Abstract
T cells are crucial for immune functions to maintain health and prevent disease. T cell development occurs in a stepwise process in the thymus and mainly generates CD4+ and CD8+ T cell subsets. Upon antigen stimulation, naïve T cells differentiate into CD4+ helper and CD8+ cytotoxic effector and memory cells, mediating direct killing, diverse immune regulatory function, and long-term protection. In response to acute and chronic infections and tumors, T cells adopt distinct differentiation trajectories and develop into a range of heterogeneous populations with various phenotype, differentiation potential, and functionality under precise and elaborate regulations of transcriptional and epigenetic programs. Abnormal T-cell immunity can initiate and promote the pathogenesis of autoimmune diseases. In this review, we summarize the current understanding of T cell development, CD4+ and CD8+ T cell classification, and differentiation in physiological settings. We further elaborate the heterogeneity, differentiation, functionality, and regulation network of CD4+ and CD8+ T cells in infectious disease, chronic infection and tumor, and autoimmune disease, highlighting the exhausted CD8+ T cell differentiation trajectory, CD4+ T cell helper function, T cell contributions to immunotherapy and autoimmune pathogenesis. We also discuss the development and function of γδ T cells in tissue surveillance, infection, and tumor immunity. Finally, we summarized current T-cell-based immunotherapies in both cancer and autoimmune diseases, with an emphasis on their clinical applications. A better understanding of T cell immunity provides insight into developing novel prophylactic and therapeutic strategies in human diseases.
Subject terms: Adaptive immunity, Tumour immunology, Infectious diseases, Immunological disorders
Introduction
T lymphocytes (T cells) are the major cell components of the adaptive immune system, responsible for mediating cell-based immune responses to keep the host healthy and prevent various types of diseases. T cells are developed from bone marrow (BM)-derived thymocyte progenitors in the thymus, and broadly grouped into CD4+ and CD8+ αβ T cells in addition to rear populations of γδ T cells and natural killer T (NKT) cells. αβ T cells recognize antigens that are presented by major histocompatibility complex (MHC) molecules on antigen-presenting cells (APCs). Upon recognition of cognate antigens (signals 1) by T cell receptor (TCR) and costimulatory molecules (signals 2) on APCs, and cytokines (signals 3), naïve CD4+ and CD8+ T cells undergo activation, clonal expansion, and differentiation to execute their effector functions of killing infected cells, producing cytokines and regulating immune responses. A small population of T cells develops into memory T cells which exhibit rapid effector functions upon reencountering the same antigens and provide the host with potent and long-term protection. In parallel, there exists a subpopulation of CD4+ T cells, named regulatory T (Treg) cells, that maintain peripheral immune tolerance. Over the past few decades, our knowledge of T cells regarding their classification, differentiation, cellular and molecular regulatory mechanisms, particularly phenotypes and functions in healthy conditions and immune-related diseases, has expanded significantly. Hence, novel strategies engaging T cell functions have been extensively developed and demonstrated unprecedented clinical efficacy in the past few decades.
In this review, we comprehensively summarize the current understandings of T cell biology and functions in both physiological and pathological settings, including the following points: (1) describe the T cell development regarding their differentiation process, T cell lineage commitment, β-selection, and CD4/CD8 lineage choice; (2) introduce major CD4+ and CD8+ T cell classification, differentiation, and the underlying regulatory mechanisms; (3) further discuss how CD8+ and CD4+ T cells respond, differentiate and contribute in infectious diseases, chronic infections and tumors, and autoimmune diseases; (4) γδ T cell development, effector subsets and function in tissue surveillance, infection, and tumor immunity; (5) T cell-based immunotherapies in cancer and autoimmune diseases and their clinical applications. Specifically, we highlight the cell signature, differentiation trajectory, regulatory mechanisms, and contributions to anti-tumor immunity of exhausted CD8+ T cells, as well as the roles of CD4+ T cells in helping CD8+ T cell responses.
T cell development
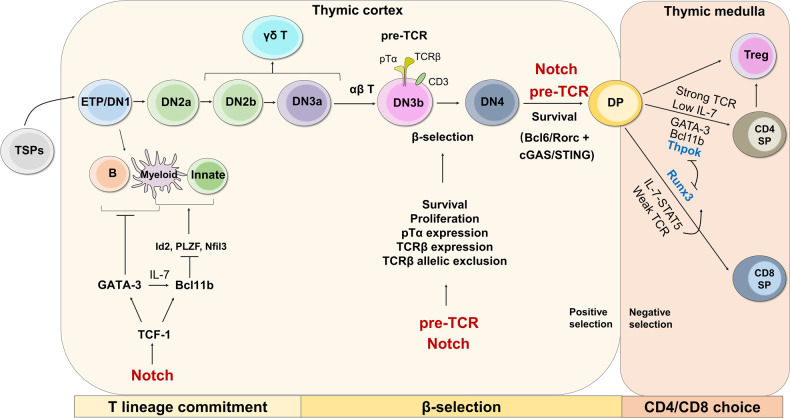
T cell development begins with BM-derived thymic seeding progenitors (TSPs) in the thymus, where T cells undergo a series of developmental stages including double negative (CD4−CD8−, DN), double positive (CD4+CD8+, DP), and single positive (CD4−CD8+ or CD4+CD8−, SP)1–3 (Fig. 1). DN thymocytes can be divided into four distinct stages from DN1 to DN4 based on CD44 and CD25 expression among lineage negative population.2,4–6 Upon Notch signaling, ETPs (DN1) acquire CD25 expression and progress into the DN2a stage, which launches the T cell lineage commitment.4,5 Bifurcation of αβ and γδ T cell lineage occurs at DN2b and DN3a stage along with upregulation of genes associated with TCRγ, TCRδ, and TCRβ rearrangement.7 A functional pre-TCR complex, consisting of CD3 protein, TCRβ and invariant pre-TCRα (pTα), drives DN3 cells to DN4, CD4+CD8− immature single positive (ISP), and DP cell development.7 Those expressing a TCRβ chain can initiate TCRα rearrangement and then form a fully functional αβTCR on the surface, which recognizes MHC I- or MHC II-peptide complexes presented by thymic APCs to become either CD8+ SP or CD4+ SP thymocytes.8 On one hand, the interaction of peptide-MHC with moderate affinity rescues DP thymocytes from apoptosis (known as positive selection) in the thymic cortex and progresses into the SP stage.8 On the other hand, recognition of self-peptide triggers immense death (known as negative selection) or skews CD4+ T cells towards Treg cells in the thymic medulla.9 The following three steps and relevant signals are required for T cell fate decision and development.
Fig. 1.
Overview of thymocyte development and regulatory mechanism. T cell development experiences three key steps: T cell lineage commitment, β-selection, and CD4/CD8 lineage choice, where T cells undergo sequential developmental stages from TSPs to DN, DP, and SP. ETPs (DN1) possess the potential to differentiate into B cells, myeloid cells, and innate-type of T cells, while DN3 can differentiate into γδ T cells. Induced by Notch signaling, transcription factors TCF-1, GATA-3, and Bcl11b play critical roles in promoting T cell lineage commitment by limiting other lineage differentiation. A pre-TCR complex consisting of TCRβ, pTα, and CD3 molecules on DN3 enforces β-selection and DN3 to DN4 development. Both pre-TCR and Notch signaling play critical roles in driving β-selection and DN to DP transition. Following positive and negative selection in the thymic cortex and medulla, respectively, DP cells differentiate into either CD4+ SP under the regulation of strong TCR and Thpok or CD8+ SP under the regulation of weak TCR and Runx3
Orchestrated trajectory for T cell lineage commitment
ETPs still possess the potential to differentiate into other immune cell lineages, such as B cells, NK cells, dendritic and myeloid cells.10,11 How ETPs commit to T cell lineage and lose the ability to convert to alternative lineages? It is well-appreciated that Notch signaling is essential for the initial commitment of T cell lineage in the thymus.12,13 Notch1 signaling induces the expression of transcription factor (TF) T cell factor 1 (TCF-1, encoded by Tcf7), which is required for the generation, survival, and proliferation of ETPs.14–16 TCF-1 promotes the upregulation of T cell-specific TFs GATA-3 and Bcl11b,15,16 and GATA-3 as well as IL-7/IL-7R signal are required for Bcl11b activation.17–19 GATA-3 suppresses both B cell and myeloid cell differentiation in TCF-1-deficient ETPs,15 whereas Bcl11b restricts the progenitor differentiation into innate lymphoid and myeloid lineages.20–22 Mechanistically, Bcl11b blocks expression of Id2, PLZF, and Nfil3 expression,21,23,24 in which Id2-repressed E protein E2A is critical for innate lymphoid cells including NK cell development,25–27 while PLZF and Nfil3 promote innate-type T cell development.28–30 Hence, enforced expression of Bcl11b can restore the DN1 to DN2 transition block resulted from TCF-1 deficiency.15 Future research needs to clarify whether GATA-3 facilitates T cell lineage and limits other lineages independent of Bcl11b. Taken together, following T cell lineage specification, the committed DN2b cells completely step on the T cell development journey.31
DN-DP transition driven by β-selection
Following the accomplishment of TCRβ rearrangement, DN3 cells expressing pre-TCR assembled with the TCRβ chain together with pTα and CD3 molecules (known as β-selection) differentiate into αβ T cells, otherwise, skew into γδ T cells.7,32,33 To date, two major signals are involved in the β-selection process: pre-TCR and Notch signaling. The pre-TCR signaling prevents thymocytes from apoptosis, stimulates their proliferation, induces allelic exclusion at the TCRβ locus in DN3b cells post-β-selection and promotes DN to DP transition.34–37 However, pre-TCR signaling alone is not sufficient for thymocyte development, as isolated DN3 thymocytes fail to differentiate into DP cells in the absence of a stromal cell-derived Notch signal.38–40 Notch signaling has been shown to promote T lineage commitment,41 thymocyte survival,42 DN to DP stage transition,42 and expression of pre-TCR components.43,44 Recently, Notch-induced endoplasmic reticulum (ER)-associated degradation (ERAD) mediates proteasomal degradation of misfolded proteins, which becomes a prerequisite for thymocyte β-selection.45 Pre-TCR and Notch signaling, by targeting ubiquitin ligase subunits Fbxl1 and Fbxl12, respectively, promote the cell cycle progression of β-selected thymocytes via accelerating degradation of cyclin-dependent kinase inhibitor Cdkn1b.46 Furthermore, β-selected thymocytes form an immunological synapse to promote proliferation, which relies on the cooperation between Notch and pre-TCR signaling.47 Interestingly, pre-TCR independent mechanisms also regulate thymocyte development. Recent studies from our and other groups demonstrated that zinc finger protein Zfp335 controlled thymocyte survival and DN to DP transition by inducing Bcl-6/Rorc expression or cGAS/STING suppression in a pre-TCR independent manner.48,49
Choice to become CD4+ or CD8+ T cells
Following positive selection, DP cells bearing MHC class I- or MHC class II-TCRs differentiate into either CD8+ or CD4+ T cells, termed as CD4/CD8 lineage choice.50,51 A well-known theory holds that DP thymocytes received positive selection signals initially terminate CD8 gene transcription and become CD4+CD8lo intermediate cells which further progress into CD4+ or CD8+ T cells depending on TCR signaling or cytokines stimulation.52–54 Persistent and strong TCR signals in intermediate thymocytes trigger differentiation into CD4+CD8- SP cells largely by inhibiting IL-7-mediated signaling, whereas transient and weak TCR signals force these cells into CD4-CD8+ SP cells, which relies on signals from IL-7 and other common gamma chain (γc) cytokines.55–57
Thpok and Runx3 are two antagonistic TFs controlling the lineage choice between CD4+ or CD8+ T cells. Thpok is highly expressed in CD4+ but not CD8+ thymocytes, and serves as a master regulator for CD4 lineage commitment.58,59 Mice with Thpok depletion or a missense mutation lack CD4+ T cells,58,60–63 whereas ectopic expression of Thpok strongly drives DP thymocytes into CD4+ SP cells.58,59 Mechanistically, Thpok represses Runx3 and CD8 lineage-related genes.61,64,65 In contrast, Runx3 facilitates CD8+ T cell development by directly downregulating CD4 and Thpok expression.62,66 In addition, Bcl11b promotes CD4 lineage commitment by directly targeting to several Thpok locus67,68 and Runx3 promoter region.67 TCR signaling-induced GATA-3 is also required for CD4 lineage commitment by enhancing Thpok expression,69,70 while the IL-7-STAT5 axis acts upstream of Runx3 to enhance its expression and promote CD8+ T cell development.71 Therefore, the balance between Thpok and Runx3 decides the lineage choice of CD4+ versus CD8+ T cells.
CD4+ T cell classification and differentaiton
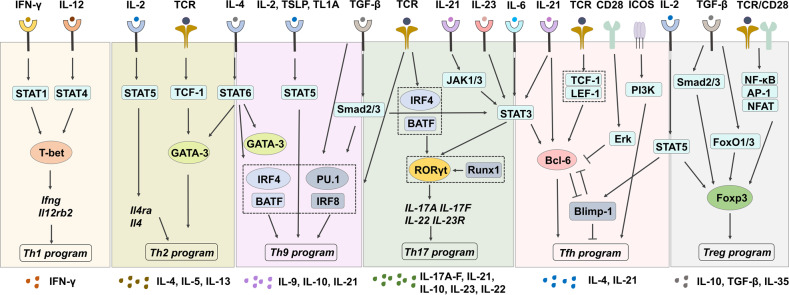
CD4+ T helper (Th) cells are a heterogeneous group of T cells playing central roles in almost all aspects of immune responses. CD4+ T cells can be activated by peptide-MHC class II complex on APCs, costimulatory stimulation, and cytokine signaling72–74 and differentiate into several subsets with a distinct expression of surface molecules, cytokines, and key TFs,75,76 such as Th1, Th2, Treg, follicular helper T (Tfh), Th17, Th9, Th22, and CD4+ cytotoxic T lymphocytes (CTLs), etc.77 Here, we will introduce six major Th subsets and the regulatory pathways of their differentiation (Fig. 2).
Fig. 2.
Cytokine signalings regulate CD4+ Th cell differentiation. Upon TCR stimulation, naïve CD4+ T cells can be differentiated into distinct effector Th subsets under different cytokines and costimulatory stimulation. IFN-γ and IL-12 drive Th1 cell differentiation by inducing the master TF T-bet expression through STAT1 and STAT4, respectively. Th2 cells are induced by TCR-stimulated TCF-1 activation and cytokine IL-2 and IL-4 signaling, expressing key TF GATA-3. Th9 cells are induced under TCR stimulation in the presence of IL-4 and TGF-β, and enhanced development by STAT5 activation. While IL-6 and TGF-β drive Th17 cell differentiation, IL-21 and IL-23 stabilize Th17 lineage by inducing RORγt. Cytokines IL-6 and IL-21 promote, while IL-2 inhibits Tfh cell differentiation. Costimulatory signaling from CD28 and ICOS play opposite roles in Tfh cell development. Treg cells can be differentiated upon TCR/CD28 stimulation in the presence of TGF-β and IL-2 through inducing Foxp3 expression. Shared cytokines are illustrated between cells: IL-4 for Th2 and Th9, TGF-β for Th9 and Th17, IL-6 for Th17 and Tfh, and IL-2 for Tfh and Treg cells. The same cytokines may induce different downstream signaling cascade and differentiation fate. For instance, IL-6-induced STAT3 activation leads to the expression of RORγt in Th17 cells but Bcl-6 in Tfh cells. Signaling complexes formed are indicated in the dashed squares
Th1 cells are the major participants in protecting hosts against intracellular bacteria and viruses by producing the pro-inflammatory cytokine IFN-γ. IL-12 and IFN-γ are two cytokines essential for Th1 differentiation.78 TCR stimulation and IFN-γ-STAT1 signaling induce the expression of T-bet (encoded by Tbx21), the major TF driving Th1 differentiation while suppressing Th2/Th17 lineages.79,80 T-bet can directly bind to the Ifng gene to increase the expression of IFN-γ80,81 and meanwhile promote the expression of IL-12Rβ2, conferring IL-12 responsiveness.82 IL-12 signaling via STAT4 activation, in turn, maintains T-bet expression.83 These feedback loops all contribute to Th1 differentiation.
Th2 cells, defined by expression of TF GATA-3 and cytokines IL-4, IL-5, and IL-13, protect the host against helminth infections, facilitate tissue repair, as well as contribute to chronic inflammation such as asthma and allergy.84 IL-4 secreted by dendritic cells (DCs) and innate lymphoid cell group 2 (ILC2) binds to IL-4R on CD4+ T cells, leading to the expression of GATA-3 through STAT6 phosphorylation and subsequent production of Th2-related cytokines.85 Autocrine production of IL-4 by activated CD4+ T cells further promotes Th2 differentiation.86 In addtion, GATA-3 mediates the repression of Th1 cell development by sliencing Th1-related genes such as Tbx21, Ifng, Stat4, and Il12rb2.87 STAT5 signaling primed by IL-2 is required for maintaining the expression of Il4ra and increasing the accessibility of Il4 chromatin.87,88 Other TFs such as NFAT1, c-Maf, IRF4, and JunB can promote Th2 program by inducing IL-4 production.87 In addition, TCF-1, activated by TCR stimulation, has been found to initiate Th2 cell differentiation by promoting GATA-3 expression.89
Th9 cells are a newly identified subset of CD4+ T cells, playing critical roles in infectious diseases, allergy, cancer, and autoimmune immunity.90–94 Th9 cells can be induced in vitro by TCR stimulation in the presence of IL-4 and TGF-β, and are characterized by expressing high levels of IL-9 and prominent TFs IRF4 and PU.1.90,95–97 Besides IL-9, IL-10, and IL-21 are also produced by Th9 cells.98 STAT6 phosphorylation mediated by IL-4 signaling induces expression of GATA-3, IRF4 and BATF to promote IL-9 transcription and Th9 cell development.99,100 Besides, TGF-β signaling activates Smads (Smad2/3), PU.1 and IRF8, contributing to Th9 cell differentiation.99,100 Furthermore, IRF4, PU.1, IRF8, and BATF form a TF complex which binds to Il9 locus and regulate Th9 differentiation.101 In addition, STAT5 phosphorylation induced by IL-2, TSLP, and TL1A promotes Th9 cell development.99 The differentiation of Th9 cells is also regulated by costimulation signaling (CD28, OX40, GITR, Notch, and DR3) and other cytokines (IL-1, IL-25, IL-7, and IL-21).91,99,100
Th17 cells, characterized by expression of featured cytokines IL-17A-F, IL-21, IL-10, IL-23, and IL-22, and steroid receptor–type nuclear receptor RORγt as the master TF,102 contribute to protection against extracellular pathogens, especially at mucosal tissue,103 as well as chronic inflammation and autoimmune diseases.104 IL-6 and TGF-β drive Th17 cell differentiation while IL-21 and IL-23 stabilize Th17 lineage.105–109 IL-6 prompts the expression of RORγt by phosphorylation of STAT3, while inhibits the expression of Foxp3 induced by TGF-β.110 RORγt induces the expression of IL-17A, IL-17F, IL-22, and IL-23R by directly targeting to their promoters.111 TGF-β signaling through Smad2/3 could sustain STAT3 activation.112 Autocrine IL-21 activates STAT3 through Janus kinase (JAK)1/3 activation, which can further increase the expression of IL-23R and confer IL-23 responsiveness of Th17 cells.113 IL-23 then enhances STAT3 activation to stabilize Th17 development.114 Recent studies have revealed a great degree of plasticity of Th17 cells depending on the presence of TGF-β. TGF-β and IL-6 induce the “classical” Th17 cells characterized by the production of IL-17, IL-21, and IL-10, whereas IL-6, IL-1β, and IL-23 induce “pathogenic” Th17 cells producing high levels of IFN-γ, GM-CSF, and IL-22.115–117 Besides RORγt, TCR signal induced transcriptional complex formed by IRF4 and BATF contributes to the initial chromatin accessibility of Th17-related genes such as Il17, Il21, Il23r, and RORc, as well as Foxp3 suppression.118–120 Runx1 enhances Th17 development through both inducing and directly interacting with RORγt.121,122 Other TFs, including RORα, c-Maf, p65, NFAT, and c-Rel, also participate in Th17 differentiation.123–127
Tfh cells are specialized CD4+ Th cells involved in supporting humoral immune responses by promoting B cell proliferation and maturation, germinal center (GC) response, and high-affinity antibody production.80,128,129 Tfh cells are featured by high expression of surface markers PD-1 and CXCR5, costimulatory receptors CD40, CD40LG, and ICOS, cytokines IL-4 and IL-21, signaling molecules SAP, as well as TF STAT3 and Bcl-6.128 Tfh cells play central roles in regulating antibody responses during infectious diseases, allergy, autoimmune diseases, and vaccination.130–132 Tfh cell development is mainly regulated by the master TF Bcl-6133 which primarily represses alternative, non-Tfh, cell fates.134–136 Bcl-6 constrains Th1, Th2 and Th17 cell differentiation by repressing their lineage-defining TFs T-bet, GATA-3, and RORγt expression.133,137,138 Suppression of B lymphocyte induced maturation protein 1 (Blimp-1, encoded by Prdm1) by Bcl-6 is also required for Tfh lineage.139 TCF-1 is involved in early induction of Bcl-6 by orchestrating with LEF-1.140,141 Other TFs, such as BATF, STAT1/3/4/5, Foxp1, KLF2, IRF4, Ets1, BACH2, Ascl2, Tox2, and Bhlhe40, have been also identified in regulating Tfh cell development.136,142–144 Additionally, Tfh cell development is regulated by costimulatory signaling in which CD28 stimulation activates ERK to suppress Tfh cell differentiation,145 whereas ICOS activates PI3K to promote and maintain Tfh cells.146 In terms of the driver cytokines for Tfh cells, IL-6 and IL-21 promote the differentiation of Tfh cells by acting STAT3 and inducing Bcl-6 expression, respectively.147,148 However, IL-2/STAT5 signaling strongly inhibits Tfh development by inducing Blimp-1 expression.149,150
Treg cells are a specialized CD4+ T cell subset for maintaining immune tolerance by suppressing an immune response. Treg cells are characterized by high expression of IL-2 receptor alpha chain (IL-2Rα, CD25), inhibitory cytokines IL-10, TGF-β, and IL-35, and master TF Foxp3.151,152 Two major subsets of Treg cells are identified based on their developmental origin: thymic Treg (tTreg) cells, also known as natural Treg (nTreg) cells that derive from thymus, and induced Treg (iTreg) cells that differentiate from conventional CD4+ T (Tconv) cells in the periphery after antigen stimulation and in the presence of TGF-β and IL-2.153,154 Given the importance of Foxp3, regulation of Foxp3 expression is critical for Treg cell development, maintenance, and function, in which both transcriptional and epigenetic mechanisms are involved.155–158 TCR/CD28 stimulation triggers Foxp3 expression by inducing bindings of NF-κB, AP-1 and NFAT to Foxp3 enhancer/promoter regions.153,159–161 In addition, TGF-β enhances Foxp3 transcription by inducing bindings of phosphorylated Smad2 and Smad3, as well as forkhead box protein O1 (FoxO1) and FoxO3 to the conserved non-coding sequences (CNSs) region of Foxp3.162 As the downstream of IL-2 signaling, STAT5 also increases the expression of Foxp3 through binding to CNS0 and CNS2.163,164 Regulation of Foxp3 stability will be further discussed in autoimmune disease section.
CD8+ T cell differentiation and regulation
CD8+ T cells play critical roles in fighting against intracellular pathogens as well as eliminating malignant cells in cancer.165 Upon antigen stimulation, naïve CD8+ T cells undergo robust expansion to give rise to effector and memory T cells. Effector CD8+ T cells, known as CD8+ CTLs, can directly induce target cell death by the interaction between Fas/Fas ligand, and secretion of cytolytic mediator perforin, which creates pores in the target cells allowing the delivery of granule serine proteases (granzymes), to induce apoptosis. Memory CD8+ T cells provide rapid and strong protection upon antigen reencounter, which is critical for effective and long-term immunity. During CD8+ T cell differentiation, heterogeneous effector and memory populations have been identified, including short-live effector CD8+ T cells (TE), exhausted CD8+ T cells (Tex), long-live memory CD8+ T cells (TM), memory precursor CD8+ T cells (TMP), central and effector memory CD8+ T cells (TCM and TEM), and tissue-resident memory (TRM) cells, which are named by their phenotype, differentiation potential and functionality.166,167 Of note, these subsets are produced at different time window and tissue location upon immune challenge, and their differentiation is under orchestrated regulation of TFs, epigenetic modification, and metabolic programs.
Key transcription factors
Several key TFs have been well-characterized to control effector versus memory CD8+ T cell differentiation in a reciprocal and antagonistic manner (Fig. 3). These TFs include T-bet versus Eomesodermin (Eomes),168,169 Blimp-1 versus Bcl-6,170–172 Id2 versus Id3,169,173,174 STAT4 versus STAT3,173,175,176 and Zeb2 versus Zeb1.177 While T-bet, Blimp-1, Id2, STAT4, and Zeb2 are predominantly expressed in TE populations and required for effector T cell lineage and/or acquisition of CTL functions, Eomes, Bcl-6, Id3, STAT3, and Zeb1 are enriched in TM populations and support memory T cell formation and maintenance. Those two sets of TFs can either enhance or antagonize each other. For example, Id2 positively regulates T-bet, which induces Zeb2 expression; STAT3 sustains Bcl-6 and Eomes expression; Blimp-1 represses Id3 expression; Bcl-6 can both repress and be repressed by Blimp-1.169,171 Currently, collective evidence has supported that the first set of TFs are activated by TCR/costimulatory signals and/or coupled with cytokine signaling (IL-2, IL-12, type I IFN, IFN-γ, IL-21, and IL-27).170,171,173 For instance, IL-2 and IL-12 drive effector CD8+ T cell differentiation by inducing expression of Blimp-1, T-bet, and Id2 expression.171 IFN-α/β stimulates the clonal expansion and production of IFN-γ in CD8+ T cells via a STAT4-dependent pathway.178 The autocrine IFN-γ further synergizes with IFN-α to promote T-bet expression.173,179 Additionally, IL-21 and IL-27 promote Blimp-1 expression in effector CD8+ T cells.180 The second set of TFs are predominantly driven by cytokine signaling (IL-7, IL-10, lL-15, and IL-21).169,173,181 TCF-1 (a downstream factor of the Wnt-signaling pathway) and FoxO1 (a factor related to metabolic pathway) are identified as indispensable TFs for memory CD8+ T cell differentiation and maintenance.182 It will be interesting to clarify how TCR and cytokine signaling sequentially activate these two sets of TFs and how the cross-regulation occurs among them.
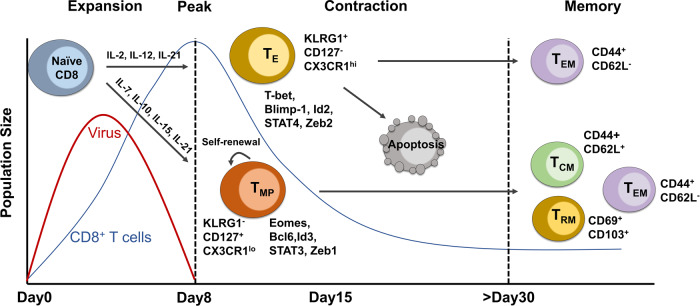
Fig. 3.
Temporal dynamics of CD8+ T cell response in acute infection. The population size of the virus (red line) and CD8+ T cells (blue line), as well as CD8+ T cell response along with the infection course, are indicated. Upon infection, CD8+ T cells undergo robust proliferation and reach the expansion peak on day 8, where the pathogens are rapidly cleared. CD8+ T cells at this stage can be separated into TE and TMP populations with distinct surface marker and differentiation potential. The differentiation of effector and memory CD8+ T cells is regulated by different transcriptional factors and cytokines. The majority of CD8+ TE cells undergo apoptosis at the contraction phase (8–15 days) and leave a subpopulation differentiating into TEM, whereas TMP cells keep self-renewal and give rise to TCM, TEM and TRM cells over 30 days post-infection
Epigenetic mechanisms
DNA methylation and histone modifications regulate chromatin accessibility of the regulatory regions of lineage-specific TFs and orchestrate the transcription of key genes to control CD8+ T cell development.183 DNA methylation, predominantly on CpG islands (CG dinucleotide-sense regions), has repressive effects on gene transcription by hindering the binding of TFs to promoters. During CD8+ T cell differentiation, DNA methylation is highly involved in regulating the transcriptional program of effector and memory CD8+ T cells.184–187 DNA methyltransferase DNMT3A catalyzes DNA methylation at sites such as the promoter of Tcf7, thus suppresses memory differentiation and supports effector differentiation.188 Methylcytosine dioxygenase TET2 induces DNA demethylation to promote effector differentiation while restrict memory T cell differentiation.189,190 In addition, histone modifications has either activating or repressive effects on gene transcription via organizing DNA into structural units termed nucleosomes.191 H3K4me3 and H3K9ac are activation-associated modifications, whereas H3K27me3 modification is associated with repressive transcription.191 TE-associated genes (Tbx21, Prdm1, Klrg1, Ifng, Gzma, Gzmb, and Prf1) and TM-associated genes (Foxo1, Klf2, Lef1, Tcf7, Il2ra, Cd27, Ccr7, and Sell) display decreased repressively but increases activating histone modifications during effector or memory lineage differentiation, respectively.184,186,187,192,193 Polycomb complex protein BMI1 and histone-lysine N-methyltransferase EZH2, components of the H3K27me3 reader complex, are induced by TCR stimulation and functionally support the expansion, survival and cytokine production of TE population.193 Similarly, PR domain zinc finger protein 1 (PRDM1) facilitates effector cell differentiation and suppresses memory lineage through recruiting repressive histone modifiers histone-lysine N-methyltransferase EHMT2 and histone deacetylase 2 (HDAC2) to the Il2ra and Cd27 loci.194 Moreover, BATF enhances effector CD8+ T cell differentiation by decreasing the expression of histone deacetylase sirtuin 1 (SIRT1) which inhibits T-bet expression though downregulating histone acetylation of the Tbx21 locus.195
Metabolic regulation
Growing evidence indicates that profound metabolic reprogramming is highly involved in CD8+ T cell differentiation. Naïve CD8+ T cells primarily depend on basal glycolysis and mitochondrial oxidative phosphorylation to meet their basal cellular processes.196–199 TE cells ensure high metabolic flux for the proliferation and functions by upregulating glycolysis197,199,200 and glutaminolysis.201 Upon TCR and costimulatory stimulation, activation of AKT-mTOR signaling in TE cells upregulates MYC expression, which induces glucose transporter type 1 (GLUT1) expression to promote glucose uptake as well as amino acid transporter SLC32A1/2 expression to increase glutamine uptake.201–203 At the same time, NFAT is also induced to upregulate GLUT1/3204 and MYC/HIF1α.197,205 TM cells differentiate and maintain the population through fatty acid oxidation fueled by long-chain and short/branched-chain fatty acids.206–208
During the process of TE towards TM differentiation, the metabolic program turns from an activated status back to a relative quiescent status. TM cells express high level of mitochondrial lipid transporter CPT1A, supporting that lipid oxidation is indispensable for memory T cell differentiation.209 In response to IL-15, TE cells upregulate CPT1A expression which mediates the transport of long-chain fatty acids into mitochondria and thereby promotes fatty acid oxidation.209 Additionally, short/branched-chain amino acid metabolism, beta-oxidation of 2-methylbutyrate, isobutyrate and isovalerate to generate ATP molecules, play a compensatory role in supporting memory T cell differentiation when long-chain fatty acids become limited.208 Upon recall stimulation, TM cells rapidly switch to glycolysis dependent on an epigenetic reprogramming controlled by TCF-1.210
Of note, there exists cross-regulations among TFs, epigenetic modification and metabolism.194,211,212 TFs and epigenetic modification co-regulate with each other, while they collaboratively regulate metabolic status.213,214 These integrated signals are involved in the fate decision and maintenance of CD8+ TE and TM populations.
T cells in acute infection and inflammation
Microbial pathogens including viruses, bacteria, fungi, and protozoa can cause acute and chronic infections in mammalian hosts, leading to various diseases even lethal damage. Owing to advances in public health management and development of vaccination, the number of deaths from pathogenic infection has reduced substantially in recent years. While infectious diseases seem faded out of the public consciousness over the past years, COVID-19 pandemic due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has caused 660 million confirmed cases and 6.6 million deaths by the end of 2022, alerting us to the danger of infectious pathogens. Though innate immune system offers the first-line defense, T cells are crucial in infectious immunity, including efficient clearance of pathogens, helping B cell response and antibody production, rapid control of reinfection, and providing long-term protection by memory formation.
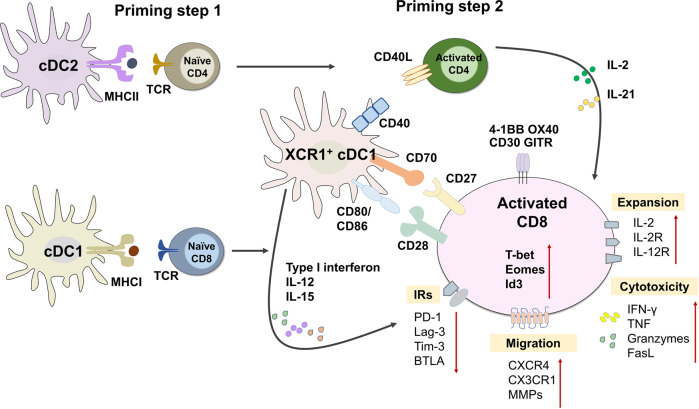
Effector CD8+ T cells contribute to protective immunity during acute infections
CD8+ T cells are main responders to viral infection but also participate in defense against bacterial and protozoal pathogens. Effector CD8+ T cells secrete pro-inflammatory cytokines such as IFN-γ and tumor necrosis factor (TNF) to inhibit viral replication,215 and express various chemokines to attract other inflammatory cells to sites of infection. Acute infections, defined as infections of only a short duration where the pathogens are eliminated rapidly after the peak of the immune response, are caused by infections of Armstrong strain of lymphocytic choriomeningitis virus (LCMV), Listeria monocytogenes (LM), influenza virus, hepatitis A virus, and vaccinia virus. The dynamics of CD8+ T cell response to acute infections has been studied extensively.216–218 The response of antigen-specific CD8+ T cells can be roughly divided into distinct stages (Fig. 3): the expansion phase (0–7 days) where CD8+ T cells are actively proliferating; the peak of expansion (day 8) where the effector CD8+ T cells reach the maximum number and stop proliferating; the contraction phase (8–15 days) where majority of effector CD8+ T cells undergo apoptosis; and the memory phase (>30 days) with only a small population of cells are survived and differentiated into distinct types of memory cells: CD44+CD62L− TEM, CD44+CD62L+ TCM, and CD69+CD103+ TRM.219 The fate decision between effector and memory T cells occurs as early as the first division of activated CD8+ T cells, in which the daughter cells with high MYC and high canonical BRG1/BRM-associated factor (cBAF) preferentially differentiate into TE cells, whereas those with low MYC and low cBAF develop into TM cells.220 At the peak of acute infection, expression of KLRG1 and CD127, the IL-7 receptor subunit-α (IL-7Rα), is used to identify short-lived terminally differentiated effector cells (TE, KLRG1+CD127-) and long-lived memory precursor cells (TMP, KLRG1-CD127+). Besides KLRG1, TE cells express a range of effector molecules including cytotoxic granzymes, perforin, cytokines (IL-2, IFN-γ, and TNF), chemokines (CCL5 and CCL3), and chemokine receptors (CX3CR1, CXCR6 and CCR5). Recently, the expression of chemokine receptor CX3CR1 on CD8+ T cells has been used to classify effector and memory T cells.221 The level of CX3CR1 on CD8+ T cells correlates with the degree of effector differentiation as CX3CR1hi subset contains the terminally differentiated effector T cells.222 The differentiation and function of effector/memory CD8+ T cells are precisely and elaborately regulated at multiple levels, which have been described in the previous section.
Overall, CD8+ T cell responses to different microbial pathogens are similar regarding to the kinetics of T cell expansion and contraction, effector function and regulation, and memory formation. However, CD8+ T cell priming, costimulatory signaling, persistence of response and intensity of the inflammation can be different in various pathogenic infections.223–227 In the acute phase of SARS-CoV-2 infection, CD8+ T cells in severe and convalescent COVID-19 patients exhibit activated phenotypes characterized by elevated expression of CD38, HLA-DR, Ki67, PD-1, perforin, and granzyme B.228–232 Comprehensive single-cell RNA-sequencing (scRNA-seq) analysis reveals that SARS-CoV-2-specific CD8+ T cells display increased “exhaustion” phenotype with high expression of inhibitory receptors (IRs) (Tim-3 and Lag-3) than influenza A virus- and Respiratory syncytial virus (RSV)-reactive CD8+ T cells. Interestingly, such “exhausted” CD8+ T cells are not dysfunctional but enriched for cytotoxicity-related genes.233 Nevertheless, SARS-CoV-2-reactive CD8+ T cells have reduced cytokine production.233 Therefore, further studies are needed to fully elucidate the function of SARS-CoV-2-specific CD8+ T cells in COVID-19 patients.
Effector CD4+ Th cells in infection
CD4+ T cells play multifaceted roles in modulating immune responses (Fig. 4), contributing to protection from a broad range of pathogenic microbes. Th1 and Th2 subsets have been long identified as crucial players in protective immunity against pathogens.234 Although effector Th cells found in vivo after infections are often heterogeneous populations, CD4+ T cells in response to viruses mainly display Th1-associated phenotypes.235 Particularly, enriched Th1 lineage is a typical feature of pulmonary infections and plays crucial roles in fighting against Mycobacterium tuberculosis (Mtb), influenza virus, Staphylococcus aureus (S. aureus), Middle East respiratory syndrome coronavirus (MERS-CoV), SARS and SARS-CoV-2.236–239 Th1 cells, characterized by expressing cytokines IFN-γ, TNF-α/β and IL-2, chemokine receptors CXCR3 and CCR5, and TFs T-bet and STAT4, mainly fight intracellular pathogens of viruses, bacteria, fungi and protozoa.76 By contrast, Th2 cells, expressing cytokines IL-2, IL-4, IL-5, IL-10, IL-13, chemokine receptors CCR3 and CCR4, and TFs GATA-3 and STAT6, are strong drivers of humoral immune reactions against extracellular helminthic parasites and allergic inflammation.240,241
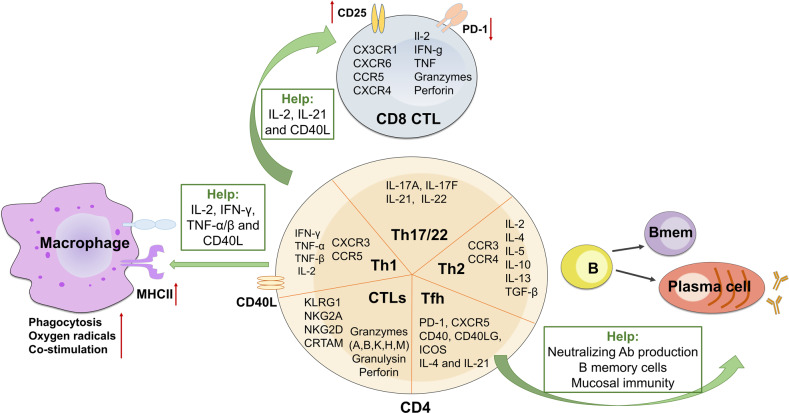
Fig. 4.
Effector CD4+ and CD8+ T cells contribute to infectious immunity. In response to infection, naïve CD8+ T cells develop into CD8+ CTLs expressing a range of chemokine receptors and effector molecules, whereas naïve CD4+ T cells develop into distinct Th1, Th2, Th17, Th22, Tfh, and CTL subsets with indicated phenotypes to exert protective functions. In addition, CD4+ T cells indirectly contribute to pathogen clearance by providing help to macrophages, CD8+ CTLs and B cell and antibody responses
Th17 response, featured by massive pro-inflammatory cytokine production, is often elicited together with Th1 cells in infections by bacterial and viral microorganisms, such as Mtb,242 S. aureus,243 MERS-CoV,244 Dengue virus,245 RSV,246 hepatitis B virus (HBV)247 and SARS-CoV-2.248 Additionally, fungal microbes, such as Pneumocystis carinii and Candida albicans can trigger strong Th17 response by inducing large amounts of IL-23 which is the key cytokine for full Th17 differentiation and function.102,249,250 Furthermore, Th22 cells are a newly identified Th subset producing IL-22 but not IFN-γ, IL-4, or IL-17.251 Th17/Th22-related cytokines can target on diverse cell types, including non-immune cell populations, such as epithelial cells, fibroblasts, and endothelium cells. Hence, Th17 and Th22 cells tend to protect against infections locally on the mucosal tissue and skin, respectively.252,253 IL-17 and IL-22 corporately augment the host immunity against infections at mucosal sites via promoting antimicrobial peptides production by mucosal epithelium and recruitment of neutrophils to eliminate bacteria and fungi.254
Moreover, CD4+ CTLs contribute to pathogen clearance through direct cytolytic activity.77,255,256 This subset of CD4+ T cells attracts much attention recently owing to their important functions in protecting against infectious disease, promoting human longevity, and mitigating tumor progression.257–259 CD4+ CTLs have been largely observed in both human and mice infected with viruses,235 such as cytomegalovirus (CMV),260 human immunodeficiency virus (HIV)-1,261 hepatitis viruses (HBV, HCV and HDV),262 Epstein–Barr virus (EBV),263 Dengue virus,264 influenza virus,265,266 and SARS-CoV-2.267 CD4+ CTLs are characterized by expression of KLRG1, natural killer group 2 (NKG2A), NKG2D, the class I-restricted T cell-associated molecule (CRTAM) and downregulated CD27/CD28.77,256 The cytotoxic activities of CD4+ CTLs attribute to the expression of pro-inflammatory cytokines, perforin, granzymes (A, B, K, H, and M), granulysin, and death receptor-dependent signaling (Fas and TRAIL).268–270 The transcriptional regulation of CD4+ CTLs is highly comparable to that of CD8+ CTLs, in which TFs T-bet, Eomes and Runx3 play critical roles in driving CD4+ CTL programming while ThPOK expression limits cytotoxic functions in CD4+ T cells.271–273 Additionally, IL-2 could drive the cytolytic phenotype of CD4+ CTLs,274 while pro-inflammatory cytokines IL-12, IL-6, and IFN-α increase granzyme B and perforin production and target killing activity.275 It remains unclear about the precursors of CD4+ CTLs or whether this population is merely the terminal differentiated Th1 cells. However, more evidence claims that CD4+ CTLs are a separate Th subset in regards to its differentiation trajectory, effector function and regulatory networks.255,276,277 Furthermore, heterogeneous populations within CD4+ CTLs have been identified in viral infection.277,278 In general, CD4+ CTLs are highly associated with antiviral immunity, however, aberrant CD4+ CTL activity has also been linked with immunopathology in some settings.279–281 For example, CD4+ CTLs contribute to the disease severity during SARS-CoV-2 infection267,282 and lung fibrosis.267,283
Accumulating evidence has suggested that more than one type of Th subsets can be triggered during the infection, and both synergy and balance among Th cells contribute to infection control. For instance, costimulation of Th1, Th2 and Th17 responses is commonly observed in various microbial infections, such as Mtb,284,285 Echinococcus multilocularis,286 Aspergillus fumigatus,287 HIV,288 SARS-CoV-2.248 Meanwhile, Treg cells can be induced during infection to prevent overstimulation of immune response and “self-attacking”.289–292 During Mtb infection, activation of macrophages induced by Th1-derived IFN-γ is crucial to control the tuberculosis. However, persistent Th1 response and pro-inflammatory cytokines can cause lung fibrosis and necrosis. Th2 cytokines IL-4, IL-10, and TGF-β are prominent to prevent pathology induced by aberrant Th1 response.293 Enhanced Th2 response during SARS-CoV-2 and influenza infection is associated with severe disease symptoms by inhibiting antiviral responses.241
Effective control of infection relies on CD4+ T cell help
CD4+ Th cells are indirectly involved in pathogen control by regulating functions of other immune cells, such as activating innate immune populations, assisting CD8+ CTL response and B cell maturation and antibody production (Fig. 4). CD4+ T cells, mainly Th1 population, are central for activation of pro-inflammatory macrophages by releasing cytokines IL-2, IFN-γ, and TNF-α/β and expressing CD40L.76 Activated macrophages augment their antimicrobial effectiveness by increasing microbial phagocytosis, production of nitric oxide (NO) and oxygen radicals, expression of MHC class II molecules and a number of costimulatory molecules for effective antigen presentation to T cells.294 Activated macrophages are also important for efficiently eliminating intracellular pathogens such as mycobacteria which grow primarily inside of macrophages and are shieled from CTLs and neutralizing antibodies.295
Furthermore, CD4+ T cell help is essential for optimal and effective CD8+ T cell response,51 although the requirement for primary CD8+ T cell response remains controversial. Some studies have shown that in the absence of CD4+ T cells, the primary CD8+ T cell expansion and cytotoxic functions during LCMV and LM infection are unaffected.296,297 However, other studies have reported that CD4+ T cells, particularly their memory subset, are required for primary effector CD8+ T cell response to herpes simplex virus (HSV) and influenza virus.298–301 The controversial effects of CD4+ T cell help for primary CD8+ T cell response are likely derived from different help-evaluation models.301 On the other hand, profound and consistent evidence indicates that CD4+ T cell help is indispensable for memory CD8+ T cell generation and their recall response to antigen restimulation.302–304 Mechanistically, CD4+ T cells support CD8+ T cell responses via cytokines IL-2 and IL-21, and CD40L signaling.301,305–307 Additionally, CD4+ T cells have been shown to help CD8+ T cells by enhancing their CD25 expression and downregulating PD-1 expression.308,309
CD4+ Tfh cells are essential for B cell responses and generating protective antibodies against viral, bacterial, parasite, and fungal pathogens in mice, non-human primates, and humans.131,310 The protective effects of Tfh cells on humoral immunity attribute to multiple mechanisms.132 First, Tfh cells help the production of protective antibodies that directly neutralize pathogens and inhibit their replication, and indirectly promote pathogen clearance through antibody opsonization. Tfh cells have long been known to highly correlate with broadly neutralizing antibodies in HIV infection.311 During SARS-CoV-2 infection, increased circulating Tfh (CCR7loPD-1+ICOS+CD38+) cells and production of neutralizing antibodies were observed in COVID-19 convalescent individuals and associated with mild symptoms.312,313 In contrast, defective Tfh cell response and delayed development of neutralizing antibodies were found in deceased patients.314 Second, Tfh cells support memory B cell formation and response, which is important for rapid humoral response upon reinfection. Thirdly, Tfh cells in mucosal-associated lymphoid tissue (MALT) can also promote IgA production and function to modulate respiratory and gastrointestinal-tract infections.315 Collectively, CD4+ T cells are crucial mediators for supporting, promoting, and regulating both humoral and cellular immunity to resolve the infections effectively.
Chronic infection and cancer: persistent antigenic stimulation
In contrast to acute infections, antigen stimulation is persistent in chronic infection and cancer. It is now well-accepted that most T cells in such circumstances adopt a unique differentiation trajectory—exhaustion.316,317 Exhausted T (Tex) cells have been identified in many high grade chronic viral infections, such as HIV, HBV, HCV, and LCMV-Clone 13 strain,318–321 and in almost every mouse and human cancer.322,323 A wealth of recent studies at single-cell level have revealed that Tex cells constitute heterogenous populations with distinct transcriptional, epigenetic and functional signatures, playing critical roles in protecting against infections and tumors. The discovery of stem-like progenitor CD8+ Tex (Tpex) cells, the main responder to immune checkpoint blockade (ICB), attracts a large attention in both preclinical and clinical research field for developing next-generation cancer immunotherapies.322,324 In this section, we will summarize current understandings of the cellular and functional features of CD8+ and CD4+ T cells in chronic infection and tumor, their developmental pathways, regulatory mechanisms, CD4+ T cell help for CD8+ CTL responses, as well as contributions to anti-tumor immunity and checkpoint blockade.
Exhausted CD8+ T cells
Exhausted CD8+ T cells represent an entirely distinct differentiation trajectory with unique cellular phenotype, heterogeneity, and functional capacity.219,325,326 Along with the exhaustion, CD8+ T cells gradually lose production of IL-2 and TNF-α, and cytotoxic function.327 Compromised IFN-γ production occurs at more later stage of exhaustion and is associated with terminally differentiated Tex.328 But terminal CD8+ Tex may retain the ability to degranulate and produce chemokines and cytokines, such as MIP1α, MIP1β, RANTES, and IL-10 329. Different from TM cells in acute infection that undergo steady homeostatic self-renewal responding to cytokines IL-7 and IL-15,330 Tex cells display defects in responsiveness to homeostatic cytokines due to impaired IL-7Rα and IL-2/15Rβ signaling pathways.331,332 Instead, persisting antigen stimulation drives a proliferative progenitor pool of Tex cells,333,334 that Tex cells adopt a self-renewing mechanism dependent on continuous TCR stimulation.333 In addition, a key hallmark of CD8+ Tex cells is the upregulated and sustained expression of multiple IRs, such as PD-1, CTLA-4, Lag-3, TIGIT, Tim-3, CD39, 2B4, CD160, etc.329,335 The extent and coexpression of IRs directly correlate with the severity of exhaustion.335,336 On the other hand, Tex cells also express costimulatory molecules which, however, favor T cell exhaustion during chronic infection and tumor. For example, costimulation of CD27 and CD28 results in an enhanced T cell exhaustion.337,338 CD28 signaling is compromised due to loss of competition to CTLA-4 for B7 family ligands.338 PD-1 signaling further suppresses T cell function by specifically inducing CD28 dephosphorylation.339
Heterogeneity and differential trajectory of CD8+ Tex cells
The exhaustion/dysfunction of CD8+ T cells in chronic infection is established progressively with sequential phases.340,341 Analysis of CD8+ cell chromatin states define two discrete dysfunctional states: early reprogrammable and late non-reprogrammable T cells that the former ones are plastic and retain the potential to form memory after adoptive transfer, whereas the latter are fixed dysfunction with massive IR expression.341,342 Regarding to Tex cell origin, it was pointed out that CD8+ Tex cells arise from the same pool of KLRG1-CD127+ TMP cells in acute infection.343 The differentiation divergence of virus-specific CD8+ T cells responding to acute and chronic viral infections occurs as early as 4.5 days post-infection.344 However, under persistent antigen stimulation, these precursors progressively lose memory potential and develop into Tex cell state.342,343 With the rapid development of single-cell technologies, extensive analysis of tumor infiltrating lymphocytes (TILs) reveal a diverse spectrum of exhausted CD8+ T cells in non-small cell lung cancer (NSCLC), melanoma, breast cancer, liver cancer, and colorectal cancer.324,345–351
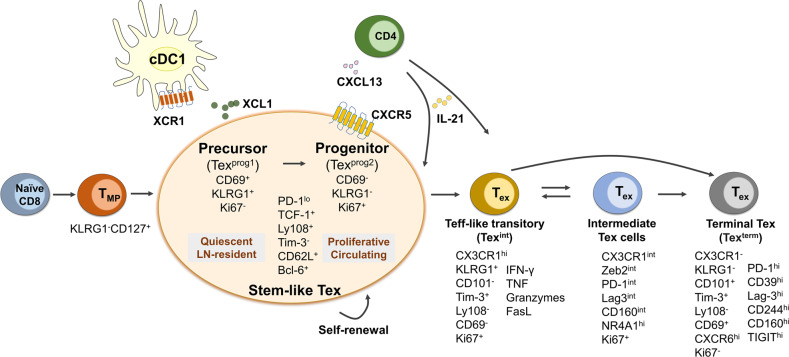
The CD8+ Tex cells being a distinct differentiation trajectory largely attributes to the identification of the stem-like, self-renewing Tpex population which is marked by expression of TCF-1 and surface profile of PD-1loTim-3-Ly108+CXCR5+.340,352 TCF-1-expressing Tpex cells are responsible for the maintenance of Tex cell populations in chronic viral infection and tumor.353,354 Tpex cells adopt a branched differentiation paradigm (Fig. 5), where they both self-renew and give rise to terminally differentiated exhausted T cells.334,344 Despite sharing similar phenotypes, the stem-like Tpex cells can be further separated into early precursor and late progenitor stages: the CD69+KLRG1+Ki67- CD8+ Tex precursors are more quiescent, lymph node (LN)-resident and having a baseline level of proliferation, whereas CD69-KLRG1-Ki67+ progenitors have robust proliferation and access to circulation.352,355 Recently, more markers are identified to define Tpex subsets. Tsui et al. reported that a small subset of TCF-1+CD62L+ Tpex cells are the stem-like population essential for long-term self-renewal, maintenance of Tex lineage and responsiveness to immunotherapy.356 In human individuals experienced latent infection such as CMV or EBV, TCF-1+ progenitors are comprised of two subsets based on PD-1 and TIGIT expression. The PD-1-TIGIT- progenitors are committed to a functional Tex differentiation, whereas PD-1+TIGIT+ progenitors are differentiated into a dysfunctional and exhausted state.357 Additionally, XCL1 is found expressed in CD8+ Tpex cells and associated with XCR1+ conventional type I dendritic cells (cDC1s).358
Fig. 5.
Heterogenous populations and differential trajectory of CD8+ Tex cells in chronic infection and tumor. Under persistent antigen stimulation, CD8+ T cells adopt an exhaustion differentiation trajectory of naïve → TMP → stem-like Tpex → effector-like transitory → intermediate → terminal Tex cells. Expression of signature markers and effector molecules at each Tex population is indicated. The stem-like Tpex cells are further divided into early precursor and late progenitor stages with discrete phenotype, proliferative status and preferential location. Tex subsets identified from different studies may use different names which are marked in the parentheses. CXCL13 and IL-21 derived from CD4+ T cells are critical for differentiation of CXCR5+ Tpex cells and CX3CR1+ Teff-like transitory Tex cells, respectively. CD8+ Tpex cells interplay with cDC1s through XCL1/XCR1 axis
Persistent antigen exposure induces downregulation of TCF-1, and drives Tpex differentiation into a “transitory” effector state and terminal exhausted T cells (Fig. 5). The transitory effector T (Teff)-like cells are critical for viral and tumor control and characterized by expression of chemokine receptor CX3CR1, producing IFN-γ, TNF and granzyme B, and enhanced cytotoxicity and cell proliferation.359,360 Generation of CX3CR1+ subset strongly depends on CD4+ T cell help and IL-21.360,361 Hudson et al. propose that Tpex differentiation follows a linear developmental trajectory where Tpex cells generate CX3CR1+Tim-3+CD101- transitory Teff-like T cells that further give rise to CX3CR1-Tim-3+CD101+ terminal Tex cells.359 Similarly, the expression of Ly108 and CD69 defines four subsets of Tex cells with a hierarchical developmental progression from Ly108+CD69+ (referred to Texprog1) to Ly108+CD69− (Texprog2) to intermediate differentiated Ly108−CD69− (Texint) cells and the most terminally differentiated Ly108−CD69+(Texterm) subset.355 Of note, Texint cells share similar transcriptional program to the CX3CR1+ Teff-like Tex cells identified in previous studies.359,360 Recently, a novel Tex subset expressing NK-associated genes (NKG2A and CD94) was uncovered within the Texint cell population.362 More evidence supporting the Tex cell differentiation trajectory comes from comprehensive analysis of antigen-specific T cells in patients with human papillomavirus (HPV)-positive head and neck cancer. Paired scRNA-seq analysis and TCR sequencing of HPV-specific CD8+ T cells sorted by MHC class I tetramers revealed that antigen-specific PD-1+TCF-1+ stem-like CD8+ T cells could proliferate and differentiate into Teff-like transitory and terminally differentiated cells.363 In addition, epigenetic landscape analysis demonstrates that the phenotypic changes of Tex cell development coincide with the chromatin accessibility of key genes.355,359 Long-term antigen stimulation leads to epigenetic reprogram which enforces the terminal exhaustion of T cells marked by high expression of IRs, diminished effector-related molecules (IFN-γ, TNF, granzymes, and T-bet) and loss of stemness and proliferation potential (TCF-1, MYB, MYC, and Ki67).219,355,359 Furthermore, in infection with chronic LCMV-Clone 13, a “bridging population” between Teff-like transitory and terminal exhausted Tex cells is characterized by intermediate expression of CX3CR1, Zeb2 and IRs, but high expression of NR4A1 (encoding NUR77), suggesting a recent activation by TCR stimulation.364 Chemokine receptors CXCR6 and CX3CR1 can be used to discriminate these three populations: Teff-like transitory cells (CX3CR1hi), intermediate Tex cells (CX3CR1int) and terminal exhausted Tex cells (CX3CR1loCXCR6hi).364 Recent high-dimensional single-cell multi-omics have revealed more heterogenous Tex clusters with distinct phenotypic, transcriptomic, epigenetic and functional patterns, which also display disease- and tissue-specificity.364–366 It is noteworthy that exhausted T cells can be also induced in acute infection with strong T cell stimulation. For instance, severe acute respiratory syndrome elicited during SARS-CoV-2 infection induces T cell exhaustion phenotypes with high level of IRs expression.229,233
Transcriptional and epigenetic regulation of CD8+ Tex cells
The differentiation of CD8+ Tex cells is tightly controlled by transcriptional and epigenetic networks. In chronic infection and tumors, TCF-1 identifies the stem-like CD8+ Tpex cells.354,367,368 Accordingly, mice with Tcf7 deficiency could not develop stem-like Tpex cells and Tex populations,353 whereas overexpression of Tcf7 led to enhanced Tpex program as well as antiviral and anti-tumor immunity.369 TCF-1 plays central roles in Tpex cells by organizing transcriptional regulatory networks.354,370 TCF-1 coordinates with FoxO1 which also acts as an upstream regulator of TCF-1 expression to promote and maintain the stemness in CD8+ T cells by augmenting pro-memory TFs Eomes, Id3, c-Myc, Bcl-2, and Bcl-6 expression while inhibiting effector-related TFs T-bet, Id2, Runx3, and Blimp-1.367,368,370–372 MYB (also known as c-Myb) is a pivotal TF for CD8+ central memory and Tpex cell generation and maintenance by acting as a transcriptional activator of Tcf7.356,373 Moreover, BACH2 promotes stem-like CD8+ T cell commitment in chronic infection and cancer by enforcing the transcriptional and epigenetic programs.374
TOX, a high-mobility group box DNA-binding protein, has recently emerged as a critical regulator for Tex cell programs.375–377 Enforced expression of TOX is sufficient to induce an exhausted T cell-associated transcriptional program with increased expression of IRs.376,378 While TOX deficiency has no impact on CD8+ T cells differentiation and effector function in acute infections, deletion of TOX in tumor-specific T cells inhibits the upregulation of IRs and augments the cytokine production, effector functions, and TCF-1 expression.375,378 Although TOX deficient T cells display a “non-exhausted” immunophenotype, those T cells remain hyporesponsive and ultimately diminish.375,378 In fact, TOX deficient CD8+ T cells fail to persist and differentiate into Tex cells, indicating that TOX-regulated exhaustion indeed protects T cells from overstimulation and activation-induced cell death.375,376,378 Additionally, TOX and nuclear receptor NR4A form positive feedback loops to impose CD8+ T cell dysfunction and exhaustion.379–381 BATF is another important TF regulating T cell exhaustion, however, its role remains controversial. Some studies report that BATF facilitates viral clearance by driving the transition from TCF-1+ Tex progenitors to CX3CR1+ effector cells during chronic viral infection.382 Moreover, BATF cooperates with IRF4 to resist exhaustion; overexpression of BATF promotes the survival and anti-tumor immunity in chimeric antigen receptor (CAR) T cells.383 However, others claim that BATF drives T cell exhaustion by directly upregulating exhaustion-associated genes, thus BATF depletion could significantly enhance T-cell resistance to exhaustion and exhibit superior efficacy against solid tumors in CAR-T cells.384–386
Intriguingly, Tex cells express certain TFs shared by T cells in acute infection, but with distinct gene transcription,387 suggesting context-dependent functions of these TFs. For instance, Eomes and T-bet are dually required for Tex cell generation.334 Eomes expression is elevated in tumor-infiltrating CD8+ T cells and high level of Eomes promotes exhaustion.388,389 But high expression of T-bet was found associated with Tpex and effector-like Tex subset.334,390,391 In addition, TF NFAT family which has a well-established role in mediating T cell activation when partners with AP-1,392 has been shown to regulate Tex cell differentiation. NFATc1 drives exhaustion program by promoting IR expression,393 whereas NFATc2 prevents the dysfunction of CD8+ Tex cells.394 The major differences of CD8+ T cells in acute and chronic infections are compared (Table 1).
Table 1.
Characteristics of CD8+ T cells in acute and chronic infections
| Infection type | Infectious gents/condition | Characteristics | Stages | Fate | Subsets | Surface marker | Key TF | Refs |
|---|---|---|---|---|---|---|---|---|
| Acute | LCMV-Armstrong, LM, influenza virus, HAV, RSV, vaccinia virus | IFN-γ, TNF, IL-2, KLRG1, Granzymes, Perforin | expansion, contraction, memory | TE, TMP, TEM, TCM, TRM | TE | KLRG1, CX3CR1, CXCR6, CCR5 | T-bet, Blimp-1, Id2, STAT4, Zeb2 | 169,173,182,193,219,223 |
| TMP | CD127, CD62L | Eomes, TCF-1, FoxO1, Bcl-6, Id3, STAT3, Zeb1 | ||||||
| Chronic | LCMV-Clone 13, HIV, HBV, HCV, CMV, EBV, SARS-CoV-2, Cancer | Loss of IL-2, IFN-γ, TNF-α; Expression of IRs (PD-1, CTLA-4, Lag-3, TIGIT, Tim-3, CD39, 2B4, CD160) | Tex precursor, Tex progenitor, Teff-like transitory, Intermediate Tex, Terminal Tex | Tex | Teff-like | KLRG1, CX3CR1, Tim-3 | T-bet, Id2, Runx3, Blimp-1 | 219,323,325,326,340,355,359,367,370,377,387 |
| Tpex | Ly108, CD62L, CXCR5, XCL1 | TCF-1, FoxO1, Eomes, Id3, c-Myc, Bcl-2, Bcl-6, MYB, BACH2 | ||||||
| Tex | PD-1, CD101, Tim-3, CXCR6 | TOX, BATF, Eomes, T-bet, NFAT |
The underlying mechanisms that govern the distinct transcriptional features of Tex cells remain poorly understood, but at least partially, are controlled by epigenetic programming. CD8+ Tex cells exhibit a unique chromatin landscape different from effector and memory T cells.342,355,362,395 The chromatin accessibility of key exhausted-associated genes such as TCR signaling, cytokines, costimulatory and coinhibitory receptors has experienced dynamically epigenetic reprogram.365,396 For instance, the gene regions around Tcf7 and Id3 are more accessible in stem-like Tpex cells while that in Prdm1, Id2, and Pdcd1 are more accessible in exhausted CD8+ T cells.397,398 Particularly, TOX acts as a crucial regulator of epigenetic programming of CD8+ Tex cells by repressing the chromatin accessibility of genes involved in effector cell differentiation. Additionally, TCF-1 regulates gene transcription by altering the three-dimensional (3D) genome organization.399,400 A prominent feature of Tpex cells is that the exhaustion commitment can be transmitted to their progeny even when adoptive transferred into new hosts received acute infection.401 The underlying mechanisms of such exhaustion inheritage are derived from epigenetic imprints which once are established, they can not be reversed by change of exogenous environment or by PD-(L)1 blockade.402–404
Tex subsets contributing to anti-tumor immunity and ICB
Tumors with high infiltration of T cells are generally considered as immune-inflamed or “hot” tumors. However, intratumoral T cells may not be tumor-reactive. TCR repertoire analysis reveals that the tumor recognizing T cells were limited to merely 10% of intratumoral CD8+ T cells.405 ICB can robustly reinvigorate Tex cell function, making it one of the most promising cancer therapies in the clinic.406–408 Antibodies targeting IRs on tumor-infiltrating T cells, such as PD-1/PD-L1 (among others), have been demonstrated impressive clinical activities across a variety of cancer types. Despite large success, ICB faces clinical challenges of low responsive rate, drug resistance, and immune-related adverse events (irAEs).409,410 Thus, it is of great significance to understand which subset of CD8+ T cells respond to ICB and how. Among heterogenous CD8+ Tex cells, it is now well-appreciated that the PD-1+TCF-1+ stem-like Tpex cell population mainly mediates tumor responses to checkpoint blockade.353,410,411 Comparison between the responder and non-responder of melanoma patients receiving ICB treatment demonstrates that the frequency of TCF-1hi tumor-infiltrating CD8+ T cells predicts positive clinical outcome.412 This CD8+ Tpex cell population has also been observed in human NSCLC, colorectal cancer, HPV-positive head and neck cancer and bladder cancer, and their number was augmented following ICB treatment.363,411,413,414 Interestingly, ICB could control tumor growth in mice depleted TCF-1-expressing T cells, indicating that later differentiated Tex cells may also be targeted by ICB.411 Indeed, comprehensive transcriptomic and TCR clonal analysis reveal that tumor/ICB-responsive CD8+ T cells including neoantigen-specific ones exhibit enhanced exhaustion compared to non-tumor-reactive bystander CD8+ T cells.415,416 Accordingly, differentiation from TCF-1+ Tpex cells into late stage of Tex cells expressing PD-1 and Tim-3 favors the tumor control.417,418 Thus, high expression of PD-1 and/or CTLA-4 on tumor infiltrating CD8+ T cells provides a predictive biomarker for responsiveness to ICB therapy.419,420
Beyond, it is also critical to address the effects of ICB on CD8+ T cell state. It has been shown that effective immunotherapies can induce remarkable remodeling of tumor environment (TME) and systemic immune activation in multiple tissues.421 Paired scRNA-seq and TCR-seq on tumor biopsies from NSCLC patients revealed that the Tpex population was accumulated in responsive tumors but not in non-responsive ones after anti-PD-1 therapy.422 The data also depicts that the increased Tpex cells are mainly derived from local expansion or replenishment from peripheral T cells with pre-existing clonotypes, a phenomenon called “clonal revival”.422 While the effect of ICB primarily relies on pre-existing state of intratumoral T cells, ICB can alter the TCR repertoire to generate novel T cell clonotypes, which is referred to as “clonal replacement”.422,423 Moreover, intratumoral exhausted T cell populations and their immunological responses to ICB exhibit features of spatial distribution.424 Studies in both mouse and human tumors have demonstrated that tumor-draining LNs (TdLNs) are the preferential reservoirs for TCF-1+ Tpex cells that remain stable regardless of the changes in TME and sustain continuous development of anti-tumor T cells.425,426 Blockade of sphingosine 1-phosphate receptor 1 (S1P1)-mediated T cell egress from TdLNs remarkably decreased the frequency of intratumoral CD8+ Tpex cells and the tumor eradication efficacy of anti-PD-1 therapy.421,426 The clonal overlapping between tumor-infiltrating CD8+ T cells and proliferating CD8+ T cells in the circulation in cancer patients following anti-PD-1 treatment highly suggests a recruitment from secondary lymphoid organs.427 A group of bona fide tumor-specific memory CD8+ T cells within TdLNs are important responders to PD-1-based ICB, highlighting their potentials in anti-tumor immunotherapy.428 Inherent in this theory, local (intratumoral, intradermal or intrapleural) administration of ICB antibodies, compared to systemic (intravenous or intraperitoneal) injection, results in enhanced tumor regression due to antibody accumulation and Tpex cell expansion within TdLNs.429,430
Complex CD4+ T helper cells
Robust and functional CD4+ T cell responses are essential for effective pathogen clearance and tumor eradication. Compared to well-defined CD8+ Tex cell differentiation, the cellular and functional signatures of CD4+ T cells in chronic disease settings are little characterized, especially with the complexity of multiple Th subsets. CD4+ T cells play multifaceted roles in chronic infection and tumor: constituting both favorable and deleterious subsets, enhancing CD8+ T cell function, and responding to ICB,427,431 which highlights potential next-generation therapeutics of harnessing CD4+ T cell function.
Are CD4+ T cells exhausted?
The effects of persistent antigenic stimulation on CD4+ T cell phenotype, differentiation and function remain less understood. Whether CD4+ T cells become “exhausted” during chronic infection remains a question for a long time. Controversial results were obtained as viral-specific CD4+ T cells lose effector function and produce reduced IFN-γ, TNF-α and IL-2 during chronic infection,432,433 but the production of IL-10 and IL-21, the important cytokines in chronic infection for sustaining CD8+ T cell and B cell responses,434–436 are increased.434,437,438 Transcriptional analysis of CD4+ T cells during chronic (LCMV-Clone 13) infection has demonstrated a unique exhaustion-associated molecular and transcriptional profile, which is distinct from CD8+ Tex cells and effector or memory CD4+ T cells in acute (LCMV-Armstrong) infection.439 In addition to reduced cytokine production, CD4+ Tex cells express markedly upregulated IRs including PD-1, CTLA-4, CD200 and BTLA, and costimulatory receptors OX40, CD27 and ICOS.439 Core TFs involved in CD4+ Tex cells include Eomes, Blimp-1, Helios, Klf4, and T-bet.439 During LCMV-Clone 13 infection, viral-specific CD4+ T cells formed multiple clusters which could be broadly grouped into Th1, Tfh and Th1/Tfh hybrid clusters at different stages, suggesting an altered Th lineage differentiation in chronic infection.431 Notably, persistent viral infection drives a progressive loss of Th1 response likely due to PD-1/PD-L1 inhibitory signaling pathway,431,440 but skews CD4+ T cells toward Th2, Th17, Treg, Tfh, and allergic CD4+ T cell lineages.439 Different from TCF-1+ CD8+ Tpex cells, TCF-1 expression in chronic virus-specific CD4+ T cells does not adequately define stem-like progenitor CD4+ T cells, rather marks and promotes Tfh cell development.431 Recently, Xia et al. identified a population of memory-like TCF-1+Bcl-6lo/− virus-specific CD4+ T cells emerged as the progenitor cells that gives rise to Teff and Tfh cells, sustaining CD4+ T cell response in chronic infection.441 Importantly, such CD4+ progenitor cells play pivotal roles in anti-tumor response preferentially at site of TdLNs.441 Hence, CD4+ T cells display exhausted yet functional phenotype in chronic infection.
CD4+ Th cell subsets
Th1 and Th2
Th1 cells predominantly exert the anti-tumor activity. The frequency of Th1 subset and IFN-γ production in TME correlate positively with better clinical outcomes in multiple tumor types including melanoma,442 breast,443,444 ovarian,445 lung,446 colorectal,447 and laryngeal cancers448 (Table 2). Th1 cells promote tumor rejection by shaping an anti-tumor immune environment and indirectly supporting effector functions of other immune cells.449,450 Th1 cells are an important CD4+ T cell subset providing help for CD8+ T cell response and function,451 which will be elaborated at the later section. The migration of effector CD8+ T cells and NK cells in TME depends on chemokine receptor CXCR3 and its ligand CXCL9 and CXCL10 which are predominantly expressed by Th1-related IFN-γ-activated macrophages, cancer-associated fibroblasts (CAFs) and tumor cells.452–454 In addition, IFN-γ and IL-2 produced by Th1 cells enhance the survival, proliferation and cytolytic function of CD8+ CTLs and NK cells.449,455 IFN-γ can significantly enhance MHC I and MHC II expression and tumor-derived antigen presentation on tumor cells.456,457
Table 2.
CD4+ T helper cell subsets in tumor immunity
| Th subset | Phenotype | Tumor immunity | Tumor types | Functions | Refs |
|---|---|---|---|---|---|
| Th1 | CXCR3, IFN-γ, TNF-α, IL-2, T-bet | anti-tumor | Melanoma, breast, ovarian, lung, colorectal and laryngeal cancers | activate macrophages, CAFs and tumor cells | 452–454 |
| enhance MHC I and MHC II expression | 456,457 | ||||
| attract NK and CD8+ T cells | 452–454 | ||||
| support effector functions of NK and CD8+ T cells | 449,455 | ||||
| Th2 | IL-3, IL-4, IL-5, IL-13, GM-CSF | anti-tumor | Plasmacytoma, melanoma, myeloma, breast cancer | activate eosinophils and M2-type macrophages | 461–463 |
| enhance NK cell cytotoxic activities | 464 | ||||
| induce cancer cell terminal differentiation | 465 | ||||
| IL-4, IL-10, TGF-β | pro-tumor | Pancreatic and breast cancer | promote breast cancer metastasis | 466 | |
| suppress Th1 response | 467,468 | ||||
| Th17 | IL-17A, IL-17B, IL-17F, IL-21, IL-22, IL-23 | anti-tumor | Chronic lymphocytic leukemia, gastric adenocarcinoma, cervical adenocarcinoma ovarian, colorectal, lung and breast cancers | induce cancer cell apoptosis | 512 |
| enhance recruitment of anti-tumor NK cells, DCs, neutrophils and macrophages | 513–516 | ||||
| attract effector CD4+ and CD8+ T cell infiltration | 474,514,517,518 | ||||
| IL-17A, IL-17D, IL-25/IL-17E | pro-tumor | Breast cancer, melanoma, bladder carcinoma, B cell acute lymphoblastic leukemia, colorectal, lung, prostate, liver, pancreatic and gastric cancers | stimulate tumor cell growth and inhibit apoptosis | 482–485 | |
| promote CSCs maintenance and activation | 486,487 | ||||
| enhance tumor invasion and metastasis | 488–490 | ||||
| promote angiogenesis | 491–493 | ||||
| promote MDSCs, TAMs and neutrophils | 494–500 | ||||
| constrain effector NK and CD8+ T cells | 501,502 | ||||
| induce terminal CD8+ Tex cell differentiation | 503 | ||||
| affect vascular endothelial cells and keratinocytes | 504–506 | ||||
| Th9 | IL-9, IL-21 | anti-tumor | Melanoma, chronic lymphocytic leukemia, non-Hodgkins lymphoma, lung, breast and colorectal cancers | direct tumor cell killing by granzymes | 521,522 |
| promote recruitment of DCs | 524,525 | ||||
| induce CD8+ CTL and NK cell responses | 98,522,523 | ||||
| elicit IFN-α/β production by monocytes | 526 | ||||
| induce mast cell activation | 521,527 | ||||
| pro-tumor | Hodgkin lymphoma, anaplastic large cell lymphoma, B and T cell lymphomas, CRC, HCC, lung, mammary, breast cancers | enhance tumor cell survival and migration | 532–536 | ||
| induce EMT and metastatic spreading | 488 | ||||
| mediate immunosuppression of mast and Treg cells | 537 | ||||
| Treg | IL-17, IFN-γ, TNF-α | anti-tumor | CRC, HNSCC, Hodgkin’s lymphoma, estrogen receptor-negative breast cancer, esophageal cancer, oral and oropharyngeal squamous cell carcinomas | suppress pro-tumor Th17 responses | 548 |
| express pro-inflammatory cytokines | 549,550 | ||||
|
CD25, ICOS, OX40, 4-1BB, GITR, PD-1, CTLA-4, Lag-3, Tim-3, TIGIT, CCR4, CCR8 IL-10, TGF-β, IL-35, IL-33, IL-37 Foxp3, FoxO1, STAT5, NFAT, T-bet, Helios, Nr4a, Foxp1 |
pro-tumor | HCC, melanoma, breast, lung, cervical, gastric, bladder, renal, endometrial and ovarian cancers | kill effector T cells, APCs and NK cells | 554,555 | |
| produce inhibitory cytokines | 556–558 | ||||
| express coinhibitory molecules | 539,559–561 | ||||
| suppress APCs function | 541,567 | ||||
| suppress NKT cell cytotoxic activity | 568 | ||||
| facilitate suppressive activity of MDSCs | 569,570 | ||||
| produce adenosine by CD73 and CD39 | 571,572 | ||||
| compete IL-2 with effector T cells | 541,573 | ||||
| produce IDO | 574,575 | ||||
| Tfh |
CXCR5, PD-1, ICOS, Bcl-6 IL-4, CXCL13, IL-21 |
anti-tumor | Melanoma, breast, colorectal and lung cancers | promote the formation of TLSs | 479,597 |
| induce pro-inflammatory cytokines | 132,598 | ||||
| activate complement cascade | 132,598 | ||||
| promote effective cytotoxic lymphocytes | 132,598 | ||||
| enhance CD8+ T cell response | 436,592,602 | ||||
| promote GC response and antibody production | 312,603,1109 | ||||
| support B cells and memory B cells | 606,607 | ||||
| respond to PD-1-based ICB | 590,608 |
The role of Th2 cells in tumor progression remains controversial with both favorable and deleterious effects458–460 (Table 2). Previously, Th2 cells have been shown to suppress tumor growth by activating eosinophils as the cytotoxic effector cells in murine plasmacytoma and melanoma.461,462 Adoptive transfer of tumor-specific Th2 cells induced massive accumulation of M2-type macrophages at the tumor site, which triggered an inflammatory immune response to eliminate myeloma cells.463 Memory Th2 cells display potent anti-tumor activity by producing IL-4 to enhance NK cell cytotoxic activities.464 Moreover, Th2 cells can directly block breast carcinogenesis by secreting IL-3, IL-5, IL-13, and GM-CSF, which induce the terminal differentiation of the cancer cells.465 However, in pancreatic cancer, thymic stromal lymphopoietin (TSLP) produced by CAFs attracts and induces Th2 cells, which correlates with reduced patient survival.459 Th2 associated IL-4 signaling in monocytes and macrophages promotes breast cancer metastasis.466 Th2 cells can also attenuate Th1-associated anti-tumor responses through IL-4 signaling.467,468 In accordance with this notion, Th1-dominant immune response—upregulation of Th1-related response while downregulation of Th2-associated response—can be used as positive prognostic indicators for certain cancers.469–471 The discrepancy of Th2-mediated tumor immunity may attribute to different tumor types and distinct Th2 cell state. For example, studies have suggested that tumor-promoting Th2 cells have high levels of IL-10 and TGF-β, whereas Th2 cells with high expression of IL-3, IL-5, IL-13, and GM-CSF exhibit pro-inflammatory and anti-tumor immunity.465,472
Th17
Th17 cells are specifically accumulated in many types of human tumors.473 Cytokine milieu formed by IL-1β, IL-6, IL-23, and TGF-β produced by tumor cells, CAFs and tumor-associated macrophages (TAMs) supports Th17 cell differentiation and expansion.474,475 However, the effects of Th17 cells and cytokine IL-17 on tumor immunity are contradictory.473,476 Therefore, the presence of Th17 cells is associated with either good or poor prognosis depending on tumor types477–479 (Table 2). The pro-tumor function of Th17 cells is attributed to both direct effects on tumor cells and indirect effects of inducing a pro-inflammatory environment.480,481 Th17 cells and IL-17 strongly stimulate tumor cell proliferation by activating growth-related kinases and TFs, while inhibit their apoptosis by acting on anti-apoptotic proteins.482–485 Th17 cells and IL-17 promote cancer stem cells (CSCs) maintenance, pro-tumorigenesis and activation.486,487 Th17 cells also enhance tumor invasion and metastasis in lung, prostate, liver, and pancreatic cancers by inducing tumor cell epithelial-mesenchymal transition (EMT), matrix metalloproteinases (MMPs) expression, and chemokine expression.488–490 A key mechanism for the pro-tumor activity of Th17 cells is that IL-17 promotes angiogenesis.491 IL-17 in TME often correlates with high vascular density and tumor overgrowth, and induces the production of angiogenic factors such as vascular endothelial growth factor (VEGF), IL-6 and IL-8 by tumor cells or stromal cells.492,493 Furthermore, Th17 cells and IL-17 indirectly shape a pro-tumor TME by recruiting and influencing other immunosuppressive cells. For instance, IL-17 promotes the development, tumor infiltration and immunosuppressive activity of myeloid derived suppressor cells (MDSCs),494,495 TAMs,496–498 and pro-tumor neutrophils.499,500 IL-17 also constrains the cytolytic activity of NK cells and CD8+ T cells by inhibiting IL-15-mediated cell maturation501 and recruiting neutrophils,502 respectively. Interestingly, IL-17 also promotes tumor progression through inducing terminal exhausted CD8+ T cell differentiation.503 Apart from immune cells, IL-17 increases vascular endothelial cells number in gastric cancer,504 triggers CAFs to produce myeloid cell stimulatory factor G-CSF,505 and promotes skin tumor formation by stimulating keratinocyte proliferation.506 Furthermore, Th17 cells secrete high level of IL-22 which enhances the tumor growth and metastasis in human colon cancer.507,508
On the contrary, Th17 cells and IL-17 are found positively associated with better prognosis and improved patient survival in various cancers474,509–511 (Table 2), indicating a tumor-protective role of Th17 cells. The underlying mechanisms for the anti-tumor activity of Th17 cells also rely on direct and indirect functions. IL-17 acts on IL-17R-expressing tumor cells and induces caspase-dependent apoptosis signaling in breast cancer.512 IL-17 enhances the recruitment and anti-tumor functions of NK cells,513 DCs,514 neutrophils,515 and pro-inflammatory macrophages.516 Th17 cells stimulate CXCL9 and CXCL10 production from tumor cells to attract effector CD4+ and CD8+ T cell infiltration, and increase IFN-γ+ T cell activity.474,514,517 Furthermore, IL-17-producing CD4+ and CD8+ T cells display improved potency to repress tumor growth.518 The multifaceted and discrepant functions of Th17 cells in the context of tumor likely derive from distinct tumor types, and more importantly, high plasticity of Th17 cells which can be transdifferentiated into other Th lineages including Th1, Th2, Tfh, and Treg cells, endowing them with discrete or opposing functions.519 Additionally, IL-17 is produced by many cell types besides Th17 cells, such as neutrophils, γδ T cells, macrophages, MDSCs, mast cells, endothelial cells, tumor cells and CAFs.519 Thus, it is important to distinguish the effects of Th17 cells and IL-17 on tumor immunity.
Th9
Th9 cells have been receiving much attention recently due to the fact that this CD4+ T cell subset and its featured cytokine IL-9 exhibit unprecedented anti-tumor immunity.100,479 High frequency of Th9 cells was found positively correlated with better prognosis in NSCLC patients.520 The potent anti-tumor activity of Th9 cells relies on both direct tumor cell killing and indirect roles in shaping anti-tumor immunity. Studies have shown that Th9 cells express high level of granzymes and display direct cytotoxic activity on melanoma cells.521,522 Th9 cells can induce robust CD8+ CTL and NK cell responses by secretion of cytokines IL-9 and IL-21.98,522,523 IL-9 may also enhance CD8+ T cell function through promoting recruitment of DCs into the tumor tissue524 and enhancing their antigen cross-presentation.525 Thus, administration of IL-9 neutralizing antibody inhibits tumor-specific CD8+ T cell responses and results in tumor progression.524 By increasing intratumor ATP, Th9 cells induce monocytes infiltration and production of IFN-α/β.526 Moreover, the anti-tumor activity of Th9 cells depends on mast cell activation.521,527 Notably, intratumoral Th9 cells are found less-exhausted and highly proliferative and cytolytic, and only Th9 cells could completely eradicate advanced tumors compared to other tumor-killing CD4+ T cell subsets such as Th1 and Th17 cells.528 Hence, Th9 cells represent an effective population of CD4+ T cells for adoptive cell therapy.526,529,530
Despite considerable evidence showing the potent anti-tumor activity of Th9 cells, pro-tumoral roles of Th9 cells have also been reported. Overexpression of IL-9 is detected in various cancers (Table 2), which is strongly associated with augmented tumorigenesis and shorter disease-free survival period.92,531,532 IL-9 can directly enhance tumor cell survival and migration through activation of JAK1 and JAK3, and STAT (STAT3 and STAT5) signaling pathways.532–534 In chronic lymphocytic leukemia (CLL) patients, an autocrine-positive feedback loop of Th9/IL-9 axis promotes malignant T cell survival.535,536 In addition, IL-9 promotes tumor progression by inducing EMT and metastatic spreading in lung cancers.488 IL-9 contributes to tumor growth by mediating immunosuppression of mast cells and Treg cells.537 IL-9 in TME functions as an immunosuppressor for adaptive immunity in which IL-9 depletion or neutralization could restore the immunological memory for effective tumor rejection.538 Given those inconsistent results, further studies are needed to fully delineate the function of Th9 cells in tumors especially their clinical relevance in human.
Treg cells
As a major immunosuppressive subset of CD4+ T cells, Treg cells are found substantially infiltrated in many solid tumors.539–541 The high frequency of Treg cells is mainly associated with worse clinical outcomes in majority of tumor types such as HCC, melanoma, breast, lung, cervical, gastric, bladder, renal, endometrial, and ovarian cancers.542–544 However, Treg infiltration may also correlate with better prognosis in CRC, HNSCC, Hodgkin’s lymphoma, estrogen receptor-negative breast cancer, esophageal cancer, and oral and oropharyngeal squamous cell carcinomas.543,545,546 This discrepancy may be related to different TME, Treg cell plasticity and their interplay with other cells. For instance, Treg cells infiltrated in CRC are enriched for less immunosuppressive Foxp3lo population rather than more immunosuppressive Foxp3hi subset.547 Th17 cell-mediated pro-inflammatory and pro-tumor responses in CRC can be attenuated by Treg cells.548 In addition, Treg cells in CRC can also be induced to express pro-inflammatory cytokines including IL-17, IFN-γ, and TNF-α, exerting an anti-tumor immunity.549,550 Therefore, high Treg cells together with a low frequency of CD8+ CTLs are better prediction for unfavorable prognosis in various types of cancer.542,551
Compared to Treg cells in non-tumor tissues, intratumoral Foxp3+ Treg cells are mostly active and highly proliferative,552 expressing elevated levels of activation markers CD25, ICOS, TNFR superfamily members OX40, 4-1BB, and GITR, various IRs, and chemokine receptors CCR4 and CCR8.542,553 Emerging evidence has revealed a variety of mechanisms contributing to Treg cell immunosuppression: (1) Treg cells can directly kill effector T cells, APCs and NK cells by expressing perforin and granzyme B, and induce cell apoptosis by FasL/Fas signaling.554,555 (2) Treg cells mediate immunosuppression through producing inhibitory cytokines, including IL-10, TGF-β, IL-35, IL-33, and IL-37.556–558 (3) Treg cells express a spectrum of high levels of coinhibitory molecules, such as CTLA-4, PD-1, Lag-3, Tim-3, and TIGIT.539,559–561 For instance, CTLA-4 competes with costimulatory receptor CD28 on effector T cells for binding to CD80/CD86 on APCs.562 CTLA-4 further downregulates CD80/CD86 expression via trans-endocytosis and trogocytosis.563–565 In addition, Treg cells maintain memory CD8+ T cell quiescence by suppressing their effector and proliferative programs through CTLA-4 signaling.566 (4) Treg cells exert immunoregulatory functions by influencing other immune cells. Engagement of CTLA-4 and Lag-3 on Treg cells with CD80/CD86 and MHC II molecules on DCs respectively, results in suppression of antigen-presenting function and subsequent activation of effector T cells.541,567 In addition, Treg cells suppress NKT cell cytotoxic activity in a cell-cell contact-dependent manner,568 while facilitate the immunosuppressive activity of MDSCs.569,570 (5) Treg cells dampen the anti-tumor immunity by shaping an immunosuppressive TME involved in suppressive metabolites. High expression of ectonucleotidase CD39 and CD73 on Treg cells can convert extracellular ATP or ADP into adenosine which induces broadly inhibitory signals in effector T cells, NK cells, and DCs.571,572 IL-2, as an essential cytokine for effector T cell activation and proliferation, is consumed by Treg cells which express high level of CD25, the high-affinity IL-2Rα.541,573 Treg cells also increase indoleamine 2, 3-dioxygenase (IDO) production which mediates tryptophan metabolism and causes effector T cell dysfunction.574,575
Another essential aspect regarding to tumor-infiltrating Treg cells is their origin. Comprehensive transcriptomic and TCR repertoire analyses have revealed both nTreg and iTreg cells serve as the cell sources,570,576,577 and tumor-infiltrating Treg cells are both recruited from the periphery or TdLNs, and expanded within the TME.578,579 A variety of chemokine receptors on Treg cells and their cognate ligands are involved in the recruitment of Treg cells, including CCR4 (CCL17 and CCL22), CCR8 (CCL1, CCL8, CCL16 and CCL18), CCR2 (CCL2), CCR5 (CCL5), CCR6 (CCL20), CCR10 (CCL28 and CCL27), CXCR3 (CXCL9, CXCL10 and CXCL11), and CXCR4 (CXCL12).345,580,581 Among distinct mechanisms, signals from tumor antigen stimulation, ICOS/ICOSL, TNFR2, 4-1BB, OX40, and GITR significantly drive Treg cell expansion and functionality.540,580,582 In addition, the nutrient-deprived TME plays critical roles in reprogramming Treg cell metabolism and activity.583 Glycolysis, fatty acid oxidation and oxidative phosphorylation are all important for the differentiation and function of tumor-infiltrating Treg cells.570,584,585 Particularly, lactic acid uptake in Treg cells promotes PD-1 expression which dampens the efficacy of anti-PD-1 immunotherapy,586,587 and uptake of free fatty acids and low-density lipoprotein via scavenger receptor CD36 is required for intratumoral Treg cell survival, amplification and suppressive function.583,588,589
Tfh and tertiary lymphoid structures (TLSs)
Tfh cells mainly support B cell responses and antibody production in infectious disease and vaccination.131,132 It is surprising to found a close link between Tfh cell response and anti-tumor immunity.131,590 Persistent antigenic stimulation during chronic viral infection and tumor redirects CD4+ T cell differentiation toward Tfh cells.131,591,592 Recent studies have revealed a positive correlation between the presence of Tfh and B cells with prolonged survival and better prognosis in a variety of human tumors, including melanoma,593 breast cancer,594 colorectal cancer,595 and lung cancers.596
The underlying mechanisms by which Tfh cells exert protective functions in infection and tumor are: (1) Tfh cell response significantly promotes the formation of TLSs which are ectopic tissue structures consisting B cells, T cells, NK cells and APCs in nonlymphoid organs under chronic inflammatory stimulation.479,597 Mature TLSs within tumors represent anti-tumor contextures with pro-inflammatory cytokines, activated complement cascade, and effective cytotoxic lymphocytes.132,598 Tumor-infiltrating Tfh cells expressing high levels of CXCL13 and IL-21 are enriched in intratumoral TLSs and strongly associated with infiltration of CD8+ T cells and B cells, as well as prolonged survival in cancer patients.599–601 (2) Tfh cells can enhance CD8+ T cell response in chronic viral infection and tumor through producing CXCL13 and IL-21,436,592,602 which will be further discussed at later section. (3) Tfh cells promote B cell and GC response and production of functional antibodies.603 Potent anti-tumor immunity requires antibody-mediated effector functions such as antibody-dependent cell cytotoxicity (ADCC), complement activation and antibody-mediated tumor cell phagocytosis.604 Tumors with high Tfh cells and mature TLSs mostly have high density and diversity of B cells and plasma cells, as well as tumor-targeting antibodies, which further induces effective anti-tumor immunity.598,605 (4) Tfh cells support the generation of memory B cells which are crucial for rapid response upon reinfection and long-term protection.606,607 (5) Tfh cells contribute to PD-1-based ICB.590,608 It is noteworthy that high PD-1 expression on Tfh cells does not indicate cell exhaustion, instead, promote Tfh cell expansion, activity and function.609,610 In clinical studies, the densities of Tfh cells, TLSs and tumor-infiltrating B cells positively correlate with the overall survival and responsiveness in patients treated with immunotherapy in various tumor types.593,611,612 The benefit of Tfh cells for anti-PD-1 therapy partially depends on their activity to recruit CD8+ T cells through CXCL13/CXCR5 signaling axis.613,614 Consistently, histological analysis confirms a spatial proximity of CXCL13+ Tfh cells, CXCR5+ CD8+ T cells and CD20+ B cells within TLSs, which enhances the efficacy of PD-1 ICB.615
CD4+ T cell help enhances anti-tumor response of CD8+ CTLs
Help mechanisms
Although CD8+ CTLs play the predominant roles in anti-tumor immunity, it is now well-appreciated that CD4+ T cells are pivotal to support the effective anti-tumor CD8+ T cell responses (Fig. 6). Growing evidence has indicated that a cooperation between CD4+ and CD8+ T cells within tumor milieu is required for effective tumor regression.449 By comparing the transcriptomic profiles of CTLs with or without CD4+ T cell help, it has been demonstrated that CD4+ T cells can help CTLs in multiple cellular processes, including priming, clonal expansion, effector function, memory formation and response to cancer immunotherapies.616,617 Full CD8+ T cell priming is a two-step process in which CD4+ T cells and CD8+ T cells first encounter antigens separately on different types of cDCs (cDC2 and cDC1 respectively) that may occur at different location of the second lymphoid tissues.617–619 In the second priming step, CD4+ T cells and CD8+ T cells recognize their antigen on the same DCs (mainly XCR1+ resident cDC1s).620–622 CD4+ T cells enhance DC activation and their antigen-presenting capability via CD40/CD40L signaling to fully prime CTL response.623,624 Therefore, eliciting CD4+ T cell response or pre-stimulating DCs with CD40 agonist are essential strategies for effective anti-tumor vaccines.625,626
Fig. 6.
CD4+ T cells support CD8+ CTL response in anti-tumor immunity. Effective CD8+ CTL priming is a two-step process dependent on CD4+ T cell help which is bridged by XCR1+ resident cDC1s. CD4+ and CD8+ T cells are activated separately by different populations of DCs. Through CD40/CD40L signaling, activated CD4+ T cells enhance the expression of CD80/CD86 and CD70 on cDC1s, which interact with CD28 and CD27 on CD8+ T cells to promote their activation. CD4+ T cell-helped cDC1s also secrete high levels of type I interferon, IL-12 and IL-15 to promote CD8+ T cell survival and effector function. CD4+ T cells can directly promote CD8+ CTL response through IL-2 and IL-21. Consequently, CD4+ T cell-helped CD8+ T cells exhibit enhanced expansion, cytotoxic activity, migratory capacity, and expression of TNFR and key transcription factors, while downregulated IRs
CD4+ T cell help also promotes the clonal expansion and effector function of CTLs. Helped CTLs have upregulated expression of IL-2, IL-2R and IL-12R to support their survival, proliferation and effector differentiation.308,627 Helped CTLs exhibit enhanced cytotoxic activities, including increased production of IFN-γ, TNF, granzymes and Fas ligand, while downregulated IRs such as PD-1, Lag-3, Tim-3, and BTLA.301,628 On the contrary, helpless CTLs display dysfunctional and exhausted phenotypes.629,630 Furthermore, CD4+ T cells help CTL migratory capacity to enter tumor tissues by upregulating their CXCR4 and CX3CR1 expression, and promote CTL extravasation at tumor site by increasing MMPs expression.628 More importantly, CD4+ T cell help is required for generating long-term memory CD8+ T cells.302,304,631 CD4+ T cell help promotes IL-15 signaling for TCM maintenance, as well as IFN-γ and granzyme B production from TEM.632 In the absence of CD4+ T cell help, memory CTLs exhibit reduced CD27 expression and IL-2 production,633 and impaired recall response likely due to massive cell apoptosis, which are associated with increased expression of the death ligand TRAIL and decreased expression of anti-apoptotic protein Bcl-2.634,635 Mechanistically, CD4+ T cells enhance the expression of key TFs for effector and memory CTLs, such as T-bet, Eomes and Id3.617,636
Help signals
The help from CD4+ T cells mostly depends on costimulatory and cytokine signals (Fig. 6). In the second step of CTL priming, CD4+ T cell help triggers upregulation of CD80/CD86 and CD70 on cDC1s, which interact with CD28 and CD27 on CD8+ T cells, respectively.637,638 CD28 costimulation is important but not sufficient to generate fully functional CTL response.639,640 Costimulation through CD70/CD27 is critical for CD8+ CTL priming, clonal expansion and differentiation into both effector and memory CTLs.641,642 Besides, other TNFR family members such as 4-1BB, OX40, CD30, and GITR may also play critical roles in mediating CD4+ T cell help.643,644 CD4+ T cell-helped cDC1s have increased expression of type I interferon, IL-12 and IL-15 to promote effector CD8+ T cell survival, differentiation and function.635,645 CD4+ T cell help augments IL-2Rα expression on primed CD8+ T cells, together with IL-2 produced by CD4+ T cells, contributing to CTL clonal expansion, effector differentiation and function.308,646 In addition, CD4+ T cell-derived IL-21 is required for CX3CR1-expressing CD8+ T cell differentiation and cytolytic function,360,602,647 promotes TCF-1+ stem-like CD8+ T cell generation and maintenance and prevents effector CD8+ T cell exhaustion.648,649 Tfh cells expressing CXCL13 attract CXCR5+ CD8+ T cell migration in chronic infection and cancer.650,651 Collectively, costimulatory and cytokine signals from CD4+ T cells collaboratively and non-redundantly support CD8+ CTL response.
T cells in autoimmune diseases
A healthy immune system is a functional network important for host homeostasis by protecting from infection while preventing self-reactivity. Disruption of this delicate immune balance causes autoimmune diseases. To date, more than 80 types of autoimmune diseases have been described, affecting approximately 5–8% of the world population.652 The autoimmune diseases can be systemic, such as systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA), or organ specific, such as multiple sclerosis (MS) and Type 1 Diabetes (T1D). Although the mechanisms underlying autoimmune disorders are complicated and poorly understood, the roles of autoreactive T cells in driving the immunopathogenesis have been characterized in various autoimmune disorders (Table 3).
Table 3.
CD4+ T helper cell subsets in autoimmune diseases
| Th subset | Mediator | Pathogenesis | Autoimmune disease | Functions | Refs |
|---|---|---|---|---|---|
| Th1 | IFN-γ | promote | MS | activate pro-inflammatory M1-like microglia | 654,655,657,673,674 |
| Th17 |
IL-17A-F, IL-21, IL-22, IFN-γ, GM-CSF |
promote | MS | activate macrophages, astrocytes, epithelial and endothelial cells and oligodendrocytes | 102,500,668,669,671–674 |
| recruit neutrophils | 102,500,668 | ||||
| disrupt BBB | 667 | ||||
| support formation of TLOs | 663,675 | ||||
| promote pathogenic myeloid cells | 683 | ||||
|
IL-17, IL-1β, TNFα, GM-CSF |
promote | RA | induce tissue-destructive enzymes, pannus growth, osteoclastogenesis and angiogenesis | 690–693 | |
| enhance proliferation of fibroblast-like synoviocytes | 694 | ||||
| stimulate GM-CSF secretion from fibroblast-like synoviocytes and ILCs | 695 | ||||
|
IL-17A, IL-17F, IL-21, IL-22, IL-23 |
promote | SLE | stimulate keratinocytes, synoviocytes, fibroblasts, macrophages and neutrophils | 704 | |
| induce the NETosis | 705 | ||||
| Th1-like Th17 |
IFN-γ, IL-17, granzymes, GM-CSF, IL-22 |
promote | MS | produce inflammatory cytokines | 707–709 |
| cross BBB | 711,712 | ||||
| promotes the neuroinflammation | 711–713 | ||||
| Th22 |
IL-22, IFN-γ, TNF-α, IL-17 |
promote |
MS, SLE, RA, psoriasis, ITP, AIH, AITD, MG, SSc |
disrupt BBB | 667 |
| affect endothelial cells | 667 | ||||
| regulate astrocytes, oligodendrocytes, Treg cells | 716,727 | ||||
| contribute to bone destruction | 731,732 | ||||
| promote fibroblasts proliferation and inflammatory responses | 731,732 | ||||
| induce osteoclast formation | 731,733 | ||||
| Th9 | IL-9 | promote |
IBD, SLE, MS, SSc, UC, RA, psoriasis, IRP, thrombocytopenia |
suppress epithelial cell proliferation | 736 |
| disrupt mucosal barrier function | 93 | ||||
| promote Th17 cell migration and differentiation | 739,746 | ||||
| induce astrocytes response | 745,747 | ||||
| promote B cell proliferation and autoantibodies production | 749,750 | ||||
| enhance MMPs production by neutrophils | 738,754 | ||||
| prevent | Gastritis, MS | dampen the pathogenic activity of Th17 cells | 755 | ||
| interfere with IL-17 and Th17 cell polarization | 756 | ||||
| maintain Treg differentiation | 757 | ||||
| Tfh |
CD40L, IL-4, IL-21, CXCL13 |
promote |
MS, RA, SLE, MG, Sjögren’s syndrome, psoriasis, AD, autoimmune thyroid, hepatitis disease, IBD and T1D |
drive autoreactive B cell response and autoantibody development | 769–773,793,794 |
| promote the inflammatory Th17 responses | 778 | ||||
| induce pathogenic CD8+ T cell responses | 784,795 | ||||
| promote osteoclasts, fibroblast-like synoviocytes, keratinocytes and synovial macrophages | 797–801 | ||||
| counteract Treg cell suppressive activity | 802,803 | ||||
| help pathogenic epitope spreading | 817,819,820 | ||||
| Treg |
CTLA-4, Lag-3, TIGIT, CD73, CD39, IL-10, TGF-β, IL-35 |
prevent |
MS, asthma, T1D, MG, RA, SLE |
prevent Tconv overactivation differentiate into Th-like Treg cells to suppress Th cells |
832,833 |
| 835–843 | |||||
| IFN-γ | promote | T1D, MS, autoimmune hepatitis, Sjögren’s syndrome | pro-inflammatory Treg: IFN-γ+Foxp3+ Th1-like Treg cells | 853–859 | |
| IL-4, IL-13 | promote |
SSc, allergy, asthma, TAK, IOI |
pro-inflammatory Treg: Foxp3+ Th2-like Treg cells | 865–871 | |
| IL-17 | promote | RA, SLE, psoriasis, mucosal autoimmunity, glomerulonephritis | pro-inflammatory Treg: IL-17+Foxp3+ Th17-like Treg cells | 841,872–875 | |
| pro-inflammatory cytokines | promote |
Diabetes, MG, MS, RA, SLE |
instability of Treg lineage: exFoxp3 cells | 883–889 | |
| impaired immunosuppressive function | |||||
| CD8 | IFN-γ, TNF, granzyme B, perforin | promote |
T1D, MS, vitiligo, Crohn disease, SLE, vasculitis, IBD |
disrupt self-tissues by cytotoxic effector molecules | 919,922,923 |
| enhance ROS production from monocytes | 919 | ||||
| presence of progenitor autoreactive T cells | 931 |
Th1, Th17, and Th1-like Th17 cells: important inflammation mediators
As a major pro-inflammatory CD4+ T cell subset, Th1 cells play critical roles in promoting pathogenesis of autoimmune diseases. MS is a chronic autoimmune disease characterized by immune dysfunction and inflammation in the central nervous system (CNS) where the immune cell infiltration triggers demyelination, axonal damage, and neurodegeneration.653 Experimental autoimmune encephalomyelitis (EAE) is the most used experimental model for MS. Th1 cells are found to be the most frequent CD4+ Th cells in the CNS of EAE and large amount of IFN-γ is detected in MS patients.654,655 Adoptive transfer of Th1 cells is sufficient to induce EAE manifestation in mouse models.656 The neuropathological roles of Th1 cells in the CNS are associated with microglia, the CNS-resident macrophages. Th1-associated factors could activate a pro-inflammatory M1-like microglia differentiation,657 and promote inflammation in EAE.657 However, later studies using IL-12p35 subunit, IL-12Rβ2 chain or IFN-γ deficient mice demonstrated that Th1 cells are not required in the pathogenesis of EAE and MS.658,659 Instead, loss of IL-23p19 subunit or IL-23R chain result in resistance to EAE.660 With the discovery of shared subunits between IL-23/IL-23R and IL-12/IL-12R, Th17 cells have been uncovered playing critical roles in autoimmune diseases.661–663 Th17 cells produce a variety of pro-inflammatory cytokines, such as IL-17A-F, IL-21 and IL-22, and pathogenic Th17 cells induced by IL-6, IL-1β, and IL-23 produce high levels of IFN-γ and GM-CSF,115 which can further act on several other cell types to amplify the inflammatory responses.
Th17 cells and IL-17 are highly involved in the pathogenesis of MS.664 In MS patients, IL-17-producing CD4+ T cells are largely found in the peripheral blood and cerebrospinal fluid.665,666 IL-17A infuses into the CNS and contributes to the disruption of blood–brain barrier (BBB).667 Pro-inflammatory cytokines produced by Th17 cells act on CNS-resident macrophages to enhance their activation, inflammatory cytokines and chemokines production, antigen-presenting activity, and recruit neutrophils into the inflammatory sites, thus promoting the axonal damage and neuroinflammation in EAE.102,500,668 Th17 cells, in cooperation with Th1 cells, affect astrocytes function by upregulation of inflammatory cytokines and chemokines while downregulation of neurotrophic factors.669 Therefore, inhibition of IL-17 signaling in astrocytes has been shown to ameliorate the EAE.670 IL-17 signaling also alters the expression of adhesion molecules on endothelial cells and actin cytoskeleton on epithelial barriers.671,672 In addition, Th17 cell- or IL-17-mediated pro-inflammatory responses inhibit the survival and maturation of oligodendrocytes whose apoptosis and dysfunction are highly associated with the demyelination and neurodegeneration in MS.673,674 Like TLSs in TME, tertiary lymphoid organs (TLOs) are observed in the chronically inflamed tissues in autoimmune diseases to sustain the local immune activation.663,675 IL-17 is required for the formation of TLOs by inducing CXCL13 and CCL19 production to recruit lymphocytes into TLOs.676,677 Furthermore, Th17 cell-derived GM-CSF has been identified as a key factor driving the inflammation during EAE development.678,679 It has been discovered that some CNS-infiltrated Th cells were IL-17A+GM-CSF+,680 and GM-CSF-producing T cells are increased in the peripheral blood and brain lesion.681,682 GM-CSF in turn enhances pathologic Th17 generation and maintenance,680 and acts on a variety of pathogenic myeloid cell types including inflammatory monocytes, monocyte-derived dendritic cells and microglia to promote EAE pathogenesis.683
RA is an autoimmune disorder characterized by the chronic inflammation in the synovial membrane. In autoimmune arthritis, Th17 cells are the dominant initiators and executors of inflammation. Increased level of IL-17 has been found in serum, synovial fluid and synovial tissue of patients with rheumatoid arthritis.684,685 Th17 activity and IL-17 correlate with the disease severity of clinical symptoms.686,687 Self-reactive T cells become activated and differentiated into CCR6+ Th17 cells in the periphery. Response to CCL20 expressed by synoviocytes, CCR6+ Th17 cells migrate to the joints to initiate inflammation by producing large amount of IL-17, IL-1β and TNFα.688,689 IL-17 contributes to the joint destruction by inducing tissue-destructive enzymes, pannus growth, osteoclastogenesis and angiogenesis.690–693 IL-17 enhances the proliferation of fibroblast-like synoviocytes through mTOR and MAPK p38 signaling.694 In addition, GM-CSF, produced directly by Th17 cells and Th17 cell-stimulated fibroblast-like synoviocytes and ILCs, is abundant in RA synovium and mediates chronic joint inflammation.695
SLE is a chronic and heterogeneous autoimmune disease featured by accumulation of autoantibodies and immune dysfunctions with systemic inflammation and tissue destruction in multiple organs such as skin, joint, kidney, brain, heart and blood.696,697 Emerging evidence has demonstrated that Th17 cells and IL-17 play essential roles in SLE pathogenesis.698,699 IL-17-producing T cells are increased in the peripheral blood and inflamed organs of SLE patients,700,701 and the IL-17 level positively correlates with the disease severity.702,703 IL-17A stimulates inflammatory cytokines and chemokines production by keratinocytes, synoviocytes, fibroblasts, macrophages and neutrophils.704 IL-17 also induces neutrophil extracellular trap formation (NETosis) which has been found promoting the pathogenesis of SLE.705 In addition, IL-23, a key cytokine for Th17 differentiation, is observed elevated in SLE patients and correlates with severe renal disease.703,706
Intriguingly, Th17 cells are highly plastic and can transdifferentiate into pathogenic Th1-like Th17 cells which are defined by producing high levels of both IFN-γ and IL-17, and co-expressing chemokine receptors CXCR3 and CCR6, as well as TFs T-bet and RORγt.707 Th1-like Th17 cells display stronger pathogenicity than Th17 cells, which may relate to the production of inflammatory cytokines GM-CSF and IL-22 and chemokine receptors CCR4, CCR6 and CXCR3.708,709 In inflammatory arthritis, both Th17 and Th1 lineage-specific TFs are highly expressed in the inflamed joints of patients. The cytokine milieu within the joints, including high levels of IL-12 but low IL-23 and TGF-β, converts Th17 cells into Th1-like cells. The direct evidence supporting the Th17 origin of Th1 cells results from the shared TCR clonality between Th1-like cells and Th17 cells.710 Th1-like Th17 cells are capable of crossing BBB and accumulate in the CNS where they promote the neuroinflammation in EAE mice and MS patients.711–713 Moreover, a CCR6+CXCR6+ cytotoxic Th17 population with expression of granzymes, IFN-γ and GM-CSF is identified to promote EAE pathology.714 Interestingly, a stem-like Th17 population is discovered by combined scRNA-seq and TCR-sequencing analysis and characterized by TCF-1 and SLAMF6 expression.715 Such Th17 progenitor cells are non-pathogenic but can give rise to GM-CSF+ and IFN-γ+ pathogenic Th17 populations under induction of IL-23, which greatly contributes to autoimmunity.715
Th22: inflammation promotors
Th22 cells and IL-22 play critical roles in promoting autoimmune diseases. The proportion of Th22 cells and IL-22 level have been found increased in the serum and/or local tissues in numerous autoimmune disorders, including MS,716 SLE,717 RA,718 psoriasis,719 ITP,720 autoimmune hepatitis (AIH),721 autoimmune thyroid diseases (AITD),722 myasthenia gravis (MG),723 and systemic sclerosis (SSc).724 The IL-22 level is dynamically changed along with the disease progression.725 High CCR6 expression facilitates Th22 cell migration into the CNS.726 IL-22R expression was upregulated in the brain tissues of MS patients and IL-22 synergized with IL-17 to disrupt BBB tight junctions by affecting endothelial cells.667 IL-22 also regulates the survival and function of astrocytes and oligodendrocytes, and inhibits Foxp3 expression in Treg cells, therefore promotes the pathogenesis of MS.716,727 In SLE, Th22 cells may represent a better prognostic marker of tissue involvement than Th17 cells.728 CCR6+ Th22 cells and IL-22 are increased in SLE patients with lupus skin diseases and significantly correlate with the SLE disease activity index (SLEDAI).729,730 The IL-22 level is also increased in the serum and kidney in patients with lupus nephritis, and treatment with anti-IL-22 monoclonal antibody could markedly reduce renal injury and inflammatory cells infiltration.717 In RA, Th22 cells positively correlate with disease activity score.718,729 High level of IL-22 in synovial tissue contributes to bone destruction and promotes fibroblasts proliferation and inflammatory responses.731,732 IL-22 also induces osteoclast formation through MAPK p38/NF-κB and JAK2/STAT3 signaling.731,733 Given the important function of Th22/IL-22 in promoting pathogenesis in many autoimmune diseases, targeting Th22/IL-22 has been considered as great therapeutic potentials.734
Th9: dual-function in autoimmune diseases
Th9 cells and IL-9 have been implicated to play pathological roles in autoimmune diseases.91 IL-9, Th9 cells and Th9 cell-associated molecular features (PU.1, IL-4, TGF-β, etc.) have been found elevated in patients with various autoimmune diseases in ulcerative colitis (UC),735 inflammatory bowel disease (IBD),736 SLE,737 RA,738 psoriasis,739 immune-related pancytopenia (IRP),740 and thrombocytopenia,741 which greatly correlates with disease severity. In IBD, Th9 cells contribute to the pathogenesis through producing IL-9 which suppresses epithelial cell proliferation and disrupts the mucosal barrier function.93,736 In MS/EAE, Th9 cells and IL-9 function in initiating disease development and promoting inflammation in CNS. Adoptive transfer of myelin oligodendrocyte glycoprotein (MOG)-specific Th9 cells into Rag1−/− mice sufficiently induces EAE more severe than transferring Th1 cells.742,743 IL-9 deficiency or neutralization exhibit attenuated EAE progression with reduced infiltration of Th17 cells and pro-inflammatory macrophages in the CNS, as well as decreased IL-17 and IFN-γ levels.744,745 Strikingly, cooperative functions of Th9 and Th17 cells have been revealed during autoimmune disorders. Th17 cells can produce IL-9 which acts as the pathogenic mediator in MS and psoriasis in animal models.739,746 In turn, IL-9 induces astrocytes to produce CCL20 which promotes Th17 cell migration into CNS and aggravates EAE development.745,747 Furthermore, the frequency of Th9 cells and serum IL-9 are positively associated with SLE disease severity.748 In murine lupus models, IL-9 is associated with increased anti-double-stranded DNA (dsDNA) antibodies via promoting B cell proliferation and autoantibody production.749,750 The enriched Th9 cell response in SLE is associated with NO751 which is elevated in SLE patients and enhances Th9 cell differentiation through TGF-β and IL-4 signaling752 and mTOR-HIF1α pathway.753 In RA patients, IL-9 and IL-9R are highly expressed in the synovial tissues, associated with synovial inflammatory infiltrates and the degree of ectopic lymphoid structures.738 Mechanistically, synovial IL-9 promotes the survival and MMPs production of neutrophils and facilitates Th17 cell differentiation.754
On the other hand, due to the complex immune microenvironment and regulatory mechanisms of autoimmune diseases, protective roles of Th9 cells are also observed. For instance, IL-9 dampens the pathogenic activity of Th17 cells in autoimmune gastritis.755 IL-9 inversely correlates with the inflammation and neurodegeneration in MS patients as high level of IL-9 interferes with IL-17 production and Th17 cell polarization.756 IL-9R deficient mice have increased Th1 and Th17 cell development but impaired Treg cell activity, which is attributed to the important role of IL-9 in modulating Th17 and Treg cell differentiation.757 Collectively, IL-9 and Th9 cells have both deleterious and protective roles in autoimmune diseases, and future comprehensive studies are required to fully delineate their functions.748
Tfh: enhance autoreactive B cell and CD8+ T cell responses
Tfh cells are strongly associated with a wide range of autoimmune diseases in both autoantibody-dependent and -independent conditions. The first evidence of dysfunctional Tfh cells promoting autoimmunity comes from a study in 2005, in which Vinuesa et al. demonstrated that Roquin gene mutation caused excessive Tfh cell differentiation and systemic autoimmunity in mice.758 Deficiency of SAP, an adapter protein required for Tfh cell–B cell interactions,759 ameliorates the autoimmune phenotype with reduced autoantibody and disease severity.760 Increased frequencies of circulating Tfh cells are observed in majority of autoimmune disorders, including MS, RA, SLE, MG, Sjögren’s syndrome, psoriasis, atopic dermatitis (AD), autoimmune thyroid and hepatitis disease, IBD, and T1D.761–763 SLE is a well-known autoantibody-mediated autoimmune disease.761,764 Activated Tfh cells, aberrant GC responses and high level of autoantibodies are frequently found in SLE murine models765,766 and in lupus nephritis patients.767,768 The autoreactive B cells in SLE patients are typically somatically mutated and the anti-dsDNA antibodies have experienced somatic hypermutation and affinity maturation, indicating that they have been “helped” by T/Tfh cells.769 Similarly, the pathological progression in RA is strongly associated with autoantibodies which are mainly Tfh cell-helped high-affinity IgG antibodies.770,771 Tfh cells are expanded in patients with active RA, which positively correlates with autoantibody titers and disease severity.772,773 In RA joints, CXCL13-expressing Tfh cells co-localize with B cells and provide their help, which further promotes ectopic lymphoid structure formation and RA pathogenesis.774,775 Hence, the decreased percentage of Tfh cells has been used as an indicative biomarker for effectiveness of autoimmune disease treatments.776,777 In mouse EAE models, CXCR5+PD1+ Tfh cells are substantially infiltrated in the CNS tissue and promote the inflammatory B cell and Th17 cell responses, contributing to the disease pathogenesis.778 Furthermore, activated-memory circulating Tfh cells (CCR7+ICOS+) are increased in patients with relapsing MS, positively correlate with the levels of autoantibodies and disease severity, but are decreased after therapeutic treatment.779 Of note, while the pathogenic autoantibodies are predominantly derived from GC response and helped by GC-Tfh cells,762,780 Tfh cells can also support extrafollicular responses and autoantibodies production.781,782 T1D is an autoantibody less-dependent autoimmune disease in which overexpression of Tfh cell-related genes such as CXCR5, ICOS, PD-1, Bcl-6, and IL-21 are also observed.783,784 T1D can be induced by transferring Tfh cells in a mouse model.783Tfh cells positively correlate with the blood glucose levels and multiple autoantibodies in T1D patients.785 The frequency of activated autoantigen-specific Tfh cells (CXCR5+PD-1+ICOS+) is increased in both patients with recently diagnosed T1D or at risk of T1D.786,787
The pathogenic activity of Tfh cells largely depends on the signature cytokine IL-21 which promotes autoimmunity through helping B cells and driving effector function of CD8+ T cells as well as other cell types.788,789 IL-21 polymorphisms and overexpression are highly associated with autoantibodies, disease pathogenesis and clinical activity in many autoimmune disorders.788,790–792 IL-21 signaling strongly drives GC response, B cell activation, plasma cell differentiation and memory B cell formation, somatic hypermutation, and antibody class switching.793,794 In addition, IL-21R is highly expressed in CD8+ T cells and IL-21 signaling induces pathogenic CD8+ T cell responses. In T1D where the destruction of pancreatic β cells is primarily mediated by CD8+ T cells, IL-21-producing Tfh cells are increased significantly784 and IL-21R expression is elevated in CD8+ T cells.795 While IL-21 overexpression drives T1D development,795 IL-21R deficiency inhibits T1D mellitus.796 The functions of autoreactive CD8+ T cell responses in autoimmunity will be discussed in later chapter. Moreover, IL-21 can promote inflammation and pathogenesis by acting on other cells, such as osteoclasts,797 fibroblast-like synoviocytes,798,799 keratinocytes800 and synovial macrophages.801 In addition, Tfh cells counteract the suppressive activity of Treg cells in autoimmune diseases through IL-21.802,803 Therefore, inhibition of Tfh cells and IL-21 signaling offers effective therapeutic strategies in autoimmune diseases.804–806 For example, treatment with steroids, immunosuppressive drugs or low-dose of IL-2, a potent inhibitor of Tfh cell differentiation,149 could significantly reduce the number of activated Tfh cells and result in improved clinical outcomes.807–809
Notably, many autoimmune diseases are likely triggered by infections due to pathogenic antigen mimics.810,811 For example, enteroviral infection has a strong association with T1D812,813; exposure to Aggregatibacter actinomycetemcomitans triggers the autoimmunity in RA814; EBV infection has a clear link with MS development815,816; autoantibodies in SLE are likely generated from response to commensal and/or environmental microbes817; patients infected with SARS-CoV-2 exhibit markedly increased autoantibodies.818 The underlying mechanisms are highly involved in Tfh cell-helped epitope spreading during infections. Specifically, self-reactive T cells cross-recognize microbial antigens and provide help to B cells bearing different specificities (bystander autoimmune B cells).817,819 For instance, influenza virus haemagglutinin-specific Tfh cells can help self-antigen MOG-specific B cells to produce autoantibodies when those B cells cocapture haemagglutinin and MOG.820 Collectively, Tfh cells potently drive the pathogenesis of autoimmune diseases through enhancing autoreactive B cell and CD8+ T cell responses.
Treg cells: critical autoimmune protectors
Autoimmune diseases are characterized as a failure of self-tolerance. As one of the most important T cell populations in maintaining immunological self-tolerance and homeostasis, Treg cells play indispensable roles in autoimmunity.821,822 Mutations in Foxp3 gene cause immunodysregulation polyendocrinopathy enteropathy X-linked syndrome (IPEX) which is a rare chromosome X-linked immunodeficiency syndrome with severe autoimmune disorders.823,824 Furthermore, mutations of Treg cell-related signature genes, such as CD25,825 CTLA-4,826,827 LRBA,828 and AIRE,829,830 result in Treg cell abnormality and severe autoimmune disorders. Depletion of Foxp3+ Treg cells indeed leads to severe autoimmunity and immunopathology which can be rescued by reconstituting Treg cells.831 By sensing IL-2 produced by autoreactive Tconv cells, Treg cells co-localize with Tconv cells to prevent their overactivation.832,833 Treg cells employ a variety of suppressive molecules for inhibitory functions, such as surface receptors CTLA-4, Lag-3, TIGIT, CD73, and CD39, and inhibitory cytokines IL-10, TGF-β, and IL-35.822,834 In addition, Treg cells are able to adapt to the environment stimuli and mirror to corresponding effector Th cells under inflammatory conditions.835 Treg cells can gain expression of signature TFs and chemokine receptors of Th1,836,837 Th2,838,839 Th17,840,841 and Tfh (known as T follicular regulatory (Tfr)) cells.842,843 By responding to different stimuli, these Th-like Treg cells migrate into the same inflammatory sites with Th effector cells, and exert stronger abilities to suppress corresponding Th cell responses.835 The change of Treg cell numbers in different autoimmune diseases has been largely studies, however, the results are strikingly inconsistent.834,844 The frequency of Treg cells seems decreased in EAE845 and asthma,846 but unaffected in T1D847 and MG.848 Nevertheless, Treg cell numbers are found either decreased,849,850 increased845,851 or unchanged in RA and SLE.845,852 Despite of inconsistence in cell number, it is well-acknowledged that the functions of Treg cells in autoimmune milieu are compromised.844
Emerging evidence has suggested that the plasticity and instability of Treg cells contribute to their dysfunction. While the Th-like Treg cells exhibit advantages for controlling host homeostasis, aberrant plasticity can affect Treg cell-mediated immunosuppression and exacerbate autoimmune diseases. It has been shown that the frequency of IFN-γ+Foxp3+ Th1-like Treg cells are increased in various autoimmune diseases, such as T1D,853 MS,854,855 autoimmune hepatitis,856 and Sjögren’s syndrome.857 Th1-like Treg cells accumulate at inflamed sites but fail to suppress effector T cell response and control the disease progression.858,859 Inflammatory cytokines TNF, IL-6, and IL-12,860–862 and PI3K-Akt-FoxO signaling pathway have been suggested to be involved in Treg cell conversion.855,863,864 Th2-like Treg cells are increased in patients with SSc,865 allergy,866 asthma,867,868 takayasu’s arteritis (TAK)869 and idiopathic orbital inflammation (IOI),870 and IL-33 derived from dermal fibroblasts contributes to Th2-like Treg transdifferentiation.871 In addition, IL-17+Foxp3+ Th17-like Treg cells are largely identified in RA,872 SLE,873 psoriasis,874 and mucosal autoimmunity,841,875 playing critical roles in disease pathogenesis. The conversion of Treg cells into Th17 cells is driven by cytokines IL-1β, IL-6, IL-4, and IL-23,862,872,876,877 Toll-like receptor 2 (TLR2) stimulation,878 pathogenic infection879 and IRF4.880 In contrast, IL-33,870 SOCS1,881 and IDO882 have been suggested to prevent Treg cell plasticity and restore their suppressive function.
Furthermore, under inflammatory or pathologic settings, instability of Treg lineage with unstable Foxp3 expression and impaired immunosuppressive function is observed.883,884 Decreased Foxp3 expression is found in Treg cells isolated from autoimmune diabetes,885 MG,886,887 MS,888 and SLE.889 Treg cells loss of Foxp3 expression (exFoxp3) exhibit activated-memory T cell phenotype and acquire effector function, such as producing pro-inflammatory cytokines and inducing autoimmune pathogenesis.890,891 Under arthritic conditions, Treg cells lose Foxp3 expression and transdifferentiate into Th17 cells (exFoxp3 Th17), which is driven by synovial fibroblast-derived IL-6. These exFoxp3 Th17 cells are more potent osteoclastogenic Th17 cells, contributing to the pathogenesis of RA.872 The mechanisms underlying Treg cell stability have been greatly associated with the expression of master regulator Foxp3. Impairment of TGF-β/IL-2 signaling leads to diminished Foxp3 expression, Treg cell function and autoimmune manifestations.885,892–894 Furthermore, the epigenic regulations of Foxp3 have been suggested playing both positive and negative roles in Treg stability.895 Current consensus suggests that Foxp3 acetylation896,897 and O-linked N-acetylglucosamine (O-GlcNAc)898 stabilize its expression and strengthen Treg stability and suppressive function, whereas methylation,899,900 phosphorylation901,902 and ubiquitination903 of Foxp3 induce instability of Treg cells. The CNSs in Foxp3 locus are critical for Foxp3 transcription and are associated with autoimmune diseases.904–906 Methylation of Treg-specific demethylation region (TSDR)—a highly conserved CpG motif within CNS2—destabilizes Foxp3 expression and disrupts the suppressive activity of Treg cells.899,907 Apart from epigenetic regulation, Treg cell stability/suppressive function are profoundly controlled at transcriptional levels. Deficiency of TFs Helios,908 Ikzf4,909 RelA,910 Smad2/Smad3,893 AP-1911 and Id3912 significantly affects the stability of Foxp3 expression. In contrast, TFs BATF3,913 IRF4,913 E47,912 and Spi-B912 repress Foxp3 expression and Treg cell induction.
Autoreactive CD8+ T cells: new players in autoimmunity
Tradition views hold that CD8+ T cells mainly participate in protection against viral infections and tumors. However, increasing evidence from recent studies implicates that excessive CD8+ T cell functionality causes self-tissue damages and autoimmune disorders.914,915 In human, autoimmune disease susceptibility is highly associated with HLA class I (human MHC I) polymorphisms, prone to autoantigen presentation to CD8+ T cells.916,917 Autoreactive CD8+ T cells have been implicated in the pathogenesis of multiple autoimmune diseases, including T1D,918 MS,919 Crohn disease,920 and vitiligo.921 Pathogenic CD8+ T cells express high levels of cytotoxic effector molecules such as IFN-γ, TNF, granzyme B and perforin.919,922,923 In the nonobese diabetic (NOD) mouse model of T1D, by 10–15 weeks of age, the pancreata exhibit severe insulitis and are largely infiltrated with CD8+ T cells recognizing NRP-V7, a peptide from the diabetes antigen IGRP. The increased frequency of NRP-V7-reactive CD8+ T cells coincides with the time of glucose intolerance, suggesting that the progression of pancreatic islet inflammation is driven by self-reactive CD8+ T cell populations.924,925 In MS, autoreactive CD8+ T cells are expanded and enriched in the CNS of patients with relapsing–remitting disease.926 In EAE models, myelin basic protein (MBP)-specific CD8+ T cells are recruited to the CNS and enhance ROS production from monocytes in the brain lesion.919 In addition, CD8+ T cells contribute to autoimmune arthritis.927 The number of CD8+ T cells is increased in active RA patients but decreased in patients in remission.928 The elevated pro-inflammatory cytokine production by CD8+ T cells positively correlates with 28-joint disease activity score (DAS28) in autoimmune arthritis.928
Recent work has revealed a great heterogeneity of autoreactive CD8+ T cells. Pathogenic CD8+ T cells in T1D, MS/EAE and vitiligo contexts are predominant effector, effector memory or resident memory cells that initiate and promote disease progression.919,922,929,930 Even though autoreactive CD8+ T cells maintain effector functions, evidence also suggests that they display exhausted features. Autoimmune CD8+ T cells in MS and T1D have upregulated expression of IRs PD-1, Lag-3, and Tim-3.919,931 The exact function of exhausted CD8+ T cells in autoimmunity is not fully understood. However, some evidence has suggested a protective role of this population since T cell exhaustion represents a hyporesponsive phenotype. For instance, exhausted CD8+ T cells in T1D and SLE patients are associated with a slow disease progression and improved prognosis.932,933 Intriguingly, TCF-1hiTOXhi stem-like progenitor CD8+ T cells have been identified in autoimmune diseases, which sustain the autoreactive T cell population.931 In T1D, this autoimmune progenitor CD8+ T cells are located at the pancreatic dLNs where they self-renew and give rise to autoimmune effector CD8+ T cells.934 Compared to the short-lived autoimmune effector cells, stem-like progenitors can induce T1D upon adoptive transfer of as few as 20 cells into recipient mice.934 Notably, the fate and functionality of self-reactive CD8+ T cells require TOX-dependent transcriptional and epigenetic reprogramming.935 Taken together, CD8+ T cells also function as autoimmune mediators, and further studies are required to better understand their cell heterogeneity, functional states and regulatory mechanisms in autoimmune diseases for developing effective therapeutic strategies.
γδ T cells
γδ T cells are a unique and rare T cell population that are mainly enriched in peripheral mucosal barriers, such as skin, lung and gut tissues, playing critical roles in both maintaining physiological homeostasis and mediating immune responses in disease conditions. During intrathymic T cell development, DN3 cells rearrange the TCR components and those expressing TCR γ and δ chains develop into γδ T lineage (known as γδ-selection).7 It has been suggested that the γδ T cell fate relies on strong and prolonged TCR signal (instructive model),7 Id3 regulation,936 Sonic hedgehog (Shh) signaling,937 CD27 costimulation, cytokine IL-7, lymphotoxin (LT) signal from αβ thymocytes (known as trans-conditioning),938 and Notch signaling.7 Nevertheless, the requirement of Notch signal for γδ T cell differentiation is controversial and varies between mouse and human. Compared to αβ T cells, γδ-lineage commitment is less Notch dependent in mice938; however, γδ T cell development in human is highly dependent on NOTCH signaling.939 TCR signals through γδ-TCR complex not only promote the survival and maturation of pre-established γδ T cells,7 but also play an instructive role in γδ T-cell lineage commitment.940 In addition, more studies have revealed that γδ T cell development is orchestrated at transcriptional,7 epigenetic941 and metabolic levels.942
γδ T cells in tissue surveillance and infection
Unlike αβ T cells that acquire effector function in the periphery, γδ T cells develop into effector cells during the development in the thymus. This early effector-programming of γδ T cells allows them to respond rapidly to pathogenic infections, inflammation, and tissue damage, endowing them with innate-cell like features. To date, two major subsets of effector γδ T cells are identified: IFN-γ producing Tγδ1 and IL-17 producing Tγδ17 cells, expressing key TFs T-bet and RORγt, respectively.938 Besides, γδ T effector cells can be distinguished by surface markers: Tγδ1 cells express CD27, CD122, NK1.1, and high level of CD45RB whereas Tγδ17 cells lack of the former three molecules but express CCR6, scavenger receptor SCART2 and low level of CD45RB.943,944 Distinct γδ T effector subpopulations have preferential Vγ usage and peripheral locations, such as IFN-γ producing cells are Vγ1+Vδ6.3+ (liver and spleen), Vγ5+ (skin), Vγ7+ (intestine), and IL-17 producing cells are mainly Vγ6+ (tongue, dermis, uterus, testis, adipose tissue, and brain) and Vγ4+ (lung, dermis, and lymph nodes).945,946 γδ T cell effector differentiation is regulated by transcriptional networks. In addition to T-bet and RORγt, TCF-1, LEF-1, Eomes, and Id3 are critical for IFN-γ producing γδ T cells, while c-Maf, Sox4, Sox13, HEB, Blk, and RelB are enriched for IL-17 producers.947,948 Of note, TCF-1 represses c-Maf/RORγt to limit Tγδ17 cells whereas c-Maf represses Tγδ1 fate by antagonizing TCF-1/LEF-1, indicating that an antagonism between c-Maf and TCF-1 controls the balance of these two γδ T effector subsets.943 Furthermore, γδ TCR signal strength impacts the effector fate, which TCR-Egr-Id3 pathway is required for IFN-γ production while TCR-E protein-TCF-1 axis supports IL-17-producing γδ T cell development.936,949 Thymic development of Tγδ1 cells requires Skint-1 signal from epithelial cells,950 while Tγδ17 cells can be differentiated in the periphery under IL-6, TGF-β, IL-1β, IL-18, and IL-23.951,952 With the advances in single-cell analysis, more insightful discoveries about the heterogeneity and developmental trajectory of tissue-specific γδ T cells have been further unveiled.953
Given the broad colonization in peripheral tissues, γδ T cells play crucial roles in tissue homeostasis and surveillance. γδ T cells sense “tissue status” by interaction with butyrophilins (BTNs) and BTN-like (BTNL) molecules which are members of the immunoglobulin superfamily.954 For example, BTNL1/BTNL6 heterodimers expressed on intestinal epithelial cells shape intestinal Vγ7+ T cells and BTNL3/BTNL8 heterodimers induce responses by colonic Vγ4+ T cells.955 γδ T cells promote wound healing and tissue repair in epithelial and mucosal barriers by producing functional factors and modulating other cells.945 In the skin, Vγ5+ dendritic epidermal T cells (DETCs) promote keratinocyte proliferation and hyaluronan production by producing keratinocyte growth factor (KGF) and insulin growth factor 1 (IGF1).956,957 Vγ7+ γδ T cells in intestines are highly associated with intestinal epithelial homeostasis through KGF1958 and IL-22.959 Gingival Vγ6+ T cells contribute to oral pathophysiology by producing IL-17 and amphiregulin.960,961 Notably, the function of Tγδ17 cells in tissue physiology can be paradoxical dependent on specific context. IL-17 producing Vγ4+ and Vγ6+ γδ T cells are found both contributing to the steady-state skin physiology962 as well as predominantly mediating the early inflammatory responses in skin diseases.963 Also, the roles of pulmonary γδ T cells can be beneficial, deleterious or dispensable in lung physiology and pathophysiology.945 Moreover, γδ T cells participate in non-barrier tissue surveillance. Vγ6+ Tγδ17 cells promote bone regeneration by stimulating the proliferation and osteoblast differentiation of mesenchymal progenitor cells.964 In the adipose tissue, γδ T cells, mainly Vγ6+ Tγδ17 subset, modulate Treg cells and adipocytes through IL-17 and TNF to promote thermogenesis.965,966 Vγ6+ Tγδ17 cells also contribute to steady-state neurophysiology967 and initiation of neuroinflammation in EAE and brain injury.676,963
γδ T cells display both innate and adaptive immune cell characteristics by expressing gene rearranged γδ TCR with limited repertoire.968 γδ T cells can recognize unprocessed peptides and various non-peptide antigens, such as lipids and the phosphoantigens without MHC restriction.969 γδ T cells constitute the first line of host defense against pathogenic infections. During the skin infection with S. aureus, IL-17 producing Vγ4+ T cells and IFN-γ/TNF producing Vγ5+ T cells enhance neutrophil recruitment and bacterial clearance.970,971 Systemic S. aureus infection led to accumulation of IL-17A+ γδ T cells in the kidney for effective infection control.972 In the infected intestinal tract, Vγ7+ γδ T cells directly kill infected cells by secreting antimicrobial peptides and cytotoxic molecules.973 In Mtb infected lung tissue, Vγ4+ γδ T cells secrete CXCL2 and TNF to promote neutrophil recruitment and Vγ4+ and Vγ6+ Tγδ17 cells contribute to granuloma formation.974,975 Moreover, γδ T cells exhibit a potent antiviral activity against a variety of viruses.976 Upon recognition of viral antigens, γδ T cells become activated and express increased pro-inflammatory cytokines (IFN-γ and TNF-α) and cytotoxic molecules (perforin and granzymes) for pathogen clearance.976 During SARS-CoV2 infection, the frequency of γδ T cells is reduced in the circulation but increased in the airway tissues.976 Both circulating and tissue-colonized γδ T cells have upregulated activation phenotypes (CD25, CD69, PD-1, IFN-γ and IL-18), suggesting an antiviral activity.976,977 Notably, given their major locations of mucosal tissues, γδ T cells have a close interaction with microbiota, which shape γδ T cell development and function in both homeostatic and pathological conditions. The crosstalk between γδ T cells and the microbiota has been reviewed previously.978 Despite the innate-like signature, γδ T cells have been recently found to have memory phenotypes that they can respond rapidly with enhanced cytokine production and pathogen clearance upon the secondary infection.979
γδ T cells in tumor immunity
The unique feature of γδ T cells in recognizing antigens without MHC restriction provides a promising application in cancer immunotherapy. Human γδ T cell subtypes are usually defined by δ chain, that Vδ1-3 are the most used gene segments and used for γδ T cell type classification.980 Vδ1 and Vδ3 T cells are less frequent γδ T cell populations and share some similarities in peripheral tissue distribution, antigen recognition and antiviral function.981,982 Vδ2 T cells—frequently paired between TCR Vδ2 and Vγ9 chains (Vγ9Vδ2 T cells)—constitute a predominant γδ T cell population in human peripheral blood after infection and malignancy.983 The phosphoantigens recognized by Vγ9Vδ2 T cells are natural products from microorganisms or generated by mammalian cells through mevalonate pathway.981 The aberrant mevalonate pathway in tumor cells leads to accumulation of phosphoantigens and Vγ9Vδ2 T cell activation and expansion in TME.984 Vγ9Vδ2 T cells recognize phosphoantigens bound by BTN3A1/BTN2A1 heterodimers.985 Therefore, phosphoantigen stimulation and agonism by targeting BTN3A1 have been shown to promote Vγ9Vδ2 T cell activation and anti-tumor activity.986,987 Non-Vγ9Vδ2 T cells, including Vδ1 and Vδ3 T cells, recognize glycolipids presented by CD1d.988 Besides, human γδ T cells express a range of natural killer receptors (NKRs), such as NKG2D, DNAM-1, NKp30, NKp44, and NKp46, which promote their cytotoxic effector functions upon recognition of cognate ligands on tumor cells.982 Moreover, γδ T cells express various TLRs and can be activated by TLR agonists to enhance cytotoxic functions.989
The function of γδ T cells in tumor immunity is versatile with both anti- and pro-tumor activities (Fig. 7). Most current evidence indicates that the presence of γδ T cells are associated with favorable outcomes in patients in CRC, breast, gastric, liver and bladder cancer, HNSCC, NSCLC and Merkel cell carcinoma.981 However, unfavorable prognosis of γδ T cells is also reported in CRC,990 gallbladder cancer,991 breast cancer,992 and acute myeloid leukemia (AML).993 Although different analysis techniques among studies could affect the results, at least, the γδ T types are likely associated with the prognostic prediction. Overall, IL-17+ γδ T cells tend to have a deleterious outcome whereas IFN-γ+ γδ T cells and NKR-expressing γδ T cells have improved outcomes.991,994 The anti-tumor activity of γδ T cells relies on multiple mechanisms995: (1) directly kill tumor cells by expression of perforin, granzymes and apoptotic receptors TRAIL and FasL;996 (2) γδ T cells upregulate CD16 (Fcγ receptor III) expression to enhance the ADCC effects of therapeutic antibodies on tumor cells;997,998 (3) γδ T cells have been shown to function as APCs that upon activation upregulate expression of MHC and costimulatory molecules and present antigens to CD4+ and CD8+ αβ T cells;999–1001 (4) γδ T cells orchestrate anti-tumor immunity through interplay with other immune cells.1002 IFN-γ production by γδ T cells exhibit an overall anti-tumor activity by increasing MHC I expression by tumor cells.1003 Vγ9Vδ2 T cells and DCs can reciprocally activate each other through both surface molecules (OX40 and BTN3A) and soluble factors (IFN-α and TNF-α).1004,1005 γδ T cells enhance NK cell activation and anti-tumor cytotoxicity via ICOS/ICOSL and 4-1BBL/4-1BB interaction.1006,1007 γδ T cells participate in humoral immunity by promoting B cell maturation, antibody production and class switching.1008 γδ T cells also modulate αβ T-cell activity indirectly through activating NK cells, DCs and B cells.1002 Intriguingly, γδ T cells are recently unveiled a critical role in mediating immune response to ICB in MHC I-deficient cancers, in which PD-1+ Vδ1 and Vδ3 T cells are the main contributors.1009
Fig. 7.
The anti- and pro-tumor immunity of γδ T cells. γδ T cells in TME play both anti- and pro-tumor activities. γδ T cells recognize phosphoantigens bound by BTN3A1/BTN2A1 heterodimers, as well as recognize glycolipids presented by CD1d. γδ T cells can directly kill tumor cells by expressing cytotoxic factors perforin and granzymes, and apoptotic receptors TRAIL and FasL. IFN-γ produced by γδ T cells enhances MHC I expression on tumor cells and their antigen presentation to CD8+ αβ T cells. γδ T cells are able to present antigens to CD4+ and CD8+ αβ T cells through MHC II and MHC I molecules, respectively. γδ T cells orchestrate the anti-tumor immunity through interacting and activating DCs, NK cells, and B cells. Expression of NKRs and TLRs promote γδ T cells activation and effector function. PD-1-expressing γδ T cells are the main responder to ICB in MHC I-deficient cancers. The pro-tumor activity of γδ T cells relies on both soluble factors and surface receptors by promoting tumor cell growth and angiogenesis, suppressing αβ T cell function, MDCSs induction, and inducing inhibitory functions
On the contrary, the pro-tumor activity of γδ T cells is largely attributed to the production of IL-17 which can promote tumor cell proliferation,1010 angiogenesis,991 accumulation of MDSCs,990 and create an immunosuppressive TME.981 In addition, pro-tumor functions of human γδ T cells may also result from expression of other mediators, such as IL-22 and amphiregulin for tumor cell growth,1011 PD-L1, galectins (Gal1 and 9), CD86, CD73, IL-10, and TLR8 for T cell suppression,1012–1014 IL-4, IL-10 and inhibitory receptors (killer Ig-like inhibitory receptors (KIRs), Ig-like transcript 2 (ILT-2), and NKG2A) for inhibitory function of Vδ T cells,1002,1015 and IL-8 and GM-CFS for MDSCs induction.990 Together, the roles of γδ T cells in the tumor milieu are complicated, and further research is required to fully elucidate the function of distinct subsets of γδ T cells to develop next-generation immunotherapies harnessing γδ T cells.
Current immunotherapies harnessing T cell immunity
Given the central roles of T lymphocytes in health and disease, novel and effective immunotherapies harnessing the T cell immunity are under extensive development. In this section, we will briefly introduce the current immunotherapies engaging T cell function in both cancer and autoimmune disease, with an emphasis on their clinical implementation and progress.
T cell-based cancer immunotherapy
Base on the biological roles and the modes of action, T cell-based immunotherapeutic approaches in cancer mainly include the following categories: immune checkpoint blockade (ICB) and costimulation, bispecific T cell engagers (TCEs) and adoptive cell therapy (ACT).
ICB and costimulation
Immunomodulation of the coinhibitory and costimulatory molecules on T cells has become a powerful and effective strategy for cancer immunotherapy. Immune checkpoint molecules refer to the inhibitory receptors expressed on the immune cells and play immunosuppressive roles upon ligand interactions to maintain self-tolerance.1016 CTLA-4 and PD-1 are so far the most potent and successful T cell immune checkpoint molecules developed for cancer therapy in the clinic.1017 Since a decade ago the first U.S. Food and Drug Administration (FDA)-approved checkpoint inhibitor Ipilimumab, a monoclonal antibody (mAb) targeting CTLA-4, seven immune checkpoint inhibitors targeting PD-1/PD-L1 and another CTLA-4 mAb Tremelimumab have been consecutively approved for multiple cancer types (Table 4). Furthermore, there are nearly 6000 clinical trials assessing anti-PD-1/PD-L1 mAbs—with majority of FDA-approved ones—as monotherapy or in combination with other therapies.1017 Besides PD-1/PD-L1, other immune checkpoint pathways have been developed in the clinic for cancer therapy, including but not limited to Lag-3, TIGIT, Tim-3, CD96, BTLA, VISTA and B7H3.1018,1019 Among them, the anti-Lag-3 mAb (Telatlimab) has been approved firstly by FDA for metastatic melanoma in combination with anti-PD-1 mAb.1020,1021 Moreover, the advanced candidates in phase III clinical trials are mAbs targeting Lag-3, TIGIT and Tim-3 (Table 5). In contrast to inhibitory checkpoints, costimulatory molecules provide critical signals for effective T cell responses and function, making them promising therapeutic targets.1022 Thus, mAbs targeting costimulatory receptors, such as GITR, 4-1BB, ICOS, CD27, CD28, and OX40, are also under evaluation in clinical trials.1023 However, agonist antibodies have not exhibited much clinical benefits.1024 So far, most of the programs targeting costimulatory pathways are in early clinical phases except for one ICOS-stimulatory mAb Feladilimab entering phase III trial (Table 5).
Table 4.
T cell-based therapies approved in the market
| Therapy type | Modality | Product name | Brand | Target | Indications | Company | Approval date |
|---|---|---|---|---|---|---|---|
| ICB | mAbs | Ipilimumab | Yervoy | CTLA-4 | Multiple cancer types | BMS | FDA 2010 |
| Tremelimumab | Imjudo | CTLA-4 | Hepatocellular carcinoma | AstraZeneca | FDA 2022 | ||
| Pembrolizumab | Keytruda | PD-1 | Multiple cancer types | Merck | FDA 2014 | ||
| Nivolumab | Opdivo | PD-1 | Multiple cancer types | BMS | FDA 2014 | ||
| Cemiplimab | Libtayo | PD-1 | Multiple cancer types | Sanofi | FDA 2018 | ||
| Dostarlimab | Jemperli | PD-1 | Multiple cancer types | GlaxoSmithKline | FDA 2021 | ||
| Atezolizumab | Tecentriq | PD-L1 | Multiple cancer types | Genentech/Roche | FDA 2016 | ||
| Avelumab | Bavencio | PD-L1 | Multiple cancer types | EMD | FDA 2016 | ||
| Durvalumab | Imfinzi | PD-L1 | Multiple cancer types | AstraZeneca | FDA 2016 | ||
| Combination | Relatlimab+Nivolumab | Opdualag+Opdivo | Lag-3+PD-1 | Metastatic melanoma | BMS | FDA 2022 | |
| bsAbs | Cadonilimab | - | PD-1xCTLA-4 | Metastatic cervical cancer | Akeso Biopharma | NMPA 2022 | |
| TCE | bsAbs | Blinatumomab | Blincyto | CD19xCD3 | r/r ALL | Amgen | FDA 2014 |
| Mosunetuzumab-axgb | Lunsumio | CD20xCD3 | Follicular lymphoma | Genentech/Roche | FDA 2022 | ||
| Teclistamab-cqyv | Tecvayli | BCMAxCD3 | r/r MM | Janssen Biotech | FDA 2022 | ||
| Elranatamab | - | BCMAxCD3 | r/r MM | Pfizer | FDA Filing Acceptance 2023 | ||
| TCR-like antibody | Tebentafusp-tebn | Kimmtrak | HLA-A*02:01/gp100 complex | Uveal melanoma | Immunocore | FDA 2022 | |
| ACT | CAR-T | Tisagenlecleucel | Kymriah | CD19 | ALL, DLBCL | Novartis | FDA 2017 |
| Axicabtagene ciloleucel | Yescarta | CD19 | NHL, DLBCL | Kite/Gilead | FDA 2017 | ||
| DLBCL | NMPA 2021 | ||||||
| Brexucabtagene autoleucel | Tecartus | CD19 | MCL, ALL | Kite/Gilead | FDA 2020 | ||
| Lisocabtagene maraleucel | Breyanzi | CD19 | DLBCL | Juno Therapeutics/BMS | FDA 2021 | ||
| Idecabtagene vicleucel | Abecma | BCMA | MM | Bluebird Bio/BMS | FDA 2021 | ||
| Ciltacabtagene autoleucel | Carvykti | BCMA | MM | Legend/Janssen Biotech | FDA 2022 | ||
| Relmacabtagene autoleucel | Carteyva | CD19 | DLBCL | JW Therapeutics | NMPA 2021 |
FDA Food and Drug Administration, NMPA National Medical Products Administration of China, r/r ALL relapsed/refractory B cell precursor acute lymphoblastic leukemia, r/r MM relapsed/refractory multiple myeloma, DLBCL diffuse large B-cell lymphoma, NHL non-Hodgkin’s lymphoma, MCL mantle cell lymphoma
Table 5.
Selected clinical-stage T cell-based immunotherapies
| Therapy type | Modality | Product name | Target | Disease | Clinical trial identifier | Sponsor | Phase |
|---|---|---|---|---|---|---|---|
| ICB | mAb | Tiragolumab | TIGIT | NSCLC | NCT04294810 | Roche | III |
| Ociperlimab | TIGIT | NSCLC | NCT04746924 | BeiGene | III | ||
| MBG453 | Tim-3 | Myelodysplastic Syndromes | NCT04266301 | Novartis Pharmaceuticals | III | ||
| Fianlimab | Lag-3 | Melanoma | NCT05608291 | Regeneron Pharmaceuticals | III | ||
| Feladilimab | ICOS | Neoplasms, Head and Neck | NCT04428333 | GlaxoSmithKline/Merck | II/III | ||
| bsAb | KN046 | PD-L1xCTLA-4 | NSCLC | NCT04474119 | Jiangsu Alphamab Biopharmaceuticals | III | |
| Tebotelimab | PD-1×Lag-3 | Gastric Cancer | NCT04082364 | MacroGenics | II/III | ||
| Lomvastomig | PD-1xTim-3 | Advanced or Metastatic ESCC | NCT04785820 | Roche | II | ||
| FS118 | PD-L1×Lag-3 | Advanced Cancer | NCT03440437 | F-star Therapeutics | I/II | ||
| XmAb22841 | CTLA-4xLag-3 | Metastatic Melanoma | NCT05695898 | Xencor | I/II | ||
| HLX301 | TIGITxPD-L1 | Advanced Tumors | NCT05390528 | Shanghai Henlius Biotech | I/II | ||
| AZD2936 | TIGITxPD-1 | NSCLC | NCT04995523 | AstraZeneca | I/II | ||
| GEN1046 | PD-L1x4-1BB | Solid Tumors | NCT03917381 | Genmab | I/II | ||
| PRS-344/S095012 | PD-L1x4-1BB | Solid Tumor | NCT05159388 | Pieris Pharmaceuticals | I/II | ||
| XmAb23104 | PD-1xICOS | Metastatic Melanoma | NCT05695898 | Xencor | I/II | ||
| Ivonescimab (AK112) | PD-1xVEGF | Advanced NSCLC | NCT05499390 | Akeso | III | ||
| PM8002 | PD-1xVEGF | NSCLC | NCT05756972 | Biotheus | II/III | ||
| Bintrafusp alfa (M7824) | PD-L1xTGFβRII | NSCLC | NCT03631706 | Merck KGaA | III | ||
| SHR-1701 | PD-L1xTGFβRII | Advanced or Metastatic NSCLC | NCT05132413 | Jiangsu Hengrui Medicine/Suzhou Suncadia Biopharmaceuticals | III | ||
| TCE | TAAxCD3 | Epcoritamab | CD20xCD3 | DLBCL | NCT04628494 | Genmab/AbbVie | III |
| Elranatamab | BCMAxCD3 | MM | NCT05317416 | Pfizer | III | ||
| Glofitamab | CD20xCD3 | DLBCL | NCT04408638 | Roche | III | ||
| Teclistamab | BCMAxCD3 | MM | NCT05083169 | Janssen Research | III | ||
| Linvoseltamab | BCMAxCD3 | MM | NCT05730036 | Regeneron Pharmaceuticals | III | ||
| Talquetamab | GPRC5DxCD3 | MM | NCT05455320 | Janssen Research | III | ||
| Catumaxomab | EpCAM x CD3 | Stomach Neoplasms | NCT04222114 | LintonPharm | III | ||
| Tarlatamab | DLL3xCD3 | SCLC | NCT05740566 | Amgen | III | ||
| CC-1 | PMSAxCD3 | Lung Cancer Squamous Cell | NCT04496674 | German Cancer Research Center | I/II | ||
| REGN4336 | PSMAXCD3 | Prostate Cancer | NCT05125016 | Regeneron Pharmaceuticals | I/II | ||
| REGN4018 | MUC16xCD3 | Ovarian Cancer | NCT03564340 | Regeneron Pharmaceuticals | I/II | ||
| EGFR BATs | EGFRxCD3 | Pancreatic Adenocarcinoma | NCT03269526 | University of Virginia | I/II | ||
| Cibisatamab | CEAxCD3 | Colorectal Cancer | NCT03866239 | Roche | I/II | ||
| Runimotamab | HER2xCD3 | HER2-expressing Solid Tumors | NCT03448042 | Genentech | I | ||
| AMG 596 | EGFRvIII and CD3 | Glioblastoma or Malignant Glioma | NCT03296696 | Amgen | I | ||
| GEM3PSCA | PSCA and CD3 | PSCA-positive solid cancers | NCT03927573 | AvenCell Europe GmbH | I | ||
| ERY974 | GPC3xCD3 | HCC | NCT05022927 | Chugai Pharmaceutical | I | ||
|
TAAx Costimulation |
REGN5668 | MUC16xCD28 | Ovarian Cancer | NCT04590326 | Regeneron Pharmaceuticals | I/II | |
| REGN5678 | PSMAxCD28 | Metastatic Castration-resistant Prostate Cancer | NCT03972657 | Regeneron Pharmaceuticals | I/II | ||
| REGN7075 | EGFRxCD28 | Advanced Solid Tumors | NCT04626635 | Regeneron Pharmaceuticals | I/II | ||
| GEN1046 | PD-L1/4-1BB | NSCLC | NCT05117242 | Genmab/BioNTech SE | II | ||
| PRS-343 | HER2/4-1BB | HER2-positive Gastric Cancer | NCT05190445 | Pieris Pharmaceuticals | II | ||
| HLX35 | EGFR×4-1BB | Advanced or Metastatic Solid Tumors | NCT05360381 | Shanghai Henlius Biotech | I | ||
| CB307 | PSMA×4-1BB | Advanced and/or Metastatic Solid Tumors | NCT04839991 | Crescendo Biologics | I | ||
| RO7122290 | FAPx4-1BB | Metastatic Colorectal Cancer | NCT04826003 | Roche | I/II | ||
| BT7480 | Nectin-4×4-1BB | Advanced Solid Tumor | NCT05163041 | BicycleTx Limited | I/II | ||
| ACT (Cancer) | CAR-T | CAR-T CD19 | CD19 | Acute Myeloid Leukemia | NCT04257175 | Sheba Medical Center | III |
| CAR-T-CD19 Cells | CD19 | DLBCL | NCT05020392 | Wuhan Union Hospital, China | III | ||
| CD19 CAR-T CELLS | CD19 | Relapsed ALL | NCT03937544 | National University of Malaysia/Gaia Science | III | ||
| BCMA CAR-T-cells | BCMA | MM | NCT04287660 | The First Affiliated Hospital of Soochow University (and 13 more) | III | ||
| JNJ-68284528 | BCMA | MM | NCT04181827 | Janssen Research & Development | III | ||
| bb2121 | BCMA | MM | NCT03651128 | Celgene | III | ||
| fhB7H3.CAR-Ts | B7H3 (CD276) | Ovarian Cancer | NCT05211557 | The Affiliated Hospital of Xuzhou Medical University | I/II | ||
| CD276 CAR-T cells | B7H3 (CD276) | Advanced Pancreatic Carcinoma | NCT05143151 | Shenzhen University General Hospital | I/II | ||
| anti-MESO CAR-T cells | Mesothelin | Ovarian Cancer | NCT03916679 | Second Affiliated Hospital, Zhejiang University | I/II | ||
| ALPP CAR-T | alkaline phosphatase | Endometrial Cancer | NCT04627740 | Xinqiao Hospital of Chongqing/TCRCure Biopharma | I/II | ||
| CNA3103 | LGR5 | Colorectal Cancer Metastatic | NCT05759728 | Carina Biotech Pty | I/II | ||
| CT041 | Claudin18.2 | Gastric Cancer/Pancreatic Cancer | NCT04404595 | CARsgen Therapeutics | I/II | ||
| RD14-01 | ROR1 | Solid Tumor | NCT05748938 | 920th Hospital of Joint Logistics Support Force of People’s Liberation Army of China | I/II | ||
| CEA CAR-T | CEA | Solid Tumor | NCT04348643 | Chongqing Precision Biotech | I/II | ||
| IVS-3001-Anti-HLA-G CAR-T | HLA-G | Solid Tumor | NCT05672459 | M.D. Anderson Cancer Center | I/II | ||
| BPX-601 | PSCA | Prostate Cancer | NCT02744287 | Bellicum Pharmaceuticals | I/II | ||
| HypoSti.CAR-HER2 T cells | HER2 | HER2-Positive Solid Tumors | NCT05681650 | Chinese PLA General Hospital | I/II | ||
| CLDN6 CAR-T | Claudin6 | Solid Tumor | NCT04503278 | BioNTech Cell & Gene Therapies GmbH | I/II | ||
| GD2-CART01 | GD2 | Neuroblastoma | NCT03373097 | Bambino Gesù Hospital and Research Institute | I/II | ||
| MUC1 CAR-T | MUC1 | Intrahepatic Cholangiocarcinoma | NCT03633773 | Second Affiliated Hospital | I/II | ||
| BOXR1030 | Glypican 3 | GPC3-Positive Solid Tumors | NCT05120271 | SOTIO Biotech | I/II | ||
| Bispecific CAR-T | bi-4SCAR CD19/22 T cells | CD19/CD22 | B Cell Malignancies | NCT05432882 | Shenzhen Geno-Immune Medical Institute | I/II | |
| bi-4SCAR CD19/70 T cells | CD19/CD70 | B Cell Malignancies | NCT05436496 | Shenzhen Geno-Immune Medical Institute | I/II | ||
| bi-4SCAR CD19/79b T cells | CD19/CD79b | B Cell Malignancies | NCT05436509 | Shenzhen Geno-Immune Medical Institute | I/II | ||
| CAR-20/19-T Cells | CD19/CD20 | B Cell Malignancies | NCT04186520 | Medical College of Wisconsin | I/II | ||
| bi-4SCAR GD2/CD70 T cells | GD2/CD70 | Cancer Disease | NCT05438368 | Shenzhen Geno-Immune Medical Institute | I/II | ||
| bi-4SCAR GD2/PSMA T cells | GD2/PSMA | Solid Tumor | NCT05437315 | Shenzhen Geno-Immune Medical Institute | I/II | ||
| bi-4SCAR PSMA/CD70 T cells | PSMA/CD70 | Cancer Disease | NCT05437341 | Shenzhen Geno-Immune Medical Institute | I/II | ||
| Dual-targeting VEGFR1 and PD-L1 CAR-T cells | VEGFR1/PD-L1 | Malignant Peritoneal Effusion | NCT05477927 | Sichuan University | I | ||
| EGFR/B7H3 CAR-T | EGFR/B7H3 | Advanced Lung Cancer/TNBC | NCT05341492 | Second Affiliated Hospital of Guangzhou Medical University | Early I | ||
| Dual-targeting HER2 and PD-L1 CAR-T cells | HER2/PD-L1 | Peritoneal Carcinoma Metastatic | NCT04684459 | Sichuan University | Early I | ||
| TCR-T | anti-MART-1 F5 T-cell receptor | MART-1 | Melanoma | NCT00509288 | National Cancer Institute (NCI) | II | |
| Anti-gp100:154-162 TCR | gp100 | Melanoma | NCT00923195 | National Cancer Institute (NCI) | II | ||
| PG13-CEA_TCR | CEA | Metastatic Cancer | NCT00923806 | National Cancer Institute (NCI) | I | ||
| WT1 TCR transduced T cells | WT1 | MDS/AML | NCT02550535 | Cell Medica | I/II | ||
| afamitresgene autoleucel | MAGE-A4 | Synovial Sarcoma | NCT04044768 | Adaptimmune Therapeutics | II | ||
| Anti-MAGE-A3-DP4 TCR PBL | MAGE-A3 | Cervical Cancer | NCT02111850 | National Cancer Institute (NCI) | I/II | ||
| autologous MC2 TCR-T cells | MAGE-C2 | Melanoma and Head and Neck Cancer | NCT04729543 | Erasmus Medical Center (and 4 more) | I/II | ||
| CD8 + T-cells, transduced with MAGE-A1 directed TCR | MAGE-A1 | Advanced Solid Tumors | NCT05430555 | knife GmbH | I/II | ||
| letetresgene autoleucel | NY-ESO-1 | Neoplasms | NCT02992743 | GlaxoSmithKline | II | ||
| NY-ESO-1c259 T cells | NY-ESO-1 | Ovarian Cancer | NCT01567891 | Adaptimmune | I/II | ||
| NY-ESO-1(TCR Affinity Enhancing Specific T cell Therapy) | NY-ESO-1 | Soft Tissue Sarcoma | NCT05549921 | Sun Yat-sen University | II | ||
| E7 TCR-T cells | HPV E7 | HPV Associated Cancers | NCT05686226 | The State University of New Jersey | II | ||
| TC-E202 cells | HPV-16 E6 | Cervical Carcinoma | NCT05357027 | TCRCure Biopharma/Fudan University | I/II | ||
| E6 TCR | HPV-16 E6 | HPV Associated Cancers | NCT02280811 | National Cancer Institute (NCI)/Kite Pharma | I/II | ||
| TCR redirected T cells | HBV | Hepatocellular Carcinoma | NCT03899415 | Beijing 302 Hospital/Lion TCR Pte | I | ||
| MCPyV-specific HLA-A02-restricted TCR T | MCPyV | Metastatic or Unresectable MCC | NCT03747484 | Fred Hutchinson Cancer Center/National Cancer Institute (NCI) | I/II | ||
| EBV-specific TCR-T | EBV | HNSCC | NCT04139057 | Xinqiao Hospital of Chongqing/TCRCure Biopharma | I/II | ||
| anti-p53 T-cell receptor transduced peripheral blood lymphocytes | Tumor protein 53 (p53) | Metastatic Cancer | NCT00393029 | National Cancer Institute (NCI) | II | ||
| Mutant KRAS G12V-specific TCR transduced autologous T cells | Mutant KRAS G12V | Pancreatic Cancer | NCT04146298 | Changhai Hospital | I/II | ||
| anti-KRAS G12D mTCR PBL | Mutant KRAS G12D | Gastrointestinal Cancer/Pancreatic Cancer | NCT03745326 | National Cancer Institute (NCI) | I/II | ||
| TC-510 T Cells | Mesothelin | Mesothelioma | NCT05451849 | TCR2 Therapeutics | I/II | ||
| Gavo-cel (TC-210) T Cells | Mesothelin | Mesothelioma | NCT03907852 | TCR2 Therapeutics | I/II | ||
| TC-110 T Cells | CD19 | NHL | NCT04323657 | TCR2 Therapeutics | I/II | ||
| TILs | Tumor Infiltrating Lymphocytes (TIL) | - | Metastatic Melanoma | NCT02278887 | The Netherlands Cancer Institute | III | |
| Lifileucel (LN-144) | - | Metastatic Melanoma | NCT02360579 | Iovance Biotherapeutics | II | ||
| LN-145 | - | Metastatic TNBC | NCT04111510 | Iovance Biotherapeutics | II | ||
| Tumor Infiltrating Lymphocytes (TIL) | - | BTC | NCT03801083 | Udai Kammula | II | ||
| Young TIL | - | Metastatic Colorectal/Pancreatic/Ovarian Cancer | NCT01174121 | National Cancer Institute (NCI) | II | ||
| Tumor Infiltrating Lymphocytes (TIL) | - | Uveal Melanoma | NCT03467516 | Udai Kammula | II | ||
| Young TIL | - | Advanced NSCLC | NCT02133196 | National Cancer Institute (NCI) | II | ||
| Tumor Infiltrating Lymphocytes (TIL) | - | Multiple advanced Solid Cancers | NCT03935893 | Udai Kammula | II | ||
| Super circulating TIL (ScTIL) | - | Gynecological Malignancies | NCT05342506 | Peking Union Medical College Hospital | II | ||
| Tumor Infiltrating Lymphocytes (TIL) | - | Metastatic Urothelial Carcinoma | NCT04383067 | Sheba Medical Center | II | ||
| Tumor Infiltrating Lymphocytes (TIL) | - | Gastrointestinal Cancer | NCT04426669 | Intima Bioscience, Inc. | I/II | ||
| Autologous tumor infiltrating lymphocytes MDA-TIL | - | Multiple advanced Solid Cancers | NCT03610490 | M.D. Anderson Cancer Center | II | ||
| ACT (Autoimmunity) | CAR-T | YTB323 | CD19 | SLE/Lupus Nephritis | NCT05798117 | Novartis Pharmaceuticals | I/II |
| CT103A cells | BCMA | Autoimmune Diseases | NCT04561557 | Tongji Hospital/Nanjing IASO Biotherapeutics | Early I | ||
| Descartes-08 | BCMA | MG | NCT04146051 | Cartesian Therapeutics | II | ||
| CD19/BCMA CAR-T-cells | CD19/BCMA | POEMS Syndrome/Amyloidosis/Autoimmune Hemolytic Anemia/Vasculitis | NCT05263817 | Zhejiang University/Yake Biotechnology | Early I | ||
| SLE | NCT05030779 | Early I | |||||
| Sjogren’s Syndrome | NCT05085431 | Early I | |||||
| Immune Nephritis | NCT05085418 | Early I | |||||
| BCMA-CD19 cCAR T cells | CD19/BCMA | Relapsed/Refractory, SLE | NCT05474885 | iCell Gene Therapeutics | I | ||
| DSG3-CAAR-T | DSG3 | Mucosal-Dominant PV | NCT04422912 | Cabaletta Bio | I | ||
| MuSK-CAAR-T | Musk | MuSK-MG | NCT05451212 | Cabaletta Bio | I | ||
| CAR-Treg | TX200-TR101 | HLA-A*02 | Kidney Transplant Rejection | NCT04817774 | Sangamo Therapeutics | I/II | |
| QEL-001 | HLA-A*02 | Rejection; Transplant, Liver | NCT05234190 | Quell Therapeutics | I/II |
NSCLC non-small cell lung cancer, ESCC esophageal squamous cell carcinoma, DLBCL diffuse large B-cell lymphoma, MM multiple myeloma, SCLC small cell lung cancer, HCC hepatocellular carcinoma, ALL B acute lymphoblastic leukemia, TNBC triple-negative breast cancer, MDS myelodysplastic syndromes, AML acute myeloid leukemia, MCC Merkel cell cancer, HNSCC head and neck squamous cell carcinoma, NHL non-Hodgkin’s lymphoma, BTC biliary tract cancer, SLE systemic lupus erythematosus, MG myasthenia gravis, PV pemphigus vulgaris
(Source: clinicaltrials.gov)
Bispecific T cell engagers (TCEs)
Emerging evidence has demonstrated that simultaneously targeting two or multiple immunomodulatory molecules display potent anti-tumor activity while reduce toxicity, leading to the revolutionary development of bispecific antibodies (bsAbs) or even trispecific antibodies (TsAbs).1025,1026 With the advances in antibody engineering, numerous formats have been exploited for bsAb design (reviewed in ref. 1026). Different from a combination of two mAbs, bsAbs can either bind to two molecules expressed on one cell (in-cis binding) or bridge two distinct cells (in-trans binding) to further enhance the therapeutic efficacy.1026 The mechanisms of action of bsAbs engaging T cells mainly include four types: (1) dual-targeting inhibitory checkpoint molecules; (2) targeting both costimulatory and inhibitory checkpoints; (3) targeting checkpoints with non-checkpoint molecules; (4) directly targeting T cells by TCE. Dual-targeting inhibitory checkpoints usually occurs between PD-1/PD-L1 and other checkpoint molecules under clinical assessment, such as CTLA-4, Lag-3, Tim-3, and TIGIT.1026,1027 Notably, Cadonilimab, a bsAb targeting PD-1×CTLA-4, is the first bsAb approved by Chinese National Medical Products Administration (NMPA) last year for treating relapsed or metastatic cervical cancer (r/mCC)1028 (Table 4). Besides, KN046 and Tebotelimab, targeting PD-L1×CTLA-4 and PD-1×LAG-3 respectively, are the most advanced bsAb candidates in late-phase clinical trials (NCT04474119 and NCT04082364) (Table 5). Other bsAbs, such as PD-1xTim-3, PD-L1×Lag-3, PD-(L)1xTIGIT, and CTLA-4xLag-3, are under evaluation in phase I/II studies (Table 5). Co-targeting checkpoint inhibitors and costimulatory molecules has a synergistic effect on enhancing T cell function and therapeutic efficacy. BsAbs in this category, including GITR×CTLA-4, 4-1BB×PD-L1,1029 OX40×PD-L1,1030 OX40×CTLA-4,1031 ICOS×PD-L1, and CD27×PD-L1,1032 are mainly under early clinical assessment. The non-checkpoint targets involved in bsAbs are mostly tumor-associated antigens (TAAs) and pro-tumor growth factors/cytokines.1027 Targeting TAAs can increase the tumor selectivity of immunomodulatory molecules and alleviate systemic toxicity, whereas inhibiting growth factors/cytokines further enhances the efficacy of tumor eradication. TAAs used for immune checkpoint targeting include EpCAM (CD40×EpCAM), EGFR (PD1×EGFR) and HER2 (PD1×HER2).1033,1034 The widely used growth factors/cytokines are pro-angiogenic VEGF and immunosuppressive TGF-β. BsAbs under late-phase clinical development are PD-1xVEGF (AK112 and PM8002) and PD-L1xTGFβRII (M7824 and SHR-1701) (Table 5). Of note, despite the rationale behind ‘trapping’ TGF-β for cancer therapy,1035 the unsatisfied clinical results of M7824 (also known as Bintrafusp alfa) in NSCLC and biliary tract cancers (BTCs)1036 raise the concern of TGF-β-targeting strategy, and further research is required to fully understand the biology of TGF-β in TME.
TCEs, also referred to bispecific T cell engagers (BiTEs), are designed bsAbs co-targeting CD3ε and specific tumor antigens to redirect cytotoxic T cells against tumor cells. Various TCE formats and platforms have been developed and reviewed elsewhere.1037,1038 TCEs activate T cells independent on MHC restriction and TCR epitope specificity and have been developed rapidly and extensively over the years, becoming a promising immunotherapy. To date, three BiTEs have been approved by FDA in the market: Blinatumomab (Blincyto; CD19×CD3; Amgen) in 2014 for patients with relapsed/refractory (r/r) B cell precursor acute lymphoblastic leukemia (ALL), Mosunetuzumab-axgb (Lunsumio; CD20xCD3; Roche) for follicular lymphoma, and Teclistamab-cqyv (Tecvayli; BCMAxCD3; Janssen Biotech) for r/r multiple myeloma (MM) in 2022. In addition, Elranatamab (BCMAxCD3; Pfizer) for r/r MM has received FDA and European Medicines Agency (EMA) filing acceptance which is expected to be approved in 2023 (Table 4). Apparently, FDA-approved TCEs and majority of the late-phase TCEs target antigens in hematological malignancies1039 (Table 5). Other hematological tumor targets in early-phase studies include CD38, CD123, CD30, CD33, FcRH5, FLT3, and CLEC12A.1026 However, compare to liquid tumors, development of TCEs against solid tumors are much challenging. Two bsAbs Catumaxomab (EpCAM×CD3) and Tarlatamab (DLL3×CD3) are so far in phase III studies, while other TCEs targeting PSMA, MUC16, EGFR, CEA, HER2, EGFRvIII, PSCA, and GPC3 are mostly in early-phase trails (Table 5). The immunological mechanisms underlying T cell response or non-response to TCEs are not fully understood. A recent clinical study in MM patients using BCMAxCD3 TCE has revealed that the pre-existing T cell landscape determines the response to TCE. Moreover, effector and naïve CD8+ T cells drive the immunological response to TCE while the exhausted CD8+ T cells are highly associated with the response failure.1040 One key challenge of CD3-TCEs in treating solid tumor is the treatment-mediated toxicity, including both cytokine release syndrome (CRS) and on-target/off-tumor toxicity.1037,1041,1042 Several strategies to overcome the adverse events of TCEs in solid tumors are under both clinical and preclinical investigations. One important approach is targeting peptide/MHC (pMHC) complexes, known as TCR mimetic antibodies. Indeed, Tebentafusp (Kimmtrak; Immunocore), a CD3 BiTE with TCR arm recognizing glycoprotein 100 (gp100) peptide presented by HLA-A*02:01, gained FDA approval in 2022 for the treatment of HLA-A*02:01-positive patients with unresectable or metastatic uveal melanoma.1043 The success of Tebentafusp has also become a major milestone for TCR-based immunotherapies. Another approach is developing conditional TCEs which are inactive prodrugs upon administration and gain activation in a tempo-spatial controlled manner within TME, such as TCEs with a masking on the binding domain.1038
In addition to CD3, alternative approaches targeting costimulatory molecules on T cells, such as CD28 and 4-1BB, have also implemented for TCE development. Engagement of costimulatory receptors mimics signal 2 for T cell activation. Costimulatory BiTEs targeting a variety of solid tumors are currently evaluated in phase I/II trials: MUC16, PSMA, EGFR, PD-L1, HER2, Nectin-4, and FAP (targeting tumor-associated fibroblasts) (Table 5). 4-1BB costimulation has been demonstrated to remarkedly improve T cell survival, activation and effector function, which occurs preferentially in CD8+ T cells.1044 TAAxCD28 BiTEs, when combined with TAAxCD3 BiTEs, could significantly enhance T cell activation and the anti-tumor activity of the CD3 BiTEs.1045 The intracellular domains of CD28 and 4-1BB are widely implemented in the CAR-T cell generation; CD28 and 4-1BB differ in both expression pattern on T cells as well as the intracellular signal cascade.1046 Further research especially results from clinical studies will help us to better understand the underlying mechanism of these costimulatory signals in cancer immunotherapy.
Adoptive cell therapy (ACT)
In addition to drugs that modulate T cell function, direct T cell adoptive transfer of autologous or allogenic T cells into patients has shown substantial promise in cancer immunotherapies. According to different T cell source and ways of antigen recognition, ACT mainly divide into three types: chimeric antigen receptor (CAR)-T cells, TCR-T cells, and tumor infiltrating lymphocyte (TIL) therapy. Generally, TIL therapy is adoptively transferring tumor-specific TILs that are isolated from tumor tissues and amplified ex vivo, whereas CAR-T cell and TCR-T cell therapies are based on T cells that are genetically engineered to express receptors recognizing antigens.
CAR-T cell therapy is one of the most prevalent and advanced types of ACT. CARs are normally engineered proteins targeting tumor antigens to enhance the tumor-killing specificity and efficacy of immune cells, such as T cells, NK cell and macrophages. A classic CAR is composed of an extracellular antigen-binding domain, a hinge, a transmembrane region, one or more costimulatory domains, and an activation domain. The antigen-binding domain consists of a single-chain variable fragment (scFv) recognizing antigens. The costimulatory domains—CD28 and/or 4-1BB—are designed to augment T cell activation, proliferation and effector function. The activation domain is usually the CD3ζ domain which transduces activation signaling for T cells.1047 The structural engineering of CAR-T cells has been gone through five generations with distinct intracellular functional domains. In addition to the basic CAR components mentioned above, the fourth and fifth generation of CAR-T cells contain cytokines or intracellular domains of cytokine receptors, which can further enhance the effector function of T cell or adaption to the immunosuppressive TME.1048
In the past two decades, CAR-T cell therapy has obtained tremendous clinical success in treating cancers particularly in patients with hematological tumors. To date, seven CAR-T products with five targeting CD19 and two for BCMA have been approved in the market (Table 4). Candidates in clinical phase III pipeline are also targeting CD19 or BCMA (Table 5). CAR-T therapies targeting antigens in solid tumors are then assessed in early-phase clinical studies, such as B7H3 (CD276), mesothelin, alkaline phosphatase, LGR5, Claudin18.2, ROR1, CEA, HLA-G, PSCA, HER2, Claudin6, GD2, MUC1, and Glypican 31049 (Table 5). Like TCEs, CAR-T therapy faces challenges in solid tumors due to multiple reasons: tumor antigen heterogeneity and escape, toxicity, inefficient tumor infiltration, poor persistency, and immunosuppressive TME.1048 Next-generation CAR-T cells for overcoming those challenges are under extensive investigations.1049,1050 For instance, to avoid tumor-antigen escape as well as off-target toxicity, dual CARs are designed to co-targeting two different tumor antigens, such as CD19/CD22, CD19/CD22, GD2/CD70, GD2/PSMA, EGFR/B7H3, etc. (Table 5). Another creative approach is applying Boolean logic to CAR-T cells, which can conditionally control T cell activity to increase T cell specificity and limit off-target toxicity.1051,1052 The logic-gates consist of OR-gate, AND-gate, NOT-gate, IF-THEN-gate and IF-BETTER-gate, and can be engineered to have constitutive expression or inducible expression.1053–1055 Most of the logic-gate CAR-T constructs have not yet been tested in the clinic except for IMPT-314, a CD19/CD20-targeted bispecific “OR-Gate” CAR-T therapy which has just gained FDA approval this year in patients with aggressive B-cell lymphoma. Some future directions for advancing CAR-T therapies include but not limit to improving CAR-T cell persistency, function and tumor infiltration, combination with other therapies, and development of allogeneic/universal CAR-T cells.1048–1050
Despite the potency, CAR-T cells target only surface antigens. In contrast, TCR-T cells can recognize intracellular antigens, which greatly increases the tumor target repertoire. TCR-T cells are much more (at least 100-fold) sensitive to antigens that a low antigen density is sufficient to activate TCR-T cells.1056,1057 In addition, TCR-T cells adopt a near-to-physiological signaling pathway compared to CAR-T cells.1056 Such enhanced sensitivity and avidity of TCR-T cells markedly improve their tumor cell recognition and killing efficacy. However, TCR-T cells recognize peptide/HLA complexes with HLA restriction, which limits their application in certain patient populations. Currently, TCR-T cell therapies have not yet been approved in the market but are assessed in early-phase clinical trials (Table 5). Given the high sensitivity of antigen detection, antigen selection is crucial for developing safe TCR-T therapies. According to the biological function, tumor antigens developed and evaluated for TCR-T therapy in the clinical trials are tissue differentiation antigens (MART-1, gp100, CEA and WT1), cancer germline antigens (MAGE-A and NY-ESO-1), viral antigens (HPV, HBV, Merkel cell polyomavirus (MCPyV), and EBV), mutation-associated neoantigens (p53, KRASG12V, and KRASG12D) as well as TAAs (mesothelin and CD19) (Table 5). TCR-T cell therapy also faces challenges such as treatment-associated toxicity, tumor antigen escape, low tumor infiltration and suppressive tumor milieu.1058 Besides, identification of tumor epitope-specific TCRs is complex. The advances of high-throughput screening using peptide libraries and barcoded tetramers and scTCR-seq facilitate the identification of antigen-specific TCRs.1059–1061
TILs, compared to non-TILs, display mostly effector memory T cell phenotype, can be activated and expanded ex vivo, and possess chemokine receptors for migration toward TME, thus severing great immunological reactivity against tumor cells.1062,1063 Although TILs can be separated from resected solid tumor tissues, the cell number is inadequate for cancer immunotherapy. High dose IL-2 exposure and nonmyeloablative lymphodepletion are key procedures to provide enough TILs for infusion and enhance the therapeutic effectiveness.1064,1065 Currently, TIL therapy has been evaluated in the clinical studies in multiple solid tumor types, such as melanoma, breast cancer, biliary tract cancer, CRC, NSCLC, gastrointestinal, and gynecological cancers (Table 5). Though no TIL therapy has been approved yet, the most advanced TIL product is lifileucel (LN-144), developed by Iovance Biotherapeutics, and has just completed its Biologics License Application (BLA) submission for unresectable or metastatic melanoma. Notably, the BLA application for lifileucel is supported by positive clinical data of a phase II study (C-144-01).1066 Besides the common challenges for T cell therapies, TIL therapy faces a key obstacle of TIL preparation. TIL therapy is the most personalized treatment; therefore, the specific TILs product must be prepared for each patient.1067 Several strategies have been developed to overcome this issue, such as CD8+ enriched young TILs,1068 rapid expansion by anti-CD3 antibody, IL-2 and feeder cells,1069 generating artificial APCs for TIL expansion,1070 and incorporation of costimulatory signals.1071 Additionally, combination of TILs with other anti-tumor therapies are also developed and tested in clinical and preclinical studies.1072
T cell-based immunotherapies in autoimmune diseases
For autoimmune diseases, traditional therapeutic drugs mainly include three classes: nonsteroid anti-inflammatory drugs (NSAIDs), steroid anti-inflammatory drugs (SAIDs), and disease-modifying antirheumatic drugs (DMARDs). While NSAIDs and SAIDs are effective for pain relief and inflammation inhibition, DMARDs are mainly reducing the tissue damages caused by severe inflammation.1073 In recent decades, biological drugs targeting inflammatory cytokines, receptors and signaling molecules have been developed and displayed great effectiveness.652,1074 Among all, Th1- and Th17-associated cytokines, such as TNF-α, IL-12, IL-6, IL-23, and IL-17, are critical for the development and pathogenesis of autoimmune diseases, thus, have been extensively studied and developed for treating multiple autoimmune diseases. A number of neutralizing antibodies or fusion proteins targeting inflammatory signaling pathways have been approved in the market: TNF-α (Infliximab, Etanercept, Adalimumab, Certolizumab, and Golimumab), IL-12/IL-23 (Ustekinumab), IL-6 (Siltuximab), IL-6R (Tocilizumab, Sarilumab, and Satralizumab), IL-23 (Guselkumab, Tildrakizumab, and Risankizumab), IL-17 (Secukinumab and Ixekizumab), and IL-17RA (Brodalumab).1075,1076 The JAK-STAT pathways, mediating the intracellular signal transduction downstream of cytokine receptors, have also been targeting by small molecule inhibitors for autoimmune diseases.1077,1078 In addition, B cell depletion by mAbs targeting various B cell types, such as anti-CD19, anti-CD20 and anti-CD22, have shown beneficial effects in autoimmune disorders.1079
CAR-T and CAAR-T cell therapy
Intriguingly, CAR-T cell-based immunotherapies have emerged increasing interest in autoimmune diseases and demonstrated promising clinical efficacy.1080,1081 Based on the recognition specificity of CARs, four strategies have been developed for CAR-T therapies in autoimmune manifestation: (1) CAR-T cells targeting autoreactive B cells; (2) Chimeric autoantibody receptor T cells (CAAR-T cells) expressing autoantigens that interact with autoantibodies on B cells; (3) CAR-T cells expressing pathogenic pMHC complexes recognized by autoreactive T cells; (4) CAR-Treg cells recognizing autoantigens and exerting immunosuppressive activity.1082,1083 B cell depletion has become an important therapeutic strategy in autoimmune diseases.1084 CAR-T cells targeting pan-B cell antigens or plasma cells, such as CD19 and BCMA, can eliminate autoantibody-producing B cells; thus, exhibit strong therapeutic effects in both preclinical1085–1087 and particularly clinical autoimmune conditions.1088–1090 Several CAR-T products targeting CD19 or BCMA or these two simultaneously are under early-phase clinical studies (Table 5). However, pan-B-cell depletion has side effect of lacking immunoglobulins.1082 To specifically target autoimmune B cells, CAAR-T cells which express autoantigens instead of traditional scFv have been developed. Hence, autoantigen recognition by autoreactive B cells leads to specific killing of pathogenic B cells by CAR-T cells.1091 A number of autoantigens have been identified highly associated with various types of autoimmune diseases.1082 CAAR-T cells expressing pemphigus vulgaris (PV) autoantigen desmoglein-3 (Dsg-3) and muscle specific kinase (MuSK) have been tested in phase I clinical trials for patients with mucosal-dominant PV and MuSK-myasthenia gravis, respectively1092,1093 (Table 5). Similarly, CAR-T cells expressing the ectodomains of pMHC complexes can specifically interact and eliminate pathogenic T cells.1094 For instance, CAR-T cells expressing I-Ag7-B:9-23 (R3) complex that the insulin B-chain peptide B:9-23 is presented by MHC II, directly target pathogenic B:9-23–specific CD4+ cells and significantly delay the onset of diabetes.1095 Likewise, genetically engineered CAR-T cells with insulin B chain peptide fused with MHC I component β2 microglobulin (β2m) could reduce the pathogenic CD8+ T cells and ameliorate diabetes in NOD mice.1096
CAR-Treg cell therapy
Given the potent immunosuppressive activity of Treg cells, therapeutic strategies harnessing Treg cell function have been proposed to restore immune tolerance in autoimmune diseases. Low-dose IL-2 therapy and engineered IL-2 with different selectivity to IL-2R (IL-2 muteins) which can preferentially induce Treg cell expansion and function without activating autoreactive Teff cells have demonstrated clinical efficacy in various autoimmune diseases.1097,1098 However, due to lacking of specificity, polyclonal Treg cells have compromised suppressive activity, whereas CAR-Treg cells with engineered CAR modules directing against autoantigens display stronger suppression of effector function.1099 CAR-Treg cells have been extensively studied in preclinical models by targeting different autoimmune antigens, including MOG for EAE,1100 2,4,6-trinitrophenyl (TNP),1101 and CEA1102 for colitis, citrullinated vimentin (CV) for RA,1103 as well as insulin for T1D.1104 In organ transplantation, HLA-A2 is commonly mismatched. CAR-Treg cells designed to express HLA-A*02 CAR have been shown to induce immunosuppression of allograft-specific effector T cells and prevent graft-versus-host disease (GVHD) in preclinical models.1105,1106 Therefore, two phase I/II clinical trials of HLA-A2-CAR-Treg cells (TX200-TR101 and QEL-001) have been registered for organ transplantation (Table 5).
Conclusions
T cells are essential for functional immune responses. In this review, we summarize the current understandings of T cell development, CD4+ and CD8+ αβ T cell and γδ T cell subsets, fate decision and regulation, functional roles in pathophysiological conditions, especially in infectious diseases, chronic infection and tumors and autoimmune diseases as well as immunotherapies harnessing T cell function in preclinical and clinical development. Cytotoxic T cells, including both CD8+ and CD4+ CTLs, can directly eliminate infected or malignant cells, while CD4+ T helper cells mainly regulate/help both innate and adaptive immune responses through costimulation and cytokine signals. Major effector T cells, including different CD4+ Th cells, effector γδ T cells and CD8+ TE cells are summarized regarding to their cellular and molecular characteristics (Table 6). Appropriate T cell immunity is essential for maintaining host homeostasis and preventing infections and malignancy, whereas aberrant T cell immune responses elicit and promote pathogenesis, tumor growth and autoimmune disorders, which may also affect its application in immunotherapy, such as CAR-T cell-induced CRS.1107
Table 6.
Effector T cell subsets and key features
| Effector T cells | Effector molecules | Surface markers | Differentiation induction | Master TF | Other regulatory TF | Functions | Refs |
|---|---|---|---|---|---|---|---|
| Th1 | IFN-γ, TNF-α/β, IL-2 | CXCR3, CCR5 | IL-12, IFN-γ | T-bet | STAT1, STAT4 |
Defense intracellular pathogens; Cell-based immunity; Pro-inflammation |
76,78,80–82,950 |
| Th2 | IL-2, IL-4, IL-5, IL-10, IL-13 | CCR3, CCR4 | IL-4 | GATA-3 |
STAT6, NFAT1, c-Maf, IRF4, JunB, TCF-1 |
Defense extracellular pathogens; Humoral immunity; Tissue repair; Allergy | 81,84,86,241 |
| Th9 | IL-9, IL-10, IL-21 |
IL-4R, TGFβR, IL-2R, OX40, GITR, Notch, DR3, TSLPR |
IL-4, TGF-β | IRF4, PU.1 | GATA-3, SMAD | Infectious diseases; Allergy; Cancer; Autoimmunity | 91–94,99,100 |
| Th17 |
IL-17A-F, IL-21, IL-10, IL-23, IL-22, IFN-γ, GM-CSF |
IL-6R, TGFβR, IL-21R, IL-23R | IL-6, TGF-β, IL-21, IL-23 | RORγt | RORα, c-Maf, p65, NFAT, c-Rel | Defense extracellular pathogens (fungi); Mucosal immunity; Autoimmunity | 102,104,108,111,117,252 |
| Tfh | IL-4, IL-21 | PD-1, CXCR5, CD40, CD40LG, ICOS, SAP | IL-6, IL-21 | Bcl-6 |
BATF, STAT1/3/4/5, Foxp1, KLF2, IRF4, Ets1, BACH2, Ascl2, Tox2, Bhlhe40, STAT5 and Blimp-1 (Inhibition) |
Humoral immunity; Autoimmunity | 130–132,136,140,145,150 |
| Treg |
IL-10, TGF-β, IL-35 |
CD25 | TGF-β, IL-2 | Foxp3 |
c-Rel, AP-1, NFAT, Smad2, Smad3, FoxO1, FoxO3, STAT5 |
Immunosuppression; Autoimmunity; Cancer | 151,152,154,156 |
| CD4 CTL | pro-inflammatory cytokines, perforin, granzymes, granulysin | KLRG1, NKG2A, NKG2D, CRTAM, Fas, TRAIL |
IL-2, IL-12, IL-6, IFN-α |
RUNX3 | T-bet, Eomes, ThPOK (Inhibition) |
Infectious diseases; Longevity; Cancer |
77,256,258,268,275,279 |
| Tγδ1 | IFN-γ | CD27, CD122, NK1.1, CD45RBhi | Skint-1 | T-bet |
TCF-1, Lef1, Eomes, Id3 |
Tissue physiology; Defense pathogenic infections; (Anti-)Cancer |
938,943,947 |
| Tγδ17 | IL-17A | CCR6, SCART2, CD45RBlo | IL-6, TGF-β, IL-1β, IL-18, IL-23 | RORγt |
c-Maf, Sox4, Sox13, HEB, Blk, RelB |
Tissue physiology and pathophysiology; Defense pathogenic infections; (Pro-)Cancer |
938,943,944,948,952 |
| CD8 TE |
IL-2, IFN-γ, TNF, perforin, granzymes, CCL5, CCL3 |
FasL, KLRG1, CX3CR1, CXCR6, CCR5 |
IL-2, IL-12, IL-21 |
T-bet |
Blimp-1, Id2, STAT4, Zeb2 |
Viral infection; Cancer |
169,173,182,193,219,223 |
T cell immunity is extremely critical but complex with significant cell heterogeneity, differentiation plasticity, functional diversity and exquisite regulatory mechanisms, which also display context-dependent features. For instance, upon acute infection, both CD4+ and CD8+ T cells differentiate into effector CD8+ T cells with robust expansion and cytotoxic functions, whereas those in chronic infection develop into exhaustion state with progressive loss of effector function and elevated inhibitory phenotype. The discrepancy of either tumor-promoting or tumor-protective effects of Th2, Th17, Th9, Treg, and Tγδ17 cells is mainly attributed to different tumor types. The differentiation plasticity of Th17 cells in tumor and autoimmune diseases is also highly dependent on the microenvironmental niche. The heterogeneity, plasticity and instability of Treg cells, such as Th-like Treg and exFoxp3 Treg cells, play important and contradictory roles in autoimmune diseases. The diverse T cell differentiation and function depend on distinct but intersected molecular regulations at transcriptional, epigenetic and metabolic levels.
Despite a comprehensive elaboration on multiple aspects of T cells, some limitations in this review are: (1) classic αβ T and γδ T cells are mainly focused here, while rare T cell populations such as mucosal-associated invariant T (MAIT) cells and NKT cells also play essential roles in immune responses. (2) Most of the current understandings on T cell immunity are derived from mouse studies, albeit highly evolutionary conservation between mouse and human, T cell response in human subjects is more clinically relevant. (3) Universal features of T cells signature and function in each disease setting are summarized. However, context-specific T cells are present in response to discrete types of pathogens or cancers. (4) We mainly summarized T cell immunity at the cellular level regarding to cell development, differentiation and functionality, whereas the molecular signaling pathways are important to understand the underlying mechanisms. For instance, TCR signaling pathway is critical for T cells in almost every aspect and contributes to human health and disease, which has been comprehensively reviewed recently.1108 Collectively, given the importance and complexity of T cell immunity, both comprehensive and delicate research are required to fully reveal T cell signature and function. Especially with the advances in single-cell technologies, future investigations need to focus on characterizing new T cell subsets, context-specific T cell heterogeneity, functional states, differential plasticity, dysfunction and programmability to provide insights into novel therapeutic strategies in human diseases.
Acknowledgements
This work was supported by the National Key Research and Development Program of China grants 2021YFA1100702 (to B.Z.), Major International (Regional) Joint Research Project grants 81820108017 (to B.Z.), National Natural Science Foundation of China grants 82271792 (to L.S.) and 32200727 (to L.S.), and Innovation Capability Support Program of Shaanxi 2021TD-38 (to B.Z.).
Author contributions
B.Z. and L.S. conceptualized and organized the review. L.S., Y. S., A.J., and X. W. wrote the manuscript. L.S. prepared the figures. All authors have read and approved the article.
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
These authors contributed equally: Lina Sun, Yanhong Su, Anjun Jiao, Xin Wang
References
- 1.Hosokawa H, Rothenberg EV. Cytokines, transcription factors, and the initiation of T-cell development. Cold Spring Harb. Perspect. Biol. 2018;10:a028621. doi: 10.1101/cshperspect.a028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 2014;14:529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosokawa H, Rothenberg EV. How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol. 2021;21:162–176. doi: 10.1038/s41577-020-00426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Q, Jeremiah Bell J, Bhandoola A. T-cell lineage determination. Immunol. Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurd N, Robey EA. T-cell selection in the thymus: a spatial and temporal perspective. Immunol. Rev. 2016;271:114–126. doi: 10.1111/imr.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta A, Zhao B, Love PE. New insights into TCR beta-selection. Trends Immunol. 2021;42:735–750. doi: 10.1016/j.it.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 9.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see) Nat. Rev. Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 11.Rothenberg EV. T cell lineage commitment: identity and renunciation. J. Immunol. 2011;186:6649–6655. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 14.Germar K, et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl Acad. Sci. USA. 2011;108:20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Perez L, et al. Functional definition of a transcription factor hierarchy regulating T cell lineage commitment. Sci. Adv. 2020;6:eaaw7313. doi: 10.1126/sciadv.aaw7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, et al. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood. 2013;122:902–911. doi: 10.1182/blood-2012-08-447839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng KK, et al. A stochastic epigenetic switch controls the dynamics of T-cell lineage commitment. Elife. 2018;7:e37851. doi: 10.7554/eLife.37851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kueh HY, et al. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat. Immunol. 2016;17:956–965. doi: 10.1038/ni.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikawa T, et al. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 23.Hosokawa H, et al. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat. Immunol. 2018;19:1427–1440. doi: 10.1038/s41590-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira ACF, et al. RORalpha is a critical checkpoint for T cell and ILC2 commitment in the embryonic thymus. Nat. Immunol. 2021;22:166–178. doi: 10.1038/s41590-020-00833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki M, et al. The E-Id protein axis specifies adaptive lymphoid cell identity and suppresses thymic innate lymphoid cell development. Immunity. 2017;46:818–834 e814. doi: 10.1016/j.immuni.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delconte RB, et al. The helix-loop-helix protein ID2 governs NK cell fate by tuning their sensitivity to interleukin-15. Immunity. 2016;44:103–115. doi: 10.1016/j.immuni.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seillet C, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 2014;211:1733–1740. doi: 10.1084/jem.20140145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Constantinides MG, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc. Natl Acad. Sci. USA. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat. Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahni H, et al. A genome wide transcriptional model of the complex response to pre-TCR signalling during thymocyte differentiation. Oncotarget. 2015;6:28646–28660. doi: 10.18632/oncotarget.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harker N, et al. Pre-TCR signaling and CD8 gene bivalent chromatin resolution during thymocyte development. J. Immunol. 2011;186:6368–6377. doi: 10.4049/jimmunol.1003567. [DOI] [PubMed] [Google Scholar]
- 34.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 35.Aifantis I, Buer J, von Boehmer H, Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 1997;7:601–607. doi: 10.1016/S1074-7613(00)80381-7. [DOI] [PubMed] [Google Scholar]
- 36.Mombaerts P, et al. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 37.Hoffman ES, et al. Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 38.Petrie HT, Hugo P, Scollay R, Shortman K. Lineage relationships and developmental kinetics of immature thymocytes: CD3, CD4, and CD8 acquisition in vivo and in vitro. J. Exp. Med. 1990;172:1583–1588. doi: 10.1084/jem.172.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/S1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 40.Ciofani M, et al. Obligatory role for cooperative signaling by pre-TCR and Notch during thymocyte differentiation. J. Immunol. 2004;172:5230–5239. doi: 10.4049/jimmunol.172.9.5230. [DOI] [PubMed] [Google Scholar]
- 41.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev. Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 42.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat. Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 43.Wolfer A, et al. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta lineage thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/S1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez-Caparros A, et al. Notch signaling controls transcription via the recruitment of RUNX1 and MYB to enhancers during T cell development. J. Immunol. 2019;202:2460–2472. doi: 10.4049/jimmunol.1801650. [DOI] [PubMed] [Google Scholar]
- 45.Liu X, et al. Notch-induced endoplasmic reticulum-associated degradation governs mouse thymocyte beta-selection. Elife. 2021;10:e69975. doi: 10.7554/eLife.69975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao B, et al. Notch and the pre-TCR coordinate thymocyte proliferation by induction of the SCF subunits Fbxl1 and Fbxl12. Nat. Immunol. 2019;20:1381–1392. doi: 10.1038/s41590-019-0469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allam AH, Charnley M, Pham K, Russell SM. Developing T cells form an immunological synapse for passage through the beta-selection checkpoint. J. Cell Biol. 2021;220:e201908108. doi: 10.1083/jcb.201908108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, et al. Zinc finger protein Zfp335 controls early T-cell development and survival through beta-selection-dependent and -independent mechanisms. Elife. 2022;11:e75508. doi: 10.7554/eLife.75508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ratiu JJ, et al. Loss of Zfp335 triggers cGAS/STING-dependent apoptosis of post-beta selection thymocytes. Nat. Commun. 2022;13:5901. doi: 10.1038/s41467-022-33610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teh HS, et al. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–233. doi: 10.1038/335229a0. [DOI] [PubMed] [Google Scholar]
- 51.Taniuchi I. CD4 helper and CD8 cytotoxic T cell differentiation. Annu. Rev. Immunol. 2018;36:579–601. doi: 10.1146/annurev-immunol-042617-053411. [DOI] [PubMed] [Google Scholar]
- 52.Brugnera E, et al. Coreceptor reversal in the thymus: signaled CD4 + 8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8 + T cells. Immunity. 2000;13:59–71. doi: 10.1016/S1074-7613(00)00008-X. [DOI] [PubMed] [Google Scholar]
- 53.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr. Opin. Immunol. 2002;14:207–215. doi: 10.1016/S0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 54.Singer A, Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 2004;83:91–131. doi: 10.1016/S0065-2776(04)83003-7. [DOI] [PubMed] [Google Scholar]
- 55.Yu Q, et al. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8 + T cells. J. Exp. Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeidan N, Damen H, Roy DC, Dave VP. Critical Role for TCR signal strength and MHC specificity in ThPOK-induced CD4 helper lineage choice. J. Immunol. 2019;202:3211–3225. doi: 10.4049/jimmunol.1801464. [DOI] [PubMed] [Google Scholar]
- 58.He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 59.Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 60.Dave VP, et al. HD mice: a novel mouse mutant with a specific defect in the generation of CD4(+) T cells. Proc. Natl Acad. Sci. USA. 1998;95:8187–8192. doi: 10.1073/pnas.95.14.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4( + ) T cells. Nat. Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luckey MA, et al. The transcription factor ThPOK suppresses Runx3 and imposes CD4(+) lineage fate by inducing the SOCS suppressors of cytokine signaling. Nat. Immunol. 2014;15:638–645. doi: 10.1038/ni.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y, et al. NuRD complex recruitment to Thpok mediates CD4( + ) T cell lineage differentiation. Sci. Immunol. 2022;7:eabn5917. doi: 10.1126/sciimmunol.abn5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/S0092-8674(02)01111-X. [DOI] [PubMed] [Google Scholar]
- 67.Kojo S, et al. Priming of lineage-specifying genes by Bcl11b is required for lineage choice in post-selection thymocytes. Nat. Commun. 2017;8:702. doi: 10.1038/s41467-017-00768-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kastner P, et al. Bcl11b represses a mature T-cell gene expression program in immature CD4(+)CD8(+) thymocytes. Eur. J. Immunol. 2010;40:2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hernandez-Hoyos G, et al. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/S1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 70.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/S1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 71.Park JH, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis MM, et al. Ligand recognition by alpha beta T cell receptors. Annu. Rev. Immunol. 1998;16:523–544. doi: 10.1146/annurev.immunol.16.1.523. [DOI] [PubMed] [Google Scholar]
- 73.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu. Rev. Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruterbusch M, Pruner KB, Shehata L, Pepper M. In vivo CD4(+) T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- 75.Geginat J, et al. Plasticity of human CD4 T cell subsets. Front. Immunol. 2014;5:630. doi: 10.3389/fimmu.2014.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4(+)T cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012:925135. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cenerenti M, Saillard M, Romero P, Jandus C. The era of cytotoxic CD4 T cells. Front. Immunol. 2022;13:867189. doi: 10.3389/fimmu.2022.867189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Meitei HT, Lal G. T cell receptor signaling in the differentiation and plasticity of CD4(+) T cells. Cytokine Growth Factor Rev. 2022;69:14–27. doi: 10.1016/j.cytogfr.2022.08.001. [DOI] [PubMed] [Google Scholar]
- 79.Dobrzanski MJ. Expanding roles for CD4 T cells and their subpopulations in tumor immunity and therapy. Front. Oncol. 2013;3:63. doi: 10.3389/fonc.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu J. T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb. Perspect. Biol. 2018;10:a030338. doi: 10.1101/cshperspect.a030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saravia J, Chapman NM, Chi H. Helper T cell differentiation. Cell Mol. Immunol. 2019;16:634–643. doi: 10.1038/s41423-019-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Afkarian M, et al. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4 + T cells. Nat. Immunol. 2002;3:549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- 83.Ylikoski E, et al. IL-12 up-regulates T-bet independently of IFN-gamma in human CD4 + T cells. Eur. J. Immunol. 2005;35:3297–3306. doi: 10.1002/eji.200526101. [DOI] [PubMed] [Google Scholar]
- 84.Walker JA, McKenzie ANJ. T(H)2 cell development and function. Nat. Rev. Immunol. 2018;18:121–133. doi: 10.1038/nri.2017.118. [DOI] [PubMed] [Google Scholar]
- 85.Jeong J, Lee HK. The role of CD4(+) T cells and microbiota in the pathogenesis of asthma. Int. J. Mol. Sci. 2021;22:11822. doi: 10.3390/ijms222111822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noben-Trauth N, Hu-Li J, Paul WE. IL-4 secreted from individual naive CD4 + T cells acts in an autocrine manner to induce Th2 differentiation. Eur. J. Immunol. 2002;32:1428–1433. doi: 10.1002/1521-4141(200205)32:5<1428::AID-IMMU1428>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 87.Spinner CA, Lazarevic V. Transcriptional regulation of adaptive and innate lymphoid lineage specification. Immunol. Rev. 2021;300:65–81. doi: 10.1111/imr.12935. [DOI] [PubMed] [Google Scholar]
- 88.Liao W, et al. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat. Immunol. 2008;9:1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu Q, et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat. Immunol. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 91.Li Y, et al. TH9 cell differentiation, transcriptional control and function in inflammation, autoimmune diseases and cancer. Oncotarget. 2016;7:71001–71012. doi: 10.18632/oncotarget.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Angkasekwinai P, Dong C. IL-9-producing T cells: potential players in allergy and cancer. Nat. Rev. Immunol. 2021;21:37–48. doi: 10.1038/s41577-020-0396-0. [DOI] [PubMed] [Google Scholar]
- 93.Vyas SP, Goswami R. A decade of Th9 cells: role of Th9 cells in inflammatory bowel disease. Front. Immunol. 2018;9:1139. doi: 10.3389/fimmu.2018.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rojas-Zuleta WG, Vasquez G. Th9 lymphocytes: a recent history from IL-9 to its potential role in rheumatic diseases. Autoimmun. Rev. 2016;15:649–655. doi: 10.1016/j.autrev.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 95.Staudt V, et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 96.Chang HC, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9 + IL-10+ Foxp3(-) effector T cells. Nat. Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.You FP, et al. Th9 cells promote antitumor immunity via IL-9 and IL-21 and demonstrate atypical cytokine expression in breast cancer. Int. Immunopharmacol. 2017;52:163–167. doi: 10.1016/j.intimp.2017.08.031. [DOI] [PubMed] [Google Scholar]
- 99.Koch S, Sopel N, Finotto S. Th9 and other IL-9-producing cells in allergic asthma. Semin. Immunopathol. 2017;39:55–68. doi: 10.1007/s00281-016-0601-1. [DOI] [PubMed] [Google Scholar]
- 100.Chen T, et al. Th9 cell differentiation and its dual effects in tumor development. Front. Immunol. 2020;11:1026. doi: 10.3389/fimmu.2020.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Humblin E, et al. IRF8-dependent molecular complexes control the Th9 transcriptional program. Nat. Commun. 2017;8:2085. doi: 10.1038/s41467-017-01070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 103.Guglani L, Khader SA. Th17 cytokines in mucosal immunity and inflammation. Curr. Opin. HIV AIDS. 2010;5:120–127. doi: 10.1097/COH.0b013e328335c2f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han L, et al. Th17 cells in autoimmune diseases. Front. Med. 2015;9:10–19. doi: 10.1007/s11684-015-0388-9. [DOI] [PubMed] [Google Scholar]
- 105.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17 + T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 106.Mangan PR, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 107.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 109.Nurieva R, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 110.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 111.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat. Rev. Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martinez GJ, et al. Smad2 positively regulates the generation of Th17 cells. J. Biol. Chem. 2010;285:29039–29043. doi: 10.1074/jbc.C110.155820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Spolski R, Leonard WJ. Interleukin-21: a double-edged sword with therapeutic potential. Nat. Rev. Drug Disco. 2014;13:379–395. doi: 10.1038/nrd4296. [DOI] [PubMed] [Google Scholar]
- 114.Chen Z, et al. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc. Natl Acad. Sci. USA. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghoreschi K, et al. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mufazalov IA, et al. IL-1 signaling is critical for expansion but not generation of autoreactive GM-CSF+ Th17 cells. EMBO J. 2017;36:102–115. doi: 10.15252/embj.201694615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Akdis M, et al. TH17 and TH22 cells: a confusion of antimicrobial response with tissue inflammation versus protection. J. Allergy Clin. Immunol. 2012;129:1438–1449. doi: 10.1016/j.jaci.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 118.Campe J, Ullrich E. T helper cell lineage-defining transcription factors: potent targets for specific GVHD therapy? Front. Immunol. 2021;12:806529. doi: 10.3389/fimmu.2021.806529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu CM, Jang SY, Fanzo JC, Pernis AB. Modulation of T cell cytokine production by interferon regulatory factor-4. J. Biol. Chem. 2002;277:49238–49246. doi: 10.1074/jbc.M205895200. [DOI] [PubMed] [Google Scholar]
- 120.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bauquet AT, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Okamoto K, et al. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464:1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- 125.Yahia-Cherbal H, et al. NFAT primes the human RORC locus for RORgammat expression in CD4(+) T cells. Nat. Commun. 2019;10:4698. doi: 10.1038/s41467-019-12680-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ruan Q, et al. The Th17 immune response is controlled by the Rel-RORγ-RORγ T transcriptional axis. J. Exp. Med. 2011;208:2321–2333. doi: 10.1084/jem.20110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang XO, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 129.Schaerli P, et al. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu. Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 131.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yu D, et al. Targeting TFH cells in human diseases and vaccination: rationale and practice. Nat. Immunol. 2022;23:1157–1168. doi: 10.1038/s41590-022-01253-8. [DOI] [PubMed] [Google Scholar]
- 133.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hatzi K, et al. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J. Exp. Med. 2015;212:539–553. doi: 10.1084/jem.20141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu X, et al. Genome-wide analysis identifies Bcl6-controlled regulatory networks during T follicular helper cell differentiation. Cell Rep. 2016;14:1735–1747. doi: 10.1016/j.celrep.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choi J, et al. Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-repressor circuits. Nat. Immunol. 2020;21:777–789. doi: 10.1038/s41590-020-0706-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kusam S, Toney LM, Sato H, Dent AL. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J. Immunol. 2003;170:2435–2441. doi: 10.4049/jimmunol.170.5.2435. [DOI] [PubMed] [Google Scholar]
- 138.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 139.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu L, et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat. Immunol. 2015;16:991–999. doi: 10.1038/ni.3229. [DOI] [PubMed] [Google Scholar]
- 141.Choi YS, et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu X, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rauschmeier R, et al. Bhlhe40 function in activated B and TFH cells restrains the GC reaction and prevents lymphomagenesis. J Exp Med. 2022;219:e20211406. doi: 10.1084/jem.20211406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu W, et al. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 2019;51:826–839.e825. doi: 10.1016/j.immuni.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 145.Wan S, et al. Costimulation molecules differentially regulate the ERK-Zfp831 axis to shape T follicular helper cell differentiation. Immunity. 2021;54:2740–2755 e2746. doi: 10.1016/j.immuni.2021.09.018. [DOI] [PubMed] [Google Scholar]
- 146.Weber JP, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 2015;212:217–233. doi: 10.1084/jem.20141432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Eto D, et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Spolski R, Leonard WJ. IL-21 and T follicular helper cells. Int. Immunol. 2010;22:7–12. doi: 10.1093/intimm/dxp112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ballesteros-Tato A, et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Johnston RJ, et al. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Plitas G, Rudensky AY. Regulatory T cells: differentiation and function. Cancer Immunol. Res. 2016;4:721–725. doi: 10.1158/2326-6066.CIR-16-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hori S. FOXP3 as a master regulator of T(reg) cells. Nat. Rev. Immunol. 2021;21:618–619. doi: 10.1038/s41577-021-00598-9. [DOI] [PubMed] [Google Scholar]
- 153.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kanamori M, et al. Induced regulatory T cells: their development, stability, and applications. Trends Immunol. 2016;37:803–811. doi: 10.1016/j.it.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 155.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 156.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4 + CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 157.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 158.Morikawa H, Sakaguchi S. Genetic and epigenetic basis of Treg cell development and function: from a FoxP3-centered view to an epigenome-defined view of natural Treg cells. Immunol. Rev. 2014;259:192–205. doi: 10.1111/imr.12174. [DOI] [PubMed] [Google Scholar]
- 159.Long M, et al. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 160.Isomura I, et al. c-Rel is required for the development of thymic Foxp3 + CD4 regulatory T cells. J. Exp. Med. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barbi J, Pardoll D, Pan F. Treg functional stability and its responsiveness to the microenvironment. Immunol. Rev. 2014;259:115–139. doi: 10.1111/imr.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maruyama T, Konkel JE, Zamarron BF, Chen W. The molecular mechanisms of Foxp3 gene regulation. Semin. Immunol. 2011;23:418–423. doi: 10.1016/j.smim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dikiy S, et al. A distal Foxp3 enhancer enables interleukin-2 dependent thymic Treg cell lineage commitment for robust immune tolerance. Immunity. 2021;54:931–946.e911. doi: 10.1016/j.immuni.2021.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chinen T, et al. An essential role for the IL-2 receptor in T(reg) cell function. Nat. Immunol. 2016;17:1322–1333. doi: 10.1038/ni.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8 + T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Joshi NS, et al. Inflammation directs memory precursor and short-lived effector CD8( + ) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kaech SM, et al. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 168.Intlekofer AM, et al. Effector and memory CD8 + T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 169.Kaech SM, Cui W. Transcriptional control of effector and memory CD8 + T cell differentiation. Nat. Rev. Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kallies A, Xin A, Belz GT, Nutt SL. Blimp-1 transcription factor is required for the differentiation of effector CD8( + ) T cells and memory responses. Immunity. 2009;31:283–295. doi: 10.1016/j.immuni.2009.06.021. [DOI] [PubMed] [Google Scholar]
- 171.Xin A, et al. A molecular threshold for effector CD8( + ) T cell differentiation controlled by transcription factors Blimp-1 and T-bet. Nat. Immunol. 2016;17:422–432. doi: 10.1038/ni.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ichii H, et al. Role for Bcl-6 in the generation and maintenance of memory CD8 + T cells. Nat. Immunol. 2002;3:558–563. doi: 10.1038/ni802. [DOI] [PubMed] [Google Scholar]
- 173.Chen Y, et al. Transcriptional and epigenetic regulation of effector and memory CD8 T cell differentiation. Front. Immunol. 2018;9:2826. doi: 10.3389/fimmu.2018.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cavalcanti E, et al. JAK3/STAT5/6 pathway alterations are associated with immune deviation in CD8 T cells in renal cell carcinoma patients. J. Biomed. Biotechnol. 2010;2010:935764. doi: 10.1155/2010/935764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schade AE, Wlodarski MW, Maciejewski JP. Pathophysiology defined by altered signal transduction pathways: the role of JAK-STAT and PI3K signaling in leukemic large granular lymphocytes. Cell Cycle. 2006;5:2571–2574. doi: 10.4161/cc.5.22.3449. [DOI] [PubMed] [Google Scholar]
- 176.Yang C, et al. STAT4: an immunoregulator contributing to diverse human diseases. Int. J. Biol. Sci. 2020;16:1575–1585. doi: 10.7150/ijbs.41852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Guan T, et al. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8( + ) T cell fates. J. Exp. Med. 2018;215:1153–1168. doi: 10.1084/jem.20171352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Curtsinger JM, et al. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J. Immunol. 2005;174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- 179.Curtsinger JM, Agarwal P, Lins DC, Mescher MF. Autocrine IFN-gamma promotes naive CD8 T cell differentiation and synergizes with IFN-alpha to stimulate strong function. J. Immunol. 2012;189:659–668. doi: 10.4049/jimmunol.1102727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang N, Bevan MJ. CD8( + ) T cells: foot soldiers of the immune system. Immunity. 2011;35:161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhao X, Shan Q, Xue HH. TCF1 in T cell immunity: a broadened frontier. Nat. Rev. Immunol. 2022;22:147–157. doi: 10.1038/s41577-021-00563-6. [DOI] [PubMed] [Google Scholar]
- 183.Franco F, et al. Metabolic and epigenetic regulation of T-cell exhaustion. Nat. Metab. 2020;2:1001–1012. doi: 10.1038/s42255-020-00280-9. [DOI] [PubMed] [Google Scholar]
- 184.Rodriguez RM, et al. Epigenetic networks regulate the transcriptional program in memory and terminally differentiated CD8 + T Cells. J. Immunol. 2017;198:937–949. doi: 10.4049/jimmunol.1601102. [DOI] [PubMed] [Google Scholar]
- 185.Scharer CD, et al. Global DNA methylation remodeling accompanies CD8 T cell effector function. J. Immunol. 2013;191:3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shin MS, et al. DNA methylation regulates the differential expression of CX3CR1 on human IL-7Ralphalow and IL-7Ralphahigh effector memory CD8 + T cells with distinct migratory capacities to the fractalkine. J. Immunol. 2015;195:2861–2869. doi: 10.4049/jimmunol.1500877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Abdelsamed HA, et al. Human memory CD8 T cell effector potential is epigenetically preserved during in vivo homeostasis. J. Exp. Med. 2017;214:1593–1606. doi: 10.1084/jem.20161760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ladle BH, et al. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8 + T-cell fate decisions following activation. Proc. Natl Acad. Sci. USA. 2016;113:10631–10636. doi: 10.1073/pnas.1524490113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Carty SA, et al. The loss of TET2 promotes CD8( + ) T cell memory differentiation. J. Immunol. 2018;200:82–91. doi: 10.4049/jimmunol.1700559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zebley CC, et al. Proinflammatory cytokines promote TET2-mediated DNA demethylation during CD8 T cell effector differentiation. Cell Rep. 2021;37:109796. doi: 10.1016/j.celrep.2021.109796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 192.He B, et al. CD8( + ) T cells utilize highly dynamic enhancer repertoires and regulatory circuitry in response to infections. Immunity. 2016;45:1341–1354. doi: 10.1016/j.immuni.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Henning AN, Roychoudhuri R, Restifo NP. Epigenetic control of CD8( + ) T cell differentiation. Nat. Rev. Immunol. 2018;18:340–356. doi: 10.1038/nri.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shin HM, et al. Epigenetic modifications induced by Blimp-1 Regulate CD8( + ) T cell memory progression during acute virus infection. Immunity. 2013;39:661–675. doi: 10.1016/j.immuni.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kuroda S, et al. Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc. Natl Acad. Sci. USA. 2011;108:14885–14889. doi: 10.1073/pnas.1105133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Krauss S, Brand MD, Buttgereit F. Signaling takes a breath-new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/S1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 197.Menk AV, et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 2018;22:1509–1521. doi: 10.1016/j.celrep.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 200.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/S1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 201.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dan HC, et al. Akt-dependent activation of mTORC1 complex involves phosphorylation of mTOR (mammalian target of rapamycin) by IkappaB kinase alpha (IKKalpha) J. Biol. Chem. 2014;289:25227–25240. doi: 10.1074/jbc.M114.554881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Gerriets VA, Rathmell JC. Metabolic pathways in T cell fate and function. Trends Immunol. 2012;33:168–173. doi: 10.1016/j.it.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Klein-Hessling S, et al. NFATc1 controls the cytotoxicity of CD8( + ) T cells. Nat. Commun. 2017;8:511. doi: 10.1038/s41467-017-00612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vaeth M, et al. Store-operated Ca(2 + ) entry controls clonal expansion of T cells through metabolic reprogramming. Immunity. 2017;47:664–679 e666. doi: 10.1016/j.immuni.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.O’Sullivan D, et al. Memory CD8( + ) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Gupta SS, et al. NIX-mediated mitophagy promotes effector memory formation in antigen-specific CD8( + ) T cells. Cell Rep. 2019;29:1862–1877 e1867. doi: 10.1016/j.celrep.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8 + T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shan Q, et al. Tcf1 preprograms the mobilization of glycolysis in central memory CD8( + ) T cells during recall responses. Nat. Immunol. 2022;23:386–398. doi: 10.1038/s41590-022-01131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yu B, et al. Epigenetic landscapes reveal transcription factors that regulate CD8( + ) T cell differentiation. Nat. Immunol. 2017;18:573–582. doi: 10.1038/ni.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Russ BE, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8( + ) T cell differentiation. Immunity. 2014;41:853–865. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Gupta SS, Wang J, Chen M. Metabolic reprogramming in CD8( + ) T cells during acute viral infections. Front Immunol. 2020;11:1013. doi: 10.3389/fimmu.2020.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kreijtz JH, Fouchier RA, Rimmelzwaan GF. Immune responses to influenza virus infection. Virus Res. 2011;162:19–30. doi: 10.1016/j.virusres.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 216.Xu X, et al. Autophagy is essential for effector CD8( + ) T cell survival and memory formation. Nat. Immunol. 2014;15:1152–1161. doi: 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kalia V, et al. Metabolic regulation by PD-1 signaling promotes long-lived quiescent CD8 T cell memory in mice. Sci. Transl. Med. 2021;13:eaba6006. doi: 10.1126/scitranslmed.aba6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Dominguez CX, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 2015;212:2041–2056. doi: 10.1084/jem.20150186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chung HK, McDonald B, Kaech SM. The architectural design of CD8 + T cell responses in acute and chronic infection: Parallel structures with divergent fates. J. Exp. Med. 2021;218:e20201730. doi: 10.1084/jem.20201730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Guo A, et al. cBAF complex components and MYC cooperate early in CD8( + ) T cell fate. Nature. 2022;607:135–141. doi: 10.1038/s41586-022-04849-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bottcher JP, et al. Functional classification of memory CD8( + ) T cells by CX3CR1 expression. Nat. Commun. 2015;6:8306. doi: 10.1038/ncomms9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Gerlach C, et al. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity. 2016;45:1270–1284. doi: 10.1016/j.immuni.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annu. Rev. Immunol. 2003;21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- 224.Perdomo-Celis F, Taborda NA, Rugeles MT. CD8( + ) T-cell response to HIV infection in the era of antiretroviral therapy. Front. Immunol. 2019;10:1896. doi: 10.3389/fimmu.2019.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Moretto MM, Harrow DI, Khan IA. Effector CD8 T cell immunity in microsporidial infection: a lone defense mechanism. Semin. Immunopathol. 2015;37:281–287. doi: 10.1007/s00281-015-0482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sung PS, Racanelli V, Shin EC. CD8( + ) T-cell responses in acute hepatitis C virus infection. Front. Immunol. 2014;5:266. doi: 10.3389/fimmu.2014.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Rha MS, Shin EC. Activation or exhaustion of CD8( + ) T cells in patients with COVID-19. Cell Mol. Immunol. 2021;18:2325–2333. doi: 10.1038/s41423-021-00750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mathew D, et al. Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:eabc8511. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sekine T, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168 e114. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Song JW, et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020;11:3410. doi: 10.1038/s41467-020-17240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kuri-Cervantes L, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5:eabd7114. doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Adamo S, et al. Profound dysregulation of T cell homeostasis and function in patients with severe COVID-19. Allergy. 2021;76:2866–2881. doi: 10.1111/all.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kusnadi A, et al. Severely ill COVID-19 patients display impaired exhaustion features in SARS-CoV-2-reactive CD8(+) T cells. Sci. Immunol. 2021;6:eabe4782. doi: 10.1126/sciimmunol.abe4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Jankovic D, Liu Z, Gause WC. Th1- and Th2-cell commitment during infectious disease: asymmetry in divergent pathways. Trends Immunol. 2001;22:450–457. doi: 10.1016/S1471-4906(01)01975-5. [DOI] [PubMed] [Google Scholar]
- 235.Swain SL, McKinstry KK, Strutt TM. Expanding roles for CD4( + ) T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Salgame P. Host innate and Th1 responses and the bacterial factors that control Mycobacterium tuberculosis infection. Curr. Opin. Immunol. 2005;17:374–380. doi: 10.1016/j.coi.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 237.Miller SM, et al. Novel lipidated imidazoquinoline TLR7/8 adjuvants elicit influenza-specific Th1 immune responses and protect against heterologous H3N2 influenza challenge in mice. Front. Immunol. 2020;11:406. doi: 10.3389/fimmu.2020.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Grifoni A, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501 e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Alhetheel A, et al. Assessment of Th1/Th2 cytokines among patients with Middle East respiratory syndrome coronavirus infection. Int. Immunol. 2020;32:799–804. doi: 10.1093/intimm/dxaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Theofilopoulos AN, Koundouris S, Kono DH, Lawson BR. The role of IFN-gamma in systemic lupus erythematosus: a challenge to the Th1/Th2 paradigm in autoimmunity. Arthritis Res. 2001;3:136–141. doi: 10.1186/ar290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Aleebrahim-Dehkordi E, et al. T helper type (Th1/Th2) responses to SARS-CoV-2 and influenza A (H1N1) virus: from cytokines produced to immune responses. Transpl. Immunol. 2022;70:101495. doi: 10.1016/j.trim.2021.101495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Shanmugasundaram, U. et al. Pulmonary Mycobacterium tuberculosis control associates with CXCR3- and CCR6-expressing antigen-specific Th1 and Th17 cell recruitment. JCI Insight. 5, e137858 (2020). [DOI] [PMC free article] [PubMed]
- 243.Bartsch P, et al. Th17 cell plasticity towards a T-bet-dependent Th1 phenotype is required for bacterial control in Staphylococcus aureus infection. PLoS Pathog. 2022;18:e1010430. doi: 10.1371/journal.ppat.1010430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Mahallawi WH, et al. MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine. 2018;104:8–13. doi: 10.1016/j.cyto.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kuczera D, et al. Isolation of dengue virus serotype 4 genotype II from a patient with high viral load and a mixed Th1/Th17 inflammatory cytokine profile in South Brazil. Virol. J. 2016;13:93. doi: 10.1186/s12985-016-0548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Schiavoni I, et al. Live attenuated B. pertussis BPZE1 rescues the immune functions of Respiratory Syncytial virus infected human dendritic cells by promoting Th1/Th17 responses. PLoS ONE. 2014;9:e100166. doi: 10.1371/journal.pone.0100166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Yan J, et al. Prevalence and clinical relevance of T-helper cells, Th17 and Th1, in hepatitis B virus-related hepatocellular carcinoma. PLoS ONE. 2014;9:e96080. doi: 10.1371/journal.pone.0096080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Gupta G, et al. Th1/Th2/Th17 cytokine profile among different stages of COVID-19 infection. Natl Acad. Sci. Lett. 2022;45:363–369. doi: 10.1007/s40009-022-01123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Rudner XL, Happel KI, Young EA, Shellito JE. Interleukin-23 (IL-23)-IL-17 cytokine axis in murine Pneumocystis carinii infection. Infect. Immun. 2007;75:3055–3061. doi: 10.1128/IAI.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J. Infect. Dis. 2004;190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 251.Trifari S, et al. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 252.Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2:403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fujita H. The role of IL-22 and Th22 cells in human skin diseases. J. Dermatol. Sci. 2013;72:3–8. doi: 10.1016/j.jdermsci.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 254.Duhen T, et al. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 255.Takeuchi A, Saito T. CD4 CTL, a cytotoxic subset of CD4( + ) T cells, their differentiation and function. Front. Immunol. 2017;8:194. doi: 10.3389/fimmu.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hoeks C, Duran G, Hellings N, Broux B. When helpers go above and beyond: development and characterization of cytotoxic CD4( + ) T cells. Front. Immunol. 2022;13:951900. doi: 10.3389/fimmu.2022.951900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hashimoto K, et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proc. Natl Acad. Sci. USA. 2019;116:24242–24251. doi: 10.1073/pnas.1907883116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Poncette L, Bluhm J, Blankenstein T. The role of CD4 T cells in rejection of solid tumors. Curr. Opin. Immunol. 2022;74:18–24. doi: 10.1016/j.coi.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Xie Y, et al. Naive tumor-specific CD4( + ) T cells differentiated in vivo eradicate established melanoma. J. Exp. Med. 2010;207:651–667. doi: 10.1084/jem.20091921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zaunders JJ, et al. Identification of circulating antigen-specific CD4 + T lymphocytes with a CCR5 + , cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 2004;103:2238–2247. doi: 10.1182/blood-2003-08-2765. [DOI] [PubMed] [Google Scholar]
- 261.Soghoian DZ, et al. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci. Transl. Med. 2012;4:123ra125. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Aslan N, et al. Cytotoxic CD4 T cells in viral hepatitis. J. Viral Hepat. 2006;13:505–514. doi: 10.1111/j.1365-2893.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 263.Choi IK, et al. Signaling by the Epstein-Barr virus LMP1 protein induces potent cytotoxic CD4(+) and CD8( + ) T cell responses. Proc. Natl Acad. Sci. USA. 2018;115:E686–E695. doi: 10.1073/pnas.1713607115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Weiskopf D, et al. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4 + T cells associated with protective immunity. Proc. Natl Acad. Sci. USA. 2015;112:E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wilkinson TM, et al. Preexisting influenza-specific CD4 + T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012;18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 266.Hua L, et al. Cytokine-dependent induction of CD4 + T cells with cytotoxic potential during influenza virus infection. J. Virol. 2013;87:11884–11893. doi: 10.1128/JVI.01461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Meckiff BJ, et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4( + ) T cells in COVID-19. Cell. 2020;183:1340–1353 e1316. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Cachot, A. et al. Tumor-specific cytolytic CD4 T cells mediate immunity against human cancer. Sci Adv. 7, eabe3348 (2021). [DOI] [PMC free article] [PubMed]
- 269.Hidalgo LG, Einecke G, Allanach K, Halloran PF. The transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4( + ) CTL, CD8( + ) CTL and NK cells. Am. J. Transpl. 2008;8:627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- 270.Canaday DH, et al. CD4(+) and CD8( + ) T cells kill intracellular Mycobacterium tuberculosis by a perforin and Fas/Fas ligand-independent mechanism. J. Immunol. 2001;167:2734–2742. doi: 10.4049/jimmunol.167.5.2734. [DOI] [PubMed] [Google Scholar]
- 271.Mucida D, et al. Transcriptional reprogramming of mature CD4(+) helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 2013;14:281–289. doi: 10.1038/ni.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Reis BS, et al. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4( + ) T cell immunity. Nat. Immunol. 2013;14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Eshima K, et al. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4(+) Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol. Lett. 2012;144:7–15. doi: 10.1016/j.imlet.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 274.Workman AM, et al. Inflammation enhances IL-2 driven differentiation of cytolytic CD4 T cells. PLoS ONE. 2014;9:e89010. doi: 10.1371/journal.pone.0089010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Brown DM, Lampe AT, Workman AM. The differentiation and protective function of cytolytic CD4 T cells in influenza infection. Front. Immunol. 2016;7:93. doi: 10.3389/fimmu.2016.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Preglej T, Ellmeier W. CD4(+) cytotoxic T cells - Phenotype, function and transcriptional networks controlling their differentiation pathways. Immunol. Lett. 2022;247:27–42. doi: 10.1016/j.imlet.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 277.Patil, V. S. et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci. Immunol. 3, eaan8664 (2018). [DOI] [PMC free article] [PubMed]
- 278.Lyu M, et al. Dissecting the landscape of activated CMV-stimulated CD4 + T cells in humans by linking single-cell RNA-seq with T-cell receptor sequencing. Front. Immunol. 2021;12:779961. doi: 10.3389/fimmu.2021.779961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Juno JA, et al. Cytotoxic CD4 T cells-friend or foe during viral infection? Front. Immunol. 2017;8:19. doi: 10.3389/fimmu.2017.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Sanchez-Martinez A, et al. Cytotoxic CD4( + ) T-cells during HIV infection: targets or weapons? J. Clin. Virol. 2019;119:17–23. doi: 10.1016/j.jcv.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 281.Tian Y, Sette A, Weiskopf D. Cytotoxic CD4 T cells: differentiation, function, and application to dengue virus infection. Front. Immunol. 2016;7:531. doi: 10.3389/fimmu.2016.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhang JY, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 283.Kaneko N, et al. Temporal changes in T cell subsets and expansion of cytotoxic CD4+ T cells in the lungs in severe COVID-19. Clin. Immunol. 2022;237:108991. doi: 10.1016/j.clim.2022.108991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Abebe F. Synergy between Th1 and Th2 responses during Mycobacterium tuberculosis infection: a review of current understanding. Int. Rev. Immunol. 2019;38:172–179. doi: 10.1080/08830185.2019.1632842. [DOI] [PubMed] [Google Scholar]
- 285.Lyadova IV, Panteleev AV. Th1 and Th17 cells in tuberculosis: protection, pathology, and biomarkers. Mediators Inflamm. 2015;2015:854507. doi: 10.1155/2015/854507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ma X, et al. Th17 cells are associated with the Th1/Th2cell balance during Echinococcus multilocularis infection. Mol. Med. Rep. 2014;10:236–240. doi: 10.3892/mmr.2014.2170. [DOI] [PubMed] [Google Scholar]
- 287.Murdock BJ, et al. Coevolution of TH1, TH2, and TH17 responses during repeated pulmonary exposure to Aspergillus fumigatus conidia. Infect. Immun. 2011;79:125–135. doi: 10.1128/IAI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Gorenec L, et al. The comparison of Th1, Th2, Th9, Th17 and Th22 cytokine profiles in acute and chronic HIV-1 infection. Micro. Pathog. 2016;97:125–130. doi: 10.1016/j.micpath.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 289.Cardona P, Cardona PJ. Regulatory T cells in Mycobacterium tuberculosis infection. Front. Immunol. 2019;10:2139. doi: 10.3389/fimmu.2019.02139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Xu Z, Jiang X, Dai X, Li B. The dynamic role of FOXP3(+) tregs and their potential therapeutic applications during SARS-CoV-2 infection. Front. Immunol. 2022;13:916411. doi: 10.3389/fimmu.2022.916411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.White MPJ, McManus CM, Maizels RM. Regulatory T-cells in helminth infection: induction, function and therapeutic potential. Immunology. 2020;160:248–260. doi: 10.1111/imm.13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wan Z, et al. Regulatory T cells and T helper 17 cells in viral infection. Scand. J. Immunol. 2020;91:e12873. doi: 10.1111/sji.12873. [DOI] [PubMed] [Google Scholar]
- 293.Infante-Duarte C, Kamradt T. Th1/Th2 balance in infection. Springe. Semin Immunopathol. 1999;21:317–338. doi: 10.1007/BF00812260. [DOI] [PubMed] [Google Scholar]
- 294.Cox N, Pokrovskii M, Vicario R, Geissmann F. Origins, biology, and diseases of tissue macrophages. Annu. Rev. Immunol. 2021;39:313–344. doi: 10.1146/annurev-immunol-093019-111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bashir S, Sharma Y, Elahi A, Khan F. Macrophage polarization: the link between inflammation and related diseases. Inflamm. Res. 2016;65:1–11. doi: 10.1007/s00011-015-0874-1. [DOI] [PubMed] [Google Scholar]
- 296.Battegay M, et al. Enhanced establishment of a virus carrier state in adult CD4 + T-cell-deficient mice. J. Virol. 1994;68:4700–4704. doi: 10.1128/jvi.68.7.4700-4704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hamilton SE, Tvinnereim AR, Harty JT. Listeria monocytogenes infection overcomes the requirement for CD40 ligand in exogenous antigen presentation to CD8( + ) T cells. J. Immunol. 2001;167:5603–5609. doi: 10.4049/jimmunol.167.10.5603. [DOI] [PubMed] [Google Scholar]
- 298.Krawczyk CM, Shen H, Pearce EJ. Memory CD4 T cells enhance primary CD8 T-cell responses. Infect. Immun. 2007;75:3556–3560. doi: 10.1128/IAI.00086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Serre K, et al. CD4 T cell help is required for primary CD8 T cell responses to vesicular antigen delivered to dendritic cells in vivo. Eur. J. Immunol. 2006;36:1386–1397. doi: 10.1002/eji.200526193. [DOI] [PubMed] [Google Scholar]
- 300.Flinsenberg TW, et al. Cognate CD4 T-cell licensing of dendritic cells heralds anti-cytomegalovirus CD8 T-cell immunity after human allogeneic umbilical cord blood transplantation. J. Virol. 2015;89:1058–1069. doi: 10.1128/JVI.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Wang JC, Livingstone AM. Cutting edge: CD4 + T cell help can be essential for primary CD8 + T cell responses in vivo. J. Immunol. 2003;171:6339–6343. doi: 10.4049/jimmunol.171.12.6339. [DOI] [PubMed] [Google Scholar]
- 302.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 303.Bourgeois C, Rocha B, Tanchot C. A role for CD40 expression on CD8 + T cells in the generation of CD8 + T cell memory. Science. 2002;297:2060–2063. doi: 10.1126/science.1072615. [DOI] [PubMed] [Google Scholar]
- 304.Janssen EM, et al. CD4 + T cells are required for secondary expansion and memory in CD8 + T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 305.Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441:890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Barker BR, et al. Critical role for IL-21 in both primary and memory anti-viral CD8 + T-cell responses. Eur. J. Immunol. 2010;40:3085–3096. doi: 10.1002/eji.200939939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Sokke Umeshappa C, et al. CD154 and IL-2 signaling of CD4 + T cells play a critical role in multiple phases of CD8 + CTL responses following adenovirus vaccination. PLoS ONE. 2012;7:e47004. doi: 10.1371/journal.pone.0047004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Obar JJ, et al. CD4 + T cell regulation of CD25 expression controls development of short-lived effector CD8 + T cells in primary and secondary responses. Proc. Natl Acad. Sci. USA. 2010;107:193–198. doi: 10.1073/pnas.0909945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Fuse S, et al. Recall responses by helpless memory CD8 + T cells are restricted by the up-regulation of PD-1. J. Immunol. 2009;182:4244–4254. doi: 10.4049/jimmunol.0802041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Huang Q, et al. Molecular basis of the differentiation and function of virus specific follicular helper CD4( + ) T cells. Front. Immunol. 2019;10:249. doi: 10.3389/fimmu.2019.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Locci M, et al. Human circulating PD-1 + CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Juno JA, et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- 313.Boppana S, et al. SARS-CoV-2-specific circulating T follicular helper cells correlate with neutralizing antibodies and increase during early convalescence. PLoS Pathog. 2021;17:e1009761. doi: 10.1371/journal.ppat.1009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kaneko N, et al. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell. 2020;183:143–157 e113. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Kato LM, Kawamoto S, Maruya M, Fagarasan S. Gut TFH and IgA: key players for regulation of bacterial communities and immune homeostasis. Immunol. Cell Biol. 2014;92:49–56. doi: 10.1038/icb.2013.54. [DOI] [PubMed] [Google Scholar]
- 316.Blank CU, et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019;19:665–674. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wherry EJ. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 318.Reignat S, et al. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. J. Exp. Med. 2002;195:1089–1101. doi: 10.1084/jem.20011723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Shankar P, et al. Impaired function of circulating HIV-specific CD8( + ) T cells in chronic human immunodeficiency virus infection. Blood. 2000;96:3094–3101. doi: 10.1182/blood.V96.9.3094.h8003094_3094_3101. [DOI] [PubMed] [Google Scholar]
- 320.Gruener NH, et al. Sustained dysfunction of antiviral CD8 + T lymphocytes after infection with hepatitis C virus. J. Virol. 2001;75:5550–5558. doi: 10.1128/JVI.75.12.5550-5558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kahan SM, Wherry EJ, Zajac AJ. T cell exhaustion during persistent viral infections. Virology. 2015;479-480:180–193. doi: 10.1016/j.virol.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Zheng L, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474. doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 323.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 324.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356 e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 325.Hashimoto M, et al. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu. Rev. Med. 2018;69:301–318. doi: 10.1146/annurev-med-012017-043208. [DOI] [PubMed] [Google Scholar]
- 326.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 2015;15:486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wherry EJ, et al. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Mackerness KJ, et al. Pronounced virus-dependent activation drives exhaustion but sustains IFN-gamma transcript levels. J. Immunol. 2010;185:3643–3651. doi: 10.4049/jimmunol.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Wherry EJ, et al. Molecular signature of CD8 + T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 330.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 331.Wherry EJ, et al. Antigen-independent memory CD8 T cells do not develop during chronic viral infection. Proc. Natl Acad. Sci. USA. 2004;101:16004–16009. doi: 10.1073/pnas.0407192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Radziewicz H, et al. Liver-infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD-1 and low levels of CD127 expression. J. Virol. 2007;81:2545–2553. doi: 10.1128/JVI.02021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Shin H, Blackburn SD, Blattman JN, Wherry EJ. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J. Exp. Med. 2007;204:941–949. doi: 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Paley MA, et al. Progenitor and terminal subsets of CD8 + T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Blackburn SD, et al. Coregulation of CD8 + T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Thommen DS, et al. Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol. Res. 2015;3:1344–1355. doi: 10.1158/2326-6066.CIR-15-0097. [DOI] [PubMed] [Google Scholar]
- 337.Penaloza-MacMaster P, et al. Opposing effects of CD70 costimulation during acute and chronic lymphocytic choriomeningitis virus infection of mice. J. Virol. 2011;85:6168–6174. doi: 10.1128/JVI.02205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Esensten JH, et al. CD28 costimulation: from mechanism to therapy. Immunity. 2016;44:973–988. doi: 10.1016/j.immuni.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Hui E, et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Philip M, Schietinger A. CD8( + ) T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 2022;22:209–223. doi: 10.1038/s41577-021-00574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Schietinger A, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 2016;45:389–401. doi: 10.1016/j.immuni.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Angelosanto JM, Blackburn SD, Crawford A, Wherry EJ. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 2012;86:8161–8170. doi: 10.1128/JVI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Yao C, et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8( + ) T cell persistence in chronic infection. Nat. Immunol. 2019;20:890–901. doi: 10.1038/s41590-019-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 346.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Lu Y, Ye C, Yuan Y. Phenotypic characteristics and T cell receptor properties in melanoma: deciphering the correlation at single-cell resolution. Signal Transduct. Target Ther. 2022;7:5. doi: 10.1038/s41392-021-00864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308 e1236. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Zhang L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564:268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 350.van der Leun AM, Thommen DS, Schumacher TN. CD8(+) T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer. 2020;20:218–232. doi: 10.1038/s41568-019-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Hudson, W. H. & Wieland, A. Technology meets TILs: deciphering T cell function in the -omics era. Cancer Cell41, 41–57. (2022). [DOI] [PMC free article] [PubMed]
- 352.Dolina JS, Van Braeckel-Budimir N, Thomas GD, Salek-Ardakani S. CD8(+) T cell exhaustion in cancer. Front. Immunol. 2021;12:715234. doi: 10.3389/fimmu.2021.715234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Utzschneider DT, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 355.Beltra JC, et al. Developmental relationships of four exhausted CD8(+) T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity. 2020;52:825–841 e828. doi: 10.1016/j.immuni.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Tsui C, et al. MYB orchestrates T cell exhaustion and response to checkpoint inhibition. Nature. 2022;609:354–360. doi: 10.1038/s41586-022-05105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Galletti G, et al. Two subsets of stem-like CD8(+) memory T cell progenitors with distinct fate commitments in humans. Nat. Immunol. 2020;21:1552–1562. doi: 10.1038/s41590-020-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Baharom F, et al. Intravenous nanoparticle vaccination generates stem-like TCF1(+) neoantigen-specific CD8(+) T cells. Nat. Immunol. 2021;22:41–52. doi: 10.1038/s41590-020-00810-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Hudson WH, et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1(+) stem-like CD8(+) T cells during chronic infection. Immunity. 2019;51:1043–1058 e1044. doi: 10.1016/j.immuni.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Zander R, et al. CD4(+) T cell help is required for the formation of a cytolytic CD8(+) T cell subset that protects against chronic infection and cancer. Immunity. 2019;51:1028–1042 e1024. doi: 10.1016/j.immuni.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kanev K, et al. Proliferation-competent Tcf1+ CD8 T cells in dysfunctional populations are CD4 T cell help independent. Proc. Natl Acad. Sci. USA. 2019;116:20070–20076. doi: 10.1073/pnas.1902701116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Giles JR, et al. Shared and distinct biological circuits in effector, memory and exhausted CD8(+) T cells revealed by temporal single-cell transcriptomics and epigenetics. Nat. Immunol. 2022;23:1600–1613. doi: 10.1038/s41590-022-01338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Eberhardt CS, et al. Functional HPV-specific PD-1(+) stem-like CD8 T cells in head and neck cancer. Nature. 2021;597:279–284. doi: 10.1038/s41586-021-03862-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Sandu I, et al. Landscape of exhausted virus-specific CD8 T cells in chronic LCMV infection. Cell Rep. 2020;32:108078. doi: 10.1016/j.celrep.2020.108078. [DOI] [PubMed] [Google Scholar]
- 365.Bengsch B, et al. Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity. 2018;48:1029–1045 e1025. doi: 10.1016/j.immuni.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Lowery FJ, et al. Molecular signatures of antitumor neoantigen-reactive T cells from metastatic human cancers. Science. 2022;375:877–884. doi: 10.1126/science.abl5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Chen Z, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51:840–855 e845. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Pais Ferreira D, et al. Central memory CD8(+) T cells derive from stem-like Tcf7(hi) effector cells in the absence of cytotoxic differentiation. Immunity. 2020;53:985–1000 e1011. doi: 10.1016/j.immuni.2020.09.005. [DOI] [PubMed] [Google Scholar]
- 369.Shan Q, et al. Ectopic Tcf1 expression instills a stem-like program in exhausted CD8(+) T cells to enhance viral and tumor immunity. Cell Mol. Immunol. 2021;18:1262–1277. doi: 10.1038/s41423-020-0436-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Zhang J, Lyu T, Cao Y, Feng H. Role of TCF-1 in differentiation, exhaustion, and memory of CD8(+) T cells: a review. FASEB J. 2021;35:e21549. doi: 10.1096/fj.202002566R. [DOI] [PubMed] [Google Scholar]
- 371.Marcel N, Hedrick SM. A key control point in the T cell response to chronic infection and neoplasia: FOXO1. Curr. Opin. Immunol. 2020;63:51–60. doi: 10.1016/j.coi.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Wu, T. et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol. 1, eaai8593 (2016). [DOI] [PMC free article] [PubMed]
- 373.Gautam S, et al. The transcription factor c-Myb regulates CD8(+) T cell stemness and antitumor immunity. Nat. Immunol. 2019;20:337–349. doi: 10.1038/s41590-018-0311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Yao C, et al. BACH2 enforces the transcriptional and epigenetic programs of stem-like CD8(+) T cells. Nat. Immunol. 2021;22:370–380. doi: 10.1038/s41590-021-00868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Alfei F, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571:265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 376.Khan O, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571:211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Sekine, T. et al. TOX is expressed by exhausted and polyfunctional human effector memory CD8(+) T cells. Sci Immunol. 5, eaba7918 (2020). [DOI] [PubMed]
- 378.Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571:270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Liu X, et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. 2019;567:525–529. doi: 10.1038/s41586-019-0979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Li F, Zhang Y. Targeting NR4As, a new strategy to fine-tune CAR-T cells against solid tumors. Signal Transduct. Target Ther. 2019;4:7. doi: 10.1038/s41392-019-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Seo H, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc. Natl Acad. Sci. USA. 2019;116:12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Chen Y, et al. BATF regulates progenitor to cytolytic effector CD8(+) T cell transition during chronic viral infection. Nat. Immunol. 2021;22:996–1007. doi: 10.1038/s41590-021-00965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Seo H, et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 2021;22:983–995. doi: 10.1038/s41590-021-00964-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Quigley M, et al. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 2010;16:1147–1151. doi: 10.1038/nm.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Wei J, et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature. 2019;576:471–476. doi: 10.1038/s41586-019-1821-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Zhang, X. et al. Depletion of BATF in CAR-T cells enhances antitumor activity by inducing resistance against exhaustion and formation of central memory cells. Cancer Cell40, 1407–1422.e7 (2022). [DOI] [PubMed]
- 387.Doering TA, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Li J, et al. High levels of Eomes promote exhaustion of anti-tumor CD8(+) T cells. Front. Immunol. 2018;9:2981. doi: 10.3389/fimmu.2018.02981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Buggert M, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10:e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Kao C, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat. Immunol. 2011;12:663–671. doi: 10.1038/ni.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.McLane LM, et al. Role of nuclear localization in the regulation and function of T-bet and Eomes in exhausted CD8 T cells. Cell Rep. 2021;35:109120. doi: 10.1016/j.celrep.2021.109120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:S67–S79. doi: 10.1016/S0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 393.Martinez GJ, et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity. 2015;42:265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Zhu L, et al. Dapl1 controls NFATc2 activation to regulate CD8(+) T cell exhaustion and responses in chronic infection and cancer. Nat. Cell Biol. 2022;24:1165–1176. doi: 10.1038/s41556-022-00942-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Abdel-Hakeem MS, et al. Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nat. Immunol. 2021;22:1008–1019. doi: 10.1038/s41590-021-00975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Scott-Browne JP, et al. Dynamic changes in chromatin accessibility occur in CD8(+) T cells responding to viral infection. Immunity. 2016;45:1327–1340. doi: 10.1016/j.immuni.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Youngblood B, et al. Chronic virus infection enforces demethylation of the locus that encodes PD-1 in antigen-specific CD8(+) T cells. Immunity. 2011;35:400–412. doi: 10.1016/j.immuni.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Jadhav RR, et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl Acad. Sci. USA. 2019;116:14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.TCF-1 mediates chromatin intermingling during T cell development. Nat. Immunol. 23, 1000–1001, (2022). [DOI] [PubMed]
- 400.Wang W, et al. TCF-1 promotes chromatin interactions across topologically associating domains in T cell progenitors. Nat. Immunol. 2022;23:1052–1062. doi: 10.1038/s41590-022-01232-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Utzschneider DT, et al. T cells maintain an exhausted phenotype after antigen withdrawal and population reexpansion. Nat. Immunol. 2013;14:603–610. doi: 10.1038/ni.2606. [DOI] [PubMed] [Google Scholar]
- 402.Belk JA, Daniel B, Satpathy AT. Epigenetic regulation of T cell exhaustion. Nat. Immunol. 2022;23:848–860. doi: 10.1038/s41590-022-01224-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Ghoneim HE, et al. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 2017;170:142–157 e119. doi: 10.1016/j.cell.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Scheper W, et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 2019;25:89–94. doi: 10.1038/s41591-018-0266-5. [DOI] [PubMed] [Google Scholar]
- 406.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Lonberg N, Korman AJ. Masterful antibodies: checkpoint blockade. Cancer Immunol. Res. 2017;5:275–281. doi: 10.1158/2326-6066.CIR-17-0057. [DOI] [PubMed] [Google Scholar]
- 409.de Miguel M, Calvo E. Clinical challenges of immune checkpoint inhibitors. Cancer Cell. 2020;38:326–333. doi: 10.1016/j.ccell.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 410.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Siddiqui I, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211 e110. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 412.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2019;176:404. doi: 10.1016/j.cell.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J. Exp. Med. 2018;215:2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Ott PA, et al. A phase Ib trial of personalized neoantigen therapy plus anti-PD-1 in patients with advanced melanoma, non-small cell lung cancer, or bladder cancer. Cell. 2020;183:347–362 e324. doi: 10.1016/j.cell.2020.08.053. [DOI] [PubMed] [Google Scholar]
- 415.Caushi JX, et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature. 2021;596:126–132. doi: 10.1038/s41586-021-03752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Oliveira G, et al. Phenotype, specificity and avidity of antitumour CD8(+) T cells in melanoma. Nature. 2021;596:119–125. doi: 10.1038/s41586-021-03704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.LaFleur MW, et al. PTPN2 regulates the generation of exhausted CD8(+) T cell subpopulations and restrains tumor immunity. Nat. Immunol. 2019;20:1335–1347. doi: 10.1038/s41590-019-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Clarke J, et al. Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J. Exp. Med. 2019;216:2128–2149. doi: 10.1084/jem.20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Thommen DS, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Daud AI, et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J. Clin. Invest. 2016;126:3447–3452. doi: 10.1172/JCI87324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Spitzer MH, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502 e415. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Liu B, et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer. 2022;3:108–121. doi: 10.1038/s43018-021-00292-8. [DOI] [PubMed] [Google Scholar]
- 423.Yost KE, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.van Pul KM, Fransen MF, van de Ven R, de Gruijl TD. Immunotherapy goes local: the central role of lymph nodes in driving tumor infiltration and efficacy. Front. Immunol. 2021;12:643291. doi: 10.3389/fimmu.2021.643291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Connolly KA, et al. A reservoir of stem-like CD8(+) T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci. Immunol. 2021;6:eabg7836. doi: 10.1126/sciimmunol.abg7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Schenkel JM, et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1(+) CD8(+) T cells in tumor-draining lymph nodes. Immunity. 2021;54:2338–2353 e2336. doi: 10.1016/j.immuni.2021.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Huang AC, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature. 2017;545:60–65. doi: 10.1038/nature22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Huang, Q. et al. The primordial differentiation of tumor-specific memory CD8(+) T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell185, 4049–4066.e25 (2022). [DOI] [PubMed]
- 429.Dammeijer F, et al. The PD-1/PD-L1-checkpoint restrains T cell immunity in tumor-draining lymph nodes. Cancer Cell. 2020;38:685–700 e688. doi: 10.1016/j.ccell.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 430.Francis, D. M. et al. Blockade of immune checkpoints in lymph nodes through locoregional delivery augments cancer immunotherapy. Sci. Transl. Med. 12, eaay3575 (2020). [DOI] [PMC free article] [PubMed]
- 431.Snell LM, et al. Dynamic CD4(+) T cell heterogeneity defines subset-specific suppression and PD-L1-blockade-driven functional restoration in chronic infection. Nat. Immunol. 2021;22:1524–1537. doi: 10.1038/s41590-021-01060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Brooks DG, Teyton L, Oldstone MB, McGavern DB. Intrinsic functional dysregulation of CD4 T cells occurs rapidly following persistent viral infection. J. Virol. 2005;79:10514–10527. doi: 10.1128/JVI.79.16.10514-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Oxenius A, Zinkernagel RM, Hengartner H. Comparison of activation versus induction of unresponsiveness of virus-specific CD4+ and CD8+ T cells upon acute versus persistent viral infection. Immunity. 1998;9:449–457. doi: 10.1016/S1074-7613(00)80628-7. [DOI] [PubMed] [Google Scholar]
- 434.Elsaesser H, Sauer K, Brooks DG. IL-21 is required to control chronic viral infection. Science. 2009;324:1569–1572. doi: 10.1126/science.1174182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. doi: 10.1126/science.1175194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Parish IA, et al. Chronic viral infection promotes sustained Th1-derived immunoregulatory IL-10 via BLIMP-1. J. Clin. Invest. 2014;124:3455–3468. doi: 10.1172/JCI66108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Frohlich A, et al. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science. 2009;324:1576–1580. doi: 10.1126/science.1172815. [DOI] [PubMed] [Google Scholar]
- 439.Crawford A, et al. Molecular and transcriptional basis of CD4(+) T cell dysfunction during chronic infection. Immunity. 2014;40:289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Snell LM, et al. Overcoming CD4 Th1 cell fate restrictions to sustain antiviral CD8 T cells and control persistent virus infection. Cell Rep. 2016;16:3286–3296. doi: 10.1016/j.celrep.2016.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Xia Y, et al. BCL6-dependent TCF-1(+) progenitor cells maintain effector and helper CD4(+) T cell responses to persistent antigen. Immunity. 2022;55:1200–1215 e1206. doi: 10.1016/j.immuni.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Mann GJ, et al. BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J. Invest. Dermatol. 2013;133:509–517. doi: 10.1038/jid.2012.283. [DOI] [PubMed] [Google Scholar]
- 443.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Ascierto ML, et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res. Treat. 2012;131:871–880. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Leffers N, et al. Identification of genes and pathways associated with cytotoxic T lymphocyte infiltration of serous ovarian cancer. Br. J. Cancer. 2010;103:685–692. doi: 10.1038/sj.bjc.6605820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Laheurte C, et al. Distinct prognostic value of circulating anti-telomerase CD4(+) Th1 immunity and exhausted PD-1(+)/TIM-3(+) T cells in lung cancer. Br. J. Cancer. 2019;121:405–416. doi: 10.1038/s41416-019-0531-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 448.Xu X, et al. Expression of Th1- Th2- and Th17-associated cytokines in laryngeal carcinoma. Oncol. Lett. 2016;12:1941–1948. doi: 10.3892/ol.2016.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Bos R, Sherman LA. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 2010;70:8368–8377. doi: 10.1158/0008-5472.CAN-10-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Dengel LT, et al. Interferons induce CXCR3-cognate chemokine production by human metastatic melanoma. J. Immunother. 2010;33:965–974. doi: 10.1097/CJI.0b013e3181fb045d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Zuazo M, et al. Systemic CD4 immunity as a key contributor to PD-L1/PD-1 blockade immunotherapy efficacy. Front. Immunol. 2020;11:586907. doi: 10.3389/fimmu.2020.586907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.House IG, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin. Cancer Res. 2020;26:487–504. doi: 10.1158/1078-0432.CCR-19-1868. [DOI] [PubMed] [Google Scholar]
- 453.Harlin H, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Wendel M, Galani IE, Suri-Payer E, Cerwenka A. Natural killer cell accumulation in tumors is dependent on IFN-gamma and CXCR3 ligands. Cancer Res. 2008;68:8437–8445. doi: 10.1158/0008-5472.CAN-08-1440. [DOI] [PubMed] [Google Scholar]
- 455.Konjevic GM, et al. The role of cytokines in the regulation of NK cells in the tumor environment. Cytokine. 2019;117:30–40. doi: 10.1016/j.cyto.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 456.Jabrane-Ferrat N, et al. Effect of gamma interferon on HLA class-I and -II transcription and protein expression in human breast adenocarcinoma cell lines. Int. J. Cancer. 1990;45:1169–1176. doi: 10.1002/ijc.2910450630. [DOI] [PubMed] [Google Scholar]
- 457.Shankaran V, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 458.Chraa D, Naim A, Olive D, Badou A. T lymphocyte subsets in cancer immunity: friends or foes. J. Leukoc. Biol. 2019;105:243–255. doi: 10.1002/JLB.MR0318-097R. [DOI] [PubMed] [Google Scholar]
- 459.De Monte L, et al. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011;208:469–478. doi: 10.1084/jem.20101876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Yoon NK, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum. Pathol. 2010;41:1794–1801. doi: 10.1016/j.humpath.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Tepper RI, Coffman RL, Leder P. An eosinophil-dependent mechanism for the antitumor effect of interleukin-4. Science. 1992;257:548–551. doi: 10.1126/science.1636093. [DOI] [PubMed] [Google Scholar]
- 462.Hung K, et al. The central role of CD4(+) T cells in the antitumor immune response. J. Exp. Med. 1998;188:2357–2368. doi: 10.1084/jem.188.12.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Lorvik KB, et al. Adoptive transfer of tumor-specific Th2 cells eradicates tumors by triggering an in situ inflammatory immune response. Cancer Res. 2016;76:6864–6876. doi: 10.1158/0008-5472.CAN-16-1219. [DOI] [PubMed] [Google Scholar]
- 464.Kitajima M, et al. Memory type 2 helper T cells induce long-lasting antitumor immunity by activating natural killer cells. Cancer Res. 2011;71:4790–4798. doi: 10.1158/0008-5472.CAN-10-1572. [DOI] [PubMed] [Google Scholar]
- 465.Boieri, M. et al. CD4+ T helper 2 cells suppress breast cancer by inducing terminal differentiation. J. Exp. Med. 219, e20201963 (2022). [DOI] [PMC free article] [PubMed]
- 466.Rodriguez-Tirado, C. et al. Interleukin 4 controls the pro-tumoral role of macrophages in mammary cancer pulmonary metastasis in mice. Cancers14, 4336 (2022). [DOI] [PMC free article] [PubMed]
- 467.Lazarski CA, et al. IL-4 attenuates Th1-associated chemokine expression and Th1 trafficking to inflamed tissues and limits pathogen clearance. PLoS ONE. 2013;8:e71949. doi: 10.1371/journal.pone.0071949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Mitchell RE, et al. IL-4 enhances IL-10 production in Th1 cells: implications for Th1 and Th2 regulation. Sci. Rep. 2017;7:11315. doi: 10.1038/s41598-017-11803-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Kusuda T, et al. Relative expression levels of Th1 and Th2 cytokine mRNA are independent prognostic factors in patients with ovarian cancer. Oncol. Rep. 2005;13:1153–1158. [PubMed] [Google Scholar]
- 470.Qin H, et al. Pan-cancer analysis identifies LMNB1 as a target to redress Th1/Th2 imbalance and enhance PARP inhibitor response in human cancers. Cancer Cell Int. 2022;22:101. doi: 10.1186/s12935-022-02467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Lee HL, et al. Inflammatory cytokines and change of Th1/Th2 balance as prognostic indicators for hepatocellular carcinoma in patients treated with transarterial chemoembolization. Sci. Rep. 2019;9:3260. doi: 10.1038/s41598-019-40078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Johansson M, Denardo DG, Coussens LM. Polarized immune responses differentially regulate cancer development. Immunol. Rev. 2008;222:145–154. doi: 10.1111/j.1600-065X.2008.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Asadzadeh Z, et al. The paradox of Th17 cell functions in tumor immunity. Cell Immunol. 2017;322:15–25. doi: 10.1016/j.cellimm.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 474.Kryczek I, et al. Phenotype, distribution, generation, and functional and clinical relevance of Th17 cells in the human tumor environments. Blood. 2009;114:1141–1149. doi: 10.1182/blood-2009-03-208249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Miyahara Y, et al. Generation and regulation of human CD4+ IL-17-producing T cells in ovarian cancer. Proc. Natl Acad. Sci. USA. 2008;105:15505–15510. doi: 10.1073/pnas.0710686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Bronte V. Th17 and cancer: friends or foes? Blood. 2008;112:214. doi: 10.1182/blood-2008-04-149260. [DOI] [PubMed] [Google Scholar]
- 477.Punt S, et al. The correlations between IL-17 vs. Th17 cells and cancer patient survival: a systematic review. Oncoimmunology. 2015;4:e984547. doi: 10.4161/2162402X.2014.984547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Wilke CM, et al. Th17 cells in cancer: help or hindrance? Carcinogenesis. 2011;32:643–649. doi: 10.1093/carcin/bgr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Ben Khelil, M. et al. Harnessing antitumor CD4(+) T cells for cancer immunotherapy. Cancers14, 260 (2022). [DOI] [PMC free article] [PubMed]
- 480.Singh N, et al. Inflammation and cancer. Ann. Afr. Med. 2019;18:121–126. doi: 10.4103/aam.aam_56_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Qianmei Y, et al. Recent advances in the role of Th17/Treg cells in tumor immunity and tumor therapy. Immunol. Res. 2021;69:398–414. doi: 10.1007/s12026-021-09211-6. [DOI] [PubMed] [Google Scholar]
- 482.Fabre, J. A. S. et al. The interleukin-17 family of cytokines in breast cancer. Int. J. Mol. Sci. 19, 3880 (2018). [DOI] [PMC free article] [PubMed]
- 483.Wang L, et al. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 2009;206:1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Bi L, et al. Increased Th17 cells and IL-17A exist in patients with B cell acute lymphoblastic leukemia and promote proliferation and resistance to daunorubicin through activation of Akt signaling. J. Transl. Med. 2016;14:132. doi: 10.1186/s12967-016-0894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Do Thi VA, Park SM, Lee H, Kim YS. The membrane-bound form of IL-17A promotes the growth and tumorigenicity of colon cancer cells. Mol. Cells. 2016;39:536–542. doi: 10.14348/molcells.2016.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Shahid A, Bharadwaj M. The connection between the Th17 cell related cytokines and cancer stem cells in cancer: Novel therapeutic targets. Immunol. Lett. 2019;213:9–20. doi: 10.1016/j.imlet.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 487.Xiang T, et al. Interleukin-17 produced by tumor microenvironment promotes self-renewal of CD133+ cancer stem-like cells in ovarian cancer. Oncogene. 2015;34:165–176. doi: 10.1038/onc.2013.537. [DOI] [PubMed] [Google Scholar]
- 488.Salazar Y, et al. Microenvironmental Th9 and Th17 lymphocytes induce metastatic spreading in lung cancer. J. Clin. Invest. 2020;130:3560–3575. doi: 10.1172/JCI124037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Li J, et al. Interleukin 17 A promotes hepatocellular carcinoma metastasis via NF-kB induced matrix metalloproteinases 2 and 9 expression. PLoS ONE. 2011;6:e21816. doi: 10.1371/journal.pone.0021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Wu HH, et al. Targeting IL-17B-IL-17RB signaling with an anti-IL-17RB antibody blocks pancreatic cancer metastasis by silencing multiple chemokines. J. Exp. Med. 2015;212:333–349. doi: 10.1084/jem.20141702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Numasaki M, et al. Interleukin-17 promotes angiogenesis and tumor growth. Blood. 2003;101:2620–2627. doi: 10.1182/blood-2002-05-1461. [DOI] [PubMed] [Google Scholar]
- 492.Pan B, et al. Interleukin-17 promotes angiogenesis by stimulating VEGF production of cancer cells via the STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci. Rep. 2015;5:16053. doi: 10.1038/srep16053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Huang Q, et al. IL-17 promotes angiogenic factors IL-6, IL-8, and Vegf production via Stat1 in lung adenocarcinoma. Sci. Rep. 2016;6:36551. doi: 10.1038/srep36551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.He D, et al. IL-17 promotes tumor development through the induction of tumor promoting microenvironments at tumor sites and myeloid-derived suppressor cells. J. Immunol. 2010;184:2281–2288. doi: 10.4049/jimmunol.0902574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Wen L, et al. Interplay between myeloid-derived suppressor cells (MDSCs) and Th17 cells: foe or friend? Oncotarget. 2016;7:35490–35496. doi: 10.18632/oncotarget.8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Mao H, et al. Feedback mechanisms between M2 macrophages and Th17 cells in colorectal cancer patients. Tumour Biol. 2016;37:12223–12230. doi: 10.1007/s13277-016-5085-z. [DOI] [PubMed] [Google Scholar]
- 497.Shen J, et al. IL-17 induces macrophages to M2-like phenotype via NF-kappaB. Cancer Manag. Res. 2018;10:4217–4228. doi: 10.2147/CMAR.S174899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Ferreira N, et al. IL-17A and IL-17F orchestrate macrophages to promote lung cancer. Cell Oncol. 2020;43:643–654. doi: 10.1007/s13402-020-00510-y. [DOI] [PubMed] [Google Scholar]
- 499.Laan M, et al. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J. Immunol. 1999;162:2347–2352. doi: 10.4049/jimmunol.162.4.2347. [DOI] [PubMed] [Google Scholar]
- 500.Pelletier M, et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood. 2010;115:335–343. doi: 10.1182/blood-2009-04-216085. [DOI] [PubMed] [Google Scholar]
- 501.Wang X, et al. IL-17 constrains natural killer cell activity by restraining IL-15-driven cell maturation via SOCS3. Proc. Natl Acad. Sci. USA. 2019;116:17409–17418. doi: 10.1073/pnas.1904125116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Dadaglio G, et al. IL-17 suppresses the therapeutic activity of cancer vaccines through the inhibition of CD8(+) T-cell responses. Oncoimmunology. 2020;9:1758606. doi: 10.1080/2162402X.2020.1758606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Kim, B. S. et al. Type 17 immunity promotes the exhaustion of CD8(+) T cells in cancer. J. Immunother. Cancer. 9, e002603 (2021). [DOI] [PMC free article] [PubMed]
- 504.Iida T, et al. Tumor-infiltrating CD4+ Th17 cells produce IL-17 in tumor microenvironment and promote tumor progression in human gastric cancer. Oncol. Rep. 2011;25:1271–1277. doi: 10.3892/or.2011.1201. [DOI] [PubMed] [Google Scholar]
- 505.Chung, A. S. et al. An interleukin-17-mediated paracrine network promotes tumor resistance to anti-angiogenic therapy. Nat. Med. 19, 1114–1123 (2013). [DOI] [PubMed]
- 506.Wu L, et al. A novel IL-17 signaling pathway controlling keratinocyte proliferation and tumorigenesis via the TRAF4-ERK5 axis. J. Exp. Med. 2015;212:1571–1587. doi: 10.1084/jem.20150204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Jiang R, et al. IL-22 is related to development of human colon cancer by activation of STAT3. BMC Cancer. 2013;13:59. doi: 10.1186/1471-2407-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Perez LG, et al. Publisher Correction: TGF-beta signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat. Commun. 2020;11:5740. doi: 10.1038/s41467-020-19685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Chen JG, et al. Intratumoral expression of IL-17 and its prognostic role in gastric adenocarcinoma patients. Int. J. Biol. Sci. 2011;7:53–60. doi: 10.7150/ijbs.7.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Lin Y, et al. Interleukin-17 is a favorable prognostic marker for colorectal cancer. Clin. Transl. Oncol. 2015;17:50–56. doi: 10.1007/s12094-014-1197-3. [DOI] [PubMed] [Google Scholar]
- 511.Punt S, et al. FoxP3(+) and IL-17(+) cells are correlated with improved prognosis in cervical adenocarcinoma. Cancer Immunol. Immunother. 2015;64:745–753. doi: 10.1007/s00262-015-1678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Furuta S, et al. IL-25 causes apoptosis of IL-25R-expressing breast cancer cells without toxicity to nonmalignant cells. Sci. Transl. Med. 2011;3:78ra31. doi: 10.1126/scitranslmed.3001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Al Omar S, Flanagan BF, Almehmadi M, Christmas SE. The effects of IL-17 upon human natural killer cells. Cytokine. 2013;62:123–130. doi: 10.1016/j.cyto.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 514.Lu L, et al. IL-17A promotes immune cell recruitment in human esophageal cancers and the infiltrating dendritic cells represent a positive prognostic marker for patient survival. J. Immunother. 2013;36:451–458. doi: 10.1097/CJI.0b013e3182a802cf. [DOI] [PubMed] [Google Scholar]
- 515.Chen CL, et al. IL-17 induces antitumor immunity by promoting beneficial neutrophil recruitment and activation in esophageal squamous cell carcinoma. Oncoimmunology. 2017;7:e1373234. doi: 10.1080/2162402X.2017.1373234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Jovanovic DV, et al. IL-17 stimulates the production and expression of proinflammatory cytokines, IL-beta and TNF-alpha, by human macrophages. J. Immunol. 1998;160:3513–3521. doi: 10.4049/jimmunol.160.7.3513. [DOI] [PubMed] [Google Scholar]
- 517.Kryczek I, et al. Endogenous IL-17 contributes to reduced tumor growth and metastasis. Blood. 2009;114:357–359. doi: 10.1182/blood-2008-09-177360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Majchrzak K, et al. Exploiting IL-17-producing CD4+ and CD8+ T cells to improve cancer immunotherapy in the clinic. Cancer Immunol. Immunother. 2016;65:247–259. doi: 10.1007/s00262-016-1797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Guery L, Hugues S. Th17 cell plasticity and functions in cancer immunity. Biomed. Res. Int. 2015;2015:314620. doi: 10.1155/2015/314620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Shen Y, et al. Fas signaling-mediated TH9 cell differentiation favors bowel inflammation and antitumor functions. Nat. Commun. 2019;10:2924. doi: 10.1038/s41467-019-10889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Purwar R, et al. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat. Med. 2012;18:1248–1253. doi: 10.1038/nm.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Vegran F, et al. The transcription factor IRF1 dictates the IL-21-dependent anticancer functions of TH9 cells. Nat. Immunol. 2014;15:758–766. doi: 10.1038/ni.2925. [DOI] [PubMed] [Google Scholar]
- 523.Wang C, et al. Th9 cells are subjected to PD-1/PD-L1-mediated inhibition and are capable of promoting CD8 T cell expansion through IL-9R in colorectal cancer. Int. Immunopharmacol. 2020;78:106019. doi: 10.1016/j.intimp.2019.106019. [DOI] [PubMed] [Google Scholar]
- 524.Lu Y, et al. Th9 cells promote antitumor immune responses in vivo. J. Clin. Invest. 2012;122:4160–4171. doi: 10.1172/JCI65459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Kim IK, et al. Glucocorticoid-induced tumor necrosis factor receptor-related protein co-stimulation facilitates tumor regression by inducing IL-9-producing helper T cells. Nat. Med. 2015;21:1010–1017. doi: 10.1038/nm.3922. [DOI] [PubMed] [Google Scholar]
- 526.Xue G, et al. Adoptive cell therapy with tumor-specific Th9 cells induces viral mimicry to eliminate antigen-loss-variant tumor cells. Cancer Cell. 2021;39:1610–1622 e1619. doi: 10.1016/j.ccell.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527.Abdul-Wahid A, et al. Induction of antigen-specific TH 9 immunity accompanied by mast cell activation blocks tumor cell engraftment. Int. J. Cancer. 2016;139:841–853. doi: 10.1002/ijc.30121. [DOI] [PubMed] [Google Scholar]
- 528.Lu Y, et al. Th9 cells represent a unique subset of CD4(+) T cells endowed with the ability to eradicate advanced tumors. Cancer Cell. 2018;33:1048–1060 e1047. doi: 10.1016/j.ccell.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Benoit-Lizon, I. et al. CD4 T cell-intrinsic STING signaling controls the differentiation and effector functions of TH1 and TH9 cells. J. Immunother. Cancer. 10, e003459 (2022). [DOI] [PMC free article] [PubMed]
- 530.Sek K, Chan CW, Beavis PA, Darcy PK. Adoptive transfer of tumor-specific Th9 cells eradicates heterogeneous antigen-expressing tumor cells. Cancer Cell. 2021;39:1564–1566. doi: 10.1016/j.ccell.2021.10.013. [DOI] [PubMed] [Google Scholar]
- 531.Gerlach, K. et al. PU.1-driven Th9 cells promote colorectal cancer in experimental colitis models through IL-6 effects in intestinal epithelial cells. J. Crohns Colitis16, 1893–1910 (2022). [DOI] [PMC free article] [PubMed]
- 532.Tan H, Wang S, Zhao L. A tumour-promoting role of Th9 cells in hepatocellular carcinoma through CCL20 and STAT3 pathways. Clin. Exp. Pharm. Physiol. 2017;44:213–221. doi: 10.1111/1440-1681.12689. [DOI] [PubMed] [Google Scholar]
- 533.Demoulin JB, et al. STAT5 activation is required for interleukin-9-dependent growth and transformation of lymphoid cells. Cancer Res. 2000;60:3971–3977. [PubMed] [Google Scholar]
- 534.Ye ZJ, et al. Differentiation and immune regulation of IL-9-producing CD4+ T cells in malignant pleural effusion. Am. J. Respir. Crit. Care Med. 2012;186:1168–1179. doi: 10.1164/rccm.201207-1307OC. [DOI] [PubMed] [Google Scholar]
- 535.Sabry SA, et al. Oxidative stress in CLL patients leads to activation of Th9 cells: an experimental and comprehensive survey. Immunol. Med. 2020;43:36–46. doi: 10.1080/25785826.2019.1700747. [DOI] [PubMed] [Google Scholar]
- 536.Kumar S, et al. The Th9 axis reduces the oxidative stress and promotes the survival of malignant T cells in cutaneous T-cell lymphoma patients. Mol. Cancer Res. 2020;18:657–668. doi: 10.1158/1541-7786.MCR-19-0894. [DOI] [PubMed] [Google Scholar]
- 537.Feng LL, Gao JM, Li PP, Wang X. IL-9 contributes to immunosuppression mediated by regulatory T cells and mast cells in B-cell non-hodgkin’s lymphoma. J. Clin. Immunol. 2011;31:1084–1094. doi: 10.1007/s10875-011-9584-9. [DOI] [PubMed] [Google Scholar]
- 538.Hoelzinger DB, Dominguez AL, Cohen PA, Gendler SJ. Inhibition of adaptive immunity by IL9 can be disrupted to achieve rapid T-cell sensitization and rejection of progressive tumor challenges. Cancer Res. 2014;74:6845–6855. doi: 10.1158/0008-5472.CAN-14-0836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Raffin C, Vo LT, Bluestone JA. Treg cell-based therapies: challenges and perspectives. Nat. Rev. Immunol. 2020;20:158–172. doi: 10.1038/s41577-019-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.McRitchie BR, Akkaya B. Exhaust the exhausters: targeting regulatory T cells in the tumor microenvironment. Front. Immunol. 2022;13:940052. doi: 10.3389/fimmu.2022.940052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat. Rev. Clin. Oncol. 2019;16:356–371. doi: 10.1038/s41571-019-0175-7. [DOI] [PubMed] [Google Scholar]
- 542.Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27:109–118. doi: 10.1038/cr.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Shang B, Liu Y, Jiang SJ, Liu Y. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci. Rep. 2015;5:15179. doi: 10.1038/srep15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Saleh R, Elkord E. FoxP3(+) T regulatory cells in cancer: prognostic biomarkers and therapeutic targets. Cancer Lett. 2020;490:174–185. doi: 10.1016/j.canlet.2020.07.022. [DOI] [PubMed] [Google Scholar]
- 545.Shan F, et al. Therapeutic targeting of regulatory T cells in cancer. Trends Cancer. 2022;8:944–961. doi: 10.1016/j.trecan.2022.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Betts G, et al. Suppression of tumour-specific CD4(+) T cells by regulatory T cells is associated with progression of human colorectal cancer. Gut. 2012;61:1163–1171. doi: 10.1136/gutjnl-2011-300970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Saito T, et al. Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 2016;22:679–684. doi: 10.1038/nm.4086. [DOI] [PubMed] [Google Scholar]
- 548.Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol. Immunother. 2011;60:909–918. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.Kryczek I, et al. IL-17+ regulatory T cells in the microenvironments of chronic inflammation and cancer. J. Immunol. 2011;186:4388–4395. doi: 10.4049/jimmunol.1003251. [DOI] [PubMed] [Google Scholar]
- 550.Yang S, et al. Foxp3+IL-17+ T cells promote development of cancer-initiating cells in colorectal cancer. J. Leukoc. Biol. 2011;89:85–91. doi: 10.1189/jlb.0910506. [DOI] [PubMed] [Google Scholar]
- 551.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 552.Gao R, Shi GP, Wang J. Functional diversities of regulatory T cells in the context of cancer immunotherapy. Front. Immunol. 2022;13:833667. doi: 10.3389/fimmu.2022.833667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Tanaka A, Sakaguchi S. Targeting Treg cells in cancer immunotherapy. Eur. J. Immunol. 2019;49:1140–1146. doi: 10.1002/eji.201847659. [DOI] [PubMed] [Google Scholar]
- 554.Cao X, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 555.Volpe E, Sambucci M, Battistini L, Borsellino G. Fas-Fas ligand: checkpoint of T cell functions in multiple sclerosis. Front. Immunol. 2016;7:382. doi: 10.3389/fimmu.2016.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Wei X, et al. Reciprocal expression of IL-35 and IL-10 defines two distinct effector Treg subsets that are required for maintenance of immune tolerance. Cell Rep. 2017;21:1853–1869. doi: 10.1016/j.celrep.2017.10.090. [DOI] [PubMed] [Google Scholar]
- 557.Sarhan D, et al. Adaptive NK cells resist regulatory T-cell suppression driven by IL37. Cancer Immunol. Res. 2018;6:766–775. doi: 10.1158/2326-6066.CIR-17-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Hatzioannou A, et al. An intrinsic role of IL-33 in Treg cell-mediated tumor immunoevasion. Nat. Immunol. 2020;21:75–85. doi: 10.1038/s41590-019-0555-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Zappasodi R, et al. CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumours. Nature. 2021;591:652–658. doi: 10.1038/s41586-021-03326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Aksoylar HI, Boussiotis VA. PD-1(+) Treg cells: a foe in cancer immunotherapy? Nat. Immunol. 2020;21:1311–1312. doi: 10.1038/s41590-020-0801-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Kurtulus S, et al. TIGIT predominantly regulates the immune response via regulatory T cells. J. Clin. Invest. 2015;125:4053–4062. doi: 10.1172/JCI81187. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 562.Wing K, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 563.Qureshi OS, et al. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332:600–603. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Gu P, et al. Trogocytosis of CD80 and CD86 by induced regulatory T cells. Cell Mol. Immunol. 2012;9:136–146. doi: 10.1038/cmi.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Tekguc, M. et al. Treg-expressed CTLA-4 depletes CD80/CD86 by trogocytosis, releasing free PD-L1 on antigen-presenting cells. Proc Natl Acad Sci USA. 118, e2023739118 (2021). [DOI] [PMC free article] [PubMed]
- 566.Kalia V, et al. Quiescence of memory CD8(+) T cells is mediated by regulatory T cells through inhibitory receptor CTLA-4. Immunity. 2015;42:1116–1129. doi: 10.1016/j.immuni.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 567.Liang B, et al. Regulatory T cells inhibit dendritic cells by lymphocyte activation gene-3 engagement of MHC class II. J. Immunol. 2008;180:5916–5926. doi: 10.4049/jimmunol.180.9.5916. [DOI] [PubMed] [Google Scholar]
- 568.Ihara F, et al. Regulatory T cells induce CD4(-) NKT cell anergy and suppress NKT cell cytotoxic function. Cancer Immunol. Immunother. 2019;68:1935–1947. doi: 10.1007/s00262-019-02417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Fujimura T, Kambayashi Y, Aiba S. Crosstalk between regulatory T cells (Tregs) and myeloid derived suppressor cells (MDSCs) during melanoma growth. Oncoimmunology. 2012;1:1433–1434. doi: 10.4161/onci.21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Li C, et al. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer. 2020;19:116. doi: 10.1186/s12943-020-01234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Young A, Mittal D, Stagg J, Smyth MJ. Targeting cancer-derived adenosine: new therapeutic approaches. Cancer Disco. 2014;4:879–888. doi: 10.1158/2159-8290.CD-14-0341. [DOI] [PubMed] [Google Scholar]
- 573.Carmenate T, et al. Blocking IL-2 signal in vivo with an IL-2 antagonist reduces tumor growth through the control of regulatory T cells. J. Immunol. 2018;200:3475–3484. doi: 10.4049/jimmunol.1700433. [DOI] [PubMed] [Google Scholar]
- 574.Moon YW, Hajjar J, Hwu P, Naing A. Targeting the indoleamine 2,3-dioxygenase pathway in cancer. J. Immunother. Cancer. 2015;3:51. doi: 10.1186/s40425-015-0094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Platten M, et al. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Disco. 2019;18:379–401. doi: 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 576.Zhang L, Zhang Z. Recharacterizing tumor-infiltrating lymphocytes by single-cell RNA sequencing. Cancer Immunol. Res. 2019;7:1040–1046. doi: 10.1158/2326-6066.CIR-18-0658. [DOI] [PubMed] [Google Scholar]
- 577.Ahmadzadeh, M. et al. Tumor-infiltrating human CD4(+) regulatory T cells display a distinct TCR repertoire and exhibit tumor and neoantigen reactivity. Sci Immunol. 4, eaao4310 (2019). [DOI] [PMC free article] [PubMed]
- 578.Oh DY, et al. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181:1612–1625 e1613. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Plitas G, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–1134. doi: 10.1016/j.immuni.2016.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Cinier, J. et al. Recruitment and expansion of Tregs cells in the tumor environment-How to target them? Cancers13, 1850 (2021). [DOI] [PMC free article] [PubMed]
- 581.Zhang Y, et al. Deep single-cell RNA sequencing data of individual T cells from treatment-naive colorectal cancer patients. Sci. Data. 2019;6:131. doi: 10.1038/s41597-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Chen Q, et al. ICOS signal facilitates Foxp3 transcription to favor suppressive function of regulatory T cells. Int. J. Med. Sci. 2018;15:666–673. doi: 10.7150/ijms.23940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Rao D, et al. Metabolic profiles of regulatory T cells in the tumour microenvironment. Cancer Immunol. Immunother. 2021;70:2417–2427. doi: 10.1007/s00262-021-02881-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Angelin A, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25:1282–1293 e1287. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Kishore M, et al. Regulatory T cell migration is dependent on glucokinase-mediated glycolysis. Immunity. 2017;47:875–889 e810. doi: 10.1016/j.immuni.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Kumagai S, et al. Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell. 2022;40:201–218 e209. doi: 10.1016/j.ccell.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 587.Watson MJ, et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature. 2021;591:645–651. doi: 10.1038/s41586-020-03045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Wang H, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020;21:298–308. doi: 10.1038/s41590-019-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Pacella I, et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc. Natl Acad. Sci. USA. 2018;115:E6546–E6555. doi: 10.1073/pnas.1720113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Basu A, et al. Differentiation and regulation of TH cells: a balancing act for cancer immunotherapy. Front. Immunol. 2021;12:669474. doi: 10.3389/fimmu.2021.669474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Fahey LM, et al. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 2011;208:987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Vella LA, Herati RS, Wherry EJ. CD4(+) T cell differentiation in chronic viral infections: the Tfh perspective. Trends Mol. Med. 2017;23:1072–1087. doi: 10.1016/j.molmed.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593.Cabrita R, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577:561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 594.Gu-Trantien C, et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Invest. 2013;123:2873–2892. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Bindea G, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 596.Cui C, et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell. 2021;184:6101–6118 e6113. doi: 10.1016/j.cell.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Chen J, Chen J, Wang L. Tertiary lymphoid structures as unique constructions associated with the organization, education, and function of tumor-infiltrating immunocytes. J. Zhejiang Univ. Sci. B. 2022;23:812–822. doi: 10.1631/jzus.B2200174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Fridman WH, et al. B cells and tertiary lymphoid structures as determinants of tumour immune contexture and clinical outcome. Nat. Rev. Clin. Oncol. 2022;19:441–457. doi: 10.1038/s41571-022-00619-z. [DOI] [PubMed] [Google Scholar]
- 599.Lin, X. et al. Follicular helper T cells remodel the immune microenvironment of pancreatic cancer via secreting CXCL13 and IL-21. Cancers13, 3678 (2021). [DOI] [PMC free article] [PubMed]
- 600.Noel, G. et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J. Clin. Invest. 131, e139905 (2021). [DOI] [PMC free article] [PubMed]
- 601.Ukita, M. et al. CXCL13-producing CD4+ T cells accumulate in the early phase of tertiary lymphoid structures in ovarian cancer. JCI Insight7, e157215 (2022). [DOI] [PMC free article] [PubMed]
- 602.Zander R, et al. Tfh-cell-derived interleukin 21 sustains effector CD8(+) T cell responses during chronic viral infection. Immunity. 2022;55:475–493 e475. doi: 10.1016/j.immuni.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Greczmiel, U. et al. Sustained T follicular helper cell response is essential for control of chronic viral infection. Sci. Immunol. 2, eaam8686 (2017). [DOI] [PubMed]
- 604.Salemme V, et al. The crosstalk between tumor cells and the immune microenvironment in breast cancer: implications for immunotherapy. Front. Oncol. 2021;11:610303. doi: 10.3389/fonc.2021.610303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Garaud S, et al. Antigen specificity and clinical significance of IgG and IgA autoantibodies produced in situ by tumor-infiltrating B cells in breast cancer. Front. Immunol. 2018;9:2660. doi: 10.3389/fimmu.2018.02660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Ma CS. Human T follicular helper cells in primary immunodeficiency: quality just as important as quantity. J. Clin. Immunol. 2016;36:40–47. doi: 10.1007/s10875-016-0257-6. [DOI] [PubMed] [Google Scholar]
- 607.Akkaya M, Kwak K, Pierce SK. B cell memory: building two walls of protection against pathogens. Nat. Rev. Immunol. 2020;20:229–238. doi: 10.1038/s41577-019-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Baumjohann, D. & Brossart, P. T follicular helper cells: linking cancer immunotherapy and immune-related adverse events. J. Immunother. Cancer9, e002588 (2021). [DOI] [PMC free article] [PubMed]
- 609.Solinas C, et al. Immune checkpoint molecules on tumor-infiltrating lymphocytes and their association with tertiary lymphoid structures in human breast cancer. Front. Immunol. 2017;8:1412. doi: 10.3389/fimmu.2017.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Shi J, et al. PD-1 controls follicular T helper cell positioning and function. Immunity. 2018;49:264–274 e264. doi: 10.1016/j.immuni.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Helmink BA, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Petitprez F, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577:556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 613.Niogret, J. et al. Follicular helper-T cells restore CD8(+)-dependent antitumor immunity and anti-PD-L1/PD-1 efficacy. J. Immunother. Cancer9, e002157 (2021). [DOI] [PMC free article] [PubMed]
- 614.Hussain M, et al. CXCL13/CXCR5 signaling axis in cancer. Life Sci. 2019;227:175–186. doi: 10.1016/j.lfs.2019.04.053. [DOI] [PubMed] [Google Scholar]
- 615.Yang, M. et al. CXCL13 shapes immunoactive tumor microenvironment and enhances the efficacy of PD-1 checkpoint blockade in high-grade serous ovarian cancer. J. Immunother. Cancer9, e001136 (2021). [DOI] [PMC free article] [PubMed]
- 616.Oosterhuis K, et al. Rational design of DNA vaccines for the induction of human papillomavirus type 16 E6- and E7-specific cytotoxic T-cell responses. Hum. Gene Ther. 2012;23:1301–1312. doi: 10.1089/hum.2012.101. [DOI] [PubMed] [Google Scholar]
- 617.Borst J, et al. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 618.Calabro S, et al. Differential intrasplenic migration of dendritic cell subsets tailors adaptive immunity. Cell Rep. 2016;16:2472–2485. doi: 10.1016/j.celrep.2016.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Gerner MY, Casey KA, Kastenmuller W, Germain RN. Dendritic cell and antigen dispersal landscapes regulate T cell immunity. J. Exp. Med. 2017;214:3105–3122. doi: 10.1084/jem.20170335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Eickhoff S, et al. Robust anti-viral immunity requires multiple distinct T cell-dendritic cell interactions. Cell. 2015;162:1322–1337. doi: 10.1016/j.cell.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Hor JL, et al. Spatiotemporally distinct interactions with dendritic cell subsets facilitates CD4+ and CD8+ T cell activation to localized viral infection. Immunity. 2015;43:554–565. doi: 10.1016/j.immuni.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 622.Bachem A, et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Bennett SR, et al. Help for cytotoxic-T-cell responses is mediated by CD40 signalling. Nature. 1998;393:478–480. doi: 10.1038/30996. [DOI] [PubMed] [Google Scholar]
- 624.Schoenberger SP, et al. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 1998;393:480–483. doi: 10.1038/31002. [DOI] [PubMed] [Google Scholar]
- 625.Schuurhuis DH, et al. Immature dendritic cells acquire CD8(+) cytotoxic T lymphocyte priming capacity upon activation by T helper cell-independent or -dependent stimuli. J. Exp. Med. 2000;192:145–150. doi: 10.1084/jem.192.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Bijker MS, et al. CD8+ CTL priming by exact peptide epitopes in incomplete Freund’s adjuvant induces a vanishing CTL response, whereas long peptides induce sustained CTL reactivity. J. Immunol. 2007;179:5033–5040. doi: 10.4049/jimmunol.179.8.5033. [DOI] [PubMed] [Google Scholar]
- 627.Schulz O, et al. CD40 triggering of heterodimeric IL-12 p70 production by dendritic cells in vivo requires a microbial priming signal. Immunity. 2000;13:453–462. doi: 10.1016/S1074-7613(00)00045-5. [DOI] [PubMed] [Google Scholar]
- 628.Ahrends T, et al. CD4(+) T cell help confers a cytotoxic T cell effector program including coinhibitory receptor downregulation and increased tissue invasiveness. Immunity. 2017;47:848–861 e845. doi: 10.1016/j.immuni.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 629.Provine NM, et al. Immediate dysfunction of vaccine-elicited CD8+ T cells primed in the absence of CD4+ T cells. J. Immunol. 2016;197:1809–1822. doi: 10.4049/jimmunol.1600591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Aubert RD, et al. Antigen-specific CD4 T-cell help rescues exhausted CD8 T cells during chronic viral infection. Proc. Natl Acad. Sci. USA. 2011;108:21182–21187. doi: 10.1073/pnas.1118450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Laidlaw BJ, Craft JE, Kaech SM. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat. Rev. Immunol. 2016;16:102–111. doi: 10.1038/nri.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Ahrends T, et al. CD4(+) T cell help creates memory CD8(+) T cells with innate and help-independent recall capacities. Nat. Commun. 2019;10:5531. doi: 10.1038/s41467-019-13438-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Matter MS, Claus C, Ochsenbein AF. CD4+ T cell help improves CD8+ T cell memory by retained CD27 expression. Eur. J. Immunol. 2008;38:1847–1856. doi: 10.1002/eji.200737824. [DOI] [PubMed] [Google Scholar]
- 634.Janssen EM, et al. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 635.Oh S, et al. IL-15 as a mediator of CD4+ help for CD8+ T cell longevity and avoidance of TRAIL-mediated apoptosis. Proc. Natl Acad. Sci. USA. 2008;105:5201–5206. doi: 10.1073/pnas.0801003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Lu YJ, et al. CD4 T cell help prevents CD8 T cell exhaustion and promotes control of Mycobacterium tuberculosis infection. Cell Rep. 2021;36:109696. doi: 10.1016/j.celrep.2021.109696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Hendriks J, Xiao Y, Borst J. CD27 promotes survival of activated T cells and complements CD28 in generation and establishment of the effector T cell pool. J. Exp. Med. 2003;198:1369–1380. doi: 10.1084/jem.20030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Feau S, et al. The CD4(+) T-cell help signal is transmitted from APC to CD8(+) T-cells via CD27-CD70 interactions. Nat. Commun. 2012;3:948. doi: 10.1038/ncomms1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Prilliman KR, et al. Cutting edge: a crucial role for B7-CD28 in transmitting T help from APC to CTL. J. Immunol. 2002;169:4094–4097. doi: 10.4049/jimmunol.169.8.4094. [DOI] [PubMed] [Google Scholar]
- 640.Curtsinger JM, Johnson CM, Mescher MF. CD8 T cell clonal expansion and development of effector function require prolonged exposure to antigen, costimulation, and signal 3 cytokine. J. Immunol. 2003;171:5165–5171. doi: 10.4049/jimmunol.171.10.5165. [DOI] [PubMed] [Google Scholar]
- 641.Bullock TN, Yagita H. Induction of CD70 on dendritic cells through CD40 or TLR stimulation contributes to the development of CD8+ T cell responses in the absence of CD4+ T cells. J. Immunol. 2005;174:710–717. doi: 10.4049/jimmunol.174.2.710. [DOI] [PubMed] [Google Scholar]
- 642.van de Ven K, Borst J. Targeting the T-cell co-stimulatory CD27/CD70 pathway in cancer immunotherapy: rationale and potential. Immunotherapy. 2015;7:655–667. doi: 10.2217/imt.15.32. [DOI] [PubMed] [Google Scholar]
- 643.Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu. Rev. Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 644.Hendriks J, et al. During viral infection of the respiratory tract, CD27, 4-1BB, and OX40 collectively determine formation of CD8+ memory T cells and their capacity for secondary expansion. J. Immunol. 2005;175:1665–1676. doi: 10.4049/jimmunol.175.3.1665. [DOI] [PubMed] [Google Scholar]
- 645.Agarwal P, et al. Gene regulation and chromatin remodeling by IL-12 and type I IFN in programming for CD8 T cell effector function and memory. J. Immunol. 2009;183:1695–1704. doi: 10.4049/jimmunol.0900592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 646.Wilson EB, Livingstone AM. Cutting edge: CD4+ T cell-derived IL-2 is essential for help-dependent primary CD8+ T cell responses. J. Immunol. 2008;181:7445–7448. doi: 10.4049/jimmunol.181.11.7445. [DOI] [PubMed] [Google Scholar]
- 647.Cui W, et al. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Snell LM, et al. CD8(+) T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity. 2018;49:678–694 e675. doi: 10.1016/j.immuni.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Yu D, Ye L. A portrait of CXCR5(+) follicular cytotoxic CD8(+) T cells. Trends Immunol. 2018;39:965–979. doi: 10.1016/j.it.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 650.Li Y, et al. CXCL13-mediated recruitment of intrahepatic CXCR5(+)CD8(+) T cells favors viral control in chronic HBV infection. J. Hepatol. 2020;72:420–430. doi: 10.1016/j.jhep.2019.09.031. [DOI] [PubMed] [Google Scholar]
- 651.Gu-Trantien, C. et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight2, e91487 (2017). [DOI] [PMC free article] [PubMed]
- 652.Fugger L, Jensen LT, Rossjohn J. Challenges, progress, and prospects of developing therapies to treat autoimmune diseases. Cell. 2020;181:63–80. doi: 10.1016/j.cell.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 653.Rodriguez Murua S, Farez MF, Quintana FJ. The immune response in multiple sclerosis. Annu. Rev. Pathol. 2022;17:121–139. doi: 10.1146/annurev-pathol-052920-040318. [DOI] [PubMed] [Google Scholar]
- 654.Voskuhl RR, et al. T helper 1 (Th1) functional phenotype of human myelin basic protein-specific T lymphocytes. Autoimmunity. 1993;15:137–143. doi: 10.3109/08916939309043888. [DOI] [PubMed] [Google Scholar]
- 655.Lock C, et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- 656.Baron JL, et al. Surface expression of alpha 4 integrin by CD4 T cells is required for their entry into brain parenchyma. J. Exp. Med. 1993;177:57–68. doi: 10.1084/jem.177.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Prajeeth CK, et al. Effector molecules released by Th1 but not Th17 cells drive an M1 response in microglia. Brain Behav. Immun. 2014;37:248–259. doi: 10.1016/j.bbi.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 658.Gran B, et al. IL-12p35-deficient mice are susceptible to experimental autoimmune encephalomyelitis: evidence for redundancy in the IL-12 system in the induction of central nervous system autoimmune demyelination. J. Immunol. 2002;169:7104–7110. doi: 10.4049/jimmunol.169.12.7104. [DOI] [PubMed] [Google Scholar]
- 659.Zhang GX, et al. Induction of experimental autoimmune encephalomyelitis in IL-12 receptor-beta 2-deficient mice: IL-12 responsiveness is not required in the pathogenesis of inflammatory demyelination in the central nervous system. J. Immunol. 2003;170:2153–2160. doi: 10.4049/jimmunol.170.4.2153. [DOI] [PubMed] [Google Scholar]
- 660.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 661.Oppmann B, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 662.Parham C, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J. Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 663.Majumder S, McGeachy MJ. IL-17 in the pathogenesis of disease: good intentions gone awry. Annu. Rev. Immunol. 2021;39:537–556. doi: 10.1146/annurev-immunol-101819-092536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Dos Passos GR, Sato DK, Becker J, Fujihara K. Th17 cells pathways in multiple sclerosis and neuromyelitis optica spectrum disorders: pathophysiological and therapeutic implications. Mediators Inflamm. 2016;2016:5314541. doi: 10.1155/2016/5314541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Brucklacher-Waldert V, et al. Phenotypical and functional characterization of T helper 17 cells in multiple sclerosis. Brain. 2009;132:3329–3341. doi: 10.1093/brain/awp289. [DOI] [PubMed] [Google Scholar]
- 666.Tzartos JS, et al. Interleukin-17 production in central nervous system-infiltrating T cells and glial cells is associated with active disease in multiple sclerosis. Am. J. Pathol. 2008;172:146–155. doi: 10.2353/ajpath.2008.070690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667.Kebir H, et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat. Med. 2007;13:1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Murphy AC, Lalor SJ, Lynch MA, Mills KH. Infiltration of Th1 and Th17 cells and activation of microglia in the CNS during the course of experimental autoimmune encephalomyelitis. Brain Behav. Immun. 2010;24:641–651. doi: 10.1016/j.bbi.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 669.Prajeeth CK, et al. Effectors of Th1 and Th17 cells act on astrocytes and augment their neuroinflammatory properties. J. Neuroinflammation. 2017;14:204. doi: 10.1186/s12974-017-0978-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Kang Z, et al. Astrocyte-restricted ablation of interleukin-17-induced Act1-mediated signaling ameliorates autoimmune encephalomyelitis. Immunity. 2010;32:414–425. doi: 10.1016/j.immuni.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 671.Setiadi AF, et al. IL-17A is associated with the breakdown of the blood-brain barrier in relapsing-remitting multiple sclerosis. J. Neuroimmunol. 2019;332:147–154. doi: 10.1016/j.jneuroim.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 672.Rahman MT, et al. IFN-gamma, IL-17A, or zonulin rapidly increase the permeability of the blood-brain and small intestinal epithelial barriers: relevance for neuro-inflammatory diseases. Biochem. Biophys. Res. Commun. 2018;507:274–279. doi: 10.1016/j.bbrc.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 673.Paintlia MK, Paintlia AS, Singh AK, Singh I. Synergistic activity of interleukin-17 and tumor necrosis factor-alpha enhances oxidative stress-mediated oligodendrocyte apoptosis. J. Neurochem. 2011;116:508–521. doi: 10.1111/j.1471-4159.2010.07136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Dulamea AO. Role of oligodendrocyte dysfunction in demyelination, remyelination and neurodegeneration in multiple sclerosis. Adv. Exp. Med. Biol. 2017;958:91–127. doi: 10.1007/978-3-319-47861-6_7. [DOI] [PubMed] [Google Scholar]
- 675.Rangel-Moreno J, et al. The development of inducible bronchus-associated lymphoid tissue depends on IL-17. Nat. Immunol. 2011;12:639–646. doi: 10.1038/ni.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 676.Pikor NB, et al. Integration of Th17- and lymphotoxin-derived signals initiates meningeal-resident stromal cell remodeling to propagate neuroinflammation. Immunity. 2015;43:1160–1173. doi: 10.1016/j.immuni.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 677.Peters A, et al. Th17 cells induce ectopic lymphoid follicles in central nervous system tissue inflammation. Immunity. 2011;35:986–996. doi: 10.1016/j.immuni.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 678.Codarri L, et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 679.Ifergan I, et al. Targeting the GM-CSF receptor for the treatment of CNS autoimmunity. J. Autoimmun. 2017;84:1–11. doi: 10.1016/j.jaut.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Sonderegger I, et al. GM-CSF mediates autoimmunity by enhancing IL-6-dependent Th17 cell development and survival. J. Exp. Med. 2008;205:2281–2294. doi: 10.1084/jem.20071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Rasouli J, et al. Expression of GM-CSF in T cells is increased in multiple sclerosis and suppressed by IFN-beta therapy. J. Immunol. 2015;194:5085–5093. doi: 10.4049/jimmunol.1403243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Carrieri PB, et al. Profile of cerebrospinal fluid and serum cytokines in patients with relapsing-remitting multiple sclerosis: a correlation with clinical activity. Immunopharmacol. Immunotoxicol. 1998;20:373–382. doi: 10.3109/08923979809034820. [DOI] [PubMed] [Google Scholar]
- 683.Croxford AL, et al. The cytokine GM-CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity. 2015;43:502–514. doi: 10.1016/j.immuni.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 684.Rosu A, et al. IL-17 patterns in synovium, serum and synovial fluid from treatment-naive, early rheumatoid arthritis patients. Rom. J. Morphol. Embryol. 2012;53:73–80. [PubMed] [Google Scholar]
- 685.Shahrara S, Huang Q, Mandelin AM, 2nd, Pope RM. TH-17 cells in rheumatoid arthritis. Arthritis Res. Ther. 2008;10:R93. doi: 10.1186/ar2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 686.Kim J, et al. Elevated levels of T helper 17 cells are associated with disease activity in patients with rheumatoid arthritis. Ann. Lab Med. 2013;33:52–59. doi: 10.3343/alm.2013.33.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Metawi SA, Abbas D, Kamal MM, Ibrahim MK. Serum and synovial fluid levels of interleukin-17 in correlation with disease activity in patients with RA. Clin. Rheumatol. 2011;30:1201–1207. doi: 10.1007/s10067-011-1737-y. [DOI] [PubMed] [Google Scholar]
- 688.Yasuda K, Takeuchi Y, Hirota K. The pathogenicity of Th17 cells in autoimmune diseases. Semin. Immunopathol. 2019;41:283–297. doi: 10.1007/s00281-019-00733-8. [DOI] [PubMed] [Google Scholar]
- 689.Hirota K, et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J. Exp. Med. 2007;204:2803–2812. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.Ito H, et al. Dual role of interleukin-17 in pannus growth and osteoclastogenesis in rheumatoid arthritis. Arthritis Res. Ther. 2011;13:R14. doi: 10.1186/ar3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Moon YM, et al. IL-32 and IL-17 interact and have the potential to aggravate osteoclastogenesis in rheumatoid arthritis. Arthritis Res. Ther. 2012;14:R246. doi: 10.1186/ar4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Pickens SR, et al. IL-17 contributes to angiogenesis in rheumatoid arthritis. J. Immunol. 2010;184:3233–3241. doi: 10.4049/jimmunol.0903271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Sato K, et al. Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J. Exp. Med. 2006;203:2673–2682. doi: 10.1084/jem.20061775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Saxena A, Raychaudhuri SK, Raychaudhuri SP. Interleukin-17-induced proliferation of fibroblast-like synovial cells is mTOR dependent. Arthritis Rheum. 2011;63:1465–1466. doi: 10.1002/art.30278. [DOI] [PubMed] [Google Scholar]
- 695.Hirota K, et al. Autoimmune Th17 cells induced synovial stromal and innate lymphoid cell secretion of the cytokine GM-CSF to initiate and augment autoimmune arthritis. Immunity. 2018;48:1220–1232 e1225. doi: 10.1016/j.immuni.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696.Tsokos GC. Systemic lupus erythematosus. N. Engl. J. Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 697.Fava, A. & Rao, D. A. Cellular and molecular heterogeneity in systemic lupus erythematosus. Semin. Immunol. 58, 101653 (2022). [DOI] [PubMed]
- 698.Koga T, Ichinose K, Kawakami A, Tsokos GC. Current insights and future prospects for targeting IL-17 to treat patients with systemic lupus erythematosus. Front. Immunol. 2020;11:624971. doi: 10.3389/fimmu.2020.624971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Burkett PR, Meyer zu Horste G, Kuchroo VK. Pouring fuel on the fire: Th17 cells, the environment, and autoimmunity. J. Clin. Invest. 2015;125:2211–2219. doi: 10.1172/JCI78085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Wong CK, et al. Hyperproduction of IL-23 and IL-17 in patients with systemic lupus erythematosus: implications for Th17-mediated inflammation in auto-immunity. Clin. Immunol. 2008;127:385–393. doi: 10.1016/j.clim.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 701.Henriques A, et al. Frequency and functional activity of Th17, Tc17 and other T-cell subsets in systemic lupus erythematosus. Cell Immunol. 2010;264:97–103. doi: 10.1016/j.cellimm.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 702.Chen XQ, et al. Plasma IL-17A is increased in new-onset SLE patients and associated with disease activity. J. Clin. Immunol. 2010;30:221–225. doi: 10.1007/s10875-009-9365-x. [DOI] [PubMed] [Google Scholar]
- 703.Zickert A, et al. IL-17 and IL-23 in lupus nephritis - association to histopathology and response to treatment. BMC Immunol. 2015;16:7. doi: 10.1186/s12865-015-0070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Waite JC, Skokos D. Th17 response and inflammatory autoimmune diseases. Int. J. Inflam. 2012;2012:819467. doi: 10.1155/2012/819467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Pisitkun P, et al. Interleukin-17 cytokines are critical in development of fatal lupus glomerulonephritis. Immunity. 2012;37:1104–1115. doi: 10.1016/j.immuni.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 706.Lubberts E. The IL-23-IL-17 axis in inflammatory arthritis. Nat. Rev. Rheumatol. 2015;11:415–429. doi: 10.1038/nrrheum.2015.53. [DOI] [PubMed] [Google Scholar]
- 707.Annunziato F, et al. Defining the human T helper 17 cell phenotype. Trends Immunol. 2012;33:505–512. doi: 10.1016/j.it.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 708.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.Kamali AN, et al. A role for Th1-like Th17 cells in the pathogenesis of inflammatory and autoimmune disorders. Mol. Immunol. 2019;105:107–115. doi: 10.1016/j.molimm.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 710.Nistala K, et al. Th17 plasticity in human autoimmune arthritis is driven by the inflammatory environment. Proc. Natl Acad. Sci. USA. 2010;107:14751–14756. doi: 10.1073/pnas.1003852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 711.Bending D, et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Paroni M, et al. Recognition of viral and self-antigens by TH1 and TH1/TH17 central memory cells in patients with multiple sclerosis reveals distinct roles in immune surveillance and relapses. J. Allergy Clin. Immunol. 2017;140:797–808. doi: 10.1016/j.jaci.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 713.Gaublomme JT, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Hou L, Yuki K. CCR6 and CXCR6 identify the Th17 cells with cytotoxicity in experimental autoimmune encephalomyelitis. Front. Immunol. 2022;13:819224. doi: 10.3389/fimmu.2022.819224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Schnell A, et al. Stem-like intestinal Th17 cells give rise to pathogenic effector T cells during autoimmunity. Cell. 2021;184:6281–6298 e6223. doi: 10.1016/j.cell.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Perriard G, et al. Interleukin-22 is increased in multiple sclerosis patients and targets astrocytes. J. Neuroinflammation. 2015;12:119. doi: 10.1186/s12974-015-0335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Yang X, et al. Increased interleukin-22 levels in lupus nephritis and its associated with disease severity: a study in both patients and lupus-like mice model. Clin. Exp. Rheumatol. 2019;37:400–407. [PubMed] [Google Scholar]
- 718.Zhang L, et al. Elevated Th22 cells correlated with Th17 cells in patients with rheumatoid arthritis. J. Clin. Immunol. 2011;31:606–614. doi: 10.1007/s10875-011-9540-8. [DOI] [PubMed] [Google Scholar]
- 719.Luan L, et al. An increased proportion of circulating Th22 and Tc22 cells in psoriasis. Cell Immunol. 2014;290:196–200. doi: 10.1016/j.cellimm.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 720.Hu Y, et al. Elevated profiles of Th22 cells and correlations with Th17 cells in patients with immune thrombocytopenia. Hum. Immunol. 2012;73:629–635. doi: 10.1016/j.humimm.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 721.Liang M, et al. The Imbalance between Foxp3(+)Tregs and Th1/Th17/Th22 Cells in Patients with Newly Diagnosed Autoimmune Hepatitis. J. Immunol. Res. 2018;2018:3753081. doi: 10.1155/2018/3753081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Vitales-Noyola M, et al. Pathogenic Th17 and Th22 cells are increased in patients with autoimmune thyroid disorders. Endocrine. 2017;57:409–417. doi: 10.1007/s12020-017-1361-y. [DOI] [PubMed] [Google Scholar]
- 723.Robat-Jazi B, et al. High frequency of Tc22 and Th22 cells in myasthenia gravis patients and their significant reduction after thymectomy. Neuroimmunomodulation. 2018;25:80–88. doi: 10.1159/000490855. [DOI] [PubMed] [Google Scholar]
- 724.Truchetet ME, et al. Increased frequency of circulating Th22 in addition to Th17 and Th2 lymphocytes in systemic sclerosis: association with interstitial lung disease. Arthritis Res. Ther. 2011;13:R166. doi: 10.1186/ar3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Muls N, et al. IL-22, GM-CSF and IL-17 in peripheral CD4+ T cell subpopulations during multiple sclerosis relapses and remission. Impact of corticosteroid therapy. PLoS ONE. 2017;12:e0173780. doi: 10.1371/journal.pone.0173780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 726.Rolla S, et al. Th22 cells are expanded in multiple sclerosis and are resistant to IFN-beta. J. Leukoc. Biol. 2014;96:1155–1164. doi: 10.1189/jlb.5A0813-463RR. [DOI] [PubMed] [Google Scholar]
- 727.Zhen J, et al. IL-22 promotes Fas expression in oligodendrocytes and inhibits FOXP3 expression in T cells by activating the NF-kappaB pathway in multiple sclerosis. Mol. Immunol. 2017;82:84–93. doi: 10.1016/j.molimm.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 728.Yang XY, et al. Th22, but not Th17 might be a good index to predict the tissue involvement of systemic lupus erythematosus. J. Clin. Immunol. 2013;33:767–774. doi: 10.1007/s10875-013-9878-1. [DOI] [PubMed] [Google Scholar]
- 729.Zhao L, et al. IL-22+CD4+ T-cells in patients with active systemic lupus erythematosus. Exp. Biol. Med. 2013;238:193–199. doi: 10.1177/1535370213477597. [DOI] [PubMed] [Google Scholar]
- 730.Zhong W, et al. Elevated levels of CCR6(+) T helper 22 cells correlate with skin and renal impairment in systemic lupus erythematosus. Sci. Rep. 2017;7:12962. doi: 10.1038/s41598-017-13344-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Miyazaki Y, et al. Th22 cells promote osteoclast differentiation via production of IL-22 in rheumatoid arthritis. Front. Immunol. 2018;9:2901. doi: 10.3389/fimmu.2018.02901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Ikeuchi H, et al. Expression of interleukin-22 in rheumatoid arthritis: potential role as a proinflammatory cytokine. Arthritis Rheum. 2005;52:1037–1046. doi: 10.1002/art.20965. [DOI] [PubMed] [Google Scholar]
- 733.Wen H, et al. Inhibitory effect and mechanism of 1,25-dihydroxy vitamin D3 on RANKL expression in fibroblast-like synoviocytes and osteoclast-like cell formation induced by IL-22 in rheumatoid arthritis. Clin. Exp. Rheumatol. 2018;36:798–805. [PubMed] [Google Scholar]
- 734.Jiang Q, et al. Role of Th22 cells in the pathogenesis of autoimmune diseases. Front. Immunol. 2021;12:688066. doi: 10.3389/fimmu.2021.688066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Nalleweg N, et al. IL-9 and its receptor are predominantly involved in the pathogenesis of UC. Gut. 2015;64:743–755. doi: 10.1136/gutjnl-2013-305947. [DOI] [PubMed] [Google Scholar]
- 736.Gerlach K, et al. TH9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat. Immunol. 2014;15:676–686. doi: 10.1038/ni.2920. [DOI] [PubMed] [Google Scholar]
- 737.Ouyang H, et al. Increased interleukin‑9 and CD4+IL-9+ T cells in patients with systemic lupus erythematosus. Mol. Med. Rep. 2013;7:1031–1037. doi: 10.3892/mmr.2013.1258. [DOI] [PubMed] [Google Scholar]
- 738.Ciccia F, et al. Potential involvement of IL-9 and Th9 cells in the pathogenesis of rheumatoid arthritis. Rheumatology. 2015;54:2264–2272. doi: 10.1093/rheumatology/kev252. [DOI] [PubMed] [Google Scholar]
- 739.Singh TP, et al. Involvement of IL-9 in Th17-associated inflammation and angiogenesis of psoriasis. PLoS ONE. 2013;8:e51752. doi: 10.1371/journal.pone.0051752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Shao Q, et al. Th9 cells in peripheral blood increased in patients with immune-related pancytopenia. J. Immunol. Res. 2020;2020:6503539. doi: 10.1155/2020/6503539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Kostic M, Zivkovic N, Cvetanovic A, Marjanovic G. CD4(+) T cell phenotypes in the pathogenesis of immune thrombocytopenia. Cell Immunol. 2020;351:104096. doi: 10.1016/j.cellimm.2020.104096. [DOI] [PubMed] [Google Scholar]
- 742.Pan HF, et al. Targeting T-helper 9 cells and interleukin-9 in autoimmune diseases. Cytokine Growth Factor Rev. 2013;24:515–522. doi: 10.1016/j.cytogfr.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 743.Jager A, et al. Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J. Immunol. 2009;183:7169–7177. doi: 10.4049/jimmunol.0901906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744.Li H, et al. IL-9 is important for T-cell activation and differentiation in autoimmune inflammation of the central nervous system. Eur. J. Immunol. 2011;41:2197–2206. doi: 10.1002/eji.201041125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Li H, et al. Neutralization of IL-9 ameliorates experimental autoimmune encephalomyelitis by decreasing the effector T cell population. J. Immunol. 2010;185:4095–4100. doi: 10.4049/jimmunol.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 746.Nowak EC, et al. IL-9 as a mediator of Th17-driven inflammatory disease. J. Exp. Med. 2009;206:1653–1660. doi: 10.1084/jem.20090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Zhou Y, et al. IL-9 promotes Th17 cell migration into the central nervous system via CC chemokine ligand-20 produced by astrocytes. J. Immunol. 2011;186:4415–4421. doi: 10.4049/jimmunol.1003307. [DOI] [PubMed] [Google Scholar]
- 748.Deng Y, et al. Th9 cells and IL-9 in autoimmune disorders: pathogenesis and therapeutic potentials. Hum. Immunol. 2017;78:120–128. doi: 10.1016/j.humimm.2016.12.010. [DOI] [PubMed] [Google Scholar]
- 749.Dantas AT, et al. Increased serum interleukin-9 levels in rheumatoid arthritis and systemic lupus erythematosus: pathogenic role or just an epiphenomenon? Dis. Markers. 2015;2015:519638. doi: 10.1155/2015/519638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Yang J, Li Q, Yang X, Li M. Interleukin-9 is associated with elevated anti-double-stranded DNA antibodies in lupus-prone mice. Mol. Med. 2015;21:364–370. doi: 10.2119/molmed.2014.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Nagy G, et al. Central role of nitric oxide in the pathogenesis of rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res. Ther. 2010;12:210. doi: 10.1186/ar3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Niedbala W, et al. Nitric oxide enhances Th9 cell differentiation and airway inflammation. Nat. Commun. 2014;5:4575. doi: 10.1038/ncomms5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Roy S, Awasthi A. ATP triggers human Th9 cell differentiation via nitric oxide-mediated mTOR-HIF1alpha pathway. Front. Immunol. 2019;10:1120. doi: 10.3389/fimmu.2019.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Chowdhury K, et al. Synovial IL-9 facilitates neutrophil survival, function and differentiation of Th17 cells in rheumatoid arthritis. Arthritis Res. Ther. 2018;20:18. doi: 10.1186/s13075-017-1505-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Stephens GL, et al. IL-9 is a Th17-derived cytokine that limits pathogenic activity in organ-specific autoimmune disease. Eur. J. Immunol. 2011;41:952–962. doi: 10.1002/eji.201040879. [DOI] [PubMed] [Google Scholar]
- 756.Ruocco G, et al. T helper 9 cells induced by plasmacytoid dendritic cells regulate interleukin-17 in multiple sclerosis. Clin. Sci. 2015;129:291–303. doi: 10.1042/CS20140608. [DOI] [PubMed] [Google Scholar]
- 757.Elyaman W, et al. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc. Natl Acad. Sci. USA. 2009;106:12885–12890. doi: 10.1073/pnas.0812530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Vinuesa CG, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 759.Qi H, et al. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.Linterman MA, et al. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Gensous N, et al. T follicular helper cells in autoimmune disorders. Front. Immunol. 2018;9:1637. doi: 10.3389/fimmu.2018.01637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nat. Immunol. 2015;16:142–152. doi: 10.1038/ni.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Walker LSK. The link between circulating follicular helper T cells and autoimmunity. Nat. Rev. Immunol. 2022;22:567–575. doi: 10.1038/s41577-022-00693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 764.Pisetsky DS. Anti-DNA antibodies–quintessential biomarkers of SLE. Nat. Rev. Rheumatol. 2016;12:102–110. doi: 10.1038/nrrheum.2015.151. [DOI] [PubMed] [Google Scholar]
- 765.He J, et al. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 766.Zhang X, et al. Circulating CXCR5+CD4+helper T cells in systemic lupus erythematosus patients share phenotypic properties with germinal center follicular helper T cells and promote antibody production. Lupus. 2015;24:909–917. doi: 10.1177/0961203314567750. [DOI] [PubMed] [Google Scholar]
- 767.Chang A, et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J. Immunol. 2011;186:1849–1860. doi: 10.4049/jimmunol.1001983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 768.Liarski VM, et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci. Transl. Med. 2014;6:230ra246. doi: 10.1126/scitranslmed.3008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Tiller T, et al. Autoreactivity in human IgG+ memory B cells. Immunity. 2007;26:205–213. doi: 10.1016/j.immuni.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 770.Lu J, et al. Follicular helper T cells: potential therapeutic targets in rheumatoid arthritis. Cell Mol. Life Sci. 2021;78:5095–5106. doi: 10.1007/s00018-021-03839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Moschovakis GL, et al. T cell specific Cxcr5 deficiency prevents rheumatoid arthritis. Sci. Rep. 2017;7:8933. doi: 10.1038/s41598-017-08935-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Zhang Y, et al. Elevated circulating Th17 and follicular helper CD4(+) T cells in patients with rheumatoid arthritis. APMIS. 2015;123:659–666. doi: 10.1111/apm.12399. [DOI] [PubMed] [Google Scholar]
- 773.Wang J, et al. High frequencies of activated B cells and T follicular helper cells are correlated with disease activity in patients with new-onset rheumatoid arthritis. Clin. Exp. Immunol. 2013;174:212–220. doi: 10.1111/cei.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774.Manzo A, et al. Mature antigen-experienced T helper cells synthesize and secrete the B cell chemoattractant CXCL13 in the inflammatory environment of the rheumatoid joint. Arthritis Rheum. 2008;58:3377–3387. doi: 10.1002/art.23966. [DOI] [PubMed] [Google Scholar]
- 775.Rao DA, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542:110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Nakayamada S, et al. Differential effects of biological DMARDs on peripheral immune cell phenotypes in patients with rheumatoid arthritis. Rheumatology. 2018;57:164–174. doi: 10.1093/rheumatology/kex012. [DOI] [PubMed] [Google Scholar]
- 777.Platt AM, et al. Abatacept limits breach of self-tolerance in a murine model of arthritis via effects on the generation of T follicular helper cells. J. Immunol. 2010;185:1558–1567. doi: 10.4049/jimmunol.1001311. [DOI] [PubMed] [Google Scholar]
- 778.Quinn JL, et al. Role of TFH cells in promoting T helper 17-induced neuroinflammation. Front. Immunol. 2018;9:382. doi: 10.3389/fimmu.2018.00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Fan X, et al. Circulating CCR7+ICOS+ memory T follicular helper cells in patients with multiple sclerosis. PLoS ONE. 2015;10:e0134523. doi: 10.1371/journal.pone.0134523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 780.Deng J, et al. T follicular helper cells and T follicular regulatory cells in rheumatic diseases. Nat. Rev. Rheumatol. 2019;15:475–490. doi: 10.1038/s41584-019-0254-2. [DOI] [PubMed] [Google Scholar]
- 781.Odegard JM, et al. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J. Exp. Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Soni C, et al. Plasmacytoid dendritic cells and type I interferon promote extrafollicular B cell responses to extracellular self-DNA. Immunity. 2020;52:1022–1038 e1027. doi: 10.1016/j.immuni.2020.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Kenefeck R, et al. Follicular helper T cell signature in type 1 diabetes. J. Clin. Invest. 2015;125:292–303. doi: 10.1172/JCI76238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Ferreira RC, et al. IL-21 production by CD4+ effector T cells and frequency of circulating follicular helper T cells are increased in type 1 diabetes patients. Diabetologia. 2015;58:781–790. doi: 10.1007/s00125-015-3509-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Xu X, et al. Inhibition of increased circulating Tfh cell by anti-CD20 monoclonal antibody in patients with type 1 diabetes. PLoS ONE. 2013;8:e79858. doi: 10.1371/journal.pone.0079858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Correction for Serr. et al. miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity. Proc. Natl Acad. Sci. USA. 2018;115:E4142. doi: 10.1073/pnas.1805675115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Viisanen T, et al. Circulating CXCR5+PD-1+ICOS+ follicular T helper cells are increased close to the diagnosis of type 1 diabetes in children with multiple autoantibodies. Diabetes. 2017;66:437–447. doi: 10.2337/db16-0714. [DOI] [PubMed] [Google Scholar]
- 788.Ren HM, Lukacher AE, Rahman ZSM, Olsen NJ. New developments implicating IL-21 in autoimmune disease. J. Autoimmun. 2021;122:102689. doi: 10.1016/j.jaut.2021.102689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 789.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Ali Abdulla A, Abdulaali Abed T, Razzaq Abdul-Ameer W. Impact of IL-21 gene polymorphisms (rs2055979) and the levels of serum IL-21 on the risk of multiple sclerosis. Arch. Razi Inst. 2022;77:81–86. doi: 10.22092/ARI.2021.356470.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.Biewenga M, et al. B-cell activating factor and IL-21 levels predict treatment response in autoimmune hepatitis. JHEP Rep. 2022;4:100460. doi: 10.1016/j.jhepr.2022.100460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Iervasi E, et al. Serum IL-21 levels from celiac disease patients correlates with anti-tTG IgA autoantibodies and mucosal damage. Autoimmunity. 2020;53:225–230. doi: 10.1080/08916934.2020.1736047. [DOI] [PubMed] [Google Scholar]
- 793.Ettinger R, et al. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J. Immunol. 2005;175:7867–7879. doi: 10.4049/jimmunol.175.12.7867. [DOI] [PubMed] [Google Scholar]
- 794.Kuchen S, et al. Essential role of IL-21 in B cell activation, expansion, and plasma cell generation during CD4+ T cell-B cell collaboration. J. Immunol. 2007;179:5886–5896. doi: 10.4049/jimmunol.179.9.5886. [DOI] [PubMed] [Google Scholar]
- 795.Sutherland AP, et al. Interleukin-21 is required for the development of type 1 diabetes in NOD mice. Diabetes. 2009;58:1144–1155. doi: 10.2337/db08-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 796.Spolski R, et al. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. Proc. Natl Acad. Sci. USA. 2008;105:14028–14033. doi: 10.1073/pnas.0804358105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Kwok SK, et al. Interleukin-21 promotes osteoclastogenesis in humans with rheumatoid arthritis and in mice with collagen-induced arthritis. Arthritis Rheum. 2012;64:740–751. doi: 10.1002/art.33390. [DOI] [PubMed] [Google Scholar]
- 798.Xing R, et al. Interleukin-21 induces migration and invasion of fibroblast-like synoviocytes from patients with rheumatoid arthritis. Clin. Exp. Immunol. 2016;184:147–158. doi: 10.1111/cei.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Xing R, et al. Interleukin-21 induces proliferation and proinflammatory cytokine profile of fibroblast-like synoviocytes of patients with rheumatoid arthritis. Scand. J. Immunol. 2016;83:64–71. doi: 10.1111/sji.12396. [DOI] [PubMed] [Google Scholar]
- 800.Caruso R, et al. Involvement of interleukin-21 in the epidermal hyperplasia of psoriasis. Nat. Med. 2009;15:1013–1015. doi: 10.1038/nm.1995. [DOI] [PubMed] [Google Scholar]
- 801.Jungel A, et al. Expression of interleukin-21 receptor, but not interleukin-21, in synovial fibroblasts and synovial macrophages of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50:1468–1476. doi: 10.1002/art.20218. [DOI] [PubMed] [Google Scholar]
- 802.Peluso I, et al. IL-21 counteracts the regulatory T cell-mediated suppression of human CD4+ T lymphocytes. J. Immunol. 2007;178:732–739. doi: 10.4049/jimmunol.178.2.732. [DOI] [PubMed] [Google Scholar]
- 803.Clough LE, et al. Release from regulatory T cell-mediated suppression during the onset of tissue-specific autoimmunity is associated with elevated IL-21. J. Immunol. 2008;180:5393–5401. doi: 10.4049/jimmunol.180.8.5393. [DOI] [PubMed] [Google Scholar]
- 804.Edo A, et al. Therapeutic effect of IL-21 blockage by gene therapy in experimental autoimmune encephalomyelitis. Neurotherapeutics. 2022;19:1617–1633. doi: 10.1007/s13311-022-01279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 805.Choi JY, et al. Disruption of pathogenic cellular networks by IL-21 blockade leads to disease amelioration in murine lupus. J. Immunol. 2017;198:2578–2588. doi: 10.4049/jimmunol.1601687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 806.Zhang M, et al. Interleukin-21 receptor blockade inhibits secondary humoral responses and halts the progression of preestablished disease in the (NZB x NZW)F1 systemic lupus erythematosus model. Arthritis Rheumatol. 2015;67:2723–2731. doi: 10.1002/art.39233. [DOI] [PubMed] [Google Scholar]
- 807.Feng X, et al. Inhibition of aberrant circulating Tfh cell proportions by corticosteroids in patients with systemic lupus erythematosus. PLoS ONE. 2012;7:e51982. doi: 10.1371/journal.pone.0051982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.He J, et al. Low-dose interleukin-2 treatment selectively modulates CD4(+) T cell subsets in patients with systemic lupus erythematosus. Nat. Med. 2016;22:991–993. doi: 10.1038/nm.4148. [DOI] [PubMed] [Google Scholar]
- 809.Rosenzwajg M, et al. Immunological and clinical effects of low-dose interleukin-2 across 11 autoimmune diseases in a single, open clinical trial. Ann. Rheum. Dis. 2019;78:209–217. doi: 10.1136/annrheumdis-2018-214229. [DOI] [PubMed] [Google Scholar]
- 810.Qiu CC, Caricchio R, Gallucci S. Triggers of autoimmunity: the role of bacterial infections in the extracellular exposure of lupus nuclear autoantigens. Front. Immunol. 2019;10:2608. doi: 10.3389/fimmu.2019.02608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.Steelman AJ. Infection as an environmental trigger of multiple sclerosis disease exacerbation. Front. Immunol. 2015;6:520. doi: 10.3389/fimmu.2015.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 812.Rodriguez-Calvo T. Enteroviral infections as a trigger for type 1 diabetes. Curr. Diab. Rep. 2018;18:106. doi: 10.1007/s11892-018-1077-2. [DOI] [PubMed] [Google Scholar]
- 813.Vehik K, et al. Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat. Med. 2019;25:1865–1872. doi: 10.1038/s41591-019-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 814.Konig MF, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci. Transl. Med. 2016;8:369ra176. doi: 10.1126/scitranslmed.aaj1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.Lanz TV, et al. Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM. Nature. 2022;603:321–327. doi: 10.1038/s41586-022-04432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 816.Bjornevik K, et al. Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis. Science. 2022;375:296–301. doi: 10.1126/science.abj8222. [DOI] [PubMed] [Google Scholar]
- 817.Zhao Z, et al. Nature of T cell epitopes in lupus antigens and HLA-DR determines autoantibody initiation and diversification. Ann. Rheum. Dis. 2019;78:380–390. doi: 10.1136/annrheumdis-2018-214125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.Wang EY, et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595:283–288. doi: 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- 819.Schwickert TA, Alabyev B, Manser T, Nussenzweig MC. Germinal center reutilization by newly activated B cells. J. Exp. Med. 2009;206:2907–2914. doi: 10.1084/jem.20091225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 820.Sanderson NS, et al. Cocapture of cognate and bystander antigens can activate autoreactive B cells. Proc. Natl Acad. Sci. USA. 2017;114:734–739. doi: 10.1073/pnas.1614472114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat. Immunol. 2018;19:665–673. doi: 10.1038/s41590-018-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 2019;50:302–316. doi: 10.1016/j.immuni.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 823.Powell BR, Buist NR, Stenzel P. An X-linked syndrome of diarrhea, polyendocrinopathy, and fatal infection in infancy. J. Pediatr. 1982;100:731–737. doi: 10.1016/S0022-3476(82)80573-8. [DOI] [PubMed] [Google Scholar]
- 824.Bacchetta R, Barzaghi F, Roncarolo MG. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Ann. N. Y Acad. Sci. 2018;1417:5–22. doi: 10.1111/nyas.13011. [DOI] [PubMed] [Google Scholar]
- 825.Caudy AA, et al. CD25 deficiency causes an immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like syndrome, and defective IL-10 expression from CD4 lymphocytes. J. Allergy Clin. Immunol. 2007;119:482–487. doi: 10.1016/j.jaci.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 826.Schubert D, et al. Autosomal dominant immune dysregulation syndrome in humans with CTLA4 mutations. Nat. Med. 2014;20:1410–1416. doi: 10.1038/nm.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827.Kuehn HS, et al. Immune dysregulation in human subjects with heterozygous germline mutations in CTLA4. Science. 2014;345:1623–1627. doi: 10.1126/science.1255904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Lo B, et al. AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349:436–440. doi: 10.1126/science.aaa1663. [DOI] [PubMed] [Google Scholar]
- 829.Yang S, et al. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Kekalainen E, et al. A defect of regulatory T cells in patients with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J. Immunol. 2007;178:1208–1215. doi: 10.4049/jimmunol.178.2.1208. [DOI] [PubMed] [Google Scholar]
- 831.Sakaguchi S, et al. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 1995;155:1151–1164. doi: 10.4049/jimmunol.155.3.1151. [DOI] [PubMed] [Google Scholar]
- 832.Liu Z, et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature. 2015;528:225–230. doi: 10.1038/nature16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 833.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 834.Zhang X, Olsen N, Zheng SG. The progress and prospect of regulatory T cells in autoimmune diseases. J. Autoimmun. 2020;111:102461. doi: 10.1016/j.jaut.2020.102461. [DOI] [PubMed] [Google Scholar]
- 835.Wing JB, Sakaguchi S. Multiple treg suppressive modules and their adaptability. Front. Immunol. 2012;3:178. doi: 10.3389/fimmu.2012.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 836.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 837.Levine AG, et al. Stability and function of regulatory T cells expressing the transcription factor T-bet. Nature. 2017;546:421–425. doi: 10.1038/nature22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 838.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 839.Wohlfert EA, et al. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J. Clin. Invest. 2011;121:4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Sefik E, et al. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Linterman MA, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat. Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 843.Chung Y, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat. Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Goschl L, Scheinecker C, Bonelli M. Treg cells in autoimmunity: from identification to Treg-based therapies. Semin. Immunopathol. 2019;41:301–314. doi: 10.1007/s00281-019-00741-8. [DOI] [PubMed] [Google Scholar]
- 845.Lin SC, et al. The quantitative analysis of peripheral blood FOXP3-expressing T cells in systemic lupus erythematosus and rheumatoid arthritis patients. Eur. J. Clin. Invest. 2007;37:987–996. doi: 10.1111/j.1365-2362.2007.01882.x. [DOI] [PubMed] [Google Scholar]
- 846.Baatjes AJ, et al. T regulatory cell phenotypes in peripheral blood and bronchoalveolar lavage from non-asthmatic and asthmatic subjects. Clin. Exp. Allergy. 2015;45:1654–1662. doi: 10.1111/cea.12594. [DOI] [PubMed] [Google Scholar]
- 847.Brusko T, et al. No alterations in the frequency of FOXP3+ regulatory T-cells in type 1 diabetes. Diabetes. 2007;56:604–612. doi: 10.2337/db06-1248. [DOI] [PubMed] [Google Scholar]
- 848.Matsui N, et al. Undiminished regulatory T cells in the thymus of patients with myasthenia gravis. Neurology. 2010;74:816–820. doi: 10.1212/WNL.0b013e3181d31e47. [DOI] [PubMed] [Google Scholar]
- 849.Lawson CA, et al. Early rheumatoid arthritis is associated with a deficit in the CD4+CD25high regulatory T cell population in peripheral blood. Rheumatology. 2006;45:1210–1217. doi: 10.1093/rheumatology/kel089. [DOI] [PubMed] [Google Scholar]
- 850.Mellor-Pita S, et al. Decrease of regulatory T cells in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 2006;65:553–554. doi: 10.1136/ard.2005.044974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Han GM, O’Neil-Andersen NJ, Zurier RB, Lawrence DA. CD4+CD25high T cell numbers are enriched in the peripheral blood of patients with rheumatoid arthritis. Cell Immunol. 2008;253:92–101. doi: 10.1016/j.cellimm.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Yates J, et al. Natural regulatory T cells: number and function are normal in the majority of patients with lupus nephritis. Clin. Exp. Immunol. 2008;153:44–55. doi: 10.1111/j.1365-2249.2008.03665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.McClymont SA, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J. Immunol. 2011;186:3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 854.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat. Med. 2011;17:673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 855.Kitz A, et al. AKT isoforms modulate Th1-like Treg generation and function in human autoimmune disease. EMBO Rep. 2019;20:e48624. doi: 10.15252/embr.201948624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 856.Arterbery AS, et al. Production of proinflammatory cytokines by monocytes in liver-transplanted recipients with de novo autoimmune hepatitis is enhanced and induces TH1-like regulatory T cells. J. Immunol. 2016;196:4040–4051. doi: 10.4049/jimmunol.1502276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 857.Yamada A, et al. Impaired expansion of regulatory T cells in a neonatal thymectomy-induced autoimmune mouse model. Am. J. Pathol. 2015;185:2886–2897. doi: 10.1016/j.ajpath.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 858.Korn T, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat. Med. 2007;13:423–431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 859.Tan TG, Mathis D, Benoist C. Singular role for T-BET+CXCR3+ regulatory T cells in protection from autoimmune diabetes. Proc. Natl Acad. Sci. USA. 2016;113:14103–14108. doi: 10.1073/pnas.1616710113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860.Yang S, et al. Differential roles of TNFalpha-TNFR1 and TNFalpha-TNFR2 in the differentiation and function of CD4(+)Foxp3(+) induced Treg cells in vitro and in vivo periphery in autoimmune diseases. Cell Death Dis. 2019;10:27. doi: 10.1038/s41419-018-1266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 861.Luo Y, Zheng SG. Hall of fame among pro-inflammatory cytokines: interleukin-6 gene and its transcriptional regulation mechanisms. Front. Immunol. 2016;7:604. doi: 10.3389/fimmu.2016.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Tarique M, et al. IL-12 and IL-23 modulate plasticity of FoxP3(+) regulatory T cells in human Leprosy. Mol. Immunol. 2017;83:72–81. doi: 10.1016/j.molimm.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 863.Ouyang W, et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864.Huynh A, et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 2015;16:188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 865.MacDonald KG, et al. Regulatory T cells produce profibrotic cytokines in the skin of patients with systemic sclerosis. J. Allergy Clin. Immunol. 2015;135:946–955 e949. doi: 10.1016/j.jaci.2014.12.1932. [DOI] [PubMed] [Google Scholar]
- 866.Noval Rivas M, et al. Regulatory T cell reprogramming toward a Th2-cell-like lineage impairs oral tolerance and promotes food allergy. Immunity. 2015;42:512–523. doi: 10.1016/j.immuni.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867.Jin HS, Park Y, Elly C, Liu YC. Itch expression by Treg cells controls Th2 inflammatory responses. J. Clin. Invest. 2013;123:4923–4934. doi: 10.1172/JCI69355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 868.Chen T, et al. The imbalance of FOXP3/GATA3 in regulatory T cells from the peripheral blood of asthmatic patients. J. Immunol. Res. 2018;2018:3096183. doi: 10.1155/2018/3096183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Gao N, et al. Contribution of Th2-like Treg cells to the pathogenesis of Takayasu’s arteritis. Clin. Exp. Rheumatol. 2020;38:48–54. [PubMed] [Google Scholar]
- 870.Chen J, et al. Increased dysfunctional and plastic regulatory T cells in idiopathic orbital inflammation. Front. Immunol. 2021;12:634847. doi: 10.3389/fimmu.2021.634847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Saigusa R, et al. Fli1-haploinsufficient dermal fibroblasts promote skin-localized transdifferentiation of Th2-like regulatory T cells. Arthritis Res. Ther. 2018;20:23. doi: 10.1186/s13075-018-1521-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 872.Komatsu N, et al. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat. Med. 2014;20:62–68. doi: 10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- 873.Jiang C, et al. Reprograming of peripheral Foxp3(+) regulatory T cell towards Th17-like cell in patients with active systemic lupus erythematosus. Clin. Immunol. 2019;209:108267. doi: 10.1016/j.clim.2019.108267. [DOI] [PubMed] [Google Scholar]
- 874.Bovenschen HJ, et al. Foxp3+ regulatory T cells of psoriasis patients easily differentiate into IL-17A-producing cells and are found in lesional skin. J. Invest. Dermatol. 2011;131:1853–1860. doi: 10.1038/jid.2011.139. [DOI] [PubMed] [Google Scholar]
- 875.Yang BH, et al. Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9:444–457. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- 876.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J. Immunol. 2007;178:6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 877.Massoud AH, et al. An asthma-associated IL4R variant exacerbates airway inflammation by promoting conversion of regulatory T cells to TH17-like cells. Nat. Med. 2016;22:1013–1022. doi: 10.1038/nm.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 878.Nyirenda MH, et al. TLR2 stimulation drives human naive and effector regulatory T cells into a Th17-like phenotype with reduced suppressive function. J. Immunol. 2011;187:2278–2290. doi: 10.4049/jimmunol.1003715. [DOI] [PubMed] [Google Scholar]
- 879.Cho SN, et al. Role of staphylococcal enterotoxin B on the differentiation of regulatory T cells in nasal polyposis. Am. J. Rhinol. Allergy. 2014;28:e17–e24. doi: 10.2500/ajra.2014.28.3995. [DOI] [PubMed] [Google Scholar]
- 880.Yu W, et al. IRF4 is correlated with the conversion to a Th17-like phenotype in regulatory T cells from the malignant pleural effusion. Int. J. Gen. Med. 2021;14:6009–6019. doi: 10.2147/IJGM.S330389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 881.Takahashi R, Nakatsukasa H, Shiozawa S, Yoshimura A. SOCS1 is a key molecule that prevents regulatory T cell plasticity under inflammatory conditions. J. Immunol. 2017;199:149–158. doi: 10.4049/jimmunol.1600441. [DOI] [PubMed] [Google Scholar]
- 882.Baban B, et al. IDO activates regulatory T cells and blocks their conversion into Th17-like T cells. J. Immunol. 2009;183:2475–2483. doi: 10.4049/jimmunol.0900986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 883.Zhou X, Bailey-Bucktrout S, Jeker LT, Bluestone JA. Plasticity of CD4(+) FoxP3(+) T cells. Curr. Opin. Immunol. 2009;21:281–285. doi: 10.1016/j.coi.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 884.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 885.Tang Q, et al. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 886.Thiruppathi M, et al. Functional defect in regulatory T cells in myasthenia gravis. Ann. N. Y Acad. Sci. 2012;1274:68–76. doi: 10.1111/j.1749-6632.2012.06840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 887.Balandina A, et al. Functional defect of regulatory CD4(+)CD25+ T cells in the thymus of patients with autoimmune myasthenia gravis. Blood. 2005;105:735–741. doi: 10.1182/blood-2003-11-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 888.Huan J, et al. Decreased FOXP3 levels in multiple sclerosis patients. J. Neurosci. Res. 2005;81:45–52. doi: 10.1002/jnr.20522. [DOI] [PubMed] [Google Scholar]
- 889.Zhang B, et al. Reduction of forkhead box P3 levels in CD4+CD25high T cells in patients with new-onset systemic lupus erythematosus. Clin. Exp. Immunol. 2008;153:182–187. doi: 10.1111/j.1365-2249.2008.03686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 890.Zhou X, et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 2009;10:1000–1007. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 891.Rubtsov YP, et al. Stability of the regulatory T cell lineage in vivo. Science. 2010;329:1667–1671. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 892.Li L, et al. Block of both TGF-beta and IL-2 signaling impedes Neurophilin-1(+) regulatory T cell and follicular regulatory T cell development. Cell Death Dis. 2016;7:e2439. doi: 10.1038/cddis.2016.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 893.Takimoto T, et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 2010;185:842–855. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- 894.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 895.Qiu R, et al. Regulatory T cell plasticity and stability and autoimmune diseases. Clin. Rev. Allergy Immunol. 2020;58:52–70. doi: 10.1007/s12016-018-8721-0. [DOI] [PubMed] [Google Scholar]
- 896.Kumar S, et al. CD4+CD25+ T regs with acetylated FoxP3 are associated with immune suppression in human leprosy. Mol. Immunol. 2013;56:513–520. doi: 10.1016/j.molimm.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 897.Geng J, et al. Publisher correction: the transcriptional coactivator TAZ regulates reciprocal differentiation of T(H)17 cells and T(reg) cells. Nat. Immunol. 2018;19:1036. doi: 10.1038/s41590-018-0055-9. [DOI] [PubMed] [Google Scholar]
- 898.Liu B, et al. The lineage stability and suppressive program of regulatory T cells require protein O-GlcNAcylation. Nat. Commun. 2019;10:354. doi: 10.1038/s41467-019-08300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 899.Alvarez Salazar EK, et al. Methylation of FOXP3 TSDR underlies the impaired suppressive function of Tregs from long-term belatacept-treated kidney transplant patients. Front. Immunol. 2017;8:219. doi: 10.3389/fimmu.2017.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 900.Miyao T, et al. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 901.Deng G, et al. Pim-2 kinase influences regulatory T cell function and stability by mediating Foxp3 protein N-terminal phosphorylation. J. Biol. Chem. 2015;290:20211–20220. doi: 10.1074/jbc.M115.638221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 902.Morawski PA, et al. Foxp3 protein stability is regulated by cyclin-dependent kinase 2. J. Biol. Chem. 2013;288:24494–24502. doi: 10.1074/jbc.M113.467704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 903.Xu, Y. et al. The E3 ligase Hrd1 stabilizes Tregs by antagonizing inflammatory cytokine-induced ER stress response. JCI Insight4, e121887 (2019). [DOI] [PMC free article] [PubMed]
- 904.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 905.Li X, et al. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 2014;158:734–748. doi: 10.1016/j.cell.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 906.Feng Y, et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–763. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 907.Wang L, et al. Mbd2 promotes foxp3 demethylation and T-regulatory-cell function. Mol. Cell Biol. 2013;33:4106–4115. doi: 10.1128/MCB.00144-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 908.Kim HJ, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350:334–339. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 909.Sharma MD, et al. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor eos. Immunity. 2013;38:998–1012. doi: 10.1016/j.immuni.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 910.Messina N, et al. The NF-kappaB transcription factor RelA is required for the tolerogenic function of Foxp3(+) regulatory T cells. J. Autoimmun. 2016;70:52–62. doi: 10.1016/j.jaut.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 911.Verrecchia F, et al. Smad3/AP-1 interactions control transcriptional responses to TGF-beta in a promoter-specific manner. Oncogene. 2001;20:3332–3340. doi: 10.1038/sj.onc.1204448. [DOI] [PubMed] [Google Scholar]
- 912.Rauch KS, et al. Id3 maintains Foxp3 expression in regulatory T cells by controlling a transcriptional network of E47, Spi-B, and SOCS3. Cell Rep. 2016;17:2827–2836. doi: 10.1016/j.celrep.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 913.Arnold PR, et al. Suppression of FOXP3 expression by the AP-1 family transcription factor BATF3 requires partnering with IRF4. Front. Immunol. 2022;13:966364. doi: 10.3389/fimmu.2022.966364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 914.Collier JL, et al. Not-so-opposite ends of the spectrum: CD8(+) T cell dysfunction across chronic infection, cancer and autoimmunity. Nat. Immunol. 2021;22:809–819. doi: 10.1038/s41590-021-00949-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 915.Walter U, Santamaria P. CD8+ T cells in autoimmunity. Curr. Opin. Immunol. 2005;17:624–631. doi: 10.1016/j.coi.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 916.Brewerton DA, et al. Ankylosing spondylitis and HL-A 27. Lancet. 1973;1:904–907. doi: 10.1016/S0140-6736(73)91360-3. [DOI] [PubMed] [Google Scholar]
- 917.Cortes A, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun. 2015;6:7146. doi: 10.1038/ncomms8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 918.Skowera A, et al. beta-cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes. 2015;64:916–925. doi: 10.2337/db14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 919.Wagner CA, et al. Myelin-specific CD8+ T cells exacerbate brain inflammation in CNS autoimmunity. J. Clin. Invest. 2020;130:203–213. doi: 10.1172/JCI132531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 920.Lee JC, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J. Clin. Invest. 2011;121:4170–4179. doi: 10.1172/JCI59255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 921.Le Gal FA, et al. Direct evidence to support the role of antigen-specific CD8(+) T cells in melanoma-associated vitiligo. J. Invest Dermatol. 2001;117:1464–1470. doi: 10.1046/j.0022-202x.2001.01605.x. [DOI] [PubMed] [Google Scholar]
- 922.Cheuk S, et al. CD49a expression defines tissue-resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity. 2017;46:287–300. doi: 10.1016/j.immuni.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 923.Amrani A, et al. Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature. 2000;406:739–742. doi: 10.1038/35021081. [DOI] [PubMed] [Google Scholar]
- 924.Garyu JW, et al. Characterization of Diabetogenic CD8+ T Cells: IMMUNE THERAPY WITH METABOLIC BLOCKADE. J. Biol. Chem. 2016;291:11230–11240. doi: 10.1074/jbc.M115.713362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 925.Han B, et al. Developmental control of CD8 T cell-avidity maturation in autoimmune diabetes. J. Clin. Invest. 2005;115:1879–1887. doi: 10.1172/JCI24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 926.Skulina C, et al. Multiple sclerosis: brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood. Proc. Natl Acad. Sci. USA. 2004;101:2428–2433. doi: 10.1073/pnas.0308689100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 927.Petrelli A, van Wijk F. CD8(+) T cells in human autoimmune arthritis: the unusual suspects. Nat. Rev. Rheumatol. 2016;12:421–428. doi: 10.1038/nrrheum.2016.74. [DOI] [PubMed] [Google Scholar]
- 928.Carvalheiro H, et al. CD8+ T cell profiles in patients with rheumatoid arthritis and their relationship to disease activity. Arthritis Rheumatol. 2015;67:363–371. doi: 10.1002/art.38941. [DOI] [PubMed] [Google Scholar]
- 929.Bender, C. et al. The healthy exocrine pancreas contains preproinsulin-specific CD8 T cells that attack islets in type 1 diabetes. Sci. Adv. 6, eabc5586 (2020). [DOI] [PMC free article] [PubMed]
- 930.Ifergan I, et al. Central nervous system recruitment of effector memory CD8+ T lymphocytes during neuroinflammation is dependent on alpha4 integrin. Brain. 2011;134:3560–3577. doi: 10.1093/brain/awr268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 931.Zakharov, P. N., Hu, H., Wan, X. & Unanue, E. R. Single-cell RNA sequencing of murine islets shows high cellular complexity at all stages of autoimmune diabetes. J. Exp. Med. 217, e20192362 (2020). [DOI] [PMC free article] [PubMed]
- 932.McKinney EF, et al. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523:612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 933.Wiedeman AE, et al. Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression. J. Clin. Invest. 2020;130:480–490. doi: 10.1172/JCI126595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 934.Gearty SV, et al. An autoimmune stem-like CD8 T cell population drives type 1 diabetes. Nature. 2022;602:156–161. doi: 10.1038/s41586-021-04248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 935.Page N, et al. Persistence of self-reactive CD8+ T cells in the CNS requires TOX-dependent chromatin remodeling. Nat. Commun. 2021;12:1009. doi: 10.1038/s41467-021-21109-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 936.Lauritsen JP, et al. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 937.Mengrelis K, et al. Sonic hedgehog is a determinant of gammadelta T-cell differentiation in the thymus. Front. Immunol. 2019;10:1629. doi: 10.3389/fimmu.2019.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 938.Ciofani M, Zuniga-Pflucker JC. Determining gammadelta versus alphass T cell development. Nat. Rev. Immunol. 2010;10:657–663. doi: 10.1038/nri2820. [DOI] [PubMed] [Google Scholar]
- 939.Van de Walle I, et al. Specific Notch receptor-ligand interactions control human TCR-alphabeta/gammadelta development by inducing differential Notch signal strength. J. Exp. Med. 2013;210:683–697. doi: 10.1084/jem.20121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 940.Scaramuzzino S, et al. Single-cell transcriptomics uncovers an instructive T-cell receptor role in adult gammadelta T-cell lineage commitment. EMBO J. 2022;41:e110023. doi: 10.15252/embj.2021110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 941.Roels J, et al. Distinct and temporary-restricted epigenetic mechanisms regulate human alphabeta and gammadelta T cell development. Nat. Immunol. 2020;21:1280–1292. doi: 10.1038/s41590-020-0747-9. [DOI] [PubMed] [Google Scholar]
- 942.Yang, K. et al. Metabolic signaling directs the reciprocal lineage decisions of alphabeta and gammadelta T cells. Sci Immunol. 3, eaas9818 (2018). [DOI] [PMC free article] [PubMed]
- 943.Parker ME, Ciofani M. Regulation of gammadelta T cell effector diversification in the thymus. Front. Immunol. 2020;11:42. doi: 10.3389/fimmu.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 944.Kisielow J, Kopf M, Karjalainen K. SCART scavenger receptors identify a novel subset of adult gammadelta T cells. J. Immunol. 2008;181:1710–1716. doi: 10.4049/jimmunol.181.3.1710. [DOI] [PubMed] [Google Scholar]
- 945.Ribot JC, Lopes N, Silva-Santos B. gammadelta T cells in tissue physiology and surveillance. Nat. Rev. Immunol. 2021;21:221–232. doi: 10.1038/s41577-020-00452-4. [DOI] [PubMed] [Google Scholar]
- 946.Narayan K, et al. Intrathymic programming of effector fates in three molecularly distinct gammadelta T cell subtypes. Nat. Immunol. 2012;13:511–518. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 947.Sagar, et al. Deciphering the regulatory landscape of fetal and adult gammadelta T-cell development at single-cell resolution. EMBO J. 2020;39:e104159. doi: 10.15252/embj.2019104159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 948.Malhotra N, et al. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 2013;38:681–693. doi: 10.1016/j.immuni.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 949.Fahl SP, et al. The E protein-TCF1 axis controls gammadelta T cell development and effector fate. Cell Rep. 2021;34:108716. doi: 10.1016/j.celrep.2021.108716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 950.Turchinovich G, Hayday AC. Skint-1 identifies a common molecular mechanism for the development of interferon-gamma-secreting versus interleukin-17-secreting gammadelta T cells. Immunity. 2011;35:59–68. doi: 10.1016/j.immuni.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 951.Sutton CE, et al. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–341. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 952.Patil RS, Bhat SA, Dar AA, Chiplunkar SV. The Jekyll and Hyde story of IL17-producing gammadeltaT cells. Front. Immunol. 2015;6:37. doi: 10.3389/fimmu.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 953.Li Z, et al. Single-cell RNA-seq and chromatin accessibility profiling decipher the heterogeneity of mouse gammadelta T cells. Sci. Bull. 2022;67:408–426. doi: 10.1016/j.scib.2021.11.013. [DOI] [PubMed] [Google Scholar]
- 954.Hayday AC, Vantourout P. The innate biologies of adaptive antigen receptors. Annu. Rev. Immunol. 2020;38:487–510. doi: 10.1146/annurev-immunol-102819-023144. [DOI] [PubMed] [Google Scholar]
- 955.Di Marco Barros R, et al. Epithelia use butyrophilin-like molecules to shape organ-specific gammadelta T cell compartments. Cell. 2016;167:203–218 e217. doi: 10.1016/j.cell.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 956.Jameson JM, et al. Gammadelta T cell-induced hyaluronan production by epithelial cells regulates inflammation. J. Exp. Med. 2005;201:1269–1279. doi: 10.1084/jem.20042057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 957.Jameson J, et al. A role for skin gammadelta T cells in wound repair. Science. 2002;296:747–749. doi: 10.1126/science.1069639. [DOI] [PubMed] [Google Scholar]
- 958.Boismenu R, Havran WL. Modulation of epithelial cell growth by intraepithelial gamma delta T cells. Science. 1994;266:1253–1255. doi: 10.1126/science.7973709. [DOI] [PubMed] [Google Scholar]
- 959.Ahlfors H, et al. IL-22 fate reporter reveals origin and control of IL-22 production in homeostasis and infection. J. Immunol. 2014;193:4602–4613. doi: 10.4049/jimmunol.1401244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 960.Wilharm A, et al. Mutual interplay between IL-17-producing gammadeltaT cells and microbiota orchestrates oral mucosal homeostasis. Proc. Natl Acad. Sci. USA. 2019;116:2652–2661. doi: 10.1073/pnas.1818812116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 961.Krishnan S, et al. Amphiregulin-producing gammadelta T cells are vital for safeguarding oral barrier immune homeostasis. Proc. Natl Acad. Sci. USA. 2018;115:10738–10743. doi: 10.1073/pnas.1802320115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 962.Spidale, N. A. et al. Neonatal-derived IL-17 producing dermal gammadelta T cells are required to prevent spontaneous atopic dermatitis. Elife. 9, e51188 (2020). [DOI] [PMC free article] [PubMed]
- 963.Papotto PH, Ribot JC, Silva-Santos B. IL-17(+) gammadelta T cells as kick-starters of inflammation. Nat. Immunol. 2017;18:604–611. doi: 10.1038/ni.3726. [DOI] [PubMed] [Google Scholar]
- 964.Ono T, et al. IL-17-producing gammadelta T cells enhance bone regeneration. Nat. Commun. 2016;7:10928. doi: 10.1038/ncomms10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 965.Hu B, et al. gammadelta T cells and adipocyte IL-17RC control fat innervation and thermogenesis. Nature. 2020;578:610–614. doi: 10.1038/s41586-020-2028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 966.Kohlgruber AC, et al. gammadelta T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol. 2018;19:464–474. doi: 10.1038/s41590-018-0094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 967.Ribeiro, M. et al. Meningeal gammadelta T cell-derived IL-17 controls synaptic plasticity and short-term memory. Sci Immunol. 4, eaay5199 (2019). [DOI] [PMC free article] [PubMed]
- 968.Holtmeier W, Kabelitz D. gammadelta T cells link innate and adaptive immune responses. Chem. Immunol. Allergy. 2005;86:151–183. doi: 10.1159/000086659. [DOI] [PubMed] [Google Scholar]
- 969.Vermijlen D, et al. gammadelta T cell responses: How many ligands will it take till we know? Semin. Cell Dev. Biol. 2018;84:75–86. doi: 10.1016/j.semcdb.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 970.Dillen CA, et al. Clonally expanded gammadelta T cells protect against Staphylococcus aureus skin reinfection. J. Clin. Invest. 2018;128:1026–1042. doi: 10.1172/JCI96481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 971.Murphy AG, et al. Staphylococcus aureus infection of mice expands a population of memory gammadelta T cells that are protective against subsequent infection. J. Immunol. 2014;192:3697–3708. doi: 10.4049/jimmunol.1303420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 972.Bertram T, et al. Kidney-resident innate-like memory gammadelta T cells control chronic Staphylococcus aureus infection of mice. Proc. Natl Acad. Sci. USA. 2023;120:e2210490120. doi: 10.1073/pnas.2210490120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 973.Shires J, Theodoridis E, Hayday AC. Biological insights into TCRgammadelta+ and TCRalphabeta+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE) Immunity. 2001;15:419–434. doi: 10.1016/S1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- 974.Nakasone C, et al. Accumulation of gamma/delta T cells in the lungs and their roles in neutrophil-mediated host defense against pneumococcal infection. Microbes Infect. 2007;9:251–258. doi: 10.1016/j.micinf.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 975.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J. Immunol. 2006;177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 976.Cimini, E. & Agrati, C. gammadelta T cells in emerging viral infection: an overview. Viruses. 14, 1166 (2022). [DOI] [PMC free article] [PubMed]
- 977.von Massow G, Oh S, Lam A, Gustafsson K. Gamma delta T cells and their involvement in COVID-19 virus infections. Front. Immunol. 2021;12:741218. doi: 10.3389/fimmu.2021.741218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 978.Papotto PH, Yilmaz B, Silva-Santos B. Crosstalk between gammadelta T cells and the microbiota. Nat. Microbiol. 2021;6:1110–1117. doi: 10.1038/s41564-021-00948-2. [DOI] [PubMed] [Google Scholar]
- 979.Sheridan BS, et al. gammadelta T cells exhibit multifunctional and protective memory in intestinal tissues. Immunity. 2013;39:184–195. doi: 10.1016/j.immuni.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 980.Kalyan S, Kabelitz D. Defining the nature of human gammadelta T cells: a biographical sketch of the highly empathetic. Cell Mol. Immunol. 2013;10:21–29. doi: 10.1038/cmi.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 981.Mensurado S, Blanco-Dominguez R, Silva-Santos B. The emerging roles of gammadelta T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol. 2023;20:178–191. doi: 10.1038/s41571-022-00722-1. [DOI] [PubMed] [Google Scholar]
- 982.Lee, D. et al. Human gammadelta T cell subsets and their clinical applications for cancer immunotherapy. Cancers. 14, 3005 (2022). [DOI] [PMC free article] [PubMed]
- 983.Willcox CR, Davey MS, Willcox BE. Development and selection of the human Vgamma9Vdelta2(+) T-cell repertoire. Front. Immunol. 2018;9:1501. doi: 10.3389/fimmu.2018.01501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 984.Gober HJ, et al. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J. Exp. Med. 2003;197:163–168. doi: 10.1084/jem.20021500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 985.Rigau, M. et al. Butyrophilin 2A1 is essential for phosphoantigen reactivity by gammadelta T cells. Science. 367, eaay5516 (2020). [DOI] [PubMed]
- 986.De Gassart A, et al. Development of ICT01, a first-in-class, anti-BTN3A antibody for activating Vgamma9Vdelta2 T cell-mediated antitumor immune response. Sci. Transl. Med. 2021;13:eabj0835. doi: 10.1126/scitranslmed.abj0835. [DOI] [PubMed] [Google Scholar]
- 987.Hoeres T, Smetak M, Pretscher D, Wilhelm M. Improving the efficiency of Vgamma9Vdelta2 T-cell immunotherapy in cancer. Front. Immunol. 2018;9:800. doi: 10.3389/fimmu.2018.00800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 988.Luoma AM, Castro CD, Adams EJ. gammadelta T cell surveillance via CD1 molecules. Trends Immunol. 2014;35:613–621. doi: 10.1016/j.it.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 989.Dar AA, Patil RS, Chiplunkar SV. Insights into the relationship between Toll like receptors and gamma delta T cell responses. Front. Immunol. 2014;5:366. doi: 10.3389/fimmu.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 990.Wu P, et al. gammadeltaT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity. 2014;40:785–800. doi: 10.1016/j.immuni.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 991.Patil RS, et al. IL17 producing gammadeltaT cells induce angiogenesis and are associated with poor survival in gallbladder cancer patients. Int. J. Cancer. 2016;139:869–881. doi: 10.1002/ijc.30134. [DOI] [PubMed] [Google Scholar]
- 992.Ma C, et al. Tumor-infiltrating gammadelta T lymphocytes predict clinical outcome in human breast cancer. J. Immunol. 2012;189:5029–5036. doi: 10.4049/jimmunol.1201892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 993.Zheng J, et al. Increased PD-1(+)Foxp3(+) gammadelta T cells associate with poor overall survival for patients with acute myeloid leukemia. Front. Oncol. 2022;12:1007565. doi: 10.3389/fonc.2022.1007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 994.Mikulak, J. et al. NKp46-expressing human gut-resident intraepithelial Vdelta1 T cell subpopulation exhibits high antitumor activity against colorectal cancer. JCI Insight4, e125884 (2019). [DOI] [PMC free article] [PubMed]
- 995.Silva-Santos B, Mensurado S, Coffelt SB. gammadelta T cells: pleiotropic immune effectors with therapeutic potential in cancer. Nat. Rev. Cancer. 2019;19:392–404. doi: 10.1038/s41568-019-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 996.Zhao Y, Niu C, Cui J. Gamma-delta (gammadelta) T cells: friend or foe in cancer development? J. Transl. Med. 2018;16:3. doi: 10.1186/s12967-017-1378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 997.Couzi L, et al. Antibody-dependent anti-cytomegalovirus activity of human gammadelta T cells expressing CD16 (FcgammaRIIIa) Blood. 2012;119:1418–1427. doi: 10.1182/blood-2011-06-363655. [DOI] [PubMed] [Google Scholar]
- 998.Tokuyama H, et al. V gamma 9 V delta 2 T cell cytotoxicity against tumor cells is enhanced by monoclonal antibody drugs-rituximab and trastuzumab. Int. J. Cancer. 2008;122:2526–2534. doi: 10.1002/ijc.23365. [DOI] [PubMed] [Google Scholar]
- 999.Brandes M, Willimann K, Moser B. Professional antigen-presentation function by human gammadelta T Cells. Science. 2005;309:264–268. doi: 10.1126/science.1110267. [DOI] [PubMed] [Google Scholar]
- 1000.Brandes M, et al. Cross-presenting human gammadelta T cells induce robust CD8+ alphabeta T cell responses. Proc. Natl Acad. Sci. USA. 2009;106:2307–2312. doi: 10.1073/pnas.0810059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001.Wang, S. et al. Human gammadelta T cells induce CD8(+) T cell antitumor responses via antigen-presenting effect through HSP90-MyD88-mediated activation of JNK. Cancer Immunol. Immunother. (2023). [DOI] [PMC free article] [PubMed]
- 1002.Chan KF, Duarte JDG, Ostrouska S, Behren A. gammadelta T cells in the tumor microenvironment-interactions with other immune cells. Front. Immunol. 2022;13:894315. doi: 10.3389/fimmu.2022.894315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1003.Riond J, et al. In vivo major histocompatibility complex class I (MHCI) expression on MHCIlow tumor cells is regulated by gammadelta T and NK cells during the early steps of tumor growth. Cancer Immun. 2009;9:10. [PMC free article] [PubMed] [Google Scholar]
- 1004.van Beek JJ, et al. Dendritic cell cross talk with innate and innate-like effector cells in antitumor immunity: implications for DC vaccination. Crit. Rev. Immunol. 2014;34:517–536. doi: 10.1615/CritRevImmunol.2014012204. [DOI] [PubMed] [Google Scholar]
- 1005.Girard P, et al. Potent bidirectional cross-talk between plasmacytoid dendritic cells and gammadeltaT cells through BTN3A, type I/II IFNs and immune checkpoints. Front. Immunol. 2020;11:861. doi: 10.3389/fimmu.2020.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1006.Cairo C, et al. Vgamma2Vdelta2 T cell costimulation increases NK cell killing of monocyte-derived dendritic cells. Immunology. 2014;144:422–430. doi: 10.1111/imm.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1007.Maniar A, et al. Human gammadelta T lymphocytes induce robust NK cell-mediated antitumor cytotoxicity through CD137 engagement. Blood. 2010;116:1726–1733. doi: 10.1182/blood-2009-07-234211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1008.Qiu, L., Zhang, Y. & Zeng, X. The function of gammadelta T cells in humoral immune responses. Inflamm. Res.72, 747–755 (2023). [DOI] [PubMed]
- 1009.de Vries NL, et al. gammadelta T cells are effectors of immunotherapy in cancers with HLA class I defects. Nature. 2023;613:743–750. doi: 10.1038/s41586-022-05593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1010.Hu Z, et al. IL-17 activates the IL-6/STAT3 signal pathway in the proliferation of hepatitis B virus-related hepatocellular carcinoma. Cell Physiol. Biochem. 2017;43:2379–2390. doi: 10.1159/000484390. [DOI] [PubMed] [Google Scholar]
- 1011.Jin C, et al. Commensal microbiota promote lung cancer development via gammadelta T cells. Cell. 2019;176:998–1013 e1016. doi: 10.1016/j.cell.2018.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1012.Daley D, et al. gammadelta T cells support pancreatic oncogenesis by restraining alphabeta T cell activation. Cell. 2020;183:1134–1136. doi: 10.1016/j.cell.2020.10.041. [DOI] [PubMed] [Google Scholar]
- 1013.Chabab G, et al. Identification of a regulatory Vdelta1 gamma delta T cell subpopulation expressing CD73 in human breast cancer. J. Leukoc. Biol. 2020;107:1057–1067. doi: 10.1002/JLB.3MA0420-278RR. [DOI] [PubMed] [Google Scholar]
- 1014.Peng G, et al. Tumor-infiltrating gammadelta T cells suppress T and dendritic cell function via mechanisms controlled by a unique toll-like receptor signaling pathway. Immunity. 2007;27:334–348. doi: 10.1016/j.immuni.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 1015.Mao Y, et al. A new effect of IL-4 on human gammadelta T cells: promoting regulatory Vdelta1 T cells via IL-10 production and inhibiting function of Vdelta2 T cells. Cell Mol. Immunol. 2016;13:217–228. doi: 10.1038/cmi.2015.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1016.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory pathways in immunotherapy for cancer. Annu. Rev. Immunol. 2016;34:539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 1017.Upadhaya S, Neftelinov ST, Hodge J, Campbell J. Challenges and opportunities in the PD1/PDL1 inhibitor clinical trial landscape. Nat. Rev. Drug Disco. 2022;21:482–483. doi: 10.1038/d41573-022-00030-4. [DOI] [PubMed] [Google Scholar]
- 1018.Wykes MN, Lewin SR. Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol. 2018;18:91–104. doi: 10.1038/nri.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1019.He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res. 2020;30:660–669. doi: 10.1038/s41422-020-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1020.Albrecht, L. J., Livingstone, E., Zimmer, L. & Schadendorf, D. The latest option: nivolumab and relatlimab in advanced melanoma. Curr. Oncol. Rep. 25, 647–657 (2023). [DOI] [PMC free article] [PubMed]
- 1021.Tawbi HA, et al. Relatlimab and nivolumab versus nivolumab in untreated advanced melanoma. N. Engl. J. Med. 2022;386:24–34. doi: 10.1056/NEJMoa2109970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1022.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat. Rev. Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1023.Mayes PA, Hance KW, Hoos A. The promise and challenges of immune agonist antibody development in cancer. Nat. Rev. Drug Disco. 2018;17:509–527. doi: 10.1038/nrd.2018.75. [DOI] [PubMed] [Google Scholar]
- 1024.Garber K. Immune agonist antibodies face critical test. Nat. Rev. Drug Disco. 2020;19:3–5. doi: 10.1038/d41573-019-00214-5. [DOI] [PubMed] [Google Scholar]
- 1025.Yao, Y., Hu, Y. & Wang, F. Trispecific antibodies for cancer immunotherapy. Immunology (2023). [DOI] [PubMed]
- 1026.Labrijn AF, Janmaat ML, Reichert JM, Parren P. Bispecific antibodies: a mechanistic review of the pipeline. Nat. Rev. Drug Disco. 2019;18:585–608. doi: 10.1038/s41573-019-0028-1. [DOI] [PubMed] [Google Scholar]
- 1027.Zhang T, Lin Y, Gao Q. Bispecific antibodies targeting immunomodulatory checkpoints for cancer therapy. Cancer Biol. Med. 2023;20:181–195. doi: 10.20892/j.issn.2095-3941.2023.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1028.Keam SJ. Cadonilimab: first approval. Drugs. 2022;82:1333–1339. doi: 10.1007/s40265-022-01761-9. [DOI] [PubMed] [Google Scholar]
- 1029.Muik A, et al. Preclinical characterization and phase I trial results of a bispecific antibody targeting PD-L1 and 4-1BB (GEN1046) in patients with advanced refractory solid tumors. Cancer Disco. 2022;12:1248–1265. doi: 10.1158/2159-8290.CD-21-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1030.Kuang Z, et al. A novel bispecific antibody with PD-L1-assisted OX40 activation for cancer tTreatment. Mol. Cancer Ther. 2020;19:2564–2574. doi: 10.1158/1535-7163.MCT-20-0226. [DOI] [PubMed] [Google Scholar]
- 1031.Kvarnhammar AM, et al. The CTLA-4 x OX40 bispecific antibody ATOR-1015 induces anti-tumor effects through tumor-directed immune activation. J. Immunother. Cancer. 2019;7:103. doi: 10.1186/s40425-019-0570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1032.Vitale LA, et al. Development of CDX-527: a bispecific antibody combining PD-1 blockade and CD27 costimulation for cancer immunotherapy. Cancer Immunol. Immunother. 2020;69:2125–2137. doi: 10.1007/s00262-020-02610-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1033.Li L, et al. Tumor-targeting anti-EGFR x anti-PD1 bispecific antibody inhibits EGFR-overexpressing tumor growth by combining EGFR blockade and immune activation with direct tumor cell killing. Transl. Oncol. 2021;14:100916. doi: 10.1016/j.tranon.2020.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1034.Gu CL, et al. Bispecific antibody simultaneously targeting PD1 and HER2 inhibits tumor growth via direct tumor cell killing in combination with PD1/PDL1 blockade and HER2 inhibition. Acta Pharm. Sin. 2022;43:672–680. doi: 10.1038/s41401-021-00683-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1035.Nixon, B. G., Gao, S., Wang, X. & Li, M. O. TGFbeta control of immune responses in cancer: a holistic immuno-oncology perspective. Nat. Rev. Immunol. (2022). [DOI] [PMC free article] [PubMed]
- 1036.Tschernia NP, Gulley JL. Tumor in the crossfire: inhibiting TGF-beta to enhance cancer immunotherapy. BioDrugs. 2022;36:153–180. doi: 10.1007/s40259-022-00521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1037.Baeuerle PA, Wesche H. T-cell-engaging antibodies for the treatment of solid tumors: challenges and opportunities. Curr. Opin. Oncol. 2022;34:552–558. doi: 10.1097/CCO.0000000000000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1038.Chen TT. Conditionally active T cell engagers for the treatment of solid tumors: rationale and clinical development. Expert Opin. Biol. Ther. 2022;22:955–963. doi: 10.1080/14712598.2022.2098674. [DOI] [PubMed] [Google Scholar]
- 1039.Esfandiari A, Cassidy S, Webster RM. Bispecific antibodies in oncology. Nat. Rev. Drug Disco. 2022;21:411–412. doi: 10.1038/d41573-022-00040-2. [DOI] [PubMed] [Google Scholar]
- 1040.Friedrich MJ, et al. The pre-existing T cell landscape determines the response to bispecific T cell engagers in multiple myeloma patients. Cancer Cell. 2023;41:711–725 e716. doi: 10.1016/j.ccell.2023.02.008. [DOI] [PubMed] [Google Scholar]
- 1041.Middelburg, J. et al. Overcoming challenges for CD3-bispecific antibody therapy in solid tumors. Cancers13, 287 (2021). [DOI] [PMC free article] [PubMed]
- 1042.Fajgenbaum DC, June CH. Cytokine storm. N. Engl. J. Med. 2020;383:2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1043.Dhillon S. Tebentafusp: first approval. Drugs. 2022;82:703–710. doi: 10.1007/s40265-022-01704-4. [DOI] [PubMed] [Google Scholar]
- 1044.Claus C, Ferrara-Koller C, Klein C. The emerging landscape of novel 4-1BB (CD137) agonistic drugs for cancer immunotherapy. MAbs. 2023;15:2167189. doi: 10.1080/19420862.2023.2167189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1045.Skokos, D. et al. A class of costimulatory CD28-bispecific antibodies that enhance the antitumor activity of CD3-bispecific antibodies. Sci. Transl. Med. 12, eaaw7888 (2020). [DOI] [PubMed]
- 1046.Cappell KM, Kochenderfer JN. A comparison of chimeric antigen receptors containing CD28 versus 4-1BB costimulatory domains. Nat. Rev. Clin. Oncol. 2021;18:715–727. doi: 10.1038/s41571-021-00530-z. [DOI] [PubMed] [Google Scholar]
- 1047.Jayaraman J, et al. CAR-T design: elements and their synergistic function. EBioMedicine. 2020;58:102931. doi: 10.1016/j.ebiom.2020.102931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1048.Mehrabadi AZ, et al. Therapeutic potential of CAR T cell in malignancies: a scoping review. Biomed. Pharmacother. 2022;146:112512. doi: 10.1016/j.biopha.2021.112512. [DOI] [PubMed] [Google Scholar]
- 1049.Yan T, Zhu L, Chen J. Current advances and challenges in CAR T-Cell therapy for solid tumors: tumor-associated antigens and the tumor microenvironment. Exp. Hematol. Oncol. 2023;12:14. doi: 10.1186/s40164-023-00373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1050.Young RM, et al. Next-generation CAR T-cell therapies. Cancer Disco. 2022;12:1625–1633. doi: 10.1158/2159-8290.CD-21-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1051.Cho JH, et al. Engineering advanced logic and distributed computing in human CAR immune cells. Nat. Commun. 2021;12:792. doi: 10.1038/s41467-021-21078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1052.Tousley AM, et al. Co-opting signalling molecules enables logic-gated control of CAR T cells. Nature. 2023;615:507–516. doi: 10.1038/s41586-023-05778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1053.Simon S, Bugos G, Salter AI, Riddell SR. Synthetic receptors for logic gated T cell recognition and function. Curr. Opin. Immunol. 2022;74:9–17. doi: 10.1016/j.coi.2021.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1054.Hamieh M, Mansilla-Soto J, Riviere I, Sadelain M. Programming CAR T cell tumor recognition: tuned antigen sensing and logic gating. Cancer Disco. 2023;13:829–843. doi: 10.1158/2159-8290.CD-23-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1055.Williams JZ, et al. Precise T cell recognition programs designed by transcriptionally linking multiple receptors. Science. 2020;370:1099–1104. doi: 10.1126/science.abc6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1056.Salter, A. I. et al. Comparative analysis of TCR and CAR signaling informs CAR designs with superior antigen sensitivity and in vivo function. Sci. Signal. 14, eabe2606 (2021). [DOI] [PMC free article] [PubMed]
- 1057.Chandran SS, Klebanoff CA. T cell receptor-based cancer immunotherapy: emerging efficacy and pathways of resistance. Immunol. Rev. 2019;290:127–147. doi: 10.1111/imr.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1058.Mao W. Overcoming current challenges to T-cell receptor therapy via metabolic targeting to increase antitumor efficacy, durability, and tolerability. Front. Immunol. 2022;13:1056622. doi: 10.3389/fimmu.2022.1056622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1059.Zhang, S. Q. et al. High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat. Biotechnol.36, 1156–1159 (2018). [DOI] [PMC free article] [PubMed]
- 1060.Lu YC, et al. An efficient single-cell RNA-seq approach to identify neoantigen-specific T cell receptors. Mol. Ther. 2018;26:379–389. doi: 10.1016/j.ymthe.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1061.Lu, Y. C. et al. Direct identification of neoantigen-specific TCRs from tumor specimens by high-throughput single-cell sequencing. J. Immunother. Cancer. 9, e002595 (2021). [DOI] [PMC free article] [PubMed]
- 1062.Lin B, et al. Tumor-infiltrating lymphocytes: warriors fight against tumors powerfully. Biomed. Pharmacother. 2020;132:110873. doi: 10.1016/j.biopha.2020.110873. [DOI] [PubMed] [Google Scholar]
- 1063.Attig S, et al. Simultaneous infiltration of polyfunctional effector and suppressor T cells into renal cell carcinomas. Cancer Res. 2009;69:8412–8419. doi: 10.1158/0008-5472.CAN-09-0852. [DOI] [PubMed] [Google Scholar]
- 1064.Dafni U, et al. Efficacy of adoptive therapy with tumor-infiltrating lymphocytes and recombinant interleukin-2 in advanced cutaneous melanoma: a systematic review and meta-analysis. Ann. Oncol. 2019;30:1902–1913. doi: 10.1093/annonc/mdz398. [DOI] [PubMed] [Google Scholar]
- 1065.Muranski P, et al. Increased intensity lymphodepletion and adoptive immunotherapy-how far can we go? Nat. Clin. Pr. Oncol. 2006;3:668–681. doi: 10.1038/ncponc0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1066.Chesney, J. et al. Efficacy and safety of lifileucel, a one-time autologous tumor-infiltrating lymphocyte (TIL) cell therapy, in patients with advanced melanoma after progression on immune checkpoint inhibitors and targeted therapies: pooled analysis of consecutive cohorts of the C-144-01 study. J. Immunother. Cancer. 10, e005755 (2022). [DOI] [PMC free article] [PubMed]
- 1067.Kumar A, Watkins R, Vilgelm AE. Cell therapy with TILs: training and taming T cells to fight cancer. Front. Immunol. 2021;12:690499. doi: 10.3389/fimmu.2021.690499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1068.Dudley ME, et al. CD8+ enriched “young” tumor infiltrating lymphocytes can mediate regression of metastatic melanoma. Clin. Cancer Res. 2010;16:6122–6131. doi: 10.1158/1078-0432.CCR-10-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1069.Sim GC, et al. Tumor-infiltrating lymphocyte therapy for melanoma: rationale and issues for further clinical development. BioDrugs. 2014;28:421–437. doi: 10.1007/s40259-014-0097-y. [DOI] [PubMed] [Google Scholar]
- 1070.Ye Q, et al. Engineered artificial antigen presenting cells facilitate direct and efficient expansion of tumor infiltrating lymphocytes. J. Transl. Med. 2011;9:131. doi: 10.1186/1479-5876-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1071.Krummel MF, Heath WR, Allison J. Differential coupling of second signals for cytotoxicity and proliferation in CD8+ T cell effectors: amplification of the lytic potential by B7. J. Immunol. 1999;163:2999–3006. doi: 10.4049/jimmunol.163.6.2999. [DOI] [PubMed] [Google Scholar]
- 1072.Kazemi MH, et al. Tumor-infiltrating lymphocytes for treatment of solid tumors: it takes two to tango? Front. Immunol. 2022;13:1018962. doi: 10.3389/fimmu.2022.1018962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1073.Li P, Zheng Y, Chen X. Drugs for autoimmune inflammatory diseases: from small molecule compounds to anti-TNF biologics. Front. Pharm. 2017;8:460. doi: 10.3389/fphar.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1074.Jung SM, Kim WU. Targeted immunotherapy for autoimmune disease. Immune Netw. 2022;22:e9. doi: 10.4110/in.2022.22.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1075.Mullard A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Disco. 2021;20:491–495. doi: 10.1038/d41573-021-00079-7. [DOI] [PubMed] [Google Scholar]
- 1076.Lai Y, Dong C. Therapeutic antibodies that target inflammatory cytokines in autoimmune diseases. Int. Immunol. 2016;28:181–188. doi: 10.1093/intimm/dxv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1077.McLornan DP, Pope JE, Gotlib J, Harrison CN. Current and future status of JAK inhibitors. Lancet. 2021;398:803–816. doi: 10.1016/S0140-6736(21)00438-4. [DOI] [PubMed] [Google Scholar]
- 1078.Hu X, et al. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct. Target Ther. 2021;6:402. doi: 10.1038/s41392-021-00791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1079.Hofmann K, Clauder AK, Manz RA. Targeting B cells and plasma cells in autoimmune diseases. Front. Immunol. 2018;9:835. doi: 10.3389/fimmu.2018.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1080.Baker DJ, June CH. CAR T therapy extends its reach to autoimmune diseases. Cell. 2022;185:4471–4473. doi: 10.1016/j.cell.2022.10.026. [DOI] [PubMed] [Google Scholar]
- 1081.Mackensen A, et al. Anti-CD19 CAR T cell therapy for refractory systemic lupus erythematosus. Nat. Med. 2022;28:2124–2132. doi: 10.1038/s41591-022-02017-5. [DOI] [PubMed] [Google Scholar]
- 1082.Su M, Zhao C, Luo S. Therapeutic potential of chimeric antigen receptor based therapies in autoimmune diseases. Autoimmun. Rev. 2022;21:102931. doi: 10.1016/j.autrev.2021.102931. [DOI] [PubMed] [Google Scholar]
- 1083.Santamaria-Alza Y, Vasquez G. Are chimeric antigen receptor T cells (CAR-T cells) the future in immunotherapy for autoimmune diseases? Inflamm. Res. 2021;70:651–663. doi: 10.1007/s00011-021-01470-1. [DOI] [PubMed] [Google Scholar]
- 1084.Lee DSW, Rojas OL, Gommerman JL. B cell depletion therapies in autoimmune disease: advances and mechanistic insights. Nat. Rev. Drug Disco. 2021;20:179–199. doi: 10.1038/s41573-020-00092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1085.Lin L, et al. Preclinical evaluation of CD8+ anti-BCMA mRNA CAR T cells for treatment of multiple myeloma. Leukemia. 2021;35:752–763. doi: 10.1038/s41375-020-0951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1086.Jin X, et al. Therapeutic efficacy of anti-CD19 CAR-T cells in a mouse model of systemic lupus erythematosus. Cell Mol. Immunol. 2021;18:1896–1903. doi: 10.1038/s41423-020-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1087.Kansal, R. et al. Sustained B cell depletion by CD19-targeted CAR T cells is a highly effective treatment for murine lupus. Sci. Transl. Med. 11, eaav1648 (2019). [DOI] [PMC free article] [PubMed]
- 1088.Muller F, et al. CD19-targeted CAR T cells in refractory antisynthetase syndrome. Lancet. 2023;401:815–818. doi: 10.1016/S0140-6736(23)00023-5. [DOI] [PubMed] [Google Scholar]
- 1089.Qin C, et al. Anti-BCMA CAR T-cell therapy CT103A in relapsed or refractory AQP4-IgG seropositive neuromyelitis optica spectrum disorders: phase 1 trial interim results. Signal Transduct. Target Ther. 2023;8:5. doi: 10.1038/s41392-022-01278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1090.Mougiakakos D, et al. CD19-targeted CAR T cells in refractory systemic lupus erythematosus. N. Engl. J. Med. 2021;385:567–569. doi: 10.1056/NEJMc2107725. [DOI] [PubMed] [Google Scholar]
- 1091.Ellebrecht CT, et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science. 2016;353:179–184. doi: 10.1126/science.aaf6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1092.Lee J, et al. Antigen-specific B cell depletion for precision therapy of mucosal pemphigus vulgaris. J. Clin. Invest. 2020;130:6317–6324. doi: 10.1172/JCI138416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1093.Huijbers MG, et al. Longitudinal epitope mapping in MuSK myasthenia gravis: implications for disease severity. J. Neuroimmunol. 2016;291:82–88. doi: 10.1016/j.jneuroim.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 1094.Kobayashi S, et al. A biomimetic five-module chimeric antigen receptor ((5 M)CAR) designed to target and eliminate antigen-specific T cells. Proc. Natl Acad. Sci. USA. 2020;117:28950–28959. doi: 10.1073/pnas.2012495117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1095.Zhang L, et al. Chimeric antigen receptor (CAR) T cells targeting a pathogenic MHC class II:peptide complex modulate the progression of autoimmune diabetes. J. Autoimmun. 2019;96:50–58. doi: 10.1016/j.jaut.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1096.Fishman S, et al. Adoptive transfer of mRNA-transfected T cells redirected against diabetogenic CD8 T cells can prevent diabetes. Mol. Ther. 2017;25:456–464. doi: 10.1016/j.ymthe.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1097.Yuan Y, et al. Therapeutic potential of interleukin-2 in autoimmune diseases. Trends Mol. Med. 2022;28:596–612. doi: 10.1016/j.molmed.2022.04.010. [DOI] [PubMed] [Google Scholar]
- 1098.Kolios AGA, Tsokos GC, Klatzmann D. Interleukin-2 and regulatory T cells in rheumatic diseases. Nat. Rev. Rheumatol. 2021;17:749–766. doi: 10.1038/s41584-021-00707-x. [DOI] [PubMed] [Google Scholar]
- 1099.Rana J, Biswas M. Regulatory T cell therapy: current and future design perspectives. Cell Immunol. 2020;356:104193. doi: 10.1016/j.cellimm.2020.104193. [DOI] [PubMed] [Google Scholar]
- 1100.Fransson M, et al. CAR/FoxP3-engineered T regulatory cells target the CNS and suppress EAE upon intranasal delivery. J. Neuroinflammation. 2012;9:112. doi: 10.1186/1742-2094-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1101.Elinav E, Waks T, Eshhar Z. Redirection of regulatory T cells with predetermined specificity for the treatment of experimental colitis in mice. Gastroenterology. 2008;134:2014–2024. doi: 10.1053/j.gastro.2008.02.060. [DOI] [PubMed] [Google Scholar]
- 1102.Blat D, et al. Suppression of murine colitis and its associated cancer by carcinoembryonic antigen-specific regulatory T cells. Mol. Ther. 2014;22:1018–1028. doi: 10.1038/mt.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1103.Raffin, C. et al. Development of citrullinated-vimentin-specific CAR for targeting Tregs to treat autoimmune rheumatoid arthritis. J. Immunol. 200, 176.117 (2018).
- 1104.Tenspolde M, et al. Regulatory T cells engineered with a novel insulin-specific chimeric antigen receptor as a candidate immunotherapy for type 1 diabetes. J. Autoimmun. 2019;103:102289. doi: 10.1016/j.jaut.2019.05.017. [DOI] [PubMed] [Google Scholar]
- 1105.Proics, E. et al. Preclinical assessment of antigen-specific chimeric antigen receptor regulatory T cells for use in solid organ transplantation. Gene Ther. 30, 309–322 (2022). [DOI] [PMC free article] [PubMed]
- 1106.Noyan F, et al. Prevention of allograft rejection by use of regulatory T cells with an MHC-specific chimeric antigen receptor. Am. J. Transpl. 2017;17:917–930. doi: 10.1111/ajt.14175. [DOI] [PubMed] [Google Scholar]
- 1107.Wei J, et al. The model of cytokine release syndrome in CAR T-cell treatment for B-cell non-Hodgkin lymphoma. Signal Transduct. Target Ther. 2020;5:134. doi: 10.1038/s41392-020-00256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1108.Shah K, Al-Haidari A, Sun J, Kazi JU. T cell receptor (TCR) signaling in health and disease. Signal Transduct. Target Ther. 2021;6:412. doi: 10.1038/s41392-021-00823-w. [DOI] [PMC free article] [PubMed] [Google Scholar]