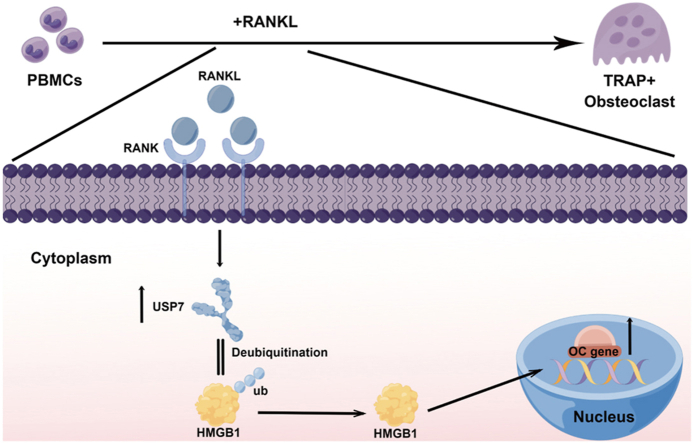
Abstract
Background
Abnormal osteoclast and osteoblast differentiation is an essential pathological process in osteoporosis. As an important deubiquitinase enzyme, ubiquitin-specific peptidase 7 (USP7) participates in various disease processes through posttranslational modification. However, the mechanism by which USP7 regulates osteoporosis remains unknown. Herein, we aimed to investigate whether USP7 regulates abnormal osteoclast differentiation in osteoporosis.
Methods
The gene expression profiles of blood monocytes were preprocessed to analyze the differential expression of USP genes. CD14+ peripheral blood mononuclear cells (PBMCs) were isolated from whole blood collected from osteoporosis patients (OPs) and healthy donors (HDs), and the expression pattern of USP7 during the differentiation of CD14+ PBMCs into osteoclasts was detected by western blotting. The role of USP7 in the osteoclast differentiation of PBMCs treated with USP7 siRNA or exogenous rUSP7 was further investigated by the F-actin assay, TRAP staining and western blotting. Moreover, the interaction between high-mobility group protein 1 (HMGB1) and USP7 was investigated by coimmunoprecipitation, and the regulation of the USP7-HMGB1 axis in osteoclast differentiation was further verified. Osteoporosis in ovariectomized (OVX) mice was then studied using the USP7-specific inhibitor P5091 to identify the role of USP7 in osteoporosis.
Results
The bioinformatic analyses and CD14+ PBMCs from osteoporosis patients confirmed that the upregulation of USP7 was associated with osteoporosis. USP7 positively regulates the osteoclast differentiation of CD14+ PBMCs in vitro. Mechanistically, USP7 promoted osteoclast formation by binding to and deubiquitination of HMGB1. In vivo, P5091 effectively attenuates bone loss in OVX mice.
Conclusion
We demonstrate that USP7 promotes the differentiation of CD14+ PBMCs into osteoclasts via HMGB1 deubiquitination and that inhibition of USP7 effectively attenuates bone loss in osteoporosis in vivo.
The translational potential of this article:The study reveals novel insights into the role of USP7 in the progression of osteoporosis and provides a new therapeutic target for the treatment of osteoporosis.
Keywords: USP7, Osteoporosis, Osteoclast differentiation, Deubiquitination
Graphical abstract
1. Introduction
Osteoporosis is a degenerative disease characterized by decreased bone mass and destruction of the bone structure, which result in a decrease in bone strength and an increase in the incidence of fractures [1]. According to recent statistics, the prevalence rate of osteoporosis in the Chinese male and female population over the age of 40 years are 5.0% and 20.6%, respectively, and the prevalence rates of vertebral fracture in these populations are 10.5% and 9.7%, respectively [2]. At present, drug therapy is the main method used to treat osteoporosis, but commonly used medications such as bisphosphonates, calcium, estrogen, and selective estrogen receptor modulators [3] have drawbacks that should not be disregarded, such as varying degrees of side effects and a lack of therapeutic targeting [[4], [5], [6]]. Therefore, it is necessary to further elucidate the specific molecular mechanism of osteoporosis to help identify new therapeutic targets.
There is an obvious imbalance in bone remodeling in osteoporosis, and bone remodeling in vivo mainly depends on the balance between the osteogenic ability of osteocytes and the bone resorption capacity of osteoclasts [7]. Human osteoclasts are mainly derived from CD14+ monocytes [8]. After stimulation with monocyte colonization stimulating factor (M-CSF) and NF-κB ligand receptor activator (RANKL), monocytes can differentiate into osteoclasts [9], and excessive activation of osteoclast differentiation plays a key role in the imbalance between osteoclasts and osteoblasts [10]. Therefore, further research on the molecular mechanism responsible for the abnormal activation of osteoclast differentiation would be helpful for understanding the occurrence and development of osteoporosis.
In recent years, posttranslational modification has been regarded as a critical factor regulating various biological activities, including the regulation of bone remodeling. As a crucial type of posttranslational modification, ubiquitin‒proteasome proteins are specifically recognized by proteasomes after binding with ubiquitin enzymes [11]. Many studies have demonstrated that the ubiquitin‒proteasome system is crucial for regulating bone remodeling [12]. We have previously revealed the mechanisms through which Smurf2 ubiquitination-mediated degradation positively regulates the osteogenic differentiation of human mesenchymal stem cells [13]. In addition, ubiquitination is a reversible process because deubiquitinating enzymes can prevent proteasome degradation by removing ubiquitin from substrate proteins [14]. As an important deubiquitinase enzyme, ubiquitin-specific peptidase 7 (USP7) was recently reported to be involved in the occurrence, progression and targeted therapy of various diseases [15]. Furthermore, emerging evidence has indicated the key role of USP7 in regulating bone remodeling. For example, USP7 regulates osteogenic differentiation by binding and deubiquitination of AXIN, a key inhibitor of the Wnt signaling pathway and a key component of the β-catenin complex [16]. However, the effect of USP7 on osteoclasts, the significance of the occurrence and progression of osteoporosis and the possibility of targeted therapy have not been clarified.
Here, we demonstrated that USP7 expression was significantly upregulated in osteoporotic osteoclasts and increased during the process of osteoclast differentiation from peripheral blood monocytes in vitro. Osteoclast differentiation could be significantly inhibited by siRNA or a specific inhibitor (P5091). Through protein spectrum analysis, we found that USP7 can bind to HMGB1 and promote osteoclast formation. Given the specific effect of USP7 on osteoclast formation and the current research results, we propose that USP7 may be a key regulatory factor in osteoclast differentiation of CD14+ PBMCs and a new target for targeted therapy of osteoporosis in the future.
2. Materials and methods
2.1. Data retrieval
The gene expression profile data in the GEO Platform dataset GSE56814, which includes blood monocytes from 42 patients with a normal bone mineral density (BMD) and 31 patients with a low BMD, and the GEO Platform dataset GSE56815, which includes blood monocytes from 40 premenopausal and 40 postmenopausal patients, were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) database via the GEOquery package (2.64.2) in R (4.2.0). The probes of the expression matrix were annotated into gene symbols through the dplyr (1.0.10) package and the GEO Platform data. Members of the USP gene family were obtained from the PathCards (https://pathcards.genecards.org/) database. Common USP genes in the above mentioned three datasets were filtered using the dplyr (1.0.10) package, and a Venn diagram was drawn with the ggVennDiagram (1.2.2) package.
2.2. Correlation analysis
Spearman's correlation coefficients of the USP genes with the menopausal status and BMD were calculated, and correlation bubble charts were plotted using ggcorrplot (0.1.4) packages.
2.3. Statistical test and box diagram preparation
The expression of USP25, USP7, USP9X, and USP16 in patients with different clinical characteristics was analyzed by a t test, and box diagrams were plotted using the ggplot2 (3.4.0) package.
2.4. Cell isolation and culture
In this study, 30 healthy donors (HDs) and 30 patients diagnosed with osteoporosis (osteoporosis patients, OPs) were recruited. The protocol was approved by the Ethics Committee of The Eighth Affiliated Hospital (Sun Yat-sen University, Guangzhou, China), and informed consent was obtained from all subjects. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood collected from the HDs and OPs. As previously described [17], density-gradient centrifugation was used to separated PBMCs. According to the manufacturer's instructions (Miltenyi Biotech), CD14+ monocytes were isolated from PBMCs labeled with magnetic-bead-conjugated anti-human CD14 mAbs via positive selection. Briefly, PBMCs were treated with fluorescein isothiocyanate (FITC)-conjugated anti-CD14 mAb (BD Biosciences) for 30 min on ice in the dark. Washed the cells with phosphate-buffered saline (PBS) three times. Then a BD Influx cell sorter (BD Biosciences) was used to identify the purity of the CD14+ monocytes. All the cells were cultured at 37 °C in 5% CO2 with α-MEM (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco).
2.5. Source of clinical bone tissue specimens
In this study, spinous process specimens were obtained from the above mentioned peripheral blood donors (including those in the HD and OP groups) who needed open lumbar surgery, and the specimens cut during the operation were subjected to decalcification, paraffin embedding, and section treatment for the next experiment. The protocol was approved by the Ethics Committee of The Eighth Affiliated Hospital (Sun Yat-sen University, Guangzhou, China), and informed consent was obtained from all subjects. Details of the donors are in Table 1.
Table 1.
Details of donors.
| OP group | ||||
|---|---|---|---|---|
| Gender | T score | Age | Diagnosis | Open surgery was performed |
| Male | −2.6 | 71 | Vertebral compression fracture (L1) | NO |
| Male | −2.7 | 69 | Vertebral compression fracture (L1) | NO |
| Male | −2.5 | 65 | Lumbar disc Herniation (L4/5) | YES |
| Male | −2.6 | 75 | Lumbar disc Herniation (L4/5) | YES |
| Male | −2.8 | 67 | Vertebral compression fracture (T12) | NO |
| Male | −2.7 | 62 | Vertebral compression fracture (T12) | NO |
| Male | −2.6 | 63 | Lumbar spinal stenosis (L4/5) | YES |
| Male | −2.6 | 65 | Lumbar spondylolisthesis (L4) | YES |
| Male | −2.7 | 65 | Lumbar disc Herniation (L5/S1) | NO |
| Male | −2.6 | 72 | Vertebral compression fracture (L1) | NO |
| Male | −2.5 | 80 | Lumbar spinal stenosis (L4/5) | YES |
| Male | −2.5 | 77 | Vertebral compression fracture (L1、L3) | YES |
| Male | −2.6 | 62 | Lumbar spinal stenosis (L4/5、L5/S1) | NO |
| Male | −2.7 | 68 | Vertebral compression fracture (T10) | NO |
| Male | −2.7 | 66 | Lumbar spinal stenosis (L5/S1) | NO |
| Female | −2.8 | 62 | Vertebral compression fracture (L1) | NO |
| Female | −2.8 | 70 | Vertebral compression fracture (L3) | YES |
| Female | −2.7 | 59 | Lumbar disc Herniation (L4/5) | YES |
| Female | −2.6 | 55 | Lumbar spinal stenosis (L4/5、L5/S1) | YES |
| Female | −2.8 | 63 | Lumbar spinal stenosis (L4/5、L5/S1) | YES |
| Female | −2.8 | 68 | Vertebral compression fracture (T8) | NO |
| Female | −2.5 | 65 | Lumbar spinal stenosis (L4/5) | NO |
| Female | −2.6 | 72 | Lumbar spinal stenosis (L5/S1) | YES |
| Female | −2.5 | 76 | Lumbar disc Herniation (L4/5) | YES |
| Female | −2.7 | 68 | Vertebral compression fracture (L1) | NO |
| Female | −2.6 | 65 | Lumbar spinal stenosis (L4/5) | YES |
| Female | −2.8 | 66 | Lumbar spinal stenosis (L4/5) | YES |
| Female | −2.6 | 67 | Vertebral compression fracture (L3) | NO |
| Female | −2.8 | 58 | Lumbar spinal stenosis (L5/S1) | YES |
| Female |
−2.7 |
69 |
Lumbar disc Herniation (L4/5) |
NO |
| HD group | ||||
| Gender |
T score |
Age |
Diagnosis |
Open surgery was performed |
| Male | −0.3 | 26 | Lumbar disc Herniation (L4/5) | NO |
| Male | 0.5 | 25 | Lumbar disc Herniation (L4/5) | NO |
| Male | 0.7 | 26 | Lumbar disc Herniation (L4/5、L5/S1) | YES |
| Male | 0.1 | 24 | Vertebral compression fracture (L3) | YES |
| Male | 0.2 | 23 | Lumbar disc Herniation (L4/5) | NO |
| Male | −0.5 | 27 | Vertebral compression fracture (L3) | YES |
| Male | −0.4 | 35 | Lumbar disc Herniation (L4/5、L5/S1) | YES |
| Male | 0.3 | 21 | Burst fracture of spine (L1) | YES |
| Male | −0.5 | 25 | Lumbar disc Herniation (L4/5) | NO |
| Male | −0.6 | 26 | Lumbar disc Herniation (L4/5、L5/S1) | NO |
| Male | −0.7 | 30 | Lumbar disc Herniation (L4/5) | YES |
| Male | 0.6 | 25 | Burst fracture of spine (T12、L1、L2) | |
| Male | 0.1 | 27 | Lumbar disc Herniation (L4/5、L5/S1) | YES |
| Male | 0 | 31 | Lumbar disc Herniation (L4/5、L5/S1) | YES |
| Male | 0.8 | 28 | Lumbar disc Herniation (L4/5、L5/S1) | NO |
| Female | −0.2 | 23 | Burst fracture of spine (L1) | YES |
| Female | −0.5 | 25 | Lumbar disc Herniation (L4/5) | NO |
| Female | −0.3 | 28 | Vertebral compression fracture (L1) | YES |
| Female | −0.1 | 29 | Lumbar disc Herniation (L4/5) | NO |
| Female | 0.6 | 27 | Vertebral compression fracture (L1) | NO |
| Female | 0.4 | 26 | Lumbar disc Herniation (L4/5) | YES |
| Female | 0.3 | 25 | Lumbar disc Herniation (L4/5) | NO |
| Female | 0.8 | 32 | Lumbar disc Herniation (L4/5) | NO |
| Female | −0.5 | 24 | Lumbar disc Herniation (L4/5、L5/S1) | NO |
| Female | −0.6 | 25 | Lumbar spondylolisthesis (L4) | YES |
| Female | 0.5 | 27 | Lumbar disc Herniation (L4/5) | NO |
| Female | 0.4 | 26 | Vertebral compression fracture (L1) | NO |
| Female | 0.6 | 24 | Lumbar disc Herniation (L4/5、L5/S1) | YES |
| Female | 0.8 | 26 | Vertebral compression fracture (L1) | NO |
| Female | −0.2 | 27 | Lumbar disc Herniation (L4/5) | NO |
2.6. H&E staining
The samples were dealkylated in xylene and hydrated with decreasing ethanol concentrations. For H&E staining, the slices were first incubated with hematoxylin for 5 min and then stained with eosin for 3 min after clearance [17]. All sections were dehydrated and observed using a microscope (Nikon).
2.7. Immunohistochemical staining
Immunohistochemical staining was performed as previously described [18]. In short, after antigen recovery, the section was sealed in 5% serum and then incubated with anti-USP7 (Abcam 264,422) at 4 °C overnight. Subsequently, the sections were incubated with the appropriate biotin-conjugated secondary antibodies and then with streptavidin solution. After staining, all the sections were dehydrated with increasing ethanol and xylene concentrations. All sections were observed using an optical microscope (Nikon).
2.8. Immunofluorescence staining
Immunofluorescence staining was performed as detailed by Xie et al. [19]. As mentioned above, the slices were dewaxed with xylene and ethanol. The cells were then fixed with 4% paraformaldehyde and incubated with 0.1% Triton X-100 at room temperature for 10 min. After blocking with goat serum, the cells were incubated with anti-USP7 (Abcam 264,422) and anti-TRAP (Abcam 191,406) antibodies overnight at 4 °C. The appropriate secondary antibody was subsequently added, and the cells were incubated at room temperature for 1 h. The sections were then restained with 4′,6-diamino-2-phenylindole (DAPI) to label the nucleus. After application of anti-fading installation medium (P0126 Magi Beyotime), the slides were covered with cover slips. An image of the sample was captured using an Axio Observer imaging system (Carl Zeiss).
2.9. Real-time quantitative reverse transcription–polymerase chain reaction (qRT‒PCR)
The experiment was performed as described by Cen et al. in detail [20]. Briefly, the cells were washed three times with PBS, and total RNA was extracted after the addition of TRIzol (Invitrogen). According to the instructions, the PrimeScript RT kit (TaKaRa) was used for the transcription of complementary DNA from 1 μg RNA. The LightCycler ®480 PCR system (Roche) and SYBR Premix Ex Taq (TaKaRa) were used for qRT‒PCR. The qRT‒PCR procedure was 95 °C for 30 s and 40 cycles of 95 °C for 5 s and 60 °C for 20 s. Each sample was analyzed in triplicate, and the average mRNA level was calculated. For confirmation of the specific magnification of the target, the melting curve of each sample was analyzed. GAPDH was used as an internal control, and the 2-delta Ct method was used. The forward and reverse primers for the target genes are shown in Table 2.
Table 2.
Primers.
| Gene symbol | Forward (5’→3′) | Reverse (3’→5′) |
|---|---|---|
| hGAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
| hUSP7 | GGAAGCGGGAGATACAGATGA | AAGGACCGACTCACTCAGTCT |
| hCTSK | GCAGAAGAACCGGGGTATTGA | GAAGGAGGTCAGGCTTGCAT |
| hACP5 | GACTGTGCAGATCCTGGGTG | GACTGTGCAGATCCTGGGTG |
| hNFATC1 | CACCGCATCACAGGGAAGAC | GCACAGTCAATGACGGCTC |
| hMMP9 | AGACCTGGGCAGATTCCAAAC | CGGCAAGTCTTCCGAGTAGT |
2.10. Western blot analysis
As described previously [21], the cells were lysed on ice for 30 min in RIPA buffer (Sigma) with 1% protease and phosphatase inhibitor (Roche). The lysate was then centrifuged at 12,000 rpm at 4 °C for 30 min, and the protein in the supernatant was quantified using a BCA Analysis Kit (Sigma) according to the manufacturer's instructions. The same amount of protein was diluted in sodium dodecyl sulfate (SDS) loading buffer, separated by 10% SDS-polyacrylamide gel electrophoresis, and electrotransferred to a polyvinylidene fluoride (EMD Millipore Billerica) membrane. The membrane was blocked in TBST with 5% skim milk at room temperature for 60 min and incubated overnight with primary antibodies against GAPDH (CST 5174), CTSK (Abcam 85,716), USP7 (CST 4833 AND Abcam 264,422) and HMGB1 (Abcam 18,256) at 4 °C. The membrane then was washed three times with TBST and incubated at room temperature with the appropriate secondary antibody against horseradish peroxidase (HRP) (diluted 1:1000, Santa Cruz Biotechnology) for 60 min. The membrane was washed three times with TBST, and the signal was detected with Immobilon Western Chemiluminescence HRP Substrate (Millipore).
2.11. TRAP staining
At days 0, 3, 6 and 9 after RANKL induction, the cells were collected and washed three times with PBS, and the TRAP activity was detected with a leucocyte acid phosphatase kit (Sigma #387A) according to the manufacturer's instructions. Paraffin sections of mouse femurs were treated as described above. TRAP-positive cells with at least three nuclei were considered osteoclasts. At least 5 visual fields covering the entire plate were evaluated, and the average number of osteoclasts was calculated [17].
2.12. F-actin assay
At days 0, 3, 6 and 9 after adding RANKL, the cells were fixed with 4% paraformaldehyde for 5 min and washed extensively with PBS. Then, the cells were stained at room temperature with FITC-conjugated phalloidin (Sigma) and 4-diamino-2-phenylindole for 40 min [22]. Subsequently, the cells were washed three times with PBS and observed with an Axio Observer fluorescence microscope (Carl Zeiss).
2.13. Coimmunoprecipitation and LC‒MS/MS
CD14+ PBMCs were inoculated into 10 cm dishes, lysed and coimmunoprecipitated using a DynabeadsTM protein G immunoprecipitation kit (Invitrogen 1007D) according to the manufacturer's instructions. The primary antibodies used in this experiment included USP7 (Abcam 264,422), HMGB1 (Abcam 18,256) and their IgG control (CST 37988). The immunoprecipitate was collected, washed and boiled in Laemmli sample buffer for 10 min. The samples were separated by SDS‒PAGE and then stained using a Coomassie blue staining kit (Beyotime Institute of Biotechnology). The different bands were collected for further LC‒MS/MS analysis of USP7-interacting proteins in PBMCs. As mentioned above, Western blotting was performed to confirm the interaction between the two proteins [23,24].
2.14. Animal studies
Twenty female C57BL/6 mice (body weight, 22.2 ± 0.13 g) were purchased from the Laboratory Animal Center of Sun Yat-sen University at two months of age and housed in public facilities at the School of Medicine of Sun Yat-sen University (Guangzhou, China). The animals were housed in pathogen-free facilities with a temperature of 25 °C, 75% humidity, a 12 h light/dark cycle, and water and food provided at will. The animal experiments were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals (8th Edition, ISBN-10:0–309-15396-4). The protocol was approved by the Committee on the Management and Use of Laboratory Animals of Sun Yat-sen University and the Ethics Committee for Laboratory Animals of Sun Yat-sen University (Ethics number: SYSU-IACUC-2023-000298).
The mice were randomly divided into different groups according to their average body weight using the random function in Excel. Mouse models of estrogen consumption were established by sham operation (n = 5) or ovariectomy (OVX, n = 20) to simulate the clinical situation of osteoporosis patients [25]. OVX mice were randomly divided into 4 groups: OVX group, OVX + P5091 group (P5091, 5 mg/kg, twice a week, intravenous injection) and OVX + P5091 group (P5091, 10 mg/kg, twice a week, caudal intravenous injection). The mice were anesthetized with isoflurane during the procedure.
The sham, OVX, and OVX + P5091 groups started treatment 3 days after surgery. The mice in the sham and OVX groups were injected with 0.9% normal saline. After 6 weeks of treatment, the mice were sacrificed by cervical dislocation after isoflurane anesthesia to relieve pain. Both femurs of the mice were removed and placed in 10% formalin solution for subsequent examination. The trabecular microstructure of the left femur was measured by microCT. The right femur was decalcified, embedded in paraffin and sectioned, and histopathologic detection was then performed with H&E staining, TRAP staining and immunohistochemical methods.
2.15. Statistical analysis
All experiments in vitro were repeated at least three times. SPSS 22.0 software (Chicago, IL, USA) was used to perform the statistical analysis. The normality of data was verified with Shapiro–Wilk test. All normally distributed values were expressed as means±SDs. Student's t test was used to compare two groups, while one-way analysis of variance (ANOVA) was performed to compare three or more groups followed by a Bonferroni test. Correlation analyses was performed with the Pearson correlation test. A P value < 0.05 was considered as statistically significance.
3. Results
3.1. The deubiquitination enzyme USP7 may be involved in the pathogenesis of osteoporosis
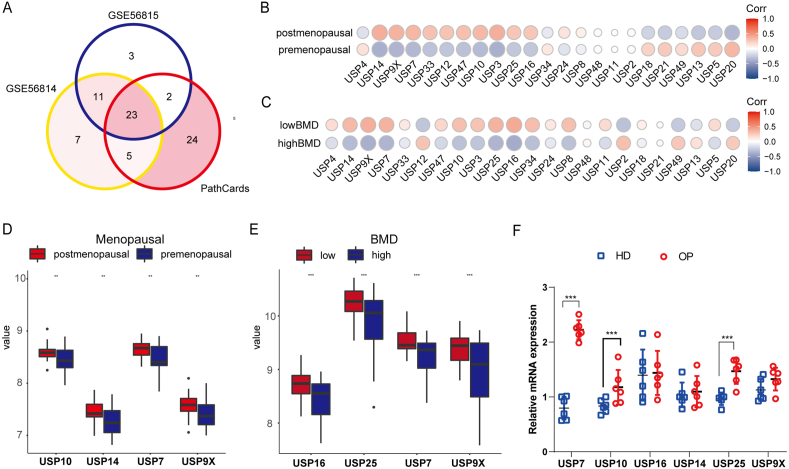
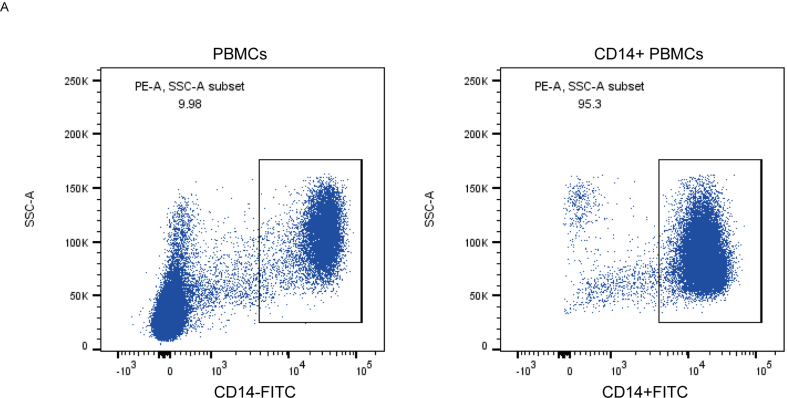
Two gene chips of human peripheral blood monocytes were obtained from the GEO database: one for premenopausal and postmenopausal women and one for human peripheral blood monocytes from patients with high and low BMD. We compared the USP family genes in the PathCards database with two gene chips. The Venn diagram showed that GSE56814 contained 47 USP genes, GSE56815 contained 39 USP genes and PathCards contained 55 USP genes and that 23 USP genes were found in the three datasets (Fig. 1A). Subsequently, the relationship between 23 USP genes and clinical characteristics was analyzed, and the results demonstrated that USP7, USP9X, USP14, USP16 and USP25 were significantly correlated with menopause status (Fig. 1B) and BMD (Fig. 1C). Moreover, the gene expression of USP7, USP9X, USP10 and USP14 was significantly upregulated in postmenopausal women compared with premenopausal women (Fig. 1D P = 0.0072, P = 0.0041, P = 0.0099 and P = 0.0022), and USP7, USP9X, USP16 and USP25 were significantly upregulated in women with a low BMD compared with women with a high BMD (Fig. 1E P = 0.00071, P = 0.00021, P = 0.00031 and P = 0.00067). We collected CD14+ peripheral blood mononuclear cells (PBMCs) from 9 patients with osteoporosis and 9 healthy donors. Flow cytometry confirmed that the purification rate of CD14+ PBMCs was >95% (Supplementary Figure S1). Subsequently, we extracted RNA according to the above mentioned steps and detected the mRNA expression of USP10, USP16, USP14, USP25, USP7 and USP9X. We found that the mRNA level of USP7 in OPs was significantly higher than that in HDs (Fig. 1F); thus, USP7 may be a key molecule in the deubiquitinase family that can affect osteoclast production.
Fig. 1.
The deubiquitination enzyme USP7 may be involved in the pathogenesis of osteoporosis. The gene chips GSE56815 and GSE56815 of premenopausal and postmenopausal women and patients with osteoporosis were obtained from the GEO database, and the datasets of USP family members were obtained from PathCards. (A) The Venn diagram shows 23 common USP genes in the three datasets. (B, D) Heatmap and box map showing that the expression of the USP7, USP9X, USP10 and USP14 genes in postmenopausal women was significantly higher than that in premenopausal women (P = 0.0072, 0.0041, 0.0099 and 0.0022). (C, E) Heatmap and box map showing that the expression of the USP7, USP9X and USP16 and USP25 genes in patients with a low BMD was significantly higher than that in patients with a high BMD (P = 0.00071, 0.00021, 0.0031 and 0.00067). (F) The CD14+ PBMCs of 9 patients with osteoporosis and healthy donors were collected, and the expression of USP7, USP10, USP16, USP14, USP25 and USP9X was measured by qRT‒PCR, which revealed that the more significant difference was found for USP7. All data are presented as the means ± standard deviations. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.2. USP7 is highly expressed in CD14+ PBMCs of osteoporosis patients
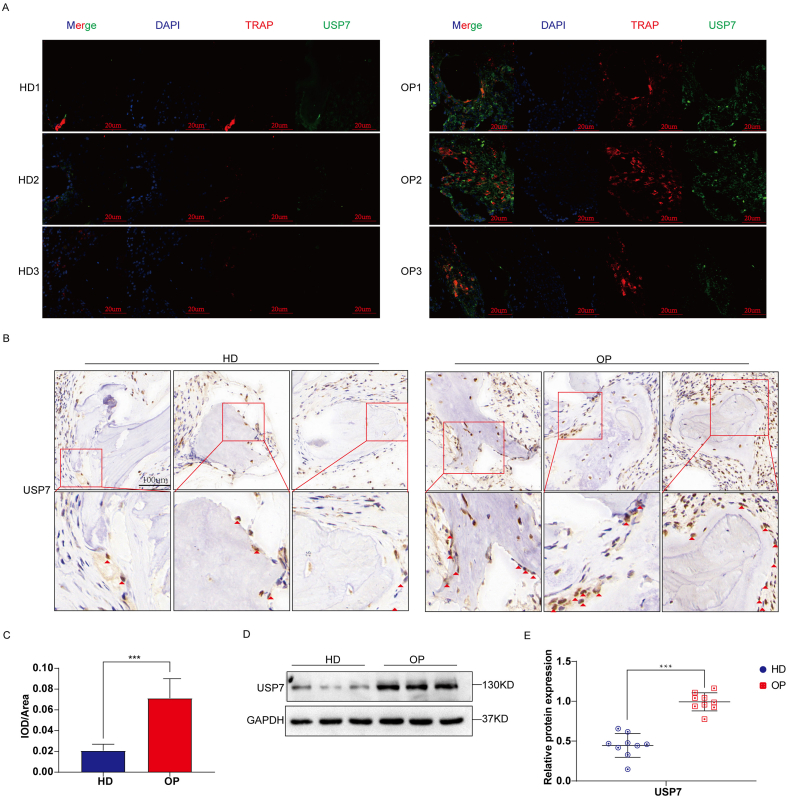
To compare the difference in USP7 expression between OPs and HDs, Western blotting, immunofluorescence, and immunohistochemistry were used to detect the specimen of OPs, HD. The immunofluorescence results showed that the expression of TRAP and USP7 in bone tissue in the OP group was significantly higher than that in the HD group (Fig. 2A). In addition, the immunohistochemistry results showed that the expression of USP7 in bone tissue was significantly increased in the OP group, and this result was semiquantitatively verified by ImageJ (Fig. 2B and C). To further explore the difference in USP7 expression between the OP group and the HD group, we collected CD14+ PBMCs from 9 OPs and HDs and extracted proteins for protein electrophoresis. The expression of the USP7 protein in the OP group was significantly higher than that in the HD group (Fig. 2D). Quantitative analysis showed that there was a significant difference in USP7 expression between the two groups (Fig. 2E). Together, our findings demonstrated that USP7 was significantly upregulated in CD14+ PBMCs of osteoporosis patients.
Fig. 2.
USP7 is highly expressed in CD14+ PBMCs of osteoporosis patients. Bone tissue from vertebrae was collected from healthy donors and osteoporotic patients (n = 3). (A) The expression and location of TRAP (red) and USP7 (green) in these tissues were detected by immunofluorescence. TRAP is considered a marker of osteoclasts. Scale bar: 20 μm (B) The expression of USP7 was detected by immunohistochemistry. Scale bar: 100 μm (C) Immunohistochemical sections were semiquantitatively detected with ImageJ. (D, E) The CD14+ PBMCs of 9 OPs and healthy donors were collected, and the level of USP7 was detected by Western blotting. All data are presented as means ± standard deviation. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. USP7 is positively correlated with the osteoclast differentiation of CD14+ PBMCs
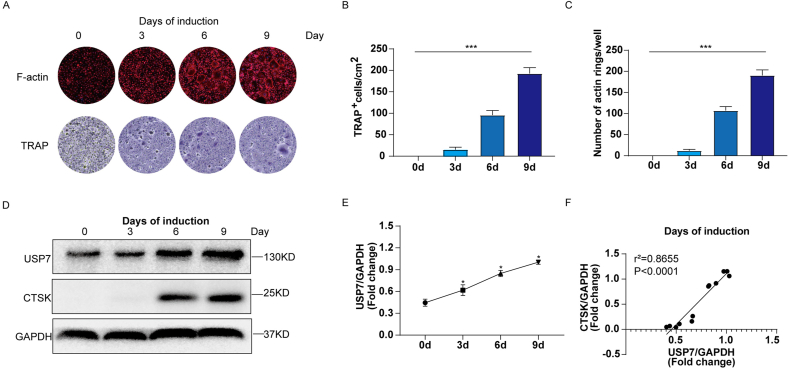
To further analyze the changes in USP7 expression during the process of osteoclast differentiation, we examined osteoclast differentiation induced by CD14+ PBMCs at days 0, 3, 6, and 9 by F-actin immunofluorescence, TRAP staining, and Western blotting. Our results showed that the osteoclasts identified in the F-actin ring and TRAP staining analyses increased gradually from day 0 to day 9 (Fig. 3A). Quantitative analysis showed that the number of osteoclasts and F-actin rings increased with increasing induction time (Fig. 3B and C). In addition, the protein level of USP7 increased with increasing induction time. Consistent with these results, the protein expression of CTSK, a key marker of osteoclast differentiation, gradually increased within 9 days during osteoclast differentiation (Fig. 3D and E). Further analysis showed that a strong positive correlation between the expression of USP7 and CTSK (Fig. 3F). In other words, USP7 is positively correlated with the osteoclast differentiation of CD14+ PBMCs.
Fig. 3.
USP7 is positively correlated with the osteoclast differentiation of CD14+ PBMCs. (A) F-actin and TRAP staining showed a gradual increase in the number of osteoclasts from day 0–9. Scale bar: 100 μm (B, C) Semiquantitative analysis showed the degree of F-actin ring formation and the number of TRAP-positive cells from day 0–9. (D, E) Western blotting showed that the protein levels of USP7 and CTSK increased gradually during osteoclast differentiation (0 d, 3 d, 6 d, 9 d). (F) The Pearson correlation test showed a strong positive correlation between USP7 expression and CTSK expression. All data are displayed as means ± standard deviations (three independent experiments and three different PBMC samples). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.4. USP7 positively regulates the osteoclast differentiation of CD14+ PBMCs in vitro
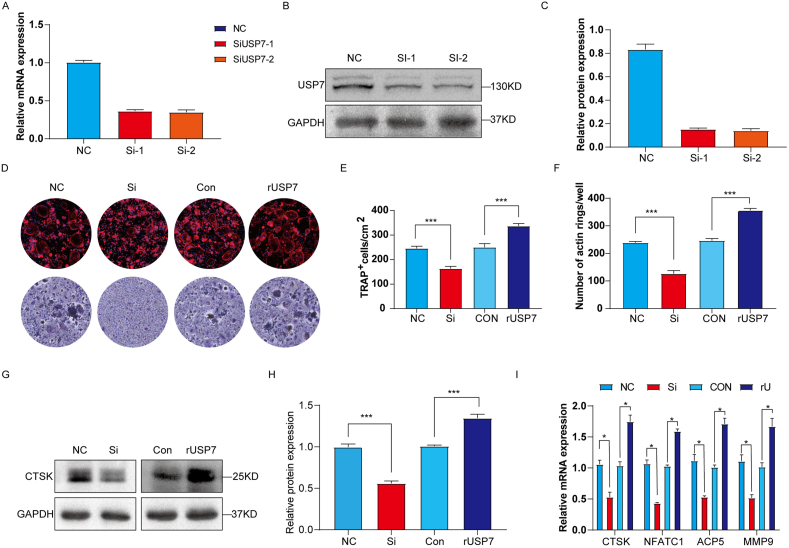
To confirm that USP7 is involved in the osteoclast differentiation of CD14+ PBMCs, we constructed siRNA to reduce the expression of USP7 and evaluated the role of USP7 in osteoclast formation. The mRNA (Fig. 4A) and protein (Fig. 4B and C) level of USP7 in the siRNA group was significantly lower than that in the Negative control (NC) group, and no significant difference in knockdown efficiency was found between the two different sequences of siRNA tested. The results showed that siRNA interfered with the production of USP7 protein, and Si-1 was chosen for the further experiments.
Fig. 4.
USP7 positively regulates the osteoclast differentiation of CD14+ PBMCs in vitro. In the knockdown group, CD14+ PBMCs were transfected with siRNA for 5 h, and the medium was then replaced with osteoclast induction medium. In the rUSP7 group, the cells were cultured with 0.5 μg/ml rUSP7 when the osteoclast culture medium was replaced and then cultured for 9 days. (A) The knockdown efficiency of two different sequences of siRNA for USP7 was measured by qRT‒PCR. (B, C) Western blotting was used to detect the changes in USP7 protein levels after USP7 knockdown. (D, E, F) F-actin and TRAP staining results showed that the knockdown group exhibited significantly decreased osteoclast production, whereas rUSP7 promoted the osteoclast differentiation of CD14+ PBMCs. Scale bar: 100 μm (G, H) Western blotting showed that the protein level of CTSK, a marker of osteoclast differentiation, was decreased in the knockdown group, whereas that in the rUSP7 group was increased. (I) The mRNA levels of osteoclast-related markers (CTSK, NFATC1, ACP5 and MMP9) in the NC group, knockdown group, control group and rUSP7 group were measured by qRT‒PCR. All data are displayed as the means ± standard deviation (three independent experiments and three different PBMC samples). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
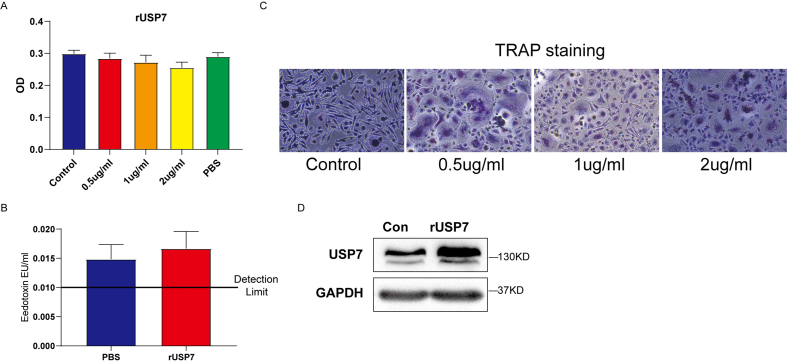
In addition, human recombinant USP7 (rUSP7) was added to constructed high-level USP7 system in CD14+PBMCs. CCK-8 demonstrated that the activity of cells was not significantly affected by 0–2 μg/mL rUSP7 (Supplementary Figure S2A). Also, the endotoxin of rUSP7 was observed and indicated that rUSP7 contained little endotoxin (Supplementary Figure S2B). Furthermore, 0.5 μg/mL rUSP7 positively induced osteoclast formation (Supplementary Figure S2C) and upregulated USP7 protein levels in CD14+ PBMC (Supplementary Figure S2D).
To further explore the function of USP7 on the osteoclast differentiation of CD14+ PBMCs, we detected the formation of the F-actin ring and the production of TRAP-positive cells in the knockdown (Si) group and rUSP7 (0.5 μg/mL) group. The results demonstrated that the formation of F-actin rings and the production of TRAP-positive cells was significantly decreased by USP7 siRNA compared to control group, suggesting USP7 knockdown inhibited the osteoclast differentiation of CD14+PBMCs. In addition, rUSP7 significantly upregulated the formation of F-actin rings and the number of TRAP-positive cells (Fig. 4D). The similar trend in F-actin and TRAP were also observed in CD14+PBMCs (Fig. 4E and F).
In addition, we detected the expression level of CTSK, a marker protein related to osteoclast differentiation. The results showed that CTSK expression was significantly downregulated by USP7 siRNA, whereas the rUSP7 resulted in significant upregulation of CTSK compared to the control group (Fig. 4G and H). Furthermore, the changes of osteoclast-related markers were detected by qRT-PCR. The mRNA levels of CTSK, NFATC1, ACP5 and MMP9 were significantly decreased by USP7 siRNA, but increased in the rUSP7 group compared to the control group (Fig. 4I). In summary, USP7 positively regulates the osteoclast differentiation of CD14+ PBMCs in vitro.
3.5. USP7 regulates osteoclast differentiation by binding to and deubiquitination of HMGB1
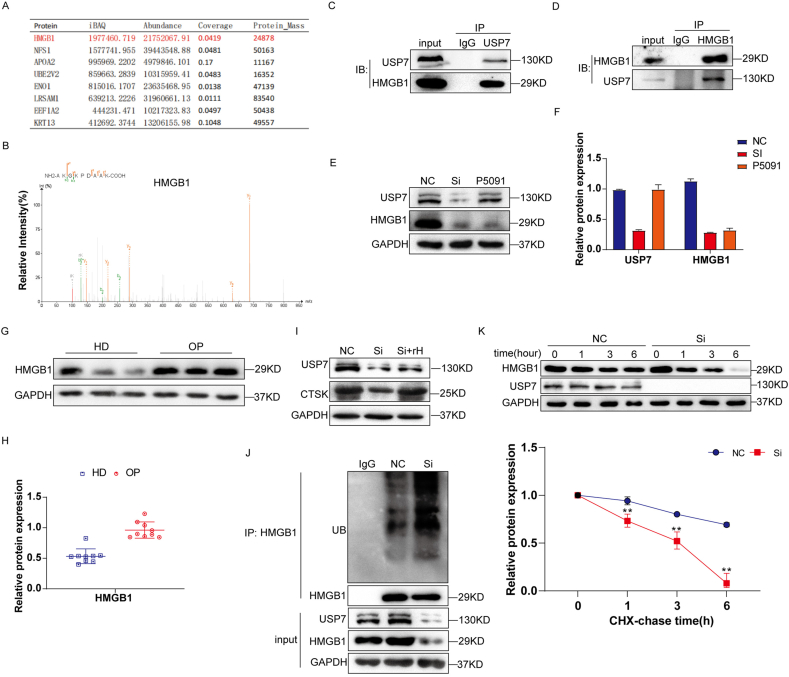
To explore the regulatory mechanism of USP7 on osteoclast formation, Co-IP and LC‒MS/MS experiments were performed to determine the interacting proteins of USP7 during osteoclast differentiation. Several proteins were identified that may interact with USP7. The iBAQ value of HMGB1 detected in the Co-IP complex was the highest, indicating that HMGB1 may be the binding partner of USP7 (Fig. 5A and B). Subsequently, mutual co-IP detection results showed that endogenous USP7 could interact with HMGB1 in CD14+ PBMCs (Fig. 5C and D).
Fig. 5.
USP7 regulates osteoclast differentiation by binding to and deubiquitination of HMGB1. (A, B) LC‒MS/MS results showed that the polypeptide sequence of HMGB1 protein was detected in the Co-IP complex. (C, D) The results of Co-IP detection showed that endogenous USP7 could interact with HMGB1 in CD14+ PBMCs. (E, F) The siRNA and USP7 inhibitor P5091 (5 μM) were used to detect the effect of USP7 on HMGB1. The Western blotting results showed that the expression of HMGB1 protein was decreased in the knockdown group and the P5091 group. (G, H) The CD14+ PMBCs of 9 patients in the OP group and HD group were collected, and the expression of HMGB1 was detected by Western blotting. (I)The expression of USP7 and CTSK in USP7 knockdown CD14+ PMBCs treated with or without rHMGB1 (rH) were detected by Western blotting. (J) After treatment with 10 nmol MG132 for 24 h, the ubiquitination level of the Co–IP complex of HMGB1 was detected in the NC group, Si group and IgG group. (K) After CHX interference of protein synthesis, the HMGB1 degradation rate of HMGB1 in the NC group and Si group was detected. All data are displayed as the means ± standard deviation (three independent experiments and three different CD14+ PBMC samples). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
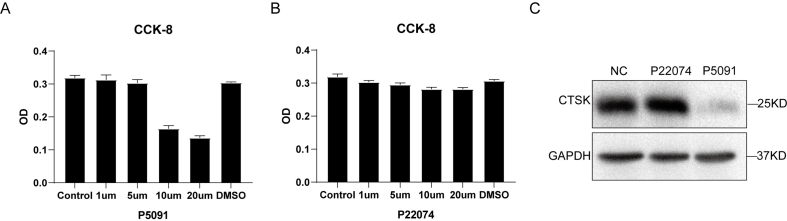
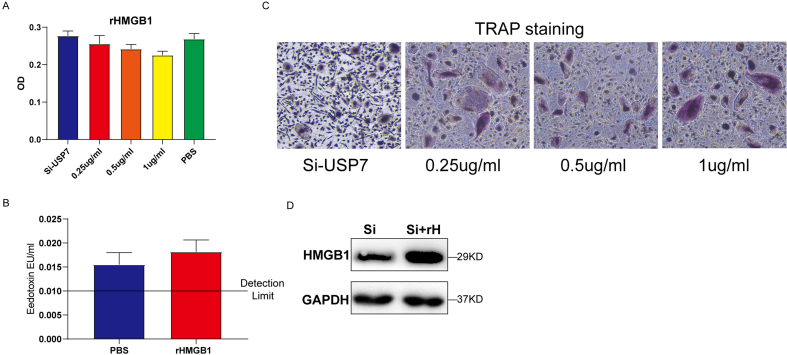
P5091 is an active inhibitor of USP7, and the CCK8 assay results suggested that 5 μM P5091 had no significant effect on cell activity (Supplementary Figure S3A). To verify the specific mechanism by which USP7 regulates HMGB1, we first compared the protein levels of HMGB1 in the NC group, knockdown group and P5091 group and found that the HMGB1 protein levels were significantly decreased in the knockdown group and inhibitor group (Fig. 5E and F). In addition, we detected the HMGB1 expression level in CD14+ PBMCs from 9 patients in the OP group and HD group, which was consistent with previous results. The level of HMGB1 protein in the OP group was significantly higher than that in the HD group (Fig. 5G and H). Similarly, human recombinant HMGB1 (rHMGB1) was used to investigate the role of HMGB1 in the USP7-mediated regulation of the osteoclast differentiation of CD14+ PBMCs. The CCK8 results showed that 0–1 μg rHMGB1 had no significant effect on cell activity (Supplementary Figure S4A). The endotoxin of rHMGB1 was not significantly different from that of PBS (Supplementary Figure S4B). TRAP staining suggested that 0.25 μg/mL rHMGB1 could induce the differentiation of an increased number of osteoclasts (Supplementary Figure S4C). Western blotting showed that the HMGB1 protein levels in the Si + rHMGB1 group were higher than those in the Si group (Supplementary Figure S4D). To further verify the previous results, we added 0.25 μg/mL exogenous rHMGB1 (rH) to the knockdown group and found that the protein level of CTSK increased (Fig. 5I).
As an important deubiquitinase enzyme, USP7 can remove the ubiquitin molecules of substrate proteins and prevent substrate proteins from being recognized and degraded by proteasomes. Therefore, we further investigated the effect of USP7 on the deubiquitination of HMGB1. MG132 (10 nmol, 24 h) was used to inhibit the function of the proteasome. The level of HMGB1 ubiquitination in the knockdown group was significantly higher than that in the NC group (Fig. 5J). In addition, we used cycloheximide (CHX) to inhibit protein synthesis and detected the protein degradation rate of the NC group and the knockdown group. The results demonstrated that the degradation rate of HMGB1 was significantly increased after USP7 knockdown (Fig. 5 K). In summary, HMGB1 is the binding companion of USP7 and plays an important regulatory role in the process of osteoclast differentiation.
3.6. P5091 attenuated bone loss in OVX mice
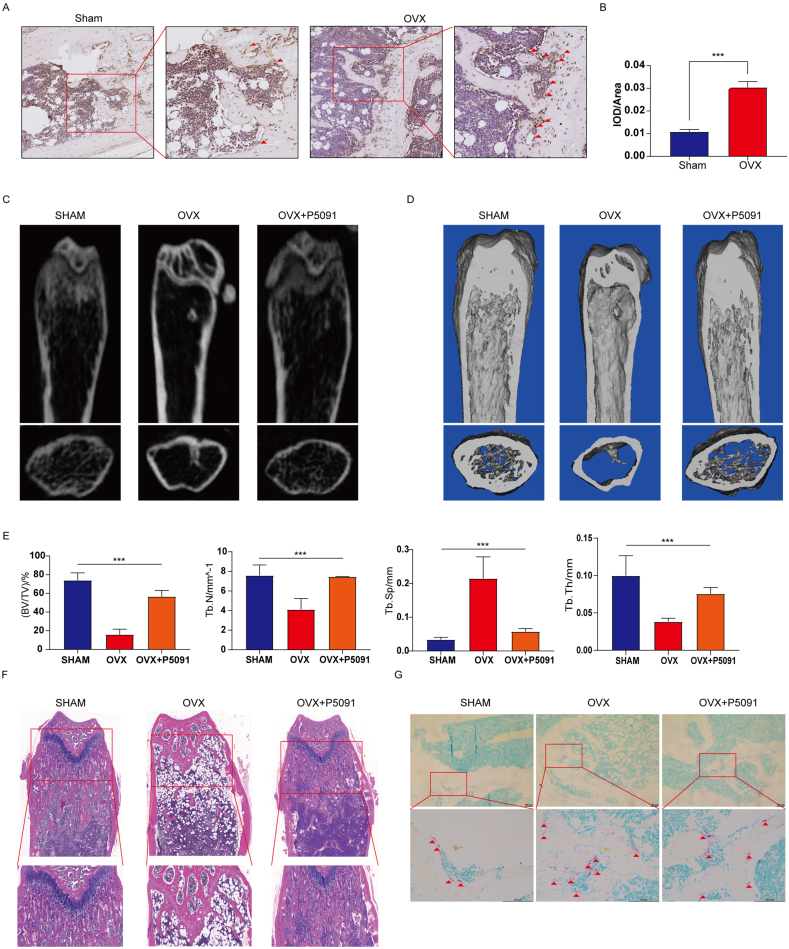
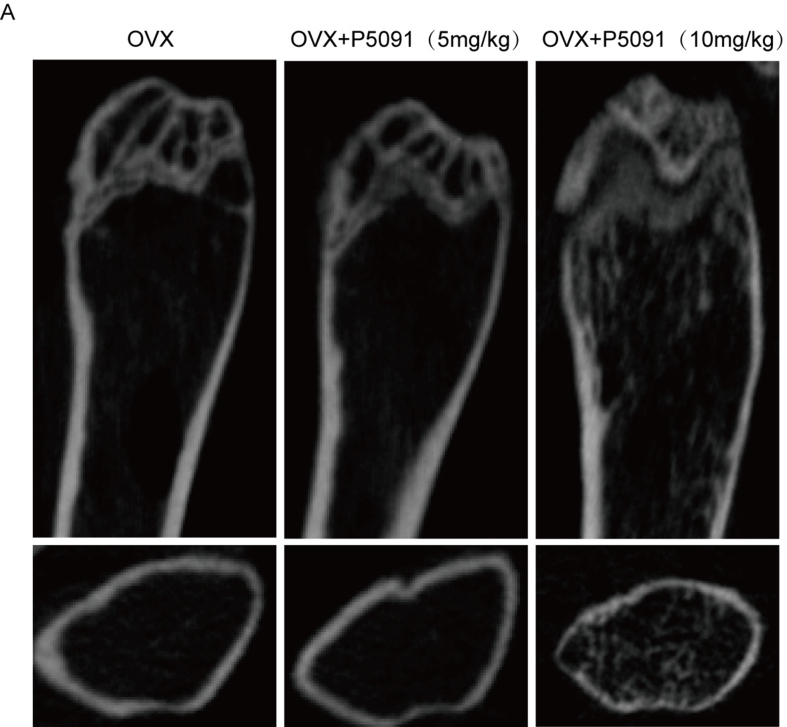
To further demonstrate the role of USP7 in osteoporosis in vivo, we investigated the effect of P5091 on bone mass in OVX mice. First, immunohistochemical determined that USP7 significantly increased the bone tissue of the distal femur of OVX mice compared with that of the Sham group (Fig. 6A and B), which is similar to the expression tend in the bone tissue of OPs. According to the instructions, three doses of P5091 were established to explore its therapeutic effect. MicroCT was used to analyze the changes in the trabeculae of the distal femur in the different model groups. The results determined that a dose of 10 mg/kg effectively attenuated bone loss in OVX mice (Supplementary Figure S5A). Furthermore, compared with the OVX group, the values of BV/TV, Tb.N and Tb. th in the OVX + P5091 group were significantly higher, and that of Tb. sp was lower. In short, P5091 significantly attenuated trabecular bone loss in OVX mice (Fig. 6C, D, E). In addition, OVX + P5091 mice lost significantly more weight than OVX mice during the whole process (Supplementary Figure S6A). Subsequently, H&E staining of femur sections was performed and demonstrated that the trabecular BMD of the distal femoral growth plate was decreased in the OVX group (Fig. 6F). Furthermore, TRAP staining of femur sections demonstrated that the number of distal osteoclasts in the OVX + P5091 group was significantly lower than that in the OVX group but was not significantly different from that in the sham group (Fig. 6G). Overall, P5091 effectively attenuated bone loss by reducing osteoclast production in OVX mice.
Fig. 6.
P5091 alleviated bone loss in OVX mice. (A, B) The expression of USP7 in the distal femurs of the sham group and OVX group was detected by immunohistochemistry. Scale bar: 100 μm (C, D, E) MicroCT was used to scan the images and three-dimensional reconstruction models of distal femoral metaphysis in the sham group, OVX group and OVX + P5091 group. The histogram shows the trabecular structure parameters of the distal femur: BV/TV, Tb.N, Tb.N, and Tb.Th. (F) Sections of distal femurs were stained with HE. Scale bar: 100 μm (G) TRAP staining of the distal femurs of mice in the Sham group, OVX group and OVX + P5091 group was performed. Scale bar: 100 μm. All data are presented as the means ± standard deviation. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
In this study, we evaluated the role of USP7 in the osteoclast differentiation of CD14+ PBMCs and observed high expression of USP7 around the bone trabeculae in OPs and osteoporotic mice. As a deubiquitinating enzyme, USP7 mediates the deubiquitination of HMGB1 and prevents it from being recognized and degraded by proteasomes, which results in positive regulation of the process of osteoclast differentiation. In addition, we evaluated the effect of P5091, a specific inhibitor of USP7, on bone mass in OVX mice and observed that P5091 could effectively attenuate bone mass loss by reducing osteoclast production in OVX mice.
USP7 has received extensive attention and been the subject of much research in recent due to its role in the pathogenesis and progression of various diseases (such as tumors and inflammation) [[26], [27], [28]]. Previous studies have clearly shown that USP7 plays an important role in the regulation of bone remodeling [29]. Ji et al. demonstrated that USP7 can stabilize AXIN1, a key protein in the Wnt pathway, and increase the degradation of β-catenin in osteoblasts, thus participating in the regulation of osteogenic differentiation [30]. However, few subsequent studies have focused on the effect of USP7 on the osteoclast differentiation of CD14+ PBMCs. It has been suggested that other deubiquitinase enzymes affect osteoclast differentiation. For example, CYLD can deubiquitinate TRAF6, a key protein downstream of RANKL, and negatively regulate osteoclast formation [31]. In addition, another member of the USP family, USP34, inhibits osteoclast differentiation by deubiquitinating the activity of IκBα, a key inhibitor of the NF-κB signaling pathway [32]. Therefore, we speculate that USP7 is also involved in the regulation of osteoclast differentiation. In this study, we evaluated the expression of USP7 in bone tissues of OPs and mice with osteoporosis, further analyzed its expression during the osteoclast differentiation in CD14+ PBMCs, and observed that manipulating the expression of USP7 significantly changed the osteoclast differentiation of CD14+ PBMCs in vitro. This result emphasizes for the first time the key role of USP7 not only in the osteoclast differentiation of CD14+ PBMCs but also in the progression of osteoporosis.
However, how USP7 regulates the osteoclast differentiation of CD14+ PBMCs remains unknown. To further explore the regulatory mechanism of USP7 in the process of osteoclast differentiation, we first screened the proteins that interact with USP7 in CD14+ PBMCs. We believe that HMGB1 is a downstream regulatory protein in this process. HMGB1 is a nonhistone nuclear protein that is released into the extracellular environment by mononuclear macrophages after cell activation [33]. Previous studies have shown that the expression of HMGB1 is upregulated during the osteoclast differentiation of CD14+ PBMCs induced by RANKL in vitro, and the recombination of the actin cytoskeleton of osteoclasts is regulated by the receptor for advanced glycation end products (RAGE) [34]. In addition, Yu et al. reported that the absence of HMGB1 led to normal trabecular arrangement and increased the bone mass in OVX mice, which further confirmed that HMGB1 promoted osteoclast formation by activating the NF-κB signaling pathway [35]. However, the upstream mechanism by which HMGB1 expression is regulated during osteoclast differentiation is unclear. In the study, we confirmed that USP7 increased in the process of osteoclast differentiation of CD14+ PBMCs and increased the expression of HMGB1 to promote the process of osteoclast differentiation. These results suggest that USP7 may be an important upstream molecule regulating the expression of HMGB1 in CD14+ PBMCs during osteoclast differentiation. These results suggest that the USP7-HMGB1 axis may promote osteoclast differentiation of CD14+ PBMCs and that USP7 increases the expression of HMGB1 and positively regulates the formation of actin rings and the production of osteoclasts. Therefore, our study complements the existing knowledge about the ubiquitination-deubiquitination regulatory network of osteoclast differentiation and contributes to a better understanding of the molecular mechanism of the osteoclast differentiation of CD14+ PBMC and the pathological process of osteoporosis.
Previous studies have shown that USP7, as an important deubiquitinase enzyme, reduces proteasome degradation of substrate proteins mainly by removing ubiquitin binding to substrate proteins [36]. Degradation is one of the most common and important types of post-translational modification and maintains physiological homeostasis by regulating the stable level of protein expression [37]. In our study, we found that the ubiquitin level of HMGB1 in the knockdown group increased significantly after the function of the proteasome was blocked with MG132. In addition, we used CHX to block protein synthesis, and the protein level of HMGB1 in the USP7 knockdown group decreased significantly. This result shows that USP7 reduces proteasome recognition and degradation by removing ubiquitin from HMGB1. Therefore, our study proved for the first time that USP7 acts as an upstream regulatory molecule of HMGB1 to regulate the osteoclast differentiation of CD14+ PBMCs.
Interestingly, our results were different from those obtained in previous studies on the regulation of osteoclast differentiation by deubiquitinase [38,39]. The following reasons could explain this discrepancy. First, the cells used in the previous studies were RAW264.7 cell lines or mouse bone marrow macrophages, whereas in this study, to better simulate the disease situation of human osteoporosis, we chose CD14+ PBMCs as the only research object, which is more suitable for the application of human life science. Second, different proteins in the same family may have different regulatory effects on the same life activity. For example, studies of the osteogenic differentiation of mesenchymal stem cells (MSCs) have proven that USP34 is essential for the differentiation and bone formation of MSCs. Conditional deletion of USP34 in MSCs weakens the activation of BMP2 by stabilizing the key osteogenic proteins Smad1 and RUNX2 in vitro [40]. Similarly, USP7 stabilizes the scaffold protein AXIN, promotes the complex destroyed by β-catenin (APC-AXIN-GSK-3 β), promotes the degradation of β-catenin, and maintains a very low concentration in normal cells, which results in inhibition of Wnt signal transduction and osteoblast formation [30,41]. However, USP15 and USP4 have related roles in osteogenesis. USP15 is a positive regulator of the BMP signaling pathway, which enhances BMP-mediated SMAD1 phosphorylation by deubiquitination of ALK3 (type I BMP receptor) [42]. In addition, USP4 enhances the Wnt signaling pathway and regulates osteogenic differentiation by deubiquitinating and stabilizing β-catenin [43]. Therefore, we hypothesize that different USP family proteins play a regulatory role in certain life processes by binding to their specific substrates and forming regulatory networks to maintain homeostasis. Generally, the USP7-HMGB1 axis is one of the members of the whole bone homeostasis regulation and plays a positive regulatory role.
Currently, targeted therapy is considered the most promising treatment for all types of inflammation and cancer. However, due to the complexity of the molecular mechanism related to osteoporosis, it is difficult to develop related targeted drugs, which hinders the further development of precision targeted therapy. At present, a number of studies have targeted osteoclast overactivation, which will broaden the idea of targeted treatment for osteoporosis in the future [[44], [45], [46]]. Therefore, our results suggest that USP7 may be a prospective target to improve the targeting efficiency to reduce the excessive activation of osteoclast differentiation of monocytes. We also found that the bone mass loss of osteoporotic mice treated with the USP7-specific inhibitor P5091 was significantly alleviated. Previous studies have shown that the abnormal USP7 expression plays an important role in the pathogenesis of multiple myeloma, gastric cancer, prostate cancer, rheumatoid arthritis and other diseases [47]. Therefore, we found abnormally high expression in OPs and mice with osteoporosis, suggesting that USP7 may play an important role in the pathogenesis of bone metabolic disorders such as osteoporosis. Further study of USP7 may provide new insights into the pathogenesis of bone metabolic disorders and reveal new therapeutic targets. However, this study has some limitations. On the one hand, The transport, intracellular localization and biological activity of recombinant proteins are all extremely concerning. our supplementary experiments cannot be excluded that rUSP7 and rHMGB1 largely reflect the non-physiological and artificial activity of the recombinant protein. On the other hand, to further explore the specific role of USP7 in osteoclast differentiation, researchers must specifically knock in or knock out USP7 in the cells of the osteoclast lineage in transgenic mice, and we are currently developing these transgenic mice for future research.
In conclusion, the study demonstrates that USP7 promotes the progression of osteoporosis via osteoclast differentiation by binding to and deubiquitinating HMGB1 in human peripheral blood monocytes. Our study reveals the significance and mechanism of USP7 in osteoporosis. In addition, P5091 effectively alleviates bone loss by reducing osteoclast production in mice with osteoporosis. These findings may provide a new and effective therapeutic target for the treatment of osteoporosis.
Funding
This work was supported by the National Natural Science Foundation of China (82172385, 82172349), Guangdong Natural Science Foundation (2023A1515010226), Futian Healthcare Research Project (FTWS2021067), and the Shenzhen Science and Technology Program (JCYJ20220530144016039).
Ethics approval and consent to participate
All human-derived cell and animal experiments were approved by the Ethics Committee of The Eighth Affiliated Hospital (Sun Yat-sen University, Guangzhou, China).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2023.05.007.
Contributor Information
Yan-Feng Wu, Email: wuyf@mail.sysu.edu.cn.
Hui-Yong Shen, Email: shenhuiy@mail.sysu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
References
- 1.Johnston C.B., Dagar M. Osteoporosis in older adults. Med Clin. 2020;104(5):873–884. doi: 10.1016/j.mcna.2020.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Wang L., Yu W., Yin X., Cui L., Tang S., Jiang N., et al. Prevalence of osteoporosis and fracture in China: the China osteoporosis prevalence study. JAMA Netw Open. 2021;4(8) doi: 10.1001/jamanetworkopen.2021.21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reid I.R., Billington E.O. Drug therapy for osteoporosis in older adults. Lancet. 2022;399(10329):1080–1092. doi: 10.1016/S0140-6736(21)02646-5. [DOI] [PubMed] [Google Scholar]
- 4.Drake M.T., Clarke B.L., Khosla S. Bisphosphonates: mechanism of action and role in clinical practice. Mayo Clin Proc. 2008;83(9):1032–1045. doi: 10.4065/83.9.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossouw J.E., Anderson G.L., Prentice R.L., LaCroix A.Z., Kooperberg C., Stefanick M.L., et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women's Health Initiative randomized controlled trial. JAMA. 2002;288(3):321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 6.Barrionuevo P., Kapoor E., Asi N., Alahdab F., Mohammed K., Benkhadra K., et al. Efficacy of pharmacological therapies for the prevention of fractures in postmenopausal women: a network meta-analysis. J Clin Endocrinol Metab. 2019;104(5):1623–1630. doi: 10.1210/jc.2019-00192. [DOI] [PubMed] [Google Scholar]
- 7.Kim J.M., Lin C., Stavre Z., Greenblatt M.B., Shim J.H. Osteoblast-osteoclast communication and bone homeostasis. Cells. 2020;9(9) doi: 10.3390/cells9092073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massey H.M., Flanagan A.M. Human osteoclasts derive from CD14-positive monocytes. Br J Haematol. 1999;106(1):167–170. doi: 10.1046/j.1365-2141.1999.01491.x. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto K., Nakashima T., Shinohara M., Negishi-Koga T., Komatsu N., Terashima A., et al. Osteoimmunology: the conceptual framework unifying the immune and skeletal systems. Physiol Rev. 2017;97(4):1295–1349. doi: 10.1152/physrev.00036.2016. [DOI] [PubMed] [Google Scholar]
- 10.Yang D., Wan Y. Molecular determinants for the polarization of macrophage and osteoclast. Semin Immunopathol. 2019;41(5):551–563. doi: 10.1007/s00281-019-00754-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pohl C., Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366(6467):818–822. doi: 10.1126/science.aax3769. [DOI] [PubMed] [Google Scholar]
- 12.Vriend J., Reiter R.J. Melatonin, bone regulation and the ubiquitin-proteasome connection: a review. Life Sci. 2016;145:152–160. doi: 10.1016/j.lfs.2015.12.031. [DOI] [PubMed] [Google Scholar]
- 13.Li J., Wang P., Xie Z., Wang S., Cen S., Li M., et al. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2019;26(12):2652–2666. doi: 10.1038/s41418-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mevissen T., Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 15.Bonacci T., Emanuele M.J. Dissenting degradation: deubiquitinases in cell cycle and cancer. Semin Cancer Biol. 2020;67(Pt 2):145–158. doi: 10.1016/j.semcancer.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji L., Lu B., Zamponi R., Charlat O., Aversa R., Yang Z., et al. USP7 inhibits Wnt/beta-catenin signaling through promoting stabilization of Axin. Nat Commun. 2019;10(1):4184. doi: 10.1038/s41467-019-12143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Wang P., Xie Z., Wang S., Ma M., Li J., et al. Abnormal inhibition of osteoclastogenesis by mesenchymal stem cells through the miR-4284/CXCL5 axis in ankylosing spondylitis. Cell Death Dis. 2019;10(3):188. doi: 10.1038/s41419-019-1448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wick M.R. The hematoxylin and eosin stain in anatomic pathology-An often-neglected focus of quality assurance in the laboratory. Semin Diagn Pathol. 2019;36(5):303–311. doi: 10.1053/j.semdp.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z., Yu W., Zheng G., Li J., Cen S., Ye G., et al. TNF-alpha-mediated m(6)A modification of ELMO1 triggers directional migration of mesenchymal stem cell in ankylosing spondylitis. Nat Commun. 2021;12(1):5373. doi: 10.1038/s41467-021-25710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cen S., Wang P., Xie Z., Yang R., Li J., Liu Z., et al. Autophagy enhances mesenchymal stem cell-mediated CD4(+) T cell migration and differentiation through CXCL8 and TGF-beta1. Stem Cell Res Ther. 2019;10(1):265. doi: 10.1186/s13287-019-1380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G., Xie Z., Wang P., Li J., Li M., Cen S., et al. Enhanced osteogenic differentiation of mesenchymal stem cells in ankylosing spondylitis: a study based on a three-dimensional biomimetic environment. Cell Death Dis. 2019;10(5):350. doi: 10.1038/s41419-019-1586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hitt A.L., Laing S.D., Olson S. Development of a fluorescent F-actin blot overlay assay for detection of F-actin binding proteins. Anal Biochem. 2002;310(1):67–71. doi: 10.1016/s0003-2697(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 23.Evans I.M., Paliashvili K. Co-Immunoprecipitation assays. Methods Mol Biol. 2022;2475:125–132. doi: 10.1007/978-1-0716-2217-9_8. [DOI] [PubMed] [Google Scholar]
- 24.Rauh M. LC-MS/MS for protein and peptide quantification in clinical chemistry. J Chromatogr, B: Anal Technol Biomed Life Sci. 2012;883-884:59–67. doi: 10.1016/j.jchromb.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 25.Komori T. Animal models for osteoporosis. Eur J Pharmacol. 2015;759:287–294. doi: 10.1016/j.ejphar.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z., Kang W., Li O., Qi F., Wang J., You Y., et al. Abrogation of USP7 is an alternative strategy to downregulate PD-L1 and sensitize gastric cancer cells to T cells killing. Acta Pharm Sin B. 2021;11(3):694–707. doi: 10.1016/j.apsb.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Eidan A., Wang Y., Skipp P., Ewing R.M. The USP7 protein interaction network and its roles in tumorigenesis. Genes Dis. 2022;9(1):41–50. doi: 10.1016/j.gendis.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Y., Huang Q., Wu Z., Chen W. Roles of protein ubiquitination in inflammatory bowel disease. Immunobiology. 2020;225(6) doi: 10.1016/j.imbio.2020.152026. [DOI] [PubMed] [Google Scholar]
- 29.Tang Y., Lv L., Li W., Zhang X., Jiang Y., Ge W., et al. Protein deubiquitinase USP7 is required for osteogenic differentiation of human adipose-derived stem cells. Stem Cell Res Ther. 2017;8(1):186. doi: 10.1186/s13287-017-0637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji L., Lu B., Zamponi R., Charlat O., Aversa R., Yang Z., et al. USP7 inhibits Wnt/beta-catenin signaling through promoting stabilization of Axin. Nat Commun. 2019;10(1):4184. doi: 10.1038/s41467-019-12143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin W., Chang M., Paul E.M., Babu G., Lee A.J., Reiley W., et al. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118(5):1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q., Wang M., Xue H., Liu W., Guo Y., Xu R., et al. Ubiquitin-specific protease 34 inhibits osteoclast differentiation by regulating NF-kappaB signaling. J Bone Miner Res. 2020;35(8):1597–1608. doi: 10.1002/jbmr.4015. [DOI] [PubMed] [Google Scholar]
- 33.Charoonpatrapong K., Shah R., Robling A.G., Alvarez M., Clapp D.W., Chen S., et al. HMGB1 expression and release by bone cells. J Cell Physiol. 2006;207(2):480–490. doi: 10.1002/jcp.20577. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Z., Han J.Y., Xi C.X., Xie J.X., Feng X., Wang C.Y., et al. HMGB1 regulates RANKL-induced osteoclastogenesis in a manner dependent on RAGE. J Bone Miner Res. 2008;23(7):1084–1096. doi: 10.1359/JBMR.080234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H., Zhou W., Zhong Z., Qiu R., Chen G., Zhang P. High-mobility group box chromosomal protein-1 deletion alleviates osteoporosis in OVX rat model via suppressing the osteoclastogenesis and inflammation. J Orthop Surg Res. 2022;17(1):232. doi: 10.1186/s13018-022-03110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pozhidaeva A., Bezsonova I. USP7: structure, substrate specificity, and inhibition. DNA Repair. 2019;76:30–39. doi: 10.1016/j.dnarep.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rousseau A., Bertolotti A. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol. 2018;19(11):697–712. doi: 10.1038/s41580-018-0040-z. [DOI] [PubMed] [Google Scholar]
- 38.Li Q., Wang M., Xue H., Liu W., Guo Y., Xu R., et al. Ubiquitin-specific protease 34 inhibits osteoclast differentiation by regulating NF-kappaB signaling. J Bone Miner Res. 2020;35(8):1597–1608. doi: 10.1002/jbmr.4015. [DOI] [PubMed] [Google Scholar]
- 39.Ang E., Pavlos N.J., Rea S.L., Qi M., Chai T., Walsh J.P., et al. Proteasome inhibitors impair RANKL-induced NF-kappaB activity in osteoclast-like cells via disruption of p62, TRAF6, CYLD, and IkappaBalpha signaling cascades. J Cell Physiol. 2009;220(2):450–459. doi: 10.1002/jcp.21787. [DOI] [PubMed] [Google Scholar]
- 40.Guo Y.C., Wang M.Y., Zhang S.W., Wu Y.S., Zhou C.C., Zheng R.X., et al. Ubiquitin-specific protease USP34 controls osteogenic differentiation and bone formation by regulating BMP2 signaling. EMBO J. 2018;37(20) doi: 10.15252/embj.201899398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song X., Wang S., Li L. New insights into the regulation of Axin function in canonical Wnt signaling pathway. Protein Cell. 2014;5(3):186–193. doi: 10.1007/s13238-014-0019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herhaus L., Al-Salihi M.A., Dingwell K.S., Cummins T.D., Wasmus L., Vogt J., et al. USP15 targets ALK3/BMPR1A for deubiquitylation to enhance bone morphogenetic protein signalling. Open Biol. 2014;4(5) doi: 10.1098/rsob.140065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yun S.I., Kim H.H., Yoon J.H., Park W.S., Hahn M.J., Kim H.C., et al. Ubiquitin specific protease 4 positively regulates the WNT/beta-catenin signaling in colorectal cancer. Mol Oncol. 2015;9(9):1834–1851. doi: 10.1016/j.molonc.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu Z., Li L., Huang Y., Shi K., Zhang L., Huang C., et al. Puerarin specifically disrupts osteoclast activation via blocking integrin-beta3 Pyk2/Src/Cbl signaling pathway. J Orthop Translat. 2022;33:55–69. doi: 10.1016/j.jot.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo W., Li H., Lou Y., Zhang Y., Wang J., Qian M., et al. Tyloxapol inhibits RANKL-stimulated osteoclastogenesis and ovariectomized-induced bone loss by restraining NF-kappaB and MAPK activation. J Orthop Translat. 2021;28:148–158. doi: 10.1016/j.jot.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang C., Zheng Y., Bai J., Shi C., Shi X., Shan H., et al. Hepatocyte growth factor overexpression promotes osteoclastogenesis and exacerbates bone loss in CIA mice. J Orthop Translat. 2021;27:9–16. doi: 10.1016/j.jot.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nininahazwe L., Liu B., He C., Zhang H., Chen Z.S. The emerging nature of Ubiquitin-specific protease 7 (USP7): a new target in cancer therapy. Drug Discov Today. 2021;26(2):490–502. doi: 10.1016/j.drudis.2020.10.028. [DOI] [PubMed] [Google Scholar]