Abstract
Background
Escherichia coli guanine-N2 (m2G) methyltransferases (MTases) RsmC and RsmD modify nucleosides G1207 and G966 of 16S rRNA. They possess a common MTase domain in the C-terminus and a variable region in the N-terminus. Their C-terminal domain is related to the YbiN family of hypothetical MTases, but nothing is known about the structure or function of the N-terminal domain.
Results
Using a combination of sequence database searches and fold recognition methods it has been demonstrated that the N-termini of RsmC and RsmD are related to each other and that they represent a "degenerated" version of the C-terminal MTase domain. Novel members of the YbiN family from Archaea and Eukaryota were also indentified. It is inferred that YbiN and both domains of RsmC and RsmD are closely related to a family of putative MTases from Gram-positive bacteria and Archaea, typified by the Mj0882 protein from M. jannaschii (1dus in PDB). Based on the results of sequence analysis and structure prediction, the residues involved in cofactor binding, target recognition and catalysis were identified, and the mechanism of the guanine-N2 methyltransfer reaction was proposed.
Conclusions
Using the known Mj0882 structure, a comprehensive analysis of sequence-structure-function relationships in the family of genuine and putative m2G MTases was performed. The results provide novel insight into the mechanism of m2G methylation and will serve as a platform for experimental analysis of numerous uncharacterized N-MTases.
Background
Most RNAs undergo a number of post-transcriptional modifications that increase the chemical diversity of the nucleotides. tRNAs are extensively modified, but rRNA, snRNA and mRNA also contain many modified nucleotides [1]. However, the functions of these nucleotides are poorly understood and few of the modifications are strongly conserved between species. Among all the modifications, only N-1 methylation of adenosine-58 in tRNAiMet was shown to be truly essential [2] and in general, modified nucleosides are of minor importance for cell growth and/or survival (reviewed in ref. [3]).
Among more than 100 naturally occurring nucleotide modifications, the simplest and most common are methylations of nucleotide bases or ribose moieties [1]. One example of this is the monomethylation of the exocyclic amine of guanine. The 16 S RNA of E. coli contains three such N2-methylguanine (m2G) residues, at positions 966, 1207, and 1516 [4]. m2G966 is located in the loop of a small stem-loop structure, which has been implicated in tRNA binding at the P site of the ribosome [5]. m2G1207 occurs in a region of the RNA that is involved in recognition of peptide chain termination codons [6], and m2G1516 is located in the 3'-terminal stem loop of 16S rRNA, which contains also two N6, N6-dimethyladenosines (m62A). In the ribosome structure, these m2G residues as well as other 7 modified residues come together forming a compact cage surrounding the location of the anticodon stem-loop structures of A and P site-bound tRNAs [7]. Nevertheless, these modifications are not essential for protein synthesis, since ribosomes constructed from totally unmodified 16S RNA are able to carry out all of the partial reactions of in vitro protein synthesis, albeit at ca. half-efficiency [8].
The enzymes responsible for the biosynthesis of m2G in 16S RNA are not completely characterized. The m2G MTases specific for G1207 and G966 (RsmC and RsmD, respectively, named for (r)ibosomal (s)mall subunit (m)ethyltransferase) have been purified [9,10] and the gene encoding the RsmC protein was cloned and characterized [10]. Recently, sequence analysis revealed a family of related proteins in Gram-negative bacteria, encompassing subfamilies typified by RsmC, the YgjO open reading frame (ORF) believed to encode the RsmD protein, and the YbiN ORF suggested to encode the m2G1516 MTase [11]. Based on similarities between the highly conserved NPPF tetrapeptide in m2G MTases and the key motif of enzymes that methylate the exocyclic amino groups of adenine (m6A) and cytosine (m4C) in nucleic acids, it was proposed that these enzymes should be classified as a single family of N-MTases [11]. It has also been hypothesized that a long non-conserved sequence in the N-terminus of m2G MTases may be implicated in recognition of the target nucleic acid molecule, in analogy to other RNA and DNA MTases [11-13]. To learn more about the evolutionary origin and the sequence-structure-function relationships in the family of RNA:m2G MTases, we attempted to infer their structural organization in a phylogenetic context.
Results
Sequence analysis
The amino-acid sequences of E. coli RsmC and RsmD were used in PSI-BLAST database searches to identify orthologous proteins (hits reported in the 1st iteration with e-values <10-40 and sequence identity > 40%; see Materials and Methods for details). The resulting multiple sequence alignment (Figure 1) was used to predict the secondary structure (separately for both protein families) and to precisely identify boundaries of the C-terminal catalytic domain. The Rsm sequences were divided into the N-terminal (variable) and C-terminal (conserved) domains (hereafter dubbed NTD and CTD) of approximately equal length. The sequences of NTD and CTD of RsmC and RsmD were used in additional PSI-BLAST searches and also submitted to the structure prediction MetaServer http://bioinfo.pl. The CTD exhibited similarity to a large class of genuine and predicted N-methyltransferases. Both the RsmC and RsmD CTD sequences showed highest similarity to each other (e-values in the range of 10-20–10-19 in the 1st iteration), but also to a family of putative MTases from Gram-positive Bacteria and Archaea, typified by the Mj0882 protein from M. jannaschii (e-value 10-19 in the 2nd iteration starting with RsmC, 10-35 in the 3rd iteration starting with RsmD). Interestingly, the crystal structure of Mj0882 has been solved as a part of a structural genomics program (1dus; deposited in the Protein Data Bank in January 2000, cited as: LW Hung, L Huang, R Kim, SH Kim: Crystal structure and functional analysis of a hypothetical protein, Mj0882, from Methanococcus jannaschii, to be published). However, neither the biochemical characterization of this protein or the analysis of its structure has been published and its function remains a mystery. More distant homologs with shorter alignments reported in the CTD-initiated search included the HemK/YfcB family of protenr:N5-glutamine MTases [14,15] and various distinct RNA and DNA MTase families (e-values > 10-16 after the RsmD-initiated search converged in the 4th iteration). Only Mj0882 and its close homologs could be aligned to the CTD of RsmC or RsmD over their entire length if the default PSI-BLAST parameters were used. Reciprocal database searches initiated with Mj0882 yielded its presumed orthologs from Archaea and Gram-positive Bacteria (detected in the 1st PSI-BLAST iteration with e-values < 10-30 and sequence identity to the query > 40%) and CTD sequences of proteobacterial m2G MTases (RsmC detected in the 2nd iteration, e-values ≥ 2*10-26 and sequence identities ≤ 26%; RsmD detected in the 3rd iteration with e values ≥ 2*10-27 and sequence identities ≤ 24%). This result of reciprocal database searches strongly suggested an orthologous relationship [16] between the RsmC/RsmD and Mj0882 families. Further support for this prediction was obtained from the threading analysis. Among structurally characterized MTases, Mj0882 was reported as the definitely best structural template for the CTD of RsmC and RsmD (confident Pcons scores 5.78 and 6.09; see Table 1 and Table 2 for the summary of the threading analysis).
Figure 1.
Multiple alignment of the Mj0882, RsmC, RsmD, and YbiN families. Sequences are denoted by the species' name and the NCBI gene identification number; sequences lacking the number were obtained from the unfinished genome data. Numbers on the left side indicate the index of the first N-terminal residue shown, numbers in parentheses indicate how many residues in an insertion were omitted for clarity. Conserved motifs are labeled according to the nomenclature described for the AdoMet-dependent MTase superfamily [12]. Conserved residues are highlighted in black; the residues with invariant physicochemical character (hydrophobic, small etc.) are highlighted in gray. Conserved carboxylates predicted to bind AdoMet are shown in black on red background; the pair of residues proposed to make van der Waals contacts to guanine is shown in green on a yellow background; the Asn residue (His in Archaeal YbiN orthologs) proposed to catalyze the methyltransfer is shown in yellow on red background; the carboxylate proposed to bind the ribose of the target nucleoside is shown in yellow on magenta background. Secondary structure elements of Mj0882, extracted from the 1dus coordinates and virtually identical for the secondary structure independently predicted for other families (see Methods), are shown above the alignment.
Table 1.
Summary of the fold recognition analysis for RsmC-CTD.
| Model number | Pcons score | Threading server reporting the model | PDB ace. # | PDB entry description |
|---|---|---|---|---|
| 1 | 5.779 | FFAS | 1dus | Mj0882 |
| 2 | 5.46 | INBGU | 1dus | Mj0882 |
| 3 | 5.085 | 3DPSSM | 1dus | Mj0882 |
| 4 | 4.701 | SAM-T99 | 1dus | Mj0882 |
| 5 | 4.326 | mGENTHREADER | 1dus | Mj0882 |
| 6 | 3.271 | FUGUE | 1dus | Mj0882 |
| 7 | 3.081 | FFAS | 1g6q | Arg MTase Hmt1 |
| 8 | 3.002 | SAM-T99 | 1g6q | Arg MTase Hmt1 |
| 9 | 2.994 | SAM-T99 | 1g6q | Arg MTase Hmt1 |
| 10 | 2.99 | SAM-T99 | 1f31 | Arg MTase Pmt3 |
Hits to the Mj0882 structure are shown in bold. Comprehensive structure prediction results are available at the URL: http://bioinfo.pl/meta/target.pl?id=3972
Table 2.
Summary of the fold recognition analysis for RsmD-CTD.
| Model number | Pcons score | Threading server reporting the model | PDB ace. # | PDB entry description |
|---|---|---|---|---|
| 1 | 6.092 | FFAS | 1dus | Mj0882 |
| 2 | 5.856 | INBGU | 1dus | Mj0882 |
| 3 | 5.43 | 3DPSSM | 1dus | Mj0882 |
| 4 | 5.132 | SAM-T99 | 1dus | Mj0882 |
| 5 | 4.893 | mGENTHREADER | 1dus | Mj0882 |
| 6 | 4.171 | PDB-Blast | 1dus | Mj0882 |
| 7 | 3.544 | FUGUE | 1dus | Mj0882 |
| 8 | 2.907 | FFAS | 2adm | DNA:m6A MTase |
| 9 | 2.906 | SAM-T99 | 1f31 | Arg MTase Prmt3 |
| 10 | 2.874 | SAM-T99 | 1g6q | Arg MTase Hmt1 |
Hits to the Mj0882 structure are shown in bold. Comprehensive structure prediction results are available at the URL: http://bioinfo.pl/meta/target.pl?id=3971
In contrast, querying PSI-BLAST with the NTD sequences did not reveal any significant similarities. The NTDs could only be aligned to each other, displaying a number of conserved residues, if the full-length RsmC or RsmD sequences were used in the PSI-BLAST search. This suggested that the NTD could represent a novel type of a variable domain fused to the common catalytic domain or a strongly diverged version of some known nucleic acid binding domain. Quite unexpectedly, all threading algorithms (see Methods) reported that the sequences of the NTD of RsmC and RsmD are compatible with the MTase fold. Moreover, the majority of servers reported the Mj0882 structure as the best template with significant scores (Table 3 and Table 4). In the rankings produced by the consensus server Pcons, the top 6 hits corresponded to the Mj0882 structure with the best score of 3.615 (RsmC NTD) and 3.805 (RsmD NTD). These results strongly suggested that the NTD and the CTD in bacterial m2G MTases are related to Mj0882 and to each other. Nevertheless, we found that the NTD lacked many residues that may be important for the enzymatic activity of Mj0882 and the CTD (see below). Hence, the NTDs could be catalytically inactive versions of the MTase domain. Therefore, the sequences of NTD were not included in subsequent studies of the m2G MTase AdoMet-binding site and the active site based on analysis of conserved residues.
Table 3.
Summary of the fold recognition analysis for RsmC-NTD.
| Model number | Pcons score | Threading server reporting the model | PDB ace. # | PDB entry description |
|---|---|---|---|---|
| 1 | 3.615 | INBGU | 1DUS | Mj0882 |
| 2 | 3.576 | FUGUE | 1DUS | Mj0882 |
| 3 | 3.266 | FFAS | 1DUS | Mj0882 |
| 4 | 3.237 | SAM-T99 | 1DUS | Mj0882 |
| 5 | 3.225 | 3DPSSM | 1DUS | Mj0882 |
| 6 | 2.624 | mGENTHREADER | 1DUS | Mj0882 |
| 7 | 2.091 | 3DPSSM | 1EIZ | RNA:ribose O-MTase |
| 8 | 1.812 | SAM-T99 | 1BHJ | glycine N-MTase |
| 9 | 1.802 | mGENTHREADER | 1DL5 | protein isoAsp-O-MTase |
| 10 | 1.673 | SAM-T99 | 1G6Q | Arg MTase Hmt1 |
Hits to the Mj0882 structure are shown in bold. Comprehensive structure prediction results are available at the URL: http://bioinfo.pl/meta/target.pl?id=3974
Table 4.
Summary of the fold recognition analysis for RsmD-NTD.
| Model number | Pcons score | Threading server reporting the model | PDB ace. # | PDB entry description |
|---|---|---|---|---|
| 1 | 3.805 | FUGUE | 1DUS | Mj0882 |
| 2 | 3.573 | INBGU | 1DUS | Mj0882 |
| 3 | 3.379 | FFAS | 1DUS | Mj0882 |
| 4 | 3.242 | 3DPSSM | 1DUS | Mj0882 |
| 5 | 2.687 | mGENTHREADER | 1DUS | Mj0882 |
| 6 | 2.262 | SAM-T99 | 1DUS | Mj0882 |
| 7 | 1.864 | SAM-T99 1 | 1F3L | Arg MTase Pmt3 |
| 8 | 1.587 | FFAS | 1G6Q | Arg MTase Hmt1 |
| 9 | 1.51 | INBGU | 1VID | catechol O-MTase |
| 10 | 1.426 | FFAS | 1D2H | glycine N-MTase |
Hits to the Mj0882 structure are shown in bold. Comprehensive structure prediction results are available at the URL: http://bioinfo.pl/meta/target.pl?id=3970
The YbiN family of predicted N-MTases has also been subjected to sequence comparisons and threading analyses. These results confirmed the previously observed similarity to sequences of RsmC and RsmD CTD [11] as well as to the Mj0882 family. PSI-BLAST reported similarity of YbiN to RsmC CTD in the 3rd iteration with the e-value of 8 * 10-5, whereas Pcons ranking of the top 4 hits corresponded to the Mj0882 structure with the best score of 3.41 (Table 5). In the PSI-BLAST searches and in sequence profile comparisons using the FFAS algorithm [17], the YbiN family showed similarity to members of the HemK/YfcB family with scores comparable to that of the RsmC/RsmD/Mj0882 family (data not shown). It has been recently shown that the HemK family comprises protein:N5-glutamine MTases, suggesting that the (D/N/S)-P-P-(F/Y/W/H) motif is characteristic for a subset of MTases that methylate -NH2 groups of various substrates, rather than only bases in nucleic acids [14,15].
Table 5.
Summary of the fold recognition analysis for YbiN.
| Model number | Pcons score | Threading server reporting the model | PDB ace. # | PDB entry description |
|---|---|---|---|---|
| 1 | 3.406 | FFAS | 1DUS | Mj0882 |
| 2 | 3.267 | 3DPSSM | 1DUS | Mj0882 |
| 3 | 3.083 | SAM-T99 | 1DUS | Mj0882 |
| 4 | 2.618 | mGENTHREADER | 1DUS | Mj0882 |
| 5 | 2.552 | PDB-Blast | 1G6Q | Arg MTase Hmt1 |
| 6 | 2.547 | INBGU | 1DUS | Mj0882 |
| 7 | 2.481 | PDB-Blast | 1DL5 | protein isoAsp-O-MTase |
| 8 | 2.467 | PDB-Blast | 1F3L | Arg MTase Pmt3 |
| 9 | 2.462 | FFAS | 1G6Q | Arg MTase Hmt1 |
| 10 | 2.4 | PDB-Blast | 1G6Q | Arg MTase Hmt1 |
Hits to the Mj0882 structure are shown in bold. Comprehensive structure prediction results are available at the URL: http://bioinfo.pl/meta/target.pl?id=3965
Although the predicted key catalytic motif of the YbiN family (NPPFH) resembles more that of bona fide guanine-N2 MTases RsmC and RsmD than of any other N-MTase family, it cannot be excluded that these proteins methylate other substrates than guanine in RNA. We hope that this analysis will facilitate the choice of representative targets for experimental characterization, which would allow more precise inference of biological function (target specificity) and relationships between various families of N-MTases.
Another piece of information that should shed light onto the broad picture of the N-MTase family is the previously unnoticed high similarity of the YbiN family to uncharacterized proteins from Eukaryota and Archaea (BLAST e-values 10-27, and 4*-04, respectively). Their close similarity (confirmed in reciprocal searches with similar e-values) indicates an orthologous relationship to bacterial YbiN proteins. This was confirmed in reciprocal searches initiated with the eukaryotic and archaeal members of the family. Interestingly, while the predicted eukaryotic YbiN orthologs possess all sequence features typical for the bacterial MTases, their archaeal counterparts lack the key Asn residue in motif IV, suggesting that they may be inactive, unless the catalytic side chain "migrated" to another position in the primary sequence (see below). Regardless of the possible loss of functional residues, the presence of common motifs in the YbiN and Mj0882 families as well as in the NTD and CTD sequences of proteobacterial m2G MTases was confirmed using the Gibbs sampling procedure [18]. Motifs I, VI and VIII were detected with probability of occurring by chance < 10-13. In addition, there is a good correlation of secondary structure elements independently predicted for all protein groups (Figure 1).
The orthologous/paralogous relationships inferred from the results of PSI-BLAST searches were confirmed by phylogenetic analysis using the neighbor-joining method of Saitou and Nei [19]. The bacterial YbiN sequences and their archaeal and eukaryotic homologs grouped together with 100% bootstrap support and the RsmC/RsmD NTD, RsmC/RsmD CTD and Mj0882 families formed groups with bootstrap support > 50% (data not shown). Taken together, these findings strongly suggest that the two domains of the proteobacterial m2G MTases RsmC and RsmD, and the predicted MTases YbiN and Mj0882 evolved from the common ancestor. An alternative scenario can be envisaged, in which the pseudodimeric structure of RsmC and RsmD results not from duplication but a fusion of the ancestral CTD with another distantly related MTase that has now degenerated. Nevertheless, fold-recognition results suggest that among various MTases of known structure, the NTD clearly shows highest similarity to Mj0882 (the archaeal counterpart of the CTD) (Table 3 and Table 4). In addition, comparison of the sequence profile of the NTD sequences with profiles of other MTase families (unpublished data) using the FFAS algorithm [17] confirmed that the Mj0882/CTD family is the best match (data not shown), as reported in the course of fold recognition searches against the profiles comprising proteins from PDB. Moreover, the scenario involving intragenic duplication of the catalytic domain in the ancestor of the RsmC/RsmD lineage, which generated a functional pseudodimer, is supported by the dimeric structure of Mj0882 in the crystal (see below). Determination of the crystal structure of a member of the RsmC/RsmD lineage will provide insight into the relationship of the inactivated NTD to known MTase structures and the mutual orientation of the two domains.
Structure-based interpretation of sequence alignment and prediction of the active site
The Mj0882 structure was solved in the absence of any ligands. Prompted by the identification of close relationships between the family of m2G MTases and Mj0882, we sought to identify the cofactor- and guanine-binding sites in the 1.8 A crystal structure of the latter protein based on comparison with structures of known N-MTases. Searching the Protein Data Bank [20] using VAST [21]http://www.ncbi.nlm.nih.gov/cgi-bin/structure/mmdbsrv?UID=1DUS and DALI [22]http://jura.ebi.ac.uk:8765/holm/qz?filename=/data/research/fssp//1dusA.fssp revealed that the Mj0882 structure is more similar to various MTases alkylating hydroxyl groups, than to other nucleic acid N-MTases. This is not surprising, since previous evolutionary analysis of MTase structures and sequences demonstrated that some nucleic acid N-MTase families do not group together in the superfamily tree. The indication is that the common motif (N/D/S)-(I/P)-P-(F/Y/W/H) evolved independently several times on the common framework [23]. It will be interesting to determine, if m2G MTases share a relatively recent common ancestor with any of the m6A or m4C MTase families and if the similarities in their active site are synapomorphic or homoplastic.
The issue of divergent or convergent evolution notwithstanding, the "catalytic loop", corresponding to motif IV, assumes a strikingly similar conformation in all N-MTase structures solved to date (Figure 2). This suggests that the interactions between the target amino group and the side chain of the first residue and the carbonyl oxygen of the second residue of the tetrapeptide are highly similar. The same conformation is retained in the Mj0882 structure, consistent with the prediction that this protein belongs to the N-MTase family. Nevertheless, in the superimposed structures there is a substantial variation in the conformation of the cofactor AdoMet or its analogs, and in their relative orientation with respect to the catalytic site (Figure 2). This variation was noted previously in the course of crystallographic analyses, and was attributed to subtle structural differences between cofactor-binding pockets of various MTases that impose distinct conformation of the same ligand [12,24] and to the fact that different ligands (AdoMet, AdoHcy, and their analogs) make different interactions and do not necessarily retain the common conformation when bound to the same MTase [24,25].
Figure 2.
Comparison of the conformations of the "catalytic loop" (motif IV) of N-MTases in stereoview. DNA:m6A MTases: M.TaqI (1g38 in PDB) [29] in yellow, M.RsrI (leg2) [55] in red, M.DpnM (2dpm) [56] in green; RNA:m6A MTase ErmC' (1qao) [24] in blue; DNA:m4C MTase M.PvuII (1boo) [57] in cyan; Mj0882 (1dus) in magenta. The ligand for each protein is shown: AdoMet for M.DpnM and ErmC', S-adenosylhomocysteine for M.PvuII, 5'-methylthioadenosine for M.RsrI, and 5'-[2-(amino)ethylthio]-5'-deoxyadenosine (cofactor analog) and the target deoxyadenosine for M.TaqI. In case of M.RsrI, ErmC' and M.PvuII, whose structures were solved in the absence of the target, the Tyr or Phe sidechain can be easily rotated to the M.TaqI-like orientation to make a face-to-face π-stacking interaction with the target base.
In order to dock the AdoMet molecule to the 1dus coordinates, we sought to identify those MTase structures, which display greatest similarity to the Mj0882 structure in the cofactor-binding region. VAST and DALI searches confirmed that the region spanning residues 55 to 117 in the Mj0882 sequence is the most similar to MTases that modify hydroxyl groups (data not shown), as well as the entire protein structure (see above). The catechol O-MTase structure (COMT) [26](1vid) was reported as the best hit by VAST (Score 7.1, RMSD 1.3). It was used as a structural template, because it was the only well-scored structure of a MTase complexed with AdoMet and not with its non-reactive analogs or the reaction product AdoHcy. Accordingly, the AdoMet moiety was copied from COMT to the Mj0882 structure based on superposition of the 1vid and 1dus coordinates. The obtained Mj0882-AdoMet complex showed no severe atomic overlaps and the cofactor seemed to fit the groove on the protein surface very well. According to AutoDock [27] the energy of the interactions between the 1dus structure and AdoMet in the docked complex is very favorable (-15.03 kcal/mol) even though it is lower from that calculated for the template COMT-AdoMet complex (-22.04 kcal/mol) in the 1vid structure [26]. From the docked model, in striking analogy to most MTase structures (reviewed in ref. [12]), the following three crucial contacts can be predicted: i) D61 from motif I coordinates the methionine amino group of AdoMet via an ordered water molecule. Even though the corresponding acidic residue is conserved in nearly all MTases analyzed to date, this contact has been identified only recently in the high-resolution structure of the RrmJ MTase [28]. ii) D84 from motif II coordinates the ribose hydroxyl groups, iii) D113 from motif III coordinates the amino group of the adenine moiety. Non-polar interactions between the side-chains of I85 and L114 and the adenine ring further contribute to the binding (Figure 3).
Figure 3.
The docking model of AdoMet and guanosine to the Mj0882 structure. Residues predicted to participate in cofactor and target binding are shown in wireframe representation and are labeled in cyan and in green, respectively.
Docking of the target guanine to the Mj0882 structure (Figure 4) was guided by superposition of the "motif IV loop" of Mj0882 and M.TaqI, the only N-MTase co-crystallized with the nucleic acid substrate [29]. Under the assumption that both enzymes bind their targets in the same plane, the N2 group of guanine could be aligned with the N6 group of the adenine amino only if the purine ring was rotated by 120 degrees (i.e. with the atoms C2 and N3 of guanine superimposed onto atoms C6 and N1 of adenine). According to AutoDock [27] the energy of the interactions between the Mj0882-AdoMet complex and the docked guanine is quite favorable and comparable with that calculated for the target adenine in the M.TaqI structure (1g38) (-6.14 kcal/mol and -7.59 kcal/mol, respectively). These values are much lower than the energy of interactions between the protein and the cofactor (see above), because the AdoMet-binding groove is very deep and hydrophobic, while the base-binding site is relatively shallow. An alternative orientation of the target guanine in complex with 1dus could be obtained if its atoms C2 and N1 were superimposed onto atoms C6 and N1 of adenine. However, the latter model resulted in steric clashes between the ribose moiety and I132 (data not shown). Even if the flexible ligand docking option of AutoDock was used, which alleviated steric clashes, energies for the guanine in the "alternative" position (a cluster of similar conformations) were significantly worse (data not shown). In the first docked model the guanine moiety fits the groove on the protein's surface surprisingly well; a residue was identified, whose side chain might coordinate the ribose hydroxyl groups, namely D41 (Figure 3). Analysis of the multiple sequence alignment (Figure 1) revealed that this residue is highly conserved in the Mj0882, RsmD, RsmC, and YbiN families, supporting its functional importance. In the "alternative" docked model, D41 is too remote from the target base to make direct contacts to the target nucleoside (data not shown). However, a substantial conformational change upon substrate binding cannot be excluded, and direct participation of D41 in catalysis as well as other modes of guanine binding can be envisaged. Identification of this conserved carboxylate as a potentially important residue will hopefully prompt site-directed mutagenesis experiments and experimental determination of its role in binding and/or catalysis.
Figure 4.
Comparison of positions of the ligands in the active site of M.TaqI and the docked model of Mj0882. M.TaqI is shown in yellow, Mj0882 is shown in magenta.
Although the presented docking model of Mj0882 complexed with AdoMet and guanine should be regarded as preliminary, it provides a useful platform for correlating the structure with sequence conservation in the Mj0882/RsmD/RsmC/YbiN family. The predicted AdoMet- and ribose-binding Asp residues (see above), as well as other invariant and highly conserved surface-exposed residues, map in the vicinity of the docked ligands, strongly supporting the model (Figure 5). The N2 group of the docked guanine is hydrogen bonded to the side chain of N129 and the carbonyl oxygen of P130. This is similar to the contacts made by the N6 group of the target adenine in the co-crystal structure of M.TaqI [29], which served as a docking template. As proposed for M.TaqI, these interactions could increase electron density of N2, change its hybridization from sp2 towards sp3, where the lone pair of N6 is no longer conjugated with the aromatic system, and thus contribute to its activation for nucleophilic attack on the activated methyl group of AdoMet. From the Mj0882 structure it is not obvious which residue may be responsible for eliminating the proton from the methylated N2 amine. The only conserved acidic residue in the vicinity of the active site corresponds to D41, and its localization is incompatible with the localization of the protons on N2 in the above-mentioned mechanism. If no massive conformational change of the m2G MTase occurs during RNA binding and catalysis, we propose that the proton is transferred directly to a bulk solvent molecule. In M.TaqI, the target base forms a face-to-face π-stacking interaction with Y108 from the NPPY tetrapeptide and a hydrophobic interaction with V21 from motif X. In Mj0882 these contacts are reversed (i.e. in motif IV Y108 is substituted by non-aromatic I and V21 is replaced by aromatic F35). Interestingly, all members of families RsmC, RsmD, and YbiN possess aromatic residues at both positions (Figure 1). It has been hypothesized that the methyltransfer reaction in N-MTases proceeds via a positively charged intermediate, which could be stabilized by cation-Ti interactions [30]. In m2G MTases, the electron-rich aromatic rings on both sides of the target could contribute to such stabilization.
Figure 5.
Molecular surface of Mj0882 colored by the conservation score. The score have been calculated using SWISS-PDB VIEWER [54] based on superposition of 1dus coordinates with homology models of YbiN and the CTD of RsmC and RsmD (all proteins from E. coli). The color scale varies from blue to yellow, with invariant residues in blue and most variable in yellow. The docked AdoMet and guanine moieties are shown in wireframe representation.
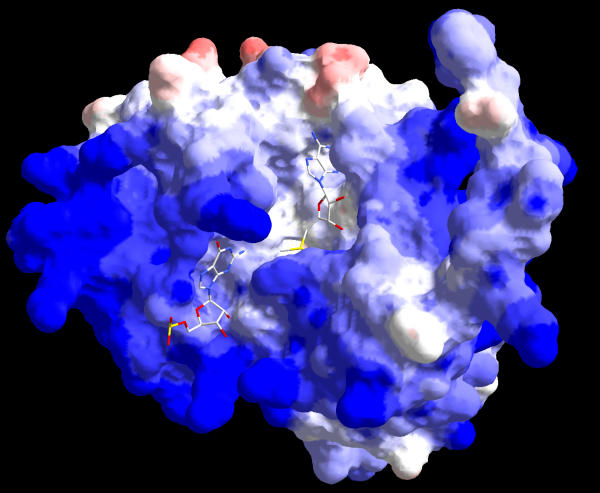
The predicted guanine-binding mode implies that the target nucleotide is "flipped out" of the rRNA into the catalytic site, similarly to the mechanism discovered for DNA: m5C MTases [31] and extended to DNA N-MTases [29,32]. In bacterial 16S rRNA G966 and G1516 occur in extrahelical regions, which should facilitate their binding in the catalytic pocket of the enzyme, but G1207 is base-paired in the helix 34 stem [7]. The m2G1207 MTase RsmC reacts with the 30S ribosomal particles but barely at all with 16S RNA, while the m2G966 MTase RsmD requires only the S7 and S19 proteins to methylate its target in the 16S RNA [10]. It has been inferred that both these nucleotides are modified after the association of several ribosomal proteins with the 16S rRNA, which is likely to assume the native secondary structure, but before assembly of the 30S subunit is complete [10]. The positively charged surface of Mj0882 in the area next to the docked guanine suggests charge complementarity to the negatively charged phosphate groups in rRNA (Figure 6). It will be interesting to study the base-flipping capabilities of RsmC and RsmD, since they may shed the light on the evolution of presumed mechanistic differences between the nucleic acid MTases that flip their target out of the double helix and those modifying extrahelical bases.
Figure 6.
Molecular surface of Mj0882 colored by the electrostatic potential. The values of surface potential are expressed as a spectrum ranging from -5 kT/e (deep red) to +5 kT/e (deep blue). The docked AdoMet and guanine moieties are shown in wireframe representation.
Analysis of the apparently "degenerated" members of the family
Analysis of the multiple sequence alignment (Figure 1) suggested that the NTD of RsmC and RsmD lack several conserved residues. However, only in connection with the docking model of Mj0882 it can be concluded, which of these residues may be involved in binding or catalysis. Specifically, all NTD sequences lack the guanine N2-coordinating Asn and the aromatic or aliphatic residue in motif IV. The NTD of all RsmC, with the exception of T. thermophilus, lacks the N-terminal extension, which includes the aromatic residue predicted to make the face-to-face π-stacking interactions with the target guanine (F35 in Mj0882). This conserved aromatic residue is also missing from the longer N-terminus of RsmD and T. thermophilus RsmC. The NTD of RsmD retained the predicted ribose-binding acidic residue at the position corresponding to D41 in Mj0882, while in the NTD of RsmC this residue is often substituted, in most cases with Ser or Thr. All NTD sequences, with the possible exception of C. crescentus and T. thermophilus RsmC, lack the AdoMet-coordinating acidic residue in motif I, corresponding to D61 of Mj0882. The NTD of RsmD retained the Asp residue corresponding to D84 in Mj0882, however in nearly all RsmC sequences this residue is substituted with various polar amino acids. A region corresponding to motif III was so diverged in the NTD of both RsmC and RsmD that it could not be aligned with confidence and the position corresponding to D113 in Mj0882 could not be identified unambiguously. It is noteworthy that the residue corresponding to D113 (motif III) is weakly conserved in the MTase superfamily, but the acidic residue corresponding to D84 (motif II) is conserved in nearly all MTase families studied to date [12](JMB, unpublished data).
In summary, the NTD of RsmC and RsmD lacks all residues, which were predicted to be a part of the active site based on the docking model and which are conserved between Mj0882 and the CTD. The NTD of RsmC also lacks most of the acidic residues predicted to coordinate AdoMet and the ribose moiety of the target nucleoside. Nevertheless, some of these residues are retained in the NTD of RsmD and in the NTD of T. thermophilus RsmC. Hence, it is inferred that the NTD of RsmC and RsmD lost the m2G MTase activity and are probably also deficient in the ability to bind the cofactor. There are known precedents of proteins with apparent severe defects in conserved MTase motifs. One example includes the yeast Kar4p protein required for expression of karyogamy-specific genes during mating [33]. Using fold recognition and homology modeling it was demonstrated that Kar4p is related to mRNA:m6A MTases and the "permuted" DNA:m6 and m4C MTases, although it lost its active site and has the AdoMet-binding site blocked by an insertion. It is quite probable that Kar4p exerts its function due to the retained ability to bind to RNA or DNA (JM Bujnicki and coworkers, manuscript submitted). Another example is the eukaryotic heterodimeric tRNA:mlA58 MTase, in which one subunit binds AdoMet and presumably carries out the catalysis, while the other is indispensable for tRNA binding [2]. It has been demonstrated that the tRNA-binding subunit is homologous to the "active" subunit, but lost the cofactor-binding and catalytic side-chains. We have also identified a single "active" tRNA:mlA58 MTase gene in Bacteria and Archaea, suggesting that the ancestral prokaryotic form was a homodimer [34]. This prediction has been recently confirmed by the crystal structure of the Rv2118c protein from Mycobacterium tuberculosis, which forms a dimer of tight homodimers [35].
Interestingly, according to the EBI Macromolecular Structure Database, the suggested biologically relevant molecule for the Mj0882 structure is a homodimer http://pdb-browsers.ebi.ac.uk/pdb-bin/macmol.pl?filename=1dus. The Mj0882 homodimer exhibits 5 interchain salt bridges within the complex and shows a loss of 2045.5 Ang**2 of solvent accessible surface area upon complex formation. However, in each subunit the active site is completely blocked by a loop from the other subunit. Nevertheless, it is tempting to speculate that the active form of Mj0882 is a homodimer that undergoes a conformational change to allow the target 16S rRNA to bind and that RsmC and RsmD MTases function as pseudodimers. If this is true, the relationship between Mj0882 and the RsmC/RsmD families would be similar to the relationships between prokaryotic and eukaryotic tRNA:mlA58 MTases.
We also found that the archaeal members of the YbiN family lack the counterpart of the conserved N129 in Mj0882. However, analysis of the multiple sequence alignment revealed that they retained all other predicted binding residues, with the possible exception of the poorly conserved D113 (Figure 1). This prompted us to build a provisional homology model of the PH0266 protein, which revealed that the "missing" Asp sidechain in motif IV is spatially replaced by a His residue from motif X (data not shown). The alignment (Figure 1) reveals that the His residue is conserved at this position in all archaeal YbiN homologs, whereas bacterial and eukaryotic YbiN proteins have Pro or Ala and the Mj0882/RsmC/RsmD lineage have Gly or Ala (G43 in Mj0882). "Migration" of catalytic and cofactor binding residues have been documented in many cases (for a review see ref. [36]), including RNA MTases that generate m5C in rRNA [37] and m7G in mRNA [38]. Nonetheless, no active nucleic acid MTase have been reported bearing the His substitution for the catalytic residue in motif IV. We hope that this analysis will stimulate experimental characterization of archaeal YbiN homologs, which may eventually lead to validation of a novel version of the methyltransfer active site.
Conclusions
Based on a comprehensive bioinformatic analysis of m2G MTases it was inferred that the prokaryotic RsmC and RsmD MTases are pseudodimers. The C-terminal catalytic domain is closely related to the structurally characterized Mj0882 protein, while the N-terminal domain lacks the cofactor-binding and catalytic side-chains. Based on analysis of sequence similarities within and between subfamilies, it is suggested that bacterial m2G MTases evolved from a duplicated Mj0882-like enzyme, although a fusion with another distantly related MTase cannot be definitely excluded. The supposed intragenic duplication or gene fusion followed by degeneration of the N-terminal domain was ancient and predated the gene duplication leading to radiation of RsmC and RsmD lineages. However, we were not able to determine the precise timing of the divergence, presumably due to the high noise-to-signal ratio in the multiple sequence alignment. This evolutionary scenario, along with the analysis of the crystal structure of Mj0882, suggests that m2G MTases function as dimers, with the one subunit carrying out catalysis and the other probably participating in recognition and binding of the target rRNA. An analogous case has been described for archaeal tRNA splicing enzymes, which exist in two forms. The M. jannaschii EndA nuclease is a homotetramer, in which two pairs of identical subunits have non-equivalent roles in tRNA binding and catalysis [39], whereas its A. fulgidus homolog is a homodimer of pseudodimers, comprising diverged domains of which only one retained the active site [40]. Analysis of the Mj0882 docking model in the light of the sequence conservation in the m2G MTase family allowed prediction of cofactor-binding and catalytic residues. In the YbiN family of predicted N-MTases (suggested to be rRNA:m2G1516 MTases, but this specificity is uncertain) new eukaryotic and archaeal members have been identified. The latter exhibited an intriguing correlated loss of a conserved Asn residue in motif IV with acquisition of an atypical His residue in motif X. Both side chains are likely to occupy a similar spatial position, suggesting a novel version of the MTase active site. All these predictions can be verified experimentally. We hope that this analysis will significantly improve the insight into the molecular mechanism of m2G methylation, facilitate further structural and functional studies on m2G MTases and contribute to better understanding of relationships between them and other nucleic acid MTases.
Methods
Sequence database searches
The position-specific, iterative (PSI-) version of BLAST [41,42] was used to search the non-redundant (nr) version of current sequence database and the publicly available complete and incomplete genome sequences available via the websites of the National Center for Biotechnology Information (Bethesda, USA; http://www.ncbi.nlm.nih.gov and the Bioinformatics Laboratory of International Institute of Molecular and Cell Biology (Warsaw, Poland; http://www.iimcb.gov.pl). All searches were initiated with stringent profile inclusion expectation (e) values (<10-20) to avoid "explosion" of hits to multiple paralogous MTase families and the cutoff was relaxed in subsequent iterations as the profile become balanced (i.e. included more orthologs of the query, which reduced the probability of assigning a significant score to a non-ortholog that exhibited a fortuitous similarity to the query, but not to the orthologs of the query). The stringent criteria for provisional assignment of orthologous relationship in PSI-BLAST searches were: the alignment spanning more than 80% of both the query and the reported hit, e-value in the 1st iteration < 10-30, sequence identity > 40%. In all searches initiated with RsmC, RsmD, and YbiN sequences, hits to the catalytic domain of HemK family members were reported before relaxation of the cutoff with e-values ranging from 10-5 to 10-18 (i.e. subsequently to the presumed members of the above-mentioned families). The choice of the relaxed cutoff was dictated by the assumption that the HemK family of protein MTases should be regarded as an outgroup and its members shall not be included in the profile. The multiple sequence alignment was retrieved from the PSI-BLAST, degapped, and used as a profile, to which all the full-length sequences were realigned using CLUSTALX [43].
Structure prediction
Protein structure predictions were carried via the MetaServer interface [44]http://bioinfo.pl/meta/ using publicly available online services for fold recognition: FFAS [17], 3DPSSM [45], BIOINBGU [46], GenThreader [47], SAM-T99 [48], and FUGUE [49], and secondary structure prediction: JPRED2 [50] and PSI-PRED [51]. The results were processed by the Pcons consensus server [52], which produced a ranking of potentially best target-template alignments and evaluated the likelihood of the models to be correct.
Protein structure analysis
Comparison of atomic coordinates with the protein structures from the Protein Data Bank [20] were carried out using VAST [21] and DALI [53]. SWISS-PDB VIEWER [54] was used for all protein structure manipulation, calculation of conservation scores and electrostatic potential distribution, and to generate the figures. Provisional homology modeling was carried out using the SWISS-MODEL/PROMOD II server http://www.expasy.ch/swissmod/SWISS-MODEL.html[54].
Authors' contributions
J.M.B. carried out the sequence and structure analysis and drafted the manuscript. L.R. provided the laboratory space and the access to the MetaServer, BIB-VIEW, local version of BLAST and sequence databases.
Contributor Information
Janusz M Bujnicki, Email: iamb@wp.pl.
Leszek Rychlewski, Email: leszek@bioinfo.pl.
Acknowledgments
We would like to thank all the genome sequencing groups that make their preliminary data publicly available, without which this work could not be done. Especially, the use of data generated at the Department of Energy Joint Genomic Institute, the Institute for Genomic Research, and the Stanford Genome Technology Center is gratefully acknowledged. We are indebted to Krzysztof Ginalski for help with AutoDock and to Richard Leach for critical reading of the manuscript. This work was supported by the Polish State Committee for Scientific Research (grants 8T11F01019 and 6P04 B00519)
References
- Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc. 2000;97:5173–5178. doi: 10.1073/pnas.090102597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog. Nucleic Acid Res. Mol. Biol. 1996;53:79–129. doi: 10.1016/s0079-6603(08)60143-9. [DOI] [PubMed] [Google Scholar]
- Raue HA, Klootwijk J, Musters W. Evolutionary conservation of structure and function of high molecular weight ribosomal RNA. Prog. Biophys. Mol. Biol. 1988;51:77–129. doi: 10.1016/0079-6107(88)90011-9. [DOI] [PubMed] [Google Scholar]
- von Ahsen U, Noller HF. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- Murgola EJ, Hijazi KA, Goringer HU, Dahlberg AE. Mutant 16S ribosomal RNA: a codon-specific translational suppressor. Proc. 1988;85:4162–4165. doi: 10.1073/pnas.85.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, demons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cunningham PR, Richard RB, Weitzmann CJ, Nurse K, Ofengand J. The absence of modified nucleotides affects both in vitro assembly and in vitro function of the 30S ribosomal subunit of Escherichia coli. Biochimie. 1991;73:789–796. doi: 10.1016/0300-9084(91)90058-9. [DOI] [PubMed] [Google Scholar]
- Weitzmann C, Tumminia SJ, Boublik M, Ofengand J. A paradigm for local conformational control of function in the ribosome: binding of ribosomal protein S19 to Escherichia coli 16S rRNA in the presence of S7 is required for methylation of m2G966 and blocks methylation of m5C967 by their respective methyltransferases. Nucleic Acids Res. 1991;19:7089–7095. doi: 10.1093/nar/19.25.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J Biol Chem. 1999;274:924–929. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM. Phylogenomic analysis of 16S rRNA:(guanine-N2) methyltransferases suggests new family members and reveals highly conserved motifs and a domain structure similar to other nucleic acid amino-methyltransferases. FASEB J. 2000;14:2365–2368. doi: 10.1096/fj.00-0076com. [DOI] [PubMed] [Google Scholar]
- Fauman EB, Blumenthal RM, Cheng X. In: In S-Adenosylmethionine-dependent methyltransferases: structures and functions. Cheng X, Blumenthal RM, editor. Singapore, World Scientific Inc.; 1999. Structure and evolution of AdoMet-dependent MTases. pp. 1–38. [Google Scholar]
- Bujnicki JM, Blumenthal RM, Rychlewski L. Sequence analysis and structure prediction of 23S rRNA:m1G methyltransferases reveals a conserved core augmented with a putative Zn-binding domain in the N-terminus and family-specific elaborations in the C-terminus. J. 2002;4:93–99. [PubMed] [Google Scholar]
- Nakahigashi K, Kubo N, Narita SS, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Rychlewski L, Jaroszewski L, Li W, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Liu JS, Lipman DJ, Lawrence CE. Extracting protein alignment models from the sequence database. Nucleic Acids Res. 1997;25:1665–1677. doi: 10.1093/nar/25.9.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr. 1996;6:377–385. doi: 10.1016/S0959-440X(96)80058-3. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM. Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol. 1999;1:1–8. http://www.bioinfo.de/isb/1999-01/0016/ [PubMed] [Google Scholar]
- Schluckebier G, Zhong P, Stewart KD, Kavanaugh TJ, Abad-Zapatero C. The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. 1999;289:277–291. doi: 10.1006/jmbi.1999.2788. [DOI] [PubMed] [Google Scholar]
- Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and sinefungin to the adenine-specific DNA methyltransferase M. TaqI. J Mol. 1997;265:56–67. doi: 10.1006/jmbi.1996.0711. [DOI] [PubMed] [Google Scholar]
- Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J. 1996;9:1–5. doi: 10.1002/(SICI)1099-1352(199601)9:1<1::AID-JMR241>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- Schluckebier G, Labahn J, Granzin J, Saenger W. M.TaqI: possible catalysis via cation-pi interactions in N-specific DNA methyltransferases. Biol. 1998;379:389–400. [PubMed] [Google Scholar]
- Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Stewart BG, Scott CF, Rose MD. The two forms of karyogamy transcription factor Kar4p are regulated by differential initiation of transcription, translation, and protein turnover. Mol. Cell Biol. 1999;19:817–825. doi: 10.1128/mcb.19.1.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM. In silico analysis of the tRNA:m1A58 methyltransferase family: homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett. 2001;507:123–127. doi: 10.1016/S0014-5793(01)02962-3. [DOI] [PubMed] [Google Scholar]
- Gupta A, Kumar PH, Dineshkumar TK, Varshney U, Subramanya HS. Crystal structure of Rv2118c: An AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J Mol. Biol. 2001;312:381–391. doi: 10.1006/jmbi.2001.4935. [DOI] [PubMed] [Google Scholar]
- Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- Liu Y, Santi DV. m5C RNA and m5C DNA methyltransferases use different cysteine residues as catalysts. Proc. 2000;97:8263–8265. doi: 10.1073/pnas.97.15.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Rychlewski L. mRNA:guanine-N7 cap methyltransferases: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. BMC Bioinformatics. 2001;2:2. doi: 10.1186/1471-2105-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Trotta CR, Abelson J. Crystal structure and evolution of a transfer RNA splicing enzyme. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- Li H, Abelson J. Crystal structure of a dimeric archaeal splicing endonuclease. J Mol. Biol. 2000;302:639–648. doi: 10.1006/jmbi.2000.3941. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM, Elofsson A, Fischer D, L Rychlewski. Structure prediction Meta Server. Bioinformatics. 2001;17:750–751. doi: 10.1093/bioinformatics/17.8.750. [DOI] [PubMed] [Google Scholar]
- Kelley LA, McCallum CM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Fischer D. Hybrid fold recognition: combining sequence derived properties with evolutionary information. Pac. 2000. pp. 119–130. [PubMed]
- Jones DT. GenTHREADER: an efficient and reliable protein fold recognition method for genomic sequences. J Mol. Biol. 1999;287:797–815. doi: 10.1006/jmbi.1999.2583. [DOI] [PubMed] [Google Scholar]
- Karplus K, Barrett C, Cline M, Diekhans M, Grate L, Hughey R. Predicting protein structure using only sequence information. Proteins. 1999;Suppl 3:121–125. doi: 10.1002/(SICI)1097-0134(1999)37:3+<121::AID-PROT16>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. Fugue: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Lundstrom J, Rychlewski L, Bujnicki JM, Elofsson A. Pcons: A neural-network-based consensus predictor that improves fold recognition. Protein Sci. 2001;10:2354–2362. doi: 10.1110/ps.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends. 1995;20:478–480. doi: 10.1016/S0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Scavetta RD, Thomas CB, Walsh MA, Szegedi S, Joachimiak A, Gumport RI, Churchill ME. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 2000;28:3950–3961. doi: 10.1093/nar/28.20.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PH, Korszun ZR, Cerritelli S, Springhorn SS, Lacks SA. Crystal structure of the DpnM DNA adenine methyltransferase from the DpnII restriction system of Streptococcus pneumoniae bound to S-adenosylmethionine. Structure. 1998;6:1563–1575. doi: 10.1016/s0969-2126(98)00154-3. [DOI] [PubMed] [Google Scholar]
- Gong W, O'Gara M, Blumenthal RM, Cheng X. Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]