Abstract
Circular RNAs (circRNAs) are a group of non-coding RNAs with a unique circular structure generated by back-splicing. It is acknowledged that circRNAs play critical roles in cardiovascular diseases. However, functional studies of circRNAs were impeded due to lack of effective in vivo silencing approaches. Since most circRNAs are produced by protein-coding transcripts, gene editing typically affects the coding activity of the parental genes. In this study, we developed a circular antisense RNA (cA-circSlc8a1) that could silence the highly expressed circRNA circSlc8a1 in the mouse heart but not its parental Slc8a1 linear mRNA. Transgenic cA-circSlc8a1 mice developed congestive heart failure resulting in a significant increase in the body weight secondary to peripheral edema and congestive hepatopathy. To further test the role of circSlc8a1, we generated transgenic mice overexpressing circSlc8a1 and observed a protective effect of circSlc8a1 in a pressure overload model. Mechanistically, we found that circSlc8a1 translocated into mitochondria to drive ATP synthesis. While establishing a transgenic murine model for antisense-mediated circRNA silencing without interfering with the parental linear RNA, our finding revealed the essential role of circSlc8a1 in maintaining heart function and may lay the groundwork of using the circular antisense RNA as a potential gene therapy approach for cardiovascular diseases.
Keywords: circular RNA, circRNA, circSlc8a1, hypertrophy, cardiac fibrosis, congenital heart disease, cardiogenic hepatopathy
Graphical abstract
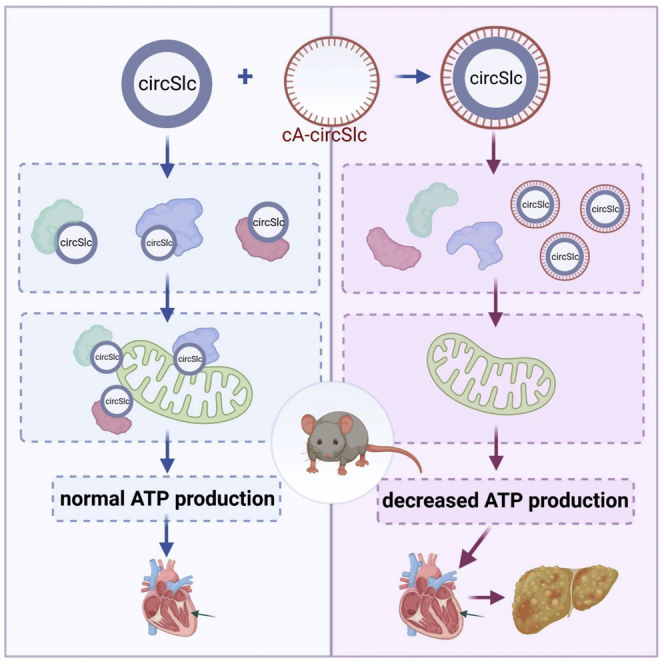
We developed a circular antisense RNA (cA-circSlc8a1) to silence the highly expressed circSlc8a1 in mouse heart. Transgenic cA-circSlc8a1 mice developed congestive heart failure phenotype. We then generated transgenic mice overexpressing circSlc8a1 and found a protective effect of circSlc8a1 on heart functions due to its mitochondrial translocation to drive ATP synthesis.
Introduction
Circular RNAs (circRNAs) are a large class of covalently linked single-stranded RNAs. Abnormal elevation of circRNA levels is associated with the onset or progression of cardiovascular diseases.1,2,3 A variety of circRNAs have been demonstrated to play critical roles in cardiovascular diseases, including cardiomyopathy, myocardial infarction, and artery diseases.4,5,6 The circRNAs have been recognized as potential therapeutic targets and biomarkers in cardiovascular diseases.7,8,9 Therefore, it is of great importance to investigate the roles of circRNAs in the development of cardiovascular diseases. Recently, some other non-coding RNAs, such as miRNAs and lncRNAs, have been successfully applied in RNA therapy in cardiovascular diseases.10,11 However, RNA therapy targeting circRNAs in cardiovascular diseases is still in the early stages of investigation.12
The circRNAs are produced by linking a 5′ splice donor with an upstream 3′ splice acceptor by back-splicing of the linear transcripts. The circRNAs are thus resistant to exonuclease treatment and more stable than the linear RNAs. The circRNAs have been identified across the genome and most of them are derived from protein-coding genes.13 Since circRNAs share a part of the exon sequences with the parental coding genes, the traditional methods for generating gene knockout mice are hardly applied to establish circRNA knockout models. The widespread use of genetic methods becomes limited in circRNA functional studies due to lack of effective tools for knocking out/down endogenous circRNAs in vivo without affecting the parental mRNAs. In the past several years, some important breakthrough progresses have been made on in vivo model studies in certain exceptional scenarios. A circRNA, Cdr1as, without detectable linear cognate mRNA was identified and the Cdr1as locus was successfully knocked out by using CRISPR-Cas9.14 A circRNA, circNfix, whose expression depends on a super-enhancer, was knocked down by mutation of its transcription factor, Meis1.5 Another study reported the knockdown of circRNAs using CRISPR-Cas13 by directly targeting the back-splicing junction (BSJ) region of the circRNA during mouse embryo preimplantation development.15 Recently, removal of intronic cassettes was also reported to knockout the expression of circZfp292 and consequently altered the endothelial flow response.16 The more popular methods used to silence a circRNA are the short hairpin RNAs and small interfering RNAs that are complementary to the BSJ of the circRNA.17,18
Here, we developed a strategy to generate a circularized antisense RNA that is complementary to the complete sequence of a circRNA. We expected that this antisense RNA was able to form a stable double-stranded RNA circle with its sense counterpart, resulting in the loss of the normal structure and functions of the sense circRNA. We first screened the circRNAs expressed in the mouse heart by RNA sequencing and selected the highest expressed circRNA, circSlc8a1, as our target. We then established a transgenic mouse model, in which the expression of circSlc8a1, but not the full-length Slc8a1 mRNA and protein, was successfully sequestered by the circular antisense against circSlc8a1 (cA-circSlc8a1) driven by a heart-specific promoter. We found that cardiac function in cA-circSlc8a1-transgenic mice was compromised, resulting in a significant increase in the body weight secondary to peripheral edema and the histological changes in the liver, including hepatic steatosis and cirrhosis. Importantly, our study demonstrates specific silencing of circRNA function. This proof-of-concept study may lay the foundation for potential clinical applications in circRNA therapy.
Results
Blocking circSlc8a1 expression by circular antisense RNA induces phenotypic changes
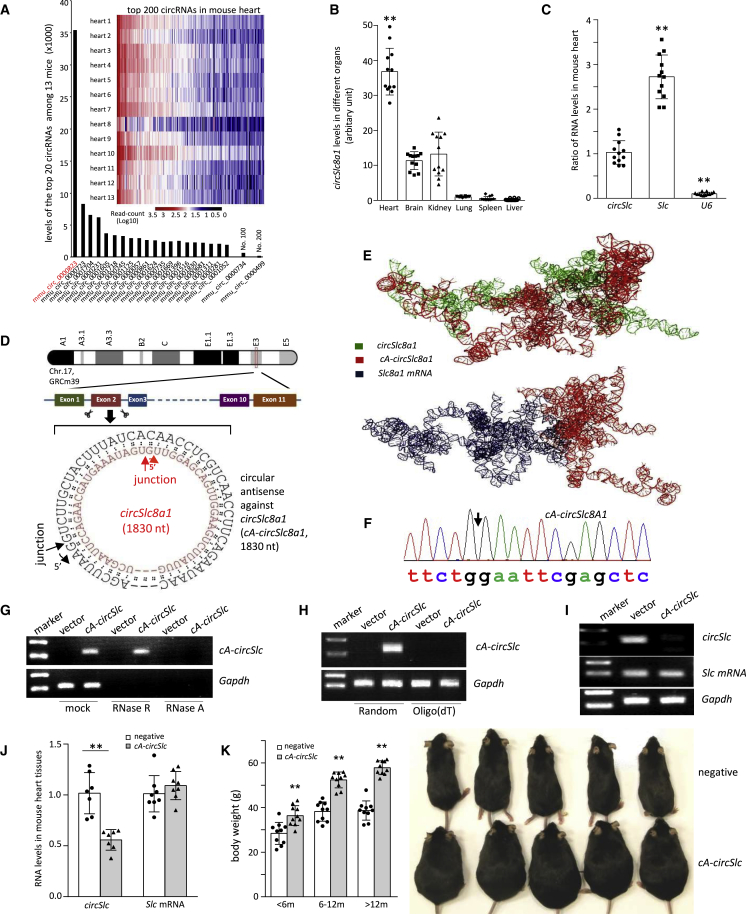
In this study, we perform high-throughput circRNA deep sequencing in myocardial tissue samples of 13 mice to search for the highest expressed circRNA in the hearts that may possess essential physiological functions. This allows us to develop an approach to silence the circRNA leading to a potent phenotype. We found that circSlc8a1 (mmu_circ_0000823) was the topmost circRNA among 27,960 circRNAs detected in the hearts. Its expression levels (sequencing readcounts) were 4.2-fold of the second highest circRNA and even more than the sum by adding the readcounts of the top 2–8 circRNAs (Figure 1A). We examined the levels of circSlc8a1 and linear Slc8a1 mRNA in six main organs and found that both circSlc8a1 and Slc8a1 were expressed the highest in the heart compared with the other organs (Figures 1B and S1A). Furthermore, we compared the levels of circSlc8a1 with Slc8a1 mRNA and the housekeeping gene U6 in the mouse heart tissues and found that the expression levels of circSlc8a1 in the heart were slightly lower than Slc8a1 mRNA but higher than the housekeeping gene U6 (Figure 1C). In addition, 2,174 ± 313.82 copies of circSlc8a1 per ng total RNA were determined in the mouse heart tissues by using droplet digital PCR. The high abundance of circSlc8a1 implied its essential physiological role in the heart. Therefore, we designed a construct to express a circular antisense RNA (cA-RNA) perfectly complementary with circSlc8a1, named cA-circSlc8a1, which had the potential to sequester circSlc8a1 by forming a sense-antisense double-stranded RNA circle (Figure 1D). The circular antisense was obtained by inserting the donor and acceptor sequences of the introns, respectively, into the antisense sequence of circSlc8a1, allowing back-splicing to form a circle. To validate whether cA-circSlc8a1 has higher affinity to the circSlc8a1 than to the linear Slc8a1 mRNA, we used the computational docking methods to predict the potential blocking ability (Figure 1E). The docking score of cA-circSlc8a1 complexed with circ-Slc8a1 was found to be −7,621.32 kCal/mole, whereas the score was −1,699.97 kCal/mole for docking cA-circSlc8a1 with the linear Slc8a1 mRNA. Our results speculated that cA-circSlc8a1 has much higher binding affinity with circSlc8a1 compared with the linear Slc8a1 mRNA.
Figure 1.
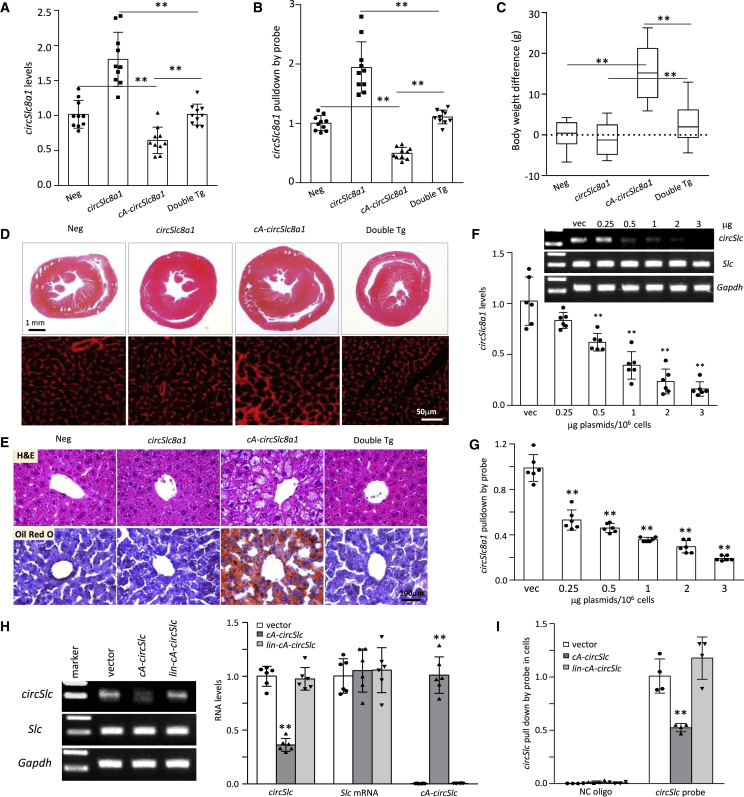
Generation of transgenic mice expressing circular antisense against circSlc8a1 (cA-circSlc8a1) knocked down cardiac circSlc8a1 and developed phenotypic changes
(A) RNA sequencing heatmap of differentially expressed circular RNA profiles in the heart of 13 mice. The highest readcount was circSlc8a1 (mmu_circ_0000823), which was more than the sum obtained by adding the readcounts of the top 2–8 circRNAs. (B) Compared with the other organs analyzed, the heart expressed the highest level of circSlc8a1. n = 12; ∗∗p < 0.01 versus other organs. (C) The ratio of circSlc8a1 (circSlc) in the mouse heart compared with Slc8a1 (Slc) mRNA and ubiquitously expressed U6. n = 12; ∗∗p < 0.01 versus circSlc. (D) A diagram showing the sequence of the circular antisense against circSlc8a1 (cA-circSlc8a1) that was perfectly complementary to circSlc8a1. (E) Graphical representation of the three-dimensional structure of cA-circSlc8a1 and circSlc8a1, or cA-circSlc8a1 and Slc8a1 mRNA docked model visualized using PyMOL. (F) The back-splice junction of the ectopic mouse cA-circSlc8a1 was validated by Sanger sequencing. (G) Representative images of cA-circSlc8a1 expression with RNase R and RNase A treatment. A cA-circSlc8a1 construct targeting mouse circSlc8a1 was transfected into human HEK293T cells. The RNAs from vector or cA-circSlc8a1-transfected cells were treated with RNase R or RNase A followed by reverse transcription. The expression of cA-circSlc8a1 was validated by mouse-specific primers against cA-circSlc8a1. n = 6. (H) Representative images of cA-circSlc8a1 expression by using random or oligo(dT) primers for reverse transcription. A cA-circSlc8a1 construct targeting mouse circSlc8a1 was transfected into human HEK293T cells. The RNAs from vector or cA-circSlc8a1-transfected cells were reverse transcribed using random or oligo(dT) primers. The expression of cA-circSlc8a1 was validated by mouse-specific primers against cA-circSlc8a1. n = 6. (I) Representative images of unbound circSlc8a1 levels after transfection of cA-circSlc8a1 in HL-1 cells. The RNAs from vector or cA-circSlc8a1-transfected cells were reverse transcribed using specific reverse primers for circSlc8a1 followed by qPCR. n = 6. (J) The levels of circSlc8a1 and Slc8a1 mRNA in the heart of cA-circSlc8a1-transgenic mice. n = 8; ∗∗p < 0.01 versus negative. (K) Left, the body weight of cA-circSlc8a1(+) mice (cA-circSlc) was measured at different time points and compared with the age-matched cA-circSlc8a1(−) counterparts (negative) litters. ∗∗p < 0.01, n = 10. Right, a photo of cA-circSlc8a1(+) mice and their age-matched negative counterparts taken at the age of 10 months, showing an increase in the body weight in cA-circSlc8a1(+) mice.
Before further studying the function of cA-circSlc8a1, we first validated the successful circularization of cA-circSlc8a1 plasmids in the cells. We transfected human HEK293 cells with the mouse cA-circSlc8a1 plasmids and measured the expression of cA-circSlc8a1 with mouse-specific divergent primers spanning the back-splice junction. The correct junction sequence that ectopically expressed in HEK293 cells was validated by Sanger sequencing as shown in Figure 1F. By pre-treatment prior to reverse transcription (RT) with RNase R (the enzyme that degrades linear RNAs but not circRNAs) and RNase A (the enzyme that degrades all RNAs), our results verified that cA-circSlc8a1 was resistant to RNase R (Figures 1G and S1B). In addition, due to lack of open ends, the circRNAs cannot be reverse transcribed with oligo(dT) primers but only with random primers. Our PCR results showed that the cA-circSlc8a1 plasmids formed circularized RNA, which could not be reverse transcribed with oligo(dT) primers (Figures 1H and S1C).
To confirm that cA-circSlc8a1 could block circSlc8a1 without affecting Slc8a1 mRNA in vitro before generating the transgenic mice, we transfected the cA-circSlc8a1 plasmids in HL-1 cells, an immortalized mouse cardiomyocytes cell line. RT using random primers for both circSlc8a1 and cA-circSlc8a1 in transfected HL-1 cells may lead to misamplification of cA-circSlc8a1 in the following qPCR analysis. To avoid this, we conducted gene-specific RT by using the reverse primer that was specifically complementary to circSlc8a1, and could not reverse transcribe cA-circSlc8a1. In addition, the reverse primer specific for circSlc8a1 could not bind to those circSlc8a1 that had already formed double-stranded RNA circle with cA-circSlc8a1 for RT. Therefore, only the free unbound circSlc8a1 could be detected by using the circRNA-specific RT with specific reverse primer followed by qPCR. Our results revealed that circSlc8a1 was blocked by cA-circSlc8a1 while the levels of Slc8a1 mRNA were not affected (Figures 1I and S1D). Taken together, these results validated that cA-circSlc8a1 plasmids successfully produced a circularized antisense RNA that blocked circSlc8a1 without affecting Slc8a1 mRNA in the cells.
To further validate the function of cA-circSlc8a1 in vivo, we established a transgenic mouse model expressing cA-circSlc8a1 driven by a heart promoter to specifically silence circSlc8a1 in the mouse heart (Figures S1E and S1F). We confirmed that the amount of free unbound circSlc8a1 was significantly decreased in the heart of cA-circSlc8a1-transgenic mice (Figure 1J). Nevertheless, the Slc8a1 mRNA and SLC8A1 protein expression in the heart of cA-circSlc8a1-transgenic mice were at the similar levels compared with the negative mice, which excluded the possibility that cA-circSlc8a1 affected the parental Slc8a1 expression (Figures 1J and S1G). Notable phenotypic changes were observed in these cA-circSlc8a1-transgenic mice compared with the litter-matched negative counterparts (Figures 1K and S1H). The body weights and the heart/tibia length ratio of cA-circSlc8a1-transgenic mice were significantly higher than those of the negative mice (Figures 1K and S2A).
Development of hepatic steatosis as a consequence of impaired heart function
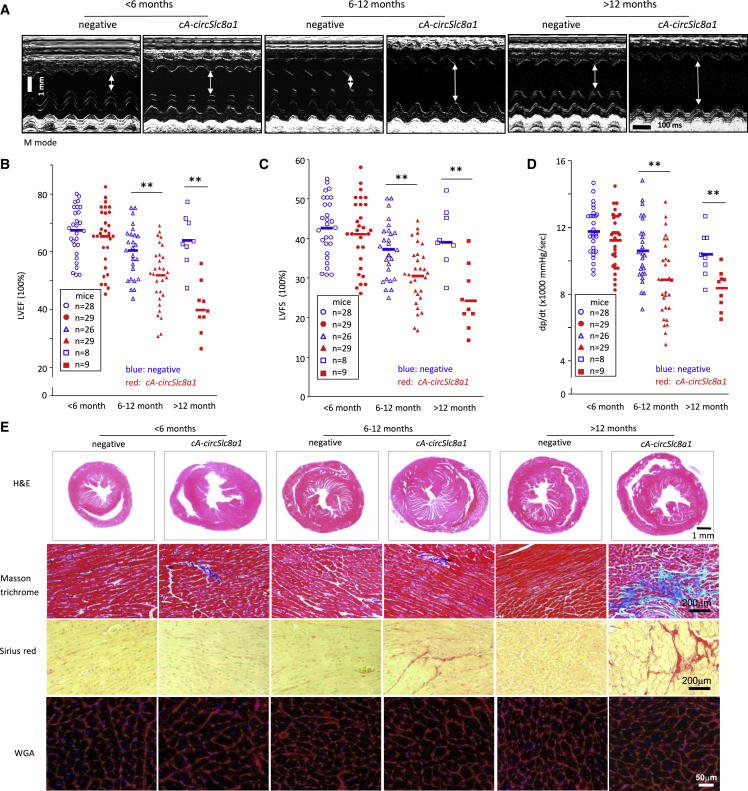
Since the transgenic cA-circSlc8a1 expression was cardiac specific, we examined whether the phenotypic changes were caused by the cA-circSlc8a1 due to depleting endogenous circSlc8a1 function in the heart. To test this, we evaluated the heart function of the cA-circSlc8a1-transgenic mice and their negative counterparts at different ages. Using the Vevo 2000 imaging system, we detected a significant malfunction in the cA-circSlc8a1-transgenic mice. Typical M-mode echocardiograms showed increased left ventricular chamber sizes and decreased ventricular contractile ability in the cA-circSlc8a1-transgenic mice compared with the litter-matched negative mice (Figure 2A). At the age of 6 months and above, cA-circSlc8a1-transgenic mice showed a significant decrease in the left ventricular ejection fraction (LVEF), left ventricular fractional shortening (LVFS), and left ventricular pressure (dp/dt) compared with the litter-matched negative mice (Figures 2B–2D). These transgenic mice showed increase in left ventricular end diastolic diameter (LVEDD) and left ventricular end systolic diameter (LVESD), leading to a decrease in LVEDD-LVESD (Figures S2B–S2D).
Figure 2.
Heart function and cardiac histological changes of cA-circSlc8a1-transgenic mice evaluated by echocardiography and staining
(A) The representative echocardiography of cA-circSlc8a1-transgenic mice at different ages. (B) Echocardiography of cA-circSlc8a1(+) mice showed reduced left ventricular ejection fraction (LVEF) compared with the litter-matched negative mice. ∗∗p < 0.01 versus negative. (C) Echocardiography of cA-circSlc8a1(+) mice showed reduced left ventricular fractional shortening (LVFS) compared with the litter-matched negative mice. ∗∗p < 0.01 versus negative. (D) Echocardiography of cA-circSlc8a1(+) mice showed reduced left ventricular pressure (dp/dt) compared with the litter-matched negative mice. ∗∗p < 0.01 versus negative. (E) Representative photographs of Masson trichrome staining and Sirius red staining showing significant development of cardiac fibrosis in cA-circSlc8a1(+) mice after age 12 months. H&E staining and WGA staining showed development of cardiac hypertrophy in cA-circSlc8a1(+) mice as early as under age 6 months.
To corroborate the effect of cA-circSlc8a1 on decreasing heart function, we further examined the morphological changes of the cardiac tissue in the transgenic mice. Hematoxylin and eosin (H&E) staining of the heart sections showed cardiac hypertrophy in the cA-circSlc8a1-transgenic mice at all time points (Figure 2E), consistent with the impaired heart functions, suggesting remodeling of the cardiac tissues. In support of this, we analyzed cardiac fibrosis by staining the heart sections with Masson’s trichrome and Sirius red to visualize the collagen deposition and detected increased fibrotic staining in the cA-circSlc8a1-transgenic mouse heart (Figures 2E and S2E). Moreover, to examine cardiac hypertrophy, we stained cardiomyocytes with Alexa Fluor 555-conjugated wheat germ agglutinin (WGA) in the cross-section of the cA-circSlc8a1-transgenic and the litter-matched negative mouse hearts and detected enlargement of cardiomyocytes sizes in the cA-circSlc8a1-transgenic mice, reaching significant levels at all time points, far before cardiac fibrosis was observed (Figures 2E and S2F). We also confirmed cardiac hypertrophy in the cA-circSlc8a1-transgenic mice by measuring the expression of ANP and BNP, the markers of myocardial hypertrophy (Figure S2G).
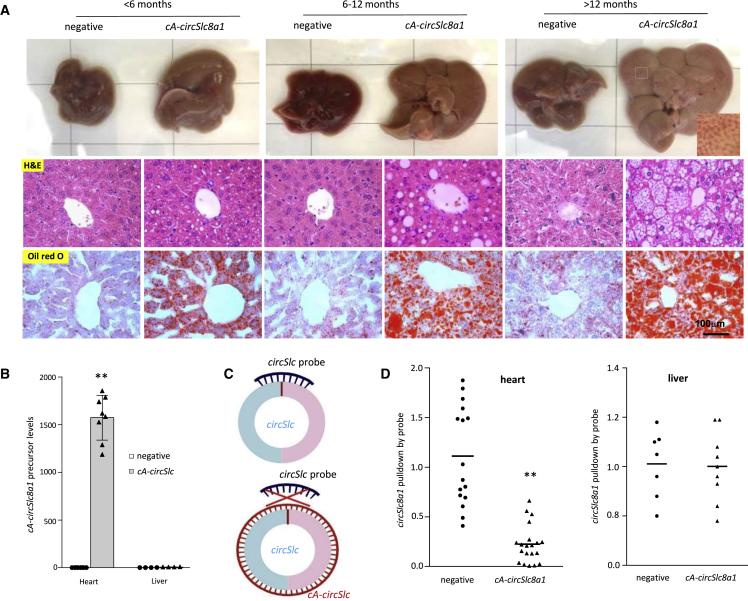
In the cA-circSlc8a1-transgenic mice, we noticed that hepatic steatosis was developed as early as 3 months old. The lipid droplets were extensively accumulated in the liver of cA-circSlc8a1-transgenic mice before the age of 6 months, visualized by both H&E and Oil Red O (ORO) staining (Figure 3A). The lipid droplets were also quantified by ImageJ scanning (Figure S3A). Moreover, 2 out of 14 cA-circSlc8a1-transgenic mice at the age of 12 months showed significant liver cirrhosis while none of the negative mice did (Figure S3B). To validate whether such hepatic steatosis was caused by the ectopic cardiac-specific expression of cA-circSlc8a1, we examined the levels of cA-circSlc8a1 precursor in different organs, including both heart and liver and detected a dramatic increase in cA-circSlc8a1 precursor levels in the heart but not in the liver of the cA-circSlc8a1-transgenic mice (Figures 3B and S3C). In addition, we also used a probe specifically targeting circSlc8a1 to pull down circSlc8a1 in the heart and liver of cA-circSlc8a1 transgenic and the litter-matched negative mice. Due to the formation of double-stranded RNA circle with cA-circSlc8a1, circSlc8a1 could not bind to its specific probe that was complementary to its BSJ region in the presence of cA-circSlc8a1 (Figure 3C). In the heart of cA-circSlc8a1-transgenic mice, significantly less circSlc8a1 was pulled down by the circSlc8a1 probe since cA-circSlc8a1 blocked the binding of the circSlc8a1 probe with the double-stranded circRNA (Figure 3D, left). However, such effects could not be observed in the liver of the cA-circSlc8a1-transgenic mice (Figure 3D, right). These findings confirmed that hepatic steatosis in the cA-circSlc8a1-transgenic mice originated from the heart dysfunction. In addition, to test the effect of the dsRNA-induced immune response, we examined the levels of multiple immune potentiated cytokines in the heart of cA-circSlc8a1-transgenic mice. Our results showed that, except for a slight increase in IL-6 expression, the expression of other cytokines in cA-circSlc8a1-transgenic mice was at similar levels to those in negative mice (Figure S3D), excluding the effect of immune response on the observed phenotype. Thus, sequestering endogenous circSlc8a1 by cA-circSlc8a1 appeared to have instigated cardiac hypertrophy and malfunction, which consequently led to hepatic steatosis and eventually cirrhosis.
Figure 3.
The cA-circSlc8a1-transgenic mice developed cardiogenic hepatic steatosis
(A) Representative photographs of gross appearance, H&E staining and Oil Red O staining of livers from cA-circSlc8a1 mice at different ages, detecting hepatic steatosis in cA-circSlc8a1(+) mice. (B) The expression levels of cA-circSlc8a1 precursor in the heart and liver of cA-circSlc8a1-transgenic mice, confirming specificity of the heart promoter. n = 8; ∗∗p < 0.01 versus negative heart. (C) A schematic illustration of the interaction of circSlc8a1 probes to circSlc8a1 with or without the presence of cA-circSlc8a1. (D) circSlc8a1 was pulled down significantly less by the circSlc8a1 probe in the heart (left) of cA-circSlc8a1(+) mice (∗∗p < 0.01 versus negative), whereas no difference was observed in the liver (right) of cA-circSlc8a1(+) mice compared with the litter-matched negative mice. n = 16–20.
Transgenic expression of circSlc8a1 prevents pressure overload-induced heart injury
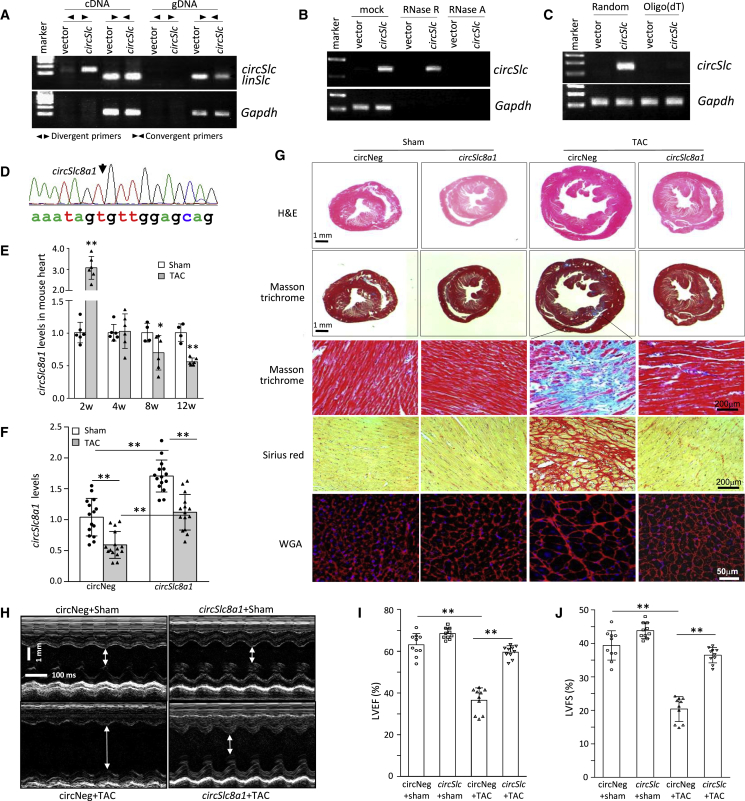
Our results from cA-circSlc8a1-transgenic mice implied that circSlc8a1 might play an essential role in maintaining the physiological function of the heart. Therefore, we constructed a circSlc8a1 plasmid for performing further functional studies on circSlc8a1. We found that circSlc8a1 could only be detected in cDNA but not in gDNA with divergent primers, while the linear form of Slc8a1 could be detected in both cDNA and gDNA with convergent primers in HL-1 cells (Figure 4A). Our results also showed that the overexpressed circSlc8a1 was resistant to RNase R treatment (Figures 4B and S4A) and could not be reverse transcribed with oligo(dT) primers (Figures 4C and S4B). In addition, the BSJ sequences of circSlc8a1 were verified in the PCR products from circSlc8a1 overexpressed HEK293 cells by Sanger sequencing (Figure 4D). All of these results validated the successful circularization of circSlc8a1 plasmid that we constructed.
Figure 4.
Transgenic expression of circSlc8a1 improved heart function against transverse aortic constriction-induced pressure overload
(A) The circSlc8a1 plasmids were transfected into HL-1 cells followed by RT-qPCR. circSlc8a1 was amplified by divergent and convergent primers in cDNA, and by convergent primers only in gDNA. (B) The circSlc8a1 plasmids were transfected into human HEK293T cells. The RNAs from vector or circSlc8a1-transfected cells were treated with RNase R or RNase A followed by reverse transcription. The expression of circSlc8a1 was validated by using mouse-specific primers against circSlc8a1. n = 4. (C) Representative images of circSlc8a1 expression by using random or oligo(dT) primers for reverse transcription. The circSlc8a1 plasmids were transfected into human HEK293T cells. The RNAs from vector or circSlc8a1-transfected cells were reverse transcribed using random or oligo(dT) primers. The expression of circSlc8a1 was validated by using mouse-specific primers against circSlc8a1. n = 6. (D) The back-splice junction of the ectopic mouse circSlc8a1 construct was confirmed by Sanger sequencing. The circSlc8a1 expression construct was used to generate transgenic mice. (E) The levels of circSlc8a1 were significantly decreased in the mouse heart tissues at 8–12 weeks after transverse aortic constriction (TAC) surgery. n = 6; ∗p < 0.05, ∗∗p < 0.01. (F) The levels of circSlc8a1 were retained at the similar levels as those in the sham group in the hearts of circSlc8a1(+)-transgenic mice 12 weeks post TAC surgery, while the levels of circSlc8a1 in the litter-matched negative mice (circNeg) were significantly decreased at the same time point after TAC surgery. n = 15; ∗∗p < 0.01. (G) Representative photographs of Masson trichrome, Sirius red, and WGA staining showed that cardiac fibrosis and cardiac hypertrophy were induced by TAC surgery in the circSlc8a1 negative (circNeg) mice. Such fibrosis and hypertrophy could be prevented in the transgenic mice overexpressing circSlc8a1. (H) The representative echocardiography of circSlc8a1-transgenic mice subjected to TAC surgery. (I and J) The circSlc8a1(+) mice subjected to TAC prevented the impairment of the heart function that was validated by (I) left ventricular ejection fraction (LVEF) and (J) left ventricular fractional shortening (LVFS). n = 10; ∗∗p < 0.01.
We then conducted transverse aortic constriction (TAC) surgery in the mice to induce pressure overload (PO) and the subsequent cardiac hypertrophy and heart failure. The expression of circSlc8a1 in the wild-type mice was increased at 2 weeks post-TAC surgery while it decreased significantly 8–12 weeks after TAC surgery in the heart of TAC mice compared with the sham mice (Figure 4E), which suggested that loss of circSlc8a1 could be associated with PO-induced heart failure. Based on these results, we established a circSlc8a1-transgenic mouse line (Figure S4C). The circSlc8a1-transgenic mice and their negative counterparts were then subjected to TAC surgery to confirm the roles of circSlc8a1 in vivo. While TAC surgery significantly decreased circSlc8a1 levels in the heart, transgenic expression of circSlc8a1 was able to maintain circSlc8a1 levels as high as those similar to negative sham mice (Figure 4F). Upon 12 weeks PO induced by TAC, the circSlc8a1-transgenic mice showed significantly less cardiac hypertrophy and fibrosis along with higher levels of circSlc8a1 in the heart compared with the litter-matched negative controls (Figures 4G, S4D, and S4E). In addition, the levels of hypertrophy markers (ANP and BNP) and fibrosis markers (collagen-I and collagen-III) were significantly lower in the hearts of the circSlc8a1-transgenic mice compared with the negative controls upon PO (Figures S5A and S5B).
Echocardiography analysis showed that TAC surgery remarkably increased the left ventricular chamber size that was prevented by transgenic expression of circSlc8a1 (Figure 4H). In cardiac functional measurements, TAC surgery significantly impaired the heart function of the negative mice, while the heart function of the circSlc8a1-transgenic mice was maintained at similar levels to the negative mice in the sham group (Figures 4I, J, and S5C–S5F). These results demonstrated the protective effects of circSlc8a1 during heart injury and indicated the importance of circSlc8a1 in maintaining physiological heart function.
The cA-circSlc8a1 induces cardiac hypertrophy and hepatic steatosis via sequestering cardiac circSlc8a1
To further validate that the phenotypic changes in cA-circSlc8a1-transgenic mice were through its specific effects on sequestering circSlc8a1, we generated a double transgenic mouse line by ectopically expressing both circSlc8a1 and cA-circSlc8a1 (Figures S6A–S6C). In the heart of double transgenic mice, the levels of circSlc8a1 were maintained at similar levels to the negative mice, which were verified by both gene-specific RT-qPCR and RNA pull-down assay (Figures 5A and 5B), while the Slc8a1 mRNA was not affected in these mice (Figure S6D). Regarding the phenotypes, we found that the body weight increase in the cA-circSlc8a1-transgenic mice was effectively reversed by re-introducing circSlc8a1 into the mice to generate double transgenic mice (Figure 5C). In addition, cardiac hypertrophy in the cA-circSlc8a1-transgenic mice was not observed in the hearts of double transgenic mice (Figures 5D and S6E). Meanwhile, hepatic steatosis that was observed in the cA-circSlc8a1-transgenic mice was not seen in the double transgenic mice (Figures 5E and S6F).
Figure 5.
The rescue of phenotypic changes in circSlc8a1 and cA-circSlc8a1 double transgenic mice
(A) The levels of unbound circSlc8a1 were detected by gene-specific RT-qPCR in circSlc8a1(+), cA-circSlc8a1(+), double transgenic mice, and the litter-matched negative mice. n = 10; ∗∗p < 0.01. (B) The levels of unbound circSlc8a1 were detected by RNA pull-down assay with circSlc8a1 probes in circSlc8a1(+), cA-circSlc8a1(+), double transgenic mice, and the litter-matched negative mice. n = 10; ∗∗p < 0.01. (C) The body weight difference in circSlc8a1(+), cA-circSlc8a1(+), double transgenic mice compared with the average of the litter-matched negative mice. n = 10; ∗∗p < 0.01. (D) Representative photographs of the whole hearts and WGA staining showed that cardiac hypertrophy was not induced in the double transgenic mice compared with the cA-circSlc8a1(+)-transgenic mice. n = 10. (E) Representative photographs of H&E and Oil Red O staining showed that hepatic steatosis was not induced in the double transgenic mice compared with the cA-circSlc8a1(+)-transgenic mice. n = 10. (F) The levels of unbound circSlc8a1 were detected by gene-specific RT-qPCR after 0.25, 0.5, 1, 2, or 3 μg cA-circSlc8a1 plasmids per 1 × 106 cells or the vectors were transfected. n = 6; ∗∗p < 0.01 versus vector. (G) The levels of unbound circSlc8a1 were detected by the RNA pull-down assay with circSlc8a1 probes after 0.25, 0.5, 1, 2, or 3 μg cA-circSlc8a1 plasmids per 1 × 106 cells or the vectors were transfected. n = 6; ∗∗p < 0.01 versus vector. (H) The unbound circSlc8a1 levels after transfection of vector, cA-circSlc8a1 plasmids and the cA-circSlc8a1 plasmids with mutated introns (lin-cA-circSlc). The levels of unbound circSlc8a1 were detected by gene-specific RT-qPCR. n = 6; ∗∗p < 0.01 versus vector. (I) The unbound circSlc8a1 levels were detected by the RNA pull-down assay after transfection with the vector, cA-circSlc8a1 plasmids and the cA-circSlc8a1 plasmids with mutated introns (lin-cA-circSlc). n = 6; ∗∗p < 0.01 versus vector.
To further verify the specific blocking of circSlc8a1 by cA-circSlc8a1, we examined the levels of free circSlc8a1 by introducing different amounts of cA-circSlc8a1 into the HL-1 cells. Our results demonstrated that the levels of circSlc8a1 were gradually decreased with transfection of elevated amounts of cA-circSlc8a1 plasmids, detected by both circRNA-specific RT-qPCR (Figure 5F) and the RNA pull-down assay (Figure 5G). In addition, to exclude the possibility that the uncircularized linear cA-circSlc8a1 precursor after transfection could also bind to and block circSlc8a1, we designed a construct with an intronic mutation which only generated linear cA-circSlc8a1 precursor but not the circular form of cA-circSlc8a1. Our results showed that the linear cA-circSlc8a1 precursor could not decrease the levels of unbound circSlc8a1, which indicated that only circularized cA-circSlc8a1 could bind with circSlc8a1 and block its function (Figures 5H and 5I). Taken together, these results demonstrated that the effects of cA-circSlc8a1 on cardiac hypertrophy and hepatic steatosis were exerted by blocking the physiological functions of circSlc8a1.
Interaction of circSlc8a1 with mitochondria-associated proteins
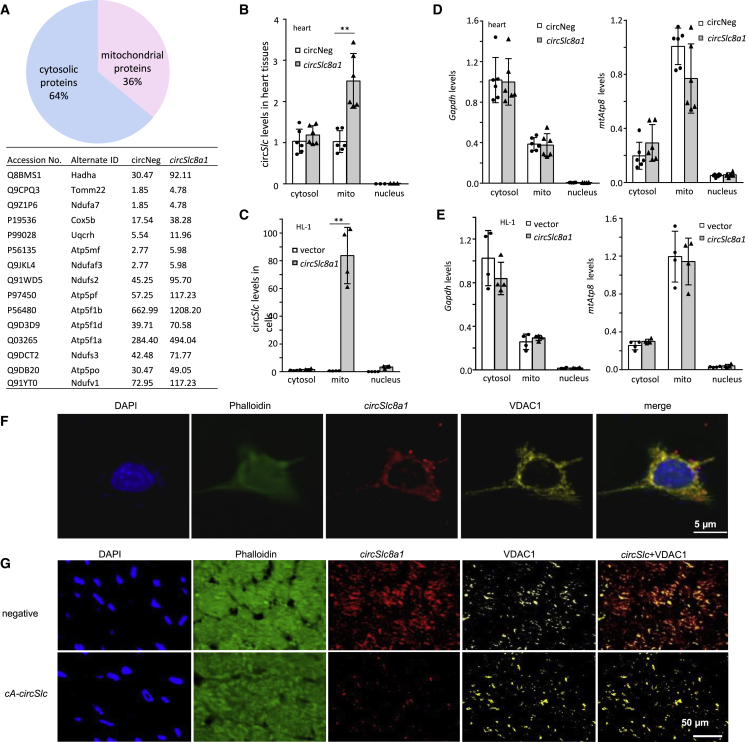
To further investigate the mechanism of the protective effects of circSlc8a1, we identified the proteins that bind with circSlc8a1 using a circSlc8a1 probe to precipitate the circSlc8a1-binding proteins followed by mass spectrometry analysis. The list showed that 36% of the total precipitated proteins were mitochondrial proteins, and 85% of these mitochondrial proteins were among the top 20 circSlc8a1-binding proteins based on the abundance in the heart tissues of the circSlc8a1-transgenic mice (Figure 6A; Table S1). To further confirm the results from the mass spectrometry analysis, we examined the subcellular localization of circSlc8a1 in the heart tissues of the circSlc8a1-transgenic mice and in HL-1 cells transfected with circSlc8a1. The results showed that circSlc8a1 levels were significantly higher in the mitochondria of the heart tissues of the circSlc8a1-transgenic mice (Figure 6B) and in the circSlc8a1-transfected HL-1 cells (Figure 6C) compared with the cytoplasm content lacking mitochondria. The purity of the mitochondria was confirmed by detecting the expression of GAPDH in the cytosol and mitochondrially encoded Atp8 (MT-ATP8) in the mitochondria (Figures 6D and 6E). We also found co-localization of circSlc8a1 with the mitochondrial marker (VDAC) in the HL-1 cells (Figure 6F) and the heart tissues of the cA-circSlc8a1-transgenic mice (Figure 6G) using fluorescence in situ hybridization (FISH) and IHF staining, which suggested that circSlc8a1 was translocated into the mitochondria to exert its function.
Figure 6.
circSlc8a1 protected the heart function via binding to mitochondrial proteins
(A) Mass spectrophotometry analysis showed that 36% of the total proteins (types) precipitated by the circSlc8a1 probe were mitochondrial proteins. (B) Subcellular localization of circSlc8a1 in the cytosol (excluding mitochondria), mitochondria, and nuclei in the mouse heart tissues. n = 6; ∗∗p < 0.01 versus circNeg. (C) Subcellular localization of circSlc8a1 in the cytosol (excluding mitochondria), mitochondria, and nuclei in HL-1 cells. n = 4; ∗∗p < 0.01 versus vector. (D) The purity of mitochondrial fraction in the mouse heart tissue was validated by examining GAPDH and the mitochondrially encoded ATP8 (MT-ATP8). n = 5. (E) The purity of mitochondrial fraction in the HL-1 cells was validated by examining GAPDH and the mitochondriallly encoded ATP8 (MT-ATP8). n = 5. (F) Representative photographs of the co-localization of circSlc8a1 and the mitochondrial marker (VDAC) in HL-1 cells. (G) Representative photographs of the co-localization of circSlc8a1 and the mitochondrial marker (VDAC) in negative and cA-circSlc8a1(+) mouse heart tissue.
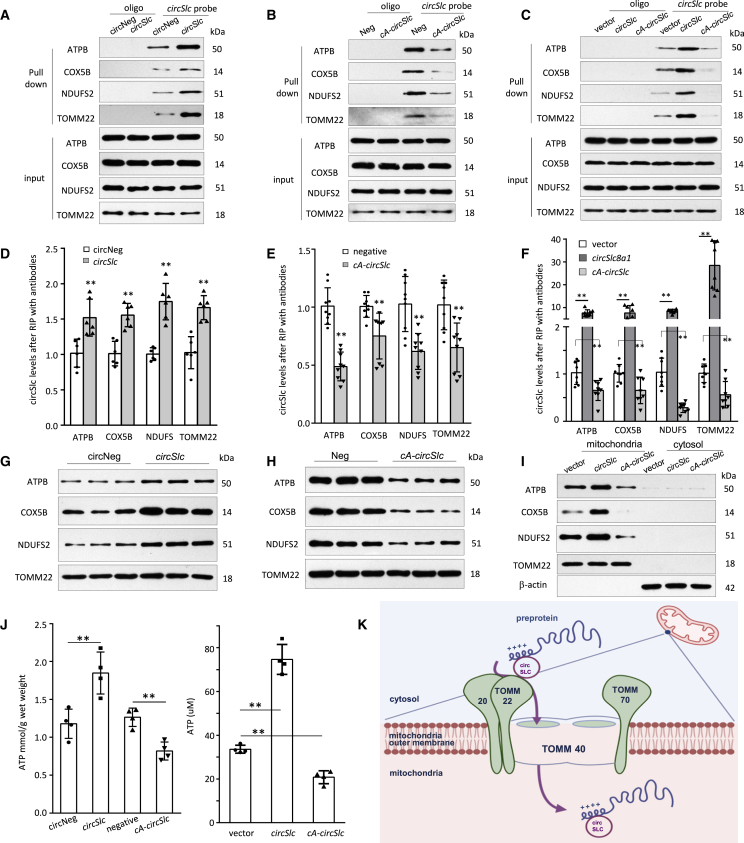
We further confirmed that circSlc8a1 could bind with the proteins of ATP synthase subunit B (ATPB), cytochrome c oxidase (COX5B), NADH dehydrogenase iron sulfur protein 2 (NDUFS2), and mitochondria import receptor subunit TOMM22 in the heart tissues of the circSlc8a1-transgenic mice (Figures 7A and S7A) or the cA-circSlc8a1-transgenic mice (Figures 7B and S7B). Such binding was also validated in the cells transfected with circSlc8a1 and cA-circSlc8a1 plasmids (Figures 7C and S7C). In accordance, the interactions between circSlc8a1 and the mitochondrial proteins, ATPB, COX5B, NDUFS2, and TOMM22, were also validated using the antibodies against these proteins to precipitate circSlc8a1 in the transgenic heart tissues expressing circSlc8a1 (Figure 7D) and cA-circSlc8a1 (Figure 7E), and the HL-1 cells transfected with circSlc8a1 and cA-circSlc8a1 plasmids (Figure 7F). We also found that mitochondrial translocation of ATPB, COX5B, and NDUFS2 was facilitated by circSlc8a1 and inhibited by cA-circSlc8a1 using western immunoblotting (Figures 7G–7I and S7D–S7F). In addition, the in situ hybridization and immunofluorescence staining results also showed that the mitochondrial translocation of these proteins was significantly suppressed in the heart tissues of cA-circSlc8a1-transgenic mice compared with their negative litter (Figure S8). This indicated that circSlc8a1 might be involved in the protein translocation to mitochondria via the translocase TOMM22 on the outer mitochondrial membrane. Since these mitochondrial proteins are associated with ATP generation in the mitochondria, we measured the ATP contents in the heart tissues of the circSlc8a1-transgenic mice and the circSlc8a1-transfected HL-1 cells. We observed a significant increase in ATP contents in the tissues and cells overexpressing circSlc8a1, which were reduced in the heart of cA-circSlc8a1-transgenic mice and cA-circSlc8a1-transfected cells (Figure 7J). These results suggested that circSlc8a1 played its cardioprotective roles by promoting the protein translocation into the mitochondria and facilitating ATP production, which are essential to maintain heart function (Figure 7K).
Figure 7.
Interaction of circSlc8a1 and the mitochondrial proteins
(A–C) Mitochondrial proteins ATPB, COX5B, NDUFS2, and TOMM22 were pulled down by circSlc8a1 probe in the heart tissues of circSlc8a1 (A) and cA-circSlc8a1 (B) transgenic mice, and in HL-1 cells transfected with circSlc8a1 or cA-circSlc8a1 plasmids (C) followed by western blotting. n = 3–4. (D and E) Antibodies against ATPB, COX5B, NDUFS2, and TOMM22 were used to precipitate circSlc8a1 from the heart tissues of circSlc8a1 (D) and cA-circSlc8a1 (E) transgenic mice. The immune-precipitated circSlc8a1 was measured by qPCR. n = 6–9; ∗∗p < 0.01 versus negative mice. (F) circSlc8a1 was immune-precipitated by ATPB, COX5B, NDUFS2, and TOMM22 antibodies in HL-1 cells transfected with circSlc8a1 or cA-circSlc8a1 plasmids followed by qPCR to detect the circSlc8a1. n = 6–9; ∗∗p < 0.01 versus vector. (G and H) Mitochondrial proteins were isolated from the heart tissue of circSlc8a1 (G) and cA-circSlc8a1 (H) transgenic mice. The mitochondrial translocation of ATPB, COX5B, and NDUFS2 proteins increased in the circSlc8a1-transgenic mice (G) but decreased in the cA-circSlc8a1-transgenic mice (H) compared with their counterpart negative litters. The outer mitochondrial membrane protein TOMM22 was not affected by circSlc8a1 expression. n = 3–4. (I) The mitochondrial and the cytosolic proteins were separated from HL-1 cells transfected with circSlc8a1 or cA-circSlc8a1 plasmids. The mitochondrial translocation of ATPB, COX5B, and NDUFS2 proteins increased in the circSlc8a1-transfected cells but decreased in the cA-circSlc8a1-transfected cells compared with the vector controls. The purity of mitochondria and cytosol was confirmed by TOMM22 and β-actin antibodies. n = 3–4. (J) ATP levels were significantly increased by circSlc8a1 overexpression and decreased by cA-circSlc8a1 in the heart tissues of the transgenic mice (left) and HL-1 cells (right). n = 4; ∗∗p < 0.01. (K) A model depicting the proposed mechanism of circSlc8a1 facilitating the translocation of pre-proteins from the cytosol into the mitochondria.
Discussion
Differential expression of circRNAs has been linked to different stages of myocardial development.19 Dysregulation of circRNAs can lead to cardiovascular diseases, and may represent potential targets for therapeutics.20,21 Usually, downregulated circRNAs could be compensated by ectopic delivery using nanoparticles or adeno-associated viruses.22,23 However, knockdown of the upregulated circRNAs may also have off-target effects impacting the parental linear mRNA. To date, several approaches have attempted to silence circRNAs without removing the linear counterparts for in vivo functional studies. 5,14,15,24 In this study, we established a simple and effective method to knockdown cardiac circRNA in vivo, which may further be applied to circRNA functional studies and gene therapy in cardiovascular diseases.
Several antisense-based therapeutic methods for linear RNAs have been thoroughly investigated in preclinical studies and have been applied clinically.25,26 The circRNAs possess unique structural conformations distinct from the linear RNA cognates,27 therefore, we engineered circularized antisense RNA to only silence circRNA. The most beneficial aspect of using circular antisense RNA is that it could possibly be used for knocking down individual circRNA depending on its own sequence or structure. By forming a complementary circle against its sense counterpart, circular antisense RNA is specific for the target circRNA of interest and does not interfere with the linear mRNA cognate. In the present study, we used the circular antisense RNA of circSlc8a1, named cA-circSlc8a1, as an example to demonstrate the effects of circular antisense RNA on sequestering its sense circRNA. The circularized antisense RNA cA-circSlc8a1 is complementary to circSlc8a1 based on the oligonucleotide sequence, which suggests that cA-circSlc8a1 and circSlc8a1 may form a double-stranded RNA circle, at least partially. While the secondary structure might be a complex, the significant high affinity between cA-circSlc8a1 and circSlc8a1 was confirmed by the computational docking method. Notably, our results demonstrated that cA-circSlc8a1 sequestered circSlc8a1 without affecting the full-length linear Slc8a1 mRNA cognate and its corresponding SLC8A1 protein in cardiomyocytes or heart tissues. One possible explanation could be that the unique circularized structure of cA-circSlc8a1 rendered it less possible to bind to and sequester the linear Slc8a1 mRNA.27 We then established a transgenic animal model with circSlc8a1 specifically silenced in the heart with cA-circSlc8a1 driven by a heart promoter. By using cA-circSlc8a1, circSlc8a1 functions in the heart were blocked. The cardiac-specific expression of cA-circSlc8a1 in mice caused dramatic phenotypic changes, including significant increase in the body weight, hepatic steatosis, and impaired heart functions. Such phenotype changes provided evidence to demonstrate that circular antisense RNA could effectively sequester the function of the targeting circRNA. With this approach, we successfully established a mouse model for studying cardiac hepatopathy and demonstrated the essential roles of circSlc8a1 in maintaining heart function. Consistent with this conclusion, another transgenic mouse model with overexpression of circSlc8a1 validated the protective effects of circSlc8a1 in a PO model. Previous studies reported that the levels of circSlc8a1 were increased in the TAC or ischemia-reperfusion mouse models.28,29 In those studies, the levels of circSlc8a1 were measured at 1 day or 3 weeks post-TAC. Our study showed that the levels of circSlc8a1 at 2 weeks post-TAC were increased in the heart of TAC mice compared with the sham mice while they were reduced significantly 8–12 weeks post-TAC. Our results are consistent with the study from Lim et al., which showed that the endogenous levels of circSlc8a1 were decreased along with the impairment of the cardiac function in the hearts of TAC mice at 5 weeks post-TAC compared with the sham mice.30 Taking the evidence together, we consider that circSlc8a1 expression may increase at the early stage of PO to compensate for the acute impairment of ejection. However, cardiac fibrosis and heart failure occurred after long-term PO (8–12 weeks post-TAC), and ectopic expression of circSlc8a1 could retain cirSlc8a1 at physiological levels, which helped to protect heart function. Our study showed the strong dynamic effects of circSlc8a1 during the progression of cardiac hypertrophy, cardiac remodeling, fibrosis, and heart failure.
Cardiac hepatopathy is associated with an increase of hepatic venous pressure and a decrease of portal vein inflow.31 Over time, liver hypoperfusion and hepatic congestion lead to the development of thrombosis and an accumulation of deoxygenated blood in the liver resulting in hepatic steatosis and fibrosis.32 Simonetto et al. developed a murine model of congestive hepatopathy by partial ligation of the inferior vena cava.33 However, the congestive hepatopathy in this model was not directly derived from heart malfunction. In addition, previous studies have also shown that the heart plays important roles in systemic metabolic homeostasis. Deletion of MED13 protein or cardiac-specific miRNA, miR-208a, in the heart, led to body weight elevation and lipid accumulation in the liver.34,35 Our study showed that genetic silencing of circSlc8a1 by cA-circSlc8a1 generated a model of heart failure leading to congestive hepatopathy. Knockdown of the cardiac circSlc8a1 by its circularized antisense RNA led to an impaired heart function, a dramatic lipid accumulation in the liver and development of hepatic fibrosis/cirrhosis at the late stage. While providing a simple mouse model for defining the circRNA function, the present proof-of-conception study may also open a novel route for gene therapy targeting circRNA. Currently, two other circular antisense circRNAs designed with the same concept as cA-circSlc8a1 are being employed for investigating circRNA functions in the heart based on their specific functions in the heart by sequestering their corresponding structures/functions in our laboratory. In our previous report, nanoparticle-conjugated plasmids were effectively delivered to the cardiac tissues and expressed well to produce circRNAs.22 In the future, delivery of circularized anti-sense RNA has the potential to be used in gene therapy for the treatment of circRNA-related disorders.
Mechanistically, a large proportion of circSlc8a1-binding proteins were identified as mitochondrial proteins in mass spectrometry analysis, such as ATP synthase, cytochrome c oxidase, NADH dehydrogenase, and outer mitochondrial membrane protein TOMM22. Co-localized expression of circSlc8a1 with mitochondrial markers in transfected cells and transgenic mice indicated that circSlc8a1 might be translocated into the mitochondria after synthesis. Most of the mitochondrial proteins are encoded by nuclear genes and synthesized on cytosolic polysomes followed by translocation into the mitochondria via a protein sorting system.36 These pre-proteins contain a consensus pre-sequence with mitochondrial targeting signal to be recognized by the mitochondrial import receptors TOMM20 and TOMM22 on the outer mitochondrial membrane.37,38 Since circSlc8a1 bound to a variety of mitochondrial proteins, we postulated that circSlc8a1 might be able to recognize and bind with the mitochondrial targeting sequence of these nuclear-encoded mitochondrial proteins after the precursor proteins are synthesized in the cytosol. Meanwhile, circSlc8a1 can also bind to TOMM22 on the outer membrane of mitochondria, which may facilitate these circSlc8a1-binding pre-proteins to interact with the outer membrane translocator complex and increase the import of these pre-proteins into the mitochondria (Figure 7K). By regulating these ATP generation-related enzymes, circSlc8a1 played an essential role in maintaining heart function. However, the binding sites and the detailed transport mechanism need further investigation. In the previous studies of the circRNAs located in the mitochondria, the circRNAs were either generated by the mitochondrial genome39 or co-localized with the mitochondrial proteins.40,41 It is of great significance to identify how the nuclear genome-derived circRNAs were translocated into the mitochondria in the future studies.
In summary, we established a circRNA knockdown mouse model by using circular antisense RNA. It is a simple and more scalable method with broad general applicability to suppress the existential circRNAs in vivo. Sequestering circSlc8a1 by its circular antisense RNA leads to malfunction of the heart, and subsequent cardiogenic hepatopathy. This in vivo circRNA knockdown model provides an effective strategy for circRNA functional studies and a potential approach for circRNA-related gene therapy.
Materials and methods
circRNA sequencing
The circRNA sequencing was conducted with the RNAs from the heart tissues obtained from 13 mice. The RNA samples were subjected to RNase R (ThemoFisher Scientific) treatment to degrade linear RNAs and depletion of rRNA. The remaining circRNAs were fragmented followed by RT using random primers. The circRNAs sequencing and analysis was performed by Novogene using HiSeq2500 System. The cDNA library was generated by the Novogene and then subjected to sequencing. Each circRNA was recorded by at least two reads spanning a head-to-tail splice junction in each sample.
Construct generation and primer design
The expression constructs of cA-circSlc8a1 and circSlc8a1 were generated according to the sequences shown in the supplemental information. The vector contains a Bluescript backbone, with one CMV promoter driving green fluorescent protein expression, and another CMV promoter driving the circRNA-forming fragments. For generating the cardiac-specific cA-circSlc8a1-transgenic mice, a heart promoter of mouse α-myosin heavy chain promoter cassette was inserted into cA-circSlc8a1 construct according to the previous report.42 For the vector control, circSlc8a1 insert sequence was replaced with a non-related random sequence. The constructs of pre-circSlc8a1 and pre-cA-circSlc8a1 were generated in which the intronic sequences for circularization signal were mutated. Thus, pre-circSlc8a1 and pre-cA-circSlc8a1 would express their linear transcripts without circularization. The sequences for all the constructs and primers used are listed in the supplemental information.
Computational docking of cA-circSlc8a1 with the circular and linear Slc8a1
To determine the secondary structures with lowest free energy, MFE algorithm implemented in Mfold (version 2.3) was used.43 Mfold, a dynamic programming algorithm, uses energy minimization to model the ensemble of possible structures by identifying optimal folding of a nucleic acid sequence within a specified energy increment. The secondary structure with a lower theoretical value of free energy was selected as a model structure for 3D structure prediction. The output file containing primary sequence and an associated secondary structure (Dot-Bracket Notation) was then submitted to 3dRNA Web Server to generate the 3D structure.44 3dRNA is an automated method of building RNA 3D structures from sequences and secondary structures by using the smallest secondary elements. Docked models were generated using a bimolecular docking server PatchDock. The main physicochemical measure that relates to binding affinity is the buried surface area (BSA). BSA = (ASA antisense-circular RNA + ASA sense-circular RNA/linear mRNA) – ASA complex. Visualization was performed by using PyMOL.
Animal models
All animal experiments were conducted in accordance with the relevant guidelines and regulations approved by the Animal Care Committee of Sunnybrook Research Institute. The cA-circSlc8a1 and circSlc8a1-transgenic mice were generated by pronuclear microinjection of DNA fragments containing either cA-circSlc8a1 with the heart promoter or circSlc8a1 into C57BL/6J, performed by the Toronto Centre for Phenogenomics. The sequences of cardiac-specific promoter (the promoter of mouse alpha myosin heavy chain gene) are shown in Figure S1E. The sequences of cA-circSlc8a1 and circSlc8a1 constructs for generating transgenic mice are listed in the supplemental information. All transgenic mice were ear tagged and genotyped after weaning. The genotyping primer sequences are listed in the Table S2.
PO-induced cardiac hypertrophy was performed by the modified TAC in mice as previously described.45,46 Successful banding was confirmed with visual confirmation of differential carotid pulpability. Success in generating PO model was confirmed by measuring the carotid artery flow velocities by Doppler. Only mice with a right carotid/left carotid flow ratio within a certain range (>5) were included for further experiments. The sham mice underwent surgery at the same time points with anesthesia and the rest of the operation, except the aortic banding.
Cell culture and transfection
The immortalized mouse cardiomyocyte cell line, HL-1 cells, were cultured in Claycomb medium supplemented with 10% FBS, 100 U/mL penicillin and streptomycin, 2 mM L-glutamine and 0.1 mM norepinephrine. The HEK293 cells were cultured in DMEM supplemented with 10% FBS and 100 U/mL penicillin and streptomycin. The cells were transfected with the mixture of plasmid and transfection reagent Polyjet (SignaGen Laboratories) for 5–12 h and collected for further examination 24–48 h after transfection.
Cardiac function assessment
Mice were anesthetized with 2% isoflurane inhalation to undergo transthoracic echocardiography. Transthoracic echocardiography was performed and analyzed in a blinded manner, using a Vevo 2100 high-resolution imaging system equipped with a 40-MHz transducer to measure LVEDD, LVESD, LVEF, LVFS, and dp/dt. A 1.4-Fr high-fidelity pressure catheter (SPR-671, Millar Instruments, Houston, TX) was inserted into the LV via the right carotid artery to evaluate left ventricular pressure (LVSP) and dp/dt using PowerLab system (AD Instruments) as described.47
Histological staining
After harvesting, the hearts were cut into two halves. The lower halves were fixed with 10% buffered formalin and embedded in paraffin, sectioned into 5 μm sections. Masson’s trichrome staining and Sirius red staining were performed to evaluate cardiac fibrosis. Masson’s trichrome stain kit (American Master Tech) was used for Masson’s trichrome staining according to the manufacturer’s instructions. In brief, the paraffin sections were deparaffinized and hydrated with xylene and absolute alcohol followed by rinsing with running tap water. The sections were then immersed into preheated Bouin’s Fluid at 56°C for 1 h followed by rinsing with running tap water until the tissues were colorless. Then, the sections were immersed in working Weigert’s hematoxylin for 5 min, in Biebrich scarlet-acid fuchsin for 15 min, in phosphomolybdic/phosphotungstic acid for 15 min, in Aniline blue stain for 10 min, and then in 1% acetic acid for 5 min. The slides were rinsed with running tap water between each step. After dehydration, the slides were mounted with permanent mounting media.
The Sirius red staining was performed according to the protocol described in the previous report.48 The mouse heart sections were de-waxed and hydrated, followed by staining with Weigert’s haematoxylin for 8 min. The sections were washed with running tap water, stained in 0.1% Picrosirius red for 1 h, and then washed in 0.1% acetic acid. After dehydration, the slides were mounted with permanent mounting media.
WGA staining was conducted to visualize the cross-section area of cardiomyocytes for evaluating cardiac hypertrophy. Alexa Fluor 555 conjugated WGA (5 μg/mL) was applied to the tissue slides and incubated at room temperature for 10 min followed by washing twice with HBSS according to the manufacturer’s instructions (ThermoFisher Scientific). Then, the slides were mounted with fluorescence mounting medium (Dako).
For ORO staining, the livers were fixed with 10% buffered formalin followed by immersion in 30% sucrose before embedded in OCT compound. Cryopreserved tissues were sectioned into 10 μm sections. The tissue slides were stained with ORO working solution (0.7%, w/v) followed by washing out the excess stain. The slides were mounted with aqueous mounting media, Glycerol Gelatin (Sigma).
The fibrosis area, cross-section area and lipid droplets area were quantified using ImageJ software.
RT-PCR and RNase treatment
Total RNAs were extracted from cells and tissues using TriRNA isolation kit (Geneaid) or TRIzol (ThermoFisher Scientific). Total RNAs were subjected to RNase R treatment to remove linear RNAs or directly used for RT to synthesize single-stranded cDNA. RNase treatment was conducted as described previously.49,50 In brief, 1 μg RNase R (Epicentre) or RNase A (QIAGEN) was added in the mixture and incubated at 37°C for 15 min prior to RT. RNA (1 μg) was subjected to RT and quantitative PCR (qPCR) using LunaScript SuperMix and LunaqPCR master mix (New England Biolabs). The small nuclear RNA U6 or GAPDH were used as internal controls to calculate the relative levels of the RNAs of interest. For examining the copy number of circSlc8a1, cDNA was prepared as described above followed by droplet digital PCR (Bio-Rad) conducted at Genomics Core Facility, Sunnybrook Research Institute. For gene-specific RT using reverse primer specific for circSlc8a1, 1 μg RNA was reverse transcribed using iScript select cDNA synthesis kit (Bio-Rad) with reverse primer specific for circSlc8a1 (5′-gtacaataagacttccaactgc-3′). The sequences of primers are listed in the supplemental information.
RNA pull-down assay
The RNA pull-down assay was performed using a biotin-labeled circRNA probe as described before.49,51 In brief, the tissues or cells were lysed in co-IP buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1 mM EDTA, 0.5% NP40 and protease inhibitor cocktail) and incubated with biotinylated DNA probes against circSlc8a1 at room temperature for 2 h. Streptavidin C1 magnetic beads (50 μL) from Invitrogen were added to each of the probe-containing reaction, and further incubated at room temperature for another 1 h. The beads were collected and washed extensively with co-IP buffer for five times. The circRNA circSlc8a1 pulled down by the probe was analyzed by RT-PCR. The proteins that bound to circSlc8a1 and pulled down by the probe were analyzed by western blotting or mass spectrometry. The biotin-oligomers for RNA pull-down of mouse circSlc8a1 were synthesized by Eurofins Genomics.
Subcellular fractionation
The subcellular fractions of the heart tissues or the cells were isolated as described.52 In brief, cultured cells or heart tissues were harvested and resuspended in 500 μL fractionation buffer (250 mM sucrose, 20 mM HEPES [pH 7.4], 10 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA, and protease inhibitor cocktail). Then, the cells were homogenized by 10 passages through a 25-G needle using a 1 mL syringe and incubated on ice for 30 min. The heart tissues were homogenized in fractionation buffer using a Dounce homogenizer on ice. The nuclear pellet was collected by centrifugation at 720 × g for 5 min. The supernatant was centrifuged again at 10,000 × g for 15 min. The pellet containing mitochondria was collected followed by further wash with fractionation buffer. The supernatant containing the cytosolic fraction was concentrated and purified with Millipore centrifugal filter units. The purity of the mitochondria was confirmed by detecting the expression of GAPDH or β-ACTIN in the cytosol and mitochondrially encoded ATP8 (MT-ATP8) in mitochondria.
FISH and immunofluorescent staining
In the FISH, cy5-labeled DNA oligo probes against circSlc8a1 were generated by fluorescence PCR labeling kit (Biolynx). The labeled probes were heated at 95°C for 2 min and chilled on ice immediately to prevent reannealing. A scramble sequence was labeled in the same way and used as a negative control. The fixed samples were dehydrated by washing for approximately 1 min each in 70%, 95%, and 100% ethanol. The dehydrated samples were air-dried and pretreated with hybridization solution in 55°C for 30 min. The pre-hybridized slides were incubated with 50 nM fluorescence-labeled DNA probes in hybridization buffer at 55°C for 2 h followed by serial washes with saline sodium citrate buffers. After the samples were blocked with TBS containing 10% goat serum for 30 min, immunofluorescence staining with mitochondrial marker, VDAC antibody (Abcam) was performed with incubation at 4°C overnight followed by Alex Fluor 647 anti-rabbit antibody (ThermoFisher Scientific). F-actin was stained with Alexa Fluor 488 phalloidin (ThermoFisher Scientific) for 2 h at room temperature. DAPI was stained with NucBlue Fixed Cell ReadyProbes Reagent (ThermoFisher Scientific). Then, the slides were mounted with fluorescence mounting medium (Dako).
Western blotting
The protein expression levels were determined by western blotting as previously described.53 In brief, proteins were isolated from tissues or cells using RIPA buffer. Proteins in the lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins were transferred onto a nitrocellulose membrane in 1× Tris/glycine buffer containing 20% methanol at 80 V and 4°C for 2 h. The membranes were blocked in TBST buffer containing 5% (w/v) non-fat dry milk powder for 1 h followed by incubation with primary antibodies at 4°C overnight. The membranes were then washed and incubated with secondary antibodies at room temperature for 2 h. After washing, the bound antibodies were visualized with an ECL detection kit (Millipore). The monoclonal antibodies against SLC8A1 and ATPB were from Abcam. The antibodies against COX5B, NDUFS2, and TOMM22 were from ABclonal. The same membranes were re-probed with rabbit anti-β-ACTIN monoclonal antibody (Proteintech) to confirm equal loading of proteins for each sample.
ATP assay
The ATP levels in the heart tissues or cultured cells were measured by ATP assay kit from Abcam according to the manufacturer’s instructions. In brief, the cultured cells were resuspended and homogenized in ATP assay buffer followed by centrifugation at 13,000 × g at 4°C for 5 min. The supernatant was collected and deproteinized by using 1 M perchloric acid (PCA). The heart tissues were homogenized in ice-cold PCA with a Dounce homogenizer and kept on ice for 30–45 min. After centrifuging at 13,000 × g for 2 min at 4°C, the supernatant was neutralized to pH 6.5–8 with KOH. Then, the supernatant, which was collected after another centrifugation, and the ATP standard were applied for fluorometric assay with the ATP probe, ATP converter and Developer Mix that was provided in the assay kit.
Mass spectrometry analysis
The mouse heart tissues were lysed in the co-IP buffer and applied for RNA pull-down assay with circSlc8a1 probe following the methods described above. The streptavidin magnetic beads were collected and washed extensively with the co-IP buffer followed by washing with PBS. The streptavidin magnetic beads were sent to SPARC Molecular Analysis Centre (The Hospital for Sick Children, Toronto) for elution, digestion and mass spectrometry analysis.
Statistical analysis
All experiments were performed in triplicates or more. The numerical data were subjected to Student’s t test with non-parametric two-tailed unpaired Mann-Whitney test for two groups or one-way ANOVA for three or more groups using GraphPad Prism. The levels of significance were set at ∗p < 0.05 and ∗∗p < 0.01.
Acknowledgements
This work was supported by a grant entitled “Regulation of ventricular remodeling by a circRNA circSlc8a1” from the Canadian Institutes of Health Research (PJT-166107) to B.B.Y.
Author contributions
B.B.Y. supervised the project. N.W. and B.B.Y. designed the experiments. N.W., F.L., W.Y., W.W.D., C.Z., J.L., S.M., K.Z., and E.E. performed the experiments and analyzed the data. F.M.A. performed computational analysis of circRNA interaction. N.W. and B.B.Y. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Ethics approval
All animal experiments were conducted in accordance with the relevant guidelines and regulations approved by the Animal Care Committee of Sunnybrook Research Institute.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.10.005.
Supplemental information
Data availability
All data and material supporting the findings of this study are presented in the paper and/or the supplemental materials. Additional data are available from the corresponding author upon request.
References
- 1.Zhang C., Huo S.T., Wu Z., Chen L., Wen C., Chen H., Du W.W., Wu N., Guan D., Lian S., Yang B.B. Rapid development of targeting circRNAs in cardiovascular diseases. Mol. Ther. Nucleic Acids. 2020;21:568–576. doi: 10.1016/j.omtn.2020.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulte C., Barwari T., Joshi A., Theofilatos K., Zampetaki A., Barallobre-Barreiro J., Singh B., Sörensen N.A., Neumann J.T., Zeller T., et al. Comparative analysis of circulating noncoding RNAs versus protein biomarkers in the detection of myocardial injury. Circ. Res. 2019;125:328–340. doi: 10.1161/CIRCRESAHA.119.314937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta S.K., Garg A., Bär C., Chatterjee S., Foinquinos A., Milting H., Streckfuß-Bömeke K., Fiedler J., Thum T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 5.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W., et al. Loss of super-enhancer-regulated circRNA nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J., et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 7.Bei Y., Yang T., Wang L., Holvoet P., Das S., Sluijter J.P.G., Monteiro M.C., Liu Y., Zhou Q., Xiao J. Circular RNAs as potential theranostics in the cardiovascular system. Mol. Ther. Nucleic Acids. 2018;13:407–418. doi: 10.1016/j.omtn.2018.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L., Han B., Zhang Z., Wang S., Bai Y., Zhang Y., Tang Y., Du L., Xu L., Wu F., et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 9.Misir S., Wu N., Yang B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–491. doi: 10.1038/s41418-022-00948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C.K., Kafert-Kasting S., Thum T. Preclinical and clinical development of noncoding RNA therapeutics for cardiovascular disease. Circ. Res. 2020;126:663–678. doi: 10.1161/CIRCRESAHA.119.315856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ylä-Herttuala S., Baker A.H. Cardiovascular gene therapy: past, present, and future. Mol. Ther. 2017;25:1095–1106. doi: 10.1016/j.ymthe.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu N., Qadir J., Yang B.B. CircRNA perspective: new strategies for RNA therapy. Trends Mol. Med. 2022;28:343–344. doi: 10.1016/j.molmed.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 14.Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P., et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 15.Li S., Li X., Xue W., Zhang L., Yang L.Z., Cao S.M., Lei Y.N., Liu C.X., Guo S.K., Shan L., et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods. 2021;18:51–59. doi: 10.1038/s41592-020-01011-4. [DOI] [PubMed] [Google Scholar]
- 16.Heumuller A.W., Jones A.N., Mourão A., Klangwart M., Shi C., Wittig I., Fischer A., Muhly-Reinholz M., Buchmann G.K., Dieterich C., et al. Locus-conserved circular RNA cZNF292 controls endothelial cell flow responses. Circ. Res. 2021;130:67–79. doi: 10.1161/CIRCRESAHA.121.320029. [DOI] [PubMed] [Google Scholar]
- 17.Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., Zeng Y., Hua J.T., Petricca J., Guo H., et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843.e22. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Pamudurti N.R., Patop I.L., Krishnamoorthy A., Ashwal-Fluss R., Bartok O., Kadener S. An in vivo strategy for knockdown of circular RNAs. Cell Discov. 2020;6:52. doi: 10.1038/s41421-020-0182-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobi T., Czaja-Hasse L.F., Reinhardt R., Dieterich C. Profiling and validation of the circular RNA repertoire in adult murine hearts. Genomics Proteomics Bioinformatics. 2016;14:216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 21.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C., et al. Author Correction: circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2020;11:2234. doi: 10.1038/s41467-020-15382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu N., Xu J., Du W.W., Li X., Awan F.M., Li F., Misir S., Eshaghi E., Lyu J., Zhou L., et al. YAP circular RNA, circYap, attenuates cardiac fibrosis via binding with tropomyosin-4 and gamma-actin decreasing actin polymerization. Mol. Ther. 2021;29:1138–1150. doi: 10.1016/j.ymthe.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F., Asokan A. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Nguyen T.M., Zhang X.O., Wang L., Phan T., Clohessy J.G., Pandolfi P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021;22:41. doi: 10.1186/s13059-021-02263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foinquinos A., Batkai S., Genschel C., Viereck J., Rump S., Gyöngyösi M., Traxler D., Riesenhuber M., Spannbauer A., Lukovic D., et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat. Commun. 2020;11:633. doi: 10.1038/s41467-020-14349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H., et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 28.Werfel S., Nothjunge S., Schwarzmayr T., Strom T.M., Meitinger T., Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Li M., Ding W., Tariq M.A., Chang W., Zhang X., Xu W., Hou L., Wang Y., Wang J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–5869. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim T.B., Aliwarga E., Luu T.D.A., Li Y.P., Ng S.L., Annadoray L., Sian S., Ackers-Johnson M.A., Foo R.S.Y. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019;115:1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 31.Samsky M.D., Patel C.B., DeWald T.A., Smith A.D., Felker G.M., Rogers J.G., Hernandez A.F. Cardiohepatic interactions in heart failure: an overview and clinical implications. J. Am. Coll. Cardiol. 2013;61:2397–2405. doi: 10.1016/j.jacc.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 32.Møller S., Bernardi M. Interactions of the heart and the liver. Eur. Heart J. 2013;34:2804–2811. doi: 10.1093/eurheartj/eht246. [DOI] [PubMed] [Google Scholar]
- 33.Simonetto D.A., Yang H.y., Yin M., de Assuncao T.M., Kwon J.H., Hilscher M., Pan S., Yang L., Bi Y., Beyder A., et al. Chronic passive venous congestion drives hepatic fibrogenesis via sinusoidal thrombosis and mechanical forces. Hepatology. 2015;61:648–659. doi: 10.1002/hep.27387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grueter C.E., van Rooij E., Johnson B.A., DeLeon S.M., Sutherland L.B., Qi X., Gautron L., Elmquist J.K., Bassel-Duby R., Olson E.N. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baskin K.K., Bookout A.L., Olson E.N. The heart-liver metabolic axis: defective communication exacerbates disease. EMBO Mol. Med. 2014;6:436–438. doi: 10.1002/emmm.201303800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vögtle F.N., Wortelkamp S., Zahedi R.P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- 37.van Wilpe S., Ryan M.T., Hill K., Maarse A.C., Meisinger C., Brix J., Dekker P.J., Moczko M., Wagner R., Meijer M., et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature. 1999;401:485–489. doi: 10.1038/46802. [DOI] [PubMed] [Google Scholar]
- 38.Pfanner N. Protein sorting: recognizing mitochondrial presequences. Curr. Biol. 2000;10:R412–R415. doi: 10.1016/s0960-9822(00)00507-8. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Q., Liu J., Deng H., Ma R., Liao J.Y., Liang H., Hu J., Li J., Guo Z., Cai J., et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell. 2020;183:76–93.e22. doi: 10.1016/j.cell.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Gong W., Xu J., Wang Y., Min Q., Chen X., Zhang W., Chen J., Zhan Q. Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct. Target. Ther. 2022;7:40. doi: 10.1038/s41392-021-00865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H., Huang S., Wei G., Sun Y., Li C., Si X., Chen Y., Tang Z., Li X., Chen Y., et al. CircRNA Samd4 induces cardiac repair after myocardial infarction by blocking mitochondria-derived ROS output. Mol. Ther. 2022 doi: 10.1016/j.ymthe.2022.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanbe A., Gulick J., Hanks M.C., Liang Q., Osinska H., Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ. Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Wang J., Huang Y., Xiao Y. 3dRNA v2. 0: an updated web server for RNA 3D structure prediction. Int. J. Mol. Sci. 2019;20:4116. doi: 10.3390/ijms20174116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X., Javan H., Li L., Szucsik A., Zhang R., Deng Y., Selzman C.H. A modified murine model for the study of reverse cardiac remodelling. Exp. Clin. Cardiol. 2013;18:e115–e117. [PMC free article] [PubMed] [Google Scholar]
- 46.Du W.W., Xu J., Yang W., Wu N., Li F., Zhou L., Wang S., Li X., He A.T., Du K.Y., et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ. Res. 2021;129:568–582. doi: 10.1161/CIRCRESAHA.120.318364. [DOI] [PubMed] [Google Scholar]
- 47.Xu J., Du W.W., Wu N., Li F., Li X., Xie Y., Wang S., Yang B.B. The circular RNA circNlgnmediates doxorubicin-inducedcardiac remodeling and fibrosis. Mol. Ther. Nucleic Acids. 2022;28:175–189. doi: 10.1016/j.omtn.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A., et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu N., Yuan Z., Du K.Y., Fang L., Lyu J., Zhang C., He A., Eshaghi E., Zeng K., Ma J., et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019;26:2758–2773. doi: 10.1038/s41418-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du W.W., Yang W., Li X., Fang L., Wu N., Li F., Chen Y., He Q., Liu E., Yang Z., et al. The circular RNA circSKA3 binds integrin beta1 to induce invadopodium formation enhancing breast cancer invasion. Mol. Ther. 2020;28:1287–1298. doi: 10.1016/j.ymthe.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Du W.W., Li X., Ma J., Fang L., Wu N., Li F., Dhaliwal P., Yang W., Yee A.J., Yang B.B. Promotion of tumor progression by exosome transmission of circular RNA circSKA3. Mol. Ther. Nucleic Acids. 2022;27:276–292. doi: 10.1016/j.omtn.2021.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 53.Li H., Chang L., Du W.W., Gupta S., Khorshidi A., Sefton M., Yang B.B. Anti-microRNA-378a enhances wound healing process by upregulating integrin beta-3 and vimentin. Mol. Ther. 2014;22:1839–1850. doi: 10.1038/mt.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and material supporting the findings of this study are presented in the paper and/or the supplemental materials. Additional data are available from the corresponding author upon request.