Abstract
Dna2 is a multifunctional enzyme in yeast that possesses endonuclease activity well suited to remove RNA–DNA primers of Okazaki fragments, raising the question of whether endonuclease activity is essential for in vivo Dna2 function. Systematic site-directed mutations of amino acid residues in Saccharomyces cerevisiae DNA2 conserved in the central region of many eukaryotic DNA2 homologs allowed us to identify mutant dna2 alleles that were divided into three groups based on the viability of the mutant cells: (i) viable; (ii) inviable only when expression was repressed; (iii) inviable. Biochemical analyses of recombinant mutant Dna2 proteins isolated from the latter two groups revealed that they possessed normal ATPase/helicase activity, but were impaired in their endonuclease activity. Cells expressing mutant Dna2 enzymes partially impaired in endonuclease activity were viable, but were unable to grow when expression of their mutant Dna2 enzymes was further reduced. Their growth was restored when the mutant Dna2 proteins decreased in nuclease activity were induced to overexpress. In contrast, mutant Dna2 proteins lacking endonuclease activity did not allow cells to grow under any conditions tested. These in vivo and in vitro results demonstrate that the endonuclease activity of Dna2 is essential for Okazaki fragment processing.
INTRODUCTION
DNA2 was originally identified by screening cell division cycle mutants of Saccharomyces cerevisiae. The gene product was shown to be essential for cell viability and to encode a 172 kDa protein that contained characteristic DNA helicase motifs (1,2). The DNA2 gene of S.cerevisiae was implicated as playing a role in chromosomal DNA replication based on results from a number of genetic and biochemical analyses (1,3–7). For example, permeabilized cells and nuclear extracts capable of replicating supercoiled plasmids were found to have reduced replication activity upon inactivation of S.cerevisiae Dna2 (ScDna2) (5). A clue to the role of Dna2 in vivo was the specific association of ScDna2 with Rad27 (3), a yeast homolog of mammalian Fen-1 that has been shown to play a crucial role in Okazaki fragment processing in conjunction with RNase HI (8–13). For this reason, DNA2 is believed to play a role in Okazaki fragment maturation in vivo. Recently, we demonstrated that Schizosaccharomyces pombe Dna2 (SpDna2) interacted genetically with two subunits of replicative DNA polymerase δ, Rad2 (the S.pombe homolog of Fen-1) and DNA ligase I, all of which are essential for lagging strand synthesis and its maturation (14). These observations strongly suggest that Dna2 participates directly in Okazaki fragment maturation during DNA replication. In support of an essential role played by Dna2 in eukaryotic DNA replication, DNA2 homologs are found throughout eukaryotes, including humans, plants, fish, fission yeast, Xenopus and nematodes (3,7,15), suggesting that its role in DNA replication may be evolutionarily conserved in all eukaryotes.
Consistent with its implied role in DNA replication, it was reported that immunoaffinity purified Dna2 fusion protein from yeast crude extracts displayed DNA-dependent ATPase activity as well as 3′→5′ DNA helicase activity and also co-purified with nuclease activity that was originally attributed to Fen-1 associated with Dna2 (1,3). Recombinant ScDna2 from insect cells, however, possessed intrinsic single-strand-specific endonuclease activity (16), suggesting that the nuclease activity that co-purified with Dna2 was not solely due to its association with Fen-1. The helicase activity associated with Dna2 was noticeable only under conditions where the nuclease activity was substantially suppressed by lowering the ratio of MgCl2 to ATP. Under these conditions, Dna2 displaced duplex DNA in a 5′→3′ direction (S.-H.Bae and Y.-S.Seo, submitted for publication), opposite to the direction claimed previously (1). We recently discovered that the endonuclease activity is not structure-specific, but prefers to cleave the free ends of single-stranded DNA (S.-H.Bae and Y.-S.Seo, submitted for publication). The unique feature of the Dna2 endonuclease activity is that this cleavage reaction, which occurred in the DNA, was stimulated by the presence of an RNA segment at the 5′-end of the flap DNA. Moreover, this cleavage event occurred efficiently within DNA in conjunction with DNA polymerases that were capable of displacement synthesis. This ensures the complete removal of the initiator RNA segment on the Okazaki fragment, providing a biochemical basis for a role of Dna2 in Okazaki fragment processing. These findings allowed us to propose a new model in which Dna2 plays a direct role in Okazaki fragment maturation in collaboration with Fen-1 (S.-H.Bae and Y.-S.Seo, submitted for publication).
A point mutation in the ATP binding motif (K1080E) of DNA2 led to inactivation of its ATPase and helicase activities and rendered the mutant cell inviable (1,16). Mutant cells expressing Dna2 K1080E, however, were still viable under certain growth conditions. They were capable of growing in medium containing lactate and glycerol instead of glucose as carbon source (7). This observation suggests that the ATPase/helicase activity of Dna2 is not constantly required for cell viability. This observation raised the possibility that the other intrinsic activity associated with Dna2, namely its endonuclease activity, is an essential function of the protein required for chromosome replication in eukaryotes.
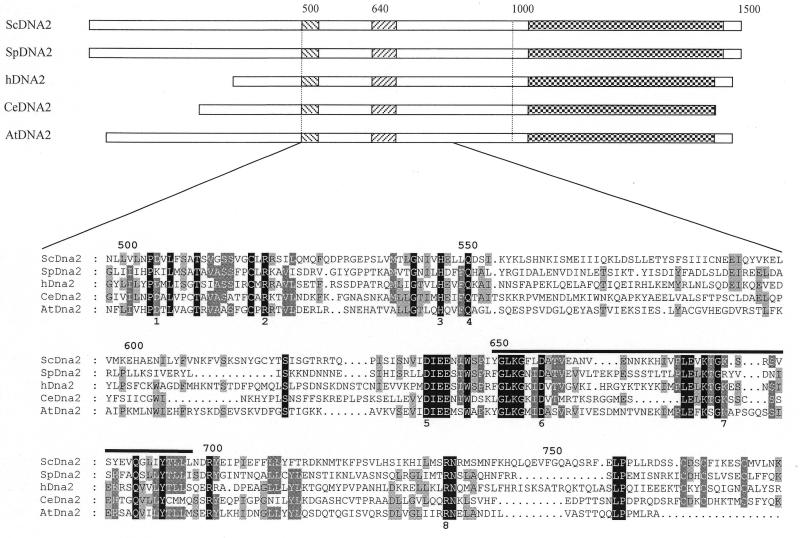
In order to test this possibility, we initiated systematic site-specific alterations of amino acid residues in the central region (amino acid positions 500–750) of yeast Dna2. This region contains sequences that show some homology between Dna2 proteins from various eukaryotic organisms (Fig. 1). In this paper, we have identified amino acid residues that are essential for endonuclease activity, but not for ATPase/helicase activity. We demonstrate that the inability of cells that are mutated in DNA2 to grow is directly related to the degree of defect in the endonuclease activity of the Dna2 protein. Our in vivo and in vitro results establish that the endonuclease activity of Dna2 plays an essential role in Okazaki fragment maturation in support of our model proposed previously (S.-H.Bae and Y.-S.Seo, submitted for publication).
Figure 1.
Comparison of the Dna2 amino acid sequences from different species and the position of ScDna2 point mutations. The most conserved parts of the Dna2 proteins from S.cerevisiae, S.pombe, human, C.elegans and A.thaliana are shown as striped regions. The amino acid residue numbers of S.cerevisiae Dna2 are indicated. The amino acid sequences of the nuclease region are aligned in the lower portions of the figure. The alignment was created by the GeneDoc program. Most conserved sequences are shown dark shadowed and the less conserved sequences are shown gray shadowed. The amino acid point mutations made in Dna2 in this study are indicated as: 1, dna2-23 (D505N); 2, dna2-15 (R521K) dna2-26 (R521E); 3, dna2-24 (H547A); 4, dna2-21 (Q551E); 5, dna2-25 (DIEE640NIQQ); 6, dna2-27 (D657A); 7, dna2-22 (K689A); 8, dna2-28 (R735A). The thick line denotes the RecB homology region (19).
MATERIALS AND METHODS
Oligonucleotides, DNA and nucleoside triphosphates and enzymes
All oligonucleotides used for the construction of various DNA substrates or for site-directed mutagenesis were synthesized commercially (Gibco BRL, Rockville, MD). Oligonucleotides longer than 50mer were gel purified prior to use. Names of mutant dna2 alleles and sequences of mutagenic primers used to construct the mutant alleles are as follows (the position of nucleotides that lead to the amino acid alteration are underlined and altered amino acids and their position in the DNA2 open reading frame are indicated in parentheses): dna2-15, 5′-GT TCA GTA GGT TGT TTA AAA CGT TCA ATT CTG CAA ATG-3′ (Q551E); dna2-21, 5′-C GTA CAC GAG TTA TTG GAA GAC TCA ATC AAA TAC-3′ (K680A); dna2-22, 5′-TTA GAA GTG AAA ACT GGA GCA TCC AGA AGC GTT TCA TAC-3′ (D505N); dna2-23, 5′-G TTG GTG CTA AAC CCT AAT GTA TTA TTT TCG GC-3′ (H547A); dna2-24, 5′-CT TTA GGC AAT ATC GTA GCC GAG TTA TTG CAA GAC-3′ (R521K); dna2-25, 5′-CT ATA TCC AAT GTG ATT AAT ATT CAA CAA AAC ATT TGG TCT CC-3′ (DIEE640NIQQ); dna2-26, 5′-CA GTA GGT TGT TTA GAA CGT TCA ATT CTG CAA ATG C-3′ (R521E); dna2-27, 5′-GGT CTT AAA GGG TTC CTA GCT GCA ACT GTT GAA GCT AAT-3′ (D657A); dna2-28, 5′-AAA CAT ATT CTC ATG TCC GCA AAT AGA ATG AGT ATG AAT-3′ (R734A).
Nucleoside triphosphates were obtained from Boehringer Mannheim (Mannheim, Germany) and [γ-32P]ATP (>5000 Ci/mmol) and [α-32P]dCTP (>6000 Ci/mmol) were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Restriction endonucleases, the Klenow fragment of Escherichia coli DNA polymerase I and polynucleotide kinase were from Promega (Madison, WI).
Strains and plasmids
The pRS series plasmids were purchased from New England Biolabs (Beverly, MA). Strain YJA1 (MATa/MATα ade1 ura3 lys1 trp1 his7 leu2 GAL+ dna2Δ::HIS3) was constructed using the following steps. The 397 bp 5′-flanking sequence of DNA2 containing the native promoter sequence was amplified by PCR using the primers described (16). The amplified fragment contained restriction sites for EcoRI and BamHI at the 5′- and 3′-ends, respectively. The 3′-flanking region (354 bp) of DNA2 was PCR amplified using the primer pair Dna2X (5′-CCG CTC GAG CTT CTC CAT AAC AGG AAA G-3′, XhoI restriction site underlined) and Dna2Y (5′-CCG GAA TTC TCC CAA GCC TCC AAG TG-3′, EcoRI restriction site underlined). The two fragments were cleaved by the appropriate restriction enzymes mentioned above and then cloned into pRS303 (His+ selection marker) cut with XhoI and BamHI to obtain pRS303-Δdna2. The chromosomal DNA2 gene was disrupted by transforming the diploid strain YPH501 (MATa/MATα ade1 ura3 lys1 trp1 his7 leu2 GAL+) with pRS303-Δdna2 linearized by EcoRI, which cut the junction between the 5′- and 3′-flanking fragments of DNA2. His+ transformants were selected and deletion of DNA2 was confirmed by both PCR and Southern blotting analyses, generating strain YJA1. YJA1 was transformed with pRS316-DNA2 (see below for construction) and sporulated. The resulting dna2Δ haploid strain harboring pRS316-DNA2 as episome was named YKH12 (MATa ade1 ura3 lys1 trp1 his7 leu2 GAL+ dna2Δ::HIS3 pRS316-DNA2).
Plasmid pDNA2, containing the entire DNA2 ORF plus its native promoter, was described previously (16). Plasmids pRS314-DNA2 (Trp+, +CEN, +ARS) and pRS304-DNA2 (Trp+, –CEN, –ARS) were constructed by subcloning EcoRI–XbaI and EcoRI–NotI fragments from pDNA2 into pRS314 and pRS304, respectively. pRS316-DNA2 was similarly constructed by subcloning the EcoRI–NotI fragment from pDNA2 into pRS316 (Ura+, +CEN, +ARS). In order to construct vectors for controlled expression of DNA2, the GAL1 promoter region was amplified in a PCR reaction. The amplified product was digested with KpnI and XhoI and cloned into pRS314 cleaved with KpnI and XhoI, and the resulting plasmid was named pRS314GU. Plasmid pRS314GU-DNA2 was constructed by subcloning the EcoRI–XbaI fragment of pDNA2 into pRS314GU.
Site-directed mutagenesis
The 1594 bp NdeI–XbaI fragment containing the putative nuclease domain of DNA2 was subcloned into pBlueScriptII-SK (Stratagene, La Jolla, CA) and mutagenized using the Quikchange system (Stratagene). Mutations of interest were confirmed by nucleotide sequencing of the entire NdeI–XbaI fragment. The altered NdeI–XbaI fragments were then swapped for the wild-type fragment in pHX-DNA2 (16), generating pHX-dna2-X (X indicates different mutant alleles). The pRS314-, pRS314GU- and pRS304-based plasmids (pRS314-dna2-X, pRS314GU-dna2-X and pRS304-dna2-X) expressing mutant dna2 alleles were constructed by replacing the BsmI fragment of pHX-dna2-X containing altered amino acids with the corresponding fragments in pRS314-DNA2, pRS314GU-DNA2 and pRS304-DNA2, respectively.
Plasmid shuffling and chromosomal integration
Strain YKH12 [MATa, ade1 ura3 lys1 trp1 his7 leu2 GAL+ dna2Δ::HIS3 pRS316-DNA2 (URA+, DNA2)] was transformed with the parent vector pRS314, pRS314-DNA2 and pRS314-dna2-X (Trp+, dna2-X mutant). Transformants were grown to saturation under Trp+ selection and then aliquots of 10-fold serial dilutions were placed on plates lacking tryptophan to determine total viability. The aliquots were also plated on medium containing 5′-fluoroorotic acid (5-FOA) to monitor cell viability upon loss of pRS316-DNA2. Growth on 5-FOA-containing plates therefore indicates the phenotype of cells containing only a mutant dna2 allele on the plasmid. For chromosomal integration, pRS304 vector only, pRS304-DNA2 and pRS304-dna2-X were digested with AatII and the linearized DNAs were used for transformation of strain YJA1. For each mutant, diploid transformants containing both Ura+ and Trp+ markers were sporulated and the resulting tetrads were dissected to examine whether each mutant allele supported cell growth. For this purpose, all viable spores were tested for the presence of His+ and Trp+ markers.
Construction of recombinant baculoviruses and overexpression and purification of the recombinant mutant Dna2 proteins in insect cells
Recombinant baculoviruses that produced mutant Dna2 proteins were constructed as recommended by the manufacturer (Gibco BRL). The recombinant baculoviruses encoded mutant Dna2 proteins with an additional 27 amino acid residues (MSYYHHHHHHDYDIPTTENLYFQGAMGS). These included six histidines (bold) fused to its N-terminal methionine to facilitate detection and purification of the recombinant Dna2 proteins. The recombinant mutant Dna2p was isolated as described previously (16) but with modifications. Briefly, infected cells (1 × 106 cells/ml, 1 l) were harvested, resuspended in 50 ml of buffer T (25 mM Tris–HCl, pH 7.5, 1 mM EDTA, 10% glycerol, 1 mM DTT, 0.1 mM PMSF, 0.15 µg/ml leupeptin and antipain) containing 100 mM NaCl and disrupted by sonication (five cycles of a 30 s pulse and a 1 min cooling interval). The extracts (5.16 mg/ml) were cleared by centrifugation at 37 000 r.p.m. for 1 h in a 45 Ti rotor (Beckman, Palo Alto, CA). The supernatants were applied directly to a heparin–Sepharose (Amersham Pharmacia Biotech) column (0.79 × 4 cm, 3.16 ml) equilibrated with buffer T plus 100 mM NaCl (buffer T100; hereafter, the number indicates the concentration of NaCl added to buffer T). The column was washed with 5 column vol of the same buffer lacking DTT and EDTA and eluted with buffer T500 (–DTT, –EDTA) plus 20 mM imidazole. The peak fractions were pooled (1.63 mg/ml, 12 ml) and loaded onto a Ni2+–NTA–agarose (Qiagen, Valencia, CA) column (1.77 × 3 cm, 5.3 ml) equilibrated with buffer T500 (–DTT, –EDTA) plus 20 mM imidazole. After washing with 2 column vol of buffer T500 (–DTT, –EDTA) plus 50 mM imidazole, the column was then eluted with 400 mM imidazole in T500 (–DTT, –EDTA). Fractions containing the mutant Dna2 protein were pooled and dialyzed for 2 h against buffer T0. The dialysate (0.26 mg/ml, 2.5 ml) was loaded onto a Hi-Trap Q column (1 ml; Amersham Pharmacia Biotech) equilibrated with buffer T100. After washing with 5 ml of buffer T100, the column was eluted with a 10 ml linear gradient of 100–500 mM NaCl in buffer T. The fractions containing the mutant Dna2 protein were pooled (0.044 mg/ml, 5 ml), concentrated 20-fold and then loaded onto a glycerol gradient (5 ml, 15–35% glycerol in buffer T500). The gradient was subjected to centrifugation for 24 h at 45 000 r.p.m. in a Beckman SW55 Ti rotor. Fractions (220 µl) were collected from the bottom of the gradient and assayed for DNA-dependent ATPase and nuclease activities.
Preparation of endonuclease substrate and enzyme assays
Standard assays for measuring DNA-dependent ATPase activity were carried out as described (16). The Y-structure partial duplex DNA substrate used to measure nuclease activity was prepared as described previously (16). The reaction mixture (20 µl) used to examine nuclease activity contained 50 mM Tris–HCl, pH 7.8, 2 mM DTT, 0.25 mg/ml BSA and the labeled DNA substrate (15 fmol). Reaction mixtures containing Dna2 enzymes were preincubated at 37°C for 5 min in the absence of MgCl2 and the reactions initiated by adding Mg2+ (2 mM or as indicated). After incubation at 37°C for 3 min, reactions were stopped with 20 µl of 2× stop solution (20 mM EDTA, pH 8.0, 95% formamide and 0.25% xylene cyanol). The reaction products were subjected to electrophoresis for 1.5 h at 150 V through a 10% native polyacrylamide gel containing 0.1% SDS in 0.5× TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA) or for 2 h at 35 W through a 20% denaturing (+ 6 M urea) polyacryamide gel in 1× TBE. The gel was dried on a DEAE–cellulose paper and subjected to autoradiography. Cleaved DNA products were quantitated using a Molecular Imager FX (Bio-Rad, Hercules, CA).
RESULTS
Mutagenesis
The fact that homologs of DNA2 are found in humans, Caenorhabditis elegans, S.pombe and Aradbidopsis suggests that this enzyme likely plays a crucial role(s) conserved in eukaryotic DNA replication. Although Dna2 possesses well-conserved DNA helicase motifs in the C-terminal one-third region (amino acid positions 1050–1522), it appeared to lack any recognizable nuclease motifs. We discovered that the N-terminal 500 amino acid sequence of yeast Dna2 is hardly conserved and deletion of the N-terminal 405 amino acids did not result in inactivation of the endonuclease and ATPase/helicase activities of Dna2 (15). This deletion, however, resulted in a temperature-sensitive phenotype (data not shown). Further deletion of the N-terminal region resulted in loss of cell viability, indicating that this region is essential for function of the Dna2 protein (data not shown). Thus, it is assumed that the endonuclease domain of ScDna2 is located somewhere between positions 405 and 1050. Furthermore, the alignment of Dna2 sequences from five different organisms, including humans (hDna2), S.pombe (SpDna2), S.cerevisiae (ScDna2), Arabidposis thaliana (ADna2), and C.elegans (CDna2) revealed a significant region of homology corresponding to amino acid positions 498–780 of ScDna2, as shown in Figure 1.
Within this region, we noted the presence of eight charged amino acid residues that are invariably conserved among the Dna2 homologs (Fig. 1). They include Arg521, His547, Gln550, Asp640, Glu642, Glu643, Asp657 and Arg734. Among these, aspartate or glutamate residues are likely to contribute to binding of the divalent Mg2+ ion (17,18). The other amino acids may play roles in substrate binding or catalysis of endonucleolytic cleavage by Dna2 and were therefore included as candidates for residues essential for endonuclease activity. The aspartic acid and lysine residues at positions 505 and 680, respectively, are not strictly conserved in all organisms. SpDna2 contained other charged amino acids (lysine and arginine, respectively) at these two positions (Fig. 1). We included these two amino acids as targets for site-directed mutagenesis, hoping that alteration of these two amino acids would lead to mutant Dna2 preparations with partially impaired endonuclease activity.
We performed site-directed mutagenesis using the mutagenic oligonucleotides and procedures described in Materials and Methods. Each candidate amino acid residue was altered singly except that the motif Asp640-Ile-Glu-Glu643, highly conserved among all eukaryotic Dna2 proteins, was changed to Asn-Ile-Gln-Gln. The Arg521 residue was mutated to either Glu or Lys. The change of Arg521 to Lys was shown previously to result in a temperature-sensitive phenotype of the mutant cells (7). We included this mutation in our study, hoping that the Dna2 protein derived from the temperature-sensitive allele would display temperature-dependent endonuclease activity. Such a property would help to demonstrate unambiguously that the endonuclease activity is intrinsic to Dna2 and that this activity is an essential function for cell growth. Thus, we constructed nine different mutant alleles of DNA2 which included (see also Table 1): R521K, Q551E, K680A, D505N, H547A, DIEE640–643NIQQ, R521E, D657A and R734A (amino acids are denoted in single letter code and the number indicates the position of the amino acid altered). Later, after we had finished constructing the mutant alleles, it was reported that the central region (amino acid positions 649–680) of Dna2 shares homology with a domain containing the nuclease active site of the RecB nuclease family (Fig. 1; 19), supporting our rationale for selection of amino acids that may cause a defect in endonuclease activity of Dna2 upon alteration.
Table 1. Spore viability and plasmid shuffling results for the DNA2 mutants.
| Mutant allele | Amino acid change | Spore viability (%) | Spore with marker (%) | pRS314 | pRS314GU + Glu | pRS314GU + Gal |
|---|---|---|---|---|---|---|
| Wild-type | 89 | 52 | +++ | +++ | +++ | |
| Group I | ||||||
| dna2-21 | Q551E | 94 | 48 | +++ | +++ | +++ |
| dna2-22 | K680A | 100 | 50 | +++ | +++ | +++ |
| Group II | ||||||
| dna2-23 | D505N | 94 | 42 | +++ | – | +++ |
| dna2-24 | H547A | 54 | 19 | +++ | – | +++ |
| dna2-15 | R521K | 90 | 44 | +++ | – | +++ |
| Group III | ||||||
| dna2-25 | DIEE640NIQQ | 46 | 0 | – | – | +/– |
| dna2-26 | R521E | 43 | 0 | – | – | – |
| dna2-27 | D657A | 46 | 0 | – | – | – |
| dna2-28 | R734A | 45 | 0 | – | – | – |
For analyses of viability of spores containing mutant DNA2 alleles, the diploid strain YJA1 (MATa/MATα DNA2/Δdna2::HIS3) was transformed with pRS304 (vector), pRS304-DNA2 (wild-type DNA2 in pRS304) and pRS304-dna2-X (X indicates the mutant allele shown above). The resulting diploid transformants were separately sporulated and tetrads were dissected. The percent spore viability was obtained by dividing the number of colonies grown by total tetrads. The viable colonies obtained were examined for the presence of both His+ and Trp+ markers. The ability of cells to grow after plasmid shuffling is shown as: –, no growth; +/–, 100- to 1000-fold less growth than wild-type; +++, growth like wild-type control. Plasmid shuffling tests were performed using plasmids in which the mutant DNA2 alleles were expressed using its native promoter (pRS314) or using the GAL1 promoter (pRS314GU) in medium containing glucose (+Glu) or galactose (+Gal).
Genetic characterization of mutant alleles revealed three different categories of mutations
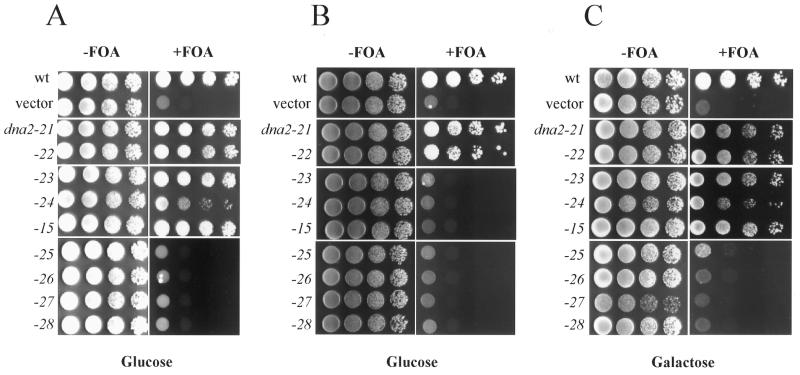
In order to search for a defect in cell growth in an isogenic background, we first introduced pRS314-dna2-X (containing a mutant dna2 allele) into YKH12 that was deleted of chromosomal DNA2, but harbors pRS316-DNA2. In this case, expression of each mutant dna2 allele was under the control of its own native promoter. As shown in Figure 2A, all of the transformants grew as efficiently as the wild-type control (–FOA), indicating that none of the mutant alleles resulted in a dominant-negative effect when expression is under these conditions. When pRS316-DNA2 was removed by growth in the presence of 5-FOA, the mutant alleles dna2-25, dna2-26, dna2-27 and dna2-28 did not support cell growth (Fig. 2A, +FOA). In contrast, the other alleles (dna2-15, dna2-21, dna2-22, dna2-23 and dna2-24) allowed cells to grow in the absence of the wild-type DNA2 allele (Fig. 2A, +FOA), although growth of dna2-24 cells was relatively poor compared to the wild-type control.
Figure 2.
Plasmid shuffling test of DNA2 mutants with its own promoter or the GAL1 promoter. (A) Strain YKH12 (YPH501 Δdna2::HIS pRS316-DNA2) was transformed with pRS314 vector only (vector), pRS314-DNA2 (wt) and pRS314-dna2-X (dna2-X) (the capital X denotes the number of the mutant allele constructed). Expression of the mutant alleles was carried out under the control of the native promoter of DNA2. Transformants were grown to saturation and serial dilutions placed on complete synthetic medium (containing glucose) lacking tryptophan in the absence (–FOA) or presence (+FOA) of 5-FOA. (B) Strain YKH12 was transformed with pRS314GU vector only (vector), pRS314GU-DNA2 (wt) and pRS314GU-dna2-X (dna2-X). Expression of the mutant alleles was carried out under control of the GAL1 promoter. Transformants were grown and plated in complete synthetic medium lacking tryptophan in the absence (–FOA) or presence (+FOA) of 5-FOA. Glucose was used as carbon source (Glucose) to repress expression of the DNA2 mutant alleles. (C) The same transformants used for (B) were plated in complete synthetic medium that contained galactose as carbon source (Galactose). They were grown in the absence (–FOA) or presence (+FOA) of 5-FOA. All plates were grown for 4 days at 30°C.
Since we expected that some of the mutant alleles could produce Dna2 with partially impaired endonuclease activity, we postulated that either repression or activation of mutant protein expression would alter the growth of the mutant cells. For this reason, we examined the relationship between the level of protein expression and the extent of cell growth. In order to carry out this experiment, each mutant allele was placed under the control of the inducible GAL1 promoter. Cells harboring plasmids containing the mutant alleles dna2-21 and dna2-22 grew efficiently regardless of the expression conditions (Fig. 2B). This suggested that the two mutant Dna2 proteins (Dna2-21p and Dna2-22p) were sufficiently functional to sustain cell growth despite significant differences in the level of enzyme produced. In marked contrast to this, cells containing the mutant alleles dna2-23, dna2-24 or dna2-15 failed to grow when their expression was repressed by growth in glucose-containing medium (Fig. 2B). However, when these mutant Dna2 proteins were strongly induced by growth in the presence of galactose, they supported cell growth to levels comparable to those observed when they were expressed from the chromosomal promoter (compare Fig. 2C, +FOA with Fig. 2A, +FOA). The cells containing the dna2-25 allele displayed only marginal ability (<100- to 1000-fold) to grow even when the mutant protein was overexpressed (Fig. 2C, +FOA). Mutant alleles dna2-26, dna2-27 and dna2-28 again did not support cell viability under any expression conditions tested (Fig. 2A–C).
Because variations in the copy number of plasmids and in their expression could also affect cell viability in the plasmid shuffling experiments described above, we replaced the chromosomal DNA2 with each mutant allele for more conclusive analyses. Mutant dna2 alleles placed under the control of its own promoter (pRS304-dna2-X) were integrated within the vector sequence in YJA1 (DNA2/dna2Δ::HIS ) in a site-specific fashion. The heterozygous diploid transformants (DNA2/dna2-X) obtained were sporulated and the tetrads dissected with a micromanipulator. After dissection, all spores were tested for the presence of the selection markers Trp+ and His+. Tetrads from mutant alleles dna2-25, dna2-26, dna2-27 and dna2-28 produced only two viable spores and none of the viable spores possessed the Trp+ marker, as shown in Table 1, demonstrating that spores with these mutant alleles were inviable. In contrast, tetrads containing the other mutant alleles all produced viable spores and the viability of total spores was >90%, except for dna2-24, which showed relatively low viability (54%) (Table 1). These results are in good agreement with those obtained from the plasmid shuffling experiments described above.
Endonuclease activity is an essential function of Dna2
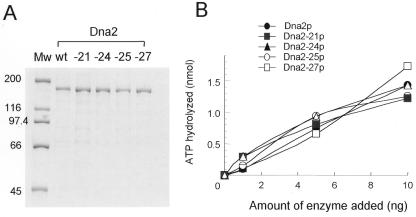
If the ability of cells to grow was related directly to the presence of functional Dna2 endonuclease activity, the above results would suggest that dna2-21 and dna2-22 yielded Dna2 products containing endonuclease activity comparable to wild-type Dna2. In contrast, mutant alleles dna2-23, dna2-24 and dna2-15 produced enzymes with partial activity that was not sufficient to support cell growth at a low protein level. Mutant alleles dna2-25, dna2-26, dna2-27 and dna2-28 are likely to produce non-functional enzymes devoid of endonuclease activity. In order to verify this hypothesis and the relationship between cell viability and the endonuclease activity of Dna2, we directly measured the endonuclease activity of mutant proteins representative of each of these groups. For this purpose, we constructed recombinant baculoviruses expressing Dna2-21p (group I, constitutively viable), Dna2-24p (group 2, lethal when expression was repressed), Dna2-25p (marginally viable only when overexpressed) and Dna2-27p (constitutively lethal). In order to facilitate detection and subsequent purification of mutant proteins, 6×His tags were fused to the N-terminal methionine of Dna2, which did not affect cell viability (16). We purified mutant Dna2 proteins to near homogeneity from extracts prepared from cells infected with baculoviruses expressing the representative mutant alleles using procedures described in Materials and Methods. As shown in Figure 3, the purity of each protein was evaluated. The mutant proteins had the same 172 kDa size as wild-type Dna2p (Fig. 3A) and were nearly as pure as wild-type Dna2p, although they were isolated using fewer steps than were used for isolation of the wild-type Dna2p (16). The authenticity of the protein was confirmed by western blot analyses using anti-Dna2 polyclonal antibodies (data not shown). For accurate measurement of enzymatic activities of the proteins purified, we assessed the concentrations of the purified mutant proteins by densitometric analyses of Coomassie blue stained protein bands. The protein concentrations were normalized with respect to that of wild-type Dna2p (data not shown).
Figure 3.
Analyses of mutant Dna2 proteins by SDS–PAGE and comparison of their ATPase activities. (A) Coomassie blue staining of a protein gel containing Dna2-wt, Dna2-21, Dna2-24, Dna2-25 and Dna2-27 proteins resolved by 8% SDS–PAGE. Each lane was loaded with ∼1 µg of the proteins, based on the value obtained in the Bradford assay (Bio-Rad). Mw indicates molecular size marker and the numbers to the left of the figure denote molecular weights (kDa). (B) Single-stranded DNA-dependent ATPase activities of mutant Dna2 enzymes were measured. The reactions, as described previously (16), were carried out in the presence of 1, 5 and 10 ng of proteins. The symbols for each mutant protein are as indicated in the figure. The amount of ATP hydrolyzed was measured as described previously (16).
Based upon protein concentrations determined as above, we first examined whether the mutant proteins were defective in their ability to hydrolyze ATP. The titration of enzyme at three different levels (1, 5 and 10 ng) resulted in linear hydrolysis of ATP. As shown in Figure 3B, the mutant proteins possessed specific ATPase activities comparable to that of the wild-type Dna2p. In this experiment, the wild-type Dna2 protein was prepared side-by-side with the mutant proteins for a more precise comparison. This result demonstrates that the mutations introduced into each protein examined did not affect their ATPase activity within the error of measurement. Since the precise quantitative comparison of DNA unwinding activity of each protein was not possible in the presence of differing levels of endonuclease (see below), we did not test this. However, the comparable levels of ATPase activities of the mutant proteins indicate that the unwinding ability might remain unchanged.
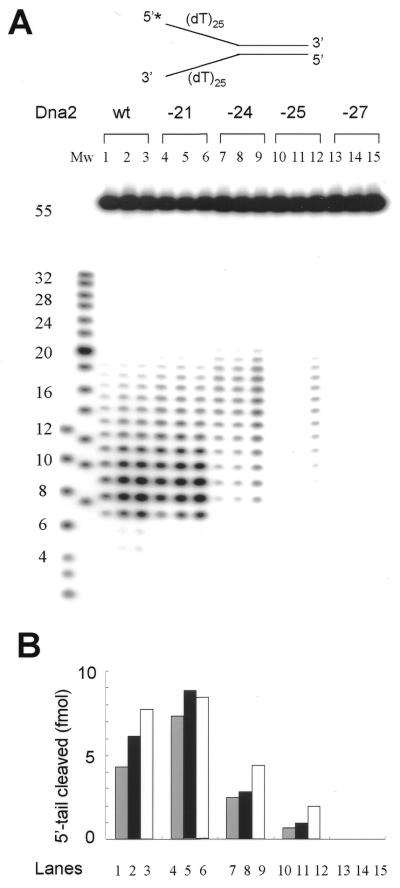
We then examined the endonuclease activity of each mutant protein as described previously (16). For this purpose, three different levels (0.25, 0.5 and 1 ng) of each enzyme preparation were analyzed (Fig. 4). The substrate used was a Y-structured substrate that contained both 5′ and 3′ oligo(dT)25 tails, but only the 5′-end of the two tails in the substrate was radiolabeled, as shown in Figure 4A. The reactions were carried out in the absence of ATP, since previous studies showed that ATP significantly inhibited the endonuclease activity of wild-type Dna2 (16). Dna2-21p displayed endonuclease activity comparable to that of wild-type Dna2p and the cleavage pattern was indistinguishable from that of wild-type Dna2p (Fig. 4A and B, compare lanes 1–3 to 4–6). In fact, the endonuclease activity of Dna2-21p was slightly (∼1.6-fold) higher than that observed with wild-type Dna2p when 0.5 ng of enzyme was added. In contrast, the endonuclease activity of Dna2-24p was slightly (2-fold) lower (Fig. 4A, lanes 7–9) than the activity detected with wild-type Dna2p. Notably, the cleaved products formed were significantly longer in length than those formed with wild-type Dna2p enzyme. Two explanations for this observation are possible. First, the decrease in endonuclease activity did not result in multiple cleavages of the already cleaved single-stranded DNA, hence increasing the final size of the products. Second, the mutation altered the binding property of the enzyme to single-stranded DNA ends, generating longer cleaved products. We prefer the latter possibility, since the cleavage pattern observed with wild-type Dna2p or mutant Dna2-24p were not altered regardless of the amount of enzyme added (Fig. 4, lanes 1–3 and 7–9). The mutant Dna2-25p enzyme displayed much weaker endonuclease activity than that observed with Dna2-24p, but with strikingly similar cleavage patterns (Fig. 4, compare lanes 7–9 and 10–12). Finally, no endonuclease activity was detected when we used Dna2-27p (Fig. 4, lanes 13–15). When we examined helicase activity with Dna2-24p, Dna2-25p and Dna2-27p, we found robust unwinding activity. This suggests that the helicase activity was not affected by the mutation (data not shown). This data supports our hypothesis proposed above that growth of cells containing a mutant Dna2 is related to the level of its endonuclease activity and indicates that the endonuclease activity intrinsic to Dna2 plays an essential role in DNA replication.
Figure 4.
Endonuclease activities of the mutant Dna2 proteins measured using a Y-structured partial duplex DNA substrate. (A) The schematic structure of the Y-structured partial duplex DNA substrate is shown at the top of the figure. An asterisk indicates the 32P label at the 5′-end of the tail. The preparation of substrate and oligonucleotides used were described previously (16). Both tails contained 25 nt oligo(dT) [(dT)25] homopolymer. Increasing amounts (0.25, 0.5 and 1 ng) of each protein were preincubated with 15 fmol of the substrate at 37°C for 5 min in the absence of MgCl2. Lanes 1–3, Dna2p (wt); lanes 4–6, Dna2-21p; lanes 7–9, Dna2-24p; lanes 10–12, Dna2-25p; lanes 13–15, Dna2-27p. The reactions were initiated by addition of MgCl2 (2 mM), incubated at 37°C for 5 min and terminated by adding 20 µl of 2× stop solution. The reaction products were analyzed on a 20% polyacrylamide gel containing 6 M urea in 1× TBE as described previously (16). Mw denotes molecular size markers prepared by labeling a synthetic oligo(dT) mixture. (B) The histogram shows the amounts of 5′-tail substrate cleaved with mutant Dna2 proteins in (A). The cleavage products were quantitated using a Molecular Imager FX (Bio-Rad). The amounts of cleaved products were plotted against each lane, which is indicated below the histogram.
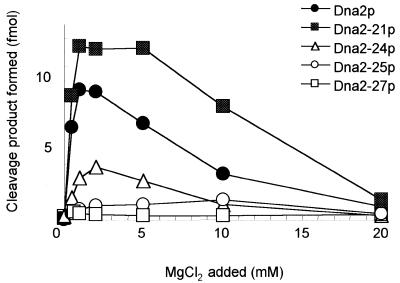
We investigated the properties of mutant Dna2 with respect to its divalent ion requirement since the amino acids altered (glutamic or aspartic acids) could have a role in divalent ion binding. As shown in Figure 5, we examined the endonuclease activities of the mutant Dna2 proteins in the presence of increasing levels (0.5, 1, 2, 5, 10 and 20 mM) of MgCl2 with the same substrate as used in Figure 4. The optimum concentration of MgCl2 for endonuclease activity of wild-type Dna2p, Dna2-21p and Dna2-24p was between 2 and 5 mM, as previously determined (16), although the extent of substrate cleavage varied with the different enzymes. In keeping with the enzyme titration results (Fig. 4), at identical protein levels Dna2-21p was reproducibly more active than wild-type Dna2p (Fig. 5). In contrast, the Dna2-25 enzyme required higher concentrations of MgCl2 (∼10 mM) for maximum cleavage activity, suggesting that the residues DIEE beginning at amino acid 640 participate in binding of Mg2+. Dna2-27p was inactive regardless of the concentrations of Mg2+ used.
Figure 5.
Requirements of Mg2+ for mutant Dna2 proteins. The reactions were carried out as described in Figure 4A, but in the presence of increasing concentrations (0.5, 1, 2, 5, 10 and 20 mM) of MgCl2. The amount of each enzyme used in the reactions was 0.2 ng. Cleavage products were analyzed by 10% native PAGE and quantitated using a Molecular Imager FX (Bio-Rad). Symbols for each protein are as indicated in the figure.
The DNA cleavage pattern observed with Dna2-24p and Dna2-25p differed from that observed with wild-type Dna2p and Dna2-21p. We examined whether this difference resulted from the alteration in the ability of the mutant proteins to bind the single-stranded DNA substrate. When we carried out electrophoretic mobility shift assays with the mutant proteins and a 5′-end-labeled oligonucleotide (98mer), all mutant proteins tested formed a complex with ssDNA with an affinity similar to that observed with wild-type Dna2p (data not shown). These findings indicate that the mutations that we introduced caused a specific defect in the catalytic function of the endonuclease activity of Dna2.
DISCUSSION
In our previous studies, we found that Dna2 contained single-strand DNA-specific endonuclease activity that appears to be an essential function of the protein (16). Subsequent characterization of Dna2p revealed that the endonuclease activity most likely plays a role in Okazaki fragment maturation (S.-H.Bae and Y.-S.Seo, submitted for publication). In this paper, we demonstrate that the endonuclease activity is essential for viability of cells by examining, both genetically and biochemically, nine different DNA2 mutants generated by site-specific mutations of invariable amino acid residues in the central region of Dna2p that are highly conserved in all eukaryotic Dna2 homologs.
Although point mutations were used in this study, changes in the region in which we introduced the site-specific mutations are likely to affect the integrity of the overall conformation of the protein. For example, the recombinant Dna2 protein lacking the N-terminal 550 amino acid residues contained no ATPase or endonuclease activity (data not shown). This finding was surprising since the truncated protein contained all of the helicase motifs. This suggests that the interaction between the nuclease and helicase domains may be crucial for both enzymatic activities. Therefore, a change in the nuclease domain could lead to inactivation of the ATPase/helicase activity of Dna2. Despite this, the inviablity of mutant cells is most likely due to the impaired endonuclease activity of Dna2 and not due to changes in the global conformation of the enzyme, which in turn may affect the protein–protein interactions necessary for Dna2 to carry out its essential function in vivo. This argument is based upon the following findings. (i) The mutations that we introduced were mostly single amino acid substitutions. (ii) The level of endonuclease activity of each mutant protein showed a strong correlation with the ability of each mutant cells to grow. (iii) Other properties of the proteins, such as single-strand DNA-dependent ATPase and substrate DNA binding activities remained unchanged.
During the course of this study, it was reported that Dna2 belongs to the RecB nuclease family (19). Sequence analyses performed using PSI-BLAST showed that the central region (amino acid positions 649–680) of Dna2 is homologous to the novel nuclease domain containing the nuclease active site of the RecB nuclease family (19). Within this region (see Fig. 1), two strictly conserved amino acid residues, corresponding to Asp657 and Glu675 in the DNA2 ORF, were found that are likely to participate in binding of Mg2+. The nuclease activity of the RecB protein was eliminated when Asp1080 (equivalent to Glu675 of Dna2) was altered to Ala, in keeping with a critical role of this residue in catalytic activity (20). In our study, the change of Asp657 to Ala (Dna2-27p) completely abolished endonuclease activity (Fig. 4), emphasizing that this residue is also critical for catalytic function. In contrast, the multiple change of the DIEE640–643 motif, which is strictly conserved in all eukaryotic Dna2 proteins, to NIQQ resulted in a mutant protein that possessed residual endonuclease activity. For this reason, it appeared that Asp657 and Glu675 play a more important role in Mg2+ binding than do Asp640 or Glu642 and Glu643 in the DIEE motif.
Within the Dna2 family, the conserved region extends into amino acid residues 500–760, as shown in Figure 1. Based on our finding that cell growth is likely to be correlated with the level of endonuclease activity, we suggest that regions (amino acids 500–648 and 681–760) of Dna2 distinct from the catalytic site conserved in RecB family proteins also contribute to Dna2 endonuclease activity. This is consistent with our finding that the mutant Dna2-24p (H547A) possessed somewhat lower endonuclease activity in vitro than the wild-type enzyme. If other regions contribute to endonuclease activity, alterations of the two strictly conserved Arg521 and Arg734 to Glu and Ala (dna2-26 and dna2-28, respectively; see Fig. 2 and Table 1) are most likely to completely abolish the endonuclease activity of Dna2. The non-conservative change of positively charged Arg521 to negatively charged Glu (dna2-26, R521E) led to cell death. In contrast, the conservative change Arg521 to Lys (dna2-15, R521K) supported cell growth, but not when expression was reduced (Table 1). This finding emphasizes the importance of the presence of a positively charged amino acid at this position and raises the possibility that Arg521 may play a role in catalytic function of the endonuclease activity. It is noteworthy that the conservative change of Arg521 to Lys resulted in temperature-sensitive growth of the mutant cells (7). This suggests that alteration of the amino acid at this position leads to a partial defect in endonuclease activity. We attempted to prepare the Dna2-15 (R521K) protein several times in order to examine whether the enzyme displayed temperature-sensitive endonuclease activity. However, we failed to purify Dna2-15p in a full-length form. Dna2-15p was found repeatedly to have suffered degradation in crude extracts of insect cells.
In addition to its role in DNA replication, Dna2 has also been implicated as playing a role in the repair of DNA damage caused by alkylating agents such as methylmethane sulfonate (MMS) (7). In order to test the possibility that the endonuclease activity plays a crucial role in this process, we focused on the group 2 mutant alleles that support cell growth in a single copy but are believed to produce enzymes partially impaired in endonuclease activity (Table 1). None of the mutant cells containing these alleles displayed any sensitivity to MMS at the concentrations used (0.01 and 0.02%), suggesting that Dna2 endonuclease activity might not play a role in MMS-induced DNA repair. In support of this, MMS-sensitive alleles of DNA2 isolated genetically by others clustered in the helicase domain, indicating that the helicase activity may be particularly important for this repair process (7). This observation was confirmed by the finding that site-directed mutations in motif I and motif II were also MMS sensitive (7). One exception to this was dna2-1 (P504S), which was highly sensitive (0.002%) to MMS (7). This suggests that dna2-1 may be defective at least in ATPase activity, although its mutation is not located within the helicase motifs. An extensive analysis with more mutant alleles will be necessary to address the issue of whether the endonuclease activity of Dna2 is required for the MMS repair process.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Jerard Hurwitz for critical reading of the manuscript. This work was supported by a grant from the Creative Research Initiatives of the Korean Ministry of Science and Technology to Y.-S.S.
REFERENCES
- 1.Budd M.E. and Campbell,J. (1995) Proc. Natl Acad. Sci. USA, 92, 7642–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budd M.E., Choe,W.-C. and Campbell,J. (1995) J. Biol. Chem., 270, 26766–26769. [DOI] [PubMed] [Google Scholar]
- 3.Budd M.E. and Campbell,J. (1997) Mol. Cell. Biol., 17, 2136–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiorentino D.F. and Crabtree,G.R. (1997) Mol. Biol. Cell, 8, 2519–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braguglia D., Heun,P., Pasero,P., Duncker,B.P. and Gasser,S.M. (1998) J. Mol. Biol., 281, 631–649. [DOI] [PubMed] [Google Scholar]
- 6.Parenteau J. and Wellinger,R.J. (1999) Mol. Cell. Biol., 19, 4143–4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formosa T. and Nittis,T. (1999) Genetics, 151, 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulian M., Richards,S.H., Heard,C.J. and Bigsby,B.M. (1990) J. Biol. Chem., 265, 19461–19471. [PubMed] [Google Scholar]
- 9.Ishimi Y., Claude,A., Bullock,P. and Hurwitz,J. (1988) J. Biol. Chem., 263, 19723–19733. [PubMed] [Google Scholar]
- 10.Waga S. and Stillman,B. (1994) Nature, 369, 207–212. [DOI] [PubMed] [Google Scholar]
- 11.Waga S. and Stillman,B. (1998) Annu. Rev. Biochem., 67, 721–751. [DOI] [PubMed] [Google Scholar]
- 12.Bambara R.A., Murante,R.S. and Hendericksen,L.A. (1997) J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- 13.Lieber M.R. (1997) Bioessays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- 14.Kang H.Y., Choi,E.J., Bae,S.H., Lee,K.H., Gim,B.S., Kim,H.D., Park,C., MacNeill,S.A. and Seo,Y.S. (2000) Genetics, 155, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q., Choe,W.C. and Campbell,J. (2000) J. Biol. Chem., 275, 1615–1624. [DOI] [PubMed] [Google Scholar]
- 16.Bae S.H., Choi,E., Park,J.S., Lee,S.H., Lee,K.H. and Seo,Y.S. (1998) J. Biol. Chem., 273, 26880–26890. [DOI] [PubMed] [Google Scholar]
- 17.Steitz T.A. and Steitz,J.A. (1993) Proc. Natl Acad. Sci. USA, 90, 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joyce C.M. and Steitz,T.A. (1994) Annu. Rev. Biochem., 63, 777–822. [DOI] [PubMed] [Google Scholar]
- 19.Aravind L., Walker,D.R. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 1223–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M., Souaya,J. and Julin,M.A. (1998) J. Mol. Biol., 283, 797–808. [DOI] [PubMed] [Google Scholar]