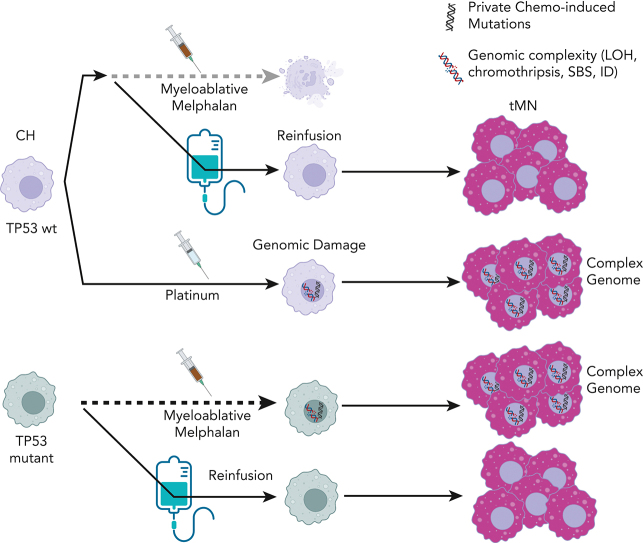
Key Points
-
•
Autologous transplantation allows clonal hematopoiesis to escape mutagenic chemotherapy and be reinfused to expand to neoplasm.
-
•
Distinct chemotherapies can promote the selection and acquisition of genomic drivers in therapy-related myeloid neoplasms.
Visual Abstract
Abstract
Patients treated with cytotoxic therapies, including autologous stem cell transplantation, are at risk for developing therapy-related myeloid neoplasms (tMN). Preleukemic clones (ie, clonal hematopoiesis [CH]) are detectable years before the development of these aggressive malignancies, although the genomic events leading to transformation and expansion are not well defined. Here, by leveraging distinctive chemotherapy-associated mutational signatures from whole-genome sequencing data and targeted sequencing of prechemotherapy samples, we reconstructed the evolutionary life-history of 39 therapy-related myeloid malignancies. A dichotomy was revealed, in which neoplasms with evidence of chemotherapy-induced mutagenesis from platinum and melphalan were hypermutated and enriched for complex structural variants (ie, chromothripsis), whereas neoplasms with nonmutagenic chemotherapy exposures were genomically similar to de novo acute myeloid leukemia. Using chemotherapy-associated mutational signatures as temporal barcodes linked to discrete clinical exposure in each patient’s life, we estimated that several complex events and genomic drivers were acquired after chemotherapy was administered. For patients with prior multiple myeloma who were treated with high-dose melphalan and autologous stem cell transplantation, we demonstrate that tMN can develop from either a reinfused CH clone that escapes melphalan exposure and is selected after reinfusion, or from TP53-mutant CH that survives direct myeloablative conditioning and acquires melphalan-induced DNA damage. Overall, we revealed a novel mode of tMN progression that is not reliant on direct mutagenesis or even exposure to chemotherapy. Conversely, for tMN that evolve under the influence of chemotherapy-induced mutagenesis, distinct chemotherapies not only select preexisting CH but also promote the acquisition of recurrent genomic drivers.
Patients receiving cytotoxic chemotherapy and autologous transplantation may develop therapy-related myeloid neoplasms (tMN). Diamond and colleagues analyzed the genomic evolution of tMN to distinguish between chemotherapy-induced mutations and evolution from previous clonal hematopoiesis. tMN arising after mutagenic chemotherapy shows a complexity of new mutations, while tMN arising after nonmutagenic chemotherapy resembles de novo acute leukemia. The trajectory of tMN may arise from the acquisition of new mutations, selection of previously mutated clones, or the expansion of mutant clones transferred from the apheresis product.
Introduction
Therapy-related myeloid neoplasms (tMN) have dismal prognoses and their incidence is predicted to increase as cancer survival rates rise.1, 2, 3 Some patients with underlying clonal hematopoiesis (CH) have a particularly high risk of tMN, especially those undergoing autologous stem cell transplantation (ASCT), a treatment with ubiquitous usage in lymphoproliferative disorders including in multiple myeloma (MM).4, 5, 6, 7, 8, 9, 10 There is evidence that anticancer therapies exert positive selective pressure on CH bearing preleukemic driver mutations (eg, TP53) such that expansion is not contingent on increased mutational burden relative to de novo acute myeloid leukemia (AML).11,12 Despite this, distinct DNA-damaging cytotoxic agents can measurably alter the mutational profile of each exposed cell, including in normal tissue, across cell lines, and in multiple malignancies.13, 14, 15, 16, 17, 18, 19
Mutational processes can be identified by examining mutations within specific nucleotide contexts. Distinct mutational signatures have been extracted and linked to intrinsic and extrinsic processes.20, 21, 22, 23, 24 The deconvolution of mutational signatures from whole-genome sequencing (WGS) data has allowed for direct quantitation of chemotherapy-mediated DNA damage and has revealed that a subset of tMN with prior platinum exposure does indeed have an increased chemotherapy-specific mutational burden compared with non–platinum-exposed tMN.25 However, it is largely unknown how chemotherapy exposure promotes CH progression to tMN. In addition to preferential selection of relatively fit CH clones among an altered and immunosuppressive bone marrow microenvironment,26, 27, 28 chemotherapy exposure may promote the acquisition of new driver events.
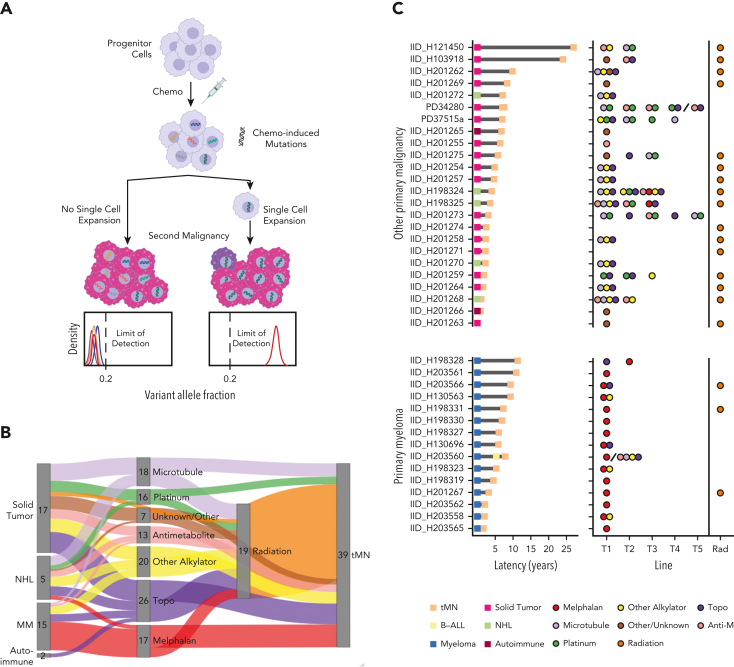
Chemotherapy-related mutational signatures are only detectable in bulk WGS after the clonal expansion of a single cell bearing a unique catalog of chemotherapy-induced mutations (ie, the single-cell expansion model, Figure 1A). The resultant mutational signature thus serves as a genomic single-cell barcode, linked to discrete clinical and temporal exposure.14,16, 17, 18,29, 30, 31 This effect is particularly significant in cells exposed to platinum and melphalan, 2 chemotherapeutics with distinct mutational signatures and highly penetrant mutagenic activity.14,18,29,31 Here, we leverage WGS and chemotherapy-induced mutational signatures as temporal molecular barcodes to measure the genomic evolution of tMN with reference to each patient’s known therapeutic history. We focused on the investigation of neoplasms that emerged after high-dose melphalan with ASCT (eg, MM) because chemotherapy is administered in a single bolus, is associated with a distinct mutational signature (SBS-MM1),29 and because the leukapheresis procedure potentially allows preleukemic clones to evade exposure to chemotherapy (supplemental Figure 1A; available on the Blood website). Overall, our data revealed 2 modes of expansion for tMN: one in which direct exposure to chemotherapy selects a preexisting CH clone and facilitates the post-therapy acquisition of distinct genomic drivers, and the other depends neither on direct chemotherapy-induced mutagenesis nor direct exposure to cytotoxic agents.
Figure 1.
Chemotherapy-induced mutagenesis and therapy-related myeloid neoplasm WGS cohort. (A) Cartoon summarizing that a single-cell expansion model is required to measure chemotherapy-associated mutational signatures in bulk WGS data. Populations that have already expanded sufficiently will develop private chemotherapy-induced mutations within multiple branching clones. With their expansion in the absence of single-cell expansion, subclonal mutations in each branch have frequencies below the bulk WGS limits of detection. (B) Sankey plot showing the therapeutic relationship between primary diagnosis and tMN. The therapy node cumulatively counts the patient’s exposure to each agent. (C) tMN for which WGS was performed with latency between the primary tumor and the second malignancy (left) and specific chemotherapy exposure (right). Forty tMN from 39 patients visually separated by the primary malignancy diagnosis are included. The line of therapy (ie, the order of treatment) is plotted on the x-axis. The backslashes separate sequential samples from the same patient. Anti-M, antimetabolite.
Methods
Study cohort
The WGS cohort was compiled using both the newly sequenced and publicly available data (supplemental Table 1). Clinical records at the Memorial Sloan Kettering Cancer Center (MSKCC) were screened to identify adult patients who developed tMN after exposure to either high-dose melphalan- or platinum-containing antineoplastic regimens. Eighteen tMN were eligible for sequencing (supplemental Tables 1-2). Eight patients were treated with melphalan as their sole cytotoxic therapy. The remaining 22 tMN genomes (from 21 patients) were imported from public datasets.12,32 21 de novo AML whole genomes were imported from TCGA33 (dbGaP: phs000178) as comparators. A total of 298 de novo AML and 22 tMN whole exomes (WES) were imported from the Beat AML data set (dbGaP: phs001657). To further gauge the effects of mutagenic chemotherapy on the genomic evolution of secondary malignancies after exposure to melphalan, we identified and sequenced 5 patients with B-ALL and 1 with transitional cell carcinoma diagnosed post-melphalan/ASCT for prior myeloma. In total, 20 patients with primary MM were included in the study, 19/20 of which received melphalan/ASCT. Two patients diagnosed with MM after exposure to platinum for ovarian and colorectal cancers were also included in the WGS (supplemental methods; supplemental Table 2). Samples and data were obtained and managed in accordance with the Declaration of Helsinki and the Institutional Review Board of MSKCC under protocols 14-276 and 15-017. A detailed description of the sequencing and analytical methods is provided in the supplemental methods.
Results
The mutational landscape of therapy-related malignancies
To measure the direct mutagenic activity of different chemotherapies and their roles in promoting tMN, we assembled a cohort of 40 tMN WGS from 39 patients with malignancies secondary to cytotoxic therapy (and/or radiation). Sixteen (40%) patients developed tMN post-melphalan/ASCT, 14 had MM, and 2 had aggressive B-cell lymphomas (Figure 1B-C; supplemental Tables 1-3; supplemental methods). Latency between the diagnosis of primary malignancy and tMN varied (median, 5.5 years; interquartile range [IQR], 2.4-7.2 years; Figure 1C), and outcomes post tMN diagnosis were expectedly poor (supplemental Figure 1B).
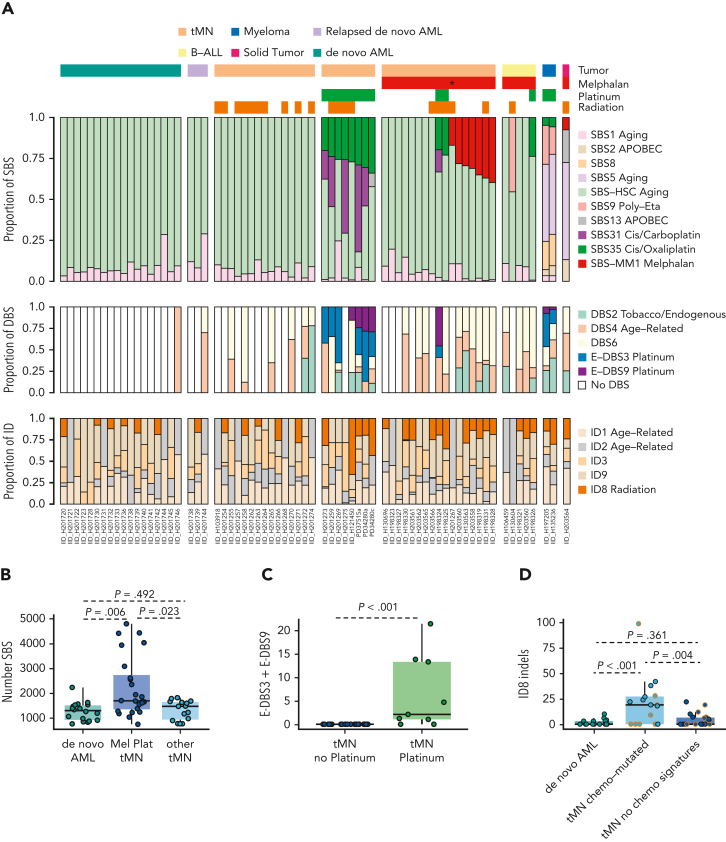
We first quantified the mutational processes that had been active in each tumor in accordance with a previously published workflow (Figure 2A; supplemental methods; supplemental Figure 2; supplemental Tables 4-8).29,32,34,35 Five known single-base substitution (SBS) mutational processes were identified in myeloid neoplasms: SBS1 and SBS-HSC, attributable to clock-like mutations that accumulate with age, observed in all hematopoietic cells25; SBS31 and SBS35, attributable to mutations induced by intercalating platinum chemotherapies14; and SBS-MM1, caused by the alkylator melphalan.18,29,30 Although SBS-MM1 has been reported in MM, lymphomas, and normal tissues previously exposed to melphalan, this is the first observation of melphalan mutagenesis in tMN. As expected, de novo AMLs and their relapsed samples bore only evidence of clock-like mutational processes, as neither of the induction agents cytarabine or anthracyclines are linked to distinct mutational signatures.17,21,36 The only antineoplastic agents that induced measurable SBS mutagenesis were platinum and melphalan, with the SBS burden in tMN having exposure to these agents being significantly higher than in either de novo AML (Wilcoxon test; P = .006) or unexposed tMN (Wilcoxon test; P = .023; Figure 2B). Though 5-fluorouracil is known to leave a distinct SBS signature (SBS17b), no evidence of it was seen in the 1 exposed sample, possibly in line with its mechanism of mutagenicity on dividing – and not quiescent – cells.25 Conversely, adducts generated by melphalan- and platinum-based agents are not dependent on cell turnover,16,25 and from previous studies are known to be highly penetrant in inducing mutagenesis in tumor and normal tissues.13,16,18,19,29, 30, 31,37 The mutational burden between de novo AML and tMN without melphalan or platinum exposure were strikingly similar (Wilcoxon test; P = .492; Figure 2B), confirming that some chemotherapies may facilitate malignancy in absence of SBS mutagenesis.25
Figure 2.
Mutational impact of chemotherapy on the whole-genome of therapy-related tumor. (A) Proportional contribution of SBS, DBS, and ID mutational signatures in each tumor sample. Each column represents a unique patient. Samples are annotated by disease histology and therapy exposure. Asterisk denotes the only patient exposed to low-dose melphalan without transplantation. (B) Boxplots for the number of SBS for platinum/melphalan-exposed tMN compared with de novo AML and other tMN. (C) Boxplot for E-DBS3 and E-DBS9 in platinum-exposed and -unexposed tMN. (D) Boxplot for ID8 in de novo AML, tMN with chemotherapy mutational signatures, and tMN without chemotherapy mutational signatures. Dots with orange borders are from patients exposed to radiation.
To further characterize the genomic impact of chemotherapy, we extracted double-base substitution (DBS) and indel (ID) signatures (supplemental methods; Figure 2A; supplemental Figures 3-4; supplemental Tables 4-6 and 9-11). Two DBS signatures known to be associated with platinum exposure14,25 were identified in 8 of 10 (80%) tMN with previous platinum exposure compared with none in unexposed samples (Wilcoxon test; P < .001; Figure 2C). Indels attributable to ID8, previously linked to double strand breaks and ionizing radiation,17,21 were enriched in tumors with chemotherapy-induced mutagenesis (Figure 2D supplemental Figure 5A), independent of prior radiotherapy, suggesting a link between ID8 and therapy-mediated genotoxic damage in tMN.21
Escape from chemotherapy-induced mutagenesis
All tMN with prior platinum exposure (n = 10) had evidence of platinum-associated SBS31 and/or SBS35 signatures, including samples with a latency from primary exposure until secondary malignancy of up to 25 years (IQR, 3.9-9.0; Figure 2A; supplemental Tables 2, 8). This complete penetrance indicates that an originating cell, with direct DNA damage from platinum chemotherapy, expanded to clonal dominance (ie, single-cell expansion model, Figure 1A; supplemental Figure 6A).14,18,29 In striking contrast, only 7 of 17 (41%) tMN with prior melphalan exposure had the SBS-MM1 melphalan signature, with neoplasms bearing SBS-MM1 containing a significantly higher tumor mutational burden than those without the signature (Wilcoxon test; P < .001; supplemental Figure 5B). All but 1 of these patients was exposed to high-dose myeloablative melphalan as a conditioning for ASCT. The absence of SBS-MM1 in these tMN can be explained by malignant progression driven by a population that has already expanded with multiple subclones in parallel (Figure 1A; supplemental Figure 6B), by the absence of direct mutagenic activity, or by a cell that has managed to escape exposure (supplemental Figure 6C).
As seen in relapsed MM, cell lines, and diffuse large B-cell lymphoma, melphalan has highly mutagenic and penetrant activity and exposure is expected to lead to mutagenesis.15,17,18,29, 30, 31,37 In fact, melphalan and platinum signatures are often seen together in patients exposed to both agents.18 Precursor escape via leukapheresis, as an explanation for the absence of SBS-MM1, is therefore the most likely explanation, and is supported by 4 lines of evidence. First, although latency between exposure to melphalan and sample collection is known to affect selection, single cell-expansion, and penetrance of SBS-MM1 detectability in MM,18,31,37 the latency between tMN and chemotherapy exposure in patients with either platinum or melphalan exposure had no influence on signature penetrance (Wilcoxon test; P = .329). Second, we sequenced 5 patients with post-melphalan/ASCT B-ALL for orthogonal support from an alternate disease biology and SBS-MM1 was not present in any, supporting a similar expansion model in an alternate secondary malignancy (Figure 2A). Third, patients with sequential exposure to platinum and then melphalan/ASCT [2 tMN (IID_H198324, IID_H198325) and 1 B-ALL (IID_H198326)] had tumors bearing only platinum-related mutational signatures, indicating that a single cell exposed to platinum, but not to melphalan, expanded to clonal dominance. Given the short latency between the 2 therapies, the absence of SBS-MM1 can only be explained by escape via leukapheresis (supplemental Figure 6D). Finally, interrogation of the WGS of tumors with precursors that did not have a possible route of escape from melphalan did indeed have SBS-MM1 signatures: 1 patient with tMN after treatment with oral melphalan without ASCT (IID_H201267), and a transitional cell carcinoma diagnosed after melphalan/ASCT (IID_H203564; supplemental methods; Figure 2A; supplemental Figure 7). The latter tumor was chosen for sequencing because unchanged melphalan is partially excreted in urine,38 thus putting the urothelium in direct contact with the mutagen. Although preleukemic clones have been demonstrated in apheresis products,7 the clone responsible for tMN progression has previously been unknown because of the categorical presence of CH within the patient, as well as in the apheresis product. Although we cannot exclude that in some samples, SBS-MM1 was below the threshold of detection (ie, absence of single-cell expansion) or that a precursor cell was somehow shielded from the exposure, these data together suggest that leukapheresis can allow for a preleukemic clone to escape exposure to melphalan and its related mutagenic activity, and then be reinfused to expand into tMN.
The driver mutations landscape of therapy-related myeloid neoplasms
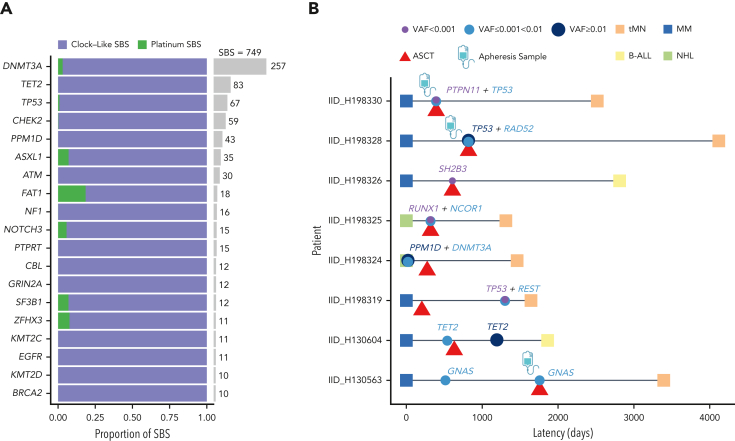
Although SNV in driver mutations for tMN and de novo AML have been extensively reported,33,39,40 it remains unclear whether exposure to chemotherapy solely selects for preexisting CH or whether it may also introduce new driver mutations. We first determined positively-selected driver genes using the dN/dS algorithm for both de novo AML and tMN, importing samples from the Beat AML data set of WES to increase power (298 de novo AML and 22 tMN; supplemental methods; supplemental Table 12).40 Only TP53 was more frequently mutated in tMN (Fisher test; P < .001; false discovery rate [FDR] = 0.009).9 Conversely, NPM1 was more frequently mutated in de novo AML (P < .001; FDR = 0.019) in line with previous findings.41 Although limited by sample size, there were no significant differences in driver gene SNVs between chemotherapy signature-positive and -negative tMN and specifically between melphalan-signature-positive and -negative cases (supplemental Figure 8; supplemental Table 13). Next, we sought to determine whether chemotherapy could introduce mutations at the driver gene level. We combined our post-platinum tMN WGS with a large number of CH mutations from a cohort of patients treated previously with platinum therapies from Bolton et al (749 mutations in 655 patients).11 Estimating the mutational signature contribution to all nonsynonymous mutations in prevalent driver genes, we observed only a minor platinum contribution (supplemental methods; Figure 3A), suggesting that the vast majority of mutations in driver genes are likely selected and not caused by chemotherapy exposure.
Figure 3.
Relationship between chemotherapy and CH mutations. (A) Per gene contribution of platinum-associated mutational signatures to 749 post-platinum mutations in 655 patients with cancer (Bolton et al).11 A small fraction (6 mutations from 5 patients) is included from clonal driver mutations in post-platinum tMN whole genomes. (B) Swimmer plot depicting the timing of CH assessment in relation to malignant diagnoses and transplant dates for tested samples in the WGS cohort. Latency reflects the time from the diagnosis of the primary tumor to the diagnosis of therapy-related malignancy. All samples were collected before melphalan administration with the exception of IID_H198319.
To expand on the relationship between mutations in driver genes and chemotherapy, we performed targeted sequencing on premelphalan blood mononuclear cells, granulocytes, and CD34+ apheresis samples from 11 of the newly sequenced hematologic malignancies from patients treated with melphalan/ASCT (supplemental methods).11,42 Knowing from the tumor WGS, the full catalog of clonal mutations, we could identify precursor mutations at lower variant allele frequencies than could be confidently called with mutation-calling algorithms, alone, in the prechemotherapy samples (supplemental methods). Eight of the 11 cases (72%), including 3 of 4 leukapheresis products, showed evidence of antecedent preleukemic clones (Figure 3B; supplemental Table 14). Notably, for IID_H198330, a case without the melphalan mutational signature, an antecedent TP53-mutated CH was detected in the apheresis product (supplemental Figure 9), supporting an evolutionary trajectory of escape from exposure to chemotherapy, reinfusion with transplant, and expansion in the absence of melphalan-induced mutagenesis. Acknowledging that gene-level signature fitting is limited by this sample size, for 10 of 21 (47.6%) driver mutations in tMN WGS not captured at prechemotherapy time points, none were in key platinum or melphalan trinucleotide contexts, suggesting that there is a low chance that they were introduced by chemotherapy (supplemental Table 15). Variants at low frequencies emphasize that CH clone size is not categorically predictive of progression to myeloid neoplasms.5,11,43, 44, 45
Copy number and structural variation in therapy-related myeloid neoplasms
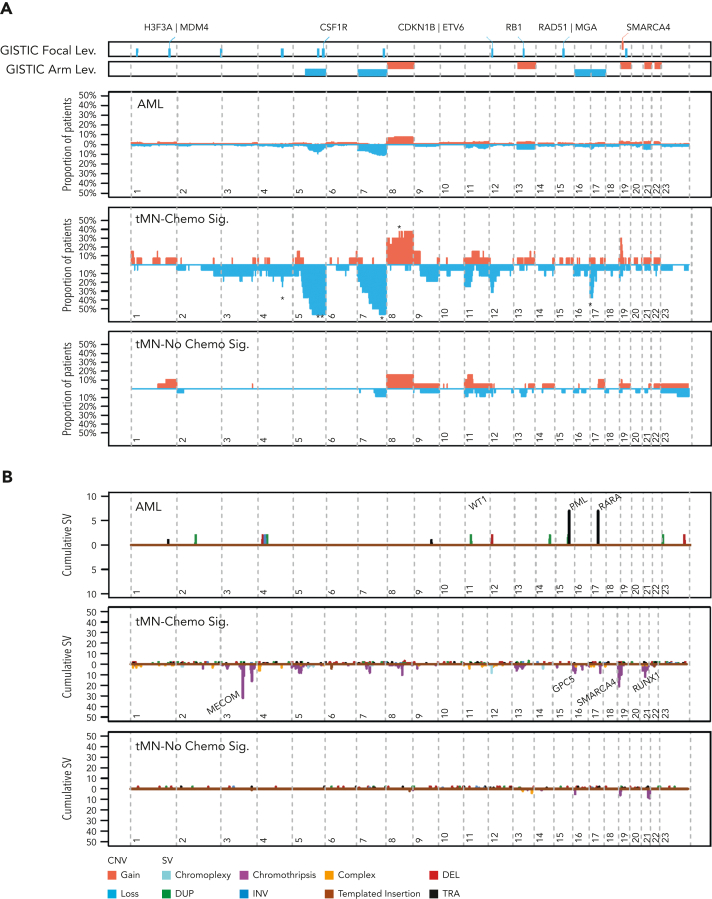
We next compared recurrent copy number aberrations (CNAs) between de novo AML and tMN, including the exomes from Beat AML (supplemental methods). We differentially detected 7 arm-level and 1 focal region (19p13.2) of amplification, and 7 arm-level and 11 focal regions of copy number loss (FDR < 0.1; supplemental Figure 10A; supplemental Table 17). The association between chromosomal aneuploidies and alkylator therapy is known however,46,47 once tMN samples were grouped by chemotherapy signature presence, a striking pattern emerged: de novo AML and tMN lacking chemotherapy-induced mutagenesis shared a similar distribution and frequency of copy number events, whereas tMN harboring chemotherapy signatures held the majority of significant CNAs, including deletions of chromosomes 5q, 7q, 17p, and focal gains in 19p13.2 (FDR < 0.1; Figure 4A; supplemental Table 17).
Figure 4.
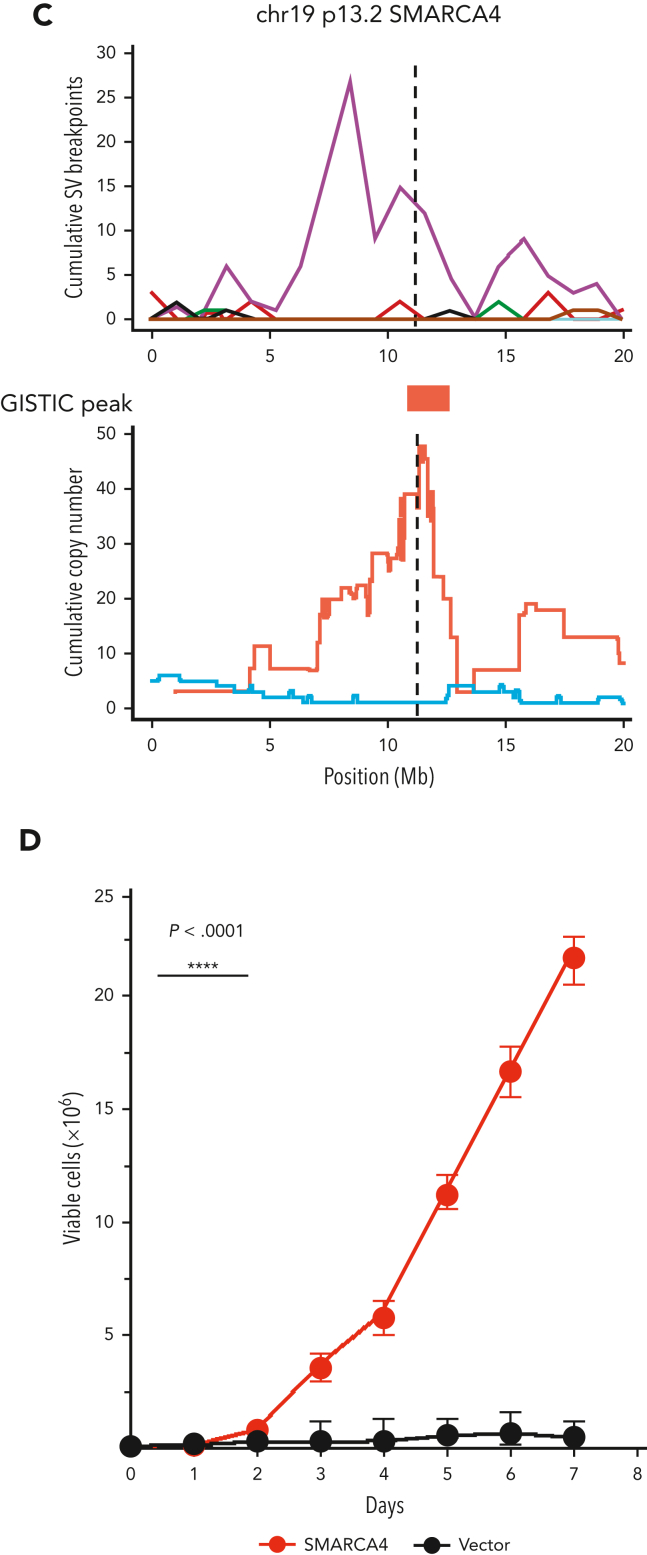
Copy number and SV landscapes in therapy-related myeloid neoplasms. (A) Cumulative copy number profile for all de novo samples (n = 316; 18 genomes from TCGA, 298 exomes from Beat AML) and 39 tMN genomes. tMN genomes are split into chemotherapy signature-positive (n = 17) and -negative (n = 22) cases. GISTIC peaks and CNA arm-level events enriched in tMN with chemotherapy signatures (P < .05; FDR < 0.1; Fisher test) are annotated with an asterisk for significance compared with: tMN without chemotherapy signatures. (B) SV landscape across de novo AML genomes from TCGA (n = 18) and tMN genomes with and without a chemotherapy-associated mutational signature. Breakpoints are binned into 1 megabase segments. Simple events point upward from the x-axis and complex events (eg, chromothripsis) point downward. (C) Plot of cumulative copy number changes and SV chromothriptic breakpoints responsible for focal multigain events in SMARCA4. (D) Growth curves of SMARCA4 transfected Ba/F3 cells versus cells transfected with the vector in the interleukin-3 cytokine independence assay. ****P < .0001.
Many previous characterizations of the tMN mutational landscape have used targeted or WES. These assays have a limited resolution for structural variants (SV) and focal CNA. We combined de novo AML and tMN WGS to explore the landscape of SVs (supplemental methods; supplemental Figure 10B). Consistent with the CNAs and known cytogenetic complexity, SVs and complex events (ie, templated insertions, chromoplexy, and chromothripsis) were significantly enriched within tMN with evidence of chemotherapy mutagenesis (Wilcoxon test; P < .001; Figure 4B; supplemental Figure 10C; supplemental Table 18). Among the complex events, chromothripsis was the most frequent and was observed in 8 of the 39 tMN cases (20.5%). This event consists of a catastrophic shattering of multiple chromosomal regions resulting in the introduction of disparate drivers.48, 49, 50, 51 Strikingly, chromothripsis involving chromosome 19p13.2 with focal amplifications (median 7; range, 5-12 copies) of the SMARCA4 locus comprised 5 of these cases, with 4 of 5 events found in chemotherapy signature-positive genomes (Figure 4C; supplemental Figure 11). In comparison, across the entire cohort of de novo AML genomes and exomes (n = 316), focal amplification of SMARCA4 was observed in only 1 case with multiple chromosomal aneuploidies (Fisher test; P < .0001; supplemental Figure 12A). Deleterious mutations in SMARCA4, generally considered a tumor suppressor gene, have been implicated in other malignancies including ovarian carcinoma, and various lymphomas.52, 53, 54 As we found it to be uniformly amplified in tMN, we sought to verify that SMARCA4 overexpression could promote leukemic cell growth. SMARCA4 transfection into Ba/F3 cells demonstrated that gain-of-function amplification could drive growth when compared with the vector with IL3 cytokine independence (Figure 4D; supplemental methods; supplemental Figure 12B). Consistent with previous evidence, the MECOM locus was also frequently involved in tMN structural variation, with a preponderance of tMN containing a chemotherapy-associated signature, further supporting its role in myeloid tumorigenesis (3 of 4 cases, Figure 4B; supplemental Figure 11).55, 56, 57 Notably, MLL aberration was found in only 1 patient exposed to melphalan without SBS-MM1 (supplemental Table 19).
Chemotherapy-related mutational signatures as molecular barcodes
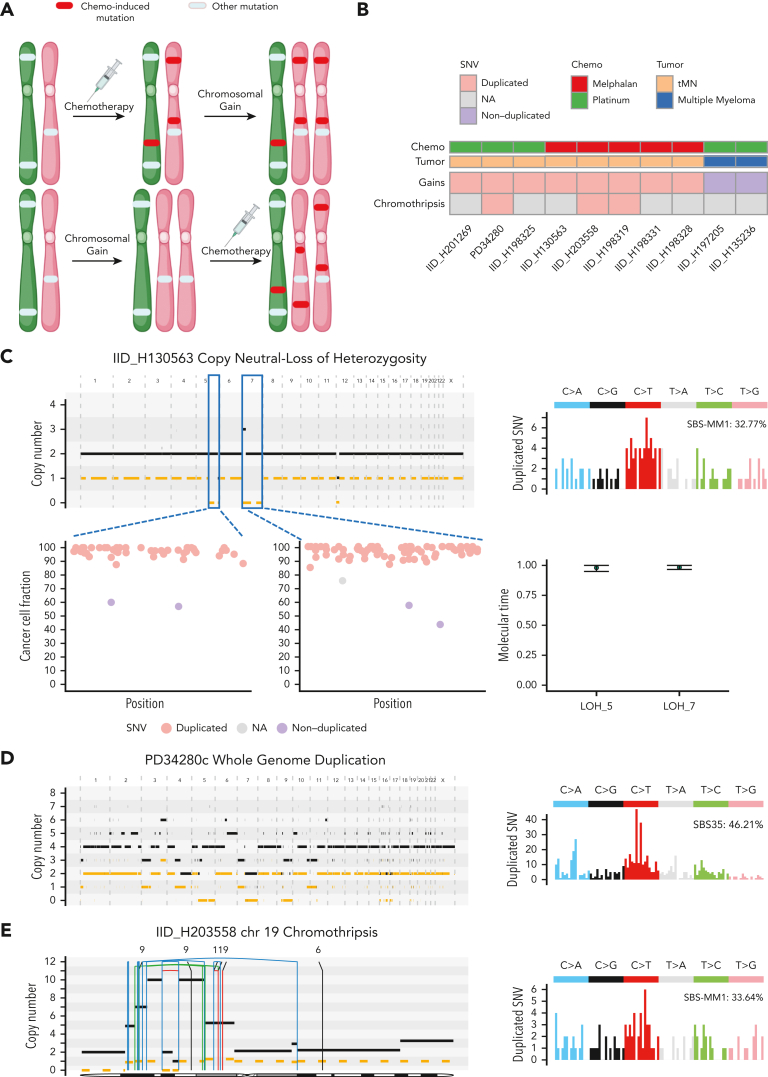
The accumulation of mutations attributable to constant, clock-like processes (SBS1, SBS5, and SBS-HSC) can be used to estimate the age at which a large chromosomal gain occurs in many malignancies.20,29,58, 59, 60 However, in tMN there is a lack of correlation between clock-like SBS and the age at diagnosis (supplemental Figure 13A).25 To overcome this limitation, we developed an approach leveraging chemotherapy-associated signatures as temporal barcodes by which to time chromosomal gains relative to chemotherapy exposure. Specifically, if a mutation is duplicated across a chromosomal gain, it must have been present before the event. Consequently, a chemotherapy-associated mutational signature present within duplicated mutations necessitates exposure before the gain. Conversely, chemotherapy-associated mutational signatures present only among nonduplicated clonal or subclonal mutations imply that the exposure occurred after the chromosomal gain (Figure 5A).61 We applied this rationale to time acquisition of CNA and SV before or after chemotherapy exposure. After collapsing together large events (eg, trisomy and copy neutral loss of heterozygosity) that occurred within the same molecular time window (supplemental methods; supplemental Figure 13B), 8 tMN with chemotherapy signatures were amenable to temporal barcoding. In all cases, melphalan or platinum signatures were detectable within duplicated clonal mutations, implying that large CNAs occurred during or after exposure to chemotherapy and late in tMN evolution (Figures 5B-D; supplemental Table 20). Importantly, 1 of the 3 patients exposed to both platinum and melphalan (IID_H198325), but only harboring platinum SBS signatures, had a post-chemotherapy trisomy 8 and clonal BCOR mutation not detected by targeted sequencing. These 2 events are clear evidence of post-chemotherapy acquisition and ongoing single-cell expansion, further supporting that a melphalan signature should also be present if not for escape in the apheresis product.
Figure 5.
Chemotherapy-associated single-base substitution signatures as temporal barcodes. (A) Cartoons summarizing the rationale used to time chromosomal duplications according to chemotherapy exposure. (B) Heatmap revealing the pattern of timing for chromosomal duplications and chromothripsis events in relation to chemotherapy for 8 tMN and 2 cases of post-chemotherapy MM. (C) Example of 2 copy neutral loss of heterozygosity (CN-LOH) acquired after chemotherapy exposure. In this case, both CN-LOH contain duplicated mutations (bottom left) consistent with late acquisition, as confirmed by molecular time analysis (bottom right), and showing large SBS-MM1 (melphalan) signature contribution within duplicated (ie, pregain) mutations (top right). (D) Example of whole-genome duplication during tMN relapse (left). Duplicated mutations (right) showed a large contribution from SBS35 (platinum), suggesting that this event was acquired after platinum exposure. (E) Chromothripsis event on chromosome 19 (SMARCA4) with multiple duplications (left). Similar to (D), the duplicated mutational signature contribution within chromothripsis-associated amplifications (right) was enriched for the SBS-MM1 contribution, implying that the chromothripsis event was acquired after melphalan exposure. In (C-E), the horizontal black line indicates the total copy number and the dashed orange line indicates the minor copy number. In (E), the vertical lines represent SVs breakpoints, color-coded based on SVs class: blue, inversion; green, tandem-duplication; red, deletion; black, translocation.
As all CNAs associated with chromothripsis occur simultaneously within a catastrophic event,62 we similarly applied this methodology to 3 tMN with large chromothriptic events characterized by amplified genomic segments with more than 40 clonal nonclustered mutations.29,60 The chemotherapy-associated signatures were present in the duplicated mutations within chromothripsis-associated gains, including the previously mentioned events in SMARCA4, supporting that these complex SVs occurred after exposure to mutagenic therapy (supplemental methods; Figures 5B,E; supplemental Table 21). These findings are the first to demonstrate that tMN development can be facilitated by the acquisition of additional drivers on a preexisting CH clone.
In the context of chemotherapy-acquired events, we pooled all somatic events among the tMN genomes (ie, SV, CNA, and SNV), and found a higher prevalence of TP53 loss (10/16, 62.5%) than in the signature-negative cases (3/23, 13%; Fisher test; P = .002). Strikingly, among those receiving melphalan/ASCT, all 6 cases with the SBS-MM1 signature (ie, not reinfused) had an event involving TP53, as compared with 2/10 (20%) without the signature (ie, reinfused; Fisher test; P = .007; supplemental Table 22). Acknowledging the limited sample size, this stark difference indicates that the loss of TP53 may allow precursors to survive direct exposure to myeloablative melphalan and accrue chemotherapy-mediated aberrations. Precursors with unperturbed TP53 may be preferentially reinfused in the transplant or selected by nonmyeloablative chemotherapy. Given the preponderance of TP53 disruption and complex genomes, we assessed the outcomes between chemotherapy-signature-positive and -negative tMN and found a nonsignificant trend toward shortened survival in the former, limited by sample size.
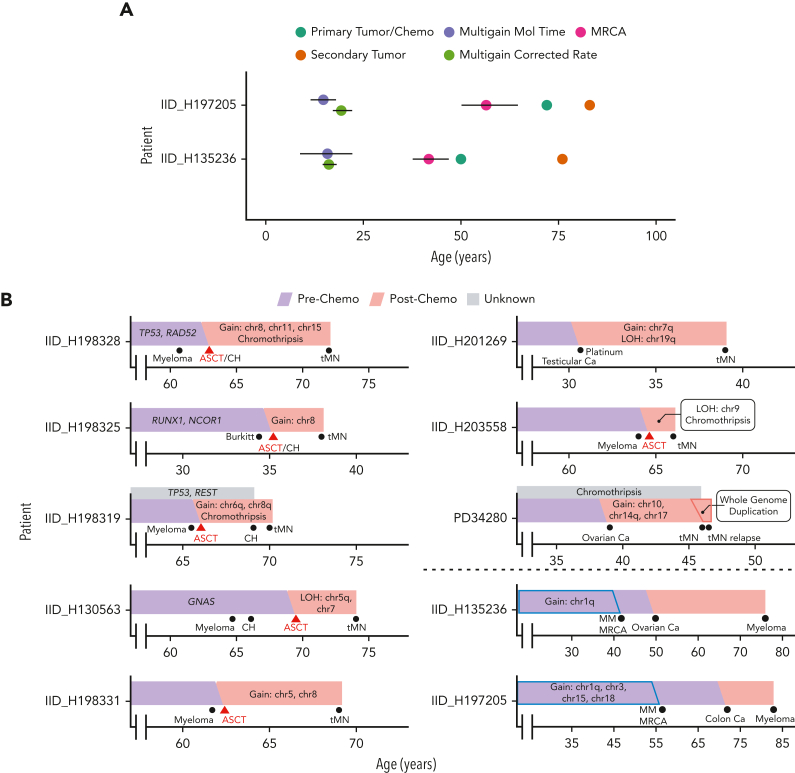
Finally, in contrast to tMN, for 2 post-platinum MM, the associated platinum signatures were seen only in subclonal (ie, nonduplicated) mutations, in agreement with previous knowledge that many chromosomal gains in MM are early events (Figure 5B).29 We sought to further validate our approach of chemotherapy-associated SBS barcoding with molecular time.29,58,60 We first calculated the individual SBS5 mutation rate per year for the 2 secondary MM and found no significant difference when compared with 77 WGS from 47 patients with primary or smoldering MM (supplemental methods; supplemental Figure 13C).29 Collapsing together large chromosomal gains occurring within the same time window, we estimated the SBS5-based molecular time to predict at which age these 2 patients acquired the first multichromosomal gain event and the emergence of the most recent common ancestor (MRCA) (supplemental methods; Figure 6A; supplemental Figure 13B). Consistent with previous reports, in both cases, the initiating gains were estimated to have occurred in the 2nd decade of life29 and MRCA arose before the diagnosis of the unrelated primary malignancy (ie, the solid tumor) and the associated platinum exposure. SBS5-based molecular time therefore aligns with the presence of platinum-associated signatures among only subclonal mutations (ie, exposure and mutagenesis after the gains) and, overall, validates chemotherapy-associated mutational signatures as temporal molecular barcodes that can be used to validate the absolute time of acquisition of copy number gains.20,58,63
Figure 6.
Evolutionary history of malignancies exposed to chemotherapy. (A) Absolute timing estimates of large chromosomal gain acquisition in 2 platinum-exposed MM with 95% confidence intervals. Multigain events for MM were timed with 2 orthogonal techniques (supplemental methods): one using SBS5-based molecular time (ie, multigain mol time) and the other using the individual SBS5 mutational rate per year estimated by a linear mixed effect model (ie, multigain corrected rate). Platinum was administered directly after the diagnosis of the primary tumor. (B) Reconstruction of tumor evolution in selected WGS cases using antecedent CH, molecular time, and duplicated chemotherapy mutation data for tMN and MM (under dotted line).
Discussion
Using WGS of tMN samples from patients exposed to a variety of antineoplastic agents and targeted sequencing of pretherapy samples, we characterized the mutational impact of chemotherapy and used chemotherapy-associated SBS signatures as temporal barcodes to reconstruct the evolution of tMN from its antecedent CH (Figure 6B). This is the first WGS profiling of tMN post–high-dose melphalan and ASCT (eg, post-MM) and the first evidence that, similar to its effects in MM and aggressive lymphomas, melphalan has direct mutagenic activity on tMN. Overall, because of the heterogeneity of the cohort and the inclusion of tMN exposed to melphalan, platinum, or other agents, our data confirmed that tMN evolving with chemotherapy-induced mutagenesis (ie, chemotherapy mutational signatures) is relatively hypermutated compared with de novo AML and tMN that develop without the direct mutational influence of chemotherapy. We also observed enrichment for distinct CNAs and SVs, including chromothripsis, among tMN with chemotherapy-induced mutagenesis. These data highlight the need for WGS to characterize the full landscape of alterations in these complex malignancies. Although we observe here in a retrospective genomic analysis that all tMN with evidence of chemotherapy-induced mutagenesis had TP53 disruption, we cannot generalize the risk of progression to TP53-variant CH. Prospective studies are needed to ascertain the individual risk of tMN, considering both variants of underlying CH and therapeutic interventions.
Prior studies have suggested that progression to tMN is driven by the chemotherapy-mediated selection of preexisting CH harboring distinct leukemic driver SNV. Here, we see supporting data in the form of the detection of antecedent CH in pretherapy samples and in the minimal contribution of chemotherapy signatures among CH mutations from platinum-exposed individuals. However, the temporal relationship of other genomic drivers, such as complex CNA and SV, with chemotherapy has been largely unexplored.25 Using a new approach based on chemotherapy signatures in duplicated SNV within chromosomal gains, we see that in contrast to driver SNV, which are almost universally selected and not caused by chemotherapy, chromothripsis and chromosomal gains are frequently acquired after mutagenic chemotherapy. These combined observations support a model in which exposure to distinct genotoxic chemotherapies can both select for and increase the genomic complexity of preexisting CH during the progression to tMN (Figure 6B). One limitation of our approach is its inability to provide estimates of deletions, for which no temporal approach has yet been developed. Importantly, chemotherapy temporal barcoding was seen here to be an orthogonal validation of previously reported molecular timing approaches for determining the age of large chromosomal gains in MM.29,58
Our investigations also characterized the disparate routes through which a preexisting CH clone can progress to tMN after high-dose melphalan and ASCT. CH clones have previously been isolated from apheresis products, indicating that they can indeed be reinfused during the ASCT procedure.4,7 Nevertheless, because the CH clone is present in both the bone marrow and apheresis products, it has not been possible to define which reservoir will be the source responsible for the progression into tMN. By leveraging mutational signatures as temporal barcodes, we were able to demonstrate that tMN may originate either from a reinfused CH clone or by an intrinsic TP53-mutant CH that survives myeloablative conditioning with high-dose melphalan.64 Although it is known that an increased chemotherapy-induced mutational burden is not requisite for tMN progression,12,25 post-ASCT tMN expansion in the absence of a melphalan signature indicates that direct cellular exposure to mutagenic chemotherapy may be required. This route of chemotherapy evasion places increased emphasis on leukemia-permissive effects of both mutagenic and nonmutagenic chemotherapy on the bone marrow compartment.11,12,65 In agreement with our findings, chemotherapy and transplantation have been shown to confer selective pressure on progenitor cells in a relatively vacant and dysfunctional marrow niche, such that hematopoiesis is reconstituted by a limited number of clones.66, 67, 68, 69 Altogether, several lines of evidence suggest that the evolution of precursor states into their malignant successors is likely driven by a complex interaction between inflammation, mutagenesis, and immune suppression.2,12,26, 27, 28,70,71 Our findings add resolution into the different routes of post-ASCT tMN progression, mediated by both selective and mutagenic aspects of therapy. As immunomodulatory and cellular therapies enter widespread clinical practice, it is becoming increasingly important to characterize their complex immune effects in the context of their ability to promote tMN evolution.
Conflict-of-interest disclosure: E.P. is a founder and equity holder and holds a fiduciary role in Isabl Inc. The remaining authors declare no competing financial interests.
Acknowledgments
This work was supported by the International Myeloma Foundation 2020 Brian D. Novis Research Award, by the Paula and Rodger Riney Foundation, by the Sylvester Comprehensive Cancer Center National Cancer Institute (NCI) Core Grant (P30 CA 240139) by the Memorial Sloan Kettering Cancer Center NCI Core Grant (P30 CA 008748), by the Conquer Cancer Young Investigator Award (16587), and by the Sylvester Comprehensive Cancer Center/American Cancer Society IRG grant. F.M. is supported by the American Society of Hematology. B.D. is supported by Myeloma Crowd, the International Myeloma Society, and the Sylvester K12 Calabresi Clinical Oncology Research Career Development Program. N.B. is supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) through investigator grant 25739.
Authorship
Contribution: F.M. designed and supervised the study, collected, and analyzed the data and wrote the manuscript; O.L. designed the study, collected the data and wrote the manuscript; B.D. and B.Z. supervised the study, collected, and analyzed the data and wrote the manuscript; K.M. collected and analyzed the data and wrote the manuscript; E.P. designed and collected the data; J. Taylor collected and analyzed the data; E.B. and G.M. collected the data and performed all the flow cytometry sorting experiments; A.M., D.J.C., and J.A.O. analyzed the data; J.J., M.A., T.M.T performed the SMARCA4 wet validation; S.U., S.N., J. Tyner, M.R., C.H., Y.Z., D.C., A.L., M.A.S., K.G., H.L., J.H.P, N.B., S.X.L., K.B., N.C., J.W., L.C. collected the data; All authors read, revised, and proofed the manuscript.
Footnotes
∗B.D. and B.Z. contributed equally to this study.
†O.L. and F.M. contributed equally to this study.
All newly sequenced whole genomes and target sequencing data used in this study have been uploaded to European Genome Archive EGAS00001006903.
Part of the data set used for this paper is derived from public sources. Twenty-two tMN genomes (from 21 patients) were imported from dbGaP phs000159 and EGAD00001005028. Twenty-one de novo AML whole genomes (including 3 relapse samples) were imported from dbGaP phs000178. Two hundred ninety-eight de novo AML and 22 tMN WES were imported from dbGaP phs001657.
Mutational calls and annotation for post-platinum CH are available on cBioPortal: http://www.cbioportal.org/study/summary?id=msk_ch_2020.
Analyses conducted using previously published code are detailed and available via github links in supplemental methods.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Majhail NS, Tao L, Bredeson C, et al. Prevalence of hematopoietic cell transplant survivors in the United States. Biol Blood Marrow Transplant. 2013;19(10):1498–1501. doi: 10.1016/j.bbmt.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNerney ME, Godley LA, Le Beau MM. Therapy-related myeloid neoplasms: when genetics and environment collide. Nat Rev Cancer. 2017;17(9):513–527. doi: 10.1038/nrc.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kayser S, Doehner K, Krauter J, et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood. 2011;117(7):2137–2145. doi: 10.1182/blood-2010-08-301713. [DOI] [PubMed] [Google Scholar]
- 4.Gibson CJ, Lindsley RC, Tchekmedyian V, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017;35(14):1598–1605. doi: 10.1200/JCO.2016.71.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai P, Mencia-Trinchant N, Savenkov O, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24(7):1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouhieddine TH, Sperling AS, Redd R, et al. Clonal hematopoiesis is associated with adverse outcomes in multiple myeloma patients undergoing transplant. Nat Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-16805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbloom B, Schreck R, Koeffler HP. Therapy-related myelodysplastic syndromes. Hematol Oncol Clin N Am. 1992;6(3):707–722. [PubMed] [Google Scholar]
- 9.Godley LA, Larson RA. Therapy-related myeloid leukemia. Semin Oncol. 2008;35(4):418–429. doi: 10.1053/j.seminoncol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coombs CC, Zehir A, Devlin SM, et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell. 2017;21(3):374–382.e374. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolton KL, Ptashkin RN, Gao T, et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat Genet. 2020;52(11):1219–1226. doi: 10.1038/s41588-020-00710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee-Six H, Olafsson S, Ellis P, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574(7779):532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 14.Pich O, Muiños F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet. 2019;51(12):1732–1740. doi: 10.1038/s41588-019-0525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain MD, Ziccheddu B, Coughlin CA, et al. Whole-genome sequencing reveals complex genomic features underlying anti-CD19 CAR T-cell treatment failures in lymphoma. Blood. 2022;140(5):491–503. doi: 10.1182/blood.2021015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplanis J, Ide B, Sanghvi R, et al. Genetic and chemotherapeutic influences on germline hypermutation. Nature. 2022;605(7910):503–508. doi: 10.1038/s41586-022-04712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucab JE, Zou X, Morganella S, et al. A compendium of mutational signatures of environmental agents. Cell. 2019;177(4):821–836.e816. doi: 10.1016/j.cell.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landau HJ, Yellapantula V, Diamond BT, et al. Accelerated single cell seeding in relapsed multiple myeloma. Nat Commun. 2020;11(1):1–10. doi: 10.1038/s41467-020-17459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore L, Cagan A, Coorens TH, et al. The mutational landscape of human somatic and germline cells. Nature. 2021;597(7876):381–386. doi: 10.1038/s41586-021-03822-7. [DOI] [PubMed] [Google Scholar]
- 20.Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet. 2015;47(12):1402–1407. doi: 10.1038/ng.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB, Kim J, Haradhvala NJ, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534(7605):47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149(5):994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pich O, Cortes-Bullich A, Muiños F, Pratcorona M, Gonzalez-Perez A, Lopez-Bigas N. The evolution of hematopoietic cells under cancer therapy. Nat Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-24858-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link D, Walter M. ‘CHIP’ping away at clonal hematopoiesis. Leukemia. 2016;30(8):1633–1635. doi: 10.1038/leu.2016.130. [DOI] [PubMed] [Google Scholar]
- 27.Stoddart A, Wang J, Fernald AA, et al. Cytotoxic therapy–induced effects on both hematopoietic and marrow stromal cells promotes therapy-related myeloid neoplasms. Blood Cancer Discov. 2020;1(1):32–47. doi: 10.1158/2643-3230.BCD-19-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zambetti NA, Ping Z, Chen S, et al. Mesenchymal inflammation drives genotoxic stress in hematopoietic stem cells and predicts disease evolution in human pre-leukemia. Cell Stem Cell. 2016;19(5):613–627. doi: 10.1016/j.stem.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Rustad EH, Yellapantula V, Leongamornlert D, et al. Timing the initiation of multiple myeloma. Nat Commun. 2020;11(1):1–14. doi: 10.1038/s41467-020-15740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maura F, Weinhold N, Diamond B, et al. The mutagenic impact of melphalan in multiple myeloma. Leukemia. 2021;35(8):2145–2150. doi: 10.1038/s41375-021-01293-3. [DOI] [PubMed] [Google Scholar]
- 31.Rasche L, Schinke C, Maura F, et al. The spatio-temporal evolution of multiple myeloma from baseline to relapse-refractory states. Nat Commun. 2022;13(1):4517. doi: 10.1038/s41467-022-32145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maura F, Degasperi A, Nadeu F, et al. A practical guide for mutational signature analysis in hematological malignancies. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-11037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Network CGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degasperi A, Amarante TD, Czarnecki J, et al. A practical framework and online tool for mutational signature analyses show intertissue variation and driver dependencies. Nat Cancer. 2020;1(2):249–263. doi: 10.1038/s43018-020-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degasperi A, Zou X, Dias Amarante T, et al. Substitution mutational signatures in whole-genome–sequenced cancers in the UK population. Science. 2022;376(6591):abl9283. doi: 10.1126/science.abl9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Kanter JK, Peci F, Bertrums E, et al. Antiviral treatment causes a unique mutational signature in cancers of transplantation recipients. Cell Stem Cell. 2021;28(10):1726–1739.e1726. doi: 10.1016/j.stem.2021.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giesen N, Paramasivam N, Toprak UH, et al. Comprehensive genomic analysis of refractory multiple myeloma reveals a complex mutational landscape associated with drug resistance and novel therapeutic vulnerabilities. Haematologica. 2022;107(8):1891–1901. doi: 10.3324/haematol.2021.279360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gouyette A, Hartmann O, Pico J-L. Pharmacokinetics of high-dose melphalan in children and adults. Cancer Chemother Pharmacol. 1986;16(2):184–189. doi: 10.1007/BF00256174. [DOI] [PubMed] [Google Scholar]
- 39.Feusier JE, Arunachalam S, Tashi T, et al. Large-scale identification of clonal hematopoiesis and mutations recurrent in blood cancers. Blood Cancer Discov. 2021;2(3):226–237. doi: 10.1158/2643-3230.BCD-20-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyner JW, Tognon CE, Bottomly D, et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562(7728):526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pich O, Reyes-Salazar I, Gonzalez-Perez A, Lopez-Bigas N. Discovering the drivers of clonal hematopoiesis. Nat Commun. 2022;13(1):1–12. doi: 10.1038/s41467-022-31878-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17(3):251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabre MA, de Almeida JG, Fiorillo E, et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature. 2022;606(7913):335–342. doi: 10.1038/s41586-022-04785-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson NA, Latorre-Crespo E, Terradas-Terradas M, et al. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. Nat Med. 2022;28(7):1439–1446. doi: 10.1038/s41591-022-01883-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abelson S, Collord G, Ng SW, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haferlach C, Dicker F, Herholz H, Schnittger S, Kern W, Haferlach T. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008;22(8):1539–1541. doi: 10.1038/leu.2008.143. [DOI] [PubMed] [Google Scholar]
- 47.Qian Z, Joslin JM, Tennant TR, et al. Cytogenetic and genetic pathways in therapy-related acute myeloid leukemia. Chem Biol Interact. 2010;184(1-2):50–57. doi: 10.1016/j.cbi.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortés-Ciriano I, Lee JJ-K, Xi R, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020;52(3):331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rustad EH, Yellapantula VD, Glodzik D, et al. Revealing the impact of structural variants in multiple myeloma. Blood Cancer Discov. 2020;1(3):258–273. doi: 10.1158/2643-3230.BCD-20-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Roberts ND, Wala JA, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578(7793):112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hadi K, Yao X, Behr JM, et al. Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell. 2020;183(1):197–210.e132. doi: 10.1016/j.cell.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jelinic P, Mueller JJ, Olvera N, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet. 2014;46(5):424–426. doi: 10.1038/ng.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Jima D, Moffitt AB, et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood. 2014;123(19):2988–2996. doi: 10.1182/blood-2013-07-517177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwartz JR, Ma J, Kamens J, et al. The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms. Nat Commun. 2021;12(1):1–11. doi: 10.1038/s41467-021-21255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertrums EJ, Huber AKR, de Kanter JK, et al. Elevated mutational age in blood of children treated for cancer contributes to therapy-related myeloid neoplasms. Cancer Discov. 2022;12(8):1860–1872. doi: 10.1158/2159-8290.CD-22-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voit RA, Tao L, Yu F, et al. A genetic disorder reveals a hematopoietic stem cell regulatory network co-opted in leukemia. Nat Immunol. 2023;24(1):69–83. doi: 10.1038/s41590-022-01370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerstung M, Jolly C, Leshchiner I, et al. The evolutionary history of 2,658 cancers. Nature. 2020;578(7793):122–128. doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fittall MW, Van Loo P. Translating insights into tumor evolution to clinical practice: promises and challenges. Genome Med. 2019;11(1):1–14. doi: 10.1186/s13073-019-0632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maura F, Bolli N, Angelopoulos N, et al. Genomic landscape and chronological reconstruction of driver events in multiple myeloma. Nat Commun. 2019;10(1):1–12. doi: 10.1038/s41467-019-11680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maura F, Ziccheddu B, Xiang JZ, et al. Molecular evolution of classic Hodgkin lymphoma revealed through whole genome sequencing of Hodgkin and Reed Sternberg cells. Blood Cancer Discov. Published online 1 February 2023. [DOI] [PMC free article] [PubMed]
- 62.Shoshani O, Brunner SF, Yaeger R, et al. Chromothripsis drives the evolution of gene amplification in cancer. Nature. 2021;591(7848):137–141. doi: 10.1038/s41586-020-03064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitchell TJ, Turajlic S, Rowan A, et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx renal. Cell. 2018;173(3):611–623.e617. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sperling AS, Guerra VA, Kennedy JA, et al. Lenalidomide promotes the development of TP53-mutated therapy-related myeloid neoplasms. Blood. 2022;140(16):1753–1763. doi: 10.1182/blood.2021014956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsu JI, Dayaram T, Tovy A, et al. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 2018;23(5):700–713.e706. doi: 10.1016/j.stem.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parmar H, Gertz M, Anderson EI, Kumar S, Kourelis TV. Microenvironment immune reconstitution patterns correlate with outcomes after autologous transplant in multiple myeloma. Blood Adv. 2021;5(7):1797–1804. doi: 10.1182/bloodadvances.2020003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dijkgraaf EM, Heusinkveld M, Tummers B, et al. Chemotherapy alters monocyte differentiation to favor generation of cancer-supporting M2 macrophages in the tumor microenvironment. Cancer Res. 2013;73(8):2480–2492. doi: 10.1158/0008-5472.CAN-12-3542. [DOI] [PubMed] [Google Scholar]
- 68.Biasco L, Pellin D, Scala S, et al. In vivo tracking of human hematopoiesis reveals patterns of clonal dynamics during early and steady-state reconstitution phases. Cell Stem Cell. 2016;19(1):107–119. doi: 10.1016/j.stem.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514(7522):322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327):842–847. doi: 10.1126/science.aag1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.