In this study, Yang et al. show that the translation initiation factor eIF4E1b protects specific maternal mRNAs and promotes their translation during oocyte-to-zygote transition. This jump-starts critical zygotic chromatin remodeling and genome activation, the loss of which results in female infertility due to developmental arrest in the two-cell embryos.
Keywords: Eif4e1b, zygotic genome activation, mouse embryo, maternal RNA translation
Abstract
Translation of maternal mRNAs is detected before transcription of zygotic genes and is essential for mammalian embryo development. How certain maternal mRNAs are selected for translation instead of degradation and how this burst of translation affects zygotic genome activation remain unknown. Using gene-edited mice, we document that the oocyte-specific eukaryotic translation initiation factor 4E family member 1b (eIF4E1b) is the regulator of maternal mRNA expression that ensures subsequent reprogramming of the zygotic genome. In oocytes, eIF4E1b binds to transcripts encoding translation machinery proteins, chromatin remodelers, and reprogramming factors to promote their translation in zygotes and protect them from degradation. The protein products are thought to establish an open chromatin landscape in one-cell zygotes to enable transcription of genes required for cleavage stage development. Our results define a program for rapid resetting of the zygotic epigenome that is regulated by maternal mRNA expression and provide new insights into the mammalian maternal-to-zygotic transition.
Terminally differentiated, transcriptionally quiescent mammalian gametes fuse at fertilization and must be reprogrammed to express embryonic genes (Ladstätter and Tachibana 2019). Maternal transcripts stored in oocytes encode proteins that directly or indirectly modify the epigenome (Zhang and Smith 2015), after which the embryonic genome orchestrates development (Tadros and Lipshitz 2009). Mechanisms controlling this maternal-to-zygotic transition (MZT) are not fully understood. The earliest transcripts from mouse zygotic genes are detected late in one-cell zygotes and are followed by a more extensive rise of gene expression in two-cell embryos. The two waves of transcription are designated minor and major zygotic genome activation (ZGA), respectively (Schulz and Harrison 2019). Translation in mammals occurs before activation of zygotic genes (Latham et al. 1991), and mouse embryos arrest at the one-cell stage if this translation is inhibited (Aoki et al. 2003; Israel et al. 2019). Why this early burst of translation is essential for embryogenesis remains unknown, but recent experiments suggest that this translation is highly selective (Alizadeh et al. 2005; Wang et al. 2010), as most maternal RNAs and proteins are rapidly cleared (Despic and Neugebauer 2018).
Considering the brief temporal window between fertilization and zygotic gene transcription, we hypothesize that maternal mRNA translation is highly regulated to ensure availability of factors for efficient zygotic genome reprogramming. Different maternal RNAs have been shown to have varied binding affinities for ribosomes, suggesting different translation efficiencies (Xiong et al. 2022; Zhang et al. 2022). However, ribosomes themselves have no RNA preference, and alternative strategies must be required to ensure translation of essential maternal RNAs in zygotes. Initiation of eukaryotic protein synthesis is controlled by the eIF4F complex (Jackson et al. 2010). eIF4F is a heterogenous set of eIF4A, eIF4E, and eIF4G. eIF4E binds directly to the 5′ 7-methylguanosine (m7G) cap structure of mRNA and is indispensable for translation initiation. The canonical eIF4E1a (also known as eIF4E) is expressed widely in different tissues and is the default eIF4E component for initiation of translation (Joshi et al. 2005). A germ cell-specific isoform of eIF4E, the eukaryotic translation initiation factor 4E family member 1b (eIF4E1b), was identified in a large-scale screen for genes that enable mouse MZT and is well conserved (Evsikov et al. 2006; Evsikov and Marin de Evsikova 2009). Using gene-edited mice in combination with multiple low-input molecular tools, we identified eIF4E1b as a regulator that is essential for pre-ZGA maternal RNA translation and the onset of preimplantation development.
Results
eIF4E1b is a germ cell-specific eIF4E homolog expressed late in oogenesis
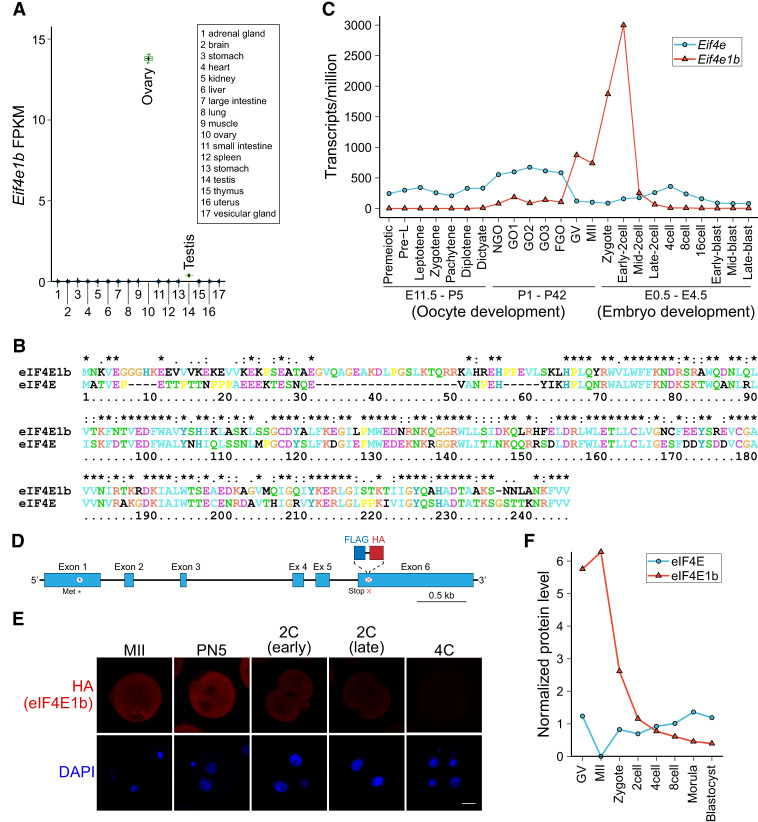
It has previously been reported that protein synthesis in fertilized mouse eggs is required for activation of zygotic genes (Wang and Latham 1997; Aoki et al. 2003), which we confirmed under our experimental conditions (Supplemental Fig. S1). To discover possible molecular mechanisms, we reviewed RNA-seq data sets of genes expressed during mouse oogenesis (Gu et al. 2019; Niu and Spradling 2020; Wu and Dean 2020) and early cleavage stage development (Deng et al. 2014). The eukaryotic translation initiation factor 4E family member 1b (Eif4e1b), a germ cell-specific (Li et al. 2017) homolog (Fig. 1A,B) of the ubiquitously expressed translation initiation factor 4E (Eif4e), is highly expressed in the latter stages of oogenesis and persists in two-cell embryos before disappearing at the four-cell stage (Fig. 1C).
Figure 1.
Eif4e1b expresses late in oogenesis and persists until two-cell embryos. (A) Abundance (FPKM [fragments per kilobase of transcript per million mapped reads]) of Eif4e1b transcripts in different tissues from published data. (B) Alignment of mouse eIF4E1b (NP_001028441.1) protein sequence with that of eIF4E (NP_031943.3). (C) Abundance (TPM [transcripts per million mapped reads]) of Eif4e1a (Eif4e) and Eif4e1b transcripts during mouse oogenesis and early embryo development. Eif4e1b transcripts are more abundant from GV to the early two-cell stage, while Eif4e is more abundant at other stages. (D) Schematic of the Eif4e1b gene locus in the Eif4e1bKI mouse line with FLAG and HA tags at the C terminus. (*) Initiator methionine, (x) stop codon. (E) Immunofluorescence of eggs and embryos derived from Eif4e1bKI female mice. Anti-HA antibody and DAPI were used to visualize the eIF4E1b fusion protein and nuclear DNA, respectively. Scale bar, 20 µm. (F) Protein levels of eIF4E and eIF4E1b from published proteomics results from GV oocyte to blastocyst stage.
eIF4E binds to the 5′ cap of poly(A) mRNAs and is critical for RNA translation (Osborne and Borden 2015). eIF4E1b maintains the 5′ cap binding ability, albeit with a lower affinity (Kubacka et al. 2015). Using a knock-in (KI) mouse line with HA and FLAG tags of the endogenous protein (Fig. 1D; Supplemental Fig. S2A,B), we detected eIF4E1b only in oocytes in mouse ovaries (Supplemental Fig. S2C), substantiating its germ cell specificity, and earlier reports suggest that it may function during MZT (Evsikov et al. 2006). We confirmed Eif4e1bKI expression in MII eggs, which persisted until late two-cell embryos after fertilization before becoming undetectable at the four-cell stage (Fig. 1E). Published quantitative proteomic data (Wang et al. 2010; Gao et al. 2017) corroborated these observations and were comparable with Eif4e1b transcript abundance (Fig. 1F).
Maternal deletion of Eif4e1b arrests embryos at the two-cell stage
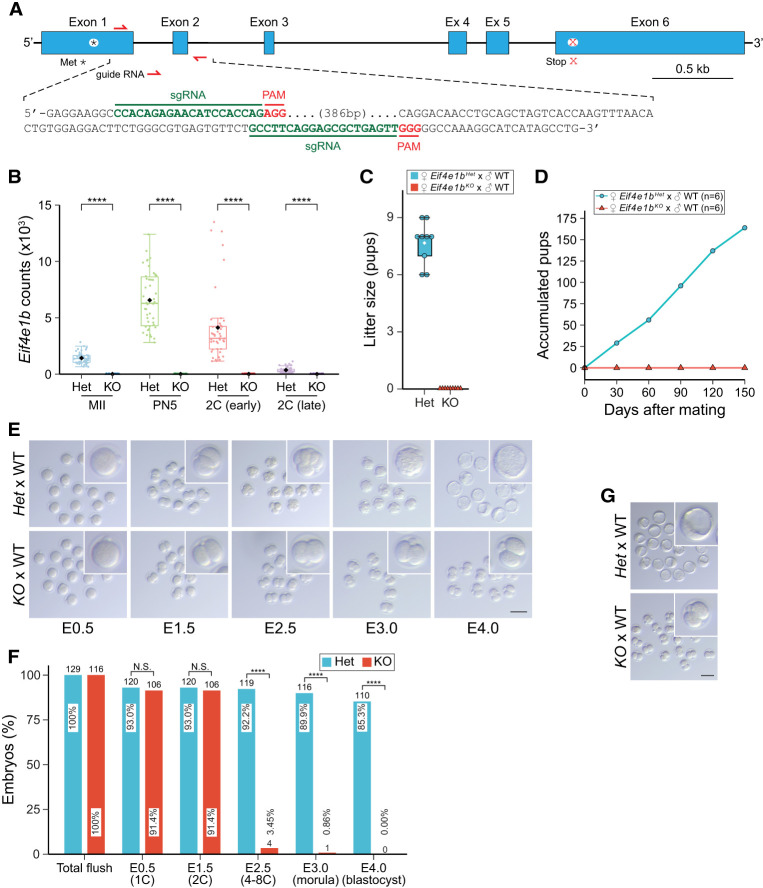
To explore Eif4e1b function, we generated null alleles of the single-copy gene in mice using CRISPR/Cas9 (Fig. 2A). After confirmation by DNA sequence, three Eif4e1b knockout mouse lines were established and designated Δ419, Δ411, and Δ329 (Supplemental Fig. S2D,E) according to the size of deletions. Unless noted, subsequent experiments were performed with the Δ419 line, which was designated Eif4e1bKO for homozygous null and designated Eif4e1bHet for heterozygous mice that were used as controls. As confirmed by single-embryo RNA-seq (Fig. 2B), Eif4e1b transcripts lost their first and second exons in oocytes and embryos from Eif4e1bKO female mice. Thus, no functional eIF4E1b protein could be translated. Homozygous null mice grew to adulthood. Adult males had normal fertility (Supplemental Fig. S3A) and testis morphology (Supplemental Fig. S3B), but the female Eif4e1bKO mice were infertile (Fig. 2C) even after >5 mo of mating (Fig. 2D).
Figure 2.
Maternal deletion of Eif4e1b leads to developmental arrest at two cells. (A) Schematic of the Eif4e1b gene (top) and sequences of sgRNAs (bottom) for generation of Eif4e1bKO mouse lines. (*) Initiator methionine, (x) stop codon. (B) Abundance of read overlaps with Eif4e1b exon 1 or 2. Residual Eif4e1b transcripts in Eif4e1bKO female mice lack exons 1 and 2 and produce no functional eIF4E1b protein. (****) P < 0.0001; two-tailed t-test. (C) Six-week-old female Eif4e1bHet (control) and homozygous Eif4e1bKO mice were mated with WT males to determine litter sizes. (D). Accumulated pups from Eif4e1bHet and Eif4e1bKO females during 5 mo of continuous mating. (E) Representative images of in vitro cultured embryos from Eif4e1bHet and Eif4e1bKO females after mating with WT males at embryonic day 0.5 (E0.5), E1.5, E2.5, E3.0, and E4.0. Inset magnification, 2.5×. Scale bar, 100 µm. (F) Quantification of embryos in E. Ratio of embryos at different stages is plotted. Total number of embryos is above each bar. (N.S.) Not significant, (****) P < 0.0001; two-sided proportion test. (G) Images of embryos flushed from Eif4e1bHet and Eif4e1bKO female reproductive tracts at E3.5 after successful in vivo mating. Inset magnification, 2.5×. Scale bar, 100 µm.
The oocytes produced by Eif4e1bKO female mice had no morphological abnormalities and were similar in size to controls (Supplemental Fig. S3C–E). More importantly, Eif4e1bKO female mice ovulate numbers of eggs comparable with controls (Supplemental Fig. S3F), and these eggs could be fertilized both in vitro and in vivo to produce normal zygotes (Supplemental Fig. S3G,H). These results suggest that Eif4e1b deletion did not affect oogenesis and oocyte quality. We thus focused on defects in embryogenesis to account for the observed Eif4e1bKO female infertility.
After mating control and Eif4e1bKO females with wild-type (WT) males, fertilized zygotes were flushed and cultured in vitro for 4 d (Fig. 2E). The ratio of embryos that developed to different stages was determined and quantified (Fig. 2F). Although Eif4e1bKO eggs were as fertile as controls and could finish the first cell division, none progressed beyond the two-cell stage, whereas control embryos became blastocysts (Fig. 2E,F). We confirmed that the two-cell arrest occurred in vivo by flushing control and Eif4e1bKO female reproductive tracts at embryonic day 3.5 (E3.5) after mating with WT males (Fig. 2G). The arrested phenotype was also observed in the Δ411 and Δ329 lines (Supplemental Fig. S2F). Together, these results confirm a role for eIF4E1b in embryo development beyond the two-cell stage.
Maternal ablation of Eif4e1b impairs zygotic gene activation
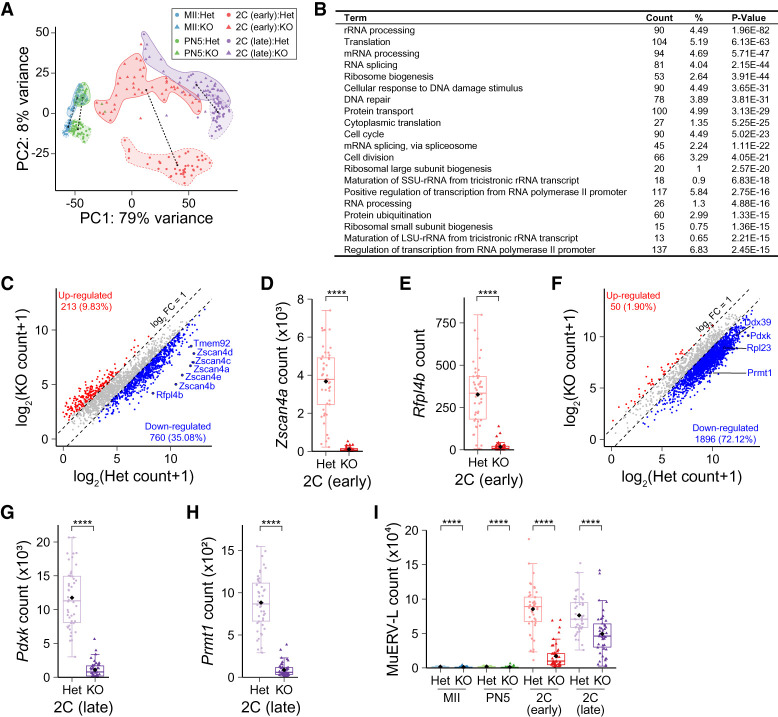
To investigate Eif4e1b function in early development, we adapted single-cell nucleosome, methylation, and transcript sequencing (scNMT-seq) (Clark et al. 2018) to single-embryo NMT-seq (seNMT-seq) (Supplemental Fig. S4A). After mating control and Eif4e1bKO female mice with WT males, zygotes were flushed for in vitro culture. Transcriptomes of these embryos, together with MII eggs and PN5 (pronuclear stage 5) zygotes, were analyzed using seNMT-seq (Supplemental Table S1). Most annotated protein-coding mRNAs and long noncoding RNAs (lncRNAs) were detected in all samples (Supplemental Fig. S4B). After quality control, principal component analysis (PCA) was performed to determine relationships among samples (Fig. 3A). From the PCA plot, we calculated Euclidian distances between the centers of the two genotypes at each developmental stage to document differences in transcriptomes between embryos from Eif4e1bKO and control female mice (Supplemental Fig. S4C). Although two-cell embryos within the same genotype exhibited considerable heterogeneity (Fig. 3A), we consistently detected more significant differences in transcriptomes of two-cell embryos derived from Eif4e1bKO and control female mice (Supplemental Fig. S4C), which could account for the observed two-cell arrest (Fig. 2E–G). In contrast, data from the two genotypes in MII eggs and PN5 zygotes were grouped closely in the PCA plot (Fig. 3A; Supplemental Fig. S4C). This may reflect the absence of developmental defects at these stages (Fig. 2E,F; Supplemental Fig. S3E–H), although down-regulated transcripts were identified after maternal Eif4e1b ablation (Supplemental Fig. S4D,E). This suggests accelerated mRNA clearance in these embryos and is consistent with the hypothesis that eIF4E1b binding protects maternal RNA from degradation. The presence of additional genes down-regulated in two-cell embryos derived from Eif4e1bKO female mice (Supplemental Fig. S4F–H) indicates that zygotic gene expression was also affected at these stages.
Figure 3.
Maternal deletion of Eif4e1b impairs ZGA. (A) Principal component analysis (PCA) plot of RNA-seq results of single embryos from Eif4e1bHet (control) or Eif4e1bKO female mice at different developmental stages. The lengths of dashed lines between cluster centers represent differences between samples. (B) Top gene ontology (GO) terms of down-regulated genes in late two-cell embryos from Eif4e1bKO females. Top 20 molecular function terms as well as the number and ratio of genes and P-values are shown. (C) Scatter plot documenting differentially expressed RNAs expected to be transcribed during minor ZGA in early two-cell embryos. mRNAs from multiple well-known minor ZGA genes are labeled in the plots. (D,E) Abundance of Zscan4a and Rfpl4b, two minor ZGA genes, at the early two-cell stage. (F) Scatter plot documenting differentially expressed RNAs expected to be transcribed during major ZGA in late two-cell embryos. mRNAs from multiple well-known major ZGA genes are labeled. (G,H) Abundance of Pdxk and Prmt1, two major ZGA genes, at the late two-cell stage. (I) Abundance of transcripts from the MuERV-L transposon in embryos from control or Eif4e1bKO female mice at different developmental stages. RNAs that have log2 fold change >2 or <−2 with adjusted P value of <0.01 are considered significantly up-regulated or down-regulated in C and F and are shown as red and blue dots, respectively. The total number of up-regulated or down-regulated RNAs is labeled in each plot. (****) P < 0.0001; two-tailed t-test.
Using gene ontology (GO) (Dennis et al. 2003), we determined that transcripts involved in RNA processing, protein translation, cell cycle, etc. were severely decreased in late two-cell embryos derived from Eif4e1bKO female mice (Fig. 3B). The minor wave of mouse ZGA is detected in late one-cell embryos ∼14 h after fertilization and continues into the early two-cell stage (Schulz and Harrison 2019). Of the 2166 reported minor ZGA transcripts (Abe et al. 2015, 2018), 1447 had reduced expression in embryos from Eif4e1bKO female mice at the early two-cell stage (Fig. 3C; Supplemental Fig. S4F; Supplemental Tables S1, S2). Protein-coding RNAs normally up-regulated during minor ZGA (Alda-Catalinas et al. 2020), including the Zscan4 cluster, Rfpl4b, and Tmem92, remained at low levels in early two-cell embryos after maternal deletion of Eif4e1b (Fig. 3D,E; Supplemental Fig. S4I).
Major ZGA follows the minor wave, and these broadly activated zygotic genes direct subsequent development to establish the blueprint of early embryos (Tadros and Lipshitz 2009). Since major ZGA is affected by the minor wave, it is not surprising that many more major ZGA genes (e.g., Prmt1, Pdxk, and Ddx39), including many epigenetic regulators, were poorly expressed in late two-cell embryos derived from Eif4e1bKO female mice (Fig. 3F–H; Supplemental Fig. S4J). Of 2629 major ZGA transcripts (Hamatani et al. 2004), 2402 were down-regulated in late two-cell embryos derived from Eif4e1bKO female mice (Fig. 3F; Supplemental Tables S1, S3), indicating devastating defects of major ZGA. Besides abnormal repression, 3087 genes were significantly up-regulated in the late two-cell embryos derived from Eif4e1bKO female mice. These results suggest that maternal ablation of Eif4e1b causes repression of gene products that should be activated (Fig. 3F), which in turn leads to abnormal gene up-regulation in late two-cell embryos (Supplemental Fig. S4G). Both activities may contribute to the two-cell embryo arrest.
Extensive activation of transposons in early mouse embryos has been reported, and long terminal repeats (LTRs) can drive gene expression during ZGA (Franke et al. 2017). MuERV-L has been used as a marker of successful zygotic genome activation (Kigami et al. 2003) and is reported to regulate pluripotency genes (Ramírez et al. 2006). In embryos from Eif4e1bKO female mice, MuERV-L abundance was decreased at the two-cell stage (Fig. 3I; Supplemental Table S4), as was global expression of LTRs (Supplemental Fig. S4K). Taken together, our results suggest that maternal Eif4e1b deletion causes systematical failure of minor ZGA, which triggers major ZGA defects, leading to developmental arrest at the two-cell stage.
Maternal eIF4E1b reprograms zygotic chromatin accessibility
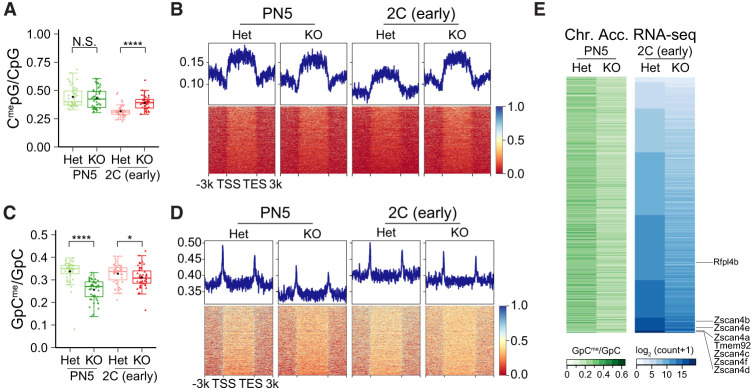
The data for DNA methylation and chromatin accessibility from seNMT-seq were sparser in each single embryo compared with that from seRNA-seq. Thus, to overcome the difficulty from low sample size, we merged the results of all single embryos with the same genotype and from the same stage together to obtain a better global view of DNA methylation and chromatin accessibility. Since abnormality of zygotic gene expression was already detected during the minor wave of ZGA in embryos from Eif4e1bKO females, we suspected that maternal ablation of Eif4e1b disrupted the activation of the earliest zygotic genes. Thus, we examined DNA methylation and chromatin accessible profiles in PN5 and early two-cell embryos collected from control and Eif4e1bKO females. Although maternal ablation of Eif4e1b slightly increased methylation of genomic DNA in early two-cell embryos, no obvious changes were detected at the earlier PN5 stage (Fig. 4A; Supplemental Fig. S5A). DNA methylation at minor ZGA gene loci also showed no significant changes between PN5 zygotes from Eif4e1bKO and control females, and hypermethylation at these regions was only detected at the early two-cell stage (Fig. 4B).
Figure 4.
Maternal eIF4E1b reprograms zygotic chromatin accessibility. (A) Ratio of endogenous methylated CpG among all CpG motifs to show global DNA methylation patterns in PN5 zygotes and early two-cell embryos from Eif4e1bHet (control) and Eif4e1bKO females. (B) DNA methylation profile around minor ZGA gene loci in PN5 zygotes and early two-cell embryos from control and Eif4e1bKO females. (N.S.) Not significant, (*) P < 0.05, (****) P < 0.0001; two-tailed t-test. (C) Same as A, but for global chromatin accessibility using an exogenous GpC methyltransferase (M.CviPI) to methylate accessible GpC. (D) Same as B, but for global chromatin accessibility. (E) Heat map showing chromatin accessibility at promoter regions (left) and expression of corresponding minor ZGA genes (right) in early two-cell embryos. Individual minor ZGA genes are annotated.
In contrast, chromatin became less accessible in both PN5 zygotes and early two-cell embryos in the absence of maternal eIF4E1b (Fig. 4C; Supplemental Fig. S5B). A significant and widespread decrease in chromatin accessibility at promoters of genes expected to express during minor ZGA was observed in PN5 zygotes derived from Eif4e1bKO female mice (Fig. 4D, left panel). The lower chromatin accessibility persisted until the early two-cell stage, albeit to a lesser extent (Fig. 4D, right panel). Analysis of multiple minor ZGA genes confirmed the overall trends observed. Hypermethylation of the DNA at their promoter regions was not observed in PN5 mutant embryos (Supplemental Fig. S5C), while lower chromatin accessibility already existed at these regions (Supplemental Fig. S5D). Similar results were confirmed at promoters of multiple transposons (Supplemental Fig. S5E,F). These results further indicate that the reduced chromatin accessibility, instead of DNA methylation, is associated with decreased expression of the earliest zygotic genes (Fig. 4E). Taken together, these results support the hypothesis that maternal deletion of Eif4e1b fails to reset zygotic chromatin to an open structure, leading to failed activation of zygotic genes.
eIF4E1b binds mRNAs selectively in MII eggs
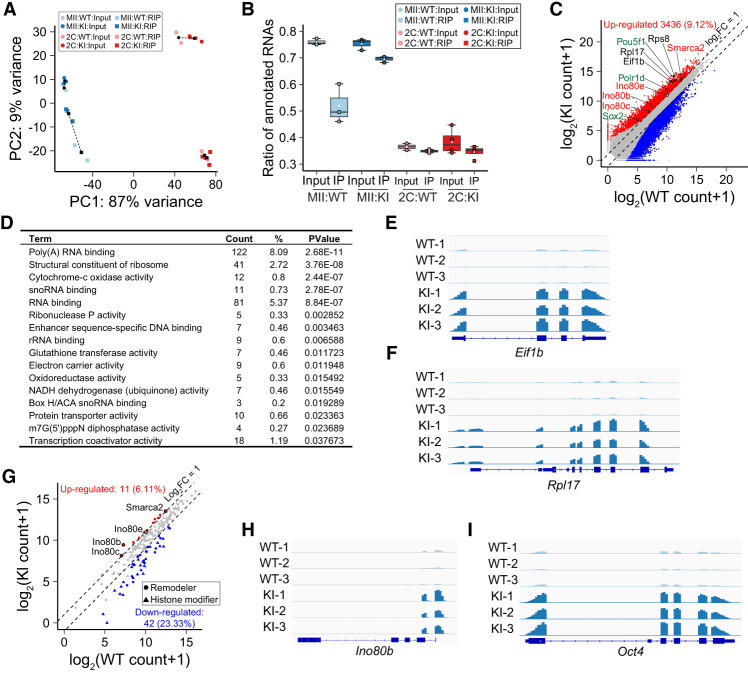
In the absence of maternal eIF4E1b, target mRNAs are not protected and quickly degrade in MII eggs and PN5 zygotes (Supplemental Fig. S4D,E). To confirm eIF4E1b binding and identify potential mRNA targets, we used MII eggs and early two-cell embryos from Eif4e1bKI and control female mice (Fig. 1D; Supplemental Fig. S2A,B) to perform RNA immunoprecipitation (RIP). There was no systematic difference in mapping input RNA to annotated genes (Fig. 5A) from the two genotypes at the same developmental stage, but eIF4E1b immunoprecipitated few annotated mRNAs in early two-cell embryos derived from either control or Eif4e1bKI female mice (Fig. 5B). This suggested that mRNAs were no longer bound by eIF4E1b at this stage of development. In contrast, eIF4E1b bound more mRNAs (Fig. 5B) transcribed from many fewer genes (Supplemental Fig. S6A) in MII eggs, consistent with specific binding to a small subset of mRNAs in MII eggs. In agreement with this result, we observed significant differences between control and Eif4e1bKI samples of the RIP-seq data from MII eggs (Supplemental Fig. S6B).
Figure 5.
eIF4E1b binds to a subset of mRNAs in MII eggs. (A) PCA of input and immunoprecipitated transcripts after eIF4E1b RIP. MII eggs or early two-cell embryos from Eif4e1bKI female mice were used, and WT eggs/embryos served as controls. (B) Ratio of input and immunoprecipitated (RIP) reads that can be mapped to annotated RNAs in RIP-seq. (C) Scatter plot documenting all differentially expressed RNAs as determined by the RIP-seq using WT and Eif4e1bKI MII eggs. Transcripts that have a log2 fold change of >1 or <−1 with adjusted P-value of <0.1 are considered significantly up-regulated and down-regulated and are shown as red and blue dots, respectively. Up-regulated RNAs represent potential Eif4e1b mRNA targets. Three groups of them are labeled in different colors (red, black, and green). (D) Top gene ontology (GO) terms of potential eIF4E1b RNA targets. Top 16 molecular function terms with P-values of <0.05 are shown. The number and ratio of genes that fall into each GO term are shown as well. (E,F) Integrated Genomics Viewer (IGV) view of eIF4E1b RIP-seq results at Eif1b and Rpl17 loci in WT and Eif4e1bKI samples. (G) Scatter plot as in C documenting differentially expressed RNAs coding all known chromatin remodeling complex subunits and histone modification enzymes as determined by RIP-seq experiments using WT and Eif4e1bKI MII eggs. Several potential eIF4E1b mRNA targets are labeled. (H,I) Same as E, but for Ino80b and Oct4 (Pou5f1) loci.
RIP data in MII eggs were reproducible within each genotype (Fig. 5A; Supplemental Fig. S6C). We identified 3436 RNAs that were more abundant in Eif4e1bKI MII eggs, representing candidate targets for eIF4E1b binding (Fig. 5C; Supplemental Table S5). We performed GO analysis using potential eIF4E1b targets (Fig. 5D). The top GO terms suggest that eIF4E1b binds to mRNAs encoding RNA binding proteins and structural units of ribosome (e.g., Eif1a, Rps8, and Rpl17), which could affect translation of maternal mRNAs (Fig. 5C–F; Supplemental Fig. S6D). There were also GO terms from multiple transcriptional factors, and we were curious whether there were additional eIF4E1b targets that could explain the altered chromatin accessibility in early embryos from Eif4e1bKO females.
Chromatin accessibility is regulated by remodeling complexes, which are affected by histone modifications (Klemm et al. 2019). We searched the RIP-seq results for the 103 known histone modifiers (Shah et al. 2020) and the 77 subunits of chromatin remodeling complexes (Hota and Bruneau 2016) in mice (Fig. 5G; Supplemental Table S6). Ten of the 77 remodeling subunits showed signification up-regulation in the RIP-seq results, while only one of the 103 histone modifiers was up-regulated. These results suggest that eIF4E1b modulates chromatin accessibility by selective regulation of subunits of remodeling complexes. We focused on multiple components of the INO80 complex (Fig. 5H; Supplemental Fig. S6E) and SMARCA2, a key member of the SWI/SNF complex (Supplemental Fig. S6F), which were potential eIF4E1b targets.
By examining genes reported to affect early embryogenesis (Supplemental Fig. S6G), we determined that Oct4 (Pou5f1), Sox2, and Polr1d transcripts were additional potential eIF4E1b targets (Fig. 5I; Supplemental Fig. S6H,I). OCT4 (POU5F1) and SOX2 are well-known pluripotency factors that regulate early embryo development (Masui et al. 2007; Pan and Schultz 2011; Lee et al. 2013). These reprogramming factors interact with multiple remodeling complexes (Wang et al. 2014; Gao et al. 2019) and may provide gene-specific recruitment during ZGA. POLR1D is an important component of RNA polymerase I, whose deletion leads to failure of embryo development (Miao et al. 2020). Together with other eIF4E1b targets controlling mRNA translation, POLR1D could facilitate translation of maternal RNAs. We searched for shared RNA motifs from potential eIF4E1b targets and identified multiple motifs that may be used by eIF4E1b for selective binding (Supplemental Fig. S6J). Taken together, our results suggest that eIF4E1b can selectively bind mRNAs encoding proteins related to the translation machinery that may further affect pre-ZGA maternal mRNA translation. eIF4E1b can also bind mRNAs of chromatin remodeling proteins and reprogramming factors in oocytes to control zygotic chromatin accessibility through regulation of these mRNA targets.
eIF4E1b controls maternal mRNA expression
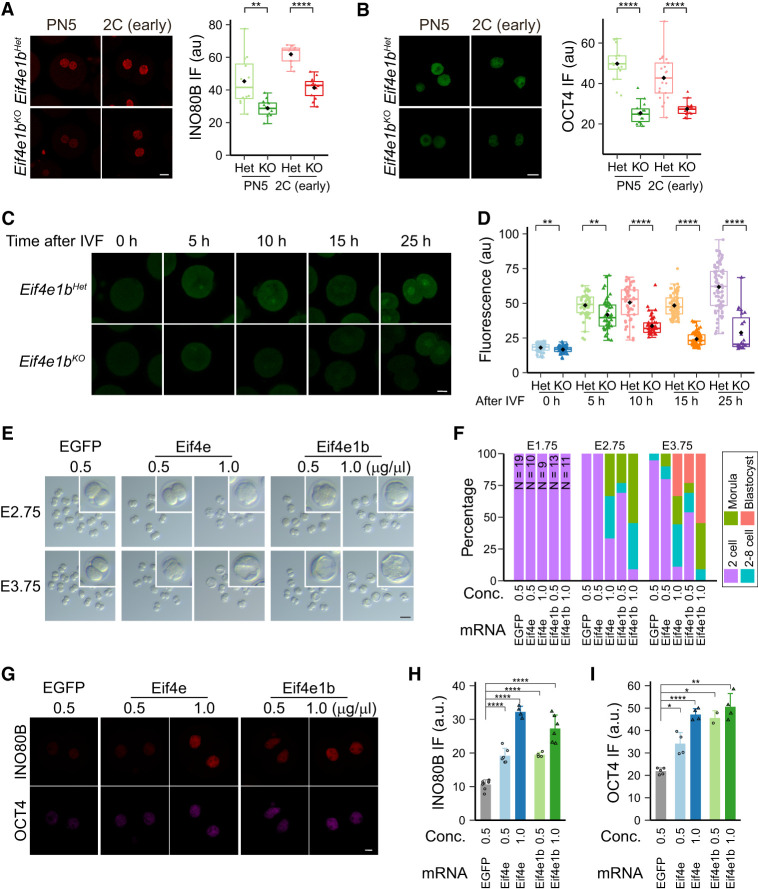
We noted that the eIF4E1b maternal RNA targets had lower abundance in zygotes and early two-cell embryos from Eif4e1bKO females (Supplemental Table S1; Supplemental Fig. S7A–J). This suggested that eIF4E1b binding protects specific maternal RNAs from rapid degradation. To determine whether eIF4E1b affects the expression of these RNAs, we performed immunofluorescence at different stages in early embryo development using INO80B-, OCT4-, and POLR1D-specific antibodies. Maternal ablation of Eif4e1b caused decreased expression of these proteins in zygotes and early two-cell embryos (Fig. 6A,B; Supplemental Fig. S7K). This is consistent with a model in which eIF4E1b binds a subset of maternal mRNAs not only to protect them from degradation but, more importantly, to promote their translation into proteins. Some eIF4E1b mRNA targets encode RNA binding proteins whose translation could further protect other maternal RNAs. We further hypothesize that translated chromatin remodeling proteins from eIF4E1b mRNA targets could then modify the zygotic genome to establish open structures to facilitate activation of early zygotic genes. Preliminary results support this hypothesis, as knocking down Smarca2 in zygotes reduced chromatin accessibility of major ZGA genes at the two-cell stage, which could further affect embryo development.
Figure 6.
eIF4E1b controls translation of maternal mRNA in mouse zygotes. (A,B) INO80B and OCT4 protein expression in embryos from Eif4e1bHet and Eif4e1bKO females at different developmental stages. Scale bar, 20 µm. The nuclear fluorescent signals are quantified at the right. (C) Imaging of nascent proteins in embryos derived from Eif4e1bHet and Eif4e1bKO females at different time points after IVF. Scale bar, 20 µm. (D) Fluorescent signal from OPP (O-propargyl-puromycin) incorporation indicating that protein translation from C was quantified. (E) Morphology of in vitro cultured embryos after microinjection of mRNAs. The number of embryos is quantified in F. Inset magnification, 2.8×. Scale bar, 100 µm. (G) Immunostaining showing expression of two eIF4E1b targets in early two-cell embryos. Scale bar, 10 µm. (H,I) Quantification of nuclear INO80B and OCT4 signals in G. (*) P < 0.05, (**) P < 0.01, (****) P < 0.0001; two-tailed t-test.
To obtain a global view of protein expression, we collected MII eggs and early two-cell embryos from control and Eif4e1bKO females for low-input mass spectrometry (Supplemental Table S7). Although no global biases (Supplemental Fig. S8A) were present due to the limited number of proteins detected, more proteins were down-regulated in early two-cell embryos derived from Eif4e1bKO than from control females (Supplemental Fig. S8B–D). Since the minor ZGA produces only a few promiscuous transcripts that are unlikely to be translated (Abe et al. 2018), the down-regulated protein expression is likely due to accumulated translation defects of maternal mRNAs at the pronuclear stage. We detected slightly lower expression of TET3 in early two-cell embryos derived from Eif4e1bKO female mice (Supplemental Fig. S8E–G), which may contribute to the modestly reduced DNA methylation in these embryos (Fig. 4B, right panel).
Mass spectrometry only reflects the total amount of proteins. To determine whether maternal ablation of Eif4e1b affected protein synthesis in zygotes, we labeled nascent proteins with O-propargyl-puromycin (OPP) in embryos after in vitro fertilization (IVF) and quantified signals at different time points. Significant reduction in protein biosynthesis was detected in zygotes and early two-cell embryos from Eif4e1bKO female mice (Fig. 6C,D). In agreement with these observations, eIF4E1b mRNA targets have an advantage in binding to ribosomes that is reduced in PN5 and early two-cell embryos after maternal ablation of eIF4E1b (Supplemental Fig. S7L). These results are consistent with eIF4E1b being essential for maternal mRNA translation in mouse zygotes. Recently published Ribo-seq data (Xiong et al. 2022) substantiated our findings, as eIF4E1b mRNA targets also bind more ribosomes compared with nontargets in embryos (Supplemental Fig. S9A). Because most eIF4E1b mRNA targets maintain binding to ribosomes at these stages, this should enable efficient translation (Supplemental Fig. S9B,C). The eIF4E1b RNA targets also have longer poly(A) tails (Supplemental Fig. S9D,E), as determined by PAIso-seq experiments (Liu et al. 2019, 2021, 2023), which could also contribute to efficient translation. Taken together, these results suggest that eIF4E1b binds to essential RNAs not only to stabilize them but also to enhance their binding to ribosomes, both of which ensure high expression of their protein products.
Microinjection of mRNA encoding eIF4E1b rescues the two-cell arrest
To determine whether the observed defects after maternal depletion of Eif4e1b could be rescued, we injected EGFP-conjugated Eif4e1b mRNA into one-cell zygotes derived from Eif4e1bKO female mice and cultured them for 3 d. Adding back ∼1.0 pL of 0.5 µg/µL Eif4e1b mRNA rescued the two-cell arrest completely in ∼25% (three out of 13) of the embryos, which progressed to blastocysts as normal. Partial rescue was observed in another ∼25% (three out of 13) of the embryos, which went beyond the two-cell stage while the rest remained as two cells. Injection of a higher dose of Eif4e1b mRNA (1.0 µg/µL) rescued the defects in all embryos, with >50% (six out of 11) becoming blastocysts and an additional 36% (four out of 11) reaching the morula stage. In contrast, embryos from Eif4e1bKO females injected with equivalent molar concentrations of 0.5 µg/µL EGFP mRNA remained at the two-cell stage (Fig. 6E,F). Using immunofluorescence, higher levels of INO80B and OCT4 were observed at the early two-cell stage in embryos with Eif4e1b mRNA injected, indicating recovered expression of maternal RNAs (Fig. 6G–I). Surprisingly, adding back mRNA encoding the canonical eIF4E protein also rescued the phenotype, although not as effectively (Fig. 6E–I). Even with doubling the amount of Eif4e mRNA, full rescue was not achieved (Supplemental Fig. S7M,N). These results suggest that eIF4E1b serves as a key component of the eIF4F translation initiation complex when eIF4E is less available at the earliest stages of embryo development and guides translation of the protein products from maternal mRNAs that are essential for embryo progression.
Discussion
After fertilization, the epigenome of mouse embryos must be reprogrammed to ensure transcription of zygotic genes (Tadros and Lipshitz 2009; Ladstätter and Tachibana 2019). This process occurs during the MZT (Schultz et al. 2018) and requires that the epigenome of parental germ cells be reprogramed for embryo development. Earlier investigations reported asymmetries in genomic DNA methylation in maternal and paternal pronuclei during reprogramming of the early zygotic epigenome. Similarly, differences of multiple histone modifications were observed between the pronuclei in early zygotes (Morgan et al. 2005). However, more recent results indicate that extensive demethylation of genome DNA occurred in both male and female pronuclei (Guo et al. 2014) and that demethylation had little gene specificity. It was also noted that maternal and paternal alleles have similar chromatin accessibility at the late one-cell (zygote) stage (Guo et al. 2017), and embryonic gene expression has no significant parental allele preference (Deng et al. 2014). Thus, differences between parental contributions to the zygotic genome do not adequately explain the requisite changes in chromatin necessary to foster gene activation. Our data suggest an alternative in which activation of early mouse embryogenesis is predicated on genetic programs predefined in female germ cells. In this scenario, essential maternal transcripts accumulate during oogenesis and, after fertilization, are translated into proteins that activate zygotic gene expression. We propose that the oocyte-specific eIF4E1b initiator factor controls selective translation of maternal transcripts that encode proteins to modify chromatin and activate transcription.
Mature eukaryotic mRNA contains a 5′ cap that is required for protein translation. The heterotrimeric eIF4F translation initiation complex (eIF4A, eIF4E, and eIF4G) binds to the cap structure and recruits ribosomes for translation and associated factors that stabilize the transcript (for review, see Pelletier and Sonenberg 2019). Oocyte-specific eIF4E1b increases dramatically in abundance late in oogenesis and binds mRNAs that encode protein translation machinery, chromatin remodelers, and reprogramming factors in MII eggs. Our analysis confirms that oocyte-specific eIF4E1b dissipates by the four-cell stage when eIF4E resumes its primary role in translation. We propose that eIF4E1b functions in a narrow window to prevent degradation of a subset of maternal RNAs and translate them into proteins that activate the zygotic genome.
The two-cell arrest of embryos derived from Eife1bKO female mice can be rescued by microinjection of mRNA encoding either the ubiquitously expressed (eIF4E) or the germ cell-specific isoform (eIF4E1b) of the initiation factor. Ubiquitously expressed eIF4E has a stronger affinity in binding mRNA caps to initiate translation (von der Haar et al. 2004; Kubacka et al. 2015) but expresses at very low levels during the MZT (Fig. 1C). However, eIF4E could not rescue the defects as efficiently as eIF4E1b, suggesting that eIF4E1b may be a specialized isoform of eIF4E that functions specifically at the earliest stage of embryogenesis. Previous reports show that eIF4E1b interacts with the CPEB1 complex in Xenopus oocytes (Minshall et al. 2007; Standart and Minshall 2008; Kubacka et al. 2015), and our results suggest that the CPEB1 complex (Duran-Arqué et al. 2022) is an important partner of mouse eIF4E1b (Supplemental Fig. S8H; Supplemental Table S8). The CPEB1 complex is reported to bind selected maternal RNA in early oocytes to repress their expression but later activates translation into protein upon resumption of meiosis (Luong et al. 2020). We propose that eIF4E1b participates in this process to protect and translate a subset of maternal RNAs after fertilization to enable normal embryo development. eIF4E1b contains a longer N terminus that is absent from eIF4E (Fig. 1B). Preliminary results suggest that this structurally flexible (Marcotrigiano et al. 1999; Jumper et al. 2021) N-terminal domain (Supplemental Fig. S10A–C) may provide eIF4E1b advantages over eIF4E in binding components of the CEPB1 complex. This N-terminal domain may facilitate eIF4E1b binding to its cofactors as well as its RNA targets, as deletion of this domain greatly reduced the function of eIF4E1b (Supplemental Fig. S10D,E). Based on our results, we further propose that after fertilization, but before ZGA, maternal eIF4E1b ensures translation of proteins required for chromatin accessibility that enables expression of early zygotic genes (Fig. 7).
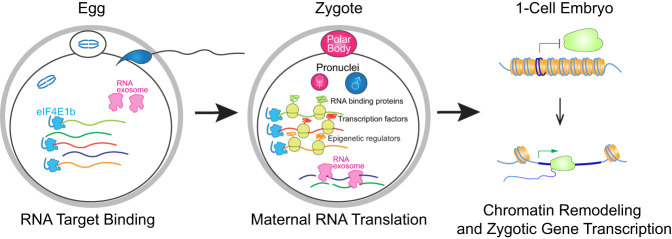
Figure 7.
Working model. eIF4E1b binds a subset of essential maternal RNAs in MII eggs to protect them from degradation. After fertilization, eIF4E1b-bound mRNAs are translated and remodel chromatin into an open state to enable transcription of the zygotic genes that establish early developmental programs.
Our results offer a possible explanation of how the pre-ZGA maternal RNA translation is regulated and why this burst of translation is essential for embryogenesis. Our model supports the hypothesis that activation of early mouse embryogenesis is based on a genetic program predefined in female germ cells. Considering the sequence conservation of eIF4E1b among different species, we suspect that this mechanism is shared among vertebrates to enable the maternal-to-zygotic transition. Further investigations will be required to determine the catalog of maternal transcripts required for zygotic gene expression and molecular mechanisms of their selection. While the cap-binding ability of eIF4E1b may be the major driver for capturing maternal RNA to regulate their translation, there could be additional selective factors, and their identification will provide better insight into the maternal regulation of early embryogenesis.
Materials and methods
Mice
All animal studies were performed in accordance with guidelines of the Animal Care and Use Committee of the National Institutes of Health under a Division of Intramural Research, National Institute of Diabetes and Digestive and Kidney Diseases-approved animal study protocol.
To generate gene-edited mouse lines, CRISPR/Cas9 crRNA XT oligonucleotides were synthesized (Integrated DNA Technologies [IDT]) and annealed with tracrRNA (IDT 1072533) to form the crRNA–tracrRNA duplex. A NEPA21 electroporator (Bulldog Bio NEPA21) was used to deliver the RNP complex into zygotes flushed from B6D2F1 female mice. A ssDNA for homologous repair was added to the RNP complex when generating the mouse line containing FLAG and HA tags fused at the C terminus of Eif4e1b. For genotyping, tail tips of mice were lysed and used for PCR. Sequences for the crRNA XT, repair oligo, and genotyping primers are in Supplemental Table S9.
To test female fertility, pairs of Eif4e1bHet (control) and Eif4e1bKO female mice were harem-mated with a WT male to determine the number and sizes of litters. Eif4e1bHet and Eif4e1bKO male mice were mated with WT females separately to determine male fertility.
Oocyte isolation and measurement
Postnatal day 14 (P14) and P28 oocytes were puncture-released from ovaries of females at corresponding ages after genotyping. MII oocytes were collect after hormone injection of 6- to 8-wk-old female mice. Egg diameters were measured by ImageJ (version 1.53k).
Histology and immunofluorescence
Histology and immunofluorescence (IF) of mouse testes and ovaries were performed as previously described (Wang et al. 2019). MII eggs and embryos were fixed in 4% PFA and washed in PBS (Invitrogen 10010023) supplemented with 0.3% polyvinylpyrrolidone (Sigma-Aldrich PVP360) for IF. Eggs/embryos were incubated in PBS with 0.3% BSA (Cell Signaling Technology [CST] 9998S) and 0.1% Tween 20 (Sigma-Aldrich P9416) for 2 h and stained overnight at 4°C with primary antibodies. Goat antimouse or rabbit antibody conjugated with Alexa Fluor was used for immunofluorescent imaging. Primary antibodies used were anti-HA (1:600; CST 3724S), anti-INO80B (1:100; Protein Tech 29488-1-AP), anti-POLR1D (1:200; Sigma-Aldrich HPA039337), and anti-OCT4 (1:60; Santa Cruz Biotechnology sc-5279).
For anti-OCT4 immunostaining, nonspecific signals from zonae pellucidae were computationally removed with a home-made Matlab script. The original images are shown in Supplemental Figure S11. All the experiments were repeated at least three times, and representative results from one replicate are presented.
Single-embryo NMT-seq
MII eggs and embryos were collected from 6- to 8-wk-old hormonally stimulated female mice and lysed in the methyltransferase reaction mix. One microliter of prediluted (1:105) ERCC spike-in (Invitrogen 4456740) was added to a single-egg/embryo lysis. cDNA from single eggs/embryos was amplified by 15 PCR cycles before dual indexing with the Nextera XT kit. The supernatants containing genomic DNA after oligo-dT capture of RNA were processed following the scNMT-seq protocol with modified adapters (sequences in Supplemental Table S9). The quality of the libraries was confirmed by Bioanalyzer 2100, and each pooled library was sequenced (150 bp paired-end) in one lane on the Illumina HiSeq4000 platform (Novogene US).
Low-input RNA immunoprecipitation (RIP) and Ribo-seq
Low-input RNA immunoprecipitation was adapted by incorporating Smart-seq2 (Picelli et al. 2014) steps into a RIP-seq protocol. For one experiment, 200–250 MII eggs or early two-cell embryos were collected from WT or Eif4e1bKI female mice. The EZ-Manga RIP kit (Millipore 17-701) was used for the RIP experiment following the product manual. Five microliters of anti-HA beads (Thermo Fisher 88836) was used for each RIP group. One-fifth of the supernatant after bead incubation was saved to extract RNA as input using RNAclean XP beads (Beckman A63987). Elution buffer containing the oligo-dT30VN primer from Smart-seq2 was added to each tube containing RNAclean XP beads (input group) or anti-HA beads (IP group) to finish elution using a thermocycler for 5 min at 55°C and 3 min at 70°C. Smart-seq2 protocol was followed to amplify cDNA in each well by 14 PCR cycles before indexing, purification, and sequencing (150 bp paired-end). Approximately 200 oocytes or zygotes or ∼100 early two-cell embryos were used as one group for low-input Ribo-seq; all embryos contained HA-tagged Rpl22 (Sanz et al. 2009). The RIP-seq protocol was reused to do the low-input Ribo-seq with minor modifications. RNase I-digested lysate was immunoprecipitated, and the libraries were finished with a smRNA-seq kit (Takara Bio 635029).
Analysis of next-generation sequencing results
The quality of all FastQ files from Illumina sequencing was analyzed and confirmed by FastQC (v0.11.8). RNA-seq reads from seNMT-seq or low-input RIP-seq were trimmed with Trimmomatic (v0.39) (Bolger et al. 2014). The trimmed RNA-seq reads were aligned by STAR (v2.7.6a) (Dobin et al. 2013) to the GRCm38 annotation plus ERCC information. StringTie (v2.1.4) was used to generate counts of genes in the GTF reference (Pertea et al. 2015). The BAM files from RIP-seq were normalized with deepTools by RPKM (Ramírez et al. 2016) and visualized with Integrative Genomics Viewer (IGV) (Robinson et al. 2011). Ribo-seq reads were processed as in RIP-seq, but only uniquely mapped reads from protein-coding genes were used for analysis.
To explore expression of transposable elements (TEs) in single embryos, TE annotation was downloaded from the Hammell laboratory (TEToolkit, Cold Spring Harbor Laboratory) and supplied to STAR for realignment of single-embryo RNA-seq data. featureCounts was used to generate the expression table of annotated TEs (Liao et al. 2014). The counting matrixes of genes or TEs from single-embryo RNA-seq were subsequently analyzed with DESeq2 (Love et al. 2014), and ERCC spike-in was used for normalization. Log2 fold change of ±2 with adjusted P-value of <0.01 was used as the cutoff for significantly up-regulated or down-regulated transcripts. ERCC normalized counts were used for box plots, and P-values were calculated by two-tailed t-test. The count matrix from RIP-seq was also analyzed by DESeq2, and log2 fold change of ±1 with adjusted P-value of <0.1 was used as the cutoff. Shared motifs of potential eIF4E1b RNA targets were searched by MEME-ChIP (Ma et al. 2014).
Single-embryo DNA-seq data for analysis of DNA methylation and chromatin accessibility were aligned using HISAT-3N (v2.2.1-3n) (Zhang et al. 2021). Picard (v2.20.5) was used to remove duplicates in the BAM files. The methylated cytosines given by HISAT-3N were annotated by a home-made C++ program to identify CG and GC dinucleotides for analysis of DNA methylation and chromatin accessibility. Results of embryos from the same stage and strain for PN5 (47 Het and 48 KO) and early two-cell (46 Het and 47 KO) embryos were merged to calculate methylation rates of detected cytosines, which were then transformed into bedGraph format. The two-sided proportion test function from R was used to calculate the P-values when methylation ratios under different experimental conditions were compared. The bedGraph files were transformed in bigwig format, and deepTools was used to generate the heat maps covering genes that were of interest (Ramírez et al. 2016). R heatmap.3 was used to generate heat map plots for expression and chromatin accessibility of genes in different samples.
Low-input mass spectrometry (MS) and IP-MS/MS
To quantify proteomic changes, two groups of ∼500 MII eggs or early two-cell embryos (after mating females to WT males) from Eif4e1bHet and Eif4e1bKO female mice were collected and treated with an encapsulated proteomic sample-processing protocol. The relative label-free quantification (LFQ) method of MaxQuant (Tyanova et al. 2016) was used to represent signal intensity of detected proteins in LC-MS/MS. A HA tag magnetic IP/co-IP kit (Pierce 88838) was used to perform IP using ovary lysates from 6-wk-old Eif4e1bKI and WT control females. The immunoprecipitated elutes were analyzed by LC-MS/MS for proteins that interact with eIF4E1b. Proteins that had log2 fold change of >1 and P-value of <0.05 were considered potential eIF4E1b cofactors.
Embryo treatment and imaging of protein synthesis
To determine effects of maternal mRNA translation on embryo development, IVF was performed with WT MII eggs. Four hours after insemination, unfertilized eggs and fertilized zygotes were washed and cultured in advanced KSOM medium supplemented with cycloheximide (CHX; Sigma-Aldrich C7698), anisomycin (Sigma-Aldrich A9789), or DMSO (Sigma-Aldrich D8418) as control for another 20 h before imaging. The Click-iT Plus OPP Alexa Fluor 488 protein synthesis assay kit (Thermo Fisher C10456) was used to determine nascent protein synthesis in each group of embryos. Nuclei were labeled with DAPI.
To quantify nascent protein synthesis in embryos, zygotes were imaged 5, 10, 15, and 25 h after insemination according to the manufacturer's instructions of the Click-iT kit. MII eggs were imaged before fertilization. All the experiments were repeated at least three times, and representative results from one replicate are presented.
mRNA in vitro transcription and microinjection
The In-Fusion cloning method was used to clone mouse Eif4e1a (Eif4e), Eif4e1b, and Eif4e1b (without the two to 62 amino acids) truncated cDNA into pCDNA3.1+ plasmid with EGFP sequence fused at their N termini. The plasmids were then used to prepare corresponding mRNAs for microinjection into mouse zygotes (Wu and Dean 2020). Approximately 1.0 pL of mRNA solution was injected into each zygote.
Quantification and statistical analysis
For all fluorescent staining experiments, the fluorescence intensity in each egg/embryo was quantified by ImageJ version 1.53k (Schneider et al. 2012) and then used for plotting in R. Gene counts from RNA-seq were normalized by ERCC spike-in for plotting. All box plots include the median and data between the 25th and 75th percentile. The black or white diamond within each box shows the average within each group. Each dot in the box plots or bar plots reflects one value. The two-tailed Student's t-test or the “two.sided” prop.test function from R was used to calculate P-values as follows: not significant (N.S.), P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). Source data for plotting are in Supplemental Table S10.
Data availability
The data in this study have been deposited at the Gene Expression Omnibus (accession no. GSE180218) and the ProteomeXchange data sets (identifiers PXD041583 and PXD041622). Scripts used in this study can be found at https://github.com/Yang-DB-Lab/Eif4e1b_Paper. All other relevant data and materials of this study are available from J.D. upon request under a material transfer agreement with the National Institutes of Health.
Supplementary Material
Acknowledgments
We thank D. Eric Anderson of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Advanced Mass Spectrometry Core for performing mass spectrometry analyses, Wen Xie of the National Heart, Lung, and Blood Institute Transgenic Core for help with microinjection, and all members of J.D.’s laboratory for their insightful comments. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) NIDDK under grant ZIAADK015603 to J.D. Part of the analyses used the computational resources of the NIH High-Performance Computing Biowulf cluster (http://hpc.nih.gov).
Author contributions: G.Y., Q.X., and J.D. conceived the project. G.Y., Q.X., and J.D. designed the experiments. G.Y. and Q.X. performed most of the experiments. I.F. helped with immunofluorescent microscopy, and D.W. performed microinjection. G.Y. analyzed results and wrote the manuscript with input from X.Q. J.D. revised the manuscript. All authors discussed and approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.350400.123.
Competing interest statement
The authors declare no competing interests.
References
- Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. 2015. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J 34: 1523–1537. 10.15252/embj.201490648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe KI, Funaya S, Tsukioka D, Kawamura M, Suzuki Y, Suzuki MG, Schultz RM, Aoki F. 2018. Minor zygotic gene activation is essential for mouse preimplantation development. Proc Natl Acad Sci 115: E6780–E6788. 10.1073/pnas.1804309115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alda-Catalinas C, Bredikhin D, Hernando-Herraez I, Santos F, Kubinyecz O, Eckersley-Maslin MA, Stegle O, Reik W. 2020. A single-cell transcriptomics CRISPR-activation screen identifies epigenetic regulators of the zygotic genome activation program. Cell Syst 11: 25–41.e9. 10.1016/j.cels.2020.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh Z, Kageyama S, Aoki F. 2005. Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol Reprod Dev 72: 281–290. 10.1002/mrd.20340 [DOI] [PubMed] [Google Scholar]
- Aoki F, Hara KT, Schultz RM. 2003. Acquisition of transcriptional competence in the 1-cell mouse embryo: requirement for recruitment of maternal mRNAs. Mol Reprod Dev 64: 270–274. 10.1002/mrd.10227 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Argelaguet R, Kapourani CA, Stubbs TM, Lee HJ, Alda-Catalinas C, Krueger F, Sanguinetti G, Kelsey G, Marioni JC, et al. 2018. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun 9: 781. 10.1038/s41467-018-03149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramsköld D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. 10.1126/science.1245316 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Despic V, Neugebauer KM. 2018. RNA tales—how embryos read and discard messages from mom. J Cell Sci 131: jcs201996. 10.1242/jcs.201996 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Arqué B, Cañete M, Castellazzi CL, Bartomeu A, Ferrer-Caelles A, Reina O, Caballé A, Gay M, Arauz-Garofalo G, Belloc E, et al. 2022. Comparative analyses of vertebrate CPEB proteins define two subfamilies with coordinated yet distinct functions in post-transcriptional gene regulation. Genome Biol 23: 192. 10.1186/s13059-022-02759-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB. 2006. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev 20: 2713–2727. 10.1101/gad.1471006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov AV, Marín de Evsikova C. 2009. Evolutionary origin and phylogenetic analysis of the novel oocyte-specific eukaryotic translation initiation factor 4E in Tetrapoda. Dev Genes Evol 219: 111–118. 10.1007/s00427-008-0268-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, Kuzman M, Fulka H, Cernohorska M, Urbanova J, et al. 2017. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res 27: 1384–1394. 10.1101/gr.216150.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Liu X, Tang B, Li C, Kou Z, Li L, Liu W, Wu Y, Kou X, Li J, et al. 2017. Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep 21: 3957–3969. 10.1016/j.celrep.2017.11.111 [DOI] [PubMed] [Google Scholar]
- Gao F, Elliott NJ, Ho J, Sharp A, Shokhirev MN, Hargreaves DC. 2019. Heterozygous mutations in SMARCA2 reprogram the enhancer landscape by global retargeting of SMARCA4. Mol Cell 75: 891–904.e7. 10.1016/j.molcel.2019.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Liu S, Wu Q, Zhang L, Guo F. 2019. Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res 29: 110–123. 10.1038/s41422-018-0125-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, et al. 2014. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15: 447–459. 10.1016/j.stem.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Guo F, Li L, Li J, Wu X, Hu B, Zhu P, Wen L, Tang F. 2017. Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res 27: 967–988. 10.1038/cr.2017.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. 2004. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6: 117–131. 10.1016/S1534-5807(03)00373-3 [DOI] [PubMed] [Google Scholar]
- Hota SK, Bruneau BG. 2016. ATP-dependent chromatin remodeling during mammalian development. Development 143: 2882–2897. 10.1242/dev.128892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel S, Ernst M, Psathaki OE, Drexler HCA, Casser E, Suzuki Y, Makalowski W, Boiani M, Fuellen G, Taher L. 2019. An integrated genome-wide multi-omics analysis of gene expression dynamics in the preimplantation mouse embryo. Sci Rep 9: 13356. 10.1038/s41598-019-49817-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi B, Lee K, Maeder DL, Jagus R. 2005. Phylogenetic analysis of eIF4E-family members. BMC Evol Biol 5: 48. 10.1186/1471-2148-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. 2003. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod 68: 651–654. 10.1095/biolreprod.102.007906 [DOI] [PubMed] [Google Scholar]
- Klemm SL, Shipony Z, Greenleaf WJ. 2019. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet 20: 207–220. 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- Kubacka D, Miguel RN, Minshall N, Darzynkiewicz E, Standart N, Zuberek J. 2015. Distinct features of cap binding by eIF4E1b proteins. J Mol Biol 427: 387–405. 10.1016/j.jmb.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladstätter S, Tachibana K. 2019. Genomic insights into chromatin reprogramming to totipotency in embryos. J Cell Biol 218: 70–82. 10.1083/jcb.201807044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D. 1991. Quantitative analysis of protein synthesis in mouse embryos. I. Extensive reprogramming at the one- and two-cell stages. Development 112: 921–932. 10.1242/dev.112.4.921 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503: 360–364. 10.1038/nature12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Qing T, Zhu J, Wen Z, Yu Y, Fukumura R, Zheng Y, Gondo Y, Shi L. 2017. A comprehensive mouse transcriptomic body map across 17 tissues by RNA-seq. Sci Rep 7: 4200. 10.1038/s41598-017-04520-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu Y, Nie H, Liu H, Lu F. 2019. Poly(A) inclusive RNA isoform sequencing (PAIso-seq) reveals wide-spread non-adenosine residues within RNA poly(A) tails. Nat Commun 10: 5292. 10.1038/s41467-019-13228-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Nie H, Zhang C, Hou Z, Wang J, Lu F. 2021. Poly(A) tail length is a major regulator of maternal gene expression during the mammalian oocyte-to-embryo transition. bioRxiv 10.1101/2021.08.29.458052 [DOI] [Google Scholar]
- Liu Y, Zhao H, Shao F, Zhang Y, Nie H, Zhang J, Li C, Hou Z, Chen ZJ, Wang J, et al. 2023. Remodeling of maternal mRNA through poly(A) tail orchestrates human oocyte-to-embryo transition. Nat Struct Mol Biol 30: 200–215. 10.1038/s41594-022-00908-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong XG, Daldello EM, Rajkovic G, Yang CR, Conti M. 2020. Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res 48: 3257–3276. 10.1093/nar/gkaa010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Noble WS, Bailey TL. 2014. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc 9: 1428–1450. 10.1038/nprot.2014.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. 1999. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716. 10.1016/S1097-2765(01)80003-4 [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. 2007. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol 9: 625–635. 10.1038/ncb1589 [DOI] [PubMed] [Google Scholar]
- Miao X, Sun T, Golan M, Mager J, Cui W. 2020. Loss of POLR1D results in embryonic lethality prior to blastocyst formation in mice. Mol Reprod Dev 87: 1152–1158. 10.1002/mrd.23427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Reiter MH, Weil D, Standart N. 2007. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J Biol Chem 282: 37389–37401. 10.1074/jbc.M704629200 [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. 2005. Epigenetic reprogramming in mammals. Hum Mol Genet 14 Spec No 1: R47–R58. 10.1093/hmg/ddi114 [DOI] [PubMed] [Google Scholar]
- Niu W, Spradling AC. 2020. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci 117: 20015–20026. 10.1073/pnas.2005570117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne MJ, Borden KL. 2015. The eukaryotic translation initiation factor eIF4E in the nucleus: taking the road less traveled. Immunol Rev 263: 210–223. 10.1111/imr.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Schultz RM. 2011. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol Reprod 85: 409–416. 10.1095/biolreprod.111.090886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 2019. The organizing principles of eukaryotic ribosome recruitment. Annu Rev Biochem 88: 307–335. 10.1146/annurev-biochem-013118-111042 [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. Stringtie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9: 171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Ramírez MA, Pericuesta E, Fernandez-Gonzalez R, Moreira P, Pintado B, Gutierrez-Adan A. 2006. Transcriptional and post-transcriptional regulation of retrotransposons IAP and MuERV-L affect pluripotency of mice ES cells. Reprod Biol Endocrinol 4: 55. 10.1186/1477-7827-4-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, Amieux PS. 2009. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci 106: 13939–13944. 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Harrison MM. 2019. Mechanisms regulating zygotic genome activation. Nat Rev Genet 20: 221–234. 10.1038/s41576-018-0087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM, Stein P, Svoboda P. 2018. The oocyte-to-embryo transition in mouse: past, present, and future. Biol Reprod 99: 160–174. 10.1093/biolre/ioy013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SG, Mandloi T, Kunte P, Natu A, Rashid M, Reddy D, Gadewal N, Gupta S. 2020. HISTome2: a database of histone proteins, modifiers for multiple organisms and epidrugs. Epigenetics Chromatin 13: 31. 10.1186/s13072-020-00354-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standart N, Minshall N. 2008. Translational control in early development: CPEB, P-bodies and germinal granules. Biochem Soc Trans 36: 671–676. 10.1042/BST0360671 [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11: 2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- von der Haar T, Gross JD, Wagner G, McCarthy JE. 2004. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 11: 503–511. 10.1038/nsmb779 [DOI] [PubMed] [Google Scholar]
- Wang Q, Latham KE. 1997. Requirement for protein synthesis during embryonic genome activation in mice. Mol Reprod Dev 47: 265–270. [DOI] [PubMed] [Google Scholar]
- Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. 2010. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci 107: 17639–17644. 10.1073/pnas.1013185107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Du Y, Ward JM, Shimbo T, Lackford B, Zheng X, Miao YL, Zhou B, Han L, Fargo DC, et al. 2014. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 14: 575–591. 10.1016/j.stem.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xu X, Li JL, Palmer C, Maric D, Dean J. 2019. Sertoli cell-only phenotype and scRNA-seq define PRAMEF12 as a factor essential for spermatogenesis in mice. Nat Commun 10: 5196. 10.1038/s41467-019-13193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Dean J. 2020. EXOSC10 sculpts the transcriptome during the growth-to-maturation transition in mouse oocytes. Nucleic Acids Res 48: 5349–5365. 10.1093/nar/gkaa249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Xu K, Lin Z, Kong F, Wang Q, Quan Y, Sha QQ, Li F, Zou Z, Liu L, et al. 2022. Ultrasensitive Ribo-seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat Cell Biol 24: 968–980. 10.1038/s41556-022-00928-6 [DOI] [PubMed] [Google Scholar]
- Zhang K, Smith GW. 2015. Maternal control of early embryogenesis in mammals. Reprod Fertil Dev 27: 880–896. 10.1071/RD14441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Park C, Bennett C, Thornton M, Kim D. 2021. Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res 31: 1290–1295. 10.1101/gr.275193.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang M, Li Y, Zhang Y. 2022. Profiling and functional characterization of maternal mRNA translation during mouse maternal-to-zygotic transition. Sci Adv 8: eabj3967. 10.1126/sciadv.abj3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data in this study have been deposited at the Gene Expression Omnibus (accession no. GSE180218) and the ProteomeXchange data sets (identifiers PXD041583 and PXD041622). Scripts used in this study can be found at https://github.com/Yang-DB-Lab/Eif4e1b_Paper. All other relevant data and materials of this study are available from J.D. upon request under a material transfer agreement with the National Institutes of Health.