ABSTRACT
Molecular mechanisms behind potentially inferior prognosis of old cholangiocarcinoma (CCA) patients are unclear. Prevalence of interventional targets and the difference between young and old CCA patients are valuable for promising precision medicine. A total of 188 CCA patients with baseline tumor tissue samples were subgrouped into the young (≤45 years) and old (>45 years) sub-cohorts. Somatic and germline mutation profiles, differentially enriched genetic alterations, and actionable genetic alterations were compared. An external dataset was used for the validation of molecular features and the comparison of overall survival (OS). Compared to young patients, KRAS alterations were more common in old patients (P = .04), while FGFR2 fusions were less frequent (P = .05). TERT promoter mutations were exclusively detected in old patients. The external dataset (N = 392) revealed no significant difference in OS between young and old patients; however, old patient-enriched KRAS (hazard ratio [HR]: 1.96, 95% confidence interval [CI]: 1.37–2.80) and TERT alterations (HR: 2.03, 95% CI: 1.22–3.38) were associated with inferior OS. Approximately 38.3% of patients were identified of actionable oncogenic mutations indicative of a potential response to targeted therapy or immunotherapy. Actionable FGFR2 fusions (P = .01) and BRAFV600E (P = .04) mutations were more frequent in young females than old patients. The enrichment of KRAS/TERT alterations in CCA patients over 45 years resulted in inferior OS. Approximately one-third of CCA patients were eligible for targeted therapy or immunotherapy given the actionable mutations carried, especially young females.
KEYWORDS: Genomic profiling, prognosis, patient age, cholangiocarcinoma
Introduction
Cholangiocarcinoma (CCA), with three subtypes including intrahepatic cholangiocarcinoma (iCCA), perihilar cholangiocarcinoma (pCCA) and distal cholangiocarcinoma (dCCA),1 is the second most common primary hepatic malignancy, accounting for approximately 15% primary liver tumors2. pCCA and dCCA are also collectively referred to as extrahepatic CCA (eCCA)3. Several well-known risk factors for CCA have been identified, such as alcohol consumption, smoking, hepatitis B/C virus infection, genetic alterations related to DNA repair, and the potential influence of obesity as well as drugs.4,5 CCA patients are usually asymptomatic in early stage,6,7 resulting disease being diagnosed in advanced stage and a poor prognosis. For CCA patients eligible for resection treatment followed by adjuvant chemotherapy, their median overall survival (mOS) and relapse-free survival (mRFS) are 51.1 and 24.4 months, respectively,8 with a high relapse rate.9,10 On the other hand, the expected mOS and median progression-free survival (mPFS) are 11.7 and 8.0 months, respectively, for CCA patients receiving palliative systemic chemotherapy due to unresectable disease.8
The influence of patient age on CCA prognosis is inconsistent across various studies, with unclear potential molecular mechanisms behind. Several previous studies have reported the negative association between patient age and clinical outcomes. In the United States, old iCCA patients ≥45 years had worse 5-year survival rates when compared to young patients under 45.11 One study focused on iCCA demonstrated that old patients had significantly inferior OS than young patients, and old age was identified as an independent prognostic factor in multivariate analyses.12 On the other hand, in a Canadian study including 200 iCCA and 191 pCCA patients, it has been revealed that similar survival benefits from surgery and palliative chemotherapy were observed irrespective of patient age.13 Similar results were reported in the sub-analysis of 13 prospective trials on advanced biliary cancer receiving palliative chemotherapy.14 A number of previous sequencing studies have been completed, discovering the hotspot IDH mutation in iCCA, identifying the actionable FGFR2 fusion mutation, as well as emphasizing the genomic complexity of CCA across subtypes.15–17 Nevertheless, few research works have comprehensively investigated genetic alterations in old and young CCA patients separately.
Molecular profiling of tumor tissue samples is able to guide the development of treatment option for CCA patients with advanced diseases, even though chemotherapy is currently recommended as the standard of care. The ABC−02 trial reported that patients with locally advanced or metastatic CCA, gallbladder cancer, or ampullary cancer receiving the combination of cisplatin and gemcitabine as the first-line palliative chemotherapy achieved significant survival advantage, compared to those treated with gemcitabine alone.18 After disease progression on first-line chemotherapy, the addition of FOLFOX (folinic acid, 5-FU and oxaliplatin) to active symptom control is recommended as the standard of care for second-line treatment, owing to superior OS as well as increased 6-month and 12-month OS rates.19 In addition to chemotherapy, targeted therapies could be options for CCA patients with locally advanced or metastatic disease, such as pemigatinib and infigratinib for patients with FGFR2 fusion or other rearrangement,20,21 ivosidenib for previously treated patients carrying IDH−1 mutations,22 etc. Immunotherapy might be another effective treatment option, while a subsequent trial was unable to confirm the efficacy of pembrolizumab in biliary tract carcinoma (BTC), with a responding rate as low as 6%.23,24 Therefore, it is meaningful to well understand the proportion of CCA patients harboring each potential intervention target and the differences between age groups.
Herein, this research aimed to comprehensively studied somatic and germline mutation profiles, as well as differentially expressed genes between young and old CCA patients. CCA prognosis data from one external data set were then explored. Prevalence of actionable mutations in young and old CCA patients was also investigated separately.
Material and methods
Patients
Participants were retrospectively included from the database of Nanjing Geneseeq Technology Inc., between August 2016 and December 2020. Main inclusion criteria were: 1) adults ≥18 years old; 2) with pathologically confirmed CCA; 3) with baseline tumor tissue samples within 90 days after initial diagnosis and prior to systemic treatment. TNM stages in CCA were determined according to the 8th edition of the American Joint Committee on Cancer classification. Demographics and clinical characteristics of participants, including age, gender, treatment history, the location of CCA, and family history of BTC and/or hepatocellular carcinoma (HCC), were obtained from the database of Nanjing Geneseeq Technology Inc. According to a previous study focused on a Chinese cohort in which a cutoff value of 45 years was used to identified young CCA patients,25 our patients whose ages at initial diagnosis ≤45 years were grouped into the young subgroup, and patients over 45 years at initial diagnosis were grouped into the old subgroup. The procedures of this study were approved by the Medical Ethics Committee of Nanjing Geneseeq Medical Laboratory (NSJB-MEC−2022-02), and each patient provided written informed consent.
DNA extraction, library preparation, and NGS data processing
Genomic profiling of baseline tumor tissue samples was performed using targeted next-generation sequencing (NGS) covering 425 cancer-related genes at a centralized Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited clinical laboratory (Nanjing Geneseeq Technology Inc). Genomic DNA from baseline formalin-fixed paraffin-embedded (FFPE) samples was extracted with QIAamp DNA FFPE Tissue Kit (QIAGEN); genomic DNA from leukocyte controls was extracted using DNeasy Blood and Tissue Kit (QIAGEN), from peripheral blood centrifuged at 1,800× g for 10 min at room temperature within 2 h after collection. Sequencing libraries were prepared using KAPA Hyper Prep Kit (KAPA Biosystems), and targeted enrichment was performed with customized xGen lockdown probes panel (Integrated DNA Technologies), Human cot−1 DNA (Life Technologies) and xGen Universal blocking oligos (Integrated DNA Technologies). Libraries were sequenced on Illumina Hiseq NGS platforms (Illumina). All procedures were conducted following the manufacturers’ instructions.
FASTQ file quality was controlled using Trimmomatic, removing leading/trailing low-quality (reading <15) or N bases.26 Sequencing data were then aligned to the reference human genome (build hg19) and processed using the Picard suite and the Genome Analysis Toolkit (GATK).27,28 A somatic mutation, filtered for common single nucleotide polymorphisms and germline mutations, was retained when it had at least 0.5% mutant allele frequency, at least three unique reads on different strands with good quality scores, and not present in public databases (Exome Variant Server, 1000 Genomes Project, and Exome Aggregation Consortium) at a population frequency > 1%. Gene fusions and copy number variations (CNV) were analyzed using FACTERA and ADTEx, respectively.29,30 and manually reviewed in Integrative Genomics Viewer Software (IGV, Broad Institute). The cutoffs of retaining CNV were 1.6 for CNV gain and 0.6 for CNV loss.
Statistical analysis
Fisher’s exact test and two-sample t-test were performed to compare the frequencies and means of variables between young and old CCA patients, respectively. For survival data, Kaplan-Meier curves for OS were generated, and log-rank tests were used to compare differences. Cox proportional hazards models were fitted to estimate hazard ratios (HR) and 95% confidence intervals (CI), and the proportionality of hazards was assessed using log(−log) survival plots. Data were analyzed using R software (version 4.0.3), and the survival package. Differential enrichment analysis for signaling pathways in which altered genes were located was conducted using KEGG pathway enrichment analysis. All quoted P-values were two-tailed, with values less than 0.05 considered to be statistically significant.
Results
Patient overview
A total of 188 eligible CCA patients with baseline tumor tissue samples were enrolled in this study, including 25 young patients ≤45 years and 163 old patients over 45 years (Figure 1). The median age of the entire study cohort was 60 (range: 23–81) years, and 56% (105/188) patients were males (Table 1). At initial diagnosis, 35% (65/188) and 19% (36/188) patients were diagnosed with stage IV and stage I–III disease, respectively; however, other 46% (87/188) patients had missing data for clinical stage. Within the panel covering 425 genes, 33% (62/188) patients were detected with tumor mutational burden (TMB) of at least 6 muts/Mb. Although treatment history records were missing in 81% (152/188) participants, it presented as a majority of patients treated with chemotherapy alone (15%, 28/188). The remaining eight patients had ever received targeted therapy (2%, 4/188) or immunotherapy (2%, 4/188). Family history of BTC/HCC were only identified in 3% patients (6/188), while patients with missing values accounted for 18% of the study cohort. The median ages of the young and old subgroups were 39 (range: 23–44) and 61 (range: 46–81) years, respectively (Table 1). Compared to the young subgroup, female patients appeared to be less frequently observed in the old subgroup (42% vs. 60%, P = .13); however, the proportions of patients diagnosed with stage I–III disease were similar between two subgroups (19% vs. 20%, P > .99). Of note, a positive relationship between patient age and TMB level at initial diagnosis was observed (≥6 muts/Mb: 36% vs. 12%, P = .02).
Figure 1.
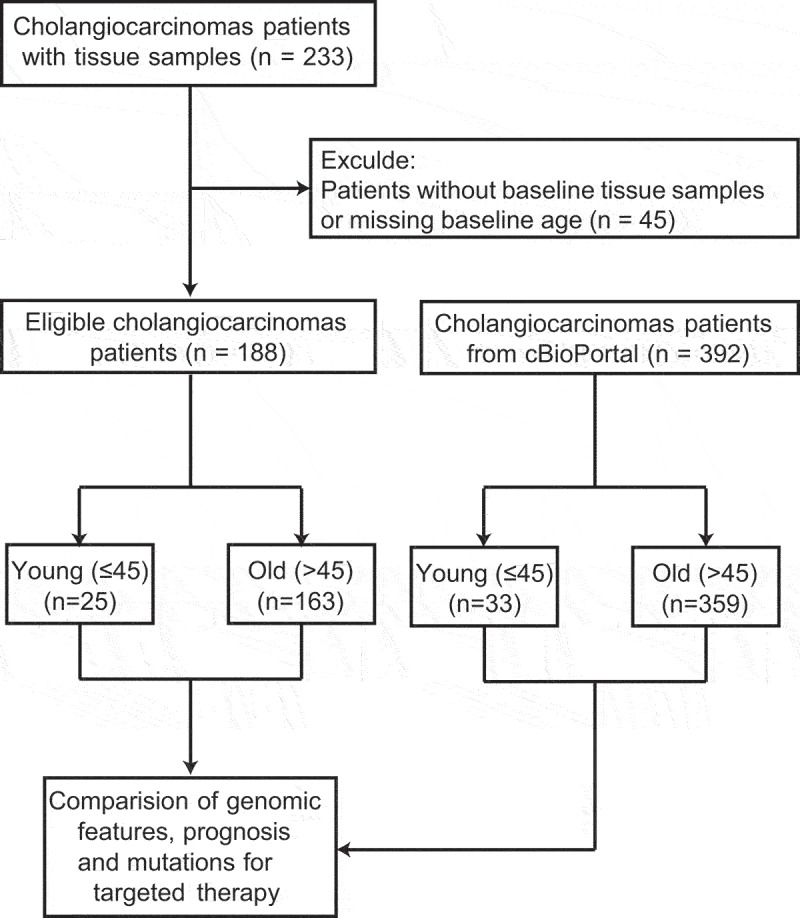
The flowchart of enrollment and analyzable patients.
A total of 188 cholangiocarcinoma patients were included in the study cohort, including 25 young patients and 163 old patients. From an external data set, 392 patients (33 young and 359 old patients) with baseline genetic alteration data and overall survival data were identified.
Table 1.
Demographics and clinical characteristics of patients.
| Characteristics | Overall (n = 188) |
Young (n = 25) |
Old (n = 163) |
P value |
|---|---|---|---|---|
| Age at initial diagnosis, median (range), y | 60 (23–81) | 39 (23–44) | 61 (46–81) | <0.001* |
| Gender, No. (%) | 0.13 | |||
| Female | 83 (44) | 15 (60) | 68 (42) | |
| Male | 105 (56) | 10 (40) | 95 (58) | |
| Clinical stage at initial diagnosis, No. (%) | >0.99 | |||
| I – III | 36 (19) | 5 (20) | 31 (19) | |
| IV | 65 (35) | 10 (40) | 55 (34) | |
| Unknown | 87 (46) | 10 (40) | 77 (47) | |
| TMB, No. (%) | 0.02* | |||
| <6 muts/Mb | 126 (67) | 22 (88) | 104 (64) | |
| ≥6 muts/Mb | 62 (33) | 3 (12) | 59 (36) | |
| Treatment, No. (%) | >0.99 | |||
| Chemotherapy | 28 (15) | 5 (20) | 23 (14) | |
| Targeted therapy | 4 (2) | 1 (4) | 3 (2) | |
| Immunotherapy | 4 (2) | 1 (4) | 3 (2) | |
| Unknown | 152 (81) | 18 (72) | 134 (82) | |
| Family history of BTC/HCC, No. (%) | 0.18 | |||
| With | 6 (3) | 2 (8) | 4 (2) | |
| Without | 149 (79) | 18 (72) | 131 (80) | |
| Unknown | 33 (18) | 5 (20) | 28 (17) |
Abbreviations: TMB, Tumor mutational burden; BTC, biliary tract carcinoma; HCC, hepatocellular carcinoma.
*Statistically significant
Distinctive mutation landscapes and development mechanisms related to patient age
For somatic alterations, the most common altered gene in the entire study cohort was TP53 (49%, 92/188), detected in 44% young patients and 50% old patients. Other frequently mutated genes included KRAS (31%, 59/188), CDKN2A (22%, 42/188), and ARID1A (16%, 31/188). We also observed CNV gain alterations of MCL1, MYC, PTK2, and MDM2, as well as CNV loss alterations of CDKN2A, CDKN2B, and PTPRD (Figure 2a). Compared to 25 young patients, mutated KRAS genes were more frequently identified in old patients (34% vs. 12%, P = .04), and TERT promoter mutations were exclusively detected in old patients (10% vs. 0%, P = .13) (Figure 2b). However, altered PTEN (16% vs. 2%, P = .01), NFE2L2 (16% vs. 2%, P = .01), and NOTCH1 (16% vs. 3%, P = .02) genes were enriched in the young subgroup. Notably, FGFR2 fusions, one type of CCA actionable mutations, were also more frequently detected in young patients than old patients (12% vs. 2%, P = .05). In addition, the prevalence of DDR2 alterations appeared to be relatively high in young patients in comparison to old patients (16% vs. 5%, P = .06). When compared iCCA with eCCA patients’ samples, TP53 (57% vs. 43%, P = .04) and SMAD4 mutations (21% vs. 12%, P = .02) were more prevalent in eCCA, whereas IDH1 (9% vs. 3%, P = .05) mutations and FGFR2 fusions (6% vs. 0%, P = .04) appeared to be more common in iCCA.
Figure 2.
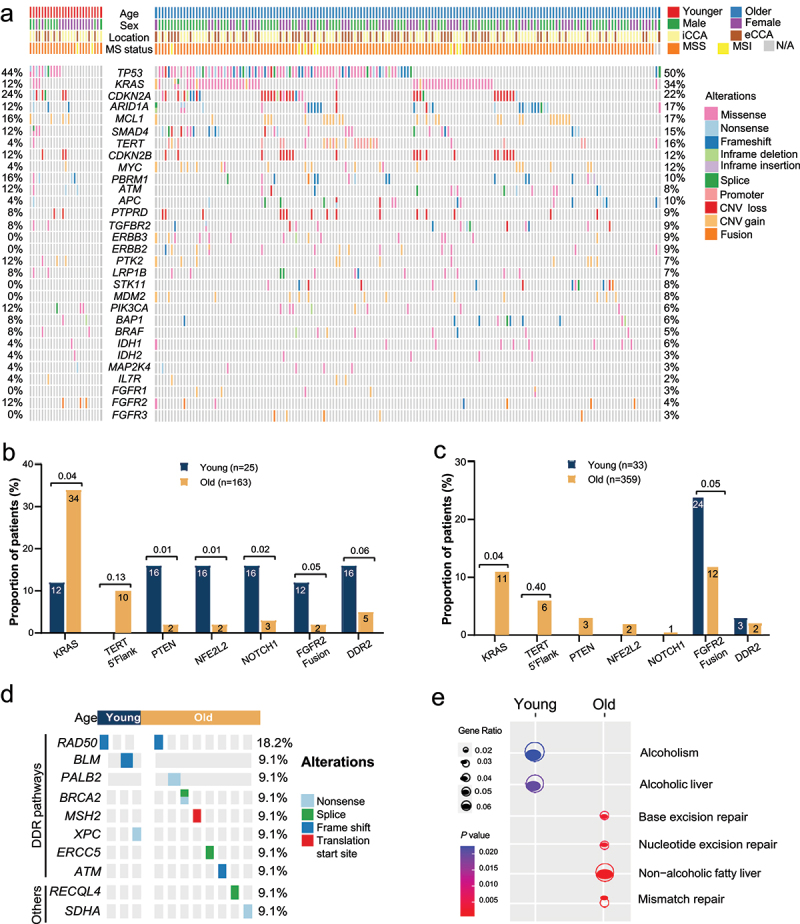
Baseline genomic profiles of young and old patients with cholangiocarcinoma.
(a) The genomic profiles of 188 patients in the study cohort. (b) Somatic alterations with different prevalence between young and old patients in the study cohort. (c) Somatic alterations with different prevalence between young and old patients in the external cohort. (d) Germline alterations of patients in the study cohort. (e) KEGG pathway enrichment analysis for altered genes.
From one bi-institutional study,31 a total of 392 iCCA patients with at least one somatic alteration and available OS data were identified for the validation of genomic features and the exploration of prognosis. Thirty-three patients aged ≤45 years were included in the young subgroup, and the remaining 359 over−45-year-old patients were included in the old subgroup. Genomic features and OS data of patients from the external data set were downloaded from cBioPortal for Cancer Genomics (https://www.cbioportal.org/study/summary?id=ihch_msk_2021). Intriguingly, in this external cohort, the proportion of patients with TMB level ≥ 6 muts/Mb was only 11%, and no significant association between patient age and TMB level was observed (Table S1). Although no KRAS alteration was detected in the young subgroup, the prevalence of KRAS alterations in old patients was only 11%, being much lower than in our study cohort (34%) (Figure 2c). TERT promoter mutations were exclusively detected in old patients, which was consistent with the finding in our study cohort. Interestingly, none of PTEN, NFE2L2, and NOTCH1 alterations were detected in the young subgroup of the external cohort, while the proportion of old patients carrying altered PTEN (3%), NFE2L2 (2%), and NOTCH1 (1%) were similar to our study cohort. The association between patient age and FGFR2 fusions detection remained significant; however, the prevalence of FGFR2 fusion in this external cohort was higher than that in our study cohort (13% vs. 3%, P < .001).
Germline mutations were identified in three (12%) young patients and eight (5%) old patients in our study cohort. Except RECQL4 and SDHA, other mutated genes were all located in the DNA damage response pathway (Figure 2d). KEGG analysis demonstrated that alcoholism and alcoholic liver relevant genes were frequently altered among young patients; however, genetic alterations related to nonalcoholic fatty liver and gene repair were enriched in the old subgroup (Figure 2e). The result of KEGG analysis could potentially reveal distinctive risk factors for CCA in young and old people.
Inferior overall survival due to genetic alterations rather than patient age
The mOS of the external cohort was 32.0 (95% CI: 26.8–36.6) months. Compared to patients with wide-type KRAS gene, patients harboring altered KRAS genes had significantly inferior OS (mOS: 17.8 vs. 33.6 months, HR: 1.96, 95% CI: 1.37–2.80, Figure 3a). Similar results were obtained in patients with mutated TERT genes (mOS: 16.9 vs. 32.7 months, HR: 2.03, 95% CI: 1.22–3.38, Figure 3b). We were not able to observe significant differences in OS across patients with different FGFR2 mutation status. The mOS of patients with FGFR2 fusions, with other types of FGFR2 alterations, and without FGFR2 alteration were 48.2, 23.8, and 30.7 months respectively. The hazard of death was 14% lower in CCA patients harboring FGFR2 fusion when compared to those without FGFR2 alteration, whereas the 95% CI (0.59–1.27) covered 1 (Figure S1a). Similarly, there was no significantly different survival outcomes between patients with IDH1 mutations and without (mOS: 35.6 vs. 30.8 months, HR: 0.81, 95% CI: 0.61–1.09, Figure S1b). TP53-mutated CCA patients displayed significantly poorer OS than those with wild-type TP53 genes (mOS: 13.7 vs. 36.6 months, HR: 2.23, 95% CI: 1.67–2.99, Figure S1c). Furthermore, a trend was observed that higher TMB level might be associated with poorer OS (mOS: 26.7 vs. 34.1 months, HR: 1.25, 95% CI: 0.98–1.60, Figure S1d).
Figure 3.
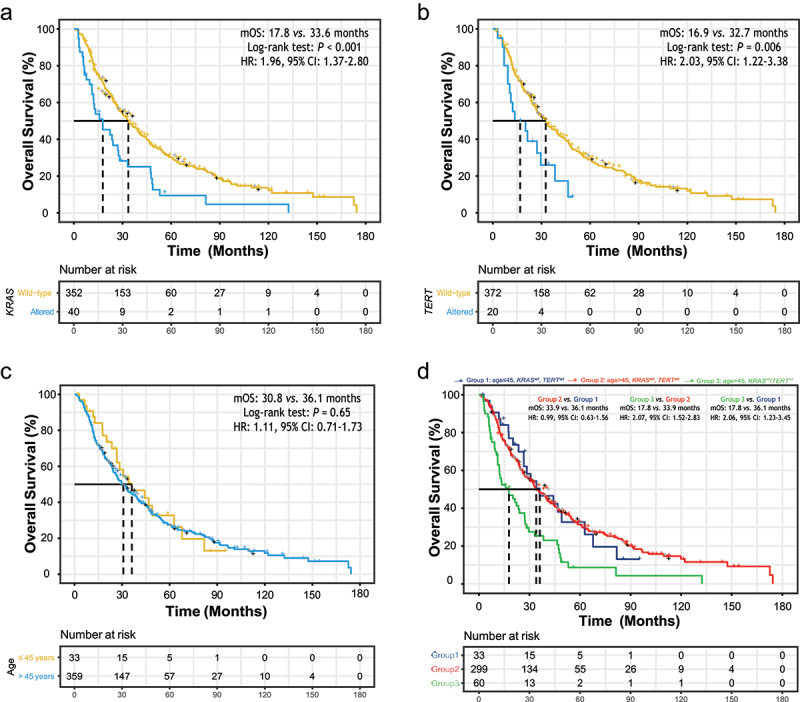
The association of overall survival (OS) with baseline clinical and genetic characteristics.
(a) Altered KRAS gene was associated with inferior OS. Median OS (mOS): 17.8 vs. 33.6 months, hazard ratio (HR): 1.96, 95% confidence interval (CI): 1.37–2.80. (b) Altered TERT gene was associated with inferior OS. mOS: 16.9 vs. 32.7 months, HR: 2.03, 95% CI: 1.22–3.38. (c) Older patient age was not significantly associated with OS. mOS: 30.8 vs. 36.1 months, HR: 1.11, 95% CI: 0.71–1.73. (d) Old patients without altered KRAS or TERT mutations had similar OS in comparison with young patients without KRAS or TERT mutations, while old patients harboring KRAS or TERT alterations had inferior OS (mOS: 17.8 vs. 33.9 months, HR: 2.07, 95% CI: 1.52–2.83; mOS: 17.8 vs. 36.1 months, HR: 2.06, 95% CI: 1.23–3.45).
We then tried confirming the previously reported association between patient age and OS. The mOS of 359 old patients and 33 young patients were 30.8 (95% CI: 26.5–36.6) months and 36.1 (95% CI: 26.7–67.8) months, respectively, without significant difference in OS between two subgroups (HR: 1.11, 95% CI: 0.71–1.73, Figure 3c). KRAS and TERT alterations, both enriched in old patients and associated with inferior OS, were potential confounders or effect modifiers of the relationship between patient age and OS. Thus, patients were further grouped according to their ages and KRAS/TERT gene alterations, and stratified analyses were conducted. For patients without altered KRAS/TERT genes, old patients (Group 2) display similar OS to young patients (Group 1) (mOS: 33.9 vs. 36.1 months, HR: 0.99, 95% CI: 0.63–1.56, Figure 3d). The association of KRAS/TERT alterations with OS remained significant when old patient with altered KRAS/TERT genes (Group 3) were compared to those without (Group 2) (mOS: 17.8 vs. 33.9 months, HR: 2.07, 95% CI: 1.52–2.83). As expected, Group 3 showed inferior OS than Group1 (mOS: 17.8 vs. 36.1 months, HR: 2.06, 95% CI: 1.23–3.45).
Therapeutic implications of patient’s actionable genomic alterations
Of 188 individuals in our study cohort, 72 (38.3%) patients were potentially eligible for receiving targeted therapy or immunotherapy, and their actionable alterations or immune-related genetic features were summarized in Table 2. Potential intervention targets mainly included IDH1/2 (7.4%), BRAFV600E (2.1%), PIK3CA (1.6%) and BRCA1/2 (4.2%) mutations, ERBB2 (3.2%) and MET (3.7%) CNV gains, FGFR2/3 fusions (5.3%), and microsatellite instability-high (5.3%) (Figure 4a). All the breakpoints of seven patients carrying FGFR2 fusions were located in FGFR2 intron 17, and the passenger fusion gene of each FGFR2 fusion was unique (Figure 4b). In addition to six passenger fusion genes having been reported, including WAC, CTNNA3, BICC1, HOOK1, RBM20, and AHCYL1, a novel passenger gene, DAAM1, was observed in our study cohort. FGFR3 fusion were observed in another three patients, with a same passenger fusion gene of TACC3, while breakpoints included FGFR3 intron 17 and intron 18. Notably, a total of 55 patients had KRAS mutations, and 76% patients with KRASG12X mutations, while KRASG12C mutations were identified in only five patients (Figure 4c), suggesting the motivation of developing treatments to tackle KRAS mutations other than KRASG12C inhibitors.
Table 2.
Actionable genomic features in cholangiocarcinoma.
| Biomarkers | No. patient | Percentage (%) |
|---|---|---|
| IDH1/2 Mutation | 14 | 7.4 |
| FGFR1–4 Fusion | 10 | 5.3 |
| BRCA1/2 Mutation | 8 | 4.3 |
| MET CNV Gain | 7 | 3.7 |
| ERBB2 CNV Gain | 6 | 3.2 |
| KRAS G12C | 5 | 2.7 |
| MSI-High | 10 | 5.3 |
| TMB-High (>10 muts/Mb) | 27 | 14.4 |
| Total | 72 | 38.3 |
Abbreviations: MSI, microsatellite instability; TMB, Tumor mutational burden.
Figure 4.
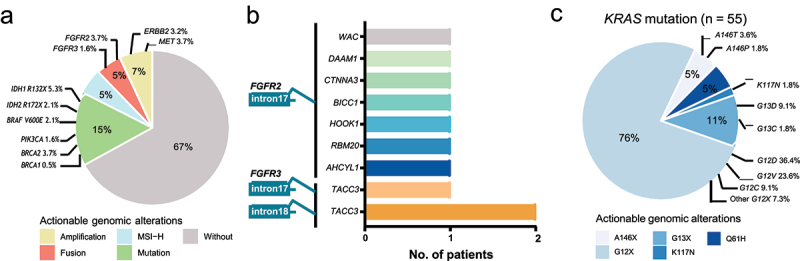
The prevalence of potentially actionable genetic alterations.
(a) The prevalence of potentially actionable genetic alterations. Approximately 33% of 188 cholangiocarcinoma patients in our study cohort were eligible for targeted therapy. (b) Seven patients carried FGFR2 fusion mutations, with all breakpoints in FGFR2 intron 17, and a novel passenger gene, DAAM1, was detected. FGFR3 fusion were observed in another three patients, with a same passenger fusion gene of TACC3. (c) KRAS mutations were identified in 55 of 188 patients, while KRASG12C mutations only accounted for 9% cases.
The proportions of patients with the potential of receiving targeted therapy were similar between young and old patients (36% vs. 27%, P = .35, Figure S2a). Females were more likely to be eligible for targeted therapy irrespective of their ages (young subgroup: 60% vs. 0%, P = .002; old subgroup: 37% vs. 20%, P = .02). Treatments targeting FGFR2 fusions (20% vs. 2%, P = .01) and BRAFV600E (13% vs. 1%, P = .04) mutations might benefit higher percentages of young females than old patients (Figure S2b). Of note, no young male CCA patients were identified with FGFR2 fusions or BRAFV600E mutations, and ERBB2 and BRCA actionable mutations were exclusively observed in the old subgroup. Although KRAS mutations were more frequently detected in old patients than in young patients, the proportions of KRASG12C inhibitors eligible patients were close between two subgroups (2% vs. 4%, P = .51).
Discussion
We addressed a comprehensive comparison of molecular features, prognosis data, and actionable mutations for target treatment between young and old CCA patients. Compared to 25 young patients whose CCA development mechanism was probably related to alcoholism consumption, CCA might attribute to DNA repair issues or nonalcoholic fatty liver in 163 old patients. Although no significant difference in OS was observed between young and old CCA patients, KRAS and TERT promoter mutations, which displayed higher prevalence in old patients, were associated with inferior OS. Nevertheless, compared to old patients, the proportion of patients who were potentially eligible for target treatment was likely to be larger in young patients, especially young females, due to more common FGFR2 fusions and BRAF mutations.
Previous studies have not drawn consistent conclusions on whether old CCA patients have poorer prognosis than young patients. Based on 11,127 iCCA patients enrolled between 1995 and 2004, higher 1-year (HR: 1.77, 95% CI: 1.56–2.02) and 5-year all-cause mortalities (HR: 1.65, 95% CI: 1.49–1.84) were observed in patients at least 45 years old, compared to patients below 45.11 Age was also identified as a separate factor predicting prognosis among gallbladder cancer and iCCA patients proceed with gemcitabine and S−1 combination chemotherapy as first-line palliative treatment.32 Similarly, patient age could serve as a prognostic factor for hilar CCA patients treated with surgical resection.33 In present study, we did not detect a significantly inferior OS among old patients, which was consistent with a couple of previous studies. For instance, in one study including 136 CCA patients, OS was not strongly associated with patient age (HR: 1.01, 95% CI: 0.99–1.03).34 Another study focused on CCA patients treated with resection neither detected an obvious relationship between prognosis and age.35 Of note, in a part of previous studies, patient age was treated as a numeric variable and included in regression models directly; however, in other studies where patient age was used as a categorical variable, different thresholds were applied over patient age to identify young and old patients. Moreover, the CCA subtype compositions differed across studies, and several studies focused on BTC also included patients with gallbladder cancers. Therefore, the heterogeneity of age group threshold and subtype of CCA cases might result in inconsistent conclusions on the association of patient age with prognosis.
In our study, KRAS and TERT alterations were enriched in old patients over 45 and inferior OS was associated with altered KRAS or TERT gene, which could partially explain the worse OS in old patients observed in some previous studies. One study where 81% of 195 total patients were diagnosed with iCCA also demonstrated that altered KRAS gene, detected in 13% patients, was negatively related to OS (P = .026). That study also showed KRAS alterations occurred with greater frequency in eCCA; however, the enrichment of KRAS alterations in old patients was unable to be confirmed due to lacking relevant data.16 In another study including 85 (85/123, 69%) iCCA patients and investigating ctDNA from their blood samples, a trend of higher prevalence of altered KRAS genes among patients ≥50 years was observed, even though the difference between early-onset (age <50 years) and old patients was not statistically significant (approximately 33% vs. 21%).36 Conversely, TERT promoter mutations appeared to be more commonly detected in early-onset patients than old patients (approximately 10% vs. 2%). In comparison with early-onset patient, TP53 mutation prevalence was relatively high in old patients (67% vs. 35%). Although we did not observe such an enrichment of mutated TP53 genes in the old subgroup of our study cohort, a strong association between TP53 mutations and poorer OS was observed. Similar negative association between TP53 mutation and clinical outcomes was also observed in BTC patients undergoing surgery treatment37 We demonstrate, for the first time to our knowledge, the potentially distinctive prognosis between young and old CCA patients might be rationalized by different prevalence of genetic alterations, whereas larger study cohorts with diverse ethnic groups are required to further confirm our findings.
Development of drugs targeting genetic alterations in CCA has got great achievements since the genomic profiling studies being launched. Ivosidenib is one potential option for iCCA patients, of whom IDH1/2 mutations are frequently identified, after chemotherapy-refractory. In one phase III randomized controlled trial including 185 post-chemotherapy iCCA patients, significantly improved PFS was observed in patients receiving ivosidenib, in comparison with placebo (HR: 0.37, 95% CI: 0.25–0.54).22 For patients harboring FGFR1/2/3 fusion, detected in approximately 5% patients in our study cohort, several FGFR inhibitors have been developed.20,38 Also, the combination of BRAF and MEK inhibitors, dabrafenib plus trametinib, showed promising treatment effects among CCA patients carrying BRAFV600E mutations.39 However, owning to the FGFR2 fusion less commonly identified among fluke-related CCA patients.40 as well as the low prevalence of BRAFV600E mutations, targeted therapy plans should be developed depending on personal genomic profiles. Moreover, over 10% patients in our study cohort harboring BRCA1/2 mutations, ERBB2 or MET CNV gain, for which potential targeted therapy might be available. It was worth noting that only approximately 10% altered KRAS genes were KRASG12C and most KRASG12X mutations were other than KRASG12C, suggesting the studies in which the efficacy of KRASG12C inhibitors could be investigated in extended cohorts. Old patients might get great benefit from these studies, as they were observed with higher KRAS mutation prevalence than young patients. For immunotherapy, a group of studies have indicated the modest efficacy in CCA, and a phase III clinical trial of pembrolizumab combined with gemcitabine and cisplatin is ongoing.41 Although chemotherapy is currently serving as the standard of care for CCA patients not eligible for surgical resection, personalized targeted therapy and immunotherapy guided by genomic profiles exhibited improved clinical outcomes when compared to routine chemotherapy (objective response rate: 87.5% vs. 25%, P < .001; mOS: not reached vs. 6.5 months, HR: 0.10, 95% CI: 0.02–0.48).42
Our study does have limitations, mainly including unknown CCA subtypes, and considerable missing data of treatment history. As a result, we were neither able to conduct stratified analyses for each CCA subtype separately nor to provide a landscape view of CCA management in real world. Another limitation was the lack of clinical stages at initial diagnosis, leading to the difficulty of controlling for the potentially confounding effect of clinical stages on OS. We had no prognosis OS data in our own study cohort, and main findings in prognosis was based on the external cohort mainly consisting of CCA cases in western countries. However, genomic landscape and etiology of CCA might vary in different countries, such as the higher proportion of microsatellite instability-high CCA patients in Asian countries,43 resulting in potentially limited generalizability of the conclusion in prognosis. Additionally, due to the lack of clinical data, we were neither able to show Carbohydrate Antigen 19–9 levels of our study cohort nor to compare the difference between young and old patients. Finally, we could not assess hepatitis B/C virus infection history because of the retrospective structure of our study, and the sample size of young patients below 45 years was relatively small. Further studies with more young patients and complete hepatitis data were needed.
Conclusions
Previously reported inferior prognosis in old CCA patients might be rationalized by more frequent KRAS/TERT alterations related to poor OS. Over 35% patients were suitable for potential personalized targeted therapy or immunotherapy, and young females were more likely to be eligible for treatment targeting FGFR fusions or BRAFV600E mutations.
Acknowledgments
The authors thank all the patients who participated in this study.
Biographies
Junhua Wang is a clinician at the Department of Biliary-pancreatic Surgery, The First People's Hospital of Foshan.
Yaoting Shi is a clinician at the Department of Medical Oncology, Beidahuang Industry Group General Hospital.
Jianbo Chen is a clinician at the Department of Medical Oncology, The First Affiliated Hospital of Xiamen University.
Juying Liu is a clinician at the Department of Radiation Oncology, Jiangsu Cancer Hospital & Jiangsu Institute of Cancer Research & The Affiliated Cancer Hospital of Nanjing Medical University.
Xiaotian Zhao is a senior research associate at Geneseeq Research Institute, Nanjing Geneseeq Technology Inc.
Jiaohui Pang is a senior research project manager at Geneseeq Research Institute, Nanjing Geneseeq Technology Inc.
Ximin Sun is a clinician at the Department of Hepatobiliary Surgery, The First Hospital Affiliated to AMU (Southwest Hospital).
Yichen Tian is a clinician at the Department of Hepatobiliary Surgery, The First Hospital Affiliated to AMU (Southwest Hospital).
Qiuxiang Ou is the leader of translational research group of Geneseeq Research Institute, and the Associate Director of R&D of Nanjing Geneseeq Technology Inc.
Feng Xia is a hepatobiliary surgery expert working at the Department of Hepatobiliary Surgery, The First Hospital Affiliated to AMU (Southwest Hospital).
Yunjie Chen is a hepatobiliary surgery expert working at the Department of Hepatopancreatobiliary Surgery, Hwa Mei Hospital, University of Chinese Academy of Sciences (Ningbo No. 2 Hospital)
Funding Statement
Key HwaMei Research Fund (Grant No. 2020HMZD17 to Yunjie Chen), Xiamen Municipal Bureau of Science and Technology (Grant No. 3502Z20189012 to Jianbo Chen).
Abbreviations
- BTC
biliary tract carcinoma
- CCA
cholangiocarcinoma
- CI
confidence interval
- CNV
copy number variation
- dCCA
distal cholangiocarcinoma
- eCCA
extrahepatic cholangiocarcinoma
- FFPE
formalin-fixed paraffin-embedded
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- iCCA
intrahepatic cholangiocarcinoma
- mOS
median overall survival
- mRFS
median relapse-free survival
- NGS
next-generation sequencing
- OS
overall survival
- pCCA
perihilar cholangiocarcinoma
- PFS
progression-free survival
- TMB
tumor mutational burden
Authors’ contributions
YC, and FX conceived and design the study. JW, YS, JC, JL, XZ, JP, XS, YT, and QO analyzed data and interpreted results. All authors wrote, revised and reviewed the manuscript. YC, and FX supervised the whole project. All authors read and approved the final manuscript.
Disclosure statement
Xiaotian Zhao, Jiaohui Pang, and Qiuxiang Ou are employees of Nanjing Geneseeq Technology Inc., China. The remaining authors have nothing to disclose.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of Nanjing Geneseeq Medical Laboratory (NSJB-MEC−2022-02), and written informed consent was provided by each participant. The work was carried out in accordance with The Declaration of Helsinki.
References
- 1.Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ.. Cholangiocarcinoma—evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15(2):95–10. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, et al. Expert consensus document: cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European network for the study of cholangiocarcinoma (ENS-CCA). Nat Rev Gastro Hepat. 2016;13(5):261–280. doi: 10.1038/nrgastro.2016.51. [DOI] [PubMed] [Google Scholar]
- 3.Khan AS, Dageforde LA. Cholangiocarcinoma. Surg Clin North Am. 2019;99(2):315–335. doi: 10.1016/j.suc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Clements O, Eliahoo J, Kim JU, Taylor-Robinson SD, Khan SA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: a systematic review and meta-analysis. J Hepatol. 2020;72(1):95–103. doi: 10.1016/j.jhep.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Xiong J, Xu W, Bian J, Huang H, Bai Y, Xu Y, Lu X, Zhao H. Aspirin use is associated with a reduced risk of cholangiocarcinoma: a systematic review and meta-analysis. Cancer Manag Res. 2018;10:4095. doi: 10.2147/CMAR.S173197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvaro D, Bragazzi MC, Benedetti A, Fabris L, Fava G, Invernizzi P, Marzioni M, Nuzzo G, Strazzabosco M, Stroffolini T, et al. Cholangiocarcinoma in Italy: a national survey on clinical characteristics, diagnostic modalities and treatment. Results from the “cholangiocarcinoma” committee of the italian association for the study of liver disease. Digestive Liver Dis. 2011;43(1):60–65. doi: 10.1016/j.dld.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Forner A, Vidili G, Rengo M, Bujanda L, Ponz‐Sarvisé M, Lamarca A. Clinical presentation, diagnosis and staging of cholangiocarcinoma. Liver Int. 2019;39(S1):98–107. doi: 10.1111/liv.14086. [DOI] [PubMed] [Google Scholar]
- 8.Banales JM, Marin JJ, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, Cardinale V, Carpino G, Andersen JB, Braconi C, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastro Hepat. 2020;17(9):557–588. doi: 10.1038/s41575-020-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koerkamp BG, Wiggers JK, Allen PJ, Besselink MG, Blumgart LH, Busch OR, Coelen RJ, D’Angelica MI, DeMatteo RP, Gouma DJ, et al. Recurrence rate and pattern of perihilar cholangiocarcinoma after curative intent resection. J Am Coll Surg. 2015;221(6):1041–1049. doi: 10.1016/j.jamcollsurg.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komaya K, Ebata T, Yokoyama Y, Igami T, Sugawara G, Mizuno T, Yamaguchi J, Nagino M. Recurrence after curative-intent resection of perihilar cholangiocarcinoma: analysis of a large cohort with a close postoperative follow-up approach. Surgery. 2018;163(4):732–738. doi: 10.1016/j.surg.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Antwi SO, Mousa OY, Patel T. Racial, ethnic, and age disparities in incidence and survival of intrahepatic cholangiocarcinoma in the United States; 1995-2014. Ann Hepatol. 2018;17(2):274–285. doi: 10.5604/01.3001.0010.8659. [DOI] [PubMed] [Google Scholar]
- 12.Cho SY, Park S-J, Kim SH, Han S-S, Kim Y-K, Lee K-W, Lee S-A, Hong EK, Lee WJ, Woo SM, et al. Survival analysis of intrahepatic cholangiocarcinoma after resection. Ann Surg Oncol. 2010;17(7):1823–1830. doi: 10.1245/s10434-010-0938-y. [DOI] [PubMed] [Google Scholar]
- 13.Horgan A, Knox J, Aneja P, Le L, McKeever E, McNamara M. Patterns of care and treatment outcomes in older patients with biliary tract cancer. Oncotarget. 2015;6(42):44995. doi: 10.18632/oncotarget.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNamara MG, Bridgewater J, Lopes A, Wasan H, Malka D, Jensen LH, Okusaka T, Knox JJ, Wagner D, Cunningham D, et al. Systemic therapy in younger and elderly patients with advanced biliary cancer: sub-analysis of ABC-02 and twelve other prospective trials. Bmc Cancer. 2017;17(1):1–9. doi: 10.1186/s12885-017-3266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farshidfar F, Zheng S, Gingras M-C, Newton Y, Shih J, Robertson AG, Hinoue T, Hoadley KA, Gibb EA, Roszik J, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH-mutant molecular profiles. Cell reports. 2017;18(11):2780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu M, Kemeny NE, O’Reilly EM, El-Dika I, Jarnagin WR, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention molecular profiling of biliary tract cancer. Clin Cancer Res. 2018;24(17):4154–4161. doi: 10.1158/1078-0432.CCR-18-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silverman IM, Murugesan K, Lihou CF, Féliz L, Frampton GM, Newton RC, Tada H, Albacker LA, Burn TC. Comprehensive genomic profiling in FIGHT-202 reveals the landscape of actionable alterations in advanced cholangiocarcinoma. Am J Clin Oncol. 2019;37(15_suppl):4080–4080. doi: 10.1200/JCO.2019.37.15_suppl.4080. [DOI] [Google Scholar]
- 18.Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, Madhusudan S, Iveson T, Hughes S, Pereira SP, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362(14):1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 19.Lamarca A, Palmer DH, Wasan HS, Ross PJ, Ma YT, Arora A, Falk S, Gillmore R, Wadsley J, Patel K, et al. Second-line FOLFOX chemotherapy versus active symptom control for advanced biliary tract cancer (ABC-06): a phase 3, open-label, randomised, controlled trial. Lancet Oncol. 2021;22(5):690–701. doi: 10.1016/S1470-2045(21)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abou-Alfa GK, Sahai V, Hollebecque A, Vaccaro G, Melisi D, Al-Rajabi R, Paulson AS, Borad MJ, Gallinson D, Murphy AG, et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21(5):671–684. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Javle M, Kelley R, Roychowdhury S, Weiss K, Abou-Alfa G, Macarulla T, Sadeghi S, Waldschmidt D, Zhu AX, Goyal L, et al. AB051. P-19. A phase II study of infigratinib (BGJ398) in previously-treated advanced cholangiocarcinoma containing FGFR2 fusions. Hepatobiliary Surg Nutr. 2019;8(S1):AB051–AB051. doi: 10.21037/hbsn.2019.AB051. [DOI] [Google Scholar]
- 22.Abou-Alfa GK, Macarulla T, Javle MM, Kelley RK, Lubner SJ, Adeva J, Cleary JM, Catenacci DV, Borad MJ, Bridgewater J, et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIdhy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21(6):796–807. doi: 10.1016/S1470-2045(20)30157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, Rugo HS, Cohen RB, O’Neil BH, Mehnert JM, et al. T-cell–inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: kEYNOTE-028. J Clin Oncol. 2019;37(4):318–327. doi: 10.1200/JCO.2018.78.2276. [DOI] [PubMed] [Google Scholar]
- 25.Feng H, Tong H, Yan J, He M, Chen W, Wang J. Genomic features and clinical characteristics of adolescents and young adults with cholangiocarcinoma. Front Oncol. 2020;9:1439. doi: 10.3389/fonc.2019.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amarasinghe KC, Li J, Halgamuge SK. CoNVEX: copy number variation estimation in exome sequencing data using HMM. BMC bioinformatics: Springer; 2013. p. 1–9. doi: 10.1186/1471-2105-14-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, Alizadeh AA. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30(23):3390–3393. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boerner T, Drill E, Pak LM, Nguyen B, Sigel CS, Doussot A, Shin P, Goldman DA, Gonen M, Allen PJ, et al. Genetic determinants of outcome in intrahepatic cholangiocarcinoma. Hepatology. 2021;74(3):1429–1444. doi: 10.1002/hep.31829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki T, Isayama H, Nakai Y, Togawa O, Kogure H, Ito Y, Yamamoto K, Mizuno S, Yagioka H, Yashima Y, et al. Prognostic factors in patients with advanced biliary tract cancer receiving chemotherapy. Cancer Chemother Pharmacol. 2011;67(4):847–853. doi: 10.1007/s00280-010-1360-7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang B-Y, Lu Y, Sun C-D, Mu P, Dong Q. Surgical treatment and prognostic analysis of 93 cases of hilar cholangiocarcinoma. Am J Med Sci. 2010;339(3):221–224. doi: 10.1097/MAJ.0b013e3181c7c8b4. [DOI] [PubMed] [Google Scholar]
- 34.Singal AG, Rakoski MO, Salgia R, Pelletier S, Welling TH, Fontana RJ, LOK AS, MARRERO JA. The clinical presentation and prognostic factors for intrahepatic and extrahepatic cholangiocarcinoma in a tertiary care centre. Aliment Pharmacol Ther. 2010;31(6):625–633. doi: 10.1111/j.1365-2036.2009.04218.x. [DOI] [PubMed] [Google Scholar]
- 35.Li S-Q, Liang L-J, Hua Y-P, Peng B-G, He Q, Lu M-D, Chen D. 2009. Long-term outcome and prognostic factors of intrahepatic cholangiocarcinoma. Chin Med J. 122(19):2286–2291. [PubMed] [Google Scholar]
- 36.Mody K, Kasi PM, Yang J, Surapaneni PK, Bekaii-Saab T, Ahn DH, Mahipal A, Sonbol MB, Starr JS, Roberts A, et al. Circulating tumor DNA profiling of advanced biliary tract cancers. JCO Precis Oncol. 2019;3(3):1–9. doi: 10.1200/PO.18.00324. [DOI] [PubMed] [Google Scholar]
- 37.Conci S, Ruzzenente A, Simbolo M, Bagante F, Rusev B, Isa G, Lawlor RT, Pedrazzani C, Iacono C, Guglielmi A, et al. Multigene mutational profiling of biliary tract cancer is related to the pattern of recurrence in surgically resected patients. Updates Surg. 2020;72(1):119–128. doi: 10.1007/s13304-020-00718-5. [DOI] [PubMed] [Google Scholar]
- 38.Javle M, Lowery M, Shroff RT, Weiss KH, Springfeld C, Borad MJ, Ramanathan RK, Goyal L, Sadeghi S, Macarulla T, et al. Phase II study of BGJ398 in patients with FGFR-altered advanced cholangiocarcinoma. J Clin Oncol. 2018;36(3):276. doi: 10.1200/JCO.2017.75.5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subbiah V, Lassen U, Élez E, Italiano A, Curigliano G, Javle M, de Braud F, Prager GW, Greil R, Stein A, et al. Dabrafenib plus trametinib in patients with BRAFV600E-mutated biliary tract cancer (ROAR): a phase 2, open-label, single-arm, multicentre basket trial. Lancet Oncol. 2020;21(9):1234–1243. doi: 10.1016/S1470-2045(20)30321-1. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z-H, Lian B-F, Dong Q-Z, Sun H, Wei J-W, Sheng Y-Y, Li W, Li Y-X, Xie L, Liu L, et al. Whole-exome mutational and transcriptional landscapes of combined hepatocellular cholangiocarcinoma and intrahepatic cholangiocarcinoma reveal molecular diversity. Biochim Et Biophys Acta (BBA) - Mol Basis Dis (BBA)-Mol Basis Of Dis. 2018;1864(6):2360–2368. doi: 10.1016/j.bbadis.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 41.Taghizadeh H, Prager GW. Immune checkpoint inhibitors for advanced biliary tract cancer. Curr Cancer Drug Targ. 2022;22(8): 639–650. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Shi J, Wang Y, Zhou H, Zhang Z, Han Z, Li G, Yang B, Cao G, Ke Y, et al. Next-generation sequencing-guided molecular-targeted therapy and immunotherapy for biliary tract cancers. Cancer Immunol Immunother. 2021;70(4):1001–1014. doi: 10.1007/s00262-020-02745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winkelmann R, Schneider M, Hartmann S, Schnitzbauer AA, Zeuzem S, Peveling-Oberhag J, Hansmann M, Walter D. Microsatellite instability occurs rarely in patients with cholangiocarcinoma: a retrospective study from a German tertiary care hospital. Int J Mol Sci. 2018;19(5):1421. doi: 10.3390/ijms19051421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


