Abstract
Magnesium ions play important roles in the structure and function of nucleic acids. Whereas the tertiary folding of RNA often requires magnesium ions binding to tight places where phosphates are clustered, the molecular basis of the interactions of magnesium ions with RNA helical regions is less well understood. We have refined the crystal structures of four decamer oligonucleotides, d(ACCGGCCGGT), r(GCG)d(TATACGC), r(GC)d(GTATACGC) and r(G)d(GCGTATACGC) with bound hexahydrated magnesium ions at high resolution. The structures reveal that A-form nucleic acid has characteristic [Mg(H2O)6]2+ binding modes. One mode has the ion binding in the deep major groove of a GpN step at the O6/N7 sites of guanine bases via hydrogen bonds. Our crystallographic observations are consistent with the recent NMR observations that in solution [Co(NH3)6]3+, a model ion of [Mg(H2O)6]2+, binds in an identical manner. The other mode involves the binding of the ion to phosphates, bridging across the outer mouth of the narrow major groove. These [Mg(H2O)6]2+ ions are found at the most negative electrostatic potential regions of A-form duplexes. We propose that these two binding modes are important in the global charge neutralization, and therefore stability, of A-form duplexes.
INTRODUCTION
DNA and RNA are highly negatively-charged polyelectrolytes, due to the negative charges associated with their phosphate groups. In solution, the charge neutralization of nucleic acids is presumably achieved through the interactions with positively-charged metal ions. Among many metal ions, magnesium ions play a particularly important role in the structure and function of nucleic acids. For example, magnesium ions play an integral role in the tertiary folding of transfer RNA (1). They are also essential in the structure and enzymatic action of ribozymes (reviewed in 2). However, it is less clear what role they play in the general structural stabilization of double helical RNA (or A-DNA).
In nucleic acid crystals, both sodium and magnesium ions have been found to interact with various sites on nucleic acids, including the phosphate oxygen and N7 of guanine. Interestingly, the interactions may either be through direct metal ion coordination or mediated through water molecules of the hydration shell around the metal ion (3,4). More recently, a number of high resolution X-ray diffraction studies of B-DNA structures revealed that hydrated magnesium ions bind near the phosphate groups (5). In addition, it has been suggested that sodium ions may directly coordinate to the A–T base pairs at the floor of the narrow minor groove of B-DNA (6), although this suggestion remains to be resolved unequivocally (5).
The RNA duplex is very different from B-DNA. RNA has a deep major groove and two anti-parallel ribose–phosphate backbones that run very close to each other, forming a narrow outer mouth as the entrance into its deep major groove. In fact, the entrance is so narrow that the major groove side of the base pairs in RNA is inaccessible to chemical probing agents (7). The A-DNA conformation is very similar to that of the RNA duplex, except for the additional 2′-OH groups bordering the two edges of the wide minor groove in RNA. The similarity was evident from the first three-dimensional structure of an RNA–DNA chimera r(GCG)d(TATACGC) (8). Therefore, many structural properties of RNA helices may be learned through the study of A-DNA structure.
Previously, we addressed questions regarding the interaction of hydrated magnesium ions with A-DNA/RNA using [Co(NH3)6]3+ as a model system. Our NMR data revealed direct NOE crosspeaks between the NH3 protons of [Co(NH3)6]3+ and the DNA protons, including GH8, GH1 (imino) and CH4 (amino) protons in DNA of (dG)n·d(C)n sequences (9). The observation suggested that [Co(NH3)6]3+ adheres to guanine bases in the deep major groove of the double helix. This is consistent with the crystal structure of d(ACCGGCCGGT) complexed with [Co(NH3)6]3+ ions (3) in which [Co(NH3)6]3+ ions are located in the major groove of the decamer A-DNA duplex at the G4pG5 and the G8pG9 steps. Such a binding mode (i.e., [Co(NH3)6]3+ ions binding at the GpG sites) has been observed in the 3 Å resolution structure of yeast phenylalanine tRNA crystals (10). Interestingly, Kieft and Tinoco have found that the [Co(NH3)6]3+ ion binds to tandem G–U mismatched wobble base pairs in the major groove of RNA duplex with good specificity (11).
In many cases the [Co(NH3)6]3+ ion behaves like the [Mg(H2O)6]2+ ion and can substitute the role played by the [Mg(H2O)6]2+ ion. For example, it is more effective in promoting the formation of the DNA four-way junction (Holliday junction) than the Mg2+ ion (12). Similarly, the activity of hammerhead ribozymes is strongly dependent on the Mg2+ concentration and [Co(NH3)6]3+ can substitute Mg2+ effectively (13). Nevertheless, one should extrapolate the observations made by the use of [Co(NH3)6]3+ ions to [Mg(H2O)6]2+ ions with caution. Therefore we wish to visualize directly the interactions between [Mg(H2O)6]2+ ions with RNA and A-DNA. We have analyzed the crystal structures of four oligonucleotides in the A-form conformation bound with [Mg(H2O)6]2+ ions. Together with our earlier analysis of [Co(NH3)6]3+ ion binding to A-DNA (3,9), we propose that there are two major characteristic [Mg(H2O)6]2+ binding modes. One involves the hydrated Mg2+ ions bound preferably at the GpG or GpT steps in the deep major groove and the other is at the exterior of the mouth of the major groove. We have also performed electrostatic potential calculations of the RNA duplex and correlated the electrostatic potential with the binding modes of [Mg(H2O)6]2+ ions.
MATERIALS AND METHODS
The DNA d(ACCGGCCGGT) and the RNA–DNA chimera r(GCG)d(TATACGC), r(GC)d(GTATACGC) and r(G)d(CGTA-TACGC) were synthesized using automated DNA synthesizers and purified by Sepharose S100 gel filtration column chromatography. Preparations of the various crystal forms of A-DNA and RNA–DNA chimera listed in Table 1 follow the procedures described previously (14). Typically, the crystallization solution contained 5 mM DNA duplex (2.5 µl), 100 mM sodium cacodylate buffer (pH 6.5) (10 µl), 20% 2-methyl-2,4-pentanediol (2-MPD) (5 µl), 15 mM spermine (2.5 µl), plus 100 mM of the MgCl2 (2.5 µl).
Table 1. Crystal and refinement data of four oligonucleotides.
| d(ACCGGCCGGT) | r(GCG)d(TATACGC) | r(GC)d(GTATACGC) | r(G)d(CGTATACGC) | |
|---|---|---|---|---|
| Cell parameters | ||||
| Space Group | P6122 | P212121 | P212121 | P212121 |
| a (Å) | 38.50 | 24.26 | 25.54 | 26.19 |
| b (Å) | 38.50 | 42.67 | 43.72 | 41.94 |
| c (Å) | 77.63 | 49.32 | 45.47 | 43.32 |
| γ (°) | 120 | |||
| Data collection | ||||
| Resolution | 1.6 | 1.4 | 1.7 | 1.7 |
| R-merge | 0.042 | 0.064 | 0.056 | 0.079 |
| R-merge (highest shell) | 0.49 | 0.18 | 0.40 | 0.23 |
| Unique reflections | 4901 | 10 433 | 5929 | 5659 |
| redundancy | 15.2 | 12.1 | 12.6 | 10.1 |
| Refinement statistics | ||||
| Refined model | half duplex | Full duplex | full duplex | full duplex |
| R-factor/R-free (1 > 2.0σ) | 0.229/0.278 | 0.193/0.239 | 0.196/0.269 | 0.194/0.240 |
| # of observables (1 > 2.0σ) | 4874 | 10 217 | 5900 | 5598 |
| R-factor/R-free (1 > 0.0σ) | 0.229/0.279 | 0.195/0.242 | 0.196/0.269 | 0.195/0.243 |
| r.m.s.d. bond length (Å) | 0.008 | 0.011 | 0.008 | 0.007 |
| r.m.s.d. bond angle (deg) | 1.7 | 1.8 | 1.6 | 1.7 |
| cations/duplex | 3 Mg(H2O)6 | 4 Mg (H2O)6 | 1 Mg (H2O)6 | 1 Mg (H2O)6 |
| water (excluding those bound with cation) | 24 | 97 | 45 | 78 |
Diffraction data, except those of r(GCG)d(TATACGC), were collected at 123 K on a Rigaku R-Axis IIc image plate area detector system. The data were processed using the software (d*trek, v.4.7) from Molecular Structure Co. Care was taken to account for intensity saturation on the image plate for strong reflections. Some frames were recollected with a faster oscillation speed in order to recover those strong (saturated) reflections.
The data for r(GCG)d(TATACGC) were measured at Structural Biology Center undulator beamline 19ID at the Advanced Photon Source using fundamental undulator harmonics (λ = 1.03321 Å) and 3×3 mosaic CCD detector (15). The experiment was carried out at 110 K from a single crystal. The complete data set was collected in one continuous scan. Crystallographic data integration and reduction was done with the program package HKL2000 (16). The quality of the new datasets is substantially better than that of the data collected previously (8,17). For example, the number of reflections for the r(GCG)d(TATACGC) and d(ACCGGCCGGT) for the new datasets is 10 217 (I > 2σ) and 4874 (I > 2σ), respectively, compared to 2521 (I > 1.5σ) and 1434 (I > 1.0σ), respectively, of the previous datasets (8,17).
All four structures were refined, first by the simulated annealing procedure incorporated in X-PLOR (18), and then by the SHELX97 (19). DNA force field parameters of Parkinson et al. (20) were used. Well-ordered water molecules and ions were located using the difference Fourier maps. After refinement, several hexahydrated magnesium ions were seen in the P6122 Mg2+-form of d(ACCGGCCGGT) and in the P212121 Mg2+-form of r(GCG)d(TATACGC). The hexahydrated magnesium ion can be easily identifed from the highly regular octahedral coordination geometry. All Mg–O(water) distances are very close to 2.0 Å. None of those ions was seen in the previous lower resolution structural analysis (8,17), thus affirming the substantial improvement of the structural quality. All metal ions have their B-factors comparable to the first shell water molecules (~35 Å2). Sample electron density maps of the hexahydrated magnesium ions of the r(GCG)d(TATACGC) and d(ACCGGCCGGT) structures, respectively, are shown in Figure 1. The atomic coordinates of the four crystal structures have been deposited at the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (accession numbers 1DNZ, 1DNO, 1DNT and 1DNX).
Figure 1.
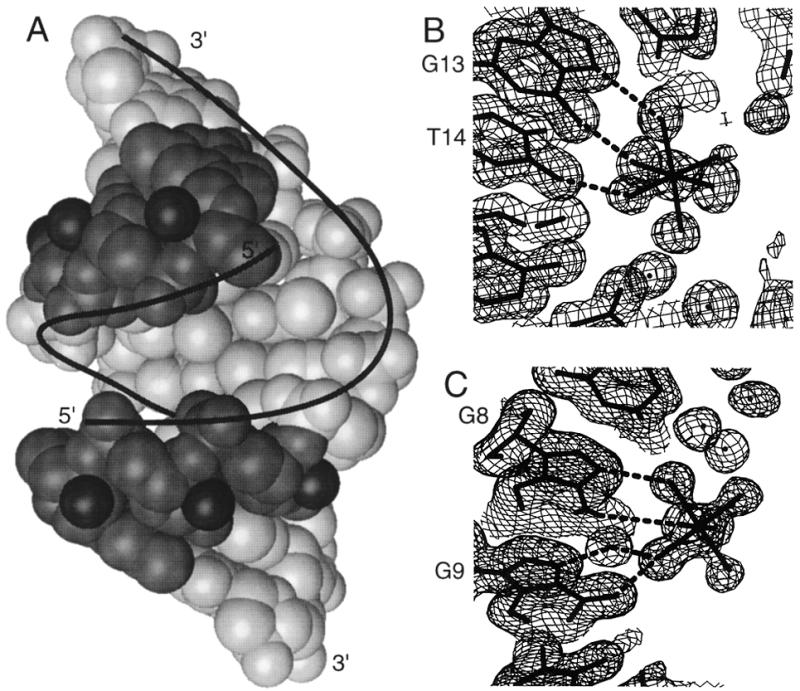
(A) The 1.4 Å resolution structure of the r(GCG)d(TATACGC) chimera. The RNA part is more darkly shaded with the 2′-hydroxyl groups depicted as dark spheres. (B) and (C) show a portion of the (2Fo-Fc) Fourier maps in which the resolved [Mg(H2O)6]2+ ions are visible. (B) The [Mg(H2O)6]2+ ion near the G13–T14 step in r(GCG)d(TATACGC) structure. (C) The [Mg(H2O)6]2+ ion near the G8–G9 step in the d(ACCCGCGGGT) A-DNA structure.
Electrostatic potential calculations were performed using the program suite of GRASP (21). Dielectric constant of the DNA parts was set at 2 with the atomic partial charge assignment according to Parkinson et al. (20) with the full negative charge option for each phosphate group. The dielectric constant of the solvent region was set at 80 and cations were explicitly added. The [Mg(H2O)6]2+ ion was assigned with a net +2 charges located on the Mg2+ ion.
RESULTS AND DISCUSSION
Conformations of A-form decamers
The crystal structures of these four DNA decamer molecules have previously been analyzed at a lower resolution. Specifically, d(ACCGGCCGGT) in the P6122 space group at 2 Å (17), r(GCG)d(TATACGC) (8) and r(GC)d(GTATACGC) (3) at 2 and 1.9 Å, respectively, in the same P212121 space group and finally, r(G)d(CGTATACGC) at 2 Å in a different P212121 space group (3,22). More recently, several RNA structures have been analyzed at significantly higher resolution, including r(CCCCGGGG) (23), the 7mer stem of tRNAAla (24), two plasmid copy control related RNA 18mer and 19mer (25). However, none of those RNA duplex crystals, obtained from high ammonium salt solutions in which no Mg2+ ion was added, contained ordered Mg2+ ions in the crystal lattice.
In general the present new structures are similar to their respective structures from earlier analysis. The most striking features of the new structures are the improved structural reliability and the observation of well-ordered magnesium ions. As most of the structural features have been discussed in detail previously, only relevant points are discussed briefly here.
As we showed before, all four oligonucleotides adopt conformations very close to the canonical A-form duplex as evident in Tables S1–S4 (see Supplementary Material available at NAR Online) which summarize the helix parameters of the four decamers in three crystal forms. The duplexes have the characteristic wide and shallow minor groove and the deep and narrow major groove. The average base pair inclination angle is in the range of 8–13° and the averaged x-displacement of the base pairs is about –4.3 Å. All sugars have the N-type (C3′-endo) pucker. Nevertheless, some small variations exist between the three crystal forms. The average rise per residue values in the P6122 and the three P212121 forms are 2.79, 2.68, 2.90 and 2.76 Å, respectively, whereas the x-displacements for the three respective crystal forms are –4.12, –4.40, –4.42 and –4.49 Å (Tables S5–S8).
However, despite the similar global conformations, the molecules show significant variations in their local structures due to crystal packing interactions. Molecules crystallized in the same space group have a more similar conformation as discussed previously (3). In the crystals, A-DNA molecules form crystal lattices by abutting their terminal base pairs onto the shallow and wide minor groove of other A-DNA duplexes. For example, in the P212121 lattice of r(GC)d(GTATACGC) (3,22), two symmetry-related terminal base pairs, G1–C10 and G11–C20, pack asymmetrically on neighboring helices with several inter-helix hydrogen bonds.
There are two independent P212121 packing arrangements for the RNA–DNA chimera decamers, with r(GCG)d(TATACGC) (9) and r(GC)d(GTATACGC) (3,22) in the same P212121 arrangement (despite quite different unit cell dimensions; see Table 1) and r(G)d(CGTATACGC) in a different P212121 arrangement (3). In both P212121 forms, since the two independent DNA strands behave differently, the helix has an asymmetric pattern of the helical twist (Ω) values for the nine unique steps. Most of them cluster around 32°, again close to that of the canonical A-DNA. Some steps are unusual. In particular, the T4pA5 step of r(GC)d(GTATACGC) has a low Ω of 27°, with a compensating larger Ω of 34° for the A5pT6 step.
The different crystal packing interactions actually affect the binding of [Mg(H2O)6]2+ to the RNA–DNA chimera in the P212121 forms. In the r(GCG)d(TATACGC) structure three ions are found in the deep major groove, whereas only one each is found in the structures of r(GC)d(GTATACGC) and r(G)d(CGTATACGC) (Table 2). In the r(GC)d(GTATACGC) structure, well-ordered [Mg(H2O)6]2+ ions at Mg I and Mg III sites are not found. This might be due to the lower resolution nature of the structure. In the r(G)d(CGTATACGC) structure, the different crystal packing results in a change in the position of one strand, causing T14 to shift away from the proper position for [Mg(H2O)6]2+ binding at a GpT step. Moreover the Mg II site (Table 2) has changed to the C2pG3 (from G3pT4) step, thus moving closer toward the Mg I site. This movement has the effect of dislodging the Mg I from its binding site.
Table 2. Interactions of Mg(H2O)62+ with base pairs in the deep groove of A-type helices.
| Sites | d(ACCGGCCGGT) | r(GCG)d(TATACGC) | r(GC)d(GTATACGC) | r(G)d(CGTATACGC) |
|---|---|---|---|---|
| Mg I | G4–G5 | G19–C18 | water at site | site interfered by Mg II |
| Mg I*a | G4*–G5* | |||
| Mg II | G8–G9 | G3–T4 | G3–T4 | C2–G3 |
| Mg II* | G8*–G9* | |||
| Mg III | G13–T14 | water at site | site lost due to T14 shift |
aThe * indicates symmetry related sites, and for d(ACCGGCCGGT), Mg I and Mg I* overlap and are each half occupied.
Two binding modes of [Mg(H2O)6]2+ to RNA and A-DNA
Four [Mg(H2O)6]2+ ions are found in the asymmetric unit of the r(GCG)d(TATACGC) crystal lattice and three of them are located in the deep major groove (Fig. 2). The hydrated Mg2+ ions are bound to the floor of the deep groove using hydrogen bonds to acceptor atoms, mostly the N7 and O6 atoms of guanine base. From Figure 2 it is clear is that even though the size of the [Mg(H2O)6]2+ ion is ~8 Å in diameter, larger than the averaged width of the narrow major groove outer mouth opening of A-DNA, the edges of the base pairs are still accessible to a bulky [Mg(H2O)6]2+ ion. Our previous results from the NMR studies of several DNA oligonucleotides, including a 34mer d(A2G30C30T2) duplex, showed that [Co(NH3)6]3+, an analog of [Mg(H2O)6]2+ ion, can enter the deep groove of A-DNA duplex (9). Paradoxically, other data suggested that the accessibility of the major groove in RNA (therefore A-DNA) is limited (7). In the case of the [Mg(H2O)6]2+ ions, it is possible that the facile exchange of the Mg2+-coordinated water molecules with the free water molecules would allow the [Mg(H2O)6]2+ ions to travel through the narrow outer mouth of the deep major groove without hindrance.
Figure 2.
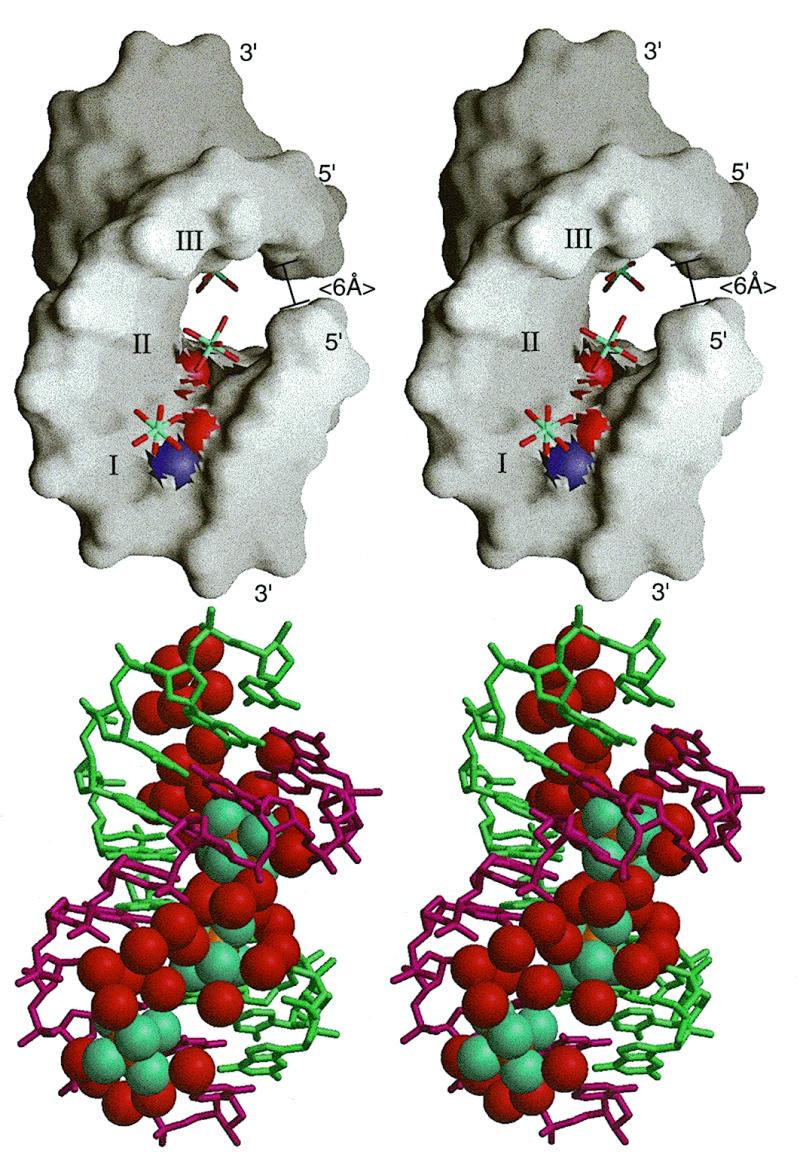
(A) Surface drawing of the RNA–DNA duplex of r(GCG)d(TATACGC) with the three [Mg(H2O)6]2+ ions (labeled I, II and III, corresponding to Mg I, Mg II and Mg III in Table 2) bound in the deep major groove of the duplex. The N7 and O6 of guanines are shown as blue and red spheres. Note that the outer mouth of the groove (~6 Å shown at right) is narrower than the size of the [Mg(H2O)6]2+ ions (~8 Å in van der Waals diameter). (B) The first shell bound waters (red) and the three [Mg(H2O)6]2+ ions (aqua) nearly fill up the major groove.
Three [Mg(H2O)6]2+ ions are found in the d(ACCGGCCGGT)-P6122 lattice. Like that in the r(GCG)d(TATACGC) decamer described above, one [Mg(H2O)6]2+ ion is bound to the edges of two adjacent intra-stranded guanines through hydrogen bonds to the N7 and O6 sites of G8 and G9 guanines. (An equivalent binding site is also located at the G8* and G9* due to the crystallographic two-fold symmetry.) Another [Mg(H2O)6]2+ ion is located at the center of the deep major groove near the G4–G5 bases of a duplex. However, due to the inherent 2-fold symmetry of the duplex (coinciding with the crystallographic 2-fold axis) the same [Mg(H2O)6]2+ ion may bind to the symmetry-related G4*–G5* bases (Fig. 3). Yet the two sites are too close to each other so that a single [Co(NH3)6]3+ ion can only bind to a GGCC:CCGG sequence at either of the two sites, but not both at the same time. This disordered binding mode is clearly discernable in the Fourier map and the two well-resolved sets of the [Mg(H2O)6]2+ ion are refined as such.
Figure 3.
The disordered [Mg(H2O)6]2+ ions located at the center of the deep major groove near the G4–G5 bases and the symmetry-related G4*–G5* bases of the d(ACCGGCCGGT) duplex.
The detailed interactions between the three different [Mg(H2O)6]2+ ions in the deep groove with the base pairs in the r(GCG)d(TATACGC) and d(ACCGGCCGGT) duplexes are shown in Figure 4. The major interactions involve the direct hydrogen bonds between three Mg2+-coordinated water molecules (from one face of the octahedron) with the hydrogen bond acceptors (principally N7 and O6 of guanine). Analogous interactions have been seen before with [Co(NH3)6]3+ ions in A-DNA (3), Z-DNA (26,28) and RNA (11).
Figure 4.
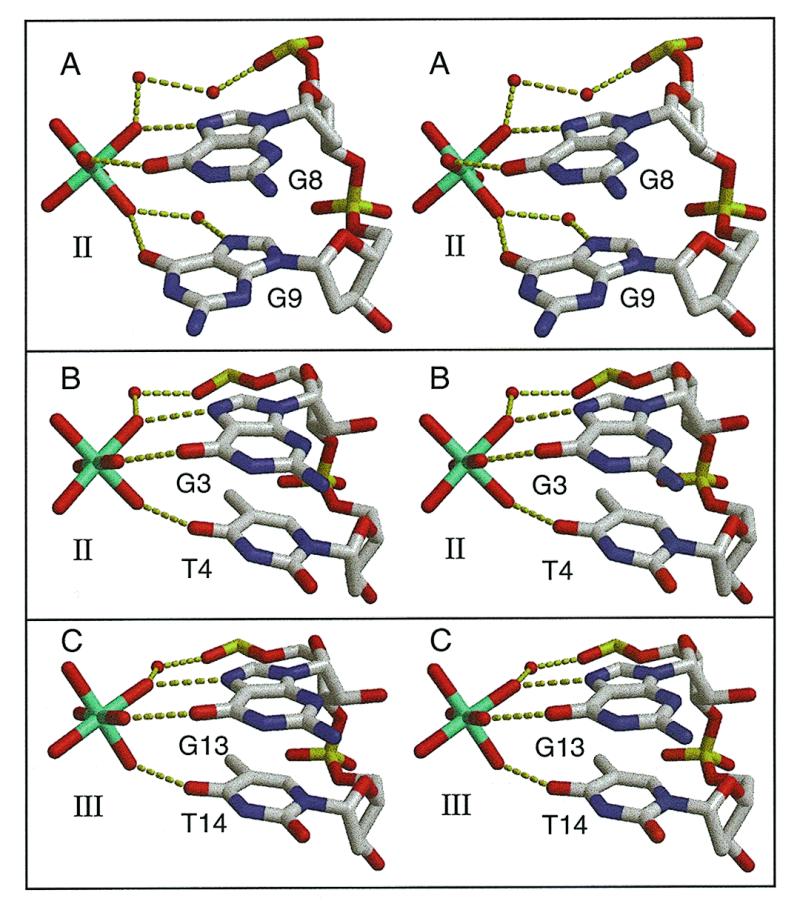
Stereoscopic views of the detailed interactions between [Mg(H2O)6]2+ ions and A-DNA. The hydrated [Mg(H2O)6]2+ ion uses three liganded water molecules from one face of the octahedron to form direct hydrogen bonds with the N7 and O6 of two adjacent guanines. (A) The G8–G9 step in the crystal structure of the P6122 form of d(ACCGGCCGGT). The hydrated [Mg(H2O)6]2+ ion (Mg II site in Table 2) adheres to the edges of two adjacent guanines. (B) The G3–G4 step (Mg II site in Table 2) in the crystal structure of r(GCG)d(TATACGC). (C) The G13–G14 step (Mg III site in Table 2) in the crystal structure of r(GCG)d(TATACGC).
Another important binding mode has been detected in our earlier analysis of the crystal structure of the d(ACCGGCCGGT) (P6122 form) in the presence of the [Co(NH3)6]3+ ions. We noted that a [Co(NH3)6]3+ ion is located midway across the narrow major groove, bridging the phosphates of C3 and C12 residues (3). In the present analysis of the Mg-form of the d(ACCGGCCGGT) structure, no [Mg(H2O)6]2+ ion could be unambiguously found at the same location of the bridging [Co(NH3)6]3+ ion due to the diffuse electron density. We believe that this type of binding mode is likely to involve diffusing [Mg(H2O)6]2+ ions, which is not surprising because this site is completely exposed to the solvent and the ion may reside at multiple locations.
Electrostatic surface potential of RNA duplex
Our high resolution crystal structural analysis of four A-DNA and RNA–DNA hybrid duplexes afforded a clear view of two important binding modes of fully-hydrated Mg2+ ions: one in the deep major groove and the other at the outer mouth of the deep groove. What may be the reasons for the positively-charged magnesium ions to be found at those locations? We address this question by analyzing the electrostatic potential of the RNA and A-DNA duplexes. Figure 5 (left panels) shows the electrostatic potential in the solvent region of a two-turn (20 bp) RNA duplex model. One notable feature of the charge distribution of the RNA duplex is that the deep groove shows the most pronounced negative potentials. Therefore, it is reasonable to expect that the deep groove of the A-form duplex is the most favorable region where the positively-charged [Mg(H2O)6]2+ ions would be located. This observation was made some time ago by Pullman et al. (28). Therefore our crystal structures with positively-charged ions found in the deep groove of RNA support such theoretical calculations.
Figure 5.
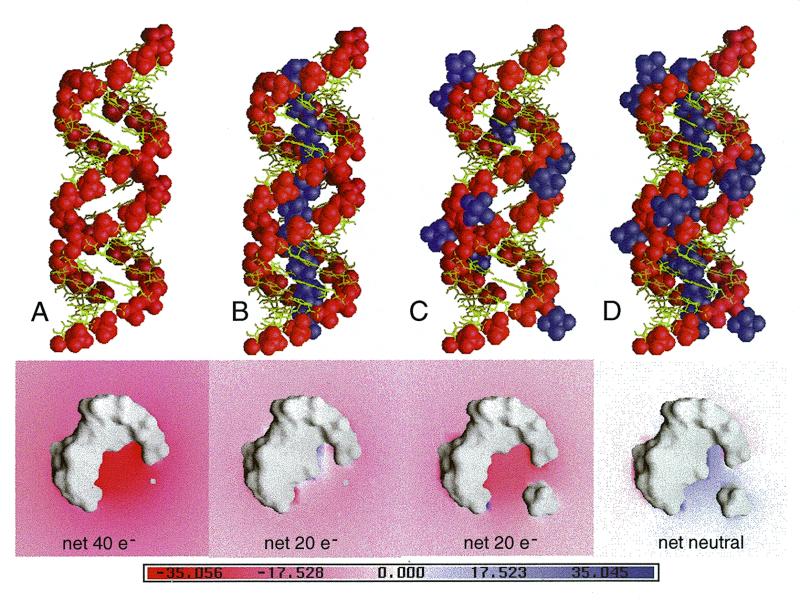
Electrostatic surface potential of an idealized RNA duplex without [(A), bottom] and with [Mg(H2O)6]2+ ions placed inside the deep groove [(B), bottom], with [Mg(H2O)6]2+ ions at the outer mouth of the groove [(C), bottom] and inside the deep groove plus at the outer mouth of the groove [(D), bottom]. The idealized poly(rG-rU) RNA model was constructed by repeating the A-form conformation of the G3pT4/A17pC18 pairs of the r(GCG)d(TATACGC) crystal structure with the idealized O2′ atoms added. The Mg II site (Table 2) is also repeated in the deep groove of the idealized helix for subsequent electrostatic calculation. The colors indicate the scale for the electrostatic potential ranging from –35 kT/e (red) to +35 kT/e (blue). The locations of the phosphates and the [Mg(H2O)6]2+ ions are indicated in the top panels as red and purple spheres, respectively. (See Materials and Methods for more details.)
It should be noted that RNA often forms complex tertiary structures in which phosphates are often clustered. In those cases, the electrostatic potential would be very different and different metal binding mode would be expected. Our discussion here addresses only the regular helical region of A-form structure.
We had demonstrated earlier that [Co(NH3)6]3+ ions bind in the deep groove of A-DNA, specifically at the edge of guanine base, using NMR spectroscopy (9). It is highly probable that [Mg(H2O)6]2+ ions would behave in a similar manner. Unfortunately, NMR cannot be used for the study of Mg2+ ions due to the rapidly-exchanging water molecules. X-ray diffraction analysis is the most direct tool for the visualization of ions in crystals. However, only well-ordered metal ions can be seen by X-ray diffraction and that usually means that the ions need to be held in a particular position with long-enough resident time. Thus favorable (i.e., strong hydrogen-bonding) interactions such as those seen in Figure 4 are required. In fact, such a binding preference toward GpU or GpG may be applied to the [Os(NH3)6]3+ ion, which is a heavier analog of the [Co(NH3)6]3+ ion. It is possible that [Os(NH3)6]3+ ions can be used as heavy atom derivatives in the structural analysis of large RNA molecules (29).
When the deep groove of an RNA duplex is filled with fully-packed [Mg(H2O)6]2+ ions (∼2 bp per ion with optimal binding sequences involving GpU or GpG) (Fig. 5B, top panel), the negative potential in the deep groove (shown in red) (Fig. 5A, bottom panel) is completely neutralized due to the densely packed [Mg(H2O)6]2+ ions (Fig. 5B, bottom panel). An important point to make here is that the deepest negative potential in A-form helix is the deep groove and [Mg(H2O)6]2+ ions are likely to be found there. We should emphasize that the fact that we have located the [Mg(H2O)6]2+ ions in the crystals does not mean that in solution they are statically bound at specific sites. Rather, they are dynamically occupying the deep groove, but with longer resident time at the favorable GpU or GpG sites so that they can be detected (e.g., by NMR).
It should be noted that residual negative potentials are found surrounding the duplex, particularly at the outer mouth of the groove. We hypothesize that additional [Mg(H2O)6]2+ ions are likely to be used at the outer mouth of the groove to fully neutralize the negative charge of phosphates (Fig. 5D, top and bottom panels).
A recent paper describing the electrostatic properties of RNA has appeared (30). It is interesting to note that the features described in our paper are also found in the 3 Å resolution structure of the loop E of 5S RNA in which possible metal ions have been identified in the major groove and were denoted as a metal zipper (31). Our high resolution structures provide support for such a ‘metal zipper’ in the deep groove of RNA.
CONCLUSION
Our high resolution structural analysis of several RNA–DNA chimera and A-DNA oligonucleotides showed that the deep groove of RNA has high negative electrostatic potential which may be neutralized with [Mg(H2O)6]2+ ions bound at certain preferred sites (like GpU or GpG steps). Additional metal ions may be bound at the outer mouth of the groove. Together these metal ions balance out the RNA negative charges which allow the opposite sugar–phosphate backbones in RNA to come close to each other without serious charge repulsion.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by a NIH grant (GM-41612) and a NSF grant (MCB98-08298) to A.H.-J.W. and by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38 to A.J.
PDB accession nos+ To whom correspondence should be addressed. Tel: +1 217 244 6637; Fax: +1 217 244 3181; Email: ahjwang@uiuc.edu 1DNZ, 1DNO, 1DNT, 1DNX
REFERENCES
- 1.Quigley G.J., Teeter,M.M. and Rich,A. (1978) Proc. Natl Acad. Sci. USA, 75, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feig A.L. and Uhlenbeck,O.C. (1999) The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 287–319.
- 3.Gao Y.-G., Sriram,M. and Wang,A.H.-J. (1993) Nucleic Acids Res., 21, 4093–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y.-G., Robinson,H., Sanishvili,R., Joachimiak,A. and Wang,A.H.-J. (1999) Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- 5.Tereshko V., Minasov,G. and Egli,M. (1999) J. Am. Chem. Soc., 121, 3590–3595. [Google Scholar]
- 6.Shui X., McFail-Isom,L., Hu,G.G. and Williams,L.D. (1998) Biochemistry, 37, 8341–8355. [DOI] [PubMed] [Google Scholar]
- 7.Weeks K.M. and Crothers,D.M. (1993) Science, 261, 1574–1577. [DOI] [PubMed] [Google Scholar]
- 8.Wang A.H.-J., Fujii,S., van Boom,J.H., van der Marel,G.A., van Boeckel,S.A.A. and Rich,A. (1981) Nature, 299, 601–604. [DOI] [PubMed] [Google Scholar]
- 9.Robinson H. and Wang,A.H.-J. (1996) Nucleic Acids Res., 24, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hingerty B.E., Brown,R.S. and Klug,A. (1982) Biochim. Biophys. Acta, 697, 78–83. [DOI] [PubMed] [Google Scholar]
- 11.Kieft J.S and Tinoco,I.,Jr (1997) Structure, 5, 713–721. [DOI] [PubMed] [Google Scholar]
- 12.Duckett D.R., Murchie,A.I.H. and Lilley,D.M.J. (1990) EMBO J., 9, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassi G.S., Mollegaard,N.-E., Murchie,A.I.H., von Kitzing,E. and Lilley,D.M.J. (1995) Nat. Struct. Biol., 2, 45–55. [DOI] [PubMed] [Google Scholar]
- 14.Wang A.H.-J. and Gao,Y.-G. (1990) Methods, 1, 91–99. [Google Scholar]
- 15.Westbrook E.M. and Naday,I. (1997) Methods Enzymol., 276, 44–68. [PubMed] [Google Scholar]
- 16.Otwinowski Z. and Minor,W. (1997) Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 17.Frederick C.A., Quigley,G.J., Teng,M.-k., Coll,M., van der Marel,G.A., van Boom,J.H., Rich,A. and Wang,A.H.-J. (1989) Eur. J. Biochem., 181, 295–307. [DOI] [PubMed] [Google Scholar]
- 18.Brünger A.T. (1992) X-PLOR 3.1, A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- 19.Sheldrick G.M. (1997) SHELX-97, Crystallographic Refinement Program. University of Gottingen, Germany.
- 20.Parkinson G., Vojtechovsky,J., Clowney,L., Brunger,A.T. and Berman,H.M. (1996) Acta Crystallogr., 52, 57–64. [DOI] [PubMed] [Google Scholar]
- 21.Nicholls A., Sharp,K. and Hoing,B. (1992) GRASP Manual. Columbia University, New York, NY.
- 22.Egli M., Usman,N. and Rich,A. (1993) Biochemistry, 32, 3221–3237. [PubMed] [Google Scholar]
- 23.Egli M., Portmann,S. and Usman,N. (1996) Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
- 24.Mueller U., Schubel,H., Sprinzl,M. and Heinemann,U. (1999) RNA, 5, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klosterman P.S., Shah,S.A. and Steitz,T.A. (1999) Biochemistry, 38, 14784–14792. [DOI] [PubMed] [Google Scholar]
- 26.Gessner R.G., Quigley,G.J., Wang,A.H.-J., van der Marel,G.A., van Boom,J.H. and Rich,A. (1985) Biochemistry, 24, 237–250. [DOI] [PubMed] [Google Scholar]
- 27.Harper A., Brannigan,J.A., Buck,M., Hewitt,L., Lewis,R.J., Moore,M.H. and Schneider,B. (1998) Acta Crystallogr., D54, 1273–1284. [DOI] [PubMed] [Google Scholar]
- 28.Lavery R. and Pullman,B. (1981) Nucleic Acids Res., 9, 4677–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cate J.H. and Doudna,J.A. (1998) Structure, 4, 1221–1229. [Google Scholar]
- 30.Chin K., Sharp,K.A., Honig,B. and Pyle,A.M. (1999) Nat. Struct. Biol., 6, 1055–1061. [DOI] [PubMed] [Google Scholar]
- 31.Correll C.C., Freeborn,B., Moore,P.B. and Steitz,T.A. (1997) Cell, 91, 705–712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.