Summary
Characterizing perturbation of molecular pathways in congenital Zika virus (ZIKV) infection is critical for improved therapeutic approaches. Leveraging integrative systems biology, proteomics, and RNA-seq, we analyzed embryonic brain tissues from an immunocompetent, wild-type congenital ZIKV infection mouse model. ZIKV induced a robust immune response accompanied by the downregulation of critical neurodevelopmental gene programs. We identified a negative correlation between ZIKV polyprotein abundance and host cell cycle-inducing proteins. We further captured the downregulation of genes/proteins, many of which are known to be causative for human microcephaly, including Eomesodermin/T-box Brain Protein 2 (EOMES/TBR2) and Neuronal Differentiation 2 (NEUROD2). Disturbances of distinct molecular pathways in neural progenitors and post-mitotic neurons may contribute to complex brain phenotype of congenital ZIKV infection. Overall, this report on protein- and transcript-level dynamics enhances understanding of the ZIKV immunopathological landscape through characterization of fetal immune response in the developing brain.
Subject areas: Immunology, Virology, Developmental neuroscience
Graphical abstract
Highlights
-
•
Multi-omics analysis revealed key molecular pathways in congenital Zika syndrome
-
•
JAK/STAT activation is a potential driver of deranged brain development
-
•
Genes associated with microcephaly were downregulated in ZIKV-infected mouse brains
-
•
ZIKV load and cell cycle-inducing protein levels showed a negative correlation
Immunology; Virology; Developmental neuroscience
Introduction
The fetal and newborn brain is highly sensitive to inflammation-induced injury1 and congenital infection is a major cause of structural abnormalities. A variety of human pathogens cause structural brain abnormalities, including “TORCH” micro-organisms (viruses, parasites, and bacteria -Toxoplasma gondii, Rubella virus, Cytomegalovirus, Herpes simplex virus, Listeria monocytogenes, Treponema pallidum; parvovirus B19, human immunodeficiency virus, and varicella-zoster virus) and, more recently, Zika virus (ZIKV).2
The acute crisis of the 2015–2016 ZIKV epidemic in the Americas led to the identification of a range of developmental CNS abnormalities because of congenital ZIKV infection, collectively known as Congenital Zika Syndrome.3,4 Affected children present with severe developmental delays, seizures, vision/hearing abnormalities, and joint contractures associated with structural brain abnormalities such as microcephaly, intracranial calcifications, gyral abnormalities, and enlarged ventricles.3,5,6 Despite significant efforts, the molecular pathways underlying the neuropathogenesis of congenital ZIKV infection remain incompletely characterized.
Systems biology is a powerful approach to define molecular and cellular signatures that correlate with clinical outcomes. In particular, multi-omics approaches, comprising genome, transcriptome, proteome, and metabolome data, or combinations thereof, provide information about thousands of genes, transcripts, proteins, and metabolites simultaneously.7,8,9 These data can now be integrated and analyzed using novel workflows developed in our lab and by others.10 Such multi-omics approaches have recently been adopted to study immune ontogeny, the immune response to infectious diseases, and microbiomes.11,12,13,14
Several systems biology studies of the pathogenesis of ZIKV in vitro have yielded important insights. A large-scale affinity purification-mass spectrometry-based proteomics study in human non-neuronal cell lines has suggested a role for ZIKV-specific interaction between non-structural protein 4A and ANKLE2, a gene linked to hereditary microcephaly.15 RNA-sequencing (RNA-Seq) analyses demonstrate dysregulation of genes associated with transcription, protein transport, stress response, cell cycle regulation, and nervous system development in ZIKV-infected cell cultures.16,17,18,19 Proteomic analyses identified the downregulation of doublecortin in ZIKV-infected primary human neural progenitor cell (hNPC) cultures20 and dysregulation of kinase cascades in ZIKV-infected neuroblastoma cultures.21 In addition, a proteo-transcriptomic analysis demonstrated that ZIKV impairs the cell cycle regulation in induced pluripotent stem cell-derived (iPSC-derived) neurospheres.22 However, to our knowledge, integrative transcription and translation studies were not done using an in vivo congenital ZIKV infection.
To gain fresh insight into the pathogenesis of ZIKV-induced neurodevelopmental disease, we devised a multi-omics approach integrating RNA-Seq and proteomics data to obtain a high-level overview of the impact of ZIKV infection in the fetal brain. We present an unbiased, proteogenomic resource characterizing the molecular events contributing to microcephaly using an established in vivo model of ZIKV infection in mice,23,24,25 highlighting the immune activation in the fetal brain. Notably, this infection model was established in an immunocompetent mouse background to study the host response to ZIKV infection in the absence of other confounding immune system perturbations. Together, these integrative systems biology approaches confirmed the activation of innate immune response pathways in the ZIKV-infected embryonic brain and suggested its role in mediating microcephaly associated with congenital infection via novel gene targets.
Results
Proteomics and gene expression analysis in the immunocompetent ZIKV microcephaly mouse model
We conducted quantitative mass spectrometry-based proteomic and RNA-Seq analyses of the embryonic brains collected from an established immunocompetent congenital ZIKV infection mouse model (Figure 1A).23,24,25
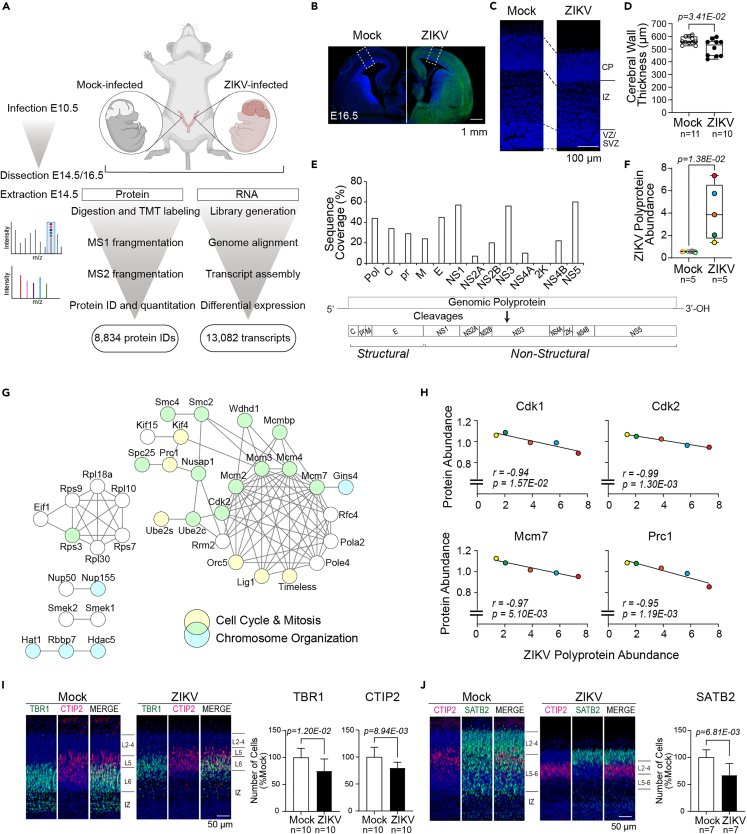
Figure 1.
Embryonic ZIKV infection recapitulates human microcephaly and shows viral load-dependent dysregulation of cell cycle-associated proteins
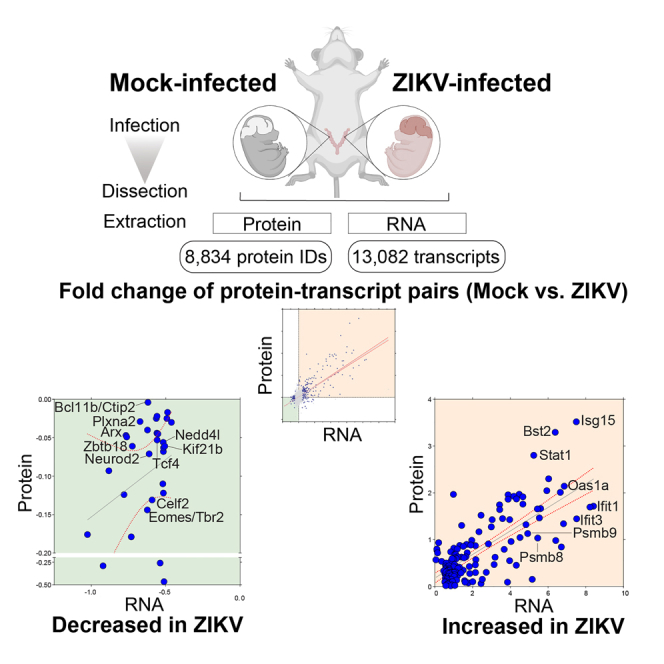
(A) Multiplexed proteomic and RNA-sequencing (RNA-Seq) analysis workflow for Mock (PBS) or Zika virus (ZIKV)-infected brains (n = 5 for proteomics and n = 3 for RNA-Seq per condition). Each placenta of the embryos received ZIKV or Mock injection at embryonic day 10.5 (E10.5). Brains were collected at E14.5 for quantitative tandem mass tag (TMT) mass spectrometry analysis and RNA-Seq, resulting in 8,834 protein IDs and 13,082 transcripts identified. Structural analysis was conducted at E16.5.
(B) Immunohistochemistry (IHC) staining of brains at E16.5, following Mock or ZIKV infection at E10.5; ZIKV Env (green), Hoechst (nuclei, blue), scale bar = 1 mm. Dotted square corresponds to the primary somatosensory area of the cerebral cortex.
(C) IHC staining of the primary somatosensory area at E16.5 showing the ventricular zone (VZ)/subventricular zone (SVZ), intermediate zone (IZ), and cortical plate (CP); Hoechst (nuclei, blue), Scale bar = 100 μm.
(D) The total thickness of the cerebral wall at E16.5. Boxplot shows the median, interquartile range, and maximum/minimum values.
(E) ZIKV polyprotein and potential processed polyprotein constituent sequence coverages for structural (C-capsid, prM-premembrane, M-membrane, E-envelope) and non-structural (NS1-5) proteins, identified by proteomics.
(F) Mean normalized abundance of ZIKV polyprotein in Mock- or ZIKV-infected brain at E14.5. Color-coding shows the embryos deriving from the same litter.
(G) Interaction network of proteins with strong negative correlation (Pearson correlation r < −0.95) with ZIKV polyprotein abundance. Node color indicates functional classification (blue, chromosome organization; yellow, cell cycle and mitosis; green, both functions); edges represent the highest confidence interactions (confidence score = 0.90) between proteins in the network based on protein-protein interaction information curated by the STRING database. Connected nodes are shown.
(H) Sum-normalized protein abundance of selected cell cycle-associated proteins, Cyclin Dependent Kinase 1 (Cdk1), Cdk2, Minichromosome Maintenance Complex Component 7 (Mcm7), and Protein Regulator of Cytokinesis 1 (Prc1), plotted against ZIKV polyprotein abundance across five ZIKV-infected brains. Color coding follows 1F. Note the negative correlation between the abundances of ZIKV polyprotein and cell cycle-inducing proteins.
(I) IHC staining of CP layers 2–6 (L2-6) and IZ from Mock- or ZIVK-infected embryonic brains, T-box Brain Protein 1 (TBR1) (green), B Cell CLL/Lymphoma 11b (CTIP2) (red) and Hoechst (nucleus, 8 blue), scale bar = 50 μm; Quantification of TBR1-positive (ZIKV-infected embryos, n = 10 from 8 litters, Mock-infected embryos, n = 10 from 6 litters) and CTIP2-positive (ZIKV-infected embryos, n = 10 from 8 litters, Mock-infected embryos, n = 10 from 6 litters) post-mitotic neurons.
(J) IHC staining of L2-6 and IZ from Mock- or ZIKV-infected embryonic brains, CTIP2 (red), Special AT-Rich Sequence-Binding Protein 2 (SATB2) (green), and Hoechst (nucleus, blue), scale bar = 50 μm; Quantification of SATB2-positive post-mitotic neurons (ZIKV-infected embryos, n = 7 from 6 litters, Mock-infected embryos, n = 7 from 5 litters). Error bars indicate standard deviation.
Pregnant wild-type CD-1 mice received an intra-placental injection of ZIKV or phosphate-buffered saline (PBS) as a control (Mock) on embryonic day 10.5 (E10.5). Each horn of the uterus was assigned for either ZIKV or Mock injection (Figure 1A). The ZIKV-injected embryos used in the experiments demonstrated smaller brains and decreased cerebral wall thickness by 9.6% (512 ± 66 vs. 566 ± 29 μm in the Mock-infected embryos, mean ± standard deviation, n = 10, p = 2.32E-02) as compared to the Mock-infected embryos at E16.5 (Figures 1B–1D and S1A–S1D). A quantitative proteomics workflow was applied to the embryonic brains on E14.5 to capture the earlier stages of molecular events leading to microcephaly. A comprehensive developmental proteome coverage of the brain was achieved, resulting in the identification of >8,000 proteins (5% false discovery rate (FDR)) and accurate quantitation of >6,800 proteins across subcellular compartments (Figure 1A). We detected ZIKV polyprotein at > 40% total sequence coverage, with peptides mapping to all ZIKV encoding structural and non-structural viral proteins except the signal peptide 2K, which does not contain trypsin-cleavable sites (Figure 1E). Although the venous outflows from the uterus of both horns merge into the inferior vena cava and thus have an overlapping circulatory system, the ZIKV abundances were significantly increased in the ZIKV-infected brains relative to the Mock-infected brains with respect to polyprotein abundances (7-fold change, Figure 1F) and virus RNA copy number (over 4000-fold change, Figure S1E).
Of interest, each embryo displayed variance in the expression of the ZIKV polyprotein despite having equal titers of virus administered, reflecting active propagation of the ZIKV within the injected embryos (Figure 1F). We leveraged the spectrum of the ZIKV polyprotein abundances within each infected brain to study the dosage response of host proteins in response to viral load. Host cell cycle progression proteins demonstrated negative abundance correlations (Pearson correlation r ≤ −0.95) with the viral polyprotein protein abundances. Notably, we identified host proteins associated with cell cycle (GO:0007049, FDR = 1.7E-08) such as Cyclin Dependent Kinase 1 (CDK1) and CDK2, chromosome organization (GO:0051276, FDR = 1.9E-11), and DNA replication (GO:0007049, FDR = 3.5E-10), components of DNA replication complexes, including the Origin of Replication Complex (ORC), Mini-Chromosome Maintenance (MCM) complex, Condensin Complex, Histone Acetylation (HAT) complexes, and the Anaphase-Promoting Complex/Cyclosome (APC/C) that were negatively correlated with ZIKV polyprotein abundances (Figures 1G and 1H, Tables 1 and S1, FDR are for gene ontology biological process (GOBP)).
Table 1.
Cell cycle-associated proteins dysregulated during ZIKV infection
| Name | Description | Functional role during mitosis | Pearson correlation |
|---|---|---|---|
| Cell cycle stage transitions/DNA replication | |||
| Cdk2 | Cyclin-dependent kinase 2 | DNA synthesis at G1/S transition; modulates G2 progression; mitotic entry | −0.989 |
| Orc5 | Origin recognition complex, subunit 5 | Origin of replication complex (ORC); pre-replication complex assembly | −0.952 |
| Anp32b | Acidic nuclear phosphoprotein 32 family, member B | G1/S transition; inhibits Caspase 3; histone chaperone | −0.981 |
| Minichromosome maintenance complex | |||
| Mcm7 | Minichromosome maintenance deficient 7 homolog | MCM complex; DNA replication initiation, elongation | −0.974 |
| Mcm4 | Minichromosome maintenance deficient 4 homolog | MCM complex; DNA replication initiation, elongation | −0.963 |
| Mcm2 | Minichromosome maintenance deficient 2 homolog | MCM complex; DNA replication initiation, elongation | −0.967 |
| Mcm3 | Minichromosome maintenance deficient 3 homolog | MCM complex; DNA replication initiation, elongation | −0.973 |
| Mcmbp | Mcm (mini-chromosome maintenance deficient) binding protein | MCM complex; binds during late S phase to promote complex disassembly; key regulator of pre-replication complex | −0.961 |
| Timeless | Timeless homolog | Binds MCM complex; regulates DNA replication/replication fork stability, circadian cycle | −0.978 |
| Wdhd1 | WD repeat and HMG-box DNA binding protein 1 | Replication initiation; MCM complex-Pola2 association | −0.963 |
| Polymerase activity | |||
| Pola2 | DNA-directed polymerase, alpha 2 | Coupling of primase complex to cellular replication machinery | −0.966 |
| Pole4 | Polymerase (DNA-directed), epsilon 4 | DNA replication and repair | −0.960 |
| Rfc4 | Replication factor C 4 | Elongation of primed DNA templates by Pold and Pole4 | −0.981 |
| Structural maintenance of chromosomes | |||
| Smc4 | Structural maintenance of chromosomes 2 | Condensin complex; chromosome assembly/segregation | −0.968 |
| Smc2 | Structural maintenance of chromosomes 4 | Condensin complex; chromosome assembly/segregation | −0.980 |
| Nsmce2 | Non-SMC element 2 homolog | E3 SUMO ligase; component of Smc5-6 complex; telomere maintenance | −0.977 |
| Histone acetylation/deacetylation | |||
| Hat1 | Histone aminotransferase 1 | DNA replication and repair; histone acetylation | −0.998 |
| Rbbp7 | Retinoblastoma binding protein 7 | Type B histone acetyltransferase (HAT) complex; post-replication chromatin assembly | −0.976 |
| Morf4l1 | Mortality factor 4-like 1 | NuA4 histone acetyltransferase (HAT) complex | −0.989 |
| Hdac5 | Histone deacetylase 5 | Histone deacetylation; midbody localization | −0.994 |
| Midzone and spindle formation/maintenance | |||
| Prc1 | Protein regulator of cytokinesis 1 | Spatiotemporal formation of midzone | −0.954 |
| Nusap1 | Nucleolar and spindle-associated protein 1 | Mitotic spindle organization | −0.977 |
| Kif4 | Kinesin family member 4 | Mitotic spindle stabilization, chromosomal positioning | −0.980 |
| Spc25 | Spindle pole body component 25 homolog/NDC80 kinetochore complex component | Spindle checkpoint activity, chromosome segregation | −0.994 |
| Haus1 | HAUS augmin-like complex, subunit 1 | Mitotic spindle assembly, centrosome integrity | −0.971 |
| Anaphase-promoting complex/cyclosome | |||
| Ube2c | Ubiquitin-conjugating enzyme E2C | Anaphase promoting complex/cyclosome (APC/C) | −0.950 |
| Ube2s | Ubiquitin-conjugating enzyme E2S | Anaphase promoting complex/cyclosome (APC/C) | −0.999 |
| Centrosome duplication/separation | |||
| Cetn3 | Centrin 3 | Microtubule organization (MTOC) center | −0.983 |
| DNA repair and additional histone modification | |||
| Rrm2 | Ribonucleotide reductase M2 | Deoxyribonucleotide biosynthesis | −0.965 |
| Lig1 | ATP-dependent DNA ligase 1 | dsDNA repair during replication, recombination | −0.977 |
| Rps3 | Ribosomal protein S3 | Endonuclease activity; DNA repair | −0.981 |
| Rybp | RING1 and YY1 binding protein | Transcriptional regulation; pro-apoptotic | −0.964 |
| Kdm3b | Lysine (K)-specific demethylase 3B | Histone demethylation | −0.957 |
| Rnf40 | Ring finger protein 40 | RNF20/40 E3 ubiquitin-protein ligase complex; histone modification | −0.970 |
Furthermore, we observed the enrichment of proteins that function as catalytic activity (GO:0003824, FDR = 2.0E-05) and nucleotide binding (GO:0000166, FDR = 7.2E-04) that positively correlate with ZIKV polyprotein abundances (Pearson correlation r ≥ 0.90, Figure S1F, FDR are for gene ontology molecular function (GOMF)). These positively correlated proteins include the Phosphoinositide 3-Kinases/Protein Kinase B/Mechanistic Target of Rapamycin (PIK3/AKT/mTOR) pathway proteins and mitochondrial proteins.
Furthermore, immunohistochemistry (IHC) staining results showed diminished numbers of neurons in the cortical plate in the ZIKV-infected embryos at E16.5. Namely, the neurons positive for TBR1, CTIP2, and SATB2 were decreased by 25% (n = 10, p = 1.2E-02), 20% (n = 10, p = 8.9E-03), and 33% (n = 7, p = 6.8E-03), respectively, in the ZIKV-infected embryos compared with the mock-infected embryos (Figures 1I and 1J).
ZIKV infection activates interferon-induced innate immunity and antigen-presenting pathways
Quantitative proteomics analysis identified 84 significantly upregulated and 16 significantly downregulated proteins in the embryonic brain on E14.5 (Figure 2A, <5% Permutation-based FDR, Table S2). GOBP analysis showed that the most enriched processes were those involved in immune response, such as innate immune response (GO:0045087, FDR = 6.91E-29), antigen processing and presentation (GO:0019882, FDR = 1.38E-11), and response to cytokine (GO:0034097, FDR = 1.83E-14) (Figure 2B). In addition, positive regulation of tumor necrosis factor production (GO:0032760; FDR = 2.63E-05) and positive regulation of apoptotic signaling pathway (GO:2001235; FDR = 6.2E-03) were also enriched in the proteomics dataset.
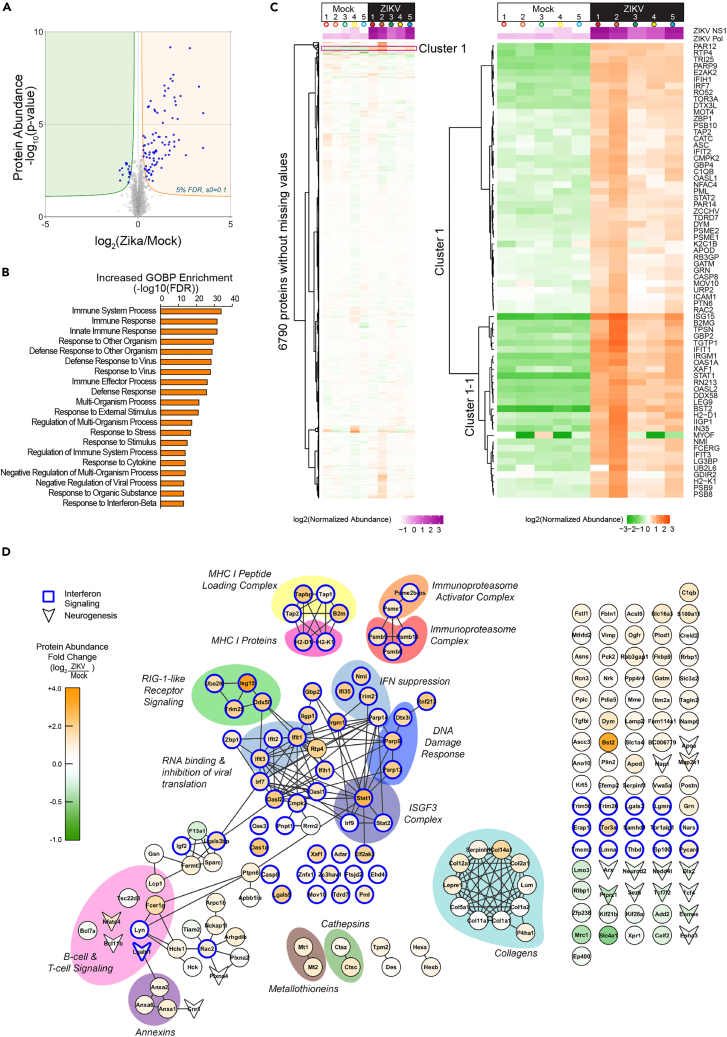
Figure 2.
Proteomics data shows the activation of interferon-induced immune pathways and antigen presentation pathways in the ZIKV-infected brain
(A) Volcano plot of protein expression fold change (log2(ZIKV/Mock)) in ZIKV vs. Mock-infected E14.5 brain tissues (n = 5 per condition) based on median-normalized protein abundance values. Significantly dysregulated proteins are highlighted in blue (5% FDR, Permutation-based). Normalized protein abundances are shown in Table S2.
(B) Top 15 significantly enriched gene ontology biological process (GOBP) terms representing significantly increased-in-abundance proteins.
(C) Hierarchical clustering of the 6790 proteins without missing values presented as low and high magnified heatmap. Cluster 1 captured the proteins that increased in abundance following ZIKV infection. Subcluster 1-1 consisted of proteins related to innate immune response and antigen presentation pathways (interaction network of subcluster 1-1 is shown in Figure S1G, results of hierarchical clustering analysis are shown in Table S3). Colored circle indicates each embryo shown in Figure 1F.
(D) The interaction network for the proteins significantly changed in abundance. Node color indicates protein-level fold change (log2(ZIKV/Mock)); edges represent high confidence interactions between proteins in the network based on protein-protein interaction information curated by the STRING database. Nodes outlined in blue indicate proteins associated with interferon signaling and innate immune response. Arrow-head node shape indicates proteins associated with neurogenesis and neurodevelopment (RNA level changes are shown in Figure S1H).
Hierarchical clustering analysis demonstrated a subset of proteins with significantly increased abundances in the ZIKV embryos (Cluster 1-1, Figures 2C and S1G, and Table S3). Of interest, the proteins in this subset were related to innate immune response and antigen processing and presentation, in accord with the GOBP analysis.
Amongst the enriched proteins, Signal Transducer and Activator of Transcription 1 (STAT1) (log2(ZIKV/Mock Fold Change) = 2.80, p = 7.52E-10), Bone Marrow Stromal Cell Antigen 2 (BST2) (log2(Fold Change) = 3.29, p = 9.06E-08), and Interferon (IFN)-Stimulated Gene 15 (ISG15) (log2(Fold Change) = 3.52, p = 2.45E-06) were the most significantly upregulated proteins (Figure 2C, Table S2, transcript level changes from RNA-Seq analysis are shown in Figures S1H and S1I), indicating ZIKV-induced activation of the IFN-induced Janus kinase (JAK)-STAT pathway (Figure 2D, blue-purple and green complex). We further captured elevated abundances of immunoproteasome complex subunits (PSMB8, 9, 10; Figure 2D, bright orange and PSME1, 2; orange), MHC-I peptide loading complex (TAP1, TAP2, TAPBP, B2M; yellow), MHC-I proteins (H2-D1, H2-K1; bright pink), and T cell and B cell signaling proteins (FCER1G, LCP1, and NFATC4; light pink). To summarize, the upregulation of the antigen presentation pathways was prominent.
ZIKV infection upregulates cytokine production downstream of the JAK/STAT pathway
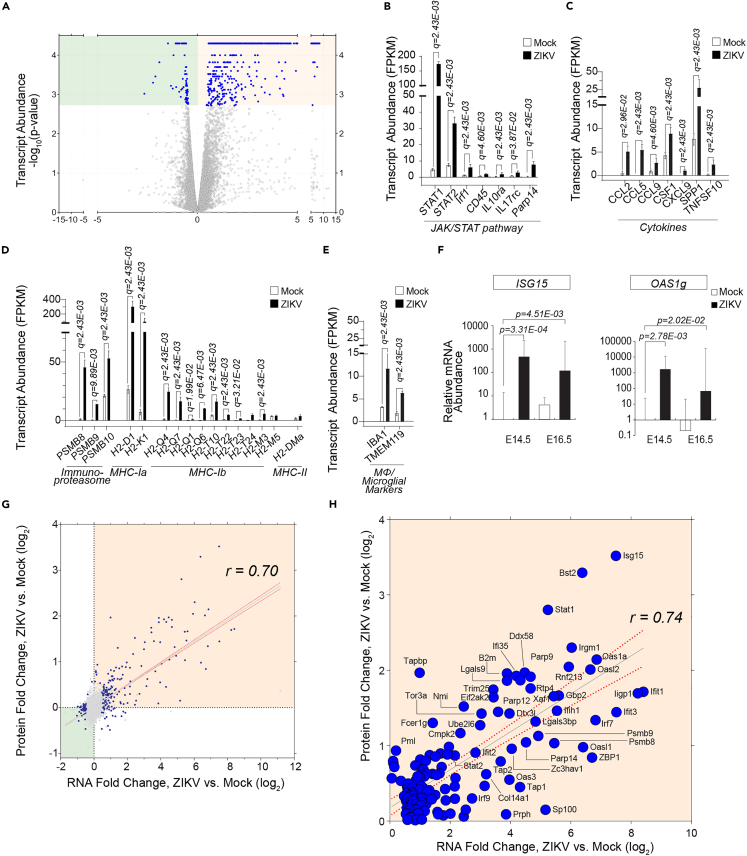
RNA-Seq data uncovered the disruption following ZIKV infection at the transcript level (560 significantly dysregulated transcripts; 491 upregulated and 69 downregulated; q value <0.05) at E14.5 (Figure 3A, Tables S4 and S5). The transcripts elevated in abundance included those involved in the IFN-induced JAK/STAT signaling pathway, such as STAT1, STAT2, IFN Regulatory Factor 1 (IRF1), and Protein Tyrosine Phosphatase Receptor Type C (Ptprc; codes CD45), which was in line with our findings in the proteomics dataset (Figures 3B and S2). In addition, we detected the elevation of cytokines induced by the JAK/STAT pathway activation, including C-C Motif Chemokine Ligand (CCL) 2, 5, 9, C-X-C Motif Chemokine Ligand (CXCL) 9; Colony Stimulating Factor (CSF) 1; Secreted Phosphoprotein (SPP) 1; and Tumor Necrosis Factor Superfamily Member (TNFSF) 10 (Figure 3C). We also detected increased abundances of transcripts involved in the antigen-presenting pathway, such as immunoproteasomes PSMB 8, 9, 10, MHC-Ia (H2-D1, H2-K1), and MHC-Ib (Figures 3D, S1H, and S1I), which was in line with the IHC staining results, showing overlapping expression of MHC-1 and PSMB 8 and 9, indicating antigen presentation via MHC-1 (Figures S3A and S3B). In addition, we captured elevated abundances of Allograft Inflammatory Factor 1 (IBA1) and Transmembrane Protein 119 (TMEM119) (Figure 3E) that indicated the recruitment of macrophages/microglia. The IHC results showed that IBA1 and MHC-1 signals were adjacent, raising a possibility that IBA1-positive cells are drawn to the region of MHC1 expression by cytokine expression either from dying infected cells or T cells activated by interactions with the MHC1 peptides (Figures S3C and S3D). Furthermore, these changes were accompanied by cleaved caspase3-positive cells indicating apoptosis within the infected brain (Figures S3E–S3G). We further conducted qPCR analysis which confirmed the increased mRNA expression level of ISG15 and OAS1g (Figure 3F).
Figure 3.
RNA-sequencing data captures interferon-induced cytokine production and macrophage/microglia infiltration in the embryonic brain
(A) Volcano plot of transcript expression (log2(ZIKV/Mock)) in ZIKV vs. Mock infected E14.5 brain tissues (n = 3 per condition) based on Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values calculated using Cufflinks/Cuffdiff. Significantly dysregulated genes are highlighted in blue (q < 0.05) (507 transcripts significantly increased; 74 transcripts significantly decreased; 581 significantly dysregulated transcripts in total). Transcripts significantly changed in abundances are listed in Tables S4 and S5.
(B) Mean relative transcript abundance (FPKM) of molecules in the Janus Kinase/Signal Transducer and Activator of Transcription (JAK/STAT) pathway based on RNA-Seq analysis. Transcript level changes mapped on interferon-induced pathways using Ingenuity Pathway Analysis are shown in Figure S2.
(C) Quantitative amount of cytokines shown in FPKM values.
(D) Quantitative amount of immunoproteasome and MHC-I and MHC-II molecules shown in FPKM values.
(E) FPKM values of macrophage/microglial markers.
(F) qPCR analysis of Interferon (IFN)-Stimulated Gene 15 (ISG15) and 2′-5′ Oligoadenylate Synthetase 1G (OAS1g).
(G) Correlation of infection-induced fold changes (log2(ZIKV/Mock)) at the protein and RNA level for 6,218 transcript-protein pairs. Dysregulated transcript-protein pairs (significant at RNA or protein level, FDR-corrected p < 0.05) are shown in blue. Transcript-protein pairs exhibiting decreased-in-abundance are highlighted in green (Quadrant III), whereas transcript-protein pairs exhibiting increased-in-abundance are highlighted in orange (Quadrant I). Quadrants II and IV are shown in Figure S4.
(H) Transcript-protein pairs with increased-in-abundance following ZIKV infection (Quadrant I of 4A). ClueGO network analysis of increased-in-abundance transcript-protein pairs is shown in Figure S1I. Error bars indicate standard deviation.
Comparison of RNA-level and protein-level dynamics of 6,218 transcript-protein pairs (Figures 3G, 3H, and 4A, Table S6) demonstrated the correspondence of transcript and protein abundances in our dataset (r = 0.70, Pearson correlation). We then filtered them for the transcript-protein pairs that showed a consistent increase in abundance, with changes that were significant at least in either the transcriptomic or proteomic dataset, resulting in 153 transcript-protein pairs that showed high correspondence of RNA-and protein-level expression increase (r = 0.74) (Figure 3H, Quadrant I of 3G, orange background). Most of the transcript-protein pairs were those involved in immune responses, as in line with our results when we analyzed proteomic and transcriptomic datasets individually.
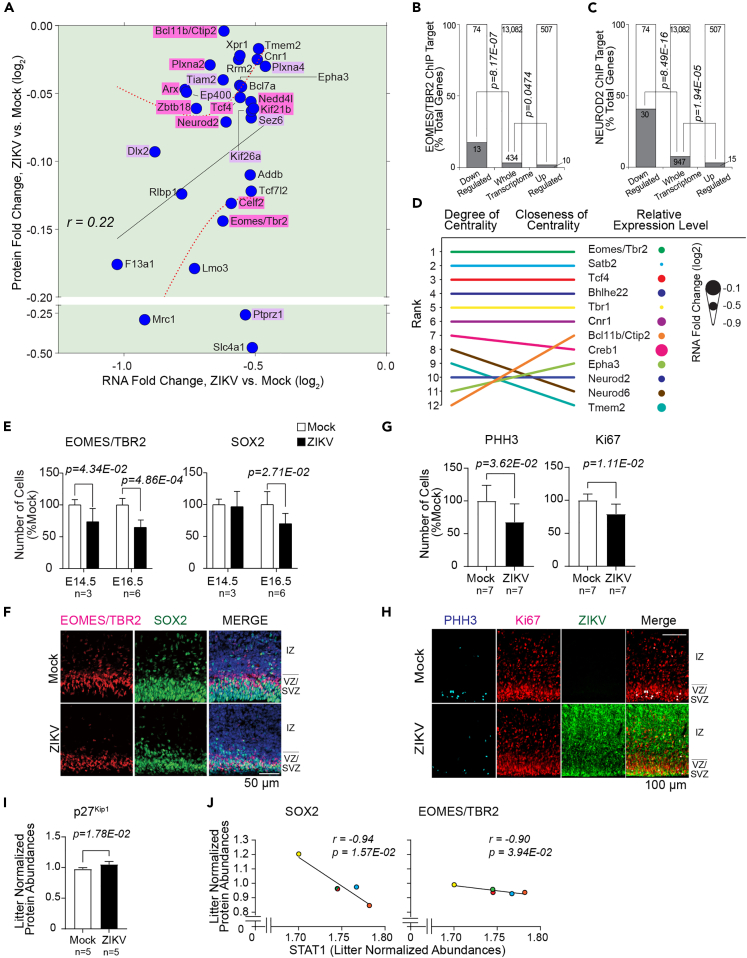
Figure 4.
ZIKV induces downregulation of critical neurodevelopmental proteins
(A) Transcript-protein pairs with decreased-in-abundance following ZIKV infection (Quadrant III of 3G). Pink; transcript-protein pairs that are known to be causative of genetic neurodevelopmental diseases in humans, Aristaless Related Homeobox (ARX), B Cell CLL/Lymphoma 11b (BCL11B or CTIP2), Eomesodermin/T-box Brain Protein 2 (EOMES/TBR2), Kinesin Family Member 21B (KIF21B), Neural Precursor Cell Expressed Developmentally Down-regulated 4-like E3 Ubiquitin Protein Ligase (NEDD4L), Neuronal Differentiation 2 (NEUROD2), Plexin A2 (PLXNA2), Transcription Factor 4 (TCF4), and Zinc Finger and BTB Domain Containing 18 (ZBTB18) (Table 2). Purple, transcript-protein pairs that are known to have neurodevelopmental functions in mice, Distal-Less Homeobox 2 (DLX2), E1A-Binding Protein P400 (EP400), Kinesin Family Member 26A (KIF26A), Plexin A4 (PLXNA4), Protein Tyrosine Phosphatase Receptor Type Z1 (PTPRZ1), Seizure Related 6 Homolog (SEZ6), and T-lymphoma invasion and metastasis-inducing protein 2 (TIAM2) (Table S7 for the functions of the highlighted transcript-protein pairs).
(B) Eomesodermin/T-box Brain Protein 2 (EOMES/TBR2) ChIP target enrichment within the down- and up-regulated subsets of the ZIKV-responsive transcriptome, relative to EOMES/TBR2 ChIP targets present in the whole transcriptome dataset; hypergeometric test p values are shown.
(C) Neuronal Differentiation 2 (NEUROD2) ChIP target enrichment within the down- and up-regulated subsets of the ZIKV-responsive transcriptome, relative to NEUROD2 ChIP targets present in the whole transcriptome dataset; hypergeometric test p values are shown.
(D) Degree and closeness of centrality and relative RNA expression level of interconnected nodes in NEUROD2/TBR2 Signaling Network. The top 10 nodes of either degree or closeness of centrality are shown.
(E) Quantification of EOMES/TBR2-positive and Sex Determining Region Y-Box 2 (SOX2)-positive cells at E14.5 and E16.5. E14.5; mock, n = 3 from 2 litters, ZIKV, n = 3 from 2 litters. E16.5; mock, n = 6 from 4 litters, ZIKV, n = 6 from 5 litters.
(F) IHC staining of VZ/SVZ and IZ of Mock- or ZIKV-infected embryonic brains, EOMES/TBR2 (red), SOX2 (green), and Hoechst (nucleus, blue), scale bar = 50 μm.
(G) Quantification of phosphoH3 (PHH3)-positive and Ki67-positive cells at E16.5; mock, n = 7 from 4 litters, zika, n = 7 from 6 litters.
(H) IHC staining of IZ and VZ/SVZ of Mock- or ZIKV-infected E16.5 brains. Mitotic marker phosphorylated Histone H3 (PHH3, cyan) and proliferation marker Ki67 (red), counterstained with ZIKV (green), Scale bar = 100 μm.
(I) Litter-normalized abundance of Cyclin-dependent Kinase Inhibitor 1B (p27Kip1) protein at E14.5.
(J) Correlation of litter-normalized abundances of neurodevelopmental genes and STAT1 abundances. Error bars indicate standard deviation.
ZIKV infection inhibits cell cycle progression and downregulates neurodevelopmental signaling cascade
We next analyzed the 30 transcript-protein pairs that showed a consistent decrease in abundances, with changes that were significant at least in either the transcriptomic or proteomic dataset. Among those transcript-protein pairs, 10 of them had been reported to cause neurodevelopmental disease following Mendelian inheritance in humans that show phenotypes overlapping with congenital Zika syndrome (Figure 4A, Quadrant III of 3G, green background, pink highlight; Table 2). These were Aristaless Related Homeobox (ARX), B Cell CLL/Lymphoma 11b (BCL11B or CTIP2), CUGBP Elav-like Family Member 2 (CELF2), Eomesodermin/T-box Brain Protein 2 (EOMES/TBR2), Kinesin Family Member 21B (KIF21B), Neural Precursor Cell Expressed Developmentally Down-regulated 4-like E3 Ubiquitin Protein Ligase (NEDD4L), Neuronal Differentiation 2 (NEUROD2), Plexin A2 (PLXNA2), TCF4, and Zinc Finger and BTB Domain Containing 18 (ZBTB18) (Table 2). In line with our findings, all except for KIF21B were reported to be loss-of-function mutations in humans (Table 2).
Table 2.
Downregulated transcript-proteins in the ZIKV-infected brains having neurodevelopmental phenotypes in humans that overlap congenital Zika syndrome
| Mouse gene symbol | Human gene symbol | Principal neurological manifestations | Effects of reported mutations | Microcephaly | References |
|---|---|---|---|---|---|
| Arx | ARX | Lissencephaly, hydranencephaly, epileptic encephalopathy, intellectual disability | Loss of function | (++) | Bienvenu et al. Kitamura et al. Kato et al.26,27,28 |
| Bcl11b | BCL11B | Intellectual disability, global developmental delay | Loss of function | (+) | Lessel et al.29 |
| Celf2 | CELF2 | Develpmental and epileptic encephalopathy, intellectual disability, autistic features | Loss of function | (−) | Gillin et al. Halgren et al. Itai et al.30,31,32 |
| Eomes/Tbr2 | EOMES | Microcephaly, polymicrogyria, callosal agenesis | Loss of function | (++) | Baala et al.33 |
| Kif21b | KIF21B | Intellectual disability | Gain of function | (+) | Asselinet al.34 |
| Nedd4l | NEDD4L | Periventricular nodular heterotopia, seizures, developmental delay | Loss of function | (+) | Broix et al.35 |
| Neurod2 | NEUROD2 | Epileptic encephalopathy | Loss of function | (+) | Segaet al.36 |
| Plxna2 | PLXNA2 | Intellectual disability | Loss of function | (−) | Altuame et al.37 |
| Tcf4 | TCF4 | Intellectual disability, hypotonia, stereotypic movements | Loss of function | (++) | Amiel et al. Zweier et al.38,39 |
| Zbtb18 | ZBTB18 | Global developmental delay, callosal hypoplasia | Loss of function | (++) | de Munnik et al.40 |
List of transcript-protein pairs downregulated in the ZIKV-infected brains (Figure 4A, highlighted in pink) that have been previously reported to be causative genes of neurodevelopmental phenotypes in humans, including microcephaly. (-), rarely or not reported; (+), sometimes reported; (++), frequently reported. Note that the majority of the mutations were loss of function mutations.
Furthermore, 7 additional downregulated transcript-protein pairs were reported to have critical functions in the developing brain (Figure 4A, Quadrant III of 3G, green background, purple highlight; Table S7), including Distal-Less Homeobox 2 (DLX2), E1A-Binding Protein P400 (EP400), Kinesin Family Member 26A (KIF26A), Plexin A4 (PLXNA4), Protein Tyrosine Phosphatase Receptor Type Z1 (PTPRZ1), Seizure Related 6 Homolog (SEZ6), and T-lymphoma invasion and metastasis-inducing protein 2 (TIAM2).
Moreover, several neurodevelopmental transcript-protein pairs, including TBR1 and Special AT-Rich Sequence-Binding Protein 2 (SATB2), were downregulated in transcript level but not in protein level, which may have reflected the long half-life of these proteins (Figure S4; Quadrants II and IV of Figure 3G).
To investigate if the downregulation of these neurodevelopmental regulators is causing the expected downstream effects in their regulatory pathways, we examined transcripts altered in abundance after ZIKV infection based on a previous genome-wide ChIPseq analysis of EOMES/TBR2-and NEUROD2-(downregulated in our dataset, shown in Figure 4A, Quadrant III of 3G) bound chromatin from E14.5 mouse cortices.41,42 In the ZIKV-infected brains, 18% of transcripts decreased in abundance were EOMES/TBR2-responsive (a 5.3-fold over-representation, p = 8.17E-07) compared to 3% within the whole transcriptome (Figure 4B, Table S8). Similarly, 40% of transcripts decreased in abundance were NEUROD2-responsive (a 5.5-fold over-representation, p = 8.49E-16) compared to 7% within the whole transcriptome (Figure 4C). To characterize downstream consequences of the NEUROD2-EOMES/TBR2 signaling cascade, we assembled a network of the NEUROD2-and EOMES/TBR2-responsive genes perturbed in transcript expression and determined connectivity among other downregulated transcripts. Among the dysregulated transcripts, EOMES/TBR2 was the highest scoring network node in terms of both degree (number of links incident on node, in this case, the neurodevelopmental genes) and closeness centrality (the average shortest path between nodes), followed by SATB2, TCF4, Basic-Helix-Loop-Helix Family Member E22 (BHLHE22), TBR1, and CTIP2 (Figure 4D), highlighting EOMES/TBR2 as a key ZIKV-perturbed molecule relevant to embryonic brain development.
In line with the transcript and protein level results, histology confirmed the decreased numbers of EOMES/TBR2-positive cells in the subventricular zone (SVZ) of the cerebral wall on E14.5 and E16.5, as well as Sex Determining Region Y-Box 2 (SOX2)-positive cells in the ventricular zone (VZ) at E16.5 (Figures 4E and 4F). In addition, the number of phospho-histone H3 (PHH3)-positive and antigen Ki67-positive cells were decreased in the ZIKV-infected brain by 32.3% (n = 7, p = 3.62E-02) and 20.7% (n = 7, p = 1.11E-02) respectively, as compared with the Mock-infected brains at E16.5 (Figures 4G and 4H). Moreover, cyclin-dependent kinase inhibitor 1B (p27Kip1) was increased in abundance in our proteomics dataset by 7.9% (n = 5, p = 1.78E-02) in the ZIKV-infected brain when the Mock- and ZIKV-infected pairs were matched for littermates (Figure 4I). Together, these findings indicate that ZIKV infection inhibited NPC cell cycle progression and proliferation, thus diminishing the NPC pool.
Of interest, STAT1 protein expression level was negatively correlated with SOX2 (r = −0.94, p = 1.57E-02) and EOMES/TBR2 (r = −0.90, p = 3.94E-02), when compared within the litter matched pairs (Figure 4J), suggesting a correlation between the JAK/STAT pathway activation and NEUROD2-EOMES/TBR2 signaling cascade suppression following ZIKV infection.
Discussion
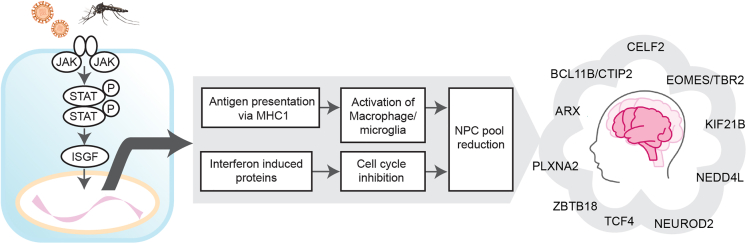
The integrated quantitative proteomic and RNA-Seq analyses conducted in the in vivo immunocompetent model of ZIKV infection provides data supporting a mechanistic model that induces microcephaly via the activation of JAK/STAT pathway leading to (1) the activation of antigen presentation programs that likely induced cell death and phagocytosis of ZIKV-infected NPCs by macrophages/microglia and (2) enhanced translation of IFN-induced proteins that suppress NPC proliferation, which together decrease the NPC pool size and neuronal output that subsequently structure a small brain (Figure 5).
Figure 5.
Model of microcephaly and neurodevelopmental impairment because of congenital ZIKV infection
JAK/STAT pathway activation decreases the NPC population in the developing brain via (1) activation of antigen presentation pathway leading to engulfment of the infected cells by macrophage and (2) increased abundances of interferon-induced proteins leading to cell-cycle arrest. Integrated proteomics and RNA-Seq analysis showed a comprehensive view of multiple neurodevelopmental proteins being downregulated by ZIKV infection in a fully immunocompetent mouse model.
Activated innate immunity via JAK/STAT pathway
Our proteomic and transcriptomic datasets showed a ZIKV-induced activation of the immune system in the murine embryonic brain, with the IFN-induced JAK/STAT pathway being the most robustly activated pathway (e.g., STAT1, 2, ISGF3). This was consistent with the increased expression of downstream transcription factors (e.g., ISG, OAS), chemokines (e.g., CCL2, CCL4, CXCL10), and IFN regulatory factors (IRFs, e.g., IRF7, IRF9), accompanied by the induction of the immunoproteasome-mediated antigen presentation pathway43 which likely results in apoptosis followed by activation of the macrophage/microglia. A previous study has shown that microglia are involved in phagocytosis and extracellular digestion during Zika virus encephalitis in 6-week-old immunodeficient mice.44 Although the macrophage/microglia are immature in the embryonic brain,45 the overlapping signals of IBA1 and MHC1 observed in our model Figures S3C and S3D) suggest that the activated macrophage/microglia have engulfed the apoptotic antigen-presenting infected cells and possibly contributed to the decrease of the neuronal population. Furthermore, the increased expression of caspase 8 and TNFR abundance at the RNA level (Table S2) indicated induction of TNF-induced apoptosis. This implication is supported by the increase in the number of cleaved caspase 3-positive cells in the brain which is mainly localized in the intermediate zone (Figures S3E–S3G). In addition, the IHC results showed that the number of SOX2-positive and TBR2-positive NPCs46,47 were decreased in the ZIKV-infected embryos (Figure 4E).
Taken together, our unbiased comprehensive approach implied that ZIKV targets NPCs that are SOX2-and/or TBR2-positive by activating the JAK/STAT pathway and antigen presentation pathway, leading to the engulfment of those NPCs by macrophages, thus reducing the proliferative population. Furthermore, our results revealed the induction of apoptosis in the intermediate zone, indicating multiple mechanisms involved in the pathophysiology of ZIKV microcephaly. However, further targeted studies are required to confirm the causal connections suggested here.
Cell cycle inhibition by JAK/STAT pathway
A major advantage of applying proteomics to in vivo models of viral infections is that direct measurements of viral proteins are possible in the context of the whole organism instead of looking at cellular level responses as in in vitro systems, thereby enabling the study of the dose-response relationships of the whole animal host proteins to virus titers. This was exemplified in our proteomics dataset, where cell cycle regulatory proteins show a negative correlation with the ZIKV polyprotein abundances (Figures 1G and 1H, Table 1). During neurodevelopment, the escape of the NPCs from the cell cycle is regulated by the expression of cell cycle regulatory proteins. Specifically, the balance between proteins that promote the cell cycle (e.g., CDK2, CDK4, cyclinD1) and inhibit the cell cycle (e.g., p27Kip1) control the numbers of NPCs and hence the endpoint number of neurons and the size of the brain.48,49,50,51 We observed a negative correlation between the abundance of cell cycle progressing proteins and ZIKV polyprotein (Figure 1H). Indeed, our quantitative proteomic analysis revealed that the amount of p27Kip1 was increased in the ZIKV-infected brain (Figure 4I). Furthermore, IHC results showed that the number of PHH3- and Ki67-positive proliferative cells were reduced in the VZ/SVZ of the ZIKV-infected embryos (Figures 4G–4H). Together, these results confirmed that ZIKV infection had suppressed the proliferation of NPCs.
Of interest, IFN has been reported to inhibit the cell cycle progression through the induction of cyclin-dependent kinase inhibitor 1 (p21Cip1/Waf1) and p27Kip1 in cultured cells.52,53 In the adult mouse brain, IFNγ suppresses the proliferation of NPCs in the SVZ by inducing STAT1, thus leading to the decrease of adult neurogenesis from the SVZ niche.54 In the developing embryonic brain, several studies have pointed out that the IFN-induced pathways have a direct biological role in neuronal development. STAT3 controls the transition of NPCs from SOX2-positive to TBR2-positive cells in mice,55 and a positive autoregulatory loop of JAK/STAT signaling controls the onset of astrogliogenesis.56 More recently, it has been reported that IFNγ treatment resulted in neurite outgrowth of neurons, accompanied by JAK/STAT pathway activation.57
To summarize, these findings showed that NPC proliferation was inhibited in the ZIKV-infected brain, accompanied by altered abundances of host cell cycle regulatory proteins. Based on previous evidence discussed above53,58,59 the underlying mechanisms could be explained by the induction of the IFN-induced proteins downstream of the JAK/STAT pathway. In combination with apoptosis and the engulfment of the NPCs by macrophages, such cell-cycle arrest would reduce the NPC population, thus leading to the reduced neuronal output and resulting in smaller brain size of the embryo.
Transcript-protein changes reflect the microcephaly phenotype
Our integrated proteo-transcript approach captured a spectrum of neurodevelopmental genes that were downregulated in the ZIKV-infected brain. Of note, these downregulated genes were significantly overrepresented among the genes known to be downstream of EOMES/TBR2 and NEUROD2 (Figures 4B and 4C). In addition, the network analysis identified EOMES/TBR2 as a key transcription factor with the highest degree and closeness of centrality among the downregulated genes (Figure 4D). EOMES/TBR2 is a critical regulator of neurogenesis in the SVZ required for differentiation and functional organization of upper cortical layer neurons.60 A previous study reported that ZIKV infection suppresses the transition of Pax6-positive NPCs in the VZ to TBR2-positive NPCs in the SVZ.45 Our results not only showed that the number of EOMES/TBR2-positive cells was decreased in the ZIKV-infected brain, but also demonstrated the prominent downstream effects of EOMES/TBR2 downregulation in ZIKV-infected brain, suggesting a critical role of this pathway in the pathogenesis of microcephaly in congenital ZIKV syndrome.
The second highest ranking gene in terms of degree and closeness of centrality was SATB2, which was decreased in abundance in the ZIKV-infected cortical plate where NEUROD2 is expressed in the post-mitotic neurons.61 NEUROD2 acts in the upstream of SATB261 and is associated strongly with its promoter region.41 In contrast to EOMES/TBR2, NEUROD2 has prominent roles in post-mitotic neurons, including axonal fasciculation and targeted axogenesis61 and synaptic maturation.62 Although EOMES/TBR2 and NEUROD2 seem to act mostly in distinct stages of cerebral cortical development, interestingly, one of the downregulated genes TCF4 is identified as a potential target of both of these transcription factors.61 This is consistent with the reported wide range of abnormalities in TCF4 mutant mice, from cell fate specification, neuronal migration, and dendrite and synapse formation.63 In total, our results suggest that, in the ZIKV-infected brain, disturbances of multiple but defined molecular pathways occur over the course of cerebral cortical development, with EOMES/TBR2 and NEUROD2 as key regulatory molecules. This likely underlies the pleomorphic neurological phenotype of congenital Zika syndrome that poses challenges in diagnosis and treatment.
Furthermore, the non-biased approach identified a broader range of neurodevelopmental genes previously not described in congenital ZIKV infection. These novel proteins were likely identified because of the robustness of the multi-omics analysis pipeline in combination with an immune-competent congenital ZIKV infection model. Among the 30 downregulated protein-transcript pairs detected, 10 were known to be causative genes of neurodevelopmental disorders in humans (ARX, BCL11B, CELF2, EOMES, KIF21B, NEDD4L, NEUROD2, PLXNA2, TCF4, ZBTB18; Figure 4A and Table 2). Though their phenotypes vary, many are associated with microcephaly and some are associated with brain malformations, such as hydranencephaly, lissencephaly and polymicrogyria, which are also seen in congenital Zika syndrome, supporting their contribution in the pathogenesis (Table 2). Furthermore, additional 7 downregulated genes were reported to have neurodevelopmental functions in mice (Table S7), underscoring the enrichment of neurodevelopmental genes and proteins in our downregulation dataset. Our results indicate a more widespread dysregulation of the developmental program of the brain than previously recognized, which may help explain the complex and varied brain malformations seen in individuals with congenital Zika syndrome. Moreover, we observed other neurodevelopmental genes that were decreased in transcript level but not in protein at E14.5 (e.g., SATB2, TBR1, NEUROD6, and DLX1). The number of SATB2- and TBR1-positive cells decreased at E16.5 (Figures 1I and 1J), and the discrepancy can reflect the long half-lives of these proteins. Taken together, our results indicate a more widespread dysregulation of the developmental program of the brain than previously recognized, which may help explain the complex and varied brain malformations seen in individuals with congenital Zika syndrome.
Integrative proteomics and transcriptomics approach to study ZIKV and other infections
Systems biology strategies such as proteomics and RNA-Seq have recently been applied to help uncover complex host-pathogen interactions. For example, transcriptomic approaches have uncovered the inflammatory host response to infection that produces pro-inflammatory cytokines and effector defense mediators via a massive gene reprogramming in host immune cells.64 However, the expression levels of transcripts and proteins can differ because of various factors such as different half-lives, post-transcriptional and translation mechanisms.9,65 Thus, the combination of transcriptomic and proteomic approaches enables a cross-referencing of findings. An in vitro study on Adenovirus type 2 has reported a low correlation between RNA- and protein-level expression of downstream genes of immune pathways during the late phase of infection (>24 h), thus highlighting the role of post-translational mechanisms.66 We improve the overall correspondence of transcript and protein abundances with relatively high statistical correlation (r = 0.70, Pearson correlation; Figure 3G), especially for the transcript-protein pairs that were increased in abundance (r = 0.74; Figure 3H), reflecting the robustness of the innate immunity activation in the embryonic brain 4 days after ZIKV exposure.
Mice are more resistant to ZIKV infection than humans as ZIKV NS5 protein has a limited ability to bind to mouse STAT2 and thus cannot antagonize the IFN response.67 In this study, however, we successfully utilized a well-established intra-placental injection model with a fully immunocompetent mouse strain23,24 which enabled us to profile the systemic immune response of the embryo that cannot be captured in immunodeficient mouse models or in in vitro studies. Moreover, we used CD-1 mice that have been repeatedly used to study embryonic neurodevelopment, hence the process of neuronal production has been described in detail.48,51 Furthermore, by matching the embryos derived from the same mother, we captured the correlation between the JAK/STAT pathway and the neurodevelopmental gene expression on E14.5; as the expression levels of such genes are highly variable in unmatched maternal progeny because of the dynamic shift of proliferation to differentiation in the NPCs (Figure 4J). One potential limitation of this model is that it entails the surgical manipulation of the mother that could potentially affect maternal immunity and embryonic brain development. This is unlikely, however, as the survival rate of the Mock-infected embryos was not reduced (Figure S1A). Moreover, hierarchical clustering analysis suggested that quantities of most proteins did not change in the ZIKV-infected brains as compared with the Mock-infected brains, indicating that only specific pathways were disturbed after the infection (Figure 2C). Alternative mouse models have been proposed, including the ZIKV infection model using humanized STAT2 mice68,69; however, neither the STAT-dependent regulation of the NPCs nor the effect of the induction of humanized STAT2 on the murine neurodevelopment have been described.
In summary, through an integrated multi-omics systems biology study design, we identified changes in innate immune and developmental pathway genes and proteins in an immunocompetent congenital ZIKV infection mouse model, many of which were not captured in prior studies using in vivo or in vitro models. This combination of a congenital infection model and multi-omics systems analysis provides important insights into the underlying mechanisms of neurodevelopmental defects where the immune system plays an essential role.
Limitations of the study
In this study, we used the placental injection model of congenital ZIKV infection. This model enabled us to study gene and protein expression changes in a wild-type, immunocompetent system to mimic the infection via the placenta; however, infection was established by a non-physiological injection of the virus. We studied a single time point for ZIKV injection and harvesting embryos for RNA-seq and mass spectrometry. Therefore, we do not have information regarding dynamic changes of gene and protein expression over time, which would be one of the focuses in our future studies. Finally, RNA-seq and mass spectrometry were performed using bulk brain tissue, and cell type-specific effects of ZIKV on gene and protein expression could not be dissected, which is also of interest for future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Activated-Caspase3 | CST | Cat#9661; RRID: AB_2341188 |
| Ctip2 | Abcam | Cat# ab18465; RRID: AB_2064130 |
| Ki67 | CST | Cat#12202; RRID: AB_2620142 |
| phospho-histone H3 | Sigma-Aldrich | Cat# H9908; RRID: AB_260096 |
| SOX2 | Santa Cruz | Cat# sc-17320; RRID: AB_2286684 |
| TBR1 | Abcam | Cat# ab31940; RRID: AB_2200219 |
| TBR2 | Abcam | Cat# ab183991; RRID: AB_2721040 |
| SATB2 | Bethyl Laboratories | Cat# A301-864A; RRID: AB_1309783 |
| ZIKV | Kerafast | Cat#EVU302 |
| PSMB9/LMP2 | Abcam | Cat#ab3328; RRID: AB_303707 |
| PSMB8/LMP7 | Abcam | Cat#ab3329; RRID: AB_303708 |
| MHC-I (rat) [ER-HR 52] | Abcam | Cat#ab15681; RRID: AB_302030 |
| MHC-I/HLA-ABC | Thermo Fisher | Cat#MA5-11723; RRID: AB_10985125 |
| NeuroD2 | Abcam | Cat#ab104430; RRID: AB_10975628 |
| IBA1 | Wako | Cat#019-19741; RRID: AB_839504 |
| Activated-Caspase3 | CST | Cat#9661S; RRID: AB_2341188 |
| Bacterial and virus strains | ||
| ZIKV strain HS-2015-BA-01 | Gift of Dr. Mauro M. Teixeira | GenBank Acc. No. KX520666 |
| Chemicals, peptides, and recombinant proteins | ||
| Fast Blue B Salt | Sigma-Aldrich | D9805 |
| PureLink RNA Mini Kit | Invitrogen | 12183018A |
| TRIzol | Invitrogen | 15596018 |
| Turbo DNA-free Kit | Invitrogen | AM1907 |
| mMESSAGE mMACHINE T7 ULTRA Transcription Kit | Ambion | AM1345 |
| RediPlate RiboGreen RNA Quantification Kit | Invitrogen | R32700 |
| agarose gel | Benchmark Scientific | A1801-LM |
| BSA | Sigma-Aldrich | A9418 |
| Triton-X-100 | Sigma-Aldrich | T9284 |
| Sodium azide | Sigma-Aldrich | S2002 |
| PBS | Invitrogen | AM9625 |
| Fluoromount-G | Southern Biotech | 0100-01 |
| Hoechst 33342 | Thermo Fisher Scientific | H3570 |
| TruSeq RNA Kit v2 | Illumina | RS-122-2001 |
| RIPA buffer | Thermo Fisher Scientific | 89901 |
| SDS | Sigma-Aldrich | L4509 |
| protease inhibitor cocktail | Roche | 5892791001 |
| phosphatase inhibitor | Roche | 4906827001 |
| Pierce™ Trypsin Protease, MS Grade | Thermo Pierce | 90057 |
| 10plex isobaric tags (TMT) | Thermo Fisher Scientific | 90110 |
| Deposited data | ||
| ProteomeXchange/PRIDE proteomics data repository | www.proteomexchange.org | ProteomeXchange Accession #: PXD040629 |
| MassIVE data repositories | https://massive.ucsd.edu/ | MassIVE Accession #: MSV000091417 |
| Gene Expression Omnibus (GEO Accession: GSE118935) | www.ncbi.nlm.nih.gov/geo/ | GEO Accession #: GSE118935 |
| Oligonucleotides | ||
| CH25H | Thermo Fisher Scientific | Cat#Mm00515486_s1 |
| EOMES | Thermo Fisher Scientific | Cat#Mm01351984_m1 |
| GAPDH | Thermo Fisher Scientific | Cat#Mm999999915_g1 |
| IFNB1 | Thermo Fisher Scientific | Cat#Mm00439552_s1 |
| MX1 | Thermo Fisher Scientific | Cat#Mm00487796_m1 |
| MX2 | Thermo Fisher Scientific | Cat#Mm00488995_m1 |
| ISG15 | Thermo Fisher Scientific | Cat#Mm01705338_s1 |
| NEUROD2 | Thermo Fisher Scientific | Cat#Mm00440465_g1 |
| NEUROD6 | Thermo Fisher Scientific | Cat#Mm01326464_m1 |
| Neurog2 | Thermo Fisher Scientific | Cat#Mm00437603_g1 |
| OAS1G | Thermo Fisher Scientific | Cat#Mm01730198_m1 |
| SOX2 | Thermo Fisher Scientific | Cat#Mm03053810_s1 |
| ZBTB18 | Thermo Fisher Scientific | Cat#Mm02343862_s1 |
| Software and algorithms | ||
| Zen lite software | Carl Zeiss Microscopy | |
| Adobe Creative Cloud software | Adobe Systems | |
| Adobe Illustrator | Adobe Systems | |
| Adobe Photoshop | Adobe Systems | |
| BioRender | BioRender | https://biorender.com/ |
| Image J software | NIH | https://imagej.nih.gov/ij/download.html |
| FastQC 0.11.3 | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Bowtie 2.2.4 (Tuxedo Suite) | Langmead et al.70 | https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.2.4/ |
| Tophat 2.1.1 (Tuxedo Suite) | Kim et al.71 | https://ccb.jhu.edu/software/tophat/index.shtml |
| Cufflinks 2.2.1 (Tuxedo Suite) | Trapnell et al.72 Trapnell et al.73 | https://github.com/cole-trapnell-lab/cufflinks |
| Picard | Broad Institute | https://broadinstitute.github.io/picard/ |
| Proteome Discoverer 2.1 | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/OPTON-30795 |
| Mascot v2.6 | Matrix Science, Perkins et al.74 | http://www.matrixscience.com/ |
| ProteoWizard | Chambers et al.75 | http://proteowizard.sourceforge.net/faq.shtml |
| Ingenuity Pathway Analysis (IPA) | Qiagen Bioinformatics | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Cytoscape | Shannon et al.76 | http://www.cytoscape.org/ |
| ClueGO | Bindea et al.77 Bindea et al.78 | http://apps.cytoscape.org/apps/cluego |
| String v10.5 | Szklarczyk et al.79 | https://string-db.org/ |
| Panther | Mi et al.80 | http://www.pantherdb.org/ |
| Perseus | http://www.coxdocs.org/doku.php?id=perseus:start | |
| Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Rstudio/R (v. 3.4.3) | Rstudio | https://cran.r-project.org/bin/windows/base/old/3.4.3/ |
| R v4.0.4 | R-project | https://cran.r-project.org/bin/windows/base/old/4.0.4/ |
| tidyverse v1.3.1 (R package) | CRAN | https://www.tidyverse.org/ |
| gridExtra v2.3 (R package) | CRAN | https://cran.r-https://cran.r-project.org/web/packages/gridExtra/index.html |
| ggdendro v0.1.22 (R package) | CRAN | https://cran.r-project.org/web/packages/gridExtra/index.html |
| reshape2 v1.4.4 (R package) | CRAN | https://cran.r-project.org/web/packages/reshape2/index.html |
| svglite v2.0.0 (R package) | CRAN | https://cran.r-project.org/web/packages/svglite/index.html |
| Other | ||
| 10 kDA MWCO Microcon spin filters | Millipore | MRCPRT010 |
| NEST C18 spin column | Nest Group | NC0194358 |
| OASIS HLB 1cc Vac Cartridges (10mg sorbent, 30μm) | Waters Corporation | 186000383 |
| Xbridge BEH C18 column (130Å, 3.5μm, 2.1mm x 100mm) | Waters Corporation | 186003022 |
| Immobiline DryStrip gels (pH 3-10, 24 cm) | GE Healthcare | 17-6002-44 |
| PicoChipGT column (BEH C18, 1.7 μm, 130A, 150μm x 100mm) | New Objectvie | 2PCH15015-100H111-FS25-GT |
| Zeiss LSM700 confocal microscope | Carl Zeiss Microscopy | |
| Axio Imager M2 | Carl Zeiss Microscopy | |
| CCD AxioCam MR | Carl Zeiss Microscopy | |
| Bioanalyzer | Agilent | |
| HiSeq 2500 Sequencing System | Illumina | |
| Pellet Pestle Motor | Kimble | |
| Infinity HPLC 1260 system | Agilent | |
| 3100 OFFGEL system | Agilent | |
| Eksigent nanoLC425 system | Sciex | |
| QExactive HF mass spectrometer | Thermo Fisher Scientific | |
Resource availability
Lead contact
Lead information and requests related to mouse model generation are best directed to and will be fulfilled by Ganeshwaran H. Mochida (ganesh.mochida@childrens.harvard.edu), and those related to multi-omics and systems analysis are best directed to Judith A. Steen (Judith.Steen@childrens.harvard.edu).
Materials availability
This study did not generate new unique reagents, mouse lines, or virus lines.
Experimental model and subject details
Ethics statement
The experiments in this study were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Mouse protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at Boston Children’s Hospital. All surgical procedures were conducted under anesthesia with isoflurane. To minimize pain following surgery, animals were administered Meloxicam. All efforts were made to minimize the number of animals required. Viral infections were performed under biosafety level 2 conditions.
Virus preparation
Viral infections were performed using the ZIKV strain HS-2015-BA-01, isolated from a viremic patient with symptomatic infection in Bahia State, Brazil, in 2015. The HS-2015-BA-01 genome is available at GenBank (accession no. KX520666). Viruses were propagated and titered, as previously described.81 ZIKV stock passage numbers are < 5. The virus isolate (a gift from Dr. Mauro Teixeira) was amplified in C6/36 cells and titered by plaque assay to generate a seed virus. Viruses for experimental use were generated from seed stocks by infecting insect C6/36 cells at a multiplicity of infection (MOI) of 0.3, followed by incubation for 96 hours. Cell supernatants were collected, filtered, and stored at -80°C. One day after freezing the virus, the titer was determined by plaque assay using a thawed aliquot.
Mouse experiments
Timed-pregnant adult CD1 mice (Charles River Laboratories) were maintained under a 12-hour light/dark schedule to acclimate for > 48 hours. Plug dates were considered to be E0.5. Pregnant CD-1 mice at specific embryonic dates were anesthetized continuously with isoflurane, and laparotomy was performed for ZIKV injections. Intrauterine placental injection of ZIKV was conducted by modifying previous ZIKV24 and CMV82 infection models. According to methods previously reported,82,83 105 flow-infectious units (FIU; similar to the plaque-forming unit (PFU)) of ZIKV suspended in 1 μl PBS were injected through the uterine wall into the labyrinth region in 50% of placentas for each pregnant mouse. The remaining 50% of placentas were injected with PBS only. To improve manipulation, 0.1% Fast Blue B Salt Dye (Sigma-Aldrich; D9805) was used to visually monitor the diffusion of solution into embryonic villi in the placenta.
Embryos of both sexes were harvested at E13.5-E16.5 time points depending on the projects. The total numbers of viable and non-viable embryos were counted for each dam. Embryonic viability was determined as the number of viable embryos divided by the total number of injected embryos in each group. The survival rates of the embryos within each litter were 63.2% (n = 6 litter) and 24.5% (n = 18 litter) in the ZIKV-infected embryos at E14.5 and E16.5, respectively, as compared with 93.3% (n = 6 litter) and 95.2% (n = 17 litter) in the Mock-infected embryos (Figure S1A). Embryos and dissected brains were photographed and weighed for analysis. The supratemporal region of the brains was collected for histological analysis. The cerebellum and adjacent brain stem were dissected for quantifying ZIKV infection by qRT-PCR. For RNA-seq and target qRT-PCR analysis, whole brains were harvested to collect total RNA.
RNA isolation and qRT-PCR for ZIKV quantification
Total RNA was extracted using PureLink RNA Mini Kit (Invitrogen) combined with TRIZOL purification and treated with Turbo DNA-free Kit (Invitrogen). ZIKV RNA copies were quantified by quantitative reverse transcriptase-PCR (qRT-PCR). Primers and probe sets were designed according to a previously reported ZIKV1162c set.84 A standard RNA covering the probe-target was synthesized from a DNA template by mMESSAGE mMACHINE T7 ULTRA Transcription Kit (Ambion) and quantified by RiboGreen RNA Quantification Kit (Invitrogen). For samples in which no qRT-PCR signal was detected, zero values were replaced with half the minimum experimentally observed value to allow log transformations. ZIKV copy number for each brain was determined relative to a standard curve from 10-fold serial dilutions of a ZIKV RNA standard. RNA copy number was calculated as previously described.85 For downstream morphometric and immunohistochemical analysis, we divided the groups into ZIKV-injected and Mock-injected and did not take into account the results of ZIKV qRT-PCR to avoid introducing biases. For and transcriptomic and proteomic analyses, Mock-injected control embryos unambiguously negative for ZIKV qRT-PCR were selected.
Method details
iFASP digestion and TMT labeling
Brain tissues from 5 litter-matched ZIKV- and Mock-infected embryos were individually lysed in RIPA buffer + 0.02% SDS supplemented with protease inhibitor cocktail and phosphatase inhibitors followed by incubation at 98°C for 2 minutes, followed by mechanical disruption using a Kimble dounce homogenizer. Protein digestion and TMT labeling were carried out using the iFASP method, as previously described.86 Briefly, 100 μg of protein lysate were incubated in 8M Urea/TEAB and 20mM DTT at 60°C with agitation on a Thermomixer (600rpm). Proteins were collected on 10 kDA MWCO Microcon spin filters (Millipore), alkylated on-filter with iodoacetamide, and digested with Trypsin (Thermo Pierce) for 18 hours at 37°C. Digested peptides were then labeled on-filter with 10plex isobaric tags (TMT, Thermo Fisher Scientific), and labeling was quenched with 5% hydroxylamine. Prior to the combination of labeled peptide samples for subsequent fractionation, 5 μL of each labeled sample were combined into a test sample to refine TMT channel mixing ratios. The test sample was desalted offline on a NEST C18 spin column, concentrated to dryness by vacuum centrifugation, and resuspended in loading buffer (5%ACN/5%FA) with sonication for 15 minutes. The test sample was analyzed by LC-MS/MS, as described below (see LC-MS/MS Analysis subheading). Raw data were searched in Proteome Discoverer (v2.1) (see MS Data Processing subheading) to assess labeling efficiency by setting TMT modification of lysine and peptide N-termini as dynamic modifications instead of as static modifications for the final analysis to allow calculation of the percentage of fully or partially labeled peptides. Labeling efficiency was confirmed to be > 99% prior to the combination of individually labeled samples 1:1 based on corrected ratios calculated from total peptide intensities per channel determined from the test sample. The combined sample was desalted on OASIS HLB 1cc Vac Cartridges (10mg sorbent, 30μm) (Waters) prior to HPLC fractionation at high pH or isoelectric focusing (Off-gel).
Chromatographic fractionation at high pH and isoelectric focusing
Peptide mixtures (300μg) were fractionated on an Agilent Infinity HPLC 1260 system by reverse-phase separation on an Xbridge BEH C18 column (130Å, 3.5μm, 2.1mm x 100mm) at high pH using a 60-minute gradient (Mobile Phase A: 20mM NH4OH/H2O, pH = 10, Mobile Phase B = 20mM NH4OH/90%ACN, pH = 10; 0-42% Mobile Phase B). Fractions (1.2 mL total volume per fraction) were collected and combined based on 280nm absorbance traces, concentrated to dryness by vacuum centrifugation, and resuspended in loading buffer (5%ACN/5%FA) with sonication for 15 minutes. For isoelectric focusing, desalted peptides were resuspended in IPG buffer (pH 3-10) with sonication and separated along with Immobiline DryStrip gels (pH 3-10, 24 cm) into 24 fractions using a 3100 OFFGEL system (Agilent). Individual fractions were collected and combined with 200μL 0.1% FA used to rinse each well for maximum peptide recovery. All fractions were desalted offline using NEST C18 spin columns and concentrated to dryness by vacuum centrifugation and resuspended in loading buffer (5%ACN/5%FA) with sonication for 15.
LC-MS/MS analysis
Samples were transferred to autosampler vials and analyzed by nLC-MS/MS on an Eksigent nanoLC425 system (Sciex) coupled to a QExactive HF mass spectrometer (Thermo Fisher Scientific). Peptides were separated by reverse-phase liquid chromatography on a heated (50°C) PicoChipGT column (BEH C18, 1.7 μm, 130A, 150μm x 100mm) at a flow rate of 1000nL/min over a 60-minute gradient (Mobile Phase A: 98%H2O, 2%ACN, 0.1%FA, Mobile Phase B = 98%ACN, 2%H2O, 0.1%FA; 2-35% Mobile Phase B). The mass spectrometer was operated in data-dependent acquisition mode with predictive AGC and dynamic exclusion enabled (repeat count 1, exclusion window = 30s). One acquisition cycle consisted of a single full-scan (m/z = 350-1400) in the Orbitrap (r = 120,000 at m/z 200), followed by HCD fragmentation of the top 20 most intense precursor ions (r = 60,000 at m/z 200). Full scan MS1 and MS2 target values were 3E6 and 8E3, respectively. Maximum IT was set to 50ms (MS1) and 100ms (MS2). HCD fragmentation was performed at an isolation width of 1.0 Th with NCE = 32, with the exclusion of singly charged ions, ions with unassigned charge states, or ions with charge states > 6. Mass spectrometry data were acquired from 5 biological replicates (5 pairs of litter-matched ZIKV- or Mock-infected embryos) analyzed together in a single 10-plex experiment using isobaric tandem mass tags. Technical replicates were not included in this study. The resulting multiplexed samples were split into two halves and fractionated by isoelectric focusing and reverse-phase liquid chromatography at high pH into 24 and 30 fractions, respectively. Individual fractions were analyzed by single-shot data-dependent mass spectrometry analysis (1-2 injections per fraction), and all resulting raw files were combined in Proteome Discoverer for protein identification and quantitation.
MS data processing
Raw files were converted into mgf files in Proteowizard75 (options: -C -k -M -T) prior to analysis in Proteome Discoverer (v2.1) using the Mascot (v.2.6.1)74 search engine (Matrix Sciences). MS/MS spectra were searched against the UniProt mouse database appended with ZIKV protein sequences (HS-2015-BA-01) and common contaminants (17,062 total sequences). During searching, the forward database was automatically reversed and concatenated to the target database for false discovery rate (FDR) determination. Spectra were filtered by imposing a minimum precursor mass limit of 600 Da and a maximum precursor mass limit of 5000 Da. Precursor mass tolerance was set to 10ppm, and the fragment mass tolerance was set to 0.01 Da. Mascot searches include the following parameters: Trypsin/P enzyme specificity allowing 2 missed cleavages; static modification of cysteine (carbamidomethylation), lysine (TMT), and peptide N-termini (TMT); and variable modification of methionine (oxidation) and protein N-termini (acetylation). Peptide identifications were refined using a target–decoy approach followed by percolation based on q-values using the built-in Proteome Discoverer Percolator node imposing a strict FDR cutoff of 0.01 and a relaxed FDR cutoff of 0.05 at the PSM and peptide level. Proteins with < 2 peptides were excluded from quantitative comparisons. Centroided reporter ion intensities (20ppm integration tolerance) with correction for isotope impurities from the Top 3 Unique and Razor peptides were used for protein abundance calculations. The reporter ion co-isolation threshold was set to 50, and the signal to noise threshold was set to 10. Intensities were normalized based on the total peptide amount and scaled based on the row averages (channel average = 100). For quantitative comparisons, confident protein IDs (< 5% protein-level FDR) were filtered to retain proteins with measured intensities in a minimum of 3 samples (i.e., 60% of total) in at least one treatment group. Protein intensities from individual samples were median-normalized, then log2 transformed prior to the calculation of protein fold changes. False discovery rates (p < 0.05) were estimated in Perseus using a stringent permutation-based method that incorporates fold-change information (s = 0.1). Spearman and Pearson correlations with ZIKV polyprotein protein abundance were calculated in Perseus87,88 based on MS-determined normalized abundance values for individual proteins identified in ZIKV-infected samples (correlations based on 5 coordinate pairs).
Selection of 5% false discovery rate for protein identification
Our rationale for selecting a more relaxed 5% FDR for protein identification was to maximize our ability to initially match proteins with corresponding gene expression data acquired by RNAseq, as the RNA dataset allowed sampling of many more genes than were detected in our MS-based protein analyses. We previously published statistical studies and tools for the robust evaluation of proteins in mass spectrometry-based studies, observing that arbitrary FDR cutoffs either overestimate or underestimate the correct group of proteins identified, while correct FDRs can be calculated using non-parametric approaches.89 By instead imposing a 1% FDR cutoff, we report the identification of 7,367 proteins (Figure 2A – High Confidence IDs), of which 6,676 were accurately quantified. Analysis of the significance of fold change differences between ZIKV-infected or Mock-infected brain tissues determined based on the subset of proteins identified using a 1% FDR cutoff yielded results highly similar to the comparison based on the subset of proteins identified using a 5% identification FDR cutoff. In the analysis reported in the paper, we report a significantly altered abundance of 102 proteins, of which all but two were identified with high confidence (1% FDR for protein identification). The proteins identified with medium confidence are NFAC4 and S4A4. Determination of abundance comparisons based on only the subset of proteins identified at a 1% FDR resulted in the identification of 113 significantly differentially abundant proteins (100 of which overlap with those identified using the 5% protein ID FDR). The additional proteins are SARAF, COT2, CATL1, FIBB, E6L1, APOB, STMN2, LGMN, CH1B2, STRP2, OSTF1, TAP1, MOV10, all of which were identified to be differentially abundant in the previous comparison (Uncorrected p < 0.05). Proteins identified only when applying a relaxed (5%) vs. stringent (1%) FDR threshold are indicated in Table S2 (Column B).
RNA sequencing (RNA-seq) analysis
Whole E14.5 brains were collected for RNA-seq analysis. Three embryos from different litters in which ZIKV copies were > 1010 copies/ug RNA were selected from the ZIKV-injected group, and 3 littermates with Mock injection were selected as controls. RNA was extracted as described above. RNA integrity number (RIN) measured by Bioanalyzer (Agilent) for all samples was above 9.0. RNA-Seq libraries were constructed using TruSeq RNA Kit (Illumina). 100bp paired-end reads were sequenced on an Illumina HiSeq 2500 platform, with a total of 21 to 31 million reads obtained for each replicate. Fastq files were evaluated with FastQC for sequence quality (Mean Phred Score > 37) and processed using the Tuxedo suite. Reads were mapped to the mouse genome (v. GRCm38.p5, (GRCm38), version M14 (Ensembl 89)) using Bowtie (v. 2.2.4)70 and Tophat (v. 2.1.1)71 with the following options enabled: library type --fr-unstranded --coverage-search --no-mixed --no-discordant with the Gencode reference GTF supplied (-G option). Following the first Tophat search, read insert size information was extracted using Picard for inclusion in a second refined Tophat search specifying --mate-inner-dist and --mate-std-dev. The resulting .bam files were assembled using Cufflinks (v. 2.2.1),72 followed by additional analysis using Cuffmerge, Cuffcompare, and Cuffdiff73 specifying two sample groups (CNTL or ZIKV) for the determination of differentially expressed genes and transcripts. For Cuffdiff analysis, FPKMs and fragment counts were scaled to the median of the geometric means of fragment counts across all libraries. Cuffdiff dispersion models were constructed using the “pooled” dispersion method, in which a single global model for all experimental conditions is generated from averaged models built from individual conditions and replicates. Gene level differential expression was determined using Cuffdiff based on a comparison of summed FPKM values for all transcripts sharing a gene_id. Expression changes were determined using the built-in Cuffdiff t-test statistic to determine p-values and Benjamini-Hochberg FDR-corrected q-values (significance threshold: q < 0.05). An FPKM expression threshold (FPKM ≥ 1 in ZIKV or Mock condition) was applied to exclude the comparison of transcripts with poor read coverage in both conditions.
Immunohistochemistry and Immunofluorescence imaging
Dissected brains were drop-fixed in 4% paraformaldehyde (PFA) overnight at 4°C. The fixed brains were washed in PBS and embedded in 4% agarose gel (Benchmark Scientific) in PBS for vibratome sectioning at 50 microns. Matched sections were blocked at 4°C in blocking buffer (3% BSA, 0.3% Triton-X-100, 0.1% sodium azide in 1xPBS) and then incubated with the primary antibody in blocking buffer at 4°C for 24-72hours. Sections were washed with PBS, followed by secondary antibody incubation in blocking buffer at 4°C overnight. Sections were rewashed with PBS before mounting with Fluoromount-G (Southern Biotech) mounting media. All samples were counterstained with Hoechst (Sigma-Aldrich).
Imaging and quantification
Images were taken with a Zeiss LSM700 confocal microscope on an upright Axio Imager M2 equipped with 405, 488, 555, and 639 nm solid-state lasers, a motorized stage, and a CCD AxioCam MR (Carl Zeiss Microscopy). Images were processed and quantified using Zen lite software (Carl Zeiss Microscopy) and Image J software (NIH), where applicable. Figures were prepared using Adobe Creative Cloud software (Adobe Systems). For qualitative assessments and image preparation, brightness and contrast adjustments were applied only to whole multi-channel images or, if not possible in the case of individual color panels, to entire single-channel images. The thickness of the cerebral wall was measured by averaging values of three points from both brain hemispheres. Crown-rump length (CRL) and brain size were measured from the images with Image J software. Cell numbers were quantified relative to the area and expressed as arbitrary units. For Ki67, pHH3, SOX2, and TBR2 quantification, only cells located within IV/SVZ and IZ were counted based on our preliminary manual counting of total cells limited to VZ/SVZ and IZ area being reasonably similar between samples. All measurements and quantification were conducted by an investigator blinded to the injection procedure.
ChIP-seq data integration for NeuroD2 and Tbr2
Decreased-in-abundance or increased-in-abundance transcripts (Cuffdiff Benjamini-Hochberg FDR-corrected q-value < 0.05) were tested for enrichment in lists of NEUROD2 or TBR2 targets identified by previous ChIP-seq analyses of E14.5 brain tissues performed by Bayam et al.41 and Sessa et al.42 relative to NeuroD2 and Tbr2 targets identified in the whole E14.5 transcriptome analysis (13,082 genes, representing the set of expressed genes after filtering Cufflinks output tables to remove pseudogene IDs (Gm#∗), Riken transcripts (∗Rik), and transcripts with no gene ID name, and to retain genes expressed with a minimum FPKM cutoff of 1 in either the ZIKV or Mock condition with “OK” test status). The significance of over-representation or under-representation was determined by a hypergeometric (phyper) test in Rstudio/R (v. 3.4.3). NeuroD2-Tbr2-responsive gene regulatory networks were constructed using Ingenuity Pathway Analysis (Qiagen Bioinformatics) to display both direct and indirect mechanistic interactions. Specifically, for NeuroD2-Tbr2 signaling pathway mapping, we retained the list of NeuroD2 ChIP and Tbr2 ChIP targets that were decreased in expression in our transcriptome dataset and imported this list into IPA to build functional networks (using direct and indirect interaction information) from genes in the list. A final network was constructed by merging all subnetworks generated from the initial gene list. We then overlaid transcriptome expression data to retain additional nodes added by IPA from the list of genes dysregulated at the transcript level in ZIKV-infected brains. Standardized degree centrality and closeness centrality scores were calculated to determine network hubs, as previously described.90 In brief, unweighted degree centrality was determined for each node based on the number of its connections to other network nodes, and closeness centrality values for each node were calculated as the inverse sum of the shortest path to all connected network nodes from a given node.
Ingenuity Pathway Analysis
The transcription expression data file was uploaded to Ingenuity Pathway Analysis (Qiagen Bioinformatics) and was analyzed with a cutoff of q < 0.05 (Benjamini-Hochberg FDR-corrected q-values). Transcript expression levels were overlayed on the canonical pathway “Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza,” which had the highest positive score of 4.243 (p = 4.19E-13) among the pathways matching with the data. The output image was imported to Adobe Illustrator (Adobe Systems) for additional visualization.
GO enrichment analysis and visualization
Gene ontology (GO) enrichment analyses were performed using multiple methods, including Panther,80 Ingenuity Pathway Analysis (Qiagen Bioinformatics),91 Perseus annotations,87,88 and String-DB (v10.5).79 Interaction networks were generated in String with additional filtering for high confidence interactions (score >0.70) and with the “textmining” option disabled for 1G, 2D, S1F, S1H, and S1G, and “textmining” option abled for S4A. Networks were imported into Cytoscape76 for additional visualization and network analysis performed using the ClueGO77,78 plugin, which draws information from the Reactome92,93 and GO databases.
Hierarchical clustering and visualization
Hierarchical clustering was performed for proteomics data in Rstudio/R (v 4.0.4) using the normalized protein abundances. Euclidean distance measure and Ward agglomeration were used for proteins. A total of 8 clusters were separated using ggdendro (v0.1.22) and visualized alongside heatmaps of intensities using reshape2 (v1.4.4), tidyverse (v1.3.1), gridExtra (v2.3), and svglite (v2.0.0).
Quantification and statistical analysis
Statistical analysis
Unless otherwise stated, statistical significance was evaluated by two-sided Student’s t-tests in which p values of 0.05 were regarded as statistically significant. For proteomics analysis, 5 ZIKV- and 5 Mock-infected embryos from the matched mothers were analyzed. For RNA-seq analysis, we analyzed 3 ZIKV- and 3 Mock-infected embryos. For statistical analysis of other experiments, five to thirteen positive embryos were analyzed for each condition (specific numbers of analyzed embryos are indicated in each figure).
Acknowledgments
This work was supported by funding from the NIH/NIGMS (1R01GM112007-01A1), Cure Alzheimer’s Fund, and Boston Children’s Hospital F.M. Kirby Neurobiology Center Kirby Innovation Grant to J.A.S. and a Developmental Neurology Institutional National Research Service Award (NRSA) T32 NS007473 grant to A.J.G. G.H.M. was supported by funding from the NIH/NINDS (R01NS35129), the Boston Children’s Hospital Office of Faculty Development Career Development Award, the Boston Children’s Hospital Research Faculty Council Pilot Award, and the David Rockefeller Center for Latin American Studies, Harvard University. K.F. was supported by Overseas Research Fellowship Grant (RRA) from the Japan Society for the Promotion of Science. T.N. was supported by fellowship grants from the Japan Society for the Promotion of Science and the Japan Foundation for Pediatric Research. A.K. was supported by NIH R21NS104633-01A1, the William Randolph Hearst Foundation, and the Charles Hood Foundation. This project was additionally supported by an NIH U19AI131135 award to L.G. We thank Dr. Maria Lehtinen, Dr. Morgan Shannon, Ms. Dilenny Gonzalez, Dr. Xiaochang Zhang, and the Animal Facility Core in Boston Children’s Hospital for technical advice on mouse manipulation, and Dr. Benjamin Croker for insightful discussions. The authors would like to thank Dr. Mauro M. Teixeira for his original gift of the ZIKV strain HS-2015-BA-01. We are grateful to Dr. Christopher A. Walsh and Dr. Timothy Yu for laboratory support for this project. We are grateful to the Boston Children’s Hospital IDDRC cores and to Harvard Medical School Research Computing for the maintenance of the Orchestra/O2 computing cluster and technical advice. OL and the Precision Vaccines Program are supported by grants and contracts from NIAID and by the Department of Pediatrics, Boston Children’s Hospital
Author contributions
G.H.M. and J.A.S. conceived of and supervised the project. G.H.M., J.A.S., T.N., and A.J.G. designed the experiments. G.H.M. supervised the mouse model work, and J.A.S. oversaw and supervised the systems biology study. A.D. and I.B. prepared viruses. L.G. contributed new reagents and supervised the preparation of the virus. T.N., A.J.G., A.M., A.K., K.F., and D.J.V. performed the experiments, collected, and analyzed the data. K.F., C.N.S., and M.K. provided additional data analysis. O.L., K.P., and S.V.H. assisted with interpreting the immunological data and editing the manuscript. All authors critically interpreted the data and contributed to the manuscript.
Declaration of interests
The authors declare no conflicts of interest.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: May 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106909.
Contributor Information
Ganeshwaran H. Mochida, Email: ganesh.mochida@childrens.harvard.edu.
Judith A. Steen, Email: judith.steen@childrens.harvard.edu.
Supplemental information
Summary of differentially expressed genes as determined by Cufflinks analysis.
Data and code availability
-
•
Raw mass spectrometry data have been deposited in the ProteomeXchange/PRIDE proteomics (ProteomeXchange Accession #: PXD040629, www.proteomexchange.org) and MassIVE data repositories: (MassIVE Accession #: MSV000091417, https://massive.ucsd.edu/) and are publicly available as of the date of publication. Raw RNAsequencing data have been deposited in the Gene Expression Omnibus (GEO Accession #: GSE118935, https://www.ncbi.nlm.nih.gov/geo/), and are publicly available as of the date of publication.
-
•
This manuscript does not contain original codes.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Strunk T., Inder T., Wang X., Burgner D., Mallard C., Levy O. Infection-induced inflammation and cerebral injury in preterm infants. Lancet Infect. Dis. 2014;14:751–762. doi: 10.1016/S1473-3099(14)70710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora N., Sadovsky Y., Dermody T.S., Coyne C.B. Microbial vertical transmission during human pregnancy. Cell Host Microbe. 2017;21:561–567. doi: 10.1016/j.chom.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda-Filho D.d.B., Martelli C.M.T., Ximenes R.A.d.A., Araújo T.V.B., Rocha M.A.W., Ramos R.C.F., Dhalia R., França R.F.d.O., Marques Júnior E.T.d.A., Rodrigues L.C. Initial description of the presumed congenital zika syndrome. Am. J. Publ. Health. 2016;106:598–600. doi: 10.2105/AJPH.2016.303115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen S.A., Jamieson D.J., Honein M.A., Petersen L.R. Zika virus and birth defects--reviewing the evidence for causality. N. Engl. J. Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 5.de Fatima Vasco Aragao M., van der Linden V., Brainer-Lima A.M., Coeli R.R., Rocha M.A., Sobral da Silva P., Durce Costa Gomes de Carvalho M., van der Linden A., Cesario de Holanda A., Valenca M.M. Clinical features and neuroimaging (CT and MRI) findings in presumed Zika virus related congenital infection and microcephaly: retrospective case series study. BMJ. 2016;353:i1901. doi: 10.1136/bmj.i1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Linden V., Filho E.L.R., Lins O.G., van der Linden A., Aragão M.d.F.V.V., Brainer-Lima A.M., Cruz D.D.C.S., Rocha M.A.W., Sobral da Silva P.F., Carvalho M.D.C.G., et al. Congenital Zika syndrome with arthrogryposis: retrospective case series study. BMJ. 2016;354:i3899. doi: 10.1136/bmj.i3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidd B.A., Peters L.A., Schadt E.E., Dudley J.T. Unifying immunology with informatics and multiscale biology. Nat. Immunol. 2014;15:118–127. doi: 10.1038/ni.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie M.D., Holzinger E.R., Li R., Pendergrass S.A., Kim D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015;16:85–97. doi: 10.1038/nrg3868. [DOI] [PubMed] [Google Scholar]
- 9.Ruggles K.V., Krug K., Wang X., Clauser K.R., Wang J., Payne S.H., Fenyö D., Zhang B., Mani D.R. Methods, tools and current perspectives in proteogenomics. Mol. Cell. Proteomics. 2017;16:959–981. doi: 10.1074/mcp.MR117.000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prabakaran S., Hemberg M., Chauhan R., Winter D., Tweedie-Cullen R.Y., Dittrich C., Hong E., Gunawardena J., Steen H., Kreiman G., Steen J.A. Quantitative profiling of peptides from RNAs classified as noncoding. Nat. Commun. 2014;5:5429. doi: 10.1038/ncomms6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee A.H., Shannon C.P., Amenyogbe N., Bennike T.B., Diray-Arce J., Idoko O.T., Gill E.E., Ben-Othman R., Pomat W.S., van Haren S.D., et al. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun. 2019;10:1092. doi: 10.1038/s41467-019-08794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckhardt M., Hultquist J.F., Kaake R.M., Hüttenhain R., Krogan N.J. A systems approach to infectious disease. Nat. Rev. Genet. 2020;21:339–354. doi: 10.1038/s41576-020-0212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard T.R., Cristea I.M. Interrogating host antiviral environments driven by nuclear DNA sensing: a multiomic perspective. Biomolecules. 2020;10:1591. doi: 10.3390/biom10121591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah P.S., Link N., Jang G.M., Sharp P.P., Zhu T., Swaney D.L., Johnson J.R., Von Dollen J., Ramage H.R., Satkamp L., et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and zika virus pathogenesis. Cell. 2018;175:1931–1945.e18. doi: 10.1016/j.cell.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M., et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X., Hua S., Chen H.R., Ouyang Z., Einkauf K., Tse S., Ard K., Ciaranello A., Yawetz S., Sax P., et al. Transcriptional changes during naturally acquired zika virus infection render dendritic cells highly conducive to viral replication. Cell Rep. 2017;21:3471–3482. doi: 10.1016/j.celrep.2017.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari S.K., Dang J., Qin Y., Lichinchi G., Bansal V., Rana T.M. Zika virus infection reprograms global transcription of host cells to allow sustained infection. Emerg. Microb. Infect. 2017;6:e24. doi: 10.1038/emi.2017.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi L., Pimentel H., Pachter L. Zika infection of neural progenitor cells perturbs transcription in neurodevelopmental pathways. PLoS One. 2017;12:e0175744. doi: 10.1371/journal.pone.0175744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang X., Dong X., Li S.H., Zhou Y.P., Rayner S., Xia H.M., Gao G.F., Yuan H., Tang Y.P., Luo M.H. Proteomic analysis of zika virus infected primary human fetal neural progenitors suggests a role for doublecortin in the pathological consequences of infection in the cortex. Front. Microbiol. 2018;9:1067. doi: 10.3389/fmicb.2018.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scaturro P., Stukalov A., Haas D.A., Cortese M., Draganova K., Płaszczyca A., Bartenschlager R., Götz M., Pichlmair A. An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature. 2018;561:253–257. doi: 10.1038/s41586-018-0484-5. [DOI] [PubMed] [Google Scholar]
- 22.Garcez P.P., Nascimento J.M., de Vasconcelos J.M., Madeiro da Costa R., Delvecchio R., Trindade P., Loiola E.C., Higa L.M., Cassoli J.S., Vitória G., et al. Zika virus disrupts molecular fingerprinting of human neurospheres. Sci. Rep. 2017;7:40780. doi: 10.1038/srep40780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vermillion M.S., Lei J., Shabi Y., Baxter V.K., Crilly N.P., McLane M., Griffin D.E., Pekosz A., Klein S.L., Burd I. Intrauterine Zika virus infection of pregnant immunocompetent mice models transplacental transmission and adverse perinatal outcomes. Nat. Commun. 2017;8:14575. doi: 10.1038/ncomms14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gladwyn-Ng I., Cordón-Barris L., Alfano C., Creppe C., Couderc T., Morelli G., Thelen N., America M., Bessières B., Encha-Razavi F., et al. Stress-induced unfolded protein response contributes to Zika virus-associated microcephaly. Nat. Neurosci. 2018;21:63–71. doi: 10.1038/s41593-017-0038-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W., Tan Y.W., Yam W.K., Tu H., Qiu L., Tan E.K., Chu J.J.H., Zeng L. In utero infection of Zika virus leads to abnormal central nervous system development in mice. Sci. Rep. 2019;9:7298. doi: 10.1038/s41598-019-43303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bienvenu T., Poirier K., Friocourt G., Bahi N., Beaumont D., Fauchereau F., Ben Jeema L., Zemni R., Vinet M.C., Francis F., et al. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum. Mol. Genet. 2002;11:981–991. doi: 10.1093/hmg/11.8.981. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura K., Yanazawa M., Sugiyama N., Miura H., Iizuka-Kogo A., Kusaka M., Omichi K., Suzuki R., Kato-Fukui Y., Kamiirisa K., et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat. Genet. 2002;32:359–369. doi: 10.1038/ng1009 ng1009. [DOI] [PubMed] [Google Scholar]
- 28.Kato M., Saitoh S., Kamei A., Shiraishi H., Ueda Y., Akasaka M., Tohyama J., Akasaka N., Hayasaka K. A longer polyalanine expansion mutation in the ARX gene causes early infantile epileptic encephalopathy with suppression-burst pattern (Ohtahara syndrome) Am. J. Hum. Genet. 2007;81:361–366. doi: 10.1086/518903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lessel D., Gehbauer C., Bramswig N.C., Schluth-Bolard C., Venkataramanappa S., van Gassen K.L.I., Hempel M., Haack T.B., Baresic A., Genetti C.A., et al. BCL11B mutations in patients affected by a neurodevelopmental disorder with reduced type 2 innate lymphoid cells. Brain. 2018;141:2299–2311. doi: 10.1093/brain/awy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilling M., Lauritsen M.B., Møller M., Henriksen K.F., Vicente A., Oliveira G., Cintin C., Eiberg H., Andersen P.S., Mors O., et al. A 3.2 Mb deletion on 18q12 in a patient with childhood autism and high-grade myopia. Eur. J. Hum. Genet. 2008;16:312–319. doi: 10.1038/sj.ejhg.5201985. [DOI] [PubMed] [Google Scholar]
- 31.Halgren C., Bache I., Bak M., Myatt M.W., Anderson C.M., Brøndum-Nielsen K., Tommerup N. Haploinsufficiency of CELF4 at 18q12.2 is associated with developmental and behavioral disorders, seizures, eye manifestations, and obesity. Eur. J. Hum. Genet. 2012;20:1315–1319. doi: 10.1038/ejhg.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itai T., Hamanaka K., Sasaki K., Wagner M., Kotzaeridou U., Brösse I., Ries M., Kobayashi Y., Tohyama J., Kato M., et al. De novo variants in CELF2 that disrupt the nuclear localization signal cause developmental and epileptic encephalopathy. Hum. Mutat. 2021;42:66–76. doi: 10.1002/humu.24130. [DOI] [PubMed] [Google Scholar]
- 33.Baala L., Briault S., Etchevers H.C., Laumonnier F., Natiq A., Amiel J., Boddaert N., Picard C., Sbiti A., Asermouh A., et al. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat. Genet. 2007;39:454–456. doi: 10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- 34.Asselin L., Rivera Alvarez J., Heide S., Bonnet C.S., Tilly P., Vitet H., Weber C., Bacino C.A., Baranaño K., Chassevent A., et al. Mutations in the KIF21B kinesin gene cause neurodevelopmental disorders through imbalanced canonical motor activity. Nat. Commun. 2020;11:2441. doi: 10.1038/s41467-020-16294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broix L., Jagline H., Ivanova E., Schmucker S., Drouot N., Clayton-Smith J., Pagnamenta A.T., Metcalfe K.A., Isidor B., Louvier U.W., et al. Mutations in the HECT domain of NEDD4L lead to AKT-mTOR pathway deregulation and cause periventricular nodular heterotopia. Nat. Genet. 2016;48:1349–1358. doi: 10.1038/ng.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sega A.G., Mis E.K., Lindstrom K., Mercimek-Andrews S., Ji W., Cho M.T., Juusola J., Konstantino M., Jeffries L., Khokha M.K., Lakhani S.A. De novo pathogenic variants in neuronal differentiation factor 2 (NEUROD2) cause a form of early infantile epileptic encephalopathy. J. Med. Genet. 2019;56:113–122. doi: 10.1136/jmedgenet-2018-105322. [DOI] [PubMed] [Google Scholar]
- 37.Altuame F.D., Shamseldin H.E., Albatti T.H., Hashem M., Ewida N., Abdulwahab F., Alkuraya F.S. PLXNA2 as a candidate gene in patients with intellectual disability. Am. J. Med. Genet. 2021;185:3859–3865. doi: 10.1002/ajmg.a.62440. [DOI] [PubMed] [Google Scholar]
- 38.Amiel J., Rio M., de Pontual L., Redon R., Malan V., Boddaert N., Plouin P., Carter N.P., Lyonnet S., Munnich A., Colleaux L. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. Am. J. Hum. Genet. 2007;80:988–993. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zweier C., Sticht H., Bijlsma E.K., Clayton-Smith J., Boonen S.E., Fryer A., Greally M.T., Hoffmann L., den Hollander N.S., Jongmans M., et al. Further delineation of Pitt-Hopkins syndrome: phenotypic and genotypic description of 16 novel patients. J. Med. Genet. 2008;45:738–744. doi: 10.1136/jmg.2008.060129. [DOI] [PubMed] [Google Scholar]
- 40.de Munnik S.A., García-Miñaúr S., Hoischen A., van Bon B.W., Boycott K.M., Schoots J., Hoefsloot L.H., Knoers N.V.A.M., Bongers E.M.H.F., Brunner H.G. A de novo non-sense mutation in ZBTB18 in a patient with features of the 1q43q44 microdeletion syndrome. Eur. J. Hum. Genet. 2014;22:844–846. doi: 10.1038/ejhg.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bayam E., Sahin G.S., Guzelsoy G., Guner G., Kabakcioglu A., Ince-Dunn G. Genome-wide target analysis of NEUROD2 provides new insights into regulation of cortical projection neuron migration and differentiation. BMC Genom. 2015;16:681. doi: 10.1186/s12864-015-1882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sessa A., Ciabatti E., Drechsel D., Massimino L., Colasante G., Giannelli S., Satoh T., Akira S., Guillemot F., Broccoli V. The Tbr2 molecular network controls cortical neuronal differentiation through complementary genetic and epigenetic pathways. Cerebr. Cortex. 2017;27:3378–3396. doi: 10.1093/cercor/bhw270. [DOI] [PubMed] [Google Scholar]
- 43.Limanaqi F., Biagioni F., Gaglione A., Busceti C.L., Fornai F. A sentinel in the crosstalk between the nervous and immune system: the (Immuno)-Proteasome. Front. Immunol. 2019;10:628. doi: 10.3389/fimmu.2019.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enlow W., Bordeleau M., Piret J., Ibáñez F.G., Uyar O., Venable M.C., Goyette N., Carbonneau J., Tremblay M.E., Boivin G. Microglia are involved in phagocytosis and extracellular digestion during Zika virus encephalitis in young adult immunodeficient mice. J. Neuroinflammation. 2021;18:178. doi: 10.1186/s12974-021-02221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C., Xu D., Ye Q., Hong S., Jiang Y., Liu X., Zhang N., Shi L., Qin C.F., Xu Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:672. doi: 10.1016/j.stem.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 46.Englund C., Fink A., Lau C., Pham D., Daza R.A.M., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fietz S.A., Kelava I., Vogt J., Wilsch-Bräuninger M., Stenzel D., Fish J.L., Corbeil D., Riehn A., Distler W., Nitsch R., Huttner W.B. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi T., Nowakowski R.S., Caviness V.S., Jr. The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J. Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarui T., Takahashi T., Nowakowski R.S., Hayes N.L., Bhide P.G., Caviness V.S. Overexpression of p27 Kip 1, probability of cell cycle exit, and laminar destination of neocortical neurons. Cerebr. Cortex. 2005;15:1343–1355. doi: 10.1093/cercor/bhi017. [DOI] [PubMed] [Google Scholar]
- 50.Lange C., Huttner W.B., Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 51.Fujimura K., Mitsuhashi T., Shibata S., Shimozato S., Takahashi T. Utero exposure to valproic acid induces neocortical dysgenesis via dysregulation of neural progenitor cell proliferation/differentiation. J. Neurosci. 2016;36:10908–10919. doi: 10.1523/JNEUROSCI.0229-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chin Y.E., Kitagawa M., Su W.C., You Z.H., Iwamoto Y., Fu X.Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 53.Xiao S., Li D., Zhu H.Q., Song M.G., Pan X.R., Jia P.M., Peng L.L., Dou A.X., Chen G.Q., Chen S.J., et al. RIG-G as a key mediator of the antiproliferative activity of interferon-related pathways through enhancing p21 and p27 proteins. Proc. Natl. Acad. Sci. USA. 2006;103:16448–16453. doi: 10.1073/pnas.0607830103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pereira L., Medina R., Baena M., Planas A.M., Pozas E. IFN gamma regulates proliferation and neuronal differentiation by STAT1 in adult SVZ niche. Front. Cell. Neurosci. 2015;9:270. doi: 10.3389/fncel.2015.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hong S., Song M.R. Signal transducer and activator of transcription-3 maintains the stemness of radial glia at mid-neurogenesis. J. Neurosci. 2015;35:1011–1023. doi: 10.1523/JNEUROSCI.2119-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He F., Ge W., Martinowich K., Becker-Catania S., Coskun V., Zhu W., Wu H., Castro D., Guillemot F., Fan G., et al. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat. Neurosci. 2005;8:616–625. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warre-Cornish K., Perfect L., Nagy R., Duarte R.R.R., Reid M.J., Raval P., Mueller A., Evans A.L., Couch A., Ghevaert C., et al. Interferon-gamma signaling in human iPSC-derived neurons recapitulates neurodevelopmental disorder phenotypes. Sci. Adv. 2020;6:eaay9506. doi: 10.1126/sciadv.aay9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chin, Y.E., Kitagawa M., Su, W.C., Su Wc Fau - You, Z.H., You Zh Fau - Iwamoto, Y., Iwamoto Y Fau - Fu, X.Y., and Fu, X.Y. Cell Growth Arrest and Induction of Cyclin-dependent Kinase Inhibitor P21 WAF1/CIP1 Mediated by STAT1 [DOI] [PubMed]
- 59.Hong, S., and Song, M.R. Signal Transducer and Activator of Transcription-3 Maintains the Stemness of Radial Glia at Mid-neurogenesis [DOI] [PMC free article] [PubMed]
- 60.Arnold S.J., Huang G.J., Cheung A.F.P., Era T., Nishikawa S.I., Bikoff E.K., Molnár Z., Robertson E.J., Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bormuth I., Yan K., Yonemasu T., Gummert M., Zhang M., Wichert S., Grishina O., Pieper A., Zhang W., Goebbels S., et al. Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J. Neurosci. 2013;33:641–651. doi: 10.1523/JNEUROSCI.0899-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilke S.A., Hall B.J., Antonios J.K., Denardo L.A., Otto S., Yuan B., Chen F., Robbins E.M., Tiglio K., Williams M.E., et al. NeuroD2 regulates the development of hippocampal mossy fiber synapses. Neural Dev. 2012;7:9. doi: 10.1186/1749-8104-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H., Zhu Y., Morozov Y.M., Chen X., Page S.C., Rannals M.D., Maher B.J., Rakic P. Disruption of TCF4 regulatory networks leads to abnormal cortical development and mental disabilities. Mol. Psychiatr. 2019;24:1235–1246. doi: 10.1038/s41380-019-0353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan M.M., Ernst O., Manes N.P., Oyler B.L., Fraser I.D.C., Goodlett D.R., Nita-Lazar A. Multi-omics strategies uncover host-pathogen interactions. ACS Infect. Dis. 2019;5:493–505. doi: 10.1021/acsinfecdis.9b00080. [DOI] [PubMed] [Google Scholar]
- 65.Haider S., Pal R. Integrated analysis of transcriptomic and proteomic data. Curr. Genom. 2013;14:91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao H., Chen M., Valdés A., Lind S.B., Pettersson U. Transcriptomic and proteomic analyses reveal new insights into the regulation of immune pathways during adenovirus type 2 infection. BMC Microbiol. 2019;19:15. doi: 10.1186/s12866-018-1375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant A., Ponia S.S., Tripathi S., Balasubramaniam V., Miorin L., Sourisseau M., Schwarz M.C., Sánchez-Seco M.P., Evans M.J., Best S.M., García-Sastre A. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gorman M.J., Caine E.A., Zaitsev K., Begley M.C., Weger-Lucarelli J., Uccellini M.B., Tripathi S., Morrison J., Yount B.L., Dinnon K.H., 3rd, et al. An immunocompetent mouse model of zika virus infection. Cell Host Microbe. 2018;23:672–685.e6. doi: 10.1016/j.chom.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wessel A.W., Kose N., Bombardi R.G., Roy V., Chantima W., Mongkolsapaya J., Edeling M.A., Nelson C.A., Bosch I., Alter G., et al. Antibodies targeting epitopes on the cell-surface form of NS1 protect against Zika virus infection during pregnancy. Nat. Commun. 2020;11:5278. doi: 10.1038/s41467-020-19096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 75.Chambers M.C., Maclean B., Burke R., Amodei D., Ruderman D.L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.H., Pagès F., Trajanoski Z., Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikolay A., Castilho L.R., Reichl U., Genzel Y. Propagation of Brazilian Zika virus strains in static and suspension cultures using Vero and BHK cells. Vaccine. 2018;36:3140–3145. doi: 10.1016/j.vaccine.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 82.Li R.Y., Tsutsui Y. Growth retardation and microcephaly induced in mice by placental infection with murine cytomegalovirus. Teratology. 2000;62:79–85. doi: 10.1002/1096-9926(200008)62:2<79::AID-TERA3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 83.Bonnin A., Goeden N., Chen K., Wilson M.L., King J., Shih J.C., Blakely R.D., Deneris E.S., Levitt P. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472:347–350. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lanciotti R.S., Kosoy O.L., Laven J.J., Velez J.O., Lambert A.J., Johnson A.J., Stanfield S.M., Duffy M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008;14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Faye O., Faye O., Diallo D., Diallo M., Weidmann M., Sall A.A. Quantitative real-time PCR detection of Zika virus and evaluation with field-caught mosquitoes. Virol. J. 2013;10:311. doi: 10.1186/1743-422X-10-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McDowell G.S., Gaun A., Steen H. iFASP: combining isobaric mass tagging with filter-aided sample preparation. J. Proteome Res. 2013;12:3809–3812. doi: 10.1021/pr400032m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 88.Tyanova S., Cox J. Perseus: a Bioinformatics platform for integrative analysis of proteomics data in cancer research. Methods Mol. Biol. 2018;1711:133–148. doi: 10.1007/978-1-4939-7493-1_7. [DOI] [PubMed] [Google Scholar]
- 89.Serang O., Paulo J., Steen H., Steen J.A. A non-parametric cutout index for robust evaluation of identified proteins. Mol. Cell. Proteomics. 2013;12:807–812. doi: 10.1074/mcp.O112.022863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Opsahl T., Agneessens F., Skvoretz J. Node centrality in weighted networks: generalizing degree and shortest paths. Soc. Network. 2010;32:245–251. [Google Scholar]
- 91.Krämer A., Green J., Pollard J., Jr., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Milacic M., Haw R., Rothfels K., Wu G., Croft D., Hermjakob H., D'Eustachio P., Stein L. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers. 2012;4:1180–1211. doi: 10.3390/cancers4041180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of differentially expressed genes as determined by Cufflinks analysis.
Data Availability Statement
-
•
Raw mass spectrometry data have been deposited in the ProteomeXchange/PRIDE proteomics (ProteomeXchange Accession #: PXD040629, www.proteomexchange.org) and MassIVE data repositories: (MassIVE Accession #: MSV000091417, https://massive.ucsd.edu/) and are publicly available as of the date of publication. Raw RNAsequencing data have been deposited in the Gene Expression Omnibus (GEO Accession #: GSE118935, https://www.ncbi.nlm.nih.gov/geo/), and are publicly available as of the date of publication.
-
•
This manuscript does not contain original codes.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.