Abstract
Preadenylated single-stranded DNA ligation adaptors are essential reagents in many next generation RNA sequencing library preparation protocols. These oligonucleotides can be adenylated enzymatically or chemically. Enzymatic adenylation reactions have high yield but are not amendable to scale up. In chemical adenylation, adenosine 5′-phosphorimidazolide (ImpA) reacts with 5′ phosphorylated DNA. It is easily scalable but gives poor yields, requiring labor-intensive cleanup steps. Here, we describe an improved chemical adenylation method using 95% formamide as the solvent, which results in the adenylation of oligonucleotides with >90% yield. In standard conditions, with water as the solvent, hydrolysis of the starting material to adenosine monophosphate limits the yields. To our surprise, we find that rather than increasing adenylation yields by decreasing the rate of ImpA hydrolysis, formamide does so by increasing the reaction rate between ImpA and 5′-phosphorylated DNA by ∼10-fold. The method described here enables straightforward preparation of chemically adenylated adapters with higher than 90% yield, simplifying reagent preparation for NGS.
Keywords: adenylation, imidazolide, formamide, RNA ligation
INTRODUCTION
RNA sequencing is a powerful and ubiquitous tool in molecular biology. Next-generation sequencing (NGS) libraries are often made by ligating preadenylated single-stranded DNA (ssDNA) adapters to RNA 3′ ends using mutant ligases that lack the ability to adenylate the ligation donor. This strategy ensures the specificity of the ligation since only the preadenylated adapter can act as a ligation donor, but necessitates either the purchase or synthesis of preadenylated ssDNA oligonucleotides (Lau et al. 2001; Viollet et al. 2011; Max et al. 2018). Adenylation of ssDNA oligonucleotides can be performed either enzymatically, using various ligases (e.g., Mth RNA Ligase or RNA Ligase 1), or chemically, using an activated adenosine 5′-phosphorimidazolide (ImpA) (Lau et al. 2001; Dai et al. 2009; Chen et al. 2012; Song et al. 2015; Lama and Ryan 2016; Max et al. 2018). In enzymatic adenylation reactions, conditions are optimized such that the first step of the ligation reaction (donor adenylation) can proceed while the second step cannot. In chemical adenylation reactions, the imidazole bound to the phosphorous of ImpA acts as a leaving group, allowing for a nucleophilic attack by the 5′ phosphate of a ssDNA oligonucleotide on the 5′ phosphate of ImpA, resulting in the formation of an adenylated ssDNA oligonucleotide. While the yield of enzymatic adenylation can be high, it is generally performed on a small scale. Chemical adenylation, on the other hand, is scalable but traditionally provides poor yields requiring labor-intensive HPLC or gel purifications post-synthesis (Max et al. 2018).
Here, we report that by performing the chemical adenylation reaction in 95% formamide instead of water, the reaction yield is increased to >90%, removing the necessity of further purification by HPLC or gel extraction. Additionally, we show that the increased yield is due to a faster reaction rate between the 5′ phosphate and ImpA in formamide than in water.
RESULTS
Formamide improves the yield of adenylation of 5′-phosphorylated DNA
Chemical adenylation of 5′-phosphorylated ssDNA oligonucleotides in solution is a common method to prepare ligation adapters used in RNA sequencing library preparations. In this reaction, ImpA is reacted with a 5′-phosphorylated ssDNA oligonucleotide to yield a 5′ adenylated product (Fig. 1A). This reaction is inefficient and requires laborious purifications to separate unreacted the 5′-phosphorylated oligonucleotide from the adenylated product.
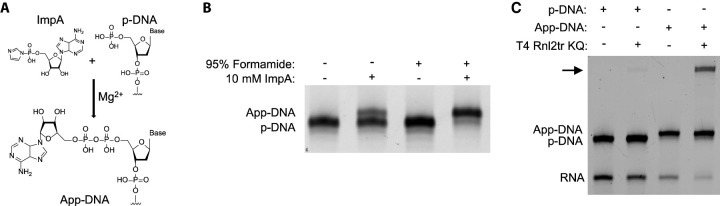
FIGURE 1.
Formamide increases the yield of chemical adenylation. (A) Schematic of the adenylation reaction. (B) Adenylation of a 5′ phosphorylated DNA oligonucleotide (ODN1) with the reaction taking place either in water or in 95% formamide. (C) Ligation of a 5′ p-DNA oligonucleotide or a 5′ App-DNA oligonucleotide (ODN1) that was adenylated in formamide to an RNA oligonucleotide (ORN1) using a mutant RNA ligase that requires a preadenylated ligation donor.
Previous studies which focused on adenylating ssDNA oligonucleotides during solid phase synthesis showed some length-dependance on adenylation yield with shorter oligonucleotides being adenylated with higher efficiency (Dai et al. 2009). We hypothesized that this length-dependency may be due to increased steric hindrance with longer oligonucleotides and that the low efficiency of the reaction in solution may, similarly, be caused by steric hindrance from oligonucleotide secondary structure. We reasoned that performing the adenylation reaction in denaturing conditions may increase yields due to decreased steric hindrance. To create denaturing conditions, we attempted the reaction in a final concentration of 95% formamide. Analysis by denaturing PAGE showed that the reaction yield was dramatically improved in the presence of formamide as compared to the reaction carried out in water (∼95% vs. ∼20%) (Fig. 1B). Mass spectrometry confirmed that the product was the adenylated oligonucleotide. To verify that the adenylation occurred on the 5′-position and produced a functional ligation donor, we carried out a ligation reaction using T4 Rnl2tr KQ, an RNA ligase that will ligate a preadenylated donor oligonucleotide to a 3′-OH RNA acceptor oligonucleotide. We observed the presence of the proper ligation product only in the presence of the preadenylated DNA donor, validating that this method yields a functional adenylated oligonucleotide (Fig. 1C).
The improved adenylation yield of formamide is generalizable
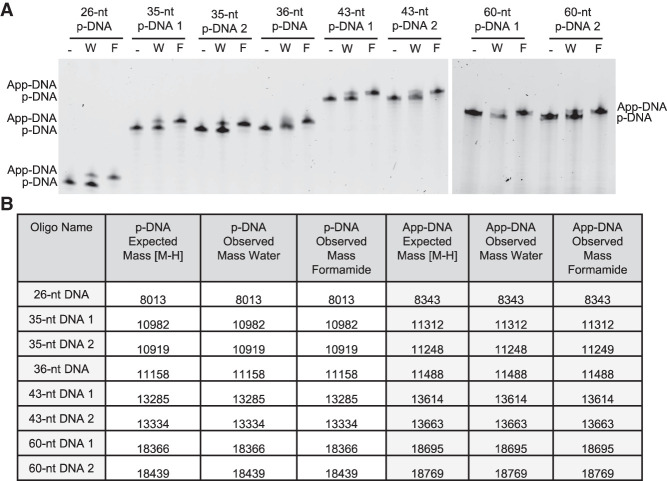
To verify that the improvements to the yield of adenylation were generalizable, we tested the adenylation efficiency of a panel of eight 5′-phosphorylated DNA oligonucleotides of varying sequence and length in either 95% formamide or water. Denaturing PAGE analysis showed that formamide drastically increased the adenylation efficiency of all eight oligonucleotides (Fig. 2A). The shift seen by denaturing PAGE was verified to be due to adenylation by mass spectrometry (Fig. 2B). Interestingly, none of the eight oligonucleotides in the panel had strong secondary structure as determined by in silico secondary structure analysis, indicating that the improvement in adenylation yield in formamide may not, in fact, be due to a reduction in secondary structure but rather through another mechanism (Supplemental Fig. 1). Intrigued by this, we tested the adenylation reaction with other additives meant to denature oligonucleotide secondary structure and found that while 50% formamide (∼50% yield) and, to a slightly lesser extent, 50% DMSO (∼40% yield) improved adenylation yield, 4 M urea did not (∼25% yield) (Supplemental Fig. 2).
FIGURE 2.
Formamide increases the yield of adenylation across multiple oligonucleotides. (A) A panel of 5′ phosphorylated DNA oligonucleotides were adenylated in either water (W) or 95% formamide (F). (B) Q-TOF LC/MS results of the adenylation reactions confirms the identity of expected products.
Additionally, when the adenylation reaction was performed in nearly pure formamide (600 mM water from MgCl2 hexahydrate solubilized in formamide), a significant fraction (∼20%) of the adenylated species had an additional +28 mass, potentially due to formylation of the oligonucleotide 5′ phosphate or ImpA (Supplemental Fig. 2; Benner et al. 2012). This additional mass is not detected when the reaction is conducted in 95% formamide with 5% water, indicating that the +28 mass is likely hydrolyzable. These results indicated to us that the formamide-induced improvement in adenylation yield is due to a mechanism other than secondary structure melting.
Formamide increases the rate of 5′-phosphorylated DNA adenylation
We reasoned that if formamide is not increasing the yield of the reaction by decreasing secondary structure, it could instead be doing so either by increasing the reaction rate between ImpA and the 5′-phosphorylated DNA or by reducing the rate of degradation of ImpA to adenosine monophosphate (pA). To test if performing the reaction in formamide affects the rate of the reaction between ImpA and 5′-phosphorylated DNA, we performed an adenylation time course. The reaction was quenched at various timepoints, resolved by denaturing PAGE, and the adenylation efficiency was measured by densitometry (Fig. 3A). Quantification of adenylation showed that the initial reaction rate increases in formamide, with the reaction in formamide proceeding ∼4.25 times faster than the reaction in water (Fig. 3B). Since adenylation of the oligonucleotide is competing with the reaction between ImpA and its hydrolysis product, pA, the increased initial rate when performing the reaction in formamide may allow more of the oligonucleotide to react with ImpA before pA accumulates, providing an explanation as to how the increased initial rate leads to a higher plateau at later timepoints. Additionally, while the reaction in water was well fit by a one-phase exponential, the reaction in formamide was better fit by a two-phase exponential potentially indicating that the adenylation reaction mechanism differs between the two conditions (Fig. 3B).
FIGURE 3.
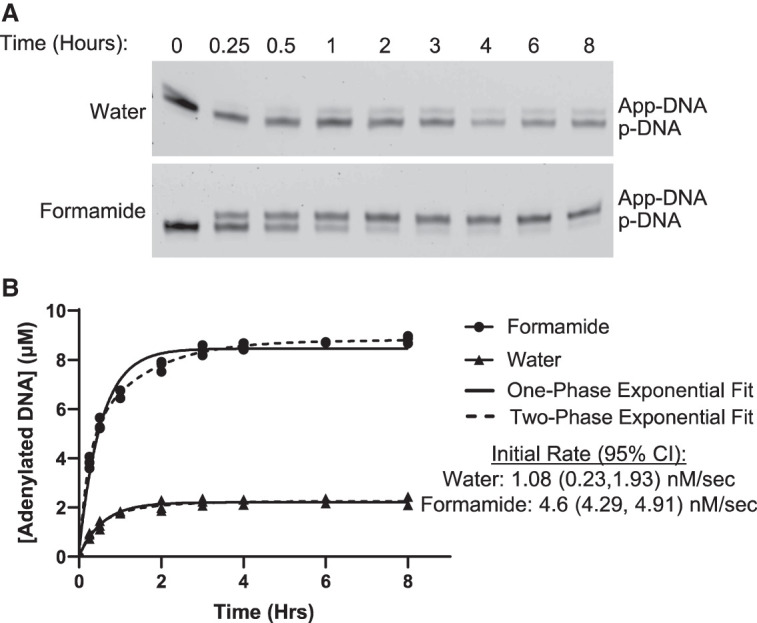
Formamide increases the rate of adenylation. (A) Time course of adenylation of a 5′ phosphorylated DNA oligonucleotide (ODN1) in either water or 95% formamide. (B) Quantification of the adenylation time course showing both one-phase exponential and two-phase exponential equations fit to the data. Mean and confidence intervals for initial rates were calculated by fitting each replicate individually and calculating the initial rate.
Formamide increases the reaction rate between ImpA and pA
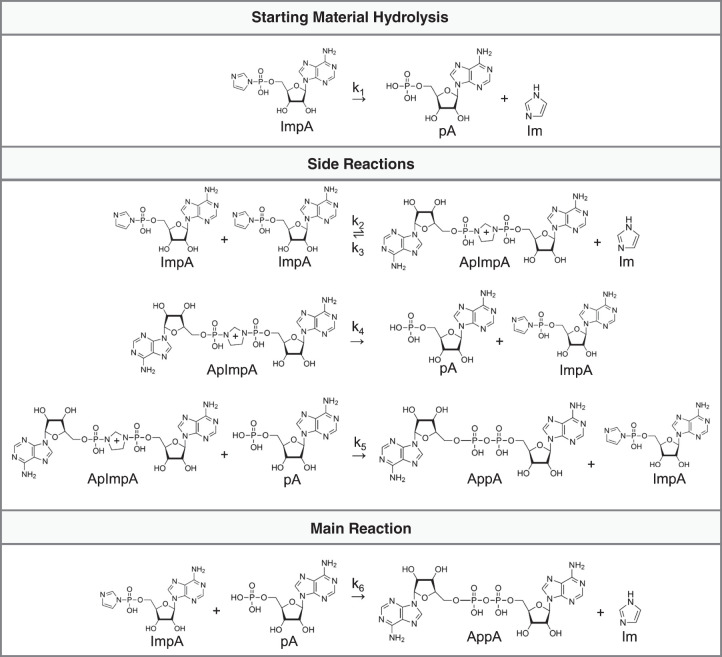
To gain more insight into the mechanism by which formamide increases the yield of the adenylation reaction, we next chose to track the reaction between mononucleotides over time by HPLC. Since ImpA will react with its hydrolysis product, pA, we are able to measure both the hydrolysis rate of ImpA into pA as well as the reaction rate between ImpA and pA (which acts as a surrogate for 5′-phosphorylated DNA oligonucleotides) to form AppA by using ImpA as the only starting material in the reaction, allowing us to gain insight into the mechanism by which formamide increases the yield of adenylation. From this data and the reaction scheme in Figure 4, a system of ordinary differential equations (ODEs) was used to fit the reaction rates. The reaction products were resolved by HPLC where clear peaks for ImpA, pA, and AppA can be seen (Fig. 5A; Supplemental Fig. 3). Plotting the measured data along with the concentrations as predicted by the ODE fit over time showed reasonable concordance with the data (Fig. 5B). In the water condition, the concentration of AppA is slightly underestimated at early timepoints, while the concentration of pA is slightly overestimated. In the formamide condition, the predicted concentrations better fit the measured concentrations although the level of pA is initially predicted to be too high and is then brought down below the level measured.
FIGURE 4.
Schematic of the reactions used to fit the system of ordinary differential equations in Figure 5.
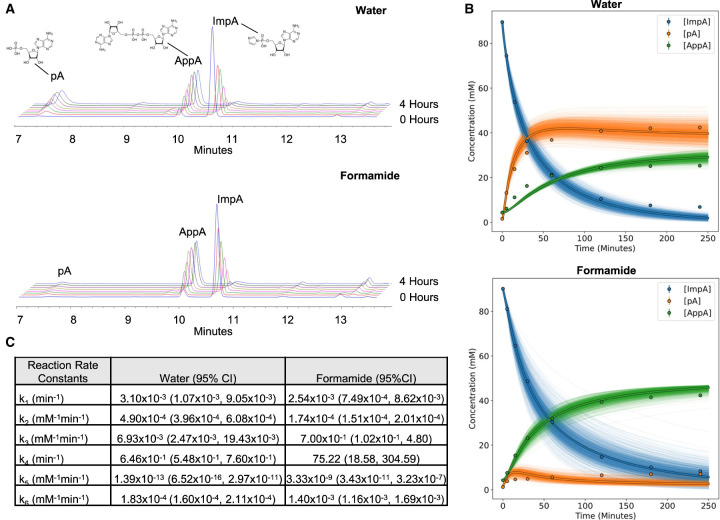
FIGURE 5.
Formamide increases the rate of the reaction between ImpA and pA. (A) Representative HPLC profiles of the reaction between ImpA and its degradation product, pA, to make AppA in either water or 95% formamide over a 4 h time course. (B) Concentrations of ImpA, pA, and AppA predicted (lines) by fitting the reaction scheme in Figure 4 to the HPLC data (points) with a system of ordinary differential equations. The bold lines are the concentrations predicted by the mean of the log-transformed fit rates, and the faint lines are the predicted concentrations from resampling the rates from their 95% confidence intervals. (C) Reaction rates for the scheme in Figure 4 fit by the system of ordinary differential equations.
The reaction rates calculated from the ODE fit showed that the fitted rate of ImpA decay was no different in formamide and water, while the predicted reaction rate between ImpA and pA was nearly ten times faster in formamide than in water (Fig. 5C), which agrees well with the oligonucleotide adenylation time course data in Figure 3. Interestingly, the difference in the concentration of AppA at the plateau is only about twofold different between the water and formamide conditions, as opposed to ∼4.5-fold in the oligonucleotide adenylation time course (Fig. 3). This is probably because the reaction in Figure 5 is only limited by the ability of ImpA to react before it is all hydrolyzed to pA whereas the oligonucleotide adenylation reaction is competing with the reaction between ImpA and pA as well. Overall, these experiments provide evidence that the main driver in the increased adenylation yield is a higher reaction rate between ImpA and 5′ phosphorylated DNA.
DISCUSSION
Chemical adenylation of DNA oligonucleotides using ImpA has long been thought of as an inefficient reaction requiring additional purification to remove unadenylated oligonucleotides. Even though the reaction on solid support has been optimized, it still yields <50% adenylated product for long DNA oligonucleotides, such as those used as adapters in RNA sequencing library preparations (Dai et al. 2009). In this work, we show that by performing the adenylation reaction in 95% formamide, the yield is improved to >90% regardless of oligonucleotide length and sequence. This improvement will make the production of 5′-adenylated oligonucleotides, a key reagent in NGS, easier and more accessible.
After verifying the generalizability of formamide for improving adenylation yields, we explored the mechanism of this improvement. The adenylation efficiency could not be explained by a reduction in secondary structure since the efficiency is improved in unstructured DNA oligonucleotides. Additionally, other agents that disrupt secondary structure do not provide the same degree of improvement as formamide. This suggested that formamide acts either to improve the reactivity between the 5′-phosphate and ImpA or the stability or ImpA, allowing more to react before it is hydrolyzed. Kinetic experiments revealed that the improved adenylation efficiency is driven by an increased reaction rate between the 5′-phosphate and ImpA in formamide as compared to water, not a change in the hydrolysis rate of ImpA.
Mass spectrometry results from oligonucleotides adenylated in pure formamide show a +28 mass, which is not detectable when performing the reaction in 95% formamide, likely corresponding to formylation of the adenylated product. Reactions between formamide and nucleotides have long been studied in the context of prebiotic chemistry, including the finding that it is a suitable solvent for the phosphorylation of nucleosides in the presence of inorganic phosphate (Schoffstall 1976; Xu et al. 2019). While it remains unclear whether formamide is increasing the reaction rate by formylating either the oligonucleotide or ImpA or through a solvent effect, the presence of this hydrolyzable +28 adduct raises the possibility that formamide is participating in the reaction as an activator. In keeping with the idea of formamide acting as an activator to increase adenylation yields, recent studies have also shown that the addition of high concentrations of 1-methyl-imidazole as an activator increases the yield of an RNA capping reaction between 7-methylguanosine 5′-diphosphate imidazolide and a 5′-phosphate RNA (Abe et al. 2022). Formamide can, however, formylate nucleotides at multiple sites, making it difficult to assess what the identity and role, if any, of this formylation may be (Benner et al. 2012).
This improved method allows for the large scale and facile synthesis of 5′-adenylated oligonucleotides, making it easier to synthesize large amounts of many adenylated oligonucleotides simultaneously without the additional step of HPLC or gel purification. This will allow researchers to spend less time and effort on reagent preparation when making RNA sequencing libraries.
MATERIALS AND METHODS
Oligonucleotides
All oligonucleotides used in this study were purchased from Integrated DNA Technologies (IDT) as 5′-phosphorylated DNA with standard desalting.
Synthesis of ImpA
Adenosine 5′-monophosphoric acid (0.5 mmol, 174 mg, 1.0 eq) in 15 mL DMF was added dropwise to a vigorously stirred solution of triphenylphosphine (1 mmol, 262 mg, 2.0 eq), 2,2′-dipyridyldisulfide (1 mmol, 220 mg, 2.0 eq), and imidazole (2.5 mmol, 170 mg, 5.0 eq) in 15 mL DMF and triethylamine (2.5 mmol, 0.9 mL, 5.0 eq). This solution was then stirred for 1.5 h at room temperature. ImpA was then precipitated adding the reaction to a stirred solution of 53 mM sodium perchlorate in 115 mL acetone and 55 mL anhydrous ethyl ether in a dropwise manner. The precipitate was then collected by passing it through a Büchner funnel and washed thoroughly with acetone. Finally, the precipitate was washed with anhydrous ethyl ether and dried under a vacuum overnight. Purity of the final product was assessed by phosphorous NMR and proton NMR (tabulation below, full spectra in Supplemental Fig. 4). ImpA was then stored under argon at −20°C.
1H NMR (DMSO-d6, 500 MHz) δppm 8.40 (1H, s), 8.14 (1H, s), 7.63 (1H, s), 7.29 (2H, brs), 7.09 (1H, brd, J = 0.8 Hz), 6.85 (1H, s) 5.89 (1H, d, J = 6.2 Hz) 5.48 (1H, d, J = 6.2 Hz), 5.30 (1H, d, J = 4.4 Hz), 4.58 (1H, dd, J = 5.8 Hz, 5.3 Hz), 4.03 (1H, d, J = 3.2 Hz), 3.94 (1H, d, J = 3.2 Hz) 3.73 (2H, m)
31P NMR (DMSO-d6, 202 MHz) δppm−10.35 (1P, s)
Oligonucleotide adenylation reaction
Adenylation reactions were carried out by mixing 100 mM MgCl2•6H2O (from a 1 M solution made either in water or formamide), 10 mM ImpA (from a 100 mM solution made either in water or deionized formamide [Invitrogen, AM9344]), and 10 µM 5′-phosphorylated DNA (from a 200 µM solution made in water) either in water or deionized formamide. This solution was then heated to 65°C for the indicated times. The reaction was then diluted 1:3 in water, ethanol precipitated, and resuspended in 95% formamide loading dye. The adenylation reactions were resolved by 7.5 M urea 15% PAGE and stained with Sybr Gold (Thermo S11494).
T4 Rnl2KQ ligation
Ligations were performed by mixing 1 µL 10 µM of DNA adapter, 1 µL 10 µM RNA oligonucleotide, 3 µL 10× RNA ligation buffer (500 mM Tris-HCl pH 7.5, 100 mM MgCl2, 100 mM DTT), 15 µL 50% PEG 8000, 3 µL Rnl2KQ (NEB M0373S), water to 30 µL. The ligations were incubated for 1 h at room temperature and quenched by boiling with 30 µL 100% formamide and 1 µL 10 mM EDTA. The reactions were resolved by 7.5 M urea 15% PAGE and stained with Sybr Gold (Thermo S11494).
Nucleotide adenylation reaction and HPLC
Reactions were carried out by mixing 1 M MgCl2, 100 mM ImpA in either water or 95% deionized formamide. The solution was heated to 65°C for the indicated times. Timepoints were quenched by adding 10 µL of the adenylation reaction to 490 µL 50 mM EDTA and flash freezing in liquid nitrogen. HPLC was performed by individually thawing the samples and analyzing them immediately. Samples were injected on a 1260 HPLC (Agilent) using a Luna Omega PS-C18 5 µm 100 Å liquid chromatography column (250 × 4.6 mm) (Phenomenex). Samples were resolved over 18 min using an isocratic elution gradient of 2% for 3 min, followed by 2%–20% over 15 min (95% acetonitrile, 5% 15 mM ammonium acetate pH 4.2) in 15 mM ammonium acetate pH 4.2 at 25°C, using a flow rate of 1.0 mL min−1. This was followed by wash steps at higher elution strengths between runs. Absorbance was recorded at 260 nm and peaks were collected manually for LCMS analysis. The order of elution was pA (∼7 min), followed by AppA (∼9.5 min), and lastly ImpA(∼10.5 min).
For high-resolution mass spectrometry (HRMS) analysis, nucleotides were first resolved using the HPLC method above, and UV peaks were manually collected in separate vials. These samples were immediately run on a 6530 Accurate-Mass Q-TOF LC/MS (Agilent) on positive ESI mode linted to a preinjection 1260 infinity HPLC (Agilent). Mass spectrometry settings to detect nucleotides were as follows: gas temperature 350°C, nebulizer gas rate 45 psig, drying gas 10 L/min, VCap 4000 V, fragmentor voltage 120 V, skimmer voltage 65 V.
Ordinary differential equation fitting for nucleotide adenylation reaction rates
To calculate the reaction rates for the nucleotide adenylation reaction, the following system of ordinary differential equations were fit to the HPLC data using the ODE45 function in Matlab R2021b:
d[ImpA]/dt = −k1[ImpA] − k2[ImpA][ImpA] + k3[ApImpA][Im] + k5[pA][ApImpA] − k6[pA][ImpA]
d[pA]/dt = k1[ImpA] + k4[ApImpA] − k5[pA][ApImpA] − k6[pA][ImpA]
d[AppA]/dt = k5[ApImpA][pA] + k6[ImpA][pA]
d[ApImpA]/dt = k2[ImpA][ImpA] − k3[ApImpA][Im] − k4[ApImpA] − k5[ApImpA][pA]
d[Im]/dt = k1[ImpA] + k2[ImpA][ImpA] − k3[ApImpA][Im] + k6[ImpA][pA]
Initial conditions were set as follows:
k1 = [0.01 0.005 0.001]
k2 = [0.01 0.005 0.001]
k3 = [0.01 0.005 0.001]
k4 = [0.01 0.005 0.001]
k5 = [0.01 0.005 0.001]
k6 = [0.01 0.005 0.001]
All combinations of initial rates were used to initialize the ODE45 optimization and the median of all fits that reached a local minimum or were stopped due to the step size being below 1 × 10−10 was calculated for each rate constant in each replicate.
The log(mean) of the fitted rates were then used to calculate confidence intervals and 1000 new rates were sampled from the 95% confidence interval for each rate constant. The mean rates as well as the resampled rates from the 95% confidence interval were used to plot the predicted reactant concentrations against the measured concentrations using the Scipy and Matplotlib packages in Python 3.7.
LC–MS analysis of oligonucleotides
The mass of oligonucleotides was measured by LC–MS analysis on an Agilent 6530 accurate mass Q-TOF using the following conditions: buffer A: 100 mM 1,1,1,3,3,3-hexafluoroisopropanol (HFIP) and 9 mM triethylamine (TEA) in LC–MS grade water; buffer B:100 mM HFIP and 9 mM TEA in LC–MS grade methanol; column, Agilent AdvanceBio oligonucleotides C18; linear gradient 0%–100% B 8 min; temperature, 60°C; flow rate, 0.5 mL/min. LC peaks were monitored at 260 nm. MS parameters: Source, electrospray ionization; ion polarity, negative mode; range, 100–3200 m/z; scan rate, 2 spectra/s; capillary voltage, 4000; fragmentor, 180 V.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We wish to thank Dr. Li Li for his invaluable advice throughout the project. This work was supported by funding from the National Institutes of Health (NIH) grants R35-GM131839 (A.K.), S10-OD020012 (A.K.), F31-NS122493 (S.R.H.), and R01-NS111990 (J.K.W.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079521.122.
REFERENCES
- Abe N, Imaeda A, Inagaki M, Li Z, Kawaguchi D, Onda K, Nakashima Y, Uchida S, Hashiya F, Kimura Y, et al. 2022. Complete chemical synthesis of minimal messenger RNA by efficient chemical capping reaction. ACS Chem Biol 17: 1308–1314. 10.1021/acschembio.1c00996 [DOI] [PubMed] [Google Scholar]
- Benner SA, Kim HJ, Carrigan MA. 2012. Asphalt, water, and the prebiotic synthesis of ribose, ribonucleosides, and RNA. Acc Chem Res 45: 2025–2034. 10.1021/ar200332w [DOI] [PubMed] [Google Scholar]
- Chen YR, Zheng Y, Liu B, Zhong S, Giovannoni J, Fei Z. 2012. A cost-effective method for Illumina small RNA-Seq library preparation using T4 RNA ligase 1 adenylated adapters. Plant Methods 8: 41. 10.1186/1746-4811-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Saikia M, Li NS, Pan T, Piccirilli JA. 2009. Efficient chemical synthesis of AppDNA by adenylation of immobilized DNA-5′-monophosphate. Org Lett 11: 1067–1070. 10.1021/ol802815g [DOI] [PubMed] [Google Scholar]
- Lama L, Ryan K. 2016. Adenylylation of small RNA sequencing adapters using the TS2126 RNA ligase I. RNA 22: 155–161. 10.1261/rna.054999.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. 10.1126/science.1065062 [DOI] [PubMed] [Google Scholar]
- Max KEA, Bertram K, Akat KM, Bogardus KA, Li J, Morozov P, Ben-Dov IZ, Li X, Weiss ZR, Azizian A, et al. 2018. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc Natl Acad Sci 115: E5334–E5343. 10.1073/pnas.1714397115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoffstall AM. 1976. Prebiotic phosphorylation of nucleosides in formamide. Orig Life 7: 399–412. 10.1007/BF00927935 [DOI] [PubMed] [Google Scholar]
- Song Y, Liu KJ, Wang TH. 2015. Efficient synthesis of stably adenylated DNA and RNA adapters for microRNA capture using T4 RNA ligase 1. Sci Rep 5: 15620. 10.1038/srep15620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollet S, Fuchs RT, Munafo DB, Zhuang F, Robb GB. 2011. T4 RNA Ligase 2 truncated active site mutants: improved tools for RNA analysis. BMC Biotechnol 11: 72. 10.1186/1472-6750-11-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Green NJ, Gibard C, Krishnamurthy R, Sutherland JD. 2019. Prebiotic phosphorylation of 2-thiouridine provides either nucleotides or DNA building blocks via photoreduction. Nat Chem 11: 457–462. 10.1038/s41557-019-0225-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.