Abstract
Interferons (IFNs) encode a family of secreted proteins involved in a number of regulatory functions such as control of cell proliferation, differentiation and regulation of the immune system. Their diverse biological actions are thought to be mediated by the products of specific but usually overlapping sets of cellular genes induced in the target cells. We have recently isolated a human cDNA encoding a new nuclear bodies-associated protein (PML-NBs), which we have termed Isg20. In this report, we describe the cloning and functional characterization of the Isg20 promoter region and the identification of sequence elements and trans-acting factors implicated in its regulation. In the absence of any recognizable TATA or CAAT elements, Isg20 promoter basal activity is dependent upon the positive transcription factors Sp-1 and USF-1. Interestingly, we demonstrate that a unique interferon stimulated response element (ISRE) mediates both IFN type I and type II Isg20 induction in the absence of functional γ-activated sequence. These inductions are strictly dependent upon of the IFN regulatory factor 1 (IRF-1). In addition, we show that the ISRE is also implicated in the constitutive transcriptional activity of Isg20 gene.
INTRODUCTION
The interferons (IFNs) are a family of secreted multifunctional proteins that exert a broad spectrum of biological activities. First characterized for their potent antiviral properties, it is now well established that they are involved in a number of regulatory functions such as control of cell proliferation, differentiation and regulation of the immune system (1). Binding of both IFN type I (IFNα/β), and IFN type II (IFNγ) to specific cell surface receptors (2) leads to activation of receptor-associated cytoplasmic Janus tyrosine kinases (JAKs) (3,4). Phosphorylation of the IFN receptor subunits by the activated JAK provides recruitment, phosphorylation and dimerization of signaling molecules that are members of the signal transducer and activator of transcription (Stats) family (recently reviewed in 3–5). The heterodimeric Stat1/Stat2 complex assembled in response to IFN type I migrates to the nuclei and associates with the p48 DNA binding protein member of the IFN regulatory factor (IRF) family to form the specific transcription complex IFN-stimulated gene factor 3 (ISGF3) (6–8). On the other hand, IFN type II leads to the formation of a homodimer Stat1/Stat1 termed the IFNγ-activation factor (GAF) (9–11). These transcription factors act at distinct cis-acting DNA elements termed the IFN-stimulatory element (ISRE) for ISGF3 and the IFNγ-activation site (GAS) for GAF and located in the promoter region of IFN-inducible genes (ISGs) (3,12). It is worth noting that these mechanisms are not restrictive since crosstalk between the signaling pathways activated by both types of IFN were described. For example, 9–27 gene-induction by IFN type II involves binding of the Stat1/Stat1 homodimer to an ISRE sequence in a p48-independent manner (13). The Stat1/Stat1 homodimer can also be induced by IFN type I (14). In addition, the phosphorylation by IFN of Stat3 (15), Stat5 (16,17) and Stat6 (16) was reported; however, their implication in IFN-signaling is not clearly understood.
The Jak/Stat signaling pathways do not require de novo protein synthesis to induce transcription of specific genes. However, the absence of induction of some ISGs after IFN treatment is observed in the presence of protein synthesis inhibitor suggesting that additional factors are involved in the IFN modulation of these genes. Some of these factors, the IRFs, act directly by binding to the ISRE sequence also designated IRF-E or IFN consensus sequence (ICS) (reviewed in 18). Other factors such CBP/p300 (19–21), USF-1 (22) or NFκB (23) could act as a co-activator. The IRF family includes seven cellular and two viral members that exert distinct biological effects (18,24). Among these factors, IRF-1 is the best characterized. IRF-1 acts as a transcriptional activator and is clearly involved in the control of cell growth and apoptosis (18,24). It was proposed as a tumor suppressor (18,24). IRF-1 is weakly expressed in most of the cells but its expression is strongly induced by virus infection (25), double-stranded RNA (26), both type of IFNs (25,27) and other cytokines such as IL-1, IL-6, tumor necrosis factor (TNF) and leukemia inhibitory factor (LIF) (reviewed in 18). Binding of IRF-1 on a sequence similar to the binding sequence of ISGF3 suggests that IRF-1 and ISGF3 might regulate an overlapping set of genes, including IFN type I and ISGs (18,24). Whether the induction of IRF-1 can alone promote efficient gene induction is not clear. Some evidences suggest that IRF-1 activity might be regulated in part by post-translational modification (28,29) and subsequent interaction with other members of the IRF family such as ICSBP (30). In particular, tyrosine phosphorylation of IRF-1 is observed in response to IFN type II treatment suggesting that like the Stats, IRF-1 activity is also modulated by tyrosine phosphorylation (29).
The biological responses of IFNs are mediated by more than 100 proteins encoded by ISGs. We have recently isolated a human cDNA encoding a new PML-nuclear bodies (PML-NBs)-associated protein which we have termed Isg20, for IFN stimulated gene product of 20 kDa (31,32). Isg20 is clearly up-regulated by both types of IFNs at the transcriptional level (31). However, the events underlying this molecular mechanism are not understood.
Here we describe the cloning and functional characterization of the Isg20 promoter region and the identification of sequence elements and trans-acting factors implicated in constitutive and IFN-induced Isg20 expression in Daudi cells. The Isg20 promoter region does not contain any recognizable TATA or CAAT elements but possesses GC-rich sequences involved in the transcription initiation of TATA-less promoters (33–35). We observed that Isg20 promoter is constitutively expressed in uninduced Daudi cells. This level of transcription is controlled by the positive factors Sp-1 and USF-1. We demonstrate that IFN type I and type II induction of Isg20, is strictly dependent of IRF-1 binding on an ISRE, in the absence of a functional GAS site. In addition, we show that the ISRE is also implicated in the transcription initiation of the Isg20 gene. Regarding all these results, Isg20 represents a unique example among the IFN-regulated genes.
MATERIALS AND METHODS
Cell cultures and antibodies
Human lymphoblastoid Daudi cells were grown in suspension in RPMI 1640 medium, supplemented with 10% (v/v) fetal calf serum (FCS). HeLa cells were grown in monolayer cultures in Dulbecco’s medium containing 10% (v/v) FCS. For IFN induction, exponentially growing cells were exposed for various times to 500 international units/ml of human IFN type I (Hu-IFNα2) obtained from Dr B. Klein (U475 INSERM, Montpellier, France) or 500 U/ml of IFN type II (a gift from Roussell-Uclaf, Romainville, France). Polyclonal rabbit antisera against Stat1α p91 (sc-345), Stat2, Stat3, IRF-1 (sc-497), USF-1 (sc-229) and monoclonal mouse antibody against c-Myc (sc-42) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Isolation of genomic Isg20 DNA
A genomic library in λ vector 2001 obtained from Dr P. Dariavach (IGMM, Montpellier, France) was screened with a full-length 32P-labeled Isg20 cDNA probe. Positive plaques were further screened with the same probe two times. The DNA of one positive clone was analyzed by Southern blot, digested by SmaI and subcloned into pUC18 vector. DNA insert was sequenced using T7 sequencing™ kit (Pharmacia Biotech, Orsay, France) and [35S]ATP. Search for sequence homologies in the EMBL and GenBank databases, as well as sequence analyses were performed by using the BISANCE facilities (36).
RNA purification and primer extension analysis
For RNA purification, the cells were pelleted, washed in phosphate-buffered saline (PBS) and total mRNAs were isolated by the guanidine thyocianate method, as previously described (37). For primer extension analysis, a synthetic oligonucleotide complementary the initiation start site of Isg20 mRNA (TCGCTCCCAGCCAT) was 5′-labeled with [γ-32P]ATP by using T4 polynucleotide kinase. Hybridization to RNAs was carried out at 65°C for 1 h in 10 µl of 40 mM Tris–HCl (pH 7.5), 60 mM KCl, 6 mM MgCl2. Then, 80 µl of a solution containing 50 mM Tris–HCl (pH 8.3), 6 mM MgCl2, 40 mM KCl, 0.5 mM of each the four deoxynucleotides triphosphates, 30 U of RNasin (Promega, France) and 2.5 U of avian myeloblastosis virus reverse transcriptase was added. Incubation was performed at 46°C for 30 min. Elongated products were phenol extracted, ethanol precipitated, and fractionated through a 6% polyacrylamide gel containing 8 M urea, before autoradiography.
Plasmid constructions
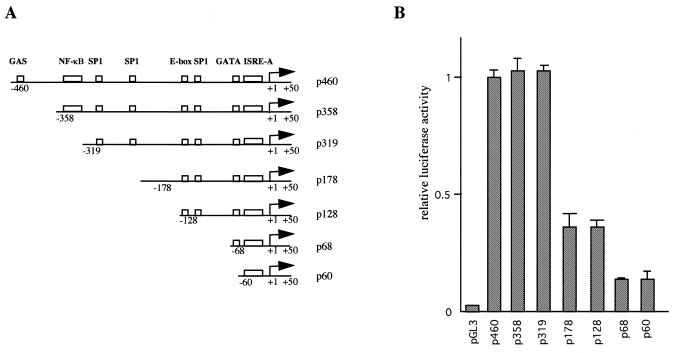
The 5′-derivative constructions of Isg20 promoter were realized by PCR amplification using various 5′-end primers located (–460,–443), (–358,–341), (–319,–302), (–178,–161), (–128,–111), (–68,–51), (–60,–43) and a 3′-end primer located (+32,+49) relative to the transcription initiation start site. The eight PCR products were verified by sequencing and cloned upstream of the luciferase reporter gene in the pGL3basic vector (Promega). These different constructs were termed p460, p358, p319, p178, p128, p68 and p60 respectively, and used for transcriptional assays.
Site-directed mutagenesis
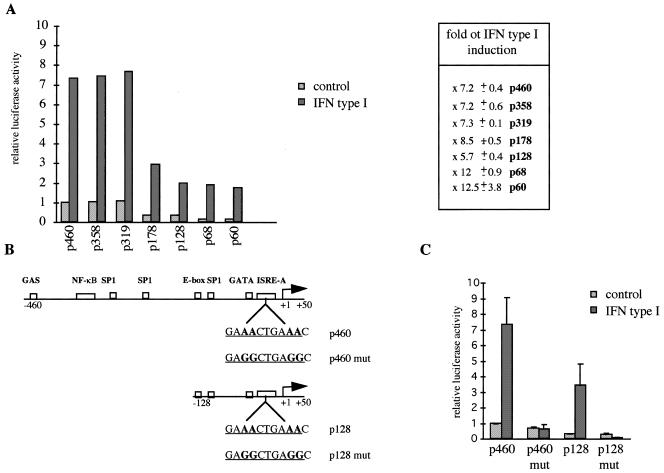
The mutagenesis was performed using the Quick Change™ Site-directed Mutagenesis Kit (Stratagene, Tebu, France). Mutagenesis of p460 and p128 constructs were realized by substitution of two conserved A residues of the ISRE (wild-type ISRE GAAACTGAAACA, mutated ISRE GAGGCTGAGGCA), previously shown to abolish IFN-responsiveness. The resulting constructs were termed p460mut and p128mut respectively.
Transient transfection experiments
Daudi cells (5 × 106) were washed twice in RPMI medium and resuspended in 800 µl RPMI serum-free at 4°C with 5 µg of ISG20 promoter constructs. The cells were electroporated at 240 V. The cells were then resuspended in 10 ml RPMI medium with 10% FCS. After 6 h of incubation, the cells were treated or not with IFN type I or type II over 16 h. The luciferase activity was measured with the Dual Luciferase™ Assay System (Promega) in a TD20 Turner Luminometer counter. HeLa cells were inoculated in 60-mm culture dishes, transfected by calcium phosphate precipitation and analyzed as described for Daudi cells.
Nuclear extract preparations
Daudi cells (2 × 106) were treated with 500 U/ml of IFN type I for various times. After washing in PBS, cells were resuspended in a buffer containing 10 mM HEPES (pH 7.8), 10 mM KCl, 2 mM MgCl2, 0,1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, a protease inhibitor cocktail from Boehringer Mannheim (Roche, Le Ulis, France), and incubated for 15 min at 4°C. Fifty microliters of 10% NP-40 were then added, and cells were vortexed and centrifuged. The pellet was resuspended in a buffer containing 50 mM HEPES (pH 7.8), 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 10% glycerol and a protease inhibitor cocktail. After an additional incubation for 20 min on ice, the suspension was cleared by centrifugation in a microfuge for 5 min and the supernatant stored at –80°C. For HeLa nuclear extract preparations, the cells were lysed in a buffer containing 20 mM HEPES (pH 7.8), 0.42 M NaCl, 0.2 mM EDTA, 1.5 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 25% glycerol and a protease inhibitor cocktail.
Oligonucleotides and probes
The oligonucleotides used in EMSA experiments were: ISRE element, 5′-TTGATAACAAACTAGAAACTGAAACAGGGTCG-3′, ISRE-MUT 1 element: 5′-TTGATAACAGCCTAGAAACTGAAACAGGGTCG-3′, E-box element: 5′-GCCCAGCCCACGTGCCCGGCCTTC-3′. Complementary oligonucleotides were annealing at 65°C and end-labeled with polynucleotide kinase (Life Technologies, France) and [γ-32P]ATP. Approximately 50 000 c.p.m. of labeled probe were used in each mobility shift assay.
Electophoretic mobility shift assays (EMSA)
Nuclear extracts were incubated for 5 min with 1 µg of poly(dI-dC) and 200 ng of unrelated single-strand oligonucleotide in 20 µl of 12 mM HEPES (pH 7.8), 5 mM MgCl2, 0.24 mM EDTA, 8% glycerol, 0.3 mM DTT, before addition of double-stranded 32P-labeled probe. The mixture was incubated for 30 min at 4°C and DNA–protein complexes were analyzed on 5% polyacrylamide gel. For competition experiments, unlabeled double-strand oligonucleotides were used in a 100-fold molar excess. For supershift experiments, nuclear extracts were pre-incubated for 20 min with specific antibodies before addition to the binding reactions.
Database access number
The nucleotide sequence reported in this paper has been submitted to the EMBL data bank with the accession number AJ249162
RESULTS
Isolation of the Isg20 5′-upstream region and mapping of the transcription start site
To identify a genomic DNA fragment containing Isg20 promoter region, a human genomic library derived from B-cells was screened with a 32P-labeled Isg20 cDNA probe. A positive clone was selected and amplified. To enhance the probability of obtaining the Isg20 regulatory region, the DNA of the clone was digested by the SmaI restriction enzyme, which is a site situated near the 5′-end of the cDNA (31), prior to subcloning in pUC18 vector. Two different clones with ~2 kb inserts were obtained and sequenced. One of them contained the first 188 bp of Isg20 cDNA with an additional 1762 bp 5′-region. Computer search for homologies in the GenBank database revealed the presence of several EST sequences identical to the Isg20 cDNA with an 80 bp longer 5′-end corresponding to the 2 kb insert sequence situated between –266 to –346 relative to the first nucleotide of the Isg20 cDNA described previously (31). These data suggest the presence of a small supplementary exon separated from the Isg20 coding region by a 265 bp intron (Fig. 1A). Therefore, the precise location of the transcription initiation site of the putative Isg20 promoter was analyzed. To this aim, the 5′-end of Isg20 mRNA from IFN-treated Daudi cells was mapped by primer extension analysis using a synthetic oligonucleotide complementary to the region spanning its translation initiation start site. A major product of 110 nt corresponding to the expected size of Isg20 mRNA plus the additional 80 nt was obtained (Fig. 1B). Interestingly, several minor initiation sites which spread over a 40 bp region were detected (Fig. 1B). This observation will be discussed in the next chapter on the basis of 5′-nucleotide sequence analysis. These data plus the comparison with some Isg20 EST present in the databases led us to localize the transcription initiation site at the nucleotide indicated +1 in Figure 1.
Figure 1.
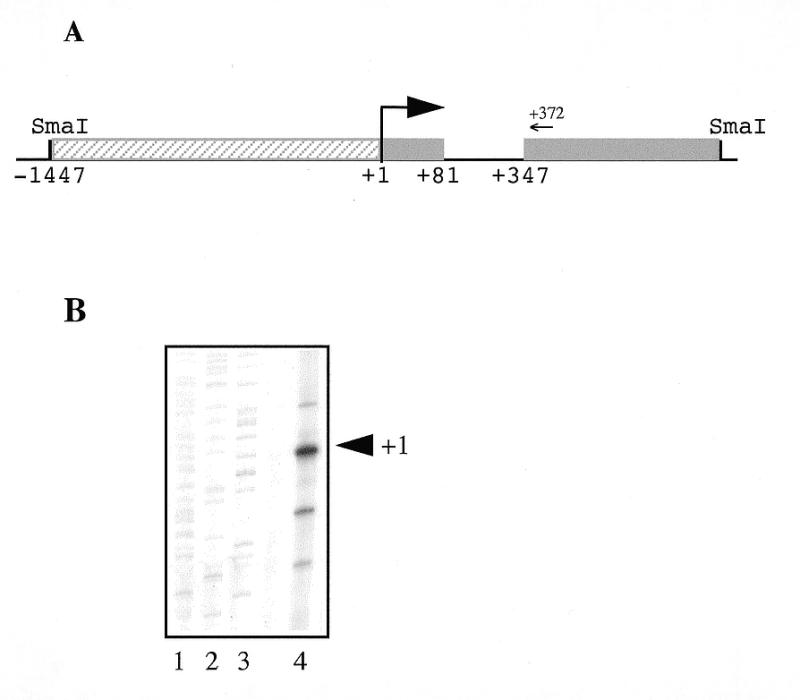
Structure of the human Isg20 promoter. (A) Schematic representation of the Isg20 promoter. The SmaI–SmaI 2 kb fragment containing the Isg20 promoter region is cloned in pUC18 cloning vector. The two first exons of Isg20 gene are represented as gray boxes and the 5′-flanking promoter region is represented as a hatched box. The arrow marks the major transcription initiation start site. The positions of the nucleotides with respect to this start site are numbered beneath the diagram. The position of the primer used for the primer extension analysis is indicated by an arrow. (B) Mapping of the transcription initiation start site by primer extension analysis. An oligonucleotide complementary to the translation initiation start site of Isg20 mRNA was 5′-end labeled and hybridized to 5 µg of total RNA extracted from IFN type I-treated Daudi cells. The cDNA was extended using avian myeloblastosis virus reverse transcriptase as described in Materials and Methods and then analyzed on a 6% polyacrylamide sequencing gel. Lanes 1–3 show a sequencing ladder used as molecular weight marker. The reverse-transcribed products are shown in lane 4. The major transcription initiation start site is indicated by +1.
Nucleotide sequence analysis of the Isg20 5′-flanking region
Analysis of Isg20 5′-proximal region (Fig. 2) reveals the absence of canonical TATA and CAAT boxes. However, like many TATA-less promoters, it contains GC-rich sequences with three binding sites for members of mammalian Sp/XKLF transcription factors family (38,39) which have been shown to make up the absence of TATA-box for transcription initiation (33–35). Consistent with a GC-rich sequence directed transcription (35), Isg20 transcription initiates at multiple clustered start sites as observed by primer extension analysis (Fig. 1B).
Figure 2.
Nucleotide sequence of Isg20 promoter. The nucleotide sequence of Isg20 extending from nucleotide –460 to +370 relative to the transcription start site. The putative cis-acting elements GAS, NFκB, GC, E-box, GATA and ISRE are boxed. The first exon and the 5′-end of the second exon of Isg20 are underlined.
In addition, the 5′-proximal region, extending from +1 to –460, shares the consensus binding sites for transcription factors, NFκB, GAS, E-box, GATA and ISRE (Fig. 2). The GAS motif (–431 5′-TTCCAATAA-3′ –422) and the ISRE motif (–51 5′-GAAACTGAAAC-3′ –42) exactly match the consensus sequence. So, the 5′-flanking region of Isg20 seems to contain all the necessary elements for constitutive and IFN-induced transcription.
Functional characterization of the Isg20 promoter
To identify the cis-acting elements involved in the Isg20 promoter activity, as well as in IFN-induction, a series of 5′-end deletion mutants was obtained by successive deletions of the consensus binding site described above. The different fragments of Isg20 promoter region extending from +80 to –50 up to –460 relative to the start site were cloned upstream the luciferase reporter gene in the pGL3-basic vector (Fig. 3A). Daudi cells were transfected by the electroporation method with these different constructs and then analyzed for luciferase activity as described in Materials and Methods. As shown Figure 3B, cells transfected with the p460 construct retaining all the described consensus binding site exhibits 40–50-fold more luciferase activity than cells transfected with pGL3 control vector. These data indicate that we have cloned the functional Isg20 promoter region. The p358 and p319 constructs corresponding to the deletion of GAS and NFκB sites, respectively, have similar luciferase activity to p460 construct indicating that neither GAS nor NFκB is implicated in the basal activity of Isg20 promoter. The constructs p178 and p128 showed a reduced luciferase activity indicating that two GC-sites located in –210 and –290 could be important for supporting high levels of Isg20 transcription in Daudi cells. In addition, the two smallest constructs p68 and p60 showed an even more reduced luciferase activity suggesting that at least the GC-site located in –80 or the E-Box located in –90 could be important for Isg20 basal activity. Interestingly, the p60 Isg20 construct retaining only the ISRE element still has a 6-fold increase in luciferase activity relative to pGL3b control vector. These data demonstrate that the GATA-box is not implicated in Isg20 promoter activity and suggest that the ISRE or downstream unidentified element can control the transcription of Isg20.
Figure 3.
Cis-acting elements implicated in Isg20 promoter activity. (A) Schematic representation of the 5′-end deletion mutants obtained by successive deletions of the consensus binding site described in Figure 2. The different constructs were cloned into the pGL3 basic vector and named p60, p68, p128, p178, p319, p358 and p460 according the length of the remaining 5′-flanking region. (B) Promoter activity of the different Isg20 promoter constructs. Daudi cells were transfected with the constructs described in Materials and Methods and analyzed after 24 h for luciferase activity. The relative luciferase activities were calculated by dividing the values measured after transfection of the different promoter constructs by the values measured after transfection of the p460 construct used as a reference. Each experiment was realized in duplicate and the standard deviations correspond to six distinct experiments.
To test the implication of the cis-acting elements in the induction of Isg20 by IFN type I, Daudi cells were transfected with the same constructs and then treated or not with IFN type I over 16 h. As shown in Figure 4A, IFN-treated Daudi cells, transfected with either constructions present a 7–12-fold increase in the level of luciferase activity as compared to untreated cells. Even p60 construct showed a greater increase in luciferase activity, similar to those observed with longer constructs when cells were treated with IFN. This result indicates that this ISRE alone is sufficient for IFN type I-induced Isg20 transcription. To confirm this result, site-directed mutagenesis into the conserved A residues of the ISRE, previously shown to abolish IFN-responsiveness, has been realized on p460 and p128 constructs and the resulting constructs were termed p460mut and p128mut respectively (Fig. 4B). As expected, the two mutated constructs have completely lost the IFN-inducibility demonstrating that only the ISRE is implicated in the induction of Isg20 by IFN type I (Fig. 4C). Interestingly, mutation in the ISRE decreases the basal activity of Isg20 promoter to about the level of pGL3b for p128mut (Fig. 4C). These data strongly suggest that in the absence of canonical initiation element in Isg20 promoter, the ISRE can direct Isg20 transcription initiation and inducibility by IFN.
Figure 4.
Cis-acting elements implicated in the modulation of Isg20 promoter by IFN type I. (A) Daudi cells transfected with p60, p68, p128, p178, p319, p358 or p460 constructs were treated or not with 500 U/ml of IFN type I for 16 h and then analyzed for luciferase activity. The promoter activity of the different Isg20 promoter constructs is presented as described in Figure 3B. Each experiment was realized in duplicate and the standard deviations correspond to at least six distinct experiments. The fold of IFN-induction for each construct is indicated at the right of the histogram. (B) Representation of p460mut and p128mut corresponding to the p460 and p128 constructs with mutation of the conserved A residues of the ISRE. (C) Contribution of the ISRE in the modulation of Isg20 with IFN type I. Daudi cells were transfected with the p460, p128, p460mut or p128mut constructs and analyzed as described in Figure 4A. Each experiment was realized in duplicate and the standard deviations correspond to four distinct experiments.
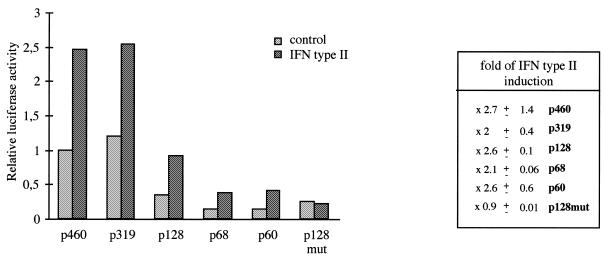
We then focused our interest on the modulation of Isg20 promoter by IFN type II. Since Daudi cells failed to respond to IFN type II, because of the lack of functional receptors, transfection experiments were performed in HeLa cells treated or not with 500 U/ml of IFN type II and analyzed for luciferase activity, as described above. According to our previously published data (31), the induction of Isg20 promoter activity was lower with IFN type II (Fig. 5). Surprisingly, p460 and p128 constructs exhibited the same 2-fold induction of luciferase activity after IFN type II treatment (Fig. 5). These data strongly suggest that the GAS element present in Isg20 promoter is not involved in IFN type II up-regulation and that the ISRE-binding site alone mediates both IFN type I and IFN type II Isg20 induction. This hypothesis was confirmed by the absence of IFN type II induction of the p128mut construct (Fig. 5).
Figure 5.
Cis-acting elements implicated in the modulation of Isg20 promoter by IFN type II. HeLa cells transfected with p60, p128, p338, p460 or p128mut constructs were treated or not with 500 U/ml IFN type II for 24 h and then analyzed for luciferase activity. The promoter activity of each Isg20 promoter construct is presented as described in Figure 3B. The fold of IFN-induction for each construct is indicated to the right of the histogram. The relative luciferase activities were calculated by dividing the values measured after transfection of the different promoter constructs by the values measured after transfection of the p460 construct used as reference.
Characterization of the proteins binding to the E-box
The presence of the E-box (CACGTG) element on the Isg20 promoter activity suggests that members of the basic-helix–loop–helix/leucine zipper (bHLH-zip) class of transcription factors including TFE3 (40), TFEB (41), USF-1 (42), USF-2 (43) and the heterodimers Myc-Max (44) and Mad-Max (45) might be involved in the regulation of Isg20 expression. Therefore, we tested whether a transcription factor binds to this E-box, using gel mobility shift assays. We used the oligonucleotide containing the Isg20 E-box and nuclear protein extracts prepared from untreated or IFN-treated Daudi cells. Three DNA–protein complexes are present with both nuclear extracts (Fig. 6). When antibodies against USF-1 were added to the binding reactions, the major DNA–protein complex completely disappeared and supershifted ones appeared (Fig. 6). On the other hand, antibodies against Myc had no effect. This result demonstrates that USF-1 binds to the E-box-Isg20 and is probably implicated in the constitutive expression of Isg20, in Daudi cells.
Figure 6.
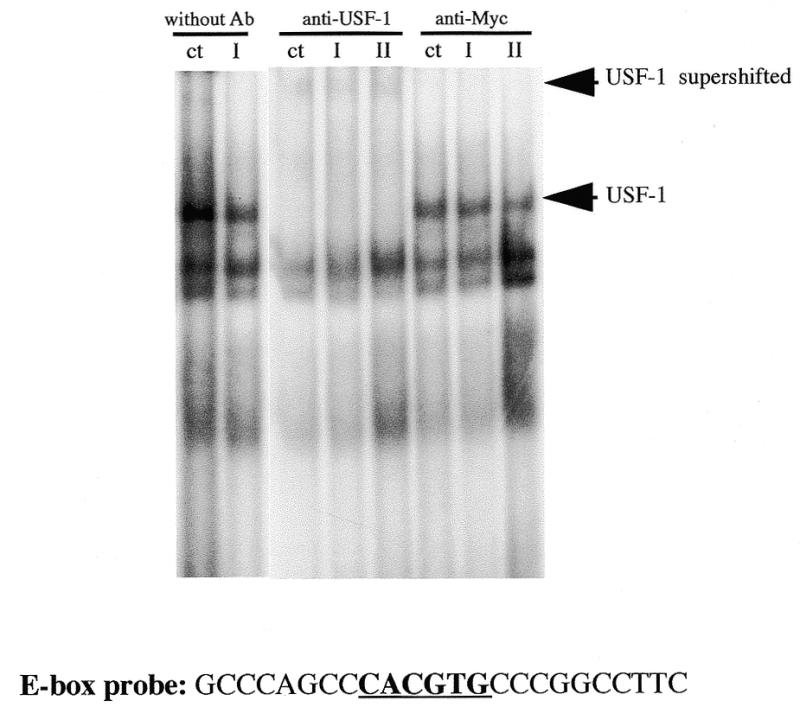
The Isg20 E-box element binds USF-1 transcription factor. Gel mobility shift assays were performed with an oligonucleotide probe containing the Isg20 E-Box element and nuclear extracts from untreated (ct) or treated Daudi cells for 2 h with either IFN type I (I) or IFN type II (II). The antibodies against USF-1 or c-Myc used for the supershift are indicated at the top of the gel. The USF-1 complex and supershifted complex is indicated. The E-box probe used is indicated beneath the gel.
Identification of proteins binding to the Isg20-ISRE
Since ISRE seems to be the only cis-acting element involved in the modulation of Isg20 by IFNs, we have investigated the transcription factors implicated in this regulation. To this aim, nuclear extracts of untreated or treated Daudi cells for various times with 500 U/ml of IFN type I, were tested in an EMSA with a 32 bp probe containing the tamdemly repeated Isg20-ISRE binding site GAAAC (Fig. 7A). Two constitutive complexes were found in untreated cells. An additional complex appears 1 h after IFN treatment and remains stable for at least 8 h (Fig. 7A). As a control of specificity, we showed that competition with unlabeled Isg20 ISRE strongly inhibited formation of the IFN-induced DNA–protein complex (data not shown). When competition assays were performed with an unlabeled Isg20 ISRE probe containing an AA→GG residue substitution in the ISRE-binding site (GAAACTGAAAC→ GAGGCTGAGGC), the IFN-induced complex formation was not altered, indicating that the ISRE binding site is important for the complex formation (data not shown). To determine the nature of the transcription factor present in the IFN-induced complex, nuclear extracts from 2 h IFN-type I-treated Daudi cells were incubated, prior EMSA analysis with specific antibodies against different known transcription factors whose activity is regulated by IFN type I and that bind to ISRE sites. As shown Figure 7B, the IFN-induced complex is completely supershifted with IRF-1 antibodies while antibodies against Stat1, Stat2 and Stat3 had no effect. According to previously published data, a 10-fold lesser supershift is observed with IRF-2 antibodies. Taken together, these experiments reveal that Isg20 induction by IFN type I is solely due to IRF-1.
Figure 7.
IFN type I induces binding of IRF-1 to the Isg20 ISRE. Gel mobility shift assays were performed with an oligonucleotide containing the tamdem repeat Isg20 ISRE and nuclear extracts from Daudi cells treated for various times with 500 U/ml IFN type I. (A) Kinetic of the IFN type I-induced complex formation to Isg20 ISRE. The 32 bp probe used is indicated beneath the gel. The time of IFN type I induction is indicated at the top of the gel. The arrow to the right indicates the IFN-induced complex. (B) Antibody supershifts for the 2 h time point of IFN-treated Daudi cells. The antibodies against Stat1, Stat2, Stat3, IRF-1 or IRF-2 used for the supershift are indicated at the top of the gel. Ct indicates mobility shift without any antibodies. The IRF-1 complex is indicated. An overexposed autoradiography of the top-right part of the gel reveals a 10-fold lesser supershifted complex with IRF-2 antibodies.
EMSA studies were then conducted with nuclear extracts prepared from HeLa cells treated or not with 500 U/ml of IFN type II. As shown in Figure 8A, IFN type II induces the formation of a specific ISRE binding complex with a delayed time as compared to that induced by IFN type I. EMSA experiments performed in the presence of specific antibodies described above led us to demonstrate that IFN type II transcriptional induction is mediated by IRF-1 transcription factor as well (Fig. 8B). An additional EMSA experiment realized with the Isg20 GAS motif did not reveal any specific IFN-induced complex formation (data not shown).
Figure 8.
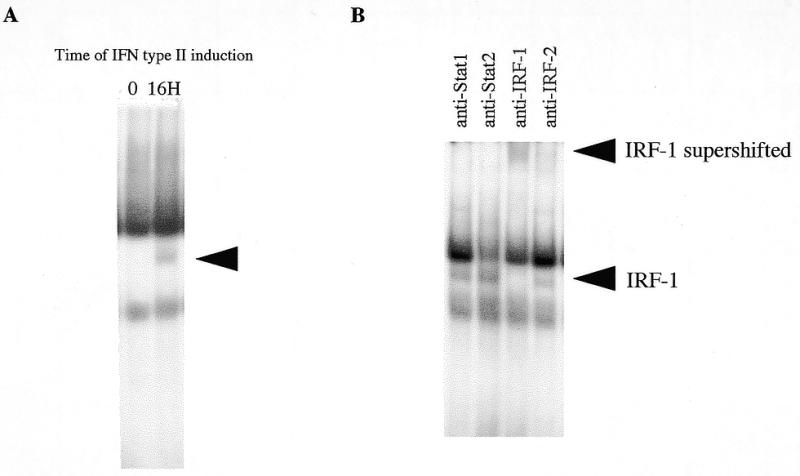
IFN type II induces binding of IRF-1 to the Isg20 ISRE. Gel mobility shift assays were performed with the oligonucleotide probe shown in Figure 7A and nuclear extracts from untreated or treated HeLa cells for 16 h with 500 U/ml IFN type II. (A) IFN type II-induced complex formation to Isg20 ISRE. The time of IFN type II induction is indicated at the top of the gel. The arrow to the right indicates the IFN-induced complex. (B) Antibody supershifts for the 16 h time point of IFN type II-treated HeLa cells. The antibodies against Stat1, Stat2, IRF-1 or IRF-2 used for the supershift are indicated at the top of the gel. The IRF-1 complex and supershifted complex are indicated.
DISCUSSION
We have recently isolated a new human cDNA strongly induced at the transcriptional level by IFN type I and to a lesser extent by IFN type II (31). This gene, which we have termed Isg20, is localized on 15q26 and encodes a PML-NBs-associated protein (31,32). In this report, we have described the isolation and functional characterization of the promoter region of the Isg20 gene, and we have examined the transcription factors involved in its regulation by the IFNs.
A 460 bp 5′-flanking region of the human Isg20 gene was shown to be sufficient to ensure high levels of expression and IFN-inducibility. Isg20 promoter region does not contain classical initiation elements such as TATA-box, CAAT-box or consensus initiation element (Inr). Although this structure has been observed primarily in housekeeping genes, some IFN-modulated genes had already been shown to lack these elements (46–49). Isg20 possesses several GC-rich motifs and an E-box element that can functionally substitute a TATA-box (33–35). In fact, transcriptional activation of TATA-less promoters may be initiated by the recruitment of the TATA-box binding protein (TBP) by a GC-rich motif, through an SP1–TBP interaction (50,51). More recently, CIF150, the human homolog of Drosophila TATA box binding protein-associated factors [dTAF(II)150] has been shown to be capable of mediating TFIID-dependent Inr activity (52). In the same way, a cooperative interaction between the bHLH-zip USF-1 transcription factor that binds the E-box element and the initiator-binding transcription initiation factor TFII-I has been shown to mediate TATA-less promoter initiation (53). TFII-I and USF-1 interact at both Inr and E-box sites (53). Therefore, we have analyzed the implication of GC-rich sequences and the E-box site in the control of Isg20 transcription. Using different Isg20 promoter–luciferase constructs, we showed that the deletion of GC-rich sites, as well as the E-box-binding site, dramatically decreased luciferase reporter gene expression in transient transfection experiments, without affecting IFN inducibility. Using EMSA experiments, we demonstrated that among the members bHLH-zip class of transcription factors only USF-1 binds on the Isg20-E-box. The USF-1 protein shares with the Myc oncoprotein, another member of the bHLH-zip family, both similar polypeptide structure and DNA-binding specificity. USF-1 and Myc play antagonistic roles in the control of mammalian cell proliferation. Although ubiquitously expressed, USF-1 has been involved in transcription of genes with tissue specificity, indicating that USF-1 can work with a specific coactivator (42,54). Recently, Stat1 and USF-1 have been shown to control the induction of the MHC class II transactivator CIITA by IFN type II, in a cooperative manner (22). Since the same E-box binding complex was obtained with nuclear extracts from IFN-treated or untreated Daudi cells, we conclude that USF-1 is not required for Isg20 modulation by IFNs.
The 5′-flanking region of Isg20 shares the consensus binding sites, for transcription factors, NFκB, GAS, ISRE, E-box and GATA. Successive 5′-end deletion in Isg20 promoter of these binding sites, led us to define a 60 bp region necessary and sufficient to promote maximal induction of transcription by both IFN type I and type II in transient transfection experiments in Daudi and HeLa cells. This region contains only the ISRE GAAACTGGAAAC motif. It is interesting to note that the GAS element located at –420 does not seem to be implicated in the modulation by IFN type II. These data were confirmed in EMSA experiments where no modulations of complex formation were detected with nuclear extracts from HeLa cells treated or untreated with IFN type II. Among the transcription factors that classically bind to ISRE, such as ISGF3 [including Stat1, Stat2, ISGF3γ (p48)], Stat3 and members of the IRF family, we showed that only IRF-1 (and at lesser extent IRF-2) transcription factor binds to the Isg20-ISRE. This complex also appears after IFN type II and remains stable for at least 8 h. As expected, mutations in the ISRE resulted in the loss of IFN type I and type II inducibility of the reporter gene. Surprisingly, these mutations strongly decrease the activity of Isg20 promoter, suggesting that the ISRE element might be involved in an IRF-1-mediated constitutive expression of Isg20-TATA-less promoter. Similar data were observed but not discussed for the regulation of the biliary glycoprotein (BGP/C-CAM-1) (55).
IFN-inducible genes could be characterized according to their dependence to IRF-1 and/or p48 (in the context of ISGF3) for induction by IFN type I and type II (56). Some of them are regulated by binding of ISGF3 or ISGF3 plus IRF-1 to ISRE and GAS sites (56). Isg20 appears to be an exceptional case since its regulation by both IFN type I and type II, involves only the binding of IRF-1 to a unique ISRE motif. The gene encoding the inducible nitric-oxide synthase (iNOS) and the mouse Gbp gene have also been shown to be regulated essentially by IRF-1. However, this was only shown in response to IFN type II or to IFN type II plus co-inducers like LPS or TNF-α (57–59). In addition, in view of the role of IRF-1 in the control of cell growth and apoptosis, it would be interesting to further study the physiological significance of the Isg20 regulation by this factor and gain information concerning the possible role of Isg20 in these processes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr P. Dariavach for providing the human genomic library. This work was supported by grants from the Association pour la Recherche contre le Cancer, the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Federation des Centres de Lutte contre le Cancer Comité du Gard, the Ligue Contre le Cancer and the Federation pour la Recherche Medicale.
DDBJ/EMBL/GenBank accession no. AJ249162
REFERENCES
- 1.Sen G.C. and Lengyel,P. (1992) J. Biol. Chem., 267, 5017–5020. [PubMed] [Google Scholar]
- 2.Pestka S. (1997) Semin. Oncol., 24 (Suppl. 9), S9-18–S9-40. [PubMed] [Google Scholar]
- 3.Schindler C. and Darnell,J.E.,Jr (1995) Annu. Rev. Biochem., 64, 621–651. [DOI] [PubMed] [Google Scholar]
- 4.Pellegrini S. and Dusanter-Fourt,I. (1997) Eur. J. Biochem., 248, 615–633. [DOI] [PubMed] [Google Scholar]
- 5.Ihle J.N. (1996) Cell, 84, 331–334. [DOI] [PubMed] [Google Scholar]
- 6.Schindler C., Fu,X.Y., Improta,T., Aebersold,R. and Darnell,J.E.,Jr (1992) Proc. Natl Acad. Sci. USA, 89, 7836–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller M., Laxton,C., Briscoe,J., Schindler,C., Improta,T., Darnell,J.E.,Jr, Stark,G.R. and Kerr,I.M. (1993) EMBO J., 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David M., Romero,G., Zhang,Z.Y., Dixon,J.E. and Larner,A.C. (1993) J. Biol. Chem., 268, 6593–6599. [PubMed] [Google Scholar]
- 9.Decker T., Lew,D.J., Mirkovitch,J. and Darnell,J.E.,Jr (1991) EMBO J., 10, 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuai K., Schindler,C., Prezioso,V.R. and Darnell,J.J. (1992) Science, 258, 1808–1812. [DOI] [PubMed] [Google Scholar]
- 11.Shuai K., Horvath,C.M., Huang,L.H., Qureshi,S.A., Cowburn,D. and Darnell,J.E.,Jr (1994) Cell, 76, 821–828. [DOI] [PubMed] [Google Scholar]
- 12.Darnell J.E. Jr, Kerr,I.M. and Stark,G.R. (1994) Science, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- 13.Bonjardim C.A. (1998) Braz. J. Med. Biol. Res., 31, 1389–1395. [DOI] [PubMed] [Google Scholar]
- 14.Haque S.J. and Williams,B.R. (1994) J. Biol. Chem., 269, 19523–19529. [PubMed] [Google Scholar]
- 15.Yang C.H., Murti,A. and Pfeffer,L.M. (1998) Proc. Natl Acad. Sci. USA, 95, 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasler-Kan E., Pansky,A., Wiederkehr,M., Battegay,M. and Heim,M.H. (1998) Eur. J. Biochem., 254, 514–519. [DOI] [PubMed] [Google Scholar]
- 17.Fish E.N., Uddin,S., Korkmaz,M., Majchrzak,B., Druker,B.J. and Platanias,L.C. (1999) J. Biol. Chem., 274, 571–573. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen H., Hiscott,J. and Pitha,P.M. (1997) Cytokine Growth Factor Rev., 8, 293–312. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J.J., Vinkemeier,U., Gu,W., Chakravarti,D., Horvath,C.M. and Darnell,J.E.,Jr (1996) Proc. Natl Acad. Sci. USA, 93, 15092–15096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horvai A.E., Xu,L., Korzus,E., Brard,G., Kalafus,D., Mullen,T.M., Rose,D.W., Rosenfeld,M.G. and Glass,C.K. (1997) Proc. Natl Acad. Sci. USA, 94, 1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhattacharya S., Eckner,R., Grossman,S., Oldread,E., Arany,Z., D’Andrea,A. and Livingston,D.M. (1996) Nature, 383, 344–347. [DOI] [PubMed] [Google Scholar]
- 22.Muhlethaler-Mottet A., Di Berardino,W., Otten,L.A. and Mach,B. (1998) Immunity, 8, 157–166. [DOI] [PubMed] [Google Scholar]
- 23.Neish A.S., Read,M.A., Thanos,D., Pine,R., Maniatis,T. and Collins,T. (1995) Mol. Cell. Biol., 15, 2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taniguchi T., Harada,H. and Lamphier,M. (1995) J. Cancer Res. Clin. Oncol., 121, 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto M., Fujita,T., Kimura,Y., Maruyama,M., Harada,H., Sudo,Y., Miyata,T. and Taniguchi,T. (1988) Cell, 54, 903–913. [DOI] [PubMed] [Google Scholar]
- 26.Fujita T., Sakakibara,J., Sudo,Y., Miyamoto,M., Kimura,Y. and Taniguchi,T. (1988) EMBO J., 7, 3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita T., Reis,L.F., Watanabe,N., Kimura,Y., Taniguchi,T. and Vilcek,J. (1989) Proc. Natl Acad. Sci. USA, 86, 9936–9940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe N., Sakakibara,J., Hovanessian,A.G., Taniguchi,T. and Fujita,T. (1991) Nucleic Acids Res., 19, 4421–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharf R., Meraro,D., Azriel,A., Thornton,A.M., Ozato,K., Petricoin,E.F., Larner,A.C., Schaper,F., Hauser,H. and Levi,B.Z. (1997) J. Biol. Chem., 272, 9785–9792. [DOI] [PubMed] [Google Scholar]
- 30.Sharf R., Azriel,A., Lejbkowicz,F., Winograd,S.S., Ehrlich,R. and Levi,B.Z. (1995) J. Biol. Chem., 270, 13063–13069. [DOI] [PubMed] [Google Scholar]
- 31.Gongora C., David,G., Pintard,L., Tissot,C., Hua,T.D., Dejean,A. and Mechti,N. (1997) J. Biol. Chem., 272, 19457–19463. [DOI] [PubMed] [Google Scholar]
- 32.Mattei M.G., Tissot,C., Gongora,C. and Mechti,N. (1997) Cytogenet. Cell Genet., 79, 286–287. [DOI] [PubMed] [Google Scholar]
- 33.Tamura T. and Mikoshiba,K. (1991) FEBS Lett., 282, 87–90. [DOI] [PubMed] [Google Scholar]
- 34.Boisclair Y.R., Brown,A.L., Casola,S. and Rechler,M.M. (1993) J. Biol. Chem., 268, 24892–24901. [PubMed] [Google Scholar]
- 35.Lu J., Lee,W., Jiang,C. and Keller,E.B. (1994) J. Biol. Chem., 269, 5391–5402. [PubMed] [Google Scholar]
- 36.Densen P., Fondrat,C.,Valencien,C. and Mugnier,C. (1980) Comput. Appl. Biosci., 6, 355–356. [DOI] [PubMed] [Google Scholar]
- 37.Mechti N., Piechaczyck,M., Blanchard,J.M., Jeanteur,P. and Lebleu,B. (1991) Mol. Cell. Biol., 11, 2832–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philipsen S. and Suske,G. (1999) Nucleic Acids Res., 27, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suske G. (1999) Gene, 238, 291–300. [DOI] [PubMed] [Google Scholar]
- 40.Beckmann H., Su,L.K. and Kadesch,T. (1990) Genes Dev., 4, 167–179. [DOI] [PubMed] [Google Scholar]
- 41.Carr C.S. and Sharp,P.A. (1990) Mol. Cell. Biol., 10, 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregor P.D., Sawadogo,M. and Roeder,R.G. (1990) Genes Dev., 4, 1730–1740. [DOI] [PubMed] [Google Scholar]
- 43.Sirito M., Walker,S., Lin,Q., Kozlowski,M.T., Klein,W.H. and Sawadogo,M. (1992) Gene Expr., 2, 231–240. [PMC free article] [PubMed] [Google Scholar]
- 44.Blackwood E.M., Kretzner,L. and Eisenman,R.N. (1992) Curr. Opin. Genet. Dev., 2, 227–235. [DOI] [PubMed] [Google Scholar]
- 45.Ayer D.E., Kretzner,L. and Eisenman,R.N. (1993) Cell, 72, 211–222. [DOI] [PubMed] [Google Scholar]
- 46.Zahedi K., Prada,A.E. and Davis,A.E. (1994) J. Biol. Chem., 269, 9669–9674. [PubMed] [Google Scholar]
- 47.Kuhen K.L., Vessey,J.W. and Samuel,C.E. (1998) J. Virol., 72, 9934–9939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grotzinger T., Jensen,K. and Will,H. (1996) J. Biol. Chem., 271, 25253–25260. [DOI] [PubMed] [Google Scholar]
- 49.George C.X. and Samuel,C.E. (1999) Gene, 229, 203–213. [DOI] [PubMed] [Google Scholar]
- 50.Smale S.T., Schmidt,M.C., Berk,A.J. and Baltimore,D. (1990) Proc. Natl Acad. Sci. USA, 87, 4509–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Shea-Greenfield A. and Smale,S.T. (1992) J. Biol. Chem., 267, 6450. [PubMed] [Google Scholar]
- 52.Kaufmann J., Ahrens,K., Koop,R., Smale,S.T. and Muller,R. (1998) Mol. Cell. Biol., 18, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy A.L., Meisterernst,M., Pognonec,P. and Roeder,R.G. (1991) Nature, 354, 245–248. [DOI] [PubMed] [Google Scholar]
- 54.Sawadogo M., Van Dyke,M.W., Gregor,P.D. and Roeder,R.G. (1988) J. Biol. Chem., 263, 11985–11993. [PubMed] [Google Scholar]
- 55.Chen C.J., Lin,T.T. and Shively,J.E. (1996) J. Biol. Chem., 271, 28181–28188. [DOI] [PubMed] [Google Scholar]
- 56.Taniguchi T., Lamphier,M.S. and Tanaka,N. (1997) Biochim. Biophys. Acta, 1333, M9–M17. [DOI] [PubMed] [Google Scholar]
- 57.Martin E., Nathan,C. and Xie,Q.W. (1994) J. Exp. Med., 180, 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spink J. and Evans,T. (1997) J. Biol. Chem., 272, 24417–24425. [DOI] [PubMed] [Google Scholar]
- 59.Briken V., Ruffner,H., Schultz,U., Schwarz,A., Reis,L.F., Strehlow,I., Decker,T. and Staeheli,P. (1995) Mol. Cell. Biol., 15, 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]