Highlights
-
•
LNP-encapsulated modified MERS-CoV RBD-mRNA presents high stability.
-
•
It induced potent neutralizing antibodies against divergent MERS-CoV strains.
-
•
Intradermal route elicited the highest anti-MERS-CoV neutralizing antibody titer.
-
•
This vaccine completely protected immunized mice against MERS-CoV infection.
Keywords: Coronavirus; MERS-CoV; Spike protein; Receptor-binding domain; mRNA vaccine; Neutralizing antibody, Protection
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV), a highly pathogenic coronavirus in the same Betacoronavirus genus and Coronaviridae family as SARS-CoV-2, continues to post a threat to human health. Mortality remains high; therefore, there is a need to develop effective vaccines to prevent MERS-CoV infection. The receptor-binding domain (RBD) within the MERS-CoV spike (S) protein is a critical vaccine target. The latest mRNA technology has enabled rapid development of much-needed vaccines with high efficiency and scalable manufacturing capacity. Here, we designed a mRNA vaccine encoding the RBD of MERS-CoV S protein (RBD-mRNA) and evaluated its immunogenicity and protective efficacy in a mouse model. The data showed that nucleoside-modified RBD-mRNA, but not RBD-mRNA lacking the nucleoside modification, was stable and elicited broadly and durable neutralizing antibody and cellular immune responses, which neutralized the original strain and multiple MERS-CoV variants. Among all immunization routes tested, the intradermal route was appropriate for this RBD-mRNA to induce strong B-cell responses and the highest neutralizing antibody titers. Importantly, injection of nucleoside-modified RBD-mRNA through the intradermal route protected immunized mice against challenge with MERS-CoV. This protection correlated with serum neutralizing antibody titers. Overall, we have developed an effective MERS-CoV RBD-based mRNA vaccine (with potential for further development) that prevents infection by divergent strains of MERS-CoV.
1. Introduction
A highly pathogenic coronavirus, Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) which causes MERS disease, was first reported in Saudi Arabia in 2012 (Zaki et al., 2012). MERS-CoV infects humans with a mortality rate of approximately 36% (936/2604) (WHO, 2023a), which is much higher than that caused by severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2, two other highly pathogenic human coronaviruses first reported in 2002 and 2019, respectively (Du et al., 2009; WHO, 2023b; Zhou et al., 2020). Unlike SARS-CoV-2, which has high human-to-human transmissibility and caused a global pandemic, MERS-CoV shows limited human-to-human transmissibility, causing sporadic outbreaks in community and healthcare-related settings (Al-Tawfiq and Memish, 2019; Drosten et al., 2014; Elkholy et al., 2020; WHO, 2023b). MERS-CoV is a zoonotic pathogen that infects humans via interaction with intermediate hosts such as dromedary camels or drinking raw camel milk (Azhar et al., 2023; Haagmans et al., 2014; Li and Du, 2019). During the 2022 FIFA World Cup in Qatar (December 2022), at least three French players were infected with MERS-CoV (Chowdhury, 2022; Azhar et al., 2023). A most recent MERS case was reported from Oman to the World Health Organization (WHO) in January 2023 (WHO, 2023c). Although MERS disease remains a significant threat to public health, there are no approved vaccines or therapeutic agents that prevent and/or treat MERS-CoV infection (Du et al., 2016b; Zhang et al., 2020).
The genome of MERS-CoV encodes four major structural proteins, among which the surface spike (S) protein plays a critical role in viral infection and pathogenesis (Du et al., 2017; Zhou et al., 2019). Similar to the S proteins of SARS-CoV-2 and SARS-CoV, the MERS-CoV S protein comprises two subunits: S1 and S2. The virus binds to a cellular receptor through the receptor-binding domain (RBD) of the S1 subunit, followed by fusion between the virus and cell membranes via the S2 subunit, resulting in virus entry into host cells (Chen et al., 2013; Du et al., 2017; Lu et al., 2013). Different from SARS-CoV-2 and SARS-CoV, both of which utilize angiotensin-converting enzyme 2 (ACE2) for virus entry, MERS-CoV uses dipeptidyl peptidase 4 (DPP4) as its cellular receptor (Du et al., 2009; Li et al., 2005; Raj et al., 2013). Therefore, the S protein, particularly the RBD, is a key target for the development of vaccines against MERS-CoV (Du and Jiang, 2015; Du et al., 2016b; Ma et al., 2014; Zhang et al., 2020, 2015).
Most of the current vaccines for MERS are in preclinical development, which are based on recombinant proteins, nanoparticles, DNA, or viral vectors (Du and Jiang, 2015; Du et al., 2016b; Zhang et al., 2020). Unlike these traditional vaccine platforms, mRNA-based vaccines can be produced quickly, at low cost, and on a large-scale; indeed, this technology enabled rapid approval of at least two mRNA vaccines to prevent Coronavirus Disease 2019 (COVID-19) caused by SARS-CoV-2 (U.S. Food and Drug Administration, 2022; Lamb, 2021; Pardi et al., 2018).
Here, we designed two MERS-CoV RBD-based mRNA (RBD-mRNA) vaccines: one with and one without a nucleoside modification. We demonstrated that the RBD-mRNA vaccine with the nucleoside modification elicited a broad neutralizing antibody response that protected mice against challenge with MERS-CoV in a mouse model.
2. Materials and methods
2.1. Construction of MERS-CoV RBD for mRNA synthesis
The RBD of MERS-CoV was constructed as described below (Shi et al., 2022b). Specifically, the DNA sequence of MERS-CoV RBD containing a N-terminal tissue plasminogen activator (tPA) signal peptide and a C-terminal His6 tag was amplified by PCR using a plasmid encoding the codon-optimized sequence of S protein of MERS-CoV (EMC2012 strain, GenBank accession number JX869059.2) as template. The purified PCR product was ligated into a pCAGGS-mCherry vector containing a N-terminal T7 promotor, 5′-untranslated region (UTR), and 3′- UTR for construction of the recombinant plasmid.
2.2. Synthesis of MERS-CoV RBD-mRNA
The RBD-mRNA of MERS-CoV was synthesized as described below (Tai et al., 2020; Wang et al., 2022). Specifically, the recombinant DNA constructed above was synthesized in vitro using MEGAscript T7 Transcription Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The RBD-mRNA-WT (without the nucleoside modification) was synthesized in the presence of ATP, GTP, CTP, and UTP nucleosides, and the RBD-mRNA-PseudoU (with the nucleoside modification) was synthesized in the presence of ATP, GTP, and CTP nucleosides, as well as modified nucleoside pseudouridine-5′-O-triphosphate (Pseudo-UTP; APExBIO). To increase stability and translation efficiency, the synthesized mRNA was capped at the N-terminus using ScriptCap™ Cap 1 Capping System (CELLSCRIPT) and tailed at the C-terminus with a poly(A) using A-Plus™ Poly(A) Polymerase Tailing Kit (CELLSCRIPT).
2.3. Formulation of the synthesized mRNA with lipid nanoparticles
The synthesized RBD-mRNA was encapsulated with lipid nanoparticles (LNPs) as described below (Tai et al., 2020; Wang et al., 2022). Specifically, PNI Formulation Buffer with or without the synthesized mRNA was combined with GenVoy-ILM (lipid mixture) (Precision Nanosystems) at 3:1 ratio using NanoAssemblr Benchtop Instrument (Precision Nanosystems). The LNP-formulated mRNA was concentrated using 10 kDa Amicon Ultra-15 Centrifugal Filters (EMD Millipore) and stored at 4 °C until use. The endotoxin level (<1 EU/ml) of each LNP-encapsulated mRNA was measured using Chromogenic LAL Endotoxin Assay Kit (GenScript), and related particle size was measured using DynaPro NanoStar II Light Scattering Detector (WYATT Technology).
2.4. Detection of protein expression
In vitro expression of MERS-CoV RBD protein in mRNA samples was detected in HEK-293T cells as described below (Tai et al., 2020; Wang et al., 2022). Specifically, cells were plated 24 h prior to the transfection, and the medium was replaced by Opti-MEM 2 h before transfection. Each purified mRNA (4 μg/well) was mixed with TransIT®-mRNA Reagent (Mirus Bio), and the mixture was added to the cells. After culture for 48 h at 37 °C, the cells were collected, fixed, and sequentially stained with MERS-CoV RBD-specific monoclonal antibody (mAb) (Mermab1; 2 μg/ml) and FITC-labeled anti-mouse antibody, followed by analysis by flow cytometry (BD LSR Fortessa 4). The percentage of FITC positive cells was used for evaluation of expression of LNP-encapsulated mRNA.
2.5. Ethical statement
Female BALB/c mice (6–8-week-old) were used in the study. The animal protocols were approved by our Institutional Animal Care and Use Committees (IACUC). The mouse-related experiments were performed according to the Guidelines for the Care and Use of Laboratory Animals of National Institutes of Health, as well as our approved protocols.
2.6. Immunization procedures and sample collection
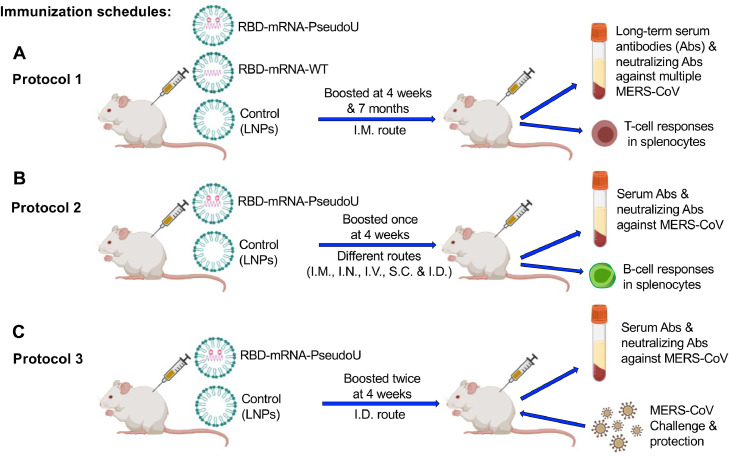
Three different immunizations were performed in mice. First, to identify vaccine-induced long-term immune responses and neutralizing antibodies, mice were immunized with each LNP-packaged RBD-mRNA (30 μg/mouse) with or without the nucleoside modification, or empty-LNP control (without mRNA), through the intramuscular (I.M.) route, and boosted twice at 4 weeks and around 7 months, respectively. Sera were collected before immunization and 10 days after each immunization, as well as monthly after the second immunization, to test specific IgG antibody responses and neutralizing antibodies. Spleens were collected 10 days after the last immunization to test specific T-cell responses. Second, to identify an effective immunization route of nucleoside-modified mRNA and test its immunogenicity at a low dose, mice were immunized with LNP-encapsulated RBD-mRNA-PseudoU (10 μg/mouse) or empty-LNP control through different immunization routes, including I.M., intranasal (I.N.), intravenous (I.V.), subcutaneous (S.C.), and intradermal (I.D.), and boosted once at 4 weeks. Splenocytes collected 10 days after the last immunization were tested for B-cell responses, and sera collected 10 days post-each dose were tested for specific antibody responses and neutralizing antibodies. Third, to evaluate protective efficacy induced by nucleoside-modified mRNA and the correlation between neutralizing antibodies and protective efficacy, mice were immunized with RBD-mRNA-PseudoU (10 μg/mouse) or empty-LNP control through the identified route (I.D.), and boosted twice at 4 weeks, followed by detection of serum neutralizing antibodies and protection against MERS-CoV infection.
2.7. ELISA
Enzyme-linked immunoassay (ELISA) was carried out to detect specific antibodies in mouse sera as described below (Tai et al., 2016a). Specifically, ELISA plates were coated with MERS-CoV S1-His-protein (1 μg/ml) overnight at 4 °C, and blocked with 2% fat-free milk in PBST for 2 h at 37 °C. The plates were washed with PBST, and incubated sequentially with mouse sera at serial dilutions and horseradish peroxidase (HRP)-conjugated anti-mouse IgG (1:5000) (Invitrogen), anti-mouse IgG1 (1:5000) (Thermo Fisher Scientific), anti-mouse IgG2a (1:5000) (Thermo Fisher Scientific), and anti-mouse IgM (1:4000) (Invitrogen) antibodies, respectively, for 1 h at 37 °C. This step was followed by sequential incubation of the plates with TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Sigma) and termination reagent (H2SO4, 1 N) to stop the reaction. Absorbance (at 450 nm) was measured by an ELISA plate reader (Tecan).
2.8. Generation of MERS-CoV pseudoviruses and pseudovirus neutralization assay
Pseudoviruses generation and subsequent pseudovirus neutralization assay were described below (He et al., 2019; Zhao et al., 2018). Specifically, a plasmid respectively expressing the S protein of wild-type or each mutant MERS-CoV and a plasmid encoding an Env-defective, luciferase-expressing HIV-1 genome (pNL4–3.luc.RE) were co-transfected into 293T cells using calcium phosphate transfection method. The medium was changed into fresh Dulbecco's Modified Eagle Medium (DMEM) containing 10% FBS 6–8 h later. Packaged pseudoviruses were collected from culture supernatants 72 h after transfection, and incubated with mouse sera at serial dilutions for 1 h at 37 °C. The serum-virus mixture was added to Huh-7 cells, which were cultured at 37 °C with 5% CO2, and fresh medium was added 24 h later. The cells were lysed in cell lysis buffer (Promega) 72 h after virus infection, incubated with luciferase substrate (Promega), and detected for relative luciferase activity using Infinite 200 PRO Luminator (Tecan). The serum neutralizing activity was reported as 50% pseudovirus neutralizing antibody titer (NT50).
2.9. Detection of cellular immune responses
B-cell responses and antigen-specific T-cell responses were determined as described below (Tai et al., 2020). For T-cell responses, splenocytes (1.0 × 106) were isolated from homogenized spleens of immunized or control mice, and stimulated with overlapping peptides covering the RBD of MERS-CoV (Genscript) for 3 days at 37 °C. Brefeldin A (BioLegend) was added to cells 6 h before staining. After stimulation, the cells were stained with Fixable Viability Dye eFluor 780 (Thermo Fisher Scientific) to distinguish live and dead cells. Cell surface markers were stained with FITC anti-mouse-CD4 and PerCP-Cy5.5 anti-mouse-CD8 antibodies (BioLegend). For intracellular cytokine staining, the cells were fixed and permed using Cytofix/Cytoperm reagent (BD Biosciences), and stained with BV 605 anti-mouse TNF-α, PE anti-mouse IFN-γ, and BV 711 anti-mouse IL-4 antibodies (BioLegend). For B-cell responses, isolated splenocytes were stained with Fixable Viability Dye eFluor 780 to distinguish live and dead cells as described above. The B-cells were then stained with PerCP-Cy5.5 anti-mouse-B220, BV421 anti-mouse-CD27, PE anti-mouse-CD138, and FITC anti-mouse-IgG antibodies (BioLegend). The stained cells were collected using BD LSRFortessa 4 Flow Cytometer. Flow cytometry data were analyzed using FlowJo Software (Tree Star Inc).
2.10. Challenge of mice with MERS-CoV
MERS-CoV challenge and subsequent sample collection were performed as described below (Channappanavar et al., 2015). Specifically, one month after last immunization, BALB/c mice were anesthetized with isoflurane, and intranasally transduced with 2.5 × 108 focus-forming unit (FFU) of adenovirus 5-human DPP4 (Ad5/hDPP4) in 75 μl of DMEM (Zhao et al., 2014). Five days post-transduction, mice were intranasally infected with authentic MERS-CoV (EMC2012 strain; 5 × 103 plaque-forming unit (PFU), a dose that caused efficient infection and lung viral replication in our preliminary studies) in 50 μl of DMEM and monitored daily. Mouse lungs were collected three days after infection, and measured for viral titer as described below. All work with authentic MERS-CoV was conducted in the Biosafety Level 3 Laboratories.
2.11. MERS-CoV plaque assay
Lung homogenate supernatants were serially diluted in DMEM, and inoculated into Vero 81 cells pre-plated in 12-well plates, which were cultured for 1 h at 37 °C and 5% CO2 with gentle rocking every 15 min. After removing the inocula, the plates were overlaid with 0.6% agarose containing 2% FBS. The agarose overlays were removed after a three-day incubation, and plaques were visualized after staining with 0.1% crystal violet (Tai et al., 2016a). Viral titer was calculated as PFU/g of tissues.
2.12. Statistical analysis
Statistical significance among different groups was calculated using GraphPad Prism statistical software. Statistical significance of neutralizing antibody responses or viral titer between RBD-mRNA-PseudoU and other groups was calculated by two-tailed Student's t-test. Antibody and neutralizing antibody responses via various immunization routes, as well as T-cell and B-cell responses, among different groups were calculated using Ordinary one-way ANOVA. Asterisks in figures indicate statistical significance among indicated groups (*, **, and *** denote P < 0.05, P < 0.01, and P < 0.001, respectively).
3. Results
3.1. Rational design and characterization of RBD-mRNA of MERS-CoV
The RBD-mRNA comprised the codon-optimized RBD of the original MERS-CoV strain, a N-terminal tPA signal peptide, and a C-terminal His6 tag. Two forms of the RBD-mRNA were synthesized: 1) RBD-mRNA-wild-type (WT) lacking a nucleoside modification, which contains ATP, GTP, CTP, and UTP, and 2) RBD-mRNA-PseudoU harboring the nucleoside modification, which contains ATP, GTP, CTP, and Pseudo-UTP (100% of the UTP was replaced with Pseudo-UTP during mRNA synthesis). Each RBD-mRNA contained a 5′-Cap, a 5′-UTR, a 3′-UTR, and a poly(A) tail, and was encapsulated by LNPs to form mRNA-LNPs for delivery (Fig. 1A). As expected, the size of the LNP-encapsulated RBD-mRNA-PseudoU particles at 25 and 37 °C was similar to that at 4 °C when stored for a period of 72 h. By contrast, the size of the RBD-mRNA-WT particles at 25 and 37 °C was different from that at 4 °C, particularly when stored for 72 h (Fig. 1B). LNP-formulated RBD-mRNA-PseudoU maintained strong integrity without obvious degradation after 72 h of storage at 4 and 25 °C, whereas significant degradation was shown in LNP-encapsulated RBD-mRNA-WT when stored at 4, 25 and 37 °C for 72 h (Fig. S1). Flow cytometry analysis revealed stronger fluorescent signals in cells incubated with RBD-mRNA-PseudoU than in cells incubated with RBD-mRNA-WT, whereas cells incubated with the empty-LNP control generated no fluorescent signals (Fig. 1C). These data demonstrate that the nucleoside-modified RBD-mRNA is more stable than the RBD-mRNA lacking the nucleoside modification, and that nucleoside-modified RBD-mRNA is expressed at higher levels than the RBD-mRNA without the nucleoside modification.
Fig. 1.
Design and characterization of MERS-CoV RBD-mRNA. (A) Schematic map of MERS-CoV spike (S) protein and design of MERS-CoV RBD-mRNA vaccines. Each RBD-mRNA consists of genes encoding signal peptide (tPA) and open reading frame of RBD flanked by the 5′ and 3′ untranslated regions (UTRs), a 5′ cap and a 3′ poly(A) tail. RBD-mRNA-wild-type (WT) was synthesized in the presence of nucleosides (UTP, CTP, ATP, and GTP), and RBD-mRNA-PseudoU was constructed by changing the UTP to modified Pseudo-UTP during mRNA synthesis. SP, signal peptide. NTD, N-terminal domain. RBD, receptor-binding domain. TM, transmembrane; CT, cytoplasmic tail. The synthesized mRNA was encapsulated with lipid nanoparticles (LNPs) and used for the subsequent experiments. (B) Characterization of the stability of each RBD-mRNA or empty-LNP (control) at different temperatures (4 °C, 25 °C, and 37 °C). Each LNP-encapsulated RBD-mRNA or LNP control was stored for 0, 24, 48, and 72 h, respectively, at the above temperatures, and the particle size (hydrodynamic diameter: nm) was measured using a Dynamic Light Scattering. (C) Representative images of flow cytometry for expression of MERS-CoV RBD protein. 293T cells transfected with each RBD-mRNA were tested for expression of RBD protein using a MERS-CoV RBD-specific monoclonal antibody (mAb, Mersmab1). 293T cells transfected with the empty-LNP were used as control. MFI, median fluorescence intensity. The data are presented as mean ± standard deviation of the mean (s.e.m.) of triple wells.
3.2. Nucleoside-modified RBD-mRNA elicited potent, broadly, and durable neutralizing antibodies
Immunization schedules were summarized in Fig. 2. To evaluate the ability of MERS-CoV RBD-mRNA with or without the nucleoside modification to elicit specific humoral immune responses, mice were immunized I.M. with each RBD-mRNA (30 μg/mouse) and then boosted twice (at 4 weeks and around 7 months). Serum samples were collected at different times after immunization, and levels of IgG antibodies specific for the S protein and neutralizing antibodies against pseudotyped MERS-CoV encoding the S protein of multiple strains were measured (Fig. 2A). Mice receiving RBD-mRNA-PseudoU generated high-titer MERS-CoV S1-specific IgG antibodies (Fig. 3A) and neutralizing antibodies against the original strain of MERS-CoV (EMC2012) (Fig. 3B), which were maintained for at least 6 months after the second immunization. Mice receiving RBD-mRNA-WT generated much lower IgG and neutralizing antibody titers (Fig. 3A and B). Notably, RBD-mRNA-PseudoU-induced antibodies neutralized multiple strains of pseudotyped MERS-CoV much more efficiently than those induced by RBD-mRNA-WT (Fig. 3C). By contrast, only background levels of IgG antibodies and neutralizing antibodies were detected in mice injected with the LNP control (Fig. 3). These data indicate that nucleoside-modified RBD-mRNA, rather than RBD-mRNA without the nucleoside modification, elicited a highly potent and durable neutralizing antibody response, with broad-spectrum activity against different MERS-CoV strains.
Fig. 2.
Immunization and challenge schedules. Mice were immunized with MERS-CoV RBD-mRNA-PseudoU, RBD-mRNA-WT, or empty-LNP (control) according to the three procedures described in Protocols 1–3 (A-C). Sera were collected to detect specific IgG antibodies (Abs), subtype (IgG1 and IgG2a) Abs, or neutralizing Abs against single or multiple strains of pseudotyped MERS-CoV. Splenocytes were collected to test specific T-cell or B-cell responses. Immunized mice were also challenged with MERS-CoV to evaluate protective efficacy of RBD-mRNA-PseudoU vaccine. I.M., intramuscular; I.N., intranasal; I.V., intravenous; S.C., subcutaneous; I.D., intradermal.
Fig. 3.
Nucleoside-modified RBD-mRNA induced potent and durable antibody responses with broadly neutralizing activity. Mice were immunized I.M. with LNP-encapsulated RBD-mRNA with or without the nucleoside modification (30 μg/mouse), or empty-LNP control, boosted twice at 4 weeks and around 7 months, respectively, and sera were collected for the tests described below. (A) Mouse sera from different time points were detected for IgG antibodies (Abs) by ELISA. ELISA plates were coated with MERS-CoV S1 protein, and the Ab titer was determined based on the endpoint serum dilution that remained positively detectable. (B) Mouse sera from different time points were measured for neutralizing Abs against infection of pseudotyped MERS-CoV encoding S protein of the original virus strain (EMC2012). (C) Mouse sera from the last immunization were measured for cross-neutralizing antibodies against multiple variants of pseudotyped MERS-CoV-2. NT50 was calculated as 50% neutralizing Ab titer against infection of the respective MERS-CoV pseudovirus in Huh-7 cells. The data are presented as mean ± s.e.m. of duplicate wells from five mice in each group. *** (P < 0.001) indicates significant difference between RBD-mRNA-PseudoU and other groups. The experiments were repeated twice, resulting in similar results.
3.3. Nucleoside-modified RBD-mRNA elicited specific T-cell responses
To evaluate the ability of MERS-CoV RBD-mRNA with or without the nucleoside modification to elicit specific T-cell responses, mice were immunized I.M. with each RBD-mRNA (30 μg/mouse) and boosted twice (at 4 weeks and around 7 months) prior to measurement of IFN-γ, TNF-α, and IL-4 cytokines secreted by CD4+ and CD8+ T cells within the splenocyte population collected after the final immunization (Fig. 2A). RBD-mRNA-PseudoU induced both CD4+ (Fig. 4A) and CD8+ (Fig. 4B) T-cell responses, resulting in secretion of MERS-CoV RBD-specific IFN-γ, TNF-α, and IL-4. The levels of these cytokines induced by RBD-mRNA-WT were significantly lower than those induced by RBD-mRNA-PseudoU (Fig. 4A-B). By contrast, the empty-LNP control elicited only background-level CD4+ and CD8+ T-cell responses (Fig. 4). These data demonstrate that nucleoside-modified RBD-mRNA elicited stronger T-cell responses than unmodified RBD-mRNA in mice.
Fig. 4.
Nucleoside-modified RBD-mRNA induced effective T-cell responses. Mice were immunized I.M. with LNP-encapsulated RBD-mRNA with or without the nucleoside modification (30 μg/mouse), or empty-LNP control, and boosted twice at 4 weeks and around 7 months, respectively. Splenocytes from the last immunization were analyzed for SARS-CoV-2 RBD-specific T-cell responses by flow cytometry. IFN-γ, TNF-α, and IL-4-secreting CD4+ (A) or CD8+ (B) T cells were stained for each cell surface marker or cytokine. The data are presented as mean ± s.e.m. of duplicate wells from five mice in each group. * (P < 0.05), ** (P < 0.01), and *** (P < 0.001) indicate significant difference between RBD-mRNA-PseudoU and other groups. The experiments were repeated twice, resulting in similar results.
3.4. An effective route and dosage for nucleoside-modified RBD-mRNA to elicit strong antibody and B-cell responses, as well as high-titer neutralizing antibodies
Next, we asked whether nucleoside-modified RBD-mRNA elicited effective neutralizing antibody responses when used at a low dose; we also identified an effective route for immunization to elicit strong B-cell responses and neutralizing antibodies. Mice were immunized with RBD-mRNA-PseudoU (10 μg/mouse) via different routes and then boosted once at 4 weeks with the same dose. We then assessed B-cell responses in splenocytes and measured serum IgG and IgM antibodies, subtype antibodies (IgG1 and IgG2a) and neutralizing antibodies against the original MERS-CoV (EMC2012) strain (Fig. 2B). We found that RBD-mRNA-PseudoU through the I.D. route significantly increased the production of B cells than other routes in the immunized splenocytes (Fig. 5). In addition, a single dose of RBD-mRNA-PseudoU was unable to induce effective IgG1 and IgG2a antibodies specific for the MERS-CoV-2 S1 protein, or relatively low titers of IgG and neutralizing antibodies against pseudotyped MERS-CoV (Fig. 6A–D); however, the boost dose caused a marked increase in production of these antibodies, regardless of the route of administration (Fig. 6E–H). Overall, the I.D. route of this RBD-mRNA elicited the highest titers of IgG, IgG1, IgG2a, and neutralizing antibodies, followed by the I.M. route (Fig. 6E–H). Notably, the I.D. route elicited higher, or significantly higher, IgG, IgG1 and IgG2a antibody levels and anti-MERS-CoV pseudovirus neutralizing antibody titers than the I.M., I.N., I.V. and S.C. routes (Fig. 6E–H). Notably, I.D. injection of RBD-mRNA-PseudoU induced a higher level of S1-specific IgM antibodies after the first dose than the boost dose (Fig. 6I and J). By contrast, the empty-LNP control induced only background IgG, IgM, IgG1 and IgG2a antibody responses, as well as neutralizing antibody and B-cell responses (Figs. 5 and 6). These data indicate that a low dose of nucleoside-modified RBD-mRNA delivered via the I.D. route elicited strong antibody and B-cell responses, as well as high-titer neutralizing antibodies against MERS-CoV.
Fig. 5.
An effective immunization route and antigen dose for nucleoside-modified RBD-mRNA to induce strong B-cell responses. Mice were immunized with LNP-encapsulated nucleoside modified RBD-mRNA (10 μg/mouse) or empty-LNP control at different routes, and boosted at 4 weeks. Splenocytes from the last immunization were analyzed for B-cell responses by flow cytometry. The cells were stained for B220+-CD27+-CD138+-IgG+ cells. The data are presented as mean ± s.e.m. of duplicate wells from five mice in each group. ** (P < 0.01) indicates significant difference between RBD-mRNA-PseudoU and other groups. The experiments were repeated twice, resulting in similar results.
Fig. 6.
An effective immunization route and antigen dose for nucleoside-modified RBD-mRNA to induce potent antibodies with neutralizing activity. Mice were immunized with LNP-encapsulated nucleoside modified RBD-mRNA (10 μg/mouse) or empty-LNP control at different routes, including I.M., I.N., I.V., S.C., and I.D., and boosted as described in Fig. 5. Sera from the first and second immunizations were detected for antibodies (Abs) and neutralizing Abs against MERS-CoV. MERS-CoV S1-specific IgG (A), IgG1 (B), IgG2a (C) Abs, and anti-MERS-CoV neutralizing Abs (D) 10 days after the first immunization, as well as MERS-CoV S1-specific IgG (E), IgG1 (F), IgG2a (G) Abs, and anti-MERS-CoV neutralizing Abs (H) 10 days after the second immunization, were evaluated by ELISA and pseudovirus neutralization assays, respectively. MERS-CoV S1-specific IgM Abs were tested by ELISA in the sera of I.D.-immunized mice from the 1st and 2nd doses (I-J). ELISA plates were coated with MERS-CoV S1 protein, and the Ab titer was determined based on the endpoint serum dilution that remained positively detectable. Neutralizing antibody activity was reported as 50% neutralizing Ab titer (NT50) against infection of pseudotyped MERS-CoV (EMC2012 strain) in Huh-7 cells. The data are presented as mean ± s.e.m. of duplicate wells from five mice in each group. ** (P < 0.01) and *** (P < 0.001) indicate significant difference of RBD-mRNA-PseudoU injected via the I.D. route and other routes. The experiments were repeated twice, resulting in similar results.
3.5. Nucleoside-modified RBD-mRNA completely protected mice from MERS-CoV challenge
Finally, we further evaluated the ability of nucleoside-modified RBD-mRNA, delivered at a low dose via the I.D. route, to protect mice against challenge with MERS-CoV. BALB/c mice were I.D. immunized with RBD-mRNA-PseudoU (10 μg/mouse) and boosted twice with the same dose at 4-week intervals. Serum was collected, and antibody responses and neutralizing antibody titers against the original MERS-CoV (EMC2012 stain) were measured. Then, mice were challenged with MERS-CoV (EMC2012 strain) after transduction with Ad5/hDPP4, and viral titers were measured in the lungs. Compared with the first boost (Fig. 6), the second boost of RBD-mRNA-PseudoU induced higher levels of IgG antibodies and a similar level of neutralizing antibodies against infection of pseudotyped MERS-CoV (Fig. 7A and B). The data from challenge studies showed that no virus was detected in the lungs of mice immunized with RBD-mRNA-PseudoU, which generated high-titer anti-MERS-CoV neutralizing antibodies (Fig. 7B and C). However, mice receiving the empty-LNP control had high titers of MERS-CoV and no detectable neutralizing antibodies (Fig. 7B and C). These data indicate that nucleoside-modified RBD-mRNA via the I.D. route completely protected mice from MERS-CoV challenge, and that the level of protection correlated with the neutralizing antibody titer.
Fig. 7.
Nucleoside-modified RBD-mRNA protected mice from MERS-CoV infection and the protection was correlated with neutralizing antibodies. Mice were immunized I.D. with LNP-encapsulated nucleoside modified RBD-mRNA (10 μg/mouse) or empty-LNP control, and boosted twice at 4 weeks. Sera collected 10 days after the last immunization were tested for MERS-CoV S1-specific IgG antibodies (Abs) by ELISA (A) and neutralizing Abs (NT50) against infection of pseudotyped MERS-CoV (EMC2012 strain) in Huh-7 cells (B). One month after the last immunization, mice were transduced with Ad5/hDPP4, and then challenged with MERS-CoV (EMC2012 strain) (5 × 103 PFU/mouse). Lungs were collected three days after challenge, and viral titer was detected from the challenged mice, which are reported as PFU/g of tissues (C). The data are presented as mean ± s.e.m. of five mice in each group. *** (P < 0.001) indicates significant difference between RBD-mRNA-PseudoU and the control group. The experiments were repeated twice, resulting in similar results.
4. Discussion and conclusions
mRNA vaccines have advantages over traditional vaccines. Unlike DNA, mRNA does not integrate into the cell genome and cause mutations, and unlike viral vectors, mRNA does not induce anti-vector immune responses (Du et al., 2022). Indeed, mRNA has a good safety profile since it does not contain any infectious components, and its stability can be improved by encapsulating it in carrier molecules such as LNPs (Du et al., 2022; Uddin and Roni, 2021). However, mRNA-based vaccines still present some challenges. For example, unmodified mRNA is recognized by germline-encoded pattern-recognition receptors such as retinoic acid–inducible gene-I-like receptors, 2′−5′-oligoadenylate synthetase, and RNA-dependent protein kinase, resulting in activation of the innate immune system and unwanted immune responses, or secretion of pro-inflammatory cytokines or chemokines (Chen and Xu, 2022; Pardi et al., 2018). Activation of innate immune sensors can be abrogated by incorporating naturally occurring, chemically modified nucleosides such as pseudouridine, 2-thiouridine, 5-methylcytidine, and 5-methoxyuridine, during the mRNA synthesis (Granados-Riveron and Aquino-Jarquin, 2021; Nance and Meier, 2021). Indeed, we showed that unmodified MERS-CoV RBD-mRNA triggers production of IL-6, IFN-α, and TNF-α, whereas replacement of the nucleoside UTP in the RBD-mRNA by 100% pseudouridine abrogated cytokine production significantly (Tai et al., 2021).
Because MERS-CoV continues to infect humans with a high fatality rate, we need to develop effective vaccines to prevent infection and reduce threats to global health. The S protein, including its RBD, is an important target for development of effective vaccines against MERS-CoV and other highly pathogenic coronaviruses such as SARS-CoV-2 and SARS-CoV (Du et al., 2009, 2016a; Tai et al., 2020; Zhou et al., 2019). SARS-CoV-2 rapidly accumulates mutations in the S protein, especially the RBD, resulting in emergence of multiple variants of concern (WHO, 2023d). The RBD or S protein of SARS-CoV-2 variants contains more than 15 or 30 amino acid mutations when compared with the original RBD or S protein. Accordingly, vaccines based on the RBD or S protein of the original SARS-CoV-2 strain may induce neutralizing antibodies with reduced titers against variants, particularly Omicron variant and its subvariants, or vice versa (Lyke et al., 2022; Shi et al., 2022a; Sievers et al., 2022). Different from SARS-CoV-2, MERS-CoV harbors fewer mutations being identified in the S protein, including the RBD. Therefore, vaccines based on the original RBD of MERS-CoV still elicit broad and potent neutralizing antibodies against MERS-CoV mutant strains (Tai et al., 2016a). Thus, it is feasible to design effective MERS vaccines that target the original RBD yet still induce broad-spectrum neutralizing activity against various MERS-CoV mutants.
Despite the number of MERS vaccines based on proteins, viral vectors, and DNAs, few mRNA-based MERS vaccines have been developed (Tai et al., 2021; Zhang et al., 2020). Here, we designed a MERS-CoV mRNA vaccine (RBD-mRNA) with or without a nucleoside modification in the mRNA component, which was based on the original RBD sequence, and then compared the ability of both vaccines to induce cellular immune responses and neutralizing antibodies against various MERS-CoV strains. We also identified an effective immunization route and antigen dosage required to elicit high neutralizing antibody titers and demonstrated protective efficacy against MERS-CoV infection.
Unlike the RBD-mRNA without the nucleoside modification (RBD-mRNA-WT), MERS-CoV RBD-mRNA with the Pseudo-UTP modification elicited long-term IgG antibodies with potent neutralizing activity and T-cell immune responses (lasting for at least 6 months). Part of the reasons for RBD-mRNA-WT in failing to induce effective immune responses might be due to its instability which potentially reduces its potency. No obvious adverse effects were noted during the injection and observation periods of the mRNA vaccine. Similar to neutralizing antibodies induced by MERS-CoV RBD proteins, neutralizing antibodies induced by the nucleoside-modified RBD-mRNA maintained broadly neutralizing activity and potency against the wild-type strain, as well as at least ten variant MERS-CoV strains. Different from RBD-based MERS-CoV subunit vaccines that generate optimal responses when delivered via S.C. or I.M. injection (Ma et al., 2014; Nyon et al., 2018; Tai et al., 2016b), we found that MERS-CoV RBD-mRNA-PseudoU delivered via the I.D. route elicited the highest neutralizing antibody titer. Of note, one or two low-dose boosts generated effective neutralizing antibody titers that protected immunized mice from MERS-CoV infection, and the level of protection correlated positively with the serum neutralizing antibody titer. Contrary to IgG antibodies, which showed an increased trend after boosts, there was a relatively higher level of IgM antibodies after the first dose than the boost. MERS-CoV-specific IgA antibodies were not detected in the immunized mouse sera (data not shown). Future studies will identify mucosal immunity of RBD-mRNA-PseudoU via various immunization routes, and compare the induced secretory IgA antibodies in the bronchoalveolar lavage. In this study, protection of the nucleoside-modified RBD-mRNA vaccine was evaluated in Ad5/hDPP4-transduced mice, with undetectable viral titers in the lungs. In the future, hDPP4-knock-in mice will be infected with mouse adapted MERS-CoV to evaluate protective efficacy of the vaccine against severe disease.
Overall, our data show that a nucleoside-modified mRNA encoding the original RBD of MERS-CoV (RBD-mRNA), but not an RBD-mRNA lacking the nucleoside modification, induced durable and potent neutralizing antibody responses that protected against MERS-CoV infection. Importantly, the vaccine elicited neutralizing antibodies with broad-spectrum neutralizing activity against multiple MERS-CoV strains. In conclusion, this RBD-mRNA vaccine has great potential for further development and use as a safe and effective method of preventing infection by diverse MERS-CoV strains as well as MERS-related coronaviruses that may emerge in the future.
Data availability
All data generated in this study are included in the published article.
CRediT authorship contribution statement
Wanbo Tai: Methodology, Data curation, Formal analysis, Validation. Jian Zheng: Data curation, Formal analysis, Writing – review & editing. Xiujuan Zhang: Data curation. Juan Shi: Methodology, Data curation. Gang Wang: Methodology, Data curation. Xiaoqing Guan: Methodology, Data curation. Jiang Zhu: Writing – review & editing. Stanley Perlman: Investigation, Writing – review & editing. Lanying Du: Conceptualization, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by NIH grants (R01AI157975, R01AI139092 and R01AI137472).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199156.
Contributor Information
Stanley Perlman, Email: stanley-perlman@uiowa.edu.
Lanying Du, Email: ldu3@gsu.edu.
Appendix. Supplementary materials
References
- Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus in the last two years: health care workers still at risk. Am. J. Infect. Control. 2019;47(10):1167–1170. doi: 10.1016/j.ajic.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhar E.I., Hui D.S., McCloskey B., El-Kafrawy S.A., Sharma A., Maeurer M., Lee S.S., Zumla A. The Qatar FIFA World Cup 2022 and camel pageant championships increase risk of MERS-CoV transmission and global spread. Lancet Glob. Health. 2023;11(2):e189–e190. doi: 10.1016/S2214-109X(22)00543-5. [DOI] [PubMed] [Google Scholar]
- Channappanavar R., Lu L., Xia S., Du L., Meyerholz D.K., Perlman S., Jiang S. Protective effect of intranasal regimens containing peptidic Middle East respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J. Infect. Dis. 2015;212(12):1894–1903. doi: 10.1093/infdis/jiv325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Xu Q. Current developments and challenges of mRNA vaccines. Annu. Rev. Biomed. Eng. 2022;24:85–109. doi: 10.1146/annurev-bioeng-110220-031722. [DOI] [PubMed] [Google Scholar]
- Chen Y., Rajashankar K.R., Yang Y., Agnihothram S.S., Liu C., Lin Y.L., Baric R.S., Li F. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J. Virol. 2013;87(19):10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury I.S., 2022. French players catch Camel flu before final match. Observer Online Report. [Cited Dec. 17, 2022]. Available from: https://www.observerbd.com/news.php?id=397883.
- Drosten C., Meyer B., Müller M.A., Corman V.M., Al-Masri M., Hossain R., Madani H., Sieberg A., Bosch B.J., Lattwein E., Alhakeem R.F., Assiri A.M., Hajomar W., Albarrak A.M., Al-Tawfiq J.A., Zumla A.I., Memish Z.A. Transmission of MERS-coronavirus in household contacts. N. Engl. J. Med. 2014;371(9):828–835. doi: 10.1056/NEJMoa1405858. [DOI] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV–a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Jiang S. Middle East respiratory syndrome: current status and future prospects for vaccine development. Expert Opin. Biol. Ther. 2015;15(11):1647–1651. doi: 10.1517/14712598.2015.1092518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Tai W., Yang Y., Zhao G., Zhu Q., Sun S., Liu C., Tao X., Tseng C.K., Perlman S., Jiang S., Zhou Y., Li F. Introduction of neutralizing immunogenicity index to the rational design of MERS coronavirus subunit vaccines. Nat. Commun. 2016;7:13473. doi: 10.1038/ncomms13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Tai W., Zhou Y., Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev. Vaccines. 2016;15(9):1123–1134. doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yang Y., Zhang X., Li F. Recent advances in nanotechnology-based COVID-19 vaccines and therapeutic antibodies. Nanoscale. 2022;14(4):1054–1074. doi: 10.1039/d1nr03831a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Yang Y., Zhou Y., Lu L., Li F., Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin. Ther. Targets. 2017;21(2):131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkholy A.A., Grant R., Assiri A., Elhakim M., Malik M.R., Van Kerkhove M.D. MERS-CoV infection among healthcare workers and risk factors for death: retrospective analysis of all laboratory-confirmed cases reported to WHO from 2012 to 2 June 2018. J. Infect. Public Health. 2020;13(3):418–422. doi: 10.1016/j.jiph.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron J.T., Aquino-Jarquin G. Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed. Pharmacother. 2021;142:111953. doi: 10.1016/j.biopha.2021.111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R., Godeke G.J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D., AlHajri M.M., Koopmans M.P. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Tai W., Li J., Chen Y., Gao Y., Li J., Sun S., Zhou Y., Du L., Zhao G. Enhanced ability of oligomeric nanobodies targeting MERS coronavirus receptor-binding domain. Viruses. 2019;11(2):166. doi: 10.3390/v11020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb Y.N. BNT162b2 mRNA COVID-19 vaccine: first approval. Drugs. 2021;81(4):495–501. doi: 10.1007/s40265-021-01480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Du L. MERS coronavirus: an emerging zoonotic virus. Viruses. 2019;11(7):663. doi: 10.3390/v11070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Lu G., Hu Y., Wang Q., Qi J., Gao F., Li Y., Zhang Y., Zhang W., Yuan Y., Bao J., Zhang B., Shi Y., Yan J., Gao G.F. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyke, K.E., Atmar, R.L., Islas, C.D., Posavad, C.M., Szydlo, D., Paul Chourdhury, R., Deming, M.E., Eaton, A., Jackson, L.A., Branche, A.R., El Sahly, H.M., Rostad, C.A., Martin, J.M., Johnston, C., Rupp, R.E., Mulligan, M.J., Brady, R.C., Frenck, R.W., Bäcker, M., Kottkamp, A.C., Babu, T.M., Rajakumar, K., Edupuganti, S., Dobrzynski, D., Coler, R.N., Archer, J.I., Crandon, S., Zemanek, J.A., Brown, E.R., Neuzil, K.M., Stephens, D.S., Post, D.J., Nayak, S.U., Suthar, M.S., Roberts, P.C., Beigel, J.H., Montefiori, D.C., Group, D.S., 2022. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant. Cell Rep. Med. 3 (7), 100679. doi: 10.1016/j.xcrm.2022.100679. [DOI] [PMC free article] [PubMed]
- Ma C., Wang L., Tao X., Zhang N., Yang Y., Tseng C.K., Li F., Zhou Y., Jiang S., Du L. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments–the importance of immunofocusing in subunit vaccine design. Vaccine. 2014;32(46):6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance K.D., Meier J.L. Modifications in an emergency: the role of N1-Methylpseudouridine in COVID-19 vaccines. ACS Cent. Sci. 2021;7(5):748–756. doi: 10.1021/acscentsci.1c00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyon M.P., Du L., Tseng C.K., Seid C.A., Pollet J., Naceanceno K.S., Agrawal A., Algaissi A., Peng B.H., Tai W., Jiang S., Bottazzi M.E., Strych U., Hotez P.J. Engineering a stable CHO cell line for the expression of a MERS-coronavirus vaccine antigen. Vaccine. 2018;36(14):1853–1862. doi: 10.1016/j.vaccine.2018.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., Thiel V., Drosten C., Rottier P.J., Osterhaus A.D., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang G., Zheng J., Verma A.K., Guan X., Malisheni M.M., Geng Q., Li F., Perlman S., Du L. Effective vaccination strategy using SARS-CoV-2 spike cocktail against Omicron and other variants of concern. NPJ Vaccines. 2022;7(1):169. doi: 10.1038/s41541-022-00580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zheng J., Zhang X., Tai W., Odle A.E., Perlman S., Du L. RBD-mRNA vaccine induces broadly neutralizing antibodies against Omicron and multiple other variants and protects mice from SARS-CoV-2 challenge. Transl. Res. 2022;248:11–21. doi: 10.1016/j.trsl.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers B.L., Chakraborty S., Xue Y., Gelbart T., Gonzalez J.C., Cassidy A.G., Golan Y., Prahl M., Gaw S.L., Arunachalam P.S., Blish C.A., Boyd S.D., Davis M.M., Jagannathan P., Nadeau K.C., Pulendran B., Singh U., Scheuermann R.H., Frieman M.B., Vashee S., Wang T.T., Tan G.S. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 2022;14(634):eabn7842. doi: 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Wang Y., Fett C.A., Zhao G., Li F., Perlman S., Jiang S., Zhou Y., Du L. Recombinant receptor-binding domains of multiple Middle East respiratory syndrome coronaviruses (MERS-CoVs) induce cross-neutralizing antibodies against divergent human and camel MERS-CoVs and antibody escape mutants. J. Virol. 2016;91(1):e01651-16. doi: 10.1128/JVI.01651-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhang X., Drelich A., Shi J., Hsu J.C., Luchsinger L., Hillyer C.D., Tseng C.K., Jiang S., Du L. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res. 2020;30(10):932–935. doi: 10.1038/s41422-020-0387-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhang X., Yang Y., Zhu J., Du L. Advances in mRNA and other vaccines against MERS-CoV. Transl. Res. 2021;242:20–37. doi: 10.1016/j.trsl.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai W., Zhao G., Sun S., Guo Y., Wang Y., Tao X., Tseng C.K., Li F., Jiang S., Du L., Zhou Y. A recombinant receptor-binding domain of MERS-CoV in trimeric form protects human dipeptidyl peptidase 4 (hDPP4) transgenic mice from MERS-CoV infection. Virology. 2016;499:375–382. doi: 10.1016/j.virol.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M.N., Roni M.A. Challenges of storage and stability of mRNA-based COVID-19 vaccines. Vaccines (Basel). 2021;9(9):1033. doi: 10.3390/vaccines9091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration, 2022. Coronavirus (COVID-19) update: FDA takes key action by approving second COVID-19 vaccine. FDA News Release. [Cited Jan. 31, 2022]. Available from: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-takes-key-action-approving-second-covid-19-vaccine.
- Wang G., Shi J., Verma A.K., Guan X., Perlman S., Du L. mRNA vaccines elicit potent neutralization against multiple SARS-CoV-2 omicron subvariants and other variants of concern. iScience. 2022;25(12):105690. doi: 10.1016/j.isci.2022.105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2023a. MERS situation update. [Cited May 2023]. Available from: https://applications.emro.who.int/docs/WHOEMCSR662E-eng.pdf?ua=1.
- World Health Organization, 2023b. WHO coronavirus (COVID-19) dashboard. [Cited Jun. 14, 2023]. Available from: https://covid19.who.int/.
- World Health Organization, 2023c. Middle East Respiratory Syndrome - Oman. [Cited Feb. 08, 2023]. Available from: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON436.
- World Health Organization, 2023d Tracking SARS-CoV-2 variants. [Cited Jun. 05, 2023]. Available from: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang N., Shang J., Li C., Zhou K., Du L. An overview of Middle East respiratory syndrome coronavirus vaccines in preclinical studies. Expert Rev. Vaccines. 2020;19(9):817–829. doi: 10.1080/14760584.2020.1813574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Tang J., Lu L., Jiang S., Du L. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res. 2015;202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., He L., Sun S., Qiu H., Tai W., Chen J., Li J., Chen Y., Guo Y., Wang Y., Shang J., Ji K., Fan R., Du E., Jiang S., Li F., Du L., Zhou Y. A novel nanobody targeting Middle East respiratory syndrome coronavirus (MERS-CoV) receptor-binding domain has potent cross-neutralizing activity and protective efficacy against MERS-CoV. J. Virol. 2018;92(18):e00837-18. doi: 10.1128/JVI.00837-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Jr, Baric R.S., Enjuanes L., Gallagher T., McCray P.B., Jr, Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. U. S. A. 2014;111(13):4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Yang Y., Huang J., Jiang S., Du L. Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. 2019;11(1):60. doi: 10.3390/v11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the published article.