Abstract
This year’s review on bioactivation and reactivity began as a part of the annual review on biotransformation and bioactivation led by Cyrus Khojasteh (see references). Increased contributions from experts in the field led to the development of a stand alone edition for the first time this year focused specifically on bioactivation and reactivity. Our objective for this review is to highlight and share articles which we deem influential and significant regarding the development of covalent inhibitors, mechanisms of reactive metabolite formation, enzyme inactivation, and drug safety. Based on the selected articles, we created two sections: (1) reactivity and enzyme inactivation, and (2) bioactivation mechanisms and safety (Table 1). Several biotransformation experts have contributed to this effort from academic and industry settings.
Beyond bioactivation
Significant world events over the past year have continued to challenge the global community – from climate disasters and the ongoing COVID-19 pandemic to the suffering of marginalized communities and war. These events strengthen our resolve to be compassionate to others and to see each other as human. We thank all those who intentionally give back, provide opportunities for the next generation, and work to make a positive difference in the world around us. A special thank you to Cyrus Khojasteh and Namandje Bumpus for being excellent examples of giving back for the next generation of scientists.
We welcome your opinions, and we extend an invitation to anyone who would like to contribute to a future edition of this review.
Klarissa Jackson, on behalf of the authors.
Table 1.
Articles covered in this review.
| Title | First author | Source | |
|---|---|---|---|
| Reactivity & enzyme inactivation | |||
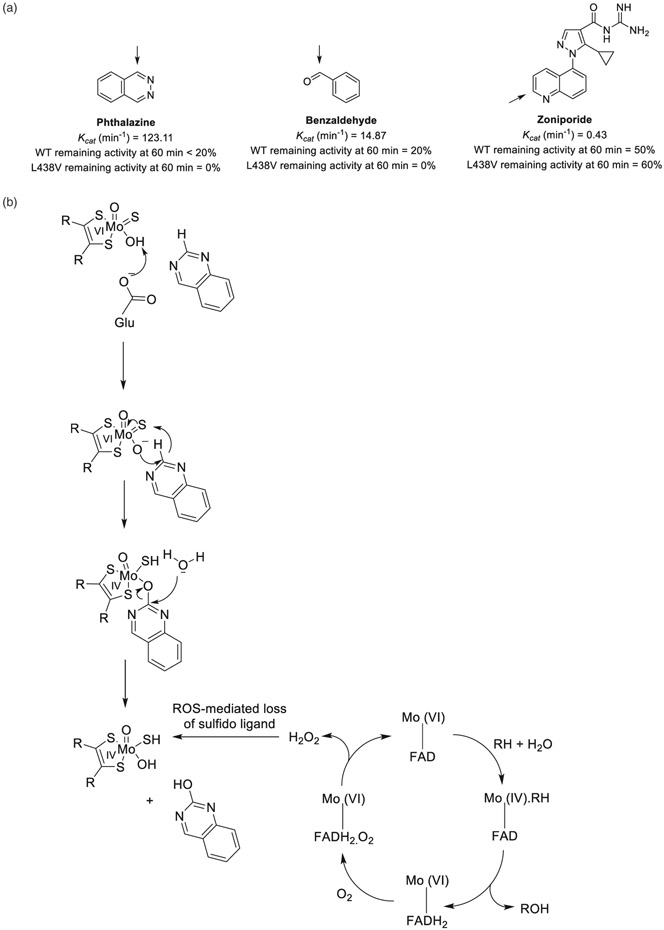
| 1 | The inactivation of human aldehyde oxidase 1 by hydrogen peroxide and superoxide. | C Garrido | Drug Metab Dispos 49:729–735, 2021 |
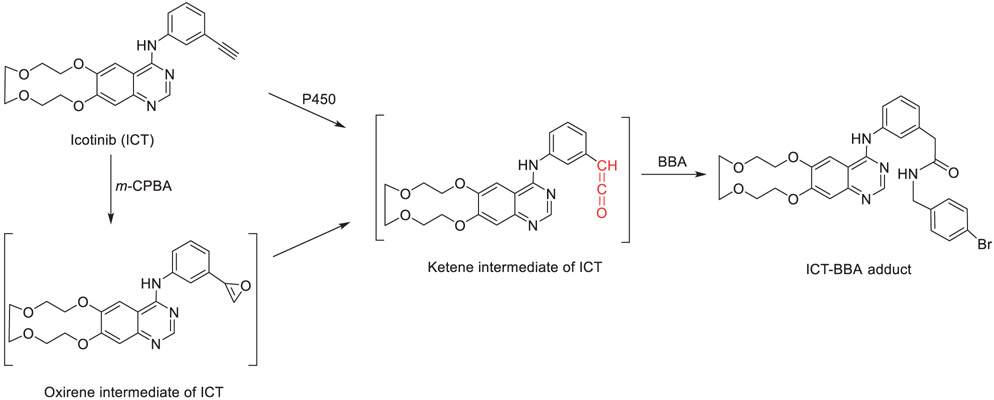
| 2 | Icotinib induces mechanism-based inactivation of recombinant human CYP3A4/5 possibly via heme destruction by ketene intermediate. | C Sun | Drug Metab Dispos 49:892–901, 2021 |
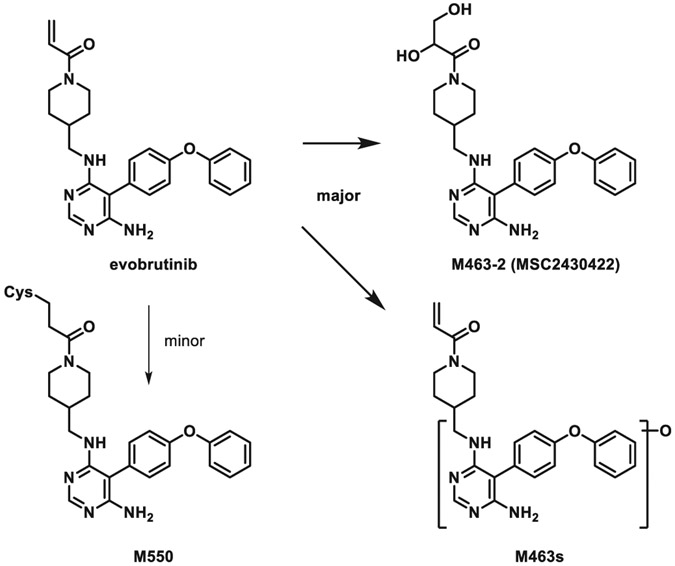
| 3 | Evobrutinib, a covalent Bruton’s tyrosine kinase inhibitor: Mass balance, elimination route, and metabolism in healthy participants. | H Scheible | Clin Transl Sci 14: 2420–2430, 2021 |
| Bioactivation mechanisms & safety | |||
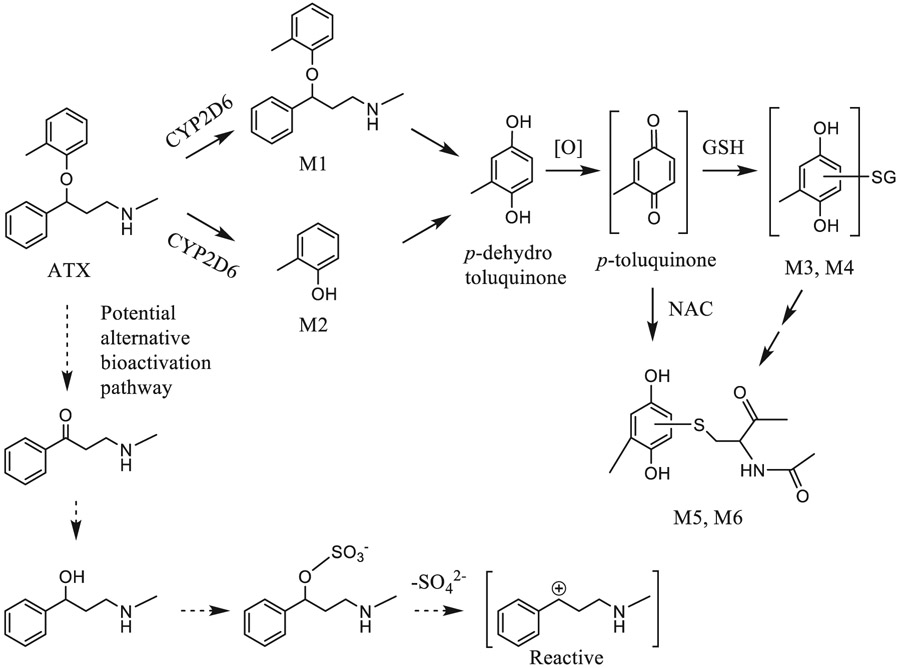
| 4 | Metabolic activation of atomoxetine mediated by cytochrome P450 2D6. | Y You | Chem Res Toxicol 34:2135–2144, 2021 |
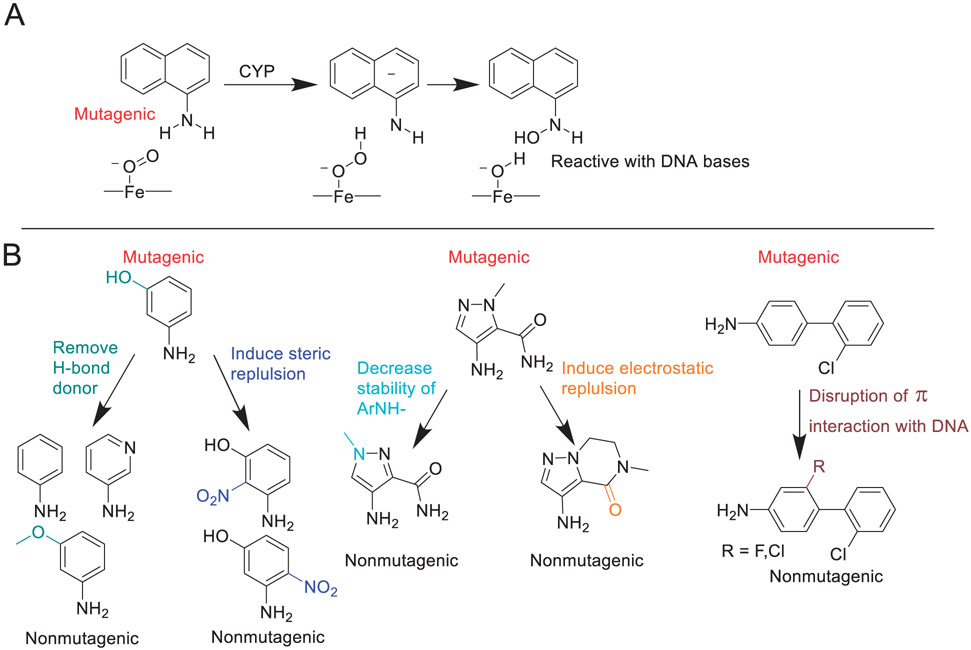
| 5 | Mechanism-based insights into removing the mutagenicity of aromatic amines by small structural alterations. | I Shamovsky | J Med Chem 64: 8545–8563, 2021 |
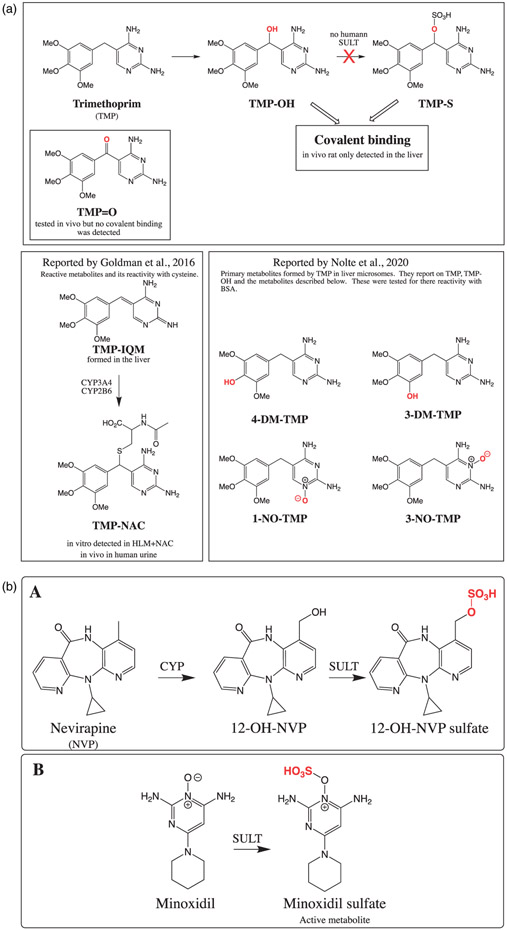
| 6 | Investigating the mechanism of trimethoprim-induced skin rash and liver injury. | Y Cao | J Toxicol Sci 180:17–25, 2021 |
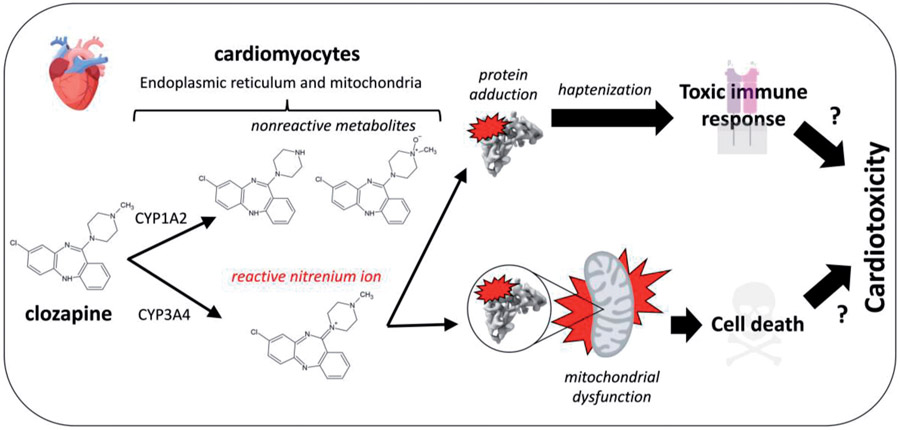
| 7 | Bioactivation of clozapine by mitochondria of the murine heart: Possible cause of cardiotoxicity. | E Arzuk | Toxicology 447: 152628, 2021 |
| 8 | CYP2C9 and 3A4 play opposing roles in bioactivation and detoxification of diphenylamine NSAIDs. | Mary Alexandra Schleiff | Biochem Pharmacol 194:114824, 2021 |
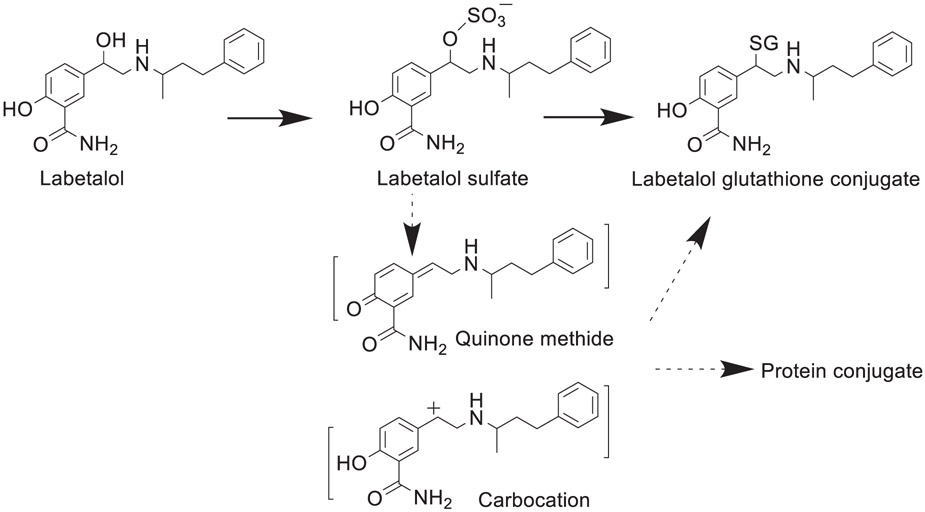
| 9 | Metabolic activation and cytotoxicity of labetalol hydrochloride mediated by sulfotransferases. | L Yang | Chem Res Toxicol 34:1612–1618, 2021 |
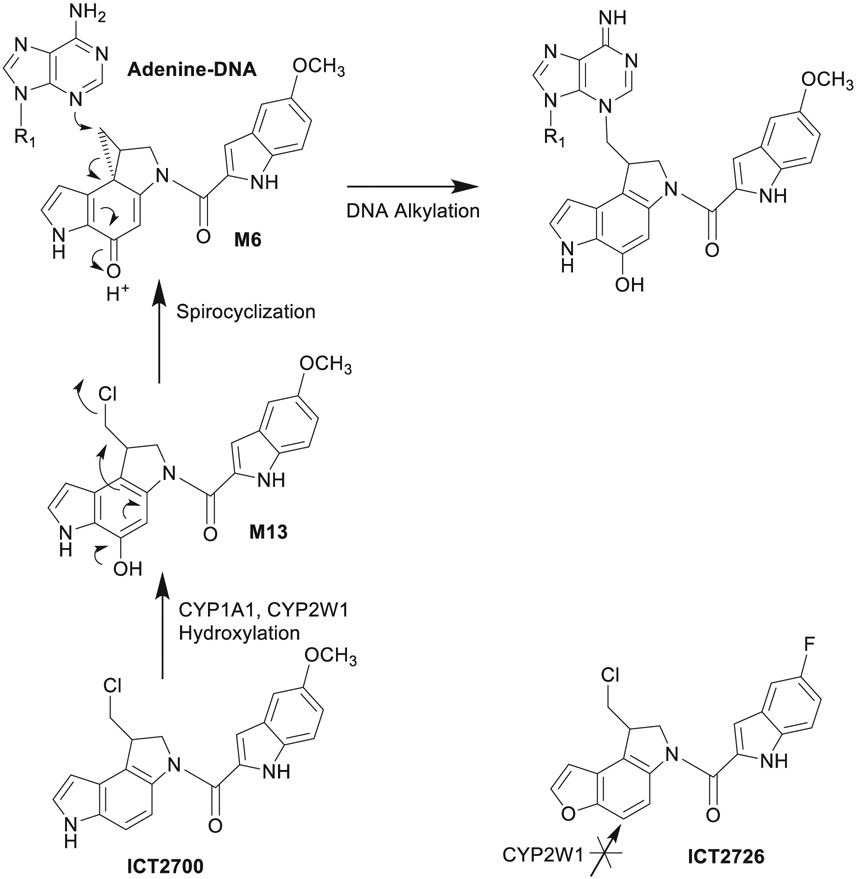
| 10 | Cytochrome P450 binding and bioactivation of tumor-targeted duocarmycin agents. | AG Bart | Drug Metab Dispos 50:49–57, 2022 |
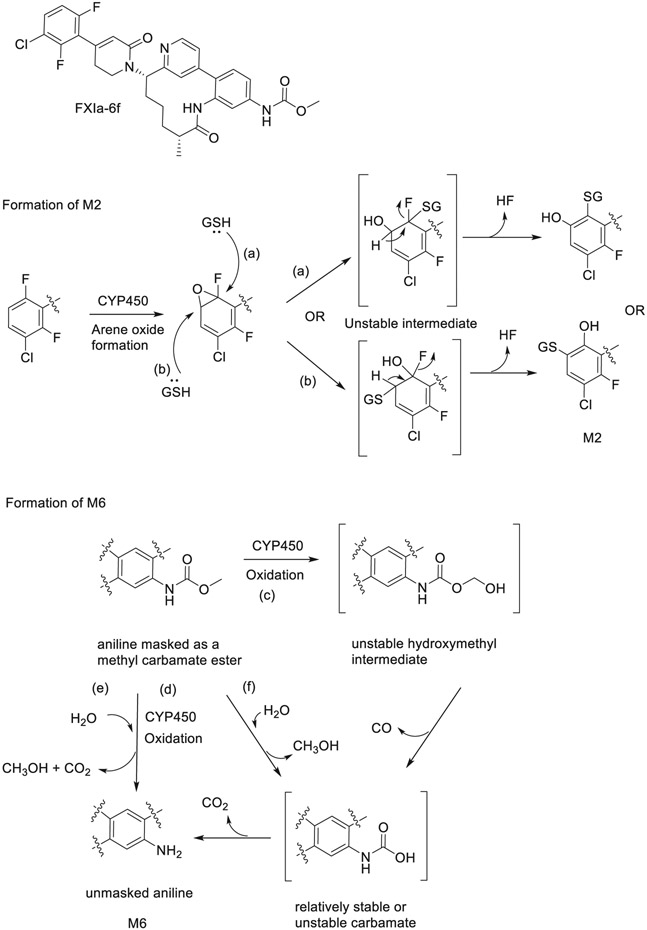
| 11 | Preclinical metabolism and disposition of an orally bioavailable macrocyclic FXIa inhibitor. | SA Chacko | Xenobiotica 51:933–948, 2021 |
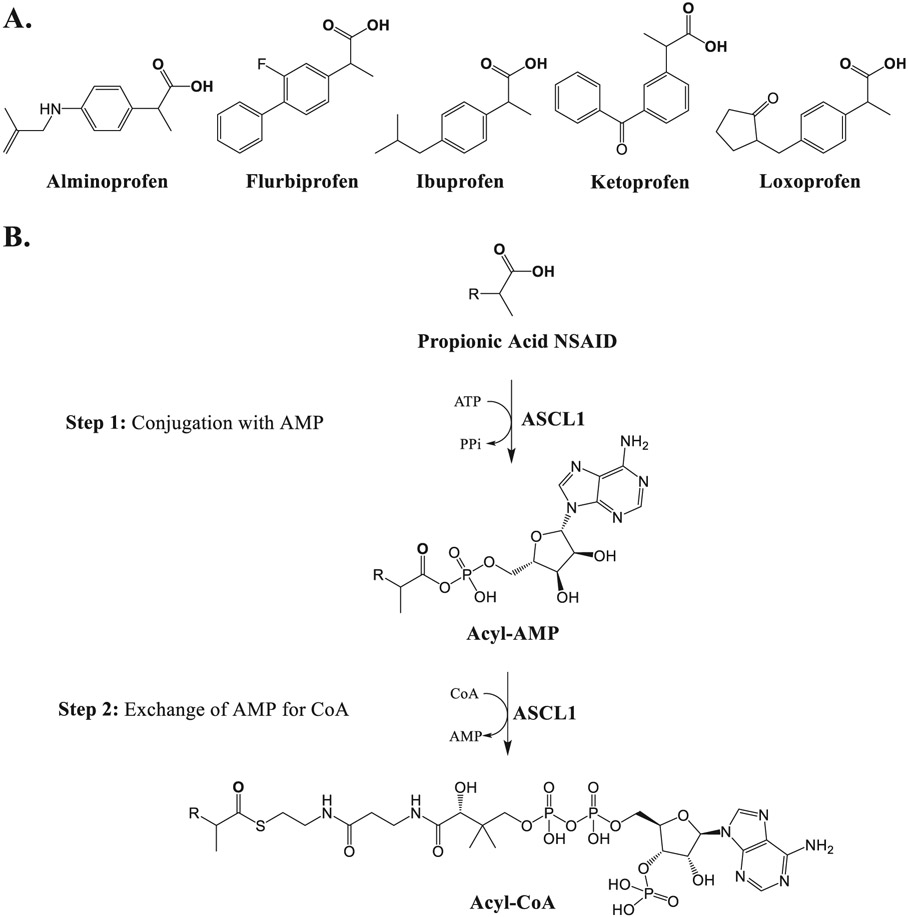
| 12 | Identification of an isoform catalyzing the CoA conjugation of nonsteroidal anti-inflammatory drugs and the evaluation of the expression levels of acyl-CoA synthetases in the human liver. | H Hashizume | Biochem Pharmacol 183:114303, 2021 |
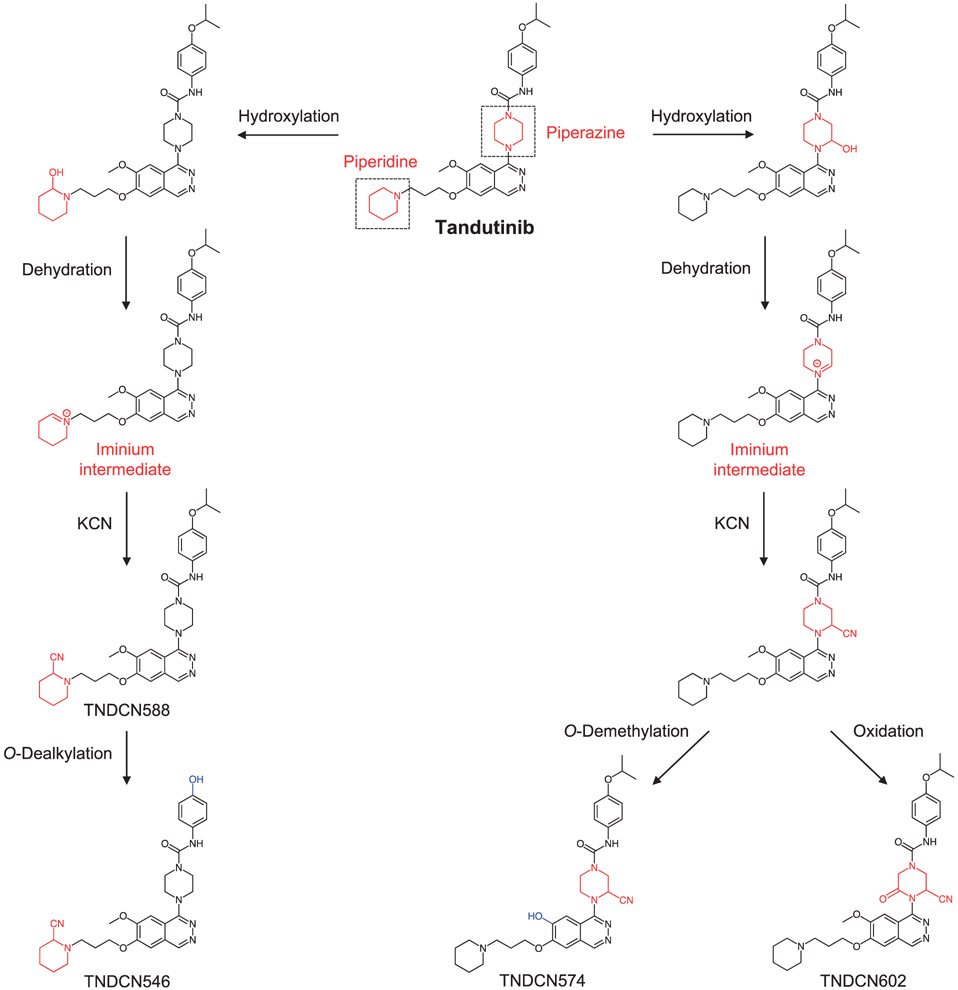
| 13 | Identification and characterization of in vitro, in vivo, and reactive metabolites of tandutinib using liquid chromatography ion trap mass spectrometry. | NS Al-Shakliah | Anal Methods 13: 399–410, 2021 |
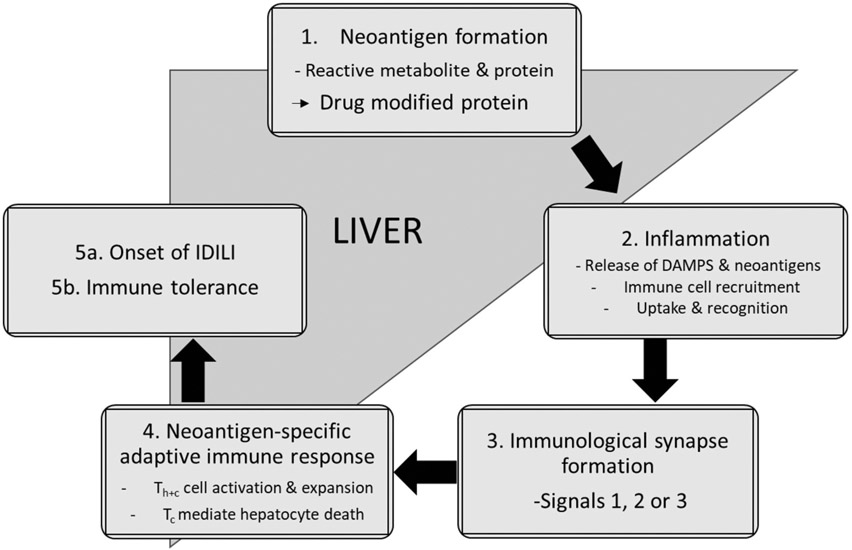
| 14 | Idiosyncratic drug-induced liver injury: Mechanistic and clinical challenges. | A Jee | Int J Mol Sci 22: 2954, 2021 |
Funding
KDJ was supported by the National Institutes of Health National Institute of General Medical Sciences [Award R35GM143044]. GPM was supported by the National Institutes of Health National Institute of General Medical Sciences [Award R01GM14063] and by the Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences (Team Science Award). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the University of Arkansas for Medical Sciences.
Footnotes
This article is dedicated to Professor Namandje Bumpus in recognition of her extensive and ongoing work in the field of bioactivation and reactivity.
Disclosure statement
Klarissa D. Jackson is co-investigator on a study funded by the Genentech Foundation. Jackson received a speaker honorarium from Genentech, Inc. to present research that is not related to this manuscript. No potential conflict of interest was reported by the author(s).
References
- Khojasteh SC, Argikar UA, Driscoll JP, Heck CJS, King L, Jackson KD, Jian W, Kalgutkar AS, Miller GP, Kramlinger V, et al. 2021. Novel advances in biotransformation and bioactivation research – 2020 year in review. Drug Metab Rev. 53(3):384–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khojasteh SC, Driscoll JP, Jackson KD, Miller GP, Mitra K, Rietjens IMCM, Zhang D 2020. Novel advances in biotransformation and bioactivation research-2019 year in review. Drug Metab Rev. 52(3):333–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khojasteh SC, Bumpus NN, Driscoll JP, Miller GP, Mitra K, Rietjens IMCM, Zhang D. 2019. Biotransformation and bioactivation reactions – 2018 literature highlights. Drug Metab Rev. 51(2):121–161. [DOI] [PubMed] [Google Scholar]
- Khojasteh SC, Miller GP, Mitra K, Rietjens IMCM. 2018. Biotransformation and bioactivation reactions – 2017 literature highlights. Drug Metab Rev. 50(3):221–255. [DOI] [PubMed] [Google Scholar]
- Khojasteh SC, Rietjens IMCM, Dalvie D, Miller G. 2017. Biotransformation and bioactivation reactions – 2016 literature highlights. Drug Metab Rev. 49(3):285–317. [DOI] [PubMed] [Google Scholar]
- Baillie TA, Dalvie D, Rietjens IMCM, Khojasteh SC. 2016. Biotransformation and bioactivation reactions – 2015 literature highlights. Drug Metab Rev. 48(2):113–138. [DOI] [PubMed] [Google Scholar]