ABSTRACT
Mycoplasma mastitis can be highly contagious, unresponsive to treatment, and cause severe economic problems in affected herds. Notable routes of Mycoplasma spp. transmissions are contaminated milking equipment and animal contact through respiratory secretions. Only a few studies report the environment as a possible source of infection. Our group studied the presence of pathogens in houseflies (Musca domestica) in a New York State dairy in the United States. Among others, a Mycoplasma spp. was found in the gut of a housefly captured in the sick pen and identified as M. arginini. Here, we characterized its genome and investigated its relatedness with eight isolates from milk, one isolate from lung tissue collected in the same dairy, and five other dairies in New York State. We applied whole-genome sequencing and phylogenetic analysis based on the sequences of the 16S rRNA gene and 76 conserved proteins. We also assessed an in silico virulence profile by considering a panel of 94 putative virulence genes. As a result of the genome analysis, the housefly M. arginini isolate was highly similar to the milk isolates; interestingly, the similarity was highest with M. arginini isolated from milk on the same dairy farm where the housefly was captured. The housefly and milk M. arginini isolates possessed 54 of the 94 pathogenicity genes considered. Our data support the hypothesis that houseflies are carriers of Mycoplasma spp. and can be considered within the possible roots of environmental transmission of infection in dairy cows. Nevertheless, M. arginini pathogenicity will need to be investigated with dedicated studies.
IMPORTANCE It is critical to control the spread of bovine mastitis caused by Mycoplasma spp., as this disease can be highly contagious and have a severe economic impact on affected dairies. A better understanding of possible transmission routes is crucial for infection control and prevention. Based on our data, the composite milk isolates are genetically similar to the housefly isolate. This provides evidence that the same Mycoplasma species found in milk and associated with mastitis can also be isolated from houseflies captured in the dairy environment.
KEYWORDS: Mycoplasma, housefly, dairy farm, mastitis, milk, cow
INTRODUCTION
Mycoplasma spp. are a significant concern for dairy farms, being associated with several diseases in cattle, including mastitis, pneumonia, otitis, arthritis, and reproductive disorders (1). Herds in the United States and many other countries have a zero-tolerance policy for cows with mycoplasma mastitis, since it could be highly contagious and unresponsive to any treatment (2). Culling animals is the most common way to control and prevent the spread of mycoplasma mastitis in the farm independently from the bacterial load and Mycoplasma species detected (3, 4). The documented transmission routes of Mycoplasma spp. are contaminated milking equipment and animal contact through respiratory secretions (5). Environmental sources of Mycoplasma spp. and their role in transmitting diseases are poorly known. To our knowledge, the few environmental sources where Mycoplasma spp. have been isolated on dairies are represented by cooling ponds, dry lots, and recycled bedding (6). Data from a study performed in Utah (7) showed that Mycoplasma spp. were found in 51% of the recycled sand bedding collected on dairy farms during outbreaks of clinical mastitis caused by Mycoplasma spp. The same study also showed that Mycoplasma spp. remained viable in recycled bedding sand for up to 8 months. Manure and other different bedding types represent the primary sources for houseflies’ nutrition and oviposition.
Houseflies represent a threat to dairy farms, resulting in significant losses attributable to the transmission of diseases, including mastitis (8, 9). To shed light on the potential role of flies in the diffusion of mycoplasmas, the present study investigated the genomic relatedness of a M. arginini isolated from a housefly captured in a dairy farm with eight M. arginini isolated from bovine milk samples and one isolated from bovine lung tissue. The M. arginini isolates of this study were obtained from different sampling periods ranging from months to years, even if specimens were collected from the same farm. In view of the uncertainty of M. arginini pathogenicity, we investigated the presence of 94 selected genes that putatively encode virulence factors (10). Since Mycoplasma bovis is the most prevalent, contagious, and clinically relevant Mycoplasma species in cattle, a set of pathogenicity genes was selected based among those already described for M. bovis, while another set was selected from the publicly available M. arginini genomic information (11).
RESULTS
Mycoplasma arginini isolation and identification.
In September 2019, we carried out a study on pathogens isolated from houseflies (Musca domestica) captured in different locations of a New York State dairy farm (12). The farm was selected based on a client list of the daily milk sample pick-up, 24h result program of Quality Milk Production Services (QMPS) and the history of cow udder pathogens isolated from milk, including S. aureus, Mycoplasma spp., Prototheca spp., and Lactococcus spp. Out of the 485 bacterial isolates obtained in that study, an isolate originating from the gut of a female housefly captured in the sick pen was identified as M. arginini by means of a multiplex-PCR assay associated with amplicon Sanger sequencing (13). Here, we present the detailed genomic analysis of this M. arginini isolate and evaluate its similarity with other isolates from bovine specimens.
Genomic DNA sequencing, assembly, and annotation of the housefly gut M. arginini isolate.
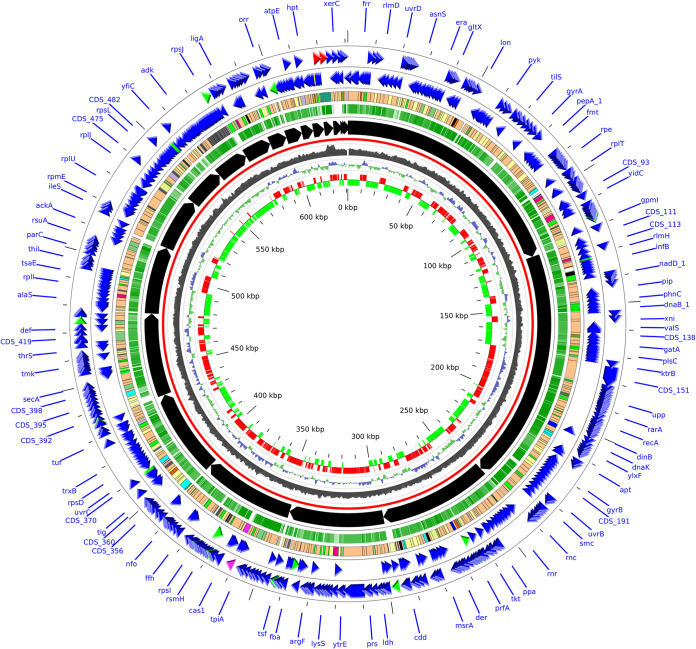
Genomic DNA sequencing and assembly of the housefly gut M. arginini isolate generated 21 scaffolds of length >500 nt, for a total of 631,023 bp; the maximum scaffold length was 122,725 bp, with 16 out of the 21 scaffolds above 5Kb and an N50 of 48,510 bp. GC content overall the scaffolds was 26.3%. Genome size and GC content were comparable to those of the reference genome HAZ145. The assembly included a total of 584 genes, including two rRNAs (16S and 23S), 32 tRNAs, 1 transfer-mRNA (tmRNA), 1 repeat region (CRISPR-associated with 32 repeat units), and 548 protein-coding genes. A hundred and eight CDS (19.7%) were annotated as “hypothetical proteins” of unknown function. Two hundred sixty-five out of the 548 protein-coding genes (48.4%) were classifiable as enzymes (e.g., nucleases, proteases, hydrolases), 14 as ATP-binding genes, 10 as lipoproteins, and 51 as ribosomal proteins. Among enzymes, the most represented were transferases (44 genes), tRNA ligases (21 genes), permeases (14 genes), nucleases (15 genes), peptidases (12 genes), and hydrolases (11 genes). Five hundred eighty-three features (99.8% of the total features) had a corresponding match on M. arginini reference strain HAZ145, with very high similarity (average identity of 99.1% ± 1.4% on a nucleotide basis) and covering nearly all the gene sequences (average coverage of 99.1% ± 7.9%) (Fig. 1).
FIG 1.
Circular plot of the draft genome of the M. arginini strain isolated from the fly gut. The 21 annotated scaffolds sum up to 631,023 bp. Each circle represents a feature of the genome: from the innermost circle, we represented: gene strand (red: positive, green: negative), the GC skew, the GC content, reference backbone (in red), scaffolds (ordered by decreasing length), the percentage of identity with M. arginini HAZ145 reference strain (opacity is proportional to % identity), CDS (colored by type; among them, light orange: enzymes, light yellow: hypothetical proteins), CDS on negative and positive strands (colors indicate: blue: proteins, red: rRNA, green: tRNA).
Five hundred seventy-eight out of the 591 CDS (97.8%) of the HAZ145 reference genome found a match in the housefly gut isolate assembly; 10 out of the 13 missing genes were annotated as hypothetical proteins, 1 was a very short peptide (a 30 aa type II restriction endonuclease) and 1 more was reported as a pseudo-gene in HAZ145.
Genomic DNA sequencing of M. arginini strains isolated from milk and lung tissue.
With the aim of understanding the relationships of the housefly gut isolate with other sources of M. arginini, we compared it to nine isolates of the same species from the same and other New York State farms and stored in our institutional repository. Specifically, we selected two milk isolates from the same farm, six milk isolates from other farms, and one bovine lung isolate. These were subjected to whole-genome sequencing. Isolates and sequencing data are reported in Table 1.
TABLE 1.
Isolates included in this study and relative sequencing data
| Isolate ID | Date | Source | Farm | Genome sizea | N. contigs | Avg. lengthb | Largest contig lengthb | N 50 | Accession no. |
|---|---|---|---|---|---|---|---|---|---|
| 20069D-01-02 | 2019-9 | Fly gut | A | 634,492 | 37 | 17,148 | 122,725 | 48,510 | JAJPGJ010000000 |
| 20069D-02-01 | 2017-3 | Milk | A | 476,969 | 602 | 792 | 6,746 | 1,025 | n.a.c |
| 20069D-03-04 | 2017-4 | Milk | A | 632,854 | 29 | 21,823 | 122,725 | 48,509 | JAPFAS000000000 |
| 20069D-02-03 | 2017-9 | Milk | B | 835,775 | 155 | 5,392 | 118,305 | 40,685 | JAPFAV000000000 |
| 20069D-03-07 | 2017-11 | Milk | B | 778,237 | 244 | 3,190 | 118,309 | 32,471 | JAPFAP000000000 |
| 20069D-03-02 | 2020-1 | Milk | C | 679,184 | 117 | 5,805 | 124,054 | 36,829 | JAPFAU000000000 |
| 20069D-03-03 | 2020-1 | Milk | C | 745,228 | 264 | 2,823 | 124,054 | 35,571 | JAPFAT000000000 |
| 20069D-03-05 | 2017-6 | Milk | D | 979,266 | 726 | 1,349 | 142,985 | 29,222 | JAPFAR000000000 |
| 20069D-03-06 | 2018-2 | Milk | E | 799,647 | 300 | 2,665 | 260,840 | 39,661 | JAPFAQ000000000 |
| 20069D-03-08 | 2018-9 | Lung tissue | F | 708,223 | 119 | 5,951 | 100,334 | 30,468 | JAPFAO000000000 |
Sum of the lengths of the individual scaffold/contigs of each assembly.
Lengths in base pairs.
See Data Availability statement.
As a result, eight out of the nine isolates (seven from milk and one from lung tissue) had an estimated genome size between 679,184 and 979,266 bp, all had similar average contig lengths (except 20069D-03-04), with an N50 comparable also to the housefly gut isolate; isolate 20069D-02-01 was an outlier, being the least sequenced one (genome size of 476,969 bp, a number of contigs three–times higher the average of the other strains, an average contig length of only 792 bp and a N50 considerably lower than the others).
Phylogenetic tree generation based on 16S rRNA.
A phylogenetic tree was generated based on 16S rRNA sequences. Adding to the newly sequenced M. arginini isolates and the reference strain, the tree included 17 additional publicly available sequences from M. arginini and related species (M. canadense, M. bovigenitalium, M. bovis, M. alkalescens, M. californicum), for a total of 27 16S rRNA sequences. The results are illustrated in Fig. 2.
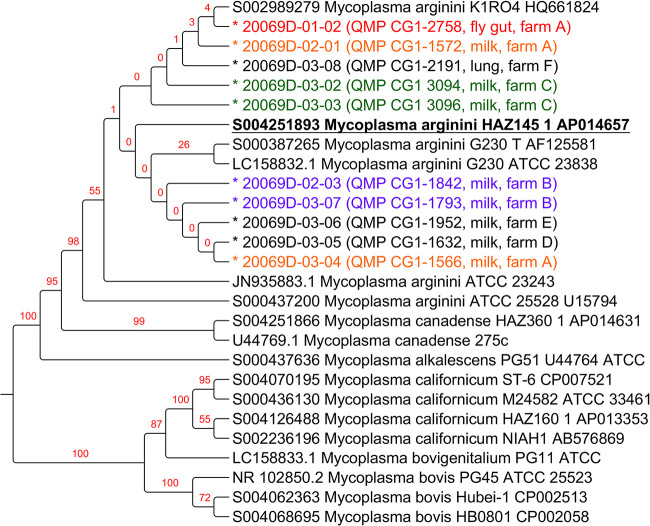
FIG 2.
Maximum likelihood phylogenetic tree among 27 16S rRNA sequences from M. arginini and related species (M. canadense, M. bovigenitalium, M. bovis, M. alkalescens, M. californicum). Values from 100 bootstraps are indicated in red over the branches. Bootstrap values indicate how many times the same branch is observed when repeating the generation of a phylogenetic tree on a resampled set of data made by artificial sequences of randomly selected sites of the original sequences with replacement; this produces a different composition of the data with the same sequence length as the original data sets. For the newly sequenced isolates (indicated by a prepending “*”), the farm and source of isolation is indicated; colors in the plot help highlighting isolates from the same farms: red = fly gut; orange = milk from the same farm as the fly gut one (i.e., farm A); Green, purple = paired isolates from the other farms. The 16S rRNA from the HAZ145 reference sequence is highlighted in bold and underlined.
All the newly sequenced strains grouped in the M. arginini branch of the tree, which, at higher levels, also encompassed M. canadense and M. alkalescens, all grouped according to their species; on the other branch of the tree, again grouped according to species, were M. californicum, M. bovigenitalium, and M. bovis. Within the M. arginini branch, some strains derived from the same farm were close one to each other, although with low bootstrap values, such as 20069D-03-02 and 20069D-03-03, isolated from farm C, in green, 20069D-02-03 and 20069D-03-07, isolated from farm B, in purple. Interestingly, the housefly gut strain 20069D-01-02, isolated in farm A, in red, and the milk isolate 20069D-02-01 from the same farm A, in orange, were close in the phylogenetic tree. The other milk isolate coming from farm A, in orange, clustered in the same branch of the tree but farther from the housefly gut isolate.
By evaluating the identity of 16S rRNA genes, all M. arginini sequences were very similar to each other with identity >98.3%. M. canadense and M. alkalescens, which were placed in the same macro-branch of the 16S rRNA tree, were very similar to M. arginini (identity >97.2%). On the other hand, similarities of M. arginini with M. californicum, M. bovigenitalium, and M. bovis were lower (<87.5%). The similarity between M. bovis, M. californicum, and M. bovigenitalium was around 94%, with the latter more similar to M. californicum (identity >97.3%) than to M. bovis (Fig. S1).
Phylogenetic classification refinement with conserved proteins in the newly sequenced strains.
The phylogenetic classification of M. arginini strains was refined by employing a subset of 49 conserved proteins in the HAZ145-1 reference genome and in all the 10 newly sequenced strains, including the housefly gut isolate to generate a maximum likelihood phylogenetic tree. The results are reported in Fig. 3.
FIG 3.
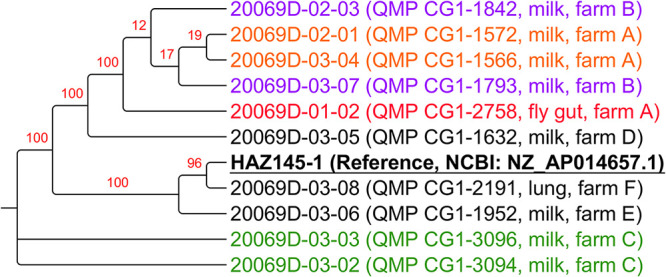
Maximum likelihood phylogenetic tree obtained creating an artificial sequence made up of 76 conserved genes among the 10 newly sequenced M. arginini strains and the HAZ145 reference genome. Values from 100 bootstraps are indicated in red over the branches. Bootstrap values indicate how many times the same branch is observed when repeating the generation of a phylogenetic tree on a resampled set of data made by artificial sequences of randomly selected sites of the original sequences with replacement; this produces a different composition of the data with the same sequence length as the original data sets. For the newly sequenced strains, the farm and source of isolation is indicated; colors in the plot help highlighting isolates from the same farms: red = fly gut strain; orange = milk-isolated strains from the same farm as the fly gut one (i.e., farm A); Green, purple = paired strains from the same farm. The HAZ145 reference sequence is highlighted in bold and underlined.
The phylogenetic tree of all 11 M. arginini strains based on the conserved protein sequences displayed four branches. In the first branch, the housefly gut isolate (20069D-01-02, red) clustered together with the two milk isolates from the same farm (A), which had been isolated from different cows 2 years earlier at different sampling times (20069D-02-01 and 20069D-03-04, orange). This branch also included the two milk isolates from farm B (20069D-02-03 and 20069D-03-07, purple). The milk isolates from farm C (20069D-03-02 and 20069D-03-03, green) clustered together on a separate branch of the tree. The milk isolate of M. arginini from farm E (20069D-03-06, black) was also distant from all the other newly sequenced isolates, clustering on the same branch with the reference strain HAZ145-1, isolated from bovine milk in Japan, and the bovine lung isolate from farm F (20069D-03-08, black). The milk isolates of farm D (20069D-03-05) fell in one branch equally distant from all the others.
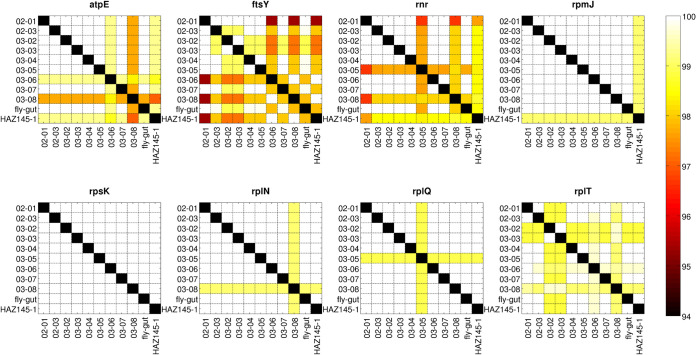
The heatmaps representing the identity percentage for the single genes among all M. arginini isolates explain their clustering behavior in the phylogenetic tree. (Fig. 4) (Fig. S2 to 9). In fact, some genes (e.g., rpsK, deoD, rpmF, upp) displayed 100% identity among all isolates, whereas others (e.g., ftsH, ftsY, mutM, and serS) had lower identity values. In the tree, the reference genome clustered separately from the majority of the newly sequenced isolates (as for example, in mnmA, mnmG, rpmJ identity plots); on the other hand, the milk isolates from farms E and F (20069D-03-06 and 20069D-03-08, respectively) clustered together with the reference genome, in line with the atpE, dnaJ, and dnaX gene identity plots; rplQ and rnr also highlighted the distance of the farm D isolate (20069D-03-05) from all the others; on the other hand, the identity patterns of the two isolates from farm C (20069D-03-02 and 20069D-03-03) were extremely similar.
FIG 4.
Representative heatmaps illustrating the percentage of identity (BLASTN value) between the 10 newly sequenced M. arginini strains and the HAZ145 reference genome. In the figure, 8 out of the 76 genes of the core genome are reported as representative of the different degree of identity among strains. The complete set of heatmaps can be found in Fig. S2 in the supplemental material −9.
Similarity analysis based on putative virulence genes.
With the purpose of investigating the virulence potential of the newly sequenced isolates, a total of 94 presumptive pathogenic genes were selected among those described in the literature (10) and aligned with all the M. arginini sequences. Of these, 35 (37%) had been previously reported for M. arginini and 59 (63%) for M. bovis.
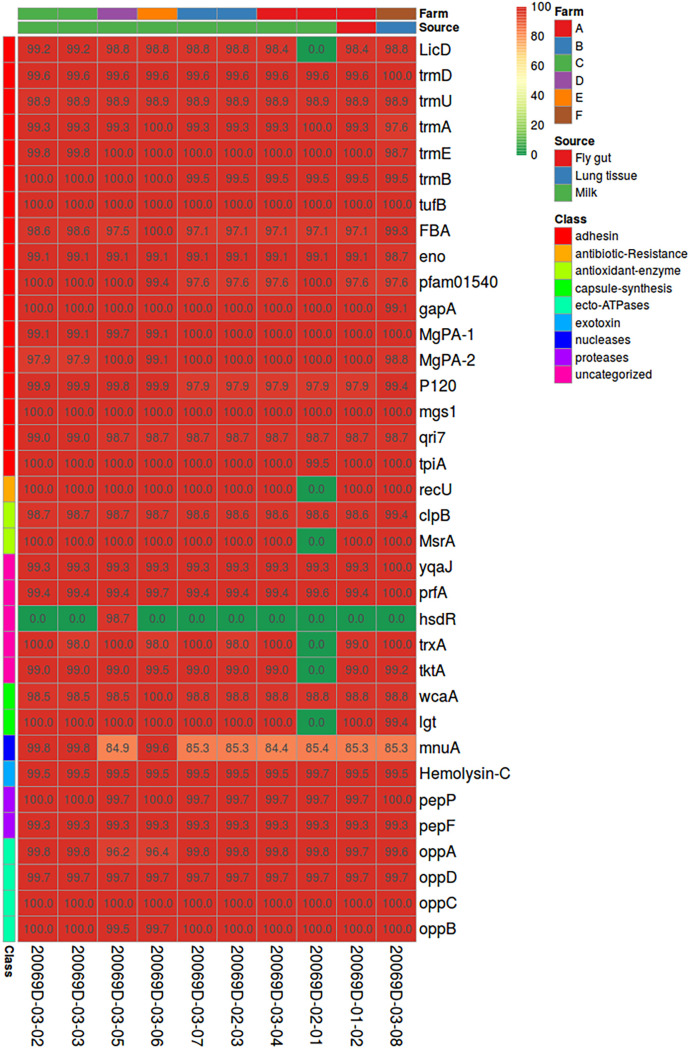
All the M. arginini isolates sequenced in this study carried 34 out of the 35 pathogenicity genes present in the M. arginini reference strain (HAZ145), all with very high protein sequence identity percentages (>98%). The only exception was hsdR (type I restriction enzyme endonuclease), missing in all but one of the newly sequenced isolates: 20069D-03-05 (milk, farm D), which showed the gene in its entirety (identity: 98.7). The lack of some genes (i.e., LicD, recU, MsrA, trxA, tktA, lgt) in 20069D-02-01 isolate (milk, farm A) was most likely due to the incomplete sequencing of this strain (Fig. 5).
FIG 5.
Heatmap representing the identity percentage (BLASTP value among translated nucleotide sequences) for a set of 35 pathogenicity-related genes found in the M. arginini reference strain HAZ145 over the 10 newly sequenced M. arginini isolates. Color is proportional to identity. Color bars over columns indicate the isolate origin and source; color bars to the left of each row represent the gene category.
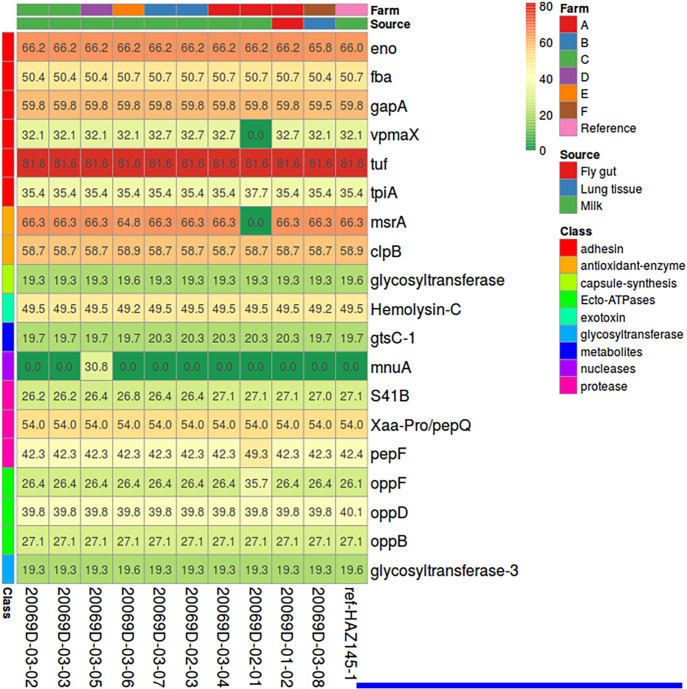
Due to the scarce information regarding M. arginini pathogenicity and the limited number of isolates already characterized, M. bovis was also selected as a comparator. As a result, the presence of M. bovis derived pathogenicity genes was dramatically lower: only 19 out of the 59 selected genes had a match with those of the newly sequenced M. arginini isolates (Fig. 6). Identity levels were generally low, in the range 19% to 82%; the genes with the highest similarities were tuf (identity of 81.6%), eno, gapA, clpB, and msrA (identities in the range of 59%–67%). Notably mnuA was lacking in all isolates with the only exception for 20069D-03-05 (milk, farm D). When present, the identity of the newly sequenced isolates with M. bovis pathogenicity genes was the same as that of the reference genome HAZ145.
FIG 6.
Heatmap representing the identity percentage (BLASTP value among translated nucleotide sequences) for a set of 59 pathogenicity-related genes found in the M. bovis strain PG45 (ATCC 25523) over the 10 newly sequenced M. arginini strains and the M. arginini reference strain HAZ145. Only 19 out of the 59 genes had a reliable match, whereas 13 others had a partial and 27 had no matches. Color is proportional to identity. Color bars over columns indicate the origin of the strain and the source the strain was extracted from; color bars to the left of each row represent the gene category.
DISCUSSION
To our knowledge, there are no previous reports of the association between bovine Mycoplasma spp. and any species of flies. On the other hand, M. arginini has been isolated from ruminant livestock’s respiratory tract and milk samples (11, 14). M. arginini is not one of the most frequent and pathogenic Mycoplasma species, but it can persist for a long time under different environmental conditions (15, 16). From other reports, no precise data suggest that M. arginini causes clinical mastitis, but it is often found in herds affected by mastitis associated with different etiologies. In a recent publication (4), M. arginini has been found to represent a 1.7% share of all mycoplasmas isolated in US dairy herds; it was isolated most frequently from bulk tank samples of herds where other Mycoplasma spp. were also found. It has been suggested that M. arginini may predispose animals to develop more severe signs of mastitis when other udder infections occur. Understanding the root of mycoplasma transmission is crucial for reducing dairy farm losses (4). To our knowledge, houseflies had never been considered possible carriers of Mycoplasma spp., contributing to their dissemination between animals or in the dairy environment. Despite the space-time interval linked to the dairy farms’ locations and to the multiple-year sampling, our results showed that the M. arginini isolated from the housefly was highly similar to the M. arginini isolated from milk samples.
The 16S rRNA-based phylogenetic tree of all Mycoplasma species sequences used for the present study identified two macro-branches. One of those located M. arginini isolates in the same macro-branch with M. canadense and M. alkalescens, similar to each other. On the other hand, M. californicum, M. bovigenitalium and M. bovis clustered in a different macro-branch distant from M. arginini. These results confirm what has been previously described by other authors concerning the phylogenesis of Mycoplasma spp. (17, 18). The same tree also placed our newly sequenced M. arginini isolates in the same branch as other published M. arginini sequences. However, our M. arginini isolates were located closer to each other, probably due to their shared country of origin (i.e., USA), whereas all other M. arginini 16S sequences in public databases were from other countries (e.g., Japan, South Africa). Within our isolates, there was also a tendency of the strains coming from the same farm to cluster together. Of particular interest was the very close clustering of the fly gut isolate with milk isolates from the same farm, even if those originated from samples collected 2 years earlier (farm A). In order to better refine the phylogenetic classification of the M. arginini isolates, we used a subset of 76 conserved proteins between the reference genome HAZ145-1 (12) and the isolates here sequenced. In this case, the housefly gut M. arginini isolate clustered together with four milk isolates originating from two of the six farms under evaluation. Most interestingly, two of them were from the same farm (farm A).
Such high similarity was especially notable since milk isolates had been obtained approximately 2 years earlier, and all the isolates from the other farms had been collected closer in time to the fly gut isolate. These findings suggest a relationship of the fly gut M. arginini isolate with the milk isolates from the same farm. The other two milk isolates clustering with the housefly gut isolate were from farm B. Milk isolates from farms C, D, and E, on the other hand, clustered in a distant branch from the fly gut isolate, and the two milk isolates from farm C clustered together on a separate branch of the tree, indicating a closer genetic makeup. The reference strain HAZ145-1, despite being described as isolated from milk, clustered with the lung isolate newly sequenced by our laboratory. The M. arginini isolated from the milk of farm E was distant from all other milk isolates but was closer to the reference strain and isolate F from the lung sample. Unfortunately, we have no data concerning exchange of cows or other factors behind the similarity between the two farms.
The 94 genes selected to characterize the M. arginini isolates of this study encoded 11 putative classes of virulence factors already described for Mycoplasma or other organisms. Instead, one group of genes was not associated with a particular class of bacterial virulence factors, despite being more related to viral or defense mechanisms (19–22) and was reported as an uncategorized class. We here evaluated the presence of genes encoding virulence factors that included (i) adhesins,(ii) antioxidant enzymes, (iii) capsule synthesis, (iv) ecto-ATPases, (v) exotoxins, (vi) nucleases, (vii) proteases, (viii) lipolytic enzymes, (ix) metabolites, (x) glycosyltransferases, and (xi) antibiotic resistance.
In the newly sequenced M. arginini isolates, we found 58% (54 out of 94) of all the putative virulence genes evaluated. The majority of these (n = 35) matched with the sequences already annotated for M. arginini HAZ145. On the other hand, only 19 genes matched with M. bovis sequences, and most of them showed a poor similarity.
The M. arginini isolated from fly gut was similar to the other milk- and lung-derived isolates in terms of presence and identity percentage to virulence factors from both M. arginini reference strain HAZ145 and M. bovis. Adhesion is the first and necessary step for both colonization and infection of the host cell (10). Thirty-seven of the 94 (39%) virulence genes considered in this study were classified as adhesins. In the genome sequences of our M. arginini isolates, we identified all the 17 putative adhesins contained in the M. arginini HAZ145 genome; these genes included LicD, trmD, trmU, trmA, trmE, trmB, tufB, FBA, eno, pfam01540, gapA, MgPA-1, MgPA-2, P120, mgs1, qri7, tpiA. A good share of these putative adhesins are metabolic enzymes reported as involved in pathogenicity with moonlighting mechanisms (23). Among these, LicD is a family of proteins involved in phosphorylcholine metabolism. Trm genes are methyltransferase groups with a possible cytoadherence function in Mycoplasma spp. (24). FBA (fructose-1,6-bisphosphate aldolase) and eno (enolase) are glycolytic enzymes with a potential role in adherence, invasion, and persistent infections as previously described for M. hyopneumoniae, M. bovis (25) and M. pneumoniae (26). gapA (Glyceraldehyde-3-phosphate) is another metabolic enzyme with a possible role as cytadhesin in M. gallisepticum (27); as mgs1, it has been described as erythrocyte-binding adhesin in M. suis (10, 28). tpiA (Triosephosphate isomerase) is a metabolic enzyme that serves as a critical interlink between glycolysis and glycerol metabolism and phospholipid synthesis. Some studies suggest that the surface-exposed TPIs also play a role in the pathogen–host interaction (29–31). It has been suggested by in vitro experiments that M. gallisepticum uses tpiA to cytoadhere the DF-1 cells (32, 33). Among the genes encoding adhesins within the M. bovis reference genome, the only one that showed high identity values (81.6%) for the newly characterized M. arginini isolates was tuf (Elongation factor TU, EF-Tu). EF-tu is one of the most abundant proteins in bacterial cells with a crucial role in protein translation (25, 34). However, EF-tu has also been reported with a cell surface localization as an important adhesin in many pathogens; it also appears to be the most immunoreactive protein and may be related to M. bovis biofilm formation (35). The biofilm growth provides M. bovis several advantages, including remarkable resistance to antimicrobials and persistence in the environment and host, which may result in the chronicity of a disease and considerable difficulty of treatment (36). EF-Tu is also a well-characterized extracellular matrix (ECM) binding adhesins for M. hyopneumoniae. The binding between EF-Tu with fibronectin contributes to the adhesion of M. hyopneumoniae in swine tracheal epithelial cells (STEC) (25). Being involved in crucial metabolic cell functions, the possible role of the above discussed housekeeping proteins in M. arginini pathogenicity through moonlighting functions will need to be investigated with specific, dedicated studies. Other proteins, however, are known for their primary role as adhesins. Based on the M. arginini HAZ145 genome annotation, pfam01540 gene codes for an adhesin lipoprotein (11). MgPA (putative MgPA-like protein) is a major adhesin mediating the attachment of the M. genitalium to the ciliated epithelium (10), resulting from a horizontal transfer from unknown origins. Both homologous MgPA-1 and MgPA-2 were found at high identity values to HAZ145 reference in all the new M. arginini isolates. P120 is a surface membrane protein described as a putative virulence factor in M. hominis (37). P120 undergoes genetic variability that could account for the ability of the microorganisms to circumvent the host immune system.
Proteases contribute to bacterial virulence for their potential to destroy functional and structural proteins of the host defense barriers (38). A total of three genes (pepP xaa-pro, pepF, and S41B) coding for proteases have been included in our analysis; all of them have been identified in the genome sequences of the new M. arginini strains characterized in this study. Other genes coding for proteins with protease functions were Xaa-Pro aminopeptidase and qri7_ Metal-dependent proteases with possible chaperone activity (39).
Four genes coding for uncategorized virulence factors (i.e., yqaJ, prfA, trxA, and tktA) were found in the characterized M. arginini isolates, including the one isolated from the fly gut. A fifth uncategorized virulence factor (hsdR) was missing in the fly gut M. arginini genome and in eight out of the other nine isolates. Another gene, recU, found in all but one the M. arginini genomes analyzed in this study, is reported to be involved in antimicrobial resistance of S. aureus (40) and Bacillus subtilis (41), and has a main role in DNA repair and homologous recombination, which is involved in the transfer of DNA within and between species and can lead to acquisition of new traits that can increase virulence or antibiotic resistance. Further studies will be needed to understand the potential role of these genes in M. arginini virulence.
In general, mycoplasmas lack the biosynthetic pathways for cholesterol, fatty acids, some amino acids, purines, and pyrimidines and, therefore, acquire these through membrane transport proteins (42). Mycoplasma genomes possess a diversity of ABC transporters predicted to be involved in the uptake of these inorganic and organic substrates (43). In the fly gut-derived M. arginini and all the other genomes analyzed in this study, the oligopeptide ABC transporter system was synthesized in the opp operon cluster genes (oppA, oppB, oppC, oppD genes). These uptake systems are mainly involved in cell nutrition; however, some might be associated with virulence (44, 45).
Notably, the in-silico virulence gene profiling highlighted that the housefly M. arginini isolate was genetically comparable to the other M. arginini isolated from bovine milk and lung samples. This is critical to define houseflies as reservoirs and carriers of M. arginini. We should consider that the contagiousness level of M. arginini is not known, and there is not much information available from the literature. Still, most dairy herds have zero tolerance for Mycoplasma spp., and the animals resulting positive are culled independently from which Mycoplasma species are isolated from milk samples. The gold standard detection methods rely on bacterial cultures showing colonies with mycoplasma morphology, and most of the time, dairy management decisions in the case of positive cows are based on culture results without going for any further speciation. At this point, it is important to reconsider the environment as a possible source of Mycoplasma spp. in dairies and in particularly houseflies as their carriers.
More in vitro or in vivo studies will be needed to confirm our findings based on in-silico analysis and to establish the mechanism of Mycoplasma transmission between houseflies and dairy cows. Since the new M. arginini strain characterized here has been isolated from the gut of a housefly and not from its exoskeleton, transmission by mechanical translocation can be excluded for the conditions of the present study. Here, transmission routes are most likely represented by flies regurgitation (bio-enhanced transmission) and or defecation (46). It is essential to consider that microbial contamination from the dairy environment also includes residues of milk, from either healthy or diseased cows, that could persist in the environment due to poor sanitation and hygiene dairy practices. Milk residue on utensils and milking equipment represent feeding elements for flies (47). Once fed and when flies find another food source or place to lay eggs, the crop fluids are either externally released by regurgitation or ingested internally to the gut to be then re-emitted into the environment by defecation. These findings reinforce the importance of mycoplasma diagnostics in cattle by performing culture on samples collected aseptically to ensure that what is detected is a natural agent of infection, not a contaminant.
Conclusion.
The data presented here support the hypothesis that houseflies could be potential carriers of Mycoplasma spp. and need to be considered within the possible roots of environmental transmission. Further studies with a higher population of flies are required to confirm our hypothesis and evaluate if other Mycoplasma spp. can be isolated from houseflies. Concerning the potential influence of M. arginini on mammary gland health, the housefly isolate possessed all the pathogenicity genes identified in the reference strain and shared a low number of virulence genes with M. bovis.
MATERIALS AND METHODS
Housefly sampling and homogenization.
The housefly that carried M. arginini isolate, the focal point of this study, was one of the specimens collected from our research survey conducted in 2019 on a commercial dairy farm in New York State. Methods for houseflies sampling and homogenization are not described in the manuscript but in the referenced paper (12).
Milk samples.
M. arginini isolates found in bovine milk originated from eight samples submitted to the Quality Milk Production Center (QMPS) at Cornell University for Mycoplasma testing. Milk samples were received from four different dairy farms in New York State. Milk samples were submitted to the QMPS labs over 3 years (2017 to 2020). Most milk isolates, 7 of 8 (87.5%), were collected from composite samples, and 1 of 8 (12.5%) was obtained from an individual quarter sample. Composite samples were tested for herd surveys or screening of fresh cows, while the quarter sample was collected from a case of clinical mastitis.
Lung tissue sample.
The M. arginini isolate found in bovine lung tissue was collected from a chronic pneumonia case submitted for Mycoplasma testing to the Bacteriology Laboratory of the Animal Health Diagnostic Center (AHDC) at Cornell University.
Mycoplasma testing.
Enrichment: The Mycoplasma enrichment step was performed only for the fly sample after being processed by the homogenization step described above. A volume of 0.5 mL from the fly’s suspension were transferred in 1 mL of Mycoplasma Broth (Hardy Diagnostics, Santa Maria, CA) and incubated for 72 h at 35°C to 38°C with 5 to 10% CO2. After the incubation, the sample was submitted for mycoplasma culture. As enrichment step, the sample was inoculated using cotton swabs on Mycoplasma Hayflick Agar (Northeast Laboratory) and incubated for 7 days at 37°C.
Culture: All samples, independently from the type of specimen, were mixed by inversion and then streaked by sterile cotton swab on Mycoplasma Hayflick Agar (Northeast Laboratory) and incubated for 7 days at 35 to 38°C with 5 to 10% CO2. Mycoplasma growth was detected by visual inspection of culture plates under an illuminated stage stereomicroscope. Mycoplasma colonies were confirmed by a PCR specific for Mycoplasma and Acholeplasma followed by Sanger sequencing for species identification (13).
Library preparation.
Library preparation and sequencing were performed by Admera Health (South Plainfield, NJ, USA). Isolated genomic DNA was quantified with Qubit 2.0 DNA HS Assay (ThermoFisher, Waltham, MA, USA) and DNA quality was evaluated by Genomic DNA ScreenTape analysis (Agilent Technologies, Santa Clara, CA, USA). Library preparation was performed using Nextera XT library kit (Illumina, San Diego, CA, USA) following the manufacturer’s recommendations while barcoding was performed using Illumina 8-nt dual-indices. The final library was evaluated by Qubit 2.0 DNA HS Assay and the quality of the library generated was examined by TapeStation HSD1000 ScreenTape (Agilent Technologies). Final library quantity was measured by KAPA SYBR FAST qPCR with QuantStudio 5 System (Applied Biosystems, Waltham, MA, USA). Equimolar pooling of libraries was performed based on qPCR QC values before sequencing on an Illumina HiSeq with a read length configuration of 2 × 150 paired end reads, obtaining 1 M PE reads per sample.
Genome assembly and annotation.
For the gut fly-isolated strain, as well as for milk- (n = 8) and lung-tissue-isolated (n = 1) strains, assembly was conducted by Admera Health using SPAdes (v. 3.14.0) with parameter “-k 127.” SPAdes was also used for generating genomic scaffolds on gut fly strain by using paired-end reads in the sequencing to concatenate contigs (48). The initial functional annotation on all sequenced strains was initially performed by prokka (v. 1.14.6) (49), and further refined by using GView Server utility (https://server.gview.ca/) (50) in PanGenome mode, employing all newly sequenced genomes and an available reference sequence (i.e., Mycoplasma arginini, strain HAZ145_1, https://www.ncbi.nlm.nih.gov/nuccore/NZ_AP014657.1) as the template (11). Finally, a second optimization was performed by performing a protein BLAST+ (v. 2.11.0) (51) search against “nr” database and limiting matches only to M. arginini-derived proteins (additional parameters: -word_size = 6. -max_target_seqs = 5, -evalue = 10e-3). Genome features were plotted by GView (50).
Phylogenetic tree building.
Phylogenetic analysis of the gut fly-isolated strain compared to other Mycoplasma species was inferred using the 16S rRNA genomic sequence as a marker. A total of 27 sequences were used for tree building: 16 M. arginini (including 10 from our assemblies and six from publicly available resources), 2 M. canadense, 1 M. alkalescens, 4 M. californicum, 1 M. bovigenitalium, and 3 M. bovis. (Table 2).
TABLE 2.
List of 16S rRNA sequences used for building the phylogenetic tree
| Mycoplasma species | Strain | Accession N. | Source |
|---|---|---|---|
| M. arginini | 20069D-01-02a | JAJPGJ000000000 b | Housefly gut |
| 20069D-02-01a | ON890822 | Bovine Composite Milk | |
| 20069D-02-03a | ON890823 | Bovine Mastitis Milk | |
| 20069D-03-02a | ON890824 | Bovine Composite Milk | |
| 20069D-03-03a | ON890825 | Bovine Composite Milk | |
| 20069D-03-04a | ON890826 | Bovine Composite Milk | |
| 20069D-03-05a | ON890827 | Bovine Composite Milk | |
| 20069D-03-06a | ON890828 | Bovine Composite Milk | |
| 20069D-03-07a | ON890829 | Bovine Composite Milk | |
| 20069D-03-08a | ON890830 | Bovine Lung | |
| G230 / ATCC 23838 | LC158832.1 | Mouse brain with sheep scrapies | |
| G230(T) | AF125581.1 | — | |
| HAZ145 | AP014657.1 | Bovine Mastitis Milk | |
| K1RO4 | HQ661824 | Vaginal swab from Dorper sheep | |
| ATCC 23243 | JN935883.1 | Sheep Lung with pneumonia | |
| ATCC 25528 | U15794 | Lion Lung | |
| M. canadense | 275c | U44769 | Bos taurus |
| HAZ360 | AP014631 | Bovine Mastitis Milk | |
| M. alkalescens | PG51 | U44764 | Bovine Nasal cavity |
| M. californicum | ATCC 33461 | M24582 | Bovine Mastitis Milk |
| NIAH1 | AB576869 | Bovine Mastitis Milk | |
| ATCC 33461T | CP007521 | Bovine Mastitis Milk | |
| HAZ160_1 | AP013353 | Bovine Mastitis Milk | |
| M. bovigenitalium | PG11 / ATCC 19852 | LC158833 | Bovine Genital Tract |
| M. bovis | ATCC 25523 | NR_102850 | Bovine Mastitis Milk |
| HB0801 | CP002058 | Bovine Lung | |
| Hubei-1 | CP002513.1 | Bovine Lung |
Indicates the M. arginini strains sequenced in the present paper; —: not available.
The sequence of 16S rRNA is contained in sequence CONTIG_17, bases 14–1531, feature id: KMZ28_00575. For each sequence, the species ID, the strain, the GenBank accession number and the source are indicated.
Identity among the 16S rRNA sequences was assessed by BLAST+. Phylogenetic reconstruction based on 76 genes conserved on all the 10 strains and the reference was performed using Phylogeny.fr utility (https://www.phylogeny.fr/) (52), using clustalO for multiple alignments, Gblocks for alignment curation, and PhyML for tree building, with 100 bootstraps. TreeGraph (v. 2.15.0-887 beta) was used to draw the final tree (53).
Check of pathogenic genes.
A total of 94 presumptive pathogenic genes were selected among those described in the literature (10); 35 were previously reported for M. arginini and 59 for M. bovis. The nucleotide sequences were retrieved from NCBI GenBank and translated into protein using utility “transeq” from EMBOSS package (54). Protein sequences of the newly sequenced M. arginini strains were those predicted by prokka. BLAST+ (-task= blastp, -evalue 10e-3) and were employed to find best matches between references and queries, selecting only those spanning at least 75% of the reference or query (whatever the shorter of the two). The percentage of identity was plotted in a heatmap using ClustVis (https://biit.cs.ut.ee/clustvis/) (55).
Data availability.
All raw data described in the paper are linked in NCBI BioProject database (accession number: PRJNA733869). Raw sequencing reads of the isolates are available within NCBI Short-Reads Archive (SRA) under accession number SRR17253962; 16S rRNA sequences are available in GenBank, under accession numbers: ON890822-ON890830. The Whole Genome Shotgun project of the fly gut-isolated M. arginini (20069D-01-02) has been deposited at DDBJ/ENA/GenBank under the accession JAJPGJ000000000. The version described in this paper is version JAJPGJ010000000. The Whole Genome Shotgun projects for the milk- and lung-derived M. arginini isolated have been deposited under accession JAPFAO000000000, JAPFAP000000000, JAPFAQ000000000, JAPFAR000000000, JAPFAS000000000, JAPFAT000000000, JAPFAU000000000, and JAPFAV000000000. Due to sequencing coverage issue, WGS for isolate 20069D-02-01 is missing; the sequences of the 76 genes used for the core genome analysis for this isolate are available in GenBank under accession numbers OQ078009-OQ078084. See also Tables 1 and 2 for more details.
ACKNOWLEDGMENTS
This study was in part funded by Quality Milk Production Service (QMPS) internal program 2019.
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
Contributor Information
P. Moroni, Email: pm389@cornell.ed.
Ryan Rego, Institute of Parasitology, Biology Centre, ASCR.
REFERENCES
- 1.Maunsell FP, Woolums AR, Francoz D, Rosenbusch RF, Step DL, Wilson DJ, Janzen ED. 2011. Mycoplasma bovis infections in cattle. J Vet Intern Med 25:772–783. doi: 10.1111/j.1939-1676.2011.0750.x. [DOI] [PubMed] [Google Scholar]
- 2.Punyapornwithaya V, Fox LK, Hancock DD, Gay JM, Alldredge JR. 2012. Time to clearance of mycoplasma mastitis: the effect of management factors including milking time hygiene and preferential culling. Can Vet J 53:1119–1122. [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholas RA, Fox LK, Lysnyansky I. 2016. Mycoplasma mastitis in cattle: to cull or not to cull. Vet J 216:142–147. doi: 10.1016/j.tvjl.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Gioia G, Addis MF, Santisteban C, Gross B, Nydam DV, Sipka AS, Virkler PD, Watters RD, Wieland M, Zurakowski MJ, Moroni P. 2021. Mycoplasma species isolated from bovine milk collected from US dairy herds between 2016 and 2019. J Dairy Sci 104:4813–4821. doi: 10.3168/jds.2020-19171. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DJ, Skirpstunas RT, Trujillo JD, Cavender KB, Bagley CV, Harding RL. 2007. Unusual history and initial clinical signs of Mycoplasma bovis mastitis and arthritis in first-lactation cows in a closed commercial dairy herd. J Am Vet Med Assoc 230:1519–1523. doi: 10.2460/javma.230.10.1519. [DOI] [PubMed] [Google Scholar]
- 6.Wilson DJ, Justice-Allen A, Goodell G, Baldwin TJ, Skirpstunas RT, Cavender KB. 2011. Risk of Mycoplasma bovis transmission from contaminated sand bedding to naive dairy calves. J Dairy Sci 94:1318–1324. doi: 10.3168/jds.2010-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Justice-Allen A, Trujillo J, Corbett R, Harding R, Goodell G, Wilson D. 2010. Survival and replication of Mycoplasma species in recycled bedding sand and association with mastitis on dairy farms in Utah. J Dairy Sci 93:192–202. doi: 10.3168/jds.2009-2474. [DOI] [PubMed] [Google Scholar]
- 8.West LS. 1951. The housefly: its natural history, medical importance, and control. Comstock Pub. Co., Ithaca, NY. [Google Scholar]
- 9.Geden CJ, Nayduch D, Scott JG, Burgess ER, IV, Gerry AC, Kaufman PE, Thomson J, Pickens V, Machtinger ET. 2021. House fy (Diptera: Muscidae): biology, pest status, current management prospects, and research needs. J Integr Pest Manag 12:39. doi: 10.1093/jipm/pmaa021. [DOI] [Google Scholar]
- 10.Yiwen CW, Yueyue WY, Qin LM, Zhu CM, You XX. 2021. Infection strategies of mycoplasmas: unraveling the panoply of virulence factors. Virulence 12:788–817. doi: 10.1080/21505594.2021.1889813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata E. 2015. Complete genome sequence of Mycoplasma arginini strain HAZ 145_1 from bovine mastitic milk in Japan. Genome Announc 3:e00265-15. doi: 10.1128/genomeA.00265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gioia G, Freeman J, Sipka A, Santisteban C, Wieland M, Gallardo VA, Monistero V, Scott JG, Moroni P. 2022. Pathogens associated with houseflies from different areas within a New York State dairy. J Dairy Sci Communications 3:285–290. doi: 10.3168/jdsc.2021-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gioia G, Werner B, Nydam DV, Moroni P. 2016. Validation of a mycoplasma molecular diagnostic test and distribution of mycoplasma species in bovine milk among New York State dairy farms. J Dairy Sci 99:4668–4677. doi: 10.3168/jds.2015-10724. [DOI] [PubMed] [Google Scholar]
- 14.Thomas A, Ball H, Dizier I, Trolin A, Bell C, Mainil J, Linden A. 2002. Isolation of Mycoplasma species from the lower respiratory tract of healthy cattle and cattle with respiratory disease in Belgium. Vet Rec 151:472–476. doi: 10.1136/vr.151.16.472. [DOI] [PubMed] [Google Scholar]
- 15.Nagatomo H, Takegahara Y, Sonoda T, Yamaguchi A, Uemura R, Hagiwara S, Sueyoshi M. 2001. Comparative studies of the persistence of animal mycoplasmas under different environmental conditions. Vet Microbiol 82:223–232. doi: 10.1016/s0378-1135(01)00385-6. [DOI] [PubMed] [Google Scholar]
- 16.Stipkovits L, Somogyi M, Asvanyi B, Toth A, Szathmary S. 2013. Short communication: role of Mycoplasma arginini in mastitis caused by Streptococcus dysgalactiae. J Dairy Sci 96:1661–1667. doi: 10.3168/jds.2012-5669. [DOI] [PubMed] [Google Scholar]
- 17.Mattsson JG, Guss B, Johansson K-E. 1994. The phylogeny of Mycoplasma bovis as determined by sequence analysis of the 16S rRNA gene. FEMS Microbiol Lett 115:325–328. doi: 10.1111/j.1574-6968.1994.tb06658.x. [DOI] [PubMed] [Google Scholar]
- 18.Weisburg WG, Tully JG, Rose DL, Petzel JP, Oyaizu H, Yang D, Mandelco L, Sechrest J, Lawrence TG, Vanetten J, Maniloff J, Woese CR. 1989. A phylogenetic analysis of the Mycoplasmas: basis for their classification. J Bacteriol 171:6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renzoni A, Klarsfeld A, Dramsi S, Cossart P. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect Immun 65:1515–1518. doi: 10.1128/iai.65.4.1515-1518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vellani TS, Myers RS. 2003. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J Bacteriol 185:2465–2474. doi: 10.1128/JB.185.8.2465-2474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domain F, Bina XR, Levy SB. 2007. Transketolase A, an enzyme in central metabolism, derepresses the marRAB multiple antibiotic resistance operon of Escherichia coli by interaction with MarR. Mol Microbiol 66:383–394. doi: 10.1111/j.1365-2958.2007.05928.x. [DOI] [PubMed] [Google Scholar]
- 22.May HC, Yu JJ, Zhang H, Wang Y, Cap AP, Chambers JP, Guentzel MN, Arulanandam BP. 2019. Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PLoS One 14:e0218505. doi: 10.1371/journal.pone.0218505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson B, Martin A. 2013. Bacterial moonlighting proteins and bacterial virulence, p 155–213. In Dobrindt U, Hacker JH, Svanborg C (ed), Between Pathogenicity and Commensalism. Springer Berlin Heidelberg, Berlin, Heidelberg. doi: 10.1007/82_2011_188. [DOI] [Google Scholar]
- 24.Khan S, Zakariah M, Rolfo C, Robrecht L, Palaniappan S. 2017. Prediction of mycoplasma hominis proteins targeting in mitochondria and cytoplasm of host cells and their implication in prostate cancer etiology. Oncotarget 8:30830–30843. doi: 10.18632/oncotarget.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, Wang H, Wang J, Feng Z, Wu M, Liu B, Xin J, Xiong Q, Liu M, Shao G. 2018. Elongation factor thermo unstable (EF-Tu) moonlights as an adhesin on the surface of Mycoplasma hyopneumoniae by binding to fibronectin. Front Microbiol 9:974. doi: 10.3389/fmicb.2018.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas C, Jacobs E, Dumke R. 2013. Characterization of pyruvate dehydrogenase subunit B and enolase as plasminogen-binding proteins in Mycoplasma pneumoniae. Microbiology (Reading) 159:352–365. doi: 10.1099/mic.0.061184-0. [DOI] [PubMed] [Google Scholar]
- 27.Indikova I, Much P, Stipkovits L, Siebert-Gulle K, Szostak MP, Rosengarten R, Citti C. 2013. Role of the GapA and CrmA cytadhesins of Mycoplasma gallisepticum in promoting virulence and host colonization. Infect Immun 81:1618–1624. doi: 10.1128/IAI.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Q, Song W, Zhang W, He L, Fang R, Zhou Y, Shen B, Hu M, Zhao J. 2018. Identification of erythrocyte membrane proteins interacting with Mycoplasma suis GAPDH and OSGEP. Res Vet Sci 119:85–90. doi: 10.1016/j.rvsc.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Furuya H, Ikeda R. 2011. Interaction of triosephosphate isomerase from Staphylococcus aureus with plasminogen. Microbiol Immunol 55:855–862. doi: 10.1111/j.1348-0421.2011.00392.x. [DOI] [PubMed] [Google Scholar]
- 30.Pereira LA, Bao SN, Barbosa MS, da Silva JLM, Felipe MSS, de Santana JM, Mendes-Giannini MJS, Soares CMD. 2007. Analysis of the Paracoccidioides brasiliensis triosephosphate isomerase suggests the potential for adhesin function. FEMS Yeast Res 7:1381–1388. doi: 10.1111/j.1567-1364.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- 31.Ramiah K, van Reenen CA, Dicks LM. 2008. Surface-bound proteins of Lactobacillus plantarum 423 that contribute to adhesion of Caco-2 cells and their role in competitive exclusion and displacement of Clostridium sporogenes and Enterococcus faecalis. Res Microbiol 159:470–475. doi: 10.1016/j.resmic.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 32.Bao SJ, Chen DQ, Yu SQ, Chen HJ, Tan L, Hu MR, Qiu XS, Song CP, Ding C. 2015. Characterization of triosephosphate isomerase from Mycoplasma gallisepticum. FEMS Microbiol Lett 362:fnv140. doi: 10.1093/femsle/fnv140. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, Yu S, Shen X, Chen D, Qiu X, Song C, Ding C. 2011. The Mycoplasma gallisepticum α-enolase is cell surface-exposed and mediates adherence by binding to chicken plasminogen. Microb Pathog 51:285–290. doi: 10.1016/j.micpath.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Weijland A, Harmark K, Cool RH, Anborgh PH, Parmeggiani A. 1992. Elongation factor Tu: a molecular switch in protein biosynthesis. Mol Microbiol 6:683–688. doi: 10.1111/j.1365-2958.1992.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, Hao H, Zhao P, Ji W, Li M, Liu Y, Chu Y. 2018. Differential immunoreactivity to bovine convalescent serum between Mycoplasma bovis biofilms and planktonic cells revealed by comparative immunoproteomic analysis. Front Microbiol 9:379. doi: 10.3389/fmicb.2018.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAuliffe L, Ellis RJ, Miles K, Ayling RD, Nicholas RAJ. 2006. Biofilm formation by mycoplasma species and its role in environmental persistence and survival. Microbiology (Reading) 152:913–922. doi: 10.1099/mic.0.28604-0. [DOI] [PubMed] [Google Scholar]
- 37.Mardassi BB, Ayari H, Béjaoui-Khiari A, Mlik B, Moalla I, Amouna F. 2007. Genetic variability of the P120' surface protein gene of Mycoplasma hominis isolates recovered from Tunisian patients with uro-genital and infertility disorders. BMC Infect Dis 7:142. doi: 10.1186/1471-2334-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lantz MS. 1997. Are bacterial proteases important virulence factors? J Periodontal Res 32:126–132. doi: 10.1111/j.1600-0765.1997.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 39.Abdullah KM, Lo RY, Mellors A. 1991. Cloning, nucleotide sequence, and expression of the Pasteurella haemolytica A1 glycoprotease gene. J Bacteriol 173:5597–5603. doi: 10.1128/jb.173.18.5597-5603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira AR, Reed P, Veiga H, Pinho MG. 2013. The Holliday junction resolvase RecU is required for chromosome segregation and DNA damage repair in Staphylococcus aureus. BMC Microbiol 13:18. doi: 10.1186/1471-2180-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen LB, Setlow P. 2000. Penicillin-binding protein-related factor A is required for proper chromosome segregation in Bacillus subtilis. J Bacteriol 182:1650–1658. doi: 10.1128/JB.182.6.1650-1658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wium M, Botes A, Bellstedt DU. 2015. The identification of oppA gene homologues as part of the oligopeptide transport system in mycoplasmas. Gene 558:31–40. doi: 10.1016/j.gene.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 43.Staats CC, Boldo J, Broetto L, Vainstein M, Schrank A. 2007. Comparative genome analysis of proteases, oligopeptide uptake and secretion systems in Mycoplasma spp. Genet Mol Biol 30:225–229. doi: 10.1590/S1415-47572007000200009. [DOI] [Google Scholar]
- 44.Cacciotto C, Cubeddu T, Addis MF, Anfossi AG, Tedde V, Tore G, Carta T, Rocca S, Chessa B, Pittau M, Alberti A. 2016. Mycoplasma lipoproteins are major determinants of neutrophil extracellular trap formation. Cell Microbiol 18:1751–1762. doi: 10.1111/cmi.12613. [DOI] [PubMed] [Google Scholar]
- 45.Cacciotto C, Addis MF, Coradduzza E, Carcangiu L, Nuvoli AM, Tore G, Dore GM, Pagnozzi D, Uzzau S, Chessa B, Pittau M, Alberti A. 2013. Mycoplasma agalactiae MAG_5040 is a Mg2+-dependent, sugar-nonspecific SNase recognised by the host humoral response during natural infection. PLoS One 8:e57775. doi: 10.1371/journal.pone.0057775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onwugamba FC, Fitzgerald JR, Rochon K, Guardabassi L, Alabi A, Kuhne S, Grobusch MP, Schaumburg F. 2018. The role of “filth flies” in the spread of antimicrobial resistance. Travel Med Infect Dis 22:8–17. doi: 10.1016/j.tmaid.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Adly E, Hegazy AA, Kamal M, Abu-Hussien SH. 2022. Midguts of Culex pipiens L. (Diptera: culicidae) as a potential source of raw milk contamination with pathogens. Sci Rep 12:13183. doi: 10.1038/s41598-022-16992-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 50.Stothard P, Grant JR, Van Domselaar G. 2019. Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform 20:1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST plus: architecture and applications. BMC Bioinform 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stover BC, Muller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. Bmc Bioinform 11. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 55.Metsalu T, Vilo J. 2015. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res 43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S9. Download spectrum.03010-22-s0001.pdf, PDF file, 3.9 MB (3.9MB, pdf)
Data Availability Statement
All raw data described in the paper are linked in NCBI BioProject database (accession number: PRJNA733869). Raw sequencing reads of the isolates are available within NCBI Short-Reads Archive (SRA) under accession number SRR17253962; 16S rRNA sequences are available in GenBank, under accession numbers: ON890822-ON890830. The Whole Genome Shotgun project of the fly gut-isolated M. arginini (20069D-01-02) has been deposited at DDBJ/ENA/GenBank under the accession JAJPGJ000000000. The version described in this paper is version JAJPGJ010000000. The Whole Genome Shotgun projects for the milk- and lung-derived M. arginini isolated have been deposited under accession JAPFAO000000000, JAPFAP000000000, JAPFAQ000000000, JAPFAR000000000, JAPFAS000000000, JAPFAT000000000, JAPFAU000000000, and JAPFAV000000000. Due to sequencing coverage issue, WGS for isolate 20069D-02-01 is missing; the sequences of the 76 genes used for the core genome analysis for this isolate are available in GenBank under accession numbers OQ078009-OQ078084. See also Tables 1 and 2 for more details.