Abstract
Purpose:
The growing size of public variant repositories prompted us to test the accuracy of pathogenicity prediction of DNA variants using population data alone.
Methods:
Under the a priori assumption that the ratio of the prevalence of variants in healthy population vs that in affected populations form 2 distinct distributions (pathogenic and benign), we used a Bayesian method to assign probability to a variant belonging to either distribution.
Results:
The approach, termed Bayesian prevalence ratio (BayPR), accurately parsed 300 of 313 expertly curated CFTR variants: 284 of 296 pathogenic/likely pathogenic variants in 1 distribution and 16 of 17 benign/likely benign variants in another. BayPR produced an area under the receiver operating characteristic curve of 0.99 for 103 functionally confirmed missense CFTR variants, which is equal to or exceeds 10 commonly used algorithms (area under the receiver operating characteristic curve range 0.54–0.99). Application of BayPR to expertly curated variants in 8 genes associated with 7 Mendelian conditions led to the assignment of a disease-causing probability of ≥80% to 1350 of 1374 (98.3%) pathogenic/likely pathogenic variants and of ≤20% to 22 of 23 (95.7%) benign/likely benign variants.
Conclusion:
Irrespective of the variant type or functional effect, the BayPR approach provides probabilities of pathogenicity for DNA variants responsible for Mendelian disorders using only the variant counts in affected and unaffected population samples.
Keywords: Bayesian analysis, Population frequency, Prevalence ratio, Variant classification, Variant interpretation
Introduction
In an era of rapidly expanding genetic testing, there is a growing challenge for clinical and research laboratories to determine the phenotypic significance of DNA variants. Many algorithms predict disease liability using population data, predictive tools, segregation analysis, novelty, allelic information, and prior reporting in expertly curated locus-specific or confederated databases.1–3 The overall trend has been to combine an ever-increasing number of tools, the most accurate being amalgams of existing prediction algorithms.4,5 This approach is fraught with the problem of circularity because many of the incorporated tools use the same variant features and training sets.6 Indeed, it is unclear whether the continued development of ever more complex algorithms will achieve greater accuracy. We therefore sought to determine if we could improve the predictive potential of 1 individual data element used to predict pathogenicity.
The widely adopted standards and guidelines devised by the American College of Medical Genetics and Genomics (ACMG)/Association of Molecular Pathology integrate population data into their criteria.7 The primary assumption when including population data is that the frequency of a disease-causing variant in a collection of affected individuals should exceed that observed in a sample of individuals without the disease of interest. However, the ACMG/AMP recommended that comparison of variant frequencies in affected and unaffected population samples using odds ratios with a somewhat arbitrary threshold is problematic when applied to Mendelian disorders with different penetrance characteristics. Furthermore, the population criterion becomes less effective as variant frequency decreases; prevalence cannot be meaningfully compared for variants that may be observed once or only a few times in a sample of diagnosed individuals or not observed in a large sample of healthy and presumably unaffected individuals. Finally, the categorical nature of assigning this information to different levels of confidence in terms of pathogenicity does not accommodate the spectrum of phenotypic consequences conferred by allelic heterogeneity.8
Bayesian methods are well suited to address this problem because they can incorporate existing sources of information to infer the likelihood of a very rare or even unobserved event.9 Sources may include orthogonal data from the subjects, prior probabilities informed by previous studies, or pooled information across individuals. Consequently, we developed and tested a population-based approach within a Bayesian framework, termed Bayesian prevalence ratio (BayPR). Using only population data, namely the prevalence of variants in samples of diagnosed and unaffected individuals, we demonstrated that most variants fall into 1 of 2 distributions under the a priori assumption that pathogenic or benign variants would self-sort into separate groups.9 Critically, the sorting would occur irrespective of functional data or other metrics of disease liability. We showed the utility of this approach using the Genome Aggregation Database (gnomAD)10 as the sample of healthy population and a worldwide collection of individuals diagnosed with cystic fibrosis (CF) (Online Mendelian Inheritance in Man 219700) (caused by variation in CFTR) as the sample of population with disease.11 We applied BayPR to expertly curated variants associated with Mendelian disorders with different inheritance patterns to demonstrate the generalizability of our approach.
Materials and Methods
Data sources
All variants were de-identified. For the control data set, aggregated genomic data were downloaded from gnomAD v2.1.1, a publicly available database consisting of 125,748 exome sequences and 15,708 whole genome sequences from unrelated individuals.10 For the CF (CFTR) data set, CFTR genotype data were provided by the Clinical and Functional Translation of CFTR (CFTR2) (https://cftr2.org) database.11 For the phenylketonuria (PKU; PAH) data set, PAH genotype data were provided by the BIOPKU database.12 For the interstitial lung disease (ABCA3) data set, data related to variants were assembled from individuals with genetic surfactant dysfunction studied in research laboratories at the Washington University School of Medicine and the Johns Hopkins University School of Medicine and from publications.13 For the X-linked adrenoleukodystrophy (ABCD1) data set, data related to ABCD1 variants were obtained from clinical testing in the Johns Hopkins Genomics DNA Diagnostic Laboratory from 2016 to 2020. For the Barth syndrome (TAFAZZIN) data set, data related to TAFAZZIN variants were obtained from the Human Tafazzin Gene Variants Database (a subdatabase of the Barth Syndrome Registry and Repository; https://www.barthsyndrome.org/research/tafazzindatabase.html).14 For the Marfan syndrome (FBN1) data set, variants were identified during clinical testing at various CLIA-approved commercial laboratories from 2006 to 2020. For the Loeys-Dietz syndrome (TGFBR1/TGFBR2) data set, variants were identified during clinical testing at various CLIA-approved commercial laboratories from 2006 to 2020. Further information concerning curation for all populations can be found in the Supplemental Material and Supplemental Figures 1-5. All genetic data (control and disease populations) and disease liability assignments for alleles can be found in the Supplemental Table 1.
Analytic method
An empirical Bayesian approach was used to analyze the counts of the variants in each disease-specific population database separately relative to the control gnomAD population using a 2-component finite mixture model. Each component modeled the number of variants arising from cases (conditional on the total number of observed variants) using a binomial likelihood, placing a beta prior on transformation of the prevalence ratio (detailed information is provided in the Supplemental Material and Supplemental Figures 1-5 section). This approach converts the proportion of variants observed in cases to the prevalence ratio of cases relative to controls. Estimates were obtained by maximizing the marginal likelihood using the expectation-maximization algorithm,15 which was implemented using the optimx package in R (R Foundation for Statistical Computing).16 A grid search was used to assess sensitivity of the starting values, which include the parameters of each mixture component and the mixing fraction for the expectation-maximization algorithm. The model allows for variation in the number of individuals successfully typed for a given variant in each database but does assume a relatively constant ratio of typed individuals in the affected database vs the control database across all variants; variation from this ratio would induce a different prior on the prevalence ratio. The R program used to generate the probabilities can be found on github (https://github.com/melishg/BayPR/). The BayPR method was compared with other predictive algorithms using nonparametric estimations of the receiver operating characteristic curves with the Bamber and Hanley confidence intervals (CIs) for the area under the receiver operating characteristic curve (AUC); the significance levels were adjusted using Sidak’s correction. Heatmaps were generated using the contour command in Stata/IC 15 (StataCorp), which uses thin plate splines for interpolation. For sensitivity and specificity analyses, score thresholds were set to determine whether a variant had a positive or negative result based on the BayPR prediction. For pathogenic/likely pathogenic (P/LP) variants, BayPR scores >90% or >80% were considered true positives. For benign/likely benign (B/LB) variants, Bayesian scores <10% or <20% were considered true negatives. The P/LP and B/LB statuses of variants were determined by independent expert review. Only genes with both P/LP and B/LB variants underwent sensitivity and specificity analysis.
Results
Predicting the disease liability of CFTR variants using prevalence ratios
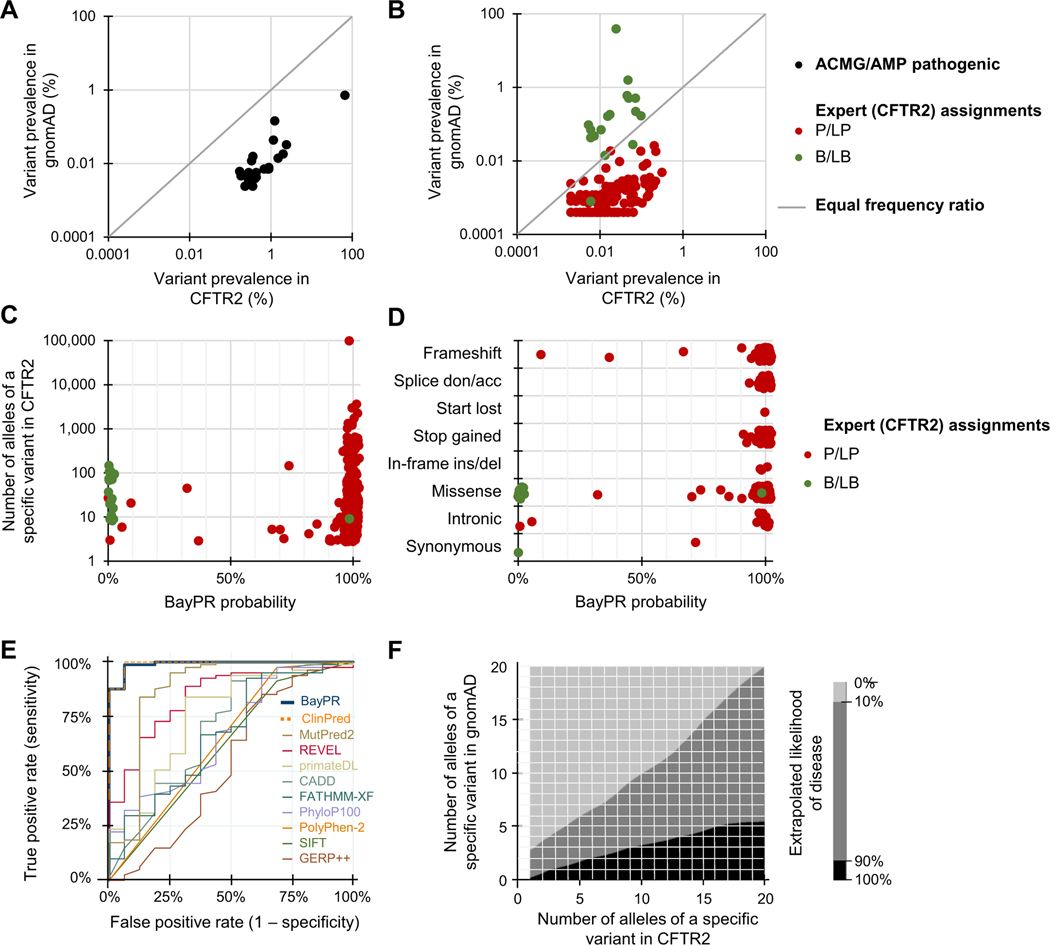
Our underlying premise is based on the differences in the prevalence of pathogenic and benign variants among healthy and affected population samples. For example, plotting 22 CFTR variants (all established to be causal for CF by an ACMG expert committee in 200417) as a ratio of their prevalence in affected and unaffected population samples reveals a clear deviation from neutrality (Figure 1A). Each variant individually generates a relative risk greater than 5.0 with CIs that do not include 1.0, thereby satisfying the ACMG/Association of Molecular Pathology criteria for pathogenicity.7 However, application of relative risk or odds ratios is problematic for many rare pathogenic variants and cannot be used to evaluate benign variants. In contrast, prevalence ratios cluster 152 common and rare expertly curated CFTR variants into distinct P/LP and B/LB groups (Figure 1B).8,11,18 To derive the probability of a variant being in one group or the other, we implemented a 2-component finite mixture model in an empirical Bayesian framework by pooling observed information across the different variants15 (a detailed explanation is provided in the Supplemental Material and Supplemental Figures 1-5). The model generates 95% credible intervals for the prevalence ratios based on observed allele counts and then groups variants according to their similarity to other variants with regard to their observed frequencies. The combining of information generates better estimates of prevalence ratio of each individual variant and allows assessment of the probability that a variant came from the population with higher- or lower-than-average prevalence ratios. The posterior probability derived from a prevalence ratio estimates the likelihood that any variant belongs in 1 of 2 distributions. We first applied our method, termed BayPR, to 313 CFTR variants interpreted using clinical, functional, and segregation information (https://cftr2.org).8,11,18 Parsing of variants based on probabilities using BayPR placed variants into 2 distinct distributions—284 out of the 296 P/LP variants placed in 1 distribution using a probability threshold of >90%, whereas 16 out of the 17 B/LB variants placed in the second distribution on the basis of having <10% probability of being assigned to the first distribution (Figure 1C, Table 1). The probability of a variant being assigned to 1 distribution or the other followed expectations for the functional mechanism. For example, 185 of 188 (98.4%) null variants (eg, splice donor/acceptor, frameshift, and nonsense variants) that can be confidently interpreted as P/LP using the ACMG/Association of Molecular Pathology variant guidelines7 had probabilities that placed them in the disease distribution (Figure 1D). Variants that may allow residual CFTR function (eg, missense and synonymous variants) appeared in both distributions, primarily in accordance with disease liability established from functional studies.8 BayPR generated an AUC of 0.99 (95% CI = 0.98–1.00) for 101 expertly interpreted missense CFTR variants (Figure 1E). This was matched only by ClinPred19 (0.99; 95% CI = 0.97–1.00), which is remarkable given that BayPR is based only on population data, whereas ClinPred uses allele frequency data and prediction scores from 16 algorithms and is trained on variants from the ClinVar database.20 The AUCs of other commonly used predictors ranged from 0.54 to 0.87 (Bonferroni corrected P values compared with those of BayPR ranged from <.001 to .03). To assess the utility of BayPR for low-frequency variants, we generated probabilities for 161 P/LP variants in CFTR2 that have counts of 20 or less. Of these, BayPR assigned >90% probability to 153 variants (95%), indicating that the method performs equally well in the case of lower-frequency variants as it does in the case of variants that arise more frequently. This attribute will be useful in a clinical situation in which a rare variant is identified. For example, if a diagnostic genetic laboratory were to identify a single novel CFTR variant not observed in gnomAD, BayPR generates a probability of 96.3%, placing it in the disease distribution; if the same variant were observed as few as 3 times in gnomAD, then the BayPR-generated probability of belonging to the disease distribution drops to 6.8% (Figure 1F).
Figure 1. Differentiating CFTR variants using BayPR.
A-D. Data are jittered for display purposes only; precise allele counts and BayPR probabilities are provided in Supplemental Table 1. A. A plot of the prevalence ratios (PRs) of 22 of the most common pathogenic CFTR variants (black dots) according to the frequencies in the affected CFTR2 and healthy (gnomAD) individuals shows an expected deviation from neutrality (gray line). The designation of each of these variants as P by the ACMG/AMP remains unchanged since their initial interpretation in 2004. B. A plot of the PRs of 135 P/LP(red dots) and 17 B/LB (green dots) variants interpreted by the CFTR2 that have been observed at least once in gnomAD reveals 2 relatively distinct populations. C. Plot of CFTR variants according to allele count and BayPR probability of being disease-causing. All 313 variants have been interpreted by CFTR2 as CF-causing P/LP(n = 296) or not CF-causing B/LB (n = 17) using clinical and functional criteria. D. BayPR probabilities of 313 CFTR variants stratified according to their functional consequence. E. Receiver operating characteristic curves were plotted for BayPR and 10 in silico prediction algorithms using data from 101 CFTR missense variants. The reference standards are annotations in the CFTR2 database based on clinical and in vitro data. F. Heatmap depicts the estimated probability of a CFTR variant to be associated with disease based on absolute counts of the variants in the CFTR2 and gnomAD databases. The area in black represents >90% probability of being associated with CF, and the area in light gray represents <10% probability. Importantly, this heatmap is only applicable to the CF data presented in this manuscript and does not apply to other genetic disorders. This heatmap was generated using the contour command in Stata/IC 15, which uses thin plate splines for interpolation. acc, acceptor; ACMG, American College of Medical Genetics; AMP, Association of Molecular Pathology; B, benign; BayPR, Bayesian prevalence ratio; CF, cystic fibrosis; CFTR2, Clinical and Functional Translation of CFTR; del, deletion; don, donor; gnomAD, Genome Aggregation Database; ins, insertion; LB, likely benign; LP, likely pathogenic; P, pathogenic.
Table 1.
Sensitivity and specificity analysis of BayPR
| Gene | Variant Interpretation | No. Variants Scored | BayPR Score Thresholda (%) | Positive n (%) | Negative n (%) | Sensitivity (95% CI) | Specificity (95% CI) | BayPR Score Thresholda | Positive n (%) | Negative n (%) | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CFTR | Pathogenic/likely pathogenic | 296 | 90 | 284 (95.9) | 12 (4.1) | 95.9 (93.0–97.9) | 94.1 (71.3–99.9) | 80 | 286 (96.6) | 10 (3.4) | 96.6 (93.9–98.4) | 94.1 (71.3–99.9) |
| Benign/likely benign | 17 | 10 | 1 (5.9) | 16 (94.1) | 20 | 1 (5.9) | 16 (94.1) | |||||
| PAH | Pathogenic/likely pathogenic | 393 | 90 | 319 (81.2) | 74 (18.8) | - | - | 80 | 385 (98.0) | 8 (2.0) | - | - |
| ABCA3 | Pathogenic/likely pathogenic | 225 | 90 | 224 (99.6) | 1 (0.4) | 99.6 (97.5–100.0) | 0.0 (0.0–52.2) | 80 | 225 (100.0) | 0 (0.0) | 100.0 (98.4–100.0) | 100.0 (47.8–100.0) |
| Benign/likely benign | 5 | 10 | 0 (0.0) | 5 (100.0) | 20 | 0 (0.0) | 5 (100.0) | |||||
| FBN1 | Pathogenic/likely pathogenic | 178 | 90 | 178 (100.0) | 0 (0.0) | - | - | 80 | 178 (100.0) | 0 (0.0) | - | - |
| TGFBR1 | Pathogenic/likely pathogenic | 23 | 90 | 2 (8.7) | 21 (91.3) | 80 | 21 (91.3) | 2 (8.7) | ||||
| TGFBR2 | Pathogenic/likely pathogenic | 55 | 90 | 2 (3.6) | 53 (96.4) | - | - | 80 | 54 (98.2) | 1 (1.8) | - | - |
| ABCD1 | Pathogenic/likely pathogenic | 61 | 90 | 61 (100.0) | 0 (0.0) | 100.0 (94.1–100.0) | 100.0 (2.5–100.0) | 80 | 61 (100.0) | 0 (0.0) | 100.0 (94.1–100.0) | 100.0 (2.5–100.0) |
| Benign/likely benign | 1 | 10 | 0 (100.0) | 1 (0.0) | 20 | 0 (100.0) | 1 (0.0) | |||||
| TAFAZZIN | Pathogenic/likely pathogenic | 143 | 90 | 41 (28.7) | 102 (71.3) | - | - | 80 | 140 (97.9) | 3 (2.1) | - | - |
BayPR, Bayesian prevalence ratio; CI, confidence interval.
Score thresholds were set to determine whether a variant had a positive or negative result from the BayPR for the purposes of sensitivity and specificity analysis. For pathogenic/likely pathogenic variants, BayPR scores >90% or >80% were considered true positives. For benign/likely benign variants, Bayesian scores <10% or <20% were considered true negatives. Pathogenic/likely pathogenic and benign/likely benign determinations were made by expert review. Only genes with both pathogenic/likely pathogenic and benign/likely benign underwent sensitivity and specificity analysis.
Predicting disease liability of unassigned CFTR variants
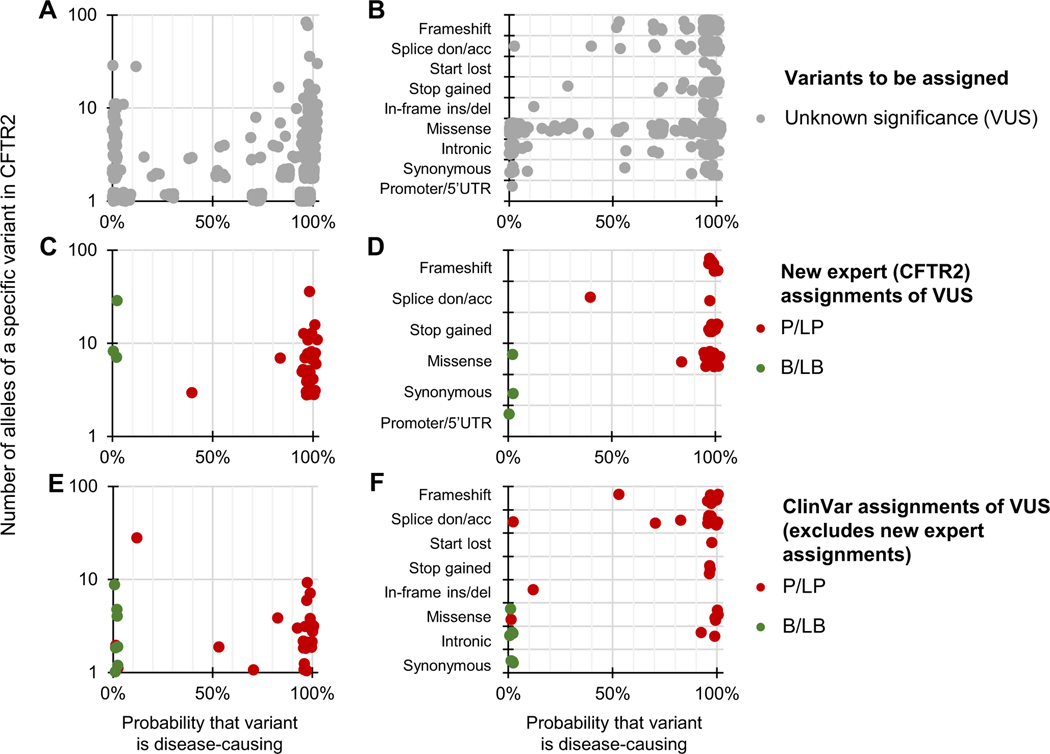
The status of 969 very rare/private CFTR variants had not been assigned at the time of this analysis (ie, variants of unknown significance [VUS]). Application of BayPR assigned probabilities exceeding 90% for 735 of these 969 VUS (75.9%), suggesting that they are likely to be disease-causing (ie, P/LP) variants, whereas 129 (13.3%) had probabilities less than 10%, indicating that they are most likely not disease-causing (ie, B/LB) variants (Figure 2A). To independently evaluate these BayPR assignments, we sorted the variants according to the predicted functional effect. BayPR correctly assigned P/LP probabilities to 378 of the 404 null variants (93.6%) (Figure 2B). To assess variants of varying functional effects (missense and others), 52 randomly selected variants were independently evaluated by the CFTR2 committee using the same criteria applied to all prior annotated variants.11 Of the 52 variants, 35 of 37 variants interpreted as P/LP had probabilities exceeding 90% and all 3 variants annotated as B/LB had 0% probability (Figure 2C). Stratification of newly interpreted variants by predicted functional consequence correlated with probability; 19 of 20 null variants and 16 of 17 missense variants newly assigned as P/LP had BayPR >90% (Figure 2D). We then compared ClinVar assignments of 41 variants that had not been expertly curated but had CF listed as a clinical phenotype, multiple submitters with no conflicts (2-star rating or higher), and a disease annotation of P/LP or B/LB to BayPR assignments (Figure 2E). In total, 28 of 34 variants with P/LP designations in ClinVar had probabilities >90%, and all 7 B/LB variants had probabilities <10%. Stratification by predicted functional effect were consistent with BayPR probabilities for the majority of variants (Figure 2F).
Figure 2. Differentiating CFTR variants of unknown significance with Bayesian prevalence ratio (BayPR).
A-F. Data are jittered for display purposes only; precise allele counts and BayPR probabilities are provided in Supplemental Table 1. A. CFTR variants that had not undergone interpretation by CFTR2 (n = 969; gray dots) cluster primarily into high (>90%; 735 variants) or low (<10%; 129 variants) probabilities. B. BayPR probabilities of unassigned CFTR variants stratified according to their functional consequence. C. A total of 52 unassigned CFTR variants were subsequently and independently assessed for disease liability by CFTR2 using clinical and functional criteria. D. BayPR probabilities of newly-assigned CFTR variants stratified according to their functional consequence. E. CFTR variants that remain without a CFTR2 interpretation and which have P/LP or B/LB interpretations in ClinVar (n = 41) cluster primarily into high (>90%; 28 variants) or low (<10%; 9 variants) probabilities. F. BayPR probabilities of ClinVar-assigned P/LP or B/LB CFTR variants without a CFTR2 annotation parsed by predicted functional effect. acc, acceptor; B, benign; CFTR2, Clinical and Functional Translation of CFTR; del, deletion; don, donor; ins, insertion; LB, likely benign; LP, likely pathogenic; P, pathogenic; UTR, untranslated region; VUS, variant of unknown significance.
Predicting disease liability of variants associated with other Mendelian disorders using BayPR
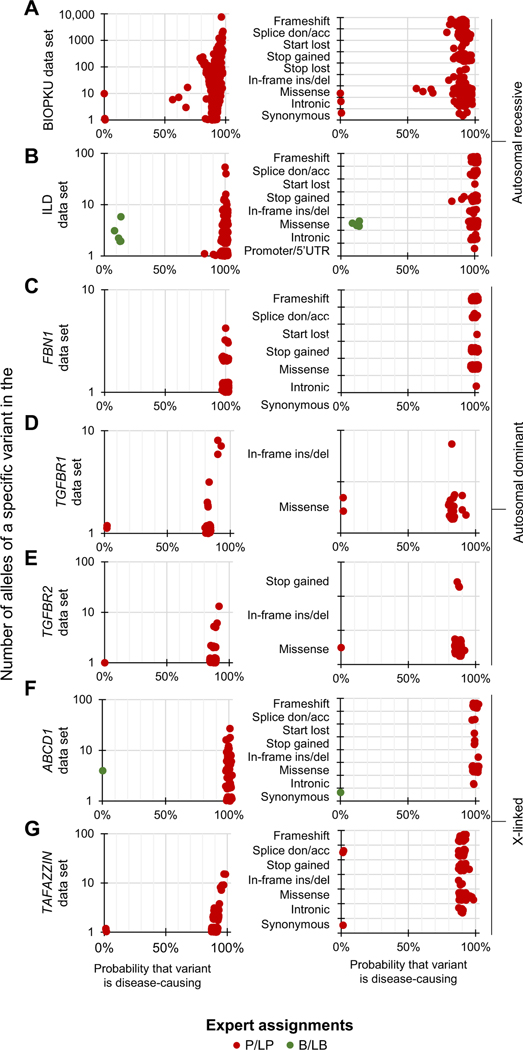
Having established the utility of BayPR for variants in CFTR, we applied this method to 2 other autosomal recessive disorders, namely PKU, caused by defects in PAH (estimated to affect 1 in 23,930 live births globally12), and interstitial lung disease, caused by dysfunction in ABCA3 (estimated to affect 1 in 3100 live births among those of European-descent to 1 in 18,000 live births among those of African-descent in the United States).21 We determined BayPR scores for 726 PAH variants that had been evaluated by an expert panel (BIOPKU).12 The BIOPKU collection differed from the CFTR2 database in that none of the PAH variants had been assigned as B/LB. Despite the absence of known negatives, BayPR assigned high disease probabilities to almost all P/LP variants. Of the 334 PAH variants associated with classic PKU, 329 had probabilities >80%, as did 56 of the 59 mild PKU-causing variants (Figure 3A, left panel). Stratifying variants according to their predicted function further demonstrated that disease assignments were consistent given that 169 of 174 missense variants assigned as P/LP had probabilities >80% (Figure 3A, right panel). Variants in ABCA3 were expertly interpreted using the ACMG/AMP criteria7 (225 P/LP; 5 B/LB; and 102 VUS). The ABCA3 variants distributed into 2 distinct groups: all 225 P/LP variants had probabilities >90%, and all 5 B/LB variants clustered in the nondisease-causing component (Figure 3B, left panel). When sorted according to predicted effect, all 75 P/LP null variants and all 135 missense variants assigned as P/LP had probabilities >90% (Figure 3B, right panel).
Figure 3. Differentiating expert assigned variants in genes associated with recessive, dominant, and X-linked disorders using BayPR.
A-G. Data are jittered for display purposes only; precise allele counts and BayPR probabilities are provided in Supplemental Table 1. Left panel. A total of 393 PAH variants are plotted by allele count and BayPR probabilities with disease assignment of either P or LP (red dots). A. Right panel. Parsing PAH variants by predicted functional effect. B. Left panel. A total of 230 ABCA3 (autosomal recessive interstitial lung disease) variants plotted by allele counts and probabilities with disease assignments of P/LP (red dots) or B/LB(green dots). B. Right panel. ABCA3 variants parsed by predicted effect. C. Left panel. A total of 178 FBN1 variants are plotted by allele count and BayPR probabilities with disease assignment of P/LP. All 178 variants have probabilities >90%. C. Right panel. FBN1 (autosomal dominant Marfan syndrome) variants parsed by predicted effect. (D) Left panel. A total of 23 TGFBR1 (autosomal dominant Loeys-Dietz syndrome) variants assigned as P/LP are plotted by allele count and BayPR probabilities. D. Right panel. TGFBR1 variants parsed by predictive effect. E. Left panel. A total of 54 TGFBR2 (autosomal dominant Loeys-Dietz syndrome) variants assigned as P/LP are plotted by allele count and BayPR probabilities. E. Right panel. TGFBR2 variants parsed by predicted effect. F. Left panel. A total of 61 ABCD1 (X-linked adrenoleukodystrophy) variants previously assigned as P/LP or B/LB are plotted by allele count and BayPR probabilities. F. Right panel. ABCD1 variants parsed by predicted effect. G. Left panel. A total of 143 TAFAZZIN (X-linked Barth syndrome) variants are plotted by allele count and BayPR probabilities, all with previous disease assignment of P/LP. G. Right panel. TAFAZZIN variants parsed by predicted effect. acc, acceptor; B, benign; BayPR, Bayesian prevalence ratio; del, deletion; don, donor; ILD, interstitial lung disease; ins, insertion; LB, likely benign; LP, likely pathogenic; P, pathogenic; UTR, untranslated region; VUS, variant of unknown significance.
We next tested the utility of BayPR for monogenic disorders with different Mendelian inheritance patterns. Marfan syndrome (caused by dysfunction of FBN1; reported prevalence of 1 in 15,400 in Denmark)22 and Loeys-Dietz syndrome (caused by dysfunction of TGFBR1 and TGFBR2; estimated prevalence of <1 in 100,000)23 are autosomal dominant monogenic disorders. We applied the BayPR analysis to 178 FBN1, 23 TGFBR1, and 55 TGFBR2 variants, all of which have been expertly curated as P/LP. All 178 FBN1 variants had probabilities >90% (Figure 3C, left panel) regardless of the predicted effect (Figure 3C, right panel). The 23 P/LP variants in TGFBR1 distributed in such a way that 2 had probabilities >90%, 19 had probabilities between 80% and 90%, and the remaining 2 had probabilities <10% (Figure 3D, left panel). All variants were either missense (n = 22) or in-frame deletions (n = 1) (Figure 3D, right panel). All but 1 of 55 TGFBR2 variants previously assigned as P/LP had probabilities >80% and 2 had >90% (Figure 3E, left and right panels). X-linked adrenoleukodystrophy (X-ALD) and X-linked Barth syndromes are monogenic disorders with an estimated baseline prevalence of 1 in 20,000 to 50,000 and 1 in 300,000 to 400,000, respectively.24,25 All 59 ABCD1 (X-ALD) variants were assigned as P/LP by a single clinical DNA testing laboratory with extensive experience in interpretation of ABCD1 variants26 and had probabilities >90%, whereas the 1 variant previously assigned as B/LB had 0% probability. Parsing by the predicted effect indicated that all 52 missense variants had probabilities >90% (Figure 3F, left and right panels). We analyzed 143 variants in tafazzin (TAFAZZIN) expertly curated as P/LP for Barth syndrome by the Human Tafazzin Gene Variants Database (https://www.barthsyndrome.org/research/tafazzindatabase.html). Although only 41 TAFAZZIN variants (28.7%) had probabilities >90%, 140 (97.9%) had probabilities >80% and 3 (2.1%) had probabilities <10% (Figure 3G, left panel). All 63 missense TAFAZZIN variants had probabilities >80%, whereas 2 of the P/LP variants with <10% probability were predicted to lead to complete loss-of-function (Figure 3G, right panel).
Sensitivity and specificity of BayPR at different probability thresholds and different size data sets
We determined the sensitivity and specificity of BayPR at different thresholds for the 3 genes (CFTR, ABCA3, and ABCD1) that harbored P/LP and B/LB variants (Table 1). For this analysis, true positives were variants annotated as P/ LP that exceeded a probability of >90% or >80%; true negatives were B/LB variants with probabilities <10% or <20%. The sensitivity of BayPR was greater than 95% for all 3 genes and for both combinations of probability. Likewise, specificity was greater than 94% for CFTR and ABCD1 at 10% and 20% probabilities, whereas specificity dramatically improved for ABCA3 at the <20% threshold when compared with the <10% threshold (100% vs 60%). For the remaining disorders, an 80% threshold achieved correct assignment of over 97% of all known P/LP variants. To test how the size of the control data set would affect BayPR probabilities, variants from all 8 genes were reanalyzed using allele counts from a subset (genomes only; ~15,000 individuals) of the full gnomAD data set (exomes and genomes of ~140,000 individuals). For CFTR, a large majority of P/LP variants moved from probabilities >90% in the full control data set to probabilities between 80% and 90% in the genomes-only control data set (Supplemental Table 1). This reduction in probability was primarily driven by rare variants (Supplemental Figure 1). In contrast, for TAFAZZIN and TGFBR2, 102 and 21 variants, respectively, moved from <90% probability in the full control data set to >90% probability in the genomes-only data set. For all other genes analyzed, variability in the control data set size had little effect on the probabilities and predictions of disease liability. Finally, inspection of the 16 P/LP variants with probabilities <50% revealed that 15 had likely explanations for the incongruence—9 occurred in populations that were underrepresented in the affected sample when compared with the gnomAD sample (6 in CFTR; 3 in PAH) and 5 had phenotype or mechanistic discrepancies (3 in TAFAZZIN and 2 in TGFBR1) (Supplemental Table 2). These results indicate that BayPR thresholds of 80% and 20% encompass the vast majority of variants assigned as P/LP and B/LB, respectively, based on the ACMG/Association of Molecular Pathology criteria.
BayPR can inform pathogenicity of variants variably associated with disease
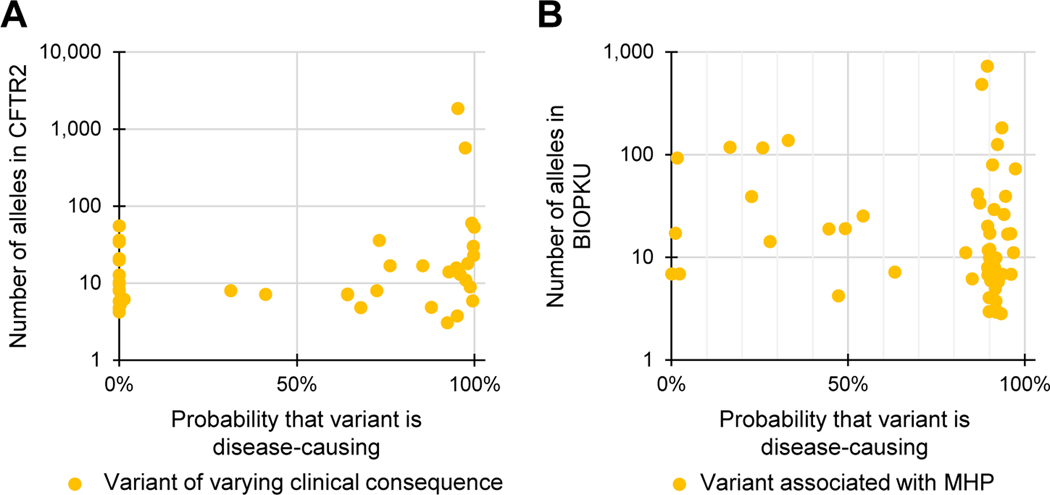
To test if BayPR can differentiate underlying causes of variable disease association, we determined the probabilities of 41 variants of varying clinical consequence (VCC) (https://cftr2.org) and observed that 17 had predicted probabilities >80% and 16 had probabilities <20%, whereas the remaining 8 variants fell between these 2 thresholds (Figure 4A). Ten variants with probabilities <20% were reported to occur in cis with other CFTR variants. The low probabilities indicate that these variants are likely benign when occurring alone but may contribute to pathogenicity when combined with other variants. Thus, variants of this type can occur in individuals with or without disease, thereby explaining their designation as VCC. CFTR variants designated as VCC with probabilities >80% could be benign or could contribute to disease but in tight linkage disequilibrium with a pathogenic variant. We also applied BayPR to variants associated with mild hyper-phenylalaninemia (MHP), which may or may not be clinically recognized. Of the 58 MHP variants, 44 produced probabilities >80%, corroborating their disease association. Only 5 variants had probabilities <20%, with the remaining 9 falling somewhere in between (Figure 4B). These findings reveal that population-based assessment can inform the interpretation of variants that can be variably associated with disease.
Figure 4. Variants associated with variable expressivity have a wide distribution of Bayesian prevalence ratio (BayPR) probabilities.
A, B. Data are jittered for display purposes only; precise allele counts and BayPR probabilities are provided in Supplemental Table 1. A. A total of 41 CFTR variants with disease assignment of varying clinical consequences are plotted by BayPR probabilities and CFTR2 allele count and show a wide distribution. Of those, 16 variants have probabilities <20%, 17 variants have probabilities >80%, and 8 have probabilities between 20% and 80%. B. A total of 58 PAH variants associated with MHP are plotted by BIOPKU allele count and BayPR probabilities and show a wide distribution. Of those, 44 have probabilities >80% and 5 variants have probabilities <10%; the remaining 9 variants fall between 20% and 80% probability of belonging to the disease-causing component. CFTR2, Clinical and Functional Translation of CFTR; MHP, mild hyperphenylaninemia.
Discussion
We show here that a component used to interpret variant pathogenicity—population frequency—has greater utility than previously recognized when used within a Bayesian framework. There are several distinct advantages to a Bayesian approach. First, probabilities can be generated for rare variants not previously observed in the general population. The rarity of such variants renders rigorous interpretation challenging because it is difficult to confirm the variant’s existence in multiple unrelated individuals, obtain adequate clinical information for phenotypic evaluation and correlation, and collect primary tissue samples or cells from a small number of affected individuals for functional assessment. Consequently, many variants interpreted using the ACMG/Association of Molecular Pathology framework remain VUS because of the paucity of data. Application of BayPR offers a way to stratify such variants as shown in Figure 2. Second, BayPR provides a substantial improvement over odds calculations because it capitalizes on the information inherent in the prevalence ratios of other pathologic and benign variants in the same gene. Third, population-based approaches to variant interpretation, such as BayPR, are independent of variant mechanism and can reveal unexpected mechanisms of pathogenicity. For example, the c.371–2A>G variant of TAFAZZIN would be predicted to cause disease (alters +2 nucleotide of splice site), but BayPR generated a probability of <10%. Further evaluation revealed that the variant interrupts a splice site at the start of exon 5, an exon excluded in the prominent RNA isoform of TAFAZZIN in humans.27 Thus, population data can provide an independent evaluation of variants that have functional testing results that are inconsistent with the associated phenotype and of variants not expected to affect the protein itself (eg, synonymous changes) but that are observed in affected individuals. Finally, based on our analysis of 8 genes, we propose that BayPR achieves excellent accuracy at probabilities >80% for pathogenic variants and <20% for benign variants. Variants with probabilities that do not fall between 20% and 80%— although potentially more difficult to annotate and interpret—may represent hypomorphic alleles with varying penetrance and/or expressivity. Identification of such variants may offer insight into the disease spectrum or represent a clinically-distinct entity, such as in the case of MHP.
There are several potential limitations to variant interpretation methods that are based on population data alone. Within the control population, size matters; larger data sets should provide a more accurate estimate of rare variant frequencies among presumably unaffected individuals. Our results support this supposition because BayPR probability predictions for CFTR variants (particularly for rare variants) improved when the full gnomAD data set (exomes and genomes) was used when compared with using genomes alone. Ongoing efforts to expand the size and diversity of healthy and affected population samples should progressively improve the accuracy and utility of population-based approaches to variant interpretation.10,28,29 It is essential to consider the ancestry composition of the affected and healthy populations even if both populations are quite diverse. Imbalance between control and affected population diversity may lead to an overestimation of the pathogenicity of variants found in underrepresented populations, whereas the converse is true for underrepresented populations in the disease population.30 For example, BayPR generated low disease probabilities for 9 pathogenic variants that occur more frequently in populations underrepresented in CFTR and PKU (Supplemental Table 2). Indeed, the prevalence of a disease may be less important than the degree of ascertainment of all affected individuals as was shown for Barth syndrome. Therefore, it is recommended that the potential effects of stratification should be considered before the use of any population-based estimator of pathogenicity,7 including BayPR.
A second consideration is phenotyping. Variants associated with mild presentation or late-onset disease could result in some affected individuals in the healthy control population. Thus, BayPR will be less effective for late-onset conditions unless appropriate age thresholds are applied to the control population. In addition, phenotypes with substantial etiologic heterogeneity owing to multiple causative genes or low heritability may have distorted prevalence ratios. However, we note that BayPR was able to accurately assign pathogenicity to variants in 2 genes responsible for Loeys-Dietz Syndrome. A third issue is applicability of prevalence ratios to de novo variants. Although our method is based on the distribution of transmitted alleles in healthy and affected individuals, BayPR generated accurate probabilities for the 2 X-linked disorders in which de novo variants account for 40% (X-ALD)31 and 13% (Barth Syndrome)14 of cases. A fourth concern was overcounting P/LP variants owing to the presence of related individuals (eg, multiple affected siblings or affected relatives from different generations) in the affected group. Overcounting, particularly of rare variants, can result in false assignment of a benign variant as disease causing because of its biased prevalence ratio. In the US CF population, there are 959 families with 1962 affected twins or siblings among a sample of 30,000 affected individuals,32 representing a potential overcounting of approximately 3.3%. Overcounting could be more problematic for rarer disorders such as Barth Syndrome because a diagnosis is more likely when 2 or more affected siblings are present; however, curation of TAFAZZIN variants requires that variants are only submitted multiple times if they are observed in unrelated individuals (https://www.barthsyndrome.org/research/tafazzindatabase.html). Conversely, a minor excess of alleles caused by the presence of siblings appeared to have no discernable decrease in the accuracy of BayPR for CF or for the rarer Mendelian disorders considered here (interstitial lung disease and Barth syndrome).
Population data provides a powerful tool for variant interpretation as recognized in the criteria developed by the ACMG/AMP and in the accuracy of interpretive algorithms that incorporate population data.7,19 Here we showed that a method based solely on variant counts can perform as good or better than popular interpretive tools using multiple data sources and algorithms (including functional predictions). We interrogated a variety of other predictive tools4,5,19,33–38 and showed that only ClinPred, which incorporates population-level data in a similar manner to our method, rivaled BayPR with regard to sensitivity and specificity. Important limitations to ClinPred and similar tools include the inability to incorporate population data if a variant does not appear in a healthy population sample and the inability to evaluate all variants. In contrast, BayPR generates probabilities for variants that are not present in control samples and for all variants in affected samples, including null variants that can provide independent evidence in situations in which it is challenging to apply the Very Strong strength level for pathogenicity in the ACMG/AMP criterion to assign pathogenicity (eg, nonsense variants that do not elicit nonsense-mediated messenger RNA decay).39 The potential of population-based data used by BayPR emphasizes the importance of disease-specific data sets, such as CFTR2 and BIOPKU, that not only collect variants observed in affected individuals but also collect the allele counts of such variants, especially those suspected to be benign. Although complete ascertainment of a disease population is ideal, we have demonstrated how multiple, current locus-specific databases have sufficient data to enable accurate interpretation of variants by BayPR. Furthermore, clinical and research laboratories could employ BayPR to generate probability scores using their own variant counts for cases in which there is reasonable confidence that the individuals tested have the phenotype of interest. As novel variants are discovered within a disease population, the affected sample can be updated and BayPR probabilities can be recalculated. Although BayPR could be a standalone predictor of pathogenicity, it could also be combined with orthogonal approaches or be incorporated into guidelines such as those developed by the ACMG/Association of Molecular Pathology and serve as a valuable tool in the classification of VUS for which limited clinical or functional information is available.
Supplementary Material
Acknowledgments
This research would not have been possible without the generous data contributions of patient and healthy subject to the Clinical and Functional Translation of CFTR project, the BIOPKU, and Genome Aggregation Database data sets and the Human TAFAZZIN Gene Variants Database.
This research was funded by the following organizations. A National Institutes of Health grant to J.M.C. under grant number R01 HL128475. This study was part of the RD-Connect initiative and was supported by an FP7-HEALTH-2021-INNOVATION-1 EU Grant to N.B. under grant number 305444. L.M.N. was supported by grants from the National Institutes of Health, American Thoracic Society, and Eudowood Foundation. G.R.C. was supported by grants from the National Institute of Diabetes and Digestive and Kidney disease (grant number R01 DK44003) and the Cystic Fibrosis Foundation under grant number CUT-TIN13A1. The funders had no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
Ethics Declaration
All data were de-identified. The Clinical and Functional Translation of CFTR project is acknowledged by the Johns Hopkins University Institutional Review Board (NA_00018599) and does not require institutional review board approval. All other data sets were obtained from publicly available resources and/or from researchers who provided de-identified variant information that meets the National Institutes of Health Exemption 4 (§46.104(d)(ii)) and does not qualify as human subject research.
Web Resources
https://gnomad.broadinstitute.org/
http://www.biopku.org/home/home.asp
https://www.barthsyndrome.org/research/tafazzindatabase.html
Additional Information
The online version of this article (https://doi.org/10.1016/j.gim.2021.08.012) contains supplementary material, which is available to authorized users.
Conflict of Interest
The rest of the authors report no conflicts of interest.
Data Availability
The data sets used for this study are available in the Supplemental Material section. The code and associated documentation generated during this study are available at https://github.com/melishg/BayPR/.
References
- 1.Kobayashi Y, Yang S, Nykamp K, Garcia J, Lincoln SE, Topper SE. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9(1):13. 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niroula A, Vihinen M. Variation interpretation predictors: principles, types, performance, and choice. Hum Mutat. 2016;37(6):579–597. 10.1002/humu.22987. [DOI] [PubMed] [Google Scholar]
- 3.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508(7497):469–476. 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ioannidis NM, Rothstein JH, Pejaver V, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh R, Oak N, Plon SE. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 2017;18(1):225. 10.1186/s13059-017-1353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raraigh KS, Han ST, Davis E, et al. Functional assays are essential for interpretation of missense variants associated with variable expressivity. Am J Hum Genet. 2018;102(6):1062–1077. 10.1016/j.ajhg.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldgar DE, Easton DF, Byrnes GB, et al. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29(11):1265–1272. 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–443. Published correction appears in Nature. 2021;590(7846):E53. 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sosnay PR, Siklosi KR, Van Goor F, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45(10):1160–1167. 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillert A, Anikster Y, Belanger-Quintana A, et al. The genetic land-scape and epidemiology of phenylketonuria. Am J Hum Genet. 2020;107(2):234–250. 10.1016/j.ajhg.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wambach JA, Casey AM, Fishman MP, et al. Genotype-phenotype correlations for infants and children with ABCA3 deficiency. Am J Respir Crit Care Med. 2014;189(12):1538–1543. 10.1164/rccm.201402-0342OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke SL, Bowron A, Gonzalez IL, et al. Barth syndrome. Orphanet J Rare Dis. 2013;8:23. 10.1186/1750-1172-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlin BP, Louis TA. Bayesian Methods for Data Analysis. 3rd ed. Chapman and Hall/CRC Press; 2008. [Google Scholar]
- 16.Nash JC, Varadhan R. Unifying optimization algorithms to aid software system users: optimx for R. J Stat Softw. 2011;43(9):1–14. 10.18637/jss.v043.i09. [DOI] [Google Scholar]
- 17.Watson MS, Cutting GR, Desnick RJ, et al. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6(5):387–391. Published correction appears in Genet Med. 2004;6(6):548. Published correction appears in Genet Med. 2005;7(4):286. 10.1097/01.gim.0000139506.11694.7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma N, Sosnay PR, Ramalho AS, et al. Experimental assessment of splicing variants using expression minigenes and comparison with in silico predictions. Hum Mutat. 2014;35(10):1249–1259. 10.1002/humu.22624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alirezaie N, Kernohan KD, Hartley T, Majewski J, Hocking TD. ClinPred: prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am J Hum Genet. 2018;103(4):474–483. 10.1016/j.ajhg.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wambach JA, Wegner DJ, Depass K, et al. Single ABCA3 mutations increase risk for neonatal respiratory distress syndrome. Pediatrics. 2012;130(6):e1575–e1582. 10.1542/peds.2012-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groth KA, Hove H, Kyhl K, et al. Prevalence, incidence, and age at diagnosis in Marfan syndrome. Orphanet J Rare Dis. 2015;10:153. 10.1186/s13023-015-0369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loughborough WW, Minhas KS, Rodrigues JCL, et al. Cardiovascular manifestations and complications of Loeys–Dietz syndrome: CT and MR imaging findings. Radiographics. 2018;38(1):275–286. 10.1148/rg.2018170120. [DOI] [PubMed] [Google Scholar]
- 24.Bezman L, Moser AB, Raymond GV, et al. Adrenoleukodystrophy: incidence, new mutation rate, and results of extended family screening. Ann Neurol. 2001;49(4):512–517. [PubMed] [Google Scholar]
- 25.Miller PC, Ren M, Schlame M, Toth MJ, Phoon CKL. A Bayesian analysis to determine the prevalence of Barth syndrome in the pediatric population. J Pediatr. 2020;217:139–144. 10.1016/j.jpeds.2019.09.074. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Busin R, Reeves C, et al. X-linked adrenoleukodystrophy: ABCD1 de novo mutations and mosaicism. Mol Genet Metab. 2011;104(1–2):160–166. 10.1016/j.ymgme.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y, Zhang S, Malhotra A, et al. Characterization of tafazzin splice variants from humans and fruit flies. J Biol Chem. 2009;284(42):29230–29239. 10.1074/jbc.M109.016642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.All of Us Research Program Investigators, Denny JC, Rutter JL, et al. The “All of Us” Research Program. N Engl J Med. 2019;381(7):668–676. 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manrai AK, Funke BH, Rehm HL, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375(7):655–665. 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp S, Pujol A, Waterham HR, et al. ABCD1 mutations and the X-linked adrenoleukodystrophy mutation database: role in diagnosis and clinical correlations. Hum Mutat. 2001;18(6):499–515. 10.1002/humu.1227. [DOI] [PubMed] [Google Scholar]
- 32.Wright FA, Strug LJ, Doshi VK, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–546. 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34(3):511–513. 10.1093/bioinformatics/btx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundaram L, Gao H, Padigepati SR, et al. Predicting the clinical impact of human mutation with deep neural networks. Nat Genet. 2018;50(8):1161–1170. Published correction appears in Nat Genet. 2019;51(2):364. 10.1038/s41588-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–121. 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP . PLoS Comput Biol. 2010;6(12):e1001025. 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abou Tayoun AN, Pesaran T, DiStefano MT, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517–1524. 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used for this study are available in the Supplemental Material section. The code and associated documentation generated during this study are available at https://github.com/melishg/BayPR/.