Abstract
Four kinds of oxidatively damaged DNA precursors, 8-hydroxydeoxyguanosine 5′-triphosphate (8-OH-dGTP), 2-hydroxydeoxyadenosine 5′-triphosphate (2-OH-dATP), 5-hydroxydeoxycytidine 5′-triphosphate (5-OH-dCTP) and 5-formyldeoxyuridine 5′-triphosphate (5-CHO-dUTP), were employed in in vitro gap-filling reactions of the supF gene conducted by the Escherichia coli DNA polymerase III holoenzyme, and these treated DNAs were transfected into various E.coli strains. When the manipulated DNAs were transfected into the repair-proficient strain, supF mutants were obtained much more frequently by the purine nucleotides than by the pyrimidine nucleotides (2-OH-dATP > 8-OH-dGTP >> 5-OH-dCTP ~ 5-CHO-dUTP). This result is in contrast to our previous observation that these four oxidatively damaged nucleotides induce chromosomal gene mutations with similar frequencies when incorporated directly into E.coli. 2-OH-dATP elicited G→T transversions, indicating the formation of G•2-OH-dATP pairs. These results demonstrate that 2-OH-dATP was highly mutagenic in this assay system containing the in vitro DNA synthesis by the E.coli replicative DNA polymerase, in addition to in the in vivo assay system reported previously. Slight increases in the mutant frequencies were observed when alkA (for 8-OH-dGTP and 2-OH-dATP) and mutY (for 2-OH-dATP) strains were used as hosts. This is the first report that clearly shows the formation of G•2-OH-dATP pairs.
INTRODUCTION
Reactive oxygen species (ROS) appear to be involved in various biological processes, including mutagenesis, carcinogenesis and aging (1,2). ROS modify DNA to form oxidative DNA damage. The formation of the oxidative DNA lesions probably alters the genetic information through their mispairing properties. In cells, various enzymatic systems work to repair DNA lesions. The base excision repair and nucleotide excision repair systems have been shown to be involved in the removal of DNA damage (3–8). Moreover, ROS modify DNA precursors to form oxidatively damaged nucleotides, such as 8-hydroxy-2′-deoxyguanosine 5′-triphosphate (8-OH-dGTP). The MutT protein and its homologue remove 8-OH-dGTP in the nucleotide pools of Escherichia coli and mammalian cells, respectively (9,10). Similar enzymes may be involved in the prevention of the mutations induced by other damaged DNA precursors.
Recently, we treated the four deoxyribonucleosides with Fenton-type reagents (Fe2+-EDTA and Fe2+-NTA) and found that 2-hydroxy-2′-deoxyadenosine, 8-hydroxy-2′-deoxyguanosine, 5-hydroxy-2′-deoxycytidine and 5-formyl-2′-deoxyuridine are formed as major products (11). Thus, the four cognate nucleotides, 2-hydroxy-2′-deoxyadenosine 5′-triphosphate (2-OH-dATP), 8-OH-dGTP, 5-hydroxy-2′-deoxycytidine 5′-triphosphate (5-OH-dCTP) and 5-formyl-2′-deoxyuridine 5′-triphosphate (5-CHO-dUTP), appear to be abundantly formed in the nucleotide pool in cells by the generation of ROS. Moreover, we studied the mutagenic potentials of the four oxidized nucleotides when they were introduced directly into E.coli and showed that they induced chromosomal gene mutations with similar frequencies in vivo (12,13). 2-OH-dATP and 8-OH-dGTP elicit G•C→T•A and A•T→C•G transversions, respectively. 5-OH-dCTP induces G•C→T•A, A•T→C•G and G•C→A•T mutations, while 5-CHO-dUTP elicits G•C→A•T, A•T→G•C and G•C→T•A mutations. In those studies, we proposed the formation of various base pairs involving the modified bases as intermediates to explain the mutations observed. However, no direct evidence has been reported for the putative mispair formation by the E.coli replicative DNA polymerase, DNA polymerase III (pol III), except for the case of 8-OH-dGTP (9). Thus, other experimental systems are required to prove the formation of the mispairs that we proposed.
In this study we prepared a gapped heteroduplex (GHD) plasmid containing the supF gene as a mutagenesis target. The E.coli pol III holoenzyme was employed in in vitro gap-filling reactions, in which an oxidized nucleotide as well as the usual nucleotides were present, in order to mimic the DNA replication in bacterial cells. Similar processes appear to occur in vivo when a damaged nucleotide is incorporated before its removal by a nucleotide pool sanitisation enzyme. The treated plasmid was transfected into repair-proficient E.coli, and mutations in the supF gene were analysed. 2-OH-dATP and 8-OH-dGTP were highly mutagenic in this assay system. 2-OH-dATP and 8-OH-dGTP elicited G→T and A→C transversions, respectively. We did not find evident mutagenic potentials of 5-OH-dCTP and 5-CHO-dUTP in this assay system.
Moreover, we transfected the treated plasmid into various repair-deficient E.coli strains, and observed slight increases in the mutant frequencies when alkA (for 8-OH-dGTP and 2-OH-dATP) and mutY (for 2-OH-dATP) strains were used as hosts.
MATERIALS AND METHODS
Materials
Oxidised deoxyribonucleotides were prepared as described previously (13–15). Unmodified nucleotides for DNA polymerase reactions were from Amersham Pharmacia Biotech. Oligonucleotides were from Hokkaido System Science (Sapporo, Japan) in purified forms. The E.coli DNA polymerase III holoenzyme and the Ligation High Kit were from Toyobo. This pol III holoenzyme was reconstituted by the addition of the β-subunit purified from an overproducing strain (16) to the pol III holoenzyme lacking the β-subunit, which was purified as described (17). The single-stranded DNA binding protein was from Amersham Pharmacia Biotech. EcoRI and HindIII were from Takara.
Bacterial strains
The E.coli strain AB1157 (F–: thr-1, leuB6, thi-1, lacY1, galK2, ara-14, xyl-5, mtl-1, proA2, his-4, argE3, rpsL31, tsx-33, supE44, flaND) was used for mutagenesis experiments. This strain was a gift from Dr Takehiko Nohmi, of the National Institute of Health Sciences, Japan. AB1157-derived, repair-deficient strains, MK603 (mutM), MK609 (mutY), MK611 (mutM/mutY) and MS23 (alkA) were from Dr Yusaku Nakabeppu of Kyushu University. KS40 [lacZ(am), CA7070, lacY1, HsdR, HsdM, Δ(araABC-leu)7679, galU, galK, rpsL, thi, gyrA]/pKY241 was from Dr Tomonari Matsuda, of Kyoto University, and was used as an indicator strain of the supF mutants.
Escherichia coli strain JM105 [endA1, supE, sbcB15, thi, rpsL, Δ(lac-proAB)/F′ (traD36, proAB+, lacIq, lacZΔM15)] was employed for the production of double-stranded (ds) and single-stranded (ss) vectors.
Preparation of the gapped heteroduplex and gap-filling by the DNA polymerase III holoenzyme
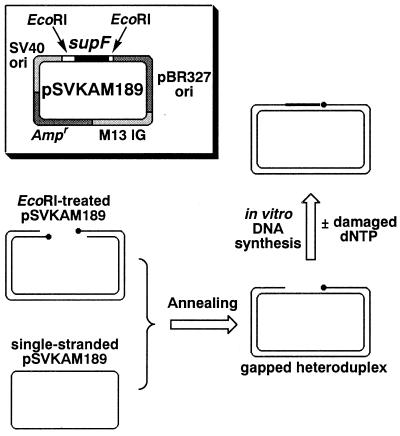
The plasmid pSVKAM189 (Fig. 1) was constructed from pMY189 (18). This new vector, designated as pSVKAM189, contains two EcoRI sites upstream and downstream from the supF gene (Fig. 1). The ss form of pSVKAM189 was obtained by superinfection of pSVKAM189-transfected JM105 with the helper phage VCS-M13 (Stratagene), and was isolated as described (19). This ss DNA was annealed with ds pSVKAM189, which was linearised by treatment with EcoRI (Fig. 1), in a buffer consisting of 6 mM Tris–HCl, 6 mM NaCl and 600 µM EDTA, pH 7.0. The GHD thus acquired was purified by high-performance liquid chromatography using a TSK-gel DNA-NPR column (φ4.6 × 75 mm, Tosoh, Japan).
Figure 1.
Structure of the parental vector, pSVKAM189 (highlighted), and experimental scheme for the gap-filling reactions.
This GHD (2.5 fmol) was used as a primed template in an in vitro DNA synthesis reaction with the E.coli DNA polymerase III holoenzyme. The reaction mixture, containing 20 mM Tris–HCl (pH 7.5), 8 mM MgCl2, 8 mM dithiothreitol, 2% glycerol, 80 µg/ml bovine serum albumin, 1 mM ATP, 1.25 µg of single-stranded DNA binding protein, 50 U of pol III, and the four deoxyribonucleoside triphosphates (20 µM each) with or without an oxidised nucleotide, was incubated at 30°C for 10 min, and the reaction was terminated by the addition of 500 mM EDTA.
The gap-filling reactions were conducted in the presence of [α-33P]dCTP to measure the efficiencies of DNA syntheses.
Mutagenesis experiments
The DNAs obtained after the gap-filling reactions by pol III were transfected into E.coli cells by electroporation using a Gene Pulser Transfection Apparatus with a Pulse Controller (Bio-Rad). An aliquot of the bacterial culture was transferred onto a Luria-Bertani agar plate and was incubated overnight at 37°C to check the transfection efficiency. The remainder of the culture was transferred into Luria-Bertani medium containing ampicillin (30 µg/ml). The amplified plasmid was isolated by the alkaline lysis method (19) from the overnight culture. The mutant frequency (MF) was calculated according to the numbers of white colonies on a Luria-Bertani agar plate containing nalidixic acid (50 µg/ml), ampicillin (150 µg/ml), chloramphenicol (30 µg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (0.008%) and isopropyl-β-d-thiogalactoside (100 µM), and of colonies on an agar plate containing ampicillin and chloramphenicol, as described (18).
The nucleotide sequences of the supF gene were analysed by sequencing using an ABI PRISM Dye Terminator Cycle Sequencing Kit (Perkin-Elmer) and an ABI model 373S DNA sequencer (Perkin-Elmer), as described previously (20).
RESULTS
Mutagenicities of damaged DNA precursors in E.coli cells
To compare the mutagenicities of 2-OH-dATP, 8-OH-dGTP, 5-OH-dCTP and 5-CHO-dUTP, we treated the GHD with DNA polymerase and one of the damaged deoxyribonucleotides in the presence of the four unmodified deoxynucleotides (Fig. 1). The pol III holoenzyme was employed to mimic the DNA replication that occurs in E.coli. The treated plasmid DNAs were then transfected into the repair-proficient AB1157 strain. After mutation fixation in the E.coli cells, the frequencies of mutations in the supF gene were measured with the indicator strain, KS40/pKY241 (21).
When only the four unmodified dNTPs (20 µM each) were present in the reaction mixture, the observed MF was ∼3 × 10–3 (Table 1). The MF increased when either 8-OH-dGTP or 2-OH-dATP was present during the DNA synthesis. The increase in the MF was detected by the addition of 0.04 µM (2 × 10–3 equivalent mole) of the damaged purine deoxyribonucleotides. With the addition of 2 µM of either 8-OH-dGTP or 2-OH-dATP, the MFs reached 1.5 and 2.3 × 10–2, respectively (Table 1). These values were >5-fold over the background (control). 2-OH-dATP appeared to be slightly more mutagenic than 8-OH-dGTP in this assay system, as well as in the in vivo assay system reported previously (12). On the other hand, the oxidatively damaged pyrimidine deoxyribonucleotides, 5-OH-dCTP and 5-CHO-dUTP, did not seem to induce mutations in the supF gene. No clear induction of mutations was observed, even when 20 µM (one equivalent mole) of either damaged pyrimidine nucleotide was present in the pol III reaction mixture. Thus, the mutagenicities of these four oxidatively damaged DNA precursors in this assay system were in the order of 2-OH-dATP > 8-OH-dGTP >> 5-OH-dCTP ~ 5-CHO-dUTP. This was in contrast with our results that the four nucleotides are similarly mutagenic when they are incorporated directly into E.coli cells (12,13).
Table 1. Frequency of supF mutants induced by damaged nucleotides in AB1157 strain.
| Damaged nucleotide addeda | Mutant frequency (× 10-3)b |
|---|---|
| None | 2.8 0.4 (1.0) |
| 0.04 µM 8-OH-dGTP | 4.4 1.8 (1.6) |
| 2.0 µM 8-OH-dGTP | 14.5 4.3 (5.2) |
| 0.04 µM 2-OH-dATP | 6.5 1.4 (2.3) |
| 2.0 µM 2-OH-dATP | 23.2 6.3 (8.3) |
| 2.0 µM 5-OH-dCTP | 0.2 0.1 (0.1) |
| 20 µM 5-OH-dCTP | 3.7 1.3 (1.3) |
| 2.0 µM 5-CHO-dUTP | 2.6 1.2 (0.9) |
| 20 µM 5-CHO-dUTP | 2.0 1.1 (0.7) |
aThe four deoxynucleotides (20 µM each) were included in the reaction mixtures.
bThe values represent the average of three or four independent experiments with standard error. Relative mutant frequencies are shown in parentheses.
We measured incorporation of radio-labeled dCTP into DNA in the presence of the other unmodified nucleotides, with and without 2 µM of an oxidatively damaged DNA precursor. The presence of the damaged nucleotides did not inhibit the gap-filling reactions.
When 2 µM of 5-OH-dCTP was present in the reaction mixture, the observed MF was 0.2 × 10–3, lower than the control (Table 1). A possible explanation is that 5-OH-dCTP inhibited the DNA synthesis. However, as described above, we did not observe the inhibition by 5-OH-dCTP. Alternatively, the presence of 5-hydroxycytosine may elicit DNA repair and, thus, MF may be reduced. However, this assumption is probably invalid because 20 µM of 5-OH-dCTP induced mutations in a level similar to the control. Further studies could resolve this phenomenon.
Mutation spectra of 2-OH-dATP and 8-OH-dGTP in E.coli
We analysed the sequences of the supF genes in the mutants induced by 8-OH-dGTP and 2-OH-dATP. We isolated the mutants obtained with the 2 µM experiments, because the MFs were >5-fold over the background. We isolated 43 colonies obtained in 29 separate transfection experiments (from 19 polymerase reactions) of plasmid treated with 2-OH-dATP. In the case of 8-OH-dGTP, 10 colonies were selected in six separate transfection experiments (from six polymerase reactions). We observed several single mutants (plasmids) contained multiple mutations. Five plasmids obtained in the 2-OH-dATP experiments possessed double substitutions. Three plasmids and one plasmid obtained by the 8-OH-dGTP treatment contained double and triple, respectively, substitutions. These phenomena may be due to independent misincorporation by pol III because high concentration (2 µM) of damaged nucleotides were present in the pol III reactions. Because some of the colonies were isolated from a single transfection experiment, the same mutant plasmid may be included in the analysis. We also analysed 12 colonies obtained in eight separate transfection experiments (from three polymerase reactions) of plasmid treated with 0.04 µM of 2-OH-dATP, and 11 colonies obtained in eight separate transfection experiments (from five polymerase reactions) of plasmid treated without any damaged nucleotide, for comparison.
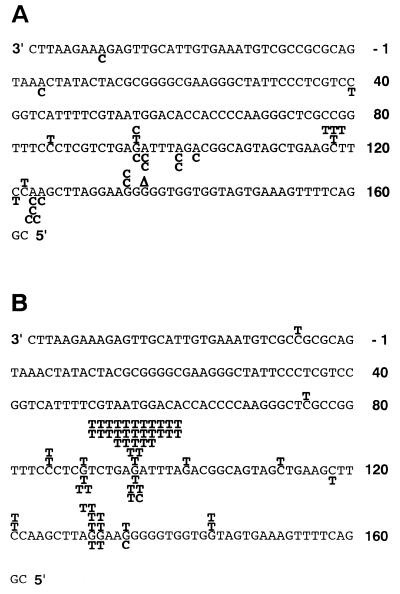
As shown in Figure 2 and Table 2, 8-OH-dGTP elicited A→C mutations, as also found in previous reports (9,12,15,22). It is very probable that this type of mutation was produced by the formation of A•8-OH-dGTP pairs. On the other hand, 2-OH-dATP induced G→T transversions, indicating that pol III inserted the oxidised dATP opposite G residues in the template DNA in vitro. This result supports our proposal that G•C→T•A mutations were induced by 2-OH-dATP in vivo, due to the formation of G•2-OH-dATP pairs (12). A major mutational hotspot was observed for 2 µM of 2-OH-dATP (at position 95, Fig. 2B). This position corresponds to the first base of the anticodon (positions 95–97) of the tRNA encoded in the supF gene. It has been reported that mutations in various positions (more than 90 sites) of the supF gene inactivate the function of the gene, thus, can be detected (18 and references therein). Moreover, the positions of hotspots observed differ depending on the kind of treatment. For example, 2-chloroacetaldehyde induces mutations at positions 85, 121 and 122 (according to the numbering system in Fig. 2) of the gene (18). In addition, no major hotspot was found in the case of 8-OH-dGTP, although an A residue is present in the anticodon position. Thus, the formation of a hotspot at position 95 by 2-OH-dATP could be interpreted as the result of preferential incorporation of 2-OH-dATP opposite G at this site by the pol III holoenzyme. When the supF mutants obtained with 0.04 µM of 2-OH-dATP were analysed, the mutations observed frequently were also G→T transversions (Table 2). The ratio of the G→T transversions at position 95 was less than that for the 2 µM of 2-OH-dATP experiment, although limited number of colonies were sequenced (Fig. 2B).
Figure 2.
The overall distribution of the point mutations detected in the supF gene. (A) The mutations obtained with the control experiments and those induced by 2 µM 8-OH-dGTP are shown above and below the sequence, respectively. The symbol Δ represents the deletion of either of the G residues located at positions 134–138. (B) The mutations induced by 2 µM 2-OH-dATP and 0.04 µM 2-OH-dATP are shown above and below the sequence, respectively. The sequence of the ss moiety of the GHD vector is shown. The anticodon corresponds to positions 95–97.
Table 2. Spectra of mutations induced by oxidatively damaged nucleotides.
| Mutations | Damaged nucleotides added | |||
|---|---|---|---|---|
| None | 0.04 µM 2-OH-dATP | 2 µM 2-OH-dATP | 2 µM 8-OH-dGTP | |
| C→T | 6 | 1 | 7 | 2 |
| G→T | 1 | 9 | 41 | 0 |
| G→C | 3 | 2 | 0 | 1 |
| A→C | 0 | 0 | 0 | 12 |
| G deletion | 1 | 0 | 0 | 0 |
| Total | 11 | 12 | 48 | 15 |
Mutagenicities in repair-deficient cells
Next we transfected the plasmid DNAs treated with pol III into various E.coli strains which one or two DNA repair genes are deficient. The mutM/mutY, mutM, mutY and alkA strains which are derived from AB1157 were employed as host strains. We used the DNAs which were treated in the presence of 0.04 µM of 8-OH-dGTP or 2-OH-dATP, or 20 µM of 5-OH-dCTP or 5-CHO-dUTP. We observed the control (no damaged nucleotide) MFs in these repair-deficient strains (in particular, in the mutM strain) were higher than in the AB1157 strain (Tables 1 and 3). These results may indicate that the substrates of MutM, MutY and AlkA were formed spontaneously and/or during the preparation of the GHD. In order to remove these effects, we calculated MF induced by a damaged nucleotide by subtracting the MF of the control.
Table 3. Frequency of supF mutants induced by damaged nucleotides in repair-deficient strains.
| Damaged nucleotide | Mutant frequency (× 10–3)b | |||
|---|---|---|---|---|
| addeda | mutM/mutY | mutM | mutY | alkA |
| None | 5.3 1.0 | 12.6 3.7 | 5.6 1.9 | 5.5 1.8 |
| 0.04 µM 8-OH-dGTP | 5.5 1.8 | 14.9 2.5 | 5.0 2.0 | 10.0 4.0 |
| 2.0 µM 8-OH-dGTP | 7.9 0.7 | NDc | 8.7 1.5 | ND |
| 0.04 µM 2-OH-dATP | 11.5 1.2 | 17.0 2.8 | 13.9 4.3 | 15.2 3.1 |
| 20 µM 5-OH-dCTP | 5.3 1.8 | ND | ND | 4.9 2.0 |
| 20 µM 5-CHO-dUTP | 5.4 1.0 | ND | ND | 5.8 3.1 |
aThe four deoxynucleotides (20 µM each) were included in the reaction mixtures.
bThe values represent the average of two to seven independent experiments with standard error.
cNot determined.
When the DNA treated with 0.04 µM 8-OH-dGTP was transfected into the mutM/mutY strain, the MF was similar to that of the control (no 8-OH-dGTP) (Table 3): 8-OH-dGTP did not elicit the mutations in this strain. Similarly, no evident mutation induction by 8-OH-dGTP was observed with the mutY strain. This effect was confirmed by using the DNA treated with 2 µM 8-OH-dGTP. The lack of the MutM protein did not affect the induced MF. These results were in agreement with the previous report by Pavlov et al. in which they observed the decrease in the MF elicited by 8-OH-dGTP when a mutM/mutY strain was used (22). The slight increase in the MF induced by 8-OH-dGTP was observed when the alkA strain was used as a host (Table 3).
Similar experiments were carried out for 2-OH-dATP. When the DNA treated in the presence of 0.04 µM 2-OH-dATP was transfected into the mutM/mutY strain, the elicited MF was slightly increased (Table 3). The mutagenesis experiments were then carried out with the mutM and mutY strains. When the mutY strain was employed, the induced MF was increased. However, the MF was not increased in the mutM strain. When the alkA strain was used as a host, the increase in the MF elicited by 2-OH-dATP was observed.
On the other hand, the MFs elicited by 5-OH-dCTP or 5-CHO-dUTP were increased in neither mutM/mutY nor alkA strains (Table 3).
DISCUSSION
It is an accepted concept that oxidative DNA lesions are formed through the incorporation of an oxidatively damaged DNA precursor by a DNA polymerase(s), as well as through the direct oxidation of a residue in DNA. In fact, it was shown that the two pathways contribute almost equally to the formation of 8-hydroxyguanine (8-OH-Gua) in DNA (23). Moreover, the presence of the MutT and MTH1 proteins, which hydrolyse the mutagenic nucleotide 8-OH-dGTP, indicates that the prevention of its incorporation into DNA is important in organisms (9,10).
The oxidatively damaged nucleotides, 8-OH-dGTP, 2-OH-dATP, 5-OH-dCTP and 5-CHO-dUTP, seem to be formed in the nucleotide pool because the cognate nucleosides are formed upon treatment of the four deoxynucleosides with Fenton-type reagents (11). Moreover, it was reported that the amounts of dATP, dCTP and dTTP in prokaryotic and eukaryotic cells were several-fold higher than that of dGTP (24–26), suggesting that other damaged nucleotides may be more abundant than 8-OH-dGTP. Thus, it is important to investigate the mutagenicities of other oxidatively damaged nucleotides.
In this study, we observed that 2-OH-dATP induced mutations slightly more frequently than 8-OH-dGTP (Table 1). This result implies that the mutagenic potential of 2-OH-dATP is as important as that of 8-OH-dGTP, when present in the nucleotide pool. We previously reported that 2-OH-dATP and 8-OH-dGTP induced chromosomal gene mutations with similar frequencies in vivo when the two nucleotides were introduced directly into E.coli (12). In our previous experiment, some of the nucleotides incorporated into cells were expected to be removed by the MutT or MutT-like enzyme, if present, and the remaining damaged nucleotides would be inserted into the DNA in E.coli cells. On the other hand, the oxidised nucleotides added into the reaction mixtures were utilised by the DNA polymerase III holoenzyme, the replicative polymerase of E.coli, without removal by the MutT-like enzyme (this study). These results predict that 2-OH-dATP is hydrolysed by an E.coli MutT-like sanitisation enzyme, because 2-OH-dATP was as mutagenic as 8-OH-dGTP in both in vitro and in vivo experiments. We recently found that 2-OH-dATP is not a substrate for the E.coli MutT protein although the human MutT homologue, MTH1, hydrolyses 2-OH-dATP more efficiently than 8-OH-dGTP (27). Thus, another protein will be involved in the removal of 2-OH-dATP from the nucleotide pool of bacteria. On the other hand, no clear mutagenic effects of 5-OH-dCTP and 5-CHO-dUTP were found (Table 1). In our previous study, these damaged nucleotides were found to be as mutagenic as 8-OH-dGTP when introduced directly into E.coli (13). This discrepancy may indicate that the hydrolysing activity specific for 5-OH-dCTP and 5-CHO-dUTP is weak, if present, in bacteria. Alternatively, effects other than mispairing properties, such as the inhibition of normal nucleotide/nucleoside metabolism, may be included in our previous results. Another explanation is that E.coli repair proteins remove these oxidised pyrimidines in a plasmid (this study) more efficiently than in chromosomal DNA (our previous study). However, this explanation seems unlikely because 2 or 20 µM of 5-OH-dCTP and 5-CHO-dUTP did not induce mutations evidently but 0.04 µM of 2-OH-dATP and 8-OH-dGTP could elicit mutations (Table 1). There appears to be qualitative difference in mutagenic potentials of the two oxidised pyrimidine nucleotides and the two damaged purine nucleotides. Further studies will resolve these interesting discrepancies.
2-OH-dATP induced G→T transversions in the supF gene (Table 2). This finding indicates that 2-OH-dATP was incorporated opposite G by pol III, and then dTTP was inserted opposite the incorporated 2-hydroxyadenine (2-OH-Ade) residue during the next round of replication (figure 3 in ref. 12). This conclusion is in contrast to our previous finding that the mammalian DNA polymerase α, another replicative DNA polymerase, inserts 2-OH-dATP opposite C residues in DNA (14). Based on our previous finding, 2-OH-dATP will elicit G•C→A•T (C→T) transitions in mammalian cells. These results suggest that misincorporation of 2-OH-dATP is DNA polymerase-specific. Interestingly, misincorporation of nucleotides opposite 2-OH-Ade in DNA also appears to depend on DNA polymerases. dCTP and dGTP are the main nucleotides incorporated opposite 2-OH-Ade by mammalian DNA polymerases α and β, and the Klenow fragment of E.coli DNA polymerase I, respectively (28). Moreover, an A→C transversion is one type of mutation induced by 2-OH-Ade in plasmid vectors in E.coli (29), whereas this type of mutation is not elicited in simian cells (30). Although the reasons for these observations are unclear, the presence of the two tautomers (2-hydroxy and 1,2-dihydro-2-oxo isomers) and of syn- and anti-conformers probably affects the base-pairing property of 2-OH-Ade (31–33). 2-OH-Ade may be able to pair with any unmodified base, in contrast to 8-OH-Gua (14,34).
The MutM glycosylase (FPG protein) cleaves the strand with oxidatively damaged purine bases (8-OH-Gua and formamidopyrimidine bases) (35–38) while the MutY glycosylase removes Ade residues opposite 8-OH-Gua (39–41). We carried out the mutagenesis experiments with the mutM/mutY, mutM and mutY strains. The MF induced by 8-OH-dGTP in the mutM strain was similar to that in the repair-proficient (AB1157) strain, and that in the mutM/mutY or mutY strain was lower than in the AB1157 strain (Table 3). Pavlov et al. reported similar results using mutM/mutY and mutM strains although they did not use a single mutY mutant strain (22). The loss of MutM may not affect the frequency of A→C mutations caused by the incorporation of 8-OH-dGTP opposite A, because 8-OH-Gua residues opposite A are the poorest substrates of the MutM protein (36,38). MutY appears to be involved in the fixation of the A→C mutations through the removal of A residues in the original DNA strand. This mutation-fixation effect of MutY was also reported by Vidmar and Cupples (42).
As shown in Table 3, the loss of MutM did not increase the MF elicited by 2-OH-dATP. Thus, MutM does not appear to excise 2-OH-Ade from DNA. On the other hand, the induced MF was higher in the mutY strain than in the AB1157 strain, suggesting that this enzyme may be involved in the repair of 2-OH-Ade. However, this result should be interpreted with care, because the degree of the increase in MF was very slight and because the basal MF was higher in the mutY strain than in the AB1157 strain.
We found that the MFs induced by 8-OH-dGTP and 2-OH-dATP were increased in the alkA strain (Table 3). The AlkA protein (3-methyladenine glycosylase II) is known to have broad substrate specificity (43–53). Thus, our results suggest that this enzyme may be involved in the prevention of the mutagenesis by the two damaged purine deoxynucleotides in vivo. It was reported that the AlkA protein removes 5-formyluracil in DNA (43–45). However, clear increase in the MF induced by 5-CHO-dUTP was not observed even in the alkA strain (Table 3).
One of our major objectives was to compare the mutagenic potentials of the four oxidized nucleotides when E.coli pol III conducts DNA synthesis in vitro, and those when these nucleotides are introduced directly into E.coli cells (our previous experiments; 12,13). It should be noted that the MutT-like activity is an important factor in the determination of the mutagenicities of damaged nucleotides in the in vivo method. We observed that the mutagenic potential of 2-OH-dATP was similar to that of 8-OH-dGTP in both the present (Table 1) and previous (12) studies. Thus, the existence of a hydrolysing activity specific for 2-OH-dATP is expected. On the other hand, prokaryotic cells may not possess sanitisation enzymes specific for 5-OH-dCTP and 5-CHO-dUTP, because these pyrimidine nucleotides did not exhibit mutagenicities in this study (Table 1).
The second major objective was to determine the mutation spectra elicited by the oxidized nucleotides. Since the chromosomal gene (in ds DNA) is the mutagenesis target in the in vivo method, we cannot show which base is opposite from the inserted damaged nucleotide. On the other hand, we could demonstrate that 2-OH-dATP was inserted opposite G in the gap-filling reactions in this study. Thus, the approach described in this study will effectively complement the in vivo method.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Takehiko Nohmi and Yusaku Nakabeppu for providing E.coli strains. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Ames B.N. (1983) Science, 221, 1256–1264. [DOI] [PubMed] [Google Scholar]
- 2.Harman D. (1981) Proc. Natl Acad. Sci. USA, 78, 7124–7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sancar A. (1996) Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 4.Demple B . and . Harrison,L (1994) Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 5.Krokan H.E., Standal,R. and Slupphaug,G. (1997) Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace S.S. (1998) Radiat. Res., 150, S60–S79. [PubMed] [Google Scholar]
- 7.Laval J., Jurado,J., Saparbaev,M. and Sidorkina,O. (1998) Mutat. Res., 402, 93–102. [DOI] [PubMed] [Google Scholar]
- 8.Croteau D.L. and Bohr,V.A. (1997) J. Biol. Chem., 272, 25409–25412. [DOI] [PubMed] [Google Scholar]
- 9.Maki H. and Sekiguchi,M. (1992) Nature, 355, 273–275. [DOI] [PubMed] [Google Scholar]
- 10.Mo J.-Y., Maki,H. and Sekiguchi,M. (1992) Proc. Natl Acad. Sci. USA, 89, 11021–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murata-Kamiya N., Kamiya,H., Muraoka,M., Kaji,H. and Kasai,H. (1997) J. Radiat. Res., 38, 121–131. [DOI] [PubMed] [Google Scholar]
- 12.Inoue M., Kamiya,H., Fujikawa,K., Ootsuyama,Y., Murata-Kamiya,N., Osaki,T., Yasumoto,K. and Kasai,H. (1998) J. Biol. Chem., 273, 11069–11074. [DOI] [PubMed] [Google Scholar]
- 13.Fujikawa K., Kamiya,H. and Kasai,H. (1998) Nucleic Acids Res., 26, 4582–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamiya H. and Kasai,H. (1995) J. Biol. Chem., 270, 19446–19450. [DOI] [PubMed] [Google Scholar]
- 15.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 16.Johanson K.O., Haynes,T.E. and McHenry,C.S. (1986) J. Biol. Chem., 261, 11460–11465. [PubMed] [Google Scholar]
- 17.Maki H., Maki,S. and Kornberg,A. (1988) J. Biol. Chem., 263, 6570–6578. [PubMed] [Google Scholar]
- 18.Matsuda T., Yagi,T., Kawanishi,M., Matsui,S. and Takebe,H. (1995) Carcinogenesis, 16, 2389–2394. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Murata-Kamiya N., Kamiya,H., Kaji,H. and Kasai,H. (1997) Nucleic Acids Res., 25, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akasaka S., Takimoto,K. and Yamamoto,K. (1992) Mol. Gen. Genet., 235, 173–178. [DOI] [PubMed] [Google Scholar]
- 22.Pavlov Y.I., Minnick,D.T., Izuta,S. and Kunkel,T.A. (1994) Biochemistry, 33, 4695–4701. [DOI] [PubMed] [Google Scholar]
- 23.Tajiri T., Maki,H. and Sekiguchi,M. (1995) Mutat. Res., 336, 257–267. [DOI] [PubMed] [Google Scholar]
- 24.Pato M.L. (1979) J. Bacteriol., 140, 518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leeds J.M., Slabaugh,M.B. and Mathews,C.K. (1985) Mol. Cell. Biol., 5, 3443–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brischwein K., Engelcke,M., Riedinger,H.J. and Probst,H. (1997) Eur. J. Biochem., 244, 286–293. [DOI] [PubMed] [Google Scholar]
- 27.Fujikawa K., Kamiya,H., Yakushiji,H., Fujii,Y., Nakabeppu,Y. and Kasai,H. (1999) J. Biol. Chem., 274, 18201–18205. [DOI] [PubMed] [Google Scholar]
- 28.Kamiya H. and Kasai,H. (1996) FEBS Lett., 391, 113–116. [DOI] [PubMed] [Google Scholar]
- 29.Kamiya H. and Kasai,H. (1997) Nucleic Acids Res., 25, 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiya H. and Kasai,H. (1997) Biochemistry, 36, 11125–11130. [DOI] [PubMed] [Google Scholar]
- 31.Seela J., Wei,C. and Kazimierczuk,Z. (1995) Helv. Chim. Acta, 78, 1843–1854. [Google Scholar]
- 32.Sepiol J., Kazimierczuk,Z. and Shugar,D. (1976) Z. Naturforsch, 31C, 361–370. [Google Scholar]
- 33.Seela J., Mertens,R. and Kazimierczuk,Z. (1992) Helv. Chim. Acta, 75, 2298–2306. [Google Scholar]
- 34.Kamiya H., Ueda,T., Ohgi,T., Matsukage,A. and Kasai,H. (1995) Nucleic Acids Res., 23, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung M.-H., Kasai,H., Jones,D.S., Inoue,H., Ishikawa,H., Ohtsuka,E. and Nishimura,S. (1991) Mutat. Res., 254, 1–12. [DOI] [PubMed] [Google Scholar]
- 36.Tchou J., Kasai,H., Shibutani,S., Chung,M.-H., Laval,J., Grollman,A.P. and Nishimura,S. (1991) Proc. Natl Acad. Sci. USA, 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boiteux S., Gajewski,E., Laval,J. and Dizdaroglu,M. (1992) Biochemistry, 31, 106–110. [DOI] [PubMed] [Google Scholar]
- 38.Castaing B., Geiger,A., Seliger,H., Nehls,P., Laval,J., Zelwer,C. and Boiteux,S. (1993) Nucleic Acids Res., 21, 2899–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaels M.L., Tchou,J., Grollman,A.P. and Miller,J.H. (1992) Biochemistry, 31, 10964–10968. [DOI] [PubMed] [Google Scholar]
- 40.Lu A-L., Tsai-Wu,J.-J. and Cillo,J. (1995) J. Biol. Chem., 270, 23582–23588. [DOI] [PubMed] [Google Scholar]
- 41.Manuel R.C. and Lloyd,R.S. (1997) Biochemistry, 36, 11140–11152. [DOI] [PubMed] [Google Scholar]
- 42.Vidmar J.J. and Cupples,C.G. (1993) Can. J. Microbiol., 39, 892–894. [DOI] [PubMed] [Google Scholar]
- 43.Bjelland S., Birkeland,N.K., Benneche,T., Volden,G. and Seeberg,E. (1994) J. Biol. Chem., 269, 30489–30495. [PubMed] [Google Scholar]
- 44.Masaoka A., Terato,H., Kobayashi,M., Honsho,A., Ohyama,Y. and Ide,H. (1999) J. Biol. Chem., 274, 25136–25143. [DOI] [PubMed] [Google Scholar]
- 45.Terato H., Masaoka,A., Kobayashi,M., Fukushima,S., Ohyama,Y., Yoshida,M. and Ide,H. (1999) J. Biol. Chem., 274, 25144–25150. [DOI] [PubMed] [Google Scholar]
- 46.Nakabeppu Y., Kondo,H. and Sekiguchi,M. (1984) J. Biol. Chem., 259, 13723–13729. [PubMed] [Google Scholar]
- 47.McCarthy T.V., Karran,P. and Lindahl,T. (1984) EMBO J., 3, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saparbaev M. and Laval,J. (1994) Proc. Natl Acad. Sci. USA, 91, 5873–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saparbaev M., Kleibl,K. and Laval,J. (1995) Nucleic Acids Res., 23, 3750–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Habraken Y., Carter,C.A., Sekiguchi,M. and Ludlum,D.B. (1991) Carcinogenesis, 12, 1971–1973. [DOI] [PubMed] [Google Scholar]
- 51.Matijasevic Z., Sekiguchi,M. and Ludlum,D.B. (1992) Proc. Natl Acad. Sci. USA, 89, 9331–9334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mattes W.B., Lee,C.S., Laval,J. and O’Connor,T.R. (1996) Carcinogenesis, 17, 643–648. [DOI] [PubMed] [Google Scholar]
- 53.Matijasevic Z., Stering,A., Niu,T.Q., Austin-Ritchie,P. and Ludlum,D.B. (1996) Carcinogenesis, 17, 2249–2252. [DOI] [PubMed] [Google Scholar]