with free access to food and water in a humidity-controlled room at 25 °C ± 1 °C under 12:12 h light/dark cycle. Focal cerebral ischemia was constructed with the middle cerebral artery occlusion (MCAO). In brief, under anesthesia with pentobarbital (30 mg/kg), mice were positioned on a heating panel at 37.0 °C during the whole procedure. Neck vessels were exposed, and branches of the right external carotid artery were carefully isolated. Then, a nylon suture blunted by silicon-coated tip was inserted into the internal carotid to occlude the origin of the middle cerebral artery. Laser Doppler flowmetry (Moor Instruments, Ltd; Devon, England) was utilized for confirming regional ischemia and reperfusion. After 24 h of reperfusion, mice were decapitated. For the control group, a sham operation without MCAO was performed.
Cell culture
Neuro-2a (N2a) cells were purchased from the National Platform of Experimental Cell Resource for Sci-Tech (Beijing, China) and cultured in DMEM (Gibco) supplemented with 1% penicillin (Gibco), 1% streptomycin (Gibco), and 10% FBS (Gibco) at 37 °C in a humidified incubator with 5% CO2. A N2a cell injury model was constructed by OGD/R. Cells were incubated in glucose-free Earle's balanced salt solution (EBSS; Life Technologies, Carlsbad, CA, USA) in a hypoxic chamber (95% N2 and 5% CO2) at 37 °C for 4 h and then in regular DMEM medium under normoxic culture conditions for 12 h for recovery. Control cells were cultured in EBSS containing glucose for 4 h and regular DMEM medium under normoxic culture conditions for 12 h.
RNA extraction and qRT-PCR
The RNAs were extracted with Trizol reagent (TakaRa Biotechnology Co., Ltd., Dalian, China) and reverse transcribed into cDNAs using M-MLV reverse transcriptase (Life Technologies) with oligo (dT) as primers. The levels of TTTY15 and IRF9 were measured with SYBR Green Real-Time PCR MM (Sigma–Aldrich, MO, USA) and normalized to GAPDH. MiRNAs were extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany) and converted to cDNAs using TaqMan MicroRNA Reverse Transcription Kit (Invitrogen, Carlsbad, CA, USA). MiR-520a-3p levels were measured with TaqMan miRNA assay kit (Invitrogen, Carlsbad, CA, USA) and normalized to U6 snRNA. Primer sequences are listed in Table 1. PCR amplifications were performed on 7900HT Fast RealTime PCR machine.
Table 1.
Sequences of primers used in qRT-PCR.
| Gene | Forward primer (5′-3′) | Reversed primer (5′-3′) |
|---|---|---|
| miR-518 b | CAAAGCGCTCCCCTTTAGAGGT | AACGCTTCACGAATTTGCGT |
| miR-1286 | TGCAGGACCAAGATGAGCCCT | GCGAGCACAGAATTAATACGAC |
| miR-382–5p | ATCCGTGAAGTTGTTCGTGG | TATGGTTGTAGAGGACTCCTTGAC |
| miR-942–5p | GCGCGCTCTTCTCTGTTTTGGC | GTGCAGGGTCCGAGGT |
| miR-494–3p | ATGAGGCTTCAGTACTTTACAG | CATAGCGTAAAAGGAGCAACA |
| miR-520a-3p | GCCACCACCATCAGCCATAC | GCACATTACTCTACTCAGAAGGG |
| miR-323a-3p | CACATTACACGGTCGACCT | AACTCAAGGTTCTTCCAGTCACG |
| U6 | CTCGCTTCGGCAGCACA | ACGCTTCACGAATTTGCGT |
| TTTY15 | TCTATGACCTGGAAGC | ATCTGATGGAACCCTA |
| IRF9 | ACAACTGAGGCCACCATTAGAGA | CACCACTCGGCCACCATAG |
| GAPDH | ACAACTTTGGTATCGTGGAAGG | GCCATCACGCCACAGTTTC |
Western blot analysis
The proteins from different groups were extracted with RIPA buffer plus containing protease inhibitors, separated by SDS-PAGE, and transferred onto PVDF membranes (BioRad, Hercules, CA). The membranes were incubated with the primary antibodies against GAPDH (1:1000, Invitrogen), cleaved caspase-3 (C-caspase-3) (1:500, Thermo), cleaved PARP (C-PARP) (1:500, Thermo), Bcl-2 (1:500, Invitrogen), Bax (1:500, Abcam, Cambridge, MA, USA), and IRF9 (1:500, Abcam) 12 h at 4 °C. After blocking for 1 h in 5% nonfat milk, the membranes were incubated with secondary antibodies at room temperature for 1 h and washed with Tris-buffered saline with Tween 20 (TBST). The signals were visualized using enhanced chemiluminescence (ECL) reagent and assessed with Image J Software 1.41.
Luciferase reporter assay
The binding sites of miR-520a-3p in IRF9 and TTTY15 were forecasted using software starBase v.3.0 and TargetScan v.7.2. A wild-type (WT) TTTY15 sequence, a mutant (MT) TTTY15 sequence, an IRF9 3′-UTR fragment (WT), and an IRF9 3′-UTR fragment (MT) including the supposed binding sites of miR-520a-3p were synthesized using Quick Change Lightning Multi Site-Directed Mutagenesis kit (Agilent Technologies, Palo Alto, CA, USA). Mutations in candidate genes of interest were validated by sequencing. After that, constructs were inserted into pmirGLO vector (Abnova, Taipei, Taiwan) to produce TTTY15-WT, IRF9-WT, TTTY15-MT, and IRF9-MT luciferase reporters. Subsequently, cells were cultured in 24-well plates and transfected with NC mimic, miR-520a-3p mimic, NC inhibitor, miR-520a-3p inhibitor, and luciferase reporter plasmids in combination or alone for 48 h. The luciferase activity was evaluated using dual-luciferase reporter assay (Promega, Madison, WI, USA) and normalized to renilla luciferase internal control.
RNA immunoprecipitation (RIP) assay
Cells were washed with cold PBS after 48 h transfection with miR-520a-3p mimic or NC mimic and then lysed with RIP lysis buffer (Amresco, Cochran, NY, USA). The cell lysates were treated with magnetic beads with mouse IgG (negative control; ThermoFisher, Waltham, MA, USA) or anti-Argonaute2 (AGO2) antibody (ThermoFisher). The beads were successively collected and washed. With the existence of proteinase K, RNAs were extracted. TTTY15 was measured using qRT-PCR.
Plasmid construction
To increase TTTY15 expression, mouse TTTY15 cDNA was inserted into pcDNA3.1 (+) mammalian expression vector (Life Technologies) between KpnI and XhoI sites to generate the pcDNA3.1-TTTY15 plasmid. Plasmids were isolated using DNA Midiprep kit (Qiagen, Germany). Cells transfected with empty pcDNA3.1 (+) vector (pcDNA3.1-NC) were used as the negative control. Cells transfected with pcDNA3.1-TTTY15 were picked with G418 (geneticin). The si-TTTY15 and si-NC were synthesized by GenePharma (Shanghai, China) for TTTY15 knockdown. The scrambled miRNAs (NC mimic), miR-520a-3p mimic, the scrambled miRNA (NC inhibitor), and miR-520a-3p inhibitor were synthesized (GenePharma, Shanghai, China) for miR-520a-3p overexpression and knockdown. The transfections were performed using Lipofectamine 2000 reagent (Life Technologies) following the manufacturer's protocol.
Cell apoptosis measurement
Cellular apoptosis was measured using the TUNEL assay kit (Abcam). Brain sections were fixed in 4% paraformaldehyde for 15 min at room temperature, washed using PBS and post-fixed at 4 °C for 5min in paraformaldehyde plus acetic acid before washed with PBS. The sections were then labeled with working strength TdT in equilibration buffer for 1 h at 37 °C. Negative controls were treated without TdT in the TUNEL reaction mixture. The percentage of TUNEL-positive cells was utilized for data presentation.
Apoptosis of cells after different treatments was measured using the Hoechst 33,258 staining kit (Sigma–Aldrich) following the manufacturer's instruction. In short, cells were fixed at room temperature with 4% paraformaldehyde for 15 min. After that, they were washed with PBS twice and stained with Hoechst 33,258 staining solution in the dark at room temperature for 10 min. Fluorescent images of the cells were visualized under a fluorescence microscope. The percentage of positive cells was used to indicate apoptotic cell numbers. Flow cytometry analysis was conducted using the flow cytometry kit (Sigma–Aldrich) following the instruction. In brief, cells were prepared as 100 cells/ml suspension and incubated with Annexin V and propidium iodide for 15 min at room temperature in the dark. Afterward, cells were analyzed on a flow cytometer, and cell cycle profile and apoptosis rate were detected using CellQuest Pro software.
Stereotaxic injection
Stereotaxic injection was operated under anesthesia. Briefly, mice were fixed on stereotaxic apparatus (Agilent Technologies) after anesthetization. A mixture of lentivirus si-NC or si-TTTY15 (5 μl) and RNAiMAX transfection reagent (ThermoFisher) were prepared and injected into the lateral ventricle (1.5 mm below brain surface, 0.2 mm posterior to bregma and 1.0 mm lateral to the midline) of mice 24 h prior to MCAO.
Infarct size and neurological deficit
Brain slices of 2-μm thickness were fixed using 10% formalin and stained with TTC for 30 min at 37 °C. Brains with subarachnoid hemorrhage or clot formation were not used. The slices were photographed, and the infarct volume was quantified.
After 24 h reperfusion, neurological status was assessed and scored as 0 for no deficit, 1 for difficulty to extend the contralateral forelimb and contralateral torso, 2 for flexion of forelimb and contralateral torso, 3 for circling to contralateral side mildly when holding the tail with feet on the floor, 4 for circling to contralateral side severely when holding the tail with feet on the floor and 5 for no spontaneous motor activity.
Statistical analysis
Quantitative data were analyzed using software SPSS 20.0. All data were presented as means ± standard deviation (SD) of at least three independent experiments. The statistical difference between two groups was analyzed using Wilcoxon rank sum test, and the difference among multiple groups was analyzed using Kruskal–Wallis followed by Dunn post hoc procedure. A p < 0.05 was considered significant statistically.
Results
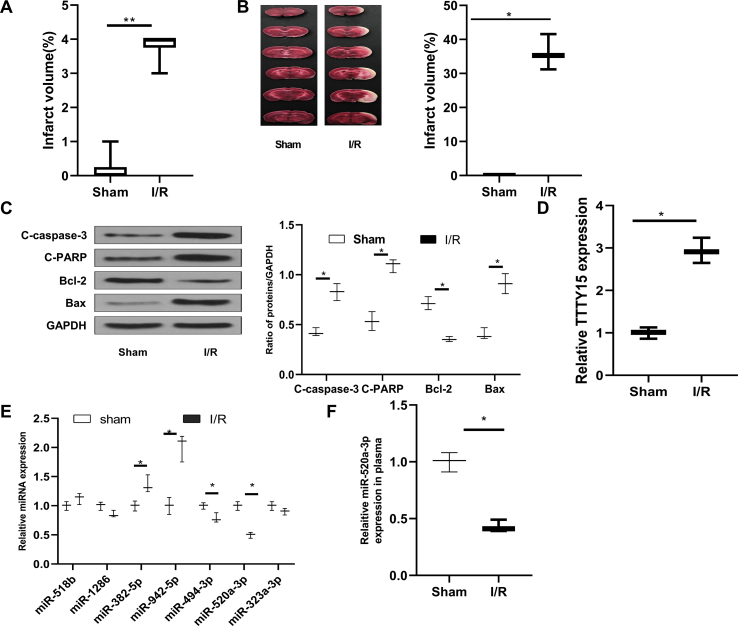
Increased TTTY15 and decreased miR-520a-3p after I/R in vivo
Significant neurologic dysfunction [Fig. 1A] and remarkable cerebral infarct volume [Fig. 1B] occurred after I/R (2 h/24 h) administration (p < 0.01). In addition, decreased Bcl-2 level and increased C-caspase-3, C-PARP, and Bax levels were also found in the brain (p < 0.01) [Fig. 1C], indicating brain cells underwent apoptosis. Moreover, TTTY15 level was upregulated following I/R administration in vivo (p < 0.01) [Fig. 1D]. Furthermore, we applied bioinformatics analysis to identify the potential TTTY15 targeting miRNAs, an important group of lncRNAs [Fig. 1E] and performed qRT-PCR. The results showed that miR-520a-3p expression was obviously reduced in brain tissues (p < 0.01) [Fig. 1E] and plasma of mice after I/R injury (p < 0.01) [Fig. 1F]. The results indicated that miR-520a-3p might be a critical target of TTTY15.
Fig. 1.
Increased TTTY15 and decreased miR-520a-3p in mice exposed to neuronal I/R in vivo. Mice (C57BL/6 J, male) experienced I/R (2 h/24 h) or sham procedure. (A) Neurobehavioral outcomes. (B) TTC staining detected infarct volume. (C) Western blot analysis. (D) TTTY15 level. (E) Level of miRNAs which were forecasted by starBase v.2.0 software. (G) Plasma miR-520a-3p expression. ∗p < 0.05, ∗∗p < 0.01. All data were exhibited as mean ± SD. n = 6.
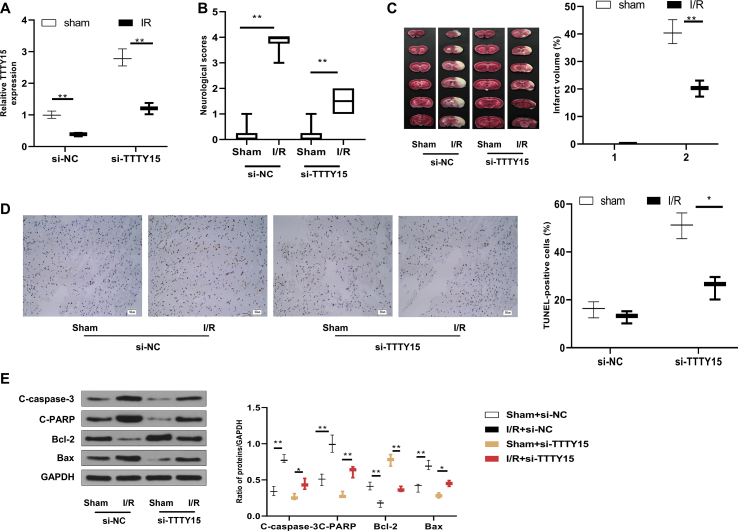
TTTY15 knockdown protects against I/R injury
We further explored the function of TTTY15 after cerebral I/R injury in vivo. Consistently, TTTY15 was upregulated in the brain of mice exposed to I/R (2 h/24 h) (p < 0.01) [Fig. 2A]. TTTY15 knockdown was achieved by injecting si-TTTY15 (p < 0.01) [Fig. 2A]. Furthermore, TTTY15 knockdown decreased neurological deficits [Fig. 2B], cerebral infarction [Fig. 2C], and TUNEL-positive cells [Fig. 2D] and reversed the expression of apoptosis-related proteins [Fig. 2E] in mice subjected to I/R (p < 0.05).
Fig. 2.
TTTY15 knockdown prevents I/R injury in vivo. si-TTTY15 or si-NC was injected operated in the lateral ventricle of mice 24 h before exposure to I/R (2 h/24 h) or sham operation. Brains were obtained for investigation. (A) Cerebral TTTY15 expression was measured by qRT-PCR. (B) Neurological deficit score. (C) TTC staining and infarct volume in brain. (D) Cell apoptosis in the cerebral cortex was analyzed with TUNEL staining ( × 200, bar = 50 μm). (E) Apoptosis-related proteins were detected using Western blot. ∗p < 0.05, ∗∗p < 0.01. Data were exhibited as mean ± SD. n = 6.
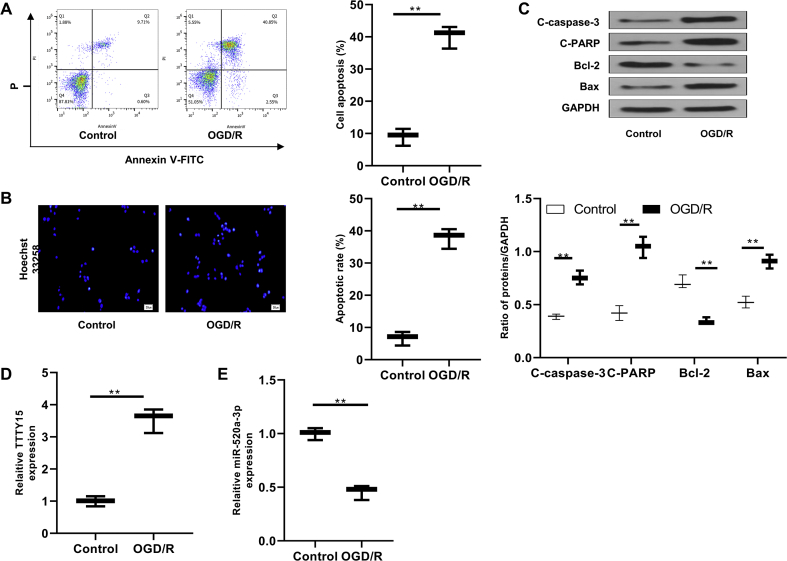
Increased TTTY15 and decreased miR-520a-3p in cells exposed to OGD/R
N2a cells exposed OGD/R (2 h/24 h) procedure in vitro underwent significant apoptosis as indicated by flow cytometry analysis [Fig. 3A], Hoechst 33,258 staining [Fig. 3B], and Western blot of apoptosis-related proteins (p < 0.01) [Fig. 3C]. Further qRT-PCR analyses showed that TTTY15 was upregulated [Fig. 3D] and miR-520a-3p was downregulated [Fig. 3E] in cells exposed to OGD/R (p < 0.01). These results indicated that TTTY15 upregulation might contribute to miR-520a-3p downregulation and neuronal injury.
Fig. 3.
TTTTY15 upregulation and miR-520a-3p downregulation N2a cells after OGD/R. Cells experienced OGD/R (2 h/24 h) treatment. (A) Apoptosis was determined by flow cytometry. (B) Apoptosis was examined using Hoechst 33,258 staining. Scale bar = 50 μm ( × 200 magnification). (C) Apoptosis-related proteins were detected using Western blot. (D, E) qRT-PCR detected TTTY15 (D) and miR-520a-3p (E) levels. ∗∗p < 0.01. Data were exhibited as mean ± SD. n = 3.
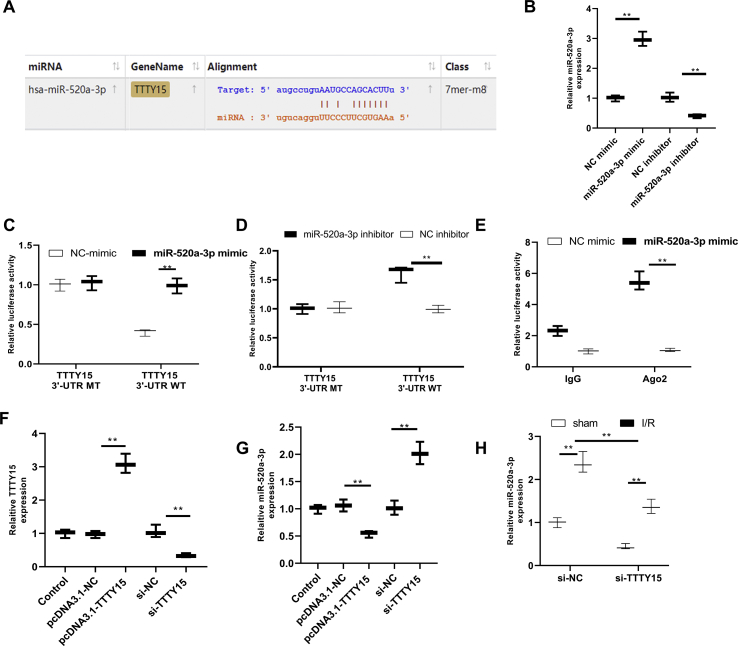
TTTY15 binds to miR-520a-3p and modulates miR-520a-3p level
Luciferase reporter systems with wild type TTTY15 (TTTY15-WT) or mutant TTTY15 with mutations at the predicted miR-520a-3p binding site (TTTY15-MT) were established to confirm the association between miR-520a-3p and TTTY15 [Fig. 4A]. MiR-520a-3p overexpression and inhibition were confirmed using qRT-PCR (p < 0.01) [Fig. 4B]. Luciferase reporter assay revealed decreased luciferase activity in TTTY15-WT co-transfected with miR-520a-3p mimic but no significant difference in TTTY15-MT co-transfected with miR-520a-3p mimic compared with that co-transfected with NC mimic (p < 0.01) [Fig. 4C]. Moreover, increased luciferase activity was exhibited in TTTY15-WT co-transfected with miR-520a-3p inhibitor, while no significance was found in TTTY15-MT co-transfected with miR-520a-3p inhibitor (p < 0.01) [Fig. 4D]. These data confirmed that TTTY15 was bound to miR-520a-3p at the predicted binding site. RNA immunoprecipitation further proved the physical relationship between TTTY15 and miR-520a-3p (p < 0.01) [Fig. 4E]. Afterward, TTTY15 overexpression and knockdown were verified with qRT-PCR (p < 0.01) [Fig. 4F]. TTTY15 overexpression downregulated miR-520a-3p level while TTTY15 knockdown increased miR-520a-3p level (p < 0.01) [Fig. 4G]. Moreover, mice subjected to I/R decreased miR-520a-3p level (p < 0.01) [Fig. 4H]. TTTY15 knockdown by si-TTTY15 injection contributed to miR-520a-3p expression elevation (p < 0.01) [Fig. 4H]. Altogether, these results indicated that TTTY15 acted as a ceRNA to downregulate miR-520a-3p via binding to miR-520a-3p.
Fig. 4.
TTTY15 regulates miR-520a-3p expression through binding to miR-520a-3p. (A) Binding sites between miR-520a-3p and TTTY15 were forecasted using StarBase v.2.0 software. (B) N2a cells were treated with miR-520a-3p mimic or miR-520a-3p inhibitor or NC mimic or NC inhibitor for 48 h, and miR-520a-3p levels were detected by qRT-PCR (n = 3). (C, D) The luciferase reporter vector with TTTY15-WT or TTTY15-MT was co-transfected with miR-520a-3p mimic or NC mimic (C) or miR-520a-3p inhibitor or NC inhibitor (D). Luciferase activity was measured 48 h after transfection (n = 3). (E) Relationship between miR-520a-3p and TTTY15 was analyzed using RNA immunoprecipitation after cells were transfected with miR-520a-3p mimic or NC mimic for 48 h (n = 3). Cells were treated by pcDNA3.1-TTTY15, si-TTTY15, pcDNA3.1-NC or si-NC. After 48 h of transfection, TTTY15 expression (F) and miR-520a-3p expression (G) were detected using qRT-PCR (n = 3). (H) Lateral ventricle of mice was treated with si-TTTY15 or si-NC. 24 h later, mice were exposed to I/R (2 h/24 h) or sham operation, and brains were sectioned for assessment. qRT-PCR was used to assess miR-520a-3p expression. ∗∗p < 0.01. Data were exhibited as mean ± SD. n = 6.
MiR-520a-3p reduces neuronal apoptosis
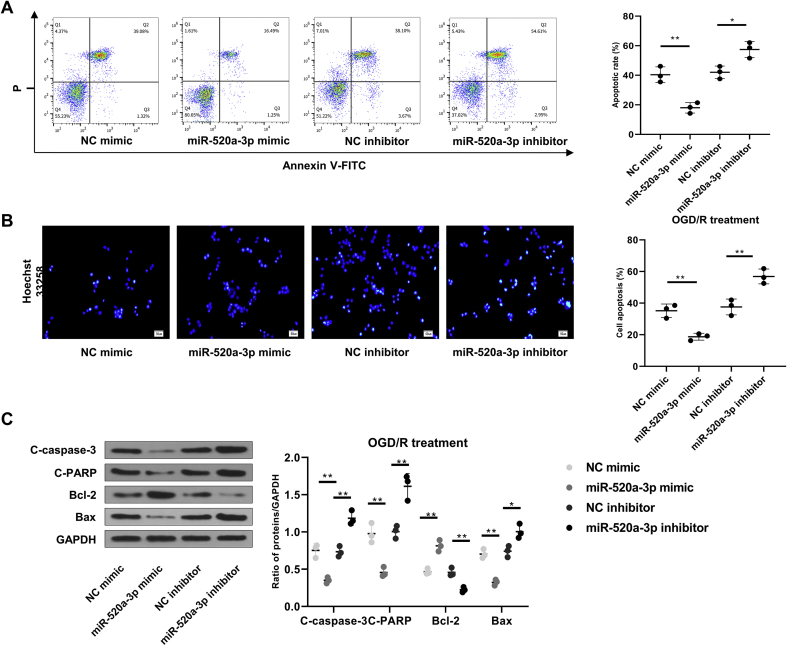
N2a cells were transfected with miR-520a-3p mimic or miR-520a-3p inhibitor to further investigate miR-520a-3p regulation in neuronal apoptosis. Flow cytometry analyses indicated that miR-520a-3p overexpression and knockdown downregulated and upregulated the OGD/R-induced apoptosis, respectively (p < 0.05) [Fig. 5A]. Hoechst 33,258 staining revealed similar results on apoptotic cell rate (p < 0.01) [Fig. 5B]. The impact of miR-520a-3p mimic or inhibitor on apoptosis following OGD/R was also evidenced by apoptosis-related proteins (p < 0.05) [Fig. 5C].
Fig. 5.
MiR-520a-3p reduces neuronal apoptosis after OGD/R exposure. miR-520a-3p mimic, NC mimic, miR-520a-3p inhibitor or NC inhibitor were transfected into N2a cells for 48 h. Subsequently, transfected cells were subjected to OGD/R. (A) Apoptosis was evaluated by flow cytometry. (B) Apoptosis was assessed using Hoechst 33,258 staining. Scale bar = 50 μm ( × 200 magnification). (C) Expression of Bcl-2, C-caspase-3, C-PARP, and Bax were measured using Western blot. ∗p < 0.05, ∗∗p < 0.01. Data were exhibited as mean ± SD. n = 3.
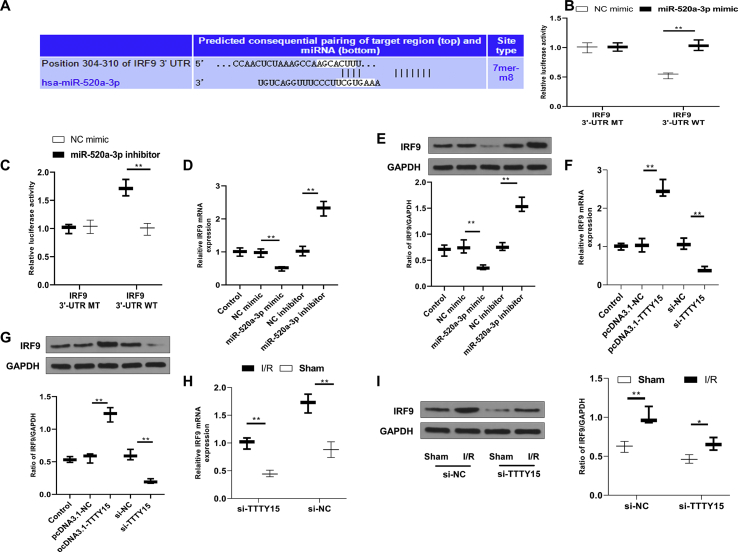
TTTY15 regulates IRF9 by serving as a ceRNA for miR-520a-3p
To confirm whether IRF9 was an actual target of miR-520a-3p, luciferase reporter systems were constructed with the wild type IRF9 3′-UTR (IRF9 3′-UTR-WT) or a mutant IRF9 3′-UTR (IRF9 3′-UTR-MT) at the binding site of miR-520a-3p [Fig. 6A]. Luciferase activity was deceased in IRF9-3′-UTR-WT co-transfected with miR-520a-3p mimic system, while no significant difference in luciferase activity was found in IRF9-3′-UTR-MT co-transfected with miR-520a-3p mimic system (p < 0.01) [Fig. 6B]. Similarly, luciferase activity was increased in IRF9-3′-UTR-WT co-transfected with miR-520a-3p inhibitor system while showed no significant difference in IRF9-3′-UTR-MT co-transfected with miR-520a-3p inhibitor system (p < 0.01) [Fig. 6C]. These results suggested that IRF9 was targeted directly by miR-520a-3p at the binding site of IRF9 3′-UTR. Then the impact of miR-520a-3p on IRF9 level was measured to further confirm that IRF9 was a target of miR-520a-3p. qRT-PCR results validated that miR-520a-3p upregulation decreased IRF9 mRNA level while miR-520a-3p inhibitor increased IRF9 mRNA level (p < 0.01) [Fig. 6D]. Western blot analyses also confirmed the findings at protein levels (p < 0.01) [Fig. 6E]. Furthermore, TTTY15 overexpression raised IRF9 expression while TTTY15 knockdown declined IRF9 expression as exhibited by qRT-PCR (p < 0.01) [Fig. 6F] and Western blot (p < 0.01) [Fig. 6G]. In addition, IRF9 level was increased after I/R (2 h/24 h) injury at both mRNA [Fig. 6H] and protein (p < 0.01) [Fig. 6I] levels. TTTY15 knockdown injected with si-TTTY15 declined IRF9 mRNA level [Fig. 6H] and protein level (p < 0.01) [Fig. 6I]. These results revealed the direct binding between IRF9 and miR-520a-3p and manifested that TTTY15 modulated IRF9 via functioning as a ceRNA for miR-520a-3p.
Fig. 6.
TTTY15 binds to miR-520a-3p competing with 3′-UTR of IRF9 mRNA. (A) Binding sites between IRF9 and miR-520a-3p forecasted in TargetScan v.7.2 software. (B, C) miR-520a-3p mimic or NC mimic (B) or miR-520a-3p inhibitor or NC inhibitor (C) was co-transfected with luciferase reporter vector carrying IRF9 3′-UTR-MT or IRF9 3′-UTR-WT in cells. 48 h after transfection, the relative activity of luciferase was measured (n = 3). (D, E) IRF9 levels were evaluated using qRT-PCR (D) and Western blot (E) in N2a cells transfected with miR-520a-3p mimic or miR-520a-3p inhibitor or NC mimic or NC inhibitor (n = 3). (F, G) IRF9 levels were evaluated using qRT-PCR (F) and Western blot (G) in cells transfected with si-TTTY15, pcDNA3.1-TTTY15, si-NC or pcDNA3.1-NC (n = 3). (H, I) The lateral ventricle of mice was administrated with si-TTTY15 or si-NC. At 24 h following the administration, mice were exposed to sham or I/R (2 h/24 h) surgery. IRF9 levels were evaluated using qRT-PCR (H) and Western blot (I). ∗p < 0.05, ∗∗p < 0.01. Data were exhibited as mean ± SD. n = 6.
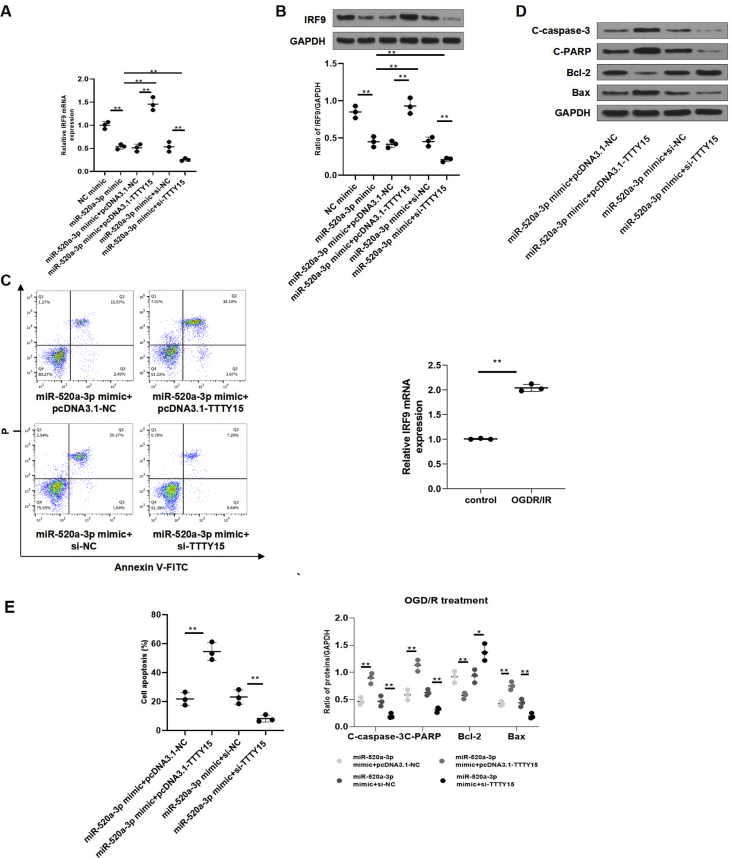
TTTY15 antagonizes the inhibitory effects of miR-520a-3p mimic on IRF9 to modulate neuronal apoptosis
To manifest TTTY15's functions in I/R injury, cells were treated with si-NC, pcDNA3.1-NC, si-TTTY15, or pcDNA3.1-TTTY15. The effects of IRF9 overexpression/knockdown on cell apoptosis upon OGD/R were shown in Figure.S1. The statistics displayed that TTTY15 overexpression restored the suppression of miR-520a-3p mimic on IRF9 while TTTY15 knockdown improved the inhibition at both mRNA level [Fig. 7A] and protein level (p < 0.01) [Fig. 7B]. The expression of IRF9 was induced by OGD/R [Fig. 7C]. Further flow cytometry analysis [Fig. 7C] and Western blot analyses of apoptosis-related proteins [Fig. 7D] exhibited that TTTY15 overexpression reversed the inhibitory effect of miR-520a-3p mimic on cell apoptosis while TTTY15 knockdown exhibited opposite effects. The statistic results of Fig. 7C and D were shown in Fig. 7E(∗p < 0.05, ∗∗p < 0.01).
Fig. 7.
TTTY15 antagonizes the inhibitory impact of miR-520a-3p on IRF9 to modulate neuronal apoptosis after OGD/R. (A, B) pcDNA3.1-TTTY15, si-TTTY15, pcDNA3.1-NC or si-NC was stably expressed in N2a cells, and miR-520a-3p mimic was transiently transfected into these cells for 48 h. In addition, miR-520a-3p mimic or mimic-NC was transiently transfected into control N2a cells for 48 h. IRF9 levels were evaluated using qRT-PCR (A) and Western blot (B). (C, D) The transfected N2a cells were co-transfected with miR-520a-3p mimic transiently for 48 h prior to OGD/R injury. (C) Apoptosis was evaluated using flow cytometry. The IRF9 expression was also enhanced by OGD/R. (E) Bcl-2, C-caspase-3, CPARP, and Bax were determined using Western blot. ∗p < 0.05, ∗∗p < 0.01. Data were exhibited as mean ± SD. n = 3.
Discussion
Normal brain function is crucial for homeostasis and remodeling. Recent studies have shown that cerebral injury caused by ischemia stroke destroys brain cell functions, resulting in neurological disorders even death [1,3]. Consequently, preventing brain cell damage is a purpose of ischemia stroke treatment to promote neurological disorder recovery and to reduce mortality. Numerous investigations have confirmed that lncRNAs target DNAs, RNAs, or proteins as sponges, signals, or guides to subsequently exert various biological functions in the pathogenesis of many human diseases [25]. But the impacts of lncRNA TTTY15 are not well understood, especially on brain cells during the progression of cerebral ischemia stroke. Our study found that TTTY15 was upregulated in cerebral injury with I/R, and TTTY15 knockdown alleviated neurological disorders, cerebral infarct, and neural apoptosis in vivo after cerebral I/R. Moreover, lncRNA TTTY15 suppressed miR-520a-3p and IRF9 expression, indicating the involvement of a TTTY15/miR-520a-3p/IRF9 axis in modulating neural apoptosis following ischemic stroke.
Many lncRNAs, such as MALAT1, MEG3, 1810034E14Rik, and N1LR, are expressed abnormally in brain cells under cerebral ischemia stroke, contributing to cell apoptosis, neurogenesis, angiogenesis, inflammation, and cell death [7,8,26]. Our study showed upregulation of a new lncRNA TTTY15 after cerebral ischemia injury in vitro and in vivo and TTTY15 knockdown alleviated neural apoptosis. A previous study has detected that TTTY15 was downregulated and TTTY15 overexpression inhibited NSCLC cell proliferation and metastasis [27]. TTTY15 level was elevated in most prostate cancer tissues and promoted prostate cancer progression [28]. TTTY15 expression was related to hypoxia, and TTTY15 attenuated cardiomyocyte injury after hypoxia [11]. Our study revealed the expression and potential function of TTTY15 in ischemic stroke. Researchers have demonstrated that lncRNAs can serve as ceRNAs, sponging specific miRNAs to modulate gene expression [29,30]. To investigate the regulation mechanism of TTTY15 in neural apoptosis following cerebral I/R, we searched for miRNAs that could bind to TTTY15 using bioinformatics. The results revealed that cerebral I/R treatment decreased miR-520a-3p level and TTTY15 negatively regulated miR-520a-3p via sponging miR-520a-3p. Additionally, we found that miR-520a-3p reduced OGD/R-induced neuronal apoptosis and downregulated its target gene IRF9, while TTTY15 antagonized the suppression impacts of miR-520a-3p on IRF9 and neuronal apoptosis after OGD/R, indicating that TTTY15 acted as a pro-apoptosis lncRNA in cerebral ischemia stroke. The interaction among lncRNAs, miRNAs and the targeted mRNAs represents a complicated regulation network of gene expression [29]. A study showed that TTTY15 targeted another miRNA (miR-455–5p) to regulate hypoxia-induced cardiomyocytes injury [11]. A previous study revealed that miR-520a-3p was also a downstream target of lncRNA NORAD and affected PI3k/Akt/mTOR signal pathway [31]. These studies revealed that the complicated networks among lncRNAs, miRNAs, and mRNA could be involved in different diseases and indicated a diverse role of the same ncRNA.
Currently, the role of miR-520a-3p in brain diseases and its impact on neural injury after cerebral I/R have not been studied. Here, we found that miR-520a-3p level decreased both in N2a cells exposed to OGD/R and in mouse brains exposed to I/R. We also showed that miR-520a-3p altered the expression of apoptosis-related proteins and prevented neural apoptosis after OGD/R treatment. Previous studies indicated that miR-520a-3p played essential roles in other diseases, such as thyroid carcinoma, osteosarcoma, and lung cancer [17,18,32]. Researchers have emphasized that miRNAs are implicated in the pathological mechanism of CNS diseases by binding to and regulating their target genes. MiR-223–5p is associated with NCKX2, and anti-miR-223–5p prevents ischemia-induced NCKX2 downregulation, consequently enhancing neuroprotection [16]. It is verified that miR-126 promotes angiogenesis and neurogenesis to improve neurobehavioral outcomes following ischemic stroke by directly inhibiting its target gene tyrosine-protein phosphatase non-receptor type 9 (PTPN9) [33]. A study has recognized Bim as a target gene of miR-124 to mediate the neuroprotection impact of miR-124, which regulates apoptosis and reduces DA neuron loss in a mouse Parkinson's disease model [34]. Subsequently, the target genes of miR-520a-3p implicated in cerebral ischemia stroke were predicted and measured. Our data indicated that miR-520a-3p targeted IRF9 to inhibit its expression at both transcriptional and translational levels.
IRF9 is a member of interferon regulatory factors (IRFs). It is expressed abnormally in CNS [21,22,35]. Studies have demonstrated that IRF9's pathological function in ischemic stroke is to negatively regulate Sirt1 transcription, resulting in cell apoptosis and cerebral injury [22]. It has been reported that in systemic lupus erythematosus, IRF9 can be targeted by miRNA-302 d to modulate IFN-induced gene expression [36]. A previous study has proven that the neuron-specific IRF9 overexpression sensitized neurons to death caused by IR and OGD/R in vivo [22]. In this study, we found that as a target gene of miR-520a-3p, IRF9 was negatively regulated by miR-520a-3p, and TTTY15 reversed this effect. Moreover, miR-520a-3p reduced IRF9 expression and alleviated OGD/R-induced cell apoptosis, and these effects were reversed by TTTY15. Furthermore, TTTY15 inhibition reduced cerebral I/R-elevated IRF9 level and improved apoptosis in vivo. Overall, our study revealed a novel TTTY15/miR-520a-3p/IRF9 pathway regulating neural apoptosis in ischemia stroke.
Conclusions
Under cerebral I/R, TTTY15 suppression attenuates neural apoptosis and neurological deficits via downregulating IRF9 expression. TTTY15 sponges miR-520a-3p as a ceRNA, promotes IRF9 expression, and thereby aggravates neural apoptosis. Therefore, TTTY15 might be an appealing target against neural apoptosis in stroke.
Ethics approval and consent to participate
Animal experiments were performed according to the American Animal Protection Legislation and approved by the Institutional Animal Care and Use Committee of The Fourth Affiliated Hospital, Harbin Medical University.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
Not applicable.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2022.04.001.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
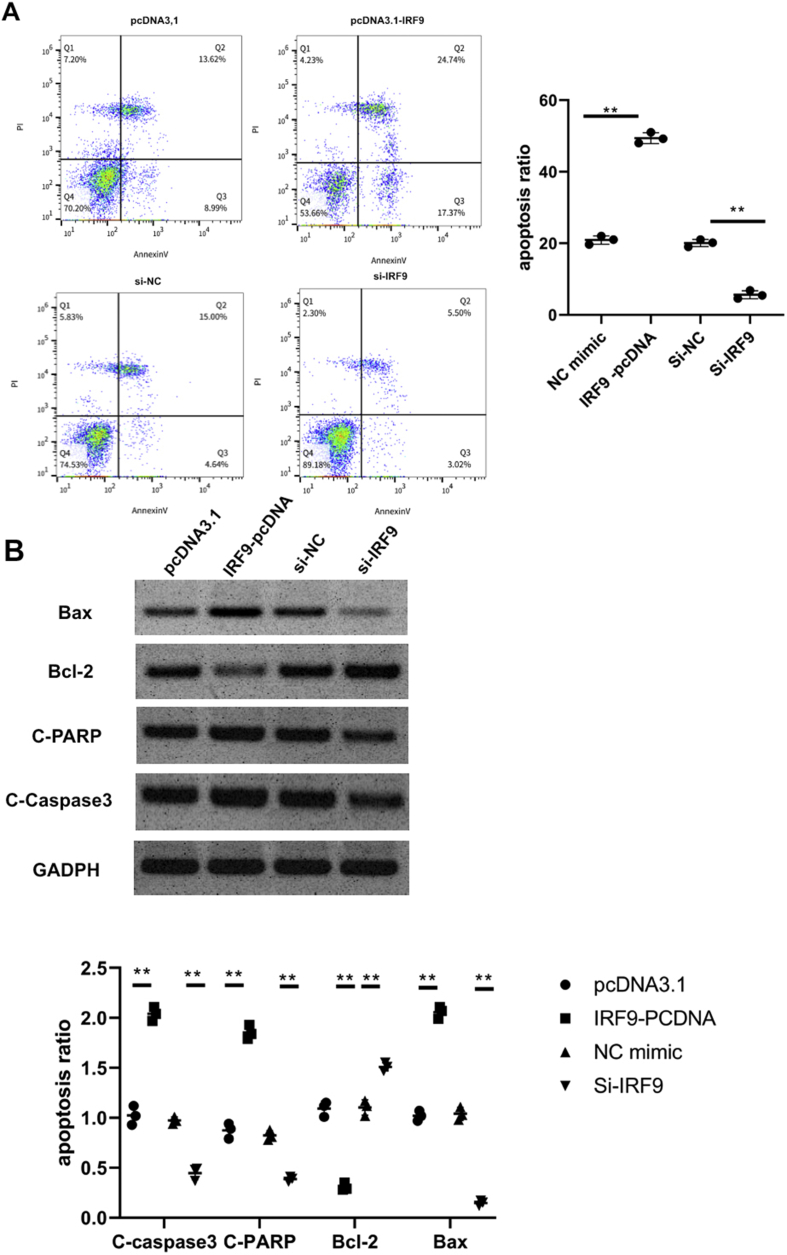
Supplementary Fig 1.
The effect of overexpression/knockdown of IRF9 on the apoptosis upon OGD/R
The transfected N2a cells were transfected with pcDNA, IRF9-PCDNA, si-NC and si-IRF9 transiently for 48h prior to OGD/R injury, respectively. (A)Apoptosis identified using Flow cytometry after OGD/R injury. (B) Bcl-2, C-caspase-3, CPARP and Bax determined with western blot. ∗P<0.05, ∗∗P<0.01. Results were exhibited as mean ± SD. n=3
References
- 1.Hankey G.J. Stroke. Lancet. 2017;389(10069):641–654. doi: 10.1016/S0140-6736(16)30962-X. [DOI] [PubMed] [Google Scholar]
- 2.Patel R.A.G., McMullen P.W. Neuroprotection in the treatment of acute ischemic stroke. Prog Cardiovasc Dis. 2017;59(6):542–548. doi: 10.1016/j.pcad.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Sekerdag E., Solaroglu I., Gursoy-Ozdemir Y. Cell death mechanisms in stroke and novel molecular and cellular treatment options. Curr Neuropharmacol. 2018;16(6):1396–1415. doi: 10.2174/1570159X16666180302115544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uzdensky A.B. Apoptosis regulation in the penumbra after ischemic stroke: expression of pro- and antiapoptotic proteins. Apoptosis. 2019;24(9-10):687–702. doi: 10.1007/s10495-019-01556-6. [DOI] [PubMed] [Google Scholar]
- 5.Chamorro A., Dirnagl U., Urra X., Planas A.M. Neuroprotection in acute stroke: targeting excitotoxicity, oxidative and nitrosative stress, and inflammation. Lancet Neurol. 2016;15(8):869–881. doi: 10.1016/S1474-4422(16)00114-9. [DOI] [PubMed] [Google Scholar]
- 6.Fok E.T., Scholefield J., Fanucchi S., Mhlanga M.M. The emerging molecular biology toolbox for the study of long noncoding RNA biology. Epigenomics. 2017;9(10):1317–1327. doi: 10.2217/epi-2017-0062. [DOI] [PubMed] [Google Scholar]
- 7.Bao M.H., Szeto V., Yang B.B., Zhu S.Z., Sun H.S., Feng Z.P. Long non-coding RNAs in ischemic stroke. Cell Death Dis. 2018;9(3):281. doi: 10.1038/s41419-018-0282-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z., Wu P., Zuo X., Yu N., Qin Y., Xu Q., et al. LncRNA-N1LR enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Mol Neurobiol. 2017;54(10):7670–7685. doi: 10.1007/s12035-016-0246-z. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Luo X., Chen F., Yuan W., Xiao X., Zhang X., et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem. 2018;119(7):5460–5472. doi: 10.1002/jcb.26705. [DOI] [PubMed] [Google Scholar]
- 10.Zheng J., Zhuo Y.Y., Zhang C., Tang G.Y., Gu X.Y., Wang F. LncRNA TTTY15 regulates hypoxia-induced vascular endothelial cell injury via targeting miR-186-5p in cardiovascular disease. Eur Rev Med Pharmacol Sci. 2020;24(6):3293–3301. doi: 10.26355/eurrev_202003_20697. [DOI] [PubMed] [Google Scholar]
- 11.Huang S., Tao W., Guo Z., Cao J., Huang X. Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced cardiomyocytes injury by targeting miR-455-5p. Gene. 2019;701:1–8. doi: 10.1016/j.gene.2019.02.098. [DOI] [PubMed] [Google Scholar]
- 12.Eyileten C., Wicik Z., De Rosa S., Mirowska-Guzel D., Soplinska A., Indolfi C., et al. MicroRNAs as diagnostic and prognostic biomarkers in ischemic stroke-A comprehensive review and bioinformatic analysis. Cells. 2018;7(12):249. doi: 10.3390/cells7120249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li G., Morris-Blanco K.C., Lopez M.S., Yang T., Zhao H., Vemuganti R., et al. Impact of microRNAs on ischemic stroke: from pre- to post-disease. Prog Neurobiol. 2018;163–164:59–78. doi: 10.1016/j.pneurobio.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Ma Q., Zhang L., Pearce W.J. MicroRNAs in brain development and cerebrovascular pathophysiology. Am J Physiol Cell Physiol. 2019;317(1):C3–C19. doi: 10.1152/ajpcell.00022.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamzei Taj S., Kho W., Riou A., Wiedermann D., Hoehn M. MiRNA-124 induces neuroprotection and functional improvement after focal cerebral ischemia. Biomaterials. 2016;91:151–165. doi: 10.1016/j.biomaterials.2016.03.025. [DOI] [PubMed] [Google Scholar]
- 16.Cuomo O., Cepparulo P., Anzilotti S., Serani A., Sirabella R., Brancaccio P., et al. Anti-miR-223-5p ameliorates ischemic damage and improves neurological function by preventing NCKX2 downregulation after ischemia in rats. Mol Ther Nucleic Acids. 2019;18:1063–1071. doi: 10.1016/j.omtn.2019.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J., Tan Q., Deng B., Fang C., Qi D., Wang R. The microRNA-520a-3p inhibits proliferation, apoptosis and metastasis by targeting MAP3K2 in non-small cell lung cancer. Am J Cancer Res. 2015;5(2):802–811. [PMC free article] [PubMed] [Google Scholar]
- 18.Bi C.L., Zhang Y.Q., Li B., Guo M., Fu Y.L. MicroRNA-520a-3p suppresses epithelial-mesenchymal transition, invasion, and migration of papillary thyroid carcinoma cells via the JAK1-mediated JAK/STAT signaling pathway. J Cell Physiol. 2019;234(4):4054–4067. doi: 10.1002/jcp.27199. [DOI] [PubMed] [Google Scholar]
- 19.Negishi H., Taniguchi T., Yanai H. The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb Perspect Biol. 2018;10:a028423. doi: 10.1101/cshperspect.a028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul A., Tang T.H., Ng S.K. Interferon regulatory factor 9 structure and regulation. Front Immunol. 2018;9:1831. doi: 10.3389/fimmu.2018.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofer M.J., Li W., Lim S.L., Campbell I.L. The type I interferon-alpha mediates a more severe neurological disease in the absence of the canonical signaling molecule interferon regulatory factor 9. J Neurosci. 2010;30(3):1149–1157. doi: 10.1523/JNEUROSCI.3711-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H.Z., Guo S., Li Z.Z., Lu Y., Jiang D.S., Zhang R., et al. A critical role for interferon regulatory factor 9 in cerebral ischemic stroke. J Neurosci. 2014;34(36):11897–11912. doi: 10.1523/JNEUROSCI.1545-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Liu X., She Z.G., Jiang D.S., Wan N., Xia H., et al. Interferon regulatory factor 9 is an essential mediator of heart dysfunction and cell death following myocardial ischemia/reperfusion injury. Basic Res Cardiol. 2014;109(5):434. doi: 10.1007/s00395-014-0434-9. [DOI] [PubMed] [Google Scholar]
- 24.Kaur H., Sarmah D., Saraf J., Vats K., Kalia K., Borah A., et al. Noncoding RNAs in ischemic stroke: time to translate. Ann N Y Acad Sci. 2018;1421(1):19–36. doi: 10.1111/nyas.13612. [DOI] [PubMed] [Google Scholar]
- 25.Qian X., Zhao J., Yeung P.Y., Zhang Q.C., Kwok C.K. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem Sci. 2019;44(1):33–52. doi: 10.1016/j.tibs.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Zhu X.L., Ji B.Y., Cao X., Yu L.J., Zhang Y., et al. LncRNA-1810034E14Rik reduces microglia activation in experimental ischemic stroke. J Neuroinflammation. 2019;16(1):75. doi: 10.1186/s12974-019-1464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai I.L., Chang Y.S., Chan W.L., Lee Y.T., Yen J.C., Yang C.A., et al. Male-specific long noncoding RNA TTTY15 inhibits non-small cell lung cancer proliferation and metastasis via TBX4. Int J Mol Sci. 2019;20:3473. doi: 10.3390/ijms20143473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao G., Yao J., Kong D., Ye C., Chen R., Li L., et al. The long noncoding RNA TTTY15, which is located on the Y chromosome, promotes prostate cancer progression by sponging let-7. Eur Urol. 2019;76(3):315–326. doi: 10.1016/j.eururo.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Yamamura S., Imai-Sumida M., Tanaka Y., Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75(3):467–484. doi: 10.1007/s00018-017-2626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandran R., Mehta S.L., Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int. 2017;111:12–22. doi: 10.1016/j.neuint.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan Y., Yao Z., Chen W., Li D. The lncRNA NORAD/miR-520a-3p facilitates malignancy in non-small cell lung cancer via PI3k/Akt/mTOR signaling pathway. OncoTargets Ther. 2020;13:1533–1544. doi: 10.2147/OTT.S230954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Xu Y., Chen X., Xiao J. Dexmedetomidine inhibits osteosarcoma cell proliferation and migration, and promotes apoptosis by regulating miR-520a-3p. Oncol Res. 2018;26:495–502. doi: 10.3727/096504017X14982578608217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu M., Pan J., Wang L., Zhou P., Song Y., Wang S., et al. MicroRNA-126 regulates angiogenesis and neurogenesis in a mouse model of focal cerebral ischemia. Mol Ther Nucleic Acids. 2019;16(3):15–25. doi: 10.1016/j.omtn.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H., Ye Y., Zhu Z., Mo L., Lin C., Wang Q., et al. MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson's disease by targeting to Bim. Brain Pathol. 2016;26(2):167–176. doi: 10.1111/bpa.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ousman S.S., Wang J., Campbell I.L. Differential regulation of interferon regulatory factor (IRF)-7 and IRF-9 gene expression in the central nervous system during viral infection. J Virol. 2005;79(12):7514–7527. doi: 10.1128/JVI.79.12.7514-7527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith S., Fernando T., Wu P.W., Seo J., Ni Gabhann J., Piskareva O., et al. MicroRNA-302d targets IRF9 to regulate the IFN-induced gene expression in SLE. J Autoimmun. 2017;79:105–111. doi: 10.1016/j.jaut.2017.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.