Summary
Background
Expansion of antimicrobial resistance monitoring and epidemiological surveillance are key components of the WHO strategy towards zero leprosy. The inability to grow Mycobacterium leprae in vitro precludes routine phenotypic drug susceptibility testing, and only limited molecular tests are available. We evaluated a culture-free targeted deep sequencing assay, for mycobacterial identification, genotyping based on 18 canonical SNPs and 11 core variable-number tandem-repeat (VNTR) markers, and detection of rifampicin, dapsone and fluoroquinolone resistance-associated mutations in rpoB/ctpC/ctpI, folP1, gyrA/gyrB, respectively, and hypermutation-associated mutations in nth.
Methods
The limit of detection (LOD) was determined using DNA of M. leprae reference strains and from 246 skin biopsies and 74 slit skin smears of leprosy patients, with genome copies quantified by RLEP qPCR. Sequencing results were evaluated versus whole genome sequencing (WGS) data of 14 strains, and versus VNTR-fragment length analysis (FLA) results of 89 clinical specimens.
Findings
The LOD for sequencing success ranged between 80 and 3000 genome copies, depending on the sample type. The LOD for minority variants was 10%. All SNPs detected in targets by WGS were identified except in a clinical sample where WGS revealed two dapsone resistance-conferring mutations instead of one by Deeplex Myc-Lep, due to partial duplication of the sulfamide-binding domain in folP1. SNPs detected uniquely by Deeplex Myc-Lep were missed by WGS due to insufficient coverage. Concordance with VNTR-FLA results was 99.4% (926/932 alleles).
Interpretation
Deeplex Myc-Lep may help improve the diagnosis and surveillance of leprosy. Gene domain duplication is an original putative drug resistance-related genetic adaptation in M. leprae.
Funding
EDCTP2 programme supported by the European Union (grant number RIA2017NIM-1847 -PEOPLE). EDCTP, R2Stop: Effect:Hope, The Mission To End Leprosy, the Flemish Fonds Wetenschappelijk Onderzoek.
Keywords: Mycobacterium leprae, Targeted next generation sequencing, Antibiotic resistance, Gene domain duplication, Diagnostics, Surveillance
Research in context.
Evidence before this study
We searched PubMed for Mycobacterium leprae genotyping and/or drug-resistance prediction methods published before January, 2023, using the following terms: ((test) OR (assay)) AND (Mycobacterium leprae) AND ((drug resistance) OR (antibiotic resistance) OR (genotyping) OR (diagnostics) OR (diagnosis)) AND (sequencing). Identified methods included phenotypic drug susceptibility and molecular testing approaches. Due to the inability to grow M. leprae in vitro, phenotypic testing requires the use of the mouse footpad model, which takes months for obtaining results. Molecular tests comprise Sanger sequencing of amplicons, real-time PCR–high-resolution melt, microarray analysis, a line probe assay (LPA) based on post-PCR reverse hybridization and multi-locus VNTR analysis performed by fragment length analysis (MLVA-FLA). These methods detect only some, predefined variants in a limited number of M. leprae genomic regions, and/or require multiple PCR reactions. Whole genome sequencing (WGS) and a targeted sequencing-based assay by Iwao et al. allow simultaneous genotyping and drug resistance prediction but they are costly and labour intensive, as they require M. leprae DNA enrichment procedures or three separate nested multiplex amplifications, respectively. The World Health Organization (WHO) has called for improving surveillance, diagnosis and monitoring of (drug resistant) leprosy.
Added value of this study
Our study describes and evaluates a test called Deeplex Myc-Lep, which can both determine M. leprae strain type and detect drug resistance-associated mutations, directly from clinical specimens and by using a single hi-plex PCR mix followed by deep DNA sequencing. The assay analyzes the entire drug resistance-determining regions of all the known gene targets associated with resistance to the WHO-recommended anti leprosy drugs, along with 29 canonical markers (SNPs/indel and VNTR) for high-resolution genotyping of M. leprae, and a target for identification of both causal agents of leprosy, M. leprae and Mycobacterium lepromatosis. Our experimental results obtained with DNA from M. leprae reference strains and from more than 300 clinical specimens from patients diagnosed with leprosy show that successful sequencing can be achieved with samples including a minimum number of genome copies in the range from 100 to 1000. Our deep sequencing data demonstrate confident detection of strain genotypes as well as resistance-associated mutations, including those carried by bacterial subpopulations, potentially causing heteroresistance, down to a 10% proportion. All SNPs detected in targets by WGS were concordantly identified by targeted deep sequencing except in a clinical sample where WGS revealed two dapsone resistance-conferring mutations instead of one by Deeplex Myc-Lep, due to partial duplication of the sulfamide-binding domain in folP1. SNPs detected uniquely by Deeplex Myc-Lep were missed by WGS due to insufficient coverage. Concordance with VNTR-FLA results was 99.4% (926/932 alleles).
Implications of all the available evidence
The Deeplex Myc-Lep assay can substantially improve the diagnosis and surveillance of (multidrug resistant) leprosy, to help reach the goal set by the WHO of 120 countries with zero new autochthonous cases and a 70% reduction in the annual number of detected incident cases by 2030. Access to this test should be favoured by the global expansion of next generation sequencing capacity as a result of the COVID-19 pandemic response, including in many high-burden countries. Our results also show that Deeplex Myc-Lep worked well on Disolol-preserved samples (at ambient temperature), facilitating surveillance in regions where fast sample transport with adequate cold chains is challenging. Furthermore, a synergy could also be expected with the progressive deployment of Deeplex Myc-TB, used in more than 30 countries to date, given the same shared technical platforms and the large prevalence of tuberculosis in most settings affected by leprosy.
Introduction
Leprosy, also called Hansen's disease, is caused by infection with Mycobacterium leprae and more rarely, M. lepromatosis.1 For several decades, the disease was treated using dapsone monotherapy, inevitably leading to emergence of resistance.2 The use of multidrug therapy recommended by the World Health Organization (WHO), with addition of rifampicin and clofazimine to dapsone,3 and of effective second-line drugs such as fluoroquinolones in case of rifampicin resistance, subsequently resulted in a decrease in the numbers of leprosy cases globally. Yet, the incidence of leprosy has plateaued since 2005,4 and the disease is still present in 120 countries, with more than 200,000 new cases reported every year.5 Emergence of (multi-) drug resistant strains of M. leprae is reported in several world regions6, 7, 8, 9 and M. leprae transmission pathways are not fully understood, nor controlled.10,11 This situation calls for new tools for diagnosis and guidance of epidemiological tracing.
A number of biological and technical challenges must be overcome in order to determine both drug resistance profiles and high-resolution genotypes of M. leprae strains. Since M. leprae cannot be cultivated on artificial media, phenotypic drug susceptibility testing requires labour, time- and cost-intensive culture in the mouse footpad model.12,13 To circumvent this, molecular tests have been developed to detect genotypic resistance to rifampicin, dapsone and fluoroquinolones directly from clinical specimens, based on known resistance mutations located in the drug resistance determining regions (DRDRs) of rpoB, folP1 and gyrA, respectively.14 In-house methods for mutation detection include Sanger sequencing,8 real-time PCR–high-resolution melt,15 and microarray analysis.16 A commercial line probe assay (LPA) is based on post-PCR reverse hybridization.17 However, these tests require multiple PCR reactions and/or identify only some, predefined high-confidence resistance mutations in these three genes.
M. leprae is moreover a clonal obligate pathogen with highly restricted genetic diversity.18 A typing system including 18 polymorphic sites,19,20 with single-nucleotide polymorphisms (SNPs) and DNA insertions/deletions (indels), canonically distinguishes M. leprae strains into four main types (1–4) and 16 subtypes (1A-4P), supported by whole genome sequencing (WGS)-based data and displaying a phylogeographical association.21,22 However, while this SNP/indel-based system is useful to identify relatively distant genetic relationships, analysis of short-range transmissions within a specific geographical setting requires markers with higher discriminatory power.23 VNTR loci present in the M. leprae genome24 exhibit higher mutation rates compared to SNPs. Therefore, multi-locus VNTR analysis (MLVA), performed by fragment-length analysis (FLA), is often used to further type M. leprae strains.9,25,26 Like for SNP typing, MLVA-FLA similarly requires multiple PCR reactions, and accurate FLA-based determination of repeat numbers (alleles) can be challenging, especially for some loci with shortest, dinucleotide repeats.
WGS, done by short read sequencing (Illumina) for M. leprae, can simultaneously capture drug resistance-associated mutations and almost all genetic variation available for subsequent epidemiological inference (except in too complex/repetitive genome regions). However, this requires the use of costly and labour intensive M. leprae DNA enrichment procedures,27,28 and frequently results in relatively limited sequencing depth, restricting genome coverage and impeding confident variant detection, especially in case of minority variants potentially reflecting drug resistance emergence (heteroresistance) or mixed strain types.
As we showed for Mycobacterium tuberculosis29, 30, 31, targeted next-generation sequencing can offer an alternative solution for combined culture-free detection of drug resistance variants and determination of strain type, also allowing for high sequencing depth and higher multiplexing of samples per sequencing run. A method was recently described for detection of resistance mutations in folP1, rpoB, gyrA and gyrB and SNP-based typing of M. leprae. This method required six PCRs, consisting of three separate nested multiplex amplifications, before amplicon sequencing.32 Here, we describe and evaluate Deeplex® Myc-Lep, a culture-free targeted deep sequencing assay based on a single 44-plex PCR, commercially available as a ready-to-use amplification kit. The targets include (i) the hsp65 gene for mycobacterial identification, (ii) 18 SNP/indel sites and 17 VNTR markers (defined as 11 core and 6 non-core markers; see below) for high-resolution genotyping of M. leprae strains, and (iii) DRDRs of folP1, rpoB, gyrA and gyrB for drug resistance prediction, as well as gene regions of ctpC, ctpI and nth. Nonsense mutations in the excision repair gene nth are linked with hypermutated genomes and drug resistance profiles in M. leprae strains.21 ctpC and ctpI are included for exploratory purposes, as it has been suggested that missense mutations in these genes are associated with resistance to rifampicin, in a strain devoid of mutation in the rpoB DRDR.33 The evaluation was performed by comparison with reference data obtained from more than 300 clinical specimens of patients affected by leprosy, DNA of four M. leprae reference strains, and of M. lepromatosis NHDP-385.
Methods
Deeplex Myc-Lep assay
The Deeplex Myc-Lep assay starts with the amplification of 43 regions of the M. leprae genome as well as of one synthetic sequence used as internal control in a single multiplex PCR step (see Results, Assay design). Amplicon libraries are prepared using the Nextera XT kit and sequenced with 150 bp or 250 bp paired-end reads in a MiSeq (Illumina, CA, USA). Sequencing data analysis is performed using a pre-parameterized automated bioinformatic pipeline.
Clinical specimens, strains and M. leprae DNA quantification
The limit of detection (LOD) of Deeplex Myc-Lep was evaluated using 213 DNA extracts from clinical specimens collected between 2017 and 2018 in the Comoros as part of the ComLep (Improved Understanding of Ongoing Transmission of Leprosy in the Hyperendemic Comoros, ITM IRB ref 1147/16) and PEOPLE (Post ExpOsure Prophylaxis for LEprosy in the Comoros and Madagascar, ITM IRB ref 1248/1834) trial studies and purified DNA from the well-characterized strains NHDP63, Thai-53, Br4923 (BEI resources) and Br14-3 (Fundação Oswaldo Cruz, Brazil) (see also Table S1 for information on the datasets used in this study). The 4 mm skin biopsies from the Comoros were inactivated directly after sampling in 1 ml of Disolol (ethanol denatured with 1% isopropanol and 1% methyl ethyl ketone) in screw cap vials at ambient temperature, and transported in batches to the Institute of Tropical Medicine (Antwerp, Belgium). The biopsies were preserved at ambient temperature up until months before analysis. Negative sampling controls, consisting of Copan FloqSwabs (Murrieta, CA, USA) that were exposed for a minimum of 1 min to air in the room where the biopsies were taken, were included each sampling day. DNA from these 213 biopsies from the Comoros were extracted as described in Braet et al., 2022,35 by using the Maxwell 16 FFPE Tissue LEV DNA Purification Kit or the Maxwell 16 FFPE Plus Tissue LEV DNA Purification Kit (Promega, WI, USA). DNA from M. leprae NHDP63, Thai-53, Br4923 was obtained from the BEI Resources Repository (VA, USA) and genomic DNA of Br14-3 was obtained from cultures of corresponding strains on mouse footpads and purified with a modified protocol using the QIAamp® DNA Microbiome Kit (Qiaqen). Evaluation of the LOD using these reference strains and these 213 biopsies was done by utilizing kits from the same Deeplex Myc-Lep production lot.
Moreover, DNA from 107 additional samples including 33 supplementary skin biopsies from the aforementioned studies as well as 74 slit skin smears (SSS) from routine leprosy diagnostics at the Fundação Oswaldo Cruz (Brazil), extracted using the Maxwell 16 FFPE Plus Tissue LEV DNA Purification Kit (Promega, WI, USA), the modified Boom method36 or the method described by van der Zanden et al.37 were included as part of a supplementary analysis of the LOD of the assay (see Table S2 for details). M. leprae DNA was quantified from all samples using quantitative PCR (qPCR) based on the M. leprae-specific repetitive element (RLEP) region.38,39
Fourteen M. leprae strains studied by WGS are detailed in Table S3. DNA was extracted from human skin biopsies or mouse footpads as described by Woods and Cole40 for crude extracts obtained by the freeze-boiling method, and by Avanzi et al.,41 including human or mouse DNA elimination for obtaining WGS quality grade DNA.
DNA from M. lepromatosis NHDP-385 and M. leprae NHDP63, used in the hsp65-based species identification experiment, was obtained from the National Hansen's Disease Program (NHDP; LA, USA) and BEI resources (VA, USA), respectively.
Determination of the limit of detection
The LOD was assessed in terms of (i) the minimum number of RLEP copies enabling correct allele detection of all Deeplex Myc-Lep core markers, (ii) the minimum proportion of detectable minority variants in mixes of a resistant (Br14-3) and a susceptible (NHDP63) M. leprae strain as well as (iii) the minimum bacterial load, expressed as the RLEP qPCR Cq value, required for the sequencing of all Deeplex Myc-Lep core markers. Dilution series of four DNA extracts (with strains NHDP63, Thai-53, Br4923 and a mix of the former two) from 3.103 to 3.106 RLEP copies representing about 80 to 80,000 M. leprae genomes and a series of mixes of a resistant (Br14-3) with a susceptible (NHDP63) strain at total 6.106 RLEP copies were prepared for the first two experiments, respectively. M. leprae was quantified by RLEP qPCR from clinical specimens. Cq values range from 13 to 30.
Species identification
Identification of mycobacterial species by Deeplex Myc-Lep was done based on amplification and sequencing of a hsp65 gene segment, followed by best-match analysis of the obtained sequences against a database of hsp65 sequences derived from Dai et al.,42 as for Deeplex Myc-TB.29
DNA from M. leprae strain NHDP63 and M. lepromatosis NHDP-385 was quantified using the Qubit dsDNA High Sensitivity assay (ThermoFisher, MA, USA) and a series of mixes of DNA from the two strains was prepared using a total of 4.5 ng of DNA in each mix.
Deeplex Myc-Lep results compared to VNTR-FLA and WGS
The ability of Deeplex Myc-Lep to correctly detect variants and VNTR marker alleles was assessed by comparing the assay's results to those of WGS and VNTR-fragment length analysis (FLA), respectively. In all cases, Deeplex Myc-Lep was performed on the same DNA extracts as those used for WGS or VNTR-FLA. Comparison to WGS was based on 11 skin biopsies with microscopy smear gradings from 2+ to 4+ collected from 2010 to 2018 as part of routine leprosy diagnostics by the National Reference Center for Mycobacteria (Paris, France) and 3 M. leprae strains cultivated in mouse footpads (Table S3). DNA extracted following the protocol published by Avanzi et al.41 was sequenced using Nextera XT DNA Library Preparation Kit and a MiSeq with 150 bp paired-end reads according to the manufacturer's instructions (Illumina, CA, USA). VNTR-FLA was performed as described by Jensen et al.43 on 89 samples, comprising 35 slit skin smears collected in Brazil as part of routine leprosy diagnostics by the Fundação Oswaldo Cruz (Fiocruz Recife), 31 skin biopsies collected in the Comoros as part of the ComLep and PEOPLE trial studies, 20 skin biopsies from diverse countries (including three from the Comoros) and 3 M. leprae cultured strains provided by the Bichat-Claude Bernard Hospital (France).
Ethics statement
The ComLep (ClinicalTrials.gov, NCT03526718) and PEOPLE studies (ClinicalTrials.gov, NCT03662022) were approved by the institutional review board of the Institute of Tropical Medicine (Antwerp, Belgium, ComLep ref 1147/16, PEOPLE ref 1248/18), the ethical committee of the University of Antwerp (Antwerp, Belgium, ComLep ref 17/05/052, PEOPLE ref 18/36/390, approved on 17/09/2018), the ethical committee on the island of Anjouan (ComLep, no ref, approved on 15/07/2017, PEOPLE ref 18-01/MSSPSPG/CNE, approved on 9/10/2018), and the Comoros national ethical committee (PEOPLE). Written informed consent was obtained from each participant, or their parent or guardian if they were younger than 18 years. Written consent was obtained for people aged 12–17 years, in addition to their parents' or guardians' consent. Participants could selectively refuse sampling if they chose to. For the control strains used in the WGS analysis and provided by the NRC France, all subjects gave written informed consent in accordance with the Declaration of Helsinki. The genotyping of slit skin smears samples was approved by the ethical committee from CPqAM/Fiocruz (ref CEP/CPqAM/FIOCRUZ 02/12). For human samples from which WGS was performed, all subjects gave written informed consent in accordance with the Declaration of Helsinki. Specimens were collected under the approval of the Centre de Ressources biologiques, Assistance publique-hôpitaux de Paris, France. DNA from mouse footpad specimens were obtained from previous work and were provided by Alexandra Aubry and Aurélie Chauffour (license number to carry out animal experiments C-75-13-01).
Role of funders
The funders had no role in study design, data collection, data analyses, interpretation, patient recruitment, or writing of this manuscript.
Results
Assay design
The gene regions and sequence positions of M. leprae genome targeted by the assay are listed in Table 1. The reaction mix comprises an internal control sequence to detect potential PCR inhibition. After amplicon sequencing on an Illumina platform with 150 bp paired-end reads, the sequencing data are automatically analysed using a proprietary, pre-parameterized bioinformatic pipeline, with integrated databases. A subsequently generated schematic representation of the results is shown in Fig. 1, comprising identification of the mycobacterial species, detection of genotypic resistance, determination of the VNTR allelic profile and of strain type based on 18 canonical SNPs. Species identification is done by best-match analysis of reference hsp65 sequences from 168 mycobacterial taxa derived from Dai et al. 2011.42 Via comparison with a proprietary reference database compiling amino acid changes reportedly associated with M. leprae antibiotic resistance, sequence variants in the relevant targets are reported as “Resistant” if known to be associated with resistance to either of the above antibiotics, or “Uncharacterised” if leading to a non-synonymous mutation not included in the current database. VNTR alleles are determined according to the numbers of repeats directly determined from the sequences amplified from the respective loci, also accounting for potential artefactual “stutter” peaks, as also seen for M. tuberculosis MIRU-VNTR markers.45 For the purpose of the analysis, markers were separated into two sets, defined as core and non-core, the latter category consisting of six VNTR markers that could be amplified only in a minority of the specimens of the test datasets using 150 bp read lengths. Of these, two VNTR markers, 18-8 and 27-5 that include longer repeat units and amplified alleles often exceeding analytic capacity with 150 bp sequencing, were recovered using longer 250 bp paired-end reads (see below).
Table 1.
M. leprae gene regions or positions targeted by Deeplex Myc-Lep, relative to the TN strain genome from MycoBrowser.44
| Target | Genomic positions | Gene positions | Codons (Gene Name) | Information | |
|---|---|---|---|---|---|
| InDel_17915 | 17915–17936 | 433–454 | pseudogene (ML0014) | Typing (SNPs/indels) | Core |
| SNP-7614 | 7614 | 297 | 99 (gyrA) | ||
| SNP-1642879 | 1642879 | 896 | pseudogene (ML1378) | ||
| SNP-2935693 | 2935693 | 753 | pseudogene (icI) | ||
| SNP-14676 | 14676 | NC | NC | ||
| SNP-8453 | 8453 | 1136 | 379 (gyrA) | ||
| SNP-313361 | 313361 | 461 | 154 (metS) | ||
| SNP-61425 | 61425 | 269 | 90 (esxA) | ||
| SNP-3102787 | 3102787 | 452 | 151 (ML2597) | ||
| SNP-1104235 | 1104235 | 239 | pseudogene (ML0934) | ||
| SNP-2751790 | 2751790 | 897 | 299 (asd) | ||
| SNP-1295195 | 1295195 | 430 | 144 (ML1119) | ||
| SNP-2312066 | 2312066 | 3 | 1 (ML1926c) | ||
| SNP-413903 | 413902 | 275 | 92 (ML0324) | ||
| SNP-20910 | 20910 | 1283 | 428 (pknA) | ||
| Ins-978589 | 978589 | 89 | 30 (ML0825c) | ||
| Del-1476522 | 1476522 | NC | NC | ||
| SNP-1527056 | 1527056 | 617 | 206 (cydD) | ||
| 6-3a | 1190305–1190395 | 518–608 | 173-203 (sigA) | Typing (VNTRsa) | Core |
| AC8a | 1531112–1531235 | 141–264 | pseudogene (cya) | ||
| AC8b | 2210951–2211090 | NC | NC | ||
| AC9 | 1452501–1452646 | NC | NC | ||
| GTA9 | 2583766–2583887 | NC | NC | ||
| GAA21 | 2785374–2785574 | NC-77 | NC-pseudogene (ML2344A) | ||
| GGT5 | 2567170–2567330 | NC | NC | ||
| 6–7 | 1816775–1816966 | 14–205 | 5-69 (ML1505) | ||
| 12–5 | 1381580–1381868 | 405–683 | 135-228 (PPE) | ||
| 21–3 | 73016–73195 | 492–671 | 164-224 (espE) | ||
| 23–3 | 2945411–2945600 | NC | NC | ||
| rpoB | 2275546–2275343 | 1267–1470 | 423–490 | Rifampicin | Core |
| folP1 | 296765–296914 | 70–219 | 24–73 | Dapsone | |
| gyrA | 7436–7638 | 119–321 | 40–107 | Fluoroquinolones | |
| gyrB | 6589–6842 | 1361–1614 | 454–538 | Fluoroquinolones | |
| ctpC | 889136–888916 | 1836–2056 | 612–686 | Exploratory | |
| ctpI | 3209132–3209379 | 4414–4661 | 1472–1554 | Exploratory | |
| nth | 2726174–2725850 | 318–642 | 106–214 | Hypermutation | |
| 18–8 | 1587513–1587860 | 188–535 | 63-179 (ML1334) | Typing (VNTRsa) | Non-core |
| 27–5 | 686961–687230 | 148–417 | 50-139 (ML0568) | ||
| TA10 | 1743996–1744180 | 872–1056 | pseudogene (ML1450A) | ||
| TA18 | 984529–984670 | 231–372 | pseudogene (ML0830c) | ||
| AT15 | 948843–949041 | NC | NC | ||
| AT17 | 2597667–2597846 | 458–637 | pseudogene (ML2183c) | ||
| hsp65 | 405683–406083 | 165–565 | 55–188 | Species identification | |
| Synthetic target | NA | NA | NA | Internal control | |
NC, non-coding.
Expected lengths of VNTR marker alleles are reported in Table S4.
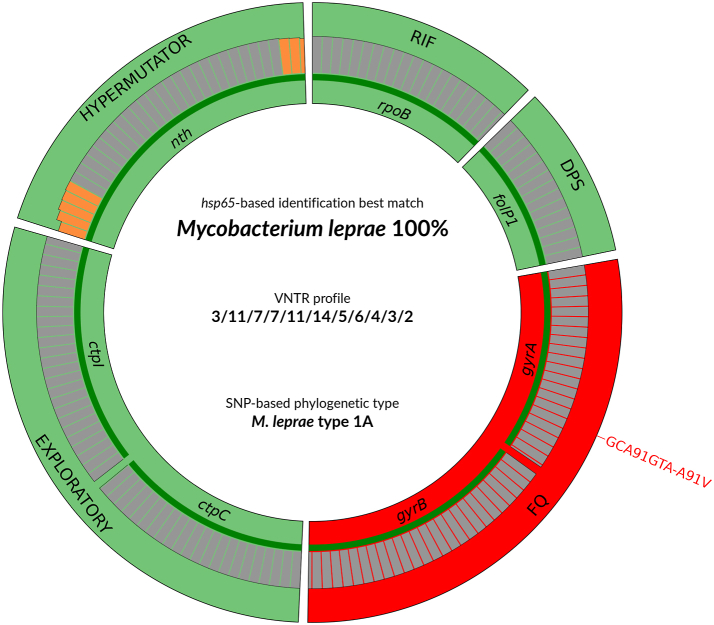
Fig. 1.
Deeplex Myc-Lep results identifying a M. leprae strain of SNP type 1A genotypically resistant to fluoroquinolones. Results are shown for a Thai53 strain derivative, mutated in gyrA (see Table S3). Information on hsp65 best match-based identification, VNTR allelic profile and SNP-based phylogenetic type is shown in the center of the circle. Information on predictions of drug susceptibility and drug resistance for anti-leprosy drugs/drug classes and on hypermutator genotype is as follows. Target gene regions are grouped within sectors in a circular map according to the prediction feature (drug resistance, hypermutation) with which they are associated. Sectors in red and green indicate targets in which resistance- or hypermutation-associated mutations or no mutations are detected, resulting in predictions of resistant or susceptible phenotypes (for rpoB, folP1, gyrA, gyrB), or hypermutator strain (nth), respectively. The ctpC and ctpI sector (and their associated drug resistance or drug susceptibility predictions) are categorized as exploratory, based on previous work suggesting an association of missense mutations in these genes with resistance to rifampicin, observed in a single strain devoid of mutation in the rpoB DRDR (see text). Green lines above gene names represent the reference sequences with coverage breadth above 95%. Limit of detection (LOD) of minority variants (resulting from subpopulations of reads bearing a mutation) depends on the read depth at each sequence position and is shown either as grey (LOD 10%) or orange zones (LOD >10%) above reference sequences. Here, LOD is >10% at the extremities of the nth target only. In the VNTR profile, VNTR markers are ordered as follows: 6-3a, AC8a, AC8b, AC9, GTA9, GAA21, GGT5, 6–7, 12-5, 21-3, 23-3. ∗RIF: Rifampicin, DPS: Dapsone, FQ: Fluoroquinolones, VNTR, variable-number tandem-repeat, SNP, single nucleotide polymorphism.
Identification of M. leprae and M. lepromatosis
The hsp65 sequencing- and best match-based system versus the reference database derived from Dai et al.42 used to identify mycobacterial species in Deeplex Myc-Lep is identical to that used in Deeplex Myc-TB (see Methods for detailed information). Its performance for species identification has previously been extensively described.29 Therefore, we evaluated here (co-)detection and distinction of M. leprae (NHDP63 strain) and M. lepromatosis (NHDP-385 strain), as the latter mycobacterium is the second causal agent of leprosy.1 For this, we applied the test on mixtures of genomic DNA from the two species at various ratios (4.5 ng total, Table 2). To note, these ratios were based on quantification of overall extracted DNA instead of specific quantification obtained by RLEP qPCR for M. leprae, since a specific qPCR was not performed for M. lepromatosis.
Table 2.
Theoretical and observed proportions of M. leprae NHDP63 and M. lepromatosis in mixes of genomic DNA from both species.
|
M. leprae |
M. lepromatosis |
||
|---|---|---|---|
| Theoretical | Detected | Theoretical | Detected |
| 100 | 99.4 | 0 | ND |
| 99 | 99.2 | 1 | ND |
| 95 | 81 | 5 | 18.9 |
| 90 | 71.3 | 10 | 28.5 |
| 80 | 64.8 | 20 | 35.1a |
| 50 | 29.1 | 50 | 70.7 |
| 20 | 24.3 | 80 | 75.5 |
| 0 | ND | 100 | 99.8 |
ND: Not detected.
Species could not be specifically identified but was reported as “Other”.
In addition to correct identification in both controls including a single species, the presence of strains from both species was explicitly detected and reported in mixtures when the minority DNA exceeded a “theoretical 5%/detected 18.9%” proportion, except in the following case. At a detected 64.8% proportion of M. leprae, M. lepromatosis was reported as “Other” at 35.1%, reflecting a large part of species-specific variants in hsp65 detected with frequencies close to 50%, making it impossible to unambiguously discriminate between hsp65 sequences of both species. Excluding the controls with a single species, detected proportions of both species differed from theoretical proportions by an average of 14.6% (SD: ±5.9), indicating semi-quantitative detection within the limits of DNA quantification accuracy indicated above. On top of this hsp65-based co-identification, the presence of M. lepromatosis in the mixes could also be inferred by detection of M. lepromatosis-specific variants in the resistance- and hypermutation-associated targets (up to 39 variants detected depending on the proportion of M. lepromatosis, Table S5).
Identification of SNPs, VNTRs and limit of detection using reference strains
Identification of SNPs and VNTR alleles, as well as the limit of detection (LOD) of the assay were first evaluated using DNA from reference M. leprae strains cultured from mouse footpads. The LOD was estimated both in terms of minimum number of genomes enabling at least 95% coverage breadth of resistance- and hypermutation-associated targets (fully comprising the DRDR region for rpoB, folP1, gyrA, and gyrB) at minimum depth of 5x (minimal threshold for base calling), correct marker allele detection and minimum proportion of detectable minority variants.
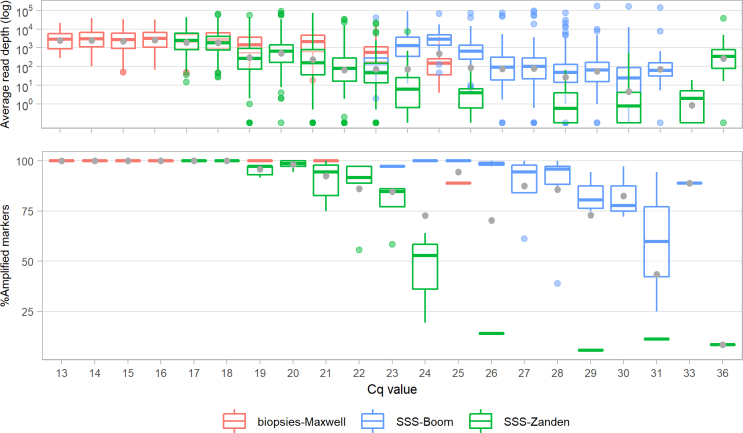
First, serial dilutions of four DNA extracts from genotypically drug susceptible strains NHDP63, Thai-53, Br4923, and an 85-15% mix of NHDP63 and Thai-53 were prepared. As estimated by RLEP qPCR, resulting amounts included per Deeplex Myc-Lep test ranged from 3.103 to 3.106 RLEP copies, representing about 80 to 80,000 genomes. While the read depth expectedly decreased with the number of genomes, all core markers (including typing SNP, VNTR and resistance- or hypermutation-associated markers) were completely covered, with a mean coverage depth of 1,718x even with 3.103 RLEP copies/80 genomes (Fig. 2), and all samples were identified as M. leprae. All expected alleles of the 18 typing SNPs and the 11 VNTR core markers were correctly called in all cases, and correct mixed SNP alleles were detected in the NHDP63/Thai-53 mix. For the 11 typing SNPs that were expected to be different between both strains, heterozygous calls were identified with the NHDP63 SNP alleles dominant as expected (see Table S6 for alleles detected in reference strains). Such detection of mixed typing SNPs was, and is, accordingly considered to report detection of mixed strain types in the sample. Further as expected, only a synonymous R99R SNP was detected in gyrA of NHDP63 and in the NHDP63/Thai-53 mixture (as a dominant allele), while no SNP was detected in any (other) resistance- or hypermutation-associated target in NHDP63, NHDP63/Thai-53, Thai-53 or Br4923. To note, because only the dominant VNTR marker alleles are called in this version of Deeplex Myc-Lep, these markers were not considered in the NHDP63/Thai-53 mixture. In contrast, less than half of the non-core VNTR alleles were called even with the highest tested numbers of genome copies, reflecting much lower read depth at these markers (average 12-108x vs 2025-5,643x for core markers).
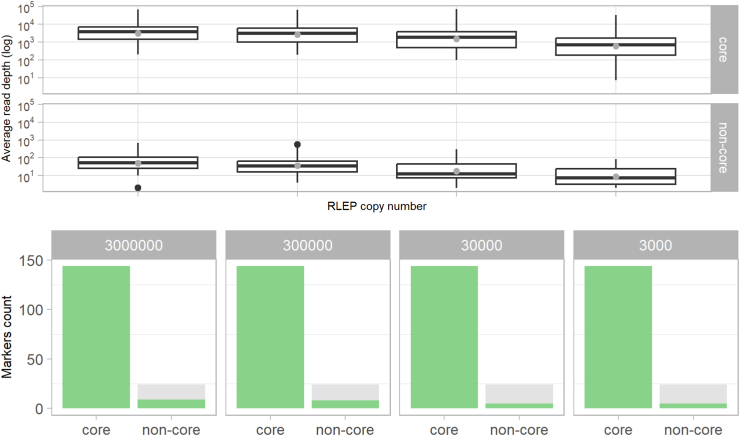
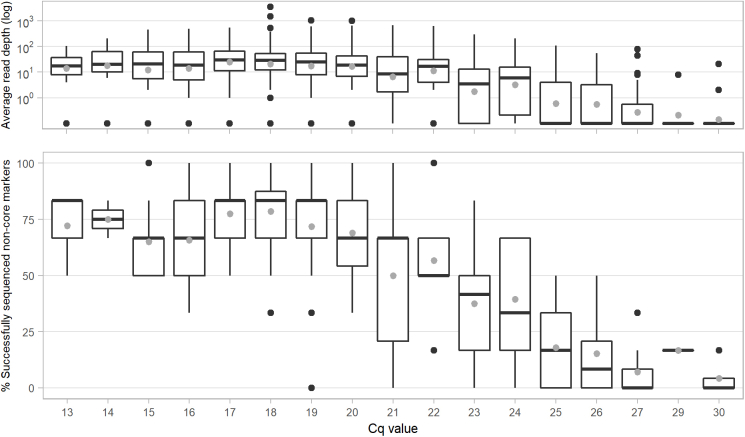
Fig. 2.
Limit of detection for correct allele calling of 42 Deeplex Myc-Lep markers. The LOD was evaluated with DNA extracts from three M. leprae strains and an 85-15% mixture of two strains. (Top) Read depth at core and non-core markers versus the number of RLEP copies. Median (line) and mean (grey dot) values as well as 25–75% quartiles are shown. (Bottom) For each serial dilution with 3.103, 3.104, 3.105 and 3.106 RLEP copies (80-80,000 M. leprae genomes), the fraction of correctly (green) and incorrectly detected or not sequenced (grey) alleles was determined for 144 core and 24 non-core alleles (36 core and six non-core markers times four DNA extracts, respectively).
Second, a series of mixes of DNA were prepared from a multidrug-resistant strain (Br14-3, known to possess resistance-conferring mutations in rpoB, gyrA, and folP1, and a stop codon in nth21) and a drug susceptible (NHDP63) strain in various proportions. In these mixes, all minority variants from Br14-3, including typing and resistance/hypermutation-conferring SNPs, were detected if the strain represented at least 10% of the input DNA (Fig. 3). Below this level, part of the expected variants were missed, while other false positive variants were detected (one at a 5% ratio and 204 at a 1% ratio were observed). The LOD of Deeplex Myc-Lep for minority variant calls was therefore set at 10%.
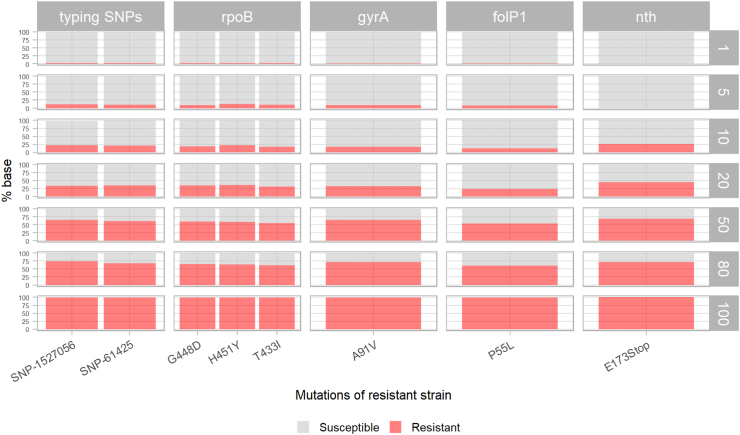
Fig. 3.
Limit of detection in terms of the minimum proportion of detectable minority variants, using M. leprae reference strains (VNTR markers not considered). Mixes of the susceptible strain NHDP63 with 1, 5, 10, 20, 50 and 80% of the resistant strain Br14-3 were used to estimate the lowest fraction of detectable variant allele. The resistant strain was also analysed alone, as a control (100%, bottom). The resistant allele is depicted in red while the susceptible allele is in grey.
Limit of detection using clinical specimens
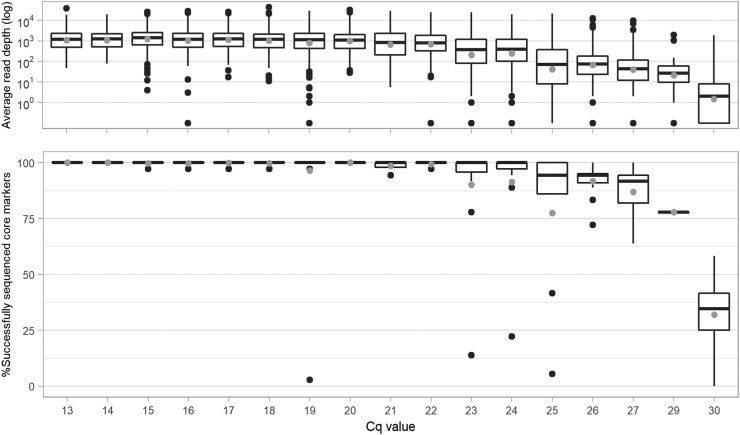
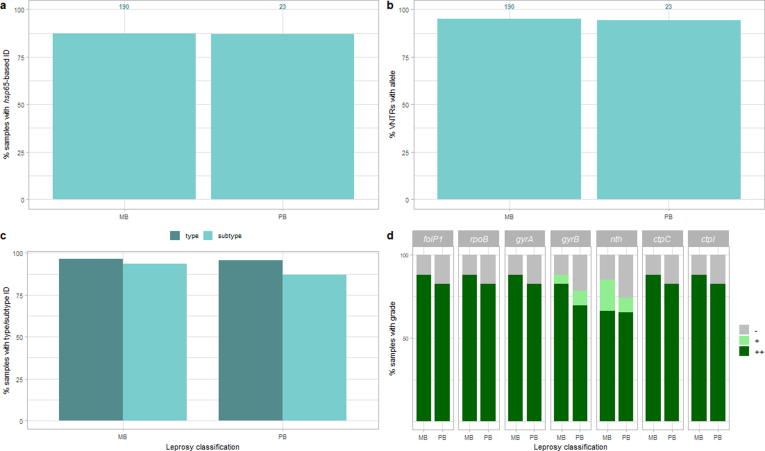
The Deeplex Myc-Lep LOD was evaluated on DNA extracted with Maxwell kits from 213 clinical specimens, consisting of skin biopsies collected from patients from Anjouan (Comoros), with leprosy diagnosed by conventional examination (Table S2). Out of these 213 samples, hsp65 sequencing results of the Deeplex confirmed the presence of M. leprae in 186 (87.3%). We first determined the coverage depth and fraction of core markers with successful sequencing results as defined above depending on the M. leprae genome numbers, as estimated by RLEP qPCR (Fig. 4). Specimens with Cq values of 24 (corresponding to 3243 genome copies, SD ± 1591) or lower had almost systematically all core markers successfully sequenced (median of 36/36 markers). With Cq values between 25 and 29 (2244-115 genome copies ±SD 3946 for Cq values of 25; a single sample was available with a Cq of 29), medians of successfully sequenced markers still ranged between 34/36 (94%) and 28/36 (78%); a marked drop in the fraction of sequenced core markers was only observed at Cq values of at least 30 (median 12/37 sequenced core markers). Regarding non-core VNTR markers, alleles were successfully detected for more than half of the markers with Cq values of 21 or lower, but complete non-core allele profiles were never obtained, even with high bacterial load, due to low overall read depths (1-86x vs 36-2,206x at core markers, Fig. S1).
Fig. 4.
Limit of detection, in terms of RLEP qPCR Cq value, for the sequencing of 36 Deeplex Myc-Lep core markers (including the species identification target), determined by using 213 biopsies from patients affected by leprosy. (Top) Read depth at Deeplex Myc-Lep core markers versus RLEP PCR Cq values 13–30. (Bottom) Proportion of successfully sequenced Deeplex Myc-Lep core markers versus RLEP PCR Cq values 13–30 (Top & Bottom) Median (line) and mean (grey dot) values as well as 25–75% quartiles are shown.
Of note, no substantial differences were seen when results were stratified by classification of samples from multi- (n = 190) or paucibacillary (n = 23) leprosy (according to WHO classification; Fig. S2), likely reflecting the limited quantitative information of this classification. Success rate for species identification and determination of core VNTR alleles were almost identical between both categories. The proportions of samples with determined SNP type/subtype and with coverage depth and breadth sufficient to detect potential variants at 10% or more (graded ++) or 80% or more frequency (+) in resistance/hypermutation targets were lower by only a few percent in the paucibacillary category.
Similar results and limits of detection were obtained in terms of RLEP Cq values when using other Deeplex Myc-Lep kit lots, on sets of DNAs extracted from 33 skin biopsies with Maxwell kits or from 74 slit skin smears with the Boom method36 and the method published in van der Zanden et al.37 Rates of successfully sequenced markers versus Cq values appeared nevertheless slightly lower on DNA extracted from slit skin smears (Fig. S3, Table S2).
Deeplex Myc-Lep versus WGS
Fourteen samples collected between 2010 and 2018 from various locations (Table S3) were sequenced using both Deeplex Myc-Lep and WGS, and variants in typing SNPs and Deeplex Myc-Lep resistance/hypermutation-associated targets were compared. Overall, 36 SNPs were concordantly detected by both methods including 3 typing SNPs and 3 resistance-associated variants (Table S7). However, 26 other SNPs, consisting exclusively of typing SNPs, were detected only by Deeplex Myc-Lep. The latter cases were straightforwardly explained by total absence of read coverage (n = 19) or coverage by a single read only (n = 7; below the threshold for confident variant calling) by WGS at the corresponding positions due to low bacillary load in the skin biopsies (Bl of 1+ and 2+). In comparison, read depths were 8-4,615x (mean 833x) at these positions by Deeplex Myc-Lep (Table S7).
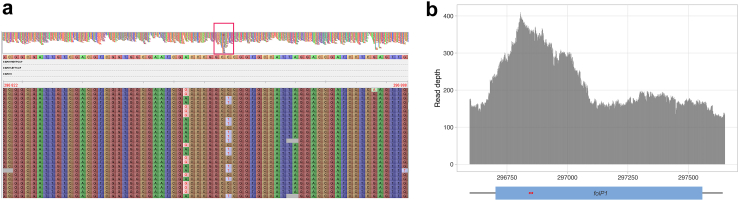
Unexpectedly, in one sample (WGS23), two variants were detected in folP1 by WGS at ∼50% (P55L and T53A) while Deeplex Myc-Lep only detected variant T53A as fixed (99.9%), even though read depths were high at both positions (>1,000x) with Deeplex Myc-Lep. Inspection of the WGS reads that mapped to folP1 in the reference genome of the TN strain shows that, despite their proximal position in the folP1 gene sequence, the two variants were systematically carried by different reads, indicating that they originate from two distinct regions in WGS23 (Fig. 5A). Moreover, the WGS coverage depth on the reference folP1 sequence was up to two times higher compared to that on flanking regions (Fig. 5B). Detailed analysis of the obtained mapping data excluded ambiguous mapping of reads from the folP2 gene paralogue (not amplified by Deeplex Myc-Lep) present in M. leprae as a potential explanation for the WGS results. Taken altogether, these observations indicate a partial duplication of folp1 in the WGS23 strain (spanning circa 350 bp, corresponding to the sulfamide-binding domain in the encoded enzyme), with T53A and P55L variants separately borne by the duplicated segments, and a possible rearrangement affecting folP1 primer regions leading to amplification of T53A only by Deeplex Myc-Lep.
Fig. 5.
WGS reads of sample WGS23 mapped to the genome of the M. leprae TN strain, around folP1. (a) Variants detected in folP1 are carried by different reads, indicating that they originate from distinct regions. The top part of the figure shows read coverage depths at the folP1 and flanking regions. A red box indicates the folP1 region with a coverage depth up to twice as high as the depth of flanking regions. The bottom part shows a zoom-in of the folP1 region showing the aligned sequence reads. Genomic positions of the extremities of the region shown are indicated on the top right and top left. Different rows represent independent sequence reads. G and T variants are never found in combination in a read, resulting in mixed wild type/variant calls with a frequency of ∼50% of the reads, at each of both variant positions. (b) Read depth at the folP1 region, with 100bp flanking sequences. Positions of the folP1 coding sequence and the two variants detected at a frequency of ∼50% by WGS are represented by a blue rectangle and red stars, respectively. Variants are 7bp apart in the reference genome.
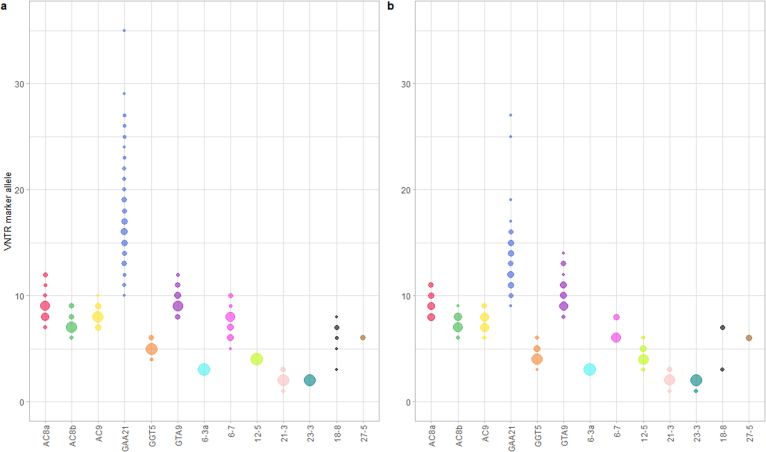
Deeplex Myc-Lep VNTR versus VNTR-FLA
The concordance of Deeplex Myc-Lep core VNTR results versus reference VNTR-FLA results was evaluated on 89 clinical specimens for which both result sets were generated. Results for two of the non-core VNTR markers (18-8 and 27-5) were also compared for a subset of 31 samples, with available 250 bp read-based sequencing data. In total, results could be obtained from both methods in 932 out of 1041 (89.5%) tested markers across the specimen set. The same allele was concordantly called in 926 markers (99.4%) by both methods (Table 3, Table S8). Three tests showed a partial match, where a same allele was identified by both methods in addition to a second allele undetected by one method. In two cases, two alleles were detected only by Deeplex Myc-Lep, each with an identical number of reads. Absence of allelic concordance between MLVA and Deeplex Myc-Lep was seen only in three tests (0.6%; for AC9, with one-repeat unit discordance in one sample, and GAA21 with one-repeat unit discordance in one sample and four-repeat unit discordance in another sample).
Table 3.
Comparison of Deeplex Myc-Lep and VNTR-FLA core and non-core VNTR analysis results from 89 M. leprae clinical samples.
| VNTR | Total Tested | Match | Partial Match | ND | Mismatch | %Match |
|---|---|---|---|---|---|---|
| 6-3a | 89 | 86 | 0 | 3 | 0 | 100 |
| AC8a | 89 | 75 | 0 | 14 | 0 | 100 |
| AC8b | 89 | 86 | 0 | 3 | 0 | 100 |
| AC9 | 89 | 82 | 0 | 6 | 1 | 98.8 |
| GTA9 | 89 | 79 | 1 | 9 | 0 | 100 |
| GAA21 | 89 | 82 | 1 | 4 | 2 | 97.6 |
| GGT5 | 89 | 86 | 0 | 3 | 0 | 100 |
| 12–5 | 89 | 84 | 0 | 5 | 0 | 100 |
| 21–3 | 89 | 76 | 1 | 12 | 0 | 100 |
| 23–3 | 89 | 70 | 0 | 19 | 0 | 100 |
| 6–7 | 89 | 65 | 0 | 24 | 0 | 100 |
| 18–8 | 31 | 26 | 0 | 5 | 0 | 100 |
| 27–5 | 31 | 29 | 0 | 2 | 0 | 100 |
| Total | 1041 | 926 | 3 | 109 | 3 | 99.5 |
Non-core VNTR markers 18-8 and 27-5 were sequenced using 250bp paired-reads on a subset of 31 samples. Match, same VNTR marker allele detected by Deeplex Myc-Lep and VNTR-FLA; partial match, two alleles detected by one of the methods, including one matching with the other method; ND, not detected by Deeplex Myc-Lep and/or VNTR-FLA; mismatch, Deeplex Myc-Lep and VNTR-FLA detected different VNTR marker alleles.
Discussion
Expansion of antimicrobial resistance monitoring and effective epidemiological surveillance are key components of the first strategic pillar of the Global Leprosy Strategy of the WHO, aiming at 120 countries with zero new autochthonous cases and a 70% reduction in the annual number of detected incident cases by 2030.46 The Deeplex Myc-Lep design is unique in that both components can be addressed in one single PCR assay, followed by NGS sequencing. This tool analyzes all the known (multi)drug resistance-associated gene targets (clofazimine resistance-associated gene(s) are as yet undetermined in M. leprae), along with 29 canonical SNPs/indel and core VNTR markers for high-resolution genotyping of M. leprae, and a mycobacterial speciation target for identification of both M. leprae and M. lepromatosis. Our results show the high degree of concordance, with an increment of superiority for some aspects as explained below, of this targeted NGS-based approach versus genome sequencing data (reference genomes and newly sequenced strains) and MLVA reference methods. We show that it can be applied directly on DNA extracts, and works best on clinical specimens with bacterial genome copies of ∼100 per test or higher as pre-quantified by RLEP qPCR. This study further uncovered an unexpected evidence of gene duplication as a source of genome plasticity and as a possible alternative mechanism of (increased) drug resistance in M. leprae.
Compared to current clinically used methods, Deeplex Myc-Lep substantially extends the diagnostic spectrum and accuracy, and additionally allows for both diagnostic and biological discovery. The commercially available LPA test specifically identifies only two common mutations in rpoB (S456L, H451Y), one in gyrA (A91V), and one in folP1 (P55L). Even if they may be less common, additional mutations in the DRDRs of these genes are known to confer resistance to rifampicin, fluoroquinolones, and dapsone.6,16,21,47 Mutations other than the four mentioned above can only be suspected in some codons within the respective DRDRs (432, 438–441, 451, and 456–458 for rpoB, 89–91 for gyrA, 53–55 for foplP1), in the absence of hybridization to “wild-type” probes, which requires additional PCR and sequencing for further assessment. Some suspected resistance mutations, such as gyrA S92A and gyrB D464N in the gyrA and gyrB DRDR segments, respectively, cannot be detected by such indirect analysis.47 Moreover, as shown for LPAs for M. tuberculosis resistance testing,48 potential synonymous or nonsynonymous mutations unrelated to resistance in these short segments could result in false inference of resistance if based on unbound wild-type probes. Microarray-based16 and high-resolution melt-based15 methods, similarly using a restricted set of pre-defined wild-type and mutant probes, or only distinguishing wild-type from mutant sequences in short DRDR segments, respectively, were exposed to the same limitations.
In contrast, Deeplex Myc-Lep analyses the entire (suspected) DRDRs of rpoB, gyrA, gyrB, and folP1 by direct sequencing, allowing to unambiguously identify all mutations conferring resistance to the current multidrug therapy validated to date in the mouse footpad model or using surrogate mycobacteria.6 The obtained mean sequencing depths of 100x or more, with RLEP qPCR Cq values of 25 or lower on DNA extracts from skin biopsies, allow extensive and highly confident detection of variants on the targets of interest. In comparison, WGS can frequently miss multiple variant positions, which were readily detected by Deeplex Myc-Lep as seen here, in case of bacillary load of 2+ or below. Our evaluation also showed that high read depths obtained by Deeplex Myc-Lep enable the detection of resistance alleles emerging within a sample, as low as a 10% heteroresistant subpopulation, which is also hard or impossible to reach with usual WGS depths or by Sanger sequencing. This deep sequencing capacity is expected to be especially useful for monitoring potential resistance emergence in the context of the anticipated scale up of preventive chemotherapy, currently done with a single dose of rifampicin, even if our preliminary data tend to be reassuring about this risk at least in the Comoros.35 In addition, targeted sequencing of relevant gene regions allows increased multiplexing of samples in a sequencing run (with, typically, 72 and 122 samples plus three controls in a single MiSeq run using 2 × 150bp or 2 × 250bp sequencing, respectively) compared to WGS, which thus reduces run cost.
Furthermore, new (candidate) resistance mutations, otherwise challenging to discover by phenotypic testing of M. leprae, could be identified as follows. Similar to WGS-based phylogenetic reconstruction,21 comparisons of the sequencing data of the rpoB, gyrA/gyrB, folP1, and the ctpC/ctpI targets with the SNP- and VNTR-based strain type information may allow detection of potential independent occurrence of the same variants in different genetic backgrounds (homoplasic mutations), indicative of positive selection likely associated with antibiotic pressure. In addition, as all hypermutated strains with nonsense nth mutations were previously found to be genotypically drug resistant, mutations detected in nth might serve as surrogate markers for inferring new candidate resistance mutations in the above targets, as well as for potential risk of treatment failure.21 Such systematic implicative relationship between nth mutations and drug resistance was not contradicted here. Indeed, nth variants were undetected in any of the 269 strains that had all resistance-associated targets successfully sequenced but showed no resistance mutation. Only one (Br14-3,21) of the four strains with confirmed resistance mutations carried a nonsense nth mutation.
Automated, direct sequencing-based allele calling of 11 core VNTR markers - extensible to 13 markers when using 250 bp read -, on top of a canonical set of typing SNPs/indels, represents an additional valuable tool for epidemiological surveillance and investigation of leprosy transmission. The observed concordance of 99.4% with VNTR-FLA results shows the high accuracy of this approach. In comparison, accurate interpretation of our VNTR-FLA data required very meticulous and tedious comparative inspections of various stutter peaks and true allelic ladders across electrophoretic profiles of many samples, which necessitated extensive expertise in VNTR typing systems (including with short sequence repeats) also developed for other mycobacteria.49,50 Although they were used in previous studies, we found that the four remaining VNTR markers (all with dinucleotide repeats) from the 17 initially tested (covering the entire repertoire of VNTR genomic loci previously used for typing the bacterium) are essentially unexploitable in most cases. Besides more difficult amplification, these loci often include apparently large numbers (well above 10) of 2-nucleotide repeats, clearly affected by too strong stutter peak effects (see Fig. 9 in Jensen et al.43) and preventing any reliable allele identification, whether by VNTR-FLA or by sequencing, as we reported for similar short sequence repeats in Mycobacterium paratuberculosis.49 Irrespective of the exclusion of non-core markers, the relative allelic diversities among the retained VNTR markers were similar between the Comoros sample set and the set of samples from other, diverse origins (Fig. S4, Table S9), suggesting similar degrees of epidemiological resolution across various settings. Moreover, in the Comoros set, the degree of genotypic resolution obtained was close to that obtained by WGS (Braet et al., in preparation).
The discovered evidence for a partial duplication in folp1 in one sample was unexpected, as the M. leprae genome is known to be otherwise prone to massive gene decay.51 The observation that each of the two partially duplicated copies carries a (different) dapsone resistance-conferring mutation known to alter the sulfamide binding site,52 with the duplication centered around the mutation positions, strongly suggests an original mechanism of domain duplication involved in (enhanced) drug resistance, reminiscent of kinase domain duplication involved in resistance of human tumoral cells to anticancer therapy.53 Of further interest, this strain neither showed resistance mutations in other resistance-associated gene targets, nor in nth, thus suggestive of a mechanism independent from hypermutation. Thus, this finding and the nth-mediated hypermutation in some other M. leprae strains21 - without known counterpart in M. tuberculosis - suggest broader capacities for genetic adaptation than could have been anticipated for a bacterium with a greatly degraded genome. This gene domain duplication was seen only in a single case among the 14 clinical samples that were successfully analysed by WGS. Therefore, knowing whether this duplication mechanism occurs relatively frequently or not, in relation with dapsone resistance in particular, will require refined (re-)analysis of M. leprae WGS data, by inspecting potential unfixed mutations and local distribution of reads and coverage depth as we did here. To note, from a diagnostic perspective, detection of only one of the two folP1 mutations was sufficient for dapsone resistance prediction by targeted sequencing.
In conclusion, based on one of the largest sample sets from a single study to date, our results show the potential of the Deeplex Myc-Lep assay to substantially improve the microbiological confirmation of the clinical diagnosis and the surveillance of (multidrug resistant) leprosy, to help reach the ambitious goals set by the WHO for this disease. Access to and use of this test should be favoured both by its availability as a commercial kit, and global expansion of next generation sequencing capacity as a result of the COVID-19 pandemic response, including in many high-burden countries. Our results also show that Deeplex Myc-Lep worked well on Disolol-preserved samples (at ambient temperature), facilitating surveillance in regions where fast sample transport with adequate cold chains is challenging. Furthermore, a synergy could also be expected with the progressive deployment of Deeplex Myc-TB, used in more than 30 countries to date, given the same shared technical platforms and the large prevalence of tuberculosis in most settings affected by leprosy. Finally, our findings reveal also a probable, previously unsuspected mechanism involved in drug resistance of M. leprae, the prevalence of which is to be further investigated.
Contributors
Conceptualisation: A.J., S.M.B, C.G., G.B., E.C., B.C.J., P.N.S., P.S.; Formal analysis: A.J., C.G., G.B., N.B, P.S.; Funding acquisition: B.C.J., Investigation: S.M.B, S.V., K.L., E.L., A.F., M.C., R.E.E.N.O.L., Y.Y.P.P.; Resources: S.M.B, C.G., A.B.F., N.L., P.R., M.M, E.H., S.G., W.A., A.S., Y.A., N.A., E.C., P.S. ; Software: A.J., Y.L. ; Supervision: E.C. B.C.J., P.N.S., P.S.; Visualisation: A.J., P.S., Writing – original draft: A.J., P.S., Writing – review and editing: A.J., S.M.B., E.C., B.C.J., P.N.S., P.S. All authors read and approved the final version of the manuscript. A.J. and P.S. verified the underlying data.
Data sharing statement
Sequence reads used in this paper were deposited in the Sequence Read Archive (SRA), National Center for Biotechnology Information (NCBI), under BioProject accession number PRJNA923280. A detailed description of the datasets is available in Table S1.
Declaration of interests
A.J., C.G., G.B., N.B., E.L., A.F., M.C. and Y.L. are employees of GenoScreen (Lille, France). P.S. is a scientific consultant for the same company. Other authors declare that they have no other competing interests.
Acknowledgements
This study is supported by a R2STOP research grant from effect:hope and The Mission To End Leprosy and the European and Developing Countries Clinical Trials program (EDCTP2) supported by the European Union (grant number RIA2017NIM-1847-PEOPLE, https://www.edctp.org/). SMB is supported by the Fonds Wetenschappelijk Onderzoek (FWO, grants 1189219N, 1189221N and V408322N, https://www.fwo.be/). E. Cambau's lab received annual grants from Fondation Raoul Follereau and Santé Publique France (French Ministry of Health). PNS was supported by grants from FAPERJ (210.877/2019) and CNPq (310418/2016-0 and 207422/2014-1). We acknowledge Aurélie Chauffour and Alexandra Aubry for providing mouse footpad specimens.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104649.
Appendix A. Supplementary data
Supplementary Figure S1.
Supplementary Figure S2.
Supplementary Figure S3.
Supplementary Figure S4.
References
- 1.Han X.Y., Seo Y.H., Sizer K.C., et al. A new Mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol. 2008;130:856–864. doi: 10.1309/AJCPP72FJZZRRVMM. [DOI] [PubMed] [Google Scholar]
- 2.Pettit J.H.S., Rees R.J.W. Sulphone resistance in leprosy: an experimental and clinical study. Lancet. 1964;2:673–674. doi: 10.1016/s0140-6736(64)92482-1. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization . 1982. Chemotherapy of leprosy for control programmes. [PubMed] [Google Scholar]
- 4.Chaptini C., Marshman G. Leprosy: a review on elimination, reducing the disease burden, and future research. Lepr Rev. 2015;86:307–315. [PubMed] [Google Scholar]
- 5.World Health Organization . 2020. Weekly epidemiological record n°36. [Google Scholar]
- 6.Aubry A., Sammarco Rosa P., Chauffour A., Fletcher M.L., Cambau E., Avanzi C. Drug resistance in leprosy: an update following 70 years of chemotherapy. Infect Dis News. 2022;52:243–251. doi: 10.1016/j.idnow.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Cambau E., Saunderson P., Matsuoka M., et al. Antimicrobial resistance in leprosy: results of the first prospective open survey conducted by a WHO surveillance network for the period 2009–15. Clin Microbiol Infect. 2018;24:1305–1310. doi: 10.1016/j.cmi.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kai M., Phuc N.H.N., Nguyen H.A., et al. Analysis of drug-resistant strains of Mycobacterium leprae in an endemic area of Vietnam. Clin Infect Dis. 2011;52 doi: 10.1093/cid/ciq217. [DOI] [PubMed] [Google Scholar]
- 9.Rosa P.S., D'Espindula H.R.S., Melo A.C.L., et al. Emergence and transmission of drug-/multidrug-resistant mycobacterium leprae in a former leprosy colony in the brazilian amazon. Clin Infect Dis. 2020;70:2054–2061. doi: 10.1093/cid/ciz570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hambridge T., Chandran S.L.N., Geluk A., Saunderson P., Richardus J.H. Mycobacterium leprae transmission characteristics during the declining stages of leprosy incidence: a systematic review. PLoS Negl Trop Dis. 2021;15 doi: 10.1371/journal.pntd.0009436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ploemacher T., Faber W.R., Menke H., Rutten V., Pieters T. Reservoirs and transmission routes of leprosy; A systematic review. PLoS Negl Trop Dis. 2020;14:1–27. doi: 10.1371/journal.pntd.0008276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy L., Ji B. The mouse foot-pad technique for cultivation of Mycobacterium leprae. Lepr Rev. 2006;77:5–24. [PubMed] [Google Scholar]
- 13.Shetty V.P., Wakade A. v., Ghate S., Pai V. v., Ganapati R., Antia N.H. Viability and drug susceptibility testing of M. LepraeUsing mouse footpad in 37 relapse cases of leprosy. Int J Lepr. 2003;71:210–217. doi: 10.1489/1544-581X(2003)71<210:VADSTO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization . 2009. Guidelines for global surveillance of drug resistance in leprosy. [Google Scholar]
- 15.Li W., Matsuoka M., Kai M., et al. Real-time PCR and high-resolution melt analysis for rapid detection of Mycobacterium leprae drug resistance mutations and strain types. J Clin Microbiol. 2012;50:742–753. doi: 10.1128/JCM.05183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuoka M., Aye K.S., Kyaw K., et al. A novel method for simple detection of mutations conferring drug resistance in Mycobacterium leprae, based on a DNA microarray, and its applicability in developing countries. J Med Microbiol. 2008;57:1213–1219. doi: 10.1099/jmm.0.2008/002600-0. [DOI] [PubMed] [Google Scholar]
- 17.Cambau E., Chauffour-Nevejans A., Tejmar-Kolar L., Matsuoka M., Jarlier V. Detection of antibiotic resistance in leprosy using GenoType LepraeDR, a novel ready-to-use molecular test. PLoS Negl Trop Dis. 2012;6 doi: 10.1371/journal.pntd.0001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schueneumann V.J., Singh P., Mendum T.A., et al. Genome-Wide comparison of medieval and modern Mycobacterium leprae. Science. 2013;341:179–183. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- 19.Truman R.W., Singh P., Sharma R., et al. Probable zoonotic leprosy in the Southern United States. N Engl J Med. 2011;364:1626–1633. doi: 10.1056/NEJMoa1010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monot M., Honoré N., Garnier T., et al. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 21.Benjak A., Avanzi C., Singh P., et al. Phylogenomics and antimicrobial resistance of the leprosy bacillus Mycobacterium leprae. Nat Commun. 2018;9 doi: 10.1038/s41467-017-02576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monot M., Honoré N., Garnier T., et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 23.Singh P., Cole S.T. Mycobacterium leprae: genes, pseudogenes and genetic diversity. Future Microbiol. 2011;6:57–71. doi: 10.2217/fmb.10.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole S.T., Supply P., Honoré N. Repetitive sequences in Mycobacterium leprae and their impact on genome plasticity. Lepr Rev. 2001;72:449–461. [PubMed] [Google Scholar]
- 25.Kimura M., Sakamuri R.M., Groathouse N.A., et al. Rapid variable-number tandem-repeat genotyping for Mycobacterium leprae clinical specimens. J Clin Microbiol. 2009;47:1757–1766. doi: 10.1128/JCM.02019-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lima L.N.C., Frota C.C., Suffys P.N., et al. Genotyping comparison of Mycobacterium leprae isolates by VNTR analysis from nasal samples in a Brazilian endemic region. Pathog Glob Health. 2018;112:79–85. doi: 10.1080/20477724.2018.1427308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefani M.M.A., Avanzi C., Bührer-Sékula S., et al. Whole genome sequencing distinguishes between relapse and reinfection in recurrent leprosy cases. PLoS Negl Trop Dis. 2017;11 doi: 10.1371/journal.pntd.0005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tió-Coma M., Avanzi C., Verhard E.M., et al. Genomic characterization of Mycobacterium leprae to explore transmission patterns identifies new subtype in Bangladesh. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jouet A., Gaudin C., Badalato N., et al. Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs. Eur Respir J. 2021;57 doi: 10.1183/13993003.02338-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makhado N.A., Matabane E., Faccin M., et al. Outbreak of multidrug-resistant tuberculosis in South Africa undetected by WHO-endorsed commercial tests: an observational study. Lancet Infect Dis. 2018;18 doi: 10.1016/S1473-3099(18)30496-1. [DOI] [PubMed] [Google Scholar]
- 31.Tagliani E., Hassan M.O., Waberi Y., et al. Culture and Next-generation sequencing-based drug susceptibility testing unveil high levels of drug-resistant-TB in Djibouti: results from the first national survey. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-17705-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iwao Y., Mori S., Ato M., Nakata N. Simultaneous determination of Mycobacterium leprae drug resistance and single-nucleotide polymorphism genotype by use of nested multiplex PCR with amplicon sequencing. J Clin Microbiol. 2021;59:814–821. doi: 10.1128/JCM.00814-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh P., Benjak A., Carat S., et al. Genome-wide re-sequencing of multidrug-resistant Mycobacterium leprae Airaku-3. Clin Microbiol Infect. 2014;20:O619–O622. doi: 10.1111/1469-0691.12609. [DOI] [PubMed] [Google Scholar]
- 34.Ortuno-Gutierrez N., Baco A., Braet S., et al. Clustering of leprosy beyond the household level in a highly endemic setting on the Comoros, an observational study. BMC Infect Dis. 2019;19 doi: 10.1186/s12879-019-4116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marijke Braet S., Jouet A., Aubry A., et al. Investigating drug resistance of Mycobacterium leprae in the Comoros: an observational deep-sequencing study. Lancet Microbe. 2022;3:e693–e700. doi: 10.1016/S2666-5247(22)00117-3. [DOI] [PubMed] [Google Scholar]
- 36.Boom R., Sol C.J.A., Salimans M.M.M., Jansen C.L., Wertheim-Van Dillen P.M.E., van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Zanden A.G.M., te Koppele-Vije E.M., Vijaya Bhanu N., van Soolingen D., Schouls L.M. Use of DNA extracts from Ziehl-Neelsen-stained slides for molecular detection of rifampin resistance and spoligotyping of Mycobacterium tuberculosis. J Clin Microbiol. 2003;41:1101–1108. doi: 10.1128/JCM.41.3.1101-1108.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez A.N., Ribeiro-Alves M., Sarno E.N., Moraes M.O. Evaluation of qPCR-Based assays for leprosy diagnosis directly in clinical specimens. PLoS Negl Trop Dis. 2011;5 doi: 10.1371/journal.pntd.0001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truman R.W., Andrews P.K., Robbins N.Y., Adams L.B., Krahenbuhl J.L., Gillis T.P. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis. 2008;2 doi: 10.1371/journal.pntd.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woods S.A., Cole S.T. A rapid method for the detection of potentially viable Mycobacterium leprae in human biopsies: a novel application of PCR. FEMS Microbiol Lett. 1989;65:305–309. doi: 10.1111/j.1574-6968.1989.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 41.Avanzi C., Lécorché E., Rakotomalala F.A., et al. Population genomics of Mycobacterium leprae reveals a new genotype in Madagascar and the Comoros. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai J., Chen Y., Lauzardo M. Web-accessible database of hsp65 sequences from Mycobacterium reference strains. J Clin Microbiol. 2011;49:2296–2303. doi: 10.1128/JCM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jensen R.W., Rivest J., Li W., Vissa V. DNA fingerprinting of Mycobacterium leprae strains using Variable number tandem repeat (VNTR) - fragment length analysis (FLA) J Vis Exp. 2011 doi: 10.3791/3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kapopoulou A., Lew J.M., Cole S.T. The MycoBrowser portal: a comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis. 2011;91:8–13. doi: 10.1016/j.tube.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 45.Supply P., Lesjean S., Savine E., Kremer K., van Soolingen D., Locht C. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J Clin Microbiol. 2001;39:3563–3571. doi: 10.1128/JCM.39.10.3563-3571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World Health Organization . 2021. Towards zero leprosy. Global leprosy (Hansen's Disease) strategy 2021–2030. [Google Scholar]
- 47.Chauffour A., Morel F., Reibel F., et al. A systematic review of Mycobacterium leprae DNA gyrase mutations and their impact on fluoroquinolone resistance. Clin Microbiol Infect. 2021;27:1601–1612. doi: 10.1016/j.cmi.2021.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Ajileye A., Alvarez N., Merker M., et al. Some synonymous and nonsynonymous gyrA mutations in Mycobacterium tuberculosis lead to systematic false-positive fluoroquinolone resistance results with the Hain GenoType MTBDRsl assays. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.02169-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thibault V.C., Grayon M., Boschiroli M.L., et al. Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of Mycobacterium avium subsp. paratuberculosis isolates. J Clin Microbiol. 2008;46:4091–4094. doi: 10.1128/JCM.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ablordey A., Swings J., Hubans C., et al. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J Clin Microbiol. 2005;43:1546–1551. doi: 10.1128/JCM.43.4.1546-1551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole S.T., Eiglmeier K., Parkhill J., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 52.Williams D.L., Gillis T.P. Drug-resistant leprosy: monitoring and current status. Lepr Rev. 2012;83:269–281. [PubMed] [Google Scholar]
- 53.Kemper K., Krijgsman O., Kong X., et al. BRAFV600E kinase domain duplication identified in therapy-refractory melanoma patient-derived xenografts. Cell Rep. 2016;16:263–277. doi: 10.1016/j.celrep.2016.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.